Abstract
Psoriasis is an immune-mediated skin disorder. Imbalance of gut microbial populations has been implicated in many diseases. We aimed to investigate whether there were differences in gut microbiota in psoriasis patients vs non-psoriasis controls and between psoriasis severity groups. 55 psoriasis patients and 27 controls were included. V3–V4 regions of the 16S rRNA gene of fecal samples were analyzed using Illumina MiSeq. Bioinformatic analysis was performed. We found changes in gut microbiome composition depending on their psoriasis status as determined by weighted unifrac (p < 0.05), in particular an increase in Firmicutes and depletion of Bacteroidetes in psoriasis patients. Additionally, the Faecalibacterium and Blautia genus were higher in psoriasis patients while Bacteroides and Paraprevotella in non-psoriasis controls (p < 0.05, LDA score > 2). Moderate-to-severe psoriasis patients had lower biodiversity than mild psoriatic patients (p = 0.049). No differences for beta-diversity were found. We developed a Psoriasis-Microbiota Index (PMI), which discriminated among psoriasis patients and controls with sensitivity: 0.78 and specificity: 0.79. Furthermore, we performed a meta-analysis with published data to validate this index. We demonstrated gut dysbiosis in psoriasis patients, suggesting a role in psoriasis pathophysiology. Furthermore, we developed a PMI with the potential to discriminate between psoriasis patients and controls across different populations, which could be used as a biomarker in the clinical practice.
Subject terms: Medical research, Next-generation sequencing
Introduction
Psoriasis is a chronic, immune-mediated inflammatory skin disease. It ranges in severity from a few scattered red, scaly plaques to involvement of almost the entire body surface1. Psoriasis is estimated to affect about 2–4% of the population in western countries, causes considerable psychosocial disability and has a major impact on patients’ quality of life2,3. Skin lesions are characterized by angiogenesis, an inflammatory reaction with recruitment of T cells into the skin, hyperproliferation of keratinocytes and altered epidermal differentiation4. Genetic and environmental factors are implicated in psoriasis, although, the exact etiology of the disease is not fully understood5,6.
The Human Microbiome Project (HMP) was initiated to fill a gap between our current understanding derived from Human Genome Project and actual physiological phenomenon. The HMP created a new view of ourselves as ‘super-organisms’ consisting of a human host and thousands of microbial symbionts7.
Imbalance of gut microbial populations or dysbiosis has important functional consequences and has been implicated in many digestive diseases, diabetes, obesity, metabolic syndrome, psoriatic arthritis, celiac disease, psychiatric disorders and others8–13.
There is a well-known relationship between psoriasis and other inflammatory diseases (obesity, inflammatory bowel disease, psoriatic arthritis, etc.)14. More importantly, bowel mucosa of active psoriasis patients without bowel symptoms show microscopic lesions, even when mucosa appeared macroscopically normal, with immune cellular infiltrates capable of producing pro-inflammatory cytokines15. Bacterial DNA translocation from the intestinal lumen has been described in patients with psoriasis suggesting that the gut microbiome may potentially act in skin diseases16,17,18.
Recent investigations point to the IL-23/Th17 axis as playing a major role in psoriasis pathogenesis19. The adhesion of specific members of gut microbiome to intestinal epithelial cells is found to be essential for the induction of Th17 cells20–22. Mice exposed to antibiotics showed inhibition of psoriasis induction by a dysregulation of gut and skin microbiota23–25.
There have been only limited studies of microbiota in psoriasis patients using molecular methods, which showed contradicting results regarding the most abundant taxa in the disease. These studies involved relatively small numbers of subjects, skin and gut microbiota and unmatched study designs6,17,26–38. Furthermore, none of existing reports evaluated changes in the gut microbiota among disease severity groups.
In the present study, we aimed to investigate whether the microbiota composition of non-treated chronic plaque psoriasis patients, as a group and divided according to disease severity, differs from non-psoriasis controls. We used strict inclusion and exclusion criteria. We included only patients with chronic plaque psoriasis and excluded patients with PsA and IBD (psoriasis comorbidities that are related to changes in the gut microbiota) and those patients under active systemic treatment, since there is evidence that methotrexate and biologic drugs induce compositional changes in the gut microbiota39–41. In addition, controls should not have family history of psoriasis in first degree relatives as genetics could also shape the gut microbiota42. Granted that there is abundant evidence that overweight or obese subjects have changes in their gut microbiota in relation to controls and that obesity and metabolic syndrome are comorbidities of psoriasis, we matched patients by sex, age and BMI43,44. Furthermore, we designed a Psoriasis-Microbiota Index (PMI) to discriminate patients against controls and performed a meta-analysis with previously published data to validate this index.
Results
Background of study cohort
This study included 55 untreated chronic plaque psoriasis patients and 27 unrelated non-psoriasis controls. The background of patients and controls are shown in Table 1. The patient group included 28 with mild disease and 27 with moderate-to-severe psoriasis. Table 2 represents the demographic data between mild and moderate-to-severe psoriasis groups, in which patients were comparable except for disease duration (longer in moderate-to-severe patients) and time since last relapse (longer for mild psoriasis).
Table 1.
Characteristics of the sample.
| Psoriasis patients | Non-psoriasis controls | p | |
|---|---|---|---|
| n: 55 | n: 27 | ||
| Age (years), mean ± SD | 44.8 (16.9) | 48.7 (18.8) | NS |
| Female (%) | 49.1 | 57.7 | NS |
| Male (%) | 50.9 | 42.3 | NS |
| Age of Psoriasis symptom onset (years), mean ± SD | 30.5 (17.5) | NA | |
| Type 1 Psoriasis (%) | 69.1 | NA | |
| Last outbreak of Psoriasis symptoms (months), mean ± SD | 4.2 (2.0) | NA | |
| Duration of Psoriasis (years), mean ± SD | 14.3 (12.0) | NA | |
| Moderate-to-severe Psoriasis (%) | 49.1 | NA | |
| Hypertension (%) | 29.1 | NA | |
| Diabetes (%) | 16.4 | NA | |
| Weight, mean ± SD | 81.8 (19.9) | 75 (15.1) | NS |
| Heigh, mean ± SD | 1.66 (0.1) | 1.63 (0.1) | NS |
| BMI, mean ± SD | 29.6 (5.5) | 28.1 (5.2) | NS |
| Metabolic syndrome (%) | 21.8 | NA | |
| Overweight (%) | 29.1 | 42.3 | NS |
| Obesity (%) | 45.5 | 30.7 | NS |
| PASI, mean ± SD | 9.9 (7.2) | NA | |
| BSA, mean ± SD | 14.5 (18.5) | NA |
Table 2.
Demographic data in mild and moderate-to-severe psoriasis patients.
| Mild psoriasis patients | Moderate-to-severe psoriasis patients | p | |
|---|---|---|---|
| n: 28 | n: 27 | ||
| Age, mean ± SD | 41 ± 14.2 | 48.6 ± 18.9 | NS |
| Female n: 27 (%) | 42.9 | 55.6 | NS |
| Male n: 28 (%) | 57.1 | 44.4 | NS |
| Age of Psoriasis symptom onset (years), mean ± SD | 31 ± 15 | 29.9 ± 19.8 | NS |
| Type 1 Psoriasis n: 38 (%) | 47.4 | 52.6 | NS |
| Last outbreak of Psoriasis symptoms (months), mean ± SD | 4.7 ± 2.1 | 3.6 ± 1.7 | 0.04 |
| Years with Psoriasis, mean ± SD | 9.9 ± 8.7 | 18.6 ± 13.4 | 0.008 |
| Hypertension n: 16 (%) | 28.6 | 29.6 | NS |
| Diabetes n: 9 (%) | 17.9 | 14.9 | NS |
| Weight, mean ± SD | 85 ± 19.5 | 79.1 ± 20.2 | NS |
| Heigh, mean ± SD | 1.68 ± 0.1 | 1.64 ± 0.1 | NS |
| BMI, mean ± SD | 29.9 ± 5.8 | 29.5 ± 5.3 | NS |
| Metabolic syndrome n: 12 (%) | 21.4 | 22.2 | NS |
| Overweight n: 16 (%) | 28.6 | 29.6 | NS |
| Obesity n: 25 (%) | 46.4 | 44.4 | NS |
| PASI, mean ± SD | 3.7 ± 1.1 | 16.3 ± 4.8 | 0.000001 |
| BSA, mean ± SD | 2 ± 1.2 | 27.5 ± 19.2 | 0.000001 |
Psoriasis vs non-psoriasis controls
Sequence analysis and comparison of microbial communities
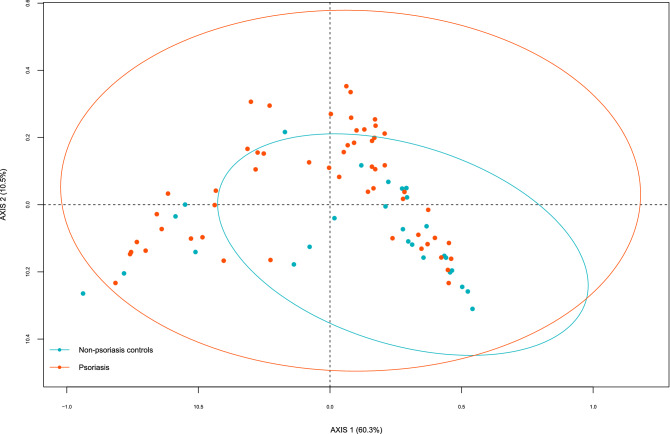
The hypervariable region V3-V4 of bacterial 16S gene was sequenced using MiSeq-Illumina system, obtaining 152,939.46 ± 18,320.34 sequences per sample. Rarefaction plots reached an asymptotic state, indicating that the sequence depth was sufficient to represent the bacterial community richness and diversity (data not shown). Therefore, when we compared species richness (Chao1 index), there were no significant differences between psoriasis patients and controls. For beta-diversity as determined by Unifrac, we found significant differences between both groups, p = 0.034 for weighted UniFrac (Fig. 1) but not for unweighted UniFrac p = 0.255 (ADONIS).
Figure 1.
PCoA of beta-diversity values (Weighted Unifrac distances). Comparison of the gut microbiota from psoriasis patients and non-psoriasis controls. Ellipses show 95% confidence intervals.
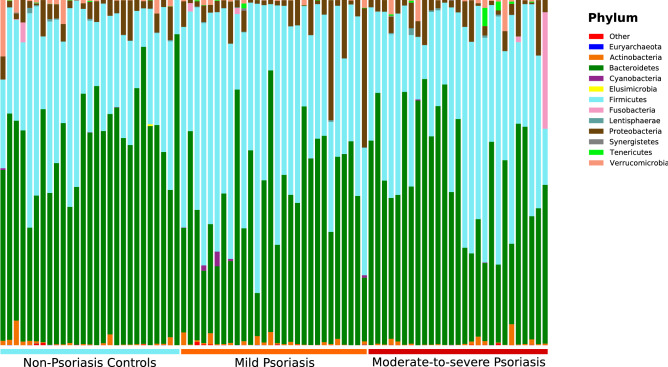
Psoriasis patients differ from controls in the observed community structure. The dominant phyla in psoriasis patients were Bacteroidetes 47.1%, Firmicutes 44.6%, Proteobacteria 5.4%, Actinobacteria 0.8% and Fusobacteria 0.7%, while the principal phyla found in controls were Bacteroidetes 59.9%, Firmicutes 33.0%, Proteobacteria 4.2%, Verrucomicrobia 1.4% and Actinobacteria 0.8% (Fig. 2).
Figure 2.
Bar plot showing the relative abundance of phyla distribution of each operational taxonomic unit (OTU) within samples.
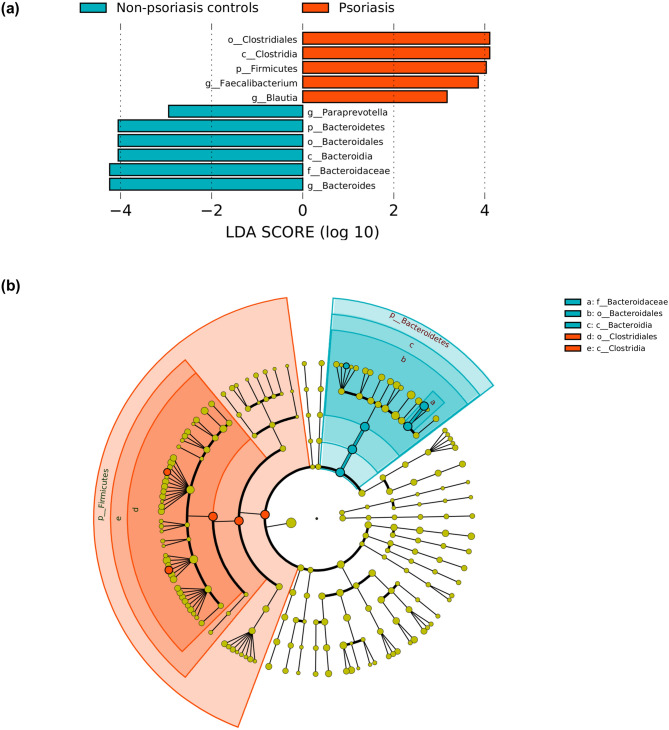
Phyla-level differences were detected between the two groups (control vs Psoriasis patients) including differences in Bacteroidetes and Firmicutes, with a Firmicutes to Bacteroidetes ratio of 0.63 ± 0.32 in non-psoriasis controls and 1.29 ± 0.81 in psoriasis patients (p = 0.0002). LefSe analysis revealed that these differences were mainly driven by changes in the Bacteroides and Paraprevotella genus which were more abundant in non-psoriasis controls while Faecalibacterium and Blautia in psoriasis patients (logarithmic LDA scores threshold was 2.0) (Fig. 3). We did not observe significant differences in gut microbiota associated with changes in age, weight and BMI.
Figure 3.
Plot from LEfSe analysis indicating enriched bacterial genus associated either with psoriasis patients (red) or non-psoriasis controls (blue). The length of the bar column represents the LDA score (a). Cladogram plotted from LEfSe analysis showing the differences in relative abundance of taxa at five levels between psoriasis patients vs non-psoriasis controls. (P < .05; LDA score 2) (b).
Mild vs moderate-to-severe psoriasis
Species richness in moderate-to-severe psoriasis patients was lower comparing with mild psoriasis patients (p = 0.049). Comparing the principal phyla detected, we did not find differences between both psoriasis groups (Supplementary Fig. S1).
We did not find differences in beta-diversity in mild vs moderate-to-severe psoriasis patients Supplementary Fig. S2. We did not observe significant differences for age, gender, age at psoriasis onset, years with psoriasis, hypertension, diabetes, weight, BMI, PASI and BSA. Only significant differences were found for metabolic syndrome (p = 0.002) in unweighted analysis.
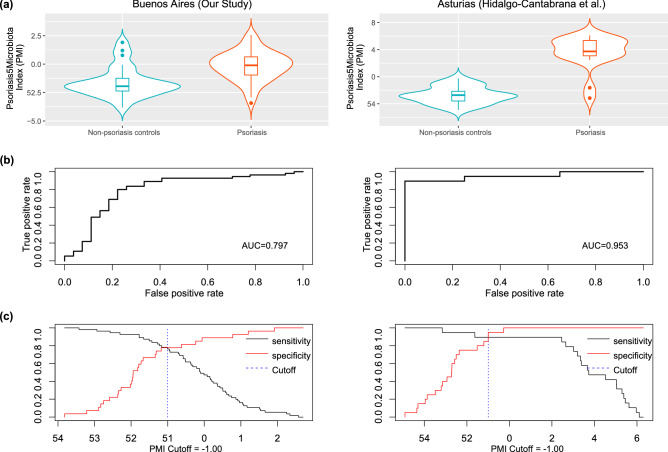
Psoriasis-Microbial index
Considering the results of relative abundance of different taxa, we generated the PMI to discriminate between psoriasis and non-psoriasis controls (Fig. 4a). We evaluated its applicability by ROC analysis. The Area Under the Curve (AUC) for the classification of Psoriasis (training dataset) was 0.797 (Fig. 4b), determining an optimal cut-off value of PMI = − 1.00 (sensitivity = 0.78 and specificity = 0.79) (Fig. 4c). When we applied the PMI according to psoriasis severity, the AUC was 0.849 and 0.743 for mild and moderate-to-severe psoriasis respectively.
Figure 4.
PMI distinguishes non psoriasis controls from psoriasis patients. Violin plot showing PMI in control and psoriasis fecal samples (a). Performance of cross-city prediction using each city-specific AD diagnosis model, as assessed via the area under the ROC curve (AUROC). The ROC curve of tenfold cross-validation was marked as blue lines and the ROC curve of the prediction as red lines. Performance of PMI, assessed via the area under the ROC curve (AUROC) (b). Sensitivity and specificity vs. PMI (Cutoff) plot in both populations (c).
Meta-analysis
We validated this PMI using datasets from previously reported literature on PubMed. We identified 7 related 16S datasets6,17,26,27,29,34,35. Only the study of Hidalgo-Cantabrana et al.34 fulfilled the inclusion criteria.
When we applied the PMI to the downloaded sequence data from Hidalgo-Cantabrana et al.(test dataset)34 (Fig. 4a), the AUC was 0.953, Sensitivity = 0.89 and Specificity = 0.90, using cut-off value obtained with our dataset (PMI = − 1.00);(Fig. 4b). Sensitivity vs Specificity curves of both datasets were plotted in Fig. 4c showing concordant results, indicating that PMI would be a powerful tool capable of discriminating between patients with psoriasis and controls from different populations.
Discussion
Intestinal dysbiosis is a possible actor in chronic inflammation, even in distant tissue sites, such as the skin. Imbalance in gut microbiota induces epithelial changes resulting in increased intestinal inflammation and altered gut permeability, which in susceptible individuals may trigger the development of different chronic disease states such as IBD, obesity, diabetes, multiple sclerosis, atopic dermatitis and cancer, among others45–50. However, until now, only a few studies have addressed this question in psoriasis6,17,26,27,29,34–36,33.
Our work demonstrates that there are differences in gut microbiota between psoriasis patients and non-psoriasis controls. We evaluated 55 untreated chronic plaque psoriasis patients (27 with moderate-to-severe psoriasis and 28 with mild disease), being according to our knowledge the study with the highest number of psoriasis patients and the first which evaluates changes in gut microbiota according to psoriasis severity based on well-defined strict criteria. We made a comparison of our study design with all the available publications on gut microbiota and psoriasis up to March 31, 2020 (Table 3).
Table 3.
Study design of the available publications included in the meta-analysis (up to March 31, 2020).
| Our study | Codoñer et al.20 | Tan et al.29 | Hidalgo-Cantabrana et al.37 | Chen et al.30 | Huang et al.6 | Scher et al.32 | Shapiro et al.38 | |
|---|---|---|---|---|---|---|---|---|
| Publication year | 2018 | 2018 | 2019 | 2018 | 2018 | 2015 | 2019 | |
| Population | Caucasian/Argentine | Caucasian/Spain | Asian/China | Caucasian/Spain | Asian/China | Asian/China | Caucasian/US | Caucasian/Israel |
| Psoriasis patients (n) | 55 | 52 | 14 | 19 | 35 | 32 | 15 Ps / 16 PsA | 24 |
| Non-Psoriasis controls (n) | 27 | 300 (from HMP) | 14 | 20 | 27 | 64 | 17 | 22 |
| Plaque psoriasis exclusive | yes | yes | yes | yes | yes | no | NA | NA |
| Matchead by | Age, sex & BMI | No | No | Age | Age, sex & BMI | No | Age & sex | Age, sex & comorbidities |
| Active systemic treatment | No | No | NA | No | Yes | Yes | No | Yes |
| Stratified by severity | Yes | No | No | No | NA | Yes | No | No |
| Concomitant PsA | no | NA | NA | NA | yes | no | yes | NA |
| 16S region analyzed | V3–V4 | V3–V4 | V4 | V2–V3 | V3–V4 | V4–V5 | V1–V2 | V4 |
| Platform | Illumina | Illumina | Illumina | Ion Chef | Illumina | Illumina | Illumina | Illumina |
| Average reads | 152,939 | ~ 85,000 | ~ 30,000 | 233,113 | ~ 85,000 | NA | NA | ~ 50,000 |
| Raw data available at | PRJNA574485 | Avaiable upon request to the corresponding author | NA | PRJNA517056 | PRJNA379878 | NA | NA | NA |
Alpha-diversity has been observed to be decreased in a dysbiotic gut51. A lower microbial diversity has been found in some psoriasis studies6,29,34 but not by other investigators17,26,27,35. Our study does not show a lower alpha-diversity.
We found that Bacteroidetes and Firmicutes were the most prevalent phyla in patients and controls. However, there were significant differences between both phyla in psoriasis patients. The Firmicutes to Bacteroidetes ratio was 1.29 ± 0.81 in psoriasis patients and 0.63 ± 0.32 in non-psoriasis controls. In line with our results, other investigations showed a high Firmicutes:Bacteroidetes ratio6,27,29,34,35.
Short Chain Fatty Acid (SCFA) like acetate, propionate and butyrate, are known to regulate not only gut specific but also distant inflammatory responses through the induction of immune cells52. An increase in Firmicutes:Bacteroidetes ratio has been implicated in a higher acetate and lower butyrate production. Butyrate is the preferred fuel for the colonic epithelial cells and the major regulator of cell proliferation and differentiation, and has important anti-inflammatory, antioxidant and anti-carcinogenic functions53. Low levels of butyrate may affect the integrity of the mucous layer compromising the gut epithelial barrier and enhance chronic colonic and systemic inflammation54.
Beta-diversity showed that genus Faecalibacterium and Blautia (both belong to the phylum Firmicutes, class Clostridia and order Clostridiales) were the most relevant genus in psoriasis patients that discriminated against non-psoriasis controls. Faecalibacterium prausnitzii (F. prausnitzii) can regulate T helper 17 cell (Th17)/regulatory T cell (Treg) differentiation and has been consistently reported as one of the main butyrate producers found in the intestine53,55. The role of F. prausnitzii in maintaining immune and physiological functions promoted this bacterium as a next generation probiotic56.
In psoriasis, a decrease in relative abundance of F. Prausnitzii has been reported in some studies30,34, but not by other investigators17,27,35. In our study the genus Faecalibacterium showed higher values in psoriasis patients. Lopez-Siles et al. determined that F. prausnitzii includes two phylogroups and recent studies suggest that other Faecalibacterium genus and species could not be ruled out53,57,58. The relative abundance, as well as which phylum and species of Faecalibacterium population are disbalanced in different diseases, makes it difficult to establish the use of a single bacteria as a general biomarker for all diseases. The use of F. prausnitzii as a gold standard of a healthy gut microbiota is limited53.
The genus Blautia includes obligate anaerobic intestinal commensal bacteria that belong to the family Lachnospiraceae and includes more than 100 different species59,60. Blautia are important members of the healthy human gut microbiota61. Jenq et al. found a lower mortality due to a graft versus-host disease after allogeneic blood/marrow transplantation among patients with high abundance of Blautia and Bajaj et al.found that Blautia was one of the bacteria associated with improved outcomes in patients with liver cirrhosis62,63. Genus Blautia has been also related to cancer. Chen et al.reported a detrimental association between lower concentrations of Blautia in the gut and colorectal cancer64. On the contrary, Luu et al. found that higher levels of Blautia were associated with poor prognosis in patients with early-stage breast cancer65. Considering the limited data available on Blautia and the huge number of species reported, we can not explain the reasons why genus Blautia was increased in our work. Additional data are required to determine their true role in human diseases.
The fact that most relevant genus in psoriasis patients that discriminated against non-psoriasis controls were Faecalibacterium and Blautia, taxa producing high levels of butyrate, contradicts the traditional association of butyrate producers observed in diseases such as IBD66,67. Therefore, results highlight the need for additional research given the observational nature and limits of 16S used in this study.
In our control group, the predominant genus were Bacteroides and Paraprevotella. These bacteria differ only in family (Bacteroideaceae for Bacteroides and Prevotellaceae for Paraprevotella)68,69. Increasing evidence proposes that Bacteroides harness complex recalcitrant glycans70. SCFAs are the major metabolic products of anaerobic fermentation of glycans by gut bacteria and have been shown to impact on the host physiology71. The beneficial effect of Bacteroides is consistent with our findings, where this genus was increased in controls and depleted in psoriasis patients.
There is evidence that age, diet, geographical location, genetics and antibiotics, among other factors, influence gut microbiota72,73. We selected unrelated controls matched by sex, age and BMI to moderate-to-severe psoriasis patients, living in the same area and with a similar diet in order to reduce those confounding factors. We did not find differences in beta-diversity according to personal features, so we postulate that changes in gut microbiota would then be dependent on psoriasis and not on other covariates.
We found that patients with moderate-to-severe psoriasis had a lower diversity (species richness) than patients with mild disease, although this difference was subtle. Only Huang et al.also studied whether the composition of the intestinal microbiota differed depending on the severity of the disease and they found that the genus Bacteroides was increased in patients with psoriasis and that it was characteristic of the subgroup with severe disease6. In our study, the genus Bacteroides was found to be diminished in patients with psoriasis but no differences were found between mild and moderate-to-severe psoriasis patients. For example, these distinctions could be due to different inclusion criteria.
When we compared whether the microbiota of patients with mild psoriasis vs patients with moderate-to-severe psoriasis was affected by age, sex, age at onset of the disease, years of illness and comorbidities such as hypertension or diabetes, we could not establish differences between both severity groups. These results could also explain that changes of the gut microbiota in psoriasis would be dependent on the presence of the disease and would not be affected by its severity.
Codoñer et al., Shapiro et al.and Hidalgo-Cantabrana et al.reported similar results to our study regarding the bacteria genus increased in psoriasis and controls17,34,35. This concordance suggests that there is probably a core gut microbiota in psoriasis patients. Unfortunately, not all the studies met the inclusion criteria for the meta-analysis. Codoñer et al. did not use a control group from the same geographic location as they used publicly available data from The Human Microbiome Project and the raw data from Shapiro et al.were not available. This serves as another example of the importance of unrestricted access to raw sequencing data, which has been already recognized by the scientific community74. Despite variations among Hidalgo-Cantabrana et al.and our study, a psoriasis model can be applied across populations from different geographical locations. The proposed PMI proved to be able to discriminate between psoriasis and controls across cities and continents with an optimal cut-off value of PMI = − 1.00.
Given that general dermatologists are able to make a diagnosis of psoriasis with a simple physical exam, the diagnostic applicability of the test will have to await further clinical experience. The PMI represents, a step forward as a combined practical, ready to use, clinical and research tool. The index will allow us to gain more knowledge on the microbial component of psoriasis and provide the possibility of increasing our understanding of the role played by the microbiome in the disease process. Moreover, as PMI was only tested in 2 cohorts of non-treated patients, we cannot exclude its role as a biomarker for evaluating treatment response. Further studies of metagenome shotgun sequencing at the species/strain levels might be useful for the update and improvement of the developed PMI.
In summary, our findings demonstrate variations in gut microbiota profiles between non-treated plaque psoriasis patients and non-psoriasis controls. This results suggest that it is likely that altered gut microbiota plays a pathophysiological role in psoriasis. However, whether modulation of gut microbiota could modify the course of the disease remains to be explored. This study is unique in being the first to propose a PMI with the ability to discriminate between psoriasis patients and age-sex-and BMI matched controls and between samples from communities of different continents. Further studies are needed to better interpret the role of the PMI as a potential biomarker test in psoriasis, and to test this index in larger and diverse populations to confirm its validity.
Methods
Study participants
This cross-sectional study recruited unrelated individuals, including consecutive chronic plaque psoriasis patients and non-psoriasis controls. Controls were matched to moderate-to-severe psoriasis patients according to sex, age (± 2 years) and Body Mass Index (BMI; ± 1). Participants were caucasian, above 18 years old and from the same geographical location. Samples were collected between October 2017 and April 2018.
Psoriasis patients were subdivided based on their severity in mild and moderate-to-severe psoriasis. Mild psoriasis was defined as actual Body Surface Area covered by psoriasis (BSA) < 10%, Psoriasis Area and Severity Index (PASI) < 10, Investigator Global Assessment (IGA) < 3 and absence of episodes of moderate-to-severe psoriasis in the past. Moderate-to-severe psoriasis was defined as BSA ≥ 10%, PASI ≥ 10 and IGA ≥ 3.
Two visits were conducted over a period of 4 weeks to take a detailed assessment of psoriasis, medical history, and a complete physical exam, including PASI, IGA and BSA involvement. Type 1 psoriasis was defined if the symptoms began on or before age 40 years; a BMI ≥ 25 was considered as excessive weight and BMI ≥ 30 as obesity.
Key exclusion criteria for psoriasis patients included concomitant diagnosis of psoriatic arthritis according to CASPAR criteria, inflammatory bowel disease (IBD), current topical treatment, systemic treatment for psoriasis (including phototherapy) 3 months previous to sample collection, assuming that immunosuppression could modify gut microbiota.
The exclusion criteria for controls were the presence of other dermatosis, family history of psoriasis in first degree relatives, immunological disorders, hypertension, fatty liver disease, diabetes mellitus, malignancy, any other serious internal disease, smoking and alcohol abuse.
Exclusion criteria applied to all groups were: antibiotic therapy 3 months previous to sample collection, extreme diet, consumption of probiotics, positive HIV test or any gastrointestinal tract surgery leaving permanent residua.
Sample collection and DNA extraction
All participants were apprised for the stool sampling collection method by receiving a standardized protocol for the collection of approximately 5 g of stool in a sterile bacteriostatic buffer tube75. Participants were asked to collect samples 24 h before the second visit. DNA extraction was performed from 200 mg of feces using QIAamp-PowerFecal DNA-Kit.
Comparison of microbial communities and sequence analysis
Hypervariable regions V3–V4 of the 16S rRNA gene were amplified with primers 337F/805R and sequenced in paired-end mode using a MiSeq sequencer (IlluminaⓇ), warranting an average of 152,939 sequences per sample.
De-multiplexed reads were quality trimmed using Trimmomatic(V0.36)76. Sequences generated were analyzed using Quantitative Insights Into Microbial Ecology (QIIME) version 1.9.1 software package77. For this purpose, the sequences obtained were compared with those from Greengenes 13_8 database78. Chimeric sequences were filtered using VSEARCH79. Operative Taxonomic Units (OTUs) were assigned to each read with an open_reference OTU picking process. SortMeRNA (v2.1)80 was used for the reference OTU picking steps (with sortmerna_coverage = 0.8) and sumaclust (v1.0.20)81 for the de novo OTU picking steps (with 10% of the failures subsampled). Low-confidence OTUs called by < 0.1% of the reads were removed using the script remove_low_confidence_otus.py82. An average of 29,872.93 ± 6,452.75 mapping high-quality sequences were obtained, leading to 455.91 ± 126.99 unique OTUs per sample. For multiple comparisons, p-values were adjusted by Bonferroni correction83. To compare microbial communities in different sample groups, we used Unifrac algorithm84. Differences on beta-diversity were assessed using ADONIS. In order to compare the relative abundance of the different taxa between groups, we performed Linear Discriminant Analysis (LDA) effect implemented in LEfSe85 .
Psoriasis-Microbiome Index development
PMI was defined as the logarithm of total abundance of organisms increased in psoriasis over total abundance of organisms decreased in psoriasis for all samples (at genus level) using the compute_taxonomy_ratio.py script86. Then, we evaluated how these PMI performed for classification subjects by psoriasis status through Receiver Operating Characteristic (ROC) analysis (training dataset). ROC analysis was performed using ROCR package (RStudio version 1.1.453)87. Cut-off value was selected as the point where the sensitivity and specificity functions intersect each other, i.e. jointly maximizing the sensitivity and specificity of PMI.
Meta-analysis
We performed a systematic literature search of PubMed databases up to March 31, 2020 using the following terms: “Psoriasis” and “gut microbiota” or “gut microbiome”. The study inclusion criteria were: Case–control studies with publicly available raw 16S data and metadata, indicating case/control status for each sample. Studies including patients with other clinical forms different from plaque psoriasis and patients under systemic treatment (DMARDS and biologics) were excluded.
Data accession
Raw sequences of 16S rRNA gene reported in this article have been deposited in NCBI Short Read Archive (SRA) and are accessible under the accession number PRJNA574485.
Ethical statement
This study received approval by the Ethics Committee of Hospital Español, Buenos Aires Argentina according to local regulations and Helsinki declaration. Written informed consent was obtained from all study participants.
Supplementary information
Acknowledgements
This research was supported by Novartis Pharma, CAIN457AAR02T. The authors thank Dr. Federico Rey for his constructive comments and suggestions and the “Centro de Investigación, Docencia y Extensión en Tecnologías de la Información y las Comunicaciones” (CIDETIC, http:/cidetic.unlu.edu.ar/), Universidad Nacional de Luján, Luján, Argentina for human and computational resources. We are grateful to Fundación H.A. Barceló, Instituto Universitario de Ciencias de la Salud, CABA, Buenos Aires, for excellent technical support.
Author contributions
I.D., F.G., H.D. and A.P.S. designed the study. I.D. performed the recruitment of the volunteers. I.D., F.G. and L.L. collected the stool samples and the extraction of fecal bacterial DNA. A.P.S. processed the raw sequences and performed the bioinformatic and statistical analysis. I.D., F.G. and A.P.S. analyzed the results. All authors wrote and reviewed the manuscript.
Competing interests
Dr Dei-Cas has received compensation as a speaker, consultant, and investigator for Novartis, Eli Lilly and Janssen. Dr Penas-Steinhardt has received compensation as a speaker for Novartis. Dr Florecina Giliberto, Dr. Leonela Luce and Dr. Hernán Dopazo declare no competing interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69537-3.
References
- 1.Griffiths CEM, Barker JN. Pathogenesis and clinical features of psoriasis. The Lancet. 2007;370:263–271. doi: 10.1016/S0140-6736(07)61128-3. [DOI] [PubMed] [Google Scholar]
- 2.Kurd SK, Gelfand JM. The prevalence of previously diagnosed and undiagnosed psoriasis in US adults: results from NHANES 2003–2004. J. Am. Acad. Dermatol. 2009;60:218–224. doi: 10.1016/j.jaad.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rapp SR, Feldman SR, Exum ML, Fleischer AB, Jr, Reboussin DM. Psoriasis causes as much disability as other major medical diseases. J. Am. Acad. Dermatol. 1999;41:401–407. doi: 10.1016/s0190-9622(99)70112-x. [DOI] [PubMed] [Google Scholar]
- 4.Baliwag J, Barnes DH, Johnston A. Cytokines in psoriasis. Cytokine. 2015;73:342–350. doi: 10.1016/j.cyto.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh S, et al. Genomic alterations driving psoriasis pathogenesis. Gene. 2019;683:61–71. doi: 10.1016/j.gene.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 6.Huang L, et al. Dysbiosis of gut microbiota was closely associated with psoriasis. Sci. China Life Sci. 2018 doi: 10.1007/s11427-018-9376-6. [DOI] [PubMed] [Google Scholar]
- 7.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landman C, Quévrain E. Gut microbiota: description, role and pathophysiologic implications. Rev. Med. Int. 2016;37:418–423. doi: 10.1016/j.revmed.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Carrera-Quintanar L, et al. The human microbiota and obesity: a literature systematic review of in vivo models and technical approaches. Int. J. Mol. Sci. 2018;19:3827. doi: 10.3390/ijms19123827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X, Devaraj S. Gut Microbiome in Obesity, Metabolic Syndrome, and Diabetes. Curr. Diab. Rep. 2018;18:129. doi: 10.1007/s11892-018-1104-3. [DOI] [PubMed] [Google Scholar]
- 11.Gulas E, Wysiadecki G, Strzelecki D, Gawlik-Kotelnicka O, Polguj M. Can microbiology affect psychiatry? A link between gut microbiota and psychiatric disorders. Psychiatr. Pol. 2018;52:1023–1039. doi: 10.12740/PP/OnlineFirst/81103. [DOI] [PubMed] [Google Scholar]
- 12.Gilis E, et al. The role of the microbiome in gut and joint inflammation in psoriatic arthritis and spondyloarthritis. J. Rheumatol. Suppl. 2018;94:36–39. doi: 10.3899/jrheum.180135. [DOI] [PubMed] [Google Scholar]
- 13.Cenit MC, Codoñer-Franch P, Sanz Y. Gut microbiota and risk of developing celiac disease. J. Clin. Gastroenterol. 2016;50(Suppl 2):S148–S152. doi: 10.1097/MCG.0000000000000688. [DOI] [PubMed] [Google Scholar]
- 14.Takeshita J, et al. Psoriasis and comorbid diseases: Epidemiology. J. Am. Acad. Dermatol. 2017;76:377–390. doi: 10.1016/j.jaad.2016.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scarpa R, et al. Microscopic inflammatory changes in colon of patients with both active psoriasis and psoriatic arthritis without bowel symptoms. J. Rheumatol. 2000;27:1241–1246. [PubMed] [Google Scholar]
- 16.Ramírez-Boscá A, et al. Identification of bacterial DNA in the peripheral blood of patients with active psoriasis. JAMA Dermatol. 2015;151:670–671. doi: 10.1001/jamadermatol.2014.5585. [DOI] [PubMed] [Google Scholar]
- 17.Codoñer FM, et al. Gut microbial composition in patients with psoriasis. Sci. Rep. 2018;8:3812. doi: 10.1038/s41598-018-22125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visser MJE, Kell DB, Pretorius E. Bacterial dysbiosis and translocation in psoriasis vulgaris. Front. Cell. Infect. Microbiol. 2019;9:7. doi: 10.3389/fcimb.2019.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blauvelt A, Chiricozzi A. The immunologic role of IL-17 in psoriasis and psoriatic arthritis pathogenesis. Clin. Rev. Allergy Immunol. 2018;55:379–390. doi: 10.1007/s12016-018-8702-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atarashi K, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163:367–380. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilck N, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017 doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flannigan KL, Denning TL. Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology. 2018 doi: 10.1111/imm.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zákostelská Z, et al. Intestinal microbiota promotes psoriasis-like skin inflammation by enhancing Th17 response. PLoS ONE. 2016;11:e0159539. doi: 10.1371/journal.pone.0159539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanvit P, et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat. Commun. 2015;6:8424. doi: 10.1038/ncomms9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stehlikova Z, et al. Crucial role of microbiota in experimental psoriasis revealed by a gnotobiotic mouse model. Front. Microbiol. 2019;10:236. doi: 10.3389/fmicb.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan L, et al. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp. Dermatol. 2018;27:144–149. doi: 10.1111/exd.13463. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y-J, et al. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp. Dermatol. 2018;27:1336–1343. doi: 10.1111/exd.13786. [DOI] [PubMed] [Google Scholar]
- 28.Chang H-W, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. 2018;6:154. doi: 10.1186/s40168-018-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scher JU, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–139. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eppinga H, et al. Similar depletion of protective Faecalibacterium prausnitziiin psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J. Crohn’s Colitis. 2016;10:1067–1075. doi: 10.1093/ecco-jcc/jjw070. [DOI] [PubMed] [Google Scholar]
- 31.Langan EA, et al. The role of the microbiome in psoriasis: moving from disease description to treatment prediction? Br. J. Dermatol. 2018;178:e360–e360. doi: 10.1111/bjd.16081. [DOI] [PubMed] [Google Scholar]
- 32.Tett A, et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes. 2017;3:14. doi: 10.1038/s41522-017-0022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakajima S, Harrison O, Merrill E, Linehan J, Belkaid Y. 648 Candida albicans colonization exacerbates skin inflammation in a murine model of psoriasis. J. Invest. Dermatol. 2017;137:S112. [Google Scholar]
- 34.Hidalgo-Cantabrana C, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br. J. Dermatol. 2019 doi: 10.1111/bjd.17931. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro J, et al. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J. Dermatol. 2019;46:595–603. doi: 10.1111/1346-8138.14933. [DOI] [PubMed] [Google Scholar]
- 36.Quan C, et al. Psoriatic lesions are characterized by higher bacterial load and imbalance between Cutibacterium and Corynebacterium. J. Am. Acad. Dermatol. 2019 doi: 10.1016/j.jaad.2019.06.024. [DOI] [PubMed] [Google Scholar]
- 37.Langan EA, et al. Combined culture and metagenomic analyses reveal significant shifts in the composition of the cutaneous microbiome in psoriasis. Br. J. Dermatol. 2019 doi: 10.1111/bjd.17989. [DOI] [PubMed] [Google Scholar]
- 38.Stehlikova Z, et al. Dysbiosis of skin microbiota in psoriatic patients: co-occurrence of fungal and bacterial communities. Front. Microbiol. 2019;10:438. doi: 10.3389/fmicb.2019.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayers E, MacGregor A, Carding SR. Drug-microbiota interactions and treatment response: relevance to rheumatoid arthritis. AIMS Microbiol. 2018;4:642–654. doi: 10.3934/microbiol.2018.4.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aden K, et al. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory bowel diseases. Gastroenterology. 2019 doi: 10.1053/j.gastro.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 41.Doherty MK, et al. Fecal microbiota signatures are associated with response to Ustekinumab therapy among Crohn’s disease patients. mBio. 2018 doi: 10.1128/mBio.02120-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeshita J, et al. Psoriasis and comorbid diseases: implications for management. J. Am. Acad. Dermatol. 2017;76:393–403. doi: 10.1016/j.jaad.2016.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nature Reviews Gastroenterology & Hepatology. 2017;14:573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maruvada P, Leone V, Kaplan LM, Chang EB. The human microbiome and obesity: moving beyond associations. Cell Host Microbe. 2017;22:589–599. doi: 10.1016/j.chom.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Patterson E, et al. Gut microbiota, obesity and diabetes. Postgrad. Med. J. 2016;92:286–300. doi: 10.1136/postgradmedj-2015-133285. [DOI] [PubMed] [Google Scholar]
- 48.Tremlett H, Bauer KC, Appel-Cresswell S, Finlay BB, Waubant E. The gut microbiome in human neurological disease: a review. Ann. Neurol. 2017;81:369–382. doi: 10.1002/ana.24901. [DOI] [PubMed] [Google Scholar]
- 49.Song H, Yoo Y, Hwang J, Na Y-C, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J. Allergy Clin. Immunol. 2016;137:852–860. doi: 10.1016/j.jaci.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 50.Saha A, Robertson ES. Microbiome and human malignancies. Microbiome Cancer. 2019 doi: 10.1007/978-3-030-4155-7_1. [DOI] [Google Scholar]
- 51.Chatelier EL, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 52.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat. Immunol. 2011;12:5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 53.Lopez-Siles M, Duncan SH, Garcia-Gil LJ, Martinez-Medina M. Faecalibacterium prausnitzii: from microbiology to diagnostics and prognostics. ISME J. 2017;11:841–852. doi: 10.1038/ismej.2016.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bach Knudsen KE, et al. Impact of diet-modulated butyrate production on intestinal barrier function and inflammation. Nutrients. 2018;10:1499. doi: 10.3390/nu10101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou L, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm. Bowel Dis. 2018 doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 56.Martín R, Bermúdez-Humarán LG, Langella P. Searching for the bacterial effector: the example of the multi-skilled commensal bacterium. Front. Microbiol. 2018;9:346. doi: 10.3389/fmicb.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fitzgerald CB, et al. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genom. 2018;19:931. doi: 10.1186/s12864-018-5313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lopez-Siles M, et al. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 2012;78:420–428. doi: 10.1128/AEM.06858-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Durand GA, et al. Blautia massiliensis sp. nov., isolated from a fresh human fecal sample and emended description of the genus Blautia. Anaerobe. 2017;43:47–55. doi: 10.1016/j.anaerobe.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 60.Website. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=572511&lvl=3&keep=1&srchmode=1&unlock [accessed on 24 August 2019].
- 61.Touyama M, Jin JS, Kibe R, Hayashi H, Benno Y. Quantification of Blautia wexlerae and Blautia luti in human faeces by real-time PCR using specific primers. Benef. Microbes. 2015;6:583–590. doi: 10.3920/BM2014.0133. [DOI] [PubMed] [Google Scholar]
- 62.Jenq RR, et al. Intestinal blautia is associated with reduced death from graft-versus-host disease. Biol. Blood Marrow Transplant. 2015;21:1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bajaj JS, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luu TH, et al. Intestinal proportion of Blautia sp. is associated with clinical stage and histoprognostic grade in patients with early-stage breast cancer. Nutr. Cancer. 2017;69:267–275. doi: 10.1080/01635581.2017.1263750. [DOI] [PubMed] [Google Scholar]
- 66.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 67.Lloyd-Price J, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taxonomy. Taxonomy browser (Bacteroides). https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=816.
- 69.Website. Taxonomy browser (Paraprevotella) [WWW Document]. URL https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=577309&lvl=3&lin=f&keep=1&srchmode=1&unlock [accessed on 24 August 2019].
- 70.Luis AS, et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018;3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh RP. Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health. Appl. Microbiol. Biotechnol. 2019 doi: 10.1007/s00253-019-10012-z. [DOI] [PubMed] [Google Scholar]
- 72.Lagier J-C, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front. Cell. Infect. Microbiol. 2012;2:136. doi: 10.3389/fcimb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Belforte FS, et al. Getting to know the gut microbial diversity of metropolitan Buenos Aires inhabitants. Front. Microbiol. 2019;10:965. doi: 10.3389/fmicb.2019.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langille MGI, Ravel J, Florian Fricke W. ‘Available upon request’: not good enough for microbiome data! Microbiome. 2018 doi: 10.1186/s40168-017-0394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gray MA, Pratte ZA, Kellogg CA. Comparison of DNA preservation methods for environmental bacterial community samples. FEMS Microbiol. Ecol. 2013;83:468–477. doi: 10.1111/1574-6941.12008. [DOI] [PubMed] [Google Scholar]
- 76.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 81.Mercier, C., Boyer, F., Bonin, A. & Coissac, E. SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. GitLabhttps://git.metabarcoding.org/obitools/sumaclust/wikis/home (2013).
- 82.Comeau AM, Douglas GM, Langille MGI. Microbiome Helper: a custom and streamlined workflow for microbiome Research. mSystems. 2017 doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 84.Lozupone CA, Knight R. The UniFrac significance test is sensitive to tree topology. BMC Bioinform. 2015;16:211. doi: 10.1186/s12859-015-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.