Abstract
Background
Abnormalities of lipid metabolism contributing to the autism spectrum disorder (ASD) pathogenesis have been suggested, but the mechanisms are not fully understood. We aimed to characterize the lipid metabolism in ASD and to explore a biomarker for clinical evaluation.
Methods
An age-matched case-control study was designed. Lipidomics was conducted using the plasma samples from 30 children with ASD compared to 30 typical developmental control (TD) children. Large-scale lipoprotein analyses were also conducted using the serum samples from 152 children with ASD compared to 122 TD children. Data comparing ASD to TD subjects were evaluated using univariate (Mann-Whitney test) and multivariate analyses (conditional logistic regression analysis) for main analyses using cofounders (diagnosis, sex, age, height, weight, and BMI), Spearman rank correlation coefficient, and discriminant analyses.
Findings
Forty-eight significant metabolites involved in lipid biosynthesis and metabolism, oxidative stress, and synaptic function were identified in the plasma of ASD children by lipidomics. Among these, increased fatty acids (FAs), such as omega-3 (n-3) and omega-6 (n-6), showed correlations with clinical social interaction score and ASD diagnosis. Specific reductions of very-low-density lipoprotein (VLDL) and apoprotein B (APOB) in serum of ASD children also were found by large-scale lipoprotein analysis. VLDL-specific reduction in ASD was correlated with APOB, indicating VLDL-specific dyslipidaemia associated with APOB in ASD children.
Interpretation
Our results demonstrated that the increases in FAs correlated positively with social interaction are due to VLDL-specific degradation, providing novel insights into the lipid metabolism underlying ASD pathophysiology.
Funding
This study was supported mainly by MEXT, Japan.
Keywords: Autism spectrum disorder, Lipid metabolism, VLDL, Social communication, Polyunsaturated fatty acid, Oxidative stress
Research in context.
Evidence before this study
To understand autism spectrum disorder (ASD), multifaceted researches, such as genetic approaches, imaging research, and animal model studies have been developed, but complete understanding has not been achieved. Recent studies demonstrated decreases in APOB100 or cholesterol high-density lipoprotein and an increase in total triglyceride in the peripheral plasma of children with ASD. Other studies also demonstrated that the Smith–Lemli–Opitz syndrome, which results in an inborn error of cholesterol synthesis, is associated with ASD. Accordingly, lipid metabolism has been suggested to have important roles in ASD aetiology. Since then, ASD metabolism has been investigated, but its roles and molecular mechanisms link to ASD remain unclear.
Added value of this study
Lipidomics was performed using peripheral blood samples from typical development controls and children with ASD. We identified 47 metabolites in ASD as novel targets involved in lipid biosynthesis and metabolism, oxidative stress, and synaptic function. Among these metabolites, we found significantly increased fatty acids, including omega-3 and omega-6 fatty acids. Interestingly, those fatty acids showed correlations with clinical social interaction score of ASD diagnosis. Performing detailed quantification of large-scale lipoproteins, we also found very-low-density lipoprotein (VLDL)–specific reductions in ASD. Our study demonstrated that increases in FAs are due to VLDL-specific degradation in ASD children.
Implications of all available evidence
Our research demonstrated the importance of lipid metabolism underlying the causes of ASD, particularly fatty acids and lipoproteins associated with social interaction. Comprehensive metabolomics information on metabolites provided may be useful for understanding ASD pathophysiology from the perspective of lipid metabolism, such as the mechanism of oxidative stress and/or nutrition for child development in ASD.
Alt-text: Unlabelled box
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder that causes pervasive abnormalities in social communication as well as repetitive behaviours and restricted interests. Worldwide, the prevalence of children diagnosed with ASD has increased significantly over recent decades [1,2]. Its aetiology is thought to involve complex, multigenic interactions, and possible environmental contributions [3]. Although several such risk factors have been implicated, including not only genetic and environmental factors, but also infectious, metabolic, and nutritional factors, the cause is known in <10% to 12% of cases [4, 5]. However, the biological mechanism underlying ASD is not fully understood. Early diagnosis and intervention improve the symptoms of ASD children [6], but scientific methods, such as biomarkers for early diagnosis, are lacking.
Lipids are essential factors in the neural development and have critical roles in neuronal migration, differentiation, morphogenesis, myelination, memory formation, and synaptic plasticity [[7], [8], [9], [10]], which are highly relevant to ASD [[11], [12], [13], [14]]. Previous studies have implicated lipid metabolism in ASD pathophysiology. For example, in the Smith–Lemli–Opitz syndrome (SLOS), which results in an inborn error of cholesterol (Chl) synthesis, approximately half of those diagnosed with SLOS also carry the diagnosis of ASD [15, 16]. SLOS is an autosomal recessive disorder caused by mutations in 7-dehydrocholesterol reductase (DHCR7) on chromosome 11q12–13, which encodes a terminal enzyme required for Chl biosynthesis [17]. Clinically, SLOS was characterised by a range of phenotypic abnormalities, including developmental delay, abnormal neural development, and abnormal peripheral lipid metabolism [16]. Cholesterol supplementation improved the symptoms of SLOS patients [18], but was less effective on the symptoms of ASD such as social communication in SLOS patients who developed ASD [16]. These studies suggest that the association between lipid metabolism and ASD, but clearly there are different mechanisms between SLOS and ASD pathophysiology.
Exploring the biomarker in ASD has been studied extensively by focusing on genes, proteins, lipids, and more using blood, urine, faeces, lymphocytes and postmortem brain from ASD children and adults. The serum proteomics study has demonstrated a reduction of apoprotein B (APOB)−100, and increases in FHR1, C1q, and FN1 in children with ASD [19]. Increased triglyceride (TG) and decreased HDL-Chl in plasma of ASD boys also have been reported, but clinical measures, such as the Autism Diagnostic Observation Schedule (ADOS) or the Autism Diagnostic Interview-Revised (ADI-R) in ASD diagnosis, were not used in this study [20]. Moreover, significant alterations of omega-3 (n-3) and omega-6 (n-6) fatty acids, including the ratio of n-3/n-6, have been reported in the plasma of ASD patients [7, 21, 22]. Imbalance of the n-3/n-6 ratio is associated with reductions of synaptic proteins expressions and synaptic vesicle density [8, 23]. A plasma FAs profiling study of ASD children using gas chromatography has shown increases in saturated FAs (SFAs), such as acetic acid and stearidonic acid, as well as decreases in polyunsaturated FAs (PUFAs), such as eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), and arachidonic acid (AA), in ASD children [24]. Thus, investigating lipid metabolism may uncover our understanding of biological mechanisms of ASD; however, previous studies had not achieved full understanding of the molecular mechanism of lipid metabolism in ASD.
On the other hand, previous studies from our group have identified inflammatory cytokines, such as transforming growth factor-β1 (TGF-β1), hepatocyte growth factor (HGF), epidermal growth factor (EGF), platelet-derived growth factor-BB (PDGF-BB), and tumor necrosis factor-α (TNF-α), in the peripheral blood of ASD children [[25], [26], [27], [28], [29]], suggesting increased inflammation in ASD children. Many findings have been reported regarding inflammation in ASD as a risk factor, which also is closely related to oxidative stress [30], for example mitochondrial dysfunction in ASD [31]. Increased oxidative protein and DNA damage have been reported to be associated with a decrease in plasma glutathione (GSH)/oxidized glutathione (GSSG) of ASD children [32]. GSH and GSH/GSSG also were decreased in the temporal cortex and cerebellum of the postmortem brain in ASD [33]. In addition, 3-nitrotyrosine, an oxidative protein damage marker, and 8-oxo-deoxyguanosine, an oxidative DNA damage marker, also were increased in the postmortem brain in ASD [33], demonstrating decreased GSH/GSSG redox/antioxidant capacity and increased oxidative stress in ASD. A study showing the relationship between oxidative stress and lipids has reported increased plasma levels of malonyldialdehyde (MDA), the end product of peroxidation of PUFA and related esters and a marker of lipid peroxidation, in ASD children [34, 35]. In addition, ASD children had reduced serum levels of key antioxidant proteins, transferrin (iron-binding protein), and ceruloplasmin (copper-binding protein) [34]. Accordingly, these studies also suggested that oxidative stress in ASD may be linked to lipid metabolism.
In our study, we characterised the mechanism of lipid metabolism underlying ASD pathology and explored a reliable biomarker for clinical evaluation, in particular for early diagnosis using peripheral blood of ASD children. Briefly, we showed metabolomic results in plasma of ASD children. We identified 48 metabolites involved in lipid biosynthesis and metabolism, oxidative stress, and synaptic function. Significantly increased FAs correlated with clinical social interaction score in ASD diagnosis (ADI-R A score) as well as lipoproteins-TG concentrations. We also analysed in detail lipoproteins using the serum of children with ASD, and identified a specific reduction of very-low-density lipoprotein (VLDL) which correlated with APOB reduction in ASD. However, we could not identify a reliable biomarker for clinical evaluation in this study. Together, our study demonstrated that the increases in FAs correlated with social interaction are due to VLDL-specific degradation, providing the novel insights into lipid metabolism in children with ASD.
2. Methods
2.1. Ethics statement
All procedures were approved by the ethics committee of the University of Fukui and the Hamamatsu University School of Medicine, and were conducted in accordance with the Ethical Guidelines for Medical and Health Research Involving Human Subjects of the Ministry of Health, Labour and Welfare of Japan. All participants were given a complete description of the study and provided written informed consent from their parent and/or legal guardian before enrolment.
2.2. Participants and diagnosis
We designed an age-matched case-control study to assess lipid metabolism in children with ASD following the Strengthening the Reporting of Observational Studies in Epidemiology guidelines for reporting case-control studies [36]. Subjects with ASD were recruited through advocacy groups in cooperation with the Asperger Society Japan (Nagoya, Japan) and the Miyagi Children's Hospital (Sendai, Japan). In contrast, we recruited age-matched TD subjects in Hamamatsu and Fukui, Japan by advertisement. According to the mean and standard deviation of serum lipid profile detected in a previous study [20], we statistically determined that the minimum sample required was 100 subjects in each group beforehand using G*Power (2 tails, effect size = 0.4, significance level = 0.05, power = 0.80) [37, 38].
At first 224 eligible cases were identified as autistic subjects and matched 1:1 with healthy control samples according to age. The reason why the sample size differed from the initial setting was that we realised we were missing height and weight data during the sample collection phase, and then we added the extra sample at that time. ASD was diagnosed by an experienced child psychiatrist based on the criteria outlined in the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision (DSM-IV-TR) based on clinical interviews and the Japanese version of the ADI-R. The ADI-R scoring was conducted by four authors (KW, TF, KM and KJT), who were experienced and reliable at diagnosing ASD with the Japanese version of the ADI-R. Then, based on these results, a DSM-IV-TR (American Psychiatric Association, 2000) diagnosis of autistic disorder was made for all subjects. The ADI-R provides a diagnostic algorithm for autism as described in the ICD-10 and DSM-IV. ADI-R is a semi-specially-formulated structured psychiatric interview with a parent, especially a mother. It is used to confirm diagnosis and evaluate the core symptoms of autism, for which we used three of its four domain scores: (1) the A domain quantifies impairment in social interaction (score, 0–32); (2) the BV domain quantifies impairment in communication (score, 0–26); and the C domain quantifies restricted, repetitive, and stereotyped behavior patterns and interests (score, 0–16). Higher scores indicate worse performance. The cutoff scores for positive symptoms in domains A, BV, and C are 10, 8, and 3, respectively. All subjects in the ASD group had scores above these thresholds. The D domain of the ADI-R was excluded because it corresponds to age of onset, and all subjects in the ASD group had onsets no later than 3 years old (D score < 1). All TD subjects underwent a comprehensive assessment of their medical history as interviewed by a physician to eliminate those with any neurologic or medical disorders. Specifically, we excluded those with fragile X syndrome, epileptic seizures, obsessive-compulsive disorder, affective disorders, and schizophrenia, together with those who had any additional psychiatric or neurologic diagnoses. All subjects were required to be drug naive including any dietary supplements and to have been free of cholesterol-lowering diets for at least 6 months before this study. The Structured Clinical Interview for DSM-IV (SCID) was used to assess for any personal or family history of past or present mental illness. We conducted the Wechsler Adult Intelligence Scale-Revised and Wechsler Intelligence Scale for Children-Third Edition of school age. We evaluated the developmental quotient (DQ) using the Kyoto Scale of Psychological Development for preschool age children. Previous reports have validated the DQ as being equivalent to the intelligence quotient (IQ) [39,40] (Supplementary Tables S1, S2).
Of 224 ASD subjects, five were excluded due to medication, four due to withdrawn consent, three due to liver dysfunction by blood test, nine due to high C-reactive protein (CRP) by blood test, and five due to no fasting at blood collection (two had high CRP and no fasting). Therefore, 200 subjects were temporarily registered in the ASD group (Fig. 1, Supplementary Tables S1, S2). Of 224 TD subjects recruited after those assessments, one was excluded due to withdrawn consent, three due to liver dysfunction by blood test, six due to high CRP by blood test, six due to no fasting at blood collection and two due to autistic finding by SCID. Therefore, 206 subjects were temporarily registered in the TD group (Fig. 1, Supplementary Tables S1, S2).
Fig. 1.
Flowcharts of trial participants for analyses.
(a, b) Flowchart illustrating the matched case-control study using peripheral blood of TD and ASD children for metabolic (a), and lipoprotein (b) profiling.
For the metabolome analysis, we randomly selected 30 ASD children from the temporarily registered 200 subjects (age, 8.27 ± 1.21 years; range, 5–11 years) and 30 TD children from the temporarily registered 206 TD subjects (age, 7.96 ± 1.44; range, 6–10 years). There also was no significant difference between the groups in age (P = 0.3154), weight (P = 0.5598), and height (P = 0.1752) as assessed by the Mann-Whitney test, but there was significant difference in the sex (P = 0.0470). The characteristics of the participants are described in Supplementary Table S1. For the large-scale lipoprotein analysis, 48 and 78 were excluded due to missing data (height and weight) from the temporarily registered 200 ASD and 206 TD subjects, respectively. Then, we enrolled 152 ASD children (age, 9.15 ± 3.96 years; range, 2–20 years) and 128 TD children (age, 9.08 ± 4.14 years; range, 2–20 years). There was no significant difference between the groups in age (P = 0.4413), weight (P = 0.2132), and height (P = 0.5156) as assessed by the Mann-Whitney test, but there was significant difference in the sex (P = 0.0375). The age information was based on the participant's date of birth at the date the blood was collected. The characteristics of the participants are summarised in Supplementary Table S2.
2.3. Blood sampling
Fasting blood samples were collected by venepuncture in a sitting position with a tourniquet between 0700 and 1200 h. Serum samples were kept at room temperature for 30 min and centrifuged at 3500 g for 10 min in a refrigerated centrifuge. Plasma samples were immediately centrifuged at 3500 g for 15 min in a refrigerated centrifuge. After centrifugation, supernatants of the samples were collected in aliquots of 200 µL and stored at −80 °C.
2.4. Blood test
Serum levels of aspartate aminotransferase (AST), alanine aminotransferase (ALT), γ-glutamyltransferase (γ-GTP), and CRP concentrations were quantified using a routine clinical biochemistry automatic analyser to exclude those with liver dysfunction or unknown inflammatory conditions that could affect lipid metabolism. All samples were analysed in triplicate and the mean value was used for analysis.
2.5. Metabolome analysis
Metabolome analysis was performed as a service by Human Metabolome Technologies, Inc. (Yamagata, Japan). Targeted 1200 free metabolites in the blood were quantified in the plasma fraction by capillary electrophoresis mass spectrometry (CE-MS) as described previously [41,42]. All samples were analysed in triplicate, and the mean value was used for analysis. Hierarchical cluster analysis and principal component analysis (PCA) analyses were performed using PeakStat and SampleStat, the proprietary statistical analysis software developed by Human Metabolome Technologies, Inc. Metabolic pathways and metabolic profiling were analysed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (available in the public domain at www.genome.jp/kegg) and the Human Metabolome Database (HMDB; hmdb.ca), respectively.
2.6. Lipoprotein analysis
Lipoprotein analysis was performed as a LipoSEARCH service by Skylight Biotech, Inc. (Akita, Japan). Briefly, Chl and TG profiles in serum lipoproteins were quantified using a gel filtration high performance liquid chromatography (GFHPLC) with patented analysing algorithm [43]. Chl and TG levels on three major classes (VLDL, high-density lipoprotein [HDL], and low-density lipoprotein [LDL]) were separated based on lipoprotein particle size. Lipoprotein particle numbers then were calculated from the data on Chl and TG levels using a newly developed program in which it was assumed that the volume ratio of the surface layer to the lipid core was equivalent for lipoproteins with the same particle size [43]. All samples were analysed in triplicate and the mean value was used for analysis.
2.7. Apoprotein analysis
Apoprotein analysis was performed as a service by Filgen, Inc. (Aichi, Japan). Serum levels of apoproteins (APOA1, APOA2, APOB, APOC3, APOE, APOH, and APOJ) were quantified using MILLIPLEX map human apolipoprotein magnetic bead panel (Cat#APOMAG-62 K; Merck Millipore, Burlington, MA, USA) and a clinical biochemistry automatic analyser (Bio-Plex 200 system; Bio-Rad Laboratories, Hercules, CA, USA). All samples were analysed in triplicate and the mean value was used for analysis.
2.8. Statistical analysis
All data are represented as means of biological independent experiments ± standard deviation (SD). The box and whisker plots show all points minimum-to-maximum value. The following statistics were used in this study. The D'Agostino and Pearson test was used to examine the normal distribution. Univariate analysis (Mann-Whitney test) was used for demographic characterization and metabolome candidate screening in the TD and ASD groups. Multivariate analysis (conditional logistic regression analysis) was used to examine the metabolome, lipoprotein, apoprotein, and liver function data between the TD and ASD groups. In conditional logistic regression analysis, diagnosis was used as the objective variable, age was considered as a matched variable, and demographic data (sex, height, weight, and body mass index [BMI]) were used as the explanatory variable. The Spearman rank correlation coefficient was used to examine the correlations among metabolome, lipoprotein, apoprotein, and/or ADI-R score. Linear regression was used only to calculate the best fit line to correlation analysis. The receiver operating characteristic (ROC) curve adjusted for age was used to determine the discrimination point between the TD and ASD groups. Discriminant analysis (linear discriminant analysis) was performed to calculate the discrimination score for distinguishing TD and ASD. The D'Agostino and Pearson test, Mann-Whitney test, Spearman rank correlation coefficient test, linear regression, and ROC curve analyses were performed using GraphPad Prism8 (GraphPad Software, San Diego, CA, USA). Conditional logistic regression analysis, and linear discriminant analysis were performed using BellCurve for Excel (Social Survey Research Information Co., Ltd., Tokyo, Japan). Asterisks indicate P values (****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05). P < 0.05 indicated statistical significance in univariate and multivariate analyses. In the correlation coefficient, r = 0.7 to 1, r = 0.4 to 0.7, r = 0.2 to 0.4, and r = 0 to 0.2 indicated a fairly strong, some, weak and almost no correlation, respectively.
3. Results
3.1. Distinct alterations of lipid metabolism in ASD
To characterize the lipid metabolism underlying ASD pathophysiology and explore a biomarker for clinical evaluation, we designed the study as a matched case-control study and recruited children with ASD and age-matched TD controls. First, 224 ASD and 224 TD subjects were evaluated for eligibility. Then, totals of 21 ASD and 18 TD children were excluded due to medication, withdrawal, liver dysfunction, and so forth (see Methods; Fig. 1). We first conducted a comprehensive metabolomic analysis targeting 1200 free metabolites in plasma samples of 30 TD children 6 to 10 years old and 30 ASD children 5 to 11 years old randomly selected from the participants (Fig. 1, Supplementary Table S1) using CE-MS, and then detected 258 metabolites.
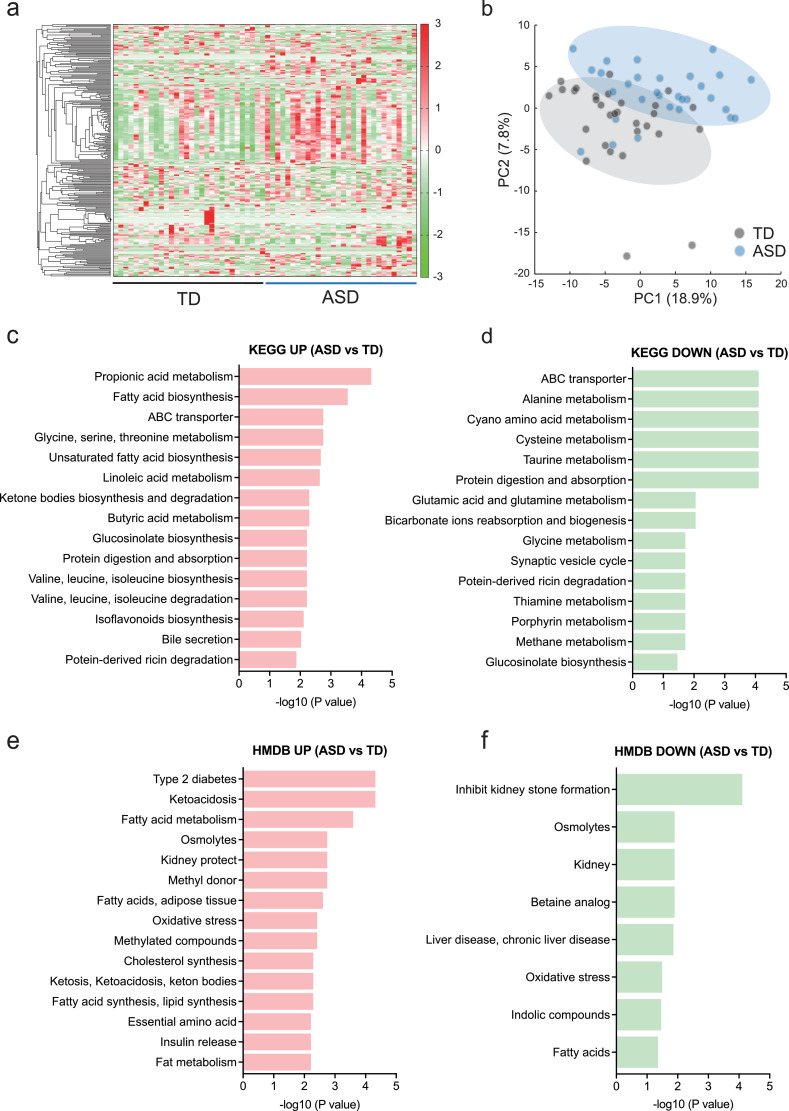
Metabolic profiles could be separated clearly between the TD and ASD groups in a heat map (Fig. 2a) and by PCA (Fig. 2b). Among 258 detected metabolites, we first identified 71 candidates altered in ASD using the Mann-Whitney test (Supplementary Fig. S1). Then, we performed multivariate analysis on these 71 candidates considering the confounders (diagnosis, sex, height, weight, and BMI) and the matched confounder (age), and identified 48 significant metabolites in plasma of ASD children compared to TD children (Table 1). Strikingly, lipid-related metabolites dominated the overall group of significant differences (Table 1). For instance, 2-aminobutyric acid (Odds ratio [OR] = 2.12, 95% confidence interval [CI] = 1.51 to 3.40, P = 0.0002, A/T ratio = 1.5), acylcarnitine (AC)(18:2) (OR = 1.03, 95% CI = 1.02 to 1.06, P = 0.0016, A/T ratio = 1.7), 2-Hydroxybutyric acid (OR = 2.17, 95% CI = 1.42 to 3.79, P = 0.0017, A/T ratio = 1.6), FA(16:1) palmitoleic acid (OR = 1.00, 95% CI = 1.00 to 1.01, P = 0.0069, A/T ratio = 2.2), FA(14:1) myristoleic acid (OR = 1.02, 95% CI = 1.01 to 1.03, P = 0.0128, A/T ratio = 1.4), and FA(20:5) eicosapentaenoic acid (EPA; OR = 1.02, 95% CI = 1.00 to 1.03, P = 0.0147, A/T ratio = 2.1) were significantly increased in the ASD group (Table 1). In contrast, alanine (Ala; OR = 0.93, 95% CI = 0.89 to 0.97, P = 0.0007, A/T ratio = 0.7), β-Ala (OR = 9.07e–4, 95% CI = 2.73e–006 to 0.106, P = 0.0084, A/T ratio = 0.9), and glutamine (OR = 2.57, 95% CI = 0.85 to 0.97, P = 0.0101, A/T ratio = 0.9) were significantly decreased in the ASD group (Table 1). These results demonstrated the abnormalities in overall metabolism centering on lipids of the patients with ASD. Especially, the increases in metabolites, such as FAs and ACs were remarkable. Moreover, increased ACs also suggested mitochondrial dysfunction and FAs hypermetabolism in ASD.
Fig. 2.
Metabolic profiles in children with ASD.
(a) Heat map visualizing metabolome results. (b) PCA showing clusters of TD and ASD. (c, d) Upregulated (c) and downregulated (d) metabolite-related biological and molecular processes using the KEGG database (genome.jp/keg) in ASD metabolome. (e, f) Upregulated (e) and downregulated (f) metabolites-related human metabolisms and diseases using the HMDB (hmdb.ca) in ASD metabolome. n = 30/condition.
Table 1.
Summary of metabolomic results.
| Compound | TD | ASD | Unit | TD vs ASD |
|||
|---|---|---|---|---|---|---|---|
| A/T ratio | P value | ||||||
| 2-Aminobutyric acid | 6.87 ± 1.60 n = 30 | 10.45 ± 2.58, n = 30 | ×103 RA | 1 0.5 | 0.0002 | *** | |
| Ala | 88.23 ± 22.02, n = 30 | 64.93 ± 19.46, n = 30 | ×103 RA | 0 0.7 | 0.0007 | *** | |
| AC(18:2) | 85.81 ± 42.82, n = 30 | 149.12 ± 44.56, n = 30 | ×106 RA | 1 0.7 | 0.0016 | ** | |
| 2-Hydroxybutyric acid | 3.46 ± 1.18, n = 30 | 5.56 ± 2.20, n = 30 | ×103 RA | 1 0.6 | 0.0017 | ** | |
| AC(16:1) | 34.41 ± 18.38, n = 28 | 76.27 ± 33.64, n = 29 | ×106 RA | 2 0.2 | 0.0017 | ** | |
| AC(14:3) | 6.06 ± 4.74, n = 29 | 12.83 ± 5.89, n = 30 | ×106 RA | 2 0.1 | 0.0025 | ** | |
| AC(16:2) | 8.46 ± 4.69, n = 28 | 18.73 ± 9.50, n = 30 | ×106 RA | 2 0.2 | 0.0026 | ** | |
| Pregnenolone sulfate | 16.03 ± 9.16, n = 30 | 27.55 ± 13.80, n = 30 | ×106 RA | 1 0.7 | 0.0027 | ** | |
| AC(14:2) | 6.062 ± 4.74, n = 29 | 98.87 ± 54.05, n = 30 | ×106 RA | 1 6.3 | 0.0029 | ** | |
| N,N-Dimethylglycine | 1.53 ± 0.37, n = 30 | 2.43 ± 1.51, n = 30 | ×103 RA | 1 0.6 | 0.0030 | ** | |
| O-Acetylcarnitine | 8.42 ± 1.67, n = 30 | 11.65 ± 3.96, n = 30 | ×103 RA | 1 0.4 | 0.0038 | ** | |
| Uric acid | 19.82 ± 4.28, n = 30 | 23.76 ± 6.51, n = 30 | ×103 RA | 1 0.2 | 0.0054 | ** | |
| Betaine | 21.55 ± 3.61, n = 30 | 25.15 ± 4.62, n = 30 | ×103 RA | 1 0.2 | 0.0055 | ** | |
| AC(14:1) | 103.85 ± 86.81, n = 29 | 217.20 ± 123.28, n = 30 | ×106 RA | 2 0.1 | 0.0065 | ** | |
| FA(16:1) Palmitoleic acid | 327.00 ± 278.79, n = 29 | 734.11 ± 477.78, n = 30 | ×106 RA | 2 0.2 | 0.0069 | ** | |
| Phenylacetaldehyde | 13.00 ± 2.78, n = 29 | 16.89 ± 3.39, n = 30 | ×106 RA | 1 0.3 | 0.0070 | ** | |
| FA(16:2) | 2.46 ± 1.35, n = 26 | 4.09 ± 1.97, n = 28 | ×106 RA | 1 0.7 | 0.0072 | ** | |
| β-Ala | 0.64 ± 0.16, n = 30 | 0.56 ± 0.14, n = 30 | ×103 RA | 0 0.9 | 0.0084 | ** | |
| FA(14:1)−1 | 6.71 ± 4.65, n = 28 | 13.78 ± 7.55, n = 28 | ×106 RA | 2 0.1 | 0. 0092 | ** | |
| Gln | 114.44 ± 8.29, n = 30 | 107.36 ± 11.17, n = 30 | ×103 RA | 0 0.9 | 0.0101 | * | |
| Daidzein | 5.39 ± 7.10, n = 18 | 13.30 ± 10.56, n = 24 | ×106 RA | 2 0.5 | 0.0102 | * | |
| FA(14:1) Myristoleic acid | 233.989 ± 65.17, n = 17 | 334.05 ± 110.85, n = 20 | ×106 RA | 1 0.4 | 0.0128 | * | |
| FA(18:2) Linoleic acid | 349.23 ± 249.76, n = 29 | 599.34 ± 332.30, n = 30 | ×106 RA | 1 0.7 | 0.0133 | * | |
| FA(18:3) Linolenic acid | 241.82 ± 179.78, n = 28 | 447.14 ± 283.38, n = 30 | ×106 RA | 1 0.8 | 0.0141 | * | |
| FA(20:5) cis-5,8,11,14,17-Eicosapentaenoic acid | 52.35 ± 47.41, n = 28 | 112.27 ± 78.90, n = 29 | ×106 RA | 2 0.1 | 0.0147 | * | |
| FA(t18:1) Ricinoleic acid | 14.70 ± 9.56 n = 28 | 25.01 ± 13.92, n = 29 | ×106 RA | 1 0.7 | 0.0148 | * | |
| FA(17:1) | 22.72 ± 16.55, n = 27 | 43.38 ± 25.97, n = 29 | ×106 RA | 1 0.9 | 0.0179 | * | |
| Benzoic acid | 0.84 ± 0.19, n = 30 | 0.67 ± 0.18, n = 21 | ×103 RA | 0 0.8 | 0.0201 | * | |
| AC(13:1) | 32.63 ± 24.25, n = 30 | 49.77 ± 24.63, n = 30 | ×106 RA | 1 0.5 | 0.0203 | * | |
| AC(20:1) | 9.42 ± 4.87, n = 28 | 13.71 ± 4.36, n = 29 | ×106 RA | 1 0.5 | 0.0208 | * | |
| Pipecolic acid | 0.51 ± 0.13, n = 30 | 0.43 ± 0.10, n = 30 | ×103 RA | 0 0.9 | 0.0222 | * | |
| Cysteine glutathione disulphide | 0.31 ± 0.15, n = 30 | 0.24 ± 0.08, n = 24 | ×103 RA | 0 0.8 | 0.0227 | * | |
| Leu | 68.18 ± 10.96, n = 30 | 76.59 ± 11.35, n = 30 | ×103 RA | 1 0.1 | 0.0250 | * | |
| FA(14:2) | 2.14 ± 0.98, n = 22 | 3.92 ± 2.43, n = 26 | ×106 RA | 1 0.8 | 0.0271 | * | |
| Loganin | 6.60 ± 2.73, n = 30 | 4.94 ± 1.69, n = 30 | ×106 RA | 0 0.7 | 0.0282 | * | |
| FA(c18:1) Oleic acid | 790.31 ± 642.27, n = 29 | 1350.17 ± 831.60, n = 30 | ×106 RA | 1 0.7 | 0.0284 | * | |
| Oleoyl ethanolamine | 24.06 ± 8. 0.47, n = 27 | 33.89 ± 14.59, n = 29 | ×106 RA | 1 0.4 | 0.0313 | * | |
| Gly | 37.93 ± 7.03, n = 30 | 34.01 ± 4.99, n = 30 | ×103 RA | 0 0.9 | 0.0325 | * | |
| FA(20:1) cis-11-Eicosenoic acid | 52.72 ± 42.86, n = 27 | 93.08 ± 60.55, n = 28 | ×106 RA | 1 0.8 | 0.0327 | * | |
| 21-Deoxycortisol | 10.24 ± 6.18, n = 30 | 21.71 ± 20.14, n = 30 | ×106 RA | 2 0.1 | 0.0328 | * | |
| 3-Hydroxybutyric acid | 5.44 ± 7.93, n = 30 | 19.26 ± 23.45, n = 30 | ×103 RA | 3 0.5 | 0.0348 | * | |
| 16α-Hydroxyestrone | 1.69 ± 0.55, n = 11 | 2.31 ± 0.53, n = 14 | ×106 RA | 1 0.4 | 0.0348 | * | |
| Urea | 382.58 ± 93.04, n = 30 | 435.65 ± 61.87, n = 30 | ×103 RA | 1 0.1 | 0.0353 | * | |
| FA(22:5) | 69.19 ± 61.03, n = 28 | 129.08 ± 91.64, n = 29 | ×106 RA | 1 0.9 | 0.0360 | * | |
| FA(20:4) Arachidonic acid | 233.86 ± 176.99, n = 29 | 393.57 ± 250.01, n = 30 | ×106 RA | 1 0.7 | 0.0395 | * | |
| FA(17:2) | 1.09 ± 0.39, n = 16 | 1.66 ± 0.67, n = 23 | ×106 RA | 1 0.5 | 0.0406 | * | |
| Trp | 17.60 ± 3.28, n = 30 | 15.97 ± 2.28, n = 30 | ×103 RA | 0 0.9 | 0.0421 | * | |
| FA(14:1)−2 | 3.76 ± 2.40, n = 27 | 7.16 ± 5.22, n = 29 | ×106 RA | 2 0.0 | 0.0453 | * | |
| N-Methylproline | 1.84 ± 1.43, n = 15 | 0.55 ± 0.26, n = 10 | ×103 RA | 0 0.3 | 0.0546 | ||
| Linoleyl ethanolamide | 8.94 ± 2.92, n = 27 | 11.87 ± 5.22, n = 29 | ×106 RA | 1 0.3 | 0.0548 | ||
| FA(20:3) Mead acid | 17.08 ± 13.78, n = 27 | 27.67 ± 16.90, n = 28 | ×106 RA | 1 0.6 | 0.0559 | ||
| Threonic acid | 3.10 ± 1.05, n = 13 | 2.14 ± 0.46, n = 8 | ×103 RA | 0 0.7 | 0.0568 | ||
| FA(16:0) Palmitic acid | 0.82 ± 0.57, n = 30 | 1.26 ± 0.77, n = 30 | ×106 RA | 1 0.5 | 0.0579 | ||
| Hypoxanthine | 0.31 ± 0.13, n = 23 | 0.39 ± 0.14, n = 27 | ×103 RA | 1 0.3 | 0.0580 | ||
| His | 21.30 ± 2.27, n = 30 | 22.74 ± 2.66, n = 30 | ×103 RA | 1 0.1 | 0.0607 | ||
| SDMA | 0.19 ± 0.03, n = 30 | 0.21 ± 0.03, n = 30 | ×103 RA | 1 0.1 | 0.0642 | ||
| 2—Hydroxyvaleric acid | 0.27 ± 0.07, n = 29 | 0.34 ± 0.11, n = 30 | ×103 RA | 1 0.2 | 0.0741 | ||
| Glycitein | 1.73 ± 0.76, n = 6 | 3.73 ± 2.56, n = 16 | ×106 RA | 2 0.2 | 0.0748 | ||
| FA(19:1) | 8.43 ± 6.06, n = 25 | 13.16 ± 7.72, n = 28 | ×106 RA | 1 0.6 | 0.0829 | ||
| AC(12:0) | 56.14 ± 49.95, n = 30 | 83.48 ± 44.28, n = 30 | ×106 RA | 1 0.5 | 0.0830 | ||
| FA(14:0) Myristic acid | 66.26 ± 46.95, n = 28 | 99.51 ± 57.13, n = 29 | ×106 RA | 1 0.5 | 0.0854 | ||
| FA(20:2) cis-11,14-Eicosadienoic acid | 39.45 ± 29.23, n = 27 | 59.91 ± 36.01, n = 29 | ×106 RA | 1 0.5 | 0.0864 | ||
| Diethanolamine | 0.25 ± 0.09, n = 21 | 0.38 ± 0.26, n = 24 | ×103 RA | 1 0.5 | 0.0906 | ||
| Cortexolone | 2.64 ± 1.25, n = 22 | 4.01 ± 1.98, n = 20 | ×106 RA | 1 0.5 | 0.1023 | ||
| Val | 95.47 ± 14.24, n = 30 | 102.46 ± 11.78, n = 30 | ×103 RA | 1 0.1 | 0.1032 | ||
| FA(20:3) cis-8,11,14-Eicosatrienoic acid | 38.45 ± 25.27, n = 27 | 55.72 ± 32.04, n = 29 | ×106 RA | 1 0.4 | 0.1074 | ||
| FA(22:4) | 40.73 ± 35.17, n = 27 | 66.27 ± 47.40, n = 29 | ×106 RA | 1 0.6 | 0.1078 | ||
| Thiaproline | 0.49 ± 0.18 n = 26 | 0.27 ± 0.06, n = 17 | ×103 RA | 0 0.5 | 0.1174 | ||
| AC(14:0) | 27.45 ± 15.53, n = 27 | 39.04 ± 17.56, n = 30 | ×106 RA | 1 0.4 | 0.1237 | ||
| FA(22:6) cis-4,7,10,13,16,19-Docosahexaenoic acid | 0.77 ± 0.71, n = 28 | 1.19 ± 0.81, n = 30 | ×103 RA | 1 0.5 | 0.1524 | ||
| Palmitoylcarnitine | 0.13 ± 0.07, n = 29 | 0.18 ± 0.06, n = 30 | ×103 RA | 1 0.3 | 0.1966 | ||
Data are represented as means (±SD). Asterisks indicate ***P < 0.001, **P < 0.01, *P < 0.05, conditional logistic regression analysis. TD, typical development; ASD, autism spectrum disorder; A/T ratio, ASD/TD ratio; AC, acylcarnitine; Ala, alanine; FA, fatty acid; Leu, leucine; Gly, glycine; Gln, glutamine; β-Ala, β-alanine; SDMA, symmetric dimethylarginine; Trp, tryptophan; Val, valine; His, histidine.
To delve further into the molecular mechanisms of lipid metabolic abnormalities in ASD, we analysed the metabolites-related biological and molecular processes using the KEGG database. We found upregulated metabolites in ASD were related to propionic acid metabolism (P < 0.0001), FA biosynthesis (P = 0.0003), unsaturated FA biosynthesis (P = 0.0020), and linoleic acid metabolism (P = 0.0022; Fig. 2c). In contrast, metabolites related to ATP-binding cassette (ABC) transporters (P < 0.0001), alanine metabolism (P < 0.0001), cysteine metabolism (P < 0.0001), glutamic acid and glutamine metabolism (P = 0.0083), and synaptic vesicle cycle (P = 0.0180; Fig. 2d) were downregulated in ASD. We also analysed the metabolites-related human metabolisms and diseases using the Human Metabolome Database (HMDB). Increased metabolites in ASD were involved in type 2 diabetes (P < 0.0001), ketoacidosis (P < 0.0001), and FAs metabolism (P = 0.0003; Fig. 2e). Moreover, metabolites related to FAs and adipose tissue (P = 0.0002), oxidative stress (P = 0.0004), and FA and lipid synthesis (P = 0.0005) were increased in the ASD group (Fig. 2e). On the other hand, decreases in metabolites involved in inhibiting kidney stone formation (P < 0.0001), osmolytes (P = 0.0119), kidney (P = 0.0119), and oxidative stress (P = 0.0030) were noted in the ASD group (Fig. 2f). Collectively, these results suggested that molecular mechanisms underlying ASD metabolism are due to hyperactivation of FAs metabolism, dysfunctions of cysteine and glutamate metabolism, and oxidative stress activation.
3.2. Increased n-3 and n-6 FAs correlated with social interaction in ASD
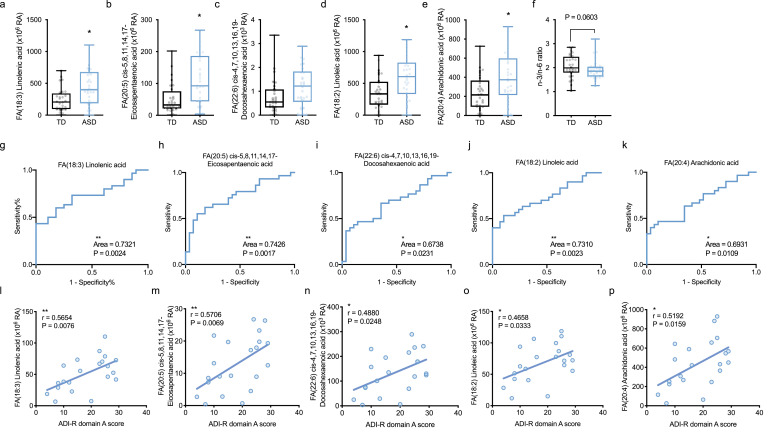
FAs are components of lipids roughly classified into SFAs and PUFAs. Notably, n-3 and n-6 FAs among PUFAs are important for nutrition and metabolism as well as central nervous system development and its function. Previous studies have reported that PUFAs are associated with ASD [21, 44]. Thus, they may be a key for uncovering the mystery of metabolism in ASD pathology. Hence, we first investigated the role of PUFAs in ASD metabolism. In n-3 FAs, we found significant increases in linolenic acid (OR = 1.00, 95% CI = 1.00 to 1.01, P = 0.0141, A/T ratio = 1.8) and EPA (OR = 1.02, 95% CI = 1.00 to 1.03, P = 0.0147, A/T ratio = 2.1), and an increased DHA (OR = 1.84, 95% CI = 0.85 to 4.56, P = 0.15, A/T ratio = 1.5; Table 1, Fig. 3a–c) in ASD by multivariate analysis (conditional logistic regression analysis). Likewise, in n-6 FAs, we also found a significant increase in linoleic acid (OR = 1.00, 95% CI = 1.00 to 1.01, P = 0.0133, A/T ratio = 1.7) and AA (OR = 1.00, 95% CI = 1.00 to 1.01, P = 0.0395, A/T ratio = 1.7; Table 1, Fig. 3d, 3e) in ASD by multivariate analysis. These results indicated that n-3 and n-6 FAs were distinctively increased in ASD, suggesting the balance of n-3/n-6 might be affected in ASD metabolism. To validate this possibility, the n-3 (sum of linolenic acid, EPA, and DHA) and n-6 (sum of linoleic acid and AA) ratio was compared between the TD and ASD groups by multivariate analysis. We found the tendency of n-3/n-6 imbalance in the ASD group (OR = 144.60, 95% CI = 0.97 to 41,489, P = 0.0603; Fig. 3f). This result suggested that the balance of n-3/n-6 may be impaired in ASD children.
Fig. 3.
Increased omega-3 and omega-6 fatty acids in ASD correlated with clinical social interaction score of ASD diagnosis.
(a–f) Increased n-3 and n-6 FAs in ASD. n-3 FAs; linolenic acid (a), EPA (b) and DHA (c) were increased in ASD. n-6 FAs; linoleic acid (d), and AA (e) also were increased in ASD. (f) The tendency of n-3/n-6 imbalance in ASD. The n-3 (sum of linolenic acid, EPA, and DHA) and n-6 (sum of linoleic acid, and AA) ratio was compared between the TD and ASD groups. Data are represented as box and whisker plots showing all points minimum-to-maximum value. Asterisk indicates *P < 0.05, conditional logistic regression analysis, n = 27–30/condition. (g–k) ROC curves of linolenic acid (g), EPA (h), DHA (i), linoleic acid (j), and AA (k). Asterisks indicate **P < 0.01, *P < 0.05, ROC adjusted for age, n = 27–30/condition. (l–p) n-3 or n-6 FAs correlated with clinical social interaction score in ASD diagnosis. Social interaction score (A score) of ADI-R was significantly correlated with linolenic acid (l), EPA (m), DHA (n), linoleic acid (o), or AA (p) in ASD. Asterisks indicate **P < 0.01, *P < 0.05, Spearman rank correlation coefficient test, n = 21/condition.
We next conducted ROC analysis adjusted for age to assess whether PUFAs are useful as biomarkers, and found that the corresponding areas under the curves (AUCs) were 0.7321 for linolenic acid (95% confidence interval [CI] = 0.60 to 0.86, P = 0.0024), 0.7426 for EPA (95% CI = 0.61 to 0.87, P = 0.0017), 0.6738 for DHA (95% CI = 0.53 to 0.81, P = 0.0231), 0.731 for linoleic acid (95% CI = 0.60 to 0.86, P = 0.0021), and 0.6931 for AA (95% CI = 0.56 to 0.83, P = 0.0109; Fig. 3g–k). These results indicated that each plasma concentration of PUFA is not a reliable biomarker in clinical evaluation.
Nevertheless, it remains interesting to examine whether PUFAs reflect the pathology of ASD. To address this, we examined the relationships between the clinical score of ASD symptom and those of FAs plasma concentrations by the Spearman rank correlation coefficient test. We used the social interaction score (A score) of the Autism Diagnostic Interview-Revised (ADI-R) as a clinical score of ASD symptom. The ADI-R A score was significantly and modestly correlated with plasma concentrations of linolenic acid (r = 0.5654, P = 0.00076), EPA (r = 0.5706, P = 0.0069), DHA (r = 0.4880, P = 0.0248), linoleic acid (r = 0.4658, P = 0.0333), and AA (r = 0.5192, P = 0.0159) in ASD (Fig. 3l–p) by the Spearman rank correlation coefficient test. We also examined whether the other FAs are correlated with ADI-R A score, and found that the majority of plasma FA concentrations, such as FA(16:0) palmitic acid (r = 0.5042, P = 0.0198) and FA(18:1) ricinoleic acid (r = 0.5784, P = 0.0060), also were correlated with ADI-R A score (Supplementary Fig. S2) by the Spearman rank correlation coefficient test. In contrast, there were no correlations between plasma AC concentrations and ADI-R A score (data not shown). Taken together, these results demonstrated that the significant increases in PUFAs in ASD, and those plasma concentrations distinctively correlated with social interaction symptoms of ASD, indicating that the increases in FAs including PUFAs are related strongly to the abnormalities of lipid metabolism in ASD.
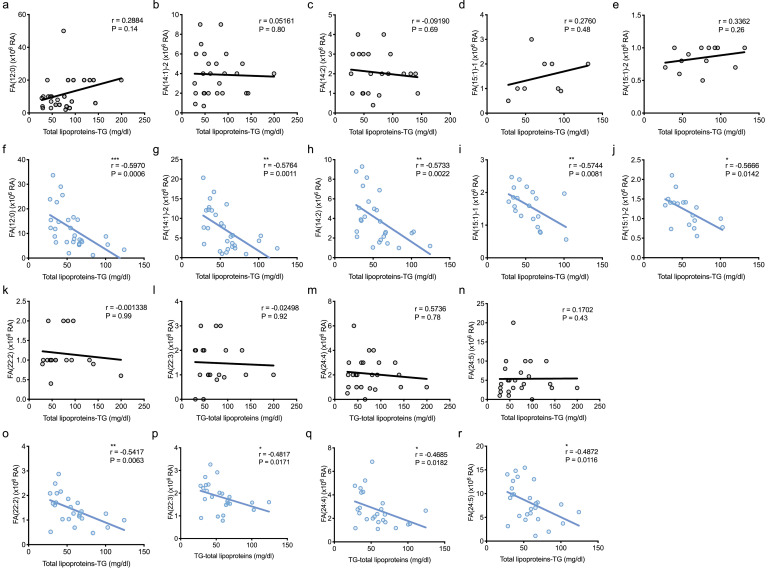
3.3. Correlations between increased FAs and lipoproteins-TG
To understand the mechanism of the increased metabolites, particularly FAs in ASD, we focused on lipoproteins, because FAs are produced by lipoprotein degradation, mainly TG. Therefore, we first analysed the correlations between FAs and total lipoproteins-TG concentrations in the TD group, but most FAs, such as FA(12:0) (r = 0.2884, P = 0.14), FA(14:1)-2 (r = 0.05161, P = 0.80, and FA(14:2) (r = −0.09190, P = 0.69), were not correlated with total lipoproteins-TG in TD (Fig. 4) by the Spearman rank correlation coefficient test. In contrast, we interestingly found that most FAs, such as FA(12:0) (r = −0.5970, P = 0.0006), FA(14:1)−2 (r = −0.5764, P = 0.0011), and FA(14:2) (r = −0.5733, P = 0.0022), were negatively and modestly correlated with total lipoproteins-TG in ASD (Fig. 4) by the Spearman rank correlation coefficient test. However, we could not observe direct correlations between total lipoproteins-TG and n-3 or n-6 FAs, such as linolenic acid (r = −0.1764, P = 0.35), EPA (r = −0.1399, P = 0.47), DHA (r = −0.1524, P = 0.42), linoleic acid (r = −0.2044, P = 0.28), and AA (r = −0.3415, P = 0.06) in the ASD group, respectively (data not shown). We also analysed the correlations of the increases in ACs in ASD, and found that most ACs also were negatively and modestly correlated with total lipoproteins-TG in ASD (Supplementary Fig. S3). Similarly, these ACs were not correlated with total lipoproteins-TG in TD as well as FAs (Supplementary Fig. S3). Together, these results indicate that the increases in FAs in ASD depended on the decreases in lipoproteins, particularly total lipoproteins-TG, suggesting the increase in FAs in ASD was due to lipoprotein degradations.
Fig. 4.
Majority of FAs were negatively correlated with lipoproteins-TG in ASD.
(a–e, k–n) No correlations of FAs were observed with total lipoproteins-TG levels in TD. FA(12:0) (a), FA(14:1)−2 (b), FA(14:2) (c), FA(15:1)−1 (d), FA(15:1)−2 (e), FA(22:2) (k), FA(22:3) (l), FA(24:4) (m), and FA(24:5) (n) were not correlated with total lipoproteins-TG. Spearman's r, n = 9–28/condition. (f–j, o–r) In contrast, increases of FAs were correlated with decreasing total lipoproteins-TG levels in ASD. FA(12:0) (f), FA(14:1)−2 (g), FA(14:2) (h), FA(15:1)−1 (i), FA(15:1)−2 (j), FA(22:2) (o), FA(22:3) (p), FA(24:4) (q), and FA(24:5) (r) were negatively correlated with total lipoproteins-TG. Asterisks indicate **P < 0.01, *P < 0.05, Spearman rank correlation coefficient test, n = 18–29/condition.
3.4. VLDL-specific reduction in ASD
To understand the role of lipoproteins in ASD, we further increased sample size and investigated in detail the serum lipoproteins using a GFHPLC [43]. After excluding the participants from the first entry, 200 ASD and 206 TD subjects were analysed for lipoprotein. Then, 48 ASD and 78 TD subjects were excluded due to missing data of height, weight, and BMI (see Methods). Therefore, we conducted a detailed lipoproteins analysis in serum samples from 128 TD and 152 ASD children 2 to 20 years old (Fig. 1, Supplementary Table S2).
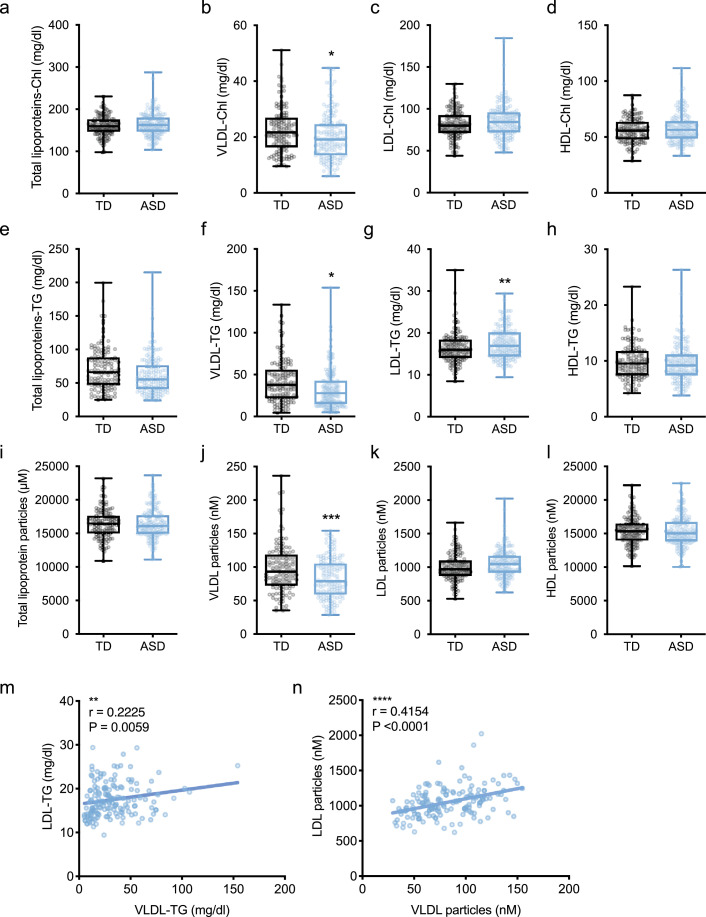
Lipoproteins are classified as VLDL, HDL, or LDL according to density or size and have Chl and TG fractions. We performed multivariate analysis on these lipoproteins considering the confounders (diagnosis, sex, height, weight, and BMI) and the matched confounder (age), and found VLDL-specific reductions in serum of ASD compared to TD children (Fig. 5). In the Chl fraction, there were no differences in total lipoprotein-Chl (OR = 1.00, 95% CI = 0.99 to 1.02, P = 0.61), LDL-Chl (OR = 1.01, 95% CI = 0.99 to 1.03, P = 0.33), and HDL-Chl (OR = 1.02, 95% CI = 0.99 to 1.05, P = 0.17; Fig. 5a, 5c, 5d). However, we found that VLDL-Chl (OR = 0.96, 95% CI = 0.93 to 1.00, P = 0.0316) was significantly decreased in the ASD group (Fig. 5b). In the TG group, we also found a significant reduction in VLDL-TG (OR = 0.99, 95% CI = 0.98 to 1.00, P = 0.0470) and a significant increase in LDL-TG (OR = 1.13, 95% CI = 1.04 to 1.22, P = 0.0045) in the ASD group (Fig. 5f, 5g), but no difference in total lipoproteins-TG (OR = 0.99, 95% CI = 0.98 to 1.00, P = 0.14) and HDL-TG (OR = 0.99, 95% CI = 0.91 to 1.08, P = 0.81; Fig. 5e, 5h). When we quantified the particulate numbers, there was a significant reduction in VLDL (OR = 0.98, 95% CI = 0.97 to 0.99, P = 0.0009; Fig. 5j), but not in total lipoproteins (OR = 1.00, 95% CI = 0.99 to 1.00, P = 0.93), LDL (OR = 1.00, 95% CI = 0.99 to 1.00, P = 0.14), and HDL (P = 0.86) particulate numbers (Fig. 5i, 5k, 5l). VLDL is formed in the liver and secreted into the blood. VLDL then is metabolised by lipoprotein lipase (LPL) to intermediate-density lipoproteins (IDL), and is eventually metabolised to LDL by hepatic lipase (HTGL). Therefore, we examined the correlations between VLDL and LDL serum concentrations in ASD by the Spearman rank correlation coefficient test, and found positive correlations between VLDL-TG and LDL-TG (r = −0.2225, P = 0.0059), as well as VLDL and LDL particulate numbers (r = −0.4154, P < 0.0001), respectively (Fig. 5m, 5n), indicating that the mechanism of VLDL degradation accelerated in ASD could not be accounted for by metabolism into LDL. These results demonstrated that a VLDL reduction is a distinct metabolic phenotype of children with ASD, but its degradation mechanism is unknown.
Fig. 5.
A VLDL-specific reduction in ASD.
(a–l) Serum concentrations of lipoprotein-cholesterols (-Chl) (a–d), triglycerides (-TG) (e–h) and particles (i–l). Significant reductions of VLDL-Chl (b), total lipoproteins-TG (e), VLDL-TG (f), and VLDL particles (j) were detected in ASD. In contrast, increased LDL-TG (g) and LDL particles (k) were detected in ASD. Data are represented as box and whisker plots showing all points minimum value to maximum value. Asterisks indicate ***P < 0.001, **P < 0.01, *P < 0.05, conditional logistic regression analysis, n = 128–152/condition. (m, n) Correlations between VLDL and LDL in ASD. VLDL-TG or the number of VLDL particles were positively correlated with VLDL-TG (m) or the number of VLDL particles (n), respectively. Asterisks indicate ****P < 0.0001, **P < 0.01, Spearman rank correlation coefficient test, n = 148/condition.
Next, we investigated whether these differences in lipoprotein concentrations would be useful as biomarkers for ASD diagnosis. We performed ROC analysis adjusted for age and found that the corresponding AUCs were 0.5977 (95% CI =0.52 to 0.67, P = 0.0128), 0.5997 (95% CI = 0.53 to 0.68, P = 0.0111), 0.6052 (95% CI = 0.53 to 0.68, P = 0.0073), and 0.6237 (95% CI = 0.55 to 0.70, P = 0.0019) for VLDL-Chl, VLDL-TG, LDL-TG, and VLDL particles, respectively (Supplementary Fig. S4). On the other hand, there were no correlations between lipoprotein concentrations and ADI-R (data not shown). These results indicated that serum levels of individual lipoproteins are not a reliable biomarker for clinical evaluation.
3.5. Lipoproteins can be an indicator of ASD pathogenesis
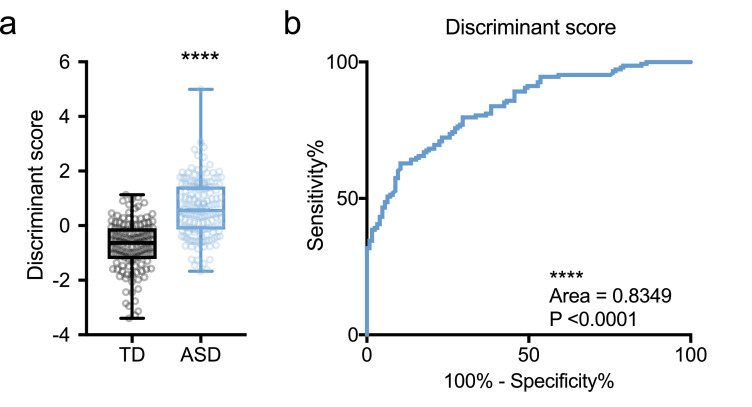
Despite the distinct phenotype of lipoprotein metabolism in ASD, each serum concentration was not useful as a reliable biomarker (Supplementary Fig. S4), but we considered comprehensive assessment of lipoproteins as a potential biomarker. We first performed discriminant analysis (linear discriminant analysis) to calculate discriminant scores of all individuals using diagnosis, sex, age, height, weight, BMI, and all lipoprotein profiles (total lipoproteins, VLDL, LDL, HDL in Chl and TG fractions, and each particulate number) information as variables. Next, we examine the normal distribution of discriminant scores by the D'Agostino and Pearson test, then examined a difference between TD and ASD by the Mann-Whitney test, because we already had considered cofounders in discriminant analysis. A significant difference in discriminant scores of serum lipoprotein profiles between TD and ASD children was found (P < 0.0001; Fig. 6a). Moreover, we recalculated the ROC curve using these discriminant scores, and found that the corresponding AUC was 0.8349 (95% CI = 0.7887 to 0.8811, P < 0.0001, sensitivity = 79.73%, specificity = 70.4%; Fig. 6b). However, there were no correlations between discriminant and ADI-R results (data not shown). These results indicated that comprehensive assessment of serum lipoprotein profiles can be an indicator, but not a reliable biomarker for clinical evaluation and ASD diagnosis.
Fig. 6.
Lipoproteins proportion potentially distinguish the children with ASD.
(a) Discriminant scores of ASD were significantly higher than TD. Those scores were calculated using sex, age, height, weight, BMI, and lipoproteins (Chl and TG fractions and particulate numbers of total lipoproteins, VLDL, LDL, and HDL) information as variables by linear discriminant analysis. Data are represented as box and whisker plots showing all points minimum-to-maximum value. Asterisk indicates ****P < 0.0001, Mann-Whitney test, n = 128–152/condition. (b) ROC curve using discriminant scores. ROC curve showing statistical significance and 83.49% AUC (sensitivity = 79.73%, specificity = 70.4%) was calculated by discriminant scores. Asterisk indicates ****P < 0.0001, ROC, n = 128–152/condition.
3.6. VLDL-associated APOB reduction in ASD
To understand the Prominent cause of VLDL-specific reduction in ASD, we first examined the liver function and other metabolic factors. Because VLDL is produced in the liver together with FAs, and both secreted into blood flow, we found that there were no differences in the serum concentration of AST (OR = 1.00, 95% CI = 0.94 to 1.07, P = 0.91), ALT (OR = 1.93, 95% CI = 0.94 to 1.04, P = 0.57), or γ-GTP (OR = 1.02, 95% CI = 0.94 to 1.10, P = 0.64) in ASD compared to TD by multivariate analysis with cofounders (diagnosis, sex, age, height, weight, and BMI; Supplementary Fig. S5). We also examined the concentration of CRP as an inflammatory marker and found no difference in serum CRP concentration in the ASD compared to the TD groups (OR = 0.05, 95% CI = 3.10e–4 to 5.57, P = 0.21; Supplementary Fig. S5). These results indicated that the VLDL-specific reduction in ASD is not due to impairment of liver function and inflammation.
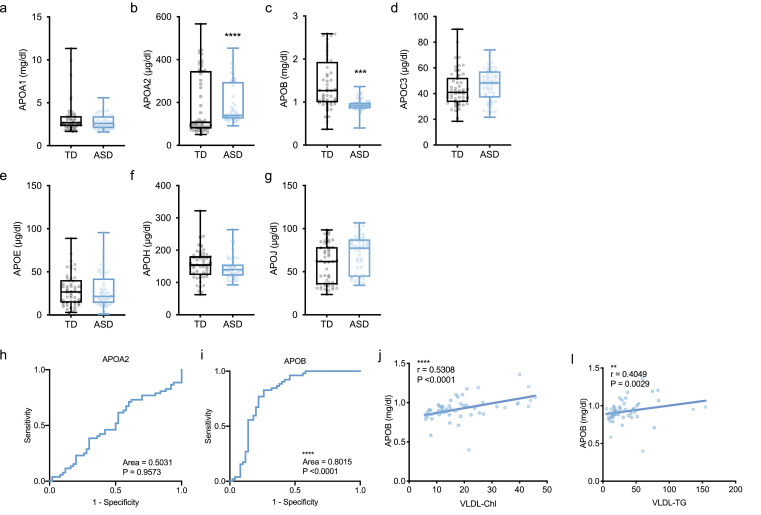
We next investigated the apoproteins that bind to lipoproteins and act as activation or coenzymes involved in lipoprotein recognition and lipid metabolism. We found no differences in serum APOA1 (OR = 0.86, 95% CI = 0.56 to 1.23, P = 0.44), APOC3 (OR = 0.99, 95% CI = 0.96 to 1.04, P = 0.96), APOE (OR = 1.02, 95% CI = 0.98 to 1.07, P = 0.29), APOH (OR = 0.99, 95% CI = 0.98 to 1.01, P = 0.29), and APOJ (OR = 1.00, 95% CI = 0.98 to 1.03, P = 0.86) in the ASD compared to the TD groups by multivariate analysis with cofounders (diagnosis, sex, age, height, weight, and BMI; Fig. 7a, 7d–g). We also found a significant increase in APOA2 (OR = 1.09, 95% CI = 1.05 to 1.14, P < 0.0001; Fig. 7b), whereas APOB (OR = 2.95e–4, 95% CI = 1.78e–6 to 9.36e–3, P = 0.0002) was significantly decreased in serum of ASD children (Fig. 7c) as reported previously [19]. The corresponding AUCs of ROC analysis adjusted for age were 0.5031 for APOA2 (95% CI = 0.39 to 0.62, P = 0.96) and 0.8015 for APOB (95% CI = 0.71 to 0.89, P < 0.0001; Fig. 7h, 7i), indicating APOB is an indicator for ASD diagnosis as previous reported [19].
Fig. 7.
Decreased APOB correlated with VLDL in ASD.
(a-g) Serum levels of APOA1 (a), APOA2 (b), APOB (c), APOC3 (d), APOE (e), APOH (f) and APOJ (g) apolipoproteins. Distinct reduction of APOB (c) was observed in ASD. On the other hand, increased APOA2 (b) and APOJ (g) also were observed in ASD. Data are represented as box and whisker plots showing all points minimum-to-maximum value. Asterisks indicate ****P < 0.0001, ***P < 0.001, conditional logistic regression analysis, n = 150–174/condition. (h–i) ROC curves of APOA2 (h) and APOB (i). Asterisks indicate ****P < 0.0001, *P < 0.05, ROC adjusted for age, n = 150–174/condition. (j, k) Correlations between APOB and VLDL-Chl and VLDL-TG. APOB was significantly correlated with VLDL-Chl (j) and VLDL-TG (k). Asterisk indicates ****P < 0.0001, Spearman rank correlation coefficient test, n = 52/condition.
APOB is a main component protein of VLDL and LDL. However, there was no decrease in LDL (Fig. 5), suggesting that a decrease in APOB could be a VLDL-specific result. Accordingly, we further investigated the correlations of APOB with VLDL-Chl and VLDL-TG in serum of ASD children by the Spearman rank correlation coefficient test. As we expected, APOB was significantly and modestly correlated with VLDL-Chl (r = 0.5308, P < 0.0001) and VLDL-TG (r = 0.4049, P = 0.0029; Fig. 7j, 7k). These results indicated that the decreased APOB correlated with VLDL in ASD. Taken together, our results demonstrated the increases of FAs in ASD are due to VLDL-specific degradation associated with APOB.
4. Discussion
In summary, we identified 48 metabolites in ASD as novel targets involved in lipid biosynthesis and metabolism, oxidative stress, and synaptic function by lipidomics. Among them, increased FAs correlated with A score of ADI-R, as well as lipoproteins-TG in ASD. We also showed that VLDL reductions depended on APOB reductions in ASD, suggesting lipoprotein proportions may potentially facilitate ASD diagnosis. We concluded that the increases in FAs are due to VLDL-specific degradation associated with ASD pathophysiology.
In our study, of the 48 metabolites identified, interestingly the majority were lipid-related molecules that showed increased concentrations in plasma of ASD children 5 to 11years old compared to TD children 6 to 10 years old (Table 1, Supplementary Table S1). Among these, FAs including n-3 and n-6, such as linolenic acid, EPA, linoleic acid, palmitic acid, oleic acid, myristic acid, ACs, 2-aminobutyric acid, and 2-hydroxybutyric acid, were listed as significant metabolites (Table 1). Consequently, we found a trend of reduction in the n-3/n-6 ratio in ASD children (Fig. 3f). Significant alterations of n-3 and n-6, including the ratio of n-3/n-6, have been reported in ASD patients [7, 21, 22, [45], [46], [47]]. In contrast to our results, a previous study showed a significant reduction in DHA and total n-3 PUFAs without n-6 PUFAs reductions in the plasma of ASD children 3 to 18 years old [21]. Moreover, increased EPA and DHA levels, and reduced AA levels also have been reported in patients with autism and Asperger's syndrome [7]. Another FAs study using gas chromatography reported increased SFAs and decreased PUFAs in plasma of ASD children (no sex information) 4 to 12 years old [24]. This plasma FAs profiling study also has shown increases in acetic, valeric, hexanoic, and stearidonic acids, as well as decreases in propionic, butyric, caprylic, decanoic, lauric, palmitic, stearic, arachidic, and linolenic acids; EPA; DHA; linoleic acid; AA, and oleic and elaidic acids [24]. A similar metabolomic study has reported increased 2-hydroxyvaleric acid and decreased myristic acid in plasma of ASD children at 4 to 6 years old [48]. Conversely, consistent with our results, several studies reported elevated PUFAs levels in ASD patients [22, 47]. An increased DHA in plasma of ASD young boys 12 to 18 years old has been reported [22]. Furthermore, elevated PUFAs and SFAs levels also were found in plasma of ASD patients [47]. Collectively, these studies demonstrate that such imbalance or abnormal amounts of n-3 and n-6 and lipid-related metabolites are the factors underlying ASD pathophysiology.
The reason for the differences in results is that the age, sex, measurement methods, and statistical analyses of the participants differed from the previous studies compared to our study. For example, several previous PUFAs studies have performed univariate analysis but not multivariate analysis as in our study. Furthermore, the differences in measurement methods and samples will greatly affect the results. Since dietary factors have ultimate effects on metabolism, we speculated that opposite metabolomic results may be derived from the differences in countries and food cultures. Notably, there is a huge difference in the intake and blood levels of PUFAs, in particular n-3 and n-6, among Western countries, Saudi Arabia, and Japan. Thus, it is believed possible that the Japanese dietary culture, such as fish intake, has raised the baseline of blood PUFAs levels in contrast to those populations [49]. However, common findings in all studies are reliable factors that PUFAs and their imbalance are definitely associated in ASD pathophysiology. In addition, it will be interesting to investigate the plasmalogens in ASD, because plasmalogens, a subgroup of ether lipids, are important membrane components involved in vesicle fusion and store PUFAs for serving antioxidants [50]. In fact, increased plasmalogen levels has been reported in plasma of ASD patients [47]. Additionally, decreased levels of total n-6, total phosphatidylethanolamine plasmalogens, and the n-6/n-3 ratio were observed in the brain of propionic acid infused ASD model rats [51]. Recent studies also demonstrate that glyceronephosphate O-acyltransferase (Gnpat) knockout mice, a model of complete deficiency in ether lipid biosynthesis including plasmalogens, displayed impaired social interaction, repetitive behavior, and hyperactivity, suggesting that plasmalogens may be involved in ASD-like behavior in mice [52, 53]. Therefore, a future study of plasmalogen also is needed to understand the molecular mechanisms underlying lipid metabolism in ASD, particularly how they behave and have a role in PUFAs.
Our results also suggested that hyperactivation of FA metabolism in ASD may result in type 2 diabetes due to dysfunction of ABC transporters, osmolytes, or kidney (Table 1, Fig. 2). In general, ABC transporters are known to transport lipids, sugars, vitamins, ions, peptides, proteins, and other metabolic substances involved in metabolism. An osmolyte is a chemical that regulates osmotic pressure in organisms. Indeed, a higher risk of type 2 diabetes in ASD has been reported in childhood to young adulthood [[54], [55], [56]], suggesting that our results also may facilitate understanding of the pathogenesis underlying this relationship.
Mitochondrial dysfunction has been reported in ASD [57, 58], and it is considered a one of causes of ASD pathology. In fact, oxidative stress is increased due to abnormalities of the mitochondrial electron transport system in the brain of ASD patients [59]. Such increased oxidative stress due to mitochondrial dysfunction has been reported by several studies, indicating strong associations with ASD pathology [31, 33, [60], [61], [62]]. As previous studies have shown GSH reduction and oxidative stress in ASD [30, [32], [33], [34], 60, 63, 64], we found a significant reduction of cysteine glutathione disulphide (OR = 1.16e–5, 95% CI = 1.90e–10 to 6.64e–2, P = 0.0227, A/T ratio = 0.8) in ASD involved in downregulations of cysteine metabolism and oxidative stress (Table 1, Fig. 2, Supplementary Fig. S1v). The major antioxidants GSH and cysteine are responsible for neutralizing the damages of oxidative stress generated. GSH is a major cellular radical scavenger and has an important role in protecting cells from exogenous and endogenous toxins, particularly in the brain [61, 64, 65]. GSH decreases have been reported in several ASD studies with increased oxidative stress and a decreased detoxification capacity [33, 61, 64, 65]. Cysteine glutathione disulphide is an oxidized form of GSH, which is produced upon oxidative stress. Thus, in general a reduction of cysteine glutathione means oxidative stress is reduced if there is a sufficient amount of GSH. However, again, decreased GSH in ASD has been reported in many studies [[32], [33], [34]]. Therefore, we speculated that the reduction of cysteine glutathione disulphide is due to baseline characteristics of GSH under ASD pathophysiology. However, we could not detect GSH using the metabolomic approach in this study. Accordingly, further study is required using an appropriate measurement method to uncover the behaviours of GSH and its related metabolites underlying oxidative stress in ASD.
Moreover, increased ACs are known indicators for mitochondrial dysfunction, in particular abnormalities in FAs metabolism responsible for energy production and conversion of ACs (Acyl-CoA) to acetylcholine (Acetyl-CoA) in the β-oxidation pathway of mitochondria. Recent studies have reported that long chain ACs, such as AC(14:1), AC(14:2), and AC(16:1), were elevated in patients with ASD [66, 67], consistent with our results (Table 1). Hence, we speculated that the increased ACs concentrations result in the possibility of mitochondrial dysfunction in ASD. Several previous studies have shown that the treatment of carnitine or co-enzyme Q10 improved ASD symptoms [57, 68]. Carnitine is involved in FA transport in the mitochondrial membrane. In this study, we detected carnitine as a one of detected 258 metabolites in metabolomic analysis, but not a significant metabolite (OR =0.99, 95% CI = 0.84 to 1.16, P = 0.86, A/T ratio = 1.1) (data not shown). On the other hand, co-enzyme Q10 is a one of key component of mitochondria, and it is biosynthesized from cholesterol. Although there was no significant difference in total lipoprotein-Chl (Fig. 5a), a significant reduction of VLDL-Chl was observed in ASD (Fig. 5b). Thus, it will be also interesting to investigate the blood level and role of co-enzyme Q10 in ASD children for understanding the association between VLDL-specific reduction and mitochondrial dysfunction of ASD in the future study. Moreover, 48 identified metabolites, such as FAs, ACs, and cysteine glutathione disulphide, have critical roles in ASD pathology, in particular generations of oxidative stress via mitochondrial dysfunction, although further investigation is needed to better understand how they behave in ASD.
Regarding reasons why lipids-related metabolites, such as FAs, were increased in ASD children, we hypothesised that it was due to lipoproteins-TG degradation. Because FAs are generated mainly in the liver, and examining the AST, ALT, and γ-GTP revealed no hepatic dysfunction, we concluded that liver function or biosynthesis was not a causal factor (Supplementary Fig. S5). Consequently, we analysed the possibility of degradation, then found that plasma FAs concentrations were negatively correlated with plasma lipoproteins-TG concentrations in the ASD group as we expected, but not in the TD group (Fig. 4). Importantly, plasma FAs concentrations indicated positive correlations between with social interaction score of ADI-R (Fig. 3). In addition, increases in betaine and 3-hydroxybutyric acid also suggested lipoproteins degradation in ASD. Together, our results demonstrated that increased FAs correlated with social interaction score of ASD diagnosis as well as lipoproteins-TG concentrations in ASD, supporting the idea that lipoproteins-TG degradation leads to increase in FAs.
A comprehensive lipoprotein analysis was conducted to determine what lipoproteins were involved. Then, we showed that VLDL was decreased in the ASD group (Fig. 5). Additionally, an increase in LDL, one of the VLDL metabolites, was observed in the ASD group, suggesting the possibility of VLDL degradation (Fig. 5). The alterations in lipoprotein composition have been reported in ASD, showing an increase in TG concentration and a decrease in HDL-Chl in plasma of ASD children 7 to 12 years old, compared to TD children 10 to 11 years old. However, those diagnoses were done without ADI-R, ADOS, and drug history in the ASD subjects [20]. In contrast, with ADI-R and drug history, we demonstrated significant reductions in VLDL-Chl and VLDL-TG as well as an increase in LDL-TG in ASD children 1 to 19 years old, compared to TD children 2 to 18 years old (Fig. 5). A recent lipidome study reported significantly decreased lipid concentrations in the prefrontal cortex of postmortem brains of ASD patients, showing a significant correlation with age 2 to 4 years [69]. These findings indicated that dyslipidaemia occurs in ASD children after birth at least, although subject age, sample and methodology for lipid quantification are different.
VLDL-specific dyslipidaemia is characterised by a decrease in serum or plasma lipoprotein and is thought to be induced by genetic and secondary factors, such as hepatic dysfunction, inflammatory condition, and malnutrition. Previous studies have shown that hyperlipidaemia is a risk factor for cardiovascular disease, but few studies have focused on hypolipidemia. To address the mechanism of VLDL-specific reduction, it was first necessary to clarify whether VLDL was decreased due to inhibited synthesis or activated degradation. VLDL is synthesised in the liver by microsomal triglyceride transfer protein (MTTP), and is degraded by LPL. We observed increased LDL, a metabolite in the degradation pathway of VLDL, which also was correlated with VLDL concentration in ASD (Fig. 5). Our results suggested that VLDL is specifically degraded in ASD.
Abetalipoproteinaemia (ABL) is an autosomal recessive disorder caused by a mutation of MTTP and is associated with a decrease in VLDL, showing specific decreases in LDL-Chl and HDL-Chl [70]. Severe deficiencies also occur in vitamin E sent to peripheral tissues via VLDL and LDL. However, to our knowledge no studies have reported VLDL-specific reductions without HDL and LDL reductions, such as the pattern observed in our study. In contrast, LPL converts VLDL into IDL by removing TG, which are taken up by macrophages and activate phagocytosis. These series of flows may uncover whether the central and peripheral phagocyte activation causes VLDL degradation. Given the characteristic phagocytosis of VLDL by macrophages [71] and activation of intracerebral microglia in ASD [72, 73], it also is possible that VLDL may be phagocytosed by macrophages.
APOB is significantly reduced in ASD children (Fig. 7), which was consistent with a previous proteomics study that demonstrated a reduction of APOB in ASD [19]. APOB has two forms, APOB100 and APOB48. VLDL, IDL, and LDL are only present on APOB100. In our study, APOB could be excluded as a causal factor for VLDL-specific reduction due to no decrease in LDL. VLDL is synthesised in the liver via the conjugation to APOB, and it only receives APOC and APOE from HDL in the blood flow, but there also were no changes in APOC3 and APOE (Fig. 7). On the other hand, APOA1 is the major protein component of HDL particles in plasm and APOA2 binds to the surface of HDL and is involved in the metabolism of HDL. Serum levels of HDL-Chl, HDL-TG, and HDL particles did not change in the TD and ASD groups (Fig. 5); however, only the APOA2 level increased in the ASD group (Fig. 7). Decreased serum APOA1 and APOA2 levels in cases of cognitive impairment have been reported to be associated with the Mini-Mental State Examination (MMSE) score in cognitive function assessment [74], suggesting that APOA2 is linked to higher brain function. However, it is unclear why only APOA2 was increased. APOA2 was correlated with serum levels of VLDL-Chl (r = −0.5105, P = 0.0001) and VLDL-TG (r = −0.4827, P = 0.0003) in ASD, but not in TD (data not shown). Thus, we speculated that an increase in APOA2 may be related to abnormal lipid metabolism in ASD. APOH (β−2-glycoprotein 1) was involved in physiologic pathways, including lipoprotein metabolism, coagulation, haemostasis and the production of antiphospholipid autoantibodies, but there was no change. An increase in APOJ in ASD was detected only by univariate analysis (P = 0.0067), but not multivariate analysis with cofounders (P = 0.86; Fig. 7). APOJ, also known as Clusterin, is involved in removal of cellular debris and apoptosis as a sensor of oxidative stress [[75], [76], [77]]. Clusterin also is implicated in oxidative stress-related disorders, such as metabolic and cardiovascular diseases, including dyslipidaemia, inflammatory diseases, aging, Alzheimer's disease, cancers, and kidney degeneration [[78], [79], [80], [81]]. Moreover, in infusion experiments into rat brain and in vitro culture systems, microglia activated by an increase in APOJ has been reported [82]. These studies suggest the possibility that APOJ is involved in the increased oxidative stress and activation of microglia. Taken together, our results suggested that VLDL-specific lipid abnormalities occur specific to ASD. However, the molecular mechanism underlying VLDL-specific dyslipidaemia in ASD remains unknown.
Lastly, the involvement of lipids in ASD also has been suggested in the study of SLOS [15, 16, 83]. In fact, DHCR7, a gene responsible for SLOS, together with variants and copy number variations also have been reported in ASD [15, 84]. Among the lipoprotein-related genes, HDL binding protein (HDLBP) [85, 86], LDL receptor related protein 2 (LRP2) [87], and LPL [88], also were reported as ASD genes. Furthermore, the deletion and duplication of 9p24.2 where the VLDL receptor (VLDLR) gene is located, also has been reported in ASD [[89], [90], [91]]. VLDLR is a known receptor for reelin (RELN) signaling [91]. RELN also are ASD genes [91, 92], having a key role in neuronal migration and layer formation during neural development. These studies also demonstrate that lipids and their metabolic pathways are essential for proper brain development and functions. In addition, whether these lipid metabolism phenotypes in ASD are congenital or acquired must be clarified by genomics and follow-up study, for example using umbilical cord, new-born or adolescent blood. This is because if it is acquired, it has the potential to become a novel therapeutic target.
On the other hand, several reports exist on dyslipidaemia in other developmental disorders as well as ASD. For example, blood levels of total Chl and LDL were elevated in adults with Asperger's disorder [93]. In the case of attention deficit hyperactivity disorder (ADHD), plasma TG and phospholipids were reduced, whereas free Chl, HDL, and APOA1 were increased in pediatric patients [94]. Interestingly, plasma EPA and DHA also were elevated in this ADHD study [94]. This result was consistent with our finding of increased plasma EPA in ASD (Fig. 3). As an opposite report, serum total Chl and LDL levels were increased in children with ADHD, but there were no correlations of clinical evaluation [95]. In Fragile X syndrome, decreases in total Chl, LDL, and HDL were reported [96]. Consequently, various abnormalities in lipid metabolism have been reported in developmental disorders, suggesting that lipid metabolism has a significant impact on brain development, function, and pathology in common.
In conclusion, we demonstrated that lipid metabolism is definitely involved in ASD pathology, particularly increases in VLDL-specific FAs in ASD correlate with social interaction signature. Our findings suggest that abnormalities of lipid metabolism, such as elevated FAs and ACs as well as VLDL-specific reduction, eventually resulting in the imbalance of n-3/n-6 and oxidative stress production via mitochondrial dysfunction, may explain the pathology of ASD. Our study also provided the comprehensive metabolic resources in the peripheral blood of children with ASD. However, we could not identify a reliable biomarker for clinical evaluation in this study. Further understanding of the role of dysfunctions of lipids metabolism in the brain should give rise to novel insights and targets for understanding the molecular mechanisms underlying ASD pathology.
We acknowledge several limitations to our study. Regarding the quantification of lipoproteins, each sample size was >100, whereas the metabolome analyses did not have a sufficient sample size. The small sample size renders the data preliminary, and a larger study with more subjects is necessary, such as a recent large-scale analysis. However, the genetic background is more uniform due to this study being composed only of Japanese subjects. Hence, we concluded that our data are free from confounders due to race, intelligence, and psychotropic drugs and reflect a certain common immunologic pathology among people with ASD. However, we acknowledge the study design and potential residual confounders, such as sex, diet, socio-economic factors, family history of developmental and psychiatric disorders, and self-selection bias. In our study, we have not recorded the family history of developmental and psychiatric disorders, educational background of parents, and economic situation of participants in both TD and ASD children. In addition, TD children were recruited by advertisement, thus we also acknowledge the limitations that there were variations in terms of educational background of parents, children's educational environment and background, and/or possible participations by parents concerned about whether their children have neurodevelopmental problems (in this regard, we have ruled out problematic participants by prior examination, see Methods). Although ASD is associated with gastrointestinal issues and problems with digestive enzymes, we have not recorded whether the participants have those problems. In addition, we have not examined the digestive enzymes secreted in the digestive tract, such as amylase, pepsin, and lipase. In the present study, we could at least rule out gastrointestinal inflammatory problems by serum CRP levels of participants. Moreover, the participants did not have any abdominal symptoms such as constipation and diarrhea at the time of blood sampling. However, we acknowledge the possibilities that the participants have problem in the gastrointestinal system. Objectively, in terms of sensitivity and specificity compared to previous studies [19, [97], [98], [99]], our current study could not provide a reliable biomarker for ASD. Accounting for this issue, further study is required to identify a truly reliable biomarker with an enormous sample size and multiple clinical evaluations. Moreover, biological validation using animal models, such as metabolic manipulated mice, genetically modified mice, and/or human-derived organoids, is essential to understand the effects of metabolic abnormalities in ASD detected in peripheral blood on brain development and its function. Overall, in terms of basic medical science, our findings definitely provide the novel insight into the lipid metabolism abnormalities at least for facilitating understanding of the neurobiologic mechanisms of ASD.
Acknowledgments
Acknowledgments
We thank all participants and their families. We also thank Chie Shimmura, Tae Takahashi, Erina Sakamoto, Mika Oyaizu, Hisako Ohba, Yasuko Hisano, Yoshimi Iwayama, Xie Min-Jue, Yoko Sasaki, Satomi Fukuhara for technical assistance and Tomoko Taniguchi for clerical supports.
Funding sources
This work was supported by the Strategic Research Program for Brain Sciences from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to H.M.; the Grant-in-Aid for Scientific Research on Innovative Areas from MEXT to H.M.; SENSHIN Medical Research Foundation to H.M. and N.U.; Hamamatsu Foundation for Science and Technology Promotion to H.M.; the Grant-in-Aid for Early-Career Scientists (18K14814) from the Japan Society for the Promotion of Science (JSPS) to N.U.; Takeda Science Foundation to N.U.; The Osaka Medical Research Foundation for Intractable Diseases to N.U.; Research Grant for Public Health Science to N.U.; Eli Lilly Japan Research Grant to N.U.; the Grant for Life Cycle Medicine from Faculty of Medical Sciences, University of Fukui to N.U..
Declaration of Competing Interest
The authors declare no competing interests.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102917.
Appendix. Supplementary materials
References
- 1.Dawson S., Glasson E.J., Dixon G., Bower C. Birth defects in children with autism spectrum disorders: a population-based, nested case-control study. Am J Epidemiol. 2009;169(11):1296–1303. doi: 10.1093/aje/kwp059. [DOI] [PubMed] [Google Scholar]
- 2.Maenner M.J., Shaw K.A., Baio J. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveill Summ. 2020;69(4):1–12. doi: 10.15585/mmwr.ss6904a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan P.F., Geschwind D.H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell. 2019;177(1):162–183. doi: 10.1016/j.cell.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grether J.K., Anderson M.C., Croen L.A., Smith D., Windham G.C. Risk of autism and increasing maternal and paternal age in a large north American population. Am J Epidemiol. 2009;170(9):1118–1126. doi: 10.1093/aje/kwp247. [DOI] [PubMed] [Google Scholar]
- 5.Network SIG . 2007. Assessment, diagnosis and clinical interventions for children and young people with autism spectrum disorders. [Google Scholar]
- 6.White S.W., Scahill L., Klin A., Koenig K., Volkmar F.R. Educational placements and service use patterns of individuals with autism spectrum disorders. J Autism Dev Disord. 2007;37(8):1403–1412. doi: 10.1007/s10803-006-0281-0. [DOI] [PubMed] [Google Scholar]
- 7.Bell J.G., MacKinlay E.E., Dick J.R., MacDonald D.J., Boyle R.M., Glen A.C.A. Essential fatty acids and phospholipase A2 in autistic spectrum disorders. Prostaglandins Leukot Essent Fatty Acids. 2004;71(4):201–204. doi: 10.1016/j.plefa.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Wurtman R.J. Synapse formation and cognitive brain development: effect of docosahexaenoic acid and other dietary constituents. Metabolism. 2008;57(Suppl 2) doi: 10.1016/j.metabol.2008.07.007. S6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salvati S., Attorri L., Avellino C., Di Biase A., Sanchez M. Diet, lipids and brain development. Dev Neurosci. 2000;22(5-6):481–487. doi: 10.1159/000017479. [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Eckel R.H. What are lipoproteins doing in the brain? Trends Endocrinol Metab. 2014;25(1):8–14. doi: 10.1016/j.tem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Torre-Ubieta L., Won H., Stein J.L., Geschwind D.H. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22(4):345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usui N., Araujo D.J., Kulkarni A. Foxp1 regulation of neonatal vocalizations via cortical development. Genes Dev. 2017;31(20):2039–2055. doi: 10.1101/gad.305037.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usui N., Co M., Harper M., Rieger M.A., Dougherty J.D., Konopka G. Sumoylation of FOXP2 regulates motor function and vocal communication through purkinje cell development. Biol Psychiatry. 2017;81(3):220–230. doi: 10.1016/j.biopsych.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heavner W.E., Smith S.E.P. Resolving the synaptic versus developmental dichotomy of autism risk genes. Trends Neurosci. 2020;43(4):227–241. doi: 10.1016/j.tins.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney E., Nwokoro N.A., Porter F.D., Freund L.S., Ghuman J.K., Kelley R.I. Behavior phenotype in the RSH/Smith-Lemli-Opitz syndrome. Am J Med Genet. 2001;98(2):191–200. doi: 10.1002/1096-8628(20010115)98:2<191::aid-ajmg1030>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Bukelis I., Porter F.D., Zimmerman A.W., Tierney E. Smith-Lemli-Opitz syndrome and autism spectrum disorder. Am J Psychiatry. 2007;164(11):1655–1661. doi: 10.1176/appi.ajp.2007.07020315. [DOI] [PubMed] [Google Scholar]
- 17.Porter F.D. Smith-Lemli-Opitz syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet EJHG. 2008;16(5):535–541. doi: 10.1038/ejhg.2008.10. [DOI] [PubMed] [Google Scholar]
- 18.Aneja A., Tierney E. Autism: the role of cholesterol in treatment. Int Rev Psychiatry. 2008;20(2):165–170. doi: 10.1080/09540260801889062. [DOI] [PubMed] [Google Scholar]
- 19.Corbett B.A., Kantor A.B., Schulman H. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry. 2007;12(3):292–306. doi: 10.1038/sj.mp.4001943. [DOI] [PubMed] [Google Scholar]
- 20.Kim E.K., Neggers Y.H., Shin C.S., Kim E., Kim E.M. Alterations in lipid profile of autistic boys: a case control study. Nutr Res (New York, NY) 2010;30(4):255–260. doi: 10.1016/j.nutres.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Vancassel S., Durand G., Barthelemy C. Plasma fatty acid levels in autistic children. Prostaglandins Leukot Essent Fatty Acids. 2001;65(1):1–7. doi: 10.1054/plef.2001.0281. [DOI] [PubMed] [Google Scholar]
- 22.Sliwinski S., Croonenberghs J., Christophe A., Deboutte D., Maes M. Polyunsaturated fatty acids: do they have a role in the pathophysiology of autism? Neuro Endocrinol Lett. 2006;27(4):465–471. [PubMed] [Google Scholar]
- 23.Bradbury J. Docosahexaenoic acid (DHA): an ancient nutrient for the modern human brain. Nutrients. 2011;3(5):529–554. doi: 10.3390/nu3050529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Ansary A.K., Bacha A.G., Al-Ayahdi L.Y. Plasma fatty acids as diagnostic markers in autistic patients from Saudi Arabia. Lipids Health Dis. 2011;10:62. doi: 10.1186/1476-511X-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makinodan M., Iwata K., Ikawa D. Tumor necrosis factor-alpha expression in peripheral blood mononuclear cells correlates with early childhood social interaction in autism spectrum disorder. Neurochem Int. 2017;104:1–5. doi: 10.1016/j.neuint.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Sugihara G., Hashimoto K., Iwata Y. Decreased serum levels of hepatocyte growth factor in male adults with high-functioning autism. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31(2):412–415. doi: 10.1016/j.pnpbp.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Okada K., Hashimoto K., Iwata Y. Decreased serum levels of transforming growth factor-beta1 in patients with autism. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31(1):187–190. doi: 10.1016/j.pnpbp.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K., Hashimoto K., Iwata Y. Decreased serum levels of epidermal growth factor in adult subjects with high-functioning autism. Biol Psychiatry. 2007;62(3):267–269. doi: 10.1016/j.biopsych.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Fujita-Shimizu A., Suzuki K., Nakamura K. Decreased serum levels of adiponectin in subjects with autism. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34(3):455–458. doi: 10.1016/j.pnpbp.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Parker W., Hornik C.D., Bilbo S. The role of oxidative stress, inflammation and acetaminophen exposure from birth to early childhood in the induction of autism. J Int Med Res. 2017;45(2):407–438. doi: 10.1177/0300060517693423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollis F., Kanellopoulos A.K., Bagni C. Mitochondrial dysfunction in Autism Spectrum Disorder: clinical features and perspectives. Curr Opin Neurobiol. 2017;45:178–187. doi: 10.1016/j.conb.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 32.Melnyk S., Fuchs G.J., Schulz E. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42(3):367–377. doi: 10.1007/s10803-011-1260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rose S., Melnyk S., Pavliv O. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Translational psychiatry. 2012;2:e134. doi: 10.1038/tp.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauhan A., Chauhan V., Brown W.T., Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin–the antioxidant proteins. Life Sci. 2004;75(21):2539–2549. doi: 10.1016/j.lfs.2004.04.038. [DOI] [PubMed] [Google Scholar]
- 35.Jain S.K. The accumulation of malonyldialdehyde, a product of fatty acid peroxidation, can disturb aminophospholipid organization in the membrane bilayer of human erythrocytes. J Biol Chem. 1984;259(6):3391–3394. [PubMed] [Google Scholar]
- 36.von Elm E., Altman D.G., Egger M., Pocock S.J., Gøtzsche P.C., Vandenbroucke J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370(9596):1453–1457. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 37.Faul F., Erdfelder E., Lang A.G., Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39(2):175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 38.Faul F., Erdfelder E., Buchner A., Lang A.G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods. 2009;41(4):1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 39.Koyama T., Osada H., Tsujii H., Kurita H. Utility of the Kyoto Scale of Psychological Development in cognitive assessment of children with pervasive developmental disorders. Psychiatry Clin Neurosci. 2009;63(2):241–243. doi: 10.1111/j.1440-1819.2009.01931.x. [DOI] [PubMed] [Google Scholar]
- 40.Kawabe K., Kondo S., Matsumoto M. Developmental quotient to estimate intelligence in autism spectrum disorder. Pediatr Int Off J Jpn Pediatr Soc. 2016;58(10):963–966. doi: 10.1111/ped.12969. [DOI] [PubMed] [Google Scholar]
- 41.Monton M.R., Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J Chromatogr A. 2007;1168(1-2):237–246. doi: 10.1016/j.chroma.2007.02.065. discussion 6. [DOI] [PubMed] [Google Scholar]
- 42.Sone M., Morone N., Nakamura T. Hybrid cellular metabolism coordinated by Zic3 and Esrrb synergistically enhances induction of naive pluripotency. Cell Metab. 2017;25(5):1103–1117. doi: 10.1016/j.cmet.2017.04.017. .e6. [DOI] [PubMed] [Google Scholar]
- 43.Okazaki M., Yamashita S. Recent advances in analytical methods on lipoprotein subclasses: calculation of particle numbers from lipid levels by gel permeation HPLC using “spherical particle model”. J Oleo Sci. 2016;65(4):265–282. doi: 10.5650/jos.ess16020. [DOI] [PubMed] [Google Scholar]
- 44.Brigandi S.A., Shao H., Qian S.Y., Shen Y., Wu B.-.L., Kang J.X. Autistic children exhibit decreased levels of essential Fatty acids in red blood cells. Int J Mol Sci. 2015;16(5):10061–10076. doi: 10.3390/ijms160510061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiest M.M., German J.B., Harvey D.J., Watkins S.M., Hertz-Picciotto I. Plasma fatty acid profiles in autism: a case-control study. Prostaglandins Leukot Essent Fatty Acids. 2009;80(4):221–227. doi: 10.1016/j.plefa.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Bell J.G., Sargent J.R., Tocher D.R., Dick J.R. Red blood cell fatty acid compositions in a patient with autistic spectrum disorder: a characteristic abnormality in neurodevelopmental disorders. Prostaglandins Leukot Essent Fatty Acids. 2000;63(1-2):21–25. doi: 10.1054/plef.2000.0186. [DOI] [PubMed] [Google Scholar]
- 47.Pastural E., Ritchie S., Lu Y. Novel plasma phospholipid biomarkers of autism: mitochondrial dysfunction as a putative causative mechanism. Prostaglandins Leukot Essent Fatty Acids. 2009;81(4):253–264. doi: 10.1016/j.plefa.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 48.West P.R., Amaral D.G., Bais P. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iso H., Sato S., Folsom A.R. Serum fatty acids and fish intake in rural Japanese, urban Japanese, Japanese American and Caucasian American men. Int J Epidemiol. 1989;18(2):374–381. doi: 10.1093/ije/18.2.374. [DOI] [PubMed] [Google Scholar]
- 50.Dorninger F., Forss-Petter S., Berger J. From peroxisomal disorders to common neurodegenerative diseases - the role of ether phospholipids in the nervous system. FEBS Lett. 2017;591(18):2761–2788. doi: 10.1002/1873-3468.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas R.H., Foley K.A., Mepham J.R., Tichenoff L.J., Possmayer F., MacFabe D.F. Altered brain phospholipid and acylcarnitine profiles in propionic acid infused rodents: further development of a potential model of autism spectrum disorders. J Neurochem. 2010;113(2):515–529. doi: 10.1111/j.1471-4159.2010.06614.x. [DOI] [PubMed] [Google Scholar]
- 52.Dorninger F., König T., Scholze P. Disturbed neurotransmitter homeostasis in ether lipid deficiency. Hum Mol Genet. 2019;28(12):2046–2061. doi: 10.1093/hmg/ddz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dorninger F., Gundacker A., Zeitler G., Pollak D.D., Berger J. Ether lipid deficiency in mice produces a complex behavioral phenotype mimicking aspects of human psychiatric disorders. Int J Mol Sci. 2019;20(16) doi: 10.3390/ijms20163929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiang A.H., Wang X., Martinez M.P., Page K., Buchanan T.A., Feldman R.K. Maternal Type 1 diabetes and risk of autism in offspring. JAMA. 2018;320(1):89–91. doi: 10.1001/jama.2018.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen M.H., Lan W.H., Hsu J.W. Risk of developing Type 2 diabetes in adolescents and young adults with autism spectrum disorder: a nationwide longitudinal study. Diabetes Care. 2016;39(5):788–793. doi: 10.2337/dc15-1807. [DOI] [PubMed] [Google Scholar]
- 56.Rivell A., Mattson M.P. Intergenerational metabolic syndrome and neuronal network hyperexcitability in autism. Trends Neurosci. 2019;42(10):709–726. doi: 10.1016/j.tins.2019.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossignol D.A., Frye R.E. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290–314. doi: 10.1038/mp.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheng N., Rho J.M., Masino S.A. Metabolic dysfunction underlying autism spectrum disorder and potential treatment approaches. Front Mol Neurosci. 2017;10:34. doi: 10.3389/fnmol.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chauhan A., Gu F., Essa M.M. Brain region-specific deficit in mitochondrial electron transport chain complexes in children with autism. J Neurochem. 2011;117(2):209–220. doi: 10.1111/j.1471-4159.2011.07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chauhan A., Chauhan V. Oxidative stress in autism. Pathophysiol. Off J Int Soc Pathophysiol. 2006;13(3):171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 61.Frustaci A., Neri M., Cesario A. Oxidative stress-related biomarkers in autism: systematic review and meta-analyses. Free Radical Biol Med. 2012;52(10):2128–2141. doi: 10.1016/j.freeradbiomed.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 62.James S.J., Rose S., Melnyk S. Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J. 2009;23(8):2374–2383. doi: 10.1096/fj.08-128926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.James S.J., Cutler P., Melnyk S. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80(6):1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- 64.Chauhan A., Audhya T., Chauhan V. Brain region-specific glutathione redox imbalance in autism. Neurochem Res. 2012;37(8):1681–1689. doi: 10.1007/s11064-012-0775-4. [DOI] [PubMed] [Google Scholar]
- 65.Geier D.A., Kern J.K., Garver C.R., Adams J.B., Audhya T., Geier M.R. A prospective study of transsulfuration biomarkers in autistic disorders. Neurochem Res. 2009;34(2):386–393. doi: 10.1007/s11064-008-9782-x. [DOI] [PubMed] [Google Scholar]
- 66.Frye R.E., Melnyk S., Macfabe D.F. Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Translational Psychiatry. 2013;3:e220. doi: 10.1038/tp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark-Taylor T., Clark-Taylor B.E. Is autism a disorder of fatty acid metabolism? Possible dysfunction of mitochondrial beta-oxidation by long chain acyl-CoA dehydrogenase. Med Hypotheses. 2004;62(6):970–975. doi: 10.1016/j.mehy.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 68.Poling J.S., Frye R.E., Shoffner J., Zimmerman A.W. Developmental regression and mitochondrial dysfunction in a child with autism. J Child Neurol. 2006;21(2):170–172. doi: 10.2310/7010.2006.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Q., He Z., Zubkov D. Lipidome alterations in human prefrontal cortex during development, aging, and cognitive disorders. Mol Psychiatry. 2018 doi: 10.1038/s41380-018-0200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tarugi P., Averna M., Di Leo E. Molecular diagnosis of hypobetalipoproteinemia: an ENID review. Atherosclerosis. 2007;195(2):e19–e27. doi: 10.1016/j.atherosclerosis.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Saraswathi V., Hasty A.H. The role of lipolysis in mediating the proinflammatory effects of very low density lipoproteins in mouse peritoneal macrophages. J Lipid Res. 2006;47(7):1406–1415. doi: 10.1194/jlr.M600159-JLR200. [DOI] [PubMed] [Google Scholar]
- 72.Vargas D.L., Nascimbene C., Krishnan C., Zimmerman A.W., Pardo C.A. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57(1):67–81. doi: 10.1002/ana.20315. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki K., Sugihara G., Ouchi Y. Microglial activation in young adults with autism spectrum disorder. JAMA psychiatry. 2013;70(1):49–58. doi: 10.1001/jamapsychiatry.2013.272. [DOI] [PubMed] [Google Scholar]
- 74.Ma C., Li J., Bao Z., Ruan Q., Yu Z. Serum levels of ApoA1 and ApoA2 are associated with cognitive status in older men. Biomed Res Int. 2015;2015 doi: 10.1155/2015/481621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Oda T., Wals P., Osterburg H.H. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1-42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136(1):22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 76.Kim J.H., Kim J.H., Jun H.O. Protective effect of clusterin from oxidative stress-induced apoptosis in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2010;51(1):561–566. doi: 10.1167/iovs.09-3774. [DOI] [PubMed] [Google Scholar]
- 77.Trougakos I.P. The molecular chaperone apolipoprotein J/clusterin as a sensor of oxidative stress: implications in therapeutic approaches - a mini-review. Gerontology. 2013;59(6):514–523. doi: 10.1159/000351207. [DOI] [PubMed] [Google Scholar]
- 78.Harold D., Abraham R., Hollingworth P. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Trougakos I.P., Gonos E.S. Regulation of clusterin/apolipoprotein J, a functional homologue to the small heat shock proteins, by oxidative stress in ageing and age-related diseases. Free Radic Res. 2006;40(12):1324–1334. doi: 10.1080/10715760600902310. [DOI] [PubMed] [Google Scholar]
- 80.Lambert J.-.C., Heath S., Even G. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 81.Park S., Mathis K.W., Lee I.K. The physiological roles of apolipoprotein J/clusterin in metabolic and cardiovascular diseases. Rev Endocr Metab Disord. 2014;15(1):45–53. doi: 10.1007/s11154-013-9275-3. [DOI] [PubMed] [Google Scholar]
- 82.Xie Z., Harris-White M.E., Wals P.A., Frautschy S.A., Finch C.E., Morgan T.E. Apolipoprotein J (clusterin) activates rodent microglia in vivo and in vitro. J Neurochem. 2005;93(4):1038–1046. doi: 10.1111/j.1471-4159.2005.03065.x. [DOI] [PubMed] [Google Scholar]
- 83.Nowaczyk M.J., Waye J.S. The Smith-Lemli-Opitz syndrome: a novel metabolic way of understanding developmental biology, embryogenesis, and dysmorphology. Clin Genet. 2001;59(6):375–386. doi: 10.1034/j.1399-0004.2001.590601.x. [DOI] [PubMed] [Google Scholar]
- 84.Fitzky B.U., Witsch-Baumgartner M., Erdel M. Mutations in the Delta7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. PNAS. 1998;95(14):8181–8186. doi: 10.1073/pnas.95.14.8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O'Roak B.J., Vives L., Girirajan S. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takata A., Miyake N., Tsurusaki Y. Integrative analyses of de novo mutations provide deeper biological insights into autism spectrum disorder. Cell Rep. 2018;22(3):734–747. doi: 10.1016/j.celrep.2017.12.074. [DOI] [PubMed] [Google Scholar]
- 87.Ionita-Laza I., Makarov V., Buxbaum J.D. Scan-statistic approach identifies clusters of rare disease variants in LRP2, a gene linked and associated with autism spectrum disorders, in three datasets. Am J Hum Genet. 2012;90(6):1002–1013. doi: 10.1016/j.ajhg.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li J., Shi M., Ma Z. Integrated systems analysis reveals a molecular network underlying autism spectrum disorders. Mol Syst Biol. 2014;10:774. doi: 10.15252/msb.20145487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krumm N., Turner T.N., Baker C. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rosenfeld J.A., Ballif B.C., Torchia B.S. Copy number variations associated with autism spectrum disorders contribute to a spectrum of neurodevelopmental disorders. Genet Med Off J Am Coll Med Genet. 2010;12(11):694–702. doi: 10.1097/GIM.0b013e3181f0c5f3. [DOI] [PubMed] [Google Scholar]
- 91.Fatemi S.H., Snow A.V., Stary J.M. Reelin signaling is impaired in autism. Biol Psychiatry. 2005;57(7):777–787. doi: 10.1016/j.biopsych.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 92.Persico A.M., D'Agruma L., Maiorano N. Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol Psychiatry. 2001;6(2):150–159. doi: 10.1038/sj.mp.4000850. [DOI] [PubMed] [Google Scholar]
- 93.Dziobek I., Gold S.M., Wolf O.T., Convit A. Hypercholesterolemia in Asperger syndrome: independence from lifestyle, obsessive-compulsive behavior, and social anxiety. Psychiatry Res. 2007;149(1-3):321–324. doi: 10.1016/j.psychres.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 94.Spahis S., Vanasse M., Belanger S.A., Ghadirian P., Grenier E., Levy E. Lipid profile, fatty acid composition and pro- and anti-oxidant status in pediatric patients with attention-deficit/hyperactivity disorder. Prostaglandins Leukot Essent Fatty Acids. 2008;79(1-2):47–53. doi: 10.1016/j.plefa.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 95.Ugur C., Uneri O.S., Goker Z., Sekmen E., Aydemir H., Solmaz E. The assessment of serum lipid profiles of children with attention deficit hyperactivity disorder. Psychiatry Res. 2018;264:231–235. doi: 10.1016/j.psychres.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 96.Berry-Kravis E., Levin R., Shah H., Mathur S., Darnell J.C., Ouyang B. Cholesterol levels in fragile X syndrome. Am J Med Genet A. 2015;167a(2):379–384. doi: 10.1002/ajmg.a.36850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fu X., Cate S.A., Dominguez M. Cysteine Disulfides (Cys-ss-X) as Sensitive Plasma Biomarkers of Oxidative Stress. Sci Rep. 2019;9(1):115. doi: 10.1038/s41598-018-35566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Goldani A.A., Downs S.R., Widjaja F., Lawton B., Hendren R.L. Biomarkers in autism. Frontiers in psychiatry. 2014;5:100. doi: 10.3389/fpsyt.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kurochkin I., Khrameeva E., Tkachev A. Metabolome signature of autism in the human prefrontal cortex. Commun Biol. 2019;2:234. doi: 10.1038/s42003-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.