Abstract
Activation of the immune costimulatory molecule cluster of differentiation 40 (CD40) in Müller glia has been implicated in the initiation of diabetes-induced retinal inflammation. Results from previous studies support that CD40 protein expression is elevated in Müller glia of diabetic mice; however, the mechanisms responsible for this increase have not been explored. Here, we evaluated the hypothesis that diabetes augments translation of the Cd40 mRNA. Mice receiving thiamet G (TMG), an inhibitor of the O-GlcNAc hydrolase O-GlcNAcase, exhibited enhanced retinal protein O-GlcNAcylation and increased Cd40 mRNA translation. TMG administration also promoted Cd40 mRNA association with Müller cell–specific ribosomes isolated from the retina of RiboTag mice. Similar effects on O-GlcNAcylation and Cd40 mRNA translation were also observed in the retina of a mouse model of type 1 diabetes. In cultured cells, TMG promoted sequestration of the cap-binding protein eIF4E (eukaryotic translation in initiation factor 4E) by 4E-BP1 (eIF4E-binding protein 1) and enhanced cap-independent Cd40 mRNA translation as assessed by a bicistronic reporter that contained the 5′-UTR of the Cd40 mRNA. Ablation of 4E-BP1/2 prevented the increase in Cd40 mRNA translation in TMG-exposed cells, and expression of a 4E-BP1 variant that constitutively sequesters eIF4E promoted reporter activity. Extending on the cell culture results, we found that in contrast to WT mice, diabetic 4E-BP1/2–deficient mice did not exhibit enhanced retinal Cd40 mRNA translation and failed to up-regulate expression of the inflammatory marker nitric-oxide synthase 2. These findings support a model wherein diabetes-induced O-GlcNAcylation of 4E-BP1 promotes Cd40 mRNA translation in Müller glia.
Keywords: diabetes, retina, cluster of differentiation 40 (CD40), O-GlcNAcylation, eukaryotic translation initiation factor 4E–binding protein 1 (eIF4EBP1), mRNA translation, post-translational modification, inflammation, thiamet G, Müller cell
Retinal inflammation plays a critical role in the pathogenesis of diabetic retinopathy (DR) (1, 2). Although inflammation is typically beneficial when resolved promptly, chronic low-grade inflammation can contribute to disease progression (3). Diabetes increases the expression of proinflammatory molecules in retinal microglia, pericytes, endothelial cells, and Müller cells (3). The conventional view of retinal inflammation is that microglia are the first responders that initiate Müller cell activation and gliosis (4). Indeed, Müller cells respond to activated microglia by increasing the expression of proinflammatory factors including interleukins 1β and 6 (5). Counter to this conventional view, Portillo et al. (6) recently demonstrated that the immune costimulatory molecule cluster of differentiation 40 (CD40) in Müller glia is specifically responsible for initiating diabetes-induced retinal inflammation. Activation of CD40 by its ligand CD154 results in increased protein nitration, retinal leukostasis, and capillary degeneration, which are all hallmark characteristics of DR (7, 8). Diabetic CD40 knockout mice fail to exhibit enhanced proinflammatory cytokine expression and are protected from both retinal leukostasis and degeneration of the retinal vasculature (6, 8). Remarkably, Müller cell-targeted add-back of CD40 to whole body CD40 knockout mice restores the retinal inflammatory response to diabetes, as well as the development of leukostasis and capillary degeneration (6).
Post-translational modification of proteins by O-linked addition of β-d-GlcNAc (O-GlcNAcylation) regulates the inflammatory response under both physiological and pathological conditions (9). Elevated retinal protein O-GlcNAcylation has been reported in rodent models of both type 1 and type 2 diabetes, and evidence supports a role in the pathogenesis of DR (10, 11). Protein O-GlcNAcylation occurs via the addition of the O-GlcNAc donor UDP-GlcNAc to the hydroxyl side chain of serine and threonine residues. Enzymatic addition of the O-GlcNAc moiety is catalyzed by O-GlcNAc transferase, whereas the modification is removed by the hydrolase O-GlcNAcase. Deficiency in protein O-GlcNAcylation inhibits T-cell proliferation and differentiation (12, 13). Similarly, O-GlcNAcylation is necessary for B-cell expansion and activation of neutrophils (14, 15). Several immune-stimulating transcription factors are O-GlcNAc modified, including NF-κB, NFAT, and SP1, resulting in enhanced transcriptional activity of these proteins and subsequent production of immunomodulatory molecules (16–20).
In the retina of diabetic mice, CD40 protein expression is elevated in Müller glia (6); however, the mechanism responsible for this increase has not been established. Expression of proinflammatory proteins is typically induced by transcription factors, which can promote inflammation by increasing the expression of cytokines and other proinflammatory molecules. Importantly, translational control also plays a critical role in the inflammatory response that is less well-explored (21). Despite evidence demonstrating widespread modification of ribosomal and other regulatory proteins, including the translational repressor 4E-BP1 (22–26), relatively little is known regarding the impact of O-GlcNAcylation on mRNA translation. Recently, we demonstrated that augmented O-GlcNAcylation causes widespread variation in retinal gene expression at the level of mRNA translation (27). In that study, the top canonical pathway identified as being translationally altered by enhanced O-GlcNAcylation was the “acute-phase response,” which is a complex early-defense system involving a range of proinflammatory cytokines. Here, we explored Cd40 as one gene candidate subject to translational regulation in retina.
The rate at which an mRNA is translated is generally limited by the recruitment of ribosomes, which can occur through either a cap-dependent or cap-independent mechanism. All eukaryotic mRNAs contain a 5′-m7GTP cap structure that is recognized by the cap-binding protein eIF4E (28). Cap-dependent recruitment of a ribosome onto an mRNA occurs via assembly of the eIF4F complex (that includes eIF4E) at the 5′-cap, followed by binding of eIF4F·mRNA to the 40S ribosomal subunit (28). Sequestration of eIF4E by the translational repressor 4E-BP1 prevents eIF4F complex assembly and represents the best-characterized mechanism for repressing cap-dependent translation initiation. As an alternative to cap-dependent translation, ∼10% of mRNAs contain sequence elements that enable translation to be initiated independent of the 5′-cap (29). These sequence elements are not conserved between genes; however, a common feature is the formation of hairpin, or stem-loop, secondary structures. Highly structured elements in the 5′-UTR of messages are thought to enable translation under physiological duress when cap-dependent translation is inhibited, such as during exposure to diabetes-induced hyperglycemia (30, 31). A number of cell surface receptors are subject to translational regulation and variation in receptor expression often reflects changes in downstream signaling (32). In this study, we evaluated the hypothesis that diabetes enhances translation of the Cd40 mRNA. Overall, the findings demonstrate that protein O-GlcNAcylation promotes cap-independent Cd40 mRNA translation via a 4E-BP1/2–dependent mechanism that involves sequences within its 5′-UTR.
Results
O-GlcNAcase inhibition enhances Cd40 mRNA translation in retina
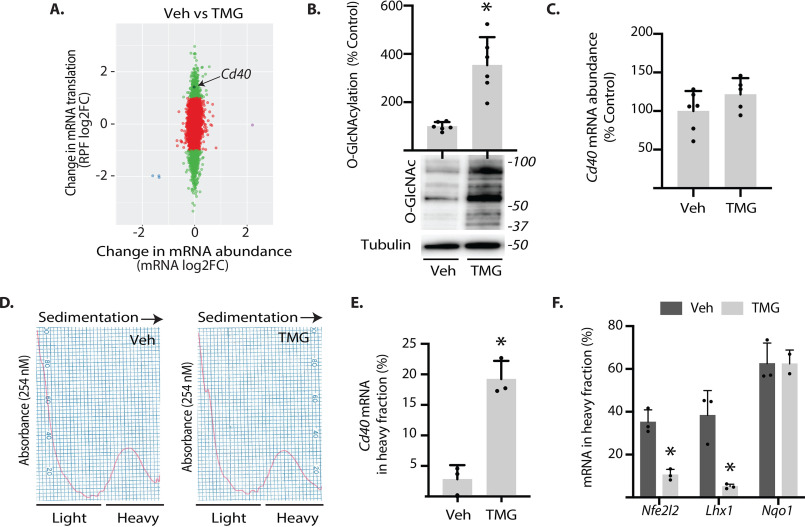
We previously evaluated the impact of O-GlcNAcase inhibition on retinal gene expression using next-generation sequencing to assess changes in mRNA abundance and ribosome-bound mRNAs undergoing translation via RNA-Seq and ribosome profiling (i.e. Ribo-Seq), respectively (27). Cd40 was identified as a candidate gene that exhibited enhanced mRNA translation in the retina of mice administered TMG as compared with PBS vehicle alone (Fig. 1A). As with the previous report (27), protein O-GlcNAcylation was enhanced in the retina of mice administered TMG as compared with PBS (Fig. 1B). Consistent with the results of ribosome profiling, Cd40 mRNA abundance was not altered in whole retinal lysates from mice administered TMG as compared with PBS (Fig. 1C). Sucrose density gradient centrifugation was used to separate retinal ribosomes into either heavy or light fractions (Fig. 1D). Poorly translated mRNAs are found in the light subpolysomal fraction, which contains both 80S monosomes, as well as individual 40S and 60S ribosomal subunits. In contrast, actively translating mRNAs associate with multiple ribosomes and form heavy polysomes. Consistent with sequencing results in Fig. 1A, the Cd40 mRNA was shifted into the heavy fraction obtained from the retina of mice administered TMG, as compared with PBS (Fig. 1E). In addition to up-regulation of Cd40 mRNA translation, mRNAs predicted by the sequencing analysis to exhibit down-regulation (Nfe2l2 and Lhx1) or no change (Nqo1) were also distributed in polysome profiles in a way that supported those observations (Fig. 1F).
Fig. 1.
O-GlcNAcase inhibition enhances translation of the Cd40 mRNA in mouse retina. Male and female mice were administered TMG or PBS vehicle (Veh). A, a recent sequencing analysis (27) revealed a TMG-induced increase in Cd40 mRNA in the ribosome-protected fragment (RPF) obtained from retina independent of a change in mRNA abundance. B, total protein O-GlcNAcylation and tubulin expression were assessed in retinal lysates by Western blotting analysis 24 h after TMG administration. Protein molecular mass (in kDa) is indicated on the right. C, Cd40 mRNA expression in whole retina was assessed by PCR. D, retinal ribosomes were separated into either light or heavy fractions via sucrose density gradient centrifugation. The distribution of mRNAs encoding Cd40 (E) or Nfe2l2, Lhx1, and Nqo1 (F) was assessed in each fraction via PCR. The results are expressed as a relative percentages of the mRNA in the heavy fraction obtained from 10 retinas. The values are means ± S.D. *, p < 0.05 versus vehicle.
Diabetes enhances Cd40 mRNA translation in retina
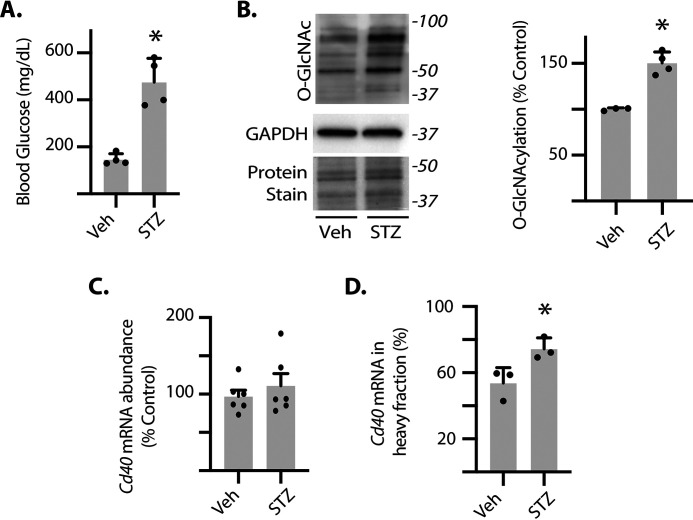
To assess whether Cd40 mRNA translation was enhanced in a model of diabetes, we generated polysome profiles from the retina of streptozotocin (STZ)–induced diabetic mice, a model of type 1 diabetes. Consistent with previous studies (11), blood glucose concentrations were significantly elevated (Fig. 2A) and retinal protein O-GlcNAcylation was enhanced in diabetic mice as compared with nondiabetic controls (Fig. 2B). To determine whether diabetes altered Cd40 transcription, we evaluated Cd40 mRNA abundance. There was no significant change in Cd40 mRNA abundance in the retina of diabetic mice as compared with nondiabetic mice (Fig. 2C). However, the relative distribution of the Cd40 mRNA was shifted into the heavy fraction obtained from the retina of diabetic mice, indicating enhanced translation (Fig. 2D).
Fig. 2.
Cd40 mRNA translation is enhanced in the retina of diabetic mice. Diabetes was induced in male mice by administration of STZ. Nondiabetic controls were administered vehicle (Veh). The retinas were collected after 3 months of diabetes. A, blood glucose concentrations were evaluated. B, protein O-GlcNAcylation and GAPDH expression were assessed in retinal lysates by Western blotting. Protein staining was used to evaluate loading. Protein molecular mass (in kDa) is indicated on the right. C, Cd40 mRNA expression was evaluated in retinal lysates by PCR. D, ribosomes from retinas were separated into either light or heavy fractions as described in the legend to Fig. 1. The distribution of the Cd40 mRNA was assessed in each fraction by PCR. The results are expressed as the relative percentage of the mRNA in the heavy fraction. The values are means ± S.D. *, p < 0.05 versus vehicle.
Diabetes and O-GlcNAcase inhibition enhance CD40 mRNA translation in retinal Müller glia
To specifically assess Cd40 mRNA translation in retinal Müller cells, we developed a RiboTag mouse (33) that expressed an epitope-tagged ribosomal protein under the control of Cre recombinase that is directed specifically toward retinal Müller cells by promoter/enhancer elements of the Pdgfra gene (34). RiboTag mice express a fully functional hemagglutinin (HA)–tagged variant of ribosomal protein L22 (Rpl22), which forms the core of the ribosomal 60S subunit and facilitates cell type–specific isolation of translationally active mRNAs (33). Hemizygous PDGFRa-Cre–positive mice were crossed with RiboTag mice, resulting in deletion of WT exon 4 in Müller cells and replacement with an exon 4 that included an HA tag (Fig. 3A). Replacement of WT exon 4 with the HA-tagged exon 4 was validated by PCR (Fig. 3B). HA-tagged Rpl22 expression was only observed in the retina of Cre-positive mice (Fig. 3C). HA-Rpl22 expression colocalized with the Müller cell–specific marker glutamine synthetase (GS) in retinal cryosections (Fig. 3D). Müller-specific HA-tagged ribosomes were immunoprecipitated from whole retinal lysates (Fig. 3E). HA-Rpl22 was detected in immunoprecipitations targeting the HA tag but not in a FLAG-tag negative control. RNA isolated by immunoprecipitation was high quality (Fig. 3F). The immunoprecipitation efficiently isolated retinal mRNAs encoding Müller cell–specific markers (CRALBP, GS, and AQP4), but not the retinal pigmented epithelium marker RPE65 (Fig. 3G). To evaluate the effect of enhanced O-GlcNAcylation on Cd40 mRNA translation, RiboTag mice were administered TMG. Cd40 mRNA association with HA-tagged ribosomes was enhanced in the retina of mice administered TMG as compared with those administered PBS, indicating up-regulated Cd40 mRNA translation (Fig. 3H). Similarly, STZ-induced diabetes also promoted Cd40 mRNA translation in retinal Müller cells (Fig. 3I).
Fig. 3.
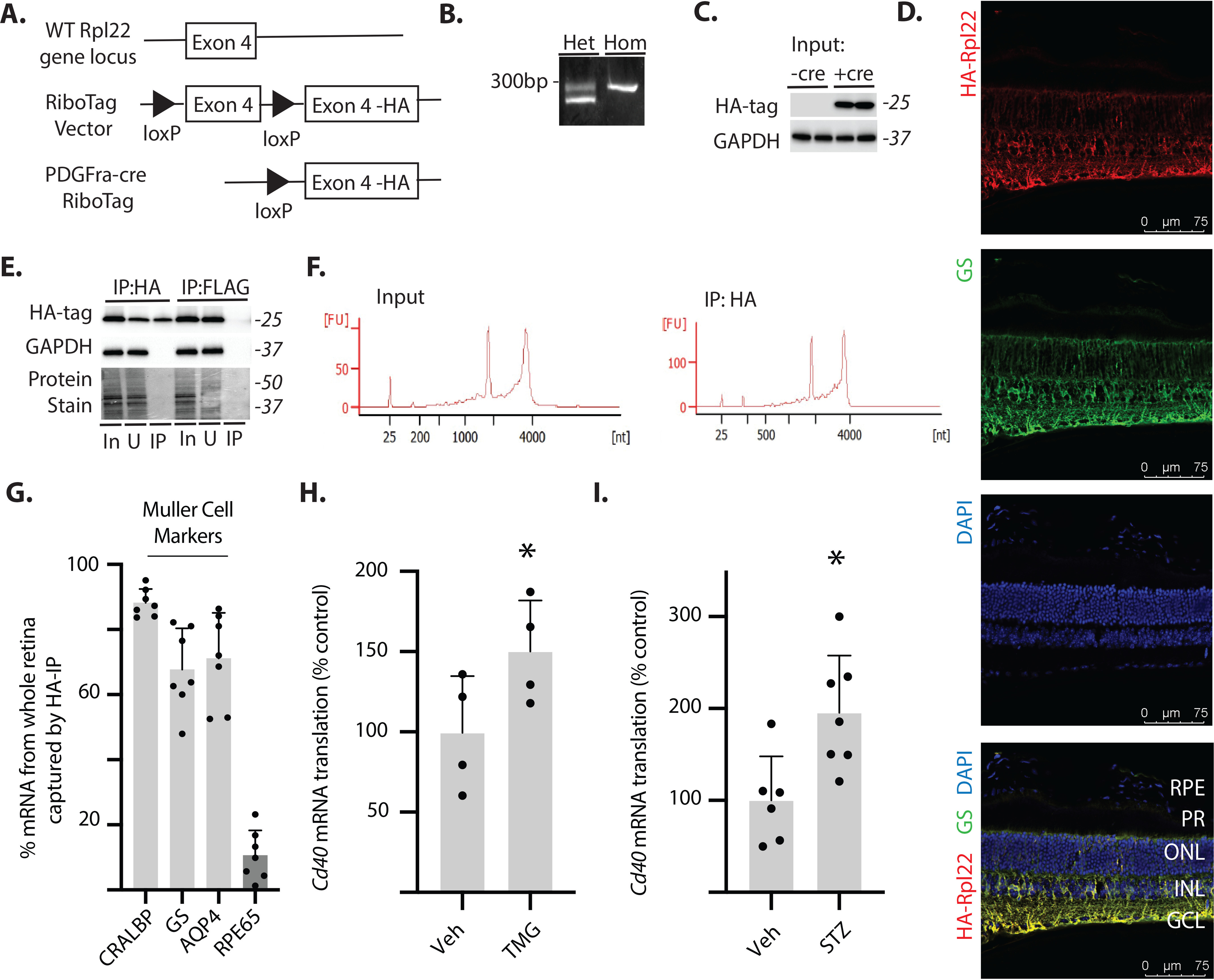
Cd40 mRNA translation is enhanced in retinal Müller glia by O-GlcNAcase inhibition or diabetes. A, Müller glia–specific expression of HA-tagged Rpl22 protein in retina was achieved by crossing WT RiboTag mice to a PDGFRa-cre recombinase–expressing mouse, resulting in deletion of the WT exon 4 in the target cell population and replacement with the Rpl22HA exon. B, PCR products were generated using oligonucleotides that amplify the loxP-containing intron sequence 5′ to the WT exon 4 of the Rpl22 gene. The WT PCR product is 260 bp, whereas the mutant PCR product is 290 bp. C, Western blotting analysis of HA-tagged Rpl22 and GAPDH in retinal lysates from WT Rpl22 (-cre) or Rpl22HA (+cre) mice. D, whole eyes were isolated, fixed, and cryosectioned into sagitally oriented longitudinal cross-sections. HA-Rpl22 (red) and the Müller glia–specific marker GS (green) was evaluated in retinal sections by immunofluorescence. The nuclei were visualized by 4′,6′-diamino-2-phenylindole (DAPI, blue). Colocalization of HA-Rpl22 and GS is shown in yellow. E, ribosomes from Rpl22HA-expressing homozygous mouse retina were isolated by immunoprecipitation. F, bioanalyzer analysis demonstrating recovery of high-quality RNA from both whole retina and following ribosome isolation (RNA Integrity Number >8.0). G, RNA from HA-tag immunoprecipitates was analyzed for recovery of Müller glia–specific markers (CRALBP, GS, and AQP4) and the retinal pigmented epithelia marker RPE65. H and I, Cd40 mRNA association with ribosomes isolated from the retina of Rpl22HA-expressing homozygous mice was determined by PCR. The retinas were isolated from male and female mice 24 h after administration of TMG (H) or from male mice 4 weeks after STZ (I). Protein molecular mass (in kDa) is indicated at right of blots in C and E. The values are means ± S.D. *, p < 0.05 versus vehicle. Het, heterozygous; Hom, homozygous; RPE, retinal pigmented epithelium; PR, photoreceptor outer segments; ONL, outer nuclear layer; INL, inner nuclear layer; IP, immunoprecipitation; GCL, ganglion cell layer; In, immunoprecipitation input; U, unbound immunoprecipitation fraction; Veh, vehicle.
Role of 4E-BP1/2 in cap-independent Cd40 mRNA translation
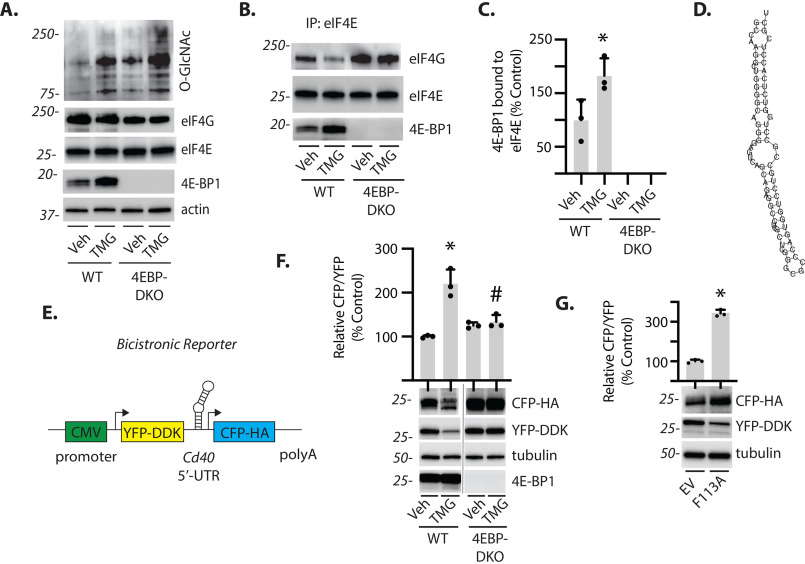
Transcript-specific translational control is generally mediated by cis-regulatory elements encoded by the UTR of the target mRNA. A recent genome-wide systematic screen indicated that sequences within the 5′-UTR of the Cd40 mRNA were capable of promoting translation independent of a 5′ mRNA cap (35). In addition, we previously provided evidence that enhanced O-GlcNAcylation causes a shift from cap-dependent to cap-independent mRNA translation that is mediated by 4E-BP1 (31). To investigate the impact of 4E-BP1 on Cd40 mRNA translation, we established a cell culture model of enhanced O-GlcNAcylation in the presence and absence of 4E-BP1. In both WT and 4E-BP1/2–deficient cells, exposure to TMG enhanced protein O-GlcNAcylation (Fig. 4A). TMG promoted 4E-BP1 coimmunoprecipitation with eIF4E, whereas the protein could not be detected in 4E-BP1/2–deficient cells (Fig. 4, B and C). Mfold web server predicted that the 5′-UTR sequence of Cd40 forms a hairpin structure (Fig. 4D). To specifically evaluate the role of the 5′-UTR of Cd40, it was cloned into a bicistronic reporter (pYIC) wherein a single mRNA encodes both yellow and cyan fluorescent protein (YFP and CFP, respectively). YFP expression was used as a reporter for cap-dependent translation, whereas CFP expression was used as a reporter for cap-independent translation driven by the Cd40 5′-UTR (Fig. 4E). In WT cells, exposure to TMG increased the CFP/YFP expression ratio, whereas there was no change in the ratio of CFP/YFP expression in 4E-BP1/2–deficient cells exposed to TMG (Fig. 4F). We also coexpressed pYIC with a 4E-BP1 F113A variant that resists mTORC1-dependent phosphorylation and therefore constitutively binds eIF4E (36) in the human MIO-M1 Müller cell line. As compared with an empty vector, 4E-BP1 F113A increased the CFP/YFP expression ratio (Fig. 4G). These data support a role for 4E-BP1 in promoting Cd40 mRNA translation.
Fig. 4.
4E-BP1 enhances cap-independent Cd40 mRNA translation. A, WT and 4E-BP1/2 double knockout (DKO) mouse embryonic fibroblasts were cultured in the presence of TMG or vehicle (Veh) for 24 h. Protein O-GlcNAcylation and expression of eIF4G, tubulin, eIF4E, and 4E-BP1 were assessed by Western blotting of retinal lysates. Protein molecular mass (in kDa) is indicated at left. B, the interaction of 4E-BP1 and eIF4G with eIF4E was examined by immunoprecipitating eIF4E from cell lysates and measuring the amount of eIF4G and 4E-BP1 in the immunoprecipitate (IP) by Western blotting. C, the interaction of 4E-BP1 with eIF4E in B was quantified. D, a hairpin structure was predicted for the Cd40 5′-UTR using the mfold web server. E, a bicistronic reporter was generated wherein a single mRNA encodes for both YFP and CFP. YFP expression is mediated by cap-dependent translation, whereas cap-independent expression of CFP is under the control of the Cd40 5′-UTR. F, WT and 4E-BP1/2 double knockout (4EBP-DKO) mouse embryonic fibroblasts expressing Cd40 reporter were cultured in the presence of TMG or vehicle for 24 h. G, human MIO-M1 Müller cell cultures were cotransfected with the Cd40 reporter, and either a plasmid that expresses an empty vector (EV) or a 4E-BP1 F113A variant that constitutively binds eIF4E. CFP, YFP, tubulin, and 4E-BP1 expression were assessed by Western blotting of cell lysates. The ratio of CFP to YFP expression was quantified from Western blots. The values are means ± S.D. A Student's t test was used to compare differences between groups in C and G. The data in E were analyzed by two-way ANOVA (Table S1). Dunnett's post hoc analysis was used to identify differences between means when significant main and interaction effects were detected. *, p < 0.05 versus vehicle; #, p < 0.05 versus WT.
4E-BP1/2 deletion prevents diabetes-induced CD40 mRNA translation
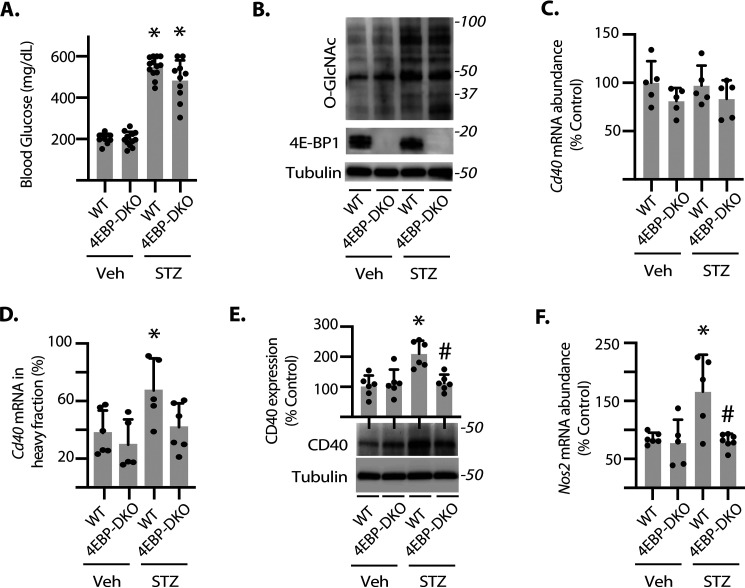
To evaluate the role of 4E-BP1 in regulating the translation of the Cd40 mRNA in the retina, diabetes was induced in WT and 4E-BP1/2 knockout mice. Blood glucose concentrations (Fig. 5A) and retinal protein O-GlcNAcylation (Fig. 5B) were significantly elevated in both WT and 4E-BP1/2–deficient diabetic mice as compared with nondiabetic mice. No significant change in total Cd40 mRNA was detected in whole retinal lysates (Fig. 5C). In polysome profiles, the Cd40 mRNA shifted into the heavy fraction obtained from the retina of WT diabetic mice as compared with WT nondiabetic mice (Fig. 5D). However, the distribution of the Cd40 mRNA in the heavy fraction obtained from the retina of 4E-BP1/2–deficient diabetic mice was similar to that observed in 4E-BP1/2–deficient nondiabetic mice (Fig. 5D). Consistent with changes in mRNA translation, CD40 protein expression was increased in the retina of WT STZ-diabetic mice, and 4E-BP1/2 deletion prevented the effect (Fig. 5E). Because CD40 is necessary for the retinal inflammatory response to diabetes, inducible nitric-oxide synthase 2 (Nos2) mRNA abundance was evaluated in retinal lysates as a marker for retinal inflammation. Diabetes enhanced Nos2 expression in the retina of WT mice; however, there was no difference in Nos2 expression in the retina of diabetic and nondiabetic 4E-BP1/2–deficient mice (Fig. 5F).
Fig. 5.
4E-BP1/2 is necessary for diabetes-induced Cd40 mRNA translation in retina. Diabetes was induced in WT and 4E-BP1/2 double knockout (4EBP-DKO) male mice using STZ. Nondiabetic controls were administered vehicle (Veh). The retinas were collected after 3 months of diabetes. A, blood glucose concentrations were evaluated. B, protein O-GlcNAcylation and expression of 4E-BP1 and tubulin were assessed by Western blotting of retinal lysates. Protein molecular mass (in kDa) is indicated at the right. C, Cd40 mRNA expression was evaluated in retinal lysates by PCR. D, ribosomes from retinas were separated into either heavy or light fractions as described in Fig. 1. The distribution of the Cd40 mRNA was assessed in each fraction by PCR. E, CD40 and tubulin protein expression were assessed in retinal lysates by Western blotting. F, Nos2 mRNA abundance was evaluated in whole retina lysates by PCR. The results are expressed as the relative percentage of the mRNA in the polysomal fraction. The values are means ± S.D. The data were analyzed by two-way ANOVA (Table S1). Dunnett's post hoc analysis was used to identify differences between means when significant main or interaction effects were detected. *, p < 0.05 versus vehicle; #, p < 0.05 versus WT.
Discussion
The TNF receptor superfamily member CD40 is a central mediator of inflammation in DR pathology (6, 8, 37). CD40 protein expression in Müller glia is elevated in the retina of diabetic rodents and is critical for the development of retinal inflammation in diabetes (6). In the present study, we demonstrated that in the retina of STZ-diabetic mice, CD40 protein expression was enhanced, independent of a change in total Cd40 mRNA abundance. Rather, the increase in CD40 protein expression in the retina of diabetic mice was associated with increased translation of the Cd40 mRNA. Using a novel RiboTag mouse model, we provide evidence that STZ-induced diabetes enhanced Cd40 mRNA translation in retinal Müller glia. In the retina of diabetic rats and in retinal Müller glia exposed to hyperglycemic conditions, the sequestration of eIF4E by 4E-BP1 is enhanced (38, 39). Our laboratory previously demonstrated that 4E-BP1 is directly O-GlcNAc–modified in response to diabetes (26, 40) and that O-GlcNAcylation promotes binding of 4E-BP1 with eIF4E (26, 31). Herein, the effect of diabetes on Cd40 mRNA translation required eIF4E sequestration, because 4E-BP1/2 ablation prevented diabetes-induced Cd40 mRNA translation in retina and normalized expression of CD40 protein, as well as the inflammatory marker NOS2. Retinal NOS2 expression is enhanced in both patients with DR and in rodent models of the disease (41, 42). Up-regulated expression of this proinflammatory marker is critical for disease progression, because NOS2-deficient mice are resistant to diabetes-induced retinal leukostasis and capillary degeneration (43, 44). Overall, the results here are consistent with a model wherein diabetes-induced O-GlcNAcylation facilitates retinal inflammation by promoting Cd40 mRNA translation (Fig. S1). An important caveat to this model is that there is likely a myriad of other signaling changes that promote the retinal inflammatory response in diabetes. Thus, it will be important for future studies to further interrogate both CD40-dependent and CD40-independent signaling events that contribute to retinal pathology in diabetes.
Portillo et al. (6–8) provided evidence for a model wherein Müller glia initiate retinal inflammation via CD40 activation, which leads to ATP release and stimulation of P2X7 purinergic receptors on microglia/macrophages. In the retina of diabetic CD40 knockout mice, NOS2 expression, leukostasis, and the development of acellular capillaries are reduced as compared with diabetic WT control (6, 8). However, Müller cell–targeted add-back of CD40 to CD40 knockout mice restores diabetes-induced retinal inflammation as assessed by inflammatory cytokine expression (e.g. NOS2) (6). Despite the in vivo work pinpointing Müller cells as a critical mediator of CD40-driven inflammation in DR, in vitro work revealed that both human and rodent Müller glia are unable to secrete cytokines in response to CD40 ligation by its ligand CD154 (6). This raised the possibility that CD40 signals to neighboring microglial cells to stimulate the production of proinflammatory cytokines. Indeed, Portillo et al. (6) found that activation of CD40 on Müller glia induced proinflammatory responses through signaling to bystander microglia. Overall, the previous findings support a model in which CD40 activation on Müller glia initiates the inflammatory response in DR. It is worth noting that other groups have not independently verified such a model. Moreover, the studies here were not designed to do so.
Herein, we explored a potential mechanism responsible for increased CD40 expression in retinal Müller glia of diabetic mice. We investigated translational regulation of the Cd40 mRNA, based on recent genome-wide screens that supported the possibility (27, 35). In the retina of diabetic mice, we found that CD40 mRNA translation was enhanced in Müller glia concomitant with an increase in retinal protein O-GlcNAcylation. In addition, Cd40 mRNA translation was enhanced in Müller glia of mice administered TMG to promote retinal protein O-GlcNAcylation.
Activation of CD40 on retinal Müller glia depends upon the presence of its ligand CD154. CD154 concentrations are enhanced in the blood of both diabetic patients and rodents, and plasma levels of soluble CD154 are positively correlated with the severity of DR (45, 46). It remains unclear how CD154 gets into the retina prior to the development of inflammation; however, it is possible that microthrombosis in the retinal capillaries of patients with DR enables activation of platelets in the retina (47). Activated platelets express surface CD154 that is rapidly shed upon ligation with CD40-expressing endothelial cells into the biologically active soluble CD154 (48). Moreover, ligation of CD154-expressing platelets with CD40-expressing endothelial cells stimulates inflammatory cascades (45). Alternatively, in advanced stages of DR when immune privilege and the blood–retinal barrier are compromised, circulating immune cells (such as CD154-expressing T cells) or serum proteins (such as soluble CD154) may infiltrate the retina and contribute to inflammation-induced neurovascular retinal damage (49).
In response to diabetes, Müller cells exhibit a range of protein expression changes that contribute to retinal inflammation, microvascular defects, and neuronal dysfunction (reviewed by Coughlin et al. in Ref. 50). Müller cells are uniquely sensitive to the metabolic environment in diabetes, in part because of their exclusive expression of the high-capacity/low-affinity GLUT2 glucose transporter (51) and predominance of the feedback inhibition–resistant glutamine-fructose-6-phosphate amidotransferase 2 isoform (52). Importantly, glutamine-fructose-6-phosphate amidotransferase is the rate-limiting enzyme of the hexosamine biosynthetic pathway. Thus, hyperglycemia potentially has dramatic effects on protein O-GlcNAcylation in Müller cells of diabetic patients. Diabetes causes Müller cells to become reactive and exhibit increased expression of the intermediate filament protein glial fibrillary acidic protein (53–55). In the retina of diabetic patients, increased glial fibrillary acidic protein expression manifests prior to development of DR, suggesting an early onset of Müller glia dysfunction (56–58).
CD40 expression is generally low under basal conditions, and increased CD40 expression is a common feature of inflammatory diseases driven by the receptor (59). A previous report supports that CD40 protein expression is elevated in Müller glia of diabetic rodents (6). Previous studies also demonstrate that TNFα and IFN-γ from microglia activate Cd40 gene transcription via cis-elements in the Cd40 promoter (60). However, cytokine-mediated Cd40 transcriptional activation is not consistent with a model wherein CD40 expression in Müller glia initiates retinal inflammation (6). Rather, transcriptional activation of CD40 through this conventional pathway would represent a model wherein retinal inflammation is initiated by gene expression changes in the microglia. In the present study, we provide new evidence that diabetes promotes Cd40 mRNA translation independent of a change in total Cd40 mRNA abundance.
We previously reported that augmented O-GlcNAcylation contributes to retinal dysfunction by causing widespread variation in the selection of mRNAs for translation, as assessed by ribosome profiling (27). CD40 was identified in that next-generation sequencing analysis as a gene candidate in which expression in retina was primarily altered at the level of mRNA translation. Translational control of CD40 expression is supported by a previous study demonstrating that a Graves' disease–associated single nucleotide polymorphism in the Cd40 5′-UTR results in increased Cd40 mRNA translation and protein expression (61). As previously mentioned, Cd40 was also identified in a genome-wide screen for sequence elements capable of promoting cap-independent translation (35). In the present study we cloned the 5′-UTR of the Cd40 mRNA into a reporter construct to evaluate cap-independent Cd40 mRNA translation. The 5′-UTR of Cd40 mRNA is not notably long at only 77 base pairs but is particularly GC rich (72%) and predicted to form a stable stem-loop structure. It is worth noting that this sequence also contains a consensus G-quadruplex (5′-…GGGCGGGGCCAAGGCTGGGGCAGGG…) (62). When located within the 5′-UTR, GC-rich secondary structures such as these inhibit cap-dependent mRNA translation (63, 64). However, these same highly structured RNA elements often facilitate preferential translation when cap-dependent translation is inhibited (30, 31). In the present study, we found that retinal Cd40 mRNA translation was enhanced by exposure to TMG. This is consistent with a relative increase in the Cd40 5′-UTR reporter that was observed in cells exposed to TMG. Importantly, TMG failed to enhance the relative activity of the Cd40 5′-UTR in the absence of 4E-BP1/2, whereas the 4E-BP1 F113A variant, which cannot be phosphorylated, was sufficient to enhance translation from the reporter. Collectively, our observations support a role for TMG-induced translational control of CD40 expression that is mediated by the Cd40 5′-UTR and requires 4E-BP1/2. Despite significant efforts, we were not able to consistently evaluate translation of endogenous Cd40 mRNA in polysomes prepared from MEFs. This is at least in part because of low Cd40 mRNA abundance in the cell culture model.
In the present study, 4-week-old mice were administered either TMG to promote O-GlcNAcylation or the toxin STZ to model type 1 diabetes. Retinal polysomes were prepared 24 h after TMG administration or 3 months after the induction of diabetes. Importantly, retinal Cd40 mRNA translation was increased by both TMG and STZ relative to their respective controls. The Cd40 mRNA was almost exclusively observed in the light subpolysomal fraction of 4-week-old mice 24 h after administration of a vehicle control. By contrast, CD40 was more evenly distributed between the light and heavy fractions obtained from the retina of nondiabetic control mice. These observations suggest that Cd40 mRNA translation may be increased in the retina of older mice. They also raise the possibility that factors other than O-GlcNAcylation potentially influence Cd40 mRNA translation.
Anti-inflammatory steroids are used clinically to treat diabetic macular edema, and the implantation of the corticosteroid dexamethasone provides a therapeutic alternative for diabetic macular edema patients refractory to the first-line anti–vascular endothelial growth factor therapies (65). However, long-acting dexamethasone implants are associated with an increased risk for elevations in intraocular pressure and cataract formation (65). Therefore, there is a need to identify alternative therapies for patients who do not respond to anti–vascular endothelial growth factor treatment. The previous body of work indicates that CD40 activation specifically on Müller glia drives the expression of multiple cytokines and chemokines, resulting in augmented inflammatory signaling and ultimately chronic retinal inflammation associated with DR. We extend the previous studies by demonstrating that diabetes and enhanced O-GlcNAcylation promote Cd40 mRNA translation via a 4E-BP1/2–dependent mechanism. Based on the data here, 4E-BP1/2–mediated control of CD40 expression in Müller glia may represent a therapeutic target for intervention to prevent retinal inflammation in DR. Notably, suppression of O-GlcNAc transferase activity by angiotensin-converting enzyme inhibition represents a potential mechanism for combating the increase in 4E-BP1 O-GlcNAcylation that is seen in diabetes (66). Regardless, further mechanistic studies delineating CD40-mediated retinal inflammation are needed to better understand the role of CD40 activation in Müller glia in DR.
Experimental procedures
Animals
At 4 weeks of age, male and female C57BL/6 mice received 50 mg/kg TMG or PBS as a control via intraperitoneal injection. The retinas were extracted 24 h after TMG injections. Alternatively, male WT or 4E-BP1/2 (Eif4ebp1; Eif4ebp2) double knockout mice were administered 50 mg/kg streptozotocin via intraperitoneal injection for 5 consecutive days to induce diabetes. Control mice received equivalent volumes of sodium citrate buffer. Diabetes was confirmed by blood glucose concentration (>250 mg/dl) in fasted animals. The retinas were extracted after 4 or 12 weeks of diabetes. All procedures were approved by the Pennsylvania State College of Medicine Institutional Animal Care and Use Committee and in accordance with the Association for Research in Vision and Ophthalmology policies on the ethical use of animals in ophthalmic research.
Protein analysis
The retinas were extracted, flash-frozen in liquid nitrogen, and later homogenized in 250 μl of extraction buffer as described previously (38). The homogenate was centrifuged at 1000 × g for 5 min at 4 °C, and the supernatant was collected for analysis. A fraction of the supernatant was added to SDS sample buffer, boiled for 5 min, and analyzed by Western blotting analysis as described previously (67). Antibodies used included O-GlcNAc (CTD110.6) (Cell Signaling Technology, catalog no. 9875), CD40 (Cell Signaling Technology, catalog no. 86165), α-tubulin (Santa Cruz, catalog no. sc-32293), GAPDH (Santa Cruz, catalog no. sc-32233), HA (Santa Cruz, catalog no. sc-805), and DDK (Origene, catalog no. TA5011-100). Preparation of the eIF4E, 4E-BP1, and eIF4G antibodies has been described previously (68, 69). Immunoprecipitations were performed by incubating cell lysates with monoclonal anti-eIF4E antibody as described previously (31).
Polysome fractionation by sucrose density gradient centrifugation and RNA isolation
Sucrose density gradient centrifugation was employed to separate the subpolysomal from the polysomal ribosome fractions as described previously (27). Two sucrose fractions representing the light and heavy portions of the gradient were collected directly into an equal volume of TRIzol reagent (Invitrogen). To improve the recovery of RNA from dense sucrose portions of the gradient, the heavy fraction was diluted twice with RNase-free water (HyClone), and the appropriate amount of TRIzol was added. RNA was extracted using the standard manufacturer's protocol and resuspended in 14 μl of RNA storage solution. An equal amount of RNA from each fraction collected was converted into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems) and subjected to quantitative real-time PCR using QuantiTect SYBR Green Master Mix (Qiagen).
RiboTag mouse model
B6J.129(Cg)-Rpl22tm1.1Psam/SjJ (RiboTag) mice carrying a targeted homozygous mutation of the ribosomal protein L22 locus to enable Cre-mediated HA epitope tagging of ribosomes from specific cell types were purchased from The Jackson Laboratory. To enable expression of HA-tagged ribosomes in Müller glia, RiboTag mice were crossed with hemizygous C57Bl/6-Tg(Pdgfra-cre)1Clc/J (PDGFRa-CRE; The Jackson Laboratory) to generate heterozygous RiboTag mice. The resulting heterozygous generation was crossed with homozygous RiboTag mice to produce offspring with a PDGFRa-cre recombinase-expressing mouse with deletion of WT exon 4 of the Rpl22 locus and replacement with the Rpl22HA exon specifically in Müller cells of the retina.
Isolation of Müller-specific ribosomes
Ribosomes were immunoprecipitated from RiboTag mice as previously described in Ref. 33. Briefly, EZview anti-HA beads (Sigma) were washed twice in immunoprecipitation buffer (50 mm Tris, pH 7.5, 100 mm KCl, 12 mm MgCl2, 1% Nonidet P-40) and blocked with 0.5% BSA, followed by two additional washes in immunoprecipitation buffer before being added to retinal homogenates. Two retinas from a single mouse were homogenized in polysome buffer (50 mm Tris, pH 7.5, 100 mm KCl, 12 mm MgCl2, 1% Nonidet P-40, 1 mm DTT, 400 units/ml Promega RNasin, 0.5 mg/ml heparin, 100 µg/ml cycloheximide, 10 µl/ml protease inhibitor mixture (Sigma) followed by centrifugation at 10,000 × g for 10 min at 4 °C. Supernatants were added to prepared beads and allowed to rotate for 16 h at 4 °C. The beads were pelleted at 8200 × g for 30 s, the supernatant was removed, and the beads were washed thrice with high-salt buffer (50 mm Tris, pH 7.5, 300 mm KCl, 12 mm MgCl2, 1% Nonidet P-40, 1 mm DTT, 100 µg/ml cycloheximide). For Western blotting analysis, 4% of the beads and 2% of the input, and supernatant were placed in Laemmli buffer. Total RNA was processed by adding 375 µl of RLT buffer (Qiagen) to the remaining beads and following manufacturer's protocol using an RNeasy Micro kit (Qiagen). RNA quantity was assessed with the NanoDrop Lite spectrophotometer (Thermo Scientific), whereas RNA quality was assessed with the RNA 2100 bioanalyzer (Agilent).
Cell culture
Cultures of WT and Eif4ebp1;Eif4ebp2 double knockout (4EBP DKO) MEFs were obtained from Dr. N. Sonenberg (McGill University) and maintained in Dulbecco's modified Eagle's medium containing 25 mm glucose (11965-084; Thermo Fisher Scientific) and supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were cultured at 37 °C and 5% CO2. Human MIO-M1 Müller cells (obtained from the UCL Institute of Ophthalmology, London, United Kingdom) were maintained in Dulbecco's modified Eagle's medium containing 5.6 mm glucose (Thermo Fisher Scientific, catalog no. 11885-084) and supplemented with 10% FBS and 1% penicillin/streptomycin. Transfections were performed with Lipofectamine 2000 (Thermo Fisher Scientific). Plasmids for expression of HA-tagged 4E-BP1-F113A were obtained from Dr. J. Blenis (Weill Cornell Medical College). pYIC was a gift from Han Htun (Addgene plasmid 18673; RRID:Addgene_18673).
Bicistronic reporter assay
A dsDNA gBlock fragment (IDT) encoding the 5′-UTR of Cd40 (5′-TTTCCTGGGCGGGGCCAAGGCTGGGGCAGGGGAGTCAGCAGAGGCCTCGCTCGGGCGCCCAGTGGTCCTGCCGCCTGGTCTCACCTCGCTATG-3′) was cloned into pCR2.1TOPO using the TOPO TA cloning kit (Invitrogen) according to the manufacturer's instructions. pYIC and TOPO plasmids were digested with EcoRI-HF (New England BioLabs) and BamHI-HF (New England BioLabs). A 4700-base pair fragment and 100-base pair fragment were gel-purified from pYIC- and TOPO-digested plasmids, respectively. The fragments were ligated and cloned into XL-1 blue supercompetent cells (Agilent). The resulting construct was digested with BstX1 (New England BioLabs) to remove the ECMV IRES. The digested construct was gel-purified, ligated, and cloned into XL-1 blue supercompetent cells. The reporter was sequence-verified using the primer ATCACATGGTCCTGCTGG.
Statistical analysis
All statistical analysis was performed with GraphPad Prism (Version 8.2.0). A Student's t test was used to compare differences between groups. In Figs. 4E and 5, where appropriate, the data were analyzed with two-way ANOVA with Dunnett's post hoc analysis (Table S1). The data are expressed as means ± S.D. Significance was defined as p < 0.05 for all analyses.
Data availability
All data for this publication are included in the article and supporting information or are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Dr. Gerald Hart (University of Georgia) and the NHLBI, National Institutes of Health P01HL107153 Core C4 at Johns Hopkins University for providing thiamet G. We thank Dr. Nahum Sonenberg (McGill University) for providing Eif4ebp1;Eif4ebp2 double knockout mice and MEFs and Dr. John Blenis (Weill Cornell Medical College) for providing the 4E-BP1 expression plasmids. We thank Scot R. Kimball and Alistair J. Barber (Penn State College of Medicine) for critically evaluating the manuscript. Parts of this study were presented in abstract form at the 79th Scientific Sessions of the American Diabetes Association, San Francisco, California, USA, in June 2019.
This article contains supporting information.
Author contributions—S. K. D. and M. D. D. conceptualization; S. K. D., A. L. T., and M. D. D. data curation; S. K. D., A. L. T., and M. D. D. formal analysis; S. K. D., A. L. T., and WPM investigation; S. K. D. and A. L. T. methodology; S. K. D. writing-original draft; S. K. D., A. L. T., S. S., and M. D. D. writing-review and editing; A. L. T. resources; WPM and M. D. D. visualization; S. S. validation; M. D. D. supervision; M. D. D. funding acquisition; M. D. D. project administration.
Funding and additional information—This work was supported by American Diabetes Association Pathway to Stop Diabetes Grant 1-14-INI-04 and NEI, National Institutes of Health Grant R01 EY029702 (to M. D. D.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- CD40
- cluster of differentiation 40
- DR
- diabetic retinopathy
- TMG
- thiamet G
- STZ
- streptozotocin
- HA
- hemagglutinin
- GS
- glutamine synthetase
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- MEF
- mouse embryonic fibroblast
- ANOVA
- analysis of variance
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Rübsam A., Parikh S., and Fort P. E. (2018) Role of inflammation in diabetic retinopathy. Int. J. Mol. Sci. 19, 942 10.3390/ijms19040942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Semeraro F., Morescalchi F., Cancarini A., Russo A., Rezzola S., and Costagliola C. (2019) Diabetic retinopathy, a vascular and inflammatory disease: therapeutic implications. Diabetes Metab. 45, 517–527 10.1016/j.diabet.2019.04.002 [DOI] [PubMed] [Google Scholar]
- 3. Tang J., and Kern T. S. (2011) Inflammation in diabetic retinopathy. Prog. Retin. Eye Res. 30, 343–358 10.1016/j.preteyeres.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang M., and Wong W. T. (2014) Microglia-Muller cell interactions in the retina. Adv. Exp. Med. Biol. 801, 333–338 10.1007/978-1-4614-3209-8_42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M., Ma W., Zhao L., Fariss R. N., and Wong W. T. (2011) Adaptive Müller cell responses to microglial activation mediate neuroprotection and coordinate inflammation in the retina. J. Neuroinflammation 8, 173 10.1186/1742-2094-8-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Portillo J. A. C., Lopez Corcino Y., Miao Y., Tang J., Sheibani N., Kern T. S., Dubyak G. R., and Subauste C. S. (2017) CD40 in retinal Müller cells induces P2X7-dependent cytokine expression in macrophages/microglia in diabetic mice and development of early experimental diabetic retinopathy. Diabetes 66, 483–493 10.2337/db16-0051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Portillo J. A., Schwartz I., Zarini S., Bapputty R., Kern T. S., Gubitosi-Klug R. A., Murphy R. C., Subauste M. C., and Subauste C. S. (2014) Proinflammatory responses induced by CD40 in retinal endothelial and Müller cells are inhibited by blocking CD40-Traf2,3 or CD40-Traf6 signaling. Invest. Ophthalmol. Vis. Sci. 55, 8590–8597 10.1167/iovs.14-15340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Portillo J. A., Greene J. A., Okenka G., Miao Y., Sheibani N., Kern T. S., and Subauste C. S. (2014) CD40 promotes the development of early diabetic retinopathy in mice. Diabetologia 57, 2222–2231 10.1007/s00125-014-3321-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y., Xie M., Men L., and Du J. (2019) O-GlcNAcylation in immunity and inflammation: An intricate system (Review). Int. J. Mol. Med. 44, 363–374 10.3892/ijmm.2019.4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurel Z., Sieg K. M., Shallow K. D., Sorenson C. M., and Sheibani N. (2013) Retinal O-linked N-acetylglucosamine protein modifications: implications for postnatal retinal vascularization and the pathogenesis of diabetic retinopathy. Mol. Vis. 19, 1047–1059 [PMC free article] [PubMed] [Google Scholar]
- 11. Kim S. J., Yoo W. S., Choi M., Chung I., Yoo J. M., and Choi W. S. (2016) Increased O-GlcNAcylation of NF-κB enhances retinal ganglion cell death in streptozotocin-induced diabetic retinopathy. Curr. Eye Res. 41, 249–257 10.3109/02713683.2015.1006372 [DOI] [PubMed] [Google Scholar]
- 12. Swamy M., Pathak S., Grzes K. M., Damerow S., Sinclair L. V., van Aalten D. M., and Cantrell D. A. (2016) Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat. Immunol. 17, 712–720 10.1038/ni.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lund P. J., Elias J. E., and Davis M. M. (2016) Global analysis of O-GlcNAc glycoproteins in activated human T cells. J. Immunol. 197, 3086–3098 10.4049/jimmunol.1502031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu J. L., Chiang M. F., Hsu P. H., Tsai D. Y., Hung K. H., Wang Y. H., Angata T., and Lin K. I. (2017) O-GlcNAcylation is required for B cell homeostasis and antibody responses. Nat. Commun. 8, 1854 10.1038/s41467-017-01677-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kneass Z. T., and Marchase R. B. (2005) Protein O-GlcNAc modulates motility-associated signaling intermediates in neutrophils. J. Biol. Chem. 280, 14579–14585 10.1074/jbc.M414066200 [DOI] [PubMed] [Google Scholar]
- 16. Krick S., Helton E. S., Hutcheson S. B., Blumhof S., Garth J. M., Denson R. S., Zaharias R. S., Wickham H., and Barnes J. W. (2018) FGF23 induction of O-linked N-acetylglucosamine regulates IL-6 secretion in human bronchial epithelial cells. Front. Endocrinol. (Lausanne) 9, 708 10.3389/fendo.2018.00708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. James L. R., Tang D., Ingram A., Ly H., Thai K., Cai L., and Scholey J. W. (2002) Flux through the hexosamine pathway is a determinant of nuclear factor κB–dependent promoter activation. Diabetes 51, 1146–1156 10.2337/diabetes.51.4.1146 [DOI] [PubMed] [Google Scholar]
- 18. Dela Justina V., Goncalves J. S., de Freitas R. A., Fonseca A. D., Volpato G. T., Tostes R. C., Carneiro F. S., Lima V. V., and Giachini F. R. (2017) Increased O-linked N-acetylglucosamine modification of NF-κB and augmented cytokine production in the placentas from hyperglycemic rats. Inflammation 40, 1773–1781 10.1007/s10753-017-0620-7 [DOI] [PubMed] [Google Scholar]
- 19. Donovan K., Alekseev O., Qi X., Cho W., and Azizkhan-Clifford J. (2014) O-GlcNAc modification of transcription factor Sp1 mediates hyperglycemia-induced VEGF-A upregulation in retinal cells. Invest. Ophthalmol. Vis. Sci. 55, 7862–7873 10.1167/iovs.14-14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Qu Y., Niu T., Wang H., and Liu K. (2017) O-GlcNAc modification of Sp1 mediates hyperglycaemia-induced ICAM-1 up-regulation in endothelial cells. Biochem. Biophys. Res. Commun. 484, 79–84 10.1016/j.bbrc.2017.01.068 [DOI] [PubMed] [Google Scholar]
- 21. Mazumder B., Li X., and Barik S. (2010) Translation control: a multifaceted regulator of inflammatory response. J. Immunol. 184, 3311–3319 10.4049/jimmunol.0903778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nandi A., Sprung R., Barma D. K., Zhao Y., Kim S. C., Falck J. R., and Zhao Y. (2006) Global identification of O-GlcNAc–modified proteins. Anal. Chem. 78, 452–458 10.1021/ac051207j [DOI] [PubMed] [Google Scholar]
- 23. Teo C. F., Ingale S., Wolfert M. A., Elsayed G. A., Not L. G., Chatham J. C., Wells L., and Boons G. J. (2010) Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat. Chem. Biol. 6, 338–343 10.1038/nchembio.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., and Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol. Cell. Proteomics 1, 791–804 10.1074/mcp.m200048-mcp200 [DOI] [PubMed] [Google Scholar]
- 25. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., and Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 10.1038/nchembio881 [DOI] [PubMed] [Google Scholar]
- 26. Dennis M. D., Schrufer T. L., Bronson S. K., Kimball S. R., and Jefferson L. S. (2011) Hyperglycemia-induced O-GlcNAcylation and truncation of 4E-BP1 protein in liver of a mouse model of type 1 diabetes. J. Biol. Chem. 286, 34286–34297 10.1074/jbc.M111.259457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dierschke S. K., Miller W. P., Favate J. S., Shah P., Imamura Kawasawa Y., Salzberg A. C., Kimball S. R., Jefferson L. S., and Dennis M. D. (2019) O-GlcNAcylation alters the selection of mRNAs for translation and promotes 4E-BP1-dependent mitochondrial dysfunction in the retina. J. Biol. Chem. 294, 5508–5520 10.1074/jbc.RA119.007494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pestova T. V., Kolupaeva V. G., Lomakin I. B., Pilipenko E. V., Shatsky I. N., Agol V. I., and Hellen C. U. (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 7029–7036 10.1073/pnas.111145798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. López-Lastra M., Rivas A., and Barría M. I. (2005) Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 38, 121–146 [DOI] [PubMed] [Google Scholar]
- 30. Holcik M., Sonenberg N., and Korneluk R. G. (2000) Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16, 469–473 10.1016/S0168-9525(00)02106-5 [DOI] [PubMed] [Google Scholar]
- 31. Dennis M. D., Shenberger J. S., Stanley B. A., Kimball S. R., and Jefferson L. S. (2013) Hyperglycemia mediates a shift from cap-dependent to cap-independent translation via a 4E-BP1–dependent mechanism. Diabetes 62, 2204–2214 10.2337/db12-1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giraud S., Greco A., Brink M., Diaz J. J., and Delafontaine P. (2001) Translation initiation of the insulin-like growth factor I receptor mRNA is mediated by an internal ribosome entry site. J. Biol. Chem. 276, 5668–5675 10.1074/jbc.M005928200 [DOI] [PubMed] [Google Scholar]
- 33. Sanz E., Yang L., Su T., Morris D. R., McKnight G. S., and Amieux P. S. (2009) Cell-type–specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. U.S.A. 106, 13939–13944 10.1073/pnas.0907143106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roesch K., Jadhav A. P., Trimarchi J. M., Stadler M. B., Roska B., Sun B. B., and Cepko C. L. (2008) The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 509, 225–238 10.1002/cne.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weingarten-Gabbay S., Elias-Kirma S., Nir R., Gritsenko A. A., Stern-Ginossar N., Yakhini Z., Weinberger A., and Segal E. (2016) Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, aad4939 10.1126/science.aad4939 [DOI] [PubMed] [Google Scholar]
- 36. Schalm S. S., Fingar D. C., Sabatini D. M., and Blenis J. (2003) TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13, 797–806 10.1016/S0960-9822(03)00329-4 [DOI] [PubMed] [Google Scholar]
- 37. Portillo J. A., Van Grol J., Zheng L., Okenka G., Gentil K., Garland A., Carlson E. C., Kern T. S., and Subauste C. S. (2008) CD40 mediates retinal inflammation and neurovascular degeneration. J. Immunol. 181, 8719–8726 10.4049/jimmunol.181.12.8719 [DOI] [PubMed] [Google Scholar]
- 38. Schrufer T. L., Antonetti D. A., Sonenberg N., Kimball S. R., Gardner T. W., and Jefferson L. S. (2010) Ablation of 4E-BP1/2 prevents hyperglycemia-mediated induction of VEGF expression in the rodent retina and in Muller cells in culture. Diabetes 59, 2107–2116 10.2337/db10-0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dennis M. D., Kimball S. R., Fort P. E., and Jefferson L. S. (2015) Regulated in development and DNA damage 1 is necessary for hyperglycemia-induced vascular endothelial growth factor expression in the retina of diabetic rodents. J. Biol. Chem. 290, 3865–3874 10.1074/jbc.M114.623058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Miller W. P., Mihailescu M. L., Yang C., Barber A. J., Kimball S. R., Jefferson L. S., and Dennis M. D. (2016) The translational repressor 4E-BP1 contributes to diabetes-induced visual dysfunction. Invest. Ophthalmol. Vis. Sci. 57, 1327–1337 10.1167/iovs.15-18719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abu El-Asrar A. M., Desmet S., Meersschaert A., Dralands L., Missotten L., and Geboes K. (2001) Expression of the inducible isoform of nitric oxide synthase in the retinas of human subjects with diabetes mellitus. Am. J. Ophthalmol. 132, 551–556 10.1016/S0002-9394(01)01127-8 [DOI] [PubMed] [Google Scholar]
- 42. Du Y., Smith M. A., Miller C. M., and Kern T. S. (2002) Diabetes-induced nitrative stress in the retina, and correction by aminoguanidine. J. Neurochem. 80, 771–779 10.1046/j.0022-3042.2001.00737.x [DOI] [PubMed] [Google Scholar]
- 43. Zheng L., Du Y., Miller C., Gubitosi-Klug R. A., Kern T. S., Ball S., and Berkowitz B. A. (2007) Critical role of inducible nitric oxide synthase in degeneration of retinal capillaries in mice with streptozotocin-induced diabetes. Diabetologia 50, 1987–1996 10.1007/s00125-007-0734-9 [DOI] [PubMed] [Google Scholar]
- 44. Leal E. C., Manivannan A., Hosoya K., Terasaki T., Cunha-Vaz J., Ambrósio A. F., and Forrester J. V. (2007) Inducible nitric oxide synthase isoform is a key mediator of leukostasis and blood–retinal barrier breakdown in diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 48, 5257–5265 10.1167/iovs.07-0112 [DOI] [PubMed] [Google Scholar]
- 45. Cipollone F., Chiarelli F., Davì G., Ferri C., Desideri G., Fazia M., Iezzi A., Santilli F., Pini B., Cuccurullo C., Tumini S., Del Ponte A., Santucci A., Cuccurullo F., and Mezzetti A. (2005) Enhanced soluble CD40 ligand contributes to endothelial cell dysfunction in vitro and monocyte activation in patients with diabetes mellitus: effect of improved metabolic control. Diabetologia 48, 1216–1224 10.1007/s00125-005-1750-2 [DOI] [PubMed] [Google Scholar]
- 46. Lamine L. B., Turki A., Al-Khateeb G., Sellami N., Amor H. B., Sarray S., Jailani M., Ghorbel M., Mahjoub T., and Almawi W. Y. (2020) Elevation in circulating soluble CD40 ligand concentrations in type 2 diabetic retinopathy and association with its severity. Exp. Clin. Endocrinol. Diabetes 128, 319–324 10.1055/a-0647-6860 [DOI] [PubMed] [Google Scholar]
- 47. Boeri D., Maiello M., and Lorenzi M. (2001) Increased prevalence of microthromboses in retinal capillaries of diabetic individuals. Diabetes 50, 1432–1439 10.2337/diabetes.50.6.1432 [DOI] [PubMed] [Google Scholar]
- 48. Henn V., Steinbach S., Büchner K., Presek P., and Kroczek R. A. (2001) The inflammatory action of CD40 ligand (CD154) expressed on activated human platelets is temporally limited by coexpressed CD40. Blood 98, 1047–1054 10.1182/blood.v98.4.1047 [DOI] [PubMed] [Google Scholar]
- 49. Xu H., and Chen M. (2017) Diabetic retinopathy and dysregulated innate immunity. Vision Res. 139, 39–46 10.1016/j.visres.2017.04.013 [DOI] [PubMed] [Google Scholar]
- 50. Coughlin B. A., Feenstra D. J., and Mohr S. (2017) Müller cells and diabetic retinopathy. Vision Res. 139, 93–100 10.1016/j.visres.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Watanabe T., Mio Y., Hoshino F. B., Nagamatsu S., Hirosawa K., and Nakahara K. (1994) GLUT2 expression in the rat retina: localization at the apical ends of Müller cells. Brain Res. 655, 128–134 10.1016/0006-8993(94)91606-3 [DOI] [PubMed] [Google Scholar]
- 52. Dai W., Dierschke S. K., Toro A. L., and Dennis M. D. (2018) Consumption of a high fat diet promotes protein O-GlcNAcylation in mouse retina via NR4A1-dependent GFAT2 expression. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3568–3576 10.1016/j.bbadis.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lieth E., Barber A. J., Xu B., Dice C., Ratz M. J., Tanase D., and Strother J. M. (1998) Glial reactivity and impaired glutamate metabolism in short-term experimental diabetic retinopathy: Penn State Retina Research Group. Diabetes 47, 815–820 10.2337/diabetes.47.5.815 [DOI] [PubMed] [Google Scholar]
- 54. Barber A. J., Antonetti D. A., and Gardner T. W. (2000) Altered expression of retinal occludin and glial fibrillary acidic protein in experimental diabetes: The Penn State Retina Research Group. Invest. Ophthalmol. Vis. Sci. 41, 3561–3568 [PubMed] [Google Scholar]
- 55. Rungger-Brändle E., Dosso A. A., and Leuenberger P. M. (2000) Glial reactivity, an early feature of diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 41, 1971–1980 [PubMed] [Google Scholar]
- 56. Sundstrom J. M., Hernández C., Weber S. R., Zhao Y., Dunklebarger M., Tiberti N., Laremore T., Simó-Servat O., Garcia-Ramirez M., Barber A. J., Gardner T. W., and Simo R. (2018) Proteomic analysis of early diabetic retinopathy reveals mediators of neurodegenerative brain diseases. Invest. Ophthalmol. Vis. Sci. 59, 2264–2274 10.1167/iovs.17-23678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vujosevic S., Micera A., Bini S., Berton M., Esposito G., and Midena E. (2016) Proteome analysis of retinal glia cells-related inflammatory cytokines in the aqueous humour of diabetic patients. Acta Ophthalmol. 94, 56–64 10.1111/aos.12812 [DOI] [PubMed] [Google Scholar]
- 58. Vujosevic S., Micera A., Bini S., Berton M., Esposito G., and Midena E. (2015) Aqueous humor biomarkers of Müller cell activation in diabetic eyes. Invest. Ophthalmol. Vis. Sci. 56, 3913–3918 10.1167/iovs.15-16554 [DOI] [PubMed] [Google Scholar]
- 59. Yellin M. J., D'Agati V., Parkinson G., Han A. S., Szema A., Baum D., Estes D., Szabolcs M., and Chess L. (1997) Immunohistologic analysis of renal CD40 and CD40L expression in lupus nephritis and other glomerulonephritides. Arthritis Rheum. 40, 124–134 10.1002/art.1780400117 [DOI] [PubMed] [Google Scholar]
- 60. Benveniste E. N., Nguyen V. T., and Wesemann D. R. (2004) Molecular regulation of CD40 gene expression in macrophages and microglia. Brain Behav. Immun. 18, 7–12 10.1016/j.bbi.2003.09.001 [DOI] [PubMed] [Google Scholar]
- 61. Jacobson E. M., Concepcion E., Oashi T., and Tomer Y. (2005) A Graves' disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology 146, 2684–2691 10.1210/en.2004-1617 [DOI] [PubMed] [Google Scholar]
- 62. Kwok C. K., Marsico G., and Balasubramanian S. (2018) Detecting RNA G-quadruplexes (rG4s) in the transcriptome. Cold Spring Harb. Perspect. Biol. 10, a032284 10.1101/cshperspect.a032284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Pelletier J., and Sonenberg N. (1985) Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- 64. Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell Biol. 9, 5134–5142 10.1128/mcb.9.11.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Urbancic M., and Gardasevic Topcic I. (2019) Dexamethasone implant in the management of diabetic macular edema from clinician's perspective. Clin. Ophthalmol. 13, 829–840 10.2147/OPTH.S206769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dierschke S. K., Toro A. L., Barber A. J., Arnold A. C., and Dennis M. D. (2020) Angiotensin-(1-7) attenuates protein O-GlcNAcylation in the retina by EPAC/Rap1-dependent inhibition of O-GlcNAc transferase. Invest. Ophthalmol. Vis. Sci. 61, 24 10.1167/iovs.61.2.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miller W. P., Ravi S., Martin T. D., Kimball S. R., and Dennis M. D. (2017) Activation of the stress response kinase JNK (c-Jun N-terminal kinase) attenuates insulin action in retina through a p70S6K1-dependent mechanism. J. Biol. Chem. 292, 1591–1602 10.1074/jbc.M116.760868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kimball S. R., Jurasinski C. V., Lawrence J. C. Jr., and Jefferson L. S. (1997) Insulin stimulates protein synthesis in skeletal muscle by enhancing the association of eIF-4E and eIF-4G. Am. J. Physiol. 272, C754–C759 10.1152/ajpcell.1997.272.2.C754 [DOI] [PubMed] [Google Scholar]
- 69. Kimball S. R., Horetsky R. L., and Jefferson L. S. (1998) Implication of eIF2B rather than eIF4E in the regulation of global protein synthesis by amino acids in L6 myoblasts. J. Biol. Chem. 273, 30945–30953 10.1074/jbc.273.47.30945 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for this publication are included in the article and supporting information or are available from the corresponding author upon request.