Abstract
The Src homology 2 (SH2) domain has a highly conserved architecture that recognizes linear phosphotyrosine motifs and is present in a wide range of signaling pathways across different evolutionary taxa. A hallmark of SH2 domains is the arginine residue in the conserved FLVR motif that forms a direct salt bridge with bound phosphotyrosine. Here, we solve the X-ray crystal structures of the C-terminal SH2 domain of p120RasGAP (RASA1) in its apo and peptide-bound form. We find that the arginine residue in the FLVR motif does not directly contact pTyr1087 of a bound phosphopeptide derived from p190RhoGAP; rather, it makes an intramolecular salt bridge to an aspartic acid. Unexpectedly, coordination of phosphotyrosine is achieved by a modified binding pocket that appears early in evolution. Using isothermal titration calorimetry, we find that substitution of the FLVR arginine R377A does not cause a significant loss of phosphopeptide binding, but rather a tandem substitution of R398A (SH2 position βD4) and K400A (SH2 position βD6) is required to disrupt the binding. These results indicate a hitherto unrecognized diversity in SH2 domain interactions with phosphotyrosine and classify the C-terminal SH2 domain of p120RasGAP as “FLVR-unique.”
Keywords: Src homology 2 domain (SH2 domain), X-ray crystallography, protein structure, phosphotyrosine, protein–protein interaction, GTPase-activating protein (GAP), RASA1, p120RasGAP, RasGAP, FLVR motif
Src homology 2 (SH2) domains are the best studied pTyr-binding protein scaffolding domains (1). They were originally identified within oncogenes of avian RNA viruses and are the second of four homology domains in Src family kinases (2–4) critical for both autoregulation and targeting (3, 5, 6). Following identification, SH2 domains rapidly revealed their presence in many kinases, phosphatases, small GTPase regulators, adaptor proteins, and transcription factors (1, 7, 8), and they are now known to integrate signals from the 90 tyrosine kinases to help regulate myriad signaling pathways and cellular processes (1, 7–9). In total, over 120 SH2 domains have been identified in the human genome distributed among ∼110 proteins (10, 11). Almost all of the members of this broad family retain a primary functional role: to bind short linear phosphotyrosine motifs (12–14). The structural conformation that allows this binding is extremely well-conserved across the family.
The SH2 domain comprises a central antiparallel seven-stranded β-sheet (βA to βG) sandwiched between two α-helices (αA and αB) (15–18) (nomenclature defined by Eck et al. (16)). This creates two binding sites—a deep pocket and a shallow cleft—within the ∼100-amino acid globular domain. Linear phosphotyrosine peptides bind perpendicular to the β-sheet in a two-pronged interaction with the pocket and cleft, a mode of binding that is reliably mirrored in cells and in the purified setting (4, 14). The deep pocket binds the pTyr residue, which is captured by electrostatic interactions with an arginine residue invariant in all pTyr-binding SH2 domains (9). In contrast, the shallow cleft binds residues C-terminal to the pTyr and varies considerably between SH2 domains allowing specificity determination (8, 13, 19, 20). This two-pronged interaction also dictates a consistent orientation of the bound phosphopeptide N to C terminus with respect to the SH2 domain (18). In v-Src, the deep pocket arginine residue is part of the Phe172-Leu173-Val174-Arg175 sequence, or “FLVR motif” (15), and arginine at this position (notated as the fifth residue on strand βB, βB5) is conserved in 117 of 120 human SH2 domains (9). Multiple lines of evidence indicate that the FLVR arginine is required for SH2–pTyr binding. To date, all structural studies of SH2 domain interactions with pTyr have demonstrated direct interactions between the FLVR arginine and pTyr. Further, the binding affinity of unphosphorylated peptides is on the order of 1000-fold weaker than phosphopeptides (15, 18). Additionally, the FLVR arginine was found to be the primary contributor of binding free energy (21, 22). Consequently, mutations of this residue are invariably used to generate a “dead” SH2 domain (23, 24).
p120RasGAP (RASA1, RasGAP, Ras GTPase-activating protein 1) was one of the first SH2 domain proteins to be identified (25–27) and was the first GTPase-activating protein (GAP) to be discovered (25, 28–30). It is ubiquitously expressed and required for life, because animals without p120RasGAP have major vascular defects (31, 32). p120RasGAP contains two SH2 domains at its N terminus that sandwich an SH3 domain, followed by PH and C2 domains for membrane recruitment and a RasGAP domain (Fig. 1A) (30, 33, 34, 84). The SH2 domains are used to directly interact with multiple phosphorylated binding partners including the p190RhoGAPs, the Eph receptor tyrosine kinases, and the Dok scaffolding proteins (32). These interactions are correlated with altered RasGAP activity and consequently with regulation of Ras signaling (35–39). The p120RasGAP SH2 domains therefore play critical roles in spatial–temporal regulation of Ras signaling by their interactions with this wide array of partners.
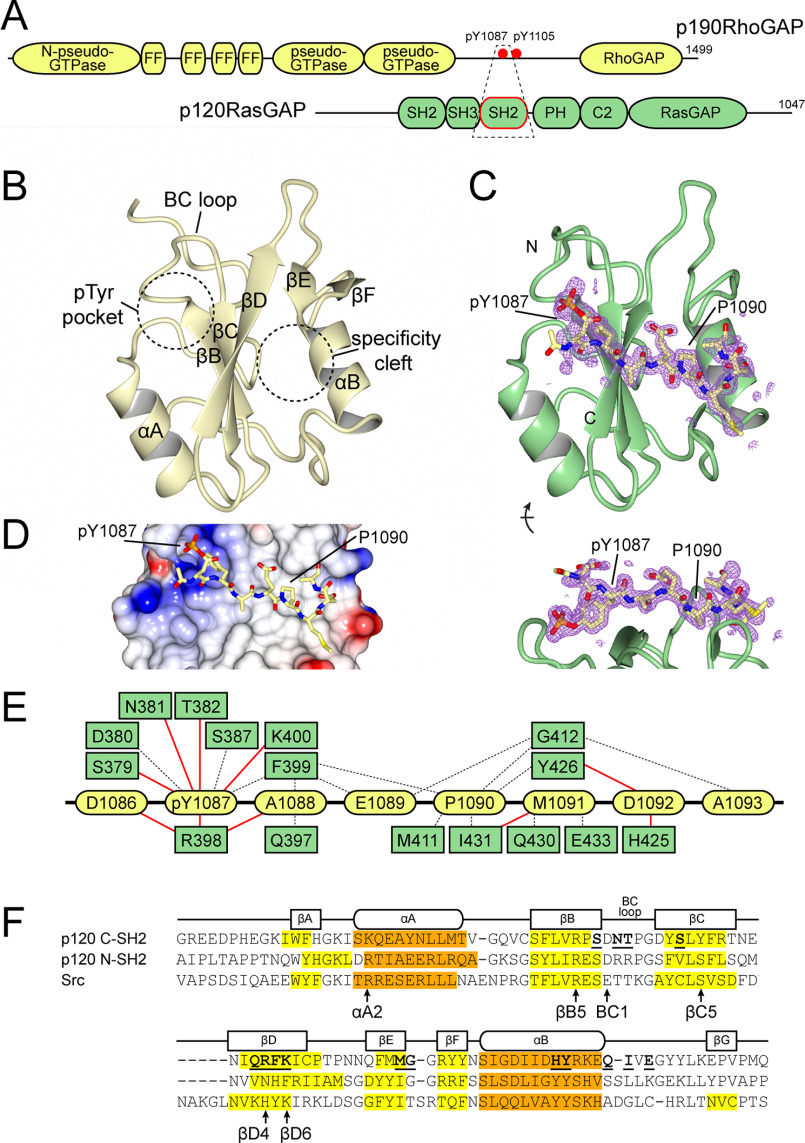
Figure 1.
Structure of p120RasGAP C-terminal SH2 domain. A, schematic of p120RasGAP interaction with p190RhoGAP. Domains are depicted, and the region co-crystallized is shown in a dashed box. B, structure of apo p120RasGAP C-SH2 indicating secondary structure elements and locations of the pTyr-binding pocket and specificity cleft. C, structure of p120RasGAP C-SH2 in complex with a synthesized phosphopeptide corresponding to p190RhoGAP-A residues 1086DpYAEPMDA1093. Simulated annealing omit Fobs − Fcalc difference map contoured at 3σ RMSD is shown. D, surface electrostatics for p120RasGAP C-SH2 with p190RhoGAP-A peptide. E, interactions observed between p120RasGAP C-SH2 (green) and p190 phosphopeptide (yellow) defined by PDBsum (84). Hydrogen bonds are shown by red lines; nonbonded elements are shown by dashed lines. F, alignment of secondary structure features for p120RasGAP C-SH2, compared with p120RasGAP N-SH2 (PDB code 6PXC) (39), and Src (PDB code 1SPS) (18). β-Strands are highlighted yellow, and α-helices are highlighted orange. Residues that contact p190 phosphopeptide as defined by PDBsum are indicated with bold and underlined text. Residues discussed in the text are indicated.
The SH2 domains of p120RasGAP are thought to be canonical SH2 domains that mediate direct interactions with pTyr residues using their FLVR arginine residue, which is conserved in both domains. We recently confirmed this to be the case for the N-terminal SH2 domain (40); however, in contrast we now demonstrate that the C-terminal SH2 domain of p120RasGAP is a novel SH2 domain. Using X-ray crystallography, we find that the βB5 FLVR arginine does not contact phosphotyrosine pTyr1087 of a bound p190RhoGAP peptide; instead, it makes a salt bridge to residue Asp380. Analysis of 715 aligned SH2 domain structures shows that this position of the FLVR arginine is rare, and point mutagenesis demonstrates that the FLVR arginine is not required for pTyr binding. Instead, pTyr binding is mediated by an alternate array of residues, including an unusual Arg398 at the βD4 position. These findings demonstrate that the C-terminal SH2 domain of p120RasGAP is “FLVR-unique.”
Results
p120RasGAP interaction with p190RhoGAP is critical for recruitment of RhoGAP activity to sites of cell adhesion and for suppression of RasGAP activity at these locations (35, 36, 41–43). Contacts between p120RasGAP and p190RhoGAP are mediated by the two SH2 domains of p120RasGAP (termed N-SH2 and C-SH2), which bind to phosphorylated tyrosine residues Tyr1087 and Tyr1105 of p190RhoGAP (44, 45); both pTyr residues reside in the preferred pYXXP sequence recognized by both SH2 domains of p120RasGAP (13, 19). We previously reported the crystal structure of N-SH2 of p120RasGAP and investigated its interaction with a p190RhoGAP pTyr1105 phosphopeptide, which revealed a canonical SH2 domain structure and pTyr peptide-binding mode with Kd of 0.3 ± 0.1 μm (40). In the present study, we aim to determine the structure and pTyr-binding mode of C-SH2 of p120RasGAP. Toward this end, we expressed and purified the isolated C-SH2 domain recombinantly and performed crystallization studies. We obtained crystals of the apo form of p120RasGAP C-SH2 and solved the structure to 1.5 Å resolution. To examine the structural basis of pTyr binding, we also solved a 1.5 Å co-crystal structure of p120RasGAP C-SH2 with a pTyr1087 p190RhoGAP phosphopeptide.
The structure of the apo form of C-SH2 (Fig. 1B and Table 1) reveals a typical overall SH2 fold that is most similar to the p120RasGAP N-SH2 (PDB code 6PXC) (40) with root-mean-squared deviation (RMSD) of ∼1.3 Å over 94 Cα atoms and 31% sequence identity; it is also highly similar to the C-terminal SH2 domain of PLCγ-1 (46) (PDB code 5TQ1; RMSD, ∼1.8 Å over 101 Cα atoms, 27% identity) and Nck2 (47) (PDB code 2CIA; RMSD, 1.1 Å over 93 Cα atoms, 25%). The two-pronged pTyr pocket and specificity cleft interaction sites (16, 18, 48) are clearly visible in p120RasGAP C-SH2 (Fig. 1B), with a strongly electropositive deep pocket and a hydrophobic shallow cleft both available to interact with a linear peptide. We then determined the 1.5 Å co-crystal structure of C-SH2 in complex with a synthetic phosphopeptide corresponding to p190RhoGAP residues 1086–1092 (1086DpYAEPMD1092) (Fig. 1C and Table 1). The structure of C-SH2 in complex with phosphopeptide is experimentally similar to the apo form (RMSDs 0.9 Å over 105 Cα atoms and 1.0 Å over 102 Cα atoms for chains A and B, respectively). As expected, the N- to C-terminal orientation of the bound phosphopeptide is consistent with other SH2 domain structures (18) and is dictated by placing pTyr1087 into the deep electrostatic pocket and Pro1090 into the shallow cleft, recapitulating canonical electrostatic and van der Waals interactions (Fig. 1D). The residues of p120RasGAP C-SH2 that contact the phosphopeptide are generally similar to previously studied SH2–phosphopeptide interactions (16, 18, 48) (Fig. 1, E and F). Curiously, however, we observed that the FLVR motif arginine residue, Arg377, is oriented differently to other SH2 domains in both the apo and peptide-bound crystal forms. We thus investigated this major structural difference in detail.
Table 1.
Data collection and refinement statistics
| p120RasGAP C-SH2 | p120RasGAP C-SH2 bound to p190RhoGAP phosphopeptide | |
|---|---|---|
| Data collection | ||
| PDB accession code | 6WAX | 6WAY |
| Wavelength (Å) | 0.97920 | 0.97920 |
| Resolution range (Å) | 50–1.50 (1.55–1.50) | 30–1.50 (1.55–1.50) |
| Space group | P 21 21 21 | C 2 2 21 |
| Cell dimensions | ||
| a, b, c (Å) | 52.5, 65.6, 71.6 | 63.1, 83.9, 54.2 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Unique reflections | 40,194 | 23,209 |
| Multiplicity | 11.7 (7.0) | 21.2 (8.6) |
| Completeness (%) | 100 (99.9) | 100 (99.8) |
| Mean I/σI | 24.1 (2.0) | 23.6 (2.0) |
| Wilson B factor (Å2) | 22.6 | 18.7 |
| Rpim (%) | 2.8 (40.7) | 3.1 (29.3) |
| CC½ | 0.993 (0.689) | 0.995 (0.806) |
| CC* | 0.998 (0.903) | 0.999 (0.945) |
| Refinement | ||
| Resolution range (Å) | 48.4–1.50 (1.54–1.50) | 27.3–1.50 (1.57–1.50) |
| Reflections used in refinement | 40,121 (2590) | 23,164 (2606) |
| Reflections used for Rfree | 1943 (126) | 1148 (130) |
| Reflections used for Rfree (%) | 4.84 | 4.96 |
| Rwork (%) | 17.9 (28.5) | 16.5 (24.8) |
| Rfree (%) | 19.7 (29.7) | 19.0 (27.8) |
| No. of non-hydrogen atoms | 2077 | 1188 |
| SH2 domains | 1800 | 921 |
| Peptide | 70 | |
| Water | 243 | 197 |
| Other solvent | 34 | |
| No. protein residues | 207 (105 chain A, 102 chain B) | 116 (106 SH2, 10 peptide) |
| Residue | ||
| p120RasGAP SH2 | 340–444, 343–444 | 339–444 |
| p190RhoGAP phosphopeptide | 1085–1094 | |
| RMSD | ||
| Bond lengths (Å) | 0.006 | 0.005 |
| Bond angles (°) | 0.872 | 0.809 |
| Ramachandran plot (%) | ||
| Favored | 98.0 | 98.1 |
| Allowed | 2.0 | 1.9 |
| Outliers | 0 | 0 |
| Rotamer outliers (%) | 1.0 | 0 |
| MolProbity clashscore | 5.27 | 2.08 |
| Average B factor (Å2) | 32.6 | 27.5 |
| SH2 domains | 30.9 | 24.4 |
| Copies A, B | 29.6, 32.3 | |
| Peptide | 34.9 | |
| Water | 42.3 | 39.3 |
In pTyr-binding SH2 domains, the conserved FLVR arginine at βB5 is required to form the base of the electropositive pocket and to hydrogen bond to pTyr (21, 22). Unexpectedly, in the p120RasGAP C-SH2/phosphopeptide co-crystal structure, the FLVR arginine Arg377 does not contact pTyr directly. Instead, Arg377 makes a salt bridge to Asp380 (at position BC1) (Fig. 2A), an interaction that is also observed in the apo C-SH2 structure (Fig. 2B). Therefore, unlike typical SH2 domains, C-SH2 Arg377 does not orient toward the phosphotyrosine-binding site in the absence of pTyr, nor does it undergo conformational change to contact pTyr directly in the presence of phosphopeptide. This unique orientation contrasts with p120RasGAP’s N-SH2 domain, which binds phosphotyrosine in a canonical fashion (Fig. 2C) (40).
Figure 2.
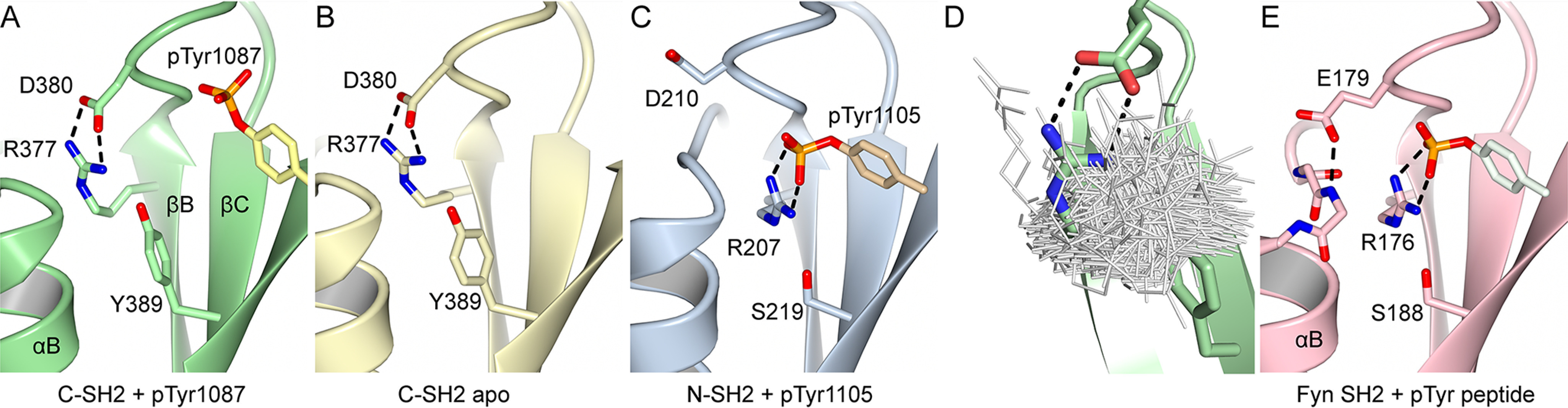
An unusual conformation for the FLVR arginine of p120RasGAP C-SH2. A, crystal structure of phosphopeptide-bound p120RasGAP C-SH2 showing the FLVR arginine Arg377, Asp380, and Tyr389 and p190RhoGAP pTyr1087. B, crystal structure of apo p120RasGAP C-SH2. C, crystal structure of phosphopeptide-bound p120RasGAP N-SH2 (PDB code 2PXC) (39) showing the equivalent residues and phosphotyrosine. D, superposition of FLVR motif arginines from 592 Dali-aligned SH2 domains. E, crystal structure of Src family kinase Fyn (PDB code 4U1P) shows residue BC1 (Glu53) caps helix αB.
To assess whether the salt-bridge engagement of FLVR Arg377 with Asp380 in p120RasGAP C-SH2 is unique among determined SH2 domains structures, we conducted in silico analysis. We find that such an orientation is rare among the 715 aligned SH2 domains identified by the DALI server (49). At the time of this analysis, there are only two other examples of FLVR arginine residues that form interactions with another residues within the SH2 domain itself: 1) in the STAT6 SH2 domain, the FLVR arginine makes salt-bridge contacts to both pTyr and Asp at position BC1 (50); and 2) in the VAV SH2 domain when bound to an unphosphorylated tyrosine, the FLVR arginine hydrogen bonds to Gln at position BC1 (51). The atypical orientation of Arg377 can be observed by superposition of aligned SH2 domains, in which the Arg377 side chain is pointed away from the pTyr-binding site, unlike the positions of a majority of the typical arginine side chains (Fig. 2D). The FLVR Arg of p120RasGAP C-SH2 is therefore unique among structures of SH2 domains.
The molecular basis of this unique FLVR arginine orientation seems to be dictated by both the salt bridge to Asp380 and by steric interference by Tyr389 (residue βC5) (Fig. 2, B and C). Tyrosine at position βC5 is not observed in any other human SH2 domain and is usually a small amino acid: over the 120 human SH2 domains, residue βC5 is observed as Ser (63 of 120 times including N-SH2 of p120RasGAP), Thr (27 times), Ala (13 times), Cys (10 times), Gly (3 times), Val (2 times), Met (1 time), and Tyr (1 time, p120RasGAP C-SH2) (1). Small amino acids at βC5 allow the FLVR arginine to engage the pTyr directly as observed in p120RasGAP N-SH2 bound to a phosphopeptide (Fig. 2C). At position BC1 (equivalent to Asp380 in p120RasGAP C-SH2), an acidic residue is frequently observed (Asp, 6 times and Glu, 38 times), but in many SH2 domains it caps helix αB (e.g. Glu179 in Fyn SH2; Fig. 2E). In contrast, in p120RasGAP C-SH2 a helix capping interaction by BC1 is not possible because the FLVR arginine guanidino group prevents close arrangement of helix αB and strand βB. The position of the FLVR arginine therefore seems to accommodate atypical orientations of other elements of the p120RasGAP C-SH2 domain.
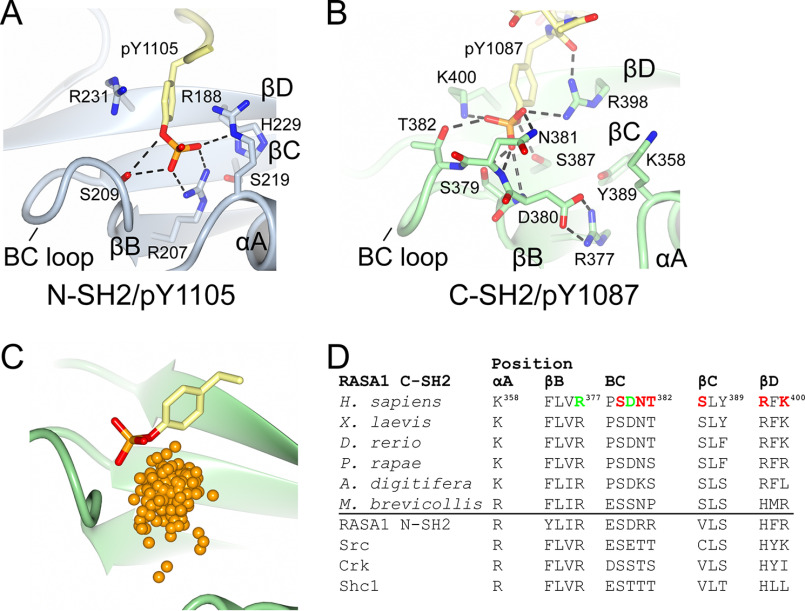
Upon inspection of the p120RasGAP C-SH2 structure in complex with phosphopeptide, we observe several additional unique features of pTyr binding in addition to the orientation of the FLVR arginine and presence of βC5 tyrosine. Across SH2 domains, three major interactions have been observed between the pTyr and its binding pocket: 1) the salt bridge between the pTyr and the FLVR motif βB5 arginine (Arg207 in N-SH2), 2) binding to a basic residue at either position αA2 (Arg188 in N-SH2) or βD6, and 3) binding to residues of the BC loop (between the βB and βC strands (Ser209 in N-SH2; Figs. 1F and 3A) (21, 40, 52). Consistent with these interactions, p120RasGAP C-SH2 uses βD6 (Lys400) and residues in the BC loop (Ser379, Asn381, and Thr382; Fig. 3B). However, in contrast, p120RasGAP C-SH2 uses additional residues βC3 (Ser387) and a basic residue at position βD4 (Arg398) to bind pTyr (Figs. 1, E and F, and 3B). Arg398 at position βD4 is unique to p120RasGAP C-SH2; this residue is most commonly a histidine (82 of 120 human SH2 domains; His229 in N-SH2; Fig. 3A) (1). The unusual binding site also manifests in the location of the phosphate—when phosphate atoms are mapped, C-SH2 binds to pTyr in a location divergent to all previously determined SH2 domains (Fig. 3C). The p120RasGAP C-SH2 pTyr-binding pocket therefore diverges from either the Src-like (where pTyr is coordinated by an Arg at position αA2) or SAP-like (where pTyr is coordinated by a basic residue at position βD6) pockets (52). Sequence alignment of human SH2 domains suggests that no other SH2 domain harbors dual basic residues at positions βD4 and βD6 (1), potentially indicating an evolution of the site to compensate for the loss of direct FLVR arginine binding. Additionally, the amino–aromatic interaction typically contributed by the arginine at αA2 (15,16) is instead fulfilled by the unique Arg398 at βD4 in C-SH2 (Fig. S1).
Figure 3.
Phosphotyrosine-binding sites of p120RasGAP. A and B, detailed binding site interactions of the phosphotyrosine to p120RasGAP N-SH2 (PDB code 2PXC) (39) (A) and p120RasGAP C-SH2 (B). C, comparison of phosphotyrosine bound to p120RasGAP C-SH2 with the location of the phosphate atom in 245 phosphotyrosine-bound SH2 domains identified by the Dali server. Locations of the phosphates are shown as orange spheres. SH2 domains identified and superposed using by the Dali server (246 contain phosphotyrosine; 2BBU not included in analysis). D, conservation of key binding site residues over evolution and in human SH2 domains. pTyr-binding residues are colored red, and the FLVR arginine–Asp380 salt bridges are colored green.
Many SH2–pTyr interactions have been studied in solution, and the measured binding affinities range in the 10−5 to 10−8 m (10 μm to 10 nm), or −7 to −11 kcal/mol range (53). We therefore used isothermal titration calorimetry (ITC) to measure the affinity of p120RasGAP C-SH2 with pTyr1087 phosphopeptide from p190RhoGAP and find the resultant Kd = 0.15 ± 0.04 μm, approximately −9 kcal/mol, to be within this expected range (Table 2 and Fig. S2). We next used ITC to assess the extent of binding of pTyr1087 phosphopeptide to C-SH2 mutants. For canonical SH2–pTyr interactions, the FLVR arginine is the primary contributor of binding free energy (21, 22), and mutation of this residue abrogates binding (23, 24). However, the unusual nature of the p120RasGAP C-SH2 suggests that the basis for its interaction with pTyr diverges from canonical SH2–pTyr interactions. We therefore tested whether Arg377 contributes to pTyr binding. Using ITC, we find that R377A mutation leads to slight reduction in affinity from WT Kd = 0.15 ± 0.04 μm to mutant Kd = 0.46 ± 0.11 μm but does not abrogate the interaction (Table 2 and Fig. S2). To our knowledge, this is the first SH2 domain studied to date that does not suffer a major loss of pTyr binding when the FLVR arginine residue is mutated. To further define the residues critical for pTyr binding, we introduced alanine point mutations at other basic residues that directly bind the pTyr: Arg398 at βD4 and Lys400 at βD6. Using ITC, we find that although the single mutants R398A and K400A only modestly impact pTyr peptide binding (Kd values of 0.31 ± 0.20 and 0.81 ± 0.15 μm, respectively), in combination, the double R398A/K400A mutant results in an ∼40-fold loss of binding (Kd of 6.24 ± 4.2 μm; Table 2 and Fig. S2). This indicates that unique among SH2 domains, the p120RasGAP C-SH2 requires multiple point mutations to abrogate pTyr binding.
Table 2.
Isothermal titration calorimetry for p120RasGAP C-SH2 with 190RhoGAP pTyr1087 peptide
Thermodynamic data for the pTyr1087 peptide–binding reactions with WT C-SH2, R377A, R398A, K400A, and R398A/K400A proteins. The errors are S.D. across multiple runs (individual runs are shown in Table S1).
| p120RasGAP C-SH2 protein | n | Kd | ΔH | ΔS | TΔS | ΔG |
|---|---|---|---|---|---|---|
| μm | kcal/mol | cal/mol*K | kcal/mol | kcal/mol | ||
| WT | 0.91 ± 0.08 | 0.16 ± 0.05 | −14.3 ± 1.6 | −16.7 ± 6.0 | −5.0 ± 1.7 | −9.3 ± 0.2 |
| R377A | 0.90 ± 0.08 | 0.46 ± 0.11 | −17.3 ± 1.5 | −29.1 ± 5.0 | −8.7 ± 1.3 | −8.7 ± 0.1 |
| R398A | 0.87 ± 0.08 | 0.19 ± 0.02 | −16.9 ± 1.3 | −26.0 ± 4.0 | −7.7 ± 1.2 | −9.2 ± 0.4 |
| K400A | 0.92 ± 0.14 | 0.81 ± 0.15 | −12.3 ± 3.0 | −13.4 ± 8.0 | −4.0 ± 2.0 | −8.3 ± 0.1 |
| R398A/K400A | 1.10 ± 0.09 | 6.24 ± 4.00 | −13.2 ± 4.0 | −20.1 ± 14.0 | −6.0 ± 4.0 | −7.2 ± 0.5 |
To assess the evolution of the p120RasGAP C-SH2 domain, we generated an alignment of 209 p120RasGAP sequences. We find that p120RasGAP is present in mammals, invertebrates, sponges, the choanoflagellates Monosiga brevicollis and Salpingoeca rosetta, the placozoa Trichoplax sp. H2, the orthonectid Intoshia linei, and in the single-celled eukaryote Capsapora owczarzaki (Figs. 1A and 3D and Fig. S3); notably, the domain architecture is preserved throughout phylogeny. We next examined the conservation of the unique phosphotyrosine-binding site. We observe that many of the phosphate-binding residues are well-conserved. However, we observe that a bulky residue at βC5 (Tyr389 in human), which sterically hinders canonical orientation of the FLVR arginine, which is retained in vertebrates and many invertebrates as Tyr or Phe, is a small residue (mostly Ser) in corals and more ancient species (Fig. S3). Similarly, the unique Arg398 at position βD4 is highly conserved in vertebrates and invertebrates but is a His in choanoflagellates and in C. owczarzaki. The evolutionary alterations in the unique pTyr-binding site therefore seem to indicate that p120RasGAP's C-terminal SH2 domain originally had a canonical binding site but that it evolved to its FLVR-unique conformation.
Discussion
Among protein domains the SH2 family is extraordinarily well-studied, making it unexpected for major divergence to have gone unidentified and even less expected that major divergence should be identified in one of the founding members. Nonetheless, we have shown that the C-terminal SH2 domain of p120RasGAP, the prototypical RasGAP and one of the earliest identified SH2 domains, is divergent from all studied SH2 domains. The lack of FLVR motif arginine contact with phosphotyrosine is unique among SH2 domains; in fact, even SH2 domains that can bind unphosphorylated partners do so in a FLVR arginine-dependent manner (e.g. SAP can bind Tyr as well as pTyr peptides) (8, 54). We therefore term the p120RasGAP C-SH2 a FLVR-unique SH2 domain.
These studies, using the isolated C-SH2 domain and single pTyr peptide, do not rule out the possibility that in the context of full-length proteins, potential domain–domain interactions within the p120RasGAP SH2–SH3–SH2 cassette (Fig. 1A) might alter the pTyr-binding site of C-SH2 to a more conventional FLVR-driven conformation. Alternatively, interdomain contacts might create an alternative unique mode of pTyr binding, similar to the tyrosine kinase ZAP70, which contains two SH2 domains separated by a short helical linker domain and binds two pTyr targets in the T-cell receptor. Specifically, the two ZAP70 SH2 domains come together to create an interface that forms one unique pTyr-binding site, whereas the other more typical pTyr-binding site is contained within a single SH2 domain (55). Future studies utilizing longer p120RasGAP constructs and dual pTyr-binding partners will help to examine these possibilities.
Classification of p120RasGAP C-SH2
Canonically, the FLVR arginine is required for interactions with pTyr peptides, contributing over 50% of their free energy of binding to the interaction (22, 56), and mutations of this residue result in an effectively “dead” SH2 domain. This residue is highly conserved, and there are only three human SH2 domains that do not contain the FLVR arginine residue (RIN has a His at this position, SH2D5 has a Trp, and Tyk2 has a Met) (9). For Tyk2, this belies the Janus kinase FERM-SH2 module mediating direct interaction with nonphosphorylated cytoplasmic tails of cytokine receptors (57, 58). The lack of diversity at the FLVR arginine position indicates conserved function, which was recently confirmed in bacteria in which a large number of SH2 domains were identified (52). Bacterial SH2 domains are classified in two groups, both of which are structurally divergent from eukaryotic SH2 domains, but even these divergent bacterial SH2 domains bind pTyr using the conserved FLVR-arginine (52). The C-terminal SH2 of p120RasGAP therefore seems to have evolved uniquely among the SH2 family of proteins.
The two-pronged pocket and cleft interaction site found in most SH2 domains (16, 18, 48) can be classified into different groups (groups IA to IE, IIA to IID, and III) that have divergent specificities, with the majority displaying a binding preference for the residue at the +2, +3, or +4 position C-terminal to the pTyr (13, 19) (although P + 1, P > 4, and regions N-terminal to the pTyr can also influence specificity (59–62)). The determinants of the specificity cleft are controlled primarily by two loops: one between the E and F β-strands (the EF loop) and one between α-helix B and the β-strand G (the BG loop). These can be thought of as analogous to the complementarity-determining regions, specificity-determining loops of antibodies (13, 19, 20). In this classification scheme, the p120RasGAP C-terminal SH2 domain is classified as a group IB SH2 domain, with specificity for hydrophobic residue at position +3 (13,19). Our crystal structures confirm the interaction site with these loops (Fig. 1).
Specificity of C-SH2 for phosphotyrosine
Two determinants of SH2 domain–binding specificity for phosphotyrosine over phosphoserine or phosphothreonine are 1) the depth of the binding pocket and 2) presence of amino–aromatic interactions (15, 16, 63–65). For p120RasGAP C-SH2 bound to a pTyr-containing peptide, we observe the typical deep, elongated pocket to accommodate the pTyr residue; the groove is lined by Arg398 and Lys400, which cradle the aromatic ring. This amino–aromatic interaction between Arg398 and the pTyr ring is similar to other typical SH2 domains such as Src and Lck (15, 16); however, Arg at the βD4 position rather than αA2 is unique to C-SH2. Taken together, these observations suggest that p120RasGAP has binding specificity for pTyr residues.
NMR solution structure of C-SH2
The NMR solution structure of p120RasGAP C-terminal SH2 domain, residues 340–446, was determined by the RIKEN Structural Genomics/Proteomics Initiative in 2006 and deposited in the Protein Data Bank (code 2GSB). The solution structure contains the 20 lowest energy models. Overall, the superposition with our crystal structures is close (RMSD 1.12 Å over 101 Cα atoms) (Fig. S4A), but in-solution conformational flexibility is observed in the EF loop, which defines +3 specificity (19), and in the BC loop, which harbors Asp380. None of the NMR structures exhibit the bidentate salt bridge between Arg377 and Asp380 (residue BC1), and both residues display extensive conformational variability, but their overall orientation is similar to our crystal structures (Fig. S4B). In contrast, Tyr389 is in a broadly similar position to our crystal structures and blocks orientation of the FLVR Arg toward the canonical pTyr-binding position. The unpublished NMR structure therefore supports our crystallographic data that p120RasGAP C-SH2 is a FLVR-unique SH2 domain.
Functional role of the FLVR-unique C-SH2
What then might the function of this highly unusual SH2 domain in p120RasGAP be? SH2 domains seem to have helped facilitate the evolutionary increase in complexity of signal transduction pathways and the transition from unicellular to multicellular organisms (as illustrated by a single SH2 domain residing in baker's yeast, Saccharomyces cerevisiae) (10). In humans there are ∼120 proteins that contain SH2 domains, but only 10 of these have dual SH2s. For p120RasGAP, the bivalent SH2 domains probably indicate that p120RasGAP has fast rebinding and long dwell times at pTyr-binding partners (66), and cooperation between the p120RasGAP domains is thought to impact signal transduction (7) with conformational changes proposed to occur on dual engagement of the SH2 domains with phosphotyrosine partners (44). However, the affinities of SH2 domain interactions with their functional phospho-partners is generally in the moderate 0.1–10 μm range (56, 67), allowing ready dissociation and thus preventing masking of the pTyr site and prevention of signal transduction (68, 69). For p120RasGAP, the combination of two tight SH2–pTyr interactions may result in extremely long dwell times that are deleterious for signal transduction. It may be that the altered pTyr-binding site in the C-terminal SH2 domain beneficially impacts p120RasGAP's signaling by tuning recruitment and retention at pTyr partners and thus prevents masking (68, 69) of these sites.
Broader implications
Disruption of SH2–pTyr interactions has theoretical clinical potential to regulate diverse signal transduction pathways, and consequently much work was conducted to achieve SH2 domain–specific inhibitors. Clinically useful selectivity has not, however, been achieved in large part because of bioavailability and the molecular similarities between SH2 domains (21). Selective picomolar binders can be designed where SH2 domains diverge as demonstrated for Grb2, which has an unusual specificity pocket that recognizes Asn at residue pY + 2 (70–72). Although it remains an outside possibility, nonetheless, it is interesting to speculate that the unique nature of p120RasGAP's C-terminal SH2 domain may represent a novel SH2-inhibitor target with the potential to beneficially alter both Ras and Rho signaling.
SH2 domains have played an important role in influencing our understanding of how cytosolic signaling occurs. Invariably these domains bind phosphotyrosine partners with their FLVR motif arginine residue acting as the linchpin of the interaction. It is striking, therefore, that one of the first identified SH2 domain proteins should diverge from this paradigm. The functional reasons for this divergence are currently unclear, but the array of bidentate binding partners for p120RasGAP and alterations in p120RasGAP's RasGAP activity on binding to phosphorylated partner proteins may hint at an evolutionary requirement to allow unmasking of the phosphotyrosine sites. The discovery of the unique nature of the C-terminal SH2 domain and how to alter its phosphotyrosine binding may therefore provide guidance to resolving how p120RasGAP functions mechanistically.
Materials and methods
Expression and purification p120RasGAP C-SH2 domain
cDNA encoding the C-terminal SH2 domain of human p120RasGAP (UniProt ID P20936; residues 340–444) was amplified by PCR and ligated into a modified pET vector, which includes an N-terminal His6 tag and TEV protease recognition site. Two native cysteine residues, Cys372 and Cys402, were mutated to serine by QuikChange mutagenesis (Agilent) to prevent formation of intermolecular disulfide bonds. The following point mutants were also generated by QuikChange mutagenesis: K377A, K398A, K400A, and K398A/K400A. Protein expression of WT and mutant C-SH2 was performed in Rosetta (DE3) cells, which were cultured in 1 liter of Luria broth at 37 °C to an A600 of 0.6–0.8 and then cooled to 18 °C, and protein expression was induced with 0.2 mm isopropyl β-d-thiogalactopyranoside. The cells were harvested by centrifugation, resuspended in lysis buffer (50 mm HEPES, pH 7.3, and 500 mm NaCl), and lysed by freeze-thaw cycles in the presence of lysozyme followed by sonication. Total cellular lysate was clarified by centrifugation at 5000 × g at 4 °C and applied to nickel–nitrilotriacetic acid–agarose resin (Qiagen) for 1 h at 4 °C to capture His6-tagged protein. Beads with bound protein were washed with 20 column volumes of wash buffer containing 50 mm HEPES, pH 7.3, 500 mm NaCl, 20 mm imidazole. The His6 tag was proteolytically removed from the C-SH2 proteins “on bead” overnight at 4 °C by the addition of TEV protease, which itself is also His6-tagged and captured by the nickel–nitrilotriacetic acid resin. The flow-through containing untagged C-SH2 protein was collected and applied to size-exclusion chromatography (Superdex 75; GE Healthcare) in buffer containing 20 mm Tris, pH 7.4, 150 mm NaCl. Finally, C-SH2 proteins were concentrated in a centrifugal filter with a molecular mass cutoff of 3,000 Da (Amicon Ultra, Millipore Sigma). 1 liter of Rosetta (DE3) cell culture resulted in a final purified protein yield of ∼20 mg for WT and up to 10 mg for mutant.
Peptide synthesis
A synthetic 7-amino acid peptide of sequence 1086DpYAEPMD1092 native to p190RhoGAP residues 1086–1092 (UniProt Q9NRY4) phosphorylated at Tyr1087 and a 15-amino acid peptide with sequence 1081GFDPSDpYAEPMDAVV1095 corresponding to residues 1081–1095 of p190RhoGAP (UniProt Q9NRY4) with N-terminal acetylation and C-terminal amidation were commercially synthesized (GenScript) and resuspended in sterile-filtered water.
Crystallization, data collection, structure determination, and refinement
p120RasGAP C-SH2 WT protein was used in crystallization trials at ∼14 mg/ml. Initial crystal screening was conducted with Index HT kit (Hampton Research) by a TTP Labtech Mosquito in sitting-drop vapor-diffusion trays at room temperature. Single crystals were obtained in Index HT position A3 containing 2.0 m ammonium sulfate, 0.1 m Bis-Tris, pH 6.5, with a 1:1 (v:v) protein:reservoir solution ratio. Optimization of this crystallization condition was achieved in hanging-drop vapor-diffusion plates, with a reservoir buffer containing 2.3 m ammonium sulfate, 0.1 m Bis-Tris, pH 6.5, and ratio of 1:1 (v:v) protein:reservoir solution. Single crystals with approximate dimensions 200 × 200 × 200 mm were harvested from the drop, cryopreserved in reservoir solution supplemented with 20% ethylene glycol, and flash-cooled in liquid nitrogen. X-ray data collection on a single crystal was performed at Northeastern Collaborative Access Team Beamline 24-ID-C at Argonne National Laboratory Advanced Photon Source. X-ray data were processed and scaled in HKL2000 (73) in space group P212121 with the unit cell dimensions a = 52.5 Å, b = 65.6 Å, c = 71.6 Å, α = β = γ = 90°, to 1.50 Å resolution. Matthews probability predicted two copies of SH2 in the asymmetric unit, and a molecular replacement solution confirmed the prediction with copies of C-SH2 obtained by Phenix Phaser (74) using the SH2 domain of SAP (PDB code 1D4T) (54) as a search model, yielding the translational function Z score of 12.7. Model building was performed in Phenix Autobuild (75), which successfully built 196 residues and correctly placed 192 in sequence (residues 344–442 of copy A and 350–442 of copy B). Manual model building was performed in Coot (76) and refinement in Phenix (77). Two sulfate ions (from the crystallization drop) and six ethylene glycol molecules (from the cryopreservative solution) were also modeled into density. Final refinement statistics are Rwork = 17.9% and Rfree = 19.7%.
For co-crystallization with the 7-amino acid p190RhoGAP pTyr1087 phosphopeptide, C-SH2 protein at 800 μm (10 mg/ml) was premixed with phosphopeptide concentrations ranging from 4 to 8 mm. Initial crystallization screens were performed with Index HT and SaltRx HT screens (Hampton Research) on a TTP Labtech Mosquito into sitting drops. Optimized co-crystals were obtained with reservoir solution containing 1.2 m sodium citrate tribasic dihydrate, 0.1 m Tris, pH 8.5. Single crystals were cryopreserved in reservoir buffer supplemented with 0.5 m sodium malonate, pH 7.0, before being flash-cooled in liquid nitrogen. X-ray data collection on a single crystal was performed at Northeastern Collaborative Access Team Beamline 24-ID-C at Argonne National Laboratory Advanced Photon Source. X-ray data were processed in HKL2000 (73) in space group C2221 with unit cell dimensions a = 63.1 Å, b = 83.9 Å, c = 54.2 Å, α = β = γ = 90° to 1.50 Å resolution and one copy per asymmetric unit. For structure solution, an all-alanine model derived from the NMR structure of p120RasGAP C-SH2 (PDB code 2GSB), truncated to residues 345–442 and B factors set to 20, was used as a search model for molecular replacement. Molecular replacement was performed by Phaser (74), which found a single solution for one copy of C-SH2 with a TFZ score of 10.6. Autobuilding was performed in Phenix (75), which removed model bias and successfully built and placed 100 residues (residues 342–442). Autobuilding also resulted in the building of five residues of the phosphopeptide, corresponding to p190RhoGAP residues 1087–1091. Manual model building was performed in Coot (76) and refinement in Phenix (77), yielding final Rwork = 16.5% and Rfree = 19.0%.
Isothermal titration calorimetry
Both C-SH2 WT and mutant proteins and pTyr1087 phosphopeptide (15 amino acids) were prepared for ITC by overnight dialysis in a common buffer with a 20 mm Tris, pH 7.4, 150 mm NaCl composition. Slide-A-Lyzer dialysis cassettes were used with a molecular mass cutoff of 3,500 Da were used for protein dialysis, and Micro Float-A-Lyzer dialysis devices with a 100–500-Da cutoff were used for peptide dialysis. The samples were retrieved from their cartridges and spun down for 10 min at 4 °C. The concentrations were measured by Nanodrop (Thermo Fisher). To determine peptide concentration, a phosphotyrosine extinction coefficient of 458.6 m−1 cm−1 at pH 7.4 was used (78). For ITC, a Nano-ITC (TA Instruments) was used. 350 μl of protein were injected into the sample cell. Both protein and peptide were degassed for 3 min prior to loading. The sample cell contents were stirred at 350 rpm at 25 °C to achieve continuous mixing. The results were analyzed using Nano-ITC Analyze software.
Evolutionary analysis
p120RasGAP sequences were identified using NCBI BLAST and searches of the FlyBase (79), WormBase (80), and MycoCosm databases (81). Sequences were aligned using the MAFFT (82) server and visualized using JalView (83).
Data availability
The coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 6WAX and 6WAY. X-ray diffraction images are available online at SBGrid Data Bank (83): doi: 10.15785/SBGRID/774 (6WAX) and doi: 10.15785/SBGRID/775 (6WAY).
Supplementary Material
Acknowledgments
We thank Mark Lemmon, Benjamin Turk, Byung Hak Ha, and Kimberly Vish for helpful comments and James Murphy for help with the ITC experiments. We also thank the staff at Northeastern Collaborative Access Team Beamline 24-ID-C at the Advanced Photon Source, Argonne National Laboratory.
This article contains supporting information.
Author contributions—R. J. C., A. L. S., and T. J. B. data curation; R. J. C., A. L. S., and T. J. B. formal analysis; R. J. C., J. W., and A. L. S. investigation; R. J. C., A. L. S., and T. J. B. visualization; R. J. C. methodology; R. J. C. and A. L. S. writing-original draft; R. J. C., J. W., A. L. S., and T. J. B. project administration; A. L. S. and T. J. B. supervision; A. L. S. and T. J. B. writing-review and editing; T. J. B. conceptualization; T. J. B. funding acquisition.
Funding and additional information—This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by National Institutes of Health Grant P41GM103403. This work used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. R. J. C. was supported in part by Mr. Morton and Mrs. Maggie Rosenfeld, the Rosenfeld Science Scholarship, and the Yale College Dean's Office. This work was also supported by National Institutes of Health Grants R01GM102262 and S10OD018007 and American Heart Association Grant 19IPLOI34740007 (to T. J. B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- SH2
- Src homology 2
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- GAP
- GTPase-activating protein
- RMSD
- root-mean-square deviation
References
- 1. Liu B. A., Jablonowski K., Raina M., Arcé M., Pawson T., and Nash P. D. (2006) The human and mouse complement of SH2 domain proteins: establishing the boundaries of phosphotyrosine signaling. Mol. Cell 22, 851–868 10.1016/j.molcel.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 2. Sadowski I., Stone J. C., and Pawson T. (1986) A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol. Cell Biol. 6, 4396–4408 10.1128/mcb.6.12.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine–SH2 domain interactions to complex cellular systems. Cell 116, 191–203 10.1016/S0092-8674(03)01077-8 [DOI] [PubMed] [Google Scholar]
- 4. Nash P. D. (2012) Why modules matter. FEBS Lett. 586, 2572–2574 10.1016/j.febslet.2012.04.049 [DOI] [PubMed] [Google Scholar]
- 5. Shah N. H., Amacher J. F., Nocka L. M., and Kuriyan J. (2018) The Src module: an ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Biol. 53, 535–563 10.1080/10409238.2018.1495173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boggon T. J., and Eck M. J. (2004) Structure and regulation of Src family kinases. Oncogene 23, 7918–7927 10.1038/sj.onc.1208081 [DOI] [PubMed] [Google Scholar]
- 7. Schlessinger J., and Lemmon M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12 10.1126/stke.2003.191.re12 [DOI] [PubMed] [Google Scholar]
- 8. Liu B. A., Engelmann B. W., and Nash P. D. (2012) The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. 586, 2597–2605 10.1016/j.febslet.2012.04.054 [DOI] [PubMed] [Google Scholar]
- 9. Kaneko T., Huang H., Cao X., Li X., Li C., Voss C., Sidhu S. S., and Li S. S. (2012) Superbinder SH2 domains act as antagonists of cell signaling. Sci. Signal. 5, ra68 10.1126/scisignal.2003021 [DOI] [PubMed] [Google Scholar]
- 10. Liu B. A., and Nash P. D. (2012) Evolution of SH2 domains and phosphotyrosine signalling networks. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2556–2573 10.1098/rstb.2012.0107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suga H., Torruella G., Burger G., Brown M. W., and Ruiz-Trillo I. (2014) Earliest holozoan expansion of phosphotyrosine signaling. Mol. Biol. Evol. 31, 517–528 10.1093/molbev/mst241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., and Lechleider R. J. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 10.1016/0092-8674(93)90404-E [DOI] [PubMed] [Google Scholar]
- 13. Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S., Li C., Roy P., Ho K., Songyang Z., Pawson T., Gao Y., and Li S. S. (2008) Defining the specificity space of the human SRC homology 2 domain. Mol. Cell. Proteomics 7, 768–784 10.1074/mcp.M700312-MCP200 [DOI] [PubMed] [Google Scholar]
- 14. Songyang Z., Shoelson S. E., McGlade J., Olivier P., Pawson T., Bustelo X. R., Barbacid M., Sabe H., Hanafusa H., and Yi T. (1994) Specific motifs recognized by the SH2 domains of Csk 3BP2, fps/fes, Grb-2, HCP, SHC, Syk and Vav. Mol. Cell Biol. 14, 2777–2785 10.1128/mcb.14.4.2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waksman G., Kominos D., Robertson S. C., Pant N., Baltimore D., Birge R. B., Cowburn D., Hanafusa H., Mayer B. J., Overduin M., Resh M. D., Rios C. B., Silverman L., and Kuriyan J. (1992) Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature 358, 646–653 10.1038/358646a0 [DOI] [PubMed] [Google Scholar]
- 16. Eck M. J., Shoelson S. E., and Harrison S. C. (1993) Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362, 87–91 10.1038/362087a0 [DOI] [PubMed] [Google Scholar]
- 17. Overduin M., Rios C. B., Mayer B. J., Baltimore D., and Cowburn D. (1992) Three-dimensional solution structure of the src homology 2 domain of c-abl. Cell 70, 697–704 10.1016/0092-8674(92)90437-H [DOI] [PubMed] [Google Scholar]
- 18. Waksman G., Shoelson S. E., Pant N., Cowburn D., and Kuriyan J. (1993) Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell 72, 779–790 10.1016/0092-8674(93)90405-F [DOI] [PubMed] [Google Scholar]
- 19. Kaneko T., Huang H., Zhao B., Li L., Liu H., Voss C. K., Wu C., Schiller M. R., and Li S. S. (2010) Loops govern SH2 domain specificity by controlling access to binding pockets. Sci. Signal. 3, ra34 10.1126/scisignal.2000796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H., Huang H., Voss C., Kaneko T., Qin W. T., Sidhu S., and Li S. S. (2019) Surface loops in a single SH2 domain are capable of encoding the spectrum of specificity of the SH2 family. Mol. Cell. Proteomics 18, 372–382 10.1074/mcp.RA118.001123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waksman G., Kumaran S., and Lubman O. (2004) SH2 domains: role, structure and implications for molecular medicine. Exp. Rev. Mol. Med. 6, 1–18 10.1017/S1462399404007331 [DOI] [PubMed] [Google Scholar]
- 22. Bradshaw J. M., Mitaxov V., and Waksman G. (1999) Investigation of phosphotyrosine recognition by the SH2 domain of the Src kinase. J. Mol. Biol. 293, 971–985 10.1006/jmbi.1999.3190 [DOI] [PubMed] [Google Scholar]
- 23. Bibbins K. B., Boeuf H., and Varmus H. E. (1993) Binding of the Src SH2 domain to phosphopeptides is determined by residues in both the SH2 domain and the phosphopeptides. Mol. Cell Biol. 13, 7278–7287 10.1128/mcb.13.12.7278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mayer B. J., Jackson P. K., Van Etten R. A., and Baltimore D. (1992) Point mutations in the abl SH2 domain coordinately impair phosphotyrosine binding in vitro and transforming activity in vivo. Mol. Cell Biol. 12, 609–618 10.1128/mcb.12.2.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trahey M., and McCormick F. (1987) A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238, 542–545 10.1126/science.2821624 [DOI] [PubMed] [Google Scholar]
- 26. Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., and Gibbs J. B. (1988) Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature 335, 90–93 10.1038/335090a0 [DOI] [PubMed] [Google Scholar]
- 27. Trahey M., Wong G., Halenbeck R., Rubinfeld B., Martin G. A., Ladner M., Long C. M., Crosier W. J., Watt K., and Koths K. (1988) Molecular cloning of two types of GAP complementary DNA from human placenta. Science 242, 1697–1700 10.1126/science.3201259 [DOI] [PubMed] [Google Scholar]
- 28. Bernards A., and Settleman J. (2004) GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14, 377–385 10.1016/j.tcb.2004.05.003 [DOI] [PubMed] [Google Scholar]
- 29. Bos J. L., Rehmann H., and Wittinghofer A. (2007) GEFs and GAPs: critical elements in the control of small G proteins. Cell 129, 865–877 10.1016/j.cell.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 30. McCormick F., Adari H., Trahey M., Halenbeck R., Koths K., Martin G. A., Crosier W. J., Watt K., Rubinfeld B., and Wong G. (1988) Interaction of ras p21 proteins with GTPase activating protein. Cold Spring Harb. Symp. Quant. Biol. 53, 849–854 10.1101/sqb.1988.053.01.097 [DOI] [PubMed] [Google Scholar]
- 31. Henkemeyer M., Rossi D. J., Holmyard D. P., Puri M. C., Mbamalu G., Harpal K., Shih T. S., Jacks T., and Pawson T. (1995) Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature 377, 695–701 10.1038/377695a0 [DOI] [PubMed] [Google Scholar]
- 32. Pamonsinlapatham P., Hadj-Slimane R., Lepelletier Y., Allain B., Toccafondi M., Garbay C., and Raynaud F. (2009) p120-Ras GTPase activating protein (RasGAP): a multi-interacting protein in downstream signaling. Biochimie (Paris) 91, 320–328 10.1016/j.biochi.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 33. Adari H., Lowy D. R., Willumsen B. M., Der C. J., and McCormick F. (1988) Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science 240, 518–521 10.1126/science.2833817 [DOI] [PubMed] [Google Scholar]
- 34. Laskowski R. A. (2001) PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 29, 221–222 10.1093/nar/29.1.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moran M. F., Polakis P., McCormick F., Pawson T., and Ellis C. (1991) Protein-tyrosine kinases regulate the phosphorylation, protein interactions, subcellular distribution, and activity of p21ras GTPase-activating protein. Mol. Cell Biol. 11, 1804–1812 10.1128/mcb.11.4.1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryant S. S., Briggs S., Smithgall T. E., Martin G. A., McCormick F., Chang J. H., Parsons S. J., and Jove R. (1995) Two SH2 domains of p120 Ras GTPase-activating protein bind synergistically to tyrosine phosphorylated p190 Rho GTPase-activating protein. J. Biol. Chem. 270, 17947–17952 10.1074/jbc.270.30.17947 [DOI] [PubMed] [Google Scholar]
- 37. Dail M., Richter M., Godement P., and Pasquale E. B. (2006) Eph receptors inactivate R-Ras through different mechanisms to achieve cell repulsion. J. Cell Sci. 119, 1244–1254 10.1242/jcs.02842 [DOI] [PubMed] [Google Scholar]
- 38. Kulkarni S. V., Gish G., van der Geer P., Henkemeyer M., and Pawson T. (2000) Role of p120 Ras-GAP in directed cell movement. J. Cell Biol. 149, 457–470 10.1083/jcb.149.2.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van der Geer P., Henkemeyer M., Jacks T., and Pawson T. (1997) Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap. Mol. Cell Biol. 17, 1840–1847 10.1128/mcb.17.4.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jaber Chehayeb R., Stiegler A. L., and Boggon T. J. (2019) Crystal structures of p120RasGAP N-terminal SH2 domain in its apo form and in complex with a p190RhoGAP phosphotyrosine peptide. PLoS One 14, e0226113 10.1371/journal.pone.0226113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang J. H., Gill S., Settleman J., and Parsons S. J. (1995) c-Src regulates the simultaneous rearrangement of actin cytoskeleton, p190RhoGAP, and p120RasGAP following epidermal growth factor stimulation. J. Cell Biol. 130, 355–368 10.1083/jcb.130.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sfakianos M. K., Eisman A., Gourley S. L., Bradley W. D., Scheetz A. J., Settleman J., Taylor J. R., Greer C. A., Williamson A., and Koleske A. J. (2007) Inhibition of Rho via Arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J. Neurosci. 27, 10982–10992 10.1523/JNEUROSCI.0793-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bradley W. D., Hernández S. E., Settleman J., and Koleske A. J. (2006) Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol. Biol. Cell 17, 4827–4836 10.1091/mbc.e06-02-0132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu K. Q., and Settleman J. (1997) Tandem SH2 binding sites mediate the RasGAP-RhoGAP interaction: a conformational mechanism for SH3 domain regulation. EMBO J. 16, 473–483 10.1093/emboj/16.3.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hernández S. E., Settleman J., and Koleske A. J. (2004) Adhesion-dependent regulation of p190RhoGAP in the developing brain by the Abl-related gene tyrosine kinase. Curr. Biol. 14, 691–696 10.1016/j.cub.2004.03.062 [DOI] [PubMed] [Google Scholar]
- 46. McKercher M. A., Guan X., Tan Z., and Wuttke D. S. (2017) Multimodal recognition of diverse peptides by the C-terminal SH2 domain of phospholipase C-γ1 protein. Biochemistry 56, 2225–2237 10.1021/acs.biochem.7b00023 [DOI] [PubMed] [Google Scholar]
- 47. Frese S., Schubert W. D., Findeis A. C., Marquardt T., Roske Y. S., Stradal T. E., and Heinz D. W. (2006) The phosphotyrosine peptide binding specificity of Nck1 and Nck2 Src homology 2 domains. J. Biol. Chem. 281, 18236–18245 10.1074/jbc.M512917200 [DOI] [PubMed] [Google Scholar]
- 48. Bradshaw J. M., Grucza R. A., Ladbury J. E., and Waksman G. (1998) Probing the “two-pronged plug two-holed socket” model for the mechanism of binding of the Src SH2 domain to phosphotyrosyl peptides: a thermodynamic study. Biochemistry 37, 9083–9090 10.1021/bi973147k [DOI] [PubMed] [Google Scholar]
- 49. Holm L., and Rosenstrom P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li J., Rodriguez J. P., Niu F., Pu M., Wang J., Hung L. W., Shao Q., Zhu Y., Ding W., Liu Y., Da Y., Yao Z., Yang J., Zhao Y., Wei G. H., et al. (2016) Structural basis for DNA recognition by STAT6. Proc. Natl. Acad. Sci. U.S.A. 113, 13015–13020 10.1073/pnas.1611228113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen C. H., Piraner D., Gorenstein N. M., Geahlen R. L., and Beth Post C. (2013) Differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain. Biopolymers 99, 897–907 10.1002/bip.22371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kaneko T., Stogios P. J., Ruan X., Voss C., Evdokimova E., Skarina T., Chung A., Liu X., Li L., Savchenko A., Ensminger A. W., and Li S. S. (2018) Identification and characterization of a large family of superbinding bacterial SH2 domains. Nat. Commun. 9, 4549 10.1038/s41467-018-06943-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ladbury J. E., Lemmon M. A., Zhou M., Green J., Botfield M. C., and Schlessinger J. (1995) Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc. Natl. Acad. Sci. U.S.A. 92, 3199–3203 10.1073/pnas.92.8.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poy F., Yaffe M. B., Sayos J., Saxena K., Morra M., Sumegi J., Cantley L. C., Terhorst C., and Eck M. J. (1999) Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol. Cell 4, 555–561 10.1016/S1097-2765(00)80206-3 [DOI] [PubMed] [Google Scholar]
- 55. Hatada M. H., Lu X., Laird E. R., Green J., Morgenstern J. P., Lou M., Marr C. S., Phillips T. B., Ram M. K., and Theriault K. (1995) Molecular basis for interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature 377, 32–38 10.1038/377032a0 [DOI] [PubMed] [Google Scholar]
- 56. Ladbury J. E., and Arold S. T. (2011) Energetics of Src homology domain interactions in receptor tyrosine kinase-mediated signaling. Methods Enzymol. 488, 147–183 10.1016/B978-0-12-381268-1.00007-0 [DOI] [PubMed] [Google Scholar]
- 57. Wallweber H. J., Tam C., Franke Y., Starovasnik M. A., and Lupardus P. J. (2014) Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nat. Struct. Mol. Biol. 21, 443–448 10.1038/nsmb.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McNally R., Toms A. V., and Eck M. J. (2016) Crystal structure of the FERM-SH2 module of human Jak2. PLoS One 11, e0156218 10.1371/journal.pone.0156218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hwang P. M., Li C., Morra M., Lillywhite J., Muhandiram D. R., Gertler F., Terhorst C., Kay L. E., Pawson T., Forman-Kay J. D., and Li S. C. (2002) A “three-pronged” binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome. EMBO J. 21, 314–323 10.1093/emboj/21.3.314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tinti M., Kiemer L., Costa S., Miller M. L., Sacco F., Olsen J. V., Carducci M., Paoluzi S., Langone F., Workman C. T., Blom N., Machida K., Thompson C. M., Schutkowski M., Brunak S., et al. (2013) The SH2 domain interaction landscape. Cell Rep. 3, 1293–1305 10.1016/j.celrep.2013.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Virdee S., Macmillan D., and Waksman G. (2010) Semisynthetic Src SH2 domains demonstrate altered phosphopeptide specificity induced by incorporation of unnatural lysine derivatives. Chem. Biol. 17, 274–284 10.1016/j.chembiol.2010.01.015 [DOI] [PubMed] [Google Scholar]
- 62. Zadjali F., Pike A. C., Vesterlund M., Sun J., Wu C., Li S. S., Rönnstrand L., Knapp S., Bullock A. N., and Flores-Morales A. (2011) Structural basis for c-KIT inhibition by the suppressor of cytokine signaling 6 (SOCS6) ubiquitin ligase. J. Biol. Chem. 286, 480–490 10.1074/jbc.M110.173526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mayer B. J. (1995) Why two heads are better. Structure 3, 977–980 10.1016/S0969-2126(01)00232-5 [DOI] [PubMed] [Google Scholar]
- 64. Eck M. J. (1995) A new flavor in phosphotyrosine recognition. Structure 3, 421–424 10.1016/S0969-2126(01)00173-3 [DOI] [PubMed] [Google Scholar]
- 65. Yaffe M. B. (2002) Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell. Biol. 3, 177–186 10.1038/nrm759 [DOI] [PubMed] [Google Scholar]
- 66. Oh D., Ogiue-Ikeda M., Jadwin J. A., Machida K., Mayer B. J., and Yu J. (2012) Fast rebinding increases dwell time of Src homology 2 (SH2)–containing proteins near the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 14024–14029 10.1073/pnas.1203397109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hause R. J. Jr., Leung K. K., Barkinge J. L., Ciaccio M. F., Chuu C. P., and Jones R. B. (2012) Comprehensive binary interaction mapping of SH2 domains via fluorescence polarization reveals novel functional diversification of ErbB receptors. PLoS One 7, e44471 10.1371/journal.pone.0044471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Haslam N. J., and Shields D. C. (2012) Peptide-binding domains: are limp handshakes safest? Sci. Signal. 5, pe40 10.1126/scisignal.2003372 [DOI] [PubMed] [Google Scholar]
- 69. Mayer B. J. (2012) Dynamics of receptor tyrosine kinase signaling complexes. FEBS Lett. 586, 2575–2579 10.1016/j.febslet.2012.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kraskouskaya D., Duodu E., Arpin C. C., and Gunning P. T. (2013) Progress towards the development of SH2 domain inhibitors. Chem. Soc. Rev. 42, 3337–3370 10.1039/c3cs35449k [DOI] [PubMed] [Google Scholar]
- 71. Morlacchi P., Robertson F. M., Klostergaard J., and McMurray J. S. (2014) Targeting SH2 domains in breast cancer. Fut. Med. Chem. 6, 1909–1926 10.4155/fmc.14.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shi Z. D., Lee K., Liu H., Zhang M., Roberts L. R., Worthy K. M., Fivash M. J., Fisher R. J., Yang D., and Burke T. R. Jr. (2003) A novel macrocyclic tetrapeptide mimetic that exhibits low-picomolar Grb2 SH2 domain-binding affinity. Biochem. Biophys. Res. Commun. 310, 378–383 10.1016/j.bbrc.2003.09.029 [DOI] [PubMed] [Google Scholar]
- 73. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 74. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., and Adams P. D. (2008) Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhang Z. Y., Maclean D., Thieme-Sefler A. M., Roeske R. W., and Dixon J. E. (1993) A continuous spectrophotometric and fluorimetric assay for protein tyrosine phosphatase using phosphotyrosine-containing peptides. Anal. Biochem. 211, 7–15 10.1006/abio.1993.1224 [DOI] [PubMed] [Google Scholar]
- 79. Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S., Marygold S. J., Matthews B. B., Millburn G., Antonazzo G., Trovisco V., Kaufman T. C., Calvi B. R., and FlyBase Consortium (2019) FlyBase 2.0: the next generation. Nucleic Acids Res. 47, D759–D765 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Harris T. W., Arnaboldi V., Cain S., Chan J., Chen W. J., Cho J., Davis P., Gao S., Grove C. A., Kishore R., Lee R. Y. N., Muller H. M., Nakamura C., Nuin P., Paulini M., et al. (2019) WormBase: a modern model organism information resource. Nucleic Acids Res. 48, D762–D767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Grigoriev I. V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., Smirnova T., Nordberg H., Dubchak I., and Shabalov I. (2014) MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–704 10.1093/nar/gkt1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Katoh K., and Standley D. M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Meyer P. A., Socias S., Key J., Ransey E., Tjon E. C., Buschiazzo A., Lei M., Botka C., Withrow J., Neau D., Rajashankar K., Anderson K. S., Baxter R. H., Blacklow S. C., Boggon T. J., et al. (2016) Data publication with the structural biology data grid supports live analysis. Nat. Commun. 7, 10882 10.1038/ncomms10882 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 6WAX and 6WAY. X-ray diffraction images are available online at SBGrid Data Bank (83): doi: 10.15785/SBGRID/774 (6WAX) and doi: 10.15785/SBGRID/775 (6WAY).