Human cytomegalovirus (HCMV) is a herpesvirus that leads to serious health consequences in neonatal or immunocompromised patients. Clinical management of infection in these at-risk groups remains a serious concern even with approved antiviral therapies available. It is necessary to increase our understanding of the cellular changes that occur during infection and their importance to virus spread. This may help to identify new targets during infection that will lead to the development of novel treatment strategies. Extracellular vesicles (EVs) represent an important method of intercellular communication in the human host. This study finds that HCMV manipulates this pathway to increase the efficiency of virus spread to uninfected cells. This finding defines a new layer of host manipulation induced by HCMV infection that leads to enhanced virus spread.
KEYWORDS: CD63, ESCRTs, GW4869, HCMV, extracellular vesicles, spread
ABSTRACT
Human cytomegalovirus (HCMV) manipulates cellular processes associated with secretory pathways within an infected cell to facilitate efficient viral replication. However, little is known about how HCMV infection alters the surrounding cellular environment to promote virus spread to uninfected cells. Extracellular vesicles (EVs) are key signaling molecules that are commonly altered in numerous disease states. Previous reports have shown that viruses commonly alter EVs, which can significantly impact infection. This study finds that HCMV modulates EV biogenesis machinery through upregulation of the endosomal sorting complex required for transport (ESCRT) proteins. This regulation appears to increase the activity of EV biogenesis, since HCMV-infected fibroblasts have increased vesicle release and altered vesicle size compared to EVs from uninfected cells. EVs generated through ESCRT-independent pathways are also beneficial to virus spread in fibroblasts, as treatment with the EV inhibitor GW4869 slowed the efficiency of HCMV spread. Importantly, the transfer of EVs purified from HCMV-infected cells enhanced virus spread. This suggests that HCMV modulates the EV pathway to transfer proviral signals to uninfected cells that prime the cellular environment for incoming infection and enhance the efficiency of virus spread.
IMPORTANCE Human cytomegalovirus (HCMV) is a herpesvirus that leads to serious health consequences in neonatal or immunocompromised patients. Clinical management of infection in these at-risk groups remains a serious concern even with approved antiviral therapies available. It is necessary to increase our understanding of the cellular changes that occur during infection and their importance to virus spread. This may help to identify new targets during infection that will lead to the development of novel treatment strategies. Extracellular vesicles (EVs) represent an important method of intercellular communication in the human host. This study finds that HCMV manipulates this pathway to increase the efficiency of virus spread to uninfected cells. This finding defines a new layer of host manipulation induced by HCMV infection that leads to enhanced virus spread.
INTRODUCTION
Human cytomegalovirus (HCMV) is a leading cause of congenital viral infection, which can result in numerous complications for the developing fetus and newborn. Infection of immunocompromised individuals is also a significant source of disease, particularly in solid organ and hematopoietic cell transplant recipients, where virus infection is commonly detected in patients (1–3). There is a need to understand the molecular mechanisms of infection to provide new therapeutic targets to combat antiviral resistance and toxicity associated with the currently approved therapies.
HCMV expresses over 200 proteins and noncoding RNAs from a large DNA genome (4, 5). This allows extensive modulation of the host to facilitate efficient replication and spread of the virus. The temporal expression of viral factors during infection supports the various stages of a prolonged replication cycle. Most studies have focused on how this regulation impacts viral replication within the infected cell. However, whether HCMV alters intercellular communication between infected and surrounding uninfected cells is less understood. HCMV causes a dramatic reorganization of cellular membranes and organelles associated with the secretory system during the formation of the cytoplasmic viral assembly compartment (cVAC), which results in altered vesicle trafficking (6–8). Viral microRNAs (miRNAs) also regulate proteins associated with secretory pathways in the cell (9). Manipulation of these pathways inevitably changes the viral and host factors that are transferred to surrounding uninfected cells. This could lead to the regulation of key pathways known to be important during HCMV infection such as metabolic activity, apoptosis, immune signaling, and cell cycle regulation. Regulation of these pathways in uninfected cells could have important implications for virus infection and spread, but the relevance to HCMV infection is largely unknown.
Cells release signaling molecules inside membrane-enclosed compartments termed extracellular vesicles (EVs) that transfer proteins and nucleic acids to recipient cells. This signaling mechanism is commonly modified during viral infection. Modulation of the content loaded into EVs during viral infection determines whether these signaling vesicles contribute to host defense or create an advantageous environment for the virus. The transfer of EVs from infected cells can activate innate antiviral activity in uninfected cells to inhibit the spread of the virus (10). Alternatively, the incorporation of viral genomes or viral proteins into EVs can enhance infection by preventing the antiviral response or through the escape from neutralizing antibody (11–14). Herpesviruses are also known to modulate this signaling pathway, as cells infected with herpes simplex virus 1 (HSV-1) release EVs that inhibit the spread of the virus (15–17). Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) both package viral miRNAs into EVs that contribute to the tumor microenvironment and also may enhance virus spread (18–21). HCMV also incorporates viral proteins into EVs, which may contribute to the transfer of viral antigen to recipient cells (22, 23). However, the EV contribution to HCMV infection has not been determined. EV biogenesis and release can occur through multiple cellular pathways. One such pathway involves the membrane scission proteins associated with the endosomal sorting complex required for transport (ESCRT) machinery. We and others have reported that ESCRT-III and VPS4 are important for efficient spread of HCMV (24, 25). Since HCMV reorganizes vesicle trafficking within infected cells and the ESCRTs play a role in infection, we hypothesized that HCMV modulates EVs to enhance virus spread.
In this study, we find that HCMV requires EVs for efficient spread. Inhibition of EV production slowed the spread of HCMV through a monolayer, which was rescued by addition of purified EVs. Furthermore, HCMV spread was enhanced with addition of EVs purified from infected fibroblasts. We found that infected fibroblasts produce more EVs, which correlates with an increase in the expression of EV biogenesis machinery. EVs produced during infection are morphologically unaltered but trend to smaller size and contain viral cargo. The functions of the viral components found in EVs are consistent with a role in preparing uninfected cells for the oncoming infection. These results uncover a mechanism by which HCMV modulates a common biological pathway to enhance viral infection.
(This research was conducted by Nicholas T. Streck in partial fulfillment of the requirements for a PhD from The Pennsylvania State University College of Medicine, Hershey, PA, 2020 [26].)
RESULTS
HCMV regulates expression of EV biogenesis machinery.
We previously found that inhibition of the ESCRT proteins hindered the spread of HCMV (24, 25). The ESCRT proteins promote membrane scission during multivesicular body (MVB) formation and represent one pathway for the formation of vesicles that can be released as EVs (27). It is known that inhibition of ESCRT activity through expression of dominant negative ESCRT subunits or RNA interference (RNAi) reduces the number of EVs released from the cell (28–30). Conversely, expression levels of ESCRT proteins are positively correlated with increased vesicle release (31).
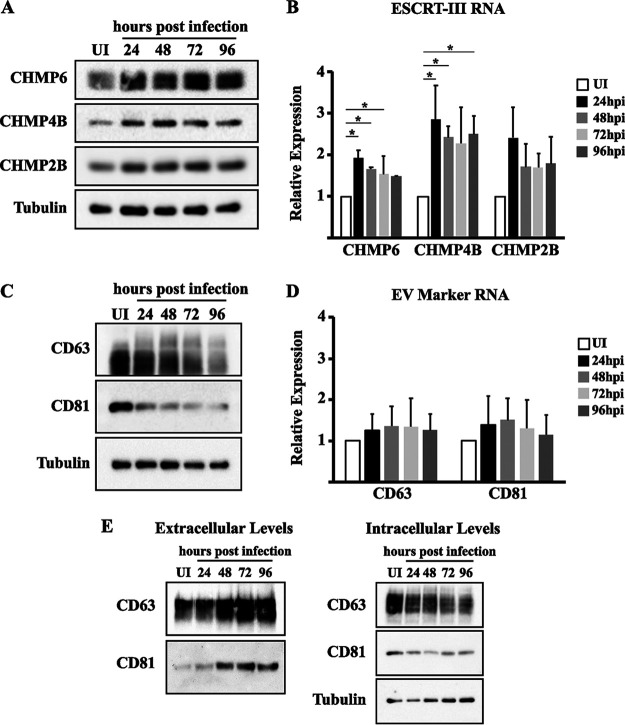
Since ESCRT-III is associated with the scission-promoting function of these complexes, we analyzed expression levels of subunits from this complex during infection of human dermal fibroblasts (HDFs). Interestingly, we found that protein levels of ESCRT-III subunits were increased throughout infection (Fig. 1A). Transcript levels of the ESCRT-III subunits tested were also increased, indicating that the increase in protein is at least partially due to increased mRNA within infected HDFs (Fig. 1B). We hypothesized that the increase in ESCRT-III protein levels could lead to an increase in EV production. To test this, we analyzed the intracellular expression levels of common EV surface markers, the tetraspanins CD63 and CD81. We found that intracellular levels of both EV markers were decreased during infection (Fig. 1C). The decrease in intracellular levels of EV surface markers does not appear to be due to changes in transcript levels, as these appear to remain relatively constant throughout infection (Fig. 1D). The decrease in intracellular EV markers could be explained by an increase in the release of EVs from the cell or an increase in protein degradation. To test the possibility that HCMV infection may cause an increase in the release of CD63 and CD81, we analyzed intracellular and extracellular levels of the proteins during infection. Interestingly, we found that extracellular levels of both proteins increased during infection (Fig. 1E). This indicates that HCMV infection alters the expression of EV biogenesis proteins and increases the release of EV markers during infection. However, this does not exclude the possibility that subsets of CD63 and CD81 are degraded during infection.
FIG 1.
HCMV infection alters EV biogenesis machinery. Uninfected and HCMV-infected (MOI = 3) fibroblasts at 24, 48, 72, and 96 hpi were harvested for protein (A and C) and RNA (B and D) analyses. (A) Western blot analysis of the ESCRT-III subunits CHMP6, CHMP4B, and CHMP2B. Tubulin was used as a loading control. (B) qPCR of CHMP6, CHMP4B, and CHMP2B. (C) Western blot of EV markers CD63 and CD81. Tubulin was used as a loading control. (D) qPCR analysis of CD63 and CD81 for a high MOI. (E) Western blot of CD63 and CD81 from HCMV-infected (MOI = 3) fibroblasts at 24, 48, 72, and 96 hpi. qPCR results are from three independent experiments. *, P < 0.05 compared to uninfected samples.
Inhibition of extracellular vesicle production slows HCMV spread.
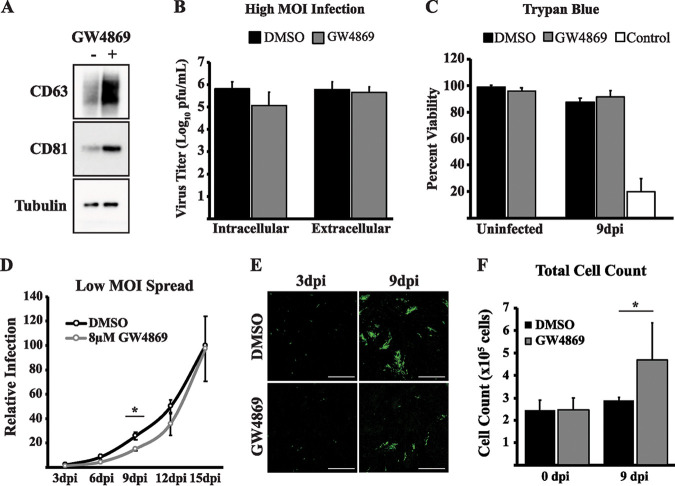
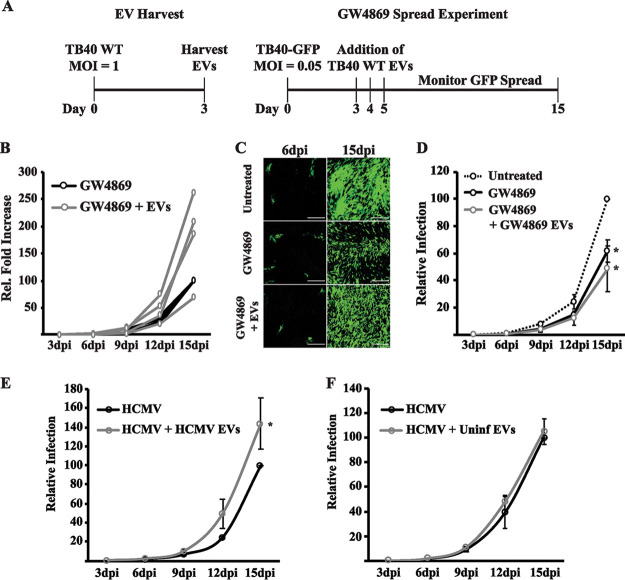
The regulation of EV-associated proteins is suggestive of an increase in EV release during infection. In addition to the ESCRT pathway, EV biogenesis occurs through the enrichment of the lipid ceramide at the endosomal membrane. Sphingomyelinases generate ceramide from a sphingomyelin precursor, and since treatment of cells with neutral sphingomyelinase (nSMase) inhibitors alters EV formation and release from the cell, these inhibitors are commonly used to disrupt EV release during diverse virus infections (10, 32–34). To test if EVs generated from this pathway also contribute to HCMV spread, we treated HCMV-infected cells with the nSMase inhibitor GW4869. The efficacy of the inhibitor was confirmed by observing an intracellular accumulation of EV markers (Fig. 2A). HCMV replication and egress are associated with cellular proteins that are also utilized in the release of EVs (35). It is possible that in addition to altering EV biogenesis, treatment with GW4869 may also inhibit the replication cycle of the virus. We therefore analyzed intracellular and extracellular virus titers at a high multiplicity of infection (MOI) in the presence of GW4869. We found that GW4869 treatment caused a slight reduction in intracellular levels of infectious virus; however, virus egress was unaffected, as extracellular virus titers were unchanged in the presence of GW4869 (Fig. 2B). GW4869 treatment, even for prolonged periods of time, did not appear to cause toxicity in the fibroblasts, because cell viabilities were similar across all treatment groups (Fig. 2C). Since we hypothesize that the spread defect observed with blocking ESCRT function is due to altered extracellular vesicle, we wanted to test whether GW4869 also altered the spread of HCMV. Thus, we hypothesized that treatment of infected cells with GW4869 would also affect the spread of HCMV. We tested the effect of GW4869 at a low MOI using a recombinant TB40/E strain that encodes green fluorescent protein (GFP) adjacent to the viral immediate early protein 2 separated by the T2A peptide (36). During infection with this virus, GFP-positive cells can be used to monitor virus spread through the monolayer after addition of the EV inhibitor GW4869 or dimethyl sulfoxide (DMSO) vehicle control. In contrast to the high MOI results, the inhibitor slowed the efficiency of virus spread through the monolayer (Fig. 2D and E). The reduction in spread following GW4869 treatment was not caused by a reduction in cell density, as total cell counts were not reduced in the treatment group (Fig. 2F). In fact, cell counts were slightly higher in GW4869-treated cells at 9 days postinfection (dpi). It is well known that HCMV infection causes cell cycle arrest and therefore prevents cell division. Since this is the time point with the largest difference in virus spread, we hypothesize that the higher cell counts in GW4869-treated cells may be caused by an increased number of uninfected cells still undergoing cell division during the experiment. Therefore, the nSMase inhibitor GW4869 slows HCMV spread without affecting virus egress or cell viability.
FIG 2.
GW4869 slows HCMV spread. (A) Western blot of intracellular CD63 and CD81 in HCMV-infected (MOI = 1) fibroblasts at 72 hpi treated with DMSO or 8 μM GW4869. Tubulin was used as a loading control. (B) Intracellular and extracellular virus titers from HCMV-infected (MOI = 3) fibroblasts at 96 hpi treated with DMSO or 8 μM GW4869. (C) Trypan blue viability for uninfected or HCMV-infected (MOI = 0.05) fibroblasts at 9 dpi. At indicated time points, cells were trypsinized and resuspended in DMEM before being mixed with trypan blue. Positive-control cells were incubated at 65°C for 10 min. (D) Virus spread in HCMV-infected (MOI = 0.05) fibroblasts treated with DMSO or 8 μM GW4869. Relative infection measured by GFP expression in the monolayer. Values for titers and virus spread are from three independent experiments. (E) Fluorescence images from low-MOI spread infection in panel D. Scale bars are 1,000 μm. (F) Total cell count of uninfected and HCMV-infected (MOI = 0.05) fibroblasts at 9 dpi treated with DMSO or 8 μM GW4869. *, P < 0.05.
Purification of EVs from HCMV-infected cells.
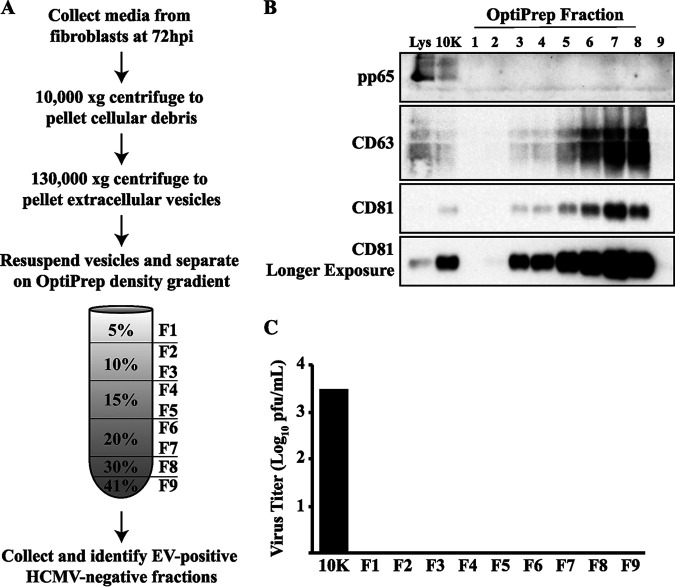
Our data show that both treatment with the EV inhibitor GW4869 and blocking ESCRT activity slow the efficiency of HCMV spread without inhibiting the viral replication cycle. These data support the hypothesis that EVs released from HCMV-infected cells are important for virus spread. To gain a better understanding of the role of EVs during infection, we purified EVs from infected or uninfected cells. We chose to purify EVs from HCMV-infected HDFs at 72 h postinfection (hpi) since this time point indicated regulation of the EV-associated proteins (Fig. 1). To characterize the EVs released from infected cells, the vesicles must be separated from viral particles that are also present in the infected media. EVs were separated from viral particles and purified using a combination of high-speed and ultracentrifugation steps with an OptiPrep gradient (Fig. 3A). Western blot analysis of the isolated fractions for CD63 and CD81 was used to identify fractions that contained EVs, and an antibody to the abundant viral tegument protein pp65 was used to identify fractions enriched with viral particles or dense bodies (Fig. 3B). CD63- and CD81-positive fractions spanned the 10% to 30% region of the gradient. Importantly, pp65 was not detected in fractions that were positive for the EV markers, suggesting that EVs were successfully separated from viral particles. To confirm this, we performed an infectious titer assay on all collected fractions. For the majority of the isolations, HCMV was pelleted at a high-speed centrifugation step labeled “10K pellet” as shown in a representative titer assay (Fig. 3C). This was unexpected, as much higher centrifugation speeds are typically used in HCMV purification protocols. However, this was the step in the purification protocol that consistently contained infectious particles. In the rare event that infectious virus was present within the gradient, these fractions were discarded and not used for any downstream analysis or experiments. Using this approach, we were able to successfully separate EVs from infectious particles.
FIG 3.
EV purification from HCMV-infected cells. (A) Protocol to purify EVs from HCMV-infected (MOI = 1) fibroblast medium. (B) Western blot analysis of the viral protein pp65 and cellular proteins CD63 and CD81 from EV purification. Cell lysate at 72 hpi (Lys), 10,000 × g centrifugation pellet (10K), and fractions from the OptiPrep density gradient (1 to 9). (C) Virus titers from a representative EV purification experiment.
HCMV infection increases EV release.
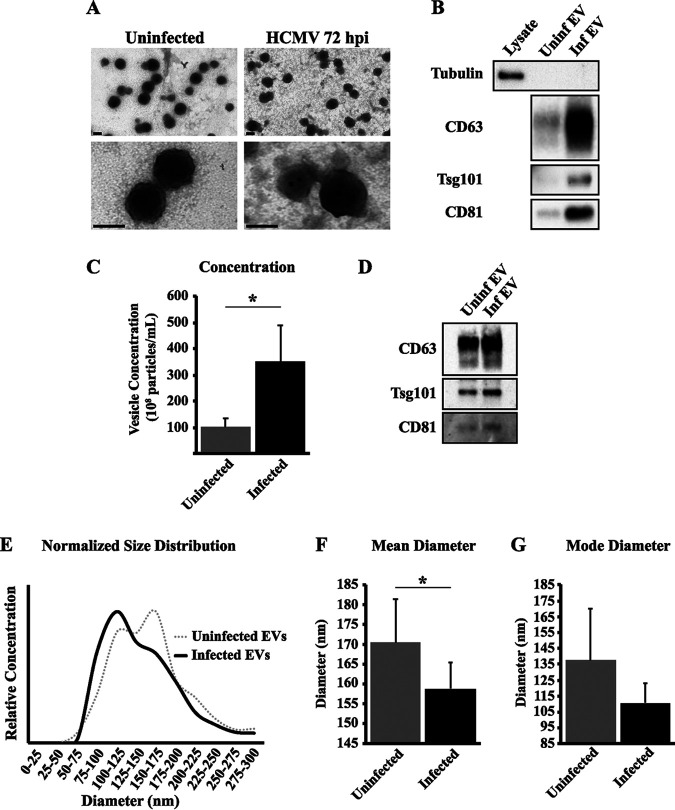
We next analyzed and compared the EV preps from infected and uninfected cells. Electron microscopy (EM) analysis of the purified EV preps revealed no differences in morphology between the samples (Fig. 4A). Importantly, we did not observe any viral particles, consistent with our infectious titer assay of vesicle fractions. Cells can release many unique vesicle populations that are characterized by origin of biogenesis, internal cargo, surface markers, and size (37). Pooled EV fractions were tested for changes in vesicle surface markers by Western blot analysis loaded in equal volumes (Fig. 4B). Both infected and uninfected EVs were positive for the vesicle markers CD63, CD81, and Tsg101 and negative for the intracellular marker α-tubulin. Strikingly, the cellular EV markers were more abundant in the infected EV samples. The increase in vesicle marker intensity could be occurring through two ways: HCMV infection could increase the number of vesicles released from the cell, or infected cells may release vesicles with a higher enrichment of these molecules per vesicle. Viral infections were previously shown to increase the release of EVs from infected cells (17, 38). To address this possibility, we analyzed the vesicle population by nanoparticle tracking analysis (NTA). Interestingly, HCMV-infected cells released 3-fold more EVs than uninfected cells, suggesting that the increase in extracellular EV markers is a result of increased release of vesicles from the cell (Fig. 4C). In support of this, Western blot analysis of EVs with equal loading of vesicle numbers showed little difference in EV marker abundance between uninfected and infected samples (Fig. 4D). In addition to increased vesicle release, HCMV infection also altered the size distribution of the purified EVs. Uninfected cells released vesicles that were categorized by their diameters into small and large EVs (Fig. 4E). Interestingly, infected cells released a more uniform population of vesicles that were smaller in size than uninfected EVs (Fig. 4E). This is supported by the fact that infected EVs have smaller median and mode vesicle diameter than uninfected EVs (Fig. 4F and G). The trend toward a smaller vesicle size is further indication that HCMV infection alters activity associated with EV pathways. Thus, HCMV-mediated regulation of the EV biogenesis machinery results in increased release of EVs and altered vesicle size.
FIG 4.
HCMV infection increases EV production. Analysis of EV populations was carried out using EV fractions from uninfected or HCMV-infected (MOI = 1) fibroblasts at 72 hpi. (A) Electron micrographs of pooled EV fractions. Scale bars are 50 nm. (B) Western blot analysis of pooled EV fractions for tubulin, CD63, Tsg101, and CD81 loaded at equal volumes. Lysate is from HCMV-infected (MOI = 1) fibroblasts at 72 hpi. (C) Particle concentration of pooled EV fractions assayed by nanoparticle tracking analysis (NTA). (D) Western blot analysis of EV fractions loaded at equal vesicle numbers. Normalized size distributions (E), mean diameters (F), and mode diameters (G) of pooled EV fractions. All NTA analyses are from seven independent EV harvests. *, P < 0.05.
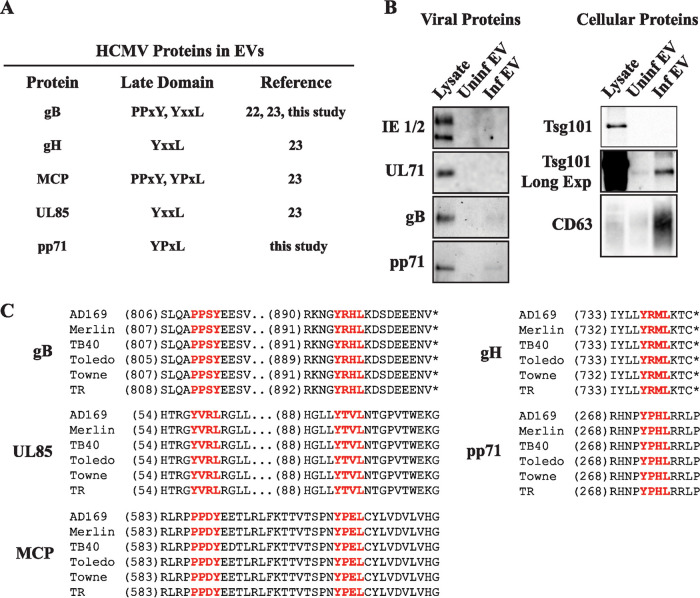
EVs from HCMV-infected cells contain viral proteins with late domain sequences.
Our data have shown that EVs released during HCMV infection appear to enhance the efficiency of virus spread. To determine how these EVs contribute to HCMV spread, it is important to identify HCMV-mediated changes in EV cargo. EVs are enriched in a number of signaling molecules, some of which include lipids, proteins, mRNAs, and noncoding RNAs such as miRNAs. These molecules can have diverse effects on the activity of the recipient cell. The genome of HCMV encodes a number of proteins that alter the activity of the infected cell to help initiate and establish a productive infection. We hypothesize that HCMV-infected cells transfer these viral factors through EVs to uninfected cells to alter the environment, which helps establish infection and enhance spread. In fact, viral proteins have been found in EVs released from HCMV-infected cells (22, 23). Upon further analysis of these previous reports, HCMV viral proteins identified in EVs contain amino acid sequences classified as late domains (Fig. 5A). Late domains were first described as amino acid sequences in viral proteins necessary for a late stage in the viral replication cycle of retroviruses and are known to be important for protein-protein interactions with ESCRT subunits, serving as a mechanism to recruit the ESCRT machinery during infection (39–41). However, late domains can also recruit proteins for incorporation into EVs through interactions with ESCRT proteins (42). The HCMV genome encodes multiple proteins with predicted late domain sequences even though the ESCRTs are not required for viral envelopment (25, 43). We analyzed our EV population for the presence of viral proteins (Fig. 5B and 3B). Consistent with previous findings, EVs were positive for the late domain-containing viral glycoprotein gB and negative for the abundant tegument protein pp65, which does not contain a late domain (22, 23). We also found the UL82 gene product pp71 in the EV population, which also contains a late domain sequence and has well-documented roles in the initiation of infection and the manipulation of the antiviral response (44–46). Interestingly, the potential late domain sequences harbored by viral proteins found in EVs were found in multiple HCMV strains (Fig. 5C). This high level of conservation suggests that the sequences are important for proper function of the proteins, perhaps due to their role in targeting the proteins for packaging into EVs. Incorporation of HCMV proteins into EVs appears to be specific, as some late domain-containing proteins (pp150) and proteins without late domains (pp65) were not found in EVs (22, 23). Together, these data suggest that HCMV proteins are selectively packaged into EVs and this may be dependent upon late domain sequences.
FIG 5.
EVs from HCMV-infected cells contain viral proteins. (A) Table of late domain sequences within viral proteins detected in infected EVs. (B) Western blot analysis of viral and cellular proteins in pooled EV fractions from uninfected or HCMV-infected (MOI = 1) fibroblasts at 72 hpi. Samples loaded at equal volumes. (C) Amino acid sequences of HCMV strains AD169 (GenBank FJ527563.1), Merlin (NCBI Reference Sequence NC_006273.2), TB40/E (GenBank EF999921.1), Toledo (GenBank GU937742.2), Towne (GenBank FJ616285.1), and TR (GenBank KF021605.1). Amino acid positions are shown in parentheses. Potential late domains are shown in red. *, stop codon.
Infected EVs do not require CD63 to enhance spread.
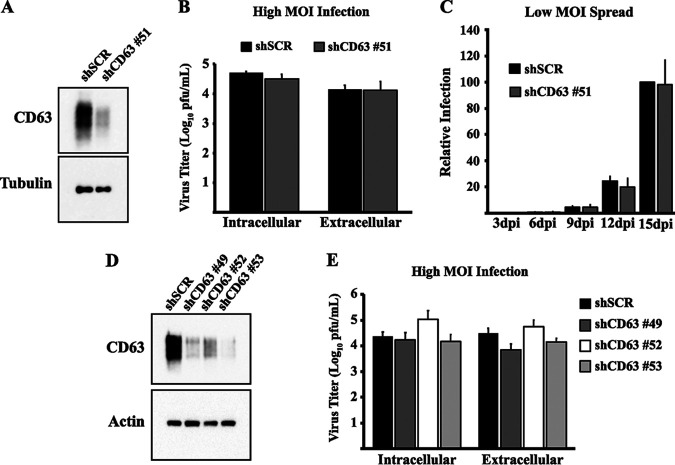
In addition to late domains, other mechanisms exist for EV cargo selection. Proteins are selectively incorporated into vesicles through posttranslational modifications or interactions with EV-associated proteins, such as CD63, that target cargo to the site of EV biogenesis (42, 47). With respect to herpesviruses, CD63 is important for incorporating the EBV latent membrane protein 1 (LMP1) into EVs (48–50). Additionally, CD63 is important for the antiviral phenotype of HSV-1 EVs, as knockdown of CD63 reduces the antiviral phenotype during HSV-1 infection (15). Conversely, overexpression of CD63 amplifies the phenotype (17). To see if CD63 was also important for HCMV spread, we used a lentiviral short hairpin RNA (shRNA) to knockdown expression of CD63 in HDFs (Fig. 6A). We found that knockdown of CD63 did not inhibit HCMV replication at a high MOI or block virus egress (Fig. 6B). Surprisingly, knockdown of CD63 also had no effect on the efficiency of HCMV spread (Fig. 6C). This appears to be in contrast to other herpesvirus infections and may represent a key difference between the mechanisms of EV manipulation. These data clearly indicate that the effector molecules necessary to enhance HCMV spread are incorporated into EVs in a CD63-independent mechanism. It was recently reported that CD63 is required for HCMV replication. Although the precise role of CD63 was not defined, evidence suggested that CD63 may be responsible for the trafficking of other membrane proteins known to be important for HCMV entry or that CD63 may be important for assembly (51). We transduced fibroblasts with three additional shRNA clones to thoroughly test whether knockdown of CD63 inhibited viral replication (Fig. 6D). While all three clones resulted in robust knockdown of CD63, none of the clones inhibited HCMV replication (Fig. 6E). A major difference between these two reported results is the duration of CD63 knockdown. In this lentiviral system, the knockdown of CD63 is for a prolonged period of time relative to that in the recent report (51). It is possible that prolonged knockdown of CD63 results in new trafficking patterns of proteins that have been linked to CD63. However, a more defined role of CD63 during this process is needed before this can be addressed. Nonetheless, this combination of data suggests that CD63 is not directly required for HCMV replication or spread.
FIG 6.
Knockdown of CD63 does not inhibit HCMV spread. (A) Western blot analysis of CD63 in uninfected fibroblasts transduced with lentivirus expressing shRNA to CD63 or scrambled control. Tubulin was used as a loading control. (B) Intracellular and extracellular virus titers from HCMV-infected (MOI = 3) transduced fibroblasts at 96 hpi. Titers are from three independent experiments. (C) Virus spread in HCMV-infected (MOI = 0.05) transduced fibroblasts. Relative infection measured by GFP expression in the monolayer. Values for low-MOI spread data represent five independent experiments. (D) Western blot analysis of CD63 in uninfected fibroblasts transduced with lentivirus expressing shRNA clones to CD63 or scrambled control. (E) Intracellular and extracellular virus titers from HCMV-infected (MOI = 3) transduced fibroblasts at 96 hpi. Titers are from three independent experiments.
Extracellular vesicles enhance HCMV infection.
We have shown that HCMV alters the number and composition of EVs released from cells and that blocking EV biogenesis impairs virus spread. To determine if HCMV-mediated changes in EVs were responsible for enhancing HCMV spread, we tested whether the addition of purified EVs from infected cells could rescue the spread defect caused by the EV inhibitor GW4869. Under physiological conditions, the number and frequency of vesicles that individual cells receive from the extracellular environment are unknown. However, using a similar approach as was used to study EVs in other herpesvirus infections, we tested whether the transfer of purified EVs could rescue the spread defect of GW4869 (15, 20) (Fig. 7A). To alleviate the concern that we may have small amounts of undetectable virus in our purified EV population, all EV isolations were performed on cells infected with a TB40/E virus that does not express any fluorescent proteins. Even if a small amount of contaminating virus is present in fractions, this would not impact the analysis of our spread experiment, which depends upon the presence of GFP to monitor virus spread. We found that treatment of cells with EVs purified from infected HDFs enhanced the spread of HCMV in the presence of the EV inhibitor (Fig. 7B). The degree of rescue was variable among the experiments, and this could be due to the quality of the purified EVs. Although addition of EVs resulted in an increase in virus spread, spread was still reduced compared to that in the untreated sample (Fig. 7C). This may be due to the number of vesicles added, frequency of vesicle addition, or when the purified vesicles were harvested. Interestingly, EVs purified from GW4869-treated cells were unable to rescue the spread phenotype of the inhibitor (Fig. 7D). This is further indication that treatment with GW4869 disrupts the EV production during HCMV infection. Since EVs rescued HCMV spread in the presence of the EV inhibitor, we next tested whether EVs could enhance the spread of an untreated infection. We found that the addition of EV to an untreated HCMV infection did in fact increase the efficiency of infection, providing further evidence that EVs released from infected cells enhance HCMV spread (Fig. 7E). Importantly, the addition of EVs purified from uninfected cells did not increase virus spread (Fig. 7F). Collectively, these data clearly state that EVs released from HCMV-infected cells transfer signals to uninfected cells that are important for efficient virus spread.
FIG 7.
EVs from HCMV-infected cells enhance virus spread. (A) Experimental outline to transfer purified EVs to HCMV-infected (MOI = 0.05) fibroblasts treated with 8 μM GW4869; 3,000 to 5,000 purified EVs were added to spread experiments at 3, 4, and 5 dpi as indicated, and GFP expression was measured at 3, 6, 9, 12, and 15 dpi. (B) Fold increases for four individual experiments showing virus spread relative to that for GW4869-treated HCMV-infected fibroblasts. (C) Fluorescence images from the spread infection in panel B. Scale bar is 250 μm. (D) Virus spread in fibroblasts treated with 8 μM GW4869 and EVs purified from cells treated with 8 μM GW4869. *, P < 0.05 compared to untreated control. (E) Virus spread in the presence of EVs purified from HCMV infection. *, P < 0.05. (F) Virus spread in the presence of EVs purified from uninfected fibroblasts.
DISCUSSION
The complex network of intercellular communication dictates the activity of cells within a given environment, and viral manipulation of this network has important implications for infection and spread. Extracellular vesicles have emerged as important players in delivering content between cells. The ESCRT proteins represent one pathway for EV biogenesis, and we and others have demonstrated that these proteins are necessary for efficient virus spread (24, 25). Since EVs are known to be important factors in the spread of other herpesvirus infections, we hypothesized that the ESCRT-dependent contribution to HCMV spread is due to its role in EV biogenesis (16, 21, 52–54). EVs are produced through both ESCRT-dependent and ESCRT-independent pathways (34). We found that the EV biogenesis inhibitor GW4869 also slowed HCMV spread and that this phenotype was partially rescued by adding purified EVs from HCMV-infected cells. Importantly, we also found that adding purified EVs to an untreated HCMV infection also enhanced the spread of the virus. This suggests that EVs released from HCMV-infected cells are important for enhancing HCMV spread.
While modulation of EV release is an intriguing finding, we sought to define the consequences of this regulation during HCMV infection. With this study, we now show that blocking ESCRT-independent EV production with the nSMase inhibitor GW4869 slows HCMV spread. The ESCRTs and ceramide production are thought to represent independent pathways of EV biogenesis, and in some cases, these pathways may even generate vesicles with distinct cargo (34, 55, 56). Since little is known about the molecules packaged into EVs released from HCMV-infected cells, it is unclear whether both pathways are generating unique EVs that contribute to the spread phenotype through mechanisms that are independent of each other. However, other virus infections are known to result in similar manipulations of both pathways (10). Future studies, including proteomic analysis of EVs under different treatment and infection conditions, will be required to understand whether vesicles released through these pathways are functionally unique.
Our investigation of proteins associated with EVs uncovered significant regulation of EV biogenesis machinery. This regulation results in HCMV-infected cells releasing 3-fold more vesicles than uninfected cells. Interestingly, EVs released from HCMV-infected cells also had an enrichment of smaller vesicles within the purified population, and the significance of this finding is unknown. Of the diverse population of EVs released from cells, exosomes are characterized by 50- to 150-nm-sized vesicles generated from endosomal membrane (37). The ESCRT proteins contribute to the formation of exosomes, and the observed increase in ESCRT protein levels during infection could be responsible for the shift in size toward the exosome population (28, 57). However, further investigation is needed to explain this difference and to understand the significance of this finding. Together, the extensive regulation of the EV biogenesis pathway results in an increase in the overall number and a shift in size distribution in EVs released from HCMV-infected cells.
In addition to modifying the size and number of EVs, HCMV also modifies EV content. Several viral proteins are incorporated into EVs, and these proteins interestingly all contain potential late domain sequences (22, 23). We used this information to direct our attempts to identify new viral proteins within these vesicles. Using this approach, we identified that the viral protein pp71 is also present in EVs. While we used potential late domain sequences to identify new viral proteins found in the EVs, it is important to note that none of the sequences have been shown to interact with ESCRT proteins. Experimental evidence is required to fully understand the role of these sequences and their link to the ESCRT machinery. The role of pp71 during the early stages of infection initiation is well documented (44–46). In fact, transfection of plasmids encoding pp71 into cells makes HCMV DNA more infectious (44). The transfer of pp71 to uninfected cells could certainly contribute to enhanced virus spread, but we do not expect that a single protein is responsible for the observed spread phenotype. We hypothesize that a combination of incoming signals from viral, and potentially host, factors is necessary to generate a proviral environment within uninfected cells.
A key component of EVs that is not addressed in this study is the impact of miRNAs on virus spread. EBV, KSHV, and HSV-1 all package viral miRNAs into EVs (16, 18, 19, 53). Within a variety of cell systems, it has been shown that miRNA incorporation is a selective process mediated by RNA binding proteins that bind to specific miRNA sequences to facilitate packaging into EVs (58–60). Two of these proteins, heterogeneous nuclear ribonuclear protein A2B1 (hnRNPA2B1) and synaptotagmin binding cytoplasmic RNA interaction protein (SYNCRIP also known as hnRNPQ), have well-described roles involving RNA transport and even antiviral defense during infection with other DNA viruses, but our understanding of their role during HCMV infection is limited (61–63). hnRNPA2B1 and SYNCRIP were recently shown to interact with the HCMV immediate early 2 IE86 protein (64). Interestingly, miRNAs harbored by HCMV contain potential binding sequences for these RNA binding proteins, and these miRNAs target pathways involving immune signaling, apoptosis, and the cell cycle (65–72). If HCMV miRNAs were incorporated into EVs, then this would add further regulation of key cellular pathways that could facilitate more efficient spread of the virus. In support of this possibility, a recent report identified several HCMV miRNAs in EVs isolated from patient samples, and the abundance of HCMV miRNAs in EVs correlated with the severity of certain pathologies associated with HCMV infection (73). A comprehensive analysis of the components found within the EVs is needed to fully understand their effects on recipient cells.
We found that EVs released from HCMV-infected cells are important for virus spread. However, it does not rule out the possibility that EVs are important during other stages of HCMV infection. Within a host, HCMV infects a variety of cells types and can undergo lytic and latent stages of viral infection. Additionally, HCMV must subvert the immune response during infection of the host, an important determinant of the severity of infection that is not present during our experiments. Thus, the enhancement in spread we observe in tissue culture may be amplified in the complex environment of the host. The content and function of EVs may also vary within a host depending on the cell type infected. It is known that infection of different cell types with the same virus can produce EVs that are functionally unique (10, 11). This study provides a system to begin to understand regulation of the EV pathway, but it will be important to understand the functional importance of EVs during infection of different cell types and during latent infection.
MATERIALS AND METHODS
Cell culture and viruses.
Primary human dermal fibroblasts (HDFs) (106-05n; Cell Applications Inc.) were used for all assays. Human lung fibroblast MRC-5 cells (ATCC CCL-171) were used to generate virus stocks as previously described (25, 74), and HEK-293TN cells (provided by David Spector) were used for lentivirus production. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS; HyClone), 2 mM GlutaMAX (Gibco), 100 U/ml penicillin, and 100 g/ml streptomycin (Corning). Culture conditions were 37°C with 5% CO2. Wild-type TB40/E or the IE-2A-GFP derivative were used for all experiments (36).
HCMV infections.
HCMV was incubated with HDFs for 3 h at 37°C. Cells were washed twice in Dulbecco’s phosphate-buffered saline (DPBS) without calcium or magnesium to remove unbound virus before replacing with culture medium. For virus titers, cells and media were harvested separately to collect intracellular and extracellular virus. Cells were scraped into culture media, sonicated, vortexed, and centrifuged at 4°C at 13,000 rpm for 10 min. Virus was flash-frozen in liquid nitrogen and stored at −80°C. Titers were calculated by diluting samples on MRC-5 cells and detecting HCMV immediate early proteins (clone D4 generated by Neil Christensen [75]) by immunofluorescence. TB40/E IE-2A-GFP was used for virus spread experiments. Unfixed HDFs in 24-well plates were imaged at the designated time points to calculate the total area of GFP-positive pixels within each image. Spread values are relative to 15-dpi control samples. For EV transfer experiments, approximately 3,000 to 5,000 EVs per cell were added at 3, 4, and 5 dpi. Images were acquired on a C2+ confocal microscope system (Nikon) and analyzed using Nikon Imaging System Advanced Research Analysis software.
Quantitative real-time PCR analysis.
RNA was isolated from HDFs infected at an MOI of 3 at designated time points using a Qiagen RNeasy minikit, and cDNA was generated using an Invitrogen SuperScript First Strand RT-PCR kit with oligo(dT) primers according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using Fast Start Universal SYBR green Master (Roche) and StepOnePlus cycler (Applied Biosystems). Samples were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and fold change was calculated compared to uninfected samples. The primers used were as follows: CHMP6-FOR, 5′-TTGGAAATGAGTGTCTGAACAAG; CHMP6-REV, 5′-CTGCGTCTCGTCCAGGAT; CHMP4B-FOR, 5′-TCCCTCTATAGCCCTACCATCA; CHMP4B-REV, 5′-TTCTCCAATTCCTTCATGTCG; CHMP2B-FOR, 5′-AAAGTGATGAATTCCCAAATGAA; CHMP2B-REV, 5′-TCTTGTTAACTGCCTGCATTGT; CD63-FOR; 5′-ACCACACTGCTTCGATCCTG; CD63-REV, 5′-TCTCCACACAGCCCTCCTTA; CD81-FOR, 5′-CCTTCCACGAGACGCTTGAC; CD81-REV, 5′-CGATCTTCTGGTGGCAGTCC; GAPDH-FOR, 5′-ACCCACTCCTCCACCTTTGAC-3′; GAPDH-REV, 5′-CTGTTGCTGTAGCCAAATTCGT.
GW4869 experiments.
HDFs were seeded at subconfluence (30% confluence) and treated for 48 h with 8 μM GW4869 (Cayman Chemical) or DMSO (Fisher) prior to infection. GW4869 or DMSO was not present during adsorption but was added back at 3 hpi. Treatment groups were spiked with additional 2.5 μM GW4869 or DMSO at 2 dpi for high-MOI experiments and at 2, 4, and 6 dpi for low-MOI experiments.
EV isolation.
For EV isolation, cells were cultured in DMEM supplemented with 10% FBS that was centrifuged at 130,000 × g at 4°C for 16 h and subsequently filtered through a 0.22-μm filter to deplete the EVs. HDFs were seeded into eight T175 culture flasks and allowed to grow to confluence for 72 h. This medium was then collected and used to isolate uninfected EVs. Confluent flasks (approximately 8.0 × 107 total HDFs) were then infected at an approximate MOI of 1, and infected cells were incubated for 72 h before collecting the medium. Medium was collected and filtered through a 0.45-μm filter, and supernatant was subsequently centrifuged at 300 × g at 4°C for 10 min, 1,200 × g at 4°C for 10 min, and 10,000 × g at 4°C for 20 min. The pellet from the 10,000 × g step was resuspended in phosphate-buffered saline (PBS) and saved as the 10K pellet. Supernatant was transferred to ultracentrifuge tubes and centrifuged at 130,000 × g at 4°C for 2 h in an SW32Ti rotor. The supernatant was discarded, and the pellet was resuspended in 1 ml of 5% OptiPrep (Sigma-Aldrich). The resuspended pellet was loaded on top of a stepwise OptiPrep gradient with 10% (1 ml), 15% (1 ml), 20% (1 ml), 30% (500 μl), and 41% (500 μl) steps and centrifuged in an SW55Ti rotor at 130,000 × g at 4°C for 16 h with no brake. Fractions were collected and brought to a final volume of 5 ml in DPBS. Diluted fractions were centrifuged in an SW55Ti rotor at 130,000 × g at 4°C for 2 h. Pellets were resuspended in DPBS and stored at −80°C.
Nanoparticle tracking analysis.
For each EV harvest, fractions positive for EV markers (CD63 and CD81) and negative for infectious virus were combined in equal volumes prior to NTA analysis. Samples were diluted to 1 ml with particle-free water. Each sample was loaded by syringe pump into the NanoSight NS300 (Malvern Instruments Ltd., Malvern, Worcestershire, UK) set in scatter mode, and five 60-s videos were captured. The flow cell was rinsed by 1 ml particle-free water 3 times between samples. The capture settings used were a screen gain of 1 and camera level of 14. The process settings used were a screen gain of 10 and detection threshold of 5. The size distribution and concentration of particles were calculated and exported using software NTA3.2 (Malvern Instruments Ltd.). To generate a normalized size distribution between infected and uninfected EVs, the sum of vesicles within the defined diameter range were divided by the total number of vesicles with a diameter of 0 to 300 nm to get a relative proportion for that range. These proportions were then plotted with a y axis from 0 to 1.
Electron microscopy.
Equal numbers of uninfected or HCMV-infected EVs were transferred to 1-nm carbon film with a 10-nm Formvar-coated grid (Electron Microscopy Sciences) and incubated with 2% uranyl acetate (Electron Microscopy Sciences) for 1 min. Images were acquired using a JEOL JEM-1400 Digital Capture transmission electron microscope.
Lentivirus production and transductions.
Lentivirus was produced in HEK-293TN cells as previously described (25). pLKO.1 scrambled shRNA was a gift from David Sabatini (Addgene plasmid no. 1864) (76). The shRNA constructs used for CD63 knockdown (pLKO.1 TRCN0000007849, TRCN0000007851, TRCN0000007852, and TRCN0000007853) were obtained from the TRC 1.0 shRNA library at Penn State College of Medicine (77). Subconfluent HDFs (30% confluence) were transduced with lentiviral vectors in DMEM containing 8 μg/ml Polybrene (Sigma-Aldrich) for 6 h at 37°C. HDFs were passaged under selection with 2 μg/ml puromycin (Thermo Fisher Scientific) before seeding for experiments.
Western blotting.
Cell lysates were harvested at the designated time points in radioimmunoprecipitation assay (RIPA) buffer supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, 0.2 mM Na3VO4, and 1 μg/ml leupeptin. For extracellular levels of CD63 and CD81 during infection, medium was collected and centrifuged at 300 × g at 4°C for 10 min. Supernatant was centrifuged at 130,000 × g at 4°C for 2 h, and the pellet was resuspended in DPBS. For Western blot analysis of EVs, aliquots were mixed 1:1 with urea lysis buffer containing 120 mM Tris-HCl (pH 6.8), 5% SDS, 8 M urea, 10 mM EDTA, and 20% glycerol. SDS-PAGE gels were transferred to polyvinylidene difluoride (PVDF) or nitrocellulose membranes and blocked with 5% nonfat dry milk. The antibodies used were against the following: CHMP6 (Proteintech), CHMP4B (Proteintech), CHMP2B (Proteintech), CD63 (TS63 [Arigo Biolaboratories] or H5C6 [Developmental Studies Hybridoma Bank]), CD81 (B-11; Santa Cruz Biotechnology), α-tubulin (DM1A; Sigma-Aldrich), HCMV pp65 (7B4; a gift from David Spector [78]), Tsg101 (Fisher Scientific), HCMV glycoprotein B (2F12; Virusys), HCMV pp71 (2H10-9; a gift from John Purdy [79]), HCMV UL71 (1G; monoclonal antibody generated by Neil Christensen [36]), HCMV IE1/2 (a gift from Jim Alwine [80]), and actin (C4; Sigma-Aldrich).
Statistical analysis.
For all statistical analyses, significance was set at a P value of less than 0.05. Data were compared using a Student's t test when comparing two samples and one-way analysis of variance (ANOVA) with Dunnett’s multiple-comparison test for data sets with more than two experimental groups. One-way ANOVA and Dunnett’s tests were performed using GraphPad Prism version 8.4.
Data availability.
We have submitted all relevant data from our experiments to the EV-TRACK knowledgebase (EV-TRACK identifier [ID] EV200015) (81).
ACKNOWLEDGMENTS
We thank Neil Christensen for the generation of monoclonal antibodies to the viral proteins UL71 and IE1/2.
This work was supported by NIH grant R01AI071286 (to N.J.B.) and NIH training grant T32CA060396 (to N.T.S.).
REFERENCES
- 1.Wreghitt TG, Abel SJ, McNeil K, Parameshwar J, Stewart S, Cary N, Sharples L, Large S, Wallwork J. 1999. Intravenous ganciclovir prophylaxis for cytomegalovirus in heart, heart-lung, and lung transplant recipients. Transpl Int 12:254–260. doi: 10.1007/s001470050219. [DOI] [PubMed] [Google Scholar]
- 2.Crocchiolo R, Bramanti S, Vai A, Sarina B, Mineri R, Casari E, Tordato F, Mauro E, Timofeeva I, Lugli E, Mavilio D, Carlo-Stella C, Santoro A, Castagna L. 2015. Infections after T-replete haploidentical transplantation and high-dose cyclophosphamide as graft-versus-host disease prophylaxis. Transpl Infect Dis 17:242–249. doi: 10.1111/tid.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bond MMK, Bond MMK, Sehn A, Dias VH, Said TL, Dos Santos CC, Finger MA, Santos AMG, Neto JMR. 2018. Cyclosporine versus tacrolimus: which calcineurin inhibitor has influence on cytomegalovirus infection in cardiac transplantation? Transplant Proc 50:809–814. doi: 10.1016/j.transproceed.2018.02.046. [DOI] [PubMed] [Google Scholar]
- 4.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci U S A 100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, Mann M, Ingolia NT, Weissman JS. 2012. Decoding human cytomegalovirus. Science 338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeltzer S, Zeltzer CA, Igarashi S, Wilson J, Donaldson JG, Goodrum F. 2018. Virus control of trafficking from sorting endosomes. mBio 9:e00683-18. doi: 10.1128/mBio.00683-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. doi: 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dreux M, Garaigorta U, Boyd B, Décembre E, Chung J, Whitten-Bauer C, Wieland S, Chisari FV. 2012. Short range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe 12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin Raj V, Jenster G, Kwekkeboom J, Tilanus HW, Haagmans BL, Baumert TF, van der Laan LJ. 2013. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A 110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Zhang L, Zhang F, Tang T, Zhou Q, Feng C, Jin Y, Wu Z. 2017. Exosome-mediated miR-146a transfer suppresses type I interferon response and facilitates EV71 infection. PLoS Pathog 13:e1006611. doi: 10.1371/journal.ppat.1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris-Love J, Gee GV, O’Hara BA, Assetta B, Atkinson AL, Dugan AS, Haley SA, Atwood WJ. 2019. JC polyomavirus uses extracellular vesicles to infect target cells. mBio 10:e00379-19. doi: 10.1128/mBio.00379-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang K, Xu S, Shi X, Xu G, Shen C, Liu X, Zheng H. 2019. Exosomes-mediated transmission of foot-and-mouth disease virus in vivo and in vitro. Vet Microbiol 233:164–173. doi: 10.1016/j.vetmic.2019.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Deschamps T, Kalamvoki M. 2018. Extracellular vesicles released by herpes simplex virus 1-infected cells block virus replication in recipient cells in a STING-dependent manner. J Virol 92:e01102-18. doi: 10.1128/JVI.01102-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalamvoki M, Du T, Roizman B. 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci U S A 111:E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dogrammatzis C, Deschamps T, Kalamvoki M. 2018. Biogenesis of extracellular vesicles during herpes simplex virus 1 infection: role of the CD63 tetraspanin. J Virol 93:e01850-18. doi: 10.1128/JVI.01850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meckes DG Jr, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. 2010. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A 107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chugh PE, Sin SH, Ozgur S, Henry DH, Menezes P, Griffith J, Eron JJ, Damania B, Dittmer DP. 2013. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog 9:e1003484. doi: 10.1371/journal.ppat.1003484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNamara RP, Chugh PE, Bailey A, Costantini LM, Ma Z, Bigi R, Cheves A, Eason AB, Landis JT, Host KM, Xiong J, Griffith JD, Damania B, Dittmer DP. 2019. Extracellular vesicles from Kaposi sarcoma-associated herpesvirus lymphoma induce long-term endothelial cell reprogramming. PLoS Pathog 15:e1007536. doi: 10.1371/journal.ppat.1007536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nanbo A, Ohashi M, Yoshiyama H, Ohba Y. 2018. The role of transforming growth factor beta in cell-to-cell contact-mediated Epstein-Barr virus transmission. Front Microbiol 9:984. doi: 10.3389/fmicb.2018.00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker JD, Maier CL, Pober JS. 2009. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J Immunol 182:1548–1559. doi: 10.4049/jimmunol.182.3.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zicari S, Arakelyan A, Palomino RAN, Fitzgerald W, Vanpouille C, Lebedeva A, Schmitt A, Bomsel M, Britt W, Margolis L. 2018. Human cytomegalovirus-infected cells release extracellular vesicles that carry viral surface proteins. Virology 524:97–105. doi: 10.1016/j.virol.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tandon R, AuCoin DP, Mocarski ES. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J Virol 83:10797–10807. doi: 10.1128/JVI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streck NT, Carmichael J, Buchkovich NJ. 2018. Nonenvelopment role for the ESCRT-III complex during human cytomegalovirus infection. J Virol 92:e02096-17. doi: 10.1128/JVI.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Streck N. 2020. Human cytomegalovirus utilizes ESCRT-III and extracellular vesicles to enhance virus spread. PhD dissertation. Pennsylvania State University, Hershey, PA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henne WM, Buchkovich NJ, Emr SD. 2011. The ESCRT pathway. Dev Cell 21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 28.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. 2013. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci 126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 29.Romancino DP, Paterniti G, Campos Y, De Luca A, Di Felice V, d'Azzo A, Bongiovanni A. 2013. Identification and characterization of the nano-sized vesicles released by muscle cells. FEBS Lett 587:1379–1384. doi: 10.1016/j.febslet.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coulter ME, Dorobantu CM, Lodewijk GA, Delalande F, Cianferani S, Ganesh VS, Smith RS, Lim ET, Xu CS, Pang S, Wong ET, Lidov HGW, Calicchio ML, Yang E, Gonzalez DM, Schlaeger TM, Mochida GH, Hess H, Lee WA, Lehtinen MK, Kirchhausen T, Haussler D, Jacobs FMJ, Gaudin R, Walsh CA. 2018. The ESCRT-III protein CHMP1A mediates secretion of sonic hedgehog on a distinctive subtype of extracellular vesicles. Cell Rep 24:973.e8–986.e8. doi: 10.1016/j.celrep.2018.06.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurwitz SN, Conlon MM, Rider MA, Brownstein NC, Meckes DG Jr. 2016. Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. J Extracell Vesicles 5:31295. doi: 10.3402/jev.v5.31295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du WL, Mutsafi Y, De Jesus-Diaz DA, Sosnovtsev SV, Levenson EA, Parra GI, Takvorian PM, Cali A, Bleck C, Vlasova AN, Saif LJ, Patton JT, Lopalco P, Corcelli A, Green KY, Altan-Bonnet N. 2018. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 24:208.e8–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Woodson M, Sherman MB, Neelakanta G, Sultana H. 2019. Exosomes mediate Zika virus transmission through SMPD3 neutral sphingomyelinase in cortical neurons. Emerg Microbes Infect 8:307–326. doi: 10.1080/22221751.2019.1578188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. 2008. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 35.Fraile-Ramos A, Cepeda V, Elstak E, van der Sluijs P. 2010. Rab27a is required for human cytomegalovirus assembly. PLoS One 5:e15318. doi: 10.1371/journal.pone.0015318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandhu P, Buchkovich NJ. 2020. HCMV decreases MHC class II by regulating CIITA transcript levels in a myeloid cell line. J Virol 94:e01901-19. doi: 10.1128/JVI.01901-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. 2019. Reassessment of exosome composition. Cell 177:428.e18–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Wu F, Liu C, Dai W, Teng Y, Su W, Kong W, Gao F, Cai L, Hou A, Jiang C. 2019. Exosomes released from rabies virus-infected cells may be involved in the infection process. Virol Sin 34:59–65. doi: 10.1007/s12250-019-00087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 40.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol 69:5455–5460. doi: 10.1128/JVI.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol 68:6605–6618. doi: 10.1128/JVI.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banfer S, Schneider D, Dewes J, Strauss MT, Freibert SA, Heimerl T, Maier UG, Elsasser HP, Jungmann R, Jacob R. 2018. Molecular mechanism to recruit galectin-3 into multivesicular bodies for polarized exosomal secretion. Proc Natl Acad Sci U S A 115:E4396–e4405. doi: 10.1073/pnas.1718921115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraile-Ramos A, Pelchen-Matthews A, Risco C, Rejas MT, Emery VC, Hassan-Walker AF, Esteban M, Marsh M. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol 9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 44.Baldick CJ Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol 71:4400–4408. doi: 10.1128/JVI.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bresnahan WA, Shenk TE. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A 97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, Shu HB. 2017. Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microbe 21:231–243. doi: 10.1016/j.chom.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Ageta H, Ageta-Ishihara N, Hitachi K, Karayel O, Onouchi T, Yamaguchi H, Kahyo T, Hatanaka K, Ikegami K, Yoshioka Y, Nakamura K, Kosaka N, Nakatani M, Uezumi A, Ide T, Tsutsumi Y, Sugimura H, Kinoshita M, Ochiya T, Mann M, Setou M, Tsuchida K. 2018. UBL3 modification influences protein sorting to small extracellular vesicles. Nat Commun 9:3936. doi: 10.1038/s41467-018-06197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG Jr. 2017. CD63 regulates Epstein-Barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-kappaB signaling. J Virol 91:e02251-16. doi: 10.1128/JVI.02251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurwitz SN, Cheerathodi MR, Nkosi D, York SB, Meckes DG Jr. 2017. Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of Epstein-Barr virus LMP1. J Virol 92:e01969-17. doi: 10.1128/JVI.01969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rider MA, Cheerathodi MR, Hurwitz SN, Nkosi D, Howell LA, Tremblay DC, Liu X, Zhu F, Meckes DG Jr. 2018. The interactome of EBV LMP1 evaluated by proximity-based BioID approach. Virology 516:55–70. doi: 10.1016/j.virol.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto Y, Sheng X, Murray-Nerger LA, Cristea IM. 2020. Temporal dynamics of protein complex formation and dissociation during human cytomegalovirus infection. Nat Commun 11:806. doi: 10.1038/s41467-020-14586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLauchlan J, Addison C, Craigie MC, Rixon FJ. 1992. Noninfectious L-particles supply functions which can facilitate infection by HSV-1. Virology 190:682–688. doi: 10.1016/0042-6822(92)90906-6. [DOI] [PubMed] [Google Scholar]
- 53.Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G. 2016. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A 113:E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bello-Morales R, Praena B, de la Nuez C, Rejas MT, Guerra M, Galán-Ganga M, Izquierdo M, Calvo V, Krummenacher C, López-Guerrero JA. 2018. Role of microvesicles in the spread of herpes simplex virus type 1 in oligodendrocytic cells. J Virol 92:e00088-18. doi: 10.1128/JVI.00088-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. 2010. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu A, Wawro P, Morgens DW, Portela F, Bassik MC, Pfeffer SR. 2018. Genome-wide interrogation of extracellular vesicle biology using barcoded miRNAs. Elife 7:e41460. doi: 10.7554/eLife.41460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. 2016. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. 2013. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. 2016. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep 17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 60.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. 2016. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 5:e19276. doi: 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoek KS, Kidd GJ, Carson JH, Smith R. 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry 37:7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 62.Mizutani A, Fukuda M, Ibata K, Shiraishi Y, Mikoshiba K. 2000. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J Biol Chem 275:9823–9831. doi: 10.1074/jbc.275.13.9823. [DOI] [PubMed] [Google Scholar]
- 63.Wang L, Wen M, Cao X. 2019. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 365:eaav0758. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- 64.Liang S, Qian D, Zhao R, Yu B, Hu M, Wang B. 2019. Human cytomegalovirus ie2 affects the migration of glioblastoma by mediating the different splicing patterns of RON through hnRNP A2B1. Neuroreport 30:805–811. doi: 10.1097/WNR.0000000000001277. [DOI] [PubMed] [Google Scholar]
- 65.Lee SH, Kalejta RF, Kerry J, Semmes OJ, O'Connor CM, Khan Z, Garcia BA, Shenk T, Murphy E. 2012. BclAF1 restriction factor is neutralized by proteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc Natl Acad Sci U S A 109:9575–9580. doi: 10.1073/pnas.1207496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim S, Seo D, Kim D, Hong Y, Chang H, Baek D, Kim VN, Lee S, Ahn K. 2015. Temporal landscape of microRNA-mediated host-virus crosstalk during productive human cytomegalovirus infection. Cell Host Microbe 17:838–851. doi: 10.1016/j.chom.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 67.Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. 2015. Human cytomegalovirus miR-UL112-3p targets TLR2 and modulates the TLR2/IRAK1/NFκB signaling pathway. PLoS Pathog 11:e1004881. doi: 10.1371/journal.ppat.1004881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hancock MH, Hook LM, Mitchell J, Nelson JA. 2017. Human cytomegalovirus microRNAs miR-US5-1 and miR-UL112-3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ. mBio 8:e00109-17. doi: 10.1128/mBio.00109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA. 2010. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog 6:e1000967. doi: 10.1371/journal.ppat.1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang YP, Qi Y, Huang YJ, Qi ML, Ma YP, He R, Ji YH, Sun ZR, Ruan Q. 2013. Identification of immediate early gene X-1 as a cellular target gene of hcmv-mir-UL148D. Int J Mol Med 31:959–966. doi: 10.3892/ijmm.2013.1271. [DOI] [PubMed] [Google Scholar]
- 71.Lau B, Poole E, Krishna B, Sellart I, Wills MR, Murphy E, Sinclair J. 2016. The expression of human cytomegalovirus microRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci Rep 6:31205. doi: 10.1038/srep31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K. 2012. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog 8:e1002577. doi: 10.1371/journal.ppat.1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang J, Huang Y, Wang Q, Ma Y, Qi Y, Liu Z, Deng J, Ruan Q. 2019. Levels of human cytomegalovirus miR-US25-1-5p and miR-UL112-3p in serum extracellular vesicles from infants with HCMV active infection are significantly correlated with liver damage. Eur J Clin Microbiol Infect Dis 39:471–481. doi: 10.1007/s10096-019-03747-0. [DOI] [PubMed] [Google Scholar]
- 74.Spector DJ, Yetming K. 2010. UL84-independent replication of human cytomegalovirus strain TB40/E. Virology 407:171–177. doi: 10.1016/j.virol.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 75.Desai D, Lauver M, Ostman A, Cruz L, Ferguson K, Jin G, Roper B, Brosius D, Lukacher A, Amin S, Buchkovich N. 2019. Inhibition of diverse opportunistic viruses by structurally optimized retrograde trafficking inhibitors. Bioorg Med Chem 27:1795–1803. doi: 10.1016/j.bmc.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 77.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 78.Kim KS, Sapienza VJ, Chen CM, Wisniewski K. 1983. Production and characterization of monoclonal antibodies specific for a glycosylated polypeptide of human cytomegalovirus. J Clin Microbiol 18:331–343. doi: 10.1128/JCM.18.2.331-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalejta RF, Bechtel JT, Shenk T. 2003. Human cytomegalovirus pp71 stimulates cell cycle progression by inducing the proteasome-dependent degradation of the retinoblastoma family of tumor suppressors. Mol Cell Biol 23:1885–1895. doi: 10.1128/mcb.23.6.1885-1895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harel NY, Alwine JC. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol 72:5481–5492. doi: 10.1128/JVI.72.7.5481-5492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.EV-TRACK Consortium, Van Deun J, Mestdagh P, Agostinis P, Akay Ö, Anand S, Anckaert J, Martinez ZA, Baetens T, Beghein E, Bertier L, Berx G, Boere J, Boukouris S, Bremer M, Buschmann D, Byrd JB, Casert C, Cheng L, Cmoch A, Daveloose D, De Smedt E, Demirsoy S, Depoorter V, Dhondt B, Driedonks TAP, Dudek A, Elsharawy A, Floris I, Foers AD, Gärtner K, Garg AD, Geeurickx E, Gettemans J, Ghazavi F, Giebel B, Kormelink TG, Hancock G, Helsmoortel H, Hill AF, Hyenne V, Kalra H, Kim D, Kowal J, Kraemer S, Leidinger P, Leonelli C, Liang Y, Lippens L, Liu S, et al. 2017. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods 14:228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We have submitted all relevant data from our experiments to the EV-TRACK knowledgebase (EV-TRACK identifier [ID] EV200015) (81).