Abstract
Changes in gene expression drive novel phenotypes, raising interest in how gene expression evolves. In contrast to the static genome, cells modulate gene expression in response to changing environments. Previous comparative studies focused on specific conditions, describing interspecies variation in expression levels, but providing limited information about variation across different conditions. To close this gap, we profiled mRNA levels of two related yeast species in hundreds of conditions and used coexpression analysis to distinguish variation in the dynamic pattern of gene expression from variation in expression levels. The majority of genes whose expression varied between the species maintained a conserved dynamic pattern. Cases of diverged dynamic pattern correspond to genes that were induced under distinct subsets of conditions in the two species. Profiling the interspecific hybrid allowed us to distinguish between genes with predominantly cis- or trans-regulatory variation. We find that trans-varying alleles are dominantly inherited, and that cis-variations are often complemented by variations in trans. Based on these results, we suggest that gene expression diverges primarily through changes in expression levels, but does not alter the pattern by which these levels are dynamically regulated.
New phenotypes emerge from mutations that change protein function or protein expression. Between closely related species, protein sequences remain highly conserved while gene regulatory regions vary, suggesting that changes in gene expression play key roles in early evolutionary divergence (King and Wilson 1975). In support of that, modifying the expression of a single, differentiation-promoting gene during organism development can give rise to novel body morphologies (Warren et al. 1994; Prud'homme et al. 2006). Although transcription programs of closely related species are in large conserved (Gilad et al. 2006; Paris et al. 2013; Wong et al. 2015), understanding the basic mechanisms of regulatory variation is of considerable interest (Yvert et al. 2003; Wittkopp et al. 2004, 2008; Landry et al. 2005; Tirosh et al. 2006, 2009; Yanai and Hunter 2009; McManus et al. 2010; Goncalves et al. 2012; Shi et al. 2012; Combs et al. 2018).
A hallmark of gene expression is its ability to adapt to changing demands. Widespread changes in gene expression occur when internal or external conditions change, allowing organisms to adapt and modify their cellular response. Therefore, expression levels measured at one condition provide limited information about expression levels at other conditions. Furthermore, determinants of expression level are often different from determinants of dynamic gene regulation. The former depends on the efficiency of the general transcriptional machinery, which is influenced, for example, by the presence of a TATA box or the nucleosome organization along the promoter (Basehoar et al. 2004; Tirosh and Barkai 2008). The latter depends on the binding of specific transcription factors (TFs) to the gene promoter. The common observation is that two genes can be expressed at a very different level, but at the same time be regulated by the same set of TFs and follow precisely the same pattern of dynamic changes when compared across conditions. Conversely, two genes can be expressed at a very similar level in most conditions, but change in expression in opposite directions owing to small environmental fluctuations, indicating the genes are subject to different regulation.
The ability to separately control expression level and expression dynamics implies that evolution can work on these two properties independently. Distinguishing between these two axes of divergence is fundamental for understanding the mechanisms of evolution. We reasoned that analysis of regulatory evolution, on a genomic scale, requires an extended comparative data set that surveys a large number of expression profiles obtained under a wide range of conditions. Following this reasoning, we profiled mRNA levels of two closely related budding yeast species and their interspecific hybrid under hundreds of conditions, which include different media and genetic perturbations, and devised computational approaches for distinguishing variations in expression levels from variations in coexpression, which resemble variations in regulatory dynamics. We further describe the cases of variation in dynamic regulation and provide insights into their mode of inheritance and the potential mechanism underlying this divergence.
Results
A compendium of comparative transcription profiles
The budding yeasts Saccharomyces cerevisiae and Saccharomyces paradoxus emerged as a model for studying interspecies variation in gene expression (Tirosh et al. 2009; Emerson et al. 2010; Artieri and Fraser 2014; McManus et al. 2014; Metzger et al. 2016; Yue et al. 2017; Weiss et al. 2018). The two species diverged approximately five million years ago, contain the same set of genes at a conserved synteny, and show 90% and 80% sequence identities at coding and noncoding regions, respectively (Scannell et al. 2011). Further, the two species can readily mate to form viable hybrids. In the context of comparative analysis, hybrids provide a key tool for classifying the genetic basis of interspecies variation: the two hybrid alleles are subject to the same trans environment so that differences in their expression must result from gene-linked differences (cis effects). Any further differences between the parents that are lost in the hybrid are attributed to variation in upstream, diffusible factors (trans effects) (Wittkopp et al. 2004, 2008; Tirosh et al. 2009, 2010; Emerson et al. 2010; McManus et al. 2010; Dori-Bachash et al. 2011; Goncalves et al. 2012; Shi et al. 2012; Artieri and Fraser 2014; Metzger et al. 2016; Combs et al. 2018).
Previous studies described interspecies variation in gene expression when compared at individual conditions. We extended this analysis by generating a large collection of comparative transcription profiles surveying a wide array of conditions (Fig. 1A; Supplemental Table S3). The larger fraction of our data set comprises a time course of transition from rich (YPD) to nitrogen-depleted media, in which we sampled wild-type (WT) strains, as well as strains that were individually deleted in 46 transcription factors (Supplemental Table S1). In addition, we profiled WT cells in three time-course experiments: progression through the cell cycle (sampled every 5 min for 3 h), transition to phosphate-depleted media (sampled every 15 min for 6 h), and progression along the growth curve on YPD (sampled every hour for a day). In total, we generated 530–570 expression profiles for each of the two species and the hybrid (Supplemental Table S3). The aforementioned experimental conditions were sampled with repeats and result in 462 samples that are comparable between species. Quality controls for the RNA-seq data set are detailed in Supplemental Note 1, Supplemental Figures S8–S10, and Supplemental Tables S4 and S5.
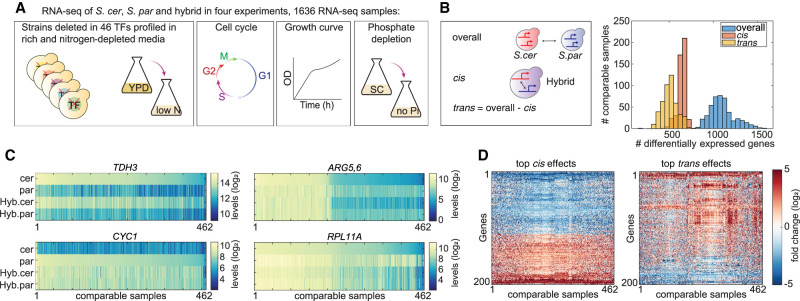
Figure 1.
Generating a compendium of comparative transcription profiles. (A) Experimental scheme. The transcription profiles of S. cerevisiae (S. cer), S. paradoxus (S. par), and the interspecific hybrid were measured in four experiments (Methods; Supplemental Table S3). Overall, 1636 samples were acquired, composed of 530–570 samples per species. The “YPD to low nitrogen” experiment was divided into three time points, each comprising 130 samples per species. (B) Hundreds of genes vary in expression level between species in each individual condition. Differentially expressed genes between species (overall), between hybrid alleles (cis), and in trans (overall-cis) were defined via DESeq2 per experiment (log2 fold change > 1, adjusted P-value < 0.05). Presented here is the distribution of genes that pass the fold change threshold in each condition and pass the P-value threshold in at least one experiment. (C) Examples of cis- and trans-varying genes: steady cis effect (TDH3), steady trans effect (CYC1), condition-dependent cis effect (ARG5,6), and condition-dependent trans effect (RPL11A). Shown are the log2-transformed expression levels of the indicated genes in the species and hybrid alleles, as measured in all samples in our data set. Samples were sorted according to expression levels in S. cerevisiae in TDH3, ARG5,6, and RPL11A, and by S. paradoxus in CYC1. (D) Consistency of cis and trans effects across conditions. Shown are log2 fold changes in cis (left) and trans (right) of the top 200 cis- and trans-affected genes.
We first aimed to evaluate global features of our data set relevant to comparative studies: the number of differentially expressed genes, the genetic basis of the variations (cis or trans), and their dependency on the environment. Consistent with previous reports (Tirosh et al. 2009; Artieri and Fraser 2014; McManus et al. 2014), about 1100 genes significantly differed in expression level between the two species at each individual condition, with fold change >2 and P-value < 0.05 (Fig. 1B; Supplemental Fig. S1A,B). A large fraction (640 of 1100, 58%) of these variations remained significant in the hybrid, indicating their origin in cis-acting mutations. To avoid errors in the estimation of the trans effect, we made the computation via a cross-replicate comparison (Supplemental Fig. S1C; Fraser 2019). On the average condition, 640 genes showed a significant greater than twofold change between the hybrid alleles (cis effect) and 500 genes showed same-magnitude trans effect. Cis and trans effects can act in the same direction, being up-regulated in a certain species and in its corresponding allele in the hybrid (reinforcing interaction) or in opposite directions (compensating interaction). We find that the two types of interactions are equally likely and relatively infrequent: on average, reinforcing interactions appeared in 60 genes and compensating interactions appeared in 70 genes (Supplemental Fig. S1D).
Some variations in expression level were consistent across conditions, whereas others were specific to a subset of conditions, as shown in Figure 1C. In general, cis effects were more reproducible across conditions (Fig. 1D; Supplemental Fig. S1B), consistent with previous reports (Smith and Kruglyak 2008; McManus et al. 2010; GTEx Consortium 2017). Genes that contain a TATA box in their promoter tend to vary in expression over evolutionary time (Tirosh et al. 2006; Landry et al. 2007; Hagai et al. 2018). We find that both cis and trans effects were highly enriched within TATA box–containing genes (Supplemental Fig. S1E). We conclude that when considering each condition individually, the pattern of expression divergence described by our data is consistent with previous reports and different subsets of genes often vary in different conditions.
Quantifying species similarity in gene regulatory dynamics by comparing coexpression patterns
Our main goal in generating this large compendium of comparative expression profiles was to assess changes in the dynamic pattern by which gene expression is regulated, that is, the pattern by which its expression varies upon genetic and environmental perturbations. To perform this analysis, we first considered the gene expression level across all conditions. Comparing this pattern directly, however, introduces differences that arise not only from changes in the transcriptional network, but from differences in environmental sensing acting upstream to the network (Supplemental Note 2; Supplemental Fig. S11). We therefore decided to measure the similarity in dynamic regulation of orthologs by comparing their coexpression pattern with all other genes in the genome, a measure we term RC (Fig. 2A). Coexpression reports on the regulatory relationships of genes within the transcriptional network. Genes that are coexpressed will show a similar pattern of induction or repression across conditions and will therefore show the same pattern of correlation with other genes in the genome. Genes that are coexpressed with a similar set of genes in the two species will receive a high RC similarity score. Indeed, coexpression is often preferred when analyzing dynamic regulation within a given species (Hughes et al. 2000; Segal et al. 2003; Ihmels et al. 2005a; Li et al. 2017) because it is less sensitive to random noise affecting the two genes being compared, or to the precise set of conditions in which expression was measured. We applied our coexpression analysis on external gene expression data sets and found that most of the genes (87%–96%) had positive RC score with our S. cerevisiae data set, with consistent coexpression of gene modules, confirming relative invariance of this measure to the precise data set used (Supplemental Note 3; Supplemental Fig. S12).
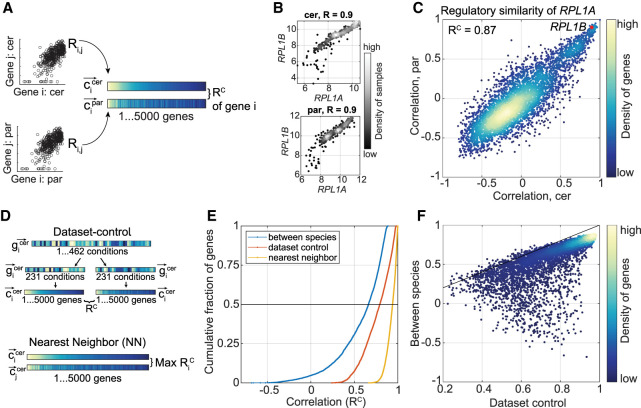
Figure 2.
Gene coexpression as a measure of regulatory similarity. (A) RC measures regulatory similarity between species. First, the pairwise correlation of gene i with all other expressed genes is measured per species. Then, the correlation of these coexpression vectors is measured, termed RC. (B) Ribosomal protein genes are highly coexpressed in both species. Shown are log2-transformed expression levels of the indicated genes in S. cerevisiae and in S. paradoxus across conditions. Pearson correlation coefficients are indicated. (C) RPL1A is regulated similarly in both species. Presented are coexpression vectors of RPL1A in S. cerevisiae (x-axis) and in S. paradoxus (y-axis), each vector comprised of 4772 genes, correlation of these (RC) is indicated. The RC value is written in the top left of the scatter plot. (D) Within data set controls. Two within data set controls were defined per species, as shown. First, data set control is obtained by splitting the set of conditions into two random subsets and measuring the gene regulatory similarity (RC) between them. Second, nearest neighbors (NN) are defined as the pair of genes with the most similar coexpression vectors. (E) Distribution of regulatory similarity scores. Shown is the cumulative distribution of the indicated measures. As NN and data set control were defined per species, here we present the mean score of the two species. (F) Control RC correlates with comparative RC. Each dot represents the RC score of a single gene, between species (y-axis) and the mean data set control of the two species (x-axis). Black line is x = y.
To test our measure of regulatory similarity, we first considered the ribosomal protein-coding genes, which both species co-induce during rapid growth. Two genes of this group, RPL1A and RPL1B, were highly coexpressed in each species in the measured conditions (Fig. 2B). Therefore, RPL1A received a high regulatory similarity score between species (RC = 0.87) (Fig. 2C). Similarly, RPL1B received RC = 0.84 (Supplemental Table S7). Examining the RC score of all orthologs, we find that half of the orthologs were assigned regulatory similarity scores higher than 0.65 (Fig. 2E). Still, the absence of genes with RC > 0.9 suggests a systematic bias caused by diverging genes; indeed, 10% of the orthologs have low to negative similarity scores (RC < 0.2 in 479/4772 genes).
We reasoned that some of these low similarity scores result from real divergence in the dynamic regulation of the respective gene between the two species, but others may result from low expression levels or inconsistent coexpression pattern of the gene within each species. To control for these possibilities, we considered two control measures for the reliability of the coexpression pattern in each species (Fig. 2D). First, we ran a bootstrapping analysis on the subset of each species separately, which we refer to as data set control. For each species, we split the expression profiles subset into two random parts, generate coexpression vectors for each gene in each half, and compute the correlation between these vectors. We repeated this process for 10 permutations, the average of these is the RC of data set control (Fig. 2D, top). Second, we measure the correlation of coexpression vectors within each species to define the nearest neighbors (NN), namely, pairs of genes with the most similar coexpression vector (Fig. 2D, bottom). As expected, these two control measures are highly correlated (R = 0.91) (Supplemental Fig. S2A). Thus, although in the between-species comparison half of the orthologs received a similarity score lower than 0.65, half of the data set control or NNs scores were above 0.79 and 0.93, respectively (Fig. 2E). Both controls and comparative measures were not dictated by the gene expression level or dynamic range, because lowly expressed genes did not typically show low similarity scores (Supplemental Fig. S2B,C). In general, genes receiving a low control score also received a low similarity score in our between-species comparison (Fig. 2F). However, a small number of genes deviate from this trend by presenting a low to negative comparative score, and a high control score (394 genes with RC between species <0.2 and RC data set control > 0.5) (Supplemental Table S7). Therefore, such cases most likely indicate regulatory divergence of these genes between species. Together, this suggests that although the majority of orthologous genes showed a high similarity score, interpreted as conserved regulation, variation can still be detected in a significant number of genes.
Variation in regulatory dynamics is distinct from variation in expression levels
Our analysis provided us with two measures of expression divergence: the common measure that quantifies the difference in expression levels, as measured in each individual condition, and another measure that quantifies differences in the pattern of gene regulatory dynamics across conditions. These two measures are related, because changes in the pattern of gene induction will modulate expression levels in respective conditions. Yet, these two parameters can also be modulated independently, for example, through regulatory mutations that affect absolute induction levels but do not alter the identity of the regulating factors. We therefore wished to characterize the extent to which these properties are related in our data set. To this end, we compared the divergence in median expression level to divergence in gene dynamic regulation. This analysis revealed that the two properties are distinct, showing no correlation (R = 0.011, P-value = 0.4) (Fig. 3A). This lack of correlation was observed for both cis- and trans-variations (R = −0.02, P-value = 0.17 [cis], R = 0.08, P-value < 1 × 10−10 [trans]) (Fig. 3B,C) and was further supported by examining specific expression modules. Stress-induced genes, for example, varied in expression levels (log2 fold change = 1.11 ± 0.8 mean ± SD) but maintained invariant regulatory dynamics in the two species (RC = 0.81 ± 0.09). In contrast, genes coding for the mitochondrial ribosomal proteins were expressed at similar levels by the two species (log2 fold change = 0.62 ± 0.34), yet they showed low regulatory similarity (RC = 0.29 ± 0.34) (Fig. 3A; Supplemental Fig. S3A).
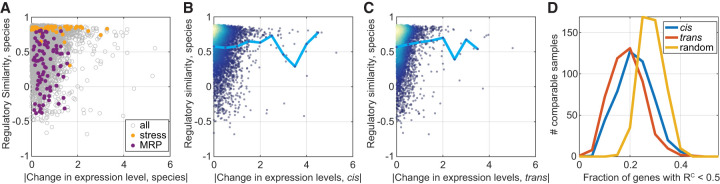
Figure 3.
Variation in regulatory dynamics is distinct from variation in expression levels. (A) Regulatory similarity is independent of changes in expression level. Presented are between-species RC scores per gene as a function of the absolute of log2 fold change in expression level between species (median of the 462 comparable samples). Expression modules: stress induced and mitochondrial ribosomal proteins (MRP) are indicated in color. Modules were defined in Ihmels et al. (2002). The correlation between the measures is R = 0.011, P-value = 0.4. (B,C) Regulatory similarity is independent of cis and trans effects. Same as in A, presented are RC as a function of the median cis effect (B) or trans effect (C). Cyan line is the mean RC value per bin. The correlation coefficients between measures are R = −0.02, P-value = 0.17 (B), and R = 0.08, P-value < 1 × 10−10 (C). Color represents density. (D) Differentially expressed genes show a conserved regulatory similarity. We considered the top 100 cis- and trans-varying genes in expression level at each comparable sample and measured the fraction that shows low regulatory similarity (<0.5). Shown are the respective distributions compared to a control in which random genes were picked per comparable sample. ANOVA test for the three distributions result in P-value = 1 × 10−113, ANOVA test for the cis- and trans-distributions result in P-value = 1 × 10−12. This analysis was repeated for different regulatory similarity thresholds as shown in Supplemental Figure S3B.
When testing individual conditions, we found no enrichment of regulatory variation within the top 100 cis or trans effects, compared with a set of 100 random genes (Fig. 3D). Together, we concluded that divergence in regulatory dynamics is largely decoupled from divergence in expression levels.
Classifying variation in dynamic regulation into cis and trans effects
We next set out to define the genetic basis of variation in regulatory dynamics. Because both cis and trans effects are likely to influence this property, we expected that within the hybrid, the two alleles would show higher similarity than between the parents, as only cis-acting effects can lead to variation in this uniform trans environment. This was indeed the case: regulatory similarity scores between the hybrid alleles are typically higher than the regulatory similarities between species (90% of the genes are above the diagonal) (Fig. 4A). This trend was not attributable to better control measures in the hybrid compared to its parents (Supplemental Fig. S4A).
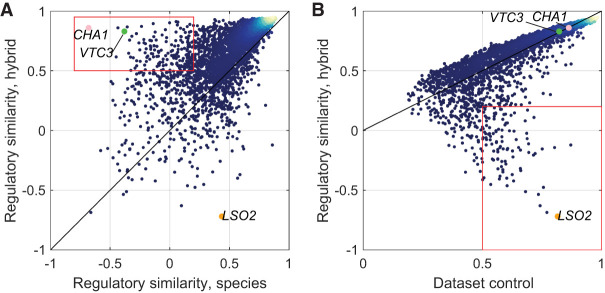
Figure 4.
Regulatory dynamics vary mainly in trans. (A) Defining trans-varying genes. Plotted are RC scores between hybrid alleles as a function of RC scores between species for all genes. Black line is x = y. Genes in the red rectangle vary in trans (184 genes). Indicated genes vary in trans (VTC3, CHA1) and in cis (LSO2) and are discussed in subsequent figures. Color represents density. (B) Defining cis-varying genes. Plotted are RC scores between hybrid alleles as a function of RC scores of the mean hybrid data set control. Genes in the red rectangle vary in cis (106 genes).
To classify dynamically varying genes into cis and trans effects, we considered both between-species and between-hybrid-alleles similarity scores. Genes that show low to negative similarity score between species (RC between species < 0.2) and high similarity score between hybrid alleles (RC hybrid > 0.5) were classified as regulatory trans effects (184 genes) (Fig. 4A). Genes with low to negative similarity between hybrid alleles (RC hybrid < 0.2) but high data set control score (RC data set control > 0.5) were classified as regulatory cis effects (106 genes) (Fig. 4B). Of note, this finding that a larger fraction of regulatory variation result from trans effects, as compared to cis effects, remained when relaxing the threshold defining regulatory varying genes (Supplemental Fig. S4B).
Having this classification of regulatory cis and trans effects, we asked whether these effects show tendency for increased variation in expression level. We first considered the median cis or trans effect on expression level across all conditions (Supplemental Fig. S4C,D). Overall, regulatory trans effects did not show tendency for higher divergence in expression level (Supplemental Fig. S4C). Regulatory cis effects did show tendency for increased cis (but not trans) variation in expression level, yet with small effect size (P-value = 3 × 10−3, rank sum statistic = 2.9) (Supplemental Fig. S4D). We observed the same result when considering the different experimental conditions: genes that diverge in cis in their expression level in at least one experiment were enriched with regulatory cis-variation (hypergeometric P-value=4 × 10−6), but the same trend was not observed for trans-variation (hypergeometric P-value = 0.07). We concluded that, overall, genes that vary in dynamic regulation are distinct from the ones that vary in expression level.
Patterns of trans-dependent regulatory variation
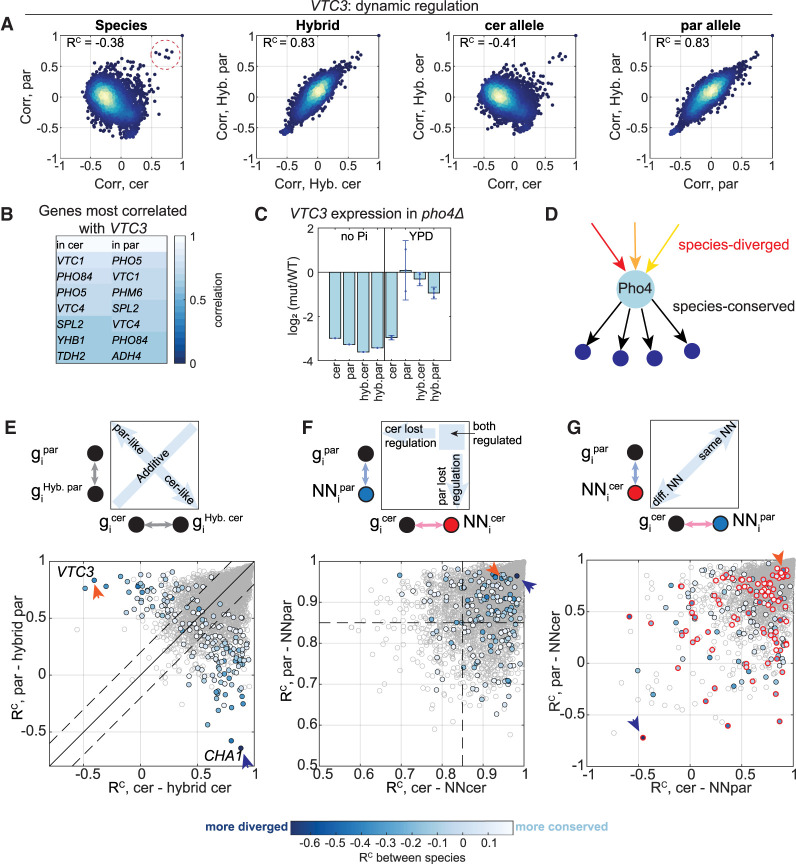
Considering the prominence of trans effects among genes of low regulatory similarity, we decided to first focus on trans-varying genes. Vacuolar transporter chaperone 3 (VTC3) is an illustrative example for a trans-varying gene. VTC3 orthologs show negative regulatory similarity, whereas the hybrid alleles remain highly similar (Fig. 5A). When tested within the hybrid, the pattern of coexpression displayed by the two alleles was similar to the one seen in S. paradoxus and different from that found in S. cerevisiae. To understand the trans-variation acting on VTC3, we examined the genes that show the highest correlation to it. These genes were all phosphate-responding genes in both species (Fig. 5B), suggesting that their direct regulator, Pho4, is still active, but differentially regulated in the two species. Indeed, PHO4 deletion affected VTC3 expression in a condition- and species-specific manner (Fig. 5C): in the absence of phosphate, Pho4 shows species-conserved activity, but in rich conditions, it is activating the expression of its targets in S. cerevisiae but not in S. paradoxus. Therefore, while Pho4 is activating a similar set of targets in both species, it is likely activated by different signals in the two species (Fig. 5D). Such differences can arise from differences in growth niche or because of the strong selection on S. cerevisiae during its domestication.
Figure 5.
Trans-variations in dynamic regulation are inherited dominantly. (A) VTC3 is an example of a trans-varying gene. Scatters of coexpression vectors between species, between hybrid alleles, between S. cerevisiae and hybrid-cerevisiae, and between S. paradoxus and hybrid-paradoxus. Each dot represents the coexpression of VTC3 with a single gene across conditions; RC scores are indicated. Genes that are highly coexpressed with VTC3 in both species are marked with a dashed red circle. (B) Pho4 activates a similar set of targets. VTC3’s highest coexpressing genes are listed. Most are shared in both species; these are known targets of Pho4 (Springer et al. 2003). (C) Pho4 is activating the expression of VTC3 in different conditions in the two species. Shown is log2 fold change (pho4Δ/WT) in gene expression of VTC3 in the indicated conditions. (D) A scheme describing Pho4 regulatory divergence. Pho4 activates the same target genes in both species, but due to different signals in each species. (E) Trans-variations in dynamic regulation are inherited dominantly. The RC scores between each species and its corresponding hybrid allele are shown, of trans-diverging genes (color, 184 genes) and of all genes (gray). Color code indicates the RC score between species. The solid black line is x = y, the dashed lines are diagonal ± 0.2, and the colored dots that pass the dashed lines represent genes that are regulated dominantly, similarly to one of the species; below the lower dashed line: cerevisiae-like (106 genes); above the upper dashed line: paradoxus-like (55 genes). Orange and blue arrow heads indicate the RC scores of VTC3 and CHA1, respectively. (F) Half the trans-diverging genes lost regulation in one of the species. The RC scores of each gene with its NN in each species are shown. The dashed line is RC = 0.85, and genes below it are referred to as genes with impaired regulation. Color code as in E. (G) Cross-species NN similarities vary between genes. Cross-species NN similarities are shown. The x-axis is the similarity of a gene in S. cerevisiae to the gene's NN as defined in S. paradoxus; the y-axis is the similarity of a gene in S. paradoxus to the NN as defined in S. cerevisiae. In the scheme on the top, the circles represent genes and the arrows represent in which species the similarity was calculated (S. cerevisiae in pink, S. paradoxus in light blue). Color code as in E. Genes that preserve high correlation with their NN (RC > 0.85 in F) are indicated with a red circle.
The high similarity of the hybrid alleles with one of the parents was surprising to us, because we expected that combining differentially regulated trans-acting factors within the same genome would result in additive or synergistic effects rather than dominant effects. Examining all trans-diverging genes, we found this dominance to be general: in the majority of cases (161/184, 87%), the hybrid alleles were either paradoxus-like or cerevisiae-like (Fig. 5E; for a list of genes, see Supplemental Table S8). Overall, cerevisiae-like regulation was more abundant, as the number of genes showing a cerevisiae-like pattern within the hybrid was double than these showing paradoxus-like dominance (106 cerevisiae-like, 55 paradoxus-like).
The widespread dominance of trans-dependent regulatory divergence suggests that allelic variants are compensated when combined in the hybrid. One case in which such compensation is expected would be when a TF becomes inactive in one species. This may result from a mutation that inactivates or impairs the regulator's activity in the conditions surveyed. In this case, both hybrid alleles will be regulated similarly to the dominant species. As a way to examine this, we used our control measure of NN similarity. This measure quantifies the degree to which a specific gene is coexpressed with (at least) one other gene in the genome. We reasoned that when a principle regulator becomes inactive, coregulation within its module will be lost as well, resulting in a low NN similarity. Consistent with our expectation, NN similarities that are lower than 0.85 were found for 51% of trans-diverging genes, compared to 23% of all genes (Fig. 5F). Further, in the vast majority of cases, the dominant species did maintain the higher NN similarity (71% of paradoxus-like genes, 91% of cerevisiae-like genes) (Supplemental Fig. S5A). Therefore, these cases of low NN similarity in the recessive species likely result from reduced activity of the direct regulator, at least in the conditions measured (for examples of specific genes, see Supplemental Fig. S5B,C).
Impaired regulation therefore appears to explain a large fraction of regulatory trans-divergence. We next examined cases of trans-divergence that are not explained by this mechanism, namely, cases in which the two orthologs are both regulated (as indicated by high NN similarity) yet show high interspecies divergence. Such cases could result from at least two scenarios. First, a gene could be regulated by the same TF in both species, but the respective TF may be activated by different signals in each species. This case appears to explain the divergence of VTC3 as discussed above. Second, the gene could be primarily regulated by one TF in one species and by another TF in the other species. To distinguish between these possibilities, we asked whether the nearest neighbors are shared between species (Fig. 5G). As expected for VTC3, the NNs are shared between species (Fig. 5G, orange arrow), as well as in other Pho4-regulated genes. Other groups showing this behavior were associated with functions such as iron transport and fatty-acid synthesis in the S. paradoxus dominance class or mitochondrial translation and cell wall maintenance in the S. cerevisiae dominance class (Supplemental Table S8). Genes that share no nearest neighbors between species are therefore expected to change their primary regulating TF, such as CHA1 that is discussed in the next section (Fig. 5G, blue arrow). To conclude, genes that preserve strong regulation in both species, and their trans effect is not explained by loss of regulation, often change their nearest neighbor and are therefore expected to change their primary regulator.
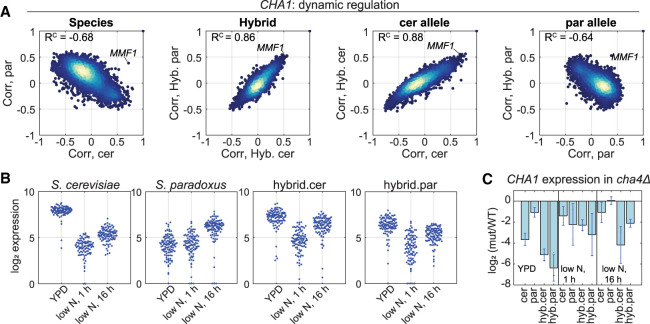
CHA1 expression is species- and condition-dependent
The most extreme variation in dynamic regulation was observed for the gene CHA1 (Fig. 4A), a catabolic L-serine and L-threonine deaminase that is known to be transcriptionaly induced in the presence of these amino acids in S. cereivisiae (Bornaes et al. 1993). This divergence in CHA1 regulation was mapped to a trans effect, because within the hybrid, regulation of the two alleles was highly similar and cerevisiae-like (Fig. 6A). We noted that in S. cereivisae, CHA1 expression is maximal in rich conditions (YPD) and is down-regulated in response to nitrogen depletion (Fig. 6B), as also seen in external data sets (Supplemental Fig. S6A). In contrast, the expression of CHA1 in S. paradoxus is basal under rich conditions (YPD) and it is not repressed upon nitrogen depletion. We find that in both species and the hybrid, the gene is mildly induced in late stages of the experiment (16 h in low nitrogen) (Fig. 6B) and on entry into stationary phase (Supplemental Fig. S6B), indicating a complex regulation. Although S. paradoxus is an autotroph, the S. cerevisiae strain used in this study (BY) is auxotroph to several amino acids. These do not include serine and threonine; therefore, we expect that our results regarding CHA1 expression are not an artifact of the strain auxotrophy.
Figure 6.
CHA1 expression is species- and condition-dependent. (A) CHA1 dynamic regulation is varying in trans. Scatters of coexpression vectors of the indicated comparisons, similar to Figure 5A. (B) CHA1 is down-regulated on nitrogen depletion in S. cerevisiae but not in S. paradoxus. Log2-transformed expression levels of CHA1 in the indicated species and conditions are shown. Expression data of all strains (deletion strains and WT) that were sampled in the indicated conditions are presented. (C) CHA1 expression is reduced upon CHA4 deletion in S. cerevisiae but not in S. paradoxus. Log2 fold change values (cha4Δ/WT) in gene expression of CHA1 in the indicated conditions are presented.
In S. cerevisiae induction of CHA1 depends on the transcription factor Cha4 (Bornaes et al. 1993; Holmberg and Schjerling 1996). Our nearest neighbor analysis (described above) suggested that the primary TF regulating CHA1 had changed between species, because the genes most correlated with CHA1 varied between the species (Fig. 5G, CHA1 is indicated by a blue arrow). We therefore hypothesized that changes in Cha4, whether in expression or activity, could account for the CHA1 regulatory variation. Indeed, deleting CHA4 caused a significant reduction in CHA1 expression in both S. cerevisiae and the hybrid. In contrast, in S. paradoxus, CHA1 expression was only mildly reduced on CHA4 deletion (Fig. 6C). In addition, the expression of CHA4 itself is twofold lower in S. paradoxus (log2 fold change = 0.8–1, P-value < 2 × 10−5) (Supplemental Fig. S6C). We therefore suspect that Cha4 is not activating the expression of CHA1 in S. paradoxus under rich conditions, perhaps owing to its lower levels. Contrary to what we expected, another Cha4 target in S. cerevisiae, MMF1, maintained a positive coexpression connection with CHA1 also in S. paradoxus (Fig. 6A). Therefore, in our interpretation, the Cha4 module did not disappear completely in S. paradoxus, but CHA1 gained an alternative regulation in S. paradoxus that is recessive in the hybrid.
Patterns of cis-dependent regulatory divergence
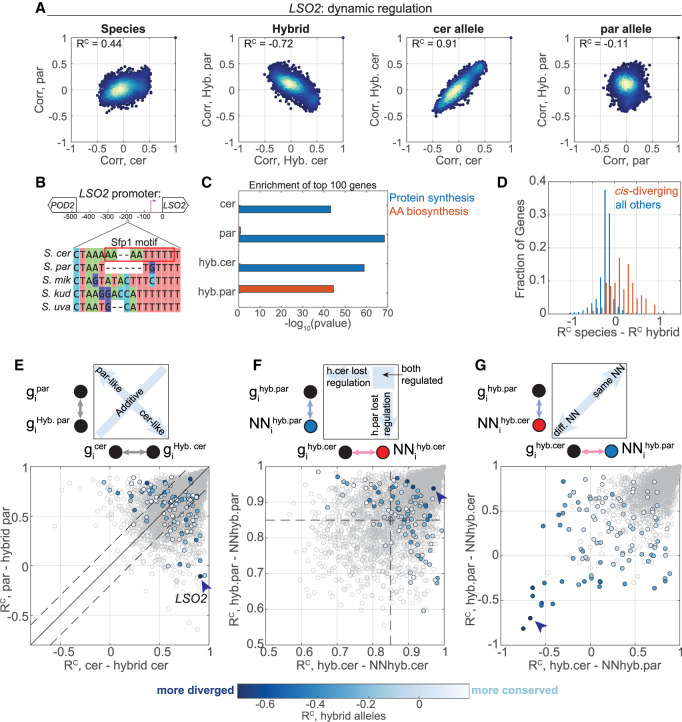
In the preceding section, we analyzed the dominant basis of regulatory divergence that is generated by trans-acting mutations. Next, we focused on the genes whose regulatory divergence was mapped to cis effects, showing low regulatory similarity between the two hybrid alleles. Illustrative examples are the genes LSO2 (Fig. 7A) and PGM1 (Supplemental Fig. S7). In S. cerevisiae, LSO2 is a ribosome-associated gene (Wang et al. 2018). Examining the promoter of LSO2, we noted that the S. cerevisiae allele has gained a binding site for Sfp1, the principle TF regulating ribosomal protein and ribosomal biogenesis genes (Fig. 7B). Consistent with the differential presence of these binding sites, we find that the S. cerevisiae LSO2 allele is coexpressed with genes coding for ribosomal proteins, whereas the LSO2 allele coming from S. paradoxus is not coexpressed with these genes (Fig. 7C) but is coexpressed with the amino acid biosynthesis module, suggesting an alternative regulation (Fig. 7C).
Figure 7.
Cis-variations in dynamic regulation are often compensated by trans effects. (A–C) LSO2 is an example for a cis-varying gene. Scatters of coexpression vectors of the indicated comparisons, similar to Figure 5A. (B) S. cerevisiae LSO2 promoter contains a Sfp1 motif. A segment of a sequence comparison of the promoters from the indicated species is shown. (C) Enrichment of ribosomal protein-coding genes, and of amino acid biosynthesis genes, within the top 100 LSO2 correlating genes in the different genetic backgrounds. (D) Cis-variations are compensated by trans-variations. Histogram of the RC difference (between species – between hybrid alleles) for all genes and for cis-varying genes (106 genes). (E) Cis-variations are not typically dominant. The RC scores of cis-diverging genes (color, 106 genes) and of all genes (gray) of the comparison between each species and its corresponding hybrid allele are shown, as in Figure 5E. Color code represents regulatory similarity between hybrid alleles. (F) Half the cis-diverging genes lost regulation in one of the alleles. The RC scores of each gene with its NN in each species are shown, similar to Figure 5F. Color code as in E. (G) Cis-variation involves a change in the nearest neighbor. Cross-species NN similarities are shown, as indicated by the scheme at the top, similar to Figure 5G. Color code as in E.
The lack of regulatory similarity between LSO2 hybrid alleles was suppressed when comparing the parental species (Fig. 7A), indicating a compensating effect acting in trans. Examining the full set of cis effects, we found compensatory trans effects to be prominent among genes showing cis-variation. Specifically, we observe that in most cases of cis effect (82 of 106 genes), regulatory similarity between the species was higher than regulatory similarity between the two hybrid alleles (Fig. 7D). Furthermore, although only ∼1% of genes showed species-hybrid difference >0.2, 50% of cis-varying genes showed such an effect. This observation is different from our observation of cis-trans variations in expression levels (Supplemental Fig. S1D) in which we find a similar number of compensating and reinforcing interactions (70 and 60 genes, respectively). Studies in mice (Goncalves et al. 2012) and flies (Landry et al. 2005; McManus et al. 2010) report that compensatory interactions are an order of magnitude more abundant than reinforcing interactions on gene expression level. Our results do not agree with previous results in the matter of cis-trans interactions on expression level but do show this trend among genes that vary in dynamic regulation. We conclude that cis-dependent dynamic regulatory variations are often accompanied by additional compensating trans effects.
Finally, we analyzed cis-diverging genes for their mode of inheritance (dominant or additive), level of regulation (correlation with their NN), and the identity of NN (common or different). Unlike trans effects, cis effects were not biased toward dominant inheritance (33 cerevisiae-like, 26 paradoxus-like, 47 additive) (Fig. 7E, Supplemental Table S9). Similar to trans effects, half the cis-diverging genes lost regulation in one of the alleles (58%) (Fig. 7F), although this did not hold for the most extreme cis-diverging genes that did maintain tight regulation of both alleles, as indicated by high NN similarities. Of note, unlike trans effects, in cis effects the identity of the NN changed (Fig. 7G), consistent with the expected change in the primary regulating TF.
Discussion
Understanding how gene expression evolves is a major challenge. In this work, we distinguished two properties of gene expression: transcript abundance and dynamic regulation. It is a common observation that within an organism, genes that are subject to a similar regulation can still be expressed at widely different levels. This ability to independently control the level and regulation of gene transcription implies that both can be subject to evolution.
Previous studies that compared gene expression between related species considered only a few conditions and therefore could only address variation in expression levels (Yvert et al. 2003; Wittkopp et al. 2004, 2008; Landry et al. 2005; Tirosh et al. 2006, 2009; Yanai and Hunter 2009; McManus et al. 2010; Goncalves et al. 2012; Shi et al. 2012; Combs et al. 2018). Whether this variation results from changes in condition-specific regulation or from differences in the capacity of the general transcriptional machinery remain unresolved. By generating a compendium of comparative data, in which we surveyed a diverse set of conditions, we distinguished variation in expression level from variation in dynamic regulation. Our major finding is that these two variation types are distinct. In particular, the majority of genes whose abundance vary between the species did maintain a conserved regulatory pattern.
We based our measure of regulatory similarity on gene coexpression. This measure is commonly used for defining coregulation within individual genomes (Hughes et al. 2000; Segal et al. 2003; Li et al. 2017) and was previously used for comparing core modules in more distant species (Ihmels et al. 2005a,b; Tsaparas et al. 2006; Thompson et al. 2013). It is beneficial in our context as it minimizes differences in environmental perception, which are upstream to the transcription network.
To gain insight on the mechanistic basis of regulatory variation, we used an interspecific hybrid. A large fraction of regulatory variation observed between the species was lost when comparing the two hybrid alleles. This indicates the prominence of trans effects in regulatory variation, which is in agreement with results from other experimental systems (Yvert et al. 2003; McManus et al. 2010; Albert et al. 2018; Liu et al. 2019; Sanchez et al. 2019). We expected that the trans effects will show additive or synergistic interactions within the hybrid. Hypothetically, if the regulatory variation was coded on a single transcription factor, activating it in a different set of conditions, the hybrid would have both types of allelic variants (and hence, both types of regulation) and would show an additive effect. In contrast to this expectation, we found that the vast majority of trans effects are being inherited in a dominant way, regulated in practically an identical pattern in the hybrid to one of the parents. We can therefore deduce that the trans-variation is coded upstream to the direct TF on possibly all regulators of the regulatory network. Dominance of trans effects was observed previously in multiple organisms, both on expression levels (Lemos et al. 2008) and transcription factor binding (Wong et al. 2017). In our case, half the trans effects are explained by limited activity of a TF in the recessive parent. This may provide a mechanistic explanation for the phenomenon of genome asymmetry that was reported before in a variety of organisms (Lemos et al. 2008; Feldman et al. 2012; Ren et al. 2019), whereby the phenotypic manifestation of some traits, including gene expression patterns, favors one genome over the other rather than being intermediate. In this sense, we found twice as many genes with a cerevisiae-like regulation than genes with a paradoxus-like regulation in the hybrid. This result is consistent with our observation that the hybrid growth phenotype is more similar to that of S. cerevisiae than S. paradoxus (Lupo et al. 2020).
We quantify regulatory similarity using a correlation-based measure, whereas variation in transcript abundance is measured in units of read counts. It is therefore difficult to conclude which of these properties change in a more prominent way. Our analysis of the data, however, suggests that cases in which genes have lost or gained regulation by specific TFs are rather rare, and that most variations result from altered activity of a common regulator. Overall, our data suggest that the massive divergence in transcript abundance masks a highly conserved transcription network.
Methods
Yeast strains
Yeast stains in this study were constructed on the background of S. cerevisiae S288c and S. paradoxus CBS432 (OS142) and their hybrid. Strains are listed in Supplemental Table S2. In this study, we expression profiled only diploid yeast cells. Transcription factors that were deleted are listed in Supplemental Table S1. All transformations were done using the standard LiAc/SS-Carrier/PEG transformation method.
Experimental conditions
Four types of time-course experiments were performed in this study: (1) transition from YPD to low-nitrogen medium, (2) cell-cycle time course, (3) growth curve, and (4) transition to phosphate starvation. These are described in full detail in the following sections.
Transition from YPD to low-nitrogen medium
In short, yeast cultures were grown to logarithmic phase in YPD, sampled for RNA purification after 6 h of growth, then washed and transferred to low-nitrogen medium. The cultures were sampled again at times of 1 h and 16 h following the transition. The experiment was performed in a 96-well plate to allow high-throughput sampling of multiple strains. In full detail, agar-grown yeast colonies were pinned to liquid starters, grown to stationary phase for 24 h, then diluted in 1.5 mL YPD in 2-mL deep-well plates. These plates contained one glass bead per well for proper mixing of the culture. In these experiments, each half plate contained a technical repeat of the same culture: one half was used for optical density (OD600) measurements, and the other was used for cell harvesting for RNA purification. The cultures were grown for 6–8 h in a 30°C shaker reaching to OD600 of 0.1–0.6 (logarithmic phase), then sampled and washed once, resuspended in low-nitrogen medium and grown in the same conditions, and sampled at 1 h and 16 h post medium shift. Cells were harvested in 0.5-mL culture, centrifuged at 4000 rpm for 30 sec; supernatant was removed using multipipette vacuum, and pellets were immediately frozen in liquid nitrogen and stored in −80°C.
Low-nitrogen medium
For the low-nitrogen medium, we used a 0.67% yeast nitrogen base without amino acids and ammonium sulfate (Bacto-YNB), 2% D-glucose, 0.05 mM ammonium sulfate, 20 mg/L Urcail, 20 mg/L Histidine, and 100 mg/L Leucine.
Cell cycle time course
Yeast starters were grown in YPD overnight at 30°C to stationary phase and were inoculated to fresh medium to OD600 of 0.005 in 100 mL in a 500-mL flask, then grown overnight. When reaching OD600 of 0.1–0.2, hydroxyurea (HU) was added to the media to a final concentration of 0.2 M for an additional 2 h. To remove HU from the media, the cells were washed twice by centrifugation (4000 rpm for 1 min) and resuspended in fresh YPD at equal volume and temperature. Then, the culture was returned to a bath orbital shaker. Cells were collected at the following time points: before HU, at 5, 10, 20, 30, 60, and 120 min in HU, and every 5 min after release for 3 h—in total, 43 time points for each strain. For RNA, samples of 1.5 mL were taken and centrifuged for 10 sec in 13,000 rpm, the supernatant was removed, and the pellets were immediately frozen in liquid nitrogen. For DNA staining (to assess proper synchronization, data shown in Lupo et al. 2020), samples of 1.5 mL were taken and centrifuged for 10 sec in 13,000 rpm and resuspended in cold 70% ethanol. Samples were kept in 4°C. This experiment was carried with two independent biological repeats for each strain.
Growth curve
Overnight starters of yeast were diluted in 50 mL YPD in 250-mL flasks and grown in a bath orbital shaker overnight (12 h) at 30°C. The time course started when the cultures reached OD600 of 0.4–0.5 and were sampled for OD600 measurements in RNA extraction at the following time points: 0, 80, 125, 170, 215, and 260 min, and 5.1, 6.1, 7.1, 8.1, 9.1, 10.1, 11.1, 23.2, and 32.5 h after start.
Transition to phosphate starvation
Overnight starters of yeast were diluted in 100 mL Synthetic Complete medium (SC) in 500-mL flasks and grown in a bath orbital shaker overnight (12 h) at 30°C. When the cultures reached OD600 of 0.2, they were sampled for RNA extraction, then washed twice in SC medium lacking Pi (phosphate-depleted) and inoculated to that media. For the next 6 h, the cultures were sampled every 15 min for RNA extraction. At the end of the time course, the three cultures (WT strains of S. cerevisiae, S. paradoxus, and the hybrid) had completed 1.5 rounds of mitotic divisions. The experiment was repeated on a small scale to include pho4Δ strains of the two species and hybrid.
RNA extraction and sequencing
RNA was extracted using a modified protocol of nucleospin 96 RNA kit. Specifically, cell lysis was done in a 96-deep-well plate by adding 450 μL of lysis buffer containing 1 M sorbitol (Sigma-Aldrich), 100 mM EDTA, and 0.45 μL lyticase (10 International units/μL). The plate was incubated for 30 min at 30°C to break the cell wall, then centrifuged for 10 min at 3000 rpm, and the supernatant was removed. From this stage, extraction proceeded as in the protocol of the nucleospin 96 RNA kit, only substituting β-mercaptoethanol with DTT. cDNA was prepared from the RNA extracts, barcoded using Tn5-mediated tagmentation protocol, and sequenced using Illumina NextSeq.
Library preparation and sequencing
Libraries were prepared as described in Voichek et al. (2018). Poly(A) RNA was selected for by reverse transcription with a barcoded poly(T) primer. The barcoded DNA–RNA hybrids were pooled and fragmented by a hyperactive variant of the Tn5 transposase (courtesy of Ido Amit). Tn5 was stripped off the DNA by treatment with SDS 0.2% followed by SPRI cleanup, and the cDNA was amplified and sequenced with Illumina NextSeq 500 with 50-bp reads.
Dual-genome alignment pipeline
Sequenced reads were mapped against the concatenated genomes of S. cerevisiae S288c and S. paradoxus CBS432 (Yue et al. 2017). Mapping was performed with STAR 2.4.2a (Dobin and Gingeras 2015) with the following parameters: ‐‐sjdbOverhang 60 ‐‐scoreGap -10. The alignments were divided into genomes based on the alignment scores (attribute AS:I in SAM format). Uniquely mapped reads were assigned to the ortholog with the better score. If there is no difference in the scores between the two genomes and the alignment is unique in the S. cerevisiae genome, then the alignment to the S. cerevisiae genome is kept and assigned as indistinguishable (and the same was done for the S. paradoxus genome). Counting was performed on the transcript end site (TES) of each gene. The TES was defined between −500 and +200 bases relative to the stop codon. The reads were counted using htseq-count, with the following parameters: ‐‐stranded yes and ‐‐mode union (https://htseq.readthedocs.io/en/master/). Count tables, divided to each of the genomes, are available in Supplemental Tables S10–S12. Mapping statistics (Supplemental Table S4) and percentage of mapping to each genome and erroneous mapping (Supplemental Table S5) are discussed in Supplemental Note 1.
Data filtering
On average, libraries were covered by 3 million reads, resulting in an average of 47 reads per gene. Samples with less than 150,000 reads were filtered out. Reads of conserved genes that were determined as indistinguishable were assigned to each sample and were divided equally between the two genomes in the hybrid samples.
Data normalization and differential expression analysis
Read counts were normalized and analyzed for differential expression via DESeq2 (Love et al. 2014) in R 3.6.3 (R Core Team 2020). Genes with reads counts fewer than five were filtered out. Read counts were given as input to DESeq2 with the following design: “∼ gb,” in which gb stands for genetic background (S. cerevisiae, S. paradoxus, hybrid-cerevisiae, hybrid-paradoxus). Supplemental Table S3 was used as the summarized experiment matrix. This design enabled differential expression analysis between species and between hybrid alleles (cis effect). Analysis of trans effect was carried out using another design: “∼ F+ species + F:species.” In this design, F represents generation, so parent samples are assigned as F0 and hybrid samples are assigned as F1, and species is either cerevisiae or paradoxus. Differential expression was determined via likelihood ratio test (test = “LRT”) focusing only on the interaction term (reduced = F + species). For both designs, results went through log2 fold change shrinkage using ashr (Stephens 2017) method. DESeq2 results are listed in Supplemental Table S6.
Regulatory similarity by coexpression (RC)
Pairwise correlation matrices for all genes in the genome were computed per data set, in which each row is referred to as a coexpression vector. Regulatory similarity is the correlation between orthologous correlation vectors as
where is the regulatory similarity of the ith gene; is the coexpression vector of the ith gene in S. cerevisiae; and similarly for . We filtered out genes that express in < 10% of the samples (resulting in 4791 genes), and coexpression vectors that compose of low correlation coefficients (|R| > 0.2 for less than 100 genes), resulting in 4772 genes in the analysis. Regulatory similarity scores are listed in Supplemental Table S7.
Data set control
Data set control is a bootstrapping analysis done to evaluate the reliability of coexpression pattern in each species. For each species, we split the expression profiles subset into two random parts, generated coexpression vectors for each gene in each half, and reported on the correlation between these vectors. We repeated this process for 10 permutations; the average of these is the RC of data set control.
Nearest neighbors
We define nearest neighbors as pairs of genes with the most similar coexpression vector. NNs were defined for each species separately. Starting from a pairwise correlation matrix, we computed the correlation between the rows of this matrix, resulting in the second correlation matrix. Nearest neighbors are the maximal value of each row of the second correlation matrix. Cross-species NNs were defined with a more relaxed criterion, in that we took the gene giving the maximal correlation value in species A out of the top five nearest neighbors of species B.
Enrichment for functional groups
Enrichment analysis was done through applying a hypergeometric test to a list of genes against functional gene groups. Functional groups include expression modules (Ihmels et al. 2002), environmental stress response (Gasch et al. 2000), GO slim, transcription factors targets (MacIsaac et al. 2006), KEGG pathways, expression levels, burst size (Newman et al. 2006), and TATA-containing promoters (Basehoar et al. 2004) OPN and DPN (Tirosh and Barkai 2008).
Promoter analysis
Promoters were defined as intergenic regions upstream of the start codon of a gene. Sequences were obtained from the genome sequences of S. cerevisiae and S. paradoxus (Yue et al. 2017). Sequences of distant species were obtained from Yeast Gene Order Browser (YGOB) (Byrne and Wolfe 2005). TF binding motifs were obtained from the YeTFaSCo database (De Boer and Hughes 2012).
Data access
The RNA-sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA592756. Tables of read counts per gene, per sample, following the initial computational pipeline, are available in Supplemental Tables S10–S12. Code used for the analysis of regulatory similarity is available at GitHub (https://github.com/GatKrieger/comparativeCoexp). All code is also available as Supplemental Code.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the Barkai laboratory members and the Levy laboratory members for helpful discussions. We thank Gil Hornung and Rotem Barzilay (The Nancy and Stephen Grand Israel National Center for Personalized Medicine [G-INCPM]) for implementing the dual-genome alignment pipeline. We thank Felix Jonas, Assaf Biran, Michal Chapal, Sagie Brodsky, Yoav Voichek, Anna Redgrave, and Patricia Wittkopp for critical reading and commenting on the manuscript. We thank Yael Maoz for proofreading. This project was supported by the U.S. National Science Foundation-U.S. Israel Binational Science Foundation-Molecular and Cellular Biosciences (NSF-BSF-MCB) (2019625), the Israel Science Foundation (ISF) (1738/15), and the Minerva center (AZ 57 46 9407 65).
Author contributions: G.K., O.L., A.A.L., and N.B. designed the research. G.K. and O.L. performed experiments and analyzed the data. All authors contributed to the writing of the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261537.120.
References
- Albert FW, Bloom JS, Siegel J, Day L, Kruglyak L. 2018. Genetics of trans-regulatory variation in gene expression. eLife 7: e35471 10.7554/eLife.35471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. 2014. Evolution at two levels of gene expression in yeast. Genome Res 24: 411–421. 10.1101/gr.165522.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699–709. 10.1016/S0092-8674(04)00205-3 [DOI] [PubMed] [Google Scholar]
- Bornaes C, Ignjatovic MW, Schjerling P, Kielland-Brandt MC, Holmberg S. 1993. A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol Cell Biol 13: 7604–7611. 10.1128/MCB.13.12.7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15: 1456–1461. 10.1101/gr.3672305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs PA, Krupp JJ, Khosla NM, Bua D, Petrov DA, Levine JD, Fraser HB. 2018. Tissue-specific cis-regulatory divergence implicates eloF in inhibiting interspecies mating in Drosophila. Curr Biol 28: 3969–3975.e3. 10.1016/j.cub.2018.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer CG, Hughes TR. 2012. YeTFaSCo: a database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res 40: D169–D179. 10.1093/nar/gkr993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. 2015. Mapping RNA-seq reads with STAR. Curr Protoc Bioinforma 51: 11.14.1–11.14.19. 10.1002/0471250953.bi1114s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori-Bachash M, Shema E, Tirosh I. 2011. Coupled evolution of transcription and mRNA degradation. PLoS Biol 9: e1001106 10.1371/journal.pbio.1001106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Hsieh LC, Sung HM, Wang TY, Huang CJ, Lu HHS, Lu MYJ, Wu SH, Li WH. 2010. Natural selection on cis and trans regulation in yeasts. Genome Res 20: 826–836. 10.1101/gr.101576.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M, Levy AA, Fahima T, Korol A. 2012. Genomic asymmetry in allopolyploid plants: wheat as a model. J Exp Bot 63: 5045–5059. 10.1093/jxb/ers192 [DOI] [PubMed] [Google Scholar]
- Fraser HB. 2019. Improving estimates of compensatory cis–trans regulatory divergence. Trends Genet 35: 3–5. 10.1016/j.tig.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245. 10.1038/nature04559 [DOI] [PubMed] [Google Scholar]
- Goncalves A, Leigh-Brown S, Thybert D, Stefflova K, Turro E, Flicek P, Brazma A, Odom DT, Marioni JC. 2012. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res 22: 2376–2384. 10.1101/gr.142281.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium. 2017. Genetic effects on gene expression across human tissues. Nature 550: 204–213. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagai T, Chen X, Miragaia RJ, Rostom R, Gomes T, Kunowska N, Henriksson J, Park JE, Proserpio V, Donati G, et al. 2018. Gene expression variability across cells and species shapes innate immunity. Nature 563: 197–202. 10.1038/s41586-018-0657-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg S, Schjerling P. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144: 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109–126. 10.1016/S0092-8674(00)00015-5 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. 2002. Revealing modular organization in the yeast transcriptional network. Nat Genet 31: 370–377. 10.1038/ng941 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Berman J, Barkai N. 2005a. Comparative gene expression analysis by a differential clustering approach: application to the candida albicans transcription program. PLoS Genet 1: e39 10.1371/journal.pgen.0010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. 2005b. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309: 938–940. 10.1126/science.1113833 [DOI] [PubMed] [Google Scholar]
- King M, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. 10.1126/science.1090005 [DOI] [PubMed] [Google Scholar]
- Landry CR, Wittkopp PJ, Taubes CH, Ranz JM, Clark AG, Hartl DL. 2005. Compensatory cis-trans evolution and the dysregulation of gene expression in interspecific hybrids of Drosophila. Genetics 171: 1813–1822. 10.1534/genetics.105.047449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Lemos B, Rifkin SA, Dickinson WJ, Hartl DL. 2007. Genetic properties influencing the evolvability of gene expression. Science 317: 118–121. 10.1126/science.1140247 [DOI] [PubMed] [Google Scholar]
- Lemos B, Araripe LO, Fontanillas P, Hartl DL. 2008. Dominance and the evolutionary accumulation of cis-and trans-effects on gene expression. Proc Natl Acad Sci 105: 14471–14476. 10.1073/pnas.0805160105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jourdain AA, Calvo SE, Liu JS, Mootha VK. 2017. CLIC, a tool for expanding biological pathways based on co-expression across thousands of datasets. PLoS Comput Biol 13: e1005653 10.1371/journal.pcbi.1005653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li YI, Pritchard JK. 2019. Trans effects on gene expression can drive omnigenic inheritance. Cell 177: 1022–1034.e6. 10.1016/j.cell.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo O, Krieger G, Jonas F, Barkai N. 2020. Mapping the architecture of regulatory variation provides insights into the evolution of complex traits. bioRxiv 10.1101/113217 [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. 2010. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res 20: 816–825. 10.1101/gr.102491.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A. 2014. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res 24: 422–430. 10.1101/gr.164996.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger BPH, Duveau F, Yuan DC, Tryban S, Yang B, Wittkopp PJ. 2016. Contrasting frequencies and effects of cis- and trans-regulatory mutations affecting gene expression. Mol Biol Evol 33: 1131–1146. 10.1093/molbev/msw011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- Paris M, Kaplan T, Li XY, Villalta JE, Lott SE, Eisen MB. 2013. Extensive divergence of transcription factor binding in Drosophila embryos with highly conserved gene expression. PLoS Genet 9: e1003748 10.1371/journal.pgen.1003748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB. 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053. 10.1038/nature04597 [DOI] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Ren L, Li W, Qin Q, Dai H, Han F, Xiao J, Gao X, Cui J, Wu C, Yan X, et al. 2019. The subgenomes show asymmetric expression of alleles in hybrid lineages of Megalobrama amblycephala × Culter alburnus. Genome Res 29: 1805–1815. 10.1101/gr.249805.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MR, Payen C, Cheong F, Hovde BT, Bissonnette S, Arkin AP, Skerker JM, Brem RB, Caudy AA, Dunham MJ. 2019. Transposon insertional mutagenesis in Saccharomyces uvarum reveals trans-acting effects influencing species-dependent essential genes. Genome Res 29: 396–406. 10.1101/gr.232330.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 1: 11–25. 10.1534/g3.111.000273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. 2003. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 34: 166–176. 10.1038/ng1165 [DOI] [PubMed] [Google Scholar]
- Shi X, Ng DWK, Zhang C, Comai L, Ye W, Jeffrey Chen Z. 2012. Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun 3: 950 10.1038/ncomms1954 [DOI] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. 2008. Gene-environment interaction in yeast gene expression. PLoS Biol 6: 810–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Wykoff DD, Miller N, O'Shea EK. 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol 1: e28 10.1371/journal.pbio.0000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M. 2017. False discovery rates: a new deal. Biostatistics 18: 275–294. 10.1093/biostatistics/kxw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Roy S, Chan M, Styczynski MP, Pfiffner J, French C, Socha A, Thielke A, Napolitano S, Muller P, et al. 2013. Evolutionary principles of modular gene regulation in yeasts. eLife 2: e00603 10.7554/eLife.00603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. 2008. Two strategies for gene regulation by promoter nucleosomes. Genome Res 18: 1084–1091. 10.1101/gr.076059.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N. 2006. A genetic signature of interspecies variations in gene expression. Nat Genet 38: 830–834. 10.1038/ng1819 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. 2009. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 324: 659–662. 10.1126/science.1169766 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. 2010. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol 6: 365 10.1038/msb.2010.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaparas P, Mariño-Ramírez L, Bodenreider O, Koonin E V, Jordan IK. 2006. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol 6: 70 10.1186/1471-2148-6-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Mittelman K, Gordon Y, Bar-Ziv R, Smit DL, Shenhav R, Barkai N. 2018. Epigenetic control of expression homeostasis during replication is stabilized by the replication checkpoint. Mol Cell 70: 1121–1133.e9. 10.1016/j.molcel.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Wang YJ, Vaidyanathan PP, Rojas-Duran MF, Udeshi ND, Bartoli KM, Carr SA, Gilbert WV. 2018. Lso2 is a conserved ribosome-bound protein required for translational recovery in yeast. PLOS Biol 16: e2005903 10.1371/journal.pbio.2005903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RW, Nagy L, Selegue J, Gates J, Carroll S. 1994. Evolution of homeotic gene regulation and function in flies and butterflies. Nature 372: 458–461. 10.1038/372458a0 [DOI] [PubMed] [Google Scholar]
- Weiss CV, Roop JI, Hackley RK, Chuong JN, Grigoriev IV, Arkin AP, Skerker JM, Brem RB. 2018. Genetic dissection of interspecific differences in yeast thermotolerance. Nat Genet 50: 1501–1504. 10.1038/s41588-018-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet 40: 346–350. 10.1038/ng.77 [DOI] [PubMed] [Google Scholar]
- Wong ES, Thybert D, Schmitt BM, Stefflova K, Odom DT, Flicek P. 2015. Decoupling of evolutionary changes in transcription factor binding and gene expression in mammals. Genome Res 25: 167–178. 10.1101/gr.177840.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Schmitt BM, Kazachenka A, Thybert D, Redmond A, Connor F, Rayner TF, Feig C, Ferguson-Smith AC, Marioni JC, et al. 2017. Interplay of cis and trans mechanisms driving transcription factor binding and gene expression evolution. Nat Commun 8: 1092 10.1038/s41467-017-01037-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Hunter CP. 2009. Comparison of diverse developmental transcriptomes reveals that coexpression of gene neighbors is not evolutionarily conserved. Genome Res 19: 2214–2220. 10.1101/gr.093815.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Lagomarsino MC, et al. 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet 49: 913–924. 10.1038/ng.3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Brem RB, Whittle J, Akey JM, Foss E, Smith EN, Mackelprang R, Kruglyak L. 2003. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet 35: 57–64. 10.1038/ng1222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.