Abstract
Landscape genomics studies focus on identifying candidate genes under selection via spatial variation in abiotic environmental variables, but rarely by biotic factors (i.e., disease). The Tasmanian devil (Sarcophilus harrisii) is found only on the environmentally heterogeneous island of Tasmania and is threatened with extinction by a transmissible cancer, devil facial tumor disease (DFTD). Devils persist in regions of long-term infection despite epidemiological model predictions of species’ extinction, suggesting possible adaptation to DFTD. Here, we test the extent to which spatial variation and genetic diversity are associated with the abiotic environment (i.e., climatic variables, elevation, vegetation cover) and/or DFTD. We employ genetic-environment association analyses using 6886 SNPs from 3287 individuals sampled pre- and post-disease arrival across the devil’s geographic range. Pre-disease, we find significant correlations of allele frequencies with environmental variables, including 365 unique loci linked to 71 genes, suggesting local adaptation to abiotic environment. The majority of candidate loci detected pre-DFTD are not detected post-DFTD arrival. Several post-DFTD candidate loci are associated with disease prevalence and were in linkage disequilibrium with genes involved in tumor suppression and immune response. Loss of apparent signal of abiotic local adaptation post-disease suggests swamping by strong selection resulting from the rapid onset of DFTD.
A central goal of molecular ecology is understanding how ecological processes generate and maintain the geographic distribution of adaptive genetic variation. Landscape genomics has emerged as a popular framework for identifying candidate loci that underlie local adaptation (Manel et al. 2010; Rellstab et al. 2015; Hoban et al. 2016; Lowry et al 2017; Storfer et al. 2018). Using genomic-scale data sets, researchers screen for loci that exhibit patterns of selection across heterogeneous environments (Haasl & Payseur 2016). One widely used method for testing for statistical associations of allele frequencies of marker loci across the genome with environmental variables is genetic-environmental associations (GEAs) (Rellstab et al. 2015; Whitlock & Lotterhos 2015; Francois et al. 2016; Hoban et al. 2016).
GEAs identify significant correlations of allele frequencies at candidate loci with abiotic environmental variables such as altitude, rainfall, and temperature. GEAs have been successful, for example, in identifying loci associated with adaptation to hypoxia in high-elevation human populations (Beall 2007, Peng et al. 2011), stress response in lichen populations along altitudinal gradients (Dal Grande et al. 2018), and leaf longevity and morphogenesis in response to aridity (Steane et al. 2014). Climatic, geographic, and fine-scale remote sensing data that explain large amounts of heterogeneity in the environment are often easily obtained (Rellstab et al. 2015). However, data on biotic factors such as life-history traits (Sun et al. 2015), community composition (Harrison et al. 2017), or disease prevalence often involve extensive fieldwork and are far more difficult and labor-intensive to collect than abiotic variables. Accordingly, few landscape genomics studies have tested for the influence of biotic variables on the spatial distribution of adaptive genetic variation.
Infectious diseases often impose strong selective pressures on their host and thereby represent key biotic variables increasingly recognized for their severe impacts on natural populations (summarized in Kozakiewicz et al. 2018, e.g., Biek & Real 2010; Wenzel et al. 2016; Leo et al. 2016; Mackinnon et al. 2016; Eoche-Bosy et al. 2017). Landscape genomic studies of the disease can help guide management programs, such as captive breeding designs and reintroductions (Hoban et al. 2016; Hohenlohe et al. 2019). Additionally, landscape genomics studies have been used to elucidate how the landscape influences the distribution and spread of the pathogen (Robinson et al. 2015; Schwabl et al. 2017). However, studies disentangling the influence of pathogen dynamics from that of other abiotic landscape features have had limited statistical power to date. For example, Wenzel et al. (2016) tested for correlations between parasite burden and genetic differentiation across the landscape but did not detect any statistically significant associations due to stochasticity in the genomic background created by dispersal. The disease may play an important role in the distribution of adaptive genetic variation across the landscape, potentially swamping signatures of local adaptation to the abiotic environment.
Tasmanian devils (Sarcophilus harrisii) and their transmissible cancer, devil facial tumor disease (DFTD), offer such an opportunity. Devils are isolated to the island of Tasmania and have been sampled across their entire geographic range. Intense mark-recapture studies and collection of thousands of genetic samples have been conducted over the past 20 years, both before and after DFTD arrival across multiple populations. This sampling effort, in addition to resources such as an assembled genome (Murchison et al. 2012; Patton et al. 2019), provides extensive data to employ GEAs to test for the relative effects of abiotic environmental factors versus DFTD.
In 1996, the first evidence of DFTD was documented in Mt. William/Wukalina National Park (McCallum 2008). In just over two decades, DFTD has spread across the majority of Tasmania with nearly a 100% case fatality rate (Hawkins et al. 2006; McCallum et al. 2009), and a second independently evolved clonal transmissible cancer (DFT2) has since emerged (Stammnitz et al. 2018; Pye et al. 2016a). Transmission appears to be largely frequency-dependent (McCallum et al. 2009; McCallum 2012), with tumors originating on the face or in the oral cavity and being transferred as allografts (Pearse et al. 2012) through biting during social interactions (Hamede et al. 2009). DFTD has a single, clonal Schwann cell origin (Murchison et al. 2010; Siddle et al. 2010) and evades host immune system detection via downregulation of host major histocompatibility complex (Siddle et al. 2013). Low overall genetic variation in devils resulting from population bottlenecks that occurred during the last glacial maximum as well as during extreme El Niño events 3000–5000 years ago (Brüniche-Olsen et al. 2014; Patton et al. 2019) has also been attributed to high susceptibility to this emerging infectious disease.
DFTD imposes extremely strong selection as it has caused local population declines exceeding 90%, and an overall species-wide decline of 80% (Jones et al. 2004; Lachish et al. 2009; Lazenby et al. 2018). However, small numbers of devils persist in areas with long-term infection likely resulting from evolutionary responses in the Tasmanian devil (Jones et al. 2008; Brüniche-Olsen et al. 2013; Epstein et al. 2016; Wright et al. 2017). Indeed, recent work has shown: (1) rapid evolution in genes associated with immune-related functions across multiple populations (Epstein et al. 2016), (2) sex-biased response in a few large-effect loci in survival following infection (Margres et al. 2018a), (3) evidence of effective immune response (Pye et al. 2016b), and (4) cases of spontaneous tumor regression (Pye et al. 2016b; Wright et al. 2017; Margres et al. 2018b). These studies, however, focused on relatively small geographic areas.
Here, we estimated allele frequencies in 6886 SNPs both randomly selected and previously shown to be associated with DFTD (Epstein et al. 2016) in 3287 devils from seven localities throughout Tasmania sampled both prior to and following disease arrival. We investigated four questions about the relative effects of abiotic environmental variables versus DFTD on the genomics of adaptation in Tasmanian devil populations: Question (1) Were there differences in the number of genetic clusters detected post-DFTD arrival? Question (2) Pre-DFTD arrival, what were the genetic-environmental associations of devil populations with abiotic variables (i.e., climatic variables, elevation, vegetation cover)? Question (3) Were statistical signals of local adaptation to abiotic variables detected pre-disease weakened following the arrival of DFTD? Question (4) Has genetic variation declined following disease arrival? Lack of significant variation in population structure (1) and genetic diversity (4) following disease arrival may suggest directional selection led to changes in allele frequencies at candidate loci rather than genetic drift owing to small surviving population sizes. We predicted that the DFTD epidemic would result in swamping of prior genetic-environmental associations with the abiotic environment (3 and 4).
Methods
TRAPPING AND SAMPLING DATA
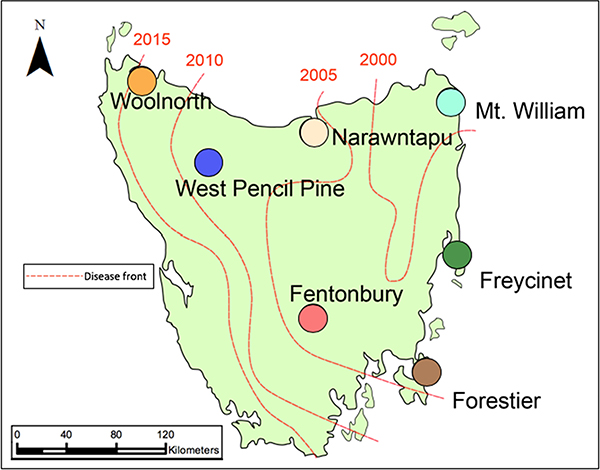
Field data and ear biopsy samples from 3287 Tasmanian devils were collected from seven different locations across their geographic range in Tasmania (Fig. 1), pre and post-DFTD, between 1999 and 2014 (Table 1). These geographic sampling locations were selected to maximize the extent of abiotic environmental heterogeneity and variation in disease prevalence across the species’ geographic range. On the eastern seaboard, coastal populations occupying narrow peninsulas were characterized by lower seasonal rainfall and moderate human activity (Freycinet and Forestier). In contrast, the coastal heathlands and dry grasslands of Mount William National Park and the coastal wetlands and lagoons that dominate the Narawntapu National Park in northeastern Tasmania experienced wetter climates. Inland, high elevation, habitats had greater annual temperature ranges and were heavily influenced by anthropogenic disturbance including increased surface area of sealed roads (Fentonbury) and commercial forestry (West Pencil Pine). While both Woolnorth and West Pencil Pine were dominated by heavy rainfall, Woolnorth had minimal human development and was primarily characterized by dry, coastal Eucalypt forests common along the northwest coast.
Figure 1.
Map of Tasmania with each sampling location. The red lines and the corresponding years indicate the first year that disease was detected in these locations.
Table 1.
Information for samples collected from each population (the acronyms for the population included in parentheses). Values listed include the number of ear biopsy samples collected per population prior to and post-disease arrival, the years the populations were sampled, and the date that DFTD was first detected in that population.
| Populations | Samples pre-disease | Samples post-disease | Years samples collected | Date of DFTD infection |
|---|---|---|---|---|
| Fentonbury (FEN) | 97 | 157 | 2004–2009 | 2005 |
| Forestier (FOR) | 137 | 382 | 2004–2010,2012–2013 | 2004 |
| Freycinet (FRY) | 511 | 562 | 1999–2014 | 2001 |
| Mt William (MTW) | N/A | 155 | 1999–2014 | 1996 |
| Narawntapu (NAR) | 258 | 153 | 1999–2000, 2003–2012 | 2007 |
| West Pencil Pine (WPP) | 55 | 357 | 2006–2014 | 2006 |
| Woolnorth (WOO) | 491 | N/A | 2006–2010, 2012 | N/A |
Five of these locations became infected with DFTD during the study; one location remained disease-free, and one location was already infected at the beginning of the study (Fig. 1, Table 1). We considered the first year of disease arrival as pre-disease in our analyses, as the generation time for Tasmanian devils is approximately two years. Tasmanian devils were trapped using standard protocols (Hamede et al. 2015) involving custom-built polypropylene pipe traps 30 cm in diameter (Hawkins et al. 2006). These traps were set and baited with meat for ten consecutive trapping nights. In each 25 km2 trapping site, at least 40 traps were set. Traps were checked daily, commencing at dawn. Each individual was permanently marked upon first capture with a microchip transponder (Allflex NZ Ltd, Palmerstone North, New Zealand). Additional specifics regarding field protocols, samples taken, and phenotypic data recorded can be found in Hawkins et al. (2006), Hamede et al. (2015), and Lazenby et al. (2018). Animal use was approved under IACUC protocol ASAF#04392 at Washington State University.
OVERVIEW OF RAD-CAPTURE ARRAY DEVELOPMENT
We used 90,000 Restriction-site Associated DNA sequencing (RAD-seq) loci generated from 430 individuals sampled across 39 sampling localities using the PstI restriction enzyme (Epstein et al. 2016; Hendricks et al. 2017) to develop a RAD-capture probe set (Ali et al. 2016). The details of the sample collection, preparation, and data processing of these original RAD loci is described in Epstein et al. (2016). Using these data, baits were designed to target a total of 15,898 RAD loci. This array included three categories of RAD loci (with some overlap among categories): (i) 7108 loci spread widely across the genome that were genotyped in more than half of the individuals, had ≤3 non-singleton SNPs, and had a minor allele frequency (MAF) ≥0.05; (ii) 6315 loci based on immune function, restricted to non-singleton SNPs genotyped in ≥ 1/3 of the individuals, in and within 50 kb of an immune-related gene; (iii) 3316 loci showing preliminary evidence of association with DFTD susceptibility with ≤5 non-singleton SNPs. Each RAD-capture locus was ≥20 kb away from other targeted loci to minimize potentially confounding effects of linkage disequilibrium. Additional details of the creation of the RAD-capture probe set have been summarized in Margres et al. (2018a).
DATA QUALITY AND FILTERING
Libraries produced from the RAD-capture arrays were constructed using 3,568 individuals from the seven distinct geographical locations across Tasmania (Fig. 1). Libraries were then sequenced on a total of 12 lanes of an Illumina platform (5 lanes on NextSeq at the University of Oregon Genomics & Cell Characterization Core Facility; seven lanes on HiSeq 4000 at the QB3 Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley). We de-multiplexed paired-end 150 bp reads, removed low quality reads, and removed potential PCR/optical duplicates using the clone_filter program (Stacks v1.21; Catchen et al. 2013). We then used Bowtie2 (Langmead & Salzberg 2012) to align reads to the reference genome (Murchison et al. 2012; Devil_ref v7.0 GCA_00189315.1 downloaded from Ensembl May 2017, N50 20.13 kilobases for contigs and 1847.19 kilobases for supercontigs). We required the entire read to align from one end to the other without trimming (-end-to-end) with sensitive and -X 900 mapping options. With the resulting bam files, we created individual GVCF files using the option to emit all sites in the aligned regions with HaplotypeCaller from GATK (McKenna et al. 2010). All GVCF files were analyzed together using the option GenotypeGVCFs that re-genotyped and re-annotated the merged records. We selected SNPs using the SelectVariants option and filtered using the following parameters: QD < 2.0 (variant quality score normalized by allele depth), FS > 60.0 (estimated strand bias using Fisher’s exact test), MQ < 40.0 (root mean square of the mapping quality of reads across all samples), MQRankSum < −12.5 (rank-sum test for mapping qualities of reference versus alternative reads), and Read-PosRankSum < −8 (rank-sum test for relative positioning of reference versus alternative alleles within reads). Using VCFtools (Danecek et al. 2011), additional filtering was performed to remove non-biallelic SNPs, indels, those with a minor allele frequency <0.05, and those missing data at more than 50% of the individuals genotyped and 40% of the sites across all individuals. To parse out the genetic-environmental associations of abiotic factors from disease, samples were divided into pre and post-disease subsets prior to analyses. After filtering, we retained 3287 Tasmanian devils (1521 before and 1765 after DFTD) and 6886 SNPs (Table 1). Post-filtering, we retained (i) 3084 SNPs (2912 unique SNPs) in RAD loci targeted for their high genotyping rate and distribution across the genome; (ii) 250 SNPs (203 unique SNPs) in RAD loci targeted for immune function; (iii) 913 SNPs (757 unique SNPs) in RAD loci targeted due to preliminary evidence of association with DFTD; and (iv) 2827 off-target SNPs widely distributed across the genome. There were 32 overlapping SNPs between categories (i) and (ii), 141 SNPs overlapping between categories (i) and (iii), and 16 SNPs overlapping between categories (ii) and (iii). However, because there was an overlap of non-unique SNPs between the overlapping categories, the numbers do not sum to the number total number of SNPs in each category.
ANALYSES OF POPULATION STRUCTURE
To examine the underlying population structure in our dataset, both pre- and post-disease, we used fastSTRUCTURE (Alexander, Novembre & Lange 2009). fastSTRUCTURE is an algorithm that estimates ancestry proportions using a variational Bayesian framework to infer population structure and assign individuals to genetic clusters. We ran both the pre- and post-disease data sets with the number of genetic clusters (K) set from 1 to 18. Genetic relationships amongst sampled populations were also evaluated using discriminant analysis of principal components (DAPC) analysis (Jombart et al. 2010) implemented in the Adegenet package in R (Jombart 2008). This method transforms genotypic data into principal components and then uses discriminant analysis to maximize between-group genetic variation and minimize within-group variation. To determine the optimal number of genetic clusters, we used the k-means clustering algorithm for increasing values of K from 1 to 20. We selected the optimal K value by selecting the visual “elbow” in the Bayesian information criterion (BIC) score, which is the lowest BIC value that also minimizes the number of components or genetic clusters retained.
GENETIC DIVERSITY
To identify signals of a potential genetic bottleneck, we tested for changes in genetic diversity pre- and post-disease emergence. We used VCFtools (Danecek et al. 2011) to calculate Weir and Cockerham’s estimator of FST (θ; Weir and Cockerham 1984) for all pairwise comparisons and all pre-post disease population comparisons. To gauge whether populations had significantly different FST values, we bootstrapped 95% confidence intervals for 10,000 iterations and tested whether the confidence intervals bounded zero. Additionally, we calculated estimated heterozygosity (Nei and Li 1979) and Tajima’s D (Tajima 1989) for each population before and after disease arrival to estimate standing levels of genetic variation (Danecek et al. 2011). Each metric was calculated by taking the average of each non-overlapping windows of 10 kilobases (kb) per population pre and post-disease. We also tested for changes in inbreeding (Fis) using the R package Adegenet (Jombart 2008) for each population pre and post-disease. Finally, we tested for changes in effective population size calculated from estimates of linkage disequilibrium among SNPs in NeEstimator v2 (Do et al. 2014).
ABIOTIC ENVIRONMENTAL VARIABLES
We used ArcGIS 10 to plot the location of every sampled Tasmanian devil. Pemberton et al. (1990) found that Tasmanian devils typically have a home range of 10–20 km2, with devils often having overlapping home ranges. A trapping area of 25 km2 thus reflects an area of overlapping home ranges for devils that has been consistently used by the Tasmanian Department of Primary Industries, Parks, Water and Environment, save the Tasmanian Devils project, University of Tasmania and other researchers for over two decades and has supported dozens of publications. Over the course of the 15 years (1999–2014; Table 1), however, there was some variation in the shape of the trapping grids at the sampling sites due to changes in land use and permissions. To account for this variation, we located the centroid of each sampling area using the calculate geometry tool in ArcGIS 10. We then drew a 25 km2 ellipse around each centroid to represent the total trapping area for each sampling location and extracted environmental data for each location. Although the Freycinet and Forestier sites were larger than other sampling sites, the centroids were in the middle of peninsulas, likely making them representatives of those sites. We collected abiotic environmental data describing variation in climate, elevation, vegetation, human development and hydrological features for this study (Table S1). The data layers for each of the climatic and vegetation variables for each site were collected and aggregated by WorldClim between 1970–2000 at 1-km spatial resolution (www.worldclim.org; Fick & Hijmans 2017). Elevation values were similarly calculated by extrapolating the centroid of the 25 km2 ellipses for each site; elevation data were collected and aggregated by Geoscience Australia between 2001–2015 at 5 m spatial resolution (www.ga.gov; Geoscience Australia 2015). The centroids for each of the climatic, elevational, and vegetative variables for each sampling location were used in downstream analyses. The only environmental variables that were not summarized as centroids were the length of sealed roads, length of public roads, and the total surface area of water. The data layers for these three variables were generated by the Land Information System Tasmanian between 2013 and 2016 at 25–500 m spatial accuracy (www.thelist.tas.gov.au). Sealed and public road lengths were calculated by cumulatively summing the total length of the sealed roads in the entire 25 km2 ellipse for each sampling location. The total surface area of water was calculated by adding the total surface area of hydrological features we hypothesized would be accessible to Tasmanian devils including “natural or dammed freshwater,” “stream,” and “watercourse” hydrological features. Table S1 includes all of the 18 abiotic environmental variables analyzed and the location of each centroid.
We extracted the values for each environmental variable at five randomly chosen locations within the ellipse generated for each trapping area to test whether there was significant heterogeneity for any of the environmental variables across the trapping area of any particular site. Using the calculate statistics tools in ArcGIS 10, we calculated the mean, standard deviation, and 95% confidence intervals for each of the environmental variables within each site. If the environmental values fell within the 95% confidence interval of the randomly selected points at each site, we deemed the environmental value as an adequate representation for that environmental variable site-wide (Table S2).
To estimate collinearity among environmental variables (Table S1), we conducted a principal component analysis (PCA) using the prcomp package in R including the environmental values for each of our seven sampling localities. The top two abiotic environmental variables that explained the greatest proportion of the variance in each of the first six principal components in the PCA were used as explanatory variables in the subsequent GEA analyses. We did not include the seventh principal component as the amount of variation in the data explained by this component was negligible (<0.001). To ensure that environmental variables were not correlated, we conducted paired correlative tests using Spearman’s ρ). Significantly correlated environmental variables were excluded (p ≤ 0.01).
BIOTIC ENVIRONMENTAL VARIABLES
To estimate disease prevalence within populations, we used trapping records compiled in a database provided by collaborators at the University of Tasmania and the Department of Primary Industries, Parks, Water and Environment from 1999 to 2014 from the six infected populations (McCallum et al. 2009; Hamede at al. 2009, 2012, 2013, 2015; Lachish et al. 2009). Field records contained data regarding every unique trapping encounter as well as phenotypic data for each devil. Likelihood of a single devil being infected with DFTD is recorded for devils in the field using a scale of 1–5 (Hawkins et al. 2006). We calculated disease prevalence using the total number of unique devils trapped with a high probability of being infected divided (DFTD score ≥ 3) by the number of unique devils captured alive that year. Infection probability scores were derived in the field based on visual infection status from 1 (no apparent signs of DFTD) to 3 (wounds and other irregularities present) to 4 (tumors present) (Lachish et al. 2007). In our GEAs, we averaged the annual disease prevalence values across years 2–4 after DFTD was confirmed at a particular sampling location. We selected these years instead of using the first year of infection of DFTD because there was variation in estimates of prevalence during the first year of DFTD detection and no guarantee that this truly was the first year of infection.
GENETIC-ENVIRONMENTAL ASSOCIATION (GEA) ANALYSES
We tested correlations of allele frequencies with abiotic and biotic environmental variables across the seven study locations using latent factor mixed models (LEA; Frichot & Francois 2015) and Bayenv2 (Gunther and Coop 2013). The R package for landscape and ecological association studies (LEAs) uses latent factors, analogous to principal components in a PCA, to account for background population structure (Frichot & Francois 2015). These latent factors serve as random effects in a linear model that tests for correlations between environmental variables and genetic variation (Frichot & Francois 2015). The number of populations sampled was used for the number of latent factors in LEA. Bayenv2 uses allelic data to generate a variance-covariance matrix, or kinship matrix, to estimate the neutral or null model for underlying demographic structure (Coop et al. 2010; Gunther and Coop 2013). Spearman’s p statistics were then calculated to provide non-parametric rankings of the strength of the association of each genotype with each environmental variable compared to the null distribution described by the kinship matrix alone (Gunther and Coop 2013). The non-parametric Spearman’s rank correlation coefficients have been shown to be more powerful in describing genetic-environmental relationships if there are extreme outliers (Whitlock and Lotterhos 2015; Rellstab et al. 2015).
CANDIDATE GENE IDENTIFICATION
Although each of the landscape genomic methods listed above detects GEAs, each program has been shown to vary in true-positive detection rate given sampling scheme, underlying demography, and population structure (Lotterhos and Whitlock 2014; Rellstab et al. 2015; Hoban et al. 2016). To reduce the false-positive rate without sacrificing statistical power, we used MINOTAUR (Lotterhos et al., 2017; Verity et al. 2017) to identify putative loci under selection. MINOTAUR takes the test statistic output from each of the GEAs and identifies the top outliers in multi-dimensional space; here, we used the Mahalanobis distance metric to ordinate our outliers. We selected this distance metric as it has been shown to have high statistical power on simulated genomic data sets (Verity et al. 2017) and because our data followed a parametric distribution. We chose our final set of candidate SNPs by selecting the top 1% of loci that had the largest Mahalanobis distance for each environmental variable. Thus, we performed seventeen separate MINOTAUR runs: eight for the eight abiotic environmental variables tested pre-DFTD arrival and nine, including the eight abiotic environmental variables and a single biotic variable, post-DFTD arrival.
We then used bedtools (Quinlan and Hall 2010) to identify genes within one kb of each candidate SNP in the devil reference genome (Murchison et al. 2010). Protein-coding genes within these windows were included in the list of candidate genes. Gene annotations were retrieved from the ENSEMBL database (Akey et al. 2002), gene IDs were derived from the NCBI GenBank database (Wheeler et al. 2007), and descriptions of putative function and gene ontologies were gathered from www.genecards.org (Fishilevich et al. 2017) and the Gene Ontology (GO) Consortium (Ashburner et al. 2000). We then tested for enrichment of GO terms in our candidate gene list using the R package SNP2GO (Szkiba et al. 2014). SNP2GO tests for over-representation of GO terms associated with genes within a specified region using Fisher’s exact test and corrects for multiple testing using the Benjamini-Hochberg and Yosef (1995) false-discovery rate (FDR). We ran this program on the pre- and post-disease candidate sets separately and used all genes within one kb of the original RAD-capture data set as our reference set. Enriched terms were those with an FDR ≤ 0.05.
FISHER′S EXACT TEST
To determine if GEAs detected pre-DFTD remained post-DFTD arrival, we calculated the change in the MINOTAUR rank of each candidate locus following disease arrival. We ranked loci by Mahalanobis distance and compared the rank of the pre-DFTD candidate loci to post-disease arrival for each of the eight abiotic environmental variables. To determine whether a greater proportion of loci detected pre-disease had reduced MINOTAUR rankings post-disease arrival than we would expect by random chance, we conducted Fisher’s exact tests using an α = 0.05. If we detected a significantly higher proportion of models in which the MINOTAUR rankings were lower post-disease than pre-disease, then we considered the molecular signal of the genetic-environmental association to be swamped by disease.
Results
POPULATION STRUCTURE AND GENETIC DIVERSITY
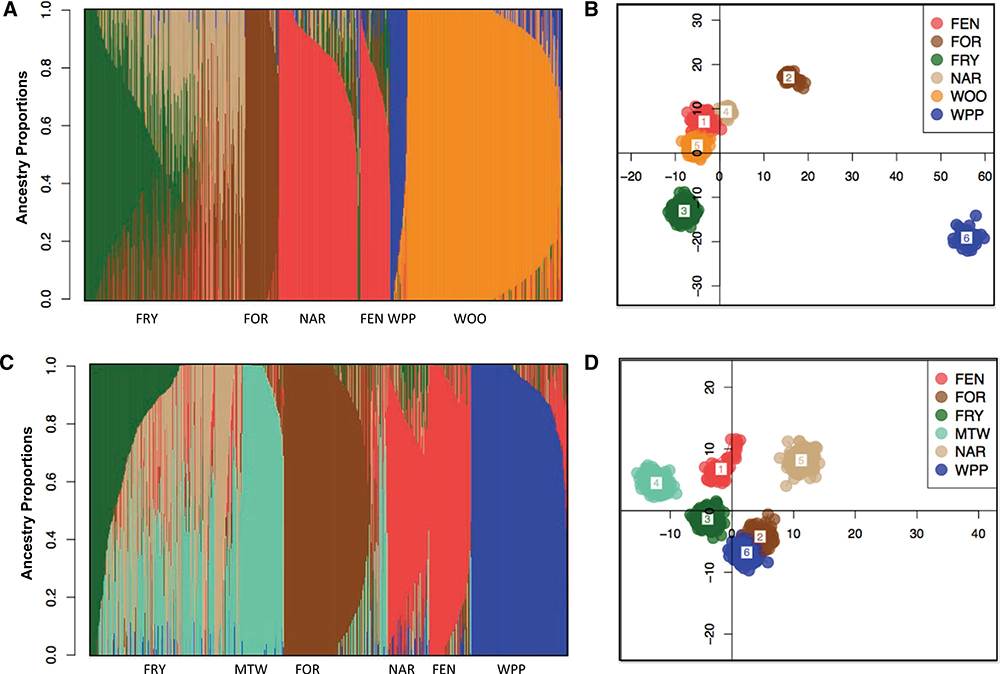
Our analyses generally supported K = 6 both pre and post-DFTD arrival reflecting the six populations sampled during each time period. However, there was uncertainty of the optimal K-value in fastSTRUCTURE. Using the “chooseK” python script recom-mended in fastSTRUCTURE, K = 9 (Fig. S1A) received the greatest support pre-DFTD and K = 8 post-DFTD (Fig. S1B). However, in both cases, the difference in the marginal likelihood between K = 4 and the selected K was very small (pre-DFTD ΔMarginal Likelihood = 0.003, post-DFTD ΔMarginal Likelihood = 0.0032; Figs. S1 and 2). For visual comparison, we also plotted K = 6 pre-DFTD (Fig. 2A) and post-DFTD (Fig. 2B), which reflects the number of populations sampled. The K value for DAPC that minimized the BIC, as well as the number of components included, was K = 6 both pre-DFTD (Fig. 2C) and post-DFTD (Fig. 2D). Pairwise FST values produced using Weir and Cockerham’s estimator would also support K = 6 as all bootstrapped confidence intervals for population comparisons did not bound zero (Table 2). There were no substantial differences between estimated heterozygosity, Tajima’s D, Nei’s Fis or effective population size (Ne) (Table S3) for any of the populations after DFTD arrived, suggesting there were no significant changes in genetic diversity following disease arrival.
Figure 2.
Population assignments computed by fastSTRUCTURE and DAPC for samples collected prior to (A and C) and post (B and D) DFTD arrival. (A and B) Each vertical bar in the fastSTRUCTURE plots represents a single individual sampled atone of the sampling locations which are abbreviated along the x-axis. Within population clusters, individuals are arranged by most common ancestry proportion. K = 6 is plotted here. Each color in all plots represents a distinct genetic cluster. (C and D) DAPC scatterplots show the first two principal components for K = 6.
Table 2.
The mean pairwise FST values ± 95% confidence intervals generated by resampling by bootstrapping 10,000 iterations. The values below the diagonal are pairwise comparisons for samples collected pre-disease and those above the diagonal are the same for post-disease. All pairwise comparisons did not bound zero and therefore were statistically significantly different from zero.
| Post-DFID |
||||||||
|---|---|---|---|---|---|---|---|---|
| FEN | FOR | FRY | MTW | NAR | WOO | WPP | ||
| Pre-DFTD | FEN | 0.049±0.028 | 0.051±0.026 | 0.062±0.035 | 0.026±0.015 | N/A | 0.066±0.037 | |
| FOR | 0.057±0.033 | 0.030±0.017 | 0.033±0.019 | 0.045±0.025 | N/A | 0.094±0.052 | ||
| FRY | 0.046±0.029 | 0.036±0.021 | 0.021±0.012 | 0.038±0.022 | N/A | 0.091±0.051 | ||
| MTW | N/A | N/A | N/A | 0.050±0.028 | N/A | 0.060 | ||
| NAR | 0.022±0.013 | 0.050±0.028 | 0.039±0.021 | N/A | N/A | 0.106±0.057 | ||
| WOO | 0.156±0.087 | 0.176±0.098 | 0.150±0.080 | N/A | 0.129±0.072 | N/A | ||
| WPP | 0.063±0.036 | 0.110±0.059 | 0.104±0.059 | N/A | 0.061±0.035 | 0.077±0.044 | ||
ENVIRONMENTAL VARIABLES
We initially considered 18 abiotic environmental variables (Table S1) that may be relevant to the distribution of genetic variation across the devil’s geographic range. Eight of these 18 variables (Table 3) explained a significant proportion (>0.99) of the variance in the environmental data as summarized in the top principal components (Table S4). Mean annual temperature and elevation were significantly correlated (mean annual = Spearman’s ρ = −0.964, p = 0.0028). We retained both of these environmental variables because we know they have a strong effect on gene flow in devil populations (Storfer et al. 2017). There were no statistically significant differences between the environmental values or any of the five randomly selected points for any of the environmental variables within each of the sampling locations (Table S2), indicating a lack of significant within-site heterogeneity. These eight abiotic environmental variables were subsequently used in the GEAs.
Table 3.
The values for the eight abiotic environmental variables and the biotic variable utilized in the GEA associations for each population. More details on the generation and calculation of these variables can be found in the methods.
| Mean Annual Temperature (°C) | Precipitation seasonality (mm) | Isothermality | Annual Temperature Range (°C) | Enhanced Vegetation Index (EVI) | Length of Sealed Roads (km) | Elevation (m) | Surface Area of Water (m2) | Average Disease Prevalence | |
|---|---|---|---|---|---|---|---|---|---|
| FEN | 10.51 | 18.24 | 49.87 | 20.07 | 34.87 | 314.16 | 232.15 | 356570.28 | 0.27 |
| FOR | 12.11 | 15.71 | 51.40 | 15.99 | 52.51 | 263.31 | 88.78 | 708797.79 | 0.22 |
| FRY | 12.67 | 14.51 | 54.09 | 17.24 | 74.01 | 61.65 | 57.17 | 4814676.77 | 0.11 |
| MTW | 13.12 | 21.61 | 49.88 | 16.4 | 52.15 | 420.78 | 43.75 | 2223617.42 | 0.19 |
| NAR | 12.48 | 31.57 | 48.07 | 18.09 | 76.58 | 671.02 | 74.31 | 6744655.41 | 0.02 |
| WPP | 7.98 | 31.17 | 47.98 | 17.11 | 61.24 | 245.23 | 703.74 | 77396.95 | 0.07 |
| WOO | 12.83 | 33.56 | 49.92 | 14.79 | 51.42 | 53.83 | 12.83 | 1066319.77 | 0.00 |
LANDSCAPE GENOMICS ANALYSES
Combining GEAs across all abiotic variables, we identified 365 unique SNPs pre-DFTD and 483 unique loci post-DFTD, of which 56 SNPs overlapped temporally, that received the greatest support from MINOTAUR as candidate loci. We identified candidate genes based on linkage disequilibrium (within one kb) to the unique loci, which resulted in 71 genes pre-disease (Table S5) and 105 genes post-disease (Table S6); twenty-four of those genes overlapped (Fig. S3, Table S5). Of the 483 unique loci post-DFTD, 59 unique loci were associated with disease prevalence. Of these 59 loci associated with disease prevalence, eight of these loci were also detected pre-disease arrival. Within one kb of these 59 SNPs, there were 13 annotated genes, two of which overlapped temporally with the pre-disease candidate set.
CANDIDATE GENE IDENTIFICATION FOR SELECTION BY THE ABIOTIC ENVIRONMENT
Mean annual temperature and annual temperature range were the two most common climatic variables significantly correlated with the allele frequencies of candidate SNPs among devil populations pre-disease arrival (Table S7). Twenty-six of the top 71 candidate genes pre-disease were associated with at least one of these two environmental variables and broadly associated with response to protein binding and response to stress (Tables S5 and S7). Specifically, genes associated with annual temperature range had GOs including cellular response to stress and RNA transport and processing. Mean annual temperature, in contrast, was correlated with genes with intracellular protein transport and protein localization GOs.
In addition to climatic variables, surface area of bodies of water, elevation, and vegetation index explained a large proportion of the observed variance in the abiotic environment across our seven sampling sites (Table S2). Surface area of bodies of water was correlated with 18 of the candidate genes pre-DFTD arrival that were associated with NOTCH signaling pathways and regulation of transcription by RNA-polymerase (Tables S5 and S7). Elevation was correlated with seven of the candidate genes with ion binding and oxidoreductase activity functions. Vegetation index was correlated with allele frequencies of 13 of the candidate genes that had GOs including transcription and cellular protein binding. Although mean elevation and mean annual temperature among sampling locations were found to be correlated, there were no shared associations between these two variables and any candidate genes (Table S7).
In our abiotic GEAs post-disease arrival, annual temperature range, mean annual temperature, and vegetation index were the top environmental variables most frequently associated with candidate SNPs (Tables S6 and S8). Similar to pre-DFTD, GOs for genes associated with these two temperature variables included a cellular response to stimulus, RNA processing and regulation, and ion binding. In contrast to pre-DFTD, mean annual temperature and annual temperature range were associated with genes with cell signaling, immune response, and apoptotic processes. Vegetation index was associated with 17 of the candidate genes post-DFTD with GOs including cellular differentiation and cell signaling pathways (Tables S6 and S8).
There was no significant enrichment of any GO category of our candidates (pre-disease p = 0.344; post-disease p = 0.297); there were commonalities in the putative functions of candidate genes pre-versus post-disease including regulation of transcription and translation and cellular response to external stress (Tables S5 and S6).
CANDIDATE GENE IDENTIFICATION FOR SELECTION BY THE BIOTIC ENVIRONMENT
Disease prevalence was significantly correlated with divergent allele frequencies among sampled devil populations (Tables S8 and S9). Thirteen of the 81 candidate genes detected uniquely post-DFTD arrival were associated with disease-prevalence (Tables S8 and S9). The associated GOs for these 13 genes included cell-cycle regulation, regulation of cell proliferation, and immune response (Table S6). All 13 genes were also associated with abiotic environmental variables. Genes TBXAS1 and FGGY were found in both the pre and post-DFTD candidate gene lists and associated with GOs including oxidoreductase activity and metabolism (Table S5).
FISHER′S EXACT TEST FOR DISEASE SWAMPING
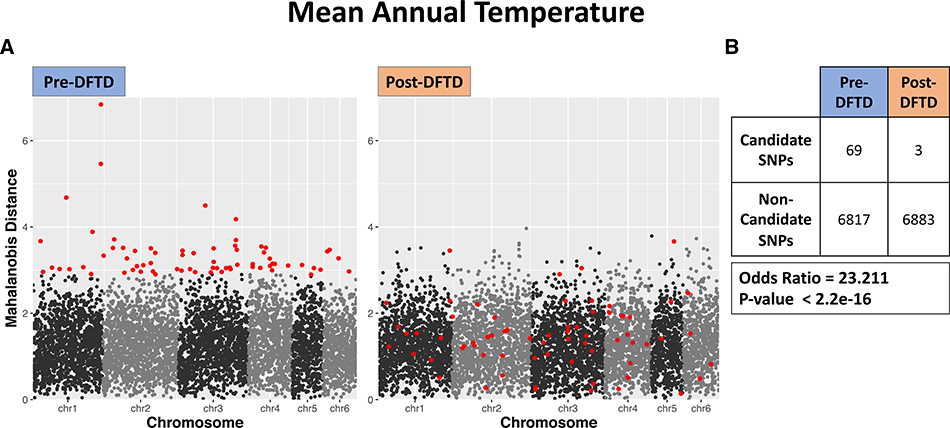
Putative GEAs found pre-DFTD were consistently undetected post-DFTD arrival (Tables S5 and S6). Using Fisher’s exact tests, fewer pre-DFTD candidate loci were detected post-DFTD arrival than expected by random chance for each MINOTAUR analysis for each of the eight abiotic environmental variables (Fig. 3; Fig. S4). For example, we only detected three of the 69 putative pre-DFTD candidate loci using MINOTAUR post-DFTD arrival for genetic associations with mean annual temperature across sampled populations (Fisher’s Exact Test Odd’s ratio = 23.211, p < 0.001; Fig. 3).
Figure 3.
Mahalanobis distances from MINOTAUR for each SNP pre-DFTD arrival (left plot) and post-DFTD arrival (right plot) for GEAs with mean annual temperature. SNPs are ordered by position along the chromosomes. The top 1% of loci with the largest Mahalanobis distance values pre-DFTD are indicated in red. (b) Post-hoc analyses found that a significantly greater number of candidate loci detected pre-DFTD arrival were not detected post-DFTD arrival. This trend was detected consistently across all eight of the abiotic environmental variables tested in genetic-environmental association analyses (output from the remaining seven variables can be found in Fig. S2).
Twenty-four of the original 71 candidate genes detected pre-disease overlapped with the 105 candidate genes post-disease arrival (Table S5). Four were associated with at least one of the same environmental variables post-disease arrival as pre-disease, and 20 were correlated with different variables (Table S9). Two of the variants linked with TBXAS1 and FGGY were found to be associated with disease prevalence post-disease arrival, but abiotic environmental variables pre-disease arrival. For example, the FGGY gene was originally associated with isothermality prior to disease arrival, but disease prevalence post-arrival. Putative functions of FGGY include carbohydrate phosphorylation and neural cell homeostasis (Singh et al. 2017; Dunckley et al. 2007; Table S5). Similarly, TBXAS1, which is putatively involved in oxidoreductase activity (Ullrich & Brugger 1994), was associated with vegetation index and mean annual temperature pre-disease arrival but was more strongly correlated with disease prevalence following disease arrival (Tables S5 and S9).
Discussion
Here, we showed Tasmanian devils are more genetically structured across Tasmania than previously documented (Miller et al. 2011; Brüniche-Olsen et al. 2014; Hendricks et al. 2017) thereby creating conditions that favor local adaptation. Indeed, GEAs showed signatures of selection across populations, suggestive of adaptation to local abiotic factors prior to disease arrival. Mean annual temperature and annual temperature range were abiotic variables most frequently associated with candidate genes (Xu et al. 2017), and these abiotic factors have been established as important for devil habitat use (Jones and Rose 1996; Jones and Barmuta 2000). Surface area of bodies of water, elevation, and vegetation index were also associated with candidate genes and correlated with the largest amount of geographic heterogeneity across the landscape (Zhao et al. 2014; Schweizer et al. 2016; Mukherjee et al. 2019). Nonetheless, most of the candidate genes detected prior to disease arrival were not detected post-DFTD emergence, reflecting the strong selection imposed by DFTD. Instead, GEAs showed evidence of significant correlations of disease prevalence with allele frequencies of several SNPs after disease arrival functionally related to immune response, apoptosis, and tumor regression. Taken together, these results suggest that DFTD swamps molecular signatures of local adaptation to abiotic variables. In contrast to previous findings (Brüniche-Olsen et al. 2016) and despite large declines in census population size across all diseased populations, no substantial changes in genetic diversity or effective population size were detected pre- versus post-disease, suggesting the detected patterns were likely not the result of genetic drift.
We found conflicting results among our demographic analyses of our sampled populations. Similar to previous studies, we detected admixture between several of the seven sampling locations using fastSTRUCTURE (Fig. 2A), which showed an optimal K value between 4 and 9. However, using DAPC (Fig. 2B), we detected distinct genetic clusters among the sampling locations, regardless of disease presence. Pairwise population FST calculations were all significantly greater than zero, indicating significant population differentiation between all populations sampled (Table 2). Regardless of the analysis employed, we identified six genetic clusters throughout the study area, representing greater amounts of genetic structure than previously detected. Two previous studies found high levels of admixture between two genetic clusters separating eastern and western Tasmania (Brüniche-Olsen et al. 2014; Hendricks et al. 2017), while another, based on mtDNA genomes, suggested three genetic clusters (Northwest, East, and Central; Miller et al. 2011). Our results differed from the previous work because we included much larger sample sizes and numbers of loci, which likely increased our power to detect population structure.
Despite extensive, range-wide population declines, we found no evidence for loss of genetic diversity following disease arrival. Devil populations had low levels of heterozygosity across the species’ geographic range, regardless of whether DFTD was present (Table S3), consistent with findings from previous studies examining levels of genetic variation among populations pre-DFTD (Jones et al. 2004; Brüniche-Olsen et al. 2014; Hendricks et al. 2017). While there was population genetic differentiation (FST) between sampled populations, there was little difference in inbreeding coefficient (FIS) among populations pre- versus post-disease arrival. Lack of a significant change in the positive Tajima’s D values across populations following disease arrival (Table S3) also suggests maintenance of standing genetic variation even after substantial population declines. One possibility for the lack of detectable changes in genetic diversity would be that simply not enough time has passed since the bottleneck. If we take into account the increased precocial breeding in diseased populations, which reduces the generation time of females from 2 to 3 years prior to disease outbreak to 1.5–2 years post-disease (Jones et al. 2008), the maximum number of generations in our data set since disease arrival would be in Freycinet for 8–9 generations (see Table 1; 2–9 generations across all populations). Detection of significant changes in genetic diversity requires substantial reduction in effective population size for several generations (Luikart et al. 2010). Following DFTD outbreak, devil populations typically decline by 90% after 5–6 years. This time lag, coupled with the short timescale, perhaps resulted in low power to detect changes in genetic diversity.
The abiotic environmental variables used in our GEAs have been shown to be important determinants of devil distribution (Rounsevell et al. 1991; Jones and Rose 1996; Jones and Barmuta 2000). Devils are distributed throughout the diverse habitats of Tasmania, but their core distribution comprises areas of low to medium rainfall which are dominated by dry, open Eucalypt forest and coastal shrubland. Devils use varying vegetation types across the environment for different functions. Devils prefer a clear understory for movement, forest-grassland edges for hunting, and generally avoid structurally complex vegetation and landscape features, such as rocky areas and steep slopes (Jones and Rose 1996; Jones and Barmuta 2000). Candidate loci linked to the CUX1 gene were strongly associated with vegetation index in devils, especially in heavily forested populations such as WPP (Table S7). CUX1, which is putatively involved in transcription regulation (Sansregret & Alain 2008) and limb development in morphogenesis (Lizarraga et al. 2002), has been detected in mammalian systems as a candidate for adaptation to environments with a complex understory (Schweizer et al. 2016; Mukherjee et al. 2019). Differential expression of this gene in muscle tissue of the bovine species Bos frontalis relative to domestic cattle was suggested to assist in reducing energetics required for navigating complex, hilly environments in India (Mukherjee et al. 2019).
Significant genetic variation among devil populations was also shown to be associated with the total surface area of bodies of fresh and marine water between sampling sites. A previous transcriptomics studies of Tasmanian devil populations showed significant enrichment of differential expression of pre-disease candidate genes associated with environmental variation between coastal and inland populations (Fraik et al. 2019). Strong genetic-environmental associations across devil populations throughout their heterogeneous geographic range provide evidence for the presence of local adaptive genetic variation.
Although we found no significant enrichment for any GO category in the 71 genes identified in strong linkage disequilibrium with the top 1% of SNPs pre-DFTD, 21 of the genes were putatively involved in the cellular response to the stimulus, and regulation of transcription and RNA polymerase II (Table S5). Climatic variables including mean annual temperature, annual temperature range, and isothermality were all important in describing observed genetic variation across the devil geographic range.
Following the arrival of DFTD, both the number of loci and proportion of variation in the allele frequencies of the pre-disease candidate loci explained by the abiotic environment were significantly reduced. The ANKYM2 gene, for example, was strongly correlated with mean annual temperature prior to disease arrival (pre-disease, MINOTAUR rank = 36), but the strength of the genetic-environmental association significantly decreased post-disease arrival and it was no longer detected as a candidate locus (post-disease, MINOTAUR rank = 2696). Only 24 of the 71 candidate genes detected pre-disease were included in the 105 genes detected post-disease, with only one gene, ARPC2 (Actin related protein 2/3 Complex Subunit 2), uniquely correlated with the same variable prior to disease arrival. FGGY, for example, was strongly correlated with mean annual temperature prior to disease arrival (pre-disease, Pearson’s correlate = −0.745, MINOTAUR rank = 25). Post-disease arrival, the strength of this relationship significantly decreased (post-disease, Pearson’s correlate = −0.082, MINOTAUR rank = 6615), and the SNP linked to this gene was more strongly correlated with disease prevalence (post-disease, MINOTAUR rank = 59). These discordant patterns of association of candidate loci with environmental variables pre-compared to post-disease arrival may possibly be explained by pleiotropy or loss of association due to DFTD.
We detected 105 candidate genes post-disease arrival associated with both abiotic environmental variables and/or disease prevalence. Although there was no significant enrichment of any GO category, most genes associated with abiotic variables had stress response GOs, whereas post-disease many genes had GOs involved in cellular processes including apoptosis, cell differentiation and cell development, similar to what has been previously detected (Epstein et al. 2016; Margres et al. 2018a; Frampton et al. 2018). Genes involved in apoptosis detected in our GEAs, including DNAJA3 in the heat shock protein family, have also been previously identified in DFTD literature as candidates for anticancer vaccines (Tovar et al. 2018) as well as for their putative involvement in immunogenicity (Graner et al. 2000) and tumor suppression (Shinagawa et al. 2008). Functionally similar to PAX3, a gene associated with apoptosis and angiogenesis (Asher et al. 1996) and devil tumor regression detected in previous studies (Wright et al. 2017), we detected genes including FLCN and BMPER post-DFTD arrival (Qi et al. 2009). We also identified MGLL and TLR6 in our post-DFTD candidate list that are involved in the inflammatory response (Epstein et al 2016; Margres et al. 2018a).
The identification of candidate genes putatively involved in immune response (e.g., RFN126 and IL9R), tumor suppression (e.g., DMBT1, FLCN, and ITFG1), signal transduction (e.g. ARHGEF37 and PPP1R12B) and cell-cycle regulation (e.g. TK2) associated with disease prevalence post-DFTD arrival provide evidence that disease may have had a strong influence on devil populations (Mollenhauer et al. 2003; Tsapogas et al. 2003; Bannert et al. 2003; Leushacke et al. 2011; Purwar et al. 2012; Tsai et al. 2014; Sun, Eriksson & Wang et al. 2014; Hasumi et al. 2015).
The lack of GO enrichment may be an artifact of the RAD-capture panel. Loci targeted herein were linked to or found in coding regions of both putatively neutral genes as well as those involved in immune response, potentially biasing our GO enrichment analysis. Another explanation for the lack of enrichment could stem from our conservative approach for identifying outliers. Using MINOTAUR, we took the top 1% of loci from each GEA as our putative candidates. Although this approach reduces false positives, it also can reduce true positives and imposes an upper bound on the number of possible associations we can ascertain between our candidate loci and disease.
Intense, long-term monitoring of Tasmanian devils coupled with an expansive temporal and geographic dataset provided the unique opportunity to test for changes in statistical signatures of GEAs. We hypothesized that DFTD would serve as an extremely selective event as it is nearly 100% lethal (Hamede et al. 2015) and can produce a rapid adaptive response (Epstein et al 2016; Wright et al. 2017; Margres et al. 2018a, b) that could swamp pre-disease allele frequency correlations with the abiotic environment detected with GEAs. Indeed, variation in allele frequencies of loci linked to candidate genes among devil populations prior to DFTD arrival appeared to be strongly associated with surface area of water among our coastal and inland populations (Tables S7–S9; Fraik et al. 2019). However, following disease arrival these GEAs were not detected. Although annual temperature range and vegetation index appeared to be important sources of selection in devil populations regardless of disease presence, most candidate genes identified pre-DFTD were not correlated with these variables after DFTD arrived (Fig. 3; Fig. S4).
The observed loss of pre-DFTD GEAs following disease arrival could also be the result of stochastic processes, such as drift operating on our populations following a significant demographic event (Lande 1976, 1993; Brüniche-Olsen et al. 2016). Following large catastrophes, locally adapted genotypes may be displaced or swamped out randomly by new genetic variation from neighboring populations via genetic rescue (Waddington 1974; Brown and Kodric-Brown 1977). If this were the case, our observation of loss of pre-DFTD candidate genes may be due to drift resulting from demographic change induced by DFTD versus the selection that DFTD imposed. In a previous study, Brüniche-Olsen and colleagues used time-series analysis and FST outlier tests to test for parallel signatures of selection in response to DFTD emergence. Using 1482 SNPs from devils sampled from six populations, they found discordance between the use of single-point and multiple timepoint selection analyses that suggested a non-conserved evolutionary response to DFTD (Brüniche-Olsen et al. 2016). However, numerous follow-up studies using significantly more loci, including another time-series analysis (Epstein et al. 2016), have provided additional evidence for a rapid evolutionary response to DFTD across devil populations, suggesting that DFTD is selecting for particular genetic variants post-DFTD arrival (summarized in Russell et al. 2018 and Storfer et al. 2018b). Additionally, despite large decreases in field- based estimates of population size, we find no detectable significant changes to genetic diversity or effective population size following disease arrival and therefore no evidence of genetic drift.
Emerging infectious diseases are increasingly recognized as significant threats to biodiversity and in extreme cases can lead to species’ range contractions and extinctions (Smith et al. 2006). Yet, few landscape genomics studies have tested for statistical correlations of allele frequencies with biotic variables such as infectious diseases. Here, we document a novel, biotic variable swamping out molecular signals of association to the abiotic environment. This finding is consistent with previous work that showed rapid evolution of Tasmanian devils in response to disease (Epstein et al. 2016; Wright et al. 2017; Margres et al. 2018a, b). Additionally, no appreciable declines in genetic diversity were detected across multiple analysis methods. Taken together, these results suggest that the observed patterns of allele frequency correlations with disease prevalence are more likely attributable to selection than genetic drift. The findings of this study demonstrate the utility of landscape genomics as a tool to explicitly test the influence of biotic factors, such as disease, on the spatial distribution of genetic variation.
Supplementary Material
Supplementary Table 1. The eighteen abiotic environmental variables used in the landscape genomic analyses as well as the easting and northing for each sampling location.
Supplementary Table 2. Mean and standard error estimates for five of the abiotic environmental variables used in the landscape genomic analyses.
Supplementary Table 3. Mean estimated heterozygosity, Tajima’s D, FIS and Ne values for the five populations in which samples were collected both prior to and post-disease arrival.
Supplementary Table 4. Summary of the PCA loading scores of the first six principal components for vegetative and geographic features for the seven sampling localities which cumulatively explain >99% of the observed variance.
Supplementary Table 5. The 71 candidate genes detected pre-disease using landscape genomics analyses.
Supplementary Table 6. The 81 candidate genes uniquely detected post-disease using landscape genomics analyses.
Supplementary Table 7. The 47 candidate genes detected uniquely pre-disease and the abiotic environmental variable(s) which the genetic-environmental association analyses detected statistically significant associations.
Supplementary Table 8. The 81 candidate genes uniquely detected post-disease and the abiotic and biotic environmental variable(s) which the genetic-environmental association analyses detected statistically significant associations.
Supplementary Table 9. The 24 candidate genes that were detected with significant genetic-environmental associations both pre- and post-DFTD and the abiotic/biotic environmental variable(s) which had statistically significant associations.
Supplementary Figure 1. Population assignments computed by fastSTRUCTURE for samples collected prior to (a,c) and post (b,d) DFTD arrival.
Supplementary Figure 2. Marginal likelihood output produced for the population assignments computed by fastSTRUCTURE pre-DFTD (a) and post- DFTD (b).
Supplementary Figure 3. Venn diagram showing the overlap of MINOTUAR candidate genes for the eight abiotic environmental variables in the pre-disease (left) and post-disease (right) candidate gene sets.
Supplementary Figure 4. (a-g) Mahalanobis distances from MINOTAUR for each SNP pre-DFTD arrival (left plot) and post-DFTD arrival (right plot) for seven of the abiotic environmental variables.
ACKNOWLEDGMENTS
We thank Omar Cornejo, and his laboratory, David Crowder, and Joanna Kelley’s laboratory for their helpful review and insightful discussion. We are grateful to the anonymous reviewers for feedback on our manuscript. This work was funded by NSF grant DEB-1316549 and NIH grant R01- GM126563 to A.S., P.A.H., M.J., and H.M. as part of the joint NSF-NIH- USDA Ecology and Evolution of Infectious Diseases program. Bioinformatics work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the NIH under grant number P30 GM103324.
Footnotes
DATA ARCHIVING
The sequence data has been deposited at NCBI under BioProject PRJNA306495 (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA306495) and BioProject PRJNA634071 (http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA634071). Code is available at github.com/jokelley/devil-landscape-genomics.
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
LITERATURE CITED
- Akey JM, Zhang G, Zhang K, Jin L, and Shriver MD. 2002. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 12:1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, and Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, Miller MR. 2016. RAD Capture (rapture): flexible and efficient sequence-based genotyping. Genetics 202:389–400. 10.1534/genetics.115.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AsherH, Sommer A, Morell R, Friedman TB. 1996. Missense mutation in the paired domain of PAX3 causes craniofacial-deafness-hand syndrome. Human Mutation 7:30–35. 10.1002/(sici)1098-1004(1996)7:1{30::aid-humu4)3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Bannert N, Vollhardt K, Asomuddinov B, Haag M, Konig H, Norley S, and Kurth R. 2003. PDZ Domain-mediated interaction of interleukin- 16 precursor proteins with myosin phosphatase targeting subunits. J. Biol. Chem. 278:42190–42199. [DOI] [PubMed] [Google Scholar]
- Beall CM 2007. Two routes to functional adaptation: Tibetan and Andean high-altitude natives. Proc. Natl. Acad. Sci. U. S. A. 104 Suppl 1:8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BenjaminiYoav Hochberg Yosef. 1995. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 57 (1): 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Biek R, and Real LA. 2010. The landscape genetics of infectious disease emergence and spread. Mol. Ecol. 19:3515–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, and Kodric-Brown A. 1977. Turnover rates in insular biogeography: effect of immigration on extinction. Ecology 58:445–449 [Google Scholar]
- Brüniche-Olsen A, Burridge CP, Austin JJ, and Jones ME 2013. Disease induced changes in gene flow patterns among Tasmanian devil populations. Biol. Conserv. 165:69–78. [Google Scholar]
- Brüniche-Olsen A, Jones ME, Austin JJ, Burridge CP, and Holland BR. 2014. Extensive population decline in the Tasmanian devil predates European settlement and devil facial tumour disease. Biol. Lett. 10:20140619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüniche-Olsen A, Austin JJ, Jones ME, Holland BR, and Burridge CP. 2016. Detecting selection on temporal and spatial scales: a genomic time-series assessment of selective responses to devil facial tumor disease. PLoS One 11:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, and Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22:3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, and Pritchard JK. 2010. Using environmental correlations to identify loci underlying local adaptation. Genetics 185:1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Grande F, Rolshausen G, Divakar PK, Crespo A, Otte J, Schleuning M, and Schmitt I. 2018. Environment and host identity structure communities of green algal symbionts in lichens. New Phytol. 217:277–289. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DoC., Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. 2014. NeEstimatorv2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14:209–214. 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, Joshipura K, Halperin RF, Stamper C, Jensen KR, Letizia D, et al. 2007. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N. Engl. J. Med. 357:775–788. [DOI] [PubMed] [Google Scholar]
- Eoche-Bosy D, Gautier M, Esquibet M, Legeai F, Bretaudeau A, Bouchez O, Fournet S, Grenier E, and Montarry J. 2017. Genome scans on experimentally evolved populations reveal candidate regions for adaptation to plant resistance in the potato cyst nematode Globodera pallida. Mol. Ecol. 26:4700–711. [DOI] [PubMed] [Google Scholar]
- Epstein B, Jones M, Hamede R, Hendricks S, McCallum H, Murchison EP, Schonfeld B, Wiench C, Hohenlohe P, and Storfer A. 2016. Rapid evolutionary response to a transmissible cancer in Tasmanian devils. Nat. Commun. 7:12684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FickStephen E, Hijmans Robert J. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37:4302–4315. 10.1002/joc.5086 [DOI] [Google Scholar]
- Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, Rosen N, Kohn A, Twik M, Safran M et al. 2017. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraik AK, Quackenbush C, Margres MJ, Comte S, Hamilton DG, Kozakiewicz CP, Jones M, Hamede R, Hohenlohe PA, Storfer A, et al. 2019. Transcriptomics of tasmanian devil (sarcophilus harrisii) ear tissue reveals homogeneous gene expression patterns across a heterogeneous landscape. Genes 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D, Schwenzer H, Marino G, Butcher LM, Pollara G, Kriston-Vizi J, Venturini C, Austin R, de Castro KF, Ketteler R, et al. 2018. Molecular signatures of regression of the canine transmissible venereal tumor. Cancer Cell 33:620–633 e626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois O, Martins H, Caye K, and Schoville SD. 2016. Controlling false discoveries in genome scans for selection. Mol. Ecol. 25:454–469. [DOI] [PubMed] [Google Scholar]
- Frichot E, and Francois O. 2015. Testing for associations between loci and environmental gradients using latent factor mixed models. Mol. Biol. Evol. 30:1687–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Australia Geoscience. 2015. Digital elevation model (DEM) of Australia derived from LiDAR 5 Metre Grid. Geoscience Australia, Canberra. http://pid.geoscience.gov.au/dataset/ga/89644 [Google Scholar]
- Graner M, Raymond A, Akporiaye E, and Katsanis E. 2000. Tumor-derived multiple chaperone enrichment by free-solution isoelectric focusing yields potent antitumor vaccines. Cancer Immunol Immunother 49:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, and Coop G. 2013. Robust identification of local adaptation from allele frequencies. Genetics 195:205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, and Payseur BA. 2016. Fifteen years of genomewide scans for selection: trends, lessons and unaddressed genetic sources of complication. Mol. Ecol. 25:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamede R, Lachish S, Belov K, Woods G, Kreiss A, Pearse AM, Lazenby B, Jones M, and McCallum H. 2012. Reduced effect of Tasmanian devil facial tumor disease at the disease front. Conserv. Biol. 26:124–134. [DOI] [PubMed] [Google Scholar]
- Hamede RK, Bashford J, McCallum H, and Jones M. 2009. Contact networks in a wild Tasmanian devil (Sarcophilus harrisii) population: using social network analysis to reveal seasonal variability in social behaviour and its implications for transmission of devil facial tumour disease. Ecol. Lett. 12:1147–1157. [DOI] [PubMed] [Google Scholar]
- ———2013. Biting injuries and transmission of Tasmanian devil facial tumour disease. J. Anim. Ecol. 82:182–190. [DOI] [PubMed] [Google Scholar]
- Hamede RK, Pearse AM, Swift K, Barmuta LA, Murchison EP, and Jones ME. 2015. Transmissible cancer in Tasmanian devils: localized lineage replacement and host population response. Proc. Biol. Sci. 282:20151468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison TL, Wood CW, Borges IL, and Stinchcombe JR. 2017. No evidence for adaptation to local rhizobial mutualists in the legume Medicago lupulina. Ecol. Evol. 7:4367–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasumi H, Baba M, Hasumi Y, Lang M, Huang Y, Oh HF, Matsuo M, Merino MJ, Yao M, Y Ito et al. 2015. Folliculin-interacting proteins Fnip1 and Fnip2 play critical roles in kidney tumor suppression in cooperation with Flcn. Proc. Natl. Acad. Sci. U. S. A. 112: E1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins CE, Baars C, Hesterman H, Hocking GJ, Jones ME, Lazenby B, Mann D, Mooney N, Pemberton D, Pyecroft S, et al. 2006. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol. Conserv. 131: 307–324. [Google Scholar]
- Hendricks S, Epstein B, Schonfeld B, Wiench C, Hamede R, Jones M, Storfer A, and Hohenlohe P. 2017. Conservation implications of limited genetic diversity and population structure in Tasmanian devils (Sarcophilus harrisii). Conserv. Genet. 18:977–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, Poss ML, Reed LK, Storfer A, and Whitlock MC. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat. 188:379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, McCallum HI, Jones ME, Lawrance MF, Hamede RK, and Storfer A. 2019. Conserving adaptive potential: lessons from Tasmanian devils and their transmissible cancer. Conserv. Genet. 20:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405 [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, and Balloux F. 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, and Barmuta LA. 2000. Niche differentiation among sympatric Australian dasyurid carnivores. J. Mammal 81:434–447. [Google Scholar]
- Jones ME, Cockburn A, Hamede R, Hawkins C, Hesterman H, Lachish S, Mann D, McCallum H, and Pemberton D. 2008. Life-history change in disease-ravaged Tasmanian devil populations. Proc. Natl. Acad. Sci. U. S. A. 105:10023–10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Paetkau D, Geffen E, and Moritz C. 2004. Genetic diversity and population structure of Tasmanian devils, the largest marsupial carnivore. Mol. Ecol. 13:2197–2209. [DOI] [PubMed] [Google Scholar]
- Jones ME, and Rose RK. 1996. Preliminary assessment of distribution and habitat associations of the spotted-tailed quoll (Dasyurus maculatus maculatus) and eastern quoll (D. viverrinus) in Tasmania to determine conservation and reservation status: Tasmanian Public Land Use Commission, Hobart, Tas. [Google Scholar]
- Kozakiewicz CP, Burridge CP, Funk WC, VandeWoude S, Craft ME, Crooks KR, Ernest HB, Fountain-Jones NM, and Carver S. 2018. Pathogens in space: Advancing understanding of pathogen dynamics and disease ecology through landscape genetics. Evol. Appl. 11:1763–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LachishShelly McCallum Hamish, Menna Jones. 2009. Demography, disease and the devil: life-history changes in a disease-affected population of Tasmanian devils (Sarcophilus harrisii). Journal of Animal Ecology 78 (2): 427–436. 10.1111/j.1365-2656.2008.01494.x. [DOI] [PubMed] [Google Scholar]
- LACHISHSHELLY JONES MENNA MCCALLUMHAMISH. 2007. The impact of disease on the survival and population growth rate of the Tasmanian devil. Journal of Animal Ecology 76 (5): 926–936. 10.1111/j.1365-2656.2007.01272.x. [DOI] [PubMed] [Google Scholar]
- Lande R 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30:314–334. [DOI] [PubMed] [Google Scholar]
- ——— 1993. Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 142:911–927. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazenby BT, Toler MW, Brown WE, Hawkins CE, Hocking GJ, Hume F, Huxtable S, Iles P, Jones ME, Lawrence C et al. 2018. Density Trends and demographic signals uncover the long-term impact of transmissible cancer in Tasmanian devils. J. Appl. Ecol. 55:1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo SS, Gonzalez A, and Millien V. 2016. Multi-taxa integrated landscape genetics for zoonotic infectious diseases: deciphering variables influencing disease emergence. Genome 59:349–361. [DOI] [PubMed] [Google Scholar]
- Leushacke M, Sporle R, Bernemann C, Brouwer-Lehmitz A, Fritzmann J, Theis M, Buchholz F, Herrmann BG, and Morkel M. 2011An RNA interference phenotypic screen identifies a role for FGF signals in colon cancer progression. PLoS One 6:e23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LizarragaGail Lichtler Alexander, Upholt William B., Kosher Robert A. 2002. Studies on the Role of Cux1 in Regulation of the Onset of Joint Formation in the Developing Limb. Developmental Biology 243 (1): 44–54. 10.1006/dbio.2001.0559. [DOI] [PubMed] [Google Scholar]
- Lotterhos KE, Card DC, Schaal SM, Wang L, Collins C, Verity B, and Kelley J. 2017. Composite measures of selection can improve the signal-to-noise ratio in genome scans. Methods in Ecology and Evolution 8:717–727. [Google Scholar]
- Lotterhos KE, and Whitlock MC. 2014. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol. Ecol. 23:2178–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Hoban S, Kelley JL, Lotterhos KE, Reed LK, Antolin MF, and Storfer A. 2017. Breaking RAD: an evaluation of the utility of restriction site-associated DNA sequencing for genome scans of adaptation. Mol. Ecol. Resour. 17:142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Ryman N, Tallmon DA, Schwartz MK, and Allendorf FW. 2010. Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conservation Genetics 11:355–373. [Google Scholar]
- Mackinnon MJ, Ndila C, Uyoga S, Macharía A, Snow RW, Band G, Rautanen A, Rockett KA, Kwiatkowski DP, and Williams TN. 2016. Environmental correlation analysis for genes associated with protection against malaria. Mol. Biol. Evol. 33:1188–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel S, Joost S, Epperson BK, Holderegger R, Storfer A, Rosenberg MS, Scribner KT, Bonin A, and Fortin MJ. 2010. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol. Ecol. 19:3760–3772. [DOI] [PubMed] [Google Scholar]
- Margres MJ, Jones ME, Epstein B, Kerlin DH, Comte S, Fox S, Fraik AK, Hendricks SA, Huxtable S, Lachish S, et al. 2018a. Large-effect loci affect survival in Tasmanian devils (Sarcophilus harrisii) infected with a transmissible cancer. Mol. Ecol. 27:4189–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margres MJ, Ruiz-Aravena M, Hamede R, Jones ME, Lawrance MF, Hendricks SA, Patton A, Davis BW, Ostrander EA, McCallum H, et al. 2018b. The genomic basis of tumor regression in tasmanian devils (Sarcophilus harrisii). Genome Biol. Evol. 10:3012–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H 2008. Tasmanian devil facial tumour disease: lessons for conservation biology. Trends Ecol Evol. 23:631–637. [DOI] [PubMed] [Google Scholar]
- ——— 2012. Disease and the dynamics of extinction. Philos Trans R. Soc. Lond B Biol Sci. 367:2828–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum H, Jones M, Hawkins C, Hamede R, Lachish S, Sinn D, Beeton N, and Lazenby B. 2009. Transmission dynamics of Tasmanian devil facial tumor disease may lead to disease-induced extinction. Ecology 90:3379–3392. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W, Hayes VM, Ratan A, Petersen DC, Wittekindt NE, Miller J, Walenz B, Knight J, Qi J, Zhao F, et al. 2011. Genetic diversity and population structure of the endangered marsupial Sarcophilus harrisii (Tasmanian devil). Proc. Natl. Acad. Sci. U. S. A. 108:12348–12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollenhauer J, Deichmann M, Helmke B, Muller H, Kollender G, Holmskov U, Ligtenberg T, Krebs I, Wiemann S, Bantel-Schaal U, et al. 2003. Frequent downregulation of DMBT1 and galectin-3 in epithelial skin cancer. Int. J. Cancer 105:149–157. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Mukherjee A, Jasrotia RS, Jaiswal S, Iquebal MA, Longkumer I, Mech M, Vupru K, Khate K, Rajkhowa C, et al. 2019. Muscle transcriptome signature and gene regulatory network analysis in two divergent lines of a hilly bovine species Mithun (Bos frontalis). Genomics 112:252–262. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C et al. 2012. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell 148:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Tovar C, Hsu A, Bender HS, Kheradpour P, Rebbeck CA, Obendorf D, Conlan C, Bahlo M, Blizzard CA et al. 2010. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science 327:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, and Li WH. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U. S. A. 76:5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton AH, Margres MJ, Stahlke AR, Hendricks S, Lewallen K, Hamede RK, Ruiz-Aravena M, Ryder O, McCallum HI, Jones ME, et al. 2019. Contemporary demographic reconstruction methods are robust to genome assembly quality: A case study in Tasmanian Devils. Mol. Biol. Evol. 36:2906–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse AM, Swift K, Hodson P, Hua B, McCallum H, Pyecroft S, Taylor R, Eldridge MD, and Belov K. 2012. Evolution in a transmissible cancer: a study of the chromosomal changes in devil facial tumor (DFT) as it spreads through the wild Tasmanian devil population. Cancer Genet. 205:101–112. [DOI] [PubMed] [Google Scholar]
- Pemberton D 1990. Organisation and Behaviour of the Tasmanian Devil, Sarcophilus harrisii. PhD thesis, Univ. Tasmania [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu B, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Purwar R, Schlapbach C, Xiao S, Kang HS, Elyaman W, Jiang X, Jetten AM, Khoury SJ, Fuhlbrigge RC, Kuchroo VK, et al. 2012. Robust tumor immunity to melanoma mediated by interleukin-9-producing T cells. Nat. Med. 18:1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye RJ, Pemberton D, Tovar C, Tubio JM, Dun KA, Fox S, Darby J, Hayes D, Knowles GW, Kreiss A, et al. 2016a. A second transmissible cancer in Tasmanian devils. Proc. Natl. Acad. Sci.USA 113.2: 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye R, Hamede R, Siddle HV, Caldwell A, Knowles GW, Swift K, Kreiss A, Jones ME, Lyons AB, and Woods GM. 2016b. Demonstration of immune responses against devil facial tumour disease in wild Tasmanian devils. Biol. Lett. 12:20160553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Zhu YT, Hu L, and Zhu YJ. 2009. Identification of Fat4 as a candidate tumor suppressor gene in breast cancers. Int. J. Cancer 124:793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rellstab C, Gugerli F, Eckert AJ, Hancock AM, and Holderegger R. 2015. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 24:4348–4370. [DOI] [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, Montgomery B, Taplin M-E, Pritchard CC, Attard G, et al. 2015. Integrative clinical genomics of advanced prostate cancer. Cell 161:1215–1228. 10.1016/jxell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounsevell D, Taylor R, and Hocking G 1991. Distribution records of native terrestrial mammals in Tasmania. Wildlife Research 18:6. [Google Scholar]
- Russell T, Madsen T, Thomas F, Raven N, Hamede R, and Ujvari B. 2018. Oncogenesis as a selective force: adaptive evolution in the face of a transmissible cancer. Bioessays 40:1700146. [DOI] [PubMed] [Google Scholar]
- SansregretLaurent Nepveu Alain. 2008. The multiple roles of CUX1: In-sights from mouse models and cell-based assays. Gene 412 (1–2): 84–94. 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Schwabl P, Llewellyn MS, Landguth EL, Andersson B, Kitron U, Costales JA, Ocana S, and Grijalva MJ. 2017. Prediction and prevention of parasitic diseases using a landscape genomics framework. Trends Parasitol. 33:264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer RM, vonHoldt BM, Harrigan R, Knowles JC, Musiani M, Coltman D, Novembre J, and Wayne RK. 2016. Genetic subdivision and candidate genes under selection in North American grey wolves. Mol. Ecol. 25:380–402. [DOI] [PubMed] [Google Scholar]
- Shinagawa N, Yamazaki K, Tamura Y, Imai A, Kikuchi E, Yokouchi H, Hommura F, Oizumi S, and Nishimura M. 2008. Immunotherapy with dendritic cells pulsed with tumor-derived gp96 against murine lung cancer is effective through immune response of CD8+ cytotoxic T lymphocytes and natural killer cells. Cancer Immunol Immunother 57:165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV, Kreiss A, Tovar C, Yuen CK, Cheng Y, Belov K, Swift K, Pearse AM, Hamede R, Jones ME, et al. 2013. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc. Natl. Acad. Sci. U. S. A. 110:5103–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddle HV,Marzec J, Cheng Y, Jones M, and Belov K.2010. MHC gene copy number variation in Tasmanian devils: implications for the spread of a contagious cancer. Proc Biol Sci. 277:2001–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C, Glaab E, and Linster CL. 2017. Molecular identification of dribulokinase in budding yeast and mammals. J. Biol. Chem. 292:1005–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KF, Sax DF, and Lafferty KD. 2006. Evidence for the role of infectious disease in species extinction and endangerment. Conserv. Biol. 20:1349–1357. [DOI] [PubMed] [Google Scholar]
- Stammnitz MR, Coorens THH, Gori KC, Hayes D, Fu B, Wang J, Martin-Herranz DE, Alexandrov LB, Baez-Ortega A, Barthorpe S et al. 2018. The origins and vulnerabilities of two transmissible cancers in tasmanian devils. Cancer Cell 33:607–619 e615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steane DA, Potts BM, McLean E, Prober SM, Stock WD, Vaillancourt RE, Byrne M. 2014. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Molecular Ecology 23:2500–2513. 10.1111/mec.12751. [DOI] [PubMed] [Google Scholar]
- Storfer A, Epstein B, Jones M, Micheletti S, Spear SF, Lachish S, Fox S. 2017. Landscape genetics of the Tasmanian devil: implications for spread of an infectious cancer. Conservation Genetics 18:1287–1297. 10.1007/s10592-017-0980-4. [DOI] [Google Scholar]
- Storfer A, Patton A, and Fraik AK. 2018. Navigating the interface between landscape genetics and landscape genomics. Front Genet. 9:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storfer A, Hohenlohe PA, Margres MJ, Patton A, Fraik AK, Lawrance M, Ricci LE, Stahlke AR, McCallum HI, and Jones ME. 2018. The devil is in the details: Genomics of transmissible cancers in Tasmanian devils. PLoS Pathog. 14:e1007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Eriksson S, and Wang L. 2014. Down-regulation of mitochondrial thymidine kinase 2 and deoxyguanosine kinase by didanosine: implication for mitochondrial toxicities of anti-HIV nucleoside analogs. Biochem. Biophys Res. Commun. 450:1021–1026. [DOI] [PubMed] [Google Scholar]
- Sun ZX, Zhai YF, Zhang JQ, Kang K, Cai JH, Fu Y, Qiu JQ, Shen JW, and Zhang WQ. 2015. The genetic basis of population fecundity prediction across multiple field populations of Nilaparvata lugens. Mol. Ecol. 24:771–784. [DOI] [PubMed] [Google Scholar]
- Szkiba D, Kapun M, von Haeseler A, and Gallach M. 2014. SNP2GO: functional analysis of genome-wide association studies. Genetics 197:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar C, Patchett AL, Kim V, Wilson R, Darby J, Lyons AB, and Woods GM. 2018. Heat shock proteins expressed in the marsupial Tasmanian devil are potential antigenic candidates in a vaccine against devil facial tumour disease. PLoS One 13:e0196469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CT, Yang P,M, Chern TR, Chuang SH, Lin JH, Klemm L, Muschen M, and Chen CC. 2014. AID downregulation is a novel function of the DNMT inhibitor 5-aza-deoxycytidine. Oncotarget 5:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsapogas P, Breslin T, Bilke S, Lagergren A, Mansson R, Liberg D, Peterson C, and Sigvardsson M. 2003. RNA analysis of B cell lines arrested at defined stages of differentiation allows for an approximation of gene expression patterns during B cell development. J. Leukoc Biol. 74:102–110. [DOI] [PubMed] [Google Scholar]
- Ullrich V, and Brugger R. 1994. Prostacyclin and thromboxane synthase: new aspects of hemethiolate catalysis. Angewandte Chemie International Edition in English 33:1911–1919. [Google Scholar]
- Verity R, Collins C, Card DC, Schaal SM, Wang L, and Lotterhos KE. 2017. minotaur: A platform for the analysis and visualization of multivariate results from genome scans with R Shiny. Mol. Ecol. Resour. 17:33–43. [DOI] [PubMed] [Google Scholar]
- Waddington CH 1974. A catastrophe theory of evolution. Ann. N Y Acad. Sci. 231:32–42. [DOI] [PubMed] [Google Scholar]
- Weir BS, and Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution. 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Wenzel MA, Douglas A, James MC, Redpath SM, and Piertney SB. 2016. The role of parasite-driven selection in shaping landscape genomic structure in red grouse (Lagopus lagopus scotica). Mol. Ecol. 25:324–341. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S et al. 2007. Database resources of the National Center for Biotechnology Information. Nucleic. Acids. Res. 35:D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, and Lotterhos KE. 2015. Reliable detection of loci responsible for local adaptation: inference of a null model through trimming the distribution of F(ST). Am. Nat. 186 Suppl 1:S24–36. [DOI] [PubMed] [Google Scholar]
- Wright B, Willet CE, Hamede R, Jones M, Belov K, and Wade CM. 2017. Variants in the host genome may inhibit tumour growth in devil facial tumours: evidence from genome-wide association. Sci. Rep. 7:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Wang YC, Liu R, Brito LF, Kang L, Yu Y, Wang DS, Wu HJ, and Liu A. 2017. Differential gene expression in the peripheral blood of Chinese Sanhe cattle exposed to severe cold stress. Genet Mol. Res. 16. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang H, Storey KB, and Chen M. 2014. RNA-seq dependent transcriptional analysis unveils gene expression profile in the intestine of sea cucumber Apostichopus japonicus during aestivation. Comp Biochem Physiol Part D Genomics Proteomics 10:30–43 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. The eighteen abiotic environmental variables used in the landscape genomic analyses as well as the easting and northing for each sampling location.
Supplementary Table 2. Mean and standard error estimates for five of the abiotic environmental variables used in the landscape genomic analyses.
Supplementary Table 3. Mean estimated heterozygosity, Tajima’s D, FIS and Ne values for the five populations in which samples were collected both prior to and post-disease arrival.
Supplementary Table 4. Summary of the PCA loading scores of the first six principal components for vegetative and geographic features for the seven sampling localities which cumulatively explain >99% of the observed variance.
Supplementary Table 5. The 71 candidate genes detected pre-disease using landscape genomics analyses.
Supplementary Table 6. The 81 candidate genes uniquely detected post-disease using landscape genomics analyses.
Supplementary Table 7. The 47 candidate genes detected uniquely pre-disease and the abiotic environmental variable(s) which the genetic-environmental association analyses detected statistically significant associations.
Supplementary Table 8. The 81 candidate genes uniquely detected post-disease and the abiotic and biotic environmental variable(s) which the genetic-environmental association analyses detected statistically significant associations.
Supplementary Table 9. The 24 candidate genes that were detected with significant genetic-environmental associations both pre- and post-DFTD and the abiotic/biotic environmental variable(s) which had statistically significant associations.
Supplementary Figure 1. Population assignments computed by fastSTRUCTURE for samples collected prior to (a,c) and post (b,d) DFTD arrival.
Supplementary Figure 2. Marginal likelihood output produced for the population assignments computed by fastSTRUCTURE pre-DFTD (a) and post- DFTD (b).
Supplementary Figure 3. Venn diagram showing the overlap of MINOTUAR candidate genes for the eight abiotic environmental variables in the pre-disease (left) and post-disease (right) candidate gene sets.
Supplementary Figure 4. (a-g) Mahalanobis distances from MINOTAUR for each SNP pre-DFTD arrival (left plot) and post-DFTD arrival (right plot) for seven of the abiotic environmental variables.