Abstract
Objective
To provide an understanding of the role of common genetic variations in colorectal cancer (CRC) risk, we report an updated field synopsis and comprehensive assessment of evidence to catalogue all genetic markers for CRC (CRCgene2).
Design
We included 869 publications after parallel literature review and extracted data for 1063 polymorphisms in 303 different genes. Meta-analyses were performed for 308 single nucleotide polymorphisms (SNPs) in 158 different genes with at least three independent studies available for analysis. Scottish, Canadian and Spanish data from genome-wide association studies (GWASs) were incorporated for the meta-analyses of 132 SNPs. To assess and classify the credibility of the associations, we applied the Venice criteria and Bayesian False-Discovery Probability (BFDP). Genetic associations classified as ‘positive’ and ‘less-credible positive’ were further validated in three large GWAS consortia conducted in populations of European origin.
Results
We initially identified 18 independent variants at 16 loci that were classified as ‘positive’ polymorphisms for their highly credible associations with CRC risk and 59 variants at 49 loci that were classified as ‘less-credible positive’ SNPs; 72.2% of the ‘positive’ SNPs were successfully replicated in three large GWASs and the ones that were not replicated were downgraded to ‘less-credible’ positive (reducing the ‘positive’ variants to 14 at 11 loci). For the remaining 231 variants, which were previously reported, our meta-analyses found no evidence to support their associations with CRC risk.
Conclusion
The CRCgene2 database provides an updated list of genetic variants related to CRC risk by using harmonised methods to assess their credibility.
Keywords: colorectal cancer
Summary box.
What is already known on this subject?
Colorectal cancer (CRC) is a global public health challenge. A large number of genetic association studies have been conducted to assess the potential correlation between common genetic variations and CRC risk.
What are the new findings?
Using an established framework for grading credibility of genetic associations, we classified 14 independent variants at 12 loci (MUTYH, SMAD7, TERT, CDH1, RHPN2, BMP2, TGFB1 and common variants tagging loci at 8q24, 8q23.3, 10p14, 11q23.1, 20p12.3) as highly credibly associated with CRC risk. A total of 63 variants at 52 loci were classified as ‘less-credible positive’ SNPs; variants of nine of these genes could be mostly highly prioritised for further investigation. For 231 variants previously reported to be associated with CRC, our meta-analyses found no evidence to support such associations.
How might it impact on clinical practice in the foreseeable future?
This database will be helpful for future research by promoting the investigation of these variants and corresponding genetic loci in populations other than of European origin, serving as a genetic basis for predicting risk estimates for population groups and providing candidate genes for further functional studies or gene-environment interaction studies.
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed malignancies and the leading cause of cancer deaths in the world, with 1.65 million new cases and about 835 000 deaths in 2015.1 The global burden of CRC is expected to increase by 60%, with more than 2.2 million new cases and almost 1.1 million deaths occurring annually by 2030.2 The distribution of CRC global burden varies widely, with more than two-thirds of all cases and about 60% of all deaths occurring in countries with a high or very high human development index, including Australia and New Zealand, Europe and North America, while incidence and mortality rates in Africa and South-Central Asia are relatively low.1 2 These geographic differences appear to be attributable to the differences in both environmental exposures and the background of genetically determined susceptibility.3
It is estimated that 15%–25% of CRC risk variance is attributed to inherited genetic factors, and the first-degree relatives of CRC patients have two to four times higher risk of developing CRC.4 5 The inherited genetic risk of CRC can be partly accounted for by a combination of rare high-penetrance mutations and large numbers of common genetic variants each conferring small risk.6 Although a number of highly penetrant mutations (eg, DNA mismatch repair genes, APC, SMAD4, LKB1/STK11, MUTYH) have been identified to influence CRC susceptibility with large effects, overall they account for only 2%–5% of incident CRC cases in the general population because these mutations are very rare.7–9 Candidate gene association studies have investigated the role of a large number of common genetic variants in CRC risk, but only a small number of them have been successfully replicated in subsequent studies.10 11
In 2012 and 2014, we reported two independently conducted series of meta-analyses to systematically evaluate associations between CRC and common variants using data from candidate gene studies and genome-wide association studies (GWASs) and identified a number of promising genetic risk variants for CRC risk.10 11 Our first field synopsis (published in 2012) reported 16 variants in 13 independent loci (MUTYH, MTHFR, SMAD7, 8q24, 8q23.3, 11q23.1, 14q22.2, 1q41, 20p12.3, 20q13.33, 3q26.2, 16q22.1, 19q13.1),10 and the second field synopsis (published in 2014) identified 8 additional variants in 5 independent loci (APC, CHEK2, DNMT3B, MLH1, MUTYH) that were strongly associated with CRC.11 These two synopses used slightly different methodologies, in that the 2012 field synopsis only included variants reported in four or more studies in meta-analyses, whereas the 2014 included variants with three or more studies, and there were some differences in the criteria applied for the evidence appraisal.10 11
In this study, we aimed to perform an updated field synopsis for CRC risk by including the most recently published genetic association studies, by following established guidelines12 13 and using harmonised methods for evidence appraisal.14–16 We systematically captured all published genetic association data on CRC for meta-analyses and subsequently incorporated data from GWAS consortia for interrogation. This study provides an up-to-date and publicly available database for CRC genetics (CRCgene2) and presents these data within a defined statistical and causal inference framework to aid interpretation of the results.13 We aimed to provide new insights into the fundamental biological mechanisms involved in colorectal carcinogenesis.
Methods
Literature search and data collection
To identify genetic association studies of CRC risk, we searched the Medline database via the Ovid gateway and the search terms comprising medical subject headings (MeSH) and keywords relating to colorectal neoplasms, the MeSH heading ‘genetic predisposition to disease’, and the keywords ‘gene$’ and ‘associate$’ were applied to terms in the entire article. The latest literature search was performed on 21 November 2018. We screened the eligibility of retrieved publications in a three-step parallel review of title, abstract and full text by following the predefined inclusion and exclusion criteria. Thus, each eligible study evaluated the association between a polymorphic genetic variant (with minor allele frequency ≥0.01 in the reference panel of the 1000 genomes) and a sporadic CRC. Studies investigating only premalignant conditions such as adenomas, polyps or dysplastic tissue were excluded. Studies investigating hereditary CRC syndromes, such as familial adenomatous polyposis, hereditary non-polyposis CRC, juvenile polyposis syndrome and Gardner’s syndrome; solely focusing on the progression or histological phenotype of CRC; or studies in animals; were excluded. Case-control, cohort and GWASs were included, while family-based studies were excluded. All included studies were published in English in a peer-reviewed journal; studies only reported as conference abstracts were excluded. Data from the eligible studies were abstracted into two standardised tables, including the key variables with regard to the study identifiers and context, study design and limitations, genotype information and outcome effects.
A list of genetic variants that were investigated in meta-analyses was summarised and data from three GWAS consortia (Scotland, Canada and Spain) were incorporated for meta-analyses, when the genotype data are available for the listed variants. In brief, the Study of CRC in Scotland (SOCCS) is a population-based case-control GWAS that includes 3417 cases and 3500 controls. The Assessment of Risk for Colorectal Tumors In Canada (ARCTIC) is a case-control GWAS database that includes 1231 cases and 1240 controls. The population-based cohort study in Spain comprised two phases (EPICOLON I and EPICOLON II) adding up to 2000 cases and 2000 controls. Restricted candidate gene genotyping data were available from both phases and GWAS data were only accessible from 881 cases and 667 controls from phase 2.17 18 More details about these GWAS datasets are present in online supplementary text.
gutjnl-2019-319313supp001.pdf (98.6KB, pdf)
Statistical analysis
Meta-analysis was performed for genetic variants with data available from at least three independent studies. Summary crude ORs and 95% CI for allelic, recessive and dominant genetic models were calculated by applying either the fixed-effect model (Mantel-Haenszel method) or the random-effects model (DerSimonian-Laird method) in case of the existence of substantial heterogeneity. The Q statistic (with a threshold of p value <0.05) and I2 metric were calculated to quantify between-study heterogeneity. Funnel plot analysis with an Egger test was conducted to test for small study effects. We also estimated the statistical power of each meta-analysis based on the significance level of α=0.05, the effect sizes and the allele frequencies of genetic variants (an integral component of the Bayesian False-Discovery Probability (BFDP) analysis).19 All statistical analysis was conducted by using R software (R x64 3.1.0).
Credibility of the identified genetic associations
We first applied the BFDP19 and the Venice criteria12 13 to assess the credibility of any observed genetic associations with p<0.05 in at least one genetic model. We then validated these associations in three additional GWAS consortia: Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO),20 Colorectal Transdisciplinary Study (CORECT, https://research.fhcrc.org/peters/en/corect-study.html) and Colon Colorectal Cancer Family Registry (CFR).21 With meta-analysis of these three GWAS datasets, we validated the observed genetic associations using data from 58 131 CRC cases and 67 347 controls (online supplementary text), and the statistical power of validation was estimated accordingly.
The BFDP assesses the noteworthiness of an observed association. The BFDP was selected rather than the false-positive report probability (FPRP) because it uses more information, defines the noteworthiness threshold explicitly in terms of the costs of false discovery and non-discovery, and does not suffer from the inferential limitations identified for the FPRP.22 We calculated BFDP values at two levels of prior probability: a medium/low prior level (0.05 to 10−3), close to what would be expected for a candidate gene, and a very low prior level (10−4 to 10−6), close to what would be expected for a random SNP. A noteworthy threshold was defined as 0.2 based on the assumption that the cost of false discovery would be four times higher than that of false non-discovery.19
According to the Venice criteria, the credibility of associations is assessed for three aspects: the amount of evidence, the extent of replication and the protection from bias.12 13 We used statistical power to assess the volume of evidence and a grade of A, B and C was assigned, respectively, when statistical power was greater than 80%, 50%–79% or less than 50%. The extent of replication was assessed by the measurement of heterogeneity (I2 criterion), and a grade of A, B and C was assigned, respectively, when I2 was less than 25%, 25%–49% or greater than 50%.12 For protection from bias, a complete assessment is difficult; instead, we considered the following aspects: (1) the phenotype definition was addressed by our inclusion criteria—namely that cases would have newly incident CRC; (2) genotyping error rates are generally low; (3) the criterion of replication across studies in part addresses potential concerns about variation in genotyping quality between studies; and (4) the magnitude of effect of population stratification appears to be small in general.23
Genetic associations were then classified into four categories based on the following criteria. Associations were classified as ‘positive’ if they: (1) were statistically significant at a p value level of 0.05 in at least two of the genetic models, (2) had a BFDP less than 0.20 at least at the p value level of 0.05, (3) had a statistical power greater than 80%, (4) had an I2 less than 50%. A class of ‘less-credible positive’, with a less-stringent threshold, was assigned to the associations (1) that were statistically significant at a p value threshold of 0.05 in at least one of the genetic models, but (2) their BFDP was greater than 0.20 or their statistical power was between 50% and 79% or had an I2 greater than 50%. Associations with p value large than 0.05 were further classified as ‘null’ or ‘negative’ by assessing if there are more than 5000 cases. After credibility assessment, genetic variants classified as ‘positive’ and ‘less-credible positive’ were sent to the CORECT coordinating centre to validate their associations with CRC risk using additive, dominant and recessive models. At this stage, ‘positive’ associations that failed to be validated (at p<0.05) were downgraded to ‘less-credible positive’. A schematic diagram is shown in figure 1 to demonstrate datasets included in each phase of the analysis.
Figure 1.
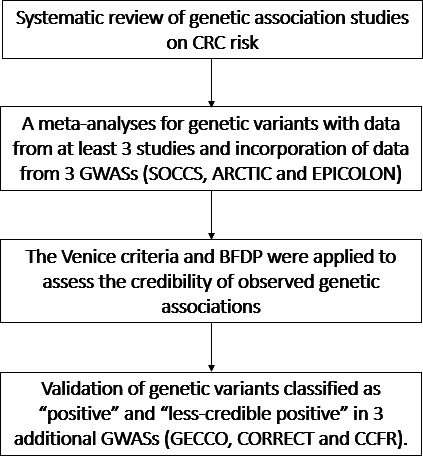
Diagram of the study design. ARCTIC, Assessment of Risk for Colorectal Tumors In Canada; BFDP, Bayesian False-Discovery Probability; CFR, Colorectal Cancer Family Registry; CORECT, Colorectal Transdisciplinary Study; CRC, colorectal cancer; EPICOLON, Gastrointestinal Oncology Group of the Spanish Gastroenterological Association; GECCO, Genetics and Epidemiology of Colorectal Cancer Consortium; GWASs, genome-wide association studies; SOCCS, Study of CRC in Scotland.
Results
Literature search and data collection
A total of 20 900 citations were identified from literature search. Of these, 6770 (32.4%) papers were published after the search period of the most recent field synopsis (31 December 2012).11 After eligibility screening, we finally included and extracted data from 869 publications (figure 2), reporting the association of CRC risk with 1063 polymorphisms in 303 different genes, of which 308 polymorphisms were reported in at least three independent studies.
Figure 2.
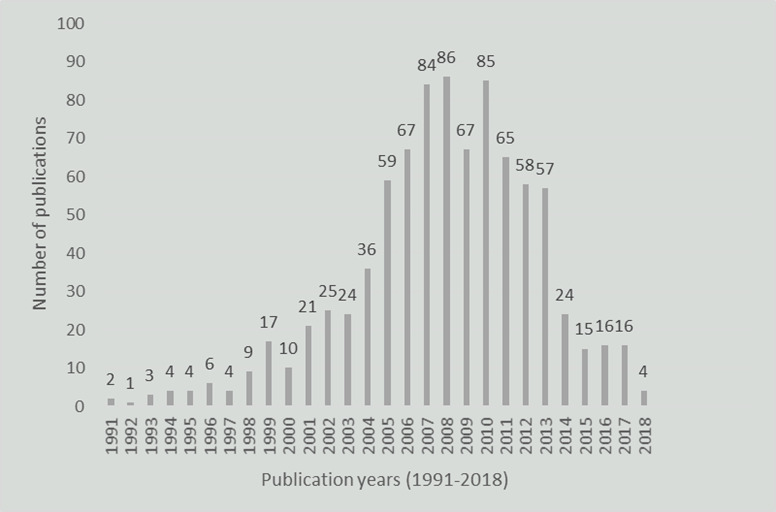
The distribution of included studies published from 1991 to 2018.
Meta-analyses
Meta-analyses were conducted for 308 polymorphisms in 158 different loci with data available in three or more candidate or GWA studies (online supplementary table 1). On average, these meta-analyses were based on 6149 CRC cases (median; IQR=2301–7334) and 7337 controls (median; IQR=2809–8885) originating from 8 (median; IQR=4–9) case–control studies. Data from the Scottish, Canadian and/ or Spanish GWAS were incorporated in the meta-analyses for 132 SNPs. Summary crude ORs and 95% CI for the allelic, dominant and recessive models are presented in table 1. Of the meta-analyses for 308 polymorphisms (tagged at different 158 loci), a total of 77 SNPs (25.6%) (tagged in 61 different loci) were identified to have a nominally statistically significant association (p value <0.05) with CRC risk in at least one of the three genetic models and were eligible for credibility assessment using the BFDP19 (online supplementary tables 2–4) and the Venice criteria12 13 and for validation in the three (GECCO, CORECT and CCFR) GWAS consortia (online supplementary tables 5–7).
Table 1.
Summary of ‘credible positive associations’ from meta-analyses and credibility assessment
| Gene | Variant | Cases vs controls (number of samples) |
Allelic model | Power | Recessive model: var/var vs wt/wt and wt/var | Dominant model: wt/var and var/var vs wt/wt | Classification | ||||||||||||||
| N | Effect size | Heterogeneity | N | Effect size | Heterogeneity | Power | N | Effect size | Heterogeneity | Power | |||||||||||
| OR (95% CI) | P value | I2 (95% CI) | P value | OR (95% CI) | P value | I2 (95% CI) | P value | OR (95% CI) | P value | I2 (95% CI) | P value | ||||||||||
| MUTYH | rs36053993 | 28 302 vs 20 935 (19*) | 19 | 1.42 (1.22 to 1.66) | 4.89E-06 | 0 (0 to 33) | 0.703 | 0.99 | na | na | na | na | na | na | 19 | 1.31 (1.12 to 1.53) | 0.001 | 0 (0 to 24) | 0.837 | 0.88 | Positive |
| MUTYH | rs34612342 | 28 180 vs 20 923 (19*) | 17 | 1.89 (1.47 to 2.42) | 5.72E-07 | 0 (0 to 62) | 0.828 | 1.00 | 7 | 3.40 (1.22 to 9.48) | 0.019 | 0 (0 to 0) | 0.998 | 1.00 | 17 | 1.80 (1.39 to 2.31) | 6.42E-06 | 0 (0 to 52) | 0.884 | 1.00 | Positive |
| SMAD7 | rs12953717 | 28 006 vs 26 539 (13*) | 13 | 1.11 (1.07 to 1.16) | 6.24E-07 | 49 (0 to 93) | 0.024 | 1.00 | 13 | 1.17 (1.08 to 1.26) | 8.34E-05 | 45 (0 to 94) | 0.041 | 1.00 | 13 | 1.14 (1.07 to 1.21) | 5.00E-05 | 54 (10 to 95) | 0.020 | 1.00 | Positive |
| SMAD7 | rs4464148 | 17 772 vs 17 356 (9*) | 9 | 1.12 (1.08 to 1.16) | 2.66E-09 | 12 (0 to 92) | 0.337 | 1.00 | 9 | 1.13 (1.00 to 1.28) | 0.054 | 49 (0 to 98) | 0.047 | 0.97 | 9 | 1.16 (1.11 to 1.21) | 2.09E-10 | 2 (0 to 86) | 0.416 | 1.00 | Positive |
| 8q24 | rs10505477 | 18 562 vs 20 132 (14) | 14 | 1.14 (1.10 to 1.19) | 2.04E-11 | 44 (0 to 82) | 0.040 | 1.00 | 14 | 1.16 (1.10 to 1.23) | 4.06E-07 | 35 (0 to 74) | 0.092 | 1.00 | 14 | 1.24 (1.17 to 1.32) | 1.04E-11 | 36 (0 to 81) | 0.084 | 1.00 | Positive |
| 20p12.3 | rs961253 | 22 971 vs 25 270 (14*) | 14 | 1.11 (1.06 to 1.16) | 1.58E-05 | 60 (28 to 91) | 0.002 | 1.00 | 13 | 1.14 (1.06 to 1.22) | 7.10E-04 | 36 (0 to 96) | 0.098 | 1.00 | 14 | 1.14 (1.07 to 1.21) | 4.75E-05 | 57 (21 to 88) | 0.004 | 1.00 | Positive |
| 8q23.3 | rs16892766 | 8351 vs 8878 (6*) | 6 | 1.24 (1.15 to 1.34) | 1.67E-08 | 4 (0 to 92) | 0.393 | 0.99 | 6 | 1.21 (0.88 to 1.67) | 0.240 | 0 (0 to 85) | 0.747 | 0.22 | 6 | 1.27 (1.17 to 1.38) | 4.00E-09 | 3 (0 to 90) | 0.400 | 1.00 | Positive |
| 10p14 | rs10795668 | 16 763 vs 18 302 (16*) | 16 | 0.88 (0.83 to 0.94) | 4.35E-05 | 67 (34 to 89) | 6.94E-05 | 1.00 | 16 | 0.83 (0.77 to 0.89) | 4.77E-07 | 11 (0 to 70) | 0.332 | 1.00 | 16 | 0.85 (0.79 to 0.93) | 2.27E-04 | 68 (34 to 87) | 3.06E-05 | 1.00 | Positive |
| 11q23.1 | rs3802842 | 22 320 vs 22 965 (20*) | 20 | 1.15 (1.11 to 1.20) | 5.21E-12 | 43 (0 to 78) | 0.024 | 1.00 | 20 | 1.24 (1.13 to 1.35) | 3.62E-06 | 45 (5 to 86) | 0.017 | 1.00 | 20 | 1.19 (1.13 to 1.25) | 7.22E-12 | 33 (0 to 70) | 0.075 | 1.00 | Positive |
| BMP2 | rs355527 | 13 539 vs 14 375 (9) | 9 | 1.12 (1.08 to 1.17) | 1.65E-10 | 0 (0 to 0) | 0.977 | 0.99 | 9 | 1.18 (1.09 to 1.27) | 3.54E-05 | 0 (0 to 94) | 0.458 | 0.99 | 9 | 1.16 (1.10 to 1.21) | 2.43E-09 | 0 (0 to 9) | 0.966 | 1.00 | Positive |
| CDH1 | rs1862748 | 17 436 vs 18 418 (11) | 11 | 0.91 (0.88 to 0.94) | 1.98E-08 | 0 (0 to 63) | 0.703 | 0.98 | 11 | 0.83 (0.77 to 0.90) | 1.59E-06 | 2 (0 to 68) | 0.427 | 1.00 | 11 | 0.91 (0.87 to 0.95) | 6.31E-06 | 0 (0 to 70) | 0.511 | 0.99 | Positive |
| RHPN2 | rs7259371 | 15 762 vs 16 700 (9*) | 9 | 0.87 (0.82 to 0.93) | 1.65E-05 | 52 (0 to 90) | 0.034 | 1.00 | 9 | 0.85 (0.73 to 0.98) | 0.027 | 17 (0 to 86) | 0.295 | 0.71 | 9 | 0.86 (0.80 to 0.92) | 3.81E-06 | 42 (0 to 87) | 0.084 | 1.00 | Positive |
| TERT | rs2736100 | 16 176 vs 18 135 (8) | 8 | 1.07 (1.04 to 1.10) | 2.95E-05 | 0 (0 to 82) | 0.526 | 0.88 | 8 | 1.06 (1.00 to 1.13) | 0.069 | 15 (0 to 86) | 0.316 | 0.64 | 8 | 1.13 (1.08 to 1.20) | 2.03E-06 | 0 (0 to 64) | 0.788 | 1.00 | Positive |
| TGFB1 | rs1800469 | 4021 vs 6024 (10) | 10 | 0.89 (0.80 to 0.99) | 0.036 | 53(2t o 92) | 0.023 | 1.00 | 10 | 0.84 (0.74 to 0.94) | 0.003 | 0 (0 to 75) | 0.527 | 1.00 | 10 | 0.84 (0.71 to 1.00) | 0.051 | 58 (15 to 92) | 0.011 | 1.00 | Positive |
*Includes GWAS data from SOCCS.
GWASs, genome-wide association studies; SOCCS, Study of CRC in Scotland.
gutjnl-2019-319313supp002.pdf (611.6KB, pdf)
Credibility assessment indicated 18 variants (5.8% of the meta-analysed SNPs) tagging 16 loci (rs36053993 and rs34612342 in MUTYH, rs2066847 in NOD2, rs12953717 and rs4464148 in SMAD7, rs1569686 in DNMT3B, rs2736100 in TERT, rs9858822 in PPAR-gamma, rs1862748 in CDH1, rs7259371 in RHPN2, rs355527 in BMP2, rs1800469 in TGFB1, rs10505477 in 8q24, rs16892766 in 8q23.3, rs3802842 in 11q23.1, rs961253 in 20p12.3, rs10795668 in 10p14, rs4951291 in 1q32.1) had the most credible associations with CRC risk and are therefore referred to as ‘positive’ SNPs (table 1, online supplementary tables 2–4). These findings are based on accrued data on 1224 to 43 652 cases and on 1381 to 60 883 controls, with a median of 17 100 cases per meta-analysis. The linkage disequilibrium (LD) between these ‘positive’ polymorphisms was checked pairwise using the Ensembl LD calculator with reference to the 1000 genomes: phase 3 CEU population and we found two pairs of SNPs (rs355527 and rs961253, rs12953717 and rs4464148) with r2 >0.20 (online supplementary tables 8). The other 59 variants (19.2% of the meta-analysed SNPs) in 49 loci with p value <0.05 were classified as ‘less-credible positive’ SNPs, given high heterogeneity, low statistical power or a high possibility of being false positive (BFDP >0.2) for their association with CRC risk (table 2, online supplementary tables 2–4). The summary findings for ‘less-credible positive’ SNPs were based on accrued data on 246 to 51 730 CRC cases and on 399 to 53 589 controls, with a median of at least 4287 CRC cases per meta-analysis.
Table 2.
Summary of ‘less-credible associations’ from meta-analyses and credibility assessment
| Gene | Variant | Cases vs controls (number of samples) |
Allelic model | Recessive model: var/var vs wt/wt and wt/var | Dominant model: wt/var and var/var vs wt/wt | Classification | |||||||||||||||
| N | Effect size | Heterogeneity | N | Effect size | Heterogeneity | Power | N | Effect size | Heterogeneity | Power | |||||||||||
| OR (95% CI) | P value | I2 (95% CI) | P value | Power | OR (95% CI) | P value | I2 (95% CI) | P value | OR (95% CI) | P value | I2 (95% CI) | P value | |||||||||
| MTHFR | rs1801133 | 43 652 vs 60 883 (94*†) | 91 | 0.96 (0.93 to 0.99) | 8.79E-03 | 53 (59 to 85) | 3.61E-09 | 0.84 | 93 | 0.87 (0.81 to 0.93) | 7.36E-05 | 56 (63 to 86) | 6.79E-11 | 1.00 | 93 | 0.98 (0.94 to 1.02) | 0.249 | 48 (52 to 82) | 1.98E-07 | 0.36 | Less credible |
| miR | rs895819 | 1322 vs 1641 (4) | 4 | 1.18 (1.06 to 1.32) | 0.003 | 4 (0 to 95) | 0.373 | 0.09 | 4 | 1.50 (1.23 to 1.83) | 6.32E-05 | 1 (0 to 96) | 0.385 | 0.44 | 4 | 1.08 (0.93 to 1.26) | 0.302 | 0 (0 to 94) | 0.474 | 0.05 | Less credible |
| ADH1B | rs1229984 | 4449 vs 6995 (9) | 9 | 1.05 (0.94 to 1.18) | 0.401 | 57 (8 to 91) | 0.018 | 0.16 | 9 | 1.23 (1.02 to 1.48) | 0.028 | 0 (0 to 70) | 0.493 | 0.68 | 9 | 1.04 (0.90 to 1.20) | 0.595 | 60 (14 to 92) | 0.010 | 0.11 | Less credible |
| XRCC1 | rs25487 | 13 017 vs 18 166 (37) | 37 | 1.11 (1.03 to 1.19) | 0.008 | 71 (68 to 90) | 8.64E-12 | 0.74 | 36 | 1.17 (1.02 to 1.34) | 0.025 | 59 (39 to 80) | 3.14E-06 | 0.91 | 37 | 1.11 (1.01 to 1.21) | 0.023 | 63 (58 to 89) | 1.44E-07 | 0.88 | Less credible |
| HFE | rs1800562 | 6547 vs 34 156 (10‡) | 10 | 1.10 (1.01 to 1.20) | 0.035 | 0 (0 to 40) | 0.850 | 0.46 | 10 | 1.15 (0.69 to 1.92) | 0.582 | 24 (0 to 84) | 0.240 | 0.12 | 10 | 1.10 (1.01 to 1.21) | 0.037 | 0 (0 to 25) | 0.937 | 0.68 | Less credible |
| MTHFR | rs1801131 | 23 523 vs 35 193 (57*) | 57 | 0.98 (0.95 to 1.01) | 0.258 | 22 (0 to 57) | 0.075 | 0.19 | 57 | 0.91 (0.84 to 0.98) | 0.010 | 23 (0 to 62) | 0.064 | 0.94 | 57 | 1.00 (0.96 to 1.04) | 0.970 | 10 (0 to 48) | 0.270 | 0.50 | Less credible |
| CYP2C9 | rs1799853 | 9588 vs 11 428 (12) | 12 | 0.95 (0.90 to 1.01) | 0.086 | 0 (0 to 73) | 0.530 | 0.26 | 12 | 1.16 (0.96 to 1.40) | 0.127 | 0 (0 to 73) | 0.667 | 0.39 | 12 | 0.93 (0.87 to 0.99) | 0.015 | 0 (0 to 72) | 0.452 | 0.65 | Less credible |
| CRP | rs1800947 | 2853 vs 3381 (3) | 2 | 1.03 (0.88 to 1.20) | 0.745 | 0 (0 to 100) | 0.334 | 0.10 | 2 | 3.95 (1.35 to 11.56) | 0.012 | 7 (0 to 99) | 0.301 | 1.00 | 3 | 0.96 (0.82 to 1.12) | 0.604 | 0 (0 to 81) | 0.911 | 0.08 | Less credible |
| CRP | rs1205 | 3037 vs 9333 (4) | 4 | 1.07 (1.00 to 1.15) | 0.065 | 0 (0 to 95) | 0.479 | 0.34 | 4 | 1.18 (1.02 to 1.37) | 0.027 | 0 (0 to 94) | 0.431 | 0.73 | 4 | 1.05 (0.96 to 1.16) | 0.288 | 0 (0 to 90) | 0.669 | 0.21 | Less credible |
| EGF | rs4444903 | 899 vs 976 (5) | 5 | 0.83 (0.64 to 1.07) | 0.143 | 70 (18 to 97) | 0.010 | 0.52 | 5 | 0.91 (0.63 to 1.31) | 0.601 | 50 (0 to 92) | 0.093 | 0.13 | 5 | 0.65 (0.44 to 0.96) | 0.030 | 67 (12 to 97) | 0.016 | 0.99 | Less credible |
| ESR2 | rs928554 | 2574 vs 2977 (3) | 3 | 1.09 (1.01 to 1.18) | 0.025 | 0 (0 to 89) | 0.743 | 0.35 | 3 | 1.12 (0.97 to 1.29) | 0.118 | 0 (0 to 93) | 0.590 | 0.36 | 3 | 1.13 (1.01 to 1.26) | 0.041 | 0 (0 to 89) | 0.876 | 0.57 | Less credible |
| HPGD | rs8752 | 3968 vs 4830 (3‡) | 3 | 1.05 (0.92 to 1.21) | 0.467 | 80 (23 to 99) | 0.007 | 0.20 | 3 | 1.12 (1.00 to 1.26) | 0.047 | 6 (0 to 97) | 0.345 | 0.53 | 3 | 1.03 (0.83 to 1.29) | 0.786 | 83 (34 to 99) | 0.003 | 0.10 | Less credible |
| LIPC | rs6083 | 7667 vs 7980 (4‡) | 4 | 0.96 (0.90 to 1.02) | 0.172 | 41 (0 to 95) | 0.167 | 0.23 | 4 | 0.89 (0.80 to 0.99) | 0.033 | 22 (0 to 94) | 0.278 | 0.68 | 4 | 0.98 (0.91 to 1.05) | 0.519 | 11 (0 to 92) | 0.340 | 0.10 | Less credible |
| miR | rs2292832 | 2355 vs 2571 (5‡) | 5 | 0.94 (0.86 to 1.02) | 0.152 | 0 (0 to 82) | 0.650 | 0.17 | 5 | 1.06 (0.89 to 1.27) | 0.522 | 0 (0 to 73) | 0.789 | 0.10 | 5 | 0.87 (0.78 to 0.98) | 0.021 | 0 (0 to 88) | 0.704 | 0.68 | Less credible |
| MSH2 | rs4608577 | 4308 vs 4011 (3‡) | 3 | 0.94 (0.86 to 1.02) | 0.112 | 0 (0 to 91) | 0.795 | 0.19 | 3 | 0.72 (0.56 to 0.92) | 0.008 | 0 (0 to 95) | 0.648 | 0.68 | 3 | 0.96 (0.88 to 1.05) | 0.406 | 0 (0 to 62) | 0.945 | 0.14 | Less credible |
| MSH3 | rs184967 | 8151 vs 10 103 (4‡) | 4 | 1.08 (1.00 to 1.16) | 0.049 | 35 (0 to 98) | 0.204 | 0.47 | 4 | 1.23 (1.02 to 1.49) | 0.031 | 0 (0 to 91) | 0.702 | 0.59 | 4 | 1.07 (0.99 to 1.16) | 0.098 | 28 (0 to 98) | 0.242 | 0.54 | Less credible |
| MSH3 | rs26779 | 6050 vs 8024 (6‡) | 6 | 1.07 (1.02 to 1.14) | 0.013 | 11 (0 to 92) | 0.343 | 0.46 | 6 | 1.13 (0.98 to 1.31) | 0.085 | 36 (0 to 93) | 0.166 | 0.59 | 6 | 1.08 (1.01 to 1.15) | 0.030 | 0 (0 to 85) | 0.682 | 0.62 | Less credible |
| MTHFD1 | rs1950902 | 9059 vs 11 358 (7‡) | 7 | 0.94 (0.89 to 0.99) | 0.026 | 3 (0 to 82) | 0.400 | 0.39 | 7 | 1.01 (0.87 to 1.18) | 0.878 | 0 (0 to 91) | 0.767 | 0.05 | 7 | 0.92 (0.86 to 0.98) | 0.015 | 15 (0 to 90) | 0.313 | 0.79 | Less credible |
| PARP1 | rs1136410 | 7002 vs 8328 (7‡) | 7 | 1.05 (0.95 to 1.17) | 0.300 | 52 (0 to 98) | 0.052 | 0.24 | 7 | 1.23 (1.05 to 1.44) | 0.012 | 0 (0 to 92) | 0.475 | 0.78 | 7 | 1.03 (0.93 to 1.15) | 0.529 | 36 (0 to 94) | 0.153 | 0.14 | Less credible |
| PGR | rs1042838 | 5232 vs 5733 (4‡) | 4 | 1.08 (1.01 to 1.16) | 0.034 | 0 (0 to 95) | 0.507 | 0.32 | 4 | 0.97 (0.77 to 1.23) | 0.819 | 0 (0 to 85) | 0.880 | 0.06 | 4 | 1.11 (1.02 to 1.21) | 0.012 | 0 (0 to 96) | 0.450 | 0.71 | Less credible |
| rs4951039 | 13 791 vs 14 288 (7‡) | 7 | 0.91 (0.81 to 1.02) | 0.106 | 81 (49 to 95) | 1.48E-05 | 0.79 | 7 | 0.78 (0.66 to 0.93) | 0.005 | 0 (0 to 68) | 0.620 | 0.82 | 7 | 0.91 (0.80 to 1.04) | 0.163 | 82 (51 to 96) | 9.30E-06 | 0.94 | Less credible | |
| SCD | rs7849 | 2011 vs 2580 (3) | 3 | 0.85 (0.73 to 0.98) | 0.025 | 29 (0 to 98) | 0.247 | 0.53 | 3 | 0.64 (0.40 to 1.02) | 0.058 | 41 (0 to 99) | 0.185 | 0.68 | 3 | 0.85 (0.71 to 1.02) | 0.084 | 33 (0 to 99) | 0.225 | 0.72 | Less credible |
| XPC | rs2228000 | 2677 vs 4253 (6) | 6 | 0.99 (0.71 to 1.36) | 0.929 | 94 (82 to 99) | 1.35E-16 | 0.05 | 6 | 0.95 (0.49 to 1.81) | 0.866 | 91 (71 to 99) | 2.18E-10 | 0.09 | 6 | 0.58 (0.41 to 0.82) | 0.002 | 92 (83 to 99) | 1.49E-11 | 1.00 | Less credible |
| VEGF | rs699947 | 4497 vs 5334 (10‡) | 10 | 1.09 (1.00 to 1.19) | 0.056 | 46 (0 to 91) | 0.053 | 0.56 | 10 | 1.14 (1.01 to 1.29) | 0.029 | 14 (0 to 89) | 0.317 | 0.74 | 10 | 1.09 (0.95 to 1.24) | 0.219 | 47 (0 to 87) | 0.047 | 0.50 | Less credible |
| IGF1 | rs35767 | 2792 vs 4988 (3) | 3 | 0.95 (0.80 to 1.14) | 0.599 | 59 (0 to 99) | 0.087 | 0.15 | 3 | 0.75 (0.62 to 0.91) | 0.003 | 0 (0 to 97) | 0.567 | 0.79 | 3 | 0.98 (0.79 to 1.21) | 0.826 | 60 (0 to 99) | 0.081 | 0.07 | Less credible |
| NAT2 | rs1799929 | 1861 vs 1952 (5*) | 5 | 0.84 (0.74 to 0.95) | 0.004 | 20 (0 to 94) | 0.289 | 0.53 | 5 | 0.68 (0.47 to 0.99) | 0.044 | 68 (14 to 98) | 0.015 | 0.81 | 5 | 0.85 (0.74 to 0.97) | 0.016 | 0 (0 to 85) | 0.872 | 0.61 | Less credible |
| NAT2 | rs1799930 | 6446 vs 7193 (8*) | 8 | 1.07 (0.99 to 1.16) | 0.099 | 44 (0 to 96) | 0.082 | 0.25 | 8 | 0.99 (0.88 to 1.12) | 0.897 | 0 (0 to 64) | 0.812 | 0.08 | 8 | 1.13 (1.00 to 1.27) | 0.043 | 51 (0 to 97) | 0.045 | 0.70 | Less credible |
| ABCB1 (MDR1) | rs9282564 | 9536 vs 9145 (6‡) | 6 | 1.07 (1.00 to 1.14) | 0.053 | 0 (0 to 77) | 0.561 | 0.29 | 6 | 0.88 (0.65 to 1.19) | 0.424 | 18 (0 to 86) | 0.299 | 0.14 | 6 | 1.09 (1.01 to 1.17) | 0.018 | 0 (0 to 70) | 0.729 | 0.65 | Less credible |
| ABCB1 (MDR1) | rs1045642 | 7537 vs 8396 (16) | 16 | 0.94 (0.88 to 1.01) | 0.108 | 46 (1 to 88) | 0.024 | 0.50 | 16 | 0.89 (0.81 to 0.97) | 0.012 | 18 (0 to 78) | 0.248 | 0.87 | 16 | 0.97 (0.85 to 1.10) | 0.608 | 57 (32 to 91) | 0.003 | 0.14 | Less credible |
| ABCB1 (MDR1) | rs1202168 | 7000 vs 6485 (5) | 5 | 1.06 (0.99 to 1.14) | 0.071 | 45 (0 to 93) | 0.124 | 0.39 | 5 | 1.04 (0.95 to 1.14) | 0.350 | 0 (0 to 81) | 0.699 | 0.14 | 5 | 1.12 (1.00 to 1.24) | 0.042 | 52 (0 to 93) | 0.080 | 0.88 | Less credible |
| ADIPOR1 | rs1342387 | 2472 vs 2848 (5‡) | 5 | 0.87 (0.78 to 0.98) | 0.018 | 48 (0 to 95) | 0.105 | 0.71 | 5 | 0.84 (0.73 to 0.96) | 0.012 | 0 (0 to 62) | 0.900 | 0.68 | 5 | 0.85 (0.69 to 1.03) | 0.098 | 63 (0 to 96) | 0.029 | 0.79 | Less credible |
| AXIN2 | rs2240308 | 4723 vs 4932 (4‡) | 4 | 1.06 (1.00 to 1.12) | 0.044 | 0 (0 to 92) | 0.397 | 0.30 | 4 | 1.07 (0.96 to 1.19) | 0.219 | 17 (0 to 95) | 0.306 | 0.29 | 4 | 1.10 (1.01 to 1.21) | 0.037 | 0 (0 to 76) | 0.758 | 0.54 | Less credible |
| MMP1 | rs1799750 | 1660 vs 2024 (10) | 9 | 0.78 (0.66 to 0.93) | 0.005 | 59 (8 to 88) | 0.012 | 0.95 | 9 | 0.78 (0.61 to 0.99) | 0.041 | 21 (0 to 68) | 0.254 | 0.75 | 10 | 0.70 (0.56 to 0.88) | 0.002 | 57 (9 to 88) | 0.013 | 1.00 | Less credible |
| CYP1A1 | rs1048943 | 9661 vs 11 774 (19*) | 19 | 1.28 (1.07 to 1.52) | 0.006 | 84 (83 to 96) | 1.15E-15 | 1.00 | 18 | 1.26 (1.04 to 1.52) | 0.016 | 0 (0 to 55) | 0.456 | 0.72 | 19 | 1.36 (1.08 to 1.72) | 0.009 | 88 (93 to 98) | 2.16E-23 | 1.00 | Less credible |
| GSTT1 | Null variant | 19 133 vs 27 821 (56) | 7 | 0.91 (0.70 to 1.20) | 0.508 | 71 (21 to 99) | 0.015 | 0.49 | 53 | 1.17 (1.07 to 1.28) | 0.001 | 70 (71 to 90) | 1.62E-15 | 1.00 | 6 | 1.00 (0.73 to 1.37) | 0.979 | 53 (0 to 98) | 0.059 | 0.05 | Less credible |
| VDR | rs1544410 | 14 789 vs 15 922 (16‡) | 16 | 0.83 (0.72 to 0.96) | 0.011 | 93 (91 to 95) | 5.59E-40 | 1.00 | 16 | 0.81 (0.67 to 0.98) | 0.033 | 85 (77 to 89) | 1.08E-14 | 1.00 | 16 | 0.82 (0.69 to 0.96) | 0.015 | 88 (83 to 91) | 2.62E-20 | 1.00 | Less credible |
| 8q24 | rs6983267 | 51 730 vs 53 589 (34‡) | 34 | 1.11 (1.06 to 1.17) | 1.75E-05 | 85 (77 to 93) | 1.93E-28 | 1.00 | 34 | 1.16 (1.08 to 1.25) | 4.92E-05 | 79 (66 to 89) | 2.80E-18 | 1.00 | 34 | 1.16 (1.08 to 1.24) | 2.98E-05 | 80 (69 to 92) | 8.68E-19 | 1.00 | Less credible |
| RAD18 | rs373572 | 6560 vs 6906 (5‡) | 5 | 1.15 (1.01 to 1.30) | 0.031 | 75 (33 to 98) | 0.003 | 0.96 | 5 | 1.23 (1.01 to 1.49) | 0.041 | 54 (0 to 95) | 0.067 | 0.93 | 5 | 1.15 (0.98 to 1.36) | 0.080 | 73 (21 to 98) | 0.005 | 0.98 | Less credible |
| NQ01 | rs1800566 | 11 183 vs 12 525 (16‡) | 16 | 1.17 (1.05 to 1.30) | 0.006 | 80 (66 to 94) | 1.17E-09 | 1.00 | 16 | 1.21 (0.99 to 1.48) | 0.060 | 55 (17 to 87) | 0.004 | 0.91 | 16 | 1.20 (1.06 to 1.37) | 0.006 | 78 (64 to 94) | 8.77E-09 | 1.00 | Less credible |
| PTGS2/COX2 | rs20417 | 7785 vs 11 371 (18) | 17 | 1.10 (1.00 to 1.2) | 0.035 | 37 (0 to 83) | 0.062 | 0.57 | 17 | 1.24 (1.04 to 1.47) | 0.016 | 0 (0 to 46) | 0.928 | 0.64 | 18 | 1.12 (1.00 to 1.26) | 0.050 | 51 (14 to 87) | 0.019 | 0.90 | Less credible |
| NOD2 | rs2066844 | 3297 vs 3088 (9) | 9 | 1.38 (1.04 to 1.84) | 0.026 | 39 (0 to 91) | 0.109 | 0.66 | 9 | 1.97 (0.55 to 7.04) | 0.299 | 0 (0 to 83) | 0.689 | 0.17 | 9 | 1.35 (1.02 to 1.78) | 0.033 | 33 (0 to 90) | 0.155 | 0.86 | Less credible |
| CCND1 | rs17852153 | 6500 vs 8885 (20) | 20 | 1.10 (1.02 to 1.19) | 0.019 | 58 (45 to 92) | 0.001 | 0.83 | 20 | 1.11 (0.97 to 1.28) | 0.128 | 63 (54 to 94) | 7.69E-05 | 0.82 | 20 | 1.15 (1.05 to 1.26) | 0.003 | 18 (0 to 76) | 0.226 | 0.94 | Less credible |
| GSTM1 | Null variant | 28 240 vs 38 880 (74) | 13 | 1.12 (0.89 to 1.40) | 0.335 | 70 (22 to 98) | 0.005 | 1.00 | 6 | 1.18 (0.90 to 1.55) | 0.234 | 67 (0 to 98) | 0.028 | 1.00 | 74 | 1.10 (1.05 to 1.16) | 0.001 | 47 (34 to 75) | 6.63E-06 | 1.00 | Less credible |
| 9p24 | rs719725 | 13 513 vs 14 999 (14) | 14 | 1.07 (1.02 to 1.11) | 0.002 | 24 (0 to 79) | 0.190 | 0.79 | 14 | 1.09 (1.02 to 1.15) | 0.006 | 29 (0 to 75) | 0.148 | 0.94 | 14 | 1.09 (1.02 to 1.17) | 0.008 | 0 (0 to 66) | 0.521 | 0.72 | Less credible |
| ERCC5 | rs17655 | 9653 vs 11 367 (14) | 13 | 1.07 (1.02 to 1.12) | 0.003 | 0 (0 to 73) | 0.626 | 0.60 | 13 | 0.98 (0.89 to 1.08) | 0.688 | 0 (0 to 77) | 0.538 | 0.07 | 14 | 1.14 (1.04 to 1.24) | 0.004 | 44 (0 to 85) | 0.038 | 1.00 | Less credible |
| GH1 | rs2665802 | 3275 vs 3848 (7) | 7 | 0.89 (0.80 to 0.99) | 0.025 | 49 (0 to 97) | 0.069 | 0.68 | 7 | 0.83 (0.71 to 0.98) | 0.028 | 37 (0 to 95) | 0.144 | 0.87 | 7 | 0.89 (0.78 to 1.02) | 0.103 | 36 (0 to 96) | 0.157 | 0.62 | Less credible |
| TP73 | G4C14 | 858 vs 1173 (4) | 4 | 1.21 (1.04 to 1.40) | 0.013 | 4 (0 to 92) | 0.371 | 0.46 | 4 | 1.75 (1.23 to 2.48) | 0.002 | 0 (0 to 94) | 0.513 | 0.91 | 4 | 1.10 (0.83 to 1.45) | 0.501 | 55 (0 to 96) | 0.083 | 0.18 | Less credible |
| TGFB1 | rs4803455 | 3747 vs 4513 (3‡) | 3 | 1.11 (1.04 to 1.18) | 0.001 | 0 (0 to 71) | 0.903 | 0.66 | 3 | 1.12 (1.01 to 1.24) | 0.027 | 0 (0 to 80) | 0.845 | 0.59 | 3 | 1.18 (1.06 to 1.30) | 0.001 | 0 (0 to 85) | 0.879 | 0.90 | Less credible |
| APC | rs1801155 | 2389 vs 4223 (5) | 5 | 1.60 (1.21 to 2.11) | 0.001 | 27 (0 to 94) | 0.244 | 0.85 | na | na | na | na | na | na | 5 | 1.62 (1.21 to 2.16) | 0.001 | 28 (0 to 94) | 0.235 | 0.99 | Less credible |
| MLH1 | rs63750447 | 937 vs 812 (4) | 3 | 2.14 (1.12 to 4.10) | 0.022 | 39 (0 to 99) | 0.194 | 0.75 | na | na | na | na | na | na | 3 | 2.17 (1.14 to 4.14) | 0.019 | 36 (0 to 98) | 0.208 | 0.96 | Less credible |
| VDR | rs11568820 | 4001 vs 4682 (5*) | 4 | 1.14 (1.05 to 1.23) | 0.001 | 0 (0 to 92) | 0.648 | 0.71 | 4 | 1.37 (1.02 to 1.85) | 0.039 | 35 (0 to 97) | 0.203 | 0.90 | 5 | 1.14 (1.04 to 1.24) | 0.004 | 0 (0 to 0) | 0.989 | 0.85 | Less credible |
| ABCB1 (MDR1) | rs1128503 | 246 vs 399 (3) | 3 | 0.70 (0.51 to 0.96) | 0.027 | 45 (0 to 98) | 0.164 | 0.59 | 3 | 0.77 (0.37 to 1.59) | 0.483 | 71 (0 to 99) | 0.031 | 0.31 | 3 | 0.52 (0.35 to 0.76) | 0.001 | 0 (0 to 96) | 0.617 | 0.93 | Less credible |
| ADIPOQ | rs2241766 | 1517 vs 1909 (5) | 5 | 1.17 (1.01 to 1.35) | 0.033 | 29 (0 to 97) | 0.231 | 0.50 | 5 | 1.04 (0.74 to 1.45) | 0.830 | 14 (0 to 98) | 0.325 | 0.06 | 5 | 1.26 (1.09 to 1.47) | 0.002 | 8 (0 to 96) | 0.364 | 0.92 | Less credible |
| CHEK2 | rs1787996 | 1687 vs 3370 (3) | 3 | 1.47 (1.12 to 1.92) | 0.006 | 0 (0 to 98) | 0.499 | 0.53 | na | na | na | na | na | na | 3 | 1.48 (1.13 to 1.95) | 0.005 | 0 (0 to 98) | 0.496 | 0.81 | Less credible |
| CHEK2 | 1100delC | 2417 vs 3615 (4) | 4 | 1.66 (1.01 to 2.73) | 0.048 | 0 (0 to 83) | 0.835 | 0.60 | na | na | na | na | na | na | 4 | 1.66 (1.01 to 2.75) | 0.047 | 0 (0 to 83) | 0.835 | 0.86 | Less credible |
| TP53 | rs1042522 | 10591 vs 12 673 (30) | 30 | 1.11 (0.99 to 1.24) | 0.077 | 85 (84 to 95) | 4.42E-26 | 0.73 | 30 | 1.21 (1.02 to 1.44) | 0.026 | 70 (55 to 88) | 2.77E-09 | 0.98 | 30 | 1.11 (0.96 to 1.28) | 0.163 | 82 (84 to 05) | 7.58E-20 | 0.64 | Less credible |
| TP53 | rs17878362 | 1812 vs 2319 (6) | 6 | 0.98 (0.76 to 1.26) | 0.859 | 70 (23 to 96) | 0.005 | 0.06 | 6 | 1.28 (0.81 to 2.03) | 0.282 | 0 (0 to 79) | 0.638 | 0.22 | 6 | 0.95 (0.71 to 1.27) | 0.734 | 71 (25 to 96) | 0.004 | 0.40 | Less credible |
| KRAS | rs712 | 1982 vs 2194 (3) | 3 | 1.22 (0.86 to 1.72) | 0.270 | 90 (55 to 100) | 6.87E-05 | 0.19 | 3 | 1.72 (0.73 to 4.06) | 0.215 | 87(44 to 100) | 3.74E-04 | 0.16 | 3 | 1.17 (0.84 to 1.65) | 0.350 | 83 (32 to 99) | 0.003 | 0.28 | Less credible |
| ARLTS1 | rs3803185 | 2281 vs 3196 (5†) | 5 | 1.09 (1.01 to 1.18) | 0.026 | 0 (0 to 76) | 0.803 | 0.35 | 5 | 1.08 (0.90 to 1.29) | 0.432 | 43 (0 to 93) | 0.133 | 0.21 | 5 | 1.18 (0.95 to 1.46) | 0.131 | 62 (0 to 96) | 0.032 | 0.77 | Less credible |
| 1q32.1 | rs4951291 | 15 835 vs 16 724 (9‡) | 9 | 0.91 (0.84 to 0.99) | 0.021 | 68 (22 to 88) | 0.002 | 0.85 | 9 | 0.74 (0.62 to 0.87) | 2.72E-04 | 0 (0 to 69) | 0.616 | 0.95 | 9 | 0.92 (0.83 to 1.01) | 0.077 | 72 (30 to 90) | 4.33E-04 | 0.91 | Less credible |
| PPAR-gamma | rs9858822 | 2152 vs 2630 (5) | 4 | 1.40 (1.19 to 1.65) | 4.69E-05 | 0 (0 to 98) | 0.697 | 0.96 | 2 | 1.69 (1.19 to 2.39) | 0.003 | 0 (0 to 98) | 0.801 | 0.54 | 4 | 1.47 (1.19 to 1.83) | 4.55E-04 | 0 (0 to 98) | 0.618 | 1.00 | Less credible |
| DNMT3B | rs1569686 | 1224 vs 1381 (5) | 4 | 0.57 (0.47 to 0.68) | 1.81E-09 | 0 (0 to 0) | 0.992 | 1.00 | 4 | 0.93 (0.54 to 1.58) | 0.775 | 0 (0 to 97) | 0.414 | 0.06 | 5 | 0.46 (0.36 to 0.59) | 2.40E-09 | 30 (0 to 95) | 0.223 | 1.00 | Less credible |
| NOD2 | rs2066847 | 4573 vs 3733 (10) | 10 | 1.39 (1.16 to 1.66) | 3.21E-04 | 0 (0 to 71) | 0.547 | 0.78 | 10 | 2.36 (0.69 to 8.13) | 0.173 | 0 (0 to 76) | 0.720 | 0.31 | 10 | 1.38 (1.14 to 1.66) | 0.001 | 0 (0 to 74) | 0.488 | 0.96 | Less credible |
*Includes GWAS data from Ontario.
†Tomlinson 2008 was based on 10 samples.
‡Includes GWAS data from SOCCS.
GWASs, genome-wide association studies; SOCCS, Study of CRC in Scotland.
Polymorphisms classified as either ‘positive’ or ‘less-credible positive’ SNPs were sent for validation in synthesised data from the GECCO, CORECT and CCFR consortia. Validation was only able to be performed for 68 out of 77 polymorphisms, as one ‘positive’ variant (rs34612342 in MUTYH [Y179C] gene) was dropped due to its low imputation quality in these GWASs; 5 ‘less-credible positive’ polymorphisms (in GSTT1, GSTM1, TP73, CHEK2 and CYP2E1) had no rs numbers; and 3 polymorphisms (in TP53 and KRAS) were not available in these GWASs; therefore, they were not able to be validated. Of the 17 ‘positive’ SNPs sent for validation, 7 polymorphisms (41.2%) in 6 different loci (rs12953717 and rs4464148 in SMAD7, rs355527 in BMP2, rs10505477 in 8q24, rs961253 in 20p12.3, rs16892766 in 8q23.3, rs3802842 in 11q23.1) reached a genome-wide statistical significance (p≤5×10−8) in at least one meta-analysis model and 6 polymorphisms (35.3%) in 6 different loci (rs7259371 in RHPN2, rs2736100 in TERT, rs10795668 in 10p14, rs36053993 in MUTYH, rs1862748 in CDH1 and rs1800469 in TGFB1) reached a nominally statistical significance (p<0.05) in at least one meta-analysis model. However, the remaining 4 polymorphisms (23.5%) in 4 different loci (rs2066847 in NOD2, rs1569686 in DNMT3B, rs9858822 in PPAR-gamma and rs4951291 in 1q32.1) failed in the GWAS validation of all genetic models (online supplementary tables 5–7) and they were therefore downgraded to ‘less-credible positive’. Of the 51 ‘less-credible positive’ SNPs sent for validation, 2 polymorphisms (3.9%) in 2 different loci (rs6983267 in 8q24 and rs1801155 in APC) reached a genome-wide statistical significance (p≤5×10−8) in at least one meta-analysis model and 11 polymorphisms (21.6%) in 11 different loci (rs1136410 in PARP1, rs11568820 in VDR, rs1342387 in ADIPOR1, rs719725 in TPD52L3, rs20417 in PTGS2/COX2, rs2665802 in GH1, rs4803455 in TGFB1, rs7849 in SCD, rs8752 in HPGD, rs36053993 in MUTYH and rs928554 in ESR2) reached a nominally statistical significance (p<0.05) in at least one meta-analysis model, whereas the remaining 38 polymorphisms (74.5%) failed in the GWAS validation of all genetic models (online supplementary tables 5–7). Overall, 26 (33.8%) out of the 77 nominally significant polymorphisms tested via meta-analysis were successfully validated in these GWASs (p<0.05).
Funnel plots were produced for all ‘credible’ (online supplementary figures 1–14) and ‘less-credible’ SNPs (online supplementary figures 15–77) for their summary estimates in allelic model. Small study effects were reported for 2 (14.3%) of the 14 ‘positive’ polymorphisms (rs36053993 in MUTYH and rs4464148 in SMAD7) and for 10 (15.9%) of 63 ‘less-credible’ polymorphisms (rs1229984 in ADH1B, rs25487 in XRCC1, rs2240308 in AXIN2, rs1799750 in MMP1, rs1048943 in CYP1A1, rs373572 in RAD18, rs1800566 in NQ01, rs2066844 in NOD2, rs2665802 in GH1, rs712 in KRAS). Therefore, their reported association with CRC risk should be interpreted with caution.
gutjnl-2019-319313supp003.pdf (17.3MB, pdf)
The remaining 231 polymorphisms (75.0%) assessed in meta-analyses were reported to have no statistically significant association (p>0.05) with CRC risk in all three genetic models (online supplementary table 9), based on accrued data on 192 to 21 929 cases and 251 to 24 054 controls, with median of at least 4017 cases per meta-analysis. Of them, 148 polymorphisms were classified as having negative associations with CRC risk because the number of cases was less than 5000 for which the null results could be due to limited statistical power, while another 83 SNPs with p>0.05 and the number of cases>5000 were classified as null variants with adequate statistical power (online supplementary table 9).
Discussion
This systematic, comprehensive field synopsis of genetic association studies on CRC updates the two previous field synopses10 11 with application of harmonised methods for evidence appraisal and further validation of the identified genetic associations in three GWAS consortia. Specifically, we extracted and collated data for 1063 polymorphisms in 303 different genes from 869 publications and performed up-to-date meta-analyses for 308 variants in 158 different genes that had data from at least three independent studies available for analysis. After credibility assessment and validation, we identified a total of 12 genetic loci credibly associated with CRC risk, of which 6 loci (MUTYH, SMAD7, 8q24, 8q23.3, 11q23.1, 20p12.3) were also classified as credibly associated with CRC risk in the previous field synopses and the other 6 loci (TGFB1, TERT, CDH1, RHPN2, BMP2 and 10p14) are novel findings as they have not been assessed or reported as credible risk loci in the previous field synopses.
We note that a synopsis was undertaken of literature on genetic associations with CRC published in the period 2012–2017.24 Our analysis differs from that study, in that it includes and updates data from our previous field synopses,10 11 includes data from three GWA studies in the meta-analysis and further validates the findings in three GWA consortia.
Similar to our previous field synopses,10 11 the present study reported two SNPs at 8q24 locus (rs6983267 and rs10505477) with strong evidence supporting significant associations with CRC risk and with these associations further replicated in three GWAS consortia. In biopsies of the rectum, sigmoid colon and cecum mucosa, proliferation has been reported to be higher among homozygotes for the risk alleles of the rs6983267 and rs10505477 variants compared with those with other genotypes in the general population.25 The rs6983267 variant has been assessed as having a highly credible association with colorectal adenomas.26 27 In fine-mapping and bioinformatic analysis performed within the GECCO-CCFR consortia, the rs6983267 variant was appraised as having a strong functional evidence.28 The rs6983267 may be a somatic target in CRC29 and may be associated with enhanced responsiveness to Wnt signalling.30 Furthermore, rs6983267 has also been found to be associated with other types of cancer, including prostate cancer.31–33 Interaction with the MYC proto-oncogene has been controversial,34–37 but in functional studies in cell lines, interaction between enhancer elements in the 8q24 locus and the MYC promoter, via transcription factor Tcf-4 binding and allele-specific regulation of MYC expression, has been demonstrated.38 Expression levels of one of these, CARLo-5, in normal colon tissue have been found to be statistically significantly correlated with rs6983267, and chromosome conformation capture analysis of genomic DNA from CRC-derived cell lines provided evidence of physical interaction between the active regulatory region of the CARLo-5 promoter and the MYC enhancer region.39 Since the end of our search period (21 November 2018), an analysis of GWAS data on 22 775 cases and 47 731 controls from 14 studies in East Asia detected a genome-wide significant association with the rs6983267 variant.40 In addition, in a combined meta-analysis of up to 58 131 cases and 67 347 controls from the GECCO, CORECT and CCFR consortia, in which imputed variants from a whole-genome sequencing analysis and Haplotype Reference Consortium panel variants were included, analysis in the 8q24.21 region conditioned on the rs6983267 and rs7013278 variants identified a genome-wide significant association with the rs4313119.41
Two genetic polymorphisms (rs12953717 and rs4464148) tagging in SMAD7 were identified to be associated with CRC with highly credible evidence. Associations with rs12953717 and rs4464148 were successfully validated in the three GWAS consortia. In fine-mapping and bioinformatics analysis performed within the GECCO-CCFR consortia, rs4464148 was appraised as having less strong functional evidence than the highly correlated rs9932005 variant, which is located within 5 kb away.28 The SMAD7 protein is an inhibitor for the TGF-ß signalling pathway.42 43 There were highly credible associations with the rs1862748 (tagging CDH1), rs355527 (BMP2), rs961253 (BMP2) and rs7259371 (RHPN2) variants, which are TGF-ß related, and replicated in the data from the GWAS consortia. In the recent East Asian analysis, the association with the rs961253 (BMP2) variant was replicated, as well as additional variants of SMAD7 (rs7229639, rs4939827), CDH1 (rs9929218), BMP2 (rs4813802) and RHPN2 (rs10411210).40 In the analysis from the GECCO, CORECT and CCFR consortia, conditioned on the rs4813802 and rs189583 variants of BMP2 as well as each other, novel associations with the BMP2 variants rs28488 and rs994308 were detected.41
Additionally, two variants (rs34612342 and rs36053993) tagging the MUTYH gene were highly credibly associated with increased CRC risk, of which rs36053993 was validated in the three GWAS consortia data, while rs34612342 was not tested due to its poor imputation quality. The MUTYH gene is known to be involved in the dysfunction of base-excision repair, which is the major pathway for repairing oxidative damage. This biological pathway in which the MUTYH gene is involved contributes to the development of multiple colorectal adenomas and carcinomas (MUTYH-associated polyposis (MAP) syndrome).44 In our analysis, the p values were nominally significant for all models for the rs34612342 variant and for the allelic and dominant models for the rs36053993 variant. For the former variant, the magnitude of effect was greatest for the recessive model, although with wide CIs, which is intriguing as MAP, in which highly penetrant mutations are implicated, and is inherited in an autosomal recessive manner.45
Highly credible associations were also reported for four variants tagging four different genes that are involved in inflammation or immune response. First, a positive association with the rs3802842 variant in the 11q23.1 was identified for CRC risk and this association was replicated in the data from the GWAS consortia and in the recent East Asian analysis.40 Fine-mapping and bioinformatics analysis performed within the GECCO-CCFR consortia support this variant with strong functional evidence.28 Fine mapping identified two genes COLCA1 and COLCA2 arranged on opposite strands and sharing a regulatory region containing rs3802842.46 It is reported that carrying the risk allele of rs3802842 is associated with the expression levels of COLCA1 and COLCA2, which is further correlated with lymphocyte infiltration of colonic lamina propia. Further, in an expression quantitative trait locus analysis in colorectal tissue, there were signals for COLCA1 and COLCA2.47 The polymorphism rs10795668 in 10p14 locus tagging GATA3 gene was reported with a highly credible association with CRC, and this association was further replicated in the GWAS consortia and in the recent East Asian analysis.40 However, in fine-mapping and bioinformatics analysis, this variant presents with weak functional evidence.28 Another less-credible positive association with the rs9858822 variant of PPARγ was identified; however, this association was not replicated in the GWAS consortia. Bioinformatics analysis showed that PPARγ and its ligands have been found to block proinflammatory genes in colon cancer cell lines, activated macrophages and monocytes.48 PPARγ is also involved in lipid metabolism, adipocyte differentiation, and glucose homeostasis and insulin sensitivity.48 A less-credible positive association also reported one variant (rs2066847) in NOD2 gene; however, this association was not replicated. Evidence from experimental studies in mice investigating the role of Nod2 in colorectal tumour risk has also been inconsistent.49–51 The recent analyses in East Asia,40 the GECCO, CORECT and CCFR consortia41 and of five UK studies and a further 10 from the COGENT consortium47 identified new associations in the major histocompatibility region.
The remaining two variants with highly credible associations with CRC risk included rs2736100 tagging TERT and rs16892766 at 8q23.3 tagging EIF3H. These variants were all validated in the GWAS consortia datasets. Fine-mapping and bioinformatics analysis performed within the GECCO-CCFR consortia supported these variants with strong functional evidence, for which polymorphism rs2736100 in TERT gene has been reported to be associated with telomere length and the risk of different types of cancer and chronic diseases other than cancer.52 For the association with the rs16892766 variant at 8q23.3 tagging EIF3H, another variant rs16888589 was previously reported with the lowest p value for the association with CRC in this locus.25 Functional analysis and chromosome conformation capture analysis in CRC cell lines have found that the genomic region harbouring rs16888589 increases EIF3H expression,53 but analysis of expression quantitative trait loci around rs16892766 suggested that UTP23 rather than EIF3H is the target of genetic variation associated with CRC in this region.54
The less credible associations with 63 variants of 52 genes involved the following pathways—adhesion (AXIN2, MMP1); alcohol metabolism (ADH1B); angiogenesis (VEGF); blood clotting (SERPINE1); DNA repair (CHEK2, ERCC5, MSH2, MSH3, PARP1, RAD18, XPC, XRCC1); hormone metabolism (ESR2, GH1, PGR); inflammation and immune response (CRP, HPGD, PTGS2/COX2); inhibition of cell growth (CCND1, EGF, TGFB1); iron metabolism (HFE); lipid metabolism (ADIPOQ, ADIPOR1, LIPC, SCD); one-carbon metabolism (MTHFR, MTFD1, MTRR); substrate metabolism (ABCB1; CYP1A1, CYP2C9, CYP2E1, GSTM1, GSTT1, NAT2, NQ01); tumour suppression (ARLTS1, miR, TP73); vitamin D metabolism (VDR)—common low penetrance variants at 1q32.1 (rs4951039, LINC00303) and 9p24 (rs719725); and the common rs1801155 (I1307K) variant of APC, for which large numbers of rare variants have been identified,55 and rs63750447 (V384D) variant of MLH1, for which rare variants confer a high risk of Lynch syndrome.56 These variants were classified as less-credible SNPs because of either the substantial heterogeneity or the high possibility of false positive; however, we would like to highlight a number of less-credible genetic loci (PARP1, MYC, VDR, ADIPOR1, APC, PTGS2/COX2, SCD, HPGD and ESR2), which were replicated in the GWAS data and for which their linked pathways are worthy of further investigation in future studies.
Updating field synopses is challenging because genetic analysis is such a fast-moving field. Recent trends highlighted by three articles published since we completed our search in November 2018 include the extension of consortia to increase statistical power to detect and replicate associations,47 the investigation of populations other than of European origin,40 and the use of whole-genome sequencing and more comprehensive reference panels to extend the range of genetic variants considered to include those that are rare or of low frequency.41 These three articles have added new loci for CRC susceptibility and indicate that most of the risk loci previously associated with CRC in populations of European origin are also associated with CRC risk in East Asian populations.
We checked whether the 14 variants we classified as ‘highly credible’ were replicated in the paper of Law et al.47 For one variant, rs34612342 MUTYH, there was poor imputation quality. However, as there was no satisfactory proxy for this, we report the available information, which supported the association (p=0.029). All the remaining 13 variants had p values for association less than 5.0×10−5 with no more than moderate heterogeneity. Such an empirical comparison was not appropriate with the data of Huyghe et al 41 because of the considerable overlap of included participants.
In summary, we have conducted a comprehensive study to capture and meta-analyse all SNP data for common genetic variants. The analysis clearly identifies 14 variants at 12 loci for which there is robust evidence of their impact on CRC risk, 63 variants at 52 loci for which further evidence through international collaboration should be generated and 231 variants for which the overall evidence does not support any association with CRC risk. With increasing availability of data from multiple SNPs, it is clear that studies to test associations must achieve very high levels of statistical stringency. Nonetheless, the analysis here provides a resource for mining available data and puts into context the sample sizes required for the identification of true associations for common genetic variants. Future resequencing studies are expected to identify rarer variants (eg, prevalence 0.05%–5%) with intermediate or perhaps even large effects, and GWAS of structural variation will likely identify deletions, amplifications and other copy number variations that may also influence CRC risk.6 This study highlights a number of common genetic variants that could be incorporated into genetic risk-prediction algorithms as further risk factors are identified and highlights the loci at which further research effort should be targeted. All data are available from the CRCgene2 database.
Acknowledgments
ET is supported by a Cancer Research UK Career Development Fellowship (C31250/A22804).
SOCCS: The work was supported by Programme Grant funding from Cancer Research UK (C348/A12076) and by funding for the infrastructure and staffing of the Edinburgh CRUK Cancer Research Centre. This work was also funded by a grant to MGD as Project Leader with the MRC Human Genetics Unit Centre Grant (U127527202 and U127527198 from 1/4/18). We are grateful to all who contribute to recruitment, data collection and data curation for the Study of Colorectal Cancer in Scotland studies. We acknowledge that these studies would not be possible without the patients and controls and their families. We acknowledge the expert support on sample preparation from the Genetics Core of the Edinburgh Wellcome Trust Clinical Research Facility.
ARCTIC: ARCTIC was funded by the Canadian Institutes of Health Research (CIHR) Team in Interdisciplinary Research on Colorectal Cancer (CTP-79845); a CIHR pilot project grant in colorectal cancer screening (200509CCS-152119-CCS-CECA-102806); the Cancer Risk Evaluation (CaRE) Program Grant from the Canadian Cancer Society Research Institute (18001).
EPICOLON: We are sincerely grateful to all patients, the Spanish National DNA Bank (BNADN) and Biobank of Hospital Clínic–IDIBAPS. SNP genotyping services were provided by the Spanish 'Centro Nacional de Genotipado' (CEGEN-ISCIII). This work was supported by grants from Fondo de Investigación Sanitaria/FEDER (PI17/00878, PI17/00509), Fundación Científica de la Asociación Española contra el Cáncer (GCB13131592CAST), PERIS (SLT002/16/00398, Generalitat de Catalunya), CERCA Programme (Generalitat de Catalunya) and Agència de Gestió d'Ajuts Universitaris i de Recerca (Generalitat de Catalunya, GRPRE 2017SGR21, GRC 2017SGR653). CIBEREHD and CIBERER are funded by the Instituto de Salud Carlos III. This article is based upon work from COST Action CA17118, supported by COST (European Cooperation in Science and Technology, www.cost.eu).
This work was also supported by grant U01 CA167551 from the National Cancer Institute and through cooperative agreements with the following Colon Cancer Family Registry sites: Australasian Colorectal Cancer Family Registry (U01 CA074778 and U01/U24 CA097735); Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783); and Seattle Colorectal Cancer Family Registry (U01/U24 CA074794). The genome-wide association studies (GWAS) were supported by grants U01 CA122839, R01 CA143237 and U19 CA148107.
Footnotes
Twitter: @ccgg_edinburgh
Contributors: JL, WZ, ET and HC conceived the study; JL, WZ, ET, HC, ZM and XL designed it; ZM, XL, ET, JL and HC wrote the article with input from other authors; ZM, XL, CN, MT and VS undertook data manipulations and statistical analysis; ZM, XL, CN, XMa, XMeng, YH and YB undertook the literature review and coordinated and/or undertook related abstraction, handling and curation of the data; SM, SC-B, CR-P, CF-R, ACarracedo, ACastells, DB, MAJ, TOK, AL, FJBvD, AW, SMF and MGD provided access to GWAS data. HC, ET, WZ and JL are joint corresponding authors.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. Fitzmaurice C, Allen C, Barber RM, et al. . Global, regional, and National cancer incidence, mortality, years of life lost, years lived with disability, and Disability-Adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol 2017;3:524–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ferlay J, Soerjomataram I, Ervik M, et al. . GLOBOCAN 2012 V1. 0, cancer incidence and mortality worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer, 2013. [Google Scholar]
- 3. Huhn S, Pardini B, Naccarati A, et al. . Ancestral susceptibility to colorectal cancer. Mutagenesis 2012;27:197–204. 10.1093/mutage/ger061 [DOI] [PubMed] [Google Scholar]
- 4. Muñoz M, Pong-Wong R, Canela-Xandri O, et al. . Evaluating the contribution of genetics and familial shared environment to common disease using the UK Biobank. Nat Genet 2016;48:980–3. 10.1038/ng.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Graff RE, Möller S, Passarelli MN, et al. . Familial risk and heritability of colorectal cancer in the Nordic twin study of cancer. Clinical Gastroenterology and Hepatology 2017;15:1256–64. 10.1016/j.cgh.2016.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tenesa A, Dunlop MG. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat Rev Genet 2009;10:353–8. 10.1038/nrg2574 [DOI] [PubMed] [Google Scholar]
- 7. de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer 2004;4:769–80. 10.1038/nrc1453 [DOI] [PubMed] [Google Scholar]
- 8. Jasperson KW, Tuohy TM, Neklason DW, et al. . Hereditary and familial colon cancer. Gastroenterology 2010;138:2044–58. 10.1053/j.gastro.2010.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lung MS, Trainer AH, Campbell I, et al. . Familial colorectal cancer. Intern Med J 2015;45:482–91. 10.1111/imj.12736 [DOI] [PubMed] [Google Scholar]
- 10. Theodoratou E, Montazeri Z, Hawken S, et al. . Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J Natl Cancer Inst 2012;104:1433–57. 10.1093/jnci/djs369 [DOI] [PubMed] [Google Scholar]
- 11. Ma X, Zhang B, Zheng W. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut 2014;63:326–36. 10.1136/gutjnl-2012-304121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ioannidis JP, Boffetta P, Little J, et al. . Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol 2008;37:120–32. 10.1093/ije/dym159 [DOI] [PubMed] [Google Scholar]
- 13. Khoury MJ, Bertram L, Boffetta P, et al. . Genome-Wide association studies, field Synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol 2009;170:269–79. 10.1093/aje/kwp119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen NC, Bagade S, McQueen MB, et al. . Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 2008;40:827–34. 10.1038/ng.171 [DOI] [PubMed] [Google Scholar]
- 15. Bertram L, McQueen MB, Mullin K, et al. . Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 2007;39:17–23. 10.1038/ng1934 [DOI] [PubMed] [Google Scholar]
- 16. Chatzinasiou F, Lill CM, Kypreou K, et al. . Comprehensive field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma. JNCI Journal of the National Cancer Institute 2011;103:1227–35. 10.1093/jnci/djr219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abulí A, Bessa X, González JR, et al. . Susceptibility genetic variants associated with colorectal cancer risk correlate with cancer phenotype. Gastroenterology 2010;139:788–96. 10.1053/j.gastro.2010.05.072 [DOI] [PubMed] [Google Scholar]
- 18. Fernandez-Rozadilla C, Cazier J-B, Tomlinson IP, et al. . A colorectal cancer genome-wide association study in a Spanish cohort identifies two variants associated with colorectal cancer risk at 1p33 and 8p12. BMC Genomics 2013;14:55 10.1186/1471-2164-14-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. The American Journal of Human Genetics 2007;81:208–27. 10.1086/519024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fehringer G, Kraft P, Pharoah PD, et al. . Cross-Cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res 2016;76:5103–14. 10.1158/0008-5472.CAN-15-2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newcomb PA, Baron J, Cotterchio M, et al. . Colon cancer family registry: an international resource for studies of the genetic epidemiology of colon cancer. Can Epidemiol Biomark Prev 2007;16:2331–43. 10.1158/1055-9965.EPI-07-0648 [DOI] [PubMed] [Google Scholar]
- 22. Lucke JF. A critique of the false-positive report probability. Genet Epidemiol 2009;33:145–50. 10.1002/gepi.20363 [DOI] [PubMed] [Google Scholar]
- 23. Freedman ML, Reich D, Penney KL, et al. . Assessing the impact of population stratification on genetic association studies. Nat Genet 2004;36:388–93. 10.1038/ng1333 [DOI] [PubMed] [Google Scholar]
- 24. Wen J, Xu Q, Yuan Y. Single nucleotide polymorphisms and sporadic colorectal cancer susceptibility: a field synopsis and meta-analysis. Cancer Cell Int 2018;18:155 10.1186/s12935-018-0656-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang B, Thyagarajan B, Gross MD, et al. . No evidence that associations of incident, sporadic colorectal adenoma with its major modifiable risk factors differ by chromosome 8q24 region rs6983267 genotype. Mol Carcinog 2014;53:E193–200. 10.1002/mc.22086 [DOI] [PubMed] [Google Scholar]
- 26. Montazeri Z, Theodoratou E, Nyiraneza C, et al. . Systematic meta-analyses and field synopsis of genetic association studies in colorectal adenomas. Int J Epidemiol 2016;45:186–205. 10.1093/ije/dyv185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abulí A, Castells A, Bujanda L, et al. . Genetic variants associated with colorectal adenoma susceptibility. PLoS One 2016;11:e0153084 10.1371/journal.pone.0153084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Du M, Jiao S, Bien SA, et al. . Fine-Mapping of common genetic variants associated with colorectal tumor risk identified potential functional variants. PLoS One 2016;11:e0157521 10.1371/journal.pone.0157521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tuupanen S, Niittymaki I, Nousiainen K, et al. . Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res 2008;68:14–17. 10.1158/0008-5472.CAN-07-5766 [DOI] [PubMed] [Google Scholar]
- 30. Tuupanen S, Turunen M, Lehtonen R, et al. . The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 2009;41:885–90. 10.1038/ng.406 [DOI] [PubMed] [Google Scholar]
- 31. Yeager M, Orr N, Hayes RB, et al. . Genome-Wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 2007;39:645–9. 10.1038/ng2022 [DOI] [PubMed] [Google Scholar]
- 32. Thomas G, Jacobs KB, Yeager M, et al. . Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 2008;40:310–5. 10.1038/ng.91 [DOI] [PubMed] [Google Scholar]
- 33. Eeles RA, Kote-Jarai Z, Giles GG, et al. . Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 2008;40:316–21. 10.1038/ng.90 [DOI] [PubMed] [Google Scholar]
- 34. Pomerantz MM, Ahmadiyeh N, Jia L, et al. . The 8q24 cancer risk variant rs6983267 shows long-range interaction with Myc in colorectal cancer. Nat Genet 2009;41:882–4. 10.1038/ng.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cicek MS, Slager SL, Achenbach SJ, et al. . Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiology Biomarkers & Prevention 2009;18:2492–500. 10.1158/1055-9965.EPI-09-0362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prokunina-Olsson L, Hall JL. No effect of cancer-associated SNP rs6983267 in the 8q24 region on co-expression of myc and TCF7L2 in normal colon tissue. Mol Cancer 2009;8:96 10.1186/1476-4598-8-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wright JB, Brown SJ, Cole MD. Upregulation of c-myc in cis through a large chromatin loop linked to a cancer risk-associated single-nucleotide polymorphism in colorectal cancer cells. Mol Cell Biol 2010;30:1411–20. 10.1128/MCB.01384-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sotelo J, Esposito D, Duhagon MA, et al. . Long-Range enhancers on 8q24 regulate c-myc. Proc Natl Acad Sci U S A 2010;107:3001–5. 10.1073/pnas.0906067107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim T, Cui R, Jeon Y-J, et al. . Long-Range interaction and correlation between Myc enhancer and oncogenic long noncoding RNA CARLo-5. Proc Natl Acad Sci U S A 2014;111:4173–8. 10.1073/pnas.1400350111 10.1073/pnas.1400350111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lu Y, Kweon S-S, Tanikawa C, et al. . Large-Scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer. Gastroenterology 2019;156:1455–66. 10.1053/j.gastro.2018.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huyghe JR, Bien SA, Harrison TA, et al. . Discovery of common and rare genetic risk variants for colorectal cancer. Nat Genet 2019;51:76–87. 10.1038/s41588-018-0286-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Loh YH, Mitrou PN, Wood A, et al. . SMAD7 and MGMT genotype variants and cancer incidence in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Cancer Epidemiol 2011;35:369–74. 10.1016/j.canep.2010.09.011 [DOI] [PubMed] [Google Scholar]
- 43. Stolfi C, Marafini I, De Simone V, et al. . The dual role of Smad7 in the control of cancer growth and metastasis. Int J Mol Sci 2013;14:23774–90. 10.3390/ijms141223774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grover S, Kastrinos F, Steyerberg EW, et al. . Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA 2012;308:485–92. 10.1001/jama.2012.8780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nielsen M, Lynch H, Infante E, et al. . MUTYH-Associated polyposis. Seattle: University of Washington, 2015. [Google Scholar]
- 46. Peltekova VD, Lemire M, Qazi AM, et al. . Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer-associated variants. International Journal of Cancer 2014;134:2330–41. 10.1002/ijc.28557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Law PJ, Timofeeva M, Fernandez-Rozadilla C, et al. . Association analyses identify 31 new risk loci for colorectal cancer susceptibility. Nat Commun 2019;10:2154 10.1038/s41467-019-09775-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang H, Taverna D, Stram DO, et al. . Genetic variation in the inflammation and innate immunity pathways and colorectal cancer risk. Can Epidemiol Biomark Prev 2013;22:2094–101. 10.1158/1055-9965.EPI-13-0694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Couturier-Maillard A, Secher T, Rehman A, et al. . Nod2-Mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest 2013;177:700–11. 10.1172/JCI62236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Robertson SJ, Zhou JY, Geddes K, et al. . Nod1 and NOD2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes 2013;4:222–31. 10.4161/gmic.24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Udden SMN, Peng L, Gan J-L, et al. . Nod2 suppresses colorectal tumorigenesis via downregulation of the TLR pathways. Cell Rep 2017;19:2756–70. 10.1016/j.celrep.2017.05.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Snetselaar R, van Oosterhout MFM, Grutters JC, et al. . Telomerase reverse transcriptase polymorphism rs2736100: a balancing act between cancer and non-cancer disease, a meta-analysis. Frontiers in Medicine 2018;5 10.3389/fmed.2018.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pittman AM, Naranjo S, Jalava SE, et al. . Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet 2010;6:e1001126 10.1371/journal.pgen.1001126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carvajal-Carmona LG, Cazier J-B, Jones AM, et al. . Fine-Mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet 2011;20:2879–88. 10.1093/hmg/ddr190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Grandval P, Blayau M, Buisine M-P, et al. . The UMD-APC database, a model of nation-wide knowledge base: update with data from 3,581 variations. Hum Mutat 2014;35:532–6. 10.1002/humu.22539 [DOI] [PubMed] [Google Scholar]
- 56. Lynch HT, Snyder CL, Shaw TG, et al. . Milestones of Lynch syndrome: 1895–2015. Nat Rev Cancer 2015;15:181–94. 10.1038/nrc3878 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319313supp001.pdf (98.6KB, pdf)
gutjnl-2019-319313supp002.pdf (611.6KB, pdf)
gutjnl-2019-319313supp003.pdf (17.3MB, pdf)


