Abstract
Salivary duct carcinoma (SDC) is a rare, aggressive malignancy with limited treatment options and poor outcome. Twenty-nine primary resected SDC, including 15 SDC de novo (SDCDN), and 14 SDC ex pleomorphic adenoma (SDCXPA) were subjected to the massive parallel sequencing assay (MSK-IMPACT™) targeting 287-468 cancer-related genes. TP53 was the most frequently altered gene (69%). TP53 mutations and ERBB2 amplification were more frequent in SDCXPA than in SDCDN (p=0.0007 and p=0.01, respectively). Potentially targetable mutations were detected in 79% (23/29) SDC involving ERBB2 (31%), PIK3CA (28%), HRAS (21%), ALK (7%) and BRAF (3%), and 22% (5/23) of those cases harbored possible primary resistance mutations involving CCNE1, NF1 and PTEN. A novel HNRNPH3-ALK rearrangement was found in one SDCDN. In another case, EML4-ALK fusion detected in the primary tumor was associated with ALK G1202R secondary resistance mutation in the post-treatment metastasis. A germline analysis of the DNA repair genes revealed a case with a pathogenic BRCA1 E23fs germline variant. SDCDN and SDCXPA are genetically distinct. Although the majority of SDC may be amenable to molecular targeted therapy, concurrent possible resistance mutations may be found in a significant minority of cases. A broad genomic profiling is necessary to ensure detection of rare but clinically actionable somatic alterations in SDC.
Keywords: salivary duct carcinoma, ALK rearrangements
1. INTRODUCTION
Salivary duct carcinoma (SDC) is an aggressive malignant epithelial tumor of the salivary glands that accounts for 9% of salivary malignancies [1]. It most commonly arises in the parotid, more frequently affect men, and has a peak incidence in those older than 50 years of age. Most patients present with a rapidly progressive disease, at clinical stage III or IV. The mainstay of therapy includes surgical removal for localized disease with or without adjuvant radiotherapy and palliative chemotherapy for metastatic disease with unresectable locally recurrent tumors. The prognosis remains poor and more than 50% of patients die within 3 to 5 years of the diagnosis [2]. SDC arises either de novo or from a pre-existing pleomorphic adenoma (PA). Except for the presence of the residual PA component in SDCXPA, the two histologic subtypes are morphologically and immunophenotypically very similar. They both usually comprise of large apocrine tumor cells forming ducts, cribriform structures and nests with comedo-type necrosis, strikingly similar to those seen in invasive mammary carcinoma with apocrine features [3]. Several genomic studies to date have shown that the majority of SDC harbor somatic genetic alterations, namely ERBB2 (HER2) amplification, and mutations in TP53, PIK3CA and HRAS [4,5]. In contrast to the prior studies focused on detection of possible druggable targets, studies exploring the potential mechanisms of resistance to targeted therapy in SDC are lacking. Here, we performed an in-depth genomic analysis of SDC aiming (1) to examine somatic genetic alterations in SDC relative to their putative cell of origin/precursor lesion and to predict their functional impact, and (2) to examine matched normal DNA for the presence of germline mutations in DNA repair genes. To achieve these aims, 15 SDCs de novo and 14 SDCs ex PA were subjected to targeted capture massively parallel sequencing (MPS) utilizing to the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT™) assay [6], which interrogates somatic genetic alterations, including select rearrangements, in 287 to 468 key cancer-related genes.
2. MATERIALS AND METHODS
2.1. Patients and tissue samples
The study was approved by the Institutional Review Board (IRB) of Memorial Sloan Kettering Cancer Center. Signed informed consents were obtained according to the approved protocol. Twenty-nine primary resected, widely invasive SDC diagnosed between December 1999 and July 2017 with sufficient formalin-fixed paraffin-embedded (FFPE) tissue were studied including 27 conventional SDC and 2 sarcomatoid variants (SDC27 and SDC28). In 15 (52%) cases, the entire tumor was processed for microscopic examination. A diagnosis of SDCXPA was made based on the presence of histologic evidence of pre-existing PA such as hypocellular hyalinized/sclerotic calcified nodule and/or presence of chondroid or myxoid areas with benign ductal elements [7]. In total, 15 (52%) SDCDN, including one case with tested primary and two subsequent metastases (SDC37), and 14 (48%) SDCXPA were analyzed. Four cases (SDC3, SDC30-32) were included in a prior study [5]. All cases were reviewed by two head and neck pathologists with interest and expertise in salivary gland pathology (SD, NK).
2.2. DNA extraction targeted capture massively parallel sequencing (MPS)
Genomic DNA was extracted from formalin-fixed paraffin embedded tissue curls cut at 10-μm using the QIAamp FFPE Tissue Kit (Qiagen) 8. Fifteen (52%) SDCDN and 14 (48%) SDCXPA were subjected to targeted capture MPS using the MSK-IMPACT™ assay. Somatic genetic alterations in 287 (7 samples), 341 (13 samples), 410 (7 samples) or 468 (4 samples) cancer-related genes were defined using our clinically validated assay as previously described [6]. A germline analysis of DNA repair genes was performed in 11 cases (SDC27-SDC37).
2.3. Clonality, pathogenicity and zygosity status of somatic mutations
The cancer cell fraction of each mutation was inferred using the number of reads supporting the reference and the alternate alleles and the segmented Log2 ratio from MPS as input for ABSOLUTE (v1.0.6) [8]. Solutions from ABSOLUTE were manually reviewed as recommended [8,9]. A mutation was classified as clonal if its clonal probability, as defined by ABSOLUTE, was >50% or if the lower bound of the 95% confidence interval of its cancer cell fraction was >90% [10]. Mutations that did not meet the above criteria were considered subclonal. The pathogenicity of mutations was determined using two methods. “Method 1” was previously described [11]. (Details are provided in Supplementary Methods). The pathogenicity/oncogenic potential of other genetic alterations such as copy number alterations, gene rearrangements and intragenic deletions was determined using OncoKB annotation (“Method 2”) available on cBioportal (www.cbioportal.org; Supplementary Table 2). The pathogenicity interpretation was compared between the two methods and alterations designated as likely pathogenic, potentially pathogenic, pathogenic, likely oncogenic, predicted oncogenic and oncogenic by either of the two methods were considered pathogenic/oncogenic. Actionability/targetability of genetic alterations and gene signaling pathways designation were determined using OncoKB. Allelic-specific copy number alterations and allelic-specific loss of heterozygosity (LOH) were defined using FACETS [12], which performs a joint segmentation of the total and allelic copy ratio and infers allele-specific copy number states. Regions of LOH were manually reviewed using plots of log ratios and B allele frequencies.
2.4. Phylogenetic tree construction
A maximum parsimony tree was built for SDC37 using binary presence/absence matrices based on the repertoire of non-synonymous and synonymous somatic mutations, gene amplifications and homozygous deletions, as described in Murugaesu et al. [13] and Guerini-Rocco et al. [14]. (Details are provided in Supplementary Methods).
2.5. Fluorescence in situ hybridization (FISH)
Formalin-fixed paraffin tumor tissue 4-μm sections were tested for ALK (SDC37) by FISH. Tissue sections were pretreated by de-paraffinizing in xylene and dehydrating in ethanol. ALK Break Apart FISH Probe Kit (Abbott Molecular, Des Plaines, IL) was used. Dual-color FISH was performed according to the protocol from Vysis/Abbott Molecular. FISH analysis and signal capture were performed on a fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany) coupled with the ISIS FISH Imaging System (Metasystems, Waltham, MA). In ALK FISH study, 100 interphase nuclei from the tumor specimen were examined and a minimum of 15% cells with the rearrangement signal was required to consider a tumor positive for ALK rearrangement.
2.6. Immunohistochemistry (IHC)
IHC studies for HER2 (Ventana, clone 4B5, antibody concentration 6ug/ml), p40 (Biocare, mouse monoclonal antibody, dilution 1:200), ALK (Cell Signaling, Danvers, MA; clone D5F3, dilution 1:250) and androgen receptor (AR; Dako, monoclonal mouse antibody clone AR-441, dilution 1:100) were performed using the standard streptavidin-biotin-peroxidase procedure with the Ventana system according to the manufacturer’s protocol (Ventana Medical Systems Inc., Tucson, AZ, United States). Appropriate positive and negative controls were used for each antibody. The immunohistochemistry results for HER2 were interpreted using the 2013 American Society of Clinical Oncologists (ASCO)/College of American Pathologists (CAP) guidelines for breast cancer [15], and cases with a score 3+ were considered HER2 positive. Androgen receptor (AR) antibody clone AR-441 (monoclonal mouse, dilution 1:100; Dako) was used to determine AR immunoreactivity. Positive nuclear labeling in at least 1% tumor cells was considered positive.
2.7. Comparison of the mutational profiles of SDC and luminal androgen receptor-positive triple negative breast cancer (LAR-TNBC)
For the comparison of mutational frequencies in SDCDN and SDCXPA with 38 LAR-TNBC reported by TCGA [16], we retrieved the publicly available mutation data from cBioPortal website (http://www.cbioportal.org/). We restricted the comparison to only 468 genes because SDC cohorts were sequenced for only this panel of genes.
2.8. Statistical analysis
Chi-square test or Fisher’s exact test for nonparametric variables and Student’s t test for continuous variables were used for statistical analyses. All tests performed were two-tailed. P values < 0.05 were considered significant. Survival analysis was performed using Log rank test. Benjamini & Hochberg (BH) method was used for p-values adjustment in multiple comparisons analysis.
3. RESULTS
3.1. Clinical characteristics of SDCDN and SDCXPA
Clinico-pathologic features of SDCDN and SDCXPA are summarized in Table 1. All but one patient was male, presenting at the median age of 67 years (range 51-82) in the SDCDN group, and at 65 years (range 31-98) in the SDCXPA group. All but one case arose in the parotid gland and all cases expressed AR. The majority of patients in both groups presented at clinical stage IV (80% and 71%) and were treated with multimodal therapy. The median follow-up was 31 months (range 5-82) and 13 months (range: 2-160) for carcinomas de novo and carcinomas ex PA, respectively. In our cohort, there were 14 deaths (48%), including 8 of 15 (53%) and 6 of 14 (43%) SDCDN and SDCXPA, respectively. Eleven patients (38%) suffered disease-related death, including 6 (50%) with SDCDN and 5 (36%) with carcinomas ex PA. A total of 17 patients (59%) had recurrence during their disease course, including 9 (60%) carcinomas de novo and 8 (57%) carcinomas ex PA cases. Survival analysis using Log rank test revealed no significant difference in overall survival, disease-specific survival and recurrence-free survival between de novo carcinomas and ex PA cases (p=0.203, 0.117 and 0.325 respectively).
Table 1.
Clinico-pathologic features of salivary duct carcinoma.
| SDCDN (N=15) | SDCXPA (N=14) | |
|---|---|---|
| Men | 14 (93%) | 11 (79%) |
| Women | 1 (7%) | 3 (21 %) |
| Age median (range), years | 67 (51-82) | 65 (31-98) |
| Smoking history | 7 (47%) | 9 (64%) |
| Primary site | ||
| Parotid | 15 (100%) | 13 (93%) |
| Submandibular gland | 0 | 1 (7%) |
| Tumor size mean (range), cm | 3.2 (1.7-5.0) | 2.5 (1.1-5.0) |
| AR immunoexpression | 15 (100%) | 14 (100%) |
| Clinical stage | ||
| I | 2 (13%) | 1 (7%) |
| II | 0 | 1 (7%) |
| III | 1 (7%) | 2 (14%) |
| IV | 12 (80%) | 10 (71%) |
| Treatment | ||
| Surgery | 15 (100%) | 14 (100%) |
| RT | 15 (100%) | 13 (93%) |
| CT | 10 (67%) | 7 (50%) |
| Anti-AR | 0 | 2 (14%) |
| Anti-HER2 | 0 | 1 (7%) |
| ALK inhibitor | 1 (7%) | 0 |
| Distant recurrence | ||
| Bone | 5 (33%) | 4 (29%) |
| Lung | 7 (47%) | 2 (14%) |
| Brain | 2 (13%) | 2 (14%) |
| Liver | 0 | 3 (21%) |
| Clinical outcome | ||
| FU period median (range), months | 31 (5-82) | 13 (2-160) |
| Death | 8 (53%) | 6 (43%) |
| Disease-related death | 6 (40%) | 5 (36%) |
| Recurrence | 9 (60%) | 9 (57%) |
Abbreviations: SDC=salivary duct carcinoma, SDCDN=salivary duct carcinoma de novo, SDCXPA=salivary duct carcinoma ex pleomorphic adenoma, AR=androgen receptor, RT=radiation therapy, CT=chemotherapy. CTRT=chemotherapy and radiation therapy, FU=follow up.
3.2. SDC displays a complex repertoire of somatic genetic alterations
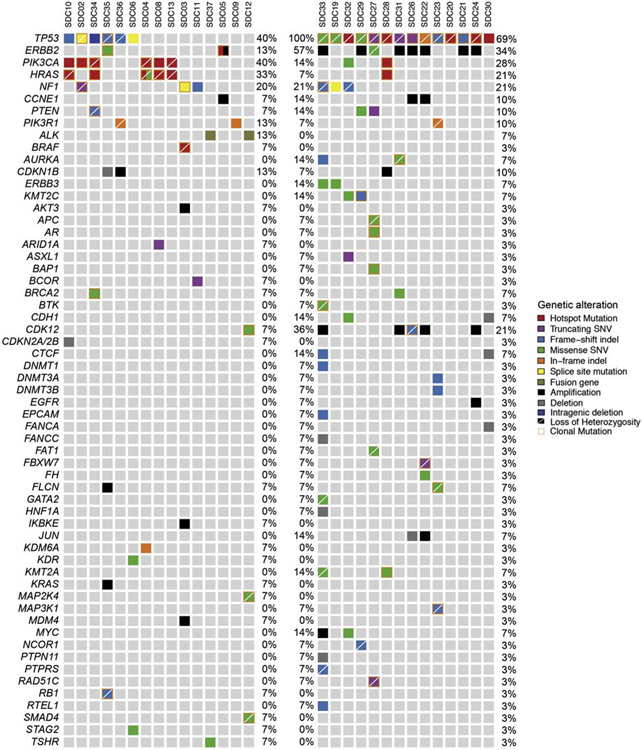
A total of 31 samples from 29 SDC (including primary and two metastases in one case) were subjected to targeted capture MPS to average read depths 539-fold (range 38-fold to 4064-fold) and 443-fold (range 44-fold to 1967-fold) for the tumor and matched normal tissue samples, respectively. Overall, 134 somatic mutations, 91 copy number alterations, and 5 structural variants including TP53 intragenic deletion (exons 6-9) and 3 fusion genes, PLAG1-CTNNB1, EML4-ALK and HNRNPH3-ALK were identified among the 29 primary tumors (Supplementary Table 1). Out of 230 unique somatic genetic alterations, 124 (54%) were defined as pathogenic/oncogenic using the two methods as described above (Supplementary Figure 1).
3.3. Genetic differences between SDCDN and SDCXPA
Genetic characteristics of SDCDN and SDCXPA are summarized in Table 2. Pathogenic/oncogenic somatic alterations were more frequent among SDCDN than SDCXPA (74% vs. 50%, p=0.0003, Fisher’s exact test, Table 2). TP53 somatic mutations were detected in 69% (20/29) cases and were significantly more frequent in SDCXPA than in carcinomas de novo (14/14, 100% vs. 6/15, 40%, p=0.0007, Fisher’s exact test). They were all designated as (likely) pathogenic, showed LOH in the majority (15/20, 75%) of cases, and were found to be clonal (i.e. present in virtually all tumor cells) in 11 (78%) carcinomas ex PA and in 2 (33%) carcinomas de novo. Interestingly, missense TP53 mutations were detected only in SDCXPA (9/14, 31% vs. 0/6, p=0.01, Fisher’s exact test) and these included 6 TP53 hotspot variants; R175H (N=3), R273H (N=1), S215G (N=1) and C238W (N=1. Table 3). ERBB2 amplification was more common in SDCXPA (7/14, 50% vs. 1/15, 7%, p=0.01, Fisher’s exact test), and tended to co-exist with TP53 mutations only in SDCXPA but not in SDCDN (7/14, 50% vs. 0/15, 0%, p=0.002, Fisher’s exact test, Table 2.
Table 2.
Genetic differences between salivary duct carcinoma de novo and salivary duct carcinoma ex pleomorphic adenoma.
| SDC (N=29) | SDCDN (N=15) | SDCXPA (N=14) | p value | |
|---|---|---|---|---|
| All somatic alterations | 230 | 85 | 145 | |
| Pathogenic/oncogenic somatic alterations | 124 (54%) | 63 (74%) | 72 (50%) | 0.0003 |
| Somatic mutations, median (range) | 135, 3 (1-19) | 49, 3 (1-6) | 85, 4 (1-19) | |
| Pathogenic/oncogenic somatic mutations | 91 (67%) | 38 (78%) | 53 (62%) | NS |
| CNA, median (range) | 91, 1 (0-22) | 32, 1 (0-15) | 59, 2 (0-22) | |
| Oncogenic CNA | 30 (33%) | 11 (34%) | 19 (32%) | NS |
| Structural variants (fusion, intragenic deletion) | 5 (17%) | 4 (27%) | 1 (7%) | |
| Oncogenic structural variants | 4 (80%) | 3 (75%) | 0 | |
| TP53 mutations | 20 (69%) | 6 (40%) | 14 (100%) | 0.0007 |
| Missense TP53 mutations | 9 (31%) | 0 | 9 (64%) | 0.01 |
| ERBB2 amplification | 8 (28%) | 1 (7%) | 7 (50%) | 0.01 |
| TP53 mutations + ERBB2 amplification | 7 (24%) | 0 | 7 (50%) | 0.002 |
Abbreviations: SDC=salivary duct carcinoma, SDCDN=salivary duct carcinoma de novo, SDCXPA=salivary duct carcinoma ex pleomorphic adenoma.
Table 3.
Spectrum of TP53 mutations in salivary duct carcinoma.
| Case | Genetic Alteration | Variant Class | OncoKB annotation (Method 2) |
OncoKB hotspot |
Zygosity status |
Clonal / Subclonal |
Pathogenicity (Method 1) |
|---|---|---|---|---|---|---|---|
| SDCDN | |||||||
| SDC02 | TP53 X307_splice | splice site | likely onc | LOH | clonal | pathogenic | |
| SDC06 | TP53 X186_splice | splice site | likely onc | subclonal | pathogenic | ||
| SDC10 | TP53 P142fs | frameshift del | likely onc | subclonal | pathogenic | ||
| SDC34 | TP53 exon 6-9 intragenic del | intragenic del | likely onc | LOH | NA | NA | |
| SDC35 | TP53 F113Qfs*5 | frameshift del | likely onc | LOH | clonal | likely pathogenic | |
| SDC36 | TP53 M340Ifs*6 | frameshift ins | likely onc | LOH | subclonal | likely pathogenic | |
| SDCXPA | |||||||
| SDC19 | TP53 A159V | missense | onc | LOH | clonal | pathogenic | |
| SDC20 | TP53 R175H | missense | onc | yes | LOH | clonal | pathogenic |
| SDC21 | TP53 A76fs*73 | frameshift | likely onc | clonal | pathogenic | ||
| SDC22 | TP53 PH177del | inframe del | likely onc | LOH | clonal | pathogenic | |
| SDC23 | TP53 Q167fs | frameshift del | likely onc | LOH | clonal | pathogenic | |
| SDC24 | TP53 R175H | missense | onc | yes | LOH | clonal | pathogenic |
| SDC26 | TP53 R306X | nonsense | likely onc | clonal | pathogenic | ||
| SDC27 | TP53 E346X | nonsense | likely onc | LOH | clonal | pathogenic | |
| SDC28 | TP53 R273H | missense | onc | yes | LOH | clonal | pathogenic |
| SDC29 | TP53 E258K | missense | likely onc | LOH | clonal | pathogenic | |
| SDC30 | TP53 R175H | missense | onc | yes | subclonal | pathogenic | |
| SDC31 | TP53 Q331* | missense | likely onc | LOH | subclonal | likely pathogenic | |
| SDC32 | TP53 S215G | missense | onc | yes | LOH | subclonal | likely pathogenic |
| SDC33 | TP53 C238W | missense | likely onc | yes | LOH | clonal | likely pathogenic |
Abbreviations: SDC=salivary duct carcinoma, SDCDN=salivary duct carcinoma de novo, SDCXPA=salivary duct carcinoma, ex pleomorphic adenoma, onc=oncogenic, LOH=loss of heterozygosity.
3.4. Targetable somatic mutations in SDC
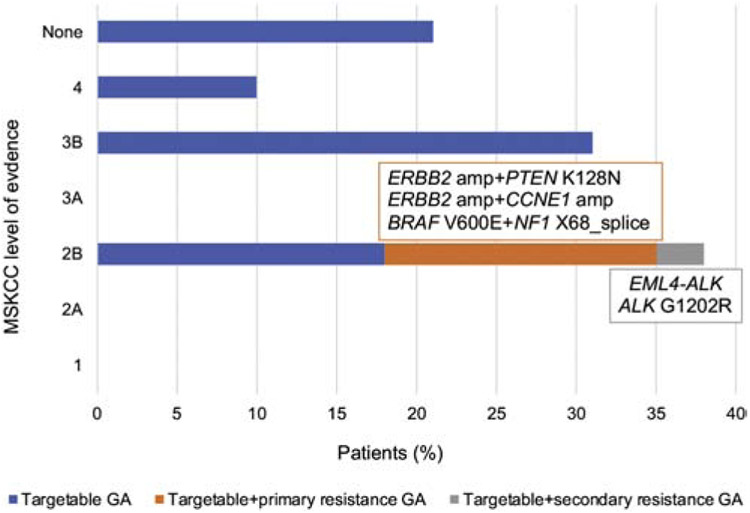
The repertoire of somatic mutations in SDC is summarized in Figure 1. Potentially targetable alterations were detected overall in 23 (79%) cases involving ERBB2 (31%), PIK3CA (28%), NF1 (21%), HRAS (17%), PTEN (10%), ALK (7%), and BRAF (3%); (Figure 2, Supplementary Table 2). Oncogenic ERBB2 alterations included ERBB2 amplification, ERBB2 V777L and ERBB2 S310F variant. The latter co-exited with ERBB2 amplification in SDC5. Interestingly, this case displayed morphologic features of invasive lobular carcinoma of the breast but was CDH1 wild-type (Supplementary Figure 2). PIK3CA activating missense mutations affected hotspots (E545K, E542K, H1047R, P104L, and D350G), co-existed with HRAS variants in 6 (21%) cases, (5 were SDCDN), and included Q61R, Q61K, G13R hotspots and one 451-2T>G splice site variant. Coexisting PIK3CA/HRAS variants were found to be both clonal in 4 (67%) cases (Supplementary Table 3). PIK3CA/HRAS mutated tumors were morphologically similar to their PIK3CA/HRAS-wild-type counterparts. Twenty-six (90%) SDC cases harbored pathogenic somatic alterations in PI3K-AKT-mTOR and/or Ras-Raf-MEK-Erk/NF1 signaling pathways. Among ERBB2 copy number neutral cases, mutations in the Ras-Raf-MEK-Erk/NF1 pathway were significantly more frequent than among ERBB2 amplified cases (15/21, 71% vs. 1/8, 13%, p=0.010, Fisher’s exact test, Supplementary Table 4).
Figure 1.
The repertoire of somatic mutations in primary SDC detected by the MSK-IMPACT™ assay. Only genes altered by at least one pathogenic/oncogenic mutation are listed. The top row lists all the cases. SDCDN are represented on the left panel with mutation frequencies displayed on the right, and SDCXPA are represented on the right panel with mutation frequencies displayed on the left. Mutation frequencies for the entire cohort, including both histologic subtypes, are on the far right.
Figure 2.
Proportion of SDC patients harboring tumors with potentially targetable genetic alterations with or without possible primary or secondary resistance mutation. MSKCC levels of evidence: 1-FDA-recognized biomarker predictive of response to an FDA-approved drug in this indication, 2A-Standard care biomarker predictive of response to an FDA-approved drug in this indication, 2B-Standard care biomarker predictive of response to an FDA-approved drug in another indication, but not standard care for this indication, 3A-Compelling clinical evidence supports the biomarker as being predictive of response to a drug in this indication, but neither biomarker and drug are standard care, 3B-Compelling clinical evidence supports the biomarker as being predictive of response to a drug in another indication, but neither biomarker and drug are standard care, 4-Compelling biological evidence supports the biomarker as being predictive of response to a drug, but neither biomarker and drug are standard care. Abbreviations: amp=amplification, GA=genetic alteration.
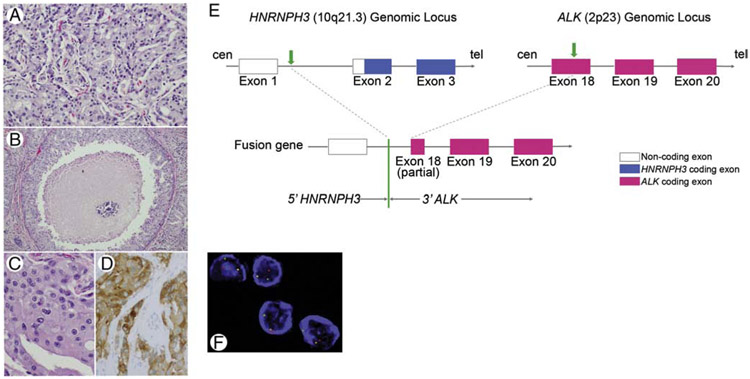
3.4.1. Novel HNRNPH3-ALK fusion in SDCDN
A novel anaplastic lymphoma kinase (ALK) gene rearrangement was detected in one de novo carcinoma. This fusion gene comprised of the 3’ALK on chromosome 2 fused to the 5’UTR of heterogeneous nuclear ribonucleoprotein H3 (2H9) (HNRNPH3), a gene ubiquitously expressed gene in human cells, located on chromosome 10, and resulted in a potentially functional in-frame product HNRNPH3-ALK. The break points mapped to exon 18 of the ALK gene and intron 1 of the HNRNPH3 gene, and the chimeric protein included the entire tyrosine kinase domain of ALK. The disruption at the ALK locus was confirmed by FISH and the presence of potentially functional fusion protein was supported by positive immunohistochemistry targeting the C-terminal portion of the tyrosine kinase domain of ALK (Figure 3).
Figure 3.
SDC12 harboring HNRHPH3-ALKfusion formed invasive nests (H&E, medium power, A), showed comedo-type necrosis (H&E, low power, B), apocrine cytology with abundant eosinophilic cytoplasm and prominent nucleoli (H&E, high power, C), and positive cytoplasmic immunostain for ALK (high power, D). Schematic diagrams of HNRNPH3 and ALK partial gene structures as well as the formation of the HNRNPH3-ALK fusion gene are depicted: HNRNPH3 is located at 10q21.3 on the plus strand of chromosome 10, and ALK is located at 2p23 on the minus strand of chromosome 2. Transparent boxes represent noncoding parts of mature transcript of HNRNPH3, blue boxes represent HNRNPH3 coding parts/exons, and red boxes represent ALK coding exons. Green arrows represent break points at HNRNPH3 locus and ALK locus, respectively. The green line represents the fusion point of HNRNPH3-ALK (E). ALK rearrangement was confirmed by FISH (3’ALK, red signal, F). Abbreviations: cen=centromere, tel=telomere.
3.5. Somatic mutations potentially associated with resistance to targeted therapy
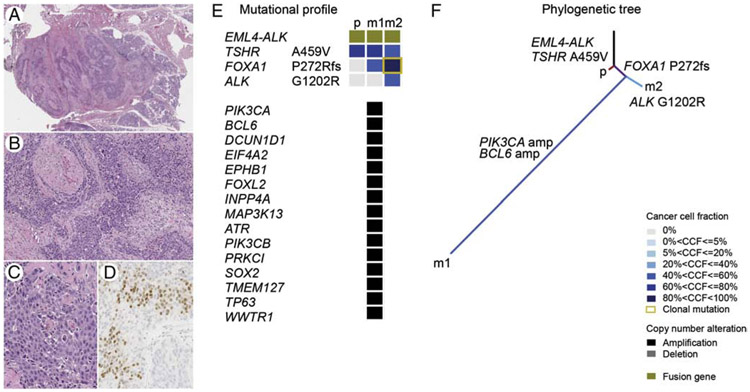
Among the 23 (79%) cases harboring potential therapeutic targets, co-existing genetic alterations potentially associated with primary resistance to targeted therapies were detected in 22% (5/23) cases i.e. in 17% cases overall (5/29, Figure 2. Supplementary Table 2). For instance, in SDC3, a BRAF V600E mutation was found in conjunction with a NF1 splice site variant associated with LOH, which may cause resistance to BRAF V600E inhibitors [17]. Recurrent amplifications in CCNE1, a mechanism of resistance to anti-HER2 agents [18] were present in 3 (14%) ERBB2 amplified cases. PTEN K128N oncogenic variant was detected in one ERBB2 amplified case and may represent a mechanism of resistance to trastuzumab [19]. The patient with EML4-ALK rearranged tumor (SDC37) was treated with an investigational ALK-inhibitor and the two subsequent metastases were also genotyped. In addition to the EML4-ALK fusion detected in each sample, metastases harbored additional genetic alterations. A phylogenetic analysis suggested that first metastasis (m1) was substantially more genetically advanced and acquired many copy number alterations than the other lesions, while second metastasis (m2) was found with an acquired ALK G1202R mutation associated with secondary resistance to select ALK-inhibitors (Figure 4).
Figure 4.
SDC37 harboring EML4-ALK fusion gene is depicted adjacent to the normal parotid showing a diffusely infiltrative growth pattern (H&E, scanning magnification, A), perineural invasion (H&E, medium power, B), large tumor cells with abundant apocrine cytoplasm and prominent nucleoli (H&E, high power, C), and positive nuclear labeling for AR (high power, D). The distinct mutational profiles with cancer cell fractions of the primary tumor and matched metastases (E), and the phylogenetic tree illustrate the clonal evolution (F).
Abbreviations: CCF=cancer cell fraction, p=primary tumor, m1=metastasis 1 (first recurrence), m2=metastasis 2 (second recurrence).
3.6. Mutational profile of SDC is similar to that of LAR-TNBC
A comparison of mutational profiles of SDC and LAR-TNBC revealed that both tumor types are significantly enriched in mutations in TP53 (66% vs. 63%, p=1, Fisher’s exact test) and PIK3CA, (28% vs. 34%, p=0.75, Fisher’s exact test, Supplementary Table 5A). However, ERBB2 amplification was found to be significantly more frequent in SDCXPA than in LAR-TNBC (50% vs. 3%, p=0.0001, Fisher’s exact test; adjusted p=0.008, Supplementary Table 5B).
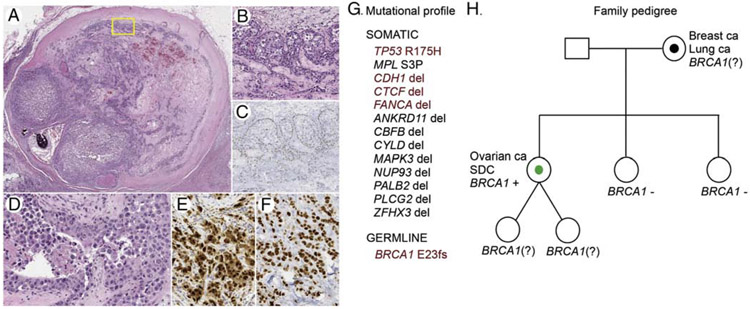
3.7. SDC patient with pathogenic germline BRCA1 mutation
Among 11 cases (SDC27-SDC37, Supplementary Table 6) evaluated for the presence of germline DNA repair genes mutations, we identified deleterious BRCA1 E23fs (c.68_69delAG) mutation in a 55-year-old woman with a prior history of treated high-grade papillary serous carcinoma of the ovary followed by a prophylactic bilateral mastectomy, and with maternal history of bilateral breast cancer and lung cancer (Figure 5, Supplementary Information). Mutational profiling of the primary parotid tumor showed multiple somatic alterations including TP53 R175H pathogenic mutation, and likely oncogenic deletions of CDH1, CTCF and FANCA genes, but no somatic BRCA1 alteration (Figure 5).
Figure 5.
Phenotype and genetics of SDC arising in a patient with a germline BRCA1 mutation. Parotid tumor with a hyalinized nodule showing features of pre-existing PA (H&E, scanning magnification, A). The tumor cells in the in situ component at the periphery (yellow frame, A) were partly surrounded by intact benign abluminal/myoepithelial cells (H&E, medium power, B), which were positive for p63 immunostain (medium power, C). The invasive component was focally necrotic and comprised of cytologically high-grade tumor cells with prominent nucleoli and ample apocrine cytoplasm (H&E, high power, D) that were positive for AR immunostain (high power, E). Immunolabeling for p53 (F) was consistent with abnormal accumulation of mutant p53 protein due to the presence of pathogenic somatic TP53 R175H variant (G). In addition, multiple somatic genetic alterations were detected in the sequenced tumor, while analysis of normal DNA revealed germline BRCA1 E23fs mutation (G; pathogenic/oncogenic alterations are highlighted dark-red). The patient’s family pedigree is depicted (H). Annotation: square=male, circle=female, diagonal line=twins, black dot=cancer diagnosis in a family member, green dot=cancer diagnosis, index case, (?)=unknown mutation status.
Abbreviations: ca=carcinoma, SDC=salivary duct carcinoma.
4. DISCUSSION
Here we analyzed a series of primary resected SDCs, including SDCDN and SDCXPA, and observed that these tumors have complex genomes, with diverse patterns of gene copy number alterations, somatic mutations and fusion genes including HNRNPH3-ALK and EML4-ALK rearrangements. Despite that the majority of SDC were found with potentially targetable alterations in genes such as ERBB2, PIK3CA, HRAS, ALK, and BRAF, a significant minority of cases harbored possible primary resistance mutations involving CCNE1, PTEN and NF, or a secondary resistance mutation ALK G1202R. Our germline analysis revealed one SDC patient harboring a pathogenic germline BRCA1 variant.
Similar to prior studies, we identified TP53 (69%), ERBB2 (31%), PIK3CA (28%), NF1 (21%), and HRAS (17%) to be the most frequently altered genes in SDC [5,20]. ERBB2 amplification (28%) was as common as previously reported [21], identifying a significant proportion of patients potentially amenable to trastuzumab therapy [22,23]. ERBB2 single nucleotide variants were also reported in SDC [4] and we identified two such variants, ERBB2 V777L and ERBB2 S310F. ERBB2 mutations are important to distinguish because distinct ERBB2 variants were found to confer different sensitivity to specific anti-HER2 agents in invasive mammary carcinomas [24,25]. For example, a breast cancer patient harboring the ERBB2 S310F in the absence of ERBB2 amplification was reported to respond to trastuzumab [26]. While our results suggest that a significant proportion of SDC patients may be eligible for anti-HER2 targeted therapy, detection of co-existing genetic alterations as a source of primary resistance to targeted therapy may need to be considered. Out of the 8 ERBB2 amplified cases 3 SDC showed concurrent CCNE1 amplification and one had the PTEN K128N variant; both were recognized as mechanisms of trastuzumab resistance in ERBB2 amplified breast cancers [18,19], further implying that SDC with similar molecular profiles may not be responsive to trastuzumab therapy. A genetic similarity between SDC and invasive mammary carcinoma was suggested earlier [5,27]. Our comparison of the mutational spectra of SDC and LAR-TNBC confirmed the remarkable genetic similarity of SDC and this breast cancer subtype. Therefore, it seems reasonable to suspect that the patterns of response or resistance to targeted therapies in SDC might mirror those observed in breast cancers with a similar genetic profile. Potential therapeutic targets were also detected in the PI3K pathway including PIK3CA hotspot variants in nearly one third of cases, and they frequently co-existed with HRAS Q61 or Q13 mutations [4,5]. SDC patients with tumors harboring mutations in PI3K pathway genes were reported to respond to PI3K/Akt/mTOR pathway inhibition [28,29]. While PIK3CA/HRAS mutated cases may be challenging to treat, development of dual-acting agents modulating MEK and PI3K signaling pathway has been in progress [30] suggesting that such treatment approach may eventually be explored in PIK3CA/HRAS mutated SDC cases.
In contrast to prior genomic studies on SDC, here we report two cases with ALK fusions. Fusions of the ALK gene with a variety of fusion partners were previously found in different cancer types, including EML4-ALK fusion in non-small cell lung carcinoma [31]. These fusions typically involving C-terminal tyrosine kinase domain of ALK, which has been a suitable target for tyrosine kinase inhibitors [32]. In the HNRNPH3-ALK fusion (SDC12), the predicted transcript of this novel fusion gene includes ALK exons 18-29 encoding a protein comprised of the intact kinase domain of ALK. Unlike most previously reported ALK fusions which typically result from a gene to gene fusion, here the fusion point maps in the 5’UTR of HNRNPH3, a gene ubiquitously expressed in human cells. The expression of 3’ALK appears to be under the regulation of the HNRNPH3 promoter while the fusion transcript does not include any HNRNPH3 coding exons. The presence of an actively translated fusion protein in the tumor cells was supported by a strong positive IHC with a monoclonal antibody targeting the C-terminal portion of the tyrosine kinase domain of ALK. The mechanism of ALK activation in HNRNPH3-ALK fusion resembles the one reported by Wiesner et al. [33]. They described a novel ALK transcript contained exons 20-29 of ALK that were preceded by ~400 base pairs of intron 19 but not exons 1 to 19. They proposed a novel mechanism of the oncogene activation, which mapped in the intronic region and was referred to as alternative transcription activation (“ATI”) [33]. HNRNPH3-ALK fusion positive SDC may belong to the same group of tumors characterized by this novel mechanism of the oncogene activation. The patient with the EML4-ALK rearrangement positive tumor (SDC37) was treated with an investigational ALK-inhibitor. He recurred with a lung metastasis harboring the ALK G1202R variant, which was recognized as secondary resistance mutation to select ALK-inhibitors in patients with EML4-ALK fusion positive lung adenocarcinoma [34]. While SDC patients with ALK rearranged tumors may benefit from ALK inhibition, the established knowledge on other ALK rearranged cancers, namely pulmonary adenocarcinomas, may help direct treatment decisions in genetically similar SDC.
In order to shed more light on the pathogenesis of SDC, we examined the differences in the mutational spectra of SDCDN and SDCXPA. To date a single study on genetic differences between SDCDN and SDCXPA using a limited gene panel has been published [20] and consistent with our results, they reported a significantly higher frequency of TP53 mutations and ERBB2 amplifications in SDCXPA then in de novo counterparts. Interestingly, we also found that certain types of TP53 alterations were nearly restricted to one or the other histologic subtype; while missense TP53 variants were limited only to SDCXPA, SDCDN harbored only TP53 frame-shift truncating mutations and intragenic deletion.
In addition to the in-depth analysis of somatic alterations in SDC and in contrast to the published literature, a subset of our cohort was also examined for the presence of germline DNA repair genes mutations. We have provided the first report of a SDC patient harboring a germline BRCA1 E23fs mutation. A single previous study has shown that the prevalence of salivary carcinoma, being salivary gland carcinoma not otherwise specified and adenoid cystic carcinoma, among BRCA1-mutated probands and likely carriers is significantly higher than the background incidence rate (0.052% vs. 0.003%, p<0.001) [35]. However, possible associations between germline BRCA1/2 mutations and SDC in particular have not been previously reported.
Our study has several limitations. First, given the rarity of SDC, our cohort size is relatively small. We intentionally selected only cases with available primary tumor resection to be able to obtain the spectrum of somatic events occurring early during SDC carcinogenesis. In addition, we included only cases with available matched normal tissue (1) to obtain accurate tumor/patient-specific somatic mutational profiles, (2) to be able to perform clonality and LOH analyses of somatic variants (by FACETS), and (3) to perform a germline analysis. Second, a lack of PLAG1/HMGA2 rearrangement status prevented us from performing additional correlations between the rearrangement status of these genes and other molecular, pathological and clinical features of SDCXPA. Third, the retrospective nature and lack of uniformity in treatment may limit the accuracy of the survival analysis. Despite the limitations several important conclusions can be drawn from the analysis made in our study. SDCDN and SDCXPA are genetically distinct. Given the complex repertoire of somatic alterations in SDC a broad genomic profiling is necessary to increase the likelihood of detection of rare but clinically actionable somatic alterations such as ALK fusions. Although the majority of SDC cases can be found with genetic alterations amenable to targeted therapy in “basket” clinical trials, somatic mutations associated with possible primary or secondary resistance to targeted therapies can occur in cancers. Further studies are warranted to evaluate the risk of SDC in BRCA1 carriers.
Supplementary Material
HIGHLIGHTS.
79% SDC harbor targetable somatic mutations.
22% SDC with targetable somatic mutations may harbor concurrent primary resistance mutations.
A novel HNRNPH3-ALK rearrangement is detected in SDC.
ALK inhibition in an EML4-ALK positive SDC can lead to ALK G1202R secondary resistance mutation.
First SDC patient harboring BRCA1 E23fs germline mutation is reported.
ACKNOWLEDGEMENTS
We wish to acknowledge the expert help of Zoran Perak and thank him for his assistance in figures preparation.
Research reported in this publication was supported by the Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute under award number P30CA008748.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement:
No conflict of interests exists for all contributory authors.
REFERENCES
- [1].Kleinsasser O, Klein HJ, Hubner G. [Salivary duct carcinoma. A group of salivary gland tumors analogous to mammary duct carcinoma]. Arch Klin Exp Ohren Nasen Kehlkopfheilkd 1968; 192: 100–105. [PubMed] [Google Scholar]
- [2].Boon E, Bel M, van Boxtel W, et al. A clinicopathological study and prognostic factor analysis of 177 salivary duct carcinoma patients from The Netherlands. Int J Cancer 2018; 143: 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Katabi N, Gomez D, Klimstra DS, et al. Prognostic factors of recurrence in salivary carcinoma ex pleomorphic adenoma, with emphasis on the carcinoma histologic subtype: a clinicopathologic study of 43 cases. Hum Pathol 2010; 41: 927–934. [DOI] [PubMed] [Google Scholar]
- [4].Chiosea SI, Williams L, Griffith CC, et al. Molecular characterization of apocrine salivary duct carcinoma. Am J Surg Pathol 2015; 39: 744–752. [DOI] [PubMed] [Google Scholar]
- [5].Dalin MG, Desrichard A, Katabi N, et al. Comprehensive Molecular Characterization of Salivary Duct Carcinoma Reveals Actionable Targets and Similarity to Apocrine Breast Cancer. Clin Cancer Res 2016; 22: 4623–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015; 17: 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].El-Naggar AK, Chan JKC, Grandis JR, et al. World Health Organization Classification of Head and Neck Tumours (4th edition). International Agency for Reseach on Cancer (IARC): Lyon, 2017. p. 173–175. [Google Scholar]
- [8].Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012; 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Piscuoglio S, Ng CK, Murray MP, et al. The Genomic Landscape of Male Breast Cancers. Clin Cancer Res 2016; 22: 4045–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ng CKY, Piscuoglio S, Geyer FC, et al. The Landscape of Somatic Genetic Alterations in Metaplastic Breast Carcinomas. Clin Cancer Res 2017; 23: 3859–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016; 44: e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Murugaesu N, Wilson GA, Birkbak NJ, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 2015; 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guerini-Rocco E, Piscuoglio S, Ng CK, et al. Microglandular adenosis associated with triple-negative breast cancer is a neoplastic lesion of triple-negative phenotype harbouring TP53 somatic mutations. J Pathol 2016; 238: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wolff AC, Hammond MEH, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 2013; 138: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Network CGA. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Whittaker SR, Theurillat JP, Van Allen E, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 2013; 3: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Scaltriti M, Eichhorn PJ, Cortés J, et al. Cyclin E amplification/overexpression is a mechanism of trastuzumab resistance in HER2+ breast cancer patients. Proceedings of the National Academy of Sciences 2011; 108: 3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].de Melo Gagliato D, Jardim DL, Marchesi MS, et al. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget 2016; 7: 64431–64446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chiosea SI, Thompson LD, Weinreb I, et al. Subsets of salivary duct carcinoma defined by morphologic evidence of pleomorphic adenoma, PLAG1 or HMGA2 rearrangements, and common genetic alterations. Cancer 2016; 122: 3136–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Glisson B, Colevas AD, Haddad R, et al. HER2 expression in salivary gland carcinomas: dependence on histological subtype. Clin Cancer Res 2004; 10: 944–946. [DOI] [PubMed] [Google Scholar]
- [22].Limaye SA, Posner MR, Krane JF, et al. Trastuzumab for the treatment of salivary duct carcinoma. Oncologist 2013; 18: 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nabili V, Tan JW, Bhuta S, et al. Salivary duct carcinoma: a clinical and histologic review with implications for trastuzumab therapy. Head Neck 2007; 29: 907–912. [DOI] [PubMed] [Google Scholar]
- [24].Bose R, Kavuri SM, Searleman AC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov 2013; 3: 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ben-Baruch NE, Bose R, Kavuri SM, et al. HER2-Mutated Breast Cancer Responds to Treatment With Single-Agent Neratinib, a Second-Generation HER2/EGFR Tyrosine Kinase Inhibitor. J Natl Compr Canc Netw 2015; 13: 1061–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Chumsri S, Weidler J, Ali S, et al. Prolonged Response to Trastuzumab in a Patient With HER2-Nonamplified Breast Cancer With Elevated HER2 Dimerization Harboring an ERBB2 S310F Mutation. J Natl Compr Canc Netw 2015; 13: 1066–1070. [DOI] [PubMed] [Google Scholar]
- [27].Di Palma S, Simpson RH, Marchiò C, et al. Salivary duct carcinomas can be classified into luminal androgen receptor-positive, HER2 and basal-like phenotypes. Histopathology 2012; 61: 629–643. [DOI] [PubMed] [Google Scholar]
- [28].Nardi V, Sadow PM, Juric D, et al. Detection of novel actionable genetic changes in salivary duct carcinoma helps direct patient treatment. Clin Cancer Res 2013; 19: 480–490. [DOI] [PubMed] [Google Scholar]
- [29].Piha-Paul SA, Cohen PR, Kurzrock R. Salivary duct carcinoma: targeting the phosphatidylinositol 3-kinase pathway by blocking mammalian target of rapamycin with temsirolimus. J Clin Oncol 2011; 29:e727–730. [DOI] [PubMed] [Google Scholar]
- [30].Van Dort ME, Hong H, Wang H, et al. Discovery of Bifunctional Oncogenic Target Inhibitors against Allosteric Mitogen-Activated Protein Kinase (MEK1) and Phosphatidylinositol 3-Kinase (PI3K). J Med Chem 2016; 59: 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007; 448: 561–566. [DOI] [PubMed] [Google Scholar]
- [32].Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. The lancet oncology 2011; 12: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wiesner T, Lee W, Obenauf AC, et al. Alternative transcription initiation leads to expression of a novel ALK isoform in cancer. Nature 2015; 526: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012; 4: 120ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shen TK, Teknos TN, Toland AE, et al. Salivary gland cancer in BRCA-positive families: a retrospective review. JAMA Otolaryngol Head Neck Surg 2014; 140: 1213–1217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.