Abstract
The first documented avian influenza virus subtype H16N3 was isolated in 1975 and is currently detectable in many countries worldwide. However, the prevalence, biological characteristics and threat to humans of the avian influenza virus H16N3 subtype in China remain poorly understood. We performed avian influenza surveillance in major wild bird gatherings across the country from 2017 to 2019, resulting in the isolation of two H16N3 subtype influenza viruses. Phylogenetic analysis showed these viruses belong to the Eurasian lineage, and both viruses presented the characteristics of inter‐species reassortment. In addition, the two viruses exhibited limited growth capacity in MDCK and A549 cells. Receptor‐binding assays indicated that the two H16N3 viruses presented dual receptor‐binding profiles, being able to bind to both human and avian‐type receptors, where GBHG/NX/2/2018(H16N3) preferentially bound the avian‐type receptor, while GBHG/NX/1/2018(H16N3) showed greater binding to the human‐type receptor, even the mice virulence data showed the negative results. Segments from other species have been introduced into the H16N3 avian influenza virus, which may alter its pathogenicity and host tropism, potentially posing a threat to animal and human health in the future. Consequently, it is necessary to increase monitoring of the emergence and spread of avian influenza subtype H16N3 in wild birds.
Keywords: avian influenza virus, evolution, H16N3, reassortant
1. INTRODUCTION
Avian influenza viruses (AIVs) belong to the genus Orthomyxoviridae. With respect to surface glycoprotein haemagglutinin (HA) and neuraminidase (NA), 16 HAs (H1 to H16) and 9 NAs (N1 to N9) have been identified in birds (Fouchier et al., 2005; Webster, Bean, Gorman, Chambers, & Kawaoka, 1992). As a natural host, wild birds play an important role in the evolution of AIVs. Many AIVs adapt to wild birds, but with the exception of some highly pathogenic H5 and H7 subtypes of AIVs, they generally cause only mild or asymptomatic infections. However, AIVs can be spread to other birds or mammals or reassort with other AIVs during the seasonal migration of wild birds, posing a potential threat to poultry farming and human public health.
Avian influenza viruses are naturally maintained in wild, aquatic waterfowl, primarily ducks and shorebirds (orders Anseriformes and Charadriiformes). The H16 subtype is generally found in gulls (Verhagen et al., 2014), shedding high concentrations of viruses through intestinal epithelial cells that are then transmitted to other hosts (Hofle et al., 2012). Based on the amino acid residues of the HA protein receptor‐binding site, H16 AIV from gulls and the low pathogenic avian influenza (LPAIV) from ducks and geese have been shown to be significantly different (Fouchier et al., 2005). The H16N3 virus is more likely to attach to the human respiratory tract, cornea and conjunctiva than other AIV subtypes, raising concerns regarding the potential spread of influenza between humans and gulls (Lindskog et al., 2013).
During October 2018, we collected faeces samples from wild bird gatherings nationwide, becoming the first to isolate two H16N3 viruses in the Ningxia Hui Autonomous Region, western China. The molecular evolution characteristics, growth properties in different cell lines and receptor‐binding properties of the viruses were investigated.
2. MATERIALS AND METHODS
2.1. Sample collection
Faecal samples from wild birds were collected in the Shahu Wetland Ningxia Hui Autonomous Region in October 2018. The bird source of the faecal samples was determined on the basis of faecal shape and colour. The collected samples were placed in a phosphate‐buffered solution (pH 7.0) that was supplemented with penicillin, streptomycin and 10% glycerinum and were stored at 4°C for transportation.
2.2. Viral isolation and identification
The samples were vortexed and then centrifuged for 5 min at 3,000 g at 4°C, after which 0.2 ml of the supernatant from each sample was inoculated into the allantoic cavity of 9‐day‐old specific pathogen‐free (SPF) chicken embryos, and the allantoic fluid was harvested after 72 hr. The haemagglutination (HA) activity of the allantoic fluid was evaluated using an HA test. Subsequently, HA subtypes of influenza A virus were identified using an HA inhibition (HI) test with a panel of H1 to H16 subtype‐positive sera, while NA subtypes were identified by N1 to N9 subtype‐specific RT‐PCR primers.
2.3. Genome sequencing
Viral RNA was extracted from the HA‐positive samples using a commercial kit (Tiangen), according to the manufacturer's instructions. cDNA was synthesized from viral RNA by reverse transcription with a 12‐bp primer (5′‐AGCAAAAGCAGG‐3′), and the influenza virus segments were amplified using H16‐specific primers. The PCR products were purified using a PCR Purification Kit (Omega) and sequenced using an Applied Biosystems DNA analyzer.
Avian influenza viruse genome amplification and sequencing were performed according to a previously described protocol (Li et al., 2005).
2.4. Phylogenetic analysis
Sequences were assembled and edited using Lasergene 8.1 (DNASTAR). The coding sequence of each gene segment was then aligned using MAFFT (PB2, 2,280 bp; PB1, 2,274 bp; PA, 2,151 bp; HA, 1,695 bp; NP, 1,497 bp; NA, 1,410 bp; MP, 982 bp; and NS, 838 bp). All duplicate submissions and incomplete sequences were removed. The randomized accelerated maximum‐likelihood (RAxML7.2.8Alpha) program under the GTRGAMMA model with 1,000 bootstrap replicates was used to construct maximum‐likelihood phylogenies. The time of most relative common ancestor (tMRCA) was estimated using the package BEAST (v1.8.4) (Drummond & Rambaut, 2007), with the SRD06 nucleotide substitution model (Shapiro, Rambaut, & Drummond, 2006), an uncorrelated lognormal relaxed clock model (Matschiner et al., 2017). All chains were run for 50,000,000 generations with a 10% burn‐in, and the effective sample size (ESS) values in the results were greater than 200. Maximum clade credibility (MCC) trees were reconstructed with a 10% burn‐in and annotated using FigTree (v1.4.4).
2.5. Viral growth characteristics in different cell lines
We evaluated the growth characteristics of the virus in human type II alveolar epithelial cells (A549), Madin‐Darby canine kidney (MDCK) cells and specific pathogen‐free (SPF) chicken embryos according to the method provided by WHO. The TCID50 (50% tissue culture infective dose) and EID50 (50% egg infective dose) values were calculated using the Reed‐Muench formula.
2.6. Receptor‐binding preference of the H16N3 influenza viruses
Receptor‐binding specificity was analysed by the use of a solid‐phase binding assay as described previously (Zhang et al., 2013), using two different glycopolymers: α‐2,3‐siaylglycopolymer [Neu5Acα2‐3Galβ1‐4GlcNAcβ1‐pAP (para‐aminophenyl)‐α‐polyglutamic acid (α‐PGA)] and α‐2,6‐sialylglycopolymer [Neu5Acα2‐6Galβ1‐4GlcNAcβ1‐pAP(para‐aminophenyl)‐α‐polyglutamic acid (α‐PGA)]. For the viruses assayed in this study, chicken antiserum against a homologous virus was used as the primary antibody. A horseradish peroxidase (HRP)‐conjugated goat anti‐chicken antibody (Sigma‐Aldrich) was used as the secondary antibody, with absorbance measured at 490 nm. Two viruses, A/chicken/Hebei/3/2013(H5N2) and A/Sichuan/1/2009(H1N1), with homological sera that bind exclusively to (‐2, 3‐ and (‐2, 6‐linked sialic acid receptors, respectively, were used as controls.
2.7. Mouse experiment
Groups of six‐week‐old female BALB/c mice (Beijing Vital River Laboratories) were anaesthetized with CO2 and inoculated intranasally (i.n.) with 106.0 EID50 of test viruses in a volume of 50 μl. The mock control group were inoculated with 50 μl PBS. A/wild bird/Tianjin/2‐300/2018(H3N8) (WB/TJ/2‐300/2018(H3N8)) was used as positive control. Three mice were euthanized on day 3 p.i., and the nasal turbinates, lungs, kidneys, spleens and brains were collected for virus titration in 9‐day‐old chicken embryos. The remaining five mice in each group were monitored daily for 14 days for weight loss and survival.
3. RESULTS
3.1. Virus isolation
In 2018, epidemiological surveillance of wild bird avian influenza was performed by collecting 248 faecal samples at the Shahu Natural Reservoir in Ningxia Hui Autonomous, which resulted in the isolation of two H16N3 viruses, yielding an AIV isolation rate of 0.8% (2 of 248). This is the first time that the H16 subtype AIV has been isolated in China, and the isolates were named A/great black‐headed gull/Ningxia/1/2018(H16N3) and A/great black‐headed gull/Ningxia/2/2018(H16N3) (abbreviated GBHG/NX/1/2018(H16N3) and GBHG/NX/2/2018(H16N3), respectively). The whole genomes of the two viruses were fully sequenced and have been uploaded to the GISAID EpiFlu database (accession numbers: EPI1604431‐EPI1604480).
3.2. Molecular characterization
The HA cleavage motif of the two H16N3 viruses was a INER↓GLFG motif, suggesting that they are LPAIVs (Horimoto, Ito, Alexander, & Kawaoka, 1995). Interestingly, although there were no mutations in 160A and 226L in HA, our results indicated that the two viruses possessed a molecular marker in 228S associated with a switch in receptor specificity from the avian type to the human type and enhanced mammalian transmissibility (Chutinimitkul et al., 2010). The deletion in the stalk‐encoding region of the NA gene enhances the lethality of AIV in mice; however, in this study, the NA genes of the two H16N3 AIVs exhibited no stalk deletions. Some amino acid substitutions in PB2, including D256G, E627K and D701N, may contribute to increased virulence and transmission of influenza viruses in mammals (Bussey, Bousse, Desmet, Kim, & Takimoto, 2010; Schat et al., 2012), but none of these substitutions were observed in the two viruses. The N30D and T215A substitutions in the M1 protein and P42S, L98F and I101M substitutions in the NS1 protein were observed in the two H16N3 viruses, which are associated with increased H5N1 virus pathogenicity in mice (Fan et al., 2009). No L26F, V27A and S31N amino acid substitutions were detected in the M2 protein, indicating that the two viral strains are sensitive to amantadine inhibitors (Table 1).
Table 1.
Molecular analysis of influenza A subtype H16 viruses from wild birds
| Protein | Amino acid position/motif | Phenotypic consequences | GBHG/NX/1/2018(H16N3) | GBHG/NX/2/2018(H16N3) |
|---|---|---|---|---|
| PB2 | D256G | Enhanced polymerase activity, mammalian host adaptation | D | D |
| E627K | Enhanced polymerase activity and increased virulence in mice | E | E | |
| D701N | Enhanced polymerase activity and increased virulence in mice | D | D | |
| PB1 | I368V | Associated with H5 transmissibility in ferrets | I | I |
| PB1‐F2 | N66S | Increased virulence, replication efficiency and antiviral response in mice | N | S a |
| PA | A515T | Increased polymerase activity in mammalian cells | T | T |
| HA | Cleavage site | Polybasic cleavage motif sequence required for high pathogenicity of avian influenza viruses | INER↓GLF | INER↓GLF |
| T160A (H3 numbering) | Associated with H5 transmissibility in ferrets | K | K | |
| Q226L (H3 numbering) | Human‐type receptor binding; associated with H5 transmissibility in ferrets | Q | Q | |
| G228S (H3 numbering) | Human‐type receptor binding; associated with H5 transmissibility in ferrets | S | S | |
| NA | Stalk deletion | Increased virulence in mice | No deletion | No deletion |
| E119A/G/V (N2 numbering) | Reduced susceptibility to zanamivir, oseltamivir and/or peramivir | E | E | |
| H274Y/R (N2 numbering) | Reduced susceptibility to oseltamivir and peramivir | H | H | |
| M1 | N30D | Increased virulence in mice | D | D |
| T215A | Increased virulence in mice | A | A | |
| M2 | L26F | Amantadine resistance | L | L |
| V27A | Amantadine resistance | V | V | |
| S31N/G | Amantadine resistance | S | S | |
| NS1 | 80‐84 deletion | Increased virulence in mice | No deletion | No deletion |
| P42S | Increased virulence in mice | S | S | |
| L98F | Increased virulence in mice | F | F | |
| I101M | Increased virulence in mice | M | M |
Underlining indicates molecular features that are associated with virulence in mammals or the transmissibility of influenza virus and were detected in the H16 isolates in this study.
3.3. Homology analysis and phylogenetic tree
To investigate the phylogenetic relationships and homology of the two viruses, we sequenced their entire genomes and constructed eight individual maximum‐likelihood phylogenetic trees, where the closest strains were identified based on nucleotide levels by alignment in the GISAID EpiFlu database. The identity of seven segments (HA, NA, PB2, PA, NP, M and NS) was higher than 98%, whereas the PB1 homology between the two viruses was 88% (Table 2). The HA genes of the two H16N3 viruses had the highest homology with A/black‐headed gull/Netherlands/37/2011(H16N3), indicating that they belong to the Eurasian lineage (Figure 1a). The NA genes of the two viruses were most likely derived from Anseriformes (duck) viruses (Figure S1). The internal genes of PB2, PA, M and NS of two viruses in the study were derived from the gull‐originated H13 virus (Figure S1). Interestingly, the PB1 gene of the GBHG/NX/2/2018(H16N3) virus was from A/duck/Hubei/ZYSYG3/2015(H6N2) and is a ternary recombinant virus from gull‐, swan‐ and duck‐originated viruses (Figure 1b).
Table 2.
Viruses with sequence homology in the GISAID EpiFlu database
| Virus | Segment | Position | Virus with the highest percentage of nucleotide identity | Segment ID | Homology (%) |
|---|---|---|---|---|---|
| GBHG/NX/1/2018(H16N3) | PB2 | 1‐2280 | A/duck/China/Weihai/2017(H13N8) | EPI1565255 | 98 |
| PB1 | 1‐2274 | A/Armenian gull/Republic of Georgia/1/2012(H13N6) | EPI617776 | 97 | |
| PA | 1‐2151 | A/black‐tailed gull/Weihai/115/2016(H13N2) | EPI1223772 | 98 | |
| HA | 1‐1695 | A/black‐headed gull/Netherlands/37/2011(H16N3) | EPI1013524 | 95 | |
| NP | 1‐1497 | A/swan/East China/SD588/2015(H13N8) | EPI1057471 | 98 | |
| NA | 1‐1410 | A/duck/Hokkaido/WZ82/2013(H16N3) | EPI531690 | 96 | |
| M | 1‐959 | A/common gull/Altai/811/2011(H13N2) | EPI467831 | 97 | |
| NS | 1‐838 | A/black‐headed gull/Netherlands/19/2009(H16N3) | EPI1014718 | 97 | |
| GBHG/NX/2/2018(H16N3) | PB2 | 1‐2280 | A/duck/China/Weihai/2017(H13N8) | EPI1565255 | 98 |
| PB1 | 1‐2274 | (A/duck/Hubei/ZYSYG3/2015(H6N2) | EPI942215 | 98 | |
| PA | 1‐2151 | A/black‐tailed gull/Weihai/115/2016(H13N2) | EPI1223772 | 99 | |
| HA | 1‐1695 | A/black‐headed gull/Netherlands/37/2011(H16N3) | EPI1013524 | 95 | |
| NP | 1‐1497 | A/swan/East China/SD588/2015(H13N8) | EPI1057471 | 98 | |
| NA | 1‐1410 | A/duck/Hokkaido/WZ82/2013(H16N3) | EPI531690 | 96 | |
| M | 1‐959 | A/common gull/Altai/811/2011(H13N2) | EPI467831 | 97 | |
| NS | 1‐838 | A/black‐headed gull/Netherlands/19/2009(H16N3) | EPI1014718 | 97 |
Figure 1.
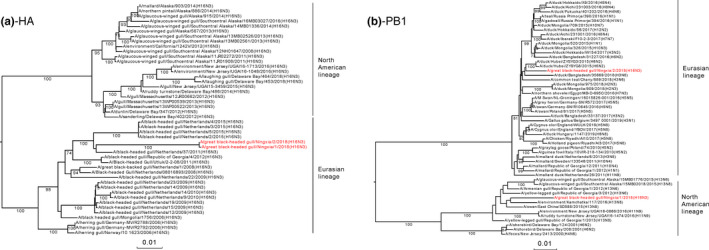
Maximum‐likelihood phylogenetic analysis of eight H16N3 virus segments: (a) HA, (b) NA, (c) PB2, (d) PB1, (e) PA, (f) NP, (g) M and (h) NS. The nucleotide sequences of the viruses listed in black were downloaded from the GISAID EpiFlu database. Reference sequences of viruses isolated from wild birds in Eurasia or the Americas are indicated with blue and red stars, respectively. The viruses listed in red were sequenced in this study [Colour figure can be viewed at wileyonlinelibrary.com]
BEAST was used to determine the tMRCA and mean evolution rate of the two H16N3 viruses in this study. The results suggested that the reassortment events that occurred to produce the two H16N3 viruses happened between July 2010 and September 2016 (Table 3, Figure 2, Figure S2); the HA genes originated from the same AIV subtype of black‐headed gull in October 2011 (Figure 2a); the NA genes were introduced from an H16N3 virus of duck origin in November 2011 (Figure S2); the two viruses obtained PB2, PA, NP and M genes from an H13 subtype virus of gull origin from mid‐2011 to late 2016 (Figure S2); and their NS genes were acquired from a black‐headed gull H16N3 virus as early as June 2010 (Figure S2). Unlike the PB1 gene of the GBHG/NX/1/2018(H16N3) virus from the H13 subtype AIV, the PB1 gene of the GBHG/NX/2/2018(H16N3) virus descended from the H6N2 virus of duck origin in March 2014 (Figure 2b). The two virus reassortant patterns and the introduction time of each segment are shown in Figure 3. From observations of the mean evolution rates of the eight gene segments of the two H16N3 viruses, the evolution of the HA gene was the fastest, while that of the PB2 gene was the most conservative and showed obvious hysteresis (Table 3).
Table 3.
Evolution rate and time of the most recent common ancestor for each of the eight segments
| Virus | Segment | Mean evolution rate (substitution/site/year) | 95% HPD interval | Most recent common ancestor | tMRCA | 95% HPD interval | Posterior probability |
|---|---|---|---|---|---|---|---|
| GBHG/NX/1/2018(H16N3) | PB2 | 6.5072E−3 | [2.9681E−3, 1.1406E−2] | A/swan/East_China/SD588/2015(H13N8) | Jun. 2016 | [Sep. 2015, Dec. 2016] | 1 |
| PB1 | 2.3845E−3 | [9.2632E−4, 3.8686E−3] | A/environment/Kamchatka/117/2016(H13N8) | Jul. 2013 | [Oct. 2014, Jun. 2012] | 0.78 | |
| PA | 1.5523E−3 | [4.951E−5, 4.4885E−3] | A/black‐tailed_gull/Weihai/115/2016(H13N2) | Sep. 2016 | [May 2016, Dec. 2016] | 0.92 | |
| HA | 1.2698E−3 | [7.152E−5, 3.2663E−3] | Netherlands black‐headed gull H16N3 stains | Oct. 2012 | [Apr. 2011, Apr. 2014] | 0.87 | |
| NP | 3.726E−3 | [2.0019E−3, 5.4001E−3] | A/swan/East_China/SD588/2015(H13N8) | Oct. 2015 | [Jun. 2015, Dec. 2015] | 1 | |
| NA | 4.3934E−3 | [1.5331E−3, 7.6825E−3] | A/duck/Hokkaido/WZ82/2013(H16N3) | Nov. 2011 | [Mar. 2011, Jul. 2012] | 1 | |
| M | 3.7627E−3 | [8.0672E−4, 8.265E−3] | A/common_gull/Altai/812/2011(H13N2) | Jun. 2011 | [Feb. 2011, Aug. 2011] | 1 | |
| NS | 2.5799E−3 | [1.6951E−4, 4.5545E−3] | A/black‐headed_gull/Utluk/2‐2‐08/2011(H16N3) | Jul. 2010 | [Nov. 2009, Apr. 2011] | 0.83 | |
| GBHG/NX/2/2018(H16N3) | PB2 | 6.5072E−3 | [2.9681E−3, 1.1406E−2] | A/swan/East_China/SD588/2015(H13N8) | Dec. 2014 | [Aug. 2013, Feb. 2016] | 1 |
| PB1 | 2.3622E−3 | [9.7416E−4, 3.8323E−3] | A/duck/Hubei/ZYSYG3/2015(H6N2) | Mar. 2014 | [Mar. 2013, Jan. 2015] | 0.98 | |
| PA | 1.5523E−3 | [4.951E−5, 4.4885E−3] | A/black‐tailed_gull/Weihai/115/2016(H13N2) | Sep. 2016 | [May. 2016, Dec. 2016] | 0.92 | |
| HA | 1.2698E−3 | [7.152E−5, 3.2663E−3] | Netherlands black‐headed gull H16N3 stains | Oct. 2012 | [Apr. 2011, Apr. 2014] | 0.87 | |
| NP | 3.726E−3 | [2.0019E−3, 5.4001E−3] | A/swan/East_China/SD588/2015(H13N8) | Oct. 2015 | [Jun. 2015, Dec. 2015] | 1 | |
| NA | 4.3934E−3 | [1.5331E−3, 7.6825E−3] | A/duck/Hokkaido/WZ82/2013(H16N3) | Nov. 2011 | [Mar. 2011, Jul. 2012] | 1 | |
| M | 3.7627E−3 | [8.0672E−4, 8.265E−3] | A/common_gull/Altai/812/2011(H13N2) | Jun. 2011 | [Feb. 2011, Aug. 2011] | 1 | |
| NS | 2.5799E−3 | [1.6951E−4, 4.5545E−3] | A/black‐headed_gull/Utlu/2‐2‐08/2011(H16N3) | Jul. 2010 | [Nov. 2009, Apr. 2011] | 0.83 |
Abbreviations: HPD, highest posterior density; tMRCA, time of most recent common ancestor.
Figure 2.
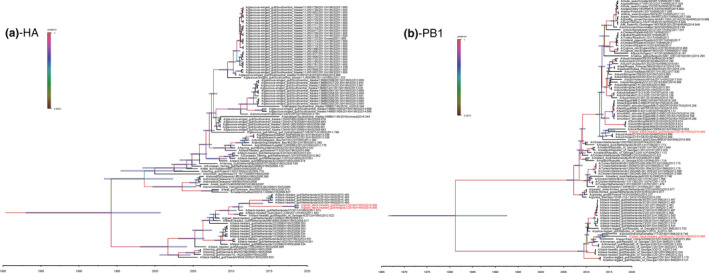
Maximum clade credibility trees of the coding sequences of eight segments. Node bars indicate 95% highest posterior density (HPD) of the node height. The two H16N3 viruses isolated in this study are coloured in red. Each branch was coloured by posterior probability. The segments shown are (a) HA, (b) NA, (c) PB2, (d) PB1, (e) PA, (f) NP, (g) M and (h) NS [Colour figure can be viewed at wileyonlinelibrary.com]
Figure 3.
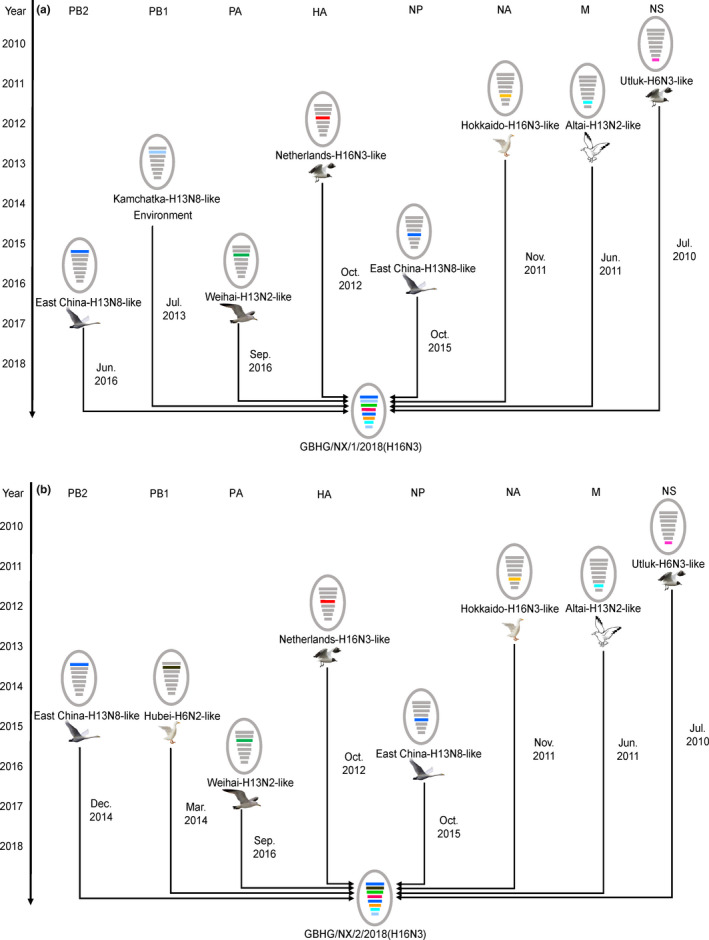
A simplified schematic showing the putative genomic composition of H16N3 viruses. The eight gene segments (from top to bottom) in each virus are polymerase basic protein 2 (PB2), polymerase basic protein 1 (PB1), polymerase acidic protein (PA), haemagglutinin (HA), nucleocapsid protein (NP), neuraminidase (NA), matrix protein (M) and non‐structural protein (NS). Each colour represents a separate virus background. The illustration is based on the nucleotide‐distance comparison and phylogenetic analysis [Colour figure can be viewed at wileyonlinelibrary.com]
3.4. Viral titration on different types of cultures
To evaluate the growth characteristics of the two viruses, they were titrated on different cell lines: A549 cells, MDCK cells and SPF chicken embryos. The TCID50 and EID50 values of the two H16N3 viruses on A549 cells, MDCK cells and SPF chicken embryos presented little difference (Table 4). The TCID50 and EID50 values of GBHG/NX/1/2018(H16N3) in all three cultures were higher than those observed for GBHG/NX/2/2018(H16N3), revealing that GBHG/NX/1/2018(H16N3) replication is more efficient. Moreover, the preferential replication in eggs of the two H16N3 viruses indicated their avian influenza properties.
Table 4.
H16N3 virus titration in different types of cultures
| Culture | Virus titration (TCID50/EID50/100μl) | |
|---|---|---|
| GBHG/NX/1/2018(H16N3) | GBHG/NX/2/2018(H16N3) | |
| MDCK | 101.50 | 101.40 |
| A549 | 101.75 | 101.50 |
| SPF chicken embryos | 106.23 | 105.68 |
3.5. Receptor binding
The data revealed that two H16N3 viruses presented dual receptor‐binding profiles, as both bound to human and avian‐type receptors, with GBHG/NX/1/2018(H16N3) preferentially binding α2,6‐siaylglycopolymer (human‐type receptor), while GBHG/NX/2/2018(H16N3) showed greater binding to α2,3‐siaylglycopolymer (avian‐type receptor) (Figure 4).
Figure 4.
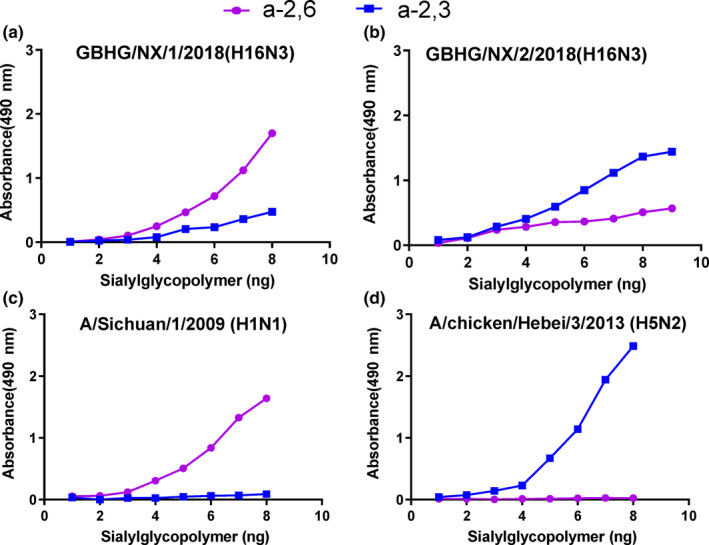
Characterization of the receptor‐binding properties of H16N3 viruses. The binding of the viruses to two different biotinylated glycans (α‐2,3 glycan, blue; α‐2,6 glycan, pink) was tested. The data shown are the means of three repeats; the error bars indicate the standard deviations. (a) GBHG/NX/1/2018(H16N3), (b) GBHG/NX/2/2018(H16N3), (c) A/chicken/Hebei/3/2013(H5N2) and (d) A/Sichuan/1/2009(H1N1) [Colour figure can be viewed at wileyonlinelibrary.com]
3.6. Mouse experiments
To evaluate the pathogenicity of A (H16N3) isolates in mammals, BALB/c mice were infected intranasally with the viruses. The results are shown in Figure 5. Weight change is shown in Figure 5a: the two H16N3 viruses in the study did not cause mice weight change; two isolates did not replicate in any investigated tissues either; as shown in Figure 5b, it implied that the viruses were none pathogenic to mice.
Figure 5.
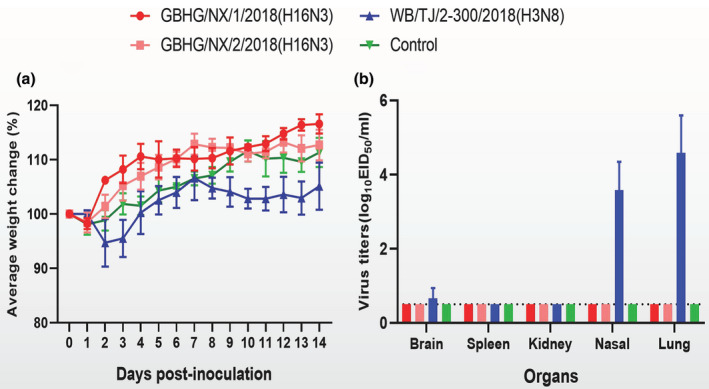
Body weight (a) and replication (b) in BALB/c mice. Body weights of mice were observed over 14 days after infection. Virus titres in organs of mice on day 3 p.i. with 106EID50 of test virus. Data shown are the mean titres from three mice; the error bars indicate the standard deviations [Colour figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
Since the H16 subtype AIV was first identified in 2005 (Fouchier et al., 2005), it has been detected in many countries worldwide, with the greatest prevalence observed in Norway and Netherlands (Figure 6). H16 subtype AIV is genetically close to H13 subtype AIV, and they are well adapted to replicate in gulls, typically causing mild or asymptomatic infections (Tonnessen, Hauge, et al., 2013; Tonnessen, Kristoffersen, et al., 2013). During the monitoring of black‐headed gulls in the Netherlands from 2006 to 2010, the prevalence of H13 and H16 AIV subtypes in birds during the second half of the breeding season in the first year was observed to be as high as 72% per week (Verhagen et al., 2014). Another study reported that H16N3 was the most common subtype of AIV in the black‐legged kittiwake, which is the most numerous gull species in the world (Toennessen et al., 2011).
Figure 6.
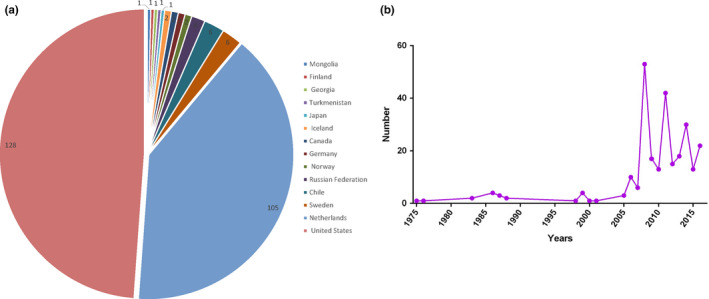
Pie chart showing all H16 viruses that have been isolated worldwide since 1975 (a) and a line chart showing the number of H16 virus isolates obtained per year (b) [Colour figure can be viewed at wileyonlinelibrary.com]
Geographically, influenza A viruses can typically be divided into Eurasian and North American lineages. Viruses of the two lineages generally exhibit little segment exchange, but these geographical limitations appear to be more easily overcome by some species that can migrate long distances, such as Charadriiformes (Dusek et al., 2014). In 2008, an H16N3 virus was isolated from a black‐backed gull in Newfoundland and was observed to contain genes from Eurasian and North American gulls (Wille, Robertson, Whitney, Bishop, et al., 2011; Wille, Robertson, Whitney, Ojkic, et al., 2011). Further study showed that the North American genes of this H16N3 virus originated from Northern Europe and Central Asia. These results demonstrated that long‐distance migrating gulls play an important role in the transfer of AIV genes from Eurasia to America (Wille, Robertson, Whitney, Bishop, et al., 2011; Wille, Robertson, Whitney, Ojkic, et al., 2011).
The H16 subtype AIV is primarily associated with gulls, which may be due to the specific characteristics of the proteins encoded by the internal genes of the virus (Tonnessen, Hauge, et al., 2013; Tonnessen, Kristoffersen, et al., 2013). However, due to intraspecific recombination, the host restrictions and pathogenicity of H16N3 may change, and there are already some reports of intraspecific recombination between wild ducks and gulls. The results of the phylogenetic analysis showed that the PB1 and NP genes of GBHG/NX/2/2018(H16N3) are related to the duck (A/duck/Hubei/ZYSYG3/2015(H6N2)) and swan (A/swan/East China/SD588/2015(H13N8)), respectively, which is seen as an evidence of inter‐species reassortment.
The titration of the virus in different cell lines indicates the growth and infection abilities of a virus. In our study, the two H16N3 viruses presented low TCID50 values in A549 and MDCK cells, likely because these cell lines are of mammalian origin. However, the replication efficiency of the two viruses in chicken embryos was high, indicating chicken embryos are habitable to these two viruses.
Receptor‐binding preference has important implications for influenza virus replication and transmission (Gao et al., 2009; Zhang et al., 2013). The change in receptor‐binding preference from α‐2,3‐linked sialic acids (avian‐type receptors) to α‐2,6‐linked sialic acids (human‐type receptors) is believed to be a prerequisite for an avian influenza virus to be transmitted between humans. In our study, the two H16N3 viruses showed dual receptor‐binding properties, showing that they have a capacity to attach to both human and avian receptors. Compared with GBHG/NX/2/2018(H16N3), GBHG/NX/1/2018(H16N3) showed preferential binding to human receptors, which may pose a potential threat to public health.
5. CONCLUSION
Although the H16 subtype avian influenza virus was isolated in 2005, its prevalence, biological characteristics and threat to humans are still poorly understood in China. In this study, for the first time, we isolated two H16N3 subtype influenza viruses from the Ningxia Hui Autonomous Region in China, with an isolation rate of 0.8% (2 of 248). None pathogenicity to mammalian model and mammalian cell‐restricted growth suggest that these viruses are less harmful towards mammals than other viruses. However, a phylogenetic analysis of the two viruses indicated the occurrence of inter‐species reassortment. Furthermore, segments from other species are continuously introduced into the H16N3 AIV, which may alter its pathogenicity and host tropism. In addition, the two H16N3 viruses showed dual receptor‐binding properties, even the data of vivo test in mammalian model showed the pathogenicity to mice was negative, GBHG/NX/1/2018(H16N3) preferentially binding the human receptor, which may pose a potential threat to animal and human health in the future. Thus, it is necessary to increase monitoring of the emergence and spread of avian influenza virus subtype H16N3 in wild birds.
ETHICAL APPROVAL
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People's Republic of China. The protocols were approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS).
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
Supporting information
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (31521005) and the Natural Science Foundation of Heilongjiang Province of China (ZD2018007). We thank the State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China, for providing a platform for the experiment. In addition, we thank all the individuals who assisted in the work described in this study.
Li Y, Li M, Tian J, et al. Characteristics of the first H16N3 subtype influenza A viruses isolated in western China. Transbound Emerg Dis. 2020;67:1677–1687. 10.1111/tbed.13511
REFERENCES
- Bussey, K. A. , Bousse, T. L. , Desmet, E. A. , Kim, B. , & Takimoto, T. (2010). PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. Journal of Virology, 84, 4395–4406. 10.1128/JVI.02642-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chutinimitkul, S. , van Riel, D. , Munster, V. J. , van den Brand, J. M. , Rimmelzwaan, G. F. , Kuiken, T. , … de Wit, E. (2010). In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. Journal of Virology, 84, 6825–6833. 10.1128/JVI.02737-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond, A. J. , & Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology, 7, 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusek, R. J. , Hallgrimsson, G. T. , Ip, H. S. , Jonsson, J. E. , Sreevatsan, S. , Nashold, S. W. , … Hall, J. S. (2014). North Atlantic migratory bird flyways provide routes for intercontinental movement of avian influenza viruses. PLoS ONE, 9, e92075 10.1371/journal.pone.0092075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, S. , Deng, G. , Song, J. , Tian, G. , Suo, Y. , Jiang, Y. , … Chen, H. (2009). Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology, 384, 28–32. 10.1016/j.virol.2008.11.044 [DOI] [PubMed] [Google Scholar]
- Fouchier, R. A. , Munster, V. , Wallensten, A. , Bestebroer, T. M. , Herfst, S. , Smith, D. , … Osterhaus, A. D. (2005). Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black‐headed gulls. Journal of Virology, 79, 2814–2822. 10.1128/JVI.79.5.2814-2822.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y. , Zhang, Y. , Shinya, K. , Deng, G. , Jiang, Y. , Li, Z. … Chen, H. (2009). Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog, 5, e1000709 10.1371/journal.ppat.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofle, U. , Van de Bildt, M. W. , Leijten, L. M. , Van Amerongen, G. , Verhagen, J. H. , Fouchier, R. A. , … Kuiken, T. (2012). Tissue tropism and pathology of natural influenza virus infection in black‐headed gulls (Chroicocephalus ridibundus). Avian Pathology, 41, 547–553. 10.1080/03079457.2012.744447 [DOI] [PubMed] [Google Scholar]
- Horimoto, T. , Ito, T. , Alexander, D. J. , & Kawaoka, Y. (1995). Cleavability of hemagglutinin from an extremely virulent strain of avian influenza virus containing a unique cleavage site sequence. Journal of Veterinary Medical Science, 57, 927–930. 10.1292/jvms.57.927 [DOI] [PubMed] [Google Scholar]
- Li, C. , Yu, K. , Tian, G. , Yu, D. , Liu, L. , Jing, B. , … Chen, H. (2005). Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology, 340, 70–83. 10.1016/j.virol.2005.06.025 [DOI] [PubMed] [Google Scholar]
- Lindskog, C. , Ellstrom, P. , Olsen, B. , Ponten, F. , van Riel, D. , Munster, V. J. , … Jourdain, E. (2013). European H16N3 gull influenza virus attaches to the human respiratory tract and eye. PLoS ONE, 8, e60757 10.1371/journal.pone.0060757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner, M. , Musilova, Z. , Barth, J. M. I. , Starostova, Z. , Salzburger, W. , Steel, M. , & Bouckaert, R. (2017). Bayesian phylogenetic estimation of clade ages supports trans‐atlantic dispersal of cichlid fishes. Systematic Biology, 66, 3–22. 10.1093/sysbio/syw076 [DOI] [PubMed] [Google Scholar]
- Schat, K. A. , Bingham, J. , Butler, J. M. , Chen, L. M. , Lowther, S. , Crowley, T. M. , … Lowenthal, J. W. (2012). Role of position 627 of PB2 and the multibasic cleavage site of the hemagglutinin in the virulence of H5N1 avian influenza virus in chickens and ducks. PLoS ONE, 7, e30960 10.1371/journal.pone.0030960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, B. , Rambaut, A. , & Drummond, A. J. (2006). Choosing appropriate substitution models for the phylogenetic analysis of protein‐coding sequences. Molecular Biology and Evolution, 23, 7–9. 10.1093/molbev/msj021 [DOI] [PubMed] [Google Scholar]
- Toennessen, R. , Germundsson, A. , Jonassen, C. M. , Haugen, I. , Berg, K. , Barrett, R. T. , & Rimstad, E. (2011). Virological and serological surveillance for type A influenza in the black‐legged kittiwake (Rissa tridactyla). Virology Journal, 8(1), 21 10.1186/1743-422X-8-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnessen, R. , Hauge, A. G. , Hansen, E. F. , Rimstad, E. , & Jonassen, C. M. (2013). Host restrictions of avian influenza viruses: In silico analysis of H13 and H16 specific signatures in the internal proteins. PLoS ONE, 8, e63270 10.1371/journal.pone.0063270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonnessen, R. , Kristoffersen, A. B. , Jonassen, C. M. , Hjortaas, M. J. , Hansen, E. F. , Rimstad, E. , & Hauge, A. G. (2013). Molecular and epidemiological characterization of avian influenza viruses from gulls and dabbling ducks in Norway. Virology Journal, 10, 112 10.1186/1743-422X-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen, J. H. , Majoor, F. , Lexmond, P. , Vuong, O. , Kasemir, G. , Lutterop, D. , … Kuiken, T. (2014). Epidemiology of influenza A virus among black‐headed gulls, the Netherlands, 2006–2010. Emerging Infectious Diseases, 20, 138–141. 10.3201/eid2001.130984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, R. G. , Bean, W. J. , Gorman, O. T. , Chambers, T. M. , & Kawaoka, Y. (1992). Evolution and ecology of influenza A viruses. Microbiological Reviews, 56, 152–179. 10.1128/MMBR.56.1.152-179.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille, M. , Robertson, G. J. , Whitney, H. , Bishop, M. A. , Runstadler, J. A. , & Lang, A. S. (2011). Extensive geographic mosaicism in avian influenza viruses from gulls in the northern hemisphere. PLoS ONE, 6, e20664 10.1371/journal.pone.0020664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille, M. , Robertson, G. J. , Whitney, H. , Ojkic, D. , & Lang, A. S. (2011). Reassortment of American and Eurasian genes in an influenza A virus isolated from a great black‐backed gull (Larus marinus), a species demonstrated to move between these regions. Archives of Virology, 156, 107–115. 10.1007/s00705-010-0839-1 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Shi, J. , Deng, G. , Guo, J. , Zeng, X. , He, X. , … Chen, H. (2013). H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science, 341, 410–414. 10.1126/science.1240532 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


