Abstract
Eukaryotic gene expression requires the cumulative activity of multiple molecular machines to synthesize and process newly transcribed pre-messenger RNA. Introns, the noncoding regions in pre-mRNA, must be removed by the spliceosome, which assembles on the pre-mRNA as it is transcribed by RNA polymerase II (Pol II). The assembly and activity of the spliceosome can be modulated by features including the speed of transcription elongation, chromatin, post-translational modifications of Pol II and histone tails, and other RNA processing events like 5′-end capping. Here, we review recent work that has revealed cooperation and coordination among co-transcriptional processing events and speculate on new avenues of research. We anticipate new mechanistic insights capable of unraveling the relative contribution of coupled processing to gene expression.
Eukaryotic pre-messenger RNA (pre-mRNA) is processed in the nuclear milieu by multiple molecular machines working in concert to affect gene expression. Pre-mRNA processing starts concurrently with transcription elongation (i.e., co-transcriptionally) and likely proceeds until the mRNA is packaged for export to the cytoplasm (Fig. 1; Beyer and Osheim 1988; Baurén and Wieslander 1994). A major component of pre-mRNA processing is RNA splicing, which excises noncoding, intervening regions (introns) from a transcript to generate mRNA. Introns can be plentiful in eukaryotic genes, and the selective removal of introns can significantly impact gene expression by altering transcript stability, coding potential, or localization. Some of the first evidence for co-transcriptional processing was the transcription-dependent recruitment of splicing factors to chromatin (Sass and Pederson 1984). In the past 10 years, global analyses have revealed that co-transcriptional removal of introns is conserved from yeast to humans (Carrillo Oesterreich et al. 2010; Ameur et al. 2011; Khodor et al. 2011, 2012; Schmidt et al. 2011; Girard et al. 2012; Tilgner et al. 2012; Windhager et al. 2012; Nojima et al. 2015; Pai et al. 2017).
Figure 1.
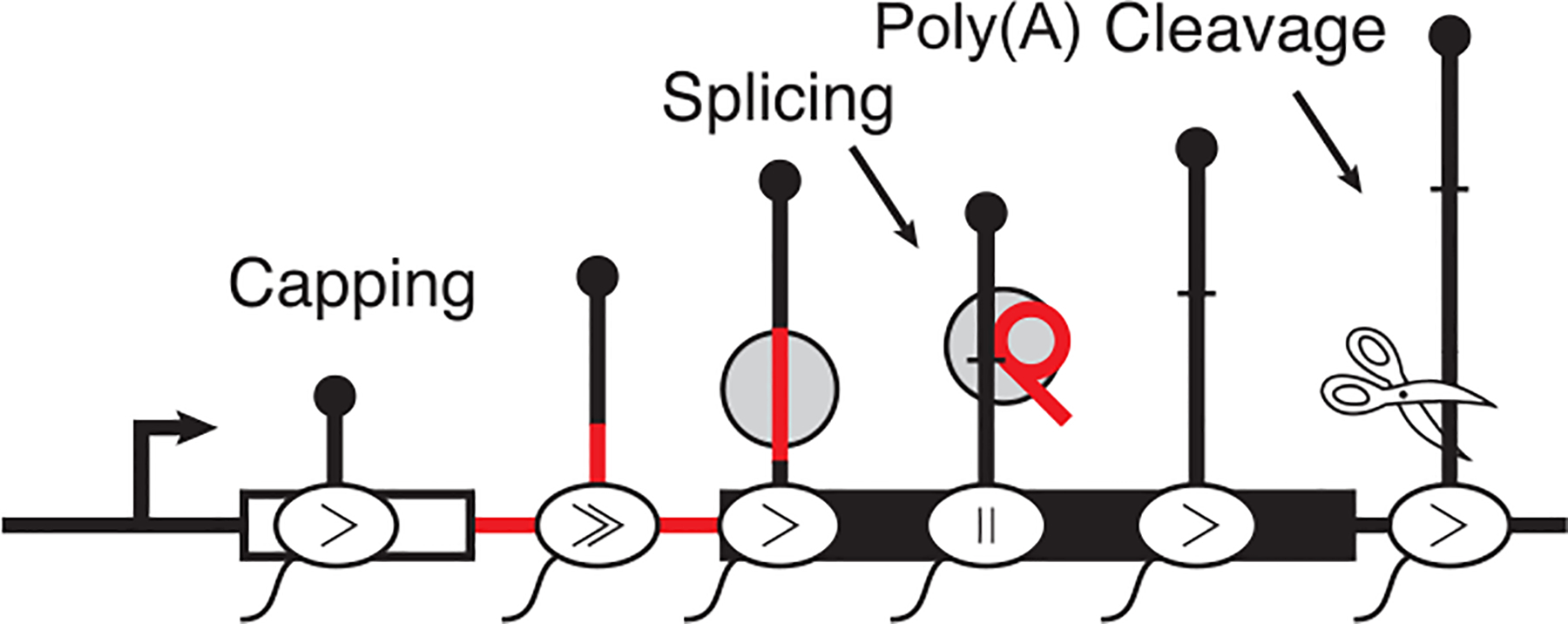
Eukaryotic mRNA is processed concurrently with transcription to modify the transcript output. Processing can include 5′-end capping with 7-methylguanosine, splicing, and polyadenylation cleavage. The rate of elongation is also non-uniform along the gene body.
An increasingly prominent theme in molecular biology is that the complexes responsible for synthesizing and modifying mRNA cross-regulate to fine-tune gene outputs. For instance, the presence of an intron in transgenes is positively correlated with transcriptional activity (Brinster et al. 1988), intron–exon boundaries are associated with active chromatin marks (Bieberstein et al. 2012), and splicing factors stimulate in vitro transcription reactions (Fong and Zhou 2001). This evidence suggests coordination between transcription and splicing. It is also well known that RNA polymerase II (Pol II) speed influences alternative splicing and intron retention in all examined species (Schor et al. 2009, 2013; Aslanzadeh et al. 2018; Saldi et al. 2018; Godoy Herz et al. 2019). Moreover, transcripts with multiple introns tend to have either all the introns removed or all the introns retained in Schizosaccharomyces pombe, suggesting coordination or cross-regulation between individual splicing events (Herzel et al. 2018). This may also be true in insect and mammalian cells, because coordination among adjacent introns has been well documented (Tilgner et al. 2015, 2018; Kim et al. 2017; Pai et al. 2017; Drexler et al. 2020). Furthermore, pre-mRNAs that are spliced are also more efficiently cleaved for polyadenylation, suggesting a relationship between the two machineries (Davidson and West 2013; Herzel et al. 2018). At the other end of the gene, we know that capping of the pre-mRNA 5′ end occurs in a rapid and coordinated fashion with transcription initiation (Hagler and Shuman 1992; Rasmussen and Lis 1993) and that promoter identity plays a role in alternative splicing and even RNA half-life (Cramer et al. 1997; Trcek et al. 2011; Fiszbein et al. 2019).
Over the years, major advances in understanding coupled processing events have emerged from new technologies. Here, we review recent progress on understanding coupled and coordinated pre-mRNA splicing focusing on the yeast Saccharomyces cerevisiae. Budding yeast is a powerful system for uncovering the mechanisms of splicing and transcription coordination, because although only 300 genes have introns, these encoded intron-containing mRNAs account for 25%–30% of the transcripts in yeast cells (Ares et al. 1999; Lopez and Seraphin 1999). Indeed, the fitness of yeast depends on having introns as the physical presence of introns in genes can promote survival in starvation conditions (Parenteau et al. 2019). The yeast spliceosome has also served as a useful model for understanding the effect of splicing factor mutations identified in human diseases including various hematopoietic malignancies and retinitis pigmentosa (Tang et al. 2016; Carrocci et al. 2017; Ruzickova and Stanek 2017).
THE MECHANISM OF PRE-mRNA SPLICING
Pre-mRNA splicing is catalyzed by the spliceosome, a single-turnover enzyme composed of the U1, U2, U4, U5, and U6 small nuclear ribonucleoproteins (snRNP) and a number of protein-only factors (Lerner et al. 1980). Early work identified several splicing factors as temperature-sensitive mutants in yeast (Hartwell 1967; Hartwell et al. 1970). The spliceosome defines the intron through recognition of conserved sequences located in the intron: the 5′ splice site (5′ss), the branchpoint sequence (bps), and the 3′ splice site (3′ss). Splice site recognition is aided by the activity of non-snRNP factors, such as Mud2 and branchpoint binding protein (BBP), which promote pairing between the 5′ss and 3′ss (Abovich and Rosbash 1997; Lacadie et al. 2006). The intronic sequences are obligate components of the splicing reaction and form part of the spliceosome active site during catalysis. Splicing occurs in two sequential trans-esterification reactions (Fig. 2; Moore and Sharp 1993). The catalytic steps of splicing are ATP-independent, but ATP is required for the activity of a number of ATPases that structurally and compositionally remodel the spliceosome during assembly and catalysis (Liu and Cheng 2015). In the first reaction, the branchpoint adenosine acts as the nucleophile to cleave the phosphodiester backbone at the 5 exon–intron boundary and generate a free 5′ exon and lariat intron–3′ exon intermediate. During the second step, the 3′OH of the 5′ exon acts as the nucleophile to excise the lariat intron and ligate the exons together.
Figure 2.
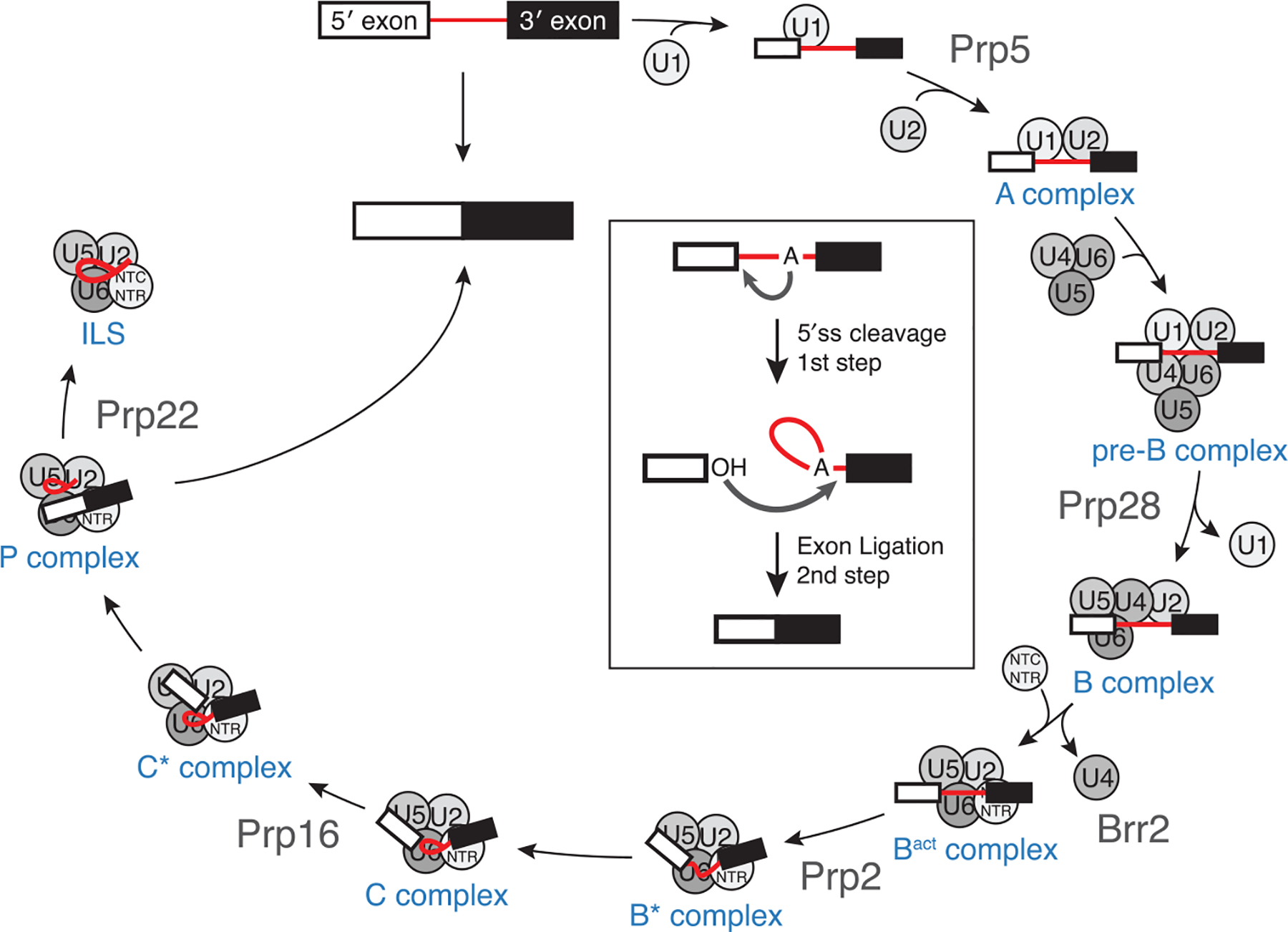
Splicing occurs in two sequential chemical steps (inset) to excise the intron (red) and ligate the coding exons. The spliceosome assembles in an ordered manner from preassembled components responsible for identifying the splice sites, forming the active site, and catalysis. The spliceosome is then released and the components recycled for the next round of splicing. Splicing complex names are shown in blue, and helicases that help mediate major transitions in the splicing cycle are shown in gray.
The spliceosome can use a variety of splice sites to varying degrees of efficiency and the conservation of splice sites differs greatly among organisms (Will and Luhrmann 2011; Qin et al. 2016). Splice sites in the yeast S. cerevisiae are very well conserved and rarely diverge from the consensus sequence, whereas metazoan splice sites are more degenerate. This is in part due to the presence of splicing regulatory proteins in metazoans that recognize splicing regulatory elements and influence splice site usage (Zhong et al. 2009). In yeast, splicing regulation has been attributed to Npl3 and Nam8. Npl3 is an SR-like protein, which is required for proper spliceosome assembly in the co-transcriptional context, and NPL3 mutations lead to intron retention (Kress et al. 2008). Interestingly, splice site usage can be further modified by the surrounding sequence context. In the context of a minigene reporter system, Wong et al. (2018) showed that the spliceosome can accommodate different 5′ss sequences depending on the origin of the intron. In yeast, Nam8 is a poly(U) binding protein that binds near the 5′ss and enhances 5′ss recognition by the U1 snRNP in a manner analogous to TIA-1 in humans (Puig et al. 1999; Förch et al. 2000; Spingola and Ares 2000; Qiu et al. 2011).
The steps of spliceosome assembly have been extensively characterized using in vitro biochemistry and genetics (Fig. 2) (Will and Luhrmann 2011). More recently, the molecular architecture of many intermediate complexes along the splicing reaction have been revealed using cryo-EM (reviewed extensively in Fica and Nagai 2017; Shi 2017; Wilkinson et al. 2019). Initially, the 5′ end of the U1 snRNA base pairs with the 5′ss found at the 5′ boundary of the intron to form the spliceosome E complex. Association of the U2 snRNP with the bps/3′ss region converts the E complex to the A complex. A complex is converted to the pre-B complex upon addition of the U4/U6.U5 tri-snRNP and subsequently to the B complex upon exchange of base pairing at the 5′ss from the U1 snRNA to the U6 snRNA and release of the U1 snRNP. Unwinding of U4/U6 base pairing and release of the U4 snRNA permits the formation of the U2/U6 catalytic active site and marks formation of the Bact complex. The spliceosome is then heavily remodeled to bring together the U2/U6 duplex and the U2/bps duplex (B* complex). Splicing factors then activate the spliceosome to promote the catalytic steps in the C and C* complexes to ligate the exons and form the postcatalytic P complex. The P complex spliceosome is then released from the mRNA and disassembled. Splicing is energetically costly and the spliceosome must assemble anew from its constituent components for every intron. This means that in mammalian cells, where genes contain on average eight introns, eight spliceosomes must assemble and disassemble for every pre-mRNA synthesized. In yeast, the majority of genes lack introns (Spingola et al. 1999); yet, the fact that 35% of transcripts must be spliced generates a strong demand for spliceosomal components. Reduced demand for splicing leads to a “hungry spliceosome,” which can act on cryptic splice sites in RNA that are not normally recognized by the spliceosome (Munding et al. 2013; Talkish et al. 2019).
CO-TRANSCRIPTIONAL ASSEMBLY OF THE SPLICEOSOME
Studies from a number of laboratories and in many species have shown that pre-mRNA splicing occurs concurrently with transcription elongation by Pol II (Carrillo Oesterreich et al. 2010; Ameur et al. 2011; Khodor et al. 2011, 2012; Schmidt et al. 2011; Girard et al. 2012; Tilgner et al. 2012; Windhager et al. 2012; Nojima et al. 2015; Pai et al. 2017). If splicing occurs co-transcriptionally, then so must spliceosome assembly. This scenario raises the possibility that components of the spliceosome would associate with the nascent RNA potentially as soon as the corresponding RNA element is synthesized (e.g., the U1 snRNP would bind the RNA as soon as Pol II has transcribed the 5′ss). Additional possibilities include the potential for Pol II itself and/or components of chromatin to help recruit spliceosomal components. The work of our laboratory and others established that splicing factors associate with gene bodies in vivo in a manner that mirrors the stepwise assembly pathway defined in vitro (Kotovic et al. 2003; Gornemann et al. 2005; Tardiff et al. 2006; Hoskins et al. 2011). Because splicing occurs co-transcriptionally, splicing factors are adjacent to the DNA axis and can cross-link to the underlying chromatin (Fig. 3A). These “splicing factor ChIP” experiments showed that U1 snRNP signal peaks in the region downstream from the 5′ exon–intron boundary and decays downstream from the intron–3′exon boundary (Fig. 3B) (Kotovic et al. 2003; Tardiff et al. 2006). Furthermore, U2 and U5 snRNPs peaked in the downstream exon, consistent with the addition of U2 and U5 subsequent to both U1 binding and synthesis of the 3′ss. ChIP data were later extended by high resolution cross-linking and immunoprecipitation (CLIP) experiments that identified the binding sites of many splicing factors on the mRNA substrate (Fig. 3C). For example, it was unclear how Mud2—similar to metazoan U2AF that binds the polypyrimidine tract at the 3′ss —might participate in gene expression in yeast, which generally lack a polypyrimidine tract. Mud2 CLIP shows that this protein binds along the entire length of the intron, displaying a different role in mRNA processing than suspected from protein homology (Baejen et al. 2014).
Figure 3.
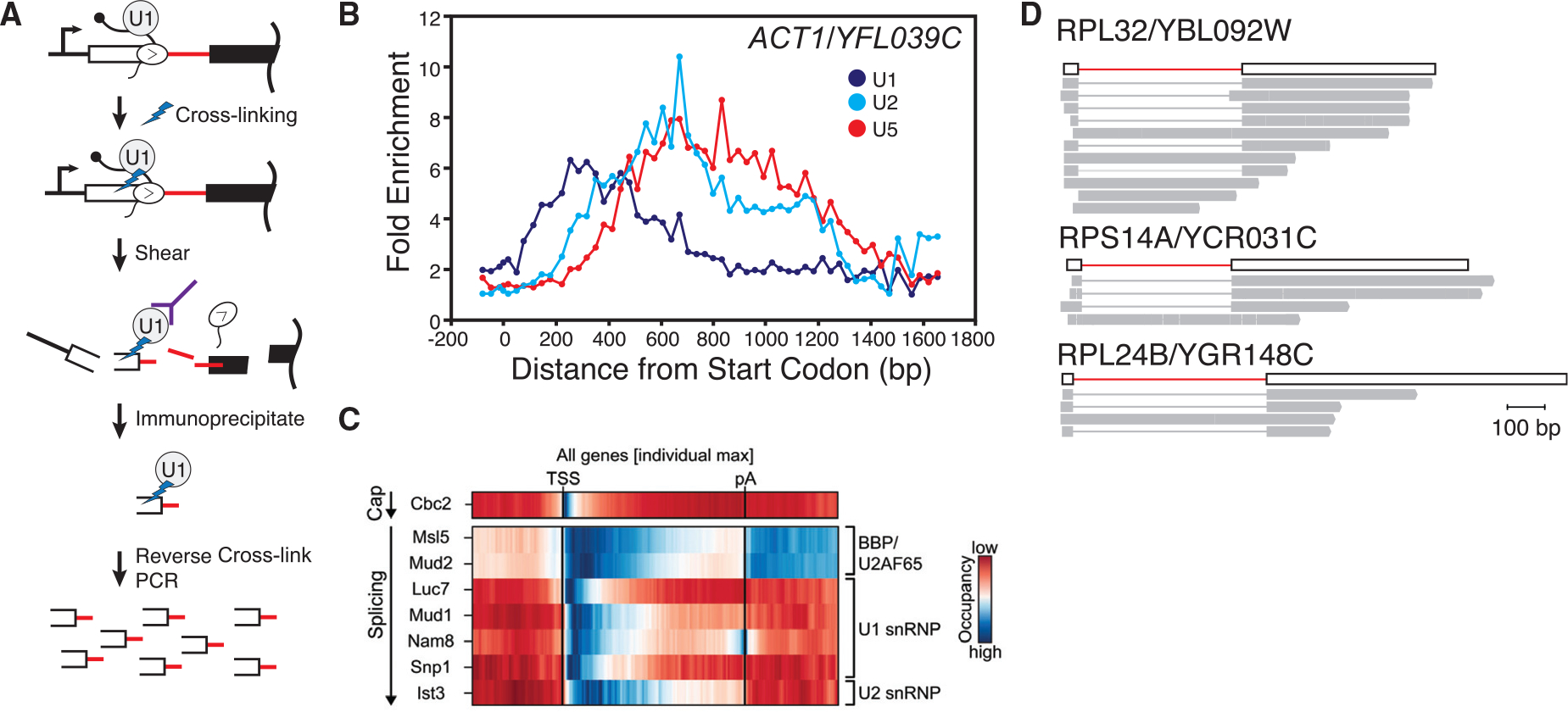
Monitoring spliceosome assembly co-transcriptionally. (A) Schematic describing a splicing factor ChIP assay. Splicing factors (i.e., U1) are formaldehyde-cross-linked to the chromatin, and chromatin is subsequently fragmented. Amplification of fragments that coimmunoprecipitated infer the association of the targeted splicing factor along the gene body. (B) Representative spliceosome assembly profile for ACT1/YFL039C. U1 associates first and departs first. U2 addition is followed by U5, consistent with the ordered assembly model. (C) Splicing factor cross-linking immunoprecipitation identifying splicing factor association with the nascent RNA. (D) Representative long reads from yeast nascent RNA sequenced on the Pacific Biosciences RSII platform showing rapid splicing of the intron while Pol II is within ~200 nt of the 3′ss. Data reproduced from Oesterreich et al. (2016). (B, Reproduced, with permission from Tardiff et al. 2006; C, reproduced, with permission, from Baejen et al. 2014.)
Broadly speaking, the ChIP and CLIP profiles in budding yeast were consistent with spliceosome assembly during transcription, but the degree to which splicing completed co-transcriptionally was contentious, with some studies concluding that exon ligation could not complete before cleavage at the poly(A) site (Tardiff et al. 2006). However, our recent work has suggested that splicing can occur rapidly relative to transcription of the 3′ss and is often upstream of previously identified ChIP peaks (Tardiff et al. 2006; Oesterreich et al. 2016; Wallace and Beggs 2017). This discrepancy is likely in part due to the low resolution of ChIP data compared to more recent custom sequencing strategies like single-molecule intron tracking and long-read sequencing, which identify the position of Pol II with single-nucleotide resolution while simultaneously determining the transcript splicing status. Spliced products can be identified while Pol II is still in close proximity to the 3′ss (Fig. 3D), suggesting Pol II and the spliceosome are physically close and corroborating earlier measurements of efficient co-transcriptional splicing with high-density tiling arrays (Carrillo Oesterreich et al. 2010). Taken together, these findings suggest that although the chemistry of RNA splicing may occur quickly, spliceosomal components may be retained in downstream gene regions because of the slow release of the post-catalytic spliceosome. This is an interesting observation because both chemical steps of splicing have been shown to be reversible in vitro (Tseng and Cheng 2008) and might suggest that an analogous complex could exist in vivo.
Simultaneous transcription and splicing afford the opportunity for cross-regulation and demand coupling for accurate gene expression. Splice site sequences are short and consensus sequences alone are insufficient to identify introns because of the presence of near-cognates in the genome. How can co-transcriptional processing aid the spliceosome in selecting the correct sites? Co-transcriptional splicing reduces the number of potential splice sites to only the subset of those in the transcript that have been transcribed at the time of spliceosome assembly. This idea led to the “first come, first served” model of splice usage which states that the splice sites that are transcribed first are used by the spliceosome (Kuhne et al. 1983). The Kornblihtt laboratory subsequently showed that “first come, first served” could refer to splicing commitment and not necessarily the completion of intron removal in the order of synthesis (de la Mata et al. 2010). Concordantly, changes in the rate of Pol II elongation can lead to changes in the outcomes of splicing even in budding yeast (Howe et al. 2003; Braberg et al. 2013; Oesterreich et al. 2016; Aslanzadeh et al. 2018). Interestingly, both faster and slower elongation leads to changes, suggesting that the transcription and splicing machinery are finely tuned to work concurrently.
Pol II elongation along gene bodies in human cells is nonuniform, with elongation rates slower over exons than introns (Veloso et al. 2014). In budding and fission yeasts, high-resolution elongation rate measurements are difficult to make and have not been accomplished; instead, a single study established an average elongation rate in budding yeast of 1.5 kb/min (Mason and Struhl 2005). Reduction in Pol II elongation (i.e., transcriptional pausing) has been proposed as an important determinant of splicing, particularly when excising introns with nonconsensus splice sites (Aslanzadeh et al. 2018). Changes in the overall rate of transcription has broad effects on splicing efficiency, indicating tuning between the rates of two processes (Oesterreich et al. 2016). Indeed, reduction in elongation rate either through mutation of Pol II or through drug treatment can rescue middle exon inclusion in multi-intron yeast genes (Howe et al. 2003). Terminal exon pausing has been identified in short endogenous genes that are spliced efficiently co-transcriptionally but the molecular features that contribute to pausing remain unclear (Carrillo Oesterreich et al. 2010). Understanding the functional consequences of pausing in terminal exons is difficult, because we currently lack the tools to manipulate pausing per se; changing the overall rate of transcription is not the same as modulating elongation at particular gene locations. Theoretically, pausing should play a role in the extent of transcripts that are spliced co-transcriptionally, but the rapid appearance of spliced products relative to transcription progress suggests that pausing may not be absolutely required for splicing. Dissecting the contribution of transcriptional pausing to efficient co-transcriptional processing will require the development of new tools to manipulate pausing on genes in their native context.
Perturbation of pre-mRNA splicing also feeds back to modify transcription. Splicing has been proposed to be a checkpoint for Pol II pausing because splicing inhibition can lead to an increase in Pol II ChIP signal over introns, further supporting communication between the two machines (Alexander et al. 2010; Chathoth et al. 2014). More recently, splicing factors, such as Mud2 and Npl3, have also been suggested to act in transcription elongation (Dermody et al. 2008; Minocha et al. 2018). Nevertheless, the molecular mechanism by which Mud2 or Npl3 contributes to efficient elongation remains unknown.
POL II MODIFICATIONS AND THE SPLICEOSOME
The major proposed mechanism of communication between elongating Pol II and complexes that modify the nascent RNA is the Pol II carboxy-terminal domain (CTD) (Custodio and Carmo-Fonseca 2016; Harlen and Churchman 2017; Wallace and Beggs 2017). The CTD is highly conserved among eukaryotes and contains a species-specific number of heptad repeats of the sequence Tyr1–Ser2–Pro3–Thr4–Ser5–Pro6–Ser7 (26 repeats in S. cerevisiae and 52 in humans). Although these repeats may act as a platform to assist in the recruitment of splicing factors to the nascent RNA, the CTD alone does not specify recruitment; for example, snRNPs are not recruited to intronless genes in yeast (Kotovic et al. 2003; Gornemann et al. 2005). Further, a CTD domain is not strictly required for splicing. Splicing occurs in the absence of transcription in vitro, and even in the context of an elongating polymerase there is no significant change in the efficiency of splicing between complexes that do and do not have a CTD (Lin et al. 1985; Natalizio et al. 2009). Eukaryotic genes including the S. pombe U6 gene that are transcribed by RNA polymerase III, which lacks a CTD, can be efficiently spliced by the spliceosome (Tani and Ohshima 1989). Nevertheless, phosphorylation of Pol II stimulates pre-mRNA splicing (Hirose et al. 1999), suggesting that splicing factor recruitment is modulated by kinases and phosphatases that dynamically post-translationally modify the CTD as Pol II transcribes along the body of the gene. Importantly, mNET-seq data from human cells has shown that Ser5P antibodies precipitate intermediates of the splicing reaction including free 5′ exons and lariat intron–3′ exon intermediates (Mayer et al. 2015; Nojima et al. 2015, 2018). This indicates that nascent RNAs that are undergoing splicing are attached to Pol II molecules with this modification. Indeed, antibodies targeting the Ser5P phospho-epitope immunoprecipitate splicing factors alongside the polymerase (Harlen et al. 2016). Additionally, pre-mRNAs transcribed by Pol II are more efficiently processed in vitro than pre-mRNAs transcribed by another polymerase lacking a CTD (Das et al. 2006). Together, these data suggest that the modified CTD may interact with the spliceosome but that other features may also significantly contribute to splicing outcomes.
CONNECTIONS TO THE mRNA CAP
The 5′ end of mRNA is capped with an inverted, methylated guanosine nucleotide that plays a major role in the life of the transcript, including mRNA splicing, 3′-end formation, export, and overall stability (Ramanathan et al. 2016). The cap is one of the first modifications installed on eukaryotic mRNA and the effects of the cap are mediated through interaction with the cap-binding complex (CBC). The CBC binds to the nascent RNA early during transcription and may help promote escape from the promoter (Lidschreiber et al. 2013). The CBC has been shown to be important, but not essential, for efficient pre-mRNA splicing in both mammalian and yeast systems (Fresco and Buratowski 1996; Schwer and Shuman 1996). Defects in mRNA capping lead to the accumulation of unspliced precursors. The CBC helps to promote E complex formation by interacting directly with U1 to recruit the snRNP and stabilize it at the 5′ss to promote spliceosome assembly (Fig. 4A,B; Lewis et al. 1996; Gornemann et al. 2005; Larson and Hoskins 2017; Li et al. 2019). Furthermore, the CBC has been proposed to play a role in the stable recruitment of the tri-snRNP (Pabis et al. 2013). Therefore, interactions with the 5′ cap of the mRNA have been proposed to be a major determinant of how the transcript will be processed. Interestingly, Ser5 phosphorylation of the Pol II CTD has also been shown to be important for RNA capping (Fabrega et al. 2003). The lethality associated with Ser5A Pol II CTD mutant in S. pombe can be bypassed by fusion of the mammalian capping enzyme to Pol II (Schwer et al. 2012). Therefore, whether the Ser5 phosphomark is directly required for splicing or whether it contributes indirectly by promoting efficient capping remains unclear.
Figure 4.
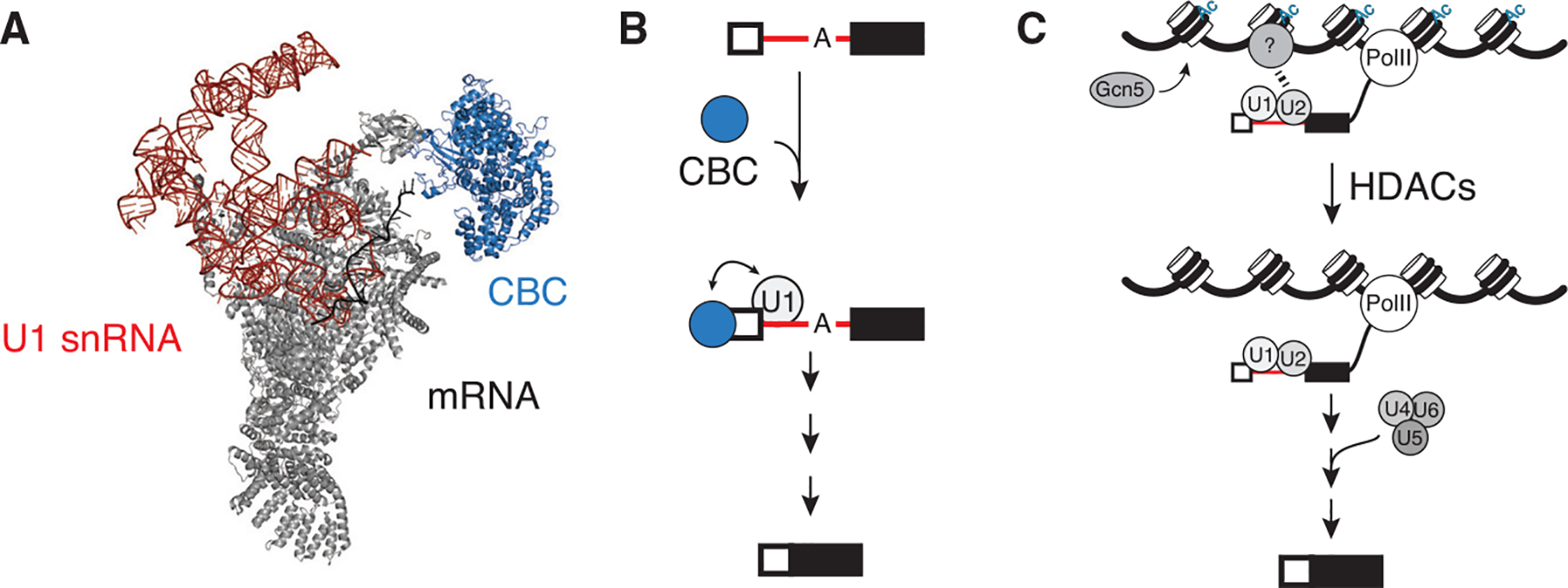
Cap-binding complex aids in U1 recruitment. (A) The structure of the spliceosome E complex showed physical interactions with CBC and the U1 snRNP component Snp1 (U1–70K in humans; pdb 6N7P) (Li et al. 2019). (B) Capping of the nascent RNA recruits CBC, which in turn can nucleate spliceosome assembly and splicing. (C) Gcn5-dependent histone acetylation recruits an unknown factor to chromatin to modulate U2 recruitment. Deacetylation facilitates subsequent steps in splicing (Gunderson et al. 2011).
CHROMATIN AND SPLICING
The co-transcriptional nature of splicing places the spliceosome in proximity to chromatin—the array of nucleosomes made up of DNA wrapped around eight histones. Some early indication of the impact of chromatin on the regulation of splicing came from work in which the integration of the adenovirus genome at different genomic locations resulted in altered splicing outcomes (Adami and Babiss 1991). In metazoans, nucleosomes are found more frequently at exons than introns, and it is proposed that they act as landmarks of exons (Kogan and Trifonov 2005; Spies et al. 2009; Tilgner et al. 2009). Nucleosome positioning can act as a transcriptional barrier that might aid in the definition of exons and subsequent assembly of the spliceosome through transient polymerase pausing (Hodges et al. 2009). Independent of nucleosome position, gene architecture—namely, the arrangement of introns and exons in any given gene—has a profound effect on the distribution of histone post-translational modifications (PTMs), which likely impacts the splicing-dependent amplification of gene output. Specifically, H3K4me3 and other active marks are best aligned to the first 5′ss in genes, even better than alignment to transcription start sites; importantly, these peaks can be moved in the gene by changing its architecture (Bieberstein et al. 2012). This suggests that components of the splicing machinery enhance gene output in part through an uncharacterized interaction with histone modifying enzymes. Consistent with this idea, perturbations of histone deacetylases (HDACs) shift alternative splicing patterns globally (Hnilicova et al. 2011). Specific and physiologically relevant examples of histone PTM alterations that determine alternative splicing patterns include the changed chromatin landscape of the NCAM gene in stimulated or differentiating neurons (Schor et al. 2009, 2013). It is very difficult to disentangle the roles of chromatin, transcription rates, and splicing, which seem to have arrows pointing in all directions. This experimental obstacle may explain why relatively few specific examples of regulation, such as the targeting of an alternative splicing regulator to chromatin sites, have emerged (Sims et al. 2007; Luco and Misteli 2011; Kfir et al. 2015). Nevertheless, these seminal studies in metazoans show the complex roles the chromatin landscape can play in splicing regulation.
Although budding and fission yeasts essentially lack alternative splicing, chromatin plays a role in splicing efficiency. The recent high-throughput method epistatic mini array profile (E-MAP) has revealed links between splicing and chromatin (Braberg et al. 2013). For example, Npl3 was found to promote H2B monoubiquitination through interactions with Bre1 in budding yeast (Moehle et al. 2012), and the chromatin remodeler SWI/SNF was linked to spliceosome activation in fission yeast (Patrick et al. 2015). Histone post-translational modifications have also been proposed to affect pre-mRNA splicing by directly modulating spliceosome assembly. The activity of the histone acetyltransferase Gcn5 promotes co-transcriptional U2 recruitment to the nascent RNA (Gunderson and Johnson 2009; Gunderson et al. 2011). More recently, genetic interactions between components of the U2 snRNP and the rare histone variant, H2A.Z, have been identified, pointing to a role in promoting splicing of introns with nonconsensus splice sites (Neves et al. 2017; Nissen et al. 2017). Furthermore, the histone methyltransferase Set2 and its corresponding H3K36 methylation mark has also been shown to play a role in the recruitment of splicing factors to the nascent transcript (Sorenson et al. 2016; Leung et al. 2019). Thus, although the molecular underpinnings of these linkages are unknown, the case for cross-regulation between chromatin and splicing is well-substantiated in the budding yeast system. Future work will be necessary to understand how this network of interactions translates into gene-specific splicing efficiencies.
OUTLOOK: PRE-mRNA SPLICING IN THE NUCLEAR LANDSCAPE
The nuclear landscape in metazoan cells is dominated by chromosomes and membraneless organelles, such as nucleoli, Cajal bodies, speckles, and others (Mao et al. 2011). Currently the field is engaged in exploring how these biomolecular condensates contribute to gene expression. It is becoming hard to imagine what nucleoplasm is, when even individual genes can be thought of as their own organelles with Pol II, RNA, and DNA- and RNA-binding proteins concentrated there (Herzel et al. 2017; Hnisz et al. 2017). Components of the transcription and splicing machinery are enriched in intrinsically disordered regions (IDRs) (Courchaine et al. 2016; Herzel et al. 2017), providing a logic for how small local condensates could form when nascent RNA is present. Indeed, SR proteins and the CTD of Pol II have some of the most extensive intrinsically disordered regions of all proteins. The concentration of SR proteins is high at transcription sites, and phosphorylation changes on the Pol II CTD during elongation could play a role in promoting splicing by recruiting snRNPs and other splicing factors, creating a high local concentration for the removal of multiple introns (Neugebauer and Roth 1997; Galganski et al. 2017; Herzel et al. 2017; Guo et al. 2019). At present, it is unclear how this understanding can be extended to budding and fission yeasts which often lack these long IDRs. Nevertheless, P-bodies were discovered in yeast (Sheth and Parker 2003), and yeast RNA has been shown to mediate condensation (Van Treeck et al. 2018). Yeast nucleoli are enormous, but speckles and Cajal bodies have not been observed. Therefore, importance of biomolecular condensation in yeast nuclei and its role in protein-coding gene expression is currently a frontier awaiting exploration.
ACKNOWLEDGMENTS
We thank Dr. Margaret Rodgers and Kirsten Reimer for their insightful comments on the manuscript. This work was supported by the National Institutes of Health (NIH) (GM134572 to T.J.C., R01 HL133406 to K.M.N., and R01 GM112766 to K.M.N.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- Abovich N, Rosbash M. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89: 403–412. doi: 10.1016/S0092-8674(00)80221-4 [DOI] [PubMed] [Google Scholar]
- Adami G, Babiss LE. 1991. DNA template effect on RNA splicing: two copies of the same gene in the same nucleus are processed differently. EMBO J 10: 3457–3465. doi: 10.1002/j.1460-2075.1991.tb04910.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander RD, Innocente SA, Barrass JD, Beggs JD. 2010. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell 40: 582–593. doi: 10.1016/j.molcel.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. 2011. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440. doi: 10.1038/nsmb.2143 [DOI] [PubMed] [Google Scholar]
- Ares M Jr, Grate L, Pauling MH. 1999. A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA 5: 1138–1139. doi: 10.1017/S1355838299991379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanzadeh V, Huang Y, Sanguinetti G, Beggs JD. 2018. Transcription rate strongly affects splicing fidelity and cotranscriptionality in budding yeast. Genome Res 28: 203–213. doi: 10.1101/gr.225615.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baejen C, Torkler P, Gressel S, Essig K, Soding J, Cramer P. 2014. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol Cell 55: 745–757. doi: 10.1016/j.molcel.2014.08.005 [DOI] [PubMed] [Google Scholar]
- Baurén G, Wieslander L. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192. doi: 10.1016/0092-8674(94)90182-1 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765. doi: 10.1101/gad.2.6.754 [DOI] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. 2012. First exon length controls active chromatin signatures and transcription. Cell Rep 2: 62–68. doi: 10.1016/j.celrep.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. 2013. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154: 775–788. doi: 10.1016/j.cell.2013.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. 1988. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci 85: 836–840. doi: 10.1073/pnas.85.3.836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo Oesterreich F, Preibisch S, Neugebauer KM. 2010. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell 40: 571–581. doi: 10.1016/j.molcel.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. 2017. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res 45: 4837–4852. doi: 10.1093/nar/gkw1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chathoth KT, Barrass JD, Webb S, Beggs JD. 2014. A splicing-dependent transcriptional checkpoint associated with prespliceosome formation. Mol Cell 53: 779–790. doi: 10.1016/j.molcel.2014.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchaine EM, Lu A, Neugebauer KM. 2016. Droplet organ-elles? EMBO J 35: 1603–1612. doi: 10.15252/embj.201593517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. 1997. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci 94: 11456–11460. doi: 10.1073/pnas.94.21.11456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M. 2016. Co-transcriptional splicing and the CTD code. Crit Rev Biochem Mol Biol 51: 395–411. doi: 10.1080/10409238.2016.1230086 [DOI] [PubMed] [Google Scholar]
- Das R, Dufu K, Romney B, Feldt M, Elenko M, Reed R. 2006. Functional coupling of RNAP II transcription to spliceosome assembly. Genes Dev 20: 1100–1109. doi: 10.1101/gad.1397406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson L, West S. 2013. Splicing-coupled 3′ end formation requires a terminal splice acceptor site, but not intron excision. Nucleic Acids Res 41: 7101–7114. doi: 10.1093/nar/gkt446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR. 2010. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912. doi: 10.1261/rna.1993510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One 3: e3273. doi: 10.1371/journal.pone.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HL, Choquet K, Churchman LS. 2020. Splicing kinetics and coordination revealed by direct nascent RNA sequencing through nanopores. Mol Cell 77: 985–998 e988. doi: 10.1016/j.molcel.2019.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrega C, Shen V, Shuman S, Lima CD. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell 11: 1549–1561. doi: 10.1016/S1097-2765(03)00187-4 [DOI] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleo-protein machine. Nat Struct Mol Biol 24: 791–799. doi: 10.1038/nsmb.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiszbein A, Krick KS, Begg BE, Burge CB. 2019. Exon-mediated activation of transcription starts. Cell 179: 1551–1565 e1517. doi: 10.1016/j.cell.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q. 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933. doi: 10.1038/414929a [DOI] [PubMed] [Google Scholar]
- Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell 6: 1089–1098. doi: 10.1016/S1097-2765(00)00107-6 [DOI] [PubMed] [Google Scholar]
- Fresco LD, Buratowski S. 1996. Conditional mutants of the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA 2: 584–596. [PMC free article] [PubMed] [Google Scholar]
- Galganski L, Urbanek MO, Krzyzosiak WJ. 2017. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res 45: 10350–10368. doi: 10.1093/nar/gkx759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Luhrmann R. 2012. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 3: 994. doi: 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- Godoy Herz MA, Kubaczka MG, Brzyzek G, Servi L, Krzyszton M, Simpson C, Brown J, Swiezewski S, Petrillo E, Kornblihtt AR. 2019. Light regulates plant alternative splicing through the control of transcriptional elongation. Mol Cell 73: 1066–1074 e1063.doi: 10.1016/j.molcel.2018.12.005 [DOI] [PubMed] [Google Scholar]
- Gornemann J, Kotovic KM, Hujer K, Neugebauer KM. 2005. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell 19: 53–63. doi: 10.1016/j.molcel.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Gunderson FQ, Johnson TL. 2009. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet 5: e1000682. doi: 10.1371/journal.pgen.1000682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson FQ, Merkhofer EC, Johnson TL. 2011. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci 108: 2004–2009. doi: 10.1073/pnas.1011982108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, et al. 2019. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572: 543–548. doi: 10.1038/s41586-019-1464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler J, Shuman S. 1992. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science 255: 983–986. doi: 10.1126/science.1546295 [DOI] [PubMed] [Google Scholar]
- Harlen KM, Churchman LS. 2017. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol 18: 263–273. doi: 10.1038/nrm.2017.10 [DOI] [PubMed] [Google Scholar]
- Harlen KM, Trotta KL, Smith EE, Mosaheb MM, Fuchs SM, Churchman LS. 2016. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep 15: 2147–2158. doi: 10.1016/j.celrep.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH. 1967. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol 93: 1662–1670. doi: 10.1128/JB.93.5.1662-1670.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, McLaughlin CS, Warner JR. 1970. Identification of ten genes that control ribosome formation in yeast. Mol Gen Genet 109: 42–56. doi: 10.1007/BF00334045 [DOI] [PubMed] [Google Scholar]
- Herzel L, Ottoz DSM, Alpert T, Neugebauer KM. 2017. Splicing and transcription touch base: co-transcriptional spliceosome assembly and function. Nat Rev Mol Cell Biol 18: 637–650. doi: 10.1038/nrm.2017.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzel L, Straube K, Neugebauer KM. 2018. Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res 28: 1008–1019. doi: 10.1101/gr.232025.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239. doi: 10.1101/gad.13.10.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnilicova J, Hozeifi S, Duskova E, Icha J, Tomankova T, Stanek D. 2011. Histone deacetylase activity modulates alternative splicing. PLoS One 6: e16727. doi: 10.1371/journal.pone.0016727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A phase separation model for transcriptional control. Cell 169: 13–23. doi: 10.1016/j.cell.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. 2009. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325: 626–628. doi: 10.1126/science.1172926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Friedman LJ, Gallagher SS, Crawford DJ, Anderson EG, Wombacher R, Ramirez N, Cornish VW, Gelles J, Moore MJ. 2011. Ordered and dynamic assembly of single spliceosomes. Science 331: 1289–1295. doi: 10.1126/science.1198830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KJ, Kane CM, Ares M Jr. 2003. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA 9: 993–1006. doi: 10.1261/rna.5390803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfir N, Lev-Maor G, Glaich O, Alajem A, Datta A, Sze SK, Meshorer E, Ast G. 2015. SF3B1 association with chromatin determines splicing outcomes. Cell Rep 11: 618–629. doi: 10.1016/j.celrep.2015.03.048 [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT II, Rosbash M. 2011. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25: 2502–2512. doi: 10.1101/gad.178962.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. 2012. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA 18: 2174–2186. doi: 10.1261/rna.034090.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Taggart AJ, Heintzelman C, Cygan KJ, Hull CG, Wang J, Shrestha B, Fairbrother WG. 2017. Widespread intra-dependencies in the removal of introns from human transcripts. Nucleic Acids Res 45: 9503–9513. doi: 10.1093/nar/gkx661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogan S, Trifonov EN. 2005. Gene splice sites correlate with nucleosome positions. Gene 352: 57–62. doi: 10.1016/j.gene.2005.03.004 [DOI] [PubMed] [Google Scholar]
- Kotovic KM, Lockshon D, Boric L, Neugebauer KM. 2003. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol 23: 5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Krogan NJ, Guthrie C. 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol Cell 32: 727–734. doi: 10.1016/j.molcel.2008.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne T, Wieringa B, Reiser J, Weissmann C. 1983. Evidence against a scanning model of RNA splicing. EMBO J 2: 727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadie SA, Tardiff DF, Kadener S, Rosbash M. 2006. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev 20: 2055–2066. doi: 10.1101/gad.1434706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JD, Hoskins AA. 2017. Dynamics and consequences of spliceosome E complex formation. Elife 6: e27592. doi: 10.7554/eLife.27592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. 1980. Are snRNPs involved in splicing? Nature 283: 220–224. doi: 10.1038/283220a0 [DOI] [PubMed] [Google Scholar]
- Leung CS, Douglass SM, Morselli M, Obusan MB, Pavlyukov MS, Pellegrini M, Johnson TL. 2019. H3K36 methylation and the chromodomain protein Eaf3 are required for proper cotranscriptional spliceosome assembly. Cell Rep 27: 3760–3769 e3764. doi: 10.1016/j.celrep.2019.05.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. 1996. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev 10: 1683–1698. doi: 10.1101/gad.10.13.1683 [DOI] [PubMed] [Google Scholar]
- Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, Shi S, Cui Y, Kappel K, Das R, et al. 2019. A unified mechanism for intron and exon definition and back-splicing. Nature 573: 375–380. doi: 10.1038/s41586-019-1523-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidschreiber M, Leike K, Cramer P. 2013. Cap completion and C-terminal repeat domain kinase recruitment underlie the initiation-elongation transition of RNA polymerase II. Mol Cell Biol 33: 3805–3816. doi: 10.1128/MCB.00361-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Newman AJ, Cheng SC, Abelson J. 1985. Yeast mRNA splicing in vitro. J Biol Chem 260: 14780–14792. [PubMed] [Google Scholar]
- Liu YC, Cheng SC. 2015. Functional roles of DExD/H-box RNA helicases in Pre-mRNA splicing. J Biomed Sci 22: 54. doi: 10.1186/s12929-015-0161-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PJ, Seraphin B. 1999. Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA 5: 1135–1137. doi: 10.1017/S135583829999091X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Misteli T. 2011. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr Opin Genet Dev 21: 366–372. doi: 10.1016/j.gde.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. 2011. Biogenesis and function of nuclear bodies. Trends Genet 27: 295–306. doi: 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PB, Struhl K. 2005. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell 17: 831–840. doi: 10.1016/j.molcel.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. doi: 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minocha R, Popova V, Kopytova D, Misiak D, Huttelmaier S, Georgieva S, Strasser K. 2018. Mud2 functions in transcription by recruiting the Prp19 and TREX complexes to transcribed genes. Nucleic Acids Res 46: 9749–9763. doi: 10.1093/nar/gky640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle EA, Ryan CJ, Krogan NJ, Kress TL, Guthrie C. 2012. The yeast SR-like protein Npl3 links chromatin modification to mRNA processing. PLoS Genet 8: e1003101. doi: 10.1371/journal.pgen.1003101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368. doi: 10.1038/365364a0 [DOI] [PubMed] [Google Scholar]
- Munding EM, Shiue L, Katzman S, Donohue JP, Ares M Jr. 2013. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell 51: 338–348. doi: 10.1016/j.molcel.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio BJ, Robson-Dixon ND, Garcia-Blanco MA. 2009. The carboxyl-terminal domain of RNA polymerase II is not sufficient to enhance the efficiency of pre-mRNA capping or splicing in the context of a different polymerase. J Biol Chem 284: 8692–8702. doi: 10.1074/jbc.M806919200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. 1997. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev 11: 1148–1159. doi: 10.1101/gad.11.9.1148 [DOI] [PubMed] [Google Scholar]
- Neves LT, Douglass S, Spreafico R, Venkataramanan S, Kress TL, Johnson TL. 2017. The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae. Genes Dev 31: 702–717. doi: 10.1101/gad.295188.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen KE, Homer CM, Ryan CJ, Shales M, Krogan NJ, Patrick KL, Guthrie C. 2017. The histone variant H2A.Z promotes splicing of weak introns. Genes Dev 31: 688–701. doi: 10.1101/gad.295287.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. 2015. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161: 526–540. doi: 10.1016/j.cell.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Rebelo K, Gomes T, Grosso AR, Proudfoot NJ, Carmo-Fonseca M. 2018. RNA polymerase II phosphorylated on CTD serine 5 interpacts with the spliceosome during cotranscriptional splicing. Mol Cell 72: 369–379 e364. doi: 10.1016/j.molcel.2018.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterreich FC, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. 2016. Splicing of nascent RNA coincides with intron exit from RNA polymerase II. Cell 165: 372–381. doi: 10.1016/j.cell.2016.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabis M, Neufeld N, Steiner MC, Bojic T, Shav-Tal Y, Neugebauer KM. 2013. The nuclear cap-binding complex interacts with the U4/U6.U5 tri-snRNP and promotes spliceosome assembly in mammalian cells. RNA 19: 1054–1063. doi: 10.1261/rna.037069.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Henriques T, McCue K, Burkholder A, Adelman K, Burge CB. 2017. The kinetics of pre-mRNA splicing in the Drosophila genome and the influence of gene architecture. Elife 6: e32537. doi: 10.7554/eLife.32537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S. 2019. Introns are mediators of cell response to starvation. Nature 565: 612–617. doi: 10.1038/s41586-018-0859-7 [DOI] [PubMed] [Google Scholar]
- Patrick KL, Ryan CJ, Xu J, Lipp JJ, Nissen KE, Roguev A, Shales M, Krogan NJ, Guthrie C. 2015. Genetic interaction mapping reveals a role for the SWI/SNF nucleosome remodeler in spliceosome activation in fission yeast. PLoS Genet 11: e1005074. doi: 10.1371/journal.pgen.1005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Seraphin B. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev 13: 569–580. doi: 10.1101/gad.13.5.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Huang L, Wlodaver A, Andrade J, Staley JP. 2016. Sequencing of lariat termini in S. cerevisiae reveals 5′ splice sites, branch points, and novel splicing events. RNA 22: 237–253. doi: 10.1261/rna.052829.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu ZR, Schwer B, Shuman S. 2011. Determinants of Nam8-dependent splicing of meiotic pre-mRNAs. Nucleic Acids Res 39: 3427–3445. doi: 10.1093/nar/gkq1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Robb GB, Chan SH. 2016. mRNA capping: biological functions and applications. Nucleic Acids Res 44: 7511–7526. doi: 10.1093/nar/gkw551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci 90: 7923–7927. doi: 10.1073/pnas.90.17.7923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzickova S, Stanek D. 2017. Mutations in spliceosomal proteins and retina degeneration. RNA Biol 14: 544–552. doi: 10.1080/15476286.2016.1191735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldi T, Fong N, Bentley DL. 2018. Transcription elongation rate affects nascent histone pre-mRNA folding and 3′ end processing. Genes Dev 32: 297–308. doi: 10.1101/gad.310896.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass H, Pederson T. 1984. Transcription-dependent localization of U1 and U2 small nuclear ribonucleoproteins at major sites of gene activity in polytene chromosomes. J Mol Biol 180: 911–926. doi: 10.1016/0022-2836(84)90263-8 [DOI] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E. 2011. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: implications for alternative splicing regulation. J Cell Biol 193: 819–829. doi: 10.1083/jcb.201009012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci 106: 4325–4330. doi: 10.1073/pnas.0810666106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR. 2013. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J 32: 2264–2274. doi: 10.1038/emboj.2013.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Shuman S. 1996. Conditional inactivation of mRNA capping enzyme affects yeast pre-mRNA splicing in vivo. RNA 2: 574–583. [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Sanchez AM, Shuman S. 2012. Punctuation and syntax of the RNA polymerase II CTD code in fission yeast. Proc Natl Acad Sci 109: 18024–18029. doi: 10.1073/pnas.1208995109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. doi: 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18: 655–670. doi: 10.1038/nrm.2017.86 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676. doi: 10.1016/j.molcel.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MR, Jha DK, Ucles SA, Flood DM, Strahl BD, Stevens SW, Kress TL. 2016. Histone H3K36 methylation regulates pre-mRNA splicing in Saccharomyces cerevisiae. RNA Biol 13: 412–426. doi: 10.1080/15476286.2016.1144009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254. doi: 10.1016/j.molcel.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spingola M, Ares M. 2000. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol Cell 6: 329–338. doi: 10.1016/S1097-2765(00)00033-2 [DOI] [PubMed] [Google Scholar]
- Spingola M, Grate L, Haussler D, Ares M. 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5: 221–234. doi: 10.1017/S1355838299981682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkish J, Igel H, Perriman RJ, Shiue L, Katzman S, Munding EM, Shelansky R, Donohue JP, Ares M Jr. 2019. Rapidly evolving protointrons in Saccharomyces genomes revealed by a hungry spliceosome. PLoS Genet 15: e1008249. doi: 10.1371/journal.pgen.1008249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, Moldon A, Xu YZ, Query CC. 2016. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev 30: 2710–2723. doi: 10.1101/gad.291872.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani T, Ohshima Y. 1989. The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature 337: 87–90. doi: 10.1038/337087a0 [DOI] [PubMed] [Google Scholar]
- Tardiff DF, Lacadie SA, Rosbash M. 2006. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell 24: 917–929. doi: 10.1016/j.molcel.2006.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001. doi: 10.1038/nsmb.1658 [DOI] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. 2012. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 22: 1616–1625. doi: 10.1101/gr.134445.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Blauwkamp T, Moshrefi A, Jaeger E, Chen F, Harel I, Bustamante CD, Rasmussen M, Snyder MP. 2015. Comprehensive transcriptome analysis using synthetic long-read sequencing reveals molecular co-association of distant splicing events. Nat Biotechnol 33: 736–742. doi: 10.1038/nbt.3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Jahanbani F, Gupta I, Collier P, Wei E, Rasmussen M, Snyder M. 2018. Microfluidic isoform sequencing shows widespread splicing coordination in the human transcriptome. Genome Res 28: 231–242. doi: 10.1101/gr.230516.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Larson DR, Moldon A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484–1497. doi: 10.1016/j.cell.2011.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC. 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784. doi: 10.1126/science.1158993 [DOI] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci 115: 2734–2739. doi: 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. 2014. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24: 896–905. doi: 10.1101/gr.171405.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace EWJ, Beggs JD. 2017. Extremely fast and incredibly close: cotranscriptional splicing in budding yeast. RNA 23: 601–610. doi: 10.1261/rna.060830.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Charenton C, Nagai K. 2019. RNA splicing by the spliceosome. Annu Rev Biochem doi: 10.1146/annurev-biochem-091719-064225 [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. doi: 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Malterer G, L’Hernault A, Schilhabel M, Schreiber S, et al. 2012. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res 22: 2031–2042. doi: 10.1101/gr.131847.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong MS, Kinney JB, Krainer AR. 2018. Quantitative activity profile and context dependence of all human 5′ splice sites. Mol Cell 71: 1012–1026 e1013. doi: 10.1016/j.molcel.2018.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10. doi: 10.1016/j.molcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]


