ABSTRACT
Synovial joint development begins with the formation of the interzone, a region of condensed mesenchymal cells at the site of the prospective joint. Recently, lineage-tracing strategies have revealed that Gdf5-lineage cells native to and from outside the interzone contribute to most, if not all, of the major joint components. However, there is limited knowledge of the specific transcriptional and signaling programs that regulate interzone formation and fate diversification of synovial joint constituents. To address this, we have performed single cell RNA-Seq analysis of 7329 synovial joint progenitor cells from the developing murine knee joint from E12.5 to E15.5. By using a combination of computational analytics, in situ hybridization and in vitro characterization of prospectively isolated populations, we have identified the transcriptional profiles of the major developmental paths for joint progenitors. Our freely available single cell transcriptional atlas will serve as a resource for the community to uncover transcriptional programs and cell interactions that regulate synovial joint development.
KEY WORDS: Synovial joint development, Interzone, Single cell RNA-Seq, Articular cartilage, Ligament, Synovium, Chondrocyte, Meniscus, Gdf5
Summary: Defining the transcriptional programs of synovial joint cells in mouse development, including progenitors of tendon, ligament, synovium, meniscus, sheath, and permanent and transient chondrocytes, using single-cell RNA-seq.
INTRODUCTION
Synovial joints are complex anatomical structures comprising diverse tissues, including articular cartilage, synovium, fibrous capsule and ligaments (Decker et al., 2014). Each of these tissues is susceptible to a range of diseases and common injuries that, collectively, have a profound global morbidity (Asahara et al., 2017; den Hollander et al., 2019). A better understanding of the inter- and intracellular networks that govern how these structures emerge during development will inform efforts to generate pluripotent stem cell derivatives for cell replacement therapy and disease modeling (Wang et al., 2019b), and efforts to elicit regeneration in situ (Johnson et al., 2012). Moreover, an improved understanding of joint development will aid in identifying putative disease-causing genes (Kelly et al., 2020).
Over the past two decades, lineage tracing has revealed much regarding the cell populations contributing to murine synovial joint development. It begins with the formation of the interzone (IZ), a region of condensed mesenchymal cells at the site of the prospective joint. In the mouse hindlimb, the IZ is initiated from a Col2a1+ Sox9+ pool of cells recruited from the mesenchymal condensation of the emerging limb bud starting at E11.5 (Hyde et al., 2008; Soeda et al., 2010). It is generally believed that chondrocytes at the presumptive joint de-differentiate (i.e. undergo a chondrocyte-to-mesenchymal transition) and begin to exhibit the flattened and layered morphology that is indicative of the IZ. A history of expressing Gdf5, a TGFβ ligand crucial to joint formation (Storm and Kingsley, 1999), marks cells that initially form the IZ or that later immigrate into it, and that subsequently go on to contribute to all of the major joint constituents, including articular chondrocytes, ligament, meniscus and synovium (Chen et al., 2016; Shwartz et al., 2016).
To gain a more comprehensive understanding of these developmental programs, bulk microarray expression profiling and RNA-Seq have been applied to the developing limb (Taher et al., 2011), to whole joints, including the elbow and knee (Pazin et al., 2012), to the meniscus (Pazin et al., 2014), to the tendon (Liu et al., 2015b), to connective tissue (Orgeur et al., 2018), and to laser-capture micro-dissected regions of the interzone (Jenner et al., 2014). Although these investigations have yielded new insights into the genetic programs underpinning limb and joint morphogenesis, they provide limited resolution of the expression states for individual cell types due to the heterogenous nature of the samples profiled. With the advent of single cell profiling, it is now possible to detect transient populations of cells, to reconstruct developmental transcriptional programs and to identify new cell populations (Guo et al., 2010; Kumar et al., 2017). For example, Feng et al. revealed molecular signatures and lineage trajectories of an interzone-related Lgr5+ population in the murine E14.5 knee joint that contribute to the formation of cruciate ligaments, synovial membrane and articular chondrocytes (Feng et al., 2019).
Here, we have applied single cell RNA-sequencing on Gdf5-lineage cells of the murine hindlimb to determine the transcriptional programs of early synovial joint development. In contrast to the recent study of Feng et al. (2019), which focused on lineage divergence of a specific Lgr5+ interzone population, we sought to characterize formation of the entire IZ and to discover the extent to which heterogeneity in the nascent interzone is resolved into the distinct lineages that are apparent later at cavitation. Therefore, we sequenced Gdf5-lineage cells from the presumptive joint of the hindlimb from E12.5 (prior to overt IZ formation) through E15.5 (coinciding with cavitation). We combined computational analytics and in situ hybridization to identify the transcriptional signatures of joint progenitors and their elaboration of the interzone into the major synovial joint lineages. To aid the community in discovering additional transcriptional programs and in inferring cell interactions that contribute to synovial joint development, we have made this data freely and easily accessible with a web application at cahanlab.org/resources/joint_ontogeny.
RESULTS
Gdf5Cre+ cells in the hindlimb from E12.5 to E15.5 Gdf5CreR26EYFP mice are primarily located in the interzone, articular cartilage, ligament, menisci and synovium, as well as in other non-joint tissues
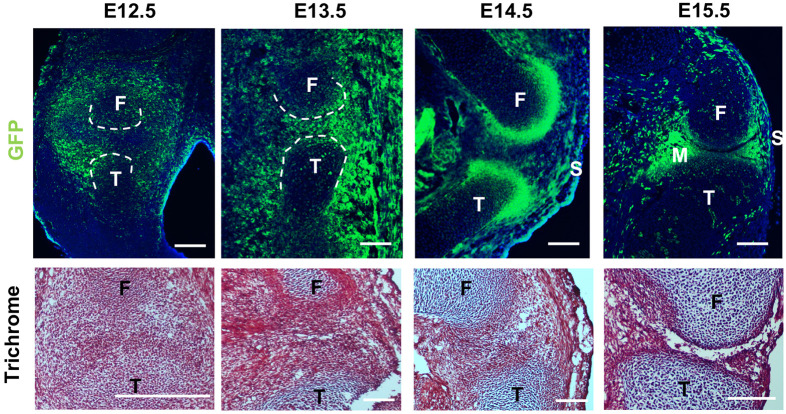
Gdf5-lineage cells contribute to several components of synovial joint, including articular cartilage, meniscus, ligaments, and synovium. To isolate joint progenitors, we crossed Gdf5 promoter-driven Cre mice with the R26 reporter mice in which loxP-flanked STOP sequence followed by the EYFP was inserted into the Gt(ROSA)26Sor locus, allowing us to identify Gdf5-lineage cells by YFP expression. We used fluorescent immunohistochemistry to determine the spatial and temporal pattern of YFP. At E12.5, YFP is mainly expressed in the presumptive joint area including part of the bone anlagen and the surrounding mesenchyme (Fig. 1). At E13.5, YFP+ cells are more centered in the interzone (IZ) and in the surrounding connective tissue; they are sparse in the anlagen of the femur and tibia. By E14.5, YFP+ staining is mainly present at the area of future articular cartilage (AC), synovium and surrounding soft tissue. YFP expression becomes obvious in menisci 1 day later. YFP+ cells are also seen in AC, epiphyseal cartilage and synovium at E15.5.
Fig. 1.
Morphology of the embryonic knee joint and localization of Gdf5-lineage cells. Top: localization of Gdf5-lineage cells in murine hindlimb. Bottom: cell density and morphology during joint formation, as shown by trichrome staining. n=6 per timepoint. F, femur; M, meniscus; S, synovium; T, tibia. Scale bars: 100 µm.
We observed ‘ectopic’ YFP expression in non-joint tissues such as the dermis and muscle, consistent with previous reports (Roelofs et al., 2017). However, because our scRNA-Seq analysis pipeline includes a ‘cell typing’ step (see below), we were able to identify these non-joint cells in silico and exclude them from our in-depth analyses that focus on the Gdf5-lineages of the joint. We refer to cells that passed our in silico filtering as Gdf5-lineage enriched (GLE) cells rather than YFP+ Gdf5-lineage cells because we cannot absolutely prove that YFP expression tracks with Gdf5-lineage in this system. Nonetheless, our staining, combined with prior reports examining Gdf5Cre cells in the limb, indicate that GLE cells are major cellular contributors to the knee joint. Therefore, determining their transcriptomes will yield insights into the genetic circuitry that accompanies IZ formation and the emergence of articular components such as ligament and tendon.
GLE cells form three distinct super clusters across two major developmental stages
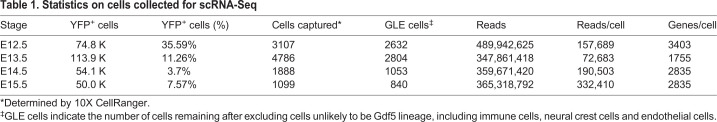
To define the transcriptional states of joint cells and their progenitors during landmark developmental events, we isolated YFP+ cells from the hind limbs of male embryos from E12.5 (the time just before overt IZ formation) to E15.5 (coinciding with cavitation). To minimize contamination with Gdf5-lineage cells from the ankle and digits, we manually dissected the region of the limb containing the presumptive joint and excluded the paw (Fig. S1A). We then collected Gdf5 lineage cells by fluorescence-activated cell sorting (FACs) of YFP+ cells after enzymatically disassociating the presumptive knee joint region (Fig. S1B). We loaded approximately 6000 cells per timepoint for single cell RNA-Seq library preparation using the 10x Genomics platform (Fig. S1C), and we sequenced the transcriptome of ∼1000 to 5000 cells at a target depth of 100,000 reads per cell (Table 1).
Table 1.
Statistics on cells collected for scRNA-Seq
Next, we performed quality control to exclude potential doublets and potential low-quality libraries. We defined doublets as the cells in the top 5% of total reads per capture, based on the estimated doublet rate of the 10x platform (Zheng et al., 2017). We labeled libraries as low-quality if they had fewer than 500 genes detected or if their total transcriptome consisted of greater than 5% of mitochondrially encoded genes. We then sought to identify the major transcriptional states in our data by clustering using the Leiden graph-based community detection algorithm (Traag et al., 2019). We found 12 clusters, many of which contained cells from multiple timepoints (Fig. S2A). To determine the cell type of each cluster, we used SingleCellNet to classify individual cells based on a well-annotated reference data set (Tan and Cahan, 2019), and we used differential gene expression to identify marker genes of cell types that are not included in current single cell reference data sets (e.g. neural crest cells and melanocytes). This approach identified six clusters made up of non-joint cell types, including myoblasts, immune and red blood cells, neural crest cells and melanocytes, and endothelial cells (Fig. S2B). After removing these non-joint cells, we re-clustered the data and performed differential gene expression analysis (Fig. S2C). All clusters had detectable levels of the osteochondral transcription factor (TF) Sox9, except one that had high levels of genes associated with dermis, including Twist2 and Irx1 (Fig. S2D). To localize the cells in this cluster, we performed in situ hybridization, confirming that they were dermal cells (Fig. S2E), and we excluded these cells from further analysis, resulting in a dataset of 7329 synovial joint GLE cells.
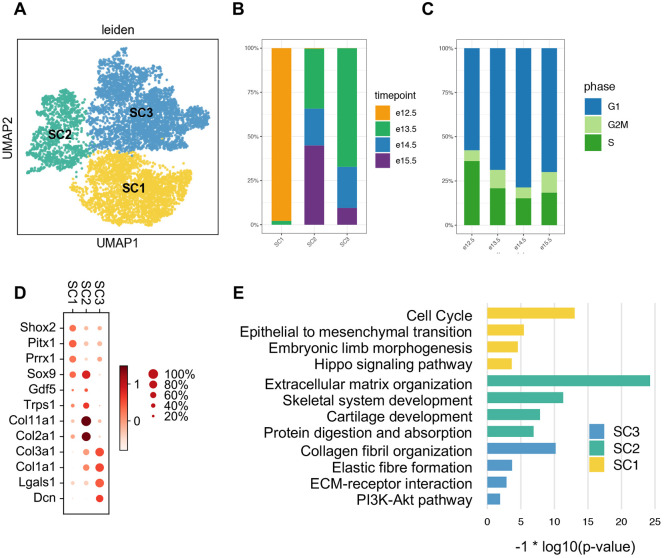
Next, we asked whether there were discernible transcriptional profiles that spanned timepoints. To address this question, we clustered all of the GLE cells and uncovered three ‘super-clusters’ (SCs), two of which contain a plurality of cells from more than a single timepoint (Fig. 2A,B). One of the clusters corresponds roughly to developmental time: SC1 is 98.2% E12.5 cells. The other two SCs are mixtures, with SC2 and SC3 predominately made up of cells from E13.5 to E15.5. We inferred the stage of cell cycle by scoring each cell based on its expression of phase-specific sets of genes. This analysis showed that, as expected, the proportion of cells in either the G2M or S stages gradually decreased over developmental time (Fig. 2C). To gain a better understanding of these SCs, we examined the expression of genes with well-established roles in limb and joint development. Prrx1 and Pitx1 are preferentially expressed in the early SC1 (Fig. 2D), consistent with their roles in specifying limb mesenchymal cells from lateral plate mesoderm (Bobick and Cobb, 2012; Marcil et al., 2003; Wang et al., 2018). Shox2, which regulates the onset of early chondrogenesis (Bobick and Cobb, 2012), has a similar expression pattern. As many cells in SC1 express Sox9 but few express Col2a1, it is likely that this supercluster comprises a mixture of progenitor cells of mesenchymal character and chondroprogenitors. SC2 is similar to SC1 in expression profile, but it also preferentially expresses Sox9, Gdf5, Col11a1 and Col2a1, suggesting that this SC is likely to contain a mixture of IZ cells and transient chondrocytes (Zhao et al., 1997). SC3 cells express the fibrous-related genes Col3a1, Col1a1, Lgals1 (Dasuri et al., 2004) and Dcn (Havis et al., 2014), indicating that SC3 largely consists of fibroblast-related cells.
Fig. 2.
scRNA-Seq of Gdf5 lineage-enriched cells during knee development. (A) Leiden clustering and UMAP embedding of the three distinct superclusters of GLE cells. (B) The proportion of cells from each timepoint varies across superclusters. (C) The proportion of mitosis phases in each timepoint. (D) Expression of genes well-characterized in limb and joint development. Size of each dot reflects the percentage of cells in which the gene is detected within the supercluster. The color indicates mean expression, including cells in which there is no detectable expression. (E) Supercluster gene set enrichment analysis, showing selected categories. Complete results are in Table S1.
Gene set enrichment analysis largely corroborated our supervised annotation of the superclusters (Fig. 2E). SC1 is enriched in limb and joint development-associated pathways, including embryonic limb morphogenesis, the Hippo signaling pathway (Bhattaram et al., 2010; Pan, 2010) and epithelial-to-mesenchymal transition. SC2 is enriched in extracellular matrix (ECM) organization, skeletal system development and cartilage development. SC3 involvement in fibrous differentiation is supported by the enrichment of collagen fibril organization and elastic fiber formation.
Taken together, this analysis has revealed three major transcriptional states of GLE cells in synovial joint development. It has also hinted at substantial heterogeneity within SCs. To more clearly define the cell types and states of GLE cells, we next analyzed each SC separately, as described in the following sections.
Two categories of early GLE cells: chondrogenic and mesenchymal
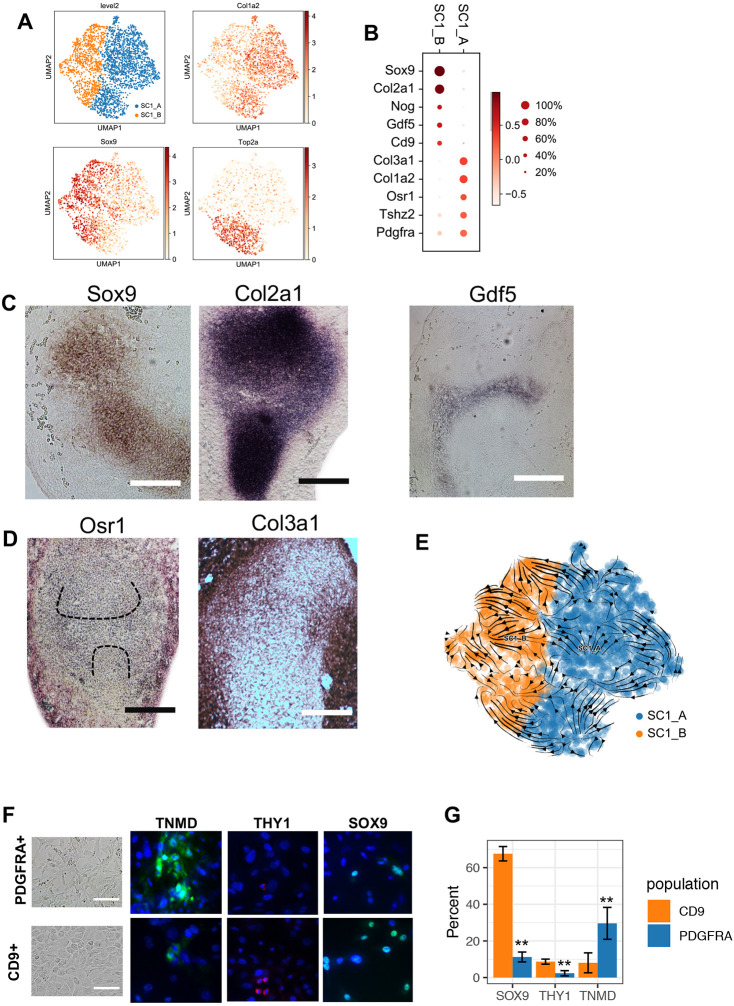
By applying Leiden clustering to only SC1, we identified two subclusters: SC1_A and SC1_B (Fig. 3A). SC1_B has high expression levels of genes associated with chondrogenesis (e.g. Sox9 and Col2a1) and the IZ (e.g. Nog and Gdf5) (Hartmann and Tabin, 2001; Ray et al., 2015; Storm and Kingsley, 1996). SC1_A exhibited high expression levels of genes associated with fibrous and mesenchymal cells, such as Col3a1 and Col1a2 (Niederreither et al., 1992), as well Osr1, which is mainly expressed in the outer mesenchyme where it promotes fibroblast differentiation and inhibits chondrogenesis (Stricker et al., 2012) (Fig. 3B). A substantial fraction of each subcluster was in S or G2M phase of cell cycle, indicating active proliferation. These results suggest that SC1 comprises chondroprogenitors and early chondrocytes of the limb anlagen, nascent IZ cells, as well as the non-chondrogenic mesenchymal cells situated outside the anlagen. We tested and confirmed this conjecture using in situ hybridization for genes indicative of each cluster (Fig. 3C,D).
Fig. 3.
SC1 is composed of chondrogenic and mesenchymal fated cells. (A) Leiden clustering and UMAP map embedding SC1 and representative gene expression patterns. (B) Dot plot expression of representative genes differentially expressed between SC1_A and SC1_B. (C,D) In situ hybridization detection for SC1_A and SC1_B representative genes. (E) RNA velocity analysis. Arrows indicate the predicted future state of SC1 cells, showing a minimal transition between SC1_A and SC1_B. (F) In vitro culture of YFP+/PDGFRA+ and YFP+/CD9+ hindlimb cells from E12.5 embryos shows distinct morphology of the cells (left). Immunofluorescence staining of the tendon and ligament marker TNMD, the fibroblast marker THY1 and the chondrocyte regulator SOX9 (right). (G) Quantification of the proportion of cells positive for each marker. Three independent experiments were conducted. Data are mean±s.d. **P<0.01 versus CD9 group (unpaired Student's t-test). Scale bars: 100 µm.
To determine the lineage relationship between these clusters, we performed RNA Velocity analysis (La Manno et al., 2018). Our results predicted that there is very little transition between SC1_A and SC1_B (Fig. 3E). To test this prediction, we prospectively isolated E12.5 YFP+ cells using antibodies specific for SC1_A (CD9) or SC1_B (PDGFRA), and measured lineage-specific marker expression after culturing the cells in vitro for 7 days. Cells from the PDGFRA+ population exhibited a mesenchymal morphology, whereas cells from the CD9+/PDGFRA− population exhibited a chondrocyte-like morphology (Fig. 3F, left). Consistent with their respective shapes and appearances, the PDGFRA+ population yielded a substantially higher proportion cells positive for the tendon and ligament marker TNMD compared with the CD9+/PDGFRA− population, and a lower proportion of cells positive for the chondrogenesis regulator SOX9 as measured by immunofluorescence [Fig. 3F (right), G]. Although the CD9+ population yielded more THY1-positive cells, neither group had a substantial fraction of positive cells. The fact that both populations were not mutually exclusive for TNMD and SOX9 expression can be explained by incomplete lineage commitment, by the imperfect ability of PDGFRA to mark SC1_A and of CD9 to mark SC1_B, and by impurity in the FACS gating. With these caveats in mind, the data support a model where the in vitro differentiation propensity of SC1_A is towards a tenocyte/ligamentocyte fate, whereas the in vitro propensity of SC1_B is towards a chondrocyte fate.
Diverse trajectories of the Sox9 high compartment of E12.5 GLE cells
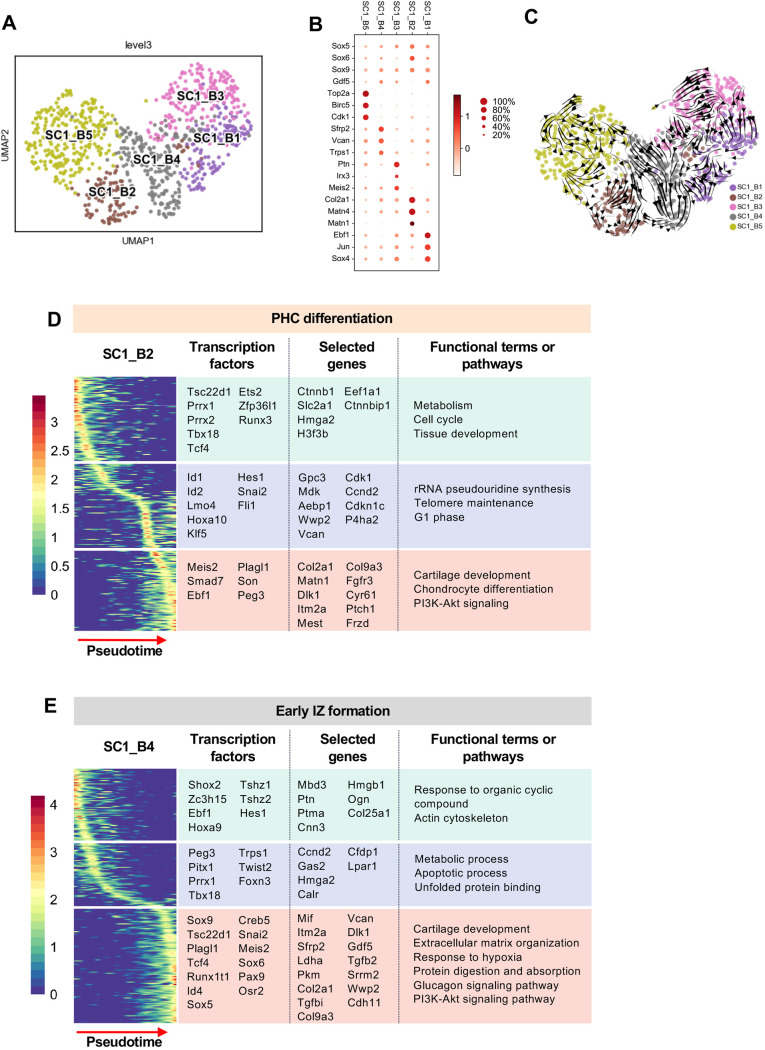
Although most SC1_B cells expressed Sox9, we noticed that they were heterogeneous in terms of IZ- and chondrocyte-related genes, suggesting that this cluster consisted of subpopulations or sub-states. To examine this further, we clustered SC1_B alone and identified five clusters: SC1_B1 to SC1_B5 (Fig. 4A). SC1_B2 was marked by high levels of Col2a1 and Matn1, indicating that it contained cells destined to become transient, epiphyseal chondrocytes (Hyde et al., 2007) (Fig. 4B). SC1_B5 was marked by high levels of cell cycle-related genes. The three other clusters expressed both chondroprogenitor transcription factors (e.g. Sox5, Sox6 and Sox9), as well as the IZ marker Gdf5. These clusters varied in the extent to which they expressed other IZ-related genes: SC1_B4 had high levels of Sfrp2, Vcan and Trps1, whereas SC1_B2 had the highest levels of Ebf1, Jun and Sox4, and SC1_B1 had the highest expression of Ptn, Irx3 and Meis2 (Fig. 4B) (Pazin et al., 2012; Choocheep et al., 2010; Kunath et al., 2002; Norris et al., 2007; Salva and Merrill, 2017).
Fig. 4.
Dynamic expression patterns of nascent interzone. (A) Leiden clustering and UMAP map embedding of SC1B. (B) Dot plot of expression of representative genes differentially expressed between SC1B subclusters. (C) RNA velocity indicates no obvious trajectories of subclusters. Pseudotime analysis identifies three stages of gene expression in PHC differentiation (D) and early IZ formation (E). Transcription factors, selected genes and functional terms or pathways from each stage are listed on the right.
To explore possible lineage relationships between these clusters, we performed RNA velocity analysis. Although there was some predicted flow from the proliferating cluster SC1_B5 to SC_B2 (PHC) and to SC1_B4 (IZ), there was otherwise very limited, consistent flow predicted between clusters, suggesting that these clusters represent distinct lineages. Next, we used pseudotime analysis to identify genes and pathways that are dynamically expressed and thus that potentially contribute to the nascent IZ and the PHC trajectories. We applied diffusion-based pseudotime on each subcluster separately, selecting a root or starting point for the trajectory based on the RNA velocity results (Fig. S3A,B) (Haghverdi et al., 2016). Then we identified genes that were significantly correlated with pseudotime along each trajectory, grouped these genes based on the period (beginning, middle or end) at which they had the highest mean expression (Street et al., 2018), and then performed gene-set enrichment.
The trajectory of PHC differentiation was characterized by initially high levels of expression of genes involved in proliferation and metabolism, as well as transcription factors (TFs) associated with early limb development, such as Prrx1, Prrx2 (ten Berge et al., 1998), Pitx1 (Nemec et al., 2017), Tbx18 (Haraguchi et al., 2015), Tcf4 (Cho and Dressler, 1998) and Ets2 (Ristevski et al., 2002) (Fig. 4D and Table S1). The detection of Ctnnbip1 (also known as ICAT), a repressor of Wnt signaling (Tago et al., 2000), at the onset of PHC pathway in cells that express both Tcf4 and β-catenin is consistent with the pre-eminent role of Wnt in regulating IZ formation (Hartmann and Tabin, 2001; Tufan and Tuan, 2001). In addition to these pathways and genes previously implicated in chondrogenesis, our analysis has identified Tsc22d1 and Zfp36l1 as potential novel regulators of this initiation phase.
Genes with diverse roles in chondrogenesis and chondrocyte differentiation were upregulated in the middle phase. The growth factor Mdk and the proteoglycan Vcan play roles in chondrogenesis via Wnt and TGFβ signaling, respectively (Choocheep et al., 2010; Haffner-Luntzer et al., 2014; Wang et al., 2014). P4ha2 encodes a component of a collagen synthesis enzyme, and mice null for this gene exhibit impaired ECM in chondrocytes (Aro et al., 2015). Gpc3, which is expressed in the regions of transient chondrocytes, controls limb patterning by interacting with BMP4, a stimulator of chondrogenesis (Miljkovic et al., 2008; Paine-Saunders et al., 2000). Aebp1 regulates collagen in ECM during chondrocytes differentiation (Blackburn et al., 2018). And Wwp2 directly interacts with Sox9 to enhance its transcriptional activity in chondrogenesis (Nakamura et al., 2011). Other genes upregulated in the middle phase do not have overt or previously described links to chondrogenesis and differentiation, but rather suggest a differentiation pause, as indicated by expression of Id1 and Id2, which interact with Hes1 to repress differentiation (Zhang et al., 2014), and by expression of several cell cycle regulators, including Cdk1, Ccnd2 and Cdkn1c.
The late stage of the PHC trajectory was marked by expression of many well-characterized players in pre-hypertrophic chondrocyte function and regulation (Fig. 4D and Table S1). Many genes encoding contributors to the ECM were upregulated in this phase: Acan, Col2a1, Col9a1, Col9a2, Col9a3, Epyc, Col7a1, Hapln1, Col11a1, Matn1 and Matn4 (Hyde et al., 2007; Lawrence et al., 2018; Mayo et al., 2009; Zhang et al., 2008). Several signaling pathways were implicated in this phase, including Fgf, Wnt, BMP/TGFβ and Hedgehog. Fgfr3, which induces terminal hypertrophic differentiation (Su et al., 2014), the Wnt antagonist Frzb (Enomoto-Iwamoto et al., 2002; Leimeister et al., 1998; Witte et al., 2009), Smad7 (Vargesson and Laufer, 2001), Cthrc1 (Kimura et al., 2008) and Dlk1 (Chen et al., 2011; Schmid et al., 2010; Taipaleenmäki et al., 2012; Wong et al., 1997), genes associated with BMP/TGFβ, and the Sonic hedgehog receptor Ptch1 were all upregulated in this phase. This analysis also revealed several TFs that have not, to our knowledge, been implicated in PHC, including Son and Peg3.
Next, we explored the early IZ trajectory of SC1_B4 cells (Fig. 4E and Table S1). They start with high expression of TFs such as Shox2, which regulates the onset of early chondrogenesis (Bobick and Cobb, 2012), the limb morphogenesis determinant Tshz1 (Coré et al., 2007) and the patterning homeobox TF Hoxa9 (Raines et al., 2015). Genes such as Cnn3 (Haag and Aigner, 2007), which is associated with the actin cytoskeleton, were also upregulated. The middle phase of early IZ formation was marked by a similarity to TFs of the early PHC trajectory (Pitx1, Prrx1, Tbx18 and Foxn3), in addition to the mesenchymal TF Twist2. In addition, Trps1 acts downstream of Gdf5 to promote chondrogenesis (Itoh et al., 2008). The late stage was characterized by a mixture of IZ- and chondrocyte-associated genes and pathways. For example, some chondrogenic-related genes and their regulators included Col2a1, Col9a3, Dlk1, Wwp2, Sulf1, Cd9 and Col11a1 (Wang et al., 2018; Chen et al., 2011; Seegmiller et al., 1971; Zhao et al., 2006), and Sox5, Sox6 and Sox9. On the other hand, IZ-associated genes such as Gdf5, Sfrp2, Osr2, Tgfbi, Tgfb2 and Pgk1 (Amarilio et al., 2007; Gao et al., 2011; Pazin et al., 2012), and Snai2 were also upregulated. Taken together, this profile suggests a complex state that is poised between chondrogenic and mesenchymal (or IZ).
IZ formation
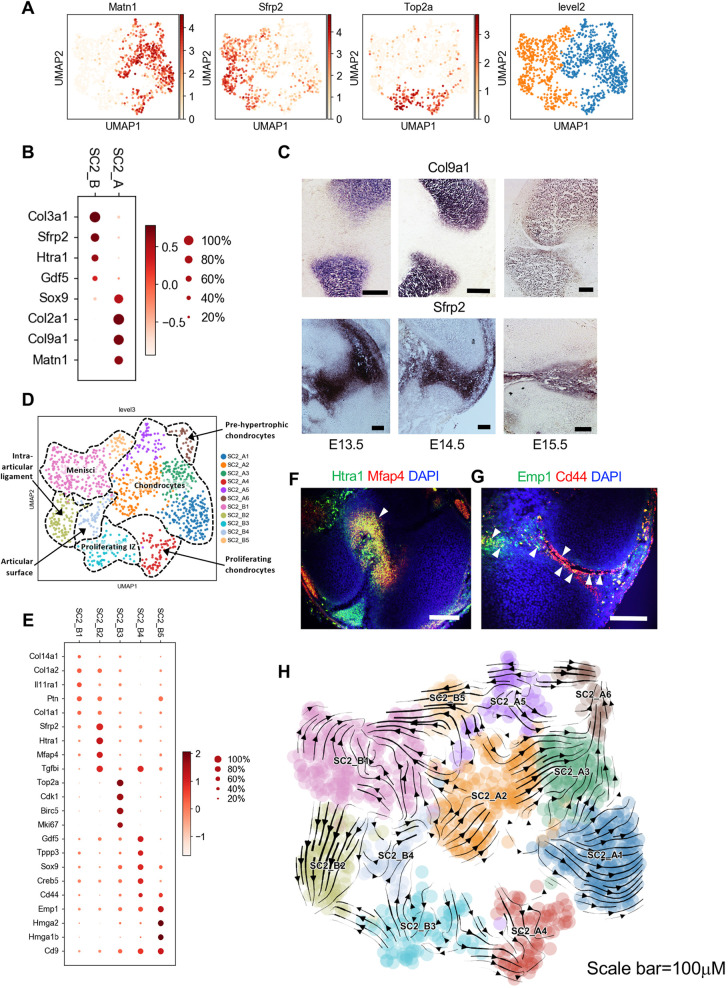
Compared with SC1, many SC2 cells had high levels of IZ-related genes such as Cd44 and Sfrp2; other SC2 cells exhibited more established chondrocyte profiles. To resolve this population heterogeneity, we performed clustering on SC2 and identified two groups (Fig. 5A). SC2_A was enriched in chondrocyte-related genes Col2a1, Col9a1, Sox9 and Matn1, whereas SC1_B expressed IZ-related genes such as Gdf5, Sfrp2 and Htra1 (Fig. 5B). We confirmed the expression localization of some of these genes by in situ hybridization (Fig. 5C and Fig. S4A).
Fig. 5.
SC2 is composed of interzone fated cells. (A) Leiden clustering and UMAP map embedding SC2 and representative gene expression patterns. (B) Dot plot expression of representative genes differentially expressed between SC2_A and SC2_B. (C) In situ hybridization detection for SC2_A (Col9a1) and SC2_B (Sfrp2) representative genes illustrating expression of Col9a1 in transient cartilage and Sfrp2 in the IZ. (D) Subclustering of SC2_A and SC2_B. Potential cell types are labeled. (E) Dot plot expression of representative genes differentially expressed between SC2_B subclusters. (F) Coronal section of E15.5 knee joint showing co-expression of Htra1 (green) and Mfap4 (red) in intra-articular ligaments. (G) Detection of Emp1 (green) and Cd44 (red) transcripts with RNAscope Hiplex probes on sagittal section of E15.5. Arrowheads indicate co-expression. (H) RNA Velocity analysis. Arrows indicate the predicted future state of SC2 cells, showing a transition between SC2_A2 and SC2_A3, SC2_A3 and SC2_A6, and SC2_B5 and SC2_B1. Scale bars: 100 µm.
Noticing heterogeneity of gene expression within SC2_A and SC2_B, we clustered them at a finer level of resolution, resulting in six and five subclusters, respectively (Fig. 5D). As in SC1_B, the chondrocyte SC2_A cluster included a subcluster (SC2_A6) that expressed Ihh, Cd200, Mef2c, and Panx3, suggesting a pre-hypertrophic state (Fig. S4B) (Etich et al., 2015). SC2_A also included clusters indicative of a proliferative state (SC2_A4), higher collagen expression patterns (SC2_A2), developmental stage (SC2_A1 and SC2_A3) and specialized chondrocyte expression signature [e.g. coordinated expression of S100a6 and S100a10, Anxa1 and Anxa2, and Timp1 in SC2_A5 (Headland et al., 2015; Yammani, 2012)]. We did not follow up on these transcriptionally distinct transient chondrocyte clusters. Rather, we chose to focus more on SC2_B because it included subclusters expressing high levels of IZ-related genes. Subcluster SC2_B1 was distinguished by higher expression of fibroblast-related collagen Col1a1, Col1a2, Ptn and Col14a1, suggesting it represents cells of the meniscus (Fig. 5E) (Hyde et al., 2008; Mittapalli et al., 2009). Further supporting this hypothesis is the fact that several genes previously reported as being more highly expressed in E16 meniscus as compared with AC or CL (i.e. Dpt, Igfbp3, Rbp1 and Ogn) were preferentially expressed in SC2_B1 (Pazin et al., 2014) (Fig. S4C). SC2_B2 expressed high levels of IZ-linked Sfrp2 and Htra1 (Oka et al., 2004), as well as Mfap4 and Tgfbi. Because Sfrp2 is expressed primarily in intra-articular ligament at later stages of development (Pazin et al., 2012), we hypothesized that SC2_B2 may include IZ cells that contribute to the presumptive intra-articular ligament. We explored this hypothesis by performing multiplex in situ hybridization using probes for Htra1 and Mfap4. We found that cells positive for both Htra1 and Mfap4 were primarily located at the nascent intra-articular ligament at E15.5, supporting the notion that SC2_B2 is a ligament associated IZ cluster (Fig. 5F).
SC2_B4 had high levels of sheath of cartilage-related and tendon-related genes Gdf5, Tppp3 (Staverosky et al., 2009), Sox9 and Cd44 (Hartmann and Tabin, 2001). This cluster also exhibited high levels of Igfbp2, which has previously been reported to be specifically expressed along the superficial aspects of the articular cavity (Pazin et al., 2014) (Fig. S4D). Based on this, we speculated that this cluster represents cells of the thin sheath of fibrous tissue that lines the surface of articular chondrocytes and the meniscus. To determine the spatial positioning of these cells, we again used multiplex in situ hybridization with probes for Cd44 and Emp1, a gene that inhibits chondrocyte differentiation (Li et al., 2011) that was also preferentially expressed in SC2_B4 and SC2_B5. We found that Cd44 was specifically expressed at the emerging articular surface (Fig. 5G), whereas Emp1 was expressed on articular cartilage, menisci and the joint capsule. We identified a small number of cells that expressed both Cd44 and Emp1 at the outermost AC layers and menisci. Taken together, these results support the notion that SC2_B4 contains synovial cavity-lining cells and that SC2_B5 comprises meniscal cells.
To understand the lineage relationship between these clusters, we performed RNA velocity analysis on SC2 cells (Fig. 5H). Overall, the minimal flow between SC2_A and SC2_B is consistent with the fact that most SC2_A cells expressed Matn1 and thus are already undergoing pre-hypertrophic chondrocyte differentiation. However, we did observe two cases in which there was flow between SC2_A and SC2_B. Approximately ten SC2_A (SC2_A5) cells had velocity towards SC2_B (SC2_B5); however, these cells did not express Matn1, suggesting that they were undergoing a chondroblast-to-mesenchymal transition (Fig. S4E). On the other hand, a small number of SC2_B (SC2_B3, SC2_B4) cells had velocity towards a Matn1-negative region of SC2_A (SC2_A2, SC2_A4), suggesting that some synovial cavity IZ cells undergo chondrocyte specification at this stage. We also discovered a consistent velocity flow into the meniscus cluster SC2_B1 from SC2_B5. SC_B5 was enriched in chondrocytes enhancers Hmga2 and Hmga1b (Richter et al., 2011), and had elevated expression levels of Cd9 (Singh et al., 2015) and Emp1, indicating that these cells might contribute to the chondrocyte-like component of the meniscus.
Development of articular fibrous components
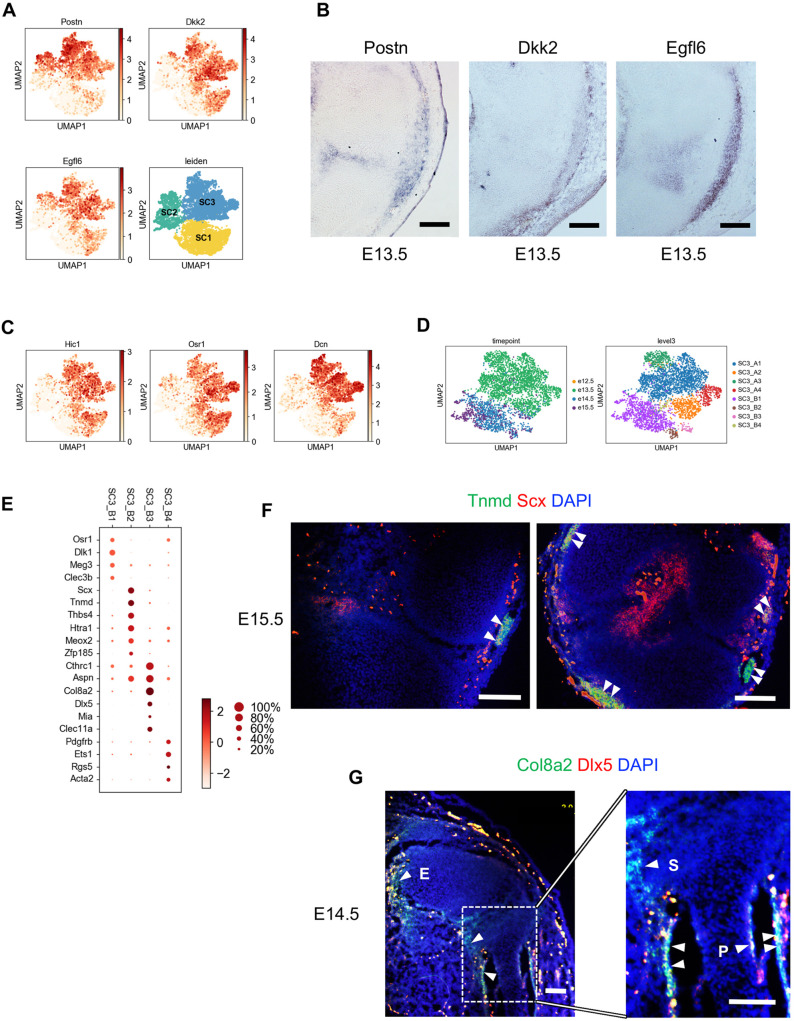
SC3 exhibited some similarities in expression profile with the IZ cells of SC2. For example, SC3 expressed high levels of Postn and Egfl6 (Fig. 6A), genes that were detected in both the IZ and flanking mesenchyme (Fig. 6B). However, SC3 specifically expressed the Wnt inhibitor Dkk2, which we detected in the flanking mesenchyme but not the IZ (Fig. 6B). SC3 specifically expressed other genes that have previously been localized to the mesenchyme adjacent to developing limb cartilage anlagen, including Hic1 (Pospichalova et al., 2011), Osr1 (Vallecillo-García et al., 2017) and Dcn (Olguin and Brandan, 2001) (Fig. 6C), suggesting that SC3 is composed of mesenchymal/fibroblast-related cells that flank the joint and developing bone. We identified other genes that distinguish SC3 from SC2, including Crabp1, Zfhx3, Ebf2, Celf2, Rspo3, Map1b and Selenop, as they were high in SC3, low or not detected in SC2, and variably expressed in SC1 (Fig. S5A).
Fig. 6.
SC3 is composed of articular fibrous component cells. (A) Gene expression patterns of three SC3 representative genes. (B) In situ hybridization detection for SC3 representative genes, showing expressions of Postn, Dkk2 and Egfl6 in the IZ region, outer tendon and fibrous tissue of E13.5 sagittal sections. (C) Distribution of genes detected in surrounding mesenchyme. (D) Subclustering of SC3 by Leiden. (E) Dot plot expression of representative genes differentially expressed among SC3_B subclusters. (F) RNAscope Hiplex for Tnmd (green) and Scx (red) of sagittal (left) and coronal (right) sections from E15.5 knee joint. Arrowheads indicate double-positive cells. (G) Sagittal sections of E14.5 knee joint showing the expression of Col8a2 (green) and Dlx5 (red). Higher magnification (right) of the boxed area (left) illustrating double-positive staining in perichondrium (P), synovium (S) and enthesis (E). Scale bars: 100 µm.
As we did for SC1 and SC2, we performed a deeper analysis of SC3 by clustering it more finely into two subclusters (Fig. S5B). SC3_A mainly comprised E13.5 cells and had high levels of genes related to cell proliferation, whereas SC3_B was made up primarily of E14.5 and E15.5 cells, and had higher levels of fibroblast- and ECM-related genes such as Postn and Col3a1 (Fig. S5C). As SC3_A and SC3_B differed mainly by developmental stage, we subclustered each to search for more subtle differences in state or lineage, resulting in four subgroups in SC3_A (Fig. 6D) and four in SC_B. Here, we focused on the subclusters of SC3_B as they appeared to have more defined and distinct signatures. SC3_B1 likely represents remaining residual undifferentiated mesenchymal progenitors, as they maintain Osr1 expression and have upregulated Dlk1 and Meg3 (Fig. 6E) imprinted genes associated with a progenitor state (Miller and Cole, 2014; Sanli et al., 2018). Cells of SC3_B2 are likely to be ligament or tendon cells based on preferential expression of Tnmd and Scx (Soeda et al., 2010; Sugimoto et al., 2013; Subramanian and Schilling, 2015), as well as other tendon-associated genes, including Thbs4 (Havis et al., 2014; Subramanian and Schilling, 2014), Htra1 (Oka et al., 2004) and Meox2 (Havis et al., 2014). Our single cell data indicated that nascent cruciate ligament cells (SC2_B2) express Scx but not Tnmd, whereas the SC3_B2 cells express both genes (Fig. S5E). We confirmed the localization of SC3_B3 to the outer tendon using multiplex in situ hybridization (Fig. 6F). SC3_B3 is likely to include synovial fibroblasts and fibrocartilage cells of the enthesis (Zelzer et al., 2014) based on the preferential expression of Cthrc1, which is produced by synovial fibroblasts (Park et al., 2004; Shekhani et al., 2016) and chondrocyte-related genes Aspn, Mia and Dlx5 (Ferrari and Kosher, 2006). We found that Col8a2 and Dlx5, which were preferentially expressed in SC3_B3, were co-expressed in synovium, enthesis and perichondrium, indicating that SC3_B3 is made up of fibrochondrocytes within these sites (Fig. 6G). Finally, the small SC3_B4 cluster is made up of pericytes/vascular smooth muscle cells based on expression of Pdgfrb, Ets1, Rgs5 and Acta2.
DISCUSSION
The synovial joint initiates from a thin layer of mesenchymal cells marked by Gdf5 expression. Through lineage tracing of Gdf5, it has become apparent that Gdf5-expressing IZ cells give rise to multiple joint lineages. The extent to which the spatial distribution of Gdf5+ descendants changes is due to cell movement, de novo expression or cell death is not precisely known; however lineage tracing indicates that cell movement and de novo expression are the primary contributors (Ito and Kida, 2000; Shwartz et al., 2016). The transcriptional programs that drive IZ formation and elaboration has remained underexplored. In this study, we applied scRNA-Seq to Gdf5 lineage cells during embryonic stages of synovial joint development to define the continuum of expression states that govern the process from IZ formation to joint cavitation. Here, we have revealed the dynamic transcriptome changes and heterogeneity in GLE cells, we have inferred the lineage trajectories of some subpopulations, and we have identified novel regulators and contributors of these developmental decisions. Several insights have emerged from our dataset and analyses that have implications for the field.
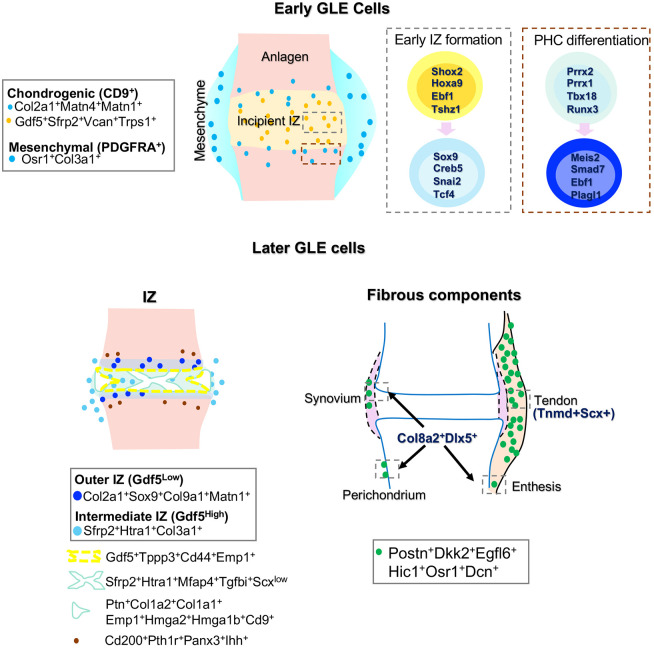
First, our results have revealed that GLE cells at E12.5 already exhibit transcriptional heterogeneity, with one cluster tending towards a more mesenchymal state (SC1_A) and one cluster tending towards a more chondroprogenitor state (SC1_B). By prospectively isolating cells using markers that distinguished these clusters, we confirmed the in vitro lineage propensity of these cell populations. The degree of commitment of these cells in vivo remains to be determined. Second, we discovered further substructure within the SC1_B group of cells, with one cluster actively proliferating (SC1_B5), one cluster expressing markers of pre-hypertrophic differentiation (SC1_B2), one cluster expressing genes indicative of spatial patterning (i.e. SC1_B3 preferentially expressed more proximal Meis2 and proximal/anterior Irx3) and one cluster expressing higher levels of IZ-associated genes (SC1_B4: Sfrp2, Vcan and Trps1). Third, we used a combination of RNA velocity and pseudotime analysis to identify known and novel regulators of the early IZ and PHC trajectories. Fourth, we have defined the expression signatures of discrete lineages within the developing synovial joint (Fig. 7) and how they emerge over time (Fig. S6).
Fig. 7.
Nascent joint development. Diagram of nascent joint development populations and markers identified here.
We note several important caveats to our results, which suggest areas for improvement and future investigation. First, we cannot exclude the possibility that certain populations of Gdf5-lineage cells were particularly vulnerable to the isolation procedure and thus underrepresented in our scRNAseq data. Another important consideration is the impact of tissue dissociation on expression state. Although we cannot exclude this latter concern, we did not detect an enrichment of immediate-early genes in any of our clusters, supporting the notion that these cells are not particularly prone to this response (van den Brink et al., 2017). Finally, it is possible that certain classes of genes are particularly sensitive to the relatively long tissue digestion necessary to achieve single cell dissociation, and that this influence may vary by cell population. We were not able to systematically assess this possibility but new technologies that enable spatial transcriptomics may illuminate the impact of this potential confounder (Ståhl et al., 2016).
In conclusion, we have made our data freely and easily accessible with a web application at www.cahanlab.org/resources/joint_ontogeny. We believe that this resource will aid the community in discovering additional transcriptional programs and in inferring cell interactions that underpin synovial joint development. Furthermore, we anticipate that these data can be used to yield improved protocols for the derivation of synovial lineages from pluripotent stem cells (Craft et al., 2015; Kawata et al., 2019; Oldershaw et al., 2010; Yamashita et al., 2015) by, for example, using it to identify candidate signaling pathways or by using the expression data as a reference against which to compare engineered cells.
MATERIALS AND METHODS
Mice
The Gdf5-Cre (Sperm Cryorecovery via IVF, FVB/NJ background) mouse strain was obtained from the Jackson laboratory. B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J (RosaEYFP) (C57BL/6J background) was gifted by the lab of Prof. Xu Cao (Johns Hopkins University, Baltimore, MD, USA). Gdf5-Cre::Rosa-EYFP mice were generated by crossing heterozygote Gdf5-Cre strain with homozygote RosaEYFP strain. The genotype of the mice was determined by PCR analyses of genomic DNA isolated from mouse tails using the following primers: Gdf5-directed Cre forward, 5′GCCTGCATTACCGGTCGATGCAACGA3′; and reverse, 5′GTGGCAGATGGCGCGGCAACACCATT3′ (protocol provided by Prof. David Kingsley, HHMI and Stanford University, CA, USA). Day 5 wild type refers to the C57/BL10 mouse. All the protocols were approved by the institutional review board of Johns Hopkins University.
Mice gender identification
We identified mouse gender by genotyping Sry Y gene using the primers: forward, 5′CTGGAAATCTACTGTGGTCTG3′; and reverse, 5′ACCAAGACCAGAGTTTCCAG3′.
Cell isolation
Mice were kept in a light-reversed room (light turns on at 10 pm and turns off at 10 am). Timing was determined by putting one male mouse and two female mice in the same cage at 9 am and separating them at 4 pm on the same day. We counted midnight as E0.5. On E12.5, E13.5, E14.5 and E15.5, the pregnant mice were sacrificed using CO2 at 3 am. The cells were isolated as previously described (Gosset et al. 2008) with modification: the embryos (usually n=6-8) were rinsed three times in PBS on ice. Two presumptive knee joints were isolated by transfemoral and transtibial division in a single 3 cm dish (Fig. 1A) and incubated in digestion solution I (3 mg/ml collagenase D, DMEM high glucose culture medium, serum free) for 45 min at 37°C, then in digestion solution II (1 mg/ml collagenase D, DMEM high glucose culture medium, serum free) for 3 h (one embryo per dish) at 37°C. During the period of incubation, the mice gender was identified by genotyping and only male samples were chosen for further processing. The tissues with medium were gently pipetted to disperse cell aggregates and filtered through a 40 µm cell strainer, then centrifuged for 10 min at 400 g. The pellet was suspended with 0.4% BSA in PBS.
Cell fractionation
All cells were fractionated by fluorescence-activated cell sorting (FACS). A MoFlo XDP sorter (Beckman Coulter) was used to collect YFP+ cells and propidium iodide was used to exclude dead cells from any observations.
Single cell RNA sequencing
GemCode Single Cell platform (10X Genomics) was used to determine the transcriptomes of single cells (Zheng et al., 2017). Cells at 1000/µl were obtained after sorting and placed on ice. Each time point, one sample was selected and profiled based on the viability and amount. A total of 6000 cells were loaded each time, followed by GEM-RT reaction and cDNA amplification. Single cell libraries were constructed by attaching P7 and P5 primer sites and sample indices to the cDNA. Single cell RNA sequencing was performed on Illumina NextSeq 500 and HiSeq 2500 to a depth ranging from 347 to 489 million reads per sample.
Analysis and visualization of scRNA seq data
CellRanger (version 2.0.0) was used to perform the original processing of single cell sequencing reads, aligning them to the mm10 reference genome. We used the command line interface of Velocyto, version 1.7.3, to count reads and attribute them as spliced, unspliced or ambiguous (La Manno et al., 2018). The resulting loom files for each sample were then concatenated and subjected to quality control processing, normalization, estimation of cell cycle phase, clustering and differential gene expression analysis using Scanpy 1.4.3 (Wolf et al., 2018). Specifically, we excluded cells in which mitochondrial gene content exceeded 5% of the total reads or in cells with fewer than 500 unique genes detected. We then excluded genes that were detected in fewer than 10 cells, resulting in a dataset of 10,124 cells and 16,352 genes. Next, we performed an initial normalization on a per cell basis followed by log transformation and scaling. We scored the phases of cell cycle using cell cycle-associated genes as previously described (Satija et al., 2015). We then identified the genes that were most variably expressed across the whole data set, and within each timepoint, resulting in 3593 genes. We performed PCA and inspected the variance ratio plots to determine the ‘elbow’, or number of PCs that account for most of the total variation in the data. We generated a graph of cell neighbors using diffusion maps (Coifman et al., 2005), and then we performed Leiden clustering (Traag et al., 2019), which we visualized with a UMAP embedding (McInnes et al., 2018). We also analyzed this with SingleCellNet (Tan and Cahan, 2019), which had been trained using the Tabula Muris dataset (Tabula Muris Consortium et al., 2018). We removed cells in clusters that were classified by SingleCellNet as ‘blood’, ‘erythroblast’ and ‘endothelial’. We also removed cells in clusters that we identified as likely to be myoblasts based on high levels of Myod1 and other muscle-specific genes, melanocyte (based on Pmel expression) and neural crest (based on Sox10 expression). Then, we repeated the pre-processing and analysis pipeline on the remaining 8378 genes. Finally, we removed cells in a cluster that we determined by in situ hybridization to consist mainly of dermis cells, resulting in final dataset of 7329 cells and 16,352 genes. Super-clusters and all subclusters were identified by following the same pipeline as described above, except that the analysis was limited to the corresponding set of cells. For example, the superclusters were identified by first finding the genes that vary across both all cells, and within each time point. A neighborhood graph was then plotted using the principal components (the number of which was decided by examining the variation ratio plot), followed by Leiden clustering and UMAP embedding, and, for some subsets of data, diffusion map embedding. Differentially expressed genes were identified using the Scanpy rank_genes_groups function. Gene set enrichment analysis was performed using GSEAPY (github.com/zqfang/GSEApy), a Python interface to enrichR (Chen et al., 2013; Kuleshov et al., 2016). The analysis pipeline of Velocyto was applied to data subsets as mentioned in the main text. We used the Velcoyto results to manually assign roots for diffusion map pseudotime analysis. Genes correlated with pseudotime were identified as described previously (Street et al., 2018).
Histochemistry, immunohistochemistry and histomorphometry
The specimens were fixed in 10% buffered formalin for 24 h at room temperature, washed with distilled water and equilibrated in 20% sucrose in PBS at 4°C overnight, then mounted in OCT and frozen at −80°C. Coronal-oriented or sagittal-oriented sections (10 μm) were cut using a cryostat.
We performed trichrome staining according to the Trichrome Stain (Connective Tissue Stain) Kit protocol (Abcam). Immunostaining was performed using a standard protocol. Sections were incubated with primary antibodies to mouse GFP (1:200) (Ishiguro et al., 2020), TNMD (1:100) (Qu et al., 2018), SOX9 (1:500) (Yu et al., 2019) and THY1 (1:100) (Chen et al., 2018) in antibody diluent (Dako) at 4°C overnight followed with three 5 min washes in TBST. The slides were then incubated with secondary antibodies conjugated with fluorescence [Ab150077, 1:200 (Wang et al., 2019a); BA1032, 1:100 (Liu et al., 2015a)] at room temperature for 1h while avoiding light followed with three 5 min washes in TBST and nuclear stained with mounting medium containing DAPI. Images were captured by Nikon EcLipse Ti-S, DS-U3 and DS-Qi2. See Table S2. Six view fields under 100 X were randomly chosen for histomophometry. The percentage of positive cells was calculated (total number of positive cells/the total number of cells under one view field×100).
The data from the histomorphometry were expressed as mean±s.d., and statistical significance was determined using an unpaired Student's t-test. The level of significance was defined as P<0.05. Data analyses were performed using SPSS 15.0 analysis software.
In situ hybridization
See Table S3 for the information of oligonucleotides used for templates for antisense RNA probes. The T7 and SP6 primer sequence were added to 5′ and 3′ ends, respectively. SP6 RNA polymerase was used for probe transcription. Probes were synthesized with digoxygenin-labeled UTP and hybridized at 68°C overnight. Results were visualized using alkaline phosphatase-conjugated anti-digoxygenin antibody and BCIP/NBT substrates.
RNAscope HiPlex
RNAscope HiPlex probes were designed by Advanced Cell Diagnostics (ACD). Assays were performed according to protocol provided by ACD. In detail, fixed frozen tissue samples were sectioned, pretreated with protease and then incubated with Hybridize RNAscope HiPlex Amp and HiPlex Fluoro probes (green). The first-round images were captured following counterstaining. The fluorophores were cleaved prior to hybridization with second round probes (red).
FACS for prospective isolation
E12.5 embryonic hindlimb cells were isolated as described in the ‘Cell isolation’ section. After filtering through a 40 µm cell strainer, cells were suspended in autoMACS rinsing solution at 1 million per ml. After spin down, E12.5 cells were then stained with PDGFRA (1 µg per 10 million cells) and CD9 (1 µg per 5 million cells) in 100 µl autoMACS rinsing solution in the dark for 30 min followed by two washes with autoMACS rinsing solution. Cells were re-suspended in autoMACS rinsing solution. A negative control without staining was used to set up the gate. The following two E12.5 populations were collected at the same time: a YFP+PDGFRA+ population and a YFP+PDGFRA−CD9+ population.
Supplementary Material
Acknowledgements
We thank Wenyan Lu for technical support with in situ hybridization probe designs. 10× Genomics single cell processing and sequencing was conducted by Melissa Olson, Kakali Sarkar and David Mohr at the Genetic Resources Core Facility, Johns Hopkins Institute of Genetic Medicine. Flow cytometry and cell sorting was conducted by Hao Zhang at the Cell Sorting Core Facility, Bloomberg School of Public Health, Johns Hopkins University.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: Q.B., P.C.; Methodology: Q.B., Y.-H.C., J.P.W., E.Y.S., D.W.K., H.W., S.Y., S.B., P.C.; Software: Y.-H.C., E.Y.S., P.C.; Formal analysis: Q.B., P.C.; Investigation: Q.B.; Resources: Q.B., P.C.; Data curation: Q.B., P.C.; Writing - original draft: Q.B.; Writing - review & editing: J.P.W., P.C.; Supervision: P.C.; Project administration: P.C.; Funding acquisition: Q.B., P.C.
Funding
This work was supported by the National Institutes of Health (R35GM124725 to P.C. and R25GM109441 to J.P.W.) and by the Maryland Stem Cell Research Fund [2017-MSCRFF-3910 (Award ID: 90074850) to Q.B.]. This work was made possible by support from the Johns Hopkins University Medicine Discovery Fund to P.C. Deposited in PMC for release after 12 months.
Data availability
Our data are freely and easily accessible with a web application at cahanlab.org/resources/joint_ontogeny. Raw data are available at GEO under accession number GSE151985.
Supplementary information
Supplementary information available online at https://dev.biologists.org/lookup/doi/10.1242/dev.185777.supplemental
Peer review history
The peer review history is available online at https://dev.biologists.org/lookup/doi/10.1242/dev.185777.reviewer-comments.pdf
References
- Amarilio R., Viukov S. V., Sharir A., Eshkar-Oren I., Johnson R. S. and Zelzer E. (2007). HIF1α regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development 134, 3917-3928. 10.1242/dev.008441 [DOI] [PubMed] [Google Scholar]
- Aro E., Salo A. M., Khatri R., Finnilä M., Miinalainen I., Sormunen R., Pakkanen O., Holster T., Soininen R., Prein C. et al. (2015). Severe extracellular matrix abnormalities and chondrodysplasia in mice lacking collagen Prolyl 4-hydroxylase isoenzyme II in combination with a reduced amount of Isoenzyme I. J. Biol. Chem. 290, 16964-16978. 10.1074/jbc.M115.662635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H., Inui M. and Lotz M. K. (2017). Tendons and ligaments: connecting developmental biology to musculoskeletal disease pathogenesis. J. Bone Miner. Res. 32, 1773-1782. 10.1002/jbmr.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattaram P., Penzo-Méndez A., Sock E., Colmenares C., Kaneko K. J., Vassilev A., Depamphilis M. L., Wegner M. and Lefebvre V. (2010). Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat. Commun. 1, 9 10.1038/ncomms1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn P. R., Xu Z., Tumelty K. E., Zhao R. W., Monis W. J., Harris K. G., Gass J. M., Cousin M. A., Boczek N. J., Mitkov M. V. et al. (2018). Bi-allelic alterations in AEBP1 lead to defective collagen assembly and connective tissue structure resulting in a variant of ehlers-danlos syndrome. Am. J. Hum. Genet. 102, 696-705. 10.1016/j.ajhg.2018.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobick B. E. and Cobb J. (2012). Shox2 regulates progression through chondrogenesis in the mouse proximal limb. J. Cell Sci. 125, 6071-6083. 10.1242/jcs.111997 [DOI] [PubMed] [Google Scholar]
- Chen L., Qanie D., Jafari A., Taipaleenmaki H., Jensen C. H., Säämänen A.-M., Sanz M. L. N., Laborda J., Abdallah B. M. and Kassem M. (2011). Delta-like 1/fetal antigen-1 (Dlk1/FA1) is a novel regulator of chondrogenic cell differentiation via inhibition of the Akt kinase-dependent pathway. J. Biol. Chem. 286, 32140-32149. 10.1074/jbc.M111.230110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Tan C. M., Kou Y., Duan Q., Wang Z., Meirelles G. V., Clark N. R. and Ma'ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Capellini T. D., Schoor M., Mortlock D. P., Reddi A. H. and Kingsley D. M. (2016). Heads, shoulders, elbows, knees, and toes: modular gdf5 enhancers control different joints in the vertebrate skeleton. PLoS Genet. 12, e1006454 10.1371/journal.pgen.1006454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhi X., Wang J. and Su J. (2018). RANKL signaling in bone marrow mesenchymal stem cells negatively regulates osteoblastic bone formation. Bone Res. 6, 34 10.1038/s41413-018-0035-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E. A. and Dressler G. R. (1998). TCF-4 binds β-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech. Dev. 77, 9-18. 10.1016/S0925-4773(98)00131-2 [DOI] [PubMed] [Google Scholar]
- Choocheep K., Hatano S., Takagi H., Watanabe H., Kimata K., Kongtawelert P. and Watanabe H. (2010). Versican facilitates chondrocyte differentiation and regulates joint morphogenesis. J. Biol. Chem. 285, 21114-21125. 10.1074/jbc.M109.096479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coifman R. R., Lafon S., Lee A. B., Maggioni M., Nadler B., Warner F. and Zucker S. W. (2005). Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc. Natl. Acad. Sci. USA 102, 7426-7431. 10.1073/pnas.0500334102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coré N., Caubit X., Metchat A., Boned A., Djabali M. and Fasano L. (2007). Tshz1 is required for axial skeleton, soft palate and middle ear development in mice. Dev. Biol. 308, 407-420. 10.1016/j.ydbio.2007.05.038 [DOI] [PubMed] [Google Scholar]
- Craft A. M., Rockel J. S., Nartiss Y., Kandel R. A., Alman B. A. and Keller G. M. (2015). Generation of articular chondrocytes from human pluripotent stem cells. Nat. Biotechnol. 33, 638-645. 10.1038/nbt.3210 [DOI] [PubMed] [Google Scholar]
- Dasuri K., Antonovici M., Chen K., Wong K., Standing K., Ens W., El-Gabalawy H. and Wilkins J. A. (2004). The synovial proteome: analysis of fibroblast-like synoviocytes. Arthritis Res. Ther. 6, R161-R168. 10.1186/ar1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker R. S., Koyama E. and Pacifici M. (2014). Genesis and morphogenesis of limb synovial joints and articular cartilage. Matrix Biol. 39, 5-10. 10.1016/j.matbio.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander W., Pulyakhina I., Boer C., Bomer N., van der Breggen R., Arindrarto W., Couthino de Almeida R., Lakenberg N., Sentner T., Laros J. F. J. et al. (2019). Annotating transcriptional effects of genetic variants in disease-relevant tissue: transcriptome-wide allelic imbalance in osteoarthritic cartilage. Arthritis Rheumatol. 71, 561-570. 10.1002/art.40748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto-Iwamoto M., Kitagaki J., Koyama E., Tamamura Y., Wu C., Kanatani N., Koike T., Okada H., Komori T., Yoneda T. et al. (2002). The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev. Biol. 251, 142-156. 10.1006/dbio.2002.0802 [DOI] [PubMed] [Google Scholar]
- Etich J., Holzer T., Pitzler L., Bluhm B. and Brachvogel B. (2015). MiR-26a modulates extracellular matrix homeostasis in cartilage. Matrix Biol. 43, 27-34. 10.1016/j.matbio.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Feng C., Chan W. C. W., Lam Y., Wang X., Chen P., Niu B., Ng V. C. W., Yeo J. C., Stricker S., Cheah K. S. E. et al. (2019). Lgr5 and Col22a1 mark progenitor cells in the lineage toward juvenile articular chondrocytes. Stem Cell Rep. 13, 713-729. 10.1016/j.stemcr.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari D. and Kosher R. A. (2006). Expression of Dlx5 and Dlx6 during specification of the elbow joint. Int. J. Dev. Biol. 50, 709-713. 10.1387/ijdb.062180df [DOI] [PubMed] [Google Scholar]
- Gao Y., Lan Y., Liu H. and Jiang R. (2011). The zinc finger transcription factors Osr1 and Osr2 control synovial joint formation. Dev. Biol. 352, 83-91. 10.1016/j.ydbio.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosset M., Berenbaum F., Thirion S. and Jacques C. (2008). Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253-1260. 10.1038/nprot.2008.95 [DOI] [PubMed]
- Guo G., Huss M., Tong G. Q., Wang C., Li Sun L., Clarke N. D. and Robson P. (2010). Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell 18, 675-685. 10.1016/j.devcel.2010.02.012 [DOI] [PubMed] [Google Scholar]
- Haag J. and Aigner T. (2007). Identification of calponin 3 as a novel Smad-binding modulator of BMP signaling expressed in cartilage. Exp. Cell Res. 313, 3386-3394. 10.1016/j.yexcr.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Haffner-Luntzer M., Heilmann A., Rapp A. E., Beie S., Schinke T., Amling M., Ignatius A. and Liedert A. (2014). Midkine-deficiency delays chondrogenesis during the early phase of fracture healing in mice. PLoS ONE 9, e116282 10.1371/journal.pone.0116282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Büttner M., Wolf F. A., Buettner F. and Theis F. J. (2016). Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845-848. 10.1038/nmeth.3971 [DOI] [PubMed] [Google Scholar]
- Haraguchi R., Kitazawa R. and Kitazawa S. (2015). Epigenetic regulation of Tbx18 gene expression during endochondral bone formation. Cell Tissue Res. 359, 503-512. 10.1007/s00441-014-2028-0 [DOI] [PubMed] [Google Scholar]
- Hartmann C. and Tabin C. J. (2001). Wnt-14 plays a pivotal role in inducing synovial joint formation in the developing appendicular skeleton. Cell 104, 341-351. 10.1016/S0092-8674(01)00222-7 [DOI] [PubMed] [Google Scholar]
- Havis E., Bonnin M.-A., Olivera-Martinez I., Nazaret N., Ruggiu M., Weibel J., Durand C., Guerquin M.-J., Bonod-Bidaud C., Ruggiero F. et al. (2014). Transcriptomic analysis of mouse limb tendon cells during development. Development 141, 3683-3696. 10.1242/dev.108654 [DOI] [PubMed] [Google Scholar]
- Headland S. E., Jones H. R., Norling L. V., Kim A., Souza P. R., Corsiero E., Gil C. D., Nerviani A., Dell'Accio F., Pitzalis C. et al. (2015). Neutrophil-derived microvesicles enter cartilage and protect the joint in inflammatory arthritis. Sci. Transl. Med. 7, 315ra190 10.1126/scitranslmed.aac5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde G., Dover S., Aszodi A., Wallis G. A. and Boot-Handford R. P. (2007). Lineage tracing using matrilin-1 gene expression reveals that articular chondrocytes exist as the joint interzone forms. Dev. Biol. 304, 825-833. 10.1016/j.ydbio.2007.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde G., Boot-Handford R. P. and Wallis G. A. (2008). Col2a1 lineage tracing reveals that the meniscus of the knee joint has a complex cellular origin. J. Anat. 213, 531-538. 10.1111/j.1469-7580.2008.00966.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K.-I., Matsuura K., Tani N., Takeda N., Usuki S., Yamane M., Sugimoto M., Fujimura S., Hosokawa M., Chuma S. et al. (2020). MEIOSIN directs the switch from mitosis to meiosis in mammalian germ cells. Dev. Cell 52, 429-445.e10. 10.1016/j.devcel.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Ito M. M. and Kida M. Y. (2000). Morphological and biochemical re-evaluation of the process of cavitation in the rat knee joint: cellular and cell strata alterations in the interzone. J. Anat. 197, 659-679. 10.1046/j.1469-7580.2000.19740659.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S., Kanno S., Gai Z., Suemoto H., Kawakatsu M., Tanishima H., Morimoto Y., Nishioka K., Hatamura I., Yoshida M. et al. (2008). Trps1 plays a pivotal role downstream of Gdf5 signaling in promoting chondrogenesis and apoptosis of ATDC5 cells. Genes Cells 13, 355-363. 10.1111/j.1365-2443.2008.01170.x [DOI] [PubMed] [Google Scholar]
- Jenner F., IJpma A., Cleary M., Heijsman D., Narcisi R., van der Spek P. J., Kremer A., van Weeren R., Brama P. and van Osch G. J. V. M. (2014). Differential gene expression of the intermediate and outer interzone layers of developing articular cartilage in murine embryos. Stem Cells Dev. 23, 1883-1898. 10.1089/scd.2013.0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Zhu S., Tremblay M. S., Payette J. N., Wang J., Bouchez L. C., Meeusen S., Althage A., Cho C. Y., Wu X. et al. (2012). A stem cell-based approach to cartilage repair. Science 336, 717-721. 10.1126/science.1215157 [DOI] [PubMed] [Google Scholar]
- Kawata M., Mori D., Kanke K., Hojo H., Ohba S., Chung U.-I., Yano F., Masaki H., Otsu M., Nakauchi H. et al. (2019). Simple and robust differentiation of human pluripotent stem cells toward chondrocytes by two small-molecule compounds. Stem Cell Rep. 13, 530-544. 10.1016/j.stemcr.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly N. H., Huynh N. P. T. and Guilak F. (2020). Single cell RNA-sequencing reveals cellular heterogeneity and trajectories of lineage specification during murine embryonic limb development. Matrix Biol. 89, 1-10. 10.1016/j.matbio.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Kwan K. M., Zhang Z., Deng J. M., Darnay B. G., Behringer R. R., Nakamura T., de Crombrugghe B. and Akiyama H. (2008). Cthrc1 is a positive regulator of osteoblastic bone formation. PLoS ONE 3, e3174 10.1371/journal.pone.0003174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A. et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90-W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Tan Y. and Cahan P. (2017). Understanding development and stem cells using single cell-based analyses of gene expression. Development 144, 17-32. 10.1242/dev.133058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath M., Lüdecke H.-J. and Vortkamp A. (2002). Expression of Trps1 during mouse embryonic development. Mech. Dev. 119 Suppl. 1, S117-S120. 10.1016/S0925-4773(03)00103-5 [DOI] [PubMed] [Google Scholar]
- La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., Lidschreiber K., Kastriti M. E., Lönnerberg P., Furlan A. et al. (2018). RNA velocity of single cells. Nature 560, 494-498. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence E. A., Kague E., Aggleton J. A., Harniman R. L., Roddy K. A. and Hammond C. L. (2018). The mechanical impact of col11a2 loss on joints; col11a2 mutant zebrafish show changes to joint development and function, which leads to early-onset osteoarthritis. Phil. Trans. R. Soc. B 373, 20170335 10.1098/rstb.2017.0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C., Bach A. and Gessler M. (1998). Developmental expression patterns of mouse sFRP genes encoding members of the secreted frizzled related protein family. Mech. Dev. 75, 29-42. 10.1016/S0925-4773(98)00072-0 [DOI] [PubMed] [Google Scholar]
- Li Z.-Y., Xiong S.-H., Hu M. and Zhang C.-S. (2011). Epithelial membrane protein 1 inhibits human spinal chondrocyte differentiation. Anat Rec (Hoboken) 294, 1015-1024. 10.1002/ar.21395 [DOI] [PubMed] [Google Scholar]
- Liu M., Jing D., Wang Y., Liu Y. and Yin S. (2015a). Overexpression of angiotensin II type 2 receptor promotes apoptosis and impairs insulin secretion in rat insulinoma cells. Mol. Cell. Biochem. 400, 233-244. 10.1007/s11010-014-2280-3 [DOI] [PubMed] [Google Scholar]
- Liu H., Xu J., Liu C.-F., Lan Y., Wylie C. and Jiang R. (2015b). Whole transcriptome expression profiling of mouse limb tendon development by using RNA-seq. J. Orthop. Res. 33, 840-848. 10.1002/jor.22886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcil A., Dumontier E., Chamberland M., Camper S. A. and Drouin J. (2003). Pitx1 and Pitx2 are required for development of hindlimb buds. Development 130, 45-55. 10.1242/dev.00192 [DOI] [PubMed] [Google Scholar]
- Mayo J. L., Holden D. N., Barrow J. R. and Bridgewater L. C. (2009). The transcription factor Lc-Maf participates in Col27a1 regulation during chondrocyte maturation. Exp. Cell Res. 315, 2293-2300. 10.1016/j.yexcr.2009.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Saul N. and Groβberger L. (2018). UMAP: uniform manifold approximation and projection for dimension reduction. J. Open Source Softw. 3, 861 10.21105/joss.00861 [DOI] [Google Scholar]
- Miljkovic N. D., Cooper G. M. and Marra K. G. (2008). Chondrogenesis, bone morphogenetic protein-4 and mesenchymal stem cells. Osteoarthr. Cartil. 16, 1121-1130. 10.1016/j.joca.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Miller A. J. and Cole S. E. (2014). Multiple Dlk1 splice variants are expressed during early mouse embryogenesis. Int. J. Dev. Biol. 58, 65-70. 10.1387/ijdb.130316sc [DOI] [PubMed] [Google Scholar]
- Mittapalli V. R., Christ B., Pröls F. and Scaal M. (2009). Pleiotrophin is expressed in avian somites and tendon anlagen. Histochem. Cell Biol. 132, 413-422. 10.1007/s00418-009-0612-2 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Yamamoto K., He X., Otsuki B., Kim Y., Murao H., Soeda T., Tsumaki N., Deng J. M., Zhang Z. et al. (2011). Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat. Commun. 2, 251 10.1038/ncomms1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec S., Luxey M., Jain D., Huang Sung A., Pastinen T. and Drouin J. (2017). Pitx1 directly modulates the core limb development program to implement hindlimb identity. Development 144, 3325-3335. 10.1242/dev.154864 [DOI] [PubMed] [Google Scholar]
- Niederreither K., D'Souza R. N. and de Crombrugghe B. (1992). Minimal DNA sequences that control the cell lineage-specific expression of the pro alpha 2(I) collagen promoter in transgenic mice. J. Cell Biol. 119, 1361-1370. 10.1083/jcb.119.5.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris R. A., Damon B., Mironov V., Kasyanov V., Ramamurthi A., Moreno-Rodriguez R., Trusk T., Potts J. D., Goodwin R. L., Davis J. et al. (2007). Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J. Cell Biochem. 101, 695-711. 10.1002/jcb.21224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka C., Tsujimoto R., Kajikawa M., Koshiba-Takeuchi K., Ina J., Yano M., Tsuchiya A., Ueta Y., Soma A., Kanda H. et al. (2004). HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development 131, 1041-1053. 10.1242/dev.00999 [DOI] [PubMed] [Google Scholar]
- Oldershaw R. A., Baxter M. A., Lowe E. T., Bates N., Grady L. M., Soncin F., Brison D. R., Hardingham T. E. and Kimber S. J. (2010). Directed differentiation of human embryonic stem cells toward chondrocytes. Nat. Biotechnol. 28, 1187-1194. 10.1038/nbt.1683 [DOI] [PubMed] [Google Scholar]
- Olguin H. and Brandan E. (2001). Expression and localization of proteoglycans during limb myogenic activation. Dev. Dyn. 221, 106-115. 10.1002/dvdy.1129 [DOI] [PubMed] [Google Scholar]
- Orgeur M., Martens M., Leonte G., Nassari S., Bonnin M.-A., Börno S. T., Timmermann B., Hecht J., Duprez D. and Stricker S. (2018). Genome-wide strategies identify downstream target genes of chick connective tissue-associated transcription factors. Development 145, dev161208 10.1242/dev.161208 [DOI] [PubMed] [Google Scholar]
- Paine-Saunders S., Viviano B. L., Zupicich J., Skarnes W. C. and Saunders S. (2000). glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev. Biol. 225, 179-187. 10.1006/dbio.2000.9831 [DOI] [PubMed] [Google Scholar]
- Pan D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491-505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y. W., Kang Y. M., Butterfield J., Detmar M., Goronzy J. J. and Weyand C. M. (2004). Thrombospondin 2 functions as an endogenous regulator of angiogenesis and inflammation in rheumatoid arthritis. Am. J. Pathol. 165, 2087-2098. 10.1016/S0002-9440(10)63259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin D. E., Gamer L. W., Cox K. A. and Rosen V. (2012). Molecular profiling of synovial joints: use of microarray analysis to identify factors that direct the development of the knee and elbow. Dev. Dyn. 241, 1816-1826. 10.1002/dvdy.23861 [DOI] [PubMed] [Google Scholar]
- Pazin D. E., Gamer L. W., Capelo L. P., Cox K. A. and Rosen V. (2014). Gene signature of the embryonic meniscus. J. Orthop. Res. 32, 46-53. 10.1002/jor.22490 [DOI] [PubMed] [Google Scholar]
- Pospichalova V., Tureckova J., Fafilek B., Vojtechova M., Krausova M., Lukas J., Sloncova E., Takacova S., Divoky V., Leprince D. et al. (2011). Generation of two modified mouse alleles of the Hic1 tumor suppressor gene. Genesis 49, 142-151. 10.1002/dvg.20719 [DOI] [PubMed] [Google Scholar]
- Qu Y., Zhou L., Lv B., Wang C. and Li P. (2018). Growth differentiation factor–5 induces tenomodulin expression via phosphorylation of p38 and promotes viability of murine mesenchymal stem cells from compact bone. Mol. Med. Rep. 17, 3640-3646. 10.3892/mmr.2017.8325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines A. M., Magella B., Adam M. and Potter S. S. (2015). Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev. Biol. 15, 28 10.1186/s12861-015-0078-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Singh P. N. P., Sohaskey M. L., Harland R. M. and Bandyopadhyay A. (2015). Precise spatial restriction of BMP signaling is essential for articular cartilage differentiation. Development 142, 1169-1179. 10.1242/dev.110940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A., Lübbing M., Frank H.-G., Nolte I., Bullerdiek J. C. and von Ahsen I. (2011). High-mobility group protein HMGA2-derived fragments stimulate the proliferation of chondrocytes and adipose tissue-derived stem cells. Eur. Cell Mater. 21, 355-363. 10.22203/eCM.v021a26 [DOI] [PubMed] [Google Scholar]
- Ristevski S., Tam P. P. L., Hertzog P. J. and Kola I. (2002). Ets2 is expressed during morphogenesis of the somite and limb in the mouse embryo. Mech. Dev. 116, 165-168. 10.1016/S0925-4773(02)00126-0 [DOI] [PubMed] [Google Scholar]
- Roelofs A. J., Zupan J., Riemen A. H. K., Kania K., Ansboro S., White N., Clark S. M. and De Bari C. (2017). Joint morphogenetic cells in the adult mammalian synovium. Nat. Commun. 8, 15040 10.1038/ncomms15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva J. E. and Merrill A. E. (2017). Signaling networks in joint development. Dev. Dyn. 246, 262-274. 10.1002/dvdy.24472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanli I., Lalevée S., Cammisa M., Perrin A., Rage F., Llères D., Riccio A., Bertrand E. and Feil R. (2018). Meg3 Non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis. Cell Rep. 23, 337-348. 10.1016/j.celrep.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Satija R., Farrell J. A., Gennert D., Schier A. F. and Regev A. (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495-502. 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid R., Schiffner S., Opolka A., Grässel S., Schubert T., Moser M. and Bosserhoff A.-K. (2010). Enhanced cartilage regeneration in MIA/CD-RAP deficient mice. Cell Death Dis. 1, e97 10.1038/cddis.2010.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegmiller R., Fraser F. C. and Sheldon H. (1971). A new chondrodystrophic mutant in mice. Electron microscopy of normal and abnormal chondrogenesis. J. Cell Biol. 48, 580-593. 10.1083/jcb.48.3.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhani M. T., Forde T. S., Adilbayeva A., Ramez M., Myngbay A., Bexeitov Y., Lindner V. and Adarichev V. A. (2016). Collagen triple helix repeat containing 1 is a new promigratory marker of arthritic pannus. Arthritis Res. Ther. 18, 171 10.1186/s13075-016-1067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz Y., Viukov S., Krief S. and Zelzer E. (2016). Joint development involves a continuous influx of Gdf5-positive cells. Cell Rep. 15, 2577-2587. 10.1016/j.celrep.2016.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Lester C., Drapp R., Hu D. Z., Glimcher L. H. and Jones D. (2015). Tetraspanin CD9 and ectonucleotidase CD73 identify an osteochondroprogenitor population with elevated osteogenic properties. Development 142, 438-443. 10.1242/dev.113571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda T., Deng J. M., de Crombrugghe B., Behringer R. R., Nakamura T. and Akiyama H. (2010). Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis 48, 635-644. 10.1002/dvg.20667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl P. L., Salmén F., Vickovic S., Lundmark A., Navarro J. F., Magnusson J., Giacomello S., Asp M., Westholm J. O., Huss M. et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78-82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Staverosky J. A., Pryce B. A., Watson S. S. and Schweitzer R. (2009). Tubulin polymerization-promoting protein family member 3, Tppp3, is a specific marker of the differentiating tendon sheath and synovial joints. Dev. Dyn. 238, 685-692. 10.1002/dvdy.21865 [DOI] [PubMed] [Google Scholar]
- Storm E. E. and Kingsley D. M. (1996). Joint patterning defects caused by single and double mutations in members of the bone morphogenetic protein (BMP) family. Development 122, 3969-3979. [DOI] [PubMed] [Google Scholar]
- Storm E. E. and Kingsley D. M. (1999). GDF5 coordinates bone and joint formation during digit development. Dev. Biol. 209, 11-27. 10.1006/dbio.1999.9241 [DOI] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R. B., Das D., Ngai J., Yosef N., Purdom E. and Dudoit S. (2018). Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S., Mathia S., Haupt J., Seemann P., Meier J. and Mundlos S. (2012). Odd-skipped related genes regulate differentiation of embryonic limb mesenchyme and bone marrow mesenchymal stromal cells. Stem Cells Dev. 21, 623-633. 10.1089/scd.2011.0154 [DOI] [PubMed] [Google Scholar]
- Su N., Jin M. and Chen L. (2014). Role of FGF/FGFR signaling in skeletal development and homeostasis: learning from mouse models. Bone Res 2, 14003 10.1038/boneres.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. and Schilling T. F. (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. Elife 3, e02372 10.7554/eLife.02372.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. and Schilling T. F. (2015). Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. Development 142, 4191-4204. 10.1242/dev.114777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y., Takimoto A., Akiyama H., Kist R., Scherer G., Nakamura T., Hiraki Y. and Shukunami C. (2013). Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140, 2280-2288. 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium, Overall coordination, Logistical coordination, Organ collection and processing, Library preparation and sequencing, Computational data analysis, Cell type annotation, Writing group, Supplemental text writing group and Principal investigators (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367-372. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K., Nakamura T., Nishita M., Hyodo J., Nagai S., Murata Y., Adachi S., Ohwada S., Morishita Y., Shibuya H. et al. (2000). Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 14, 1741-1749. [PMC free article] [PubMed] [Google Scholar]
- Taher L., Collette N. M., Murugesh D., Maxwell E., Ovcharenko I. and Loots G. G. (2011). Global gene expression analysis of murine limb development. PLoS ONE 6, e28358 10.1371/journal.pone.0028358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipaleenmäki H., Harkness L., Chen L., Larsen K. H., Säämänen A.-M., Kassem M. and Abdallah B. M. (2012). The crosstalk between transforming growth factor-β1 and delta like-1 mediates early chondrogenesis during embryonic endochondral ossification. Stem Cells 30, 304-313. 10.1002/stem.792 [DOI] [PubMed] [Google Scholar]
- Tan Y. and Cahan P. (2019). SingleCellNet: a computational tool to classify single cell RNA-Seq data across platforms and across species. Cell Syst. 9, 207-213; e2. 10.1016/j.cels.2019.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge D., Brouwer A., Korving J., Martin J. F. and Meijlink F. (1998). Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development 125, 3831-3842. [DOI] [PubMed] [Google Scholar]
- Traag V. A., Waltman L. and van Eck N. J. (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufan A. C. and Tuan R. S. (2001). Wnt regulation of limb mesenchymal chondrogenesis is accompanied by altered N-cadherin-related functions. FASEB J. 15, 1436-1438. 10.1096/fj.00-0784fje [DOI] [PubMed] [Google Scholar]
- Vallecillo-García P., Orgeur M., Vom Hofe-Schneider S., Stumm J., Kappert V., Ibrahim D. M., Börno S. T., Hayashi S., Relaix F., Hildebrandt K. et al. (2017). Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nat. Commun. 8, 1218 10.1038/s41467-017-01120-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink S. C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C. S., Robin C. and van Oudenaarden A. (2017). Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935-936. 10.1038/nmeth.4437 [DOI] [PubMed] [Google Scholar]
- Vargesson N. and Laufer E. (2001). Smad7 misexpression during embryonic angiogenesis causes vascular dilation and malformations independently of vascular smooth muscle cell function. Dev. Biol. 240, 499-516. 10.1006/dbio.2001.0481 [DOI] [PubMed] [Google Scholar]
- Wang W., Rigueur D. and Lyons K. M. (2014). TGFβ signaling in cartilage development and maintenance. Birth Defects Res. C Embryo Today 102, 37-51. 10.1002/bdrc.21058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. S., Infante C. R., Park S. and Menke D. B. (2018). PITX1 promotes chondrogenesis and myogenesis in mouse hindlimbs through conserved regulatory targets. Dev. Biol. 434, 186-195. 10.1016/j.ydbio.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kumar M., Deng Q., Wang X., Liu M., Gong Z., Zhang S., Ma X., Xu-Monette Z. Y., Xiao M. et al. (2019a). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces peripheral blood abnormalities and plasma cell neoplasms resembling multiple myeloma in mice. Cancer Lett. 440-441, 135-144. 10.1016/j.canlet.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Nimkingratana P., Smith C. A., Cheng A., Hardingham T. E. and Kimber S. J. (2019b). Enhanced chondrogenesis from human embryonic stem cells. Stem Cell Res. 39, 101497 10.1016/j.scr.2019.101497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte F., Dokas J., Neuendorf F., Mundlos S. and Stricker S. (2009). Comprehensive expression analysis of all Wnt genes and their major secreted antagonists during mouse limb development and cartilage differentiation. Gene Expr. Patterns 9, 215-223. 10.1016/j.gep.2008.12.009 [DOI] [PubMed] [Google Scholar]
- Wolf F. A., Angerer P. and Theis F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M., Kireeva M. L., Kolesnikova T. V. and Lau L. F. (1997). Cyr61, product of a growth factor-inducible immediate-early gene, regulates chondrogenesis in mouse limb bud mesenchymal cells. Dev. Biol. 192, 492-508. 10.1006/dbio.1997.8766 [DOI] [PubMed] [Google Scholar]
- Yamashita A., Morioka M., Yahara Y., Okada M., Kobayashi T., Kuriyama S., Matsuda S. and Tsumaki N. (2015). Generation of scaffoldless hyaline cartilaginous tissue from human iPSCs. Stem Cell Rep. 4, 404-418. 10.1016/j.stemcr.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yammani R. R. (2012). S100 proteins in cartilage: role in arthritis. Biochim. Biophys. Acta 1822, 600-606. 10.1016/j.bbadis.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Dawson L. A., Yan M., Zimmel K., Lin Y.-L., Dolan C. P., Han M. and Muneoka K. (2019). BMP9 stimulates joint regeneration at digit amputation wounds in mice. Nat. Commun. 10, 424 10.1038/s41467-018-08278-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E., Blitz E., Killian M. L. and Thomopoulos S. (2014). Tendon-to-bone attachment: from development to maturity. Birth Defects Res. C Embryo Today 102, 101-112. 10.1002/bdrc.21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Pritchard M. R., Middleton F. A., Horton J. A. and Damron T. A. (2008). Microarray analysis of perichondral and reserve growth plate zones identifies differential gene expressions and signal pathways. Bone 43, 511-520. 10.1016/j.bone.2008.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Yantiss R. K., Nam H.-S., Chin Y., Zhou X. K., Scherl E. J., Bosworth B. P., Subbaramaiah K., Dannenberg A. J. and Benezra R. (2014). ID1 is a functional marker for intestinal stem and progenitor cells required for normal response to injury. Stem Cell Rep. 3, 716-724. 10.1016/j.stemcr.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Eberspaecher H., Lefebvre V. and De Crombrugghe B. (1997). Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 209, 377-386. [DOI] [PubMed] [Google Scholar]
- Zhao W., Sala-Newby G. B. and Dhoot G. K. (2006). Sulf1 expression pattern and its role in cartilage and joint development. Dev. Dyn. 235, 3327-3335. 10.1002/dvdy.20987 [DOI] [PubMed] [Google Scholar]
- Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J. et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.