Abstract
Background
Recently, it has emerged from the international scientific literature that quorum sensing (QS) is a promising way for the effective treatment of diseases caused by pathogenic bacteria. One of the crucial proteins in the QS system of Gram-positive bacteria is the pheromone. Some research has reported secondary metabolites from natural products capable of attenuating bacteria through the interruption of the quorum sensing system. One of the Indonesian herbal plants containing bioactive compounds is Sarang Semut (Myrmecodia pendans). A phenolic compound, dibenzo-p-dioxin-2,8-dicarboxylic acid, has been isolated from this plant which had antibacterial activity against Enterococcus faecalis. However, the molecular mechanism of it has not been known.
Aim
The study in question aimed to predict the molecular action of the compound M. pendans against some proteins that act as a signal in the mediated QS of Gram-positive bacteria, called pheromones, including PrgQ, PrgX, PrgZ, and CcfA.
Materials and Methods
The methods used in this in silico study were ligand-protein docking and virtual screening that were performed by some software and programs. The compound 1 and some positive controls act as ligand were subject binding to PrgQ, PrgX, PrgZ, and CcfA as proteins target, the ligands were free for blind docking. A framework was presented potency of phenolic compounds to inhibit the protein’s target from its affinity binding scores.
Results
It was found thatcompound 1 was potential to inhibit all of the tested protein and gave the highest binding affinity to PrgX (−9.2 kcal.mol−1; the site at Phe59B, Phe59B, Asn63A, and Asn63B residue) and PrgZ (−7.4 kcal.mol−1; the site at Leu4B, Thr65A, Thr82A. Gln81A, and Val5B residue).
Conclusion
It is proposed that compound 1 has a good activity to inhibit E. faecalis through its peptide pheromones in the QS system.
Keywords: oral bacteria, quorum sensing, peptide pheromones, PrgQ, PrgX, PrgZ, CcfA
Introduction
Dental caries is the most common oral disease among the population. It is a kind of serious disease that increases the quality of life. The most prevalent cause of this problem is demineralization dental hard tissues that cause a lot of people to lose their teeth.1 Several factors cause this disease, one kind of bacteria that contribute to oral disease is E. faecalis. It is the main bacteria that cause caries, endodontic infection, peri-implantitis, chronic periodontitis, and failed root canal treatment involving chronic apical periodontitis.2,3
The E. faecalis is a Gram-positive, nonmotile, commensal member of the gut microbiota and spherical bacteria. These bacteria attend about 90% infection in humans that causes the enterococci group. It is very often found in root canals treated teeth, causes failed endodontic treated teeth, and it has emerged a multidrug resistance such as some strain E. faecalis are resistant rifampicin, erythromycin, and azithromycin.4–6 The resistance of pathogenic bacteria against antibiotics is a serious problem for the future.7 An alternative approach is needed such as finding new modes of the action of novel antibiotics.8 One of the solutions to finding the novel antibiotic is a computational approach. Virtual screening is a valuable method to identify the effectiveness of lead compound activity.9 With the computational and docking methods, the prediction provided by virtual screening aims the researcher to find the most appropriate set of a compound that has required bioactivity from a natural resource, in a very effective and efficient way.10,11
The antibiotic treatment eliminates vulnerable bacteria from the bacterial population, leaving resistant bacteria to grow and multiply. In the E. faecalis, acquired elements, including antibiotic resistance genes, are estimated to represent over 25% of its genome. Acquired and intrinsic resistance mean that E. faecalis shows resistance to a variety of antibiotics. Virulence-specific therapeutics could avoid the selective pressure posed by antibiotics. Therefore, alternative anti-virulence therapeutic strategies, such as inhibition of the quorum-sensing system, could be sought to target this opportunistic pathogen.12
Quorum-sensing, termed QS, is the way bacteria communicate with each other or each cell which uses specific signals to coordinate population behaviors. Its regulate some process that occurs in the cell or synchrony requirement members in their community, such as biofilm formation and dispersal, virulence factor regulation, competence development, sporulation, and many more.13,14
Pheromone is a small peptide that acts as signals in mediate of QS in Gram-positive bacteria. These signal peptides regulate a wide array of processes, including many related to host-microbe interaction, and thus many provide novel targets for therapies that interfere with communication to disrupt bacterial infection. The pCF10 is a conjugative plasmid that occurs in the conjugation system of E. faecalis. Some precursors and regulators that occur in this system are PrgQ, PrgX, PrgY, PrgZ, and CcfA.15
The pCF10 is divided into two types of peptide pheromones, they are cCF10 (the amino acid sequence is LVTLVFV) and iCF10 (the amino acid sequence is AITLIFI). The cCF10 encoded by CcfA on the chromosome, while the iCF10 encoded by PrgQ on the plasmid.15,16 In pCF10 peptide regulation, the positive conjugation regulated by cCF10, act as an inducer of pCF10 conjugation genes from a donor cell, while negative conjunction is regulated by iCF10, act as inhibitor peptide which represses their expression to avoid self-induction by the endogenous pheromone from the recipient cell. The resected cCF10 activities neutralized by iCF10, in the donor cell, the induction state depends on the ratio cCF10/iCF10.16 The importation of the mature cCF10 peptide into the cytosol is facilitated by PrgZ.17 Then, both of mature cCF10 and iCF10 compete for binds to PrgX, the master regulator of conjugation peptide from RNPP (Rap, NprR, PlcR, and PrgX) regulators family. PrgX dimer specifically binds pCF10 at two promoters in plasmid pCF10. The iCF10 stabilizes PrgX tetramer, forming a DNA loop restricting access to RNA polymerase. The cCF10 competes with iCF10 for the binding pocket of PrgX. When cCF10 binds to PrgX, it disrupts the tetramers and eventually releases; dimer binding to DNA promoters allow access of RNA polymerase.18 There is no extended region of high sequence similarity between PgrX and any other known protein, including the TraA proteins.19
In therapy and drug development, effectiveness, and safety are an important point to use by humans. So, finding a lead compound and treatment method that give no side effect is needed. Natural products can be an option as a source of the active compound.20 Some plants such as Austroeupatorium inulaefolium, Leoheo domatiophorus Chaowasku, Paramignya trimera (Oliv.) Guillaum and Limnocitrus littoralis (Miq.) Swingle contains an essential oil that has various biological activities including antibacterial, antiviral, antimycotic, and antitrichomonas activities.21–23 Another potential plant is Sarang Semut (Myrmecodia pendans) or commonly known as ant nests plant is one of Rubiciae family which associated with ants. This epiphytic plant is spread in tropical forests, such as Malay Peninsula, Philippines, Cambodia, Cap York, Solomon Island, Sumatra, Borneo, Toraja, Java, and especially in Papua islands.24,25 Some Papuan peoples consume M. pendans as traditional medicine by boil dried parts of this plant, like consume tea.
Some therapeutic activity from M. pendans had been reported treating a variety of systemic diseases such as leukemia, heart diseases, tuberculosis, kidney, and prostate dysfunction, various allergies, migraines, rheumatism, hemorrhoid, infection diseases,26,27 especially the infection that causes dental caries.27 Several compounds that were isolated from M. pendans is a derivate of phenolic compound, and it was reported that some derivates of phenolic compounds had good antibacterial activity, especially against Enterococcus faecalis.28,29
In this study, it is supposed to give information about a prediction of the drug ability of antibacterial and anti-QS compounds from M. pendans and the prediction of the potential target proteins to which these compounds bind. Furthermore, it is hoped to be a guide for discovering novel antipathogenic agents.
Materials and Methods
The Ligand for this research is a phenolic compound that isolated from Sarang Semut (M. pendans), it was characterized as dibenzo-p-dioxin-2,8-dicarboxylic Acid30 while the positive controls were amoxicillin (CID 33613), ampicillin (CID 6249), enalapril (CID 5388962), and esomeprazole (CID 9568614).31 The chemical structure and molecular formula of this ligand retrieved from the PubChem compound database (https://www.ncbi.nlm.nih.gov/pccompound). Some proteins were involved in quorum sensing of E. faecalis was used as a protein target. The protein targets were PrgQ (2grm), prgX (2AW6), PrgZ (4FAJ), and CcfA (YidC) were retrieved from UniProt knowledgebase. (http://www.uniprot.org/).
In silico Characterization of the Compound 1
The phenolic characteristics were confirmed using two online programs. The chemical structures of the Sarang Semut phenolic were drawn again using PubChem Sketcher V2.4 – Mozilla Firefox online to obtain their canonical SMILE. The SMILE for dibenzo-p-dioxin-2,8-dicarboxylic acid was retrieved from the PubChem Compound database.32 Those SMILEs were used to convert the chemical structure into 3D using OPEN BABEL 2.4.1 program, in PDB format.33 The SMILEs of the phenolic were used further in the prediction of bioactivity. The 3-D structure model of proteins target was build using the SWISS-MODEL server (https://swissmodel.expasy.org/) based on its canonical sequence. The model obtained fitted to the template of the 3-D structure database, PDB entry – 4n0f. This model was in PDB format.34
Molecular Docking Between PgrX/PgrQ/PgrZ/ccfA with Compound 1 and Positive Controls
The ligand-protein docking and virtual screening were performed using Autodock Vina in open source software PyRx 0.8.35 The phenolic compound act as ligands were subject for binding to PrgQ, PrgZ, prgX, and CcfA as protein target; the ligands were free for blind docking. The most favorable free binding energy for the molecular interaction is the one that had a binding energy score that less than 1.0Å in positional root-mean-square deviation (RMSD).36
Complex PgrX/PgrQ/PgrZ/CcfA with Compound 1 and Positive Controls Visualization and Analysis
The docking results were visualized using pymol and then analyzed by software protein plus.37,38 Pymol showed docking poses and ligand-residue interaction in the 3D molecular picture. To show which residues bind to a ligand, proteins.plus program analyzes the protein-ligand results complex file, and then the picture of molecular interactions come out in 2D structure. For the best visualization, those molecular interactions were showed in the 3D molecular picture.
Results
Refer to the in vitro test, it was known that dibenzo-p-dioxin-2,8-dicarboxylic acid from M. pendans (hereinafter referred to as compound 1) had antibacterial activity against E. faecalis with the inhibition zone was 8.05 mm at in concentration of 1000 μg.mL−1.30 Some positive controls which had in vitro test were amoxicillin, ampicillin, enalapril, and esomeprazole gave antibacterial activity with their inhibition zones were 16±0.46, 11.5±0.12, 13±0.5 and 17.3±0.53 mm, respectively, with the different concentration about 5–25 μg.mL−1.31
In this docking process, we used several proteins involved in QS, more precisely in the peptide pheromone signaling process, that known as PrgX, PrgQ, PrgZ dan CcfA/CcfA.5 The result of docking showed in Tables 2 and 3.
Table 2.
Prediction of Antibacterial Activity of Compounds from M. pendans and Positive Controls (in vitro Study)
| Ligands | Binding Affinity of Ligand-Protein Complex (Kcal.mol−1) | |||
|---|---|---|---|---|
| PrgX | PrgQ | PrgZ | CcfA | |
| Compound 1 | −9.2 | −6.7 | −8.6 | −7.8 |
| Amoxycillin | −7.7 | −7.9 | −7.3 | −8.3 |
| Ampicillin | −8.3 | −6.1 | −6.5 | −8.3 |
| Enalapril | −5.9 | −6.8 | −6.4 | −6.9 |
| Esomeprazole | −8.4 | −6.7 | −7.3 | −7.3 |
Table 3.
Hydrogen Bond in Protein-Ligand from M. pendans
| Ligands | Residues Binding at Ligand-Protein Complex | |||
|---|---|---|---|---|
| PrgX | PrgQ | PrgZ | CcfA | |
| Compound 1 | Asn63A Asn63B Phe59B Phe59B | Asn161B Ser275B | Leu4B Thr65A Thr82A Gln81A Val5B | Ser240A Arg72A |
| Amoxycillin | Arg154A Ser169A Lys173A Ser170B | Arg145A Ser169A Lys173A Ser170B | Pro200A Ala474A Asp475A Trp511A | Arg72 A Ser240A Gln243A Gly137A Met196A |
| Ampicillin | Asn73B Asn63B Asp185B | Glu278A Asn161A | Ser72A Thr77A | Gly137A Gln243A Ser240A Arg72A |
| Enalapril | Leu4E Asn161B Phe86B | Tyr186A Gln216A Phe182A Val179A | Glu160A Glu484A Ala172A Val159A | Arg72A Ala1A Ser240A Ser83A Gly79A Ala1A |
| Esomeprazole | Asn73A Asn68A Asp185A | Gly150B Val179B | Leu503A Lys504A | Tyr188A |
Note: The BOLD text indicates amino residue to form hydrophobic bond with ligands, while the regular text indicates amino acid residue to form hydrophilic bond with ligands.
Discussion
Bioavailability and Antibacterial Activity Prediction of Compound 1 and Positive Controls Through Molecular Interaction with PrgX, PrgQ, PrgZ and CcfA
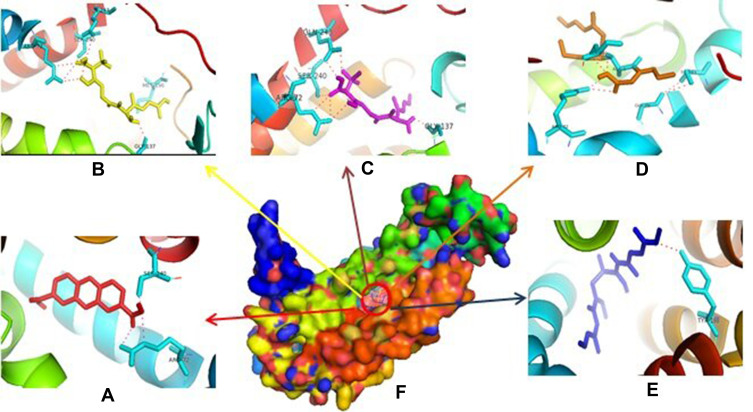
Based on Table 1, it showed that compound 1 was given higher activity against PrgX and PrgZ than other positive controls with binding affinity value was −9.2 kcal.mol−1 and −7.4 kcal.mol−1, respectively. The highest binding affinity against PgrQ was given from amoxicillin (−7.9 kcal.mol−1), while the highest binding affinity against CcfA was given from amoxicillin and ampicillin (both of them show the binding affinity value −8.3 kcal.mol−1). The active site protein which binds with the compound showed in Table 2. Based on data from Table 2, majority compounds were competitive inhibitor against CcfA proteins binding to Ser240A dan Arg72A (Figure 1). Although the esomeprazole was bound to the Tyr188A site located in the same place with other compounds in CcfA proteins, the esomeprazole is not a competitive inhibitor because it is bind to a different site than other compounds.
Table 1.
Antibacterial Activity of Compounds from M. pendans and Positive Controls (in vitro Study)
| Samples | Inhibition Zone (mm) | Disc Diffusion (mm in diameter) |
|---|---|---|
| Compound 1 | 8.05 | 6.0 |
| Amoxycillin | 16.0±0.46 | 15.4 ± 0.50 |
| Ampicillin | 11.5±0.12 | 10.0 ± 0.50 |
| Enalapril | 13.0± 0.50 | 11.0 ± 0.40 |
| Esomeprazole | 17.3± 0.53 | 16.0 ± 0.50 |
Figure 1.
Binding site of compound 1 (A), amoxycillin (B), ampicillin (C), enalapril (D), esomeprazole (E) against amino acid residue from CcfA (F) proteins.
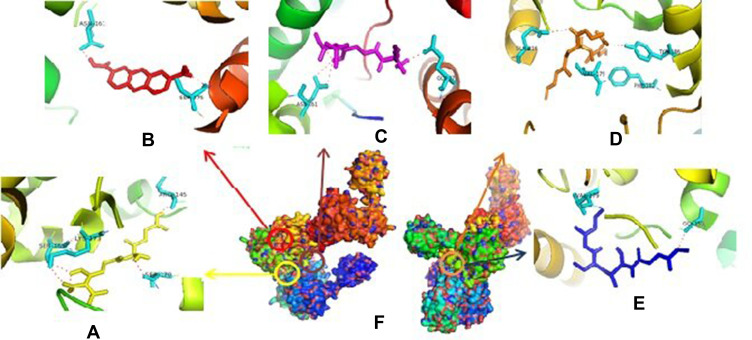
The inhibition of CcfA by compounds will certainly prevent the cCF10 leading decrease in cCF10 production.39 If the formation of pCF10 is inhibited, the induction system will be interrupted so that the interaction between bacterial cells are interrupted. In PrgQ, all compound that was tested show their activity with the different binding site that means they were noncompetitive inhibitor against PrgQ. The highest activity against PrgQ showed by amoxicillin with its binding affinity was −7.9 kcal.mol−1 and bond at Arg145A, Ser169A, Lys173A, and Ser170B residue, while a value of binding affinity compound 1 against PrgQ was −6.7 kcal.mol−1 and bond at Asn161B dan Ser275B residue (Figure 2).
Figure 2.
Binding site of amoxycillin (A), compound 1 (B), ampicillin (C), enalapril (D), esomeprazole (E) against amino acid residue from PrgQ proteins (F).
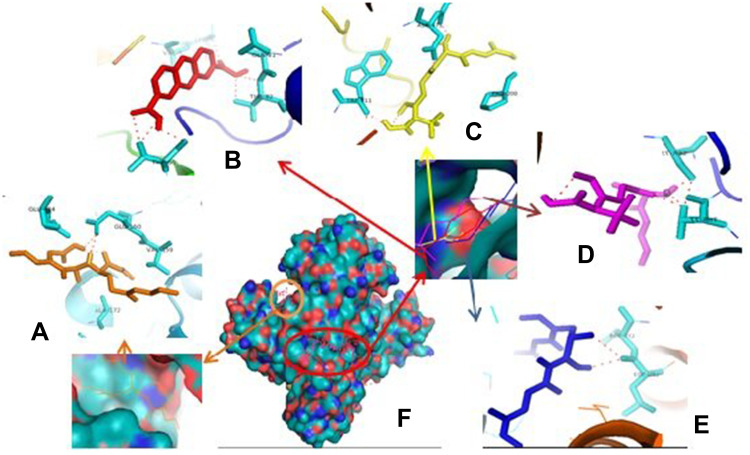
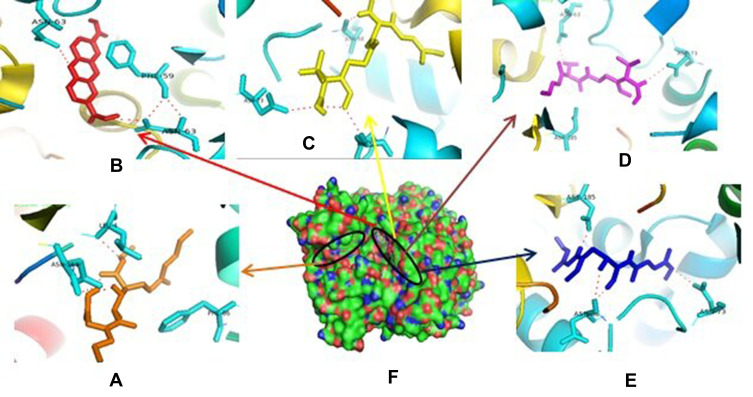
Therefore, if there is an inhibition of PrgQ (iCF10 producing) the function of the conjugative precursor inhibition will be inhibited so that the spread of genes throughout the bacterial population becomes uncontrolled. Both of PrgZ and PrgX proteins, all compounds were noncompetitive inhibitor because they were bond at the different sites against the protein, and the highest affinity value was showed by compound 1 with its scores −8.6 kcal.mol−1 and bond to Leu4B, Thr65A, Thr82A. The Gln81A and Val5B residue against PrgZ (Figure 3) and it was −9.2 kcal.mol−1 and bond to Phe59B, Phe59B, Asn63A, and Asn63B residue against PrgX (Figure 4).
Figure 3.
Binding site of enalapril (A), compound 1 (B), amoxycillin (C), ampicillin (D), esomeprazole (E) against amino acid residue from PrgZ (F) proteins.
Figure 4.
Binding site of enalapril (A), compound 1 (B), amoxycillin (C), ampicillin (D), esomeprazole (E) against amino acid residue from PrgX proteins (F).
In pCF10 regulation, the PrgZ acts as a facilitator in the importing process of peptide signals from plasmid to cytosol. Meanwhile, the PrgX acts as a master regulator that establishes complex with the peptide signal in the cytosol. Therefore, inhibition both of PrgZ or PrgZ will obstruct the main pCF10 regulation process and the QS system will be blocked.
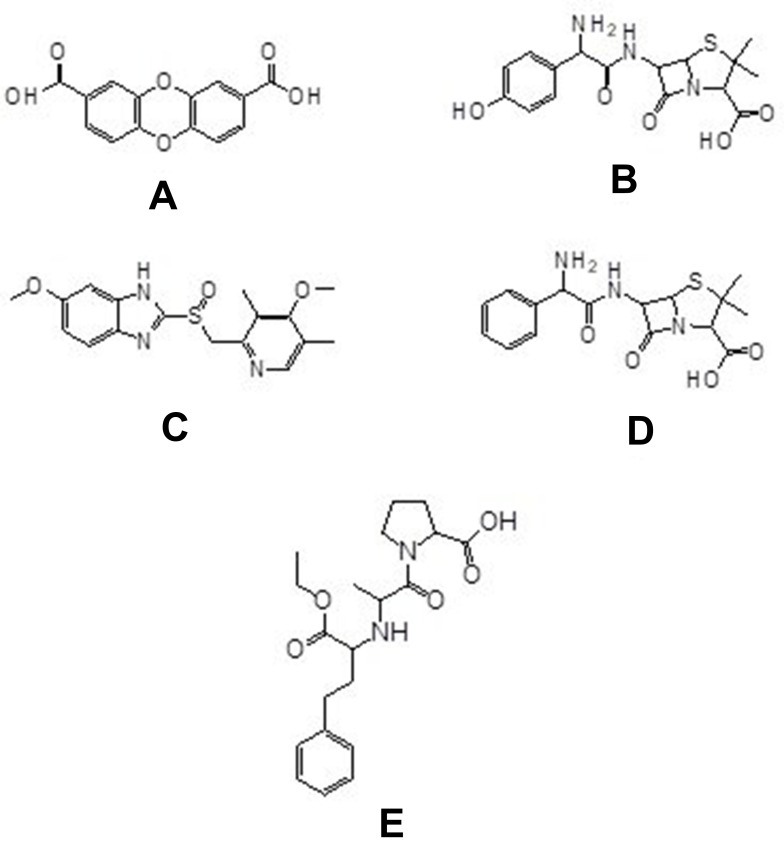
Based on the data, it is obtained that compound 1 has higher activity against PrgX and PrgZ proteins than all positive controls, while the amoxicillin has the highest activity against PrgQ and CcfA proteins. The different activity and binding site from all compounds is suggested by the different structure as follow in Figure 5.
Figure 5.
Chemical structure of compounds of dibenzo-p-dioxin-2,8-dicarboxylic acid (A) from M. pendans; positive control: amoxycillin (B), enalapril (C), ampicillin (D), and esomeprazole (E).
Chemical Structure Relationship on Antibacterial Activities of Compound 1 and Positive Controls
Based on the result and the structural comparison of all compound test, it suggests that the existence of carboxyl groups give the most contribute to the activity.40 Compound 1 has the most activity because it has two carboxylic acid groups while amoxicillin, ampicillin, and esomeprazole have one carboxylic group, respectively. Even though the amoxicillin and ampicillin have a similar structure, in this case, they have a different activity. The activity of amoxicillin is better than ampicillin’s activity because there is a hydroxyl group of amoxicillin which suggests increasing the activity of its compound.41
Besides, pheromone inhibitors isolated from bacteria such as iPD1, iAD1, iCF10, and iAM373 have a linear peptide structure. They have a similar characteristic in which the structure of them contains hydrophobic amino acid-like Leu, Phe, Gly, Thr, and Ile that match with the part of pheromone structure.42 It could be the same analogy for the interaction of compound 1 and positive control with the protein (pheromone). The part of compound 1 and the positive control structure is hydrophobic alkyl includes phenyl, methyl, etc.
Comparison Between the Result of in vitro and in silico
The in vitro study showed that compound 1 has the lowest of inhibition zone (8.8 mm) while in silico study suggested it has the highest binding affinity against PrgX and PrgZ (−9.2 kcal.mol−1 and −7.4 kcal.mol−1). Difference result from in vitro and in silico methods are cannot be separated from the complexity of biological system factors which cannot be fully represented by a computer program. Moreover, as a nonlinear system, biological entities is also showing ‘screwd up behavior’43
Conclusion
The dibenzo-p-dioxin-2,8-dicarboxylic acid (compound 1) from Sarang semut (Myrmecodia pendans) has a good activity to inhibit Enterococcus faecalis through its peptide pheromones, which cause the existence of its carboxyl groups, or in another word, compound 1 has antibacterial and anti-QS activity. The result of this study can be guidance in advance research including in vitro, in vivo, and trial clinical.
Acknowledgments
The authors are grateful to Universitas Padjadjaran. This research was supported by Academic Leadership Grant 2019–2020 from Universitas Padjadjaran, Sumedang Indonesia.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Veiga N, Aires D, Douglas F, et al. Dental caries: a review. J Dent Oral Health. 2016;2(5):1–3. [Google Scholar]
- 2.Wang QQ, Zhang CF, Chu CH, et al. Prevalence of Enterococcus faecalis in saliva and filled root canals of teeth associated with apical periodontitis. Int J Oral Sci. 2012;4(1):19–23. doi: 10.1038/ijos.2012.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.E Y K, Lapesqueur LSS, Yassuda CG, et al. Enterococcus species in the oral cavity: prevalence, virulence factors, and antimicrobial susceptibility. PLoS One. 2016;11(9):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flanangan D. Enterococcus faecalis and dental implants. J Oral Implantol. 2017;43(1):8–11. doi: 10.1563/aaid-joi-D-16-00069 [DOI] [PubMed] [Google Scholar]
- 5.Tyne DV, Martin MJ, Gilmore MS. Virulence effect of Enterococcus faecalis structure, function, and biology of the Enterococcus faecalis cytolysin. Toxins. 2013;5(5):895–911. doi: 10.3390/toxins5050895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golob M, Pete M, Kusar D, et al. Antimicrobial resistance and virulence genes in Enterococcus faecium and Enterococcus faecalis from humans and retail red meat. Biomed Res Int. 2019;2019:1–12. doi: 10.1155/2019/2815279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seethaler M, Hertlein T, Wecklein B, et al. Novel small-molecule antibacterial against Gram-positive pathogens of Staphylococcus and Enterococcus species. Antibiotics. 2019;8(4):1–9. doi: 10.3390/antibiotics8040210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usai D, Donadu M, Bua A, et al. Enhancement of antimicrobial activity of pump inhibitors associating drugs. J Infect Dev Ctries. 2019;13(2):162–164. doi: 10.3855/jidc.11102 [DOI] [PubMed] [Google Scholar]
- 9.Tang YT, Marshall GR. Virtual screening for lead discovery In: Satyanarayanajois SD, editor. Drug Design and Discovery: Method and Protocols. New York: Springer Science; 2011:1–22. [DOI] [PubMed] [Google Scholar]
- 10.Shen J, Xu X, Cheng F, et al. Virtual screening on natural products for discovering active compound and target information. Curr Med Chem. 2003;10(21):2327–2342. doi: 10.2174/0929867033456729 [DOI] [PubMed] [Google Scholar]
- 11.Lavecchia A, Giovanni CD. Virtual screening strategies in drug discovery: A critical review. Curr Med Chem. 2013;20(23):2839–2860. doi: 10.2174/09298673113209990001 [DOI] [PubMed] [Google Scholar]
- 12.Ali L, Goraya MU, Arafat Y, et al. Molecular mechanism of quorum-sensing in Enterococcus faecalis: its role in virulence and therapeutic approaches. Int J Mol Sci. 2017;18(5):1–19. doi: 10.3390/ijms18050960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang Q, Chen J, Yang C, et al. Quorum sensing: a prospective therapeutic target for bacterial diseases. Biomed Res Int. 2019;2019:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook LC, Federle MJ. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol Ecol. 2014;38(3):473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Bandyopadhyay A, Kozlowicz BK. Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. Microbiology Open. 2017;6(4):1–13. doi: 10.1002/mbo3.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antiporta MH, Dunny GM. CcfA, The genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184(4):1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi K, Brown CK, Gu ZY, et al. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proceedings of the National Academy of Sciences. 2005;102(51):18596–18601. doi: 10.1073/pnas.0506163102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estarda JR, Diez AE, Guarneros G, et al. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl Microbiol Biotechnol. 2010;87(3):913–923. doi: 10.1007/s00253-010-2651-y [DOI] [PubMed] [Google Scholar]
- 19.Kozlowicz BK, Shi K, Gu YZ, et al. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol. 2006;62(4):1–19. doi: 10.1111/j.1365-2958.2006.05434.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomford NE, Senthebane DA, Rowe A, et al. Natural product for drug discover in the 21st century: innovetions for novel drug discovery. Int J Mol Sci. 2018;19(6):1–29. doi: 10.3390/ijms19061578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le NT, Ho DV, Doan TQ, et al. In vitro antimicrobial activity of essential oil extracted from leaves of Leoheo domatiophorus chaowasku, D.T. Ngo and H.T. Le in Vietnam. Plants. 2020;453(9):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le NT, Ho DV, Doan TQ, et al. Biological activities of essential oil from leave of Paramigya trimera (Oliv.) Guillaum and Limnocitrus littoralis (Miq.) Swingle. Antibiotics. 2020;207(9):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bua A, Usai D, Donadu MG, et al. Antimicrobial activity of Austroeupatorium inulaefolium (H.B.K.) against intracellular and extracellular organism. Nat Prod Res. 2018;23(32):2869–2871. [DOI] [PubMed] [Google Scholar]
- 24.Bashari MH, Hidayat S, Ruswandi YAR, et al. The n-hexane fraction of Myrmecodia pendans inhibits cell survival and poliferation in colon cancer cell line. Int J Pharm Pharm Sci. 2018;10(1):108–112. doi: 10.22159/ijpps.2018v10i1.21882 [DOI] [Google Scholar]
- 25.Gartika M, Pramesti HT, Kurnia D, et al. A terpenoid isolated from Sarang Semut (Myrmecodia pendans) bulb and it’s potential for the inhibition and eradication of Streptococcus mutans biofilm. BMC Complement Altern Med. 2018;18(151):1–8. doi: 10.1186/s12906-018-2213-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudiono J, Oka CT, Trisfilha P. The scientific base of Myrcomedia pendans as herbal remedies. Br J Med Med Res. 2015;8(3):230–237. doi: 10.9734/BJMMR/2015/17465 [DOI] [Google Scholar]
- 27.Gartika AM, Wartadewi, Mariam MS, et al. Antibacterial of terpenoid A from Sarang Semut (Myrmrcodia pendans) against Streptococcus mutans. Int J Chemtech Res. 2018;11(1):228–233. [Google Scholar]
- 28.Engida AM, Faika S, Thi BTN, et al. Analysis of major antioxidants from extracts of Myrmecodia pendans by UV/visible spectrophotometer, liquid chromatography/tandem mass spectrometry, and high-performance liquid chromatography/UV techniques. J Food Drug Anal. 2015;23(2):303–309. doi: 10.1016/j.jfda.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pimia RH, Nohynek L, Meier C, et al. Antimicrobial properties of phenolic compounds from Berries. J Appl Microbiol. 2001;90(4):494–507. doi: 10.1046/j.1365-2672.2001.01271.x [DOI] [PubMed] [Google Scholar]
- 30.Kurnia D, Sumiarsa D, Dharsono HAD, et al. Bioactive compound isolated from Indonesian epiphytic plan of Sarang Semut and their antibacterial activity against pathogenic oral bacteria. Nat Prod Commun. 2017;12(8):1201–1204. doi: 10.1177/1934578X1701200814 [DOI] [Google Scholar]
- 31.Selvaraj C, Sivakamavalli J, Vaseeharan B, et al. Structural elucidation of SrtA enzyme in Enterococcus faecalis: an emphasis on screening of potential inhibitors against the biofilm formation. Mol Biosyst. 2014;10(7):1775–1789. doi: 10.1039/C3MB70613C [DOI] [PubMed] [Google Scholar]
- 32.Ihlenfeldt WD, Bolton EE, Bryant SH. The PubChem chemical structure sketcher. J Cheminformat. 2009;1(20):1–9. doi: 10.1186/1758-2946-1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Boyle NM, Banck M, James CA, et al. Open babel: an open chemical toolbox. J Cheminform. 2011;3(33):1–14. doi: 10.1186/1758-2946-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse A, Bertoni M, Bienert S, et al. Sarang Semut (Myrmrcodia pendans) against Streptococcus mutans. Int J Chemtech Res. 2018;11(1):228–233. [Google Scholar]
- 35.Dallakyan S, Oslon AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. [DOI] [PubMed] [Google Scholar]
- 36.Azam SS, Abbasi SW. Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor Biol Med Model. 2013;10(63):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLano WL, Bromberg S. Pymol User’s Guide. USA: DeLano Scientific LLC; 2004. [Google Scholar]
- 38.Rauf MA, Zubair S, Azhar A. Ligand docking and binding site analysis with Pymol and Autodock/Vina. Int J Basic Appl Sci. 2015;4(2):168–177. doi: 10.14419/ijbas.v4i2.4123 [DOI] [Google Scholar]
- 39.Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also act as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci. 2005;102(43):15617–15622. doi: 10.1073/pnas.0505545102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varadwaj PK, Lahiri T. Functional group based ligand binding affinity scoring function at atomic enviromental level. Bioinformation. 2009;3(6):268–274. doi: 10.6026/97320630003268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaushik AC, Kumar S, Wei DQ, et al. Structure based virtual screening studies to identify novel potential compound for GPR142and their relative dynamic analysis for study of type 2 diabetes. Front Chem. 2018;6(23):1–14. doi: 10.3389/fchem.2018.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama J, Ono Y, Suzuki A. Isolation and structure of the sex pheromone inhibitor, iAM373, of Enterococcus faecalis. Biosci Biotech Biochem. 1995;59(7):1358–1359. doi: 10.1271/bbb.59.1358 [DOI] [PubMed] [Google Scholar]
- 43.Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: applications to targets and beyond. Br J Pharmacol. 2007;152(1):21–37. doi: 10.1038/sj.bjp.0707306 [DOI] [PMC free article] [PubMed] [Google Scholar]