Abstract
Graft versus host disease (GVHD) is a post-transplant pathology in which donor-derived T cells present in the Peyer’s patches target the cell-surface alloantigens of the recipient, causing host tissue damages. Therefore, the GVHD has long been considered only a purely immunological process whose prevention requires an immunosuppressive treatment. However, since the early 2010s, the impact of gut microbiota on GVHD has received increased attention. Both a surprising fall in gut microbiota diversity and a shift toward Enterobacteriaceae were described in this disease. Recently, unexpected results were reported that further link GVHD with changes in bacterial composition in the gut and disruption of intestinal epithelial tight junctions leading to abnormal intestinal barrier permeability. Patients receiving allogenic hematopoietic stem cell transplant (allo-HCT) as treatment of hematologic malignancies showed a decrease of the overall diversity of the gut microbiota that affects Clostridia and Blautia spp. and a predominance of lactic acid bacteria (LAB) of the Enterococcus genus, in particular the lactose auxotroph Enterococcus faecium. The reduced microbiota diversity (likely including Actinobacteria, such as Bifidobacterium adolescentis that cross feed butyrogenic bacteria) deprives the butyrogenic bacteria (such as Roseburia intestinalis or Eubacterium) of their capacity to metabolize acetate to butyrate. Indeed, administration of butyrate protects against the GVHD. Here, we review the data highlighting the possible link between GVHD and lactase defect, accumulation of lactose in the gut lumen, reduction of Reg3 antimicrobial peptides, narrower enzyme equipment of bacteria that predominate post-transplant, proliferation of En. faecium that use lactose as metabolic fuels, induction of innate and adaptive immune response against these bacteria which maintains an inflammatory process, elevated expression of myosin light chain kinase 210 (MLCK210) and subsequent disruption of intestinal barrier, and translocation of microbial products (lactate) or transmigration of LAB within the liver. The analysis of data from the literature confirms that the gut microbiota plays a major role in the GVHD. Moreover, the most recent publications uncover that the LAB, butyrogenic bacteria and bacterial cross feeding were the missing pieces in the puzzle. This opens new bacteria-based strategies in the treatment of GVHD.
Keywords: microbiota, butyrate, lactase, lactose, cross-feeding, graft versus host disease, hematopoietic cell transplant
The Gut Microbiota
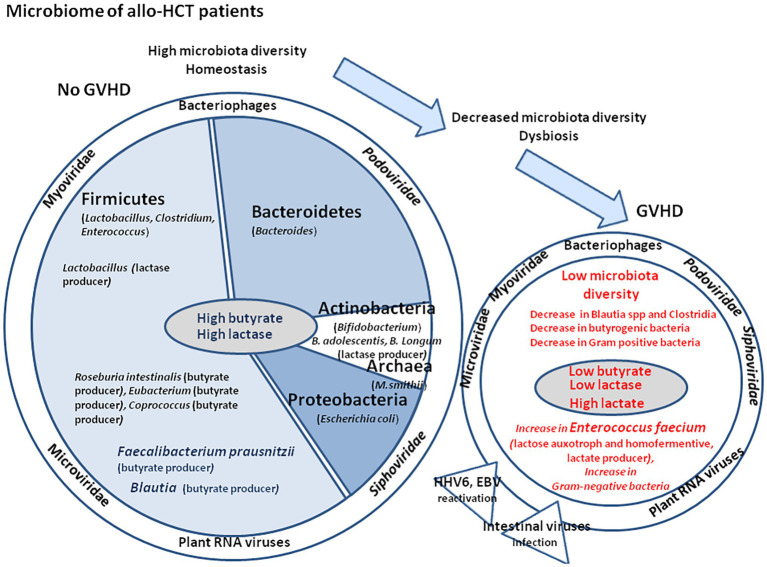
About 100 trillion bacteria present in the intestinal lumen (especially the colon) compose the human gut microbiota. During the last two decades, the advanced methods of high throughput sequencing and culturomics (Lagier et al., 2016) have highlighted the enormous diversity of bacteria found in humans. With its considerable bacterial genetic diversity, over 1,000 species and 7,000 strains identified, the human gut microbiota is a quite complex ecosystem, in which the phyla Firmicutes (species such as Lactobacillus, Enterococcus, and Clostridium) and Bacteroidetes (species such as Bacteroides) account for the majority of species. Other phyla including Proteobacteria (Escherichia coli), Actinobacteria (Bifidobacteria), Cyanobacteria, Fusobacteria, and Verrucomicrobia are also present in lower abundance (Figure 1; Eckburg et al., 2005; Qin et al., 2010). Microorganism DNA represents about 90% of the total DNA found within human bodies (Human Microbiome Project Consortium, 2012). The capacity to rapidly identify a large number of bacterial species in human microbiota opened the way to compare the gut bacterial composition in cohorts of individuals with metabolic or infectious diseases to healthy controls in search of beneficial and non-beneficial bacteria (Million et al., 2016; Tidjani Alou et al., 2017; Elkrief et al., 2019).
Figure 1.
Schematic representation of oral intake including milk that contains water, vitamins, minerals, and biologically active molecules such as adipokines, leptin, adiponectin, and growth factors such as epidermal growth factor and fibroblast growth factor 21 (FGF21). The nutriments first meet the stomach microenvironment. The figure illustrates the variations in pH with marked changes from acidic in the stomach to almost neutral or slightly alkaline in the intestine. The diversity of microbiota increases from the stomach to duodenum, small intestine, and colon, where the concentration of bacteria is the highest. The abundance of microbes is about 101–103 colony forming unit (CFU) per gram in the stomach and duodenum, 104–107 CFU/g in the small intestine, and 1010–1011 CFU/g in the colon. The middle panel illustrates the microbiome and virome diversity in the intestinal microenvironment. The right panel illustrates the physiological organization of the intestinal epithelium during homeostasis. The epithelium (composed of epithelial cells) is protected by a mucus layer synthesized by goblet cells which secrete the mucins (e.g., MUC2 – mucin gel). The mucus layer allows nutrients to transport to epithelial cells but prevents bacteria attachment. The commensal intestinal microbiota is limited to the epithelium-distal mucus layer, while the epithelium-proximal mucus is largely devoid of bacteria. Intestinal bacteria such as Bacteroides and Clostridia ferment lactose into gases, hydrogen carbon dioxide, and methane when the microbiota contains an abundance of Archaea.
During the embryo development (between the first and third trimester of pregnancy), there are shifts in maternal microbiota composition, which likely provide advantages to the fetus survival (Koren et al., 2012). It is usually admitted that the microbial colonization occurs first in the amniotic fluid and placenta, and then in the maternal gut microbiota which supports the development of a prenatal microbiota in the fetus (Collado et al., 2016). When the in utero development is achieved, the fetus migration through the vagina favor a bacterial transfer between the microbiota colonizing the birth canal of the mother and the fetus. In favor of an early bacterial colonization of the fetus, it was observed that Es. coli, Enterococcus faecalis, and Staphylococcus epidermidis were isolated from the meconium of healthy neonates (Nicholson et al., 2012). The neonate survival and growth next depends on mother milk feeding that is his/her essential source of nutrients and that contributes to shape his/her gut microbiota. During the early period of breastfeeding, the infant’s gut is characterized by a low microbiota diversity and become colonized by beneficial bacteria such as Bifidobacterium, Lactobacillus, and Prevotella (Rautava et al., 2012; Arrieta et al., 2014). The mother milk contains about 70 g/L (7%) of lactose produced by the mammary gland (lactose synthesis requires the enzyme galactosyl transferase, which combines activated uridine di-phosphate galactose with glucose), as well as numerous biologically active factors including growth factors. The lactose is well digested by newborns whose small intestinal brush border enterocytes produce lactase in abundance. Dietary intake and bile acids (steroid acids produced in the liver and whose main function is to facilitate the absorption of fat-soluble vitamins and cholesterol) determine at least in part the microbiota assembled during the first few years of life and a shift is observed in favor of anaerobic bacteria that contribute to produce many metabolites of fermentation such as production of short-chain fatty acids (SCFAs; e.g., acetate, propionate, and butyrate; Shanahan et al., 2017). By a mechanism of cross feeding, the intestinal symbiotic microbiota contributes to maintain the production of butyrate by butyric acid bacteria; they also participate to the inhibition of pathogens growth by competing consumption of nutriments and allow to prevent toxin translocation by decomposing metabolic carbohydrates to obtain SCFAs. These SCFAs act on enteroendocrine cells of the gut through heterotrimeric guanine nucleoside-binding protein (G-protein)-coupled receptors that secrete a variety of bioactive compounds.
Lactase production decrease in the majority of the world population after weaning and most healthy adults (67–75%) produce less, sometimes very little lactase (about 10% of the concentration found of neonatal levels), whereas 25–33% retain the ability to digest lactose into adulthood (Troelsen et al., 1997). The study of ileostomy effluent samples from adult patients provided evidence that the small intestine metagenome is enriched in genes related to carbohydrate metabolism compared to the fecal metagenome (Zoetendal et al., 2012), suggesting that carbohydrate metabolism is a central function of the small intestine with lactase and propionate fermentation activities encoded by many taxa from the ileal effluent, in particular Streptococcus that help the growth of secondary fermenters (e.g., Veillonella and Clostridium). It is also admitted that the small intestine microbiota is phylogenetically less diverse but more dynamic than that of the colon (Booijink et al., 2010). Many bacteria living in the intestinal tract such as Lactobacillus sp., Bifidobacterium sp., Bacillus sp., and Es. coli produce lactase and play a major function in lactose absorption in the colon (Rhimi et al., 2009; Juajun et al., 2011). Dietary fibers (non starch polysaccharides) escape digestion in the human small intestine and are then broken down into simple sugars by anaerobic bacteria in the caecum and colon. The colon is more equipped to degrade complex carbohydrates and its main function is absorption of water and electrolytes and storage of fecal matter before expulsion (Turnbaugh et al., 2010). Diet degradation and absorption is under control of metabolic processes that largely depend on expression of bacterial enzymes, which are a direct reflection of the gut microbiota. The intestinal microbiota produces essential vitamins such as vitamin K, B1, B6, B9 (folic acid), and B12. Bacteria from the Firmicutes, Bacteroidetes, and Actinobacteria phyla play a role in bile acids metabolism through bile salt hydrolase activity, which catalyzes the deconjugation of conjugated bile acids (cholesterol derivatives synthesized in the liver) to liberate free primary bile acids, upregulates mucosal defenses, and controls the cholesterol homeostasis (Jones et al., 2008).
It is currently well established that homeostasis and dysbiosis are largely influenced by the composition of gut microbiota and the balance existing between different strains of bacteria (Hooper and Macpherson, 2010; Littman and Pamer, 2011). Environmental selection pressure (e.g., over‐ or under-nutrition, and antibiotics; World bank, 2017; Vieco-Saiz et al., 2019) and competitive exclusion between bacteria (e.g., probiotics and pathogenic bacteria) are expected to be the major driving forces that shape the bacterial composition of the human gut microbiota (Walter and Ley, 2011; Devaux and Raoult, 2018). Here, we provide evidence that the microbiome (the microbiota and the bacteria enzymatic equipment-driven metabolome) determine the outcome of graft versus host disease (GVHD).
Impact of the Gut Microbiota on GVHD
If, for certain human diseases, the pathological process linked to change in the composition of the gut microbiota begins to be understood, for others it remains to be elucidated. Hematopoietic stem cell (HSC) transplantation remains indispensable for the treatment of several malignant disorders (Korbling and Freireich, 2011). Before the graft infusion, most protocols require killing of malignant cells (by chemotherapeutic drugs and/or radiotherapy myelosuppressive treatment) that cause a cytotoxic burst of tumor and normal immune cells associated with a pro-inflammatory status (Blazar et al., 2012). Then, cellular reconstitution is achieved by transplantation of peripheral blood stem cells that is the preferred source of allogenic HSC in adults in order to replace the hematopoiesis of the recipient by that of the donor. Next, it is of major importance to avoid graft rejection (mediated by the recipient immune cells) using immunosuppressive drugs. The differentiated immune cells from donor origin colonize the body (including the gut) with a risk of GVHD. The immunosuppressive therapies practiced in these patients incompletely control GVHD and increase susceptibility to infections. GVHD is considered to be mediated by activated CD8+ cytotoxic T-cell (CTL) from the graft donor after these cells had met graft recipient alloantigens in the context of antigen presenting cells (APCs) in the subepithelial dome of gut Peyer’s patches, the major sites where immune response is set up against luminal antigens and microorganisms (Murai et al., 2003). About 40–50% patients experience severe gastrointestinal damages from acute GVHD that turn to be fatal in about 15% of allo-transplant recipients refractory to standard steroid therapy (Blazar et al., 2012; Köhler and Zeiser, 2019). The follow-up of chimerism in patients after allo-HCT allows quantification of the donor or recipient origin of cells obtained from blood or bone marrow samples, and the chimerism is considered complete when 95% of cells are phenotypically of donor origin.
Beside the immunological activation that has long been studied in allo-transplantation, the impact of gut microbiota on GVHD has received increasing attention over the recent years. Indeed, bacterial lipopolysaccharide (LPS) released from injured gut during the condition regiment taken by the patient was considered as responsible for initiating an innate immune response through activation of toll-like receptors (TLRs) and production of cytokines, serving as a breeding ground for the onset of GVHD (Hill et al., 2000). In humans, the treatment of hematologic malignancies by allo-HCT was characterized by a marked decrease of the overall diversity of the gut microbiota, Enterococcus domination, and the patients with the lowest gut microbiota diversity were those with the higher mortality outcomes (Taur et al., 2012, 2014; Andermann et al., 2018). Thus, the pathophysiological mechanisms implemented in the context of a post-transplant GVHD accompanied by a modification of the gut microbiota deserve further investigation. Early administration of large spectrum antibiotics depleting Blautia spp. (and at a lesser extend Clostridia) was associated with increased GVHD and higher mortality, whereas increased abundance in Blautia spp. improves survival (Jenq et al., 2015). It was next confirmed that the abundance of Clostridia decreased in the microbiota of allo-transplant patients that experienced GHVD and was accompanied by alteration in gastrointestinal microbiota-derived butyrate (Mathewson et al., 2016). Metabolites, such as 3-indoxyl sulfate that originates from the degradation of dietary protein-derived tryptophan to indole by the tryptophanase of intestinal commensal bacteria and that is known to enhance epithelial barrier integrity and to reduce inflammation (Bansal et al., 2010), may serve as a urine marker for monitoring GVHD since microbiota perturbation in patients with GVHD is associated with lower urine levels of 3-indoxyl sulfate (Weber et al., 2015, 2017). Although bacterial populations such as Streptococcus, Gemella, and Veillonella considered as genera defining the core oral microbiota were little affected by the allo-HCT, at the bacterial species level, it was reported that the oral microbiota was affected in patients who developed respiratory complication after allo-HCT (e.g., decrease in Streptococcus infantis and increase in Veillonella parvula; Ames et al., 2019). Since microbiota is connected and dysbiosis is likely to be generalized. The oral microbiota could therefore be considered as a new biomarker to monitor the allo-HCT evolution.
Recently, three independent research groups decipher the molecular mechanism linking GVHD to changes in the species composition of the gut microbiota (Golob et al., 2019; Nalle et al., 2019; Stein-Thoeringer et al., 2019). The analysis of the gut microbiota of a cohort of patients with allo-HCT revealed an increase in Enterococcus faecium, a lactose auxotroph bacteria (which requires lactose for its in vitro growth), in the patient’s gut microbiota along with inflammation and intestine damages (Stein-Thoeringer et al., 2019). A decrease in Clostridium spp. accompanied by a significant reduction of fecal butyrate was also reported in the allo-HCT patients. This corroborate the results from Golob et al. (2019) who reported that a decrease in butyrogenic bacteria (e.g., Blautia spp.) in allo-HCT patients favor the GVHD, whereas administration of butyrate provide protection against GVHD.
Stein-Thoeringer et al. (2019) found that the sugar metabolism of the allo-HCT patients was impaired with an over-expression of enzymes involved in the degradation of lactose and galactose and that patients bearing a genetic polymorphism which decreases lactase expression (an enzyme also named β-d-galactosidase that is synthesized by enterocytes of the small intestine that break down the disaccharide d-lactose into d-galactose and d-glucose monomers) suffer from lactose absorption failure. It was hypothesized that lactic acid bacteria (LAB) En. faecium may possibly mediate the pro-inflammatory process (Zitvogel and Kroemer, 2019). Although patients with GVHD have increased intestinal permeability, the distribution of zonula occludens-1 (ZO-1) and actin were found unaltered and, in most cases, epithelial damages are limited to apoptosis of crypt epithelial cells associated with an over-expression of myosin light chain kinase 210 (MLCK210) and increased myosin II regulatory light chain phosphorylation (Nalle et al., 2019). Non-muscle myosin II (NMII), notably NMIIA, a key Rho kinase target, plays a role in epithelial cell-cell adhesion by controlling the local E-cadherin accumulation at the cell-cell contact (Priya et al., 2015). Moreover, E-cadherin is known for being used as target receptor for several bacteria and cleavage of E-cadherin by sheddases followed by the release of soluble E-cadherin is a mechanism frequently involved in disruption of the intestinal epithelium and invasive bacteria transmigration (Devaux et al., 2019). A higher risk of blood stream infection caused either by vancomycin-resistant Enterococcus or Gram-negative bacteria (e.g., Es. coli or Klebsiella pneumoniae) was reported, and post-engraftment vancomycin-resistant Enterococcus colonization was associated with increased mortality (Ford et al., 2017; Tamburini et al., 2018; Zama et al., 2020).
The Butyrate Pathway
Recently, Golob et al. (2019) reported that the butyrogenic bacteria Blautia spp. were less abundant in allo-HCT who experienced GVHD, whereas administration of butyrate provided protection against GVHD. A decrease of fecal butyrate was also reported by Stein-Thoeringer et al. (2019) in the allo-HCT patients who experienced GVHD (Figure 2).
Figure 2.
The butyrate pathway. Most of the dominant species of human colon microbiota produce acetate during the fermentation of carbohydrates. Acetate, propionate, and butyrate are the main short-chain fatty acids (SCFAs) found in humans. The main pathway for acetate formation is the oxidative decarboxylation of pyruvate leading to the synthesis of an ATP molecule. In the human colon, propionate is mainly produced by the succinate decarboxylation pathway. Propionate is mainly synthesized by the species of the genus Bacteroides, Propionibacterium, and Veillonella. However, the synthesis of propionate via lactate (not shown), could also be important, especially in the case of diets rich in non-digestible oligosides. Clostridium, Eubacterium, Faecalibacterium, Roseburia, and Butyrivibrio have been shown to be capable, in vitro, of producing butyrate by the condensation of two molecules of acetyl-CoA with synthesis of one molecule of ATP. Acetyl-CoA is also the precursor of the synthesis of ethanol, produced in vitro by many species such as those of the genera Bacteroides, Lactobacillus, or Bifidobacterium. Faecalibacterium prauznitzii was found able to use acetate as a source of butyrate. Butyrate is a major metabolite in colonic lumen arising from bacterial fermentation of dietary fiber and has been shown to be a critical mediator of the colonic inflammatory response. Butyrate is a source of energy and it plays a critical role in homeostasis through its ability to counteract inflammation-mediated ulcerative colitis. Patients receiving allo-HCT as treatment for hematologic malignancies and who have an abundance of butyrate-producing bacteria (left) have the highest probability to get reed of carbon-dioxide (GVHD), whereas those with low abundance of butyric acid bacteria (right) are more often faced to GVHD.
Butyrate, considered a protective molecule against inflammation is the end-product of anaerobic bacteria fermentation of non-digestible carbohydrates and also a component of dairy products (e.g., butter, milk, and cheese). The butyric acid bacteria are ubiquitously present in the gut microbiota of healthy humans. Oxidative stress sensitive bacteria include the butyrate producers, which mainly belong to the Firmicutes, Lachnospiraceae, Ruminococcaceae (e.g., F. prausnitzii, Roseburia intestinalis, Eubacterium spp., and Coprococcus spp.), and mucosa associated symbionts such as Lactobacillus spp. or B. adolescentis. These bacteria contribute to absorb dietary fats and lipid-soluble vitamins and facilitate lipid absorption and can digest polysaccharide dietary fiber and oxidize sugar to pyruvate. Different functionally distinct groups of butyrate-producing bacteria are present in the human large intestine: strains of R. intestinalis and F. prausnitzii possess butyryl CoA: acetate CoA transferase and acetate kinase activities but lack butyrate kinase, whereas the strain L2–50 of Coprococcus sp., possesses butyryl CoA: acetate CoA transferase, acetate kinase, and butyrate kinase (Duncan et al., 2002). This observation corroborates old studies performed on anaerobic butyric acid bacteria of cattle rumen that already demonstrated that among 48 strains, about 50% were able to hydrolyze starch and the number of strains that fermented certain carbohydrates was glucose, 48; esculin, 46; xylose, maltose, and cellobiose, 44; sucrose and salicin, 43; fructose, 42; lactose and inulin, 40; dextrin, 37; xylan, 34; trehalose, 16; and mannitol, 1, indicating major strain differences in genes coding for their enzyme equipment (Bryant and Small, 1956).
During the fermentation process, they produce SCFAs: acetate (C2), propionate (C3), and butyrate (C4). The SCFAs concentration is 10 times higher in the colon than in the ileum (Cummings et al., 1987). Acetate, propionate, and butyrate represent between 85 and 95% of total SCFAs found in the host and the molar ratio acetate/propionate/butyrate is about 60/20/20 (Cummings and Macfarlane, 1991; Topping and Clifton, 2001). About 95% of the SCFAs produced in the colon are absorbed from the colonic epithelium and 5–10% of the SCFAs are excreted by the feces (Hijova and Chmelarova, 2007). Acetate and propionate are transported through the blood to a variety of organs, where they are substrates for oxidation, lipid synthesis, and energy metabolism. Acetate is mainly metabolized by liver cells (50–70%), but also by the heart and skeletal muscles and the brain, making it an important source of energy. Propionate is also metabolized by the liver and used by hepatocytes for glucogenesis (Cummings et al., 1987; Nicholson et al., 2012). Butyrate is used by the cells of the colon and liver and targeted to their mitochondrial compartment for energy purposes (Clausen and Mortensen, 1994; Basson et al., 2000; Della Ragione et al., 2001).
Most of the dominant species of human colon microbiota produce acetate during the fermentation of carbohydrates. The main pathway for acetate formation is the oxidative decarboxylation of pyruvate leading to the synthesis of an ATP molecule. In the human colon, propionate is mainly produced by the succinate decarboxylation pathway (Miller and Wolin, 1979). Propionate is mainly synthesized by the species of the genus Bacteroides, Propionibacterium, and Veillonella. However, the synthesis of propionate via lactate could also be important, especially in the case of diets rich in non-digestible oligosides. Butyrate is mainly produced by distinct families within the Firmicutes, Ruminococcaceae, and Lachnospiraceae. But members of other phyla including Bacteroidetes, Actinobacteria, Fusobacteria, and Proteobacteria are potential butyrate producers (Vital et al., 2014; Zhang et al., 2019). Bacteria that belong to Clostridium, Eubacterium, Faecalibacterium, Roseburia, and Butyrivibrio have been shown to be capable, in vitro, of producing butyrate by the condensation of two molecules of acetyl-CoA with synthesis of one molecule of ATP. Most of the bacterial diversity in bacteria that produce butyrate by the acetyl-CoA pathway is associated with Ruminococcaceae and Lachnospiraceae, whereas other bacteria such as Bacteroidetes mainly use other (Glutarate, Lysine) pathways (Vital et al., 2014). Acetyl-CoA is also the precursor of the synthesis of ethanol, produced in vitro by many species such as those of the genera Bacteroides, Lactobacillus, or Bifidobacterium. Pyruvate is a central compound of the butyrate pathway and in turn can be oxidized to acetyl CoA with production of hydrogen, methane, and carbon dioxide gases (CO2 increases the pH) and energy, which the bacteria require for growth and production of many molecules. Part of acetyl CoA is converted into acetic acid with ATP production and butyryl CoA that is metabolized into butyrate with ATP production. In acetate-rich diet, where acetate was delivered by resistant starch directly into the gastrointestinal tract, butyric acid bacteria can convert both exogenous and gut microbiota-produced acetate into butyrate (a reaction that requires acetate CoA transferase and butyrate kinase). One of the rare known bacteria that can achieve this enzymatic reaction is F. prausnitzii (Duncan et al., 2002), thereby contributing to the protection of the intestinal mucosa barrier and to the promotion of Bifidobacterium and Lactobacillus improving intestinal function (Si et al., 2018). F. prausnitzii grows poorly in culturomic medium that does not contain acetate and this requirement likely explains its dependence on rumen fluid. SCFAs are able to bind and activate the G-protein coupled cell surface receptors (including FFA2/GPR43 and FFA3/GPR41) expressed by enteroendocrine I, K, and/or L epithelial cells that are known to play an important role in the regulation of glucose homeostasis and appetite (Jorsal et al., 2018; Priyadarshini et al., 2019). In a murine model, it was shown that FFA2/GPR43 is also strongly expressed in a large population of leukocytes in the lamina propria; that FFA3/GPR41 is expressed in subpopulations of ghrelin and gastrin cells in the stomach, in secretin cells of the proximal small intestine, in GLP-1, YY, and neurotensin cells of the distal small intestine and proximal colon; and that a gradient of FFA3/GPR41 expression exists among the somatostatin cells from less than 5% in the stomach to more than 95% in the rectum (Nohr et al., 2013). FFA2/GPR43 has similar affinity for acetate, propionate, and butyrate whereas FFA3/GPR41 has preferential affinity for propionate, although it also binds butyrate. In a murine model of GVHD, mice orally force-fed with GPR43 antagonist GLPG0974 (10 mg/kg/day) demonstrated significant higher GVHD than control, indicating a protective effect against GVHD when butyrate can bind FFA2/GPR43 (Fujiwara et al., 2018). Beside being a major source of energy for the intestinal epithelium (this allow energy-deprived cells to escape autophagy) and source of nutriment for microbes such as Desulfotomaculum spp. (Kuwahara, 2014), butyrate acts on the epigenetic regulation of genes by inhibiting an histone deacetylase (HDAC; Waldecker et al., 2008) and as an agonist for peroxisome proliferator-activated receptors (PPARs) that control both lipid metabolism and inflammation (Varga et al., 2011). Butyrate increases expression of proteins (such as junctional adhesion molecules/JAM/occludin) involved in the stability of tight junctions in colon epithelia; it regulates the neutrophil function and migration and inhibits inflammatory cytokine-induced expression of vascular cell adhesion molecule-1 (V-CAM1). It can pass through the enterocytes into the circulation. Butyrate and retinoic acid (RA) co-operate to regulate the innate immune response. Vitamin A is taken from food in the form of retinol, retinoic acid, or beta-carotene. In the gut, dendritic cells (DCs) metabolize vitamin A in RA, and RA co-operates with butyrate to induce mucosal-like CD103+DCs differentiation required to trigger the differentiation and intestinal recruitment of FoxP3+ T regulator (T reg) cells, IgA antibody secretion, and reduce inflammation (Qiang et al., 2017). The butyrate receptors, FFA2/GPR43 and FFA3/GPR41, are all found expressed on liver cells. At high concentration, butyrate is expected to promote anti-tumor effect by optimizing the effector function of CD8+ T cells that produce IFN-γ (Luu et al., 2018).
Radiotherapy, Antibiotics/Chemotherapy, and Oxidative Stress-Induced Loss of Cross Feeding Bacteria
Radiotherapy and antibiotic/chemotherapy treatments that are practiced to kill fast dividing cancer cells and as treatment of complications of allo-HCT are known to induce oxidative stress, where high levels of reactive oxygen species (ROS) and reactive nitrogen species (NOS) are generated. The free oxygen radicals hydroxyl radical (OH), superoxide anion (O2−), hydrogen peroxide (H2O2) trigger the up-regulation of cyclooxygenases (COX), nitric oxide synthase, lipoxygenases, and nicotinamide adenine dinucleotide phosphate oxidase, leading to DNA damage, inflammation (radiation-induced enteritis/radiation-induced bowel injury), cell apoptosis, and also modify the microbiota homeostasis (Saha et al., 2017; Deleemans et al., 2019; Severyn et al., 2019). In a non-human primate model, ionizing radiations were also reported to induce up-regulation of tumor necrosis factor alpha (TNFα) and metalloprotease MMP7 (Zheng et al., 2015), likely affecting the intestinal epithelium barrier integrity and bacterial infiltration.
Radiation enteritis and dysbiosis were reported in patients who received a radiotherapy treatment. The dysbiosis was characterized by a relative higher abundance of Proteobacteria and Gammaproteobacteria and lower abundance of Bacteroides. A deeper analysis found an increase in oxidative stress resistant Enterobacteriaceae, Phyllobacteriaceae, and Beijerinckiaceae, whereas oxidative stress sensitive Bacteroidaceae and Ruminococcaceae were decreased (Figure 3; Wang et al., 2019). It confirms previous observation from radiation-induced intestinal chronic inflammation (Kumagai et al., 2018). Within the microbiota, some bacteria are resistant to oxidative stress (bacteria that synthesize the anti-oxidant enzyme superoxide dismutase, SOD, which neutralize O2−, and catalase, and CAT, which neutralize H2O2), whereas others are highly sensitive. Bifidobacterium that are essential to fermentation are not able to produce butyrate although they are associated to a butyrogenic effect due to cross-feeding between Bifidobacterium and butyrate producing colon bacteria (Falony et al., 2009; De Vuyst and Leroy, 2011). Bifidobacterium are preferentially stimulated to growth in the presence of fructose oligosaccharides (FOS), inulin-type fructans (ITF), and xylo-oligosaccahride (XOS; Tuohy et al., 2001; Roberfroid, 2007; Lecerf et al., 2012). Cross-feeding between starch-degrading Bifidobacteria and lactate-converting, butyrate‐ producing colon bacteria (e.g., Eubacterium hallii and Anaerostipes caccae) has been demonstrated (Duncan et al., 2004). Indeed, Eu. hallii L2-7 and A. caccae L1-92 failed to grow on starch in pure culture, but in co-culture with B. adolescentis L2-32, butyrate was formed, indicating cross-feeding of metabolites to the lactate user bacteria (Belenguer et al., 2006). Obligate cross-feeding was also reported between B. longum BB53 and A. caccae DSM 14662 and R. intestinalis DSM 14610, B. longum acting as acetate producer for the butyrogenic bacteria (Falony et al., 2006), and in co-cultures of B. adolescentis L2-32 with F. prausnitzii S3/L3 or F. prausnitzii A2-165 with FOS as carbon source, resulting in acetate decrease and butyrate increase (Rios-Covian et al., 2015). Metabolic cross-feeding can occur via intercellular nanotubes among bacteria (Pande et al., 2015).
Figure 3.
Cross-feeding that take place between Bifidobacterium spp. and species of butyrate-producing colon bacteria. Within the microbiota, the bacteria equipped with superoxide dismutase and catalase anti-oxidant enzymes can be resistant to the free radical produced after radiotherapy/chemotherapy, whereas bacteria that lack these enzymes are highly sensitive to oxidative stress. Bifidobacterium that are essential to fermentation are not able to produce butyrate but produce acetate to cross-feed the butyrate producing colon bacteria. Under oxidative stress, the Bifidobacterium are killed and no longer play their role as cross-feeding bacteria with direct consequences on the production of butyrate by the butyrogenic bacteria.
In the presence of carbohydrates, Bifidobacterium cross feed the butyrogenic bacteria that in turn produce butyrate (Rivière et al., 2016). However, Bifidobacterium strains such as B. adolescentis are highly sensitive to oxidative stress. It is therefore possible that during the chemotherapy that is practiced in patients with allo-HCT, such bacteria are lost. In methotrexate chemotherapy of lymphoblastic leukemia, a significant reduction of Bifidobacterium, Lactobacillus, and Es. coli was reported (Huang et al., 2012). Without cross-feeding, the production of butyrate will be drastically reduced. Treatment of patients with melatonin (N-acetyl-5-methoxytryptamine know to exhibit antioxidant activity) post-irradiation significantly increased both SOD and CAT, suggesting that the treatment restore the oxidative stress resistant bacteria (Musa et al., 2019).
Taken together, these results suggest that digestive bacteria sensitive to oxidative stress, capable of catabolizing lactose and increasing butyrate production are lost in GVHD. It was reported that the use of Lactobacillus rhamnosus (a butyric bacteria species) could prevent the occurrence of diarrhea in patients receiving radiotherapy (Delia et al., 2002). Other studies reported that use of Lactobacillus acidophilus and Bifidobacterium bifidum were beneficial to the patients, yet, the results remain controversial (Kumagai et al., 2018; Wei et al., 2018), likely because these bacteria display phenotypic variation on strain level and each clone is different in terms of enzymatic equipment. More recently, it was reported that fecal microbiota transplant (FMT) improves the outcomes of an allo-HCT with GVHD (Severyn et al., 2019).
The Lactase Pathway
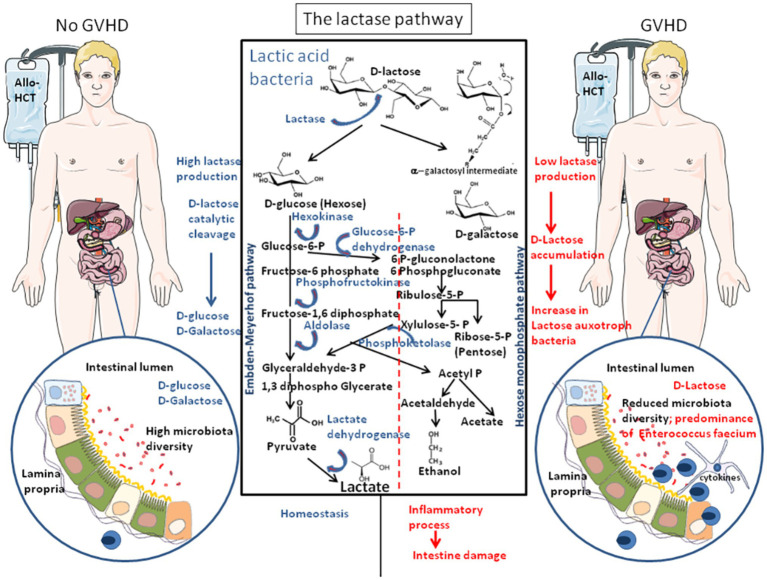
Recently, Stein-Thoeringer et al. (2019) reported that patients with allo-HCT who experienced GVHD were characterized by an increase in the lactose auxotroph (which require lactose for their in vitro growth) bacteria En. faecium in their gut microbiota that was accompanied by inflammation and intestine damages. It was hypothesized that LAB En. faecium may possibly mediate the pro-inflammatory process (Figure 4; Zitvogel and Kroemer, 2019).
Figure 4.
The lactase pathway. The FGF21 was found to enhance MAPkinases (ERK1/2) phosphorylation in intestinal explants and to stimulate lactase production and glucose homeostasis. The lactic acid bacteria (LAB) of the intestinal microbiota are the main source of lactase in adult. Bacterial lactase genes in the intestinal microbiota derived from Actinobacteria, Proteobacteria, and Firmicutes. The bacterial species that transform the carbohydrates into lactate mainly belong to the genera Bifidobacterium and Lactobacillus, as well as Streptococcus and Enterococcus. Lactate is produced by reduction of pyruvate. LAB can be divided in two metabolic classes: first, the homofermentative bacteria that produce lactate includes some Lactobacilli and most species of Pediococcus, Vagococci, Streptococci, Tetragenococci, and Enterococci (including Enterococcus faecium), and the heterofermentative species include Leuconostocs, Oenococci, Weissella species, and some lactobacilli that produce lactate, carbon-dioxide (CO2), acetate, and/or ethanol. Patients receiving allo-HCT as treatment of hematologic malignancies and who have an abundance of lactase-producing bacteria (left) produce lactate from lactose and are less sensitive to GVHD. Patients with allo-HCT, who carry a microbiota that produces low lactase, accumulate lactose that increases the abundance of lactose auxotroph bacteria (En. faecium) with a concomitant decrease of the microbiota diversity (right), triggering pro-inflammatory responses, and serving as a breeding ground for GVHD.
The intestinal microbiota is the main source of lactase in adult. Bacterial lactase genes in the intestinal microbiota mainly derived from Actinobacteria, Proteobacteria, and Firmicutes (Long et al., 2018). It is excluded to draw general conclusions on a metabolism picture that would be common for all LAB. Each species and possibly each clone may behave differently depending on their/its enzymatic equipment. LAB are Gram-positive bacteria, acid-tolerant, low G+C content in the DNA, that belong to Lactobacillus, Enterococcus, Streptococcus, Pediococcus, Lactococcus, and Oenococcus genus have the capability to modify the environment in which they are delivered (Makarova et al., 2006). LAB genomes code between 1,700 and 2,500 proteins. The growth optimum for LAB is at pH 5.5–5.8, and these bacteria have complex nutritional requirements for carbohydrates and other compounds such as fatty acids, peptides, vitamins, amino acids, peptides, nucleotide bases, and minerals. LAB produce lactase, an enzyme that catalyze the cleavage of carbohydrates (lactose, glucose, sucrose, or galactose) into lactic acid (Rhimi et al., 2009; Russo et al., 2017). Lactase is an important enzyme associated with lactose absorption. Dysbacteriosis associated to inhibition of intestinal lactase activity cause body diarrhea by affecting body’s absorption of nutriments. A variety of diarrhea can be treated by supplementing oral intake with LAB or lactase (Luo et al., 2016).
Based on sugar fermentation patterns, LAB can be divided in two metabolic classes: first, the homofermentative bacteria that include some Lactobacilli (such as Lactobacillus casei, Lactococcus lactis, and Lactobacillus plantarum), most species of Pediococcus (such as Pediococcus pentasaceus and Pediococcus acidilactici), Enterococci (such as En. faecium), Streptococci, Tetragenococci, and Vagococci that ferment hexoses by the Embden-Meyerhof pathway. It was reported that Streptococcus faecalis, that produce glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase, regulate the metabolism of glucose by the specific inhibitory interaction of the Embden-Meyerhof intermediate fructose-1,6-diphosphate with 6-phosphogluconate dehydrogenase (Brown and Wittenberger, 1971). In the presence of homofermentative LAB, lactate is the primary product with a production of two moles of lactate from one mole of glucose (homofermentative LAB are bacteria common to the dairy industry); second, the heterofermentative species include Leuconostocs, Oenococci, Weissella species, and some Lactobacilli (such as Lactobacillus buchneri). The heterofermentative LAB species produce one mole of lactate from one mole of glucose, as well as CO2 and acetate and/or ethanol. High amount of ethanol was shown to be produced by Lactobacillus fermentum by glucose metabolism and Weissella confusa by fructose metabolism, whereas addition of pyruvate reduced their production of ethanol with a shift to acetate production (Elshaghabee et al., 2016). LAB can also synthesize different compounds such as bacteriocins (ribosomally synthesized anti-microbial peptides), H2O2, and enzymes capable to facilitate nutriment acquisition (such as protease, amylase,…), and other enzymes (such as β-galactosidase, galactose mutarotase, l-lactate dehydrogenase,…; Liao and Nyachoti, 2017). The apparent difference between the homofermentative and heterofermentative classes of LAB is the presence or absence of the fructose-1,6-diphosphate and/or phosphoketolases. Bacteria that produce xylose isomerase and xylulokinase can convert xylose, a pentose, to xylulose 5-phosphate. In many bacteria, such as Es. coli, xylulose 5-phosphate is further catabolized to form glyceraldehyde-3 phosphate by the transketolase and transaldolase enzyme of the pentose phosphate pathway. Another xylose catabolic pathway used by heterofermentative bacteria such as Clostridium acetobutylicum, involves phosphoketolases that cleave xylulose 5-phosphate into acetyl-phosphate and glyceraldehyde-3-phosphate (Liu et al., 2012). In Bifidobacteria, phosphoketolases are key enzymes to convert fructose-6-phosphate to acetyl-phosphate and erythrose-4-phosphate (Grill et al., 1995). Recently, a Collinsella aerofaciens subspecies that uses butyric acid kinase and phosphatase butyryltransferase enzyme to metabolize sugars was described (Qin et al., 2019).
When produced at high concentration, lactate may in turn inhibit proline oxidase, thereby regulating the bioavailability of proline (a markedly reduced rate of proline degradation) in the liver (Kowaloff et al., 1977). A pathologic hyperprolinemia could be related to the hyperlactacidemia found in acquired lactic acidosis and alcoholic cirrhosis, and there are evidence in the literature that in infants with plasma concentration of lactate 8–10-fold normal, their plasma proline was two to three-fold the normal concentration and that patients with lactic acidosis showed a plasma concentration of proline five to six-fold the normal level. Since, proline can be a source for gluconeogenesis (a metabolic pathway that results in the generation of glucose from glucogenic amino acids, triglycerides, glycerol, pyruvate, and lactate), increased lactate concentration in the liver may trigger acute hepatic gluconeogenesis that acts as metabolic fuels.
Several reports indicate that LAB induce adaptive immune responses (Kiczorowska et al., 2017; Han et al., 2018). Feeding piglets with L. rhamnosus prevented acute infectious diarrhea by triggering the lamina propria CD3+/CD4+ T cells activation (Zhu et al., 2014). L. plantarum was shown to stimulate anti-Salmonella immune response in pigs (Tran et al., 2016). In chickens, feed supplementation with L. acidophilus increases the production of CD3+/CD4+ and CD3+/CD8+ T cells in their gastrointestinal tract and peripheral blood (Asgari et al., 2016). LAB (mainly Lactobacillus) are currently considered as a possible probiotics and they are intensively studied to select the most valuable strains for commercial use (Aristimuño Ficoseco et al., 2018). Thus, Lactobacilli are mainly grouped in the growth promoter beneficial bacteria.
Possible Relationship Between Pathogenic Lactic Acid Enterococcus Genus and GVHD
Although Enterococci are part of the normal intestinal microbiota, several species have a notable clinical implication, in particular En. faecalis and En. faecium. These Enterococci species cause a variety of diseases, including endocarditis, urinary tract infections, prostatitis, and cellulitis (Noris et al., 2010; Paganelli et al., 2016; Tien et al., 2017; Beganovic et al., 2018; Reissier et al., 2018). About 3% of the En. faecalis and En. faecium genomes encode enzymes involved in lactose and galactose metabolism (Figure 5; Stein-Thoeringer et al., 2019). En. faecium, is known for its ability to co-metabolize citrate and lactose and to produce high amounts of lactate (by conversion of pyruvate into lactate; Cabral et al., 2007). Risk factors for colonization and infection must be recognized, in particular prior treatment with antibiotics such as cephalosporins or quinolones. The veterinary use of vancomycin and avoparcin as growth factor in the feed supplementation of farm animals is likely one of the reasons for the selection of VanA strains that resist both vancomycin and avoparcin (a resistance that is uncommon except in Enterococci; Bortolaia et al., 2015). En. faecalis was reported as capable of producing extracellular superoxide and H2O2 that damage colonic epithelial cell DNA (Huycke et al., 2002). Unlike most commensal strains, the genome of multidrug-resistant (MDR) strains of En. faecalis clinical isolates are rich in mobile genetic elements and lack genome defense system composed by the clustered regularly interspaced short palindromic repeat (CRISP) and the CRISP-associated protein (CAS; Hullahalli et al., 2018). Moreover, En. faecalis incorporates to its membrane host-derived fatty acids found in human serum that protect the bacteria against membrane-damaging antibiotics (e.g., daptomycin; Saito et al., 2018). Another species of Enterococci, Enterococcus gallinarum was found to expand in mice treated with proton pump inhibitors (PPIs; Llorente et al., 2017). This strain is known to trigger pro-inflammatory pathways, to alter gut-barrier related molecules in the small intestine, and to translocate to the mesenteric lymph node, spleen and liver, inducing systemic autoimmunity both in mice and humans (Manfredo Vieira et al., 2018). En. gallinarum was also found to cause endocarditis (Angelos et al., 2018).
Figure 5.
Microbiome and virome in patients with allo-HCT. Many bacteria living in the intestinal tract (left panel) such as Lactobacillus sp., Bifidobacterium sp., Bacillus sp., and Escherichia coli produce lactase and play a major function in lactose absorption in the colon. Bacteria from the Firmicutes, Bacteroidetes, and Actinobacteria phyla play a role in bile acids metabolism and up-regulate mucosal defenses. Faecalibacterium prausnitzii is one of the rare bacteria that can convert acetate into butyrate, thereby contributing to the protection of the intestinal mucosa barrier and to the promotion of Bifidobacterium and Lactobacillus improving intestinal function. The presence of F. prausnitzii and Blautia spp. in the patient microbiota is a favorable prognosis mark for the patient. During GVHD (right panel), an increase in the lactose auxotroph bacteria En. faecium is observed along with inflammation and intestine damages. A decrease in Clostridium spp. and Blautia spp. (butyrogenic bacteria) is accompanied by a significant reduction of fecal butyrate in GVHD. Gram-positive Lactobacilli, Clostridia, Bifidobacteria, and Bacillus spp. were less abundant in GVHD. In contrast, the proportion of Gram-negative bacteria (Enterobacteriaceae, Enterococci, and Bacteroides/Prevotella spp.) increased, as well as release of lipopolysaccharide (LPS) that trigger toll-like receptor (TLR) induction and low-grade chronic inflammation. During GVHD reactivation of viruses [e.g., human herpes virus 6 (HHV-6) or EBV] is frequently observed.
Microbiota and the Innate Immunity: Relationship to GVHD
Interactions between the gut microbiota and the host immune system begin at birth and these two systems are involved in a complex interplay. The crosstalk that takes place determines the host immune inflammation status. Several studies reported that innate immune cells (e.g., neutrophils and inflammatory monocytes) are recruited to the gut shortly after allo-HCT (Schwab et al., 2014).
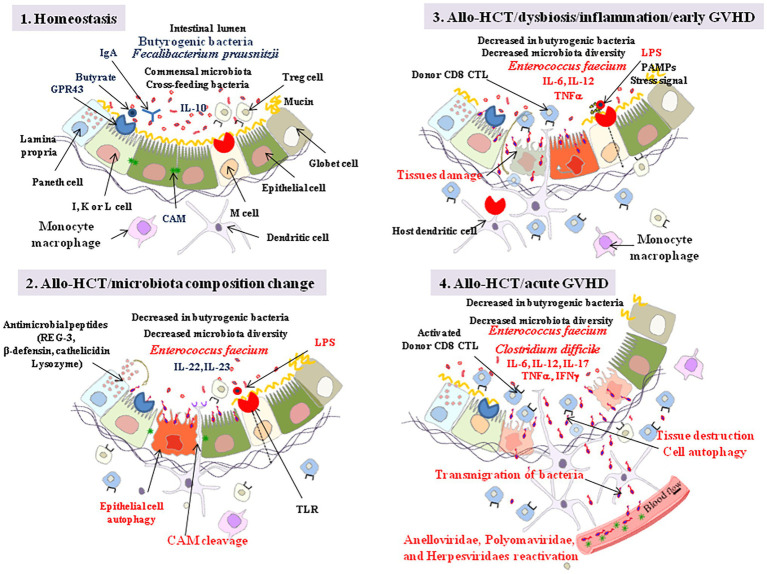
The gut microbiota shapes the gut mucosal immune system and the intestinal immunoglobulin A (IgA) produced by B cells predominantly target the commensal bacteria that reside in the small intestine, yet more IgA-producing B cells are recruited during inflammatory processes (Million et al., 2018). Gut homeostasis does not just rely on IgA production following microbial immune priming. Vitamin A derived RA together with butyrate were shown to maintain gut immune homeostasis through the induction of CD103+ DC cells (in the mesenteric lymph nodes and colonic lamina propria) and recruitment of FoxP3+ T reg cells (Qiang et al., 2017). The butyric acid bacteria F. prausnitzii (one of the most abundant Firmicutes in the gut) that are enzymatically equipped for the conversion of acetate into butyrate (it express acetate CoA transferase and butyrate kinase; Duncan et al., 2002), were found to exhibit anti-inflammatory properties through the stimulation of anti-inflammatory cytokine IL-10 expression and reduction of the pro-inflammatory cytokine IL-8 expression (Tremorali and Backhed, 2012; Heinken et al., 2014). Butyrate was also found to suppress expression of proinflammatory cytokines interleukin 6 (IL-6) and tumor necrosis factor (TNF)-α to reduce IL-12 expression, and butyrate-treated DCs showed a decreased capability in priming CD4+ T cell proliferation. However, SFCAs at high concentration can also induce Th1 and Th17 cells upon immunological challenge and, therefore, also have the potential to induce inflammatory responses (production of IL-17 and IFNγ; Park et al., 2020). Moreover, the induction of the pro-inflammatory cytokine IL-23 in DCs by butyrate is linked to its role as a HDAC inhibitor for epigenetic modification of genes. Through binding to the retinoic acid-receptor -γt, IL-23 induces the production of the epithelial cell regenerative factor IL-22. Altogether, these observations indicate that CD103+ DC cells and FoxP3+ T reg cells, therefore, regulate the balance between gut tolerance and intestinal inflammation (Figure 6).
Figure 6.
Allo-HCT: from homeostasis to acute GVHD. Step 1: intestinal lumen microenvironment characteristic of homeostasis with high butyrate, presence of FoxP3 T reg cells, and anti-inflammatory interleukin 10 (IL-10). Butyrate controls the homeostasis through interaction with GPR43 expressed by enteroendocrine I, K, and/or L cells. The engagement of GPR43 triggers ERK signaling and subsequent NOD-LRR‐ and pyrin domain-containing protein 3 (NLRP3) inflammasome activation which promotes protection of intestinal epithelial cells and repairs by increasing IL-18 secretion. Butyrate also acts as a histone deacetylase inhibitor, thereby increasing expression of anti-apoptotic genes (e.g., BCL-B, a member of the BCL-2 family). The epithelial cells maintain a barrier through intercellular junctions that involve CAM, such as E-cadherin. Step 2: following allo-HCT, when the gut microbiota suffers quantitative and qualitative changes in bacteria composition (e.g., predominance of En. faecium), the release of bacterial LPS is considered as responsible for initiating an innate immune response through activation of TLRs at the surface of villous microfold cells (M cells). These cells deliver the luminal antigens to the underlying immune system to set up a whole arsenal of anti-bacterial innate immunity. Indole transitorily stimulates the production of IL-22 which protects the intestinal epithelial cells. Host DCs present recipient allo-antigens to donor CD4+ and CD8+ T cells that produce pro-inflammatory cytokines, serving as a breeding ground for GVHD. Through their MR1 molecules, dendritic cells (DCs) also activate host mucosal associated invariant T (MAIT) cells to secrete IL-17A, which enhances the intestinal barrier integrity. The Paneth cells produce antimicrobial proteins (e.g., C-type lectin REG3γ, β-defensins, cathelicidins, and lysozyme). Sheddases produced by bacteria or by eukaryotic cells through bacterial signaling initiate the cleavage of cell adhesion molecule (CAM; e.g., cleavage of E-cadherin), creating damages in the intestinal barrier. Step 3: the sequential detection of stress signals, tissue damages, and pathogen-associated molecular patterns (PAMPs; e.g., lipopolysaccharide, flagellin, peptidoglycan, lipoproteins, and unique bacterial nucleic acid structures) by the host antigen presenting cells activates the immune system. KLRG1+ dendritic cells and monocytes/macrophages, CD103+ T-cells, KLRG1+ T-cells, and other immune cell subpopulations, colonize the lamina propria. To challenge the dysbiosis, epithelial cell autophagic clearance and dead cells renewal accelerates but remains unsuccessful due to aggravation of the pro-inflammatory process that combine activation of anti-recipient host alloantigens donor CD8+ cytotoxic T cells (CTL) and proinflammatory cytokines (IL-6, IL-12, TNFα) production. Alloreactive T cells from the donor attack healthy tissues in the recipient allo-HCT patient. Step 4: the inflammatory process develop accompanied with transmigration of bacteria outside the intestinal lumen (Clostridium difficile has been frequently found associated with severe GVHD), reactivation of viruses, further tissue destruction and global aggravation (spread to other tissues) of the disease with characteristic symptoms including abdominal cramping and diarrhea, and biological markers of abnormal liver function (elevation of alkaline phosphatase and bilirubin). GVHD can be also monitored by skin, liver, and gut biopsis. GVHD is also associated with lower urine levels of 3-indoxyl sulfate.
A high fat diet is known to alter the composition of the intestinal microbiota, notably decreasing the number of the Gram-positive Bifidobacteria bacteria and increasing the proportion of Gram-negative bacteria in the gut, and, hence, increase the release and plasma concentration of LPS and trigger TLR induction that generates low-grade chronic inflammation (Wagnerberger et al., 2012). Indeed, the TLR+ villous microfold cells (M cells) present in the intestinal epithelium deliver luminal antigens to the underlying immune system either by transcytosis or microvesicle uptake, while alternatively M cell apoptosis and generation of transcellular pores through which DCs can gain access to the intestinal lumen may also allow capture of antigens and antigen presentation (Million et al., 2018). In mice model with deleted LPS receptors and CD14 mutants (CD14 is the co-receptor for the LPS-receptor TLR4), a hypersensitivity to insulin was reported, suggesting that high-fat diet-induced metabolic dysfunction occurs through the LPS/CD14 signaling pathway (Cani et al., 2007). LPS is a marker of Gram-negative bacteria death. High-fat diets have been shown to induce the passage of LPS from the intestine into the mesenteric lymphatic system (possibly through binding of chylomicrons, formed from dietary triglycerides, to LPS), leading to inflammation (Vreugdenhil et al., 2003). Production of chylomicrons was also reported capable to promote production the (TNF)-α proinflammatory cytokine. In mice models, cross linking of TLR9 by bacterial DNA stimulate GVHD whereas mutations in the TLR9 and TLR4 genes preventing bacterial LPS recognition were found to reduce GVHD (Calcaterra et al., 2008; Imado et al., 2010). Polymorphism of nucleotide-binding oligomerization domain 2 (NOD2) gene that encodes a receptor for the pathogen-associated molecular patterns (PAMPs) bacterial associated molecules is linked with a higher incidence of GVHD in HSC transplant recipients (Penack et al., 2010). The TLR5 agonist fagellin extracted from bacterial flagella was shown to reduce GVHD and preserve long post-transplant immune reconstitution characterized by more Fop-3+ T regulator (T reg) cells (Hossain et al., 2011). Studying the role for TLR9 and its downstream signaling adaptor MYD88 in an intestine GVHD model, Heimessat et al. (2010) observed a shift towards Enterobacteriaceae, Enterococci, and Bacteroides/Prevotella spp. being elevated in the colon lumen during GVHD, whereas Lactobacilli, Clostridia, Bifidobacteria and Bacillus spp. were less abundant. In fine, GVHD is mediated by CD8+ CTLs that express anti-host specificities. In a murine model, it was found that GVHD may be inhibited by preventing CD8+ CTL migration into the Peyer’s patch either by disrupting the gene encoding the CCR5 chemokine receptor or by blocking the integrin α4β7-mucosal vascular addressin (MAdCAM-1) interaction into the gut Peyer’s patches (Murai et al., 2003). Since mutants in the integrin α4β7 can also affect the integrin binding to E-cadherin (Higgins et al., 2000), it remains possible that such mutants might also affect the anti-bacterial CTL immune response through aberrant homing signals that involve E-cadherin, soluble E-cadherin, CD103, and KLRG1, as previously suggested (Devaux et al., 2019).
Is there Room for Viruses in GVHD?
Immense populations of viruses, among which bacteriophages, are present in the human gut, and lysogenic or temperate phages are able to integrate their chromosome into the bacterial genome, sometimes altering the phenotype of host bacteria. A number of different eukaryotic viruses have also been found in the human gut virome with a predominance of the pepper mild mottle virus, a plant-infecting RNA virus derived from diet (Zhang et al., 2006). A very elegant study by Minot et al. (2011) reported the presence of Myoviridae, Siphoviridae, Podoviridae, Tectiviridae, Inoviridae, and Microviridae in human stools from healthy donors. Among double stranded DNA viruses found in the human gut, a predominance of Podoviridae followed by Siphoviridae and Myoviridae was reported in healthy humans while Microviridae dominated the single strand DNA viruses (Kim et al., 2011). Kim et al. (2011) also reported the rare presence of picornaviruses, herpesviruses, and poxviruses. Many viruses (e.g., rotaviruses, caliciviruses, astroviruses, enteric adenoviruses, toroviruses, and parechoviruses) are known to induce gastroenteritis in humans, the leading cause of diarrheal disease being rotaviruses, a Reoviridae (Clark and McKendrick, 2004). Therefore, it cannot be ignored that the virome could influence GVHD just as easily as the microbiome. Although eukaryotic cell-borne viruses are minor components of the human gut virome compared to the bacteriophages, latent viral infections can be reactivated in patients with allo-HCT who experience a phase of immunosuppression. Norovirus induced gastroenteritis was found to be a major threat to patients to allo-HCT (Schwartz et al., 2011). Reactivation of human herpes virus 6 (HHV-6) was reported to be a predictive factor for acute GVHD (Pichereau et al., 2012). The HHV-6 reactivation reported in 35% of patients who received an allo-HCT (sometimes associated with a subsequent cytomegalovirus reactivation) was found predictive of mortality after allo-HCT (Zerr et al., 2012). Feghoul et al. (2015) reported that human adenovirus infections constitute a major cause of morbidity in pediatric allo-HCT patients. The incidence rate at day 100 was 35.9% for the adenovirus digestive infections and 24.0% for the systemic infections. Infection with herpes simplex virus-1 and 2 is mostly seen during the pre-engraftment phase, whereas cytomegalovirus and HHV-6 are mainly found during the post-engraftment phase, and Epstein-Barr virus and varicella-zoster virus infections are often observed after the 100th day post-transplant (Sahin et al., 2016).
Yang et al. (2016) reported unexpected results indicating that mice treated with antiviral cocktail display more severe dextran sulfate sodium-induced colitis than untreated mice suggesting that gut resident viruses may insure maintenance of gut homeostasis. DCs isolated from colon of inflamed mice produced interferon-β in a TLR-dependent manner. When mice were reconstituted with toll-like receptors or rotavirus, colitis symptoms were significantly ameliorated, suggesting that enteric rotaviruses damped gut inflammation via toll-like receptors TLR-3 and TLR-7-mediated interferon β production. Consequently, it could not be excluded that human gut eukaryotic viruses can also be beneficial to patients in limiting GVHD. Legoff et al. (2017) reported that within a cohort of patients who received allo-HCT, patients who experienced enteric GVHD had both decreased richness of virome and higher abundance of Microviridae. Anelloviridae, Polyomaviridae, and Herpesviridae were found in immunocompromised allo-HCT patients likely related to viral reactivation. Recently, it was reported that myxoma virus, a Poxviridae that exhibits oncolytic activity against various hematologic malignancies like multiple myeloma or acute myeloid leukemia could be used, has a tool for ex-vivo treatment of allo-HCT with evidence of possible GVHD abrogation without impairing graft-versus-tumor effects against residual cancer cells (Villa and McFadden, 2018).
Discussion
Allo-HCT is a potentially curative treatment of hematologic malignancies. However, the effectiveness of this treatment remains limited by the high incidence of acute GVHD, which is the principal cause of death in allo-HCT (Riwes and Reddy, 2018). Pioneers’ experimental studies carried out in the 1970s already indicated less acute GVHD in mice which received allo-HCT in germ-free conditions or experimental group receiving gut decontamination antibiotics (van Bekkum et al., 1974). The influence of intestinal bacteria on the development of acute GVHD was further confirmed in patients receiving allo-HCT as treatment of hematologic malignancies. More than 20 years ago, evidence was obtained that antimicrobial chemotherapy (metronidazole and ciprofloxacin) targeted to intestinal anaerobic bacteria in marrow transplant recipients significantly reduced the severity of acute GVHD (Beelen et al., 1999). Although it was hypothesized that allogenic GVHD is driven by initial interaction between APCs that encounter donor T lymphocytes that in turn release pro-inflammatory cytokines recruiting allo-reactive T cells, the molecular crosstalk that account for allo-HCT tolerance or GVHD was largely ignored (Magenau et al., 2016). Indeed, it is very likely that during the set up of post-transplant GVHD, the control of the innate immune response is dependent on both the bacteria which produce butyrate and those which produce lactase. The precise quantitative and qualitative adjustment of these bacterial populations is probably the key to the balance between immune tolerance and inflammatory process/GVHD.
A decrease in SCFAs (butyrate) was found to be associated with an increased mortality from GVHD following allo-HCT transplantation (Mathewson et al., 2016). Next, the same research group reported that the GVHD protective effect of SCFAs requires GPR43-mediated intracellular signaling that triggers ERK phosphorylation, ERK-dependent activation of IL-18, and activation of the NOD-LRR‐ and pyrin domain-containing protein 3 (NLRP3) inflammasome (Fujiwara et al., 2018). GPR43 is expressed by intestinal epithelial cells, as well as by antigen presenting cells (APCs, such as macrophages and DCs). Fujiwara et al. (2018), reported that macrophages and DCs isolated from ileum and colon of allo-HCT recipients show greater expression of GPR43 (whereas in vivo GPR43 expression on donor T cells is weak), but only the expression of GPR43 on non-hematopoietic cells (presumably intestinal epithelial cells) was involved in protective effect of SCFAs against GVHD. Moreover, a diet supplemented with butyrate mitigates GVHD. According to Golob et al. (2019), the beneficial response to butyrate would be biphasic with the first phase corresponding to the prevention of acute GVHD and the second phase dealing with the recovery, once colitis is durably established. It was also reported that the abundance of Clostridia decreased in the microbiota of allo-transplant patients that experienced GHVD and was accompanied by alteration in gastrointestinal microbiota-derived butyrate (Mathewson et al., 2016). The central role played by butyrate in the prevention of GVHD indicates that the presence of butyrogenic bacteria is essential for GVHD prevention. Butyrate is the main microbiota-derived regulator of gut mucosal immunity. Butyrogenic bacteria are considered oxidative stress sensitive. Most of these bacteria do not synthesize butyrate in the absence of cross-feeding bacteria that are oxidative stress sensitive. Under oxidative stress, the presence of butyrogenic bacteria in the microbiota decrease and oxidative stress-resistant strains (that encode enzymes such as catalase or superoxide dismutase required to deal with oxidative stress) are selected and trigger secretion of pro-inflammatory cytokines by immune response cells (Million et al., 2018).
In human, lactase persistence is associated with enhanced milk intake, higher waist circumference, and cardiometabolic abnormalities. Higher dairy intake was associated with higher body mass index supporting a causal link between lactose ingestion and body weight (Huang et al., 2018). When consuming dairy products, a person with lactase intolerance (the most common cause of the disease being a genetic mutation in the promoter of the gene that codes for lactase resulting in loss of intestinal lactase) may experience the symptoms of lactose intolerance which includes nausea, abdominal pain, gas, bloating, and diarrhea (Misselwitz et al., 2019; Kastl et al., 2020). Beside lactose, dairy products also provide numerous biologically active factors including growth factors, including the fibroblast growth factor FGF21 (mainly produced in the liver, brown and white adipocyte tissues, and pancreas) that was found to enhance MAPkinases (ERK1/2) phosphorylation in intestinal explants and to stimulate lactase production and lactose absorption (Gavaldà-Navarro et al., 2015). Gavaldà-Navarro et al. (2015) reported that mice feed with milk from FGF21 knockout mice showed decreased expression of lactase and maltase glucoamylase in the ileum. FGF21 binds to the FGFreceptor in complex with βKlotho to trigger cell signaling (Kurosu et al., 2007), yet the mode of FGF21 regulation of lactase expression by intestinal cells remains to be explored. This growth factor is known for stimulating the oxidation of fatty-acids, the production of ketone bodies, and inhibition of lipogenesis, thus regulating the glucose-lipid metabolism (Tezze et al., 2019). Recently, it was reported that feeding allo-transplanted mice with lactose free diet mitigated GVHD and reduced post-transplant Enterococcus proliferation (the optimal growth of which depends on lactose availability), whereas in mice feed with a dairy products diet, the lactase expression declined in the duodenum during the course of transplantation and induced a pathological state resembling lactose intolerance (Stein-Thoeringer et al., 2019).
Holler et al. (2014) reported a shift toward Enterococci after allo-HCT with a decrease in the obligate Firmicutes anaerobic bacteria that was much pronounced in the patients treated with antibiotics. Stein-Thoeringer et al. (2019) hypothesized that intestinal mucosal damages caused by irradiation or allo-reactive T cells may reduce the production of lactase from the small intestine enterocytes, that duodenal lactase progressively decline allowing undigested lactose to reach the lower intestinal tract where it serves as metabolic fuels for the growth of Enterococcus (e.g., En. faecium in human, En. faecalis in mice). Nalle et al. (2019) recently reported that MLCK210-deficient mice exhibited limited GVHD and were protected from epithelial barrier damages and CD8+ T cells proliferation. These authors discriminate the initiation of GVHD considered to be tight junction-independent from GVHD propagation that is MLCK210-dependent. Stein-Thoeringer et al. (2019) reported that En. faecium (only recently considered a human pathogen) dominate in patients with allo-HCT between 3-week and 3-month post-transplantation and that fecal domination by Enterococcus in the early post-transplant period (the first 3-week post-transplant) was associated with increased GVHD and overall mortality. They also found the VanA operon in a subset (37.4%) of patients. In an animal model, Stein-Thoeringer et al. (2019) reported a transient expansion of En. faecalis in GVHD mice and administration of En. faecalis aggravated the GVHD, thus demonstrating a direct link between the Enterococcus predominance and the GVHD. A major breakthrough came along with the attempt to demonstrate that the post-transplant defect of mucosal defense mechanisms facilitate the expansion of Enterococcus. They found that intestinal antimicrobial peptides of the Reg3 family, known to suppress the growth of vancomycin resistant Enterococci (VRE bacteria) that exploit innate immune deficits (Brandl et al., 2008), were reduced in the ileum of GVHD mice. Obviously, the primary strategy to prevent GVHD after allo-HCT is immunosuppression, but such treatment may increase the risk of enteropathogenic bacteria invasion. Stein-Thoeringer et al. (2019) also provided an elegant demonstration that En. faecium-dominated microbiota observed after HCT were enriched in bioactive compounds involved in lactose and galactose degradation pathway (with similar result in En. faecalis-dominated microbiota in mice). En. faecalis expresses the gelatinase (GelE) a matrix metalloprotease, so-called sheddase, that cleaves E-cadherin (Steck et al., 2011) and activates the protease-activated receptor 2 (PARP2), a transmembrane G-protein-coupled receptor (Maharshak et al., 2015). These molecules contribute to the disruption of intestinal barriers and inflammation. According to the UniProt database1, En. faecalis also encodes a predicted htrA protein serine protease, another candidate sheddase (Devaux et al., 2019).
In mice animal model, it was observed that PPIs (namely omeprazole) administered to mice cause reduced gastric secretion and favor the expansion of Enterococcus species including En. faecalis (Llorente et al., 2017). This favor bacterial translocation in mesenteric lymph nodes and the liver with subsequent inflammation, hepatocytes death, and non-alcoholic steatohepatosis. Mice that lack expression of the TLR2 (a cell membrane receptor that recognizes peptidoglycan from Gram-positive bacteria) or the myeloid differentiation primary response 88, MYD88 (intracellular adaptor molecule for TLRs), lack innate immune response and were protected from En. faecalis-induced inflammation, steatosis, and liver injury. En. faecalis was found capable of suppressing the innate immune response of macrophages through repression of NF-κB signaling (Tien et al., 2017). In a human cohort, administration of omeprazole for preclinical evaluation, the frequency of Enterococcus in the fecal samples was increased after 2-week of PPI treatment. This corroborates previous observation indicating that an omeprazole therapy in cirrhosis demonstrated a shift in fecal microbiota composition (Bajaj et al., 2014; Merli et al., 2015). Innate immune response that follows the engagement of TLRs is involved in the process of GVHD. The TLR5 agonist fagellin was shown to reduce GVHD and preserve long post-transplant immune reconstitution characterized by more Fop-3+ T regulator (T reg) cells (Hossain et al., 2011). Although it remains to be documented in humans suffering from ten-eleven translocation (TET) methylcytosine dioxygenase 2 (TET2)-deficiency, in an animal model it was reported that mutations in TET2, which encodes an epigenetic modifier enzyme that influence the activation of regulatory CD4+ T cells (T reg cells) via FoxP3 (Yue et al., 2016), can be associated with dysfunction of the small intestine barrier, bacterial translocation and IL-6 production known to be a critical activator of myelopoiesis in response to systemic bacterial dissemination (Meisel et al., 2018).
Clostridium difficile has also been implicated as a potential trigger of immune reaction that can contribute to the development of GVHD. Working with a cohort of 75 allo-HCT patients, Chakarbati et al. (2000), found that C. difficile was frequently associated with severe GVHD (grade 3–4) and the presence of the bacteria in stool worsened the pathophysiology of GVHD in 60% of patients. The relationship between the presence of C. difficile and GVHD was also reported by Dubberke et al. (2007) who reported that patients with C. difficile showed a higher propensity to develop new-onset GVHD and severe forms of GVHD. Similar observations were reported from the investigation of a cohort of 822 allo-HCT patients regarding the risk to develop severe GVHD at day 60 and day 100 after allo-HCT when C. difficile was present in the patients (Trifilio et al., 2012). The incidence was more than 20% in patients who were older than 60 years and carried a vancomycin-resistant (VRE) C. difficile. Indeed, C. difficile positive diagnosis was found to precede GVHD diagnosis in 85.7% of allo-HCT patients who developed gut GVHD while the overall 1-year incidence of C. difficile was 9.2% among a cohort of 999 allo-HCT patients (Alonso et al., 2012; Alonso and Marr, 2013). Although the presence of VRE C. difficile was not investigated in the main papers discussed in this review, it can be hypothesized that the decreased microbiota diversity (butyrogenic bacteria) facilitates the clonal expansion C. difficile strains in the gut of these patients (Hocquart et al., 2018), which in turn represents an aggravating factor regarding the risk of mortality by GVHD of allo-HCT patients.
CTLA-4, currently known as a target for immune checkpoint inhibitor antibodies, was originally described as a T cell surface molecule that negatively influence immune response by competing with CD28 for binding to ligands on APCs. In patients with severe alcoholic hepatitis, it was reported that PD-1 immune checkpoint-receptor inhibition restored the adaptive antibacterial T cell response which otherwise was defective (Markwick et al., 2015). The approval of immune checkpoint inhibitors (such as CTLA-4 or PD1/PD-L1 inhibitor antibodies) in the treatment of cancers has changed the outcome of several of such severe diseases and highlighted the strong molecular crosstalk between the microbiota and the immune system (Zitvogel et al., 2018; Gong et al., 2019). Defects in the microbiota can compromise the therapeutic efficacy or have secondary side effect on the gut. In melanoma therapy, anti-PD1 and anti-CTLA-4 antibodies have been approved. However, immune-mediated colitis in patients with melanoma after dual checkpoint inhibitors treatment was described (Thalambedu et al., 2019). A variety of genera can reduce the immune checkpoint inhibitor-induced colitis possibly by limiting the inflammation through expansion of CD4+/FoxP3+ T reg cells and/or production of anti-inflammatory cytokines (Dubin et al., 2016). In a murine model of melanoma, CD8+ T cell activation in response to PD-L1 inhibitor correlated with mice, which received fecal transplantations from patients abundant in bacteria from the Ruminococcaceae family and Faecalibacterium spp., whereas nonresponders were characterized by elevated presence of CD4+/FoxP3+ T reg cells and received stools which were abundant in Bacteroidales (Gopalakrishnan et al., 2018).
The experience accumulated in the field of cancer therapy with immune checkpoint inhibitors provides evidence that the microbiota govern the balance between CD8+ T cell activation and inflammation on one side and CD4+/FoxP3+ T reg cells and lack of inflammation on the other side. It can be extrapolated that high butyrate allows CD4+/FoxP3+ T reg cells expansion and prevents inflammation, and GHVD reaction whereas low lactase induces lactic acid bacteria expansion, CD8+ T cell activation, inflammation and GVHD. However, we are just beginning to understand what could be the role of the microbiota and the impact of cross feeding in GVHD. First, variation in the composition of the microbiota between allo-HCT patients that lack GVHD and those who unfortunately experience GVHD is currently well established. Second, the role played by butyrogenic and lactic acid bacteria begun to be understood. However, it is now necessary to enter more deeply into the characterization of the bacterial species that distinguish homeostasis from dysbiosis in non-GVHD versus GVHD patients and to study their enzymatic equipment which can vary both at the level of the bacterial strain and at the clone level. There is currently evidence that the adverse outcomes of irradiation and antibiotics/chemotherapy treatments (oxidative stress) on gut microbiota could be avoided by using probiotics and/or fecal microbiota transplant, opening new perspective for the prevention of GVHD in allo-HCT patients (Delia et al., 2002; Kaito et al., 2018; Severyn et al., 2019; Shouval et al., 2019). The next challenge is to offer transplant patients a beneficial combination of easy-to-consume probiotics, which should prevent the occurrence of GVHD. Strict anaerobic bacteria are not the easiest to cultivate and produce in large quantity for therapeutic application. We have recently reported that oxygen-sensitive bacteria can maintain butyrate production despite the presence of oxygen when treated with antioxidants (Million et al., 2020). Among lactic acid bacteria, the Bifidobacterium seem to be the best candidates as anti-GVHD probiotics. In contrast, the Enterococci which also metabolize lactose are suspected of being deleterious and responsible for the production of noxious substances. If this therapeutic cocktail must contain cross feeding bacteria and butyrogenic bacteria, it will be necessary to exclude those which are known for their deleterious side effects (i.e., Clostridium butyricum, associated with enterocolitis, or Mediterraneibacter gnavus, associated with obesity) and to investigate the beneficial cooperative effects of bacteria such as B. adolescentis, B. longum, Eu. hallii, R. intestinalis, and particularly F. prausnitzii.
Author Contributions
CD, MM, and DR contributed to the conception of the manuscript. CD wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Figures were designed using the Servier Medical Art supply of images available under a Creative Commons CC BY 3.0 license.
Funding. This study was supported by IHU Méditerranée Infection, University of Marseille, and CNRS, France. This work has benefited from the French State support, managed by the “Agence Nationale de la Recherche,” including the “Programmed’Investissement d’Avenir” under the reference Méditerranée Infection 10-1AHU-03.
References
- Alonso C. D., Marr K. A. (2013). Clostridium difficile infection among hematopoietic stem cell transplant recipients: beyond colitis. Curr. Opin. Infect. Dis. 26, 326–331. 10.1097/QCO.0b013e3283630c4c, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C. D., Treadway S. B., Hanna D. B., Huff C. A., Neofytos D., Carroll K. C., et al. (2012). Epidemiology and outcomes of Clostridium difficile infections in hematopoietic stem cell transplant recipients. Clin. Infect. Dis. 54, 1053–1063. 10.1093/cid/cir1035, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames N. J., Sulima P., Ngo T., Barb J., Munson P. J., Paster B. J., et al. (2019). A characterization of the oral microbiome in allogeneic stem cell transplant patients. PLoS One 7:e47628. 10.1371/journal.pone.0047628, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andermann T., Peled J., Ho C., Reddy P., Riches M., Storb R., et al. (2018). The microbiome and hematopoietic cell transplantation: past, present, and future. Biol. Blood Marrow Transplant. 24, 1322–1340. 10.1016/j.bbmt.2018.02.009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelos T., Nikolaos S., Nikolaos K., Nektarios P., Evdokia P. (2018). A rare case of tricuspid valve endocarditis caused by Enterococcus gallinarum in a patient with recurrent urinary tract infection. Cardiovasc. Hematol. Disord. Drug Targets 18, 162–164. 10.2174/1871529X18666180302113428, PMID: [DOI] [PubMed] [Google Scholar]
- Aristimuño Ficoseco C., Mansilla F. I., Maldonado N. C., Miranda H., Nader-Macias M. E. F., Vignolo G. M. (2018). Safety and growth optimization of lactic acid bacteria isolated from feedlot cattle for probiotic formula design. Front. Microbiol. 9:2220. 10.3389/fmicb.2018.02220, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta M. C., Stiemsma L. T., Amenyogbe N., Brown E. M., Finlay B. (2014). The intestinal microbiome in early life: health and disease. Front. Immunol. 5:427. 10.3389/fimmu.2014.00427, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgari F., Madjid Z., Falak R., Bahar M. A., Nasrabadi M. H., Raiani M., et al. (2016). Probiotic feeding affects T cell populations in blood and lymphoid organs in chickens? Benef. Microbes 7, 669–675. 10.3920/BM2016.0014, PMID: [DOI] [PubMed] [Google Scholar]
- Bajaj J. S., Cox I. J., Betrapally N. S., Heuman D. M., Schubert M. I., Ratneswaran M., et al. (2014). System biology analysis of omeprazole therapy in cirrhosis demonstrates significant shift in gut microbiota composition and function. Am. J. Physiol. Gastrointest. Liver Physiol. 307, G951–G957. 10.1152/ajpgi.00268.2014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T., Alaniz R. C., Wood T. K., Jayaraman A. (2010). The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U. S. A. 107, 228–233. 10.1073/pnas.0906112107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. D., Liu Y. W., Hanly A. M., Emenaker N. J., Shenoy S. G., Gould Rothberg B. E. (2000). Identification and comparative analysis of human colonocyte short-chain fatty acid response genes. J. Gastrointest. Surg. 4, 501–512. 10.1016/S1091-255X(00)80093-1, PMID: [DOI] [PubMed] [Google Scholar]
- Beelen D. W., Elmaagacli A., Müller K. D., Hirche H., Schaefer U. W. (1999). Influence of intestinal bacterial decontamination using metronidazole and ciprofloxacin or ciprofloxacin alone on the development of acute graft-versus-host disease after marrow transplantation in patients with hematologic malignancies: final results and long-term follow-up of an open-label prospective randomized trial. Blood 93, 3267–3275. PMID: [PubMed] [Google Scholar]
- Beganovic M., Luther M. K., Rice L. B., Arias C. A., Rybak M. J., LaPlante K. L. (2018). A review of combination antimicrobial therapy for Enterococcus faecalis bloodstream infections and infective endocarditis. Clin. Infect. Dis. 67, 303–309. 10.1093/cid/ciy064, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A., Duncan S. H., Calder G., Holtrop G., Louis P., Lobley G. E., et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 72, 3593–3599. 10.1128/AEM.72.5.3593-3599.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazar B. R., Murphy W. J., Abedi M. (2012). Advances in graft-versus-host disease biology and therapy. Nat. Rev. Immunol. 12, 443–458. 10.1038/nri3212, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booijink C. C., El-Aidy S., Rajilic-Stojanovic M., Heilig H. G., Troost F. J., Smidt H., et al. (2010). High temporal and inter-individual variation detected in the human ileal microbiota. Environ. Microbiol. 12, 3213–3227. 10.1111/j.1462-2920.2010.02294.x, PMID: [DOI] [PubMed] [Google Scholar]
- Bortolaia V., Mander M., Jensen L. B., Olsen J. E., Guardabassi L. (2015). Persistence of vancomycin resistance in multiple clones of Enterococcus faecium isolated from Danish broilers 15 years after the ban of avoparcin. Antimicrob. Agents Chemother. 59, 2926–2929. 10.1128/AAC.05072-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl K., Plitas G., Mihu C. N., Ubeda C., Jia T., Fleisher M., et al. (2008). Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807. 10.1038/nature07250, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. T., Wittenberger C. L. (1971). Mechanism for regulating the distribution of glucose carbon between the Embden-Mayerhof and hexose-monophosphate pathways in Streptococcus faecalis. J. Bacteriol. 106, 456–467. 10.1128/JB.106.2.456-467.1971, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Small N. (1956). The anaerobic monotrichous butyric acid-producing curved ros-shaped bacteria of the rumen. J. Bacteriol. 72, 16–21. 10.1128/JB.72.1.16-21.1956, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral M. E., Mukdsi M. A. C., Medina de Figueroa R. X., Gonzalez S. N. (2007). Citrate metabolism by Enterococcus faecium and Enterococcus durans isolated from goat’s and ewe’s milk: influence of glucose and lactose. Can. J. Microbiol. 53, 607–615. 10.1139/W07-011, PMID: [DOI] [PubMed] [Google Scholar]
- Calcaterra C., Sfondrini L., Rossini A., Sommariva M., Rumio C., Ménard S., et al. (2008). Critical role of TLR9 in acute graft-versus-host disease. J. Immunol. 181, 6132–6139. 10.4049/jimmunol.181.9.6132, PMID: [DOI] [PubMed] [Google Scholar]
- Cani P. D., Amar J., Iglesias M. A., Poggi M., Knauf C., Bastelica D., et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. 10.2337/db06-1491, PMID: [DOI] [PubMed] [Google Scholar]
- Chakrabarti S., Lees A., Jones S. G., Milligan D. W. (2000). Clostridium difficile infection in allogeneic stem cell transplant recipients is associated with severe graft-versus-host disease and nonrelapse mortality. Bone Marrow Transplant. 26, 871–876. 10.1038/sj.bmt.1702627, PMID: [DOI] [PubMed] [Google Scholar]
- Clark B., McKendrick M. (2004). A review of viral gastroenteritis. Curr. Opin. Infect. Dis. 17, 461–469. 10.1097/00001432-200410000-00011, PMID: [DOI] [PubMed] [Google Scholar]
- Clausen M. R., Mortensen P. B. (1994). Kinetic studies on the metabolism of short-chain fatty acids and glucose by isolated rat colonocytes. Gastroenterology 106, 423–432. 10.1016/0016-5085(94)90601-7, PMID: [DOI] [PubMed] [Google Scholar]
- Collado M. C., Rautava S., Aakko J., Isolauri E., Salminen S. (2016). Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6:23129. 10.1038/srep23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J. H., Macfarlane G. T. (1991). The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70, 443–459. 10.1111/j.1365-2672.1991.tb02739.x, PMID: [DOI] [PubMed] [Google Scholar]
- Cummings J. H., Pomare E. W., Branch W. J., Naylor C. P. E., MacFarlane G. T. (1987). Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227. 10.1136/gut.28.10.1221, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vuyst L., Leroy F. (2011). Cross-feeding between Bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int. J. Food Microbiol. 149, 73–80. 10.1016/j.ijfoodmicro.2011.03.003, PMID: [DOI] [PubMed] [Google Scholar]
- Deleemans J. M., Chleilat F., Reimer R. A., Henning J. W., Baydoun M., Piedalue C. A., et al. (2019). The chemo-gut study: investigating the long-term effects of chemotherapy on gut microbiota, metabolic, immune, psychological and cognitive parameters in young adult cancer survivors; study protocol. BMC Cancer 19:1243. 10.1186/s12885-019-6473-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delia P., Sansotta G., Donato V., Messina G., Frosina P., Pergolizzi S., et al. (2002). Prevention of radiation-induced diarrhea with the use of VSL#3, a new high-potency probiotic preparation. Am. J. Gastroenterol. 97, 2150–2152. 10.1111/j.1572-0241.2002.05946.x, PMID: [DOI] [PubMed] [Google Scholar]
- Della Ragione F., Criniti V., Della Pietra V., Borriello A., Oliva A., Indaco S., et al. (2001). Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Lett. 499, 199–204. 10.1016/S0014-5793(01)02539-X, PMID: [DOI] [PubMed] [Google Scholar]
- Devaux C. A., Mezouar S., Mege J. L. (2019). The E-cadherin cleavage associated to pathogenic bacteria infections can favor bacterial invasion and transmigration, dysregulation of the immune response and cancer induction in humans. Front. Microbiol. 10:2598. 10.3389/fmicb.2019.02598, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C., Raoult D. (2018). The microbiological memory, an epigenetic regulator governing the balance between good health and metabolic disorders. Front. Microbiol. 9:1379. 10.3389/fmicb.2018.01379, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubberke E. R., Sadhu J., Gatti R., Reske K. A., DiPersio J. F., Devine S. M., et al. (2007). Severity of Clostridium difficile-associated disease (CDAD) in allogeneic stem cell transplant recipients: evaluation of a CDAD severity grading system. Infect. Control Hosp. Epidemiol. 28, 208–211. 10.1086/511792, PMID: [DOI] [PubMed] [Google Scholar]
- Dubin K., Callahan M. K., Ren B., Khanin R., Viale A., Ling L., et al. (2016). Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7:10391. 10.1038/ncomms10391, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Barcenilla A., Stewart C. S., Pryde S. E., Flint H. J. (2002). Acetate utilization and butyryl coenzyme A (CoA): acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 68, 5186–5190. 10.1128/AEM.68.10.5186-5190.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Louis P., Flint H. J. (2004). Lactate-utilizing bacteria, isolated from human faeces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70, 5810–5817. 10.1128/AEM.70.10.5810-5817.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. 10.1126/science.1110591, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkrief A., Derosa L., Zitvogel L., Kroemer G., Routy B. (2019). The intimate relationship between gut microbiota and cancer immunotherapy. Gut Microbes 10, 424–428. 10.1080/19490976.2018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshaghabee F. M. F., Bockelmann W., Meske D., Vrese M. D., Walte H. G., Schrezenmeir J., et al. (2016). Ethanol production by selected intestinal microorganisms and lactic acid bacteria growing under different nutritional conditions. Front. Microbiol. 7:47. 10.3389/fmicb.2016.00047, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G., Calmeyn T., Leroy F., De Vuyst L. (2009). Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl. Environ. Microbiol. 75, 2312–2319. 10.1128/AEM.02649-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G., Vlachou A., Verbrugghe K., De Vuyst L. (2006). Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 72, 7835–7841. 10.1128/AEM.01296-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghoul L., Chevret S., Cuinet A., Dalle J. H., Ouachée M., Yacouben K., et al. (2015). Adenovirus infection and disease in paediatric haematopoietic stem cell transplant patients: clues for antiviral pre-emptive treatment. Clin. Microbiol. Infect. 21, 701–709. 10.1016/j.cmi.2015.03.011, PMID: [DOI] [PubMed] [Google Scholar]
- Ford C. D., Gazdik M. A., Lopansri B. K., Webb B., Mitchell B., Coombs J., et al. (2017). Vancomycin-resistant Enterococcus colonization and bacteremia and hematopoietic stem-cell transplantation outcomes. Biol. Blood Marrow Transplant. 23, 340–346. 10.1016/j.bbmt.2016.11.017, PMID: [DOI] [PubMed] [Google Scholar]
- Fujiwara H., Docampo M. D., Riwes M., Peltier D., Toubai T., Henig I., et al. (2018). Microbial metabolite sensor GPR43 controls severity of experimental GVHD. Nat. Commun. 9:3674. 10.1038/s41467-018-06048-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavaldà-Navarro A., Hondares E., Giralt M., Mampel T., Iglesias R., Villarroya F. (2015). Fibroblast growth factor 21 in breast milk controls neonatal intestine function. Sci. Rep. 5:13717. 10.1038/srep13717, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golob J. L., DeMeules M. M., Loeffelhoz T., Quinn Z. Z., Dame M. K., Silvestri S. S., et al. (2019). Butyrogenic bacteria after acute graft-versus-host disease (GVHD) are associated with the development of steroid-refractory GVHD. Blood Adv. 3, 2866–2869. 10.1182/bloodadvances.2019000362, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Chehrazi-Raffle A., Placencio-Hickok V., Guan M., Hendifar A., Salgia R. (2019). The gut microbiome and response to immune checkpoint inhibitors: preclinical and clinical strategies. Clin. Transl. Med. 8:9. 10.1186/s40169-019-0225-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C. N., Nezi L., Reuben A., Andrews M. C., Karpinets T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. 10.1126/science.aan4236, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill J. P., Crociani J., Ballongue J. (1995). Characterization of fructose 6 phosphate phosphoketolases purified from Bifidobacterium species. Curr. Microbiol. 31, 49–54. 10.1007/BF00294634, PMID: [DOI] [PubMed] [Google Scholar]
- Han J., Wang Y., Song D., Lu Z., Dong Z., Miao H., et al. (2018). Effects of Clostridium butyricum and Lactobacillus plantarum on growth performance, immune function and volatile fatty acid level of caecal digesta in broilers. Food Agric. Immunol. 29, 797–807. 10.1080/09540105.2018.1457013 [DOI] [Google Scholar]
- Heimesaat M. M., Nogai A., Bereswill S., Plickert R., Fischer A., Loddenkemper C., et al. (2010). MyD88/TLR9 mediated immunopathology and gut microbiota dynamics in a novel murine model of intestinal graft-versus-host disease. Gut 59, 1079–1087. 10.1136/gut.2009.197434, PMID: [DOI] [PubMed] [Google Scholar]
- Heinken A., Khan M. T., Paglia G., Rodionov D. A., Harmsen H. J. M., Thiele I. (2014). Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J. Bacteriol. 196, 3289–3302. 10.1128/JB.01780-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J. M., Cemadas M., Tan K., Irie A., Wang J., Takada Y., et al. (2000). The role of alpha and beta chains in ligand recognition by beta 7 integrins. J. Biol. Chem. 275, 25652–25664. 10.1074/jbc.M001228200, PMID: [DOI] [PubMed] [Google Scholar]
- Hijova E., Chmelarova A. (2007). Short chain fatty acids and colonic health. Bratisl. Lek. Listy 108, 354–358. PMID: [PubMed] [Google Scholar]
- Hill G. R., Teishima T., Rebel V. I., Krijanovski O. I., Cooke K. R., Brinson Y. S., et al. (2000). The p55 TNF-α receptor plays a critical role in T cell alloreactivity. J. Immunol. 164, 656–663. 10.4049/jimmunol.164.2.656, PMID: [DOI] [PubMed] [Google Scholar]
- Hocquart M., Lagier J. C., Cassir N., Saidani N., Eldin C., Kerbai J., et al. (2018). Early fecal microbiota transplantation improves survival in severe Clostridium difficile infections. Clin. Infect. Dis. 66, 645–650. 10.1093/cid/cix762, PMID: [DOI] [PubMed] [Google Scholar]
- Holler E., Butzhammer P., Schmid K., Hundsrucker C., Koestler J., Peter K., et al. (2014). Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol. Blood Marrow Transplant. 20, 640–645. 10.1016/j.bbmt.2014.01.030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L. V., Macpherson A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10, 159–169. 10.1038/nri2710, PMID: [DOI] [PubMed] [Google Scholar]
- Hossain M. S., Jaye D. L., Pollack B. P., Farris A. B., Tselanyane M. L., David E., et al. (2011). Flagellin, a TLR5 agonist, reduces graft-versus-host disease in allogeneic hematopoietic stem cell transplantation recipients while enhancing antiviral immunity. J. Immunol. 187, 5130–5140. 10.4049/jimmunol.1101334, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T., Ding M., Bergholdt H. K. M., Wang T., Heianza Y., Sun D., et al. (2018). Dairy consumption and body mass index among adults: mendelian randomization analysis of 184802 individuals from 25 studies. Clin. Chem. 64, 183–191. 10.1373/clinchem.2017.280701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang W., Liu H., Duan J., Zhang Y., Liu M., et al. (2012). Effect of high-dose methotrexate chemotherapy on intestinal Bifidobacteria, Lactobacillus and Escherichia coli in children with acute lymphoblastic leukemia. Exp. Biol. Med. (Maywood) 237, 305–311. 10.1258/ebm.2011.011297, PMID: [DOI] [PubMed] [Google Scholar]
- Hullahalli K., Rodrigues M., Nguyen U. T., Palmer K. (2018). An attenuated CRISP-Cas system in Enterococcus faecalis permits DNA acquisition. mBio 9, e00414–e00418. 10.1128/mBio.00414-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. 10.1038/nature11234, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke M. M., Abrams V., Moore D. R. (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536. 10.1093/carcin/23.3.529, PMID: [DOI] [PubMed] [Google Scholar]
- Imado T., Iwasaki T., Kitano S., Satake A., Kuroiwa T., Tsunemi S., et al. (2010). The protective role of host toll-like receptor-4 in acute graft-versus-host disease. Transplantation 90, 1063–1070. 10.1097/TP.0b013e3181f86947, PMID: [DOI] [PubMed] [Google Scholar]
- Jenq R. R., Taur Y., Devlin S. M., Ponce D. M., Goldberg J. D., Ahr K. F., et al. (2015). Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol. Blood Marrow Transplant. 21, 1373–1383. 10.1016/j.bbmt.2015.04.016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B. V., Begley M., Hill C., Gahan C. G., Marchesi J. R. (2008). Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 105, 13580–13585. 10.1073/pnas.0804437105, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorsal T., Rhee N. A., Pedersen J., Wahlgren C. D., Mortensen B., Jepsen S. L., et al. (2018). Endocrine K and L cells in healthy and type 2 diabetic individuals. Diabetologia 61, 284–294. 10.1007/s00125-017-4450-9, PMID: [DOI] [PubMed] [Google Scholar]
- Juajun O., Nguyen T. H., Maischberger T., Iqbal S., Haltrich D., Yammabhai M. (2011). Cloning, purification, and characterization of β-galactosidase from Bacillus licheniformis DSM 13. Appl. Microbiol. Biotechnol. 89, 645–654. 10.1007/s00253-010-2862-2, PMID: [DOI] [PubMed] [Google Scholar]
- Kaito S., Toya T., Yoshifugi K., Kurosawa S., Inamoto K., Takeshita K., et al. (2018). Fecal microbiota transplantation with frozen capsules for a patient with refractory acute gut graft-versus-host disease. Blood Adv. 2, 3097–3101. 10.1182/bloodadvances.2018024968, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastl A. J., Jr., Terry N., Wu G. D., Albenberg L. G. (2020). The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell. Mol. Gastroenterol. Hepatol. 9, 33–45. 10.1016/j.jcmgh.2019.07.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiczorowska B., Samolinska W., Mustafa Al-Yasiry A. R., Kiczorowski P., Winiarska-Mieczan A. (2017). The natural feed additives as immunostimulants in monogastric animal nutrition—a review. Ann. Anim. Sci. 17, 605–625. 10.1515/aoas-2016-0076 [DOI] [Google Scholar]
- Kim M. S., Park E. J., Roh S. W., Bae J. W. (2011). Diversity and abundance of single-stranded DNA viruses in human feces. Appl. Environ. Microbiol. 77, 8062–8070. 10.1128/AEM.06331-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler N., Zeiser R. (2019). Intestinal microbiota influence immune tolerance post allogeneic hematopoietic cell transplantation and intestinal GVHD. Front. Immunol. 9:3179. 10.3389/fimmu.2018.03179, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbling M., Freireich E. J. (2011). Twenty-five years of peripheral blood stem cell transplantation. Blood 117, 6411–6416. 10.1182/blood-2010-12-322214, PMID: [DOI] [PubMed] [Google Scholar]
- Koren O., Goodrich J. K., Cullender T. C., Spor A., Laitinen K., Bäckhed H. K., et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480. 10.1016/j.cell.2012.07.008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowaloff E. M., Phang J. M., Granger A. S., Downing S. J. (1977). Regulation of proline oxidase activity by lactate. Proc. Natl. Acad. Sci. U. S. A. 74, 5368–5371. 10.1073/pnas.74.12.5368, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai T., Rahman F., Smith A. M. (2018). The microbiome and radiation induced-bowelinjury: evidence for potential mechanistic role in disease pathogenesis. Nutrients 10:1405. 10.3390/nu10101405, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H., Choi M., Ogawa Y., Dickson A. S., Goetz R., Eliseenkova A. V., et al. (2007). Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282, 26687–26695. 10.1074/jbc.M704165200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara A. (2014). Contributions of colonic short-chain fatty acid receptors in energy homeostasis. Front. Endocrinol. 5:144. 10.3389/fendo.2014.00144, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier J. C., Khelaifia S., Alou M. T., Ndongo S., Dione N., Hugon P., et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1:16203. 10.1038/nmicrobiol.2016.203, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lecerf J. M., Dépeint F., Clerc E., Dugenet Y., Niamba C. N., Rhazi L., et al. (2012). Xylo-oligosaccharide (XOS) in combination with inulin modulates both the intestinal environment and immune status in healthy subjects, while XOS alone only shows prebiotic properties. Br. J. Nutr. 108, 1847–1858. 10.1017/S0007114511007252, PMID: [DOI] [PubMed] [Google Scholar]
- Legoff J., Resche-Rigon M., Bouquet J., Robin M., Naccache S. N., Mercier-Delarue S., et al. (2017). The eukaryotic gut virome in hematopoietic stem cell transplantation: new clues in enteric graft-versus-host disease. Nat. Med. 23, 1080–1085. 10.1038/nm.4380, PMID: [DOI] [PubMed] [Google Scholar]
- Liao S. F., Nyachoti C. M. (2017). Using probiotics to improve swine gut health and nutrient utilization. Anim. Nutr. 3, 331–343. 10.1016/j.aninu.2017.06.007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman D. R., Pamer E. G. (2011). Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe 10, 311–323. 10.1016/j.chom.2011.10.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang L., Tang W., Gu Y., Hua Q., Yang S., et al. (2012). Phosphoketolase pathway for xylose catabolism in Clostridium acetobutylicum revealed by 13C metabolic flux analysis. J. Bacteriol. 194, 5413–5422. 10.1128/JB.00713-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente C., Jepsen P., Inamine T., Wang L., Bluemel S., Wang H. J., et al. (2017). Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat. Commun. 8:837. 10.1038/s41467-017-00796-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Liu Y., He L., Tan Q., Yu Z., Xiao N., et al. (2018). Bacterial lactase genes diversity in intestinal mucosa of mice with dysbacterial diarrhea induced by antibiotics. Biotech 8:176. 10.1007/s13205-018-1191-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W. X., Xie M., Gao L. W. (2016). Observation on the therapeutic effect of lactase on lactose intolerance in infants with diarrhea. Strait Pharm. J. 28, 153–154. [Google Scholar]
- Luu M., Weigand K., Wedi F., Breidenbend C., Leister H., Pautz S., et al. (2018). Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 8:14430. 10.1038/s41598-018-32860-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenau J., Runaas L., Reddy P. (2016). Advances in understanding the pathogenesis of graft-versus-host disease. Br. J. Haematol. 173, 190–205. 10.1111/bjh.13959, PMID: [DOI] [PubMed] [Google Scholar]
- Maharshak N., Hub E. Y., Paiboonrungruang C., Shanahan M., Thurlow L., Herzog J., et al. (2015). Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect. Immun. 83, 2762–2770. 10.1128/IAI.00425-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K., Slesarev A., Wolf Y., Sorokin A., Mirkin B., Koonin E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. U. S. A. 103, 15611–15616. 10.1073/pnas.0607117103, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredo Vieira S., Hiltensperger M., Kumar V., Zegarra-Ruiz D., Dehner C., Khan N., et al. (2018). Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 360, 1156–1161. 10.1126/science.aar7201, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwick L. J. L., Riva A., Ryan J. M., Cooksley H., Palma E., Tranah T. H., et al. (2015). Blockade of PD1 and TIM3 restores innate and adaptive immunity in patients with acute alcoholic hepatitis. Gastroenterology 148, 590–602. 10.1053/j.gastro.2014.11.041, PMID: [DOI] [PubMed] [Google Scholar]
- Mathewson N. D., Jenq R., Mathew A. V., Koenigsknecht M., Hanash A., Toubai T., et al. (2016). Gut microbiome derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513. 10.1038/ni.3400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel M., Hinterleitner R., Pacis A., Chen L., Earley Z. M., Mayassi T., et al. (2018). Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficeint host. Nature 557, 580–584. 10.1038/s41586-018-0125-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merli M., Lucidi C., Di Gregorio V., Giannelli V., Giusto M., Ceccarelli G., et al. (2015). The chronic use of beta-blockers and proton pump inhibitors may affect the rate of bacterial infections in cirrhosis. Liver Int. 35, 362–369. 10.1111/liv.12593, PMID: [DOI] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J. (1979). Fermentations by saccharolytic intestinal bacteria. Am. J. Clin. Nutr. 32, 164–172. 10.1093/ajcn/32.1.164, PMID: [DOI] [PubMed] [Google Scholar]
- Million M., Armstrong N., Khelaifia S., Guilhot E., Richez M., Lagier J. C., et al. (2020). The antioxidants glutathione, ascorbic acid and uric acid maintain butyrate production by human gut Clostridia in the presence of oxygen. Sci. Rep. 10:7705. 10.1038/s41598-020-64834-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M., Tidjani Alou M., Khelaifia S., Bachar D., Lagier J. C., Dione N., et al. (2016). Increased gut redox and depletion of anaerobic and methanogenic prokaryotes in severe acute malnutrition. Sci. Rep. 6:26051. 10.1038/srep26051, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Million M., Tomas J., Wagner C., Lelouard H., Raoult D., Gorvel J. P. (2018). New insights in gut microbiota and mucosal immunity of the small intestine. Hum. Microb. J. 7-8, 23–32. 10.1016/j.humic.2018.01.004 [DOI] [Google Scholar]
- Minot S., Sinha R., Chen J., Li H., Keilbaugh S. A., Wu G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. 10.1101/gr.122705.111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misselwitz B., Butter M., Verbeke K., Fox M. R. (2019). Update on lactose malabsorption and intolerance: pathogenesis, diagnosis and clinical management. Gut 68, 2080–2091. 10.1136/gutjnl-2019-318404, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai M., Yoneyama H., Ezaki T., Suematsu M., Terashima Y., Harada A., et al. (2003). Peyer’s patch is the essential site in initiating murine acute and lethal graft-versus-host reaction. Nat. Immunol. 4, 154–160. 10.1038/ni879, PMID: [DOI] [PubMed] [Google Scholar]
- Musa A. E., Shabeeb D., Alhilfi H. S. Q. (2019). Protective effect of melatonin against radiotherapy-induced small intestinal oxidative stress: biochemical evaluation. Medicina 55:308. 10.3390/medicina55060308, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalle S. C., Zuo L., Ong M. L. D. M., Singh G., Worthylake A. M., Choi W., et al. (2019). Graft-versus-host disease propagation increased intestinal epithelial thight junction permeability. J. Clin. Invest. 129, 902–914. 10.1172/JCI98554, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. 10.1126/science.1223813, PMID: [DOI] [PubMed] [Google Scholar]
- Nohr M. K., Pedersen M. H., Gille A., Egerod K. L., Engelstoft M. S., Husted A. S., et al. (2013). GPR41/FFAR3 and GPR43/FFAR2 as cosensors for short-chain fatty acids in enteroendocrine cells vs FFAR3 in enteric neurons and FFAR2 in enteric leukocytes. Endocrinology 154, 3552–3564. 10.1210/en.2013-1142, PMID: [DOI] [PubMed] [Google Scholar]
- Noris R. D., Mahamed A. Z., Martin J. M., Docimo S. G. (2010). Enterococcus faecalis cellulitis/fasciitis after hypospadias surgery. Urology 76, 107–108. 10.1016/j.urology.2009.11.053, PMID: [DOI] [PubMed] [Google Scholar]
- Paganelli F. L., Huebner J., Singh K. V., Zhang X., van Schaik W., Wobser D., et al. (2016). Genome-wide screening identifies phosphotransferases system permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J. Infect. Dis. 214, 189–195. 10.1093/infdis/jiw108, PMID: [DOI] [PubMed] [Google Scholar]
- Pande S., Shitut S., Freund L., Westermann M., Bertels F., Colesie C., et al. (2015). Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat. Commun. 6:6238. 10.1038/ncomms7238, PMID: [DOI] [PubMed] [Google Scholar]
- Park J., Goergen C. J., HogenEsch H., Kim C. H. (2020). Chronically elevated levels of short-chain fatty acids induce T cell-mediated ureteritis and hydronephrosis. J. Immunol. 196, 2388–2400. 10.4049/jimmunol.1502046, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penack O., Holler E., van den Brink M. R. (2010). Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood 115, 1865–1872. 10.1182/blood-2009-09-242784, PMID: [DOI] [PubMed] [Google Scholar]
- Pichereau C., Desseaux K., Janin A., Scieux C., Peffault de Latour R., Whaard A., et al. (2012). The complex relationship between human herpesvirus 6 and acute graft-versus-host disease. Biol. Blood Marrow Transplant. 18, 141–144. 10.1016/j.bbmt.2011.07.018, PMID: [DOI] [PubMed] [Google Scholar]
- Priya R., Gomez G. A., Budnar S., Verma S., Cox H. L., Hamilton N. A., et al. (2015). Feedback regulation through myosin II confers robustness on RhoA signalling at E-cadherin junctions. Nat. Cell Biol. 17, 1282–1293. 10.1038/ncb3239, PMID: [DOI] [PubMed] [Google Scholar]
- Priyadarshini M., Kotlo K. U., Dudeja P. K., Layden B. T. (2019). Role of short chain fatty acid receptors in intestinal physiology and pathology. Compr. Physiol. 8, 1091–1115. 10.1002/cphy.c170050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang Y., Xu J., Yan C., Jin H., Xiao T., Yan N., et al. (2017). Butyrate and retinoic acid imprint mucosal-like dendritic cell development synergistically from bone marrow cells. Clin. Exp. Immunol. 189, 290–297. 10.1111/cei.12990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. 10.1038/nature08821, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P., Zou Y., Dai Y., Luo G., Zhang X., Xiao L. (2019). Characterization a novel butyric acid-producing bacterium Collinsella aerofaciens subsp. Shenzhenensis subsp. Nov. Microorganisms 7:78. 10.3390/microorganisms7030078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautava S., Luoto R., Salminen S., Isolauri E. (2012). Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 9, 565–576. 10.1038/nrgastro.2012.144, PMID: [DOI] [PubMed] [Google Scholar]
- Reissier S., Saleh-Mghir A., Guerin F., Massias L., Ghout I., Sinel C., et al. (2018). In vivo daptomycin efficacy against experimental vancomycin-resistant Enterococcus faecium endocarditis. J. Antimicrob. Chemother. 73, 981–986. 10.1093/jac/dkx499, PMID: [DOI] [PubMed] [Google Scholar]
- Rhimi M., Aghajari N., Jaouadi B., Juy M., Boudebbouze S., Maquin E., et al. (2009). Exploring the acidotolerance of betagalactosidase from Lactobacillus delbrueckii subsp. bulgaricus: an attractive enzyme for lactose bioconversion. Res. Microbiol. 160, 775–784. 10.1016/j.resmic.2009.09.004, PMID: [DOI] [PubMed] [Google Scholar]
- Rios-Covian D., Gueimonde M., Duncan S. H., Flint H. J., de los Reyes-Gavilan C. G. (2015). Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol. Lett. 362:fnv176. 10.1093/femsle/fnv176, PMID: [DOI] [PubMed] [Google Scholar]
- Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. 10.3389/fmicb.2016.00979, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riwes M., Reddy P. (2018). Microbial metabolites and graft versus host disease. Am. J. Transplant. 18, 23–29. 10.1111/ajt.14443, PMID: [DOI] [PubMed] [Google Scholar]
- Roberfroid M. B. (2007). Inulin-type fructans: functional food ingredient. J. Nutr. 137, 2493S–2502S. 10.1093/jn/137.11.2493S, PMID: [DOI] [PubMed] [Google Scholar]
- Russo P., Pia M., Fiocco D., Capozzi V., Drider D., Spano G. (2017). Lactobacillus plantarum with broad antifungal activity: a promising approach to increase safety and shelf-life of cereal-based products. Int. J. Food Microbiol. 247, 48–54. 10.1016/j.ijfoodmicro.2016.04.027, PMID: [DOI] [PubMed] [Google Scholar]
- Saha S. K., Lee S. B., Won J., Choi H. Y., Kim K., Yang G. M., et al. (2017). Correlation between oxidative stress, nutrition, and cancer initiation. Int. J. Mol. Sci. 18:1544. 10.3390/ijms18071544, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Toprak S. K., Atilla P. A., Atilla E., Demirer T. (2016). An overview of infectious complications after allogenic hematopoeitic stem cell transplantation. J. Infect. Chemother. 22, 505–514. 10.1016/j.jiac.2016.05.006, PMID: [DOI] [PubMed] [Google Scholar]
- Saito H. E., Harp J. R., Fozo E. M. (2018). Enterococcus faecalis responds to individual exogenous fatty acids independently of their degree of saturation or chain length. Appl. Environ. Microbiol. 84, e01633–e01717. 10.1128/AEM.01633-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab L., Goroncy L., Palaniyandi S., Gautam S., Triantafyllopoulou A., Mocsai A., et al. (2014). Neutrophil granulocytes recruited upon translocation of intestinal bacteria enhance graft-versus-host disease via tissue damage. Nat. Med. 20, 648–654. 10.1038/nm.3517, PMID: [DOI] [PubMed] [Google Scholar]
- Schwartz S., Vergoulidou M., Schreier E., Loddenkemper C., Reinwald M., Schmidt-Hieber M., et al. (2011). Norovirus gastroenteritis causes severe and lethal complications after chemotherapy and hematopoietic stem cell transplantation. Blood 117, 5850–5856. 10.1182/blood-2010-12-325886, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severyn C. J., Brewster R., Andermann T. M. (2019). Microbiota modification in hematology: still at the bench or ready for the bedside? Blood Adv. 3, 303–314. 10.1182/bloodadvances.2019000365, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F., van Sinderen D., O’Toole P. W., Stanton C. (2017). Feeding the microbiota: transducer of nutrient signals for the host. Gut 66, 1709–1717. 10.1136/gutjnl-2017-313872, PMID: [DOI] [PubMed] [Google Scholar]
- Shouval R., Geva M., Nagler A., Youngster I. (2019). Fecal microbiota transplantation for treatment of acute graft-versus-host disease. Clin. Hematol. Int. 1, 28–35. 10.2991/chi.d.190316.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si X., Shang W., Zhou Z., Strappe P., Wang B., Bird A., et al. (2018). Gut microbiome-induced shift of actetate to butyrate positively manages dysbiosis in high fat diet. Mol. Nutr. Food Res. 62:1700670. 10.1002/mnfr.201700670, PMID: [DOI] [PubMed] [Google Scholar]
- Steck N., Hoffmann M., Sava I. G., Kim S. C., Hahne H., Tonkonogy S. L., et al. (2011). Enteroccus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology 141, 959–971. 10.1053/j.gastro.2011.05.035, PMID: [DOI] [PubMed] [Google Scholar]
- Stein-Thoeringer C. K., Nichols K. B., Lazrak A., Docampo M. D., Slingerland A. E., Slingerland J. B., et al. (2019). Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science 366, 1143–1149. 10.1126/science.aax3760, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini F. B., Andermann T. M., Tkachenko E., Senchyna F., Banaei N., Bhatt A. S. (2018). Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat. Med. 24, 1809–1814. 10.1038/s41591-018-0202-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y., Jenq R. R., Perales M. A., Littmann E. R., Morjaria S., Ling L., et al. (2014). The effects of intestinal tract bacterial diversity on mortality following allogenic hematopoietic stem cell transplantation. Blood 124, 1174–1182. 10.1182/blood-2014-02-554725, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y., Xavier J. B., Lipuma L., Ubeda C., Goldberg J., Gobourne A., et al. (2012). Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914. 10.1093/cid/cis580, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezze C., Romanello V., Sandri M. (2019). FGF21 as modulator of metabolism in health and disease. Front. Physiol. 10:419. 10.3389/fphys.2019.00419, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalambedu N., Khan Y., Zhang Q., Khanal S., Ashfaq A. (2019). Immune-mediated colitis from dual checkpoint inhibitors. Cureus 11:e6233. 10.7759/cureus.6233, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidjani Alou M., Million M., Traore S. I., Mouelhi D., Khelaifia S., Bachar D., et al. (2017). Gut bacteria missing in severe acute malnutrition, can we identify potential probiotics by culturomics? Front. Microbiol. 8:899. 10.3389/fmicb.2017.00899, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien B. Y. Q., Goh H. M. S., Chong K. K. L., Bhaduri-Tagore S., Holec S., Dress R., et al. (2017). Enterococcus faecalis promotes innate immune suppression and polymicrobial catheter-associated urinary tract infection. Infect. Immun. 85. pii: e00378–e00417. 10.1128/IAI.00378-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping D. L., Clifton P. M. (2001). Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81, 1031–1064. 10.1152/physrev.2001.81.3.1031, PMID: [DOI] [PubMed] [Google Scholar]
- Tran T. H. T., Everaert N., Bindelle J. (2016). Review on the effects of potential prebiotics on controlling intestinal enteropathogens Salmonella and Escherichia coli in pig production. J. Anim. Physiol. Anim. Nutr. 102, 17–32. 10.1111/jpn.12666 [DOI] [PubMed] [Google Scholar]
- Tremorali V., Backhed F. (2012). Functional interaction between the gut microbiota and host metabolism. Nature 489, 242–249. 10.1038/nature11552, PMID: [DOI] [PubMed] [Google Scholar]
- Trifilio S. M., Pi J., Mehta J. (2012). Changing epidemiology of Clostridium difficile-associated disease during stem cell transplantation. Biol. Blood Marrow Transplant. 19, 405–409. 10.1016/j.bbmt.2012.10.030, PMID: [DOI] [PubMed] [Google Scholar]
- Troelsen J. T., Mitchelmore C., Spodsberg N., Jensen A. M., Noren O., Sjöström H. (1997). Regulation of lactase-phlorizin-hydrolase gene expression by the caudal-related homoeodomain protein Cdx-2. Biochem. J. 322, 833–838. 10.1042/bj3220833, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuohy K. M., Kolida S., Lustenberger A. M., Gibson G. R. (2001). The prebiotic effects of biscuits containing partially hydrolysed guar gum and fructo-oligosaccharides—a human volunteer study. Br. J. Nutr. 86, 341–348. 10.1079/BJN2001394, PMID: [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Quince C., Faith J. J., McHardy A. C., Yatsunenko T., Niazi F., et al. (2010). Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc. Natl. Acad. Sci. U. S. A. 107, 7503–7508. 10.1073/pnas.1002355107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bekkum D. W., Roodenburg J., Heidt P. J., van der Waaij D. (1974). Mitigation of secondary disease of allogeneic mouse radiation chimeras by modification of the intestinal microflora. J. Natl. Cancer Inst. 52, 401–404. 10.1093/jnci/52.2.401, PMID: [DOI] [PubMed] [Google Scholar]
- Varga T., Czimmerer Z., Nagy L. (2011). PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 1812, 1007–1022. 10.1016/j.bbadis.2011.02.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieco-Saiz N., Belguesmia Y., Raspoet R., Auclair E., Gancel F., Kempf I., et al. (2019). Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 10:57. 10.3389/fmicb.2019.00057, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N. Y., McFadden G. (2018). Virotherapy as potential adjunct therapy for graft-vs-host disease. Curr. Pathobiol. Rep. 6, 247–263. 10.1007/s40139-018-0186-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M., Chuang Howe A., Tiedje J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. mBio 5:e00889. 10.1128/mBio.00889-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreugdenhil A. C., Rousseau C. H., Hartung T., Greve J. W., van’tVeer C., Buurman W. A. (2003). Lipopolysaccharide (LPS)-binding protein mediates LPS detoxification by chylomicrons. J. Immunol. 170, 1399–1405. 10.4049/jimmunol.170.3.1399, PMID: [DOI] [PubMed] [Google Scholar]
- Wagnerberger S., Spruss A., Kanuri G., Volynets V., Stahl C., Bischoff S. C., et al. (2012). Toll-like receptor 1-9 are elevated in livers with fructose-induced hepatic steatosis. Br. J. Nutr. 107, 1727–1738. 10.1017/S0007114511004983, PMID: [DOI] [PubMed] [Google Scholar]
- Waldecker M., Kautenburger T., Daumann H., Busch C., Schrenk D. (2008). Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 19, 587–593. 10.1016/j.jnutbio.2007.08.002, PMID: [DOI] [PubMed] [Google Scholar]
- Walter J., Ley R. (2011). The human gut microbiome: ecology and recent evolutionary changes. Annu. Rev. Microbiol. 65, 411–429. 10.1146/annurev-micro-090110-102830, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang Q., Wang X., Zhu L., Chen J., Zhang B., et al. (2019). Gut microbial dysbiosis is associated with development and progression of radiation enteritis during pelvic radiotherapy. J. Cell. Mol. Med. 23, 3747–3756. 10.1111/jcmm.14289, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D., Jenq R. R., Peled J. U., Taur Y., Hiergeist A., Koestler J., et al. (2017). Microbiota disruption induced by early use of broad-spectrum antibiotics is an independent risk factor of outcome after allogeneic stem cell transplantation. Biol. Blood Marrow Transplant. 23, 845–852. 10.1016/j.bbmt.2017.02.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber D., Oefner P. J., Hiergeist A., Koestler J., Gessner A., Weber M., et al. (2015). Low urinary indoxyl sulfate levels early after transplantation reflect a disrupted microbiome and are associated with poor outcome. Blood 126, 1723–1728. 10.1182/blood-2015-04-638858, PMID: [DOI] [PubMed] [Google Scholar]
- Wei D., Heus P., van de Wetering F. T., van Tienhoven G., Verleye L., Scholten R. J. P. M. (2018). Probiotics for the prevention or treatment of chemotherapy or radiotherapy-related diarrhoea in people with cancer. Cochrane Database Syst. Rev. 8. Art.No: CD008831. 10.1002/14651858.CD008831.pub3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Bank (2017). Drug-resistant infections: a threat to our economic future (Vol. 2): final report (English) number 114679, disclosure date April 08, 2017. HNP/Agriculture Global Antimicrobial Resistance Initiative. Washington, DC: World Bank Group. Available at: http://documents.worldbank.org/curated/en/323311493396993758/final-report
- Yang J. Y., Kim M. S., Kim E., Cheon J. H., Lee Y. S., Kim Y., et al. (2016). Enteric viruses ameliorate gut inflammation via toll-like receptor 3 and toll-like receptor 7-mediated interferon-b production. Immunity 44, 889–900. 10.1016/j.immuni.2016.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- Yue X., Trifari S., Aijö T., Tsagaratou A., Pastor W. A., Zepeda-Martinez J. A., et al. (2016). Control of Foxp3 stability through modulation of TET activity. J. Exp. Med. 213, 377–397. 10.1084/jem.20151438, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zama D., Bossu G., Leardini D., Muratore E., Biagi E., Prete A., et al. (2020). Insights into the role of intestinal microbiota in hematopoietic stem-cell transplantation. Ther. Adv. Hematol. 11, 1–19. 10.1177/2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr D. M., Boeckh M., Delaney C., Martin P. J., Xie H., Adler A. L., et al. (2012). HHV-6 reactivation and associated sequelae after hematopoietic cell transplantation. Biol. Blood Marrow Transplant. 18, 1700–1708. 10.1016/j.bbmt.2012.05.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W. H., Run J. Q., Wei C. L., Soh S. W., et al. (2006). RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. 10.1371/journal.pbio.0040003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Song L., Wang Y., Liu C., Zhang L., Zhu S., et al. (2019). Beneficial effect of butyrate-producing Lachnospiraceae on stress-induced visceral hypersensitivity in rats. J. Gastroenterol. Hepatol. 34, 1368–1376. 10.1111/jgh.14536, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wang J., Pouliot M., Authier S., Loose D. S., Hauer-Jensen M. (2015). Gene expression profiling in non-human primate jejunum, ileum and colon after total-body irradiation: a comparative study of segment-specific molecular and cellular responses. BMC Genomics 16:984. 10.1186/s12864-015-2168-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. H., Li X. Q., Zhang W., Zhou D., Liu H. Y., Wang J. F. (2014). Dose-dependent effects of Lactobacillus rhamnosus on serum interleukin-17 production and intestinal T-cell responses in pig challenged with Escherichia coli. Appl. Environ. Microbiol. 80, 1787–1798. 10.1128/AEM.03668-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Kroemer G. (2019). Immunostimulatory gut bacteria. Science 366, 1077–1078. 10.1126/science.aaz7595, PMID: [DOI] [PubMed] [Google Scholar]
- Zitvogel L., Ma Y., Raoult D., Kroemer G., Gajewski T. F. (2018). The microbiome in cancer immunotherapy: diagnostic tools and therapeutic stragies. Science 359, 1366–1370. 10.1126/science.aar6918, PMID: [DOI] [PubMed] [Google Scholar]
- Zoetendal E. G., Raes J., van den Bogert B., Arumugam M., Booijink C. C., Troost F. J., et al. (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426. 10.1038/ismej.2011.212, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]