Abstract
Purpose:
Poor outcomes for ovarian cancer patients relate to dormant, drug resistant cancer cells that survive following primary surgery and chemotherapy. Ovarian cancer (OvCa) cells persist in poorly vascularized scars on the peritoneal surface and depend upon autophagy to survive nutrient deprivation. We have sought drugs that target autophagic cancer cells selectively to eliminate residual disease.
Experimental Design:
Using unbiased siRNA screens, we found that knockdown of anaplastic lymphoma kinase (ALK) reduced survival of autophagic OvCa cells. Small molecule ALK inhibitors were evaluated for their selective toxicity against autophagic OvCa cell lines and xenografts. Autophagy was induced by re-expression of DIRAS3 or serum starvation and evaluated with western blot analysis, fluorescence imaging and transmission electron microscopy. Signaling pathways required for crizotinib-induced apoptosis of autophagic cells were explored with flow cytometric analysis, western blot analysis, shRNA knockdown of autophagic proteins, and small molecule inhibitors of STAT3 and BCL-2.
Results:
Induction of autophagy by re-expression of DIRAS3 or serum starvation in multiple OvCa cell lines significantly reduced the IC50 of crizotinib and other ALK inhibitors. In two human OvCa xenograft models, the DIRAS3 expressing tumors treated with crizotinib had significantly decreased tumor burden and long-term survival in 67-79% of mice. Crizotinib treatment of autophagic cancer cells further enhanced autophagy and induced autophagy-mediated apoptosis by decreasing p-STAT3 and BCL-2 signaling.
Conclusions:
Crizotinib might eliminate dormant, autophagic drug resistant OvCa cells that remain after conventional cytoreductive surgery and combination chemotherapy. A clinical trial of ALK inhibitors as maintenance therapy following second look operations should be seriously considered.
Keywords: Autophagy, Crizotinib, Anaplastic Lymphoma Kinase (ALK), dormancy, autophagy, ovarian cancer, DIRAS3
INTRODUCTION
Five-year survival for advanced stage OvCa has risen over the last four decades to ~50% due to advances in cytoreductive surgery and combination chemotherapy (2). Despite improvements in treatment, the disease still proves lethal in >60% of cases (1, 2). One of the most important factors contributing to poor patient outcomes is the ability of metastatic, drug-resistant OvCa cells to survive initial therapy, often remaining dormant on the peritoneal surface for years before growing progressively to kill the patient.
Dormancy of metastatic cancer cells has been attributed to the persistence of small deposits of metastatic cancer cells that are 1) either not cycling or undergoing balanced cell proliferation and death, 2) failing to develop microvasculature, and 3) evading the host immune response (3-4). In OvCa, second look surveillance operations have been performed in the past following primary surgery and chemotherapy where a complete clinical remission has been documented with negative imaging and normalization of the serum biomarker CA125. Small deposits of metastatic residual disease have been found in poorly vascularized nodules on the surface of the parietal or visceral peritoneum in approximately half of the patients (5). Previous studies reported that the outgrowth of clinically apparent cancer will occur in 85% of patients with positive second look operations within a median of 16 months, but disease can recur after intervals as long as 10 years (5). Given the years required to detect recurrence in some patients after positive second look operations, the drug resistant OvCa cells that remain after surgery and chemotherapy evidently do not grow exponentially and at least a fraction can be considered dormant.
Survival of dormant cancer cells in a hypovascular, nutrient-poor environment can depend upon autophagy. In greater than 80% of positive second look operations, persistent drug resistant OvCa cells exhibit widespread autophagy evidenced by punctate immunostaining for LC3 (microtubule-associated protein 1A/1B light chain 3) in autophagosomes (6). By contrast, autophagy is observed in only about 20% of well-vascularized primary cytoreductive surgical specimens obtained from the same patients prior to chemotherapy (6). Autophagy, a natural process which recycles worn and damaged organelles or proteins, has long been implicated in cancer cell survival (6-7). Upregulation of autophagy provides energy through catabolism of amino acids and fatty acids that allows cancer cells to survive in a nutrient poor environment (6-7). Interestingly, a similar fraction of positive second look specimens contain punctate deposits of DIRAS3 (ARHI) which has been shown to regulate autophagy and to co-localize with LC3 in the membrane of autophagosomes (6).
We have previously demonstrated that DIRAS3 induces both autophagy and tumor dormancy in human OvCa xenografts (6). DIRAS3 is an imprinted tumor suppressor gene that is downregulated in 60% of primary ovarian cancers and its downregulation is associated with shortened progression free survival (6, 8-19). Re-expression of DIRAS3 in OvCa cells upregulates autophagy and induces tumor dormancy by inhibiting proliferation, motility, angiogenesis and xenograft growth (6, 8-19). In earlier studies with a DIRAS3-inducible model for OvCa dormancy, we had found that functional inhibition of autophagy with chloroquine markedly delayed outgrowth of dormant, autophagic human OvCa xenografts after downregulation of DIRAS3, consistent with a role for autophagy in facilitating survival of dormant cancer cells in a nutrient-poor sparsely-vascularized environment.
Drugs that target autophagic cancer cells might eliminate residual disease, particularly in OvCa where cancer cells destined to recur are undergoing autophagy. Using unbiased siRNA screens, we found that knockdown of anaplastic lymphoma kinase (ALK) reduced survival of autophagic OvCa cells. Induction of autophagy by upregulation of DIRAS3 or serum starvation in multiple OvCa cell lines significantly enhanced the activity of crizotinib and other ALK inhibitors. In two human OvCa xenograft models, treatment of dormant, autophagic grafts with crizotinb produced long-term survival in a fraction of mice. Crizotinib treatment of dormant, autophagic cancer cells further enhanced autophagy and induced apoptosis by decreasing p-STAT3 and BCL-2 signaling.
RESULTS
Knockdown of ALK with siRNA kills autophagic OvCa cells selectively.
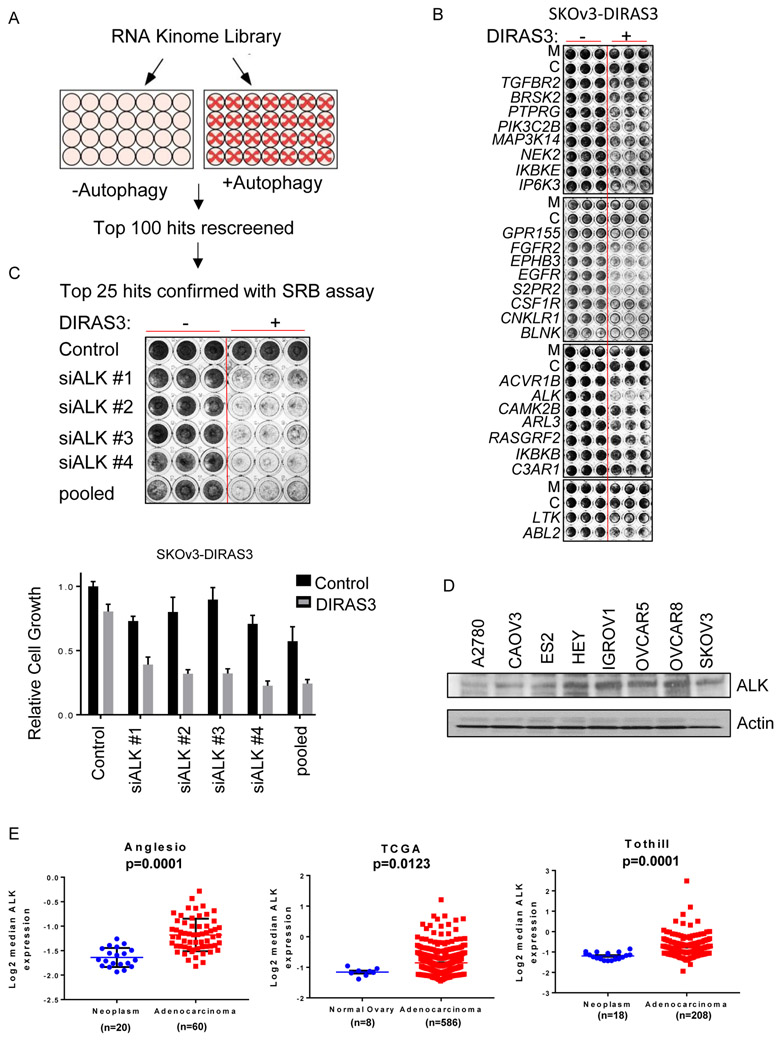
A high throughput screen was performed in triplicate using a small interfering RNA (siRNA) library that targeted 570 kinases and G-protein coupled receptors to identify targets that regulate the survival of SKOv3 OvCa cells which are undergoing autophagy induced by the re-expression of DIRAS3 or by amino acid starvation (Figure 1A). This library includes a pool of 4 distinct siRNA targeting each gene. This screen was repeated twice in an unbiased manner. The 100 targets which significantly (p<0.05) reduced viability selectively in the autophagic cancer cells were rescreened. Cytotoxicity of the top 25 targets was confirmed and the outcome validated by deconvoluting each pool and testing each of the siRNAs independently in a sulforhodamine B (SRB) assay in SKOV3 and OVCAR8 Cells (Figure 1B, Supplemental Figure 1A) (20). Consistent with the high throughput screen results, the siRNAs targeting ALK resulted in the greatest cytotoxicity for autophagic cells, when compared to non-autophagic cells of the same line. Four unique siRNAs targeting ALK each induced cytotoxicity in autophagic cancer cells, confirming that ALK is responsible for the selective cytotoxicity (Figure 1C).
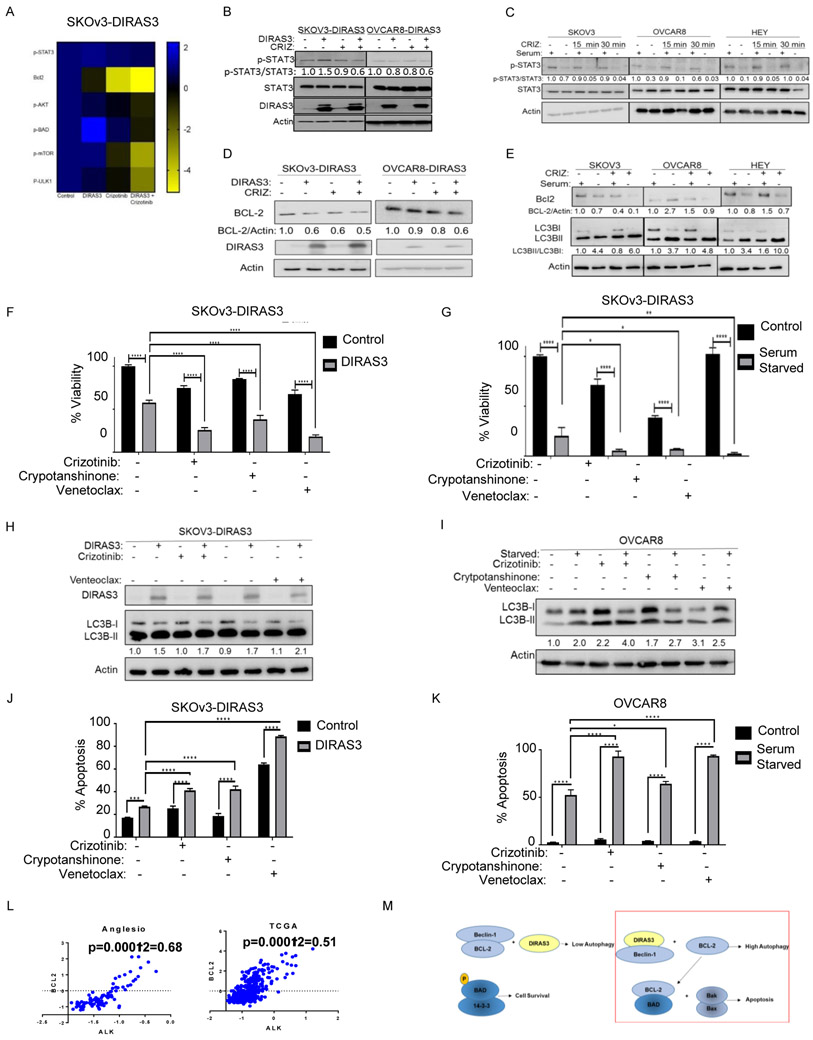
Figure 1. ALK selectively eliminates autophagic OvCa cells.
A. High throughput screen using an siRNA library that targeted 570 kinases and G-protein coupled receptors to identify proteins that regulate the survival of SKOv3 cells that are undergoing autophagy. After 24 hrs transfection, ± doxycycline (1 μg/μl) was added for 72 hrs to SKOV3-DIRAS3 cells. B. The top 25 hits were confirmed in an independent SRB assay using SKOv3-DIRAS3. Cells were inversely transfected with siRNA, allowed to adhere overnight then treated ± doxycycline (1 ug/mL). After 72 hrs, an SRB assay was performed. C. Four unique siRNAs targeting ALK were used to determine the specificity of the pooled knockdown. D. Lysates from OvCa cell lines were probed from endogenous ALK expression on western blot analysis. E. Analysis of ALK mRNA expression in ovarian cancer data sets. Error bars represent mean −/+ SEM of log2-median centered ALK expression levels. n= number of patients.
ALK is expressed in several OvCa cell lines.
Greater than 27% of serous ovarian carcinomas express ALK with reports of gene copy number gain, aberrant phosphorylation, and a novel fusion protein (FN1-ALK) (21, 22). Similarly, we were able to detect full length ALK (220 kDa) expression in multiple OvCa cell lines by WB and mRNA expression by qPCR analysis (Figure 1D, Supplemental Figure 2). Analysis of publicly available data sets shows higher expression of ALK mRNA in ovarian adenocarcinomas when compared to controls (Figure 1E). Neither the inducible nor the parental OvCa cells we used for this paper had ALK translocations nor obvious amplifications (Supplemental Figure 3).
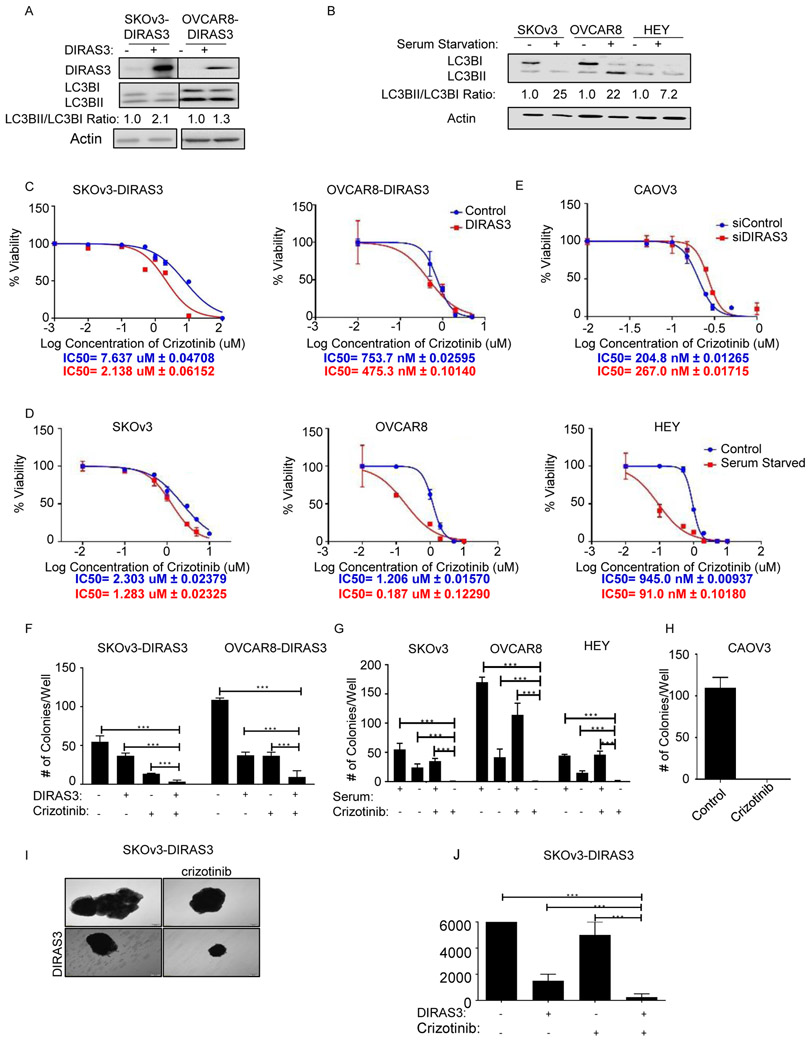
The IC50 of crizotinib is significantly reduced in autophagic OvCa cells that express DIRAS3 or are deprived of serum.
Either inducible re-expression of DIRAS3 (SKOv3-DIRAS3 or OVCAR8-DIRAS3) or serum starvation (SKOv3, OVCAR8, or HEY) in OvCa cell lines results in conversion of LC3B-I to LC3B-II, a surrogate marker for autophagy (Figure 2A-B). OvCa cells undergoing DIRAS3-induced autophagy or serum starvation-induced autophagy were treated with crizotinib. The IC50s for crizotinib were significantly lower for OvCa cell lines undergoing autophagy than for the same cancer cell lines not undergoing autophagy (Figure 2C-D). Conversely, CAOV3 has high basal autophagy related to higher endogenous expression of DIRAS3. Knockdown of DIRAS3 using siRNAs in CAOV3 cells raised the IC50 of crizotinib (Figure 2E). Crizotinib appeared to be on target, decreasing viability by inhibiting ALK in SKOv3-DIRAS3 inducible OvCa cells (Supplemental Figure 4). Knockdown of ALK with siRNA decreased the mass of proteins, a surrogate for viability, in the SRB assay to a greater extent in DIRAS3-induced autophagic cells than in control cells. In DIRAS3-induced autophagic cells with ALK knockdown, the addition of crizotinib had no greater effect than knockdown of ALK alone in DIRAS-induced autophagic cells. (Supplemental Figure 4A). Knockdown of c-met with siRNA produced less cell killing than knockdown of ALK in DIRAS3 (P<0.001) induced autophagic cells, but still decreased the fraction of viable cells when compared to controls (P=0.0006) (Supplemental Figure 4B). Other ALK inhibitors, which do not target c-MET, including alectinib, ceritinib, and lorlatinib inhibit growth of OvCa cells expressing DIRAS3 and undergoing autophagy more than cells from the same lines that were not undergoing autophagy (Supplemental Figure 5) (23-25).
Figure 2. Autophagy increases crizotinib sensitivity.
A. DIRAS3-inducible cells were treated ± doxycycline (1 ug/mL) for 48 hrs before western blot analysis. B. OvCa cells were grown ± serum for 48 hrs. C and D. IC50 of crizotinib in autophagic and non-autophagic OvCa cells by upregulation of DIRAS3 (C) or serum starvation (D) using SRB assays. Cells were plated (5,000 cells/well) and after 24 hrs, treated ± doxycycline (C), or (D) were grown ± serum and ± crizotinib. Cells were treated a second time C) ± doxycycline and C-D) ± crizotinib after 48 hrs. After 6 days, SRB assays were performed and the IC50 was calculated using Graphpad prism. E. CAOV3 cells were inversely transfected with siRNAs targeting DIRAS3 or a scrambled control sequence, plated (10,000 cells/well) and 24h later treated with crizotinib. Cell viability was measured with an SRB assay after 72 hrs. F. After 24 hrs, cells were treated ± doxycycline and ± crizotinib (1uM). G. After 24 hrs cells were washed and grown in ± serum. Cells were then treated ± crizotinib (1uM). H. CAOv3 cells were treated ± 500nM crizotinib. H. After 12 days, cells were stained with 0.5% methylene blue and colonies were scored. I. Spheroids of SKOV3-DIRAS3 cells grown in suspension were treated with ±doxycycline and ± crizotinib (1uM). J. After 2 weeks, cell viability was assessed with trypan blue dye. All experiments were repeated in triplicate and subjected to ANOVA analysis. * p<0.05, ** p<0.01, *** p<0.005, **** p<0.001
Crizotinib reduces colony formation and inhibits the growth of 3-D spheroids containing OvCa cells undergoing autophagy.
Clonogenic assays were performed with DIRAS inducible sublines of SKOv3 and OVCAR8 (SKOv3-DIRAS3 and OVCAR8-DIRAS3). Treatment of DIRAS3-expressing, autophagic OvCa cells lines with 1μM crizotinib reduced colony formation more than treatment of cells with 1uM crizotinib alone or induction of DIRAS3 alone (Figure 2F and Supplemental Figure 6). When OvCa cell lines were incubated in the presence or absence of serum to induce autophagy, treatment with crizotinib significantly reduced clonogenic growth of autophagic OvCa (Figure 2G and Supplemental Figure 6). In each of these cases, crizotinib alone reduced the size of the colonies. CAOV3 cells that endogenously express elevated levels of DIRAS3 and exhibit high basal autophagy were unable to form any colonies in the presence of crizotinib (Figure 3H and Supplemental Figure 6).
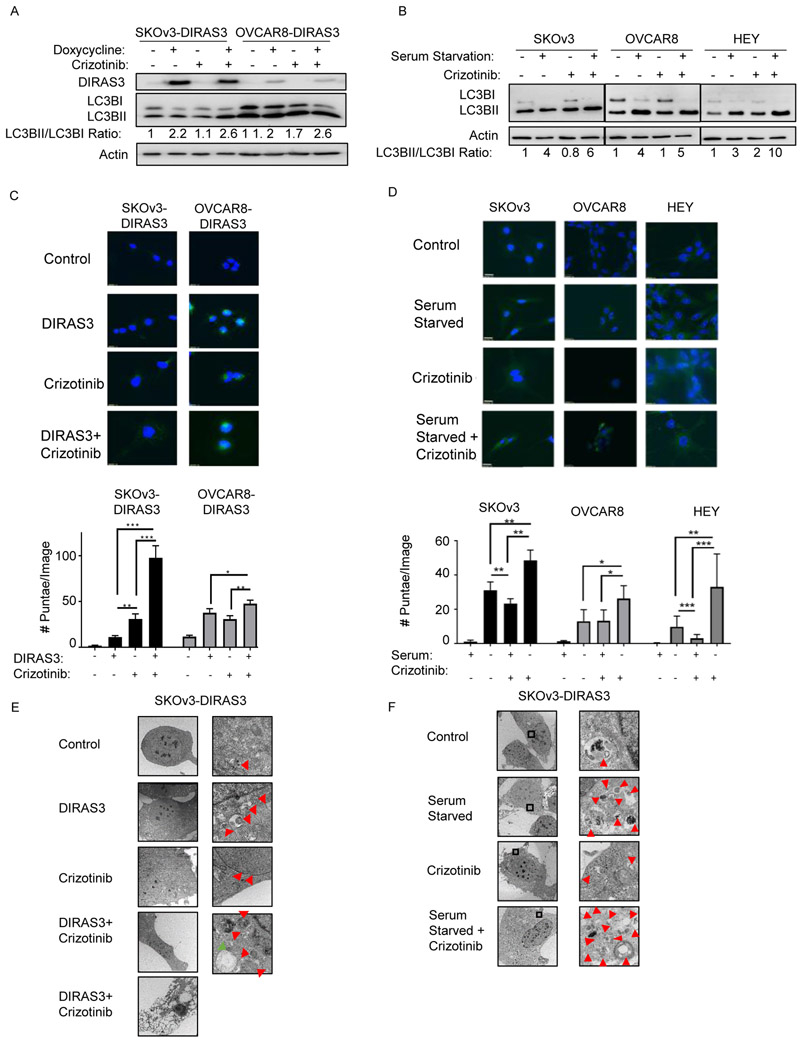
Figure 3. Crizotinib further increases autophagy.
A-B. SKOv3-DIRAS3 and OVCAR8-DIRAS3 cells were treated with +/− doxycycline (A), and SKOv3, OVCAR8, and HEY cells were serum-starved (B) during treatment with crizotinib for 72 hrs before western blot analysis of MAP-LC3 conversion. C-D. Effect of crizotinib on autophagy induced by DIRAS3 (C) or serum starvation (D). Cells treated as described in A and B were fixed and stained for LC3B by immunofluorescence. Quantification of LC3B punctae are graphed below. All experiments were repeated in triplicate and subjected to ANOVA analysis. * p<0.05, ** p<0.01, *** p<0.005, **** p<0.001. Autophagy was further visualized with transmission electron microscopy in E. ± DIRAS3 ± crizotinib treated SKOv3-DIRAS3 cells and F. ± serum starved ± crizotinib treated SKOv3 cells at 2 magnifications (3000x and 5000x). Red arrows: autophagosomes, green arrow: autophagolysosomes.
At the time of initial surgery for advanced stage disease, clumps of OvCa cells are found suspended in ascites fluid and found within established metastases on the peritoneal surface (26). 3D cancer cell spheroids might provide a better model than monolayer cultures for the behavior of OvCA growing within the peritoneal cavity (27). Biocompatible nanoparticles were used to magnetize, levitate, and grow 3D spheroids from inducible SKOv3-DIRAS3 and OVCAR8-DIRAS3 ovarian cancer cells. Spheroids grown in suspension for 2 weeks with crizotinib and with doxycycline to induce DIRAS3 expression were remarkably smaller in size than spheroids treated with crizotinib or doxycycline alone. When spheroids were dissociated, dual treatment yielded the smallest number of viable cells (Figure 2I-J and Supplemental Figure 7).
Crizotinib treatment further increases autophagy in autophagic ovarian cancer cells.
Treatment with crizotinib increased LC3B-I to LC3B-II conversion in OvCa cells already undergoing autophagy induced by DIRAS3 re-expression (Figure 3A) or by serum starvation (Figure 3B). Cleaved and lipidated LC3B-II is associated with autophagosomes and can be detected as punctate staining with immunofluorescence microscopy (28). LC3B-I, which is not associated with the autophagosome, can be found throughout the cytoplasm. Induction of autophagy by re-expressing DIRAS3 or by serum starvation significantly increased LC3B–II punctae compared to control cells (Figure 3C-D). Treatment with crizotinib in these already autophagic cells, further increased the number of LC3B-II punctae per cell, indicating an increase in autophagy. Moreover, transmission electron microscopy (TEM) revealed a greater number of autophagosomes in SKOv3-DIRAS3 OvCa cells expressing DIRAS3 and treated with crizotinib. Similar results were obtained in SKOv3 cells that were serum starved and treated with crizotinib (Figure 3E-F).
Crizotinib induces apoptosis in autophagic OvCa cells.
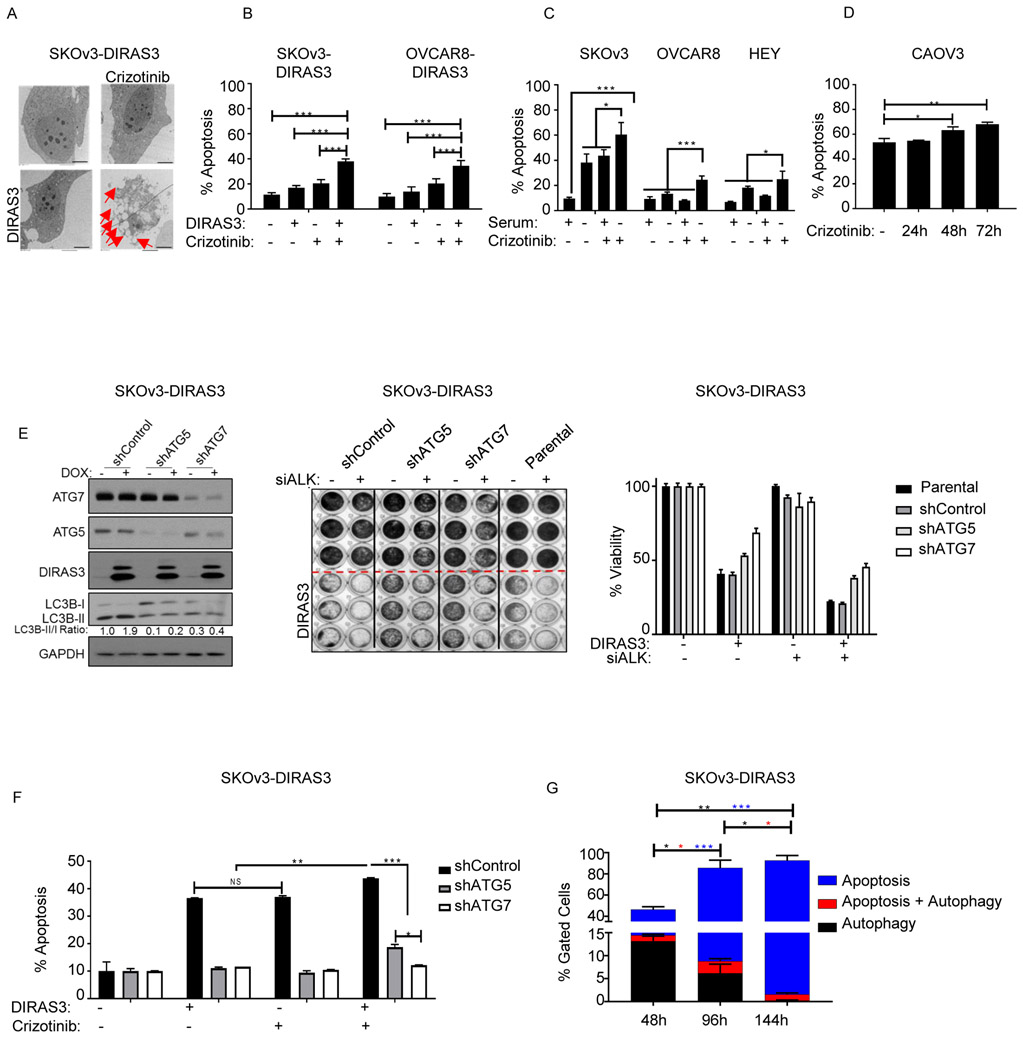
TEM of the SKOv3-DIRAS3 cells expressing DIRAS3 and treated with crizotinib revealed not only cells with increased autophagic vesicles but also cells with apoptotic bodies (Figure 3E). At a later time point, 6 days after expression of DIRAS3 and treatment with crizotinib, TEM revealed that the few cells which remained were filled with apoptotic bodies (Figure 4A).
Figure 4. Crizotinib induces apoptosis in autophagic OvCa cells.
A. Measurement of induction of apoptosis with Transmission electron microscopy (TEM) in SKOv3-DIRAS3 treated ± doxycycline and ± crizotinib for 6 days. The red arrows shows the increased in apoptotic bodies. B. SKOV3-DIRAS3 and OVCAR8-DIRAS3 cells expressing DIRAS3 and treated ±crizotinib and C. SKOv3, OVCAR8, and HEY cells were +/− serum starved and treated ± crizotinib for 6 days. D. CAOV3 cells treated with 500nM crizotinib for 24, 48, or 72 hrs were stained for annexinV/PI and subjected to flow cytometry. E. SKOV3-DIRAS3 cells stably expressing shControl, shATG5, or shATG7 were inversely transfected with a pool of 4 siRNAs targeting a scrambled control or ALK and plated (100,000 cells/well: lysate) (5,000 cells/well: SRB). After 24h, cells were treated ±doxycycline. After 72hrs, lysates were collected for western blot analysis or SRB assay. F. SKOV3-DIRAS3 cells stably expressing shControl, shATG5, or shATG7 were treated ± doxycycline and ± crizotinib (1uM) for 6 days, stained for annexin v/pi and subjected to flow cytometry. G. SKOv3-DIRAS3 cells were treated with ± doxycycline and ± crizotinib (1uM) for 48 hrs, 96 hrs, or 144 hrs, stained for autophagy with CYTO-ID® Autophagy detection kit and for apoptosis with annexin v-apc and subjected to flow cytometry. Only the brightest florescent cells as compared to chloroquine treatment were gated and labeled as “autophagic.” All experiments were repeated in triplicate and subjected to ANOVA analysis. * p<0.05, ** p<0.01, *** p<0.005, **** p<0.001
The role of apoptosis in crizotinib-mediated cytotoxicity for cells undergoing autophagy was further evaluated with annexin V/pi staining followed by flow cytometric analysis. Crizotinib induced substantially greater apoptosis in cells undergoing DIRAS3-induced or serum starvation-induced autophagy than in non-autophagic OvCa cells (Figure 4B-C). SK-N-SH, a neuroblastoma cell line, which has already been reported to undergo apoptotic cell death following crizotinib treatment was used as a positive control. SK-N-SH cells had a significantly higher fraction of apoptotic cells after treatment with crizotinib, but doxycycline treatment did not affect the fraction of annexin V+/pi+ cells (Supplemental Figure 8). CAOV3 cells which have high basal levels of autophagy, underwent apoptosis in a time dependent manner during treatment with crizotinib (Figure 4D).
Autophagy is required for apoptotic cell death.
Stable knockdown of key components of autophagy, ATG5 or ATG7, with shRNAs reduced autophagy in cancer cells and partially rescued DIRAS3 expressing cells transfected with pooled siALK (Figure 4E). We hypothesized that cell death could occur in each of two subpopulations of cells: one undergoing Type I apoptotic cell death and the other undergoing Type II autophagic cell death. An alternative hypothesis was that autophagy was a precursor of apoptotic cell death, as previously reported (29). To distinguish these alternatives, we established sublines of SKOv3-DIRAS3 that expressed shRNAs against ATG5, ATG7 or a scrambled control. Knockdown of key genes required for autophagy, ATG5 or ATG7, dramatically reduced the percentage of apoptotic cells detected following re-expression of DIRAS3 and treatment with crizotinib, consistent with the hypothesis that autophagy was a required precursor for the apoptosis that occurred in this setting (Figure 4F). To test this possibility further, we stained cells for biomarkers of both autophagy and apoptosis and measured the percentage of cells undergoing autophagy or apoptosis in each treatment group (Figure 4G, Supplemental Figure 9). SKOv3-DIRA3 cells treated for 4 or 6 days had significantly (P<0.005) more apoptosis than cells treated for only 2 days. Concurrently, the fraction of cells undergoing autophagy decreased significantly with each time point after treatment with a mean of less than 0.3% of cells undergoing autophagy alone at day 6 (P<0.05). The greatest proportion of cells that were both autophagic and apoptotic was on day 4 (P<0.05).
Crizotinib induces autophagy and apoptosis in autophagic OvCa cells by downregulating p-STAT3.
Using reverse phase protein arrays (RPPAs), p-STAT3(Tyr705), BCL-2, p-AKT(Ser473), p-BAD, p-mTOR, and p-ULK1 were found to be decreased in DIRAS3 expressing, crizotinib-treated OvCa cells (Figure 5A, Supplemental Figure 10). Within a short interval (15-60m) p-STAT3 (Tyr705) was dramatically reduced in autophagic OvCa cells treated with crizotinib (Figure 5B-C). BCL-2 is a well characterized transcriptional target of p-STAT3(30). Since we saw a dramatic reduction in BCL-2 expression in the RPPA, we hypothesized that BCL-2 would be reduced in autophagic OvCa cells following crizotinib treatment (Figure 5D-E). Using specific inhibitors of p-STAT3 at Tyr705, cryptotanshinone (Tanshinone c), and BCL-2, venetoclax (ABT-199, VENCLEXTA®), we were able to mimic the significant reductions in viability in the autophagic cells that we observe with crizotinib (Figure 5F-G, Supplemental Figure 11). Both cryptotanshinone and venetoclax increased autophagy and apoptosis, similar to crizotinib, in autophagic OvCa cells (Figure 5H-K, Supplemental Figure 12). Moreover, in OvCa patient samples available on TCGA, ALK expression was positively correlated with BCL2 expression (Figure 5L).
Figure 5. Crizotinib targets autophagic OvCa cells by downregulating p-STAT3.
A. Heatmap of proteins changed based on reverse phase protein array analysis. B and C. DIRAS3-inducible cells were treated ± doxycycline (1 ug/mL) for 24 hrs and C) SKOv3, OVCAR8, and HEY cells ± serum for 30 minutes prior to treatment with 1uM crizotinib. D and E. DIRAS-inducible cells were treated ±doxycycline for 24 hrs (D) and SKOv3, OVCAR8, and HEY cells were serum starved € for 30 minutes prior to treatment with crizotinib for 72 hrs. Lysates were collected for western blot (WB) analysis and band density was quantified with image j software. F - I. SKOv3-DIRAS3 (F) and OVCAR8 cells (G) were plated (5,000 cells/well) in a 96-well plate. After 24 hrs cells were treated ± doxycycline (F) or grown ± serum (G) followed by treatment with inhibitors at 24 hrs and 96 hrs. Cell viability was measured with an SRB assay 72 hrs after the second treatment. IC50 was calculated using GraphPad Prism. SKOv3-DIRAS3 cells were plated and 24 hrs later treated ± doxycycline (H). After an additional 24 hrs, cells were treated with inhibitors for 72 hrs. OVCAR8 cells were serum starved 24 hrs after plating for 1h followed by treatment with inhibitors for 72 hrs (I). Lysates were collected for WB. J. SKOV3-DIRAS3 expressing DIRAS3 and K. OVCAR8 cells were serum starved and treated ± inhibitors two times over 6 days and then stained for annexin v/pi and subjected to flow cytometry. All experiments were repeated in triplicate and subjected to ANOVA analysis. * p<0.05, ** p<0.01, *** p<0.005, **** p<0.001; 1uM crizotinib, 1uM cryptotanshinone, 250nM venetoclax. L. Analysis of ovarian cancer datasets to determine the correlation between ALK and BCL2 mRNA levels. The scale for the expression of ALK and BCL2 is log2 median-centered ratio. The Pearson-correlation p-values and r2-values were calculated using GraphPad prism. M. Schematic of proposed mechanism.
We also observed decreases in p-AKT, but not p-ERK1/2, in autophagic SKOv3-DIRAS3 ovarian cells treated with crizotinib (Supplemental Figure 13). This decrease in p-AKT was associated with a decrease p-mTOR and p-ULK1, all of which can contribute to an increase in autophagy. Decreased p-AKT also correlated with a decrease in its downstream effector, p-BAD (Supplemental Figure 13). Taken together, a decrease in p-BAD would permit BAD to sequester any remaining BCL-2 present in the autophagic OvCa cells treated with crizotinib, permitting Bax and Bak to oligomerize and apoptosis to occur (Figure 5M). Since Bcl-XL is also a target of STAT3 we performed a western blot analysis and observed a decreased in autophagic SKOV3-DIRAS3 and OVCAR8-DIRAS3 cells treated with crizotinib (Supplemental Figure 14).
Crizotinib significantly prolongs survival in dormant xenograft models that express DIRAS3 and are undergoing autophagy.
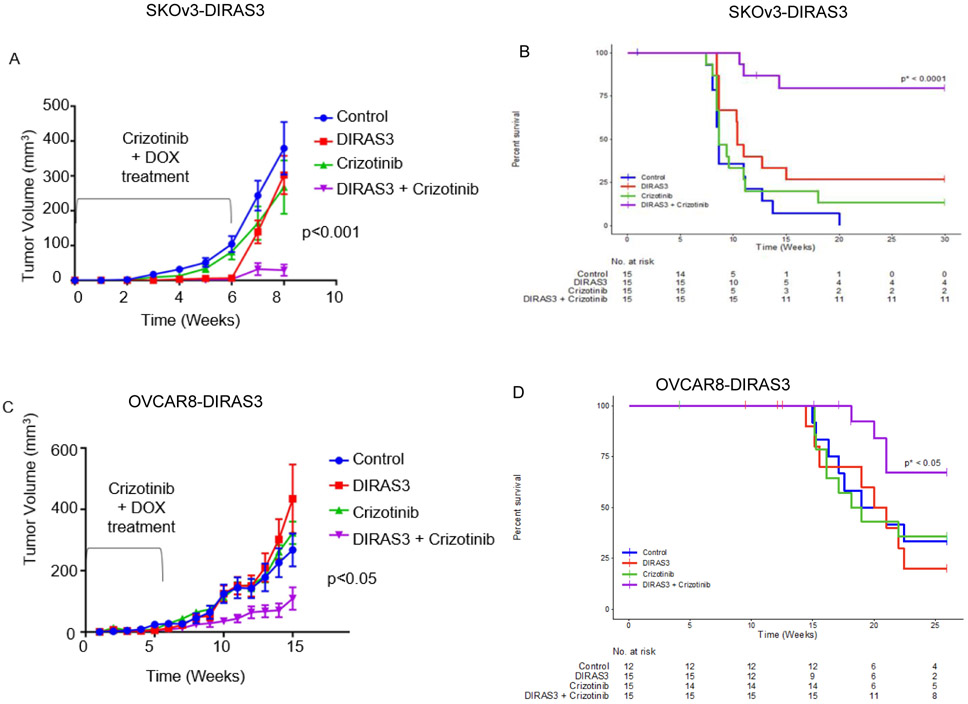
In in vivo models of dormancy, mice bearing SKOv3-DIRAS3 or OVCAR8-DIRAS3 OvCa subcutaneous xenografts, are given doxycycline in the sucrose drinking water for 6 weeks (Supplemental Figure 15). In earlier studies (6), persistent induction of DIRAS3 driven by continued administration of DOX suppressed xenograft growth for months in most animals. Ovarian cancer cells in the dormant xenografts expressed DIRAS3 and underwent autophagy, similar to the ovarian cancer cells found at second look operations (31). When doxycycline was removed after six weeks, tumors grew progressively (Figure 6A, green line), mimicking tumor growth in control mice (blue line) and the mice on sucrose drinking water and gavaged with crizotinib daily during the same 6 week interval (red line). Mice that were both given DOX and crizotinib p.o. daily for 6 weeks had fewer tumors grow out after decreasing DIRAS3 expression and smaller average tumor volumes (purple line). Impressively, treatment of crizotinib in mice bearing DIRAS3 expressing SKOV3-DIRAS3 tumors significantly prolonged survival in 79% of mice beyond 30 weeks (Figure 6B and Supplemental Table 1). Similar significant trends were observed in mice with OVCAR8-DIRAS3 subcutaneous xenografts where 67% of mice survived at 25 weeks (Figure 6C-D and Supplemental Table 1).
Figure 6. Crizotinib significantly prolongs survival in dormant xenograft models that express DIRAS3 and are undergoing autophagy.
5.5 x 106 cells were injected subcutaneously; next day nude mice began p.o. treatment of diluent or 20 mg/kg crizotinib q.d. for 5 days/week for 6 weeks. Mice were given 5% sucrose or 5% sucrose + 0.02% doxycycline water. After 6 weeks, mice bearing A. SKOv3-DIRAS3 or C. OVCAR8-DIRAS3 DIRAS3 expressing tumors treated with crizotinib (purple line) had significantly decreased tumor burden compared to controls (blue line), the DIRAS3 expressing tumors without receiving crizotinib (red line) and the non-DIRAS3 expressing tumors treated with crizotinib (green line).This decrease in tumor burden prolonged the survival (to ethical endpoint). Figure 6B. Kaplan-Meier curves for SKOV3. The p-value p* is for the hypothesis test comparing Crizotinib and DIRAS+Crizotinib. Figure 6D. Kaplan-Meier curves for OVCAR8. The p-value p* is for the hypothesis test comparing Crizotinib and DIRAS+Crizotinib
Discussion
Less than 40% of OvCa patients can be cured, due in large part to the persistence of dormant, drug resistant cancer cells in the peritoneal cavity (1, 2). When second look operations are performed for patients with a complete clinical remission after primary cytoreductive surgery and chemotherapy, small deposits of OvCa cells are found in poorly-vascularized collagenous nodules on the surface of the peritoneal cavity. In more than 80% of cases, persistent cancer cells are undergoing autophagy, providing a unique opportunity for anti-autophagic therapy (6). Using an unbiased siRNA screen of the targets for FDA-approved drugs, we have shown for the first time that autophagic OvCa cells are selectively vulnerable to ALK inhibition. While ALK has not proven to be useful for primary ovarian cancer, only 20% of primary cases have significant numbers of autophagic OvCa cells. Having confirmed that knockdown of ALK reduced the viability of OvCa cells in which autophagy had been induced by expression of DIRAS3 or serum starvation, we found that several FDA-approved inhibitors of ALK exhibited dramatically lower IC50s in autophagic OvCa cells. Crizotinib, an ALK inhibitor, dramatically reduced the outgrowth of two different dormant, autophagic OvCa xenografts and significantly increased survival.
Dormant, autophagic OvCa cells required ALK signaling to remain viable. The oncogenic role of ALK and its frequent translocations has been well characterized in many hematopoietic and solid tumors (32-34). Crizotinib, the first FDA-approved drug used to treat lung cancer with ALK rearrangements, is a small molecule inhibitor of ALK with some ability to inhibit c-ros (ROS1) and c-MET (34). In preclinical reports, crizotinib was highly selective for ALK and effectively inhibited cell proliferation which was associated with increased apoptosis. Crizotinib has also been reported to induce autophagy via inhibition of the STAT3 signaling in NSCLC, although in this case the autophagy was cytoprotective (35-36). Clinical trials conducted in diverse patient populations indicated that crizotinib was simultaneously effective and safe; it was well tolerated with rare grade 3/4 treatment-related adverse events (37).
Other approaches have been taken to target autophagic cancer cells (7, 38-39). When autophagy is seen to be cytoprotective, inhibitors such as chloroquine and hydroxychloroquine (HCQ), have been used to block autophagic flux, inducing cell death often through apoptosis (38-39). In clinical trials, a subset of patients with various cancers treated with HCQ in combination with other anti-cancer agents had partial responses and stable disease; however, few patients in any of these studies experienced an objective response (38-39). At least one recent study documented a dramatic partial response when HCQ was added to trametinib in a patient with refractory pancreatic cancer and suggested that drug dose could be a critical factor (40). Clinical trials of anti-autophagy agents with greater potency and better safety profiles are underway (39).
While most clinical studies have attempted to inhibit autophagy, an alternative approach would enhance autophagy, inducing type II cell death. Our group recently reported that OvCa cells undergoing DIRAS-3 induced autophagy depend critically on at least three survival factors produced by the cancer cells or host microenvironment to avoid type II autophagic death, including VEGF, IGF and IK-8 (41). Neutralizing these factors or blocking their receptors with monoclonal antibodies during DIRAS3 induced dormancy and autophagy, prolonged survival of mice with DIRAS3 inducible xenografts.
Treatment with crizotinib further increased the level of autophagy that had been induced by expression of DIRAS3 or serum starvation, resulting in apoptosis. Knockdown of autophagic genes reduced crizotinib-induced apoptotic cell death in OvCa cells. Several groups have outlined a mechanism in which an intrinsic stress signal can activate a death-inducing signaling complex (DISC)-like complex that is assembled on autophagosomal membranes (29, 41-42). The activation of this DISC-like complex then activates cleavage of caspase 8 which can be recruited to the autophagosome through direct interaction with p62/LC3B or through an interaction with FADD and ATG5(29, 41-42). Our data and data from others have noted that in certain instances cell death occurs by one integrated cell death mechanism that initially requires autophagic components and later requires apoptotic components (29, 42-43).
Our data suggest that ALK inhibition by crizotinib reduces the viability of dormant, autophagic OvCa cells primarily by decreasing p-STAT3 levels. STAT3 in its non-phosphorylated state is typically found in the cytoplasm, whereas phosphorylated STAT3 can translocate to the nucleus and induce transcription of target genes (44). More than 70% of OvCa have high nuclear STAT3 staining which can be correlated with poor prognosis (45). Previously, we have found that DIRAS3 interacts with STAT3, sequestering it in the cytoplasm (10). Our work here has demonstrated that treatment with an ALK inhibitor in DIRAS3 expressing cells results in even more dramatic concurrent reduction of p-STAT3. Others have shown that activation of the STAT3 signaling pathway helps cells to escape from the dormant state and that decreased p-STAT3 signaling pushes dormant cells toward death (3-4). While neither STAT3 nor its downstream effector, BCL-2, were a part of our initial siRNA screen, we were able to replicate the decrease in viability in vitro with a selective p-STAT3 inhibitor, cryptotanshinone, and a selective BCL-2 inhibitor, venetoclax (46-47). Targeting either STAT3 or BCL-2 could provide to be an alternative method for targeting dormant, autophagic OvCa cells (48-49).
Taken together, we have sought to exploit vulnerabilities of autophagic cancer cells by targeting kinases required for their survival in nutrient poor settings where DIRAS3 is upregulated (49). Our pre-clinical data suggest that crizotinib and other ALK inhibitors might eliminate dormant, autophagic drug resistant OvCa cells that remain after conventional cytoreductive surgery and combination chemotherapy. A clinical trial of ALK inhibitors as maintenance therapy following second look operations should be seriously considered.
Materials and Methods
Kinome Screen.
To identify targets whose knockdown would selectively eliminate autophagic cancer cells, an unbiased screen was conducted with a short interfering (si) RNA library that targeted 570 kinases and G-protein receptors. siRNAs were transfected into SKOv3-DIRAS3 and OVCAR8-DIRAS3 OvCa cells. After 24 hours of transfection, doxycycline (DOX) in tissue culture medium (1 μg/μl) or tissue culture medium alone was added to multiwall plates to induce DIRAS3 expression. After 72 hours incubation, cell mass was measured using a sulforhodamine B assay (SRB)(30). In confirmatory studies with amino acid starvation after 72 hours of transfection of parental SKOv3 OvCa cells, culture medium was replaced with amino acid deficient medium for 24 hours or 48 hours before assay with an SRB assay (20).
Flow Cytometry.
Annexin V-488/PI was performed as previously described (8). Double staining of autophagy and apoptosis was done by collecting all attached and floating cells after indicated length of treatment, washing in PBS, incubating in the CYTO-ID® Green Autophagy Stain Solution (ENZO) for 30m in the dark at room temperature, washing again in PBS, incubating in Annexin V-APC (Life Technologies) for 15m, adding Sytox Blue, and sorting on the BCI Gallios Analyzer.
Human OvCa Xenografts in Nude Mice.
Experiments using female athymic nu/nu mice were reviewed and approved by IACUC (ID: 00001195-RN00; MD Anderson Cancer Center, Houston, TX). Baytril water was given prophylactically for 2 weeks to prevent dry skin. SKOv3-DIRAS3 and OVCAR8-DIRAS3 cells were injected subcutaneously. At the time of injection, mice were given fresh 5% sucrose water or 5% sucrose + 0.02% doxycyline water, which was changed every other day for 6 weeks. Crizotinib was provided by Pfizer Pharmaceuticals (Groton, CT) administrated orally 20 mg/kg/day 5 days per week for 6 weeks. At the time of injection, the mice were randomly assigned to treatment groups (n = 15 mice per group): (i) 5% sucrose + control gavage; (ii) 5% sucrose + crizotinib; (iii) 5% sucrose + 0.02% Doxycyline + control gavage; (iv) 5% sucrose + 0.02%Doxycyline + crizotinib. Tumors were measured weekly. Once the tumor burden reached 1.5cm, mice were sacrificed by CO2.
Statistical Methods
Pearson’s Correlation Coefficients were calculated to measure the correlations between ALK and BCL2 mRNA levels. ANOVAs were applied to test the difference among multiple groups for continuous variables and t-tests were used for two-group comparisons. Overall survival distributions were estimated using the Kaplan-Meier method and compared between groups using the log-rank tests. P-values less than 0.05 were considered statistically significant. Statistical analyses were conducted in GraphPad Prism 8.0 and R 3.6.1.
Please see the supplementary Materials and Methods for additional information.
Supplementary Material
Table 1.
Median survival and survival rate at 25 weeks for each arm.
| Arm | Median survival (week) | Survival rate (%) at 25 weeks | ||
|---|---|---|---|---|
| SKOV3 | OVCAR8 | SKOV3 | OVCAR8 | |
| Control | 8.6 | 20 | 0 | 33.3 |
| DIRAS3 | 10.4 | 20.5 | 26.7 | 20.0 |
| Crizotinib | 8.6 | 18.6 | 13.3 | 35.7 |
| DIRAS3 + Crizotinib | > 14.3 | > 21 | 79.4 | 67.1 |
ACKNOWLEDGEMENTS
The authors thank Mr. Kenneth Dunner Jr. in the High-Resolution Electron Microscopy Facility and all of the staff at the Flow Cytometry and Cellular Imaging Core Facility. “EE was supported by the CPRIT Research Training Program (RP170067)”
Financial Support: This work was supported by National Cancer Institute (NCI) R01 CA135354 P30CA016672, MD Anderson SPORE in Ovarian Cancer NCI P50 CA 83639 and 217685, National Foundation for Cancer Research, Anne and Henry Zarrow Foundation, Mossy Foundation, Roberson Endowment, and Stuart and Gaye-Lynn Zarrow. A.M.B. is an MDACC Odyssey Fellow and is supported by the CFP Foundation. Crizotinib was provided by Pfizer Pharmaceuticals.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to report.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh AC, Ramaswamy S. Mechanisms of cancer cell dormancy--another hallmark of cancer? Cancer Res. 2015;75:5014–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greer BE, Bundy BN, Ozols RF, et al. Implications of second-look laparotomy in the context of optimally resected stage III OvCa: a non-randomized comparison using an explanatory analysis: a Gynecologic Oncology Group study. Gynecol Oncol. 2005;99:71–79. [DOI] [PubMed] [Google Scholar]
- 6.Lu Z, Luo RZ, Lu Y, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Z, Yang H, Sutton MN, et al. ARHI (DIRAS3) induces autophagy in ovarian cancer cells by downregulating the epidermal growth factor receptor, inhibiting PI3K and Ras/MAP signaling and activating the FOXo3a-mediated induction of Rab7. Cell Death Differ. 2014;21:1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y, Luo R, Lu Z, et al. Biochemistry and biology of ARHI (DIRAS3), an imprinted tumor suppressor gene whose expression is lost in ovarian and breast cancers. Methods Enzymol. 2006;407:455–468. [DOI] [PubMed] [Google Scholar]
- 10.Badgwell DB, Lu Z, Le K, et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene. 2012;31:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bao JJ, Le XF, Wang RY, et al. Reexpression of the tumor suppressor gene ARHI induces apoptosis in ovarian and breast cancer cells through a caspase-independent calpain-dependent pathway. Cancer Res. 2002;62:7264–7272. [PubMed] [Google Scholar]
- 12.Chen J, Shi S, Yang W, Chen C. Over-expression of ARHI decreases tumor growth, migration, and invasion in human glioma. Med Oncol. 2014;31:846. [DOI] [PubMed] [Google Scholar]
- 13.Dalai I, Missiaglia E, Barbi S, et al. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia. 2007;9:181–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Cui G, Sun L, et al. ARHI overexpression induces epithelial ovarian cancer cell apoptosis and excessive autophagy. Int J Gynecol Cancer. 2014;24:437–443. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z, Bast RC Jr. The tumor suppressor gene ARHI (DIRAS3) inhibits ovarian cancer cell migration through multiple mechanisms. Cell Adh Migr. 2013;7:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z, Luo RZ, Peng H, et al. Transcriptional and posttranscriptional down-regulation of the imprinted tumor suppressor gene ARHI (DRAS3) in ovarian cancer. Clin Cancer Res. 2006;12:2404–2413. [DOI] [PubMed] [Google Scholar]
- 17.Luo RZ, Fang X, Marquez R, et al. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene. 2003;22:2897–2909. [DOI] [PubMed] [Google Scholar]
- 18.Washington MN, Suh G, Orozco AF, et al. ARHI (DIRAS3)-mediated autophagy-associated cell death enhances chemosensitivity to cisplatin in ovarian cancer cell lines and xenografts. Cell Death Dis. 2015;6:e1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Y, Xu F, Peng H, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A. 1999; 96:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Chang IS, Lin W, et al. ARHI (DIRAS3), an imprinted tumour suppressor gene, binds to importins and blocks nuclear import of cargo proteins. Biosci Rep. 2009;30:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. [DOI] [PubMed] [Google Scholar]
- 22.Sutton MN, Huang GY, Zhou J, et al. Amino acid deprivation-induced autophagy requires upregulation of DIRAS3 through reduction of E2F1 and E2F4 transcriptional repression. Cancers (Basel). 2019;11:E603. doi: 10.3390/cancers110506039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang S, Yang F, Du X, Lu Y, Zhang L, Zhou X. Aberrant expression of anaplastic lymphoma kinase in ovarian carcinoma independent of gene rearrangement. Int J Gynecol Pathol. 2016;35:337–347. [DOI] [PubMed] [Google Scholar]
- 24.Ren H, Tan ZP, Zhu X, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–3323. [DOI] [PubMed] [Google Scholar]
- 25.Anglesio MS, Arnold JM, George J, Tinker AV, Tothill R, Waddell N, Simms L, Locandro B, Fereday S, Traficante N, Russell P, Sharma R, Birrer MJ, deFazio A, Chenevix-Trench G, Bowtell DDL. Mutation of ERBB2 Provides a Novel Alternative Mechanism for the Ubiquitous Activation of RAS-MAPK in Ovarian Serous Low Malignant Potential Tumors. Molecular Cancer Research. 2008; 6(11) 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM: ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 6:1–6. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, Traficante N, Fereday S, Hung JA, Chiew Y-E, Haviv I, Gertig D, deFazio A, Bowtell DDL. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clinical Cancer Research. 2008; 14(16) 5198–5208. [DOI] [PubMed] [Google Scholar]
- 28.McKeage K. Alectinib: a review of its use in advanced ALK-rearranged non-small cell lung cancer. Drugs. 2015;75:75–82. [DOI] [PubMed] [Google Scholar]
- 29.Shaw AT, Kim DW, Mehra R, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorlatinib is active in drug-resistant NSCLC. Cancer Discov. 2016;6:OF1. [DOI] [PubMed] [Google Scholar]
- 31.Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haisler WL, Timm DM, Gage JA, Tseng H, Killian TC, Souza GR. Three-dimensional cell culturing by magnetic levitation. Nat Protoc. 2013;8:1940–1949. [DOI] [PubMed] [Google Scholar]
- 33.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter RL, Lo HW.STAT3 target genes relevant to human cancers. Cancers(Basel).2014:6:897–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Z, Baquero M, Yang H, et al. ARHI (DIRAS3) regulates the autophagosome initiation complex and is expressed in autophagic ovarian cancer cells at second look surgery. Autophagy. 2014;10:1071–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prokoph N, Larose H, Lim MS, Burke GAA, Turner SD. Treatment options for paediatric anaplastic large cell lymphoma (ALCL): current standard and beyond. Cancers (Basel). 2018;10:E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Inamura K. Translocation renal cell carcinoma: an update on clinicopathological and molecular features. Cancers (Basel). 2017;9:E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trigg RM, Turner SD. ALK in Neuroblastoma: biological and therapeutic implications. Cancers (Basel). 2018;10:E113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Millett RL, Elkon JM, Tabbara IA. Directed Therapies in anaplastic lymphoma kinase-rearranged non-small cell lung cancer. Anticancer Res. 2018;38:4969–4975. [DOI] [PubMed] [Google Scholar]
- 41.You L, Shou J, Deng D, et al. Crizotinib induces autophagy through inhibition of the STAT3 pathway in multiple lung cancer cell lines. Oncotarget. 2015;6:40268–40282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandhi L, Janne PA. Crizotinib for ALK-rearranged non-small cell lung cancer: a new targeted therapy for a new target. Clin Cancer Res. 2012;18:3737–3742. [DOI] [PubMed] [Google Scholar]
- 43.Verbaanderd C, Maes H, Schaaf MB, et al. Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience. 2017;11:781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chude CI, Amaravadi RK. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int J Mol Sci. 2017;18:E1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinsey CG, Camolotto SA, Boespflug AM, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mao W, Peters HL, Sutton MN, et al. The role of vascular endothelial growth factor, interleukin 8, and insulinlike growth factor in sustaining autophagic DIRAS3-induced dormant ovarian cancer xenografts. Cancer. 2019;125:1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubinstein AD, Kimchi A. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci. 2012;125(pt 22):5259–5268. [DOI] [PubMed] [Google Scholar]
- 49.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. [DOI] [PubMed] [Google Scholar]
- 50.Min H, Wei-hong Z. Constitutive activation of signal transducer and activator of transcription 3 in epithelial ovarian carcinoma. J Obstet Gynaecol Res. 2009;35:918–925. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Lu Y, Chen G, Huang S. Molecular evidence of cryptotanshinone for treatment and prevention of human cancer. Anticancer Agents Med Chem. 2013;13:979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gentile M, Petrungaro A, Uccello G, et al. Venetoclax for the treatment of chronic lymphocytic leukemia. Expert Opin Investig Drugs. 2017;26:1307–1316. [DOI] [PubMed] [Google Scholar]
- 53.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genentech. Venclexta® tablets. Important Safety Information. Accessed November 27, 2018 https://www.venclexta.com/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.