Abstract
Objective:
To reliably and quickly diagnose children with posterior urethral valves (PUV), we developed a multi-instance deep learning method to automate image analysis.
Methods:
We built a robust pattern classifier to distinguish 86 children with PUV from 71 children with mild unilateral hydronephrosis based on ultrasound images (3504 in sagittal view and 2558 in transverse view) obtained during routine clinical care.
Results:
The multi-instance deep learning classifier performed better than classifiers built on either single sagittal images or single transverse images. Particularly, the deep learning classifiers built on single images in the sagittal view and single images in the transverse view obtained area under the receiver operating characteristic curve (AUC) values of 0.796±0.064 and 0.815±0.071, respectively. AUC values of the multi-instance deep learning classifiers built on images in the sagittal and transverse views with mean pooling operation were 0.949±0.035 and 0.954±0.033, respectively. The multi-instance deep learning classifiers built on images in both the sagittal and transverse views with a mean pooling operation obtained an AUC of 0.961±0.026 with a classification rate of 0.925±0.060, specificity of 0.986±0.032, and sensitivity of 0.873±0.120, respectively. Discriminative regions of the kidney located using classification activation map demonstrated that the deep learning techniques could identify meaningful anatomical features from ultrasound images.
Conclusion:
The multi-instance deep learning method provides an automatic and accurate means to extract informative features from ultrasound images and discriminate infants with PUV from male children with unilateral hydronephrosis.
Keywords: Multi-instance deep learning, posterior urethral valves, ultrasound images, kidney
Congenital abnormalities of the kidney and urinary tract (CAKUT) are the most common cause of end-stage renal disease (ESRD) in childhood.1 Although many children with CAKUT are diagnosed prenatally,2 postnatal ultrasound remains the cornerstone for characterizing and diagnosing congenital kidney abnormalities3 and in surveilling the kidneys of children with CAKUT diagnosed prenatally. The diagnosis of CAKUT based on ultrasound imaging relies on multiple anatomical measures, such as kidney size, symmetry of the kidneys, hydronephrosis, and echogenicity of the kidney parenchyma.4 However, these anatomical measures are typically obtained manually and exhibit moderate inter-observer variation, which decreases reliability of diagnosis.5 Automated image analysis that accurately identify congenital kidney disease could improve the efficiency and reliability of diagnosis of children with CAKUT early in life.
Pattern recognition models built on imaging features of diagnostic imaging data have demonstrated promising performance to aid diagnosis of kidney diseases.6, 7 However, most features of kidney images were empirically defined and therefore may not fully harness the full discriminative power of the ultrasound images. Recent deep learning studies using diagnostic imaging have demonstrated promising performance in learning imaging features to aid diagnosis and predict clinical outcomes.8, 9 Particularly, convolutional neural networks (CNNs) have been widely adopted to learn informative imaging features in order to achieve optimal pattern recognition performance, including using a single 2D ultrasound images to classify normal kidneys and those with CAKUT.10, 11 A single 2D ultrasound image provides partial anatomic information of the kidney and the same kidney’s appearance in 2D ultrasound images varies in angle and orientation. Consequently, the classification models built upon single 2D ultrasound images might not be robust enough to different views of ultrasound images of the same kidney. On the other hand, multiple 2D ultrasound scans of the same kidney in different views are routinely collected in clinical practice. A classification model built upon different views of ultrasound images of the same kidney might be able to achieve robust classification performance. Such a classification model could be built using multiple instance learning (MIL) that treats all 2D ultrasound images of the same individual together to yield an overall classification score.12
In the present study, we develop a multi-instance deep learning method to learn discriminative features of kidney ultrasound images and determine its performance in discriminating children with CAKUT from children with isolated hydronephrosis.
Materials and Methods
Imaging data and Participants
Ultrasound kidney images were obtained from 86 infants with CAKUT and 71 children with mild unilateral hydronephrosis (controls) at a local hospital for children. The individuals with CAKUT all had posterior urethral valves (PUV), the most common CAKUT diagnosis. These were randomly sampled from patients enrolled in the Registry of Urologic Disease, a comprehensive patient registry that includes 90% of patients seen in the Urology clinic since 2000. The controls were male children with unilateral mild hydronephrosis (Society of Fetal Urology grade I-II). The children with CAKUT had varying degrees of increased cortical echogenicity, decreased corticomedullary differentiation, and hydronephrosis. In order to decrease selection bias, all cases and controls were selected without knowledge of the appearance of the kidneys on ultrasound.
The first ultrasound scans after birth were used and all identifying information was removed. For each individual, multiple 2D ultrasound images in sagittal and transverse views were collected during routine clinical care using Philips, Siemens, or General Electric ultrasound scanners with an abdominal transducer. All the images had 1024×768 pixels with pixel size ranging from 0.08×0.08mm2 to 0.12×0.12mm2. The subjects had varied numbers of 2D ultrasound images in different views.
The work described has been carried out in accordance with the Declaration of Helsinki. The study has been reviewed and approved by the Institutional Review Board.
Problem formulation using multiple instance learning
We formulated the diagnosis of CAKUT based on ultrasound images as a pattern classification problem. Given a subject X with multiple 2D kidney images xi (i = 1, …, I), we built a classifier in a MIL setting to classify X as either a CAKUT patient with a positive label or a control with a negative label. In the MIL setting, each 2D image is referred to as an instance, and all instances of the same subject constitute a bag. In this study, we adopt the standard MIL assumption that all instances of a negative bag are all negative (normal) and a positive (abnormal) bag has at least one positive instance. The classification of a bag X is achieved by a scoring function P(X) that is permutation-invariant to its elements of X in the following form13
where f and g are suitable transformation functions. In our method, f is implemented as a deep learning model at an instance-level to obtain a probability of CAKUT for each 2D image xi, i.e., pi = f(xi), and g is implemented as a mean or max pooling operation.
Deep learning for CAKUT diagnosis at an instance-level based on transfer learning
Instead of building an instance-level deep learning classification model on raw ultrasound kidney images, we adopted a transfer learning strategy to extract informative image features from the images as a starting point, as we have successfully adopted the transfer learning in studies of ultrasound image classification and segmentation.11, 14, 15 Our transfer learning is based on a deep learning model, referred to as VGG16, which was developed for image classification and achieved a top-5 test-accuracy of 92.7% in ImageNet Challenge 2014.16
We trained a deep learning model by adopting the VGG16 model for CAKUT classification at an instance-level. We also compared the VGG16 model with other top-performing deep learning networks of natural image classification, including the ResNet17 and the Inception network.18 Implementation details are presented in the Supplement.
Computer aided classification of CAKUT based on multi-instance pooling
Once a deep learning model was trained for CAKUT classification at an instance-level, we applied the model to each individual’s kidney images for computing an overall bag-level classification score.
Image preprocessing and data augmentation
We manually identified all kidney images in the sagittal view and the transverse view for each individual. Then, all the images were resized to have the same size of 425×321 with cropping but without changing the width to height ratio, and each image’s intensity values were normalized to [0, 255]. In the training stage, we adopted the following data augmentation strategies, including random cropping of the training images by 10 pixels in horizontal directions or 5 pixels in in vertical directions, random rotation of the training images in [−45°, 45°], and left-right flipping of the training images. After data augmentation, the number of images per view and per patient for training was the number of the original training images multiplied by 42042 (21×11×91×2).
Implementation and evaluation of the deep learning model
The deep learning method was implemented based on Python 3.7.0 and TensorFlow r1.11. We trained deep learning models with a minibatch of 8 images using Adam stochastic optimization with a learning rate of 10−5. The maximum number of iteration steps was set to 50000. A TITAN XP graphics processing unit (GPU) on a Linux workstation was used to train the deep learning model.
We trained 2 different deep learning models. The first one was trained based on images in the sagittal view, referred to as S training; and the second one was trained based on images in the transverse view, referred to as T training. Since the deep learning models were trained to classify individual kidney images, we refer to the models as single-instance deep learning (SIL) models. Based on the SIL models, corresponding MIL models were built. Furthermore, a multi-view MIL model was built to utilize images in both the sagittal view and the transverse view by applying the SIL models to images in their corresponding views.
The deep learning models were evaluated in terms of their classification performance, estimated using a 5-fold cross-validation. We first randomly split the whole dataset into 5 subsets. Then, we selected one subset as testing data to evaluate and optimize classification performance of a deep learning model trained based on the other 4 subsets. This process was repeated until each subset had been used as the testing set. The classification performance was measured by area under receiver operating characteristic curve (AUC). We also reported classification accuracy (ACC), specificity, and sensitivity.
We first evaluated the classification performance of the SIL models in order to estimate the diagnostic performance based on a single 2D ultrasound kidney image. We also adopted the gradient-weighted class activation mapping (CAM) [31] to visualize how different kidney regions contributed to the kidney classification.
We then evaluated the classification performance of the MIL models that were built on images in the sagittal view, the transverse view, or both views. We also compared the MIL models built using the mean and max pooling operators. Table S1 of supplementary data summarizes all evaluation in terms of classification models trained and tested using images in different views.
RESULTS
Data characteristics
Table 1 summarizes demographic data and numbers of 2D ultrasound images in different views of all the participants. Consistent with the diagnosis of PUV, the cases and sex-matched controls were all male and had similar ages at time of imaging. However, they differed by race and gestational age, although the gestational age differences would likely not be clinically important (37.1 vs 38.5 weeks). Participants had varied numbers of 2D ultrasound images in different views. No significant difference was observed between the patients with PUV and controls in their numbers of 2D images of different views.
Table 1.
Demographic information and numbers of 2D kidney images in different views.
| Cases (N = 86) | Controls (N = 71) | p-value | ||
|---|---|---|---|---|
| Characteristics | ||||
| Sex n (%) | N/A | |||
| Male | 86 (100) | 71 (100) | ||
| Age at first US (months) Mean (Std) | 0.91 (1.89) | 1.48 (2.32) | 0.09 | |
| Gestational age (weeks)* Mean (Std) | 37.1 (2.8) | 38.5 (2.3) | 0.004 | |
| Race n (%) | ||||
| White | 28 (32.6) | 40 (56.3) | 0.0006 | |
| Black | 34 (39.5) | 8 (11.3) | ||
| Asian | 3 (3.5) | 4 (5.6) | ||
| Other | 21 (24.4) | 19 (26.8) | ||
| CAKUT n (%) | PUV | 86 (100) | N/A | N/A |
| number of 2D images in sagittal view/individual | 22.7±9.4 | 21.9±7.6 | 0.546 | |
| number of 2D images in transverse view/individual | 15.8±6.1 | 16.9±6.5 | 0.285 | |
N used to calculate gestational age statistics is lower due to lack of existing data; N = 75 for cases and N = 42 for controls. Difference in numbers of 2D kidney images of individuals was assessed using two-sample t-test.
Classification performance and computational cost of the SIL models and the MIL models
Table 2 summarizes classification performance of different SIL and MIL models on the testing datasets. All the SIL models obtained AUC values were around 80%, and the SIL models trained and tested on images in the transverse view had slightly better performance than those trained and tested on images in the sagittal view. The Multi-View MIL model had the best classification accuracy and its AUC value was slightly better than the T (T training) MIL model’s AUC value.
Table 2.
Five-fold cross-validation results of SIL models and MIL models on test datasets.
| Models | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| S (S training) SIL | 0.796±0.064 | 0.838±0.047 | 0.811±0.112 | 0.875±0.042 |
| T (T training) SIL | 0.815±0.071 | 0.852±0.053 | 0.841 ±0.092 | 0.863±0.070 |
| S (Straining) MIL | 0.949±0.035 | 0.912±0.036 | 0.873±0.090 | 0.960±0.060 |
| T (T training) MIL | 0.954±0.033 | 0.904±0.038 | 0.868±0.073 | 0.945±0.093 |
| Multi-view MIL | 0.961±0.026 | 0.925±0.060 | 0.873±0.120 | 0.986±0.032 |
ROC curves of different classification models on all subjects under comparison are shown in Figure 1. The ROC curves further demonstrated that the MIL models had better performance than the SIL models and the multi-view MIL model with mean pooling had the best performance.
Figure 1.
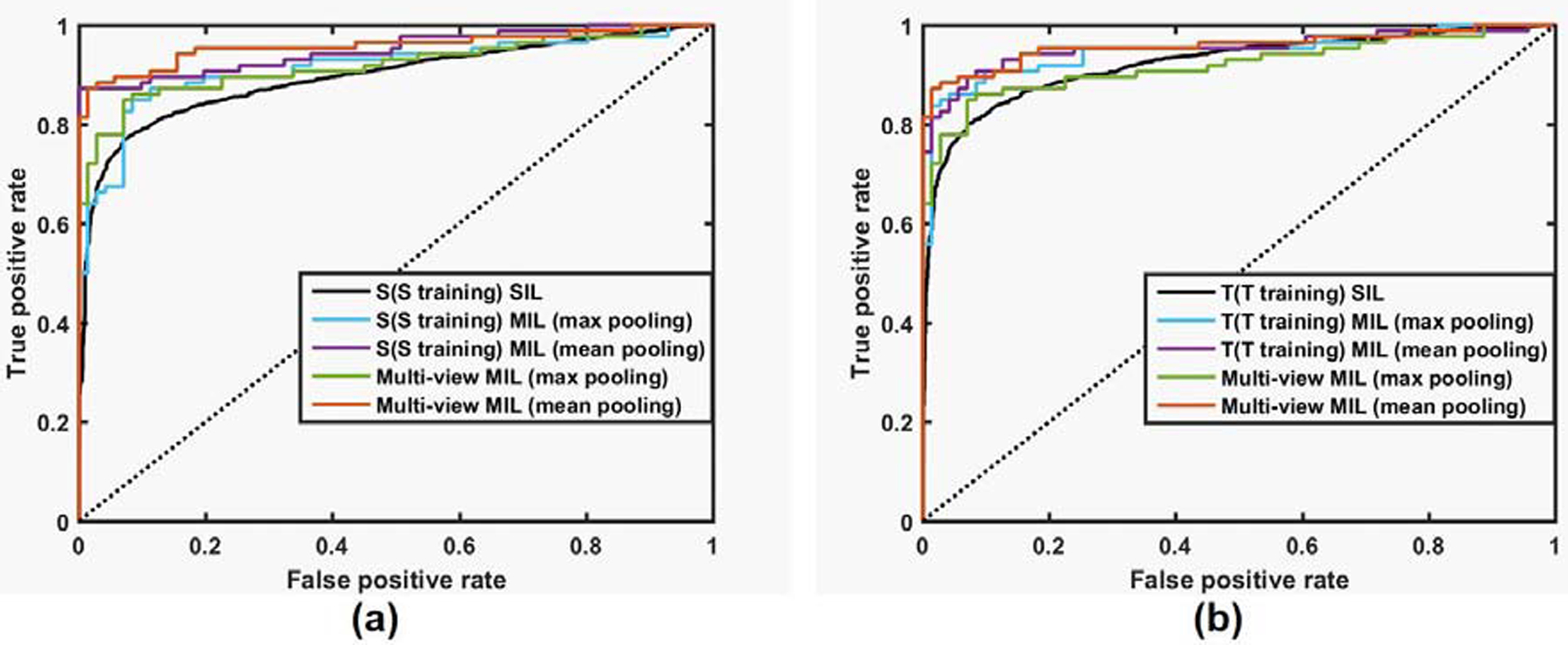
ROC curves of different classification models under comparison. (a) ROC curves of S (S training) SIL, S (S training) MIL models, and multi-view MIL models; (b) ROC curves of T (T training) SIL, T (T training) MIL models and Multi-view MIL models.
On average, the SIL’s computational time was 0.189s/image, the S (S training) MIL’s computational time was 4.22s/subject, the T (T training) MIL’s computational time was 3.019s/subject, and the Multi-view MIL’s computational time was 7.236s/subject.
Kidney regions contributed to the classification
Figure 2 show class activation mapping results of randomly selected kidney images obtained using the SIL models trained on images in different views. These results indicated that discriminative regions were located in both the renal parenchyma and the collecting system.
Figure 2.
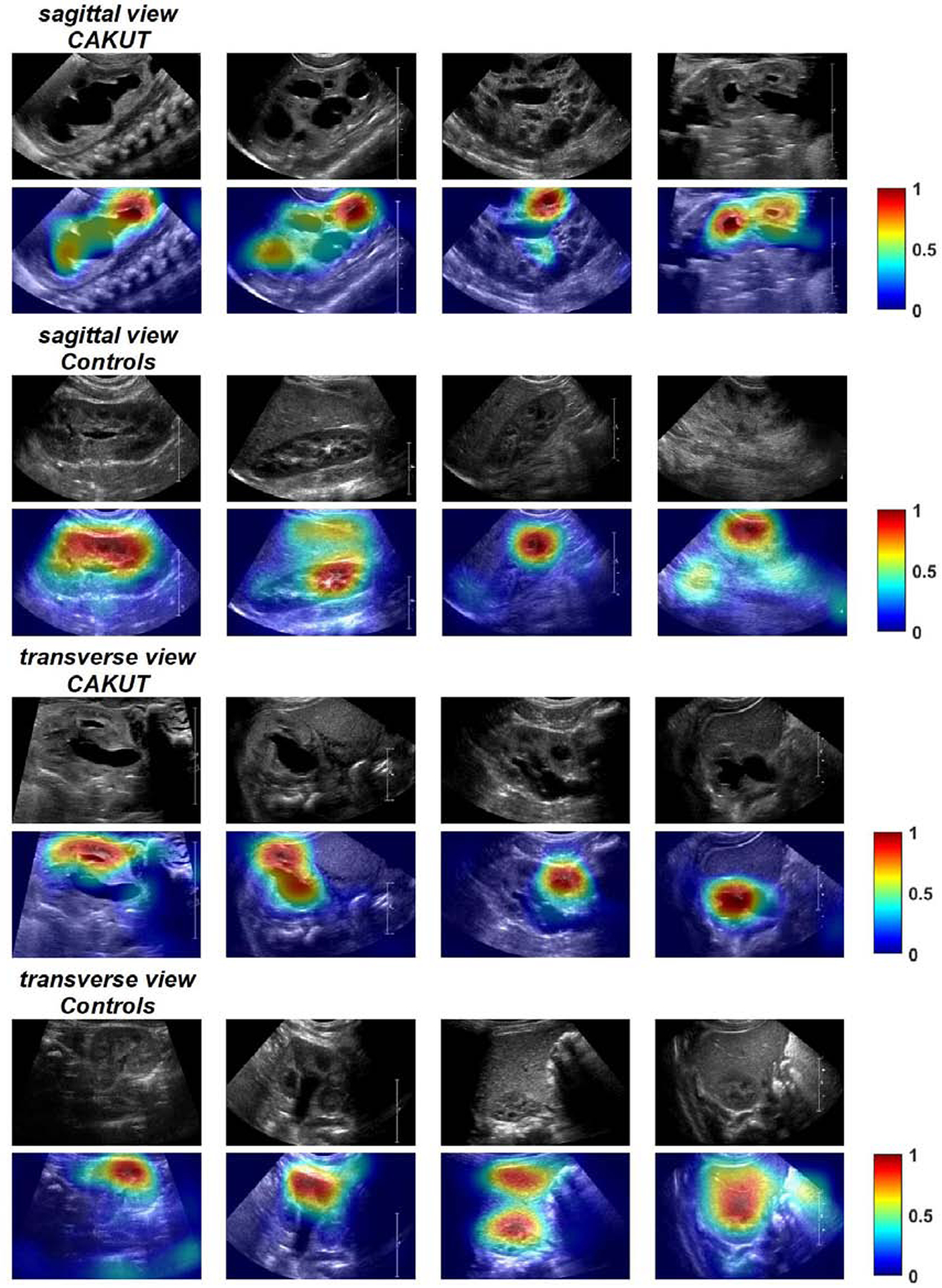
Class activation mapping results of randomly selected CAKUT and control kidney images obtained by the SIL model trained on images in sagittal view and transverse view. Kidney regions in warm color contributed more to the classification than those in cold color. The color bar indicates relative activation.
Comparison with other deep learning methods
Supplemental Table 3 summarizes classification accuracy of different classification models on the testing datasets. The classification models trained with Inception_Resnet v2 obtained the best classification accuracy for classifying images in sagittal view and multiple views, and the classification models trained with the VGG16 obtained the best classification accuracy for classifying images in in transverse view.
DISCUSSION
In this study, we have demonstrated that multi-instance deep learning could achieve excellent performance to rapidly classify infants with PUV based on kidney ultrasound images. These results indicate that the multi-view multi-instance deep learning model built on kidney images in multiple views could obtain higher classification accuracy than deep learning models built on individual kidney images and kidney images in one single view, either sagittal or transverse. The relatively small standard deviations of the classification performance measures across different runs of the cross-validation experiments further demonstrated that the classification performance was relatively stable across different testing datasets. The high computational efficiency (8 seconds/individual) indicated that the deep learning classification system could be adopted in clinical practice.
We compared two different fusion strategies for integrating classification scores of individual kidney images, including the max pooling and the mean pooling (supplementary data). The experimental results indicated that the mean pooling operator led to better classification performance, highlighting that integrating multiple kidney images could improve the diagnosis since individual 2D ultrasound kidney images only provide partial anatomic information of the kidney. The experimental results suggested that the deep learning models built on images in transverse view had slightly better classification performance than those built on images in sagittal view. Moreover, the integration of images in both sagittal and transverse views led to higher classification accuracy.
As illustrated in Supplementary Figure 4, representative CAKUT images misclassified by the deep learning classifiers are visually normal, especially those in top 2 rows. These misclassified images highlighted the fact that single 2D images only provide partial information of the kidney under study and an individual 2D image of a CAKUT kidney could be similar to images of children with mild hydronephrosis both in appearance and in the deep leaning feature space.
The deep learning classification models also automatically identified discriminative image regions for the classification of CAKUT and normal kidneys. The discriminative image regions were located in both the parenchyma of the kidney and the collecting system. These results indicated that the deep learning models extracted informative imaging features since the deep learning models were built upon ultrasound images without telling the algorithm where the kidney is in the images. This finding is important since controls had mild unilateral hydronephrosis; despite the dilation of the collecting system, the deep learning models could discriminate severity of dilation and associated parenchymal changes.
This study did not determine the clinical utility of deep learning. Specifically, these results do not suggest replacing clinical decision making or current standards of care. A much greater body of evidence, including prospective comparative effectiveness studies, is needed before making any recommendations that have such high consequences. However, these findings suggest that deep learning might be able to guide whether subsequent diagnostic imaging, such as voiding cystourethrogram, should be obtained for those infants with kidneys that have features of PUV. In addition, these findings raise the possibility that the discriminative image regions might be associated with severity of underlying kidney disease and clinical outcomes such as chronic kidney disease progression. The deep learning results provide complementary discriminative imaging information to anatomical kidney features, such as renal parenchymal area19, 20 and texture features.6, 7 However, no kidney segmentation is needed to compute the anatomical measures for the deep learning method. In fact, the deep learning models built upon the whole kidney images had better performance than a classification model built upon deep transfer learning features and texture features of the kidney in ultrasound images alone.11 We speculate that the refinement of parameters of a pretrained VGG16 model adopted in our study improved discriminative power of the deep learning imaging features and the multi-view and multi-instance learning further improved the classification performance.
We compared three different pre-trained deep learning models to initialize our kidney image classification networks, including the VGG16, the ResNet, and the Inception network. The experimental results indicated that they yielded similar classification accuracy for both single image classification and multi-instance image classification. However, class activation mapping results obtained by the ResNet and the Inception network also highlighted regions outside of the kidney, as illustrated in Supplementary Figure 2 and Supplementary Figure 3.
One limitation of the present study is that the classification performance was estimated using cross-validation based on ultrasound data collected at a single institution. However, ultrasound images were obtained with a variety of ultrasound machines with different technologists performing the scans. We will validate the deep learning method based on external data that contains other CAKUT diagnoses in order to obtain a more accurate and generalizable estimation of the classification. Second, although our method is fully automatic and does not require kidney segmentation, it did require manually selecting images in sagittal view or transverse view. A fully automatic method is needed to automate the selection of images in different views. Furthermore, our deep learning models did not include the kidney size or other clinical measures that are potentially informative for the disease diagnosis.21 It merits further investigations if the kidney size or other clinical information could provide additional discriminative information to the learned deep learning imaging features for diagnosis of CAKUT, but beyond the scope of this report since we focused on the ultrasound kidney images. In addition, we only included postnatal ultrasounds, but there is clinical value in examining antenatal ultrasounds when the diagnosis is unknown. Future studies should also evaluate the clinical utility of advanced image analysis for children with kidney disease.
Supplementary Material
Funding:
This work was supported in part by the National Institutes of Health (R21DK117297, P50DK114786, and K23DK106428); the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR001878); the National Natural Science Foundation of China (61772220 and 61473296); the Key Program for International S&T Cooperation Projects of China (2016YFE0121200); the Hubei Province Technological Innovation Major Project (2017AAA017 and 2018ACA135); the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics, and the China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare that they have no relevant financial interests.
References
- 1.Dodson JL, Jerry-Fluker JV, Ng DK, et al. Urological disorders in chronic kidney disease in children cohort: clinical characteristics and estimation of glomerular filtration rate. The Journal of urology. 2011;186:1460–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiesel A, Queisser-Luft A, Clementi M, Bianca S, Stoll C. Prenatal Detection of Congenital Renal Malformations by Fetal Ultrasonographic Examination: An Analysis of 709,030 Births in 12 European Countries. European Journal of Medical Genetics. 2005;48:131–144. [DOI] [PubMed] [Google Scholar]
- 3.Richter-Rodier M, Lange AE, Hinken B, et al. Ultrasound Screening Strategies for the Diagnosis of Congenital Anomalies of the Kidney and Urinary Tract. Ultraschall in Med. 2012;33:E333–E338. [DOI] [PubMed] [Google Scholar]
- 4.Hálek J, Flögelová H, Michálková K, et al. Diagnostic accuracy of postnatal ultrasound screening for urinary tract abnormalities. Pediatric Nephrology. 2009;25:281. [DOI] [PubMed] [Google Scholar]
- 5.Nelson CP, Lee RS, Trout AT, et al. Interobserver and Intra-Observer Reliability of the Urinary Tract Dilation Classification System in Neonates: A Multicenter Study. Journal of Urology. 2019;201:1186–1192. [DOI] [PubMed] [Google Scholar]
- 6.Sharma K, Virmani J. A Decision Support System for Classification of Normal and Medical Renal Disease Using Ultrasound Images: A Decision Support System for Medical Renal Diseases. Int J Ambient Comput. 2017;8:52–69. [Google Scholar]
- 7.Attia MW, Abou-Chadi FEZ, Moustafa HE, Mekky N. Classification of Ultrasound Kidney Images using PCA and Neural Networks. Int J Adv Comput Sc. 2015;6:53–57. [Google Scholar]
- 8.Li H, Zhong H, Boimel PJ, Ben-Josef E, Xiao Y, Fan Y. Deep convolutional neural networks for imaging based survival analysis of rectal cancer patients. International Journal of Radiation Oncology • Biology • Physics. 2017;99:S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Habes M, Wolk DA, Fan Y. A deep learning model for early prediction of Alzheimer’s disease dementia based on hippocampal magnetic resonance imaging data. Alzheimer’s & dementia. 2019;15:1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng Q, Tasian G, Fan Y. Transfer learning for diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data. Proceedings of IEEE 15th International Symposium on Biomedical Imaging 2018:1487–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Q, Furth SL, Tasian GE, Fan Y. Computer-aided diagnosis of congenital abnormalities of the kidney and urinary tract in children based on ultrasound imaging data by integrating texture image features and deep transfer learning image features. Journal of Pediatric Urology. 2019;15:75.e71–75.e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amores J Multiple instance classification: Review, taxonomy and comparative study. Artificial intelligence. 2013;201:81–105. [Google Scholar]
- 13.Zaheer M, Kottur S, Ravanbakhsh S, Poczos B, Salakhutdinov RR, Smola AJ. Deep sets. Proceedings of Advances in neural information processing systems2017:3391–3401. [Google Scholar]
- 14.Yin S, Peng Q, Li H, et al. Automatic kidney segmentation in ultrasound images using subsequent boundary distance regression and pixelwise classification networks. Medical Image Analysis. 2020;60:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin S, Zhang Z, Li H, et al. Fully-automatic segmentation of kidneys in clinical ultrasound images using a boundary distance regression network. Proceedings of IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) Venice, Italy: 2019:1741–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karen Simonyan AZ. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv:1409.1556 2014.
- 17.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. Proceedings of Computer Vision and Pattern Recognition2016:770–778. [Google Scholar]
- 18.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, inception-ResNet and the impact of residual connections on learning. Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence2017:4278–4284. [Google Scholar]
- 19.Pulido JE, Furth SL, Zderic SA, Canning DA, Tasian GE. Renal Parenchymal Area and Risk of ESRD in Boys with Posterior Urethral Valves. Clinical Journal of the American Society of Nephrology. 2014;9:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rickard M, Lorenzo AJ, Braga LH, Munoz C. Parenchyma-to-hydronephrosis Area Ratio Is a Promising Outcome Measure to Quantify Upper Tract Changes in Infants With High-grade Prenatal Hydronephrosis. Urology. 2017;104:166–171. [DOI] [PubMed] [Google Scholar]
- 21.O’Neill WC. Renal relevant radiology: use of ultrasound in kidney disease and nephrology procedures. Clinical journal of the American Society of Nephrology : CJASN. 2014;9:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


