Abstract
Dominant mutations of ATP1A3, a neuronal Na,K-ATPase α subunit isoform, cause neurological disorders with an exceptionally wide range of severity. Several new mutations and their phenotypes are reported here (p.Asp366His, p.Asp742Tyr, p.Asp743His, p.Leu924Pro, and a VUS, p.Arg463Cys). Mutations associated with mild or severe phenotypes [rapid-onset dystonia-parkinsonism (RDP), alternating hemiplegia of childhood (AHC), or early infantile epileptic encephalopathy (EIEE)] were expressed in HEK-293 cells. Paradoxically, the severity of human symptoms did not correlate with whether there was enough residual activity to support cell survival. We hypothesized that distinct cellular consequences may result not only from pump inactivation but also from protein misfolding. Biosynthesis was investigated in four tetracycline-inducible isogenic cell lines representing different human phenotypes. Two cell biological complications were found. First, there was impaired trafficking of αβ complex to Golgi apparatus and plasma membrane, as well as changes in cell morphology, for two mutations that produced microcephaly or regions of brain atrophy in patients. Second, there was competition between exogenous mutant ATP1A3 (α3) and endogenous ATP1A1 (α1) so that their sum was constant. This predicts that in patients, the ratio of normal to mutant ATP1A3 proteins will vary when misfolding occurs. At the two extremes, the results suggest that a heterozygous mutation that only impairs Na,K-ATPase activity will produce relatively mild disease, while one that activates the unfolded protein response could produce severe disease and may result in death of neurons independently of ion pump inactivation.
Keywords: Dystonia, ataxia, epilepsy, neurodegeneration, mutation validation, phenotype-genotype relationship, cytopathology
Introduction
Since the discovery of ATP1A3 mutations in rapid-onset dystonia-parkinsonism, mutations of this gene have been found to produce a range of neurological problems (Rosewich et al., 2017; Carecchio et al., 2018). Alternating hemiplegia of childhood (AHC) emerges in infancy (0-18 months) (Heinzen et al., 2014; Rosewich et al., 2017). Early infantile epilepsy and encephalopathy (EIEE) with or without apnea is more severe (Paciorkowski et al., 2015). Milder ATP1A3 mutations manifest in childhood or adulthood, often after a trigger. Syndromes include rapid-onset dystonia-parkinsonism (RDP), cerebellar ataxia, areflexia, pes cavus, optic nerve atrophy, and sensorineural deafness (CAPOS), relapsing encephalopathy with cerebellar ataxia (RECA) and fever-induced paroxysmal weakness and encephalopathy (FIPWE) (de Carvalho Aguiar et al., 2004; Demos et al., 2014; Dard et al., 2015; Yano et al., 2017; Brashear et al., 2018). Most known patients fit into discrete syndromes, and some are on a phenotypic continuum. To date, all disease causing ATP1A3 mutations are heterozygous, and when tested usually had loss of function or altered kinetic properties (Holm et al., 2016).
In the CNS, ATP1A3 is expressed only in neurons, and has critical electrophysiological functions (McGrail et al., 1991; Dobretsov and Stimers, 2005; Benarroch, 2011). Experimental work has shown that Na,K-ATPase not only restores ion gradients after activity, but powers afterhyperpolarizations (Kim et al., 2007; Picton et al., 2017); impacts burst firing (Vaillend et al., 2002); and distributes to dendritic spines (Shiina et al., 2011; Blom et al., 2011). ATP1A3 appears to be highly expressed in inhibitory neurons, particularly fast-spiking interneurons (Richards et al., 2007; Anderson et al., 2010; Paciorkowski et al., 2015). In many but not all neurons it is co-expressed with ATP1A1 (Wetzel et al., 1999). An important functional feature is that α3 has lower intrinsic affinity for sodium than α1, and this allows α3 to respond to transient sodium elevation (Soga et al., 2001; Dobretsov and Stimers, 2005; Azarias et al., 2013).
The wide range of disease severity seen with ATP1A3 mutations is not yet understood. Many laboratory studies have shown the successful expression of protein with no detectable ATP hydrolysis or pump current, i.e. the pump is inactive. Other mutations of ATP1A3 and of ATP1A2 have detectable activity, and have been extensively studied in Xenopus oocytes, in Sf9 insect cells, and in mammalian cell lines (Friedrich et al., 2016; Holm et al., 2016; Clausen et al., 2017). Often a reduction of affinity for sodium was seen, which would have little effect on survival of mammalian cell lines when α1 is inhibited, but serious consequences for neuronal physiology when intracellular sodium is elevated (Azarias et al., 2013; Swarts et al., 2013; Toustrup-Jensen et al., 2014). Studies on neuron-differentiated iPSCs from ATP1A3 patients showed membrane depolarization (Simmons et al., 2018). Some mutations impair affinity for potassium, the capacity for conformation change, or voltage-dependent properties and proton leaks (Friedrich et al., 2016; Holm et al., 2016; Tranebjaerg et al., 2018). Dominant-negative effects have been documented (Li et al., 2015). Reductions in expression level have also been reported, albeit with some variability between laboratories (de Carvalho Aguiar et al., 2004; Heinzen et al., 2012; Holm et al., 2016).
Examination of the locations of mutations in crystal structures of Na,K-ATPase in the sodium-bound or potassium-bound conformations has shown clustering of ~70% of the AHC mutations close to the ion binding sites buried in the membrane domain, whereas few mild phenotypes are associated with mutations there (Sweadner et al., 2019). For most mutations, however, there was no simple correlation between location and phenotype. The aim here was to investigate mutations for their effects on biosynthesis, using isogenic cell lines to facilitate comparisons. Three converging sources of impairment or cytopathology contribute to the wide range of pathogenicity of ATP1A3 mutations.
Materials and methods
Patients
Written, informed consent was obtained from all patients, and research protocols were approved by the Institutional Review Boards of Wake Forest School of Medicine; La Pitié-Salpêtrière University Hospital; the Icahn School of Medicine at Mount Sinai; Children’s Hospital Colorado & Denver Health Medical Center, University of Colorado; and the Massachusetts General Hospital. This work did not entail use of human subjects apart from clinical evaluation and DNA testing; those procedures were in accordance with The Code of Ethics of the World Medical Association.
Mutagenesis, transient transfection, and the survival assay
pCMV6-XL5 plasmid containing ATP1A3 cDNA with two mutations conferring ouabain resistance (WT-OR) (de Carvalho Aguiar et al., 2004) was used as template for missense mutations identified in patients (Mutagenex, Inc., Hillsborough, NJ). Transient transfection was performed in HEK-293T, HEK-293, or Flp-In™ T-REX™ 293 cells (Life Technologies) as described (de Carvalho Aguiar et al., 2004; Paciorkowski et al., 2015). 24 h post-transfection, cells were treated with ouabain at concentrations ranging from just sufficient to kill the host cells that express only endogenous ATP1A1 (0.5 μM) to well - over saturating for ATP1A1 (3-10 μM). The expressed ATP1A3 was ouabain-resistant, and so cell survival depended on whether it had enough activity. Cell viability was monitored daily. The assays were performed at least 3 times.
Establishment of stable cell lines
Flp-In™ T-REX™ 293 cells (Life Technologies) were used to generate stable lines expressing mutations of ATP1A3, following manufacturer’s protocols. Induction of stably expressed mutant forms of ATP1A3 was with tetracycline (Sigma-Aldrich). The major advantage of the system is that the defined recombination site and isogenic background allows direct comparison of different mutants.
Rates of growth
Flp-In lines were seeded into poly-lysine coated 38-mm2 wells of 96-well flat-bottomed tissue culture plates (103 cells per well) in quadruplicate and allowed to adhere overnight. Cell proliferation was assayed over the following 2–4 days using WST-1 (Roche Diagnostics). Quantification was with a microtiter plate reader (Beckman Coulter DTX 880 Multimode Detector) at 450 nm. Data from daily measurements were normalized to the averaged 24 h readings for each individual cell line. Semi -log plots were generated with GraphPad Prism 7 software with data from at least 3 replicates.
Sample preparation for SDS gel analysis
Cell lysates of Flp-In lines were obtained with a RIPA buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na-EDTA, 1 mM Na-EGTA, 1% NP-40, 1% sodium deoxycholate, and protease inhibitor cocktail (Complete Mini, Roche Diagnostics, Mannheim, Germany). Lysed cells were scraped from plastic dishes, homogenized with a pellet pestle motor homogenizer (Kimble Chase), and centrifuged at 5,000 x g for 10 min at 4°C. The pellets were discarded, and the supernatants were assayed for protein using the Lowry method.
Western blots
Gel electrophoresis was in Nu-Page 4-12% gradient Bis-Tris NuPage gels (Life Technologies) followed by transfer to nitrocellulose. Mouse pan-specific antibody 9A7 (gift of Dr. Maureen McEnery, Case Western Reserve University, Cleveland, OH) was employed to detect the total amount of alpha subunit isoforms. Monoclonal antibodies 6F (Developmental Studies Hybridoma Bank) or M17-P5-F11 (gift of Dr. W. James Ball, University of Cincinnati) were used to detect human α1 and β2 subunits of Na,K-ATPase, respectively. α3 subunit of Na,K-ATPase was detected with the goat peptide-directed antibody C16 (Santa Cruz Biotechnology). Actin-HRP antibody was from Santa Cruz Biotechnology. Secondary antibodies were HRP-conjugated, and final detection was with chemiluminescence (WesternBright ECL, Advansta, CA). An LAS 4000 imager (GE Healthcare Life Sciences) with ImageQuant software was used for the analysis of Western blots. All analyses were performed with a minimum of 3 biological replicates, and actin was used as a loading control for all samples for quantification. When protein induction is large compared to the basal level, the accuracy of blot quantification is limited by the accuracy of background subtraction: three different ImageQuant automatic background subtraction methods were employed, and the results averaged for a best-estimate of expression differences.
Deglycosylation
PNGase F (Peptide:N-glycosidase F) digestion of N-glycans was performed on Flp-In cell lysate in RIPA buffer (above) with the kit from New England Biolabs following the manufacturer’s protocol. Incubation was for 1 h at 37°C. Western blots were stained with M17-P5-F11 antibody against β1.
Biotinylation detection of surface proteins
Biotinylation was performed largely as described (Vagin et al., 2008). Briefly, Flp-In lines were grown until confluent in 12-well plates. Before treatment, plates were put on ice, cells were washed twice with ice-cold Dulbecco’s PBS containing 1mM CaCl2and MgCl2, pH 7.4 (D-PBS), and then treated with either biotinylation reagent (EZ-Link-Sulfo-NHS-SS-Biotin, 1mg/ml in D-PBS) or D-PBS for 30 min at 4°C on ice. Quenching was with 50mM NH4Cl for 15 min on ice followed by two washes with ice-cold D-PBS. Lysis was with RIPA buffer (above) for 1 hour on ice. Cells were scraped, homogenized, and the suspension was centrifuged at 15,000 xg for 10 min at 4°C. The pellets were discarded, and the supernatants were incubated with rotation with 50 μl streptavidin-bound agarose beads (Vector Laboratories, Burlingame, CA) in buffer A containing 15 mM Tris-base, 0.5% Triton X-100, 4 mM EGTA, and 150 mM NaCl, pH 8.0. After 1 hour, the beads were washed with buffer A with low salt, 150 mM NaCl, and then high salt, 0.5M NaCl. Low ionic strength elution was performed with buffer A containing no NaCl. Western blots were stained with antibodies against β1.
Immunofluorescence
Flp-In lines were grown in 8-chamber glass slides (Nunc or CellTreat Scientific) until confluent, fixed with 4% PLP (paraformaldehyde/lysine/periodate), treated with 1% SDS in PBS for antigen retrieval, and blocked in 1% bovine serum albumin in PBS supplemented with 0.2% Triton X-100. The 6F, M17-P5-F11, and C16 antibodies for Na,K-ATPase mentioned above were also employed for immunofluorescence. Incubation with primary antibodies was overnight at 4°C. Detection was with Alexa Fluor-conjugated secondary antibodies (Life Technologies). Images were collected with a Nikon Eclipse E800 fluorescence microscope using Nikon NIS-Elements imaging software. Conventional microscopy was used rather than confocal because it captured more of the observed gross structural abnormalities in cells that tend to mound and present an irregular surface.
qPCR
Total RNA from 106 α3WT Flp-In cells with and without tet induction was isolated with the RNAeasy kit (Qiagen), and single strand DNA was synthesized with the iScript Advanced cDNA kit for RT-qPCR (Bio-Rad) in accordance with manufacturer protocols. The primers were the following: α1FWD GAACAGACTTGAGCCGGGGA; α1REV CCCCAAAGAGCTGCCGACAA; α3FWD TCACCCAAGAAGAACAAGGGCA; α3REV TCCGGCAGACCTCTTCCACT. qPCR analysis was performed on the Applied Biosystems StepOnePlus Real-Time PCR System. Actin mRNA was used as the internal control.
Results
New ATP1A3 mutations and clinical phenotypes
The new mutations reported here include two cases of RDP; one case with cerebellar atrophy that is ataxia-predominant but also has symptoms of RDP; one case of severe EIEE with apnea; and one case suspicious of late-onset RDP but with a variant that is typically not predicted to be disease causing because of its frequency in controls [i.e. a variant of unknown significance (VUS)].
p.Asp366His.
The patient was a male of European ancestry, 55 years old at examination, with a history typical of RDP (Haq et al., 2019). He reported onset of symptoms at 16 years of age triggered by running on his high school track team, where running became slower and more labored. Psychological stress was also present at onset. Speech and left arm and leg were affected with dystonia first. Symptoms gradually stabilized 1-6 months after onset. Speech, swallowing, left arm and leg, neck and both hands were affected at the time of interview, and UPDRS score for parkinsonian symptoms was 54. No second episodes or learning difficulty were reported. He ambulated independently, although he reported some falls. MRI was normal. Sanger sequencing showed ATP1A3 c.1096G>C, p.Asp366His (the transcript used is NM_152296, ENST00000302102). During ATP hydrolysis the terminal phosphate of ATP is transferred to this exposed aspartate in the active site, forming a labile high-energy phosphoester bond essential for ion transport. Experimental mutations of this site have been made in a number of labs, and Na,K-ATPase activity and ability to be phosphorylated during turnover have always been abolished, even with the conservative substitution of glutamate for aspartate (Ohtsubo et al., 1990; Liang et al., 2006). There is a Hailey-Hailey disease-related mutation of the corresponding aspartate in the SERCA Ca2+-ATPase ATP2C1, c.1049A>T, p.Asp350Val (Cheng et al., 2010).
p.Asp742Tyr.
The patient was a male of European ancestry, 45 years old at examination. Based on detailed parental notes, he had psychomotor delay, learning disabilities, and showed ataxic gait as soon as he started walking. He did not experience acute neurological episodes of any kind, and he was stable during childhood and adolescence with mildly disabling symptoms. In adulthood, his gait worsened due to a mild worsening of cerebellar functions, the onset of mild pyramidal signs, and mild dystonic postures that progressed over several years. He was able to walk without aid and work in a protective environment. On exam, ataxia, mild dystonic postures, bradykinesia and a masked face were observed, consistent with ataxia-RDP. MRI showed severe cerebellar atrophy (Supplement Figure S1), similar to a p.Gly316Ser ataxia-RDP patient previously described (Sweadner et al., 2016). Exome and Sanger sequencing showed de novo ATP1A3 c.2224G>T, p.Asp742Tyr. There is a recent report of an unrelated female patient with the same mutation (c.2224G>T, p.Asp742Tyr) but with onset of paroxysmal motor symptoms at 6 weeks of age, seizures, apnea, and severe developmental delay. MRI at age 16 in that case showed parietal temporal atrophy and a short corpus callosum (Marzin et al., 2018).
p.Asp743His/p.Ser729Tyr.
This 38 year old woman of European ancestry proved to have two de novo mutations in ATP1A3. Symptoms were characteristic of RDP with gradual progression. Age at onset was 22 years with neck stiffness that evolved to frank posturing. One to two months later she developed dysarthria, restriction in mouth opening, and deterioration of handwriting with micrographia. After 3 months, she started dragging her right foot. Her walking worsened 2 years after onset. Over the next 3-4 years she developed dystonic symptoms in both hands. She received botulinum toxin injection for blepharospasm, lower cranial dystonia, especially jaw closure, cervical dystonia, and bilateral distal arm dystonia. Sinemet and Artane were without benefit; there was some response to clonazepam. At last exam there was prominent cranial-cervical dystonia that was generalized to involve both legs, and only minimal parkinsonism. Sanger sequencing showed de novo c.2186C>A, p.Ser729Tyr and c.2227G>C, p.Asp743His.
Both variants are in the same exon of ATP1A3. To test for compound heterozygosity, the region of interest was recovered by PCR from patient genomic DNA and inserted in plasmids for transformation of E. coli. Eight isolated subclone colonies were picked and sequenced. Four had both variants and four had neither variant, demonstrating that both variants are on the same strand. Each variant was tested for function separately in the survival assay, as detailed below (Table 1), and the results indicated that p.Asp743His is pathogenic. The survival of cells transfected with p.Ser729Tyr is inconclusive, but it was not studied further because it is at the surface in the Na,K-ATPase structures.
Table 1.
Ability of ATP1A3 mutations to support cell survival in HEK-293.
| mutation | [ouab] μM |
survival/ growth |
phenotypes | age at onset |
recurrence | ref. for survival assay |
1st ref. for patients |
|
|---|---|---|---|---|---|---|---|---|
| α3 WTor | 0.5-10 | +++ | − | − | − | (1) | ||
| R463C | 3 | +++ | (VUS) | dystonia, tremor, parkinsonism | 60 y | 130 in gnomAD | this work | (2) |
| I274T | 10 | ++ | mild | RDP | 37 y | no, but I274N | (1) | |
| E277K“ | 10 | + | mild intermed. | RDP late AHC | 14-26 y 2.6 y | yes | (1) | (1,3) |
| G316S | 0.5 | +/− | mild | ataxia-RDP | 19 y | no | (4) | |
| G358V | 0.5 | − | severe | EIEE | 4 h | no | (5) | |
| I363N | 0.5 | − | severe | EIEE | 6 w | no | (5) | |
| D366H | 3 | − | mild | RDP | 16 y | no | this work | |
| C596R | 0.5 | − | severe | AHC | 3 w | no | this work | (6) |
| T613M | 10 | + | mild | RDP | 8-28 y | yes | (1) | |
| S729Y* | 0.5 | +++ | (VUS) | − | no | this work | ||
| D742Y“ | 3 | − | (severe) severe | ataxia-RDP paroxysmal | 1 y 6 w | yes | this work | (7) |
| D743H* | 3 | − | mild | RDP | no | this work | ||
| I758S | 10 | + | mild | RDP | 14-45 y | no, but I758F | (1) | |
| F780L | 10 | + | mild | RDP | 16-35 y | no | (1) | |
| D801Y“ | 10 | +/− | mild severe | RDP AHC | 12-23 y 14 mo | yes | (1) | (1,6) |
| E815K | 3 | − | severe | AHC | 1 w-18 m | yes | this work | (8) |
| D923N““ | 3 | + | severe intermed. mild | AHC late AHC RDP | 2 m,8 m 10 @ 2-4y 2 @ 20 y | yes | this work | (9-11) |
| L924P | 3 | ++ | severe | EIEE | 3 d | no | this work | |
| G947R | 0.5 | +/− | severe | AHC | 1 w-18 m | yes | this work | (8) |
Survival was scored as follows. No survival, −. Survival of some cells but no cell division, +/−. Slow, intermediate, and normal growth in ouabain, +, ++, and +++ respectively. The clinical syndrome most closely matching the phenotype seen is given, and phenotypes were categorized as severe (onset under 18 months of age), intermediate (2-4 years of age here) and mild (onset from 8 years to middle age). The use of different concentrations of ouabain depended on the investigator; all concentrations were sufficient, and some were in excess. Note that while four mutations presenting with severe phenotypes showed no survival in ouabain, another two mutations presenting with mild phenotypes also did not survive (boldface). L924P is a mutation with severe phenotype (fatal in this case) yet cells survived. Independent recurrence of each mutation is stated; one family is one occurrence. VUS: variant of unknown significance.
These two variants occurred in the same patient. WTOR, ouabain-resistant wild type ATP1A3. References: 1) (de Carvalho Aguiar et al., 2004), 2) (Yang et al., 2014), 3) (Boelman et al., 2014), 4) (Sweadner et al., 2016), 5) (Paciorkowski et al., 2015), 6) (Viollet et al., 2015), 7) (Marzin et al., 2018), 8) (Heinzen et al., 2012), 9) (Roubergue et al., 2013), 10) (Anselm et al., 2009), 11) (Zanotti-Fregonara et al., 2008)
p.Leu924Pro.
The patient was a male child of mixed ancestry born at 36 weeks. Symptoms of apnea appeared at 3 days. EEG was monitored for apneic spells, and EEG abnormalities were noted at 36 days. Epileptifom discharges were seen from 2.5 months, eventually including focal status epilepticus. MRI without contrast at 1 month was normal, but he had microcephaly with head circumference around the 1st centile at all ages. He died during an apparent apneic spell following seizure at age 2 years. Whole exome sequencing revealed de novo ATP1A3 c.2771T>C, p.Leu924Pro. The symptoms resembled the previous cases described for G358V and I363N (Paciorkowski et al., 2015), however neither of those mutations had enough activity to support cells in the survival assay, but L924P did (Table 1 below). Because of the uncommon clinical presentation, an extended case description is in the Supplement.
p.Arg463Cys.
The patient, a male of European ancestry, developed a right leg tremor at age 61, followed by right-hand rest tremor. Two days after tremor onset he developed flexion dystonia in the extremities of the right side. He did not report any potential triggers. Over months his dystonia fluctuated in severity but grew more painful, and episodic spasmodic dysphonia developed. MRI of the brain was not informative. At initial exam mild symmetric hyperreflexia without Babinski signs was observed, and he was hypophonic with mild dysphonia. He demonstrated a typical parkinsonian 6-8 Hz resting tremor in his right arm and hand as well as limited dystonic posturing. Two years after symptom onset his symptoms had worsened subjectively but remained fairly constant on exam (UPDRS-III of 43 in 2009, 46 in 2011), with neck flexion and posturing now of his left leg and foot, and a moderate tremor limited to the right hand and leg. His exam was otherwise normal. He was initially significantly levodopa responsive (50% on 200 mg levodopa 2 years after onset, 22% 4 years after onset) and had a bilaterally and asymmetrically abnormal DaT scan.
Sanger sequencing showed c.1387C>T, p.Arg463Cys. The same variant appears in gnomAD 127 times, giving it a frequency of 4.489e-4 overall and a frequency of 7.74e-4 among Europeans. It has been seen in at least three other unpublished ATP1A3 candidate AHC or RDP patients that we are aware of. Although it is predicted to affect protein function by SIFT, the arginine is exposed at the surface where pathogenicity seems less likely, and it supported cell life in the survival assay (Table 1 below). Although R463C lacks experimental support as a causative mutation, possibilities such as increased sensitivity to oxidative stress due to the introduction of a cysteine at the protein surface cannot be ruled out. Because of the RDP-like presentation, an extended case description is in the Supplement.
Screening variants for functional impairment
The survival assay is a useful test of the functional capability of Na,K-ATPase variants expressed in live mammalian cells. When mammalian cells (HEK 293, HeLa or COS) are treated with ouabain to inhibit the endogenous α1 Na, K-ATPase, acidification of the medium due to metabolism stops, and death of all of the cells follows within 48 h. Prior transfection with a ouabain-resistant Na, K-ATPase (ATP1A1, ATP1A2, or ATP1A3 modified with two benign mutations near the ouabain site) (Price and Lingrel, 1988) rescues cell survival and growth. Impairment of survival or of growth rate is validating evidence for mutation pathogenicity, however survival of cells and normal growth may require as little as 5-15% of normal ATPase activity (Jewell-Motz and Lingrel, 1993; Holm et al., 2016). Many pathogenic ATP1A1, ATP1A2, and ATP1A3 mutations have been shown to allow no cell survival at all, but others support growth and have functional modifications.
Table 1 shows the survival of human ATP1A3 mutations that have been investigated in our laboratories using transient transfection of HEK 293, with the corresponding patient phenotypes. Mutations were selected to represent a wide range of phenotype severity including RDP, AHC, and EIEE. Also included was the VUS R463C that has a 4.5/10,000 incidence in gnomAD. There are two important takeaways from Table 1. The simple prediction that mutations with low or no activity (no survival) in the survival assay should correlate with phenotype severity, does not hold up. Mutations supporting no survival at all can be found in a range of phenotypes: mild RDP cases (D366H, D743H), the most common AHC mutation (D801N), more severe AHC cases with seizures (E815K), and fatal EIEE (G358V). In contrast, the novel mutation found in the fatal EIEE case described above (L924P) did support survival.
Isogenic stable cell line growth rate
The lack of correlation between cell survival and the severity of patient phenotype le us to consider the cell biological consequences of mutations, such as misfolding and trafficking. For uniformity, stable isogenic cell lines were produced in Flp-In T-REX 293 cells for expression of tetracycline-inducible, ouabain-resistant ATP1A3 cDNAs with and without mutations, each inserted at a fixed FRT recombination site. The cell lines included α3WT; D366H; two pairs of adjacent amino acids associated with contrasting phenotypes, D742Y, D743H, and D923N, L924P; E815K (a representative AHC mutation); and the VUS, R463C.
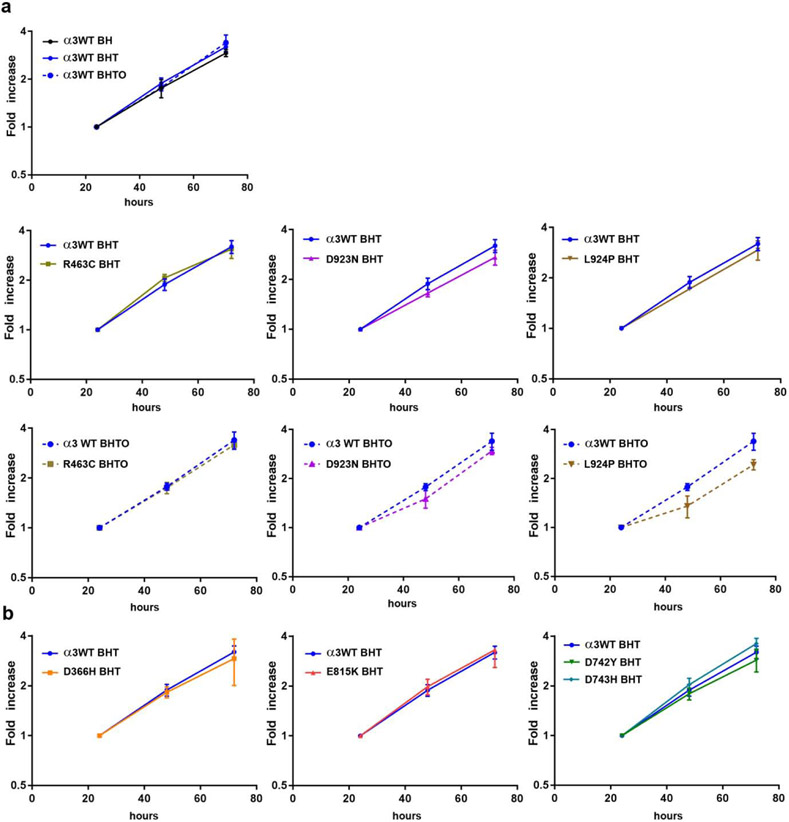
After propagation of the ouabain-surviving lines for 5 or more passages with or without 3 μM ouabain, logarithmic growth rates were compared using the cell proliferation reagent WST-1 (Fig. 1). The first graph in Fig. 1a shows the Flp-In line with wild type ouabain-resistant α3 subunit in three conditions: without tet induction of α3WT, with tet induction, and with tet induction and enough ouabain to inhibit the endogenous α1. Growth rate was not significantly altered. Then the growth rates of three mutations that do have enough activity to support cell survival are compared with and without ouabain in Fig. 1a. Growth rates were not significantly different in the absence of ouabain or in its presence for the VUS R463C, but D923N and L924P grown chronically in ouabain showed transient lags followed by recovery to the rates shown by α3WT. The lags were statistically significant by 1-tailed t test with equal variance. The lags suggest a slower rate of recovery from trypsinization and replating when the cells depend on the activity of exogenous mutant α3. This explained our observation that those lines were slower to grow during routine cell culture. Fig. 1b shows the growth rates of Flp-In lines for D366H, E815K, D742Y and D743H, which do not survive in the presence of ouabain. Their growth rates, necessarily dependent on endogenous α1, were not significantly different from α3WT.
Figure 1. Growth rates of isogenic cell lines expressing wild type or mutated ATP1A3.
In all cases, cells had been maintained in the indicated mixture of drugs. BH: blasticidin and hygromycin, antibiotics used to ensure the stability of the constructs. In this condition only endogenous ATP1A1 is expressed. BHT: BH with tetracycline to induce the exogenous ATP1A3 construct. BHTO: BHT with 3 μM ouabain to inhibit the activity of endogenous ATP1A1. Cells were trypsinized and replated at time zero. (a) These four cell lines all had enough ATP1A3 activity to support life in the survival assay and were grown with and without ouabain. R463C cells had a growth rate similar to WT, but D923N and L924P cells both consistently exhibited lags in growth after plating. (b) These four cell lines died when treated with ouabain, and so they were grown without it so that endogenous α1 was active during growth. [color optional if redone with distinctive lines]
Representative pairs of mutations
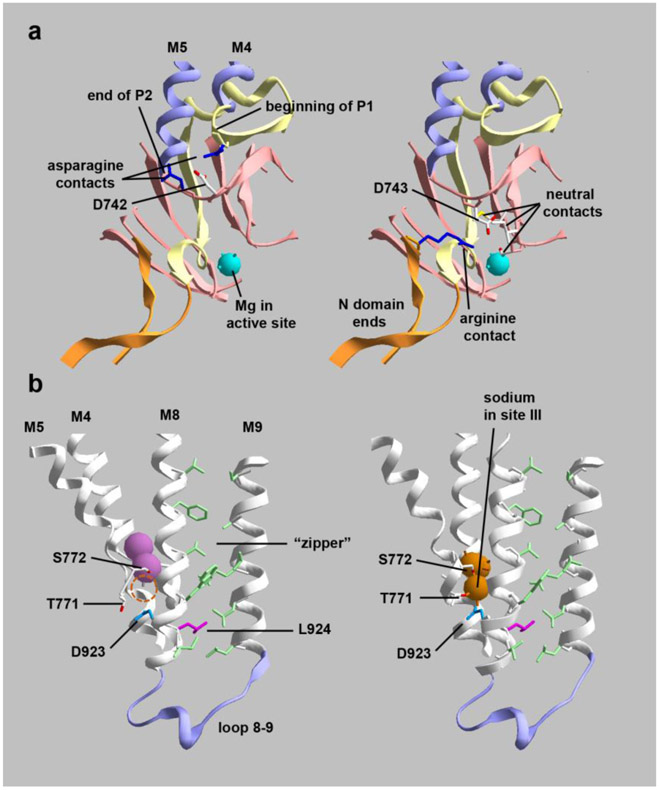
We selected four mutants to investigate further. Two were from patients with relatively mild symptoms without abnormal brain MRI (D923N and D743H), and two were from patients with symptom-associated regional atrophy or microcephaly (D742Y and L924P). The mutations selected are also pairs of adjacent residues, one associated with neuron loss and the other not. Figures 2 illustrates how the side chains of each member of a pair point in different directions and have different structural roles despite their proximity.
Figure 2. Cutaway images of crystal structures showing the positions and interactions of mutated residues.
(a) D742 and D743 are adjacent aspartates at a junction between protein domains. They are on an unstructured link between the second part of the P domain (P2) and transmembrane span M5. (left) D742 (colored in CPK) has polar interactions with two asparagines (dark blue), one at the beginning of P1 and one at the end of P2. Substitution with a bulky tyrosine (D742Y) may impede the folding of the two parts of the P domain. (right) D743 (also colored in CPK) points down toward the active site where Mg+2 is positioned. D743 interacts with 3 local residues in P2 and with an arginine (dark blue) located at the beginning of the N domain; D743 and R586 do not appear to form a strong salt bridge. Substitution with hydrophilic histidine may not affect protein folding but may impact the active site. (b) D923N (blue) and L924P (magenta) are adjacent residues that also point in different directions. (left) K+-bound conformation (2 pink spheres); (right) Na+-bound conformation (3 orange spheres). D923 on M8 is close to T771 and S772 on M5, residues that bind the third sodium ion. The empty sodium binding site is marked with a dashed circle in the K+ conformation. T771 and S772 produce AHC when mutated, while D923N produces RDP or AHC. D923N is well-known to have residual activity and to reduce affinity for sodium (Einholm et al., 2010; Holm et al., 2016). L924 (magenta) has a distinct structural role. While most transmembrane spans in polytopic membrane proteins intersect each other at angles, M8 and M9 are very parallel. They are held together by a zipper-like structure of opposing residues (light green). L924 (magenta) interacts with two opposing leucines in the zipper near its base where the M8-M9 hairpin establishes the position of the 8-9 loop, an important part of the structure. Substitution of L924 with a proline will disrupt the helix at that position, so it is likely to cause misfolding and may displace both D923 and the 8-9 loop. The structures are PDB codes 2ZXE (Shinoda et al., 2009) and 3WGU (Kanai et al., 2013). [color essential]
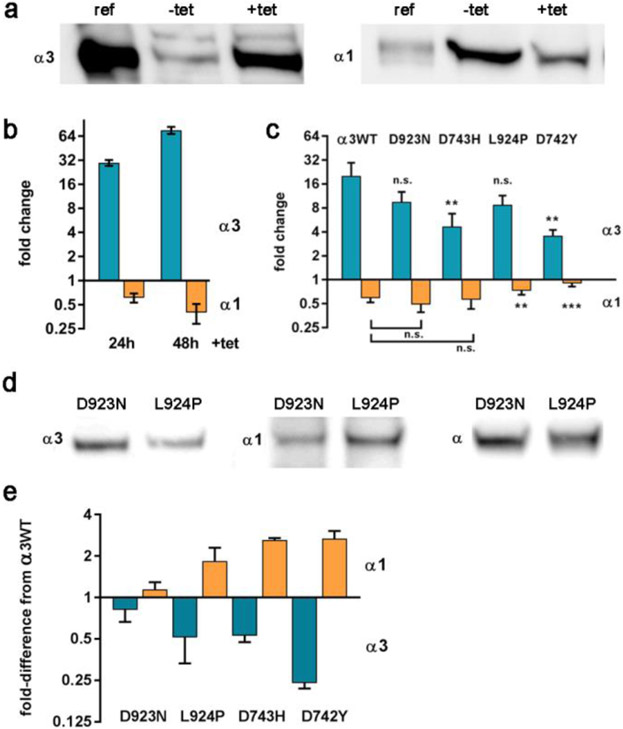
Induction of the stable α3WT transfectant cell line by tetracycline increased ATP1A3 mRNA levels 13.1-fold ± 5.1 , as calculated by qPCR with the ΔΔCτ method, but did not affect the level of ATP1A1 mRNA (1.02-fold ± 0.47) (5 independent biological replicates). This rules out an effect of the exogenous ATP1A3 on the mRNA level of the endogenous ATP1A1. In similar qPCR experiments (3 biological replicates), the relative amounts of ATP1A3 mRNA showed no significant differences by 2-tailed t-test for α3WT, D923N, L924P, D743H, and D742Y (average 1.43 ± 0.36 S.D., P = 0.3).
Competition between α subunits
When tetracycline was added to induce expression of ATP1A3, the level of α1 protein was reduced both acutely and at steady state. Fig. 3a shows a representative blot illustrating the increase in expression of α3 protein and an accompanying reduction in α1 upon tet induction for 24 and 48 h. Fold-changes in protein expression are quantified in Fig. 3b. The reduction in α1 is an expected feature of Na,K-ATPase biosynthesis: newly synthesized α and β subunits must assemble into an αβ protomer before leaving the endoplasmic reticulum (Geering, 2001; Gatto et al., 2001; Tokhtaeva et al., 2009). It is known that the total amount of Na,K-ATPase alpha subunit stays fixed when an exogenous subunit is introduced to compete with it (Devarajan et al., 1992; Rajasekaran et al., 2004; Clifford and Kaplan, 2009). This can be readily understood if the nascent α subunits co-translationally compete for β subunits. Blots stained with a pan-specific α antibody showed no change in total α: the average change in protein was 1.04 ± 0.085 for 3 biological replicates each of α3WT and all four mutants.
Figure 3. Competition between exogenous and endogenous Na,K-ATPase α subunits in isogenic cell lines.
(a-c) Acute addition of tetracycline produced changes in expression of α3 and α1 as estimated by western blot quantification. (a) A representative blot where ref is a reference sample of mouse brain microsomes, and −tet and +tet are lysates of α3WT-expressing cells. (b) Quantification of replicates for the response of α3WT over 24 and 48 h, expressed as fold-change. The 30- and 65- fold increase of α3 is relative to a low level of leakiness of the promoter or the endogenous ATP1A3 gene, and α1 in turn dropped by ~30 and 60%. (c) 24 h induction experiments where α3WT, D923N, D743H, L924P, and D742Y were compared (averaged from 3-5 experiments). Significance by two-tailed t-test of <0.01, **; of <0.001, ***. Error bars show standard deviations. The reduction of α1 was not significantly different for WT, D923N, and D743H. (d, e) Chronic addition of tet produced reduced α3 and increased α1 at steady state, i.e. after >5 passages in tet-containing medium. (d) A representative blot showing that the level of total α subunit was equal in D923N cells and L924P cells, as detected by a pan -specific antibody 9A7, while the two cell lines differed in the levels of α3 and α1. (e) The steady state results for the four isogenic cell lines were normalized to the levels of α3 and α1 in α3WT (= 1.0). Shown are averages of 3-5 biological replicates in chronic tetracycline. [Color optional]
Fig. 3c shows the semi-quantitative relationship after 24 h of tet induction between the expression of α3 and the suppression of expression of α1 in the isogenic Flp-In cell lines for α3WT and the four mutations. The mutation with the least expression also had the least impact on α1 protein level.
Even after prolonged culture of ATP1A3 mutations in tet-containing media, competition for β subunit affected the expression level of α1. Fig. 3d shows representative blots for two isogenic cell lines grown for >5 passages with tet induction. D923N expressed more α3 than L924P and the opposite was found for α1, but the total detected by the pan-specific α antibody was unchanged. Fig. 3e summarizes average changes in the levels of α3 and α1 at steady state in tet for the four mutant cell lines. The values obtained in the control α3WT were taken as 1, and the chart shows the increases or decreases in each line relative to that standard. D923N was well-expressed with ~15% lower α3 than WT and with ~15% higher α1. The mutations L924P and D743H were similar in showing half the level of α3 than α3WT and double the level of α1 than α3WT. D742Y had the lowest expression of α3 but the increase in α1 was not significantly higher than with the other mutants, suggesting that a maximum may have been reached.
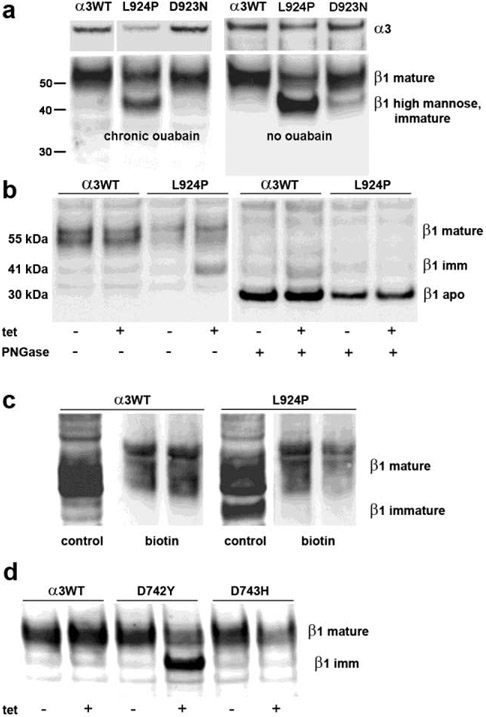
Impairments of post-translational processing of beta subunit
When expression of mutated α3 was turned on with tet, altered glycosylation of β subunit was observed with some mutations and not others. The nascent β subunit has a molecular weight of 32 kDa, but N-glycans are added in the ER at three intraluminal (future extracellular)sites. The N-glycans serve as binding ligands for the lectin-like chaperones calnexin and calreticulin and play a critical role in protein folding and trafficking (Tokhtaeva et al., 2010). N-glycan addition in the ER is a high-mannose form, and the β subunit in this compartment has an apparent molecular weight of ~40 kDa on SDS gels. Subsequently the N-glycans are modified in the Golgi apparatus, forming β subunits migrating at higher molecular weight due to differences in the extent of glycan branching and to the negative charge of sialylation. The mature N-glycan β traffics to the cell surface with its α subunit partner.
When cells were induced by tetracycline to steady state and grown continuously with or without ouabain, most of the β subunit of α3WT and D923N was in the mature, fully glycosylated form, although traces of nascent apo-β and a weak band corresponding to immature glycosylation could sometimes be seen (Fig. 4a). In contrast, more than half of the β stain in L924P cells migrated to where the immature form is expected, again with or without ouabain. Scans of blots showed that the sum of the two major bands was equal to the mature β band in α3WT. The lower molecular weight band was not increased in L924P cells until acute induction of α3 with tet (Fig. 4b). The epitope for the anti - β antibody is at approximately amino acid 200 (out of 305) (Sun and Ball, Jr., 1994), and so it was possible that the new band in L924P cells was due to proteolytic cleavage at either end of the protein. To test this, two experiments were performed. First, preparations isolated from α3WT and L924P cultures with and without tet induction were treated with PNGase F (peptide-N-glycosidase F) to remove all N-glycans. Fig. 4b shows that all β bands were reduced to the size of nascent, unglycosylated β, demonstrating that the β subunit in L924P was still full-length. The data also illustrate that the amount of β was not changed by tet induction of α3WT or L924P. Second, β subunits at the plasma membrane were biotinylated by impermeable biotinylation reagent, and the biotinylated proteins were collected with streptavidin beads and analyzed by western blot. Fig. 4c shows that the band at ~40 kDa was not biotinylated, demonstrating that its trafficking to the plasma membrane was blocked.
Figure 4. Retention of β in endoplasmic reticulum.
(a) Cells were grown continuously in tet with or without continuous ouabain to inhibit α1. Blots were cut, and blot pieces were stained separately for α3 and β1. The expression of α3 was reduced for L924P relative to α3WT and D923N, as also seen in Fig. 2. The majority of β subunit migrated at 55 kDa in the α3WT and D923N cells, while more than half migrated at ~40 kDa in L924P, the position of β with high-mannose N-glycans in the ER. (b) On the left, L924P cells did not accumulate immature β until tet induction of mutant α3. On the right, removal of all N-glycans with PNGase F reduced both the mature and immature N-glycan forms of β to the mobility of the intact apoprotein, 30 kDa. (c) When cells were biotinylated with an impermeable reagent, only the mature form of β was recovered with streptavidin beads, not the immature N-glycan form, consistent with its residence in the ER. (d) D742Y but not D743H accumulated immature β when α3 was induced with tet. [gray scale]
L924P cells had a significant proportion of β in fully-glycosylated form. This most likely reached the plasma membrane partly with endogenous α1 and partly with an unknown fraction of α3 that trafficked correctly. We know that some L924P α3 trafficked correctly because of the cell survival assay data of Table 1 and the growth rates of Fig. 1a, when α1 was inhibited by ouabain. We surmised that L924P trafficking was impaired but not abolished, and that detection of the immature form of β is a marker of this impairment. The expression of D742Y was also accompanied by the appearance of a high proportion of immature β, while D743H behaved like α3WT(Fig. 4d). Because all experiments with both D742Y and D743H were necessarily performed without ouabain, β subunit was partially retained in D742Y despite the availability of α1 to support cell growth. This is consistent with misfolding and ER retention as the cause, the expected consequence of the unfolded protein response (UPR) (Karagoz et al., 2019). Not shown is that three other mutations from Table 1 and Figure 1, D366H, R463C, and E815K, did not result in accumulation of immature β. D366H is inactive because of the aspartate’s role in catalysis.
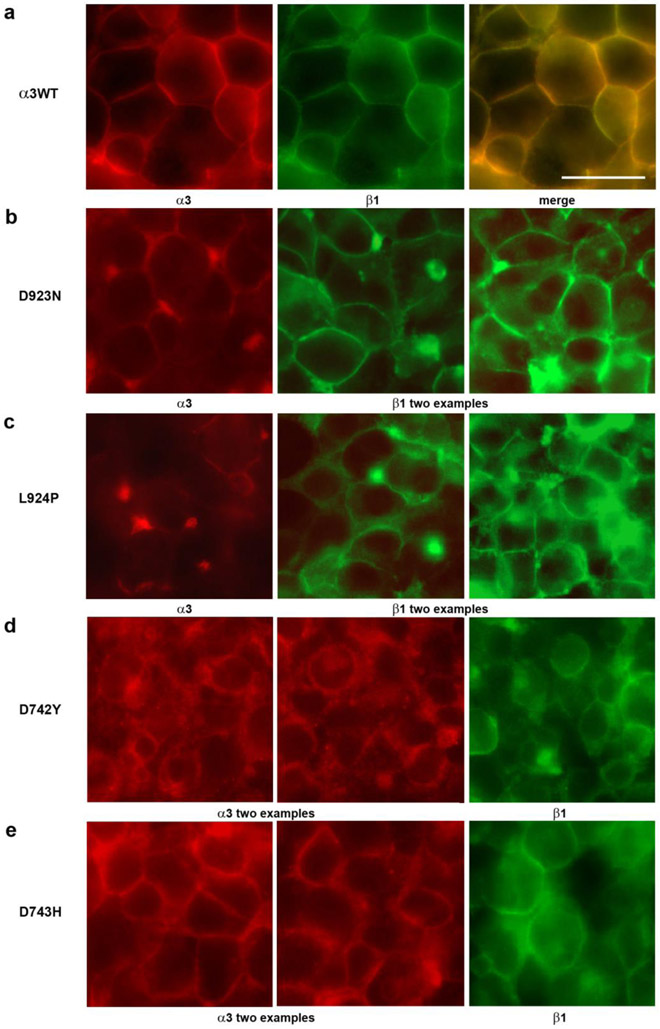
Cytopathology and aberrant trafficking
Light microscopy with immunofluorescence for α3 and β1 was used to investigate the distribution of Na, K-ATPase subunits when ATP1A3 was mutated. The microscopy also revealed cytopathology that differed between mutations. Fig. 5a shows cells expressing α3WT under tet induction. The double -label stain for α3 and β1 was largely coincident as expected. The cells tended to be in smooth, closely-packed monolayers or mounds. A distinctive polygonal appearance results from the tight apposition of healthy adjacent cells. Prior work in other cell lines has shown that this is due to β subunit -mediated intercellular adhesion (Vagin et al., 2008).
Figure 5. Cytopathology in Flp-In cells expressing ATP1A3 mutations.
(a) α3WT cells, like the parent cell line, packed tightly when confluent and adopted a polygonal appearance. Both α3 and β1 stain was mainly at the surface. (b and e) Both D923N cells and D743H cells were often almost as healthy as α3WT, but also showed some blebs and mislocalization. (c) In L924P cells, stain for both subunits was more irregular, with blebs and occasional cytoplasmic localization. (d) In D742Y cells there was deterioration of the packing of cells accompanied by intracellular localization of stain and blebbing. The length bar is 20 μm. [Color essential]
Fig. 5b-e show representative examples of the cell morphology and distribution of subunits for D923N, L924P, D742Y and D743H. D923N and D743H resembled α3WT in that there was a polygonal cell appearance and a majority of both subunits had a plasma membrane distribution. Nonetheless there was also some intracellular stain and there were bright concentrations of stain in some cells, possibly aggregates, and occasional large blebs at cell interfaces. L924P had relatively faint plasma membrane stain for α3 and prominent accumulations of stain for β subunit in cytoplasm; its blebs contained both α3 and β1 (not shown). D742Y had almost all of the α3 stain located intracellularly, while β subunit was both at the surface (presumably with endogenous α1) and intracellular. D742Y was notable for having lost most of the polygonal interaction planes between cells. The cytopathology was qualitatively consistent with the observation that some mutations (D923N and D743H) had mostly mature β subunit, while others (L924P and D742Y) had substantial immature β subunit. L924P clearly had some α3 stain that trafficked correctly, while D742Y apparently failed to traffic.
Discussion
To date there are no recessive mutations in ATP1A3, and so either haploinsufficiency or dominant effects are assumed to be causative. The significance of the findings reported here is that misfolding of α subunit and allele competition for β subunit are factors distinct from changes in Na,K -ATPase activity or kinetics for the potential to cause disease. This means that for each mutation, there are interdependent forces contributing to a sliding scale of deleterious impact at the cellular level. Any given mutation may produce a combination of effects on activity, misfolding, and the competition of WT and mutant alleles for β. This predicts a heterogeneity of pathogenicity that may go a long way toward explaining the wide range of human phenotypes observed with different ATP1A3 mutations.
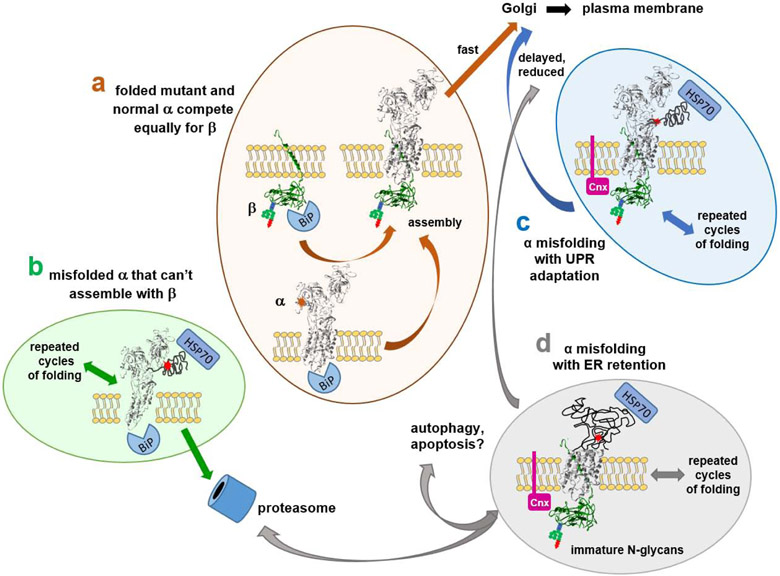
Some predictions can be made based on this framework: there are at least four ways that things can go badly. First, a mutated protein that folds as well as the normal allele should be trafficked to the plasma membrane in a 50:50 ratio with the normal protein (Fig. 6a). That would be the worst outcome in the event of a gain-of-toxic-function mutation like ion leakage, but not if a mutation was simply inactive. Second, a mutation that disrupts folding of the C-terminal third of the protein (the location of the obligatory β binding sites) might have reduced impact: the α from the mutant allele will not compete with the normal allele if it is incapable of interacting with β (Fig. 6b). This could make more β available to the nascent α chains from the good allele for a better-than-haploinsufficient result, such as reported for some Na,K-ATPase mutations in Drosophila (Ashmore et al., 2009). In the ER, chaperones like BiP will assist in extruding the unfolded mutant alpha from ER to cytoplasm, followed by ubiquitination and proteasome degradation (ERAD), a system that is constitutively active to deal with the normal background of errors in translation and folding. Third, a misfolding mutation that does complex with beta will engage in calnexin or calreticulin-mediated cycles of refolding, and it has a chance of succeeding and trafficking to the Golgi and beyond, even though defective (Fig. 6c). α subunit does not have the N-glycans required for association with those chaperones. In the event of an increased burden of misfolded protein, the unfolded protein response (UPR) will expand the ER and upregulate proteins that facilitate folding adaptively (Karagoz et al., 2019). This could impact the effect of gain-of-toxic-function mutations. Aggregates of misfolded protein will activate autophagy as another protective UPR response. Fourth, if expansion of the ER and autophagy are insufficient, the UPR will induce apoptosis, leading to neuronal cell death (Fig. 6d). In extreme cases, that likely leads to pre- and post-natal microcephaly, and in less extreme cases may account for the progressive regional atrophy detected by MRI. All of these possibilities are types of biosynthesis defects, and they will impact the competition of WT and mutant alleles, further influencing the clinical outcome.
Figure 6. Alternative fates of ATP1A3 mutations.
There are four ways that Na,K-ATPase mutations can navigate the biosynthetic pathway: (a) unimpaired folding and biosynthesis; (b) irremediable misfolding followed by proteasomal degradation (ERAD); (c) successful chaperone-assisted folding, which delays biosynthesis; and (d) failed chaperone-assisted folding. The chaperones illustrated (BiP/GRP-78, calnexin, and HSP70) are just representative of different chaperone classes. The factor that makes the consequences of Na,K-ATPase mutations difficult to predict is the competition that must occur between mutant and normal ATP1A3 alleles for β subunit. In (a), well-folded mutant and normal α subunits should compete equally well for β and achieve a 50:50 ratio in patient neurons. In (b), misfolded α3 incapable of binding to β will be degraded by ERAD. In this case only WT α3 subunits will succeed in reaching the plasma membrane, leading to a mild phenotype. Mutant α3 degradation may also increase the pool of β subunit available to nascent chains from the normal allele. In (c), association of mutant α3 with β will lead to delayed folding (Tokhtaeva et al., 2010) and potentially to a less-than-50:50 ratio of mutant to normal α3 at the membrane. This could lessen the effect of gain-of-toxic-function mutations. In (d), prolonged retention of β subunit in the ER could effectively sequester β subunit and lead to further reduction of Na,K-ATPase expression, protein aggregation, and the triggering of autophagy and apoptosis. Any given mutation could lie between these four extremes. Superimposed on the outcomes for (a, c, and d) will be any functional effects of mutation on the activity or kinetics of the folded and trafficked protein. [Color essential]
In these experiments, D742Y and L924P showed β retention and the lowest expression. The two known D742Y patients both exhibited atrophy in the brain, although in different locations, parietal temporal cortex and a short corpus callosum in one case and cerebellum in the other. Atrophy in the patients is consistent with the D742Y cytopathology seen here if we postulate that the UPR was driven to the point of apoptosis. The patients differed in severity of symptoms, possibly related to the location of the atrophy. The adjacent mutation D743H showed much less evidence of cytopathology in culture and no retention of β subunit, and it was associated with a milder, RDP phenotype. L924P had α3 levels as low as D742Y and correspondingly high α1. Proline substitutions often lead to misfolding because they force a change in backbone direction, and that is particularly a problem when they are introduced into an α-helix as here, because it will change the rotation of its nearest neighbors. In the Flp-In cell line, however, a fraction of the expressed L924P did fold and traffic to the membrane with enough activity to keep the cells alive in the survival assay. Nonetheless, the patient was severely affected and had microcephaly. From this we postulate that the defect in biosynthesis was toxic on its own, by inducing apoptosis. That would not necessarily be obvious in cell lines with 24 h doubling times, but in vivo it should accrue in developing or post-mitotic neurons. The adjacent residues D923N and L924P also had contrasting properties. D923N, which has been well -studied and reduces Na+ affinity by its effect on the third ion binding site (Holm et al., 2016), appeared to be almost as well-expressed as the normal allele; was accompanied by strong competition with α1; and displayed no retention of β. However, the cells also showed a lag in growth and routinely took longer to propagate when grown with ouabain to inhibit α1.
Other members of the broader P-type ATPase family have some human mutations subject to misfolding, including ATP1A2 (familial hemiplegic migraine) (Coppi and Guidotti, 1997), SERCA2C1 (Darier disease) (Wang et al., 2011), ATP7A (Menkes disease) (Vonk et al., 2012), ATP7B (Wilson’s disease) (Dmitriev et al., 2011) and ATP8B1 (intrahepatic cholestasis) (Hegde et al., 2017). However, of these, only the paralog ATP1A2 and the lipid flippase ATP8B1 also have a β-like subunit obligatory for ER exit (Paulusma et al., 2008; Tokhtaeva et al., 2012). like ATP1A3, ATP8B1 mutations also manifest in severe and mild forms, and it is interesting to predict that allele competition and the degree of misfolding will contribute to severity there as well.
There are additional potential sources of patient phenotype variability between mutations. One is that the burden of misfolding and the UPR may impact different classes of neurons to different degrees, based on baseline rates of Na,K-ATPase biosynthesis, size, and axon and dendrite targeting. The normal relative proportions of α1 and α3 differ between types of neurons as well, with the result that in many neurons, competition between WT and mutant alleles of ATP1A3 will also be complicated by a reduction of α3 relative to α1 (Azarias et al., 2013). And of course, a source of variability between individuals with the same mutation is that genetic background may make some people less resilient to misfolding mutations than others. An intriguing example is the cerebellar atrophy of a patient with ATP1A3 p.Gly316Ser who also had a de novo mutation in UBQLN4, an adaptor protein for ubiquitin ligases (Sweadner et al., 2016).
This conceptual framework cannot answer every question, not the least because there are cases where identical ATP1A3 mutations have produced different clinical outcomes, as shown in Table 1. The experiments here implicate the UPR, however more investigation is needed to understand which of its arms are activated, how the responses differ between mutations, and how it impacts neurons. Understanding the involvement of the UPR, and whether its actions are beneficial or toxic for each ATP1A3 mutation, is relevant to the development of treatments for ATP1A3 diseases because of the potential for using chemical chaperones or innovative methods to promote protein folding (Hegde et al., 2017).
Supplementary Material
Highlights.
Mutations in ATP1A3 cause disease ranging widely from newborn- to adult-onset
Loss of activity cannot explain the severity of phenotype
Protein misfolding and ER retention did correlate with clinical severity here
Misfolding also affected the ratio between good and bad alleles, by competition
Acknowledgements
We thank the patients for their participation in research; W.J. Ball, University of Cincinnati and M.W. McEnery, Case Western Reserve University, for monoclonal antibodies; J.H. Kaplan, University of Illinois, and J.K. Young, Massachusetts General Hospital for plasmids. We thank Prof. C. Toyoshima, Univ. of Tokyo, for useful discussions.
The research was funded by NIH grant NS058949 to A.B.. The microscopy core was supported by P30EY003790 to Pablo Argueso. The funding sources had no involvement in the design, performance, or publication of the work.
Footnotes
Declaration of interest
E.A., T.L., F.M., P.F., C.S., J.F.C., S.D., L.J.O., and K.J.S. declare no competing interests. I.U.H. has performed research for Allergan, Boston Scientific, Great Lakes Neurotechnology, and Pfizer. His conflicts of interest are managed by Wake Forest School of Medicine. R.S-P. has support from the NIH (U01-NS094148), the Bigglesworth Family Foundation, and consults for Denali. S.B. has support from the Michael J. Fox Foundation and consults for Denali. A.B. performs research funded by NINDS, Revance, and Ipsen, and consults for Revance and Ipsen. Her conflicts of interest are managed by Wake Forest School of Medicine.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson TR, Huguenard JR, Prince DA, 2010. Differential effects of Na+-K+ ATPase blockade on cortical layer V neurons. Journal of Physiology 588, 4401–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselm IA, Sweadner KJ, Gollamudi S, Ozelius LJ, Darras BT, 2009. Rapid-onset dystonia-parkinsonism in a child with a novel ATP1A3 gene mutation. Neurology 73, 400–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashmore LJ, Hrizo SL, Paul SM, Van Voorhies WA, Beitel GJ, Palladino MJ, 2009. Novel mutations affecting the Na, K ATPase alpha model complex neurological diseases and implicate the sodium pump in increased longevity. Hum.Genet 126, 431–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarias G, Kruusmagi M, Connor S, Akkuratov EE, Liu X-L, Lyons D et al. , 2013. A specific and essential role for Na,K-ATPase α3 in neurons co-expressing α1 and α3. Journal of Biological Chemistry 288, 2734–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benarroch EE, 2011. Na+,K+-ATPase: Functions in the nervous system and involvement in neurologic disease. Neurology 76, 287–293. [DOI] [PubMed] [Google Scholar]
- Blom H, Ronnlund D, Scott L, Spicarova Z, Widengren J, Bondar A et al. , 2011. Spatial distribution of Na+-K+-ATPase in dendritic spines dissected by nanoscale superresolution STED microscopy. BMC Neurosci. 12, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelman C, Lagman-Bartolome AM, MacGregor DL, McCabe J, Logan WJ, Minassian BA, 2014. Identical ATP1A3 mutation causes alternating hemiplegia of childhood and rapid-onset dystonia parkinsonism phenotypes. Pediatr.Neurol 51, 850–853. [DOI] [PubMed] [Google Scholar]
- Brashear A, Sweadner KJ, Cook JF, Swoboda KJ, Ozelius LJ, 2018. ATP1A3-related neurologic disorders. Gene Reviews http://www.ncbi.nlm.nih.gov/books/NBK1115/. [Google Scholar]
- Carecchio M, Zorzi G, Ragona F, Zibordi F, Nardocci N, 2018. ATP1A3-related disorders: An update. Eur.J.Paediatr. Neurol 22, 257–263. [DOI] [PubMed] [Google Scholar]
- Cheng TS, Ho KM, Lam CW, 2010. Heterogeneous mutations of the ATP2C1 gene causing Hailey-Hailey disease in Hong Kong Chinese. J.Eur.Acad.Dermatol.Venereol 24, 1202–1206. [DOI] [PubMed] [Google Scholar]
- Clausen MV, Hilbers F, Poulsen H, 2017. The structure and function of the Na,K-ATPase isoforms in health and disease. Front Physiol 8, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford RJ, Kaplan JH, 2009. Regulation of Na,K-ATPase subunit abundance by translational repression. Journal of Biological Chemistry 284, 22905–22915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppi MV, Guidotti G, 1997. Intracellular localization of Na,K-ATPase alpha2 subunit mutants. Archives of Biochemistry and Biophysics 346, 312–321. [DOI] [PubMed] [Google Scholar]
- Dard R, Mignot C, Durr A, Lesca G, Sanlaville D, Roze E et al. , 2015. Relapsing encephalopathy with cerebellar ataxia related to an ATP1A3 mutation. Dev.Med.Child Neurol 57, 1183–1186. [DOI] [PubMed] [Google Scholar]
- de Carvalho Aguiar P, Sweadner KJ, Penniston JT, Zaremba J, Liu L, Caton M et al. , 2004. Mutations in the Na,K-ATPase α3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron 43, 169–175. [DOI] [PubMed] [Google Scholar]
- Demos MK, van Karnebeek CDM, Ross CJD, Adam S, Shen Y, Zhan SH et al. , 2014. A novel recurrent mutation in ATP1A3 causes CAPOS syndrome. Orph.J.Rare Dis 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Gilmore Hebert M, Benz EJ Jr., 1992. Differential translation of the Na,K-ATPase subunit mRNAs. Journal of Biological Chemistry 267, 22435–22439. [PubMed] [Google Scholar]
- Dmitriev OY, Bhattacharjee A, Nokhrin S, Uhlemann EM, Lutsenko S, 2011. Difference in stability of the N-domain underlies distinct intracellular properties of the E1064A and H1069Q mutants of copper-transporting ATPase ATP7B. Journal of Biological Chemistry 286, 16355–16362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov M, Stimers JR, 2005. Neuronal function and alpha3 isoform of the Na/K-ATPase. Frontiers in Bioscience 10, 2373–2396. [DOI] [PubMed] [Google Scholar]
- Einholm AP, Toustrup-Jensen MS, Holm R, Andersen JP, Vilsen B, 2010. The rapid-onset dystonia parkinsonism mutation D923N of the Na+,K+-ATPase α3 isoform disrupts Na+ interaction at the third Na+ site. Journal of Biological Chemistry 285, 26245–26254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich T, Tavraz NN, Junghans C, 2016. ATP1A2 mutations in migraine: seeing through the facets of an ion pump onto the neurobiology of disease. Front.Physiol 7, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto C, McLoud SM, Kaplan JH, 2001. Heterologous expression of Na+-K+-ATPase in insect cells: intracellular distribution of pump subunits. American Journal of Physiology 281, C982–C992. [DOI] [PubMed] [Google Scholar]
- Geering K, 2001. The functional role of β subunits in oligomeric P-type ATPases. Journal of Bioenergetics and Biomembranes 33, 425–438. [DOI] [PubMed] [Google Scholar]
- Haq IU, Snively BM, Sweadner KJ, Suerken CK, Cook JF, Ozelius LJ et al. , 2019. Revising Rapid-Onset Dystonia-Parkinsonism: broadening indications for ATP1A3 testing. Movement Disorders in press. DOI: 10.1002/mds.27801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RN, Subramanian A, Pothukuchi P, Parashuraman S, Luini A, 2017. Rare ER protein misfolding-mistrafficking disorders: Therapeutic developments. Tissue Cell 49, 175–185. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Arzimanoglou A, Brashear A, Clapcote SJ, Gurrieri F, Goldstein DB et al. , 2014. Distinct neurological disorders with ATP1A3 mutations. Lancet Neurol. 13, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen EL, Swoboda KJ, Gurrieri F, Nicole S, de Vries B, Tiziano FD et al. , 2012. De novo mutations in ATP1A3 cause alternating hemiplegia of childhood. Nat.Genet 44, 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm R, Toustrup-Jensen MS, Einholm AP, Schack VR, Andersen JP, Vilsen B, 2016. Neurological disease mutations of alpha3 Na+,K+-ATPase: Structural and functional perspectives and rescue of compromised function. Biochim.Biophys.Acta 1857, 1807–1828. [DOI] [PubMed] [Google Scholar]
- Jewell-Motz EA, Lingrel JB, 1993. Site-directed mutagenesis of the Na,K-ATPase: consequences of substitutions of negatively-charged amino acids localized in the transmembrane domains. Biochemistry 32, 13523–13530. [DOI] [PubMed] [Google Scholar]
- Kanai R, Ogawa H, Vilsen B, Cornelius F, Toyoshima C, 2013. Crystal structure of a Na+-bound Na+,K+-ATPase preceding the E1P state. Nature 502, 201–206. [DOI] [PubMed] [Google Scholar]
- Karagoz GE, Acosta-Alvear D, Walter P, 2019. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb.Perspect.Biol in press doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sizov I, Dobretsov M, von Gersdorff H, 2007. Presynaptic Ca2+ buffers control the strength of a fast post-tetanic hyperpolarization mediated by the α3 Na+/K+-ATPase. Nat.Neurosci 10, 196–205. [DOI] [PubMed] [Google Scholar]
- Li M, Jazayeri D, Corry B, McSweeney KM, Heinzen EL, Goldstein DB et al. , 2015. A functional correlate of severity in alternating hemiplegia of childhood. Neurobiol.Dis 77, 88–93. [DOI] [PubMed] [Google Scholar]
- Liang M, Cai T, Tian J, Qu W, Xie Z-J, 2006. Functional characterization of Src-interacting Na/K-ATPase using RNA interference assay. Journal of Biological Chemistry 281, 19709–19719. [DOI] [PubMed] [Google Scholar]
- Marzin P, Mignot C, Dorison N, Dufour L, Ville D, Kaminska A et al. , 2018. Early-onset encephalopathy with paroxysmal movement disorders and epileptic seizures without hemiplegic attacks: About three children with novel ATP1A3 mutations. Brain Dev. 40, 768–774. [DOI] [PubMed] [Google Scholar]
- McGrail KM, Phillips JM, Sweadner KJ, 1991. Immunofluorescent localization of three Na,K-ATPase isozymes in the rat central nervous system: both neurons and glia can express more than one Na,K-ATPase. Journal of Neuroscience 11, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Noguchi S, Takeda K, Morohashi M, Kawamura M, 1990. Site-directed mutagenesis of Asp-376, the catalytic phosphorylation site, and Lys-507, the putative ATP-binding site, of the α-subunit of Torpedo californica Na+/K+-ATPase. Biochimica et Biophysica Acta 1021, 157–160. [DOI] [PubMed] [Google Scholar]
- Paciorkowski AR, McDaniel SS, Jansen LA, Tully H, Tuttle E, Ghoneim DH et al. , 2015. Novel mutations in ATP1A3 associated with catastrophic early life epilepsy, episodic prolonged apnea, and postnatal microcephaly. Epilepsia 56, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulusma CC, Folmer DE, Ho-Mok KS, de Waart DR, Hilarius PM, Verhoeven AJ. et al. , 2008. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology 47, 268–278. [DOI] [PubMed] [Google Scholar]
- Picton LD, Nascimento F, Broadhead MJ, Sillar KT, Miles GB, 2017. Sodium pumps mediate activity-dependent changes in mammalian motor networks. Journal of Neuroscience 37, 906–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, Lingrel JB, 1988. Structure-function relationships in the Na,K-ATPase alpha subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry 27, 8400–8408. [DOI] [PubMed] [Google Scholar]
- Rajasekaran SA, Gopal J, Willis D, Espineda C, Twiss JL, Rajasekaran AK, 2004. Na,K-ATPase β1-subunit increases the translation efficiency of the α1-subunit in MSV-MDCK cells. Molec.Biol.Cell 15, 3224–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KS, Bommert K, Szabo G, Miles R, 2007. Differential expression of Na+/K+-ATPase α-subunits in mouse hippocampal interneurons and pyramidal cells. Journal of Physiology 5852, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosewich H, Sweney MT, DeBrosse S, Ess K, Ozelius L, Andermann E et al. , 2017. Research conference summary from the 2014 International Task Force on ATP1A3-Related Disorders. Neurol.Genet 3, e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubergue A, Roze E, Vuillaumier-Barrot S, Fontenille MJ, Meneret A, Vidailhet M et al. , 2013. The multiple faces of the ATP1A3-related dystonic movement disorder. Mov Disord. 28, 1457–1459. [DOI] [PubMed] [Google Scholar]
- Shiina N, Yamaguchi K, Tokunaga M, 2011. RNG105 deficiency impairs the dendritic localization of mRNAs for Na+/K+ ATPase subunit isoforms and leads to the degeneration of neuronal networks. Journal of Neuroscience 30, 12816–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C, 2009. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature 459, 446–450. [DOI] [PubMed] [Google Scholar]
- Simmons CQ, Thompson CH, Cawthon BE, Westlake G, Swoboda KJ, Kiskinis E et al. , 2018. Direct evidence of impaired neuronal Na/K-ATPase pump function in alternating hemiplegia of childhood. Neurobiol.Dis 115, 29–38. [DOI] [PubMed] [Google Scholar]
- Soga TM, Nakayama T, Inoue N, 2001. Expression and regulation of Na pump isoforms in cultured cerebellar granule cells. NeuroReport 12, 829–832. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ball WJ Jr., 1994. Identification of antigenic sites on the Na,K-ATPase β-subunit: their sequences and the effects of thiol reduction upon their structure. Biochimica et Biophysica Acta 1207, 236–248. [DOI] [PubMed] [Google Scholar]
- Swarts HG, Weigand KM, Venselaar H, van den Maagdenberg AM, Russel FG, Koenderink JB, 2013. Familial hemiplegic migraine mutations affect Na,K-ATPase domain interactions. Biochimica et Biophysica Acta 1832, 2173–2179. [DOI] [PubMed] [Google Scholar]
- Sweadner KJ, Arystarkhova E, Penniston JT, Swoboda KJ, Brashear A, Ozelius LJ, 2019. Genotype-structure-phenotype relationships diverge in paralogs ATP1A1, ATP1A2, and ATP1A3. Neurol.Genet, 5:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweadner KJ, Toro C, Whitlow CT, Snively BM, Ozelius LJ, Markello TC et al. , 2016. ATP1A3 mutation in adult rapid-onset ataxia. PLOS One 11, (3): e0151429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E, Clifford RJ, Kaplan JH, Sachs G, Vagin O, 2012. Subunit isoform selectivity in assembly of Na,K-ATPase alpha-beta heterodimers. Journal of Biological Chemistry 287, 26115–26125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E, Munson K, Sachs G, Vagin O, 2010. N-glycan-dependent quality control of the Na,K-ATPase β2 subunit. Biochemistry 49, 3116–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokhtaeva E, Sachs G, Vagin O, 2009. Assembly with the Na,K-ATPase α1 subunit is required for export of β1 and β2 subunits from the endoplasmic reticulum. Biochemistry 48, 11421–11431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toustrup-Jensen MS, Einholm AP, Schack VR, Nielsen HN, Holm R, Sobrido MJ et al. , 2014. Relationship between intracellular Na+ concentration and reduced Na+ affinity in Na+,K+-ATPase mutants causing neurological disease. Journal of Biological Chemistry 289, 3186–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranebjaerg L, Strenzke N, Lindholm S, Rendtorff ND, Poulsen H, Khandelia H et al. , 2018. The CAPOS mutation in ATP1A3 alters Na/K-ATPase function and results in auditory neuropathy which has implications for management. Hum.Genet 137, 111–127. [DOI] [PubMed] [Google Scholar]
- Vagin O, Tokhtaeva E, Yakubov I, Shevchenko E, Sachs G, 2008. Inverse correlation between the extent of N-glycan branching and intercellular adhesion in epithelia. Contribution of the Na,K-ATPase β1 subunit. Journal of Biological Chemistry 283, 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillend C, Mason SE, Cuttle MF, Alger BE, 2002. Mechanisms of neuronal hyperexcitability caused by partial inhibition of Na+-K+-ATPases in the rat CA1 hippocampal region. Journal of Neurophysiology 88, 2963–2978. [DOI] [PubMed] [Google Scholar]
- Viollet L, Glusman G, Murphy KJ, Newcomb TM, Reyna SP, Sweney M et al. , 2015. Alternating Hemiplegia of Childhood: Retrospective genetic study and genotype-phenotype correlations in 187 subjects from the US AHCF Registry. PLOS One 10, e0127045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk WI, de BP, Wichers CG, van den Berghe PV, van der Plaats R, Berger R et al. , 2012. The copper-transporting capacity of ATP7A mutants associated with Menkes disease is ameliorated by COMMD1 as a result of improved protein expression. Cell Mol.Life Sci 69, 149–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bruce AT, Tu C, Ma K, Zeng L, Zheng P et al. , 2011. Protein aggregation of SERCA2 mutants associated with Darier disease elicits ER stress and apoptosis in keratinocytes. Journal of Cell Science 124, 3568–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel RK, Arystarkhova E, Sweadner KJ, 1999. Cellular and subcellular specification of Na,K-ATPase α and β isoforms in the postnatal development of mouse retina. Journal of Neuroscience 19, 9878–9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Gao H, Zhang J, Xu X, Liu X, Wu X et al. , 2014. ATP1A3 mutations and genotype-phenotype correlation of alternating hemiplegia of childhood in Chinese patients. PLOS One 9, e97274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano ST, Silver K, Young R, DeBrosse SD, Ebel RS, Swoboda KJ et al. , 2017. Fever-Induced paroxysmal weakness and encephalopathy, a new phenotype of ATP1A3 mutation. Pediatr.Neurol 73, 101–105. [DOI] [PubMed] [Google Scholar]
- Zanotti-Fregonara P, Vidailhet M, Kas A, Ozelius LJ, Clot F, Hindie E et al. , 2008. [123l]-FP-CIT and [99mTc]-HMPAO single photon emission computed tomography in a new sporadic case of rapid-onset dystonia-parkinsonism. Journal of Neurological Science 273, 148–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.