Abstract
Lanthanide elements have been recently recognized as “new life metals” yet much remains unknown regarding lanthanide acquisition and homeostasis. In Methylorubrum extorquens AM1, the periplasmic lanthanide-dependent methanol dehydrogenase XoxF1 produces formaldehyde, which is lethal if allowed to accumulate. This property enabled a transposon mutagenesis study and growth studies to confirm novel gene products required for XoxF1 function. The identified genes encode an MxaD homolog, an ABC-type transporter, an aminopeptidase, a putative homospermidine synthase, and two genes of unknown function annotated as orf6 and orf7. Lanthanide transport and trafficking genes were also identified. Growth and lanthanide uptake were measured using strains lacking individual lanthanide transport cluster genes, and transmission electron microscopy was used to visualize lanthanide localization. We corroborated previous reports that a TonB-ABC transport system is required for lanthanide incorporation to the cytoplasm. However, cells were able to acclimate over time and bypass the requirement for the TonB outer membrane transporter to allow expression of xoxF1 and growth. Transcriptional reporter fusions show that excess lanthanides repress the gene encoding the TonB-receptor. Using growth studies along with energy dispersive X-ray spectroscopy and transmission electron microscopy, we demonstrate that lanthanides are stored as cytoplasmic inclusions that resemble polyphosphate granules.
Subject terms: Microbiology, Bacteria, Cellular microbiology, Microbial genetics
Introduction
Lanthanide (Ln) metals have long been recognized for their magnetic and superconductive properties and have facilitated the advancement of our communication, green energy, and medical technologies1–4. However, it has been less than a decade since an inherent role for Ln in methylotrophic bacteria was described5–7. Since these initial reports, bacterial strains that are not considered methylotrophs such as Pseudomonas putida and Bradyrhizobium sp. have been shown to similarly utilize Ln as cofactors in alcohol dehydrogenase (ADH) enzymes, suggesting the impact of Ln on microbial metabolism may be more widespread than initially thought8–13.
To use methanol as a carbon and energy source, many Gram-negative methylotrophic bacteria first oxidize methanol in the periplasmic space using pyrroloquinoline quinone (PQQ)-dependent ADH enzymes. MxaFI is a two-subunit Ca2+-dependent ADH that was long considered to be the predominant methanol dehydrogenase (MeDH) in nature until Ln-dependent XoxF enzymes were first described in 20115. Since their role in methanol oxidation became apparent, XoxF enzymes have been classified into five phylogenetically distinct clades (Type 1 to 5)14–16. In 2016, a Ln-dependent ethanol dehydrogenase was described and named as ExaF based on homology with the Ca2+-dependent Exa ethanol dehydrogenases from Pseudomonas and Rhodopseudomonas strains, and homology with XoxF- and MxaF-type MeDHs17,18.
When incorporated into the active site, Ln act as potent Lewis acids to facilitate a hydride transfer from the alcohol to the catalytic PQQ cofactor to prompt alcohol oxidation19–21. All methylotrophic PQQ-ADHs are periplasmic enzymes associated with a cytochrome cL (MxaG, XoxG, and ExaG, respectively) that transfers electrons from PQQ to additional cytochromes in the electron transport chain. In addition to the partnering cytochrome cL, operons or genomic clusters that encode xoxF and exaF genes often contain homologs of mxaJ (xoxJ and exaJ genes respectively), which encode periplasmic binding proteins that have been suggested to function in the activation of the ADHs 22. mxa operons encode additional proteins that are suggested to function in Ca2+ insertion, facilitate interactions between MxaFI MeDH and its cytochrome, and are required for regulation of the mxa operon expression23.
Methylorubrum extorquens AM1 (formerly Methylobacterium extorquens AM1) produces a MxaFI-type MeDH (encoded by mxaF: MexAM1_META1p4538 and mxaI: MexAM1_META1p4535), two XoxF (type 5) MeDHs (encoded by xoxF1: MexAM1_META1p1740 and xoxF2: MexAM1_META1p2757), and an ExaF-type ethanol dehydrogenase (encoded by exaF: MexAM1_META1p1139)17,18. The XoxF enzymes from M. extorquens AM1 share 90% amino acid similarity, and are named as XoxF1 and XoxF2 to distinguish them from one another24. Phenotypic studies suggest M. extorquens AM1 XoxF1 and XoxF2 have redundant function18. However, in the laboratory, XoxF1 appears to be the dominant XoxF-MeDH as xoxF2 is expressed at low levels18,25. A fifth putative PQQ-ADH encoded by MexAM1_META1p4973 does not contribute to methanol growth under the conditions tested17. When Ln are absent, MxaFI is the only known contributor to methanol oxidation in M. extorquens AM1. When Ln are available, the XoxF enzymes produce formaldehyde from methanol whereas ExaF oxidizes methanol further to formate25.
In addition to catalysis, Ln serve as a signal for a transcriptional response called “the Ln-switch” or “rare earth-switch,” though the mechanism of Ln signal sensing is not completely understood18,26–29. When Ln are available, the mxa operon (mxaFJGIRSACKLDEHB) is downregulated and transcript levels of the xox1 operon genes (xoxF1GJ) are upregulated18,25–28. In M. extorquens AM1 and closely related PA1 strain, the MxbDM two-component system along with the xoxF1 and xoxF2 genes themselves have been shown to be required for operation of the Ln-switch24,30,31. However, suppressor mutations in the mxbD sensor kinase encoding gene can arise, which bypass the need for XoxF1 and XoxF2, presumably by constitutively activating the MxbM response regulator24,29,31. Though much progress has been made regarding the catalytic and regulatory roles of Ln in methylotrophic bacteria, relatively little is known about how these Ln are acquired and incorporated into the enzymes that use them.
The machinery necessary for Ln transport is in the early stages of characterization and is predicted to be analogous to siderophore-mediated iron transport4,31 with the siderophore-like molecule referred to as a lanthanophore32. In M. extorquens AM1, ten genes predicted to encode proteins involved in Ln transport and utilization are clustered together in the genome (MexAM1_META1p1778–MexAM1_META1p1787) and encode an ABC-type transporter, four hypothetical periplasmic proteins of unknown function, a periplasmic protein that binds Ln (encoded by MexAM1_META1p1781), a TonB-dependent transporter, and lanmodulin (encoded by MexAM1_META1p1786)4,33. In closely related M. extorquens strain PA1, Ochsner et al. characterized the Ln transport cluster by generating deletions spanning multiple genes in the predicted transport system31. Their results showed that a TonB-dependent transporter and a putative ABC transporter were necessary for Ln transport, suggesting Ln transport into the cytosol. A detailed analysis of the contribution of each gene from the transport cluster is still lacking and Ln transport has not been quantified. Subsequently, Mattocks et al. demonstrated that in strain AM1 the hypothetical periplasmic protein encoded by MexAM1_META1p1781 efficiently binds lighter Ln and that similar to strain PA1, Ln are transported into the cytosol4.
In this study, we describe new pieces necessary to complete the Ln puzzle: the identification of novel genes that contribute to Ln metabolism and methanol oxidation, and the discovery and visualization of Ln storage in M. extorquens AM1. Detailed growth studies for strains lacking each component of the first eight genes in the Ln transport cluster are described, including the ability of these strains to mutate or acclimate (phenotypic change that is inducible and reversible) to allow Ln transport through a predicted secondary mechanism. Ln uptake is also quantified for several transport mutant strains. Finally, we show that M. extorquens AM1 stores Ln with phosphate as crystalline cytoplasmic deposits.
Results
A genetic study identifies gene products that contribute to methanol oxidation
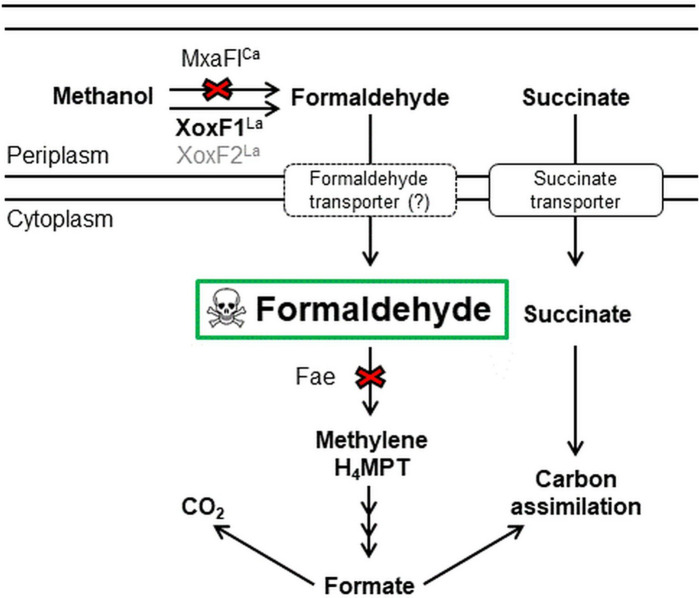
A transposon mutagenesis study was designed to take advantage of the in vivo formaldehyde production capability of XoxF1 and XoxF2 to identify genes required for XoxF-dependent methanol oxidation. The strain used to conduct the mutant hunt contained a mutation in mxaF to make cells dependent on the exogenously provided La3+ for formaldehyde production, and a second mutation in fae, which would result in formaldehyde accumulation and cell death when methanol is oxidized to formaldehyde by XoxF1 and XoxF2 (Fig. 1). Transposon insertions that reduced or eliminated formaldehyde production allowed survival and colony formation on media containing both methanol and succinate, since methanol resistant strains could use succinate for growth. In addition to genes required for XoxF1 and XoxF2 function, this mutant hunt had the potential to identify a hypothetical formaldehyde import system, disruption of which might reduce formaldehyde levels in the cytoplasm (Fig. 1). As ExaF oxidizes methanol directly to formate25, exaF and genes specific to ExaF function were not expected to be identified though this genetic study.
Figure 1.
Schematic representation of the metabolic processes relevant to the mxaF fae transposon mutagenesis study. XoxF1 and XoxF2 oxidize methanol to formaldehyde, which accumulates to lethal levels in the fae mutant strain. If a process required for XoxF-dependent methanol oxidation is disrupted by a transposon insertion, formaldehyde is reduced or eliminated, and cells use succinate for growth. A dashed line around the formaldehyde transporter is to indicate that this function has not been demonstrated.
Over six hundred transposon mutants were isolated, and their insertion locations mapped to the M. extorquens AM1 genome. As it is likely that a portion of these transposon mutants became methanol-resistant due to spontaneous second-site suppressor mutations and not the transposon insertion, only genes that were independently identified four or more times were considered for further analysis and are listed in Table 1. From the twenty eight genes identified in our transposon mutagenesis study (Table 1), mutations in twenty three genes were constructed in mxaF and/or wild-type strain backgrounds and methanol growth in the presence of La3+ was assessed (Tables 2, 3).
Table 1.
Genes identified four or more times via transposon mutagenesis.
| Gene designation | Gene name | Predicted function |
|---|---|---|
| MexAM1_META1p0863 | LysR-type regulator | |
| MexAM1_META1p1292 | cycL | c-type cytochrome biogenesis |
| MexAM1_META1p1293 | cycK | Heme lyase |
| MexAM1_META1p1294 | cycJ | Periplasmic heme chaperone |
| MexAM1_META1p1740 | xoxF1 | Ln-dependent methanol dehydrogenase |
| MexAM1_META1p1741 | xoxG | Cytochrome c |
| MexAM1_META1p1742 | xoxJ | Periplasmic binding protein |
| MexAM1_META1p1746 | orf6 | Unknown |
| MexAM1_META1p1747 | orf7 | Unknown |
| MexAM1_META1p1748 | pqqE | PQQ biosynthesis |
| MexAM1_META1p1749 | pqqCD | PQQ biosynthesis |
| MexAM1_META1p1750 | pqqB | PQQ biosynthesis |
| MexAM1_META1p1771 | MxaD homolog | |
| MexAM1_META1p1778 | lutA | ABC transporter-periplasmic binding component |
| MexAM1_META1p1779 | lutB | Exported protein |
| MexAM1_META1p1782 | lutE | ABC transporter-ATP binding component |
| MexAM1_META1p1783 | lutF | ABC transporter-membrane component |
| MexAM1_META1p1784 | lutG | Exported protein |
| MexAM1_META1p1785 | lutH | TonB-dependent receptor |
| MexAM1_META1p2024 | hss | Homospermidine synthase |
| MexAM1_META1p2330 | pqqF | Protease |
| MexAM1_META1p2331 | pqqG | Protease |
| MexAM1_META1p2359 | ABC transporter—fused ATPase and transmembrane components | |
| MexAM1_META1p2732 | ccmC | Heme export |
| MexAM1_META1p2734 | ccmG | c-type cytochrome biogenesis |
| MexAM1_META1p2825 | ccmB | Heme export |
| MexAM1_META1p2826 | ccmA | Heme export |
| MexAM1_META1p3908 | Leucyl aminopeptidase | |
Table 2.
Growth parameters for strains grown in methanol medium with La3+.
| Strain | Growth rate (h−1)a in MeOH + La3+ |
|---|---|
| Growth of control strains | |
| Wild type | 0.16 ± 0.01 |
| mxaF | 0.16 ± 0.01 |
| xoxF1 | 6–9 h lag, 0.07 ± 0.00 |
| xoxF1 xoxF2 | 6 h lag, 0.04 ± 0.01 |
| mxaF xoxF1 xoxF2 | 6 h lag, 0.04 ± 0.00 |
| Growth of strains lacking novel genes identified in genetic selection | |
| MexAM1_META1p0863 | 0.14 ± 0.00 |
| MexAM1_META1p1771 | 0.11 ± 0.01 |
| orf6 | 0.03 ± 0.00 |
| orf7 | 9 h lag, 0.08 ± 0.01 |
| MexAM1_META1p2359 | No growth |
| hss | 6 h lag, 0.10 ± 0.01 |
| MexAM1_META1p3908 | 15 h lag, 0.07 ± 0.00 |
| Growth of strains lacking genes with known function | |
| pqqBCDE | No growth |
| pqqF | No growth |
| cycK | No growth |
| ccmB | No growth |
| ccmC | No growth |
| Growth of strains lacking lanthanide utilization and transport genes | |
| lutA | 0.02 ± 0.01, 96 h; 0.08 ± 0.01 |
| mxaF lutA | 0.02 ± 0.00 |
| lutB | 0.02 ± 0.00, 87 h; 0.05 ± 0.01 |
| mxaF lutB | 0.02 ± 0.00 |
| lutC | 9 h lag, 0.11 ± 0.01 |
| mxaF lutC | 9 h lag, 0.11 ± 0.01 |
| lutE | No growth |
| mxaF lutE | No growth |
| lutF | No growth |
| mxaF lutF | No growth |
| lutG | 12 h lag, 0.03 ± 0.00 |
| mxaF lutG | 12 h lag, 0.03 ± 0.00 |
| lutH | 0.16 ± 0.00 |
| mxaF lutH | No growth |
aData for a minimum of three biological replicates are reported.
±Indicates standard deviations.
Table 3.
Growth parameters for xox strains grown in methanol medium with and without La3+.
| Strain | Growth rate (h−1)a | |
|---|---|---|
| MeOH | MeOH + La3+ | |
| Wild type | 0.14 ± 0.01 | 0.16 ± 0.01 |
| xoxF1 xoxF2 | No growth | 6 h lag, 0.04 ± 0.01 |
| xoxG | 3 h lag, 0.11 ± 0.01 | 0.04 ± 0.00 |
| xoxJ | 21 h lag, 0.13 ± 0.00 | 0.04 ± 0.01 |
aData for a minimum of three biological replicates are reported.
±Indicates standard deviations.
Growth phenotypes of strains lacking genes identified in the transposon mutagenesis study
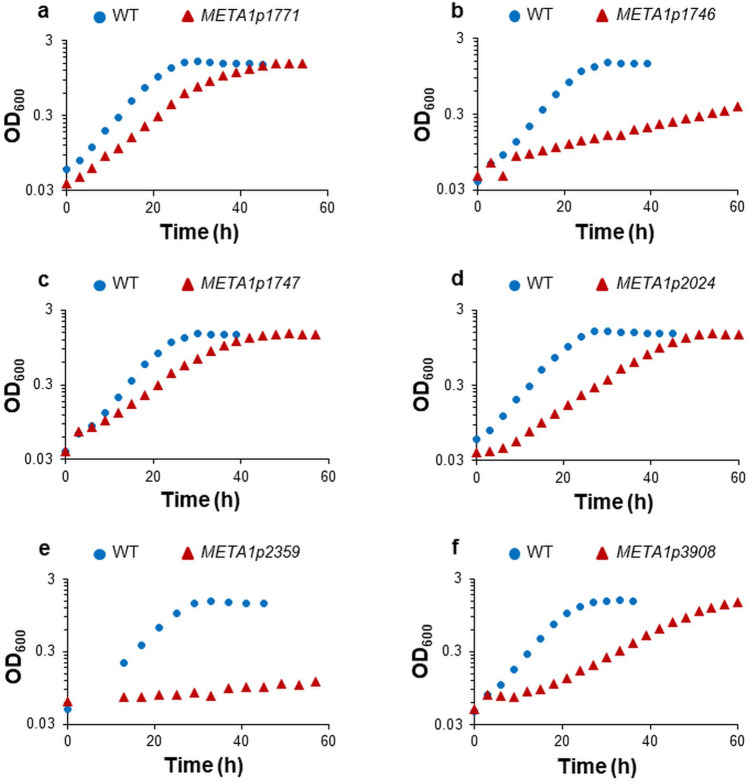
Novel genes were identified in the transposon mutagenesis study which impacted methanol growth with La3+ when deleted from the M. extorquens AM1 genome (Table 1). The identified genes encode a LysR-type transcriptional regulator (MexAM1_META1p0863), two proteins of unknown function annotated as orf6 and orf7 (MexAM1_META1p1746 and MexAM1_META1p1747 respectively), an MxaD homolog (MexAM1_META1p1771), a putative homospermidine synthase (MexAM1_META1p2024), an ABC-type transporter (MexAM1_META1p2359), and an aminopeptidase (MexAM1_META1p3908). Growth rates for strains lacking these genes are shown in Table 2 and growth curves for mutant strains that had a 30% or greater reduction in growth rate are shown in Fig. 2a–f. Among these identified genes, loss of the MexAM1_META1p2359 ABC-type transporter, putative homospermidine synthase, and lysR-type regulator resulted in similar growth defects in the absence of La3+ (Table S1) which suggests an involvement in both XoxF1- and MxaFI-facilitated methanol growth. The identification of these genes and initial growth studies lay the foundation to explore the specific roles and requirements for these gene products in methanol oxidation.
Figure 2.
Growth of mutant strains lacking genes identified through transposon mutagenesis in medium containing methanol and La3+. Growth of M. extorquens AM1 wild-type is represented by blue circles and depicted mutant strains (red triangles) have the following genes deleted: (a) MexAM1_META1p1771, mxaD homolog; (b) MexAM1_META1p1746, orf6; (c) MexAM1_META1p1747, orf7; (d) MexAM1_META1p2024, homospermidine synthase; (e) MexAM1_META1p2359, ABC transporter of unknown function; (f) MexAM1_META1p3908, aminopeptidase. Representative data from biological triplicates are shown. One-way analysis of variance (ANOVA) determined that growth rate differences between mutant and parent strains are significantly different (p < 0.005).
Also included in the list of genes identified in the transposon mutagenesis study were those that encode proteins with described roles in methanol oxidation (PQQ biosynthesis, xoxF1, xoxG, xoxJ) and those with predicted functions based on sequence similarity (cytochrome synthesis and Ln transport31,34). Deletion of PQQ biosynthesis genes (ΔpqqBCDE; ΔpqqF) and genes in the three identified cytochrome c biogenesis and heme export clusters34 (cycK; ccmB; ccmC) eliminated methanol growth in the presence and absence of La3+ as expected (Tables 2, S1).
XoxF1 and XoxF2 catalytically contribute to methanol oxidation and growth when methanol and Ln are provided 17,18, and exert a regulatory role in the absence of Ln as production of XoxF1 or XoxF2 is required for expression of the Ca2+-dependent MxaFI-MeDH18,24. Growth analysis and transcriptional reporter fusion studies were employed to see if strains lacking xoxG or xoxJ had similar effects on growth and mxa operon expression. In methanol La3+ medium, loss of either xoxG or xoxJ was equivalent to loss of both xoxF1 and xoxF2 (Fig. 3; Table 3), which is consistent with XoxG and XoxJ being essential for XoxF-dependent methanol oxidation as suggested by recent biochemical studies22,35,36.
Figure 3.
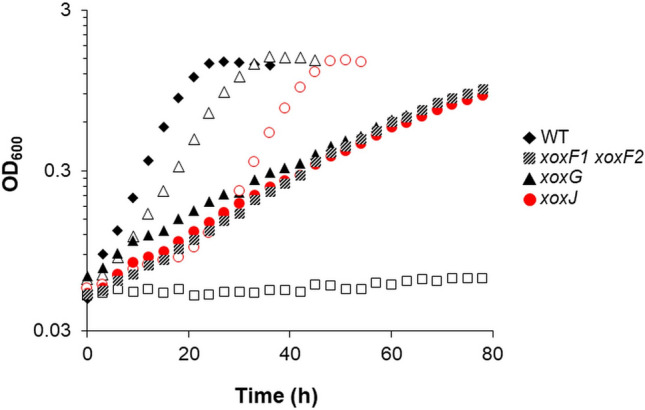
Growth of xoxG and xoxJ mutant strains in the presence (filled symbols) and absence (open symbols) of La3+. Representative data from biological triplicates are shown. One-way analysis of variance (ANOVA) determined that growth rate differences between mutant and parent strains are significantly different (p < 0.005).
Unlike the xoxF1 xoxF2 double mutant strain, transcriptional reporter fusion studies that assessed expression from the mxa promoter determined that the growth phenotypes observed for the xoxG and xoxJ mutants grown in the absence of La3+ were not due to impaired mxa expression (Table 4). Taken together, these phenotypes suggest XoxG and XoxJ may play a broader role in methanol metabolism in M. extorquens AM1 independent from facilitating XoxF1 and XoxF2 catalytic and regulatory functions.
Table 4.
Expression from mxa and xox1 promoters (RFU/OD600) in wild type and mutant strains grown in methanol media with or without La3+ (Data for a minimum of three biological replicates is reported).
| Promoter | MeOH | MeOH + La3+ | ||
|---|---|---|---|---|
| mxa | xox1 | mxa | xox1 | |
| Strain | ||||
| Wild type | 323 ± 63 | 44 ± 3 | 61 ± 10 | 206 ± 11 |
| mxaF lutH | ND | ND | 363 ± 11 | 53 ± 7 |
| mxaF lutH acclimated | ND | ND | 82 ± 14 | 240 ± 17 |
| xoxF1 xoxF2 | 25 ± 4 | ND | ND | ND |
| xoxG | 365 ± 24 | ND | ND | ND |
| xoxJ | 566 ± 55 | ND | ND | ND |
ND not determined.
Detailed phenotypic characterization of Ln utilization and transport cluster mutant strains.
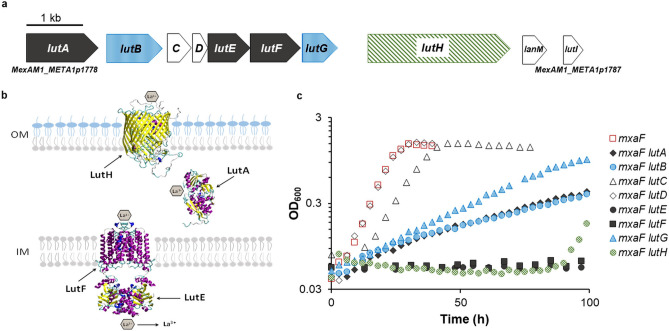
Consistent with previous reports for strain PA1, our transposon mutagenesis studies suggest that genes encoding homologs of the TonB- and ABC-dependent Fe3+ scavenging systems play a role in methanol metabolism when Ln are present31. In addition to the transport system homologs, two of six hypothetical periplasmic proteins encoded in the Ln transport cluster were identified in the transposon mutagenesis study (Fig. 4a). We expanded Ochsner’s study by dissecting the contribution of individual genes (MexAM1_META1p1778 through MexAM1_META1p1785). Based upon work detailed within, we propose to name the Ln transport cluster genes as lut, for Ln utilization and transport.
Figure 4.
Characterization of the lut cluster. (a) Genomic map of the lut genes (MexAM1_META1p1778 to MexAM1_META1p1787). Black, genes encoding the ABC transport system; green, gene encoding the TonB-dependent transporter; blue, genes encoding putative periplasmic proteins identified by transposon mutagenesis; white, lanmodulin and additional genes encoding periplasmic proteins not identified by transposon mutagenesis. (b) Model for Ln transport. The three-dimensional structures of monomers of LutH and LutA were predicted using homology modeling (HHpredserver and MODELLER)37 and homodimers of the ABC transporter were predicted using GalaxyHomomer38. (c) Growth of mxaF and mxaF lut mutant strains with 2 μM LaCl3. Graphs depict representative data from three biological replicates. Growth rate averages and standard deviations are shown in Table 2.
As reported by Ochsner et al.31, loss of lutH (Mextp1853 in strain PA1) alone did not result in a growth defect in medium containing methanol and La3+, which is consistent with the hypothesis that the TonB-dependent transporter is needed for transport of La3+ into the periplasm. If La3+ does not enter the periplasm, the mxaF and mxaI genes are likely expressed and used for methanol oxidation. Loss of both mxaF and lutH arrested growth, but after 90–120 h, growth of the mxaF lutH double mutant strain occurred in approximately 60% of the 24 cultures tested (Fig. 4c; Tables 2, 5). Consistent with an acclimation process and not a suppressor mutation, the mxaF lutH double mutant strain lost the ability to grow in methanol medium with La3+ after passage onto medium containing succinate. To determine if acclimated growth was due to production of XoxF1, expression from the mxa and xox1 promoters was measured before and after acclimation (Table 4). Before acclimation, mxa promoter expression occurred at levels comparable to those in media lacking La3+ in the wild-type strain while xox1 expression was repressed as the cells could not transport La3+ to promote the Ln-switch. Once acclimation occurred and the mxaF lutH strain began to grow, expression from the mxa promoter was repressed and expression from the xox1 promoter was induced, suggesting the cells were able to uptake La3+ through the outer membrane via an unknown mechanism.
Table 5.
Growth parameters of suppressor and acclimation events.
| Strain | MeOH + La3+ | |
|---|---|---|
| Time of S/A | Growth ratea | |
| (h) | (h−1) | |
| MexAM1_META1p2359 | Suppressor: 85 | 0.10 ± 0.01 |
| lutE | Suppressor: 75–90 | 0.07 ± 0.01 |
| mxaF lutE | Suppressor: 200–220 | 0.02 ± 0.00 |
| lutF | Suppressor: 79–91 | 0.09 ± 0.02 |
| mxaF lutF | Suppressor: 145–157 | 0.02 ± 0.01 |
| mxaF lutH | Acclimation: 90–120 | 0.14 ± 0.02 |
aData for a minimum of three biological replicates is reported.
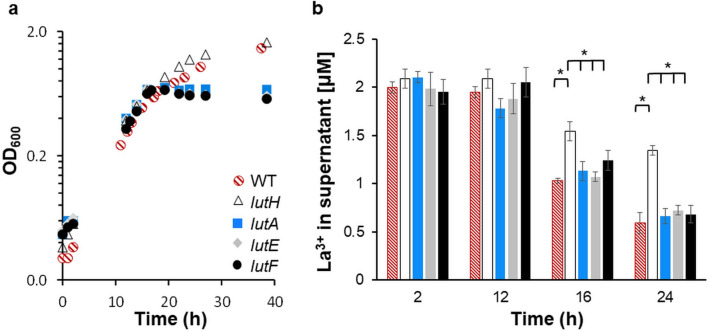
LutA, a periplasmic binding protein, is predicted to traffic lanthanophore bound Ln through the periplasm to the inner membrane transporter system (encoded by lutE and lutF) for transport into the cytoplasm (Fig. 4b). Strains lacking lutE or lutF in the mxaF mutant strain background were unable to grow in medium containing methanol and La3+ (Fig. 4c, Table 2). However, after 200 h and 150 h respectively, second-site suppressor mutations arose which facilitated growth albeit 88% slower than that of the wild-type strain (Table 5). In contrast, loss of lutA in the absence of mxaF still allowed La3+-dependent growth but at a reduced rate (Fig. 4c). Genes lutB and lutG, encoding hypothetical periplasmic proteins, were also identified in the transposon mutagenesis study. Growth of the mxaF lutB and mxaF lutG double mutant strains was similar to that of the mxaF lutA double mutant strain, suggesting an equally important but non-essential function. The growth observed for the lutA, lutB, and lutG mutant strains in the absence of mxaF was not identified as acclimation or second-site suppression. To ensure that observed phenotypes were not due to polarity, mutants lacking individual transport cluster genes (lutABEFG) were complemented by expressing the respective gene in pCM6239 and growth similar to the wild-type strain was restored in each case (Fig. S1).
Genes encoding LutC, LutD, LanM, and LutI were not identified in the transposon study but LutD and LanM have been shown to bind Ln4,33. Loss of lutD did not result in a significant growth defect in the mxaF mutant and wild-type strain backgrounds while loss of lutC resulted in a small growth defect when La3+ was provided (Fig. 4c; Table 2).
Quantification of La3+ uptake
To assess how loss of the Ln transport system affects La3+ uptake, La3+ levels were quantified from the spent media at different phases of growth (Fig. 5). Strains were grown in methanol medium containing 2 µM LaCl3 and limiting succinate (3.75 mM) as lutA, lutE, and lutF mutant strains are unable to grow or grow poorly with methanol as a sole carbon source (Fig. 4c; Table 2). While significant decreases in La3+ levels were not observed for any strain during early- to mid-exponential phase (Fig. 5a,b), as cultures continued to grow, and succinate was likely depleted, significant differences in La3+ levels became apparent. In the wild-type strain, La3+ uptake continued into stationary phase (Fig. 5a,b). Growth of the lutE and lutF mutants (encoding ABC-transporter components) was arrested at an approximate OD600 of 0.7 (Fig. 5a), yet La3+ concentrations in the spent media continued to decrease to 0.7 µM eight hours into stationary phase (Fig. 5b). Taken together with the growth phenotypes, these data suggest that in the absence of the Lut-ABC-transporter system, Ln are likely transported through the outer membrane into the periplasm but cannot enter the cytoplasm. Levels of La3+ in the supernatants from the lutH TonB-dependent transporter mutant were significantly higher than wild type in mid-exponential and stationary phases (Fig. 5b). However, a small decrease in La3+ content from the lutH mutant strain culture supernatants was observed.
Figure 5.
Growth studies and measurement of La3+ uptake suggest La3+ storage capability. (a) Growth of wild type (red dashed circles), lutH (white triangles), lutA (blue squares), lutE (gray diamonds), and lutF (black circles) strains grown with limiting succinate (3.75 mM), methanol (125 mM), and 2 μM LaCl3. Data are the average of three biological replicates. Standard deviations between biological replicates were less than ± 0.02. (b) La3+ concentrations (μM) in culture supernatants of lut mutant strains depicted in same color code as in (a). Data depict the average of three biological replicates with error bars showing the standard deviation. One-way analysis of variance (ANOVA) followed by a t-test was used to represent statistical significance. *The p-value is < 0.005.
Expression from the lutH promoter is repressed by La3+
Genes found within the lut cluster are likely not expressed as a single transcript as suggested by the spacing between genes such as lutG and lutH, and by multiple promoters predicted by the bacterial promoter prediction tool, BPROM40. To test if expression of the of Ln uptake genes is regulated by Ln, a fluorescent transcriptional reporter fusion was used to monitor expression from the predicted lutH promoter region in methanol media. Addition of exogenous La3+ repressed expression fourfold (RFU/OD600: 210 ± 7 without La3+ and 47 ± 5 with La3+) showing that when Ln are in excess, transport is down-regulated. Similar repression has been reported for transcription of the TonB-dependent transporter of Methylotuvimicrobium buryatense 5GB1C41.
La3+ is stored as cytoplasmic crystalline deposits
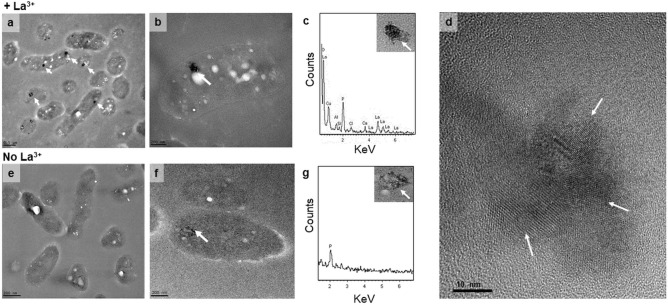
Transmission electron microscopy (TEM) coupled with energy dispersive X-ray spectroscopy (EDS) has been used to determine the elemental composition of cellular inclusions40,42 while La3+ has been widely used as an intracellular and periplasmic stain for electron microscopy43–45. Here, we show that La3+ can be directly identified by TEM if accumulated inside M. extorquens AM1 cells. Electron-dense deposits were observed in the cytoplasm from M. extorquens AM1 cells grown with exogenous LaCl3 (Fig. 6a,b). Samples were analyzed using EDS and corroborated that the electron dense deposits contained La3+ (Fig. 6c). When grown without La3+, only a few cells showed smaller electron dense areas (Fig. 6e, f); however, La3+ was not detected in EDS analysis in these cases (Fig. 6g). These data demonstrate that La3+ can be stored in M. extorquens as metal deposits. Moreover, EDS analysis of electron dense areas from the wild-type strain grown with La3+ determined a content of lanthanum (22.2 ± 1.0 weight %), phosphorus (15.1 ± 2.1 weight %), and oxygen (51.1 ± 1.9 weight %), suggesting La3+ is complexed with phosphates (Fig. S2). Traces of chloride (3.0 ± 1.0 weight %), calcium (2.2 ± 0.6 weight %), and aluminum (3.4 ± 0.6 weight %) ions were also detected. The copper, carbon, and silicon ion content from the support grids and embedding medium were not considered for metal content calculations. High-resolution transmission electron microscopy (HRTEM) images of the La3+ deposits showed an atomic lattice with a Moiré fringes pattern, indicating a crystalline nature46 (Fig. 6d). Together, these results suggest that La3+ is embedded in inorganic phosphate crystals, which form the electron dense deposits observed in the cytoplasm.
Figure 6.
Visualization of La3+ storage. TEM of ultrathin sections of wild-type cells harvested at OD600 of ~ 0.6 after growth with 3.75 mM succinate and 125 mM methanol containing (a, b) and lacking (e, f) 20 µM La3+. (c, g) Elemental analysis of electron dense deposits from cells grown with (c) and without (g) La3+. (d) High-resolution transmission electron microscopy analysis of the wild-type strain shows an atomic lattice structure in electron-dense areas suggesting that La3+ is embedded in cytoplasmic crystals.
Visualization of La3+ accumulation in lut transporter mutants
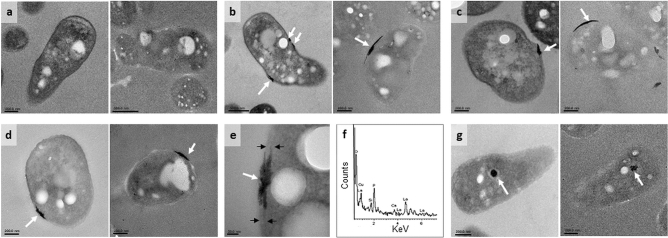
To determine if La3+ could be visualized in strains lacking the Lut-TonB-ABC transport system, TEM and EDS were employed for the analysis of lutA, lutE, lutF, and lutH mutant strains. Strains were grown in methanol medium containing La3+ and limiting succinate. Samples stained with OsO4 and 2% uranyl acetate allowed the outer and inner membrane of the bacterial cells to be distinguished (Fig. 7a–e and g left subpanels), while visualization without staining enabled metal content analysis by removing the interaction between Os8+ and phosphate, which interferes with La3+ measurements (Fig. 7a–e and g right subpanels). Mutants lacking the TonB-dependent transporter (encoded by lutH) did not display La3+ deposits (Fig. 7a compared to Fig. 7g). In contrast, localized La3+ deposits were visualized in the periplasmic space in mutant strains lacking the ABC-transporter components (encoded by lutA, lutE, and lutF) as shown in Fig. 7b–e. EDS microanalyses confirmed these electron dense periplasmic deposition areas contained La3+ (Fig. 7f). Taken together, these findings directly demonstrate a role for the TonB-dependent and ABC transporters in Ln transport.
Figure 7 .
Visualization of La3+ localization using TEM. Thin sections of (a) lutH, (b) lutE, (c) lutF, (d) lutA, and (g) wild-type strains grown with 3.75 mM succinate, 125 mM methanol, and 20 µM LaCl3. White arrows indicate deposits of electron scattering material in the periplasm. Cells were fixed with 2.5% glutaraldehyde and stained with OsO4 and uranyl acetate to detect cell membranes (left subpanel) or left unstained for elemental analysis (right subpanel). (e) Magnification of the La3+-deposits localized in the periplasmic space from the lutA mutant strain; black arrows indicate the boundaries of the outer membrane and inner membrane of the lutA mutant strain. (f) Elemental analysis of the electron-dense deposits observed inside the periplasm.
Discussion
The Ln-dependent XoxF1-MeDH produces formaldehyde in vivo25. Here, we took advantage of this property which allowed lethal levels of formaldehyde to accumulate when methanol was oxidized by XoxF1 and fae was deleted from the genome. These phenotypes enabled a genetic selection to identify gene products required for or involved in XoxF1-mediated methanol oxidation.
Ln must be transported into the cell and incorporated into the XoxF1 active site25,47 but is not yet known if incorporation occurs in the cytoplasm or periplasmic space. Our growth, transport, TEM, and EDS analyses are consistent with LutH facilitating Ln transport into the periplasm and the Ln-ABC transport system facilitating Ln transport into the cytoplasm. La3+ concentrations found in the supernatant from strain variants lacking transport system components suggest that once in the periplasm, significant concentrations of La3+ do not go back outside of the cell as ABC transporter mutant strains showed uptake of La3+ from the medium similar to the wild-type strain. Intriguingly, TEM and EDS studies with ABC transporter mutant strains demonstrated localized accumulation of La3+ in the periplasmic space. It is not yet clear how or why La3+ accumulates in specific areas rather than appears diffused throughout the periplasm.
In M. extorquens, the MxbDM two-component system has been proposed to sense periplasmic Ln either directly or indirectly to facilitate differential regulation of the mxa and xox1 operons18,31. Results from our growth, transport, and visualization studies support the hypothesis by Oschner et al.31 that the regulatory network controlling differential expression may be more complex than initially thought. Consistent with Ochsner et al. and Mattocks et al., our data suggest Ln must enter the cytoplasm for xoxF1 to be expressed4,31; lut ABC transporter mutants accumulated La3+ in the periplasmic space yet were unable to grow. It is also possible that xoxF1 is expressed and produced in the lut ABC transporter mutants, yet periplasmic La3+ is not incorporated into the active site of XoxF1 as it is bound by lanthanophores. It is not yet known if Ln release from the lanthanophore occurs in the periplasm, or if Ln are first released in the cytoplasm then exported back into the periplasmic space for insertion into XoxF1. While a LysR-type transcriptional regulator was identified multiple times in this genetic study, no severe growth defect was observed when disrupted.
Our data also suggest periplasmic Ln may be enough to prevent mxa expression since the lut ABC transporter mutants could not grow when mxaF was intact in the genome. An analogous system in P. putida KT2440 exists where expression of the pedH and pedE genes, which encode Ln- and Ca-dependent ADHs respectively, are differentially regulated by Ln through the PedS2/R2 two-component system10. Unlike M. extorquens, Ln-ABC transport mutants of P. putida KT2440 express pedH when Ln are present, suggesting periplasmic Ln are enough to facilitate pedH expression48.
The phenotypes for the lutH outer membrane transporter mutant were distinct from the ABC transporter mutant phenotypes. Surprisingly, an approximate 0.6 µM decrease in La3+ content from the lutH mutant strain culture supernatants was observed. However, since methanol growth did not occur in the lutH strain when mxaF was also deleted from the genome (Fig. 4c), this suggests that significant quantities of La3+ do not enter the cell to trigger xox1 expression. Lack of La3+ transport into the periplasm would explain why the lutH mutant was able to grow when mxaF was not deleted from the cell; the cell would not sense periplasmic La3+ and the mxa operon would be expressed. The decrease of La3+ from the media likely reflects adsorption of La3+ onto the surface of the cells or interaction of La3+ with lipopolysaccharide, a phenomenon observed for other metals in different bacterial species49.
A discovery from this work is that the Ln transport system can be bypassed by suppression or acclimation. Acclimation of the mxaF lutH double mutant strain allowed xoxF1 expression and rapid growth. It may be that Ln can leak into the periplasm slowly over time and once a threshold is reached, xoxF1 is expressed. A second possibility is that an alternative outer membrane transport system is expressed in the acclimated cultures.
The requirement for the Ln-ABC transport system was bypassed by suppression which allowed growth at reduced rate. Suppressor mutations may facilitate Ln uptake through an alternative system, either by increased expression of such system or by amino acid changes in a different metal import system to facilitate Ln transport. In Pseudomona putida KT2440, growth in the absence of the analogous Ln-ABC transporter still occurs if exogenous Ln3+ concentrations are high (100 µM) and/or Fe2+/3+ concentrations are low. These results suggest that in P. putida KT2440, Ln are in competition with metals such as Fe3+ and can be transported by other metal (Fe3+) transport systems when in excess48. Intriguingly, Gu and Semrau saw differential expression of multiple genes encoding ABC-type and TonB-dependent transporters in the methanotroph, Methylosinus trichosporium OB3b, when cerium was added to the growth medium50. However, in M. extorquens AM1, Good et al. did not observe a similar upregulation of metal transport systems with the addition of La3+25.
We did not identify transposon insertions in the lanM or lutD genes though the proteins they encode have been shown to bind Ln4,33. These findings are consistent with studies in strain PA1 and suggest a non-essential or redundant role for these gene products31.
Notably, our transposon mutagenesis study did not identify obvious gene candidates for lanthanophore biosynthesis though over 600 insertions were mapped to the genome. This may indicate that more than one lanthanophore is produced or that the lanthanophore has an essential role that is not yet understood.
Our genetic study identified processes and gene products previously known or predicted to be required for XoxF function such as PQQ and cytochrome synthesis, heme export, and the XoxF, XoxJ, and XoxG proteins themselves. Recent biochemical and structural analyses of XoxG suggest that this cytochrome is tuned specifically for light Ln (lanthanum to samarium), while XoxJ interacts with and may activate XoxF122. Here we show that loss of either xoxG or xoxJ results in growth that mirrors the xoxF1 xoxF2 double mutant strain, consistent with XoxG and XoxJ as essential for the activity of XoxF-enzymes. However, using growth and transcriptional reporter fusion studies, we show that unlike XoxF1, XoxG and XoxJ are not required for expression of the mxa genes yet loss of the xoxG and xoxJ genes impacts growth in the absence of La3+. In the methanotroph Methylomonas sp. strain LW13, loss of xoxG also results in a growth defect in methanol medium lacking Ln suggesting an unknown role in metabolism in addition to functioning as a cytochrome for XoxF-mediated methanol oxidation36. Our results are in contrast to previous reports for M. extorquens PA1 and AM1, where loss of xoxG and xoxJ did not result in a growth phenotype in methanol medium lacking Ln31,51. Notably, the previous AM1 studies were carried out on agar plates where subtle growth defects may not be apparent.
Strains lacking novel genes were also identified that only displayed a requirement if La3+ was provided and include an mxaD homolog, orf6 and orf7. MxaD is a 17-kDa periplasmic protein that directly or indirectly stimulates the interaction between the MxaFI-MeDH and cytochrome cL 52 . Before the existence of Ln-dependent MeDHs was known, it was concluded that orf6 and orf7 gene products did not have a role in C1-metabolism though orf6 and orf7 are proximal to other methylotrophy genes51. Results here suggest that orf6 and orf7 contribute to Ln-dependent methylotrophy.
Our genetic study identified novel gene products that facilitate methanol growth regardless of La3+ presence or absence. An ABC-type transporter of unknown function was identified as essential. Lack of a periplasmic binding component suggests an export function rather than import. One possible function for META1p2359 could be the export of PQQ into the periplasm for incorporation into XoxF1 and MxaFI. This hypothesis is consistent with structural analysis of this exporter and the essential role for META1p2359 in methanol oxidation. An aminopeptidase was also identified which may be involved in processing one or more proteins required for XoxF1 and MxaFI function. Alternatively, the aminopeptidase could function in processing PQQ, as PQQ is peptide based and not all PQQ processing proteins have been identified53,54.
Lastly, our TEM and EDS analyses demonstrate that M. extorquens AM1 stores Ln in the cytoplasm in crystal form. For many bacteria, biomineralization is a mechanism used to cope with toxicity of different metals, manage waste products, sense and change orientations in accordance with geomagnetic fields, and store important cations for growth55–61. It has been reported that some bacteria store cations like Mg2+ and Ca2+ complexed to polyphosphate in the form of volutin (also known as metachromatic granules) or acidocalcisomes62–65. It is not yet known if M. extorquens AM1 stores La3+ complexed to polyphosphate, however, the ratios of P, O, and La3+ detected in our studies are consistent with La3+ phosphates. A gene encoding a putative homospermidine synthase (hss) was identified. Homospermidine synthases function in polyamine biosynthesis66,67. Polyamines have diverse roles but have been suggested to complex with polyphosphate to reduce spermidine toxicity in P. aeruginosa and stabilize polyphosphate granule formation in Escherichia coli68,69. Detailed studies are necessary to define the exact chemical structure of Ln storage deposits in M. extorquens AM1 and if these granules are membrane or lipid bound. Our current findings bring exciting implications for Ln metabolism and for the development of bioremediation and biometallurgy strategies for Ln recovery.
Methods
Bacterial strains and cultivation
Strains and plasmids used in this study are listed in Table S2. E. coli strains were cultivated in Lysogeny Broth (LB) medium70 (BD, Franklin Lakes, NJ) at 37 °C. M. extorquens AM1 strains were grown in Methylobacterium PIPES [piperazine-N,N’-bis(2-ethanesulfonic acid)] (MP) media71 supplemented with succinate (15 mM) and/or methanol (125 mM) as described18 unless otherwise stated. Conjugations took place on Difco Nutrient Agar (Thermo Fisher Scientific, Waltham, MA). Liquid cultures were grown at 29 °C and shaken at 200 and 180 rpm in New Brunswick Innova 2,300 and Excella E25 shaking incubators (Eppendorf, Hauppauge, NY), respectively. LaCl3 was supplemented to a final concentration of 2 or 20 μM when indicated. When necessary, antibiotics were added at the following concentrations: rifamycin (Rif, 50 µg/mL), tetracycline (Tc, 10 µg/mL for LB, 5 µg/mL for MP or 10 µg/mL when used together with Rif), kanamycin (Km, 50 µg/mL), ampicillin (Ap, 50 µg/mL).
Plasmid and strain construction
Primers used for plasmid and strain construction are listed in Table S3. The allelic exchange plasmid pHV2 was constructed by cloning the sacB gene from pCM43372 into the PscI site of pCM1873 in the same orientation as the Tc resistance gene. Insertion and orientation of sacB was confirmed by colony PCR. The lutH transcriptional reporter fusion was constructed by cloning the promoter region of lutH into the AclI and EcoRI sites upstream of a promoter-less venus gene in pAP524. To create overexpression constructs for complementation studies, individual genes in the lut operon (lutA, lutB, lutE, lutF, and lutG) were cloned into the KpnI and SacI sites downstream of a Plac promoter in pCM6239. Diagnostic PCR was used to confirm successful integration of inserts. Plasmids were maintained in E. coli TOP10 (Invitrogen, Carlsbad, CA). Gene deletions were constructed using pCM184 or pHV2 as previously described18 except 5% sucrose was added for counter selection against single crossovers72 when using pHV2. Plasmids were conjugated into M. extorquens AM1 via biparental mating using E. coli S17-174 or triparental mating using E. coli TOP10 (Invitrogen, Carlsbad, CA) and E. coli harboring the conjugative plasmid pRK2013 as described18. When indicated, the Km resistance cassette was resolved using pCM157 to achieve marker-less deletions73.
Transposon mutagenesis
Suicide vector pCM639 carrying a mini transposon ISphoA/hah-T75 was conjugated into the mxaF fae strain background via triparental mating as described18,76. Dilutions of the mating mixtures were plated onto MP succinate (15 mM) plus methanol (50 mM) La3+ medium containing 10 µg/mL Tc to select for successful integration of the mini transposon into the M. extorquens AM1 genome and 50 µg/mL Rif to counter select against E. coli strains bearing pCM639 or pRK2013. Plates were incubated for 5–7 days at 29 °C. Transposon mutant colonies were streaked onto MP succinate methanol La3+ Tc medium for downstream studies.
Location of transposon insertions
To identify the transposon insertion sites, genomic DNA was isolated using a Qiagen DNeasy UltraClean Microbial Kit (Qiagen, Germantown, MD). Degenerate nested PCR was performed as described76,77 with the following exceptions: PCR reactions contained 1 µM of each primer, 0.05 U/µL Dream Taq (Thermo Fisher Scientific, Waltham, MA), and 5% dimethyl sulfoxide. Modifications to the PCR amplification parameters included 2 min for the initial denaturation at 95 °C, 6 cycles of annealing at 40 °C followed by 25 cycles of annealing at 65 °C for the first PCR reaction, and 30 cycles of annealing at 65 °C for the second PCR reaction. PCR products were purified using a Qiagen QIAquick 96 PCR Purification Kit (Germantown, MD). Sequence analysis was performed using TransMapper, a Python-based program developed in-house to identify transposon insertion locations and map them to the M. extorquens AM1 genome for visualization using SnapGene Viewer (GSL Biotech LLC, Chicago, IL).
Phenotypic analyses
Growth phenotypes were determined on solid or in liquid MP media using a minimum of three biological replicates. On solid media, colony size was scored after four days. Growth curve experiments were conducted at 29 °C in an Excella E25 shaking incubator (New Brunswick Scientific, Edison, NJ) using a custom-built angled tube rack holder as previously described18. Optical density (OD600) was measured at 600 nm using a Spectronic 20D spectrophotometer (Milton Roy Company, Warminster, PA). For strains with extended growth lags, suppression and acclimation was assessed. Strains from the growth curves were streaked onto methanol La3+ medium after they reached stationary phase. If the parent stock strain did not grow on methanol La3+ medium and the strain post-growth curve grew, acclimation versus suppression was tested. Strains were passaged from the methanol medium plate to a succinate medium plate. After colonies grew on succinate medium, they were streaked back onto methanol La3+ medium. If strains retained the ability to grow on methanol medium, it was concluded that growth was due to a suppressor mutation. If strains lost the ability to grow on methanol medium after succinate passage, it was concluded growth was due to acclimation and not a genetic change.
Transcriptional reporter fusion assays
M. extorquens AM1 strains carrying mxa, xox1, and lutH transcriptional reporter fusions (Table S2) which use venus78 as a fluorescent reporter were grown in MP media supplemented with methanol only or methanol and succinate with and without La3+ as indicated in the text. Once cells reached an OD600 of 0.6, expression was measured as relative fluorescent units (RFU) using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) and normalized to OD600 as previously described18. For measurements of xox1 and mxa expression before and after acclimation in the mxaF lutH strain, measurements were taken every three hours before after sub-culturing until stationary phase. Expression after acclimation is reported in Table 4 from cells harvested at an OD600 of 0.6.
La3+ depletion during M. extorquens AM1 growth
Overnight cultures of wild type, lutA, lutE, lutF, and lutH mutant strains were inoculated 1:50 into 250 mL polycarbonate flasks (Corning Inc., Corning, NY) containing 75 mL of MP medium 71. Succinate (3.75 mM) and methanol (125 mM) were added as carbon sources with 2 μM LaCl3. Flasks were incubated at 28 °C at 200 rpm in Innova 2,300 shaking incubators (Eppendorf, Hauppauge, NY) for 44 h. To monitor La3+ depletion during M. extorquens AM1 cultivation, the Arsenazo III assay was used79. 5 mL samples were collected at four different time points (2, 12, 16, and 24 h) and the concentration of La3+ remaining in the supernatant was calculated using the calibration curve prepared as previously described79. A control of three uninoculated flasks containing MP medium with 2 μM LaCl3 were considered to determine La3+ adsorption by the flasks which was subtracted from the culture measurements. The initial concentration of La3+ in the media (before growth) was measured using the Arsenazo III assay in the same way as described above. Significant differences between depletion of La3+ by different strains were calculated using One-way ANOVA followed by a t-test.
Cellular locations of Ln visualized using transmission electron microscopy (TEM)
Sample preparation for TEM: wild type, lutA, lutE, lutF, and lutH mutant strains were grown in MP medium containing 125 mM methanol and 3.75 mM succinate as carbon sources with or without the addition of 20 μM LaCl3 until they reached an OD600 of ~ 0.6. 3 mL of cells was harvested by centrifugation for 3 min at 1,500×g at room temperature and fixed for 30 min in 1 mL of 2.5% (v/v) glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in 0.1 M cacodylate buffer (Electron Microscopy Sciences, Hatfield, PA). After fixation, cells were pelleted by centrifugation for 3 min at 1,500×g and washed with 1 mL of 0.1 M cacodylate buffer. Cell pellets were embedded in 2% (w/v) agarose and washed three times with 0.1 M cacodylate buffer. When indicated, pellets in agarose blocks were stained for 30 min in 1% osmium tetroxide in 0.1 M cacodylate buffer. Samples were washed three times with 0.1 M cacodylate buffer, dehydrated in acetone, and embedded in Spurr resin (Electron Microscopy Sciences, Hatfield, PA). Blocks were polymerized at 60ºC for 48 h. 70 nm sections were obtained with a Power Tome XL ultramicrotome (RMC Boeckeler Instruments, Tucson AZ), deposited on 200 mesh carbon coated grids, and stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA). To assess the presence of La3+ by EDS, sections were left unstained. To image the distribution of cellular La3+, a TEM JOEL 1,400 Flash (Japan Electron Optics Laboratory, Tokyo, Japan) was used. Detection of La3+ in the cells and high-resolution imaging were done with a JEOL 2200FS (Japan Electron Optics Laboratory, Tokyo, Japan) operated at 200 kV. X-ray energy dispersive spectroscopy was performed using an Oxford Instruments INCA system (Abingdon, United Kingdom).
Supplementary information
Acknowledgements
We would like to thank Dr. Lena Daumann and Dr. Nathan Good for critical review of this manuscript. We would like to thank SJSU General Microbiology students who isolated transposon mutants and purified genomic DNA for sequencing as part of a class research project. The mutant hunt in this study was inspired by Dr. Elizabeth Skovran’s undergraduate research mentor, Dr. Marc Rott at the University of Wisconsin-LaCrosse. We would like to thank Timothy Andriese for assistance with sequencing of the transposon mutant DNA and all Skovran lab members for assistance with growth curves and transcriptional reporter fusion assays. TEM work was done at the Center for Advanced Microscopy, MSU. We would like to thank Dr. Alicia Withrow for invaluable assistance with TEM experiments and Dr. Xudong Fan for his expertise using TEM-EDS. This material is based upon work supported by the National Science Foundation under Grant No. 1750003 and by a California State University Program for Education and Research in Biotechnology (CSUPERB) Joint Venture Grant. P.R-J. was supported by the National Science Foundation under Grant No. 1750003. E.M.A and F.Y. were supported by the National Science Foundation Research Initiative for Scientific Enhancement (RISE) award under Grant No. R25GM071381. F.Y. was also supported by the National Institute of Health Maximizing Access to Research Careers Undergraduate Student Training in Academic Research (MARC U-STAR) award under Grant No. 4T34GM008253. Funding for transposon DNA isolation, PCR, and sequencing was provided by San José State University through the Department of Biological Sciences.
Author contributions
E.S., N.C.M-G., P.R-J., and H.N.V. directed experiments. N.C.M-G., and E.S. wrote the manuscript. P.R-J. conducted microscopy and Ln transport experiments. H.N.V, G.A.S, J.C., and E.C carried out transposon mutagenesis studies. H.N.V., G.A.S., R.C., J.C., C.R., E.M.A., E.C., N.F.L., and F.Y. constructed strains and conducted growth experiments. R.C., C.H., J.P.W., and G.A.S. conducted expression studies. R.T.N created TransMapper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Paula Roszczenko-Jasińska and Huong N. Vu.
Contributor Information
Norma C. Martinez-Gomez, Email: mart1754@msu.edu
Elizabeth Skovran, Email: elizabeth.skovran@sjsu.edu.
Supplementary information
is available for this paper at 10.1038/s41598-020-69401-4.
References
- 1.Zepf, V., Reller, A., Rennie, C., Ashfield, M. & Simmons, J. Materials Critical to the Energy Industry. An Introduction. (BP, 2014).
- 2.Martinez-Gomez NC, Vu HN, Skovran E. Lanthanide chemistry: from coordination in chemical complexes shaping our technology to coordination in enzymes shaping bacterial metabolism. Inorg. Chem. 2016;55:10083–10089. doi: 10.1021/acs.inorgchem.6b00919. [DOI] [PubMed] [Google Scholar]
- 3.Teo RD, Termini J, Gray HB. Lanthanides: applications in cancer diagnosis and therapy. J. Med. Chem. 2016;59:6012–6024. doi: 10.1021/acs.jmedchem.5b01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattocks JA, Ho JV, Cotruvo JA. A selective, protein-based fluorescent sensor with picomolar affinity for rare earth elements. J. Am. Chem. Soc. 2019;141:2857–2861. doi: 10.1021/jacs.8b12155. [DOI] [PubMed] [Google Scholar]
- 5.Hibi Y, et al. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J. Biosci. Bioeng. 2011;111:547–549. doi: 10.1016/j.jbiosc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 6.Pol A, et al. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol. 2014;16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa T, et al. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS ONE. 2012;7:e50480. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitriyanto NA, et al. Ce 3+ -induced exopolysaccharide production by Bradyrhizobium sp. MAFF211645. J. Biosci. Bioeng. 2011;111:146–152. doi: 10.1016/j.jbiosc.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J. Functional role of lanthanides in enzymatic activity and transcriptional regulation of pyrroloquinoline quinone-dependent alcohol dehydrogenases in Pseudomonas putida KT2440. MBio. 2017;8:e00570–e617. doi: 10.1128/mBio.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wehrmann M, Berthelot C, Billard P, Klebensberger J. The PedS2/PedR2 two-component system is crucial for the rare earth element switch in Pseudomonas putida KT2440. mSphere. 2018;3:e00376–e003718. doi: 10.1128/mSphere.00376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L, et al. Lanthanide-dependent methanol dehydrogenase from the legume symbiotic nitrogen-fixing bacterium Bradyrhizobium diazoefficiens strain USDA110. Enzyme Microb. Technol. 2019;130:109371. doi: 10.1016/j.enzmictec.2019.109371. [DOI] [PubMed] [Google Scholar]
- 12.Skovran E, Martinez-Gomez NC. Just add lanthanides. Science. 2015;348:862–863. doi: 10.1126/science.aaa9091. [DOI] [PubMed] [Google Scholar]
- 13.van Teeseling MCF, et al. Expanding the verrucomicrobial methanotrophic world: description of three Novel Species of Methylacidimicrobium gen nov. Appl. Environ. Microbiol. 2014;80:6782–6791. doi: 10.1128/AEM.01838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, et al. Rare earth element alcohol dehydrogenases widely occur among globally distributed, numerically abundant and environmentally important microbes. ISME J. 2019;13:2005–2017. doi: 10.1038/s41396-019-0414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keltjens JT, Pol A, Reimann J, Op Den Camp HJM. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 2014;98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 16.Chistoserdova L. Lanthanides: new life metals? World J. Microbiol. Biotechnol. 2016;32:138. doi: 10.1007/s11274-016-2088-2. [DOI] [PubMed] [Google Scholar]
- 17.Good NM, et al. Pyrroloquinoline quinone-containing ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multi-carbon substrates. J. Bacteriol. 2016;198:3109–3118. doi: 10.1128/JB.00478-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vu HN, et al. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J. Bacteriol. 2016;198:1250–1259. doi: 10.1128/JB.00937-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prejanò M, Iziana Marino T, Russo N, Marino T, Russo N. How can methanol dehydrogenase from Methylacidiphilum fumariolicum work with the alien Ce III ion in the active center? A theoretical study. Chem. A Eur. J. 2017;23:8652–8657. doi: 10.1002/chem.201700381. [DOI] [PubMed] [Google Scholar]
- 20.McSkimming A, Cheisson T, Carroll PJ, Schelter EJ. functional synthetic model for the lanthanide-dependent quinoid alcohol dehydrogenase active site. J. Am. Chem. Soc. 2018;140:1223–1226. doi: 10.1021/jacs.7b12318. [DOI] [PubMed] [Google Scholar]
- 21.Lumpe H, Pol A, Op Den Camp H, Daumann L. Impact of the lanthanide contraction on the activity of a lanthanide-dependent methanol dehydrogenase: a kinetic and DFT study and DFT study. Dalt. Trans. 2018;47:10463–10472. doi: 10.1039/c8dt01238e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Featherston ER, et al. Biochemical and structural characterization of XoxG and XoxJ and their roles in lanthanide-dependent methanol dehydrogenase activity. ChemBioChem. 2019;20:2360–2372. doi: 10.1002/cbic.201900184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chistoserdova L, Chen S-W, Lapidus A, Lidstrom ME. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J. Bacteriol. 2003;185:2980–2987. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skovran E, Palmer AD, Rountree AM, Good NM, Lidstrom ME. XoxF is required for expression of methanol dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 2011;193:6032–6038. doi: 10.1128/JB.05367-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Good NM, Moore RS, Suriano CJ, Martinez-Gomez NC. Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci. Rep. 2019;9:4248. doi: 10.1038/s41598-019-41043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu F, Beck DAC, Lidstrom ME. MxaY regulates the lanthanide-mediated methanol dehydrogenase switch in Methylomicrobium buryatense. PeerJ. 2016;4:e2435. doi: 10.7717/peerj.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semrau J. Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium. OB. 2016;3:1–20. doi: 10.1093/femsle/fnw129. [DOI] [PubMed] [Google Scholar]
- 28.Masuda S, et al. Lanthanide-dependent regulation of methylotrophy in Methylobacterium aquaticum Strain 22A. mSphere. 2018;3:e00462-17. doi: 10.1128/mSphere.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skovran E, Raghuraman C, Martinez-Gomez NC. Lanthanides in methylotrophy. Methyl. Methyl. Communit. 2019;1:101–116. doi: 10.21775/cimb.033.101. [DOI] [PubMed] [Google Scholar]
- 30.Springer AL, Morris CJ, Lidstrom ME. Molecular analysis of mxbD and mxbM, a putative sensor-regulator pair required for oxidation of methanol in Methylobacterium extorquens AM1. Microbiology. 1997;143:1737–1744. doi: 10.1099/00221287-143-5-1737. [DOI] [PubMed] [Google Scholar]
- 31.Ochsner AM, et al. Use of rare-earth elements in the phyllosphere colonizer Methylobacterium extorquens PA1. Mol. Microbiol. 2019;111:1152–1166. doi: 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daumann LJ. Essential and ubiquitous: the emergence of lanthanide metallobiochemistry. Angew. Chem. Int. Ed. 2019;58:12795–12802. doi: 10.1002/anie.201904090. [DOI] [PubMed] [Google Scholar]
- 33.Cotruvo, Jr., J. A., Featherston, E. R., Mattocks, J. A., Ho, J. V. & Laremore, T. N. Lanmodulin: a highly selective lanthanide-binding protein from a lanthanide-utilizing bacterium. J. Am. Chem. Soc. 140, 15056–15061 (2018). [DOI] [PubMed]
- 34.Sawyer EB, Barker PD. Continued surprises in the cytochrome c biogenesis story. Protein Cell. 2012;3:405–409. doi: 10.1007/s13238-012-2912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Versantvoort W, et al. Characterization of a novel cytochrome c as the electron acceptor of XoxF-MDH in the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV. Biochim. Biophys. Acta. 2019;1867:595–603. doi: 10.1016/j.bbapap.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Zheng C, Huang Y, Zhao J, Chistoserdova F. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. MBio. 2018;9:e02430–e2517. doi: 10.1128/mBio.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek M, Park T, Heo L, Park C, Seok C. GalaxyHomomer: a web server for protein homo-oligomer structure prediction from a monomer sequence or structure. Nucleic Acids Res. 2017;45:W320–W324. doi: 10.1093/nar/gkx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou H-H, Marx CJ. Optimization of gene expression through divergent mutational paths. Cell Rep. 2012;1:133–140. doi: 10.1016/j.celrep.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klaus T, Joerger R, Olsson E, Granqvist C-G. Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. 1999;96:13611–13614. doi: 10.1073/pnas.96.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Groom JD, Ford SM, Pesesky MW, Lidstrom ME. A mutagenic screen identifies a TonB-dependent receptor required for the lanthanide metal switch in the type I methanotroph Methylotuvimicrobium buryatense 5GB1C. J. Bacteriol. 2019;201:e00120-19. doi: 10.1128/JB.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maleke M, et al. Biomineralization and bioaccumulation of europium by a thermophilic metal resistant bacterium. Front. Microbiol. 2019;10:1–10. doi: 10.3389/fmicb.2019.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leeson TS, Higgs G. Lanthanum as an intracellular stain for electron microscopy. Histochem. J. 1982;14:553–560. doi: 10.1007/BF01011888. [DOI] [PubMed] [Google Scholar]
- 44.Bayer ME, Bayer MH. Lanthanide accumulation in the periplasmic space of Escherichia coli B. J. Bacteriol. 1991;173:141–149. doi: 10.1128/jb.173.1.141-149.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merroun ML, Ben Chekroun K, Arias JM, Alez-Munoz MTGA. Lanthanum fixation by Myxococcus xanthus: cellular location and extracellular polysaccharide observation. Chemosphere. 2003;52:113–120. doi: 10.1016/S0045-6535(03)00220-0. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto H, Mannami MNT. Dynamical theory of electron diffraction for the electron microscopic image of crystal lattices. II. Image of superposed crystals (Moiré pattern) Philos. Trans. R. Soc. 1961;253:490–516. [Google Scholar]
- 47.Good, N. M. et al. Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function. J. Biol. Chem. 295, 8272–8284 (2020) [DOI] [PMC free article] [PubMed]
- 48.Wehrmann M, Berthelot C, Billard P, Klebensberger J. Rare earth element (REE)-dependent growth of Pseudomonas putida KT2440 relies on the ABC-transporter Peda1A2BC and is influenced by iron availability. Front. Microbiol. 2019;10:2494. doi: 10.3389/fmicb.2019.02494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langley S, Beveridge TJ. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl. Environ. Microbiol. 1999;65:489–498. doi: 10.1128/aem.65.2.489-498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gu W, Semrau JD. Copper and cerium-regulated gene expression in Methylosinus trichosporium OB3b. Appl Microbiol Biotechnol. 2017;101:8499–8516. doi: 10.1007/s00253-017-8572-2. [DOI] [PubMed] [Google Scholar]
- 51.Chistoserdova L, Lidstrom ME. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology. 1997;143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 52.Toyama H, Inagaki H, Matsushita K, Anthony C, Adachi O. The role of the MxaD protein in the respiratory chain of Methylobacterium extorquens during growth on Methanol. Biochim. Biophys. Acta. 2003;1647:372–375. doi: 10.1016/s1570-9639(03)00097-9. [DOI] [PubMed] [Google Scholar]
- 53.Martins AM, et al. A two-component protease in Methylorubrum extorquens with high activity toward the peptide precursor of the redox cofactor pyrroloquinoline quinone. J. Biol. Chem. 2019;294:15025–15036. doi: 10.1074/jbc.RA119.009684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bultreys A, Gheysen I. Production and comparison of peptide siderophores from strains of distantly related Pathovars of Pseudomonas syringae and Pseudomonas viridiflava LMG 2352. Appl. Environ. Microbiol. 2000;66:325–331. doi: 10.1128/aem.66.1.325-331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhan G, Li D, Zhang L. Aerobic bioreduction of nickel(II) to elemental nickel with concomitant biomineralization. Appl. Microbiol. Biotechnol. 2012;96:273–281. doi: 10.1007/s00253-011-3827-9. [DOI] [PubMed] [Google Scholar]
- 56.Bai HJ, Zhang ZM, Guo Y, Yang GE. Biosynthesis of cadmium sulfide nanoparticles by photosynthetic bacteria Rhodopseudomonas palustris. Colloids Surf. B. 2009;70:142–146. doi: 10.1016/j.colsurfb.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 57.Sousa T, Chung A-P, Pereira A, Piedade AP, Morais PV. Aerobic uranium immobilization by Rhodanobacter A2–61 through formation of intracellular uranium-phosphate complexes. Metallomics. 2013;5:390–397. doi: 10.1039/c3mt00052d. [DOI] [PubMed] [Google Scholar]
- 58.Couradeau E, et al. An early-branching microbialite cyanobacterium forms intracellular carbonates. Science. 2012;336:459–462. doi: 10.1126/science.1216171. [DOI] [PubMed] [Google Scholar]
- 59.Debieux CM, et al. A bacterial process for selenium nanosphere assembly. Proc. Natl. Acad. Sci. 2011;108:13480–13485. doi: 10.1073/pnas.1105959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahn-Lee L, Komeili A. The magnetosome model: Insights into the mechanisms of bacterial biomineralization. Front. Microbiol. 2013;4:352. doi: 10.3389/fmicb.2013.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat. Rev. Microbiol. 2017;15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kulaev I. The Biochemistry of Inorganic Polyphosphates. Hoboken: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 63.Friedberg I, Avigad G. Structures containing polyphosphate in Micrococcus lysodeikticus. J. Bacteriol. 1968;96:544–553. doi: 10.1128/jb.96.2.544-553.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pallerla SR, et al. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 2005;243:133–140. doi: 10.1016/j.femsle.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 65.Widra A. Metachromatic granules of microorganisms. J. Bacteriol. 1959;78:664–670. doi: 10.1128/jb.78.5.664-670.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krossa S, Faust A, Ober D, Scheidig AJ. Comprehensive structural characterization of the bacterial homospermidine synthase-an essential enzyme of the polyamine metabolism. Sci. Rep. 2015;6:19501. doi: 10.1038/srep19501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wortham BW, Patel CN, Oliveira M. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv. Exp. Med. Biol. 2007;603:106–115. doi: 10.1007/978-0-387-72124-8_9. [DOI] [PubMed] [Google Scholar]
- 68.Peng YC, Lu CY, Li G, Eichenbaum Z, Lu CD. Induction of the pho regulon and polyphosphate synthesis against spermine stress in Pseudomonas aeruginosa. Mol. Microbiol. 2017;104:1037–1051. doi: 10.1111/mmi.13678. [DOI] [PubMed] [Google Scholar]
- 69.Motomura K, Takiguchi N, Ohtake H, Kuroda A. Polyamines affect polyphosphate accumulation in Escherichia coli. J. Environ. Biotechnol. 2006;6:41–46. [Google Scholar]
- 70.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delaney NF, et al. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS ONE. 2013;8:e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marx CJ. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marx CJ, Lidstrom ME. Broad-host-range Cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques. 2002;33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 74.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1983;1:784–791. [Google Scholar]
- 75.Jacobs MA, et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 2003;100:14339–14344. doi: 10.1073/pnas.2036282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marx CJ, O’Brien BN, Breezee J, Lidstrom ME. Novel methylotrophy genes of Methylobacterium extorquens AM1 identified by using transposon mutagenesis including a putative dihydromethanopterin reductase. J. Bacteriol. 2003;185:669–673. doi: 10.1128/JB.185.2.669-673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manoil C, Traxler B. Insertion of in-frame sequence tags into proteins using transposons. Methods. 2000;20:55–61. doi: 10.1006/meth.1999.0905. [DOI] [PubMed] [Google Scholar]
- 78.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 79.Hogendoorn C, et al. Facile arsenazo III-based assay for monitoring rare earth element depletion from cultivation media for methanotrophic and methylotrophic bacteria. Appl. Environ. Microbiol. 2018;84:e02887–e2917. doi: 10.1128/AEM.02887-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.