Abstract
During normal T cell development in the thymus, αβ T cell receptors (TCR) signal immature thymocytes to differentiate into mature T cells by binding to peptide-MHC ligands together with CD4/CD8 coreceptors. Conversely, in MHC and CD4/CD8 coreceptor-deficient mice, the thymus generates mature T cells expressing MHC-independent TCRs that recognize native conformational epitopes rather than linear antigenic-peptides presented by MHC. To this date, no structural information of MHC-independent TCRs is available and their structural recognition of non-MHC ligand remains unknown. Here, we determined the first structures of two murine MHC-independent TCR (A11 and B12A) that bind with high nanomolar affinities to mouse adhesion receptor CD155. Solution binding demonstrated Vαβ-domain is responsible for MHC-independent B12A recognition of its ligand. Analysis of A11 and B12A sequences against various MHC-restricted and -independent TCR sequence repertoires showed that individual V-genes of A11 and B12A did not exhibit preference against MHC-restriction. Likewise, CDR3 alone did not discriminate against MHC binding, suggesting VDJ recombination together with Vα/Vβ pairing determine their MHC-independent specificity for CD155. The structures of A11 and B12A TCR are nearly identical to those of MHC-restricted TCR, including the conformations of CDR1 and 2. Mutational analysis, together with negative-staining electron microscopy images, showed that the CDR regions of A11 and B12A recognized epitopes on D1 domain of CD155, a region also involved in CD155 binding to poliovirus and Tactile in human. Taken together, MHC-independent TCRs adopt canonical TCR structures to recognize native antigens, highlighting the importance of thymic selection in determining TCR ligand specificity.
Introduction
After the initial activation by evolutionarily conserved innate immunity components, the immune systems of humans and other mammals further develop robust, specific and memory-type responses against various pathogens. These acquired immunities bifurcate into two equally important and mutually reacting arms, B cell mediated humoral responses and T cell mediated cellular responses. To confront a large array of potential pathogens, B and T cells utilize similar gene recombination machineries to generate diverse number of antigen receptors. While B-cell receptors (BCR) recognize both linear and 3-dimensional antigenic epitopes, peripheral T cells normally recognize only linear peptides presented by major histocompatibility antigens (MHCs), a feature referred to as “MHC-restriction”. A wealth of structural studies has shown that αβTCRs adopt a common binding geometry on peptide-MHC with germline derived CDR1 and CDR2 loops primarily interacting with MHC helices, and CDR3 engaging the peptide. The fixed TCR docking geometry on MHC was interpreted as co-evolution between TCR and MHC. However, in CD4/CD8 coreceptor-deficient mice that are also MHC-deficient QuadKO mice (B2m−/−H-2Ab1−/−CD4−/−CD8a−/− ), the thymus generates mature T cells expressing MHC-independent TCRs that bind to conformational epitopes on native ligands independently of MHC. Recently, the molecular and sequence signatures of the MHC-independent TCR repertoire from QuadKO mice was found to be distinct from the MHC-restricted TCR repertoire of conventional B6 mice, especially in their hypervariable CDR3 regions(1). For example, both TCRα and β sequences of the QuadKO showed increased Cys frequency in their hypervariable CDR3 regions compared to their MHC-sufficient littermates. In addition, the average CDR3α and CDR3β are longer in the QuadKO repertoires than those of MHC-restricted repertoires. Whether the T cells from QuadKO mice are genuine non-MHC restricted T cells that lack specific binding to peptide-MHC and instead bind specifically to non-MHC ligands, or they are a subset of MHC-restricted T cells exhibiting cross reactivities to non-MHC ligands, is intensely debated as their ligands are mostly unknown. To further characterize MHC-independent T cell receptors, we cloned two TCR from the QuadKO T cells, A11 and B12A, and identified CD155 as their selecting ligand (2). To date, there is no structural information available on MHC-independent TCR and how they recognize non-MHC ligands remains unknown.
Here, we report the first crystal structures of two MHC-independent αβTCR and characterized their conformational epitopes on CD155. Specifically, we attempted to address if MHC-independent TCR possess unique structural features and determined their binding epitopes on CD155.
Materials and methods
Stimulation of A11 and B12A transduced T cells
Full length TCR cDNA for TCRα and TCRβ were cloned with PCR primers specific to sequences 5’ of the start and 3’ of the stop codon of rearranged TCR chains. TCR expression constructs were cloned into murine stem cell virus (MSCV)-based retroviral plasmids with pMX-IRES-GFP (provided by R. Bosselut, NCI) for TCRβ and MSCV-IRES-tNGFR (provided by W. Pear, University of Pennsylvania) for TCRα, and transfected into PlatE cells separately to produce retrovirus containing supernatants. TCR-negative 4G4 T cells were retrovirally transduced with both TCRα and TCRβ-containing retroviruses in the presence of 2 µg/ml of polybrene. The resulting B12A-4G4 or A11–4G4 cells (5x104 cells/well) were incubated overnight at 37℃ with B6 or MHCKO stimulator spleen cells that had been treated overnight with LPS (10µg/ml), irradiated with 3000R, and T-depleted. Purified B10.A lymph node T cells that were alloreactive against H-2b stimulators were used as control responder T cells. MHC blocking antibodies used were a mixture of monoclonal antibodies specific for MHC class I anti-H-2Kb/H-2Db (28–8-6) and MHC-II Y3P (I-A), M5/114 (I-A/I-E) at 10 µg/ml each. Culture supernatants of stimulated B12A- and A11–4G4 cells were collected and IL-2 concentrations were measured by ELISA (R&D Systems).
Protein expression and purification
Chimeric DNA encoding TCR A11 or B12A alpha and beta variable regions and corresponding human constant regions (Cα and Cβ) were synthesized and cloned into pET30a vectors (Genescript Inc., Piscataway, NJ), as previously described. The soluble TCR A11 and B12A were produced as disulfide-linked heterodimers by a rapid dilution refolding procedure as previously described (3). In brief, each receptor chain was expressed separately as inclusion bodies in BL21 (DE3) cells. Then, purified inclusion bodies of TCR α and β chains were mixed at ~1:1 molar ratio and dissolved in 8M urea, 20mM Tris (pH8.0), 1.5M Guanidine-HCl, 5mM EDTA, and 5mM sodium acetate supplemented with 5mM DTT. The dissolved inclusion bodies were injected into the refolding buffer containing 0.4M arginine, 100mM Tris, pH8.5, 5mM reduced glutathione, 0.5mM oxidized glutathione, 2mM EDTA at 4°C. Two days later, refolding mixture was dialyzed against 10mM Tris, pH8.0 and 100mM urea. Soluble TCRs were purified by anion-exchange chromatography (HiTrap Q FF, GE Healthcare), followed by size exclusion chromatography (Superdex200 16/60, GE Healthcare) in 10mM Hepes, pH7.4 and 0.15M NaCl. The V-domain only single chain construct of B12A consists of amino acids 1–116 from Vα, 1–116 from Vβ with a (GGGGS)3 linker between the variable regions and a C-terminal six-histidine tag. The inclusion body of and B12A single-chain variable domains were refolded and dialyzed the same as described for the heterodimers. Soluble monomeric single chain variable domains were purified by Ni-NTA affinity chromatography and size exclusion chromatography.
Crystallization, Structure determination and refinement
TCR A11 and B12A proteins were concentrated to an A280nm of 12.2 and 10.5, respectively. TCRA11 was crystallized in 20% PEG 4000, 0.1M Tris, pH8.5, 10mM MgCl2, and optimized by streak seeding. TCR B12A was grown in 25% PEG 4000, 0.1M sodium acetate, pH4.6, and 0.2M (NH4)2SO4. The crystals were immersed in cryoprotectant containing the respective mother liquor plus 15% glycerol prior to flash-cooling in liquid nitrogen. All X-ray data were collected at SER-CAT beamlines and processed with HKL2000 (4)(Table 2). The structures of TCR A11 and B12A were solved by a molecular replacement method with the program Phaser(5) in CCP4 packages(6) using the TCR B3K506 structure (PDB ID: 3C5Z) as the search model. The structure refinement was subsequently carried out by autoBUSTER(7) with repeated cycles of rebuilding in COOT(8). Data collection and model statistics are summarized in Table 2. Figures were prepared with PyMOL(9). The structural numbering of residues does not include the signal peptide region of TCR, and thus is different from the sequencing numbering. In particular, CDR3β is between Cys 89-Phe 100 for A11, and Cys 90-Phe 105 for B12A, and Cys 104- Phe 118 for all repertoire sequences.
Table 2.
X-ray Data collection and refinement statistics
| Data collection | ||
|---|---|---|
| A11 | B12A | |
| Space group | P212121 | C2 |
| Unit cell dimension (Å) | a=89.2, b=102.6, c=118.5, α=90.0, β=90.0, γ=90.0 | a=128.0, b=43.6, c=98.4, α=90.0, β=125.9, γ=90.0 |
| Resolution range (Å) | 50.0–2.60 (2.65–2.60)* | 40.0–2.43 (2.47–2.43)* |
| Unique reflections | 32,187(1262) | 16,078(603) |
| Average redundancy | 9.9(4.0) | 6.6(3.5) |
| Rmerge(%)a | 9.5(48.7) | 12.5(38.5) |
| I/σ(I) | 23.0(2.1) | 13.6(2.1) |
| Completeness (%) | 94.7 (68.4) | 95.0 (74.8) |
| Refinement statistics | ||
| A11 | B12A | |
| Refinement resolution (Å) | 34.0–2.60(2.67–2.60)* | 39.7–2.43(2.59–2.43)* |
| Rcryst (%)b | 20.5(26.4) | 20.5(23.9) |
| Rfree (%) | 25.8(39.1) | 25.0(31.5) |
| Protein atoms | 6,993 | 3,520 |
| Water molecules | 142 | 48 |
| R.m.s.d from ideal values | ||
| Bond length (Å) | 0.01 | 0.01 |
| Bond angle (°) | 1.29 | 1.26 |
| Mean B-factor (Å2) | 71.6 | 62.3 |
| Wilson plot B-factor (Å2) | 69.5 | 59.4 |
| Ramachandran statistics | ||
| Most favored region | 93.2% | 94.5% |
| Additionally allowed | 6.8% | 5.5% |
Rcryst = ∑|| Fo| - |Fc|| / ∑|Fo| calculated from working data set. Rfree is calculated from 6.0% of data randomly chosen not to be included in refinement.
Values for the highest resolution shell in data collection and refinement are listed in the parenthesis.
Surface plasmon resonance (BIAcore) experiment
Surface plasmon resonance measurements were performed using a BIAcore 3000 instrument and analyzed with BIAevaluation 4.1 software (Biacore AB). Murine CD155 and human CD155 were obtained from Sino Biological, Inc. and R&D systems, respectively. To measure the affinity, TCRs or CD155 proteins were immobilized on carboxylated dextran CM5 chips (Biacore AB) to 200–500 response units (RU) using a primary amine-coupling and 5 μl/min flow rate in 10mM sodium acetate (pH 4.0). The analyte consisted of serial dilutions of TCRs or CD155 between 0.3 and 2.5 µM in a buffer containing 10mM Hepes (pH7.4) and 0.15M NaCl. The dissociation constants were obtained by kinetic curve fitting using BIAevaluation 4.1 (BIAcore Inc.).
Negative-stain electron microscopy
Recombinant TCR B12A and CD155(Sino biological, Inc.) proteins were dialyzed against Hepes buffer (10 mM HEPES, 150 mM NaCl pH 7.5), respectively. To prepare TCR B12A-CD155 complex samples, equal volumes of TCR B12A (4mg/ml) and CD155(4mg/ml) were mixed and chemically cross-linked with 0.04% glutaraldehyde for 30 minutes on ice. The cross-linking reaction was quenched by addition of Tris buffer(pH8) to a final concentration of 10mM. The mixture was subsequently subjected to size-exclusion chromatography (Superdex200 10/300, GE Healthcare) and proteins corresponding to molecular weight of ~100kDa were collected to fractions of 0.5ml. To prepare samples for negative-stained electron microscopy, each fraction was adsorbed to plasma-cleaned (Solarus Model 950 cleaner, Gatan) electron microscope grids coated with continuous carbon film, which were subsequently washed with Hepes buffer and stained with 0.75% uranyl formate. Images were collected using EPU software (FEI) on a Tecnai T12 electron microscope (FEI) fitted with a 4K charge-coupled device camera (Gatan) at an effective pixel size of 0.18 nm in the specimen plane with the magnification of 110K or 67K.
T cell activation assay
Wild-type or chimeric CD155 constructs were synthesized (GeneScript) and cloned into SPORT6 or pIRES2-ZsGreen1 expression vectors. All constructs (300–400 ng of DNA) were transfected in 293T cells (105 cells/well) using Lipofectamine 2000 (Invitrogen) in 96-well flat-bottom plates and left overnight. Mouse CD155 surface expression levels were analyzed 24 hours post-transfection by flow cytometry. Transduced 4G4 responder cells (105 cells/well) transduced with B12A αβ chain as described previously were cocultured with transfected 293T cells for 24 hours (2) and mIL-2 amounts in the supernatants were subsequently determined by ELISA (R&D).
Data availability
The structural coordinates of MHC-independent TCRs A11 and B12A have been deposited in the Protein Data Bank (PDB, www.rcsb.org) under the accession 6C68 and 6C61, respectively. The sequence data described in the manuscript are freely accessible through ImmuneAccess [http://clients.adaptivebiotech.com/pub/lu-2019-natcomms] with doi [10.21417/JL2019] and are available upon request.
Results
A11 and B12A recognized MHC-independent ligands
In contrast to MHC deficient animals that are devoid of mature αβT cells (10–13), Quad deficient mice developed ~25% of mature αβT cells, which was further increased to that similar to B6 in the presence of Bcl2 transgene (14). Two of these T hybridomas, A11 and B12A, recognized mouse CD155, a murine poliovirus receptor homolog (2). Moreover, transgenic T cells expressing A11 or B12A failed to proliferate in CD155−/− but MHC sufficient host while developed normally in CD155+/+ host mice (15), suggesting these TCR utilized CD155 as their selection ligand during thymic development. To further confirm that A11 and B12A TCRs have no intrinsic MHC reactivity, we retrovirally expressed each αβTCR in a TCR-negative 4G4 T cell line (16). Both A11 and B12A expressing 4G4 cells responded to anti-TCR stimulation as measured by their IL-2 secretion (Fig 1), demonstrating a functional reconstitution of their αβTCR on 4G4 cells. However, unlike MHC-restricted B10.A T cells which reacted only against H-2b expressing but not against MHC-deficient stimulators (Fig 1A), A11 and B12A T cells responded to both B6 and MHCKO(b2m−/−Abb−/−) splenocytes (Fig 1B–C). Comparable amount of IL-2 were secreted in the presence and absence of MHC, demonstrating that the reactivity of A11 or B12A-expressing 4G4 T cells was independent of MHC. To further rule out if their response to MHCKO stimulators resulted from cross reactivity of TCR to non-MHC ligands, we stimulated B12A-expressing 4G4 T cells in the presence of MHC-blocking antibodies. The results showed that while blocking MHC abolished the B6 stimulation of MHC-restricted B10.A lymph node T cells, it did not affect the B12A and A11 response to B6 stimulation (Fig 1B–C). Thus, these TCR have no intrinsic MHC reactivity and are functional independent of MHC.
FIGURE 1.
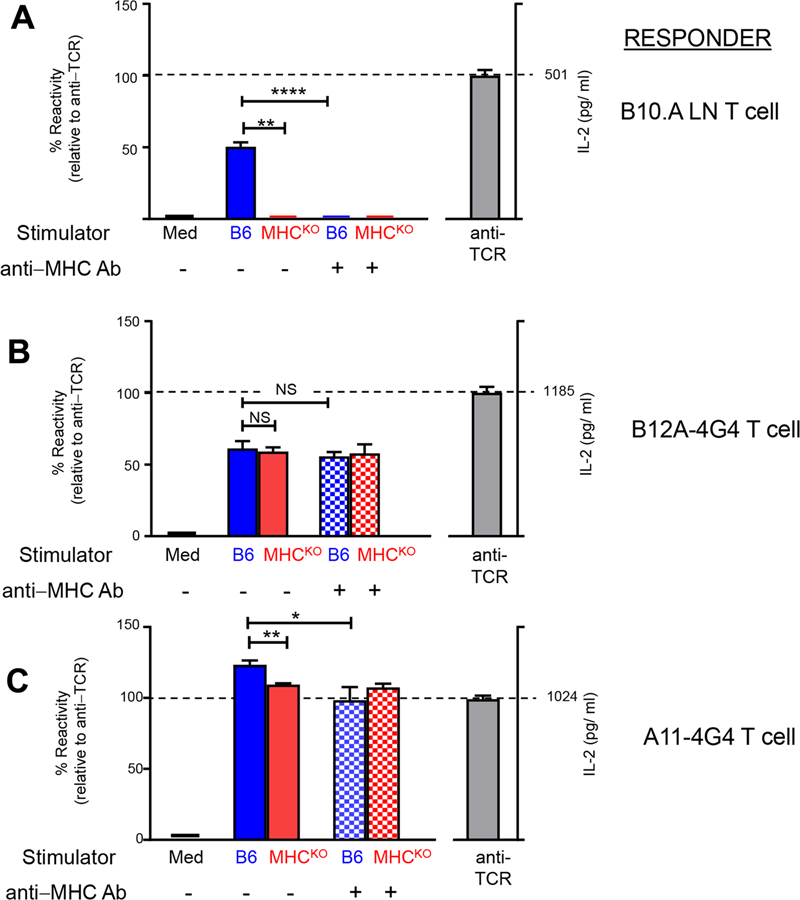
B12A and A11 TCRs display MHC-independent reactivity. TCR− 4G4 T cells were retrovirally transduced with plasmids encoding TCRα and TCRβ sequences as previously described(2). A) Purified B10.A lymph node T cells that were alloreactive against H-2b stimulators were used as control responder T cells. B)5x104 B12A-4G4 or C)A11–4G4 cells were stimulated in vitro overnight with B6 or MHCKO stimulator spleen cells that had been LPS-treated (10μg/ml), irradiated with 3000R, and T-depleted. Where indicated a cocktail of blocking mAbs specific for all H-2b class I and II proteins response cultures were added to the response cultures (10 μg/ml each of Y3P, M5–114, and anti-H-2Db). IL-2 secreted into culture supernatants were measured by ELISA (R&D Systems). IL-2 amounts (pg/ml) secreted in response to platebound anti-TCRβ mAb (H57–597) stimulation is shown (right axis), whereas IL-2 amounts secreted in response to stimulator cells is shown relative to the amount stimulated by platebound anti-TCRβ which was set as 100% (left axis). NS, not significant; ****, p<0.0001; **, p<0.01; *, p<0.05.
Variable domain of MHC-independent TCR is responsible for CD155 recognition
Previously, we showed that the solution binding affinity of B12A to CD155 was 230 nM, much higher than a typical TCR-MHC binding (2). We also measured the binding affinity of A11 to recombinant soluble CD155 and obtained 280 nM dissociation constant KD (Fig 2A). Thus, both A11 and B12A have high affinity binding of CD155. Conventional αβ TCRs recognize short peptidyl- or glyco-antigens presented by MHC or MHC-like molecules. Their antigenic interactions are mediated by TCR variable CDR regions (17, 18). Unlike the conventional MHC-restricted αβ TCRs, both A11 and B12A bound native CD155 without MHC. However, the structural requirement for TCR recognition of non-MHC dependent antigen remains unknown. A previous mutational work of A11 TCR showed the involvement Tyr 46 and Tyr 48 of CDR2β in recognition of CD155 (2). To further address if the recognition of CD155 is mediated by the receptor variable domain, we generated a V-domain only single chain construct of B12A and obtained its solution binding affinity of 400 nM to soluble CD155, slightly less but similar to that of V-C two domain B12A (Fig 2B). Thus, B12A V-domain alone is sufficient to recognize CD155.
FIGURE 2.
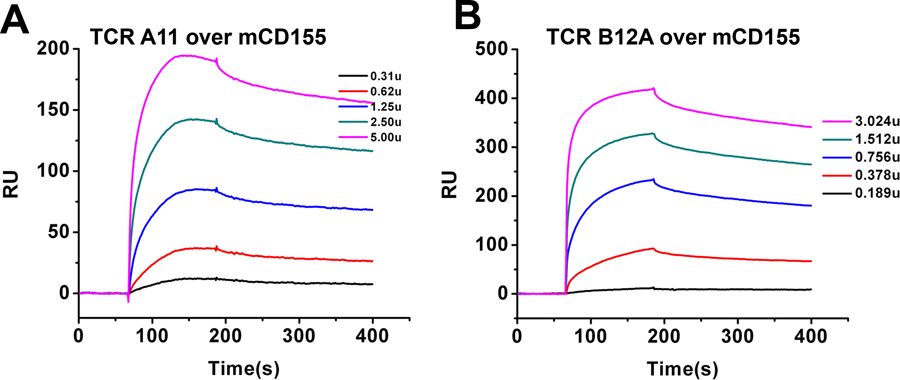
BIAcore binding of soluble murine CD155 by A) A11 and B) B12A V-domain single chain TCRs.
Structures of CD155-specific αβ T cell receptors A11 and B12A
While both A11 and B12A recognize CD155, their TCR sequences are derived from different variable gene segments. The V-region of A11 consists of TRAV12–1 (Vα8) and TRBV13–3*3 (Vβ8.1) alleles; and B12A is composed of TRAV6D-6*02 (Vα4) and TRBV26*01 (Vβ3) segments (Table 1). The CDR3α and β of A11 contain 13 and 10 amino acids while those of B12A contain 13 and 14 residues, respectively. To investigate the structures of MHC-independent TCRs and their recognition of CD155, we expressed soluble αβ TCR A11 and B12A heterodimers, fused to the constant region of human TCR, with an engineered interchain disulfide bond between Cα146 and Cβ154 as described for MHC-restricted αβ TCRs (19). The crystal structures of A11 and B12A were determined to resolutions of 2.6Å and 2.4Å, respectively (Table 2 and Fig 3A–B). All CDR regions of A11 and B12A are well ordered. The overall structure of A11 is nearly identical to that of B12A with a 0.5 Å root-mean-square deviation (rmsd) among all Cα carbons (Fig 3C). In fact, both A11 and B12A exhibit canonical TCR structures with a root-mean-square deviation (rmsd) of 1 Å when superimposed to the structure of an MHC-restricted TCR (3PQY) (Fig 3D).
Table 1.
TCR A11 and B12A V, D, and J segments
| TCR | V-gene | CDR1 | CDR2 | J | CDR3 | |
|---|---|---|---|---|---|---|
| A11 | α | Vα8/TRAV12–01 | TAYSDVA | SSTDNK | J40–01 | CALSPHNTGNYKYVF |
| β | Vβ8.1/TRBV13–03*3 | NNHDY | SYVADS | J1–02 | CASSQTNSDYTF | |
| B12A | α | Vα4/TRAV6D-6*02 | ATSIAYPN | VITAGQ | J37–01 | CALGIITGNTGKLIF |
| β | Vβ3/TRBV26–01 | KGHPV | FQNQEV | J2–02 | CASSPGQGAATGQLYF | |
FIGURE 3.
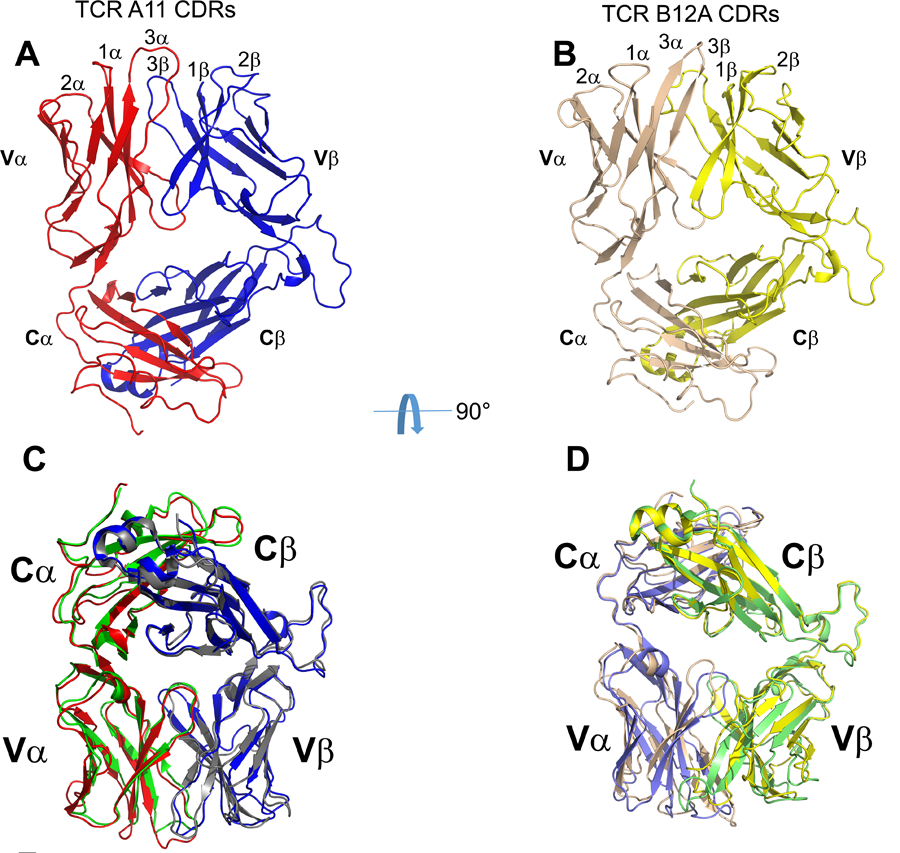
The structures of A11 and B12A TCR. A) and B) Crystal structures of TCR A11 and B12A. C) Structure superposition of TCR A11 and B12A, TCR alpha and beta chain of A11 and B12A were respectively shown by red, blue, green and gray cartoons. D) Structure superposition of TCRB12A and MHC-restricted TCR (PDB ID:3PQY), whose alpha and beta chain were colored in purple and green, respectively.
CDR regions of CD155-specific TCR
As both A11 and B12A exhibit canonical TCR structures, we then examined their CDR sequences for possible reasons of discriminating against MHC binding. Despite recognizing the same CD155 ligand, A11 and B12A use different V-gene segments. A11 and B12A α-chains CDR1&2 are derived from TRAV12–1 (Vα8) and TRAV6D-6*2 (Vα4), respectively, while their β-chains are derived from TRBV13–3 and TRBV26–1, respectively. There is no sequence conservation in A11 and B12A CDR1, CDR2 (Table 1). To address if these V-genes are favored for MHC-independent ligand binding, we compared the usage frequencies of these V-genes between MHC-restricted (B6 and its Bcl2 transgenic littermate B6Bcl2tg) and MHC-independent (QuadKO and its Bcl2 transgenic littermate QuadKOBcl2tg) TCR sequence repertoires(1). The results showed that while the frequencies of both A11 and B12A Vα-genes were slightly higher in MHC-independent TCR sequences, their Vβ-genes frequencies were similar or less in MHC-independent TCR sequences (Fig 4A). Indeed, the same Vα and Vβ-genes of A11 are also found in the structures of MHC-restricted TCR 1LP9 and 1NFD, respectively (Fig 4B–C). Structure comparison showed that both CDR1 and 2 of A11 α and β chains adopted near identical conformations as those of MHC-restricted ones (Fig 4B–C). Similarly, the CDR1 and 2 conformations of Vβ-gene observed in B12A are also in close agreement with those of MHC-restricted (Fig 4D–E). Thus, A11 and B12A V-genes do not discriminate against MHC-restricted ligand.
FIGURE 4.
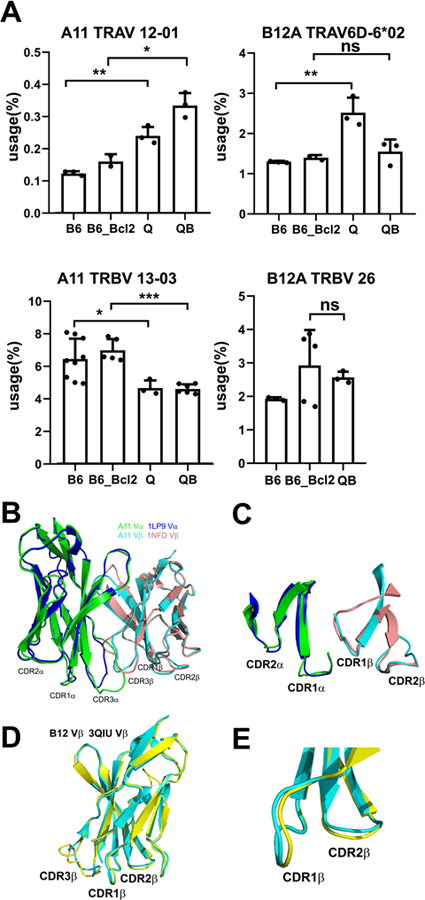
A) Cognate Vα and Vβ -gene usage of TCR A11 and B12A in MHC-restricted (B6 and B6_Bcl2) and MHC-independent (Q and QB) TCRβ repertoires. B) the structural superposition of TCR A11 beta and alpha chains to their cognate germline V gene structures (PDB ID: 1LP9 and 1NFD) showed nearly identical conformations of germline CDR1 and CDR2 loops. C) Enlarged view of germline CDR1 and CDR2 loop structures of TCR A11 as indicated in the superposition in B). D) the structural superposition of TCR B12A beta chain to its cognate germline V gene structure (PDB ID: 3QIU). E) Enlarged view of germline CDR1 and CDR2 loop structures of TCR B12A as superposed in D).
Conventional MHC-restricted TCR favor CDR3 length of 8–13 amino acids (1). To further address if the length of A11 and B12A CDR3 is less optimal for binding to MHC, we compiled CDR3α and CDR3β length distributions from 132 published TCR-MHC complex structures in protein data bank (PDB). Majority of their CDR3α and β are 9–12 amino acids long with average lengths of 11.5 and 11.7 amino acids for CDR3α and β, respectively (Fig 5A). While the lengths of A11 CDR3α and β are within the optimal length of those MHC-restricted TCRs, those of B12A are slightly longer than most of MHC-restricted TCRs (Fig 5A). In the cases of TCR contained a longer CDR3α or β, the longer CDR3 often results in decreased MHC contacts from CDR1 and 2 (1). The longer CDR3 found in CD155-specific B12A, however, is consistent with those from γδTCR as well as those present in the pre-selection repertoire of MHC-restricted animals (20).
FIGURE 5.
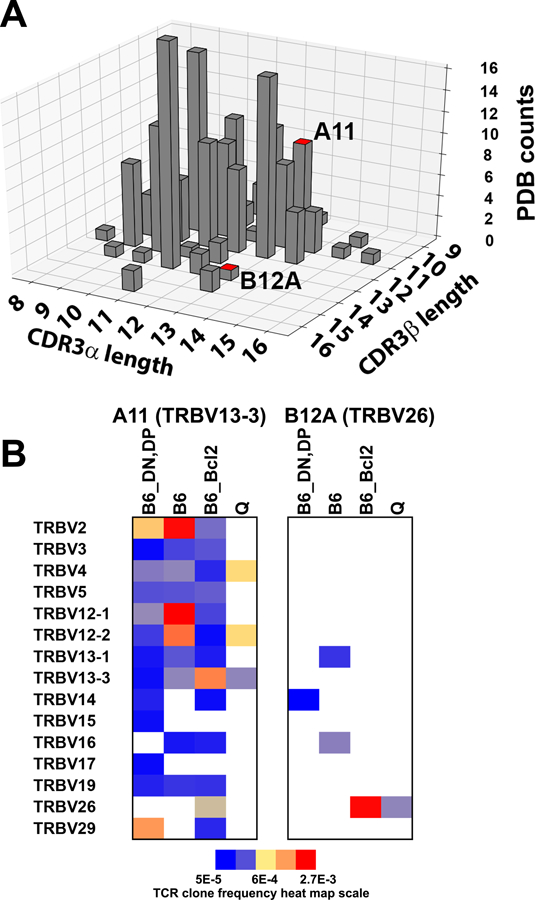
A) paired CDR3α and CDR3β length distribution of MHC-restricted αβ TCRs structures from Protein Data Bank (PDB). The bars showing the equivalent CDR3α and CDR3β length of TCR A11 and B12a were highlighted in red on top. B) heatmap showing the association of A11 or B12A CDR3β sequences with various Vβ genes in pre-selection (B6_DN, DP), MHC-restricted (B6 and B6_Bcl2) and MHC-independent (Q) TCRβ sequence repertoires.
To further assess the uniqueness of A11 and B12A CDR3 sequences, we examined the occurrence of A11 and B12A CDR3 sequences in various TCR sequence repertoires, including pre-selection (B6_DN,DP), mature MHC-restricted (B6) and mature MHC-independent sequences (QuadKO). The 10-residue A11 CDR3β has an optimum length of MHC-restricted CDR3 and its sequence is found in all three repertoires as well as in B6Bcl-2tg repertoires (Fig 5B)(1). In fact, A11 CDR3β occurs rather frequently in B6 with some in the top 1000 sequences out of ~300000 sequences, albeit it is sometimes associated with different V-genes. In addition, the intact TCRβ of A11, the recombination of TRBV13–3 with CDR3β (CDR1, 2 and 3β), can be found in all four repertoires (Fig 5B). Thus, TCRβ of A11 is not unique to MHC-independent TCR. However, TCRα of A11 is not found in any of the MHC-restricted repertoires, suggesting the non-MHC ligand specificity of A11 is derived from the unique pairing between its α/β chains during thymic selection. In contrast, the length of the 14-residue B12A CDR3β is less optimum for MHC-restricted TCR (1), and its sequence occurred rarely in various repertoires but is observed in B6 repertoire (Fig 5B). The TCRβ of B12A (recombination of TRBV26 with its CDR3β), however, is only found in B6Bcl2tg repertoire but not in B6. Consistent with non-MHC ligand specificity is derived from the pairing between TCR α/β chains, the α- but not β-chain of B12A were found in MHC-restricted repertoires.
A11 and B12A TCR recognize two overlapping epitopes on CD155 D1 domain
Previous domain swapping of murine and human CD155 showed that the activation of A11 and B12A required D1-D2 domains of CD155 (2). To further assess the mode of TCR-CD155 recognition, we acquired negative staining electron microscopy images of recombinant B12A in complex with soluble CD155. B12A TCR alone appears as a donut shaped structure and the TCR-CD155 complex resembles the shape of a tadpole with TCR as its head and CD155 as its tail in negatively stained EM images (Fig 6A). While the resolution of the EM images is not sufficient to determine the precise molecular interface, the structural images are best fit with a binding mode in which the TCR is orientated with its CDR regions docked onto the D1 domain of CD155 (Fig 6B). Since both A11 and B12A responded to murine but not human CD155, we used mutational approach to further map the TCR binding region on CD155 D1-domain. In all, there are 25 variant surface residues on CD155 D1 domain between human and mouse (Supplemental Fig S1A). We generated 12 cluster mutations based on the crystal structure of human CD155 (PDB entry 3UDW) to replace mouse CD155 residues with either human counterparts or alanine residues (Fig 6C, Supplemental Fig S1B). The activation of A11 or B12A-transduced 4G4 T cells was measured by IL-2 production in the presence of wildtype or mutant CD155 expressing HEK293T cells (2). Both the mutants and wild type exhibited similar surface expressions of CD155 (Supplemental Fig S2). While majority mutations stimulated either A11 or B12A TCR to produce comparable level of IL-2 as the wild-type CD155, two separate cluster mutations resulted significantly reduced IL-2 production from either A11 or B12A-transduced T cells despite their similar surface expressions of CD155 as the wild-type (Fig 7A–B). Specifically, cluster mutations 6 and 7, located at C’-strand and C’C”-loop impaired A11 activation and mutations 5 and 6, located at CC’-loop abolished B12A activation. Clusters 6 and 7, and 5 and 6 define two continuous surface patches on CD155 structure. These mutational results show that A11 and B12A TCR recognize two closely related but different epitopes on D1 domain of CD155. To further confirm the mutational result, we aggregated the five activating mutations, 2,4,7, 9 and 12, to generate a penta-cluster (Y36T/N57E/K83T/K84Q/E92K/Q124L) mutation. The penta-cluster mutant activated both A11 and B12A like wild-type CD155 (Fig 7A–B). Interestingly, clusters 6 and 7 are located in human CD155 region identified for poliovirus and Tactile (CD96) binding (21, 22). The docking of B12A on CD155 showed the close contact of both CDR2 and CDR3 of B12A TCR with the ligand. This is consistent with previous Ala mutation of A11 CDR2β, that resulted in decreased IL-2 production in response to CD155 (2). The involvement of CDR3 is further supported by the failure of B12F TCR to respond to CD155 stimulation (2). B12F differs from B12A only in its CDR3α residues. Thus, unlike conventional TCRs that only recognize short peptide fragments presented by MHC molecules, A11 and B12A recognize distinct native surface patches on D1 domain of CD155, consistent with their different surface electrostatic potential distributions of their CDR loop regions (Fig 8). The combination of high affinity binding and different surface shape of A11 and B12A epitopes are characteristic of antibody recognition. It suggests non-MHC restricted TCR may recognize a broad spectrum of conformational antigens.
FIGURE 6.
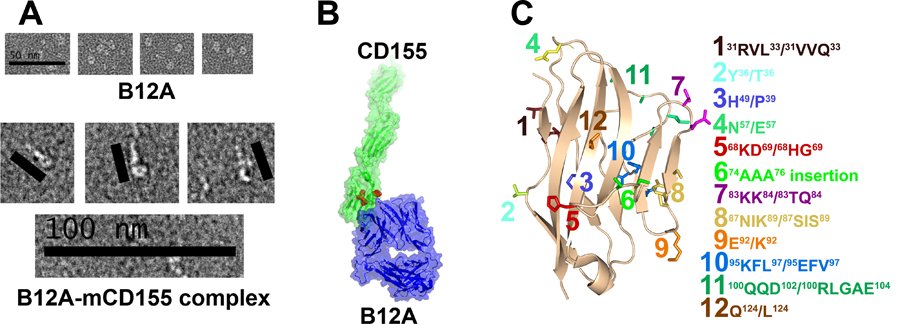
A) representative micrographs of negative-stain electron microscopy of TCR B12A and Glutaraldehyde-crosslinked TCR B12A and CD155 complex. B) The surface representations of TCR B12A(blue) and human CD155 (green) structures. They are positioned based on the micrographs in A) to model the binding of TCR B12A to CD155. C) schematic showing CD155 mutation sites based on hCD155 structure. Among them, site 5 and 6 were highlighted in red in panel B).
FIGURE 7.
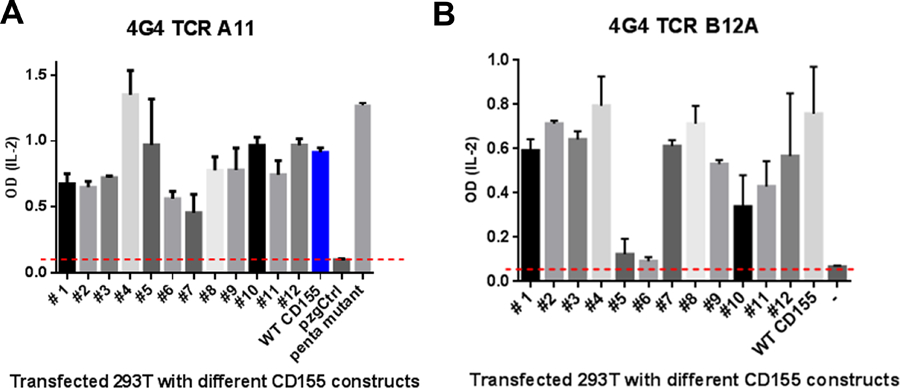
IL-2 production of TCR B12A transduced 4G4 cells activated by different CD155 mutant-transfected HEK293 cells.
FIGURE 8.
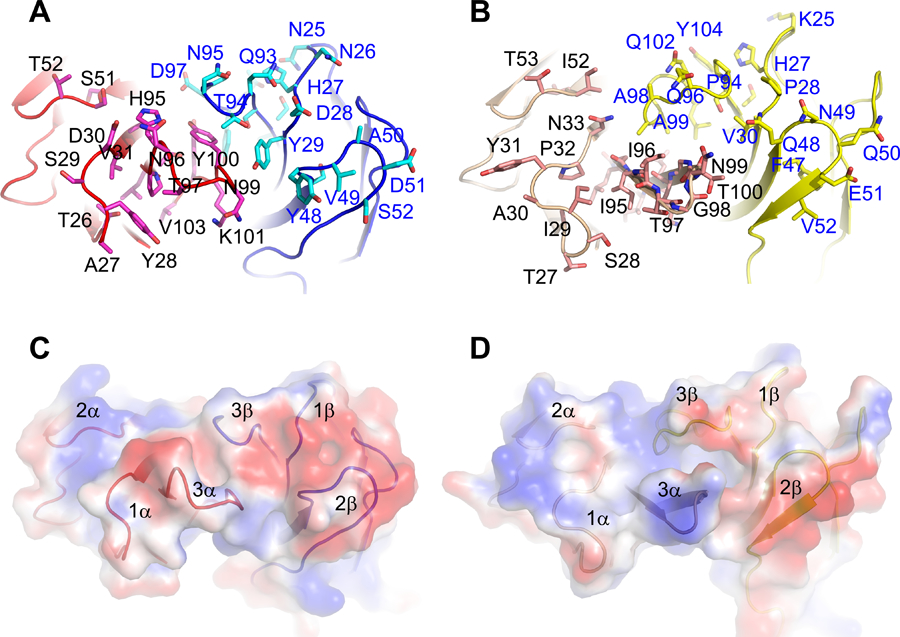
A) and B) CDR loops of A11 and B12A TCR. The CDR residues on A11 and B12A were respectively represented by magenta, cyan, pink and yellow sticks. The numbering of CDR residues was based on signal peptide-cleaved TCR alpha and beta chains. C)and D) Electrostatic surfaces of MHC-independent A11 (A) and B12A (B) TCR showed different charge distribution at the CDR loops of A11 and B12A.
Discussion
We showed previously that TCRs can recognize non-MHC ligands (2). Here, we showed A11 and B12A TCR produced IL-2 in response to MHC-deficient stimulators and blocking of MHC did not affect the IL-2 production from MHC-sufficient stimulators (B6) (Fig 1), demonstrating the functional independence of A11 and B12A to MHC-expression. We determined the first structure of these MHC-independent TCR and mapped their binding epitope on CD155. The structures of A11 and B12A resemble those of MHC-restricted TCR. We showed here that unlike conventional αβTCR recognizing short peptide fragments presented by MHC molecules, A11 and B12A TCRs recognize distinct native surface patches on D1 domain of CD155, consistent with their distinct amino acid and surface electrostatic potential distributions of their CDR loop regions (Fig 8). The combination of high affinity binding and different surface shape of A11 and B12A epitopes are characteristic of antibody recognition.
However, MHC-independent TCRs A11 and B12A bear normal germline genes that are also utilized in MHC-restricted αβTCRs. Through deep sequencing, we could identify their respective Vβ region including CDR3β among preselected, MHC-restricted and MHC-independent mature T cell repertoires, suggesting individual V genes were not sufficient to pre-determine the ligand specificity of TCRs. Recent publication of Krovi et al. showed that the stimulation of unselected αβTCRs by MHC-expressing antigen presenting cells was largely inhibited by anti-MHC blockade (23). It is not clear, however, if these randomized unselected TCRs would fail to respond to MHC-deficient stimulation. While the unselected T cells necessarily contained many MHC-restricted T cells, it is not clear if they are sufficient for the response. The presence of a minor population of non-MHC-restricted T cells in the pool of unselected T cells could not be ruled out without testing their response to MHC-deficient antigen stimulation. Here, both A11 and B12A responded to MHC-deficient antigen presenting cells, while MHC-restricted B10.A allogenic T cells did not (Fig 1). Furthermore, unlike the TCR response from Krovi et al., the response of B12A-4G4 T cells was not affected in the presence of MHC-blockade. Indeed, substantial αβTCRs underwent positive selection and maturation in quad-deficient mice (B2m−/−H-2Ab1−/−CD4−/−CD8a−/− ), demonstrating αβTCRs do not necessarily have intrinsic activity to MHC molecules. This also highlights the importance of thymic selection and its intricate selection mechanism in determining the ligand specificity of αβTCRs.
Our previous study also revealed that transgenic TCRs A11 and B12A could only initiate positive selection through CD155 in the thymus but MHC-independent TCR B12F that was identical to B12A except the CDR3α residues could not undergo positive selection through CD155, demonstrating that MHC-independent reactivity was TCRαβ pair specific. Through VDJ recombination, the adaptive immune system generates huge numbers of TCR α and β chains. Although structural studies have glinted important information for individual α or β chain in the contribution of MHC and antigen recognition, the functional role of their pairing remains to be further investigated. A recent study using TCRβ chain (Vβ8.2 or TRBV13–2) of IAb-3k-reactive YAe62 TCR as a transgene demonstrated that TCRs paired with this transgene and different Vα chains resulted in exclusive MHC II specificity, or cross-reactivity with classical or non-classical MHC-I ligands upon thymic selection on different MHC background, revealing a role of differential TCR αβ pairing in directing MHC ligand specificity (24). The influence of TCR αβ pairing on the ligand specificity of TCR repertoire was also examined by high-through put single-cell sequencing, which showed little overlap between CD4 and CD8 TCR repertoires (25). Therefore, αβ pairing showed strong association with T cell lineage and their corresponding MHC-antigen specificities. In our previous deep-sequencing based repertoire comparisons, we showed that MHC-restricted and MHC-independent TCR repertoires display different molecular constraints as MHC-restricted repertoires have shorter CDR3 length and limit the usage of positively charged and hydrophobic amino acids in CDR3β. In current study, we showed that two MHC-independent TCRs A11 and B12A form canonical TCR structures, whose CDR3s can also be found in pre-selection and MHC-restricted repertoire, but often were recombined with other germline V genes (Fig 5B). Interestingly, each MHC-restricted animal may contain either matched α- or β- but not both chains of A11 and B12A TCR sequences, suggesting the pairing of A11 and B12A is unique for MHC-independent ligand recognition. With the emerging high throughput TCR pairing based on single cell RNA sequencing, the functional importance of TCRαβ pairing in directing their ligand specificity would expand our current understanding of T cell repertoires at different states such as infection, tumor and autoimmunity.
Our current study showed that TCRs may recognize a broad spectrum of conformational antigens other than MHC molecules, which could expand our current dogma of T cell mediated biology. A recent example showed the presence of tumor specific αβT cells in a patient with renal cancer recognized intact TRAIL-DR4 antigen independent of class I and II MHC on human renal cell carcinomas (26, 27). The recognition of a newly formed conformational tumor antigen by a naturally occurring TCR exemplifies that TCR could have broader ligand specificity not as expected. In addition, the antibody like recognition by MHC-independent A11 and B12A could transform our view of how T cell receptors interact with their antigens. As T cells from QuadKOBcl-2tg animals displayed activation or memory phenotype with some animals exhibited autoreactive T cell infiltrations in lung and pancreas (14), it is intriguing to speculate that self-reactive MHC-independent T cells are present in the preselection stage but are normally deleted in thymus. Failure to remove them, however, may result in autoimmunity. Recently a diverse of human γδ T cells were found to be autoreactive to the monomorphic MHC-related protein 1(MR1) in peripheral blood and tissues. Instead of recognizing the MR1-antigen binding groove, these γδ T cells bind underneath the antigen-binding platform of MR1 like antibodies (28). Ignoring the potential link between MHC-independent T cell-antigen recognition and autoimmune diseases may impede our search for cure. Therefore, the current study not only broadens our knowledge of TCR ligand specificity but also highlights an intriguing question whether TCRs recognizing non-MHC ligand would be of clinical implications.
Supplementary Material
Key points:
αβT cell receptors, A11 and B12A, recognize CD155 independent of MHC presentation.
CDR3 of α or β-chain alone is insufficient to discriminate against MHC binding.
This is the first structure of αβTCR in complex with a non-MHC ligand.
Acknowledgement
We thank Kira A. Podolsky and Dr. Siriram Subramaniam for the support of negative staining electron microscopy at the Center for Cancer Research, National Cancer Institute, National Institutes of Health. The funding for this work is from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, and National Cancer Institute.
The funding of this work is provided by the Intramural Research Program (IRP) of National Cancer Institute (NCI) and National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health.
Reference
- 1.Lu J, Van Laethem F, Bhattacharya A, Craveiro M, Saba I, Chu J, Love NC, Tikhonova A, Radaev S, Sun X, Ko A, Arnon T, Shifrut E, Friedman N, Weng NP, Singer A, and Sun PD 2019. Molecular constraints on CDR3 for thymic selection of MHC-restricted TCRs from a random pre-selection repertoire. Nature communications 10: 1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tikhonova AN, Van Laethem F, Hanada K, Lu J, Pobezinsky LA, Hong C, Guinter TI, Jeurling SK, Bernhardt G, Park JH, Yang JC, Sun PD, and Singer A 2012. alphabeta T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity 36: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clements CS, Kjer-Nielsen L, MacDonald WA, Brooks AG, Purcell AW, McCluskey J, and Rossjohn J 2002. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta crystallographica. Section D, Biological crystallography 58: 2131–2134. [DOI] [PubMed] [Google Scholar]
- 4.Otwinowski Z, and Minor W 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276: 20. [DOI] [PubMed] [Google Scholar]
- 5.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ 2007. Phaser crystallographic software. Journal of applied crystallography 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, and Wilson KS 2011. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bricogne G, B. E., Brandl M, Flensburg C, Keller P, Paciorek W, and S. A. Roversi P, Smart OS, Vonrhein C, Womack TO 2011. BUSTER 2.11.2. United Kingdom: Global Phasing Ltd. [Google Scholar]
- 8.Emsley P, Lohkamp B, Scott WG, and Cowtan K 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC. [Google Scholar]
- 10.Grusby MJ, Auchincloss H Jr., Lee R, Johnson RS, Spencer JP, Zijlstra M, Jaenisch R, Papaioannou VE, and Glimcher LH 1993. Mice lacking major histocompatibility complex class I and class II molecules. Proceedings of the National Academy of Sciences of the United States of America 90: 3913–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, and Fugger L 1999. Mice lacking all conventional MHC class II genes. Proceedings of the National Academy of Sciences of the United States of America 96: 10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koller BH, Marrack P, Kappler JW, and Smithies O 1990. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science 248: 1227–1230. [DOI] [PubMed] [Google Scholar]
- 13.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, and Jaenisch R 1990. Beta 2-microglobulin deficient mice lack CD4–8+ cytolytic T cells. Nature 344: 742–746. [DOI] [PubMed] [Google Scholar]
- 14.Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, Adoro S, Adams A, Sharrow SO, Feigenbaum L, and Singer A 2007. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity 27: 735–750. [DOI] [PubMed] [Google Scholar]
- 15.Van Laethem F, Tikhonova AN, Pobezinsky LA, Tai X, Kimura MY, Le Saout C, Guinter TI, Adams A, Sharrow SO, Bernhardt G, Feigenbaum L, and Singer A 2013. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell 154: 1326–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong SC, Chelouche A, Lin RH, Shaywitz D, Braunstein NS, Glimcher L, and Janeway CA Jr. 1992. An MHC interaction site maps to the amino-terminal half of the T cell receptor alpha chain variable domain. Cell 69: 999–1009. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y, Li Y, and Mariuzza RA 2012. Structural basis for self-recognition by autoimmune T-cell receptors. Immunol Rev 250: 32–48. [DOI] [PubMed] [Google Scholar]
- 18.Garcia KC, and Adams EJ 2005. How the T cell receptor sees antigen--a structural view. Cell 122: 333–336. [DOI] [PubMed] [Google Scholar]
- 19.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, and Jones EY 2003. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol 4: 657–663. [DOI] [PubMed] [Google Scholar]
- 20.Fahl SP, Coffey F, Kain L, Zarin P, Dunbrack RL Jr., Teyton L, Zuniga-Pflucker JC, Kappes DJ, and Wiest DL 2018. Role of a selecting ligand in shaping the murine gammadelta-TCR repertoire. Proceedings of the National Academy of Sciences of the United States of America 115: 1889–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Y, Mueller S, Chipman PR, Bator CM, Peng X, Bowman VD, Mukhopadhyay S, Wimmer E, Kuhn RJ, and Rossmann MG 2003. Complexes of poliovirus serotypes with their common cellular receptor, CD155. J Virol 77: 4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deuss FA, Watson GM, Fu Z, Rossjohn J, and Berry R 2019. Structural Basis for CD96 Immune Receptor Recognition of Nectin-like Protein-5, CD155. Structure 27: 219–228 e213. [DOI] [PubMed] [Google Scholar]
- 23.Krovi SH, Kappler JW, Marrack P, and Gapin L 2019. Inherent reactivity of unselected TCR repertoires to peptide-MHC molecules. Proceedings of the National Academy of Sciences of the United States of America 116: 22252–22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadinski BD, Trenh P, Smith RL, Bautista B, Huseby PG, Li G, Stern LJ, and Huseby ES 2011. A role for differential variable gene pairing in creating T cell receptors specific for unique major histocompatibility ligands. Immunity 35: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter JA, Preall JB, Grigaityte K, Goldfless SJ, Jeffery E, Briggs AW, Vigneault F, and Atwal GS 2019. Single T Cell Sequencing Demonstrates the Functional Role of alphabeta TCR Pairing in Cell Lineage and Antigen Specificity. Front Immunol 10: 1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanada K, Wang QJ, Inozume T, and Yang JC 2011. Molecular identification of an MHC-independent ligand recognized by a human {alpha}/{beta} T-cell receptor. Blood 117: 4816–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang QJ, Hanada K, Feldman SA, Zhao Y, Inozume T, and Yang JC 2011. Development of a genetically-modified novel T-cell receptor for adoptive cell transfer against renal cell carcinoma. J Immunol Methods 366: 43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Nours J, Gherardin NA, Ramarathinam SH, Awad W, Wiede F, Gully BS, Khandokar Y, Praveena T, Wubben JM, Sandow JJ, Webb AI, von Borstel A, Rice MT, Redmond SJ, Seneviratna R, Sandoval-Romero ML, Li S, Souter MNT, Eckle SBG, Corbett AJ, Reid HH, Liu L, Fairlie DP, Giles EM, Westall GP, Tothill RW, Davey MS, Berry R, Tiganis T, McCluskey J, Pellicci DG, Purcell AW, Uldrich AP, Godfrey DI, and Rossjohn J 2019. A class of gammadelta T cell receptors recognize the underside of the antigen-presenting molecule MR1. Science 366: 1522–1527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structural coordinates of MHC-independent TCRs A11 and B12A have been deposited in the Protein Data Bank (PDB, www.rcsb.org) under the accession 6C68 and 6C61, respectively. The sequence data described in the manuscript are freely accessible through ImmuneAccess [http://clients.adaptivebiotech.com/pub/lu-2019-natcomms] with doi [10.21417/JL2019] and are available upon request.


