The genus Flavivirus includes major human pathogens, as well as animal-infecting viruses with zoonotic potential. In order to counteract the threats these viruses represent, it is important to understand their basic biology to develop universal attenuation strategies. Here, we demonstrate that five different flaviviruses use cap-independent translation, indicating that the phenomenon is probably common to all members of the genus. The mechanism used for flavivirus cap-independent translation was found to be different from that of IRES-mediated translation and dependent on both 5′ and 3′ UTRs that act in cis. As cap-independent translation was also observed in mosquito cells, its role in flavivirus infection is unlikely to be limited to the evasion of consequences of the shutoff of host translation. We found that the inhibition of cap-independent translation results in decreased viral proliferation, indicating that the strategy could be applied to produce attenuated variants of flaviviruses as potential vaccine candidates.
KEYWORDS: duck Tembusu virus, cap-independent translation, secondary structure, untranslated regions
ABSTRACT
Duck Tembusu virus (DTMUV) (genus Flavivirus) is a causative agent of duck egg drop syndrome and has zoonotic potential. The positive-strand RNA genomes of flaviviruses are commonly translated in a cap-dependent manner. However, dengue and Zika viruses also exhibit cap-independent translation. In this study, we show that RNAs containing 5′ and 3′ untranslated regions (UTRs) of DTMUV, mosquito-borne Tembusu virus (TMUV), and Japanese encephalitis virus can be translated in a cap-independent manner in mammalian, avian, and mosquito cells. The ability of the 5′ UTRs of flaviviruses to direct the translation of a second open reading frame in bicistronic RNAs was much less than that observed for internal ribosome entry site (IRES) encephalomyocarditis virus, indicating a lack of substantial IRES activity. Instead, cap-independent translation of DTMUV RNA was dependent on the presence of a 3′ UTR, RNA secondary structures located in both UTRs, and specific RNA sequences. Mutations inhibiting cap-independent translation decreased DTMUV proliferation in vitro and delayed, but did not prevent, the death of infected duck embryos. Thus, the 5′ and 3′ UTRs of DTMUV enable the virus to use a cap- and IRES-independent RNA genome translation strategy that is important for its propagation and virulence.
IMPORTANCE The genus Flavivirus includes major human pathogens, as well as animal-infecting viruses with zoonotic potential. In order to counteract the threats these viruses represent, it is important to understand their basic biology to develop universal attenuation strategies. Here, we demonstrate that five different flaviviruses use cap-independent translation, indicating that the phenomenon is probably common to all members of the genus. The mechanism used for flavivirus cap-independent translation was found to be different from that of IRES-mediated translation and dependent on both 5′ and 3′ UTRs that act in cis. As cap-independent translation was also observed in mosquito cells, its role in flavivirus infection is unlikely to be limited to the evasion of consequences of the shutoff of host translation. We found that the inhibition of cap-independent translation results in decreased viral proliferation, indicating that the strategy could be applied to produce attenuated variants of flaviviruses as potential vaccine candidates.
INTRODUCTION
Duck Tembusu virus (DTMUV) is a causative agent of acute egg drop syndrome in ducks (1). DTMUV is related to mosquito-borne Tembusu virus (TMUV) and belongs to the genus Flavivirus (family Flaviviridae), which also includes human pathogens, such as yellow fever virus (YFV), Japanese encephalitis virus (JEV), West Nile virus (WNV), Zika virus (ZIKV), and dengue virus (DENV) (2). DTMUV replicates well in many types of nonavian cells, and its RNA has been frequently detected in human oral swabs, raising public concern regarding the transmission of DTMUV to humans (3). DTMUV also replicates in mosquito cells, indicating the possibility of mosquito transmission.
Like other flaviviruses, DTMUV has an approximately 11-kb-long positive-strand RNA genome that has a 5′ cap and lacks a 3′ poly(A) tail. The genome comprises a single large open reading frame (ORF) encoding a polyprotein that is processed into 3 structural proteins and 7 nonstructural proteins (4). The ORF is flanked by untranslated regions (UTRs), and for the DTMUV strain CQW1, the 5′ and 3′ UTRs have lengths of 95 and 619 nucleotides (nt), respectively. The genomes of flaviviruses contain a number of highly conserved RNA secondary structures, including stem-loops (SLs) A (SLA) and B (SLB) in the 5′ UTR and a 5′ cyclization sequence (5′ CS) in the region encoding the capsid protein. The 3′ UTR contains two dumbbell (DB) structures, a 3′ cyclization sequence (CS), a 3′ short hairpin structure (sHP), pseudoknots (PKs), and several SLs, including one at the extreme 3′ end of the genome (3′ SL). These structures are crucial for flaviviral RNA translation, replication, and packaging and to counteract the innate immune responses of hosts (5).
Protein synthesis is one of the most important physiological activities in cells. Cap-dependent translation is dependent on a 5′ cap structure that is used to bind the eukaryotic initiation factor 4F (eIF4F) complex (eIF4E, -4G, and -4A) and eIF4B. This binding results in mRNA activation, recruitment of the 43S preinitiation complex, and mRNA scanning in the 5′-to-3′ direction until a start codon in a suitable context is reached. Then, the 80S ribosome is assembled, and polypeptide synthesis is initiated (6). Viruses lack their own translation apparatus and must therefore rely on cellular translation machinery. Many viruses have adopted the use of the canonical 5′ cap-dependent translation initiation mechanism. However, some viruses use different cap-independent translation initiation strategies that rely on the use of internal ribosome entry sites (IRESs), 5′ cap-independent translation enhancers (5′ CITEs), 3′ CITEs, or N6-methyladenosine (m6A) modification-mediated translation (7, 8). The IRES of encephalomyocarditis virus (EMCV) interacts with eIFs via determinants located in a specific domain that mimics the 5′ cap-binding eIF4F complex. The use of an IRES allows EMCV to discriminate between its own genome and cellular mRNAs and, by inhibiting cap-dependent translation, to generate conditions that are advantageous for the translation of the former molecules (9). The folding of the hepatitis C virus (HCV) IRES depends on binding of liver-specific miR-122 (10). and it directly recruits eIFs to initiate the assembly of a translation complex (11). 3′ CITEs have been identified in many plant viruses, including satellite tobacco necrosis virus (12), barley yellow dwarf virus (13), and turnip crinkle virus (14). In cooperation with 5′ UTRs, 3′ CITEs recruit components of the translation machinery using RNA secondary structures, resulting in positioning of ribosome subunits at the 5′ end of the corresponding mRNA (15).
The translation of flavivirus genomes that have 5′ cap structures is commonly initiated by cap-dependent ribosomal scanning. Unexpectedly, Edgil et al. demonstrated that DENV genotype 2 (DENV2) RNA can be translated in either a cap-dependent or cap-independent manner and that the latter process is not mediated by an IRES (16). Subsequently, the ability of DENV2 and ZIKV to resist cap-dependent translation inhibition was confirmed (17). It was also proposed that the 5′ UTR of DENV2 and that of ZIKV harbor IRES functions. Thus, the data regarding the mechanism of cap-independent initiation of translation of flavivirus genomes are contradictory. Furthermore, it is unknown whether this strategy is used by many flaviviruses or represents a specific property of the closely related ZIKV and DENV2.
In this study, we report that a cap-independent translation strategy is used by a number of flaviviruses. The cap-independent translation of DTMUV, TMUV, and JEV RNAs was observed in mammalian, avian, and mosquito cells. We provide evidence that the IRES activity of the DTMUV 5′ UTR, if it exists at all, is a minor contributor to cap-independent translation. Instead, to be effective, the 5′ end of the DTMUV 5′ UTR must be free and the 3′ UTR of DTMUV must be present in the same RNA molecule. This finding confirms the crucial role of 3′ UTRs in cap-independent translation in flaviviruses, a conclusion that is further supported by the observation that all individual RNA secondary structures of 3′ UTRs are essential for this process. The repression of cap-independent translation decreased DTMUV proliferation in vitro and delayed the death of infected duck embryos. Thus, the repression of cap-independent translation represents a new and promising strategy for viral attenuation.
RESULTS
DTMUV infection suppresses the translation of cellular mRNAs.
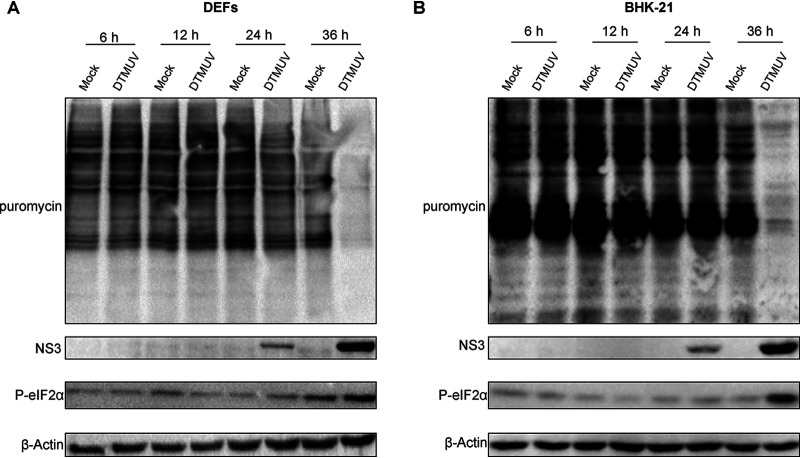
In vertebrate cells, many flaviviruses have been shown to cause the shutoff of translation of cellular mRNAs. To analyze if this phenomenon also occurs in DTMUV-infected avian and mammalian cells, a puromycin incorporation assay was performed. At 6, 12, and 24 h postinfection (p.i.), puromycin incorporation rates were identical in DTMUV-infected and mock-infected duck embryo fibroblasts (DEFs) and BHK-21 cells. However, at 36 h p.i., a notable difference between the infected and mock-infected cells was observed (Fig. 1A and B). The shutdown of cellular translation coincided with an increase in eIF2α phosphorylation, suggesting that DTMUV infection inhibited the initiation of cellular mRNA translation. Importantly, in both cell types, the shutoff of cellular translation also coincided with an accumulation of viral NS3 protein (Fig. 1). These results demonstrate that DTMUV selectively inhibited the translation of cellular mRNAs, while the synthesis of viral proteins remained active.
FIG 1.
DTMUV infection results in cellular mRNA translation shutoff. DEFs (A) and BHK-21 cells (B) were infected with DTMUV, treated with 1 μg/ml (for DEFs) or 10 μg/ml (for BHK-21 cells) puromycin for 30 min at the indicated time points, harvested, lysed, and analyzed using Western blotting. Puromycin incorporation was detected using a mouse anti-puromycin monoclonal antibody, synthesis of viral proteins was detected using a mouse anti-DTMUV-NS3 polyclonal antibody, and eIF2α phosphorylation was detected using a corresponding rabbit monoclonal antibody. β-Actin was detected as the loading control.
DTMUV has a cap-independent translation strategy.
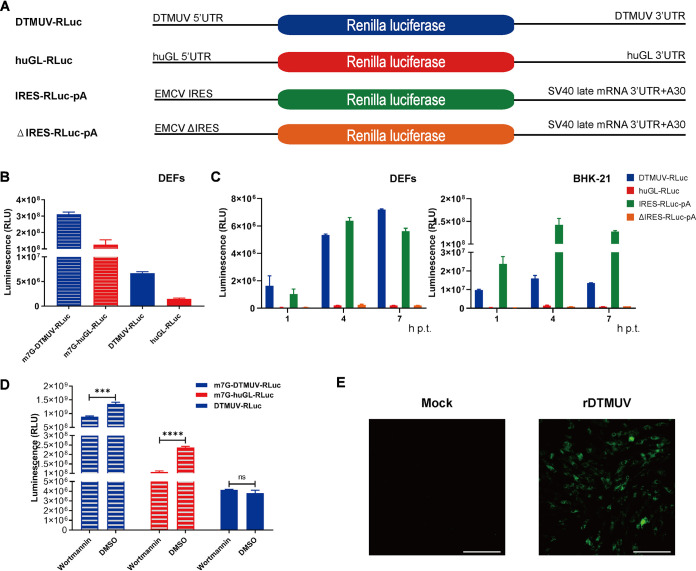
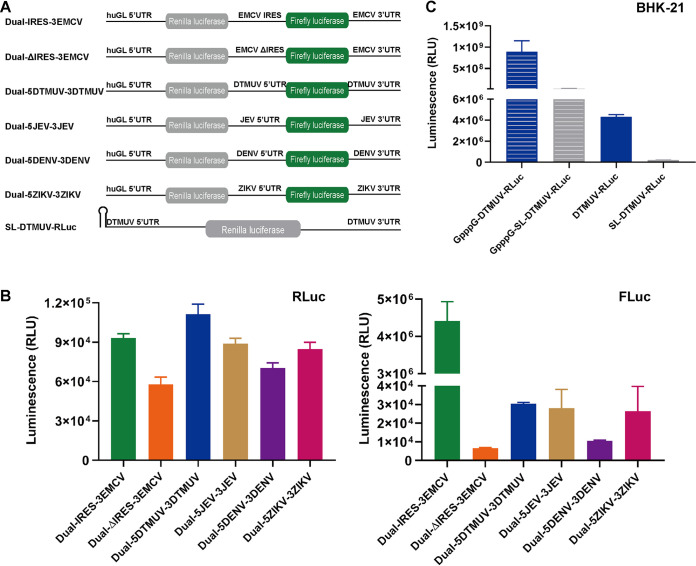
The ability of DTMUV to maintain high levels of viral protein synthesis in the presence of phosphorylated eIF2α suggests that the virus may use noncanonical strategies of translation initiation. To simplify the analysis of DTMUV translation, an RNA reporter was constructed in which a sequence encoding the Renilla luciferase (RLuc) marker was flanked by 5′ and 3′ UTRs of DTMUV. In a control reporter construct, RLuc was placed between the UTRs of the efficiently translated human β-globin (huGL) mRNA (Fig. 2A). In IRES-RLuc-pA, the 5′ UTR was replaced by the EMCV IRES, while in the ΔIRES-RLuc-pA construct, the EMCV IRES activity was eliminated by a deletion.
FIG 2.
DTMUV has a cap-independent translation initiation strategy. (A) Schematic presentation of monocistronic reporters. The diagrams are not to scale. (B) DEFs were transfected with capped/uncapped DTMUV-RLuc or huGL-RLuc, and RLuc activities were measured at 4 h p.t. (C) Uncapped DTMUV-RLuc, huGL-RLuc, IRES-RLuc, and ΔIRES-RLuc were used to transfect DEFs (left) or BHK-21 cells (right). The time course detection of RLuc activities were measured at 1 h, 4 h, and 7 h p.t. RLU, relative light units. (D) BHK-21 cells transfected with capped DTMUV-RLuc, with capped huGL-RLuc, or with uncapped DTMUV-RLuc were treated with 1 μM wortmannin or with the appropriate amount of DMSO. RLuc activities were measured at 4 h p.t. (B to D) The values obtained are presented as means plus standard deviations (SD) (n = 3). ns, not significant; ***, P < 0.001; ****, P < 0.0001 (Student's t test). (E) Supernatants from BHK-21 cells transfected with uncapped transcripts from the DTMUV icDNA clone pACYC FL-CQW1 were used to infect naive BHK-21 cells. At PID 10, the presence of rDTMUV was assessed using a mouse anti-DTMUV polyclonal antibody. The scale bar corresponds to 100 μm.
In DEFs, capped DTMUV-RLuc and huGL-RLuc were efficiently translated, resulting in similar RLuc activities. At 4 h, the translation of corresponding uncapped RNAs was less efficient (Fig. 2B), with observed reductions in RLuc expression of ∼46-fold for DTMUV-RLuc and ∼85-fold for huGL-RLuc. Uncapped DTMUV-RLuc produced ∼5-fold more RLuc than uncapped huGL-RLuc, indicating that its translation is less dependent on a 5′ cap than the translation of cellular mRNA. In DEFs, the natural host cells for DTMUV, translation rates of uncapped DTMUV-RLuc and IRES-RLuc-pA were comparable at 4 h and 7 h posttransfection (p.t.) (Fig. 2C, left). Efficient translation of uncapped DTMUV-RLuc was also observed in BHK-21 cells, although the translation of RNA containing the EMCV IRES in these cells was ∼10-fold higher (Fig. 2C, right) at 4 h and 7 h posttransfection. The translation of uncapped huGL-RLuc was inefficient in both cell types (Fig. 2C). BHK-21 or DEFs were transfected with irrelevant RNA, which was transcribed from pCDNA3.1 (with no RLuc gene), for 4 h, and RLuc activities were detected as the limit of detection. The number was about 1,000, which was too low to be included in the figure.
To confirm the use of a cap-independent translation strategy by DTMUV, BHK-21 cells were transfected with capped huGL-RLuc or capped or uncapped DTMUV-RLuc and treated with wortmannin, an inhibitor of cap-dependent translation that inhibits phosphatidylinositol 3-kinase (PI3K) and leads to the dephosphorylation of 4e binding protein 1, causing eIF4E to be isolated from eIF4F, thereby inhibiting cap-dependent translation. As expected, this treatment significantly reduced RLuc expression from capped mRNAs: ∼30% for DTMUV-RLuc and >50% for huGL-RLuc. In contrast, wortmannin treatment had no negative effect on the translation of uncapped DTMUV-RLuc (Fig. 2D), confirming that DTMUV has a cap-independent strategy of translational initiation.
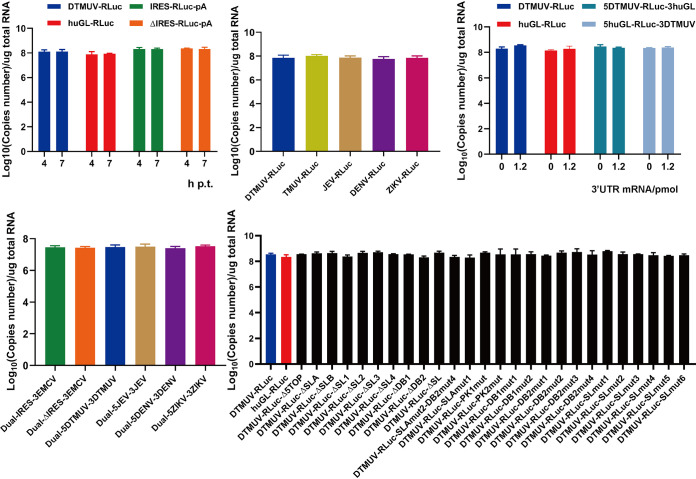
It has been shown that DENV2 can be rescued from uncapped full-length genome RNAs (17). To assess whether this is the case for DTUMV, BHK-21 cells were transfected with uncapped transcripts of DTMUV infectious cDNA (icDNA). The cell culture supernatant, harvested at 5 days p.t., was used to infect naive BHK-21 cells. At postinfection day (PID) 10, the immunofluorescence assay (IFA) results revealed large numbers of DTMUV-infected cells (Fig. 2E), confirming that uncapped RNAs corresponding to the DTMUV genome are infectious. All of these reporter RNA stabilities were analyzed by quantitative PCR (qPCR), which showed no significant difference (see Fig. 9).
FIG 9.
RNA stability analysis of RNA reporters. BHK-21 cells transfected with RNA reporters were lysed, and total RNA was extracted. RT-qPCRs that detected RLuc were used to measure the RNA copy numbers.
Cap-independent translation initiation strategies are common among flaviviruses.
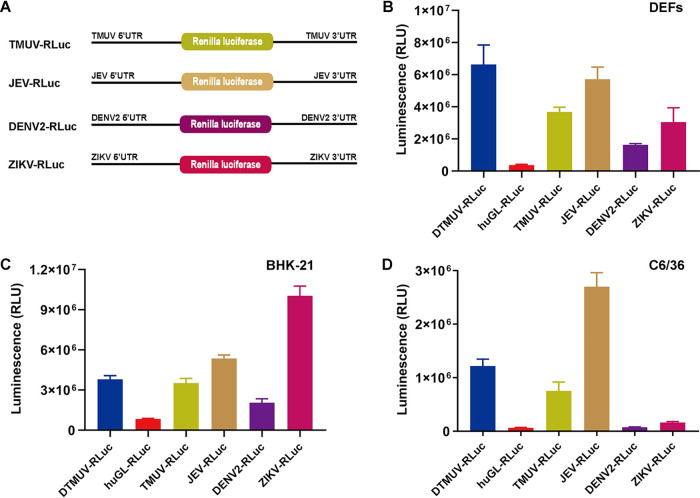
To assess whether the cap-independent translation strategy is also used in other flaviviruses, monocistronic reporters were constructed for TMUV, JEV, DENV2, and ZIKV (Fig. 3A); the reporters were analyzed in avian, mammalian, and mosquito cells. In DEFs, the highest level of cap-independent translation was observed for a reporter based on duck-borne DTMUV. However, this construct was closely followed by that based on swine-borne JEV and those based on human-borne ZIKV and TMUV isolated from mosquitoes. Only the reporter based on human-borne DENV2 showed a somewhat lower level of cap-independent translation in DEFs (Fig. 3B). The DENV2 reporter also exhibited the least efficient cap-independent translation in BHK-21 cells. The translation of the DTMUV, TMUV, and JEV reporters was somewhat more efficient, while the ZIKV-based reporter clearly outperformed the others (Fig. 3C). Again, no clear correlation between the original host of the virus and the efficiency of cap-independent translation was observed.
FIG 3.
Cap-independent translation initiation is a strategy shared by different flaviviruses. (A) Schematic presentation of monocistronic reporters for TMUV, JEV, DENV2, and ZIKV. The diagrams are not to scale. (B to D) DEFs (B) and BHK-21 (C) and C6/36 (D) cells were cultured in 48-well plates. When the cells reached ∼70 to 90% confluence, they were transfected with the indicated uncapped reporters. The activities of RLuc measured at 4 h p.t. are presented as means plus SD (n = 3).
In contrast to vertebrate cells, substantial differences between reporters based on different flaviviruses were observed in C6/36 mosquito cells. In these cells, the translation of uncapped DENV2 and ZIKV reporters was inefficient and comparable with that of the huGL-RLuc used as a negative control (Fig. 3D). In contrast, uncapped reporters based on the other three flaviviruses were efficiently translated, and the efficiencies were comparable to those observed in vertebrate cells (compare Fig. 3D with Fig. 3B and 4C). Thus, in mosquito cells, the uncapped RNAs of ZIKV and DENV2 were translated much less efficiently that those of DTMUV, TMUV, and JEV.
FIG 4.
The 5′ UTR of DTMUV lacks substantial IRES activity. (A) Schematic presentation of bicistronic reporters. The diagrams are not to scale, and the poly(A) tails of Dual-IRES-3EMCV and Dual-ΔIRES-3EMCV are not shown. (B) BHK-21 cells transfected with uncapped bicistronic reporters were lysed at 4 h p.t., and RLuc and FLuc activities were measured. (Left) Translation of the upstream cistron (RLuc). (Right) Translation of the downstream cistron (FLuc). The RLuc and FLuc activities are presented as means plus SD (n = 3). (C) BHK-21 cells were transfected with capped/uncapped DTMUV-RLuc or SL-DTMUV-RLuc, and RLuc activities were measured at 4 h p.t. (C) BHK-21 cells transfected with capped/uncapped DTMUV-RLuc or SL-DTMUV-RLuc were lysed at 4 h p.t., and RLuc activities were measured.
The 5′ UTR of DTMUV lacks substantial IRES activity.
In DEFs, the translation of uncapped DTMUV-RLuc was similar to that of the EMCV IRES-based IRES-RLuc-pA reporter (Fig. 2C, left). Based on these data, we questioned whether the DTMUV 5′ UTR has IRES activity. To address this question, a panel of bicistronic reporter constructs, including those based on ZIKV, DENV, and JEV sequences, was constructed (Fig. 4A) and analyzed.
In BHK-21 cells, all the uncapped reporters expressed the upstream cistron (RLuc) with similarly low efficiencies (Fig. 4B, left). The EMCV IRES-driven expression of the second cistron (firefly luciferase [FLuc]) occurred at very high levels (Fig. 4B, right). The translation of FLuc from the 5′ UTR of any flavivirus was at least 100-fold lower than the translation of FLuc from the EMCV IRES. Nevertheless, FLuc expression, presumably mediated by 5′ UTRs of flaviviruses, did exceed that mediated by ΔIRES by up to 4-fold (Fig. 4B, right). The correlation between the expression levels of RLuc and FLuc (Fig. 4B) indicates that the differences between the flavivirus reporters were, at least in part, caused by different stabilities of the corresponding RNAs. Thus, the 5′ UTR of DTMUV and those of other flaviviruses either do not have IRES competence or their IRES-like activities are several orders of magnitude lower than that of the EMCV IRES (Fig. 4B), indicating that an IRES activity of the 5′ UTR is highly unlikely to be the primary contributor to the cap-independent translation of flavivirus RNAs. To further study whether DTMUV cap-independent translation is 5′-end dependent, SL-DTMUV-RLuc, with a stem-loop that can inhibit 5′-end translation, was used (18); capped and uncapped SL-DTMUV-RLuc translation activities were over 30-fold lower than those of capped/uncapped DTMUV-RLuc. These results clearly indicate that efficient DTMUV cap-independent translation is highly dependent on a free 5′ end, which may allow the translation complex to initiate at the 5′ terminus and scan along the 5′ UTR (Fig. 4C).
Cap-independent translation of DTMUV RNA requires the presence of 5′ and 3′ UTRs in cis.
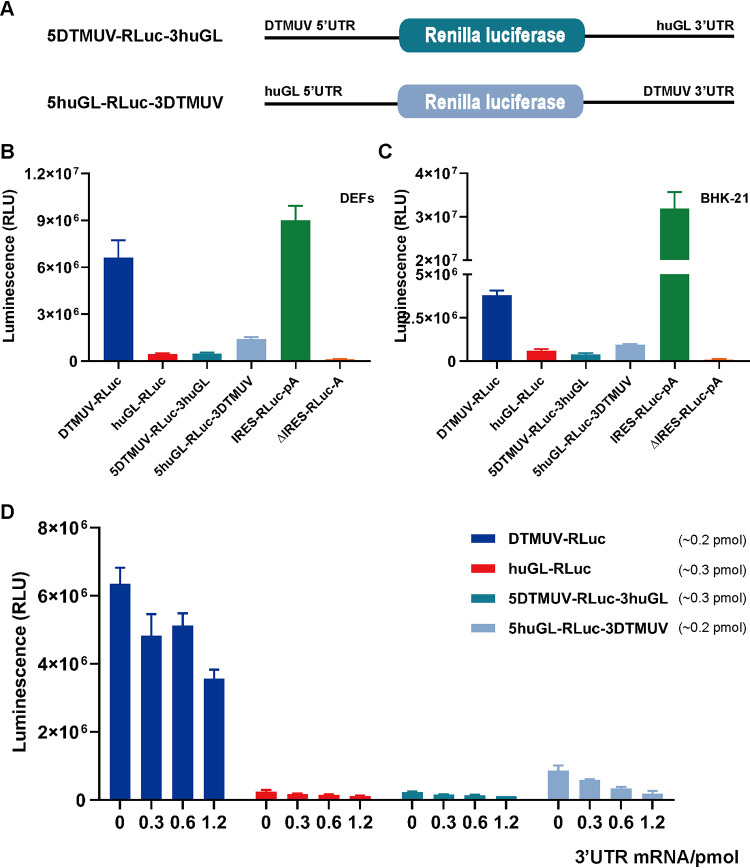
To elucidate the role of the DTMUV 5′ and 3′ UTRs in cap-independent translation, two chimeric reporters were constructed and analyzed (Fig. 5A). The replacement of the 5′ UTR in the huGL-RLuc construct with that of DTMUV did not result in any increase of cap-independent RLuc expression. The increase in RLuc expression resulting from the replacement of the 3′ UTR of huGL-RLuc was detectable but still very modest (Fig. 5B and C) and may represent a consequence of the stabilization of reporter RNA by the DTMUV 3′ UTR. These results clearly indicated that both the 5′ and 3′ UTRs of DTMUV are crucial for efficient cap-independent translation.
FIG 5.
The presence of both UTRs in cis is essential for cap-independent translation of DTMUV RNA. (A) Schematic presentation of chimeric monocistronic reporters. The diagrams are not to scale. (B and C) DEFs (B) and BHK-21 cells (C) transfected with the indicated uncapped reporters were lysed at 4 h p.t., and RLuc activities were measured. (D) DEFs were cotransfected with 120 ng of uncapped DTMUV-RLuc, huGL-RLuc, 5DTMUV-RLuc-3huGL, or 5huGL-RLuc-3DTMUV reporters and increasing amounts (0, 60, 120, and 240 ng) of RNA corresponding to the DTMUV 3′ UTR. A constant amount of total RNA (360 ng/well) used for each transfection was obtained by adding the appropriate amount (240, 120, 60, or 0 ng) of irrelevant RNA obtained by transcription of plasmid pCDNA3.1. The cells were lysed at 4 h p.t., and RLuc activities were measured. (B to D) RLuc activities are presented as means plus SD (n = 3).
To determine if these elements need to be present in the same RNA molecule (in cis) or can also function in trans, DEFs were cotransfected with DTMUV-RLuc, huGL-RLuc, 5DTMUV-RLuc-3huGL, and 5huGL-RLuc-3DTMUV and with increasing amounts of the DTMUV 3′ UTR RNA. This RNA, provided in trans, did not increase the translation of any uncapped reporter. Instead, it inhibited the translation of the uncapped DTMUV-RLuc and huGL-RLuc-3DTMUV reporters in a dose-dependent manner. The presence of increasing amounts of DTMUV 3′ UTR RNA had little effect on the translation of the uncapped huGL-RLuc and 5DTMUV-RLuc-3huGL reporters (Fig. 5D), either because the reporter RNAs lacked the 3′ UTR of DTMUV or because the low level of RLuc expression from the reporters hampered the detection of negative effects resulting from the presence of the DTMUV 3′ UTR RNA. The presence of the DTMUV 3′ UTR did not affect the RNA stabilities of the reporters (see Fig. 9). Taken together, these data clearly show that the 3′ UTR of DTMUV participates in cap-independent translation and that it must be present in cis.
Cap-independent translation of DTMUV RNA depends on sequences and RNA secondary structures present in both UTRs.
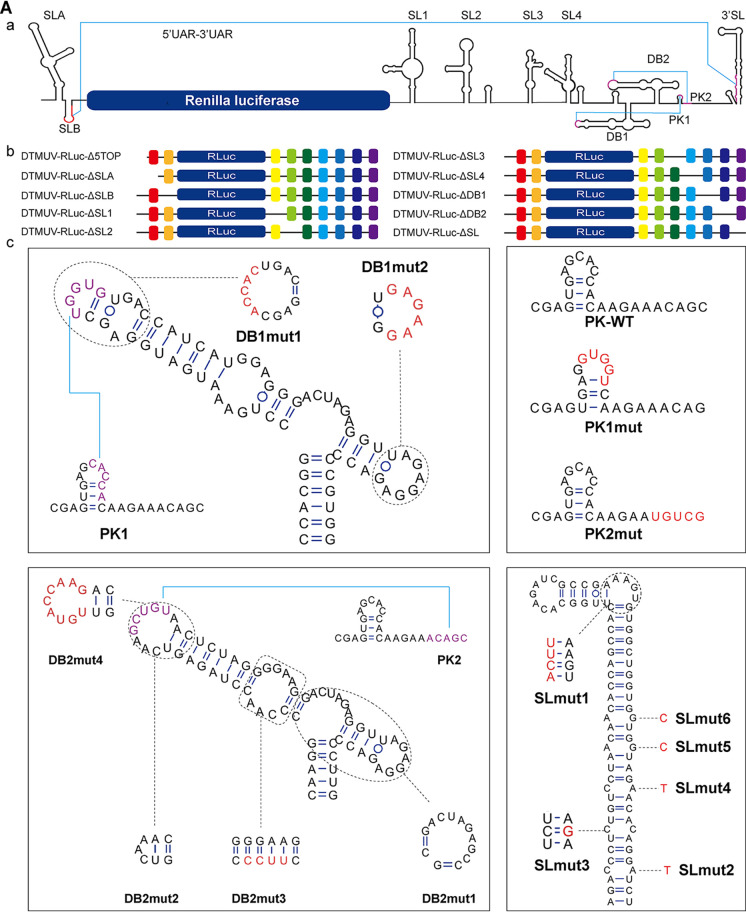
Cap-independent initiation of translation commonly depends on high-order RNA structures. The secondary structures present in the 5′ and 3′ UTRs of DTMUV (Fig. 6A, a) predicted using the RNAfold Web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) were similar to those that were experimentally identified in the genomes of DENV and other flaviviruses (5, 19). To identify the potential impact of each of these structures on DTMUV cap-independent translation, deletions and substitutions resulting in the removal, alteration, or preservation of predicted secondary-structure elements (Table 1 and Fig. 6A, b and c) were introduced into DTMUV-RLuc. To increase the sensitivity of the analysis, a huGL-FLuc construct was used for normalization.
FIG 6.
DTMUV 5′ and 3′ UTR sequences and RNA secondary-structural elements are crucial for cap-independent translation. (A) Schematic presentation of the reporters used in this study. The diagrams are not to scale. Detailed information regarding the introduced mutations is presented in Table 1. (a) Predicted secondary structures in the UTRs of the DTMUV-RLuc reporter. UARs (i.e., upstream of AUG) and the DB pseudoknot are in purple. (b) Schematic presentation of reporter mRNAs harboring a deletion at the 5′ end or deletions of predicted secondary-structural elements. (c) Predicted native (wild-type [WT]) secondary structures of elements located in the 5′ UTR/3′ UTR and structures resulting from introduced mutations. Introduced substitutions are shown in red. (B) Translation efficiencies of DTMUV-RLuc reporters containing mutations shown in panel A. BHK-21 cells were cotransfected with the indicated uncapped reporters and uncapped huGL-FLuc control. The cells were lysed at 4 h p.t., and the activities of RLuc and FLuc were measured. The activities of RLuc expressed by different reporters were normalized to the activity of FLuc expressed by the transfection control. The relative luciferase activities (RLuc/FLuc) are presented as means plus SD (n = 3). ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (Student’s t test).
TABLE 1.
Mutations introduced into DTMUV-RLuc
| Reporter | Nucleotides | Region | Mutation |
|---|---|---|---|
| DTMUV-RLuc-Δ5TOP | 1–5 | 5′ UTR | Deleted |
| DTMUV-RLuc-ΔSLA | 6–75 | 5′ UTR | Deleted |
| DTMUV-RLuc-ΔSLB | 76–95 | 5′ UTR | Deleted |
| DTMUV-RLuc-ΔSL1 | 1–79 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔSL2 | 80–153 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔSL3 | 184–247 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔSL4 | 248–310 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔDB1 | 349–417 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔDB2 | 426–514 | 3′ UTR | Deleted |
| DTMUV-RLuc-ΔSL | 525–619 | 3′ UTR | Deleted |
| DTMUV-RLuc-DB1mut1 | 371–375 | 3′ UTR | UGGUG→ACCAC |
| DTMUV-RLuc-DB1mut2 | 402–407 | 3′ UTR | AGAGGA→GAGAAG |
| DTMUV-RLuc-DB2mut1 | 474–485 | 3′ UTR | Deleted |
| DTMUV-RLuc-DB2mut2 | 447–452 | 3′ UTR | Deleted |
| DTMUV-RLuc-DB2mut3a | 432–434 | 3′ UTR | CCA→UUCC |
| DTMUV-RLuc-DB2mut4 | 444–452 | 3′ UTR | CAAGCUGTA→UGUACCAAG |
| DTMUV-RLuc-PK1mut | 500–504 | 3′ UTR | CACCA→GUGGU |
| DTMUV-RLuc-PK2mutb | 511–515 | 3′ UTR | ACAGC→UGUCG |
| DTMUV-RLuc-SLmut1 | 569–570 | 3′ UTR | CU→CACUUU |
| DTMUV-RLuc-SLmut2 | 616 | 3′ UTR | A→U |
| DTMUV-RLuc-SLmut3 | 612 | 3′ UTR | C→G |
| DTMUV-RLuc-SLmut4 | 608 | 3′ UTR | A→U |
| DTMUV-RLuc-SLmut5c | 604 | 3′ UTR | G→C |
| DTMUV-RLuc-SLmut6 | 601 | 3′ UTR | G→C |
The same mutation is also present in pCMV-DTMUV-Rep-NLuc-DB2mut3 and in pCMV-icDTMUV-DB2mut3.
The same mutation is also present in pCMV-DTMUV-Rep-NLuc-PK2mut.
The same mutation is also present in pCMV-DTMUV-Rep-NLuc-SLmut5 and in pCMV-icDTMUV-SLmut5.
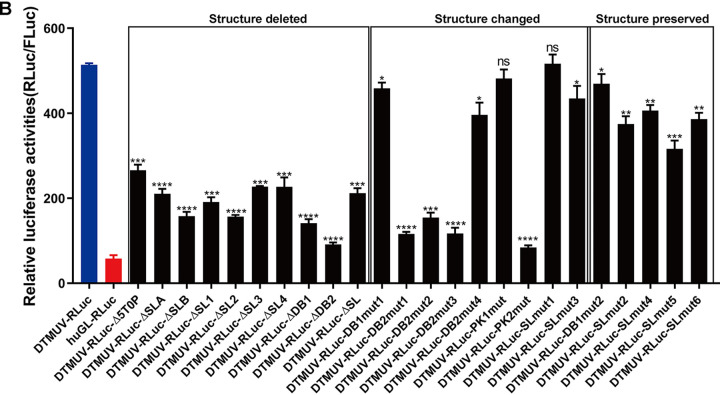
The deletion of 5 nucleotides from the 5′ end of DTMUV-RLuc, as well as the deletion of any predicted secondary-structural element, significantly reduced the cap-independent translation of reporter RNAs. The most substantial effects were caused by deleting the DB1 or DB2 structure (Fig. 6B). To exclude the possibility that the observed effect was caused by reduced stability of the reporter RNA harboring UTR deletions, the amounts of DTMUV-RLuc and mutant reporters at 4 h p.t. were determined using qPCR (see Fig. 9). No significant differences were observed, confirming that these deletions had no effect on reporter RNA stability. Thus, the observed differences in the RLuc/FLuc ratio originated from different efficiencies in cap-independent translation.
Deletions of entire secondary-structural elements may impact the high-order structure of the entire reporter RNA. Therefore, the roles of the DTMUV UTR sequences and RNA structures were further analyzed using smaller mutations (Table 1) designed to either alter the predicted secondary-structural elements or to preserve the original secondary structures (Fig. 6A, c). With exception of SLmut1 and PK1mut, mutations leading to changes in the predicted RNA secondary structure significantly reduced the cap-independent translation of the corresponding reporters. However, this effect was substantial for only three deletions in DB2, indicating that the structure of the DB2 element is especially important for cap-independent translation. In contrast, the introduction of mutations designed to preserve predicted secondary structure in DB1 or the SL caused only a minor, albeit significant, reduction in cap-independent translation (Fig. 7B). Thus, while the 3′ SL structure itself is important for cap-independent translation, smaller changes within the structure were tolerated, demonstrating that cap-independent translation is not only structure dependent, but also sequence dependent.
FIG 7.
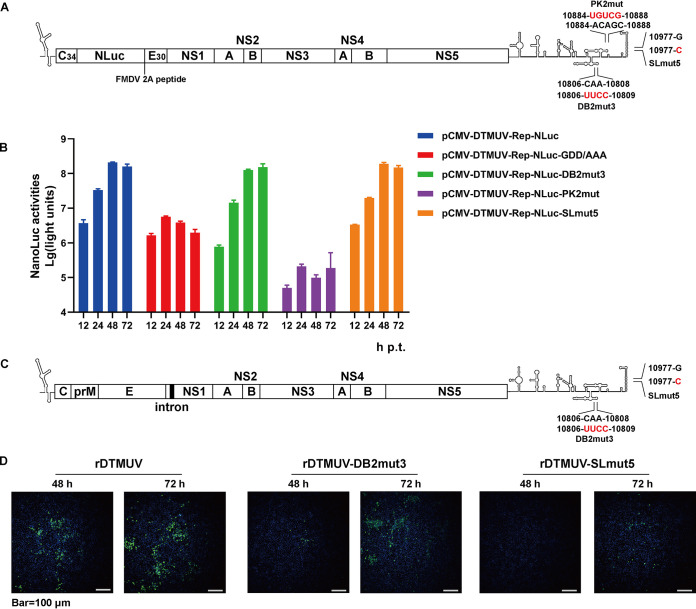
Defects in cap-independent translation attenuate DTMUV replicons and viruses. (A and C) Schematic presentation of DTMUV replicons (A) and genomes (C) harboring mutations in the 3′ UTR. WT residues and their positions in the genome of DTMUV are shown; mutated residues are indicated in red. C34, region encoding a truncated capsid protein; E30, region encoding 30 C-terminal residues of E glycoprotein; FMDV, foot-and-mouth disease virus; NLuc, region encoding the nanoluciferase reporter. The intron shown in panel C is present only in the icDNA plasmid and is removed by splicing upon viral rescue. (B) BHK-21 cells were transfected with the indicated plasmids. At 12, 24, 48, and 72 h p.t., the cells were lysed, and NLuc activities were measured. The data are presented as means plus SD (n = 3). (D) BHK-21 cells were transfected with pCMV-icDTMUV, pCMV-icDTMUV-DB2mut3, or pCMV-icDTMUV-SLmut5. The rescue of the recombinant viruses at 48 and 72 h p.t. was assessed using a mouse anti-DTMUV polyclonal antibody.
Inhibition of cap-independent translation reduces DTMUV proliferation in cell cultures but does not eliminate virulence in embryonated duck eggs.
The DTMUV replicon and icDNA clones were used to assess the impacts of mutations that reduce cap-independent translation on DTMUV RNA replication, viral multiplication, and pathogenicity. For this analysis, three relatively small substitutions (Table 1 and Fig. 7A and C) with a substantial (PK2mut and DB2mut3) or moderate but highly significant (SLmut5) effect on cap-independent translation (Fig. 6B) were chosen.
The transfection of BHK-21 cells with pCMV-DTMUV-Rep-NLuc-PK2mut resulted in very low levels of nanoluciferase (NLuc) activity that did not increase after 24 h p.t., indicating that the PK2mut substitution abolished DTMUV RNA replication. Interestingly, the levels of NLuc produced by the replicon were considerably lower than those produced by the replicon harboring a GDD-to-AAA mutation in the active site of the NS5 polymerase (Fig. 7B), indicating a reduced level of translation of cellular RNA polymerase II-generated pCMV-DTMUV-Rep-NLuc-PK2mut transcripts. Thus, PK2mut also affects the cap-dependent translation of DTMUV RNA, resulting in a lack of RNA replication. In contrast, the expression levels of NLuc in cells transfected with pCMV-DTMUV-Rep-NLuc-DB2mut3, pCMV-DTMUV-Rep-NLuc-SLmut5, and pCMV-DTMUV-Rep-NLuc were similar (Fig. 7B), indicating that DB2mut3 and SLmut5 did not have a negative effect on viral RNA replication.
Next, mutations that were tolerated in the context of the DTMUV replicon were introduced into the icDNA clone of DTMUV (Fig. 7C). Corresponding recombinant viruses, designated rDTMUV-DB2mut3 and rDTMUV-SLmut5, were rescued in BHK-21 cells (Fig. 7D). The rescue of rDTMUV-SLmut5 was substantially delayed, and no or little spread of the virus was observed, which was a phenotype that contrasted with the essentially uncompromised RNA replication of the corresponding replicon (Fig. 7B). Thus, in addition to diminished cap-independent translation, rDTMUV-SLmut5 harbored another substantial functional defect that excluded its use in subsequent experiments.
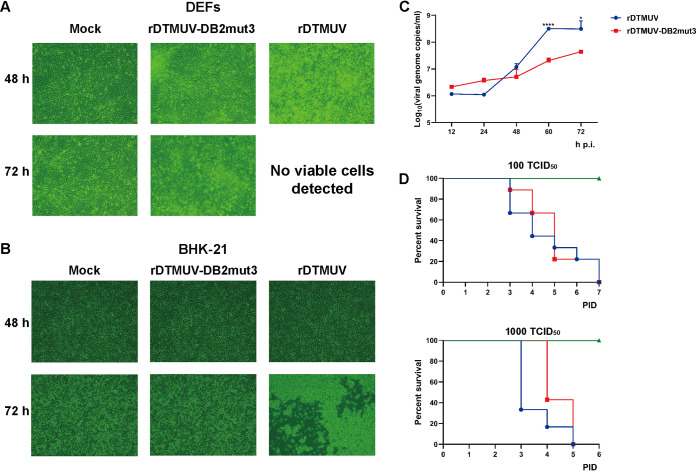
Despite DB2mut3 having been designed to alter the structure of the DB2 element (Fig. 6A) and to have a strong impact on cap-independent translation (Fig. 6B), the corresponding recombinant virus was successfully rescued and detected by IFA (Fig. 7D). Next, rDTMUV-DB2mut3 was compared to rDTMUV. The progeny rDTMUV-DB2mut3 (p1, i.e., rescued recombinant virus was passaged in BHK-21 cells once) which was determined by sequencing not to have new mutations, infected DEFs or BHK-21 cells. In rDTMUV-infected DEFs, cytopathic effects (CPE) were observed at 48 h p.i. (Fig. 8A); the development of CPE in BHK-21 cells was observed at 72 h p.i. (Fig. 8B). In rDTMUV-DB2mut3-infected cells, CPE was not detected even at 72 h p.i. (Fig. 8A and B), indicating that DB2mut3 attenuated the growth and/or reduced the cytotoxic properties of DTMUV. The growth curve experiment performed using DEFs confirmed that, compared to rDTMUV, the propagation of rDTMUV-DB2mut3 was delayed and occurred at a lower level. However, while the titers of rDTMUV plateaued at 60 h p.i., those of rDTMUV-DB2mut3 continued to increase (Fig. 8C).
FIG 8.
Replication of rDTMUV-DB2mut3 is attenuated in cultured vertebrate cells. DEFs (A) or BHK-21 cells (B) grown in 12-well plates were infected with 1,000 TCID50 of rDTMUV or rDTMUV-DB2mut3, while control cells were mock infected. The development of CPE was observed at 48 and 72 h p.i. using a Nikon 80i fluorescence microscope. (C) DEFs grown in 12-well plates were infected with recombinant viruses at 300 TCID50 per well. At the indicated time points, the supernatants were harvested, and the copy number of viral genomic RNA was determined using RT-qPCR. The data are presented as means plus SD (n = 3). *, P < 0.05; ****, P < 0.0001 (Student’s t test). (D) Groups of 9-day-old duck embryos (n = 7) were mock infected or infected with rDTMUV or rDTMUV-DB2mut3 by allantoic-cavity inoculation. The viruses were used at 100 TCID50 (top) or 1,000 TCID50 (bottom) per embryo. The duck embryos were incubated at 37°C and assessed for survival daily.
Finally, the virulence of rDTMUV and rDTMUV-DB2mut3 was evaluated by infecting 9-day-old duck embryos via allantoic-cavity inoculation. At 100 50% tissue culture infective doses (TCID50), both viruses caused the death of all infected embryos by PID 7. At 1,000 TCID50, all infected embryos died by PID 5. Notably, the death of rDTMUV-infected embryos occurred earlier than that of the rDTMUV-DB2mut3-infected embryos, although the difference did not reach statistical significance (Fig. 8D). Taken together, these results indicate that mutations inhibiting cap-independent translation delay/decrease rDTMUV proliferation in cell culture but do not eliminate DTMUV virulence in duck embryos.
DISCUSSION
All viruses are dependent on the translation machinery of the host cell. Since a positive-strand RNA genome acts as an mRNA, it needs to compete with the large excess of host mRNAs in the early stages of infection. At the late stage of infection, many viruses hijack cellular resources to produce large amounts of structural proteins necessary for the assembly of progeny virions. For these purposes, viruses have developed various strategies to inhibit the synthesis of host proteins and/or to enhance the translation of their own RNAs. Well-known examples include use of noncanonical mechanisms of initiation of translation combined with cleavage of eIF4G, the induction of eIF2α phosphorylation, and other mechanisms leading to a shutoff of host cell translation (19–21). In this study, we report that DTMUV infection represses host protein synthesis in both avian and mammalian cells and that this activity is accompanied by induction of eIF2α phosphorylation. The same has been previously observed for JEV- and WNV-infected cells, but not DENV-infected cells (22–24). The repression of host mRNA translation, whether caused by eIF2α phosphorylation or not, suggests the existence of a noncanonical translation strategy of DTMUV genomic RNA in vertebrate cells.
Arbovirus members of the genus Flavivirus do not cause major CPE in invertebrate cells. However, interestingly, both DENV and ZIKV RNAs have been shown to be capable of cap-independent translation in mosquito cells (17). In this study, cap-independent translation was observed for DTMUV, TMUV, and JEV RNAs and was more efficient for these viruses than for DENV2 or ZIKV (Fig. 3D). Thus, cap-independent translation in mosquito cells is a common phenomenon for many, if not all, flaviviruses. However, the biological implications of the phenomenon are not obvious. It is possible that sequences and secondary structures of flavivirus UTRs simply function the same way in both vertebrate and invertebrate cells. However, it is more likely that cap-independent translation is important to overcome a problem(s) common to both host cell types. Indeed, the functional significance of cap-independent translation may not be limited to facilitating the synthesis of viral proteins upon the shutoff of host cell translation. It has recently been shown that replicases of distantly related alphaviruses (family Togaviridae) synthesize large amounts of uncapped transcripts during the late stages of infection. Such RNAs are important for alphaviruses, as an increase in capping efficiency has a negative effect on virion formation and attenuates viral growth and pathogenicity (25, 26). To our knowledge, it is not known whether this or similar phenomena also occur in flaviviruses. However, intriguingly, similar phenotypes were observed for an alphavirus with increased capping efficiency and for rDTMUV-DB2mut3 harboring a specific defect in cap-independent translation (Fig. 6 and 8). Taking into account that flaviviruses use Xrn1 nuclease-mediated degradation of full-length RNAs to generate short subgenomic flavivirus RNAs (sfRNAs) (27), it is plausible that these viruses also synthesize uncapped full-length transcripts that may have an important role(s) in the infection process.
Cap-independent translation of DTUMV and other flaviviruses is different from IRES-mediated translation. Consistent with a previous report (16), the “IRES-like” activities of the flavivirus 5′ UTRs in bicistronic constructs did not exceed 1% of that of the EMCV IRES (Fig. 4). The superior efficiency of the EMCV IRES-driven translation was also observed by Song et al., although the difference between the translation by the EMCV IRES and DENV 5′ UTR was less pronounced (17). The conclusion of these authors that the 5′ UTRs of DENV and ZIKV function as IRES elements was primarily based on the comparison of translation from these 5′ UTRs with that from the HCV IRES and its defective variant. The HCV IRES is weaker than that of EMCV, and its correct folding depends on a liver-specific miR-122 (10), the presence of which in BHK-21 cells has not been reported. The use of different controls is the only notable difference between our study and that of Song et al., since the designs of the bicistronic reporters were similar and the 5′ UTRs of the ZIKV strains FSS1302 and BeH819015 are identical. Consistent with their study, we also observed modestly increased translation of the second reporter in the bicistronic construct when it was placed under the control of the 5′ UTR of the flavivirus genome (Fig. 4). However, comparison with the strong EMCV IRES led us to conclude that the observed translation efficiency was much closer to that of the negative control. The use of the HCV IRES and its defective version most likely generated an entirely different context, leading researchers to different interpretations of otherwise similar data.
While our data do not exclude the existence of low IRES activity of flavivirus 5′ UTRs, they do unequivocally demonstrate that the key to efficient translation of uncapped flavivirus RNAs lies in the functional interaction between elements located within the 5′ and 3′ UTRs. Furthermore, it was observed that the 5′ and 3′ UTRs must be present in cis, as providing the 3′ UTR in trans resulted in a reduction in translation efficiency (Fig. 5). Interestingly, in both vertebrate and mosquito cells, flaviviruses produce large amounts of noncoding and highly structured sfRNAs that are important for many biological functions, including pathogenicity and vector transmission (28, 29). The sfRNAs roughly correspond to the 3′ UTR of the flavivirus genome and are therefore similar to the RNA that was shown to suppress cap-independent translation (Fig. 5D). If this is the case in infected cells, then sfRNAs may also serve as tools to regulate the translation of the viral genome. The observed inhibition of translation may result from the 3′ UTR provided in trans interacting with sequences/structures within the 5’ UTR and preventing the 3′ UTR present in cis from doing the same. Another possibility is that the cap-independent translation depends on interaction of 3′ UTR and key protein factor and that the provided 3′ UTR competitively binds this key factor. The cap-independent translation of 5huGL-RLuc-3DTMUV, in which DTMUV 5′ UTR does not exist and thus potential interactions between DTMUV 5′ UTR and the provided 3′ UTR are avoided, was also inhibited by providing 3′ UTR. It was recently demonstrated that circularization of the flavivirus genome, which on one hand is mediated by the 5′ UTR, downstream AUG region, and 5′ cyclization sequence and on the other hand is mediated by the 3′ UTR, inhibits cap-dependent translation (30).
Because any genomic-RNA translation is incompatible with the use of the same RNA molecule as a template for RNA polymerase, it is logical to assume that the same conformational change should inhibit cap-independent translation, as well. This assumption is supported by the finding that the SLB structure, which is essential for cap-independent translation (Fig. 6B), is not present in the circular form of the flavivirus genome (31). As our reporter RNAs lacked 5′ circularization sequences, they should have a linear conformation, which has also been shown to be a dominant form of the flavivirus genome in infected cells (32). Thus, the interaction between the 5′ and 3′ UTRs, which is crucial for cap-independent translation, is unlikely to be mediated by the formation of circular structures and more likely involves host cell factor-mediated interactions. Indeed, two DB structures in the DENV2 3′ UTR have been shown to interact with poly(A)-binding protein, and this interaction is crucial for cap-dependent translation (33).
The finding that deletions of these elements and mutations altering the structure of DB2 had the biggest impact on the efficiency of cap-independent translation (Fig. 6B) indicates that the same interactions also affect cap-independent translation. However, in contrast to their negative effect on cap-independent translation, SLmut5 and DB2mut3 did not affect the replication and cap-dependent translation of DTMUV replicons (Fig. 7B). Thus, the overlap of sequences and/or higher-order structures responsible for cap-dependent and cap-independent translation of flavivirus genomes is considerable, but not complete.
The inefficient rescue and spread of rDTMUV-SLmut5 clearly contrast with the uncompromised replication of the corresponding replicon, indicating the presence of a defect(s) in functions that are required for the virus. Unfortunately, multiple overlapping functions of sequences and RNA structures located within the 3′ UTRs of flaviviruses makes it difficult to attribute the biological effects of SLmut5 to a single compromised function. It is conceivable that rDTMUV-SLmut5 has a defect in the release of the viral genome from the capsid, in the initiation of genome translation, or in the switch from translation to RNA replication, although the observation that DB2mut3 results in a virus with different biological properties argues against these possibilities. Therefore, it is more likely that SLmut5 affects the late stages of infection. Diminished virion formation can be caused by reduced synthesis of structural proteins and/or by affecting the packaging of viral genomes. Although the former possibility is consistent with the proposed role of cap-independent translation (i.e., maintaining the production of viral proteins in cells where cap-independent translation is inhibited), it is unlikely to be the primary reason. Because the DB2mut3 mutation has a more substantial effect on cap-independent translation than does SLmut5 (Fig. 6B) but does not cause similar attenuation, it is more likely that the primary effect of SLmut5 on the recombinant virus is due to inhibition of RNA packaging.
The cap-independent translation of DTMUV and other flavivirus genomes was proposed to be, at least in part, an evasion strategy to resist host translation shutoff. While we cannot exclude the possibility that DB2mut3 has multiple mechanisms of action, the phenotype of the corresponding virus supports this assumption. The lack of a detectable defect in RNA replication most likely indicates that sufficient numbers of replicase complexes are formed at the earlier stages of infection and/or that their formation is less sensitive to a reduction in viral protein expression. In contrast, a deficit of structural proteins should result in a delay in virion formation, leading to delayed spread of infection and slower virion accumulation, which is exactly what was observed for rDTMUV-DB2mut3 (Fig. 8C and D). Interestingly, these defects delayed but did not abolish the death of infected duck embryos. The phenotype was expected, as the mutation in the 3′ UTR only reduces and/or delays accumulation of viral proteins and does not change their structures or functions. The observed virulence of rDTMUV-DB2mut3 may also be a consequence of the high viral dose used and/or the high susceptibility of embryos used for virus infection assays. Thus, cap-independent translation is important for flavivirus propagation and also affects viral infection in vivo.
Collectively, these data affirm the existence of cap-independent translation in DTMUV, TMUV, DENV2, ZIKA, and JEV. This phenomenon is probably common to all members of the genus Flavivirus. The efficiency of cap-independent translation of RNAs of flaviviruses varies with the virus species and host cell type. This is the first report to show that for cap-independent translation in DTMUV (and presumably other flaviviruses), 5′ and 3′ UTRs act in cis and should also be present in their native locations. The mechanism by which these regions activate cap-independent translation remains unknown, although it clearly depends on both secondary structures and primary sequences within UTRs. As cap-independent translation was also observed in mosquito cells, its role in flavivirus infection is unlikely to be limited to the evasion of consequences of the shutoff of host translation. We demonstrated that the inhibition of cap-independent translation can be used to decrease viral proliferation. Although this approach only delayed the death of infected duck embryos, it may be sufficient to eliminate virulence in adults, which are less susceptible to flavivirus infection than embryos or neonatal animals. If so, the inhibition of cap-independent translation could be used to produce attenuated variants of DTMUV and other flaviviruses that represent potential vaccine candidates.
MATERIALS AND METHODS
Viruses and cells.
The DTMUV strain CQW1 (NCBI accession no. KM233707.1) was provided by the Research Center of Avian Disease, Sichuan Agricultural University, Chengdu, Sichuan, China. The JEV strain SA14 (NCBI accession no. KU323483.1) was a gift from the Research Center of Swine Disease, Sichuan Agricultural University. DEFs were isolated from 9-day-old duck embryos and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% newborn calf serum (NBCS) at 37°C under a 5% CO2 atmosphere. Baby hamster kidney 21 (BHK-21) cells (ATCC CCL-10) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) at 37°C under a 5% CO2 atmosphere. Aedes albopictus C6/36 cells were cultured in minimal essential medium (MEM) supplemented with 10% FBS at 28°C with no additional CO2.
Sequences used in reporter constructs.
All fragments based on the genome of DTMUV were amplified from the plasmid pACYC FL-DTMUV, which contains the icDNA of DTMUV (34). Sequences corresponding to the 5′ and 3′ UTRs of TMUV strain MM1775 (NCBI accession no. MH414569) and those of human β-globin mRNA without a poly(A) tail (NCBI accession no. NM_000518.5) and the EMCV 3′ UTR with a poly(A) tail were obtained as synthetic DNAs (Sangon Biotech, China). EMCV IRES cDNA was synthesized by GenScript. Sequences encoding RLuc and FLuc were PCR amplified from the vectors pRL-TK and pGL3 (Promega), respectively. Sequences corresponding to the 5′ and 3′ UTRs of DENV2 were PCR amplified from an icDNA clone of DENV2 strain New Guinea C (NCBI accession no. AF038403.1), which was a gift from Suresh Mahalingam (Griffith University, Australia). The 5′ and 3′ UTRs of ZIKV strain BeH819015 (NCBI accession no. KU365778.1) were PCR amplified from the corresponding icDNA clone (35).
Construction of reporters.
DNA corresponding to mono- and bicistronic reporter RNAs was cloned into the vector pMD19-T (TaKaRa, Japan). Then, a promoter for bacteriophage T7 RNA polymerase was placed upstream, and an XhoI restriction site was placed downstream of the sequences corresponding to the ends of the reporter. Monocistronic reporters containing a sequence encoding RLuc flanked by flavivirus 5′ and 3′ UTRs were designated DTMUV-RLuc, DENV2-RLuc, ZIKV-RLuc, JEV-RLuc, and TMUV-RLuc. SL-DTMUV-RLuc consisted of DTMUV-RLuc and a stem-loop (GGGGGGGGGGTTCGCCCCCCCCCC) that was located at the 5′ end of DTMUV-RLuc. The control reporter huGL-RLuc had the same design except that the 5′ and 3′ UTRs were obtained from human β-globin mRNA (Fig. 2A and 4A); in the huGL-FLuc reporter, the coding sequence of RLuc in huGL-RLuc was replaced with that of FLuc. The positive control IRES-RLuc-pA contained the EMCV IRES, RLuc, and the 3′ UTR of simian virus 40 (SV40) late mRNAs followed by a poly(A) tail consisting of 30 A residues (Fig. 2A). In the negative control, designated ΔIRES-RLuc-pA, residues 430 to 551 of the EMCV IRES were deleted. Reporters harboring heterologous combinations of DTMUV and human β-globin UTRs were designated 5DTUMV-RLuc-3huGL and 5huGL-RLuc-3DTMUV (Fig. 6A).
Bicistronic reporters followed a similar basic design, where the T7 promoter was followed by sequences corresponding to the 5′ UTR of the mRNA of human β-globin, and RLuc was used as an upstream reporter. A cassette for the expression of FLuc, used as a downstream reporter, contained 5′ and 3′ UTRs of different flaviviruses. The resulting constructs were designated Dual-5DTMUV-3DTMUV, Dual-5DENV-3DENV, Dual-5ZIKV-3ZIKV, and Dual-5JEV-3JEV (Fig. 5A). In the control constructs, designated Dual-IRES-3EMCV and Dual-ΔIRES-3EMCV, the 5′ UTR of the flavivirus was replaced with the EMCV IRES or ΔIRES, respectively, while the 3′ UTR of the downstream reporter was that of EMCV with a poly(A) tail consisting of 30 A residues.
Introduction of mutations into the DTUMV-RLuc reporter.
A plasmid expressing DTMUV-RLuc was modified using a Fast Mutagenesis System kit (Transgen Biotech, China) to obtain plasmids expressing the reporters shown in Fig. 7A. In the DTMUV-RLuc-Δ5TOP construct, the first 5 nucleotides of the 5′ UTR (AGAAG) were deleted. Reporters harboring deletions of predicted RNA secondary-structural elements were designated DTMUV-RLuc-ΔSLA (nucleotides 6 to 75 of the 5′ UTR deleted), DTMUV-RLuc-ΔSLB (nucleotides 76 to 95 of the 5′ UTR deleted), DTMUV-RLuc-ΔSL1 (nucleotides 1 to 79 of the 3′ UTR deleted), DTMUV-RLuc-ΔSL2 (nucleotides 80 to 153 of the 3′ UTR deleted), DTMUV-RLuc-ΔSL3 (nucleotides 184 to 247 of the 3′ UTR deleted), DTMUV-RLuc-ΔSL4 (nucleotides 248 to 310 of the 3′ UTR deleted), DTMUV-RLuc-ΔDB1 (nucleotides 349 to 417 of the 3′ UTR deleted), DTMUV-RLuc-ΔDB2 (nucleotides 426 to 514 of the 3′ UTR deleted), and DTMUV-RLuc-ΔSL (nucleotides 525 to 619 of the 3′ UTR deleted). Reporters with mutations designed to alter the secondary structures present in the DTMUV 3′ UTR were designated DTMUV-RLuc-DB2mut1 (nucleotides 474 to 485 of the 3′ UTR deleted), DTMUV-RLuc-DB2mut2 (nucleotides 447 to 452 of the 3′ UTR deleted), DTMUV-RLuc-DB1mut1 (nucleotides 371 to 375 of the 3′ UTR altered), DTMUV-RLuc-DB2mut3 (nucleotides 432 to 434 of the 3′ UTR altered), DTMUV-RLuc-PK1mut (nucleotides 500 to 504 of the 3′ UTR altered), DTMUV-RLuc-SLmut1 (nucleotides ACTT inserted between residues 569 and 570 of the 3′ UTR), and DTMUV-RLuc-SLmut3 (nucleotide 612 of the 3′ UTR altered). Reporters containing mutations designed to preserve secondary structures in the DTMUV 3′ UTR were designated DTMUV-RLuc-DB2mut4 (nucleotides 444 to 452 of the 3′ UTR altered), DTMUV-RLuc-DB1mut2 (nucleotides 402 to 407 of the 3′ UTR altered), DTMUV-RLuc-PK2mut (nucleotides 511 to 515 of the 3′ UTR altered), DTMUV-RLuc-SLmut2 (nucleotide 616 of the 3′ UTR altered), DTMUV-RLuc-SLmut4 (nucleotide 608 of the 3′ UTR altered), DTMUV-RLuc-SLmut5 (nucleotide 604 of the 3′ UTR altered), and DTMUV-RLuc-SLmut6 (nucleotide 601 of the 3′ UTR altered). All the mutated sites are shown in Fig. 6A, and details of the mutations are provided in Table 1.
In vitro transcription, RNA transfection, and analysis of luciferase activity and RNA stabilities.
Plasmids containing cDNAs of mono- and bicistronic reporters were linearized using the XhoI endonuclease (New England Biolabs) and transcribed in vitro using a TranscriptAid T7 high-yield transcription kit to obtain uncapped RNAs or using a mMessage mMachine kit (Thermo Fisher Scientific) to obtain RNAs with 5′ m7G cap structures. In some experiments, the reporter RNAs containing viral sequences were mixed with huGL-FLuc RNA at a ratio of 20:1 and used as described below (Fig. 9).
DEFs or BHK-21 or C6/36 cells were seeded into 48-well plates and incubated at 37°C (for DEFs and BHK-21 cells) or 28°C (for C6/36 cells) until they reached ∼70 to 90% confluence, after which the cells were transfected with RNA transcripts (250 ng/well) using Lipofectamine MessengerMax reagent (Invitrogen) and incubated at 37°C or 28°C, respectively. In experiments involving the use of wortmannin, an inhibitor of cap-dependent translation, BHK-21 cells grown on a 12-well plate were treated with 1 μM wortmannin or a corresponding volume of vehicle control (dimethyl sulfoxide [DMSO]) and transfected with reporter mRNAs (250 ng/well). At 4 h p.t., the cells were harvested and lysed, after which the activities of RLuc and FLuc were detected using a dual-luciferase reporter assay system (Promega). DEFs or BHK-21 cells were transfected with DTMUV-RLuc, huGL-RLuc, IRES-RLuc-pA or ΔIRES-RLuc-pA for 1 h, 4 h, and 7 h, and then RLuc activities were detected.
BHK-21 cells were transfected with reporter RNA for 4 h (7 h for DTMUV-RLuc, huGL-RLuc, IRES-RLuc-pA, and ΔIRES-RLuc-pA), cells were harvested, and total RNA was isolated. Reverse transcription (RT)-qPCR, which detected the RLuc gene, was used to measure the copy numbers of reporter RNAs.
Construction of mutant DTMUV replicon and virus cDNAs.
The plasmids pCMV-DTMUV-Rep-NLuc and pCMV-icDTMUV, in which the transcription of DTMUV cDNA is driven by an early promoter of human cytomegalovirus (CMV), were used to obtain cDNAs of DTMUV replicons and genomes containing mutations in the 3′ UTR sequences. The plasmids obtained were designated according to the introduced mutations (Table 1) as follows: pCMV-DTMUV-Rep-NLuc-DB2mut3, pCMV-DTMUV-Rep-NLuc-PK2mut, and pCMV-DTMUV-Rep-NLuc-SLmut5 (Fig. 8A) or pCMV-icDTMUV-DB2mut3 and pCMV-icDTMUV-SLmut5 (Fig. 8C).
Replication assay and rescue of mutant viruses.
For replication assays, BHK-21 cells were transfected with 1.6 μg of pCMV-DTMUV-Rep-NLuc, pCMV-DTMUV-Rep-NLuc-DB2mut3, pCMV-DTMUV-Rep-NLuc-PK2mut, and pCMV-DTMUV-Rep-NLuc-SLmut5 using TransIntro EL transfection reagent (TransGen Biotech, China) according to the manufacturer’s instructions. At 12, 24, 48, and 72 h p.t., the cells were lysed, and NLuc activities were measured using the Nano-Glo luciferase assay system (Promega).
For recombinant-virus rescue, BHK-21 cells were transfected with 1.6 μg of pCMV-icDTMUV, pCMV-icDTMUV-DB2mut3, or pCMV-icDTMUV-SLmut5 as described above. The rescue of viruses designated rDTMUV, rDTMUV-DB2mut3, and rDTMUV-SLmut5 was assessed using an IFA as described below.
An icDNA clone of DTMUV pACYC FL-CQW1, where the viral icDNA is placed under the control of a bacteriophage T7 RNA polymerase promoter (34), was in vitro transcribed using a TranscriptAid T7 high-yield transcription kit. The uncapped transcripts corresponding to the DTMUV genome obtained were used to transfect BHK-21 cells grown in 12-well plates. The transfected cells were then incubated in DMEM supplemented with 2% FBS for 5 days. Subsequently, the supernatant was harvested and used to infect naive BHK-21 cells, which were then incubated for 10 days and analyzed for the presence of rDTMUV using a mouse anti-DTMUV polyclonal antibody.
Western blotting and indirect immunofluorescence assay.
DEFs or BHK-21 cells grown on 12-well plates were infected with 10,000 TCID50 of DTMUV and cultured at 37°C. After incubating for 6, 12, 24, or 36 h, the cells were treated with 1 μg/ml (for DEFs) or 10 μg/ml (for BHK-21 cells) puromycin for 30 min. Then, the cells were harvested and analyzed by Western blotting. A mouse anti-puromycin monoclonal antibody (Merk-Millipore, Germany), a mouse anti-DTMUV-NS3 polyclonal antibody, a mouse anti-β-actin antibody (Ruiying Biological, China) and a rabbit anti-phospho-IF2α monoclonal antibody (CST) were used as primary antibodies. The membranes were then incubated with appropriate secondary antibodies conjugated to horseradish peroxidase, and proteins were visualized using Clarity Western ECL substrate (Bio-Rad).
For indirect IFA, infected or transfected cells were washed with phosphate-buffered saline (PBS)-Tween 20 (PBST), fixed with 4% paraformaldehyde for 1 h, permeabilized with 0.25% Triton X-100 in PBS for 30 min, and blocked with 0.5% bovine serum albumin (BSA) for 1 h at 37°C. After three rounds of washing with PBST, the cells were incubated with mouse anti-DTMUV polyclonal antibody as the primary antibody overnight at 4°C. The cells were then washed and treated with secondary goat anti-mouse Alexa 568 (Life Technologies) antibody for 1 h, after which nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Solarbio, China) for 15 min at room temperature. Subsequently, the cells were imaged using an 80i fluorescence microscope (Nikon, Japan).
Analysis of growth kinetics and virulence of the recombinant viruses.
rDTMUV-DB2mut3 was propagated in BHK-21 (p1) cells, and the virus genome RNA was isolated, reverse transcribed, and sequenced. This rDTMVU-DB2mut3 (p1) was used in the subsequent experiments. To analyze the CPE of rDTMUV and rDTMUV-DB2mut3 infection, BHK-21 cells or DEFs grown in 12-well plates were infected using 300 TCID50 of the virus (p1) per well. Infected cells were assessed for CPE and imaged at 48 and 72 h p.i.
To analyze the growth kinetics of the recombinant viruses, DEFs grown in 12-well plates were infected with rDTMUV or rDTMUV-DB2mut3 using 300 TCID50 per well. At 12, 24, 48, 60, and 72 h p.i., supernatants were harvested, and viral genomic RNA was extracted using a DNA/RNA isolation kit (Tiangen, China). For RT-qPCR, viral RNA was reverse transcribed using HiScript III RT SuperMix for qPCR (plus genomic-DNA [gDNA] wiper) (Vazyme, China) according to the manufacturer’s protocol. The protocols used for quantitative PCR and construction of the rDTMUV and rDTMUV-DB2mut3 growth curves were previously described (36).
To analyze the virulence of the recombinant viruses, 9-day-old embryonated duck eggs were infected by allantoic-cavity inoculation with 100 or 1,000 TCID50 of rDTMUV or rDTMUV-DB2mut3, while the mock-infection group received 100 μl of DMEM. The injected embryos were incubated at 37°C and assessed for signs of infection every day until PID 7.
Statistical analysis.
The significance of the observed differences was assessed using a two-tailed, unpaired, and unequal variant of Student's t test with GraphPad Prism 8.0. P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
We are grateful to Sanjie Cao (Research Center of Swine Disease, Sichuan Agricultural University) for the Japanese encephalitis virus strain SA14 (NCBI accession no. KU323483.1) and to Suresh Mahalingam (Institute for Glycomics, Griffith University) for the DENV2 strain New Guinea C cDNA clone.
This work was supported by grants from the National Key Research and Development Program of China (2017YFD0500800), Sichuan-International Joint Research for Science and Technology (2018HH0098), the China Agricultural Research System (CARS-42-17), the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and the Integration and Demonstration of Key Technologies for Goose Industrial Chain in Sichuan Province (2018NZ0005).
REFERENCES
- 1.Cao Z, Zhang C, Liu Y, Liu Y, Ye W, Han J, Ma G, Zhang D, Xu F, Gao X, Tang Y, Shi S, Wan C, Zhang C, He B, Yang M, Lu X, Huang Y, Diao Y, Ma X, Zhang D. 2011. Tembusu virus in ducks, China. Emerg Infect Dis 17:1873–1875. doi: 10.3201/eid1710.101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W, Chen S, Mahalingam S, Wang M, Cheng A. 2017. An updated review of avian-origin Tembusu virus: a newly emerging avian Flavivirus. J Gen Virol 98:2413–2420. doi: 10.1099/jgv.0.000908. [DOI] [PubMed] [Google Scholar]
- 3.Tang Y, Gao X, Diao Y, Feng Q, Chen H, Liu X, Ge P, Yu C. 2013. Tembusu virus in human, China. Transbound Emerg Dis 60:193–196. doi: 10.1111/tbed.12085. [DOI] [PubMed] [Google Scholar]
- 4.Zhu K, Huang J, Jia R, Zhang B, Wang M, Zhu D, Chen S, Liu M, Yin Z, Cheng A. 2015. Identification and molecular characterization of a novel duck Tembusu virus isolate from Southwest China. Arch Virol 160:2781–2790. doi: 10.1007/s00705-015-2513-0. [DOI] [PubMed] [Google Scholar]
- 5.Ng WC, Soto-Acosta R, Bradrick SS, Garcia-Blanco MA, Ooi EE. 2017. The 5′ and 3′ untranslated regions of the flaviviral genome. Viruses 9:137. doi: 10.3390/v9060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brocard M, Ruggieri A, Locker N. 2017. m6A RNA methylation, a new hallmark in virus-host interactions. J Gen Virol 98:2207–2214. doi: 10.1099/jgv.0.000910. [DOI] [PubMed] [Google Scholar]
- 8.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2019. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 11:a033001. doi: 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman MA, Palmenberg AC. 1995. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol 69:4399–4406. doi: 10.1128/JVI.69.7.4399-4406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chahal J, Gebert LFR, Gan HH, Camacho E, Gunsalus KC, MacRae IJ, Sagan SM. 2019. miR-122 and Ago interactions with the HCV genome alter the structure of the viral 5′ terminus. Nucleic Acids Res 47:5307–5324. doi: 10.1093/nar/gkz194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto GA, Puglisi JD. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 12.Danthinne X, Seurinck J, Meulewaeter F, Van Montagu M, Cornelissen M. 1993. The 3′ untranslated region of satellite tobacco necrosis virus RNA stimulates translation in vitro. Mol Cell Biol 13:3340–3349. doi: 10.1128/mcb.13.6.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L, Allen EM, Miller WA. 2001. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol Cell 7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- 14.Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholson BL, White KA. 2011. 3′ Cap-independent translation enhancers of positive-strand RNA plant viruses. Curr Opin Virol 1:373–380. doi: 10.1016/j.coviro.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Edgil D, Polacek C, Harris E. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol 80:2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Y, Mugavero J, Stauft CB, Wimmer E. 2019. Dengue and Zika virus 5′ untranslated regions harbor internal ribosomal entry site functions. mBio 10:e00459-19. doi: 10.1128/mBio.00459-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terenin IM, Smirnova VV, Andreev DE, Dmitriev SE, Shatsky IN. 2016. A researcher’s guide to the galaxy of IRESs. Cell Mol Life Sci 74:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrasco L, Sanz MA, González-Almela E. 2018. The regulation of translation in alphavirus-infected cells. Viruses 10:70. doi: 10.3390/v10020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belsham GJ, McInerney GM, Ross-Smith N. 2000. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol 74:272–280. doi: 10.1128/jvi.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras A-C, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci U S A 93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Courtney SC, Scherbik SV, Stockman BM, Brinton MA. 2012. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J Virol 86:3647–3657. doi: 10.1128/JVI.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, Stoecklin G, Ruggieri A. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. mBio 8:e02150-16. doi: 10.1128/mBio.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu Y-C, Yu C-Y, Liang J-J, Lin E, Liao C-L, Lin Y-L. 2012. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J Virol 86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPointe AT, Moreno-Contreras J, Sokoloski KJ. 2018. Increasing the capping efficiency of the Sindbis virus nsP1 protein negatively affects viral infection. mBio 9:e02342-18. doi: 10.1128/mBio.02342-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sokoloski K, Haist K, Morrison T, Mukhopadhyay S, Hardy R. 2015. Noncapped alphavirus genomic RNAs and their role during infection. J Virol 89:6080–6092. doi: 10.1128/JVI.00553-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Melian EB, Edmonds J, Dong H, Shi P-Y, Khromykh AA. 2010. RNA structures required for production of subgenomic flavivirus RNA. J Virol 84:11407–11417. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi P-Y, Hall RA, Khromykh AA. 2008. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 29.Göertz GP, van Bree JWM, Hiralal A, Fernhout BM, Steffens C, Boeren S, Visser TM, Vogels CBF, Abbo SR, Fros JJ, Koenraadt CJM, van Oers MM, Pijlman GP. 2019. Subgenomic flavivirus RNA binds the mosquito DEAD/H-box helicase ME31B and determines Zika virus transmission by Aedes aegypti. Proc Natl Acad Sci U S A 116:19136–19144. doi: 10.1073/pnas.1905617116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanford TJ, Mears HV, Fajardo T Jr, Locker N, Sweeney TR. 2019. Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res 47:9789–9802. doi: 10.1093/nar/gkz686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, Qin CF. 2016. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 5:e17636. doi: 10.7554/eLife.17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P, Wei Y, Mei M, Tang L, Sun L, Huang W, Zhou J, Zou C, Zhang S, Qin C-F, Jiang T, Dai J, Tan X, Zhang QC. 2018. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe 24:875–886. doi: 10.1016/j.chom.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 33.Polacek C, Friebe P, Harris E. 2009. Poly(A)-binding protein binds to the non-polyadenylated 3′ untranslated region of dengue virus and modulates translation efficiency. J Gen Virol 90:687–692. doi: 10.1099/vir.0.007021-0. [DOI] [PubMed] [Google Scholar]
- 34.Chen S, He Y, Zhang R, Liu P, Yang C, Wu Z, Zhang J, Wang M, Jia R, Zhu D, Mafeng L, Qiao Y, Ying W, Anchun C. 2018. Establishment of a reverse genetics system for duck Tembusu virus to study virulence and screen antiviral genes. Antiviral Res 157:120–127. doi: 10.1016/j.antiviral.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Mutso M, Saul S, Rausalu K, Susova O, Zusinaite E, Mahalingam S, Merits A. 2017. Reverse genetic system, genetically stable reporter viruses and packaged subgenomic replicon based on a Brazilian Zika virus isolate. J Gen Virol 98:2712–2724. doi: 10.1099/jgv.0.000938. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Wang T, Liu P, Yang C, Wang M, Jia R, Zhu D, Liu M, Yang Q, Wu Y, Xinxin Z, Anchun C. 2019. Duck interferon regulatory factor 7 (IRF7) can control duck Tembusu virus (DTMUV) infection by triggering type I interferon production and its signal transduction pathway. Cytokine 113:31–38. doi: 10.1016/j.cyto.2018.06.001. [DOI] [PubMed] [Google Scholar]