Abstract
Exposure of airborne particulate matter (PM) with an aerodynamic diameter less than 2.5 μm (PM2.5) is epidemiologically associated with lung dysfunction and respiratory symptoms, including pulmonary fibrosis. However, whether epigenetic mechanisms are involved in PM2.5-induced pulmonary fibrosis is currently poorly understood. Herein, using a PM2.5-induced pulmonary fibrosis mouse model, we found that PM2.5 exposure leads to aberrant mRNA 5-methylcytosine (m5C) gain and loss in fibrotic lung tissues. Moreover, we showed the m5C-mediated regulatory map of gene functions in pulmonary fibrosis after PM2.5 exposure. Several genes act as m5C gain-upregulated factors, probably critical for the development of PM2.5-induced fibrosis in mouse lungs. These genes, including Lcn2, Mmp9, Chi3l1, Adipoq, Atp5j2, Atp5l, Atpif1, Ndufb6, Fgr, Slc11a1, and Tyrobp, are highly related to oxidative stress response, inflammatory responses, and immune system processes. Our study illustrates the first epitranscriptomic RNA m5C profile in PM2.5-induced pulmonary fibrosis and will be valuable in identifying biomarkers for PM2.5 exposure-related lung pathogenesis with translational potential.
Keywords: PM2.5 exposure, mRNA m5C, Pulmonary fibrosis, Inflammation, Immune response
Introduction
Exposure to airborne particulate matter with aerodynamic diameter less than 2.5 μm (PM2.5) has been epidemiologically associated with respiratory diseases [1], [2]. Harmful PM2.5 pollutants are released into the pulmonary surfactant and then attached to pulmonary epithelial cells [2]. Thus, it can increase the risk of multiple airway illnesses, including chronic obstructive pulmonary disease (COPD) [3], bronchitis [4], asthma [5], and idiopathic pulmonary fibrosis (IPF) [6]. Pulmonary fibrosis is characterized by excessive deposition of collagen in the lungs, leading to chronically impaired gas exchange and death [6]. It has been verified that PM2.5 and PM10 exposure accelerates functional decline in IPF patients determined by linear multivariable mixed-effects model [7].
Inflammatory responses [8], [9], immune cell activation [9], [10], [11], and oxidative stress [12], [13] are highly associated with pulmonary fibrosis pathogenesis. Oropharyngeal aspiration of PM2.5 induces significant collagen deposition and increases in the levels of the inflammatory markers interleukin 1-β (IL-1β) and transforming growth factor (TGF-β1), in mouse lungs after 21 days of treatment [8]. Anti-inflammatory cytokine IL-13 mediates fibrogenesis by regulating TGF-β1 expression and recruiting leukocytes into the lesion site [9]. Tumor necrosis factor-α (TNF-α) can also activate macrophages to secrete TGF-β1, leading to extracellular matrix (ECM) deposition [11]. In addition, high levels of oxidant stress have been detected in IPF patients compared with controls [14], and oxidant–antioxidant imbalances in the lower respiratory tract are strongly associated with IPF pathogenesis [12]. A recent study has unraveled that exposure to PM2.5 promotes fibrotic response by increasing the expression of Col1a1, Col3a1, Cox-4, and TGF-β1, activating smad3 expression, as well as generating reactive oxygen species (ROS) [15]. These findings further support the critical role of oxidative stress in fibrogenesis.
Epigenetic mechanisms, especially aberrant transcriptome induced by altered DNA methylation, have been shown to be associated with PM2.5 exposure and possible pathogenesis [16], [17]. RNA methylation, another epigenetic mechanism that regulates gene expression, has been shown to participate in various disease processes [18]. For example, as the most abundant internal RNA modification, N6-methyladenosine (m6A) is involved in acute myelocytic leukemia (AML), metabolic disorders, and nervous system disorders [18]. 5-Methylcytosine (m5C) is another prevalent RNA modification in mammals [19], [20] and plants [21], [22]. m5C modification is catalyzed by NOP2/Sun RNA methyltransferase family member 2 (NSUN2) in humans, mice, and human immunodeficiency viruses (HIV) [19], [20], [23], [24], [25], [26]. NUSN2 has been shown to mediate RNA transport by m5C reader of the nuclear protein Aly/REF export factor (ALYREF) [20], [27]. m5C participates in the regulation of RNA processing [24], [28], RNA stability [29], [30], [31], [32], and translation [33], [34]. Moreover, it plays critical roles in several important biological pathways in mice and plants [19], [22]. Recent advances in high-throughput sequencing technologies have facilitated m5C site identification at single-base resolutions [19], [20], [22], [23]. These findings suggest that m5C modification serves as a key posttranscriptional regulatory factor. However, whether the m5C methylome is altered upon PM2.5 exposure and further participates in the pathogenesis of pulmonary fibrogenesis is not well understood.
In our previous study, we have uncovered that PM2.5 directly affects macrophage polarization and induces the expression of the pro-inflammatory cytokines granulocyte–macrophage colony stimulating factor (GM-CSF), TNF-β, IL-1β, and IL-6 in mice [10]. In this study, the m5C methylome was profiled in the pathogenesis of lung fibrosis. We identified a possible novel role of altered m5C modification in mediating PM2.5-induced lung fibrosis through posttranscriptional regulation.
Results and discussion
PM2.5 exposure leads to pulmonary inflammation and fibrosis
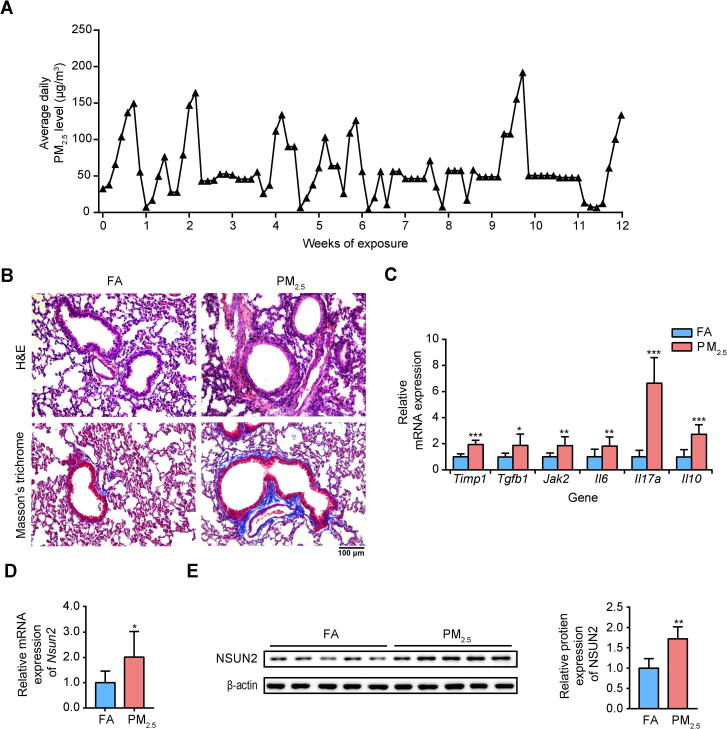
To mimic human exposure as accurately as possible, mice were maintained in ambient airborne PM2.5 chamber (Table S1). As shown in Figure 1A, the weekly average PM2.5 concentrations during the exposure period varied from 35.66 μg/m3 to 101.60 μg/m3. The mean airborne PM2.5 concentration to which the mice were exposed during the study period was ∼59.77 μg/m3. Histopathological examination of lung sections using hematoxylin and eosin (H&E) and Masson staining showed that compared to filter air (FA)-exposed mice, PM2.5-exposed mice exhibited severe lung injury and fibrosis (Figure 1B). Exposure caused intense inflammatory cell infiltration and thickened alveolar walls in the lungs (Figure 1B, top); it also caused excessive collagen deposition around the bronchi (Figure 1B, bottom). Moreover, the expression levels of the inflammatory markers Timp1, Tgfb1, Jak2, Il6, Il17a, and Il10 were higher in the PM2.5-exposed group than control group (P < 0.05, two-sided Wilcoxon and Mann–Whitney tests, Figure 1C). These results are consistent with previous reports demonstrating that PM2.5-induced disorder of inflammatory cytokine networks may lead to the death of lung epithelial cells and fibroblasts [35], [36], [37]. Importantly, we found that PM2.5 exposure increased NSUN2 mRNA and protein levels in the lung (Figure 1D and E). NSUN2 has been shown to methylate a variety of mRNAs and to promote or inhibit cell growth and proliferation [38]. It has been reported that the protein expression of NSUN2 is increased in human breast cancer [39]. As NSUN2 is a well-validated m5C methyltransferase [19], [20], [23], [24], [25], it is possible that altered expression of Nsun2 could disturb basic biological functions by affecting the number or methylation level of m5C sites. Thus, we hypothesize that PM2.5 may contribute to altered mRNA m5C methylomes during pulmonary fibrosis development.
Figure 1.
PM2.5 exposure induced pulmonary inflammation and fibrosis
A. The average daily PM2.5 concentrations in the exposure chamber during the study period. B. Representative images of H&E and Masson’s trichrome staining in lungs of FA- and PM2.5-exposed mice. Scale bar = 100 μm. C. RT-qPCR analysis showing the relative expression levels (compared to FA) of inflammatory and fibrotic genes in mouse lungs. D. The mRNA expression of Nsun2 in mouse lungs examined by RT-qPCR. E. The protein expression of NSUN2 in mouse lungs examined by Western blotting. The P values were determined using two-tailed Student’s t-tests. *, P < 0.05; **, P < 0.01; ***, P < 0.01. FA, filtered air; PM2.5, particulate matter with an aerodynamic diameter less than 2.5 μm; H&E, hematoxylin and eosin; Timp1, tissue inhibitor of metalloproteinase 1; Tgfb1, transforming growth factor, beta 1; Jak2, Janus kinase 2; Il, interleukin; NSUN, NOL1/NOP2/Sun domain family protein.
PM2.5 exposure triggers m5C modification in pulmonary fibrosis
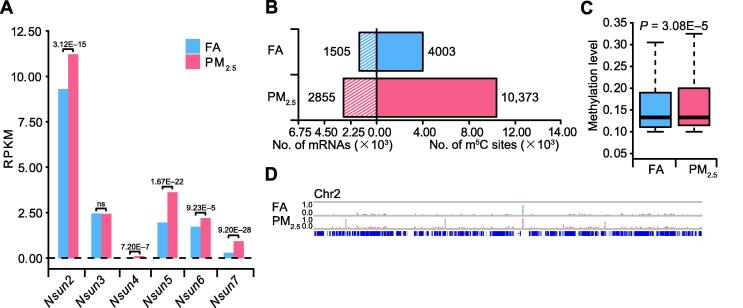
To define the potential mechanism of PM2.5-induced pulmonary fibrosis, we performed RNA bisulfite sequencing (RNA-BisSeq) and RNA sequencing (RNA-Seq) for the lung samples from FA- or PM2.5-exposed mice (Table S2). We found that the mRNA levels of most Nsun family members were upregulated in lung samples from PM2.5-exposed group with fold change (FC) > 1.2 and false positive rate (FDR) < 9.23E−5. Of all the Nsuns members, Nsun2 showed the highest expression level (Figure 2A). These results are consistent with the results of Western blot and RT-qPCR analyses (Figure 1D and E). Similarly, we observed that expression of Sftpc, Lcn2, Dbp, Mmp9, Ace2, and Hist1h2ad was upregulated in PM2.5-induced pulmonary fibrosis, whereas expression of Pink1, Jak1, Tgfb, and other nine pulmonary fibrosis-related genes was downregulated (Figure S1A) [40], [41], [42], [43]. Interestingly, 5/6 of the upregulated genes (Lcn2, Dbp, Mmp9, Ace2, and Hist1h2ad) had more m5C sites when comparing ling samples from PM2.5-exposed to those from FA-exposed mice samples. The results support a reasonable speculation that m5C probably participates in the regulation of pulmonary fibrosis development.
Figure 2.
Distribution profile of m5C in the lung tissues of FA- and PM2.5-exposed mice
A. Bar chart illustrating the expression levels of NOP2/Sun RNA methyltransferase family protein members in lungs of FA- and PM2.5-exposed mice, as examined by RNA-Seq. FDR values calculated using Benjamini–Hochberg method were labeled on top of the genes. B. Bar chart showing the numbers of m5C sites and m5C-modified mRNAs lung samples of FA- and PM2.5-exposed mice. C. Boxplots showing the overall distributions of mRNA m5C levels in lung samples of FA- and PM2.5-exposed mice using meRanTK. The P values were calculated using two-sided Wilcoxon and Mann–Whitney tests. D. IGV tracks displaying the distributions of m5C methylation level (in the range of 0.0–1.0) on chromosome 2 in lung samples of FA- and PM2.5-exposed mice. FDR, false positive rate; IGV, Integrative Genomics Viewer.
NSUN2 upregulation is probably the main reason for m5C alterations. It has been demonstrated that PM2.5 can elicit oxidative stress by inducing ROS generation and disrupting intracellular redox balance [17]. Moreover, oxidative stress (H2O2 exposure) has been shown to increase NSUN2 protein expression [44]. We found that transcription factors (TFs) encoded by 12 genes, including Egr1, Egr2, Hlf, Hoxa3, Hsf1, Hsf2, Irf7, Max, Nfe2, Pax6, Pparg, and Stat4 (P < 0.001, hypergeometric test), can potentially bind to the promoter sequence of Nsun2 [45]. Expression of these genes was upregulated in PM2.5-exposed group (FC > 1.2, FDR < 0.05). Thus, we speculate that PM2.5 induces oxidative stress and increases expression of TFs, which are involved in the upregulation of Nsun2 and consequently the increase in m5C level. However, how PM2.5 induces pulmonary fibrogenesis through modulating NSUN2 and TFs warrants further investigation.
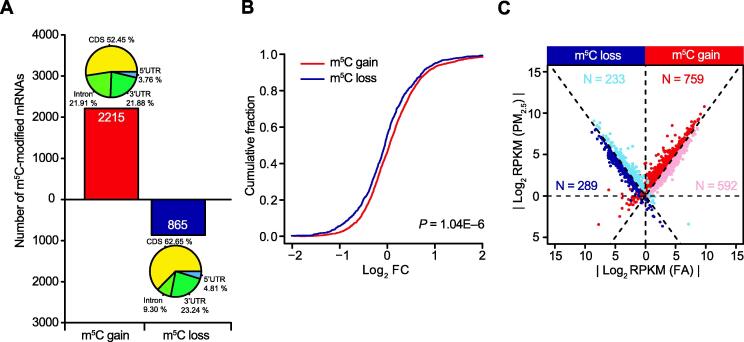
We next compared the m5C features between PM2.5-exposed and control FA groups. We found consistent m5C distribution in different regions and conserved m5C motif (Figure S1B and C). Intriguingly, the numbers of m5C sites and modified mRNAs increased markedly in PM2.5-treated group; 6370 additional m5C sites in 1350 mRNAs were detected in the PM2.5-treated group (Figure 2B). The global methylation level was also increased (P = 3.08E−5, two-sided Wilcoxon and Mann–Whitney tests, Figure 2C). In addition, we examined the number of modified mRNAs among different chromosomes. Almost all the chromosomes underwent m5C gain, except mitochondrial and Y chromosomes, most likely due to their shorter genome lengths (Figure S1D). The Integrative Genomics viewer (IGV) tracks also displayed a wide range of m5C gain across the whole chromosome 2 (Figure 2D). We then identified 2215 mRNAs with m5C gain and 865 mRNAs with m5C loss in lungs of PM2.5-exposed mice compared to those of FA-exposed mice (Figure 3A). The expression of genes with m5C gain was upregulated compared to that of genes with m5C loss overall (P = 1.04E−6, two-sided Wilcoxon and Mann–Whitney tests, Figure 3B). To exclude the potential impact caused by gene expression, we compared the coverage of m5C-modified genes based on the RNA-BisSeq data for the m5C gain and loss groups between lung samples of FA and PM2.5-exposed mice RNA-BisSeq (Figure S2A). High Pearson correlation coefficients between FA- and PM2.5-exposed mice for gene coverage were obtained in both the m5C gain (R = 0.89, P < 2.20E−16) and loss groups (R = 0.99, P < 2.20E−16). This result indicates that the m5C global changes are not affected by gene expression.
Figure 3.
mRNA m5C gain or loss is associated with aberrant transcriptome
A. Number of mRNAs with m5C gain (red) and loss (dark blue) in lung samples from PM2.5-exposed mice, compared to those from FA-exposed mice. The pie chart shows the distributions of m5C sites in different genic regions. B. Cumulative distribution displaying the expression level change of genes with m5C gain (red) and loss (dark blue) from RNA-Seq data in lung samples from PM2.5-exposed mice, compared to those from FA-exposed mice. The P values were calculated using two-sided Wilcoxon and Mann–Whitney tests and are shown in the bottom-right corner. C. Scatter plot showing the distributions of genes with significant RNA abundance changes (FC > 1.5, FDR < 0.05) with regard to both m5C gain/loss and gene expression levels in lung samples from PM2.5-exposed mice, compared to those from FA-exposed mice. The dots in red, pink, blue, and dark blue represent upregulated genes with m5C gain, downregulated genes with m5C gain, upregulated genes with m5C loss and downregulated genes with m5C loss, respectively. FC, fold change.
We further identified 12,595 differentially expressed genes (DEGs) in PM2.5-exposed group (FC > 1.2, FDR < 0.05), including 8964 upregulated DEGs and 3631 downregulated DEGs (Figure S2B). Functional enrichment analysis showed that these DEGs were highly enriched in cellular metabolic processes, RNA processing, immune responses, anatomical structure morphogenesis, system development, and cell differentiation (Figure S2C and D). To delineate the impact of mRNA m5C on the transcriptome profile in mouse pulmonary fibrosis, we compared the alterations in RNA abundance of gene sets with m5C gain or loss. We divided the gene sets into four groups (see details in Materials and methods): upregulated genes with m5C gain (n = 759), upregulated genes with m5C loss (n = 233), downregulated genes with m5C gain (n = 592), and downregulated genes with m5C loss (n = 289) (Figure 3C). Importantly, over half of the genes with m5C gain exhibited upregulated expression levels (759 out of 1351, 56.18%). We found hundreds of mRNAs whose expression was abundant and probably affected by m5C gain or loss in lungs of PM2.5-exposed mice. Altogether, m5C modification may regulate expression of the modified genes that are associated with the pathogenesis of PM2.5-induced pulmonary fibrosis.
m5C modification regulates PM2.5-induced immune and fibrotic responses
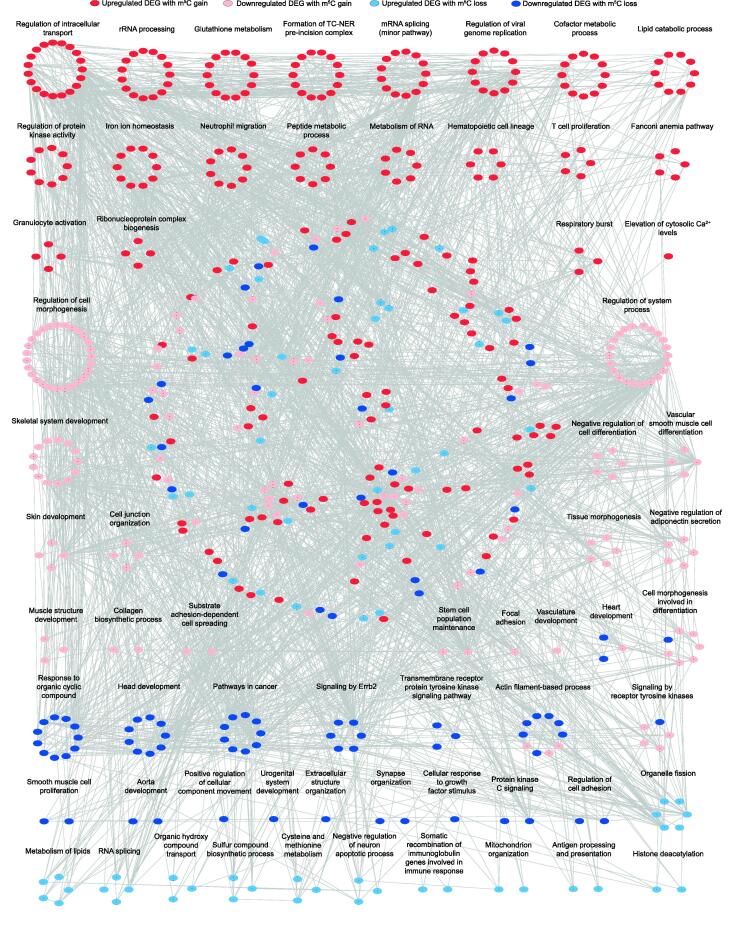
To further determine the biological significance of the m5C-modified DEGs in the lungs of mice exposed to PM2.5, gene ontology (GO) and pathway analyses were carried out. A protein–protein interaction (PPI) network analysis of the proteins encoding these DEGs indicates that both up- and downregulated DEGs are involved in multiple processes (Figures 4 and S3; Tables S3 and S4). As shown in Figure 4, for example, upregulated genes with m5C gain were enriched in neutrophil migration, granulocyte activation, as well as mRNA splicing and metabolism. In contrast, downregulated genes with m5C loss were involved in cancer, cell adhesion regulation, and other processes. These results suggest that PM2.5 exposure may regulate mRNA processing in lung lesions, leading to the recruitment and activation of immune cells. In particular, mRNA m5C gain negatively affected normal metabolic activity by upregulating expression levels of some genes in the lung. Therefore, we speculate that mRNA m5C modification may function as a pivotal factor regulating the pathogenesis of pulmonary fibrosis.
Figure 4.
GO enrichment map of m5C-modifed DEGs
Groups of upregulated genes with m5C gain (red), downregulated genes with m5C gain (pink), upregulated genes with m5C loss (blue), and downregulated genes with m5C loss (dark blue) are marked in the network (P < 0.01, hypergeometric test; for details, see Tables S3 and S4). The dots in the middle of the network with no functional labels represent genes that participate in multiple biological processes. GO, Gene Ontology; PPI, protein–protein interaction; DEG, differentially expressed gene.
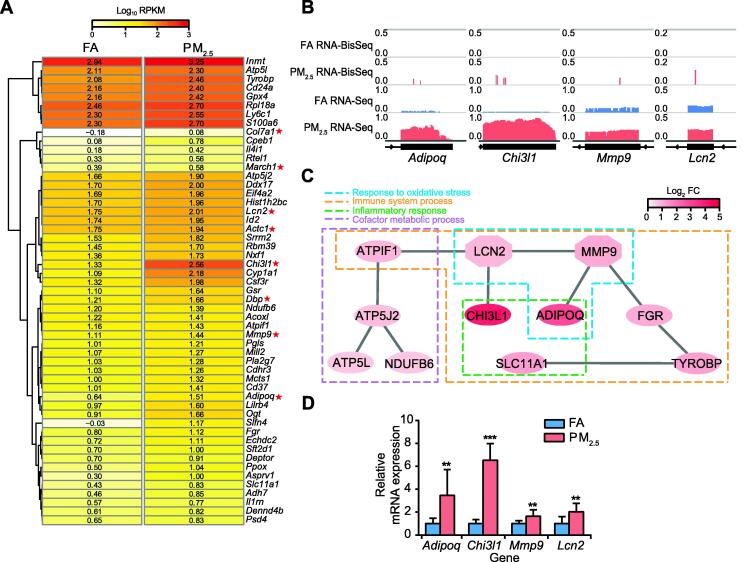
GO enrichment map of these gene sets showed that upregulated genes with m5C gain were mainly associated with RNA metabolism, oxidative stress responses, inflammatory responses, and immune system process (Figure 4). Thus, we selected top 53 upregulated DEGs with m5C gain that are highly associated with these processes to explore the potential influence of m5C in the lungs of mice with pulmonary fibrosis (Figure 5A). The IGV tracks of Adipoq, Chi3l1, Mmp9, and Lcn2 are shown in Figure 5B as examples of alterations in m5C levels and gene expression. The tracks clearly exhibit m5C gain sites and the higher expression levels of these genes in PM2.5-exposed mice than in FA-exposed mice. Moreover, large changes in RNA abundance mediated by acquisition of m5C modifications were associated with pulmonary fibrosis induction (Figure 5C). Among the important genes selected (Figure 5A), Lcn2 and Mmp9 have been reported to participate in pulmonary fibrosis development [46], [47]. Chi3l1 plays a major role in inflammation and is a potential target for prevention and treatment [48], whereas Adipoq is highly associated with the incidence of COPD [49]. The expression of these four key genes was also validated by RT-qPCR (Figure 5D).
Figure 5.
Upregulated DEGs with m5C gain were associated with immune and inflammatory responses in pulmonary fibrosis
A. Heatmap illustrating the expression levels of the top 53 upregulated DEGs with m5C gain (ranked by FDR) in mouse lungs (FC > 1.5, FDR < 0.05). The red asterisks mark the important DEGs with validated functions in oxidative stress responses, inflammatory responses, and immune system processes. B. IGV tracks displaying m5C and RNA-Seq read distributions in the FA (blue) and PM2.5-exposed (watermelon red) samples (from left to right: Adipoq, Chi3l1, Mmp9, and Lcn2). C. PPI network map of 11 pulmonary fibrosis-related upregulated genes with m5C gain. The color bar represents the mRNA log2 fold change between the two groups. D. RT-qPCR results showing the increased expression of four representative genes in lung samples from FA-exposed (bars in blue) and PM2.5-exposed (bars in watermelon red) mice. P values were determined using two-tailed Student’s t-tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ADOPOQ, adiponectin, C1Q and collagen domain-containing; CHI3L1, chitinase-like 1; MMP9, matrix metallopeptidase 9; LCN2, lipocalin 2; ATPIF1, ATPase inhibitory factor 1; ATP5J2, ATP synthase, H+-transporting mitochondrial F0 complex subunit F2; ATP5L, ATP synthase, H+-transporting mitochondrial F0 complex subunit G; NDUFB6, NADH: ubiquinone oxidoreductase subunit B6; SLC11A1, solute carrier family 11; TYROBP, TYRO protein tyrosine kinase binding protein.
These observations are consistent with a previous report showing the important role of Lcn2 and Mmp9 in inflammation and fibrosis through the IL-17 signaling pathway (Figure S4) [48], [49]. IL-17 induces inflammation by activating fibroblasts, endothelial cells, and epithelial cells [50]; it also enhances neutrophil recruitment to sites of inflammation [51]. In the present study, upregulation of Mmp9 and Lcn2 expression through m5C gain upon PM2.5 exposure may be critical in m5C-mediated fibrosis. Furthermore, m5C modification is potentially implicated in the pathogenesis of PM2.5-induced pulmonary fibrosis.
Conclusion
In this study, we report the first epitranscriptomic RNA m5C profile in PM2.5-induced pulmonary fibrosis. Our study offers insight into the relationship between PM2.5-induced pulmonary fibrosis and an altered mRNA m5C methylome. Moreover, mRNA m5C gain probably negatively affects normal metabolic activity, including RNA metabolism, oxidative stress responses, inflammatory responses, and immune system processes by upregulating gene expression levels in lungs of PM2.5-exposed mice. The results suggest that mRNA m5C modification may play a critical role in the pathogenesis of PM2.5-induced pulmonary fibrosis. Altogether, the results provides valuable evidence for elucidating the mechanism of PM2.5 exposure-related lung pathogenesis and identifying novel potential biomarkers.
Materials and methods
Experimental animals
Four-week-old male C57BL/6 mice were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). The mice were randomly assigned into the FA control group (n = 5) or PM2.5-exposed group (n = 5). Animals were either exposed to ambient PM2.5-polluted air or FA for 12 h/day, 7 days/week for 12 weeks (October 2017–January 2018) in a whole-body PM2.5 exposure system at the Zhongguancun Campus of the University of Chinese Academy of Sciences (UCAS), Beijing, China (N39°57′39.83″E116°20′10.97″). The exposure system included two separate chambers. In the FA chamber, ambient microparticles were removed by high-efficiency particulate air filter (Shanghai Liancheng Purification Equipment, Shanghai, China). In the PM2.5 chamber, ambient PM2.5 were collected with a swirler as previously reported [52]. The PM2.5 levels in the chamber were continually monitored and sampled daily. During the exposure period, the animals were maintained in exposure chamber with 12 h light/dark cycle (temperature: 21 °C–22 °C; humidity: 40%–60%) and given standard laboratory chow and sterile deionized water ad libitum.
PM2.5 sampling and preparation
To evaluate the PM2.5 levels in the PM2.5 chamber, ambient PM2.5 was sampled during the exposure period every day. The distance between the sampling inlets and the PM2.5 chamber was 30 m. The PM2.5 particles were collected by Teflon coated filters (diameter = 90 mm, Whatman, St. Louis, MO) with medium-volume samplers (TH-150D II, Tianhong, Wuhan, China) at 100 l/min flow rate for 12 h (08:00–20:00) every day. Before and after sampling, the filters were equilibrated for 48 h at 30% relative humidity and room temperature (25 °C). Then the filters were weighed for calculation of ambient PM2.5 levels. The sampled PM2.5 was extracted by sonication into deionized water (18 MΩ·cm) according to the method we reported previously [53]. In brief, PM2.5 samples were sonicated for 0.5 h with the sonicator (Catalog No. KQ-700 V, Shumei, Kunshan, China). The PM2.5 suspension was stored at −80 °C prior to analysis.
The chemical properties of the PM2.5 were analyzed as described previously [53]. Briefly, the metal elements were extracted by acid digestion (HNO3: HF = 7:3). Then the solution measured by inductively coupled plasma mass spectrometry (ICP-MS, Elemental X7, Thermo Fisher Scientific, Waltham, MA). The water-soluble inorganic components were measured with ion chromatography (Dionex-600, Thermo Fisher Scientific). Organic carbon (OC) and elemental carbon (EC) were determined by thermal-optical analyzer (Sunset Laboratories, Tigard, OR). The concentrations of metal elements, inorganic components, and carbon of the PM2.5 samples are listed in Table S1.
Tissue processing
After exposing to PM2.5, mice were anesthetized and sacrificed. The lung tissues were fixed using 4% paraformaldehyde solution, then embedded in paraffin. Lung sections (6 μm) were stained by H&E (Catalog No. G1005, Wuhan Servicebio Technology, Wuhan, China) or with a Masson trichrome stain kit (Catalog No. DC0033, Beijing Leagene, Beijing, China).
Construction of the RNA-Seq and RNA-BisSeq libraries
Total RNA was extracted from mouse lungs with an RNAsimple Total RNA Kit (Catalog No. DP419, Tiangen, Beijing, China), and mRNA was isolated using a Dynabeads® mRNA Purification Kit (Catalog No.61006, Ambion, Waltham, MA). TURBO™ DNase (Catalog No. 61006, Ambion) treatment was used to eliminate DNA contamination at 37 °C for 30 min. Then, the mRNA was purified by ethanol precipitation. RNA extracted from the group of FA-exposed mice and that of PM2.5-exposed mice was pooled separately and used for construction of the RNA-Seq and RNA-BisSeq libraries.
The RNA-Seq libraries were constructed with a KAPA Stranded mRNA-Seq Kit (Catalog No. 07962207001, KAPA, Wilmington, MA). The RNA-BisSeq libraries were constructed based on the methods in a previous study with minor optimization [20]. Briefly, in vitro- transcribed mouse Dhfr mRNA, a methylation conversion control, was mixed with approximately 200 ng of purified mRNA at a ratio of 1:300. Then, the RNA was fragmented into ∼100-nt fragments in 10× RNA Fragmentation Reagent (Catalog No. AM8740, Ambion) at 90 °C (1 min), which was terminated by 10× RNA stop solution (Catalog No. AM8740, Ambion). After precipitation with 100% ethanol, the RNA pellet was resuspended in 100 µl of a bisulfite solution containing a 100:1 mixture of 40% sodium bisulfite (Catalog No. 13438, Sigma, St. Louis, MO) and 600 µM hydroquinone (pH 5.1; catalog No. H9003, Sigma), and heat-incubated at 75 °C for 4.5 h. Nanosep columns with 3 K Omega membranes (Catalog No. OD003C35, PALL Corporation, Port Washington, NY) were used to desalt the reaction mixture with centrifugation. The RNA pellet was washed with nuclease-free water followed by centrifugation for five times. Finally, the RNA was re-suspended in 75 μl of nuclease-free water and then incubated at 75 °C for 1 h with equal volume of 1 M Tris-HCl (pH 9.0) for desulfonation. The RNA was dissolved in 11 µl of RNase-free water after ethanol precipitation. After reverse transcription with SuperScript II Reverse Transcriptase (Catalog No. 18064014, Invitrogen, Waltham, MA) and ACT random hexamers, a KAPA Stranded mRNA-Seq Kit (Catalog No. 07962207001, KAPA) was used to perform the subsequent procedures according to the manufacturer’s instructions.
Quantitative real-time PCR
Total RNA was isolated with an RNAsimple Total RNA Kit (Catalog No. DP419, Tiangen). cDNA was synthesized using a GoScript Reverse Transcription System (Catalog No. A5001, Promega, Madison, WI) according to the manufacturer’s instructions. The primer pairs used in this study are listed in Table S5. The gene expression levels were normalized to that of ACTB. RT-qPCR was performed as previously described [54]. P values were determined using two-tailed Student’s t-test. A difference with P < 0.05 was defined as significant.
Western blotting analysis
Protein was extracted from the mouse lung tissues using RIPA lysis buffer containing PMSF (Catalog No. 97064-672, Amresco, Radnor, PA) and protease and phosphatase inhibitor cocktails (Catalog Nos. B14001 and B15001, Bimake, Houston, TX). The solution was centrifuged at 12,000g and 4 °C for 10 min, the supernatant was collected for further analysis. The expression of NSUN2 and β-actin in whole-cell lysates was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Immunoreactive bands were detected with ECL reagents (Catalog No. 1705061, Bio-Rad, Berkeley, CA) according to the manufacturer’s instructions. The following antibodies were used: rabbit anti-NSUN2 (Catalog No. 20854-1-AP, Proteintech, Wuhan, China) and mouse β-actin (Catalog No. AF0003, Beyotime, Shanghai, China).
Data analysis for high-throughput sequencing
RNA-Seq and RNA-BisSeq were performed using an Illumina HiSeq 2500 platform with a paired-end read length of 150 bp. The adaptor sequences were trimmed off and low-quality bases were removed using Trimmomatic (version 0.33) [55]. For RNA-Seq, the remaining reads with lengths greater than 35 nt were used for the alignment with the mouse reference genome (version mm10) with TopHat (version 2.1.1, default parameters) [56]. Uniquely mapped reads (q ≥ 20) were used for the downstream analysis (Table S2). DEGs were detected with the DEGseq package in the R language [57]. Genes with FC > 1.2 and FDR < 0.05 were defined as significantly up- or downregulated genes. For RNA-BisSeq, reads that contained more than three sequential Cs were removed to reduce the number of false positives [58], [59], and the resulting reads were then aligned to reference genomes with meRanT align (meRanTK version 1.2.0) [60] with parameters: -fmo -mmr 0.01. The m5C sites were called by meRanCall (meRanTK, version 1.2.0) [60] with parameters: -mBQ 20 -mr 0 and only sites with coverage depth ≥ 10, methylation level ≥ 0.1, methylated cytosine depth ≥ 2 were considered credible. BEDTools’ intersectBed (version 2.26.0) [61] was used to annotate the m5C sites. mRNAs with m5C gain or loss were defined as mRNAs that were specifically modified in the PM2.5 exposure or control samples. IGV (IGVTools, version 2.3.8) [62] was used for visualization.
To explore the essential role of m5C modification in PM2.5-induced pulmonary fibrosis in mice, DEGs were separated into four groups based on m5C gain or loss and upregulation or downregulation of gene expression (FC > 1.2, FDR < 0.05). Only genes with reads per kilobase per million reads (RPKM) >1 in at least one sample and mRNA with FC > 1.5 were subjected to downstream analysis. The signaling network in lungs of mice exposed to PM2.5 was assessed with KEGG Mapper (https://www.kegg.jp/kegg/mapper.html). DAVID (version 6.8, http://david.abcc.ncifcrf.gov/) and Metascape (http://metascape.org) were used to perform GO analysis. GO terms with P < 0.05 (hypergeometric test) were considered statistically significant (Table S3). DEGs from the four groups mentioned above were used to perform PPI network analysis. PPI network was generated using the Search Tool for the Retrieval of Interacting Genes (STRING) database [63] (Table S4) and visualized with Cytoscape (version 3.6.0) [64].
Promoter is defined as the region ±2 kb around the transcription start site [45]. Prediction of TFs that potentially bind to the promoter sequence of Nsun2, was performed on https://biogrid-lasagna.engr.uconn.edu/lasagna_search/.
Ethical statements
The animal studies were performed under the guidance of laboratory animal care (NIH publication no. 85-23, revised 1985) and approved by the University of Chinese Academy of Sciences Animal Care and Use Committee.
Data availability
The RNA-Seq and RNA-BisSeq data have been uploaded to the Gene Expression Omnibus database (GEO: GSE122493), and to the Genome Sequence Archive [65] at the National Genomics Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences/China National Center for Bioinformation (GSA: CRA001230 with BioProject ID: PRJCA001116), and are publicly accessible at https://bigd.big.ac.cn/gsa/.
Authors’ contributions
WD and YGY conceived this project. HL, XH, ZZ, and WY performed the experiments and analyzed the data. CW and FZ contributed to animal care and technical supports. XL performed the physiochemical characterization of PM2.5 analysis. XH and BS performed bioinformatics analysis. XH, HL, ZZ, YZ, and GJ wrote the manuscript with the help of all authors. All authors read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
This work was supported by the State Key Program of the National Natural Science Foundation of China (Grant No. 91643206), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB14030300), and the Chinese Academy of Sciences/State Administration of Foreign Experts Affairs (CAS/SAFEA) International Partnership Program for Creative Research Teams of China.
Handled by Chengqi Yi
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.11.005.
Contributor Information
Yun-Gui Yang, Email: ygyang@big.ac.cn.
Wenjun Ding, Email: dingwj@ucas.ac.cn.
Supplementary material
The following are the Supplementary data to this article:
Distribution features of mRNA m5C in the lungs of FA and PM2.5-exposed mice A. Heatmap illustrating the mRNA expression levels and m5C modification of several pulmonary fibrosis-related genes in the lung. The color bar represents the normalized RPKM of the genes. The blue and white boxes represent m5C-modified and unmodified genes, respectively. B. Pie chart showing the transcriptome-wide distribution of mRNA m5C sites. C. Sequence frequency logo for the sequences proximal to mRNA m5C sites (generated using WebLogo). D. RNA-BisSeq results revealing the numbers of m5C-modified mRNAs among different chromosomes. MT, mitochondria.
PM2.5 exposure leads to altered gene expression in mouse lungs A. Scatter plots showing the coverage of genes with m5C gain and loss in lung samples from FA-exposed and PM2.5-exposed mice. The Pearson correlation coefficient (R) is shown in the top left corner. B. Volcano plots displaying enrichment of DEGs in lung samples from PM2.5-exposed mice compared to FA-exposed mice. The numbers of upregulated (blue) or downregulated (purple) DEGs (FC > 1.2, FDR < 0.05) are shown. The vertical dashed lines indicate the FC cut-off (1.2), whereas the horizontal dashed lines indicate the FDR cut-off (0.05). C. Representative GO biological process categories enriched for the upregulated genes. D. Representative GO biological process categories enriched for the downregulated genes. The color bar represents the −log10P value.
Bubble diagram of GO biological process categories enriched for DEGs with m5C gain or loss The pink circle represents the −log10P value.
Signaling network of m5C-modified Mmp9 and Lcn2 in pulmonary fibrosis mice The network was produced using KEGG Mapper (). Mmp9 and Lcn2 were marked in red and yellow, respectively.
References
- 1.Sese L., Nunes H., Cottin V., Sanyal S., Didier M., Carton Z. Role of atmospheric pollution on the natural history of idiopathic pulmonary fibrosis. Thorax. 2018;73:145–150. doi: 10.1136/thoraxjnl-2017-209967. [DOI] [PubMed] [Google Scholar]
- 2.Feng S., Gao D., Liao F., Zhou F., Wang X. The health effects of ambient PM2.5 and potential mechanisms. Ecotox Environ Safe. 2016;128:67–74. doi: 10.1016/j.ecoenv.2016.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Sint T., Donohue J.F., Ghio A.J. Ambient air pollution particles and the acute exacerbation of chronic obstructive pulmonary disease. Inhal Toxicol. 2008;20:25–29. doi: 10.1080/08958370701758759. [DOI] [PubMed] [Google Scholar]
- 4.Hertz-Picciotto I., Baker R.J., Yap P.S., Dostal M., Joad J.P., Lipsett M. Early childhood lower respiratory illness and air pollution. Environ Health Persp. 2007;115:1510–1518. doi: 10.1289/ehp.9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li N., Hao M., Phalen R.F., Hinds W.C., Nel A.E. Particulate air pollutants and asthma: a paradigm for the role of oxidative stress in PM-induced adverse health effects. Clin Immunol. 2003;109:250–265. doi: 10.1016/j.clim.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Johannson K.A. Air pollution exposure and IPF: prevention when there is no cure. Thorax. 2018;73:103–104. doi: 10.1136/thoraxjnl-2017-210808. [DOI] [PubMed] [Google Scholar]
- 7.Winterbottom C.J., Shah R.J., Patterson K.C., Kreider M.E., Panettieri R.A., Jr, Rivera-Lebron B. Exposure to ambient particulate matter is associated with accelerated functional decline in idiopathic pulmonary fibrosis. Chest. 2018;153:1221–1228. doi: 10.1016/j.chest.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng R., Tao L., Jian H., Chang Y., Cheng Y., Feng Y. NLRP3 inflammasome activation and lung fibrosis caused by airborne fine particulate matter. Ecotox Environ Safe. 2018;163:612–619. doi: 10.1016/j.ecoenv.2018.07.076. [DOI] [PubMed] [Google Scholar]
- 9.Borthwick L.A., Barron L., Hart K.M., Vannella K.M., Thompson R.W., Oland S. Macrophages are critical to the maintenance of IL-13-dependent lung inflammation and fibrosis. Mucosal Immunol. 2016;9:38–55. doi: 10.1038/mi.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q.J., Chen H., Yang T., Rui W., Liu F., Zhang F. Direct effects of airborne PM2.5 exposure on macrophage polarizations. Biochim Biophys Acta. 2016;1860:2835–2843. doi: 10.1016/j.bbagen.2016.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Lemos D.R., Babaeijandaghi F., Low M., Chang C.K., Lee S.T., Fiore D. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nat Med. 2015;21:786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 12.Kinnula V.L., Fattman C.L., Tan R.J., Oury T.D. Oxidative stress in pulmonary fibrosis: a possible role for redox modulatory therapy. Am J Resp Crit Care. 2005;172:417–422. doi: 10.1164/rccm.200501-017PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kliment C.R., Oury T.D. Oxidative stress, extracellular matrix targets, and idiopathic pulmonary fibrosis. Free Radical Biol Med. 2010;49:707–717. doi: 10.1016/j.freeradbiomed.2010.04.036. [DOI] [PubMed] [Google Scholar]
- 14.Cantin A.M., North S.L., Fells G.A., Hubbard R.C., Crystal R.G. Oxidant-mediated epithelial cell injury in idiopathic pulmonary fibrosis. J Clin Invest. 1987;79:1665–1673. doi: 10.1172/JCI113005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin G., Xia J., Zhang Y., Guo L., Chen R., Sang N. Ambient fine particulate matter exposure induces reversible cardiac dysfunction and fibrosis in juvenile and older female mice. Part Fibre Toxicol. 2018;15:27. doi: 10.1186/s12989-018-0264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng L., Liu J., Li B., Liu S., Li X., Tu H. Cigarette smoke-induced hypermethylation of the GCLC gene is associated with COPD. Chest. 2016;149:474–482. doi: 10.1378/chest.14-2309. [DOI] [PubMed] [Google Scholar]
- 17.Wei H., Liang F., Meng G., Nie Z., Zhou R., Cheng W. Redox/methylation mediated abnormal DNA methylation as regulators of ambient fine particulate matter-induced neurodevelopment related impairment in human neuronal cells. Sci Rep. 2016;6:33402. doi: 10.1038/srep33402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X., Su R., Weng H., Huang H., Li Z., Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amort T., Rieder D., Wille A., Khokhlova-Cubberley D., Riml C., Trixl L. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017;18:1. doi: 10.1186/s13059-016-1139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y. 5-Methylcytosine promotes mRNA export – NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui X., Liang Z., Shen L., Zhang Q., Bao S., Geng Y. 5-Methylcytosine RNA methylation in Arabidopsis Thaliana. Mol Plant. 2017;10:1387–1399. doi: 10.1016/j.molp.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 22.David R., Burgess A., Parker B., Li J., Pulsford K., Sibbritt T. Transcriptome-wide mapping of RNA 5-methylcytosine in Arabidopsis mRNAs and noncoding RNAs. Plant Cell. 2017;29:445–460. doi: 10.1105/tpc.16.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain S., Sajini A.A., Blanco S., Dietmann S., Lombard P., Sugimoto Y. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khoddami V., Cairns B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat Biotechnol. 2013;31:458–464. doi: 10.1038/nbt.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Courtney D.G., Tsai K., Bogerd H.P., Kennedy E.M., Law B.A., Emery A. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019;26:217–227. doi: 10.1016/j.chom.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominissini D., Rechavi G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res. 2017;27:717–719. doi: 10.1038/cr.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan S., Tang H., Xing J., Fan X., Cai X., Li Q. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34:3630–3641. doi: 10.1128/MCB.00243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y.F., Zhang X.D., Shi J.C., Tuorto F., Li X., Liu Y.S. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20:535–540. doi: 10.1038/s41556-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Li A., Sun B.F., Yang Y., Han Y.N., Yuan X. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat Cell Biol. 2019;21:978–990. doi: 10.1038/s41556-019-0361-y. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y., Wang L., Han X., Yang W.L., Zhang M., Ma H.L. RNA 5-methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol Cell. 2019;75:1188–1202. doi: 10.1016/j.molcel.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Tang H., Fan X.Q., Xing J.Y., Liu Z.Y., Jiang B., Dou Y. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging. 2015;7:1143–1158. doi: 10.18632/aging.100860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing J.Y., Yi J., Cai X.Y., Tang H., Liu Z.Y., Zhang X.T. NSun2 promotes cell growth via elevating cyclin-dependent kinase 1 translation. Mol Cell Biol. 2015;35:4043–4052. doi: 10.1128/MCB.00742-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S., Zhang W., Zhang F., Roepstorff P., Yang F., Lu Z. TMT-based quantitative proteomics analysis reveals airborne PM2.5-induced pulmonary fibrosis. Int J Environ Res Pub Heal. 2018;16 doi: 10.3390/ijerph16010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z., Li Z., Liao Z., Gao S., Hua L., Ye X. PM2.5 induced pulmonary fibrosis in vivo and in vitro. Ecotox Environ Safe. 2018;171:112–121. doi: 10.1016/j.ecoenv.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 37.Zehender A., Huang J., Gyorfi A.H., Matei A.E., Trinh-Minh T., Xu X. The tyrosine phosphatase SHP2 controls TGF beta-induced STAT3 signaling to regulate fibroblast activation and fibrosis. Nat Commun. 2018;9:3259. doi: 10.1038/s41467-018-05768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W. mRNA methylation by NSUN2 in cell proliferation. Wiley Interdiscip Rev RNA. 2016;7:838–842. doi: 10.1002/wrna.1380. [DOI] [PubMed] [Google Scholar]
- 39.Yi J., Gao R., Chen Y., Yang Z., Han P., Zhang H. Overexpression of NSUN2 by DNA hypomethylation is associated with metastatic progression in human breast cancer. Oncotarget. 2017;8:20751–20765. doi: 10.18632/oncotarget.10612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cottin V., Reix P., Khouatra C., Thivolet-Bejui F., Feldmann D., Cordier J.F. Combined pulmonary fibrosis and emphysema syndrome associated with familial SFTPC mutation. Thorax. 2011;66:918–919. doi: 10.1136/thx.2010.151407. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y., Nunes F.A., Berencsi K., Gonczol E., Engelhardt J.F., Wilson J.M. Inactivation of E2a in recombinant adenoviruses improves the prospect for gene therapy in cystic fibrosis. Nat Genet. 1994;7:362–369. doi: 10.1038/ng0794-362. [DOI] [PubMed] [Google Scholar]
- 42.Wynn T.A. Fibrotic disease and the TH1/TH2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhal B.D., Li X., Piasecki C.C., Molina-Molina M. Angiotensin signalling in pulmonary fibrosis. Int J Biochem Cell Biol. 2012;44:465–468. doi: 10.1016/j.biocel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X., Liu Z., Yi J., Tang H., Xing J., Yu M. The tRNA methyltransferase NSun2 stabilizes p16INK(4) mRNA by methylating the 3′-untranslated region of p16. Nat Commun. 2012;3:712. doi: 10.1038/ncomms1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreos R., Ambrosini G., Perier R.C., Bucher P. The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 2015;43:D92–D96. doi: 10.1093/nar/gku1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wree A., McGeough M.D., Inzaugarat M.E., Eguchi A., Schuster S., Johnson C.D. NLRP3 inflammasome driven liver injury and fibrosis: roles of IL-17 and TNF in mice. Hepatology. 2018;67:736–749. doi: 10.1002/hep.29523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amatya N., Garg A.V., Gaffen S.L. IL-17 signaling: the Yin and the Yang. Trends Immunol. 2017;38:310–322. doi: 10.1016/j.it.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y., Peng H., Sun H., Peng X., Tang C., Gan Y. Chitinase 3-like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med. 2014;6:240ra76. doi: 10.1126/scitranslmed.3007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan Y., Jiang H., Kuang J., Hou X., Feng Y., Su Z. Genetic variations in ADIPOQ gene are associated with chronic obstructive pulmonary disease. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwakura Y., Ishigame H., Saijo S., Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Roussel L., Houle F., Chan C., Yao Y., Berube J., Olivenstein R. IL-17 promotes p38 MAPK-dependent endothelial activation enhancing neutrophil recruitment to sites of inflammation. J Immunol. 2010;184:4531–4537. doi: 10.4049/jimmunol.0903162. [DOI] [PubMed] [Google Scholar]
- 52.Wang H., Shen X., Tian G., Shi X., Huang W., Wu Y. AMPKalpha2 deficiency exacerbates long-term PM2.5 exposure-induced lung injury and cardiac dysfunction. Free Radic Biol Med. 2018;121:202–214. doi: 10.1016/j.freeradbiomed.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Rui W., Guan L., Zhang F., Zhang W., Ding W. PM2.5-induced oxidative stress increases adhesion molecules expression in human endothelial cells through the ERK/AKT/NF-kappaB-dependent pathway. J Appl Toxicol. 2016;36:48–59. doi: 10.1002/jat.3143. [DOI] [PubMed] [Google Scholar]
- 54.Zhao C., Li T., Han B., Yue W., Shi L., Wang H. DDAH1 deficiency promotes intracellular oxidative stress and cell apoptosis via a miR-21-dependent pathway in mouse embryonic fibroblasts. Free Radic Biol Med. 2016;92:50–60. doi: 10.1016/j.freeradbiomed.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L.K., Feng Z.X., Wang X., Wang X.W., Zhang X.G. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 58.Edelheit S., Schwartz S., Mumbach M.R., Wurtzel O., Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m(5)C within archaeal mRNAs. PLoS Genet. 2013;9:e1003602. doi: 10.1371/journal.pgen.1003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaefer M. RNA 5-methylcytosine analysis by bisulfite sequencing. Methods Enzymol. 2015;560:297–329. doi: 10.1016/bs.mie.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Rieder D., Amort T., Kugler E., Lusser A., Trajanoski Z. meRanTK: methylated RNA analysis ToolKit. Bioinformatics. 2016;32:782–785. doi: 10.1093/bioinformatics/btv647. [DOI] [PubMed] [Google Scholar]
- 61.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y.Q., Song F.H., Zhu J.W., Zhang S.S., Yang Y.D., Chen T.T. GSA: Genome Sequence Archive. Genomics Proteomics Bioinformatics. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution features of mRNA m5C in the lungs of FA and PM2.5-exposed mice A. Heatmap illustrating the mRNA expression levels and m5C modification of several pulmonary fibrosis-related genes in the lung. The color bar represents the normalized RPKM of the genes. The blue and white boxes represent m5C-modified and unmodified genes, respectively. B. Pie chart showing the transcriptome-wide distribution of mRNA m5C sites. C. Sequence frequency logo for the sequences proximal to mRNA m5C sites (generated using WebLogo). D. RNA-BisSeq results revealing the numbers of m5C-modified mRNAs among different chromosomes. MT, mitochondria.
PM2.5 exposure leads to altered gene expression in mouse lungs A. Scatter plots showing the coverage of genes with m5C gain and loss in lung samples from FA-exposed and PM2.5-exposed mice. The Pearson correlation coefficient (R) is shown in the top left corner. B. Volcano plots displaying enrichment of DEGs in lung samples from PM2.5-exposed mice compared to FA-exposed mice. The numbers of upregulated (blue) or downregulated (purple) DEGs (FC > 1.2, FDR < 0.05) are shown. The vertical dashed lines indicate the FC cut-off (1.2), whereas the horizontal dashed lines indicate the FDR cut-off (0.05). C. Representative GO biological process categories enriched for the upregulated genes. D. Representative GO biological process categories enriched for the downregulated genes. The color bar represents the −log10P value.
Bubble diagram of GO biological process categories enriched for DEGs with m5C gain or loss The pink circle represents the −log10P value.
Signaling network of m5C-modified Mmp9 and Lcn2 in pulmonary fibrosis mice The network was produced using KEGG Mapper (). Mmp9 and Lcn2 were marked in red and yellow, respectively.
Data Availability Statement
The RNA-Seq and RNA-BisSeq data have been uploaded to the Gene Expression Omnibus database (GEO: GSE122493), and to the Genome Sequence Archive [65] at the National Genomics Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences/China National Center for Bioinformation (GSA: CRA001230 with BioProject ID: PRJCA001116), and are publicly accessible at https://bigd.big.ac.cn/gsa/.