Significance
Molecular studies of plant immune responses have mainly focused on qualitative resistance, a form of immunity determined by a few large effect genes. In contrast, very limited information exists about quantitative disease resistance (QDR), although it is extensively observed in wild and crop species. We used systems biology approaches to describe this form of immunity in Arabidopsis thaliana. On the basis of gene regulation studies and search for protein–protein interactions, we report the reconstruction of a highly interconnected and distributed network, organized in five modules with differential robustness to genetic mutations. These studies revealed key functions of QDR, mainly distinct from those previously identified for plant immunity, and shed some light on the complexity of this plant immune response.
Keywords: plant pathogen interactions, immunity, quantitative disease resistance, systems biology, regulatory networks
Abstract
Quantitative disease resistance (QDR) represents the predominant form of resistance in natural populations and crops. Surprisingly, very limited information exists on the biomolecular network of the signaling machineries underlying this form of plant immunity. This lack of information may result from its complex and quantitative nature. Here, we used an integrative approach including genomics, network reconstruction, and mutational analysis to identify and validate molecular networks that control QDR in Arabidopsis thaliana in response to the bacterial pathogen Xanthomonas campestris. To tackle this challenge, we first performed a transcriptomic analysis focused on the early stages of infection and using transgenic lines deregulated for the expression of RKS1, a gene underlying a QTL conferring quantitative and broad-spectrum resistance to X. campestris. RKS1-dependent gene expression was shown to involve multiple cellular activities (signaling, transport, and metabolism processes), mainly distinct from effector-triggered immunity (ETI) and pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) responses already characterized in A. thaliana. Protein–protein interaction network reconstitution then revealed a highly interconnected and distributed RKS1-dependent network, organized in five gene modules. Finally, knockout mutants for 41 genes belonging to the different functional modules of the network revealed that 76% of the genes and all gene modules participate partially in RKS1-mediated resistance. However, these functional modules exhibit differential robustness to genetic mutations, indicating that, within the decentralized structure of the QDR network, some modules are more resilient than others. In conclusion, our work sheds light on the complexity of QDR and provides comprehensive understanding of a QDR immune network.
In nature, plants are continuously exposed to various pathogenic microbes and have evolved multiple layers of immune responses in order to protect themselves. A large body of data has been produced to unravel the molecular mechanisms underlying qualitative resistance that is determined by a few genes conferring an almost complete resistance such as nucleotide-binding domain leucine-rich repeat (NLR) receptors. In contrast, quantitative disease resistance (QDR) remains poorly understood (1–4) despite the fact that it represents the predominant form of resistance in natural populations and crops (5). The complexity of the genetic architecture underlying natural variation of QDR, which involves many QTLs with minor to moderate effects, has limited the analysis of the underlying molecular mechanisms. However, several genes with various functions have been cloned in recent years (6). For example, an ABC (adenosine triphosphate [ATP]-binding cassette) transporter encoded by the gene Lr34 was identified and confers resistance against multiple fungi in wheat (7). A caffeoyl-CoA O-methyltransferase associated with lignin production is shown to confer QDR to diverse maize necrotrophic diseases (8). Interestingly, kinases have been shown in several cases to play key roles in QDR. One of the best studied cases is the maize wall-associated kinase, ZmWAK1, conferring QDR to northern corn leaf blight, and for which the molecular function has been associated with the biosynthesis of secondary metabolites (9, 10). In some cases, NLR genes can also confer QDR (1, 11). Thus, the molecular functions underlying QDR seem to be more diverse than those responsible for qualitative resistance. However, for these few identified genes, the identity of the downstream components of QDR and the corresponding gene networks remain largely unknown.
QDR is determined by multiple QTLs of minor to major effects and can be hypothetically defined as a complex network integrating diverse pathways in response to multiple pathogenic determinants (2). As other responses to pathogen attack, QDR may require the coordinated and early expression of plant defense mechanisms including several defense-associated metabolites, proteins, or genes potentially involved in other plant immunity pathways. While many transcriptomic approaches were used to characterize other forms of immunity (12), to date, comparative transcriptome analyses of plant–pathogen interactions showing QDR remain scarce. In some cases, differential gene expression was studied in near-isogenic lines (NILs), or susceptible versus tolerant lines (13, 14). Transcriptomic analysis of NILs differing for ZmWAK-RLK1 (10) showed a close association of WAK-mediated QDR with changes in expression of genes involved in the biosynthesis of benzoxazinoids, thereby revealing a new role for this metabolism in immunity to fungal pathogens. This demonstrates that transcriptome approaches on focused QDR genes can help to understand some components of the complex responses associated with this form of resistance. However, one major limitation is that an individual analysis of an identified pathway might lead to the possible underestimation of multiple and coordinated pathways acting together to mount the QDR response.
In this context, genome-wide modular network analysis has been recently used to decipher such biological complexity and first applied to the plant immune responses ETI (effector-triggered immunity) and PTI (PAMP-triggered immunity) (12, 15–17). Comparison at different scales indicated a densely connected network shared between ETI and PTI (12). Important interactions between these two immune responses were identified, one signaling subnetwork mediating the hypersensitive response being inhibited by PTI signaling (18). QDR, considered as the result of an interplay between multiple pathogen effectors, toxins, or other metabolites and diverse plant targets and defense responses, presents a high complexity and therefore prompted us to use systems biology approaches (19). Indeed, these approaches allow the study of network properties referred to as “emergent property” of complex biological networks by combining molecular network modeling and experimental investigation. Among them, robustness is a key property “that allows a system to maintain its functions against internal and external perturbations” and involved in immune systems and biological network evolution (20). In addition, QDR genes confer broad-spectrum resistance and have been associated with durable resistance in many cases (4). Consequently, the QDR plant immune system should have evolved to acquire a particularly high robustness and tunability. These properties were proposed for PTI to occur through cell surface receptor complexes (21) and multiple paths of signaling networks (17). The extent to which these properties are associated with the QDR immune network, together with the identification of the network components, remain open questions.
The bacterial vascular pathogen Xanthomonas campestris (Xc) causes the black rot disease, possibly the most important disease of crucifers, and is one of the most prevalent bacterial pathogens in natural populations of A. thaliana (22–24). We identified by map-based cloning and nested genome-wide association mapping the Arabidopsis gene RKS1, which confers quantitative and broad-spectrum resistance to Xc (25). RKS1 encodes an atypical kinase, for which no kinase activity has been detected so far. Interestingly, atypical kinases (or pseudokinases) are known to be regulators of signaling networks (26), acting as molecular scaffolds for assembly of protein complexes or modulation of the activity of a catalytically active enzyme (27). RKS1 constitutes a starting point for exploration of the signaling pathways leading to QDR to Xcc in Arabidopsis thaliana. Recently, RKS1 has also been shown to be a component of an NLR resistosome (28, 29). These results suggest that RKS1 might be part of a signaling platform in response to recognition of various pathogen determinants and a key element to coordinate gene expression of the plant immune system. Here we show, by a combination of omics, network reconstruction, and mutational approaches, that the RKS1-dependent gene network reveals unexpected and multiple regulatory modules controlling QDR to Xc in A. thaliana, and exhibiting differential robustness.
Results
Transcriptome Analysis Reveals that RKS1 Controls Multiple Cellular Activities.
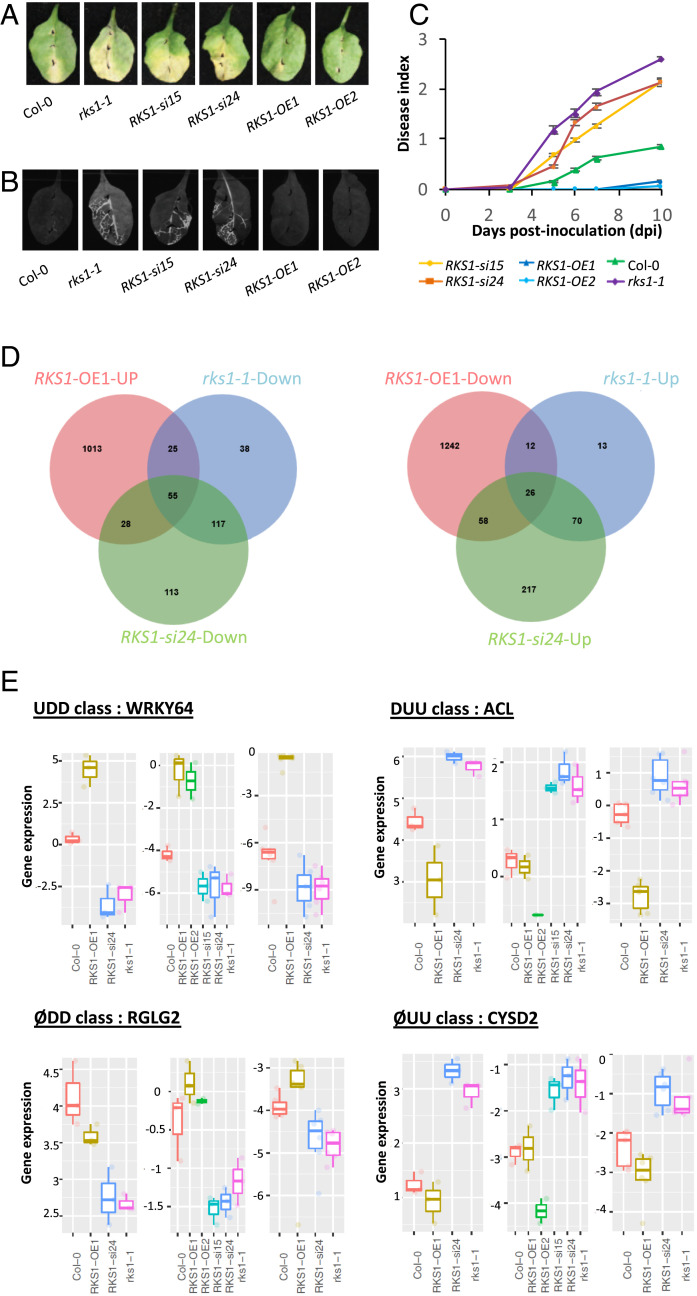
To investigate QDR regulatory pathways and potential cell reprogramming specifically controlled by RKS1, a transcriptomic analysis was performed using RKS1 deregulated lines expressing contrasted phenotypes (Fig. 1). The rks1-1 mutant and amiRNA lines for RKS1 (RKS1-si/Col-0) were previously characterized and showed a susceptible phenotype (25). Transgenic lines overexpressing RKS1 were generated (RKS1-OE/Col-0; SI Appendix, Fig. S1), and all presented increased resistance to Xcc568, the Xc strain used for the identification of RKS1 (Fig. 1 A–C and SI Appendix, Fig. S2A). This phenotype was associated with an absence of leaf bacterial invasion as shown by using an Xcc568 strain harboring the Photorhabdus luminescens lux operon (30) and by in planta bacterial growth (Fig. 1B and SI Appendix, Fig. S2B).
Fig. 1.
Global gene expression analyses in RKS1-deregulated transgenic lines after X. campestris pv. campestris inoculation led to the identification of 268 differentially expressed genes. (A) Visual observations of the disease symptoms 10 d after inoculation with a bacterial suspension adjusted to 109 cfu/mL. (B) Luminescence imaging illustrates quantitative and spatial aspects of leaf colonization by Xcc568. Photos were taken with a CCD camera under light (exposure time 10 ms) and dark conditions (exposure time 10 s) 7 d postinoculation with the Xcc568-Lux reporter strain. From these pictures, an overlay image was generated. (C) Time course evaluation of disease index after inoculation with Xcc568 with a bacterial suspension adjusted to 109 cfu/mL. Means and SEs were calculated from 18 plants in 3 independent experiments. (D) Venn diagrams showing the distribution of genes up- or down-regulated in the different RKS1 transgenic or mutant lines, as compared to the wild type accession Col-0. (E) Quantitative RT-PCR validation of the expression profile for four specific genes, each belonging to an expression class: WRKY64 as an example of the class UDD (up-regulated in RKS1-OE1 line, down-regulated in RKS1-si24 and in rks1-1 lines), RGLG2 as ØDD class (not affected in RKS1-OE1 line, down-regulated in RKS1-si24 and in rks1-1 lines), ACL as DUU class (down-regulated in RKS1-OE1 line, up-regulated in RKS1-si24 and in rks1-1 lines), and CYSD2 as ØUU class (not affected in RKS1-OE1 line, up-regulated in RKS1-si24 and in rks1-1 lines). From left to right and for each class: expression pattern on RNA-seq datasets, qPCR datasets using initial transcriptome samples, and independent experiment. Statistical analyses for qPCR were performed with the Wilcoxon test.
For transcriptome analysis, we harvested leaves after inoculation with the bacterial strain Xcc568 and focused on early responses (0, 1.5, 3, and 6 h postinoculation [hpi]). Three independent experiments were performed, and RNA was sequenced with an average of 20.6 million reads per sample. The major driver of differential gene expression was the time (hpi/time 0, accounting for 78.3% of the total variance; SI Appendix, Fig. S3). The line/time effect accounted for 13.7% of total variance and revealed that significant differences among lines were mainly observed at 6 hpi (SI Appendix, Table S1), i.e., these effects greatly exceeded the effects from biological replicates. A total of 6,733 differentially expressed genes (DEGs) between inoculated lines and mock Col-0 samples were identified, corresponding to genes with significant line or hpi effect. We then selected the genes significantly deregulated at least for one time point in one transgenic line as compared to the wild type. Most of these 3,412 genes were found to be deregulated 6 hpi, among which 1,645 were up-regulated and 1,954 down-regulated as compared to the wild type (SI Appendix, Fig. S4). To identify DEGs specifically associated with RKS1, we focused on genes found at 6 hpi to be up- or down-regulated in the RKS1-OE line and in the opposite way in the RKS1-si24 line and the rks1-1 mutant. We identified a total of 268 DEGs (Dataset S1), which belong to four classes of gene regulation: up-regulated in the RKS1-OE1 (up) line and down-regulated in the rks1-1 mutant and RKS1-si24 lines (down-down), DEGs with the opposite profile (down-up-up), DEGs affected similarly in the rks1-1 mutant and RKS1-si24 line (down-down or up-up), and DEGs not affected in the RKS1-OE1 line (Figs. 1D and 2A and Dataset S1). To confirm patterns of gene expression after Xcc inoculation, we analyzed the expression profile of ∼10% of the DEGs randomly selected from each regulation class by RT-qPCR, using the RKS1 deregulated lines used for the transcriptomic analysis. Under these conditions, we found that about 80% of the genes showed an expression profile similar to the one obtained by RNA-seq data (SI Appendix, Fig. S5A), and a highly significant correlation was found between the RNA-seq and RT-qPCR data (Pearson’s rho = 0.86; SI Appendix, Fig. S5B). By using two additional lines (RKS1-OE2 and RKS1-si15) and another independent experiment, 56% of the genes were still validated (SI Appendix, Fig. S5A). The expression pattern of selected genes for each regulation class (Fig. 1E) indicates that the differential expression pattern of RNA-seq datasets, qPCR datasets using initial transcriptome samples, and independent experiments were consistent.
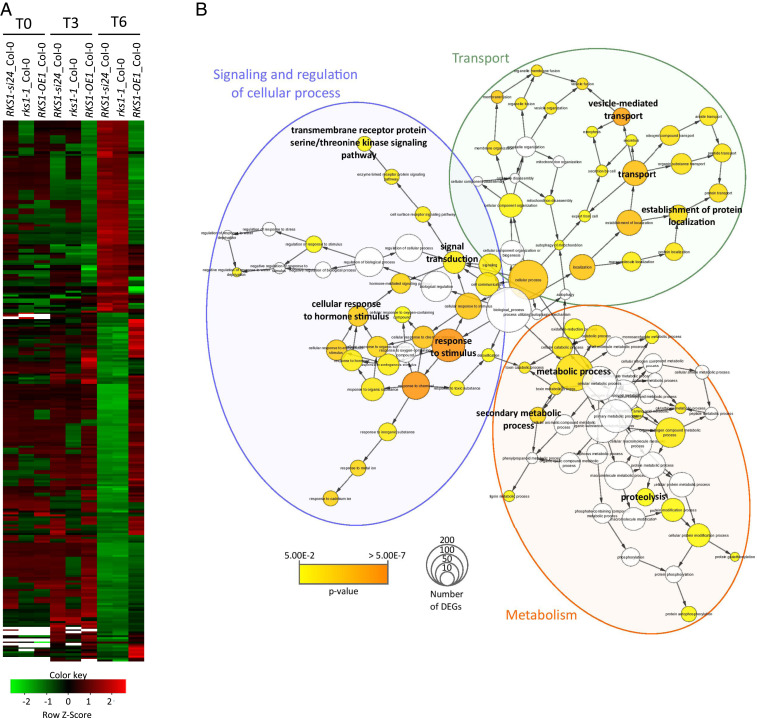
Fig. 2.
Clustering and Gene Ontology analyses of the 268 differentially expressed genes (DEGs) reveal multiple gene functional modules. (A) Clustering of the 268 DEGs whose expression is shared by the different RKS1 deregulated lines (P values < 0.05 corrected by BH statistical analysis). Each line was compared with Col-0 at 0, 3, and 6 hpi. Green indicates down-regulated values, red indicates up-regulated values, black indicates unchanged values, and white indicates missing values. (B) Gene Ontology (GO) enrichment analysis of the 268 DEGs was plotted by using BINGO module from Cytoscape software. GO process annotation was recovered for 232 of 268 DEGs. Enrichment was calculated using BINGO and plotted on GO hierarchy using Cytoscape. Circle size represents gene number in one GO, and the colors represent GO enrichment (GO white is not enriched, and color increased with P value < 0.05).
To document the nature of the transcriptional changes dependent on RKS1, a Gene Ontology (GO) enrichment analysis was performed on the 268 DEGs (Fig. 2B and SI Appendix, Fig. S6). This analysis revealed an enrichment in three main cellular activities, including cellular responses, transport, and metabolism. Functions related to transport and metabolism were strongly enriched in the class of genes whose expression was down-regulated in rks1-1 and RKS1-si24 lines. These results show that RKS1-dependent gene expression involves different cellular activities in the early stages of infection, in line with the complex nature of the QDR response. We then examined whether the identified DEGs include genes already associated with other forms of immunity (SI Appendix, Fig. S7). Interestingly, very few genes were found in common with previously identified ETI- and/or PTI-associated genes in A. thaliana during the early stages of pathogen infection (12, 18, 31): 42 genes related to ETI (15.6% of QDR DEGs, 1.8% of ETI genes) and 35 genes related to PTI (13.1% of QDR DEGs, 1.6% of PTI genes). Therefore, we conclude that RKS1 controls multiple QDR pathways that are mainly distinct from ETI and PTI responses in A. thaliana.
Protein–Protein Interaction Network Reconstitution Revealed a Highly Connected and Distributed RKS1-Dependent Network.
Transcriptomic analysis revealed that RKS1-dependent gene expression in the early stages of infection involves different cellular activities, and many of them, such as signaling pathways, protein metabolism, and trafficking, are known to involve protein–protein interaction networks. Hence, to decipher the signaling pathways downstream of RKS1 and the putative protein–protein interaction (PPI) network controlled by RKS1, we investigated the functional protein association network depending on RKS1 using three strategies. First, we assessed the subcellular localization of RKS1 to reveal where potential interactions take place. Second, we experimentally searched for proteins interacting with RKS1 by using the yeast two-hybrid (Y2H) system. Third, we reconstructed a model of PPI network by looking for known interactors of the proteins encoded by the 268 DEGs and of the RKS1 putative interactors identified in this study.
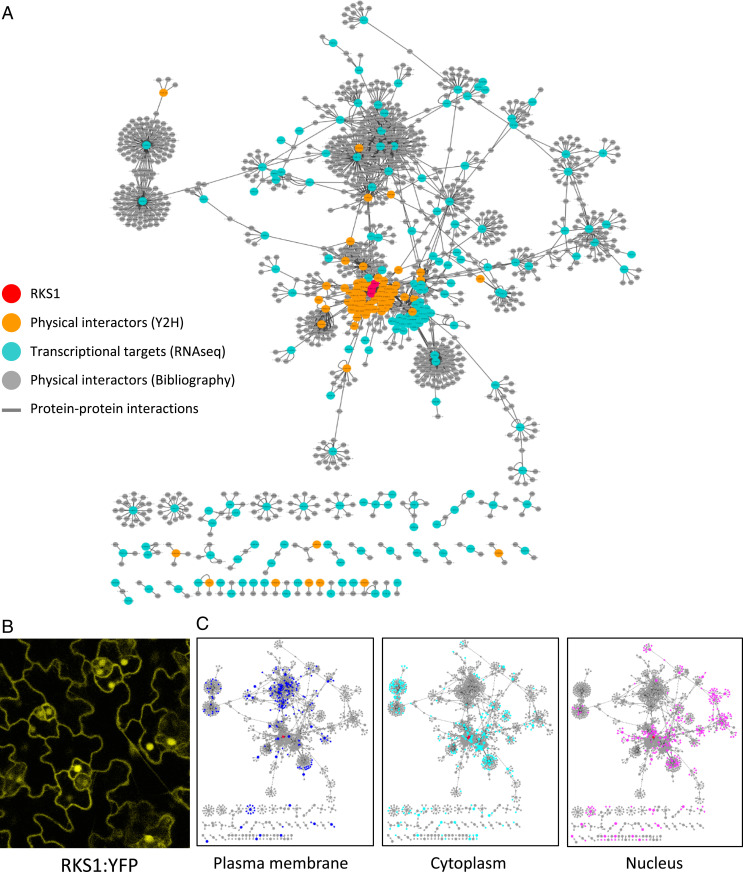
Using confocal laser-scanning microscopy, we observed leaves of overexpressing lines containing an RKS1–yellow fluorescent protein (YFP) fusion construct (Fig. 3B). The YFP signal was observed in the plasma membrane, in cytoplasmic tracks, and in the nucleus. The same observation was done after transient expression of a RKS1-GFP fusion construct in A. thaliana seedlings and confirmed by coexpression with different subcellular markers (SI Appendix, Fig. S8). Although we cannot rule out a possible side effect of the fluorescent tag, these results indicate that the RKS1 protein localization was observed in multiple compartments including the plasma membrane, the cytoplasm, and the nucleus. Moreover, a mutated version of RKS1 (RKS1D191A) was used as bait to screen a Y2H Arabidopsis cDNA library generated from mRNAs isolated from leaf tissue after bacterial inoculation (32). Forty-one prey cDNA clones were identified in two independent screening rounds (SI Appendix, Table S2). In parallel, we collected information based on Biological General Repository for Interaction Datasets (BioGRID) data, a database collecting experimental evidences for protein–protein interactions, in order to identify primary interactors of the 268 DEGs and the 41 putative interacting proteins. Each of the 2,294 interactions found were manually curated with data available in UniProt Knowledgebase (https://www.uniprot.org/) and using the published articles related to the proteins of interest (Dataset S2). The final PPI network is composed of 1,330 nodes (proteins) corresponding to 1,047 genes involved in 1,876 interactions and integrates 49% of the DEGs. A total of 906 proteins (87% of the proteins) are linked to RKS1 via protein–protein interactions (Fig. 3A). Such features indicate that RKS1 might be involved in an intricated protein–protein interaction network, a part of this network corresponding to genes whose expression is RKS1-dependent.
Fig. 3.
Protein–protein interaction network reconstitution reveals a highly interconnected and distributed RKS1-dependent network. (A) The RKS1 protein–protein interaction network plotted with Cytoscape showing the components used for its construction: RKS1 (in red), the physical interactors of RKS1 identified by yeast two-hybrid screening (in orange), the proteins corresponding to the 268 DEGs (in blue), and the proteins identified in the bibliography as experimentally interacting with the proteins corresponding to the 268 DEGs (in gray). (B) Confocal scanning microscopy observation of Arabidopsis leaves of RKS1-OE lines. RKS1 localizes to multiple subcellular compartments: the nucleus, plasma membrane, and cytoplasmic tracks. (C) Display of the subcellular localization of the network components in the compartments where RKS1 has been observed: plasma membrane, cytoplasm, and nucleus.
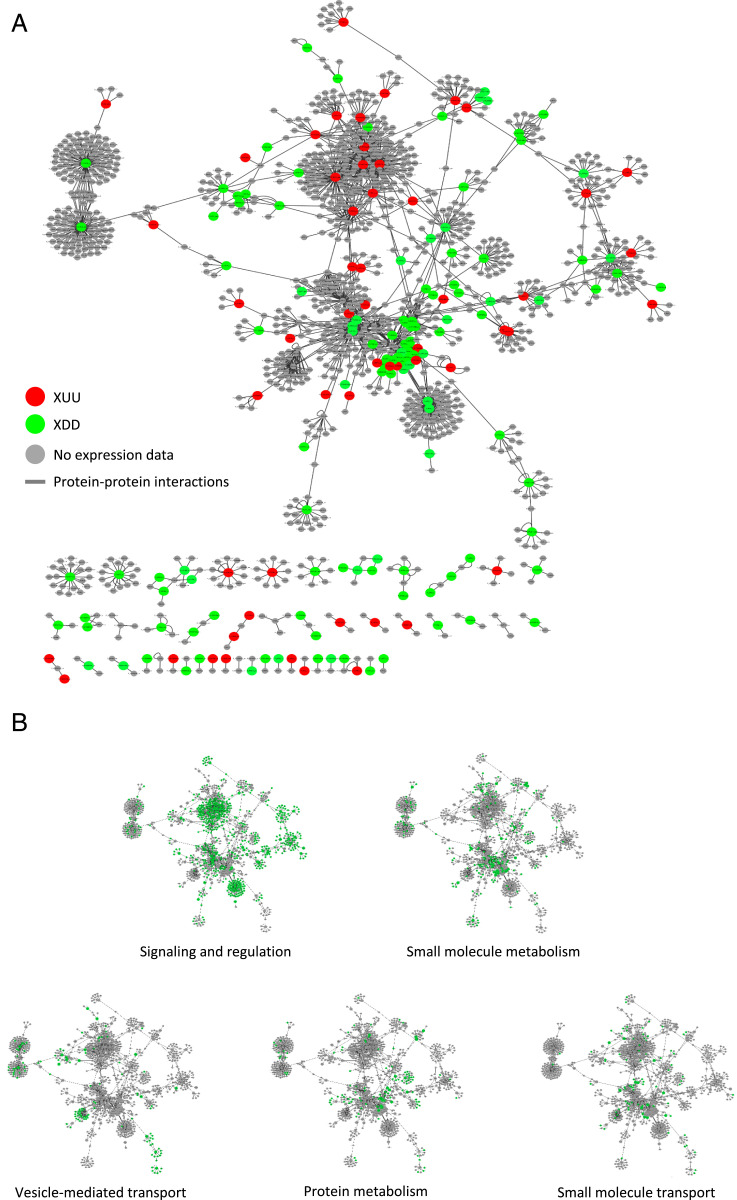
Analysis of the protein localization reported for the network components revealed that the three most abundant localization sites are in the plasma membrane (198 components; 14.8%), the cytoplasm (259; 19.3%), and the nucleus (273; 20.4%; Fig. 3C). Taking in account that RKS1 has been observed in these compartments, this proportion indicates that the PPI network contains components able to establish interactions and which might participate in signaling from the plasma membrane to the nucleus. Thus, this network structure corroborates the role of RKS1 in triggering rapid gene expression reprogramming at an early stage of infection. Interestingly, expression classes were found distributed within the network, indicating that the RKS1-dependent network presents a decentralized structure (Fig. 4A). In addition, the network was distributed in five main modules of different biological functions, i.e., signaling and regulation of cellular process, vesicle-mediated and small molecule transport, and protein and small molecule metabolism (Fig. 4B). Proteins assigned to the different biological functions are highly interconnected and distributed in the RKS1-dependent network. Indeed, analysis of the network connectivity reveals that 33% (346 of 1,047) of the proteins are connected with at least two partners and 7% (n = 73) are connected with five or more partners (SI Appendix, Fig. S9). Proteins related to signaling and regulatory functions are the more connected proteins, with 40% of the proteins connected. RKS1 appears as one of the main hubs, with 44 interacting partners. In summary, we established the RKS1-dependent network and its highly connected nature and decentralized topology is in agreement with the hypothesis about the complex and multigenic nature of quantitative disease resistance.
Fig. 4.
Expression profiles and functions of components of the RKS1 protein–protein interaction network. (A) The RKS1 protein–protein interaction network plotted with Cytoscape showing the components for which the gene expression pattern has been evaluated: in red, XDD DEGs (classes UDD and ØDD); in green, XUU DEGs (classes DUU and ØUU). (B) Display on the RKS1 protein–protein interaction network of the functional annotation of the different proteins. In green are proteins assigned to the functional group indicated.
Differential Robustness of Subnetworks of the QDR Response Revealed by Mutant Analysis.
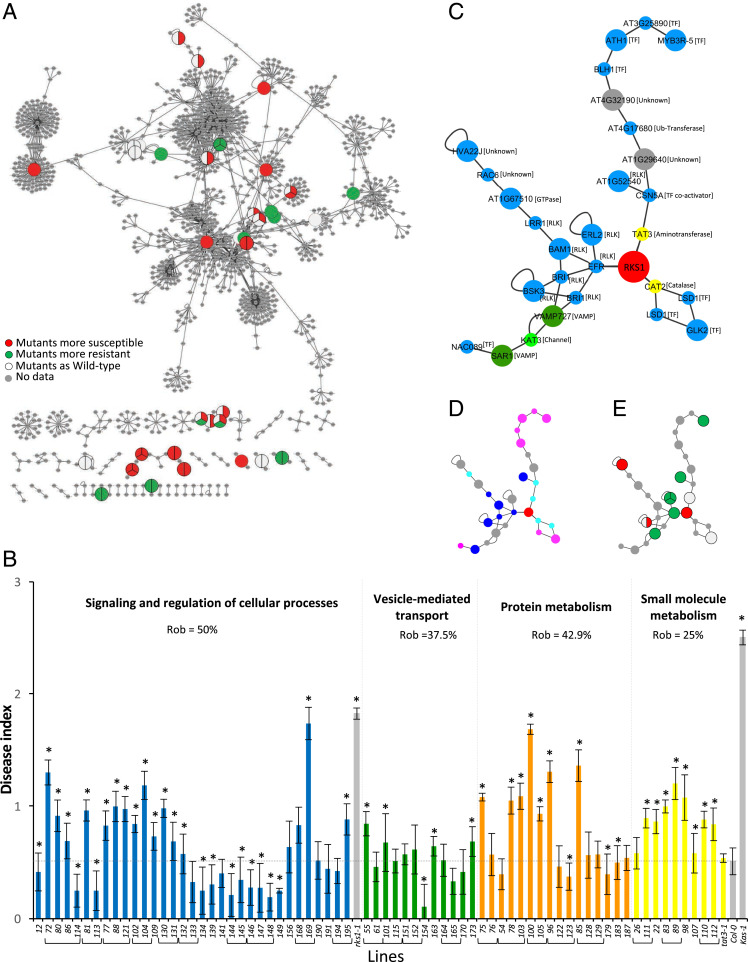
In order to validate the complex and multigenic nature of the RKS1-dependent QDR, we hypothesized that, if the five functional gene classes identified in the reconstructed PPI network play key roles in resistance to Xc, we should expect that mutants affecting genes belonging to these modules present alteration of their phenotype in response to Xcc568. Seventy-one T-DNA mutants in the Col-0 background were collected, first characterized by evaluating gene expression and sequencing the T-DNA insertion site, and phenotyped after inoculation with Xcc568 (SI Appendix, Fig. S10). The disease scores were plotted on the corresponding genes within the reconstructed RKS1-dependent network (Fig. 5A). From the mutants tested, 49 T-DNA mutants were significantly affected for their response to Xcc: 35 were found more susceptible, 14 more resistant (SI Appendix, Fig. S10). The mutant disease scores were quantitatively distributed between those of Col-0 and rks1-1, suggesting that each gene/gene module participates partially to RKS1 mediated resistance. Interestingly, these mutations do not similarly affect the different functional modules of the network, with the metabolism module being particularly affected (Fig. 5B). Indeed, we calculated each module’s robustness and found that the protein metabolism module including genes related to ubiquitination/proteasome was strongly impacted (100% of the genes mutated lead to resistance alteration). However, the resistance robustness to mutational perturbation remains at 0.43, i.e., only 57% of the mutants display a resistance decrease. The small molecule metabolism also presented a significant perturbation: 75% of the genes mutated led to resistance alteration, and, for almost all of them, to a reduction of resistance (robustness 0.25). In the opposite, the vesicle-mediated transport module is the less impacted sector of the network (62% of the genes affected and robustness of 0.38). The signaling and regulation of cellular process sector for which a particularly high number of genes was tested because of the putative signaling function of RKS1 showed a distinct pattern of response to mutation. Out of 20 genes tested using 34 mutants, 16 (80%) were found to exert a role in resistance, as the corresponding mutants were significantly affected as compared to the wild type. Interestingly, for half of these genes, the corresponding mutants were exhibiting an increased level of resistance, suggesting their implication in the negative regulation of resistance to Xcc. This module displays the highest level of robustness for resistance (0.50). Thus, the functional subnetworks of the QDR response to Xcc show differential robustness, indicating that, within the decentralized structure of the QDR network, some modules are more resilient than others, suggesting that the genes implicated in these different modules can compensate for each other.
Fig. 5.
Evaluation of robustness of the RKS1-dependent network by phenotypic characterization of mutants corresponding to some network components. (A) The RKS1 protein–protein interaction network plotted with Cytoscape showing the components for which mutants have been phenotyped in response to Xcc. Each circle represents the mutant phenotype corresponding to the protein component of the network: red indicates mutants significantly more susceptible than the wild type accession, green more resistant, and white not affected. No mutant was tested for genes represented in gray. (B) Disease index at 7 dpi after inoculation with a bacterial suspension adjusted to 2.108 cfu/mL of mutants corresponding to genes belonging to the different functional groups. Mutants belonging to “Signaling and regulation of cellular process” (blue), “Vesicle-mediated transport” (green), “Protein metabolism (ubiquitination/proteasome)” (orange), and “Small molecule metabolism” (yellow) are identified by numbers (SI Appendix, Table S3 shows correspondence to gene accession numbers). Mutants corresponding to the same gene have been grouped. Means were calculated from 4 to 24 plants (*Kinetic modeling deference with Col-0 time course, 0 = P value > 0.05 and 1 = P value ≤ 0.05). (C–E) A signaling subnetwork was extracted from the RKS1 protein–protein interaction network. (C) Subnetwork with protein functional groups (blue, signaling and regulation; yellow, small molecule metabolism; green, small molecule transport; dark green, vesicle-mediated transport; grey, others) and the protein molecular functions. Large circles indicate proteins encoded by genes from the 268 DEGs, and little circles indicate proteins from Y2H or BioGRID candidates. (D) Subnetwork with the protein subcellular localization (dark blue, plasma membrane; pale blue, cytoplasm; pink, nucleus; grey, others). (E) Subnetwork with the insertional mutant phenotypes (red, mutants more susceptible than the wild-type; green, mutants more resistant than the wild-type; white, mutants as the wild-type; grey, not determined).
A subnetwork including RKS1 and composed of 27 proteins (29 nodes) is presented in Fig. 5C. Interestingly, this subnetwork comprises essential signaling and regulatory proteins, located in the plasma membrane, nucleus, and cytoplasm (Fig. 5D) and including seven receptor-like kinases and a kinase (in close connection with RKS1), seven transcriptional factors, and two proteins related to metabolism (TAT3 and CAT2). Twelve insertional mutants corresponding to nine of these genes were analyzed, demonstrating that seven of them have an impact on resistance to Xcc (Fig. 5E). These findings suggest an essential role of this subnetwork in RKS1-dependent QDR and open avenues for functional studies.
Discussion
In recent years, considerable progress has been made in understanding the organization and dynamics of the plant immune system by using system-level approaches allowing integration of high-throughput genomic data. For instance, ETI and PTI are organized in diverse and interconnected defense modules, including receptor subnetworks, explaining the relative robustness of the plant immune system to pathogen perturbations (12, 17, 21). Communication between these immune networks and some pathogen molecules (effectors) is now being explored, revealing contact points (33, 34). However, while ETI and PTI are controlled by large effect genes that are mainly involved in pathogen perception and signaling, quantitative disease resistance is mediated by multiple functions, extending beyond pathogen perception (3). Despite its preeminence in natural plant populations and crops, the comprehension of its mechanistics has been hampered by its biological complexity (4). While many QDR genes have been recently identified, complete description and functional analysis of the gene networks underlying this immune response are still lacking. Using transcriptome and protein interactome analyses, systems biology, and functional validation by mutational approaches, we provide clues on the molecular networks acting downstream and possibly upstream of the RKS1 gene that regulates QDR in response to Xcc.
Previous studies of QDR by RNA-seq approaches indicated multifaceted defense responses according to the resistant genotype studied and the different QTLs involved (35, 36). However, a major limitation associated with these studies is the genetic diversity of the plant lines used. Here, we used RKS1 deregulated lines in order to precisely unravel the molecular mechanism underlying this resistance QTL. Among the 268 DEGs identified 6 hpi, 23% are associated with signaling and regulation of cellular processes, including 15 kinases or kinase-like proteins; 16% are related to vesicular and small molecule transport; and finally, 41% are involved in metabolic processes. This demonstrates that 1) genes associated with the early steps of QDR belong to diverse functional classes, in line with the idea that QDR does not involve simply perception of pathogenic molecules (3); 2) the major functional class is related to metabolic processes; and 3) these genes are mostly distinct from PTI and ETI gene networks already described (12, 18, 31) (SI Appendix, Fig. S7). A similar observation was reported by comparative transcriptomic analyses on barley in response to stem rust, indicating that 25% of barley genes are altered in response to infection, but only very few of these genes are controlled by the R locus (37). In line with these observations, we found a small proportion (6%) of genes putatively associated with response to hormones, none of them related to defense phytohormones such as ethylene (ET), jasmonate (JA), or salicylic acid (SA) (31, 38). However, previous studies have indicated that a four-sector network including ET, JA, SA, and PAD4 (PhytoAlexin Deficient 4) was required for PTI and ETI (18, 38, 39). Therefore, we tested single, double, triple, and quadruple mutants of ein2-1, pad4-1, sid2-2, and dde2-2. Most of these mutants were either more resistant or not affected in response to Xcc (SI Appendix, Fig. S11), indicating that 1) the QDR gene modules are mainly distinct from PTI and ETI networks found in response to Pseudomonas syringae, in good agreement with our findings by transcriptomic data comparison (SI Appendix, Fig. S7), and 2) a negative regulatory impact of the four hormonal sector network controlling ETI and PTI was observed on resistance to Xcc, revealing a balance between the different forms of immune responses. This finding is in agreement with the ETI-mediating and PTI-inhibited sector (EMPIS) mechanism identified, which tunes the immune response in response to P. syringae, and specifically the ETI and HR responses when the PTI is effective (18). Taking in account that ZAR1 forms a complex with RKS1 to perceive the effector XopAC from Xcc (30), these findings also suggest that the immune response triggered by this perception event is probably part of (or corresponds to) the immune response studied here and/or includes components mainly distinct from those already identified for ETI, as previously suggested (40). Therefore, to explore this hypothesis, we used three complementary strategies: 1) we studied the resistance phenotypes of the different RKS1 lines in response to Xcc568 and Xcc568ΔXop-AC, 2) we analyzed the expression of a subset of genes from our RKS1-dependent gene network in these lines in response to the same bacterial strains and in the zar1-2 mutant as compared to the wild type and rks1-1 mutant, and 3) we analyzed the resistance phenotypes of two mutants corresponding to genes of the RKS1-dependent network in response to the same bacterial strains. As previously shown with the Xanthomonas strain 8004 (28), the two mutants rks1-1 and zar1-2 were susceptible to Xcc568 and Xcc568ΔXopAC to a similar extent (SI Appendix, Fig. S12A). In contrast, the RKS1-OE1 line, which was more resistant than Col-0 to Xcc568 (129% at 7 dpi), was also more resistant to Xcc568ΔXopAC (41% at 7 dpi). These results demonstrate that the resistance phenotype observed in RKS1-OE1 is partially XopAC-independent. This observation was reinforced by testing the expression profile of a subset of genes (30 tested) from the RKS1-dependent gene network. Several genes showed an expression independent of the presence of XopAC in the RKS1-OE1 line (SI Appendix, Fig. S12B). Similarly, several genes showed a differential expression in the mutants zar1-2 and rks1-1 as compared to the wild type (SI Appendix, Fig. S12C). A similar behavior was observed for the resistance phenotype of two mutants corresponding to genes of the RKS1-dependent network (SI Appendix, Fig. S13). All these experiments taken together suggest that RKS1-dependent resistance is only partially dependent on XopAC and/or ZAR1, suggesting that QDR probably results from integration of different perception and signaling pathways, each of them representing a part of the resistance phenotype. This suggests also that ZAR1-dependent ETI differs from already characterized ETI pathways (40).
The most intriguing question about how QDR response is mounted at early time points of the interaction is whether multiple recognition and/or signaling/regulatory pathways are activated in parallel and then interconnected to produce a coherent response to the pathogen attack. In this context, we identified five different subnetworks that are different players in mediating QDR. Targeted protein localization and transport are essential for every stage of plant response to environmental constraints, especially during interactions with pathogenic microbes (4), and commonly associated with QDR (41, 42). Thus, VAMPs, syntaxins, and ABC transporters have been found among the DEGs, but most of them have not been described as associated with plant immunity yet. In the same line, proteasome proteolytic pathway and ubiquitination play major roles in plant immunity (43, 44). With the exception of RIN2 (RPM1 interacting protein 2) (45), 19 genes encoding ubiquitin-ligases or proteasome subunits have been identified in our study, most of them not previously described as involved in plant immunity. An important part of 268 DEGs (28%) was identified as part of small molecule metabolism with various functions, including carbon and secondary metabolism, confirming the large panel of functions usually described in QDR. Interestingly, we identified not less than 8 kinases and 10 receptor kinases in the signaling gene module. Considering that RKS1 encodes an atypical kinase, possibly a pseudokinase, kinases or receptor kinases constitute good candidates as potential coregulators. In agreement with previously described functions for pseudokinases (27, 46), RKS1 might act as a scaffolding protein to modulate their activity by activation or change in substrate specificity. The subnetwork extracted from the RKS1 network (Fig. 5C) supports such a hypothesis, as RKS1 interacts directly or indirectly with diverse RLKs. Moreover, 5 phosphatases and 16 transcription factors were also found within the DEGs, most of them not previously described in the context of plant immunity. In line with this central role of RKS1 in immune signaling pathways, RKS1 appears as a hub (44 connections) at the center of a dense network of components from plasma membrane to the cytoplasm and nucleus. Further studies, including transcriptomic studies at later timepoints, would help to get a dynamic comprehension of the RKS1 network and of its placement in relation with other forms of immunity.
Robustness is a key property of biological networks, especially of immune systems (20), and potentiates the evolvability of biological systems, which is a cornerstone in plant–pathogen interactions. We investigated the robustness of the RKS1-dependent network to mutational perturbations. Such loss-of-function perturbations may mimic gene function disruption triggered by pathogen effectors. Most mutants (69%, representing 76% of genes) showed phenotypic alteration but with only a moderate effect on resistance. This observation suggests a decentralized network because none of the gene disruption exerts a drastic and complete loss of resistance. However, most of the genes have a quantitatively significant effect, indicating their active role and the absence or low level of redundant/compensatory pathways replacing their loss of function. Interestingly, the potentially upstream functions, related to signaling and regulation, display the highest level of robustness (0.50), whereas the putatively more downstream processes, like small molecule metabolism, display low robustness (0.25). It should be noted that, for more than half of the genes related to signaling and regulation, mutations do not lead to a loss of resistance. This is mainly because a large part of these genes might be involved in mitigating resistance rather than in mounting resistance, with mutations in these genes leading to increased resistance. Hence our analysis shed light on the network structure of QDR and provides explanation of its high robustness to pathogen breakdown (4). Indeed, QDR depends on the contribution of many components involved in various and multiple defense mechanisms, with low functional redundancy. In contrast, in the case of qualitative resistance, robustness is due to functional redundancy of NLRs or decoy resistance proteins as well as compensatory downstream pathways (21, 31, 47, 48). In an attempt to compare the different forms of immune systems and the observed phenotypes (quantitative and qualitative resistance), qualitative phenotypes are expected to depend on upstream network components through which pathogen recognition is almost essential for triggering signaling and defense mechanisms. In such case, functional redundancy may be the major source of robustness (21). Also, our analysis did not reveal a high level of compensatory downstream pathways in the case of QDR even if more deep analysis of the pathways would be required to identify such complex mechanisms. This feature may also differentiate QDR from the qualitative resistance. Quantitative phenotypes are expected to depend on early, intermediary, and downstream components, the contribution to the information flow toward mounting resistance being distributed through the whole network. In such case, decentralized distribution of information seems the major source of robustness.
Analysis of multiple and combined disruptions of apparently unrelated genes (different functional categories) will further help to decipher the defense regulators and their distributed contributions to QDR. This should reveal if defense mechanisms operate in an additive or synergistic manner.
Material and Methods
Full and detailed methods are described in SI Appendix, Material and Methods.
Plant Growth and Bacterial Inoculation.
A. thaliana Col-0 accession was grown on Jiffy pots in a growth chamber at 22 °C with a 9-h photoperiod at 192 µmol m−2 s−1. Four-week-old plants were used for experiments. The inoculation tests were done with the strain LMG568/ATCC33913 (Xcc568) (49) carrying the LUX operon of Photorhabdus luminescens (50).
Fluorescence Microscopy.
Fluorescence images were acquired using a Leica SP8 confocal microscope equipped with a water immersion objective lens (×25, numerical aperture 1.20; PL APO).
Yeast Two-Hybrid.
To identify proteins interacting with RKS1, a mutated version of RKS1 (RKS1D191A) was used to screen a cDNA library made from A. thaliana leaves infected with the strain Xcc147 (32).
Transcriptomic Analyses.
Arabidopsis plants mis-expressing RKS1 [At3g57710; rks1-1, RKS1-si24 (25) and RKS1-OE1 lines] were used. Three independent experiments were performed at four time points (0, 1.5, 3, and 6 h postinoculation). Samples were sequenced by Fasteris on an Illumina HiSEq 2500 instrument. Reads were mapped on Col-0 genome downloaded from TAIR (https://www.arabidopsis.org/) using the glint software (http://lipm-bioinfo.toulouse.inrae.fr/download/glint/; release glint-1.0.rc6). Raw and normalized RNA-seq data have been deposited in the SRA database (accession number SRP233656).
Network Reconstruction.
Interactors of the 268 coregulated genes and the 41 potential interactors of RKS1D191A identified by yeast two-hybrid screens were recovered from Arabidopsis BioGRID protein interaction datasets version 3.5.179 (51). Protein–protein interactions were plotted with Cytoscape software v3.7.2. GO annotation analysis was done using BINGO module from Cytoscape. Network connectivity was calculated using Cytoscape.
Supplementary Material
Acknowledgments
We thank Aurélie Le Ru for technical assistance in cell imaging; Jérôme Gouzy, David Rengel, Richard Berthomé, Adelin Barbacci and Sarah Ranty-Roby for their help in bioinformatic and statistical analyses; and Sylvain Raffaële and Stéphane Genin for critically reading the manuscript. We also wish to acknowledge Benoît Lefebvre and Laurent Noël (LIPM, Toulouse, France) for providing biological material. This work was supported by a French Agence Nationale de la Recherche (ANR) Grant (RIPOSTE ANR-14-OE19-0024-01) and the French Laboratory of Excellence Project TULIP (ANR-10-LABX-41). F.D. is funded by a grant from Région Occitanie and the Plant Health & Environment Division of INRAE.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data of RNA sequencing experiments have been deposited in the National Center for Biotechnology Information Sequence Read Archive database (accession number SRP233656).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2000078117/-/DCSupplemental.
Data Availability.
All data necessary for replication are included in the submission and/or publicly available. The readers will be able to access the data, associated protocols, code, and materials in the paper, including the raw data of RNA sequencing experiments, which have been deposited in the SRA database at https://www.ncbi.nlm.nih.gov/sra (accession number SRP233656).
References
- 1.Poland J. A., Balint-Kurti P. J., Wisser R. J., Pratt R. C., Nelson R. J., Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 14, 1360–1385 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Roux F. et al., Resistance to phytopathogens e tutti quanti: Placing plant quantitative disease resistance on the map. Mol. Plant Pathol. 15, 427–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corwin J. A., Kliebenstein D. J., Quantitative resistance: More than just perception of a pathogen. Plant Cell 29, 655–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson R., Wiesner-Hanks T., Wisser R., Balint-Kurti P., Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 19, 21–33 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Bartoli C., Roux F., Genome-wide association studies in plant pathosystems: Toward an ecological genomics approach. Front Plant Sci 8, 763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.French E., Kim B. S., Iyer-Pascuzzi A. S., Mechanisms of quantitative disease resistance in plants. Semin. Cell Dev. Biol. 56, 201–208 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Krattinger S. G. et al., A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Yang Q. et al., A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 49, 1364–1372 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Hurni S. et al., The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang P. et al., Fungal resistance mediated by maize wall-associated kinase ZmWAK-RLK1 correlates with reduced benzoxazinoid content. New Phytol. 221, 976–987 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Barbacci A., et al. , Rapid identification of an Arabidopsis NLR gene conferring susceptibility to Sclerotinia sclerotiorum using time-resolved automated phenotyping. bioRxiv:10.1101/488171 (6 December 2018). [DOI] [PMC free article] [PubMed]
- 12.Dong X., Jiang Z., Peng Y. L., Zhang Z., Revealing shared and distinct gene network organization in Arabidopsis immune responses by integrative analysis. Plant Physiol. 167, 1186–1203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim M., Hyten D. L., Niblack T. L., Diers B. W., Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci. 51, 934 (2011). [Google Scholar]
- 14.Kebede A. Z., Johnston A., Schneiderman D., Bosnich W., Harris L. J., Transcriptome profiling of two maize inbreds with distinct responses to Gibberella ear rot disease to identify candidate resistance genes. BMC Genomics 19, 131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katagiri F., A global view of defense gene expression regulation–a highly interconnected signaling network. Curr. Opin. Plant Biol. 7, 506–511 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Sato M. et al., Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6, e1001011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillmer R. A. et al., The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 13, e1006639 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatsugai N. et al., A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J. 36, 2758–2769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyraud R. et al., Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J. 90, 720–737 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitano H., Biological robustness. Nat. Rev. Genet. 5, 826–837 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Wu C.-H., Derevnina L., Kamoun S., Receptor networks underpin plant immunity. Science 360, 1300–1301 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Williams P. H., Black rot: A continuing threat to world crucifers. Plant Dis. 64, 736–742 (1980). [Google Scholar]
- 23.Kniskern J. M., Traw M. B., Bergelson J., Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol. Plant Microbe Interact. 20, 1512–1522 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Bartoli C. et al., In situ relationships between microbiota and potential pathobiota in Arabidopsis thaliana. ISME J. 12, 2024–2038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huard-Chauveau C. et al., An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 9, e1003766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reiterer V., Eyers P. A., Farhan H., Day of the dead: Pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol. 24, 489–505 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Blaum B. S. et al., Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 186, 112–121 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Wang G. et al., The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285–295 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Wang J. et al., Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, 44 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Meyer D., Lauber E., Roby D., Arlat M., Kroj T., Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 6, 327–333 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Mine A. et al., The defense phytohormone signaling network enables rapid, high-amplitude transcriptional reprogramming during Effector-Triggered Immunity. Plant Cell 30, 1199–1219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Froidure S. et al., AtsPLA2-alpha nuclear relocalization by the Arabidopsis transcription factor AtMYB30 leads to repression of the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 107, 15281–15286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed H. et al., Network biology discovers pathogen contact points in host protein-protein interactomes. Nat. Commun. 9, 2312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brauer E. K. et al., Integrative network-centric approach reveals signaling pathways associated with plant resistance and susceptibility to Pseudomonas syringae. PLoS Biol. 16, e2005956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y. et al., Transcriptome dynamics associated with resistance and susceptibility against fusarium head blight in four wheat genotypes. BMC Genomics 19, 642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu Y. et al., Transcriptome analysis reveals the molecular mechanisms of the defense response to gray leaf spot disease in maize. BMC Genomics 19, 742 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscou M. J., Lauter N., Steffenson B., Wise R. P., Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PLoS Genet. 7, e1002208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F., Network properties of robust immunity in plants. PLoS Genet. 5, e1000772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim Y. et al., Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host Microbe 15, 84–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis J. D., Wu R., Guttman D. S., Desveaux D., Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6, e1000894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gu Y., Zavaliev R., Dong X., Membrane trafficking in plant immunity. Mol. Plant 10, 1026–1034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niks R. E., Qi X., Marcel T. C., Quantitative resistance to biotrophic filamentous plant pathogens: Concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53, 445–470 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Sorel M., Mooney B., de Marchi R., Graciet E., UbiquitinPproteasome system in plant pathogen responses. Annu. Plant Rev. 2, 65–116 (2019). [Google Scholar]
- 44.Furniss J. J. et al., Proteasome-associated HECT-type ubiquitin ligase activity is required for plant immunity. PLoS Pathog. 14, e1007447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawasaki T. et al., A duplicated pair of Arabidopsis RING-finger E3 ligases contribute to the RPM1- and RPS2-mediated hypersensitive response. Plant J. 44, 258–270 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Hammarén H. M., Virtanen A. T., Silvennoinen O., Nucleotide-binding mechanisms in pseudokinases. Biosci. Rep. 36, e00282 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis J. G., Integrated decoys and effector traps: How to catch a plant pathogen. BMC Biol. 14, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cui H., Tsuda K., Parker J. E., Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511 (2015). [DOI] [PubMed] [Google Scholar]
- 49.da Silva A. C. et al., Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417, 459–463 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Winson M. K. et al., Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163, 193–202 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Oughtred R. et al., The BioGRID interaction database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary for replication are included in the submission and/or publicly available. The readers will be able to access the data, associated protocols, code, and materials in the paper, including the raw data of RNA sequencing experiments, which have been deposited in the SRA database at https://www.ncbi.nlm.nih.gov/sra (accession number SRP233656).