Abstract
Based on genome-scale loss-of-function screens we discovered that Topoisomerase III-β (TOP3B), a human topoisomerase that acts on DNA and RNA, is required for yellow fever virus and dengue virus-2 replication. Remarkably, we found that TOP3B is required for efficient replication of all positive-sense-single stranded RNA viruses tested, including SARS-CoV-2. While there are no drugs that specifically inhibit this topoisomerase, we posit that TOP3B is an attractive anti-viral target.
Keywords: Topoisomerase III-ß (TOP3B), Positive stranded RNA viruses, Host factor, Dengue, SARS-CoV-2
Highlights
-
•
Topoisomerase III-ß (TOP3B) is a host factor for all single stranded positive strand RNA viruses.
-
•
TOP3B is not a host factor for Ebola and influenza viruses, which are negative strand RNA viruses.
-
•
TOP3B acts directly on the viral genome of DENV2.
-
•
TOP3B could be a target for antiviral development.
Acute viral infections, particularly those caused by RNA viruses, are dangerous threats to public health. This has been made evident by the ongoing pandemic of severe acute respiratory syndrome (SARS), officially named COVID-19, caused by the betacoronavirus SARS-CoV-2. When confronted with emerging viruses we are left with few countermeasures that can be deployed to specifically prevent or treat the diseases they cause. Understanding the molecular mechanisms required for viral replication for known viruses can reveal potential vulnerabilities for related novel viruses. These vulnerabilities could be based on viral targets, such as the RNA-dependent RNA polymerases, or dependency on host encoded pro-viral activities. To identify such host pro-viral factors we embarked on genome-scale screens for diverse families of viruses and identified scores of required host factors (Barrows et al., 2019; Le Sommer et al., 2012; Sessions et al., 2009; Anderson et al., 2019). Some of these host factors are attractive drug targets.
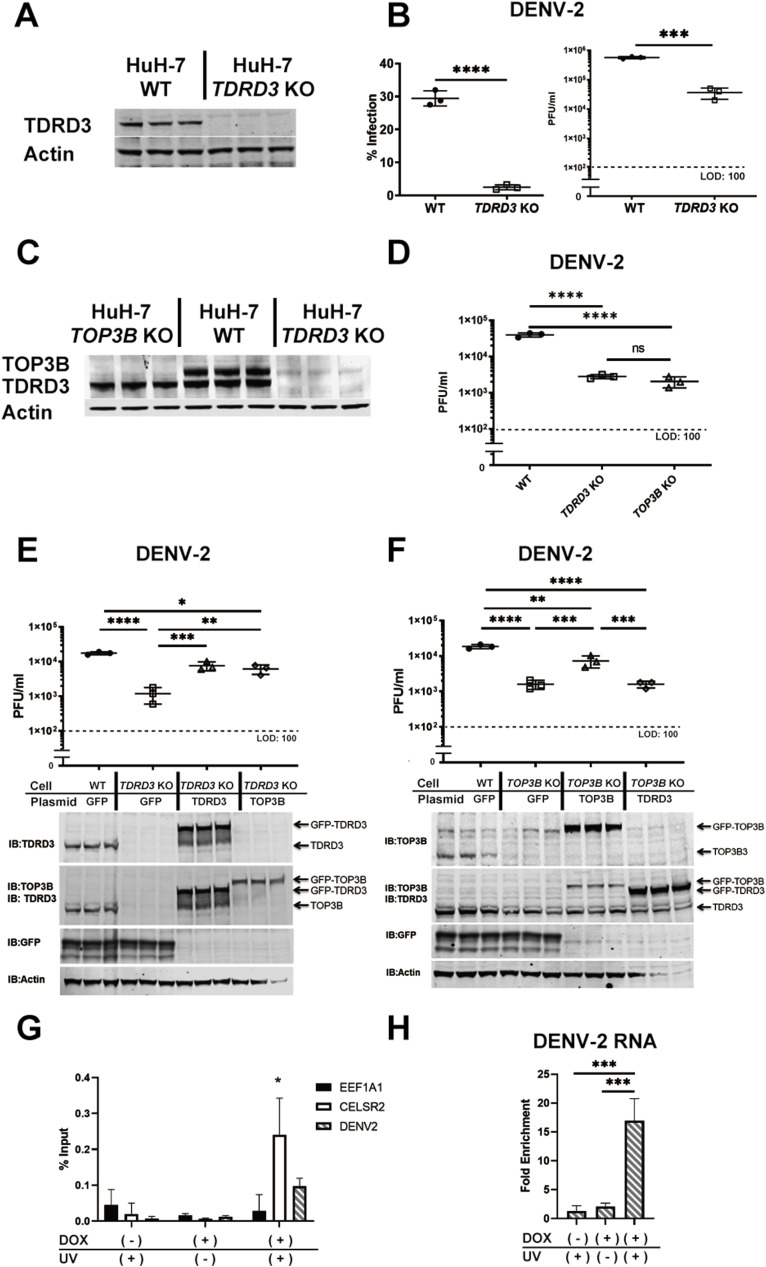
A meta-analysis of RNAi-based loss-of-function screens for yellow fever virus-17D (YFV) and dengue virus-2 (DENV-2) host factors revealed 274 common candidates (Barrows et al., 2019). TDRD3, a Tudor domain containing protein that interacts with methylated arginine motifs (Cote and Richard, 2005), was identified as a candidate host factor required for both DENV-2 and YFV. In YFV screens TDRD3 ranked 66th of over 21,500 gene products in terms of how well its knockdown decreased YFV (adjusted p value = 0.0006). CRISPR-Cas9 mediated knockout of TDRD3 in HuH-7 cells confirmed that this protein was required for efficient DENV-2 replication (Supplementary Fig. 1, Fig. 1 A and B).
Fig. 1.
TOP3B is required for efficient replication of DENV-2. (A) TDRD3 levels assessed by western blotting with TDRD3 specific antibody from lysates of WT and TDRD3 KO HuH-7 cells (B) TDRD3 KO inhibits DENV-2 propagation; percentage of DENV-2 infected HuH-7 cells was determined by immunofluorescence microscopy using an antibody to the envelope protein (left); comparison of viral titers in the media of WT and TDRD3 KO cell cultures infected with DENV-2 (right). (C) TOP3B and TDRD3 levels assessed by western blotting with TOP3B and TDRD3 specific antibodies respectively from lysates of WT, TOP3B KO and TDRD3 KO HuH-7 cells. (D) Comparison of viral titers in the supernatants of WT HuH-7, TDRD3 KO and TOP3B KO cells infected with DENV-2. (E) TOP3B overexpression rescues DENV-2 infection in TDRD3 KO cells. WT and TDRD3 KO HuH-7 cells transfected with plasmids expressing GFP-TDRD3, GFP-TOP3B or a control plasmid (GFP) were infected with DENV-2. The supernatants were harvested 72 hpi and the viral titers were determined by plaque assay. (F) TDRD3 overexpression does not rescue DENV-2 infection in TOP3B KO cells. WT and TOP3B KO HuH-7 cells transfected with plasmids expressing GFP-TOP3B, GFP-TDRD3, or a control plasmid (GFP) were infected with DENV. The supernatants were harvested 72 hpi and the viral titers were determined by plaque assay. In the western blots shown in E and F in the panels probed for both TOP3B and TDRD3 we could not distinguish the endogenous TOP3B band. (H) Crosslinking and immunoprecipitation (CLIP) indicates binding of FLAG-TOP3B in HEK-293 cells to cellular and DENV-2 RNAs. (G) Crosslinking of FLAG-TOP3B to DENV-2 RNA depended on UV irradiation and induction of FLAG-TOP3B expression. *: p < 0.05, **: p < 0.01, ***: p < 0.001 and ****: p < 0.0001.
A well-known function of TDRD3 is to bind and stabilize Topoisomerase III-β (TOP3B) (Xu et al., 2013; Stoll et al., 2013; Yang et al., 2014), a type IA topoisomerase and the only human topoisomerase known to act on both DNA and RNA (Xu et al., 2013; Siaw et al., 2016). We confirmed that in HuH-7 cells TDRD3 interacted with TOP3B (Supplementary Fig. 2) and stabilized it, since knockout of TDRD3 led to very low levels of TOP3B (Fig. 1C). Knockout of TOP3B, which does not alter TDRD3 levels, resulted in the same dramatic decrease in DENV-2 as knockout of TDRD3 (Supplementary Fig. 1A and 1C & D), which suggested that a major, or perhaps the only role of TDRD3 in viral replication was to stabilize TOP3B. Indeed, TOP3B overexpression rescued DENV-2 infection in TDRD3 KO cells (Fig. 1E), while the reverse was not true (Fig. 1F). Therefore, we conclude that TOP3B is a proviral host factor for DENV-2. Not surprisingly, TOP3B was also required for efficient replication of YFV and Zika virus (Cambodian strain) (Supplementary Fig. 3), leading us to suggest that TOP3B is required for diverse flaviviruses.
The genetic approaches we utilized above do not distinguish between direct and indirect modes of action. To address whether or not TOP3B directly interacted with DENV-2 genomes we used UV crosslinking followed by RNA immunoprecipitation (CLIP). We carried out CLIP assays using a HEK-293T cell line that expressed a FLAG-tagged TOP3B upon doxycycline treatment and anti-FLAG antibodies to carry out the immunoprecipitation. FLAG-TOP3B preferentially crosslinked to CELSR2 RNA, which is known to bind this topoisomerase (Xu et al., 2013), relative to EEF1A1 RNA, which we use as a negative control (Fig. 1G). Importantly, TOP3B crosslinked DENV-2 RNA (Fig. 1G and H), strongly suggesting that TOP3B acts directly on the viral genome.
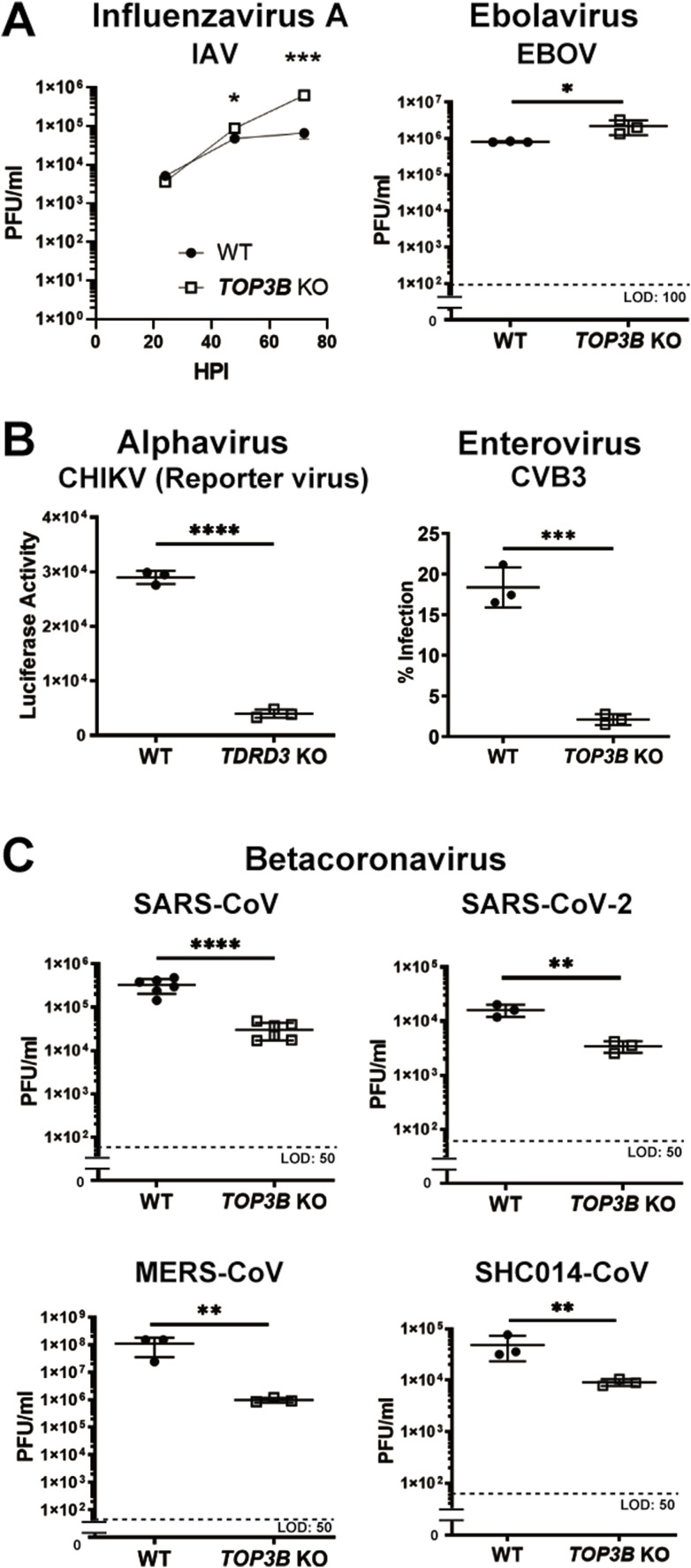
Since TOP3B was required for DENV-2, YFV and ZIKV replication, we asked whether or not this topoisomerase was required for replication of other viruses. Influenza A virus, which has a negative-sense segmented RNA genome, replicated significantly better in TOP3B KO cells than in the parental HuH-7 cells (Fig. 2 A, left panel), suggesting that TOP3B plays an anti-viral role for influenza A virus. We also tested Ebola virus, which has a negative sense RNA genome and again noted that Ebola virus replicated slightly better in TOP3B KO cells than in the parental HuH-7 cells (Fig. 2A, right panel). We thus suggest that TOP3B is not required for influenza A virus or Ebola virus replication.
Fig. 2.
TOP3B is required for efficient replication of a diverse group of (+) ss RNA viruses. (A) Influenza A virus (IAV) and Ebola virus (EBOV) replicate efficiently in TOP3B KO cells. A time course of viral titers in the media of WT and TDRD3 KO cultures infected with IAV (left); HPI, hours post infection. Viral titers in the media of WT and TDRD3 KO cultures infected with EBOV (right). (B) TDRD3 KO inhibits luciferase expression by a reporter chikungunya virus (left); TOP3B KO inhibits percentage of CVB3 infected HuH-7 cells (right). (C) TOP3B KO inhibits viral titers in the media of SARS-CoV, SARS-CoV-2, MERS-CoV, and SCH 1 014-CoV infected cell cultures. *: p < 0.05, **: p < 0.01, ***: p < 0.001 and ****: p < 0.0001.
We tested a series of positive-sense single-stranded RNA viruses ((+) ss RNA viruses). A recombinant chikungunya virus (CHIKV), an alphavirus, was sensitive to TDRD3 knockout (Fig. 2B, left panel), and coxsackievirus B3 (CVB3), an enterovirus, was dependent on TOP3B or TDRD3 for efficient replication (Fig. 2B, right panel, & Supplementary Fig. 4). Most important in the context of the COVID-19 pandemic, four betacoronaviruses, SARS-CoV, SARS-CoV-2, MERS-CoV, and SCH 1 014-CoV, a bat coronavirus, were significantly crippled by TOP3B KO (Fig. 2C). These results indicated that TOP3B is required for efficient replication of a diverse group of (+) ss RNA viruses.
Among host factors required for diverse groups of RNA viruses are components of the translation machinery and the proteasome, in addition to factors that mediate membrane transactions required for entry and exit of viruses. The understanding of the broad requirement for TOP3B among (+) ss RNA viruses will likely provide important insights into heretofore understudied steps in their lifecycles. While the human topoisomerase TOP1, a type IB topoisomerase, has been shown to exacerbate inflammation induced by RNA virus infection (Rialdi et al., 2016), the direct action of an RNA topoisomerase in viral replication is unprecedented. A requirement for a RNA topoisomerase as a critical cofactor for (+) ss RNA viruses is reasonable since viral genomes, because of their complex structures and size, especially the large coronaviruses, may lend themselves to topological problems (Ahmad et al., 2016; Pommier et al., 2016).
Could TOP3B be considered an antiviral drug target? It must be clear that we ask this suggestion responsibly with full understanding that our work is an early step in what must be a rigorous process to identify effective antivirals. We also note that the level of reduction in viral propagation caused by TOP3B KO varied depending on the virus studied between 10-fold (e.g., DENV-2) to approximately 100-fold (e.g., for MERS-CoV), and this may not be enough to alter infection in vivo. Nonetheless, several facts support an exploration of TOP3B as an antiviral drug target. TOP3B−/- mice are viable (Kwan and Wang, 2001) and humans with a homozygous 240-kb deletion of chromosome 22q11.22 spanning the TOP3B gene are also viable (Stoll et al., 2013). Although both homozygous null mice and humans have abnormal phenotypes, the enzyme is not essential. Topoisomerase inhibitors are well known drugs used to treat cancer (e.g. doxorubicin) (Tacar et al., 2013) and bacterial infections (e.g., ciproflaxin) (Zhanel et al., 2006). Unfortunately, we are not aware of any approved TOP3B drugs and prior to this report there was no evidence that inhibiting TOP3B could be a useful in treating acute viral infections. In fact other than the ACE-2 receptor, and perhaps furin, required for SARS-CoV-2 attachment and entry (Walls, 2020), TOP3B is the only other known required host factor. We hope this brief communication will spur interactions between individuals studying TOP3B and those working on positive-sense RNA viruses, including SARS-CoV-2.
Author contributions
KRP did the majority of the experimental work in Fig. 1 and coordinated the work in Fig. 2, MH did the CLIP experiments in Fig. 1, analyzed the data for all figures and prepared final figures. CS, XX and PYS made the reporter CHIKV and provided essential reagents for flavivirus work. MGB wrote the first draft of the manuscript, except the online methods, which was drafted by MH and KRP. MTB provided critical reagents and expertise needed to start the work on this project. WSF, SSB and MGB supervised the work in the Garcia-Blanco laboratory. AH performed and analyzed, and RR supervised work on IAV. CAP performed and AB supervised work with EBOV.
Declaration of competing interest
MTB is cofounder of EpiCypher. Others declare no competing interests.
Acknowledgements
We thank members of our laboratories at the University of Texas Medical Branch for comments and suggestions. We also thank Dr. Yuk-Ching Tse Dinh (Florida International University), Dr. Yves Pommier (National Cancer Institute), Dr. Weidong Wang (National Institute on Aging), and Dr. Phillip A. Sharp (MIT) for important discussions and comments. We acknowledge support from the Uehara Foundation Fellowship (MH), NIH R01 CA204806, Vacek Chair, and UTMB startup package (MGB), NIH R01 GM126412 (MTB), NIH AI142759, and the Sealy & Smith, Kleberg, John S. Dunn, Amon G. Carter, Gilson Longenbaugh, and Summerfield Robert Foundations (PYS), NIH/NIAID T32 AI007526 (AH), R21 AI26012 and R01 AI134907 (RR), UTMB start up funds (VM).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2020.104874.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmad M. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016;44:6335–6349. doi: 10.1093/nar/gkw508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.E. Comparative loss-of-function screens reveal ABCE1 as an essential cellular host factor for efficient translation of paramyxoviridae and pneumoviridae. mBio. 2019;10 doi: 10.1128/mBio.00826-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows N.J. Dual roles for the ER membrane protein complex in flavivirus infection: viral entry and protein biogenesis. Sci. Rep. 2019;9:9711. doi: 10.1038/s41598-019-45910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote J., Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- Kwan K.Y., Wang J.C. Mice lacking DNA topoisomerase IIIbeta develop to maturity but show a reduced mean lifespan. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5717–5721. doi: 10.1073/pnas.101132498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sommer C., Barrows N.J., Bradrick S.S., Pearson J.L., Garcia-Blanco M.A.G. protein-coupled receptor kinase 2 promotes flaviviridae entry and replication. PLoS Neglected Trop. Dis. 2012;6 doi: 10.1371/journal.pntd.0001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y., Sun Y., Huang S.N., Nitiss J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016;17:703–721. doi: 10.1038/nrm.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rialdi A. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352 doi: 10.1126/science.aad7993. aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions O.M. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaw G.E., Liu I.F., Lin P.Y., Been M.D., Hsieh T.S. DNA and RNA topoisomerase activities of Top3beta are promoted by mediator protein Tudor domain-containing protein 3. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E5544–E5551. doi: 10.1073/pnas.1605517113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013;16:1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- Walls A.C. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 Apr 16;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. Epub 2020 Mar 9. PMID: 32155444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013;16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell. 2014;53:484–497. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel G.G. A review of new fluoroquinolones : focus on their use in respiratory tract infections. Treat. Respir. Med. 2006;5:437–465. doi: 10.2165/00151829-200605060-00009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.