Abstract
The world is currently facing a serious health burden of waterborne diseases, including diarrhea, gastrointestinal diseases, and systemic illnesses. The control of these infectious diseases ultimately depends on the access to safe drinking water, properly managed sanitation, and hygiene practices. Therefore, ultrasensitive, rapid, and specific monitoring platforms for bacterial pathogens in ambient waters at the point of sample collection are urgently needed. We conducted a literature review on state-of-the-art research of rapid in-field aquatic bacteria detection methods, including cell-based methods, nucleic acid amplification detection methods, and biosensors. The detection performance, the advantages, and the disadvantages of the technologies are critically discussed. We envision that promising monitoring approaches should be automated, real-time, and target-multiplexed, thus allowing comprehensive evaluation of exposure risks attributable to waterborne pathogens and even emerging microbial contaminants such as antibiotic resistance genes, which leads to better protection of public health.
Keywords: waterborne pathogens, exposure risk assessment, detection methods, rapidity, portability
Access to adequate water, sanitation, and hygiene (WASH) has long been a significant public health concern and an international development policy. According to the World Health Organization, global mortality attributable to waterborne diseases is estimated to be > 2.2 million per year, among which about 1.4 million are children, resulting in nearly $12 billion per year of economic loss worldwide [1]. It is estimated that diarrhea alone amounts to 842 000 deaths per year due to unsafe WASH and includes 361 000 deaths of children < 5 years of age, mostly in low-income countries [2]. Ultrasensitive, rapid, and specific monitoring platforms for bacterial pathogens in ambient waters at the point of sample collection are essential for timely water quality surveillance and microbial risk assessment. Therefore, the development of such platforms plays a key role in predicting and assessing the risk of disease outbreaks and providing quality care in healthcare settings such as improving the effectiveness of vaccine distribution.
Microbial detection techniques are usually classified into phenotypic methods and molecular methods. Culture-based methods as the mainstream of phenotyping have the advantages of cost-effectiveness and simplicity, and remain the gold standard for bacterial monitoring and identification. However, it requires days for culture-based methods to provide conclusive results, which greatly hampers their applications in water quality monitoring [3]. Molecular analyses including conventional polymerase chain reaction (PCR)–based methods, immunology-based methods, etc, however, require lengthy processes of sample pretreatment (eg, concentration, cell lysis, purification), expensive equipment, and trained personnel in centralized laboratory facilities. The demanding requirements of molecular methods represent a major disadvantage for their application in resource-limited communities [4, 5]. In addition, the majority of currently available molecular techniques have low precision (~20%) and are poorly suited for absolute quantification, thus having limited application in low-concentration pathogen detection [6]. To tackle this problem, in addition to enhancing specificity and mitigating competitive side reactions, researchers have also been exploring the “digital detection” concept. It realizes absolute quantification through separating the sample into sufficient partitions followed by individual molecular reaction and endpoint counting of positive and negative signals in each reaction [6, 7]. In addition, biosensor is also a promising technique for future waterborne pathogen monitoring systems. Biosensor generally provides more reliable results from real-time measurements and allows rapid analysis without the requirement of complicated pretreatment steps such as the target enrichment process, which still has a lot of room to be developed [8–10].
Overall, microbial pathogen detection is urged to be ultrasensitive, rapid, simple, low-cost, field-deployable, and easily operable by undertrained individuals for applications in environmental surveillance. Over the past years, numerous research advances have been made in such integrated platform for detection and identification of bacterial pathogens including but not limited to Salmonella enterica serovar Typhi (S. typhi) in water. Here, we review representative technologies categorized into cell-based methods, nucleic acid amplification methods, and biosensors. We also further discuss the needs of future developments on microbial monitoring platforms in the underdeveloped parts of the world.
CELL-BASED DETECTION METHODS
Compared to molecular-based detection platforms that target specific nucleic acids or proteins, cell-based detection methods offer direct identification and measurement with relative simple workflows [11]. Utilization of commercial instruments simplifies the construction of cell-based detection platforms. Recent development of miniaturized analysis systems has further promoted the efficiency and portability of cell-based detection methods, thus enabling complex diagnostics or monitoring procedures.
Miniaturized cell cultivation techniques based on microfluidic devices and Lab-on-a-Chip technologies consume less fluid, take less volume, and usually have higher tolerance toward ambient conditions, thus reducing the total cost and time for bacterial analysis [12]. One example of miniaturized cell cultivation is a palm-size device developed by Futai et al utilizing Braille display, monolithic surface, modified culture media and transparent heater [13]. This device was successfully used to culture highly carbon dioxide (CO2)–dependent cells in nonpreferable growing environment with limited CO2, humidity and a non-37°C temperature. Even for uncultivatable microbial species in various environments, an isolation chip with miniature diffusion chambers was developed to achieve parallel cultivation and isolation [14]. However, these miniaturized cell cultivation routines unavoidably take a long time, can usually be labor intensive, and require skilled operators.
Compared to cell culture, flow cytometers (FCMs) for direct cell counting enable fast quantification of the total bacterial community in the environment with high reproducibility and relatively small standard deviation. More importantly, many commercial FCMs are available for adaptations and the setup of FCM is suitable for automation, making FCM a great candidate for online routine bacterial monitoring [15]. Besmer et al used an automated in situ FCM analysis platform to help characterize the temporal variation of dynamic aquatic environments enabled by a commercial FCM (C6 flow cytometer, BD Accuri, San Jose California) coupled with a fully automated staining robot [16, 17]. Going one step further, Props et al combined the use of real-time FCM and advanced fingerprinting metrics, which aided the detection and characterization of microbial dynamic changes with a high temporal resolution of 10–30 seconds [18]. Nevertheless, FCM techniques have some major drawbacks, including difficulties in distinguishing between live and dead cells and specific strains of bacteria, and in discriminating bacterial aggregates and clusters. Incorporating microscopic imaging to FCM could boost the specificity of this detection platform. For example, an automatic imaging FCM was developed with a deep learning–based phase-recovery and holographic-reconstruction framework to generate pictures of micro-object in water samples without fluorescence triggering, and the pictures generated could be used for characterization [19]. However, current holograms taken by the microscopy and reconstructed images do not have a resolution high enough for specific bacterial pathogen characterization and thus further research is needed.
Besides miniature cell cultivation and FCM, other online cell-based sensing methods have also been developed. A real-time sensor using multiangle light scattering (MALS) technology was developed by Sherchan et al. By comparing the light scattering patterns after using a laser beam to strike particulates in water (including organic particles and microbial cells) with light scattering patterns in the computerized database, data obtained was characterized and the load of injected Escherichia coli was back-calculated [20]. Due to the existence of fluorophores in bacterial cells such as tryptophan, phenylalanine, or nucleic acids, which emit fluorescence light after excited by ultraviolet light, Simões and Dong developed an optical microfluidic sensor based on tryptophan intrinsic fluorescence with 3D-printing prototyping [21]. Furthermore, direct 3D image recognition for online pathogen detection was enabled by the combination of a sample-holding flow cell and a field imaging system (including a light source, a magnifying lens, and a camera). An image analysis system was developed to analyze 59 parameters of the images obtained and was able to distinguish between bacteria and abiotic particles. 3D image recognition analysis also provides quantification results, which correlates well with actual bacterial counts [22]. Tables 1 and 2 summarize specific detection parameters and comments on the application and detection parameters of the above-mentioned cell-based technologies.
Table 1.
Pathogen Detection Methods and Their Samples Studied
| Detection Method | Phenotypic or Genetic | Waterborne Microbial Agent Tested | Complex Sample Matrices Tested | Treated Volume, mL | ||
|---|---|---|---|---|---|---|
| A. Cell-based | A1. Isolation chip | Phenotypic | Total bacteria | Seawater and soil | NA | |
| A2. Online flow cytometry | Phenotypic | Total bacteria | Drinking water, river water, and groundwater | 0.015 | ||
| A3. Real-time flow cytometry | Phenotypic | Total bacteria | Nonchlorinated municipal drinking water, river water, and pond water | 0.016/min | ||
| A4. MALS sensor | Phenotypic | Escherichia coli | Distilled and tap water | 600 | ||
| A5. Optical microfluidic sensor based on tryptophan intrinsic fluorescence | Phenotypic | E. coli and Legionella | Distilled water | NA | ||
| A6. Novel optical sensor | Phenotypic | Total particles | Nonchlorinated water and water from cattle slaughterhouse | 200 | ||
| B. NAA | PCR-based | B1. Coaxial channel-based DNA extraction and microfluidic PCR | Genetic | E. coli | Milk | 10 |
| LAMP-based | B2. Self-contained microfluidic gLAMP | Genetic | Proteus hauseri | Serum | NA | |
| Vibrio parahaemolyticus | ||||||
| Salmonella subsp enterica | ||||||
| E. coli | ||||||
| B3. Centrifugal microfluidic automatic wireless endpoint LAMP | Genetic | E. coli | Chicken meat | NA | ||
| Salmonella spp | ||||||
| Vibrio cholerae | ||||||
| B4. One-step single-layer membrane for digital LAMP | Genetic | E. coli | Culture media | NA | ||
| Salmonella Typhi | ||||||
| Enterococcus faecalis | ||||||
| B5. Asymmetric double-layer membrane for digital LAMP | Genetic | E. coli | Unprocessed environmental water | 10 | ||
| Salmonella Typhi | ||||||
| B6. In-gel LAMP | Genetic | MS2 | Culture media | NA | ||
| C. Biosensor | C1. MOF-bacteriophage biosensor | Phenotypic | Staphylococcus aureus | Pastry cream | 0.6 | |
| C2. Impedimetric paper-based biosensor | Phenotypic | Cultures from sewage sludge | Synthetic wastewater | NA | ||
| C3. Immunomagnetic separation and colorimetric paper-based device | Phenotypic | Salmonella Typhimurium | Bird feces and whole milk | 1 | ||
| C4. Real-time amperometric immunoassay amplified by nanomaterial | Phenotypic | E. coli | Water | 0.2 | ||
| C5. Phage-mediated separation with quantitative PCR detection | Combined | E. coli O157:H7 | Agricultural water and city water | 1 | ||
| C6. Carbon nanotube multilayer biosensors and on-chip LAMP | Combined | E. coli O157:H7 | Juice and milk | 1 | ||
Abbreviations: LAMP, loop-mediated isothermal amplification; MALS, multiangle light scattering; MOF, metal-organic framework; NA, not available; NAA, nucleic acid analysis; PCR, polymerase chain reaction.
Table 2.
Pathogen Detection Methods and Their Technical Characteristics
| Detection Method | Limit of Detection | Recovery Efficiency, % | Dynamic Range | Time to Answer, h | Absolute or Relative Quantification | Trained Personnel Required | Tests at Species Level | Ready for Field Test | Reference |
|---|---|---|---|---|---|---|---|---|---|
| A1 | NA | Up to 50% | ~500 cells | 2 wk | Relative | Yes | No | No | [14] |
| A2 | 103 cells/mL−1 | NA | 103–106 cells/mL−1 | 0.25 | Absolute | No | No | Yes | [16, 17] |
| A3 | 103 cells/mL−1 | NA | ~103 cells/mL−1 | 0.25 | Absolute | No | No | Yes | [18] |
| A4 | 103 CFU/mL−1 | NA | 103–106 CFU/mL−1 | 0 | Relative | Yes | No | No | [20] |
| A5 | 1.4 × 103 CFU/mL−1 | NA | 7 × 105 to 1 × 104 CFU/mL−1 | 0 | Relative | Yes | No | Yes | [21] |
| A6 | 1.6 × 102 particles/mL−1 | NA | 1.6 × 102–5 × 106 particles/mL−1 | 10 | Relative | No | No | Yes | [22] |
| B1 | 12 CFU/mL−1 | 97.4–100.6 | NA | 1.5 | Relative | No | Yes | No | [23] |
| B2 | 3 copies/μL−1 | NA | 3–3000 copies/μL−1 | 1.2 | Relative | No | Yes | Yes | [24] |
| 3 copies/μL−1 | 3–3000 copies/μL−1 | ||||||||
| 2 copies/μL−1 | 2–2000 copies/μL−1 | ||||||||
| 3 copies/μL−1 | 3–3000 copies/μL−1 | ||||||||
| B3 | 3 × 10−5 ng/μL−1 or 2.7 × 104 CFU/mL−1 | NA | 3 × 10−5–3 × 100 ng/μL−1 | 1 | Relative | No | Yes | Yes | [25] |
| B4 | 11 copies/μL−1 | NA | 11–1.1 × 105 copies/μL−1 | 1 | Absolute | No | Yes | Yes | [26] |
| B5 | 0.3 cells/mL−1 | 99.9 | 0.3–10 000 cells/mL−1 | 1 | Absolute | No | Yes | Yes | [27] |
| 3 cells/mL−1 | NA | 3–10 000 cells/mL−1 | |||||||
| B6 | 0.7 PFU per reaction | NA | 1–1000 PFU per reaction | 0.5 | Absolute | No | Yes | Yes | [28] |
| C1 | 31 CFU/mL−1 | 96–104 | 40–4 × 108 CFU/mL−1 | 0.33 | Relative | No | Yes | Yes | [29] |
| C2 | 1.9 × 103 CFU/mL−1 | NA | 103–106 CFU/mL−1 | 0.75 | Relative | Yes | No | Yes | [30] |
| C3 | 102 CFU/mL−1 | 8.84–21.3 | NA | 1.5 | Relative | Yes | Yes | Yes | [31] |
| C4 | 50 CFU/mL−1 | NA | 50–107 CFU/mL−1 | 0.53 | Relative | No | Yes | Yes | [32] |
| C5 | 102 CFU/mL−1 | 45.4–80.2 | 102–106 CFU/mL−1 | 2 | Relative | Yes | Yes | Yes | [33] |
| C6 | 1 CFU/mL−1 | 101–112.1 | 5–105 CFU/mL−1 | 2 | Relative | Yes | Yes | Yes | [34] |
Abbreviations: CFU, colony-forming units; NA, not available; PFU, plaque-forming units.
Many methods mentioned in the section have been successfully implanted for days or even months with full automation, and can be constructed easily with commercial instruments. However, the sensitivity of these methods can be easily influenced by different environmental factors and the detection limit is relatively high. Moreover, it is challenging to identify specific pathogens solely based on cell-level analysis, not to mention their genetic information. Therefore, further molecular level detections are needed to secure higher sensitivity and specificity.
NUCLEIC ACID AMPLIFICATION DETECTION PLATFORMS
Compared to phenotyping methods, molecular methods typically based on the quantification and identification of specific genomic segments of the pathogen’s genomes allow rapid, highly specific, and more sensitive detection, which better fit the expectations of timely monitoring and effective surveillance of aquatic pathogens in a range of water environmental settings. In this section, advances in monitoring methods based on PCR and loop-mediated isothermal amplification (LAMP) are respectively discussed.
PCR-BASED METHODS
The major drawback of PCR-related methods usually lies in their long response time and limited portability, since they rely on fussy thermal cycling and require additional equipment to detect the amplification products [35]. Another drawback is that trained personnel with experimental skills are needed to perform the assays, thus making the PCR-based systems impractical in resource-limited settings [4, 5]. Therefore, there is an urgent demand for a mobile and automated PCR-based device to monitor water microbial quality. Microfluidics have been demonstrated to provide a higher surface-to-volume ratio and a higher rate of mass and heat transfer, thus offering better performance than conventional systems due to significantly reduced reaction time [36]. Zhang et al reported a microfluidic PCR system integrated with the sample pretreatment technique of coaxial channel-based DNA extraction that was able to detect E. coli in milk matrix [23]. Detailed information about this system can be found in Tables 1 and 2. Some companies have directly tackled the mobility issue of PCR systems by developing handheld PCR instruments as shown in Table 3. Nguyen et al investigated the feasibility of using the Biomeme handheld quantitative PCR (qPCR) system for rapid (< 50 minute) on-site detection and monitoring of Flavobacterium psychrophilum in filtered water samples [37]. The study showed a close match between the results of the Biomeme handheld qPCR system and those of traditional bench qPCR, highlighting the feasibility of field-based qPCR systems in rapidly detecting and timely monitoring bacterial pathogens in water.
Table 3.
Summary of Commercially Available Handheld Quantitative Polymerase Chain Reaction Systems
| Company | Item | Weight, kg | Footprint, cm2 |
|---|---|---|---|
| Chai | Open quantitative PCRa | 4 | 28.0 × 24.0 |
| Ubiquitome | Freedom 4b | Not available | 10.2 × 20.3 |
| Ubiquitome | Liberty 16c | 3.2 | 21.2 × 11.0 |
| Amplyus | miniPCRb | 0.45 | 12.7 × 5.1 |
| Biomeme | Franklinb | 0.91 | About the size of a soda can |
Abbreviation: PCR, polymerase chain reaction.
aProduct information is from https://www.chaibio.com/openqpcr.
bProduct information is from Reference 35.
cProduct information is from https://insights.ubiquitomebio.com/liberty16-personal-qpcr- machine.
LAMP-BASED METHODS
LAMP is one of the most commonly used isothermal amplification methods [23, 38] and has attracted the most attention due to its high specificity, high amounts of amplification product, and superior tolerance to inhibitors [39]. Moreover, LAMP can be carried out at a constant temperature, so that it does not require a thermal cycler, which simplifies the detection procedure and allows better portability compared to PCR-based methods. Chen et al introduced a self-contained microdevice to in-gel LAMP (gLAMP) for multiplexed pathogen detection in complex clinical samples such as serum [24]. Escherichia coli, Proteus hauseri, Vibrio parahemolyticus, and Salmonella subspecies were simultaneously detected with high selectivity and sensitivity, as shown in Tables 1 and 2. Another major merit of the detection system was that the microchip preloaded with agarose solution containing LAMP reagents could maintain activity for 30 days when stored at 4°C, allowing the long-term storage and transportation of LAMP reagents, which is essential for LAMP-based point-of-use applications [24]. Sayad et al developed a wireless automatic endpoint detection system using centrifugal microfluidics for food safety examination. Foodborne pathogenic bacteria including E. coli, Salmonella species, and Vibrio cholerae in chicken meat were successfully detected with the sample-to-response time of < 1 hour [25]. Moreover, since this system is performed in an entirely automated way with the help of Bluetooth wireless technology, it is accessible for field application in environmental water samples. However, for the methods described above, the adaptability to environmental water matrix rather than food or blood needs further investigation and validation; in addition, the above methods were semiquantitative and not suitable for absolute quantification. Hoffmann’s laboratory has done a lot of work on developing rapid microbial pathogen detection systems based on digital LAMP (dLAMP) for absolute quantification in environmental waters [26–28]. Lin et al demonstrated that 1-step LAMP can be successfully performed on single-layer commercial polycarbonate membrane to achieve absolute quantification of the genome DNA of E. coli, Enterococcus faecalis, and S. Typhi [26]. Lin et al further reported the development and validation of the simpler and more robust double-layer membrane for dLAMP of bacterial pathogens in complex environmental waters. Absolute quantification of E. coli and S. Typhi spiked in unprocessed pond water and seawater could be completed within 1 hour with the sensitivity down to single cell [27]. Huang et al developed a gLAMP system enabling absolute quantification of microbial pathogens in environmental waters within 30 minutes at a very low cost of $5 per test. Bacteria (E. coli and S. Typhi) and viruses (bacteriophage MS2) were immobilized with LAMP reagents in polyethylene glycol hydrogel matrix and were then amplified [28]. Although the authors demonstrated that the above system could also be used for absolute quantification of bacterial targets including E. coli and S. Typhi, relevant detection limits were not reported, which needs further validation. More detailed information about all of the above-mentioned LAMP-based systems can be found in Tables 1 and 2. In this emerging field, a range of rapid and easy-to-operate platforms have been developed for low-concentration pathogen detection. It has great potential for future application in point-of-sample detection in field upon proper modification of consumables such as reagents and microchips.
BIOSENSORS
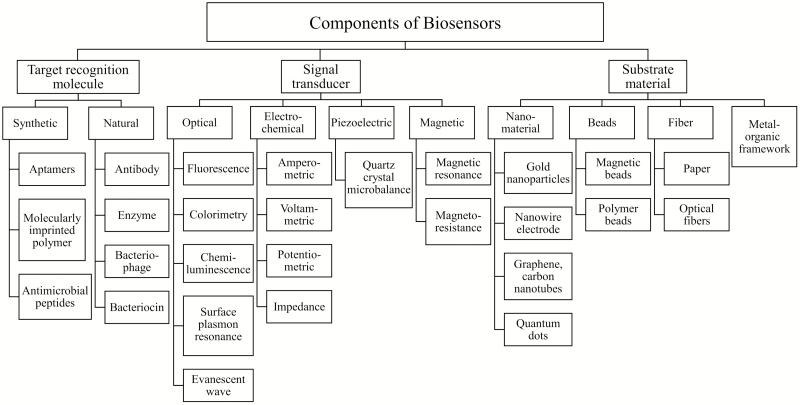
Biosensors are analytical devices that consist of target recognition molecules and signal transducers to detect the interaction between the recognition molecules and the specific target. Innovations in recognition molecules and signal transduction methods, as summarized in Figure 1, are emerging to achieve sensitive, rapid, and specific pathogen detection. We note that thorough reviews are available on various types of recognition molecules and signal transducers applicable to waterborne bacterial pathogen detection [40, 41]. Below we highlight novel biosensors that are portable for in-field applications or hold promise for online water quality monitoring.
Figure 1.
Recent developments in biosensors for bacterial pathogen detection. Widely used or innovative target recognition molecules, signal transducers, and substrate materials are summarized based on Justino et al [40], Kumar et al [41], and Vikesland and Wigginton [42].
The novel combination of target recognition molecules and new substrates for their immobilization has been demonstrated to boost the sensitivity of biosensors and the applicability in-field applicability. For example, Bhardwaj et al conjugated bacteriophage onto metal-organic framework (MOF) for specific quantification of Staphylococcus aureus [29]. The MOF, NH2-MIL-53(Fe), functioned as a water-dispersible and stable matrix, and also as an optical transducer whose reduction in photoluminescence was proportional to target bacterial concentration. This type of stable and economical biosensor with notable quantification performance could be an attractive solution to scale up for point-of-sample-collection detection. However, it should be noted that such target-specific bacteriophage is not available for every bacterial pathogen. As an alternative class of recognition molecules, aptamers (synthetic single-stranded oligonucleotides) can fold into designed 3D structure to bind specific targets. The sequence of the aptamers can be selected in vitro through systematic evolution of ligands by exponential enrichment, and the easily synthesized aptamers have high stability, specificity, and affinity to the targets [43].
Integration of nanomaterials with paper microfluidics has led to development of convenient portable biosensor devices. Commercial test strips, such as RapidCheK and Watersafe, are available for environmental detection of E. coli and Salmonella Typhimurium. However, these commercial kits mainly use colorimetric detection based on nanoparticle aggregation caused by antibody–antigen reaction, which takes hours to give qualitative results [44]. Using an alternative detection approach, Rengaraj et al conjugated concanavalin A, the recognition molecule binding saccharide on bacterial cell surfaces, onto commercial hydrophobic paper with screen-printed conductive carbon ink for impedance measurement. This device has potential in-field applicability in terms of portable instrumentation and relative assay stability against environmental disturbance [30]. However, as typical to capillary force-driven paper microfluidics, the sample size at microliters is too small to be relevant for environmental pathogen monitoring without a prior sample concentration step. To overcome this limitation, Srisa-Art et al adopted an approach combining immunomagnetic separation using anti-Salmonella coated Dynabeads and paper-based sandwich immunoassay using the detection enzyme β-galactosidase, which forms a red-violet product with chlorophenol red galactopyranoside for colorimetric detection. The immunomagnetic separation enabled species-specific capture and enrichment from a 1-mL sample [31]. Although the detection device is paper-based, laboratory equipment such as vortex and pipette was still required for the immunomagnetic separation step. To adapt paper microfluidics for in-field environmental detection, the integration of pathogen-specific separation with biosensors represents both an opportunity and a challenge.
For automated and low-cost bacterial pathogen monitoring, immunoassay-based electrochemical biosensors are approaching commercialization, owing to the consistent assay performance and easily automated instrumentation [45]. For example, based on an electrochemical biosensor, Altintas et al developed a fully automated portable system for real-time amperometric measurements of E. coli–specific immunoassay on a microfluidic chip [32]. The instrument prototype with programmed fluid manipulation, electrochemical measurements, and user interface was also developed and tested, thus showing great promise for commercialization. However, since the protein-based recognition reaction is intrinsically weak and susceptible to matrix effect, the majority of these novel biosensors are still limited in sensitivity and specificity compared to nucleic acid analysis (NAA) methods. One solution would be to employ biosensors for target capture utilizing the specific target recognition, while using a nucleic acid–based method to amplify target DNA or RNA for detection. Wang et al demonstrated this approach with bacteriophage-coated Dynabeads for magnetic separation of pathogenic E. coli followed by qPCR detection of total bacterial DNA [33]. Li et al combined antibody-coated carbon nanotube multilayer biosensors for specific capture of E. coli and microfluidic chip-based LAMP detection [34]. The latter study achieved single cell detection in 1 mL complex samples such as juice and milk [33, 34]. More detailed information on above biosensors can be found in Tables 1 and 2. With the automated platforms available for LAMP and PCR, these studies demonstrated that coupled biosensor-NAA would be a promising approach for further development of a fully automated environmental pathogen detection system.
CONCLUSIONS
Portable systems for rapid, ultrasensitive, and specific environmental pathogen monitoring are essential in risk assessment, outbreak prevention, and vaccine distribution for low-resource settings. Recent advances in cell-based, nucleic acid–based, and biosensor-based platforms are reviewed here, with a focus on promising solutions for bacterial pathogen detection in ambient waters at the point of sample collection. Among the reviewed technologies, miniaturized PCR instruments is the most well-developed and commercialized method that is readily deployable in field for sensitive and specific pathogenic bacterial detection, as summarized in Table 3. For biosensors, the combination of biosensor and NAA-based detection holds promise for improved detection efficiency and thus deserves further research and commercial development. Overall, future research should focus on Lab-on-a-Chip pretreatment approaches that can be integrated with subsequent detection [46], entirely automated devices with preloaded reagents, multiplex detection systems, and online real-time monitoring. Such platforms would benefit further comprehensive and timely hazard identification, exposure risk assessment, and pollution control and management. For example, to cope with the global health crisis caused by widespread and fast-evolving antibiotic resistance genes (ARGs), point-of-sample-collection gene sequencing [47] has been developed. This technique provides information on hundreds of ARG subtypes and toxin genes for a range of water environments. Acquiring this information in-field is a pressing need not only for pathogen source tracking, but also for preventing ARG dissemination across various environments.
Notes
Financial support. This work was supported by the Bill & Melinda Gates Foundation (grant number OPP1111252 and OPP1192379).
Supplement sponsorship.This supplement is funded with support from the Coalition against Typhoid Secretariat, housed at the Sabin Vaccine Institute in Washington, DC and made possible by a grant from the Bill & Melinda Gates Foundation.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Ramírez-Castillo FY, Loera-Muro A, Jacques M, et al. Waterborne pathogens: detection methods and challenges. Pathogens 2015; 4:307–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prüss-Ustün A, Bartram J, Clasen T, et al. Burden of disease from inadequate water, sanitation and hygiene in low- and middle-income settings: a retrospective analysis of data from 145 countries. Trop Med Int Health 2014; 19:894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang D, Bi H, Liu B, Qiao L. Detection of pathogenic microorganisms by microfluidics based analytical methods. Anal Chem 2018; 90:5512–20. [DOI] [PubMed] [Google Scholar]
- 4. Gunda NSK, Mitra SK. Rapid water quality monitoring for microbial contamination. Electrochem Soc Interface 2017; 25:73–78. [Google Scholar]
- 5. Song J, Mauk MG, Hackett BA, Cherry S, Bau HH, Liu C. Instrument-free point-of-care molecular detection of Zika virus. Anal Chem 2016; 88:7289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heyries KA, Tropini C, Vaninsberghe M, et al. Megapixel digital PCR. Nat Methods 2011; 8:649–51. [DOI] [PubMed] [Google Scholar]
- 7. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999; 96:9236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deisingh AK, Thompson M. Strategies for the detection of Escherichia coli O157:H7 in foods. J Appl Microbiol 2004; 96:419–29. [DOI] [PubMed] [Google Scholar]
- 9. Vikesland PJ, Wigginton KR. Nanomaterial enabled biosensors for pathogen monitoring—a review. Environ Sci Technol 2010; 44:3656–69. [DOI] [PubMed] [Google Scholar]
- 10. Yilmaz E, Majidi D, Ozgur E, Denizli A. Whole cell imprinting based Escherichia coli sensors: a study for SPR and QCM. Sensors Actuators B Chem 2015; 209:714–21. [Google Scholar]
- 11. Mairhofer J, Roppert K, Ertl P. Microfluidic systems for pathogen sensing: a review. Sensors (Basel) 2009; 9:4804–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coluccio ML, Perozziello G, Malara N, et al. Microfluidic platforms for cell cultures and investigations. Microelectron Eng 2019; 208:14–28. [Google Scholar]
- 13. Futai N, Gu W, Song JW, Takayama S. Handheld recirculation system and customized media for microfluidic cell culture. Lab Chip 2006; 6:149–54. [DOI] [PubMed] [Google Scholar]
- 14. Nichols D, Cahoon N, Trakhtenberg EM, et al. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol 2010; 76:2445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Van Nevel S, Koetzsch S, Proctor CR, et al. Flow cytometric bacterial cell counts challenge conventional heterotrophic plate counts for routine microbiological drinking water monitoring. Water Res 2017; 113:191–206. [DOI] [PubMed] [Google Scholar]
- 16. Besmer MD, Weissbrodt DG, Kratochvil BE, Sigrist JA, Weyland MS, Hammes F. The feasibility of automated online flow cytometry for in-situ monitoring of microbial dynamics in aquatic ecosystems. Front Microbiol 2014; 5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Besmer MD, Epting J, Page RM, Sigrist JA, Huggenberger P, Hammes F. Online flow cytometry reveals microbial dynamics influenced by concurrent natural and operational events in groundwater used for drinking water treatment. Sci Rep 2016; 6:38462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Props R, Rubbens P, Besmer M, et al. Detection of microbial disturbances in a drinking water microbial community through continuous acquisition and advanced analysis of flow cytometry data. Water Res 2018; 145:73–82. [DOI] [PubMed] [Google Scholar]
- 19. Gӧrӧcs Z, Tamamitsu M, Bianco V, et al. A deep learning-enabled portable imaging flow cytometer for cost-effective, high-throughput, and label-free analysis of natural water samples. Light Sci Appl 2018; 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sherchan S, Miles S, Ikner L, Yu HW, Snyder SA, Pepper IL. Near real-time detection of E. coli in reclaimed water. Sensors (Switzerland) 2018; 18:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simões J, Dong T. Continuous and real-time detection of drinking-water pathogens with a low-cost fluorescent optofluidic sensor. Sensors (Switzerland) 2018; 18:2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Højris B, Christensen SCB, Albrechtsen HJ, Smith C, Dahlqvist M. A novel, optical, on-line bacteria sensor for monitoring drinking water quality. Sci Rep 2016; 6:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang H, Huang F, Cai G, Li Y, Lin J. Rapid and sensitive detection of Escherichia coli O157:H7 using coaxial channel-based DNA extraction and microfluidic PCR. J Dairy Sci 2018; 101:9736–46. [DOI] [PubMed] [Google Scholar]
- 24. Chen C, Liu P, Zhao X, Du W, Feng X, Liu BF. A self-contained microfluidic in-gel loop-mediated isothermal amplification for multiplexed pathogen detection. Sensors Actuators B Chem 2017; 239:1–8. [Google Scholar]
- 25. Sayad A, Ibrahim F, Mukim Uddin S, Cho J, Madou M, Thong KL. A microdevice for rapid, monoplex and colorimetric detection of foodborne pathogens using a centrifugal microfluidic platform. Biosens Bioelectron 2018; 100:96–104. [DOI] [PubMed] [Google Scholar]
- 26. Lin X, Huang X, Urmann K, Xie X, Hoffmann MR. Digital loop-mediated isothermal amplification on a commercial membrane. ACS Sens 2019; 4:242–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin X, Huang X, Zhu Y, Urmann K, Xie X, Hoffmann MR. Asymmetric membrane for digital detection of single bacteria in milliliters of complex water samples. ACS Nano 2018; 12:10281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang X, Lin X, Urmann K, et al. Smartphone-based in-gel loop-mediated isothermal amplification (gLAMP) system enables rapid coliphage MS2 quantification in environmental waters. Environ Sci Technol 2018; 52:6399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhardwaj N, Bhardwaj SK, Mehta J, Kim KH, Deep A. MOF-bacteriophage biosensor for highly sensitive and specific detection of Staphylococcus aureus. ACS Appl Mater Interfaces 2017; 9:33589–98. [DOI] [PubMed] [Google Scholar]
- 30. Rengaraj S, Cruz-Izquierdo Á, Scott JL, Di Lorenzo M. Impedimetric paper-based biosensor for the detection of bacterial contamination in water. Sensors Actuators B Chem 2018; 265:50–8. [Google Scholar]
- 31. Srisa-Art M, Boehle KE, Geiss BJ, Henry CS. Highly sensitive detection of Salmonella Typhimurium using a colorimetric paper-based analytical device coupled with immunomagnetic separation. Anal Chem 2018; 90:1035–43. [DOI] [PubMed] [Google Scholar]
- 32. Altintas Z, Akgun M, Kokturk G, Uludag Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens Bioelectron 2018; 100:541–8. [DOI] [PubMed] [Google Scholar]
- 33. Wang Z, Wang D, Kinchla AJ, Sela DA, Nugen SR. Rapid screening of waterborne pathogens using phage-mediated separation coupled with real-time PCR detection. Anal Bioanal Chem 2016; 408:4169–78. [DOI] [PubMed] [Google Scholar]
- 34. Li T, Zhu F, Guo W, et al. Selective capture and rapid identification of E. coli O157:H7 by carbon nanotube multilayer biosensors and microfluidic chip-based LAMP. RSC Adv 2017; 7:30446–52. [Google Scholar]
- 35. Marx V. PCR heads into the field. Nat Methods 2015; 12:393–7. [DOI] [PubMed] [Google Scholar]
- 36. Foudeh AM, Fatanat Didar T, Veres T, Tabrizian M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012; 12:3249–66. [DOI] [PubMed] [Google Scholar]
- 37. Nguyen PL, Sudheesh PS, Thomas AC, Sinnesael M, Haman K, Cain KD. Rapid detection and monitoring of Flavobacterium psychrophilum in water by using a handheld, field-portable quantitative PCR System. J Aquat Anim Health 2018; 30:302–11. [DOI] [PubMed] [Google Scholar]
- 38. Zhang C, Xu J, Ma W, Zheng W. PCR microfluidic devices for DNA amplification. Biotechnol Adv 2006; 24:243–84. [DOI] [PubMed] [Google Scholar]
- 39. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 2000; 28:E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Justino CIL, Duarte AC, Rocha-Santos TAP. Recent progress in biosensors for environmental monitoring: a review. Sensors (Switzerland) 2017; 17:2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kumar N, Hu Y, Singh S, Mizaikoff B. Emerging biosensor platforms for the assessment of water-borne pathogens. Analyst 2018; 143:359–73. [DOI] [PubMed] [Google Scholar]
- 42. Vikesland PJ. Nanosensors for water quality monitoring. Nat Nanotechnol 2018; 13:651–60. [DOI] [PubMed] [Google Scholar]
- 43. Zhao YW, Wang HX, Jia GC, Li Z. Application of aptamer-based biosensor for rapid detection of pathogenic Escherichia coli. Sensors (Switzerland) 2018; 18:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ge X, Asiri AM, Du D, Wen W, Wang S, Lin Y. Nanomaterial-enhanced paper-based biosensors. Trends Anal Chem 2014; 58:31–9. [Google Scholar]
- 45. Furst AL, Francis MB. Impedance-based detection of bacteria. Chem Rev 2019; 119:700–26. [DOI] [PubMed] [Google Scholar]
- 46. Zhu Y, Huang X, Xie X, et al. Propidium monoazide pretreatment on a 3D-printed microfluidic device for efficient PCR determination of “live: versus dead” microbial cells. Environ Sci Water Res Technol 2018; 4:956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu YOO, Ndegwa N, Alneberg J, et al. Stationary and portable sequencing-based approaches for tracing wastewater contamination in urban stormwater systems. Sci Rep 2018; 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]