Abstract
Circular RNAs (circRNAs) belong to a diverse class of stable RNAs expressed in all cell types. Their proposed functions include sponging of microRNAs (miRNAs), sequestration and trafficking of proteins, assembly of multimeric complexes, production of peptides, and regulation of transcription. Backsplicing due to RNA structures formed by an exceptionally high number of Alu repeats lead to the production of a vast repertoire of circRNAs by human Survival Motor Neuron genes, SMN1 and SMN2, that code for SMN, an essential multifunctional protein. Low levels of SMN due to deletion or mutation of SMN1 result in spinal muscular atrophy (SMA), a major genetic disease of infants and children. Mild SMA is also recorded in adult population, expanding the spectrum of the disease. Here we review SMN circRNAs with respect to their biogenesis, sequence features, and potential functions. We also discuss how SMN circRNAs could be exploited for diagnostic and therapeutic purposes.
Keywords: spinal muscular atrophy, SMA; Survival Motor Neuron, SMN; backsplicing; circRNA; Alu elements; microRNA
1. Introduction
Circular RNAs (circRNAs) are a diverse class of RNAs expressed in all organisms from bacteria to humans [1–4]. While multiple mechanisms account for their biogenesis [1–4], the primary mechanism of circRNA production in humans is backsplicing, which will be the focus of this review [4]. Initially referred to as scrambled exons, circRNAs were first thought to be accidental byproducts of pre-mRNA splicing [5,6]. With the advent of next-generation sequencing, thousands of circRNAs began to be identified in diverse cell types, some of which represent the predominant RNA isoform of their host genes [4]. Soon, several studies outlining the biogenesis, characteristics, and potential functions of circRNA started to emerge, reinforcing the status of circRNAs as a bona fide category of functional RNAs [7–9].
The identified functions of circRNAs include sponging of microRNAs (miRNAs) and sequestration of RNA-binding proteins (RBPs) [7,8,10–13]. Some circRNAs are translated into proteins or peptides [14]. Similar to linear long non-coding RNAs (lncRNAs) that serve as protein scaffolds [15,16] or regulate chromatin modification [17,18], circRNAs can have critical non-coding functions. While exonic circRNAs are primarily localized in the cytoplasm [19], EIciRNAs that harbor both exonic and intronic sequences are localized in the nucleus [20]. EIciRNAs promote transcription of their host genes through interactions with U1 snRNP [20]. Several circRNAs formed by intron-only sequences have been found to be localized in the nucleus and implicated in the regulation of transcription [21].
Backsplicing that generates most circRNAs in humans requires pairing of a downstream 5′ splice site (5′ss) with an upstream 3′ splice site (3′ss) [4]. Such splice site pairing is often achieved through an RNA secondary structure formed by inverted repeat sequences, such as Alu elements in humans and B1 elements in mice [9,22]. RBPs also promote backsplicing by interacting with flanking intronic sequences upstream and downstream of the 5′ss and 3′ss, respectively [12,23]. With the help of RNA structures and/or RBPs, backsplicing can occur within a lariat intermediate harboring skipped exon(s) [24]. Intron-only lariat intermediates themselves can form stable circRNAs by escaping the debranching reaction following splicing [25].
Alu elements are bipartite primate-specific repeats derived from the 7SL RNA portion of the signal recognition particle [26]. Alu-like sequences occupy ~10% of the human genome and are more conserved than older repeat families such as MIR elements [27–29]. Upon transcription, inverted Alu repeats can form stable RNA:RNA duplex structures through base pairing of the complementary sequences. Depending on the context, such structures may modulate alternative pre-mRNA splicing [22,30] as well as backsplicing [9,22]. Also, intronic Alu elements give rise to novel exons [31]. Exonization of an Alu sequence is favored by splice-site-like motifs located within the Alu element itself [32]. Currently, it is not known whether Alu-derived exons are more common in circRNAs than in linear mRNAs.
The SMN genes (SMN1 and SMN2) code for the survival motor neuron (SMN) protein that is involved in multiple cellular processes including pre-mRNA splicing, transcription, translation, stress granule formation, signal transduction, and macromolecular trafficking [33]. Low expression of SMN due to deletion of or mutation in SMN1 results in spinal muscular atrophy (SMA), a leading genetic disease of infants and children [34,35]. The SMN locus generates a variety of transcripts [36]. The major transcript produced from the SMN1 gene, SMN1 mRNA, codes for the full-length SMN; it contains 9 exons, i.e., exons 1, 2A, 2B, 3, 4, 5, 6, 7 and 8 [36]. The SMN2 gene cannot compensate for the loss of SMN1 due to predominant skipping of exon 7 during SMN2 pre-mRNA splicing [37,38], leading to production of a truncated and highly unstable protein, SMNΔ7 [39–41]. However, correction of SMN2 exon 7 splicing holds the promise for cure [42–44]. Indeed, the first approved therapy for SMA, Spinraza (Nusinersen), is based on this approach [45,46]. Spinraza is an antisense oligonucleotide (ASO) that promotes exon 7 inclusion by blocking intronic splicing silencer N1 (ISS-N1), a negative cis-element located within intron 7 of SMN genes [46,47]. Two small compounds currently in clinical trials also act through correction of SMN2 exon 7 splicing [35,48,49]. In addition to exon 7, SMN exons 3 and 5 undergo alternative splicing [50,51]. Skipping of other SMN exons is triggered by either oxidative stress or depletion of U1 snRNP [50,52,53]. It has been shown that an Alu-derived sequence within intron 6 (referred to as exon 6B) can be exonized, resulting in an SMN isoform with an altered C-terminus [40].
The SMN genes are highly enriched in intronic Alu elements (Figure 1) [54]. Therefore, they are an ideal source for the production of circRNAs. Consistently, recent reports independently confirm the generation of a vast repertoire of exonic circRNAs by the SMN genes [55,56]. Every single internal exon of the SMN genes can become incorporated into circRNAs and several of the SMN circRNAs contain portions of exons 1 or 8 due to usage of novel splice sites [55,56]. Some of the SMN circRNAs include exon 6B and/or novel exons derived from portions of intron 1 and intergenic sequences downstream of exon 8 [55]. At least two SMN circRNAs contain exons from other genes, suggesting cooccurrence of backsplicing and trans-splicing for some of the SMN transcripts [55]. In this review, we describe the nature of the broad spectrum of SMN circRNAs and propose mechanisms for their generation. We also discuss potential functions of SMN circRNAs.
Figure 1. Organization of the SMN genes.
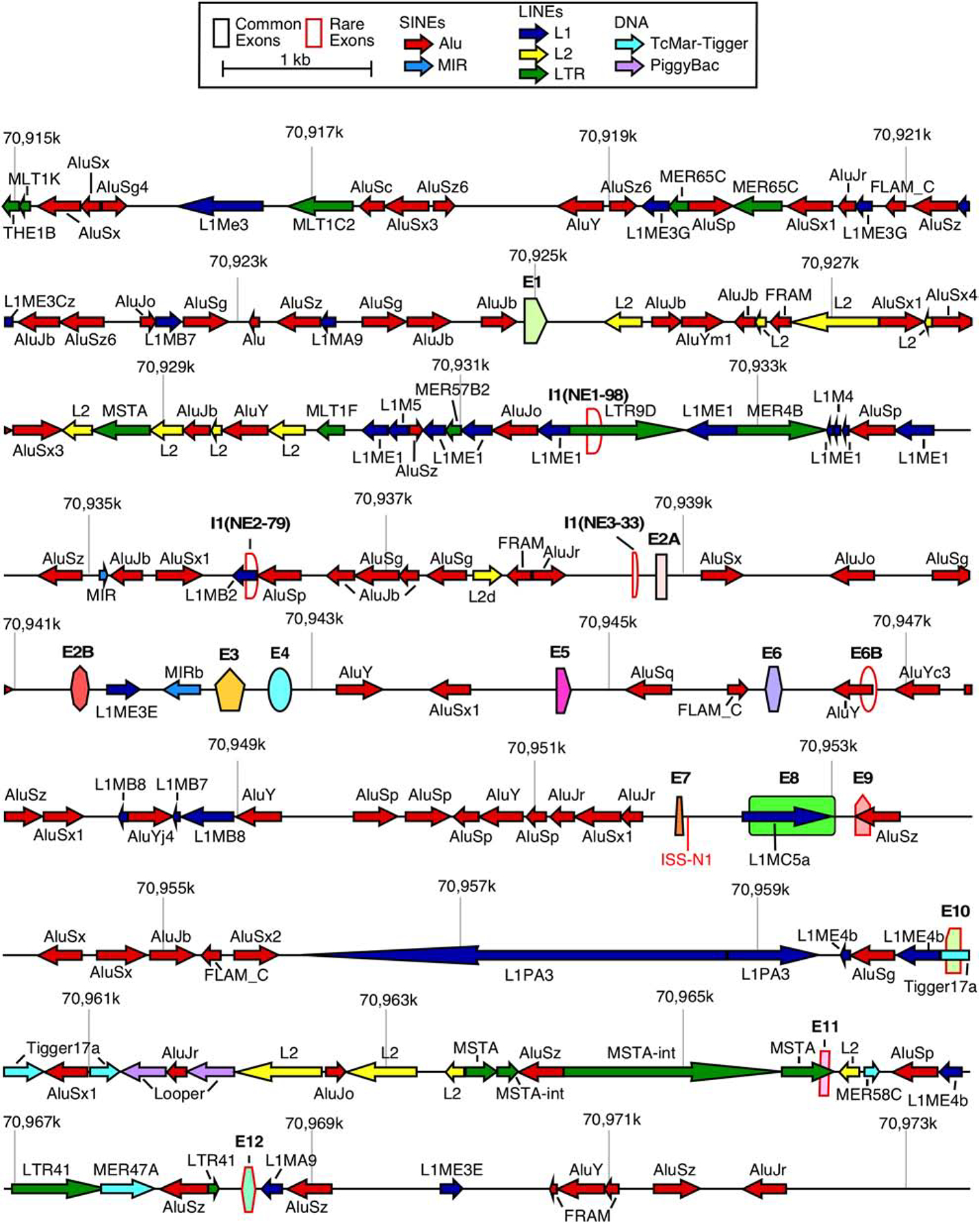
A scale depiction of the SMN1 gene (SMN2 has an identical overall structure) and flanking sequences are shown. Exons are depicted by colored shapes. Common exons (≥10% of total SMN RNA) are outlined in black, rare exons (<10% of total SMN RNA) are outlined in red. Repeat sequences as identified by Repeatmasker are depicted by colored arrows. Arrow direction indicates the orientation of the repeat sequence. ISS-N1, a critical splicing regulatory sequence in intron 7, is shown in red. Numbers indicate position within chromosome 5 of the GRCh38 human genome build. Abbreviations: SINEs, short interspersed nuclear elements; LINEs, Long interspersed nuclear elements. Scale and color coding of exons and repeat sequences are given in the boxed region.
2. Identification and classification of SMN circRNAs
The first evidence that human SMN genes could produce circRNAs came from early surveys in human cell lines. As reported in circBase, RNA-Seq of Hs68 fibroblasts identified reads supporting backsplicing of SMN exon 4 to exon 2B [9]. Another RNA-Seq study performed in H1 human embryonic stem cells identified reads supporting backsplicing of exon 6 to exon 5 [57]. The vast majority of SMN circRNAs were identified by two independent studies using a candidate-based approach [55,56]. Both studies identified circRNAs by reverse transcription and PCR (RT-PCR) employing divergent primers annealing to either a single exon or one primer annealing to the backsplice junction [55,56]. To prevent mis-identification of linear products and/or RT-PCR artifacts, both studies used RNA samples treated with RNase R that selectively degrades linear transcripts [55,56]. While minor differences in experimental approach and primer design led to differences in the apparent relative expression of some SMN circRNAs, many of the predominant isoforms identified by each were identical in exon content and backsplice site usage [55,56]. In addition to PCR using divergent primers, the expression of five of the most highly expressed circRNAs was independently confirmed by RNase protection assay [55].
SMN circRNAs are broadly categorized into four subtypes (Figure 2). Type 1 circRNAs are generated exclusively from the early transcribed region of SMN, encompassing a combination of the first 5 canonical exons of SMN. Three of the most highly expressed type 1 circRNAs utilize the 5′ss of exon 4 for backsplicing (Figure 2), indicating that sequences or RBPs binding in the vicinity of the 5′ss of exon 4 are highly conducive for backsplicing. Some of the type 1 circRNAs contain one of three novel exons, I1(NE1–98), I1(NE2–76), and I1(NE3–33), derived from sequences within intron 1 (Figure 2). None of these exonized sequences are derived from Alu elements; instead, I1(NE1–98) is derived from an LTR9D element, I1(NE2–76) is derived from an L1 repeat, and I1(NE3–33) is derived from a non-repeat sequence (Figure 1).
Figure 2. SMN genes generate a vast repertoire of circRNAs.
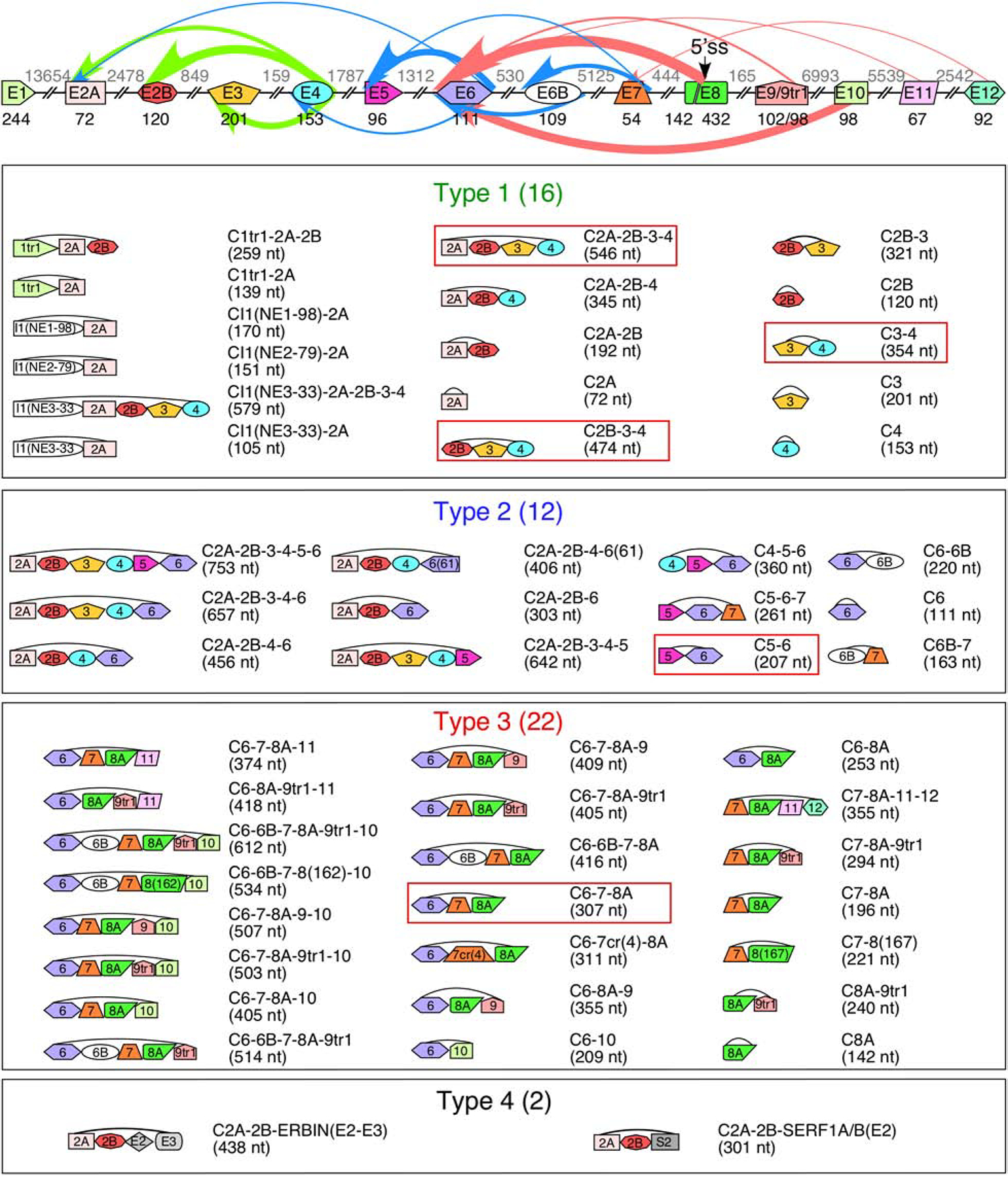
The genomic layout of the SMN genes and prominent backsplicing events is given at the top. Exons are shown as colored shapes, introns as broken lines. Exon sizes are given in black below each exon, intron sizes are given in gray above each intron. A cryptic 5′ss in exon 8 is indicated with an arrow. Colored arrows represent backsplicing events. The relative thickness of each arrow represents backsplice site usage frequency; arrow color represents type of circRNA (green for type 1, blue for type 2, and red for type 3). Bottom 4 panels show the total catalog of each type of SMN circRNA. The exon content of each circRNA is indicated graphically. The name of each circRNA is given at the right with total size in parentheses. circRNAs with the highest expression level are boxed.
Type 2 circRNAs are more varied in exon composition and are broadly characterized as containing at least one “middle” exon, i.e., exons 5, 6, and 7 (Figure 2). All type 3 circRNAs contain exon 8A with or without the downstream exons derived from the intergenic sequences. While the 3′ss of exon 6 is utilized for the production of most type 3 circRNAs, the 5′ss usage varies. Type 3 circRNAs are diverse, incorporating 8 exons in various combinations, including exon 6B and four novel exons generated from intergenic sequences located downstream of exon 8 (Figure 2) [55]. Of note, while exon 6B was originally identified in linear SMN mRNA as a rare exon [40], it appears to be much more commonly included in circRNAs [55].
The four novel exons generated from the intergenic sequences are referred to as exons 9, 10, 11, and 12. Of these four, the two most proximal exons, 9 and 10, are much more frequently incorporated into circRNAs [55]. Exon 9 is derived from the right arm of an antisense AluSz element located immediately (<200 nt) downstream of exon 8. Of note, exon 9 has two alternative 3′ss separated by 4 nt. The shorter form, 9tr1, appears to be the predominant variant [55]. Exon 10 is located ~7 kb downstream of exon 9 and is derived from a Tigger17a repeat element (Figure 1). Exon 11 is located ~5.5 kb downstream of exon 10 and is derived from an MSTA element (Figure 1). Exon 12 is located ~2.6 kb downstream of exon 11 and is derived from a non-repeat sequence (Figure 1).
3. Biogenesis of SMN circRNAs
All SMN circRNAs reported thus far contain at least one exon. In general, an exonic circRNA is generated by a backsplicing event catalyzed by the spliceosome, when a given 5′ss preferentially pairs with an upstream 3′ss. Such pairing of the splice sites could be facilitated by unique contexts described below.
3a. Backsplicing mediated by RNA structure
In several instances, an RNA:RNA duplex formed between complementary sequences located upstream and downstream of the 3′ss and 5′ss, respectively, facilitate backsplicing. In humans, such RNA:RNA duplex is often formed by inverted Alu repeats [9,22,58]. Non-repeat sequences capable of forming an RNA:RNA duplex by long-distance interactions can also facilitate backsplicing, although such duplexes are difficult to predict due to the presence of alternative structures in long transcripts [59,60]. SMN has an exceptionally high Alu content (~40%) within its introns (Figure 1) [54]. Therefore, it is likely that structures associated with the Alu elements are the prime drivers of SMN circRNA generation [61]. For example, intron 4 contains two Alu elements, an AluY and an AluSx1 (Figure 3A). Production of C2A-2B-3–4 requires pairing of the 5′ss of exon 4 with the 3′ss of exon 2A. Consistently, intron 1 contains numerous Alu elements that the intron 4 elements could pair with (Figure 1). Specifically, near the 3′ss of exon 2A, there are three antisense Alu elements that can each pair with the AluY located immediately downstream of exon 4 (Figure 3A). Alternatively, a sense AluJr element upstream of the 3′ss of exon 2A can pair with the antisense AluSx1 element located in the middle of intron 4 (Figure 3A). In order for C2B-3–4 to be produced, the 5′ss of exon 4 must pair with the 3′ss of exon 2B. Consistently, intron 2A contains three Alu elements, of which the AluSx and AluJo are particularly well-situated to pair with both the AluY and AluSx1 in intron 4 (Figure 3A). Unlike introns 1 and 2A, intron 2B does not contain an Alu element (Figures 1 and 3A). Despite this, C3–4 is one of the most abundant circRNAs produced by the SMN genes [55]. Since intron 3 is relatively short (159 nt), it is possible that pairing of the 5′ss of exon 4 and 3′ss of exon 3 is reinforced by the same Alu:Alu base pairing that is used for the production of C2B-3–4 (Figure 3A). Another possibility is that a non-Alu RNA:RNA duplex and/or RBPs facilitate the required splice site pairing.
Figure 3. Potential Alu:Alu pairing leading to exon circularization.
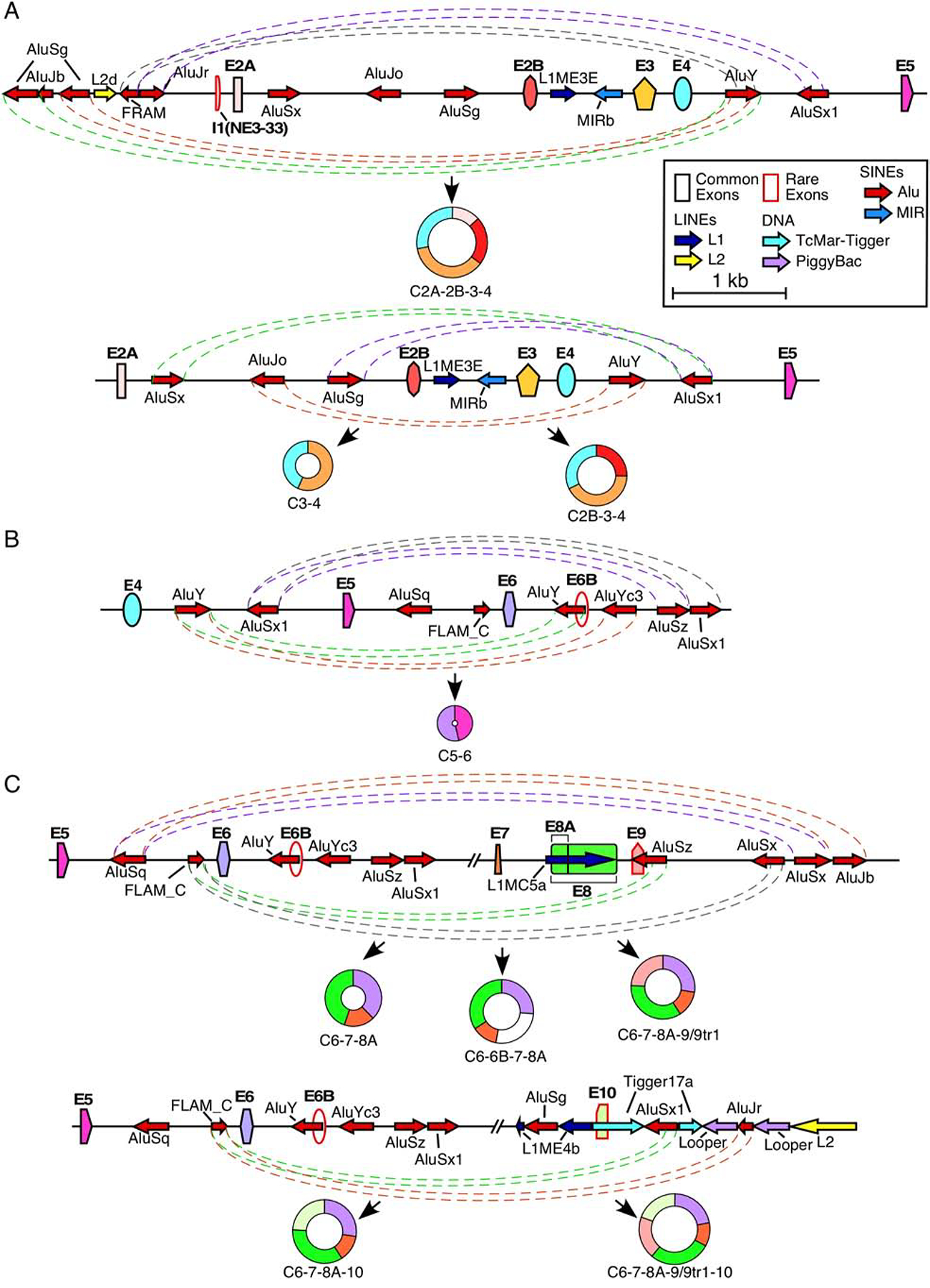
(A) Alu-mediated backsplicing of major type 1 circRNAs. Top panel shows the genomic overview of the region from ~2kb upstream of exon 2A to exon 5 and production of C2A-2B-3–4, bottom panel shows the region from exon 2A to exon 5 and production of C2B-3–4 and C3–4. Exons are shown as colored shapes and labeled in bold. Introns are represented by black lines. Repeat elements are represented by colored arrows, the direction of each arrow indicating the orientation of the repeat. Dashed colored lines denote potential pairings between complementary Alu elements. Coloring is arbitrary and is meant to aid in distinguishing between pairings. The circRNA(s) predicted to result from Alu:Alu base pairing is indicated below. Scale and color coding of exons and repeat sequences is given in the boxed region. (B) Alu-mediated backsplicing of C5–6, the major type 2 circRNA. The genomic overview of the region from exon 4 to ~2 kb downstream of exon 6 is given. Coloring and labeling are the same as in (A). (C) Alu-mediated backsplicing of major type 3 circRNAs. The top panel shows the genomic overview of the region from exon 5 to ~2 kb downstream of exon 9 and production of C6–7–8A, C6–6B–7–8A, and C6–7–8A–9/9tr1. The bottom panel shows the genomic overview of the region from exon 5 to ~2kb downstream of exon 10 and production of C6–7–8A–10 and C6–7–8A–9/9tr1–10. Coloring and labeling are the same as in (A).
The predominant type 2 circRNA, C5–6, is produced by pairing of the 5′ss of exon 6 with the 3′ss of exon 5. This splice site pairing is likely facilitated by RNA:RNA duplex formed between inverted Alu repeats located within introns 4 and 6. Of note, numerous Alu elements present within intron 6 can base pair with one of the two Alu elements present within intron 4 (Figure 1). In particular, one of the two antisense AluYs in the vicinity of 5′ss of exon 6 can potentially pair with the sense AluY located upstream of exon 4 (Figure 3B).
Almost all type 3 circRNAs utilize the 3′ss of exon 6 for backsplicing. Therefore, Alu sequences within intron 5 are likely to be critical for the formation of structures facilitating their backsplicing events. Intron 5 contains two Alu elements, an AluSq and a FLAM (Figure 3C). FLAM elements are ancestral Alu elements that contain only the left arm of the Alu consensus sequence. Interestingly, all of the downstream introns involved in the formation of type 3 circRNAs (e.g. introns 8A, 9, 10, 11, and 12) possess an antisense Alu element within 1 kb distance of the 5′ss involved (Figure 3C) [55]. Therefore, it is likely that in order to enable backsplicing the sense FLAM element forms an RNA:RNA duplex by base pairing with these antisense Alu elements.
3b. Backsplicing mediated by RNA binding proteins
Many RBPs have self-interacting domains, allowing them to dimerize or oligomerize on their target RNA molecules and as a consequence bring distantly located sequences in close proximity. Based on the type of looped out sequences due to self-dimerization/multimerization of proteins bound to distant motifs, skipping or inclusion of specific exons could be enabled [62]. Looping out can also result in backsplicing when RBPs bind near a 5′ss and the partner 3′ss comes from the upstream sequence. RBPs known to influence backsplicing include MBL, DHX9, FUS, Sam68, hnRNP L, and QKI [12,22,23,56,63,64].
As of now, DHX9 and Sam68 are the only two RBPs known to modulate backsplicing of specific SMN exons [55,56]. DHX9 is a ubiquitously expressed RNA helicase that unwinds Alu:Alu base pairing, thus inhibiting Alu-associated production of circRNAs [12]. Under conditions of DHX9 knockdown, some SMN exons, especially exons 3, 4, and 5, are skipped in linear transcripts producing the splice isoforms SMNΔ3–5 and SMNΔ3–5,7 [55]. At the same time, biogenesis of some circRNAs is strongly induced, with the biggest relative increase observed for C3–4 [55,56]. Since both exons 3 and 4 are included in the lariat products, it is likely that the increase in C3–4 is due to backsplicing occurring within the lariat intermediate containing exons 3, 4, and 5. Unlike DHX9 that inhibits generation of SMN circRNAs, Sam68 facilitates generation of specific circRNAs of SMN [56]. Knockdown of Sam68 results in a reduction of type 1 circRNAs as well as C6–7–8A–9tr1. In addition, deletion of potential Sam68 binding sites drastically reduces the circularization of C6–7–8A-9tr1 in a SMN minigene [56]. Similar results were also obtained in Sam68 knockout mice harboring the human SMN2 transgene [56].
3c. Backsplicing through a lariat intermediate
Lariats containing skipped exons may provide a constrained structural context favorable for backsplicing. However, it is not easy to prove that a lariat containing skipped exons was indeed the origin of a specific circRNA since the corroborating evidence, which is a linear byproduct(s) that lacks these very same exons, can be “destroyed” by nonsense-mediated decay (NMD). In mice carrying the human SMN2 transgene, SMN2 exons 3–5 and 5–7 can independently undergo co-skipping, producing lariats containing all exons found in C3–4 and C5–6, respectively [40]. In addition, all exons from 3 to 7 can also be skipped together under the conditions of oxidative stress [40]. Such skipping events could be conducive for the formation of specific circRNAs, including C3–4 and C5–6, although this possibility requires further investigation. As discussed above, the best evidence for SMN circRNA arising from backsplicing within a lariat containing skipped exons comes from DHX9 knockdown experiments described earlier [55].
4. Exon 8A splicing, downstream transcription, and type 3 circRNAs
There are two distinct features uniquely inherent to many of the type 3 circRNAs. Firstly, all type 3 circRNAs utilize a cryptic 5′ss located at the 142nd position of exon 8 [55]. Activation of this splice site produces a novel exon, i.e., exon 8A [55]. Secondly, many type 3 circRNAs contain exons located downstream of the canonical SMN cleavage and polyadenylation site (PAS) located within exon 8. An Alu element predicted to participate in exon 8A backsplicing to exon 6 is also located downstream of the canonical PAS. This implies that the transcription of intergenic sequences may be required for the generation of all type 3 circRNAs. The low levels of linear transcripts harboring intergenic exons may suggest their degradation by NMD [55]. It is also likely that the intergenic exons are predominantly incorporated into circRNAs rather than linear mRNA.
Recognition of the 5′ss by U1 snRNP is a critical early step during pre-mRNA splicing [65]. Recognition is achieved by base pairing of the 5′ss with the 5′ end of the U1 snRNA (5′ss:U1 base pairing) [66]. Recruitment of U1 snRNP to the 5′ss can be modulated by RNA secondary structure and/or RBPs interacting in the vicinity of the 5′ss. The 5′ss of exon 8A can form a moderately strong 5′ss:U1 duplex comprised of five canonical base pairs and an additional two wobble base pairs (Figure 4). Such a level of base pairing was previously found to activate the cryptic 5′ss (Cr1) located within intron 7 [53]. Therefore, it is likely that U1 snRNA binding in the vicinity of the 5′ss of exon 8A induces its activation in a similar manner [53,67]. Future studies will determine if the recognition of the 5′ss of exon 8A is modulated by RBPs interacting with nearby cis-elements.
Figure 4. Recognition of exon 8A 5′ss by U1 snRNA.
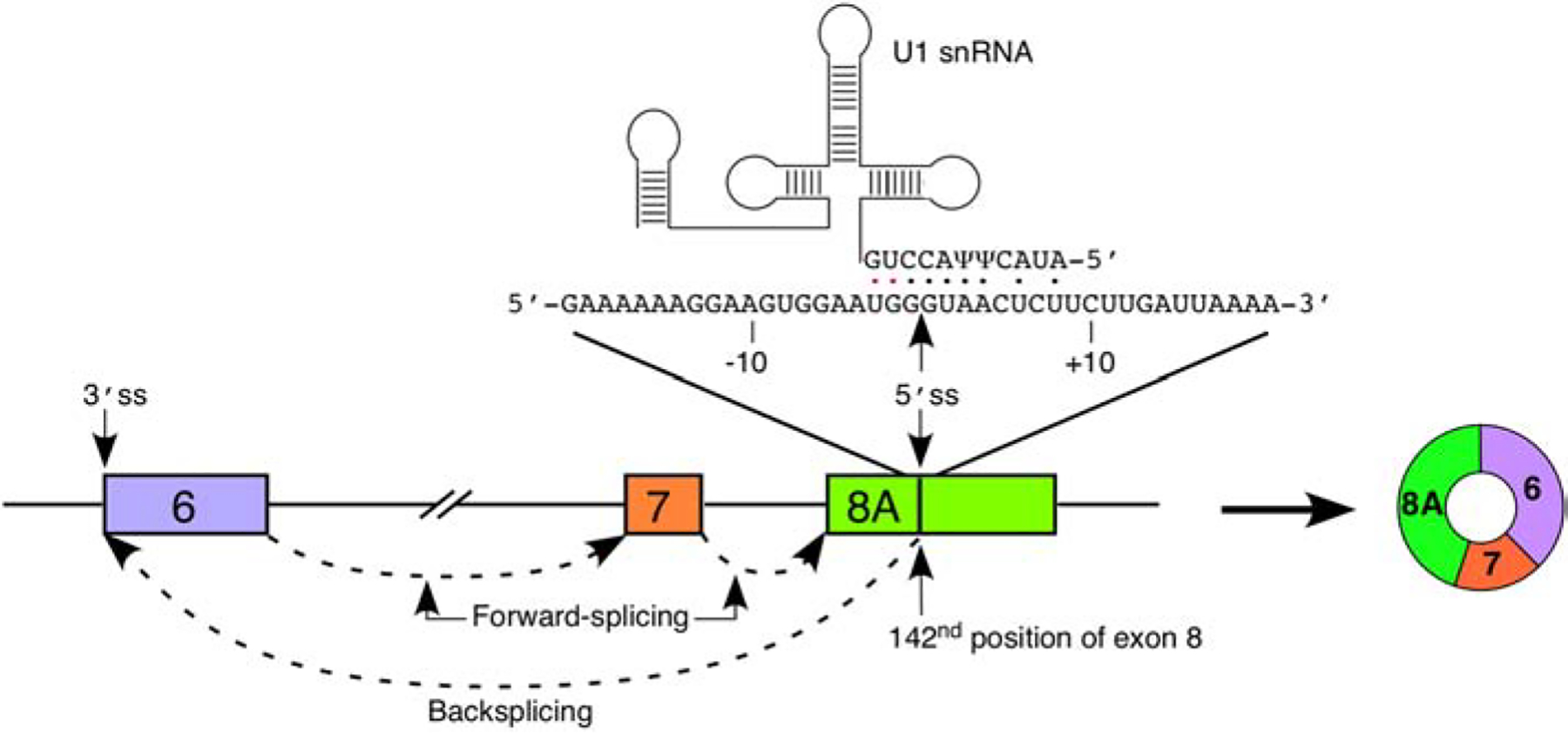
A diagram of exons 6 through 8 and intervening is given. Exons are shown as colored boxes, introns as lines/broken lines. Splicing events are indicated by dashed arrows. The sequence of exon 8 flanking the 8A 5′ss is given above the exon diagram. Numbering is given relative to the 8A 5′ss. The location of the 5′ss is indicated by an arrow. U1 snRNA is shown above. Base pairs between the 5′ portion of U1 snRNA and the 5′ss are indicated by dots; black dots indicate canonical base pairs and red dots indicate wobble base pairs. The primary splicing product of exon 8A recognition, C6–7–8A, is shown to the right. Of note, other circRNAs including additional exons are also produced downstream of recognition of the 5′ss of exon 8A (See Figures 2 and 3).
Currently, it is not known why a fraction of SMN transcripts bypass the canonical PAS. It may be a stochastic event, occurring at a baseline level due to a simple failure of the cleavage and polyadenylation machinery to assemble on the nascent transcript. It also may be due to a competing binding of RBP(s) to the 3′ untranslated region (3′UTR) of SMN that leads to inhibition of PAS recognition. Several RBPs, including Gemin5, U1A and HuR, have been proposed to bind to the 3′UTR of SMN [68–70]. It remains to be seen if an interaction of any of these factors with the 3′UTR of SMN is required for the intergenic transcription. U1 snRNP is recruited at more sites than are used for pre-mRNA splicing; this recruitment was shown to repress the usage of a potential PAS by a process known as “telescripting” [71,72]. Hence, it is likely that binding of U1 snRNP to the 5′ss of exon 8A inhibits usage of the canonical PAS and promotes intergenic transcription.
5. Functions of SMN circRNAs
The proposed functions of circRNAs fall into multiple categories: sponging of miRNAs or RBPs, assembly and recruitment of RBPs into complexes, generation of polypeptides and direct regulation of transcription. We briefly discuss each of these potential functions of SMN circRNAs.
5a. Sponging of miRNAs
CDR1as is one of the most studied circRNAs for its role in miRNA sponging as it encompasses 63 potential binding sites for miR-7 [7]. Overexpression of CDR1as recapitulates the effects of a miR-7 knockdown, suggesting a positive role of this circRNA in translation of mRNAs targeted by miR-7 [7]. CircSry is another circRNA implicated in miRNA sponging due to the presence of 16 target sites for miR-138 [8]. At the same time, circMTO1 contains only a single target site for miR-9 but still acts as its sponge inhibiting cell proliferation in hepatocellular carcinoma [11]. This finding underscores the important role of circRNAs in miRNA sponging independent of the number of binding sites of a given miRNA. However, arguments have also been made that just a few miRNA binding sites on the low abundant circRNAs may not capture sufficient numbers of miRNA molecules to have a significant biological effect [73].
As per the publicly available circInteractome database [74], two SMN circRNAs, C5–6 and C2B-3–4, potentially interact with AGO1 [36], which is a surrogate marker for miRNA binding [36,74]. Figure 5A shows the nine most highly expressed SMN circRNAs along with the miRNA target sites identified by two complementary miRNA binding site prediction tools [75,76]. MiR-510–3p, a miRNA linked to azoospermia [77], has a predicted binding site in the 5′ portion of exon 4 (Figure 5A) as well as the 3′UTR of SMN [36]. Thus, this miRNA could potentially interact with all type 1 circRNAs (Figure 5A), competing with binding to the SMN mRNA and likely impacting its translation. Notably, low SMN expression has been linked to the male infertility of SMA mouse models as well as mild SMA patients [78,79]. Another miRNA, miR-339–5p, which potentially targets the junction between exons 6 and 6B in C6–6B-7–8A (Figure 5A), is associated with Alzheimer’s disease [80]. MiR-2110 and miR-496 that have predicted binding sites within exons 3 and 10, respectively (Figure 5A), have been shown to regulate neuronal differentiation [81,82].
Figure 5. Predicted miRNA binding to SMN circRNAs.
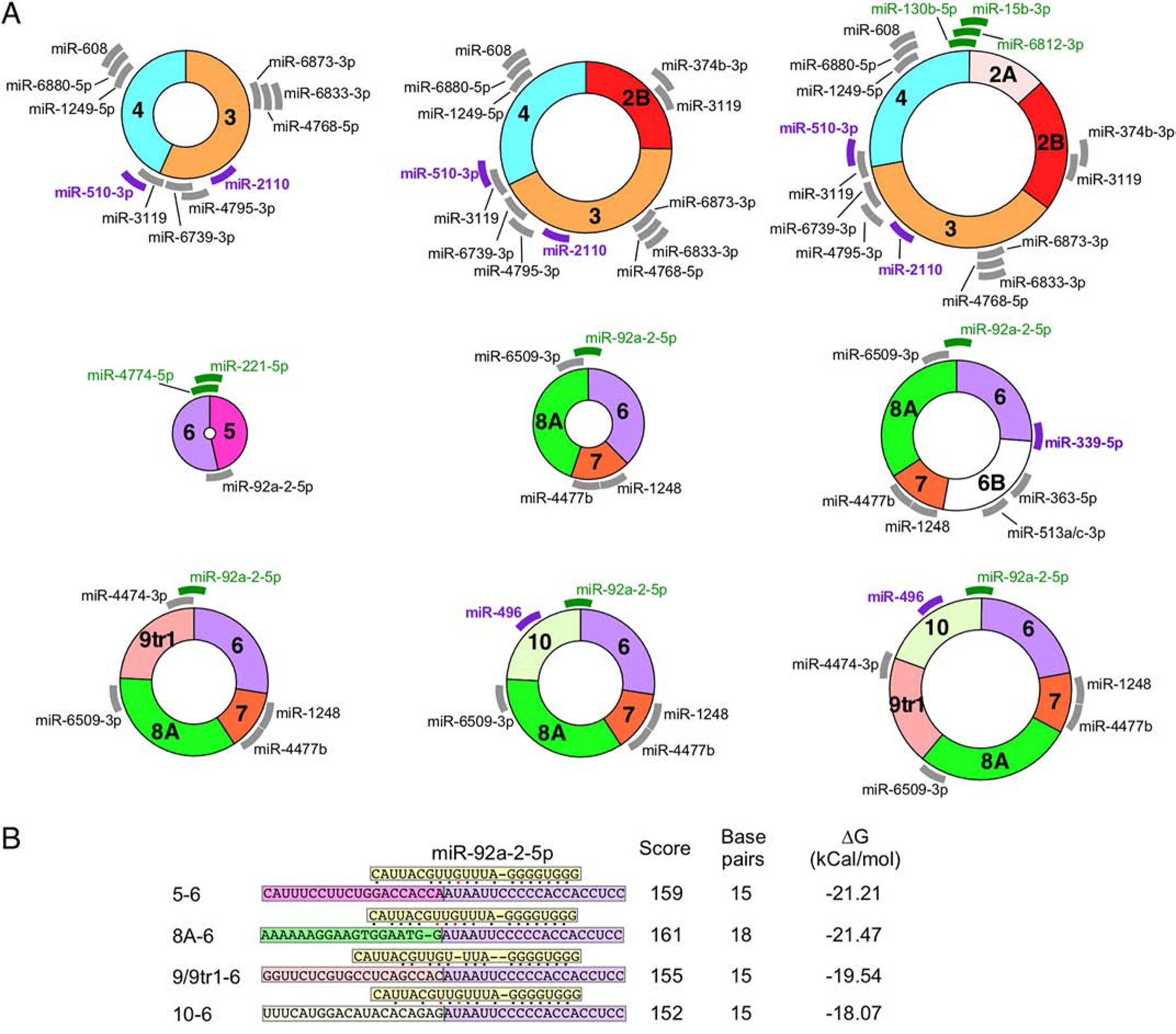
(A) The 9 SMN circRNAs with the highest predicted expression levels are indicated graphically. Each exon is shown in different colors. miRNA with predicted binding sites located in SMN circRNA are indicated with thick lines with the position representing the predicted binding site. Gray color represents miRNAs of unknown function. Purple color represents miRNAs with identified functions related to SMN biology and/or neurodegeneration. Green color represents miRNAs whose targets are located across the backsplice junction. Thus, these miRNAs bind differentially to circRNA as compared to their linear mRNA counterparts. (B) The predicted binding strength of miR-92a-2–5p targeting different SMN RNAs. miR-92a-2–5p (boxed in yellow) is shown binding to different splice junctions (boxed in exon-specific colors). Base pairs are indicated with dots; black dots indicate canonical base pairs and red dots indicate wobble base pairs. Locations of bulging bases are indicated by dashes. The identity of each junction is indicated at the left. The miRanda score, number of base pairs, and free energy change of binding are given at the right.
The backsplice junction provides a unique miRNA target specific to a given circRNA. For instance, miR-130b-5p, miR-6812–3p and miR-15b-3p are predicted to bind the backsplice junction between exons 4 and 2A in C2A-2B-3–4, which happens to be one of the abundantly expressed SMN circRNAs (Figure 5A). The backsplice junction between exons 6 and 5 creates potential binding sites for miR-4774–5p and miR-221–5p (Figure 5A). Interestingly, miR-92a-2–5p is predicted to interact with the backsplice junctions of multiple type 3 circRNAs as well as with the linear SMN transcript (Figures 5A and 5B). However, the strongest predicted binding of miR-92a-2–5p occurs in the context of C6–7–8A that furnishes 18 potential base pairs (between the miRNA and the target sequence) with a free energy change of −21.47 kCal/mol (Figure 5B). Contexts of linear SMN and other type 3 circRNAs are predicted to form 15 base pairs with free energy change between −18.07 and −21.21 kCal/mol, suggesting a slightly weaker interaction with miR-92a-2–5p (Figure 5B). Future experiments will reveal if slight variations in the free energy of miRNA interactions with their target sequences coupled with the changes in the relative expressions of type 3 circRNAs would have any physiological significance.
5b. Sponging and/or assembly of RBPs
RBP interactions with circRNAs have been less studied than miRNA sponging. This is likely due to the fact that sequence complementarity alone is enough to decide the fate of a miRNA binding to its target, therefore, miRNA targets are much easier to predict. However, a few clear examples of RBP sponging do exist. For example, circMBL contains multiple binding sites for the protein product of its host gene, MBL, presumably as part of a feedback loop to control MBL activity [12]. Sequestration of HuR, a multifunctional RBP, by circPABPN1 reduces the translation of HuR mRNA, supporting a point of view that circRNAs could play an important role in executing feedback loops [13]. In addition to the simple sequestration of RBPs, circRNAs could provide a much complex function such as acting as a scaffold for the assembly of multimeric complexes. For example, circFOXO3 binds both CDK2 and p21 in order to regulate cell cycle progression [83].
Due to extensive studies on SMN exon 7 splicing regulation, much is known about RBPs that bind to this exon (Table 1) [51,61,84–92]. Since exon 7 is present in nearly all type 3 circRNAs, it is likely that type 3 circRNAs indirectly regulate SMN exon 7 splicing by competing for RBP binding in the nucleus. It is also possible that these circRNAs shuttle some RBPs to the cytoplasm. Outside of exon 7, a few RBPs have been shown to bind to the 3′UTR of SMN [68–70]. However, the majority of relevant studies was focused on the 3′-most portion of exon 8, which is not present in circRNAs. Several RBPs are predicted by UV crosslinking and immunoprecipitation (CLIP) to interact with SMN RNA sequences, including all but one exon (exon 2A) present in SMN circRNAs (Table 1). Some of these RBPs, including hnRNP A1, FUS, TAF15, and TARDBP/TDP-43, are associated with neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) [93,94]. Some of these proteins (FUS and TDP43) directly interact with SMN, and perturbations of SMN function and/or localization have been linked to ALS and FTD [94–97]. Hence, it is likely that SMN circRNAs “cross-talk” with both SMN and SMN-interacting proteins implicated in ALS and FTD. SMN2 gene deletion and atypical SMN1 copy number are suggested risk factors for ALS [98]. It remains to be seen if the perturbations in the relative expression of SMN circRNAs due to an aberrant copy number of the SMN1 and/or SMN2 genes are pathogenic.
Table 1. RNA binding proteins predicted to interact with SMN circRNAs.
(A)Experimentally validated RBPs shown to interact with SMN sequences present in circRNAs. RBP name, SMN exon bound, and major circRNA isoforms that contain the target exon are given. Original studies identifying the RBP-SMN interaction are given in the ‘Reference’ column. (B)RBPs identified by CLIP and reported in public databases. The first three columns are the same as in (A). The fourth column lists the database(s) reporting CLIP tags.
| Table 1A. Experimentally validated RNA binding proteins | |||
|---|---|---|---|
| Protein | Exon | circRNAs | Reference |
| SF2/ASF (SRSF1) | 7 | Type 3 (all) | [84] |
| hnRNP A1/A2B1 | 7 | Type 3 (all) | [85] |
| Sam68 | 7 | Type 3 (all) | [86] |
| hnRNP Q | 7 | Type 3 (all) | [87] |
| Tra2-β | 7 | Type 3 (all) | [88,89] |
| hnRNP G | 7 | Type 3 (all) | [90] |
| hnRNP M | 7 | Type 3 (all) | [91] |
| PSF | 7 | Type 3 (all) | [92] |
| Table 1B. RNA binding proteins identified by CLIP | |||
| Protein | Exon | circRNAs | Database |
| FUS | 2B | C2A–2B–3–4, C2B–3–4 | CircInteractome, starBase |
| TAF15 | 2B, 7 | C2A–2B–3–4, C2B–3–4, Type 3 (all) | starBase |
| IGF2BP2 | All exons | All circRNA | starBase |
| AGO1 | 3, 5 | Type 1 (all), Type 2 (all) | CircInteractome |
| SF2/ASF (SRSF1) | 3, 4, 5, 7, 8A | Type 1 (all), Type 2 (all) | starBase |
| SRSF3 | 3, 4, 5, 8A | Type 1 (all), Type 2 (all) | starBase |
| MOV10 | 3, 8A | Type 1 (all), Type 3 (all) | starBase |
| U2AF2 | 4, 5, 7 | Type 1 (all), Type 2 (all) | starBase |
| YTHDC1 | 4, 8A | Type 1 (all), Type 3 (all) | starBase |
| EIF4A3 | 5 | Type 2 (all) | circInteractome, starBase |
| LIN28 | 6 | Type 2 (all), Type 3 (all) | circInteractome, starBase |
| hnRNP U | 6, 7 | Type 2 (all), Type 3 (all) | starBase |
| TARDBP | 6, 7 | Type 2 (all), Type 3 (all) | starBase |
| ILF3 | 6 | Type 2 (all), Type 3 (all) | starBase |
| KHDRBS1 | 7 | Type 3 (all) | starBase |
CircRNAs are known to have an altered profile of N6-adenosine methylation (m6A) modifications as compared to their linear counterparts [99]. YTHDC1, an m6A “reader” protein, participates in the recognition, binding, and nuclear export of m6A containing transcripts, along with SRSF3 [100]. YTHDC1 is predicted to bind to SMN exons 4 and 8A, and SRSF3 is predicted to bind to SMN exons 3, 4, 5, and 8A (Table 1). Future studies will determine whether some of the SMN circRNAs are m6A modified. IGF2BP2 (also known as IMP2) is predicted to bind extensively throughout both mRNA and circRNAs of SMN (Table 1). The IMP family of RBPs plays critical roles in mRNA stabilization and localization, and IGF2BP2 specifically is associated with a risk for type 2 diabetes [101]. The related protein, IMP1, interacts with SMN during the trafficking of cytoplasmic mRNP granules [102]. It is possible that SMN circRNAs play an important regulatory role by sequestering IMP1, IMP2, and IMP-containing complexes. EIF4A3 is predicted to interact with SMN exon 5, which is present in most type 2 circRNAs (Table 1). EIF4A3 is a multifunctional protein, which participates in the formation of the exon junction complex [103], circRNA generation [104], NMD [105], and ribosome biogenesis [106]. Of particular interest to SMN function, EIF4A3 plays a regulatory role in selenocysteine incorporation [107], a process in which SMN participates as well [108]. LIN28B protein is predicted to interact with exon 6, suggesting that it may bind both type 2 and type 3 circRNAs (Table 1). The canonical function of LIN28 proteins is to regulate let-7 family miRNAs, but it also directly binds to thousands of mRNAs [109].
5c. Potential translation of SMN circRNAs
CircRNAs have been reported to be translated, presumably through an internal ribosome entry site (IRES)-mediated process [14]. Due to diverse structural features, it is currently difficult to predict the presence of an IRES; therefore, any AUG (start codon) within circRNA could be considered as a potential translation start site in the context of a presumed IRES. Table 2 lists all AUGs that correspond to translation open reading frames (ORFs) of at least 10 amino acids of length. Every major SMN circRNA can potentially generate at least 6 peptides of various sizes. Interestingly, several circRNAs contain AUGs that are in-frame with the SMN ORF, meaning that they can replicate several domains of SMN. One such AUG lies in the 3′ portion of exon 4 and is present in all type 1 circRNAs. For C3–4 and C2B-3–4, rolling-circle translation would create a multimer containing the entire Tudor domain and its nearby sequences, while for C2A-2B-3–4, the multimer would additionally contain an RNA binding domain (Table 2) [110–112]. There are three in-frame AUGs in exon 6. For C5–6, the primary type 2 circRNA, rolling-circle translation would produce multiple copies of a proline-rich region of SMN, which serves as a binding site for Profilin, an actin cytoskeleton regulatory protein [113,114]. For type 3 circRNAs, the same AUGs would produce peptides that contain the YG box, an essential domain for the multimerization of SMN [115]. One could speculate that such peptides could either compete with full-length SMN for self-interaction, or alternatively interact with monomeric SMN to stabilize it, as monomeric SMN is highly unstable [41].
Table 2. Predicted translation products of SMN circRNAs.
All AUGs resulting in peptides of at least 10 amino acids are reported. The first column gives the circRNA name. Start and stop exons give the location of AUG start codon and UAG/UAA stop codons, respectively. Start position refers to the position of the first nucleotide of AUG within the start exon. Stop position refers to the position of the last nucleotide of UAG/UAA within the stop exon. Peptide length refers to the predicted size in amino acids of the translation product. The last column refers to whether the circRNA ORF contains portions of the primary in-frame SMN ORF. N/A: There is no predicted stop codon, resulting in rolling-circle translation. ∞: Due to rolling-circle translation, the predicted translation product is of indeterminate length.
| circRNA | Start exon | Start Position | Stop exon | Stop position | Peptide length | In frame? |
|---|---|---|---|---|---|---|
| C3–4 | 3 | 18 | 3 | 77 | 19 | No |
| 3 | 116 | 3 | 172 | 18 | No | |
| 3 | 191 | 3 | 172 | 111 | No | |
| 4 | 2 | 3 | 172 | 107 | No | |
| 4 | 8 | 3 | 172 | 105 | No | |
| 4 | 29 | 3 | 172 | 98 | No | |
| 4 | 93 | 3 | 77 | 45 | No | |
| 4 | 124 | N/A | N/A | ∞ | Yes | |
| C2B–3–4 | 2B | 14 | 3 | 172 | 92 | No |
| 3 | 18 | 3 | 77 | 19 | No | |
| 3 | 116 | 3 | 172 | 18 | No | |
| 3 | 191 | 2B | 10 | 57 | No | |
| 4 | 2 | 2B | 10 | 53 | No | |
| 4 | 8 | 2B | 10 | 51 | No | |
| 4 | 29 | 2B | 10 | 44 | No | |
| 4 | 93 | 2B | 20 | 26 | No | |
| 4 | 124 | N/A | N/A | ∞ | Yes | |
| C2A–2B–3–4 | 2A | 5 | 2A | 37 | 10 | No |
| 2A | 47 | 2B | 10 | 11 | No | |
| 2B | 14 | 3 | 172 | 92 | No | |
| 3 | 18 | 3 | 77 | 19 | No | |
| 3 | 116 | 3 | 172 | 18 | No | |
| 3 | 191 | 2A | 37 | 66 | No | |
| 4 | 2 | 2A | 37 | 62 | No | |
| 4 | 8 | 2A | 37 | 60 | No | |
| 4 | 29 | 2A | 37 | 53 | No | |
| 4 | 93 | 2A | 8 | 22 | No | |
| 4 | 124 | N/A | N/A | ∞ | Yes | |
| C5–6 | 5 | 17 | 6 | 4 | 27 | No |
| 5 | 63 | 6 | 41 | 24 | No | |
| 6 | 64 | N/A | N/A | α | Yes | |
| 6 | 75 | 6 | 41 | 57 | No | |
| 6 | 82 | N/A | N/A | ∞ | Yes | |
| 6 | 109 | N/A | N/A | ∞ | Yes | |
| C6–7–8A | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 6 | 41 | 14 | No | |
| C6–7–8A–9 | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 6 | 41 | 48 | No | |
| C6–7–8A–9trl | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 7 | 46 | 87 | Yes | |
| C6–7–8A–10 | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 10 | 32 | 11 | No | |
| 10 | 62 | 6 | 41 | 25 | No | |
| 10 | 83 | 6 | 41 | 18 | No | |
| C6–6B–7–8A | 6 | 64 | 6B | 51 | 32 | Yes |
| 6 | 75 | 6B | 20 | 18 | No | |
| 6 | 82 | 6B | 51 | 26 | Yes | |
| 6 | 109 | 6B | 51 | 17 | Yes | |
| 6B | 93 | 7 | 46 | 20 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 6 | 41 | 48 | No | |
| C6–7–8A–9–10 | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 10 | 32 | 45 | No | |
| 10 | 62 | 6 | 41 | 25 | No | |
| 10 | 83 | 6 | 41 | 18 | No | |
| C6–7–8A–9trl-10 | 6 | 64 | 7 | 46 | 32 | Yes |
| 6 | 75 | 8A | 26 | 38 | No | |
| 6 | 82 | 7 | 46 | 26 | Yes | |
| 6 | 109 | 7 | 46 | 17 | Yes | |
| 8A | 27 | 8A | 68 | 13 | No | |
| 8A | 139 | 10 | 45 | 48 | No | |
| 10 | 62 | 6 | 41 | 25 | No | |
| 10 | 83 | 6 | 41 | 18 | No |
6. Conclusions
The SMN protein carries out surprisingly diverse cellular functions [33]. Likewise, alternative splicing, transcription initiation, and 3′ end processing result in a stunning array of transcripts, both linear and circular, from the SMN1 and SMN2 loci [36]. The SMN genes produce a very large number of circRNAs, containing sequences from every one of annotated SMN exons as well as exons never before detected in linear SMN mRNA [55,56]. Although no single circRNA is expressed at high levels compared to the major linear SMN transcripts (e.g. full-length and exon 7-skipped transcripts), the sheer number of unique SMN circRNAs results in a total sum of expression that is likely to be substantial.
The strongest driver of SMN circRNA production appears to be the high content of Alu-derived sequences within SMN introns. Sam68 enhances circularization by interacting with the intronic sequences in the vicinity of some Alu elements [56]. Several other proteins likely promote backsplicing by binding to intronic sequences close to the splice sites of SMN exons. Low SMN levels are linked to alternative transcription termination by two distinct mechanisms. Firstly, SMN interacts with the C-terminal domain of polymerase II (PolII CTD) and Senataxin to mediate the resolution of RNA:DNA hybrids known as R-loops at the 3′ end of genes [116]. Secondly, SMN is required for the proper assembly of U1 snRNP that participates in telescripting in which recruitment of U1 snRNP at random locations on the elongating transcripts inhibits cleavage and polyadenylation at the nearby PASs [72,117]. Therefore, it is likely that the production of type 3 SMN circRNAs that contain sequences downstream of the canonical PAS can be directly linked to SMN functions through a variety of feedback mechanisms. In fact, several proteins whose functions are tied to SMN are predicted to bind to exons contained in SMN circRNAs, highlighting a possible regulatory relationship.
One of the major hopes for circRNA research is that, due to their high stability and persistent presence in biological fluids such as blood, saliva, and urine, they may serve as disease biomarkers and/or outcome measures for the therapeutic treatments [118]. As new therapies for SMA are being developed and the existing treatment regimens continue to evolve over time, there is a growing need for non-invasive, unbiased outcome measures. Specific SMN circRNAs could serve as such outcome measures if their relative expressions against other circRNAs are firmly established. Considering SMN circRNAs are likely to be part of the feedback mechanism that regulates SMN function, they could potentially serve as therapeutic targets for SMA as well. Some of the SMN circRNAs, including C2A-2B-3–4, C4 and C5–6–7, are present in both human and mouse, supporting their conserved function in mammals [55]. However, several of the SMN circRNAs, such as C2B-34, C3–4 and all type 3 circRNAs, appear to be specific to primates, supporting their evolutionary significance. Now that the generation of the vast repertoire of SMN circRNAs is independently confirmed, future studies will reveal if SMN circRNAs are as integral to cellular functions as the SMN protein.
Acknowledgements
Authors acknowledge members of the Singh lab for critical reading of the manuscript and for valuable discussions and suggestions. While authors have attempted to include most contributions on SMN transcript diversity, they regret not being able to include several related references due to the lack of space.
Funding: This work was supported by grants from the National Institutes of Health (R01 NS055925).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures and competing interests: ISS-N1 target (US patent # 7,838,657) mentioned in this review was discovered in the Singh lab at UMASS Medical School (Worcester, MA, USA). Inventors, including RNS and UMASS Medical School, are currently benefiting from licensing of ISS-N1 target to IONIS Pharmaceuticals/Biogen, which is marketing Spinraza™ (Nusinersen), the FDA-approved drug, based on ISS-N1 target. RNS is co-founder of RNACorrect, Inc., an Iowa-based small business engaged in research and development.
References
- [1].Mance LG, Mawla I, Shell SM, Cahoon AB, Mitochondrial mRNA fragments are circularized in a human HEK cell line. Mitochondrion. 51 (2020) 1–6. [DOI] [PubMed] [Google Scholar]
- [2].LaRoche-Johnston F F, Monat C, Coulombe S, Cousineau B B, Bacterial group II introns generate genetic diversity by circularization and trans-splicing from a population of intron-invaded mRNAs, PLoS Genet 21(14) (2018) 11:e1007792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J, Circular RNA Is Expressed across the Eukaryotic Tree of Life, Plos One 9(3) (2014) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO, Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types, Plos One 7(2) (2012) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B, Scrambled exons, Cell 64(3) (1991) 607–13. [DOI] [PubMed] [Google Scholar]
- [6].Cocquerelle C, Mascrez B, Hétuin D, Bailleul B, Mis-splicing yields circular RNA molecules, FASEB J 7(1) (1993) 155–60. [DOI] [PubMed] [Google Scholar]
- [7].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N, Circular RNAs are a large class of animal RNAs with regulatory potency, Nature 495(7441) (2013) 333–338. [DOI] [PubMed] [Google Scholar]
- [8].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, Natural RNA circles function as efficient microRNA sponges, Nature 495(7441) (2013) 384–388. [DOI] [PubMed] [Google Scholar]
- [9].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu JZ, Marzluff WF, Sharpless NE, Circular RNAs are abundant, conserved, and associated with ALU repeats, Rna-a Publication of the Rna Society 19(2) (2013) 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zheng QP, Bao CY, Guo WJ, Li SY, Chen J, Chen B, Luo YT, Lyu DB, Li Y, Shi GH, Liang LH, Gu JR, He XH, Huang SL, Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs, Nature Communications 7 (2016) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han D, Li JX, Wang HM, Su XP, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang MY, Li N, Zhou WP, Yu YZ, Cao XT, Circular RNA circMTO1 Acts as the Sponge of MicroRNA-9 to Suppress Hepatocellular Carcinoma Progression, Hepatology 66(4) (2017) 1151–1164. [DOI] [PubMed] [Google Scholar]
- [12].Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S, circRNA Biogenesis Competes with Pre-mRNA Splicing, Molecular Cell 56(1) (2014) 55–66. [DOI] [PubMed] [Google Scholar]
- [13].Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M, Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1, Rna Biology 14(3) (2017) 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S, Translation of CircRNAs, Molecular Cell 66(1) (2017) 9-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY, Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes, Science 329(5992) (2010) 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yoon JH, Abdelmohsen K, Kim J, Yang XL, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M, Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination, Nature Communications 4 (2013) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].d’Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ, The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy, Neuron (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Woo CJ, Maier VK, Davey R, Brennan J, Li GD, Brothers J, Schwartz B, Gordo S, Kasper A, Okamoto TR, Johansson HE, Mandefro B, Sareen D, Bialek P, Chau BN, Bhat B, Bullough D, Barsoum J, Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy, Proceedings of the National Academy of Sciences of the United States of America 114(8) (2017) E1509–E1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zaghlool A, Ameur A, Wu CL, Westholm JO, Niazi A, Manivannan M, Bramlett K, Nilsson M, Feuk L, Expression profiling and in situ screening of circular RNAs in human tissues, Scientific Reports 8 (2018) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li ZY, Huang C, Bao C, Chen L, Lin M, Wang XL, Zhong GL, Yu B, Hu WC, Dai LM, Zhu PF, Chang ZX, Wu QF, Zhao Y, Jia Y, Xu P, Liu HJ, Shan G, Exon-intron circular RNAs regulate transcription in the nucleus, Nature Structural & Molecular Biology 22(3) (2015) 256–264. [DOI] [PubMed] [Google Scholar]
- [21].Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL, Circular intronic long noncoding RNAs. Mol Cell. 51 (2013) 792–806. [DOI] [PubMed] [Google Scholar]
- [22].Aktas T, Ilik IA, Maticzka D, Bhardwaj V, Rodrigues CP, Mittler G, Manke T, Backofen R, Akhtar A, DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome, Nature 544(7648) (2017) 115-+. [DOI] [PubMed] [Google Scholar]
- [23].Errichelli L, Modigliani SD, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, De Santis R, Scarfo R, Peruzzi G, Lu L, Caffarelli E, Shneider NA, Morlando M, Bozzoni I, FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons, Nature Communications 8 (2017) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Barrett SP, Wang PL, Salzman J, Circular RNA biogenesis can proceed through an exon-containing lariat precursor, Elife 4 (2015) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Talhouarne GJS, Gall JG, Lariat intronic RNAs in the cytoplasm of vertebrate cells, Proceedings of the National Academy of Sciences of the United States of America 115(34) (2018) E7970–E7977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Deininger P, Alu elements: know the SINEs, Genome Biology 12(12) (2011) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].International Human Genome Sequencing Consortium, Initial sequencing and analysis of the human genome, Nature 409(6822) (2001) 860–921. [DOI] [PubMed] [Google Scholar]
- [28].Kapitonov V, Jurka J, The age of Alu subfamilies, Journal of Molecular Evolution 42(1) (1996) 59–65. [DOI] [PubMed] [Google Scholar]
- [29].de Koning APJ, Gu WJ, Castoe TA, Batzer MA, Pollock DD, Repetitive Elements May Comprise Over Two-Thirds of the Human Genome, Plos Genetics 7(12) (2011) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G, Intronic Alus Influence Alternative Splicing, Plos Genetics 4(9) (2008) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shen SH, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, Sato S, Davidson BL, Xing Y, Widespread establishment and regulatory impact of Alu exons in human genes, Proceedings of the National Academy of Sciences of the United States of America 108(7) (2011) 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sorek R, Lev-Maor G G, Reznik M, Dagan T, Belinky F, Graur D, Ast G, Minimal conditions for exonization of intronic sequences: 5’ splice site formation in alu exons. Mol Cell. 14 (2004) 221–31. [DOI] [PubMed] [Google Scholar]
- [33].Singh RN, Howell MD, Ottesen EW, Singh NN, Diverse role of survival motor neuron protein, Biochim Biophys Acta 1860(3) (2017) 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ahmad S, Bhatia K, Kannan A, Gangwani L, Molecular Mechanisms of Neurodegeneration in Spinal Muscular Atrophy, Journal of Experimental Neuroscience 10 (2016) 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wirth B, Karakaya M, Kye MJ, Mendoza-Ferreira N, Twenty-Five Years of Spinal Muscular Atrophy Research: From Phenotype to Genotype to Therapy, and What Comes Next, Annu Rev Genomics Hum Genet (2020). [DOI] [PubMed] [Google Scholar]
- [36].Singh NN, Ottesen EW, Singh RN (2020) A survey of transcripts generated by spinal muscular atrophy genes. Biochim Biophys Acta Gene Regul Mech. 2020 (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lorson CL, Hahnen E, Androphy EJ, Wirth B, A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy, Proceedings of the National Academy of Sciences of the United States of America 96(11) (1999) 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AHM, McPherson JD, A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2, Human Molecular Genetics 8(7) (1999) 1177–1183. [DOI] [PubMed] [Google Scholar]
- [39].Cho SC, Dreyfuss G, A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity, Genes & Development 24(5) (2010) 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN, A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein, Scientific Reports 6 (2016) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gray KM, Kaifer KA, Baillat D, Wen Y, Bonacci TR, Ebert AD, Raimer AC, Spring AM, ten Have S, Glascock JJ, Gupta K, Van Duyne GD, Emanuele MJ, Lamond AI, Wagner EJ, Lorson CL, Matera AG, Self-oligomerization regulates stability of survival motor neuron protein isoforms by sequestering an SCFSlmb degron, Molecular Biology of the Cell 29(2) (2018) 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Seo J, Howell MD, Singh NN, Singh RN, Spinal muscular atrophy: An update on therapeutic progress, Biochimica Et Biophysica Acta-Molecular Basis of Disease 1832(12) (2013) 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sivanesan S, Howell MD, DiDonato CJ, Singh RN, Antisense oligonucleotide mediated therapy of spinal muscular atrophy, Translational Neuroscience 4(1) (2013) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Howell MD, Singh NN, Singh RN, Advances in therapeutic development for spinal muscular atrophy, Future Medicinal Chemistry 6(9) (2014) 1081–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ottesen EW, ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy, Translational Neuroscience 8 (2017) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Singh NN, Howell MD, Androphy EJ, Singh RN, How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy, Gene Therapy 24(9) (2017) 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Singh NK, Singh NN, Androphy EJ, Singh RN, Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron, Molecular and Cellular Biology 26(4) (2006) 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang JX, Schultz PG, Johnson KA, Mechanistic studies of a small-molecule modulator of SMN2 splicing, Proceedings of the National Academy of Sciences of the United States of America 115(20) (2018) E4604–E4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cheung AK, Hurley B, Kerrigan R, Shu L, Chin DN, Shen YP, O’Brien G, Sung MJ, Hou Y, Axford J, Cody E, Sun R, Fazal A, Fridrich C, Sanchez CC, Tomlinson RC, Jain M, Deng L, Hoffmaster K, Song C, Van Hoosear M, Shin Y, Servais R, Towler C, Hild M, Curtis D, Dietrich WF, Hamann LG, Briner K, Chen KS, Kobayashi D, Sivasankaran R, Dales NA, Discovery of Small Molecule Splicing Modulators of Survival Motor Neuron-2 (SMN2) for the Treatment of Spinal Muscular Atrophy (SMA), Journal of Medicinal Chemistry 61(24) (2018) 11021–11036. [DOI] [PubMed] [Google Scholar]
- [50].Singh NN, Seo J, Rahn SJ, Singh RN, A Multi-Exon-Skipping Detection Assay Reveals Surprising Diversity of Splice Isoforms of Spinal Muscular Atrophy Genes, Plos One 7(11) (2012) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Singh NN, Howell MD, Singh RN, Transcriptional and Splicing Regulation of Spinal Muscular Atrophy Genes, in: Charlotte SJ, Paushkin S, Ko C-P (Eds.), Spinal Muscular Atrophy: Disease Mechanisms and Therapy, Elsevier Inc; 2016. [Google Scholar]
- [52].Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN, Oxidative Stress Triggers Body-Wide Skipping of Multiple Exons of the Spinal Muscular Atrophy Gene, Plos One 11(4) (2016) 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Singh NN, del Rio-Malewski JB, Luo D, Ottesen EW, Howell MD, Singh RN, Activation of a cryptic 5 ‘ splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene, Nucleic Acids Research 45(21) (2017) 12214–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ottesen EW, Seo J, Singh NN, Singh RN, A Multilayered Control of the Human Survival Motor Neuron Gene Expression by Alu Elements, Frontiers in Microbiology 8 (2017) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ottesen EW, Luo D, Seo J, Singh NN, Singh RN, Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs, Nucleic Acids Research 47(6) (2019) 2884–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pagliarini V, Jolly A, Bielli P, Di Rosa V, De la Grange P, Sette C, Sam68 binds Alu-rich introns in SMN and promotes pre-mRNA circularization, Nucleic Acids Research 48(2) (2020) 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO, Cell-Type Specific Features of Circular RNA Expression, Plos Genetics 9(9) (2013) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Welden JR, Stamm S, Pre-mRNA structures forming circular RNAs, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1862(11–12) (2019) 6. [DOI] [PubMed] [Google Scholar]
- [59].Pervouchine DD, Circular exonic RNAs: When RNA structure meets topology, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1862(11–12) (2019) 7. [DOI] [PubMed] [Google Scholar]
- [60].Andrews RJ, Moss WN, Computational approaches for the discovery of splicing regulatory RNA structures, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1862(11–12) (2019) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Singh NN, Singh RN, How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1862(11–12) (2019) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Martinez-Contreras R, Fisette JF, Nasim FH, Madden R, Cordeau M, Chabot B, Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing, Plos Biology 4(2) (2006) 172–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fei T, Chen YW, Xiao TF, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG, Wu Q, Feng FY, Loda M, He HSH, Liu XS, Brown M, Genomewide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing, Proceedings of the National Academy of Sciences of the United States of America 114(26) (2017) E5207–E5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ, The RNA Binding Protein Quaking Regulates Formation of circRNAs, Cell 160(6) (2015) 1125–1134. [DOI] [PubMed] [Google Scholar]
- [65].Matlin AJ, Clark F, Smith CWJ, Understanding alternative splicing: Towards a cellular code, Nature Reviews Molecular Cell Biology 6(5) (2005) 386–398. [DOI] [PubMed] [Google Scholar]
- [66].Roca X, Krainer AR, Eperon IC, Pick one, but be quick: 5 ‘ splice sites and the problems of too many choices, Genes & Development 27(2) (2013) 129–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Singh RN, Singh NN, A novel role of U1 snRNP: Splice site selection from a distance, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1862(6) (2019) 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Farooq F, Balabanian S, Liu XJ, Holcik M, MacKenzie A, p38 Mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR, Human Molecular Genetics 18(21) (2009) 4035–4045. [DOI] [PubMed] [Google Scholar]
- [69].Workman E, Veith A, Battle DJ, U1A Regulates 3 ‘ Processing of the Survival Motor Neuron mRNA, Journal of Biological Chemistry 289(6) (2014) 3703–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Workman E, Kalda C, Patel A, Battle DJ, Gemin5 Binds to the Survival Motor Neuron mRNA to Regulate SMN Expression, Journal of Biological Chemistry 290(25) (2015) 15662–15669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Oh JM, Di C, Venters CC, Guo JN, Arai C, So BR, Pinto AM, Zhang ZX, Wan LL, Younis I, Dreyfuss G, U1 snRNP telescripting regulates a size-function-stratified human genome, Nature Structural & Molecular Biology 24(11) (2017) 993-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Venters CC, Oh JM, Di C, So BR, Dreyfuss G, U1 snRNP Telescripting: Suppression of Premature Transcription Termination in Introns as a New Layer of Gene Regulation, Cold Spring Harbor Perspectives in Biology 11(2) (2019) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Thomson DW, Dinger ME, Endogenous microRNA sponges: evidence and controversy, Nature Reviews Genetics 17(5) (2016) 272–283. [DOI] [PubMed] [Google Scholar]
- [74].Dudekulay DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M, CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs, Rna Biology 13(1) (2016) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS, MicroRNA targets in Drosophila, Genome Biology 5(1) (2004) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Liu WJ, Wang XW, Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data, Genome Biology 20 (2019) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ji J, Qin YF, Zhou R, Zang RJ, Huang ZY, Zhang Y, Chen MJ, Wu W, Song L, Ling XF, Shen HB, Hu ZB, Xia YK, Lu CC, Wang XR, X chromosome-wide identification of SNVs in microRNA genes and non-obstructive azoospermia risk in han chinese population, Oncotarget 7(31) (2016) 49122–49129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ottesen EW, Howell MD, Singh NN, Seo J, Whitley EM, Singh RN, Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy, Scientific Reports 6 (2016) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lipnick SL, Agniel DM, Aggarwal R, Makhortova NR, Finlayson SG, Brocato A, Palmer N, Darras BT, Kohane I, Rubin LL, Systemic nature of spinal muscular atrophy revealed by studying insurance claims, Plos One 14(3) (2019) 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Long JM, Ray B, Lahiri DK, MicroRNA-339–5p Down-regulates Protein Expression of beta-Site Amyloid Precursor Protein-Cleaving Enzyme 1 (BACE1) in Human Primary Brain Cultures and Is Reduced in Brain Tissue Specimens of Alzheimer Disease Subjects*, Journal of Biological Chemistry 289(8) (2014) 5184–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhao ZZ, Partridge V, Sousares M, Shelton SD, Holland CL, Pertsemlidis A, Du LQ, microRNA-2110 functions as an oncosuppressor in neuroblastoma by directly targeting Tsukushi, Plos One 13(12) (2018) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rago L, Beattie R, Taylor V, Winter J, miR379–410 cluster miRNAs regulate neurogenesis and neuronal migration by fine-tuning N-cadherin, Embo Journal 33(8) (2014) 906–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Du WW, Yang WN, Liu E, Yang ZG, Dhaliwal P, Yang BB, Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2, Nucleic Acids Research 44(6) (2016) 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Cartegni L, Krainer AR, Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1, Nat Genet 30(4) (2002) 377–84. [DOI] [PubMed] [Google Scholar]
- [85].Kashima T, Rao N, Manley JL, An intronic element contributes to splicing repression in spinal muscular atrophy, Proceedings of the National Academy of Sciences of the United States of America 104(9) (2007) 3426–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C, The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy, Embo Journal 29(7) (2010) 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY, The RNA Binding Protein hnRNP Q Modulates the Utilization of Exon 7 in the Survival Motor Neuron 2 (SMN2) Gene, Molecular and Cellular Biology 28(22) (2008) 6929–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B, Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2), Proceedings of the National Academy of Sciences of the United States of America 97(17) (2000) 9618–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Chen YC, Chang JG, Jong YJ, Liu TY, Yuo CY, High Expression Level of Tra2-beta 1 Is Responsible for Increased SMN2 Exon 7 Inclusion in the Testis of SMA Mice, Plos One 10(3) (2015) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Moursy A, Allain FHT, Clery A, Characterization of the RNA recognition mode of hnRNP G extends its role in SMN2 splicing regulation, Nucleic Acids Research 42(10) (2014) 6659–6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Cho S, Moon H, Loh TJ, Oh HK, Williams DR, Liao DJ, Zhou JH, Green MR, Zheng XX, Shen HH, PSF contacts exon 7 of SMN2 pre-mRNA to promote exon 7 inclusion, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1839(6) (2014) 517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cho S, Moon H, Loh TJ, Oh HK, Choy HE, Song WK, Chun JS, Zheng X, Shen H, hnRNP M facilitates exon 7 inclusion of SMN2 pre-mRNA in spinal muscular atrophy by targeting an enhancer on exon 7, Biochimica Et Biophysica Acta-Gene Regulatory Mechanisms 1839(4) (2014) 306–315. [DOI] [PubMed] [Google Scholar]
- [93].Weishaupt JH, Hyman T, Dikic I, Common Molecular Pathways in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia, Trends in Molecular Medicine 22(9) (2016) 769–783. [DOI] [PubMed] [Google Scholar]
- [94].Chi BK, O’Connell JD, Iocolano AD, Coady JA, Yu Y, Gangopadhyay J, Gygi SP, Reed R, The neurodegenerative diseases ALS and SMA are linked at the molecular level via the ASC-1 complex, Nucleic Acids Research 46(22) (2018) 11939–11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Yamazaki T, Chen S, Yu Y, Yan BA, Haertlein TC, Carrasco MA, Tapia JC, Zhai B, Das R, Lalancette-Hebert M, Sharma A, Chandran S, Sullivan G, Nishimura AL, Shaw CE, Gygi SP, Shneider NA, Maniatis T, Reed R, FUS-SMN Protein Interactions Link the Motor Neuron Diseases ALS and SMA, Cell Reports 2(4) (2012) 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Sun SY, Ling SC, Qiu JS, Albuquerque CP, Zhou Y, Tokunaga S, Li HR, Qiu HY, Bui A, Yeo GW, Huang EJ, Eggan K, Zhou HL, Fu XD, Lagier-Tourenne C, Cleveland DW, ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP, Nature Communications 6 (2015) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Groen EJN, Fumoto K, Blokhuis AM, Engelen-Lee J, Zhou YP, van den Heuvel DMA, Koppers M, van Diggelen F, van Heest J, Demmers JAA, Kirby J, Shaw PJ, Aronica E, Spliet WGM, Veldink JH, van den Berg LH, Pasterkamp RJ, ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN, Human Molecular Genetics 22(18) (2013) 3690–3704. [DOI] [PubMed] [Google Scholar]
- [98].Butchbach ME, Copy Number Variations in the Survival Motor Neuron Genes: Implications for Spinal Muscular Atrophy and Other Neurodegenerative Diseases, Front Mol Biosci 3 (2016) 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang JK, Van Wittenberghe N, Xing Y, Giallourakis CC, Mullen AC, Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs, Cell Reports 20(9) (2017) 2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Roundtree IA, Luo GZ, Zhang ZJ, Wang X, Zhou T, Cui YQ, Sha JH, Huang XX, Guerrero L, Xie P, He E, Shen B, He C, YTHDC1 mediates nuclear export of N-6 - methyladenosine methylated mRNAs, Elife 6 (2017) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Christiansen J, Kolte AM, Hansen TVO, Nielsen FC, IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes, Journal of Molecular Endocrinology 43(5–6) (2009) 187–195. [DOI] [PubMed] [Google Scholar]
- [102].Fallini C, Rouanet JP, Donlin-Asp PG, Guo P, Zhang HL, Singer RH, Rossoll W, Bassell GJ, Dynamics of Survival of Motor Neuron (SMN) Protein Interaction with the mRNA-Binding Protein IMP1 Facilitates Its Trafficking into Motor Neuron Axons, Developmental Neurobiology 74(3) (2014) 319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chan CC, Dostie J, Diem MD, Feng WQ, Mann M, Rappsilber J, Dreyfuss G, eIF4A3 is a novel component of the exon junction complex, Rna 10(2) (2004) 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wang RJ, Zhang S, Chen XY, Li N, Li JW, Jia RC, Pan YQ, Liang HQ, EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis, Molecular Cancer 17 (2018) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Isken O, Maquat LE, The multiple lives of NMD factors: balancing roles in gene and genome regulation, Nature Reviews Genetics 9(9) (2008) 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Alexandrov A, Colognori D, Steitz JA, Human eIF4AIII interacts with an eIF4G-like partner, NOM1, revealing an evolutionarily conserved function outside the exon junction complex, Genes & Development 25(10) (2011) 1078–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Budiman ME, Bubenik JL, Miniard AC, Middleton LM, Gerber CA, Cash A, Driscoll DM, Eukaryotic Initiation Factor 4a3 Is a Selenium-Regulated RNA-Binding Protein that Selectively Inhibits Selenocysteine Incorporation, Molecular Cell 35(4) (2009) 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Gribling-Burrer AS, Leichter M, Wurth L, Huttin A, Schlotter F, Troffer-Charlier N, Cura V, Barkats M, Cavarelli J, Massenet S, Allmang C, SECIS-binding protein 2 interacts with the SMN complex and the methylosome for selenoprotein mRNP assembly and translation, Nucleic Acids Research 45(9) (2017) 5399–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Hafner M, Max KEA, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T, Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition, Rna 19(5) (2013) 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S, The RNA-binding properties of SMN: deletion analysis of the zebrafish orthologue defines domains conserved in evolution, Human Molecular Genetics 8(5) (1999) 775–782. [DOI] [PubMed] [Google Scholar]
- [111].Lorson CL, Androphy EJ, The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding, Human Molecular Genetics 7(8) (1998) 1269–1275. [DOI] [PubMed] [Google Scholar]
- [112].Ottesen EW, Singh NN, Luo D, Singh RN, High-affinity RNA targets of the Survival Motor Neuron protein reveal diverse preferences for sequence and structural motifs, Nucleic Acids Research 46(20) (2018) 10983–11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Giesemann T, Rathke-Hartlieb S, Rothkegel M, Bartsch JW, Buchmeier S, Jockusch BM, Jockusch H, A role for polyproline motifs in the spinal muscular atrophy protein SMN - Profilins bind to and colocalize with SMN in nuclear gems, Journal of Biological Chemistry 274(53) (1999) 37908–37914. [DOI] [PubMed] [Google Scholar]
- [114].Nolle A, Zeug A, van Bergeijk J, Tonges L, Gerhard R, Brinkmann H, Al Rayes S, Hensel N, Schill Y, Apkhazava D, Jablonka S, O’Mer J, Srivastav RK, Baasner A, Lingor P, Wirth B, Ponimaskin E, Niedenthal R, Grothe C, Claus P, The spinal muscular atrophy disease protein SMN is linked to the rho-kinase pathway via profilin, Human Molecular Genetics 20(24) (2011) 4865–4878. [DOI] [PubMed] [Google Scholar]
- [115].Martin R, Gupta K, Ninan NS, Perry K, Van Duyne GD, The Survival Motor Neuron Protein Forms Soluble Glycine Zipper Oligomers, Structure 20(11) (2012) 1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhao DY, Gish G, Braunschweig U, Li Y, Ni ZY, Schmitges FW, Zhong GQ, Liu K, Li WG, Moffat J, Vedadi M, Min JR, Pawson TJ, Blencowe BJ, Greenblatt JF, SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination, Nature 529(7584) (2016) 48-+. [DOI] [PubMed] [Google Scholar]
- [117].Paushkin S, Gubitz AK, Massenet S, Dreyfuss G, The SMN complex, an assemblyosome of ribonucleoproteins, Current Opinion in Cell Biology 14(3) (2002) 305–312. [DOI] [PubMed] [Google Scholar]
- [118].Memczak S, Papavasileiou P, Peters O, Rajewsky N, Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood, Plos One 10(10) (2015) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]


