Summary
The emergence of the novel coronavirus disease 2019 (COVID-19) is placing an increasing burden on healthcare systems. Although the majority of infected patients experience non-severe symptoms and can be managed at home, some individuals develop severe symptoms and require hospital admission. Therefore, it is critical to efficiently assess the severity of COVID-19 and identify hospitalization priority with precision. In this respect, a four-variable assessment model, including lymphocyte, lactate dehydrogenase, C-reactive protein, and neutrophil, is established and validated using the XGBoost algorithm. This model is found to be effective in identifying severe COVID-19 cases on admission, with a sensitivity of 84.6%, a specificity of 84.6%, and an accuracy of 100% to predict the disease progression toward rapid deterioration. It also suggests that a computation-derived formula of clinical measures is practically applicable for healthcare administrators to distribute hospitalization resources to the most needed in epidemics and pandemics.
Keywords: COVID-19, hospitalization priority, learning-based model
Graphical Abstract
Highlights
-
•
A model was developed to evaluate hospitalization priority in COVID-19 pandemics
-
•
This model used easily accessible biomarkers to evaluate the risk of severe COVID-19
-
•
The evaluation can be rapidly processed using an online program
-
•
Performance of different algorithms in evaluation of COVID-19 severity was explored
The Bigger Picture
The COVID-19 pandemic is threatening millions of lives and putting medical systems under stress worldwide. Although the infection growth in some areas has ceased, there is a risk of a second wave. Therefore, a sustainable strategy to defend against a pandemic using the current limited but effective healthcare resources is in high demand. Our study aims to find a solution that triages patients to hospitalization by identifying their severity progression. In this study, a model that used four easily accessible biomarkers to assess the risk of severe COVID-19 was successfully developed. This model is easy to use, and it eliminates the dependence on expensive equipment to make a decision. It was found to be effective in identifying the risk of severe COVID-19. Thus, it is practically applicable for general practitioners to effectively assess the infection and allocate inpatient care to the cases who need it most. Our study is expected to have a prolonged social impact under the current circumstances.
The authors propose a learning-based model to assist clinicians in quick and efficient triage of patients in places where medical resources are limited in COVID-19 pandemics. This model used four easily accessible biomarkers to assess the severity of COVID-19 and was found to be effective in identifying the risk of severe COVID-19. It enables healthcare administrators to distribute hospitalization resources where they are most needed.
Introduction
The novel coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was first reported in December 2019 in China and rapidly spread across the world, affecting over 16 million people worldwide and killing more than a half million infected patients at time of writing.1, 2, 3 Even worse, the global pandemic of COVID-19 is expected to continue growing, as no effective vaccines have been officially approved for prophylaxis of this disease.4 Although the growth in detected infections has declined in East Asia and Europe, the number of infections in the US, South America, and places in Africa is continuing to grow.3 Moreover, the fear of a second wave pandemic outbreak still remains.5
In a pandemic, a nation's healthcare system bears extraordinary burdens. However, most patients infected with SARS-CoV-2 generally have non-severe symptoms and can be safely managed at home and recover under limited and basic medical care.6 For infections with severe symptoms or progression toward rapid deterioration, immediate admission to hospital for close monitoring and intensive treatment has been proven to be effective in reducing complications and mortality.7 Therefore, a way of identifying COVID-19 patients at high risk of severe illness and prioritizing them for immediate admission to hospitals is needed urgently, especially in nations and territories where the healthcare systems are insufficient to deal with all infections and suspicions of infection.
Some studies have reported how to predict deterioration and mortality during hospitalization,8, 9, 10, 11, 12, 13, 14, 15 and even predict the probability of SARS-CoV-2 infections, which enables the timely quarantine of high-risk infections and prevents spread.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 But none of these studies provide a solution for rational triage of patients in places where medical resources are limited.
In light of this unmet need in efficient triage of COVID-19 cases, this study aims to develop and validate a learning-based model that evaluates patients' priority for admission to hospital care due to their appearance or susceptibility to severe COVID-19. The model, provided with a simple user interface, can efficiently assess the severity of COVID-19, and predict disease progression, with high rates of accuracy. Our study is expected to have a prolonged social impact under the current circumstances, when the simple and practical model will be accepted to assist clinicians in quick and efficient triage of COVID-19 patients. This study was approved by the Guizhou Provincial People's Hospital Ethics Committee.
Results
Clinical Characteristics of COVID-19 Cases
The patient cohorts enrolled in this study comprised data of 134 COVID-19 cases retrieved from the World Health Organization (WHO) COVID-19 database26 (Figure S1; Table S1) and 467 COVID-19 cases recruited from a multi-center dataset in China. Of the 601 patients, 25.4% developed severe symptoms on admission and 6.5% presented with non-severe symptoms on admission but later developed severe symptoms after admission. The minimal, medium, and maximal time from hospital admission to severe disease progression were less than 1, 5, and 12 days, respectively. The prevalence of underlying comorbidities was 24.8%. Hypertension (11%) was the most common comorbidity, followed by endocrine diseases (6.7%). The medium age was 48 years. Fever was the most common initial symptom (63.1%), followed by a cough (50.1%), fatigue (21.3%), and dyspnea (9.3%). Table 1 shows the baseline laboratory results obtained on or soon after admission. All of the patients were cases of laboratory-confirmed COVID-19 and the severity of COVID-19 was stratified into severe and non-severe categories according to the criteria shown in Table 2.
Table 1.
Baseline Characteristics of Patients Enrolled to the Study
| Clinical Features | Overall |
|---|---|
| Age, mean ± SD, years | 47.63 ± 15.6 |
| Gender, n (%) | |
| Male | 319 (53) |
| Female | 280 (47) |
| Underlying comorbidities, n (%) | 149 (24.8) |
| Hypertension | 67 (11) |
| Endocrine disease | 40 (6.7) |
| Cardiovascular disease | 18 (3) |
| Chronic lung disease | 9 (1.5) |
| Digestive disease | 20 (3.3) |
| Renal disease | 7 (1.2) |
| Tumor | 3 (0.5) |
| Cerebrovascular/nervous disease | 7 (1.2) |
| Immune disorder | 8 (1.3) |
| Others | 35 (5.8) |
| Signs or Symptoms, n (%) | |
| Fever | 379 (63.1) |
| Cough | 301 (50.1) |
| Expectoration | 6 (1.0) |
| Hemoptysis | 2 (0.3) |
| Dyspnea | 56 (9.3) |
| Catarrh | 9 (1.5) |
| Fatigue | 128 (21.3) |
| Anorexia | 3 (0.5) |
| Nausea/emesis | 14 (2.3) |
| Myalgia | 47 (7.8) |
| Dizziness/headache | 37 (6.2) |
| Pharyngalgia | 9 (1.5) |
| Abdominal pain/diarrhea | 7 (0.2) |
| Laboratory Findings, mean ± SD | |
| White blood cell count, 109/L | 6.02 ± 15.98 |
| Lymphocyte count, 109/L | 1.25 ± 1.22 |
| Neutrophil count, 109/L | 3.64 ± 2.73 |
| Erythrocyte sedimentation rate, mm/h | 43.75 ± 28.86 |
| C-reactive protein, mg/L | 25.24 ± 28.92 |
| Procalcitonin, ng/mL | 0.91 ± 7.58 |
| D-dimer, μg/mL | 68.26 ± 515.42 |
| Alanine aminotransferase, U/L | 25.54 ± 15.47 |
| Aspartate aminotransferase, U/L | 29.5 ± 16.15 |
| Total bilirubin, μmol/L | 13.56 ± 8.09 |
| Albumin, g/L | 39.75 ± 9.28 |
| Lactate dehydrogenase, U/L | 229.5 ± 118.41 |
| Blood urea nitrogen, mmol/L | 5.29 ± 3.34 |
| Serum creatinine, μmol/L | 62.52 ± 34.39 |
| Prothrombin time, s | 13.27 ± 3.34 |
| Lactic acid, mmol/L | 271.37 ± 359.03 |
| Creatine kinase, U/L | 110.23 ± 118.38 |
| SpO2, % | 93.93 ± 14.69 |
Table 2.
Classification of the COVID-19 Severity
| Classifications | Definitions |
|---|---|
| Non-severe COVID-19 | Patients have non-specific symptoms, such as fever, cough, fatigue, myalgia, pharyngalgia, but have no signs of dehydration, sepsis, or shortness of breath. The radiological examination shows no signs of severe pneumonia. |
| Severe COVID-19 | Adult cases meeting any of the following criteria in the quiescent state: (1) Respiratory distress (respiratory rate ≥ 30 breaths/min); (2) Peripheral capillary oxygen saturation (SpO2) ≤ 93%; (3) Arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤ 300 mmHg; (4) Pulmonary lesion progression exceeds 50% in 24–48 h (5) Respiratory failure that requires mechanical ventilation; (6) Shock; (7) Organ failure that requires to be managed in intensive care unit. |
Identification of Critical Variables for Model Establishment
The clinical variables of most patients were measured multiple times across different days during hospitalization to assess the prognosis. As this study aimed to identify hospitalization priority according to the prehospital assessment of severe COVID-19 risk, only clinical data obtained on admission were used to evaluate the importance of clinical variables in identification of severe or potentially severe cases. Given that various data were missing on different clinical variables and different patients, a strategy was adopted to set a threshold value alpha to remove these missing data, minimizing their impact on data analysis. It was found that, as the threshold alpha increased, available variables decreased, while available observations, i.e., the available COVID-19 cases, increased (Figure S2). A threshold alpha of 350 was selected to remove the missing data, and to obtain as many clinical variables and observations as possible. Hence, a total of 29 clinical variables and 214 patients with non-missing variable values were used for subsequent analysis.
Next, a univariate analysis was performed to investigate the difference in the 29 clinical variables between the severe and non-severe groups. As shown in Table S2, a total of 12 clinical variables were significantly different between the two groups, including age, fever, dyspnea, lymphocyte, neutrophil, C-reactive protein (CRP), lactic dehydrogenase (LDH), creatine kinase (CK), D-dimer, alanine aminotransferase (ALT), aspartate aminotransferase (AST), and albumin. These 12 clinical variables could be used to discriminate between the severe and non-severe COVID-19 cases.
Development of Risk Assessment Models
Extreme Gradient Boosting (XGBoost), which is a high-performance machine learning algorithm and works with a sequence of decision trees where the latter tree tries to minimize the net error from previous trees, was used to generate the risk assessment model. In addition to XGBoost classifier, other representative algorithms, including linear discriminant analysis (LDA), logistic regression, support vector machine (SVM), and random forest and decision trees, were benchmarked as the baselines.
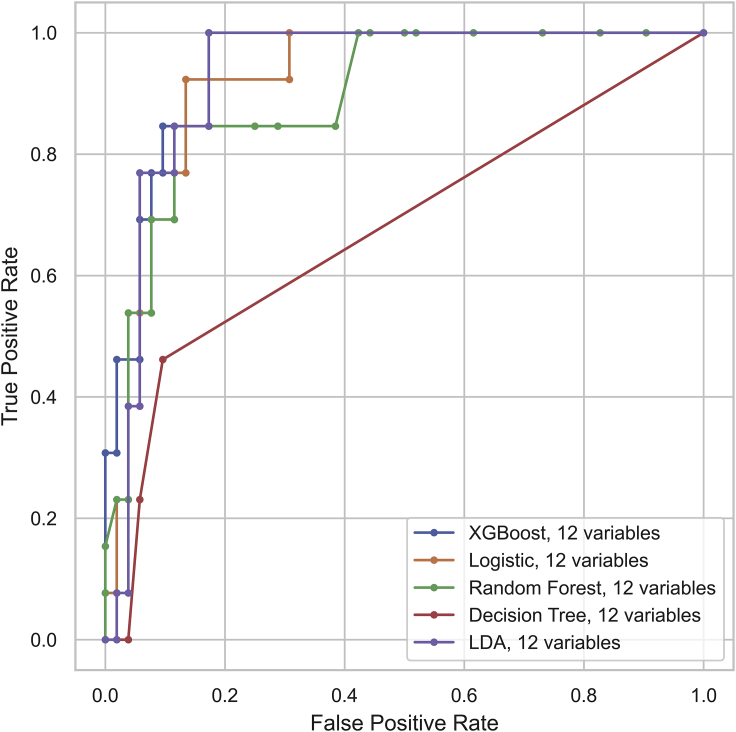
A sample of 65 randomly chosen cases from the total of 214 cases served as a holdout testing set (Figure S3). The remaining 149 cases were used for training and cross-validation. Then, the 12 significant variables in the previous univariant analysis were included to construct the risk assessment models based on the XGBoost classifier as well as other classifiers. They were first trained in the training set of 149 cases and then evaluated in the holdout testing set of 65 cases, by comparing the values of accuracy, F1 score, sensitivity, specificity, and the area under curve (AUC) score of the receiver operating characteristic (ROC) curve. The definition of these evaluation metrics is shown in Table S3. The 12-variable XGBoost classifier proved to have an accuracy of 89.2% in discriminating severe COVID-19 cases from their non-severe counterparts (Table 3). Moreover, it outperformed other classifiers in the evaluations listed above, with the exception of specificity (Table 3; Figure 1). Our study was in agreement with a reported conclusion that the XGBoost algorithm had a high discriminative performance,9 and thus could be used to assess hospitalization priority with precision.
Table 3.
The Performance of 12-Variable Models for Identification of Severe COVID-19 on Admission
| LDA | Logistic Regression | Random Forest | Decision Tree | SVM | XGBoost | |
|---|---|---|---|---|---|---|
| AUC macro | 0.929 | 0.917 | 0.903 | 0.676 | – | 0.953 |
| F1 weighted | 0.891 | 0.854 | 0.848 | 0.769 | 0.848 | 0.896 |
| Accuracy | 0.892 | 0.862 | 0.862 | 0.800 | 0.862 | 0.892 |
| Sensitivity | 0.692 | 0.538 | 0.462 | 0.231 | 0.462 | 0.846 |
| Specificity | 0.942 | 0.942 | 0.962 | 0.942 | 0.962 | 0.904 |
Figure 1.
Receiver Operating Characteristic Curve for the Performance of 12-Variable Models in Discriminating the Severe COVID-19 Cases
The 12 variables included age, fever, dyspnea, lymphocyte, neutrophil, C-reactive protein, lactic dehydrogenase, creatine kinase, D-dimer, alanine aminotransferase, aspartate aminotransferase, and albumin.
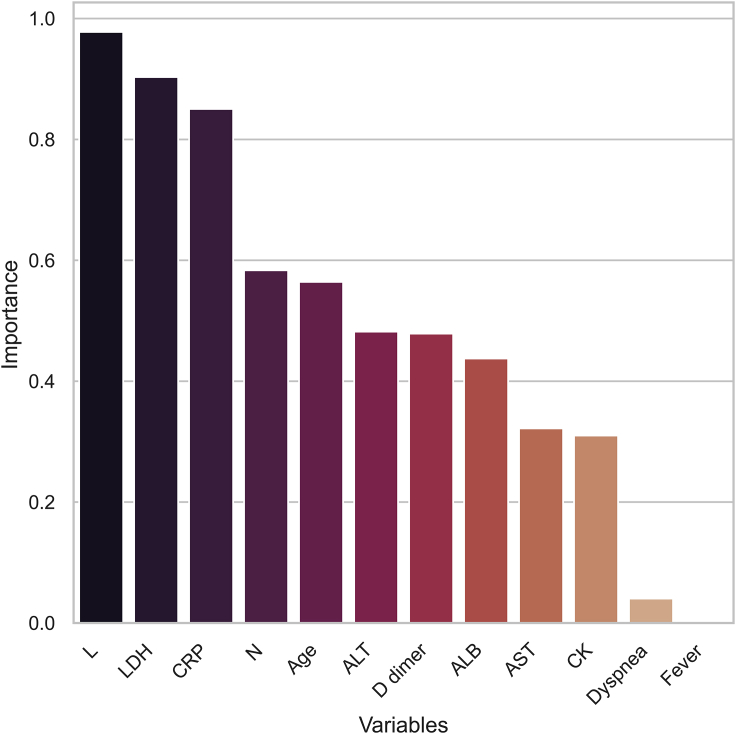
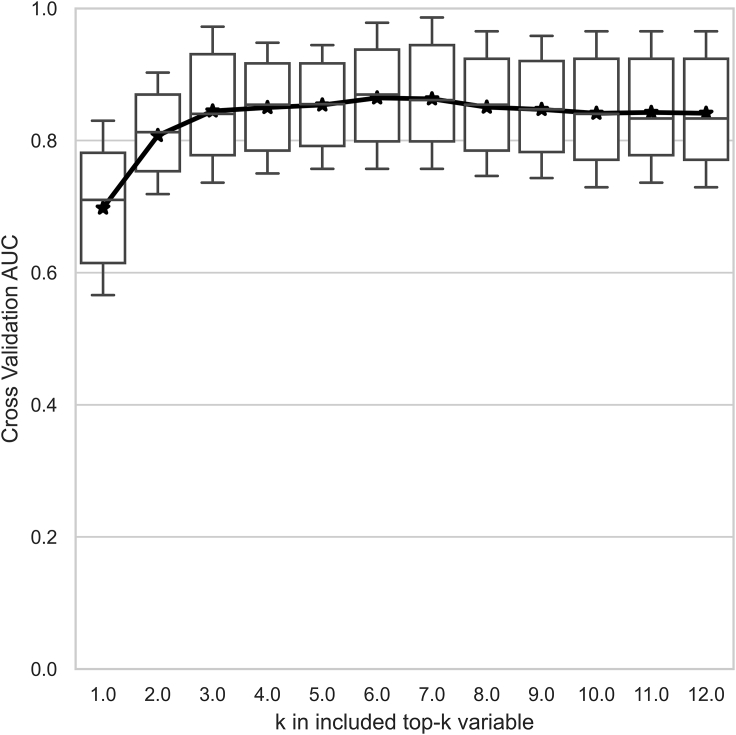
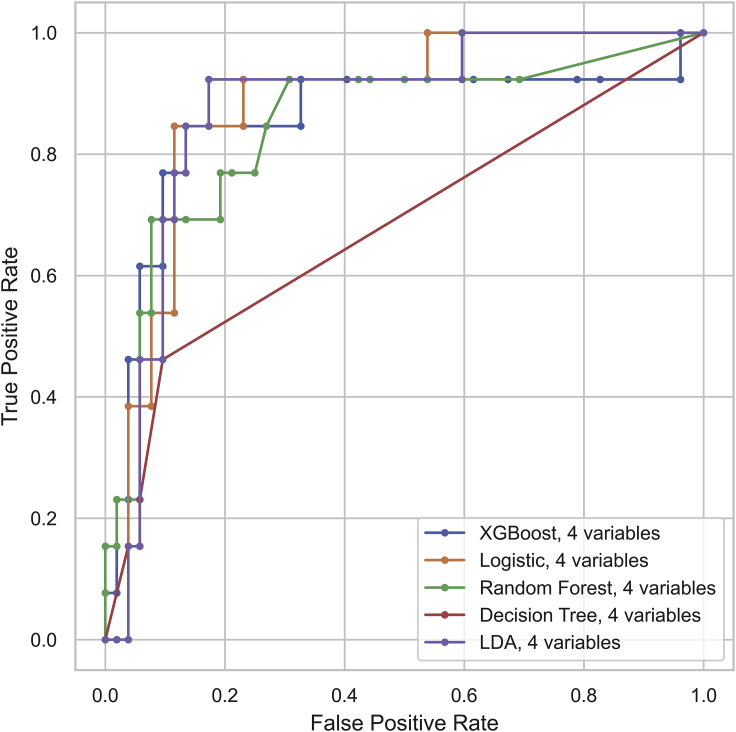
Next, the assembly of variables was minimized to make clinical use easier. For this purpose, a sequential variable selection approach was used to find the optimal variable set based on its assessment performance. In brief, important variables ranked by XGBoost (Figure 2) were sequentially assembled in an individualized manner to investigate their incremental effects in terms of AUC scores by cross-validation. The AUC scores ceased to grow when the count of assembled variables increased to 4 (Figure 3). Thus, the previous 12-variable models were shrunk to the selected 4-variable models, where the XGBoost classifier achieved an accuracy of 84.6% in the identification of severe COVID-19 cases. Table 4 compares the performance of various classifiers in the 4-variable models. The AUC score of XGBoost was slightly decreased compared with other models, but XGBoost achieved the highest F1 score and accuracy among the classifiers (Table 4; Figure 4). A greater than 80% accuracy indicated that the 4-variable XGBoost model could play a crucial role in distinguishing most cases that required immediate medical attention. Overall, the 4-variable XGBoost model was evaluated to be the most competitive and easy-to-use tool compared with other prevalent models.
Figure 2.
Top Key Clinical Variables That Are Ranked According to Their Importance in the Multi-Tree XGBoost Algorithm
Abbreviations: L, lymphocyte; LDH, lactic dehydrogenase; CRP, C-reactive protein; N, neutrophil; ALT, alanine aminotransferase; ALB, albumin; CK, creatine kinase; AST, aspartate aminotransferase.
Figure 3.
Important Variables Ranked by the XGBoost Algorithm Were Sequentially Assembled to Investigate Their Incremental Effects on the Model Performance
Abbreviation: AUC, area under the receiver operating characteristic curve.
Table 4.
The Performance of 4-Variable Models for Identification of Severe COVID-19 on Admission
| LDA | Logistic Regression | Random Forest | Decision Tree | SVM | XGBoost | |
|---|---|---|---|---|---|---|
| AUC macro | 0.876 | 0.879 | 0.864 | 0.680 | – | 0.859 |
| F1 weighted | 0.815 | 0.802 | 0.815 | 0.769 | 0.815 | 0.856 |
| Accuracy | 0.831 | 0.815 | 0.831 | 0.800 | 0.831 | 0.846 |
| Sensitivity | 0.385 | 0.385 | 0.385 | 0.231 | 0.385 | 0.846 |
| Specificity | 0.942 | 0.923 | 0.942 | 0.942 | 0.942 | 0.846 |
Figure 4.
Receiver Operating Characteristic Curve for the Performance of 4-Variable Models in Discriminating the Severe COVID-19 Cases
The four variables were lymphocyte, lactic dehydrogenase, C-reactive protein, and neutrophil.
Finally, the study investigated whether the 4-variable XGBoost model was effective to predict the risk of deterioration in patients who presented with non-severe symptoms on admission. For this purpose, a total of 39 patients who had non-severe COVID-19 on admission but experienced a deterioration during hospitalization were enrolled as an external testing set for analysis (Figure S3). The 4-variable XGBoost model achieved 100% accuracy in predicting the risk of rapid deterioration (Table 5). For 17 patients who suffered exacerbation and had complete time course of exacerbation, the minimal, medium, and maximal prediction horizons were less than 1, 5, and 12 days, respectively, suggesting that our model could predict the risk of disease deterioration up to 12 days before its occurrence.
Table 5.
The Performance of the 4-Variable XGBoost Model for Prediction of COVID-19 Deterioration
| LDA | Logistic Regression | Random Forest | Decision Tree | SVM | XGBoost | |
|---|---|---|---|---|---|---|
| Accuracy | 1.000 | 1.000 | 0.974 | 0.974 | 1.000 | 1.000 |
To test whether a clinically operable single-tree XGBoost classifier based on the lymphocyte count, CRP level, and LDH level as reported by Yan et al.9 was able to accurately identify the risk of severe symptoms of the disease on admission, we performed the single-tree XGBoost to identify severe COVID-19 cases as well as to predict the risk of in-hospital deterioration from non-severe to severe symptoms of the disease. Table S4 shows that the single-tree XGBoost had an 80% accuracy in identifying severe COVID-19 cases on admission, but only a 38.5% accuracy in predicting the risk of in-hospital deterioration. This suggests that models used for other purposes or reported in other works do not fulfill our goal in this study, which is to identify hospitalization priority for COVID-19 infections.
Collectively, the 4-variable XGBoost model is the first computation model used to assess hospitalization priority that enables rational triage of infected patients and prioritizes hospitalization to those who need it most.
Model Interpretation
SHAP (SHapley Additive exPlanations), a game theoretic approach that interpreted the impact of each input variable toward the model output, has been relied upon for model interpretation. In Figure S4, each dot corresponds to an individual case in the study. Different colors encode different values of input variables, while the SHAP value represents the impact of each variable on the prediction outcome. The risk of severe COVID-19 was found to be associated with a decrease in lymphocyte count, and an increase in LDH levels, CRP levels, and neutrophil count.
Subsequently, the t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm, a technique for dimensionality reduction, was used to project the 4D data (lymphocyte, LDH, CRP, neutrophil) into a 3D feature space for visualization.27 It enabled the visualization of different features among the three groups of patients, including severe cases, non-severe cases, and cases who showed severe progression. The cases who showed severe progression were patients with non-severe symptoms on admission but who subsequently developed severe symptoms. There was a clear separation between the non-severe (distributed in the core) and severe cases (distributed in the periphery), whereas the cases who showed severe progression were distributed between the core and the periphery (Figure S5; Video S1).
Discussion
The worldwide epidemic of COVID-19 is placing increasing stress on healthcare systems in many countries and territories as no effective vaccines have been approved to protect the general population from this highly contagious disease.4,5 This, therefore, requires decision-makers to efficiently distribute medical resources according to the need in those places where healthcare resources are rare.
To date, many studies have reported the detection of COVID-19 in patients suspected of infection.15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25 For instance, Menni et al.16 reported the efficiency of self-reported symptoms in early identification of potential COVID-19. This allows medical providers to timely admit all potential COVID-19 cases to hospitals in countries and territories where medical resources are relatively adequate, such as China.5 However, in places where medical resources are limited for all infected patients, identifying patients at a higher risk of severe COVID-19 and prioritizing them to hospitalization are crucial. Recent studies that assessed the outcome of COVID-19 patients does not allow the distribution priority of high-risk infections to hospital admission. For instance, a machine-learning model developed by Yan et al.,9 and a big data analysis performed by Williamson et al.,8 focused on early identification of COVID-19 mortality during hospital stay, rather than the timely identification or prediction of severe disease. The poor performance of the mortality identification model in assessing severity suggests that the previously reported prediction models are not suitable for the identification of hospitalization priority for severe or potentially severe COVID-19 cases (Table S4).
More recently, a nomogram developed by Gong et al.10 has been used to assist in the early identification of severe COVID-19 cases with a sensitivity of 77.5% and a specificity of 78.4%; however, this study was published prematurely before all participants had reached the final outcome. Therefore, our study is the first to focus on the development of an easy-to-use model that provides an insightful solution for doctors to triage patients, achieving a relatively high sensitivity and specificity, without negatively affecting its performance by involving participants still in treatment.
In this study, the clinical features of COVID-19 were screened to identify a total of 12 critical variables that were found to be associated with the risk of severe COVID-19 (Figure 2). To make clinical use easier, a model comprising four clinical variables was established. The 4-variable XGBoost model is effective in the identification of nearly 85% of severe cases who require immediate medical attention (Table 4). Importantly, it accurately predicts the risk of progression toward rapid deterioration for up to 12 days before its occurrence (Table 5).
Our study is in line with previous studies that showed that increased inflammatory responses, as demonstrated by an increase in neutrophil count and CRP level, and impaired antiviral capacity, such as lymphopenia, were associated with severe disease.11, 12, 13,28, 29, 30, 31 A high LDH level associated with tissue breakdown in various diseases also indicates severe disease.9
This machine-learning-based model is expected to be of clinical use in the context of the current COVID-19 pandemic. The four indices included in the model are easily accessible even in rural and less-developed situations; evaluation can be rapidly processed using the online program with a user-friendly interface (Figure S6; link: covid-19.zyh.science:8888). The model has general applicability, as the training and testing datasets were retrieved from the datasets of different hospitals in different cities in China as well as the WHO database, with participants who had experienced the disease with varying levels of severity.
There are some limitations in this study. First, this is a respective study with a relatively small sample size. A prospective study should be performed to validate our findings. Second, the datasets of different resources suffer various ratios of missing values that may affect the data analysis. The principle of cases and variables inclusion is a trade-off between the number of cases and the number of included variables (Figure S2). In this way, the impact of missing values on data analysis can be reduced. Third, there was no significant difference in the prevalence of comorbidities between severe and non-severe COVID-19 cases in our datasets (Table S2). This is probably because the number of patients with comorbidities is not enough to identify any difference in this factor between the two groups. It is unclear if the inclusion of comorbidity or other potential risk factors can improve the performance of the model. But, most recently, researches did not find critical roles of comorbidity in improving the accuracy of the model in predicting COVID-19 prognosis.9,10 However, as more data become available, we can easily update our findings and generate a more accurate model using the same procedure.
In summary, we developed this machine-learning-based model to contribute to healthcare management during the current pandemic. It is practically applicable for healthcare decision-makers and professionals to efficiently assess the infections and allocate inpatient care to those who need it most, and to contribute to the global battle against the spread of this coronavirus.
Experimental Procedures
Resource Availability
Lead Contact
Shaohua Ma; ma.shaohua@sz.tsinghua.edu.cn.
Materials Availability
No new unique reagents were generated in this study.
Data and Code Availability
The clinical data used in this study were obtained from the WHO COVID-19 database (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov), Guizhou Provincial People's Hospital, Affiliated Hospital of Zunyi Medical University, Jiangjunshan Hospital of Guizhou Province, Zhongnan Hospital of Wuhan University, and the Radiology Quality Control Center database of Hunan province. The code is available at GitHub (https://github.com/cow8/Covid-19_Severity).
Method Details
Patient Enrollment
Data on COVID-19 cases were retrieved from the WHO COVID-19 database,26 the Radiology Quality Control Center database of Hunan province, and the datasets of four major hospitals (Guizhou Provincial People's Hospital, Affiliated Hospital of Zunyi Medical University, Jiangjunshan Hospital of Guizhou Province, Zhongnan Hospital of Wuhan University).
A strategy to retrieve studies that reported individual data on clinical characteristics, clinical types, and prognosis of COVID-19 cases is shown in Figure S1. In brief, the publication list was first screened to exclude reviews, opinions, guidelines, corrections, epidemiological and pharmacological studies, and other nonclinical or irrelevant investigations, and identify clinical studies reporting both the clinical characteristics and patient outcomes. To enroll COVID-19 cases from the multi-center dataset in China, the medical records were examined to identify patients who were admitted to hospitals for SARS-CoV-2 infection from January to March, 2020.
All patients recruited to this study were laboratory-confirmed COVID-19 cases. Patients who were pregnant or younger than 18 years were excluded. The demographic information, clinical characteristics, laboratory findings, and prognosis of patients were extracted from these datasets. By the time of data collection, all patients had experienced the final outcome (e.g., death, or recovery and discharge).
Disease Assessment
All patients enrolled in this study were tested for SARS-COV-2 before or after being admitted to hospital. Oropharyngeal swab, nasopharyngeal swab, sputum, serum, feces, endotracheal aspirate, or bronchoalveolar lavage were collected from each patient to detect the SARS-CoV-2 RNA or anti-SARS-CoV-2 IgM/IgG if feasible. The COVID-19 cases were confirmed upon the detection of unique sequences of SARS-CoV-2 RNA by real-time RT-PCR assay or viral genome sequencing, or anti-SARS-CoV-2 IgM/IgG in the paired serum specimens, following the clinical guidelines.28,32 All of the patients enrolled in the study were also assessed for disease severity on admission and were repeatedly evaluated for progression during hospitalization. The severity of COVID-19 was stratified into non-severe and severe categories using the criteria that were published by WHO and the National Health Commission of China,28,31 with minor modifications (Table 2). Specifically, a severe case of COVID-19 was defined by the presence of any of the following conditions in the quiescent state, such as an increased respiratory rate of ≥ 30 breaths/minute, decreased oxygenation index of ≤ 300 mmHg, or decreased peripheral capillary oxygen saturation (SpO2) of ≤ 93%. Moreover, patients who developed shock, multiple organ failure that required to be admitted to the intensive care unit, or respiratory failure that warranted mechanical ventilation, were stratified into the severe category in this study. Finally, patients with pulmonary lesions that showed rapid progression of over 50% within 24–48 h were considered to have severe disease.
Critical Variables Inclusion
Clinical variables that were widely available in the clinic and were previously demonstrated to be closely associated with the severity of COVID-19 were obtained for data analysis in this study. They include but are not limited to age, gender, underlying comorbidity, symptoms (i.e., dyspnea), the hematological and biochemical parameters of lymphocyte, neutrophil, CRP, procalcitonin, D-dimer, erythrocyte sedimentation rate, ALT, AST, bilirubin, albumin, LDH, serum creatinine, blood urea nitrogen, prothrombin time, lactic acid, CK, oxygenation index, and SpO2.10, 11, 12, 13,28, 29, 30, 31,33, 34, 35 All these hematological and biochemical variables were detected using standard automated laboratory methods or commercially available kits following the manufacturers' protocols. All the clinical variables of patients were determined on admission or soon after hospitalization. The critical variables chosen to establish the model were measures easily accessible by practitioners from most countries and territories, and were common to all databases across different institutions.
Statistical Analysis
Data Analysis and Visualization Were Implemented Using Python
Categorical variables were described as number and frequency, while the continuous variables were expressed as mean, standard deviation (SD), interquartile range (IQR) where appropriate.
As there were various missing data in the datasets, a threshold alpha was set to remove these missing data (Figure S2). First, the clinical variables with missing data that exceeded the threshold alpha were removed (bottom figure). Second, the observations (that were COVID-19 cases) with missing data for any of the resulting clinical variables were removed. An optimal threshold alpha should be selected to obtain as many clinical variables and observations as possible.
Next, COVID-19 cases with non-missing variable values were grouped into severe and non-severe categories according to the criteria in Table 2. The difference in the clinical variables between the two groups was identified via the univariate descriptive statistics (Table S2). A p value of < 0.05 was considered statistically significant and was used as a threshold to identify key clinical variables for model development. These key clinical variables were assembled to generate the risk-assessment models based on the XGBoost classifier as well as other classifiers, such as LDA, logistic regression, SVM, random forest, and decision trees.
Then, the COVID-19 cases were randomly split into the training set and holdout testing set at a ratio of 7:3. The different models were trained in the training set and evaluated in the holdout testing set by comparing the values of accuracy, F1 score, sensitivity, specificity, and AUC score.
Subsequently, the assembly of key clinical variables was minimized to generate the simplified models. For this purpose, all the key clinical variables were ranked according to the importance calculated by XGBoost (Figure 2), followed by the sequential variable selection approach (Figure 3). This was to minimize the variable set while optimizing the model performance. The simplified models based on the minimized variable set were trained and evaluated in accordance with a method mentioned above. To assess the effectiveness of models in early prediction of severe progression, patients who presented with non-severe symptoms on admission but developed severe disease during hospitalization were enrolled as an external testing set for analysis. The performance of models was reflected by accuracy (Table 5). In comparison, the performance of a previously validated single-tree XGBoost model9 in identification of severe COVID-19 risk was assessed in our datasets (Table S4).
Finally, the SHAP values were plotted to visualize the impact of each input variable toward the model output and the t-SNE algorithm was used for dimensionality reduction that enabled to visualize the difference in features among different groups of patients.
Acknowledgments
We thank the above-mentioned cooperating hospitals for kindly sharing the data with us, in accordance with the Declaration of Helsinki. The work was supported by the National Natural Science Foundation of China (grant nos. 61722209 and 61971255), the fund from the Shenzhen Science and Technology Innovation Committee (grant no. KQJSCX20180327143623167), and the Guizhou Science and Technology Project (grant no.QKHZC[2020]4Y002).
Author Contributions
S.M. and L.F. conceived the project. Y. Zheng, Y. Zhu, and M.J. designed and performed the experiments and data analysis. Y. Zheng and Y. Zhu innovated and implemented the algorithm. R.W., X.L., and M.Z. contributed the data. S.M., L.F., and R.W critically reviewed the manuscript. All co-authors contributed to writing of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: August 3, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.patter.2020.100092.
Contributor Information
Lu Fang, Email: fanglu@sz.tsinghua.edu.cn.
Shaohua Ma, Email: ma.shaohua@sz.tsinghua.edu.cn.
Supplemental Information
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronavirus disease 2019 (COVID-19) Situation Report – 189. 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
- 4.Yang P., Wang X. COVID-19: a new challenge for human beings. Cell Mol. Immunol. 2020;17:555–557. doi: 10.1038/s41423-020-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., Chen Q., Feng L., Rodewald L., Xia Y., Yu H., Zhang R., An Z., Yin W., Chen W. Active case finding with case management: the key to tackling the COVID-19 pandemic. Lancet. 2020 doi: 10.1016/S0140-6736(20)31278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infection prevention and control in the household management of people with suspected or confirmed coronavirus disease (COVID-19) 2020. https://www.ecdc.europa.eu/sites/default/files/documents/Home-care-of-COVID-19-patients-2020-03-31.pdf
- 7.Sun Q., Qiu H., Huang M., Yang Y. Lower mortality of COVID-19 by early recognition and intervention: experience from Jiangsu Province. Ann. Intensive Care. 2020;10:33. doi: 10.1186/s13613-020-00650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson E., Walker A.J., Bhaskaran K.J., Bacon S., Bates C., Morton C.E., Curtis H.J., Mehrkar A., Evans D., Inglesby P. OpenSAFELY: factors associated with COVID-19-related hospital death in the linked electronic health records of 17 million adult NHS patients. medRxiv. 2020 doi: 10.1101/2020.05.06.20092999. [DOI] [Google Scholar]
- 9.Yan L., Zhang H.-T., Goncalves J., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Zhang M. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. [Google Scholar]
- 10.Gong J., Ou J., Qiu X., Jie Y., Chen Y., Yuan L., Cao J., Tan M., Xu W., Zheng F. A tool to early predict severe corona virus disease 2019 (COVID-19): a multicenter study using the risk nomogram in Wuhan and Guangdong, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal. Transduct Target Ther. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y., Yu X., Zhao H., Wang H., Zhao R., Sheng J. Host susceptibility to severe COVID-19 and establishment of a host risk score: findings of 487 cases outside Wuhan. Crit. Care. 2020;24:108. doi: 10.1186/s13054-020-2833-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan M., Yin W., Tao Z., Tan W., Hu Y. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One. 2020;15:e0230548. doi: 10.1371/journal.pone.0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynants L., Van Calster B., Bonten M.M.J., Collins G.S., Debray T.P.A., De Vos M., Haller M.C., Heinze G., Moons K.G.M., Riley R.D. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., Ganesh S., Varsavsky T., Cardoso M.J., El-Sayed Moustafa J.S. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020 doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diaz-Quijano F.A., Silva J.M.N.d., Ganem F., Oliveira S., Vesga-Varela A.L., Croda J. A model to predict SARS-CoV-2 infection based on the first three-month surveillance data in Brazil. medRxiv. 2020 doi: 10.1101/2020.04.05.20047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv. 2020 doi: 10.1101/2020.02.25.20021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng Z., Wang M., Song H., Guo S., Zhou Y., Li W., Zhou Y., Li M., Song X., Zhou Y. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv. 2020 doi: 10.1101/2020.03.18.20035816. [DOI] [Google Scholar]
- 21.Song C.-Y., Xu J., He J.-Q., Lu Y.-Q. COVID-19 early warning score: a multi-parameter screening tool to identify highly suspected patients. medRxiv. 2020 doi: 10.1101/2020.03.05.20031906. [DOI] [Google Scholar]
- 22.Martin A., Nateqi J., Gruarin S., Munsch N., Abdarahmane I., Knapp B. An artificial intelligence-based first-line defence against COVID-19: digitally screening citizens for risks via a chatbot. bioRxiv. 2020 doi: 10.1101/2020.03.25.008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Weng J., Li Z., Hou R., Zhou L., Ye H., Chen Y., Yang T., Chen D., Wang L. Development and validation of a diagnostic nomogram to predict COVID-19 pneumonia. medRxiv. 2020 doi: 10.1101/2020.04.03.20052068. [DOI] [Google Scholar]
- 24.Wu J., Zhang P., Zhang L., Meng W., Li J., Tong C., Li Y., Cai J., Yang Z., Zhu J. Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results. medRxiv. 2020 doi: 10.1101/2020.04.02.20051136. [DOI] [Google Scholar]
- 25.Mao X., Liu X.-P., Xiong M., Yang X., Jin X., Li Z., Zhou S., Chang H. Development and validation of chest CT-based imaging biomarkers for early stage COVID-19 screening. medRxiv. 2020 doi: 10.1101/2020.05.15.20103473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Global research on coronavirus disease (COVID-19) 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov
- 27.Maaten L.v. d., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 28.Diagnosis and treatment of novel coronavirus pneumonia (trial version 7) 2020. http://www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm [DOI] [PMC free article] [PubMed]
- 29.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ai J., Li Y., Zhou X., Zhang W. COVID-19: treating and managing severe cases. Cell Res, 2020;30:370–371. doi: 10.1038/s41422-020-0329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical management of severe acute respiratory infection when COVID-19 is suspected: Interim guidance V 1.2, (2020). https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected.
- 32.Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, (2020). https://apps.who.int/iris/handle/10665/331329.
- 33.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao W., Li T. COVID-19: towards understanding of pathogenesis. Cell Res. 2020 doi: 10.1038/s41422-020-0327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The clinical data used in this study were obtained from the WHO COVID-19 database (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov), Guizhou Provincial People's Hospital, Affiliated Hospital of Zunyi Medical University, Jiangjunshan Hospital of Guizhou Province, Zhongnan Hospital of Wuhan University, and the Radiology Quality Control Center database of Hunan province. The code is available at GitHub (https://github.com/cow8/Covid-19_Severity).