Abstract
The emergence and perseverance of drug resistant strains of Mycobacterium tuberculosis (Mtb) ensures that drug discovery efforts remain at the forefront of tuberculosis research. There are numerous different approaches that can be employed to lead to the discovery of anti-tubercular agents. In this work, we endeavored to optimize the anthraquinone chemical scaffold of a known drug, rhein, converting it from a compound with negligible activity against Mtb, to a series of compounds with potent activity. Two compounds exhibited low toxicity and good liver microsome stability and were further progressed in attempts to identify the biological target. Whole genome sequencing of resistant isolates revealed inactivating mutations in a monoglyceride lipase. Over-expression trials and an enzyme assay confirmed that the designed compounds are prodrugs, activated by the monoglyceride lipase. We propose that rhein is the active moiety of the novel compounds, which requires chemical modifications to enable access to the cell through the extensive cell wall structure. This work demonstrates that re-engineering of existing antimicrobial agents is a valid method in the development of new anti-tubercular compounds.
Keywords: Rhein, Mycobacterium, Lipase, Drug discovery
1. Introduction
Mycobacterium tuberculosis (Mtb), the etiological agent of tuberculosis (TB), is the ninth leading cause of death worldwide and is the primary cause of death from a single infectious agent. In 2018, an estimated 10 million people were diagnosed with the disease, which caused an estimated 1.4 million deaths. Efforts to reduce the incidence and mortality rates are being sought by the World Health Organization’s End TB Strategy and the United Nations’ Sustainable Development Goals, but it is apparent that progress is not sufficient to meet targets to ultimately end the global TB epidemic (World Health Organisation, 2019). The standard six-month treatment regimen for TB has remained unchanged over several decades, facilitating the development of drug resistance with an increasing prevalence of multi-drug resistant (MDR) and extensively-drug resistant (XDR) strains of Mtb. Currently there are 23 drugs in Phase I, II or III clinical trials, including new and repurposed drugs (World Health Organisation, 2019). In addition to early diagnosis, it is essential that new treatment strategies are developed to preserve the existing treatment program and to combat drug resistance.
Drug resistance can manifest by a number of different mechanisms including mutations to the target (Abrahams et al., 2016, Abrahams et al., 2012, Abrahams et al., 2017, Gurcha et al., 2014), transcriptional regulation (for example down-regulation of drug importers or up-regulation of exporters (Milano et al., 2009)), chemical modification (inactivating an inhibitor (Kasik, 1965) or preventing the conversion of a prodrug into an active compound (Rouse and Morris, 1995)). In order to establish the effectiveness of a drug, if possible, it is important to not only establish the target of the inhibitor but also to determine any associated resistance mechanisms that could be used to make the compound ineffective in a clinical setting. Thereby, decisions can be made as to whether to progress a compound in medicinal chemistry efforts or to employ alternative strategies to combat the resistance mechanism. For example, until recently, β-lactam antibiotics were considered to be ineffective against Mtb due to the expression of a β-lactamase. However, combination therapy with a β-lactam and a β-lactamase inhibitor has been shown to be bactericidal against replicating and non-replicating Mtb (Hugonnet et al., 2009, Rullas et al., 2015).
The continuing emergence of drug resistant strains of Mtb threatens to render current treatment programs ineffective, reinforcing the urgent requirement of new anti-tubercular agents. Innovative strategies are being sought to broaden the potential of developing or discovering the next front-line antibiotic. These include: re-purposing of existing approved drugs (Harbut et al., 2015, Maitra et al., 2015), such as known antibacterials; high-throughput phenotypic screening of extensive compound libraries (Ballell et al., 2013); target based screening (Moreira et al., 2015); optimizing the chemical scaffold of known drugs (Nigam et al., 2014). Given the global efforts in the search for new anti-tubercular agents, together with diversity of these approaches employed, it is anticipated that more compounds will make it into clinical trials for progression into the treatment regimen.
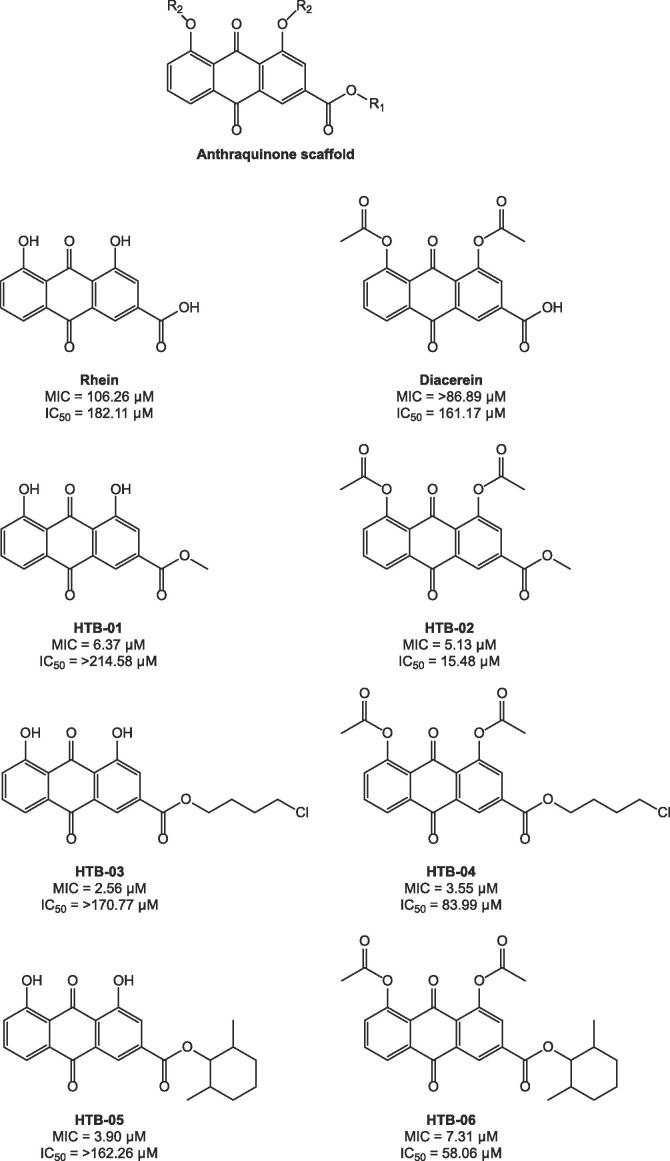
Re-purposing of existing antibacterial drug classes and structural modifications based on natural products with antibacterial activity are undoubtedly effective approaches to discover new anti-tubercular agents. In this work, we endeavored to optimize the chemical scaffold of the natural plant product rhein (Fig. 1). First isolated in 1895 from Chinese rhubarb, rhein has since been extracted from other plant species such as Cassia reticulata (Anchel, 1949). It has been shown to exhibit a multitude of medicinal properties including anti-cancer, anti-inflammatory, anti-diabetic and anti-microbial effects (Zhou et al., 2015). Consequently, rhein has an impact on a number of metabolic processes, examples of which are electron transport (Floridi et al., 1990), ATP production (Castiglione et al., 1990), and oxidative phosphorylation (Floridi et al., 1990) in eukaryotic cells. With relevance to its antimicrobial activity, rhein has been shown to inhibit Helicobacter pylori (Chung et al., 1998) and Staphylococcus aureus (Yu et al., 2008), although the mode of action is yet to be confirmed. Rhein does not exhibit anti-tubercular activity (Gajadeera et al., 2015). However, owing to the pharmacological potential rhein, we investigated whether structurally optimizing the anthraquinone core structure could convert an essentially inactive compound into a potent inhibitor against Mtb.
Fig. 1.
Chemical structures of the anthraquinone rhein, diacerein and synthesized derivatives. The Mtb MIC and cytotoxicity (Vero, IC50) of each compound is illustrated.
Herein, we show that the anthraquinone rhein and diacerein derivatives, HTB-03 and HTB-04, with a middle size 4-chloro-1-butyl ester group, demonstrated potent anti-TB activity, low toxicity and good liver microsome stability. Furthermore, we have determined that these compounds are prodrugs, which require activation by the monoglyceride lipase, Rv0183. Mycobacterium bovis BCG spontaneous resistant mutants raised against these compounds exhibit amino acid substitutions, deletion and STOP mutations in the Rv0183 homologue BCG_0220, thereby reducing or inactivating the enzyme activity, leading to the identification of a novel resistance mechanism. This is informative for the development of these compounds and for future hit optimization studies through focused medicinal chemistry efforts.
2. Materials and methods
2.1. HTB compounds
To a magnetically stirred solution of rhein/diacerein (0.2 mmol) in CH2Cl2 (2 mL), thionyl chloride (1 mL) was added. The reaction mixture was heated to reflux for 4 h under an atmosphere of argon. After cooling to room temperature, the reaction mixture was evaporated under reduced pressure. The residue was dissolved in anhydrous CH2Cl2 (4 mL) and 4-dimethylaminopyridine (DMAP) (0.04 mmol) was added and cooled with an ice bath. The mixture of selected alcohol (0.25 mmol) and CH2Cl2 (2 mL) were slowly added to the reaction mixture cooled with an ice bath. The reaction mixture was stirred for 3–6 h at room temperature. The solvent was evaporated under reduced pressure and the residue was purified by column chromatography (EtOAc: petroleum ether = 1:5) to give the title compounds. The general experimental information and structural characterization are provided in the Supplementary Material.
2.2. Minimum inhibitory concentration (MIC) and cytotoxicity assay
All target compounds were evaluated for their anti-mycobacterial activities against Mtb H37Rv or isolated clinical strains using the Microplate Alamar Blue Assay (MABA) (Collins and Franzblau, 1997). The MIC was defined as the lowest concentration effecting a reduction in fluorescence of ≥90% relative to the mean of replicate bacterium-only controls. All target compounds were further tested for their cytotoxicity against a mammalian cell line (Vero cells) using methyl-thiazolyldiphenyl-tetrazolium bromide (MTT) as a read out for cell viability and measured by the concentration required for inhibiting 50% cell growth (IC50) as compared to the no-treatment control (Lu et al., 2011).
2.3. Liver microsome stability assay
The assay was performed with liver microsomes from male CD-1 mouse (Xenotech) and pooled human (Bioreclamation). Compounds HTB-03, HTB-04 and HTB-05 were tested at 1 μM with a final concentration of microsomal protein of 1 mg/mL. The reaction was initiated by the addition of NADPH (1 mM), and samples were incubated for up to 60 min at 37 °C in a shaking incubator. The reaction was terminated at 0, 5, 15 and 30 min by the addition of ice-cold ACN/MeOH (50:50) spiked with internal standard. Samples were centrifuged at 4000 rpm at 4 °C for 15 min and the supernatants analyzed by LC-MS/MS. The assay evaluated the metabolic stability of compounds by measuring the amount of parent remaining to test compounds with or without NADPH cofactor. LC-MS/MS: AB Sciex 4500 Qtrap; Kinetex 2.6 μm C18 100 Å column (3.0 mm × 30 mm); mobile phase A, 0.1% formic acid in H2O; mobile phase B, 0.1% formic acid in ACN; flow rate, 0.8 mL/min; room temperature.
2.4. Culturing of mycobacteria
M. bovis BCG strain Pasteur and derivatives were cultured in static liquid media (Middlebrook 7H9 (Difco), 0.05% (v/v) Tween-80, 10% (v/v) Middlebrook ADC and 0.25% (v/v) glycerol) or on solid media (Middlebrook 7H11 agar (Difco) with 10% (v/v) Middlebrook OADC and 0.5% (v/v) glycerol) at 37 °C, 5% CO2. Where appropriate, 25 μg/mL kanamycin was added to the media to select for mycobacterial plasmids.
2.5. Generation of spontaneous resistant mutants and whole genome sequencing
M. bovis BCG was cultured to mid-log phase (OD600nm 0.4–0.8) and 1 × 108 cells were plated on solid media containing 50× and 100× MIC HTB-03 and 20× MIC HTB-04 (based on MICs for M. tuberculosis H37Rv). Potential resistant mutants were cultured and reselected on 100× MIC HTB-03 and 20× MIC HTB-04 to confirm the resistant phenotype. The genomic DNA (gDNA) from the mutants was purified following standard methods and the whole genome sequencing (WGS) and bioinformatics were performed by MicrobesNG, University of Birmingham. Briefly, barcoded DNA libraries were generated from genomic DNA, the fragments were purified and quantified before size evaluation and sequencing. High frequency, statistically relevant single nucleotide polymorphisms (SNPs) were identified by aligning the mutant sequence reads to the M. bovis BCG Pasteur 1173P2 (accession: NC_008769.1) genome sequence.
2.6. Preparation of plasmid expression constructs
The plasmids pVV16 and pET28a were used for over-expression studies in M. bovis BCG and Escherichia coli respectively, both of which encode kanamycin resistance for selection. Vector-encoded poly-histidine tags were exploited from both pVV16 (C-terminal) and pET28a (N-terminal). Primers were designed to clone WT BCG_0220 (equivalent to Rv0183 from Mtb H37Rv) and the mutant genes, BCG_0220-S172P and BCG_0220-VAAQD137D, from M. bovis BCG genomic DNA (isolated as described above) into the multiple cloning sites of the plasmids, exploiting the restriction sites NdeI and BamHI, and are listed in Table 1. The Rv0183 gene was amplified using Q5 DNA polymerase (New England Biolabs) following the manufacturer’s instructions. The PCR products were purified (QIAquick PCR Purification Kit, Qiagen), and along with the plasmids, digested with restriction enzymes (FastDigest, Thermo Fisher Scientific) and the products were extracted from a 1% agarose gel (QIAquick Gel Extraction Kit, Qiagen). The digested PCR products and vectors were ligated using T4 DNA ligase (New England Biolabs). Following transformation into E. coli TOP10 cells, the constructs in positive clones were confirmed by DNA sequencing (Source Bioscience).
Table 1.
Primers used for the generation of over-expression constructs. The restriction site is identified in bold type.
| Primer | Plasmid | Sequence (5′-3′) |
|---|---|---|
| Rv0183 sense | pVV16 | GATCGACTCATATGACTACCACCCGGACTGAACG |
| pET28a | ||
| Rv0183 Anti-sense | pVV16 | GATCGATCGGATCCCCAACCGCTCGGTGAGCCAG |
| pET28 | GATCGATCGGATCCCTACAACCGCTCGGTGAGC |
2.7. Electroporation of mycobacteria
Electrocompetent M. bovis BCG (HTB-04 mutant 1, BCG_0220-VAAQD137D and HTB-04 mutant 2, BCG_0220-S172P) were prepared by pelleting and subsequently washing a mid-log culture with decreasing volumes of room temperature 10% (v/v) glycerol. The cells were incubated at 37 °C with 1 μg plasmid DNA (pVV16 or pVV16-Rv0183) for 10 min in a 2 mm electrode gap electroporation cuvette. A single pulse of 2.5 kV was applied, and the cells were immediately recovered in liquid media and incubated overnight at 37 °C. Positive clones were selected on solid medium containing the 25 μg/mL kanamycin.
2.8. Liquid IC50 determination
The impact of HTB-04 on M. bovis BCG mutant strains over-expressing Rv0183 was analyzed in liquid media. Using a 96-well plate (flat, black bottom, polystyrene), 1 μL compound (100% DMSO) at 100× desired concentration (0.14–21.8 μM) was aliquoted in triplicate into each well. The different strains were cultured to OD600nm 0.4–0.8 and diluted to 1 × 106 colony-forming units (CFU)/mL, before adding 100 μL to each well. The plate was incubated at 37 °C, 5% CO2 for 7 days. 30 μL 0.02% (w/v) resazurin and 12.5 μL 20% (v/v) Tween 80 was added to each well and incubated for a further 24 h. Cell viability was determined by measuring the fluorescence with a λ excitation at 530 nm and λ emission at 590 nm using a POLARstar Omega plate reader. GraphPad Prism was used to fit the data using non-linear regression
2.9. Expression and purification of recombinant Rv0183
Recombinant WT BCG_0220 and the mutants BCG_0220-S172P and BCG_0220-VAAQD137D (equivalent to and henceforth referred to as Rv0183 variants as described) were over-expressed in E. coli SHuffle T7 (K12) cells and E. coli BL21 (DE3) cells, respectively, from the pET28a construct. An overnight culture was used to inoculate 1 L Terrific broth and incubated at 37 °C, 180 rpm, until OD600nm 0.4–06 was reached. The cultures were cooled to 16 °C before protein production was induced using 1 mM IPTG. Cells were cultured overnight at 16 °C before harvesting the following day and the resulting cell pellet was frozen at −20 °C.
The cell pellet was resuspended in 50 mM sodium phosphate, pH 7.5, 0.5 M NaCl and 25 mM imidazole and DNAse and a Complete protease inhibitor cocktail tablet (Roche) were added. Cells were lysed by sonication, 30 s on, 30 s off, for 12 cycles and the insoluble material was pelleted at 14000×g for 45 min. The recombinant protein was purified from the clarified lysate using immobilized metal affinity chromatography (IMAC) using a 1 mL nickel-charged IMAC column. The column was washed and the recombinant protein was eluted using a step gradient of 25, 50, 100, 150, 200, 300 and 1000 mM imidazole. Here, fractions containing the purified Rv0183-S172P and Rv0183-VAAQD137D recombinant proteins were dialyzed into 50 mM Tris pH 7.5, 500 mM NaCl, 10% (v/v) glycerol. The WT Rv0183 recombinant protein was desalted into 50 mM Tris pH 7.5, 10% (v/v) glycerol and was further purified by anion exchange using a 1 mL Q HP anion exchange column with an increasing NaCl gradient. Purified proteins were stored at −20 °C.
2.10. TLC analysis of Rv0183 activity
Each reaction was performed in 50 mM Tris, pH 7.5, 200 mM NaCl, with a final volume of 50 μL. 10 μM Rv0183 (WT and mutants) was incubated with 5 μg compound (HTB-03, HTB-04, rhein, diacerein) with a final DMSO concentration of 2%. Enzyme-free and compound-free controls were also performed. The reactions were incubated at 37 °C for 1 h before being dried by vacuum centrifugation. The samples were resuspended in methanol and loaded onto a TLC plate (silica gel 60) and developed in either 40:10:1 (v/v) chloroform:methanol:ammonia (for separation of rhein and diacerein) or 80:20 pet-ether (60–80):ethyl acetate (for separation of HTB-03 and HTB-04). Compounds were visualised using UV radiation.
3. Results
3.1. Anti-mycobacterial activities, toxicity and stability of anthraquinone analogues
To investigate the therapeutic potential of anthraquinone analogues as Mtb inhibitors we designed and synthesized two series of esters (Fig. 1) through esterification of rhein and diacetyl rhein (diacerein, a marketed drug for the treatment of osteoarthritis) with representative different chain length alcohols, such as methanol, 2,6-dimethylcyclohexanol and 4-chloro-1-butanol. All six compounds together with rhein and diacerein (Fig. 1) were evaluated for their anti-mycobacterial activities against Mtb H37Rv using the Microplate Alamar Blue Assay (MABA) and further tested for their cytotoxicity against a mammalian cell line (Vero cells). As shown in Fig. 1, HTB compounds displayed potent in vitro anti-mycobacterial activities with MICs in the range from 2.56 to 7.31 μM compared to their parent compounds rhein and diacerein. Interestingly, compounds esterified from rhein exhibited low cytotoxicity with IC50 > 162 μM, whereas compounds esterified from diacerein displayed some toxicity with IC50 in the range of 15.48 to 83.99 μM.
To identify the metabolic liabilities of the novel HTB compounds, three compounds (HTB-03, HTB-04 and HTB-05) with more potent anti-mycobacterial activities were investigated for their stability in mouse liver microsome (MLM) and human liver microsome (HLM). As summarized in Table 2, Table 3, compounds HTB-03 and HTB-04 with the flexible side chain R1 exhibited good to excellent stability in MLM or HLM, while the compound with rigid cyclic side chain R1 (HTB-05) showed a relatively low microsomal stability both in MLM and HLM, exemplified by the substrate remaining of HTB-05 in MLM as 42.9% while the substrate remaining of HTB-03 and HTB-04 as 66.3% and 79.8% respectively. Therefore, compounds HTB-03 and HTB-04 were further tested against two drug-resistant Mtb clinical isolates. The two compounds displayed potent in vitro activity not only against drug-susceptible but also drug-resistant Mtb clinical strains (Table 2). These compounds could be promising novel scaffolds for TB drug discovery against drug-resistant strains.
Table 2.
Anti-tubercular activity and liver microsome stability of the selected compounds. The MICs of the selected HTB compounds against different Mtb strains are shown. aMtb resistant to isoniazid (INH), rifampicin (RFP), streptomycin (SM), ethambutol (EMB), rifapentine (RFT), rifabutin (RFB) and paza-aminosalicylate (PAS). bMtb resistant to INH, EMB, RFP, RFB, RFT, amikacin (AMK) and capreomycin (CPM). Compound stability was analyzed in liver microsomes. cSubstrate concentrations were determined in incubations with NADPH after 30 min and normalized to concentrations at time zero. MIC, minimum inhibitory concentration; MLM, mouse liver microsome; HLM, human liver microsome.
| Compounds | MIC (μM) |
IC50 (Vero) (μM) | Substrate remaining (%)c |
|||
|---|---|---|---|---|---|---|
| H37Rv | 12611a | 14231b | MLM | HLM | ||
| HTB-03 | 2.56 | 2.64 | 2.40 | >170.77 | 66.3 | 92.8 |
| HTB-04 | 3.55 | 3.66 | 2.04 | 83.99 | 79.8 | 82.4 |
| HTB-05 | 3.90 | – | – | >162.26 | 42.9 | 67.8 |
| INH | 0.26 | 141.97 | >291.67 | – | – | – |
Table 3.
Whole genome sequencing results of M. bovis BCG spontaneous resistant mutants raised against HTB-03 and HTB-04. High frequency single nucleotide polymorphisms resulting in changes in the amino acid sequence are reported. Genomic positions of the SNP are relative to M. bovis BCG strain Pasteur 1173P2. The base changes (deletions, insertions or substitutions) are capitalized. The amino acid changes are documented, where *1, *2, *3 and *4 represent the generation of a stop codon, amino acid deletions, insertion of a base leading to an amino acid change and a subsequent frameshift, and amino acid insertions. Spontaneous resistant mutants are numbered and the MIC at which they were generated is shown. Dots represent areas where sequence coverage was not sufficient for analysis. Blank spaces indicate the absence of a base change relative to the reference strain.
| Gene | Genome position of SNP | Codon change | Amino acid changes | Frequency of SNP | HTB-03 Mutants |
HTB-04 Mutants |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 50 × MIC |
100 × MIC |
20 × MIC |
||||||||||
| 1 | 2 | 1 | 2 | 3 | 1 | 2 | 3 | |||||
| BCG_0220 | 244,283 | taT/taG | Y99*1 | 98.86 | G | |||||||
| 244,393 | GTGGCGGCACAGgac/gac | VAAQD137D*2 | 97.09 | GTGGCGGCACAG | GTGGCGGCACAG | GTGGCGGCACAG | GTGGCGGCACAG | |||||
| 244,500 | Tct/Cct | S172P | 100 | C | ||||||||
| BCG_0404c | 475,202 | Gcc/Acc | A143T | 100 | A | A | A | A | A | A | ||
| Non-coding region | 1,344,671 | C/G | – | 100 | G | G | G | G | G | G | G | G |
| 1,344,672 | G/C | – | 100 | C | C | C | C | C | C | C | C | |
| PE_PGRS28 | 1,661,298 | Gcc/Acc | A651T | 100 | . | A | . | . | . | A | . | . |
| 1,661,300 | gTc/gGc | V650G | 100 | . | G | . | . | . | G | . | . | |
| 1,661,303 | aTc/aAc | I649N | 100 | . | A | . | . | . | A | . | . | |
| 1,661,304 | Atc/Gtc | I649V | 100 | . | G | . | . | . | G | . | . | |
| 1,661,306 | aTc/aGc | I648S | 100 | . | G | . | . | . | G | . | . | |
| 1,661,307 | Atc/Gtc | I648V | 100 | . | G | . | . | . | G | . | . | |
| 1,661,309 | gAc/gGc | D647G | 100 | . | G | . | . | . | G | . | . | |
| pks12 | 2,282,655 | Ggt/Agt | G2662S | 100 | ||||||||
| 2,282,656 | tgG/tgA | W2661*1 | 100 | |||||||||
| proB | 2,702,724 | Gcc/Tcc | A232S | 100 | T | |||||||
| fadD28 | 3,237,397 | ttt/ttAt | F25L*3 | 100 | TA | TA | TA | TA | TA | TA | ||
| BCG_3499c | 3,833,488 | gcc/gcGGCc | A433AA*4 | 100 | GGC | GGC | GGC | GGC | GGC | GGC | GGC | |
| cut3 | 3,854,969 | ggc/gGTCgc | G261GR*4 | 100 | GTC | GTC | GTC | GTC | GTC | GTC | GTC | |
| PE_PGRS53 | 3,907,860 | Aac/Gac | N599D | 100 | G | G | G | G | G | G | G | G |
| PE_PGRS57 | 3,927,720 | Acc/Gcc | T598A | 100 | G | . | . | . | . | G | . | . |
| BCG_3742 | 4,098,093 | gaC/gaA | D188E | 100 | A | A | A | A | A | A | ||
| glpK | 4,111,717 | ttG/ttC | L385F | 100 | C | C | C | C | C | C | ||
3.2. Efforts to identify the molecular target of the anthraquinone analogues through the generation of resistant isolates
In drug discovery, elucidating the molecular target of an inhibitory compound is a primary step, and can provide valuable information to direct medicinal chemistry efforts to improve compound efficacy and toxicity for the progression into clinical drug candidates. Without prior information of the target, whole genome sequencing of spontaneous resistant mutants generated against an inhibitor is a validated approach to begin de-convoluting its molecular mode of action (Abrahams et al., 2016, Abrahams et al., 2012, Abrahams et al., 2017, Gurcha et al., 2014). This methodology was employed to probe the target of HTB-03 and HTB-04, and ultimately rhein, which could open up new opportunities for discovering novel anti-TB drug candidates through structural modification.
Given the minimal inhibitory concentrations (MICs) of HTB-03, HTB-04 and rhein determined in Mtb (Fig. 1; 2.56, 3.55 and 106.26 μM, respectively), spontaneous resistant mutants were generated in the Mtb model organism, M. bovis BCG, at 50× and 100× MIC of HTB-03 and 20× MIC of HTB-04 with frequencies of resistance of 83 × 10-8, 100 × 10-8 and 85 × 10-8, respectively. Following confirmation of the resistant phenotype of five mutants raised against HTB-03 and three mutants raised against HTB-04, the gDNA was extracted and the whole genome was sequenced. Through comparison to the wild-type (WT) M. bovis BCG reference strain (Genbank accession number NC_008769.1), a number of statistically relevant SNPs were identified (Table 3). Three out of the five mutants generated against HTB-03 and all three mutants generated against HTB-04 had mutations in the gene BCG_0220, suggesting a role in the resistance mechanism. It is noteworthy that there are a number of SNPs in other genes with high frequencies of occurrence within the population sequenced. These genes were not considered to be involved in the resistance phenotype either because they are commonly observed during in vitro spontaneous resistant mutant generation, or because they occurred only in a minority of the mutants sequenced. The BCG_0220 gene has 100% sequence identity to Rv0183, which has been characterized as a monoglyceride lipase (Cotes et al., 2007). Although classified by Himar1-based transposon mutagenesis as non-essential (Sassetti et al., 2003), Rv0183 is highly conserved and present in the minimal genome of Mycobacterium leprae. Additionally, its activity is maintained in dormant Mtb (Tallman et al., 2016), suggesting its essential role in the survival and pathogenicity of Mtb, warranting further investigation for its role in resistance against the HTB-03 and HTB-04 compounds.
3.3. Over-expression of Rv0183 in HTB resistant strains increases sensitivity to the anthraquinone analogues
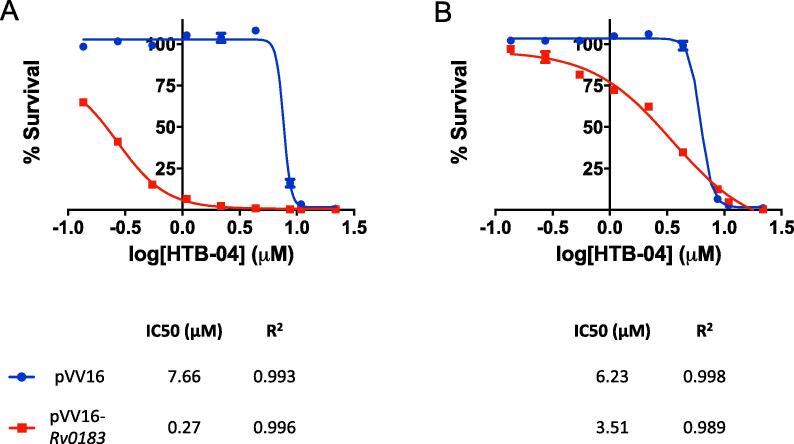
From the WGS results, the SNPs in BCG_0220 are codon deletions, STOP mutations or changes to a conserved residue. These mutations suggest that the reduction or inactivation of BCG_0220 activity results in resistance and from this we can infer that Rv0183 is not the target of the HTB compounds. To assess the involvement of Rv0183 in the resistance mechanism, the impact on the MIC of HTB-04 was investigated using two resistant mutant strains (HTB-04 mutant 1, BCG_0220-VAAQD137D and HTB-04 mutant 2, BCG_0220-S172P (mutation of a conserved residue (Cotes et al., 2007)); mutant nomenclature taken from Table 3). The mycobacterial expression vectors, pVV16 (empty vector) and pVV16-Rv0183 (over-expressing WT Rv0183 (BCG_0220)) were electroporated into the mutant strains and the liquid MIC was analyzed as shown in Fig. 2. In both mutant strains, the over-expression of Rv0183 markedly reduced the IC50 of HTB-04 compared to the empty vectors strain (mutant 1, 7.66 to 0.27 μM; mutant 2, 6.23 to 3.51 μM), indicating a restoration of sensitivity and suggesting a role of the mutated forms of Rv0183 in the resistance mechanism.
Fig. 2.
The impact on survival of HTB resistant mutants over-expressing WT Rv0183. The mycobacterial expression constructs pVV16 (blue) and pVV16-Rv0183 (red) were electroporated into the HTB-04 resistant mutants (A) 1 (BCG_0220-VAAQD137D) and (B) 2 (BCG_0220-S172P) and the percentage survival was analyzed using increasing concentrations of HTB-04. Each point shows the mean with bars representing the standard error based on triplicate data. GraphPad Prism was used to fit the data using non-linear regression. IC50 and the corresponding R2 values are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Rv0183 hydrolyzes the chlorinated acyl chain of the anthraquinone analogues
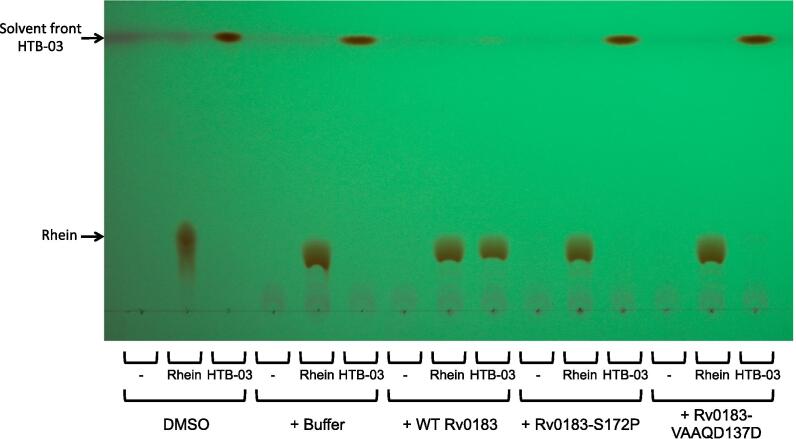
After establishing the involvement of Rv0183 in the resistance to HTB compounds, the mechanism was investigated. Relating the structure of the compounds with the characterized activity of Rv0183, it was plausible that the enzyme hydrolyzed the ester linkage, releasing the chlorinated carbon chain. To validate this hypothesis, recombinant WT BCG_0220, BCG_0220-S172P and BCG_0220-VAAQD137D (equivalent to and henceforth referred to as Rv0183 variants as described) were incubated with HTB-03 and HTB-04 and the compounds were analyzed by thin layer chromatography (TLC). As controls, the migration of the HTB-03 derivative, rhein, and the HTB-04 derivative, diacerein, (without the chlorinated carbon chain) were analyzed alongside the HTB compounds. In the absence of enzyme, HTB-03 and HTB-04 migrated with the solvent front (Fig. 3, Supplementary Fig. 1A). Fig. 3 shows that upon incubation with WT Rv0183, HTB-03 was converted to rhein. However, the mutants Rv0183-VAAQD137D and Rv0183-S172P showed minimal and no activity with HTB-03, respectively. The enzymes had no apparent activity on rhein. The activities of the enzymes with HTB-04 were inconclusive (Supplementary Fig. 1). In addition to the limited solubility of HTB-04, it was evident that the R2 acyl groups on diacerein were hydrolyzed in the buffer system used, leading to the production of rhein (Supplementary Fig. 1A). This was also apparent for the R2 acyl groups on HTB-04, where partial degradation of the compound led to the generation of various products, one of which coincided with HTB-03 (Supplementary Fig. 1B). Upon the incubation of HTB-04 with Rv0183, a band representing rhein appeared. Despite this apparent activity, it was not possible to confirm whether Rv0183-dependent hydrolysis of the chlorinated carbon tail occurred before or after the enzyme-independent hydrolysis of the R2 acyl groups on the diacerein moiety. Also, due to the instability of diacerein in the assay conditions, it was also not possible to determine whether Rv0183 was able to hydrolyze the acyl groups in addition to the spontaneous degradation. Regardless of limited information gained from the enzyme activity with HTB-04, the hydrolysis of the chlorinated acyl chain of HTB-03 implicates rhein as the active moiety of the HTB compounds.
Fig. 3.
TLC analysis of HTB-03 compound modification by Rv0183. Rv0183, WT and mutant variants, were incubated with Rhein and HTB-03 and the samples were analyzed by TLC (40:10:1 (v/v) chloroform:methanol:ammonia) and exposed under UV light. For reference, compounds in DMSO were loaded directly onto the TLC and were not subjected to the assay conditions. Compound only in buffer and enzyme only incubations were used as controls. The solvent front and the positions of HTB-03 and rhein are indicated.
4. Discussion
4.1. Derivatives of rhein as suitable compounds to treat Mtb
The natural product, rhein, has a number of pharmacological activities, and its known antimicrobial properties directed this investigation as to whether rhein derivatives could be used to treat Mtb. It has, however, previously been demonstrated that rhein does not inhibit Mtb (Gajadeera et al., 2015). Mycobacteria possess an extensive lipophilic mycobacterial cell wall, and it is this feature that could prevent entry of rhein into the cell and imparting its effect. Therefore, we envisaged that optimizing the chemical structure to one that was more lipophilic, could aid entry into the cell. From this, HTB-03 and HTB-04 (derivatives of rhein and diacerein, respectively), which have flexible chlorinated carbon chains, were synthesized. Both compounds exhibit anti-tubercular activities as well as low toxicity and good liver microsome stability. This preliminary result revealed that there are more opportunities to identify promising leads for the anti-TB drug discovery with novel scaffolds.
4.2. Mutations in the lipase, Rv0183, conferred resistance to the HTB compounds
To further progress the HTB compounds, their mode of action was investigated, firstly via WGS of resistant mutants. Although the target was not elucidated, we have identified that mutations in BCG_0220 are responsible for resistance against the HTB compounds. From this, we have deduced that the HTB compounds are prodrugs, requiring activation by the Mtb homologue Rv0183. Resistance associated with prodrug activation is a common problem associated with the first-line drug in TB treatment, isoniazid, where mutations in katG prevent compound activation and the generation of the isoniazid-NADH adduct, which exhibits anti-tubercular activity. In clinical MDR isolates and INH-resistant strains generated within the laboratory, the frequency of mutations relating to the actual target, inhA, are low in comparison to katG (Jagielski et al., 2014, Tseng et al., 2015), indicating the potential difficulties with identifying the target of the HTB compounds through whole genome sequencing of resistant isolates. In this regard, we attempted to generate spontaneous resistant mutants against the hypothesized active moiety of the HTB compounds, rhein, but were unsuccessful despite testing a broad range of concentrations.
4.3. The role of Rv0183 in prodrug activation
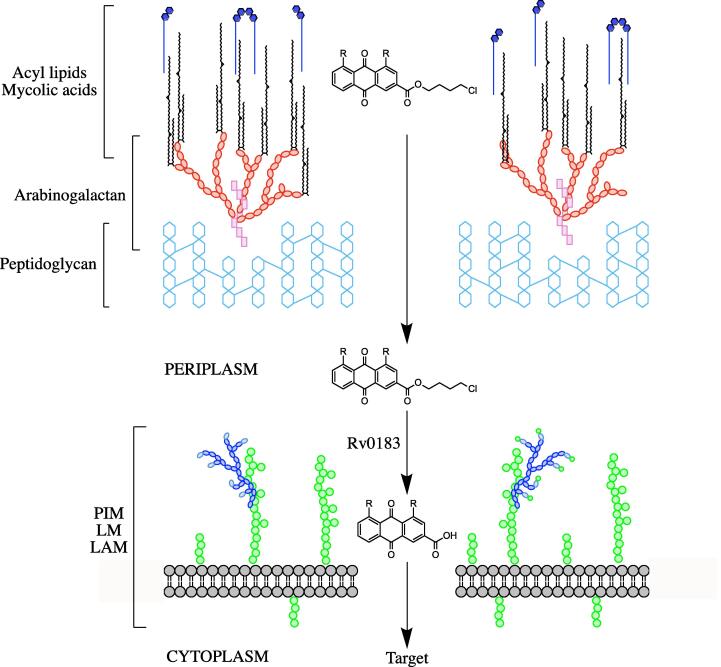
In this work, we have discovered that Rv0183 restores sensitivity to HTB mutants in a whole cell assay, and that recombinant Rv0183 hydrolyzes the ester linkage on the rhein derivative HTB-03, in vitro. Unfortunately, we were unable to establish enzyme activity on the diacerein derivative HTB-04 due to compound insolubility and the apparent instability of the acyl groups in our buffer system. Therefore, it is not possible to conclude whether diacerein can function as the active molecule, or whether further modifications by other lipases are necessary prior to or post Rv0183 activity. The higher MIC of HTB-04 compared to HTB-03 could indicate the latter is plausible. Interestingly, due to the high MIC exhibited by rhein in Mtb, the chlorinated 4-carbon chain of the HTB compounds must contribute to their potency. Previous localization studies have revealed that Rv0183 exists in the cell wall and extracellular space (Cotes et al., 2007). From these considerations, we propose a model in which the HTB compounds traverse the lipid rich mycobacterial cell wall by exploiting the carbon chain, where the compounds interact with Rv0183 in the periplasmic space, releasing rhein, which can pass through the cytoplasmic membrane, where it can be further modified, if necessary, and reach the target (depicted in Fig. 4). In this model, the activity of Rv0183 would have to be regulated; activity exterior to the cell wall would convert the compounds into rhein or similar, which is essentially inactive against Mtb. It has been suggested that Rv0183 is bound to large particles on the outside of the cell wall (Cotes et al., 2007), the role of which could control the activity of the lipase.
Fig. 4.
Model of HTB entry and activation into the mycobacterial cell. The HTB-03 or HTB-04 compound is shown to traverse the lipid layer, exploiting the lipophilic chain. In the periplasm, Rv0183 hydrolyzes the ester linkage, releasing 4-chlorobutan-1-ol. We propose that rhein (or diacerein) can be imported through the cytoplasmic membrane into the cell to exert its effect. R, —OH (HTB-03) or —COOH (HTB-04). PIM, phosphatidyl-myo-inositol mannoside; LM, lipomannan; LAM, lipoarabinomannan.
4.4. Rv0183 as a direct target for anti-tubercular drugs
The activity of lipases is important during the survival and pathogenicity of Mtb, where lipid metabolism is key to providing a carbon source during infection, dormancy and reactivation. It has recently been established that Rv0183, along with CaeA (esterase and protease) and Rv1683 (long-chain acyl-CoA synthase and lipase), are active during all stages of TB disease (Tallman et al., 2016). Furthermore, disruption of the ortholog of Rv0183 in Mycobacterium smegmatis has been shown to impact drug susceptibility (Dhouib et al., 2010). These features make these enzymes, along with their accessible location, suitable targets for anti-tubercular drugs, which are currently being pursued (Aschauer et al., 2018, Point et al., 2013, Point et al., 2012, Saravanan et al., 2012). As we have proposed that Rv0183 is involved in the activation of the HTB compounds, inhibition of this enzyme would be undesirable in the context of the potential treatment with compounds of this class. Dormant Mtb has a limited enzymatic activity, and enzymes that remain active are expected to be essential to sustain survival, especially during latency. In this regard, inactivating mutations in Rv0183, although causing resistance against the HTB compounds, would likely be prohibitory for survival. Additionally, due to the activity of Rv0183 during dormancy, the activated HTB compounds maintain the potential (target dependent) to inhibit latent Mtb infections. To conclude, the HTB compounds and other rhein derivatives could be conceivably progressed into clinical drug candidates, and this will be further advanced by the elucidation of the molecular target.
5. Author statements
5.1. Ethics statement
All experiments were approved by the University of Birmingham, Institute of Materia Medica, and Beijing Tuberculosis and Thoracic Tumor Research Institute ethical committees where required and there are no ethical issues to report.
CRediT authorship contribution statement
Katherine A. Abrahams: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization. Wei Hu: Conceptualization, Methodology, Validation, Investigation. Gang Li: Conceptualization, Methodology, Validation, Formal analysis, Investigation. Yu Lu: Conceptualization, Methodology, Validation, Investigation. Emily J. Richardson: Formal analysis, Investigation. Nicholas J. Loman: Formal analysis, Investigation. Haihong Huang: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration. Gurdyal S. Besra: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing - original draft, Writing - review & editing, Visualization, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thanks Albel Singh and Natacha Veerapen for their technical support. GSB acknowledges support in the form of a Personal Research Chair from Mr. James Bardrick, a Royal Society Wolfson Research Merit Award, and the Medical Research Council, UK (MR/R001154/1 and MR/S000542/1). HH acknowledges support from the CAMS Innovation Fund for Medical Sciences (CAMS-2016-I2M-1-010).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tcsw.2020.100040.
Contributor Information
Haihong Huang, Email: joyce@imm.ac.cn.
Gurdyal S. Besra, Email: g.besra@bham.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abrahams K.A., Chung C.W., Ghidelli-Disse S., Rullas J., Rebollo-Lopez M.J., Gurcha S.S., Cox J.A., Mendoza A., Jimenez-Navarro E., Martinez-Martinez M.S., Neu M., Shillings A., Homes P., Argyrou A., Casanueva R., Loman N.J., Moynihan P.J., Lelievre J., Selenski C., Axtman M., Kremer L., Bantscheff M., Angulo-Barturen I., Izquierdo M.C., Cammack N.C., Drewes G., Ballell L., Barros D., Besra G.S., Bates R.H. Identification of KasA as the cellular target of an anti-tubercular scaffold. Nat. Commun. 2016;7:12581. doi: 10.1038/ncomms12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams K.A., Cox J.A., Spivey V.L., Loman N.J., Pallen M.J., Constantinidou C., Fernandez R., Alemparte C., Remuinan M.J., Barros D., Ballell L., Besra G.S. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahams K.A., Cox J.A.G., Futterer K., Rullas J., Ortega-Muro F., Loman N.J., Moynihan P.J., Perez-Herran E., Jimenez E., Esquivias J., Barros D., Ballell L., Alemparte C., Besra G.S. Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface. Sci. Rep. 2017;7:9430. doi: 10.1038/s41598-017-09642-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchel M. Identification of the antibiotic substance from Cassia reticulata as 4,5-dihydroxyanthraquinone-2-carboxylic acid. J. Biol. Chem. 1949;177:169–177. [PubMed] [Google Scholar]
- Aschauer P., Zimmermann R., Breinbauer R., Pavkov-Keller T., Oberer M. The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition. Sci. Rep. 2018;8:8948. doi: 10.1038/s41598-018-27051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballell L., Bates R.H., Young R.J., Alvarez-Gomez D., Alvarez-Ruiz E., Barroso V., Blanco D., Crespo B., Escribano J., Gonzalez R., Lozano S., Huss S., Santos-Villarejo A., Martin-Plaza J.J., Mendoza A., Rebollo-Lopez M.J., Remuinan-Blanco M., Lavandera J.L., Perez-Herran E., Gamo-Benito F.J., Garcia-Bustos J.F., Barros D., Castro J.P., Cammack N. Fueling open-source drug discovery: 177 small-molecule leads against tuberculosis. ChemMedChem. 2013;8:313–321. doi: 10.1002/cmdc.201200428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglione S., Paggi M.G., Delpino A., Zeuli M., Floridi A. Inhibition of protein synthesis in neoplastic cells by rhein. Biochem. Pharmacol. 1990;40:967–973. doi: 10.1016/0006-2952(90)90481-y. [DOI] [PubMed] [Google Scholar]
- Chung J.G., Tsou M.F., Wang H.H., Lo H.H., Hsieh S.E., Yen Y.S., Wu L.T., Chang S.H., Ho C.C., Hung C.F. Rhein affects arylamine N-acetyltransferase activity in Helicobacter pylori from peptic ulcer patients. J. Appl. Toxicol. 1998;18:117–123. doi: 10.1002/(sici)1099-1263(199803/04)18:2<117::aid-jat486>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Collins L., Franzblau S.G. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob. Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotes K., Dhouib R., Douchet I., Chahinian H., de Caro A., Carriere F., Canaan S. Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem. J. 2007;408:417–427. doi: 10.1042/BJ20070745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouib R., Laval F., Carriere F., Daffe M., Canaan S. A monoacylglycerol lipase from Mycobacterium smegmatis involved in bacterial cell interaction. J. Bacteriol. 2010;192:4776–4785. doi: 10.1128/JB.00261-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floridi A., Gentile F.P., Bruno T., Castiglione S., Zeuli M., Benassi M. Growth inhibition by rhein and lonidamine of human glioma cells in vitro. Anticancer Res. 1990;10:1633–1636. [PubMed] [Google Scholar]
- Gajadeera C., Willby M.J., Green K.D., Shaul P., Fridman M., Garneau-Tsodikova S., Posey J.E., Tsodikov O.V. Antimycobacterial activity of DNA intercalator inhibitors of Mycobacterium tuberculosis primase DnaG. J. Antibiot. (Tokyo) 2015;68:153–157. doi: 10.1038/ja.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcha S.S., Usha V., Cox J.A., Futterer K., Abrahams K.A., Bhatt A., Alderwick L.J., Reynolds R.C., Loman N.J., Nataraj V., Alemparte C., Barros D., Lloyd A.J., Ballell L., Hobrath J.V., Besra G.S. Biochemical and structural characterization of mycobacterial aspartyl-tRNA synthetase AspS, a promising TB drug target. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbut M.B., Vilcheze C., Luo X., Hensler M.E., Guo H., Yang B., Chatterjee A.K., Nizet V., Jacobs W.R., Jr., Schultz P.G., Wang F. Auranofin exerts broad-spectrum bactericidal activities by targeting thiol-redox homeostasis. Proc. Natl. Acad. Sci. USA. 2015;112:4453–4458. doi: 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugonnet J.E., Tremblay L.W., Boshoff H.I., Barry C.E., 3rd, Blanchard J.S. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–1218. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagielski T., Bakula Z., Roeske K., Kaminski M., Napiorkowska A., Augustynowicz-Kopec E., Zwolska Z., Bielecki J. Detection of mutations associated with isoniazid resistance in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J. Antimicrob. Chemother. 2014;69:2369–2375. doi: 10.1093/jac/dku161. [DOI] [PubMed] [Google Scholar]
- Kasik J.E. The nature of Mycobacterial Penicillinase. Am. Rev. Respir. Dis. 1965;91:117–119. doi: 10.1164/arrd.1965.91.1.117. [DOI] [PubMed] [Google Scholar]
- Lu Y., Zheng M., Wang B., Fu L., Zhao W., Li P., Xu J., Zhu H., Jin H., Yin D., Huang H., Upton A.M., Ma Z. Clofazimine analogs with efficacy against experimental tuberculosis and reduced potential for accumulation. Antimicrob. Agents Chemother. 2011;55:5185–5193. doi: 10.1128/AAC.00699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A., Bates S., Kolvekar T., Devarajan P.V., Guzman J.D., Bhakta S. Repurposing-a ray of hope in tackling extensively drug resistance in tuberculosis. Int. J. Infect. Dis. 2015;32:50–55. doi: 10.1016/j.ijid.2014.12.031. [DOI] [PubMed] [Google Scholar]
- Milano A., Pasca M.R., Provvedi R., Lucarelli A.P., Manina G., Ribeiro A.L., Manganelli R., Riccardi G. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis (Edinb) 2009;89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Moreira W., Ngan G.J., Low J.L., Poulsen A., Chia B.C., Ang M.J., Yap A., Fulwood J., Lakshmanan U., Lim J., Khoo A.Y., Flotow H., Hill J., Raju R.M., Rubin E.J., Dick T. Target mechanism-based whole-cell screening identifies bortezomib as an inhibitor of caseinolytic protease in mycobacteria. MBio. 2015;6:e00253–e315. doi: 10.1128/mBio.00253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam A., Almabruk K.H., Saxena A., Yang J., Mukherjee U., Kaur H., Kohli P., Kumari R., Singh P., Zakharov L.N., Singh Y., Mahmud T., Lal R. Modification of rifamycin polyketide backbone leads to improved drug activity against rifampicin-resistant Mycobacterium tuberculosis. J. Biol. Chem. 2014;289:21142–21152. doi: 10.1074/jbc.M114.572636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Point V., Malla R.K., Carriere F., Canaan S., Spilling C.D., Cavalier J.F. Enantioselective inhibition of microbial lipolytic enzymes by nonracemic monocyclic enolphosphonate analogues of cyclophostin. J. Med. Chem. 2013;56:4393–4401. doi: 10.1021/jm4000787. [DOI] [PubMed] [Google Scholar]
- Point V., Malla R.K., Diomande S., Martin B.P., Delorme V., Carriere F., Canaan S., Rath N.P., Spilling C.D., Cavalier J.F. Synthesis and kinetic evaluation of cyclophostin and cyclipostins phosphonate analogs as selective and potent inhibitors of microbial lipases. J. Med. Chem. 2012;55:10204–10219. doi: 10.1021/jm301216x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D.A., Morris S.L. Molecular mechanisms of isoniazid resistance in Mycobacterium tuberculosis and Mycobacterium bovis. Infect. Immun. 1995;63:1427–1433. doi: 10.1128/iai.63.4.1427-1433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullas J., Dhar N., McKinney J.D., Garcia-Perez A., Lelievre J., Diacon A.H., Hugonnet J.E., Arthur M., Angulo-Barturen I., Barros-Aguirre D., Ballell L. Combinations of beta-lactam antibiotics currently in clinical trials are efficacious in a DHP-I-deficient mouse model of tuberculosis infection. Antimicrob. Agents Chemother. 2015;59:4997–4999. doi: 10.1128/AAC.01063-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanan P., Dubey V.K., Patra S. Potential selective inhibitors against Rv0183 of Mycobacterium tuberculosis targeting host lipid metabolism. Chem. Biol. Drug Des. 2012;79:1056–1062. doi: 10.1111/j.1747-0285.2012.01373.x. [DOI] [PubMed] [Google Scholar]
- Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Tallman K.R., Levine S.R., Beatty K.E. Small-molecule probes reveal esterases with persistent activity in dormant and reactivating Mycobacterium tuberculosis. ACS Infect. Dis. 2016;2:936–944. doi: 10.1021/acsinfecdis.6b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.T., Tai C.H., Li C.R., Lin C.F., Shi Z.Y. The mutations of katG and inhA genes of isoniazid-resistant Mycobacterium tuberculosis isolates in Taiwan. J. Microbiol. Immunol. Infect. 2015;48:249–255. doi: 10.1016/j.jmii.2013.08.018. [DOI] [PubMed] [Google Scholar]
- World Health Organisation . World Health Organisation; Geneva, Switzerland: 2019. Global tuberculosis Report. [Google Scholar]
- Yu L., Xiang H., Fan J., Wang D., Yang F., Guo N., Jin Q., Deng X. Global transcriptional response of Staphylococcus aureus to rhein, a natural plant product. J. Biotechnol. 2008;135:304–308. doi: 10.1016/j.jbiotec.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Zhou Y.X., Xia W., Yue W., Peng C., Rahman K., Zhang H. Rhein: a review of pharmacological activities. Evid. Based Complement Alternat. Med. 2015;2015 doi: 10.1155/2015/578107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.