Abstract
NAMD is a molecular dynamics program designed for high-performance simulations of very large biological objects on CPU- and GPU-based architectures. NAMD offers scalable performance on petascale parallel supercomputers consisting of hundreds of thousands of cores, as well as on inexpensive commodity clusters commonly found in academic environments. It is written in C++ and leans on Charm++ parallel objects for optimal performance on low-latency architectures. NAMD is a versatile, multipurpose code that gathers state-of-the-art algorithms to carry out simulations in apt thermodynamic ensembles, using the widely popular CHARMM, AMBER, OPLS, and GROMOS biomolecular force fields. Here, we review the main features of NAMD that allow both equilibrium and enhanced-sampling molecular dynamics simulations with numerical efficiency. We describe the underlying concepts utilized by NAMD and their implementation, most notably for handling long-range electrostatics; controlling the temperature, pressure, and pH; applying external potentials on tailored grids; leveraging massively parallel resources in multiple-copy simulations; and hybrid quantum-mechanical/molecular-mechanical descriptions. We detail the variety of options offered by NAMD for enhanced-sampling simulations aimed at determining free-energy differences of either alchemical or geometrical transformations and outline their applicability to specific problems. Last, we discuss the roadmap for the development of NAMD and our current efforts toward achieving optimal performance on GPU-based architectures, for pushing back the limitations that have prevented biologically realistic billion-atom objects to be fruitfully simulated, and for making large-scale simulations less expensive and easier to set up, run, and analyze. NAMD is distributed free of charge with its source code at www.ks.uiuc.edu.
I. INTRODUCTION
Grasping the function of very large biological objects, such as those of the cell machinery, necessitates at its very core not only the structural knowledge of these organized systems but also their dynamical signature. However, despite formidable advances on the experimental front, the intrinsic limitations of conventional approaches have often thwarted access to the missing microscopic detail of these complex, dynamic molecular constructs, restricting their observation to static pictures. The so-called computer revolution, which began over 40 years ago, considerably modified the perspectives, paving the road to structural biology investigations by means of numerical simulations from first principles. Such simulations form the central idea of the computational microscope,1,2 an emerging instrument for cell biology at atomic resolution, which the molecular dynamics (MD) program NAMD embodies.
A. The NAMD philosophy
The goal of NAMD development since its beginning has been to enable practical supercomputing for biomedical research. This goal of practicality is reflected first by the pursuit of affordable hardware such as workstation clusters in the 1990s, Linux clusters in the 2000s, and GPU acceleration in the 2010s—viewed as another computer revolution. Practicality is more deeply and enduringly reflected by the attitude of the NAMD development community that the target user of the program is the experimentalist or their collaborator, not the programmer or computer expert, or even the method developer.
In pursuit of this goal, NAMD has been designed to be a single program available across all platforms, preserving the knowledge of the users as their science grows from reproducing tutorials and case studies on a laptop, to production science on departmental commodity clusters, to large and multi-copy simulations on leadership-class supercomputers. NAMD is distributed free of charge for both academic and private-sector use as both the source code and pre-compiled binaries for most platforms. As cutting-edge biomolecular simulations are never truly routine, user extensions that are portable without recompilation across both platforms and NAMD releases are supported via the Tcl and Python scripting languages.
NAMD development relies on symbiotic relationships with multiple stakeholders. The oldest longstanding relationship is with the computer scientist developers of the Charm++ parallel programming system (see Sec. II), with which the NAMD developers have shared both a 2002 Gordon Bell Prize and a 2012 IEEE Fernbach Award. As the most popular Charm++ application, NAMD provides the Charm++ developers with real-world feedback from a broad community of users and drives access and support for Charm++ on leadership platforms. In return, Charm++ supplies enhancements that address performance, usability, and programmability issues faced by both NAMD users and developers.
The second indispensable relationship is between the NAMD and Charm++ developers and the high-performance computing technology providers, such as Intel, NVIDIA, AMD, IBM, Cray, and Mellanox. These corporations provide critical insights into current and upcoming technology, as well as software engineering expertise and code contributions to improve the performance of both NAMD algorithms and Charm++ high-speed network communication.
The third relationship that drives NAMD development is with computing resource providers, at both the various National Science Foundation (NSF) centers and Department of Energy (DOE) national laboratories that lead the United States exascale program. Early development and science access to leadership platforms ensures NAMD users of a smooth transition to new technologies and biomolecular applications of a generous share of high-performance computing resources, which could be readily consumed by other fields of science with less potential impact on human health and well-being. In return, the centers can promote the early scientific impacts of their machines, such as the all-atom model of the HIV-1 virus capsid3 solved during the early science period of the National Center for Supercomputing Applications (NCSA) Blue Waters machine.
The final symbiotic relationship driving NAMD is with the broad community of computational scientists who are ever expanding and furthering the scope of simulation methods. The NAMD core developers in Urbana together with other NAMD contributors scattered across the world maintain a strong intellectual connection with this community. The team of core developers and contributors meet yearly to coordinate ongoing efforts, discuss new methodological advances, and plan future changes to the program. In recent years, these joint efforts have facilitated the implementation of important advanced techniques, including integration algorithms,4 polarizable force fields,5 free-energy calculations, and enhanced-sampling strategies.6,7 Nevertheless, some tensions are to be expected between performance and innovation—NAMD was never designed to serve as a virtual laboratory platform for method development, and hence, sacrifices in modularity and modifiability have been made in the interest of scalability and performance. Regardless, NAMD offers today a mature and complete set of simulation capabilities, incorporating many features that have proven of general utility and continue to be extended and improved in response to both feedback from the NAMD user community and innovation by the method developers, who appear as co-authors on this new reference paper.
B. Perspective
To this date, NAMD has been downloaded by over 110 000 registered users, over 30 000 of whom have downloaded multiple releases. The 2005 NAMD reference paper8 has been cited over 13 000 times, and the NAMD user email list has over 1000 subscribers. NAMD is also typically reported as one of, if not the most utilized program at the NSF supercomputing centers. For a code developed for over two decades to enable leading-edge simulations on emerging platforms, NAMD serves a surprisingly large community of researchers. This can be attributed to the leadership of the late Klaus Schulten, who sought to share not only his scientific achievements with the world but also the tools with which they were accomplished and gathered a team of software and method developers that shared this vision.
Most users obtain NAMD by downloading a pre-compiled binary from www.ks.uiuc.edu. Current and multiple previous NAMD releases are available for download, along with the most recent nightly build. The download process requires creating an account by completing a brief registration form, which provides the NAMD developers with user statistics to justify future funding. The download process also asks the user to agree to a license that prohibits redistribution and requires attribution and citation of NAMD in publications resulting from its use. The standard license allows NAMD to be employed by both academic and private-sector researchers but prohibits commercialization of the program. Derived work may utilize up to 10% of the NAMD source code without restriction when combined with an equal amount of original code, allowing for substantial reuse by the biomolecular simulation community. All releases include the source code for both NAMD and the specific version of the Charm++ parallel runtime with which it was built. The NAMD and the Charm++ source code are also available via separate public Git repositories, and binaries for both releases and recent builds are maintained publicly on several NSF and DOE supercomputers. Official NAMD releases occur approximately annually and are each preceded by a series of beta releases to aid in bug discovery. The NAMD source code is intended to be of production quality at all times, and hence, bug fixes implemented after the beta period are not back-ported to the previous release.
The size of the NAMD user community both necessitates and enables a community support model. Basic training in MD simulation concepts and their application in NAMD is provided in a series of hands-on workshops, which can be attended in person or streamed at any time, and the pedagogical material, i.e., tutorials and case studies, used in the workshops is available for download. The tutorials require only a laptop, allowing them to be done without access to external resources, and possibly continued after the workshop. The pedagogical team of the workshops is formed of faculty members and teaching assistants, who are experienced NAMD users. Questions regarding the use of the program are directed to the public NAMD mailing list both because the collective experience of the user community will generally produce more useful and varied responses than the developers could and to provide a publicly searchable record of previous questions and answers. End-users are encouraged to search the mailing list archives along with the manual and other training materials before asking their questions on the mailing list. Reports of suspected bugs along with logs and ideally a reproducing input set for the latest NAMD release are regularly sent to the developer mailing list. Furthermore, high-level personal support is provided for selected driving projects that are testing or co-developing new NAMD capabilities.
NAMD software engineering practices are based on two decades of experience in ensuring correctness with minimal overhead. Both NAMD and Charm++ use Git for revision control, with Charm++ also employing formal issue tracking due to its larger numbers of both active developers and minor enhancement requests. NAMD has a smaller number of developers, with more differentiated expertise and goals, and this separation of responsibilities makes formal centralized issue tracking less beneficial. Moreover, due to the dynamic nature of development and changing priorities, schedules of planned enhancements are not reliable and are not advertised. It is better that the user make progress with the currently available version of the code than delay work based on the promise of an unproven feature. Separate developer documentation is avoided with the intent that the source code itself be legible and discoverable on its own.
NAMD development is guided by a small set of driving projects, which are scientifically important and require enhanced NAMD capability. While the driving project provides the essential eager end-user to test and provide feedback on the implementation of the new capability, it is important to note that no single-user or single-project features are implemented in NAMD itself. Instead, the goal of the development process is either to extend an existing capability to enable the driving project or, if necessary, to implement some new general-purpose and scalable feature that will enable both the specific driving project and a larger class of related simulations. Necessary project-specific code may be segregated outside of the NAMD code base through a Tcl scripting interface. Capabilities enabled via scripting in NAMD include top-level protocols such as equilibration, annealing, and replica-exchange or multiple-copy strategies, as well as application of long-range steering forces onto small numbers of atoms and application of independent boundary forces to each atom in the molecular system. While an optional Python scripting interface is available, Tcl remains the recommended choice due to its stable interface, compact and embeddable interpreter, simple and flexible syntax, and shell-like appearance, which appeals to non-programmers.
NAMD quality assurance practices are adapted to the unique challenges of feature-rich scientific software running at scale on a wide variety of platforms. All-platform builds and installs are automated, and record both default and optional modules loaded during compilation, as well as the specific version of Charm++ utilized. Extensive testing of each build on different platforms is prohibitively complex to manage and expensive in terms of computer time. Still, the most likely sources of observed defects in reviewed and merged code are compiler bugs and rare unanticipated edge cases in new end-user input sets. For this reason, NAMD contains multiple internal consistency checks, which are active at all times, including in production runs. The goal of consistency checks is not so much to prevent crashes, which are often easily diagnosed and relatively harmless, but to avoid silently generating flawed simulation outputs, which would waste both computer and human time and leave bad science in their wake. Consistency-check failures raise fatal errors, both in order to halt the program without wasting future cycles and because, in our experience, end-users ignore warning messages—and often barely read error messages.
C. Key features
NAMD supports classical MD simulations, most commonly of full atomistic nature, in explicit solvent, with periodic boundary conditions and full particle mesh Ewald (PME) electrostatics (see Sec. V) in a variety of thermodynamic ensembles (see Sec. IV), including constant–pH (see Sec. XI), although coarse-grained models, implicit solvent, and non-periodic or semi-periodic boundary conditions can also be employed. CHARMM9 and similar academic force fields, e.g., AMBER,10 OPLS–AA,11 and GROMOS,12 are supported, including the CHARMM Drude polarizability model.5,13 A flexible interface for quantum-mechanical/molecular-mechanical (QM/MM) calculations connects NAMD to external quantum-chemistry codes (see Sec. XII), namely, ORCA14 and MOPAC,15 thereby allowing physical phenomena that are not captured by classical models to be captured, e.g., chemical reactions involving bond formation and rupture. NAMD also supports alchemical free-energy calculations (see Sec. IX).16
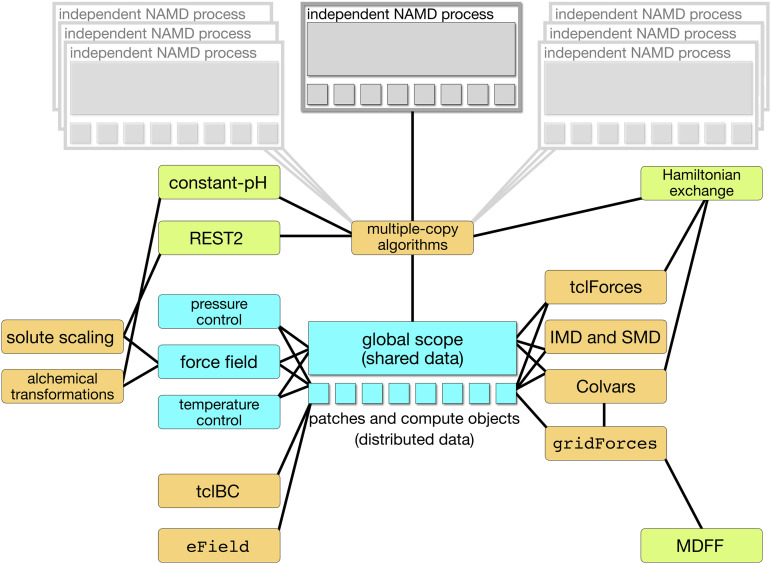
Built on this foundation are a variety of features to add external forces in the simulations, including the Colvars module,6 which allows the end-user to define collective variables (CVs) as control parameters for biased MD and free-energy calculations (see Sec. VI). Flexible fitting17 of structures to electron density maps from cryo-electron microscopy (cryo-EM) can be performed via grid potentials18 (see Sec. XIII). Methods for accelerating sampling include user-customizable multiple-copy algorithms (MCAs) (see Sec. X) for both parallel-tempering strategies and free-energy calculations of geometrical and alchemical nature.16 The workflow architecture of a NAMD simulation is summarized in Fig. 1.
FIG. 1.
Workflow architecture of a NAMD simulation. The blue boxes denote the core physical details of the simulation, the orange boxes denote the features that are embedded in the code (written in C++), and the green boxes denote the features that are fully implemented through user scripts (written in Tcl or Python).
Support for setup, analysis, and visualization of NAMD simulations is implemented in the co-developed program VMD.19 In VMD, molecular structures can be assembled either with the low-level psfgen module or via the QwikMD plugin,20 which provides a graphical interface and guides the user through the stages of the standard MD workflow (see Sec. XV). A large number of VMD plugins are available to aid in analysis tasks. Guidance on the use of NAMD and VMD is provided by a variety of tutorials and case studies available on the www.ks.uiuc.edu website.
NAMD supports most computational platforms, ranging from MacOS–and Windows–operated laptops to leadership-class supercomputers. NVIDIA GPU acceleration has been implemented since 2009 (see Sec. III),21 and support for AMD and Intel GPUs is being developed as part of readiness programs for the Oak Ridge Frontier and Argonne Aurora exascale machines. Simulation sizes of up to 2 × 109 atoms are possible when the program is compiled in memory-optimized mode, which is recommended above 2 × 106 atoms.
Most users will have no need to compile NAMD from the source code. NAMD achieves cross-platform extensibility through scripting in Tcl, a language familiar to the end-users, notably from its use in VMD, allowing custom simulation protocols to be implemented without recompiling the program. Pre-compiled binaries are available for laptops, desktops, and clusters, both with or without high-speed InfiniBand networks. No MPI library is required, but the standard mpiexec program may be leveraged to simplify launching NAMD within the batch environment of the cluster.
II. THE PARALLEL, OBJECT-ORIENTED PROGRAMMING LANGUAGE CHARM++
NAMD is implemented on top of Charm++,22,23 an adaptive, asynchronous, distributed, message-driven, task-based parallel programming model using C++.
A. Underpinnings of Charm++
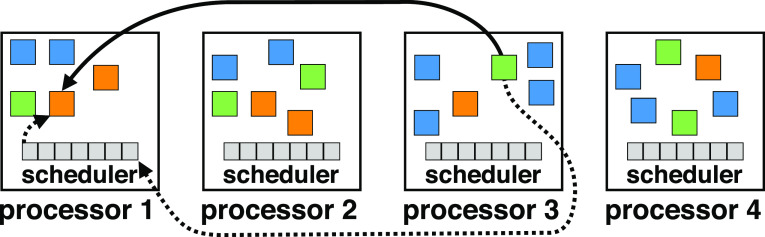
In Charm++, computations are partitioned into migratable objects called chares, each with its own data. Chares communicate by sending messages that invoke methods on other objects, which can be located locally or remotely, on a different node than the sender. An example of chare layout and messaging is shown in Fig. 2. Charm++ also features a powerful introspective runtime system (RTS), which measures and tunes the performance of applications at runtime. Put together, these properties allow for dynamic load balancing, in which the runtime relocates chares to different processors to evenly distribute computational load. These aspects of Charm++ allow high performance and scalability to be achieved on a wide variety of applications and large-scale computer architectures. Charm++ has also been carefully crafted to maximize portability so that applications can be executed on virtually any hardware, from laptops to supercomputers.
FIG. 2.
Overview of a Charm++ application on four processor elements (PEs). The solid line indicates an object on PE 3 sending a message to an object on PE 1, and the dashed line shows how the message is sent via per-PE schedulers.
1. Over-decomposition
Most parallel programming paradigms partition a problem into the number of processors that are being used to execute the application. This approach tries to minimize overhead while placing the onus on the programmer to provide an even distribution of work to the processors. However, in Charm++, applications are intended to be over-decomposed or broken into many more pieces than there are processors in the system. Using this finer decomposition may increase overhead, but it also provides scope for the runtime system to perform optimizations outweighing it, especially for dynamic, irregular applications. One virtue of this approach is communication-computation overlap. Since there are many small objects on a processor, as opposed to fewer large objects, the RTS can schedule the computation of an object while it awaits communication required by another. In doing so, Charm++ more effectively uses both the processors and network, thereby reducing starvation of processors due to network delays.
2. Load balancing
Another key facet of the Charm++ model is dynamic load balancing.24 Over-decomposition creates many high-granularity objects, giving the RTS scope to finely control distribution of load via chare migration. The Charm++ RTS actively measures the per-object load at runtime, which it then feeds into one of many different, customizable algorithms to determine a new and improved distribution of chares based on these empirical data. This design implies that load balancing retains efficacy even for applications with load properties that are difficult to predict a priori. Native support for general load balancing in the Charm++ RTS constitutes a dramatic improvement over frameworks such as MPI, which require end-users to write application-specific load-balancing code. In general, load balancing is critical to achieve high scalability, as the likelihood of significantly overloading some processor grows as more processors are added to a job.
3. Modern Charm++ features
While currently unused by NAMD, Charm++ has many other features to tune applications. Remaining within certain power and temperature budgets has become vital as supercomputers grow ever denser and larger. Charm++ includes modules that work with the processor and system controls over these properties while still retaining high performance.25 For instance, it can leverage the load balancer to migrate away chares from overheated processors to avoid slowdowns when the system underclocks them to reduce their temperatures. Charm++ features a checkpoint-restart facility, which makes applications fault tolerant.26 In the case of system-level hardware or software failures, rather than crashing, potentially wasting hours of computer time, the RTS can automatically recover and continue execution using the non-failed parts of the system. Charm++ also has a module to integrate GPU data and task management with the RTS.27 Programming models that are built on top of Charm++ also exist, such as Adaptive MPI, which allows MPI applications to execute atop Charm++ and gain several of its advantages,28 as well as charm4py, a Python interface to Charm++.29
B. Parallel structure of NAMD and its use of Charm++
The various features of Charm++ described previously have contributed to the success of NAMD in achieving high performance, scalability, and portability.30–32 NAMD and Charm++ share a long history, having been synergistically codeveloped since 1994. In fact, NAMD was the first large, driving application for Charm++, and many features and design decisions of Charm++ were inspired by the needs and challenges presented by NAMD. The architecture of NAMD maps very naturally to Charm++, allowing the structure and parallelism of the code to be easily and cleanly expressed without sacrificing speed.
MD simulations involve calculating and applying forces, bonded and nonbonded, that simulated atoms exert onto each other. NAMD takes as input a molecular structure, iteratively computes these bonded and nonbonded forces, and integrates the equations of motion to update atomic positions and velocities. NAMD implements this process using a unique hybrid approach, decoupling the distribution of data from the distribution of computation. The simulation space is spatially divided into small boxes called “patches”—objects containing simulation data, while force calculations are done by objects called “computes,” which operate on data received from patches. After the force calculation is completed by the computes, the forces are sent back to patches, which update their constituent atoms. Hydrogen atoms are stored on the same patch as the heavy atom they are bonded to, and atoms are reassigned to different patches as they move in space.
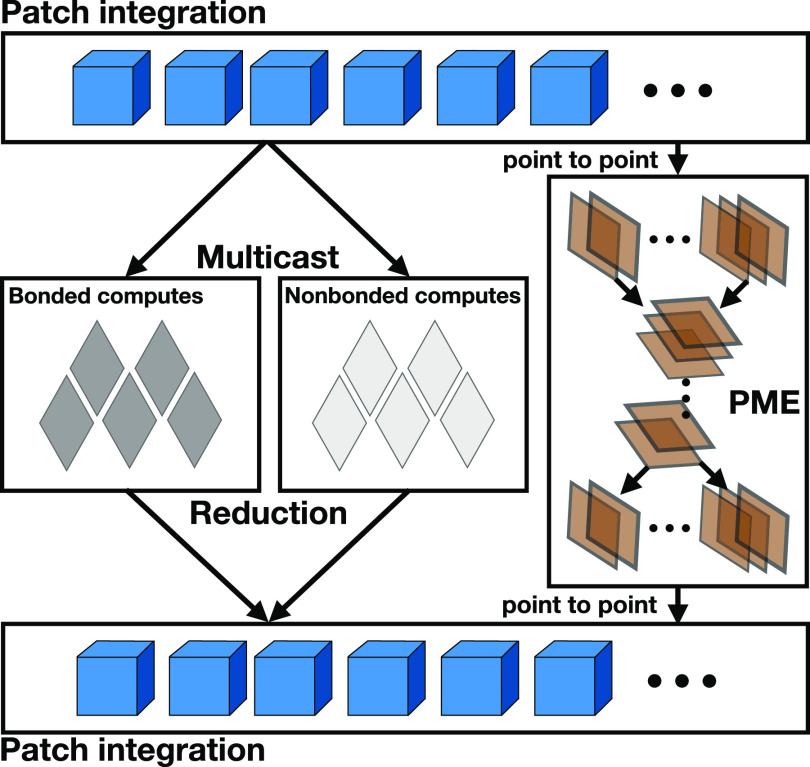
An overview of the parallel structure of NAMD is depicted in Fig. 3. There are three different kinds of computes, namely, bonded, nonbonded, and PME, respectively, responsible for bonded forces, short-range nonbonded forces, and long-range nonbonded electrostatic forces using the PME algorithm (see Sec. V). Naïve computation of all nonbonded forces would result in a computational complexity in , where N stands for the number of particles, whereas the PME approach of only computing pairwise nonbonded forces explicitly for atoms within some cutoff radius and interpolating the reciprocal-space Ewald sums for more distant atoms improves the overall complexity to .33 Patches are sized such that only the 26 neighbors of the three-dimensional patch are involved in the bonded and short-range nonbonded interactions or, formally, that non-neighboring patches are separated by at least the cutoff radius. Each nonbonded compute is responsible for calculating the forces between a given pair of neighboring patches, including self-pairs, and, correspondingly, each patch sends its data to the twenty-seven non-bonded computes that use its atoms. PME computes are done using a two-dimensional pencil decomposition of the charge grid and consist of three different subtypes, x, y, z, one for each of the three directions of the Cartesian space. This step involves performing several fast Fourier transforms (FFTs) and transpositions between the different dimensions, making PME relatively light in terms of computation but heavy in terms of communication. It is noteworthy that Charm++ supports message priorities, which are set to high in the case of PME, since communication is crucial here. Bonded computes are also relatively light computationally, since they only operate on the bonded components, so the majority of the computational load comes from the nonbonded computes.
FIG. 3.
NAMD software architecture. “Computes” are objects responsible for force calculations, operating on data received from “patches,” small boxes generated by the spatial decomposition of the computational assay. Long-range electrostatic forces are handled by the PME algorithm.
At runtime, these compute objects in NAMD are distributed throughout the available processors for the job. Notably, only nonbonded computes are relocatable as they represent most of the load. Patches and the other computes are statically assigned to the available processors at startup. Loads and communication patterns of the various objects, whether relocatable or not, are empirically measured and fed to the load balancer, which redistributes the nonbonded computes to minimize communication and equalize load between processors.
Generally, the above descriptions are equally valid for the CPU-based and GPU-based versions of NAMD. However, there are a few extra considerations in the GPU version, chiefly that of aggregating data and work requests. GPUs are computationally extremely powerful, but since they are physically separated from the CPUs and, thus, have high communication latency, performing many small transfers of data, or starting many small kernels, is relatively expensive. In order to avoid this bottleneck, NAMD aggregates data and work requests for the GPU, sending the data of several patches or invoking a kernel corresponding to several computes at once. While this strategy limits in some way the ability of Charm++ to perform optimizations, it also allows NAMD to leverage the immense power of GPUs without inordinate penalties due to latency, making it a worthwhile trade-off.
III. GPU CAPABILITIES AND PERFORMANCE
Historically, NAMD was designed for optimal performance on large arrays of computing units interconnected by low-latency, high-bandwidth networks. As use of GPUs in high-performance computing gained steam, NAMD has been adapted to novel, GPU-based architectures. In this section, we first review the performance of the code on a variety of platforms, prior to describing ongoing efforts toward more fully utilizing GPU acceleration.
A. NAMD performance
From its inception, NAMD was designed for high-performance classical simulations of large atomic models of biomolecular systems, often comprising 100 000 atoms or more. Through pervasive use of parallel computing technologies, the system size scales and simulation timescales amenable to NAMD have grown by orders of magnitude, as the program evolved from being able to handle a few nanoseconds of simulated time on hundreds of thousands of atoms34 to a hundred nanoseconds for millions of atoms,3 finally arriving at where we stand today, namely, being able to simulate hundreds of nanoseconds on hundred-million atom systems.35
The growth in performance achieved by NAMD can be primarily attributed to its ability to harness the computational power of tens of thousands to millions of processing elements in parallel. Over the past decade, computer architectures have undergone a paradigm shift toward hardware designs that favor parallelism as the primary mechanism for improved performance, e.g., by increasing core counts from one processor generation to the next. NAMD supports diverse computing hardware architectures, including multicore and many-core CPUs with wide single-instruction-multiple-data (SIMD) vector arithmetic units and heterogeneous computing platforms containing massively parallel GPU accelerators. NAMD decomposes MD simulation algorithms into very fine-grained data-parallel operations, which can be executed by the available pool of computing resources, maximizing the use of hardware-optimized functions, or so-called “kernels,” on CPU vector units or GPU accelerators. These kernels parallelize operations on individual atoms using a hardware-specific approach, thereby maximizing arithmetic performance and simulation throughput for the target hardware.
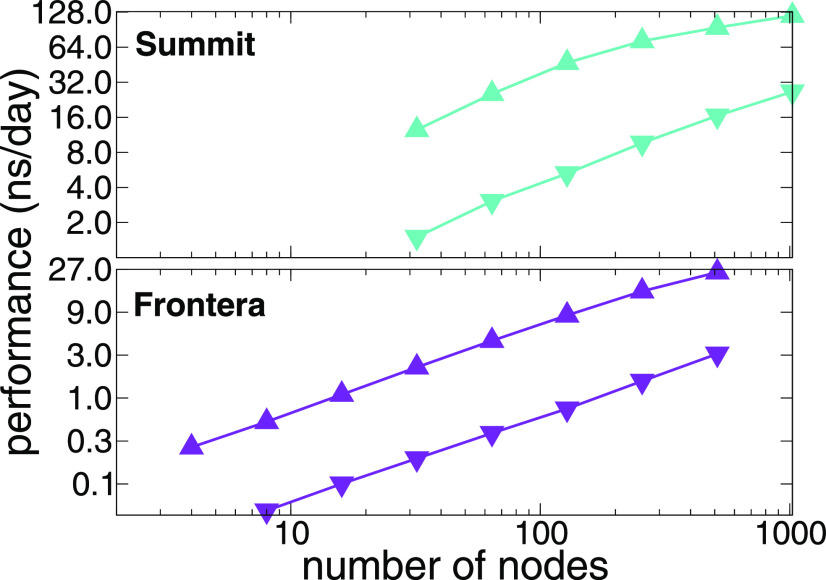
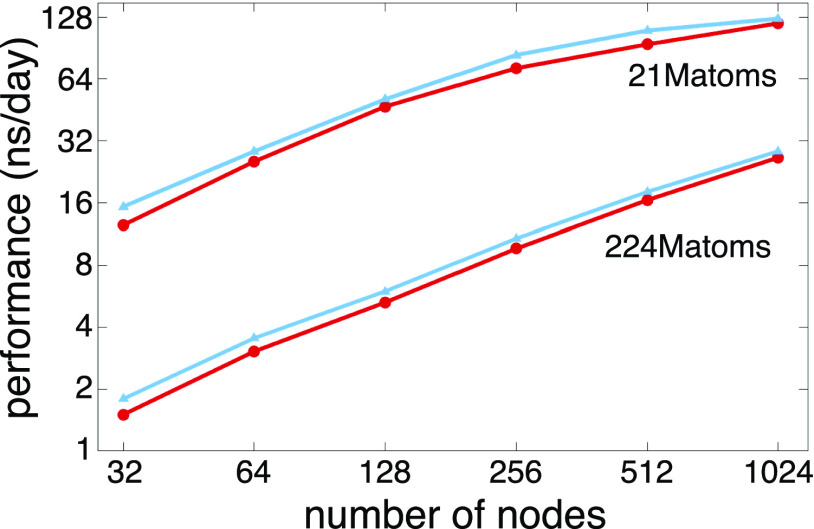
NAMD distributes work to shared memory CPU cores and distributed memory compute nodes using a three-dimensional decomposition of interactions among patches (see Sec. II), spatial subvolumes sized such that interactions with the 26 nearest neighbor patches embrace all of the bonded, van der Waals, and short-range electrostatics force contributions. Decomposition of the simulation into patch–patch interactions affords a large amount of parallelism for shared- and distributed-memory parallelism and enables NAMD to maintain load balance by shifting work from overloaded to underloaded cores or nodes over the course of the simulation. Building on these design points, extensive use of asynchronous message passing techniques and Charm++ runtime system features have allowed state-of-the-art NAMD simulations to run at high performance on petascale supercomputers,36–40 as well as pre-exascale supercomputers.32 Figure 4 summarizes the distributed-memory scaling performance of NAMD simulating two benchmark systems, each consisting of a tiled array of a 1 × 106-atom satellite tobacco mosaic virus (STMV),41 on two contemporary leadership supercomputers with CPU-based (Frontera) and GPU-accelerated (Summit) hardware platforms.
FIG. 4.
NAMD distributed memory parallel scaling benchmarks performed on satellite tobacco mosaic virus (STMV) in water on leadership supercomputers, Summit, using up to 1024 nodes containing 6144 GPUs (upper panel), and Frontera, using up to 512 CPU-based nodes (lower panel). Two benchmark systems were constructed as a tiled array of a periodic 1.067M-atom system, a 5 × 2 × 2 tiling for 21M atoms (▴), and a 7 × 6 × 5 tiling for 224M atoms (▾).
B. GPU acceleration
One of the biggest changes to the high-performance computing ecosystem in the past decade has been the emergence of GPUs as the dominant type of accelerator for scientific applications, leading to their rapid adoption and widespread use in computational chemistry applications.42 NAMD was the first fully featured MD package to exploit GPU acceleration,21 and it was also a pioneer in supporting GPU-accelerated clusters.43,44 NAMD development has evolved to encompass two main computing approaches, namely, (i) large-scale distributed memory parallel computing (continuing past NAMD 2.x approach) and (ii) GPU-resident computing to support new and emerging platforms that provide dense, tightly coupled GPU accelerators, with shared memory among GPUs and hosts (newly added in NAMD 3.x).
The initial use of GPUs in NAMD accelerated calculation of the short-range non-bonded forces, the biggest computational workload at each time step. The GPU kernels themselves, similar to the CPU, interpolate force interactions from a table but make use of fast-texture memory lookup and automatic linear interpolation, which avoids calculation of square-root and exponential functions required for the PME algorithm33 (see Sec. V) and eliminates conditionals needed to support switching functions for van der Waals forces. Energies are similarly computed with table lookup and only when required for output. Enhancements were introduced to improve performance by streaming the force calculations back to the patches, enabling each patch to proceed asynchronously with time stepping as soon as all of its forces are computed.21 GPU acceleration was next applied to the PME long-range electrostatics calculation, specifically to the patch-based calculations involving the spreading of charges from atoms to grid points and the gathering of forces from grid points to atoms.45 Parallel scalability is improved by doubling the PME grid spacing, together with increasing the interpolation order from 4 to 8 to maintain the same accuracy, thereby reducing the communication bandwidth by a factor of 8 while increasing the intensity of arithmetic offloaded to the GPU.
Since then, NAMD has evolved to allow all force terms to be computed on the GPU. With the 2.12 release, NAMD incorporated new CUDA kernels for both short-range, nonbonded forces and long-range electrostatics calculations, which yield better performance and scaling in general. The new short-range non-bonded kernels compute pairwise interactions between two sets of atoms subdivided into tiles of 32 × 32, producing tile lists that can be executed by any CUDA thread block. To further improve performance, the kernels also benefit from Newton’s third law, avoiding duplicated calculation between atom pairs and eliminating the need for synchronization between thread blocks, which allows CUDA warps to execute independently.46 This new scheme also introduced generalized Born implicit solvent (GBIS) neighbor list calculation kernels for existing GPU-accelerated GBIS functions.47 The revised implementation of PME now offloads the reciprocal-space calculation as well to GPU and uses the NVIDIA cuFFT library for calculating forward and inverse FFTs,46 although the scalability of this approach is limited to no more than four nodes. The 2.13 release of NAMD added new CUDA kernels for calculating the bonded forces and the correction for excluded interactions.32 The 2.14 release yields better performance on modern GPU architectures and contains a more stable pair list generation scheme for large domain decomposition cycles. NAMD 2.14 performance results reported in this contribution represent the outcome from runs using the traditional distributed memory NAMD design.
NAMD 3.0 maintains the traditional large-scale distributed memory computing paradigm but is the first version to pioneer the new GPU-resident computing approach. The NAMD 3.0 benchmarks reported here represent the currently achieved performance using the new GPU-resident computing scheme, applied to single-node GPU-accelerated hardware platforms. We note that since NAMD 3.0 and its new GPU-resident computing approach are still actively in development at the time of writing this article, we expect to achieve even higher performance by the time it is finalized.
Based on a Charm++ parallel object paradigm, NAMD over-decomposes the total work into small, easily migratable tasks at startup, which are consequently distributed across the available allocated CPU resources for a particular run (see Sec. II). This scheme is effective on large parallel computers with a myriad of CPU cores as tasks are small enough to allow for dynamic load balancing at runtime and better scaling overall. However, fine-grained decompositions that are appropriate for large CPU core counts often result in task sizes that are insufficient to saturate GPU with work, as is necessary to approach peak GPU performance. Moreover, running large batches of small tasks typically requires excessive host-device memory transfers as they have disjoint memory spaces. Thus, for launching GPU tasks efficiently, NAMD usually performs an aggregation step, whereby patches are gathered together into contiguous memory spaces, and their corresponding atoms are rearranged in order to coarsen task grain size to improve GPU utilization and reduce overhead. It is also possible for the end-user to control which force terms are offloaded onto the GPU, as force calculations are scheduled independently, for best combined use of hybrid CPU and GPU computing resources.
The improvements to existing GPU kernels and more effective GPU offloading schemes have allowed NAMD to benefit not only from thousands of CPU cores but also from thousands of discrete GPUs. Figure 4 depicts NAMD benchmarks occupying up to a quarter of ORNL Summit, a GPU-dense supercomputer with 4600 compute nodes, each containing two IBM POWER9 CPUs and six NVIDIA Volta V100 GPUs. Two different tiled replications of the 1.067 × 106 atom, freely distributed, STMV computational assays were employed as benchmark systems—a 5 × 2 × 2 replication with 21 × 106 atoms and a 7 × 6 × 5 replication with 224 × 106 atoms. The results demonstrate the fact that NAMD is able to benefit from the thousands of GPUs available in Summit, delivering approximately eight times improved performance compared to CPU-only runs.
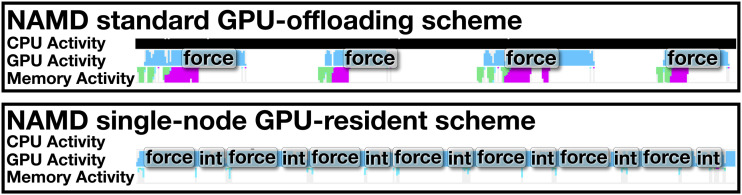
The rapid pace of performance gains on state-of-the-art computing architectures has tipped the balance of computing power even more dramatically in favor of GPUs. The traditional GPU offloading scheme in NAMD overlaps CPU and GPU work, with forces being calculated on the GPU and the integration of the Newton equations of motion being calculated on the CPU as soon as the forces are processed by the GPU. However, in-depth NAMD performance analysis reveals that current high-end GPUs are idling for a large fraction of the simulation time, since integration is a critical step and must be performed before the next launch of GPU force kernels. Ongoing development efforts have begun to ameliorate this imbalance, whereby a new GPU-resident computing scheme maintains data in-place on the GPU throughout force calculations and integration of atomic coordinates, with drastically reduced involvement from host CPUs and minimal host-device memory transfers. Kernels for the Verlet integration (see Sec. IV), rigid bond constraints, and Langevin dynamics have been introduced, without any need for data transfer between the CPU and GPU. Figure 5 depicts a timeline profile of a simulation of apolipoprotein 1 (ApoA1) in the microcanonical ensemble, with roughly 92 224 atoms, on a NVIDIA Titan-V GPU, demonstrating that the standard offloading scheme is not capable of fully saturating the GPU with work as there are large gaps between the GPU force calculations with integration tasks running on the CPU.
FIG. 5.
Standard GPU offload approach compared against new GPU-resident execution scheme for a single-node NAMD simulation of apolipoprotein 1 (ApoA1) in water, consisting of 92 224 atoms. The light blue line tracks GPU activity, while the black strip tracks CPU activity. GPU force calculations are labeled “force,” and GPU integration calculations are labeled “int.”
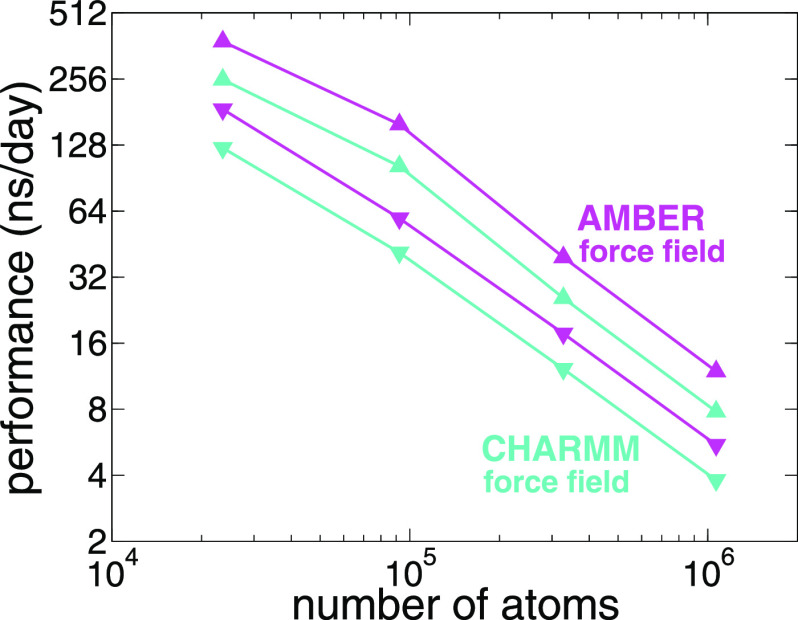
The new, GPU-resident computing scheme is able to effectively saturate the GPU with work, showing almost no gaps between kernel calls. Figure 6 shows preliminary benchmarks of this GPU-resident scheme for simulations in the microcanonical ensemble, with a 2-fs time step and the standard CHARMM force-field parameters, on a NVIDIA Titan V GPU and an Intel Xeon E5-2650, for four independent computational assays of increasing size.
FIG. 6.
Comparison of NAMD GPU computing schemes for simulations with increasing atom counts, namely, (i) reductase enzyme (DHFR) with 23 558 atoms, (ii) apolipoprotein 1 (ApoA1) with 92 224 atoms, (iii) F1-ATPase with 327 506 atoms, and (iv) STMV with 1 066 628 atoms for the GPU-resident scheme in NAMD 3.0 (▴) and for standard GPU offloading scheme in NAMD 2.14 (▾). The simulations were performed in the microcanonical ensemble, using the CHARMM9 (turquoise line) and AMBER-like10 (magenta line) cutoff schemes: 12 Å and 8 Å, respectively.
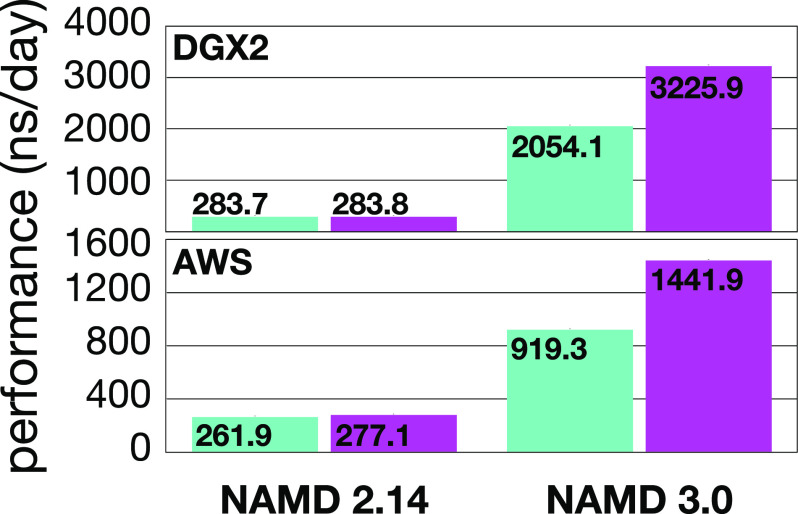
The new GPU-resident computing scheme, using only a single CPU core, outperforms the original GPU offload scheme by approximately a factor of 2, despite the original offload scheme’s use of all 16 CPU cores. In addition, since most of the computational bottlenecks have been removed from the CPU, it is possible to achieve perfectly linear scaling when running independent replica simulations on single-node multi-GPU platforms, with one replica per GPU. This approach can produce aggregate simulation times of microseconds per day, as shown in Fig. 7.
FIG. 7.
NAMD performance compared on two platforms, NVIDIA DGX-2 (upper panel) and Amazon Web Services (AWS) P3.16xlarge (lower panel), using the standard GPU offloading scheme in NAMD 2.14 and the GPU-resident scheme in NAMD 3.0. The DGX-2 and AWS P3.16xlarge platforms consisted, respectively, of 16× NVIDIA Tesla V100 GPUs and 8× NVIDIA Tesla V100 GPUs associated with a 48-core CPU. The benchmarks were conducted on replica simulations of apolipoprotein 1 (ApoA1) in water, representing 92 224 atoms, in the microcanonical ensemble, with one independent replica running on each GPU. The benchmarks were carried out with the CHARMM9 (turquoise bars) and AMBER-like10 (magenta bars) cutoff schemes: 12 Å and 8 Å, respectively.
IV. PROPAGATORS
One of the features that has made NAMD a widely popular MD engine is its ability to generate trajectories in apt thermodynamic ensembles, with minimal approximation and corner-cutting. This section reviews the numerical schemes implemented in NAMD to integrate the equations of motion, together with the algorithms utilized to maintain the temperature and the pressure constant.
A. Numerical integration
It is useful to distinguish the two ways whereby MD simulations might be carried out. For actual dynamics, the dynamical parameters, e.g., the mass and the thermostat and barostat coupling parameters, are to be given physically realistic values. For sampling, however, parameters can be chosen to reduce autocorrelation times, thereby increasing the number of independent samples. In both cases, it is, however, normally expected that a numerical integrator retains the following property of the dynamics: if an ensemble of trajectories has initial positions and velocities chosen from a given distribution, e.g., the canonical ensemble, this distribution is preserved as the trajectories evolve. This is a useful property even for sampling, in which only a single, long trajectory is generated.
1. Verlet and symplectic integrators
NAMD provides both deterministic and stochastic equations of motion. The deterministic model rests upon the Newton equations of motion, and the basic integrator is provided by the Verlet algorithm.48 This method happens to be symplectic, which ensures that numerical trajectories possess properties similar to those of the analytical Hamiltonian dynamics. In particular, Hamiltonian dynamics preserves volume in phase space and conserves the energy exactly, which, together, ensure the preservation of any distribution, such as the Boltzmann distribution, that is a function of the Hamiltonian alone. A symplectic integrator also preserves volume in phase space but conserves a so-called “shadow” energy, which differs from the actual energy by a modest . Actually, this is not precisely correct—there is a “very small” drift in the conservation of the energy over “very long” timescales.49 The very small error shrinks dramatically, and the very long timescales expand dramatically as Δt is reduced. In a plot of the actual energy, the fluctuations indicate the error in the shadow energy, and an insignificant secular drift indicates a sound integration. In practice, for a symplectic integrator, it is adequate that the time step be not much smaller than that needed to avoid drift. For one thing, the greatest error in the shadow Hamiltonian occurs in the less important high-frequency modes. Second, temporal discretization error is typically much smaller than that due to limited sampling. Of any integrator that employs only full force evaluations, the Verlet integrator allows the largest stable time step for a given computational effort.50
2. Multiple time stepping
Attaining larger time steps with a symplectic integrator is possible if the energy function is partitioned based on timescales and the forces corresponding to shorter timescales are evaluated more frequently. This numerical scheme,51 known variously as r-RESPA52 and the impulse method, is implemented in NAMD with up to three levels of multiple time stepping, namely, bonded, short-range nonbonded, and long-range nonbonded, e.g., 1 fs, 2 fs, and 4 fs. There is a limit to the largest time step due to possible resonances between the frequencies of the long-range impulses and natural frequencies of the bonded interactions.53
3. Constrained dynamics
For constrained dynamics, NAMD uses an extension of Verlet known as SHAKE.54 This method is dynamically equivalent to RATTLE,55 which is symplectic.56 More specifically, RATTLE performs a post-processing on velocities, which is purely cosmetic, in the sense that it affects only the output velocities, but has no effect on the trajectory.57 In the NAMD implementation, only covalent bonds to hydrogen atoms are allowed to be constrained, thereby reducing the frequencies of the fastest bonded interactions while avoiding any additional parallel communication, since each cluster of atoms to be constrained is in close enough proximity to be kept together on a single processing element. A consequence of removing these hard degrees of freedom by means of holonomic constraints is the possibility to utilize longer time steps to integrate the equations of motion. Generally, the use of SHAKE requires an iterative process to satisfy all constraints. To rigidify water molecules, the constrained equations of motion are solved analytically, employing a formulation known as SETTLE.58
4. Stochastic dynamics
Commonly used to simulate a heat bath, the Langevin dynamics implementation of NAMD is based on a second-order accurate extension of Verlet, known as the Brooks–Brünger–Karplus (BBK) scheme.59 A less computationally demanding approach incorporated in NAMD and referred to as stochastic velocity rescaling60 leans on a stochastic process to infer the rescaling parameter. NAMD also employs Langevin dynamics to control piston fluctuations for controlling pressure in the context of the Langevin piston method.61
B. Thermostats and barostats
NAMD provides mechanisms to control temperature and pressure in a way that generates the correct ensemble distribution. For isothermal simulations, thermostat control is provided by Langevin dynamics or, alternatively, by stochastic velocity rescaling.60 Langevin dynamics is advantageous for parallel scaling since no additional communication is required. Moreover, the friction term that appears in the Langevin equation tends to enhance dynamic stability. The method also lends itself to a great deal of flexibility, where different parts of the computational assay, e.g., the solute and the solvent, can be handled using different coupling coefficients defined on a per-atom basis. However, the computational cost of the integration is increased with respect to the basic Verlet implementation due to the need for a Gaussian-distributed random number for each degree of freedom and at every time step.
NAMD provides a less costly alternative in the form of the stochastic velocity rescaling method,60 which is an inexpensive stochastic extension of the weak-coupling Berendsen thermostat,62 requiring just two Gaussian-distributed random numbers for every rescaling of the velocities. The method has the virtue of being less disruptive to the underlying dynamics than Langevin dynamics, conserving both holonomic constraints and linear momentum. However, each rescaling involves a global broadcast of the rescaling parameter. Rescaling should not be done more often than the largest time step employed in the simulation and can in practice be done less often, corresponding to the update of the domain decomposition of the cell, which typically occurs every 20 time steps. The overall reduction in computational effort results in up to a 20% performance improvement for parallel, GPU-accelerated simulations, as demonstrated by the parallel scaling benchmarks on the Summit supercomputer at Oak Ridge National Laboratory depicted in Fig. 8.
FIG. 8.
NAMD scaling on the Summit supercomputer shows the performance advantage of using stochastic velocity rescaling (blue line) over the Langevin damping thermostat (red line), up to 20% faster for the same number of nodes.
Isothermal–isobaric simulations are handled in NAMD with an implementation of the Langevin piston algorithm,61 which combines the Hoover constant-pressure equations of motion63,64 with piston fluctuations controlled by Langevin dynamics.61 (Reference 64 details the difference between the Hoover formulation and the original Nosé–Andersen equations.65,66) The resulting equations of motion were independently proposed in another work, which proved that the correct ensemble is generated.67 To perform MD simulations in the isothermal–isobaric ensemble, the Langevin piston barostat must be used in conjunction with either of the aforementioned thermostats.
V. ELECTROSTATICS
Evaluation of electrostatic interactions represents a significant component of the computational effort in MD simulations. In this section, we review the algorithms implemented in NAMD to handle electrostatic forces, in particular, their long-range nature.
A. Periodic electrostatics
NAMD supports periodic boundary conditions, or PBCs, with periodicity in one, two, or three directions, as well as nonperiodic simulations. Periodicity gives rise to forces resulting from infinite sums of image charges. For periodicity in two or three directions, these forces are not well defined, the net force depending on the ordering of the terms. To have a well defined force, it is necessary to be more precise about the limiting process. The best such construction envisions a sphere of complete periodic cells and considers the limit as its radius goes to infinity. The result is the classic Ewald model68 plus a dipole term, the coefficient of which depends on the dielectric constant of the surroundings. Such a term seems inadvisable for solvated biomolecules, so a so-called “tin foil” boundary is assumed to ensure that it is equal to zero. For a system with a nonzero net charge, the limiting process remains valid—provided that a neutralizing uniform background charge is introduced, which has no effect on forces but introduces a constant correction to the electrostatic energy per periodic cell.69
B. Ewald summation
The energy and forces associated with the Ewald sum represent a significant part of the computational effort in an MD simulation. Ewald summation decomposes the Coulomb interaction kernel into a short-range part plus a long-range part, based on the error function erf, 1/r = erfc(βr)/r + erf(βr)/r, where erfc is the complementary error function. The short-ranged first term gives rise to a real-space summation between nearby pairs of atoms. The long-ranged and bounded second term gives rise to a summation of interactions between the charge densities of the unit cell and all of its periodic images, which converges rapidly in reciprocal space after applying a Fourier transformation. The parameter β is chosen to minimize the computational effort, yielding an operation count proportional to N3/2.70
1. Smoothed particle–mesh Ewald
NAMD reduces the operation count to 33 by employing the smoothed algorithm.71 PME achieves this speedup by replacing the complex exponential in the reciprocal space sum with a B-spline interpolant, which yields an approximation on a grid amenable to the use of the FFT.
2. Implementation of particle–mesh Ewald
To enhance the performance of PME, NAMD tabulates quantities used in the PME energy calculation and interpolates from these quantities, thereby avoiding the calculation of expensive transcendental functions, erfc and exp, during the simulation. The exact details, however, depend on the computer. In the CPU version of NAMD, cubic Hermite interpolation results in an energy function with continuous first derivatives, and its gradient is used to calculate forces. Conversely, the GPU version utilizes a linear interpolation of the force and an additional linear interpolation of the energy when needed for output. Having continuous forces is important for minimization and crucial for dynamics. Furthermore, having a force that is a gradient is equally crucial for Hamiltonian dynamics.
3. Performance of particle–mesh Ewald
The FFTs calculated by PME pose a challenge to parallel scaling due to communication requirements. A one-dimensional pencil decomposition of the three-dimensional FFT improves the scaling for larger assemblies of atoms over the two-dimensional slab decomposition originally supported by earlier versions of NAMD, where the FFTW library is employed to calculate the constituent FFTs on CPU cores. GPUs can be used with PME to calculate the spreading of charge from atoms to grid points and the gathering of the force from grid points to atoms. This use of GPU acceleration can outperform the CPU version when one doubles the grid spacing and increases the order of interpolation from 4 to 8, which maintains the expected accuracy and simultaneously increases the arithmetic intensity of the GPU while reducing the communication bandwidth by a factor of 8. There is also a version of PME for single-node MD simulations, which implements all kernels on the GPU and employs the NVIDIA cuFFT library for the FFT calculation. The performance of PME in the context of the adaptive, asynchronous programming model Charm++ is discussed in Sec. II.
4. Conservation of momentum
The best known fast methods for electrostatics are all based on a gridded approximation72 and, therefore, compute energies that are not translation-independent. Consequently, the total linear momentum is not expected to be conserved. NAMD, however, provides a simple device73 that conserves momentum without incurring energy drift of any significance (comparing to Sec. IV of Ref. 71).
C. Multilevel summation method
For simulations that are not periodic or periodic in only two directions, the use of FFT is less efficient. Additionally, FFT does not parallelize very well for very large molecular objects simulated on a large array of processors. These drawbacks are overcome by the multilevel summation method (MSM), which generalizes the basic idea of PME by decomposing the Coulomb interaction kernel into two or more parts of increasing length scale. The intermediate parts are approximated on meshes/grids of increasing coarseness, and these intermediate computations are performed in real space rather than in reciprocal space, exploiting the finite range of the intermediate parts of the kernel. The long part, of infinite range, is on a grid coarse enough for the computation to be carried out efficiently by an FFT or even directly. In practice, instead of erf(βr)/r, a softened kernel is employed,74 which has the virtue of creating short-range and intermediate-range kernels that are exactly zero beyond a given cutoff. NAMD implements MSM based on the use of piecewise polynomial interpolation having continuous first derivatives.75
D. Treatment of induced polarization
In addition to the simpler, pairwise additive force fields with fixed electric charges featured in the program, NAMD offers an extension that models induced electronic polarization using a classical Drude-oscillator approach.5,13 Potential functions that represent electrostatic interactions in terms of fixed effective partial charges are clearly limited by their ability to provide a realistic and accurate representation of both microscopic and thermodynamic properties, most notably when induced electronic polarization is expected to play a significant role. One approach to incorporate these effects is the classical oscillator model introduced by Drude,76 which addresses induction phenomena by introducing an auxiliary charged particle attached to each polarizable atom by means of a harmonic spring.13 One noteworthy advantage of this model is that it preserves the simple particle–particle Coulomb electrostatic interaction employed in pairwise additive force fields, allowing an implementation that is computationally simple and effective. The development and parametrization of the Drude force field over the last 15 years now includes water, ions, proteins, lipids, nucleic acids, and carbohydrates.77,78 To avoid the computationally prohibitive self-consistent field (SCF) procedure, which is normally required to treat induced polarization, an extended Lagrangian with a dual Langevin thermostat scheme applied to the Drude-nucleus pairs has been developed.5,13 This approach enables the efficient generation of classical trajectories that are near the SCF limit. To achieve SCF-like dynamics, it is critical that the cold thermostat act on the atom-Drude oscillator pairs rather than on the Drude particles directly.13 The implementation in NAMD is achieved by separating the dynamics of each atom-Drude pair with coordinates in terms of the global motion of the center of mass and the relative internal motion of the oscillator. This implementation has made possible the efficient simulation of very large molecular systems accounting for through-space induction phenomena.5
E. Generalized Born implicit solvation
NAMD features alternatives to an explicit description of the environment, chief among which is the generalized Born implicit solvent (GBIS) model.79 GBIS is a fast, albeit approximate, model for the calculation of electrostatic interactions within a solvent described as a dielectric continuum by means of the Poisson–Boltzmann equation. It allows large molecular objects to be simulated at a fraction of the cost of a model that would include explicit solvent molecules and is available in both the CPU and GPU versions of NAMD.47,80
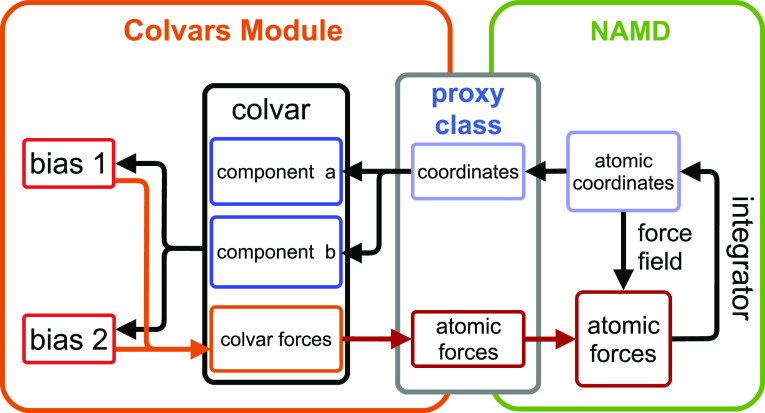
VI. THE COLLECTIVE VARIABLES MODULE
The collective variables module, or Colvars, is a contributed software library, which supports enhanced-sampling methods in a space of reduced dimensionality.6 Since its introduction as part of NAMD version 2.7, Colvars has provided the computational platform for most of the enhanced-sampling methods listed in Sec. VII and other derived methods.81 It is written primarily in C++ and included in the official source code as well as precompiled NAMD builds.
A. Principles of a biased simulation with Colvars
Simulations using methods implemented by Colvars require the end-user to choose and define two entities, namely, (i) at least one collective variable (CV) (colvar), which is a function of atomic coordinates, and (ii) a method that modifies the dynamics of the CV, i.e., a bias. There can be of course multiple variables or biases defined simultaneously. The Colvars module offers many choices of both variables and biasing methods, ranging from massively parallel compiled code to arbitrary functions chosen by the user at runtime; both are described hereafter (Fig. 9).
FIG. 9.
Graphical representation of a Colvars configuration.
B. Using VMD to prepare or analyze simulations
Setting up Colvars calculations with NAMD is now facilitated by the interactive Colvars dashboard in VMD, which provides helpers for preparing a configuration describing the CVs. The configuration can be saved to a file that is directly readable by NAMD for biased simulations. The Colvars dashboard also facilitates the analysis of NAMD simulations, regardless of whether these were carried out with Colvars. It is distributed with VMD, starting with version 1.9.4.
C. Parallel performance
When NAMD is run in parallel on multiple nodes, only one instance of the Colvars module is run on the first, master node. This requires all-to-one communication of selected atomic coordinates, or their partial contributions to the CVs, and one-to-all communication of the biasing forces on the atoms. Updating the Colvars module is executed asynchronously alongside the force calculation by NAMD, resulting in efficient latency hiding.
Two types of parallelization schemes are implemented in Colvars/NAMD, addressing, respectively, cases of CPU and communication bottlenecks. On multicore platforms that support shared-memory parallelism, calculation of multiple CV components and biases is distributed over all cores of the first node. This is helpful when several computationally expensive variables or components are defined. Separately, when CVs depend on atomic coordinates only via the centers of mass of groups of atoms, the centers of massare calculated in parallel using NAMD functions. There, a single biasing force is computed for each group and is distributed onto the constituent atoms only within each node that carries them. This arrangement achieves parallelism without communicating the entire molecular system over all nodes, preserving the capability of NAMD to treat very large computational assays. Similarly, biasing forces can also be applied indirectly to atoms via volumetric maps, as will be detailed hereafter.
D. Scripting interface
Colvars takes advantage of the NAMD embedded scripting interpreters to enable custom extensions without the need to write and compile C++ codes. These custom extensions may take two forms: (i) directives in the main NAMD script and (ii) callbacks to user-defined functions. Scripting directives are typically employed to control simulation workflows based on CVs, forming the basis for the implementation of numerical schemes such as the string method with swarms of trajectories82 or the adaptive multilevel splitting algorithm.83,84 The primary scripting language of Colvars in NAMD is Tcl. Colvars can also be called from a Python script indirectly through the Tcl interface. In response to a growing demand from a broad community of users, Python objects will be made available in the near future, in conjunction with improved Python support by recent NAMD builds.
Complementary to workflow control, user-defined functions, i.e., callbacks, can also be used by the Colvars library. Such scripted functions provide the framework for the implementation of custom-tailored CVs and biasing algorithms without the necessity to recompile NAMD. Compared with tclForces and tclBC, Colvars-based callbacks carry the advantage that custom variables and biases are typically calculated in low-dimensional spaces. They have, therefore, minimal performance overhead because atom-level processing is done by C++ functions of Colvars and NAMD. A simpler but slightly less flexible way to define variables as custom mathematical functions of existing components is provided using the lightweight expression parsing library Lepton written by Eastman.85 Using Lepton requires no knowledge of programming at all as new variables can be expressed in conventional mathematical notation. Gradients of such custom-tailored variables are calculated transparently using automatic differentiation. The example configuration below defines a CV based on two components, namely, the scalar distance, d, between two atom groups and the vector distance, r, between the same groups. The value of the CV is the unit vector joining the two groups, the individual scalar components of which are calculated by three custom functions.
| colvar { |
| name myUnitVectorColvar |
| # Uses two predefined basis functions, |
| # scalar and vector distance |
| distance { |
| name d |
| # This quantity is referred to by its |
| # name ’d’ in custom functions |
| group1 { atomNumbers 1 2 3 4 } |
| group2 { atomNumbers 5 6 7 8 } |
| } |
| distanceVec { |
| name r |
| # Scalar components of the vector r are accessed |
| # as ′r1′, ′r2′, and ′r3′ |
| group1 { atomNumbers 1 2 3 4 } |
| group2 { atomNumbers 5 6 7 8 } |
| } |
| # Together, the 3 instances of customFunction |
| # define a 3-vector colvar |
| customFunction r1 / d |
| customFunction r2 / d |
| customFunction r3 / d |
| } |
E. Projecting atomic forces on collective variables
A key feature of Colvars is the projection of total atomic forces onto specific CVs, forming the basis of the classic thermodynamic integration (TI) free-energy estimator.86 This estimator is typically used in the adaptive biasing force (ABF) method.87–89 Starting with NAMD 2.13, it can also be used in combination with other methods, including steered MD (SMD),90 umbrella sampling,91 and metadynamics92 (see Sec. VII).6
Because the projection of total forces requires the fulfillment of certain mathematical conditions,88,93 the TI estimator cannot be used directly in some cases. However, variables can still be coupled to an extended Lagrangian system, the forces of which approximate then the true total forces of the variables for estimating the free energy, as is done, for instance, in the extended-system ABF algorithm,94 implemented in Colvars.95 The free energy can then be recovered from unbiased estimators.95,96 In two- and three-dimensional cases, the free energy is now automatically computed based on estimates of its gradients. To work around the problem that the inexact numerical estimate of the gradients is not generally the gradient of a scalar field,97 the free-energy landscape is obtained as the solution of a Poisson equation, stating that the Laplacian of the free energy is equal to the divergence of the gradient estimator, subject to appropriate boundary conditions (periodic or non-periodic depending on the CVs in use).
The collection of total forces requires that Colvars computations are carried out at the end of the force calculation by NAMD, which would introduce an additional latency. To circumvent this shortcoming, total forces computed at the previous time step, rather than the current one, are utilized by Colvars. This approach is specific to NAMD and is not currently used by Colvars in other MD engines. The one-step lag is inconsequential for methods that rely on force averages over many time steps. This detail must, however, be kept in mind when the time series of total forces is important.
A second noteworthy detail is that the total forces computed by NAMD include contributions both from the force field and any externally applied forces. Colvars automatically subtracts its own biasing forces under the most typical scenarios, when the TI free-energy estimator is employed by a single enhanced-sampling method, e.g., ABF. Otherwise, it should be noted that any external restraints will be accounted for by the TI free-energy estimator,86,93 thereby potentially introducing a bias.
F. New and notable coordinates
The basis set of coordinates provided by the C++ Colvars library has been extended with spherical polar coordinates, employed, for instance,98 in a restraint scheme for standard binding free-energy calculations,99 dipole-moment magnitude and direction,100 and geometric path-based variables.101 Independently, path-collective variables102 are available in Tcl as scripted functions. The variable that calculates coordination numbers (coordNum) has quadratic complexity in the number of atoms involved, with a potentially high computational cost. It can now be computed at a tunable level of approximation using pair lists, drastically improving its performance.
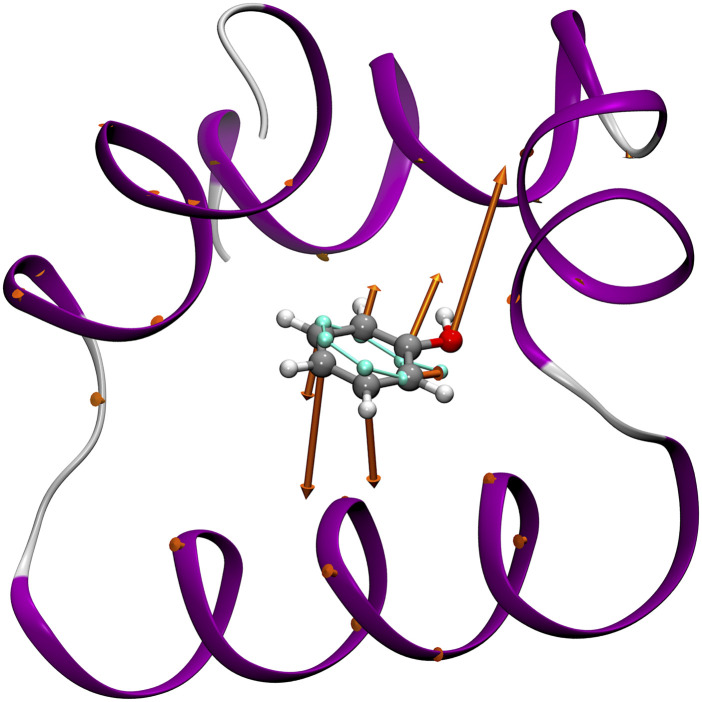
Any CV implemented by Colvars can be calculated in a moving frame of reference tied to a set of atoms in the molecular system. This way, any external degree of freedom can be turned into an internal degree of freedom of the relevant subsystem (the reader is referred to Ref. 6 for further details). This approach facilitates the description of the relative motion of molecular objects and is the foundation of the Distance to Bound Configuration (DBC) coordinate for absolute binding free-energy calculations, which measures the deviation of translational, rotational, and conformational degrees of freedom of the ligand (see Fig. 10).81
FIG. 10.
Gradients of the Distance to Bound Configuration (DBC) coordinate for a phenol molecule bound to T4 lysozyme. The reference pose of phenol heavy atoms is shown in light cyan. The arrows are proportional to the derivatives of the CV with respect to atomic Cartesian coordinates. The small gradient contributions on protein Cα carbons illustrate the purely roto-translational counter-forces exerted on the receptor when biasing forces are applied to a DBC coordinate.
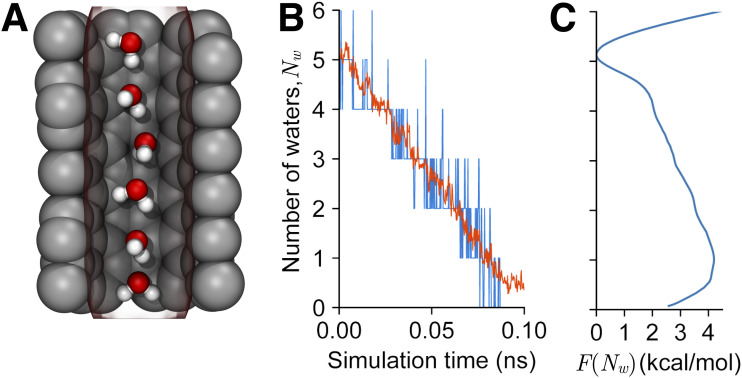
Certain phenomena, such as protein–solvent interaction or membrane dynamics, require dynamical selections of the relevant atoms. Toward this end, volumetric maps (see Sec. XIII) may be used to define CVs that, for instance, “count” the number of atoms within an arbitrary region of space. The recently introduced Multi-Map variable103 utilizes this approach to study the deformation of lipid membranes over biologically relevant scales, as well as solvent-density fluctuations in confined cavities (see Fig. 11). This functionality will also serve as the basis for new sampling methods based on electron density maps (see Sec. XIV).
FIG. 11.
CVs for solvent reorganization based on volumetric maps. Snapshot of a hydrophobic cavity containing a varying number of water molecules (a). A volumetric map (red transparent contour) is used to define a continuous variable measuring the number of molecules in the cavity. Trajectory of a short SMD simulation on (red), compared to the number of molecules counted with VMD (blue) (b). PMF of , computed over ∼400 ns. The use of gridForces maps (see Sec. XIII) in Colvars allows the development of methods for enhanced sampling of fully dynamic aggregates, such as water densities and lipid membranes103 (c).
G. New and notable biasing methods
In addition to the biasing methods originally described in the Colvars reference,6 other notable methods have been introduced more recently, particularly in the context of introducing experimental constraints into the simulation. Leveraging tools added to NAMD for other replica-exchange algorithms (see Sec. X), the multiple-walker (MW) version of ABF104 has been introduced for free-energy calculation encumbered by hidden barriers (see Sec. VII).105 Two new methods target directly a certain probability distribution via harmonic restraints on the probability distribution computed over multiple copies of certain atoms106 or by the more general ensemble-biased metadynamics method.107 Alternatively, experimental restraints may be enforced following the maximum-entropy principle as a constant-force, linear restraint.108 The magnitude of the force can be learned automatically within the simulation, such as in the adaptive linear bias (ALB) method109 and in the restrained-average dynamics (RAD) method,110 which further reduces nonequilibrium effects by incorporating the experimental uncertainty in the biasing forces.
VII. ENHANCED SAMPLING METHODS
NAMD provides a large set of methods and algorithms to enhance, boost, and accelerate the natural molecular motions during MD simulations. Depending upon the context and implementation, several of these methods may be used to enhance conformational sampling while remaining consistent with a Boltzmann equilibrium distribution. A broad set of approaches, referred to as Hamiltonian tempering,111 aim at enhancing configurational sampling via a modification of the underlying potential energy function of the system. Those include accelerated MD112,113 (aMD) and its Gaussian variant114 (GaMD), which, as will be detailed in Sec. VIII, attempt to parametrically “lift” the energy floor of the potential function to make the energy wells more shallow yet without perturbing the energy barriers. Another method is solute tempering (REST2),115 which aims at enhancing sampling by scaling the solute–environment interaction energy to lower the barriers that separate conformational states. Hamiltonian tempering methods effectively attempt to smooth the potential energy surface in order to enhance sampling. Hamiltonian tempering methods can generate Boltzmann-distributed configurations either via a post-hoc re-weighting analysis or by generating the simulation within a Hamiltonian tempering replica-exchange scheme (see Sec. X).
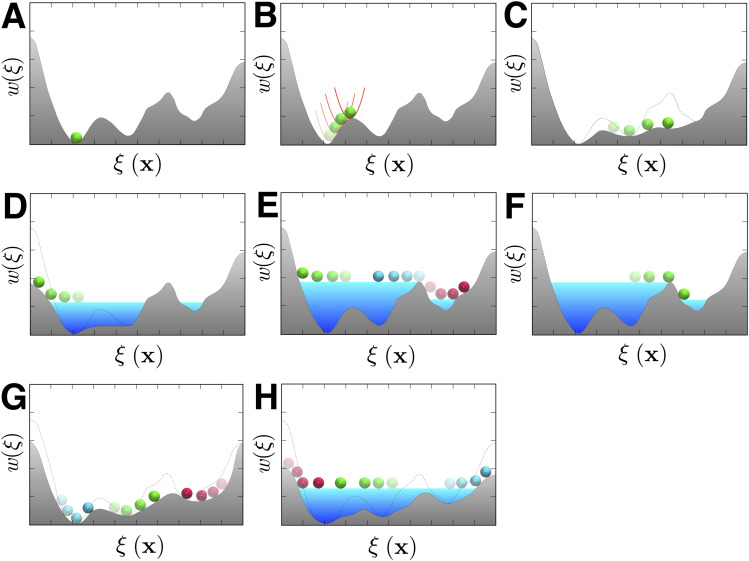
Studies of complex conformational transitions occurring on long timescales often proceed by identifying a geometric transformation associated with a general-extent parameter,116 , often referred to as the reaction-coordinate model117 and formed by collective variables.88 Several enhanced-sampling algorithms16,118,119 aimed at encouraging the exploration of relevant regions of configurational space along such a user-chosen reaction-coordinate model are available in NAMD. One of the most direct choices to boost the motion of a system along this reaction-coordinate model consists in applying a time-dependent non-equilibrium perturbation via SMD.90 In principle, such non-equilibrium SMD trajectories can be used to determine the equilibrium free energy via post-hoc analysis based on the Jarzynski identity,120 although reaching convergence may be challenging in practice. By far, the most widely used approaches to map the free-energy landscape along the reaction-coordinate model of complex biomolecular systems rely on configurational sampling in the presence of some biasing potential, following the general statistical technique of importance sampling,16,118,119 which seeks to estimate a particular distribution by sampling from a different distribution. The Colvars module,6 described in Sec. VI, serves as a repository for these importance-sampling approaches, as well as a toolkit for the implementation of new numerical schemes. Among the widely used algorithms associated with geometric transformations that are available in Colvars are umbrella sampling91,121 (US), metadynamics92,122,123 (MtD), well-tempered metadynamics124 (WT-MtD), and ABF87,89 and its different variants95,96,125,126 (see Fig. 12).
FIG. 12.
Sampling of a rugged free-energy landscape. Boltzmann sampling favors low-energy regions (a). Free-energy barriers can be overcome by depositing harmonic potentials, as is done in US (b); by applying a time-dependent bias that yields a Hamiltonian bereft of an average force along ξ(x), as is done in ABF, or its extended-Lagrangian variant, eABF (c); or by flooding valleys by means of Gaussian potentials, or hills, as is done in MtD (d). Combination of MtD and eABF, meta-eABF, concurrently shaves free-energy barriers and floods valley (e). To enhance ergodic sampling, a multiple-walker extension of MtD (f), ABF or eABF (g), and meta-eABF (h) has been implemented.
Importance sampling approaches such as US, MtD, and ABF, which promote configurational exploration by means of a bias along a chosen direction ξ(x), remain plagued by slow degrees of freedom in directions orthogonal to ξ(x).127 For instance, misrepresentation of the true conformational transformation associated with the molecular transition in terms of a naive reaction-coordinate model, ξ(x), consisting of a single collective variable, may often result in severe sampling nonuniformity.118,119 While it is never possible to formally guarantee a complete ergodic sampling, multiple-walker (MW) strategies, e.g., MW-MtD128 or MW-ABF,104,105 are available to address this issue. A common denominator is the combination of information accrued by the different walkers, namely, the Gaussian-potential weights and widths in MW-MtD, and the free-energy gradients in MW-ABF. Those algorithms, directly embedded in Colvars,6 can be brought to a higher level of sophistication by means of Tcl scripting. For instance, Darwinian selection rules clone good walkers that cover large stretches of ξ(x) while eliminating the less efficient, kinetically trapped ones.105 A number of replica-exchange strategies built on the powerful multiple-copy algorithm129 (MCA) infrastructure of NAMD may be used (see Sec. X). These strategies include multi-canonical temperature and Hamiltonian tempering replica-exchange MD111,130–132 (REMD) and bias-exchange window-swapping umbrella sampling133 (BEUS). Available MCA sampling algorithms may be extended via the Tcl scripting interface.
All the sampling methods described above rely on a standard microscopic isothermal MD propagator to evolve the atomic configuration of the system according to the Newton classical equations of motion. Another type of isothermal propagator, which leans on a combination of Metropolis–Hastings Monte Carlo (MC), with proposed moves generated by non-equilibrium MD (neMD) switches134 may be employed to further enhance conformational sampling. This hybrid neMD-MC propagator may be combined with any number of enhanced sampling functionalities of NAMD, e.g., ABF.135 At the heart of the hybrid propagator is the neMD switch, during which the molecular system is evolved under the influence of a time-dependent Hamiltonian that is perturbed for a brief period of time. The resulting configuration at the end of the neMD trajectory is then accepted or rejected according to a Metropolis criterion. A symmetric two-end momentum reversal prescription is used at the end of the neMD trajectories to guarantee that the algorithm obeys microscopic detailed balance to yield the proper equilibrium Boltzmann distribution.134 The hybrid neMD-MC sampling propagator with the Hamiltonian tempering scheme aMD and REST2115 is available in NAMD in the form of Tcl scripting. Additional variants of neMD-MC involving the exchange of molecular species—e.g., lipid swapping in membrane models, may be implemented through the alchemical free-energy perturbation (FEP) module and Tcl scripting.
As an illustration of an enhanced-sampling application with NAMD, we have simulated the permeation by a drug-like molecule, 2′, 3′–dideoxyadenosine (DDA), of a membrane model consisting of 1–palmitoyl–2–oleoyl-phosphatidylcholine:cholesterol mixture at a 2:1 ratio, employing both the ABF and its recent variant that combines MtD92,122,123 and the extended-Lagrangian version of ABF,95,96 coined meta-eABF.7,126 The potential of mean force (PMF) characterizing the permeation events was obtained by discretizing the reaction-coordinate model, chosen as the Euclidian distance between the center of mass of DDA and that of the bilayer projected onto its normal, z, into bins 0.1 Å wide, wherein samples of the local force were accrued. To improve the efficiency of the free-energy calculation, the reaction pathway spanning −45 Å ≤ z ≤ +45 Å was stratified into nonoverlapping windows. However, whereas nine windows were utilized with the ABF algorithm, the aggressive sampling of MtD in meta-eABF simulation allows a coarser stratification scheme to be employed, with only three windows in this particular instance. Details of the simulations can be found in Refs. 136 and 126. Compared to Ref. 136, sampling has been increased to 4.5 μs to reduce to less than 0.4 kcal/mol the hysteresis in the PMF between the upper and the lower leaflets. The meta-eABF free-energy profile was obtained in 1.5 µs, though as early as 0.8 µs, the hysteresis was equal to kBT. As shown in Fig. 13, the two PMFs almost perfectly overlap, differing only by 0.1 kcal/mol at the barrier (z = 0) and by 0.2 kcal/mol in the humps of the interfacial region (z = ±24 Å). The virtual absence of difference between the PMFs determined with ABF and meta-eABF underscores the efficiency of the latter algorithm, being able to map a complex free-energy landscape 3–6 times faster than the former.7,126 It is also worth noting that this class of enhanced-sampling methods based on time-dependent biases acting on CVs allows kinetic models to be built, obviating the need for additional simulations. For further details, the reader is referred to Refs. 137 and 138.
FIG. 13.
Potential of mean force characterizing the permeation of a 1–palmitoyl–2–oleoyl-phosphatidylcholine:cholesterol model membrane by 2′, 3′–dideoxyadenosine, obtained using ABF (red line) and meta-eABF (blue line) (a). The reaction-coordinate model is the projection onto the z-axis of the distance separating the center of mass of the permeant from that of the lipid bilayer (b).
VIII. IMPLEMENTATION OF ACCELERATED MD IN NAMD
Accelerated MD (aMD) pertains to the family of enhanced-sampling methods discussed in Sec. VII and smoothens the potential energy landscape of a system through adding a boost potential, Uboost(x), to the original potential, U(x), whenever the latter falls below a certain threshold E. In the original version of aMD,139 Uboost(x) takes the following form when U(x) < E: , where α is a user-defined acceleration factor that fine-tunes the modified potential energy landscape, U*(x). The smaller the α, the more flattened the U*(x) becomes, while as α increases, U*(x) asymptotically approaches the original potential, U(x). The ensemble average of an observable can be recovered through reweighting: . Here, β = 1/kBT, where kB is the Boltzmann constant and T is the temperature, and ⟨⋯⟩* represents the ensemble average with the modified potential.
Available since the 2.8 release of NAMD,113 the above version of aMD has been applied to investigate a range of biological objects, such as lipid mixtures,140 the maltose binding protein,141 the ubiquitin ligase,142 and the dopamine transporter.143 The overhead for performing an aMD simulation, relative to a standard MD run, is only ∼10% on average.113,114 However, a major limitation has been the convergence of the reweighted ensemble average—the numerical evaluation of equations like the previous one is known to be challenging due to the strong non-linearity of the exponential terms.144,145 Much effort has been invested to address this reweighting problem in aMD.146,147 Starting with version 2.12, the Gaussian accelerated MD (GaMD) method148 has been implemented in NAMD.114 GaMD differs from the original aMD version in three main aspects: First, the boost potential in GaMD adopts a harmonic form when U(x) < E: , where k is a force constant. Second, GaMD employs a second-order cumulant expansion to perform reweighting, which is more accurate than an exponential average when Uboost(x) follows a near-Gaussian distribution.146 Third, unlike in the original version of aMD, where the end-user must provide all parameters, GaMD automatically determines key parameters based on statistics collected from a series of short preparative simulations. The user only needs to specify the length of the preliminary runs and σ0, the upper limit for the standard deviation of the boost potential. A smaller σ0 tends to enforce a more stringent requirement on the normality of the boost potential distribution.
In both the original aMD and GaMD implementations, three modes for applying the boost potential are available: (i) boosting the dihedral potential, (ii) boosting the total potential, and (iii) boosting both, i.e., the so-called “dual-boost,” with separate parameters controlling the boost level for each term. Modes (i) and (iii), in which the dihedral potential is singled out to enable direct manipulation of the torsional degrees of freedom, are used most frequently. Overall, the dual-boost mode affords the highest boost potential, which may produce the greatest effect of enhanced sampling, but simultaneously poses the biggest challenge for reweighting. With the GaMD implementation, the dual-boost mode has been applied on molecular objects ranging from short peptides to G-protein coupled receptors, with normality tests revealing minimal anharmonicity of the resulting boost potential distributions.148
Unlike many other enhanced sampling methods, aMD does not require a predefined reaction-coordinate model. This feature confers to it the flexibility to explore the available conformational space in a somewhat “unrestrained” manner. In particular, it renders the method suitable for complex objects without knowledge or obvious choices of collective variables. However, this very feature also means that the sampling effort in aMD may be less focused along a given reaction-coordinate model, which, in turn, hampers convergence of the resulting free-energy profile. In order to address this issue, a promising and actively pursued direction is the combination of aMD with other enhanced-sampling methods, which can be applied either sequentially149,150 or simultaneously.135,151
IX. ALCHEMICAL FREE-ENERGY CALCULATIONS
Alchemical free-energy calculations16,118,119 are aimed at transforming chemical species through a virtual process involving intermediate species that are unphysical. Many types of calculations are possible and supported by NAMD, depending on the transformation process linking the end states, altering either the intermolecular interaction as a whole or the individual force field parameters within a single-152 or dual-topology153 framework (see Fig. 14). Dual-topology indicates that the covalent structure of the two alchemical end states is intact and present at all stages of the calculations, while single-topology is meant to indicate that a covalent scaffold common to the two end states is being used. The choice of one or both of these frameworks can have significant effects on statistical efficiency, and one or the other might intuitively seem better suited for a particular problem.118 Indeed, for computational efficiency, the transformation is often carried out on a hybrid molecule formed by a single-topology common scaffold attached to two distinct dual-topology moieties. Nonetheless, by virtue of free energy being a state function, both the single- and dual-topology paradigms are valid approaches. While the single-topology framework generally seems to imply a lesser perturbation, its setup and implementation require particular care compared to its dual-topology counterpart.
FIG. 14.
Single- and dual-topology frameworks for alchemical transformations in NAMD. When the reference and the target states are chemically different and their respective charge distributions are distinct, a minimum common substructure is sought in the dual-topology paradigm [(a) and (c)]. The two topologies coexist in a hybrid compound, albeit do not interact, either directly (through nonbonded contributions) or indirectly (through bonded contributions). Unchanged (black), incoming (blue), and outgoing (magenta) atoms are defined in three partitions. In some extreme cases, when the end states are chemically distinct and no common substructure can be found, the two topologies are introduced independently, and geometric restraints are enforced to prevent them from drifting apart from each other. In the single-topology paradigm [(b) and (d)], holonomic constraints are applied to the common substructure (black), and dummy particles are introduced to describe the alternate state. As the transformation proceeds, these dummy particles become real, interacting atoms. To avoid the creation of an unphysical net force, the alternate state is only partially chemically bonded to the rest of the molecule.
NAMD supports a wide variety of methodologies for computing the free-energy difference between alchemical end states. The two most conventional approaches are thermodynamic integration86 (TI) and energy difference methods based on so-called free-energy perturbation154,155 (FEP). In FEP, the free-energy change between the reference state, 0, and the target state, 1, writes, . Here, ΔU is the perturbation, which is the difference between the potential energy of the reference state and that of the target perturbed state, and ⟨⋯⟩0 denotes an ensemble average over configurations representative of the reference state. In practice, even for modest perturbations, the transition between the end states is stratified into nonphysical intermediates by means of a transformation parameter λ to reduce the variance of the free-energy estimate. Convergence of the free-energy calculation is intimately related to the number of intermediates, ensuring suitable overlap of the underlying configurational ensembles. In TI, the free-energy change between the end states is expressed as an integral, , which, in practice, is also determined using finite increments of the coupling parameter. Here, the number of points is a trade-off between the accuracy of the numerical integral and the computational cost. To circumvent singularities arising from particles appearing in a locus already occupied, for instance, by solvent molecules, the Lennard-Jones potential is simultaneously shifted and scaled,156 resulting in a smooth transition between the chemical states. From a practical standpoint, the alchemical transformation can be carried out in a sequential fashion from 0 to 1, or in the opposite direction, by means of Tcl scripting, avoiding a restart of NAMD between intermediate states. Under these premises, the end-user takes advantage of NAMD performance on parallel architectures to handle the computational assay at a given value of the coupling parameter λ.
An alternate view to FEP is to note that ΔU is essentially the instantaneous work required to carry out particular perturbation in question.157 Following Jarzynski,120 it is possible instead to define a protocol such that the perturbation is carried out over a certain amount of time in a fashion that disrupts the equilibrium. In this case, the FEP equation154,155 still holds, but ΔU is instead replaced by the observed work required to carry out the transformation and the ensemble average represents initial conditions for multiple perturbations—which is still an equilibrium ensemble. The potential advantage of this approach is that it eliminates the need for stratified intermediate states. However, the accuracy and efficiency are highly contingent on the intrinsic timescales needed to carry out the transformation effectively.
Assuming suitable computational resources, it is also possible to run the different intermediate states concurrently within the multiple copy algorithm129 (MCA) infrastructure of NAMD (see Sec. X). Optimally, each replica is handled by an array of computing cores commensurate with the system size. The data generated in an alchemical transformation, either sequentially or concurrently for all strata, are generally post-processed to obtain a better estimator of the free energy than that of FEP, e.g., Bennett acceptance ratio158 based on bidirectional free-energy calculations,157 as well as estimates of the associated statistical and systematic errors. For performance purposes, it might be desirable to carry out the forward and the backward transformations concomitantly. One possible option provided by NAMD is the interleaved double-wide sampling algorithm,81 which switches the target value of lambda on the fly to supply energy differences in both directions of change, which are necessary to calculate statistically optimal free-energy estimators, in a single simulation. An interesting characteristic of importance-sampling algorithms such as FEP and TI is that they can be melded seamlessly with numerical schemes aimed at improving ergodic sampling.118,119 Running simultaneously the different intermediate states of the alchemical transformation, it is natural to combine the free-energy method with a replica-exchange algorithm, e.g., Hamiltonian-exchange,131 temperature-exchange,130 or both.
REMD111,130–132 has proven to improve the convergence of FEP calculations involving considerable reorganization of the surroundings. The Hamiltonian-exchange or FEP/(λ, )–REMD algorithm has been introduced, primarily for ligand binding, wherein replicas along the alchemical thermodynamic coupling axis, λ, are spawned as a series of Hamiltonian–boosted copies along a second axis to form a two-dimensional replica-exchange exchange map (see Fig. 15).132 Aiming to achieve a similar performance at a lower computational cost, a modified version of this algorithm has been implemented, in which only the end states along the alchemical axis are augmented by boosted replicas. The reduced FEP/(λ, )–REMD numerical scheme, with a one-dimensional unbiased alchemical thermodynamic coupling axis, λ, is introduced in the context of the generic MCA chassis of NAMD (see Sec. X). Different Hamiltonian-tempering boosting schemes could be employed to accelerate the convergence of the free-energy calculation, e.g., a potential-energy rescaling of a selected subset of the computational assay with REST2115 and the introduction of biases flattening the torsional free-energy barriers.
FIG. 15.
Hamiltonian-exchange FEP calculations or FEP/()–REMD (see Sec. X). The computational assay is cloned into replicas corresponding to the different strata of the alchemical transformation whereby the interaction of a small molecule with its environment is progressively turned off. The configurations of adjacent λ-states, i and j, are swapped periodically, and the proposed move is accepted following a Metropolis–Hastings criterion, .
Historically, alchemical transformations in NAMD were introduced through the dual-topology framework,159 but a number of applications, e.g., in drug discovery and in constant–pH MD,160 have provided an impetus for the introduction of an alternate scheme, now available in the free-energy module of the code. An effective hybrid single-dual topology protocol has been designed for the calculation of relative binding affinities of small ligands to a receptor.161 The protocol has been developed as an expansion of NAMD, which hitherto exclusively supported a dual-topology framework for relative alchemical FEP calculations. In this protocol, the alchemical end states are represented as two separate molecules sharing a common substructure identified through maximum structural mapping. Within the substructure, an atom-to-atom correspondence is established, and each pair of corresponding atoms is holonomically constrained to share identical coordinates at all time throughout the simulation. The forces are projected and combined at each step for propagation. To enhance sampling of the dual-topology region, the FEP calculations can be carried out within a REMD strategy supported by the MCA framework of NAMD (see Sec. X), with periodic attempted swapping of the thermodynamic coupling parameter, λ, between neighboring states. This hybrid single-dual topology scheme combines the conceptual simplicity of the dual-topology paradigm with the advantageous sampling efficiency of the single-topology approach.
X. MULTIPLE COPY/REPLICA ALGORITHMS
Numerical schemes that couple the dynamical propagation of a set of copies of the computational assay of interest, referred here to as “replicas,” offer powerful and flexible strategies to characterize complex molecular processes. Such MCAs can be employed to enhance sampling, compute reversible work and free energies, as well as refine transition pathways. Widely used examples of MCAs include temperature and Hamiltonian-tempering REMD, i.e., T–REMD and –REMD;130,131,133,162 alchemical FEP with λ–replica-exchange, i.e., FEP/(λ, )–REMD;132,163 umbrella sampling with Hamiltonian replica exchange, i.e., US/–REMD;164 and the string method with swarms of trajectories (SMwST)82 to sample conformational transition pathways.
In Sec. IX, we have mentioned the use of MCAs in the context of FEP/(λ, )–REMD calculations with NAMD (see Fig. 15). Here, we outline how MCAs can be employed for SMwST simulations, whereby a putative transition path between basins of the conformational free-energy landscape can be refined. This pathway, or “string,” consists of a set of m discrete conformations or “images.” A set of CVs is introduced to reduce the dimensionality of the process at hand (see Sec. VI). The swarms result from n MD trajectories started from a single image, combined to yield an average drift and requiring communication between the n copies of that image. The average drift for the different images is utilized to update the string, following a redistribution of these images to remove any drift tangential to the path. This re-parameterization step also implies communication between the m images that form the string. The images are then prepared for the next optimization cycle by performing restrained simulations with the CVs to their updated values. The MCA implementation of this method utilizes m × n independent replicas that intercommunicate once at each iteration.
A robust and general infrastructure has been built upon the parallel programming system Charm++ upon which NAMD rests to enable the implementation of a suite of MCAs for MD simulations (see Fig. 16). Multiple concurrent NAMD instances are launched with internal partitions of Charm++ and located continuously within a single-communication world. Messages between NAMD instances are passed by low-level, point-to-point communication functions, accessible through the Tcl scripting interface of NAMD. The communication-enabled Tcl scripting provides a sustainable application interface for the end-users to set up generalized MCAs without the explicit need to modify the source code, thereby conferring to the present implementation both versatility and massive scalability.
FIG. 16.
Generic implementation of MCA in the Charm++ RTS. Multiple NAMD instances run concurrently, and each of them is executed by an independent Charm++ RTS (gray vertical lines). Replica exchange/inter-copy communications (red double arrows) are implemented through a Tcl scripting interface. comm_local denotes a local internal Charm++ partition, dedicated to be the communication layer of a single trajectory run/NAMD instance. comm_cross denotes a global Charm++ communication layer across all local partitions. A scripting interface of replica exchange is implemented with communication-enabled Tcl functions built on top of comm_cross.
Representative applications of MCAs with fine-grained, inter-copy communication structure, including global λ–exchange in FEP/(λ, )–REMD, window-swapping US/–REMD in multidimensional order-parameter space, and SMwST simulations, have been implemented with Tcl scripting on top of Charm++. Once Charm++ is initialized, each Charm++ processing element can be logically mapped onto a designated local partition. When Charm++/NAMD enters the inter-copy communication phase, all localized processing elements are mapped back to a global state. Furthermore, on leadership supercomputers, such internal partitions can benefit from performance gains from the low-level machine-specific communication library, such as the parallel active messaging interface (PAMI) on IBM Blue Gene/Q165 or the user generic network interface (uGNI) on Cray XK7.166
The Tcl interface of NAMD is intended to offer maximum flexibility to the end-user for the implementation of MCAs without touching the source code. For instance, through Tcl scripting, variables and expressions utilized initially to define options can be changed dynamically in the course of the simulation. Said differently, once the user-friendly Tcl communication commands are built on top of the low-level, point-to-point communication functions, the end-user can design generic MCAs without modifying or adding a single line of C++. Within an MCA Tcl script, inter-copy communications are executed by the Tcl replicaSend/Recv/Sendrecv functions on top of the Charm++ communication layer, and the end-user only needs to designate the communication partners for each copy. A significant advantage of the present Tcl-based MCA scheme is that the end-user can define virtually any type of MCA through simple Tcl scripting as all the parameters and biasing energy terms are already registered in the Tcl-scripting interface of NAMD. For instance, in an absolute free-energy calculation, the nonbonded scaling parameters for different atom types can be wrapped into a single-parameter unit, which will be exchanged along the entire alchemical reaction path. Similarly, multiple orthogonal order parameters can be alternatively exchanged in the context of a multidimensional US Hamiltonian exchange.
XI. CONSTANT–pH MOLECULAR DYNAMICS
In constant-pH MD simulations, selected ionizable sites are allowed to spontaneously change their protonation state as a function of time in response to the environment and the imposed pH. This is an increasingly popular approach for directly probing different protonation states in biomolecules. Indeed, several methods and implementations have been reported160,167–172 and employed to study pH effects on molecular conformation,173 ligand binding,174,175 as well as enzymatic activity.176 In practice, how these calculations are carried out and subsequently analyzed can depend rather significantly on the specific methodology being used. The implementation in NAMD retains as many characteristics of conventional MD as possible and, with only minor additions, shares all of the same system preparation tools and output formats. With only a few exceptions, which will be discussed hereafter, pH-dependent mechanical observables can be estimated directly from trajectory averages.
The method implemented in NAMD is based on a hybrid scheme,177 which consists of carrying out short nonequilibrium MD (neMD) switching trajectories to generate physically plausible configurations with changed protonation states that are subsequently accepted or rejected according to a Metropolis Monte Carlo (MC) criterion.160,172 The constant-pH neMD-MC method is essentially an elaboration of conventional MD, which is still utilized to sample new molecular configurations in an explicit solvent model (this functionality is not currently compatible with implicit solvent models). By using a symmetric momentum reversal prescription134 and a generalized Metropolis–Hastings criterion,178 the neMD-MC hybrid procedure rigorously captures the proper Boltzmann statistics while allowing the environment, e.g., the solvent, to relax according to its natural dynamics,179 i.e., the protonation states and configurations are sampled jointly. This combination builds upon the concept of “stochastic titration”180 and a switching protocol.177 NAMD also utilizes efficiency improvements in both the switching procedure and the way MC candidate states are selected.134,160,172 In particular, sampling can be improved by iteratively updating an estimate of the pKa of each titratable group, namely, an adjustable parameter referred to as the “inherent” pKa.160,172 These estimates need not be exact in practice, and a reference estimate from a single residue in solution is often suitable to guide sampling away from improbable protonation-state changes.
The constant-pH MD analog of atomic coordinates is the protonation site “occupation vector,” a sequence of ones and zeros indicating which protons are physically coupled to the system and which are simply modeled as dummy atoms. Usage of dummy atoms is a purely bookkeeping measure and does not impact the thermodynamics of the molecular objects at play in any way.160 By adding extra placeholder atoms, constant-pH MD trajectories can be easily visualized and analyzed with the myriad tools available for fixed particle count simulations, chief among which is VMD.19 The only caveat to this process is that the occupation vector must be considered as additional data in each trajectory frame used to filter out the cases in which a given proton is either interacting or non-interacting.
The occupation vector can also be utilized to construct simple and intuitive observables. For instance, the number of simulation frames in which a proton occupies a site can be directly related to the protonated fraction of a given state, i.e., the fraction of simulation time in which that state is occupied. A protonation state of interest can often be composed of the occupations from multiple sites, possibly on multiple residues. Since these occupations are discrete, there is no ambiguity in doing so. Ultimately, the protonated fraction or, more generally, the state occupation at multiple pH values constitutes a titration curve that can be analyzed to yield a pKa value, usually by fitting to a model like a Hill curve (see Fig. 17). The output from NAMD simulations at multiple pH values is also amenable to use with reweighting schemes such as the weighted histogram analysis method.160,183
FIG. 17.
Sample titration curves computed from a constant-pH MD trajectory of the BBL miniprotein181,182 reveal a range of protonation states over a modest pH range. Solid lines represent fits to a Hill curve, from which pKa values and Hill coefficients, n, are derived. The present results were obtained using the CHARMM36 force field and standard settings (e.g., rigid bonds with hydrogen, PME). An MC step was performed every ps, and a uniform switch time of 15 ps was used for each move (particular residues requiring longer/shorter times can also be specified). About 20 000 moves were attempted at seven pH values, leading to an aggregate of about 2 µs of MD.
XII. HYBRID QM/MM MOLECULAR DYNAMICS SIMULATIONS
Classical MD simulations, which rely on molecular mechanics (MM) force fields, are well suited for tackling most computational biophysics problems, from folding184 to the study of large macromolecular complexes,185 but fail when the electrons play an important role in the investigated phenomena.186 In particular, charge redistribution and creation or rupture of chemical bonds are two frequent problems that have limited the treatment at the MM level.187 In such cases, quantum mechanical (QM) calculations provide a much more detailed view of the chemical process at hand.188 The cost for a more detailed treatment offered by QM methods is a significant increment in the computational complexity, even for the smallest of proteins, making an ns-long full-QM MD simulation still impractical or outright prohibitive.
Partitioning a biomolecular system in MM and QM regions has become a common approach to balance precision and efficiency.189 Hybrid QM/MM simulations do just that by focusing the computational resources on atoms that play a significant role in the process of interest.190 In practice, the QM/MM approach augments the MM force field, restricting the electronic structure calculation at the QM level to a small part of the system, while the remaining atoms serve as an environment that can affect the electronic distribution in the QM region.189,191 The classical MD integrator then combines the QM potential and the MM force field potential to dictate the movement of the atoms over time, keeping the dynamics of the system at the classical mechanics level in the Born–Oppenheimer approximation.192
For most applications, hybrid QM/MM protocols are well-established and have been employed broadly to investigate a variety of biomolecular problems, most notably in enzymatic activity, including the mechanism of HIV integrases, glutamine synthetases, glycoside hydrolases, dioxygenases, lipases, dehydrogenases, catalases, glycosyltransferases, and nearly every other class of enzymes with known structure.193–199 The large list of enzymatic mechanisms that have been explored showcases the significant contribution of QM/MM methods to the development of enzymology.200 In addition, polarization effects have also been the focus of QM/MM simulations.201 By studying both lipids and drugs at the QM level, QM/MM approaches have been momentous in understanding how small drugs interact with lipid membranes, particularly on how local anesthetics are stabilized by dipalmitoylphosphatidylcholine (DPPC) lipids at water/lipid interfaces.202 More recently, the QM/MM implementation of NAMD was used to investigate a key step in the translation of the genetic code,203 revealing details of the aminoacylation mechanism of glutamyl-tRNA synthetase (GluRS). In this investigation, two QM regions were simulated simultaneously over multiple replicas of the same system: one for the active site of GluRS and the other for its anticodon recognition domain. This combination of biological system and software infrastructure made possible a unique analysis of the information transferred between the QM and MM regions, indicating that both levels of calculation were closely integrated to render a cohesive view of the biological object.
Despite the increase in interest and widespread applications, QM/MM simulations remain difficult to set up, most notably when a combination of methods is needed, e.g., QM/MM plus enhanced sampling (see Sec. VII). The philosophy behind the development of the NAMD QM/MM interface was to make hybrid methods available to all structural biologists while reducing the typical learning barrier and making this interface flexible and easily adaptable. The NAMD QM/MM suite includes an interface to multiple QM codes that can be combined with NAMD, orbital visualization tools within the program VMD,19 and the user-friendly QwikMD20 module (see Sec. XV) for preparing, running, and analyzing QM/MM simulations.203 The NAMD QM/MM interface also allows a large number of independent QM regions to be simulated concurrently, with a full integration of the vast collection of methods featured in NAMD, including SMD, enhanced-sampling, and free-energy estimation methods (see Sec. VII). Moreover, a reaction-oriented biased simulation method was introduced, providing a quick start for the study of reaction mechanisms.203
In hybrid QM/MM simulations, NAMD offloads part of its force and energy calculations to a quantum-chemistry package, referred here to as “QM software.”203 The QM software receives positions and elements of all atoms in the QM region and then returns partial charges, forces, and total energy. Using simple keywords, a user can direct NAMD to provide the positions and charges of the classical atoms that surround the QM region, allowing the QM calculation to be carried out in an electrostatic embedding. In case a covalent bond connects a QM and an MM atom, a link atom, typically a hydrogen atom,204 is introduced to cap the QM atom, and the classical partial charge from the MM atom is distributed over surrounding classical atoms (see Fig. 18). Different keywords control various aspects of the electrostatic interactions between the MM and QM regions, as well as the charge redistribution around the QM–MM bonds. All keywords are used in a regular NAMD configuration file, essentially expanding the classical MD simulation.
FIG. 18.
Schematic representation of a hybrid QM/MM MD step in NAMD. Two trialanines, each containing one QM region, are simulated through simultaneous and independent executions of the chosen QM software (ORCA). In a QM/MM simulation, positions and elements of QM atoms are sent by NAMD to the QM software, along with positions and partial charges representing the local MM environment within a given cutoff (dashed magenta or purple lines, top-left panel). NAMD expects from the QM software forces, total energy, and partial charges for all QM atoms, and forces acting on MM partial charges due to electrostatic interactions. All atoms are moved based on the calculated force gradients. Different atoms will have their resulting force gradient computed either by the QM software, by NAMD, or a combination thereof (top-right panel). If selected, long-range electrostatics can be calculated by NAMD for all atoms using PME, utilizing the updated partial charges calculated by the QM software. For additional details, the reader is referred to Ref. 203.
Taking advantage of the advanced state of current quantum-chemistry packages, NAMD can efficiently carry out a QM/MM MD simulation using a memory-backed temporary filesystem (“RAM disk”) to exchange input and output files with the chosen QM software. Most QM packages use “state files” to store the results of a single-point calculation and are optimized to be called sequentially, quickly loading state files from a previous iteration and using that information to achieve convergence faster. The underlying assumption is that atoms will move only slightly between two consecutive MD steps so that the previous result makes a good initial guess for the current calculation. The choice of using files in memory to communicate with the QM software has two main advantages: First, the time spent with file reading and writing is negligible, compared to a single-point calculation in an MD step. Second, it facilitates the integration with multiple QM software. To that effect, the NAMD QM/MM interface was built to communicate natively with MOPAC205,206 and ORCA207 and to provide a flexible standardized interface that allows virtually any quantum chemistry package to be wrapped in a script and used by NAMD.
The QM/MM implementation in NAMD has been extensively tested for accuracy, stability, and performance. Both MOPAC and ORCA were used to validate results, and all tests were thoroughly described in Ref. 203. Energy conservation was observed using a variety of simulation protocols, and trajectory stability was assessed by conserving energy in simulations up to 100 ns long. Moreover, tests with NAMD/ORCA at the PM3, HF, and density functional theory (DFT) levels of approximation revealed that long-range electrostatics could be safely integrated into the simulation using PME (see Sec. V) with little impact on energy conservation. As expected, benchmarks indicated that performance depends heavily on the chosen QM method and the size of the QM region. For very small systems, i.e., 12 QM atoms, our benchmarks showed a performance of up to 10 ns/day of QM/MM simulation when employing NAMD/MOPAC with the PM7 semi-empirical method208 and running on a 4.2-GHz Intel Core i7-7700K CPU, with 64 GB of RAM.
XIII. GRID-BASED POTENTIALS
While most MD simulations operate with particles and particle-based interactions, there is a world of continuum models used throughout all branches of basic science and engineering. The gridForces module of NAMD makes it possible to couple a particle-based MD simulation to external grid-based potentials, opening exciting possibilities for multi-scale and multi-physics simulations of biomolecular objects and offering an extreme flexibility for user customization of external forces in an MD simulation.
The gridForces module of NAMD was initially developed to facilitate simulation of single-stranded DNA (ssDNA) passage through the nanopore of α-hemolysin,18 a water-filled transmembrane channel [see Fig. 19(a)]. In experiment,212 ssDNA passage through the nanopore is driven by a gradient of an electric potential that varies dramatically at the entrance and within the nanopore [see Fig. 19(b)]. The ms-timescale of the translocation process rules out a brute force MD approach, whereas conventional accelerated dynamics approaches, such as constant force pulling or SMD, fail to produce realistic translocation events, owing to ssDNA stretching under the force.18 The solution—referred to as grid-steered MD (G-SMD)—was to first determine the average distribution of the electrostatic potential within the nanopore and then use this distribution as an external potential, magnified by a custom factor, to guide DNA through the nanopore. Using the G-SMD protocol, multiple complete translocation events of ssDNA through α-hemolysin were obtained within just tens of ns while introducing minimal distortions to the DNA conformations. In this particular example, the gridForces module was employed to enable simulations at, effectively, very high transmembrane biases without producing high electric-field artifacts, such as membrane rupture. A step-by-step guide to using G-SMD for nanopore transport simulation can be found in a dedicated tutorial on nanopore modeling.213,214
FIG. 19.
Using grid-based potentials to steer an MD simulation. (a) A cut-away view of an all-atom simulation system featuring a single DNA strand (orange) passing through an α-hemolysin nanopore (blue) embedded in a lipid membrane (green). Water and ions are not shown for clarity. (b) Electrostatic potential contour map in and around an α-hemolysin nanopore determined using all-atom MD methods.209 Coupling this potential to atoms of ssDNA only through the gridForces module of NAMD dramatically accelerates the simulation of the DNA translocation process.18 (c) The cryo-EM density (white wireframe surface) is used as a steering potential for docking the all-atom structure of an α/β (red/blue) tubulin dimer to build an all-atom model of a microtubule.210 (d) All-atom MD simulation of plasmonic trapping. Electric field-driven translocation of DNA (blue and orange) through a nanopore in a solid-state membrane (gray) is halted by the plasmonic field (semi-transparent red surface) generated by the gold bowtie nanostructure (yellow). The arrows illustrate the magnitude and direction of plasmonic forces acting on the DNA molecules.211 Water and ions are not shown for clarity.
Being able to apply position-dependent forces according to a custom three-dimensional potential in an all-atom MD simulation became a game changer for the field cryogenic electron microscopy (cryo-EM) reconstruction,215 where crystallography-resolved all-atom models of proteins or nucleic acids are modified to match a three-dimensional density determined using cryo-EM [see Fig. 19(c)]. The molecular dynamics flexible fitting (MDFF) method,17 which will be detailed in Sec. XIV, uses the experimentally determined density as an external potential in a G-SMD simulation of a crystallographic structure to obtain the best match between the atomic coordinates and the electron density. Originally developed for implicit-solvent docking,216 the MDFF protocol can also be employed in an explicit solvent simulation, as was the case, for instance, in the construction of a complete all-atom structure of a biological microtubule.210
The gridForces module of NAMD offers limitless possibilities with regard to building initial models for all-atom simulations or coupling an all-atom simulation to external continuum models. For example, a nanoscale Swarovski sculpture can be built by melting a silicon dioxide crystal and then annealing the molten atoms in the presence of a steric repulsive potential that acts as a kind of a nano-imprint mask.217 Similarly, atomically precise repulsive potentials can be used to create pockets of vacuum in water218 or a lipid membrane35 prior to insertion of folded biomolecules, thereby eliminating steric clashes. Multi-scale modeling is enabled by coupling an MD simulation to external potentials obtained by solving continuum models, such as the COMSOL Multiphysics® program, or any other three-dimensional solver. Examples of such simulations include the study of thermophoretic stretching of ssDNA in a locally heated solid-state nanopore,219,220 simulations of plasmonic trapping of DNA molecules [see Fig. 19(d)],211 and coarse-grained simulations of double-nanopore systems.221
At present, the gridForces module is activated using a set of standard NAMD keywords. The reader is referred to the NAMD user’s guide and the user-defined forces tutorial218 for a detailed description. The information about grid-force potentials is provided to NAMD in the form of .dx formatted file, the header of which specifies the dimensions and location of the grid. For each gridForce potential, which could be multiple, the end-user must supply a pdb file with a flag on those atoms or particles that will be subject to the external potential. Optionally, the user can specify custom factors that couple the potential to individual atoms, as well as a global coupling factor, which can be set separately for each of the Cartesian axes. During a NAMD simulation, each of the flagged atoms will be subject to an external force, the magnitude and direction of which are determined by (i) the instantaneous coordinates of the atoms, (ii) the eight values of the potential at the nearby nodes of the grid, and (iii) additional user-defined scaling factors. The value of the force is computed as the negative gradient of the local potential, which in turn is determined using either linear or cubic interpolation schemes. If the physical dimensions of the grid do not match the physical dimension of the computational assay, i.e., the simulation cell, special care must be taken in defining conditions at the boundary of the grid to avoid interpolation artifacts, such as forces produced by an abrupt change in the potential at the edge of the grid or the unintended application of the grid potential over the periodic boundary of the simulation cell. Additional care should be taken when using cubic interpolation for the computation of grid forces in the case of rapidly varying (digitized) potentials, as such an interpolation is prone to produce local attractive potentials. We strongly recommend to examine the shape, the location, and the profile of the external grid-based potentials, e.g., visually in VMD or through analysis scripts, prior to running production simulations.
Among current limitations of the gridForces module is the static nature of the external potentials, that is, once the potential has been activated, its spatial location, orientation, and values cannot be changed. While the framework for modifying external potentials in the course of an MD simulation is in the works, several workarounds are already available. A trivial one consists in splitting one continuous NAMD run into several ones, replacing the potential file between consecutive runs, which is a convenient method to enable a self-consistent multi-scale simulation. Another possibility consists in using multiple grid potentials and changing their global scaling factors via scripting commands in the NAMD configuration file or via external forces applied by Colvars onto one or more grid potentials, for example, via the Multi-Map variable.103 The grid-force potentials themselves can be prepared using third-party software, such as Matlab, COMSOL Multiphysics, or APBS,222 obtained using the VolMap plugin of the visualization program VMD19 or generated from scratch using custom Tcl, Perl, or Python scripts.
XIV. MOLECULAR DYNAMICS FLEXIBLE FITTING
While the most widely used method for acquiring biomolecular structures is x-ray crystallography, crystallization of very large biomolecules, macromolecular complexes, and membrane proteins is extremely challenging. In response, cryo-EM, which obviates the difficult crystallization step and makes imaging possible under physiologically relevant conditions, is increasingly becoming a mainstream approach for structure determination of biomolecular systems.
Historically, computational methods were required to bridge the resolution gap between crystallography and cryo-EM to produce atomic-resolution models of biomolecular objects. One such method is MDFF,17,215,223,224 which is a feature of NAMD. It has proven to be an extremely successful refinement method as evidenced by its numerous applications215,225 ranging from the intricate ribosomal machinery216,226–228 to a host of non-enveloped viruses.3
A. The original MDFF algorithm
The essence of MDFF is, given an initial all-atom structure and a corresponding cryo-EM density, to match the structure to the density by means of an MD simulation (see Fig. 20). Toward this end, the structure is first docked rigidly into the density. Then, flexible fitting is performed by applying to the structure an external biasing potential, in addition to the classical force field. This biasing potential is derived by inverting the cryo-EM density and bounding the resultant map from below a threshold to remove noise. Quality of the fitting procedure is controlled by a user-defined factor, gscale, which scales the biasing potential relative to the force field.
FIG. 20.
A simple MDFF example of fitting a protein structure into a density map. (a) The x-ray structure of an adenylate kinase protein (PDB:1AKE). (b) The protein structure rigid body docked to the density map. (c) The protein structure inside the density map after running an MDFF simulation. (d) The overall quality of fit (cross correlation) during an MDFF simulation.
In practice, the potential energy function used for fitting is defined on a three-dimensional grid and incorporated into the MD simulation using the gridForces feature of NAMD8,18 (see Sec. XIII). Forces are computed from the added potential and applied, in addition to the intrinsic forces, to each atom as a function of its position on the grid using an interpolation scheme. The computed density-derived forces drive the atoms into regions of high density, producing an atomic-resolution structure in the conformation captured in the cryo-EM density. The MD-based nature of MDFF allows for flexibility and sampling while maintaining a realistic structural geometry through incorporation of the molecular mechanical force fields. Restraints imposed during the simulation help preserve the secondary structure and stereochemical correctness (using the extraBonds feature of NAMD229), as well as any symmetry230 of the protein investigated. Traditional MDFF is applicable with cryo-EM densities in the resolution range of 5 Å–15 Å. For density maps between 3 Å–5 Å resolution, the refinement steps get entrapped into biologically irrelevant energy minima, usually yielding incorrect models. This limitation of MDFF has been overcome by introducing a density-based simulated annealing protocol called cascade MDFF and, subsequently, in a more automated replica-exchange algorithm referred to as ReMDFF.231
B. xMDFF: MDFF for low-resolution x-ray crystallography
Although originally developed for fitting crystal structures into cryo-EM densities, MDFF has been extended to refine structures from x-ray crystallographic diffraction data, leading to an algorithm coined xMDFF. This methodology employs a real-space refinement scheme, which flexibly fits atomic models into a density map through an iterative updating. The performance of xMDFF has been demonstrated for the refinement of protein crystallographic data at resolutions ranging from 3.6 Å to 7 Å.232 For abiological macromolecules, xMDFF refinements were performed up to 1.6 Å resolution.233
xMDFF uses model-phased density maps, which incorporate the phases from a search model and the amplitudes from the x-ray diffraction data. These density maps are created utilizing tools in the Phenix software suite.234 The density map is used as a potential for steering the search model into the appropriate locations by means of MDFF forces. Once the search model is fitted into the density, it provides new phases to be used with the observed amplitudes of the x-ray diffraction data to generate an updated density. This model is then fitted into the new map using MDFF, and the process proceeds iteratively until a sufficiently low reliability factor for assessing possible over-modeling of the data, or Rfree-value, is obtained.
Mirroring the strengths of MDFF, xMDFF also benefits from the equilibrium and enhanced-sampling capabilities of NAMD. This sampling capability enables the determination of multiple occupancies in biological objects.235 Conventional MD simulations generate ensembles of atomic structures under constraints, such as constant pressure, volume, temperature, and number of particles. Generally, xMDFF is compatible with any such ensemble-generation scheme, e.g., isobaric–isothermal or canonical, as well as microenvironmental conditions, such as vacuum or explicit or implicit solvent, all achievable within typical NAMD simulations.
XV. USABILITY, REPRODUCIBILITY, AND EXTENSIBILITY
An MD simulation in NAMD is typically configured using one or multiple files, referred to as configuration files. These files set the values of many tunable parameters, e.g., temperature and pressure, and include a series of instructions and manipulations to drive the simulation following a well-defined workflow. Such a workflow can be as simple as a temperature ramp, where the temperature increases every so many steps, or a more sophisticated sequence of runs, analysis, and manipulations, as would be the case in constant-pH160 simulations and in a number of replica-exchange strategies.129 Written either in Tcl or in Python, these configuration files are a resourceful platform that allows the user to adapt the NAMD wealth of features to the specificities of each and every molecular object and computational assay. Since these workflows are written in commonly used scripting languages, end-users can exploit the scripting interface that connects NAMD with their favorite analysis and modeling tools to perform more complex operations guiding the simulation.
Method development at the scripting level, e.g., constant-pH, requiring minimal or no manipulation at all of the source code, has many strategic advantages, most notably in terms of code maintenance. Furthermore, as a result of their flexibility and accessibility, scripting languages such as Tcl and Python enjoy a broad community of developers who have developed over the years a vast library of modules, packages, and scripts, often critical for non-programming researchers. To formalize the interface as a development platform, we have expanded the NAMD Tcl (8.5 and above) and Python (3.7 and above) interpreters to support plugin/module utilization and distribution. Establishing a formal plugin engine, similar to the VMD19 plugin system, allows the developers not only to develop their methods more conveniently but also to distribute them to a growing community of users. Beyond improving the usability of NAMD, formalization of the plugin distribution also ensures reproducibility of the simulations by generalizing the common functionalities between different modules while confining the end-user intervention to the manipulation of variables and user-defined functions, as opposed to the duplication and manipulation of entire modules.
Irrespective of the architecture where NAMD is executed, the same configuration file can be used to drive a simulation on a laptop, a local workstation, or a supercomputer—differences between these architectures appearing at the level of execution commands and the internals of NAMD and Charm++. This commonality has allowed for the development of a number of tools to set up MD simulations with NAMD, such as QwikMD,20,203 MDFF graphical user interface, the binding free-energy estimator (BFEE),236 and Colvars6 dashboard, all distributed in the visualization code VMD,19 as well as web tools, such as CHARMM-GUI.237
In general, preparation tools for MD simulations manage the end-user input information on the computational assay, the structures of the molecular objects at hand, and the simulation parameters, and generate the necessary files toward execution of NAMD on virtually any architecture. When using QwikMD and MDFF-GUI, the simulation can be performed on the same machine where the files were prepared, and the end-user can interact with the simulation as it runs. Interactivity is supported by the interactive molecular dynamics (IMD) module, implemented in both VMD and NAMD, and allowing visualization, analysis, and manipulation of the simulation during its execution. Ability to interact with the simulation has proven particularly useful in structure refinement with MDFF-GUI as the user can manually promote structure fitting into regions of the density map and, more generally, in any MD simulation to detect abnormal events at an early stage. Moreover, the IMD module is often used for educational purposes to visualize the dynamics of the molecular object in the course of an MD simulation, a feature largely exploited in QwikMD and its training material.
QwikMD is an MD toolkit available in VMD, which guides the end-user through the main steps toward the setup, the execution, and the analysis of MD simulations. The user-friendly GUI of QwikMD facilitates the preparation of MD simulations in a point-and-click fashion, using preset parameters for unbiased, steered-MD, MDFF, or hybrid QM/MM computations,203 and is fully integrated with the molecular visualization interface of VMD. Beyond assisting the end-user at the setup stage, this tool ensures the reproducibility of the simulations by recording and logging all parameters and steps taken during the preparation and analysis process. The results of the simulations are then readily analyzed in QwikMD, employing a wide variety of tools available in VMD.19
The development of QwikMD was facilitated by the existence of other modeling VMD plugins that are utilized in a very well-defined workflow, chief among which is the protein structure file (psf) generator, or PSFGEN, the common structure preparation tool for simulations with NAMD. This tool can either be employed in a standalone mode or as a VMD plugin to map the input user-defined structure into the CHARMM force field,9 generating the psf that describes the identity and topology of the molecular object, e.g., atom types and charges, as well as bonds, valence angles, dihedral angles, and cross terms. PSFGEN was recently expanded to support the recent developments in both the additive9 and non-additive77 CHARMM36 force fields, being now able to generate and manipulate Drude particles and lone pairs. Improvements in the performance and implementation of the hydrogen-mass repartition238 now allow the end-user to set up faster and simulate larger molecular objects with longer time steps.
XVI. CONCLUDING REMARKS AND PERSPECTIVE
Empowered with continuing advances in hardware technology on both commodity and large supercomputing platforms enabling faster and larger simulations, with development of more efficient algorithms allowing for better exploitation of the computing architectures available to computational scientists (see Secs. II and III) and with more effective interaction between scientists of different disciplines, we can only expect broader and more sophisticated applications of molecular simulations to more complex biomolecular objects and processes. As a result, the scope of molecular modeling and simulation will continue to extend into a broader range of biomolecular phenomena of strong biological and biomedical relevance.
One particular area that will certainly keep its already fast pace of progress is that of modeling and simulating cellular-scale phenomena. Both inspired and enabled by advances in structural biology on the experimental side, biomolecular modelers will be able to aim at modeling complex subcellular organelles and cellular behavior.3,35,239 The resulting structural models will provide scientists with crucial information to start thinking in the context of realistic, concrete molecular models and to pose relevant molecular questions more rationally. Since its inception, NAMD has always regarded large-scale simulations as a key area in biomolecular modeling and, thus, continues to invest heavily to allow researchers to model and simulate realistic cellular systems. Recent modifications of the code have enabled the program to be benchmarked for the first time on molecular assemblies of up to 2 × 109 atoms (see Fig. 21).
FIG. 21.
Continuous development effort over the years toward simulating with NAMD realistic biological objects of increasing complexity from a small, solvated protein, on the thousand-atom size scale, in the early 1990s, to a full protocell, on the billion-atom size scale, nowadays.
Notably, in the context of cellular-scale molecular modeling and simulation, a major aspect, prominent in recent studies, has been the degree to which the modelers make an effort to maximize the biological realism by including as extensively as possible available experimental data into the computational model and to match the simulation conditions with the experimental ones.17,215,223,224 While unavailability of some experimental details may limit the scope of cellular modeling to some degree, in many areas, computational techniques offer alternative solutions to build biological models, e.g., construction of reliable models of a biological membrane with heterogeneous lipid composition. In this regard, MD simulations will continue to provide a powerful tool to build detailed molecular models of systems of interest. At the same time, there is a need for new modeling tools streamlining the nontrivial process of setting up such complex models (see Sec. XV).20,237 Successful projects employing MD simulations will not only benefit from experimental verification but also be empowered by incorporation of reliable experimental data into the model, an area in which biomolecular modelers have made tremendous progress particularly over the last decade. While we have developed already advanced methodologies to integrate a broad range of experimental information to guide our models and simulations, there is clearly a need to develop the methodology and the software tools to allow for better, more accurate, more comprehensive, and preferably automated ways of doing so. Such molecular modeling efforts are, therefore, essential to pave the way toward modeling cell-scale simulations in meaningful times.
Another major area that has and continues to largely benefit from advances in molecular simulation, in general, and free-energy calculations, in particular, is computational drug design.16,240–242 Supported by progress on the hardware front, most prominently, with the availability of fast GPUs on a variety of platforms, as well as on the software front, with optimally designed tools (see Secs. IX and XV),81,99,129,159,161,236 one can now screen large numbers of small molecules for potential pharmacological effects on specific biological targets within timeframes compatible with industrial requirements and with meaningful, quantitative free energies, e.g., binding affinities.243–245 In this regard, development of more accurate force fields, such as polarizable force fields aimed at capturing induction phenomena more faithfully, as well as machine-learning protocols such as the accurate neural network engine for molecular energies (ANAKIN-ME or ANI for short),246 is a critical advance in order to provide reliable quantitative scales. Modern MD engines have to be conscious and aware of such developments and ready to incorporate them efficiently and in a time-bound fashion. Furthermore, in cases where more complex electronic effects might be important to describe ligand–protein interactions, one can resort to more expensive levels of theory, most notably in the form of a QM description amid a classical representation of the rest of the macromolecular system—a methodology known as the QM/MM calculation already successfully implemented in NAMD203 (see Sec. XII).
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
ACKNOWLEDGMENTS
NAMD core development has been supported by the National Institutes of Health (NIH), Grant No. P41-GM104601 (to E.T., K.S., A.A., L.V.K., and Z.L.-S.). Additional support for specific algorithm/software development was provided by NIH R01-GM072558 (to B.R.), and the National Science Foundation, Grant Nos. PHY-1430124 (to Z.L.-S., K.S., and A.A.), MCB-1616590 (to Z.L.-S. and R.C.B.), MCB-1517221 (to B.R.), and MCB-1942763 (to A.S.), and the Agence Nationale de la Recherche grant ProteaseInAction. NAMD development and testing and simulation of specific molecular systems were enabled by NSF resources provided through multiple XSEDE and PRAC allocations at the National Center for Supercomputing Applications (NCSA) and the Texas Advanced Computing Center (TACC), as well as resources at DOE National Laboratories through leadership allocations (INCITE) and early science and application readiness projects at Oak Ridge National Laboratory (ORNL), Argonne National Laboratory (ANL), and the National Energy Research Scientific Computing Center (NERSC). Significant technical and development assistance for NAMD and Charm++ was contributed by the NCSA, ORNL, and ANL leadership computing centers as well as by the technology companies NVIDIA, IBM, Intel, AMD, Mellanox, and Cray.
Note: This paper is part of the JCP Special Topic on Classical Molecular Dynamics (MD) Simulations: Codes, Algorithms, Force fields, and Applications.
Contributor Information
Christophe Chipot, Email: .
Emad Tajkhorshid, Email: .
REFERENCES
- 1.Shaw D. et al. , “Anton, a special-purpose machine for molecular dynamics simulation,” ACM SIGARCH Comput. Archit. News 35, 1–12 (2007). 10.1145/1273440.1250664 [DOI] [Google Scholar]
- 2.Lee E. H., Hsin J., Sotomayor M., Comellas G., and Schulten K., “Discovery through the computational microscope,” Structure 17, 1295–1306 (2009). 10.1016/j.str.2009.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao G. et al. , “Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics,” Nature 497, 643–646 (2013). 10.1038/nature12162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leimkuhler B. and Matthews C., “Rational construction of stochastic numerical methods for molecular sampling,” Appl. Math. Res. eXpress 2013, 34–56. 10.1093/amrx/abs010 [DOI] [Google Scholar]
- 5.Jiang W. et al. , “High-performance scalable molecular dynamics simulations of a polarizable force field based on classical Drude oscillators in NAMD,” J. Phys. Chem. Lett. 2, 87–92 (2011). 10.1021/jz101461d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fiorin G., Klein M. L., and Hénin J., “Using collective variables to drive molecular dynamics simulations,” Mol. Phys. 111, 3345–3362 (2013). 10.1080/00268976.2013.813594 [DOI] [Google Scholar]
- 7.Fu H., Shao X., Cai W., and Chipot C., “Taming rugged free-energy landscapes using an average force,” Acc. Chem. Res. 52, 3254–3264 (2019). 10.1021/acs.accounts.9b00473 [DOI] [PubMed] [Google Scholar]
- 8.Phillips J. C. et al. , “Scalable molecular dynamics with NAMD,” J. Comput. Chem. 26, 1781–1802 (2005). 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. D. MacKerell, Jr. et al. , “All–atom empirical potential for molecular modeling and dynamics studies of proteins,” J. Phys. Chem. B 102, 3586–3616 (1998). 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 10.Cornell W. D. et al. , “A second generation force field for the simulation of proteins, nucleic acids, and organic molecules,” J. Am. Chem. Soc. 117, 5179–5197 (1995). 10.1021/ja00124a002 [DOI] [Google Scholar]
- 11.Jorgensen W. L., Maxwell D. S., and Tirado-Rives J., “Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids,” J. Am. Chem. Soc. 118, 11225–11236 (1996). 10.1021/ja9621760 [DOI] [Google Scholar]
- 12.Oostenbrink C., Villa A., Mark A. E., and van Gunsteren W. F., “A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force–field parameter sets 53A5 and 53A6,” J. Comput. Chem. 25, 1656–1676 (2004). 10.1002/jcc.20090 [DOI] [PubMed] [Google Scholar]
- 13.Lamoureux G. and Roux B., “Modeling induced polarization with classical Drude oscillators: Theory and molecular dynamics simulation algorithm,” J. Chem. Phys. 119, 3025–3039 (2003). 10.1063/1.1589749 [DOI] [Google Scholar]
- 14.Neese F., “The ORCA program system,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2, 73–78 (2012). 10.1002/wcms.81 [DOI] [Google Scholar]
- 15.Stewart J. J. P., MOPAC: Stewart Computational Chemistry, 2016.
- 16.Chipot C., “Frontiers in free-energy calculations of biological systems,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 4, 71–89 (2014). 10.1002/wcms.1157 [DOI] [Google Scholar]
- 17.Trabuco L. G., Villa E., Mitra K., Frank J., and Schulten K., “Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics,” Structure 16, 673–683 (2008). 10.1016/j.str.2008.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells D. B., Abramkina V., and Aksimentiev A., “Exploring transmembrane transport through α-hemolysin with grid-steered molecular dynamics,” J. Chem. Phys. 127, 125101 (2007). 10.1063/1.2770738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Humphrey W., Dalke A., and Schulten K., “VMD—Visual molecular dynamics,” J. Mol. Graphics 14, 33–38 (1996). 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 20.Ribeiro J. V. et al. , “QwikMD—Integrative molecular dynamics toolkit for novices and experts,” Sci. Rep. 6, 26536 (2016). 10.1038/srep26536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stone J. E. et al. , “Accelerating molecular modeling applications with graphics processors,” J. Comput. Chem. 28, 2618–2640 (2007). 10.1002/jcc.20829 [DOI] [PubMed] [Google Scholar]
- 22.Kalé L. et al. (2020), “UIUC-PPL/charm,” Zenodo. 10.5281/zenodo.3370873. [DOI]
- 23.Kalé L. V. and Zheng G., “The Charm++ programming model,” in Parallel Science and Engineering Applications: The Charm++ Approach, 1st ed., edited by Kale L. V. and Bhatele A. (CRC Press, Inc., Boca Raton, FL, USA, 2013), Chap. 1, pp. 1–16. [Google Scholar]
- 24.Brunner R. K. and Kalé L. V., “Handling application-induced load imbalance using parallel objects,” in Parallel and Distributed Computing for Symbolic and Irregular Applications (World Scientific Publishing, 2000), Vol. 1, pp. 167–218. [Google Scholar]
- 25.Acun B. et al. , “Power, reliability, and performance: One system to rule them all,” Computer 49, 30–37 (2016). 10.1109/mc.2016.310 [DOI] [Google Scholar]
- 26.Meneses E., Ni X., Zheng G., Mendes C. L., and Kalé L. V., “Using migratable objects to enhance fault tolerance schemes in supercomputers,” IEEE Trans. Parallel Distrib. Syst. 26, 2061–2074 (2015). 10.1109/tpds.2014.2342228 [DOI] [Google Scholar]
- 27.Robson M. P., Buch R., and Kalé L. V., “Runtime coordinated heterogeneous tasks in Charm++,” in 2016 Second International Workshop on Extreme Scale Programming Models and Middlewar (ESPM2) (IEEE, 2016), pp. 40–43. [Google Scholar]
- 28.Kalé L. V. and Zheng G., “Charm++ and AMPI: Adaptive runtime strategies via migratable objects,” in Advanced Computational Infrastructures for Parallel and Distributed Applications, edited by Parashar M. (Wiley-Interscience, 2009), pp. 265–282. [Google Scholar]
- 29.Galvez J. J., Senthil K., and Kalé L., “CharmPy: A python parallel programming model,” in 2018 IEEE International Conference on Cluster Computing (CLUSTER) (IEEE, 2018), pp. 423–433. [Google Scholar]
- 30.Phillips J. C. et al. , “Scalable molecular dynamics with NAMD,” in Parallel Science and Engineering Applications: The Charm++ Approach, 1st ed., edited by Kale L. V. and Bhatele A. (CRC Press, Inc., Boca Raton, FL, USA, 2013), Chap. 4, pp. 59–75. [Google Scholar]
- 31.Acun B., Buch R., Kalé L. V., and Phillips J. C., “NAMD: Scalable molecular dynamics based on the Charm++ parallel runtime system,” in Exascale Scientific Applications: Scalability and Performance Portability (Chapman & Hall, 2017). [Google Scholar]
- 32.Acun B. et al. , “Scalable molecular dynamics with NAMD on the summit system,” IBM J. Res. Dev. 62, 4-1 (2018). 10.1147/jrd.2018.2888986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Darden T., York D., and Pedersen L., “Particle mesh Ewald: An N log(N) method for Ewald sums in large systems,” J. Chem. Phys. 98, 10089–10092 (1993). 10.1063/1.464397 [DOI] [Google Scholar]
- 34.Villa E., Balaeff A., and Schulten K., “Structural dynamics of the lac repressor–DNA complex revealed by a multiscale simulation,” Proc. Natl. Acad. Sci. U. S. A. 102, 6783–6788 (2005). 10.1073/pnas.0409387102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singharoy A. et al. , “Atoms to phenotypes: Molecular design principles of cellular energy metabolism,” Cell 179, 1098–1111.e23 (2019). 10.1016/j.cell.2019.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar S. et al. , “Scalable molecular dynamics with NAMD on the IBM Blue Gene/L system,” IBM J. Res. Dev. 52, 177–188 (2008). 10.1147/rd.521.0177 [DOI] [Google Scholar]
- 37.Mei C. et al. , “Enabling and scaling biomolecular simulations of 100 million atoms on petascale machines with a multicore-optimized message-driven runtime,” in Proceedings of the 2011 ACM/IEEE Conference on Supercomputing (IEEE, Seattle, WA, 2011), Vol. 61, pp. 1–11. [Google Scholar]
- 38.Sun Y. et al. , “Optimizing fine-grained communication in a biomolecular simulation application on Cray XK6,” in Proceedings of the 2012 ACM/IEEE Conference on Supercomputing (IEEE Press, Salt Lake City, UT, 2012), pp. 1–11. [Google Scholar]
- 39.Phillips J. C., Sun Y., Jain N., Bohm E. J., and Kalé L. V., “Mapping to irregular torus topologies and other techniques for petascale biomolecular simulation,” in Proceedings of the International Conference on High Performance Computing, Networking, Storage and Analysis, SC’14 (IEEE Press, 2014), pp. 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips J. C. et al. , “Petascale Tcl with NAMD, VMD, and Swift/T,” in SC’14 Workshop on High Performance Technical Computing in Dynamic Languages, SC’14 (IEEE Press, 2014), pp. 6–17. [Google Scholar]
- 41.Freddolino P. L., Arkhipov A. S., Larson S. B., McPherson A., and Schulten K., “Molecular dynamics simulations of the complete satellite tobacco mosaic virus,” Structure 14, 437–449 (2006). 10.1016/j.str.2005.11.014 [DOI] [PubMed] [Google Scholar]
- 42.Stone J. E., Hardy D. J., Ufimtsev I. S., and Schulten K., “GPU-accelerated molecular modeling coming of age,” J. Mol. Graphics Modell. 29, 116–125 (2010). 10.1016/j.jmgm.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Phillips J. C., Stone J. E., and Schulten K., “Adapting a message-driven parallel application to GPU-accelerated clusters,” in SC’08: Proceedings of the 2008 ACM/IEEE Conference on Supercomputing (IEEE Press, Piscataway, NJ, USA, 2008), p. 9. [Google Scholar]
- 44.Kindratenko V. et al. , “GPU clusters for high performance computing,” in 2009 IEEE International Conference on Cluster Computing and Workshops, CLUSTER’09 (IEEE, 2009), pp. 1–8. [Google Scholar]
- 45.Hardy D. J. et al. , “Fast molecular electrostatics algorithms on GPUs,” in GPU Computing Gems, Applications of GPU Computing Series (Elsevier, 2011), pp. 43–58. [Google Scholar]
- 46.Stone J. E., Hynninen A.-P., Phillips J. C., and Schulten K., “Early experiences porting the NAMD and VMD molecular simulation and analysis software to GPU-accelerated OpenPOWER platforms,” High Perform. Comput. 9945, 188–206 (2016). 10.1007/978-3-319-46079-6_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanner D. E., Phillips J. C., and Schulten K., “GPU/CPU algorithm for generalized Born/solvent-accessible surface area implicit solvent calculations,” J. Chem. Theory Comput. 8, 2521–2530 (2012). 10.1021/ct3003089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verlet L., “Computer “experiments” on classical fluids. I. Thermodynamical properties of Lennard-Jones molecules,” Phys. Rev. 159, 98–103 (1967). 10.1103/physrev.159.98 [DOI] [Google Scholar]
- 49.Hairer E. and Lubich C., “The life-span of backward error analysis for numerical integrators,” Numer. Math. 76, 441–462 (1997). 10.1007/s002110050271 [DOI] [Google Scholar]
- 50.Chawla M. M., “On the order and attainable intervals of periodicity of explicit Nyström methods for y′′ = f(t, y),” SIAM J. Numer. Anal. 22, 127–131 (1985). 10.1137/0722009 [DOI] [Google Scholar]
- 51.Grubmüller H., Heller H., Windemuth A., and Schulten K., “Generalized Verlet algorithm for efficient molecular dynamics simulations with long-range interactions,” Mol. Simul. 6, 121–142 (1991). 10.1080/08927029108022142 [DOI] [Google Scholar]
- 52.Tuckerman M., Berne B. J., and Martyna G. J., “Reversible multiple time scale molecular dynamics,” J. Chem. Phys. 97, 1990–2001 (1992). 10.1063/1.463137 [DOI] [Google Scholar]
- 53.Schlick T., Mandziuk M., Skeel R. D., and Srinivas K., “Nonlinear resonance artifacts in molecular dynamics simulations,” J. Comput. Phys. 140, 1–29 (1998). 10.1006/jcph.1998.5879 [DOI] [Google Scholar]
- 54.Ryckaert J.-P., Ciccotti G., and Berendsen H. J. C., “Numerical integration of the Cartesian equation of motion of a system with constraints: Molecular dynamics of n-alkanes,” J. Comput. Phys. 23, 327–341 (1977). 10.1016/0021-9991(77)90098-5 [DOI] [Google Scholar]
- 55.Andersen H. C., “Rattle: A ‘velocity’ version of the shake algorithm for molecular dynamics calculations,” J. Comput. Phys. 52, 24–34 (1983). 10.1016/0021-9991(83)90014-1 [DOI] [Google Scholar]
- 56.Leimkuhler B. J. and Skeel R. D., “Symplectic numerical integrators in constrained Hamiltonian systems,” J. Comput. Phys. 112, 117–125 (1994). 10.1006/jcph.1994.1085 [DOI] [Google Scholar]
- 57.Allen M. P. and Tildesley D. J., Computer Simulation of Liquids (Clarendon Press, Oxford, 1987). [Google Scholar]
- 58.Miyamoto S. and Kollman P. A., “SETTLE: An analytical version of the SHAKE and RATTLE algorithm for rigid water molecules,” J. Comput. Chem. 13, 952–962 (1992). 10.1002/jcc.540130805 [DOI] [Google Scholar]
- 59.Brünger A., Brooks C. L., and Karplus M., “Stochastic boundary-conditions for molecular dynamics simulations of ST2 water,” J. Chem. Phys. 105, 495–500 (1984). 10.1016/0009-2614(84)80098-6 [DOI] [Google Scholar]
- 60.Bussi G., Donadio D., and Parrinello M., “Canonical sampling through velocity rescaling,” J. Chem. Phys. 126, 014101 (2007). 10.1063/1.2408420 [DOI] [PubMed] [Google Scholar]
- 61.Feller S. E., Zhang Y., Pastor R. W., and Brooks B. R., “Constant pressure molecular dynamics simulation: The Langevin piston method,” J. Chem. Phys. 103, 4613–4621 (1995). 10.1063/1.470648 [DOI] [Google Scholar]
- 62.Berendsen H. J. C., Postma J. P. M., Van Gunsteren W. F., DiNola A., and Haak J. R., “Molecular dynamics with coupling to an external bath,” J. Chem. Phys. 81, 3684–3690 (1984). 10.1063/1.448118 [DOI] [Google Scholar]
- 63.Hoover W. G., “Canonical dynamics: Equilibrium phase-space distributions,” Phys. Rev. A 31, 1695–1697 (1985). 10.1103/physreva.31.1695 [DOI] [PubMed] [Google Scholar]
- 64.Hoover W. G., “Constant-pressure equations of motion,” Phys. Rev. A 34, 2499–2500 (1986). 10.1103/physreva.34.2499 [DOI] [PubMed] [Google Scholar]
- 65.Andersen H. C., “Molecular dynamics simulations at constant pressure and/or temperature,” J. Chem. Phys. 72, 2384–2393 (1980). 10.1063/1.439486 [DOI] [Google Scholar]
- 66.Nosé S., “A molecular dynamics method for simulations in the canonical ensemble,” Mol. Phys. 52, 255–268 (1984). 10.1080/00268978400101201 [DOI] [Google Scholar]
- 67.Quigley D. and Probert M. I. J., “Langevin dynamics in constant pressure extended systems,” J. Chem. Phys. 120, 11432–11441 (2004). 10.1063/1.1755657 [DOI] [PubMed] [Google Scholar]
- 68.Ewald P. P., “Die Berechnung optischer und elektrostatischer Gitterpotentiale,” Ann. Phys. 64, 253–287 (1921). 10.1002/andp.19213690304 [DOI] [Google Scholar]
- 69.Figueirido F., Del Buono G. S., and Levy R. M., “On finite-size effects in computer simulations using the Ewald potential,” J. Chem. Phys. 103, 6133–6142 (1995). 10.1063/1.470721 [DOI] [Google Scholar]
- 70.Perram J. W., Petersen H. G., and de Leeuw S. W., “An algorithm for the simulation of condensed matter which grows as the 3/2 power of the number of particles,” Mol. Phys. 65, 875–889 (1988). 10.1080/00268978800101471 [DOI] [Google Scholar]
- 71.Essmann U. et al. , “A smooth particle mesh Ewald method,” J. Chem. Phys. 103, 8577–8593 (1995). 10.1063/1.470117 [DOI] [Google Scholar]
- 72.Toukmaji A. Y. and J. A. Board, Jr., “Ewald summation techniques in perspective: A survey,” Comput. Phys. Commun. 95, 73–92 (1996). 10.1016/0010-4655(96)00016-1 [DOI] [Google Scholar]
- 73.Skeel R. D., “An alternative construction of the Ewald sum,” Mol. Phys. 114, 3166–3170 (2016). 10.1080/00268976.2016.1222455 [DOI] [Google Scholar]
- 74.Brandt A. and Venner C. H., “Multilevel evaluation of integral transforms with asymptotically smooth kernels,” SIAM J. Sci. Comput. 19, 468–492 (1998). 10.1137/s106482759528555x [DOI] [Google Scholar]
- 75.Hardy D. J. et al. , “Multilevel summation method for electrostatic force evaluation,” J. Chem. Theory Comput. 11, 766–779 (2015). 10.1021/ct5009075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Drude P., Lehrbuch der Optik. 1. Ausgabe (Verlag von S. Hirzel, Leipzig, 1900). [Google Scholar]
- 77.Lemkul J. A., Huang J., Roux B., and MacKerell A. D., “An empirical polarizable force field based on the classical Drude oscillator model: Development history and recent applications,” Chem. Rev. 116, 4983–5013 (2016). 10.1021/acs.chemrev.5b00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin F.-Y. et al. , “Further optimization and validation of the classical Drude polarizable protein force field,” J. Chem. Theory Comput. 16, 3221–3239 (2020). 10.1021/acs.jctc.0c00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Onufriev A., Bashford D., and Case D. A., “Exploring protein native states and large-scale conformational changes with a modified generalized Born model,” Proteins: Struct., Funct., Genet. 55, 383–394 (2004). 10.1002/prot.20033 [DOI] [PubMed] [Google Scholar]
- 80.Tanner D. E., Chan K.-Y., Phillips J. C., and Schulten K., “Parallel generalized Born implicit solvent calculations with NAMD,” J. Chem. Theory Comput. 7, 3635–3642 (2011). 10.1021/ct200563j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salari R., Joseph T., Lohia R., Hénin J., and Brannigan G., “A streamlined, general approach for computing ligand binding free energies and its application to GPCR-bound cholesterol,” J. Chem. Theory Comput. 14, 6560–6573 (2018). 10.1021/acs.jctc.8b00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pan A. C., Sezer D., and Roux B., “Finding transition pathways using the string method with swarms of trajectories,” J. Phys. Chem. B 112, 3432–3440 (2008). 10.1021/jp0777059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teo I., Mayne C. G., Schulten K., and Lelièvre T., “Adaptive multilevel splitting method for molecular dynamics calculation of benzamidine-trypsin dissociation time,” J. Chem. Theory Comput. 12, 2983–2989 (2016). 10.1021/acs.jctc.6b00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lopes L. J. S. and Lelièvre T., “Analysis of the adaptive multilevel splitting method on the isomerization of alanine dipeptide,” J. Comput. Chem. 40, 1198–1208 (2019). 10.1002/jcc.25778 [DOI] [PubMed] [Google Scholar]
- 85.Eastman P. and Pande V. S., “OpenMM: A hardware independent framework for molecular simulations,” Comput. Sci. Eng. 12, 34–39 (2015). 10.1109/MCSE.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirkwood J. G., “Statistical mechanics of fluid mixtures,” J. Chem. Phys. 3, 300–313 (1935). 10.1063/1.1749657 [DOI] [Google Scholar]
- 87.Darve E. and Pohorille A., “Calculating free energies using average force,” J. Chem. Phys. 115, 9169–9183 (2001). 10.1063/1.1410978 [DOI] [Google Scholar]
- 88.Hénin J., Fiorin G., Chipot C., and Klein M. L., “Exploring multidimensional free energy landscapes using time-dependent biases on collective variables,” J. Chem. Theory Comput. 6, 35–47 (2010). 10.1021/ct9004432 [DOI] [PubMed] [Google Scholar]
- 89.Comer J. et al. , “The adaptive biasing force method: Everything you always wanted to know but were afraid to ask,” J. Phys. Chem. B 119, 1129–1151 (2015). 10.1021/jp506633n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Izrailev S., Stepaniants S., Balsera M., Oono Y., and Schulten K., “Molecular dynamics study of unbinding of the avidin–biotin complex,” Biophys. J. 72, 1568–1581 (1997). 10.1016/s0006-3495(97)78804-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Torrie G. M. and Valleau J. P., “Monte Carlo study of phase separating liquid mixture by umbrella sampling,” J. Chem. Phys. 66, 1402–1408 (1977). 10.1063/1.434125 [DOI] [Google Scholar]
- 92.Laio A. and Parrinello M., “Escaping free energy minima,” Proc. Natl. Acad. Sci. U. S. A. 99, 12562–12565 (2002). 10.1073/pnas.202427399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ciccotti G., Kapral R., and Vanden-Eijnden E., “Blue moon sampling, vectorial reaction coordinates, and unbiased constrained dynamics,” ChemPhysChem 6, 1809–1814 (2005). 10.1002/cphc.200400669 [DOI] [PubMed] [Google Scholar]
- 94.Lelièvre T., Rousset M., and Stoltz G., “Computation of free energy profiles with parallel adaptive dynamics,” J. Chem. Phys. 126, 134111 (2007). 10.1063/1.2711185 [DOI] [PubMed] [Google Scholar]
- 95.Lesage A., Lelièvre T., Stoltz G., and Hénin J., “Smoothed biasing forces yield unbiased free energies with the extended-system adaptive biasing force method,” J. Phys. Chem. B 121, 3676–3685 (2017). 10.1021/acs.jpcb.6b10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fu H., Shao X., Chipot C., and Cai W., “Extended adaptive biasing force algorithm. An on-the-fly implementation for accurate free-energy calculations,” J. Chem. Theory Comput. 12, 3506–3513 (2016). 10.1021/acs.jctc.6b00447 [DOI] [PubMed] [Google Scholar]
- 97.Alrachid H. and Lelièvre T., “Long-time convergence of an adaptive biasing force method: Variance reduction by Helmholtz projection,” SMAI J. Comput. Math. 1, 55–82 (2015). 10.5802/smai-jcm.4 [DOI] [Google Scholar]
- 98.Contributions to the Colvars module. Geometric variables for standard binding free-energy calculations by Haohao Fu. Dipole-moment magnitude and direction by Alejandro Bernardin. Geometric path-based variables by Haochuan Chen. Path-based variables as Tcl scripts by Chris Chipot. Coordination numbers by Joshua Vermaas.
- 99.Fu H., Cai W., Hénin J., Roux B., and Chipot C., “New coarse variables for the accurate determination of standard binding free energies,” J. Chem. Theory Comput. 13, 5173–5178 (2017). 10.1021/acs.jctc.7b00791 [DOI] [PubMed] [Google Scholar]
- 100.Garate J. A. et al. , “Orientational and folding thermodynamics via electric dipole moment restraining,” J. Phys. Chem. B 123, 2599–2608 (2019). 10.1021/acs.jpcb.8b09374 [DOI] [PubMed] [Google Scholar]
- 101.Díaz Leines G. and Ensing B., “Path finding on high-dimensional free energy landscapes,” Phys. Rev. Lett. 109, 020601 (2012). 10.1103/physrevlett.109.020601 [DOI] [PubMed] [Google Scholar]
- 102.Branduardi D., Gervasio F. L., and Parrinello M., “From A to B in free energy space,” J. Chem. Phys. 126, 054103 (2007). 10.1063/1.2432340 [DOI] [PubMed] [Google Scholar]
- 103.Fiorin G., Marinelli F., and Faraldo-Gómez J. D., “Direct derivation of free energies of membrane deformation and other solvent density variations from enhanced sampling molecular dynamics,” J. Comput. Chem. 41, 449–459 (2020). 10.1002/jcc.26075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Minoukadeh K., Chipot C., and Lelièvre T., “Potential of mean force calculations: A multiple-walker adaptive biasing force approach,” J. Chem. Theory Comput. 6, 1008–1017 (2010). 10.1021/ct900524t [DOI] [Google Scholar]
- 105.Comer J., Phillips J. C., Schulten K., and Chipot C., “Multiple-replica strategies for free-energy calculations in NAMD: Multiple-walker adaptive biasing force and walker selection rules,” J. Chem. Theory Comput. 10, 5276–5285 (2014). 10.1021/ct500874p [DOI] [PubMed] [Google Scholar]
- 106.Shen R. et al. , “Structural refinement of proteins by restrained molecular dynamics simulations with non-interacting molecular fragments,” PLoS Comput. Biol. 11, e1004368 (2015). 10.1371/journal.pcbi.1004368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Marinelli F. and Faraldo-Gómez J. D., “Ensemble-biased metadynamics: A molecular simulation method to sample experimental distributions,” Biophys. J. 108, 2779–2782 (2015). 10.1016/j.bpj.2015.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pitera J. W. and Chodera J. D., “On the use of experimental observations to bias simulated ensembles,” J. Chem. Theory Comput. 8, 3445–3451 (2012). 10.1021/ct300112v [DOI] [PubMed] [Google Scholar]
- 109.White A. D. and Voth G. A., “Efficient and minimal method to bias molecular simulations with experimental data,” J. Chem. Theory Comput. 10, 3023–3030 (2014). 10.1021/ct500320c [DOI] [PubMed] [Google Scholar]
- 110.Marinelli F. and Fiorin G., “Structural characterization of biomolecules through atomistic simulations guided by DEER measurements,” Structure 27, 359–370.e12 (2019). 10.1016/j.str.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swendsen R. H. and Wang J.-S., “Replica Monte Carlo simulation of spin glasses,” Phys. Rev. Lett. 57, 2607–2609 (1986). 10.1103/physrevlett.57.2607 [DOI] [PubMed] [Google Scholar]
- 112.Voter A. F., “Hyperdynamics: Accelerated molecular dynamics of infrequent events,” Phys. Rev. Lett. 78, 3908–3911 (1997). 10.1103/physrevlett.78.3908 [DOI] [Google Scholar]
- 113.Wang Y., Harrison C. B., Schulten K., and McCammon J. A., “Implementation of accelerated molecular dynamics in NAMD,” Comput. Sci. Discovery 4, 015002 (2011). 10.1088/1749-4699/4/1/015002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pang Y. T., Miao Y., Wang Y., and McCammon J. A., “Gaussian accelerated molecular dynamics in NAMD,” J. Chem. Theory Comput. 13, 9–19 (2017). 10.1021/acs.jctc.6b00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang L., Friesner R. A., and Berne B. J., “Replica exchange with solute scaling: A more efficient version of replica exchange with solute tempering (REST2),” J. Phys. Chem. B 115, 9431–9438 (2011). 10.1021/jp204407d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zwanzig R., Nonequilibrium Statistical Mechanics (Oxford University Press, 2001). [Google Scholar]
- 117.Bolhuis P. G., Dellago C., and Chandler D., “Reaction coordinates of biomolecular isomerization,” Proc. Natl. Acad. Sci. U. S. A. 97, 5877–5882 (2000). 10.1073/pnas.100127697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Free Energy Calculations. Theory and Applications in Chemistry and Biology, edited by Chipot C. and Pohorille A. (Springer-Verlag, Berlin, Heidelberg, NY, 2007). [Google Scholar]
- 119.Lelièvre T., Stoltz G., and Rousset M., Free Energy Computations: A Mathematical Perspective (Imperial College Press, 2010). [Google Scholar]
- 120.Jarzynski C., “Nonequilibrium equality for free energy differences,” Phys. Rev. Lett. 78, 2690–2693 (1997). 10.1103/physrevlett.78.2690 [DOI] [Google Scholar]
- 121.Roux B., “The calculation of the potential of mean force using computer simulations,” Comput. Phys. Commun. 91, 275–282 (1995). 10.1016/0010-4655(95)00053-i [DOI] [Google Scholar]
- 122.Huber T., Torda A. E., and van Gunsteren W. F., “Local elevation: A method for improving the searching properties of molecular dynamics simulation,” J. Comput.-Aided Mol. Des. 8, 695–708 (1994). 10.1007/bf00124016 [DOI] [PubMed] [Google Scholar]
- 123.Grubmüller H., “Predicting slow structural transitions in macromolecular systems: Conformational flooding,” Phys. Rev. E 52, 2893–2906 (1995). 10.1103/physreve.52.2893 [DOI] [PubMed] [Google Scholar]
- 124.Barducci A., Bussi G., and Parrinello M., “Well-tempered metadynamics: A smoothly converging and tunable free-energy method,” Phys. Rev. Lett. 100, 020603 (2008). 10.1103/physrevlett.100.020603 [DOI] [PubMed] [Google Scholar]
- 125.Chipot C. and Lelièvre T., “Enhanced sampling of multidimensional free-energy landscapes using adaptive biasing forces,” SIAM J. Appl. Math. 71, 1673–1695 (2011). 10.1137/10080600x [DOI] [Google Scholar]
- 126.Fu H. et al. , “Zooming across the free-energy landscape: Shaving barriers, and flooding valleys,” J. Phys. Chem. Lett. 9, 4738–4745 (2018). 10.1021/acs.jpclett.8b01994 [DOI] [PubMed] [Google Scholar]
- 127.Zheng L., Chen M., and Yang W., “Random walk in orthogonal space to achieve efficient free-energy simulation of complex systems,” Proc. Natl. Acad. Sci. U. S. A. 105, 20227–20232 (2008). 10.1073/pnas.0810631106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Raiteri P., Laio A., Gervasio F. L., Micheletti C., and Parrinello M., “Efficient reconstruction of complex free energy landscapes by multiple walkers metadynamics,” J. Phys. Chem. B 110, 3533–3539 (2006). 10.1021/jp054359r [DOI] [PubMed] [Google Scholar]
- 129.Jiang W. et al. , “Generalized scalable multiple copy algorithms for molecular dynamics simulations in NAMD,” Comput. Phys. Commun. 185, 908–916 (2014). 10.1016/j.cpc.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sugita Y. and Okamoto Y., “Replica-exchange molecular dynamics method for protein folding,” Chem. Phys. Lett. 314, 141–151 (1999). 10.1016/s0009-2614(99)01123-9 [DOI] [Google Scholar]
- 131.Woods C. J., Essex J. W., and King M. A., “The development of replica-exchange-based free-energy methods,” J. Phys. Chem. B 107, 13703–13710 (2003). 10.1021/jp0356620 [DOI] [Google Scholar]
- 132.Jiang W. and Roux B., “Free energy perturbation Hamiltonian replica-exchange molecular dynamics (FEP/H–REMD) for absolute ligand binding free energy calculations,” J. Chem. Theory Comput. 6, 2559–2565 (2010). 10.1021/ct1001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sugita Y., Kitao A., and Okamoto Y., “Multidimensional replica–exchange method for free–energy calculations,” J. Chem. Phys. 113, 6042–6051 (2000). 10.1063/1.1308516 [DOI] [Google Scholar]
- 134.Chen Y. and Roux B., “Efficient hybrid non-equilibrium molecular dynamics - Monte Carlo simulations with symmetric momentum reversal,” J. Chem. Phys. 141, 114107 (2014). 10.1063/1.4895516 [DOI] [PubMed] [Google Scholar]
- 135.Suh D., Radak B. K., Chipot C., and Roux B., “Enhanced configurational sampling with hybrid non-equilibrium molecular dynamics/Monte Carlo propagator,” J. Chem. Phys. 148, 014101 (2018). 10.1063/1.5004154 [DOI] [PubMed] [Google Scholar]
- 136.Tse C. H., Comer J., Wang Y., and Chipot C., “Link between membrane composition and permeability to drugs,” J. Chem. Theory Comput. 14, 2895–2909 (2018). 10.1021/acs.jctc.8b00272 [DOI] [PubMed] [Google Scholar]
- 137.Comer J., Chipot C., and González-Nilo F. D., “Calculating position-dependent diffusivity in biased molecular dynamics simulations,” J. Chem. Theory Comput. 9, 876–882 (2013). 10.1021/ct300867e [DOI] [PubMed] [Google Scholar]
- 138.Chipot C. and Comer J., “Subdiffusion in membrane permeation of small molecules,” Sci. Rep. 6, 35913 (2016). 10.1038/srep35913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hamelberg D., Mongan J., and McCammon J. A., “Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules,” J. Chem. Phys. 120, 11919–11929 (2004). 10.1063/1.1755656 [DOI] [PubMed] [Google Scholar]
- 140.Wang Y., Markwick P. R. L., de Oliveira C. A. F., and McCammon J. A., “Enhanced lipid diffusion and mixing in accelerated molecular dynamics,” J. Chem. Theory Comput. 7, 3199–3207 (2011). 10.1021/ct200430c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bucher D., Grant B. J., and McCammon J. A., “Induced fit or conformational selection? The role of the semi-closed state in the maltose binding protein,” Biochemistry 50, 10530–10539 (2011). 10.1021/bi201481a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Caulfield T. R. et al. , “Phosphorylation by PINK1 releases the UBL domain and initializes the conformational opening of the E3 ubiquitin ligase parkin,” PLoS Comput. Biol. 10, e1003935 (2014). 10.1371/journal.pcbi.1003935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cheng M. H. and Bahar I., “Molecular mechanism of dopamine transport by human dopamine transporter,” Structure 23, 2171–2181 (2015). 10.1016/j.str.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hummer G., “Fast-growth thermodynamic integration: Error and efficiency analysis,” J. Chem. Phys. 114, 7330–7337 (2001). 10.1063/1.1363668 [DOI] [Google Scholar]
- 145.Park S., Khalili-Araghi F., Tajkhorshid E., and Schulten K., “Free energy calculation from steered molecular dynamics simulations using Jarzynski’s equality,” J. Chem. Phys. 119, 3559–3566 (2003). 10.1063/1.1590311 [DOI] [Google Scholar]
- 146.Miao Y. et al. , “Improved reweighting of accelerated molecular dynamics simulations for free energy calculation,” J. Chem. Theory Comput. 10, 2677–2689 (2014). 10.1021/ct500090q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Doshi U. and Hamelberg D., “Towards fast, rigorous and efficient conformational sampling of biomolecules: Advances in accelerated molecular dynamics,” Biochim. Biophys. Acta, Gen. Subj. 1850, 878–888 (2015). 10.1016/j.bbagen.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 148.Miao Y., Feher V. A., and McCammon J. A., “Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation,” J. Chem. Theory Comput. 11, 3584–3595 (2015). 10.1021/acs.jctc.5b00436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wereszczynski J. and McCammon J. A., “Nucleotide-dependent mechanism of Get3 as elucidated from free energy calculations,” Proc. Natl. Acad. Sci. U. S. A. 109, 7759–7764 (2012). 10.1073/pnas.1117441109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sibener L. V. et al. , “Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding,” Cell 174, 672–687 (2018). 10.1016/j.cell.2018.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang Y.-m. M., McCammon J. A., and Miao Y., “Replica exchange Gaussian accelerated molecular dynamics: Improved enhanced sampling and free energy calculation,” J. Chem. Theory Comput. 14, 1853–1864 (2018). 10.1021/acs.jctc.7b01226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jorgensen W. L. and Ravimohan C., “Monte Carlo simulation of differences in free energies of hydration,” J. Chem. Phys. 83, 3050–3054 (1985). 10.1063/1.449208 [DOI] [Google Scholar]
- 153.Gao J., Kuczera K., Tidor B., and Karplus M., “Hidden thermodynamics of mutant proteins: A molecular dynamics analysis,” Science 244, 1069–1072 (1989). 10.1126/science.2727695 [DOI] [PubMed] [Google Scholar]
- 154.Landau L. D., Statistical Physics (Clarendon Press, Oxford, 1938). [Google Scholar]
- 155.Zwanzig R. W., “High–temperature equation of state by a perturbation method. I. Nonpolar gases,” J. Chem. Phys. 22, 1420–1426 (1954). 10.1063/1.1740409 [DOI] [Google Scholar]
- 156.Zacharias M., Straatsma T. P., and McCammon J. A., “Separation-shifted scaling, a new scaling method for Lennard-Jones interactions in thermodynamic integration,” J. Chem. Phys. 100, 9025–9031 (1994). 10.1063/1.466707 [DOI] [Google Scholar]
- 157.Pohorille A., Jarzynski C., and Chipot C., “Good practices in free-energy calculations,” J. Phys. Chem. B 114, 10235–10253 (2010). 10.1021/jp102971x [DOI] [PubMed] [Google Scholar]
- 158.Bennett C. H., “Efficient estimation of free energy differences from Monte Carlo data,” J. Comput. Phys. 22, 245–268 (1976). 10.1016/0021-9991(76)90078-4 [DOI] [Google Scholar]
- 159.Dixit S. B. and Chipot C., “Can absolute free energies of association be estimated from molecular mechanical simulations? The biotin–streptavidin system revisited,” J. Phys. Chem. A 105, 9795–9799 (2001). 10.1021/jp011878v [DOI] [Google Scholar]
- 160.Radak B. K. et al. , “Constant-pH molecular dynamics simulations for large biomolecular systems,” J. Chem. Theory Comput. 13, 5933–5944 (2017). 10.1021/acs.jctc.7b00875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jiang W., Chipot C., and Roux B., “Computing relative binding affinity of ligands to receptor: An effective hybrid single-dual-topology free-energy perturbation approach in NAMD,” J. Chem. Inf. Model. 59, 3794–3802 (2019). 10.1021/acs.jcim.9b00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sindhikara D., Meng Y., and Roitberg A. E., “Exchange frequency in replica exchange molecular dynamics,” J. Chem. Phys. 128, 024103 (2008). 10.1063/1.2816560 [DOI] [PubMed] [Google Scholar]
- 163.Jiang W., Hodoscek M., and Roux B., “Computation of absolute hydration and binding free energy with free energy perturbation distributed replica-exchange molecular dynamics (FEP/REMD),” J. Chem. Theory Comput. 5, 2583–2588 (2009). 10.1021/ct900223z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jiang W., Luo Y., Maragliano L., and Roux B., “Calculation of free energy landscape in multi-dimensions with Hamiltonian-exchange umbrella sampling on petascale supercomputer,” J. Chem. Theory Comput. 8, 4672–4680 (2012). 10.1021/ct300468g [DOI] [PubMed] [Google Scholar]
- 165.Kumar S. et al. , “PAMI: A parallel active message interface for the Blue Gene/Q supercomputer,” in 2012 IEEE 26th International Parallel and Distributed Processing Symposium (IEEE, 2012), pp. 763–773. [Google Scholar]
- 166.Sun Y., Zheng G., Kalé L. V., Jones T. R., and Olson R., “A uGNI-based asynchronous message-driven runtime system for Cray supercomputers with gemini interconnect,” in 2012 IEEE 26th International Parallel and Distributed Processing Symposium (IPDPS) (IEEE, 2012), pp. 751–762. [Google Scholar]
- 167.Wallace J. A. and Shen J. K., “Continuous constant pH molecular dynamics in explicit solvent with pH-based replica exchange,” J. Chem. Theory Comput. 7, 2617–2629 (2011). 10.1021/ct200146j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Donnini S., Tegeler F., Groenhof G., and Grubmüller H., “Constant pH molecular dynamics in explicit solvent with λ-dynamics,” J. Chem. Theory Comput. 7, 1962–1978 (2011). 10.1021/ct200061r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Swails J. M., York D. M., and Roitberg A. E., “Constant pH replica exchange molecular dynamics in explicit solvent using discrete protonation states: Implementation, testing, and validation,” J. Chem. Theory Comput. 10, 1341–1352 (2014). 10.1021/ct401042b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lee J., Miller B. T., Damjanović A., and Brooks B. R., “Constant pH molecular dynamics in explicit solvent with enveloping distribution sampling and Hamiltonian exchange,” J. Chem. Theory Comput. 10, 2738–2750 (2014). 10.1021/ct500175m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang Y., Chen W., Wallace J. A., and Shen J., “All-atom continuous constant pH molecular dynamics with particle mesh Ewald and titratable water,” J. Chem. Theory Comput. 12, 5411–5421 (2016). 10.1021/acs.jctc.6b00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chen Y. and Roux B., “Constant–pH hybrid nonequilibrium molecular dynamics–Monte Carlo simulation method,” J. Chem. Theory Comput. 11, 3919–3931 (2015). 10.1021/acs.jctc.5b00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Sarkar A., Gupta P. L., and Roitberg A. E., “pH-dependent conformational changes due to ionizable residues in a hydrophobic protein interior: The study of L25K and L125K variants of SNase,” J. Phys. Chem. B 123, 5742–5754 (2019). 10.1021/acs.jpcb.9b03816 [DOI] [PubMed] [Google Scholar]
- 174.Ellis C. R., Tsai C.-C., Hou X., and Shen J., “Constant pH molecular dynamics reveals pH-modulated binding of two small-molecule BACE1 inhibitors,” J. Phys. Chem. Lett. 7, 944–949 (2016). 10.1021/acs.jpclett.6b00137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Harris R. C., Tsai C.-C., Ellis C. R., and Shen J., “Allostery modulates the inhibitor selectivity for β-secretase,” J. Phys. Chem. Lett. 8, 4832–4837 (2017). 10.1021/acs.jpclett.7b02309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dissanayake T., Swails J. M., Harris M. E., Roitberg A. E., and York D. M., “Interpretation of pH-activity profiles for acid-base catalysis from molecular simulations,” Biochemistry 54, 1307–1313 (2015). 10.1021/bi5012833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stern H. A., “Molecular simulation with variable protonation states at constant pH,” J. Chem. Phys. 126, 164112 (2007). 10.1063/1.2731781 [DOI] [PubMed] [Google Scholar]
- 178.Chen Y. and Roux B., “Generalized Metropolis acceptance criterion for hybrid non-equilibrium molecular dynamics–Monte Carlo simulations,” J. Chem. Phys. 142, 024101 (2015). 10.1063/1.4904889 [DOI] [PubMed] [Google Scholar]
- 179.Radak B. K. and Roux B., “Efficiency in nonequilibrium molecular dynamics Monte Carlo simulations,” J. Chem. Phys. 145, 134109 (2016). 10.1063/1.4964288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Baptista A. M., Teixeira V. H., and Soares C. M., “Constant-pH molecular dynamics using stochastic titration,” J. Chem. Phys. 117, 4184–4200 (2002). 10.1063/1.1497164 [DOI] [Google Scholar]
- 181.Arbely E., Rutherford T. J., Sharpe T. D., Ferguson N., and Fersht A. R., “Downhill versus barrier-limited folding of BBL 1: Energetic and structural perturbation effects upon protonation of a histidine of unusually low pKa,” J. Mol. Biol. 387, 986–992 (2009). 10.1016/j.jmb.2008.12.055 [DOI] [PubMed] [Google Scholar]
- 182.Arbely E. et al. , “Carboxyl pKa values and acid denaturation of BBL,” J. Mol. Biol. 403, 313–327 (2010). 10.1016/j.jmb.2010.08.052 [DOI] [PubMed] [Google Scholar]
- 183.Kumar S., Rosenberg J. M., Bouzida D., Swendsen R. H., and Kollman P. A., “The weighted histogram analysis method for free energy calculations on biomolecules. I. The method,” J. Comput. Chem. 13, 1011–1021 (1992). 10.1002/jcc.540130812 [DOI] [Google Scholar]
- 184.Lindorff-Larsen K., Piana S., Dror R. O., and Shaw D. E., “How fast-folding proteins fold,” Science 334, 517–520 (2011). 10.1126/science.1208351 [DOI] [PubMed] [Google Scholar]
- 185.Perilla J. R. et al. , “Molecular dynamics simulations of large macromolecular complexes,” Curr. Opin. Struct. Biol. 31, 64–74 (2015). 10.1016/j.sbi.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kirchner B., Wennmohs F., Ye S., and Neese F., “Theoretical bioinorganic chemistry: The electronic structure makes a difference,” Curr. Opin. Chem. Biol. 11, 134–141 (2007). 10.1016/j.cbpa.2007.02.026 [DOI] [PubMed] [Google Scholar]
- 187.Boulanger E. and Harvey J. N., “QM/MM methods for free energies and photochemistry,” Curr. Opin. Struct. Biol. 49, 72–76 (2018). 10.1016/j.sbi.2018.01.003 [DOI] [PubMed] [Google Scholar]
- 188.Senn H. M. and Thiel W., “QM/MM methods for biomolecular systems,” Angew. Chem., Int. Ed. Engl. 48, 1198–1229 (2009). 10.1002/anie.200802019 [DOI] [PubMed] [Google Scholar]
- 189.Field M. J., Bash P. A., and Karplus M., “A combined quantum mechanical and molecular mechanical potential for molecular dynamics simulations,” J. Comput. Chem. 11, 700–733 (1990). 10.1002/jcc.540110605 [DOI] [Google Scholar]
- 190.Jindal G. and Warshel A., “Exploring the dependence of QM/MM calculations of enzyme catalysis on the size of the QM region,” J. Phys. Chem. B 120, 9913–9921 (2016). 10.1021/acs.jpcb.6b07203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Warshel A. and Levitt M., “Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lysozyme,” J. Mol. Biol. 103, 227–249 (1976). 10.1016/0022-2836(76)90311-9 [DOI] [PubMed] [Google Scholar]
- 192.Lev B., Roux B., and Noskov S. Y., “Relative free energies for hydration of monovalent ions from QM and QM/MM simulations,” J. Chem. Theory Comput. 9, 4165–4175 (2013). 10.1021/ct400296w [DOI] [PubMed] [Google Scholar]
- 193.Queens M. G., Borowski T., and de Visser S. P., “Quantum mechanics/molecular mechanics modeling of enzymatic processes: Caveats and breakthroughs,” Chemistry 22, 2562–2581 (2016). 10.1002/chem.201503802 [DOI] [PubMed] [Google Scholar]
- 194.Kaila V. R. I., Wikström M., and Hummer G., “Electrostatics, hydration, and proton transfer dynamics in the membrane domain of respiratory complex I,” Proc. Natl. Acad. Sci. U. S. A. 111, 6988–6993 (2014). 10.1073/pnas.1319156111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Świderek K., Tuñón I., and Moliner V., “Predicting enzymatic reactivity: From theory to design,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 4, 407–421 (2014). 10.1002/wcms.1173 [DOI] [Google Scholar]
- 196.Kamerlin S. C. L. and Warshel A., “The empirical valence bond model: Theory and applications,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 1, 30–45 (2011). 10.1002/wcms.10 [DOI] [Google Scholar]
- 197.Rovira C., “The description of electronic processes inside proteins from Car–Parrinello molecular dynamics: Chemical transformations,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 3, 393–407 (2013). 10.1002/wcms.1153 [DOI] [Google Scholar]
- 198.Sousa S. F. et al. , “Application of quantum mechanics/molecular mechanics methods in the study of enzymatic reaction mechanisms,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 7, e1281 (2017). 10.1002/wcms.1281 [DOI] [Google Scholar]
- 199.Cen Y. et al. , “Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution,” Nat. Commun. 10, 3198 (2019). 10.1038/s41467-019-11155-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.van der Kamp M. W. and Mulholland A. J., “Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology,” Biochemistry 52, 2708–2728 (2013). 10.1021/bi400215w [DOI] [PubMed] [Google Scholar]
- 201.Du S., Fu H., Shao X., Chipot C., and Cai W., “Addressing polarization phenomena in molecular machines containing transition metal ions with an additive force field,” J. Chem. Theory Comput. 15, 1841–1847 (2019). 10.1021/acs.jctc.8b00972 [DOI] [PubMed] [Google Scholar]
- 202.Bernardi R. C. and Pascutti P. G., “Hybrid QM/MM molecular dynamics study of benzocaine in a membrane environment: How does a quantum mechanical treatment of both anesthetic and lipids affect their interaction,” J. Chem. Theory Comput. 8, 2197–2203 (2012). 10.1021/ct300213u [DOI] [PubMed] [Google Scholar]
- 203.Melo M. C. R. et al. , “NAMD goes quantum: An integrative suite for hybrid simulations,” Nat. Methods 15, 351–354 (2018). 10.1038/nmeth.4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Amara P. and Field M. J., “Evaluation of an ab initio quantum mechanical/molecular mechanical hybrid-potential link-atom method,” Theor. Chem. Acc. 109, 43–52 (2003). 10.1007/s00214-002-0413-3 [DOI] [Google Scholar]
- 205.Stewart J. J. P., “MOPAC: A semiempirical molecular orbital program,” J. Comput.-Aided Mol. Des. 4, 1–103 (1990). 10.1007/bf00128336 [DOI] [PubMed] [Google Scholar]
- 206.Maia J. D. C. et al. , “GPU linear algebra libraries and GPGPU programming for accelerating MOPAC semiempirical quantum chemistry calculations,” J. Chem. Theory Comput. 8, 3072–3081 (2012). 10.1021/ct3004645 [DOI] [PubMed] [Google Scholar]
- 207.Neese F., “Software update: The ORCA program system, version 4.0,” Wiley Interdiscip. Rev.: Comput. Mol. Sci. 8, e1327 (2018). 10.1002/wcms.1327 [DOI] [Google Scholar]
- 208.Stewart J. J. P., “Optimization of parameters for semiempirical methods VI: More modifications to the NDDO approximations and re-optimization of parameters,” J. Mol. Model. 19, 1–32 (2013). 10.1007/s00894-012-1667-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Aksimentiev A. and Schulten K., “Imaging α-hemolysin with molecular dynamics: Ionic conductance, osmotic permeability and the electrostatic potential map,” Biophys. J. 88, 3745–3761 (2005). 10.1529/biophysj.104.058727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wells D. B. and Aksimentiev A., “Mechanical properties of a complete microtubule revealed through molecular dynamics simulation,” Biophys. J. 99, 629–637 (2010). 10.1016/j.bpj.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Belkin M., Chao S.-H., Jonsson M. P., Dekker C., and Aksimentiev A., “Plasmonic nanopores for trapping, controlling displacement, and sequencing of DNA,” ACS Nano 9, 10598–10611 (2015). 10.1021/acsnano.5b04173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kasianowicz J. J., Brandin E., Branton D., and Deamer D. W., “Characterization of individual polynucleotide molecules using a membrane channel,” Proc. Natl. Acad. Sci. U. S. A. 93, 13770–13773 (1996). 10.1073/pnas.93.24.13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Comer J., Wells D. B., and Aksimentiev A., “Modeling nanopores for sequencing DNA,” in DNA Nanotechnology, Methods and Protocols, edited by Zuccheri G. and Samori B. (Humana Press, New York, 2011), Vol. 749, Chap. 22, pp. 317–358. [DOI] [PubMed] [Google Scholar]
- 214.See http://bionano.physics.illinois.edu/tutorials/modeling-nanopores-sequencing-dna for Modeling Nanopores for Sequencing DNA Tutorial.
- 215.McGreevy R., Teo I., Singharoy A., and Schulten K., “Advances in the molecular dynamics flexible fitting method for cryo-EM modeling,” Methods 100, 50–60 (2016). 10.1016/j.ymeth.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Villa E. et al. , “Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis,” Proc. Natl. Acad. Sci. U. S. A. 106, 1063–1068 (2009). 10.1073/pnas.0811370106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Aksimentiev A., Brunner R., Cruz-Chu E., Comer J., and Schulten K., “Modeling transport through synthetic nanopores,” IEEE Nanotechnol. Mag. 3, 20–28 (2009). 10.1109/mnano.2008.931112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.See http://bionano.physics.illinois.edu/tutorials/user-defined-forces-namd for User-Defined Forces in NAMD Tutorial.
- 219.Belkin M., Maffeo C., Wells D. B., and Aksimentiev A., “Stretching and controlled motion of single-stranded DNA in locally heated solid-state nanopores,” ACS Nano 7, 6816–6824 (2013). 10.1021/nn403575n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Belkin M., Chao S.-H., Giannetti G., and Aksimentiev A., “Modeling thermophoretic effects in solid-state nanopores,” J. Comput. Electron. 13, 826–838 (2014). 10.1007/s10825-014-0594-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Pud S. et al. , “Mechanical trapping of DNA in a double-nanopore system,” Nano Lett. 16, 8021–8028 (2016). 10.1021/acs.nanolett.6b04642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A., “Electrostatics of nanosystems: Application to microtubules and the ribosome,” Proc. Natl. Acad. Sci. U. S. A. 98, 10037–10041 (2001). 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Trabuco L. G., Villa E., Schreiner E., Harrison C. B., and Schulten K., “Molecular dynamics flexible fitting: A practical guide to combine cryo-electron microscopy and X-ray crystallography,” Methods 49, 174–180 (2009). 10.1016/j.ymeth.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Singharoy A., Barragan A. M., Thangapandian S., Tajkhorshid E., and Schulten K., “Binding site recognition and docking dynamics of a single electron transport protein: Cytochrome c2,” J. Am. Chem. Soc. 138, 12077–12089 (2016). 10.1021/jacs.6b01193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Goh B. C. et al. , “Atomic modeling of an immature retroviral lattice using molecular dynamics and mutagenesis,” Structure 23, 1414–1425 (2015). 10.1016/j.str.2015.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Trabuco L. G. et al. , “Applications of the molecular dynamics flexible fitting method,” J. Struct. Biol. 173, 420–427 (2011). 10.1016/j.jsb.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Frauenfeld J. et al. , “Cryo-EM structure of the ribosome-SecYE complex in the membrane environment,” Nat. Struct. Mol. Biol. 18, 614–621 (2011). 10.1038/nsmb.2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wickles S. et al. , “A structural model of the active ribosome-bound membrane protein insertase YidC,” eLife 3, e03035 (2014). 10.7554/eLife.03035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schreiner E., Trabuco L. G., Freddolino P. L., and Schulten K., “Stereochemical errors and their implications for molecular dynamics simulations,” BMC Bioinf. 12, 190 (2011). 10.1186/1471-2105-12-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chan K.-Y. et al. , “Symmetry-restrained flexible fitting for symmetric EM maps,” Structure 19, 1211–1218 (2011). 10.1016/j.str.2011.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Singharoy A. et al. , “Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps,” eLife 5, e16105 (2016). 10.7554/elife.16105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.McGreevy R. et al. , “xMDFF: Molecular dynamics flexible fitting of low-resolution X-ray structures,” Acta Crystallogr., Sect. D: Biol. Crystallogr. 70, 2344–2355 (2014). 10.1107/s1399004714013856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Singharoy A. et al. , “Macromolecular crystallography for synthetic abiological molecules: Combining xMDFF and PHENIX for structure determination of cyanostar macrocycles,” J. Am. Chem. Soc. 137, 8810–8818 (2015). 10.1021/jacs.5b04407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Adams P. D. et al. , “PHENIX: A comprehensive Python-based system for macromolecular structure solution,” Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 213–221 (2010). 10.1107/s0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li Q. et al. , “Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain,” Nat. Struct. Mol. Biol. 21, 244–252 (2014). 10.1038/nsmb.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Fu H. et al. , “BFEE: A user-friendly graphical interface facilitating absolute binding free-energy calculations,” J. Chem. Inf. Model. 58, 556–560 (2018). 10.1021/acs.jcim.7b00695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Jo S., Kim T., Iyer V. G., and Im W., “CHARMM-GUI: A web-based graphical user interface for CHARMM,” J. Comput. Chem. 29, 1859–1865 (2008). 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- 238.Balusek C. et al. , “Accelerating membrane simulations with hydrogen mass repartitioning,” J. Chem. Theory Comput. 15, 4673–4686 (2019). 10.1021/acs.jctc.9b00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Durrant J. D. et al. , “Mesoscale all-atom influenza virus simulations suggest new substrate binding mechanism,” ACS Cent. Sci. 6, 189–196 (2020). 10.1021/acscentsci.9b01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Shirts M. R., Mobley D. L., and Chodera J. D., “Alchemical free energy calculations: Ready for prime time?,” in Annual Reports in Computational Chemistry (Elsevier, The Netherlands, 2007), Vol. 3, pp. 41–59. [Google Scholar]
- 241.Chodera J. D. et al. , “Alchemical free energy methods for drug discovery: Progress and challenges,” Curr. Opin. Struct. Biol. 21, 150–160 (2011). 10.1016/j.sbi.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Cournia Z., Allen B., and Sherman W., “Relative binding free energy calculations in drug discovery: Recent advances and practical considerations,” J. Chem. Inf. Model. 57, 2911–2937 (2017). 10.1021/acs.jcim.7b00564 [DOI] [PubMed] [Google Scholar]
- 243.Chipot C., Rozanska X., and Dixit S. B., “Can free energy calculations be fast and accurate at the same time? Binding of low–affinity, non–peptide inhibitors to the SH2 domain of the src protein,” J. Comput.-Aided Mol. Des. 19, 765–770 (2005). 10.1007/s10822-005-9021-3 [DOI] [PubMed] [Google Scholar]
- 244.Gumbart J. C., Roux B., and Chipot C., “Standard binding free energies from computer simulations: What is the best strategy?,” J. Chem. Theory Comput. 9, 794–802 (2013). 10.1021/ct3008099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rizzi A. et al. , “The SAMPL6 SAMPLing challenge: Assessing the reliability and efficiency of binding free energy calculations,” J. Comput.-Aided Mol. Des. 34, 601–633 (2020). 10.1007/s10822-020-00290-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Smith J. S., Isayev O., and Roitberg A. E., “ANI-1: An extensible neural network potential with DFT accuracy at force field computational cost,” Chem. Sci. 8, 3192–3203 (2017). 10.1039/c6sc05720a [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.