Abstract
The technology of organ-on-a-chip tries to mimic the complexity of native tissues in vitro. Important progress has been made recently in using this technology to study the gut with and without microbiota. These in vitro models can serve as an alternative to animal models for studying physiology, pathology, and pharmacology. While these models have greater physiological relevance compared to two-dimensional (2D) cell systems in vitro, endocrine and immunological functions in gut-on-a-chip models are still poorly represented. Furthermore, the construction of complex models, in which different cell types and structures interact, remains a challenge. Generally, gut-on-chip models have the potential to advance our understanding of the basic interactions found within the gut and lay the foundation for future applications in understanding pathophysiology, developing drugs, and personalizing medical treatments.
Keywords: Gut, Microfluidics, Organ-on-a-chip, Microphysiological systems, Tissue engineering
1. INTRODUCTION
Though the gut is primarily responsible for facilitating nutrient digestion and absorption, mounting evidence suggests it also plays a central role in the proper function of other organs [1, 2] as well as the etiopathology of many diseases [3–7]. Fundamentally, its barrier function limits the transport of compounds and microbes in and out of the digestive system and protects the body from the passage of unwanted substances and the expansion of pathogenic organisms [7, 8]; however, the gut is also an important part of the immune [3, 9] and endocrine systems [2, 10–13]. Defective digestive, immune, or endocrine function in the gut can lead to the onset of disease in various organs beyond the gut itself [3,4]. Despite the integral role of the gut in maintaining human health, there are still major gaps in our knowledge regarding the mechanisms of its influence [14, 15]. These gaps are the result of ineffective study methods and the poor availability of effective models [15–17].
Animal models have been the primary mechanism for studying the gut [18]; however, these models are limited by their failure to faithfully represent human physiology [19]. In many cases, particularly in studies of drug response, animal models are not able to predict the responses observed in humans [20]. Drug toxicity often differs significantly between species and compounds that are toxic in humans may not be so in animals and vice versa [21]. In addition to their inadequate representative ability, animal experimentation is costly [22] and comes with ethical issues [23]. Moreover, there are several ongoing efforts to modernize toxicity testing [24]. For example, a committee of experts in different fields produced the report titled Toxicity Testing in the 21st Century - A Vision and a Strategy (Tox21c). The committee proposed to decrease using animals and instead use in vitro toxicity tests employing human cells or cell lines, high-throughput screening and quantitative parameters [25]. Efforts like these have driven interest in developing alternative testing models in the past decade [26].
Beyond animal models, in vitro models have also been used extensively to study the gut [20]. Like animal models, these current systems also lack significant representation of physiological processes. Most in vitro models rely upon two-dimensional (2D) cell culture which is inherently limited as it cannot recapitulate the three-dimensional (3D) structure and interactions found in native tissue [27, 28]. In order to properly study gut physiology, pathology, or pharmacology, 3D models and dynamic culture systems must be used [29].
Recent advances in microfabrication techniques and microfluidics [30–32] have facilitated the development of lab-on-a-chip systems that can be used for biological analyses [17, 33]. Systems that more closely represent the 3D structure and physiological microenvironment of native tissues could be created by incorporating live cells into these microfluidic platforms. Furthermore, the cell culture systems could be designed to recapitulate essential organ functions thus, allowing the investigation of such functions in vitro [34, 35]. These culture systems can be made dynamic by controlling fluid flow and introducing new compounds (metabolites, drugs, nutrients, etc.) [36]. Such microphysiological systems, also called organ-on-a-chip (OoC) systems, are considered the next generation of in vitro tools capable of recreating in vivo-like, physiologically-relevant microenvironments needed for 3D tissue culture as well as tissue barriers as they are capable of monitoring the biological responses of cells and tissues to changes in the homeostasis of the device [37]. These platforms have the potential to supplement many 2D culture methods and animal experiments to build a more accurate understanding of the basic biological function and physiological response of intestinal ecosystems [38, 39].
Furthermore, these systems can facilitate new capabilities that were not previously possible using 2D or animal models. Integration with sensors and real-time analytic instrumentation [40] is one way in which the systems can provide insight that is otherwise unattainable [40–42]. With proper design, it is possible to closely monitor cell-level and tissue-level events [43], simulate and reverse pathological situations [44], and study the effect of etiological factors on tissue function [45–47]. The modular nature of on-a-chip devices makes it possible to connect several chips to create multi-organ-on-a-chip (MoC) systems, also referred to as a “body-on-a-chip” [40, 48–50]. These MoC systems can be useful to study inter-organ communication, systemic pathology, pharmacology, and pathogen invasion and distribution [49]. This is particularly useful for studying secondary toxicity that results from the metabolism of drugs in organs such as the gut and liver [8, 51, 52]. While OoC systems can be useful independently, each organ model plays a distinct role in contributing to a physiologically relevant body-on-a-chip model [53].
Gut-on-a-chip systems offer a new and powerful in vitro platform for studying human gut physiology, pathology, and pharmacology [54]. These OoC systems will help advance the understanding and treatment of prevalent diseases such as inflammatory bowel disease (IBD) [55] and colorectal cancer [56]. The use of patient-derived stem cells [57, 58] in OoC systems can also contribute to the development of personalized medicine and drug screening technologies [59]. The development and recent progress made in gut-on-a-chip technology are discussed in this review. The role of these systems in studying pathophysiology and drug development are highlighted. In addition, the current challenges and future perspectives associated with the development and further use of gut platforms are discussed.
2. THE FUNDAMENTALS OF GUT-ON-A-CHIP MODELS
In order to effectively design gut-on-a-chip devices, one must understand the key structures and functions of the organ. The primary functions of the gut are to absorb and transport nutrients, electrolytes and drugs from the digestive system to the vasculature for distribution throughout the rest of the body [20]. The secondary involvement in both the immune and endocrine systems arise from the presence of specialized human cells and the microbiome [60]. The gut is characterized by its enormous surface area, achieved through the presence of folded microstructures known as the intestinal villi and microvilli. These finger-like protrusions of epithelial and specialized cells facilitate the multiple functions of the gut. In addition, the intestines harbor important microorganisms that aid in digestion, immune regulation, and protection from foreign pathogens [60]. These mutualistic microorganisms are able to survive due to the unique hypoxic environment found in the intestines [61]. In addition to the physical and chemical environment in the intestines, mechanical stress is applied by the characteristic peristaltic motion which stretches and squeezes the tissue to propel the contents of the bowel along the gastrointestinal tract (GIT) [62].
2.1. Common chip features
While a wide array of OoC devices and systems have been designed, many share similar characteristics (Figure 1). The body of the chip houses the channels or chambers and any other embedded elements such as sensors, electrodes, or valves. The chips are generally optically transparent and gas permeable (see further discussion in the Materials section) to allow for facile imaging and observation as well as to facilitate the diffusion of gases such as oxygen (O2) and carbon dioxide (CO2). Most models of the gut involve some form of two microchannels that are separated by a porous and flexible membrane. This arrangement is common as it is used to simulate the barrier between the intestinal lumen and the draining vasculature. Cells are initially seeded into channels under static culture conditions to ensure cell adherence and later, media is pumped through the channels using syringe, pressure, or peristaltic pumps to simulate the dynamic microenvironment found in vivo. Generally, one of the channels represents the lumen of the gut and is lined with gut epithelial cells, while the other channel represents a blood vessel and is lined with vascular endothelial cells. The membrane that can be made of an array of different materials with a variety of pore sizes. While the materials may differ, the membranes’ core function remains the same – to allow the transport of soluble molecules between the simulated intestine and blood vessel. Although most gut-on-a-chip systems use membranes (due to their ease of use and facile manufacturing), recent work has eliminated the membrane to improve the similarity to native tissue [63].
Figure 1.
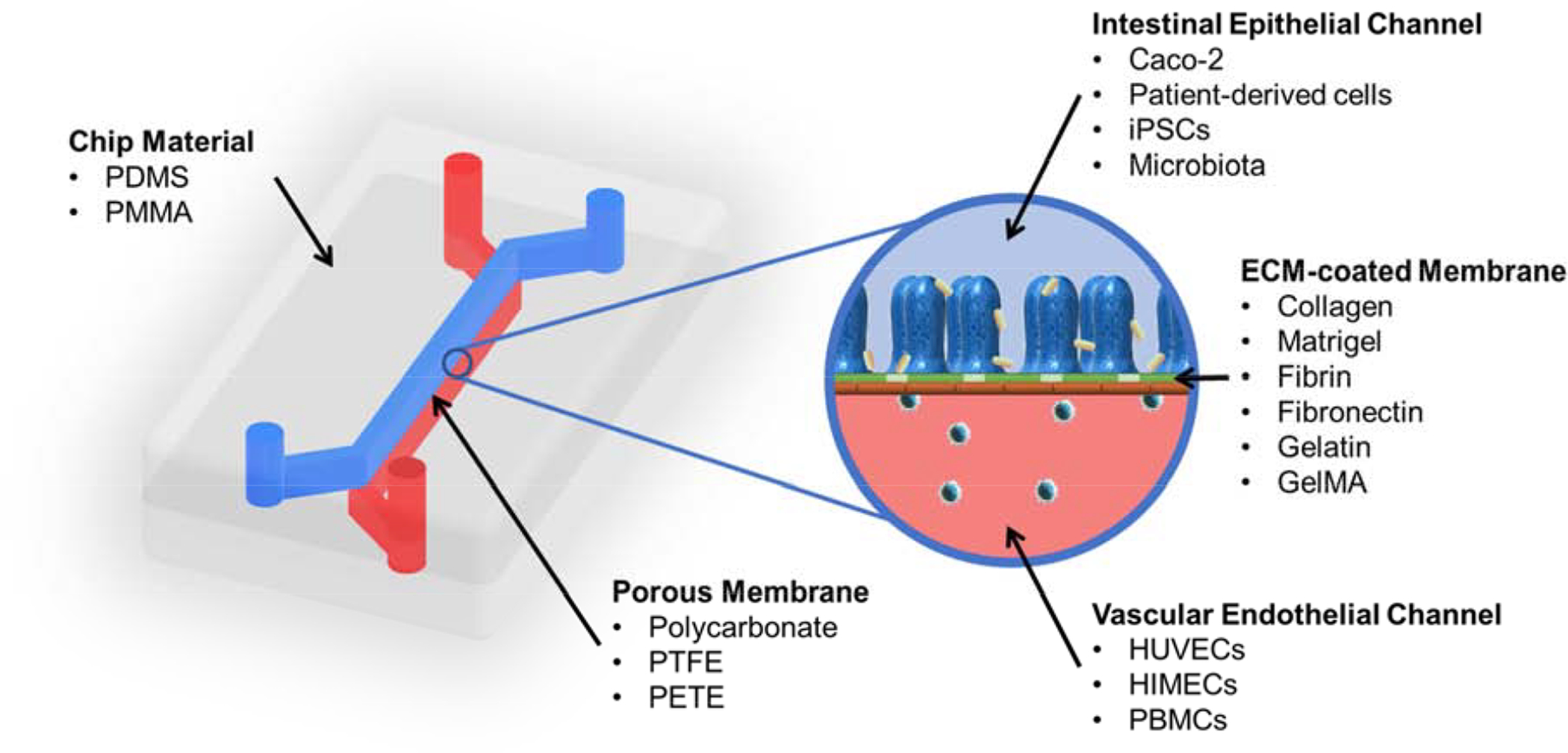
Common features and components of gut-on-chip microphysiological systems.
Another conserved feature amongst in vitro gut models is the cellular structures within the devices. In most cases, the channel representing the vasculature is lined on all sides with a confluent monolayer of vascular endothelial cells. Though simple, these channels have the ability to recreate the barrier properties of native vascular tissue making them important components for studying the absorption and transport of small molecules [64–66].
The analytical methods used to characterize the cells and tissues within the device are also similar across many models. Optically transparent chip materials allow for analysis with light, fluorescent, and confocal microscopy to elucidate cell organization and observe characteristics like polarization [41, 67]. Additional characterization can be performed on the effluent from the device. The used media from both the intestinal and vascular channels can be assayed for dissolved O2, pH, metabolites, signaling molecules, and drug concentration [41, 68].
Another common characterization technique is transepithelial electrical resistance (TEER) measurement. TEER is a non-invasive method for characterizing the barrier function of formed layers of cells inside the OoC. It works by assessing the electrical resistance across a cell layer and it is taken as a relative indicator of the permeability and integrity of this layer. Importantly, TEER measurements must be analyzed in the context of each individual system, as a higher value is not necessarily indicative of a superior barrier. Many factors beyond the integrity of the cell layer are also accounted for in this measurement, including the membrane material physical placement of electrodes, and resistance of the media. In many gut-on-chip systems in which cellular barriers are created represent in vivo interfaces, TEER measurements can be performed with electrodes placed on opposite sides of the barrier [69, 70]. Srinivasan et al. have extensively reviewed variations in TEER values due to factors such as temperature, medium formulation, passage number of cells, and measurement techniques. Their review also analyzes the optimal parameters for TEER measurements and discusses the strengths and weaknesses associated with the various measurement methods [71]. Odijk et al. reemphasized that additional consideration must be given when comparing TEER values across platforms by comparing measurements from an organ-on-chip and Transwell model (Figure 2) [18]. In their comparison, they noted that the TEER measurements taken in the gut-on-chip model were consistently higher compared to a static control (Figure 2C). This trend was constant throughout the 120 hour duration of the study, however, the shear-induced formation of complex 3D architecture in the microfluidic device caused the resistance of the tissue to increase in comparison with the confluent 2D monolayer formed in the Transwell system. Additionally, the group presented and validated a mathematical model that reconciles the differences in TEER values between Transwell devices and organ-on-a-chip systems during the first three days of culture when both systems contain a monolayer of cells. However, this model fails once the villi-like structures are formed in the microfluidic chip. Though TEER is a frequently used measurement for the characterization of barrier integrity due to its simplicity and minimally invasive form, the structure of the model and the maturity of the cultured tissue must be considered when comparing measurements across systems.
Figure 2.
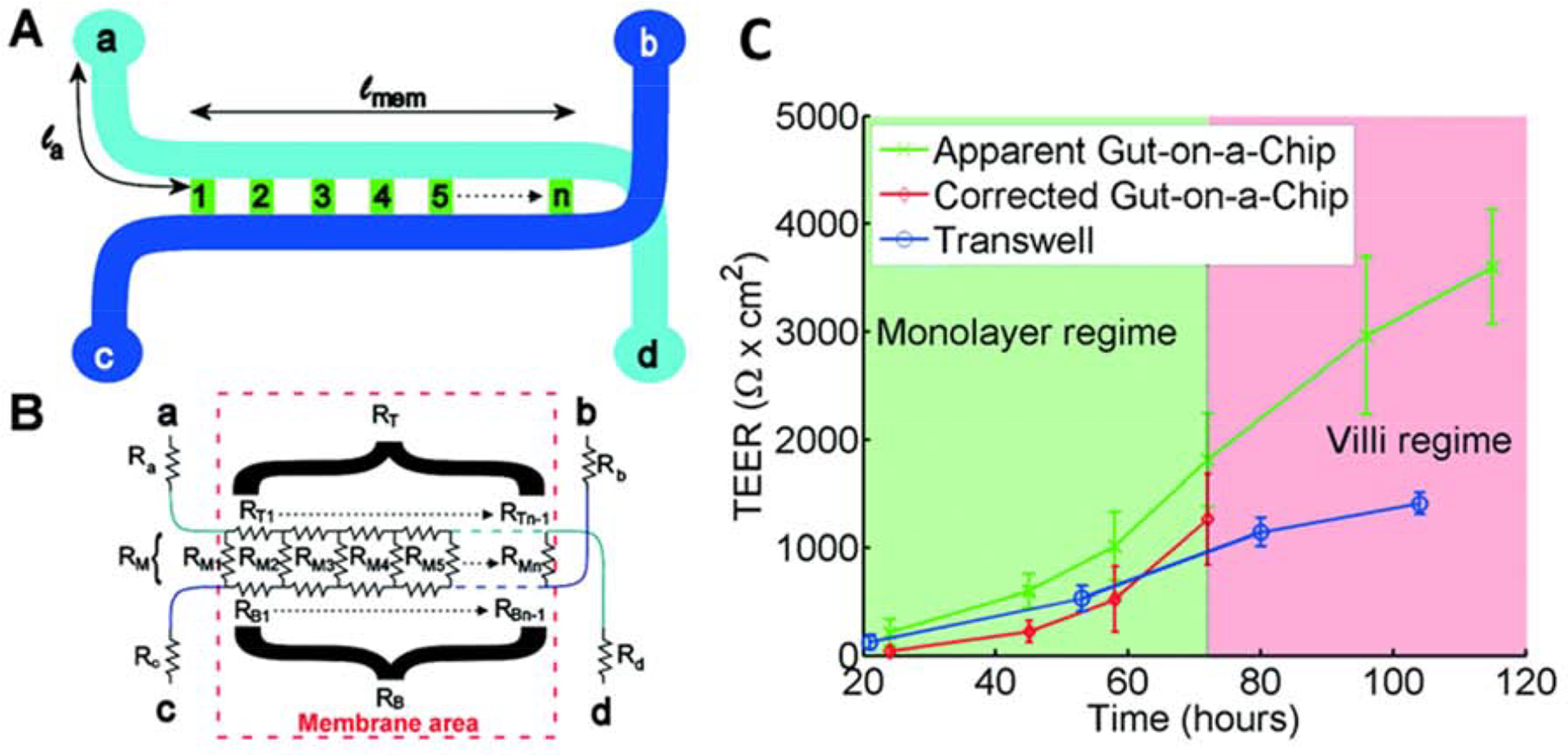
Transepithelial electrical resistance (TEER) measurement in gut-on-a-chip platform: A) Chip layout, B) equivalent electrical circuit for the chip, C) Comparison of TEER measurements values for the gut-on-a-chip (green line) and Transwell (blue line) human intestinal epithelial Caco-2 cells. The corrected gut-on-a-chip line is indicated as red line. Reproduced from [18], with permission from the Royal Society of Chemistry.
2.2. Materials and Fabrication
The material most often used to create the microfluidic systems is polydimethylsiloxane (PDMS). PDMS is the dominant material used for OoC systems because of its cell- and researcher-friendly properties [72]. For cells, PDMS provides a biologically inert, gas-permeable, non-toxic surface with low adhesion [73, 74]. This generally leads to more cell-cell interactions in the devices compared to cell-substrate interactions in traditional 2D culture models. Additionally, PDMS is inexpensive and has many desirable qualities for the creation of microfluidic bioreactors in the laboratory setting. The optical transparency of the material facilitates visual analysis of the enclosed tissue by standard light microscopes. The ability to observe the cells within the device is important as cell viability, activity, and morphology can be tracked in real-time. Because PDMS begins as a viscous liquid prior to curing, it is ideal for casting onto molds to create the customized features required for the fabrication of organ-on-chip devices. The major drawbacks of PDMS used for gut-on-chip models are its hydrophobicity, which can lead adsorption of lipophilic compounds to the walls of the device [75–78], and its gas permeability. Several strategies have been developed to mitigate both concerns. To reduce nonspecific adsorption of hydrophobic compounds, several groups have developed surface modification protocols and other material modifications that are discussed in detail in these recommended review articles [79, 80]. Additionally, while most OoC devices desire gas permeability to ensure sufficient transfer of O2 and CO2 to the cells within the device, gas permeation is ill-suited for the creation of the hypoxic environment found in the intestines. Water vapor can also pass through the PDMS which leads to small amounts of water loss along microfluidic channels, especially at higher temperatures [81]. Other materials used for the creation of gut-on-chip systems include polymethyl methacrylate (PMMA) [40, 82] and glass [83]. While they offer benefits like reduced drug adsorption [84], they are used much less frequently due to increased manufacturing difficulty.
As for the membranes used to separate the channels in gut-on-chip models, they are composed of flexible, porous materials such as polycarbonate, PDMS, and polyester (PET) [72, 83, 85], or biological materials, like collagen [86]. The material is often selected by its pore size which can be modulated to control the translocation of molecules between the two chambers of the device and the availability of nutrients. One study, conducted by Guo et al. observed increased production of sucrase-isomaltase and villin in Caco-2 cells cultured on a porous (0.45μm pore size) non-porous membrane compared to a non-porous PDMS membrane [87]. This suggests that porous substrates may play a role in upregulating cell activity in vitro. Additionally, to increase the biocompatibility of certain materials, chambers and membranes can be coated with natural polymers to facilitate cell attachment, migration, and representative function. Collagen [86, 88], Matrigel [88], gelatin [89], gelatin methacryloyl [90], fibrin [82], and fibronectin [91] are all common coatings for the inner surfaces of the devices.
In some works, topographically patterned scaffolds were also included to help villus formation. Wang et al. implemented a microfabricated topographic analog of intestinal crypts formed in PDMS (Figure 3) [91]. Fibronectin was used to coat the PDMS surface on which Caco-2 cells were seeded and cultured for two weeks. Cell spreading, metabolic activity and differentiation were found to be affected by the topography of the substrate. Specifically, the Caco-2 cells seeded on the patterned substrate showed elevated mitochondrial activity and lower alkaline phosphatase activity at early stages compared to cells seeded on flat substrates. In another study Shim et al. used collagen scaffold with protrusions to seed Caco-2 cells on. The group used photolithography to create a villi mold and casted alginate over the surface to yield a dissolvable mold. Collagen was cast over the alginate to yield the final patterned scaffold. Though the scaffold helped create microvilli structures within the chip, the study concluded that both patterning and fluid flow were necessary to produce in vivo-like enzymatic activity and barrier permeability (Figure 4) [72].
Figure 3.
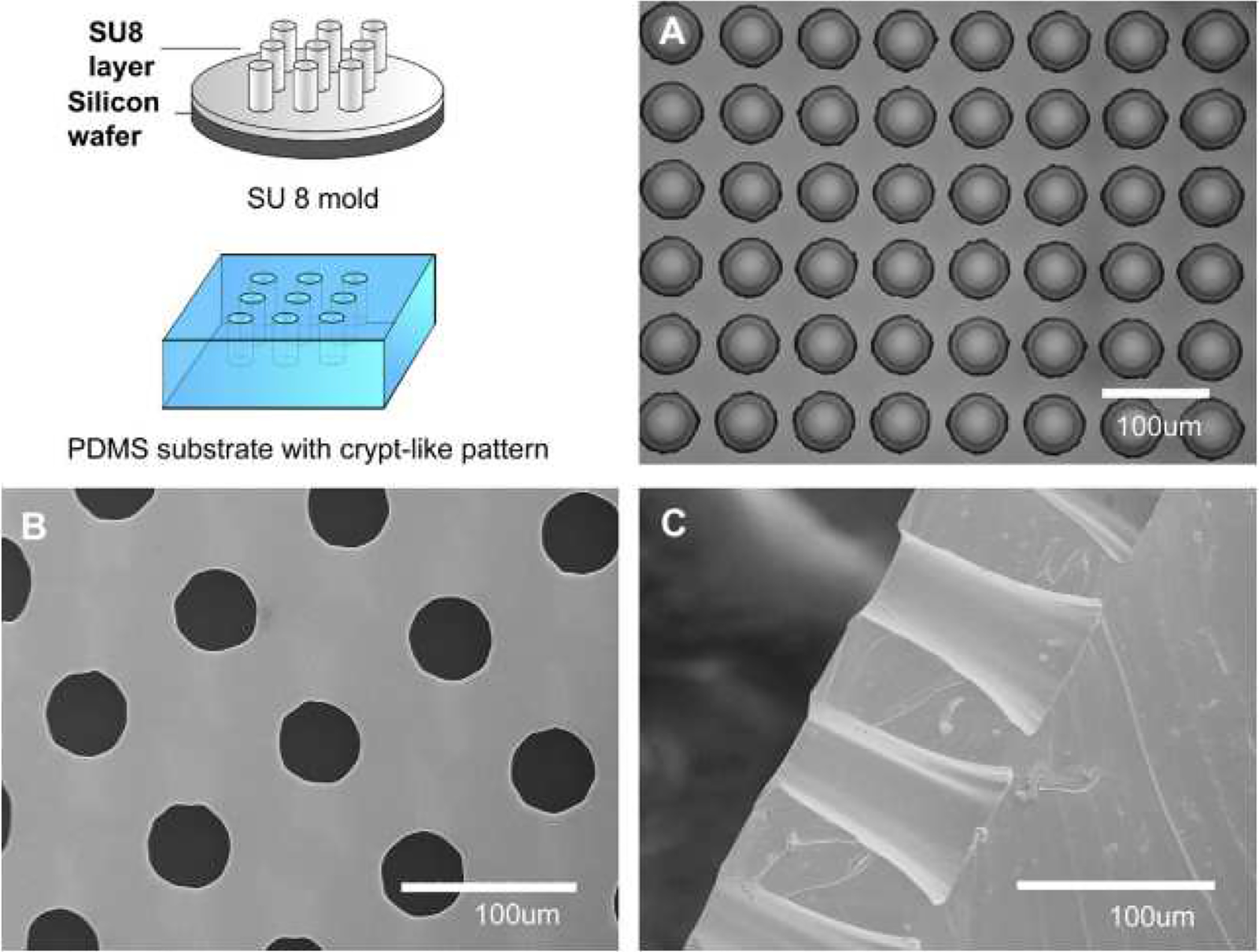
Design and fabrication of a SU-8 master mold and a PDMS negative replicate. (A) Related microscopic images of SU-8 mold. B) Top view SEM images of PDMS scaffolds with micro-well structures, and C) Cross-sectional SEM image of PDMS scaffolds. Reproduced from [91] with permission from Elsevier.
Figure 4.
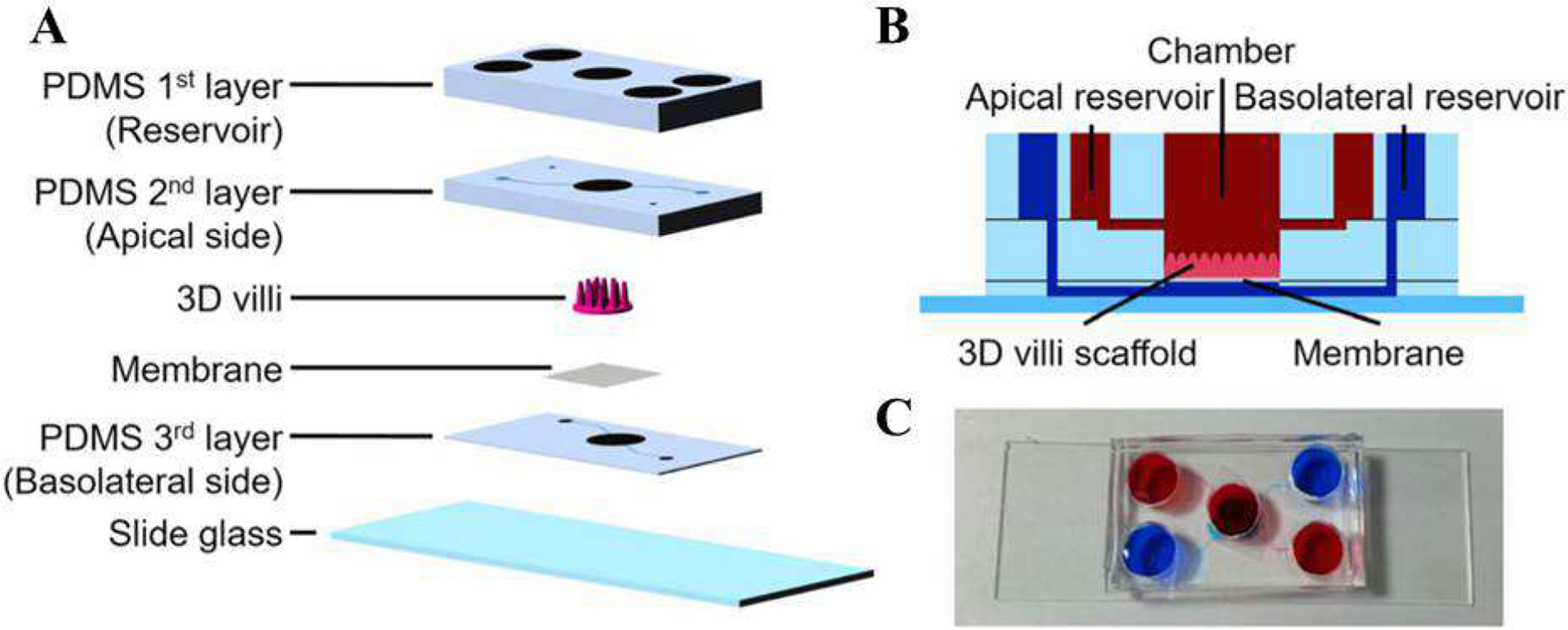
A) Schematics of different component of a gut-on-a-chip with villi structure. B) Side-view of the platform. C) Picture of a platform showing two sets of reservoirs for the apical (red) and basolateral sides (blue). Reproduced from [72], with permission from Springer.
A recent study employed collagen type I-based gel (instead of membrane) to separate two channels of the system (commercially available as OrganoPlate) [63]. One side of the gel was seeded with Caco-2 cells, resulting in the formation of a confluent epithelium with tight junctions and brush borders. To study barrier leakage, 4.4kDa TRITC-dextran and 150kDa FITC-dextran were used. Cells were exposed to staurosporine and aspirin to investigate their effect on membrane permeability. The system showed that concentrations as low as 0.36μM of staurosporine and 40mM of aspirin significantly reduced cell viability and increased permeability of FITC-dextran and TRITC-dextran through the epithelial barrier.
Most microfluidic devices, including OoC systems, are fabricated by creating molds using soft photolithography and casting PDMS over the surface. Typically, the master mold is composed of photoresists (like SU-8) and patterned with photomasks. PDMS is composed of two components, the polymer and its curing agent, which are combined with a weight ratio of 10:1, respectively. The combined PDMS is poured over the master mold and placed in a desiccator to degas the solution and remove any air bubbles. The polymer is cured by placing in an oven set to 80°C for approximately two hours. The cured PDMS can then be peeled from the master mold and it will recreate the features of the mold with micron-scale fidelity. The fluidic ports can be created in the device using flat tip needles or biopsy punches. Once all of the layers needed for fabricating the device are prepared, the multiple components of the device can be assembled with the semipermeable membrane. This is typically done by plasma treating the surface of the PDMS and other silicon substrates (like glass) to expose hydroxyl groups. Upon contact the hydroxyl groups can create stable bonds to form covalent Si-O-Si linkages [72, 83]. While this is the most common fabrication processes, recent work has developed similar systems through 3D printing [32].
2.3. Cells
Multiple cell types have been used within gut-on-a-chip devices and the primary differentiating factor is their source. For cells modeling the vasculature, vascular endothelial cell lines such as human umbilical vascular endothelial cells (HUVECs) [92] and human intestinal microvascular endothelial cells (HIMECs) [88, 93] are commonly used. As for the intestinal cells, a larger variety of cells have been used. Many studies incorporate immortalized cell lines, most notably Caco-2 cells. These cells were derived from human colon carcinoma, but have been the standard for studying the intestines for decades [85, 94–97]. These cells are robust, easily accessible, and have the ability to spontaneously form crypt and villi structures in under five days [95] while differentiating into polarized epithelial cells [97]. Though Caco-2 cells can perform the basic functions of native intestinal epithelial cells (including differentiation), they are severely limited by their inability to produce a significant mucosal layer [98]; this sheet of mucous greatly impacts the permeability of drugs and molecules as it heavily influences the solubility of compounds at the surface of the cells. As a result, several studies have moved beyond immortalized cell lines due to their lack of key features of the typical physiological microenvironment [94, 97].
Primary cells derived from both humans and animals have been incorporated into devices as well. While these cells have increased physiological relevance due to the lack of genetic modifications, they tend to be less robust, require exogenous growth factors, and are less accessible. One study utilized primary human duodenal cells [88]. In these experiments, organoids derived from endoscopic biopsies of healthy tissue were used to create the models. These crypts were then expanded in vitro over 5 to 25 passages and introduced into the device after disaggregation (Figure 5). Within the device, the cells were cultured on a porous, PDMS membrane coated with type I collagen and Matrigel. In the lower vascular channel, HIMECs were loaded. With stimulation from fluidic flow and lateral mechanical strain, the cells were able to occupy the device and form a coherent cell layer within eight to 12 days. Villi-like projections formed in their chip from polarized epithelial cells with multi-lineage differentiation. Transcriptomic analysis, SEM imaging, and immunostaining showed that this system recapitulates the entire human duodenum better than static culture of the organoids from which the cells were sourced.
Figure 5.
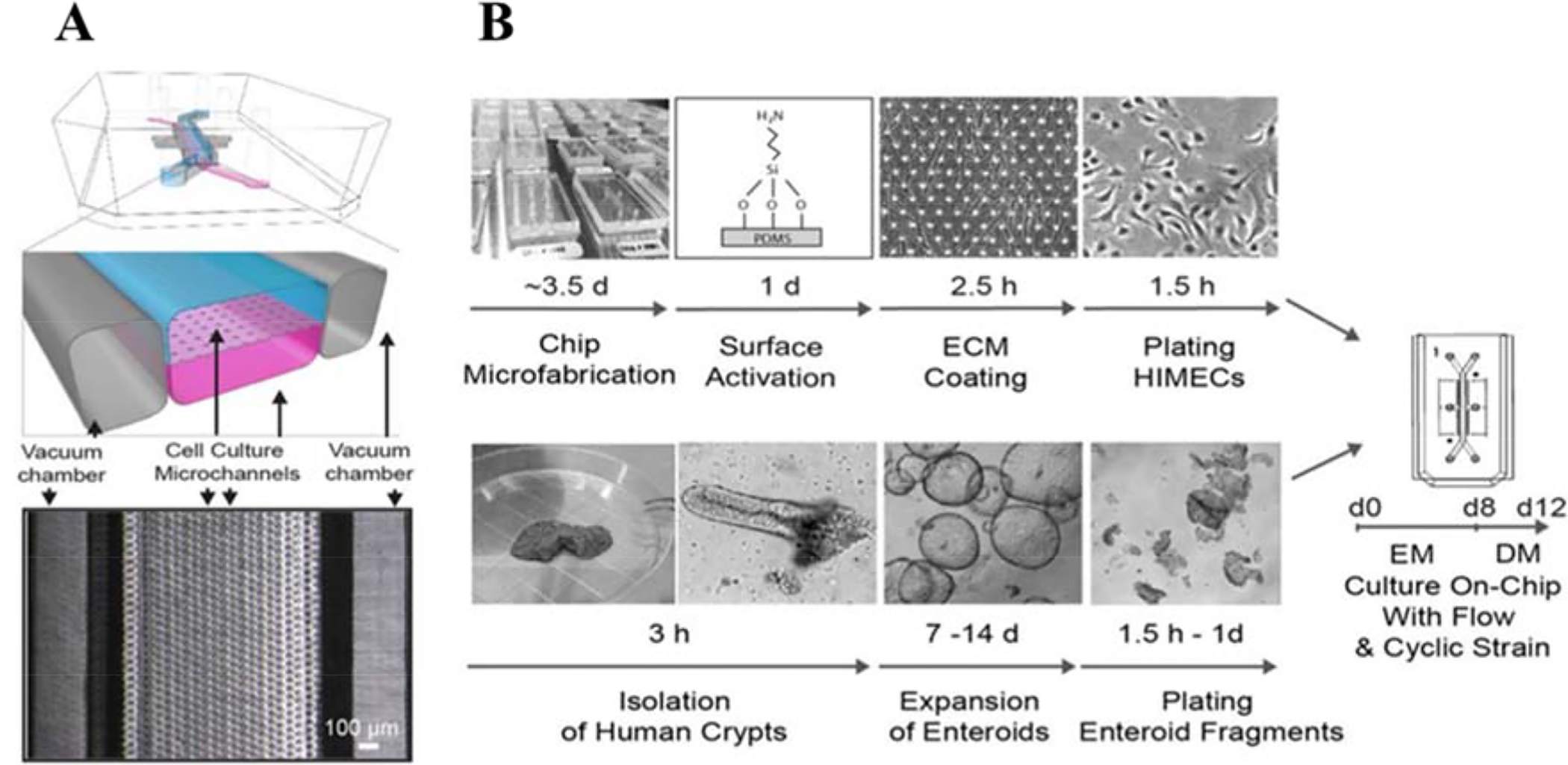
Primary human Intestine Chip platform. (A) A schematic illustration of the chip and a top view phase contrast micrograph of the chip. Vacuum chambers were incorporated to facilitate peristalsis-like mechanical deformation of the tissue, which has been shown to aid in formation and maturation of 3D tissue structures. (B) Schematic illustration of the step-by-step procedure for the establishment of on-chip co-cultures of primary human intestinal epithelium and intestinal microvascular endothelium. Reproduced from [88], with permission from Nature Publishing Group.
Another system developed by Yissachar et al. integrated a resected part of intestine from perinatal mice into an OoC device [99]. It was thought that this helps to preserve the natural microstructure as well as the different cell type composition of the intestine. Models containing whole unmodified tissue segments have the potential to facilitate the investigation of complex interactions between the gut, immune and nervous system by preserving native physiological structure and cellular diversity.
The rise of stem cells has been also translated to OoC models. OoC systems have included an array of stem cell types, including multipotent mesenchymal stem cells (MSCs) [100] and pluripotent embryonic stem cells (ESCs) [101] and induced pluripotent stem cells (iPSCs) [100]. Stem cells have the potential to revolutionize gut-on-chip systems by augmenting their physiological representation through differentiation into the cellular subtypes that are found throughout the intestines, and ultimately develop personalized medicine [102].
2.4. Stimuli
Multiple techniques have been implemented in order to stimulate cells to create mature tissue in vitro. Perhaps the most important of the stimuli is the presence of fluid flow to provide biomimetic shear stress to cells. One of the first studies regarding the importance of mechanically stimulating the cells was published by Chi et al., in which mucin-2 (MUC-2) and actin expression were upregulated under flow in comparison with a Transwell model [103]. Flow also was shown to significantly reduce the number of adherent E. coli in the system. A study by Kim and Ingber showed the ability to induce Caco-2 cell polarization, morphogenesis, and differentiation of into complex villi by simulating luminal flow [95]. The epithelial cells differentiated into different cell types (absorptive, mucus-secretory, enteroendocrine, and Paneth cells) to yield a physiological intestinal tissue model. In comparison to stationary monolayer Caco-2 cell culture, differentiated 3D intestinal structure led to enhanced drug metabolism. In addition to fluid flow, peristalsis-like strain on cells was also found to induce morphogenesis into 3D intestinal villi [95]. Beyond the formation of the villi structures, the cells also demonstrated expression of tight junctions, presentation of a brush layer, and production of mucous. In another gut model studying the remodeling of ECM, Caco-2 cells were co-cultured with subepithelial myofibroblasts to generate a full-thickness model of the intestine [104]. Their findings suggest that the presence of flow accelerates the ability of the cells to remodel the surrounding microenvironment by producing and altering collagen networks, thus contributing to a more physiologically relevant model due to the self-assembly of ECM networks. A recent study extensively characterized the influence of flow on the creation of 3D tissue architecture. Maurer et al. directly compared the morphology and protein expression of Caco-2 cells cultured on a PET membrane in an organ-on-chip system compared to cells in a Transwell system [105]. The variable culture conditions yielded drastically different cellular constructs after seven days (Figure 6A–F) indicating that shear stress induced villi formation. Staining for surface markers on the anatomical structures indicated that the intestinal epithelial cells showed polarized expression of carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1), villin, and tight-junction proteins ZO-1 and occludin (Figure 6G–M). The vascular endothelial cells on the opposite side of the PET membrane showed strong expression of the von Willebrand factor (vWF) (Figure 6I). Differentiation of the epithelial cells was also confirmed by the presence of cytochrome P450 3A4 (CYP3A4), a protein responsible for drug metabolism; α-defensin, a marker of Paneth cells; and MUC-2, a marker of goblet cells (Figure 6N–Q).
Figure 6.
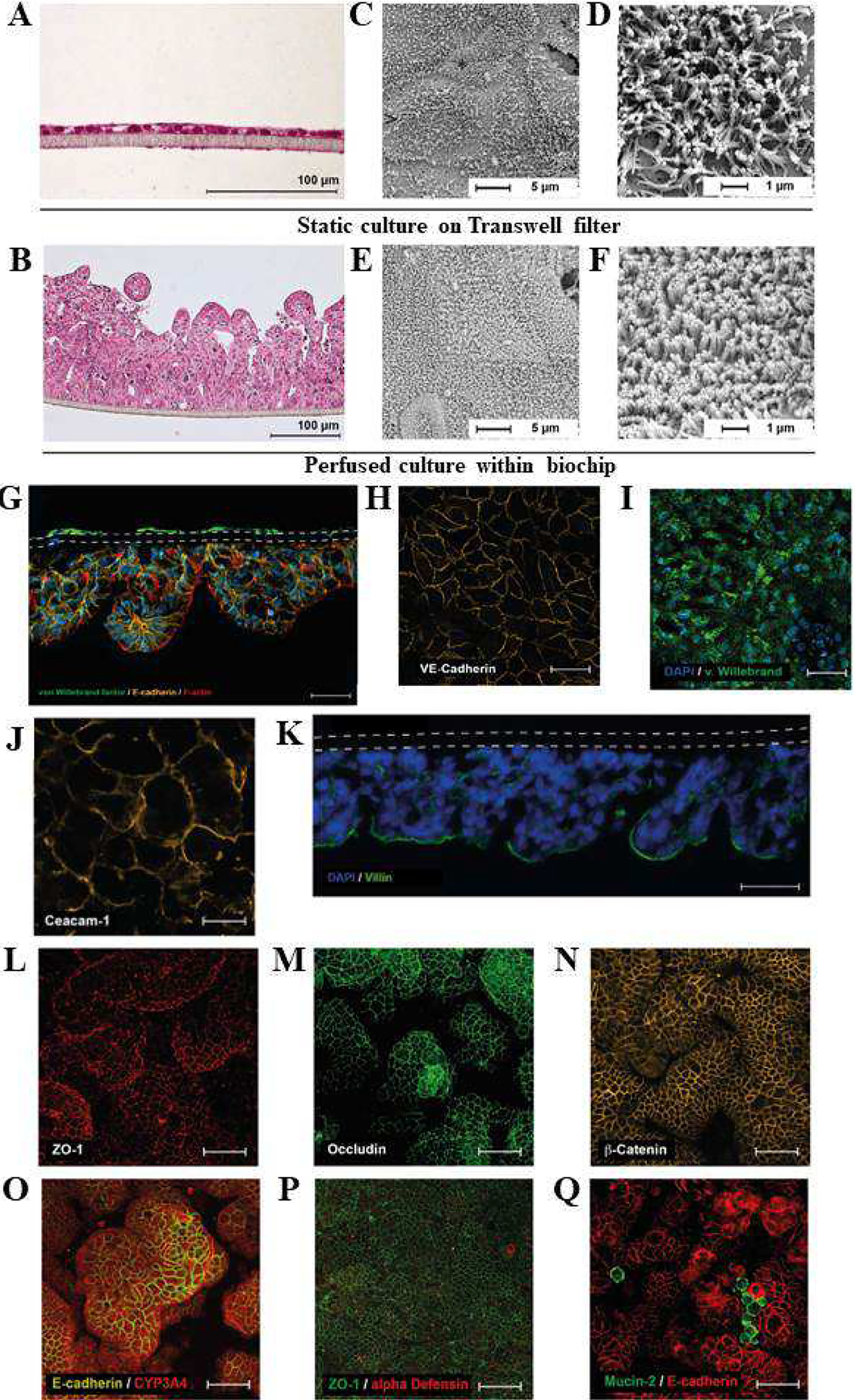
Mechanically stimulated cells form crypt and villus-like structures and express endothelial and intestinal epithelial markers. A, B) Histological H&E staining of Caco-2 cell layers cultured A) statically in the Transwell, and B) under perfusion in the biochip. C-F) Scanning electron microscopy of Caco-2 cell layers cultured for 7 days under C, D) static conditions on Transwell filters, and E, F) under perfusion in the biochip. G) Cross-section of the three-dimensional intestinal model perfused at 50μl/min (in both channels): endothelial cells express von Willebrand factor (green), epithelial cells express E-cadherin (orange), and F-actin (red). Both cell layers are separated by a porous membrane (dashed line). Actin filaments are stained with phalloidin (red). Scale bar 100 μm. Nuclei were stained with DAPI (blue). H-I) Endothelial cells form a confluent monolayer and express H) VE-cadherin (orange) and I) von Willebrand factor (green). J-Q) Epithelial cell layer: Expression of J) CEACAM-1 (orange); K) villin (green) (DAP blue, dashed lines marks membrane); L) ZO-1 (red); M) occludin (green); N) β-catenin (orange); O) E-cadherin (orange); CYP3A4 (red); P) α-defensin (red); ZO-1 (green); Q) mucin-2 (green); E-cadherin (red); Nuclei were stained with DAPI (blue). Figure reproduced from Ref. [105], with permission from Elsevier.
Mechanical stimuli not only impact the intestinal epithelial cells, but also resident microorganisms. Grassart et al. exploited organ-chip technology to study the effect of mechanical forces on Shigella infection [106]. In the 3D microenvironment, Shigella was observed to infect enterocytes from the apical side, using the crypt-like invaginations for early colonization. In the presence of simulated intestinal peristalsis, invasion increased. They concluded that their intestinal model revealed that Shigella leverages the intestinal microarchitecture and mechanical forces to invade the tissue.
2.5. Applications
Thus far, gut-on-a-chip development has been motivated by the need to understand the basic functions of the gut and how they are influenced by environmental conditions, drugs, and other cells. Absorption and barrier function have been extensively studied with these microfluidic in vitro models. One study by Pocock et al. characterized absorption of products of the chemotherapeutic agent SN38 [39]. When comparing the cells cultured under microfluidic conditions compared to the static Transwell model, the device demonstrated superior biological relevance as the cells self-assembled microvilli and produced significantly more F-actin.
Investigating the permeability of the intestine was another early research objective of gut-on-a-chip studies [86]. The most common methods of testing barrier function of tissue within a device include tracking diffusion of marker molecules [63, 86] and TEER [18]. Though TEER is simple and quantifiable, studying the transport of specific molecules often provides better insight into the properties of the molecules dictating their transport across the barrier. The primary factors limiting passage are molecular weight and hydrophobicity. These are tested using an array of compounds, but high molecular weight substances, such as dextran [63], or fluorescent molecules, like Lucifer yellow [86], are commonly used. Other molecules such as curcumin are also used due to the ease of detection using mass spectrometry [107].
Another interesting application of the in vitro gut models is study of the effects of radiation therapy. A model was created to study human intestinal injury resulting from exposure to γ-radiation. The device was used to evaluate the efficacy of novel radioprotective drugs. The device had a PDMS membrane that was lined by human intestinal epithelial cells on one side and by vascular endothelial cells on the other side. Within the device, the team observed generation of reactive O2 species, fragmentation of DNA, cytotoxicity, apoptosis, disruption of tight junctions, villus blunting, and compromised intestinal barrier integrity. Using the device as a screening platform, dimethyloxaloylglycine was identified as an effective prophylactic countermeasure, suppressing all of the adverse effects of radiation therapy [108].
3. MICROBIOME-ON-A-CHIP
The human body hosts at least 100 trillion (1014) microbial cells [109]. These microorganisms are present primarily in the intestine and on the outside of the body, on the skin. Collectively these organisms constitute our microbiome. This complex community interacts with each other and with the host, strongly impacting human physiology [110]. However, the role of the microbiome in influencing human health and etiology of disease is still not fully understood. This knowledge gap has stimulated research as hundreds of studies on the subject have already been published and many others are currently being performed [111].
Alteration in the normal microbiome, i.e. dysbiosis, is commonly linked to a variety of modern-world health problems [112]. Dysbiosis is implicated in the pathology of many diseases, such as metabolic syndrome, obesity, diabetes, autoimmune disorders, allergies, inflammatory bowel disease, hepatic inflammatory disease and some forms of cancer [17]. Additionally, alterations in the gut microbiome can leave the body susceptible to the expansion and invasion of opportunistic pathogens such as Candida albicans and Clostridium difficile [113–115].
Because the microbiome plays such a significant role in human health, the development of new technology to identify and characterize the bacteria colonizing the body is essential. Gut-on-chip systems provide a unique opportunity to study these communities ex vivo. Microbiome composition in organ-on-chip systems can be determined using traditional metagenomic sequencing [116, 117]; however, novel lab-on-chip systems have been developed [118, 119].
Beyond the use of microfluidics for lab-on-a-chip platforms with microbiome applications, gut-on-chip technology can also be employed to investigate host-microbiome interactions and provide useful tools for elucidating the relationship between the composition of gut microbiome and health in humans. The investigation of interactions between microbiota and host in vivo is limited to studies employing analyses of fecal samples or to those using animal models. However, animal models that are used for studying human microbiome are not representative to human physiology [120]. Regulatory pressure to shift away from animal testing, the desire to reduce costs incurred during drug development, and concerns related to the relevance of using animal models [121] are the main driving forces toward developing alternative in vitro models.
In vitro models that deal with investigating microbiome may allow the representation of luminal [122, 123] and adherent microbiota [124] but they generally do not include the evaluation of host response. Traditionally, host response have been evaluated by using bacteria-free supernatants added to cultured human cells [125]. Alternatively, direct-contact co-cultures using Transwell systems [126], beads [127], or gut organoid models [128, 129] were used.
Microscale engineering technologies, such as microfabrication and microfluidics, have enabled to precise control of the cellular microenvironment including the exposure of cells to certain mechanical and biochemical signals. In this section, we present studies of the microbiome that have been carried out using microfluidic approaches. Marzorati et al. developed a host-microbiota interaction (HMI) microfluidic system (Figure 7A), where the response of Caco-2 cells to bacterial products can be investigated [130]. The HMI model consists of two compartments separated, one containing mixed microbiota and the other enterocytes. By using the HMI module, the authors tried to recreate physiologically relevant GIT conditions consisting of: i) a mucosal area for bacterial adherence with shear stress; ii) bilateral transport of low molecular weight metabolites; and iii) microaerophilic conditions. The HMI model has several notable benefits when compared to other systems. First, is the possibility to simulate bacterial adhesion to the wall of the gut and the indirect effect on cells, combined in one system. The potential to perform studies over longer periods of time, of up to 48 hours, with a complex representative microbiota is also a key advantage. The results from their study demonstrated that both host and bacterial cells can be maintained viable in the HMI for 48 hours of co-culture. Mucus-associated microbiota were different from the luminal species and the composition of the microbial community was affected by administered treatment a fermentation product, known to have anti-inflammatory properties. The treatment led to reduced production of the proinflammatory IL-8 between 24 and 48 hours.
Figure 7.
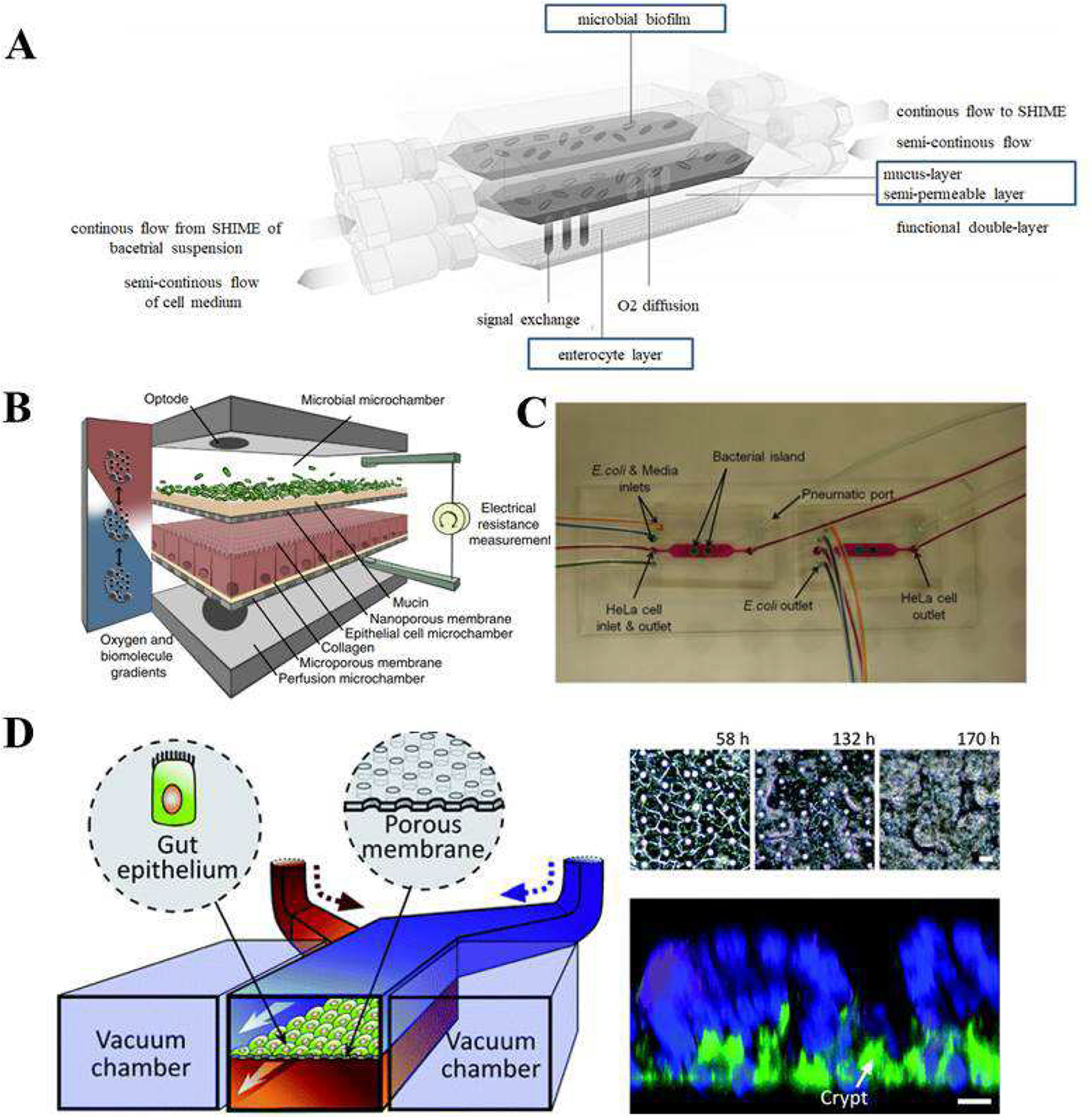
Gut Microbiota-on-a-Chip devices. A) Scheme of the HMI module for long-term studies of the host-microbiota interaction in the GIT. Reproduced from Marzorati et al. [130], under the Creative Commons Attribution 4.0 International License. B) Conceptual diagram of the HuMiX model for the representative co-culture of human epithelial cells with gastrointestinal microbiota. Reproduced from Shah et al. [131] with permission form Nature. C) Microfluidic model for co-culture of epithelial cells and bacteria. Reproduced from Kim et al. [132] with permission from the Royal Society of Chemistry. D) Schematic of the gut-on-a-chip device showing the flexible porous ECM-coated membrane lined by gut epithelial cells cross horizontally through the middle of the central microchannel, and full height vacuum chambers on both sides (left); and Spontaneous formation of intestinal villi by Caco-2 cells cultured in the gut-on-a-chip (right). Reproduced from Kim et al. [134] with permission from the Royal Society of Chemistry.
Using a similar approach in which the bacteria are physically separated from eukaryotic cells, Shah et al. developed a microbiome-gut-on-chip device, which has three chambers, one for epithelial cells, and one for microbial cells and one for medium perfusion (Figure 7B) [131]. The microchambers were separated by a nanoporous membrane. The device had also O2 sensors to monitor the concentration of dissolved O2. TEER was used to assess cell growth and barrier function by using electrodes in the channels. Cells and bacteria were obtained from human colon. Cell differentiation was evaluated by using TEER and fluorescent microscopical evaluation of the expression of Occludin (tight junction protein). Although the system was developed with the intention to study on molecular host–microbe interactions, it may also be applied for drug screening and nutritional studies.
Another model developed by Kim et al. used the epithelial and bacterial cell co-culture for investigating their interactions with the aim of studying the development of GIT infection [132]. Their system was comprised two chambers, one for epithelial cells and the other for bacteria (E. coli BW25113 strain representing commensal bacteria) (Figure 7C). The pathogenic E. coli strain O157:H7 was then added to the commensal bacterial region to mimic the process of intestinal infection. Interaction of pathogenic bacteria with commensals led to reduced degree of infection, as it was indicated by the viability of intestinal cells. This study verified the notion that commensal biofilms in the human intestines are a key component of controlling infectivity and virulence of pathogens.
Just as bacteria can be detrimental to healthy tissue, they can also play an important role in remediation. A study by Kim et al. used an OoC device to investigate the role bacteria plays in the repair of inflammation-damaged gut epithelium [133]. Probiotic bacteria were introduced to the OoC to treat inflammation resulting from pathogenic E. coli. Models like these, while good for studying the negative effects of pathogens, may also be used to develop new therapies for tissues damaged by chronic inflammatory conditions.
To further improve the physiological relevance of in vitro systems, other researchers have co-cultured epithelial cells and bacteria in the same chamber. Kim et al. cultured Lactobacillus rhamnosus GG, on Caco-2 cell monolayer (Figure 7D) [134]. The system was compared with Transwell chambers under static conditions. Caco-2 cells were cultured first for 4–5 days to develop intestinal barrier function, and then the antibiotic-free medium was used 12 hours prior to LGG cell seeding. Results showed that bacterial microcolonies remained tightly adherent to the Caco-2 monolayer for 96 hours, (Figure 7D). Additionally, Caco-2 cells remained viable after co-culture with bacteria. The authors emphasized that the intestinal cell monolayer was able to maintain normal barrier functions in the microfluidic device.
In vitro investigation of direct interactions between intestinal tissue and the microbiome is challenging, because even commensal bacteria can overgrow and kill human cells when grown on culture dishes. A particularly challenging aspect of microbiome models is that most commensal microbes in the intestines are anaerobic [60], thus requiring low O2 conditions which can damage human cells. Recently, Ingber’s team developed a gut-on-a-chip to study direct interactions between the microbiome and intestinal tissue by co-culturing human intestinal epithelial cells with gut microbiota (aerobic and anaerobic) by generating controlled O2 gradients (Figure 8A and B) [93]. Six O2-quenched fluorescent particles positioned in the inlets, centers, and outlets of the upper and lower channels were used to characterize the O2-rich regions of the device (Figure 8C). In the device, physiologically relevant anaerobic conditions (<0.5%) were possible to create by flowing nitrogen at the rate of 243 ml min−1 (Figure 8D). The chips were lined with Caco-2 cells and which were observed to form villi containing polarized cells that were connected by tight junctions (Figure 8E). Sensors in the system indicated that hypoxic culture conditions can be maintained for up to seven days. It was also possible to provide different O2 concentrations at different locations within the device (Figure 8F).
Figure 8.
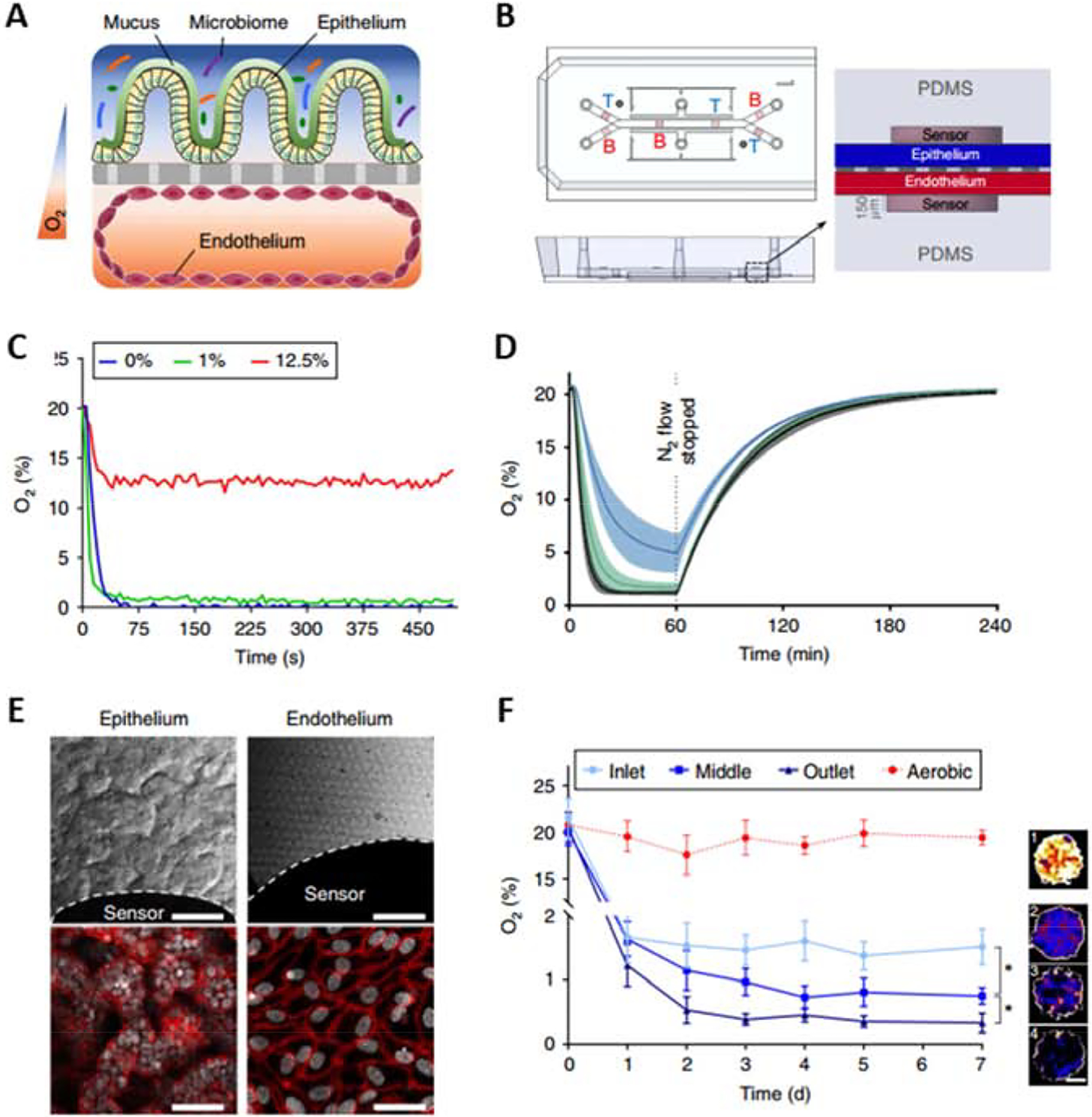
Oxygen-sensitive human Intestine Chip. A) A schematic of the two-channel Chip device with an oxygen gradient. The human intestinal epithelium, which is overlaid with its own mucus layer and complex gut biota, is located over an extracellular matrix-coated porous and flexible membrane. The vascular endothelium lies under the porous membrane. B) A schematic representation of the Intestine Chip with six oxygen-quenched fluorescent particles embedded in the inlets, middles, and outlets of the top and bottom channels. C) A sensitivity analysis of oxygen spots located in the Intestine Chip in response to defined, standard oxygen concentrations. D) Anaerobic chamber validation at various N2 inflow pressures. E) Microscopy images showing the villus morphology of human Caco2 intestinal epithelium (top left; scale bar, 100 μm) and vascular endothelium (top right; scale bar, 100 μm) cultured for 6 days in the microfluidic chip under anaerobic conditions, white dashed lines indicate the border of the oxygen sensor spot. F) Oxygen concentration profiles in aerobically and anaerobically cultured Intestine Chips. Representative pseudocolor insets indicate the average oxygen concentration in the aerobic chip (1) and the inlet (2), middle (3) and outlet (4) of the anaerobically cultured epithelium channel, at day 7 of culture. Scale bar, 200 μm. Reproduced from Jalili-Firoozinezhad et al. [93], with permission from Springer Nature Publishing.
In another study by Gumuscu et al., Caco-2 cells were co-cultured with intestinal bacteria (E. coli) under continuous perfusion in a microfluidic device to conduct preliminary evaluation of drug effects [135]. Antibiotic efficacy was studied in vitro by treating the co-culture with 34μg ml−1 chloramphenicol over 36 hours. The Caco-2 cells were cultured in compartments with varied geometries containing small and large hydrogel interfaces were used to create differences in cell spreading and proliferation of Caco-2 cells. This compartmentalized cell culture system may be adapted to significantly improve the in vitro throughput of drug screening.
4. GUT-IMMUNE INTERACTIONS-ON-A-CHIP
Another physiological realm that can be investigated by employing gut-on-a-chip models is the interaction between the gut and the immune system. The microbiome plays an important role early on as the maternal microbiota is passed on to the newborn to colonize the gut [136]. The initial exposure is responsible for forming the immune system through the activation of the innate immune system and training of the adaptive system. This process is essential as it shapes the mutualistic relationship between the host and microorganisms by initiating responses and desensitizing the immune system to certain microorganisms. While the development period of the immune system occurs early on, it has lasting implications on metabolism [137] and susceptibility to disease [136]. Kim et al. have developed a human gut-on-a-chip platform specifically for the purpose of studying gut-immune interactions [133]. In the device, a porous and flexible membrane was coated with ECM and Caco-2 cells were cultured on its upper surface. With culture medium constantly perfused at 30 μL/h, yielding a stress of 0.02 dyne/cm2, and cyclic mechanical deformation (10% strain, 0.15 Hz), the Caco-2 cells spontaneously formed villi after ~100 h of culture. To study inflammatory activation in the presence of flowing medium with cyclic mechanical deformation, human capillary endothelial cells or human lymphatic microvascular endothelial cells were cultured on the other side of the membrane to model blood vessel walls (Figure 9). The host–microbe co-culture method was the same as described in their previous work [134] as VSL#3, green fluorescent protein-labeled E. coli (GFP-EC), or enteroinvasive E. coli (EIEC) were seeded into the upper chamber. When the non-pathogenic bacteria (GFP-EC strain) were allowed to attach to the luminal surface of the villi, under static conditions, they colonized crypts between the villi. Pathogenic bacteria (serotype O124: NM strain of EIEC) were then introduced into the luminal channel. They were first localized between villi, but then grew over the surface of villi within 24 hours. As a control, pathogenic E. coli derived lipopolysaccharide (LPS) endotoxin was added (15 μg/mL) to the system and the response was compared to the device having EIEC in co-culture. The response to the toxin reproduced in vitro results that were reported in previous studies [105, 138, 139] and animal models [140], as the compound alone was unable to disrupt the on-a-chip intestinal barrier. On the other hand, when the apical surface of epithelium was exposed to EIEC cells, within a short time period of 24–36 hours, normal villus architecture and intestinal barrier function were entirely lost. In patients with intestinal inflammatory diseases, such as IBD, additional immune cells are recruited from the lamina propria as part of the inflammatory response [141]. The authors introduced isolated human peripheral blood mononuclear cells (PBMCs) into the lower capillary channel of the device and allowed them to interact with the lumen without flow for 2 h. PBMCs contain a mixed population of innate (e.g., monocytes and granulocytes) and adaptive (e.g., lymphocytes) immune cells. The production and secretion of inflammatory cytokines is another key property of intestinal inflammatory diseases. Analysis revealed that immune cells and LPS together stimulated epithelial cells to produce four proinflammatory cytokines (IL-8, IL-6, IL-1β, and TNF-α) that induce villus injury and compromise intestinal barrier function. The ongoing inflammation in the intestine resulting from the pathophysiological recruitment of circulating immune cells is regulated by activation of the underlying vascular endothelium. To analyze this organ-level inflammatory response, a monolayer of human microvascular endothelial cells or lymphatic endothelial cells was cultured on the opposite side of the porous ECM-coated membrane. To induce intestinal inflammatory responses, LPS or TNF-α was added to the upper epithelial channel for 24 h, after which the PBMCs were added to the vascular channel for 1 h without flow. Treatment with both LPS (or TNF-α) and PBMCs resulted in the activation of intercellular adhesion molecule-1 (ICAM-1) expression on the surface of the endothelium and a significant increase in the number of PBMCs that adhered to the surface of the capillary endothelium.
Figure 9.
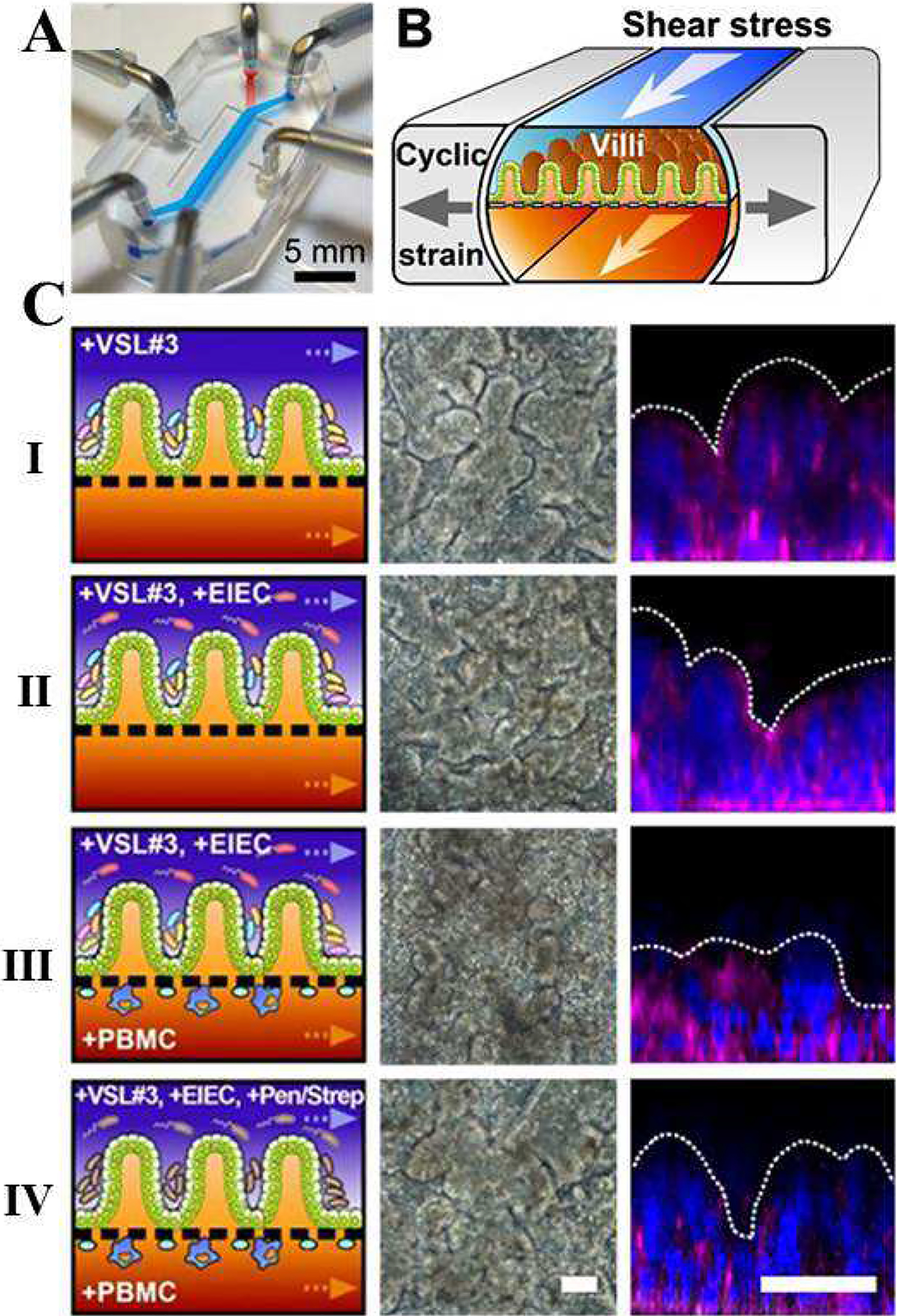
The human gut-on-a-chip microfluidic device for investigation of contributions of the microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation. Probiotic VSL#3 protect against EIEC-induced, immune cell-associated intestinal injury on-chip. (A) A photograph of the device. (B) A schematic of a 3D cross-section of the device showing how repeated suction to side channels (gray arrows) exerts peristalsis-like cyclic mechanical strain and fluid flow (white arrows) generates a shear stress in the perpendicular direction. (C) Morphological analysis of intestinal villus damage under the exposure to (I) probiotic VSL#3 bacteria alone or (II) to the coculture with EIEC (+VSL#3 +EIEC), (III) PBMCs (+VSL#3 +PBMC), or (IV) all cells (+VSL#3 +EIEC +PBMC). The effect of the antibiotic mixture (mixture of penicillin and streptomycin) was tested before the addition of PBMCs (+VSL#3 +EIEC +Pen/Strep +PBMC; open magenta diamonds). The left, middle, and right columns show schematics, phase contrast images (taken at 57 h), and fluorescence confocal micrographs (vertical cross-sectional views) of villi recorded at 83 h after staining for F-actin (magenta), and nuclei (blue). Reproduced from Kim et al. [133], under the Creative Commons Attribution 4.0 International License.
Another study by Shin and Kim developed a similar immune model to observe the onset of gut inflammation [142]. Their system, containing a model of the intestinal epithelium in the upper (apical) channel and the vascular endothelium in the lower (basolateral), was used to examine the intercellular crosstalk between Caco-2BBE cells and peripheral blood mononuclear cells (PBMC) in response to insult by dextran sodium sulfate (DSS), lipopolysaccharide endotoxin, probiotic VSL#3, and non-pathogenic E. coli. Unlike comparable animal models, this system facilitated the isolation of each component of the signaling cascade to identify the root cause of the inflammatory response. It was determined that a sequential disruption of the intestinal epithelium (by the application of DSS) followed by the addition of LPS or bacteria led to elevated secretion of proinflammatory cytokines by the PBMC. Without prior disruption to the gut epithelium, the presence of the LPS and bacteria failed to activate the immune component of the model. Immune-competent models of the gut, such as the one described above, provide unique opportunities to elucidate pathways of dysfunction that may otherwise be obscured using complex animal models.
Ramadan et al. investigated the immuno-modulatory function of dairy food by using a microfluidic system, NutriChip [143]. The chip contained a permeable membrane that separated a confluent layer of Caco-2 cells, which allows the application of in vitro-digested food on its apical side, from a basolateral culture of a monocytic cell line (U937 cells) differentiated into macrophages. U937 is a histiocytic lymphoma cell line with monoblastic characteristics, the authors differentiated the U937 cells into macrophage-like cells after treatment with phorbol esters. This arrangement mimicked the interactions found within the human GIT and facilitated the study of nutrient passage and immune activation. A significant increase in IL-6 secretion was observed in the basolateral media after treating the apical side of Caco-2 cells with LPS and TNF-α for 24 h. No cell de-sensitization was observed after LPS and TNF-α treatment. Furthermore, their results showed a significant IL-6 concentration increase after treating the macrophages with LPS, which demonstrates the possibility of quantifying the induced cytokine production using an on-chip immunomagnetic assay. The group continued to study the impact of immune modulators such as LPS, milk on the production of cytokines and the activation of immune cells.
Models of the gut have also been used to study various types of infection. Earlier studies prior to the widespread use of on-chip systems culminated in the use of fermenters for studying growth and expansion of pathogens [144, 145], however, the use of gut-on-chip models has facilitated investigation into the role of tissue structure in infection. Motivated by the challenges of studying enterovirus infection in animal models, Villenave et al. created a chip to study coxsackievirus B1 (CBV1) in vitro [146]. This model was particularly needed as animals express different virus receptors from those seen in humans, and 2D culture models fail to recapitulate the human gut complexity. The cultured epithelium cells spontaneously formed undulating villus-like structures exhibiting a tight epithelial barrier when analyzed with fluorescent inulin. This was achieved by culturing Caco-2 cells for six days under continuous perfusion (~0.02 dyne/cm2) and cyclic mechanical strain (10%; 0.2Hz), mimicking fluid flow and physiological peristaltic motions of the human intestine, respectively. CVB1 was seeded into the epithelium-lined chamber in the device. To characterize the interaction between CVB1 and the gut-on-a-chip, the authors analyzed the concentration of IP-10 and IL-8 released into the basal and apical effluents. Both cytokine levels were elevated in the infected chips. However, it was consistently much higher in the apical effluent, suggesting a polarized release of cytokines to the luminal side of the epithelium independent of the route of virus entry. The authors observed that it was possible to induce infection when the virus was introduced via the luminal chamber. However, when the virus was delivered through the blood vessel-representing chamber of the device, to simulate a basal route of infection, there was lower viral titers, cytopathic effects, and caspase-3 activation. Additional studies have also investigated the bacterial and fungal infections. Grassart, et al. used an on-chip model to study the role of mechanical force on Shigella infection [147]. The group found that lateral deformation of the epithelium leads to distinct differences in the bacterial invasion compared to unstimulated and 2D Transwell models. Another chip developed by Maurer, et al. was used to study the role of L. rhamnosus in moderating the fungal load of the opportunistic pathogen, C. albicans [105]. Using their model consisting of endothelial cells, Caco-2 cells, and PBMCs, the study demonstrated two key components of native gut physiology: immunotolerance of the bacteria on the apical surface of an intact gut epithelium, reduced colonization of C. albicans in the presence of probiotic L. rhamnosus. Thus far, researchers have significantly advanced knowledge surrounding the role of the immune system in the GIT; however, these microphysiological systems have greater potential in the characterization of the role of immune cells in microbial immunity, food allergies, and pathogen invasion.
5. MULTI-ORGANS-ON-A-CHIP
The incorporation of multiple cell types from several organs is a prerequisite to the creation of in vitro systems that faithfully reproduce relevant biological responses [148]. Though many preclinical models test toxicity on a model of the target organ, advanced multi-organ platforms are required to evaluate the systemic impacts of a compound [149]. Induced inflammatory phenotypes and drug metabolism are factors that must be studied in multi-organ systems to understand biological responses [150, 151]. To address this, several systems have been developed with the goal of studying multi-organ interactions. Vernetti et al. created integrated biological systems recreating the drug digestion process ranging from compound absorption using primary jejunal enteroid cultures; metabolism with hepatocytes, Kupffer, and Stellate cells; and clearance by primary human proximal tubular epithelial cells. These digestion systems were applied to skeletal muscle cultures (primary human myocytes) and neurovascular models composed of iPSC-derived human neurons and astrocytes. Media flowed sequentially from one organ to the other as the system was composed of three microfluidic devices (modeling the liver, kidney, and brain) and one Transwell model (gut) [50]. Another study by Maschmeyer et al. presented a MoC model consisting of primary human small intestinal epithelial cells (intestine model), primary human hepatic stellate cells co-cultured with immortalized HepaRG cells (liver organoid), human skin biopsies, and RPTEC/TERT1 cells (kidney organoid) for studying multi-organ toxicity, drug absorption, distribution, metabolism, and excretion (ADME) [152]. Viability and function of the tissues were maintained concurrently over 28 days and were verified by immunostaining and mRNA expression. Li et al. proposed another system that integrated Caco-2 cells as a model of the intestines in conjunction with rat primary glomerular endothelial cells (simulating the kidney) in a compartmentalized micro-chamber of a microfluidic system [153]. The system was then used to demonstrate that digoxin induced cell death and endothelial permeability, which is consistent with clinical digoxin nephrotoxicity.
The intestine and liver are the main barriers in first pass metabolism for orally delivered compounds. Their ability to modulate drug transport greatly influences the available concentrations of the drug in vivo and modulates drug efficacy and the presence of side effects. The multi-organ nature of first pass metabolism is difficult to study in current in vitro models using conventional cell culture approaches. To address this issue, Choe et al. created a gut-liver platform on-a-chip that can mimic the dynamics of first pass metabolism [8]. Caco-2 cells were used as a model of the gut epithelium while HepG2 cells were used as the liver model. To recreate the process of absorption and metabolism, fluid passed from the gut chamber into the liver chamber. Notably, co-culture of the cell lines led to changes in the physiological function of both cell types. The permeability of the Caco-2 cells was drastically decreased in the presence of shear stress and both cell lines exhibited increased activity of cytochrome P450 metabolic activities significantly increased. The significant alterations in the behavior of both cell types affirms the need for the development of co-culture systems to increase the physiological relevance of in vitro models. Other models of the gut-liver relationship have been developed to focus on the management of specific therapeutics including the intestine’s ability to shield the liver from nanoparticles [154] and the variable metabolism of epirubicin, irinotecan, and cyclophosphamide by a lung-liver-gut microfluidic system [155]. Chen et al. created a liver-gut system that was used to demonstrate the gut’s ability to genetically manipulate the production of bile by the liver. Additionally, under inflammatory conditions, the two organs communicate to amplify the inflammatory response [156]. Tsamandouras et al. proposed another MoC in vitro system to investigate the different pharmacokinetic processes related to oral drug administration in humans including intestinal permeability and hepatic metabolism [45]. Their findings showed that interorgan communication can upregulate hepatic metabolism. Midwoud et al. proposed one of the first attempts to study gut-liver axis in a chip-based system [157]. They studied the applicability of a microfluidic device for the perfusion of precision-cut intestinal slices and the sequential perfusion of intestinal and liver slices in a co-culture perfusion system, to mimic in vivo first pass metabolism. The use of tissue slices maintained the physiological tissue structure found in the body. Another study by Bricks et al. used a gut-liver coculture system to show uptake of phenacetin by the intestinal epithelium and subsequent metabolism by the liver cells into bioactive paracetamol [158]. Their proposed bioreactor showed higher metabolic performance compared to coculture in a static petri dish. These approaches that integrate multi-cellular tissue constructs, multiple MPS, and quantitative mechanistic modeling, are powerful tools that have broad applications in pre-clinical drug development.
In order to demonstrate the potential of GIT–liver models for predicting human response in preclinical studies, Chen et al. designed an integrated system culturing primary human intestinal cells (hIECs) and 3D liver tissue together [159]. The hIECs were made immortal by transducing the cells with hTERT, and following seeding, they were connected with the liver tissue by gravity-driven media flow between the respective chambers. Both hIECs and HepG2 C3A liver cells demonstrated excellent viability after 14 days of co-culture. The study continued to compare the permeability of the intestinal model with a Transwell model using Caco-2 cells by testing the uptake of caffeine, mannitol, and propranolol. The gut-liver-on-chip system was tested over 2 weeks and yielded consistent metabolic rates as urea and albumin production was uniform throughout the system. When compared to single OoC systems, metabolizing enzyme CYP activity in the co-culture GI tract–liver system was elevated significantly [159].
A final interesting application of MoC platforms is the study of colon cancer. Skardal et al. developed a metastasis-on-a-chip platform that used fluorescent tracking of colon cancer cells from a hydrogel gut construct to observe invasion of a downstream liver construct using a circulating fluidic device [160]. Metastatic tumor foci grew and eventually disseminated from the intestinal construct, entering circulation, and subsequently reaching the liver construct. While gut-liver systems have been primarily used to study metabolism and interorgan communication, these models also have promise to provide insight into the behavior of invasive and metastatic cancers.
6. COMMERCIAL USE AND PERSONALIZED MEDICINE
New drugs, cosmetics, and chemicals must be evaluated for safety prior to clinical use. For decades, 2D cell culture and animal models served as the gold standard for preclinical safety and efficacy tests [161]. Though animal trials allow the study of compounds within complex organisms, these studies are time-consuming, resource-intensive, and ethically controversial, while they often yield species-specific information that does not accurately predict human toxicity [162, 163]. The failure of these models to faithfully represent the in vivo response of human organs has contributed to delayed observation of organ toxicity that can avoid identification until phase III and phase IV clinical trials [164, 165]. The limitations of traditional toxicity models have produced interest in gut-on-chip systems to serve as a supplementary models to study compound toxicity.
Though many academic labs have devoted their resources to the design and development of gut models, relatively few have tailored their systems for large scale commercial use. Like other microphysiological systems, gut-on-a-chip devices have been extensively used to study specific cellular and tissue-level interactions and relationships in vitro; however, the objectives of commercial systems would primarily focus on efficacy and toxicity tests for pharmaceuticals, chemicals, and other materials. Many organizations are looking to shift away from animal models (due to validity and ethical concerns) as the demand for early screening methods rises. To satisfy industrial users, commercial platforms must be inexpensive, easy-to-use systems that provide reproducible data with minimal employee training and capital investment. Recently, several companies such as Emulate, Mimetas, and StemoniX have entered the commercial market to bring the cutting-edge microfluidic systems to pharmaceutical and cosmetics companies such as Roche, Takeda, Merck, and Johnson & Johnson [166–168].
One of the primary questions that must be addressed for many pharmaceuticals and ingested small molecules is their permeability through the gut epithelium. For orally delivered compounds, poor permeability or high susceptibility to degradation can significantly inhibit absorption, thus restricting distribution throughout the rest of the body [169]. Early gut models studied the permeability of hydrophobic and hydrophilic drugs through simulated epithelial barriers [107, 170]; however, MoC models are needed to increase the physiological relevance. The liver is especially relevant for drug metabolism studies as it is responsible for clearance of endogenous and exogenous molecules, like drugs, from the blood by metabolizing the compounds to become more hydrophilic [171]. Due to the chemical modifications that are made, the solubility, absorption, and chemical interactions of the molecule can be altered and lead to drastic changes in efficacy and toxicity. Because of the role of the liver in metabolism, Tsamandouras, et al. presented a complex microfluidic system that houses both gut and liver tissues to create microphysiological system for preclinical drug screening [45]. Their aim was to investigate the different pharmacokinetic processes accompanying oral drug administration and metabolism of diclofenac and hydrocortisone. The system demonstrated that the tissues communicated through soluble signals to modulate hepatic metabolism. While the system could faithfully emulate interorgan signaling, the fluidic platform used was complex and would likely require additional equipment to operate on an industrial scale. Another device developed by Bricks et al. also housed monolayers of intestine and liver tissues [172]. This study focused on the transport and metabolism of phenacetin, an analgesic taken orally that is metabolized into paracetamol, from the apical side of the intestinal epithelium into the liver. They demonstrated that microfluidic circulation of the fluidic greatly increased liver metabolism of the drug compared to controls in a static culture. Chen et al. proposed an integrated gut-liver microfluidic system to co-culture primary human intestinal and hepatic cells [159]. When comparing the behavior of the cells independently and in co-culture, the activity of CYP (a metabolic enzyme) had notably enhanced in the GI–liver co-culture system, suggesting that simple single organ-on-chip models may be insufficient to predict efficacy. One notable aspect of the system developed by Chen et al. is that the device is pump-free and uses gravity-driven flow. Commercial systems can greatly benefit from pump-free devices as they reduce the capital cost required to adopt the model platforms in addition to reducing the training required to use the devices. While there has been remarkable progress towards developing multi-organ digestive models, a greater push must be made to make the systems simple and cost-effective to move these systems into commercial use.
Though the first commercial applications of gut-on-a-chip platforms will likely be for toxicology, these chips have the potential to improve patient care through the realization of personalized medicine. These devices could be used to culture patient-derived cells that emulate the biology and physiology of the individual. Once a relevant model has been established, health care providers could screen potential therapies with the goal of identifying the optimal treatment plan prior to administration to the patient. Several academic groups have worked towards this goal by studying the inclusion of patient-derived cells and organoids in gut-on-chip devices. A recent study by Kasendra et al. fabricated models of the small intestine by incorporating epithelial cells harvested from biopsies [88]. In the presence cyclic strain and fluid flow, the primary cells remained viable and formed the complex villi structures native to the GI tract. While the study demonstrated that primary cells can be used, the cells were not seeded in the device until several weeks after the initial biopsy. The extensive culture time required prior to their use may be acceptable for some models; however, for many clinical applications, relevant data must be obtained within days to effectively inform treatment decisions. Jalili-Firoozinezhad et al. built on the previous success by incorporating patient-derived microbiota into a gut-on-chip device [93]. Fecal samples were collected from infants and cultured in anaerobic conditions in a microfluidic channel with patient-derived intestinal cells. Under anaerobic conditions, the device sustained over 200 unique bacterial types representing 11 different genera. While additional work needs to be done to facilitate the in vitro culture of a more representative microbiome, the device used is an excellent example of balancing device simplicity and physiological relevance. Additionally, the essential role of the intestinal microbiome in digestion and gastrointestinal health warrants its inclusion in in vitro models of the gut, especially for personalized medicine [4, 173, 174]. Another strategy for the development of personalized in vitro models include the use of iPSCs to generate the tissue models [100, 175], however, these methods are currently time-consuming and complex. While the foundation for the use of gut-on-chip devices for personalized medicine applications has been laid, additional work is required to study the validity of current models and to develop new devices the better represent the in vivo behavior of the human intestine.
7. CHALLENGES AND FUTURE DIRECTIONS
While extensive work has already been conducted in the pursuit of representative in vitro models of the human intestines, the field has significant challenges it must overcome to improve the utility and implementation of these devices in industrial and clinical settings. In general, new systems must balance complexity with relevance in order to optimize cost, ease-of-use, and the output of meaningful data using technologies such as machine learning [176]. The current primary challenge is the incorporation of physiologically relevant cells. Traditionally, animal-derived or immortalized cell lines with poor biological relevance to human cells found in vivo have been used in microfluidic models [94]. The widespread adoption of iPSC-derived cells from humans has facilitated the generation of more relevant terminally differentiated human cells [100]. Intestinal organoids derived from human pluripotent stem cells (hPSCs) can be another valuable source of cells to recreate physiological responses in vitro with high fidelity [177] and will play an important role in drug development [178]. Of primary importance for preclinical drug development models are gut-liver systems. The co-culture of human liver spheroids with human intestine in a MoC platform is a promising approach for recreating the fundamental drug metabolism mechanisms in vitro [152, 179]. Beyond improvements in cell sourcing, incorporating stimuli-responsive materials [180, 181] and biosensors [182] in the systems can assist in better directing and monitoring stem cell differentiation into the desired cell types [183]. Better control over the terminal differentiation of cells will further improve our models of GI physiology, GI disease, and drug toxicity screening.
One major obstacle for organ-on-chips is the limited life span of cells in the devices [184]. This limitation is exacerbated when using primary—as opposed to immortalized—cells in the systems. The limited ability of current immortalized cell lines to faithfully mimic in vivo biology is also a primary concern with these systems. Chip systems aimed at studying drug toxicity have all been hampered by inadequate function of luminal drug transporters and metabolic enzymes [184], and as a result, current models can only achieve partial recreation of normal physiology. One cause of this may be the use of inadequate biomaterials in the microfluidic systems. Clinically useful systems must use materials that are biocompatible, inert, non-adsorbent, and non-leaching (to avoid introducing by-products of the material unintentionally [185]. As discussed in last sections, for instance in the application of PDMS in microfluidic OoC devices it should be considered that for a wide range of cellular assays, PDMS can adsorb small and hydrophobic molecules, also some drugs or fluorescent marker that incubated with cells can be diffused into the PDMS walls devices, which reduce the reproducibility of concentration of solute molecules in solution [76, 77], so modification of the PDMS surface by some alternative polymeric or ECM-based materials can solve these drawbacks [40].
The challenges associated with creating in vivo-like tissue structures with relevant cell lineages and organization within these devices may be addressed by three-dimensional bioprinting technologies. These methods facilitate precise, reproducible production of complex and multiplexed structures [186–188]. Multimaterial and multicomponent bioinks development can also improve cell-cell and tissue-tissue interaction by the use of materials that can be metabolized and help in the formation of extracellular matrix [189–191]. This strategy has also been used by researchers engaged in the generation of droplets for cells encapsulation [192]. These strategies, if applied in the area of OoC, can facilitate the development of platforms with greater tissue-tissue interaction, besides contributing with greater spatial organization of different cell types. Newer printing technologies such as four-dimensional (4D) bioprinting have also been developed [193–195] to create tissues with higher-level, dynamic functions that [196] and they can potentially be integrated into microfluidic platforms.
Aside from improving the cellular and biological contents of the microfluidic systems, monitoring the tissues with integrated sensors and improved microscopy techniques will help generate more data to gain a better understanding of the [197, 198] cellular behavior in the device [199]. While TEER measurements derived from embedded electrodes have been widely used for quantifying epithelial barrier integrity [200], additional analytical methods such as the integration of mass spectrometric analysis [201] can facilitate identification and quantification of genetic, proteomic, and metabolomic signatures in response to specific compounds and stimuli [185, 202]. The inclusion of data communication tools in the sensor suite will also lead to new capabilities such as real-time data analysis from remote computation centers. Advances in imaging technologies to better capture the 3D tissue structures maintained within the devices have the potential to yield additional information, especially when observing multi-cellular constructs. While these improvements will not enhance the biological behavior of the cells, sensors, imaging, and communication tools will facilitate the implementation of more complex computational and analytical methods.
One of the primary goals of using organ-on-chip systems in preclinical settings is the profiling of drug ADME characteristics. While extensive preliminary data suggests that some systems may serve as viable tools, system validation, design standardization, and integration with standard laboratory tools will be prerequisite to large scale adoption of these technologies [185]. The currently available devices are not sufficient for comprehensive ADME studies and, as a result, no single system has gained widespread acceptance. The first devices to successfully predict toxicity will be significant advances in biomedical science; however, the current systems are limited by insufficient tissue-tissue interactions and absent lymphatic and nervous components.
8. CONCLUSIONS
In the past few decades advances in the field of microfluidics and microfabrication have contributed to the development of dynamic cell culture systems and in vitro models recapitulating the human gut. These on-chip systems can be useful tools in the prediction of drug toxicity and for modeling various disease states in vitro. They hold promise to reduce the occurrence of unpredicted side-effects to medications, and to cut costs of drug development leading improved health care practices with more accessible pharmaceuticals and fewer problems. By developing advanced in vitro models of the human intestines, gut-on-a-chip technology can accelerate research into the physiology, pathology, and pharmacology of gastrointestinal diseases.
Acknowledgements
The authors also acknowledge funding from the National Institutes of Health (R01AR066193, R01AR057837, and R01EB021857).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure
No financial interest in the subject or funds received in the preparation of this work. No conflict of interest reported.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REEFRENCES
- [1].Compare D, Coccoli P, Rocco A, Nardone OM, De Maria S, Cartenì M, Nardone G, Gut–liver axis: The impact of gut microbiota on non alcoholic fatty liver disease, Nutrition, Metabolism and Cardiovascular Diseases 22(6) (2012) 471–476. [DOI] [PubMed] [Google Scholar]
- [2].Lyte M, Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior, PLoS Pathog 9(11) (2013) e1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Moser G, Fournier C, Peter J, Intestinal microbiome-gut-brain axis and irritable bowel syndrome, Wien Med Wochenschr 168(3–4) (2018) 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C, Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment, Genome Biol 13(9) (2012) R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J, The neuroactive potential of the human gut microbiota in quality of life and depression, Nature Microbiology (2019). [DOI] [PubMed] [Google Scholar]
- [6].Raimondi MT, Albani D, Giordano C, An Organ-On-A-Chip Engineered Platform to Study the Microbiota–Gut–Brain Axis in Neurodegeneration, Trends in Molecular Medicine (2019). [DOI] [PubMed] [Google Scholar]
- [7].Schaik W.v., The human gut resistome, Philosophical Transactions of the Royal Society B: Biological Sciences 370(1670) (2015) 20140087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Choe A, Ha SK, Choi I, Choi N, Sung JH, Microfluidic Gut-liver chip for reproducing the first pass metabolism, Biomed Microdevices 19(1) (2017) 4. [DOI] [PubMed] [Google Scholar]
- [9].Fung TC, Olson CA, Hsiao EY, Interactions between the microbiota, immune and nervous systems in health and disease, Nat Neurosci 20(2) (2017) 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jameson KG, Hsiao EY, Linking the gut microbiota to a brain neurotransmitter, Trends Neurosci 41(7) (2018) 413–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, Dinan TG, Minireview: Gut microbiota: the neglected endocrine organ, Molecular endocrinology (Baltimore, Md.) 28(8) (2014) 1221–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cani PD, Knauf C, How gut microbes talk to organs: The role of endocrine and nervous routes, Mol Metab 5(9) (2016) 743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Latorre R, Sternini C, De Giorgio R, Greenwood-Van Meerveld B, Enteroendocrine cells: a review of their role in brain-gut communication, Neurogastroenterol Motil 28(5) (2016) 620–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Conlon MA, Bird AR, The impact of diet and lifestyle on gut microbiota and human health, Nutrients 7(1) (2014) 17–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Guinane CM, Cotter PD, Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ, Therap Adv Gastroenterol 6(4) (2013) 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Karl JP, Hatch AM, Arcidiacono SM, Pearce SC, Pantoja-Feliciano IG, Doherty LA, Soares JW, Effects of Psychological, Environmental and Physical Stressors on the Gut Microbiota, Front Microbiol 9 (2018) 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Trujillo-de Santiago G, Lobo-Zegers MJ, Montes-Fonseca SL, Zhang YS, Alvarez MM, Gut-microbiota-on-a-chip: an enabling field for physiological research, Microphysiological Systems 1 (2018) 1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Odijk M, van der Meer AD, Levner D, Kim HJ, van der Helm MW, Segerink LI, Frimat JP, Hamilton GA, Ingber DE, van den Berg A, Measuring direct current trans-epithelial electrical resistance in organ-on-a-chip microsystems, Lab Chip 15(3) (2015) 745–52. [DOI] [PubMed] [Google Scholar]
- [19].Shanks N, Greek R, Greek J, Are animal models predictive for humans?, Philosophy, Ethics, and Humanities in Medicine 4(1) (2009) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DEJC, m. gastroenterology, hepatology, Microfluidic organ-on-a-chip models of human intestine, 5(4) (2018) 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR, Can animal models of disease reliably inform human studies?, PLoS Med 7(3) (2010) e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Malcolm W, The Potential of Organ on Chip Technology for Replacing Animal Testing, Brill, Leiden, The Netherlands, 2019, pp. 639–653. [Google Scholar]
- [23].Doke SK, Dhawale SC, Alternatives to animal testing: A review, Saudi Pharm J 23(3) (2015) 223–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ashammakhi N, Darabi MA, Çelebi-Saltik B, Tutar R, Hartel MC, Lee J, Hussein SM, Goudie MJ, Cornelius MB, Dokmeci MR, Khademhosseini A, Therapeutic Nanomaterials: Microphysiological Systems: Next Generation Systems for Assessing Toxicity and Therapeutic Effects of Nanomaterials (Small Methods 1/2020), Small Methods 4(1) (2020) 2070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krewski D, Acosta D Jr., Andersen M, Anderson H, Bailar JC 3rd, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S Jr., Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L, Toxicity testing in the 21st century: a vision and a strategy, Journal of toxicology and environmental health. Part B, Critical reviews 13(2–4) (2010) 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Adler AI, Boyko EJ, Ahroni JH, Smith DG, Lower-extremity amputation in diabetes. The independent effects of peripheral vascular disease, sensory neuropathy, and foot ulcers, Diabetes Care 22(7) (1999) 1029. [DOI] [PubMed] [Google Scholar]
- [27].Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E, Davenport-Huyer L, Lai B, Zhang B, Zhao Y, Mandla S, Korolj A, Radisic M, Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies, Adv Healthc Mater 7(2) (2018). [DOI] [PubMed] [Google Scholar]
- [28].Zhang B, Korolj A, Lai BFL, Radisic M, Advances in organ-on-a-chip engineering, Nature Reviews Materials 3(8) (2018) 257–278. [Google Scholar]
- [29].Hartung T, Toxicology for the twenty-first century, Nature 460(7252) (2009) 208–12. [DOI] [PubMed] [Google Scholar]
- [30].Whitesides GM, The origins and the future of microfluidics, Nature 442(7101) (2006) 36873. [DOI] [PubMed] [Google Scholar]
- [31].Shamloo A, Abdorahimzadeh S, Nasiri R, Exploring contraction-expansion inertial microfluidic-based particle separation devices integrated with curved channels, AIChE Journal (2019). [Google Scholar]
- [32].Fetah K, Tebon P, Goudie MJ, Eichenbaum J, Ren L, Barros N, Nasiri R, Ahadian S, Ashammakhi N, Dokmeci MR, Khademhosseini A, The emergence of 3D bioprinting in organ-on-chip systems, Progress in Biomedical Engineering 1(1) (2019) 012001. [Google Scholar]
- [33].Nasiri R, Shamloo A, Ahadian S, Amirifar L, Akbari J, Goudie MJ, Lee K, Ashammakhi N, Dokmeci MR, Di Carlo D, Khademhosseini A, Microfluidic-based approaches in targeted cell/particle separation based on physical properties: Fundamentals and applications, Small (2020). [DOI] [PubMed] [Google Scholar]
- [34].Ashammakhi N, Elkhammas E, Hasan A, Translating advances in organ-on-a-chip technology for supporting organs, Journal of biomedical materials research. Part B, Applied biomaterials 107(6) (2019) 2006–2018. [DOI] [PubMed] [Google Scholar]
- [35].Tian C, Tu Q, Liu W, Wang J, Recent advances in microfluidic technologies for organ-on-a-chip, TrAC Trends in Analytical Chemistry 117 (2019) 146–156. [Google Scholar]
- [36].Shuler ML, Advances in organ-, body-, and disease-on-a-chip systems, Lab on a chip 19(1) (2019) 9–10. [DOI] [PubMed] [Google Scholar]
- [37].Kratz SRA, Höll G, Schuller P, Ertl P, Rothbauer M, Latest Trends in Biosensing for Microphysiological Organs-on-a-Chip and Body-on-a-Chip Systems, Biosensors (Basel) 9(3) (2019) 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bhatia SN, Ingber DE, Microfluidic organs-on-chips, Nature Biotechnology 32 (2014) 760. [DOI] [PubMed] [Google Scholar]
- [39].Pocock K, Delon L, Bala V, Rao S, Priest C, Prestidge C, Thierry B, Intestine-on-a-Chip Microfluidic Model for Efficient in Vitro Screening of Oral Chemotherapeutic Uptake, ACS biomaterials science & engineering 3(6) (2017) 951–959. [DOI] [PubMed] [Google Scholar]
- [40].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Sanati Nezhad A, Manbohi A, De Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A, Multisensorintegrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors, Proc Natl Acad Sci U S A 114(12) (2017) E2293–E2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kilic T, Navaee F, Stradolini F, Renaud P, Carrara S, Organs-on-chip monitoring: sensors and other strategies, Microphysiological Systems; Vol 2 (September 2018): Microphysiological Systems (2018). [Google Scholar]
- [42].Christenson C, Baryeh K, Ahadian S, Nasiri R, Dokmeci MR, Goudie M, Khademhosseini A, Ye JY, Enhancement of label-free biosensing of cardiac troponin I, International Society for Optics and Photonics, p. 112512J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS, Ingber DE, Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro, Cell Rep 21(2) (2017) 508–516. [DOI] [PubMed] [Google Scholar]
- [44].Sosa-Hernández J, Villalba-Rodríguez A, Romero-Castillo K, Aguilar-Aguila-Isaías M, García-Reyes I, Hernández-Antonio A, Ahmed I, Sharma A, Parra-Saldívar R, Iqbal HJM, Organs-on-a-chip module: a review from the development and applications perspective, 9(10) (2018) 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M, Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies, AAPS J 19(5) (2017) 1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lasli S, Kim HJ, Lee K, Suurmond CAE, Goudie M, Bandaru P, Sun W, Zhang S, Zhang N, Ahadian SJAB, A Human Liver-on-a-Chip Platform for Modeling Nonalcoholic Fatty Liver Disease, 1900104. [DOI] [PMC free article] [PubMed]
- [47].Ceri-Anne SL Suurmond E, van den Dolder Floor W., Ung Aly, Kim Han-Jun, Bandaru Praveen, Lee KangJu, Ahadian Samad, Ashammakhi Nureddin, Dokmeci Mehmet R., Lee Junmin, Khademhosseini Ali., In vitro human liver model of nonalcoholic steatohepatitis by co culturing hepatocytes, endothelial, and Kupffer cells, Biomaterials (2019). [DOI] [PubMed] [Google Scholar]
- [48].Skardal A, Murphy SV, Devarasetty M, Mead I, Kang H-W, Seol Y-J, Shrike Zhang Y, Shin S-R, Zhao L, Aleman J, Hall AR, Shupe TD, Kleensang A, Dokmeci MR, Jin Lee S, Jackson JD, Yoo JJ, Hartung T, Khademhosseini A, Soker S, Bishop CE, Atala A, Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform, Scientific Reports 7(1) (2017) 8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang C, Zhao Z, Abdul Rahim NA, van Noort D, Yu H, Towards a human-on-chip: culturing multiple cell types on a chip with compartmentalized microenvironments, Lab Chip 9(22) (2009) 3185–92. [DOI] [PubMed] [Google Scholar]
- [50].Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL, Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle, Sci Rep 7 (2017) 42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Webb SPD, CELLS IN THE THIRD DIMENSION, Biotechniques 62(3) (2017) 93–98. [DOI] [PubMed] [Google Scholar]
- [52].Singh D, Cho WC, Upadhyay G, Drug-Induced Liver Toxicity and Prevention by Herbal Antioxidants: An Overview, Front Physiol 6 (2015) 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, Platt V, Bridges R, Cai Y, Santhanam N, Berry B, Najjar S, Akanda N, Guo X, Martin C, Ekman G, Esch MB, Langer J, Ouedraogo G, Cotovio J, Breton L, Shuler ML, Hickman JJ, Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs, Sci Rep 6 (2016) 20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dongeun Huh HJK, Fraser Jacob P, Shea Daniel E, Khan Mohammed, Bahinski Anthony, Hamilton Geraldine A & Ingber Donald E, Microfabrication of human organs-on-chips, protocol (2013). [DOI] [PubMed] [Google Scholar]
- [55].Kim DH, Cheon JH, Pathogenesis of Inflammatory Bowel Disease and Recent Advances in Biologic Therapies, Immune Netw 17(1) (2017) 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Shin MK, Kim SK, Jung H, Integration of intra- and extravasation in one cell-based microfluidic chip for the study of cancer metastasis, Lab on a Chip 11(22) (2011) 3880–3887. [DOI] [PubMed] [Google Scholar]
- [57].Musah S, Mammoto A, Ferrante TC, Jeanty SSF, Hirano-Kobayashi M, Mammoto T, Roberts K, Chung S, Novak R, Ingram M, Fatanat-Didar T, Koshy S, Weaver JC, Church GM, Ingber DE, Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip, Nat Biomed Eng 1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AMJN, Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro, 470(7332) (2011) 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lee J, Choi JH, Kim HJ, Human gut-on-a-chip technology: will this revolutionize our understanding of IBD and future treatments?, Expert Rev Gastroenterol Hepatol 10(8) (2016) 883–5. [DOI] [PubMed] [Google Scholar]
- [60].Donaldson GP, Lee SM, Mazmanian SK, Gut biogeography of the bacterial microbiota, Nat Rev Microbiol 14(1) (2016) 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schmidt TM, Kao JY, A little O2 may go a long way in structuring the GI microbiome, Gastroenterology 147(5) (2014) 956–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hall JJG, Hall PE Textbook of Medical Physiology 13th ed. Philadelphia, General principles of gastrointestinal function-motility, nervous control, and blood circulation, (2016). [Google Scholar]
- [63].Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Kustermann S, Roth A, Hankemeier T, Moisan A, Vulto P, Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes, Nature Communications 8(1) (2017) 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yeste J, Illa X, Alvarez M, Villa R, Engineering and monitoring cellular barrier models, Journal of Biological Engineering 12(1) (2018) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kasendra M, Luc R, Yin J, Manatakis DV, Kulkarni G, Lucchesi C, Sliz J, Apostolou A, Sunuwar L, Obrigewitch J, Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model, eLife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sakolish CM, Esch MB, Hickman JJ, Shuler ML, Mahler GJ, Modeling Barrier Tissues In Vitro: Methods, Achievements, and Challenges, EBioMedicine 5 (2016) 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].McDonald JC, Whitesides GM, Poly (dimethylsiloxane) as a material for fabricating microfluidic devices, Accounts of chemical research 35(7) (2002) 491–499. [DOI] [PubMed] [Google Scholar]
- [68].Bhatia SN, Ingber DE, Microfluidic organs-on-chips, Nature biotechnology 32(8) (2014) 760. [DOI] [PubMed] [Google Scholar]
- [69].van der Helm MW, Odijk M, Frimat JP, van der Meer AD, Eijkel JCT, van den Berg A, Segerink LI, Fabrication and Validation of an Organ-on-chip System with Integrated Electrodes to Directly Quantify Transendothelial Electrical Resistance, J Vis Exp (127) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hyun Jung Kim JL, Choi Jin-Ha, Bahinski Anthony, Ingber Donald E., Co-culture of Living Microbiome with Microengineered Human Intestinal Villi in a Gut-on-a-Chip Microfluidic Device, Journal of Visualized Experiments (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Srinivasan B, Kolli AR, Esch MB, Abaci HE, Shuler ML, Hickman JJ, TEER measurement techniques for in vitro barrier model systems, J Lab Autom 20(2) (2015) 107–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Shim KY, Lee D, Han J, Nguyen NT, Park S, Sung JH, Microfluidic gut-on-a-chip with three-dimensional villi structure, Biomed Microdevices 19(2) (2017) 37. [DOI] [PubMed] [Google Scholar]
- [73].Mata A FA, Roy S, Characterization of polydimethylsiloxane (PDMS) properties for biomedical micro/nanosystems, Biomed Microdevices 7 (2005) 281–293. [DOI] [PubMed] [Google Scholar]
- [74].van Poll ML, Zhou F, Ramstedt M, Hu L, Huck WT, A self-assembly approach to chemical micropatterning of poly(dimethylsiloxane), Angew Chem Int Ed Engl 46(35) (2007) 6634–7. [DOI] [PubMed] [Google Scholar]
- [75].Duffy DC, McDonald JC, Schueller OJA, Whitesides GM, Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane), Analytical Chemistry 70(23) (1998) 4974–4984. [DOI] [PubMed] [Google Scholar]
- [76].Toepke MW, Beebe DJ, PDMS absorption of small molecules and consequences in microfluidic applications, Lab Chip 6(12) (2006) 1484–6. [DOI] [PubMed] [Google Scholar]
- [77].Wang JD, Douville NJ, Takayama S, ElSayed M, Quantitative analysis of molecular absorption into PDMS microfluidic channels, Ann Biomed Eng 40(9) (2012) 1862–73. [DOI] [PubMed] [Google Scholar]
- [78].Halldorsson S, Lucumi E, Gomez-Sjoberg R, Fleming RMT, Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices, Biosens Bioelectron 63 (2015) 218–231. [DOI] [PubMed] [Google Scholar]
- [79].Wolf MP, Salieb-Beugelaar GB, Hunziker P, PDMS with designer functionalities—Properties, modifications strategies, and applications, Progress in Polymer Science 83 (2018) 97–134. [Google Scholar]
- [80].Gokaltun A, Yarmush ML, Asatekin A, Usta OB, Recent advances in nonbiofouling PDMS surface modification strategies applicable to microfluidic technology, Technology (Singap World Sci) 5(1) (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Xu L, Lee H, Jetta D, Oh KW, Vacuum-driven power-free microfluidics utilizing the gas solubility or permeability of polydimethylsiloxane (PDMS), Lab Chip 15(20) (2015) 3962–79. [DOI] [PubMed] [Google Scholar]
- [82].Sriram G, Alberti M, Dancik Y, Wu B, Wu R, Feng Z, Ramasamy S, Bigliardi PL, Bigliardi-Qi M, Wang Z, Full-thickness human skin-on-chip with enhanced epidermal morphogenesis and barrier function, Materials Today 21(4) (2018) 326–340. [Google Scholar]
- [83].Kulthong K, Duivenvoorde L, Mizera BZ, Rijkers D, Dam G.t., Oegema G, Puzyn T, Bouwmeester H, van der Zande M, Implementation of a dynamic intestinal gut-on-a-chip barrier model for transport studies of lipophilic dioxin congeners, RSC Advances 8(57) (2018) 32440–32453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nge PN, Rogers CI, Woolley AT, Advances in microfluidic materials, functions, integration, and applications, Chem Rev 113(4) (2013) 2550–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo SS, Vincent PA, Dai G, Creating perfused functional vascular channels using 3D bio-printing technology, Biomaterials 35(28) (2014) 8092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Imura Y, Asano Y, Sato K, Yoshimura E, A microfluidic system to evaluate intestinal absorption, Analytical sciences : the international journal of the Japan Society for Analytical Chemistry 25(12) (2009) 1403–7. [DOI] [PubMed] [Google Scholar]
- [87].Guo Y, Li Z, Su W, Wang L, Zhu Y, Qin J, A Biomimetic Human Gut-on-a-Chip for Modeling Drug Metabolism in Intestine, Artificial Organs 42(12) (2018) 1196–1205. [DOI] [PubMed] [Google Scholar]
- [88].Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, Scholl W, Zhang C, Rickner H, Richmond CA, Li H, Breault DT, Ingber DE, Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids, Sci Rep 8(1) (2018) 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ahn S, Ardona HAM, Lind JU, Eweje F, Kim SL, Gonzalez GM, Liu Q, Zimmerman JF, Pyrgiotakis G, Zhang Z, Beltran-Huarac J, Carpinone P, Moudgil BM, Demokritou P, Parker KK, Mussel-inspired 3D fiber scaffolds for heart-on-a-chip toxicity studies of engineered nanomaterials, Anal Bioanal Chem 410(24) (2018) 6141–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen MB, Srigunapalan S, Wheeler AR, Simmons CA, A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell-cell interactions, Lab Chip 13(13) (2013) 2591–8. [DOI] [PubMed] [Google Scholar]
- [91].Wang L, Murthy SK, Fowle WH, Barabino GA, Carrier RL, Influence of micro-well biomimetic topography on intestinal epithelial Caco-2 cell phenotype, Biomaterials 30(36) (2009) 6825–34. [DOI] [PubMed] [Google Scholar]
- [92].Jain RK, Au P, Tam J, Duda DG, Fukumura D, Engineering vascularized tissue, Nature Biotechnology 23(7) (2005) 821–823. [DOI] [PubMed] [Google Scholar]
- [93].Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, Levy O, Gregory KE, Breault DT, Cabral JMS, Kasper DL, Novak R, Ingber DE, A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip, Nature Biomedical Engineering 3(7) (2019) 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chow ECY, Liu S, Du Y, Pang KS, The Caco-2 cell monolayer: usefulness and limitations AU - Sun, Huadong, Expert Opinion on Drug Metabolism & Toxicology 4(4) (2008) 395–411. [DOI] [PubMed] [Google Scholar]
- [95].Kim HJ, Ingber DE, Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation, Integr Biol (Camb) 5(9) (2013) 1130–40. [DOI] [PubMed] [Google Scholar]
- [96].Hidalgo IJ, Raub TJ, Borchardt RT, Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability, Gastroenterology 96(2) (1989) 736–749. [PubMed] [Google Scholar]
- [97].Sambuy Y, De Angelis I, Ranaldi G, Scarino M, Stammati A, Zucco F, The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics, Cell biology and toxicology 21(1) (2005) 1–26. [DOI] [PubMed] [Google Scholar]
- [98].Lea T, Caco-2 Cell Line, in: Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A, Requena T, Swiatecka D, Wichers H (Eds.), The Impact of Food Bioactives on Health: in vitro and ex vivo models, Springer International Publishing, Cham, 2015, pp. 103–111. [PubMed] [Google Scholar]
- [99].Yissachar N, Zhou Y, Ung L, Lai NY, Mohan JF, Ehrlicher A, Weitz DA, Kasper DL, Chiu IM, Mathis D, Benoist C, An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk, Cell 168(6) (2017) 1135–1148.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Cimetta E, Sirabella D, Yeager K, Davidson K, Simon J, Moon RT, Vunjak-Novakovic G, Microfluidic bioreactor for dynamic regulation of early mesodermal commitment in human pluripotent stem cells, Lab on a chip 13(3) (2013) 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wan C.-r., Chung S, Kamm RD, Differentiation of embryonic stem cells into cardiomyocytes in a compliant microfluidic system, Annals of biomedical engineering 39(6) (2011) 1840–1847. [DOI] [PubMed] [Google Scholar]
- [102].Ashammakhi NA, Elzagheid A, Organ-on-a-Chip: New Tool for Personalized Medicine, Journal of Craniofacial Surgery 29(4) (2018). [DOI] [PubMed] [Google Scholar]
- [103].Chi M, Yi B, Oh S, Park D-J, Sung JH, Park S, A microfluidic cell culture device (μFCCD) to culture epithelial cells with physiological and morphological properties that mimic those of the human intestine, Biomedical microdevices 17(3) (2015) 58. [DOI] [PubMed] [Google Scholar]
- [104].De Gregorio V, Corrado B, Sbrescia S, Sibilio S, Urciuolo F, Netti PA, Imparato G, Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation, Biotechnology and Bioengineering 117(2) (2020) 556–566. [DOI] [PubMed] [Google Scholar]
- [105].Maurer M, Gresnigt MS, Last A, Wollny T, Berlinghof F, Pospich R, Cseresnyes Z, Medyukhina A, Graf K, Gröger M, Raasch M, Siwczak F, Nietzsche S, Jacobsen ID, Figge MT, Hube B, Huber O, Mosig AS, A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies, Biomaterials 220 (2019) 119396. [DOI] [PubMed] [Google Scholar]
- [106].Grassart A, Malardé V, Gobaa S, Sartori-Rupp A, Kerns J, Karalis K, Marteyn B, Sansonetti P, Sauvonnet N, Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection, Cell Host & Microbe 26(4) (2019) 565. [DOI] [PubMed] [Google Scholar]
- [107].Gao D, Liu H, Lin JM, Wang Y, Jiang Y, Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device, Lab Chip 13(5) (2013) 978–85. [DOI] [PubMed] [Google Scholar]
- [108].Jalili-Firoozinezhad S, Prantil-Baun R, Jiang A, Potla R, Mammoto T, Weaver JC, Ferrante TC, Kim HJ, Cabral JMS, Levy O, Ingber DE, Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip, Cell Death Dis 9(2) (2018) 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Whitman WB, Coleman DC, Wiebe WJ, Prokaryotes: The unseen majority, Proceedings of the National Academy of Sciences 95(12) (1998) 6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Clemente JC, Ursell LK, Parfrey LW, Knight R, The impact of the gut microbiota on human health: an integrative view, Cell 148(6) (2012) 1258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Abdelmannan D, Farooqi M, Ashammakhi N, Beshyah S, Expanding the research on gut microbiota in health and disease, Ibnosina Journal of Medicine and Biomedical Sciences 10(6) (2018) 181–183. [Google Scholar]
- [112].Karkman A, Lehtimäki J, Ruokolainen L, The ecology of human microbiota: dynamics and diversity in health and disease, 1399(1) (2017) 78–92. [DOI] [PubMed] [Google Scholar]
- [113].Bertolini M, Ranjan A, Thompson A, Diaz PI, Sobue T, Maas K, Dongari-Bagtzoglou A, Candida albicans induces mucosal bacterial dysbiosis that promotes invasive infection, PLOS Pathogens 15(4) (2019) e1007717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Boĭkov SS, Moroz AF, Babaeva EE, Association of Candida albicans fungi with some opportunistic microorganisms in intestinal dysbiosis in patients of different age groups, Zh Mikrobiol Epidemiol Immunobiol (2) (2005) 65–69. [PubMed] [Google Scholar]
- [115].Zuo T, Wong SH, Cheung CP, Lam K, Lui R, Cheung K, Zhang F, Tang W, Ching JYL, Wu JCY, Chan PKS, Sung JJY, Yu J, Chan FKL, Ng SC, Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection, Nature Communications 9(1) (2018) 3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Tringe SG, Rubin EM, Metagenomics: DNA sequencing of environmental samples, Nature Reviews Genetics 6(11) (2005) 805–814. [DOI] [PubMed] [Google Scholar]
- [117].Jovel J, Patterson J, Wang W, Hotte N, O’Keefe S, Mitchel T, Perry T, Kao D, Mason AL, Madsen KL, Wong GK-S, Characterization of the Gut Microbiome Using 16S or Shotgun Metagenomics, Front Microbiol 7(459) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bjerketorp J, Ng Tze Chiang A, Hjort K, Rosenquist M, Liu W-T, Jansson JK, Rapid lab-on-a-chip profiling of human gut bacteria, Journal of Microbiological Methods 72(1) (2008) 8290. [DOI] [PubMed] [Google Scholar]
- [119].Tottey W, Denonfoux J, Jaziri F, Parisot N, Missaoui M, Hill D, Borrel G, Peyretaillade E, Alric M, Harris HMB, Jeffery IB, Claesson MJ, O’Toole PW, Peyret P, Brugère J-F, The human gut chip “HuGChip”, an explorative phylogenetic microarray for determining gut microbiome diversity at family level, PloS one 8(5) (2013) e62544–e62544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Nguyen TLA, Vieira-Silva S, Liston A, Raes J, How informative is the mouse for human gut microbiota research?, Disease models & mechanisms 8(1) (2015) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jud C, Clift MJ, Petri-Fink A, Rothen-Rutishauser B, Nanomaterials and the human lung: what is known and what must be deciphered to realise their potential advantages?, Swiss medical weekly 143 (2013) w13758. [DOI] [PubMed] [Google Scholar]
- [122].Molly K, Vande Woestyne M, Verstraete W, Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem, Applied Microbiology and Biotechnology 39(2) (1993) 254–258. [DOI] [PubMed] [Google Scholar]
- [123].Macfarlane S, Quigley ME, Hopkins MJ, Newton DF, Macfarlane GT, Polysaccharide degradation by human intestinal bacteria during growth under multi-substrate limiting conditions in a three-stage continuous culture system, 26(3) (1998) 231–243. [Google Scholar]
- [124].Cinquin C, Fliss I, Le Blay G, Lacroix C, New three-stage in vitro model for infant colonic fermentation with immobilized fecal microbiota, FEMS Microbiology Ecology 57(2) (2006) 324–336. [DOI] [PubMed] [Google Scholar]
- [125].Venema K, de Ligt RAF, Doornbos RP, van Nuenen MHMC, Kuipers EJ, van der Woude JCJ, The influence of microbial metabolites on human intestinal epithelial cells and macrophages in vitro, FEMS Immunology & Medical Microbiology 45(2) (2005) 183–189. [DOI] [PubMed] [Google Scholar]
- [126].Parlesak A, Haller D, Brinz S, Baeuerlein A, Bode C, Modulation of Cytokine Release by Differentiated CACO-2 Cells in a Compartmentalized Coculture Model with Mononuclear Leucocytes and Nonpathogenic Bacteria, 60(5) (2004) 477–485. [DOI] [PubMed] [Google Scholar]
- [127].Höner zu Bentrup K, Ramamurthy R, Ott CM, Emami K, Nelman-Gonzalez M, Wilson JW, Richter EG, Goodwin TJ, Alexander JS, Pierson DL, Pellis N, Buchanan KL, Nickerson CA, Three-dimensional organotypic models of human colonic epithelium to study the early stages of enteric salmonellosis, Microbes and Infection 8(7) (2006) 1813–1825. [DOI] [PubMed] [Google Scholar]
- [128].Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, Roeselers G, Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids, MBio 5(4) (2014) e01438–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Marzorati M, Vanhoecke B, De Ryck T, Sadaghian Sadabad M, Pinheiro I, Possemiers S, Van den Abbeele P, Derycke L, Bracke M, Pieters J, Hennebel T, Harmsen HJ, Verstraete W, Van de Wiele TJBM, The HMI™ module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro, 14(1) (2014) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Marzorati M, Vanhoecke B, De Ryck T, Sadaghian Sadabad M, Pinheiro I, Possemiers S, Van den Abbeele P, Derycke L, Bracke M, Pieters J, Hennebel T, Harmsen HJ, Verstraete W, Van de Wiele T, The HMI™ module: a new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro, BMC Microbiology 14(1) (2014) 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jäger C, Seguin-Devaux C, Zenhausern F, Wilmes P, A microfluidics-based in vitro model of the gastrointestinal human–microbe interface, Nature Communications 7 (2016) 11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kim J, Hegde M, Jayaraman A, Co-culture of epithelial cells and bacteria for investigating host-pathogen interactions, Lab Chip 10(1) (2010) 43–50. [DOI] [PubMed] [Google Scholar]
- [133].Kim HJ, Li H, Collins JJ, Ingber DE, Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip, Proc Natl Acad Sci U S A 113(1) (2016) E7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kim HJ, Huh D, Hamilton G, Ingber DE, Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow, Lab Chip 12(12) (2012) 2165–74. [DOI] [PubMed] [Google Scholar]
- [135].Gumuscu B, Albers HJ, van den Berg A, Eijkel JCT, van der Meer AD, Compartmentalized 3D Tissue Culture Arrays under Controlled Microfluidic Delivery, Sci Rep 7(1) (2017) 3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].El Aidy S, van den Bogert B, Kleerebezem M, The small intestine microbiota, nutritional modulation and relevance for health, Curr Opin Biotechnol 32 (2015) 14–20. [DOI] [PubMed] [Google Scholar]
- [137].El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, Reijngoud D-J, Kleerebezem M, The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation, Gut 62(9) (2013) 1306–1314. [DOI] [PubMed] [Google Scholar]
- [138].Van De Walle J, Hendrickx A, Romier B, Larondelle Y, Schneider YJ, Inflammatory parameters in Caco-2 cells: effect of stimuli nature, concentration, combination and cell differentiation, Toxicology in vitro : an international journal published in association with BIBRA 24(5) (2010) 1441–9. [DOI] [PubMed] [Google Scholar]
- [139].Haller D, Holt L, Parlesak A, Zanga J, Bauerlein A, Sartor RB, Jobin C, Differential effect of immune cells on non-pathogenic Gram-negative bacteria-induced nuclear factor-kappaB activation and pro-inflammatory gene expression in intestinal epithelial cells, Immunology 112(2) (2004) 310–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Chelvarajan RL, Collins SM, Van Willigen JM, Bondada S, The unresponsiveness of aged mice to polysaccharide antigens is a result of a defect in macrophage function, Journal of leukocyte biology 77(4) (2005) 503–12. [DOI] [PubMed] [Google Scholar]
- [141].Thompson-Chagoyán OC, Maldonado J, Gil A, Aetiology of inflammatory bowel disease (IBD): Role of intestinal microbiota and gut-associated lymphoid tissue immune response, Clinical Nutrition 24(3) (2005) 339–352. [DOI] [PubMed] [Google Scholar]
- [142].Shin W, Kim HJ, Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip, Proceedings of the National Academy of Sciences 115(45) (2018) E10539–E10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ramadan Q, Jafarpoorchekab H, Huang C, Silacci P, Carrara S, Koklü G, Ghaye J, Ramsden J, Ruffert C, Vergeres G, Gijs MAM, NutriChip: nutrition analysis meets microfluidics, Lab on a Chip 13(2) (2013) 196–203. [DOI] [PubMed] [Google Scholar]
- [144].Baines SD, O’Connor R, Saxton K, Freeman J, Wilcox MH, Activity of vancomycin against epidemic Clostridium difficile strains in a human gut model, Journal of Antimicrobial Chemotherapy 63(3) (2008) 520–525. [DOI] [PubMed] [Google Scholar]
- [145].Baines SD, Crowther GS, Freeman J, Todhunter S, Vickers R, Wilcox MH, SMT19969 as a treatment for Clostridium difficile infection: an assessment of antimicrobial activity using conventional susceptibility testing and an in vitro gut model, Journal of Antimicrobial Chemotherapy 70(1) (2014) 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, Ferrante TC, Bahinski A, Elkins CA, Kulka M, Ingber DE, Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro, PLoS One 12(2) (2017) e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Grassart A, Malarde V, Gobaa S, Sartori-Rupp A, Kerns J, Karalis K, Marteyn B, Sansonetti P, Sauvonnet N, Bioengineered Human Organ-on-Chip Reveals Intestinal Microenvironment and Mechanical Forces Impacting Shigella Infection, Cell Host Microbe 26(3) (2019) 435–444 e4. [DOI] [PubMed] [Google Scholar]
- [148].Cho S, Yoon JY, Organ-on-a-chip for assessing environmental toxicants, Curr Opin Biotechnol 45 (2017) 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Ashammakhi N, Darabi MA, Çelebi-Saltik B, Tutar R, Hartel MC, Lee J, Hussein SM, Goudie MJ, Cornelius MB, Dokmeci MR, Khademhosseini A, Microphysiological Systems: Next Generation Systems for Assessing Toxicity and Therapeutic Effects of Nanomaterials, Small Methods 4(1) (2020) 1900589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Mackay SM, Funke AJ, Buffington DA, Humes HD, Tissue Engineering of a Bioartificial Renal Tubule, ASAIO Journal: Artificial Organ Research and Development 44(3) (1998). [DOI] [PubMed] [Google Scholar]
- [151].Sanechika N, Sawada K, Usui Y, Hanai K, Kakuta T, Suzuki H, Kanai G, Fujimura S, Yokoyama TA, Fukagawa M, Terachi T, Saito A, Development of bioartificial renal tubule devices with lifespan-extended human renal proximal tubular epithelial cells, Nephrology Dialysis Transplantation 26(9) (2011) 2761–2769. [DOI] [PubMed] [Google Scholar]
- [152].Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hubner J, Lindner M, Drewell C, Bauer S, Thomas A, Sambo NS, Sonntag F, Lauster R, Marx U, A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents, Lab Chip 15(12) (2015) 2688–99. [DOI] [PubMed] [Google Scholar]
- [153].Li Z, Su W, Zhu Y, Tao T, Li D, Peng X, Qin J, Drug absorption related nephrotoxicity assessment on an intestine-kidney chip, Biomicrofluidics 11(3) (2017) 034114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Esch MB, Mahler GJ, Stokol T, Shuler ML, Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury, Lab on a chip 14(16) (2014) 3081–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Kimura H, Ikeda T, Nakayama H, Sakai Y, Fujii T, An on-chip small intestine-liver model for pharmacokinetic studies, Journal of laboratory automation 20(3) (2015) 265–73. [DOI] [PubMed] [Google Scholar]
- [156].Chen WLK, Edington C, Suter E, Yu J, Velazquez JJ, Velazquez JG, Shockley M, Large EM, Venkataramanan R, Hughes DJ, Stokes CL, Trumper DL, Carrier RL, Cirit M, Griffith LG, Lauffenburger DA, Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk, Biotechnol Bioeng 114(11) (2017) 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].van Midwoud PM, Merema MT, Verpoorte E, Groothuis GMM, A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices, Lab on a Chip 10(20) (2010) 2778–2786. [DOI] [PubMed] [Google Scholar]
- [158].Bricks T, Paullier P, Legendre A, Fleury M-J, Zeller P, Merlier F, Anton PM, Leclerc E, Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines, Toxicology in Vitro 28(5) (2014) 885–895. [DOI] [PubMed] [Google Scholar]
- [159].Chen HJ, Miller P, Shuler ML, A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells, Lab Chip 18(14) (2018) 2036–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Skardal A, Devarasetty M, Forsythe S, Atala A, Soker S, A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening, Biotechnol Bioeng 113(9) (2016) 2020–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Modeling Physiological Events in 2D vs. 3D Cell Culture, Physiology (Bethesda, Md.) 32(4) (2017) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [162].Severance EG, Prandovszky E, Castiglione J, Yolken RH, Gastroenterology issues in schizophrenia: why the gut matters, Curr Psychiatry Rep 17(5) (2015) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wang B, Gray G, Concordance of Noncarcinogenic Endpoints in Rodent Chemical Bioassays, Risk Anal 35(6) (2015) 1154–66. [DOI] [PubMed] [Google Scholar]
- [164].Redfern WS, Bialecki R, Ewart L, Hammond TG, Kinter L, Lindgren S, Pollard CE, Rolf M, Valentin J-P, Impact and prevalence of safety pharmacology-related toxicities throughout the pharmaceutical life cycle, Journal of Pharmacological and Toxicological Methods 62(2) (2010). [Google Scholar]
- [165].Naughton C, Drug-induced nephrotoxicity, Am. Fam. Physician 78 (2008) 743–750. [PubMed] [Google Scholar]
- [166].Mullin R, Roche and Takeda try organ on a chip, Chemical & Engineering News, American Chemical Society, 2018, p. 15. [Google Scholar]
- [167].Hale C, StemoniX rakes in $14.4M for micro-organ drug testing chips built from stem cells, FierceBiotech, Questex, 2019. [Google Scholar]
- [168].Balijepalli A, Sivaramakrishan V, Organs-on-chips: research and commercial perspectives, Drug Discovery Today 22(2) (2017) 397–403. [DOI] [PubMed] [Google Scholar]
- [169].Rouge N, Buri P, Doelker E, Drug absorption sites in the gastrointestinal tract and dosage forms for site-specific delivery, International Journal of Pharmaceutics 136(1) (1996) 117–139. [Google Scholar]
- [170].Sjöberg Å, Lutz M, Tannergren C, Wingolf C, Borde A, Ungell A-L, Comprehensive study on regional human intestinal permeability and prediction of fraction absorbed of drugs using the Ussing chamber technique, European Journal of Pharmaceutical Sciences 48(1) (2013) 166–180. [DOI] [PubMed] [Google Scholar]
- [171].Almazroo OA, Miah MK, Venkataramanan R, Drug Metabolism in the Liver, Clinics in Liver Disease 21(1) (2017) 1–20. [DOI] [PubMed] [Google Scholar]
- [172].Bricks T, Paullier P, Legendre A, Fleury MJ, Zeller P, Merlier F, Anton PM, Leclerc E, Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines, Toxicology in vitro : an international journal published in association with BIBRA 28(5) (2014) 885–95. [DOI] [PubMed] [Google Scholar]
- [173].Gilbert JA, Blaser MJ, Caporaso JG, Jansson JK, Lynch SV, Knight R, Current understanding of the human microbiome, Nat Med 24(4) (2018) 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Lyte M, The microbial organ in the gut as a driver of homeostasis and disease, Medical hypotheses 74(4) (2010) 634–8. [DOI] [PubMed] [Google Scholar]
- [175].Workman MJ, Gleeson JP, Troisi EJ, Estrada HQ, Kerns SJ, Hinojosa CD, Hamilton GA, Targan SR, Svendsen CN, Barrett RJ, Enhanced Utilization of Induced Pluripotent Stem Cell–Derived Human Intestinal Organoids Using Microengineered Chips, Cellular and Molecular Gastroenterology and Hepatology 5(4) (2018) 669–677.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Fetah KL, DiPardo BJ, Kongadzem E-M, Tomlinson JS, Elzagheid A, Elmusrati M, Khademhosseini A, Ashammakhi N, Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration, Small 15(50) (2019) 1901985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Nadkarni RR, Abed S, Cox BJ, Bhatia S, Lau JT, Surette MG, Draper JS, Functional Enterospheres Derived In Vitro from Human Pluripotent Stem Cells, Stem Cell Reports 9(3) (2017) 897–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Kretzschmar K, Clevers H, Organoids: Modeling Development and the Stem Cell Niche in a Dish, Dev Cell 38(6) (2016) 590–600. [DOI] [PubMed] [Google Scholar]
- [179].Maschmeyer I, Hasenberg T, Jaenicke A, Lindner M, Lorenz AK, Zech J, Garbe LA, Sonntag F, Hayden P, Ayehunie S, Lauster R, Marx U, Materne EM, Chip-based human liver-intestine and liver-skin co-cultures--A first step toward systemic repeated dose substance testing in vitro, Eur J Pharm Biopharm 95(Pt A) (2015) 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Ashammakhi N, Kaarela O, Stimuli-Responsive Biomaterials: Next Wave, Journal of Craniofacial Surgery 28(7) (2017). [DOI] [PubMed] [Google Scholar]
- [181].Lu Y, Aimetti AA, Langer R, Gu Z, Bioresponsive materials, Nature Reviews Materials 2(1) (2016) 16075. [Google Scholar]
- [182].Hasan A, Nurunnabi M, Morshed M, Paul A, Polini A, Kuila T, Al Hariri M, Lee YK, Jaffa AA, Recent advances in application of biosensors in tissue engineering, Biomed Res Int 2014 (2014) 307519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Jenkinson SE, Chung GW, van Loon E, Bakar NS, Dalzell AM, Brown CD, The limitations of renal epithelial cell line HK-2 as a model of drug transporter expression and function in the proximal tubule, Pflugers Arch 464(6) (2012) 601–11. [DOI] [PubMed] [Google Scholar]
- [184].Ashammakhi N, Wesseling-Perry K, Hasan A, Elkhammas E, Zhang YS, Kidney-on-a-chip: untapped opportunities, Kidney Int 94(6) (2018) 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R, Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening, Trends Biotechnol 34(2) (2016) 156–170. [DOI] [PubMed] [Google Scholar]
- [186].Ma X, Qu X, Zhu W, Li YS, Yuan S, Zhang H, Liu J, Wang P, Lai CS, Zanella F, Feng GS, Sheikh F, Chien S, Chen S, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc Natl Acad Sci U S A 113(8) (2016) 2206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Liu J, Hwang HH, Wang P, Whang G, Chen S, Direct 3D-printing of cell-laden constructs in microfluidic architectures, Lab Chip 16(8) (2016) 1430–8. [DOI] [PubMed] [Google Scholar]
- [188].Tumbleston SD JR, Ermoshkin N, et al. , Additive manufacturing. continuous liquid interface production of 3D objects, Science (New York, N.Y.) 347 (2015). [DOI] [PubMed] [Google Scholar]
- [189].Ali M, Pr AK, Yoo JJ, Zahran F, Atala A, Lee SJ, A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation, Advanced healthcare materials 8(7) (2019) e1800992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Ashammakhi N, Ahadian S, Xu C, Montazerian H, Ko H, Nasiri R, Barros N, Khademhosseini A, Bioinks and Bioprinting Technologies to Make Heterogeneous and Biomimetic Tissue Constructs, Materials Today Bio (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Seyedmahmoud Rasoul S Çelebi, Barros, Nasiri, Banton, Shamloo, Ashammakhi, Dokmeci, Ahadian, Three-Dimensional Bioprinting of Functional Skeletal Muscle Tissue Using GelatinMethacryloyl-Alginate Bioinks, Micromachines 10(10) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Yu Y, Moncal KK, Li J, Peng W, Rivero I, Martin JA, Ozbolat IT, Three-dimensional bioprinting using self-assembling scalable scaffold-free “tissue strands” as a new bioink, Sci Rep 6 (2016) 28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Tibbits S, 4D Printing: Multi-Material Shape Change, Architectural Design 84(1) (2014) 116–121. [Google Scholar]
- [194].Ashammakhi N, Ahadian S, Zengjie F, Suthiwanich K, Lorestani F, Orive G, Ostrovidov S, Khademhosseini A, Advances and Future Perspectives in 4D Bioprinting, Biotechnology Journal 13(12) (2018) 1800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Li Y-C, Zhang YS, Akpek A, Shin SR, Khademhosseini A, 4D bioprinting: the next-generation technology for biofabrication enabled by stimuli-responsive materials, Biofabrication 9(1) (2016) 012001. [DOI] [PubMed] [Google Scholar]
- [196].Li YC, Zhang YS, Akpek A, Shin SR, Khademhosseini A, 4D bioprinting: the next-generation technology for biofabrication enabled by stimuli-responsive materials, Biofabrication 9(1) (2016) 012001. [DOI] [PubMed] [Google Scholar]
- [197].Anna SL, Droplets and Bubbles in Microfluidic Devices, Annual Review of Fluid Mechanics 48(1) (2016) 285–309. [Google Scholar]
- [198].Stone HA, Stroock AD, Ajdari A, Engineering Flows in Small Devices, Annual Review of Fluid Mechanics 36(1) (2004) 381–411. [Google Scholar]
- [199].Shin SR, Kilic T, Zhang YS, Avci H, Hu N, Kim D, Branco C, Aleman J, Massa S, Silvestri A, Kang J, Desalvo A, Hussaini MA, Chae SK, Polini A, Bhise N, Hussain MA, Lee H, Dokmeci MR, Khademhosseini A, Label-Free and Regenerative Electrochemical Microfluidic Biosensors for Continual Monitoring of Cell Secretomes, Adv Sci (Weinh) 4(5) (2017) 1600522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Henry OYF, Villenave R, Cronce MJ, Leineweber WD, Benz MA, Ingber DE, Organs-on-chips with integrated electrodes for trans-epithelial electrical resistance (TEER) measurements of human epithelial barrier function, Lab on a Chip 17(13) (2017) 2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Oedit A, Vulto P, Ramautar R, Lindenburg PW, Hankemeier T, Lab-on-a-Chip hyphenation with mass spectrometry: strategies for bioanalytical applications, Curr Opin Biotechnol 31 (2015) 79–85. [DOI] [PubMed] [Google Scholar]
- [202].Maoz BM, Herland A, Henry OYF, Leineweber WD, Yadid M, Doyle J, Mannix R, Kujala VJ, FitzGerald EA, Parker KK, Ingber DE, Organs-on-Chips with combined multi-electrode array and transepithelial electrical resistance measurement capabilities, Lab Chip 17(13) (2017) 2294–2302. [DOI] [PubMed] [Google Scholar]


