Abstract
Hepatocellular carcinoma (HCC) is the most commonly diagnosed primary liver malignancy. The limited success with relapse of the disease in HCC therapy is frequently associated with the acquired resistance to anticancer drugs. To develop a strategy and design for overcoming the resistance of HCC cells to TNF-related apoptosis inducing ligand (TRAIL)-induced cell death, we evaluated the efficacy of a non-steroidal anti-inflammatory drug (NSAID) in combination with TRAIL against TRAIL-resistant HCC cells expressing a high level of CD44. We revealed by MTT and western blotting, respectively, that celecoxib (CCB), an NSAID, and 2,5-dimethyl celecoxib (DMC), a non-cyclooxygenase (COX)-2 inhibitor analog of CCB, were able to sensitize TRAIL-resistant HCC cells to TRAIL, implicating a COX-independent mechanism. CCB dose-dependently enhanced LC3-II and reduced p62 levels through AMPK activation and inhibition of the Akt/mTOR pathway and upregulated expression of ATF4/CHOP, leading to activation of endoplasmic reticulum (ER) stress-dependent autophagy. The TRAIL sensitization capacity of CCB in TRAIL-resistant HCC cells was abrogated by an ER stress inhibitor. In addition, we also revealed by flow cytometry and western blotting, respectively, that accelerated downregulation of TRAIL-mediated c-FLIP expression, DR5 activation and CD44 degradation/downregulation by NSAID resulted in activation of caspases and poly(ADP-ribose) polymerase (PARP), leading to the sensitization of TRAIL-resistant HCC cells to TRAIL and thereby reversal of TRAIL resistance. From these results, we propose that NSAID in combination with TRAIL may improve the antitumor activity of TRAIL in TRAIL-resistant HCC, and this approach may serve as a novel strategy that maximizes the therapeutic efficacy of TRAIL for clinical application.
Keywords: hepatocellular carcinoma, TRAIL, non-steroidal anti-inflammatory drug, autophagy, CD44, c-FLIP, endoplasmic reticulum stress
Introduction
The most common type of liver cancer is hepatocellular carcinoma (HCC), and the prognosis of patients with advanced HCC is poor due to acquired resistance to current chemotherapeutic regimens through the de-regulation of signaling pathways governing cell proliferation and survival (1). Resistance to apoptosis of HCC cells is a critical obstacle in cancer treatment. Among the diverse modalities inducing apoptosis in cancer cells including HCC cells, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), a death receptor ligand is one of the promising anticancer agents due to its capability to induce apoptosis selectively in cancer cells but not in most normal cells (2). However, most primary cancer cells show resistance to TRAIL monotherapy. Therefore, combination therapies are required for reduced development of drug resistance, better effectiveness, and reduced toxicity. TRAIL combinations have been studied to induce synergism or sensitize TRAIL-resistant cancer cells (3), and identification of effective combination that synergize with TRAIL to kill HCC cells is needed for a more extensive and successful application of TRAIL-based therapies in the future.
TRAIL-induced apoptosis occurs through the binding of TRAIL to its cognate surface receptors. Following the binding of TRAIL to the death receptor TRAIL-R1 (DR4) and/or TRAIL-R2 (DR5), the activated receptors recruit the adapter protein FAS-associated death domain (FADD) and the effector capase-8, resulting in the assembly of the death-inducing signaling complex (DISC). After binding the DISC, caspase-8 undergoes cleavage and promotes apoptosis by activating the downstream effector caspase-3 and the mitochondrial apoptotic pathway (2). The cellular-FLICE inhibitory protein (c-FLIP), which consists of two isoforms, FLIPL and FLIPS, resembles an initiator procaspase, except in the absence of a proteolytic domain. Following the recruitment of c-FLIP to the DISC, this protein competes with procaspases-8 and −10, blocking the processing and activation of these procaspases and inhibiting DR4- and DR5-mediated cell death. Therefore, c-FLIP hinders apoptosis by inhibiting the activation of caspase-8 and accordingly the inhibition of c-FLIP enhances TRAIL-induced apoptosis in cancer cells (4).
It has been shown that several cancer cell lines including HCC cells are resistant to TRAIL (5). An overexpression of c-FLIP, an endogenous antiapoptotic factor which inhibits procaspase-8 in DISC complex, may represent an important mechanism for resistance to apoptosis in cancer cells (6). In addition, the downregulation of antiapoptotic proteins involving c-FLIP and/or upregulation of death receptors, and the activation of C/EBP homologous protein (CHOP) can overcome TRAIL resistance in cancer cells (7). CHOP, which is induced during the unfolded protein response, mediates the transcriptional control during endoplasmic reticulum (ER) stress-induced apoptosis (8). c-FLIPL is a CHOP control target, and CHOP downregulates c-FLIPL expression at the post-transcriptional level (9).
It has been known that an interplay of autophagy and apoptosis, which are interconnected in their signaling pathways, greatly affects cell death during stress responses. An insufficient activity of autophagy may trigger apoptosis due to accumulation of aberrant proteins and defective organelles, while excessive activity of autophagy can also lead to cell death, even in the insufficient stimuli of apoptosis (10). Therefore, the interconnection of signaling pathways of both autophagy and apoptosis is not surprising, and may modulate sensitivity to anticancer drugs. Autophagy is associated with improvement of TRAIL sensitivity of cancer cells through both upregulation of DR5 and c-FLIP degradation (11,12). It has been suggested that the addition of autophagy-inducing agents to some apoptosis-inducing therapeutic agents could be a useful therapeutic approach to overcome resistance of cancers (13).
Nonsteroidal anti-inflammatory drugs (NSAIDs), which are a structurally diverse group of drugs, are widely used to treat inflammation, fever and pain, and are currently known to have diverse effects in cancer. Celecoxib (CCB), a non-cyclooxygenase (COX)-2 selective NSAID, exhibits therapeutic effects in tumor cells in combination with chemotherapy and radiotherapy in preclinical investigations (14). Ibuprofen, a commonly used NSAID, was also found to enhance the anticancer effect of cisplatin in lung cancer cells (15). The antitumor effects of NSAIDs seems to be at least in part related to their autophagy-modulating effects, and the use of NSAIDs in combination with anticancer drugs may be helpful for the treatment of drug-resistant tumors (16).
In this study, we investigated whether NSAID could potentiate TRAIL cytotoxicity of TRAIL-resistant HCC cells, and the molecular mechanism underlying the combined effect of NSAID and TRAIL on sensitization of TRAIL-resistant HCC cells to TRAIL for reversal of TRAIL resistance by inducing autophagic cell death as well as apoptosis.
Materials and methods
Cell culture and reagents
Hepatocellular carcinoma (HCC) cell lines (SNU-354, SNU-423, SNU-449 and SNU-475) derived from HCC tissues of patients were purchased from the Korea Cell Line Bank (17). TRAIL-resistant SNU-475/TR subline was isolated from SNU-475 cells by stepwise increases in concentrations of TRAIL, starting with 10 ng/ml and reaching 200 ng/ml. After approximately 2 month of drug exposure, SNU-475/TR cells were able to grow in the presence of 200 ng/ml TRAIL, and this phenotype remained stable through passages. SNU-475/TR cells were at least 100-fold more resistant than SNU-475 cells to TRAIL. Cells were maintained in RPMI-medium (Welgene) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS; Welgene), 100 U/ml penicillin and 100 µg/ml streptomycin in a 5% CO2 humidified incubator at 37°C. Recombinant human soluble TRAIL was obtained from R&D systems. Celecoxib (CCB), 2,5-dimethyl celecoxib (DMC), cycloheximide (CHX), 3-methyladenine (3-MA), chloroquine (CQ), LY294002 (LY), and 4-phenylbutyric acid (4-PBA) were purchased from Sigma-Aldrich; Merck KGaA.
Cell proliferation assay
Cell proliferation was measured by counting viable cells using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric dye-reduction method. Exponentially growing cells (1×104 cells/well) were plated in a 96-well plate and incubated in growth medium treated with the indicated concentration of TRAIL in the absence or presence of the NSAID at 37°C. After 96 h, the medium was removed using centrifugation (500 × g for 10 min), and MTT-formazan crystals solubilized in 100 µl DMSO. The optical density (OD) of each sample at 570 nm was measured using an ELISA reader. The optical density of the medium was proportional to the number of viable cells. Inhibition of proliferation was evaluated as a percentage of control growth (no drug in the medium). All experiments were repeated in at least two experiments in triplicate.
Western blot analysis
Cells were lysed by RIPA cell lysis buffer (1X) with EDTA (GenDEPOT, Inc.) and the Bradford protein (Bio-Rad Laboratories, Inc.) assay was used for protein determination. The protein samples from each group were resolved by 10–15% SDS-PAGE (20 µg protein per lane) and transferred to PVDF membrane (EMD Millipore). Membranes were blocked with 5% non-fat milk in TBST at room temperature (RT) for 1 h, and then incubated at 4°C overnight with the primary antibodies: Caspase-8 (dilution 1:1,000, cat. no. 9746), caspase-9 (dilution 1:1,000, cat. no. 9508), caspase-3 (dilution 1:1,000, cat. no. 9662), c-FLIP (dilution 1:1,000, cat. no. 56343), ALDH1 (dilution 1:1,000, cat. no. 36671), Nanog (dilution 1:1,000, cat. no. 3580), CHOP (dilution 1:1,000, cat. no. 5554), CD44 (dilution 1:1,000, cat. no. 5640), β-tubulin (dilution 1:10,000, cat. no. 2146), AMPK (dilution 1:1,000, cat. no. 2532), phosphorylated (p)-AMPK (Thr172) (dilution 1:1,000, cat. no. 2535), Akt (dilution 1:1,000, cat. no. 9272), p-Akt (Thr308) (dilution 1:1,000, cat. no. 9271), mTOR (dilution 1:1,000, cat. no. 2972), p-mTOR (Ser2448) (dilution 1:1,000, cat. no. 2971), p70S6K (dilution 1:1,000, cat. no. 9202), p-p70S6K (Thr389) (dilution 1:1,000, cat. no. 9205), 4E-BP1 (dilution 1:1,000, cat. no. 9644) and p-4E-BP1 (Thr37/46) antibodies (dilution 1:1,000, cat. no. 2855) (all above were purchased from Cell Signaling Technology, Inc.) ATF4 (dilution 1:1,000, cat. no. ab34034), DR5 (dilution 1:1,000, cat. no. ab16942; Abcam), PARP (dilution 1:10,000, cat. no. sc-7150; Santa Cruz Biotechnology, Inc.), β-actin (dilution 1:10,000, cat. no. A5316; Sigma-Aldrich; Merck KGaA), LC3B (dilution 1:5,000, cat. no. NB100-2220), p62 (dilution 1:1,000, cat. no. NBP1-31381; Novus) CD133 (dilution 1:1,000, cat. no. PAB12663; Abnova). Membranes were washed with TBST three times (15 min per time), incubated with horseradish peroxidase-conjugated secondary antibody (dilution 1:10,000, cat. no. 7074; dilution 1:50,000, cat. no. 7076; Cell Signaling Technology, Inc.) and washed thrice with TBST. Target proteins were visualized using ECL and detected with an enhanced chemiluminescence (ECL) detection system (NEL103E001EA, PerkinElmer Inc.), and the film (Agfa RADIOMAT™) that detected chemiluminescence signals from Western blot was developed by JP-33 Automatic X-Ray Film Processor (JPI America, Inc.) in a darkroom.
Apoptosis analysis by flow cytometry
Cell apoptosis was detected using an Annexin V/propidium iodide (PI) apoptosis detection kit according to the manufacturer's protocols. Cells were treated with TRAIL in the presence or absence of CCB (or DMC). After 24 h, the cells were harvested by trypsinization and washed in PBS. The cells were centrifuged (400 × g for 5 min) and resuspended in 100 µl binding buffer, and then 5 µl FITC Annexin V and 3 µl PI was added. The cells were gently vortexed and incubated for 15 min at RT in the dark. After that 400 µl of binding buffer was added to each tube. The samples were analyzed using a CANTO II (BD Biosciences) flow cytometer and the data were analyzed using FlowJo (ver.10; Tress Star).
Fluorescence-activated cell sorting (FACS) analysis
Expression profiles of cell surface molecules on HCC cell lines were conducted using FITC mouse anti-human CD44 (dilution 1:50, cat. no. 555478; BD Biosciences), APC mouse anti-human CD133 (dilution 1:100, cat. no. 130-090-854; Miltenyi Biotec) and FITC mouse anti-human DR5 (dilution 1:10, cat. no. MA1-19759; Invitrogen; Thermo Fisher Scientific, Inc.) monoclonal antibodies (mAbs). Cells (5×105 cells/well) were centrifuged at 500 × g and resuspended in 500 µl PBS. Cells were then incubated for 1 h on ice with 5 µl of mouse IgG, anti-CD44 or anti-CD133 mAb for 30 min. After washing with PBS, PE-conjugated rabbit anti-mouse IgG (dilution 1:500, cat. no. IC002P; R&D Systems) was added to the cell suspensions, incubated for 30 min on ice, and washed with PBS. After rinsing, samples were analyzed by flow cytometry using a FACS Calibur flow cytometer (BD Biosciences). The data were analyzed using CellQuest software (version 3.3; BD Biosciences). Cell surface expression of CD44 or DR5 on HCC cells treated with or without NSAID was determined by flow cytometry. Briefly, cells were washed once at the time of harvesting with PBS/0.1% sodium azide and aliquoted into polystyrene tubes. Cells were stained with FITC-labeled anti-CD44 mAb or anti-DR5 mAb. Autofluorescence and isotype (IgG2b)-matched control Abs (dilution 1:500, cat. no. MA5-14447; BD Biosciences) were included. Data was collected using BD FACSCanto II or BD FACSCalibur flow cytometer (BD Biosciences) and subsequently analyzed with FlowJo software (version 10; Tree Star).
Statistical analysis
Statistical analyses were performed using SPSS ver. 24.0 (IBM Corp.). One-way analysis of variance (ANOVA) with Tukey's post hoc test was performed to analyze multiple groups, and unpaired t-test was used to analyze differences in data between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
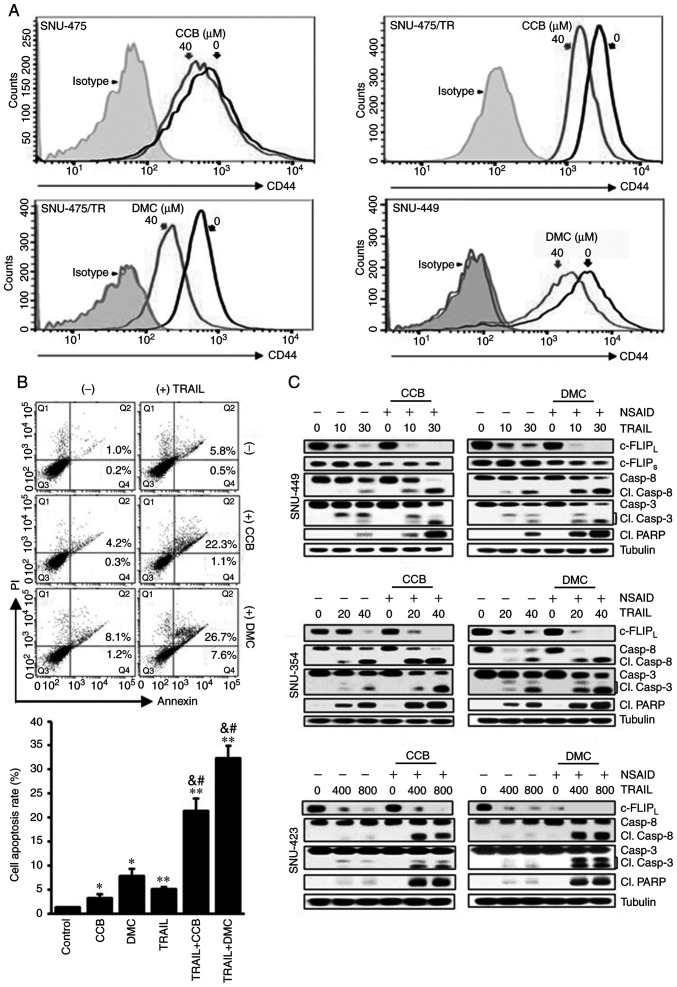
NSAID sensitizes TRAIL-resistant CD44-overexpressing HCC cells to TRAIL
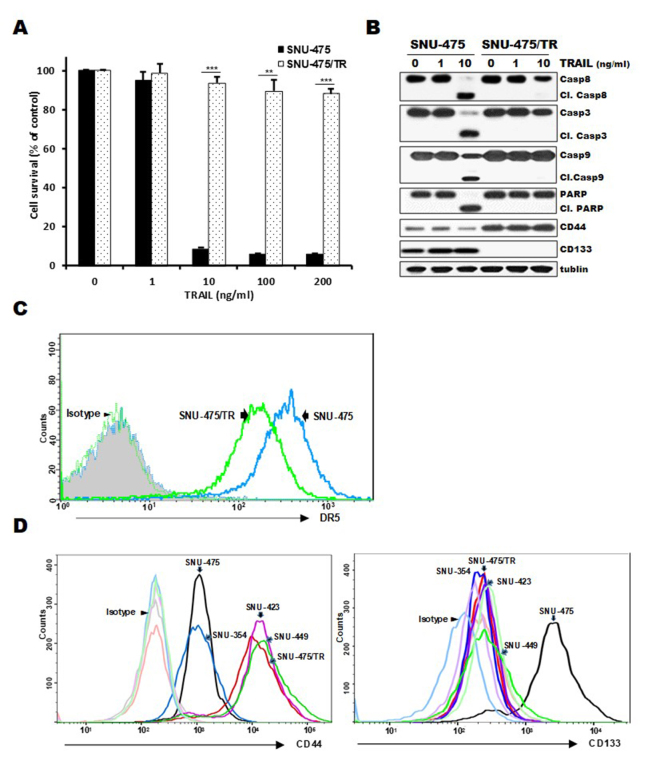
Expression of CD44 and CD133 as markers of liver cancer stem cells can contribute to hepatocellular carcinoma (HCC) development and poor responsiveness of HCC towards chemotherapy and radiotherapy (18). Since high susceptibility to TRAIL of HCC cells is associated with CD133 expression (19), we therefore investigated whether the acquisition of resistance to TRAIL is associated with distribution of CD44/CD133 expression in HCC cells. When both SNU-475/TR cells exhibiting high-level resistance to TRAIL and the parental TRAIL-sensitive SNU-475 cells were treated with increasing doses of TRAIL, marked activation of caspases (caspase-8, −9 and −3) and PARP, showing cleavage of caspases and PARP, occurred in the SNU-475 cells but not in the SNU-475/TR cells, indicating resistance of SNU-475/TR cells to TRAIL (Fig. 1A and B). Moreover, SNU-475/TR cells exhibited relatively high basal expression of CD44 but undetectable expression of CD133, as compared to those in SNU-475 cells, and also the upregulated CD44 level of SNU-475/TR cells was not modulated by TRAIL treatment (Fig. 1B), suggesting that the expression of CD44 in HCC cells might be linked to the acquisition of the TRAIL-resistance phenotype. We next compared cell surface expression of DR5, a major predictor of TRAIL sensitivity, between SNU-475/TR and SNU-475 cells by FACS analysis. As expected, the cell surface expression of DR5 in SNU-475/TR cells was significantly decreased when compared with that in SNU-475 cells (Fig. 1C). We therefore evaluated the expression of both CD44 and CD133 cell-surface markers in four HCC cell lines and SNU-475/TR subline (Fig. 1D). The surface expression of CD44 in SNU-423, SNU-449, SNU-475/TR, SNU-423 and SNU-449 cells was significantly higher than that in the SNU-475 and SNU353 cells whereas the surface expression of CD133 in SNU-353, SNU-423, SNU-449 and SNU-475/TR cells was significantly lower than in SNU-475 cells. Indeed, we previously reported that CD133-high SNU475 cells were more sensitive to TRAIL than CD133-low SNU-423, SNU-449 and SNU-354 cells (19). These results indicate that the expression ratio of CD44/CD133 plays a critical role in HCC cells for TRAIL sensitivity, and downregulation of CD44 is required for enhancing the TRAIL sensitivity of HCC cells.
Figure 1.
Comparison of cytotoxicity, activity of caspases and surface expression of DR5, CD44 and CD133 in SNU-475/TR cells and other HCC cells in the absence and presence of TRAIL. (A) SNU-475/TR cells and the parental SNU-475 cells were treated with various concentrations of TRAIL (0. 1. 10. 100 and 200 ng/ml). Percentage of cell survival was determined after 96 h of incubation using MTT assay. Each bar represents the mean ± SD of triplicate experiments. **P<0.01, ***P<0.001 compared to the SNU-475 cells. (B) SNU-475/TR and SNU-475 cells were treated with TRAIL (1 or 10 ng/ml) for 24 h, and the level of activation by the cleavage (Cl.) of pro-caspase-8 (Casp8), −3 (Casp3), −9 (Casp9) and PARP and the expression of CD44/CD133 were determined by western blot analysis. Tubulin was used as a loading control. (C) Both cell lines were stained with anti-DR5 antibody to determine the surface expression of DR5 by a flow cytometer. (D) Four HCC and SNU-475/TR cell lines were stained with anti-CD133 or anti-CD44 antibody to determine the surface expression of CD44 or CD133 by a flow cytometer. HCC, hepatocellular carcinoma; TRAIL, TNF-related apoptosis inducing ligand; DR4, death receptor TRAIL-R1; PARP, poly(ADP-ribose) polymerase.
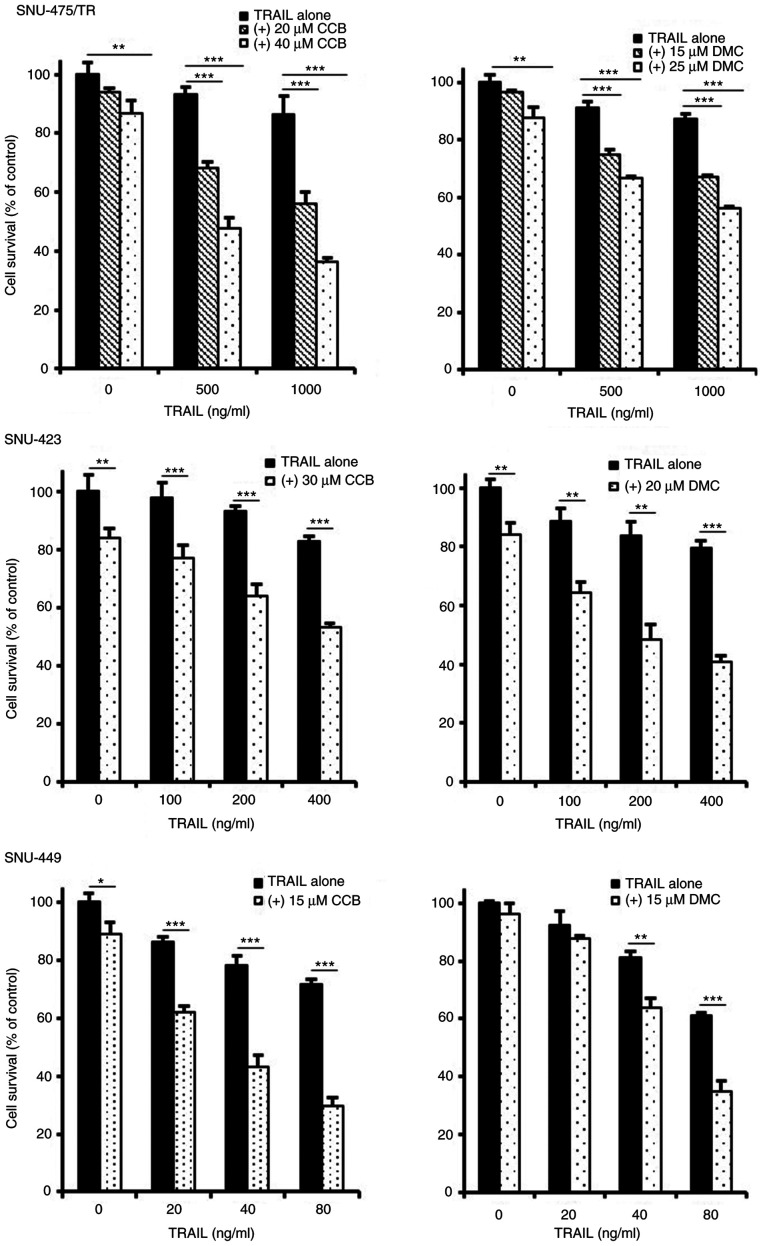
To enhance the TRAIL-mediated cytotoxicity and thus to overcome TRAIL resistance of TRAIL-resistant HCC cells, it is important to develop novel TRAIL sensitizing agent in combination with TRAIL. Indeed, combining TRAIL with single-drug therapy such as TRAIL-sorafenib combination has shown potential in colon cancer cells (20). In the present study, we investigated the combined effect of NSAID as TRAIL a sensitizing agent with TRAIL for obtaining sensitization of TRAIL-resistant HCC cells to TRAIL. To evaluate the effectiveness of NSAID as a new modulator of TRAIL sensitivity, the changing susceptibility of TRAIL-resistant HCC cells to TRAIL by NSAIDs such as celecoxib (CCB) and 2,5-dimethyl celecoxib (DMC), a non-COX-2 inhibitory analog of celecoxib was determined by MTT assay (Fig. 2). TRAIL-resistant CD44-overexpressing SNU-475/TR, SNU-423 and SNU-449 cells were treated with various concentrations of TRAIL in the presence or absence of CCB or DMC. SNU-475/TR cells showed increased susceptibility to TRAIL by combined treatment with CCB and TRAIL than TRAIL alone, indicating potentiation of TRAIL sensitivity in SNU-475/TR cells by CCB. We also determined whether DMC could enhance susceptibility of SNU-475/TR cells to TRAIL. DMC significantly potentiated TRAIL cytotoxicity in SNU-475/TR cells, suggesting that DMC could exert TRAIL-potentiating effect, regardless of COX-2 activity. Similar results were observed in other TRAIL-resistant SNU-423 and SNU-449 cells co-treated with TRAIL and CCB (or DMC). SNU-354 cells also showed that both CCB and DMC potentiated TRAIL cytotoxicity, and another non-selective COX inhibitor ibuprofen also potentiated TRAIL cytotoxicity in TRAIL-resistant HCC cells (data not shown). These results strongly suggest that NSAID could be a promising candidate for a new class of TRAIL sensitizers in HCC cells.
Figure 2.
Enhancement of TRAIL cytotoxicity by NSAID in TRAIL-resistant HCC cells. SNU-475/TR, SNU-423 and SNU-449 cells were treated with various concentrations of TRAIL in the absence or presence of celecoxib (CCB) or dimethyl celecoxib (DMC). The percentage of cell survival was determined after 96 h of incubation using MTT assay. Each bar represents the mean ± SD of triplicate experiments. *P<0.05, **P<0.01, ***P<0.001, compared with the TRAIL alone treated cells. NSAID, non-steroidal anti-inflammatory drug; TRAIL, TNF-related apoptosis inducing ligand; HCC, hepatocellular carcinoma.
NSAID modulates the expression of DR5/c-FLIP and CSC markers through ATF4/CHOP signaling axis in TRAIL-resistant HCC cells
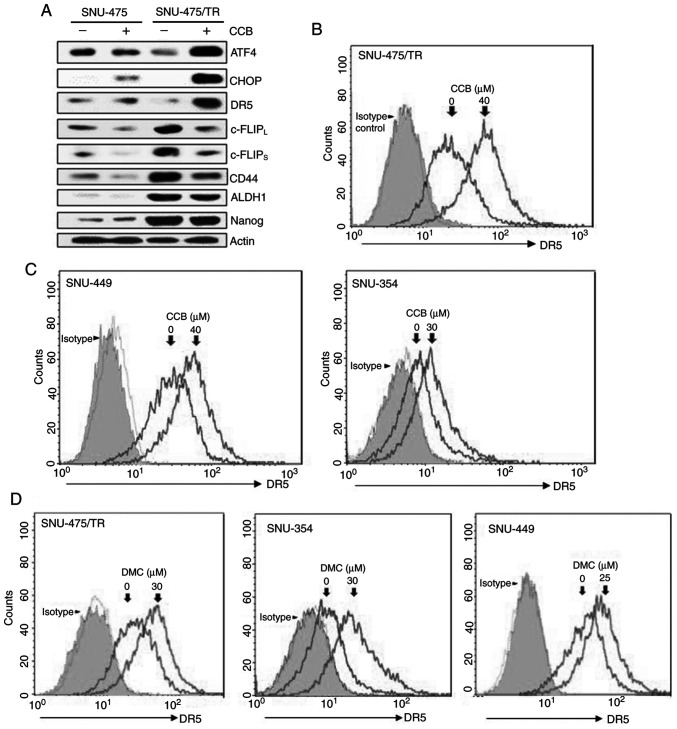
Since enhancement of DR5 expression is mediated through upregulation of ER stress-induced major transcriptional factor CHOP (8,21), we determined whether CCB could induce upregulation of DR5 via activation of ATF4 and CHOP, one of the direct ATF4 target genes, in SNU-475/TR cells in comparison to parental SNU-475 cells. CCB-induced ATF4/CHOP-dependent DR5 upregulation was observed in SNU-475/TR cells but not in SNU-475 cells (Fig. 3A, upper rows). It has been reported that the level of c-FLIP is a key regulator of TRAIL, and upregulation of c-FLIP induces TRAIL resistance (22). Downregulation of c-FLIP plays a critical role in overcoming TRAIL resistance and is caused by facilitating autophagy-mediated c-FLIP degradation, leading to apoptosis (23). We found that CCB-induced c-FLIPL/S downregulation in SNU-475/TR cells with high basal level of c-FLIPL/S was more prominent than SNU-475 cells, and concurrently the expression of CSC markers CD44, ALDH1 and Nanog that are overexpressed in SNU-475/TR cells was decreased by CCB (Fig. 3A, bottom rows). To investigate the mechanisms underlying enhancement of TRAIL-induced cytotoxicity by NSAID, modulation of DR5 cell surface expression by NSAID in HCC cells was investigated by FACS analysis since DR5 plays a critical role in TRAIL sensitization via the extrinsic pathway. In SNU-475/TR cells, the cell surface expression of DR5 was significantly increased by CCB treatment (Fig. 3B). Next, we determined whether NSAID could induce DR5 activation in other TRAIL-resistant HCC cells. SNU-449 and SNU-354 cells also showed that DR5 surface expression was increased by CCB treatment (Fig. 3C). Similar results were obtained in DMC-treated with HCC cells. The cell surface expression of DR5 was significantly increased in SNU-475/TR, SNU-354 and SNU-449 cells by DMC treatment (Fig. 3D). These results indicate that NSAID-induced DR5 activation might contribute to sensitization of TRAIL-resistant HCC cells to TRAIL.
Figure 3.
CCB-induced regulation of DR5/c-FLIP and CSC markers through the ATF/CHOP signaling axis in SNU-475/TR cells, and the effect of NSAID on the cell surface expression of DR5 in HCC cells. (A) The altered levels of ATF4, CHOP, DR5 (mature form), c-FLIPL/S, CD44, ALDH1 and Nanog in SNU-475 and SNU-475/TR cells by CCB treatment (40 µM for 24 h) were determined by western blot analysis. Actin was used as a loading control. (B-D) HCC cell lines were treated with the indicated dose of CCB or DMC for 24 h. The cells were stained with control IgG or anti-DR5 antibody and subsequently labeled with PE-conjugated secondary antibodies to determine the surface expression of DR5, and the cell surface expression were measured by a flow cytometer. CCB, celecoxib; DR5, death receptor TRAIL-R2; c-FLIP, cellular-FLICE inhibitory protein; CSC, cancer stem cell; ATF, activating transcription factor; CHOP, C/EBP homologous protein; ALDH1, aldehyde dehydrogenase 1; NSAID, non-steroidal anti-inflammatory drug; HCC, hepatocellular carcinoma; DMC, 2,5-dimethyl celecoxib.
NSAID induces degradation of c-FLIP and upregulation of DR5 through activation of ER stress-dependent autophagy
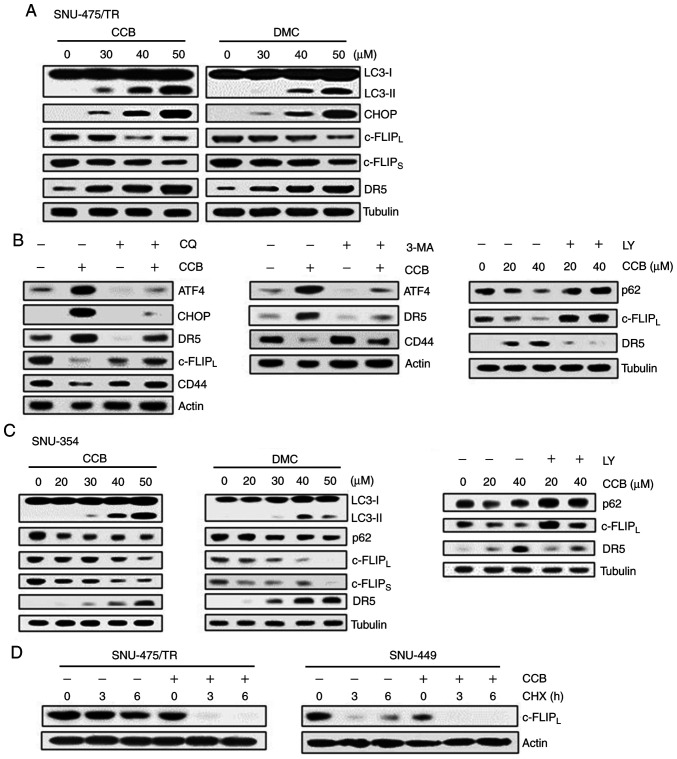
The protein c-FLIP was found to be degraded by autophagy and competes with microtubule-associated protein light chain 3 (LC3) for Atg3 binding (12,24). LC3 is widely used to monitor autophagy. The amount of LC3 conversion (LC3-I to LC3-II) is clearly correlated with the number of autophagosomes, and an increase in endogenous LC3-II may be regarded as a marker for autophagy (25). In addition, ER stress-inducible transcription factors ATF4/CHOP signaling has been shown to play significant role in the regulation of autophagy (26). In SNU-475/TR cells, we therefore examined whether CCB (or DMC) treatment could induce ER stress-dependent autophagy and lead to the modulation of DR5 and c-FLIP expression (Fig. 4A). CCB dose-dependently upregulated both LC3-II and CHOP, resulting in downregulation of c-FLIPL/S and upregulation of DR5. These results indicate CCB-induced autophagic degradation of c-FLIP and CHOP-mediated upregulation of DR5. Similarly, DMC treatment of SNU-475/TR cells also resulted in an increase in LC3-II, CHOP and DR5 levels and a decrease in c-FLIPL/S levels. We further confirmed the ability of CCB to induce autophagy related to the degradation of CD44 through ATF4/CHOP-dependent autophagy in SNU-475/TR cells since the degradation of CD44 in cancer cells is mediated by autophagy (27). When SNU-475/TR cells were treated with CCB in the presence of autophagy inhibitor, we determined whether CCB-induced modulation of DR5 and c-FLIPL/CD44 was blocked via autophagy inhibition-mediated ATF/CHOP inactivation (Fig. 4B). Autophagy inhibitor chloroquine (CQ) that blocks the final steps of autophagic degradation prevented CCB-induced upregulation of both ATF4/CHOP and DR5 and downregulation of c-FLIPL and CD44 in SNU-475/TR cells. Similar results were observed when SNU-475/TR cells were treated with CCB in the presence of another autophagy inhibitor 3-methyladenine (3-MA) or LY294002 (LY) that suppresses autophagy via inhibition of class III PI3K. CCB-induced upregulation of ATF4 and DR5 and downregulation of CD44 was significantly attenuated by 3-MA treatment in SNU-475/TR cells. In addition to LC3, degradation of p62 is another widely used marker to monitor autophagic activity and thus the reduced level of p62 can be used to monitor autophagic flux (25). The CCB-induced reduction of p62 level and subsequent downregulation of c-FLIPL and upregulation of DR5 were blocked by LY treatment. These results indicate that ATF4/CHOP-induced autophagy induction might be associated with degradation/downregulation of c-FLIP and CD44. As shown in Fig. 4C, SNU-354 cells also demonstrated that treatment of CCB or DMC resulted in a significant increase in LC3-II and decrease in p62 level and concurrent downregulation of c-FLIPL/S, and upregulation of DR5 in a dose-dependent manner, which were prevented by LY treatment, indicating that NSAID positively regulates autophagy and TRAIL activity in HCC cells. Since autophagy-mediated c-FLIP degradation in lung adenocarcinoma A549 cells has been reported (23), we examined the CCB-mediated c-FLIPL degradation in HCC cells in the presence of protein synthesis inhibitor cycloheximide (CHX) after treatment with CCB (Fig. 4D). When the half-life of c-FLIPL protein was determined in SNU-475/TR cells treated with or without CCB by performing a CHX chase assay, degradation of c-FLIP protein in CCB-treated cells was accelerated in the presence of CHX compared with CCB-untreated cells. Similarly, the half-life of c-FLIPL protein in SNU-449 cells was also significantly reduced by CCB treatment. These results suggest that CCB causes reduction of c-FLIP level in TRAIL-resistant HCC cells possibly through autophagic degradation.
Figure 4.
Effect of NSAID on expression of CHOP/DR5 and c-FLIP and autophagic degradation of c-FLIP in HCC cells. (A) SNU-475/TR cells were treated with serial doses of CCB or DMC for 24 h. (B) SNU-475/TR cells were pretreated with 5 µM chloroquine (CQ), 10 mM 3-methyladenine (3-MA) or 5 µM LY294002 (LY) for 3 h followed by treatment of 40 µM CCB for 24 h. (C) SNU-354 cells were treated with serial doses of CCB or DMC for 24 h or 5 µM LY for 3 h followed with treatment of 40 µM CCB for 24 h. Subsequently, altered levels of the indicted molecules in these cells were determined by western blot analysis. (D) SNU-475/TR and SNU-449 cells were treated with or without 50 µM CCB for 24 h, and were collected at 0, 3 and 6 h following treatment with 20 µg/ml cycloheximide (CHX) and the level of c-FLIPL was determined by western blot analysis. TRAIL, TNF-related apoptosis inducing ligand; NSAID, non-steroidal anti-inflammatory drug; CHOP, C/EBP homologous protein; DR5, death receptor TRAIL-R2; c-FLIP, cellular-FLICE inhibitory protein; HCC, hepatocellular carcinoma; CCB, celecoxib; DMC, 2,5-dimethyl celecoxib.
NSAID reduces CSC marker CD44 surface expression and accelerates TRAIL-mediated downregulation of c-FLIP and activation of caspases and PARP
Since TRAIL-resistance phenotype of SNU-475/TR cells was closely linked to high expression of CD44, we determined whether CCB could modulate surface expression of CD44 by FACS analysis (Fig. 5A). SNU-475/TR cells expressing a high level of CD44 showed that CCB significantly reduced CD44 surface expression when compared to SNU-475 cells. Similarly, inhibition of CD44 surface expression by DMC treatment occurred in SNU-475/TR and SNU-449 cells. These results demonstrated that treatment of HCC cells with DMC as well as CCB induced reduction of CD44 surface expression and suggest that NSAID could suppress the expression of CD44, one of the major causes of TRAIL resistance, and overcome TRAIL resistance in HCC cells. We next determined the effect of TRAIL and NSAID, singly and in combination, on apoptosis induction in HCC cells. SNU-449 cells showed an increase in the induction of apoptosis after co-treatment with TRAIL and CCB (or DMC) compared to the individual drug treatments (Fig. 5B). The increase in apoptosis after the combination treatment of TRAIL and NSAID was observed in other HCC cells (data not shown). We also investigated the effects of NSAID on TRAIL-mediated c-FLIP expression and caspase activity in various HCC cells. We found that NSAID accelerated TRAIL-mediated c-FLIP downregulation, and subsequent caspase and PARP activation in HCC cells (Fig. 5C). CCB or DMC promoted TRAIL-mediated c-FLIPL/S downregulation and subsequent activation of caspase-8 and −3 and PARP cleavage in SNU-449 cells, indicating acceleration of TRAIL-mediated cell death by NSAID. Similarly, TRAIL-mediated c-FLIPL downregulation and caspase-8 and −3 and PARP activation were significantly accelerated by combined treatment of TRAIL and CCB in SNU-423 and SNU-354 cells compared to the results of TRAIL treatment alone. Our data suggest that sensitization of TRAIL-resistant HCC cells to TRAIL by NSAID is partially caused by degradation/downregulation of c-FLIP and CD44 via ATF4/CHOP-induced autophagy.
Figure 5.
Acceleration of TRAIL-mediated caspase and PARP activities by CCB in HCC cells through inhibition of CD44 surface expression and downregulation of c-FLIP. (A) SNU-475, SNU-475/TR and SNU-449 cells treated with 40 µM CCB or DMC for 24 h were stained with anti-CD44 antibody to determine the surface expression of CD44 by a flow cytometer. (B) Apoptosis assay. SNU-449 cells treated with TRAIL (30 ng/ml) in the presence or absence of NSAID (20 µM CCB or DMC). Annexin V/PI double staining by flow cytometry (upper image). Statistical analysis of the cell apoptosis assay (lower histogram). The number of apoptotic cells is the sum of Q2 and Q4. Data are mean ± SD, n=3. *P<0.01, **P<0.001 vs. the control; &P<0.001 vs. TRAIL alone; #P<0.001 vs. CCB or DMC alone, respectively. (C) SNU-449, SNU-354 and SNU-423 cells treated with the indicated doses of TRAIL (ng/ml) in the presence or absence of NSAID (20 µM CCB or DMC) for 24 h. The levels of c-FLIP and activity of caspase and PARP in these cells were determined by western blot analysis. Cl., cleaved; TRAIL, TNF-related apoptosis inducing ligand; PARP, poly(ADP-ribose) polymerase; CCB, celecoxib; DMC, 2,5-dimethyl celecoxib; HCC, hepatocellular carcinoma; NSAID, non-steroidal anti-inflammatory drug; c-FLIP, cellular-FLICE inhibitory protein.
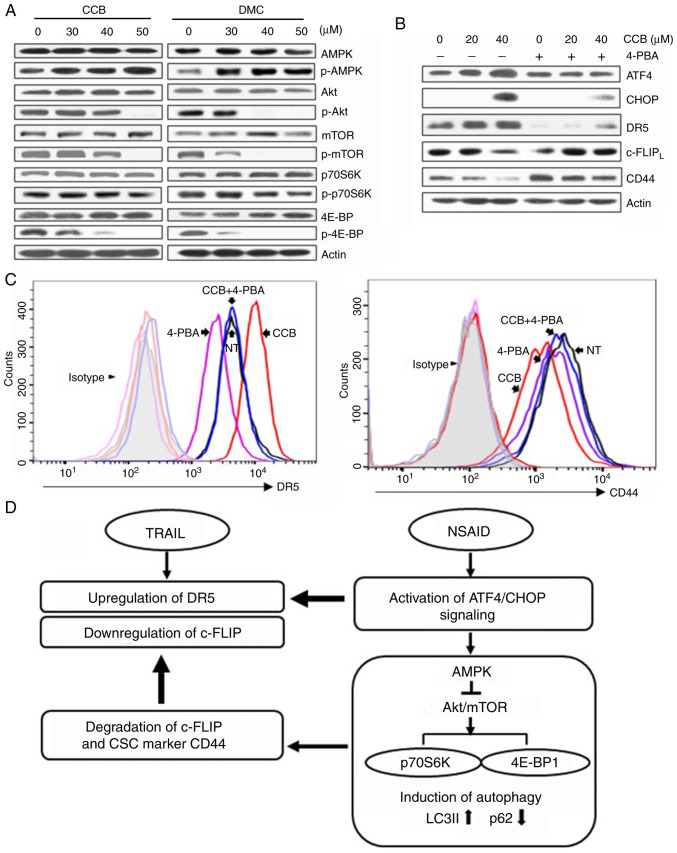
NSAID activates AMPK but inhibits Akt/mTOR signaling pathway through ER-mediated ATF4/CHOP pathway
It has been reported that autophagy is promoted by AMPK and inhibited by mTOR, and mTOR phosphorylates downstream targets such as ribosomal protein S6 kinase (p70S6K) and eukaryotic initiation factor 4E-binding protein (4E-BP1) (28). We investigated both total and phosphorylated forms of AMPK and key proteins of Akt/mTOR signaling pathway. Treatment of SNU-475/TR cells with CCB (or DMC) resulted in an increase in phosphorylated AMPK (p-AMPK) and a decrease in phosphorylated Akt (p-Akt), mTOR (p-mTOR), p70S6K (p-p70S6K) and 4E-BP1 (p-4E-BP1) without affecting total protein levels of them, indicating that activation of AMPK phosphorylation and inhibition of Akt/mTOR/p70S6K/4E-BP1 phosphorylation by CCB participates in the event of CCB-induced autophagy (Fig. 6A). These results indicate the possibility that autophagic response triggered by NSAID via AMPK activation and inhibition of Akt/mTOR signaling may be linked to activation of ATF/CHOP signaling in HCC cells. Since ATF4/CHOP is the key signal for autophagy induced by ER stress, and the above data showed that CCB-induced up-regulation of DR5 and downregulation of c-FLIP and CD44 occurred through ATF4/CHOP pathway, we determined whether ER stress inhibitor 4-phenylbutyric acid (4-PBA) could modulate the CCB-induced expression of ATF4/CHOP, DR5, c-FLIP and CD44 in SNU-475/TR cells (Fig. 6B). Our data showed that blocking ER stress with 4-PBA suppressed CCB-induced upregulation of ATF4 and CHOP and subsequent upregulation of DR5 and also blocked downregulation of c-FLIP and CD44. We further determined whether treatment of 4-PBA could block CCB-induced surface expression of DR5 and CD44 by FACS analysis (Fig. 6C). In SNU-475/TR cells, CCB-mediated increase of DR5 and decrease of CD44 surface expression was significantly attenuated by 4-PBA. These results indicate that CCB triggers activation of ER stress-dependent autophagy, leading to promote upregulation of DR5 and degradation/downregulation of c-FLIP and CD44.
Figure 6.
Activation of AMPK and inhibition of Akt/mTOR signaling by NSAID and inhibition of NSAID-activated ATF4/CHOP pathway by ER stress inhibitor. (A) SNU-475/TR cells were treated with the indicated doses of CCB or DMC for 24 h or (B) the cells were pretreated with or without 1.5 mM 4-phenylbutyric acid (4-PBA) for 24 h before treatment of serial doses of CCB for 24 h, and the altered levels of indicated molecules were determined by western blot analysis. (C) SNU-475/TR cells were additionally treated with CCB (40 µM for 24 h) after pretreatment of 4-PBA (1.5 mM for 24 h) and stained with anti-DR5 or anti-CD44 antibody to determine the cell surface expression of DR5 or CD44 by a flow cytometer. (D) Proposed scheme showing how NSAID enhances TRAIL activity. NSAID induces ER stress via activation of ATF4/CHOP signaling, which upregulates TRAIL-mediated DR5 expression and also induces autophagy via AMPK activation and inhibition of Akt/mTOR signaling. NSAID-induced autophagy causes degradation of c-FLIP/CD44 that can accelerate TRAIL-mediated c-FLIP downregulation. NSAID, non-steroidal anti-inflammatory drug; ATF, activating transcription factor; ER, endoplasmic reticulum; CCB, celecoxib; DMC, 2,5-dimethyl celecoxib; DR5, death receptor TRAIL-R2; CHOP, C/EBP homologous protein; TRAIL, TNF-related apoptosis inducing ligand; CSC, cancer stem cell; LC3, microtubule-associated protein light chain 3.
In summary, we propose that the autophagic response triggered by NSAID could be attributed to activation of ATF/CHOP signaling, which leads to upregulation of TRAIL-mediated DR5 expression, and also NSAID-induced autophagy via AMPK activation and inhibition of Akt/mTOR signaling. This consequently leads to increased LC3B-II and decreased p62 levels resulting in degradation of c-FLIP and TRAIL resistance-related CSC marker CD44, which accelerates TRAIL-mediated c-FLIP downregulation, and reduces CSC marker protein in TRAIL-resistant HCC cells (Fig. 6D).
Discussion
Resistance to TNF-related apoptosis inducing ligand (TRAIL) remains a major issue in the treatment of cancer, and overcoming resistance to TRAIL is desirable for TRAIL-based therapy. In the present study, to sensitize TRAIL-resistant hepatocellular carcinoma (HCC) cells to TRAIL, we examined the cytotoxic effect of TRAIL alone and in combination with a non-steroidal anti-inflammatory drug (NSAID) such as celecoxib (CCB) or 2,5-dimethyl celecoxib (DMC), a non-cyclooxygenase (COX)-2 inhibitor analog of CCB in TRAIL-resistant HCC cells involving SNU-475/TR, SNU-423 and SNU-449 cells with high levels of CD44. We propose NSAID as a new sensitizer for TRAIL in TRAIL-resistant HCC cells. NSAID significantly potentiated the sensitivity of TRAIL-resistant HCC cells to TRAIL and thus could overcome TRAIL resistance in a COX-2-independent manner.
In addition to the anti-inflammatory effects, some NSAIDs including CCB are effective in the treatment and prevention of cancer, and the anticancer effects of NSAIDs include their ability to induce apoptosis (29). Although CCB-mediated apoptosis has not been fully elucidated, it appears to be associated with induction of endoplasmic reticulum (ER) stress involving upregulation of CHOP and downregulation of the anti-apoptotic protein survivin (30). It has been reported that several signaling pathways are involved in the complex cross-talk between apoptosis and autophagy, and autophagy has been closely linked to ER stress/unfolded protein response pathway (31). ATF4 and CHOP have been shown to play a significant role in the regulation of autophagy (32). Both CHOP and ATF4 are increased upon treatment with CCB or other agents, and CCB induced CHOP and DR5 expression through an ATF4-dependent mechanism. ATF4 can directly upregulate CHOP expression through binding to the the promoter region of CHOP, and subsequently CHOP upregulates a number of autophagy genes (33).
Autophagy has been known to affect the apoptotic process, serving either a pro-survival or pro-death function (10). Indeed, TRAIL has been shown to induce autophagy as well as apoptosis in cancer cell lines. TRAIL induces autophagy in lung adenocarcinoma cells and activates a Fas-associated death domain pathway mediated by both autophagy and apoptosis (34). In addition, gefitinib, ginsenoside and compound K were also found to increase DR5 expression through induction of autophagy, and inhibition of autophagy reduced DR5 expression in human colon cancer cells (11,35). We showed that CCB and DMC increased ATF4 and subsequent CHOP expression in parallel with upregulation of DR5 in TRAIL-resistant HCC cells. Treatment of SNU-475/TR cells with NSAID resulted in a dose-dependent increase in LC3-II and decrease in p62 levels, leading to autophagic degradation/downregulation of c-FLIP and CD44 through activation of the ATF/CHOP pathway, which contributes to enhance TRAIL-mediated apoptosis in TRAIL-resistant HCC cells. Insight into the complex network of TRAIL-induced apoptosis and autophagy contributes to the development of novel chemosensitizers to reverse TRAIL resistance in HCC cells. Our data showed that treatment of HCC cells with NSAID resulted in the increased expression of DR5 and concurrent decreased level of c-FLIP and CD44, major causes of TRAIL resistance, leading to accelerate TRAIL-induced cell death. Therefore, NSAIDs may represent promising chemosensitizers with which to reverse TRAIL resistance in CD44-overexpressing TRAIL-resistant HCC cells by inducing apoptotic and autophagic cell death to enhance TRAIL sensitivity. We previously reported that NSAIDs exert their autophagy-inducing effect through activation of AMPK and inhibition of the Akt/mTOR/p70S6K/4EBP signaling axis in CD44high K562 cells (36). Similarly, we showed that autophagic responses triggered by CCB could be attributed to activation of the ATF/CHOP signaling axis, which leads to induction of autophagy in TRAIL-resistant HCC cells via AMPK activation and inhibition of Akt/mTOR/p70S6K/4EBP1 signaling pathway.
Since cancer stem cells (CSCs), which are resistant to anticancer therapy, are usually characterized by a dysregulation of autophagy, it is crucial to investigate their relationship (37). Notably, nigericin induced autophagy and subsequently suppressed CSC properties in glioma cells (38), and induction of autophagy by rottlerin was found ot lead to apoptosis in breast CSCs (39). These reports indicate a possibility that induction of autophagy could finally suppress stemness and lead to cell death in CSCs, and thus, autophagy is a potential target to decrease the resistance of CSCs to anticancer therapy. The CSC population in HCC can be responsible for metastasis, recurrence and chemoresistance of HCC (40). In HCC, markers most frequently used for CSC are CD44 and CD133 (18), and particularly CD44 expression was reported to be correlated with high HCC histologic grades, vascular invasion, and reduced survival outcomes (41). Another potential CSC marker is CD133 that is released from the plasma membrane to the cytoplasm, and overexpression of CD133 constitutively activates autophagy via inhibition of mTORC1 and mTORC2 signaling and potentiation of autophagy flux (42). In the present study, we revealed a pivotal role of CD44/CD133 expression for HCC resistance towards TRAIL. CD133−/CD44high SNU475/TR cells are significantly resistant to TRAIL compared with CD133high/CD44− SNU475 cells. In SNU-475/TR cells, the expression of CD44 on the cell surface was significantly enhanced while the CD133 level was markedly reduced as compared to those in the parental TRAIL-sensitive SNU-475 cells. Other CD133−/CD44high HCC cells such as SNU-423 and SNU-449 cells were also resistant to TRAIL, indicating that upregulation of CD44 may be associated with acquisition of TRAIL resistance in HCC cells, and thus a high ratio of CD44/CD133 relative expression is an important marker for TRAIL resistance.
It has been reported that CD44 protein degradation in human breast and pancreatic cancer cells can occur through the lysosomal/autophagic pathway (43). In addition, CD44 degradation in head and neck squamous cell carcinoma cells was blocked by autophagy inhibitor CQ not by pretreatment with the proteasome inhibitor MG132 (27). We also showed that CCB inhibited cell surface expression of CD44, and pre-treatment of autophagy inhibitor 3-MA or CQ blocked CCB-induced reduction of CD44 level in SNU-475/TR cells, indicating CCB-induced autophagic degradation of CD44 in HCC cells. In addition, the expression of stemness-related makers ALDH1 and Nanog as well as CD44 in SNU-475/TR cells was downregulated by CCB. Indeed, ALDH1high cells were found to demonstrate higher resistance to common chemotherapeutic reagents, and ALDH1A3 protein, one of the ALDH1 family members, was downregulated by autophagy (44), and the expression of Nanog as well as CD44 in gastric cancer stem cells was also decreased by ibuprofen (45), indicating the possibility of NSAID-induced autophagic degradation of CSC marker proteins CD44, ALDH1 and Nanog in SNU-475/TR cells. Recently, most studies have focused on CSCs due to their abilities to cause tumorigenicity, drug resistance, and cancer recurrence. NSAIDs are among the most commonly used medications worldwide and inexpensive. Here, we showed the effectiveness of NSAIDs on TRAIL-resistant HCC cells overexpressing CSC markers to perform effective cancer therapy. The combination of NSAID and TRAIL resulted in a greater cytotoxicity than TRAIL alone by NSAID-induced acceleration of TRAIL-mediated c-FLIP downregulation and DR5 activation via ER stress ATF4/CHOP-mediated autophagic cell death pathway, leading to subsequent activation of caspases and PARP and finally NSAID-mediated potentiation of TRAIL-induced cell death in TRAIL-resistant HCC cells. Taken together, our findings highlight a novel mechanism underlying the combination effect of NSAID and TRAIL, and thus NSAID can be considered as a novel chemosensitizer of TRAIL for effective treatment of HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Pusan National University Research Grant, 2019.
Availability of data and materials
The datasets used during the present study are available from the corresponding authors upon reasonable request.
Authors' contributions
SHK and CDK conceived and designed the study. SHL, HJM and YSL performed the experiments and the statistical analysis and participated in the writing of the manuscript. YSL participated in the revision of the manuscript critically for important intellectual content. All authors approved the manuscript and agree to be accountable for all aspects of the work in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019;460:1–9. doi: 10.1016/j.canlet.2019.114428. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W, Wu DB, Fu SY, Chen EQ, Tang H, Zhou TY. Insight into the role of TRAIL in liver diseases. Biomed Pharmacother. 2019;110:641–645. doi: 10.1016/j.biopha.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Trivedi R, Mishra DP. Trailing TRAIL resistance: Novel targets for TRAIL sensitization in cancer cells. Front Oncol. 2015;5:69. doi: 10.3389/fonc.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Yang X, Xu T, Kong Q, Zhang Y, Shen Y, Wei Y, Wang G, Chang KJ. Overcoming resistance to TRAIL-induced apoptosis in solid tumor cells by simultaneously targeting death receptors, c-FLIP and IAPs. Int J Oncol. 2016;49:153–163. doi: 10.3892/ijo.2016.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koehler BC, Urbanik T, Vick B, Boger RJ, Heeger S, Galle PR, Schuchmann M, Schulze-Bergkamen H. TRAIL-induced apoptosis of hepatocellular carcinoma cells is augmented by targeted therapies. World J Gastroenterol. 2009;15:5924–5935. doi: 10.3748/wjg.15.5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Seo SU, Min KJ, Woo SM, Nam JO, Kubatka P, Kim S, Park JW, Kwon TK. Garcinol enhances TRAIL-induced apoptotic cell death through up-regulation of DR5 and down-regulation of c-FLIP expression. Molecules. 2018;23:1614. doi: 10.3390/molecules23071614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeon MY, Min KJ, Woo SM, Seo SU, Choi YH, Kim SH, Kim DE, Lee TJ, Kim S, Park JW, Kwon TK. Maritoclax enhances TRAIL-induced apoptosis via CHOP-mediated ipregulation of DR5 and miR-708-mediated downregulation of cFLIP. Molecules. 2018;23:3030–3043. doi: 10.3390/molecules23113030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubillos-Ruiz JR, Bettigole SE, Glimcher LH. Tumorigenic and Immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noh HJ, Lee SJ, Sung EG, Song IH, Kim JY, Woo CH, Kwon TK, Lee TJ. CHOP down-regulates cFLIP(L) expression by promoting ubiquitin/proteasome-mediated cFLIP(L) degradation. J Cell Biochem. 2012;113:3692–3700. doi: 10.1002/jcb.24242. [DOI] [PubMed] [Google Scholar]
- 10.Sheng J, Qin H, Zhang K, Li B, Zhang X. Targeting autophagy in chemotherapy-resistant of hepatocellular carcinoma. Am J Cancer Res. 2018;8:354–365. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Meng Y, Sun Q, Zhang Z, Guo X, Sheng X, Tai G, Cheng H, Zhou Y. Ginsenoside compound K sensitizes human colon cancer cells to TRAIL-induced apoptosis via autophagy-dependent and -independent DR5 upregulation. Cell Death Dis. 2016;7:e2334. doi: 10.1038/cddis.2016.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Xu X, Shi S, Wang Q, Saxton B, He W, Gou X, Jang JH, Nyunoya T, Wang X, et al. Autophagy-mediated degradation of IAPs and c-FLIP(L) potentiates apoptosis induced by combination of TRAIL and Chal-24. J Cell Biochem. 2016;117:1136–1144. doi: 10.1002/jcb.25397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy A, Le J, Sausville E, Emadi A. Autophagy modulation: A target for cancer treatment development. Cancer Chemother Pharmacol. 2015;75:439–447. doi: 10.1007/s00280-014-2637-z. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Chen F, Shang L. Advances in antitumor effects of NSAIDs. Cancer Manag Res. 2018;10:4631–4640. doi: 10.2147/CMAR.S175212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endo H, Yano M, Okumura Y, Kido H. Ibuprofen enhances the anticancer activity of cisplatin in lung cancer cells by inhibiting the heat shock protein 70. Cell Death Dis. 2014;5:e1027. doi: 10.1038/cddis.2013.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu C, Li WB, Liu JB, Lu JW, Feng JF. Autophagy: Novel applications of nonsteroidal anti-inflammatory drugs for primary cancer. Cancer Med. 2018;7:471–484. doi: 10.1002/cam4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku JL, Park JG. Biology of SNU cell lines. Cancer Res Treat. 2005;37:1–19. doi: 10.4143/crt.2005.37.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozeik MS, Hammam OA, Ali AI, Magdy M, Khalil H, Anas A, Abo El Hassan AA, Rahim AA, El-Shabasy AI. Evaluation of CD44 and CD133 as markers of liver cancer stem cells in Egyptian patients with HCV-induced chronic liver diseases versus hepatocellular carcinoma. Electron Physician. 2017;9:4708–4717. doi: 10.19082/4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SH, Hyun SK, Kim HB, Kang CD, Kim SH. Potential role of CD133 expression in the susceptibility of human liver cancer stem-like cells to TRAIL. Oncol Res. 2016;24:495–509. doi: 10.3727/096504016X14685034103950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennarun B, Kleibeuker JH, Boersma-van Ek W, Kruyt FA, Hollema H, de Vries EG, de Jong S. Targeting FLIP and Mcl-1 using a combination of aspirin and sorafenib sensitizes colon cancer cells to TRAIL. J Pathol. 2013;229:410–421. doi: 10.1002/path.4138. [DOI] [PubMed] [Google Scholar]
- 21.Lee JY, Jung KH, Morgan MJ, Kang YR, Lee HS, Koo GB, Hong SS, Kwon SW, Kim YS. Sensitization of TRAIL-induced cell death by 20(S)-ginsenoside Rg3 via CHOP-mediated DR5 upregulation in human hepatocellular carcinoma cells. Mol Cancer Ther. 2013;12:274–285. doi: 10.1158/1535-7163.MCT-12-0054. [DOI] [PubMed] [Google Scholar]
- 22.Hassanzadeh A, Farshdousti Hagh M, Alivand MR, Akbari AAM, Shams Asenjan K, Saraei R, Solali S. Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL. J Cell Physiol. 2018;233:6470–6485. doi: 10.1002/jcp.26585. [DOI] [PubMed] [Google Scholar]
- 23.Nazim UM, Jeong JK, Park SY. Ophiopogonin B sensitizes TRAIL-induced apoptosis through activation of autophagy flux and downregulates cellular FLICE-like inhibitory protein. Oncotarget. 2017;9:4161–4172. doi: 10.18632/oncotarget.23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He MX, He YW. c-FLIP protects T lymphocytes from apoptosis in the intrinsic pathway. J Immunol. 2015;194:3444–3451. doi: 10.4049/jimmunol.1400469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang P, Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Su M, Mei Y, Sinha S. Role of the Crosstalk between autophagy and apoptosis in Cancer. J Oncol. 2013;2013:102735. doi: 10.1155/2013/102735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanbu T, Umemura N, Ohkoshi E, Nanbu K, Sakagami H, Shimada J. Combined SN-38 and gefitinib treatment promotes CD44 degradation in head and neck squamous cell carcinoma cells. Oncol Rep. 2018;39:367–375. doi: 10.3892/or.2017.6105. [DOI] [PubMed] [Google Scholar]
- 28.Matter MS, Decaens T, Andersen JB, Thorgeirsson SS. Targeting the mTOR pathway in hepatocellular carcinoma: Current state and future trends. J Hepatol. 2014;60:855–865. doi: 10.1016/j.jhep.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umar A, Steele VE, Menter DG, Hawk ET. Mechanisms of nonsteroidal anti-inflammatory drugs in cancer prevention. Semin Oncol. 2016;43:65–77. doi: 10.1053/j.seminoncol.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Jendrossek V. Targeting apoptosis pathways by Celecoxib in cancer. Cancer Lett. 2013;332:313–324. doi: 10.1016/j.canlet.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Song S, Tan J, Miao Y, Li M, Zhang Q. Crosstalk of autophagy and apoptosis: Involvement of the dual role of autophagy under ER stress. J Cell Physiol. 2017;232:2977–2984. doi: 10.1002/jcp.25785. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto H, Miyazaki S, Matsuyama S, Takeda M, Kawano M, Nakagawa H, Nishimura K, Matsuo S. Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open. 2013;2:1084–1090. doi: 10.1242/bio.20135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.B'chir W, Chaveroux C, Carraro V, Averous J, Maurin AC, Jousse C, Muranishi Y, Parry L, Fafournoux P, Bruhat A. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal. 2014;26:1385–1391. doi: 10.1016/j.cellsig.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y, Zhou X, Qiao J, Bao A. Autophagy is a regulator of TRAIL-induced apoptosis in NSCLC A549 cells. J Cell Commun Signal. 2017;11:219–226. doi: 10.1007/s12079-016-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Meng Y, Guo X, Sheng X, Tai G, Zhang F, Cheng H, Zhou Y. Gefitinib enhances human colon cancer cells to TRAIL-induced apoptosis of via autophagy- and JNK-mediated death receptors upregulation. Apoptosis. 2016;21:1291–1301. doi: 10.1007/s10495-016-1287-5. [DOI] [PubMed] [Google Scholar]
- 36.Moon HJ, Park SY, Lee SH, Kang CD, Kim SH. Nonsteroidal anti-inflammatory drugs sensitize CD44-overexpressing cancer cells to Hsp90 inhibitor through autophagy activation. Oncol Res. 2019;27:835–847. doi: 10.3727/096504019X15517850319579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F. Autophagy and cancer stem cells: Molecular mechanisms and therapeutic applications. Cell Death Differ. 2019;26:690–702. doi: 10.1038/s41418-019-0292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegazy AM, Yamada D, Kobayashi M, Kohno S, Ueno M, Ali MA, Ohta K, Tadokoro Y, Ino Y, Todo T, et al. Therapeutic strategy for targeting aggressive malignant gliomas by disrupting their energy balance. J Biol Chem. 2016;291:21496–21509. doi: 10.1074/jbc.M116.734756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar D, Shankar S, Srivastava RK. Rottlerin-induced autophagy leads to the apoptosis in breast cancer stem cells: Molecular mechanisms. Mol Cancer. 2013;12:171. doi: 10.1186/1476-4598-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji J, Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 2012;39:461–472. doi: 10.1053/j.seminoncol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Y, Tan Y. Prognostic value of CD44 expression in patients with hepatocellular carcinoma: Meta-analysis. Cancer Cell Int. 2016;16:47. doi: 10.1186/s12935-016-0325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharya S, Yin J, Winborn CS, Zhang Q, Yue J, Chaum E. Prominin-1 is a novel regulator of autophagy in the human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2017;58:2366–2387. doi: 10.1167/iovs.16-21162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haakenson JK, Khokhlatchev AV, Choi YJ, Linton SS, Zhang P, Zaki PM, Fu C, Cooper TK, Manni A, Zhu J, et al. Lysosomal degradation of CD44 mediates ceramide nanoliposome-induced anoikis and diminished extravasation in metastatic carcinoma cells. J Biol Chem. 2015;290:8632–8643. doi: 10.1074/jbc.M114.609677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu W, Schecker J, Würstle S, Schneider F, Schönfelder M, Schlegel J. Aldehyde dehydrogenase 1A3 (ALDH1A3) is regulated by autophagy in human glioblastoma cells. Cancer Lett. 2018;417:112–123. doi: 10.1016/j.canlet.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 45.Akrami H, Moradi B, Borzabadi Farahani D, Mehdizadeh K. Ibuprofen reduces cell proliferation through inhibiting Wnt/β catenin signaling pathway in gastric cancer stem cells. Cell Biol Int. 2018;42:949–958. doi: 10.1002/cbin.10959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding authors upon reasonable request.