Bacteria exhibit distinct swim modes, thereby facilitating robust and efficient chemotaxis performance in complex habitats.
Abstract
Bacterial chemotaxis—a fundamental example of directional navigation in the living world—is key to many biological processes, including the spreading of bacterial infections. Many bacterial species were recently reported to exhibit several distinct swimming modes—the flagella may, for example, push the cell body or wrap around it. How do the different run modes shape the chemotaxis strategy of a multimode swimmer? Here, we investigate chemotactic motion of the soil bacterium Pseudomonas putida as a model organism. By simultaneously tracking the position of the cell body and the configuration of its flagella, we demonstrate that individual run modes show different chemotactic responses in nutrition gradients and, thus, constitute distinct behavioral states. On the basis of an active particle model, we demonstrate that switching between multiple run states that differ in their speed and responsiveness provides the basis for robust and efficient chemotaxis in complex natural habitats.
INTRODUCTION
Bacterial swimming is one of the most widespread forms of cellular locomotion that plays a key role in many biological processes, such as the spreading of infections or the formation of biofilms (1). Bacterial swimmers propel themselves by rotating helical flagella that are attached to their cell body. During continuous rotation of the flagellar motors, cells propagate in straight runs. Such episodes of smooth swimming are typically interrupted by sharp turning events that are initiated if motors interrupt or change their sense of rotation. In the classical case of Escherichia coli that carries several flagella distributed across the cell body (peritrichous flagellation), a smooth run requires that all motors rotate in a counterclockwise (CCW) direction to form a coherent flagellar bundle. As soon as one or several motors change direction, the bundle falls apart and the cell undergoes a tumble event that results in an erratic change of the swimming direction (2). Species that carry only one flagellum (monotrichous flagellation) may interrupt their runs by pausings in the motor rotation, such as in the case of Rhodobacter sphaeroides (3), or they may change the sense of motor rotation to reverse their direction of motion and switch from a pushing to a pulling mode, such as many marine bacteria (4) or the soil bacterium Azospirillum brasilense (5).
One of the prime challenges of bacterial swimmers is to purposefully navigate in their complex natural habitats to find food or flee from poisons (6). For E. coli, it is well known that cells bias their swimming pattern toward sources of food by decreasing the tumbling frequency during runs that are oriented in the direction of increasing nutrient concentration. While this paradigmatic chemotaxis strategy has been confirmed in several species that exhibit a single swimming mode, much less is known about the chemotactic behavior of bacteria that rely on a combination of several run modes. Here, we will study the chemotaxis strategy of the soil bacterium Pseudomonas putida KT2440, a well-established model organism with three different types of runs (7) P. putida propels itself with a tuft of helical flagella attached to one of the cell poles (lophotrichous flagellation). It swims in straight runs that are interrupted by sharp reversals in the swimming direction and occasional stop events (8). Moreover, the swimming speed can change between runs by a factor of two on average (9). On the basis of high-speed imaging of fluorescently tagged flagella, we could recently show that P. putida may switch between pushing and pulling modes by synchronously reversing the rotation of its motors (7). In addition, P. putida may wrap its flagellar bundle around the cell body to swim in a slow corkscrew-type configuration, a previously unidentified type of swimming motility that we call wrapped mode, which was recently found in several bacterial species (10–12).
Relying on P. putida as an example, we will assess the role of the corkscrew configuration in bacterial chemotaxis and, at the same time, shed light onto the chemotaxis strategies of multimode swimmers beyond the classical paradigm of E. coli. Using combined fluorescence (F-L) and phase-contrast (P-C) imaging, we simultaneously tracked cell positions and flagellar configurations to infer run times, turning angles, and transition probabilities between the different modes in the presence of a chemoattractant gradient from experimental data. On the basis of this dataset, we propose an active particle model for swimmers with multimode motility that reproduces, on the one hand, the short time dynamics of the bacterial swimmers inferred from experiments and, furthermore, allows to perform numerical simulations and analytical calculations to predict quantities that describe the long-time dynamics such as the chemotactic drift velocity and the diffusion coefficient. In this way, we can reliably predict the long-time chemotactic response, thereby bridging the gap between experimental short-time observations and chemotactic motion in the long time limit. Furthermore, our model provides the basis for a critical assessment of the efficiency and robustness of chemotaxis strategies of bacteria with multimode motility in general, both in the bulk fluid and in disordered environments.
RESULTS
Simultaneous tracking of cell body and flagellar configuration
The motility pattern of P. putida is composed of persistent runs that are interrupted by stops and turn events (Fig. 1). Three different run types can be distinguished depending on whether cells swim in the push, pull, or wrapped mode (7). To capture the full swimming statistics, cell tracking thus requires the simultaneous recording of both the trajectory of the cell body and the configuration of its flagella to distinguish between the different swimming modes (13). To achieve this goal, a dedicated imaging setup was designed and constructed to record a sequence of alternating P-C and F-L images at a frame rate of 20 s−1 (see Materials and Methods for technical details). The cell body was tracked on the basis of the P-C images (see Fig. 1A and movie S1 for a typical trajectory). Reversal and stop events, which separate individual runs, were identified by an automatized tumble recognition algorithm based on the time series of cell speed and angular velocity. Reversals of the direction of motion caused sharp spikes in the angular velocity time series combined with a drop in the swimming speed, whereas stops were characterized by a decrease in the speed without abrupt changes in the direction of motion (see Fig. 1, E and F, for examples). All results were cross-checked manually (see the Supplementary Materials for details regarding the experiment and data analysis).
Fig. 1. A combined F-L/P-C microscopy technique enables tracking of the multimode swimmer P. putida with simultaneous information on cell position and orientation of the flagellar bundle.
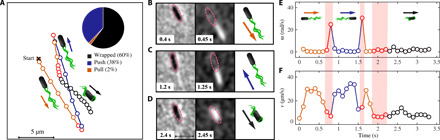
(A) A typical trajectory containing multiple changes of the flagellar configuration. Cell body positions are displayed by circular markers at 0.1-s intervals. The three run modes are highlighted in color: pull (orange), push (blue), and wrapped (black). A schematic of the flagellar bundle configuration is placed close to the corresponding part of the trajectory (not drawn to scale). The arrows indicate the swimming direction. The transition events, identified by the tumble analysis, are shown in red. The frequency of observations of the different swimming modes is represented as an inset (built from 2642 runs in total). The panels in the middle (B to D) represent the corresponding image series of P-C (left) and F-L (middle) with the time delay of 0.05 s between two consecutive P-C and F-L images. The swim modes are symbolized by cartoons on the right. The pink ellipses are the position of the cell body, obtained by linear interpolation between adjacent P-C images. (E and F) Time series of the absolute value of the rotational velocity ω (E) and the swimming speed v (F) [same color scheme as in (A)]. Scale bars, 5 μm.
The flagella were stained with a fluorescent dye and displayed in the F-L frames of our recordings. By aligning adjacent P-C and F-L images, the position of the flagellar bundle with respect to the cell body and the direction of motion could be determined, thus allowing for a clear assignment of pushing, pulling, and wrapped filament configurations to individual runs. In Fig. 1A, an example of such an assignment is displayed, together with selected P-C and F-L images in Fig. 1 (B to D). The P-C images are used to track the cell body, whereas F-L images contain information about the state of the flagellar bundle. The first row (Fig. 1B) displays the cell in the pull mode with the flagellar bundle placed in front of the cell body, the second row shows the push mode (Fig. 1C) characterized by a long tight bundle located behind the cell body, and the third row (Fig. 1D) is the wrapped mode identified by an oval-shaped bundle covering the cell body. In total, 1799 trajectories were recorded in the absence of a chemoattractant and 1378 in a chemoattractant gradient.
The majority of runs in our recordings displayed wrapped or pushing flagellar configurations (see the inset in Fig. 1A); only a small number of swimmers with a stable pulling bundle were observed in contrast to earlier data (7). Please note that the recordings reported in (7) were taken close to the microscope coverslip surface due to the much smaller working distance of the 60× and 100× objectives as compared with the 20× objective used here. Hydrodynamic surface interactions may, however, crucially influence the dynamics and stability of the flagellar bundle. In our present work, we therefore decided to take advantage of the larger working distance of the 20× objective and tracked the bacteria in the bulk to exclude all hydrodynamic wall effects on the flagellar dynamics. On the basis of our present data, the pulling mode appears as a short-lived transient within the flagellar reconfiguration from push to wrapped mode that was well captured in our earlier high-speed recordings but is practically invisible at the lower temporal resolution used in this study (cf. movie S2 and corresponding fig. S2 for an example illustrating a push-wrap transition). Nevertheless, we note that also at high temporal resolution direct transitions from push to wrapped mode can be observed (movies S3 and S7 and figs. S3 and S7A), which is in agreement with the behavior reported for Burkholderia sp. RPE64, whose flagellation pattern is similar to P. putida (12). Because of the low number of observations of the pull mode, it is considered irrelevant for the chemotaxis strategy of P. putida; the subsequent analysis rather focuses on the push and wrapped mode.
Turn angle distributions
Bacteria can change their direction of motion either by stopping the flagellar motors or by changing their sense of rotation. As a consequence, persistent runs are interrupted by stop events or by abrupt turns in the swimming direction (see the Supplementary Materials for movies and figs. S2 to S7 showing exemplary trajectories). For P. putida, the distribution of the turn angle ψ, i.e., the change in swimming direction from the end of one run to the beginning of the next run, has been reported to be bimodal, with peaks at ψ ≃ 0° and ψ ≃ 180° corresponding to stop and reversal events, respectively (8, 9). Knowing the configuration of the flagellar bundle for every run now enabled us to extract separate turn angle distributions for the different transition scenarios, as shown in Fig. 2. When runs in the push mode were interrupted by pauses in motor rotation, the flagellar bundle gradually disassembled and we observed a stop event with only marginal changes of the direction of motion (see Fig. 2A for the turn angle distribution and movie S4 and fig. S4 for an example). The turn angle distributions for transitions from push to wrapped mode and vice versa (movies S2 and S6 and figs. S2 and S6) show that these changes in flagellar configuration induce reversals in the swimming direction, as can be seen from a pronounced peak at around ψ ≃ 180° in Fig. 2 (C and D). Last, transitions from wrapped to wrapped mode (movie S5 and fig. S5) are characterized by a broad turn angle distribution with a peak at ψ ≈ 60° (Fig. 2B). They may be triggered by an interruption of the bundle rotation (movie S8 and fig. S7B) or by a single filament leaving the coherent bundle (movie S9 and fig. S7C). At a phenomenological level, the motility pattern of P. putida thus combines elements of the run-and-tumble motility of E. coli (2), the run-and-stop motion of R. sphaeroides (3), and the run-and-reverse pattern that is known from many marine bacteria (4, 14). The duration of turn or stop events (Δt = 0.3 s) is typically one order of magnitude shorter than the average run duration in both push and wrapped modes.
Fig. 2. Statistics of the turn angles of P. putida in the bulk for transitions from one run mode to another.
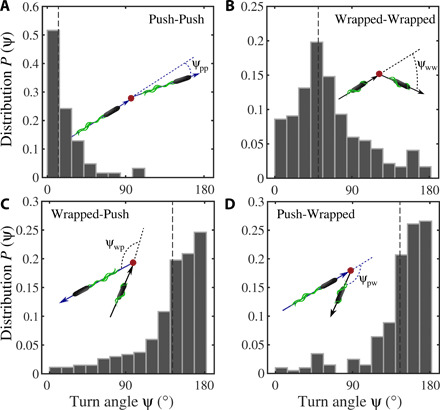
(A) Narrow turn angle distribution centered around zero degree corresponding to motor stops during push runs; (B) broad distribution of turn angles with a characteristic peak around 60° for transitions from wrapped to wrapped mode; (C) and (D) reveal that reversals of the direction of motion (peak at 180°) occur whenever the swimming mode changes from push to wrapped and vice versa. Dashed lines represent the median values. Distributions are built from 59, 415, 266, and 199 events for the histograms (A to D), respectively.
Chemotaxis of P. putida relies on wrapped swimming mode
To elucidate how P. putida uses its multimode swimming pattern to navigate toward a source of chemoattractant, we imaged bacteria in a linear casamino acid gradient and identified turn events and run types, as described above. Most trajectories contained zero, one, or two turn events. Runs in push and wrapped modes were analyzed separately. In both cases, two subsets of the data were created that contained all runs oriented up or down the gradient direction, respectively. For all four datasets, the sojourn probabilities s(t) were determined, i.e., the probabilities of observing a run that is longer than a given time t. In Fig. 3, the results are displayed for the runs in push mode (A) and wrapped mode (B). The sojourn probabilities s(t) were estimated on the basis of a nonparametric maximum likelihood approach proposed in (15), which correctly takes incomplete runs—those that do not begin or end with a turn event because bacteria enter or leave the field of view—into account as censored data.
Fig. 3. Run-time bias of P. putida in a linear concentration gradient.
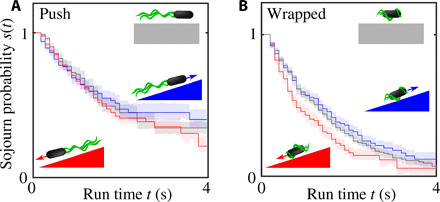
The plots show the sojourn probabilities s(t) that a run is longer than a given time t for runs in the push mode (A) and wrapped mode (B). Upgradient runs are shown in blue and downgradient runs in red. The control experiments in the absence of a gradient are plotted in gray for comparison. The error bars indicate the 1σ interval estimated by bootstrapping (34). The run-time bias depends on the swim mode: Whereas the run-time statistics up- and downgradient is practically indistinguishable for bacteria swimming in the push mode, there is a significant run-time bias in the wrapped mode; bacteria in the wrapped mode tend to decrease the downgradient run time. Each curve includes information from at least 900 runs.
Comparing upgradient and downgradient runs in the wrapped mode, we observe a clear run-time bias, i.e., the average run time depends on the direction of motion with respect to the gradient direction. This is reflected in a difference of the sojourn probabilities of up- and downgradient runs, so that runs toward the source of the chemoattractant are, on average, longer than runs in the opposite direction (see Fig. 3B). Unexpectedly, for runs in the push mode, no such run-time bias is observed; the statistics of swimming durations up and down the gradient are virtually indistinguishable (see Fig. 3A). Thus, the push and wrapped swimming modes differ in their chemotactic responsiveness. Note that the wrapped and push modes are associated with opposite senses of flagellar rotation (7). Thus, the time that a flagellum rotates in a clockwise (CW) direction (wrapped mode) is affected by the chemoattractant, whereas the time of CCW rotation (push mode) is not. This asymmetry is reminiscent of the different responses of CW and CCW rotation times observed in Vibrio alginolyticus (14, 16) in response to a stepwise temporal increase in chemoattractant (17).
Another remarkable feature of the chemotaxis strategy of P. putida is revealed by comparing the run-time statistics in a linear gradient to the control experiment, where this gradient is absent: While the sojourn probability of upgradient runs is practically identical to the control experiment, downgradient runs are significantly shorter (Fig. 3B). This is markedly different from the well-studied case of E. coli, where bacteria extend their run duration when heading in the direction of the chemical gradient (2).
A parametric maximum likelihood parameter inference based on a Weibull distribution, which substantiates our findings discussed above, is presented in the Supplementary Materials. In particular, we show that differences in the up- and downgradient run-time distributions are statistically not significant (they correspond to a 1.6σ fluctuation) in the case of swimmers in the push mode, whereas in the case of bacteria swimming in the wrapped mode, they are significant (3.1σ fluctuation). Consequently, we conclude that the wrapped mode is the relevant swimming state for chemotaxis of P. putida.
Modeling active particles with multimode motility
We propose an active particle model for the multimode swimmer P. putida based on the experimentally observed motility states and the statistics of the corresponding transitions between them (see Fig. 4). The two essential run modes are push (p) and wrap (w); since the pull mode is rarely observed, it is not taken into account here. Neglecting the pull runs does not change any of the results discussed in the following because the relative fraction of time that bacteria swim in the pull mode is small (2%, as shown in Fig. 1); a more detailed discussion of this aspect is presented in the Supplementary Materials. During run phases, bacteria move at characteristic speeds—denoted by vp and vw for push and wrapped modes, respectively—which are assumed to be approximately constant. The finite persistence length of runs is taken into account by rotational noise with the intensities Dp and Dw. For simplicity, we assume all run, stop, and transition times to be exponentially distributed, such that they are characterized by average durations or, equivalently, by effective transition rates denoted by κi in Fig. 4. Using parametric maximum likelihood parameter inference based on the experimental data, we tested that the run-time distributions are reasonably well described by an exponential distribution (see the Supplementary Materials).
Fig. 4. Model representation of the motility pattern of P. putida: The two essential run states, push and wrap, are interrupted by stops of the flagellar driving or their reconfiguration with respect to the cell body inducing transitions from one run mode to another accompanied by turns.
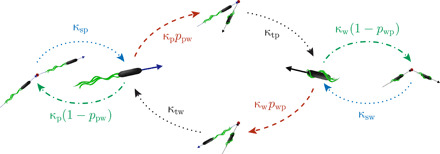
The mean duration of all states is parameterized by inverse transition rates, denoted by κi. The parameter ppw reflects the probability that the push mode is followed by the wrapped mode (and vice versa for pwp).
During stop and transition phases, there is no substantial spatial displacement of the cell body. The bacterial swimmers initiate these maneuvers by changes in flagellar activity. However, we do not focus on the mechanistic details of flagellar dynamics here but describe the turning events in terms of their probabilistic turn angle distributions (Fig. 2). Last, the parameters ppw and pwp, accounting for the probabilities that a push (wrap) run is followed by a run in the wrapped (push) mode, complete the model. The full temporal dynamics is mathematically described by a set of master equations, defining the motility in each state and the transition statistics between them (see the Supplementary Materials for mathematical details).
We stress that all parameters of the active particle model for the short-time dynamics were estimated on the basis of our experimental data. The details on the parameter inference techniques are listed in the Supplementary Materials. In the following, we will mostly focus on the parameter regime relevant for P. putida. However, we will also use our model to explore regimes that do not correspond to the experimentally observed parameter values in order to address the question why the motility pattern of P. putida has developed in the way we observed it. Is it optimized to perform chemotaxis or to navigate specific environments? The possibility to vary model parameters is crucial to assess the chemotactic performance of our model swimmer and also to characterize the consequences of multimode swimming for bacterial chemotaxis in general.
Cells that swim in a chemical gradient may experience a run-time bias, i.e., the average run times depend on the direction of motion with respect to the chemical gradient. We account for this on a phenomenological level via gradient-dependent transition rates (18, 19)
| (1) |
where the unit vector e denotes the direction of motion. Without a chemical gradient, the transition rates are given by λp,w. The coupling of the run times to the chemical gradient is determined by the parameters ηp,w: The run times are enhanced upgradient (taxis toward a chemoattractant) for ηp,w > 0, implying that the corresponding transition rates are decreased.
The actual run-time bias ηp,w depends on the way in which the bacterium senses extracellular concentrations and on the internal signaling cascade that controls flagellar activity. For P. putida, we observed ηp ≪ ηw (Fig. 3). As we do not address the biochemistry of the chemotaxis pathway in this work, we do not explain the origin of the run-time bias ηp,w but rather consider it as a parameter. Note in this context that the bias of run times may, in principle, depend on other motion characteristics, such as the speed in the respective run state. This may occur, for example, if the underlying model for the run-time bias relies on a memory kernel (16, 20–24), but it is also observed in alternative models describing navigation of active particles in concentration gradients (25).
Keller-Segel–type transport in the long time limit
What is the effective long-time chemotactic response of a bacterium with a given motility pattern and a specific run-time bias? We answer this question using the active particle model introduced above, which, on the one hand, reproduces the short-time dynamics of the bacterial swimmers and, furthermore, allows us to perform numerical simulations and analytical calculations to derive quantities that characterize the long-time dynamics, such as the chemotactic drift velocity and the diffusion coefficient. In this way, we can reliably predict the long-time chemotactic response even though experimental observations of individual bacteria are restricted to short-time intervals. For this purpose, we generalized the coarse-graining approaches established in (18, 22, 23, 25) to a theoretical framework that enables us to derive long-term transport properties analytically for arbitrary underlying multimode motility patterns [see also (16, 20, 21) for different theoretical approaches]. By reducing the full dynamics to a Keller-Segel–type equation (26) for the particle density ρ via a mode expansion
| (2) |
we demonstrate that, on large spatial scales and to first order in gradients of the chemoattractant ∇c, the long-time dynamics of P. putida in an external chemical gradient is given by an upgradient drift (the actual chemotactic response), superimposed by diffusion; as we assumed that run-time distributions decay exponentially for large run times, anomalous diffusion as discussed in (27) is not expected in this model. The mean chemotactic drift velocity vd = μ∇c and the long-time diffusion coefficient D are functions of the underlying motility pattern, the motion characteristics of individual runs, and the run-time bias. On the basis of this derivation of the effective transport properties from a descriptive model of the motility—bridging the gap between short-time and long-time dynamics—it is possible to predict the chemotactic response even though experimental observations of individual bacteria are restricted to short-time intervals (cf. the Supplementary Materials for the details on the derivation).
DISCUSSION
The complete expressions for the transport coefficients μ and D are rather involved; they are explicitly given in the Supplementary Materials. Here, we will particularly discuss the chemotactic drift coefficient μ, with respect to efficiency and robustness of P. putida’s chemotactic performance. To first order in gradients of the chemoattractant ∇c, the chemotactic drift vd = μ∇c is determined by the response functions μp, w, which are defined via
| (3) |
for a given run-time bias ηp,w. Below, we address how these response functions μp,w depend on the motion characteristics of a bacterial swimmer. The duration of stop and turn events is neglected here, as their duration is at least one order of magnitude smaller than the average run time according to our experimental results.
Chemotaxis in uniform bulk fluid: Swimming speed versus sensing capacity
Our experiments revealed that out of the two accessible swimming modes, only the wrapped mode, which exhibits the slower swimming speed, is chemotactically responsive. How is P. putida’s chemotactic efficiency affected by this choice? An intuitive answer is obtained considering the expression for the drift coefficient in the limit of vanishing rotational diffusion, i.e., for straight runs in between stops or turns (Dp,w → 0)
| (4) |
where Pp is the probability to find a bacterium in the push mode (Pw = 1 − Pp) and d denotes the spatial dimension. Equivalently, this means that μp,w = Pp,wvp,w/(λp,wd) in this limit (see Eq. 3). The efficiency of the run modes is set by a combination of the run speed vi and the corresponding relative run-time bias ϵi = ηi/λi, which reflects the sensing capacity. Thus, the ratio of the products of speed and relative run-time bias (ϵpvp)/(ϵwvw) reveals which of the swim modes is more efficient. If out of two accessible run modes only one is chemotactic, then the chemotactic performance is optimized if the chemotactic mode also exhibits the larger speed vi. However, in the case of P. putida, the chemotactic wrapped mode is the slower one with vw/vp ≈ 0.5. We thus conclude that the chemotaxis strategy of P. putida is not optimized for chemotactic motion in uniform bulk liquid.
Mixed chemotaxis strategies: Efficiency and robustness
Our experiments have shown that for P. putida, there is no significant run-time bias in the push mode (ηp ≈ 0); only the wrapped mode is chemotactically active (ηw > 0) (see Fig. 3). Is it, nevertheless, beneficial to use a mixed chemotaxis strategy, where the bacterium occasionally moves in the push mode even though it does not sense the chemoattractant during those phases? Or would it be more efficient to remain in the wrapped mode all the time, interrupted only by turn events?
To answer this question, we take a closer look at how the response function μw depends on the probability pwp that a wrapped run is followed by a push run (to reduce the number of parameters, we set ppw = 1 − pwp). In the limit of pwp = 0, the bacterium performs runs in the wrapped mode, which are intermittently interrupted by turns similar to the run-and-tumble pattern of E. coli (2). In the opposite limit of pwp = 1, the bacterium performs runs in the push mode interrupted by stop events reminiscent of the navigation strategy of R. sphaeroides (3). Note that the chemotactic drift coefficient μ tends to zero in this limit as the bacterium swims in the push mode only, but it is not responding to the chemoattractant in this state (ηp ≈ 0). The full dependence of the response function μw on pwp between those limiting cases is shown in Fig. 5A. The dependence of μw on pwp is not monotonic: There may exist a maximum for intermediate values of pwp. Thus, the chemotactic drift may be optimized by a mixed chemotaxis strategy, where the chemotactic wrapped mode is interrupted by runs in the nonresponsive push mode. The existence of this maximum depends on the mean cosine of the turn angle ⟨cos ψww⟩ for events that interrupt runs in the chemotactic wrapped mode. If turn angles are small on average, then it is beneficial to switch to the push mode from time to time since the corresponding flagellar reconfigurations are accompanied by a reversal of the direction of motion (Fig. 2). In particular in cases where the bacterium is moving downgradient, such reversals may be beneficial to reorient in the upgradient direction, thereby maximizing the chemotactic drift. In short, mixed chemotaxis strategies may outperform single-mode motility if reorientations in the chemotactic wrapped mode are not efficient enough, even though push runs are episodes of blind flight, during which bacteria do not respond to the chemoattractant. Note that in the case of P. putida, the maximum in μw at intermediate pwp values does not occur over the entire range of experimentally observed ⟨cos ψww⟩ values, indicated by the solid lines in Fig. 5A. However, even though this effect may only play a secondary role for P. putida, it highlights an interesting feature of chemotaxis in multimode swimmers in general.
Fig. 5. Quantitative prediction and parameter study of the long-time chemotaxis response of P. putida.
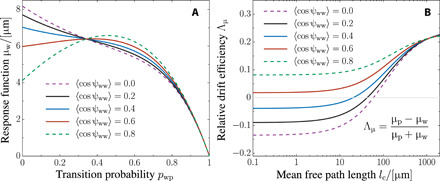
(A) Response function μw as a function of the transition probability pwp from the wrapped to the push mode (we set ppw = 1 − pwp). For pwp = 0, bacteria stay in the wrapped mode and perform run-and-turn motility. In the opposite limit (pwp = 1), they perform push runs that are occasionally interrupted by stops. The dependence of μw on pwp is not monotonic, and all curves intersect around pwp ≈ 0.35, reflecting a robust chemotactic response independent of the turn statistics ⟨cos ψww⟩. Parameters: Average turn angles ⟨cos ψpp⟩ = 0.9, ⟨cos ψpw⟩ = − 0.9, and ⟨cos ψwp⟩ = − 0.9; speeds vp = 25 μm/s and vw = 13 μm/s; average reorientation rates λp = 0.3 s−1 and λw = 0.4 s−1; rotational diffusion Dp = 0.03 s−1 and Dw = 0.13 s−1; and spatial dimension d = 3. (B) Chemotaxis in heterogeneous environments. The plot shows the relative difference Λμ of the response functions μp and μw as a function of the mean free path length in a disordered environment. If the obstacle density is low (large mean free path length), then the faster run mode is the beneficial one. However, as collisions become more frequent, i.e., if the mean free path is comparable to the average run times, both run modes are equally efficient. In crowded environments, where lc is smaller than the average run length, the wrapped mode may even become the more efficient one (Λμ < 0). Parameters: Mean turn angles for collision with obstacles ⟨cos ψc⟩ = − 0.3, ppw = 0.7, and pwp = 0.4; other parameters as in (A). On the basis of the data, we estimated 0.2 ≤ ⟨cos ψww⟩ ≤ 0.6 for P. putida, represented by solid lines in both panels.
Figure 5A reveals another interesting aspect regarding the robustness of the chemotactic response: All curves for different mean turn angles intersect for an intermediate value of pwp ≈ 0.35. The reorientation angles are highly fluctuating quantities, as they depend on the shape and structure of the bacterial cell body and the shape of the flagella and their dynamics during stop events. Bacteria can nevertheless achieve a robust chemotactic drift by a mixed chemotaxis strategy, as this drift becomes independent of the reorientation angle in a certain range of pwp values. This reveals a way of ensuring robustness of bacterial chemotaxis, unique to swimmers with multiple run modes. We believe that this type of robustness of the chemotactic performance is an intriguing feature of bacteria with multimode motility in general, beyond the specific case of P. putida, as it can ensure survival under changing environmental conditions, i.e., independent of specific parameter values of the motility pattern.
Chemotaxis in disordered environments
Our theoretical estimates have shown that a mixed strategy, where chemotactic motion is interrupted by runs in a nonresponsive swimming mode, may optimize the overall chemotactic performance of P. putida. However, it remains unclear why chemotactic sensing of P. putida relies on the wrapped mode, which exhibits a lower swimming speed than the nonresponsive push mode—a choice that is clearly not optimal for chemotaxis in uniform fluid environments. This question can only be addressed by taking into account that P. putida’s natural habitat is densely packed, disordered soil. We approximate this heterogeneous environment by a disordered array of obstacles of size σ at a density ρo. During run phases, the trajectories of bacteria are fairly straight because the characteristic run time, which is on the order of 1 s, is smaller than the persistence time. In this regime, the random arrangement of obstacles determines the mean free path lc = 1/(ρoσ) that bacteria traverse ballistically on average before colliding with an obstacle (28). In the following, we discuss how the chemotactic response in disordered environments depends on the mean free path or, equivalently, on the obstacle density. The collision rates in the push and wrapped modes, and , depend on the speed of the respective run mode. We include these collisions into the active particle model, assuming that bacteria turn by a random angle ψc upon a collision (see the Supplementary Materials for technical details), and find that the presence of obstacles, implying a finite mean free path, effectively renormalizes the rotational diffusion
| (5A) |
| (5B) |
In the limit of large mean free paths (lc → ∞), the presence of obstacles yields only a minor correction to the rotational noise. The relative increase in rotational diffusion is furthermore proportional to the speed in the respective run state and, thus, higher in the push mode as collisions are more likely to occur for large speeds in a given time interval. For high obstacle densities—if the mean free path is small—we find, in contrast, the following expressions for the response functions
| (6A) |
| (6B) |
to first order in lc. In this limit, the response functions are independent of speed and ultimately vanish for very dense environments (lc → 0). In this regime, the chemotactic drift is rather determined by purely geometric properties that characterize turn events—the expression in the numerator of Eq. 6A is the weighted average cosine of the reorientation angle given a wrapped run ends. Thus, not the speed but the ability to escape from traps determines the chemotactic efficiency in heterogeneous environments. This is illustrated in Fig. 5B, where the relative difference in the response functions Λμ = (μp − μw)/(μp + μw) is shown as a function of the mean free path. For the motility parameters of P. putida, we find that the wrapped mode may become the more efficient one (Λμ < 0) for performing chemotaxis in crowded environments if the mean free path length is of the same order (or smaller) than the mean run length. This can be understood by keeping in mind that wrap-wrap transitions allow for large turn angles, whereas stop events during push runs are small and, thus, do not enable effective navigation through the soil (cf. the turn angle distributions in Fig. 2).
CONCLUSIONS
We have performed a detailed analysis of the chemotaxis strategy of a bacterial swimmer that exhibits more than one run mode. As a representative multimode swimmer, we have chosen for our study the soil bacterium P. putida that exhibits run phases with pushing, pulling, and wrapped filament configurations. While the pull mode can be largely neglected, the two remaining dominant run modes showed distinctly different chemotactic responses: For runs in the wrapped mode (CW rotation of the flagellar motors), we observed a clear run-time bias, whereas runs in the push mode (CCW motor rotation) were not affected by the presence of a chemoattractant gradient. Here, we did not address the mechanistic origin of this asymmetric response at the level of the intracellular signaling pathway. For details on the chemosensory system and signaling pathway of P. putida, we refer the reader to (29) and references therein. This remains an open question and will be the subject of future studies. Instead, we developed an active particle model of multimode motility to analyze the impact of this asymmetry on the chemotactic motion of the bacterial swimmers in the long time limit. On the basis of our model, we could show that the presence of a nonresponsive run mode may still be beneficial to ensure an efficient and robust chemotactic performance of the swimmers. Moreover, our model suggests that the lower swimming speed during the chemoresponsive wrapped mode, as compared with the nonresponsive push mode, enhances the chemotactic efficiency in dense, crowded environments.
Our study can be seen as a first step to understand more complex chemotaxis strategies of bacterial swimmers beyond the well-known paradigm of E. coli. In particular, over the past decade, it became increasingly clear how different arrangements of the flagella across the cell body result in different motion patterns, such as, for example, the run-reverse-flick cycle of V. alginolyticus (14, 30). How these more complex multimode swimmers navigate in chemical gradients is an open question that we have started addressing in this work, choosing P. putida as a representative example (7). Along with the experimental data, the active particle model that we propose offers a systematic approach to explore different chemotaxis strategies beyond the specific case that we investigated experimentally, so that their performance can be critically assessed and compared. Our findings for the soil bacterium P. putida also demonstrate that a given movement strategy can be fully understood only if the bacterial swimmer is considered in the context of its natural habitat. We thus expect that future experimental studies will increasingly focus on motility and chemotaxis in complex environments, such as porous, granular materials or viscous filamentous matrices that mimic typical bacterial habitats, such as soil or tissue.
MATERIALS AND METHODS
Cell culturing
P. putida KT2440 FliCS267C (P. putida FliC) is a genetically modified strain from the wild-type P. putida KT2440 (31). The genetic modification causes a single cysteine substitution in the flagellar filament protein as described in (7). P. putida FliC was grown in 50 ml of tryptone broth [tryptone (10 g liter−1; AppliChem) and NaCl (5 g liter−1)] overnight, being shaken at 300 rpm and 30°C. The F-L staining of flagella with Alexa Fluor 488 followed the protocol of (7). Both the chemotaxis and the bulk experiments were performed in a μ-Slide Chemotaxis three-dimensional device (ibidi; see fig. S1A). The overnight cell culture that contained the highly motile bacteria was directly used for further experiments (cf. the Supplementary Materials for additional technical details).
Microscopy setup
All digital images were obtained using an IX71 inverted microscope (Olympus) equipped with a high-speed camera (ORCA-Flash4.0, Hamamatsu) and two light sources (cf. fig. S1B). For the combined imaging, a 470-nm light-emitting diode (LED) (Prizmatix, UHP-T-LED-460) was used to visualize the stained flagellar filaments in F-L imaging mode, and a white LED (Prizmatix, UHP-LED-White) was the light source for the P-C imaging part. Images were captured with a 20× UPLFLN-PH objective (Olympus) at a frame rate of 20 frames per second (fps). The microscope was controlled by μManger (http://micro-manager.org/). The high-speed F-L images were captured with a 100× UPLFLN-PH objective (Olympus) and the 470-nm LED at 100 to 400 fps.
Combined P-C/F-L imaging
In order to track the position of bacteria and visualize the state of the flagellar bundle simultaneously, alternating sequences of P-C and F-L images were generated. Accordingly, the combined image series was P-C1/F-L2/P-C3/.../F-LN for a sequence of N images. The P-C images were processed to track the cell body; afterward, F-L recordings were checked to determine the orientation of flagella by visual inspection. Since the flagellar bundle is stable during the whole run, the time delay of 0.05 s between sequential P-C/F-L images is negligible. To generate these image series, a devise controller, Triggerscope (Advanced Research Consulting), was used. The Arduino microcontroller board in Triggerscope established the communication among the computer, the light sources, and the camera. The trigger output signals in the ORCA-Flash4.0 camera provide an option by which the camera and the external devices get synchronized. The camera signals are sent out at exposure time during each frame acquisition. In particular, the camera trigger signals, TTLs (transistor-transistor logic), are sent out over a BNC cable to the Triggerscope. As a result, the Triggerscope controlled the light sources, which generated the combined image series. Upon each pulse, the LEDs are set to the correct states by the Triggerscope and therefore perform a flip/flop illumination, i.e., the two LEDs are switched on and off alternately (see fig. S1B for the devices communication scheme).
Event recognition algorithm
In order to detect events in the recorded trajectories, an automatized event recognition algorithm was used, as introduced by Masson et al. (32) and modified by Theves et al. (9). The idea is to identify large fluctuations in the direction of motion or sudden drops in speed [cf. (19)]. First, the trajectories were smoothed using a second-order Savitzky-Golay filter (33) with a window size of five data points corresponding to 500 ms. Afterward, the instantaneous speed v, direction of propagation φ, and angular velocity ω were computed from these smoothed tracks.
In order to identify turn events from the time series of the speed, all local minima of the speed are identified first. Let v(t1) and v(t2) be the adjacent maxima next to a minimum v(tm). The depth of a minimum is defined via Δv = max [v(t1) − v(tm), v(t2) − v(tm)]. We consider a speed drop as significant if Δv/v(tm) > 2; the event duration is determined by the condition v(t) ≤ v(tm) + 0.45Δv. In addition, further turn events may be detected from the time series of the absolute value of the angular speed. Let a maximum exist at tM with two adjacent minima at times t1,2. If the total angular reorientation is larger than with and Dr = 0.1 s−1, then we consider it a turn event whose duration is calculated according to ∣ω(tM) − ω(t)∣ ≤ 0.85Δω, where Δω = max [ω(tM) − ω(t1), ω(tM) − ω(t2)]. If an instant of time in the time series meets one of the two criteria above, then we consider it an event. The event detection was verified by visual inspection, taking advantage, in particular, of the F-L information on the configuration of the flagellar bundle with respect to the cell body. In this way, the threshold and event duration parameters given above were adjusted.
Supplementary Material
Acknowledgments
We thank A. Blanco for technical help concerning Triggerscope. We are grateful to C. Elster for valuable insights on data analysis and uncertainty quantification for censored data. Funding: Z.A., V.P., M.H., and C.B. acknowledge funding by Deutsche Forschungsgemeinschaft (DFG) in the framework of research training group GRK1558. C.B. acknowledges further financial support by Deutsche Forschungsgemeinschaft (DFG) via SFB1294. Author contributions: C.B. designed the research; Z.A., V.P., and M.H. performed the experiments; Z.A. and R.G. analyzed the data; Z.A. contributed new analytic tools; R.G. developed theory and performed modeling; and Z.A., R.G., and C.B. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz6153/DC1
REFERENCES AND NOTES
- 1.H. C. Berg, E. coli in Motion (Springer, 2004). [Google Scholar]
- 2.Berg H. C., Brown D. A., Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239, 500–504 (1972). [DOI] [PubMed] [Google Scholar]
- 3.Armitage J. P., Macnab R. M., Unidirectional, intermittent rotation of the flagellum of Rhodobacter sphaeroides. J. Bacteriol. 169, 514–518 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansen J. E., Pinhassi J., Blackburn N., Zweifel U. L., Hagström Å., Variability in motility characteristics among marine bacteria. Aquat. Microb. Ecol. 28, 229–237 (2002). [Google Scholar]
- 5.Mukherjee T., Elmas M., Vo L., Alexiades V., Hong T., Alexandre G., Multiple CheY homologs control swimming reversals and transient pauses in Azospirillum brasilense. Biophys. J. 116, 1527–1537 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raina J.-B., Fernandez V., Lambert B., Stocker R., Seymour J. R., The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hintsche M., Waljor V., Großmann R., Kühn M. J., Thormann K. M., Peruani F., Beta C., A polar bundle of flagella can drive bacterial swimming by pushing, pulling, or coiling around the cell body. Sci. Rep. 7, 16771 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy K. J., Ford R. M., Turn angle and run time distributions characterize swimming behavior for Pseudomonas putida. J. Bacteriol. 179, 1428–1430 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theves M., Taktikos J., Zaburdaev V., Stark H., Beta C., A bacterial swimmer with two alternating speeds of propagation. Biophys. J. 105, 1915–1924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kühn M. J., Schmidt F. K., Eckhardt B., Thormann K. M., Bacteria exploit a polymorphic instability of the flagellar filament to escape from traps. Proc. Natl. Acad. Sci. U.S.A. 114, 6340–6345 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Constantino M. A., Jabbarzadeh M., Fu H. C., Shen Z., Fox J. G., Haesebrouck F., Linden S. K., Bansil R., Bipolar lophotrichous Helicobacter suis combine extended and wrapped flagella bundles to exhibit multiple modes of motility. Sci. Rep. 8, 14415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinosita Y., Kikuchi Y., Mikami N., Nakane D., Nishizaka T., Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME J. 12, 838–848 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner L., Ping L., Neubauer M., Berg H. C., Visualizing flagella while tracking bacteria. Biophys. J. 111, 630–639 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie L., Altindal T., Chattopadhyay S., Wu X.-L., Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 108, 2246–2251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vardi Y., Nonparametric estimation in renewal processes. Ann. Statist. 10, 772–785 (1982). [Google Scholar]
- 16.Altindal T., Xie L., Wu X.-L., Implications of three-step swimming patterns in bacterial chemotaxis. Biophys. J. 100, 32–41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie L., Lu C., Wu X.-L., Marine bacterial chemoresponse to a stepwise chemoattractant stimulus. Biophys. J. 108, 766–774 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnitzer M. J., Theory of continuum random walks and application to chemotaxis. Phys. Rev. E 48, 2553–2568 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Pohl O., Hintsche M., Alirezaeizanjani Z., Seyrich M., Beta C., Stark H., Inferring the chemotactic strategy of P. putida and E. coli using modified Kramers-Moyal coefficients. PLOS Comput. Biol. 13, e1005329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Gennes P.-G., Chemotaxis: The role of internal delays. Eur. Biophys. J. 33, 691–693 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Locsei J. T., Persistence of direction increases the drift velocity of run and tumble chemotaxis. J. Math. Biol. 55, 41–60 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Celani A., Vergassola M., Bacterial strategies for chemotaxis response. Proc. Natl. Acad. Sci. U.S.A. 107, 1391–1396 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taktikos J., Stark H., Zaburdaev V., Correction: How the motility pattern of bacteria affects their dispersal and chemotaxis. PLOS ONE 8, e81936 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seyrich M., Palugniok A., Stark H., Traveling concentration pulses of bacteria in a generalized Keller–Segel model. New J. Phys. 21, 103001 (2019). [Google Scholar]
- 25.Nava L. G., Großmann R., Peruani F., Markovian robots: Minimal navigation strategies for active particles. Phys. Rev. E 97, 042604 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Keller E. F., Segel L. A., Model for chemotaxis. J. Theor. Biol. 30, 225–234 (1971). [DOI] [PubMed] [Google Scholar]
- 27.Thiel F., Schimansky-Geier L., Sokolov I. M., Anomalous diffusion in run-and-tumble motion. Phys. Rev. E 86, 021117 (2012). [DOI] [PubMed] [Google Scholar]
- 28.E. M. Lifshitz, L. P. Pitaevskii, Physical Kinetics, vol. 10 of Course of Theoretical Physics (Pergamon Press, 1981). [Google Scholar]
- 29.López-Farfán D., Reyes-Darias J. A., Matilla-Vazquez M. A., Krell T., Concentration dependent effect of plant root exudates on the chemosensory systems of Pseudomonas putida KT2440. Front. Microbiol. 10, 78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Son K., Guasto J. S., Stocker R., Bacteria can exploit a flagellar buckling instability to change direction. Nat. Phys. 9, 494–498 (2013). [Google Scholar]
- 31.Belda E., van Heck R. G. A., José Lopez-Sanchez M., Cruveiller S., Barbe V., Fraser C., Klenk H.-P., Petersen J., Morgat A., Nikel P. I., Vallenet D., Rouy Z., Sekowska A., Martins dos Santos V. A. P., de Lorenzo V., Danchin A., Médigue C., The revisited genome of Pseudomonas putida KT2440 enlightens its value as a robust metabolicchassis. Environ. Microbiol. 18, 3403–3424 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Masson J.-B., Voisinne G., Wong-Ng J., Celani A., Vergassola M., Noninvasive inference of the molecular chemotactic response using bacterial trajectories. Proc. Natl. Acad. Sci. U.S.A. 109, 1802–1807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Savitzky A., Golay M. J. E., Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964). [Google Scholar]
- 34.B. Efron, R. J. Tibshirani, An introduction to the bootstrap (Chapman and Hall/CRC, 1994). [Google Scholar]
- 35.Zengel P., Nguyen-Hoang A., Schildhammer C., Zantl R., Kahl V., Horn E., μ-Slide Chemotaxis: A new chamber for long-term chemotaxis studies. BMC Cell Biol. 12, 21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapur J. N., Sahoo P. K., Wong A. K. C., A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. 29, 273–285 (1985). [Google Scholar]
- 37.Crocker J. C., Grier D. G., Methods of digital video microscopy for colloidal studies. J. Coll. Interfat. Sci. 179, 298–310 (1996). [Google Scholar]
- 38.Großmann R., Peruani F., Bär M., A geometric approach to self-propelled motion in isotropic & anisotropic environments. Eur. Phys. J. Spec. Top. 224, 1377–1394 (2015). [Google Scholar]
- 39.Godrèche C., Luck J. M., Statistics of the occupation time of renewal processes. J. Stat. Phys. 104, 489–524 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/22/eaaz6153/DC1


