Here, Reizel et al. investigated the FoxA factor's role in maintaining the regulatory network needed for liver development, and ablated all FoxA genes in the adult mouse liver. They found that loss of FoxA caused rapid and massive reduction in the expression of critical liver genes, and that FoxA proteins are be required for maintaining enhancer activity, chromatin accessibility, nucleosome positioning, and binding of HNF4α.
Keywords: pioneer factor, transcriptional network, winged helix protein
Abstract
The FoxA transcription factors are critical for liver development through their pioneering activity, which initiates a highly complex regulatory network thought to become progressively resistant to the loss of any individual hepatic transcription factor via mutual redundancy. To investigate the dispensability of FoxA factors for maintaining this regulatory network, we ablated all FoxA genes in the adult mouse liver. Remarkably, loss of FoxA caused rapid and massive reduction in the expression of critical liver genes. Activity of these genes was reduced back to the low levels of the fetal prehepatic endoderm stage, leading to necrosis and lethality within days. Mechanistically, we found FoxA proteins to be required for maintaining enhancer activity, chromatin accessibility, nucleosome positioning, and binding of HNF4α. Thus, the FoxA factors act continuously, guarding hepatic enhancer activity throughout adult life.
Developmental programs are composed of multiple successive signals that induce accurate and robust patterns of gene expression necessary for organ formation. The role of pioneer factors is to initiate transcriptional activity through nucleosome repositioning at cis-regulatory sequences (Zaret and Mango 2016). Nucleosome repositioning is essential for the recruitment of the full complement of cell type-specific transcription factors to a specific set of enhancers (Zaret and Mango 2016). Once full transcriptional activation is achieved in terminally differentiated cells, pioneer factors are considered unnecessary for the maintenance of transcriptional execution, either because they are replaced by other transcription factors or because these regulatory sequences are fully activated (Spitz and Furlong 2012). For example, PU.1 is essential for the establishment of hematopoiesis in embryonic life and in the maintenance of stem and progenitor cells in the adult (Scott et al. 1994; Iwasaki et al. 2005). However, it is not required for the function of mature B cells (Iwasaki et al. 2005). Similarly, Sox9 is essential for the establishment of hair follicles during development (Vidal et al. 2005) and for stem cell maintenance in the adult, but is not required in terminally differentiated cells (Kadaja et al. 2014). Likewise, in Drosophila melanogaster, Zelda is another pioneer factor required for transcriptional activation during early embryonic development and not at later stages, when it is replaced by other transcription factors occupying the same regulatory sequences (Liang et al. 2008).
The FoxA gene family of DNA-binding transcription factors consists of the three closely related FoxA1, FoxA2, and FoxA3 genes (originally known as hepatocyte nuclear factor 3 α, β, and γ, respectively). These genes play multiple important roles in organ development and function including in pancreas and gut development (Gao et al. 2007, 2008; Ye and Kaestner 2009) and are critical for the induction of the liver primordium from foregut endoderm (Lee et al. 2005; Friedman and Kaestner 2011). The FoxA proteins were previously defined as “paradigm pioneer factors” (Lee et al. 2005; Li et al. 2012; Iwafuchi-Doi et al. 2016) as they can bind to nucleosomal DNA both in vitro and in vivo, occupy cis-regulatory sequences before activation of their associated genes, and recruit ATP-dependent chromatin remodelers (Li et al. 2012; Iwafuchi-Doi et al. 2016). Circumstantial evidence suggests that FoxA proteins, similar to Zelda, Sox9, and PU.1, are not essential for maintaining the gene network of terminally differentiated cells. First, after FoxA proteins execute their pioneering activity during the prehepatoblast stage, additional transcription factors are progressively activated and often bind to the same enhancers already engaged by FoxA proteins, creating a highly complex network thought to be resistant to loss of binding by any specific hepatic transcription factor (Kyrmizi et al. 2006). The resilience of the regulatory network in the mature liver is evidenced by the fact that while depletion of HNF4α during fetal liver development results in lethality, ablation of the same transcription factor during adulthood does not cause death (Kyrmizi et al. 2006; Bonzo et al. 2012). A similar phenomenon was observed for C/EBPα, for which only its early ablation causes lethality (Inoue et al. 2004). Second, while FoxA1/A2 are critical for the formation of the liver primordium (Lee et al. 2005) and embryonic survival, their removal at later stages of development has a much milder phenotype that includes cholestasis under specific conditions (Li et al. 2009). Third, recently, Thakur et al. (2019) analyzed the epigenetic function of HNF4α and FoxA2 in the adult liver and concluded that FoxA2 has no significant role in maintaining enhancer activity. Fourth, although the FoxA proteins are abundant in the adult liver, FoxA1 and FoxA2 transcript levels are lower than they are in embryonic life (Supplemental Fig. 1A). Thus, the prevailing view suggests that FoxA proteins function mainly as competency factors to establish a stable enhancer network during embryonic development. In this study, we evaluated this concept by removing all FoxA proteins in the adult mouse liver and assaying network stability.
Results
FoxA proteins are required to maintain liver homeostasis
We depleted all three FoxA genes in 8-wk-old mice by injecting adeno-associated virus 8 (AAV8) carrying the gene for Cre recombinase under the control of the hepatocyte-specific thyroid-binding globulin (Tbg) promoter to adult FoxA1L/L/FoxA2L/L/FoxA3−/− mice (“FoxA triple null” below for short) (Fig. 1A) and validated FoxA depletion (Supplemental Fig. 1B,C). As controls, we injected adult FoxA1L/L/FoxA2L/L/FoxA3−/− mice with AAV8 expressing GFP also driven by the Tbg promoter (“control” in all figures unless specified alternatively). FoxA-triple-null mice started losing weight 15 d after virus injection. Twenty percent of these mice died within the next 4 d, and the remainder had to be sacrificed following ethical guidance (Fig. 1B). Livers were pale and severely damaged (Fig. 1C; Supplemental Fig. 2A) and plasma levels of the hepatocyte enzymes alanine aminotransferase and alkaline phosphatase were dramatically elevated, indicating severe liver injury (Fig. 1D). Acute midzonal coagulative necrosis was present in individual or clustered hepatocytes as well as multifocal hepatocellular swelling and clearing (Fig. 1E; Supplemental Fig. 2B) indicating internal liver failure. In addition, we observed multifocal random mixed lymphocyte infiltration, demonstrating that inflammation followed cell destruction and not the opposite. Interestingly, in liver sections of FoxA-triple-null mice taken 10 d after AAV-Cre injection, there was only mild multifocal hepatocellular atrophy with centrilobular hepatic cord hypercellularity, corresponding to a higher Ki67 labeling index at this stage (Fig. 1E; Supplemental Figs. 2B, 3). Overall, the striking phenotype following complete FoxA depletion in the mature liver demonstrates its necessity for liver homeostasis, indicating that the critical role of FoxA proteins is not restricted to early organ development.
Figure 1.
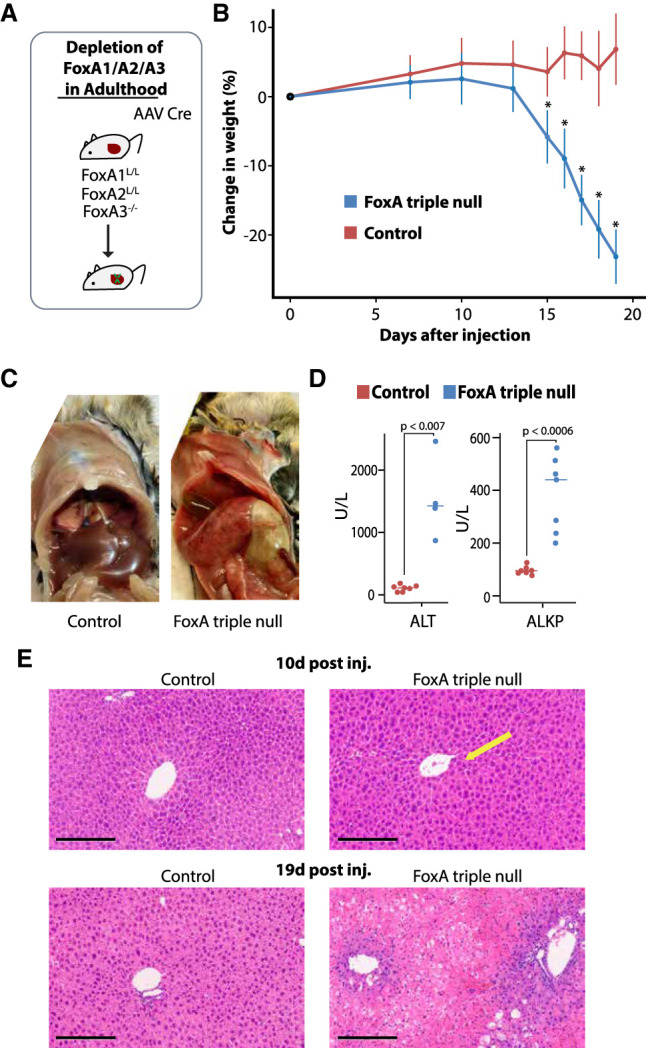
Mice deficient for FoxA proteins succumb to liver failure. (A) Description of gene ablation model. (B) Rapid weight loss after acute FoxA deletion. (*) P < 0.0005, Wilcoxon-Mann-Whitney test, n ≥ 8. (C) Liver anatomy of FoxA-triple-null mutants and controls. (D) Blood levels of ALT and ALKP (P < 0.007 for all comparisons, Wilcoxon-Mann-Whitney test, n ≥ 4). (E) Liver histology demonstrates a mild phenotype 10 d after AAV-Cre injection in the FoxA triple nulls compared with controls, but massive necrosis 19 d following injection. Scale bar, 200 μm. Arrow indicates hypercellularity.
FoxA ablation in adult mice eliminates expression of key liver genes
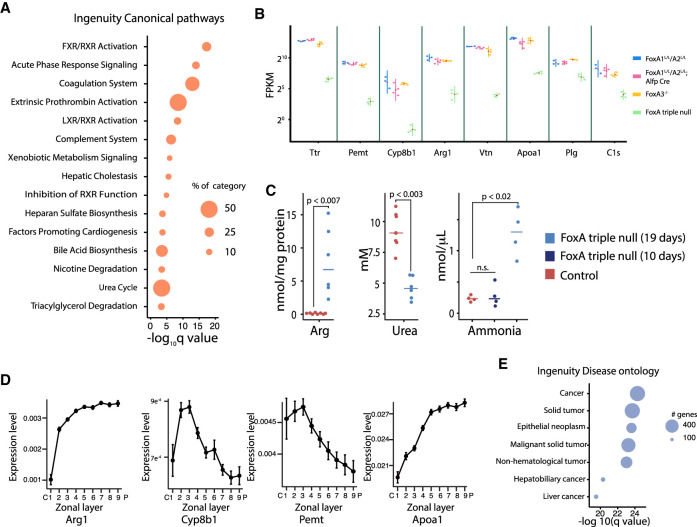
Next, we compared hepatocyte gene expression levels of FoxA-triple-null livers with controls or with mice lacking only FoxA1 and FoxA2 in the liver established previously (Li et al. 2009). We performed this analysis 7 d after AAV injection before hepatic necrosis had begun to capture only the most relevant and direct FoxA targets. We discovered 663 genes specifically down-regulated in the FoxA-triple-null livers but not in partially FoxA-depleted models (Supplemental Fig. 4A). Gene ontology analysis identified key liver processes associated with the down-regulated genes such as lipid metabolism, acute phase response, coagulation, and complement system, metabolism of xenobiotics, bile acid synthesis, and the urea cycle (Fig. 2A). Many key liver genes like albumin (Alb) and afamin (Afm) displayed reduced expression in the FoxA-null liver (Fig. 2B; Supplemental Fig. 4B), and a subset of these showed near total loss of steady state mRNA, with decreases of >30-fold (Fig. 2B). Among these was the transport protein transthyretin (Ttr) that carries thyroid hormone and retinol. The Ttr promoter contains the FoxA binding site that was used to purify the FoxA proteins (then termed “hepatocyte nuclear factor 3”) >30 yr ago (Costa et al. 1988), but only now is shown to be dependent on FoxA for gene activation (Fig. 2B). Among the FoxA-controlled genes, Arginase 1 (Arg1) is most relevant to the pathophysiology of the mutant mice. Arg1 catalyzes the last step of the urea cycle and is required for the hydrolysis of arginine to ornithine and urea. Importantly, its expression is reduced 40-fold in FoxA mutant mice (Fig. 2B). Complete liver-specific deficiency for Arg1 is lethal due to hyperammonemia and liver damage (Iyer et al. 2002; Sin et al. 2013). Consistent with the observed dramatic reduction of Arg1 expression, FoxA-triple-null mice also exhibit hyperargininemia, hypouremia, and hyperammonemia, similar to pathologies present in Arg1-null mice. Therefore, we attribute the lethality of the FoxA-triple-null mice to this defect in Arg1 expression (Fig. 2C; Supplemental Fig. 5D). Moreover, no elevation in ammonia levels was observed 10 d after Cre injection when gene expression was already perturbed but no phenotype was observed, indicating that elevated ammonia levels are associated with the severe phenotype that becomes evident at later stages (Fig. 2C). We found additional key metabolic genes with massively reduced expression such as Pemt, which encodes an enzyme that converts phosphatidylethanolamine to phosphatidylcholine; Apoa1, which is a major component of HDL particles in the plasma (Li et al. 2019); and Cyp8b1, which controls the balance between cholic acid and chenodeoxycholic acid (Fig. 2B). The fact that additional key metabolic genes controlling cholesterol homeostasis are also strongly FoxA dependent is reflected in significantly altered lipid and carnitine levels, which likely contribute to the severe liver damage in FoxA deficient mice (Supplemental Fig. 5). Interestingly, we found that the midzone of the hepatic lobule, which displays the most significant pathology (Fig. 1E; Supplemental Fig. 2B), exhibits the highest expression levels of genes completely dependent on FoxA, including Arg1, Pemt, Cyp8b1, and Apoa1, as determined by zonation gene expression profiling (Fig. 2D; zonal expression data were reused with permission from Halpern et al. (2017), suggesting contribution of these key metabolic genes to the severe midzonal damage.
Figure 2.
Key liver gene expression is dependent on FoxA proteins in the adult liver. (A) Canonical pathways associated with down-regulated genes in the FoxA-triple-null mutants. (B) Examples of key hepatic genes down-regulated in the FoxA-triple-null liver. Note the log scale (red triangle represents the mean, adjusted P-value < 10−40 for all comparisons between FoxA partial depletion or controls and FoxA triple nulls). (C) Hepatic levels of arginine and urea and blood levels of ammonia 10 and 19 d after Cre injection in FoxA triple nulls and controls (Wilcoxon-Mann-Whitney test, n ≥ 4). (D) Zonal distribution of four key genes down-regulated in the FoxA-triple-null mutants showing high expression in the midzonal region (data reused with permission from Halpern et al. 2017). (E) Gene ontology of combined up-regulated and down-regulated genes using ingenuity pathway analysis.
We also found 513 genes to be up-regulated in FoxA-triple-null livers (Supplemental Fig. 4C,D). Interestingly, gene ontology analysis of the up-regulated genes indicated that many of these are associated with cancer and related proliferative pathways (Fig. 2E). This gene signature explains the hepatic hypercellularity and increased proliferation rate immediately following AAV-Cre administration reported above (Fig. 1E; Supplemental Fig. 3) and likely reflects the attempt of the injured liver to restore functional tissue mass. Of these up-regulated genes, only 23% were highly expressed during early development, and gene ontology analysis suggests that these genes are regulated by transcription factors associated with proliferation (Supplemental Fig. 6A–D). Activation of the proliferative response is likely a secondary effect to the loss of functional liver mass as a result of ongoing liver injury. The remaining 77% of up-regulated genes are not developmentally controlled and are associated with nuclear receptors. Thus, most of the up-regulated genes likely represent an indirect effect of FoxA depletion (Supplemental Fig. 6A–D).
FoxA proteins maintain the activation state of a set of key liver genes
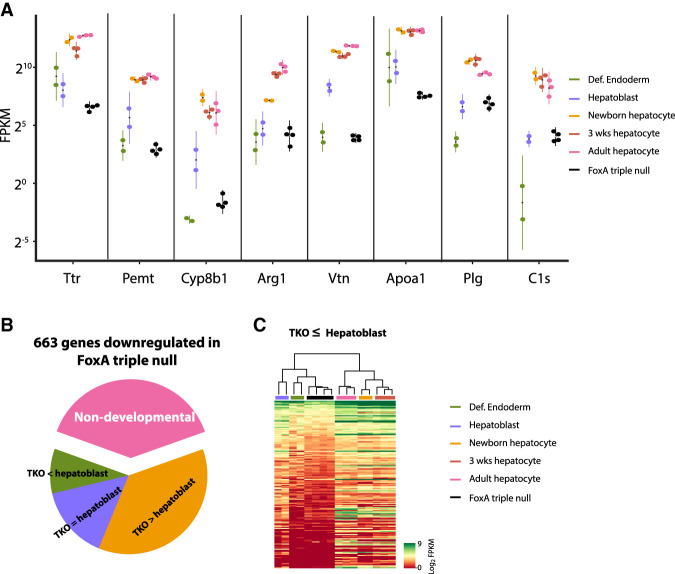
Because FoxA proteins are central to the induction of gene expression during early prehepatic endoderm development (Lee et al. 2005), we asked how many of the genes down-regulated in the FoxA-triple-null model are developmentally induced. Strikingly, we found that all genes with a >30-fold decrease in the mutants (Fig. 2B) are massively activated during liver development, and in most cases, their expression levels in the adult FoxA-triple-null liver are as low as they are normally only in the prehepatic endodermal stage (gene expression of endodermal and hepatoblast were obtained from Nicetto et al. [2019] and early postnatal hepatocytes from Reizel et al. [2018]) (Fig. 3A). When we extended this analysis to all 663 significantly down-regulated genes, we found that 63% of these are developmentally induced and 24% are deactivated in the triple-null mutants back to fetal levels (Fig. 3B,C). Overall, our transcriptome analysis demonstrates that the FoxA proteins are continuously required to maintain the expression of a specific set of crucial developmentally induced genes.
Figure 3.
Adult ablation of FoxA factors reduces expression of key liver genes back to early developmental stages. (A) Expression of FoxA target genes is reduced back to the levels of early fetal stages following acute FoxA ablation. (Def.) Definitive. All gene expression levels are significantly lower in the FoxA-triple-null compared with hepatoblasts (adjusted P-value < 0.001) except for Arg1, Plg, and C1s. (B) The majority of FoxA-dependent genes in the adult liver are induced during liver development, and many are deactivated in the FoxA-triple-null liver back to total hepatoblast levels (adjusted P-value < 0.01) (for details, see the Materials and Methods). (TKO) FoxA-triple-null mutants. (C) Heat map demonstrating genes down-regulated back to hepatic or prehepatic stage in the FoxA-triple-null mutants.
FoxA-binding sites are associated with down-regulated genes
To determine the association between FoxA-binding sites and down-regulated genes in FoxA-triple-null mice, we focused on FoxA3-binding sites in liver deficient for FoxA1 and FoxA2 (for explanation, see the Materials and Methods; Supplemental Fig. 7A–C, for convenience we symbolized these sites as FoxA3*, ChIP-seq data was obtained from Iwafuchi-Doi et al. (2016) (GSE57559). We found enrichment of Foxa3* binding in the promoters of genes down-regulated in the FoxA-triple-null livers (Supplemental Fig. 7B,D). We also show a significant association between FoxA3* distal regulatory regions and the down-regulated genes using the HiC map of liver chromatin generated by Kim et al. (2018) (GSE104129) (Supplemental Fig. 7E,F).
HNF4α binding is dependent on the presence of FoxA at cobound sites
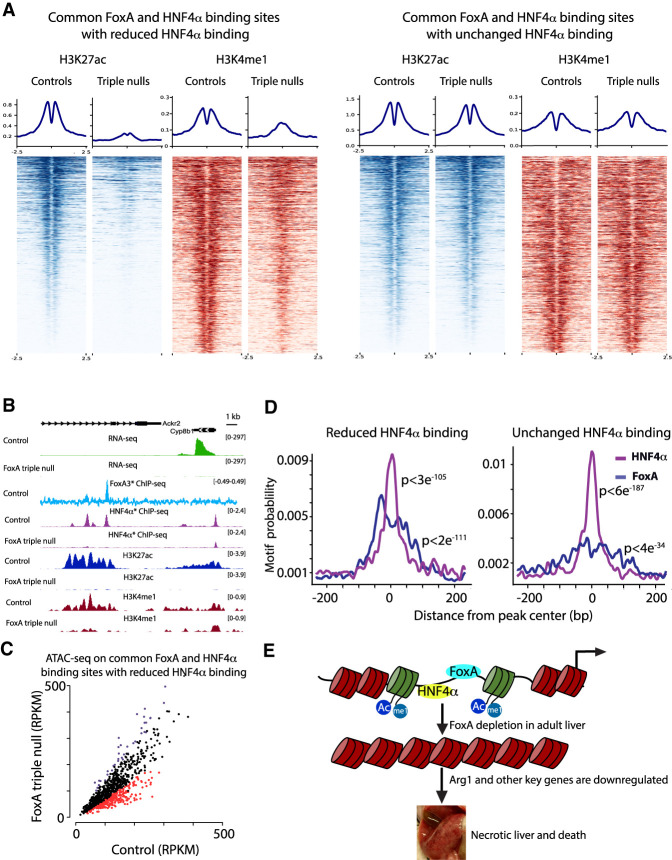
Since FoxA proteins play a key role in chromatin remodeling during development (Li et al. 2012; Zaret and Mango 2016), we investigated whether the FoxA proteins act only as pioneer factors during fetal organogenesis or whether they are continuously required for the hepatic gene regulatory network at the terminally differentiated state. We focused on the effects of FoxA depletion on HNF4α binding because HNF4α is known to have a key role in hepatic maintenance and binds to most hepatic enhancers. Indeed, 61% of FoxA-binding sites (FoxA3*) are also occupied by HNF4α (Fig. 4A) and 37% of genes down-regulated in the FoxA-triple-null livers are also HNF4α-dependent (Fig. 4B; Supplemental Fig. 8A). In order to examine the effect of FoxA depletion in the adult liver on HNF4α binding, we performed ChIP-seq for HNF4α on FoxA-triple-null and control livers. We note that HNF4α protein levels are maintained in FoxA-triple-null livers, excluding the possibility that reduced protein levels explains the loss of HNF4α binding (Supplemental Fig. 8B). We found HNF4α occupancy at binding sites not shared with FoxA binding sites to be similar between mutants and controls, as expected (Fig. 4C). However, at FoxA sites (FoxA3*) cobound by HNF4α, HNF4α occupancy was dramatically reduced at 40.6% percent of the sites, especially those with relatively weak HNF4α binding (Fig. 4D). Thus, FoxA is required not only during the establishment of the hepatic primordium for the generation of the hepatic enhancer landscape, but also for maintaining the adult hepatic regulatory network by constantly enabling HNF4α binding.
Figure 4.
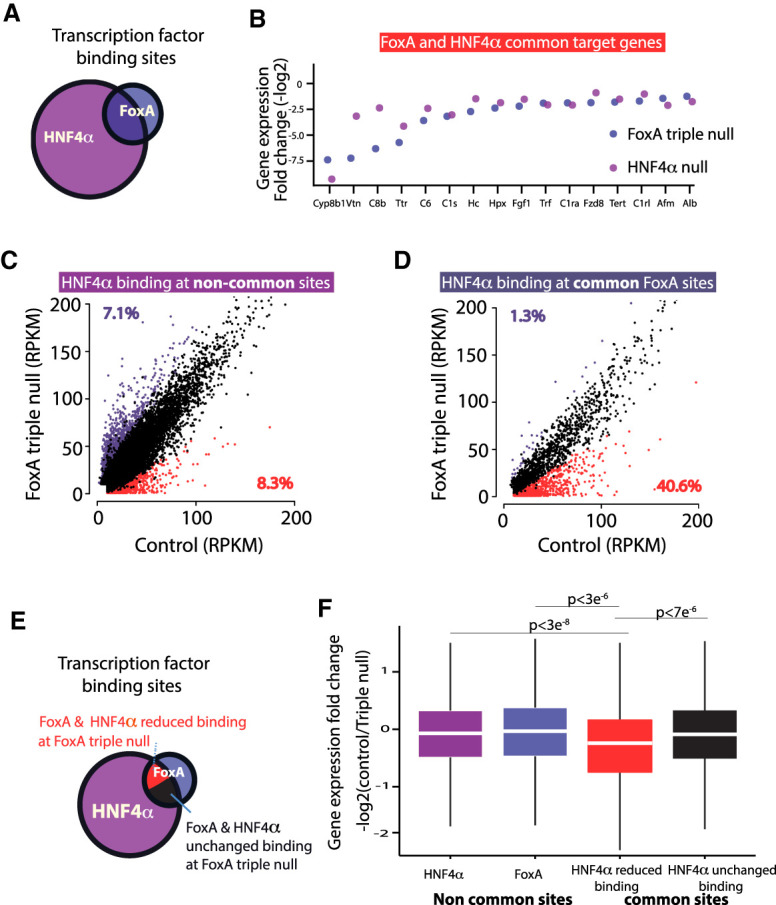
HNF4α binding is dependent on FoxA proteins at cobound sites. (A) Venn diagram of FoxA (FoxA3*) and HNF4α-binding sites in the adult liver. (B) Transcript levels of key liver genes down-regulated in the FoxA-triple-null mutants or in adult livers depleted for HNF4α. Presented is fold change of mutants compared with controls for both models. (C) Scatter plot of HNF4α binding values ([RPKM] reads per kilobase per million) of FoxA triple nulls compared with controls at HNF4α sites that do not overlap with FoxA. (D) Scatter plot of HNF4α binding values at sites with FoxA occupancy. For C and D, increased or decreased binding is purple or red, respectively. FDR < 0.05. Each group is the mean of four samples. (E) Venn diagram showing the proportion of four groups of binding sites: noncommon sites of either HNF4α or FoxA proteins, common sites that show no loss of HNF4α occupancy, and common sites with loss of HNF4α binding. (F) Box plot demonstrating fold change of the nearest gene in the FoxA triple nulls compared with controls for each of the groups indicated in E (Wilcoxon-Mann-Whitney test).
Next, we examined the association between the reduction of HNF4α occupancy at cobound sites and gene expression. For this purpose, we divided the FoxA- and HNF4α-binding sites into four classes: noncommon sites bound only by HNF4α, noncommon sites bound only by FoxA, common sites that show no loss of HNF4α occupancy, and common sites with loss of HNF4α binding (Fig. 4E). Strikingly, only common sites with reduced HNF4α binding showed a statistically significant decrease in expression of nearest genes (Fig. 4F). This finding demonstrates that FoxA generates its strongest effects on gene expression in the adult liver at enhancers also bound by HNF4α.
FoxA maintains enhancer activity, nucleosome positioning, and chromatin accessibility at associated enhancers
Next, we performed ChIP-seq on H3K27ac and H3K4me1, the two main chromatin markers associated with enhancer activity, on the livers of FoxA-triple-null mutants and controls. We focused on HNF4α- and FoxA-cobound sites and divided those into two categories: sites with loss of HNF4α binding and sites with unchanged HNF4α binding in FoxA-triple-null mutants. Strikingly, we found that 69.5% of FoxA/HNF4α-cobound regions with loss of HNF4α exhibit a dramatically reduced H3K27ac signal, while only 8% of cobound sites with unchanged HNF4α occupancy display reduced H3K27ac signal (FDR < 0.05) (Fig. 5A,B; Supplemental Fig. 9). In addition, we showed that 20.1% of cobound sites with loss of HNF4α binding have a reduced signal for the H3K4me1 modification. Strikingly, the H3K4me1 mark, clearly present on two nucleosomes flanking the transcription factor binding sites in the control liver as expected for primed or active enhancers (Fig. 5A), collapsed onto a single nucleosome in the vast majority of the FoxA/HNF4α cobound enhancers (Fig. 5A), demonstrating a shift in nucleosome positioning. However, of the cobound sites with unchanged HNF4α binding, only 1.9% displayed a reduced H3K4me1 signal, and no change in nucleosome positioning was observed (FDR < 0.05) (Fig. 5A,B; Supplemental Fig. 9). Next, we performed ATAC-seq for the mapping of accessible chromatin in order to address the mechanism by which FoxA maintains enhancer activity. We found reduced accessibility at 34.4% of FoxA- and HNF4α-cobound sites that lost HNF4α binding in the mutants (FDR < 0.05) (Fig. 5C; Supplemental Fig. 10A–C), while only 0.4% of sites with no decrease in HNF4α binding displayed reduced accessibility (Supplemental Fig. 10A–C). Interestingly, the very minor reduction in accessibility was associated with complete loss of the H3K27ac signal (Supplemental Fig. 10B). Thus, the FoxA proteins are continuously required to maintain nucleosome positioning and active enhancer marks at regulatory sequences of key genes necessary for liver viability, establishing the FoxA factors as active enhancer guards.
Figure 5.
FoxA is required to maintain active enhancers, nucleosome positioning, and chromatin accessibility at cobound HNF4α/FoxA-binding sites. (A) Heat map and quantification of H3K27ac and H3K4me1 signal at HNF4α/FoxA common binding sites that show no loss of HNF4α occupancy, and common sites with loss of HNF4α binding (n = 2, for controls or mutants). (B) HNF4α/FoxA-cobound site next to the Cyp8b1 gene in which loss of FoxA binding results in loss of expression, loss of HNF4α binding, and altered H3K27ac and H4K4me1 signals. (C) Scatter plot of ATAC-seq RPKM values in FoxA-triple-null livers compared with controls at cobound HNF4α/FoxA sites that show loss of HNF4α binding. Decreased or increased accessibility sites are indicated as red or purple dots, respectively. FDR < 0.05 (n = 3 for controls or mutants). (D) Motif frequency as a function of distance from peak center (base pairs) of the binding sites indicated in A using Centrimo. (E) Model for the role of FoxA proteins in maintaining enhancer activity, chromatin accessibility, and gene expression in the adult liver.
Since we observed a strong effect of FoxA on H3K27 acetylation and HNF4α binding at certain sites but not in others, we performed motif frequency analysis using Centrimo, as we hypothesized that binding site density could be a determining factor differentiating between enhancers that are sensitive to FoxA loss versus those that are resistant. Indeed, we found that the stable FoxA/HNF4α enhancers showed a much higher frequency for the HNF4α motif over the FoxA motif (Fig. 5D), while those that had lost HNF4α binding and enhancer marks displayed equal frequencies of the two binding elements.
Finally, we wondered what additional transcription factor binding motifs are enriched at FoxA regulated enhancers. Therefore, we compared all FoxA3* sites that in FoxA-triple-null mutants lost or maintained chromatin accessibility as determined by ATAC-seq. We found higher enrichment for CEBP, NF1, and Erra consensus binding sites at FoxA3* sites that lost chromatin accessibility in the mutants, whereas PPARα and RAR binding sites were enriched at FoxA3* sites with unchanged accessibility in the triple-null livers (Supplemental Fig. 11A). Likewise, similar factors were identified when we focused our analysis only on FoxA3*/HNF4α-cobound sites that lost H3K27ac mark in the mutants and performed motif enrichment on subgroups either with reduced or unchanged accessibility (Supplemental Fig. 11B). Interestingly, CEBPβ binding was previously shown to be sensitive to nearby FoxA occupancy (Iwafuchi-Doi et al. 2016), in agreement with the data reported here.
Discussion
In this study, we discovered a set of critical metabolic pathways and genes that are fully dependent on FoxA proteins in terminally differentiated hepatocytes and that are necessary for liver function and survival. Many of these key genes, including Arg1, Plg, and Pemt, were never before described as FoxA targets. Thus, the FoxA proteins are not acting exclusively as competency factors required during liver development, but also continuously maintain critical hepatic functions in the adult. We also established that the hepatic gene regulatory network is not resilient to FoxA loss. We suggest a model in which FoxA proteins are continuously required to maintain enhancer activity and nucleosome positioning at enhancers with a high density of FoxA motifs. FoxA binding enables occupancy by other transcription factors and without FoxA proteins many critical enhancers collapse, leading to near complete loss of gene activity (Fig. 5E).
FoxA is known to play a key role in the development of many organs including the pancreas, intestine, and lung (Wan et al. 2005; Gao et al. 2008; Ye and Kaestner 2009). It is possible that in each of these organs removal of the FoxA proteins will result in lethality and organ degradation. If correct, this work demonstrates a phenomenon that is applicable for many biological systems and could identify additional metabolic pathways and gene networks regulated by FoxA proteins and required for organ function. Moreover, in the present study, we investigated the role of FoxA in normal homeostasis using complete deletion. Partial depletion of FoxA was shown to increase fibrosis and steatosis in the human liver (Moya et al. 2012; Dong et al. 2017; Wang et al. 2017). It would be of interest to examine the effects of complete FoxA depletion on these pathologies and hepatocellular carcinoma.
Our finding that the paradigm pioneer factor is continuously required to maintain chromatin accessibility in the mature organ suggests that this could be the case for other mammalian transcriptional regulators. This is unlike Zelda, which acts exclusively to restrict enhancer activation at early developmental stages flies, or the mammalian Sox9 and PU.1 pioneer factors that are dispensable for the function of fully differentiated tissues. Thus, FoxA proteins function not only as pioneers to establish cell type-specific patterns of gene activation, but also as “settlers” that continuously maintain the enhancer landscape.
Materials and methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pennsylvania. The derivation of the FoxA1loxP/loxP;FoxA2 loxP/loxP;FoxA3−/− as well as HNF4αloxP/loxP mice has been reported previously (Kaestner et al. 1998; Sund et al. 2000; Parviz et al. 2002; Gao et al. 2008). Male mice at ages 8–12 wk were jugular vein-injected with 1011 particles of AAV expressing either Cre recombinase (Addgene 107787-AAV8) or GFP (Addgene 105535-AAV8) under the control of the hepatocyte-specific thyroid-binding globulin (Tbg) promoter. Mice were genotyped by PCR of tail DNA as described in the above-mentioned papers. Validation of HNF4α depletion was shown by Armour et al. (2017).
Metabolite measurements
Either plasma samples and/or a neutralized perchloric acid (PCA) extract prepared from liver were used for metabolite measurements. The concentration of amino acids was determined by the Agilent 1260 HPLC system, using precolumn derivatization with o-phthalaldehyde (Nissim et al. 2012, 2014). Organic acid levels were determined by the isotope dilution approach and GC-MS system (Weinberg et al. 2000). Carnitine, Acyl-Carnitine, and b-OH-butyrate levels were determined by the Agilent LC/MS 6410 Triple Quad system as described (Nissim et al. 2012). Total plasma urea level was determined as described in Nissim et al. (2014) and protein levels in liver extracts were determined as detailed in Nissim et al. (2012, 2014). Hypercellularity was determined by a veterinary pathologist blinded to the genotype of the mice.
Western blot analysis
About 30 mg of frozen liver was added to 500 μL of RIPA buffer (150 mM NaCl, 50 mM Tris-HCl at pH 8.0, 1% Triton X-100, 1 mM PMSF, 0.5% sodium deoxycholate, 0.1% SDS, 1× cOmplete protease inhibitor cocktail). Tissue was homogenized in a Tissue Lyser II (Qiagen) using 5-mm stainless steel beads (Qiagen) at 20 Hz for 4 min. Following homogenization, the samples were sonicated using a standard Bioruptor (Diagenode UCD-200) with the following parameters: intensity = high (H), multitimer = off: 30 sec, on: 30 sec, total timer = 2.5 min. Next, the samples were incubated for 30 min at 4°C with rotation. Following incubation, samples were centrifuged at 15,000 RCF for 15 min and the supernatant was transferred to a new tube. Then, protein quantification using BCA protein assay kit (Thermo Fisher) was completed. Fifty micrograms of protein was brought up to 6.5 μL with DDW and added to 2.5 μL of 4× LDS sample buffer (Thermo Fisher) and 1 μL of Nupage reducing agent (Thermo Fisher). Samples were denatured for 10 min at 70°C and loaded onto a precast 4%–12% mini bis-tris gel (Thermo Fisher) using the XCell SureLock Mini-Cell system (Thermo Fisher). Samples were run with 1× MOPS SDS running buffer (Thermo Fisher) with 500 μL of antioxidant (Thermo Fisher). The gel was run at a constant 200 V for 50 min. Proteins were transferred to a PVDF membrane (Bio-Rad) using a semiwet method. One liter of transfer buffer was prepared with 50 mL of 20× Nupage transfer buffer (Thermo Fisher), 100 mL of methanol, 1 mL of antioxidant, and 849 mL of H2O. Gels were transferred using XCell SureLock minicell system at 30 V for 1 h. Following the transfer, the membrane was washed with TBST (1× TBS [Bio-Rad], 0.1% Tween-20) for 1 h and subsequently blocked with 5% nonfat dry milk (Labscientific) in TBST for 1 h. Following blocking, the membrane was washed in TBST for 30 min and incubated with the primary antibody diluted at a ratio of 1:1000 in 5% milk in TBST overnight at 4°C. The following antibodies were used: anti-FOXA1 (Abcam ab23738), anti-FoxA2/HNF3β (D56D6) XP Rabbit mAb (Cell Signaling Technology 8186), and anti-HNF4a (Abcam ab181604). The following day, the membrane was washed in TBST for 30 min and then incubated with the secondary antibody diluted at a ratio of 1:5000 in 5% nonfat dry milk in TBST for 1 h at room temperature. Then, the membrane was washed in TBST for 30 min, and protein signal was measured by chemiluminescence with the application of West Dura extended duration substrate (Thermo Fisher). Following development, the membrane was washed in TBST for 30 min, changed to fresh TBST, and stripped in Western blot stripping buffer (Thermo Fisher) for 45 min at room temperature. The membrane was then reprobed with an anti-α Tubulin (Santa Cruz Biotechnologies sc-23948) antibody following the same protocol.
Immunohistochemistry
Paraformaldehyde-fixed liver tissues were paraffin-embedded and sectioned. To deparaffinize tissue, slides were incubated for 15 min at 56°C and then submerged twice in xylene for 5 min each. Tissue sections were rehydrated, quickly dipped in H2O, and then incubated in PBS for 5 min. For antigen retrieval, slides were submerged in 1× R-buffer A (Electron Microscopy Sciences) and loaded into a 2100 Antigen Retriever (Proteogenix). After reaching the target temperature, slides were allowed to cool for 2 h. Slides were rinsed with running water for 5 min and then incubated in PBST (1× PBS, 0.1% Tween-20) for 5 min. Slides were then incubated three times in PBS for 5 min each. Slides were submerged in 3% H2O2 for 15 min to quench peroxidases, rinsed with gentle running water for 5 min, and placed in PBS. To block the tissue, Avidin D blocking reagent (Vector Laboratories) was applied to the slide for 15 min at room temperature. Following the incubation, slides were placed in PBS. Biotin blocking reagent (Vector Laboratories) was applied to the tissue, and slides were again incubated for 15 min at room temperature and then washed in PBS. CAS-Block (Thermo Fisher) was applied to the tissue, and the slides were incubated for 10 min at room temperature. Ki-67 antibody (ThermoFisher RM-9106) was diluted in CAS Block, applied to the tissue, and incubated overnight at 4°C in a humidified chamber. The following day, slides were incubated in PBST for 5 min and then twice in PBS for 5 min. Secondary antibody was diluted at a ratio of 1:200 in CAS Block, applied to the tissue for 30 min at 37°C, and then washed twice in PBS for 5 min. ABC HRP reagent (Vector Laboratories PK-7100) was applied to tissue, and slides were incubated for 30 min in a humidified chamber at 37°C. Slides were washed in PBS twice for 5 min. Using the DAB substrate kit for peroxidase (Vector Laboratories), the substrate working solution was applied to tissue and signal development was monitored under a microscope. Slides were rinsed with running water for 10 min to stop the reaction. To counterstain tissue, slides were dipped in hematoxylin solution (Sigma GHS216) and then rinsed with running water for 5 min. Tissue sections were dehydrated and then mounted using Cytoseal XYL (Thermo Fisher).
RNA extraction and library preparation
A maximum of 500,000 hepatocytes was homogenized in TRIzol reagent (Thermo Fisher) and incubated at room temperature. Next, chloroform purification (Millipore Sigma C2432) was performed followed by ethanol precipitation. RNA was then purified using the RNeasy minikit (Qiagen) and eluted with DDW. RNA was quantified using a NanoDrop-1000 (Thermo Fisher), and RNA quality was measured using a BioAnalyzer RNA Nano Chip (Agilent). Libraries were prepared using the Ultra RNA library preparation kit for Illumina (New England BioLabs) with 150 ng of input RNA. Libraries were sequenced on the Illumina HiSeq 4000. RNA was extracted from the following genotypes: FoxA1loxP/loxP; FoxA2loxP/loxP, FoxA1loxP/loxP; FoxA2loxP/loxP; Cre-Alfp, FoxA1loxP/loxP; FoxA2loxP/loxP; FoxA3−/− injected with AAV GFP, and FoxA1loxP/loxP; FoxA2loxP/loxP; FoxA3−/− injected with AAV Cre.
ChIP-seq
About 130 mg of frozen liver was minced in 2 mL of cold 1× DPBS (Thermo Fisher 14080055). The volumes of liver samples were brought up to 10 mL of 1% formaldehyde in 1× DPBS. The samples were incubated for 8 min at room temperature with rotation. To quench the reaction, the sample was brought to 0.125 M glycine and incubated for 5 min at room temperature with rotation. Samples were centrifuged at 2500 RCF for 2 min at 4°C, the supernatant was discarded, and the pellet was washed once with 1× DPBS. The pellet was resuspended in 1 mL of cold ChIP cell lysis buffer (10 mM Tris-HCl at pH 8.0, 10 mM NaCl, 3mM MgCl2, 0.5% Igepal CA-630, 1× cOmplete protease inhibitor cocktail) and transferred to a 10-mL tissue grinder on ice. The samples were homogenized with a smooth Teflon pestle 20 times and incubated for 5 min at 4°C. Following incubation, the samples were centrifuged at 17,000 RCF for 5 min at 4°C to pellet nuclei. The supernatant was discarded, and nuclei were resuspended in 1 mL of cold ChIP nuclear lysis buffer (50 mM Tris-HCl at pH 8.0, 5 mM EDTA, 1% SDS, 1× cOmplete protease inhibitor cocktail). The samples were sonicated for two rounds using the Standard Bioruptor (Diagenode UCD-200) with the following parameters: intensity = high (H), multitimer = off: 30 sec, on: 30 sec, total timer = 7.5 min with maintaining 4°C. The Bioruptor was cooled down for 15 min between rounds. Following sonication, the samples were centrifuged at 17,000 RCF for 10 min at 4°C, and the supernatant containing sheared chromatin was recovered.
To determine the amount of material required for immunoprecipitation, we isolated DNA from sonicated chromatin. DNA was quantified using a NanoDrop-1000 (Thermo Fisher) and run on a BioAnalyzer high-sensitivity DNA chip. Following quantification, 10 μg of sonicated chromatin was added to 1 mL of ChIP dilution buffer (16 mM Tris-HCl at pH 8.0, 167 mM NaCl, 0.01% SDS, 1.1% Triton-X100, 1× cOmplete protease inhibitor cocktail). Three micrograms of the following antibodies was added to the samples: H3K4me1(Abcam ab8895), H3K27ac (Active Motif 39133), and HNF4a (Abcam ab181604). The samples were incubated overnight at 4°C with rotation. Forty microliters of recombinant Protein G agarose beads (Thermo Fisher) was washed three times with 1 mL of ChIP dilution buffer. Following the washes, the beads were resuspended in 75 μL of ChIP dilution buffer with 5 μL of BSA (New England BioLabs) and incubated overnight at 4°C with rotation for blocking. Next, chromatin samples were added to the blocked beads and the samples were incubated for 1 h at 4°C with rotation. Samples were centrifuged at 100 RCF for 30 sec, and the supernatant was discarded. The beads were then washed sequentially with each of the following buffers by resuspending the beads in 1 mL of buffer, incubating beads for 5 min at room temperature with rotation, pelleting beads, and discarding supernatant: TSE I (20 mM Tris-HCl at pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), TSE II (20 mM Tris-HCl at pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), ChIP buffer III (10 mM Tris-HCl at pH 8.0, 0.25 M LiCl, 1 mM EDTA, 1% IGEPAL CA-630, 1% sodium deoxycholate), and TE (10 mM Tris-HCl at pH 8.0, 1 mM EDTA). Following the washes, the beads were resuspended in 100 μL of elution buffer (0.1 M NaHCO3, 1% SDS) and incubated for 15 min at room temperature with rotation. Four microliters of 5 M NaCl was added to the supernatant, and the samples were then incubated overnight at 65°C. DNA was then purified from the samples.
Enrichment of IP samples was measured through qPCR using the following program: 3 min at 95°C followed by 40 cycles of 5 sec at 95°C and 20 sec at 60°C, with the primers indicated below. Then, libraries were prepared using the NEBNext Ultra II DNA library preparation kit for Illumina. Libraries were sequenced on the Illumina HiSeq 4000.
The HNF1α promoter used was F-GCACTTGCAAGGCTGAAGTC and R-ATTGGAGCTGGGGAAATTCT. The 40S used was F-AGCGAGCTGTGCTGAAGTTT and R-AGGCTGCTTGGATCTGGTTA.
ATAC-seq
About 50 mg of frozen livers was minced in 1 mL of cold swelling buffer (10 mM Tris-HCl at pH 7.5, 2 mM MgCl2, 3 mM CaCl2). Tissue was homogenized with a Dounce tissue grinder (DWK Life Sciences 357544) using the loose pestle 10 times and was then incubated for 20 min at 4°C. Following incubation, the sample was homogenized 20 times with the tight pestle. Three milliliters of cold swelling buffer was added to the lysate, and the sample was passed through a 70-μm cell strainer (Corning). The lysate was centrifuged at 600 RCF for 20 min at 4°C. The supernatant was discarded, and the pellet was resuspended in 1.8 mL of swelling buffer and 200 μL of glycerol. While vortexing, 2 mL of lysis buffer (1× swelling buffer, 10% glycerol, 1% Igepal CA-630) was added drop by drop to the lysate, and the lysate was incubated for 5 min at 4°C. Following incubation, 5 mL of lysis buffer was added to the sample, and the sample was centrifuged at 700 RCF for 13 min at 4°C. The supernatant was discarded, and the nuclei were resuspended in 2 mL of 1× DPBS (Thermo Fisher 14080055). Nuclei were counted using a hemocytometer, 135,000 nuclei were added to a new 1.5-mL microcentrifuge tube, and nuclei were pelleted by centrifugation at 900 RCF for 10 min at 4°C. The supernatant was discarded, and nuclei were resuspended in 75 μL of 2× TD buffer (Illumina), 49.5 μL of 1× DPBS, 7.5 μL Tn5 Transposase (Illumina), 1.5 μL of 10% Tween-20, 1.5 μL of 1% digitonin, and 15 μL of DDW. Nuclei were incubated for 30 min at 37°C. Following incubation, DNA was purified using the MinElute reaction cleanup kit (Qiagen), and eluted in 10 μL of EB. Twenty-five microliters of high-fidelity 2× PCR master mix (New England BioLabs), 2.5 µL of indexing primer, and 10 μL of DDW were added to the 10 μL of purified DNA and the DNA was amplified five cycles using the following PCR program: 5 min at 72°C, 30 sec at 98°C, and five cycles of 10 sec at 98°C, 30 sec at 63°C, and 1 min at 72°C. Next, 5 μL of the partially amplified library was removed and used to determine the appropriate number of additional cycles by qPCR with the following mix: 3.85 μL of DDW, 0.5 μL of indexed primer, 0.15 μL of SYBR Green I nucleic acid gel stain diluted 1:100 (Thermo Fisher), and 5 μL of High-Fidelity 2× PCR master mix. We used the following qPCR program: 30 sec at 98°C, 20 cycles of 10 sec at 98°C, 30 sec at 63°C, and 1 min at 72°C. Additional cycles were determined as one-third of the plateau. The remaining 45 μL of the partially amplified library was further amplified accordingly. Double-sided size selection was performed using Ampure XP beads (Beckman Coulter) by adding 22.5 μL of beads to the library and keeping the supernatant. The supernatant was transferred to another tube and then 58.8 μL of beads was added. The beads were kept and resuspended in 20 μL of DDW, from which the final library was eluted. Libraries were sequenced on the Illumina HiSeq 4000.
Data analysis
References for FoxA-binding sites indicated in this study
While FoxA1 and FoxA2 are nearly identical in amino acid sequence and in their cistromes, FoxA3 has diverged during vertebrate evolution, which is reflected by a partially divergent hepatic cistrome (Iwafuchi-Doi et al. 2016). Thus, FoxA3 has both unique binding sites and many others shared with FoxA1/A2. Interestingly, mice lacking only hepatic FoxA1/A2 have a much milder phenotype compared with the FoxA-triple-null mutants (Li et al. 2009). We reasoned that FoxA3 must be able to partially compensate for loss of FoxA1/A2. Indeed, the number of FoxA3 binding sites doubles in the FoxA1/A2 mutant mice through occupancy of sites normally bound by FoxA1/2 (from 1998 to 3732), indicative of enhancer switching (for convenience, FoxA3-binding sites occupied in FoxA1/A2-null livers are symbolized as FoxA3*) (FoxA3 ChIP-seq data are from Zaret and Mango 2016). Here, we determined that the FoxA-triple-null mutant phenotype is lethal and identify a set of down-regulated genes that is specific to this model and not down-regulated in partially depleted FoxA models. Thus, enhancers still bound by FoxA3 in FoxA1/A2-null livers are likely critical for hepatic survival and are the most relevant for studying the relationship between FoxA binding and down-regulated genes in the FoxA-triple-null mutants. Indeed, we found a positive association between FoxA3*-binding sites and down-regulated genes in the FoxA-triple-null mutants (as shown in Supplemental Fig. 6). Therefore, we report FoxA3*-binding sites as the most relevant binding sites to the observed phenotype. Thus, the low number of FoxA-binding sites reported here are due to our strategy of focusing only on FoxA3* sites.
RNA-seq
RNA-seq reads were mapped to the mouse genome (mm9) using STAR version 2.5.2a (Dobin et al. 2013) with the flags “–readFilesCommand zcat –outSAMtype BAM SortedByCoordinate –outSAMstrandField intronMotif.” SAMtools view version 1.1 was used to remove duplicate reads with the flag “-bhq 255.”
Cufflinks v2.2.1 (Trapnell et al. 2013) was used for gene expression quantification. We used the following flags: “-max-mle-iterations 500000 ‐‐max-bundle-frags 100000000.” The reads were mapped against GENCODE vM1. Then, we ran the DESeq2 package on R in order to estimate differential expression significance. We kept genes with FPKM >1. Differentially expressed genes were defined as genes with adjusted P-value <0.01 and fold change >1.5. RNA-seq data of hepatoblasts and endoderm were derived from Nicetto et al. (2019), and newborn and 20-d-old hepatocyte data were derived from Reizel et al. (2018). Heat maps for either up-regulated or down-regulated genes were produced with ggplot2 using Heatmap.2. Log2 of all FPKM was calculated, and for better visualization, log2FPKM values >9 or <0 were attributed as 9 or 0, respectively. To analyze gene ontology, a list of either down-regulated or up-regulated genes and their log2 fold change compared with controls was input into Ingenuity Pathway Analysis (Qiagen). Motif frequency analysis was completed with CentriMo (Bailey and Machanick 2012). Gene example figures were made with ggplot2 on log2FPKM values. Zonal expression data were derived from Halpern et al. (2017).
ChIP-seq
Single-end sequencing reads were trimmed with Cutadapt and aligned to the mouse genome (assembly mm9) using Bowtie2 version 2.3.4.1. PCR duplicates were removed using picard.jar tool and the command MarkDuplicates with the following flags: “REMOVE_DUPLICATES = true VALIDATION_STRINGENCY = LENIENT.” Peak calling was done with MACS2 version 2.1.1.20160309, and peak calling for HNF4α was done on united files of all control samples using default MACS2 parameters. In order to compare differential binding between controls and FoxA-triple-null mutants, we used DiffBind version 2.10.0 (Ross-Innes et al. 2012). Peaks were defined as ±250 bp from peak center. Normalization was done with DBA_SCORE_RPKM_FOLD and DB.DESeq2 was used to find statistically significant differences between groups. We took only binding sites with an RPKM average >10 in controls or FoxA-triple-null mutatns, and counted as significant only sites with FDR <0.05. The same parameters were used for HNF4α-binding comparison, H3K27ac, H3K4me1, and ATAC-seq signal.
Bam files were converted to Bedgraph files after normalizing to 1 million reads per library using BEDtools (“genomecov”) and then converted to bigwig format using UCSC toolkit (“bedGraphToBigWig”). For control and FoxA-triple-null groups, an average bigwig file was also generated from the replicates (“bigWigMerge“). Bigwig files were loaded on the WashU browser for visualization. Deeptools (version 2.5.7, “computeMatrix” and “plotHeatmap”) was used to generate enrichment heat maps surrounding selected regions. Motif frequency analysis was performed using Centrimo version 1.5.0. (Bailey and Machanick 2012).
ATAC-seq
Paired-end sequencing reads were aligned to the mouse genome (assembly mm9) using Bowtie2 allowing ends to be soft clipped. Only properly paired alignments were retained for downstream analysis. PCR duplicates were removed using SAMtools (“rmdup”). Reads mapped to mitochondria and ENCODE blacklist regions were removed. All reads aligning to the positive strand were offset by +4 bp, and all reads aligning to the negative strand were offset −5 bp. Comparison between controls and FoxA-triple-null mutants was conducted using DiffBind in a similar way to ChIP-seq processing. Quality control metrics were evaluated before alignment using FastQC (version 0.11.8) and after alignment using QualiMap (version 2.2) to ensure comparable quality of sequencing libraries.
Nearest genes analysis
To analyze association between FoxA/HNF4α-binding sites and down-regulated genes in the FoxA-triple-null mutants, we identified the nearest genes using Homer AnnotatePeaks.pl. Then, we filtered out genes with a distance >1 Mb and genes with RPKM values <1. We chose 1 Mb as a cutoff because HiC analysis showed that at this distance there is still enrichment between FoxA3*-binding sites and the promoters of down-regulated genes. For each gene, −log2 of fold change between FoxA-triple-null mutants over controls was calculated. Identical amounts of genes were randomly sampled for each group.
HiC analysis
In order to determine the association between FoxA-binding sites and down-regulated genes in the FoxA-triple-null mutants using HiC maps, we took advantage of liver HiC maps generated by Kim et al. (2018). We computed the signal between FoxA binding sites and the TSS of the above-mentioned genes using the following approach. We binned the genome into 5-kb tiles, each with specific interaction scores, measuring interaction with other tiles on the same chromosome. We took all score values that connect tiles overlapping with FoxA3* with tiles of promoters of down-regulated or unchanged genes. Statistical tests were calculated on all possible interactions.
Distribution of materials, data, and code
Distribution of materials
Mouse strains are available from the Mutant Mouse Resource and Research Center.
Deep sequencing data
All data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE140423. A secure token has been created to allow the review of record GSE140423 while it remains in private status (ehyvssccprgbxub).
Supplementary Material
Acknowledgments
We thank Dr. M. Lazar for critical suggestions on the manuscript and the Lazar laboratory for the RNA-seq data from HNF4α-deficient livers. We thank J. Liu and additional members of our laboratory for technical help and fruitful discussions, and H.M. Kolev for critical reading of the manuscript. We thank Dr. Itzhak Nissim, Y. Daikhin, O. Horyn, and Dr. Ilana Nissim for performing the analysis of urea, protein, amino and organic acids, carnitine, and acyl-carnitine levels at the Metabolomics Core Facility at the Children's Hospital of Philadelphia. We thank Dr. C.A. Assenmacher for pathology analysis of liver sections and providing relevant images. We acknowledge Dr. J. Schug for his advice on bioinformatics analysis, and thank Nicholas J. Hand for fruitful discussions. This work was supported by R01 DK102667. We thank the University of Pennsylvania Diabetes Research Center for the use of the Functional Genomics Core (P30 DK19525) and the Center for Molecular Studies in Digestive and Liver Diseases (P30 DK050306) for the use of the Molecular Pathology and Imaging Core.
Author contributions: Y.R. and K.H.K. conceived the study and wrote the manuscript. Y.R., A.M., and E.L.W. performed the methodology. Y.R. and A.M. were responsible for the resources. Y.R., L.G., Y.L., E.M., and K.H.K. curated and visualized the data. K.H.K. and A.W.W. supervised the study. K.H.K. acquired the funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.337691.120.
References
- Armour SM, Remsberg JR, Damle M, Sidoli S, Ho WY, Li Z, Garcia BA, Lazar MA. 2017. An HDAC3–PROX1 corepressor module acts on HNF4α to control hepatic triglycerides. Nat Commun 8: 549 10.1038/s41467-017-00772-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Machanick P. 2012. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. 2012. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem 287: 7345–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RH, Lai E, Grayson DR, Darnell JE. 1988. The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol Cell Biol 8: 81–90. 10.1128/MCB.8.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Yang Y, Shen Z, Zheng C, Jin Z, Huang Y, Zhang Z, Zheng S, Chen G. 2017. Forkhead box A3 attenuated the progression of fibrosis in a rat model of biliary atresia. Cell Death Dis 8: e2719 10.1038/cddis.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. 2011. On the origin of the liver. J Clin Invest 121: 4630–4633. 10.1172/JCI59652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, White P, Doliba N, Golson ML, Matschinsky FM, Kaestner KH. 2007. Foxa2 controls vesicle docking and insulin secretion in mature β cells. Cell Metab 6: 267–279. 10.1016/j.cmet.2007.08.015 [DOI] [PubMed] [Google Scholar]
- Gao N, Lelay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. 2008. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev 22: 3435–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern KB, Shenhav R, Matcovitch-Natan O, Tóth B, Lemze D, Golan M, Massasa EE, Baydatch S, Landen S, Moor AE, et al. 2017. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542: 352–356. 10.1038/nature21065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ. 2004. Disruption of hepatic C/EBPα results in impaired glucose tolerance and age-dependent hepatosteatosis. J Biol Chem 279: 44740–44748. [DOI] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62: 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Somoza C, Shigematsu H, Duprez EA, Iwasaki-Arai J, Mizuno S, Arinobu Y, Geary K, Zhang P, Dayaram T, et al. 2005. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood 106: 1590–1600. 10.1182/blood-2005-03-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RK, Yoo PK, Kern RM, Rozengurt N, Tsoa R, O'Brien WE, Yu H, Grody WW, Cederbaum SD. 2002. Mouse model for human arginase deficiency. Mol Cell Biol 22: 4491–4498. 10.1128/MCB.22.13.4491-4498.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaja M, Keyes BE, Lin M, Pasolli HA, Genander M, Polak L, Stokes N, Zheng D, Fuchs E. 2014. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev 28: 328–341. 10.1101/gad.233247.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestner KH, Hiemisch H, Schütz G. 1998. Targeted disruption of the gene encoding hepatocyte nuclear factor 3γ results in reduced transcription of hepatocyte-specific genes. Mol Cell Biol 18: 4245–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Marhon SA, Zhang Y, Steger DJ, Won K-J, Lazar MA. 2018. Rev-erbα dynamically modulates chromatin looping to control circadian gene transcription. Science 359: 1274–1277. 10.1126/science.aao6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. 2006. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20: 2293–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Friedman JR, Fulmer JT, Kaestner KH. 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947. 10.1038/nature03649 [DOI] [PubMed] [Google Scholar]
- Li Z, White P, Tuteja G, Rubins N, Sackett S, Kaestner KH. 2009. Foxa1 and Foxa2 regulate bile duct development in mice. J Clin Invest 119: 1537–1545. 10.1172/JCI38201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151: 1608–1616. 10.1016/j.cell.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Xu Y, Jadhav K, Zhu Y, Yin L, Zhang Y. 2019. Hepatic forkhead box protein A3 regulates ApoA-I (Apolipoprotein A-I) expression, cholesterol efflux, and atherogenesis. Arterioscler Thromb Vasc Biol 39: 1574–1587. 10.1161/ATVBAHA.119.312610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya M, Benet M, Guzmán C, Tolosa L, García-Monzón C, Pareja E, Castell JV, Jover R. 2012. Foxa1 reduces lipid accumulation in human hepatocytes and is down-regulated in nonalcoholic fatty liver. PLoS One 7: e30014 10.1371/journal.pone.0030014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, et al. 2019. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 363: 294–297. 10.1126/science.aau0583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim I, Horyn O, Nissim I, Daikhin Y, Wehrli SL, Yudkoff M, Matschinsky FM. 2012. Effects of a glucokinase activator on hepatic intermediary metabolism: study with 13C-isotopomer-based metabolomics. Biochem J 444: 537–551. 10.1042/BJ20120163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim I, Horyn O, Daikhin Y, Chen P, Li C, Wehrli SL, Nissim I, Yudkoff M. 2014. The molecular and metabolic influence of long term agmatine consumption. J Biol Chem 289: 9710–9729. 10.1074/jbc.M113.544726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F, Li J, Kaestner KH, Duncan SA. 2002. Generation of a conditionally null allele ofhnf4α. Genesis 32: 130–133. [DOI] [PubMed] [Google Scholar]
- Reizel Y, Sabag O, Skversky Y, Spiro A, Steinberg B, Bernstein D, Wang A, Kieckhaefer J, Li C, Pikarsky E, et al. 2018. Postnatal DNA demethylation and its role in tissue maturation. Nat Commun 9: 2040 10.1038/s41467-018-04456-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265: 1573–1577. 10.1126/science.8079170 [DOI] [PubMed] [Google Scholar]
- Sin YY, Ballantyne LL, Mukherjee K, St. Amand T, Kyriakopoulou L, Schulze A, Funk CD. 2013. Inducible Arginase 1 deficiency in mice leads to hyperargininemia and altered amino acid metabolism. PLoS One 8: e80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Furlong EEM. 2012. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics 13: 613–626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Sund NJ, Ang SL, Sackett SD, Shen W, Daigle N, Magnuson MA, Kaestner KH. 2000. Hepatocyte nuclear factor 3β (Foxa2) is dispensable for maintaining the differentiated state of the adult hepatocyte. Mol Cell Biol 20: 5175–5183. 10.1128/MCB.20.14.5175-5183.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A, Wong JCH, Wang EY, Lotto J, Kim D, Cheng JC, Mingay M, Cullum R, Moudgil V, Ahmed N, et al. 2019. Hepatocyte nuclear factor 4-α is essential for the active epigenetic state at enhancers in mouse liver. Hepatology 70: 1360–1376. 10.1002/hep.30631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal VP, Chaboissier MC, Lützkendorf S, Cotsarelis G, Mill P, Hui CC, Ortonne N, Ortonne JP, Schedl A. 2005. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr Biol 15: 1340–1351. 10.1016/j.cub.2005.06.064 [DOI] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang S-L, Wert S, Stahlman MT, Whitsett JA. 2005. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem 280: 13809–13816. 10.1074/jbc.M414122200 [DOI] [PubMed] [Google Scholar]
- Wang W, Yao LJ, Shen W, Ding K, Shi PM, Chen F, He J, Ding J, Zhang X, Xie WF. 2017. FOXA2 alleviates CCl4-induced liver fibrosis by protecting hepatocytes in mice. Sci Rep 7: 15532 10.1038/s41598-017-15831-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg JM, Venkatachalam MA, Roeser NF, Nissim I. 2000. Mitochondrial dysfunction during hypoxia/reoxygenation and its correction by anaerobic metabolism of citric acid cycle intermediates. Proc Natl Acad Sci 97: 2826–2831. 10.1073/pnas.97.6.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye DZ, Kaestner KH. 2009. Foxa1 and Foxa2 control the differentiation of goblet and enteroendocrine L- and D-cells in mice. Gastroenterology 137: 2052–2062. 10.1053/j.gastro.2009.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Mango SE. 2016. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev 37: 76–81. 10.1016/j.gde.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.