Abstract
Lipid asymmetry in biological membranes is essential for various cell functions, such as cell polarity, cytokinesis, and apoptosis. P4-ATPases (flippases) are involved in the generation of such asymmetry. In Saccharomyces cerevisiae, the protein kinases Fpk1p/Fpk2p activate the P4-ATPases Dnf1p/Dnf2p by phosphorylation. Previously, we have shown that a blue-light-dependent protein kinase, phototropin from Chlamydomonas reinhardtii (CrPHOT), complements defects in an fpk1Δ fpk2Δ mutant. Herein, we investigated whether CrPHOT optically regulates P4-ATPase activity. First, we demonstrated that the translocation of NBD-labelled phospholipids to the cytoplasmic leaflet via P4-ATPases was promoted by blue-light irradiation in fpk1Δ fpk2Δ cells with CrPHOT. In addition, blue light completely suppressed the defects in membrane functions (such as endocytic recycling, actin depolarization, and apical-isotropic growth switching) caused by fpk1Δ fpk2Δ mutations. All responses required the kinase activity of CrPHOT. Hence, these results indicate the utility of CrPHOT as a powerful and first tool for optogenetic manipulation of P4-ATPase activity.
Subject terms: Biological techniques, Cell biology, Molecular biology
Introduction
In eukaryotic cells, lipid bilayers exhibit asymmetric phospholipid distributions. This is particularly evident in the plasma membrane: the cytoplasmic leaflet has an abundance of phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and its derivatives, whereas the exoplasmic leaflets are rich in phosphatidylcholine (PC), sphingomyelin (SM) and glycosphingolipids1,2. These lipid asymmetries across membranes play a crucial role in many cell functions, such as vesicular transport3, cytokinesis4,5, cell signalling6, apoptosis7, cell migration8, and the immune response9. The asymmetry of phospholipids in biological membranes is established and maintained by lipid transporters10. A family of such transporters is P4-ATPase (i.e., flippase), which transports specific lipids from the exoplasmic leaflet to the cytoplasmic leaflet using the energy of ATP hydrolysis10. Most P4-ATPases interact with a noncatalytic subunit, Cdc50 family protein, for proper subcellular localization and ATPase activity11.
The P4-ATPase subfamily is widely conserved in eukaryotes12. To date, many studies in mammals, Caenorhabditis elegans, Saccharomyces cerevisiae, Arabidopsis thaliana and others have been reported11,13,14,15. As an example, mammalian studies have reported that knockdown of a P4-ATPase ATP8A1 disrupts the asymmetric distribution of PS in recycling endosomes (REs) and suppresses vesicle transport from endosomes16. The involvement of P4-ATPases in vesicular transport has been reported in various organisms and is considered to be a general function of P4-ATPases3,17. Probes that specifically bind to PS (e.g. LactC2-GFP18 and GFP-evt2-2XPH19) enable direct observation of the PS distribution in the cytosolic leaflet16. Analysis using a biotinylated PE probe (Ro09-0198 20) in yeast cells showed that PE is specifically exposed on the surface of polarized ends such as the bud site and bud neck21. Mutation of P4-ATPase genes disrupts these PE polarized distributions in yeast cells21. As described above, genetic analysis using mutants and overexpression reveals many physiological functions of P4-ATPases in various organisms. Cell biological analysis using specific lipid probes reveals the location of intracellular phospholipids. In contrast, many issues regarding the spatiotemporal regulation of P4-ATPases and translocated phospholipids remain. To elucidate them, the development of a new tool is required.
The budding yeast S. cerevisiae, has five P4-ATPases, Drs2p, Dnf1p, Dnf2p, Dnf3p, and Neo1p. Neo1p is essential, while the others are redundantly essential for viability22. Dnf1p and Dnf2p have 69% amino acid sequence identity and are localized in the plasma membrane and inner membranes (trans-Golgi network (TGN), early endosomes and transport vesicles)22,23,24,25. Dnf1p/Dnf2p transport PE, PC, their lyso-forms, and monosaccharide glycosphingolipids like glucosylceramide (GlcCer) and galactosylceramide (GalCer). GlcCer is primarily transported by Dnf2p13,23,26,27,28. Both Dnf1p and Dnf2p require interaction with Lem3p, a member of the Cdc50p family, for subcellular localization and function; hence, lem3∆ and dnf1∆ dnf2∆ are phenocopies29. In both dnf1∆ dnf2∆ and lem3∆ mutants at a late mitotic phase, translocation of PE to the inner leaflet, actin depolarization at the bud tip and switching from apical to isotropic growth are significantly delayed compared to those in wild-type cells23,30,31,32. These results indicate that Dnf1p/Dnf2p-Lem3p are involved in the maintenance of cell polarity.
Dnf1p/Dnf2p-Lem3p are also involved in endocytic recycling of the vesicle-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (v-SNARE) Snc1p. Mutation of another P4-ATPase, Drs2p, or its partner subunit Cdc50p causes an internal accumulation of Snc1p, and a combination of dnf1∆ dnf2∆ (or lem3∆) and drs2∆ (or cdc50∆) mutations results in severe vesicle-transport abnormalities22,29,32,33. Hence, Dnf1p/Dnf2p-Lem3p and Drs2p-Cdc50p are redundant in the regulation of vesicle transport in this pathway.
The only factors regulating the activity of Dnf1p/Dnf2p are the Ser/Thr protein kinase Fpk1p and its paralog, Fpk2p. The FPK1 gene was identified as a gene responsible for a synthetic lethal mutation with cdc50∆, and the fpk1Δ fpk2∆ mutant shows almost the same phenotypes as the dnf1Δ dnf2Δ or the lem3Δ mutant32. Moreover, Fpk1p phosphorylates Dnf1p/Dnf2p in vivo and in vitro32,34,35,36; thus, Fpk1p/Fpk2p are Dnf1p/Dnf2p-activating kinases. Fpk1p and Fpk2p share the highest sequence homology in yeast with the AGCVIII kinase domain of phototropins (PHOTs), plant specific blue-light (BL) photoreceptors32,37. We have previously introduced a PHOT from Chlamydomonas reinhardtii (CrPHOT) into an fpk1Δ fpk2Δ mutant with a conditional Cdc50p mutant (PGAL1-CDC50 fpk1Δ fpk2Δ. As a result, we have shown that CrPHOT complements the growth defect of the fpk1Δ fpk2Δ mutant in a BL-dependent manner37. This result indicates the possibility that CrPHOT controls P4-ATPases in a light-dependent manner. Therefore, we examined whether this system could be used as a new optogenetic technology.
In this study, we reintroduced CrPHOT into the fpk1Δ fpk2Δ mutant or the PGAL1-CDC50 fpk1Δ fpk2Δ mutant and biochemically assessed the lipid translocation activity to show that it was indeed regulated by BL irradiation. Furthermore, Dnf1p/Dnf2p-dependent cellular processes, such as vesicle transport, were regulated in these cells by BL. Our results suggest the potential of CrPHOT as an optogenetic tool to regulate membrane functions.
Results
Optical control of yeast cell growth by Chlamydomonas PHOT
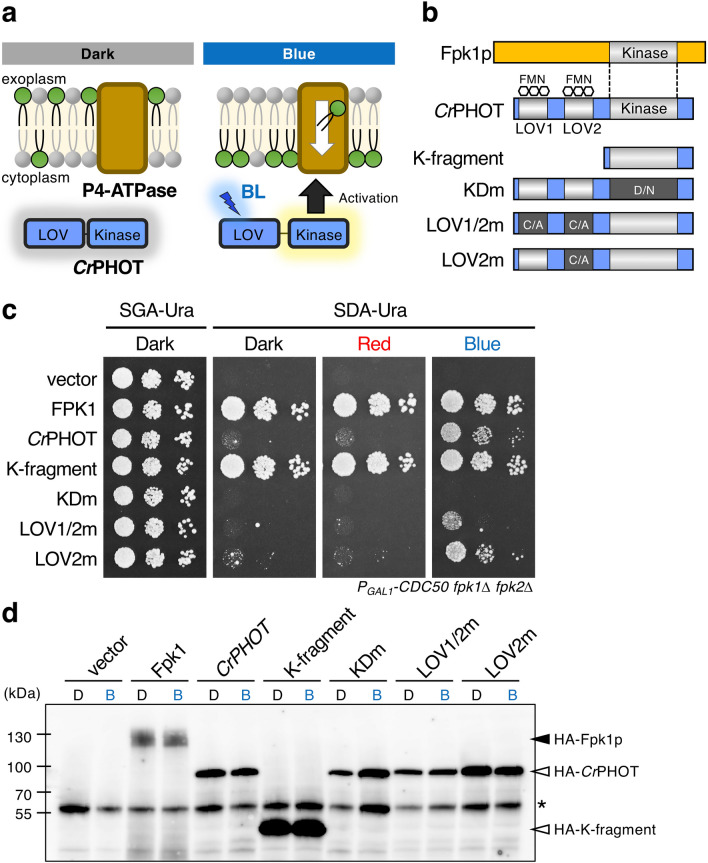
To establish a system to control P4-ATPase (flippase) activity by light (Fig. 1a), we attempted to prove that CrPHOT regulates P4-ATPases at the molecular level. On the basis of the highest sequence homology among the kinase domains of Fpk1p/Fpk2p and CrPHOT32, we have shown that CrPHOT complements the synthetic lethality of a PGAL1-CDC50 fpk1Δ fpk2Δ yeast mutant in a BL-dependent manner37. In this study, we introduced CrPHOT and its derivatives (Fig. 1b) into the mutant and examined its growth activity again. Consequently, we reconfirmed that CrPHOT suppresses the growth defect of the PGAL1-CDC50 fpk1Δ fpk2Δ yeast mutant in glucose-containing medium under BL but not under red light (RL) or in darkness (Fig. 1c, CrPHOT). Furthermore, a K-fragment with constitutive kinase activity suppressed the growth defect regardless of the light condition, and KDm with a kinase-dead mutation, failed to restore the growth even under BL (Fig. 1b, c)37. The substitution of a conserved cysteine (Cys) to an alanine (Ala) in the light-oxygen-voltage (LOV) domain is known to decrease the extent of light activation38. LOV2m, which has this mutation in the LOV2 domain, was used as a mutant to reduce light sensitivity in previous research37. LOV2m certainly reduced but did not completely abolish the ability to restore growth under BL37 (Fig. 1b, c). Therefore, we newly constructed LOV1/2 m, which has Cys to Ala mutations in two LOV domains (LOV1 and LOV2; Fig. 1b). The growth activity of LOV1/2 m under BL was considerably lower than that of LOV2m (Fig. 1c). Hence, LOV1/2 m was used as a mutant to reduce photosensitivity instead of LOV2m in this study.
Figure 1.
Light control of yeast cell growth by CrPHOT. (a) A schematic of BL-induced activation of flippases through CrPHOT in yeast. (b) Schematic illustration of Fpk1p and CrPHOT and its derivatives. K-fragment, kinase domain fragment; KDm, kinase-dead mutant with D549N; LOV1/2 m and LOV2m, mutants with a decreased extent of light activation by substitution of conserved Cys in LOV1 and LOV2 domain or LOV2 domain only, respectively, to Ala. Flavin mononucleotide (FMN) is a chromophore of phototropin. (c) Growth of the yeast conditional mutant PGAL1-CDC50 fpk1Δ fpk2Δ carrying pRS416-CrPHOT or its derivatives. Yeast cells were serially diluted and spotted onto plates containing galactose (SGA-Ura) or glucose (SDA-Ura), which were incubated in darkness (Dark) or under 10 μmol m−2 s−1 RL (Red) or BL (Blue) irradiation at 28 °C for 3 days. The experiments were performed more than three times with different biological samples. A representative image is shown in (c). (d) The protein level of CrPHOT and its derivatives is not affected by light conditions. HA-fused Fpk1p and CrPHOT and its derivatives were extracted from the yeast transformants (same strains shown in c) cultured in darkness (d) or under 10 μmol m−2 s−1 BL irradiation (b) at 28 °C for 16 h, which were then resolved by SDS-PAGE and analysed by immunoblotting with anti-HA antibodies. Asterisk indicates non-specific bands.
We then examined the expression level of CrPHOT and derived proteins in yeast with an anti-HA tag antibody (Fig. 1d). The level of CrPHOT protein was not altered with light conditions as in previous analysis37. The amount of each CrPHOT derivative was also not changed by BL irradiation, and their levels differed little. These results reconfirmed that complementation by CrPHOT depends on its photoactivation and furthermore indicate that the degree of growth activity by the derivatives depends on their biochemical properties rather than the protein amount.
CrPHOT and Dnf1p/Dnf2p P4-ATPases are localized in similar subcellular compartments
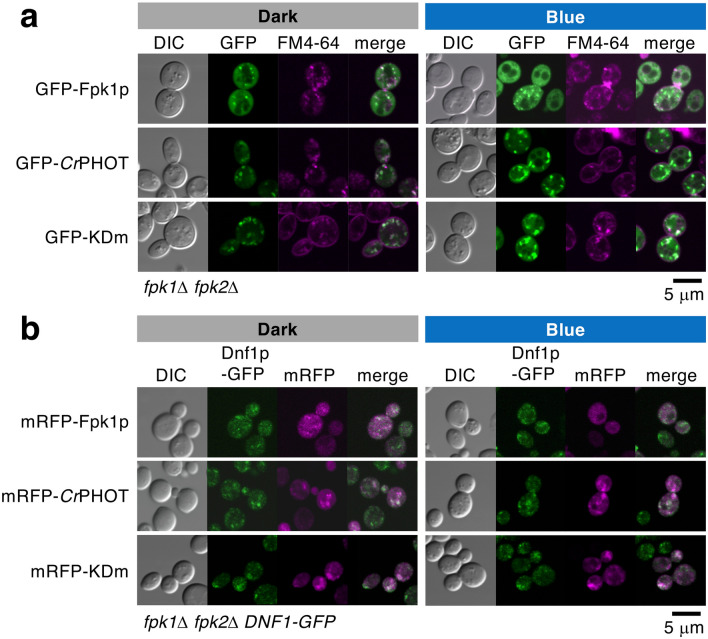
Fpk1p/Fpk2p, which regulate P4-ATPase activity, are mainly distributed throughout the cytoplasm but are partially localized in the early endosome/TGN and plasma membrane25,32,39. To clarify the control of P4-ATPases by CrPHOT, we compared the intracellular localization of fluorescent protein-tagged CrPHOT with that of Fpk1p. Before analysis, we confirmed that the tagged proteins were functional by a growth assay of the PGAL1-CDC50 fpk1Δ fpk2Δ mutant (Supplementary Fig. S1). An fpk1Δ fpk2Δ mutant expressing GFP-Fpk1p or GFP-CrPHOT cultured in darkness was briefly stained with a lipophilic dye, FM4-6432,40, which was employed as a marker for the plasma membrane and endosomal/TGN compartments (Fig. 2a). GFP-CrPHOT was occasionally observed at the plasma membrane but primarily localized to intracellular punctate structures, which mostly merged with the fluorescence of FM4-64 (64.4% of the GFP-CrPHOT speckles were merged, n = 163; Fig. 2a). The localization was similar to that of GFP-Fpk1p (66.0% of the GFP-Fpk1p speckles were merged, n = 152; 67.5% in previous analysis32; Fig. 2a). The speckles of GFP-CrPHOT, as with GFP-Fpk1p, were colocalized with another TGN marker, Sec7p-mRFP32 (Supplementary Fig. S2). These results suggest that GFP-CrPHOT is primarily localized to endosomal/TGN compartments in yeast cells.
Figure 2.
Intracellular localization of CrPHOT is similar to that of FPK1p regardless of BL irradiation. (a) Punctate structures of GFP-CrPHOT and GFP-KDm stained with FM4-64 were similar to that of GFP-FPK1p. Yeast fpk1Δ fpk2Δ cells carrying pRS416-GFP-FPK1, -GFP-CrPHOT or -GFP-KDm were incubated at 25 °C with FM4-64 under each light condition, followed by confocal microscopic observation. (b) Confocal images of KKT332 (fpk1Δ fpk2Δ DNF1-GFP) cells carrying pKT1639 (pRS416-mRFP-FPK1), pRS416-mRFP-CrPHOT or -mRFP-KDm. In (a) and (b), yeast cells were grown to logarithmic phase in darkness (Dark) or under 10 μmol m−2 s−1 BL irradiation (Blue) at 18 °C. Images were merged to compare the two signal patterns. Scale bars = 5 μm.
We then examined the effect of BL on the subcellular localization of GFP-CrPHOT. This was because BL irradiation changes phototropin intracellular localization in plants41,42. The localization analysis by FM4-64 staining in yeast cells was performed under BL irradiation. The results showed that the localization of GFP-CrPHOT under BL was the same as that in the dark and that most of its speckles merged with FM4-64 (62.5%, n = 120; Fig. 2a). To investigate the effect of kinase activity on CrPHOT localization, we observed the localization of GFP-KDm. The localization pattern of GFP-KDm was the same as that of GFP-CrPHOT regardless of the light condition (68.0% in the dark and 57.9% under BL, n = 166 and n = 164, respectively; Fig. 2a). These results suggest that the localization of CrPHOT in yeast cells is not dependent on either BL or its own kinase activity.
Under normal culture conditions, Dnf1p and Dnf2p are primarily localized to early endosomal/TGN compartments and partially to the plasma membrane of the bud and bud neck22,23,25,29,33. Fpk1p is known to colocalize with Dnf1p/Dnf2p at endosomal/TGN compartments and the plasma membrane32. We then examined the colocalization of mRFP-CrPHOT and Dnf1p-GFP in yeast cells. The results showed that Dnf1p-GFP was localized to punctate structures distributed throughout the cell, and many but not all of them colocalized with mRFP-CrPHOT (Fig. 2b). The influence of BL or the introduction of mRFP-KDm on the localization of Dnf1p-GFP was hardly observed (Fig. 2b). The same results were found for the localization of Dnf2p-GFP (Supplementary Fig. S2). These results suggest that CrPHOT, as is the case with Fpk1p, constitutively colocalizes with Dnf1p/Dnf2p in endosomal/TGN compartments.
Optical control of phospholipid translocation by CrPHOT in a kinase activity-dependent manner
At the plasma membrane, Dnf1p/Dnf2p are involved in flipping phospholipids (mainly PC and PE) and glycosphingolipids13,21,23,26,28,29,30. Fpk1p/Fpk2p regulates phospholipid uptake through Dnf1p/Dnf2p across the plasma membrane32,34,39,43. Since CrPHOT was able to complement Fpk1p/Fpk2p function in yeast cell growth37 (Fig. 1b), BL irradiation likely activates CrPHOT to control phospholipid uptake in the same way as Fpk1p/Fpk2p.
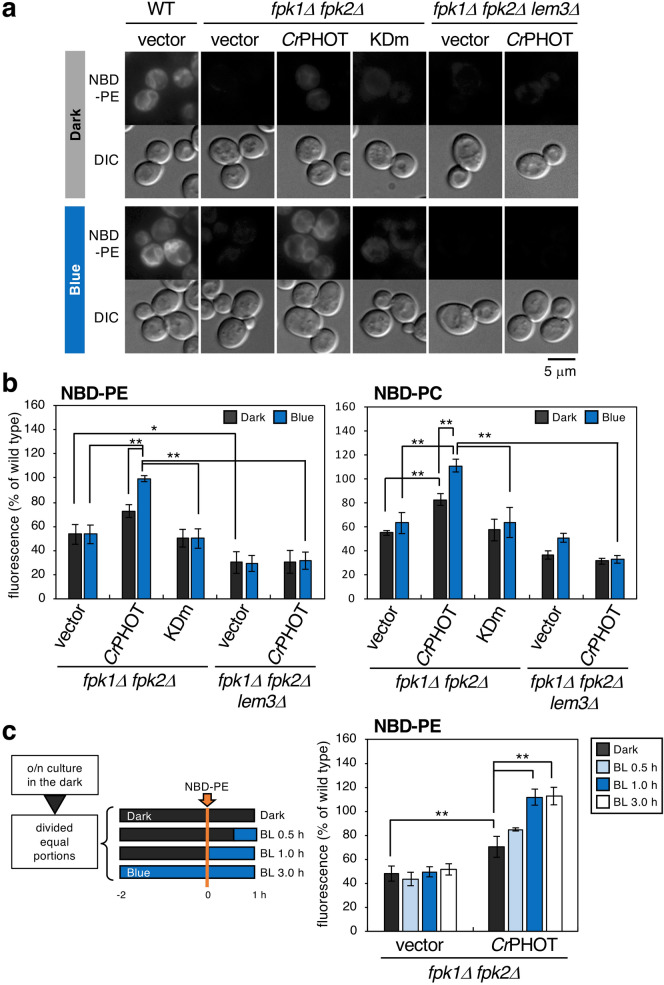
We thus examined whether CrPHOT controls the uptake of nitrobenzoxadiazole (NBD)-labelled phospholipids (NBD-PE and NBD-PC) in the fpk1Δ fpk2Δ mutant. NBD-phospholipids taken up by P4-ATPases are transported mainly to the endoplasmic reticulum (ER) in a short time, and strong fluorescence at the ER membrane is observed30,44. When NBD-PE was added, fluorescence was observed at the ER membrane in wild-type cells but not in the fpk1Δ fpk2Δ mutant (Fig. 3a, WT + vector and fpk1Δ fpk2Δ + vector). The internalized NBD-PE was quantified by flow cytometry of the cells (Fig. 3b). When the fpk1Δ fpk2Δ mutant harboured empty vector, the amount of NBD-PE was significantly decreased as described previously32 (53 ± 8% in the dark and 53 ± 8% under BL relative to wild-type levels, Fig. 3b). When the fpk1Δ fpk2Δ mutant harboured CrPHOT plasmids, intracellular fluorescence was hardly observed and showed a low value in the dark (Fig. 3a; 73 ± 5%, Fig. 3b) but was observed to the same extent as that in the wild type under BL (Fig. 3a; 99 ± 2%, Fig. 3b). In contrast, no promotion of uptake by CrPHOT under BL was observed in the fpk1Δ fpk2Δ lem3Δ background strain (Fig. 3a; 31 ± 9% in the dark and 32 ± 7% under BL for NBD-PE, Fig. 3b). This indicates that NBD-PE uptake promoted by CrPHOT depends on Dnf1p/Dnf2p-Lem3p. The same results were obtained when NBD-PC was added (Fig. 3b, Supplementary Fig. S3). These results suggest that P4-ATPase-mediated phospholipid uptake can be photo-controlled via CrPHOT. In addition, when the fpk1Δ fpk2Δ mutant harboured KDm plasmids, intracellular fluorescence was hardly observed even under BL (Fig. 3a; 50 ± 8% for NBD-PE and 64 ± 13% for NBD-PC under BL, Fig. 3b; Supplementary Fig. S3), suggesting that control by CrPHOT is dependent on its own kinase activity.
Figure 3.
CrPHOT promotes NBD-phospholipid flipping by flippases in a BL-dependent manner. (a) NBD-PE internalization by CrPHOT under BL. Wild type (WT) harbouring vector plasmid, fpk1Δ fpk2Δ mutant harbouring vector, CrPHOT or KDm plasmids, and KKT274 (fpk1Δ fpk2Δ lem3Δ) harbouring vector or CrPHOT plasmids were grown in SC medium in darkness or under 10 μmol m−2 s−1 BL at 30 °C and treated with NBD-PE. A representative cell image obtained by microscopic observation is shown. Scale bar = 5 μm. (b) Quantification of internalized NBD-labelled phospholipids. Cells grown in SDA-U medium in the dark at 30 °C were labelled with NBD-PE or -PC for 60 min in the dark or under 10 μmol m−2 s−1 BL, and then washed with SD containing 2.5% BSA before flow cytometry. (c) Time course of NBD-labelled PE internalization by BL. Cells grown in the dark were incubated for a total of 3 h in the indicated light condition (dark, BL 0.5 h, BL 1.0 h, or BL 3.0 h), in which the last 1 h was incubated with the NBD-PE. The internalized NBD-phospholipids were quantitated by flow cytometry. Data are presented as the average percentage ± SD relative to wild-type measurements of three independent experiments (10,000 cells per sample) in each light condition (*p < 0.05, **p < 0.01; Tukey’s test).
These results were consistent with the altered sensitivity of the fpk1Δ fpk2Δ mutant expressing CrPHOT or its derivatives to duramycin (Supplementary Fig. S4)37. Duramycin is a peptide toxin that specifically binds to PE in biological membranes45,46. The fpk1Δ fpk2Δ mutant is sensitive to duramycin because PE is not enriched in the inner leaflet and is exposed on the outer leaflet of the plasma membrane32,34,39,43. CrPHOT suppressed the sensitivity of the fpk1Δ fpk2Δ mutant to duramycin in a BL-dependent manner, and the K-fragment suppressed it regardless of light conditions. In contrast, the fpk1Δ fpk2Δ mutant harbouring KDm showed increased sensitivity to duramycin even under BL (Supplementary Fig. S4)37. Under BL, the resistance activity by LOV1/2 m remained moderate, although it decreased the suppression compared to that of CrPHOT (Supplementary Fig. S4). These results reconfirm the suggestion of controlling phospholipid flipping by light as described above.
We next investigated the time-course of phospholipids uptake in response to BL. Cells grown in the dark were further incubated for a total of 3 h. During incubation, cells were irradiated with different lengths of BL (0, 0.5, 1, 3 h) towards the end of the period. NBD-PE was added 1 h before the end of incubation time (Fig. 3c). As a result of quantification by flow cytometry, the internalization of NBD-PE reached the same level as that of the wild-type in 1 h BL irradiation (112 ± 7% relative to wild-type levels, Fig. 3c), although the effect of 0.5 h BL was not clear. These data demonstrate that this CrPHOT system results in a superior, light-induced control of flipping activity.
Light regulates both actin depolarization and switching of apical-isotropic growth
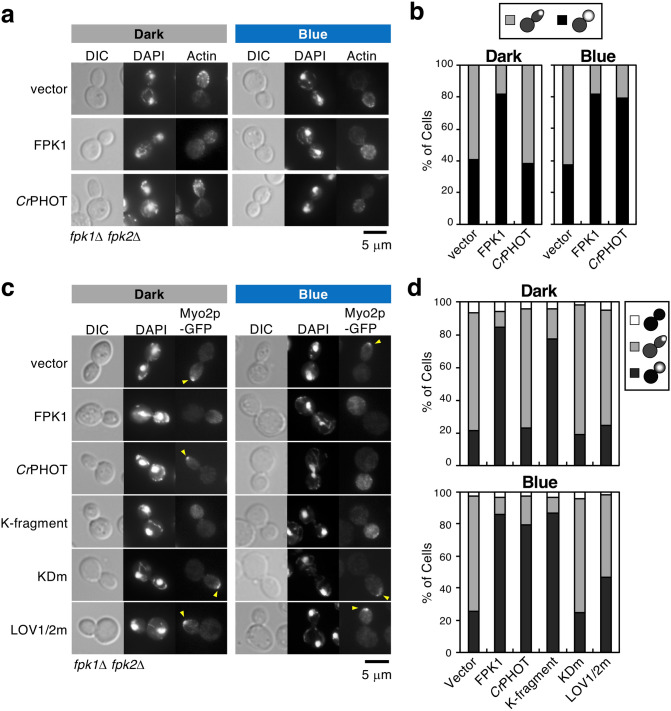
In small-budded cells (at G2/early mitotic phase), cortical actin patches (small assemblage of actin filaments) are polarized at the tip of the bud, and daughter cells exhibit apical growth47. In large-budded cells (at a late mitotic phase), the actin patches are randomly distributed, and the cells switch to isotropic growth47. Dnf1p/Dnf2p-Lem3p and Fpk1p/Fpk2p are involved in actin depolarization and switching from apical to isotropic growth. Therefore, defects in these genes result in prolonged polarization of actin patches at the tip and extended elongation of the bud even at the late mitotic phase31,32. We confirmed this phenotype in the fpk1Δ fpk2Δ mutant. Cells cultured in each light condition were stained with phalloidin-TRITC (tetramethylrhodamine B isothiocyanate peptide) to visualize actin and with DAPI (4′,6-diamidino-2-phenylindole) to confirm the cell cycle stage. As a result, the polarized actin patches and elongated bud shape were most prominent in large-budded cells regardless of light conditions (Fig. 4a, vector), and the ratio of cells with dispersed actin patches was much lower than that in FPK1p-expressing cells (Fig. 4a, vector; 40.8% in the dark and 37.5% under BL, Fig. 4b, vector). If light controls P4-ATPase activity, it will also restore both switching of actin positioning and proper growth direction. We next examined the localization of actin patches and bud morphology in the fpk1Δ fpk2Δ mutant expressing CrPHOT. In the dark, a low number of cells had depolarized actin (Fig. 4a; 38.5%, Fig. 4b), and the bud morphology remained tapered in large-budded cells. When cells were irradiated with BL, actin patches were distributed throughout the daughter cells, and the buds exhibited a round shape similar to FPK1p-expressing cells (Fig. 4a; 79.3%, Fig. 4b). These results suggest that CrPHOT is able to control both actin depolarization and switching of cell growth in a BL-dependent manner.
Figure 4.
Optical control of actin depolarization associated with apical-isotropic growth switching. (a, b) Optical control of F-actin distribution by CrPHOT. Yeast fpk1Δ fpk2Δ cells carrying pKT1639 (pRS416-FPK1) or pRS416-CrPHOT were cultured in darkness (Dark) or under 10 μmol m-2 s-1 BL irradiation (Blue) at 18 °C, followed by staining with phalloidin-TRITC and DAPI to visualize actin and nuclei, respectively. (a) A representative cell image by microscopic observation. Scale bar = 5 μm. (b) Large-budded cells with divided nuclei were classified as showing actin polarized to the bud tip (grey) or its distribution in whole daughter cells (black). n = 155–253. (c, d) Optical control of Myo2p-GFP localization in a kinase-dependent manner. KKT353 (fpk1Δ fpk2Δ MYO2-GFP) cells carrying pKT1639 (pRS416-FPK1) or pRS416-CrPHOT or its derivatives were cultured in YPDA medium in darkness (Dark) or under BL irradiation (Blue) at 18 °C, followed by staining with DAPI to visualize nuclei. (c) A representative cell image obtained by microscopic observation. Arrowheads indicate Myo2p-GFP polarized to the bud tip. Scale bar = 5 μm. (d) Large-budded cells with divided nuclei were classified as showing Myo2p-GFP distributed in whole daughter cells (black), polarized to the bud tip (grey) or delocalized (white). n = 96–180.
The same results were obtained in the localization analysis of Myo2p-GFP (Fig. 4c, d). Type V myosin Myo2p transports polarity proteins and is mostly localized at the bud tip during bud formation and dispersed throughout the cell at the late mitotic phase48. In the lem3Δ or the fpk1Δ fpk2Δ mutant, Myo2p-GFP remains polarized at the bud tip even at the late mitotic phase31,32. We thus investigated the photo-regulation of Myo2p-GFP localization by CrPHOT. In the fpk1Δ fpk2Δ mutant expressing CrPHOT, Myo2p-GFP was polarized in the dark, but it was distributed throughout the cells under BL as FPK1 was (Fig. 4c; 23.0% in the dark and 79.6% under BL, ratio of cells with dispersed Myo2p-GFP, Fig. 4d). Depolarization of Myo2p-GFP was observed in the cells expressing the K-fragment regardless of light irradiation (77.2% in the dark and 86.7% under BL), while prolonged polarization was observed even under BL in KDm- (Fig. 4c; 19.3% in the dark and 25.0% under BL, Fig. 4d) or LOV1/2 m-expressing cells (Fig. 4c; 24.7% in the dark and 46.6% under BL; Fig. 4d). These results suggest that light can regulate cell polarity switching in a CrPHOT kinase activity-dependent manner.
Light regulates endocytic recycling to the TGN in the CrPHOT system
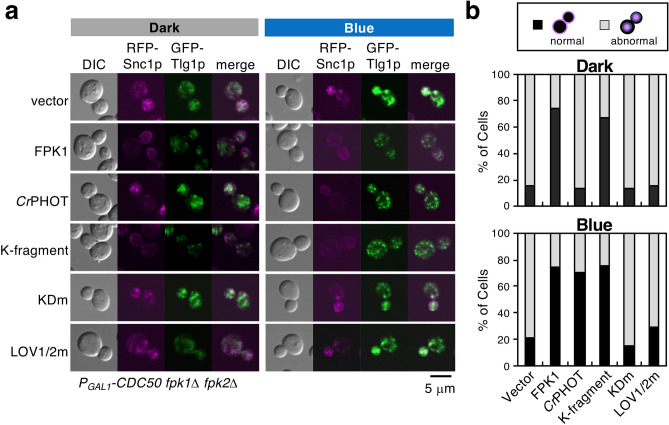
Dnf1p/Dnf2p-Lem3p and Fpk1p/Fpk2p, redundantly with Drs2p-Cdc50p, regulate endocytic recycling. The lem3Δ or fpk1Δ fpk2Δ mutant with Cdc50p-depletion exhibits severe defects in the retrieval pathway from early endosomes to the TGN22,32,33. We thus investigated whether CrPHOT can regulate this pathway in a light-dependent manner. An exocytic vesicle-SNARE Snc1p is recycled from the plasma membrane via early endosomes to the TGN by this pathway49. mRFP-Snc1p is primarily localized at the plasma membrane of daughter cells during bud formation in partially Cdc50p-depleted fpk2Δ cells (Fig. 5a, FPK1)32. A target-SNARE, Tlg1p, is recycled between the TGN and early endosomes, and thus, Tlg1p-GFP was observed in punctate fluorescence, indicating endosome/TGN localization in this cell (Fig. 5a, FPK1)50. When the retrieval pathway from early endosomes to the TGN is inhibited, Snc1p and Tlg1p accumulate in abnormal structures in the cell (Fig. 5a, vector)32,33.
Figure 5.
Optical control of endocytic recycling from early endosomes to the TGN. YTM2110 (PGAL1-CDC50 fpk1Δ fpk2Δ mRFP-SNC1) cells carrying pKT1651 (pRS315-GFP-TLG1) and pKT1639 (pRS416-FPK1) or pRS416-CrPHOT or its derivatives were cultured in darkness (Dark) or under 10 μmol m−2 s−1 BL irradiation (Blue) at 18 °C for 12 h to deplete Cdc50p in YPDA medium, followed by fixation with 0.5% formaldehyde. (a) A representative cell image by confocal microscopic observation. Scale bar = 5 μm. (b) Cells were classified as showing mRFP-Snc1p localized in the plasma membrane (normal, black) or in abnormal intracellular structures (abnormal, grey). n = 155–315.
Plasmids encoding CrPHOT or its derivatives were introduced into the Cdc50p-depleted fpk1Δ fpk2Δ mutant expressing mRFP-Snc1p and GFP-Tlg1p and cultured in the dark or under BL. After fixing the cells with formaldehyde, the localization of those marker proteins was observed using a microscope. In CrPHOT-expressing cells, mRFP-Snc1p and GFP-Tlg1p were observed in the abnormal aggregates in the dark; cells in which mRFP-Snc1p was normally localized in the plasma membrane were hardly observed (Fig. 5a; 13.3%, Fig. 5b, CrPHOT) as in the case of vector (15.0% in the dark, 20.9% under BL). Under BL irradiation, GFP-Tlg1p was localized in the normal punctate structures, and mRFP-Snc1p was localized in the cell periphery (Fig. 5a; 70.7%, Fig. 5b, CrPHOT), as in the case of the cells expressing Fpk1p (Fig. 5a; 74.5% in the dark, 74.2% under BL, Fig. 5b, FPK1). LOV1/2 m exhibited abnormal aggregation of both marker proteins and failed to restore them to normal localization even under BL irradiation (Fig. 5a; 15.5% in the dark, 29.2% under BL, Fig. 5b, LOV1/2 m). These results suggest that the endocytic recycling pathway can be photo-controlled by CrPHOT. We then investigated the necessity for kinase activity in the photo-control of endocytic recycling. As a result, both mRFP-Snc1p and GFP-Tlg1p showed normal localization in cells harbouring the K-fragment regardless of light irradiation (Fig. 5a; 67.1% in the dark and 75.0% under BL, Fig. 5b), but failed for KDm even under BL (Fig. 5a; 13.9% in the dark and 15.5% under BL, Fig. 5b). These results suggest that the control of endocytic recycling by light requires CrPHOT kinase activity.
Discussion
In this study, we succeeded in photo-controlling phospholipid (such as PC and PE) flipping and biological membrane functions (actin depolarization and endocytic recycling) in yeast. To date, some optogenetic techniques have been established to transiently control the amounts of phosphatidylinositol 4,5- bisphosphate (PI (4,5) P2) and its metabolites in cells using PI-metabolizing enzymes51. The tool we proposed is the first technique to optically control the intracellular distribution of non-PI phospholipids.
The basic mechanism of this new tool is that CrPHOT light-controls the activity of select P4-ATPases in S. cerevisiae. The P4-APases localized in the plasma membrane of yeast are mainly Dnf1p/Dnf2p-Lem3p22,23,24,25. In the NBD-phospholipid uptake analysis, NBD-PC and NBD-PE added to the medium were taken up into the cell in a light-dependent manner in the presence of CrPHOT, and the lem3Δ mutation impaired this uptake (Fig. 3). These results provide direct evidence that CrPHOT optically regulates the flipping of PE and PC through activation of Dnf1p/Dnf2p-Lem3p in the plasma membrane. Furthermore, we showed that NBD-PE uptake by CrPHOT reached that of the wild-type cells at least within one hour of BL irradiation (Fig. 3c). Therefore, the activation itself of flippase by BL is presumed to occur in a short time. However, the fpk1Δ fpk2Δ mutant harbouring CrPHOT plasmids showed slightly higher NBD-phospholipids uptake even in the dark (Fig. 3b,c). In the case of KDm, the uptake activity was suppressed to the same level as that of empty vector (Fig. 3b), hence the CrPHOT activity within yeast cells is presumed to leak in the dark. Improvements of CrPHOT molecules to strictly inhibit activity in the dark are awaited. Although detailed analysis of photo-reversibility is also needed in the future, we provide the first basis of a tool for optic controlling lipid uptake in this study.
How does CrPHOT regulate P4-ATPases? All P4-ATPase-related responses analysed in this study were dependent on the kinase activity of CrPHOT (Figs. 1, 3, 4, 5, Supplementary Fig. S3 and S4). Yeast Fpk1p phosphorylates Dnf1p/Dnf2p P4-ATPases in vivo and in vitro32,35,36. The kinase activity of Fpk1p and phosphorylation of Dnf1p are required for the optimal function of Dnf1p32,35. The control of P4-ATPase activity by Fpk1p/Fpk2p and control of P4-ATPase activity by CrPHOT under BL irradiation were remarkably consistent; therefore, we conclude that CrPHOT also regulates Dnf1p/Dnf2p by phosphorylation. Some phosphorylation sites of Dnf1p by Fpk1p have been identified, and the combination of mutations at those sites within Dnf1p reduces PE flipping in yeast cells35. The molecular mechanism underlying P4-ATPase regulation by kinase-related phosphorylation is still unknown, but this system that switches the activity of P4-ATPases by light will be quite useful for elucidating it in the future.
Next, it is necessary to consider the effect of CrPHOT on factors other than Dnf1p/Dnf2p, e.g., other P4-ATPases. In microscopic observation, localization of CrPHOT to TGN/endosomes was mainly observed (Fig. 2, Supplementary Fig. S2). Drs2p and Dnf3p are P4-ATPases mainly localized to the TGN/endosomes. Furthermore, their function in the endocytic recycling pathway is partially redundant with that of Dnf1p/Dnf2p22,23,24,33. Drs2p and Dnf3p are also phosphorylated by Fpk1p in vitro32,36, and Dnf3p is isolated as a phosphorylation substrate for Fpk1p/Fpk2p in vivo34,35. However, the necessity of Fpk1p/Fpk2p activity to the functions of Drs2p and Dnf3p is currently unknown. Further analysis is needed to determine whether CrPHOT regulates the functions of other P4-ATPases, such as Drs2p and Dnf3p.
Another is the effects of Fpk1p/Fpk2p on factors other than P4-ATPases. The protein kinases Ypk1p and Akl1p are known as phosphorylation substrates for Fpk134,35,36,43. Target of rapamycin complex 2 (TORC2) serves as a sensor and regulator for plasma membrane status and is involved in actin-cytoskeleton regulation, sphingolipid synthesis, and endocytosis in S. cerevisiae52,53. Under plasma membrane stresses, TORC2 phosphorylates and negatively regulates Fpk1p through activation of Ypk1p34. Fpk1p activated under normal conditions suppresses the upstream inhibitor Ypk1p by phosphorylation and promotes the endocytic pathway from the plasma membrane through inhibition of Akl1p by phosphorylation, independent of Dnf1p/Dnf2p activation35,36.
To investigate the involvement of CrPHOT in this endocytic pathway, CrPHOT or FPK1 was introduced into the PGAL1-CDC50 fpk1Δ fpk2Δ strain, and endocytosis from the plasma membrane was observed by FM4-64 staining. Unfortunately, the PGAL1-CDC50 fpk1Δ fpk2Δ cells failed to show any FM4-64 uptake delay even when the corresponding empty vector was introduced, despite experiments at various temperatures (4–24 °C) and depression conditions (data not shown). This observation indicated that the endocytic pathway functioned properly even in the absence of Dnf1p/Dnf2p-Lem3p and Drs2p-Cdc50p in our experimental conditions. Therefore, the involvement of CrPHOT in endocytosis could not be investigated. We would need to investigate whether CrPHOT regulates this pathway, including the phosphorylation of Akl1p and Ypk1p.
RSK3 (belonging to the p90-S6K subfamily) and Ca2+-dependent protein kinase C (PKC) are known as kinases that would be involved in phosphorylation of P4-ATPases in mammals. RSK3 has been identified as a functional counterpart of Fpk1p/Fpk2p in yeast screening43, but P4-ATPase regulation and phosphorylation in mammalian cells are not understood. PKC controls the endocytosis of the P4-ATPase ATP11C (relates to B-cell maturation, erythrocyte shape, anaemia and hyperbilirubinemia), and phosphorylation of the ATP11C C-terminal region is required for endocytosis54. Hence, P4-ATPases are likely regulated by kinases in many eukaryotes, and CrPHOT may also function in mammalian cells. Although an analysis of photo-reversibility and improvement in controllability are required, this study is able to propose the basis of a P4-ATPase light control system.
Methods
Media and growth conditions
Yeast strains were cultured in YPDA-rich medium (1% yeast extract [BD Biosciences, San Jose, CA], 2% bacto-peptone [BD], 2% glucose [Nacalai Tesque, Kyoto, Japan], and 0.01% adenine [FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan]). Strains carrying plasmids were selected in synthetic medium (SD) containing the required nutritional supplements55. When appropriate, 0.5% casamino acids [BD] were added to SD medium without uracil [FUJIFILM Wako] (SDA-Ura). For induction of the GAL1 promoter, 3% galactose [FUJIFILM Wako] and 0.2% sucrose [Nacalai] were used as carbon sources instead of glucose (YPGA and SGA-Ura). Blue (peak at 470 nm) and red (peak at 660 nm) light-emitting diode panels [SL-150X150 series; CCS, Tokyo, Japan] were used as light sources. In all analyses, BL and RL were used at an intensity of 10 μmol m−2 s−1 unless otherwise noted. When grown on the agar medium, the yeast cells were irradiated with light vertically from a height of about 10 cm above the plate. When cultured in a liquid medium, the entire test tube was irradiated with light from a distance of about 10 cm from the side of the tube. Escherichia coli strains were cultured in LB medium [Nacalai] containing appropriate antibiotics as needed. The lithium acetate method was used to introduce plasmids into yeast cells56,57.
Strains and plasmids
The S. cerevisiae strains used in this study are listed in Table 1. Yeast strains carrying monomeric red fluorescent protein (mRFP)-tagged SNC1 were constructed by integrating linearized pRS306-mRFP-SNC1 into the URA3 locus, followed by a marker change from URA3 to TRP1. Strain carrying SEC7-mRFP was constructed by PCR-based procedures as described58,59. E. coli strain DH5α was used for the construction and amplification of plasmids.
Table 1.
Yeast strains used in this study.
| Straina | Relevant genotype | Derivation/source |
|---|---|---|
| BY4743 | MATa/α LYS2/lys2Δ0 ura3Δ0/ura3Δ0 his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 | Brachmann et al., 199863 |
| YEF473 | MATa/α lys2-810/ lys2-810 ura3-52/tura3-52 his3Δ-200/his3Δ-200 trp1Δ-63/trp1Δ-63 leu2Δ-1/leu2Δ-1 | Bi and Pringle, 199664 |
| KKT330 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 HIS3MX6::PGAL1-3HA-CDC50 fpk1Δ::HphMX4 fpk2Δ::KanMX6 (designated here as PGAL1-CDC50 fpk1Δ fpk2Δ) | Nakano et al., 200832 |
| KKT268 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 fpk1Δ::HphMX4 fpk2Δ::KanMX6 (designated here as fpk1Δ fpk2Δ) | Nakano et al., 200832 |
| KKT353 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 MYO2-GFP::HIS3MX6 fpk1Δ::HphMX4 fpk2Δ::KanMX6 | Nakano et al., 200832 |
| YKT905 | MATa ura3-52 his3Δ-200 trp1Δ-63 leu2Δ-1 lys2-801 SEC7-mRFP::TRP1 | Sakane et al., 200665 |
| KKT492 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 HIS3MX6::PGAL1-3HA-CDC50 fpk1Δ::HphMX4 fpk2Δ::KanMX6 ura3::TRP1::mRFP-SNC1 | This study |
| KKT332 | MATa lys2Δ0 ura3Δ0 his3Δ1 leu2Δ0 met15Δ0 DNF1-GFP::HIS3MX6 fpk1Δ::HphMX4 fpk2Δ::KanMX6 | Nakano et al., 200832 |
| KKT336 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 DNF2-GFP::HIS3MX6 fpk1Δ::HphMX4 fpk2Δ::KanMX6 | Nakano et al., 200832 |
| KKT274 | MATa LYS2 ura3Δ0 his3Δ1 leu2Δ0 MET15 lem3Δ::KanMX6 fpk1Δ::HphMX4 fpk2Δ::HIS3MX6 | This study |
aKKT strains are isogenic derivatives of BY4743. YKT strains are isogenic derivatives of YEF473.
The plasmids used in this study are listed in Table 2. pRS416-GFP-CrPHOT, pRS416-mRFP-CrPHOT, pRS416-GFP-KDm and pRS416-mRFP-KDm were constructed as follows. GFP- or mRFP-tagged CrPHOT or CrPHOT(D549N)37 was constructed by megaprimer PCR-based procedures60 and cloned into the BamHI/SalI site of pRS41661. pRS416-LOV1/2 m was generated using a QuikChange site-directed mutagenesis kit [Agilent Technologies, Santa Clara, CA] with pRS416-LOV2m37. The genes inserted into pRS416 were constitutively expressed in a form tagged with two N-terminal tandem repeats of the influenza virus haemagglutinin epitope (2HA) under control of the TPI1 promoter. All regions constructed by PCR-based procedures were verified by DNA sequencing.
Table 2.
Plasmids used in this study.
| Plasmid | Characteristics | Derivation/source |
|---|---|---|
| YCplac111 | LEU2 CEN4 | Gietz and Sugino, 198866 |
| YEplac181 | LEU2 2 μm | Gietz and Sugino, 198866 |
| YEplac195 | URA3 2 μm | Gietz and Sugino, 198866 |
| pKO10 | PGAL1-HA URA3 2 μm | Kikyo et al., 199967 |
| pRS416 | URA3 CEN6 | Sikorski and Hieter, 198961 |
| pRS315 | LEU2 CEN6 | Sikorski and Hieter, 198961 |
| pKT1634 [pRS416-GFP-FPK1] | PTPI1-GFP-FPK1 URA3 CEN6 | Nakano et al., 200832 |
| pKT1638 [pRS416-mRFP-FPK1] | PTPI1-mRFP-FPK1 URA3 CEN6 | Nakano et al., 200832 |
| pKT1639 [pRS416-HA-FPK1] | PTPI1-HA-FPK1 URA3 CEN6 | Nakano et al., 200832 |
| pKT1651 [pRS315-GFP-TLG1] | PTPI1-GFP-TLG1 LEU2 CEN6 | This study |
|
pKT2177 [pRS306-mRFP-SNC1] pRS416-CrPHOT |
PTPI1-mRFP-SNC1 URA3 PTPI1-HA-CrPHOT URA3 CEN6 |
This study Aihara et al., 201237 |
| pRS416-K-fragment | PTPI1-HA-CrPHOTΔN URA3 CEN6 (identical to Kinase-fragment in Aihara et al., 201237) | Aihara et al., 201237 |
| pRS416-KDm | PTPI1-HA-CrPHOT(D549N) URA3 CEN6 (identical to Kinase-dead (D549N) in Aihara et al., 201237) | Aihara et al., 201237 |
| pRS416-LOV2m | PTPI1-HA-CrPHOT(C250A) URA3 CEN6 (identical to (C250A) in Aihara et al., 201237) | Aihara et al., 201237 |
| pRS416-LOV1/2 m | PTPI1-HA-CrPHOT(C57A, C250A) URA3 CEN6 | This study |
| pRS416-GFP-CrPHOT | PTPI1-HA-GFP-CrPHOT URA3 CEN6 | This study |
| pRS416-mRFP-CrPHOT | PTPI1-HA-mRFP-CrPHOT URA3 CEN6 | This study |
| pRS416-GFP-KDm | PTPI1-HA-GFP-CrPHOTΔN URA3 CEN6 | This study |
| pRS416-mRFP-KDm | PTPI1-HA-mRFP-CrPHOTΔN URA3 CEN6 | This study |
Yeast growth assay
The yeast cells were grown at 28 °C in liquid SGA-Ura medium to an A600 of 0.6–0.8 and diluted with sterile water to an A600 of 0.1. Ten-fold serial dilutions (10–1, 10–2, 10–3, 10–4) were made in sterile water. A 10-μl aliquot of each of the diluted cell suspensions was then spotted on a plate with SGA-Ura or SDA-Ura medium. The plates were then placed at 28 °C under different light conditions for 3 days. For growth sensitivity to duramycin, the yeast cells grown in liquid SDA-Ura were diluted as above, and 2 μl of each of the diluted cell suspensions were examined on YPDA plates containing 20 μM duramycin [Sigma-Aldrich, St. Louis, MO] at 28 °C.
Immunoblot analysis
Yeast cells were grown to logarithmic phase in YPGA medium at 28 °C under different light conditions. Protein extraction from yeast cells was performed as described62. The extracted proteins (40 μg) were separated in a 4–15% SDS-polyacrylamide gel [Bio-Rad Laboratories, Hercules, CA] and blotted onto polyvinylidene difluoride (PVDF) membrane [Bio-Rad], and immunoblotting was performed using mouse anti-HA monoclonal antibody [MBL, Nagoya, Japan] and horseradish peroxidase (HRP)-conjugated anti-mouse IgG [Promega, Madison, WI]. The signals were detected using an LAS-3000 imaging system [FUJIFILM, Tokyo, Japan]. The raw image is presented in Supplementary Fig. S5, which includes an unprocessed original data that was used to prepare Fig. 1d.
Microscopic observations
Microscopic observations were performed as previously described32. Yeast cells were grown to early-midlogarithmic phase in YPDA liquid medium at 18 °C in darkness or under BL irradiation. All experiments requiring dark conditions were performed in a dark room with the aid of green safe light. Most GFP- or mRFP-tagged proteins were observed in living cells, which were observed using a Fluoview FV1000 confocal microscope [Olympus Corporation, Tokyo, Japan]. The lipophilic styryl dye FM4-64 [Thermo Fisher Scientific, Waltham, MA, USA] was used to visualize endosomal structures. Cells grown in the logarithmic phase were harvested by centrifugation, washed twice with ice-cold SC medium, resuspended in 100 μl of ice-cold SC medium containing 4 μl of 1 mM FM4-64 in dimethyl sulfoxide (DMSO) (40 μM final concentration), and then incubated at 25 °C for 20 min. Labelled cells were immediately observed using a confocal microscope. Localization of mRFP-Snc1p and GFP-Tlg1p or Sec7p-mRFP was observed in fixed cells. Cells grown in the logarithmic phase were fixed by the addition of formaldehyde (3.7% final concentration) [FUJIFILM Wako] into the medium and incubated at 18 °C for 10 min and then 30 °C for 10 min. After fixation, cells were washed twice with phosphate-buffered saline (PBS) and immediately observed using a confocal microscope. To observe Myo2p-GFP in cells with divided nuclei, cells fixed as above were stained with DAPI (0.5 μg ml-1 final concentration) [Sigma-Aldrich] in 100 μl of water at room temperature for 10 min. To visualize F-actin, cells were fixed by the addition of formaldehyde (5.0% final concentration) into the medium and incubated at 18 °C for 10 min and then 25 °C for 30 min. After fixation, the cells were labelled with phalloidin-TRITC (0.1925 μM final concentration) [Sigma-Aldrich] in PBS at room temperature for 30 min, washed three times with PBS, and then stained with DAPI as described above. Cells stained with DAPI were washed three times with water and immediately observed using a BX51 biological microscope [Olympus] with the appropriate fluorescence filter sets [Olympus]. Images were acquired with an ORCA-fusion digital CMOS camera [C14440-20UP; Hamamatsu Photonics, Hamamatsu, Japan].
Internalization of fluorescence-labelled phospholipids into yeast cells
Large unilamellar vesicles containing NBD-phospholipids were prepared as described29. 1-palmitoyl-2-(6-NBD-aminocaproyl)-PE (NBD-PE), 1-palmitoyl-2-(6-NBD-aminocaproyl)-PC (NBD-PC), and dioleoylphosphatidylcholine (DOPC) were obtained from Avanti Polar Lipids (Alabaster, AL). Fluorescently labelled phospholipid internalization experiments were performed as described29,30. Briefly, cells were grown to early logarithmic phase in SDA-U medium at 30 °C in the dark. After dilution to 0.35 A600 ml-1, cells were incubated for 60 min at 30 °C with liposomes containing 40% NBD-phospholipid and 60% DOPC at a final concentration of 20 μM in the dark or under BL. Cells were then suspended in cold SD containing 20 mM sodium azide and 2.5% bovine serum albumin (BSA), incubated for 20 min, and washed with PBS. Flow cytometry of fluorescently labelled cells was performed on a FACSCanto II cytometer [BD]. For investigation of time response, overnight cultures were diluted to 0.2 A600 ml-1 and incubated for a total of 3 h in the indicated condition (dark, BL 0.5 h, BL 1.0 h, or BL 3.0 h), in which the last 1 h was incubated with the NBD-PE liposome. The experiments were performed in three biological replicates consisting of 10,000 cells per sample. Significance for Fig. 3 was determined using a one-way analysis of variance with a Tukey’s test.
Supplementary information
Acknowledgements
We are grateful to N. Mochizuki (Kyoto University, Japan) for helpful discussion, S. Kawamura and Y. Naito for technical support. This work was supported by JST, PRESTO Grant Number JPMJPR1786, Japan.
Author contributions
T.S. conceived and conducted most of the experiments and data analysis and wrote the paper. T.M. and K.T. advised on most of experiments, and designed and conducted the lipid incorporation analysis. A.N. supervised the project and edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tomomi Suzuki, Email: tomo@physiol.bot.kyoto-u.ac.jp.
Akira Nagatani, Email: nagatani@physiol.bot.kyoto-u.ac.jp.
Supplementary information
is available for this paper at 10.1038/s41598-020-69459-0.
References
- 1.Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993;294:1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pomorski T, Hrafnsdóttir S, Devaux PF, van Meer G. Lipid distribution and transport across cellular membranes. Semin. Cell Dev. Biol. 2001;12:139–148. doi: 10.1006/scdb.2000.0231. [DOI] [PubMed] [Google Scholar]
- 3.Muthusamy B-P, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim. Biophys. Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emoto K, et al. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA. 1996;93:12867–12872. doi: 10.1073/pnas.93.23.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo J, et al. Phosphatidylethanolamine Is Required for Normal Cell Morphology and Cytokinesis in the Fission Yeast Schizosaccharomyces pombe. Eukaryot. Cell. 2009;8:790–799. doi: 10.1128/EC.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kay JG, Fairn GD. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun. Signal. 2019;17:126. doi: 10.1186/s12964-019-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato U, et al. Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. J. Biol. Chem. 2013;288:4922–4934. doi: 10.1074/jbc.M112.402701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donnell VB, Rossjohn J, Wakelam MJO. Phospholipid signaling in innate immune cells. J. Clin. Investig. 2018;128:2670–2679. doi: 10.1172/JCI97944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montigny C, Lyons J, Champeil P, Nissen P, Lenoir G. On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim. Biophys. Acta. 2016;1861:767–783. doi: 10.1016/j.bbalip.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Andersen JP, et al. P4-ATPases as phospholipid flippases—structure, function, and enigmas. Front. Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmgren M, Østerberg JT, Nintemann SJ, Poulsen LR, López-Marqués RL. Evolution and a revised nomenclature of P4 ATPases, a eukaryotic family of lipid flippases. Biochim. Biophys. Acta. 2019;1861:1135–1151. doi: 10.1016/j.bbamem.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Panatala R, Hennrich H, Holthuis JCM. Inner workings and biological impact of phospholipid flippases. J. Cell Sci. 2015;128:2021–2032. doi: 10.1242/jcs.102715. [DOI] [PubMed] [Google Scholar]
- 14.Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG. P4-ATPases: lipid flippases in cell membranes. Pflüg. Arch. 2014;466:1227–1240. doi: 10.1007/s00424-013-1363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nintemann SJ, Palmgren M, López-Marqués RL. Catch you on the flip side: a critical review of flippase mutant phenotypes. Trends Plant Sci. 2019;24:468–478. doi: 10.1016/j.tplants.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Lee S, et al. Transport through recycling endosomes requires EHD 1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Best JT, Xu P, Graham TR. Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr. Opin. Cell Biol. 2019;59:8–15. doi: 10.1016/j.ceb.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung T, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–213. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 19.Uchida Y, et al. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc. Natl. Acad. Sci. USA. 2011;108:15846–15851. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aoki Y, Uenaka T, Aoki J, Umeda M, Inoue K. A novel peptide prove for studying the transvilayer movement of phosphatidylethanolamine. J. Biochem. (Tokyo) 1994;116:291–297. doi: 10.1093/oxfordjournals.jbchem.a124522. [DOI] [PubMed] [Google Scholar]
- 21.Iwamoto K, et al. Local exposure of phosphatidylethanolamine on the yeast plasma membrane is implicated in cell polarity: polarized PE exposure on plasma membrane. Genes Cells. 2004;9:891–903. doi: 10.1111/j.1365-2443.2004.00782.x. [DOI] [PubMed] [Google Scholar]
- 22.Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-Type ATPases is required for protein trafficking between golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomorski T, et al. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K, Hua Z, Nepute JA, Graham TR. Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell. 2007;18:487–500. doi: 10.1091/mbc.E06-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sartorel E, Barrey E, Lau RK, Thorner J. Plasma membrane aminoglycerolipid flippase function is required for signaling competence in the yeast mating pheromone response pathway. Mol. Biol. Cell. 2015;26:134–150. doi: 10.1091/mbc.E14-07-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baldridge RD, Xu P, Graham TR. Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 2013;288:19516–19527. doi: 10.1074/jbc.M113.476911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riekhof WR, et al. Lysophosphatidylcholine Metabolism in Saccharomyces cerevisiae: the role of p-type ATPases in transport and a broad specificity acyltransferase in acylation. J. Biol. Chem. 2007;282:36853–36861. doi: 10.1074/jbc.M706718200. [DOI] [PubMed] [Google Scholar]
- 28.Roland BP, et al. Yeast and human P4-ATPases transport glycosphingolipids using conserved structural motifs. J. Biol. Chem. 2019;294:1794–1806. doi: 10.1074/jbc.RA118.005876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito K, et al. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-Type atpase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato U, et al. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- 31.Saito K, et al. Transbilayer Phospholipid Flipping Regulates Cdc42p Signaling during Polarized Cell Growth via Rga GTPase-Activating Proteins. Dev. Cell. 2007;13:743–751. doi: 10.1016/j.devcel.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell. 2008;19:1783–1797. doi: 10.1091/mbc.E07-07-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Endocytic Recycling in Yeast Is Regulated by Putative Phospholipid Translocases and the Ypt31p/32p–Rcy1p Pathway. Mol. Biol. Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roelants FM, Baltz AG, Trott AE, Fereres S, Thorner J. A protein kinase network regulates the function of aminophospholipid flippases. Proc. Natl. Acad. Sci. USA. 2010;107:34–39. doi: 10.1073/pnas.0912497106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roelants FM, et al. TOR complex 2-regulated protein kinase Fpk1 stimulates endocytosis via inhibition of Ark1/Prk1-related protein kinase Akl1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2017;37:e00627. doi: 10.1128/MCB.00627-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourgoint C, et al. Target of rapamycin complex 2–dependent phosphorylation of the coat protein Pan1 by Akl1 controls endocytosis dynamics in Saccharomyces cerevisiae. J. Biol. Chem. 2018;293:12043–12053. doi: 10.1074/jbc.RA117.001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aihara Y, et al. Mutations in N-terminal flanking region of blue light-sensing Light-Oxygen And Voltage 2 (LOV2) domain disrupt its repressive activity on kinase domain in the Chlamydomonas Phototropin. J. Biol. Chem. 2012;287:9901–9909. doi: 10.1074/jbc.M111.324723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuoka D, Tokutomi S. Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. USA. 2005;102:13337–13342. doi: 10.1073/pnas.0506402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roelants FM, et al. Protein kinase Gin4 negatively regulates flippase function and controls plasma membrane asymmetry. J. Cell Biol. 2015;208:299–311. doi: 10.1083/jcb.201410076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vida TA. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakamoto K, Briggs WR. Cellular and subcellular localization of phototropin 1. Plant Cell. 2002;14:1723–1735. doi: 10.1105/tpc.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong S-G, et al. Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 2006;45:994–1005. doi: 10.1111/j.1365-313X.2006.02667.x. [DOI] [PubMed] [Google Scholar]
- 43.Yamane-Sando Y, et al. Fpk1/2 kinases regulate cellular sphingoid long-chain base abundance and alter cellular resistance to LCB elevation or depletion. MicrobiologyOpen. 2014;3:196–212. doi: 10.1002/mbo3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanson PK, Grant AM, Nichols JW. NBD-labeled phosphatidylcholine enters the yeast vacuole via the pre-vacuolar compartment. J. Sci. 2002;115:2725–2733. doi: 10.1242/jcs.115.13.2725. [DOI] [PubMed] [Google Scholar]
- 45.Navarro J, et al. Interaction of duramycin with artificial and natural membranes. Biochemistry. 1985;24:4645–4650. doi: 10.1021/bi00338a025. [DOI] [PubMed] [Google Scholar]
- 46.Noji T, et al. Mutational analysis of the Lem3p-Dnf1p putative phospholipid-translocating P-type ATPase reveals novel regulatory roles for Lem3p and a carboxyl-terminal region of Dnf1p independent of the phospholipid-translocating activity of Dnf1p in yeast. Biochem. Biophys. Res. Commun. 2006;344:323–331. doi: 10.1016/j.bbrc.2006.03.095. [DOI] [PubMed] [Google Scholar]
- 47.Pruyne D, Bretscher A. Polarization of cell growth in yeast II. The role of the cortical actin cytoskeleton. J. Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 48.Schott D, Huffaker T, Bretscher A. Microfilaments and microtubules: the news from yeast. Curr. Opin. Microbiol. 2002;5:564–574. doi: 10.1016/s1369-5274(02)00369-7. [DOI] [PubMed] [Google Scholar]
- 49.Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol. Biol. Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siniossoglou S. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Idevall-Hagren O, De Camilli P. Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta. 2015;1851:736–745. doi: 10.1016/j.bbalip.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem. Sci. 2009;34:620–627. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Weisman R, Cohen A, Gasser SM. TORC 2—a new player in genome stability. EMBO Mol. Med. 2014;6:995–1002. doi: 10.15252/emmm.201403959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takatsu H, et al. Phospholipid flippase ATP11C is endocytosed and downregulated following Ca2+-mediated protein kinase C activation. Nat. Commun. 2017;8:1423. doi: 10.1038/s41467-017-01338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 56.Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 57.Gietz RD, Woods RA. Transformation of yeast by lithium/acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzym. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 58.Longtine MI, Demarini S, Wach B, Philippsen P. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 59.Goldstein AL. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 60.Barik S. Mutagenesis and Gene Fusion by Megaprimer PCR. Berlin: Springer; 1997. pp. 173–182. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces ceratisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;9:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 63.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 64.Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sakane H, Yamamoto T, Tanaka K. The functional relationship between the Cdc50p-Drs2p putative aminophospholipid translocase and the Arf GAP Gcs1p in vesicle formation in the retrieval pathway from yeast early endosomes to the TGN. Cell Struct. Funct. 2006;31:87–108. doi: 10.1247/csf.06021. [DOI] [PubMed] [Google Scholar]
- 66.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 67.Kikyo M, et al. An FH domain-containing Bnr1p is a multifunctional protein interacting with a variety of cytoskeletal proteins in Saccharomyces cerevisiae. Oncogene. 1999;18:7046–7054. doi: 10.1038/sj.onc.1203184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.