Abstract
Zinc is required for the regulation of proliferation, metabolism, and cell signaling. It is an intracellular second messenger, and the cellular level of ionic, mobile zinc is strictly controlled by zinc transporters. In mammals, zinc homeostasis is primarily regulated by ZIP and ZnT zinc transporters. The importance of these transporters is underscored by the list of diseases resulting from changes in transporter expression and activity. However, despite numerous structural studies of the transporters revealing both zinc binding sites and motifs important for transporter function, the exact molecular mechanisms regulating ZIP and ZnT activities are still not clear. For example, protein phosphorylation was found to regulate ZIP7 activity resulting in the release of Zn2+ from intracellular stores leading to phosphorylation of tyrosine kinases and activation of signaling pathways. In addition, sequence analyses predict all 24 human zinc transporters to be phosphorylated suggesting that protein phosphorylation is important for regulation of transporter function. This review describes how zinc transporters are implicated in a number of important human diseases. It summarizes the current knowledge regarding ZIP and ZnT transporter structures and points to how protein phosphorylation seems to be important for the regulation of zinc transporter activity. The review addresses the need to investigate the role of protein phosphorylation in zinc transporter function and regulation, and argues for a pressing need to introduce quantitative phosphoproteomics to specifically target zinc transporters and proteins involved in zinc signaling. Finally, different quantitative phosphoproteomic strategies are suggested.
Keywords: Zinc signaling, Protein phosphorylation, ZIP, ZnT
Introduction
Zinc has been implicated as a factor in the development and progression of many pathological conditions such as cancer, inflammation, diabetes as well as in neurological and psychiatric diseases [1–14]. An estimated 3000 proteins interact with zinc, representing 10% of the genome, and zinc is a known regulator of gene expression through metal-responsive transcription factor-1 [15–19]. Furthermore, zinc has been identified as an intracellular second messenger involved in regulating various pathways, adding an extra dimension to its role in cellular regulation [20–22]. The basal level varies in different cell types. It ranges from tens to hundreds of pM free zinc, and it is strictly controlled as deviations from the normal cellular level may be cytotoxic [23, 24]. As zinc flux is primarily controlled by zinc transporters, we expect zinc-related dysfunction is not likely to result from dietary zinc deficiency or abundance alone, but rather from deviations in the function of proteins regulating zinc homeostasis.
The review gives a brief introduction to the role of zinc transporters in human diseases as well as evidence linking protein phosphorylation of zinc transporters to zinc signaling. The case is made that quantitative phosphoproteomics is an important approach to now understand more fully the role of phosphorylation in regulating zinc homeostasis. Finally, different quantitative phosphoproteomic strategies are suggested. In addition to zinc ions, ZIP and ZnT transporter are capable of transporting other cations such as iron, manganese, and cadmium. The mechanisms for transport of these ions and their link to different diseases is described elsewhere, and will not be discussed in this review as our focus will be on zinc transport [25–33].
Zinc transporters
Four major zinc transporter families have been identified: (1) P-type ATPases; (2) RND (resistance, nodulation and division) multidrug efflux transporters; (3) the Zrt-, Irt-like protein (ZIP) family (Slc39A); and (4) the superfamily of cation diffusion facilitators (CDF) that includes the ZnT family of zinc transporters. The P-type ATPases are identified in bacteria and plants [34, 35], whereas the RND transporters only exist in a few Gram-negative bacteria [36]. In humans, the uptake of zinc is regulated by zinc transporters of the ZIP family (Slc39A) located in membranes such as the plasma membrane, the endoplasmic reticulum, and Golgi [37]. These transporter proteins import zinc from the extracellular environment or from organelles to increase the concentration of cytosolic zinc [37–39]. On the other hand, the mammalian CDF family and the Zn transporter (ZnT) family (Slc30A) mediates the export of zinc from the cytosol into organelles or out of the cell [37, 40]. ZnT transporters are found in membranes of intracellular organelles except for ZnT1 which is located in the plasma membrane [1, 41–43]. These two transporter families account for 14 human ZIP proteins and 10 human ZnT proteins and are responsible for controlling zinc homeostasis. In addition, zinc binding proteins such as metallothioneins assist in regulating the level of free zinc in the cell.
Zinc transporters in diseases
Changes in the expression and activity of different zinc transporters have been directly linked to both systemic and central nervous system diseases, and to rare diseases such as acrodermatitis enteropathica (AE) [44] as well as lifestyle diseases and conditions affecting large numbers of people worldwide [45–58].
Diabetes
ZnT8 is the zinc transporter best studied in diabetes. It is expressed in pancreatic beta cells and functions as target autoantigen in patients with type 1 diabetes [59]. A common W325R variant in the ZnT8 large C-terminal domain (CTD) has been associated with changed autoantibody specificity in type 1 diabetes as well as increased risk of developing type 2 diabetes [60]. ZIP14 and ZIP4 were also found to be involved in diabetes as altered zinc trafficking in Zip14−/− mice resulted in a phenotype with defects in glucose homeostasis [45], and in the murine pancreatic beta cell line MIN6, overexpression of ZIP4 leads to increased granular zinc content and glucose-stimulated insulin secretion [50].
Neurological and psychiatric diseases
In the nervous system, both transporters controlling zinc influx as well as zinc efflux play key roles in cellular regulation. The increase in cytosolic zinc mediated by ZIP12 leads to CREB phosphorylation and activation which is important for neuronal differentiation [61]. A mutation in the SLC39A8 gene encoding the zinc and manganese transporter ZIP8 results in low levels of both Zn2+ and Mn2+ in the blood and increased levels in urine due to increased renal wasting in the affected patients [62]. The resulting autosomal-recessive disorder is characterized by intellectual disability, developmental delay, hypotonia, strabismus, cerebellar atrophy, and variable short stature [62]. ZnT3 has been identified as critical for transport of zinc into synaptic vesicles of a subset of glutamatergic neurons [63–65], and ZnT3 expression is reduced in patients with Alzheimer’s disease [66, 67] and Parkinson’s disease-related dementia [68]. Moreover, ZnT3 expression decreases with age suggesting a role in the prevention of aging-related cognitive loss [69]. Recently, Whitfield et al. proved a link between reduced ZnT3 protein level and depression in patients with dementia [14]. In a study in postmortem brain tissue, Rafalo-Ulinska et al. found a reduction in the ZnT3 protein level as well as a significant increase in the level of ZnT1, ZnT4, ZnT5 protein in the prefrontal cortex of subjects diagnosed with major depressive disorder (MDD) and in non-diagnosed suicide victims, relative to control subjects suggesting that zinc transporters are important in the pathophysiology of MDD and suicide [12]. ZnT3 is also inked to increased risk of febrile seizures [63, 70]. Changes in the level of ZnT transporers have also been identified in rats subjected to olfactory bulbectomy (OB), which is a model of depression [13].
Cancer
Dysregulation of zinc homeostasis is critical in a variety of cancers. ZIP4 is found to be upregulated in several types of cancer cells [53, 54, 58]. ZIP6 also plays an important role in numerous cancers, particularly breast cancer [47, 49, 56, 57, 71]. Overexpression of ZnT2 is found to decrease invasive phenotypes of breast cancer cells [72]. In addition, reduced ZnT4 expression is observed in the progression from benign to invasive prostate cancer [73].
Immune response
ZIP10 is important for B-cell survival and function and along with ZIP6 is involved in antigen presentation to T cells [46, 52]. Dendritic cells are important for immune response as they are involved in antigen presentation to T cells [74]. When dendritic cells are exposed to lipopolysaccharides, it results in the fragmentation of a ligand for toll-like receptor 4 (TLR4) from the outer membrane of Gram-negative bacteria. The TLR4–TRIF (Toll/Il-1 receptor domain-containing adapter protein-inducing interferon β) axis is activated leading to the downregulation of ZIP6 and ZIP10, and upregulation of ZnT1 and ZnT6 resulting in a significant drop in the intracellular zinc level [52]. ZIP8 is highly involved in inflammation [55], and was recently reported to be central to the development of osteoarthritis [51]. ZnT5 is required for the mast cell-mediated delayed-type allergic response by playing a role in Fc epsilonRI-induced translocation of protein kinase C (PKC) to the plasma membrane and in the nuclear translocation of nuclear factor kappaB [75].
ZnT and ZIP structures
No three-dimensional structures of full-length human ZnT or ZIP proteins have been solved. Instead, most structural information on these transporters comes from sequence analysis and prediction studies as well as data on the crystal structures of prokaryotic homologs [76, 77]. The current knowledge on zinc transporter structure and function is described in detail in recent reviews [46, 69, 78].
ZnT transporters
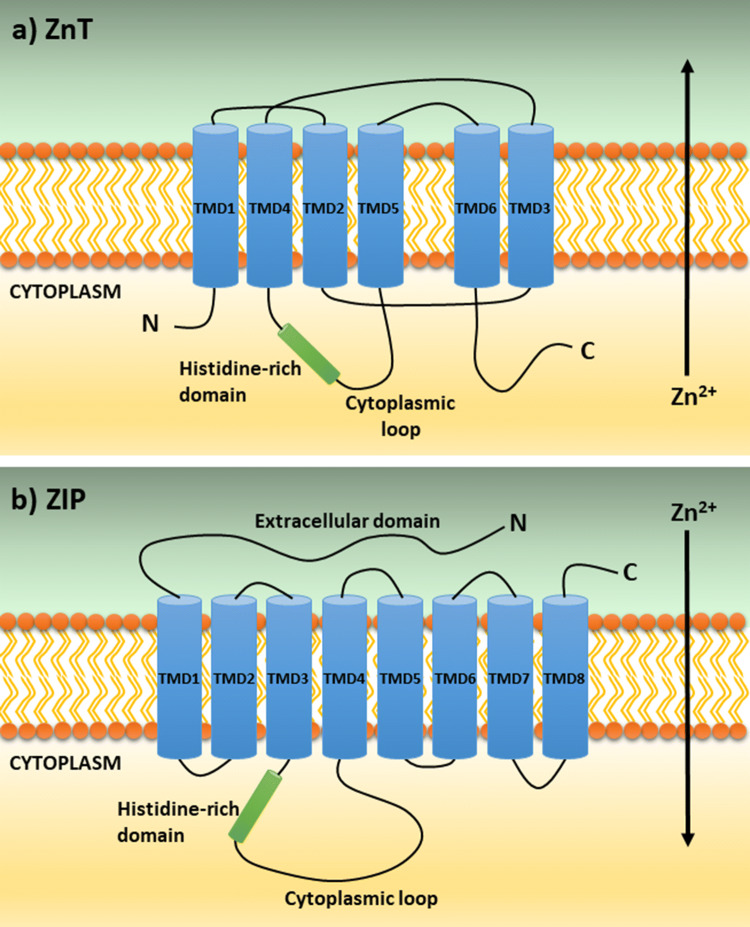
Based on the three-dimensional structure of Escherichia coli homolog YiiP, the ZnT transporters have been predicted to contain six transmembrane domains (TMDs) with intracellular NH2 and COOH termini (Fig. 1a). The exception is ZnT5, which contains seven TMDs [78]. The literature presents different predictions for the location of the TMDs but overall, the ZnTs are described with a histidine/serine-rich domain in a cytosolic loop of varying lengths between TMD4 and TMD5, and a large CTD with an overall structural similarity with the copper metallochaperone Hah1, although the ZnT CTDs do not share sequence homology with Hah1 [77] (Fig. 1a).
Fig. 1.
Predicted structures of ZnT and ZIP transporter proteins. a Predicted structure of members of the Zn transporter (ZnT) family (Slc30A) adapted from [79]. b Predicted structure of members of the Zrt-, Irt-like protein (ZIP) family (Slc39A). The long extracellular domain at the N-terminus is unique for members of the LIV-1 subfamily
Based on studies of YiiP, TMD1, TMD2, TMD4, and TMD5 are thought to form a compact four-helix structure in which four conserved hydrophilic coordination residues in TMD2 and TDM5 form a intramembranous zinc-binding site [69]. Mutations of these conserved residues have been shown to inhibit zinc transport activity [80]. Three of these coordination residues are conserved between bacteria and human, whereas the fourth site is a histidine residue in ZnT but alternates between a histidine and a aspartate residue in YiiP [81]. This enables ZnT, in contrast to YiiP, to discriminate between Zn2+ and the toxic ion Cd2+ [81]. Other studies point to the cytoplasmic domain as important for transporter activity as described for ZnT8 [60], as well as in ZnT10 where a L349P missense mutation in the CTD affects transporter activity, and a synonymous mutation (M250P) in the prokaryotic CDF protein, MamM, was found to be critical for the CTD fold illustrating the importance of the CTD [82]. The identification of a CDF superfamily lacking the CTD, however, points to additional strategies for zinc transport by ZnT transporters [43]. Furthermore, a di-proline motif in the luminal loop 2 conserved in ZnT5 and ZnT7 was recently found to be important for activation of the secretory and membrane-bound zinc-requiring enzyme, tissue-non-specific alkaline phosphatase (TNAP) [83].
ZIP transporters
Based on sequence similarity, the ZIP family is divided into subfamily ZIP I, ZIP II, gufA, and LIV-1 (Liverpool-1) [84–86]. The ZIP1, ZIP2, and ZIP3 proteins belong to the ZIP II subfamily, ZIP9 to the ZIP I subfamily and ZIP11 to the gufA subfamily. The remaining nine ZIP proteins are part of the LIV-1 subfamily. These transporters are related to the estrogen-regulated gene, LIV-1 and have primarily been identified in mammals [85]. The ZIP transporters contain eight putative transmembrane domains (TMDs) with extracellular NH2 and COOH termini (Fig. 1b) [38, 69, 78, 79, 87]. They all have a very short COOH terminus and a large cytoplasmic loop between TMD3 and TMD4 [46, 85, 88]. The cytoplasmic loop contains a histidine-rich motif with metal-binding properties [85]. In addition, members of the LIV-1 subfamily have a HEXXH motif in their TMD5 expected to function as a metal binding site and their sequences include a large NH2-terminal extracellular domain (ECD) not found in other ZIP subfamilies [85]. The ECD contains a highly conserved proline–alanine–leucine (PAL) motif, and for several of the LIV-1 subfamily members, these highly variable ECDs contain a high degree of histidine residues [69, 78, 85]. The expression of ZIP transporters at the cell surface is generally found to be upregulated upon dietary zinc deficiency (except for ZIP5) underscoring the tight connection between expression/function of the transporters and zinc signaling [89–91]. However, limited experimental data hamper our understanding of the regulation of zinc transport and zinc signaling in humans.
The ZIP4 protein is the best characterized of the ZIP transporters. ZIP4 is the ZIP transporter responsible for dietary zinc uptake as it is the exclusive transporter expressed on the apical surface of the intestinal epithelium [89]. Reduced zinc uptake caused by mutations in the ZIP4 gene results in the rare autosomal recessive disorder acrodermatitis enteropathica (AE) [44]. Fifteen missense mutations in ZIP4 cause AE, of these, seven are found in the ECD emphasizing the central role of this domain [92, 93]. Recently, the first crystal structure of a mammalian ZIP4-ECD was reported [94]. The study revealed two independent subdomains; a helix-rich domain and a PAL motif containing domain connected by a short loop and stabilized by four disulfide bonds. The study showed how two ECDs form a dimer centered at the PAL motif. As some of the AE-causing mutations eliminate the first and the fourth disulfide bond and downregulate ZIP4 glycosylation in mice, the two disulfide bonds are found to be critical for ZIP4 folding [94].
ZIP4 is found to be important for zinc transport activity, transporter processing and trafficking [94–96]. During zinc deficiency, the ZIP4-ECD is cleaved to form a ~ 35 kDa ZIP4 peptide. This peptide accumulates as a peripheral membrane protein, whereas the remaining ~ 37-kDa ZIP4 COOH-terminal processed peptide accumulates as an integral membrane protein [96]. Two AE mutations are found to block ECD cleavage and other mutations have been shown to diminish cleavage suggesting that proteolytic processing of ECD is important for ZIP4 function in regulating zinc homeostasis [96]. ECD cleavage has been found to occur in both ZIP4, ZIP6, and ZIP10 [97].
Dempski and co-workers investigated the function of the large human ZIP4 (hZIP4) cytoplasmic loop between TMD3 and TMD4 (M3M4) [98]. Combining site-directed mutagenesis, metal binding affinity assays, and X-ray absorption spectroscopy demonstrated how two Zn2+ ions bind sequentially to two different sites in the cytosolic loop due to different zinc affinities [98]. First, one Zn2+ ion binds to a CysHis3 site with a nanomolar binding affinity. Then a second Zn2+ ion binds to a His4 site with a weaker affinity, suggesting a form of zinc sensing role of the M3M4 domain, perhaps involved in the regulation of the ZIP4 level at the plasma membrane according to the need for cytosolic zinc [98].
Studies of hZIP4 and the mouse homologue (mZIP4) have shown how the cytosolic concentration of zinc ions regulate ZIP4 surface expression [88]. The level of murine ZIP4 mRNA in enterocytes and yolk sac is increased upon zinc limitation and suppressed upon zinc repletion [89, 99], and mZIP4 protein expression at the plasma membrane is reduced when zinc-deficient mice are fed a zinc-replete diet [89]. High-cytosolic Zn2+ concentrations were found to reduce mZIP4 expression at the plasma membrane through Zn2+-dependent endocytosis [100]. Kim et al. demonstrated that both mZIP4 and hZIP4 protein accumulate at the plasma membrane during zinc deficiency and undergo endocytosis when cells are exposed to low zinc concentrations (~ 1 µM Zn) leading to reduced zinc uptake through ZIP4 [100]. Furthermore, Mao and co-workers showed how a histidine-rich cluster in the cytoplasmic loop mediates ubiquitination and proteasomal degradation of hZIP4 at higher concentrations of zinc to protect against zinc cytotoxity [101].
It is clear that the regulation of cellular zinc is both stringent and complex and that various strategies are used to control the level and activity of ZIP4, and other zinc transporters as well [102].
General overview of protein phosphorylation
Protein phosphorylation is another post-translational modification (PTM) involved in zinc transport and signaling. It is a transient PTM that enables the cell to change the conformation, activity, and interaction of target proteins within a very short timeframe. It is a reversible modification, and a complex interplay between specific protein kinases and protein phosphatases keeps a strict temporal and spatial control of the phosphorylation and dephosphorylation of target proteins at specific sites. This enables the cell to quickly transduce extracellular signals into intracellular signals through signal transduction pathways in which a large range of target proteins are affected (Fig. 2). Protein phosphorylation is one of the most widespread regulatory mechanisms in nature, acting as an important modulator of intracellular biological processes such as proliferation, differentiation, cell survival, transcription, and translation [103–107]. Genomic sequencing indicates that 2–3% of all eukaryotic genes are likely to code for protein kinases [105], and at present, protein phosphorylation is the most studied and best understood post-translational modification [108, 109].
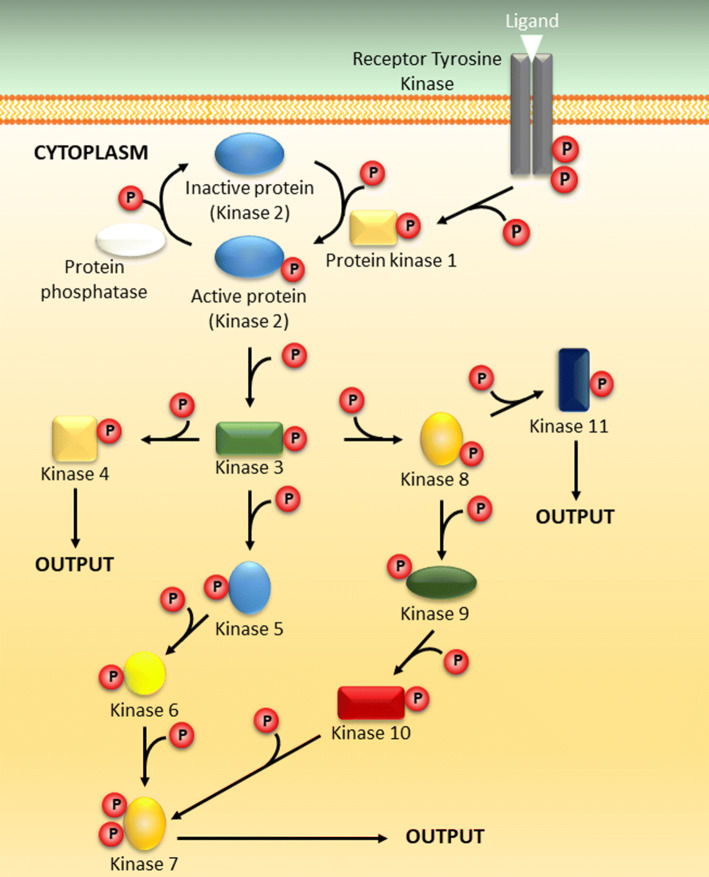
Fig. 2.
Signal transduction pathway. The figure illustrates the principle of a protein phosphorylation cascade. Protein phosphorylation of target proteins is catalysed by protein kinases, whereas dephosphorylation is catalysed by protein phosphatases. As an example, binding of a ligand to a receptor tyrosine kinase results in dimerization and autophosphorylation of the receptor. This leads to the phosphorylation and activation of kinase 2 initiating a phosphorylation cascade affecting several protein kinases. Through signal transduction pathways, the cell can propagate and enhance the cellular output. The cellular output depends on the specific pathway. Red P circle indicates protein phosphorylation. Double red P circles indicate protein phosphorylation at multiple sites. White figure—protein phosphatase. Coloured figures—different protein kinases
Protein phosphorylation is linked to zinc transporters and zinc signaling
Zinc and protein tyrosine phosphatases
A strong collaboration between zinc signaling and protein phosphorylation is clear from the actions of zinc as a second messenger in different cell types leading to the activation of various phosphorylation cascades, and the inhibition of protein tyrosine phosphatase (PTP) activity [20, 21]. PTPs play central roles in cellular function as key regulators of protein phosphorylation, directly affecting the activity of a great number of phosphoproteins and tyrosine kinase pathways [110–114]. PTPs contain a substrate binding site (in the P-loop) binding the phosphate ester group of the substrate and an active site (in the protein loop known as the WPD-loop due to the conserved tryptophan–proline–aspartic acid sequence). The WPD-loop exists in an open conformation in which the conserved aspartic acid is positioned 8–10 Å from the active site [115]. Substrate binding results in a conformational change in the WPD-loop leading to a closure of the loop placing it over the active site and bringing the conserved aspartic acid close to the bound substrate [115, 116]. Upon binding of a substrate, the nucleophilic Cys215 thiol in the P-loop attacks the substrate phosphate ester group. The leaving group is protonated by Asp181 in the WPD-loop forming a phospho-enzyme intermediate and releasing peptidyl tyrosine [115, 116]. Subsequent hydrolysis of the phospho-enzyme intermediate releases inorganic phosphate and regenerates the PTP enzyme with the WPD-loop in the open conformation [117, 118].
PTPs are regulated by protein phosphorylation, redox signaling, dimerization, and proteolysis. We suspect that zinc may inhibit PTP activity by affecting these processes directly or using other mechanisms. The PTP regulating the insulin and leptin signaling pathway, PTP1B (PTPN1) is one of the most extensively studied PTPs. Bellomo et al. optimised an assay for investigating zinc inhibition of PTPs using PTP1B [118]. They found the binding of zinc to PTP1B to require activation in the presence of a substrate as it only binds to the catalytic pocket of the closed PTP1B phospho-intermediate [118]. The analysis of three PTP1B 3D structures (PDB id: 2CM2, 3I80, and 1A5Y) revealed putative zinc binding sites and supported the hypothesis of inhibition only of the phospho-intermediate. Furthermore, they showed how PTP1B inhibition can also result from an alternative mechanism possibly initiated by the binding of zinc to the PTP1B surface. As PTP1B and the zinc transporter ZIP7 are both localized on the ER membrane, zinc influx through ZIP7 may play an important role in the regulation of PTP1B activity [118].
Zinc and receptor tyrosine kinases
Zinc is found to induce phosphorylation of a variety of receptor tyrosine kinases including the insulin receptor (IR), IGF-1R, and EGFR resulting in the activation of the PI3K/Akt and Erk signaling cascades [119, 120]. This affects insulin-stimulated glucose metabolism and cell survival. Zinc also initiates phosphorylation and activation of Erk1/2 in neurons leading to neurotoxicity [121–124]. In 2002, Du et al. showed that a brief exposure to methylisothiazolinone, a widely used industrial and household biocide, resulted in a zinc-dependent activation of Erk1/2 via a 12-lipoxygenase-mediated pathway leading to neuronal death in neurons in culture [122, 123]. Later, Ho and co-workers found zinc accumulation resulting from oxidative stress to selectively inhibit Erk1/2-directed phosphatases either by increased degradation or reduced enzyme activity leading to increased kinase activity in the murine hippocampal neuronal cell line, HT22 cells, and immature neurons [121].
The neurotrophin receptor, TrkB, is required for neuronal survival and differentiation as well as synaptic structure, function, and plasticity [125, 126]. As a consequence, dysregulation of TrkB activity and signaling is involved in various neurological and psychiatric disorders [127–129]. TrkB is a receptor tyrosine kinase activated by the binding of neurotrophins leading to dimerization, increased kinase activity, and autophosphorylation of the receptor resulting in the activation of downstream phosphorylation cascades [130]. Zinc, however, is found to activate TrkB in a neurotrophin-independent manner [131, 132]. Activation results from increased activity of the Src family of protein tyrosine kinases. The TrkB activation by zinc has been shown to potentiate the hippocampal mossy fiber-CA3 pyramid synapse [132]. Also, Zn2+ induces the activation of Trk signaling pathways leading to multi-phosphorylation of the striatal-enriched phosphatase (STEP) as well as phosphorylation and activation of Erk2, a STEP substrate involved in Zn2+-dependent neurotoxicity [133].
Zinc and NFκB
NFκB (NF-kappa B, nuclear factor kappa-light-chain-enhancer of activated B cells) is a major transcription factor also regulating immune responses to infection, stress, free radicals etc. NFκB is also important for synaptic signaling is NFκB [134]. In unstimulated cells, NFκB is located in the cytosol inactivated by the binding to inhibitory protein IκB (inhibitory proteins of NFκB). As a response to infection, the binding of pro-inflammatory cytokines stimulates the phosphorylation of IκB proteins by specific kinases such as IκB kinase (IKK). This leads to the ubiquitination and degradation of IκB. NFκB is subsequently released and free to translocate to the nucleus to stimulate the transcription of specific target genes by binding to their promoter regions [135–137]. The research group of Sarkar found zinc to activate NFκB in HUT-78 cells [138]. They also found zinc to be required for gene expression of both interleukin-2 (IL-2) and interleukin-2 receptor alpha and beta (IL-R2α and IL-R2β) which was also seen in vivo [139, 140]. Subsequently they showed that the upregulation of NFκB activity was a result of zinc stimulating IκB phosphorylation [141]. Increased protein tyrosine kinase (PTK) activity and reduced PTP activity caused by increased oxidative stress during aging was found to result in the phosphorylation of NFκB, NIK/IKK, and MAPKs (Erk, p38, and JNK) leading to NFκB activation [142, 143]. As a general point, zinc appears to play a key role in the regulation of PTK/PTP balance, by the activation of phosphorylation as a result of inhibition of PTPs or by direct activation of kinases.
Zinc signaling in cancer cells
Zinc transporters have also been linked to protein phosphorylation and signaling in cancer cells. In MDA-MB-468 breast cancer cells and PC-3 prostate cancer cells transfected with ZIP9 (PC3-ZIP9) cells, ZIP9 was found to act as a specific Gi-coupled membrane androgen receptor (mAR) binding testosterone and initiating MAPK and zinc signaling leading to apoptosis [144, 145].
In the estrogen receptor-positive human breast cancer cell line MCF-7, the ability of cells to spread to the lymph nodes was found to be a result of ZIP6-mediated zinc influx [71]. Signal transducer and activator of transcription 3 (STAT3) and oestrogen induce transcription of ZIP6. Upon transcription, ZIP6 is proteolytically cleaved at the N-terminus to translocate to the plasma membrane where it mediates Zn2+ influx. The increase in cellular zinc then results in the phosphorylation and inactivation of glycogen synthase kinase-3beta (GSK-3β) possibly through insulin-mimetic actions of zinc or indirectly through activation of Akt [71, 146, 147]. The inhibition of GSK-3β prevents it from phosphorylating transcription factor Snail, which is then retained in the nucleus acting as a transcriptional repressor of the E-cadherin gene, CDH1 (epithelial cadherin), CDH1, causing cell rounding, and detachment [71].
In 2012, it was shown in MCF-7 cells that the activity of human endoplasmic reticulum zinc channel hZIP7 was regulated by protein phosphorylation at Ser275 and Ser276 upon zinc stimulation [20]. Phosphorylation of hZIP7 was linked to the release of Zn2+ ions from intracellular stores which further led to the phosphorylation of tyrosine kinases such as Erk1/2 and Akt, and the subsequent activation of signaling pathways within the cell [20, 148]. Two additional phosphorylation sites at S294 and T294 have also been found to be important for maximal hZIP7 activity [113].
Besides being important for the regulation of cell signaling as mentioned above, protein phosphorylation is also likely to be central to the regulation of zinc transporter activity and regulation as in silico analysis predict all human ZnT and ZIP transporter proteins to be phosphorylated (Table 1 and Table 2). This underscores the importance of characterizing protein phosphorylation in zinc transporter proteins.
Table 1.
Phosphorylation sites in human ZnT transporter proteins
| Zinc transporter | #P-sites (predicted) | #P-sites (published) | P-sites (published) | References |
|---|---|---|---|---|
| ZnT1 | 39 | 15 | S29, S167, S172, S173, T196, S426, S429, T439, T449, T462, S466, S468, S473, S505, S506 | [149–156] |
| ZnT2 | 22 | 2 | T239, S247 | |
| ZnT3 | 34 | 3 | S32, S38, T66, S311 | [157] |
| ZnT4 | 40 | 4 | S12, S75, S313, S412 | |
| ZnT5 | 59 | 12 | Y5, T30, Y32, T39, T88, S364, S378, T382, Y385, Y757, T762, Y763 | [153, 158] |
| ZnT6 | 38 | 6 | S122, T376, S381, S382, S388, T391, | [152, 159, 160] |
| ZnT7 | 29 | 2 | Y11, S31 | |
| ZnT8 | 25 | 2 | T7, Y8, | |
| ZnT9 | 53 | 3 | T222, S230, S355 | |
| ZnT10 | 36 | 7 | S187, S189, T192, S197, S402, S469, Y479 | [161] |
Number of predicted phosphorylation sites (#P-sites) in human ZnT transport proteins according to the NetPhos 3.1 Server is listed along with phosphorylation sites that have been experimentally identified and published according to UniProt and PhosphoSitePlus [162, 163]. Phosphorylation sites listed in the NetPhos 3.1 Server with a prediction score above 0.5 are included. References listed in the table only account for research articles. References for phosphorylation sites found on curated info pages are not listed in the table.
Table 2.
Phosphorylation sites in human ZIP transporter proteins
| Zinc transporter | #P-sites (predicted) | #P-sites (published) | P-sites (published) | References |
|---|---|---|---|---|
| ZIP1 | 14 | 0 | ||
| ZIP2 | 23 | 0 | ||
| ZIP3 | 26 | 3 | S125, S129, Y147 | [150, 159, 164] |
| ZIP4 | 57 | 2 | T137, S490 | [165, 166] |
| ZIP5 | 4 | 1 | S336 | [167] |
| ZIP6 | 34 | 11 | S471, Y473, S475, S478, T479, T486, T490, Y493, Y528, Y531, S583 | [152, 160, 168–172] |
| ZIP7 | 8 | 4 | S275, S276, S293, T294 | [113, 154, 173] |
| ZIP8 | 1 | 4 | S275, S278, S288, T424 | [172–174] |
| ZIP9 | 1 | 0 | ||
| ZIP10 | 20 | 12 | S323, T536, S539, T540, S546, T553, S556, S570, S573, T583, S591, Y596 | [149, 152, 155, 158, 159, 174–179] |
| ZIP11 | 4 | 2 | S125, S153 | [152, 153] |
| ZIP12 | 5 | 0 | ||
| ZIP13 | 3 | 3 | S39, T42, T44 | |
| ZIP14 | 9 | 5 | S256, Y258, S260, S309, S311 | [167, 169, 180, 181] |
Number of predicted phosphorylation sites (#P-sites) in human ZIP transport proteins according to the NetPhos 3.1 Server is listed along with phosphorylation sites that have been experimentally identified and published according to UniProt and PhosphoSitePlus [162, 163]. Phosphorylation sites listed in the NetPhos 3.1 Server with a prediction score above 0.5 are included. References listed in the table only account for research articles. References for phosphorylation sites found on curated info pages are not listed in the table.
Mass spectrometry and quantitative phosphoproteomics for studying zinc signaling
Despite the ubiquitous role of protein phosphorylation, phosphoproteins have low abundance within the cell due to the transient nature of phosphorylation. In addition, phosphate groups are easily lost during sample handling. These problems make the identification and characterization of phosphorylated proteins challenging, and for years, the study of protein phosphorylation was hampered by the lack of efficient and specific methods. Traditional molecular approaches frequently focus on the analysis of a single phosphoprotein and entire signaling pathways are studied protein by protein. One often has to work from the assumption or prior knowledge that specific proteins are phosphorylated and investigate this using approaches such as cloning, 1D or 2D gel electrophoresis combined with Pro-Q Diamond staining [182–190], radioactive labelling using 32P or 33P [182, 183], or directed antibody-based strategies limited by their low specificity and high cost. Furthermore, detection provides no information of the specific sites of phosphorylation unless antibodies have been generated against specific phosphorylation sites.
Mass spectrometry (MS) is a powerful tool for discovery-based analysis of proteins and phosphoproteins where no prior knowledge is required. In MS, the protein or peptide samples are ionized, separated according to their mass-to-charge ratios before the ions are registered by a detector. The measurement of the mass-to-charge ratio of a sample molecule provides exact details on the mass, sequence and PTMs which in turn can give useful information about the structure, interaction and regulation of the particular molecule. The analysis of protein phosphorylation by MS, termed phosphoproteomics, requires selective phospho-specific enrichment methods in combination with MS strategies. Phosphorylation-specific antibodies have been used at both the protein and peptide levels. However, the poor specificity and sensitivity of anti-phosphoserine and anti-phosphothreonine antibodies have limited their use leaving the more specific anti-phosphotyrosine antibodies as the preferred method for antibody-based enrichment [191–198]. In nature, however, the vast majority of protein phosphorylation is on serine and threonine residues [199].
For many years, the primary method for phosphopeptide enrichment prior to MS analysis was immobilized metal ion affinity chromatography (IMAC) using the affinity of negatively charged phosphate groups on proteins and peptides towards positively charged metal ions (Fe3+, Al3+, Ga3+, or Co2+) chelated to nitriloacetic acid (NTA) or iminodiacetic acid (IDA)-coated beads [125, 200–208]. IMAC, however, has a limited selectivity when used on complex samples, and requires an O-methylesterification step for improved selectivity adding more complexity to the sample [209]. Titanium dioxide (TiO2) was found to have affinity for phosphate ions in aqueous solutions [210–216]. This initiated great progress in the development of new and highly sensitive strategies for phospho-specific enrichment before MS analysis [214, 217–220], and TiO2 chromatography quickly replaced all other strategies as the method of choice for large-scale phosphoproteomic studies [176, 179, 214, 217–228]. The reason for the popularity of the method is the high selectivity toward phosphopeptides but also due to the setup being simple and fast, and that TiO2 beads, unlike IMAC, are extremely tolerant toward most buffers and salts used in cell biology laboratories [229]. Later, the strengths of both IMAC and TiO2 chromatography were combined in the SIMAC method (sequential elution from IMAC) improving not only the number of phosphorylation sites detected from complex samples, but also significantly increasing the detection of multiply phosphorylated peptides from low amounts of material [230, 231]. These strategies allow for large-scale phosphoproteomic screening of various cell lines and tissues [214, 217–220, 230]. Indeed, all but one phosphorylation site identified in human ZnT and ZIP transporters have been identified in large-scale phosphoproteomic studies (Tables 1, 2).
The use of phosphoproteomic strategies can be very powerful. By combining these methods with quantitation strategies and highly efficient MS technology the comparison of proteins and PTMs in different cells, disease stages, treatments etc. is possible. Common quantitative strategies include the introduction of different isotopic labels to proteins or peptides from samples to be compared. Subsequent MS analysis then reveals differences in the abundance of labelled species originating from the different samples. Labelling can be introduced at the protein level using metabolic labelling such as 14/15 N-labeling or stable isotope labelling by amino acids in cell culture (SILAC) where different isotopic tags are added to the media and incorporated into proteins during cell culturing [232]. Chemical modification is introduced either during protein digestion (O18-labelling) or at peptide level using isobaric peptide tags for relative and absolute quantification (iTRAQ), tandem mass tags (TMT) or dimethyl labelling introduced during sample preparation [233–238]. In addition, MS offers the use of label-free strategies where quantitation is performed by the comparison of liquid chromatography (LC)-MS or MS/MS data obtained in sequential experiments [239, 240]. The different strategies within phosphoproteomics and quantitative phosphoproteomics are described in various reviews [241–244].
Conclusions and future perspectives
Despite the power of these strategies, phosphoproteomic approaches have barely been used in the study of zinc homeostasis and zinc signaling. Except for one phosphorylation site, all sites identified in the ZnT and ZIP transporters were discovered in large-scale screenings to characterize phosphorylation profiles of different cell types or conditions or during the development of new MS strategies to identify even more phosphorylation sites [149–161, 164–181]. None of the studies were designed to investigate zinc homeostasis or zinc signaling. Instead the ZnT and ZIP protein phosphorylation sites were all identified by chance. The time is ripe for using these sensitive strategies to specifically target zinc transporters and proteins involved in zinc signaling. Quantitative phosphoproteomic approaches have already proven their potential in biological studies [245–249].
The low-abundance and highly hydrophobic nature of membrane proteins adds additional challenge to the identification of phosphorylation sites in zinc transporter proteins which could explain why the sites identified primarily come from large-scale studies using large amounts of starting material. Over-expression of specific zinc transporters combined with membrane enrichment strategies would improve identification in discovery-based studies. The identification of novel phosphorylation sites in ZnT and ZIP proteins will enable the investigation of possible cytosolic binding partners in pull-down experiments using phospho-specific antibodies directed against these novel sites.
In addition, these approaches will be useful for understanding zinc physiology from studies of over-expressing or knocking out zinc transporters, time and dose dependent zinc stimulation, or studying specific diseases linked to aberrant zinc homeostasis. Combining such data with molecular methodology will lead to a deeper understanding of the molecular mechanisms regulating zinc homeostasis and shed light on the signaling pathways affected by zinc stimulation.
Acknowledgements
This work was supported by a research grant (10115) from Villum Foundation (TT; Project no. 10115); National Institutes of Health (US) (EY024481, EY027881, HD018655).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Costello LC, Fenselau CC, Franklin RB. Evidence for operation of the direct zinc ligand exchange mechanism for trafficking, transport, and reactivity of zinc in mammalian cells. J Inorg Biochem. 2011;105(5):589–599. doi: 10.1016/j.jinorgbio.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haase H, Rink L. The immune system and the impact of zinc during aging. Immun Ageing. 2009;6:9. doi: 10.1186/1742-4933-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prasad AS. Impact of the discovery of human zinc deficiency on health. J Am Coll Nutr. 2009;28(3):257–265. doi: 10.1080/07315724.2009.10719780. [DOI] [PubMed] [Google Scholar]
- 4.Sensi SL, Paoletti P, Bush AI, Sekler I. Zinc in the physiology and pathology of the CNS. Nat Rev Neurosci. 2009;10(11):780–791. doi: 10.1038/nrn2734. [DOI] [PubMed] [Google Scholar]
- 5.Sladek RRG, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 6.Taylor KMVP, Jordan N, Hiscox S, Hendley R, Nicholson RI. ZIP7-mediated intracellular zinc transport contributes to aberrant growth factor signaling in antihormone-resistant breast cancer Cells. Endocrinology. 2008;149(10):4912–4920. doi: 10.1210/en.2008-0351. [DOI] [PubMed] [Google Scholar]
- 7.Vallee BL, Falchuk KH. The biochemical basis of zinc physiology. Physiol Rev. 1993;73(1):79–118. doi: 10.1152/physrev.1993.73.1.79. [DOI] [PubMed] [Google Scholar]
- 8.Prasad AS. Discovery of human zinc deficiency and studies in an experimental human model. Am J Clin Nutr. 1991;53(2):403–412. doi: 10.1093/ajcn/53.2.403. [DOI] [PubMed] [Google Scholar]
- 9.Hambidge M. Human zinc deficiency. J Nutr. 2000;130(5S Suppl):1344S–1349S. doi: 10.1093/jn/130.5.1344S. [DOI] [PubMed] [Google Scholar]
- 10.Fraker PJ, King LE. Reprogramming of the immune system during zinc deficiency. Annu Rev Nutr. 2004;24:277–298. doi: 10.1146/annurev.nutr.24.012003.132454. [DOI] [PubMed] [Google Scholar]
- 11.Devirgiliis C, Zalewski PD, Perozzi G, Murgia C. Zinc fluxes and zinc transporter genes in chronic diseases. Mutat Res. 2007;622(1–2):84–93. doi: 10.1016/j.mrfmmm.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Rafalo-Ulinska A, Piotrowska J, Kryczyk A, Opoka W, Sowa-Kucma M, Misztak P, Rajkowska G, Stockmeier CA, Datka W, Nowak G, Szewczyk B. Zinc transporters protein level in postmortem brain of depressed subjects and suicide victims. J Psychiatr Res. 2016;83:220–229. doi: 10.1016/j.jpsychires.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafalo A, Zadrozna M, Nowak B, Kotarska K, Wiatrowska K, Pochwat B, Sowa-Kucma M, Misztak P, Nowak G, Szewczyk B. The level of the zinc homeostasis regulating proteins in the brain of rats subjected to olfactory bulbectomy model of depression. Prog Neuropsychopharmacol Biol Psychiatry. 2017;72:36–48. doi: 10.1016/j.pnpbp.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Whitfield DR, Vallortigara J, Alghamdi A, Hortobagyi T, Ballard C, Thomas AJ, O'Brien JT, Aarsland D, Francis PT. Depression and synaptic zinc regulation in Alzheimer disease, dementia with lewy bodies, and Parkinson disease dementia. Am J Geriatr Psychiatry. 2015;23(2):141–148. doi: 10.1016/j.jagp.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Hogstrand C, Zheng D, Feeney G, Cunningham P, Kille P. Zinc-controlled gene expression by metal-regulatory transcription factor 1 (MTF1) in a model vertebrate, the zebrafish. Biochem Soc Trans. 2008;36(Pt 6):1252–1257. doi: 10.1042/BST0361252. [DOI] [PubMed] [Google Scholar]
- 16.Laity JH, Andrews GK. Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1) Arch Biochem Biophys. 2007;463(2):201–210. doi: 10.1016/j.abb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Giedroc DP, Chen X, Apuy JL. Metal response element (MRE)-binding transcription factor-1 (MTF-1): structure, function, and regulation. Antioxid Redox Signal. 2001;3(4):577–596. doi: 10.1089/15230860152542943. [DOI] [PubMed] [Google Scholar]
- 18.Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc-proteins encoded in the human genome. J Proteome Res. 2006;5(1):196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 19.Passerini A, Andreini C, Menchetti S, Rosato A, Frasconi P. Predicting zinc binding at the proteome level. BMC Bioinform. 2007;8:39. doi: 10.1186/1471-2105-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor KM, Hiscox S, Nicholson RI, Hogstrand C, Kille P. Protein kinase CK2 triggers cytosolic zinc signaling pathways by phosphorylation of zinc channel ZIP7. Sci Signal. 2012;5(210):ra11. doi: 10.1126/scisignal.2002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamasaki S, Sakata-Sogawa K, Hasegawa A, Suzuki T, Kabu K, Sato E, Kurosaki T, Yamashita S, Tokunaga M, Nishida K, Hirano T. Zinc is a novel intracellular second messenger. J Cell Biol. 2007;177(4):637–645. doi: 10.1083/jcb.200702081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase H, Maret W. Intracellular zinc fluctuations modulate protein tyrosine phosphatase activity in insulin/insulin-like growth factor-1 signaling. Exp Cell Res. 2003;291(2):289–298. doi: 10.1016/s0014-4827(03)00406-3. [DOI] [PubMed] [Google Scholar]
- 23.Maret W. Zinc in cellular regulation: the nature and significance of "zinc signals". Int J Mol Sci. 2017 doi: 10.3390/ijms18112285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hara T, Takeda TA, Takagishi T, Fukue K, Kambe T, Fukada T. Physiological roles of zinc transporters: molecular and genetic importance in zinc homeostasis. J Physiol Sci. 2017;67(2):283–301. doi: 10.1007/s12576-017-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor CA, Hutchens S, Liu C, Jursa T, Shawlot W, Aschner M, Smith DR, Mukhopadhyay S. SLC30A10 transporter in the digestive system regulates brain manganese under basal conditions while brain SLC30A10 protects against neurotoxicity. J Biol Chem. 2019;294(6):1860–1876. doi: 10.1074/jbc.RA118.005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukhopadhyay S. Familial manganese-induced neurotoxicity due to mutations in SLC30A10 or SLC39A14. Neurotoxicology. 2018;64:278–283. doi: 10.1016/j.neuro.2017.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy M, Elkoshi N, Barber-Zucker S, Hoch E, Zarivach R, Hershfinkel M, Sekler I. Zinc transporter 10 (ZnT10)-dependent extrusion of cellular Mn(2+) is driven by an active Ca(2+)-coupled exchange. J Biol Chem. 2019;294(15):5879–5889. doi: 10.1074/jbc.RA118.006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalton TP, He L, Wang B, Miller ML, Jin L, Stringer KF, Chang X, Baxter CS, Nebert DW. Identification of mouse SLC39A8 as the transporter responsible for cadmium-induced toxicity in the testis. Proc Natl Acad Sci USA. 2005;102(9):3401–3406. doi: 10.1073/pnas.0406085102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girijashanker K, He L, Soleimani M, Reed JM, Li H, Liu Z, Wang B, Dalton TP, Nebert DW. Slc39a14 gene encodes ZIP14, a metal/bicarbonate symporter: similarities to the ZIP8 transporter. Mol Pharmacol. 2008;73(5):1413–1423. doi: 10.1124/mol.107.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He L, Girijashanker K, Dalton TP, Reed J, Li H, Soleimani M, Nebert DW. ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol Pharmacol. 2006;70(1):171–180. doi: 10.1124/mol.106.024521. [DOI] [PubMed] [Google Scholar]
- 31.Zogzas CE, Aschner M, Mukhopadhyay S. Structural elements in the transmembrane and cytoplasmic domains of the metal transporter SLC30A10 are required for its manganese efflux activity. J Biol Chem. 2016;291(31):15940–15957. doi: 10.1074/jbc.M116.726935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Himeno S, Yanagiya T, Fujishiro H. The role of zinc transporters in cadmium and manganese transport in mammalian cells. Biochimie. 2009;91(10):1218–1222. doi: 10.1016/j.biochi.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Yang ZB, You JF, Yang ZM. Manganese uptake and transportation as well as antioxidant response to excess manganese in plants. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao. 2007;33(6):480–488. [PubMed] [Google Scholar]
- 34.Williams LE, Mills RF. P(1B)-ATPases—an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005;10(10):491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Sitsel O, Meloni G, Autzen HE, Andersson M, Klymchuk T, Nielsen AM, Rees DC, Nissen P, Gourdon P. Structure and mechanism of Zn2+-transporting P-type ATPases. Nature. 2014;514(7523):518–522. doi: 10.1038/nature13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hantke K. Bacterial zinc transporters and regulators. Biometals. 2001;14(3–4):239–249. doi: 10.1023/a:1012984713391. [DOI] [PubMed] [Google Scholar]
- 37.Sekler I, Sensi SL, Hershfinkel M, Silverman WF. Mechanism and regulation of cellular zinc transport. Mol Med. 2007;13(7–8):337–343. doi: 10.2119/2007-00037.Sekler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichten LA, Cousins RJ. Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr. 2009;29:153–176. doi: 10.1146/annurev-nutr-033009-083312. [DOI] [PubMed] [Google Scholar]
- 39.Kambe T, Hashimoto A, Fujimoto S. Current understanding of ZIP and ZnT zinc transporters in human health and diseases. Cell Mol Life Sci. 2014;71(17):3281–3295. doi: 10.1007/s00018-014-1617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura T, Kambe T. The functions of metallothionein and ZIP and ZnT Transporters: an overview and perspective. Int J Mol Sci. 2016;17(3):336. doi: 10.3390/ijms17030336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu YY, Kirschke CP, Huang L. Immunohistochemical analysis of ZnT1, 4, 5, 6, and 7 in the mouse gastrointestinal tract. J Histochem Cytochem. 2007;55(3):223–234. doi: 10.1369/jhc.6A7032.2006. [DOI] [PubMed] [Google Scholar]
- 42.Qin Y, Thomas D, Fontaine CP, Colvin RA. Silencing of ZnT1 reduces Zn2+ efflux in cultured cortical neurons. Neurosci Lett. 2009;450(2):206–210. doi: 10.1016/j.neulet.2008.11.069. [DOI] [PubMed] [Google Scholar]
- 43.Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T. Cation diffusion facilitator family: structure and function. FEBS Lett. 2015;589(12):1283–1295. doi: 10.1016/j.febslet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Kury S, Dreno B, Bezieau S, Giraudet S, Kharfi M, Kamoun R, Moisan JP. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet. 2002;31(3):239–240. doi: 10.1038/ng913. [DOI] [PubMed] [Google Scholar]
- 45.Aydemir TB, Chang SM, Guthrie GJ, Maki AB, Ryu MS, Karabiyik A, Cousins RJ. Zinc transporter ZIP14 functions in hepatic zinc, iron and glucose homeostasis during the innate immune response (endotoxemia) PLoS ONE. 2012;7(10):e48679. doi: 10.1371/journal.pone.0048679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bin BH, Seo J, Kim ST. Function, structure, and transport aspects of ZIP and ZnT zinc transporters in immune cells. J Immunol Res. 2018;2018:9365747. doi: 10.1155/2018/9365747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowanadisai W, Lonnerdal B, Kelleher SL. Zip6 (LIV-1) regulates zinc uptake in neuroblastoma cells under resting but not depolarizing conditions. Brain Res. 2008;1199:10–19. doi: 10.1016/j.brainres.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Costello LC, Franklin RB, Zou J, Naslund MJ. Evidence that human prostate cancer is a ZIP1-deficient malignancy that could be effectively treated with a zinc ionophore (clioquinol) approach. Chemotherapy (Los Angel) 2015 doi: 10.4172/2167-7700.1000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grattan BJ, Freake HC. Zinc and cancer: implications for LIV-1 in breast cancer. Nutrients. 2012;4(7):648–675. doi: 10.3390/nu4070648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hardy AB, Prentice KJ, Froese S, Liu Y, Andrews GK, Wheeler MB. Zip4 mediated zinc influx stimulates insulin secretion in pancreatic beta cells. PLoS ONE. 2015;10(3):e0119136. doi: 10.1371/journal.pone.0119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, Jeon J, Shin M, Won Y, Lee M, Kwak JS, Lee G, Rhee J, Ryu JH, Chun CH, Chun JS. Regulation of the catabolic cascade in osteoarthritis by the zinc-ZIP8-MTF1 axis. Cell. 2014;156(4):730–743. doi: 10.1016/j.cell.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Kitamura H, Morikawa H, Kamon H, Iguchi M, Hojyo S, Fukada T, Yamashita S, Kaisho T, Akira S, Murakami M, Hirano T. Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat Immunol. 2006;7(9):971–977. doi: 10.1038/ni1373. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang SM, Cousins RJ, Fisher WE, Brunicardi FC, Logsdon CD, Chen C, Yao Q. Aberrant expression of zinc transporter ZIP4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci USA. 2007;104(47):18636–18641. doi: 10.1073/pnas.0709307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y, Chen Y, Wang Y, Yang J, Zhu VF, Liu Y, Cui X, Chen L, Yan W, Jiang T, Hergenroeder GW, Fletcher SA, Levine JM, Kim DH, Tandon N, Zhu JJ, Li M. ZIP4 is a novel molecular marker for glioma. Neuro Oncol. 2013;15(8):1008–1016. doi: 10.1093/neuonc/not042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu MJ, Bao S, Galvez-Peralta M, Pyle CJ, Rudawsky AC, Pavlovicz RE, Killilea DW, Li C, Nebert DW, Wewers MD, Knoell DL. ZIP8 regulates host defense through zinc-mediated inhibition of NF-kappaB. Cell Rep. 2013;3(2):386–400. doi: 10.1016/j.celrep.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lue HW, Yang X, Wang R, Qian W, Xu RZ, Lyles R, Osunkoya AO, Zhou BP, Vessella RL, Zayzafoon M, Liu ZR, Zhau HE, Chung LW. LIV-1 promotes prostate cancer epithelial-to-mesenchymal transition and metastasis through HB-EGF shedding and EGFR-mediated ERK signaling. PLoS ONE. 2011;6(11):e27720. doi: 10.1371/journal.pone.0027720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Unno J, Satoh K, Hirota M, Kanno A, Hamada S, Ito H, Masamune A, Tsukamoto N, Motoi F, Egawa S, Unno M, Horii A, Shimosegawa T. LIV-1 enhances the aggressive phenotype through the induction of epithelial to mesenchymal transition in human pancreatic carcinoma cells. Int J Oncol. 2009;35(4):813–821. doi: 10.3892/ijo_00000394. [DOI] [PubMed] [Google Scholar]
- 58.Weaver BP, Zhang Y, Hiscox S, Guo GL, Apte U, Taylor KM, Sheline CT, Wang L, Andrews GK. Zip4 (Slc39a4) expression is activated in hepatocellular carcinomas and functions to repress apoptosis, enhance cell cycle and increase migration. PLoS ONE. 2010 doi: 10.1371/journal.pone.0013158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawasaki E. ZnT8 and type 1 diabetes. Endocr J. 2012;59(7):531–537. doi: 10.1507/endocrj.ej12-0069. [DOI] [PubMed] [Google Scholar]
- 60.Parsons DS, Hogstrand C, Maret W. The C-terminal cytosolic domain of the human zinc transporter ZnT8 and its diabetes risk variant. FEBS J. 2018;285(7):1237–1250. doi: 10.1111/febs.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chowanadisai W, Graham DM, Keen CL, Rucker RB, Messerli MA. Neurulation and neurite extension require the zinc transporter ZIP12 (slc39a12) Proc Natl Acad Sci USA. 2013;110(24):9903–9908. doi: 10.1073/pnas.1222142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boycott KM, Beaulieu CL, Kernohan KD, Gebril OH, Mhanni A, Chudley AE, Redl D, Qin W, Hampson S, Kury S, Tetreault M, Puffenberger EG, Scott JN, Bezieau S, Reis A, Uebe S, Schumacher J, Hegele RA, McLeod DR, Galvez-Peralta M, Majewski J, Ramaekers VT, Care4Rare Canada C. Nebert DW, Innes AM, Parboosingh JS, Abou Jamra R. Autosomal-Recessive Intellectual Disability with Cerebellar Atrophy Syndrome Caused by Mutation of the Manganese and Zinc Transporter Gene SLC39A8. Am J Hum Genet. 2015;97(6):886–893. doi: 10.1016/j.ajhg.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McAllister BB, Dyck RH. Zinc transporter 3 (ZnT3) and vesicular zinc in central nervous system function. Neurosci Biobehav Rev. 2017;80:329–350. doi: 10.1016/j.neubiorev.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Palmiter RD, Cole TB, Quaife CJ, Findley SD. ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc Natl Acad Sci USA. 1996;93(25):14934–14939. doi: 10.1073/pnas.93.25.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole TB, Wenzel HJ, Kafer KE, Schwartzkroin PA, Palmiter RD. Elimination of zinc from synaptic vesicles in the intact mouse brain by disruption of the ZnT3 gene. Proc Natl Acad Sci USA. 1999;96(4):1716–1721. doi: 10.1073/pnas.96.4.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adlard PA, Parncutt JM, Finkelstein DI, Bush AI. Cognitive loss in zinc transporter-3 knock-out mice: a phenocopy for the synaptic and memory deficits of Alzheimer's disease? J Neurosci. 2010;30(5):1631–1636. doi: 10.1523/JNEUROSCI.5255-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bjorklund NL, Reese LC, Sadagoparamanujam VM, Ghirardi V, Woltjer RL, Taglialatela G. Absence of amyloid beta oligomers at the postsynapse and regulated synaptic Zn2+ in cognitively intact aged individuals with Alzheimer's disease neuropathology. Mol Neurodegener. 2012;7:23. doi: 10.1186/1750-1326-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitfield DR, Vallortigara J, Alghamdi A, Howlett D, Hortobagyi T, Johnson M, Attems J, Newhouse S, Ballard C, Thomas AJ, O'Brien JT, Aarsland D, Francis PT. Assessment of ZnT3 and PSD95 protein levels in Lewy body dementias and Alzheimer's disease: association with cognitive impairment. Neurobiol Aging. 2014;35(12):2836–2844. doi: 10.1016/j.neurobiolaging.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 69.Kambe T, Tsuji T, Hashimoto A, Itsumura N. The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol Rev. 2015;95(3):749–784. doi: 10.1152/physrev.00035.2014. [DOI] [PubMed] [Google Scholar]
- 70.Hildebrand MS, Phillips AM, Mullen SA, Adlard PA, Hardies K, Damiano JA, Wimmer V, Bellows ST, McMahon JM, Burgess R, Hendrickx R, Weckhuysen S, Suls A, De Jonghe P, Scheffer IE, Petrou S, Berkovic SF, Reid CA. Loss of synaptic Zn2+ transporter function increases risk of febrile seizures. Sci Rep. 2015;5:17816. doi: 10.1038/srep17816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hogstrand C, Kille P, Ackland ML, Hiscox S, Taylor KM. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3) Biochem J. 2013;455(2):229–237. doi: 10.1042/BJ20130483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bostanci Z, Alam S, Soybel DI, Kelleher SL. Prolactin receptor attenuation induces zinc pool redistribution through ZnT2 and decreases invasion in MDA-MB-453 breast cancer cells. Exp Cell Res. 2014;321(2):190–200. doi: 10.1016/j.yexcr.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Henshall SM, Afar DE, Rasiah KK, Horvath LG, Gish K, Caras I, Ramakrishnan V, Wong M, Jeffry U, Kench JG, Quinn DI, Turner JJ, Delprado W, Lee CS, Golovsky D, Brenner PC, O'Neill GF, Kooner R, Stricker PD, Grygiel JJ, Mack DH, Sutherland RL. Expression of the zinc transporter ZnT4 is decreased in the progression from early prostate disease to invasive prostate cancer. Oncogene. 2003;22(38):6005–6012. doi: 10.1038/sj.onc.1206797. [DOI] [PubMed] [Google Scholar]
- 74.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106(3):255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 75.Nishida K, Hasegawa A, Nakae S, Oboki K, Saito H, Yamasaki S, Hirano T. Zinc transporter Znt5/Slc30a5 is required for the mast cell-mediated delayed-type allergic reaction but not the immediate-type reaction. J Exp Med. 2009;206(6):1351–1364. doi: 10.1084/jem.20082533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang T, Liu J, Fellner M, Zhang C, Sui D, Hu J. Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci Adv. 2017;3(8):e1700344. doi: 10.1126/sciadv.1700344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu M, Fu D. Structure of the zinc transporter YiiP. Science. 2007;317(5845):1746–1748. doi: 10.1126/science.1143748. [DOI] [PubMed] [Google Scholar]
- 78.Fukada T, Kambe T. Molecular and genetic features of zinc transporters in physiology and pathogenesis. Metallomics. 2011;3(7):662–674. doi: 10.1039/c1mt00011j. [DOI] [PubMed] [Google Scholar]
- 79.Kambe T. Molecular architecture and function of ZnT transporters. Curr Top Membr. 2012;69:199–220. doi: 10.1016/B978-0-12-394390-3.00008-2. [DOI] [PubMed] [Google Scholar]
- 80.Wei Y, Fu D. Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J Biol Chem. 2006;281(33):23492–23502. doi: 10.1074/jbc.M602254200. [DOI] [PubMed] [Google Scholar]
- 81.Hoch E, Lin W, Chai J, Hershfinkel M, Fu D, Sekler I. Histidine pairing at the metal transport site of mammalian ZnT transporters controls Zn2+ over Cd2+ selectivity. Proc Natl Acad Sci USA. 2012;109(19):7202–7207. doi: 10.1073/pnas.1200362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barber-Zucker S, Uebe R, Davidov G, Navon Y, Sherf D, Chill JH, Kass I, Bitton R, Schuler D, Zarivach R. Disease-homologous mutation in the cation diffusion facilitator protein MamM causes single-domain structural loss and signifies its importance. Sci Rep. 2016;6:31933. doi: 10.1038/srep31933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujimoto S, Tsuji T, Fujiwara T, Takeda TA, Merriman C, Fukunaka A, Nishito Y, Fu D, Hoch E, Sekler I, Fukue K, Miyamae Y, Masuda S, Nagao M, Kambe T. The PP-motif in luminal loop 2 of ZnT transporters plays a pivotal role in TNAP activation. Biochem J. 2016;473(17):2611–2621. doi: 10.1042/BCJ20160324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor KM. LIV-1 breast cancer protein belongs to new family of histidine-rich membrane proteins with potential to control intracellular Zn2+ homeostasis. IUBMB Life. 2000;49(4):249–253. doi: 10.1080/15216540050033087. [DOI] [PubMed] [Google Scholar]
- 85.Taylor KM, Nicholson RI. The LZT proteins; the LIV-1 subfamily of zinc transporters. Biochim Biophys Acta. 2003;1611(1–2):16–30. doi: 10.1016/s0005-2736(03)00048-8. [DOI] [PubMed] [Google Scholar]
- 86.Gaither LA, Eide DJ. Eukaryotic zinc transporters and their regulation. Biometals. 2001;14(3–4):251–270. doi: 10.1023/a:1012988914300. [DOI] [PubMed] [Google Scholar]
- 87.Jeong J, Eide DJ. The SLC39 family of zinc transporters. Mol Aspects Med. 2013;34(2–3):612–619. doi: 10.1016/j.mam.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kasana S, Din J, Maret W. Genetic causes and gene-nutrient interactions in mammalian zinc deficiencies: acrodermatitis enteropathica and transient neonatal zinc deficiency as examples. J Trace Elem Med Biol. 2015;29:47–62. doi: 10.1016/j.jtemb.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 89.Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem. 2003;278(35):33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- 90.Gaither LA, Eide DJ. Functional expression of the human hZIP2 zinc transporter. J Biol Chem. 2000;275(8):5560–5564. doi: 10.1074/jbc.275.8.5560. [DOI] [PubMed] [Google Scholar]
- 91.Lichten LA, Ryu MS, Guo L, Embury J, Cousins RJ. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS ONE. 2011;6(6):e21526. doi: 10.1371/journal.pone.0021526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kury S, Kharfi M, Schmitt S, Bezieau S. Clinical utility gene card for: acrodermatitis enteropathica. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmitt S, Kury S, Giraud M, Dreno B, Kharfi M, Bezieau S. An update on mutations of the SLC39A4 gene in acrodermatitis enteropathica. Hum Mutat. 2009;30(6):926–933. doi: 10.1002/humu.20988. [DOI] [PubMed] [Google Scholar]
- 94.Zhang T, Sui D, Hu J. Structural insights of ZIP4 extracellular domain critical for optimal zinc transport. Nat Commun. 2016;7:11979. doi: 10.1038/ncomms11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang F, Dufner-Beattie J, Kim BE, Petris MJ, Andrews G, Eide DJ. Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J Biol Chem. 2004;279(23):24631–24639. doi: 10.1074/jbc.M400680200. [DOI] [PubMed] [Google Scholar]
- 96.Kambe T, Andrews GK. Novel proteolytic processing of the ectodomain of the zinc transporter ZIP4 (SLC39A4) during zinc deficiency is inhibited by acrodermatitis enteropathica mutations. Mol Cell Biol. 2009;29(1):129–139. doi: 10.1128/MCB.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ehsani S, Salehzadeh A, Huo H, Reginold W, Pocanschi CL, Ren H, Wang H, So K, Sato C, Mehrabian M, Strome R, Trimble WS, Hazrati LN, Rogaeva E, Westaway D, Carlson GA, Schmitt-Ulms G. LIV-1 ZIP ectodomain shedding in prion-infected mice resembles cellular response to transition metal starvation. J Mol Biol. 2012;422(4):556–574. doi: 10.1016/j.jmb.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bafaro EM, Antala S, Nguyen TV, Dzul SP, Doyon B, Stemmler TL, Dempski RE. The large intracellular loop of hZIP4 is an intrinsically disordered zinc binding domain. Metallomics. 2015;7(9):1319–1330. doi: 10.1039/c5mt00066a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dufner-Beattie J, Kuo YM, Gitschier J, Andrews GK. The adaptive response to dietary zinc in mice involves the differential cellular localization and zinc regulation of the zinc transporters ZIP4 and ZIP5. J Biol Chem. 2004;279(47):49082–49090. doi: 10.1074/jbc.M409962200. [DOI] [PubMed] [Google Scholar]
- 100.Kim BE, Wang F, Dufner-Beattie J, Andrews GK, Eide DJ, Petris MJ. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem. 2004;279(6):4523–4530. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- 101.Mao X, Kim BE, Wang F, Eide DJ, Petris MJ. A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J Biol Chem. 2007;282(10):6992–7000. doi: 10.1074/jbc.M610552200. [DOI] [PubMed] [Google Scholar]
- 102.Bowers K, Srai SKS. The trafficking of metal ion transporters of the Zrt- and Irt-like protein family. Traffic. 2018;19(11):813–822. doi: 10.1111/tra.12602. [DOI] [PubMed] [Google Scholar]
- 103.Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther. 1999;82(2–3):111–121. doi: 10.1016/s0163-7258(98)00056-4. [DOI] [PubMed] [Google Scholar]
- 104.Hunter T. Signaling—2000 and beyond. Cell. 2000;100(1):113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 105.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 106.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103(2):211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 107.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 108.Morandell S, Stasyk T, Grosstessner-Hain K, Roitinger E, Mechtler K, Bonn GK, Huber LA. Phosphoproteomics strategies for the functional analysis of signal transduction. Proteomics. 2006;6(14):4047–4056. doi: 10.1002/pmic.200600058. [DOI] [PubMed] [Google Scholar]
- 109.Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005;5(16):4052–4061. doi: 10.1002/pmic.200401289. [DOI] [PubMed] [Google Scholar]
- 110.Pan E, Zhang XA, Huang Z, Krezel A, Zhao M, Tinberg CE, Lippard SJ, McNamara JO. Vesicular zinc promotes presynaptic and inhibits postsynaptic long-term potentiation of mossy fiber-CA3 synapse. Neuron. 2011;71(6):1116–1126. doi: 10.1016/j.neuron.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sindreu C, Palmiter RD, Storm DR. Zinc transporter ZnT-3 regulates presynaptic Erk1/2 signaling and hippocampus-dependent memory. Proc Natl Acad Sci USA. 2011;108(8):3366–3370. doi: 10.1073/pnas.1019166108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brautigan DL, Bornstein P, Gallis B. Phosphotyrosyl-protein phosphatase. Specific inhibition by Zn. J Biol Chem. 1981;256(13):6519–6522. [PubMed] [Google Scholar]
- 113.Nimmanon T, Ziliotto S, Morris S, Flanagan L, Taylor KM. Phosphorylation of zinc channel ZIP7 drives MAPK, PI3K and mTOR growth and proliferation signaling. Metallomics. 2017;9(5):471–481. doi: 10.1039/c6mt00286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wilson M, Hogstrand C, Maret W. Picomolar concentrations of free zinc(II) ions regulate receptor protein-tyrosine phosphatase beta activity. J Biol Chem. 2012;287(12):9322–9326. doi: 10.1074/jbc.C111.320796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hengge AC. Isotope effects in the study of phosphoryl and sulfuryl transfer reactions. Acc Chem Res. 2002;35(2):105–112. doi: 10.1021/ar000143q. [DOI] [PubMed] [Google Scholar]
- 116.Brandao TA, Johnson SJ, Hengge AC. The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B. Arch Biochem Biophys. 2012;525(1):53–59. doi: 10.1016/j.abb.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brandao TA, Hengge AC, Johnson SJ. Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps. J Biol Chem. 2010;285(21):15874–15883. doi: 10.1074/jbc.M109.066951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bellomo E, Massarotti A, Hogstrand C, Maret W. Zinc ions modulate protein tyrosine phosphatase 1B activity. Metallomics. 2014;6(7):1229–1239. doi: 10.1039/c4mt00086b. [DOI] [PubMed] [Google Scholar]
- 119.Ohashi K, Nagata Y, Wada E, Zammit PS, Shiozuka M, Matsuda R. Zinc promotes proliferation and activation of myogenic cells via the PI3K/Akt and ERK signaling cascade. Exp Cell Res. 2015;333(2):228–237. doi: 10.1016/j.yexcr.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 120.Sun W, Yang J, Wang W, Hou J, Cheng Y, Fu Y, Xu Z, Cai L. The beneficial effects of Zn on Akt-mediated insulin and cell survival signaling pathways in diabetes. J Trace Elem Med Biol. 2018;46:117–127. doi: 10.1016/j.jtemb.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 121.Ho Y, Samarasinghe R, Knoch ME, Lewis M, Aizenman E, DeFranco DB. Selective inhibition of mitogen-activated protein kinase phosphatases by zinc accounts for extracellular signal-regulated kinase 1/2-dependent oxidative neuronal cell death. Mol Pharmacol. 2008;74(4):1141–1151. doi: 10.1124/mol.108.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Du S, McLaughlin B, Pal S, Aizenman E. In vitro neurotoxicity of methylisothiazolinone, a commonly used industrial and household biocide, proceeds via a zinc and extracellular signal-regulated kinase mitogen-activated protein kinase-dependent pathway. J Neurosci. 2002;22(17):7408–7416. doi: 10.1523/JNEUROSCI.22-17-07408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang Y, Aizenman E, DeFranco DB, Rosenberg PA. Intracellular zinc release, 12-lipoxygenase activation and MAPK dependent neuronal and oligodendroglial death. Mol Med. 2007;13(7–8):350–355. doi: 10.2119/2007-00042.Zhang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.He K, Aizenman E. ERK signaling leads to mitochondrial dysfunction in extracellular zinc-induced neurotoxicity. J Neurochem. 2010;114(2):452–461. doi: 10.1111/j.1471-4159.2010.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McAllister AK, Katz LC, Lo DC. Neurotrophins and synaptic plasticity. Annu Rev Neurosci. 1999;22:295–318. doi: 10.1146/annurev.neuro.22.1.295. [DOI] [PubMed] [Google Scholar]
- 126.Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2(1):24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 127.Coull JA, Beggs S, Boudreau D, Boivin D, Tsuda M, Inoue K, Gravel C, Salter MW, De Koninck Y. BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain. Nature. 2005;438(7070):1017–1021. doi: 10.1038/nature04223. [DOI] [PubMed] [Google Scholar]
- 128.McNamara JO, Huang YZ. Leonard AS (2006) Molecular signaling mechanisms underlying epileptogenesis. Sci STKE. 2006;356:re12. doi: 10.1126/stke.3562006re12. [DOI] [PubMed] [Google Scholar]
- 129.Chao MV, Rajagopal R, Lee FS. Neurotrophin signaling in health and disease. Clin Sci (Lond) 2006;110(2):167–173. doi: 10.1042/CS20050163. [DOI] [PubMed] [Google Scholar]
- 130.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 131.Hwang JJ, Park MH, Choi SY, Koh JY. Activation of the Trk signaling pathway by extracellular zinc. Role of metalloproteinases. J Biol Chem. 2005;280(12):11995–12001. doi: 10.1074/jbc.M403172200. [DOI] [PubMed] [Google Scholar]
- 132.Huang YZ, Pan E, Xiong ZQ, McNamara JO. Zinc-mediated transactivation of TrkB potentiates the hippocampal mossy fiber-CA3 pyramid synapse. Neuron. 2008;57(4):546–558. doi: 10.1016/j.neuron.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 133.Poddar R, Rajagopal S, Shuttleworth CW, Paul S. Zn2+-dependent activation of the Trk signaling pathway induces phosphorylation of the brain-enriched tyrosine phosphatase STEP: molecular basis for Zn2+-induced ERK MAPK activation. J Biol Chem. 2016;291(2):813–825. doi: 10.1074/jbc.M115.663468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6(10):1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 135.Chang TP, Vancurova I. NFkappaB function and regulation in cutaneous T-cell lymphoma. Am J Cancer Res. 2013;3(5):433–445. [PMC free article] [PubMed] [Google Scholar]
- 136.Henkel TMT, Alkalay I, Krönke M, Ben-Neriah Y, Baeuerle PA. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365(6442):182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 137.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 138.Prasad AS, Bao B, Beck FW, Sarkar FH. Zinc activates NF-kappaB in HUT-78 cells. J Lab Clin Med. 2001;138(4):250–256. doi: 10.1067/mlc.2001.118108. [DOI] [PubMed] [Google Scholar]
- 139.Prasad AS, Bao B, Beck FW, Sarkar FH. Zinc enhances the expression of interleukin-2 and interleukin-2 receptors in HUT-78 cells by way of NF-kappaB activation. J Lab Clin Med. 2002;140(4):272–289. doi: 10.1067/mlc.2002.127908. [DOI] [PubMed] [Google Scholar]
- 140.Prasad AS, Bao B, Beck FW, Sarkar FH. Correction of interleukin-2 gene expression by in vitro zinc addition to mononuclear cells from zinc-deficient human subjects: a specific test for zinc deficiency in humans. Transl Res. 2006;148(6):325–333. doi: 10.1016/j.trsl.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 141.Bao B, Prasad AS, Beck FW, Sarkar FH. Zinc up-regulates NF-kappaB activation via phosphorylation of IkappaB in HUT-78 (Th0) cells. FEBS Lett. 2007;581(23):4507–4511. doi: 10.1016/j.febslet.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 142.Jung KJ, Lee EK, Yu BP, Chung HY. Significance of protein tyrosine kinase/protein tyrosine phosphatase balance in the regulation of NF-kappaB signaling in the inflammatory process and aging. Free Radic Biol Med. 2009;47(7):983–991. doi: 10.1016/j.freeradbiomed.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 143.Schulze-Osthoff K, Ferrari D, Riehemann K, Wesselborg S. Regulation of NF-kappa B activation by MAP kinase cascades. Immunobiology. 1997;198(1–3):35–49. doi: 10.1016/s0171-2985(97)80025-3. [DOI] [PubMed] [Google Scholar]
- 144.Thomas P, Pang Y, Dong J. Membrane androgen receptor characteristics of human ZIP9 (SLC39A) zinc transporter in prostate cancer cells: androgen-specific activation and involvement of an inhibitory G protein in zinc and MAP kinase signaling. Mol Cell Endocrinol. 2017;447:23–34. doi: 10.1016/j.mce.2017.02.025. [DOI] [PubMed] [Google Scholar]
- 145.Thomas P, Pang Y, Dong J, Berg AH. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology. 2014;155(11):4250–4265. doi: 10.1210/en.2014-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ilouz R, Kaidanovich O, Gurwitz D, Eldar-Finkelman H. Inhibition of glycogen synthase kinase-3beta by bivalent zinc ions: insight into the insulin-mimetic action of zinc. Biochem Biophys Res Commun. 2002;295(1):102–106. doi: 10.1016/s0006-291x(02)00636-8. [DOI] [PubMed] [Google Scholar]
- 147.Lee S, Chanoit G, McIntosh R, Zvara DA, Xu Z. Molecular mechanism underlying Akt activation in zinc-induced cardioprotection. Am J Physiol Heart Circ Physiol. 2009;297(2):H569–575. doi: 10.1152/ajpheart.00293.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Taylor KM, Kille P, Hogstrand C. Protein kinase CK2 opens the gate for zinc signaling. Cell Cycle. 2012;11(10):1863–1864. doi: 10.4161/cc.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Beli P, Lukashchuk N, Wagner SA, Weinert BT, Olsen JV, Baskcomb L, Mann M, Jackson SP, Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46(2):212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang H, Haar Petersen M, Ibanez-Vea M, Lassen PS, Larsen MR, Palmisano G. Simultaneous enrichment of cysteine-containing peptides and phosphopeptides using a cysteine-specific phosphonate adaptable tag (CysPAT) in combination with titanium dioxide (TiO2) chromatography. Mol Cell Proteom. 2016;15(10):3282–3296. doi: 10.1074/mcp.M115.054551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci Signal. 2011;4(179):rs5. doi: 10.1126/scisignal.2001497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, Kawaler E, Mundt F, Krug K, Tu Z, Lei JT, Gatza ML, Wilkerson M, Perou CM, Yellapantula V, Huang KL, Lin C, McLellan MD, Yan P, Davies SR, Townsend RR, Skates SJ, Wang J, Zhang B, Kinsinger CR, Mesri M, Rodriguez H, Ding L, Paulovich AG, Fenyo D, Ellis MJ, Carr SA, Nci C. Proteogenomics connects somatic mutations to signaling in breast cancer. Nature. 2016;534(7605):55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 155.Ruse CI, McClatchy DB, Lu B, Cociorva D, Motoyama A, Park SK, Yates JR., 3rd Motif-specific sampling of phosphoproteomes. J Proteome Res. 2008;7(5):2140–2150. doi: 10.1021/pr800147u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Shiromizu T, Adachi J, Watanabe S, Murakami T, Kuga T, Muraoka S, Tomonaga T. Identification of missing proteins in the neXtProt database and unregistered phosphopeptides in the PhosphoSitePlus database as part of the Chromosome-centric Human Proteome Project. J Proteome Res. 2013;12(6):2414–2421. doi: 10.1021/pr300825v. [DOI] [PubMed] [Google Scholar]
- 157.Imami K, Sugiyama N, Imamura H, Wakabayashi M, Tomita M, Taniguchi M, Ueno T, Toi M, Ishihama Y. Temporal profiling of lapatinib-suppressed phosphorylation signals in EGFR/HER2 pathways. Mol Cell Proteom. 2012;11(12):1741–1757. doi: 10.1074/mcp.M112.019919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sharma K, D'Souza RC, Tyanova S, Schaab C, Wisniewski JR, Cox J, Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8(5):1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 159.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31(3):438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 160.Zhou H, Di Palma S, Preisinger C, Peng M, Polat AN, Heck AJ, Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12(1):260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 161.Boeing S, Williamson L, Encheva V, Gori I, Saunders RE, Instrell R, Aygun O, Rodriguez-Martinez M, Weems JC, Kelly GP, Conaway JW, Conaway RC, Stewart A, Howell M, Snijders AP, Svejstrup JQ. Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 2016;15(7):1597–1610. doi: 10.1016/j.celrep.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40((Database issue)):D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294(5):1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 164.Tzouros M, Golling S, Avila D, Lamerz J, Berrera M, Ebeling M, Langen H, Augustin A. Development of a 5-plex SILAC method tuned for the quantitation of tyrosine phosphorylation dynamics. Mol Cell Proteom. 2013;12(11):3339–3349. doi: 10.1074/mcp.O113.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5(2):204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, Chen HY, Nesvizhskii AI, Ishihama Y, Chen YJ. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. doi: 10.1038/ncomms7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, Sun D, Wang L, Ye M, Zou H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteom. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 168.Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR, 3rd, Lee JD. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69(6):2663–2668. doi: 10.1158/0008-5472.CAN-08-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, Levine DA, Townsend R, Erdmann-Gilmore P, Snider JE, Davies SR, Ruggles KV, Fenyo D, Kitchens RT, Li S, Olvera N, Dao F, Rodriguez H, Chan DW, Liebler D, White F, Rodland KD, Mills GB, Smith RD, Paulovich AG, Ellis M, Carr SA. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteom. 2014;13(7):1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Palacios-Moreno J, Foltz L, Guo A, Stokes MP, Kuehn ED, George L, Comb M, Grimes ML. Neuroblastoma tyrosine kinase signaling networks involve FYN and LYN in endosomes and lipid rafts. PLoS Comput Biol. 2015;11(4):e1004130. doi: 10.1371/journal.pcbi.1004130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu F, Wang P, Zhang J, Young LC, Lai R, Li L. Studies of phosphoproteomic changes induced by nucleophosmin-anaplastic lymphoma kinase (ALK) highlight deregulation of tumor necrosis factor (TNF)/Fas/TNF-related apoptosis-induced ligand signaling pathway in ALK-positive anaplastic large cell lymphoma. Mol Cell Proteom. 2010;9(7):1616–1632. doi: 10.1074/mcp.M000153-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81(11):4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 173.Kim JE, Tannenbaum SR, White FM. Global phosphoproteome of HT-29 human colon adenocarcinoma cells. J Proteome Res. 2005;4(4):1339–1346. doi: 10.1021/pr050048h. [DOI] [PubMed] [Google Scholar]
- 174.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods. 2013;10(7):634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3(136):ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 177.Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol Cell Proteom. 2009;8(12):2796–2808. doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4(164):rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 179.Thingholm TE, Larsen MR, Ingrell CR, Kassem M, Jensen ON. TiO(2)-based phosphoproteomic analysis of the plasma membrane and the effects of phosphatase inhibitor treatment. J Proteome Res. 2008;7(8):3304–3313. doi: 10.1021/pr800099y. [DOI] [PubMed] [Google Scholar]
- 180.Klammer M, Kaminski M, Zedler A, Oppermann F, Blencke S, Marx S, Muller S, Tebbe A, Godl K, Schaab C. Phosphosignature predicts dasatinib response in non-small cell lung cancer. Mol Cell Proteom. 2012;11(9):651–668. doi: 10.1074/mcp.M111.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M, Seo HC, Nagiec M, Luna RE, Reinherz EL, Blenis J, Gygi SP, Wagner G. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci USA. 2014;111(21):E2182–2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Su HC, Hutchison CA, 3rd, Giddings MC. Mapping phosphoproteins in Mycoplasma genitalium and Mycoplasma pneumoniae. BMC Microbiol. 2007;7:63. doi: 10.1186/1471-2180-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Eymann C, Becher D, Bernhardt J, Gronau K, Klutzny A, Hecker M. Dynamics of protein phosphorylation on Ser/Thr/Tyr in Bacillus subtilis. Proteomics. 2007;7(19):3509–3526. doi: 10.1002/pmic.200700232. [DOI] [PubMed] [Google Scholar]
- 184.Chitteti BR, Peng Z. Proteome and phosphoproteome dynamic change during cell dedifferentiation in Arabidopsis. Proteomics. 2007;7(9):1473–1500. doi: 10.1002/pmic.200600871. [DOI] [PubMed] [Google Scholar]
- 185.Clarke SJ, Khaliulin I, Das M, Parker JE, Heesom KJ, Halestrap AP. Inhibition of mitochondrial permeability transition pore opening by ischemic preconditioning is probably mediated by reduction of oxidative stress rather than mitochondrial protein phosphorylation. Circ Res. 2008;102(9):1082–1090. doi: 10.1161/CIRCRESAHA.107.167072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park SH, Kim JW, Yun SH, Leem SH, Kahng HY, Kim SI. Characterization of beta-ketoadipate pathway from multi-drug resistance bacterium, Acinetobacter baumannii DU202 by proteomic approach. J Microbiol. 2006;44(6):632–640. [PubMed] [Google Scholar]
- 187.Baik JY, Joo EJ, Kim YH, Lee GM. Limitations to the comparative proteomic analysis of thrombopoietin producing Chinese hamster ovary cells treated with sodium butyrate. J Biotechnol. 2008;133(4):461–468. doi: 10.1016/j.jbiotec.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 188.Masaki S, Yamada T, Hirasawa T, Todaka D, Kanekatsu M. Proteomic analysis of RNA-binding proteins in dry seeds of rice after fractionation by ssDNA affinity column chromatography. Biotechnol Lett. 2008;30(5):955–960. doi: 10.1007/s10529-007-9619-8. [DOI] [PubMed] [Google Scholar]
- 189.Cole JN, Aquilina JA, Hains PG, Henningham A, Sriprakash KS, Caparon MG, Nizet V, Kotb M, Cordwell SJ, Djordjevic SP, Walker MJ. Role of group A Streptococcus HtrA in the maturation of SpeB protease. Proteomics. 2007;7(24):4488–4498. doi: 10.1002/pmic.200700626. [DOI] [PubMed] [Google Scholar]
- 190.Yue QX, Cao ZW, Guan SH, Liu XH, Tao L, Wu WY, Li YX, Yang PY, Liu X, Guo DA. Proteomics characterization of the cytotoxicity mechanism of ganoderic acid D and computer-automated estimation of the possible drug target network. Mol Cell Proteom. 2008;7(5):949–961. doi: 10.1074/mcp.M700259-MCP200. [DOI] [PubMed] [Google Scholar]
- 191.Gronborg M, Kristiansen TZ, Stensballe A, Andersen JS, Ohara O, Mann M, Jensen ON, Pandey A. A mass spectrometry-based proteomic approach for identification of serine/threonine-phosphorylated proteins by enrichment with phospho-specific antibodies: identification of a novel protein, Frigg, as a protein kinase A substrate. Mol Cell Proteom. 2002;1(7):517–527. doi: 10.1074/mcp.m200010-mcp200. [DOI] [PubMed] [Google Scholar]
- 192.Pandey A, Fernandez MM, Steen H, Blagoev B, Nielsen MM, Roche S, Mann M, Lodish HF. Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J Biol Chem. 2000;275(49):38633–38639. doi: 10.1074/jbc.M007849200. [DOI] [PubMed] [Google Scholar]
- 193.Collins MO, Yu L, Choudhary JS. Analysis of protein phosphorylation on a proteome-scale. Proteomics. 2007;7(16):2751–2768. doi: 10.1002/pmic.200700145. [DOI] [PubMed] [Google Scholar]
- 194.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 195.Kasyapa C, Gu TL, Nagarajan L, Polakiewicz R, Cowell JK. Phosphorylation of the SSBP2 and ABL proteins by the ZNF198-FGFR1 fusion kinase seen in atypical myeloproliferative disorders as revealed by phosphopeptide-specific MS. Proteomics. 2009;9(16):3979–3988. doi: 10.1002/pmic.200800852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhang Y, Yoshida Y, Nameta M, Xu B, Taguchi I, Ikeda T, Fujinaka H, Magdeldin S, Tsukaguchi H, Harita Y, Yaoita E, Yamamoto T. Glomerular proteins related to slit diaphragm and matrix adhesion in the foot processes are highly tyrosine phosphorylated in the normal rat kidney. Nephrol Dial Transplant. 2010;25(6):1785–1795. doi: 10.1093/ndt/gfp697. [DOI] [PubMed] [Google Scholar]
- 197.Shomori K, Ochiai A, Akimoto S, Ino Y, Shudo K, Ito H, Hirohashi S. Tyrosine-phosphorylation of the 12th armadillo-repeat of beta-catenin is associated with cadherin dysfunction in human cancer. Int J Oncol. 2009;35(3):517–524. doi: 10.3892/ijo_00000363. [DOI] [PubMed] [Google Scholar]
- 198.Duran MC, Chan HL, Timms JF. Identification of oxidative stress-induced tyrosine phosphorylated proteins by immunoprecipitation and mass spectrometry. Methods Mol Biol. 2009;527(33–45):ix. doi: 10.1007/978-1-60327-834-8_3. [DOI] [PubMed] [Google Scholar]
- 199.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77(3):1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Andersson L, Porath J. Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem. 1986;154(1):250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 201.Neville DC, Rozanas CR, Price EM, Gruis DB, Verkman AS, Townsend RR. Evidence for phosphorylation of serine 753 in CFTR using a novel metal-ion affinity resin and matrix-assisted laser desorption mass spectrometry. Protein Sci. 1997;6(11):2436–2445. doi: 10.1002/pro.5560061117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51(5):931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Michel HP, Bennett J. Identification of the phosphorylation site of an 8.3 kDa protein from photosystem II of spinach. FEBS Lett. 1987;212:103–108. [Google Scholar]
- 204.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteom. 2005;4(3):310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 205.Figeys D, Gygi SP, McKinnon G, Aebersold R. An integrated microfluidics-tandem mass spectrometry system for automated protein analysis. Anal Chem. 1998;70(18):3728–3734. doi: 10.1021/ac980320p. [DOI] [PubMed] [Google Scholar]
- 206.Nuhse TS, Stensballe A, Jensen ON, Peck SC. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol Cell Proteom. 2003;2(11):1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 207.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71(14):2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 208.Li S, Dass C. Iron(III)-immobilized metal ion affinity chromatography and mass spectrometry for the purification and characterization of synthetic phosphopeptides. Anal Biochem. 1999;270(1):9–14. doi: 10.1006/abio.1999.4060. [DOI] [PubMed] [Google Scholar]
- 209.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 210.Ikeguchi Y, Nakamura H. Determination of organic phosphates by column-switching high performance anionexchange chromatography using on-line preconcentration on titania. Anal Sci. 1997;13:479–483. [Google Scholar]
- 211.Ikeguchi Y, Nakamura H. Selective enrichment of phospholipids by titania. Anal Sci. 2000;16:541. [Google Scholar]
- 212.Jiang ZT, Zuo YM. Synthesis of porous titania microspheres for HPLC packings by polymerization-induced colloid aggregation (PICA) Anal Chem. 2001;73(3):686–688. doi: 10.1021/ac001008u. [DOI] [PubMed] [Google Scholar]
- 213.Kawahara M, Nakamura H, Nakajima T. Titania and zirconia: possible new ceramic microparticulates for high-performance liquid chromatography. J Chromatogr A. 1990;515:149–158. [Google Scholar]
- 214.Sano A, Nakamura H. Chemo-affinity of titania for the column-switching HPLC analysis of phosphopeptides. Anal Sci. 2004;20(3):565–566. doi: 10.2116/analsci.20.565. [DOI] [PubMed] [Google Scholar]
- 215.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA. 2007;104(14):5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Connor PA, McQuillan AJ. Phosphate adsorption onto TiO2 from aqueous solutions: an in situ internal reflection infraredspectroscopicstudy. Langmuir. 1999;15:2916–2921. [Google Scholar]
- 217.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteom. 2005;4(7):873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 218.Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ. Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem. 2004;76(14):3935–3943. doi: 10.1021/ac0498617. [DOI] [PubMed] [Google Scholar]
- 219.Sano A, Nakamura H. Titania as a chemo-affinity support for the column-switching HPLC analysis of phosphopeptides: application to the characterization of phosphorylation sites in proteins by combination with protease digestion and electrospray ionization mass spectrometry. Anal Sci. 2004;20(5):861–864. doi: 10.2116/analsci.20.861. [DOI] [PubMed] [Google Scholar]
- 220.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1(4):1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 221.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4(9):709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 222.Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Slijper M, Menke FL. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteom. 2007;6(7):1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 223.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4(3):231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 224.Dengjel J, Akimov V, Olsen JV, Bunkenborg J, Mann M, Blagoev B, Andersen JS. Quantitative proteomic assessment of very early cellular signaling events. Nat Biotechnol. 2007;25(5):566–568. doi: 10.1038/nbt1301. [DOI] [PubMed] [Google Scholar]
- 225.Hopf C, Bantscheff M, Drewes G. Pathway proteomics and chemical proteomics team up in drug discovery. Neurodegener Dis. 2007;4(2–3):270–280. doi: 10.1159/000101851. [DOI] [PubMed] [Google Scholar]
- 226.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA. 2007;104(7):2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Pinkse MW, Mohammed S, Gouw JW, van Breukelen B, Vos HR, Heck AJ. Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J Proteome Res. 2008;7(2):687–697. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 228.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7(3):1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 229.Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Spectrom. 2007;21(22):3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 230.Thingholm TE, Jensen ON, Robinson PJ, Larsen MR. SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol Cell Proteom. 2008;7(4):661–671. doi: 10.1074/mcp.M700362-MCP200. [DOI] [PubMed] [Google Scholar]
- 231.Thingholm TE, Larsen MR. Sequential elution from IMAC (SIMAC): an efficient method for enrichment and separation of mono- and multi-phosphorylated peptides. Methods Mol Biol. 2016;1355:147–160. doi: 10.1007/978-1-4939-3049-4_10. [DOI] [PubMed] [Google Scholar]
- 232.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteom. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 233.Mirgorodskaya OA, Kozmin YP, Titov MI, Korner R, Sonksen CP, Roepstorff P. Quantitation of peptides and proteins by matrix-assisted laser desorption/ionization mass spectrometry using (18)O-labeled internal standards. Rapid Commun Mass Spectrom. 2000;14(14):1226–1232. doi: 10.1002/1097-0231(20000730)14:14<1226::AID-RCM14>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 234.Stewart II, Thomson T, Figeys D. 18O labeling: a tool for proteomics. Rapid Commun Mass Spectrom. 2001;15(24):2456–2465. doi: 10.1002/rcm.525. [DOI] [PubMed] [Google Scholar]
- 235.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73(13):2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 236.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteom. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 237.Choe L, D'Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7(20):3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 239.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 240.Steen H, Jebanathirajah JA, Springer M, Kirschner MW. Stable isotope-free relative and absolute quantitation of protein phosphorylation stoichiometry by MS. Proc Natl Acad Sci USA. 2005;102(11):3948–3953. doi: 10.1073/pnas.0409536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Palmisano G, Thingholm TE. Strategies for quantitation of phosphoproteomic data. Expert Rev Proteom. 2010;7(3):439–456. doi: 10.1586/epr.10.19. [DOI] [PubMed] [Google Scholar]
- 242.Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9(6):1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 243.von Stechow L, Francavilla C, Olsen JV. Recent findings and technological advances in phosphoproteomics for cells and tissues. Expert Rev Proteom. 2015;12(5):469–487. doi: 10.1586/14789450.2015.1078730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chan CY, Gritsenko MA, Smith RD, Qian WJ. The current state of the art of quantitative phosphoproteomics and its applications to diabetes research. Expert Rev Proteom. 2016;13(4):421–433. doi: 10.1586/14789450.2016.1164604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2(84):rs46. doi: 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 246.Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339(6125):1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 247.Ye J, Zhang H, He W, Zhu B, Zhou D, Chen Z, Ashraf U, Wei Y, Liu Z, Fu ZF, Chen H, Cao S. Quantitative phosphoproteomic analysis identifies the critical role of JNK1 in neuroinflammation induced by Japanese encephalitis virus. Sci Signal. 2016;9(ra448):98. doi: 10.1126/scisignal.aaf5132. [DOI] [PubMed] [Google Scholar]
- 248.Rusin SF, Schlosser KA, Adamo ME, Kettenbach AN. Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells. Sci Signal. 2015;8(398):rs12. doi: 10.1126/scisignal.aab3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332(6035):1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]