Abstract
Genome-derived noncoding RNAs (ncRNAs), including microRNAs (miRNAs), small interfering RNAs (siRNAs), and long noncoding RNAs (lncRNAs), play an essential role in the control of target gene expression underlying various cellular processes, and dysregulation of ncRNAs is involved in the pathogenesis and progression of various diseases in virtually all species including humans. Understanding ncRNA biology has opened new avenues to develop novel RNA-based therapeutics. Presently, ncRNA research and drug development is dominated by the use of ncRNA mimics that are synthesized chemically in vitro and supplemented with extensive and various types of artificial modifications and thus may not necessarily recapitulate the properties of natural RNAs generated and folded in living cells in vivo. Therefore, there are growing interests in developing novel technologies for in vivo production of RNA molecules. The two most recent major breakthroughs in achieving an efficient, large-scale, and cost-effective fermentation production of recombinant or bioengineered RNAs (e.g., tens of milligrams from 1 L of bacterial culture) are (1) using stable RNA carriers and (2) direct overexpression in RNase III-deficient bacteria, while other approaches offer a low yield (e.g., nano- to microgram scales per liter). In this article, we highlight these novel microbial fermentation-based technologies that have shifted the paradigm to the production of true biological ncRNA molecules for research and development.
Keywords: Noncoding RNA, MicroRNA, siRNA, Bioengineering, Biotechnology
Introduction
Noncoding DNA sequences account for approximately 98.5% of the genome in humans, while they are less than 50% in prokaryotes and simple eukaryotes, and over 50% among plants and animals (Mattick 2004). Indeed, around 75% of the whole human genome is clearly transcribed into a diverse array of RNA molecules at variable levels and different localizations during development (Djebali et al. 2012). Besides the well-known protein-coding mRNAs that are translated into functional proteins, a large percentage of the transcripts has been annotated as noncoding RNAs (ncRNAs), including long noncoding RNAs (lncRNAs), ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), microRNAs (miRNAs or miRs), small interfering RNAs (siRNAs), transfer RNAs (tRNAs), enhancer RNAs (eRNAs), and other forms of short or small RNAs (sRNAs) which may be directly involved in the pathogenesis and progression of various diseases among virtually all species including humans (Ambros 2004; Cech and Steitz 2014; Dahlberg 1989; Dupuis-Sandoval et al. 2015; Kopp and Mendell 2018; Rak et al. 2018). The recognition of the importance of functional ncRNAs in physiology and diseases, especially the discovery of miRNA- and siRNA-mediated posttranscriptional gene regulation and RNA interference (RNAi) mechanisms, has changed the traditional definition of a gene and revolutionized life sciences and biomedical research. This has also propelled the development of novel RNA-based therapeutics for the treatment of human diseases (Burnett and Rossi 2012; Yu et al. 2019).
While ncRNAs are important for basic research and drug development, nonviral or viral-based ncRNA expression systems have been commonly used for related in vitro and in vivo studies (Liu and Berkhout 2011). Since these expression systems are DNA-based, the actual doses or expression levels of target ncRNA products are widely variable and even unknown or undetermined at all. Furthermore, expression of ncRNA products does not necessarily correlate with the amount of DNA plasmid administered since the generation of target ncRNAs is dependent upon the transfection and infection efficiencies, as well as the RNA biogenesis machineries within the hosts. Meanwhile, chemically engineered ncRNA “mimics” made in vitro (Bramsen and Kjems 2012; Khvorova and Watts 2017; Lundin et al. 2013) are frequently used at quantifiable doses or concentrations. These chemo-engineered ncRNA mimics actually lack important posttranscriptional modifications occurring in natural RNAs produced in cells (Ho and Yu 2016; Yu et al. 2019). Rather, they have an array of artificial modifications at different locations that are expected to increase chemical diversity and metabolic stability (Yu et al. 2019). Therefore, chemo-engineered ncRNA mimics are basically different RNA molecules with their own structures, physicochemical properties, and biological activities and thus may not represent the actions of natural ncRNA molecules transcribed from the genome and folded in living cells. In addition, although chemical synthesis of RNAs has been automated, chemo-engineered ncRNA mimics remain costly, especially with increased length and the requirement for precise purification to reagent, pharmaceutical or analytical grades of purity. The tradition of using chemo-engineered RNA mimics for biological research is also in sharp contrast to standard highly successful approaches for protein research and drug development (Burley et al. 2019; Usmani et al. 2017) which employ bioengineered or recombinant proteins produced and folded in living cells instead of polypeptides or proteins synthesized chemically in vitro. Moreover, even small-molecule biochemical compounds with stereostructures, while being readily prepared by stereosynthesis, may benefit from fermentation-based in vivo production (Kim and Park 2019; Min et al. 2015; Willke 2014).
Besides in vitro enzymatic production, great efforts have been made to develop new biotechnologies for the in vivo manufacture of true biological ncRNA molecules in living cells (see recent reviews (Ho and Yu 2016; Yu et al. 2019)). The use of stable RNA scaffolds or carriers (e.g., hybrid tRNA/pre-miRNA) to assemble various types of target RNAs (e.g., aptamers, miRNAs, siRNAs, sRNAs) has proven to be the most efficient approach to achieve a large-scale production of novel ncRNA molecules (e.g., tens of milligrams of pure ncRNAs from 1 L of bacterial culture) for research and development (Chen et al. 2015; Ho et al. 2018; Petrek et al. 2019). Direct heterogenous expression in particular bacterial strains remains a challenge, yielding only nano- to micrograms of target RNAs per liter of fermentation (Kikuchi and Umekage 2018), until the very recent demonstration of efficient overexpression of two specific RNAs in RNase III-deficient bacteria (Hashiro et al. 2019a; Hashiro et al. 2019b). Therefore, following a brief introduction of in vitro enzymatic production approaches, we focus on the review and discussion of novel fermentation-based in vivo production of ncRNAs, including the use of particular RNAs as carriers and unique bacterial strains to avoid degradation and achieve efficient overexpression and accumulation. These recombinant or bioengineered ncRNA molecules should better represent natural ncRNAs for research and development as both are produced and folded in living cells.
In vitro enzymatic reactions
In vitro transcription is a reliable enzymatic approach to produce target RNA molecules (Milligan et al. 1987), including RNAi agents (Beckert and Masquida 2011). This approach relies on the utilization of a DNA template (e.g., linearized plasmid, PCR product, or synthetic oligonucleotides) to specifically biosynthesize target RNA molecules under a particular promoter in vitro using the corresponding RNA polymerase. The greatest advantage of in vitro transcription is the ease and versatility of this method in producing a wide variety of RNA molecules covering both mRNAs and ncRNAs, and there are now many commercial kits available. Nevertheless, the challenge lies in the reliability of T7 RNA polymerase that may relapse as the length of transcript increases, usually resulting in the addition of untemplated nucleotides to the 3′ end and/or heterogeneity at 3′ or 5′ ends (Gardner et al. 1997; Pleiss et al. 1998). Previous studies have also revealed a significant level of template-dependent transcriptional infidelity by using the enzyme T7 RNA polymerase that is commonly used for in vitro transcription (Wons et al. 2015). Furthermore, the RNAs made by in vitro transcription lack the necessary posttranscriptional modifications due to the absence of posttranscriptional modification machineries. In addition, it remains costly to produce larger quantities (e.g., milligrams) of target RNA molecules using this method.
In vitro transcription has been successfully used alone or in combination with other enzymatic reactions for the production of various types of RNAi agents. On the one hand, complementary single-stranded RNAs (ssRNAs) can be made by in vitro transcription and then annealed to offer target double-stranded (dsRNAs) that can be directly utilized for RNAi studies (Wianny and Zernicka-Goetz 2000; Yang et al. 2002). On the other hand, recombinant RNase Dicer can be employed to process the in vitro transcribed dsRNAs into target siRNAs in vitro, which are biologically active in knocking down target genes in mammalian cells (Myers et al. 2003). The combination of in vitro transcription with Dicer-mediated cleavage has also been established as a single-pot enzymatic reaction for the production of functional siRNA agents (Guiley et al. 2012). Actually, chemically synthesized RNAs can be directly subjected to enzymatic reactions, e.g., cleavage by recombinant bacterial RNase III (Yang et al. 2002), for the production of target siRNAs. While enzymatic reactions were proven to be a robust means to produce functional RNAi molecules in these studies, the yield and purity of RNA products were surprisingly not reported or demonstrated.
Stable RNA carriers for fermentation production in vivo
Transfer RNA scaffold
The tRNA has been proven to be a simple and useful scaffold (Fig. 1a) for the production of recombinant RNA molecules in bacteria in vivo (Nelissen et al. 2012; Ponchon et al. 2009; Ponchon and Dardel 2007; Ponchon and Dardel 2011). This discovery shifted the paradigm of RNA production to in vivo fermentation-based (Table 1). This method is grounded on the rationale that tRNA scaffold would be recognized by cellular machinery as an endogenous molecule, enabling overexpression and accumulation, and further supported by the evidence that the bacterial tRNAMet and the tRNA-like domains of chimeric transfer-messenger RNA (tmRNA) with tRNA and mRNA properties may be recombinantly expressed in bacteria and accumulated to approximately 5% of total bacterial tRNAs (Gaudin et al. 2003; Meinnel et al. 1988). The tRNA scaffold is specifically exploited to produce recombinant RNAs by replacing the anticodon with RNAs of interest (Fig. 1a), while the other segments are retained to maintain the cloverleaf structure and overall stability. Production of recombinant RNA is driven by the Escherichia coli murein lipoprotein (lpp) promoter (Ponchon et al. 2009; Ponchon and Dardel 2007) while the T7 promoter can be utilized as well (Nelissen et al. 2012) (Table 1). When successfully overexpressed, the recombinant RNA may be isolated by affinity, filtration, or ion exchange chromatography methods to offer up to tens of milligrams of pure RNAs. In addition, the target RNA species could be released on demand by corresponding RNases or ribozyme for further studies. As the tRNA scaffold has been used for the successful production of various forms of target RNAs, e.g., viral RNAs and aptamers (Ponchon et al. 2009; Ponchon and Dardel 2007), hammerhead riboswitch RNAs (Nelissen et al. 2012), pre-miRNAs (Li et al. 2015; Li et al. 2014; Wang et al. 2015), and snoRNAs (Peng et al. 2014), the efficiency in heterogenous expression truly depends upon the structure of chimeric RNAs and resistance to bacterial RNase (Chen et al. 2015; Ho et al. 2018; Li et al. 2014).
Fig. 1.
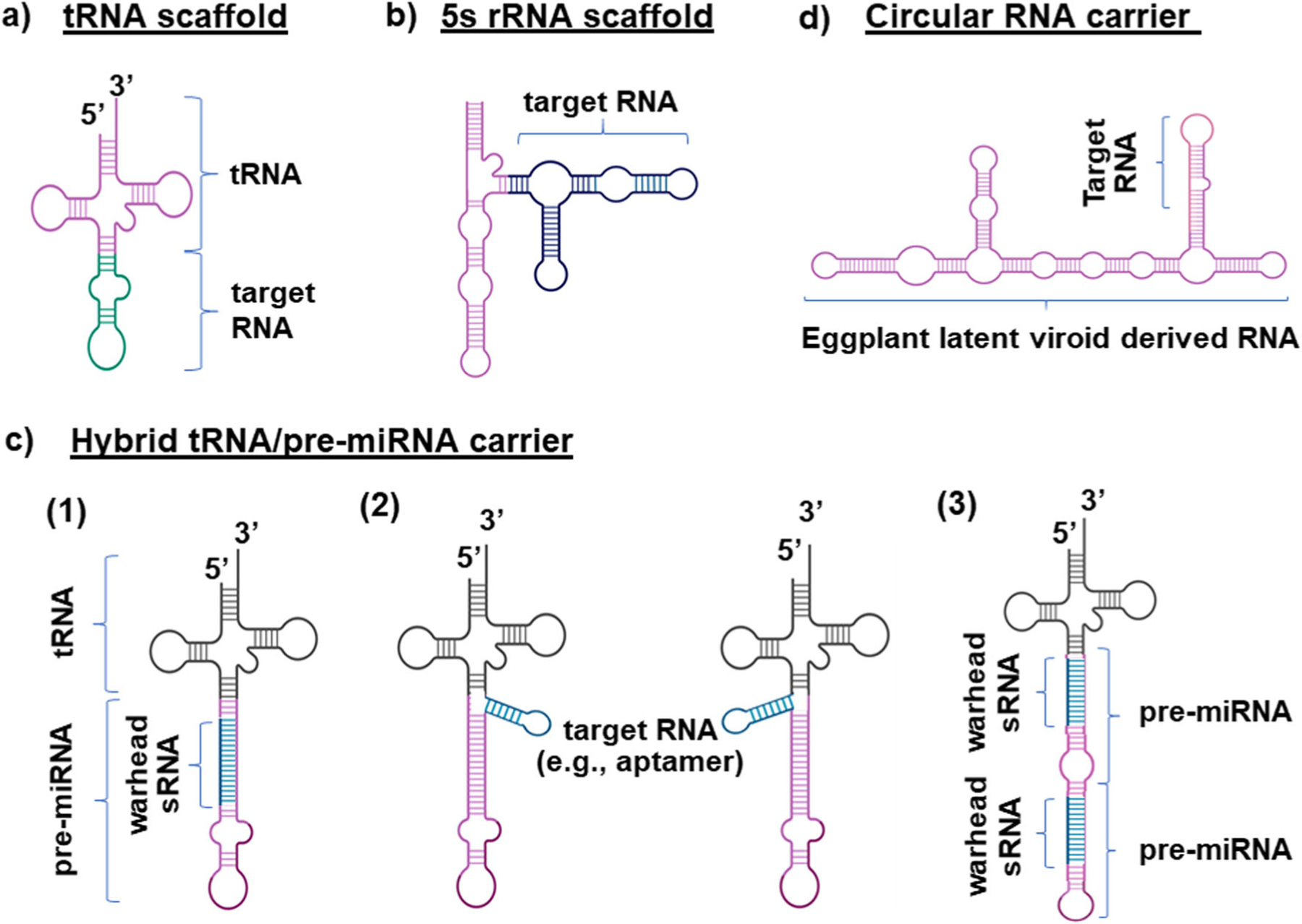
Various carriers have been used to achieve efficient production of recombinant or biologic RNA molecules through cost-effective microbial fermentation. a The tRNA scaffold where the anticodon is substituted by a target RNA. b The 5S rRNA scaffold allows the insertion or substitution of desired RNAs into stems II and III. c Stable hybrid tRNA/pre-miRNA molecules have been identified as novel carriers to accommodate various types of RNAs including (1) substitution of the authentic miRNA with target miRNA, siRNA, or sRNA, (2) insertion of target RNA aptamers or sRNAs at the 5′ or 3′ of pre-miRNA, and (3) addition of another pre-miRNA (or shRNA) to adopt multiple warhead sRNAs. d A stable circular RNA carrying target RNA aptamer has been created by expressing the eggplant latent viroid (ELVd)-derived RNAs in E. coli, along with the eggplant tRNA ligase to complete circularization
Table 1.
Overview of novel approaches having been established for microbial production of RNA molecules
| Approach | Target RNA | Bacterial strain | Vector | Promoter | Purification method | Yield (mg/L culture) | References |
|---|---|---|---|---|---|---|---|
| tRNA scaffold | Various | E. coli JMlOlTr E. coli BL21 (DE3) | pBSTNAV derivative or pET23b | lpp or T7 | PAGE, affinity or ion exchange FPLC | Tens | Nelissen et al. 2012; Ponchon et al. 2009; Ponchon and Dardel 2007 |
| rRNA scaffold | Aptamers, sRNAs | E. coli JM109 (DE3) | pCV and pCR derivatives | rrnB P1/P2 | Gel electrophoresis | 2–8 mg/g cells | Liu et al. 2010 |
| tRNA/pre-miRNA carrier | Various | E. coli HST08 and DH5a | pBSTNAV | lpp | Spin column, FPLC methods, etc. | Tens | Chen et al. 2015; Ho et al. 2018; Li et al. 2015; Petrek et al. 2019 |
| ELVd RNA carrier | Aptamer, sRNA | E. coli BL21 or derivative | P15tRnlSm andpLELVd-BZB | lpp | PAGE, FPLC | ~150 (estimated; total ELVd RNAs) | Daros et al. 2018 |
| siRNA binding protein (pl9) | siRNA | E. coli T7 Express Iq (BL21derivative) | pRSF-GST-p19-His and pCDF-GST-p19-His | tac (p19) and T7 (siRNA) | Affinity chromatograph and anion exchange HPLC | ~0.04 | Huang et al. 2013 |
| Using RNase III-deficient C. glutamicum | U1A*-RNA diapl *-dsRNA | C. glutamicum 2256LΔrnc | pVH2 | F1 | (Not reported or demonstrated) | Up to 75–300 | Hashiro et al. 2019a; Hashiro et al. 2019b |
Ribosomal RNA scaffold
The rRNAs are the most abundant RNA species in cells, among which the 5S rRNA (Fig. 1b) has been established as a reliable carrier to achieve microbial fermentation production of target RNAs, including aptamers and sRNAs (Table 1). Initially, the 5S rRNA was tagged with various small RNAs as “identifiers” for the monitoring of microorganisms (D’Souza et al. 2003; Pitulle et al. 1995), which were indeed stably expressed. As such, the 5S rRNA has been employed to accommodate target RNAs within its stem II and stem III sites (Fig. 1b) (Zhang et al. 2009), while other sites may yet be explored. Specifically, a 71 nt artificial 3xpen RNA was inserted into stem II while the stem III loop B and C regions were completely replaced. Expression of chimeric RNAs is driven by two rRNA gene promoters, rrnB P1 and P2, followed by rrnB T1 and T2 transcription terminators (Table 1). Chimeric RNAs (160 nt) overexpressed in bacteria may be purified by practical preparative polyacrylamide gel electrophoresis (Zhang et al. 2009). Furthermore, with the addition of DNAzyme-specific sequences, a selective release of the target RNA from the purified chimeric RNA has been demonstrated (Liu et al. 2010). This approach has been shown to offer as much as 2.5–7.5 mg of purified RNA from 1 g of bacteria (Table 1). Although the rRNA carrier is relatively less explored, its potential for the efficient production of recombinant RNAs should not be underestimated.
Hybrid tRNA/pre-miRNA carrier
The hybrid tRNA/pre-miRNA molecules have recently been identified and established as novel ncRNA carriers to achieve a consistent high-yield large-scale production of bioengineered or biologic RNA agents (BERAs) through a cost-effective microbial culture (Chen et al. 2015; Ho et al. 2018). With direct heterogenous expression in bacteria, human pre-miRNAs did not accumulate to an adequate level for purification, and even when using the tRNA scaffold, there were huge variations in RNA expression levels with a low overall success rate of high-level accumulation of recombinant RNAs (e.g., 10% of total bacterial RNA) (Chen et al. 2015; Li et al. 2015; Li et al. 2014; Wang et al. 2015). Therefore, a hybrid tRNA/pre-miRNA molecule showing overexpression in bacteria was reasoned to serve as a ncRNA carrier to assemble target RNAs (Fig. 1c). Indeed, the hybrid tRNAMet/pre-miR-34a stably being overexpressed and accumulated up to 15% of total bacterial RNA (Chen et al. 2015) was proven to be a versatile carrier for the production of various types of biologic RNAi molecules of interest, albeit offering an overall success rate of less than 30% (Ho et al. 2018). Optimization of human pre-miR-34a sequence revealed that the pre-miR-34a G138U/139ΔG derivative fused to the tRNAMet was superior to the first-generation wild-type tRNAMet/pre-miR-34a carriers by offering much higher levels of expression (e.g., >30% of bacterial RNA) and success rate (e.g., 80%) for the production of target BERAs (Ho et al. 2018) (Table 1).
A wide variety of target RNAs may be accommodated to the hybrid tRNA/pre-miRNA carrier in different ways (Fig. 1c). First, the miRNA (e.g., miR-34a-5p) duplexes within the tRNA/pre-miRNA (e.g., tRNAMet/pre-miR-34a) carrier can be directly replaced by target miRNA, siRNA, or sRNA, along with its complementary sequence. Second, a target RNA aptamer or sRNA may be directly inserted at the 5′ or 3′ of pre-miRNA. Third, the tRNA/pre-miRNA carrier can accommodate another pre-miRNA (or shRNA) to carry multiple warhead sRNAs. Following the design, the BERA coding sequence is cloned into a derivative of pBSTNAV vector with a lpp promoter (Fig. 2). After transformation and fermentation, expression of target BERA is readily verified by RNA gel electrophoresis which indicates a remarkably high-level accumulation in bacteria (>30% of total bacterial RNAs) (Ho et al. 2018; Jilek et al. 2019; Li et al. 2018). Spin column and fast protein liquid chromatography (FPLC)-based methods have been respectively established for small- and large-scale purification of target BERA from bacterial total RNA (Ho et al. 2018). The purity of the final BERA product can be quantitatively determined by HPLC (Ho et al. 2018; Wang et al. 2015), and the endotoxin level was examined with commercially available kits. Most BERAs purified by a general anion exchange FPLC method in a single run exhibited a high degree of homogeneity (e.g., >97%) and low endotoxin activity (e.g., <1 EU/μg RNA) while some might require re-purification (Ho et al. 2018; Petrek et al. 2019) as the FPLC method was not optimized for each BERA. This platform allows consistent and efficient production of tens of milligrams of target BERAs from 1 L of bacterial culture (Table 1).
Fig. 2.
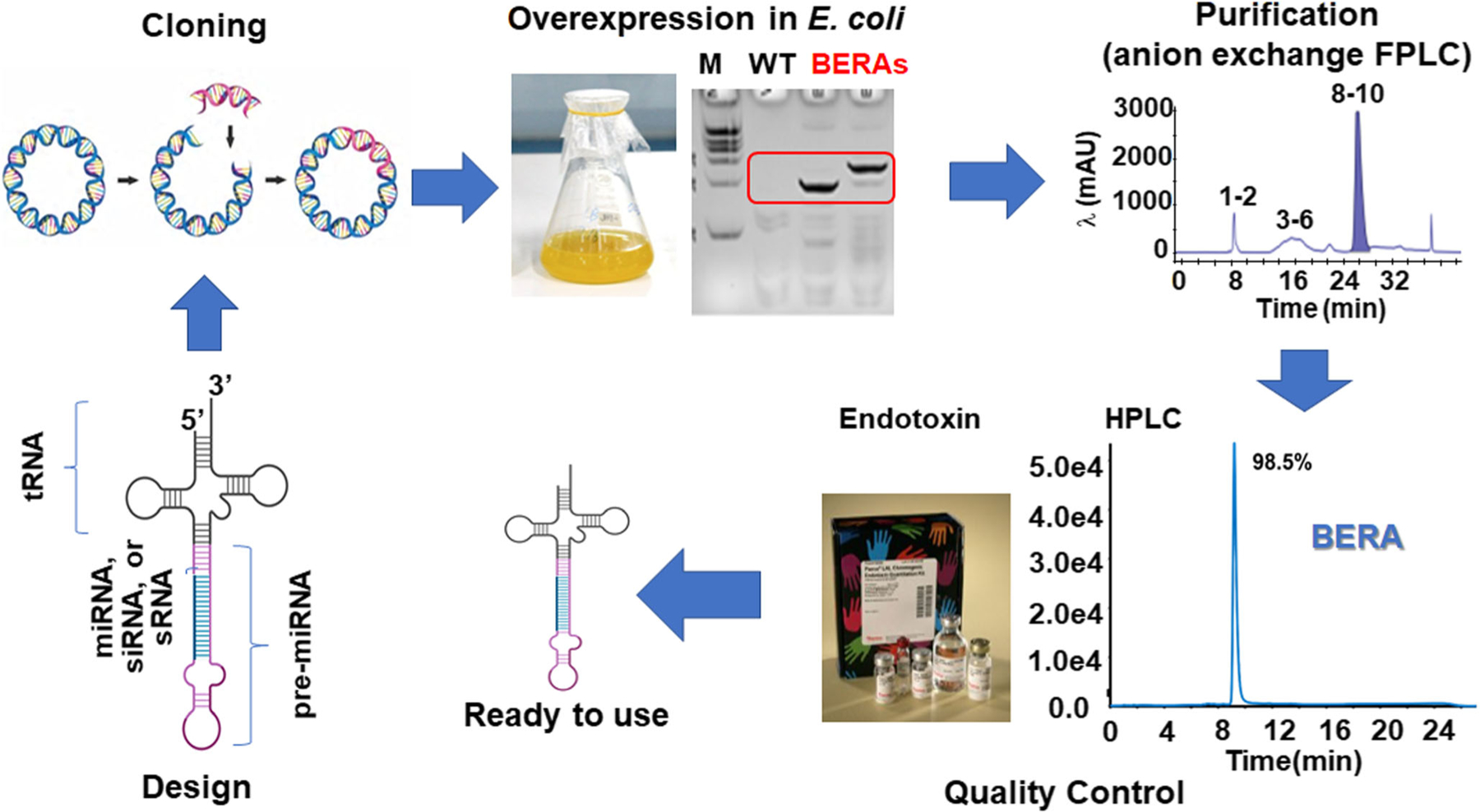
Streamlined workflow for fermentation production of biologic RNAi agent or BERA using the tRNA/pre-miRNA carrier. After a BERA is designed to accommodate the miRNA/siRNA/sRNA of interest, the coding sequence is cloned into a vector. Overexpression of the target BERA is easily verified by gel electrophoresis of total bacterial RNA. After the BERA is purified (e.g., with anion exchange FPLC method), its purity and endotoxin level are verified by HPLC method and commercially available endotoxin assay kits, respectively. These BERAs should better recapitulate the properties of natural RNAs as both are produced and folded in living cells. Therefore, BERAs are a novel class of biologic RNA molecules for research and development
BERAs carry no or minimal posttranscriptional modifications, are biologically active in regulating target gene expression and cellular processes, and are well tolerated in animal models (Alegre et al. 2018; Chen et al. 2015; Ho et al. 2018; Jian et al. 2017; Jilek et al. 2017; Jilek et al. 2019; Li et al. 2015; Li et al. 2018; Tu et al. 2019; Umeh-Garcia et al. 2019; Xu et al. 2019; Zhang et al. 2018; Zhao et al. 2016; Zhao et al. 2015), resembling cellular RNAi mechanisms (Fig. 3). While endogenous miRNA biogenesis starts with the production of a pri-miRNA transcript within the nucleus, followed by the generation of pre-miRNA by Drosha and transport into the cytoplasm by Exportin-5, recombinant BERAs likely overlap with pre-miRNA during processing and action in the cytoplasm (Fig. 3) after crossing the cellular barrier through the endocytosis pathway and endosome release (Dominska and Dykxhoorn 2010; Johannes and Lucchino 2018). In contrast, chemo-engineered miRNA mimics or siRNAs presumably enter into the processes in later steps (Fig. 3), following endosome release and cytosolic translocation (Johannes and Lucchino 2018). These different agents converge at the posttranscriptional gene expression through translation inhibition or mRNA degradation mechanisms (Fig. 3). In addition, we cannot exclude the possibility that exogenously introduced BERAs, siRNAs and miRNA mimics, could be processed into the multivesicular body and incorporated into exosomes or extracellular vesicles for intercellular signaling and communications (Abels and Breakefield 2016; Hessvik and Llorente 2018).
Fig. 3.
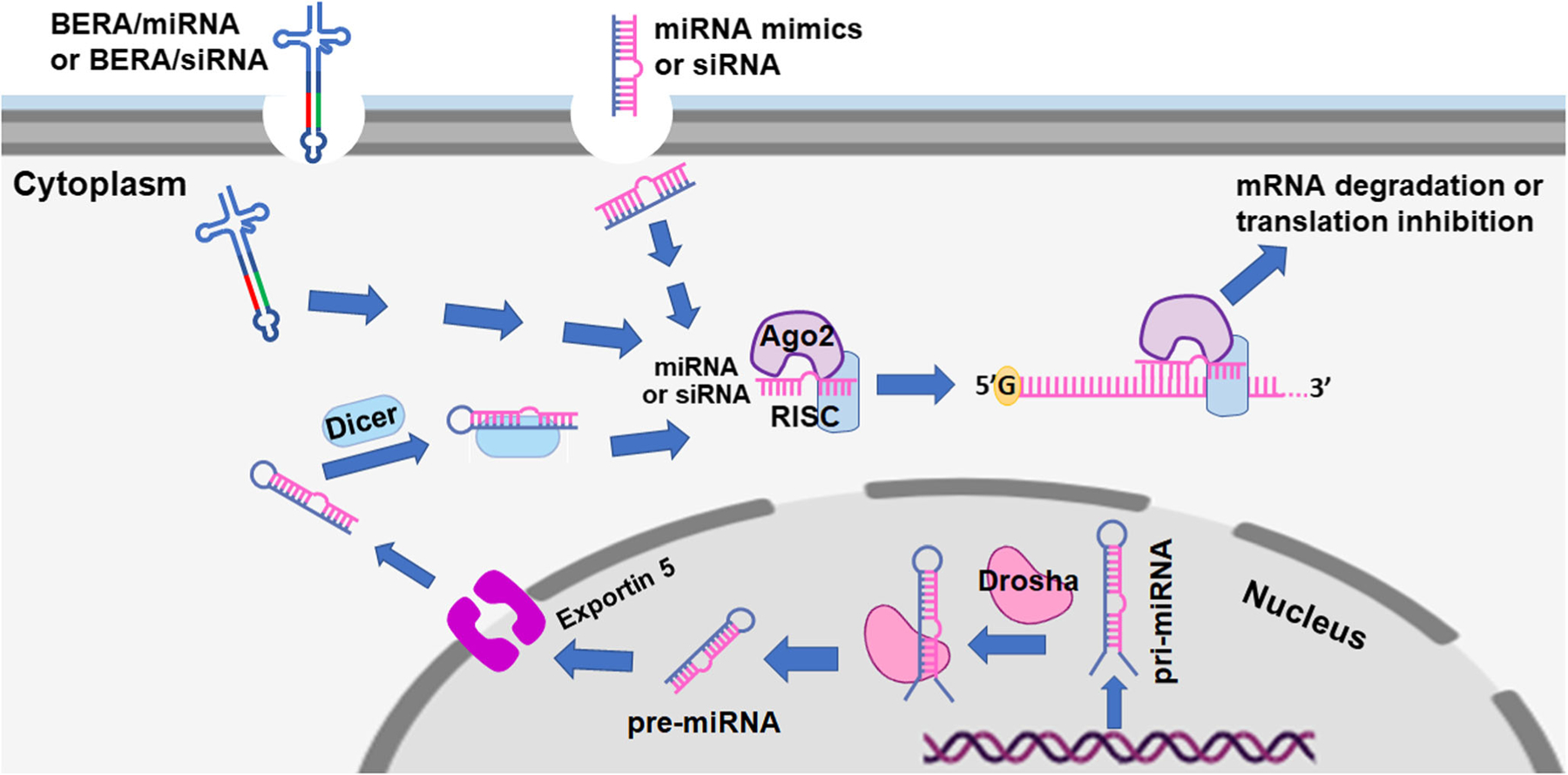
Overview of the miRNA biogenesis pathway and the mechanistic actions of BERA/miRNAs and BERA/siRNAs as well as chemo-engineered miRNA mimics and siRNAs. The biogenesis of miRNA starts with the formation of a long primary miRNA (pri-miRNA) transcript from the genome within the nucleus. The microprocessor complex, comprised of Drosha, cleaves the pri-miRNA to offer the precursor miRNA (pre-miRNA). The pre-miRNA is exported to the cytoplasm in an Exportin-5/RanGTP-dependent manner and thus is processed to miRNA duplex in a Dicer-dependent or independent manner. Finally, either the 5p or 3p dominant strand mature miRNA is loaded onto the Argonaute (AGO) family of proteins to form a miRNA- or siRNA-induced silencing complex (RISC). This leads to a functional RISC complex where it binds to corresponding target transcripts to induce either translational inhibition or mRNA degradation. After getting into human cells through the endocytosis pathway and undergoing endosome release and cytosolic translocation, BERA/miRNA or BERA/siRNA is specifically processed by the intrinsic miRNA machinery to target miRNA or siRNA molecule in a Dicer-dependent or independent manner, while chemo-engineered miRNA mimic or siRNA may be involved in later steps, to exert posttranscriptional gene expression
It is noteworthy that RNA sequencing studies have demonstrated a selective release of target miRNA (and siRNA) molecules from recombinant BERA/miRNAs (or BERA/siRNAs) in human cells (Chen et al. 2015; Ho et al. 2018; Umeh-Garcia et al. 2019), which, rather surprisingly, may (miR-34a-5p) or may not (miR-124–3p) depend on Dicer although the same tRNA/pre-miRNA carrier was used (Ho et al. 2018). The release of target miRNA from BERA/miRNA consequently altered the miRNome in human cells, as well as the transcriptome (Ho et al. 2018). The selectivity of biologic or recombinant miRNAs in target gene regulation was supported by the fact that miRNA enrichment analyses identified corresponding miRNAs (e.g., miR-34a-5p) underlying those downregulated transcripts (Ho et al. 2018). The tRNA/pre-miRNA-based RNA bioengineering technology has been proven to be the most robust, versatile, and efficient means to achieve a consistent high-yield production of a wide variety of RNA agents including miRNAs, siRNAs, aptamers, and other types of sRNAs (Fig. 1c), and BERAs are a novel class of RNA molecules for research and development (Duan and Yu 2016; Ho and Yu 2016; Yu et al. 2019).
Viroid-derived circular RNA cargo
A new viroid-derived system was reported very recently to produce large amounts of recombinant RNAs (Fig. 1d) in E. coli (Daros et al. 2018). Viroids are a special class of infectious agents of higher plants that specifically contain short, single-stranded, noncoding circular RNA (circRNA) molecules without any proteins (Branch et al. 1988; Branch and Robertson 1984). In the host cells, the viroid RNA is transcribed by the host RNA polymerase II or the chloroplastic nuclear-encoded RNA polymerase (Daros et al. 1994). Viroid RNA depends on the host type-III RNase for its processing. In some viroids with embedded ribozymes, self-cleavage of RNA intermediates during replication results in a viroid monomer that is subsequently circularized by either the host DNA ligase 1 or the chloroplastic isoform of tRNA ligase (Nohales et al. 2012a; Nohales et al. 2012b) to offer circRNA.
The use of viroid-derived circRNA for the efficient production of recombinant RNAs has been established through the co-expression of eggplant latent viroid (ELVd)-derived RNAs in E. coli and the eggplant tRNA ligase essential for RNA circularization (Daros et al. 2018). As such, this approach consists of two plasmids, pLELVd-BZB with the lpp promoter to express target RNA-bearing ELVd RNA and p15LtRnlSm to produce eggplant tRNA ligase (Table 1). The ELVd transcripts self-cleave efficiently in non-host E. coli cells through the embedded hammerhead ribozymes. The resulting viroid monomers can be stably circularized to generate target circular ELVd RNA chimera, which is indeed dependent on the co-expression of eggplant tRNA ligase. The utility of this approach was demonstrated by the successful production of an ELVd-spinach RNA chimera (Daros et al. 2018). The monomeric circular and linear ELVd RNAs, with a circular-to-linear ratio of 3:1, were estimated to accumulate up to 150 mg per liter of bacterial culture, and circRNAs could be purified by gel electrophoresis or FPLC methods (Daros et al. 2018). However, the yield and purity of isolated circRNAs were somehow not reported. Although it has not been explored whether this ELVd circRNA-based cargo could assemble RNAi agents for functional studies, this novel approach holds great promise for RNA bioengineering.
Co-expression with RNA-binding protein
Another approach for the production of fully processed siRNA molecules has been established via co-expression of corresponding shRNAs with a RNA-binding protein, p19, in bacteria (Huang et al. 2013). The plant RNA virus tombusvirus encodes a 19 kD protein, namely p19, which selectively binds to siRNAs of ~21 nt in length and suppresses siRNA function. Since p19 is known to selectively bind to double-stranded siRNAs with high affinity, p19 has been utilized for the isolation/detection or stabilization of siRNAs/miRNAs (Qiu et al. 2002; Silhavy et al. 2002). Through co-expression of p19 with siRNA-embedded shRNA (Huang et al. 2013), the target siRNA can be first generated from shRNA by bacterial RNase and then form a stable complex with p19. Thus, when total protein was extracted, the siRNA-p19 complex consisting of a histidine tag was readily purified by nickel affinity chromatography. SDS treatment allowed for the separation of the siRNA from the complex which was followed by isolation of target siRNA species by anion exchange HPLC (Huang et al. 2013). While this method was utilized for the successful production of a few functional siRNAs, the overall yield is very low (tens of micrograms per liter of bacterial culture; Table 1), and it is unknown whether similarly sized bacterial sRNAs are completely separated from target siRNAs.
Direct expression using particular bacteria strains
Purple bacteria Rhodovulum sulfidophilum
R. sulfidophilum is a marine phototrophic bacterium that is able to generate extracellular nucleic acids in nature (Kikuchi and Umekage 2018; Nagao et al. 2015; Pereira et al. 2017). Since this bacterium does not produce any RNases in the culture medium, a target RNA might be produced and retained in the bacterial culture medium, without requiring any additional means to protect against degradation by nucleases. R. sulfidophilum has been employed for direct production of various species of RNAs, including tRNAs, rRNAs, aptamers, (Ando et al. 2006; Nagao et al. 2014; Suzuki et al. 2010) as well as human pre-miRNAs (Pereira et al. 2016a; Pereira et al. 2016b; Pereira et al. 2016c). However, the levels of recombinant RNAs, occurring either at extracellular or intracellular locations, are extremely low, from nanogram to microgram ranges per liter of culture (Kikuchi and Umekage 2018; Pereira et al. 2017). This might be due to an intrinsic low transcription efficiency of this bacterial strain, the presence of intracellular RNases to which the recombinant RNAs are immediately susceptible, and ineffective diffusion or efflux of the RNAs into the culture medium. Therefore, the potential of direct production of biologic RNAs using R. sulfidophilum remains to be explored.
RNase III-deficient Corynebacterium glutamicum strain
A most recent advance is the development and application of RNase III-deficient C. glutamicum bacterium for in vivo production of target RNA molecules (Table 1) (Hashiro et al. 2019a; Hashiro et al. 2019b). C. glutamicum has been widely used in industrial settings for large-scale production of various biochemical compounds such as amino acids and lipids (Kalinowski et al. 2003). To avoid degradation and achieve overexpression of target RNA products, the RNase III-encoding gene rnc was disrupted to offer a new bacterial strain, namely C. glutamicum 2256LΔrnc. A strong F1 promoter was utilized to drive the expression of recombinant RNA (Fig. 4). In one case, an RNA molecule, namely U1A*-RNA, about 160 nt in length, was evaluated (Hashiro et al. 2019b), which consists of a stem/loop II (SLII, hairpin II) structure from U1 small nuclear RNA (snRNA) (Fig. 4a), and binds to U1A protein to form a U1 sn-ribonucleoprotein critical for pre-mRNA splicing. A prominent expression of RNA in the C. glutamicum 2256LΔrnc corresponding to the U1A*-RNA transcript was evident, while a truncated form was also notable (Hashiro et al. 2019b). In another case, a diap1*-dsRNA was designed as a pesticide through suppressing the expression of the essential gene diap1 (encoding death-associated inhibitor of apoptosis protein 1) in the pest Henosepilachna vigintioctopunctata. Overexpression of diap1*-dsRNA was achieved in C. glutamicum 2256LΔrnc strain by convergent transcription under two strong F1 promoters (Fig. 4b) (Hashiro et al. 2019a). The target diap1*-dsRNA and U1A*-RNA was estimated to reach up to 75 and 300 mg per liter of bacterial fermentation, respectively (Table 1), although the purification and overall yield were not reported. This new system may serve as an efficient platform for in vivo preparation of biologic RNAs while its applicability to other specific RNAs warrants further evaluation.
Fig. 4.
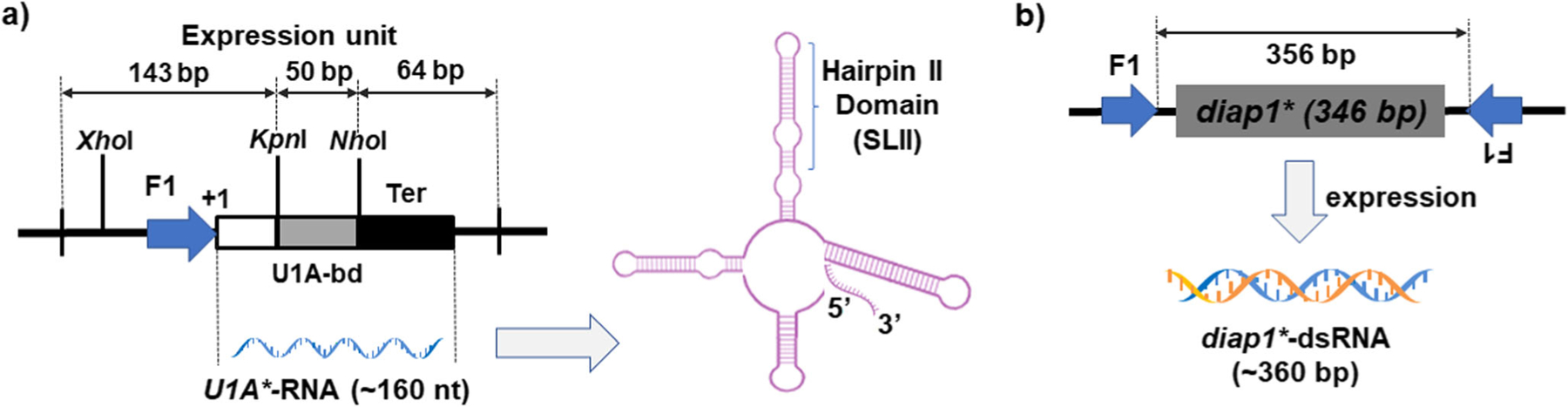
Direct overexpression of diap1*-dsRNA and U1A*-RNA in C. glutamicum strains 2256LΔrnc deficient in RNase III. a Schematic illustration of the expression unit of U1A*-RNA consisting of the F1 promoter and a transcription terminator (Ter). The predicted secondary structure of recombinant U1A*-RNA product includes the hairpin II domain that is also named SLII. b A convergent transcription-based expression of diap1*-dsRNA, among which the sense and antisense strands of the RNA are driven by two F1 promoters
Conclusions and perspectives
Given the presence of many more ncRNA-coding genes than protein-coding genes in the human genome and the recognition of a variety of ncRNA-controlled gene regulation mechanisms in essentially all species, it is crucial to understand the biological functions of ncRNAs. Their roles in normal physiology and disease should be fully understood towards the development of new therapeutic or agricultural strategies. While chemo-engineered ncRNA mimics continue to dominate ncRNA research and development, recent efforts have led to breakthroughs in novel biotechnologies for fermentation production of RNA molecules (Chen et al. 2015; Hashiro et al. 2019a; Hashiro et al. 2019b; Ho et al. 2018; Ponchon et al. 2009; Ponchon and Dardel 2007). Among them, the tRNA/pre-miRNA carrier-based technology has proven to be a robust and versatile platform to achieve a consistent, efficient, large-scale, and cost-effective production of a wide variety of biologic RNA agents, including miRNAs, siRNAs, aptamers, and sRNAs, all of which are biologically active in vitro and in vivo. Direct overexpression in RNase III-deficient bacterial strains has been demonstrated very recently by the production of two specific RNAs. Bioengineered or recombinant RNAs are a novel class of RNA molecules that should better recapitulate the properties of natural RNAs as both are made and folded in living cells. Future research shall help to challenge and refine these approaches, explore novel strategies, and critically assess the utilities of bioengineered ncRNAs.
Acknowledgments
This study was supported by National Cancer Institute (grant No. R01CA225958) and National Institute of General Medical Sciences (R01GM113888), National Institutes of Health.
Footnotes
Compliance with ethical standards
Ethical statement The authors confirm that the article does not contain any studies with human participants or animals.
Conflict of interest
The authors are named inventors of patent applications related to RNA bioengineering technology and utilities that are owned by the UC Davis, and Dr. Yu is a founder of AimRNA, Inc. which has an agreement to license the intellectual property.
References
- Abels ER, Breakefield XO (2016) Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 36(3):301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre F, Ormonde AR, Snider KM, Woolard K, Yu AM, Wittenburg LA (2018) A genetically engineered microRNA-34a prodrug demonstrates anti-tumor activity in a canine model of osteosarcoma. PLoS One 13(12):e0209941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355 [DOI] [PubMed] [Google Scholar]
- Ando T, Suzuki H, Nishimura S, Tanaka T, Hiraishi A, Kikuchi Y (2006) Characterization of extracellular RNAs produced by the marine photosynthetic bacterium Rhodovulum sulfidophilum. J Biochem 139(4):805–811 [DOI] [PubMed] [Google Scholar]
- Beckert B, Masquida B (2011) Synthesis of RNA by in vitro transcription. Methods Mol Biol 703:29–41 [DOI] [PubMed] [Google Scholar]
- Bramsen JB, Kjems J (2012) Development of therapeutic-grade small interfering RNAs by chemical engineering. Front Genet 3:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Benenfeld BJ, Robertson HD (1988) Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci U S A 85(23):9128–9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Robertson HD (1984) A replication cycle for viroids and other small infectious RNA’s. Science 223(4635):450–455 [DOI] [PubMed] [Google Scholar]
- Burley SK, Berman HM, Bhikadiya C, Bi C, Chen L, Di Costanzo L, Christie C, Dalenberg K, Duarte JM, Dutta S, Feng Z, Ghosh S, Goodsell DS, Green RK, Guranovic V, Guzenko D, Hudson BP, Kalro T, Liang Y, Lowe R, Namkoong H, Peisach E, Periskova I, Prlic A, Randle C, Rose A, Rose P, Sala R, Sekharan M, Shao C, Tan L, Tao YP, Valasatava Y, Voigt M, Westbrook J, Woo J, Yang H, Young J, Zhuravleva M, Zardecki C (2019) RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res 47(D1):D464–D474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ (2012) RNA-based therapeutics: current progress and future prospects. Chem Biol 19(1):60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA (2014) The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157(1):77–94 [DOI] [PubMed] [Google Scholar]
- Chen QX, Wang WP, Zeng S, Urayama S, Yu AM (2015) A general approach to high-yield biosynthesis of chimeric RNAs bearing various types of functional small RNAs for broad applications. Nucleic Acids Res 43(7):3857–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza LM, Larios-Sanz M, Setterquist RA, Willson RC, Fox GE (2003) Small RNA sequences are readily stabilized by inclusion in a carrier rRNA. Biotechnol Prog 19(3):734–738 [DOI] [PubMed] [Google Scholar]
- Dahlberg AE (1989) The functional role of ribosomal RNA in protein synthesis. Cell 57(4):525–529 [DOI] [PubMed] [Google Scholar]
- Daros JA, Aragones V, Cordero T (2018) A viroid-derived system to produce large amounts of recombinant RNA in Escherichia coli. Sci Rep 8(1):1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daros JA, Marcos JF, Hernandez C, Flores R (1994) Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci U S A 91(26):12813–12817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue C, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu Y, Ruan X, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan Y, Wold B, Carninci P, Guigo R, Gingeras TR (2012) Landscape of transcription in human cells. Nature 489(7414):101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominska M, Dykxhoorn DM (2010) Breaking down the barriers: siRNA delivery and endosome escape. J Cell Sci 123(Pt 8):1183–1189 [DOI] [PubMed] [Google Scholar]
- Duan Z, Yu AM (2016) Bioengineered non-coding RNA agent (BERA) in action. Bioengineered 7(6):411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Sandoval F, Poirier M, Scott MS (2015) The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA 6(4):381–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LP, Mookhtiar KA, Coleman JE (1997) Initiation, elongation, and processivity of carboxyl-terminal mutants of T7 RNA polymerase. Biochemistry 36(10):2908–2918 [DOI] [PubMed] [Google Scholar]
- Gaudin C, Nonin-Lecomte S, Tisné C, Corvaisier S, Bordeau V, Dardel F, Felden B (2003) The tRNA-like domains of E. coli and A. aeolicus transfer–messenger RNA: structural and functional studies. J Mol Biol 331(2):457–471 [DOI] [PubMed] [Google Scholar]
- Guiley KZ, Pratt AJ, MacRae IJ (2012) Single-pot enzymatic synthesis of Dicer-substrate siRNAs. Nucleic Acids Res 40(5):e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, Chikami Y, Kawaguchi H, Niimi T, Yasueda H (2019a) Construction of Corynebacterium glutamicum cells as containers encapsulating dsRNA overexpressed for agricultural pest control. Appl Microbiol Biotechnol 103(20):8485–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiro S, Mitsuhashi M, Yasueda H (2019b) Overexpression system for recombinant RNA in Corynebacterium glutamicum using a strong promoter derived from corynephage BFK20. J Biosci Bioeng 128(3):255–263 [DOI] [PubMed] [Google Scholar]
- Hessvik NP, Llorente A (2018) Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 75(2):193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Duan Z, Batra N, Jilek JL, Tu MJ, Qiu JX, Hu Z, Wun T, Lara PN, DeVere White RW, Chen HW, Yu AM (2018) Bioengineered non-coding RNAs selectively change cellular miRNome profiles for cancer therapy. J Pharmacol Exp Ther 365(3):494–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PY, Yu AM (2016) Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip Rev RNA 7(2):186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jin J, Deighan P, Kiner E, McReynolds L, Lieberman J (2013) Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat Biotechnol 31(4):350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian C, Tu MJ, Ho PY, Duan Z, Zhang Q, Qiu JX, DeVere White RW, Wun T, Lara PN, Lam KS, Yu AX, Yu AM (2017) Co-targeting of DNA, RNA, and protein molecules provides optimal outcomes for treating osteosarcoma and pulmonary metastasis in spontaneous and experimental metastasis mouse models. Oncotarget 8(19):30742–30755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Tian Y, Yu AM (2017) Effects of MicroRNA-34a on the pharmacokinetics of cytochrome P450 probe drugs in mice. Drug Metab Dispos 45(5):512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilek JL, Zhang QY, Tu MJ, Ho PY, Duan Z, Qiu JX, Yu AM (2019) Bioengineered let-7c inhibits orthotopic hepatocellular carcinoma and improves overall survival with minimal immunogenicity. Mol Ther Nucleic Acids 14:498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, Lucchino M (2018) Current challenges in delivery and cytosolic translocation of therapeutic RNAs. Nucleic Acid Ther 28(3): 178–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Kramer R, Linke B, McHardy AC, Meyer F, Mockel B, Pfefferle W, Puhler A, Rey DA, Ruckert C, Rupp O, Sahm H, Wendisch VF, Wiegrabe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol 104(1–3):5–25 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Watts JK (2017) The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 35(3):238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Umekage S (2018) Extracellular nucleic acids of the marine bacterium Rhodovulum sulfidophilum and recombinant RNA production technology using bacteria. FEMS Microbiol Lett 365(3) [DOI] [PubMed] [Google Scholar]
- Kim SK, Park YC (2019) Biosynthesis of omega-hydroxy fatty acids and related chemicals from natural fatty acids by recombinant Escherichia coli. Appl Microbiol Biotechnol 103(1):191–199 [DOI] [PubMed] [Google Scholar]
- Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172(3):393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Addepalli B, Tu MJ, Chen QX, Wang WP, Limbach PA, LaSalle JM, Zeng S, Huang M, Yu AM (2015) Chimeric microRNA-1291 biosynthesized efficiently in Escherichia coli is effective to reduce target gene expression in human carcinoma cells and improve chemosensitivity. Drug Metab Dispos 43(7):1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MM, Wang WP, Wu WJ, Huang M, Yu AM (2014) Rapid production of novel pre-microRNA agent hsa-mir-27b in Escherichia coli using recombinant RNA technology for functional studies in mammalian cells. Drug Metab Dispos 42(11):1791–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PC, Tu MJ, Ho PY, Jilek JL, Duan Z, Zhang QY, Yu AX, Yu AM (2018) Bioengineered NRF2-siRNA is effective to interfere with NRF2 pathways and improve chemosensitivity of human cancer cells. Drug Metab Dispos 46(1):2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Stepanov VG, Strych U, Willson RC, Jackson GW, Fox GE (2010) DNAzyme-mediated recovery of small recombinant RNAs from a 5S rRNA-derived chimera expressed in Escherichia coli. BMC Biotechnol 10:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YP, Berkhout B (2011) miRNA cassettes in viral vectors: problems and solutions. Biochim Biophys Acta 1809(11–12):732–745 [DOI] [PubMed] [Google Scholar]
- Lundin KE, Hojland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J, Smith CI (2013) Biological activity and biotechnological aspects of locked nucleic acids. Adv Genet 82:47–107 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2004) RNA regulation: a new genetics? Nat Rev Genet 5(4): 316–323 [DOI] [PubMed] [Google Scholar]
- Meinnel T, Mechulam Y, Fayat G (1988) Fast purification of a functional elongator tRNAmet expressed from a synthetic gene in vivo. Nucleic Acids Res 16(16):8095–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J, Groebe D, Witherell G, Uhlenbeck O (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15(21):8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min K, Park K, Park DH, Yoo YJ (2015) Overview on the biotechnological production of L-DOPA. Appl Microbiol Biotechnol 99(2):575–584 [DOI] [PubMed] [Google Scholar]
- Myers JW, Jones JT, Meyer T, Ferrell JE Jr (2003) Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat Biotechnol 21(3):324–328 [DOI] [PubMed] [Google Scholar]
- Nagao N, Hirose Y, Misawa N, Ohtsubo Y, Umekage S, Kikuchi Y (2015) Complete genome sequence of Rhodovulum sulfidophilum DSM 2351, an extracellular nucleic acid-producing bacterium. Genome Announc 3(2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao N, Suzuki H, Numano R, Umekage S, Kikuchi Y (2014) Short hairpin RNAs of designed sequences can be extracellularly produced by the marine bacterium Rhodovulum sulfidophilum. J Gen Appl Microbiol 60(6):222–226 [DOI] [PubMed] [Google Scholar]
- Nelissen FH, Leunissen EH, van de Laar L, Tessari M, Heus HA, Wijmenga SS (2012) Fast production of homogeneous recombinant RNA—towards large-scale production of RNA. Nucleic Acids Res 40(13):e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Flores R, Daros JA (2012a) Viroid RNA redirects host DNA ligase 1 to act as an RNA ligase. Proc Natl Acad Sci U S A 109(34):13805–13810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Molina-Serrano D, Flores R, Daros JA (2012b) Involvement of the chloroplastic isoform of tRNA ligase in the replication of viroids belonging to the family Avsunviroidae. J Virol 86(15):8269–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Yu G, Tian S, Li H (2014) Co-expression and co-purification of archaeal and eukaryal box C/D RNPs. PLoS One 9(7):e103096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P, Pedro AQ, Queiroz JA, Figueiras AR, Sousa F (2017) New insights for therapeutic recombinant human miRNAs heterologous production: Rhodovolum sulfidophilum vs Escherichia coli. Bioengineered 8(5):670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira P, Pedro AQ, Tomas J, Maia CJ, Queiroz JA, Figueiras A, Sousa F (2016a) Advances in time course extracellular production of human pre-miR-29b from Rhodovulum sulfidophilum. Appl Microbiol Biotechnol 100(8):3723–3734 [DOI] [PubMed] [Google Scholar]
- Pereira P, Queiroz JA, Figueiras A, Sousa F (2016b) Affinity approaches in RNAi-based therapeutics purification. J Chromatogr B Analyt Technol Biomed Life Sci 1021:45–56 [DOI] [PubMed] [Google Scholar]
- Pereira PA, Tomas JF, Queiroz JA, Figueiras AR, Sousa F (2016c) Recombinant pre-miR-29b for Alzheimer’s disease therapeutics. Sci Rep 6:19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrek H, Batra N, Ho PY, Tu MJ, Yu AM (2019) Bioengineering of a single long noncoding RNA molecule that carries multiple small RNAs. Appl Microbiol Biotechnol 103(15):6107–6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulle C, Hedenstierna KO, Fox GE (1995) A novel approach for monitoring genetically engineered microorganisms by using artificial, stable RNAs. Appl Environ Microbiol 61(10):3661–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Derrick ML, Uhlenbeck OC (1998) T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4(10):1313–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coli using a tRNA scaffold. Nat Protoc 4(6):947–959 [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4(7):571–576 [DOI] [PubMed] [Google Scholar]
- Ponchon L, Dardel F (2011) Large scale expression and purification of recombinant RNA in Escherichia coli. Methods 54(2):267–273 [DOI] [PubMed] [Google Scholar]
- Qiu W, Park JW, Scholthof HB (2002) Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant-Microbe Interact 15(3):269–280 [DOI] [PubMed] [Google Scholar]
- Rak R, Dahan O, Pilpel Y (2018) Repertoires of tRNAs: the couplers of genomics and proteomics. Annu Rev Cell Dev Biol 34:239–264 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21(12):3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Ando T, Umekage S, Tanaka T, Kikuchi Y (2010) Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl Environ Microbiol 76(3):786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu MJ, Ho PY, Zhang QY, Jian C, Qiu JX, Kim EJ, Bold RJ, Gonzalez FJ, Bi H, Yu AM (2019) Bioengineered miRNA-1291 prodrug therapy in pancreatic cancer cells and patient-derived xenograft mouse models. Cancer Lett 442:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeh-Garcia M, Simion C, Ho PY, Batra N, Berg A, Carraway III K, Yu AM, Sweeney C (2019) Suppressing the growth and metastatic potential of triple negative breast cancer cells using novel bioengineered microRNA-127 prodrug. Cancer res:(in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usmani SS, Bedi G, Samuel JS, Singh S, Kalra S, Kumar P, Ahuja AA, Sharma M, Gautam A, Raghava GPS (2017) THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One 12(7): e0181748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WP, Ho PY, Chen QX, Addepalli B, Limbach PA, Li MM, Wu WJ, Jilek JL, Qiu JX, Zhang HJ, Li T, Wun T, White RD, Lam KS, Yu AM (2015) Bioengineering novel chimeric microRNA-34a for prodrug cancer therapy: high-yield expression and purification, and structural and functional characterization. J Pharmacol Exp Ther 354(2):131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M (2000) Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol 2(2):70–75 [DOI] [PubMed] [Google Scholar]
- Willke T (2014) Methionine production—a critical review. Appl Microbiol Biotechnol 98(24):9893–9914 [DOI] [PubMed] [Google Scholar]
- Wons E, Furmanek-Blaszk B, Sektas M (2015) RNA editing by T7 RNA polymerase bypasses InDel mutations causing unexpected phenotypic changes. Nucleic Acids Res 43(8):3950–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sun J, Ho PY, Luo Z, Ma W, Zhao W, Rathod SB, Fernandez CA, Venkataramanan R, Xie W, Yu AM, Li S (2019) Creatine based polymer for codelivery of bioengineered microRNA and chemodrugs against breast cancer lung metastasis. Biomaterials 210: 25–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM (2002) Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc Natl Acad Sci U S A 99(15):9942–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu AM, Jian C, Yu AH, Tu MJ (2019) RNA therapy: are we using the right molecules? Pharmacol Ther 196:91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, Ho PY, Tu MJ, Jilek JL, Chen QX, Zeng S, Yu AM (2018) Lipidation of polyethylenimine-based polyplex increases serum stability of bioengineered RNAi agents and offers more consistent tumoral gene knockdown in vivo. Int J Pharm 547(1–2):537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Potty AS, Jackson GW, Stepanov V, Tang A, Liu Y, Kourentzi K, Strych U, Fox GE, Willson RC (2009) Engineered 5S ribosomal RNAs displaying aptamers recognizing vascular endothelial growth factor and malachite green. J Mol Recognit 22(2):154–161 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tu MJ, Wang WP, Qiu JX, Yu AX, Yu AM (2016) Genetically engineered pre-microRNA-34a prodrug suppresses orthotopic osteosarcoma xenograft tumor growth via the induction of apoptosis and cell cycle arrest. Sci Rep 6:26611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Tu MJ, Yu YF, Wang WP, Chen QX, Qiu JX, Yu AX, Yu AM (2015) Combination therapy with bioengineered miR-34a prodrug and doxorubicin synergistically suppresses osteosarcoma growth. Biochem Pharmacol 98(4):602–613 [DOI] [PMC free article] [PubMed] [Google Scholar]


