Abstract
The coronavirus disease 2019 (COVID-19) pandemic has elicited a swift response by the scientific community to elucidate the pathogenesis of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2)-induced lung injury and develop effective therapeutics. Clinical data indicate that severe COVID-19 most commonly manifests as viral pneumonia-induced acute respiratory distress syndrome (ARDS), a clinical entity mechanistically understood best in the context of influenza A virus-induced pneumonia. Similar to influenza, advanced age has emerged as the leading host risk factor for developing severe COVID-19. In this review we connect the current understanding of the SARS-CoV-2 replication cycle and host response to the clinical presentation of COVID-19, borrowing concepts from influenza A virus-induced ARDS pathogenesis and discussing how these ideas inform our evolving understanding of COVID-19-induced ARDS. We also consider important differences between COVID-19 and influenza, mainly the protean clinical presentation and associated lymphopenia of COVID-19, the contrasting role of interferon-γ in mediating the host immune response to these viruses, and the tropism for vascular endothelial cells of SARS-CoV-2, commenting on the potential limitations of influenza as a model for COVID-19. Finally, we explore hallmarks of ageing that could explain the association between advanced age and susceptibility to severe COVID-19.
Short abstract
Review of viral ARDS pathogenesis, how it informs evolving models of COVID-19, and how hallmarks of ageing explain the age-related morbidity and mortality of severe COVID-19 https://bit.ly/39Ca0c0
Main novel ideas and hypotheses
Our review explores influenza A virus-induced acute respiratory distress syndrome (ARDS) as a paradigm for understanding coronavirus disease 2019 (COVID-19)-induced ARDS pathogenesis and ageing as a risk factor for severe disease. Integrating established knowledge of influenza A virus-induced ARDS pathophysiology, we discuss how shared clinical findings frame influenza as only an approximate model for COVID-19. We argue that the impaired interferon-I and -III response of severe COVID-19 is reminiscent of severe acute respiratory syndrome-coronavirus (SARS-CoV) and influenza pathobiology, suggesting conserved virulence mechanisms among these viruses. We also review clinically apparent differences in the immune responses elicited by these viruses according to the latest clinical data and recommend that investigators note these differences in ongoing efforts to elucidate the pathogenicity of SARS-CoV-2. Specifically, we argue that the hypercoagulable and hyperinflammatory state of severe COVID-19 is a consequence of the expanded tropism of SARS-CoV-2, which allows it to infect vascular endothelial cells, and that cytokine storm physiology contributes to a lesser degree. Moreover, we discuss the deleterious effect of interferon-γ activity in influenza infection and how this observation contrasts with findings in severe COVID-19. We also explore the potential aetiologies of the lymphopenia associated with severe COVID-19: the virus' expanded tropism, elevated serum cytokines (particularly interleukin-6 and tumour necrosis factor-α), and excessive lymphocyte recruitment to the lungs. Finally, we discuss how certain hallmarks of ageing (epigenetic alterations, mitochondrial dysfunction, telomere attrition, cellular senescence, and altered intercellular communication) predispose the ageing population to severe COVID-19. We speculate that the ability of SARS-CoV-2 to affect T-lymphocyte and myeloid cell physiology coupled with age-related maladaptive biological phenomena explain the strong association between advanced age and increased risk of COVID-19-related morbidity and mortality.
Introduction
The spread of a novel coronavirus (severe acute respiratory syndrome-coronavirus-2, or SARS-CoV-2) has challenged the capacity of healthcare systems and the public health policies of governments worldwide [1]. As of July 2020, the coronavirus disease 2019 (COVID-19) pandemic has resulted in over 12 million cases and over 550 000 confirmed deaths, although the implementation of social distancing measures and wearing masks have allayed the spread of the virus in many parts of the world [2]. Nevertheless, its high transmissibility portends further waves of contagion, warranting continued efforts to fully elucidate its pathogenesis and identify putative therapeutic targets. Mortality attributable to SARS-CoV-2 infection occurs mainly through the development of viral pneumonia-induced acute respiratory distress syndrome (ARDS), a clinical entity best understood in the context of influenza A virus (influenza) infection [2, 3]. While the exact mechanisms through which SARS-CoV-2 causes ARDS and how certain host factors confer an increased risk of developing severe disease remain unclear, one factor has emerged as a dominant predictor of disease severity and risk of mortality, which is age [4, 5]. In fact, reports from Italy and China early in the pandemic noted case-fatality rates of 15–20% among patients aged >80 years compared with <1% in patients aged <50 years. In this review we examine clinically relevant mechanisms of influenza-induced ARDS pathophysiology, discuss their relevance to COVID-19 pathophysiology and how they contrast with COVID-19-specific clinical findings, and explore hallmark biological ageing processes that may underlie the age-related susceptibility to severe COVID-19.
Viral replication cycle and clinical presentation
SARS-CoV-2 (an enveloped, positive-sense, single-stranded RNA virus) belongs to the Betacoronavirus genus. It shares 79% of its RNA sequence with the pathogen that caused the 2003–2004 SARS epidemic, SARS-CoV [6]. Structurally, both viruses contain a spike (S)-protein on their envelope, which, following its enzymatic activation via host proteases and subsequent binding to the human ACE2 receptor, mediates viral fusion and endocytosis [7]. Once inside the cell, the viral genome is transcribed by the viral RNA-dependent RNA polymerase and then translated by host ribosomes to synthesise viral proteins. Finally, mature virions are assembled in the cytoplasm, where they are primed for exocytosis [8].
SARS-CoV has tropism for ciliated airway epithelial cells and type II pneumocytes, which is conferred by its dependence on both the human ACE2 receptor, as well as the host membrane serine protease TMPRSS2 for S-protein cleavage and subsequent activation [7]. While SARS-CoV-2 shares this tropism and invasion mechanism, it has also acquired a furin cleavage site that allows S-protein fusion domain exposure by ubiquitous furin proteases [9]. These furin proteases are found on the membrane of myriad cells across the body that also express ACE2, including vascular endothelial cells [10]. This expanded tropism ultimately translates to increased infectivity as well as novel clinical consequences relative to SARS-CoV infection.
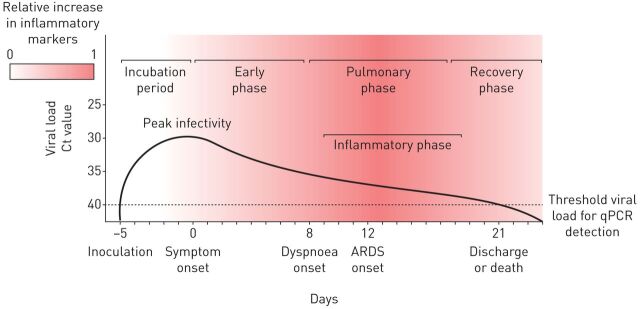
Current models of SARS-CoV-2 transmissibility have established a mean incubation period of 5 days, with viral loads peaking before symptom onset and monotonically declining thereafter. Viral loads are detectable on posterior naso/oropharyngeal swabs for a minimum of 8 days and a median of 20 days from symptom onset, according to latest reports [11–14]. While the maximum duration of viral shedding remains unclear, it likely wanes by day 40 post-infection in most cases. There is significant variability in the symptoms reported during the early symptomatic phase of infection, ranging from mild constitutional symptoms such as fever, fatigue, dry cough and diarrhoea, to sore throat and loss of taste; some cases remain entirely asymptomatic in this phase [15–17]. In many patients these prodromal symptoms are followed by the pulmonary phase of infection, characterised by respiratory symptoms and findings including dyspnoea, radiographic pulmonary infiltrates and hypoxaemia [14]. Most morbidity and mortality related to COVID-19 occurs in the following inflammatory phase, characterised by a dysregulated immune response and hypercoagulable state that is associated with life-threatening complications, including cardiac and renal failure, cerebrovascular disease, and ARDS [18, 19]. Finally, among survivors, there follows a recovery phase, in which inflammation resolves and damaged lung tissue is repaired, ultimately restoring organ system homeostasis (figure 1). Of the life-threatening sequelae associated with the inflammatory phase, ARDS has been documented as the most commonly occurring, representing a substantial proportion of COVID-19 patients requiring intensive care unit (ICU) admission [15].
FIGURE 1.
Stereotypical clinical course of coronavirus disease 2019 complicated by acute respiratory distress syndrome (ARDS). Mean incubation period and viral load threshold cycle (Ct) values at days 0, 7, 14 and 21 according to He et al. [12]. Median days from symptom onset to dyspnoea (interquartile range 4–9 days), ARDS (interquartile range 8–15 days), and discharge or death (interquartile range 17–25 days) according to Zhou et al. [14].
Current understanding of viral ARDS and host immune responses
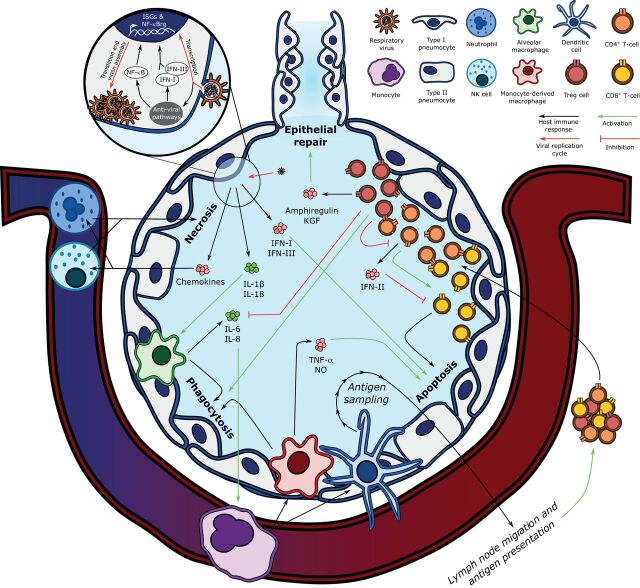
The Berlin Criteria define ARDS by acute hypoxaemic respiratory failure following an acute event (such as a respiratory viral infection) that presents as bilateral pulmonary infiltrates on lung imaging in the absence of a purely cardiogenic or hydrostatic aetiology [20]. ARDS is a heterogeneous clinical syndrome, encompassing a range of endotypes that require further characterisation for the development of effective therapeutics [21]. Much progress has been made in understanding the pathophysiology of viral pneumonia-induced ARDS, particularly in the context of influenza [3]. The canonical understanding of influenza-induced ARDS begins with the presence of its negative-sense, single-stranded RNA genome in the cytoplasm of respiratory epithelial cells (figure 2). Cytoplasmic viral RNA triggers the secretion of type-I and -III interferons (IFN-I and -III) and the pro-inflammatory cytokines interleukin (IL)-1β and IL-18 through the activation of well-conserved intracellular immune pathways such as Toll-like receptors, mitochondria-associated anti-viral signalling proteins, and the NOD-, LRR- and pyrin domain-containing protein 3 inflammasome [22, 23]. The induction of these anti-viral pathways results in the upregulation of interferon-stimulated genes and nuclear factor-κB-regulated genes, as well as the recruitment of effector and regulatory immune cells. Alveolar macrophages respond to these cues by phagocytosing infected and apoptotic epithelial cells, promoting viral clearance, and secreting more pro-inflammatory and chemotactic cytokines including IL-6 and IL-8, characteristic of a pro-inflammatory macrophage response [24]. These lung epithelial cell and alveolar macrophage responses work synergistically to recruit other immune cell types, inducing the invasion of neutrophils and natural killer (NK) cells and the differentiation of circulating monocytes into monocyte-derived macrophages and dendritic cells.
FIGURE 2.
Key components of the pathogenesis of influenza-induced acute respiratory distress syndrome. Influenza's replication cycle requires the presence of viral RNA in the host epithelial cell's cytoplasm, which is recognised by anti-viral pathways such as Toll-like receptors, mitochondria-associated anti-viral signalling, and the NLRP3 inflammasome. The host cell responds by upregulating nuclear factor (NF)-κB-regulated genes (NF-κBrg) and secreting type-I and -III interferons (IFN-I and IFN-III), which stimulate the transcription of interferon-stimulated genes (ISGs). Infected epithelial cell-derived IFN-I and IFN-III induce epithelial cell apoptosis, and interleukin (IL)-1β and IL-18 secretion activates alveolar macrophages, which phagocytose apoptotic cells. Alveolar macrophages also secrete IL-6 and IL-8, which recruit the central immune system, including monocytes, to the lung parenchyma. Monocytes differentiate into monocyte-derived macrophages and dendritic cells. Monocyte-derived macrophages also contribute to infected epithelial cell phagocytosis and secrete tumour necrosis factor (TNF)-α and nitric oxide (NO) to activate epithelial cell apoptosis. Dendritic cells link the innate and adaptive immune responses by sampling alveolar antigens and migrating to lymph nodes, where they present viral antigens to naïve T-cells. After undergoing antigen-specific clonal expansion, T-lymphocytes are mobilised to the lung. CD8+ T-cells exert their cytotoxicity on viral antigen-presenting cells when co-stimulated by CD4+ T-cells, which also secrete type-II interferon (IFN-II), a negative regulator of CD8+ T-cell activity. In turn, CD4+ T-cells are negatively regulated by regulatory T-cells (Treg), which also promote epithelial repair via the secretion of amphiregulin and keratinocyte growth factor (KGF), and attenuate inflammation by inhibiting alveolar macrophage-derived IL-6 and IL-8 secretion while potentiating their phagocytic activity (efferocytosis). NK: natural killer.
As part of the innate immune response, neutrophils and NK cells are recruited to the lung parenchyma in response to the secretion of chemotactic cytokines, including CCL2, CCL5, CXCL8 and CXCL10 [25, 26]. Neutrophils induce nonspecific epithelial cell necrosis via the secretion of many effector compounds, including neutrophil extracellular traps. While these processes exert protective effects against influenza virus and other pathogens, they also contribute to lung injury when they remain unchecked [27]. Similarly, NK cells are actively recruited during influenza infection and exert both pro-necrotic and -apoptotic effects via the secretion of cytotoxic granzymes and perforins; indeed, excessive NK cell-mediated cytotoxicity is associated with lethal influenza infection from uncontrolled lung injury [28]. Infiltrating monocyte-derived macrophages and dendritic cells further contribute to the immune response by secreting other pro-inflammatory molecules including TNF-α and nitric oxide, known to drive influenza clearance and alveolar injury through the induction of epithelial cell apoptosis [29, 30]. Dendritic cells also bridge the innate and adaptive immune systems by sampling viral antigens from lung alveoli and migrating to draining lymph nodes. There, they act as antigen-presenting cells for naïve CD8+ and CD4+ T-cells, activating them to expand and mature in an antigen-specific manner, after which they are mobilised to the lung [31]. Once in the lung, CD8+ T-cells induce the lysis of cells presenting viral antigens, and CD4+ T-cells modulate the inflammatory response in a myriad of ways, including the secretion of additional cytokines such as IFN-II [32, 33].
These immune processes result in the active clearance of the influenza virus at the expense of severe lung injury from epithelial cell destruction and prolonged inflammation, manifesting as ARDS in a subset of patients. Interestingly, post mortem studies of patients that succumbed to influenza induced-ARDS showed undetectable viral loads in most patients, suggesting that the direct cytotoxic effects of the virus are not the only drivers of mortality; the host's inability to dampen inflammation and repair damaged lung tissue contribute to a great degree [34]. These findings have generated substantial interest in the physiologic processes that mediate the resolution of inflammation and repair of lung injury and how they contribute to viral pneumonia-induced ARDS-related morbidity and mortality. Many host components responsible for mediating the recovery phase have been identified, including FOXP3+ regulatory T-cells (Treg) [35–42]. Treg cells are well known for their role in promoting immune self-tolerance through the negative regulation of CD4+ T cells, but they also serve other homeostatic functions, mediating resolution of inflammation by inhibiting the pro-inflammatory effects of macrophages and enhancing their phagocytic activity [39, 42, 43]. Treg cells also promote lung epithelial cell proliferation via the secretion of amphiregulin, a ligand of the epidermal growth factor receptor, and keratinocyte growth factor [44, 45]. These processes modulate the host immune response and help to restore lung parenchymal homeostasis. Consequently, their potential malfunction partially explains the wide spectrum of outcomes among patients with influenza virus-induced ARDS.
Insights into COVID-19-induced ARDS
The heterogeneity associated with the clinical presentation of COVID-19 has prompted the conceptualisation of novel paradigms of respiratory disease in an effort to explain the observed variability and individualise clinical management of COVID-19 [46]. Nevertheless, a recent cohort study reported that as many as 85% of ICU patients with COVID-19 meet the Berlin Criteria definition of ARDS and that well-established supportive interventions for ARDS, such as low tidal volumes and prone ventilation, resulted in significant improvement in oxygenation and lung compliance [47]. It is therefore reasonable to explore other causes of viral pneumonia-induced ARDS to glean insights into severe COVID-19 while we await more disease-focused data. Accordingly, there is evidence suggesting both parallels and contrasts between influenza and SARS-CoV-2 infections [48].
Global immune signature of SARS-CoV-2
Early work performed to characterise the host immune response of COVID-19 suggested an immune signature consisting of elevated serum cytokines (particularly IL-1β, IL-6 and tumour necrosis factor (TNF)-α), impaired interferon responses, and peripheral lymphopenia as markers of severe disease; other associated inflammatory serum markers include elevated levels of ferritin, lactate dehydrogenase, d-dimer, C-reactive protein, and coagulation factors [4, 14, 49]. Furthermore, transcriptional profiling of lung epithelial cells following SARS-CoV-2 and influenza infections in vitro revealed similar dampening of IFN-I and -III signalling in the host response to both of these viruses and a shared cytokine signature (including IL-6 and TNF-α). In contrast, the SARS-CoV-2-specific immune signature identified in vitro was characterised by high IFN-II signalling and chemokine expression relative to influenza [50].
Impaired IFN-I and -III responses
Both COVID-19 and influenza are associated with an impaired IFN-I and -III host response relative to other viruses, and COVID-19 severity correlates with the degree of impairment [50, 51]. The influenza and SARS-CoV genomes code for the nonstructural protein 1 (NS1), which antagonises IFN-I and -III signalling; similarly, the SARS-CoV-2 proteins ORF6, ORF8 and its nucleocapsid also inhibit IFN-I signalling in vitro [52–54]. The conserved nature of NS1, combined with the associated COVID-19 IFN-I and -III impairment, suggest that SARS-CoV-2 has shared virulence factors with SARS-CoV that are also recapitulated in influenza infection. IFN-I modulates the pro-inflammatory response of macrophages, making it reasonable to hypothesise that downregulation of IFN-I could explain the hyperinflammatory state associated with COVID-19 [55]. However, pharmacologic inhibition of IFN-I signalling in SARS-CoV-2-infected cells in vitro did not increase cytokine production, suggesting that impaired IFN-I secretion is insufficient to explain the severe hyperinflammatory state of COVID-19 [50]. IFN-III (or IFN-λ) shares similar functions with IFN-I but is more potent in promoting influenza clearance without inducing excessive inflammation [56]. IFN-III is essential in mediating the initial response to influenza, holding non-redundant roles in regulating the tissue-destructive properties of neutrophils as well as in potentiating CD8+ memory T-cell immunity against influenza by inducing the migration of dendritic cells to draining lymph nodes [57, 58]. These observations have made pharmacologic IFN-λ a candidate for the management of COVID-19, although murine studies of the deleterious effect of IFN-λ on the host's susceptibility to secondary bacterial infections raise concerns about its safety [59]. Nevertheless, given the observation that these pro-inflammatory molecules are able to limit SARS-CoV-2 replication in vitro, ongoing clinical trials are ascertaining the therapeutic benefit of exogenous IFN-I and IFN-III administration for COVID-19 (clinicaltrials.gov/ NCT04343768 and NCT04354259, respectively) [60].
Cytokine storm as driver of COVID-19 severity
The overlap in secreted cytokines in response to SARS-CoV-2 and influenza can be explained by the presence of viral RNA in the host cell's cytoplasm during the replication cycle of both viruses, which likely induces the activation of similar intracellular anti-viral pathways and subsequent recruitment of similar immune cells to the respiratory epithelium. The pro-inflammatory immune signature of SARS-CoV-2 has been likened to macrophage-activation syndrome (MAS), a life-threatening clinical entity observed in autoimmune diseases and mimicked in many viral infections, including influenza [61, 62]. MAS is associated with impaired cytolytic activity of NK cells and specific CD8+ T-cell subpopulations, which are tasked with lysing infected host cells to prevent prolonged secretion of inflammatory cytokines by compromised cells [63]. Such impairment in cell-mediated lysis is driven by high levels of IL-6, establishing a vicious cycle of cytokine-driven pro-inflammatory cytokine secretion. Therefore, elevated IL-6 levels and their association with severe disease are thought to reflect an over-exuberant inflammatory response that lacks proper regulation and resolution, mimicking an MAS-like pathologic state.
Vasculopathy of COVID-19 and implications for the cytokine storm hypothesis
SARS-CoV-2's expanded tropism poses a substantial challenge when attempting to use the influenza associated cytokine storm to derive conclusions about the potential virulence mechanisms of COVID-19. Importantly, available data from patients who succumbed to COVID-19 suggest that SARS-CoV-2 infects endothelial cells to cause inflammation (endothelialitis) [64]. This observed endothelialitis supports the idea that SARS-CoV-2 has tropism for vascular endothelial cells, which express the ACE receptor [10]. Moreover, viral cytotoxicity could be playing a larger role in mediating severe COVID-19 than in influenza, since post mortem detection of replicating virus is less frequent in the latter [34, 65]. These findings could also explain the multi-system organ failure and hypercoagulable state associated with severe COVID-19, since local pulmonary endothelialitis would result in activation of the coagulation cascade and exuberant production of endothelium-derived pro-inflammatory cytokines without the need to invoke an MAS-like pathologic state. Moreover, reported plasma IL-6 levels in COVID-19 patients appear to be significantly lower on average (10- to 40-fold) when compared with those reported in other non-COVID-19 ARDS cohorts that display signs of a cytokine storm [66]. These observations lend less credibility to the hypothesis that elevated serum cytokines are driving the unprecedented morbidity and mortality observed in severe COVID-19, suggesting instead that they are consequences of local vasculopathy. Of note, a clinical trial exploring the therapeutic benefit of monoclonal IL-6 receptor antibodies for COVID-19 was recently discontinued in the absence of a detectable benefit (clinicaltrials.gov/ NCT04315298).
The dichotomous role of interferon-II
There are important differences between influenza and COVID-19 regarding the role of IFN-II (or IFN-γ) in driving disease severity. Previous studies on the role of IFN-γ in the host response to influenza revealed that attenuation of IFN-γ signalling during the late phases of inflammation can improve clinically relevant outcomes by promoting survival of CD8+ T-cells [67, 68]. This protective effect of low IFN-γ in influenza differs with the inverse correlation observed between levels of IFN-γ produced by CD4+ T-cells and the severity of COVID-19 [4]. Moreover, SARS-CoV-2 was shown to induce a stronger IFN-γ response in vitro compared with influenza, suggesting that IFN-γ might play a larger role in the host immune response to COVID-19 [50]. Some have suggested that the IL-6-to-IFN-γ ratio could be used as a clinical tool to stratify patients by their relative risk of developing severe COVID-19, and preliminary analyses of this ratio as a risk-stratifying measure for severe COVID-19 showed promising results [69]. Nevertheless, a better understanding of the role of IFN-γ in driving severe disease and the validation of the IL-6-to-IFN-γ ratio in larger cohorts will be required before its deployment as a reliable clinical tool.
COVID-19-associated lymphopenia
Severe COVID-19 is associated with lymphopenia that disproportionately affects the T- rather than B-cell compartment [4]. Similar changes in lymphocyte counts occur in other viral infections via mechanisms such as TNF-α-mediated inhibition of lymphocyte recirculation [70]. In fact, IL-6 inhibition improved the lymphopenia of severe disease in a subset of COVID-19 patients, suggesting a role for elevated serum cytokines in depleting peripheral T-lymphocytes [71]. Nevertheless, recent immune profiling of COVID-19 patients revealed that the T-cell compartment, although more strongly affected in severe versus moderate COVID-19, is still significantly diminished in the peripheral blood of COVID-19 patients compared with cases of non-COVID-19 pneumonia [72]. This finding suggests that cytokine excess is not the only causal factor for the lymphopenia of COVID-19. Another potential explanation for COVID-19-associated lymphopenia is the excessive recruitment and sequestration of lymphocytes in the lung. In support of this hypothesis, post mortem examination of independent cohorts of COVID-19 patients identified pulmonary lymphoid infiltrates in a subset of cases, and similar observations were made in bronchoalveolar lavage fluid samples from patients with both mild and severe COVID-19 [64, 73, 74]. Finally, some hypothesise this lymphopenia to be a direct consequence of the expanded tropism of SARS-CoV-2, which could confer cytotoxicity to T-cell populations; however, such tropism remains unverified. As lymphopenia is being considered for patient risk stratification, elucidating the aetiology of COVID-19-associated lymphopenia is crucial for our understanding of COVID-19 pathogenesis and efforts to identify patients at risk of developing severe disease [75].
Transmissibility profile and host risk factors for severe disease
Current models of the transmissibility of COVID-19 recapitulate an influenza-like profile of viral shedding, suggesting that contact precautions for influenza could prove efficacious in the context of COVID-19 [12]. Importantly, there are key differences between these two viruses with respect to identified host risk factors for severe disease. For example, influenza-induced ARDS carries a female predominance, while COVID-19-induced ARDS is mostly associated with male sex, suggesting that they exploit different host susceptibilities to enhance their pathogenicity [76, 77]. Regardless, advanced age still remains the main predictor for the development of severe disease associated with both pathogens, signifying that our knowledge of advanced age as a risk factor for influenza-induced ARDS can inform our understanding of age in predisposing to severe COVID-19.
Potential mechanisms through which ageing drives severe COVID-19
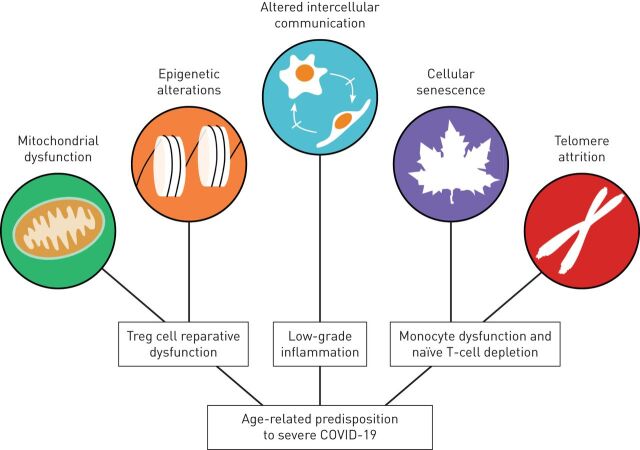
Advanced age is associated with an increased risk of developing life-threatening infections [78]. Indeed, almost 90% of influenza-related mortality and a disproportionate fraction of COVID-19-related mortality occurs in individuals aged >65 years [4, 5, 79, 80]. This age-related susceptibility to severe disease does not arise solely because of prolonged exposure to environmental and host risk factors, such as tobacco smoke or primary hypertension, but also through the development of maladaptive physiological processes that affect an individual's ability to maintain homeostasis when faced with a stressor. These processes have been coined the hallmarks of ageing and have been broadly associated with an age-related susceptibility to stress, also known as homeostenosis [81, 82]. The increasingly older global population and its myriad of age-related comorbidities have generated substantial interest in understanding the pathophysiology of these hallmarks of ageing, particularly as they relate to the risk of severe COVID-19. While their influence has not been elucidated in the context of SARS-CoV-2 infection, our current knowledge of their role in influenza provides an insight into why ageing is a risk factor for the development of severe COVID-19 (figure 3).
FIGURE 3.
Examples of the contribution of hallmarks of ageing to the age-related predisposition to severe coronavirus disease 2019 (COVID-19). Age-related mitochondrial dysfunction can induce epigenetic changes in regulatory T-cells (Treg), which impair their pro-recovery functions to hinder proper resolution of inflammation and repair of lung injury. Monocytes and naïve T-lymphocytes undergo cellular senescence following telomere attrition from sustained replication, impairing the host's ability to mount an efficient immune response to a viral challenge or create a memory T-cell response to vaccines. Finally, altered intercellular communication underlies the low-grade inflammation associated with ageing, which contributes to the development of age-related comorbidities.
Epigenetic alterations and mitochondrial dysfunction
Advanced age is associated with changes in the epigenetic and metabolic landscape of immune cells that mediate the host response to viral pneumonia-induced ARDS, including Treg cells [39]. The pro-repair function of Treg cells is associated with DNA hypomethylation of Treg cell lineage-specific loci. Indeed, inhibition of DNA methyltransferases in Treg cells accelerates lung injury resolution in a murine influenza model, suggesting that DNA methylation is required to maintain the Treg cell lineage over the lifespan but impairs Treg cell reparative function after lung injury [40]. While alterations in Treg cell function and numbers have been shown to occur with ageing, the mechanisms through which these changes occur remain unclear; however, recent studies suggest they could be mediated by the accumulation of toxic metabolites and reactive oxygen species caused by the mitochondrial dysfunction associated with ageing [83]. For example, intracellular accumulation of the metabolite L-2-hydroxyglutarate from its synthesis during hypoxia and other states of mitochondrial dysfunction induces DNA hypermethylation and impairs suppressive function in Treg cells [84]. Such metabolite accumulation could ultimately inhibit Treg cell-dependent ARDS recovery by repressing the expression of key loci associated with Treg cell pro-resolution and pro-repair pathways. Given the age-related phenomenon of mitochondrial dysfunction, irrespective of its proximate cause, it is plausible that similar phenomena are occurring across other cell types also required for viral ARDS resolution.
Telomere attrition and cellular senescence
Ageing is associated with the erosion of chromosomal telomeres from successive cellular replication (telomere attrition) and the concomitant cell cycle arrest of somatic cells (cellular senescence), including monocytes and lymphocytes. Monocytes from aged mice suffer from an inability to properly phagocytose bacterial pathogens, while producing significantly higher levels of TNF-α relative to young mice. These findings correlate with the telomere length of monocytes, which are shorter in the elderly, suggesting an age-related impairment in pathogen clearance and excessive TNF-α-mediated apoptosis that is in part mediated by telomere attrition [85]. These age-related phenomena also affect naïve T- and B-cell numbers, manifesting as low peripheral blood counts of these cells in older hosts [86]. Coupled with the process of thymic involution, age-related depletion of naïve T cell counts poses a substantial impediment for the adaptive immune system's ability to generate antigen-specific memory lymphocytes [87]. Accordingly, a recent genome-wide association study of genetic factors linked to severe COVID-19 susceptibility identified a risk allele associated with decreased levels of CXCR6 expression, which is a chemokine receptor expressed in tissue resident memory T-cells [88]. This finding supports the hypothesis that impaired memory T-cell function predisposes to developing severe COVID-19. Age-related naïve T-cell deficiency is also hypothesised to play a role in the prolonged hospital stays and increased susceptibility to severe disease in the ageing population affected by influenza-induced ARDS [89]. Finally, naïve T-cell depletion also underlies the decreased efficacy of seasonal influenza vaccines observed with increasing age, restricting preventive therapeutic alternatives for older adults [80].
Altered intercellular communication
The replicative senescence related to ageing affects a variety of somatic cells and is associated with a shift toward a basal, chronic secretion of pro-inflammatory cytokines, a phenomenon termed the senescence-associated secretory phenotype (SASP) [82]. According to the senescence hypothesis, accumulation of these SASP-like cells throughout ageing results in the persistent recruitment and activation of effector immune cells that perturb the local communication of the pro- and anti-inflammatory arms of the immune system, inducing tissue damage and impeding tissue repair [90]. Accordingly, selective depletion of stress-induced senescent cells in vivo attenuated the SASP-related transcriptional signal following acute lung injury and improved physical function in mice, suggesting that the accumulation of senescent alveolar epithelial cells ultimately compromises the host's ability to properly recover from acute lung injury [91]. Such pro-inflammatory states may also explain the predisposition to many age-associated pathologies, including primary hypertension, the comorbidity most strongly correlated with an individual's risk for developing severe COVID-19 [92].
Conclusion
Our evolving understanding of COVID-19 suggests that the pathogenesis of influenza provides insights into COVID-19, particularly with regard to the impaired IFN-I and -III host response. However, findings in influenza fall short in explaining the distinct clinical course in severe COVID-19, endothelialitis, the role of IFN-II, and the lymphopenia associated with severe COVID-19. We suggest that such differences between these two infections represent SARS-CoV-2-specific virulence mechanisms that, if elucidated, could inform the development of novel therapeutic strategies. Therefore, we contend that these differences need to be accounted for and should be a focus for ongoing research efforts. We also speculate that the severe COVID-19-specific immune signature, coupled with age-related maladaptive phenomena, could impede the host's ability to dampen the inflammatory phase of severe COVID-19 and resolve lung injury, contributing to the protracted ICU care episodes and the attendant morbidity and mortality associated with prolonged ICU stays among older adults. Given the interdependent and synergistic nature of these hallmarks of ageing, we suggest that these hallmarks' influence on relative susceptibility to developing severe COVID-19 should be studied in concert. Ultimately, the mechanistic associations between the biology of ageing and the relative susceptibility to severe viral pneumonia-induced ARDS carry substantial clinical implications that require careful consideration in future efforts to develop population-specific preventive and therapeutic interventions for COVID-19.
Shareable PDF
Footnotes
Conflict of interest: M.A. Torres Acosta has nothing to disclose.
Conflict of interest: B.D. Singer has a patent US Patent App. 15/542,380, “Compositions and Methods to Accelerate Resolution of Acute Lung Inflammation” pending.
Support statement: M.A. Torres Acosta was supported by NIH award T32GM008152. B.D. Singer was supported by NIH awards K08HL128867, U19AI135964, R01HL149883, and P01AG049665. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with Pneumonia in China, 2019. N Engl J Med 2020; 382: 727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Coronavirus disease (COVID-19) pandemic. www.who.int/emergencies/diseases/novel-coronavirus-2019 Date last accessed: 10 July 2020.
- 3.Herold S, Becker C, Ridge KM, et al. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur Respir J 2015; 45: 1463–1478. doi: 10.1183/09031936.00186214 [DOI] [PubMed] [Google Scholar]
- 4.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020; 130: 2620–2629. doi: 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Onder G, Rezza G, Brusaferro S. Case-fatality rate and characteristics of patients dying in relation to COVID-19 in Italy. JAMA 2020; 323: 1775–1776 [DOI] [PubMed] [Google Scholar]
- 6.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses . The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020; 5: 536–544. doi: 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–280.doi: 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jain J, Gaur S, Chaudhary Y, et al. The molecular biology of intracellular events during Coronavirus infection cycle. Virusdisease 2020; 31: 1–5. doi: 10.1007/s13337-020-00591-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 2020; 181: 281–292. doi: 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamming I, Timens W, Bulthuis ML, et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 2004; 203: 631–637. doi: 10.1002/path.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 2020; 20: 565–574. doi: 10.1016/S1473-3099(20)30196-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 2020; 26: 672–675. doi: 10.1038/s41591-020-0869-5 [DOI] [PubMed] [Google Scholar]
- 13.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382: 1177–1179. doi: 10.1056/NEJMc2001737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020; 395: 1054–1062. doi: 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Morales AJ, Cardona-Ospina JA, Gutierrez-Ocampo E, et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Travel Med Infect Dis 2020; 34: 101623. doi: 10.1016/j.tmaid.2020.101623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. Sci China Life Sci 2020; 63: 706–711. doi: 10.1007/s11427-020-1661-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Felice FG, Tovar-Moll F, Moll J, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the central nervous system. Trends Neurosci 2020; 43: 355–357. doi: 10.1016/j.tins.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 2020; 8: 475–481. doi: 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ARDS Definition Task Force , Ranieri VM, Rubenfeld GD, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA 2012; 307: 2526–2533. [DOI] [PubMed] [Google Scholar]
- 21.Calfee CS, Delucchi K, Parsons PE, et al. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2014; 2: 611–620. doi: 10.1016/S2213-2600(14)70097-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durbin RK, Kotenko SV, Durbin JE. Interferon induction and function at the mucosal surface. Immunol Rev 2013; 255: 25–39. doi: 10.1111/imr.12101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pothlichet J, Meunier I, Davis BK, et al. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog 2013; 9: e1003256. doi: 10.1371/journal.ppat.1003256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Nikrad MP, Travanty EA, et al. Innate immune response of human alveolar macrophages during influenza A infection. PLoS One 2012; 7: e29879. doi: 10.1371/journal.pone.0029879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol 2017; 8: 550. doi: 10.3389/fimmu.2017.00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlin LE, Hemann EA, Zacharias ZR, et al. Natural killer cell recruitment to the lung during influenza A virus infection is dependent on CXCR3, CCR5, and virus exposure dose. Front Immunol 2018; 9: 781. doi: 10.3389/fimmu.2018.00781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narasaraju T, Yang E, Samy RP, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol 2011; 179: 199–210. doi: 10.1016/j.ajpath.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdul-Careem MF, Mian MF, Yue G, et al. Critical role of natural killer cells in lung immunopathology during influenza infection in mice. J Infect Dis 2012; 206: 167–177. doi: 10.1093/infdis/jis340 [DOI] [PubMed] [Google Scholar]
- 29.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 2012; 122: 787–795. doi: 10.1172/JCI59643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.AldridgeJR, Jr., Moseley CE, Boltz DA, et al. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci USA 2009; 106: 5306–5311. doi: 10.1073/pnas.0900655106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.GeurtsvanKessel CH, Willart MA, van Rijt LS, et al. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J Exp Med 2008; 205: 1621–1634. doi: 10.1084/jem.20071365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Topham DJ, Tripp RA, Doherty PC. CD8(+) T cells clear influenza virus by perforin or Fas-dependent processes. J Immunol 1997; 159: 5197–5200. [PubMed] [Google Scholar]
- 33.Kim TS, Sun J, Braciale TJ. T cell responses during influenza infection: getting and keeping control. Trends Immunol 2011; 32: 225–231. doi: 10.1016/j.it.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shieh WJ, Blau DM, Denison AM, et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol 2010; 177: 166–175. doi: 10.2353/ajpath.2010.100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal NR, Tsushima K, Eto Y, et al. Immunological priming requires regulatory T cells and IL-10-producing macrophages to accelerate resolution from severe lung inflammation. J Immunol 2014; 192: 4453–4464. doi: 10.4049/jimmunol.1400146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Alessio FR, Craig JM, Singer BD, et al. Enhanced resolution of experimental ARDS through IL-4-mediated lung macrophage reprogramming. Am J Physiol Lung Cell Mol Physiol 2016; 310: L733–L746. doi: 10.1152/ajplung.00419.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath-Morrow SA, Ndeh R, Helmin KA, et al. DNA methylation regulates the neonatal CD4(+) T-cell response to pneumonia in mice. J Biol Chem 2018; 293: 11772–11783. doi: 10.1074/jbc.RA118.003589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mock JR, Garibaldi BT, Aggarwal NR, et al. Foxp3+ regulatory T cells promote lung epithelial proliferation. Mucosal Immunol 2014; 7: 1440–1451. doi: 10.1038/mi.2014.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer BD, Chandel NS. Immunometabolism of pro-repair cells. J Clin Invest 2019; 129: 2597–2607. doi: 10.1172/JCI124613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singer BD, Mock JR, Aggarwal NR, et al. Regulatory T cell DNA methyltransferase inhibition accelerates resolution of lung inflammation. Am J Respir Cell Mol Biol 2015; 52: 641–652. doi: 10.1165/rcmb.2014-0327OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter JM, Helmin KA, Abdala-Valencia H, et al. Multidimensional assessment of alveolar T cells in critically ill patients. JCI Insight 2018; 3: e123287. doi: 10.1172/jci.insight.123287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Alessio FR, Tsushima K, Aggarwal NR, et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J Clin Invest 2009; 119: 2898–2913. doi: 10.1172/JCI36498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proto JD, Doran AC, Gusarova G, et al. Regulatory T cells promote macrophage efferocytosis during inflammation resolution. Immunity 2018; 49: 666–677.doi: 10.1016/j.immuni.2018.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arpaia N, Green JA, Moltedo B, et al. A Distinct function of regulatory T cells in tissue protection. Cell 2015; 162: 1078–1089. doi: 10.1016/j.cell.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dial CF, Tune MK, Doerschuk CM, et al. Foxp3(+) regulatory T cell expression of keratinocyte growth factor enhances lung epithelial proliferation. Am J Respir Cell Mol Biol 2017; 57: 162–173. doi: 10.1165/rcmb.2017-0019OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gattinoni L, Chiumello D, Caironi P, et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med 2020; 46: 1099–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziehr DR, Alladina J, Petri CR, et al. Respiratory pathophysiology of mechanically ventilated patients with COVID-19: a cohort study. Am J Respir Crit Care Med 2020; 201: 1560–1564. doi: 10.1164/rccm.202004-1163LE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang X, Du R, Wang R, et al. Comparison of hospitalized patients with ARDS caused by COVID-19 and H1N1. Chest 2020; 158: 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goshua G, Pine AB, Meizlish ML, et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol 2020; 7: e575–e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020; 181: 1036–1045. doi: 10.1016/j.cell.2020.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe 2020; 27: 883–890. doi: 10.1016/j.chom.2020.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia D, Rahbar R, Chan RW, et al. Influenza virus non-structural protein 1 (NS1) disrupts interferon signaling. PLoS One 2010; 5: e13927. doi: 10.1371/journal.pone.0013927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zust R, Cervantes-Barragan L, Kuri T, et al. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 2007; 3: e109. doi: 10.1371/journal.ppat.0030109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li JY, Liao CH, Wang Q, et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res 2020; 286: 198074. doi: 10.1016/j.virusres.2020.198074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Channappanavar R, Fehr AR, Vijay R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 2016; 19: 181–193. doi: 10.1016/j.chom.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidson S, McCabe TM, Crotta S, et al. IFNlambda is a potent anti-influenza therapeutic without the inflammatory side effects of IFNalpha treatment. EMBO Mol Med 2016; 8: 1099–1112. doi: 10.15252/emmm.201606413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemann EA, Green R, Turnbull JB, et al. Interferon-lambda modulates dendritic cells to facilitate T cell immunity during infection with influenza A virus. Nat Immunol 2019; 20: 1035–1045. doi: 10.1038/s41590-019-0408-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galani IE, Triantafyllia V, Eleminiadou EE, et al. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 2017; 46: 875–890. doi: 10.1016/j.immuni.2017.04.025 [DOI] [PubMed] [Google Scholar]
- 59.Broggi A, Ghosh S, Sposito B, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 2020; 369: 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Felgenhauer U, Schoen A, Gad HH, et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J Biol Chem 2020; in press [ 10.1074/jbc.AC120.013788]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulert GS, Zhang M, Fall N, et al. Whole-exome sequencing reveals mutations in genes linked to hemophagocytic lymphohistiocytosis and macrophage activation syndrome in fatal cases of H1N1 influenza. J Infect Dis 2016; 213: 1180–1188. doi: 10.1093/infdis/jiv550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lerkvaleekul B, Vilaiyuk S. Macrophage activation syndrome: early diagnosis is key. Open Access Rheumatol 2018; 10: 117–128. doi: 10.2147/OARRR.S151013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crayne CB, Albeituni S, Nichols KE, et al. The immunology of macrophage activation syndrome. Front Immunol 2019; 10: 119. doi: 10.3389/fimmu.2019.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med 2020; 383: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020; 395: 1417–1418. doi: 10.1016/S0140-6736(20)30937-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med 2020; 180: 1152–1154. [DOI] [PubMed] [Google Scholar]
- 67.Nicol MQ, Campbell GM, Shaw DJ, et al. Lack of IFNgamma signaling attenuates spread of influenza A virus in vivo and leads to reduced pathogenesis. Virology 2019; 526: 155–164. doi: 10.1016/j.virol.2018.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prabhu N, Ho AW, Wong KH, et al. Gamma interferon regulates contraction of the influenza virus-specific CD8 T cell response and limits the size of the memory population. J Virol 2013; 87: 12510–12522. doi: 10.1128/JVI.01776-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lagunas-Rangel FA, Chavez-Valencia V. High IL-6/IFN-gamma ratio could be associated with severe disease in COVID-19 patients. J Med Virol 2020; 92: 1789–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shiow LR, Rosen DB, Brdickova N, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 2006; 440: 540–544. doi: 10.1038/nature04606 [DOI] [PubMed] [Google Scholar]
- 71.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 2020; 27: 992–1000.doi: 10.1016/j.chom.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang X, Du RH, Wang R, et al. Comparison of hospitalized patients with ARDS caused by COVID-19 and H1N1. Chest 2020; 158: 195–205. doi: 10.1016/j.chest.2020.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menter T, Haslbauer JD, Nienhold R, et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020; 77: 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voiriot G, Fajac A, Lopinto J, et al. Bronchoalveolar lavage findings in severe COVID-19 pneumonia. Intern Emerg Med 2020; in press [ 10.1007/s11739-020-02356-6]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan L, Wang Q, Zhang D, et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther 2020; 5: 33. doi: 10.1038/s41392-020-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mertz D, Kim TH, Johnstone J, et al. Populations at risk for severe or complicated influenza illness: systematic review and meta-analysis. BMJ 2013; 347: f5061. doi: 10.1136/bmj.f5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angus DC, Linde-Zwirble WT, Lidicker J, et al. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 2001; 29: 1303–1310. doi: 10.1097/00003246-200107000-00002 [DOI] [PubMed] [Google Scholar]
- 79.Thompson W, Comanor L DKS. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis 2006; 194: S82–S91. doi: 10.1086/507558 [DOI] [PubMed] [Google Scholar]
- 80.Kostova D, Reed C, Finelli L, et al. Influenza illness and hospitalizations averted by influenza vaccination in the United States, 2005–2011. PLoS One 2013; 8: e66312. doi: 10.1371/journal.pone.0066312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lopez-Otin C, Blasco MA, Partridge L, et al. The hallmarks of aging. Cell 2013; 153: 1194–1217. doi: 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khan SS, Singer BD, Vaughan DE. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell 2017; 16: 624–633. doi: 10.1111/acel.12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morales-Nebreda L, McLafferty FS, Singer BD. DNA methylation as a transcriptional regulator of the immune system. Transl Res 2019; 204: 1–18. doi: 10.1016/j.trsl.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weinberg SE, Singer BD, Steinert EM, et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 2019; 565: 495–499. doi: 10.1038/s41586-018-0846-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hearps AC, Martin GE, Angelovich TA, et al. Aging is associated with chronic innate immune activation and dysregulation of monocyte phenotype and function. Aging Cell 2012; 11: 867–875. doi: 10.1111/j.1474-9726.2012.00851.x [DOI] [PubMed] [Google Scholar]
- 86.Yan J, Greer JM, Hull R, et al. The effect of ageing on human lymphocyte subsets: comparison of males and females. Immun Ageing 2010; 7: 4. doi: 10.1186/1742-4933-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Palmer DB. The effect of age on thymic function. Front Immunol 2013; 4: 316. doi: 10.3389/fimmu.2013.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med 2020; in press [ 10.1056/NEJMoa2020283]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moitra VK, Guerra C, Linde-Zwirble WT, et al. Relationship between ICU length of stay and long-term mortality for elderly ICU survivors. Crit Care Med 2016; 44: 655–662. doi: 10.1097/CCM.0000000000001480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coppe JP, Desprez PY, Krtolica A, et al. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol 2010; 5: 99–118. doi: 10.1146/annurev-pathol-121808-102144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cai Y, Zhou H, Zhu Y, et al. Elimination of senescent cells by beta-galactosidase-targeted prodrug attenuates inflammation and restores physical function in aged mice. Cell Res 2020; 30: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsioufis C, Dimitriadis K, Selima M, et al. Low-grade inflammation and hypoadiponectinaemia have an additive detrimental effect on aortic stiffness in essential hypertensive patients. Eur Heart J 2007; 28: 1162–1169. doi: 10.1093/eurheartj/ehm089 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-02049-2020.Shareable (373.5KB, pdf)