Abstract
Mycobacteria use type VII secretion systems (T7SSs) to translocate a wide range of proteins across their diderm cell envelope. These systems, also called ESX systems, are crucial for the viability and/or virulence of mycobacterial pathogens, including Mycobacterium tuberculosis and the fish pathogen Mycobacterium marinum. We have previously shown that the M. tuberculosis ESX‐5 system is unable to fully complement secretion in an M. marinum esx‐5 mutant, suggesting species specificity in secretion. In this study, we elaborated on this observation and established that the membrane ATPase EccC5, possessing four (putative) nucleotide‐binding domains (NBDs), is responsible for this. By creating M. marinum‐M. tuberculosis EccC5 chimeras, we observed both in M. marinum and in M. tuberculosis that secretion specificity of PE_PGRS proteins depends on the presence of the cognate linker 2 domain of EccC5. This region connects NBD1 and NBD2 of EccC5 and is responsible for keeping NBD1 in an inhibited state. Notably, the ESX‐5 substrate EsxN, predicted to bind to NBD3 on EccC5, showed a distinct secretion profile. These results indicate that linker 2 is involved in species‐specific substrate recognition and might therefore be an additional substrate recognition site of EccC5.
Keywords: chimeras, ESX, membrane ATPase, mycobacterium, substrate specificity, type VII secretion
One of the major virulence factors of Mycobacterium tuberculosis and other pathogenic mycobacteria are the type VII secretion systems. Here, we provide an important insight into the mechanism of substrate recognition by these systems by identifying a putative second substrate recognition site on the central type VII secretion membrane ATPase EccC.
1. INTRODUCTION
Type VII secretion systems (T7SSs) are crucial virulence determinants for pathogenic mycobacteria, such as Mycobacterium tuberculosis and Mycobacterium marinum (Groschel et al., 2016). Pathogenic mycobacteria can have up to five T7SSs, named ESX‐1 to ESX‐5 (Houben et al., 2014), of which three, i.e., ESX‐1, ESX‐3 and ESX‐5, have been shown to be functional (Pym et al., 2002; Stanley et al., 2003; Abdallah et al., 2006; Siegrist et al., 2009; Simeone et al., 2012). These secretion systems are paramount for diverse processes, such as the utilization of nutrients and iron, and completion of the macrophage infection cycle. In pathogenic mycobacteria, ESX‐1 is crucial for intracellular survival by mediating phagosomal membrane rupture (van der Wel et al., 2007; Simeone et al., 2012). The importance of this secretion system is further substantiated by the fact that the lack of a large part of the esx‐1 gene cluster is the decisive factor in the attenuation of the live vaccine strain Mycobacterium bovis BCG (Pym et al., 2002; Simeone et al., 2012). Both ESX‐3 and ESX‐5 systems are essential for in vitro growth and have been linked to iron and fatty acid uptake respectively (Serafini et al ., 2009; Siegrist et al., 2009; Ates et al., 2015).
In mycobacteria, T7SSs secrete a diverse array of substrates, which includes monomeric as well as heterodimeric protein pairs. Most well known are the Esx substrates, which are small proteins forming heterodimeric complexes (Renshaw et al., 2005). The first described proteins of this family are the ESX‐1 substrate EsxA (also named ESAT‐6) and its secretion partner EsxB (also called CFP‐10). Dimerization of Esx proteins is mediated by two helix‐turn‐helix structures and in mycobacteria one of the partner proteins harbors a C‐terminal conserved secretion motif YxxxD/E (Poulsen et al., 2014). Two major classes of other T7SS substrates, called the PE and PPE proteins, also form stable heterodimers (Strong et al., 2006; Chen et al., 2017). The PE proteins, named after a conserved proline (P) and glutamic acid (E) motif located N‐terminally, have a conserved N‐terminal domain of approximately 110 residues. This domain forms a helix‐turn‐helix structure, followed by an YxxxD/E motif, similar to Esx proteins (Strong et al., 2006; Daleke, et al., 2012a; Chen et al., 2017). PPE proteins, named after a similar conserved motif with an additional proline (P) residue, have a larger conserved N‐terminal domain of ~180 amino acids. Part of this conserved domain forms a helix‐turn‐helix structure involved in the dimerization of the PPE with its PE partner (Strong et al., 2006; Chen et al., 2017). The N‐terminal PPE domain furthermore contains a so‐called helical‐tip domain that does not interact with the partner protein (Strong et al., 2006; Chen et al., 2017). Both PE and PPE proteins can have additional C‐termini that are highly variable and might make up the functional domain of the substrate (Mishra et al., 2008; Daleke et al., 2011; Burggraaf et al., 2019). The majority of the PE and PPE proteins are secreted by the ESX‐5 system (Abdallah et al., 2009; Ates et al., 2015). ESX‐5 is the most recently evolved mycobacterial T7SS and is present only in the so‐called slow‐growing mycobacteria, which includes important pathogens such as M. tuberculosis and Mycobacterium leprae. A large portion of the substrates that are secreted by the ESX‐5 system belong to the subfamily of the PE_PGRS proteins, named after the polymorphic GC‐rich repetitive sequence motifs in their genes. Although this subfamily contains many members in M. tuberculosis, their function is not very clear. The M. tuberculosis PE_PGRS30 protein has been shown to be involved in virulence (Bottai et al., 2012; Iantomasi et al., 2012; Fishbein et al., 2015; Deng et al., 2017) and PE_PGRS33 has been shown to interact with TLR2 (Basu et al., 2007). However, a recent study implicated that a M. tuberculosis strain not secreting any PE_PGRS proteins due to a spontaneous ppe38 deletion showed in fact (moderate) hypervirulence at later time points (Ates et al., 2018). Due to the same mutation this strain was also deficient in the secretion of the so‐called PPE_MPTR proteins.
The different mycobacterial T7SSs contain a set of conserved components, including two cytosolic and five membrane proteins. The cytosolic chaperone EspG has been shown to be involved in substrate recognition (Daleke et al., 2012b; Ekiert and Cox, 2014; Korotkova et al., 2014; Phan et al., 2017). EspG interacts specifically with PE/PPE heterodimers and helps to keep these dimers soluble by binding to a hydrophobic patch on the helical‐tip domain of the PPE protein (Daleke et al., 2012b; Ekiert and Cox, 2014; Korotkova et al., 2014). By swapping the helical‐tip domain of PPE substrates of different systems, these substrates can be redirected from one system to another (Phan et al., 2017), showing that this domain is involved in determining system specificity. Four of the conserved membrane components (EccBCDE) assemble into a hexameric complex of approximately 2 MDa (Houben et al., 2012). While a first low‐resolution image of a full T7SS membrane complex, i.e., of a hexameric ESX‐5 membrane complex, has been provided by negative stain electron microscopy (EM) (Beckham et al., 2017), very recently two high‐resolution structures of a dimeric complex of ESX‐3 have been solved by cryo‐EM (Famelis et al., 2019; Poweleit et al., 2019). Although the quaternary structures differ between the ESX‐3 and ESX‐5 complex, ESX‐3 has also been observed to form higher order multimers, suggesting the dimeric structure is a subcomplex (Famelis et al., 2019; Poweleit et al., 2019). Another discrepancy between the negative stain structure and the cryo‐EM structures is the reported EccBCDE stoichiometry of 1:1:1:1 and 1:1:2:1 respectively. This variation might be due to the highly hydrophobic and thereby aggregation‐prone nature of EccD, which could result in its underrepresentation in stoichiometric measurements (Beckham et al., 2017; Famelis et al., 2019). The fifth conserved and essential membrane component, the subtilisin‐like protease mycosin or MycP, interacts only transiently with this complex and is involved in complex stabilization (van Winden et al., 2016). This component is also involved in cleaving specific substrates (Ohol et al., 2010).
A central component of T7SS is EccC, which is a membrane‐associated ATPase and most likely the motor protein of the membrane complex (Houben et al., 2012). Importantly, EccC is the only conserved membrane protein in the more distantly related T7SSs of Firmicutes (Abdallah et al., 2007). The ATPase contains two predicted N‐terminal transmembrane domains, three nucleotide binding domains (NBDs) and an extra flexible domain of unknown function (DUF) between the ATPase and transmembrane domains (Figure 1b,c) (Rosenberg et al., 2015; Beckham et al., 2017). The recent cryo‐EM structure of ESX‐3 revealed that the DUF domain exhibits an ATPase fold, similar to the previously described NBDs of EccC (Rosenberg et al., 2015; Famelis et al., 2019; Poweleit et al., 2019). All three NBDs as well as the DUF domain of EccC are part of a family of so‐called P‐loop NTPases that show strong similarities to FtsK/SpoIIIE proteins. Proteins in this family use the energy released from ATP hydrolysis to drive the translocation of macromolecules (Burton and Dubnau, 2010). Whereas the activity of NBD2 and NBD3 of EccC has been shown to be partially dispensable for secretion, it is NBD1, normally held in an auto‐inhibited state, that is crucial for T7SS activity (Ates et al., 2015; Rosenberg et al., 2015). The EccC protein of the ESX‐1 system has the unique feature that it is split up in two subunits, i.e., EccCa1 and EccCb1. The C‐terminal 7 amino acids of EsxB have been shown by yeast‐two‐hybrid and pulldown analysis to interact with the EccCb1 subunit (Champion et al., 2006; Rosenberg et al., 2015). Subsequently, structural analyses of co‐crystals of EccC from Thermonospora curvata (Rosenberg et al., 2015) and NBD3 of EccCb1 of M. tuberculosis (Wang et al., 2019) with a peptide mimicking the C‐terminal domain of cognate EsxB and full‐length M. tuberculosis EsxB, respectively, revealed again that the last seven amino acids of the peptide were bound to NBD3 of EccC. However, substrate binding to EccC has only been shown for Esx proteins and it therefore remains unclear whether and how the other substrate classes, in particular the major substrate group of PE and PPE proteins, bind to this membrane ATPase. In addition, to what extent EccC is able to recognize substrates in a system‐specific fashion is still very poorly understood.
FIGURE 1.
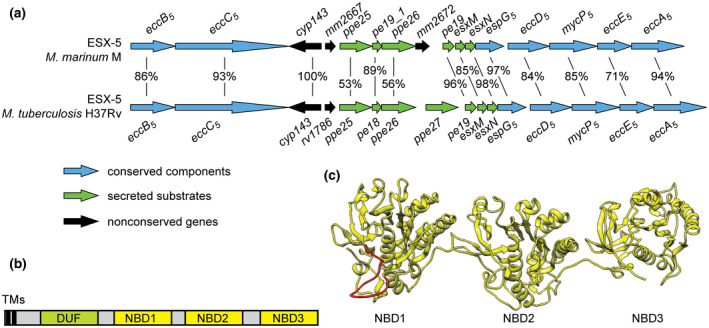
(a) Genetic organization of the esx‐5 loci in M. marinum and in M. tuberculosis. The shared identities of the orthologous proteins encoded by the genes are indicated. (b) General domain architecture of mycobacterial EccC ATPases. The constructs contain: TM, transmembrane domains; DUF, domain of unknown function, shown to adopt an ATPase fold for EccC3 (Famelis et al., 2019; Poweleit et al., 2019); NBD, nucleotide binding domain. (c) Structural model of the M. marinum EccC5 ATPase domains (residues 432–1388), generated with Phyre2 using the structure of T. curvata EccC as a template. The missing 65 residues of the linker 2 region that could not be modeled are indicated in red
Previous results from our group have shown that the ESX‐5 system is essential for growth of the fish pathogen M. marinum (Ates et al., 2015). Strikingly, this essentiality can be circumvented by increasing the permeability of the mycobacterial outer membrane, e.g., by introducing an outer membrane porin from Mycobacterium smegmatis, called MspA (Ates et al., 2015). Expression of the homologous esx‐5 operon from M. tuberculosis also allowed for the successful deletion of the entire operon in M. marinum (Ates et al., 2015). Although the ESX‐5 system of M. tuberculosis is able to mediate growth of the M. marinum ∆esx‐5 mutant, the complementation is only partial as the M. tuberculosis system is unable to mediate the secretion of many M. marinum ESX‐5 substrates, most of which are M. marinum‐specific PE_PGRS proteins (Ates et al., 2015). This result was unexpected, because the components of the two systems have an overall amino acid identity of 78% (Figure 1a). We speculated that the observed secretion defects are caused by the fact that many M. marinum substrates are not recognized by the M. tuberculosis ESX‐5 system. If this assumption is correct, we expected that the responsible proteins could be the two conserved components shown to be involved in substrate recognition, i.e., EspG5 and EccC5. In this study, we tested this hypothesis by investigating the species‐specific roles of EspG5 and EccC5 in ESX‐5‐mediated secretion.
2. RESULTS
2.1. EspG5mtub complements secretion of an M. marinum ∆espG5 mutant
EspG is a dedicated T7SS chaperone present in four of the five ESX systems, which binds PE/PPE proteins in a system‐specific fashion (Daleke et al., 2012b; Korotkova et al., 2014). As we hypothesized that the inability of the M. marinum ∆esx‐5::esx‐5mtub to secrete most M. marinum PE/PPE substrates is due to the species‐specific recognition of these substrates, EspG5 was a prime candidate for causing this effect. To test this, we used an M. marinum ∆espG5 knock‐out strain that expresses MspA to circumvent the essentiality of ESX‐5 for growth (Phan et al., 2018). As previously demonstrated, this mutant showed no PE_PGRS secretion, as determined by analyzing cell surface fractions extracted by the mild detergent Genapol X‐080 (Figure S1). Also the ESX‐5 substrate EsxN was not secreted by this mutant (Figure S1). This reduction in secretion in the espG5 mutant did not result in increased amounts of ESX‐5 substrates in the cell, which is consistent with previous observations made for esx‐5 mutants in M. marinum (Houben et al., 2012; Ates et al., 2015; Ates et al., 2016; Ates et al., 2018; van Winden et al., 2019). In these previous studies, it was shown that this instability phenotype is not linked to regulatory effects on a transcriptional level (Abdallah et al., 2009; Bottai et al., 2012; Houben et al., 2012; Ates et al., 2018). PE_PGRS protein secretion could be restored to WT levels upon complementation with the M. marinum espG5 gene, although EsxN secretion was only partially restored (Figure S1). Importantly, introduction of the M. tuberculosis espG5 gene restored secretion to similar levels (Figure S1). We therefore conclude that EspG5mtub is fully functional in M. marinum and not the cause for the M. marinum ∆esx‐5::esx‐5mtub species‐specific secretion defect of PE_PGRS proteins.
2.2. EccC5mtub complements essentiality but not PE_PGRS secretion in an M. marinum ∆eccC5 mutant
The central membrane ATPase EccC5 is the second T7SS protein that has been shown to bind substrates (Stanley et al., 2003; Rosenberg et al., 2015; Wang et al., 2019). We therefore examined whether EccC was responsible for species‐specific secretion. First, we checked whether eccC5mtub was able to rescue the essentiality phenotype of an M. marinum ∆eccC5 strain by introducing the same plasmid into a previously described ∆eccC5::eccC5mmar strain that lacks MspA (Ates et al., 2015). This strain bears an integrative plasmid containing both eccC5mmar and a hygromycin resistance marker, which was exchanged with a kanamycin‐resistant integrative plasmid harboring eccC5mtub. Multiple colonies appeared that showed kanamycin resistance and hygromycin sensitivity, demonstrating that the plasmid exchange was successful and therefore that eccC5mtub is able to mediate growth of the M. marinum ∆eccC5 mutant.
Next, we tested the secretion profile of an M. marinum ∆eccC5 mutant expressing MspA, which was complemented with eccC5 from either M. marinum or M. tuberculosis. As expected, this MspA‐expressing ∆eccC5 mutant showed no secretion of PE_PGRS or EsxN substrates (Figure 2c). While secretion could be restored to WT levels upon complementation with the M. marinum eccC5 gene, introduction of a plasmid containing eccC5 from M. tuberculosis showed no visible PE_PGRS proteins in the cell surface‐enriched protein fraction (Figure 2c). Surprisingly, the culture supernatant fraction of this strain showed only one PE_PGRS protein band, which was similar to the previously observed PE_PGRS secretion phenotype of M. tuberculosis (Figures 2c and 3a) (Houben et al., 2012). In addition, no EsxN secretion was observed in this strain. The difference in secretion is surprising, as the overall sequence identity between the two eccC5 orthologues is 93% (Figure 1a). In addition, the residues present in the pocket of NBD3, which have been predicted to be crucial for Esx substrate binding (Rosenberg et al., 2015; Wang et al., 2019) are conserved between the two species. On the other hand, the most C‐terminal residues of EsxM, the partner protein of EsxN, containing the C‐terminal YxxxD/E motif and predicted to be involved in EccC binding, are not well conserved between M. marinum and M. tuberculosis (Figure S4b). We also checked the colony phenotype for the mutant and the complemented strains. Opposed to the WT and the ∆eccC5::eccC5mmar strain, which showed smooth colony morphology and monodispersed growth in liquid cultures, the ∆eccC5 as well as the ∆eccC5::eccC5mtub strains showed a nondispersed growth phenotype in culture and flat and dry colonies on plate (Figure S2). Together, these results show that eccC5mtub is unable to fully complement the eccC5 mutation in M. marinum due to species‐specific functioning.
FIGURE 2.
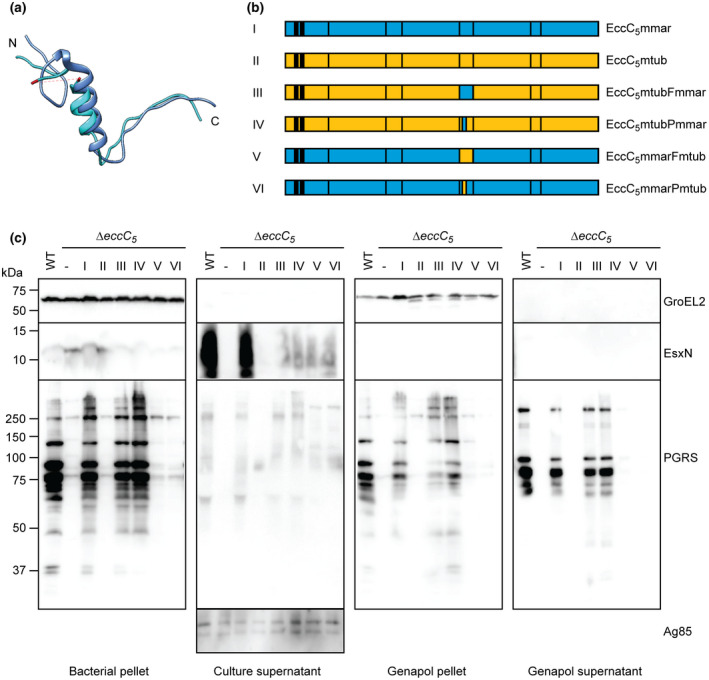
Role of linker 2 in substrate specificity in M. marinum ESX‐5. (a) Superimposition of T. curvata linker 2 (bright blue) and linker 3 (dark blue). The 43 residues that were not resolved in the crystal structure are indicated in red. (b) Schematic overview of M. marinum and M. tuberculosis EccC5 as well as the chimeric constructs used to complement eccC5 mutants of these species. See Figure S4a for the exchanged sequences. (c) Secretion analysis of M. marinum ∆eccC5 complemented with WT eccC5mmar, eccC5mtub and chimeric constructs depicted in b. Proteins were visualized by SDS‐PAGE and immunoblotting using antibodies against EsxN and PE_PGRS proteins (ESX‐5 substrates), GroEL2 (lysis and whole‐cell loading control) and Ag85 (secreted fraction loading control)
FIGURE 3.
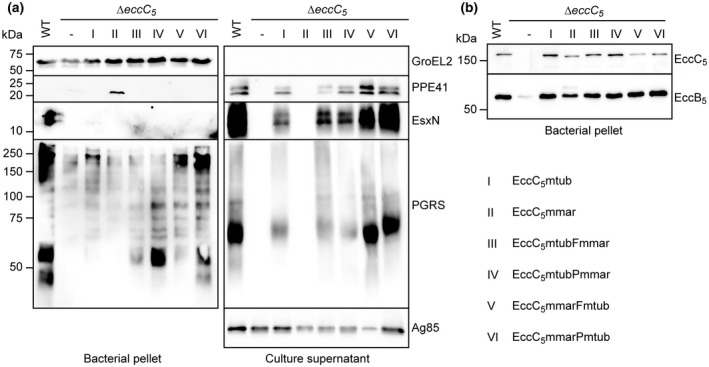
Role of linker 2 in substrate‐specificity in M. tuberculosis ESX‐5. (a) Secretion analysis of a M. tuberculosis EccC5 transposon mutant complemented with WT eccC5mmar, eccC5mtub and chimeric constructs. Roman numerals indicate the constructs as depicted on the bottom right (see Figure 2b for a schematic overview of the EccC5 proteins). Proteins were visualized by SDS‐PAGE and immunoblotting using antibodies against EsxN, PE_PGRS proteins and PPE41 (ESX‐5 substrates), GroEL2 (lysis and whole‐cell loading control) and Ag85 (secreted fraction loading control). (b) SDS‐PAGE and immunoblot analysis of whole cells of M. tuberculosis WT, ∆eccC5 and ∆eccC5 complemented with constructs depicted in Figure 2b. Proteins were visualized using antibodies against EccB5 and EccC5 (membrane components of ESX‐5)
We next investigated whether the observed secretion defects were due to the unsuccessful incorporation of EccC5mtub in the ESX‐5 membrane complex. We have shown previously that the ESX‐5 membrane complex of ~2 MDa can be visualized using BN‐PAGE and western blot analysis of DDM solubilized cell envelopes using antibodies against any of the four complex components (Houben et al., 2012). Using the same approach, the ∆eccC5 strain showed reduced expression of the ESX‐5 membrane components EccB5 and EccE5 and membrane complex formation was abrogated (Figure S3a). Complex formation was restored upon complementation with either the M. marinum or M. tuberculosis eccC5 containing plasmid (Figure S3a). Similarly, expression of the EccB5 and EccE5 components were restored to WT levels (Figure S3b). We therefore conclude that the lack of PE_PGRS secretion by the M. marinum ∆eccC5::eccC5mtub was probably not caused by any defect in the assembly of the ESX‐5 membrane complex. Because the secretion phenotype of the ∆eccC5::eccC5mtub strain was similar to that of the ∆esx‐5::esx‐5mtub complementation strain, we conclude that EccC is the key component responsible for this distinct secretion defect.
2.3. EccC5 linker 2 domain is involved in species‐specific secretion in M. marinum
Although EccC5mtub is properly integrated in the ESX‐5 membrane complex and rescues essentiality of an M. marinum ∆eccC5 mutant, this ATPase was unable to restore secretion of all substrates. A sequence alignment of the EccC5 proteins of M. marinum and M. tuberculosis showed high overall conservation, but also revealed some variations (Figures S4a and S6). Aligning the two proteins with the sequence of the T. curvata EccC, for which the crystal structure has been solved (Rosenberg et al., 2015), revealed that the amino acids that were shown to be important for ATPase activity and substrate binding are highly conserved (Figures S4a and S6). In particular, the interacting amino acids lining the substrate‐binding pocket on NBD3, i.e., E1237, L1253, I1282 for eccC5mtub and E1234, L1250, I1279 for eccC5mmar (I1163, I1179 and L1208 in the T. curvata system), are all conserved (Figure S6). In addition, also critical residues within the linker 2 region are conserved, i.e., the tryptophan (W810mtub and W807mmar) and glutamine (Q811mtub and Q808mmar (L763 in T. curvata)) that keep the critical first NBD in an inhibited state (Rosenberg et al., 2015). However, there are key differences in the same linker 2 region (Figure S4a). As this domain blocks ATPase activity of NBD1, it has been speculated that a, yet unknown, event might lead to the displacement of linker 2 from the pocket of NBD1, allosterically regulating its activity (Rosenberg et al., 2015). Interestingly, a significant part (41 residues) of this linker 2 domain is disordered and therefore not present in the crystal structure of EccC of T. curvata (Figure 2a). This disordered region also revealed the lowest sequence identity between the EccC proteins of M. tuberculosis and M. marinum. Compared to EccC of T. curvata, the linker 2 region is considerably larger for the EccC5 proteins, with an additional 31 residues for EccC5mtub and 27 residues for EccC5mmar.
Taking these observations into consideration, we reasoned that this disordered region in linker 2 might play a crucial role in the (in)activation of NBD1 through regulating substrate binding and/or specificity. In order to test this, we made two chimeric eccC5 constructs where the backbone originates from M. tuberculosis and the linker 2 region from M. marinum. The linker 2 portion covered either the entire region after NBD1 until just after the two amino acids WQ that interact with the pocket of NBD1 (named full—EccC5mtubFmmar) or only a small part of the linker 2 that shows the most sequence divergence between the two and also aligns broadly with the disordered region (named partial—EccC5mtubPmmar) (Figures 2b and S4A). Both constructs could rescue the essentiality of the M. marinum eccC5 knockout in the absence of MspA. Subsequently, we examined the expression of ESX‐5 membrane components and the presence of the ESX‐5 membrane complex by SDS‐ and BN‐PAGE immunoblot analysis, which showed that both proteins were incorporated (Figure S3a,b). Finally, while expression of the original EccC5mtub in the eccC5 mutant resulted in flat and dry colonies, this phenotype was reversed to the WT situation upon exchange of the linker 2 in the EccC5mtubFmmar or EccC5mtubPmmar plasmids (Figure S2).
We next checked if these constructs could alleviate the secretion defect caused by the EccC5mtub complementation. Remarkably, although EsxN was not fully restored, secretion of PE_PGRS proteins was restored back to WT levels with both full and partial swapped constructs (Figure 2c). This is intriguing, as only 19 amino acids were different between the swapped region in the partial construct and the WT eccC5mtub gene. These data confirmed our hypothesis that linker 2 is involved in substrate specificity and/or (in)activation of NBD1.
Subsequently, we wondered if conversely, we could repress secretion of the ∆eccC5mmar::eccC5mmar complementation by swapping the linker 2 of EccC5mmar with that of EccC5mtub. Importantly, although the linker 2 region originated from EccC5mtub, the rest of the gene was M. marinum WT, thus keeping all other (un)known potential interaction sites. These constructs were named EccC5mmarFmtub and EccC5mmarPmtub (Figures 2b and S4a). Although only 25 residues for the full linker 2 and 19 residues for partial linker 2 were different, PE_PGRS secretion with the chimeric constructs was completely abolished, thus substantiating our initial findings. Conversely, EsxN was present in the supernatant, but only in low amounts, similar to the reciprocal chimeric constructs. Both constructs could rescue the essentiality of the M. marinum ∆eccC5 mutant and showed a somewhat intermediate phenotype between the smooth WT colonies and the rough and dry colonies of the ∆eccC5 and ∆eccC5::eccC5mtub (Figure S2). From this, we conclude that the linker 2 domain of EccC5 is involved in species‐specific secretion of PE_PGRS substrates. In addition, as EsxN secretion was only partially recovered by all chimeric constructs, the optimal secretion of EsxN is not only dependent on NBD3 but is regulated by multiple domains of or interactions with EccC.
2.4. EccC5 linker 2 is involved in substrate specificity in M. tuberculosis
We next tested whether the drastic effects upon exchanging only a small part of the linker 2 of EccC5, observed in M. marinum, could also apply to other species. For this, we introduced the same chimeric constructs as analyzed in M. marinum in a previously described M. tuberculosis eccC5 transposon mutant (Figures 2b and S4a) (Houben et al., 2012).
The different constructs were efficiently expressed in the M. tuberculosis mutant strain (Figure 3b). In addition, while expression of EccB5 was strongly affected in the mutant strain, its expression was restored to WT levels in the presence of all constructs. From this, we conclude that the chimeric EccC proteins were again able to stabilize other components of the ESX‐5 membrane complex, suggesting these constructs were properly integrated in the membrane complex (Figure 3b). Next, the secretion of PE_PGRS proteins and EsxN was analyzed. It should be noted that surface‐associated PE_PGRS proteins are not extractable in M. tuberculosis (Houben et al., 2012; Ates et al., 2018), while in our M. marinum experiments only this subset of substrates was completely dependent on the cognate linker 2. Similar to M. marinum, in M. tuberculosis secretion of PE_PGRS proteins and EsxN into the culture supernatant was completely abolished in the absence of eccC5, and this could be restored by the WT M. tuberculosis gene, albeit to a slightly lower level (Figure 3a). Importantly, the M. marinum gene was not able to complement both PE_PGRS and EsxN secretion, identical to the reciprocal experiment in M. marinum. Both chimeric EccC5 proteins, i.e., M. tuberculosis EccC5 with the linker 2 of the M. marinum protein and the M. marinum protein with the linker 2 of M. tuberculosis, showed restored PE_PGRS and EsxN secretion, again similar to what was observed for the culture supernatant fractions of M. marinum (Figure 3a). From this, we conclude that the linker 2 domain has a similar role in M. tuberculosis, as described for M. marinum ESX‐5.
3. DISCUSSION
ESX‐5 is the most recently evolved T7SS in mycobacteria and is responsible for the secretion of the majority of PE and PPE proteins, among which are most or even all members of the large family of PE_PGRS proteins. Previous studies have shown that introduction of the ESX‐5 system of M. tuberculosis is able to take over the essential role of the ESX‐5 system in M. marinum (Ates et al., 2015). However, this system is only marginally able to restore secretion of M. marinum ESX‐5‐dependent substrates, suggesting that substrate recognition is at least partially species specific. This study revealed that a highly specific domain in the central membrane ATPase of the ESX‐5 system is the determining factor for the specifies‐specific secretion of PE_PGRS proteins.
To identify the component responsible for the observed species‐specific secretion of PE_PGRS proteins, we used individual esx‐5 component mutants and complemented these with the corresponding gene from either M. marinum or M. tuberculosis. Because PE_PGRS proteins were not secreted by the M. marinum ∆esx‐5::esx‐5mtub and have been widely used as model ESX‐5 substrates, we decided to use these proteins as a readout for ESX‐5 functionality. Additionally, by assessing PE_PGRS protein secretion, we could monitor a whole set of substrates and not just individual proteins. Notably, blocking ESX‐5 secretion also leads to lower cellular levels of PE_PGRS proteins in M. marinum, which has been observed previously for more esx‐5 mutants, even when specific substrates were controlled by constitutive promoters (Abdallah et al., 2009; Ates et al., 2018).
EspG was our initial most prominent candidate, as this chaperone has been shown to bind PE/PPE heterodimers and the corresponding EspG‐binding domain is a determining factor in the system‐specific secretion of these substrates. However, ∆espG5 complementations did not show any marked differences in secretion between EspG5 of M. marinum and M. tuberculosis. As the conservation between the two proteins is very high, i.e., 97% identity (Figure 1a), both of them can probably serve each other's function in the opposite species.
The only other component known to recognize substrates is EccC, although only Esx substrates have been shown to interact with this central ATPase component (Stanley et al., 2003; Rosenberg et al., 2015; Wang et al., 2019). Indeed, complementation of M. marinum ∆eccC5 with eccC5mtub was able to restore growth, but not the presence of PE_PGRS proteins on the cell surface or EsxN secretion in the culture supernatant. Importantly, using M. marinum‐M. tuberculosis EccC5 chimeras, we showed that surface‐localization of PE_PGRS proteins strictly depends on the presence of the native linker 2 domain, irrespective of the origin of the EccC5 backbone. The absence of surface‐localized PE_PGRS proteins is also linked to a distinct flat and dry colony morphology. Interestingly, while the presence of EccC5mtub showed only a single PE_PGRS protein in the secreted fraction by western blotting, M. marinum EccC5 with linker 2 of M. tuberculosis was able to mediate secretion of multiple PE_PGRS proteins into the culture supernatant, although the PE_PGRS protein pattern was distinct from that of the WT strain.
Significantly, we obtained similar results in M. tuberculosis. The only problem in studying this process in M. tuberculosis is that surface‐associated PE_PGRS proteins are not extractable in this species (Houben et al., 2012; Ates et al., 2018). Analysis of the PE_PGRS substrates in the secreted fraction, however, shows an identical profile as in M. marinum. Whereas secretion analysis of an eccC5 mutant showed abolished secretion of PE_PGRS and EsxN in both species, complementation with the native gene restores this back to WT levels. Importantly, introduction of a plasmid containing eccC5 from the opposite species was unable to restore secretion into the culture supernatant, while expression of an EccC5 chimera, which contains the native linker 2 and the remainder of the protein from the opposite species does recover secretion in both M. marinum and M. tuberculosis. This further strengthens the hypothesis that the EccC5 linker 2 domain is involved in determining substrate‐specificity for the secretion of PE_PGRS substrates.
Although the interface between NBD1 and NBD2 is similar to that between NBD2 and NBD3, the interdomain linkers are variable. Both these linkers form a main α‐helix that potentially mimics the C‐terminal tail of EsxB‐like proteins, which binds to NBD3. However, there is a region of variability between linkers 2 and 3 in sequence and size immediately N‐terminal from of this α‐helix, i.e., this region is significantly larger in the linker 2 interdomain (Figure S5). Highly intriguing is that a large part of this variable region of linker 2 is disordered in the only available structure containing all three NBDs of an EccC homologue from T. curvata (Rosenberg et al., 2015), suggesting flexibility (Figure 2a). Sequence alignments show that this is also the most variable region between the two EccC5 proteins of M. tuberculosis and M. marinum (Figure S6). An alignment of the EccC ATPases from all five mycobacterial ESX systems shows that this region of the linker 2 domain is extended in ESX‐2 and ESX‐5 systems, i.e., the most recently evolved systems (Figure S7). ESX‐5 most likely evolved through a duplication of the esx‐2 cluster (Gey van Pittius et al., 2006). This duplication event is followed by the vast expansion of pe and ppe genes, in particular the most recently evolved PE_PGRS proteins and the so‐called PPE_MPTR proteins, of which at least a major portion are secreted through the ESX‐5 system (Abdallah et al., 2009; Bottai et al., 2012). Previous mass spectrometry analysis of an ∆esx‐5::esx‐5mtub strain showed that majority of PE/PPE proteins, that are not secreted by this strain, are specific for M. marinum (Ates et al., 2015). In contrast, different ESX‐5‐dependent PE/PPE heterodimers that are conserved between M. tuberculosis and M. marinum showed no notable secretion difference in M. marinum ∆eccC5::eccC5mmar and ∆eccC5::eccC5mtub (our unpublished results). These findings indicate that PE/PPE proteins that are more recently evolved in M. marinum are not recognized by (the linker 2 of) EccC5mtub. Therefore, the variable extension of the linker 2 domain might have co‐evolved with the expansion of PE/PPE proteins, especially the PE_PGRS proteins, to allow the recognition of this vast group of substrates.
Importantly, the secretion of EsxN is also species‐specific, as both the M. marinum and M. tuberculosis eccC5 mutants could only be restored to WT levels upon complementation with the cognate eccC5 gene. This is in contrast to the conservation found in the putative binding pocket of Esx substrates on NBD3. Notably, EsxM, i.e., the partner protein of EsxN carrying the predicted C‐terminal secretion signal, has a stop codon in M. tuberculosis. However, both EsxN and EsxM have multiple highly homologous paralogs in M. marinum and M. tuberculosis, which probably have redundant functions. Although there is high conservation between the different EsxM homologs from M. marinum and M. tuberculosis, the most C‐terminal four amino acids are divergent. As both for the T. curvata EccC and EccCb1 of M. tuberculosis the last seven amino acids of EsxB are involved in binding to NBD3 (Rosenberg et al., 2015; Wang et al., 2019), this provides an explanation for the species‐specific secretion of EsxN. However, our observation that all chimeric EccC5 proteins with exchanged linker 2 regions, irrespective of the origin of NBD3, showed intermediate levels of EsxN secretion in M. marinum and full secretion in M. tuberculosis is more difficult to explain. As T7SS substrates have been shown to be interdependent on each other for secretion (Fortune et al., 2005; Champion et al., 2009; Ates et al., 2018; Damen et al., 2020), it might be possible that PE/PPE protein(s) and EsxM/EsxN are secreted in a concerted fashion. Indeed, it has recently been shown that the ESX‐1 secreted heterodimer EsxB_1/EsxA_1 is efficiently secreted only when PE35/PPE68_1, which are encoded by the same operon as the Esx proteins, are co‐expressed and secreted (Damen et al., 2020). While substrate interdependency for secretion is a not yet understood phenomenon in T7SS, these data together suggest that secretion‐specificity of Esx heterodimers is not only dependent on the interaction with the cognate EccC NBD3.
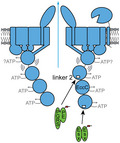
Based on our current results and previous data we propose a working model for substrate recognition by EccC in mycobacterial T7SSs (Figure 4). In this model, the EccBCDE membrane complex is a stable complex in the mycobacterial inner membrane. Without substrate binding, EccC is hexameric via its transmembrane regions, while its cytosolic domain is highly flexible through the N‐terminal DUF domain (Beckham et al., 2017). Interaction of the C‐terminal NBD3 with the C‐terminal tail of specific (Esx) substrates drives multimerization, but NBD1 remains inactive through the pocket 1/linker 2 connection (Rosenberg et al., 2015). The final secretion activation step takes place upon linker 2 displacement from pocket 1, which at least in the mycobacterial ESX‐5 system, is triggered by binding of PE/PPE substrates or a (yet unknown) chaperone for these proteins. While the role of linker 2 of EccC in keeping the highly important NBD1 in an inactive state has previously been described, we present here a new role for this domain of EccC ATPases in substrate specificity.
FIGURE 4.
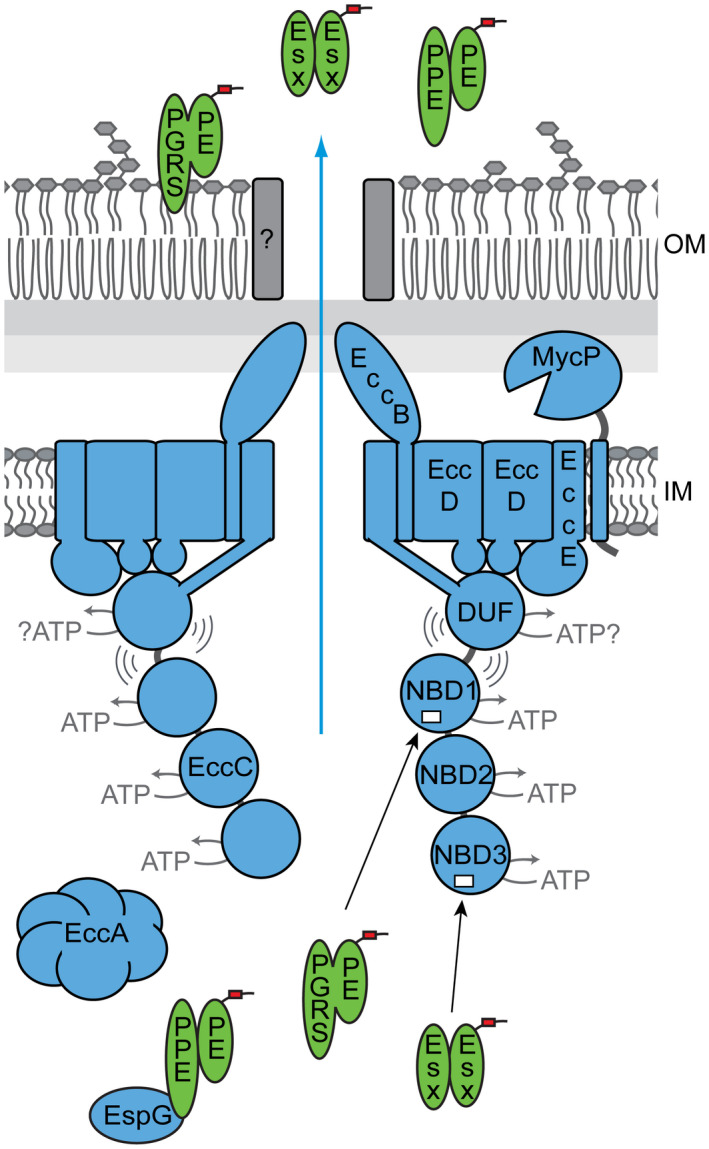
Model for ESX‐5 mediated secretion. Substrate recognition occurs at two separate sites on the EccC5 ATPase. The third NBD interacts with the C‐terminus of specific Esx proteins, leading to the multimerization of the soluble domain of EccC. A second site of substrate recognition is located in the linker 2 domain at the first NBD. As the interaction between NBD1 and this linker has an inhibitory effect on the activity of NBD1, binding of PE_PGRS proteins and perhaps also other PE and PPE proteins disrupts this interaction and activates this crucial ATPase domain
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and culture conditions
M. marinum M and M. tuberculosis CDC155 were used for all experiments involving EccC5 and M. marinum E11 was used for the EspG5 analysis. M. marinum was grown at 30°C on 7H10 agar with 10% Middlebrook OADC (BD Biosciences) or in 7H9 liquid medium with 10% Middlebrook ADC and 0.05% Tween 80 (Merck). M. tuberculosis was grown at 37°C under similar conditions. Culture medium was supplemented with the necessary antibiotics at the following concentrations: kanamycin, 25 µg/ml; hygromycin, 50 µg/ml; streptomycin, 30 µg/ml.
4.2. Molecular cloning
All cloning was performed in Escherichia coli DH5α, with restriction enzymes from New English Biolabs and PCR amplifications with Iproof (BioRad). Difficult ligations or ligations that included more than two fragments were performed with In‐Fusion (TakaraBio) with 15 bp homologies coded in the primer sequence. Two identical pMV361 vectors cut with XmnI and HindIII and coding for HygR or KanR were used (Ates et al., 2015). The esxM–esxN–espG5 region or eccC5 was amplified from M. tuberculosis H37Rv genomic DNA using anchored primers 1 and 2 or 3 and 4, respectively (XmnI, HindIII, Table S2), and ligated in two pMV361 vectors coding for HygR or KanR (Ates et al., 2015) using XmnI and HindIII, resulting in the plasmids pMV‐espG5mtub‐hygR and pMV‐eccC5mtub‐kanR. Primers 5 and 6 were used to PCR amplify the partial linker 2 domain (between F744 and V786) from the pMV‐eccC5mmar plasmid. This PCR product and the pMV‐eccC5mtub plasmid were both cut with FspAI and MunI and ligated, resulting in the plasmid pMV‐eccC5mtubPmmar.
Primers 7 and 8 were used to amplify the full linker 2 domain of eccC5mmar (between residues A680 and D819) and primers 9 and 10 were used to PCR amplify the rest of the eccC5mtub gene, downstream of the linker 2, which was cut out in the process. Plasmid pMV‐eccC5mtub was cut with SfiI and HindIII and ligated with both PCR products via In‐Fusion cloning, resulting in plasmid pMV‐eccC5mtubFmmar. Primers had a 15bp overlap for In‐Fusion cloning.
pMV‐eccC5mmarFmtub and pMV‐eccC5mmarPmtub were cloned in a similar fashion. Due to a lack of proper restriction sites, these plasmids were cloned from scratch using In‐Fusion cloning. The eccC5mmar region upstream of the linker 2 domain was amplified from the pMV‐eccC5mmar plasmid with primers 11 and 12 for the full domain (PCR1) and primers 11 and 17 for the partial domain (PCR2). The eccC5mtub linker 2 domain was amplified from the corresponding plasmid with primers 13 and 14 for the full (between residues A679 and D822—PCR3) and 18 and 19 for the partial domain (between residues F743 and T789—PCR4). The eccC5mmar region downstream of the linker 2 domain was PCR amplified from pMV‐eccC5mmar with primers 15 and 16 for the full domain—PCR5—and primers 16 and 20 for the partial domain—PCR6. A pMV‐kanR plasmid cut with the XmnI and HindIII restriction sites was In‐Fusion ligated with PCR products 1, 3 and 5 to result in the plasmid pMV‐eccC5mmarFmtub and with PCR products 2, 4 and 6 to result in the plasmid pMV‐eccC5mmarPmtub.
4.3. Protein secretion and western blot analysis
For protein secretion, M. marinum strains were grown in 7H9 liquid medium with 10% Middlebrook ADC, 0.05% Tween 80 and appropriate antibiotics until mid‐log phase. Cells were harvested, washed and inoculated at an OD600 of 0.4–0.5 in 7H9 liquid medium with 0.2% dextrose, 0.2% glycerol, 0.05% Tween 80 and appropriate antibiotics. After overnight growth, cells were pelleted at an OD600 of 0.8–1. Supernatants were passed through an 0.2 µm filter and precipitated with trichloroacetic acid (TCA) (culture supernatant fraction). Cell pellets were split in two and half was treated with 0.5% Genapol X‐080 (Fluka) for 30 min, head over head at room temperature, after which cells were spun down and supernatant was collected (Genapol surface‐extracted fraction and Genapol‐treated cells). Both whole cell samples, treated or not with Genapol‐X080, were lysed by bead‐beating. SDS loading buffer was added and samples were boiled and loaded on SDS‐PAGE gels (10%–16%, depending on the size of the proteins of interest), transferred to nitrocellulose membranes and stained with appropriate antibodies. For experiments involving M. tuberculosis, the procedure was similar, but cells and culture supernatants were heat inactivated for 30 min at 80°C after harvesting. The antibodies that were used were anti‐GroEL2 (CS44; John Belisle, NIH, Bethesda, MD, USA), anti‐EsxN (Mtb9.9a), anti‐PE_PGRS (Abdallah et al., 2006), anti‐Ag85 (Bei Resources), anti‐PPE41 (Abdallah et al., 2006), anti‐EspG5 (Houben et al., 2012), anti‐EccB5 (Houben et al., 2012), anti‐EccC5 (Houben et al., 2012), anti‐EccE5 (Houben et al., 2012) and anti‐FtsH (Houben et al., 2012).
4.4. Cell envelope isolation
For cell envelope isolations, M. marinum was grown in liquid 7H9 media with 10% Middlebrook ADC, 0.05% Tween 80 and appropriate antibiotics to an OD600 of 1.2–1.5. Cells were washed in PBS and resuspended in CE buffer (20 mM Tris‐HCl, 300 mM NaCl and 10% glycerol). Cells were lysed by passing through a One‐Shot Cell disruptor (Constant Systems Ltd.) and unbroken cells were pelleted at 5,000× g. Cell envelopes (CE) were separated from the soluble fraction by ultracentrifugation at 150.000× g for 90 min. After ultracentrifugation, supernatant was discarded, pellets were washed in CE buffer and resuspended in CE buffer.
5. BN‐PAGE
For BN‐PAGE analysis of membrane complexes, cell envelopes were solubilized with 0.25% DDM for 1 hr at 4°C. Nonsolubilized material was pelleted by centrifugation at 100.000× g for 20 min at 4°C. NativePage 5% G‐250 Sample Additive (Invitrogen) was added to the resulting supernatant fraction and samples were run on a 3%–12% NativePage Bis‐Tris Protein Gel (Invitrogen). Gels were blotted to a PVDF membrane and stained with appropriate antibodies.
CONFLICT OF INTEREST
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: C.M.B., W.B., E.N.G.H. Performed the experiments: C.M.B., R.U. Analyzed the data: C.M.B., W.B., E.N.G.H. Wrote the initial draft C.M.B. Manuscript finalization: C.M.B., W.B., E.N.G.H.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by a VIDI grant (864.12.006; to CMB and ENGH) from the Netherlands Organization of Scientific Research. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Bunduc CM, Ummels R, Bitter W, Houben ENG. Species‐specific secretion of ESX‐5 type VII substrates is determined by the linker 2 of EccC5 . Mol Microbiol. 2020;114:66–76. 10.1111/mmi.14496
REFERENCES
- Abdallah, A.M. , Gey van Pittius, N.C. , Champion, P.A. , Cox, J. , Luirink, J. , Vandenbroucke‐Grauls, C.M. , et al (2007) Type VII secretion–mycobacteria show the way. Nature Reviews Microbiology, 5, 883–891. [DOI] [PubMed] [Google Scholar]
- Abdallah, A.M. , Verboom, T. , Hannes, F. , Safi, M. , Strong, M. , Eisenberg, D. , et al (2006) A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Molecular Microbiology, 62, 667–679. [DOI] [PubMed] [Google Scholar]
- Abdallah, A.M. , Verboom, T. , Weerdenburg, E.M. , Gey van Pittius, N.C. , Mahasha, P.W. , Jimenez, C. , et al (2009) PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX‐5. Molecular Microbiology, 73, 329–340. [DOI] [PubMed] [Google Scholar]
- Ates, L.S. , Dippenaar, A. , Ummels, R. , Piersma, S.R. , van der Woude, A.D. , van der Kuij, K. , et al (2018) Mutations in ppe38 block PE_PGRS secretion and increase virulence of Mycobacterium tuberculosis . Nature Microbiology, 3, 181–188. [DOI] [PubMed] [Google Scholar]
- Ates, L.S. , Ummels, R. , Commandeur, S. , van de Weerd, R. , Sparrius, M. , Weerdenburg, E. , et al (2015) Essential role of the ESX‐5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genetics, 11, e1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates, L.S. , van der Woude, A.D. , Bestebroer, J. , van Stempvoort, G. , Musters, R.J. , Garcia‐Vallejo, J.J. , et al (2016) The ESX‐5 system of pathogenic mycobacteria is involved in capsule integrity and virulence through its substrate PPE10. PLoS Pathogens, 12, e1005696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, S. , Pathak, S.K. , Banerjee, A. , Pathak, S. , Bhattacharyya, A. , Yang, Z. , et al (2007) Execution of macrophage apoptosis by PE_PGRS33 of Mycobacterium tuberculosis is mediated by Toll‐like receptor 2‐dependent release of tumor necrosis factor‐alpha. Journal of Biological Chemistry, 282, 1039–1050. [DOI] [PubMed] [Google Scholar]
- Beckham, K.S. , Ciccarelli, L. , Bunduc, C.M. , Mertens, H.D. , Ummels, R. , Lugmayr, W. , et al (2017) Structure of the mycobacterial ESX‐5 type VII secretion system membrane complex by single‐particle analysis. Nature Microbiology, 2, 17047. [DOI] [PubMed] [Google Scholar]
- Bottai, D. , Di Luca, M. , Majlessi, L. , Frigui, W. , Simeone, R. , Sayes, F. , et al (2012) Disruption of the ESX‐5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Molecular Microbiology, 83, 1195–1209. [DOI] [PubMed] [Google Scholar]
- Burggraaf, M.J. , Speer, A. , Meijers, A.S. , Ummels, R. , van der Sar, A.M. , Korotkov, K.V. , et al (2019) Type VII secretion substrates of pathogenic mycobacteria are processed by a surface protease, mBio, 10, e01951‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, B. and Dubnau, D. (2010) Membrane‐associated DNA transport machines. Cold Spring Harbor Perspectives in Biology, 2, a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion, P.A. , Champion, M.M. , Manzanillo, P. and Cox, J.S. (2009) ESX‐1 secreted virulence factors are recognized by multiple cytosolic AAA ATPases in pathogenic mycobacteria. Molecular Microbiology, 73, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion, P.A. , Stanley, S.A. , Champion, M.M. , Brown, E.J. and Cox, J.S. (2006) C‐terminal signal sequence promotes virulence factor secretion in Mycobacterium tuberculosis . Science, 313, 1632–1636. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Cheng, H.F. , Zhou, J. , Chan, C.Y. , Lau, K.F. , Tsui, S.K. , et al (2017) Structural basis of the PE‐PPE protein interaction in Mycobacterium tuberculosis . Journal of Biological Chemistry, 292, 16880–16890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , Cascioferro, A. , de Punder, K. , Ummels, R. , Abdallah, A.M. , van der Wel, N. , et al (2011) Conserved Pro‐Glu (PE) and Pro‐Pro‐Glu (PPE) protein domains target LipY lipases of pathogenic mycobacteria to the cell surface via the ESX‐5 pathway. Journal of Biological Chemistry, 286, 19024–19034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , Ummels, R. , Bawono, P. , Heringa, J. , Vandenbroucke‐Grauls, C.M. , Luirink, J. , et al (2012a) General secretion signal for the mycobacterial type VII secretion pathway. Proceedings of the National Academy of Sciences of the United States of America, 109, 11342–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke, M.H. , van der Woude, A.D. , Parret, A.H. , Ummels, R. , de Groot, A.M. , Watson, D. , et al (2012b) Specific chaperones for the type VII protein secretion pathway. Journal of Biological Chemistry, 287, 31939–31947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damen, M.P.M. , Phan, T.H. , Ummels, R. , Rubio‐Canalejas, A. , Bitter, W. and Houben, E.N.G. (2020) Rerouting of an Esx substrate pair from the ESX‐1 type VII secretion system to ESX‐5 by modifying a PE/PPE substrate pair. bioRxiv. 10.1101/2020.01.15.906685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W. , Long, Q. , Zeng, J. , Li, P. , Yang, W. , Chen, X. , et al (2017) Mycobacterium tuberculosis PE_PGRS41 enhances the intracellular survival of M. smegmatis within macrophages via blocking innate immunity and inhibition of host defense. Scientific Reports, 7, 46716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert, D.C. and Cox, J.S. (2014) Structure of a PE‐PPE‐Esp G complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proceedings of the National Academy of Sciences of the United States of America, 111, 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famelis, N. , Rivera‐Calzada, A. , Degliesposti, G. , Wingender, M. , Mietrach, N. , Skehel, J.M. , et al (2019) Architecture of the mycobacterial type VII secretion system. Nature, 576, 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein, S. , van Wyk, N. , Warren, R.M. and Sampson, S.L. (2015) Phylogeny to function: PE/PPE protein evolution and impact on Mycobacterium tuberculosis pathogenicity. Molecular Microbiology, 96, 901–916. [DOI] [PubMed] [Google Scholar]
- Fortune, S.M. , Jaeger, A. , Sarracino, D.A. , Chase, M.R. , Sassetti, C.M. , Sherman, D.R. , et al (2005) Mutually dependent secretion of proteins required for mycobacterial virulence. Proceedings of the National Academy of Sciences of the United States of America, 102, 10676–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey van Pittius, N.C. , Sampson, S.L. , Lee, H. , Kim, Y. , van Helden, P.D. and Warren, R.M. (2006) Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT‐6 (esx) gene cluster regions. BMC Evolutionary Biology, 6, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel, M.I. , Sayes, F. , Simeone, R. , Majlessi, L. and Brosch, R. (2016) ESX secretion systems: mycobacterial evolution to counter host immunity. Nature Reviews Microbiology, 14, 677–691. [DOI] [PubMed] [Google Scholar]
- Houben, E.N.G. , Bestebroer, J. , Ummels, R. , Wilson, L. , Piersma, S.R. , Jimenez, C.R. , et al (2012) Composition of the type VII secretion system membrane complex. Molecular Microbiology, 86, 472–484. [DOI] [PubMed] [Google Scholar]
- Houben, E.N.G. , Korotkov, K.V. and Bitter, W. (2014) Take five—Type VII secretion systems of Mycobacteria. Biochimica et Biophysica Acta, 1843, 1707–1716. [DOI] [PubMed] [Google Scholar]
- Iantomasi, R. , Sali, M. , Cascioferro, A. , Palucci, I. , Zumbo, A. , Soldini, S. , et al (2012) PE_PGRS30 is required for the full virulence of Mycobacterium tuberculosis . Cellular Microbiology, 14, 356–367. [DOI] [PubMed] [Google Scholar]
- Korotkova, N. , Freire, D. , Phan, T.H. , Ummels, R. , Creekmore, C.C. , Evans, T.J. , et al (2014) Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25‐PPE41 dimer. Molecular Microbiology, 94, 367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, K.C. , de Chastellier, C. , Narayana, Y. , Bifani, P. , Brown, A.K. , Besra, G.S. , et al (2008) Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. Infection and Immunity, 76, 127–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol, Y.M. , Goetz, D.H. , Chan, K. , Shiloh, M.U. , Craik, C.S. and Cox, J.S. (2010) Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX‐1 secretion and virulence. Cell Host & Microbe, 7, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.H. , Ummels, R. , Bitter, W. and Houben, E.N.G. (2017) Identification of a substrate domain that determines system specificity in mycobacterial type VII secretion systems. Scientific Reports, 7, 42704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, T.H. , van Leeuwen, L.M. , Kuijl, C. , Ummels, R. , van Stempvoort, G. , Rubio‐Canalejas, A. , et al (2018) EspH is a hypervirulence factor for Mycobacterium marinum and essential for the secretion of the ESX‐1 substrates EspE and EspF. PLoS Pathogens, 14, e1007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen, C. , Panjikar, S. , Holton, S.J. , Wilmanns, M. and Song, Y.H. (2014) WXG100 protein superfamily consists of three subfamilies and exhibits an alpha‐helical C‐terminal conserved residue pattern. PLoS ONE, 9, e89313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poweleit, N. , Czudnochowski, N. , Nakagawa, R. , Trinidad, D.D. , Murphy, K.C. , Sassetti, C.M. , et al (2019) The structure of the endogenous ESX‐3 secretion system, eLife, 8, e52983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym, A.S. , Brodin, P. , Brosch, R. , Huerre, M. and Cole, S.T. (2002) Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti . Molecular Microbiology, 46, 709–717. [DOI] [PubMed] [Google Scholar]
- Renshaw, P.S. , Lightbody, K.L. , Veverka, V. , Muskett, F.W. , Kelly, G. , Frenkiel, T.A. , et al (2005) Structure and function of the complex formed by the tuberculosis virulence factors CFP‐10 and ESAT‐6. EMBO Journal, 24, 2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, O.S. , Dovala, D. , Li, X. , Connolly, L. , Bendebury, A. , Finer‐Moore, J. , et al (2015) Substrates control multimerization and activation of the multi‐domain ATPase motor of type VII secretion. Cell, 161, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini, A. , Boldrin, F. , Palu, G. and Manganelli, R. (2009) Characterization of a Mycobacterium tuberculosis ESX‐3 conditional mutant: essentiality and rescue by iron and zinc. Journal of Bacteriology, 191, 6340–6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist, M.S. , Unnikrishnan, M. , McConnell, M.J. , Borowsky, M. , Cheng, T.Y. , Siddiqi, N. , et al (2009) Mycobacterial Esx‐3 is required for mycobactin‐mediated iron acquisition. Proceedings of the National Academy of Sciences of the United States of America, 106, 18792–18797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone, R. , Bobard, A. , Lippmann, J. , Bitter, W. , Majlessi, L. , Brosch, R. , et al (2012) Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathogens, 8, e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley, S.A. , Raghavan, S. , Hwang, W.W. and Cox, J.S. (2003) Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proceedings of the National Academy of Sciences of the United States of America, 100, 13001–13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong, M. , Sawaya, M.R. , Wang, S. , Phillips, M. , Cascio, D. and Eisenberg, D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proceedings of the National Academy of Sciences of the United States of America, 103, 8060–8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel, N. , Hava, D. , Houben, D. , Fluitsma, D. , van Zon, M. , Pierson, J. , et al (2007) M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell, 129, 1287–1298. [DOI] [PubMed] [Google Scholar]
- van Winden, V.J. , Ummels, R. , Piersma, S.R. , Jimenez, C.R. , Korotkov, K.V. , Bitter, W. , et al (2016) Mycosins are required for the stabilization of the ESX‐1 and ESX‐5 Type VII secretion membrane complexes, mBio, 7:e01471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Winden, V.J.C. , Damen, M.P.M. , Ummels, R. , Bitter, W. and Houben, E.N.G. (2019) Protease domain and transmembrane domain of the mycosin protease determine system‐specific functioning in mycobacteria. Journal of Biological Chemistry, 294, 4806–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Zhou, K. , Yang, X. , Zhang, B. , Zhao, Y. , Xiao, Y. , et al (2019) Structural insights into substrate recognition by the type VII secretion system. Protein Cell, 11, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


