Abstract
Increasing awareness about germline predisposition and the widespread application of unbiased whole exome sequencing contributed to the discovery of new clinical entities with high risk for the development of haematopoietic malignancies. The revised 2016 WHO classification introduced a novel category of “myeloid neoplasms with germline predisposition” with GATA2, CEBPA, DDX41, RUNX1, ANKRD26 and ETV6 genes expanding the spectrum of hereditary myeloid neoplasms (MN). Since then, more germline causes of MN were identified, including SAMD9, SAMD9L, and ERCC6L2. This review describes the genetic and clinical spectrum of predisposition to MN. The main focus lies in delineation of phenotypes, genetics and management of GATA2 deficiency and the novel SAMD9/SAMD9L-related disorders. Combined, GATA2 and SAMD9/SAMD9L (SAMD9/9L) syndromes are recognized as most frequent causes of primary paediatric myelodysplastic syndromes, particularly in setting of monosomy 7. To date, ~550 cases with germline GATA2 mutations, and ~130 patients with SAMD9/9L mutations had been reported in literature. GATA2 deficiency is a highly penetrant disorder with a progressive course that often rapidly necessitates bone marrow transplantation. In contrast, SAMD9/9L disorders show incomplete penetrance with various clinical outcomes ranging from spontaneous haematological remission observed in young children to malignant progression.
Keywords: Germline predisposition, Hereditary MDS, GATA2, SAMD9, SAMD9L
Introduction: germline predisposition in myeloid neoplasms
Nearly a century ago the first classical inherited bone marrow failure (BMF) syndrome predisposing to myeloid neoplasms (MN) had been reported by the paediatrician Guido Fanconi, and later named Fanconi Anemia (FA) [1]. Since then, a number of additional inherited BMF syndromes with risk for the development of myelodysplastic syndrome (MDS) and leukemia have been discovered, including severe congenital neutropenia (SCN), Shwachman Diamond syndrome (SDS), telomere biology disorders/dyskeratosis congenita, Down syndrome, RASopathies, and DNA repair disorders [[2], [3], [4], [5], [6], [7], [8], [9], [10]]. These disorders are usually straightforward to diagnose because of preexisting dysmorphologies and haematological symptoms arising from abnormalities of haematopoietic stem and progenitor cells. The characteristics of recurrently occurring genetic syndromes are outlined in Table 1 . Although BMF manifests early in life, MDS and leukemia develop as a secondary disease after a latency of years to decades from diagnosis of the underlying condition [5,[11], [12], [13], [14], [15], [16]]. Clonal evolution is usually associated with a number of recurrent somatic mutations, for example RUNX1 in FA, CSF3R and RUNX1 in SCN, TP53 in SDS, GATA1 short and cohesin genes in Down syndrome [5,7,10,12,[16], [17], [18], [19], [20], [21], [22], [23]]. Additional mutations can involve typical leukemia drivers including RAS pathway genes, SETBP1, ASXL1, EZH2, and others. Recurrent karyotype abnormalities are also found, particularly loss of chromosome 7 (monosomy 7 (−7), del(7q), der(1; 7), isochromosome 7q), trisomy 8 (+8), or trisomy 21 (+21). While most of the somatic mutations are known leukemia drivers (RUNX1, RAS pathway, TP53), others might represent benign adaptive responses (EIF6, CSF3R). In addition, revertant somatic events including back mutations or uniparental disomy have been reported in FA and TERT/TERC mutated telomere biology disorders [[24], [25], [26], [27]].
Table 1.
Germline syndromes predisposing to myeloid neoplasms.
| Disease/Gene | Risk for MN | Age of MN onset, years * | Population at High Risk for MN | Reported somatic mutations | Reported karyotypes | Congenital anomalies | Immune deficiency |
|---|---|---|---|---|---|---|---|
| Germline predisposition to myelodysplastic syndromes/acute myeloid leukemia (MDS/AML) | |||||||
| GATA2 | High | 0.4–78 (~20) | Children – Adults | SETBP1, ASXL1, RUNX1, PTPN11, NRAS, KRAS, CBL, EZH2, ETV6, STAG2, JAK3, IKZF1, CRLF2, IDH2, TP53 | −7, der(1; 7), +8, +21 | ++ | ++ |
| SAMD9, SAMD9L | Moderate | Paediatric age, not yet defined | Children | SETBP1, ASXL1, RUNX1, PTPN11, KRAS, CBL, EZH2, ETV6, BRAF, RAD21 | −7, del(7q), del12p13.2, UPD7q | ++ | – |
| Somatic revertant mosaicism (cis SAMD9/9L, UPD7q) | |||||||
| RUNX1 | High | 6-77 (~33) | Children – Adults | RUNX1 (trans mutation or duplication via LOH), ASXL1, BCOR, DNMT3A, PHF6, WT1, GATA2, FLI1, JMJD5, KDM6B, CDC25C | +21, +8, −7 | – | – |
| CEBPA | High | 2-50 (~25) | Children – Adults | CEBPA (trans mutation at 3′ end), GATA2, WT1, EZH2, TET2, SMC3, NRAS, DDX41, CSF3R | – | – | |
| ETV6 | Moderate (mostly ALL) | 8–82 | Adults | BCOR, RUNX1, NRAS | – | – | |
| DDX41 | Moderate | 6-93 (~55) | Adults | DDX41 (trans p.Arg525His mutation, p.Ala255Asp, p.Glu247Lys, p.Pro321Lys) | del(20q), del(7q), −7, +8 | – | – |
| ANKRD26 | Low | >30 | Adults | – | – | ||
| TET2 (1 family with p.K1363fs mutation) | Not known | 53-61 (60) | Adults | TET2 (p.His863fs), BRAF, ZRSR2, SRSF2, JAK2, GATA2 | – | – | – |
| Classical inherited bone marrow failure syndromes | |||||||
| Fanconi Anemia | |||||||
| 22 FA genes | High | 0.1–49 (13) | Children | Somatic revertant mosaicisms (back mutations), | del(7q), dup(1q), dup(3q), complex | ++ | – |
| RUNX1 | |||||||
| Severe Congenital Neutropenia ELANE, G6PC3, HAX1, JAGN1, GFI1, VPS45A, TCRG1 |
High | 2-49 (12) | Children – Adults | CSF3R, RUNX1, RAS genes | −7, del(7q), +21 | + | – |
| Shwachman Diamond Syndrome SBDS, ELF1, SRP54, DNAJC21 |
Variable | MDS: 5–42 (8) | Children – Adults | TP53 (>50% of cases with SBDS germline mutation), EIF6, PRPF8, CSNK1A1, U2AF1, IDH1, RUNX1, SETBP1, NRAS, KRAS, BRAF, DNMT3A, TET2, ASXL1 | isochromosome 7q, −7, del(20q) |
++ | (+) |
| Telomere Biology Disorders DC: TERC, TERT, DKC1, RTEL1, TINF2, ACD, CTC1, NHP2, NOP10, NPM1, PARN, WRAP53 Pulmonary fibrosis: POT1, ZCCHC8, NAF1 CLL/Melanoma: POT1 |
Moderate (mostly adults) | 19-61 (35) | Adults | Somatic revertant mosaicism (UPD of TERT/TERC allele, or activating TERT promoter mutations – very rare); | + | + | |
| Leukemia mutations uncommon | |||||||
| Down syndrome and rasopathies | |||||||
| Trisomy 21 | Moderate | 1-4 (1.5) | Children | GATA1 short, cohesin (RAD21, STAG2, NIPBL, SMC1A, SMC3), CTCF, EZH2, KANSL1, BCOR, WT1, DCAF7 TP53, NRAS, KRAS, PTPN11, JAK2, JAK3, SH2B3 | MLL gene rearrangements, complex | ++ | (+) |
| Rasopathies NF1, CBL, PTPN11, NRAS, KRAS |
Moderate | CBL: 0.1–3.6 (1.1) | Children | Duplicaton of mutant allele (via UPD), additional RAS pathway mutations | −7 | ++ | – |
| DNA repair syndromes | |||||||
| ERCC6L2 | High | 14-65 (38) | Adults | TP53, IDH1 | −7, +20, −18, del(5q) | + | – |
| Xeroderma pigmentosum C (XP-C) | Low | 7-29 (25) | Adults | TP53, CSF3R, TET2, RAD21 | −7, del(5q), complex | + | – |
| Other | |||||||
| TP53, CMMRD, Werner/Bloom syndrome, NBS, AT, Ligase IV deficiency | MN are rare | All ages | Children – Adults | Chromothripsis | −7, complex | + | + |
Abbreviations: DC, dyskeratosis congenita; CMMRD, Constitutional Mismatch Repair Deficiency; NBS, Nijmegen Breakage Syndrome; AT, Ataxia telangiectasia; MN, myeloid neoplasms.
+, present; (+), possibly present; ++, commonly present; -, absent.
*Approximate age range (median) assessed from literature reports; CLL chronic lymphocytic leukemia; ALL, acute lymphoblastic leukemia; UPD, uniparental disomy; LOH, loss of heterozygosity.
The identification of germline mutations in RUNX1 (1999) and CEBPA (2004) initiated a new era for the discovery of monogenic disorders predisposing to MN [28,29]. For the most part these “new” syndromes result from heterozygous mutations in haematopoietic transcription factors or regulators (GATA2, RUNX1, CEBPA, ETV6, DDX41, SAMD9, SAMD9L, and TET2, Table 1) [31], [32], [33], [34], [35], [36], [37], [52], [147]. These syndromes often pose unique diagnostic challenges. First, the full spectrum of clinical and genetic manifestations is not yet fully defined, forcing us to adopt the “expect the unexpected“ approach for the workup of such cases. Second, unlike the classical inherited BMF syndromes which are often diagnosed based on medical history, these new entities can often manifest with MN without preceding clinical problems. And finally, many patients have negative family history (even in families with multiple mutated individuals) which can be attributed to incomplete penetrance/variable expressivity and a considerable rate of de novo germline mutations. Despite these challenges, there are genetic and phenotypic patterns that can serve as diagnostic red flags. These can include certain types of mutations that are identified on a somatic sequencing panel, for example variants with allelic frequency nearly 50% in a patient without significant blast increase, DDX41 somatic hotspot mutation that co-occurs with germline DDX41 alterations, bi-allelic CEBPA mutation with one mutation positioned at 3′-end, and finally the domain-specific localization such as missense GATA2 mutations in zinc finger 2. In addition, certain clinical signs can be syndrome-specific, i.e. lymphedema, hydrocele, and congenital deafness in GATA2, ataxia in SAMD9L, adrenal hypoplasia in SAMD9, or preceding thrombocytopenia in individuals with RUNX1/ETV6/ANKRD26 mutations.
GATA2 and SAMD9/SAMD9L (SAMD9/9L) syndromes are the most frequent predisposing conditions in children and adolescents with primary MDS and are associated with the development of −7 karyotype: collectively they account for at least 50% of paediatric MDS with −7 [37], [38], [39], [52]. The following text will discuss in detail the clinical and genetic spectrum of GATA2 and SAMD9/9L syndromes.
GATA2 deficiency
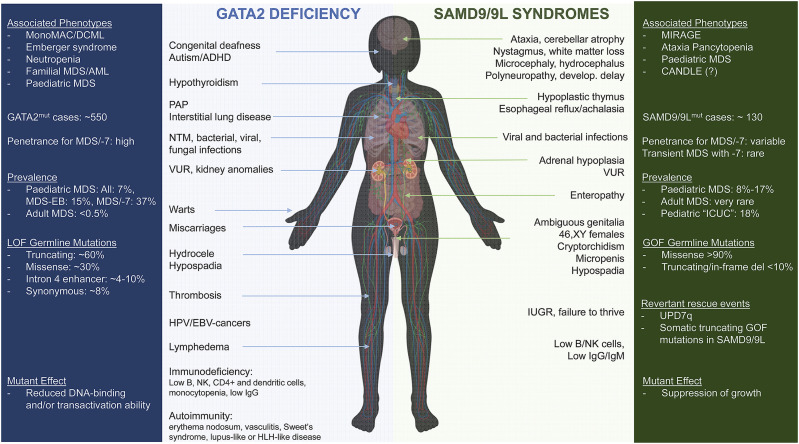
GATA2 is a key transcription factor critical for ontogenesis of haematopoietic system, including haematopoietic stem cell (HSC) activity and self-renewal, myeloid and myelo-erythroid progenitor cell differentiation, and erythroid precursor cell maintenance [[40], [41], [42], [43], [44]]. In the past decade, heterozygous germline GATA2 mutations have been identified in a number of cohorts with cellular deficiencies (immunodeficiency syndromes initially referred to as MonoMAC syndrome, DCML deficiency, Emberger syndrome, chronic neutropenia) [[45], [46], [47], [48]], and in patients with familial MDS and acute myeloid leukemia (AML), as well as paediatric MDS [[47], [48], [49], [50], [51], [52], [53]]. To date, approximately 150 unique GATA2 germline mutations have been identified in roughly 550 patients (Fig. 1 ).
Fig. 1.
Clinical characteristics of GATA2 deficiency and SAMD9/9L syndromes.
Key information referring to phenotypes, prevalence and genetics are summarized in the outside coloured boxes; clinical manifestations are depicted in the middle. Abbreviations: MonoMAC, Monocytopenia and Mycobacterium avium complex infection syndrome; DCML, Dendritic cell, Monocytes, B/NK Lymphocytes deficiency; MDS, Myelodysplastic Syndrome; MDS-EB, MDS with Excess of Blasts; AML, Acute Myeloid Leukemia; −7, monosomy 7; LOF, loss of function; ADHD, Attention Deficit Hyperactivity Disorder; PAP, Pulmonary Alveolar Proteinosis; NTM, Nontuberculous mycobacteria; VUR, Vesicoureteral Reflux; IUGR, Intrauterine Growth Restriction; MIRAGE, Myelodysplasia, Infection, growth Restriction, Adrenal hypoplasia, Genital phenotypes, and Enteropathy; CANDLE, Chronic Atypical Neutrophilic Dermatosis with Lipodystrophy and Elevated temperaturę; ICUC, idiopathic cytopenia of unknown cause (denotes children with unclear cytopenia and suspected bone marrow failure); GOF, gain of function.
Penetrance and prevalence
GATA2 deficiency follows an autosomal dominant inheritance pattern with the majority (up to 80%) of cases arising de novo [49,[52], [53], [54], [55]]. Although the lifetime penetrance for the development of MN is very high, incomplete penetrance is possible, as suggested by the presence of several asymptomatic mutation carriers of various ages within affected families [50,52,53,[56], [57], [58]]. Recently, distinct patterns of GATA2 promoter methylation leading to disbalance in allelic expression have been identified in 2 patients and proposed as a mechanism for reduced clinical expressivity [59,60].
Summarizing published cohort studies and smaller case series, approximately 75% of GATA2 mutation carriers develop MN at an estimated median age of 20 years (Table 1). The spectrum of MN includes primary paediatric MDS, AML, chronic myelomonocytic leukemia (CMML) and myeloproliferative neoplasms [47,[49], [50], [51], [52], [53], [54], [55],61]. In children and adolescents with primary MDS, GATA2 deficiency is a predominant germline predisposition accounting for 7% of all MDS cases, 15% of patients with advanced MDS, and 37% of patients with MDS and −7 karyotype [52,62]. Among children, the prevalence of GATA2 deficiency increases with age, and 2/3 of adolescents with MDS and monosomy 7 carry germline GATA2 mutations. In adult MDS, GATA2 deficiency is rare and present in less than 0.5% of individuals [52], however the true prevalence in various age groups has yet to be defined.
Clinical presentation
The initial haematological presentation in patients with GATA2 deficiency can be very variable, ranging from cytopenias and hypocellular BMF-like picture, severe immunodeficiency to myeloid neoplasms. Many patients often lack family history of MDS and exhibit mild initial symptoms with preceding cellular deficiencies [49,52,62]. However, MDS can also manifest as a stand-alone diagnosis without preceding cytopenia. GATA2-deficient patients often suffer from preexisting monocytopenia, B-cell and NK-cell lymphopenia, reduction/lack of CD56bright NK cells and dendritic cells, inverted ratio of CD4:CD8 cells, and chronic neutropenia [48,53,58,[61], [62], [63], [64], [65]]. Immune deficiency is typically recorded as a consequence of profound cytopenias and loss of functional stem cells [66]. When compared to other marrow failure conditions in children, reduction of progenitor and mature B-cells are the most constant feature of GATA2 deficiency [62]. Notably, monocytosis has been observed at diagnosis in GATA2 patients with MDS and is likely attributed to disease progression [52,53], although transient monocytosis in infancy has been also observed (unpublished own observations). The immunological phenotypes are heterogeneous. Many patients can initially present with HPV-related infections (warts, generalized verrucosis, cervical intraepithelial neoplasia), disseminated nontuberculous mycobacterial, as well as systemic bacterial and fungal infections [50,53,67]. Recurrent respiratory tract infections can result in a development of pulmonary alveolar proteinosis (PAP) or interstitial lung disease [50,53]. Moreover, compromised function of the immune system may contribute to malignant transformation of HPV- or EBV-related neoplasia and increased occurrence of other solid tumors [47,50,53,68]. Furthermore, GATA2 mutated patients have been repeatedly shown to suffer from autoimmune dysregulation such as autoimmune cytopenia, arthritis, panniculitis, erythema nodosum, psoriasis, lupus-like syndrome and autoimmune hepatitis [53,56,63,[69], [70], [71]].
GATA2-MDS patients demonstrate heterogeneous morphological features involving hypocellular marrow with cytopenias, or normal to hypercellular marrow in case of advanced MDS. Frequently recognized dysplasia is seen in megakaryocyte lineage, but other lineages are also affected [50,63].
Apart from haematological and immunological symptoms, at least 50% of GATA2-deficient patients present with constitutional abnormalities affecting different organ systems (Fig. 1). In addition to lymphedema, hydrocele and congenital deafness, abnormalities of pulmonary, cardiovascular, urogenital and neurological systems have been repeatedly observed [48,50,52,53,56,62,63]. This includes i.e. PAP, thrombosis, pulmonary embolism, vesicoureteral reflux, hypospadias, hydrocele, developmental delay, or behavioral disorders/ADHD. The presence of both immune deficiency and typical constitutional features, especially in context of MDS should trigger genetic workup for GATA2 mutations.
GATA2 germline mutations
De novo or inherited heterozygous loss of function mutations have been identified as the genetic basis of GATA2 deficiency. These mutations are thought to lead to loss of function of GATA2 protein (through loss of one allele or malfunctioning protein) specifically abolishing the DNA-binding function of the C-terminal zinc finger (ZF2) [39], [47], [49], [72], [73], [74], [75], [76], [77], [79]. Overall, 3 main mutation categories can be distinguished i.e. null (frameshift, nonsense, splice site and whole gene deletions) located prior or within ZF2 and accounting for around two thirds of all reported variants, missense substitutions clustered within ZF2 representing one third of GATA2 mutations and intronic alterations affecting +9.5 kb enhancer element (EBOX-GATA-ETS) detected in ~4–10% of cases (Fig. 1). Additionally, rare changes including in-frame deletions/insertions and missense mutations downstream of ZF2 were found [39]. Most recently, we identified 5 heterozygous synonymous GATA2 mutations (p.Thr117Thr, p.Leu217Leu, p.Gly327Gly, p.Ala341Ala, p.Pro472Pro) in 9 patients with GATA2 deficiency that led to selective loss of mutated copy of GATA2 mRNA [81]. The hotspot p.Thr117Thr mutation has been also described by others [77,82] and mechanistically was found to introduce a new splice donor resulting in premature translation termination associated with nonsense-mediated decay.
Experimental studies found impaired ability of mutant GATA2 protein to bind DNA and activate transcription of target genes. This was shown for several mutations (p.Arg330X, p.Ala345delinsALLVAALLAA, p.Thr354Met, p.Thr355del, p.Thr358Asn, p.Arg361Leu, p.Cys373Arg, p.Arg396Gln, p.Arg396Leu, p.Arg398Trp) [47,49,[73], [74], [75], [76],79]. Moreover, some of the mutations were shown to affect proliferation, differentiation and apoptosis in haematopoietic cells [49,74]. Variable expressivity of GATA2 ZF2 germline mutations is common. The most recurrent mutations p.Thr354Met, p.Arg396Gln and p.Arg398Trp all predispose to myeloid malignancies, however p.Thr354Met was shown to be associated with MDS as initial presentation, while p.Arg396Gln and p.Arg398Trp mutations were suggested to correlate with a phenotype of immunodeficiency manifesting prior to malignant transformation [75]. Although, reduced DNA-binding/transactivation ability has been shown to be causal of GATA2 haploinsufficiency, this does not explain the genotype/phenotype correlation for individual ZF2 mutations. One possible explanation could be the altered interaction between GATA2 and other proteins, as shown for GATA2 p.Thr354Met and p.Cys373Arg mutations that compared to wild type GATA2 protein bind more strongly to the haematopoietic differentiation factor PU.1 [75]. Hence, detailed mechanistic studies are paramount to understand these phenotypic differences and functional consequences of germline GATA2 mutations. Genotype-phenotype correlative analyses have thus far been unsuccessful and potential associations seen in some studies were not validated in other cohorts. In a recent study encompassing 79 French patients with GATA2 deficiency, patients harboring missense mutations (14 out of 38) were more likely to develop leukemia than patients with frameshift mutations (2 out of 28; p = 0.007) [53]. However, an analysis conducted by our group in a large cohort of paediatric MDS (N = 137) did not confirm this association (own unpublished observations).
Somatic GATA2 mutations compared to germline mutations show different localization within the protein and are associated with other haematological phenotypes. Of the roughly 50 reported somatic mutations, the majority are found within the boundaries of N-terminal zinc finger (ZF1), but some mutations also occur in ZF2 [83]. These mutations are generally rare and were identified in paediatric and adult AML (predominantly accompanied by biallelic CEBPA mutations), chronic myeloid leukemia in blast crisis, as well as acute erythroid leukemia [[84], [85], [86], [87], [88], [89], [90], [91], [92], [93]]. In contrast to germline variants, acquired GATA2 mutations were characterized as either LOF (p.Pro304His, p.Leu321Val, p.Leu321Pro, p.Arg330Gln, p.Arg362Gln, p.Ala341_Gly346del) or gain-of-function (GOF; p.Gly320Asp, p.Leu359Val). It is interesting to note that certain somatic GATA2 ZF2 mutations in adults with MN can “phenocopy” symptoms of GATA2 germline disorder, i.e. immunodeficiency (monocytopenia, low B-/NK-cells, recurrent infections) accompanied by lymphedema or PAP [94,95].
Acquired cytogenetic and genetic aberrations
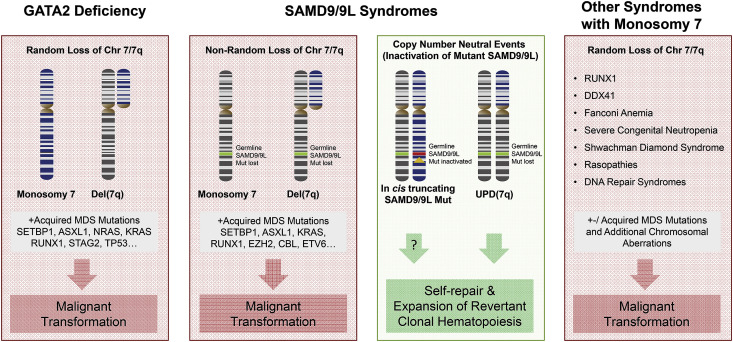
The most common karyotype abnormalities are monosomy 7 or der(1; 7) that can occur in up to 80% of GATA2-related MDS patients, with average estimate across all published studies of ~41% (Fig. 2 ) [39]. Trisomy 8 is the second most common aberration identified in up to 40% of patients in single cohorts, and an average of 15% across published studies. Additional common abnormality is trisomy 21, while complex karyotypes are generally not encountered. GATA2-related MDS is also associated with acquired MDS/leukemia driver mutations (Table 1). Recurrent oncogenic alterations were identified in genes ASXL1, SETBP1, RUNX1, STAG2, CBL, EZH2, NRAS, KRAS, JAK3, PTPN11. [51,54,58,[96], [97], [98], [99], [100]] Furthermore, single GATA2-MDS cases were also reported to harbor somatic mutations in IKZF1, CRLF2, HECW2, GATA1, GATA2, ATRX, BRCA2, GPRC5A, IDH2, TP53, WT1 [97,99,101].
Fig. 2.
Monosomy 7 driven mechanisms of clonal evolution in MDS predisposition syndromes.
Malignant transformation of GATA2 deficiency (outside left box), SAMD9/9L disorders (middle boxes) and other predisposing syndromes (outside right box) is associated with loss of whole chromosome 7 or its long (7q) arm followed by acquisition of somatic mutations in leukemia-related genes. Loss of chromosome 7 is non-random in SAMD9/9L syndromes and leads to disappearance of germline SAMD9/9L mutation. Revertant clonal haematopoietic in SAMD9/9L syndromes arise from UPD7q or acquired truncating SAMD9/9L mutation (middle right box). Abbreviations: MDS, myelodysplastic syndrome; AML, acute myeloid leukemia; UPD, uniparental disomy.
Therapeutic considerations
There are no consensus guidelines on management of GATA2 deficiency and the surveillance strategies are individually tailored. Most patients are being followed by haematologists, immunologists or transplant physicians and general recommendations include periodic assessment of complete blood counts and immune status, yearly bone marrow evaluation with cytogenetics and somatic mutational testing, as well as screening for HPV-related cancers and pulmonary symptoms. Haematopoietic stem cell transplantation (HSCT) is the only curative treatment with reported outcomes ranging from 54% (4 year overall survival (OS)) in adults transplanted for MDS/AML or immunodeficiency [50], to 66% (5 year OS) in children transplanted for MDS with -7 [52], or 62% (5 year OS) in a French GATA2 cohort [53], and 86% (2 year disease-free survival) in young adults with MDS [68]. Because −7 karyotype is associated with a high risk of progression to more advanced MDS, patients with this cytogenetic category should undergo HSCT as soon as possible [102]. While myeloablative conditioning is preferred in MDS with -7 (independent of blasts) as well as advanced MDS/AML, reduced intensity conditioning might be preferred option in patients with hypocellular MDS without high risk somatic alterations, as well as patients with immunodeficiency alone. HSCT was shown to reverse HPV-related lesions as well as respiratory problems (PAP) [68,[103], [104], [105]]. Patients with stable disease course, without relevant infections, bone marrow dysplasia and transfusion-dependency might qualify for a watch & wait strategy [106,107]. However, it can be assumed that most GATA2-deficient patients show progressive disease and even with careful watching the best opportunity for low risk HSCT might be missed. Currently, transplant indications include progressing immunodeficiency with recurrent infections, respiratory complications (PAP), transfusion-dependency, and MDS [50,52,104,105,108]. The optimal point in time for performing HSCT would be the stage of hypocellular MDS or immunodeficiency prior development of MDS/leukemia evolution or severe organ dysfunction.
SAMD9 and SAMD9L syndromes
Sterile alpha motif domain-containing protein 9 (SAMD9) and the paralogue gene SAMD9-like (SAMD9L) are located side-by-side on chromosome 7q21. Initial study describing acquired microdeletions of 7q21 in patients with MN drew preliminary attention to these poorly characterized genes [113]. SAMD9/9L are IFN and TNF-α responsive proteins that were shown to play a role in antiviral response [[109], [110], [111], [112]], tumor suppression [113,114], inflammation [115,116], development [[117], [118], [119]] and endosomal fusion [118,120]. Samd9l-deficient mice develop myeloid disease resembling human MDS with −7 [120]. The first link to human disease was a description of biallelic LOF SAMD9 mutations (p.K1495E and p.R344X) in several consanguineous Jewish-Yemenite families with normophosphatemic familial tumor calcinosis (NFTC), however no further NFTC cases with SAMD9 mutations were found [115,116]. In 2016, heterozygous missense SAMD9 mutations with GOF effect were linked to a severe early-onset condition with Myelodysplasia, Infections, Restriction of growth, Adrenal hypoplasia, Genital phenotypes and Enteropathy (MIRAGE) [118,119]. At the same time, missense GOF mutations in SAMD9L were described as a cause of a syndrome with progressive neurological phenotype, pancytopenia and hypocellular bone marrow (Ataxia Pancytopenia (ATXPC)) [117,121]. Both conditions have a unifying phenotype of early onset myelodysplasia with monosomy 7. Following the first discoveries in syndromic cohorts, a number of studies reported germline missense SAMD9 and SAMD9L mutations in paediatric cohorts with MDS (often without syndromic manifestation) [[122], [123], [124], [125]]. Recently, germline frameshift SAMD9L mutations were also discovered in several children presenting with autoinflammatory panniculitis resembling Chronic Atypical Neutrophilic Dermatosis with Lipodystrophy and Elevated Temperatures (CANDLE) syndrome [126].
Penetrance and prevalence
SAMD9/9L syndromes are autosomal dominant syndromes resulting from GOF, mostly missense mutations with variable penetrance. SAMD9 mutations are associated with a high rate of de novo cases with very high penetrance especially in pedigrees presenting with MIRAGE syndrome. For SAMD9L mutations, the penetrance for haematological disease is incomplete and has been estimated at 70% [[123]]. Similarly, low penetrance for neurological phenotypes (that might increase with age) has been observed [117].
Germline SAMD9/9L mutations account for a large proportion of primary childhood MDS and the prevalence was shown to range from 8% (43/548) in a large multi-institutional consecutive cohort of the European Working Group of Childhood MDS (EWOG-MDS) [37] to 17% (8/46) in a single institution report [122]. Interestingly, in a French cohort of patients with idiopathic cytopenia of unknown cause (“ICUC”), 18.6% (16/86) of patients carried germline SAMD9/9L mutations [125].
Clinical presentation
Multiple organ systems can be affected in SAMD9/9L syndromes, with predominant haematologic, immunologic, endocrine, genital and neurologic involvement (Fig. 1). Initial clinical presentation is heterogeneous and can range from severe disease with high infant mortality, to transient cytopenia and immune dysfunction. Patients with MIRAGE (SAMD9) phenotype present with early onset adrenal hypoplasia and primary adrenal insufficiency, intrauterine growth restriction, genital phenotypes (46XY females, bifid shawl scrotum, testicular dysgenesis, intra-abdominal or inguinal testes, clitoromegaly), gastrointestinal issues (enteropathy, reflux, achalasia), severe systemic infections, as well as thrombocytopenia and anaemia at birth - which in some patients can be self-limiting during infancy [37,118,119,[128], [129], [130], [131], [132], [133], [134], [135], [136], [137], [138], [139]]. Patients with SAMD9L mutations might show disease-specific neurological findings with very variable age of onset and dynamics of progression. Severe cerebellar ataxia is observed in some but not all cases [117]. Some patients might show cerebellar atrophy, dysmetria, nystagmus, white matter abnormalities, and loss of Purkinje cells [117,140,141].
Of note, haematological phenotype is the “common denominator” of both syndromes manifesting in the majority of patients [117,[121], [122], [123],125,142]. The haematological spectrum involves single lineage cytopenia (mostly thrombocytopenia) or pancytopenia with hypocellular marrow, and MDS with −7 or del(7q). In a large paediatric MDS cohort the majority (90%) of SAMD9/9L-mutated patients presented with refractory cytopenia of childhood (RCC), while MDS with excess blasts was found in 10% of cases [37]. In a small subset of patients advanced leukemic disease (AML, CMML) can be diagnosed [37,[122], [123], [124]]. The median age at diagnosis in paediatric MDS with SAMD9/9L was shown to be 9.6 years (0.2–17.6) which is comparable with GATA2-related MDS [37]. The most widespread aberrant karyotype is −7/del(7q) and shows a unique non-random pattern where the chromosome 7 with mutant SAMD9/9L copy is always lost, which by itself can be considered a pathognomonic sign of SAMD9/9L syndromes (Fig. 2).
Immune dysfunction is not well defined and shows varying phenotype and severity. Mostly in patients with MIRAGE phenotype (SAMD9) but also in several SAMD9L-mutated patients, severe invasive infections have been described. The causative organisms were bacteria (pseudomonas aeruginosa, Clostridium difficile, Staphylococcus, Serratia marcescens, Enterococcus faecium, Escherichia coli, Klebsiella pneumoniae, Stenotrophomonas maltophilia, Streptococcus pyogenes), viruses (MRSA, CMV, EBV, rhinovirus, coronavirus, varicella), and fungi (aspergillus, candida) causing sepsis, meningitis, otitis, sinusitis, laryngitis, hepatitis, bronchiolitis, pneumonia, neonatal necrotizing enterocolitis, pancolitis, gastroenteritis, enteropathy, urinary tract infections, otitis media, ecthyma gangrenosum, warts, dental abscesses, and urethritis [118,119,121,124,125,[128], [129], [130], [131], [132], [133],[136], [137], [138], [139],141,[143], [144], [145]]. However, the majority of non-syndromic SAMD9/9L-MDS patients generally do not appear to have high risk to develop immune dysfunction and severe infections. Decreased peripheral B/NK-cells, low IgG and IgM or increased TNF-alpha and IL-6 levels were documented in cases with SAMD9/9L mutations [121,125,128,131,135,143]. Other rare dysmorphic features documented in single patients include skeletal abnormalities (scoliosis, joint contracture at wrist and ankles), hearing loss, dysmorphic facial features, camptodactyly, arachnodactyly, glomerular proteinuria, dysautonomia and speech delay [129,131].
SAMD9/9L genetics
Thus far 38 distinct SAMD9 and 26 SAMD9L mutations have been identified in 110 symptomatic patients as mutually exclusive events [118,119,121,122,124,126,[129], [130], [131], [132], [133], [134],[140], [141], [142], [143],145,146]. The majority of these patients exhibit haematological phenotype with cytopenias, and/or MDS with −7. Most germline mutations are missense and occur predominantly in the second half (C-terminus) of SAMD9/9L proteins, encompassing the predicted P-LOOP_NTPase domain. Six cases with truncating germline mutations in SAMD9L were also reported in children with CANDLE phenotype [126].
Thus far, all functionally evaluated germline missense SAMD9/9L mutations were shown to inhibit cell growth in 293 cellular overexpression assay [118], [119], [123]. Of note, many mutated amino acids show moderate or weak conservation across species, thus posing a risk of being scored as ‘likely benign’ based on in silico predictors. The current state-of-art for assessment of pathogenicity includes functional validation on a research basis. However, such testing must be interpreted with caution since sensitivity and specificity have not been defined and various groups utilize different readouts [118], [119], [122], [123].
Clonal evolution and somatic reversion
The unique aspect of SAMD9/9L disease mechanism is ‘adaptation by aneuploidy’ that is achieved by the non-random loss chromosome 7 (−7/del(7q)) which contains mutated SAMD9/9L gene copy (Fig. 2)[118], [119], [121], [122], [123], [124], [125], [129], [134], [135], [141], [142], [143], [144], [145]. The decrease of mutant allele in haematopoiesis poses a diagnostic challenge, since germline SAMD9/9L mutations show decreased variant allele frequency (VAF), with VAF even below 5% (own observations), necessitating germline validation in non-haematopoietic specimens, i.e. fibroblasts.
Several case reports documented complete disappearance of −7 clones, thus far seen exclusively in young children, a phenomenon previously referred to as transient monosomy 7 syndrome [121], [122], [123], [124], [141], [142], [143]. However, −7 is a high risk lesion with malignant propensity (i.e. due to loss of several tumor suppressor genes such as EZH2). Clonal evolution to advanced MDS/AML is a recurrent complication in SAMD9/9L-related MDS with −7 and was shown to be accompanied by somatic driver mutations in SETBP1, ASXL1, RUNX1, PTPN11, KRAS, CBL, EZH2, ETV6, BRAF, and RAD21 [[122], [123], [124],142].
Somatic revertant mosaicism with expansion of benign, corrected haematopoiesis represents another unique feature of SAMD9/9L syndromes (Fig. 2). The two mechanisms observed so far are the acquisition of truncating SAMD9/9L mutations or an independent uniparental disomy of 7q (UPD7q). Somatic SAMD9/9L mutations are acquired in cis (on the same allelle) and are thought to exert a LOF effect and “neutralize” the GOF germline mutation, as documented in cellular growth assays [119,121,123,124,131,134,142]. Missense somatic SAMD9/9L are rarely encountered but were also shown to modify germline mutant function [121]. A true genetic reversion with replacement of the mutant SAMD9/9L allele via UPD7q has been thus far reported in 11 patients who experienced spontaneous remission [121], [122], [123], [124], [125], [141], [142], [143]. This reversion likely arises through non-allelic homologous recombination in a del(7q) clone, where the wild type 7q arm is duplicated. Strikingly, the reversion seems to be definitive, as shown in patients who normalized their blood counts and marrow cellularity with normal findings up to over 20 years after diagnosis [123,125]. UPD7q can be considered a protective mechanism against the development of MDS, but it is not clear how it arises and how it outcompetes −7/del(7q) clones.
Therapeutic considerations
SAMD9/9L are newly described MDS predisposition syndromes where clinical outcome data is derived from retrospective studies and no guidelines exist on prospective management. The current practice for patients mild haematological phenotypes is guided by the morphological subtype (as recommended by the EWOG-MDS working group). For example, patients with RCC without severe neutropenia and no transfusion dependency can be followed with a watch & wait strategy with periodic assessment of blood counts and yearly marrow evaluation aiming at detection of high-risk somatic changes (−7, somatic driver mutations). A very careful consideration must be given to patients with SAMD9/9L syndromes and −7, where not enough data exists to deviate from the general recommendation for paediatric MDS with −7 where HSCT is performed in a timely manner [102]. Children with severe multi-organ involvement in context of MIRAGE syndrome who received HSCT were shown to have rather poor outcome complicated by syndrome-related comorbidities [128,145]. On the other hand, children with SAMD9/9L germline mutations who were transplanted for MDS had satisfactory outcomes with a 5 year OS of 85% [37]. Going forward one might speculate that young children with clinically stable disease (MDS and −7 without severe cytopenias and without somatic leukemia mutations) might benefit from careful watching with repeated molecular studies to document loss of monosomy 7 clone and emergence of revertant UPD7q clones. At the same time however, the patients might be exposed to the risk of clonal evolution to a more advanced disease where HSCT outcome might be inferior compared to initial disease state.
Practice points
-
•
Recently described autosomal dominant syndromes predisposing to myeloid neoplasms often manifest without preexisting features or family history and show variable expressivity and incomplete penetrance
-
•
GATA2 and SAMD9/SAMD9L syndromes are most common germline drivers of paediatric MDS and account for at least half of paediatric MDS with monosomy 7
-
•
HSCT is indicated in patients with transfusion dependency, neutropenia, immunodeficiency, morphologically advanced disease, and high-risk cytogenetic and genetic lesions
-
•
GATA2 deficiency is a highly penetrant disease with progressive course necessitating HSCT, while SAMD9/SAMD9L syndromes can show diverse outcomes ranging from spontaneous remission (in young children) to clonal progression.
Research agenda
-
•
Prospective monitoring of patients with hereditary predisposition to MN might reveal risk factors for clonal evolution
-
•
It remains to be answered if a careful watch & wait strategy in stable patients with SAMD9/9L-related MDS and monosomy 7 might identify patients with spontaneous genetic reversion and disappearance of monosomy 7, and eventually become a standard approach.
-
•
Collaborative studies are required to address the question of incomplete penetrance in syndromes predisposing to MN.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by ALSAC/St. Jude Children's Research Hospital, Fritz-Thyssen Foundation 10.17.1.026MN (to MWW), Deutsche Kinderkrebsstifung DKS 2017.03, ERAPerMed by German Federal Ministry of Education and Research (BMBF) 2018-123/01KU1904, and Deutsche Krebshilfe 109005 (to MWW), Baden-Wuerttemberg LGFG stipend (to EJK), BMBF MyPred Network for young individuals with syndromes predisposing to myeloid malignancies (to MWW). The authors thank the members of the Wlodarski lab and the St. Jude Bone Marrow Failure/MDS program, as well as Prof. Charlotte Niemeyer and the EWOG-MDS Coordinating Study Center Freiburg for helpful discussions.
References
- 1.Böhles H. Historische Fälle aus der Medizin: erstbeschreibungen von der Ahornsiruperkrankung bis zum Pfeifferschen Drüsenfieber. Springer; Berlin, Heidelberg: 2020. Die perniziöse Fanconi-Anämie, 1927; pp. 249–259. Berlin Heidelberg. [Google Scholar]
- 2.Skokowa J., Dale D.C., Touw I.P., Zeidler C., Welte K. Severe congenital neutropenias. Nat Rev Dis Primers. 2017;3:17032. doi: 10.1038/nrdp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boocock G.R., Morrison J.A., Popovic M., Richards N., Ellis L., Durie P.R., et al. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 4.Niewisch M.R., Savage S.A. An update on the biology and management of dyskeratosis congenita and related telomere biology disorders. Expet Rev Hematol. 2019;12:1037–1052. doi: 10.1080/17474086.2019.1662720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida K., Toki T., Okuno Y., Kanezaki R., Shiraishi Y., Sato-Otsubo A., et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 6.Niemeyer C.M., Kang M.W., Shin D.H., Furlan I., Erlacher M., Bunin N.J., et al. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niemeyer C.M. RAS diseases in children. Haematologica. 2014;99:1653–1662. doi: 10.3324/haematol.2014.114595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altmuller F., Lissewski C., Bertola D., Flex E., Stark Z., Spranger S., et al. Genotype and phenotype spectrum of NRAS germline variants. Eur J Hum Genet. 2017;25:823–831. doi: 10.1038/ejhg.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneider M., Chandler K., Tischkowitz M., Meyer S. Fanconi anaemia: genetics, molecular biology, and cancer - implications for clinical management in children and adults. Clin Genet. 2015;88:13–24. doi: 10.1111/cge.12517. [DOI] [PubMed] [Google Scholar]
- 10.Tummala H., Dokal A.D., Walne A., Ellison A., Cardoso S., Amirthasigamanipillai S., et al. Genome instability is a consequence of transcription deficiency in patients with bone marrow failure harboring biallelic ERCC6L2 variants. Proc Natl Acad Sci U S A. 2018;115:7777–7782. doi: 10.1073/pnas.1803275115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman M.H., Bonilla M.A., Fier C., Bolyard A.A., Scarlata D., Boxer L.A., et al. Myelodysplasia syndrome and acute myeloid leukemia in patients with congenital neutropenia receiving G-CSF therapy. Blood. 2000;96:429–436. [PubMed] [Google Scholar]
- 12.Germeshausen M., Ballmaier M., Welte K. Incidence of CSF3R mutations in severe congenital neutropenia and relevance for leukemogenesis: results of a long-term survey. Blood. 2007;109:93–99. doi: 10.1182/blood-2006-02-004275. [DOI] [PubMed] [Google Scholar]
- 13.Shimamura A., Alter B.P. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cave H., Caye A., Strullu M., Aladjidi N., Vignal C., Ferster A., et al. Acute lymphoblastic leukemia in the context of RASopathies. Eur J Med Genet. 2016;59:173–178. doi: 10.1016/j.ejmg.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Peffault de Latour R., Soulier J. How I treat MDS and AML in Fanconi anemia. Blood. 2016;127:2971–2979. doi: 10.1182/blood-2016-01-583625. [DOI] [PubMed] [Google Scholar]
- 16.Douglas S.P.M., Siipola P., Kovanen P.E., Pyorala M., Kakko S., Savolainen E.R., et al. ERCC6L2 defines a novel entity within inherited acute myeloid leukemia. Blood. 2019;133:2724–2728. doi: 10.1182/blood-2019-01-896233. [DOI] [PubMed] [Google Scholar]
- 17.Quentin S., Cuccuini W., Ceccaldi R., Nibourel O., Pondarre C., Pages M.P., et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- 18.Klimiankou M., Mellor-Heineke S., Zeidler C., Welte K., Skokowa J. Role of CSF3R mutations in the pathomechanism of congenital neutropenia and secondary acute myeloid leukemia. Ann N Y Acad Sci. 2016;1370:119–125. doi: 10.1111/nyas.13097. [DOI] [PubMed] [Google Scholar]
- 19.Germeshausen M., Kratz C.P., Ballmaier M., Welte K. RAS and CSF3R mutations in severe congenital neutropenia. Blood. 2009;114:3504–3505. doi: 10.1182/blood-2009-07-232512. [DOI] [PubMed] [Google Scholar]
- 20.Olofsen P.A., Touw I.P. RUNX1 mutations in the leukemic progression of severe congenital neutropenia. Mol Cell. 2020;43:139–144. doi: 10.14348/molcells.2020.0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy A.L., Myers K.C., Bowman J., Gibson C.J., Camarda N.D., Furutani E., et al. Distinct genetic pathways define pre-leukemic and compensatory clonal hematopoiesis in Shwachman-Diamond syndrome. bioRxiv. 2020;2020 doi: 10.1038/s41467-021-21588-4. 06.04.134692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bezzerri V., Cipolli M. Shwachman diamond syndrome: molecular mechanisms and current perspectives. Mol Diagn Ther. 2019;23:281–290. doi: 10.1007/s40291-018-0368-2. [DOI] [PubMed] [Google Scholar]
- 23.Jongmans M.C., van der Burgt I., Hoogerbrugge P.M., Noordam K., Yntema H.G., Nillesen W.M., et al. Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. Eur J Hum Genet. 2011;19:870–874. doi: 10.1038/ejhg.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lo Ten Foe J.R., Kwee M.L., Rooimans M.A., Oostra A.B., Veerman A.J., van Weel M., et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5:137–148. [PubMed] [Google Scholar]
- 25.Gross M., Hanenberg H., Lobitz S., Friedl R., Herterich S., Dietrich R., et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98:126–135. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- 26.Asur R.S., Kimble D.C., Lach F.P., Jung M., Donovan F.X., Kamat A., et al. Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype. Mol Genet Genomic Med. 2018;6:77–91. doi: 10.1002/mgg3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jongmans M.C.J., Verwiel E.T.P., Heijdra Y., Vulliamy T., Kamping E.J., Hehir-Kwa J.Y., et al. Revertant somatic mosaicism by mitotic recombination in dyskeratosis congenita. Am J Hum Genet. 2012;90:426–433. doi: 10.1016/j.ajhg.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song W.J., Sullivan M.G., Legare R.D., Hutchings S., Tan X., Kufrin D., et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 29.Smith M.L., Cavenagh J.D., Lister T.A., Fitzgibbon J. Mutation of CEBPA in familial acute myeloid leukemia. N Engl J Med. 2004;351:2403–2407. doi: 10.1056/NEJMoa041331. [DOI] [PubMed] [Google Scholar]
- 31.Galera P., Dulau-Florea A., Calvo K.R. Inherited thrombocytopenia and platelet disorders with germline predisposition to myeloid neoplasia. Int J Lab Hematol. 2019;41(Suppl 1):131–141. doi: 10.1111/ijlh.12999. [DOI] [PubMed] [Google Scholar]
- 32.Tawana K., Wang J., Renneville A., Bodor C., Hills R., Loveday C., et al. Disease evolution and outcomes in familial AML with germline CEBPA mutations. Blood. 2015;126:1214–1223. doi: 10.1182/blood-2015-05-647172. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M.Y., Churpek J.E., Keel S.B., Walsh T., Lee M.K., Loeb K.R., et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polprasert C., Schulze I., Sekeres M.A., Makishima H., Przychodzen B., Hosono N., et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Canc Cell. 2015;27:658–670. doi: 10.1016/j.ccell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duployez N., Goursaud L., Fenwarth L., Bories C., Marceau-Renaut A., Boyer T., et al. Familial myeloid malignancies with germline TET2 mutation. Leukemia. 2020;34:1450–1453. doi: 10.1038/s41375-019-0675-6. [DOI] [PubMed] [Google Scholar]
- 36.Brown A.L., Hahn C.N., Scott H.S. Secondary leukemia in patients with germline transcription factor mutations (RUNX1, GATA2, CEBPA) Blood. 2020;136:24–35. doi: 10.1182/blood.2019000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahoo SS, Pastor VP, Panda PK, Szvetnik EK, Kozyra EJ, Voss R, et al. SAMD9 and SAMD9L germline disorders in patients Enrolled in studies of the European working group of MDS in childhood (EWOG-MDS): prevalence, outcome, phenotype and functional characterisation. Blood. 2018;132(1):613. doi: 10.1182/blood-2018-99-118389. [DOI] [Google Scholar]
- 38.Hirabayashi S., Wlodarski M.W., Kozyra E., Niemeyer C.M. Heterogeneity of GATA2-related myeloid neoplasms. Int J Hematol. 2017;106:175–182. doi: 10.1007/s12185-017-2285-2. [DOI] [PubMed] [Google Scholar]
- 39.Wlodarski M.W., Collin M., Horwitz M.S. GATA2 deficiency and related myeloid neoplasms. Semin Hematol. 2017;54:81–86. doi: 10.1053/j.seminhematol.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai F.Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 41.Persons D.A., Allay J.A., Allay E.R., Ashmun R.A., Orlic D., Jane S.M., et al. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood. 1999;93:488–499. [PubMed] [Google Scholar]
- 42.Lim K.C., Hosoya T., Brandt W., Ku C.J., Hosoya-Ohmura S., Camper S.A., et al. Conditional Gata2 inactivation results in HSC loss and lymphatic mispatterning. J Clin Invest. 2012;122:3705–3717. doi: 10.1172/JCI61619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Pater E., Kaimakis P., Vink C.S., Yokomizo T., Yamada-Inagawa T., van der Linden R., et al. Gata2 is required for HSC generation and survival. J Exp Med. 2013;210:2843–2850. doi: 10.1084/jem.20130751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Menendez-Gonzalez J.B., Vukovic M., Abdelfattah A., Saleh L., Almotiri A., Thomas L.A., et al. Gata2 as a crucial regulator of stem cells in adult hematopoiesis and acute myeloid leukemia. J Stem Cell Res Rev Rep. 2019;13:291–306. doi: 10.1016/j.stemcr.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsu A.P., Sampaio E.P., Khan J., Calvo K.R., Lemieux J.E., Patel S.Y., et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickinson R.E., Griffin H., Bigley V., Reynard L.N., Hussain R., Haniffa M., et al. Exome sequencing identifies GATA-2 mutation as the cause of dendritic cell, monocyte, B and NK lymphoid deficiency. Blood. 2011;118:2656–2658. doi: 10.1182/blood-2011-06-360313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostergaard P., Simpson M.A., Connell F.C., Steward C.G., Brice G., Woollard W.J., et al. Mutations in GATA2 cause primary lymphedema associated with a predisposition to acute myeloid leukemia (Emberger syndrome) Nat Genet. 2011;43:929–931. doi: 10.1038/ng.923. [DOI] [PubMed] [Google Scholar]
- 48.Pasquet M., Bellanne-Chantelot C., Tavitian S., Prade N., Beaupain B., Larochelle O., et al. High frequency of GATA2 mutations in patients with mild chronic neutropenia evolving to MonoMac syndrome, myelodysplasia, and acute myeloid leukemia. Blood. 2013;121:822–829. doi: 10.1182/blood-2012-08-447367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hahn C.N., Chong C.E., Carmichael C.L., Wilkins E.J., Brautigan P.J., Li X.C., et al. Heritable GATA2 mutations associated with familial myelodysplastic syndrome and acute myeloid leukemia. Nat Genet. 2011;43:1012–1017. doi: 10.1038/ng.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spinner M.A., Sanchez L.A., Hsu A.P., Shaw P.A., Zerbe C.S., Calvo K.R., et al. GATA2 deficiency: a protean disorder of hematopoiesis, lymphatics, and immunity. Blood. 2014;123:809–821. doi: 10.1182/blood-2013-07-515528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West R.R., Hsu A.P., Holland S.M., Cuellar-Rodriguez J., Hickstein D.D. Acquired ASXL1 mutations are common in patients with inherited GATA2 mutations and correlate with myeloid transformation. Haematologica. 2014;99:276–281. doi: 10.3324/haematol.2013.090217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wlodarski M.W., Hirabayashi S., Pastor V., Starý J., Hasle H., Masetti R., et al. Prevalence, clinical characteristics, and prognosis of GATA2-related myelodysplastic syndromes in children and adolescents. Blood. 2016;127:1387–1397. doi: 10.1182/blood-2015-09-669937. [DOI] [PubMed] [Google Scholar]
- 53.Donadieu J., Lamant M., Fieschi C., de Fontbrune F.S., Caye A., Ouachee M., et al. Natural history of GATA2 deficiency in a survey of 79 French and Belgian patients. Haematologica. 2018;103:1278–1287. doi: 10.3324/haematol.2017.181909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bodor C., Renneville A., Smith M., Charazac A., Iqbal S., Etancelin P., et al. Germ-line GATA2 p.THR354MET mutation in familial myelodysplastic syndrome with acquired monosomy 7 and ASXL1 mutation demonstrating rapid onset and poor survival. Haematologica-the Hematology Journal. 2012;97:890–894. doi: 10.3324/haematol.2011.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holme H., Hossain U., Kirwan M., Walne A., Vulliamy T., Dokal I. Marked genetic heterogeneity in familial myelodysplasia/acute myeloid leukaemia. Br J Haematol. 2012;158:242–248. doi: 10.1111/j.1365-2141.2012.09136.x. [DOI] [PubMed] [Google Scholar]
- 56.Dickinson R.E., Milne P., Jardine L., Zandi S., Swierczek S.I., McGovern N., et al. The evolution of cellular deficiency in GATA2 mutation. Blood. 2014;123:863–874. doi: 10.1182/blood-2013-07-517151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brambila-Tapia A.J.L., Garcia-Ortiz J.E., Brouillard P., Nguyen H.L., Vikkula M., Rios-Gonzalez B.E., et al. GATA2 null mutation associated with incomplete penetrance in a family with Emberger syndrome. Hematology. 2017;22:467–471. doi: 10.1080/10245332.2017.1294551. [DOI] [PubMed] [Google Scholar]
- 58.McReynolds L.J., Yang Y., Yuen Wong H., Tang J., Zhang Y., Mule M.P., et al. MDS-associated mutations in germline GATA2 mutated patients with hematologic manifestations. Leuk Res. 2019;76:70–75. doi: 10.1016/j.leukres.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al Seraihi A.F., Rio-Machin A., Tawana K., Bodor C., Wang J., Nagano A., et al. GATA2 monoallelic expression underlies reduced penetrance in inherited GATA2-mutated MDS/AML. Leukemia. 2018;32(11):2502–2507. doi: 10.1038/s41375-018-0134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim N., Choi S., Kim S.M., Lee A.C., Im K., Park H.S., et al. Monozygotic twins with shared de novo GATA2 mutation but dissimilar phenotypes due to differential promoter methylation. Leuk Lymphoma. 2019;60:1053–1061. doi: 10.1080/10428194.2018.1516039. [DOI] [PubMed] [Google Scholar]
- 61.Kazenwadel J., Secker G.A., Liu Y.J., Rosenfeld J.A., Wildin R.S., Cuellar-Rodriguez J., et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood. 2012;119:1283–1291. doi: 10.1182/blood-2011-08-374363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Novakova M., Zaliova M., Sukova M., Wlodarski M., Janda A., Fronkova E., et al. Loss of B cells and their precursors is the most constant feature of GATA-2 deficiency in childhood myelodysplastic syndrome. Haematologica. 2016;101:707–716. doi: 10.3324/haematol.2015.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang M.Y., Keel S.B., Walsh T., Lee M.K., Gulsuner S., Watts A.C., et al. Genomic analysis of bone marrow failure and myelodysplastic syndromes reveals phenotypic and diagnostic complexity. Haematologica. 2015;100:42–48. doi: 10.3324/haematol.2014.113456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganapathi K.A., Townsley D.M., Hsu A.P., Arthur D.C., Zerbe C.S., Cuellar-Rodriguez J., et al. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood. 2015;125(1):56–70. doi: 10.1182/blood-2014-06-580340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maciejewski-Duval A., Meuris F., Bignon A., Aknin M.L., Balabanian K., Faivre L., et al. Altered chemotactic response to CXCL12 in patients carrying GATA2 mutations. J Leukoc Biol. 2016;99:1065–1076. doi: 10.1189/jlb.5MA0815-388R. [DOI] [PubMed] [Google Scholar]
- 66.Dotta L., Badolato R. Primary immunodeficiencies appearing as combined lymphopenia, neutropenia, and monocytopenia. Immunol Lett. 2014;161:222–225. doi: 10.1016/j.imlet.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 67.Calvo K.R., Vinh D.C., Maric I., Wang W., Noel P., Stetler-Stevenson M., et al. Myelodysplasia in autosomal dominant and sporadic monocytopenia immunodeficiency syndrome: diagnostic features and clinical implications. Haematologica. 2011;96:1221–1225. doi: 10.3324/haematol.2011.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parta M., Shah N.N., Baird K., Rafei H., Calvo K.R., Hughes T., et al. Allogeneic hematopoietic stem cell transplantation for GATA2 deficiency using a busulfan-based regimen. Biol Blood Marrow Transplant. 2018;24:1250–1259. doi: 10.1016/j.bbmt.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bigley V., Haniffa M., Doulatov S., Wang X.N., Dickinson R., McGovern N., et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011;208:227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb G., Chen Y.Y., Li K.K., Neil D., Oo Y.H., Richter A., et al. Single-gene association between GATA-2 and autoimmune hepatitis: a novel genetic insight highlighting immunologic pathways to disease. J Hepatol. 2016;64:1190–1193. doi: 10.1016/j.jhep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fakhri B., Cashen A.F., Duncavage E.J., Watkins M.P., Wartman L.D., Bartlett N.L. Fifty Shades of GATA2 mutation: a case of plasmablastic lymphoma, nontuberculous mycobacterial infection, and myelodysplastic syndrome. Clin Lymphoma, Myeloma & Leukemia. 2019;19:e532–e535. doi: 10.1016/j.clml.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 72.Hsu A.P., Johnson K.D., Falcone E.L., Sanalkumar R., Sanchez L., Hickstein D.D., et al. GATA2 haploinsufficiency caused by mutations in a conserved intronic element leads to MonoMAC syndrome. Blood. 2013;121:3830–3837. doi: 10.1182/blood-2012-08-452763. S1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hahn C.N., Brautigan P.J., Chong C.E., Janssan A., Venugopal P., Lee Y., et al. Characterisation of a compound in-cis GATA2 germline mutation in a pedigree presenting with myelodysplastic syndrome/acute myeloid leukemia with concurrent thrombocytopenia. Leukemia. 2015;29:1795–1797. doi: 10.1038/leu.2015.40. [DOI] [PubMed] [Google Scholar]
- 74.Cortes-Lavaud X., Landecho M.F., Maicas M., Urquiza L., Merino J., Moreno-Miralles I., et al. GATA2 germline mutations impair GATA2 transcription, causing haploinsufficiency: functional analysis of the p.Arg396Gln mutation. J Immunol. 2015;194:2190–2198. doi: 10.4049/jimmunol.1401868. [DOI] [PubMed] [Google Scholar]
- 75.Chong C.E., Venugopal P., Stokes P.H., Lee Y.K., Brautigan P.J., Yeung D.T.O., et al. Differential effects on gene transcription and hematopoietic differentiation correlate with GATA2 mutant disease phenotypes. Leukemia. 2018;32:194–202. doi: 10.1038/leu.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sologuren I., Martinez-Saavedra M.T., Sole-Violan J., de Borges de Oliveira E., Jr., Betancor E., Casas I., et al. Lethal influenza in two related adults with inherited GATA2 deficiency. J Clin Immunol. 2018;38(4):513–526. doi: 10.1007/s10875-018-0512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wehr C., Grotius K., Casadei S., Bleckmann D., Bode S.F.N., Frye B.C., et al. A novel disease-causing synonymous exonic mutation in GATA2 affecting RNA splicing. Blood. 2018;132:1211–1215. doi: 10.1182/blood-2018-03-837336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cavalcante de Andrade Silva M., Katsumura K.R., Mehta C., Velloso E., Bresnick E.H., Godley L.A. Breaking the spatial constraint between neighboring zinc fingers: a new germline mutation in GATA2 deficiency syndrome. Leukemia. 2020;Apr 14(Online ahead of print) doi: 10.1038/s41375-020-0820-2. [DOI] [PubMed] [Google Scholar]
- 81.Kozyra E.J., Pastor V.B., Lefkopoulos S., Sahoo S.S., Busch H., Voss R.K., et al. Synonymous GATA2 mutations result in selective loss of mutated RNA and are common in patients with GATA2 deficiency. Leukemia. 2020;Jun 18(Online ahead of print) doi: 10.1038/s41375-020-0899-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fox L.C., Tan M., Brown A.L., Arts P., Thompson E., Ryland G.L., et al. A synonymous GATA2 variant underlying familial myeloid malignancy with striking intrafamilial phenotypic variability. Br J Haematol. 2020;Jun 3(Online ahead of print) doi: 10.1111/bjh.16819. [DOI] [PubMed] [Google Scholar]
- 83.Tien F.M., Hou H.A., Tsai C.H., Tang J.L., Chiu Y.C., Chen C.Y., et al. GATA2 zinc finger 1 mutations are associated with distinct clinico-biological features and outcomes different from GATA2 zinc finger 2 mutations in adult acute myeloid leukemia. Blood Canc J. 2018;8:87. doi: 10.1038/s41408-018-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang S.J., Ma L.Y., Huang Q.H., Li G., Gu B.W., Gao X.D., et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2008;105:2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Greif P.A., Dufour A., Konstandin N.P., Ksienzyk B., Zellmeier E., Tizazu B., et al. GATA2 zinc finger 1 mutations associated with biallelic CEBPA mutations define a unique genetic entity of acute myeloid leukemia. Blood. 2012;120:395–403. doi: 10.1182/blood-2012-01-403220. [DOI] [PubMed] [Google Scholar]
- 86.Luesink M., Hollink I.H., van der Velden V.H., Knops R.H., Boezeman J.B., de Haas V., et al. High GATA2 expression is a poor prognostic marker in pediatric acute myeloid leukemia. Blood. 2012;120:2064–2075. doi: 10.1182/blood-2011-12-397083. [DOI] [PubMed] [Google Scholar]
- 87.Niimi K., Kiyoi H., Ishikawa Y., Hayakawa F., Kurahashi S., Kihara R., et al. GATA2 zinc finger 2 mutation found in acute myeloid leukemia impairs myeloid differentiation. Leuk Res. 2013;2:21–25. doi: 10.1016/j.lrr.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fasan A., Eder C., Haferlach C., Grossmann V., Kohlmann A., Dicker F., et al. GATA2 mutations are frequent in intermediate-risk karyotype AML with biallelic CEBPA mutations and are associated with favorable prognosis. Leukemia. 2013;27:482–485. doi: 10.1038/leu.2012.174. [DOI] [PubMed] [Google Scholar]
- 89.Green C.L., Tawana K., Hills R.K., Bodor C., Fitzgibbon J., Inglott S., et al. GATA2 mutations in sporadic and familial acute myeloid leukaemia patients with CEBPA mutations. Br J Haematol. 2013;161:701–705. doi: 10.1111/bjh.12317. [DOI] [PubMed] [Google Scholar]
- 90.Shiba N., Funato M., Ohki K., Park M.J., Mizushima Y., Adachi S., et al. Mutations of the GATA2 and CEBPA genes in paediatric acute myeloid leukaemia. Br J Haematol. 2014;164:142–145. doi: 10.1111/bjh.12559. [DOI] [PubMed] [Google Scholar]
- 91.Hou H.A., Lin Y.C., Kuo Y.Y., Chou W.C., Lin C.C., Liu C.Y., et al. GATA2 mutations in patients with acute myeloid leukemia-paired samples analyses show that the mutation is unstable during disease evolution. Ann Hematol. 2015;94(2):211–221. doi: 10.1007/s00277-014-2208-8. [DOI] [PubMed] [Google Scholar]
- 92.Theis F., Corbacioglu A., Gaidzik V.I., Paschka P., Weber D., Bullinger L., et al. Clinical impact of GATA2 mutations in acute myeloid leukemia patients harboring CEBPA mutations: a study of the AML study group. Leukemia. 2016;30:2248–2250. doi: 10.1038/leu.2016.185. [DOI] [PubMed] [Google Scholar]
- 93.Ping N., Sun A., Song Y., Wang Q., Yin J., Cheng W., et al. Exome sequencing identifies highly recurrent somatic GATA2 and CEBPA mutations in acute erythroid leukemia. Leukemia. 2017;31:195–202. doi: 10.1038/leu.2016.162. [DOI] [PubMed] [Google Scholar]
- 94.Sekhar M., Pocock R., Lowe D., Mitchell C., Marafioti T., Dickinson R., et al. Can somatic GATA2 mutation mimic germ line GATA2 mutation? Adv Blood Grouping. 2018;2:904–908. doi: 10.1182/bloodadvances.2017012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alfayez M., Wang S.A., Bannon S.A., Kontoyiannis D.P., Kornblau S.M., Orange J.S., et al. Myeloid malignancies with somatic GATA2 mutations can be associated with an immunodeficiency phenotype. Leuk Lymphoma. 2019;60:2025–2033. doi: 10.1080/10428194.2018.1551535. [DOI] [PubMed] [Google Scholar]
- 96.Loyola V.B.P., Hirabayashi S., Pohl S., Kozyra E.J., Catala A., De Moerloose B., et al. Somatic genetic and epigenetic architecture of myelodysplastic syndromes arising from GATA2 deficiency. Blood. 2015;126(23) doi: 10.1182/blood.V126.23.299.299. 299–299. [DOI] [Google Scholar]
- 97.Wang X.A., Muramatsu H., Okuno Y., Sakaguchi H., Yoshida K., Kawashima N., et al. GATA2 and secondary mutations in familial myelodysplastic syndromes and pediatric myeloid malignancies. Haematologica. 2015;100:E398–E401. doi: 10.3324/haematol.2015.127092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ding L.W., Ikezoe T., Tan K.T., Mori M., Mayakonda A., Chien W., et al. Mutational profiling of a MonoMAC syndrome family with GATA2 deficiency. Leukemia. 2017;31:244–245. doi: 10.1038/leu.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fisher K.E., Hsu A.P., Williams C.L., Sayeed H., Merritt B.Y., Elghetany M.T., et al. Somatic mutations in children with GATA2-associated myelodysplastic syndrome who lack other features of GATA2 deficiency. Adv Blood Grouping. 2017;1:443–448. doi: 10.1182/bloodadvances.2016002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pastor V., Hirabayashi S., Karow A., Wehrle J., Kozyra E., Nienhold R., et al. Mutational landscape in children with myelodysplastic syndromes is distinct from adults: specific somatic drivers and novel germline variants. Leukemia. 2017;31:759. doi: 10.1038/leu.2016.342. [DOI] [PubMed] [Google Scholar]
- 101.Fujiwara T., Fukuhara N., Funayama R., Nariai N., Kamata M., Nagashima T., et al. Identification of acquired mutations by whole-genome sequencing in GATA-2 deficiency evolving into myelodysplasia and acute leukemia. Ann Hematol. 2014;93:1515–1522. doi: 10.1007/s00277-014-2090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niemeyer C.M. In: The EBMT handbook: hematopoietic stem cell transplantation and cellular therapies. th Carreras E., Dufour C., Mohty M., Kroger N., editors. 2019. Pediatric MDS including refractory cytopenia and juvenile myelomonocytic leukemia; pp. 557–560. Cham (CH) [PubMed] [Google Scholar]
- 103.Cuellar-Rodriguez J., Gea-Banacloche J., Freeman A.F., Hsu A.P., Zerbe C.S., Calvo K.R., et al. Successful allogeneic hematopoietic stem cell transplantation for GATA2 deficiency. Blood. 2011;118:3715–3720. doi: 10.1182/blood-2011-06-365049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Maeurer M., Magalhaes I., Andersson J., Ljungman P., Sandholm E., Uhlin M., et al. Allogeneic hematopoietic cell transplantation for GATA2 deficiency in a patient with disseminated human Papillomavirus disease. Transplantation. 2014;98:e95–e96. doi: 10.1097/TP.0000000000000520. [DOI] [PubMed] [Google Scholar]
- 105.Tholouli E., Sturgess K., Dickinson R.E., Gennery A., Cant A.J., Jackson G., et al. In vivo T-depleted reduced-intensity transplantation for GATA2-related immune dysfunction. Blood. 2018;131:1383–1387. doi: 10.1182/blood-2017-10-811489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simonis A., Fux M., Nair G., Mueller N.J., Haralambieva E., Pabst T., et al. Allogeneic hematopoietic cell transplantation in patients with GATA2 deficiency-a case report and comprehensive review of the literature. Ann Hematol. 2018;97:1961–1973. doi: 10.1007/s00277-018-3388-4. [DOI] [PubMed] [Google Scholar]
- 107.Bogaert D.J., Laureys G., Naesens L., Mazure D., De Bruyne M., Hsu A.P., et al. GATA2 deficiency and haematopoietic stem cell transplantation: challenges for the clinical practitioner. Br J Haematol. 2020;188:768–773. doi: 10.1111/bjh.16247. [DOI] [PubMed] [Google Scholar]
- 108.Grossman J., Cuellar-Rodriguez J., Gea-Banacloche J., Zerbe C., Calvo K., Hughes T., et al. Nonmyeloablative allogeneic hematopoietic stem cell transplantation for GATA2 deficiency. Biol Blood Marrow Transplant. 2014;20(12):1940–1948. doi: 10.1016/j.bbmt.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu J., Wennier S., Zhang L., McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol. 2011;85:3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meng X., Krumm B., Li Y., Deng J., Xiang Y. Structural basis for antagonizing a host restriction factor by C7 family of poxvirus host-range proteins. Proc Natl Acad Sci U S A. 2015;112:14858–14863. doi: 10.1073/pnas.1515354112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nounamo B., Li Y., O'Byrne P., Kearney A.M., Khan A., Liu J. An interaction domain in human SAMD9 is essential for myxoma virus host-range determinant M062 antagonism of host anti-viral function. Virology. 2017;503:94–102. doi: 10.1016/j.virol.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meng X., Zhang F., Yan B., Si C., Honda H., Nagamachi A., et al. A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Asou H., Matsui H., Ozaki Y., Nagamachi A., Nakamura M., Aki D., et al. Identification of a common microdeletion cluster in 7q21.3 subband among patients with myeloid leukemia and myelodysplastic syndrome. Biochem Biophys Res Commun. 2009;383:245–251. doi: 10.1016/j.bbrc.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 114.Wu L., Pu X., Wang Q., Cao J., Xu F., Xu L.I., et al. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Topaz O., Indelman M., Chefetz I., Geiger D., Metzker A., Altschuler Y., et al. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet. 2006;79:759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chefetz I., Ben Amitai D., Browning S., Skorecki K., Adir N., Thomas M.G., et al. Normophosphatemic familial tumoral calcinosis is caused by deleterious mutations in SAMD9, encoding a TNF-alpha responsive protein. J Invest Dermatol. 2008;128:1423–1429. doi: 10.1038/sj.jid.5701203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chen D.H., Below J.E., Shimamura A., Keel S.B., Matsushita M., Wolff J., et al. Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am J Hum Genet. 2016;98:1146–1158. doi: 10.1016/j.ajhg.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Narumi S., Amano N., Ishii T., Katsumata N., Muroya K., Adachi M., et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48:792–797. doi: 10.1038/ng.3569. [DOI] [PubMed] [Google Scholar]
- 119.Buonocore F., Kuhnen P., Suntharalingham J.P., Del Valle I., Digweed M., Stachelscheid H., et al. Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest. 2017;127(5):1700–1713. doi: 10.1172/JCI91913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nagamachi A., Matsui H., Asou H., Ozaki Y., Aki D., Kanai A., et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Canc Cell. 2013;24:305–317. doi: 10.1016/j.ccr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 121.Tesi B., Davidsson J., Voss M., Rahikkala E., Holmes T.D., Chiang S.C., et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS and neurological symptoms. Blood. 2017;129(16):2266–2279. doi: 10.1182/blood-2016-10-743302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schwartz J.R., Ma J., Lamprecht T., Walsh M., Wang S., Bryant V., et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8:1557. doi: 10.1038/s41467-017-01590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pastor V.B., Sahoo S.S., Boklan J., Schwabe G.C., Saribeyoglu E., Strahm B., et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica. 2018;103:427–437. doi: 10.3324/haematol.2017.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wong J.C., Bryant V., Lamprecht T., Ma J., Walsh M., Schwartz J., et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 2018:3. doi: 10.1172/jci.insight.121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bluteau O., Sebert M., Leblanc T., Peffault de Latour R., Quentin S., Lainey E., et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131:717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 126.de Jesus A.A., Hou Y., Brooks S., Malle L., Biancotto A., Huang Y., et al. Distinct interferon signatures and cytokine patterns define additional systemic autoinflammatory diseases. J Clin Invest. 2020;130(4):1669–1682. doi: 10.1172/JCI129301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sarthy J., Zha J., Babushok D., Shenoy A., Fan J.M., Wertheim G., et al. Poor outcome with hematopoietic stem cell transplantation for bone marrow failure and MDS with severe MIRAGE syndrome phenotype. Adv Blood Grouping. 2018;2:120–125. doi: 10.1182/bloodadvances.2017012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jeffries L., Shima H., Ji W., Panisello-Manterola D., McGrath J., Bird L.M., et al. A novel SAMD9 mutation causing MIRAGE syndrome: an expansion and review of phenotype, dysmorphology, and natural history. Am J Med Genet. 2018;176:415–420. doi: 10.1002/ajmg.a.38557. [DOI] [PubMed] [Google Scholar]
- 130.Kim Y.M., Seo G.H., Kim G.H., Ko J.M., Choi J.H., Yoo H.W. A case of an infant suspected as IMAGE syndrome who were finally diagnosed with MIRAGE syndrome by targeted Mendelian exome sequencing. BMC Med Genet. 2018;19:35. doi: 10.1186/s12881-018-0546-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shima H., Koehler K., Nomura Y., Sugimoto K., Satoh A., Ogata T., et al. Two patients with MIRAGE syndrome lacking haematological features: role of somatic second-site reversion SAMD9 mutations. J Med Genet. 2018;55:81–85. doi: 10.1136/jmedgenet-2017-105020. [DOI] [PubMed] [Google Scholar]
- 132.Shima H., Hayashi M., Tachibana T., Oshiro M., Amano N., Ishii T., et al. MIRAGE syndrome is a rare cause of 46,XY DSD born SGA without adrenal insufficiency. PLoS One. 2018;13 doi: 10.1371/journal.pone.0206184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mengen E., Kucukcongar Yavas A. A rare Etiology of 46, XY disorder of Sex development and adrenal insufficiency: a case of MIRAGE syndrome caused by mutations in SAMD9 gene. J Clin Res Pediatr Endocrinol. 2019;12(2):206–211. doi: 10.4274/jcrpe.galenos.2019.2019.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Perisa M.P., Rose M.J., Varga E., Kamboj M.K., Spencer J.D., Bajwa R.P.S. A novel SAMD9 variant identified in patient with MIRAGE syndrome: further defining syndromic phenotype and review of previous cases. Pediatr Blood Canc. 2019;66 doi: 10.1002/pbc.27726. [DOI] [PubMed] [Google Scholar]
- 135.Formankova R., Kanderova V., Rackova M., Svaton M., Brdicka T., Riha P., et al. Novel SAMD9 mutation in a patient with immunodeficiency, neutropenia, impaired anti-CMV response, and severe gastrointestinal involvement. Front Immunol. 2019;10:2194. doi: 10.3389/fimmu.2019.02194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y., Zhang Y., Zhang V.W., Zhang C., Ding H., Yin A. Mutations in both SAMD9 and SLC19A2 genes caused complex phenotypes characterized by recurrent infection, dysphagia and profound deafness - a case report for dual diagnosis. BMC Pediatr. 2019;19:364. doi: 10.1186/s12887-019-1733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Roucher-Boulez F., Mallet D., Chatron N., Dijoud F., Gorduza D.B., Bretones P., et al. Reversion SAMD9 Mutations Modifying Phenotypic Expression of MIRAGE Syndrome and Allowing Inheritance in a Usually de novo Disorder. Front Endocrinol. 2019;10:625. doi: 10.3389/fendo.2019.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Onuma S., Wada T., Araki R., Wada K., Tanase-Nakao K., Narumi S., et al. MIRAGE syndrome caused by a novel missense variant (p.Ala1479Ser) in the SAMD9 gene. Hum Genome Var. 2020;7:4. doi: 10.1038/s41439-020-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Viaene A.N., Harding B.N. The neuropathology of MIRAGE syndrome. J Neuropathol Exp Neurol. 2020;79:458–462. doi: 10.1093/jnen/nlaa009. [DOI] [PubMed] [Google Scholar]
- 140.Thunstrom S., Axelsson M. Leukoencephalopathia, demyelinating peripheral neuropathy and dural ectasia explained by a not formerly described de novo mutation in the SAMD9L gene, ends 27 years of investigations - a case report. BMC Neurol. 2019;19:89. doi: 10.1186/s12883-019-1319-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cheah J.J.C., Brown A.L., Schreiber A.W., Feng J., Babic M., Moore S., et al. A novel germline SAMD9L mutation in a family with ataxia-pancytopenia syndrome and pediatric acute lymphoblastic leukemia. Haematologica. 2019;104:e318–e321. doi: 10.3324/haematol.2018.207316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schwartz J.R., Wang S., Ma J., Lamprecht T., Walsh M., Song G., et al. Germline SAMD9 mutation in siblings with monosomy 7 and myelodysplastic syndrome. Leukemia. 2017;31:1827–1830. doi: 10.1038/leu.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Csillag B., Ilencikova D., Meissl M., Webersinke G., Laccone F., Narumi S., et al. Somatic mosaic monosomy 7 and UPD7q in a child with MIRAGE syndrome caused by a novel SAMD9 mutation. Pediatr Blood Canc. 2019;66 doi: 10.1002/pbc.27589. [DOI] [PubMed] [Google Scholar]
- 144.Hockings C., Gohil S., Dowse R., Hoade Y., Mansour M.R., Gale R.E., et al. In trans early mosaic mutational escape and novel phenotypic features of germline SAMD9 mutation. Br J Haematol. 2020;188:e53–e57. doi: 10.1111/bjh.16322. [DOI] [PubMed] [Google Scholar]
- 145.Ahmed I.A., Farooqi M.S., Vander Lugt M.T., Boklan J., Rose M., Friehling E.D., et al. Outcomes of hematopoietic cell transplantation in patients with germline SAMD9/SAMD9L mutations. Biol Blood Marrow Transplant. 2019;25:2186–2196. doi: 10.1016/j.bbmt.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Amano N., Narumi S., Hayashi M., Takagi M., Imai K., Nakamura T., et al. Genetic defects in pediatric-onset adrenal insufficiency in Japan. Eur J Endocrinol. 2017;177:187–194. doi: 10.1530/EJE-17-0027. [DOI] [PubMed] [Google Scholar]
- 147.Wlodarski MW, Niemeyer CM. Introduction: Genetic syndromes predisposing to myeloid neoplasia. Semin Hematol. 2017;54(2):57–59. doi: 10.1053/j.seminhematol.2017.05.001. [DOI] [PubMed] [Google Scholar]