Abstract
The Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) family mediates membrane fusion during membrane trafficking and autophagy in all eukaryotic cells, with a number of SNAREs having cell type-specific functions. The endosome-trans-Golgi network (TGN) localized SNARE, Vesicle transport through interaction with t-SNAREs 1A (Vti1a), is unique among SNAREs in that it has numerous neuron-specific functions. These include neurite outgrowth, nervous system development, spontaneous neurotransmission, synaptic vesicle and dense core vesicle secretion, as well as a process of unconventional surface transport of the Kv4 potassium channel. Furthermore, the human VT11A gene is known to form fusion products with neighboring genes in cancer tissues, and VT11A variants are associated with risk in cancers, including glioma. In this review, I highlight VTI1A's known physio-pathological roles in brain neurons, as well as unanswered questions in these regards.
Keywords: Neuron, Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE), VAMP4, VAMP7, Vti1a, Neuroscience, Cellular neuroscience, Molecular neuroscience, Membrane, Gene mutation
Neuron, Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE), VAMP4, VAMP7, Vti1a, Neuroscience, Cellular neuroscience, Molecular neuroscience, Membrane, Gene mutation.
1. Introduction
Vesicle fusion in eukaryotic membrane (or vesicular) traffic is mediated by N-ethylmaleimide-sensitive factor (NSF), Soluble NSF attachment proteins (SNAPs) and SNAP receptors (SNAREs) (Söllner et al., 1993; Weber et al., 1998). These form a distinct physical complex in vitro – the 20S complex (Hohl et al., 1998; Zhou et al., 2015). A SNARE protein is membrane associated via a C-terminal transmembrane domain or lipid anchor, and is thus largely cytoplasmically oriented. SNAREs harbor one or two α-helical coiled-coil signature motif known as the SNARE domain (Weimbs et al., 1998). SNARE domains are composed of repeated hydrophobic residues that is interrupted by a charged arginine (R) or glutamine (Q), and thus the basis of classification of SNAREs into R- or Q-SNAREs (Fasshauer et al., 1998; Kloepper et al., 2007). In membrane fusion, SNARE domains belonging to those that are either transport vesicle-associated (v-SNAREs) or target membrane-localized (t-SNAREs) interact spontaneously to form complexes in trans with the right composition and stoichiometry (Katz and Brennwald, 2000). In completion of the pairing of the SNARE domain α-helixes, or the formation of multiple ` SNAREpins’ (Weber et al., 1998), the energetically favorable SNAREpin formation and clustering at the fusion site provides the biophysical or entropic force required to overcome the electrostatic repulsion and bring about fusion of two negatively charged lipid bilayers (Mostafavi et al., 2017).
In this regard, the fusion between synaptic vesicles and the pre-synaptic plasma membrane during synaptic vesicle exocytosis in neurons offers a classic example (Südhof, 2014). This action potential-triggered process is highly regulated (Rizo and Xu, 2015), and canonical synaptic vesicle fusion is driven by a trans-SNARE complex formed between the plasma membrane associated Qa-SNARE Syntaxin 1 (STX1A/B) and the Qb-SNARE Synaptosome Associated Protein 25 (SNAP25) with the vesicle-bound R-SNARE Synaptobrevin 2 (Syb2)/Vesicle-associated membrane protein 2 (VAMP2) (Pevsner et al., 1994; Südhof, 2014). Structural analyses indicate that this synaptic exocytic SNARE complex takes the form of a 4-helix bundle, with leucine-zipper-like layers formed by the SNARE domains at the center (Sutton et al., 1998; Poirier et al., 1998; Katz et al., 1998). Embedded within these leucine-zipper layers is the ionic `zero’ layer, consisting of the 1 R (from VAMP2) and 3 Q (1 from STX1A/B and 2 from SNAP25) residues contributed by each of the four SNARE domain α-helices. On the other hand, other SNARE complexes, such as that responsible for endoplasmic reticulum (ER)-Golgi transport, has its 3 Qs contributed by 3 different t-SNAREs (Xu et al., 2000). This particular `1R + 3Q′ stoichiometric arrangement provides topological constraint (Parlati et al., 2000) and compositional specificity (Yang et al., 2008) for functional membrane fusion, and is likely conserved for all SNARE-mediated membrane fusion in the cell. The unique ionic `zero’ layer was shown to be required for eventual dissociation of formed SNARE complex by NSF/α-SNAP (Scales et al., 2001), although mutational analyses in yeast have suggested that the R could also be functionally replaced by a Q (Katz and Brennwald, 2000).
Neurons are polarized cells with rather specific membrane trafficking needs and processes that are exemplified by, but not limited to, synaptic vesicle exocytosis. SNAREs are required in other membrane fusion processes in neurons (Wang and Tang, 2006), including those that are neuron-specific such as neurite outgrowth and neuroendocrine secretion. In this regard, a subset of R-SNAREs known as the longins (as opposed to the brevins) (Filippini et al., 2001; Rossi et al., 2004), have been quite extensively implicated in neuron-specific functions (Rossi et al., 2004; Daste et al., 2015). The longin domain with a profilin-like fold (Gonzalez et al., 2001; Rossi et al., 2004) can be found in the R-SNAREs Sec22b (Gonzalez et al., 2001), Ykt6 (Tochio et al., 2001) and VAMP7 (Proux-Gillardeaux et al., 2007). The multi-functional Ykt6 complexes with several other SNAREs and is shown to be involve in ER-Golgi transport (McNew et al., 1997), Golgi transport (Xu et al., 2002), vacuole targeting (Kweon et al., 2003) and more recently in autophagosome-lysosome fusion (Yong and Tang, 2019; Kriegenburg et al., 2019). However, Ykt6 appears to be specialized for the trafficking of a poorly characterized unique neuronal membrane compartment (Hasegawa et al., 2003). The mammalian Sec22b is involved in ER-Golgi exocytic transport (Hay et al., 1997; Zhang et al., 1999), secretory autophagy (Kimura et al., 2017), ER-plasma membrane (PM) apposition and PM expansion (Petkovic et al., 2014), and has also been implicated in ER-phagosome antigen cross presentation in dendritic cells (Cebrian et al., 2011; Alloatti et al., 2017; Cruz et al., 2020). VAMP7 mediates late endosome-lysosome transport (Advani et al., 1999), TGN-late endosome transport (Pols et al., 2013) and the unconventional process of lysosomal secretion (Sato et al., 2011; Wang et al., 2018). In neurons, VAMP7 is also known to mediate neurite outgrowth (Arantes and Andrews, 2006; Burgo et al., 2009) and spontaneous neurotransmission (Bal et al., 2013).
Orthologue of the SNARE, Vesicle transport through interaction with t-SNAREs 1A (Vti1a), is first identified in yeast (as Vti1p), with functions as an endo-lysosomal SNARE in Golgi retrograde (Lupashin et al., 1997) and Golgi to vacuole transport (Fischer von Mollard and Stevens, 1998). In yeast, Vti1p is the only SNARE of the Qb.IIIb subtype (Kloepper et al., 2007), and two Qb SNAREs with the closest homology to Vti1p in mammals are Vti1a and Vti1b (Fischer von Mollard and Stevens, 1998). The mammalian Vti1a and Vti1b appear to have distinct subcellular localizations, cognate SNARE partners (Kreykenbohm et al., 2002) and trafficking functions. For example, Vti1a functions in insulin-stimulated glucose transport (Bose et al., 2005) and regulated exocytosis in adrenal chromaffin cells, but VTI1b null cells show no secretion defects (Walter et al., 2014). Deletion of Vti1b is non-lethal, but rather specifically resulted in reduced amounts of Syntaxin 8 due to enhanced degradation of the latter (Atlashkin et al., 2003). However, they are also likely to have key overlapping functions, as knockout of either of them did not affect viability in mice (Atlashkin et al., 2003; Kunwar et al., 2011), but a double-knockout of both Vti1a and Vti1b resulted in perinatal lethality (Kunwar et al., 2011).
Early works have implicated Vti1a in both anterograde (Xu et al., 1998) and retrograde (Mallard et al., 2002) traffic, as well as its enrichment in small synaptic vesicles (Antonin et al., 2000). Vti1a and VAMP4 partners with a number of SNAREs associated with the trans-Golgi network (TGN) and the endosome to mediate connecting traffic between the exocytic and endocytic pathways. A SNARE complex formed between Vti1a, Syntaxin 6 (STX6) (Wendler and Tooze, 2001), Syntaxin 16 (STX16) (Simonsen et al., 1998; Tang et al., 1998) and either VAMP3/cellubrevin (McMahon et al., 1993) or VAMP4 (Steegmaier et al., 1999) mediates early/recycling endosome transport of Shiga toxin and TGN46 (Mallard et al., 2002; Kreykenbohm et al., 2002; Laufman et al., 2011). Another related complex, consisting of Vti1a, Syntaxin 10 (STX10) (Wang et al., 2005; Ganley et al., 2008), STX16, and VAMP3, works in mediating Mannose 6-phosphate receptors (MPRs) transport from the endosomes to the Golgi (Ganley et al., 2008). Vti1a, VAMP4 and STX6, in conjunction with Syntaxin 13 (STX13) (Prekeris et al., 1998), forms another complex that mediate homotypic fusion of early endosomes (Brandhorst et al., 2006).
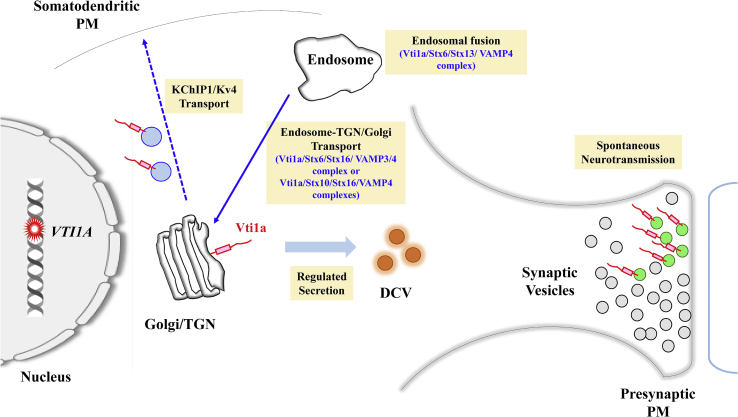
Importantly, Vti1a appears to have specific roles in neuronal processes (Ramirez and Kavalali, 2012; Emperador-Melero et al., 2019), including neuronal development (Kunwar et al., 2011), non-canonical neurotransmission processes (Ramirez et al., 2012; Emperador-Melero et al., 2018), dense core granule secretion (Walter et al., 2014; Emperador-Melero et al., 2018), unconventional transport in neuron (Flowerdew and Burgoyne, 2009), and perhaps also brain malignancy (Kinnersley et al., 2015; Wang et al., 2017a, Wang et al., 2017b; Davidsen et al., 2018). In the paragraphs below, Vti1a's involvement in neuronal development, physiology and pathological processes (summarized in Figure 1) shall be highlighted and discussed.
Figure 1.
Schematic diagram illustrating the roles of Vti1a in neurons. As in non-neuronal cells, Vti1a is part of a SNARE complexes that mediate endosome-trans-Golgi network (TGN) transport. Vti1a also mediates regulated secretion of dense core vesicles (DCV) and marks a pool of spontaneously fusing synaptic vesicles (green) that recycles at rest and fuse with the axonal presynaptic plasma membrane (PM). Vti1a and VAMP7 (VAMP7) are also involved in Potassium channel interacting protein 1 (KChIP1)/voltage-gated K+ channels (Kv4) to the somatodendritic plasma membrane. Mutations/variants and gene fusion of VTI1A (red star) in neuronal or glia progenitor cells could be oncogenic. See text for more details.
2. Vti1a/b in neural development
In 2011, von Mollard's group has generated Vti1a−/− Vti1b−/− double-knockout (DKO) mouse embryos and found somewhat surprisingly that fibroblasts immortalized from embryos exhibit neither obvious aberrance in plasma or endo-lysosomal membrane morphology, nor overt defects in cargo trafficking (Kunwar et al., 2011). However, major neuronal projection tracts and commissures were either absent or reduced in size in E18.5 DKO mice, indicating severely impaired axonal growth. In the DKO embryos, the peripheral ganglia also exhibited varying degrees of neurodegeneration, likely due to neurotrophic deprivation resulting from a lack of axonal targeting and innervation. The DKO animals thus have neuronal trafficking processes that are crucial for axonal growth that was severely defective with the loss of both Vti1a and Vti1b. That defects do not occur in single knockout or double heterozygous mice indicate that Vti1a and Vti1b could compensate for the loss of each other. Notably, the axonal growth phenotype of Vti1a/b DKO in mice is much more severe than those of VAMP7 (Sato et al., 2011; Danglot et al., 2012), although the latter has been implicated in a major neurite outgrowth mechanism involving specific structures known as enlargeosomes (Racchetti et al., 2010; Meldolesi, 2011; Colombo et al., 2014). A more recent study by Verhage's group also showed that Vti1a/Vti1b DKO neurons have diminished viability in culture, are smaller and form fewer synapses (Emperador-Melero et al., 2018). Superficially, the findings would suggest that Vti1a and Vti1b function in a redundant manner in axonal or neurite growth, with the presence of either being sufficient for postnatal survival.
3. Vti1a in spontaneous neurotransmission
Other than action potential-evoked plasma membrane fusion of synaptic vesicles, all synapses exhibit a low background of spontaneous vesicle fusion and action potential-independent miniature neurotransmission (Sutton et al., 2006). Although it has been argued that spontaneous and evoked transmission arise from the same synaptic vesicle pools (Groemer and Klingauf, 2007; Hua et al., 2010; Wilhelm et al., 2010), the notion is controversial and evidence exists for spontaneous fusion being the result of a separate pool of vesicles recycling spontaneously (Sara et al., 2005; Fredj and Burrone, 2009; Andreae et al., 2012; Ramirez et al., 2012). Kavalali's group showed that some synaptic vesicles do recycle when neurons are at rest, and identified by differential tagging of synaptic vesicles a `reluctantly releasable’ or `reserved pool’ (Sara et al., 2005). Spontaneous, but not evoke transmission, is affected by neurotransmitter depletion at rest rather selectively from the spontaneously fusing vesicles. Also, the pools of activity-dependent and spontaneously recycling vesicles could become indistinguishable in the absence of Syb2 (Sara et al., 2005).
Using Syb2, VAMP7 or Vti1a tagged with different fluorescent probes to examine the trafficking and fusion of labeled vesicles both at rest and when stimulated, the group found a population of Vti1-containing vesicles that appear to traffic preferentially at rest (Ramirez et al., 2012). Furthermore, endogenous levels of Vti1a in live neurons correlated with the degree of spontaneous vesicle recycling at individual synaptic termini, and silencing of Vti1a selectively impaired the generation of signals from spontaneous, but not the stimuli-evoked transmission. Conversely, expression of an N-terminally truncated version of Vti1a with a likely disinhibition of Vti1a's SNARE activity, augmented spontaneous neurotransmission. Importantly, this apparently Vti1a-dependent spontaneous vesicle trafficking and fusion still occurs in Syb2-knockout neurons. This latter point is important as it suggests the possibility that Syb2, which is critical for the canonical synaptic vesicle fusion SNARE complex, is not required for functional fusion of the Vti1a-containing vesicles. Vti1a therefore appears to be important for the trafficking and recycling of a rather unique spontaneously-fusing vesicle pool at rest. The questions is whether Vti1a actually participates in the SNARE complex at the final plasma membrane fusion step remains unclear. At first look this notion would appear untenable as the canonical synaptic fusion machinery typically uses the Qb-SNARE SNAP25 (Emperador-Melero et al., 2018), but a non-exclusive role for Vti1a in this regard could not yet be completely ruled out. Notably, the R-SNARE VAMP4, which forms a SNARE complex with Vti1a in mediating endosome-TGN transport (Mallard et al., 2002), also has a pre-synaptic role in maintaining bulk Ca2+-dependent asynchronous neurotransmission that is independent of Syb2-mediated synchronous neurotransmission (Raingo et al., 2012). The role of VAMP4 in this regard is also unclear. VAMP4 has been identified as a component of synaptic vesicle (Raingo et al., 2012; Ramirez and Kavalali, 2012), but there is yet no functional evidence to confirm that VAMP4 directly mediates synaptic vesicle fusion.
In a more recent analysis, Verhage's group found that both synaptic vesicle and dense-core vesicle (DCV) number and secretion were greatly reduced in Vti1a/Vti1b DKO mouse neurons (Emperador-Melero et al., 2018). Whole cell patch clamp recordings indicate that evoked neurotransmitter release upon stimulation was decreased by 80–90%, while frequency of spontaneous fusion events was reduced by 65%. These authors also showed that the Vti1a/b DKO neurons have much reduced levels of proteins important for synaptic vesicle fusion, such as the Qb-SNARE SNAP25, Munc13-1 and Synaptotagmin. There is in general a reduced protein flux into axons and the presynaptic termini, while the somatic Golgi apparatus exhibit distended cisternae and cargo accumulation. These findings indicate that loss of both Vti1a and Vti1b in neurons severely impairs the more upstream event of TGN exocytosis. Vti1a/b DKO neurons also have impaired endosome-Golgi transport as marked by Cholera Toxin subunit-B (Emperador-Melero et al. 2018, 2019). The secretory defects noted in Vti1a/b DKO neurons were rescued almost completely by exogenous expression of Vti1a alone, but some of which (such as total DCV pool and retrograde transport) could only be partially rescue by the expression of Vti1b alone. These findings of Vti1a associated upstream defects makes deciphering of any distinct synaptic role of Vtia difficult. Surprisingly, spontaneous transmission is also not completely abolished in the Vti1a/b DKO neurons, which would suggest that at least some spontaneously fusing vesicles at rest could fuse independently of Vti1a.
4. Vti1a/b and secretory granule exocytosis
The first hint that Vti1a could be involved in regulated transport or secretion came from the report which showed that Vti1a regulates insulin-stimulated glucose transport and the secretion of the hormone Adipocyte complement related protein of 30 kD (Acrp30), or adiponectin, in 3T3-L1 adipocytes (Bose et al., 2005). Two studies have since confirmed a role for Vti1a in dense-core vesicle biogenesis and secretion in the endocrine and neuronal systems, respectively (Walter et al., 2014; Emperador-Melero et al., 2018). In the earlier report, Walter and colleagues showed that Vti1a is localized to a subdomain of TGN that appears to be negative for the TGN marker TGN38, but harbors the SNARE Syntaxin 6 (Walter et al., 2014). Analysis of the adrenal glands of Vti1a-null mice showed a reduction in Syb2 levels. The Syb2-positive large dense-core vesicles (LDCVs) in adrenal chromaffin cells are devoid of Vti1a. However, the Ca2+ channel abundance and stimuli-evoked LDCV exocytosis are reduced in Vti1a-null cells. With manipulation of exocytic stimulus, it is shown that despite the secretion defect and the decrease in the number of secretory granules, LDCV secretion in the Vti1a-null cells are unchanged in terms of kinetics and Ca2+ sensitivity. The LDCV secretory phenotype is not exhibited by Vti1b-null cells and not exacerbated by a Vti1a/Vti1b DKO, indicating that Vti1b is not involved in, and could not complement Vti1a's role in regulated exocytosis of adrenal chromaffin cells.
As mentioned in the section above, Emperador-Melero and colleagues discovered that, as with synaptic vesicles, DCV number and secretion were also greatly reduced in Vti1a/Vti1b DKO mouse neurons (Emperador-Melero et al., 2018). In the case of neuronal DCV, Vti1b expression could rescue the DCV secretion, but only partially rescue the reduction in total DCV pool. Therefore there appears to be a difference between DCV secretion in neurons and LDCV secretion in adrenal chromaffin cells in terms of Vti1b's ability to compensate for the loss of Vti1a. These results should be interpreted with some caution as compensatory mechanisms that could be activated or engaged by the cells may be different in the case of losing one of the Vti protein as compared to losing both (Villarreal et al., 2013). Born without either Vti1a or Vti1b, different cell types may find different ways to cover for the secretory defect and survive the deficiency.
5. Vti1a in neuronal unconventional exocytic transport
In neurons, the trafficking of voltage-gated Kv4 potassium (K+) channels to the plasma membrane is dependent on the Potassium channel interacting proteins (KChIPs) (Jerng and Pfaffinger, 2014). The somatodendritic A-type K+ current underlies neuronal excitability, and KChiPs-modulated Kv4 trafficking is thus important for homeostatic excitability (Wang et al., 2013). KChIP1 is targeted via an N-terminal myristoylation to vesicles that appear to be trafficking intermediates from the ER to the Golgi, but these differ from those conveying conventional ER-Golgi traffic of the classical exocytic cargo marker, Vesicular stomatitis virus G protein (VSVG) (Tang et al., 2005). These require Coat protein I (COPI), but are not COPII-coated and not inhibited by a GTP-locked Sar1 mutant (Hasdemir et al., 2005). These KChIP1-postive vesicles do not have components of the usual ER-Golgi SNARE complexes, but are instead positive for both Vti1a and VAMP7. Silencing of either Vti1a or VAMP7 inhibited the transport of Kv4/KChIP1 to the plasma membrane in HeLa and Neuro2A cells (while not affecting VSVG transport), but not Kv4 transport stimulated by KChIP2 (Flowerdew and Burgoyne, 2009).
Kv4/KChIP1 trafficking in neuron may thus occur via a somewhat unconventional exocytic route that is dependent on Vti1a and VAMP7. The precise nature and other membrane trafficking requirements of this route is still unclear. Several routes of unconventional exocytosis in non-neuronal cells have been described (Rabouille et al., 2012; Chua et al., 2012; Ng and Tang, 2016; Rabouille, 2017). However, the SNARE-dependence of KChIP1 vesicle transport does not appear to fit directly with any of these other described modes of unconventional exocytosis. Furthermore, while the trafficking of KChIP1 vesicles does not appear to involve COPII, its requirement of COPI may nonetheless indicate a Golgi-dependent exocytic process. Further work would be required to better decipher the exocytic itinerary and the mechanism of Vti1a-dependent Kv4/KChIP1 surface transport in more detail.
6. Vti1a′s possible connection with brain malignancy
Vti1a's potential involvement in human malignancy is first indicated by the identification of a VTI1A gene fusion with a neighboring gene encoding Transcription factor 7-like 2 (TCF7L2)/T-cell transcription factor 4 (TCF4) (Bass et al., 2011). TCF7L2/TC4 is known to co-operate with β-catenin in colorectal carcinogenesis (Morin et al., 1997), but the fusion product lacks the TCF4 β-catenin-binding domain. A more recent analysis has indicated that the Vti1a-TCF4 fusion protein acts as a dominant-negative regulator of Wnt-β-catenin signaling (Davidsen et al., 2018) but the oncogenic role of the fusion protein remains unclear. The genomic region around VTI1A appears rather prone to the production of fusion products. Another report has, however, suggested that the high frequency of fusion transcripts between TCF7L2 and its neighboring genes including VTI1A, even in non-cancerous tissues, could be due largely to transcription induced chimeras that are expressed at low levels (Nome et al., 2014).
More recent Genome-wide association study (GWAS)-based analysis have also identified colorectal cancer (Wang et al., 2014) and lung cancer (Su et al., 2015) susceptibility locus in VTI1A. Moreover, GWAS-based analysis have shown that the VTI1A gene harbors risk locus for glioma in a European population (Kinnersley et al., 2015), and the VTI1A-associated single nucleotide polymorphism (SNP) variant rs11196067 is significantly associated with glioma risk in a Chinese population (Wang et al., 2017a, Wang et al., 2017b). A summary of VTI1A's association with cancer is provided in Table 1. A recent meta-analysis of variants reported in the VTI1A-TCF7L2 region and potential function of these variants using data from the Encyclopedia of DNA Elements (ENCODE) has indicated that the VTI1A-TCF7L2 region does have a significant role in cancer pathogenesis (Zhang et al., 2018). The generation of oncogenic fusion gene product via chromosomal translocation is well-known, but fusion gene products involving SNAREs are uncommon. In this regard, the fusion of VAMP2 and Neuregulin 1 (NRG1) genes has been found in non-small-cell lung carcinoma and is predicted to be oncogenic (Jung et al., 2015). Likewise, STX16's fusion with Aminopeptidase Like 1 (NPEPL1) has been identified in gastrointestinal stromal tumors (Kang et al., 2016).
Table 1.
A summary of VTI1A and other SNARE family members with known genetic association with various cancers. VTI1A - Vesicle transport through interaction with T-SNAREs 1A (Vti1a); TCF7L2 - Transcription factor 7-like 2; CFAP46 - Cilia and flagella associated protein 46; VAMP2 – Vesicle-associated membrane protein 2; STX16 – Syntaxin 16; NRG1 – Neuregulin 1; NPEPL1 – Aminopeptidase-like 1.
| Cancer and data type | Vti1a mutation/variant | Reference |
|---|---|---|
| Colorectal cancer (CRC) (primary tumor sample and matched adjacent tissues) | VTI1A-TCF7L2 gene fusion | Bass et al., 2011 |
| CRC cell lines and tissues | Splice variants of VTI1A-TCF7L2 fusion transcripts | Nome et al., 2014 |
| CRC genome wide association studies (GWAS) | Risk locus at 10q25 (rs12241008, intronic to VTI1A) | Wang et al., 2014 |
| Non-small cell lung cancer association | Allele A of VTI1A SNP rs7086803 | Su et al., 2015 |
| Glioma GWAS | Risk locus at 10q25.2 (rs11196067), near the VTI1A gene locus | Kinnersley et al., 2015 |
| CRC GWAS | Risk locus at 10q25.2 (rs10506868) | Zeng et al., 2016 |
| Glioma association | Risk locus rs11196067 | Wang et al., 2017a, Wang et al., 2017b |
| Associations between variants in the VTI1A-TCF7L2 region and cancer susceptibility | 8 common variants of VTI1A and TCF7L2 associated with various cancers | Zhang et al., 2018 |
| Hepatocellular carcinoma (HCC) RNA sequencing | VTI1A-CFAP46 fusion transcript (from a genomic DNA VTI1A-CFAP46 translocation event) | Tsuge et al., 2019 |
| Non-small-cell lung adenocarcinoma, whole-transcriptome sequencing | VAMP2-NRG1 gene fusion | Jung et al., 2015 |
| Gastrointestinal stromal tumors, exome and transciptome sequencing | STX16-NPEPL1 gene fusion | Kang et al., 2016 |
Our understanding of involvement of membrane trafficking components in human cancer is largely based on the oncogenic roles of Rabs (Chia and Tang, 2009; Tang and Ng, 2009; Wang et al., 2017a, Wang et al., 2017b; Shaughnessy and Echard, 2018). At the moment it is difficult to determine with any certainty how dysregulated or mutated Vti1a could be oncogenic. Given the role of Wnt signaling in brain tumors (McCord et al., 2017; Rajakulendran et al., 2019), it is conceivable that the Vti1a-TCF7L2/TCF4 fusion product may perturb oncogenic signaling in the brain. Furthermore, it is speculatively plausible that the membrane trafficking role of Vti1a may impact on growth receptor signaling, or in some yet undefined manner facilitates migration or metastasis in cancer cells.
7. Future questions and research directions
In this brief review, the unusually wide and unique roles for the endosomal-TGN SNARE Vti1a in neuron-specific processes are highlighted and discussed. Vti1a appears to function in axonal/neurite outgrowth and thus nervous system development, spontaneous neurotransmission, dense core vesicle exocytosis, and may speculatively have a role in brain malignancy. Our understanding of these Vti1a functions in neurons are in most cases still lacking in mechanistic details, and many important research questions remained. A prominent question would be whether Vti1a could participate directly in a synaptic vesicle SNARE fusion complex, perhaps replacing SNAP25 as a Qb-SNARE. The SNARE complexes formed by VTI1A with other SNAREs in neurons, as well as biochemical and biophysical aspects of their functions in the neuronal processes discussed above, have remained underexplored. Another important question is how does Vti1a's role in endosome-TGN retrograde traffic align with, or explains its apparent role in regulated exocytosis. Little is known about the COPII-independent route of KChIP1 that involves Vti1a. In this regard, does Vti1a have more roles in an unconventional exocytosis involving other cargoes in neurons? Given VTI1A's deciphered role in nervous system development and neurotransmission, it is likely that VTI1A mutations and variants could result in nervous system dysfunctions and neurological disorders. This possibility should also be further explored.
With regards to the oncogenic potential of the VTI1A locus, an important question would be how do VTI1A variants or mutants, in conjunction with gene fusion products or on their own, drive oncogenesis, particularly those associated with the brain? Forthcoming answers to these questions would undoubtedly enrich our overall fundamental understanding of neuronal physiology and pathology.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Advani R.J., Yang B., Prekeris R., Lee K.C., Klumperman J., Scheller R.H. VAMP-7 mediates vesicular transport from endosomes to lysosomes. J. Cell Biol. 1999;146:765–776. doi: 10.1083/jcb.146.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alloatti A., Rookhuizen D.C., Joannas L., Carpier J.M., Iborra S., Magalhaes J.G. Critical role for Sec22b-dependent antigen cross-presentation in antitumor immunity. J. Exp. Med. 2017;214:2231–2241. doi: 10.1084/jem.20170229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae L.C., Fredj N.B., Burrone J. Independent vesicle pools underlie different modes of release during neuronal development. J. Neurosci. 2012;32:1867–1874. doi: 10.1523/JNEUROSCI.5181-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Riedel D., von Mollard G.F. The SNARE Vti1a-beta is localized to small synaptic vesicles and participates in a novel SNARE complex. J. Neurosci. 2000;20:5724–5732. doi: 10.1523/JNEUROSCI.20-15-05724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arantes R.M.E., Andrews N.W. A role for synaptotagmin VII-regulated exocytosis of lysosomes in neurite outgrowth from primary sympathetic neurons. J. Neurosci. 2006;26:4630–4637. doi: 10.1523/JNEUROSCI.0009-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlashkin V., Kreykenbohm V., Eskelinen E.L., Wenzel D., Fayyazi A., Fischer von Mollard G. Deletion of the SNARE vti1b in mice results in the loss of a single SNARE partner, syntaxin 8. Mol. Cell Biol. 2003;23:5198–5207. doi: 10.1128/MCB.23.15.5198-5207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal M., Leitz J., Reese A.L., Ramirez D.M.O., Durakoglugil M., Herz J. Reelin mobilizes a VAMP7-dependent synaptic vesicle pool and selectively augments spontaneous neurotransmission. Neuron. 2013;80:934–946. doi: 10.1016/j.neuron.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass A.J., Lawrence M.S., Brace L.E., Ramos A.H., Drier Y., Cibulskis K. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat. Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A., Guilherme A., Huang S., Hubbard A.C., Lane C.R., Soriano N.A. The v-SNARE Vti1a regulates insulin-stimulated glucose transport and Acrp30 secretion in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:36946–36951. doi: 10.1074/jbc.M508317200. [DOI] [PubMed] [Google Scholar]
- Brandhorst D., Zwilling D., Rizzoli S.O., Lippert U., Lang T., Jahn R. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2701–2706. doi: 10.1073/pnas.0511138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgo A., Sotirakis E., Simmler M.C., Verraes A., Chamot C., Simpson J.C. Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 2009;10:1117–1124. doi: 10.1038/embor.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 2011;147:1355–1368. doi: 10.1016/j.cell.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Chia W.J., Tang B.L. Emerging roles for Rab family GTPases in human cancer. Biochim. Biophys. Acta. 2009;1795:110–116. doi: 10.1016/j.bbcan.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Chua C.E.L., Lim Y.S., Lee M.G., Tang B.L. Non-classical membrane trafficking processes galore. J. Cell. Physiol. 2012;227:3722–3730. doi: 10.1002/jcp.24082. [DOI] [PubMed] [Google Scholar]
- Colombo F., Racchetti G., Meldolesi J. Neurite outgrowth induced by NGF or L1CAM via activation of the TrkA receptor is sustained also by the exocytosis of enlargeosomes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:16943–16948. doi: 10.1073/pnas.1406097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F.M., Colbert J.D., Rock K.L. The GTPase Rab39a promotes phagosome maturation into MHC-I antigen-presenting compartments. EMBO J. 2020;39 doi: 10.15252/embj.2019102020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danglot L., Zylbersztejn K., Petkovic M., Gauberti M., Meziane H., Combe R. Absence of TI-VAMP/Vamp7 leads to increased anxiety in mice. J. Neurosci. 2012;32:1962–1968. doi: 10.1523/JNEUROSCI.4436-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daste F., Galli T., Tareste D. Structure and function of longin SNAREs. J. Cell Sci. 2015;128:4263–4272. doi: 10.1242/jcs.178574. [DOI] [PubMed] [Google Scholar]
- Davidsen J., Larsen S., Coskun M., Gögenur I., Dahlgaard K., Bennett E.P. The VTI1A-TCF4 colon cancer fusion protein is a dominant negative regulator of Wnt signaling and is transcriptionally regulated by intestinal homeodomain factor CDX2. PloS One. 2018;13 doi: 10.1371/journal.pone.0200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperador-Melero J., Huson V., van Weering J., Bollmann C., Fischer von Mollard G., Toonen R.F. Vti1a/b regulate synaptic vesicle and dense core vesicle secretion via protein sorting at the Golgi. Nat. Commun. 2018;9:3421. doi: 10.1038/s41467-018-05699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emperador-Melero J., Toonen R.F., Verhage M. Vti proteins: beyond endolysosomal trafficking. Neuroscience. 2019;420:32–40. doi: 10.1016/j.neuroscience.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R.B., Brunger A.T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini F., Rossi V., Galli T., Budillon A., D'Urso M., D'Esposito M. Longins: a new evolutionary conserved VAMP family sharing a novel SNARE domain. Trends Biochem. Sci. 2001;26:407–409. doi: 10.1016/s0968-0004(01)01861-8. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G., Stevens T.H. A human homolog can functionally replace the yeast vesicle-associated SNARE Vti1p in two vesicle transport pathways. J. Biol. Chem. 1998;273:2624–2630. doi: 10.1074/jbc.273.5.2624. [DOI] [PubMed] [Google Scholar]
- Flowerdew S.E., Burgoyne R.D. A VAMP7/Vti1a SNARE complex distinguishes a non-conventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels. Biochem. J. 2009;418:529–540. doi: 10.1042/BJ20081736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredj N.B., Burrone J. A resting pool of vesicles is responsible for spontaneous vesicle fusion at the synapse. Nat. Neurosci. 2009;12:751–758. doi: 10.1038/nn.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., Espinosa E., Pfeffer S.R. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J. Cell Biol. 2008;180:159–172. doi: 10.1083/jcb.200707136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez L.C., Weis W.I., Scheller R.H. A novel snare N-terminal domain revealed by the crystal structure of Sec22b. J. Biol. Chem. 2001;276:24203–24211. doi: 10.1074/jbc.M101584200. [DOI] [PubMed] [Google Scholar]
- Groemer T.W., Klingauf J. Synaptic vesicles recycling spontaneously and during activity belong to the same vesicle pool. Nat. Neurosci. 2007;10:145–147. doi: 10.1038/nn1831. [DOI] [PubMed] [Google Scholar]
- Hasdemir B., Fitzgerald D.J., Prior I.A., Tepikin A.V., Burgoyne R.D. Traffic of Kv4 K+ channels mediated by KChIP1 is via a novel post-ER vesicular pathway. J. Cell Biol. 2005;171:459–469. doi: 10.1083/jcb.200506005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H., Zinsser S., Rhee Y., Vik-Mo E.O., Davanger S., Hay J.C. Mammalian ykt6 is a neuronal SNARE targeted to a specialized compartment by its profilin-like amino terminal domain. Mol. Biol. Cell. 2003;14:698–720. doi: 10.1091/mbc.E02-09-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J.C., Chao D.S., Kuo C.S., Scheller R.H. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hohl T.M., Parlati F., Wimmer C., Rothman J.E., Söllner T.H., Engelhardt H. Arrangement of subunits in 20 S particles consisting of NSF, SNAPs, and SNARE complexes. Mol. Cell. 1998;2:539–548. doi: 10.1016/s1097-2765(00)80153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Sinha R., Martineau M., Kahms M., Klingauf J. A common origin of synaptic vesicles undergoing evoked and spontaneous fusion. Nat. Neurosci. 2010;13:1451–1453. doi: 10.1038/nn.2695. [DOI] [PubMed] [Google Scholar]
- Jerng H.H., Pfaffinger P.J. Modulatory mechanisms and multiple functions of somatodendritic A-type K (+) channel auxiliary subunits. Front. Cell. Neurosci. 2014;8:82. doi: 10.3389/fncel.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Yong S., Kim P., Lee H.Y., Jung Y., Keum J. VAMP2-NRG1 fusion gene is a novel oncogenic driver of non-small-cell lung adenocarcinoma. J. Thorac. Oncol. 2015;10:1107–1111. doi: 10.1097/JTO.0000000000000544. [DOI] [PubMed] [Google Scholar]
- Kang G., Yun H., Sun C.H., Park I., Lee S., Kwon J. Integrated genomic analyses identify frequent gene fusion events and VHL inactivation in gastrointestinal stromal tumors. Oncotarget. 2016;7:6538–6551. doi: 10.18632/oncotarget.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Brennwald P. Testing the 3Q:1R "rule": mutational analysis of the ionic "zero" layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell. 2000;11:3849–3858. doi: 10.1091/mbc.11.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., Hanson P.I., Heuser J.E., Brennwald P. Genetic and morphological analyses reveal a critical interaction between the C-termini of two SNARE proteins and a parallel four helical arrangement for the exocytic SNARE complex. EMBO J. 1998;17:6200–6209. doi: 10.1093/emboj/17.21.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Jia J., Kumar S., Choi S.W., Gu Y., Mudd M., Dupont N., Jiang S., Peters R., Farzam F., Jain A., Lidke K.A., Adams C.M., Johansen T., Deretic V. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36:42–60. doi: 10.15252/embj.201695081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnersley B., Labussière M., Holroyd A., Di Stefano A.L., Broderick P., Vijayakrishnan J. Genome-wide association study identifies multiple susceptibility loci for glioma. Nat. Commun. 2015;6:8559. doi: 10.1038/ncomms9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepper T.H., Kienle C.N., Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol. Biol. Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreykenbohm V., Wenzel D., Antonin W., Atlachkine V., von Mollard G.F. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 2002;81:273–280. doi: 10.1078/0171-9335-00247. [DOI] [PubMed] [Google Scholar]
- Kriegenburg F., Bas L., Gao J., Ungermann C., Kraft C. The multi-functional SNARE protein Ykt6 in autophagosomal fusion processes. Cell Cycle. 2019;18:639–651. doi: 10.1080/15384101.2019.1580488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunwar A.J., Rickmann M., Backofen B., Browski S.M., Rosenbusch J., Schöning S. Lack of the endosomal SNAREs vti1a and vti1b led to significant impairments in neuronal development. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2575–2580. doi: 10.1073/pnas.1013891108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon Y., Rothe A., Conibear E., Stevens T.H. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole. Mol. Biol. Cell. 2003;14:1868–1881. doi: 10.1091/mbc.E02-10-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O., Hong W., Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J. Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin V.V., Pokrovskaya I.D., McNew J.A., Waters M.G. Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol. Biol. Cell. 1997;8:2659–2676. doi: 10.1091/mbc.8.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F., Tang B.L., Galli T., Tenza D., Saint-Pol A., Yue X. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord M., Mukouyama Y.S., Gilbert M.R., Jackson S. Targeting WNT signaling for multifaceted glioblastoma therapy. Front. Cell. Neurosci. 2017;11:318. doi: 10.3389/fncel.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon H.T., Ushkaryov Y.A., Edelmann L., Link E., Binz T., Niemann H. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature. 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- McNew J.A., Sogaard M., Lampen N.M., Machida S., Ye R.R., Lacomis L. Ykt6p, a prenylated SNARE essential for endoplasmic reticulum-Golgi transport. J. Biol. Chem. 1997;272:17776–17783. doi: 10.1074/jbc.272.28.17776. [DOI] [PubMed] [Google Scholar]
- Meldolesi J. Neurite outgrowth: this process, first discovered by Santiago Ramon y Cajal, is sustained by the exocytosis of two distinct types of vesicles. Brain Res. Rev. 2011;66:246–255. doi: 10.1016/j.brainresrev.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Mostafavi H., Thiyagarajan S., Stratton B.S., Karatekin E., Warner J.M., Rothman J.E. Entropic forces drive self-organization and membrane fusion by SNARE proteins. Proc. Natl. Acad. Sci. U.S.A. 2017;114:5455–5460. doi: 10.1073/pnas.1611506114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng F., Tang B.L. Unconventional protein secretion in animal cells. Methods Mol. Biol. 2016;1459:31–46. doi: 10.1007/978-1-4939-3804-9_2. [DOI] [PubMed] [Google Scholar]
- Nome T., Hoff A.M., Bakken A.C., Rognum T.O., Nesbakken A., Skotheim R.I. High frequency of fusion transcripts involving TCF7L2 in colorectal cancer: novel fusion partner and splice variants. PloS One. 2014;9 doi: 10.1371/journal.pone.0091264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., McNew J.A., Fukuda R., Miller R., Söllner T.H., Rothman J.E. Topological restriction of SNARE-dependent membrane fusion. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- Petkovic M., Jemaiel A., Daste F., Specht C.G., Izeddin I., Vorkel D., Verbavatz J.M., Darzacq X., Triller A., Pfenninger K.H., Tareste D., Jackson C.L., Galli T. The SNARE Sec22b has a non-fusogenic function in plasma membrane expansion. Nat. Cell Biol. 2014;16:434–444. doi: 10.1038/ncb2937. [DOI] [PubMed] [Google Scholar]
- Pevsner J., Hsu S.C., Braun J.E., Calakos N., Ting A.E., Bennett M.K. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Poirier M.A., Xiao W., Macosko J.C., Chan C., Shin Y.K., Bennett M.K. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat. Struct. Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- Pols M.S., van Meel E., Oorschot V., ten Brink C., Fukuda M., Swetha M.G. hVps41 and VAMP7 function in direct TGN to late endosome transport of lysosomal membrane proteins. Nat. Commun. 2013;4:1361. doi: 10.1038/ncomms2360. [DOI] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y.A., Scheller R.H. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proux-Gillardeaux V., Raposo G., Irinopoulou T., Galli T. Expression of the Longin domain of TI-VAMP impairs lysosomal secretion and epithelial cell migration. Biol. Cell. 2007;99:261–271. doi: 10.1042/BC20060097. [DOI] [PubMed] [Google Scholar]
- Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27:230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- Rabouille C., Malhotra V., Nickel W. Diversity in unconventional protein secretion. J. Cell Sci. 2012;125:5251–5255. doi: 10.1242/jcs.103630. [DOI] [PubMed] [Google Scholar]
- Racchetti G., Lorusso A., Schulte C., Gavello D., Carabelli V., D'Alessandro R. Rapid neurite outgrowth in neurosecretory cells and neurons is sustained by the exocytosis of a cytoplasmic organelle, the enlargeosome. J. Cell Sci. 2010;123:165–170. doi: 10.1242/jcs.059634. [DOI] [PubMed] [Google Scholar]
- Raingo J., Khvotchev M., Liu P., Darios F., Li Y.C., Ramirez D.M.O. VAMP4 directs synaptic vesicles to a pool that selectively maintains asynchronous neurotransmission. Nat. Neurosci. 2012;15:738–745. doi: 10.1038/nn.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakulendran N., Rowland K.J., Selvadurai H.J., Ahmadi M., Park N.I., Naumenko S. Wnt and Notch signaling govern self-renewal and differentiation in a subset of human glioblastoma stem cells. Genes Dev. 2019;33:498–510. doi: 10.1101/gad.321968.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez D.M.O., Kavalali E.T. The role of non-canonical SNAREs in synaptic vesicle recycling. Cell. Logist. 2012;2:20–27. doi: 10.4161/cl.20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez D.M.O., Khvotchev M., Trauterman B., Kavalali E.T. Vti1a identifies a vesicle pool that preferentially recycles at rest and maintains spontaneous neurotransmission. Neuron. 2012;73:121–134. doi: 10.1016/j.neuron.2011.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Xu J. The synaptic vesicle release machinery. Annu. Rev. Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- Rossi V., Banfield D.K., Vacca M., Dietrich L.E.P., Ungermann C., D'Esposito M. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem. Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Sara Y., Virmani T., Deák F., Liu X., Kavalali E.T. An isolated pool of vesicles recycles at rest and drives spontaneous neurotransmission. Neuron. 2005;45:563–573. doi: 10.1016/j.neuron.2004.12.056. [DOI] [PubMed] [Google Scholar]
- Sato M., Yoshimura S., Hirai R., Goto A., Kunii M., Atik N. The role of VAMP7/TI-VAMP in cell polarity and lysosomal exocytosis in vivo. Traffic. 2011;12:1383–1393. doi: 10.1111/j.1600-0854.2011.01247.x. [DOI] [PubMed] [Google Scholar]
- Scales S.J., Yoo B.Y., Scheller R.H. The ionic layer is required for efficient dissociation of the SNARE complex by alpha-SNAP and NSF. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14262–14267. doi: 10.1073/pnas.251547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy R., Echard A. Rab35 GTPase and cancer: linking membrane trafficking to tumorigenesis. Traffic. 2018;19:247–252. doi: 10.1111/tra.12546. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Bremnes B., Rønning E., Aasland R., Stenmark H. Syntaxin-16, a putative Golgi t-SNARE. Eur. J. Cell Biol. 1998;75:223–231. doi: 10.1016/S0171-9335(98)80116-7. [DOI] [PubMed] [Google Scholar]
- Söllner T., Whiteheart S.W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Steegmaier M., Klumperman J., Foletti D.L., Yoo J.S., Scheller R.H. Vesicle-associated membrane protein 4 is implicated in trans-Golgi network vesicle trafficking. Mol. Biol. Cell. 1999;10:1957–1972. doi: 10.1091/mbc.10.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W.M., Chen Z.H., Zhang X.C., Su J., Xie Z., Yan H.H. Single nucleotide polymorphisms in VTI1A gene contribute to the susceptibility of Chinese population to non-small cell lung cancer. Int. J. Biol. Markers. 2015;30:e286–e293. doi: 10.5301/jbm.5000140. [DOI] [PubMed] [Google Scholar]
- Südhof T.C. The molecular machinery of neurotransmitter release (Nobel lecture) Angew Chem. Int. Ed. Engl. 2014;53:12696–12717. doi: 10.1002/anie.201406359. [DOI] [PubMed] [Google Scholar]
- Sutton M.A., Ito H.T., Cressy P., Kempf C., Woo J.C., Schuman E.M. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Tang B.L., Low D.Y., Lee S.S., Tan A.E., Hong W. Molecular cloning and localization of human syntaxin 16, a member of the syntaxin family of SNARE proteins. Biochem. Biophys. Res. Commun. 1998;242:673–679. doi: 10.1006/bbrc.1997.8029. [DOI] [PubMed] [Google Scholar]
- Tang B.L., Ng E.L. Rabs and cancer cell motility. Cell Motil Cytoskeleton. 2009;66:365–370. doi: 10.1002/cm.20376. [DOI] [PubMed] [Google Scholar]
- Tang B.L., Wang Y., Ong Y.S., Hong W. COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta. 2005;1744:293–303. doi: 10.1016/j.bbamcr.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Tsuge S., Saberi B., Cheng Y., Wang Z., Kim A., Luu H., Abraham J.M., Ybanez M.D., Hamilton J.P., Selaru F.M., Villacorta-Martin C., Schlesinger F., Philosophe B., Cameron A.M., Zhu Q., Anders R., Gurakar A., Meltzer S.J. Detection of novel fusion transcript VTI1A-CFAP46 in hepatocellular carcinoma. Gastrointest. Tumors. 2019;6:11–27. doi: 10.1159/000496795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio H., Tsui M.M., Banfield D.K., Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Villarreal A.M., Adamson S.W., Browning R.E., Budachetri K., Sajid M.S., Karim S. Molecular characterization and functional significance of the Vti family of SNARE proteins in tick salivary glands. Insect Biochem. Mol. Biol. 2013;43:483–493. doi: 10.1016/j.ibmb.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A.M., Kurps J., de Wit H., Schöning S., Toft-Bertelsen T.L., Lauks J. The SNARE protein vti1a functions in dense-core vesicle biogenesis. EMBO J. 2014;33:1681–1697. doi: 10.15252/embj.201387549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Nola S., Bovio S., Bun P., Coppey-Moisan M., Lafont F. Biomechanical control of lysosomal secretion via the VAMP7 hub: a tug-of-war between VARP and LRRK1. iScience. 2018;4:127–143. doi: 10.1016/j.isci.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Burnett T., Kono S., Haiman C.A., Iwasaki M., Wilkens L.R. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat. Commun. 2014;5:4613. doi: 10.1038/ncomms5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.G., He X.P., Li Q., Madison R.D., Moore S.D., McNamara J.O. The auxiliary subunit KChIP2 is an essential regulator of homeostatic excitability. J. Biol. Chem. 2013;288:13258–13268. doi: 10.1074/jbc.M112.434548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Deng Z., Wang M., Li R., Xu G., Bao G. Additional evidence supports association of common genetic variants in VTI1A and ETFA with increased risk of glioma susceptibility. J. Neurol. Sci. 2017;375:282–288. doi: 10.1016/j.jns.2017.02.013. [DOI] [PubMed] [Google Scholar]
- Wang S., Hu C., Wu F., He S. Rab25 GTPase: functional roles in cancer. Oncotarget. 2017;8:64591–64599. doi: 10.18632/oncotarget.19571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tai G., Lu L., Johannes L., Hong W., Tang B.L. Trans-Golgi network syntaxin 10 functions distinctly from syntaxins 6 and 16. Mol. Membr. Biol. 2005;22:313–325. doi: 10.1080/09687860500143829. [DOI] [PubMed] [Google Scholar]
- Wang Y., Tang B.L. SNAREs in neurons--beyond synaptic vesicle exocytosis (Review) Mol. Membr. Biol. 2006;23:377–384. doi: 10.1080/09687860600776734. [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Weimbs T., Mostov K., Low S.H., Hofmann K. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- Wendler F., Tooze S. Syntaxin 6: the promiscuous behaviour of a SNARE protein. Traffic. 2001;2:606–611. doi: 10.1034/j.1600-0854.2001.20903.x. [DOI] [PubMed] [Google Scholar]
- Wilhelm B.G., Groemer T.W., Rizzoli S.O. The same synaptic vesicles drive active and spontaneous release. Nat. Neurosci. 2010;13:1454–1456. doi: 10.1038/nn.2690. [DOI] [PubMed] [Google Scholar]
- Xu D., Joglekar A.P., Williams A.L., Hay J.C. Subunit structure of a mammalian ER/Golgi SNARE complex. J. Biol. Chem. 2000;275:39631–39639. doi: 10.1074/jbc.M007684200. [DOI] [PubMed] [Google Scholar]
- Xu Y., Wong S.H., Tang B.L., Subramaniam V.N., Zhang T., Hong W. A 29-kilodalton Golgi soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Vti1-rp2) implicated in protein trafficking in the secretory pathway. J. Biol. Chem. 1998;273:21783–21789. doi: 10.1074/jbc.273.34.21783. [DOI] [PubMed] [Google Scholar]
- Xu Y., Martin S., James D.E., Hong W. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Mol. Biol. Cell. 2002;13:3493–3507. doi: 10.1091/mbc.E02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.J., Nakanishi H., Liu S., McNew J.A., Neiman A.M. Binding interactions control SNARE specificity in vivo. J. Cell Biol. 2008;183:1089–1100. doi: 10.1083/jcb.200809178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong C.Q.Y., Tang B.L. Another longin SNARE for autophagosome-lysosome fusion-how does Ykt6 work? Autophagy. 2019;15:352–357. doi: 10.1080/15548627.2018.1532261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Matsuda K., Jia W.H., Chang J., Kweon S.S., Xiang Y.B., Shin A., Jee S.H., Kim D.H., Zhang B., Cai Q., Guo X., Long J., Wang N., Courtney R., Pan Z.Z., Wu C., Takahashi A., Shin M.H., Matsuo K., Matsuda F., Gao Y.T., Oh J.H., Kim S., Jung K.J., Ahn Y.O., Ren Z., Li H.L., Wu J., Shi J., Wen W., Yang G., Li B., Ji B.T., Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) Brenner H., Schoen R.E., Küry S., Colorectal Transdisciplinary (CORECT) Study. Gruber S.B., Schumacher F.R., Stenzel S.L., Colon Cancer Family Registry (CCFR) Casey G., Hopper J.L., Jenkins M.A., Kim H.R., Jeong J.Y., Park J.W., Tajima K., Cho S.H., Kubo M., Shu X.O., Lin D., Zeng Y.X., Zheng W. Identification of susceptibility loci and genes for colorectal cancer risk. Gastroenterology. 2016;150:1633–1645. doi: 10.1053/j.gastro.2016.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Tang M., Fang Y., Cui H., Chen S., Li J. Cumulative evidence for relationships between multiple variants in the VTI1A and TCF7L2 genes and cancer incidence. Int. J. Canc. 2018;142:498–513. doi: 10.1002/ijc.31074. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wong S.H., Tang B.L., Xu Y., Hong W. Morphological and functional association of Sec22b/ERS-24 with the pre-Golgi intermediate compartment. Mol. Biol. Cell. 1999;10:435–453. doi: 10.1091/mbc.10.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Huang X., Sun S., Li X., Wang H.W., Sui S.F. Cryo-EM structure of SNAP-SNARE assembly in 20S particle. Cell Res. 2015;25:551–560. doi: 10.1038/cr.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]