Abstract
Direct infusion-based shotgun lipidomics is one of the most powerful and useful tools in comprehensive analysis of lipid species from lipid extracts of various biological samples with high accuracy/precision. However, despite of many advantages, the classical shotgun lipidomics suffers some general dogmas of limitations, such as ion suppression, ambiguous identification of isobaric/isomeric lipid species, and ion source-generated artifacts, restraining the applications in analysis of low abundance lipid species, particularly those less ionizable or isomers that yield almost identical fragmentation patterns. This article reviews on the strategies (such as modifier addition, prefractionation, chemical derivatization, charge feature utilization, etc.) that have been employed to improve/eliminate these limitations in modern shotgun lipidomics approaches (e.g., high mass resolution mass spectrometry-based and multi-dimensional mass spectrometry-based shotgun lipidomics). Therefore, with the enhancement of these strategies for shotgun lipidomics, comprehensive analysis of lipid species including isomeric/isobaric species can be achieved in a more accurate and effective manner, greatly substantiating the aberrant lipid metabolism, signaling trafficking, and homeostasis under pathological conditions.
Keywords: In-source fragmentation, Ion suppression, Isobaric/isomeric species, Multi-dimensional mass spectrometry, Shotgun lipidomics
1. Introduction
1.1. Complication of lipids
Lipids play many crucial roles in organism. Aside as essential constituents of cellular membranes, lipids play key roles in numerous important biological processes, including signaling transduction, energy storage, cell growth, differentiation, and survival [1]. Moreover, with the development of other omics (i.e., genomics, transcriptomics, and proteomics) and molecular biology, new functions of lipids are being discovered [2]. It has been demonstrated that aberrant metabolism of lipids involves the pathogenesis of many human diseases, such as obesity and diabetes, cancer, cardiovascular, neurodegenerative, and autoimmune diseases [3]. Thus, determination of lipid alteration under different conditions could greatly contribute to the understanding of disease pathogenesis, development of potential biomarkers, and discovery of novel drug targets.
However, identification and quantification of all lipids is an enormously challenge: 1) the number of cellular lipid species existing in a cell, an organ, or a biological system, is extremely huge (Tens to hundreds of thousands of possible lipids as predicted are present in cellular lipidomes at the levels of amol/mg to nmol/mg protein [4]); 2) the content and composition of these lipids are dynamically changing with life cycle, environmental conditions, or physiological and/or pathological perturbation [1, 5]; and 3) in general, cellular lipids are highly diverse as they consist of different polar head groups, backbones, and various aliphatic chains. Meanwhile, the lipids within one class/subclass have some similarities [6]. They have an identical polar head group and backbone, but various aliphatic chains different in length (i.e., different number of carbon atoms), degrees of unsaturation, locations of double bonds (e.g., n-7/n-9/n-12 18:1 fatty acid (FA) isomers) [7], potential branches, linkages connecting to backbone (e.g., 18:1_16:0 and 16:0_18:1 phosphatidylcholine (PC) isomers), etc. To overcome the challenge, lipidomics was proposed in early 2000 as an independent disciplinary field to analyze all lipid species in a large scale and at the levels of intact molecular species [8].
1.2. Shotgun lipidomics
The progress of lipidomics is greatly accelerated by a series of modern analytical technologies, such as nuclear magnetic resonance, fluorescence spectroscopy, high performance liquid chromatography (HPLC), microfluidic technology, especially mass spectrometry (MS) [9]. Nowadays, MS-based approaches, possessing very high sensitivities and specificity, are the most powerful and remarkable ones in lipidomics. On the basis of whether the lipid solution delivered to the ion source chamber is under a condition of constant concentration during the period of lipid analysis, the MS-based approaches can roughly classified into two main categories: one is LC-MS based lipidomics, the other is directly infusion-based lipidomics (also termed “shotgun lipidomics”), which is the topic reviewed in the present article [6].
The basic principles of shotgun lipidomics are to exploit the unique chemical and physical properties inherent in discrete lipid classes, subclass, and individual lipids species to analyze lipid species directly from organic extracts of biological samples under a constant concentration of lipid solution [6]. Analysis of lipids after direct infusion was demonstrated at the earliest time when electrospray ionization mass spectrometry (ESI-MS) was applied for lipid analysis [10–12]. Identification of individual lipid species in these studies was conducted after tandem MS in the product ion mode. All these studies served as the protype of shotgun lipidomics for high mass resolution MS-based shotgun lipidomics.
Each lipid species of a polar lipid class has a common head group, which can yield one or more characteristic fragment ion(s) after collision-induced dissociation (CID). Through detection of the characteristic fragment ion(s) in the precursor-ion scanning (PIS) or neutral-loss scanning (NLS) mode using a tandem mass spectrometer, all individual species of such a class can be “specifically” detected. In 1997, Brugger et al. [13] developed a method to “isolate” the individual species of a class of interest through specific PIS or NLS with a triple quadrupole mass spectrometer after direct infusion. In the method, the signal-to-noise (S/N) ratio of spectra was greatly improved after the double filtering process of MS/MS. This method has been widely used for profiling of biological samples (e.g., plant samples) due to the great advantages, including simplicity, efficiency, ease of management, and less expensive instrumental requirement [14]. The tandem MS-based methodology becomes the choice of lipidomics profiling at the early stage of lipidomics field and thus is also termed as classical shotgun lipidomics. This classical one lays the foundation for development of multi-dimensional MS-based shotgun lipidomics (MDMS-SL).
With the rapid development of MS instrumentations and the joint efforts of investigators focusing on shotgun lipidomics, other two modern shotgun lipidomics platforms have been developed and well documented in the literature according to the features and the mass spectrometers employed. One is high mass resolution MS-based shotgun lipidomics as commercially available high mass resolution/accuracy mass spectrometers (i.e., quadrupole time-of-flight (Q-TOF), orbitrap, and Fourier-transform ion cyclotron resonance (FTICR)) have been advanced and widely used [15]. The other is MDMS-SL [16, 17]. The former takes the advantage of the resolving power of the mass spectrometer [15]. The latter maximally exploits the unique chemical and physical properties of lipids, including hydrophobicity, stability, and reactivity inherent in different lipid classes or subclasses for analysis of individual lipid species, even achievable for those of less ionizable lipid classes or in very low abundance [16].
1.3. General dogmas of limitations in classical shotgun lipidomics
Compared with LC-MS based approaches, one of the features of shotgun lipidomics is allowing unlimited time to perform detailed tandem MS analyses in different fragmentation modes, such as PIS, NLS, and product ion analysis. Therefore, in theory, the tandem MS-based shotgun lipidomics approach (i.e., classical shotgun lipidomics) could be used to detect individual species of any targeted lipid classes at the level of instrumentation sensitivity directly from lipid extracts of biological samples, which makes it still as one of the most widely used approaches in the current lipidomics studies [16].
However, this classical shotgun lipidomics suffers a few inherent limitations as mostly being criticized, which restrict its utilizations in lipidomics. The mostly criticized one is ion suppression, which may affect ion formation (thereby dynamic range and the limitation of detection), detection precision, and quantification accuracy. The existence of ion suppression may limit the analysis of some lipid classes, such as those present in low abundance and/or less ionizable. It is well known that the mass resolution and mass accuracy of a triple quadrupole mass spectrometer is not high (~1000 mass resolution and 100–1500 ppm mass accuracy). The ion suppression present in shotgun lipidomics makes the signals of the low abundance and/or less ionizable lipid species essentially buried in baselines, especially in the analysis by the full mass scan.
The second issue present in the classical shotgun lipidomics is the difficulties to resolve the isobaric/isomeric mass overlap between individual lipid species, which limits unambiguous lipid identification even between lipid classes/subclasses. For instance, it is difficult to provide unambiguous identification of isobaric (i.e., same nominal) lipid species of a class of interest present in a lipid extract, such as PC 30:1 and PC O-31:1 (“O” indicates an alkyl ether linkage at the sn-1 position). Moreover, since the classical shotgun lipidomics only focuses on the class-specific head group fragments, the FA substituents of lipid species are not identified. Thus, it does not allow researchers to distinguish isomeric lipid species that have different fatty acid chains or different linkages connecting to the backbone (e.g., PC 18:2_20:4 and PC 16:0_22:6; and PC 18:1/16:0 and PC 16:0/18:1).
Finally, the classical shotgun lipidomics may also be problematic to identify ion source-generated artifactual peaks in the specific spectrum. In-source fragmentation is always present in ESI-MS regardless of that the ESI source is believed to be soft. When a characteristic fragment of a class is detected to filter the potential species present in the lipid class of interest, some ion source-generated artifactual peaks could be displayed in the acquired mass spectrum. So far, these artifactual peaks cannot be distinguished by this approach.
Due to these aforementioned limitations, some people hold incorrect concepts/bias on shotgun lipidomics. In turn, the general dogmas of limitations lead to argument that lipids cannot be quantified by using shotgun lipidomics approaches. In the following sections, we would dissect these limitations and further summarize the strategies employed for improvement/elimination of these problems in modern shotgun lipidomics approaches.
2. Strategies for reduction/improvement of ion suppression in modern shotgun lipidomics technologies
2.1. Ion suppression
The term “ion suppression” usually means ionization efficiency or intensity of a compound or a group of compounds is significantly reduced owing to the presence of other compounds, changes in the matrix components, or due to the dramatic changes of the concentration of the compound itself [18]. The reduction of ionization efficiency of targeted compounds is largely due to the changes of the efficiency of droplet formation or droplet desolvation owing to the presence of other compounds/changes in the concentrations of themselves. Although the mechanism of ion suppression remains unclear, some factors are known to induce ion suppression, such as high concentration, mass overlapping, basicity, and elution in the same retention windows as the analyte of interest (in the case of LC-MS analysis) [19]. Therefore, ion suppression is present in both shotgun lipidomics and LC-MS analysis.
Generally, it is not difficult to imagine that multiple components at high concentrations compete for either space or charge in the process of ionization, as it occurs in shotgun lipidomics where directly infused lipid extracts of biological samples are extremely complex. Such competition leads to a lower signal for each component. Therefore, someone could dogmatically believe that lipids cannot be quantified by using a shotgun lipidomics approach, as ion suppression is present in all the approaches after direct infusion. In fact, this concept only does hold true for the analysis of lipids in the high-concentration region, in which lipids have aggregated, leading to differential ionization efficiencies. However, it has been demonstrated that the ionization efficiencies of individual lipid species of a polar lipid class are nearly identical in the low concentration range (e.g., pmol/μL or lower) due to these ionization efficiencies predominantly depend on the charge property of the same polar head group [11, 20]. In other words, although ion suppression is present, shotgun lipidomics approaches can be employed to quantify individual lipid species in comparison to the coexisting standard(s) under certain conditions.
Although ion suppression sometimes refers to as the matrix effect in LC-MS analysis, there are two types of phenomena related to ion suppression in shotgun lipidomics. The first one is intra-class ion suppression. It means that the low abundance species is suppressed by abundant species in a lipid class. The ionization efficiencies of individual lipid species within one lipid class are essentially identical, their concentrations in lipid extracts, however, are huge different from amol/mg to nmol/mg protein as aforementioned. The intra-class ion suppression results in even lower signals of the low abundance species, leading to difficult determination of the low or very low abundance lipid species. The second one is inter-class ion suppression. As its name indicates, ionization efficiencies of low abundance lipid class(es) are suppressed in the presence of the other abundance lipid class. Although the intensity ratios of individual species of low abundance lipid class(es) is essentially kept unchanged, the inter-class ion suppression causes lower signal intensities of lipid species of the whole class(es), greatly influencing the linear dynamic range of quantification for these types of lipid class(es).
2.2. Improvement of ion suppression by high mass accuracy-based shotgun lipidomics
In the high mass accuracy-based shotgun lipidomics platform, Q-TOF or orbitrap mass spectrometers offer not only excellent duty cycle, but also high mass resolution and mass accuracy (e.g., >10,000 mass resolution and <2–5 ppm mass accuracy) in comparison with the conventional triple quadrupole mass spectrometers [15, 21]. The improved duty cycle can greatly reduce the baseline noise and increase the detection sensitivity, thus directly improving the S/N ratio of spectra. The high mass resolution and mass accuracy could partially resolve at least two problems including peak broadening and peak shift, thus reducing the interferences from neighboring peaks. Then the enhancement of the S/N ratio of a selected ion is also achieved. Collectively, the FTICR mass spectrometer shows a high sensitivity as it possesses much higher resolution and mass accuracy (e.g., >100,000 mass resolution and <1–2 ppm mass accuracy), largely due to the reduction of the baseline noise [15]. Compared with the Q-TOF or orbitrap mass spectrometers, however, the FTICR mass spectrometer has lower duty cycle, needing longer cycle time for every scanning event. It obviously restricts its applications in LC-MS based lipidomics approaches. However, the potentials of a FTICR mass spectrometer for lipidomics could be fully explored in shotgun lipidomics if the cost is not a consideration since shotgun lipidomics offers unlimited time to conduct mass acquisition and perform every detailed tandem MS analyses. In summary, ion suppression in this platform is improved through reduction of the baseline noise, thus broadening the linear dynamic range of quantification for lipid class(es) and individual species.
2.3. Reduction of ion suppression by multi-dimensional MS-based shotgun lipidomics
Unlike high mass accuracy-based shotgun lipidomics, MDMS-SL approach minimizes ion suppression by improving the signals of targeted molecular ions of lipid class(es) in a spectrum. Many strategies are adapted to enhance molecular ions in this platform (Fig. 1).
Fig. 1.
A summary of the strategies for reduction of ion suppression in MDMS-SL. Many strategies have been exploited to reduce ion suppression through enhancement of the signals of molecular ions of selected lipid class(es) in MDMS-SL analysis. These include, but are not limited to, (1) adding a suitable modifier to enhance ionization responses of non-polar lipids; (2) exploiting more sensitive fragments to detect lipid species of a class or a subclass; (3) developing different derivatization methods for different moieties present in different lipid classes to selectively enhance ionization efficiency and/or to generate informative, sensitive, and specific fragment ions; and (4) using unique charge properties or prefractionation to analyze a particular lipid class(es) (e.g., cardiolipin as doubly-charged ions or lysolipids with prefractionation).
1). To use an appropriate modifier
Modifiers (e.g., ammonium acetate, ammonium formate, and lithium chloride) are used to enhance ionization responses of non-polar lipids, such as glycerolipids (including triacylglycerol (TAG) and diacylglycerol (DAG)) and cholesterol ester species. Usually, the protonated ions or deprotonated ions of different lipids are used to detect in the positive-ion mode or the negative-ion mode of ESI-MS analysis, respectively, in shotgun lipidomics. However, glycerolipids cannot readily be ionized by protonation from nonaqueous solutions, even in the presence of organic acids under an ESI condition since a prominent charge site is absent in this lipid class [10]. In contrast, these species can be easily ionized as small cation adducts, such as their ammonium, lithium, or sodium counterparts in the positive-ion mode, to display very abundant ion peaks in the spectra. Thus, an appropriate modifier can be added to the lipid solution to form one of these adducts for analysis of these lipids sensitively [10, 22]. Similarly, it is able to profile individual molecular species of cholesterol esters by PIS of m/z 369.3 (corresponding to a sterol derivative) from ammoniated molecular ions in the presence of an ammonium salt as a modifier in the lipid solution [23].
2). More sensitive fragments are defined
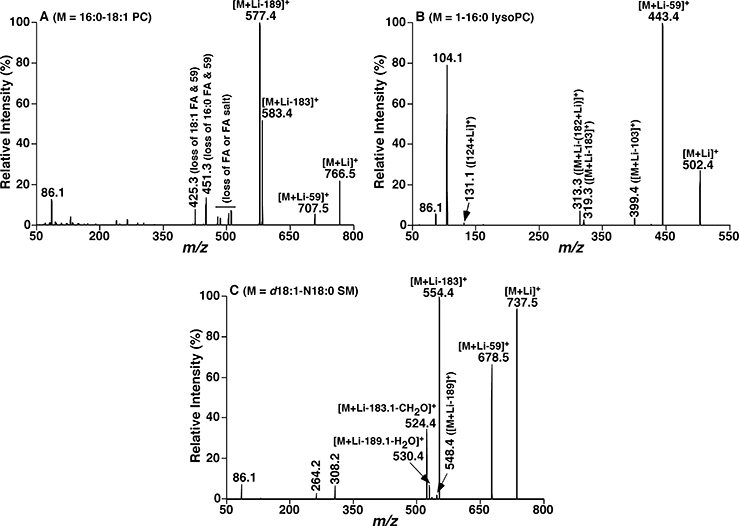
We take MDMS-SL analysis of phosphocholine-containing lipid species as an example. These lipids include PC, lysophosphatidylcholine (lysoPC), and sphingomyelin (SM) species. They all contain a quaternary amine in the form of zwitterions with the phosphate. The head group and the backbone of both PC and lysoPC species are the same, while lysoPC species have only a single FA chain connected to the glycerol backbone at either sn-1 or −2 position in the mass range from m/z 500–600. Both PC and SM species largely fall into the mass range between m/z 650 and 900. Under an acidic condition or an ammonium salt as a modifier added, protonated molecular species are readily formed in the positive-ion mode for all of these lipids. A fragment ion at m/z 184, corresponding to a phosphocholine ion is predominant in the product-ion spectra yielding from the protonated molecules of these lipid species, while other structurally informative fragment ions corresponding to the losses of FA chain(s) are in very low abundance. It is well known that the PC species are the most abundant phospholipids in most biological samples, such as plasma, the liver, kidneys, the heart, etc. [24] Thus, there exists obvious ion suppression, i.e., the SM and lysoPC species are suppressed by PC species and the low abundance PC species are suppressed by the abundant PC species when PIS of 184 Da is conducted as evidenced in classical shotgun lipidomics analysis [13]. However, when alkaline ions are available in the lipid solution, these lipid species can be readily ionized as alkaline adducts (alk = Li, Na, K) and yield different fragment ions in the positive-ion mode after low-energy CID. If no modifier carrying a cation is added to the lipid solution, sodium adducts are readily formed. In the case of lithium adduct (Fig. 2), three fragment ions resulting from the head group could be displayed in the product-ion spectrum of PC species: neutral losses of 59 Da (i.e., trimethylamine), 183 Da (i.e., choline phosphate), and 189 Da (i.e., lithium choline phosphate)). Additionally, fragments ions related to FA chains are also present in the product-ion spectra of lithiated PC species. The fragmentation pattern of lysoPC species is similar to PC species. In the MS/MS spectra of lithiated SM species, in addition to the three fragment ions from the PC head group as aforementioned, there also exist specific fragment ions to SM species: one is NLS of 213 Da (i.e., lithium choline phosphate plus methyl aldehyde); the second one is the neutral loss of phosphocholine and part of the sphingosine (e.g., NLS of 429 Da from SM species containing d18:1 sphingoid backbone, i.e., sphingosine); the last one is yielded from the backbone of the sphingoid base in SM (i.e., a fragment ion of m/z 246.2 from those SM species containing sphingosine). Therefore, lysoPC and SM species could be sensitively detected by NLS of 59 Da with the mass range from 500–600 and NLS of 213 Da, respectively. The low abundance PC species could be “isolated” and determined by NLS of 59 plus the contained FAs (e.g., odd-numbered fatty acids) [25]. Other fragment ions could be used for further validation in MDMS-SL analysis. Similar approaches could be applied to reduce ion suppression for other lipid classes by MDMS-SL analysis as long as these lipid species can be ionized to a certain degree and yield sensitive and specific fragment ions under the selected experimental condition. It should be mentioned that distinguishing ether-containing PC species from diacyl PC species can be readily achieved with the distinct ratios of the intensities of ions at NLS 59 and NLS 183 as previously described in detail [26].
Fig. 2.
Representative product-ion ESI-MS spectra of lithiated PC, lysoPC and SM species after CID. Product-ion ESI-MS mass spectra of lithiated PC 16:0/18:1 (a), lithiated 1-palmitoyl-sn-3-phosphocholine (b), and lithiated SM d18:1/18:0 (c) were performed on a QqQ mass spectrometer. Collision activation was conducted with collision energy of 32, 22, and 32 eV, respectively, and gas pressure at 1.0 mTorr.
3). Chemical derivatization is one of the most powerful methods to minimize ion suppression
Although many strategies have been exploited to reduce ion suppression, it is still difficult to analyze the species of some lipid classes because they are in very low concentrations, or unstable, or suppressed by other co-existing lipids, or no characteristic fragment ion(s). Chemical derivatization is the most attractive and effective strategy to resolve these difficulties and to selectively enhance the targeted analysis of a lipid class of interest by shotgun lipidomics [15, 17, 25, 27]. Some papers published by our group have summarized the major strategies recently exploited to enhance the MDMS-SL analysis in detail [17, 25, 27].
Briefly, these strategies could enhance MDMS-SL analysis in the following multiple ways. First, chemical derivatization could selectively enhance ionization efficiency of a selected lipid class which is unionizable through attaching an ionizable moiety, converting the less ionizable lipid species into readily ionized derivatives of lipid species of interest and eliminating the inter-class ion suppression. Moreover, introduction of a polar group could make the ionization efficiency of the selected lipid class mainly depend on the tagged group, indicating that the ionization efficiencies of lipid species within one non-polar class become essentially identical under an appropriately experimental condition. This approach can greatly improve the quantification accuracy of the non-polar lipid species. Furthermore, new fragmentation ion(s) can be generated from the derivatives of the tagged lipid species after CID, leading to more sensitive and specific identification of these lipids. Finally, the molecular masses of lipid species of interest can shift from the overlapped region to a new region without overlaps [25].
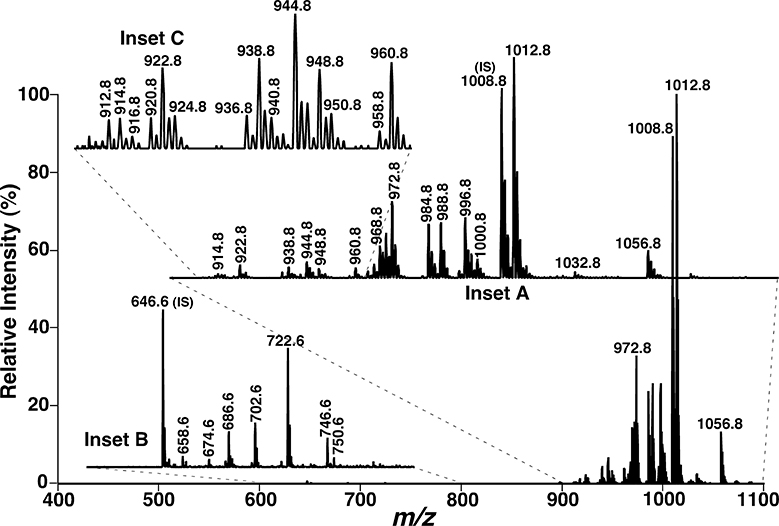
The analysis of phosphatidylethanolamine (PE) and lysophosphatidylethanolamine (lysoPE) molecular species derivatized with fluorenylmethoxylcarbonyl chloride (Fmoc-Cl) can also be used as an example to evidence the reduction of ion suppression between the classes. It is known that PE and lysoPE species carry a weakly zwitterionic head group and become anionic under an alkaline condition. Thus, these lipid species are usually analyzed after addition of a small amount of LiOH in the negative-ion mode. However, due to ion suppression of lysoPE and low abundance PE species by abundant PE species, the signals of many low-abundance PE species and almost the entire lysoPE class are buried in the baseline noise under experimental conditions. Moreover, these PE species containing long fatty acids can potentially overlap with lipid species from other lipid classes (i.e., phosphatidylinositol), bringing difficulties for identification. Our group has developed a strategy to improve the analysis of these lipids through derivatization of the primary amino group with Fmoc-Cl [28]. After CID, a prominent product ion resulting from the neutral loss of 222.2 Da (i.e., Fmoc moiety) is detected from all derivatives of Fmoc-PE and Fmoc-lysoPE species. The fragment feature can be employed to screen the presence of these lipids. Compared with the previous method, the detection limitation for PE and lysoPE analysis using this method has been improved by at least 100-fold with a >15,000-fold dynamic range (Fig. 3). Except the improvement of detection limitation, the reaction also shifts the PE species to a higher m/z region compared to the underived counterparts, greatly reducing overlaps with other endogenous lipids in the lipid extracts. Therefore, this approach can be used to unambiguously identify and quantify very low-abundance PE and lysoPE species.
Fig. 3.
Representative tandem mass spectrum of Fmoc-derivatized phoshoethanolamine-containing lipids in the lipid extract of mouse retinas by NLS of the Fmoc moiety. Tandem mass spectra of Fmoc-PE (inset A) and Fmoc-lysoPE (inset B) were acquired by NLS of 222.2 Da (i.e., Fmoc moiety) on a QqQ mass spectrometer with collision energy of 30 eV and collision gas pressure of 1.0 mTorr. Inset C shows the presence of many very low-abundance PE species in the region. IS denotes internal standard [28].
Similar methods have been exploited to reduce ion suppression mainly through improving the ionization efficiencies of lipid classes to enhance MDMS-SL platform for analyses of these lipid species [7, 29, 30].
4). Unique physical/chemical properties are exploited to reduce ion suppression
Although the MDMS-SL platform possesses more than 4 orders of magnitude dynamic range inherent in MS, it is still difficult to analyze the extremely low abundance lipids with the sensitivities of commercially available mass spectrometers due to ion suppression. Thus enrichment is necessary in some cases. In MDMS-SL analysis, different prefractionation methods are conducted to enrich a particular lipid class(es) depending on the unique physical/chemical properties of these lipids. Prefractionation herein means some methods to fractionate lipid categories, such as different solvent extraction, recovery of polar lipids from aqueous phase, separation by liquid chromatography or a solid phase extraction column, and others. There are some examples of prefractionation for shotgun lipidomics.
Non-polar lipids (e.g., TAGs) are predominant in some lipid extracts of biological samples (e.g., adipose tissue, plasma, plant oil, etc.), affecting the analyses of other lipids in shotgun lipidomics. A large amount of non-polar lipids dramatically reduces the concentration of lipid mixture in the infusion solution, whereas the upper limit of this concentration is to avoid formation of aggregation as previously described [31]. Even though a similar concentration of mixture could be achieved, the presence of excess non-polar lipids in the infusion solution may destabilize the ionization current, change the efficiency of droplet formation or droplet desolvation, and then reduce the ionization efficiency based on the principles of ESI-MS. Therefore, it is very helpful to remove, even though partially, these non-polar lipids for comprehensive analysis of other lipids present in the lipid extracts. To this end, hexane extraction could be used to washout the excess non-polar lipid prior to the subsequent analysis of other lipids since non-polar could be readily extracted with hexane against methanol [32].
Comprehensive analysis of lysophospholipids by shotgun lipidomics is another representative instance using a prefractionation method. Although most lipid species are extractable with organic solvents [33], some lipid classes (e.g., lysophospholipids) tend to be present in the aqueous phase after organic solvent extraction since lysophospholipids are composed of only a single FA chain and a polar head group. Recovery of these lipid classes from aqueous phase could enhance shotgun lipidomics analysis of lysophospholipids. Usually, besides lysoPE and lysoPC species, most of other lysophospholipids are present at very low concentrations in lipid extracts. Recently, to remove ion suppression resulting from phospholipids interferences, especially PC and PE species, a type of HybridSPE column has been used for preparation of biological samples prior to LC-MS analysis [34]. The working mechanism of HybridSPE is founded on the Lewis acid-base interaction between HybridSPE zirconia ions and phospholipids. Typically, the phosphate group inherent all phospholipids acts as a very strong Lewis base that interacts strongly with zirconia atoms coated on the particle surface in the solid phase extraction column under acidic conditions. The bounded lysophospholipids then could be eluted with a basic solution through varied pH conditions. Following the line of reasoning, our group has established a MDMS-SL method for comprehensive analysis of lysophospholipid species founded on the principles of shotgun lipidomics in combination with a novel strategy of sample preparation that recovers and enriches lysophospholipids from aqueous solution after solvent extraction using a HybridSPE column [35].
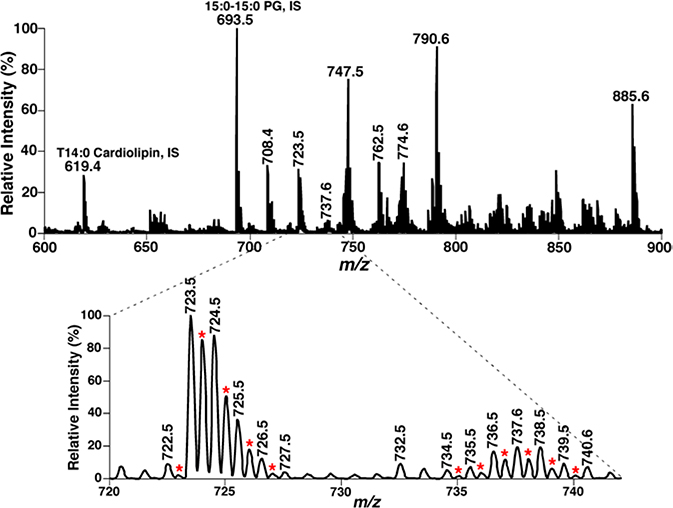
The special charge character (i.e., doubly-charged ions) of a lipid class could also be exploited by the MDMS-SL platform for selective and sensitive analysis of lipids. The majority of cellular lipids are only singly-charged ions under common experimental conditions. However, some classes of lipids, including cardiolipin, some gangliosides, polyphosphoinositides, etc. could be readily ionized as doubly-charged molecular ions. This charge character is unique to these lipids and has been used by MDMS shotgun lipidomics for analysis of these lipids (e.g., cardiolipin). Specifically, the majority of the formed doubly-charged ions of cardiolipin species and their isotopologues (i.e., [M-2H]2− and [M-2H+2]2−) fall into the mass region of m/z 600–800. Although the [M-2H]2− and [M-2H+2]2− peaks of cardiolipin species could overlap with the other lipid species, the plus-one isotopologue peaks (e.g., [M-2H+1]2− of doubly-charged cardiolipin molecular species) are very unique in lipid analysis of these species and could be easily recognized [36]. Fig. 4 shows an example of mass spectral analysis on lipid extracts of mouse myocardium by a triple quadrupole mass spectrometer. Through searching the plus-one isotopologue peaks of doubly-charged cardiolipin ions (indicated with red asterisks in the inset of Fig. 4), over 25 baseline-resolved doubly charged ion peaks could be easily recognized, corresponding to cardiolipin species present in the lipid extracts of mouse myocardium. Their FA constituents and structural isomers could be further identified and quantified in MDMS-SL analysis as previously demonstrated [36, 37].
Fig. 4.
Negative ion ESI-MS analysis of a lipid extract of mouse myocardium with a Thermo Scientific TSQ ESI mass spectrometer. The expanded mass spectrum clearly shows the presence of many doubly-charged ions corresponding to cardiolipin molecular species. IS, internal standard.
In addition to the strategies aforementioned, many other approaches, such as acid/base hydrolysis and the paired rule, are also exploited by the MDMS-SL platform to reduce ion suppression [25, 38].
3. Strategies for resolving isobaric/isomeric lipid species in modern shotgun lipidomics technologies
3.1. Isobaric and isomeric lipid species
The term “isomer” is widely used to describe the molecules that have identical formulas but distinct structures. Three major kinds of isomers are present in lipidomics: 1) lipids within different classes/subclasses but with the same elemental compositions, such as PC 30:1 and PE 33:1, PC O-31:1 and PC P-31:0 (“P” indicates a vinyl ether linkage at the sn-1 position); 2) isomeric lipid species within the same class, subclass, and sum composition, but differing in FA chain compositions, or in sn-1 and −2 regiochemistry (e.g., PC 18:2_20:4 vs. PC 16:0_22:6, PC 18:1/16:0 vs. PC 16:0/18:1); and 3) structurally defined isomers resulting from the different double bond locations of FA substituents (e.g., PE 16:0/18:1(n-7) vs. PE 16:0/18:1(n-9)) or different locations of one or two phosphate moieties in polyphosphoinositides (PPI).
Another phenomenon that these lipid species have a same nominal molecular mass with the different elemental composition (termed “isobaric” lipids) should be mentioned in lipidomics. For example, the protonated monoisotopic precursor ions of PC 30:1 (m/z 704.5225), PC O-31:1 (m/z 704.5589), and the 13C1 isotope ion of SM 34:1 (m/z 704.5887) all have a nominal mass of 704 and are displayed with one ion peak when a quadrupole-type mass spectrometer is used. LC-MS based lipidomics could resolve these isobaric mass overlaps of lipid species after LC separation. However, it is difficult to unambiguously identify them in classical shotgun lipidomics since all of them can yield a “characteristic” phosphocholine fragment ion at m/z 184 in the positive-ion mode after CID. The similar issues could also exist in analyses of other lipid classes using the classical shotgun lipidomics approach. Therefore, the presence of isobaric/isomeric lipid species within complex lipid extracts further complicates the mass spectrum, making identification and quantification of individual lipid species as a huge challenge.
3.2. Strategies to resolve isobaric and isomeric species by high resolution MS-based shotgun lipidomics
High mass accuracy-based shotgun lipidomics resolving isobaric mass overlaps of lipids strongly depend on the mass resolution and mass accuracy of the employed instrument. The mass difference of the isobaric pairs between the two-13C-atom-containing isotopologue of a species M (i.e., M+2 isotopologue) and the ion of a species with one less double bond than M is 0.009 Da, which is one of the most prominent mass differences in lipidomics as all lipid species are potentially affected by it. Computational modeling suggests that a mass analyzer with 100,000 mass resolution and sub-ppm mass accuracy could yield recognized individual signals in conventional spectral processing software, which can easily achieve the identification and quantification of these lipids, although these isobaric mass overlaps are not completely solved [15]. However, higher mass resolution (i.e., exceeding 500,000) might make little sense for identification of the isobaric lipids [39].
The fragmentation patterns of each lipid classes are employed to characterize isomeric lipid species by product-ion analysis in high resolution MS-based shotgun lipidomics. After CID, the fragmentation pattern of a lipid class depends on its intrinsic chemical structure and charge properties. Thus, in theory, the majority of lipid classes have a unique fragmentation pattern, including the fragment ions that relate to/reflect the head groups and the FA constituents of lipid class of interest. Therefore, in analysis by high mass resolution MS-based shotgun lipidomics, identification of isomeric lipid species is achieved by the fragments corresponding to either FA constituents or head group moieties recorded in the product-ion mass spectra using special software programs. Incidentally, compared with a triple quadrupole mass spectrometer, high resolution MS/MS could display completely resolved peaks of isobaric fragments (e.g., 0.1 amu or higher), greatly eliminating many possibilities of false-positive identification. However, some types of isomers which possess a similar/identical fragmentation pattern cannot be identified by this approach. Then other solutions are needed, including chromatographic fractionation, differential or multistep lipid extraction, intrasource separation and selective ionization during MS analysis, or derivatization [15].
3.3. Strategies to resolve isobaric and isomeric species by multi-dimensional MS-based shotgun lipidomics
In the MDMS-SL platform, the unique fragmentation patterns of each lipid classes are fully exploited to resolve isobaric and isomeric species. Specifically, the fragment ions or the neutrally-lost fragments are recorded by using two powerful tandem MS techniques (i.e., NLS and PIS) in a mass ramp format after CID. Then, the informative fragmentation(s) from either the headgroup or resulted from the neutral loss of headgroup are used to identify the lipid class of interest, and PIS and/or NLS of FA chains can be used to definitely the individual molecular species present within the class. Collectively, individual lipids species, including isobaric and isomeric species, as well as regioisomers, can be effectively and thoroughly identified using this approach.
For example, protonated PC ions yield an abundant fragment at m/z 184, whereas fragment ions corresponding to the losses of FA chain(s) are low abundance, and even undetectable in many cases. In contrast, in addition to the fragment ions resulting from the NLS of 59, 183, and 189 Da related the headgroup, one or two fragment ions corresponding to neutral losses of FA chain(s) plus 59 Da are also present in the product-ion spectra of alkaline adducts of PC species (Fig. 2). Therefore, NLS of 59, 183, and 189 Da of the lipid extract allows identification of the ion peaks in the full MS scan belonging to the PC class/subclasses. The spectra are also acquired from NLS of 59 Da plus all naturally occurring FAs in MDMS-SL. Then mapping of all of these spectra constitutes a two-dimensional mass spectrum. Individual PC species including regioisomers could be identified by using the two-dimensional mass spectrometric format. For example, this method has been achievable in analysis of lipid extracts of rabbit myocardium and 46 PC molecular species were identified and quantified [26].
A similar approach is developed in the analysis of TAG species by MDMS-SL. Due to the presence of multiple complicate isomeric ion peaks and severe mass overlaps between TAG and PC molecular species, identification and quantification of TAG species directly from the lipid extract of a biological sample is a huge challenge. As mentioned in Section 2.2, TAG species can be readily ionized in the positive-ion ESI-MS as their lithium adducts if an according modifier is present. Tandem MS analysis of lithiated TAG species shows a unique fragmentation pattern of fragment ions corresponding to the neutral losses of paired free FA(s) and their lithium salt(s). Thus, the number of abundant fragment ions present in a product-ion mass spectrum of lithiated TAG species is based on the number of different FA chains present in the TAG species. Individual TAG species including isomeric species, except the isomers due to double-bond locations and the regiospecific position, could be identified by exploiting this fragmentation pattern. Specifically, a full MS scan plus all NLS of naturally occurring FA chains (approximately 30 types of FAs) are sequentially acquired and used to construct a two-dimensional MS mapping. In the constructed mass spectrum, the cross peaks of a given precursor ion in the full MS scan with the NLS spectra present in the second dimension represent the FA chains that constitute the isomeric TAG species corresponding the selected precursor ion. Therefore, these isomeric TAG species could be readily determined from the number and intensities of these underlying neutral-loss fragment peaks in combination with the m/z value of TAG molecular ion [22].
However, without prior extensive separation by HPLC, it is usually impractical to distinguish these isobaric/isomeric lipid species that have similar/identical fragmentation patterns by shotgun lipidomics. In order to overcome these problems, chemical derivatization is also routinely used in the MDMS-SL approach, turning identical fragmentation patterns into different ones. In the past few years, our group has developed many methods in this respect [7, 29, 40, 41].
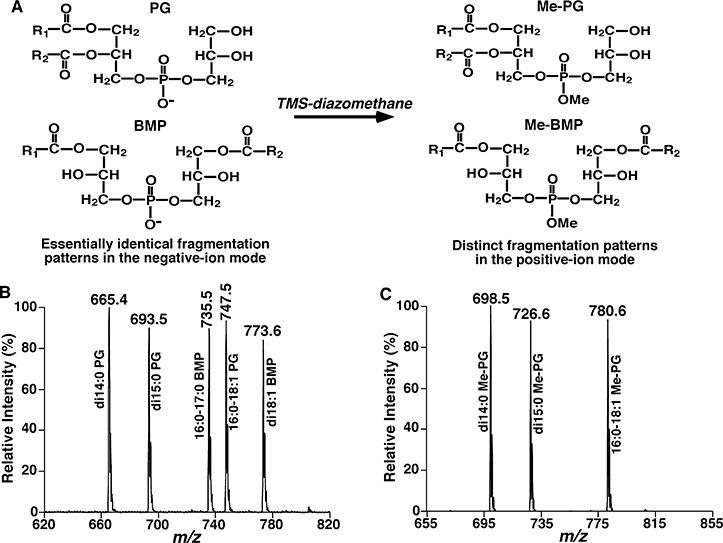
We take the analysis of isomeric bis(monoacylglycero)phosphate (BMP) and phosphatidylglycerol (PG) species as an example. BMP, playing important roles in glycosphingolipid degradation and cholesterol transport [42], is a class of negatively-charged glycerophospholipids with unusual sn-1/sn-1’ structural configuration (Fig. 5). While, the structural isomers of BMP, PG species also involve in many key cellular processes, accounting for ~1–2 % of phospholipids in most animal tissues [43]. Both of them have an identical fragmentation pattern in tandem MS spectra and could not be distinguished in shotgun lipidomics without extra solution treated. Moreover, HPLC-MS based studies on lipids are usually not comprehensive because of time restriction for total identification of FA substituents. On the principle of the derivative products of selected lipid species generating new fragmentation patterns, our group has successfully exploited a strategy for quantitative analysis of these isomeric lipid species using shotgun lipidomics through a methylation reaction [41]. After the methylation reaction with trimethylsilyl-diazomethane (TMS-diazomethane), the fragmentation patterns of ammoniated/lithiated Me-PG and Me-BMP species are obviously different, such as a dominant ion resulting from the neutral loss of 203 Da (methylated glycerophosphate derivative) is present in the product ion mass spectra of all ammoniated Me-PG species, whereas this peak is very minimal (absent) in those of Me-BMP species, in which display only one or two fragmentation ions corresponding to the losses of FA chains [41]. Typically, individual PG and BMP species as well as their potential mixtures present in lipid extracts of biological samples are identified by separate product ion analyses of individual ions and their total content is quantified compared with the prior added internal standard by high mass accuracy/resolution MS. Then after a methylation reaction, NLS 203 spectra of the methylated lipid extracts are acquired to identify and quantify Me-PG species. Finally, identification and quantification of individual Me-BMP species can be derived from these analyses. The principle of the approach for analysis of isomeric PG and BMP species is shown in Fig. 5.
Fig. 5.
Schematic illustration of the methylation reaction with TMS-diazomethane for analysis of isomeric PG and BMP species. Representative structures of PG and BMP with and without methylation (Panel A). Full-scan mass analysis (Panel B) and neutral loss scan of 203 Da after methylation (Panel C) [41].
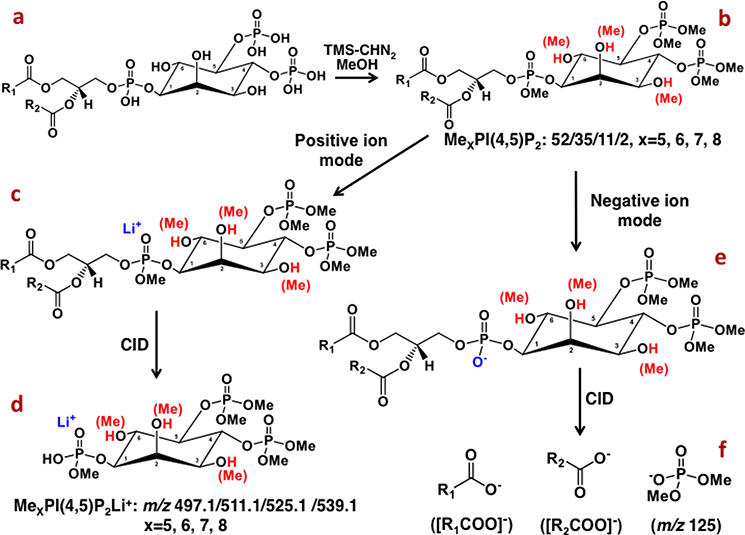
Analysis of PPI species with the MDMS-SL approach is another representative sample. PPI species, involving in numerous cellular processes, are a family of important cellular lipids [44]. However, the structures of isomeric PPI species, such as PIP (i.e., PIP[3’], PIP[4’], and PIP[5’]), and PIP2 (PIP2[3’,4’], PIP2[3’,5’], PIP2[4’,5’]) are very similar, leading to almost identical fragmentation patterns in tandem MS analysis. Recently, the methylation reaction with TMS-diazomethane has further been exploited to achieve a comprehensive analysis of individual PPI isomers using MDMS-SL approach [40]. For instance, the mass analyses of lithiated derivatives of PIP2 classes revealed that individual PIP2 classes have class-specific methylation patterns, that is, methylation of five sites was mainly in the classes of PIP2[3’,4’] and PIP2[4’,5’], while methylation of six groups in PIP2[3’,5’] was predominant. The lithiated species of PIP2 class containing the same number of methyl groups yield identical fragment ions (i.e., m/z 497.1, 511.1, 525.1, and 539.1) corresponding to lithiated polymethylphosphoinsitol (Fig. 6). Thus PIS analysis of these different methylated species in the positive-ion mode can be used to detect these lipids and quantify the total mass levels of the ions (including isomers) in the presence of an IS. Quantification of individual PIP2 species can be achieved through simulation of the mixtures of isomers based on the class-specific methylation patterns of PIP2 [40]. In addition, there exist abundant fragment ions resulting from FA constituent(s) and an ion at m/z 125 corresponding to dimethylphosphate in product-ion mass spectra of PIP2 species in the negative-ion mode (Fig. 6). These features can be used for identification of FA substituent(s) of individual species. Therefore, global identification of individual PIP2 species including phosphorylation isomers and FA chains is achieved. The procedures for analysis of other PPI species (e.g., PIP and PIP3) are similar [40].
Fig. 6.
Representative scheme of the methylation reaction of PIP2 with TMS-diazomethane and their resultant ions in MS and tandem MS analyses.
Although many approaches reviewed in this paper are used the MDMS-SL platform to reduce ion suppression or resolve isobaric and isomeric species as an example, the strategies can also be used in high mass accuracy-based shotgun lipidomics platform to overcome the same problems. Furthermore, there are many special approaches that are developed to enhance the analysis of the high mass accuracy-based shotgun lipidomics platform [15, 45], which have been reviewed previously [17].
4. Identification of existence of ion source-generated artifacts in modern shotgun lipidomics technologies
In-source fragmentation, a well-known phenomenon in ESI-MS, is a fragmentation process that occurs in the ion source during an MS analysis. The occurrence of this process largely depends on the analyte structure and harsh ionization conditions (e.g., high ionization temperature, high ionization voltage, very short distance between the spray tip and the inlet, leading to an extremely high electrical capacity, inappropriate setting of the gate voltages, etc.). In lipidomics, this phenomenon can be problematic, especially if the ion source-generated artifacts that correspond to other endogenous lipids. It has been demonstrated that the ions of lysoPC and PC species fragment in-source to produce ions with the same mass as lysoPE or PE species, respectively [46]; phosphatidic acid (PA) species could be generated from in-source fragmentation of their phosphatidylserine (PS) counterparts. Meantime, the observation of lysophosphatidic acid (lysoPA) species could also be resulted from in-source fragmentation of lysoPC species [47]. The in-source fragmentation could greatly impact on the lipid identification and subsequent quantitation in complex biological samples, especially in shotgun lipidomics. For example, the presence of both precursors and ion source-generated artifacts increase spectral complexity and complicate data analysis. Moreover, in-source fragmentation could reduce ionization sensitivity and alter their quantitation due to the loss of a certain amount of lipids in this process, producing falsely low concentrations. Finally, if ion source-generated artifacts have the same mass as endogenous lipids, these fragment ions might be identified as true lipids, leading to a falsely high concentration of the corresponding endogenous lipids.
Therefore, recognition of in-source fragmentation and identification of the resulted artifacts are of importance during the method development stage for a selected category of lipids. In modern shotgun lipidomics, identification of this type of ion source-generated artifacts is usual through addition of standards, which are absent endogenously. For example, a particular standard of PS species should be added to see the in-source produced PA species, which should be not overlapped with that endogenously-presented. Then the ion source-generated artifacts could be easily distinguished from the endogenous lipids. If the peaks of ion source-generated artifacts are abundant in the mass spectrum, a standard solution of lipids should be used to further tune the instrument to minimize/eliminate the in-source fragmentation process with a less harsh ionization condition. A suitable ionization condition can be examined with the minimal existence of some ion source-generated artifacts.
In summary, in-source fragmentation has been recognized and ion source-generated artifacts can also be distinguished with addition of standards in modern shotgun lipidomics approaches. Meanwhile, to reduce these types of artifacts, some measures could be taken to eliminate/minimize the in-source fragmentation during the development of an analytical method.
5. Conclusion
The strategies exploited/developed to reduce ion suppression, resolve isobaric/isomeric lipids, and identify/reduce ion source-generated artifacts in modern shotgun lipidomics approaches have been reviewed. Through these enhancements, shotgun lipidomics has been one of the most powerful analytical approaches in global analysis of cellular lipidomes directly from lipid extracts of biological samples with much more accuracy/precision. Currently, MDMS-SL could identify and quantify thousands of individual lipid species (including regioisomers) of >50 lipid classes in an automated and relatively high-throughput manner [27, 48]. It is anticipated that shotgun lipidomics can make greater contributions to the research on lipid metabolism and translational medicine with enhancement for comprehensive analysis of lipid species.
Acknowledgements
The work was partially supported by National Natural Science Foundation of China (No. 81803861), National Key R&D Program of China (2018YFC1705500), NIH/NIA (RF1 AG061872), intramural institutional research funds from the University of Texas Health Science Center at San Antonio (UT Health SA), the Mass Spectrometry Core Facility of UT Health SA, and the Methodist Hospital Foundation endowment.
Abbreviations
- BMP
bis(monoacylglycero)phosphate
- CID
collision-induced dissociation
- DAG
diacylglycerol
- FA
fatty acyl or fatty acid
- FTICR
Fourier-transform ion cyclotron resonance
- Fmoc-Cl
fluorenylmethoxylcarbonyl chloride
- HPLC
high performance liquid chromatography
- IS
internal standard
- lysoPA
lysophosphatidic acid
- lysoPC
lysophosphatidylcholine
- lysoPE
lysophosphatidylethanolamine
- MS
mass spectrometry
- MDMS-SL
multi-dimensional MS-based shotgun lipidomics
- NLS
neutral-loss scanning
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PIS
precursor-ion scanning
- PPI
polyphosphoinositide
- PS
phosphatidylserine
- Q-TOF
quadrupole time-of-flight
- SM
sphingomyelin
- TMS
diazomethane, trimethylsilyl-diazomethane
- TAG
triacylglycerol
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- [1].Han X, Nat. Rev. Endocrinol 2016, 12, 668. [DOI] [PubMed] [Google Scholar]
- [2].Schmitt S, Castelvetri LC, Simons M, Biochim. Biophys. Acta 2015, 1851, 999; A. C. Kendall, M. Kiezel-Tsugunova, L. C. Brownbridge, J. L. Harwood, A. Nicolaou, Biochimica et biophysica acta. Biomembranes 2017, 1859, 1679; A. Laganowsky, E. Reading, T. M. Allison, M. B. Ulmschneider, M. T. Degiacomi, A. J. Baldwin, C. V. Robinson, Nature 2014, 510, 172. [DOI] [PubMed] [Google Scholar]
- [3].Hu C, Zhou J, Yang S, Li H, Wang C, Fang X, Fan Y, Zhang J, Han X, Wen C, Free Radical Biol. Med 2016, 101, 475; H. Cheng, S. Guan, X. Han, J. Neurochem. 2006, 97, 1288; X. Han, D. M Holtzman, D. J. McKeel, J. Kelley, J. Morris, J. Neurochem. 2002, 82, 809. [DOI] [PubMed] [Google Scholar]
- [4].Shevchenko A, Simons K, Nat. Rev. Mol. Cell Biol 2010, 11, 593; X. Han, X. Jiang, Eur. J. Lipid Sci. Technol. 2009, 111, 39. [DOI] [PubMed] [Google Scholar]
- [5].Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, Kraut-Cohen J, Wang M, Han X, Asher G, Cell Metab. 2014, 19, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Han X, Gross RW, Mass Spectrom. Rev 2005, 24, 367. [DOI] [PubMed] [Google Scholar]
- [7].Wang M, Han RH, Han X, Anal. Chem 2013, 85, 9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Han X, Gross RW, Lipid Res J 2003, 44, 1071; M. Lagarde, A. Géloën, M. Record, D. Vance, F. Spener, BBA-Mol Cell Biol L 2003, 1634, 61. [DOI] [PubMed] [Google Scholar]
- [9].Armstrong D, Lipidomics, Vol. 1, Humana Press, 2009. [Google Scholar]
- [10].Duffin KL, Henion JD, Shieh JJ, Anal. Chem 1991, 63, 1781. [DOI] [PubMed] [Google Scholar]
- [11].Han X, Gross R, Proc Natl Acad Sci U S A 1994, 91, 10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kerwin JL, Tuininga AR, Ericsson LH, J. Lipid Res 1994, 35, 1002. [PubMed] [Google Scholar]
- [13].Brügger B, Erben G, Sandhoff R, Wieland F, Lehmann W, Proc Natl Acad Sci U S A 1997, 94, 2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X, J. Biol. Chem 2002, 277, 31994. [DOI] [PubMed] [Google Scholar]
- [15].Ryan E, Reid GE, Acc. Chem. Res 2016, 49, 1596. [DOI] [PubMed] [Google Scholar]
- [16].Han X, Yang K, Gross RW, Mass Spectrom. Rev 2012, 31, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu C, Wang C, He L, Han X, TrAC, Trends Anal. Chem 2018, 10.1016/j.trac.2018.11.028. [DOI] [Google Scholar]
- [18].Furey A, Moriarty M, Bane V, Kinsella B, Lehane M, Talanta 2013, 115, 104. [DOI] [PubMed] [Google Scholar]
- [19].Han X, Lipidomics: Comprehensive Mass Spectrometry of Lipids, Wiley: Hoboken, NJ, USA: 2016. [Google Scholar]
- [20].Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P, Lipid Res J 2001. 42, 663; X. Han, K. Yang, J. Yang, K. N. Fikes, H. Cheng, R. W. Gross, J. Am. Soc. Mass. Spectrom. 2006, 17, 264. [PubMed] [Google Scholar]
- [21].Wang J, Wang C, Han X, Anal. Chim. Acta 2019, 10.1016/j.aca.2019.01.043. [DOI] [Google Scholar]
- [22].Han X, Gross RW, Anal. Biochem 2001, 295, 13. [DOI] [PubMed] [Google Scholar]
- [23].Duffin K, Obukowicz M, Raz A, Shieh J, Anal. Biochem 2000, 279, 10. [DOI] [PubMed] [Google Scholar]
- [24].Cole VJ LK, Vance DE, Biochim. Biophys. Acta 2012, 1821, 8. [Google Scholar]
- [25].Wang C, Han X, Oil Crop Science 2017, 2, 195. [Google Scholar]
- [26].Yang K, Zhao Z, Gross RW, Han X, J. Chromatogr. B 2009, 877, 2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang M, Wang C, Han RH, Han X, Prog. Lipid Res 2016, 61, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Han X, Yang K, Cheng H, Fikes KN, G. R. W, J. Lipid Res 2005, 46, 1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang M, Hayakawa J, Yang K, Han X, Anal. Chem 2014, 86, 2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang M, Fang H, Han X, Anal. Chem 2012, 84, 4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang M, Wang C, Han X, Mass Spectrom. Rev 2017, 36, 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun G, Yang K, Zhao Z, Guan S, Han X, Gross R, Anal. Chem 2008, 80, 7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cruz M, Wang M, Frisch-Daiello J, Han X, Lipids 2016, 51, 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rahman M, Ahmad S, Gupta A, Hussain A, Kalra H, Raut B, Journal of Pharmacy and Bioallied Sciences 2012, 4, 267; D. Neville, R. Houghton, S. Garrett, Bioanalysis 2012, 4, 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang C, Wang M, Han X, Anal. Chem 2015, 87, 4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Han X, Yang K, Yang J, Cheng H, Gross RW, J. Lipid Res 2006, 47, 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Han X, Yang J, Yang K, Zhao Z, Abendschein D, Gross R, Biochemistry 2007, 46, 6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang K, Zhao Z, Gross R, Han X, PLoS One 2007, 2, e1368; X. Jiang, H. Cheng, K. Yang, R. W. Gross, X. Han, Anal. Biochem. 2007, 371, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang M, Huang Y, Han X, Rapid Commun. Mass Spectrom 2014, 28, 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang C, Palavicini JP, Wang M, Chen L, Yang K, Crawford PA, Han X, Anal. Chem 2016, 88, 12137. [DOI] [PubMed] [Google Scholar]
- [41].Wang M, Palavicini JP, Cseresznye A, Han X, Anal. Chem 2017, 89, 8490. [DOI] [PubMed] [Google Scholar]
- [42].Akgoc Z, Sena-Esteves M, Martin DR, Han X, d’Azzo A, Seyfried TN, J. Lipid Res 2015, 56, 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Mejiaa Edgard M., Nguyen Hieu, Hatch Grant M., Chem. Phys. Lipids 2014, 179, 6; H. Hirai, S. Natori, K. Sekimizu, Arch. Biochem. Biophys. 1992, 298, 458. [DOI] [PubMed] [Google Scholar]
- [44].Ketel K, Krauss M, Nicot AS, Puchkov D, Wieffer M, Muller R, Subramanian D, Schultz C, Laporte J, Haucke V, Nature 2016, 529, 408. [DOI] [PubMed] [Google Scholar]
- [45].Schuhmann K, Thomas H, Ackerman JM, Nagornov KO, Tsybin YO, Shevchenko A, Anal. Chem 2017, 89, 7046. [DOI] [PubMed] [Google Scholar]
- [46].Gathungu RM, Larrea P, Sniatynski MJ, Marur VR, Bowden JA, Koelmel JP, Starke-Reed P, Hubbard VS, Kristal BS, Anal. Chem 2018, 90, 13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhao Z, Xu Y, J Chromatogr B Analyt Technol Biomed Life Sci 2009, 877, 3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yang K, Cheng H, Gross RW, Han X, Anal. Chem. 2009, 81, 4356; C. Hu, M. Wang, X. Han, Redox Biol. 2017, 12, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]