Abstract
Acetate-utilizing methanogens are responsible for approximately two-thirds of the one billion metric tons of methane produced annually in Earth’s anaerobic environments. Methanosarcina acetivorans has emerged as a model organism for the mechanistic understanding of aceticlastic methanogenesis and reverse methanogenesis applicable to understanding the methane and carbon cycles in nature. It has the largest genome in the Archaea, supporting a metabolic complexity that enables a remarkable ability for adapting to environmental opportunities and challenges. Biochemical investigations have revealed an aceticlastic pathway capable of fermentative and respiratory energy conservation that explains how Ms. acetivorans is able to grow and compete in the environment. The mechanism of respiratory energy conservation also plays a role in overcoming endothermic reactions that are key to reversing methanogenesis.
Keywords: global warming, archaea, methane, ecology, evolution, biochemistry, acetate, enzymology
Introduction
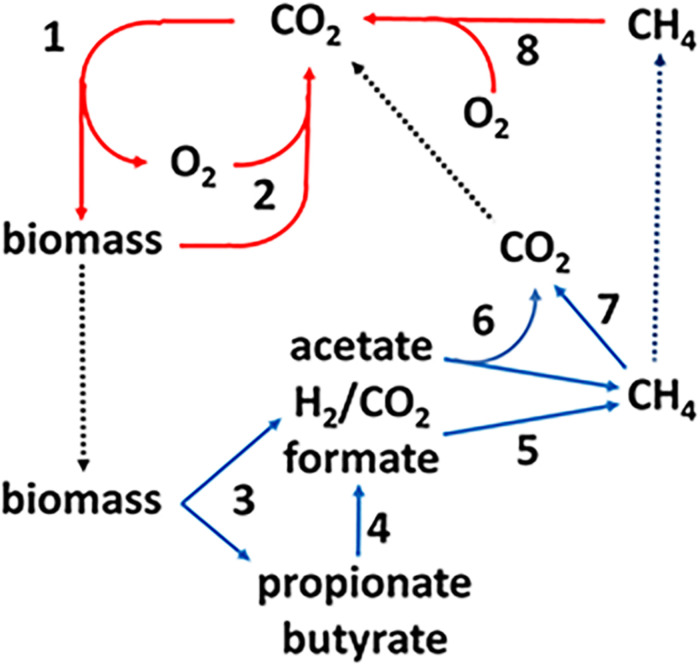
The production and consumption of CH4, the methane cycle, is an important link in the global carbon cycle (Figure 1). The complex biomass produced by photosynthetic plants and microbes is hydrolyzed and oxidized in aerobic habitats by O2-respiring microbes producing CO2 that re-enters the carbon cycle (steps 1, 2). A fraction of the biomass enters diverse anoxic environments where it is metabolized by microbial food chains comprized of fermentative, acetogenic, and methanogenic anaerobes (steps 3–6) producing an estimated one billion tons of methane (Thauer, 1998). The complex biomass is hydrolyzed and metabolized by fermentative anaerobes that produce primarily acetate plus other higher volatile fatty acids (VFA), H2 and formate. The VFA are oxidized to acetate and either formate or H2 by acetogens. Thus, acetate is the major metabolite in the food chain that acetotrophic methanogens convert to CH4 and CO2 (Mah et al., 1977). The balance of global methane production derives primarily from methanogens that oxidize H2 or formate and reduce CO2 to CH4. Methylotrophic methanogens produce minor, although significant, amounts of methane from methyl-containing compounds such as methanol and methylated amines. The CH4 produced in anaerobic environments is oxidized to CO2 by reversal of methanogenic pathways (step 7). The CO2 and residual CH4 diffuses into aerobic zones where O2 respiring methanotrophs oxidize CH4 to CO2 thereby closing the carbon cycle (step 8). However, not all the CH4 is oxidized and the remaining escapes to the upper atmosphere.
FIGURE 1.
The global carbon cycle. Solid lines indicate aerobic (red) and anaerobic (blue) steps in the cycle and dotted lines indicate transfer of material between aerobic and anaerobic environments. See text for explanation of numbered steps. Not shown are lesser amounts of methane produced from methylotrophic substrates such as methanol and methylamines. Reproduced (Yan and Ferry, 2018).
Methane is a greenhouse gas with a global warming potential approximately 20-fold greater than CO2 (Ramaswamy et al., 2001). The CH4 cycle (production and oxidation) plays an important role in controlling Earth’s climate (Valentine, 2002; Rhee et al., 2009). Indeed, Earths greatest mass extinction is attributed in part to the evolution of acetotrophic methanogens that produced a methanogenic burst in the end-Permian carbon cycle that contributed to a sharp increase in global warming (Rothman et al., 2014). Anthropogenic CH4 emissions to the atmosphere have increased sharply since 2007 raising awareness of the potential consequences (Nisbet et al., 2019). A mechanistic biochemical understanding of the CH4 cycle is paramount to a deeper understanding necessary to predict and control CH4 emissions. Although the understanding of aerobic methanotrophic microbes is well developed, mechanistic understanding of anaerobic CH4 oxidation (AOM) is in the early stages.
This review features relevant and recent mechanistic understanding of the aceticlastic pathway and reverse methanogenesis for which Methanosarcina acetivorans has emerged as a model.
Aceticlastic Pathways
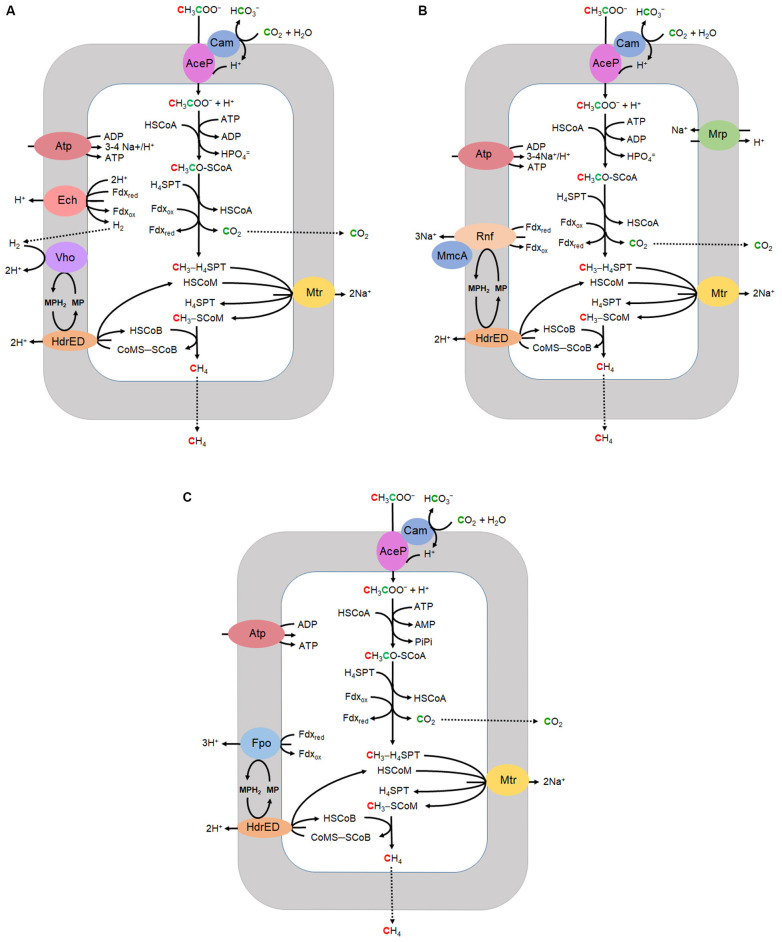
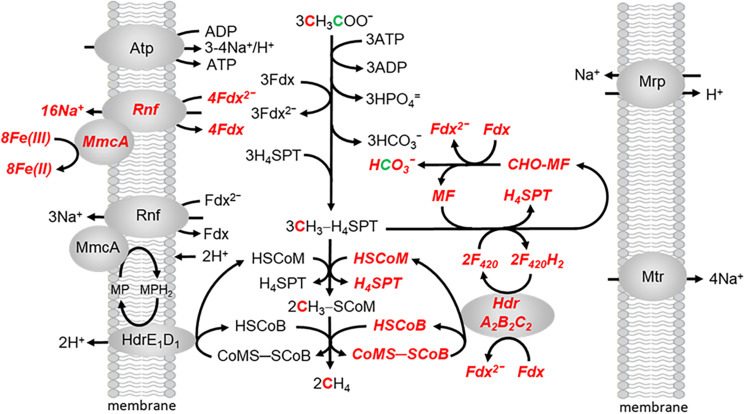
Most CH4 produced in Earth’s diverse anaerobic environments derives from acetate although only two genera, Methanosarcina and Methanothrix (formerly Methanosaeta) are known to grow with acetate and produce CH4. Acetotrophic methanogens utilize three variations of the aceticlastic pathway of which two are typical of the genus Methanosarcina (Ms.) while the third is characteristic of the genus Methanothrix (Mt.) (Figure 2). All three have in common the transport of acetate, activation to acetyl-CoA, decarbonylation of acetyl-CoA, and one-carbon reactions transforming the methyl group to CH4. The variations diverge in the mechanisms of electron transport and energy conservation. Most investigations have centered on Methanosarcina for which there are two divergent electron transport pathways, H2 dependent and H2 independent. The H2 dependent pathway (Figure 2A) is well established for Methanosarcina barkeri and Methanosarcina mazei (Welte and Deppenmeier, 2014). However, the pathway of several acetotrophic Methanosarcina species is independent of H2 and instead contains the Rnf complex for which Ms. acetivorans has emerged as the model (Figure 2B). The Rnf complex is also encoded in all sequenced genomes of diverse methylotrophic genera that includes Methanosarcina1. Isolated from marine sediment, Ms. acetivorans has the largest genome among all methanogens and amenable to robust genetic manipulation (Sowers et al., 1984a; Galagan et al., 2002; Nayak and Metcalf, 2017). The Rnf-dependent aceticlastic pathway of Ms. acetivorans (Figure 2B) is supported by transcriptomic, proteomic and modeling investigations (Li et al., 2005a, b; Li et al., 2007; Satish Kumar et al., 2011; Benedict et al., 2012; Peterson et al., 2014).
FIGURE 2.
Aceticlastic pathways. (A) H2 dependent Methanosarcina. (B) H2 independent Methanosarcina. (C) Methanothrix. CoA, coenzyme A; H4SPT, tetrahydrosarcinapterin; Fdx, ferredoxin; HSCoM, coenzyme M; HSCoB, coenzyme B; MP, methanophenazine; Cam, gamma carbonic anhydrase; AceP, acetate permease; Mrp, multisubunit sodium/proton antiporter; Atp, ATP synthase; Rnf, homolog of rhodobacter nitrogen fixation complex; MmcA, multiheme c-type cytochrome; HdrED, membrane bound heterodisulfide reductase; Mtr, CH3-H4SPT:HSCoM methyltransferase; Ech, energy-converting ferredoxin-dependent hydrogenase; Vho, F420-nonreactive membrane-bound hydrogenase; Fpo, F420H2 dehydrogenase multi-subunit complex. Adapted (Smith and Ingram-Smith, 2007).
Acetate Transport and Activation
AceP from Ms. acetivorans was shown to transport acetate by a proton symport mechanism (Ribas et al., 2018). A homolog of AceP was shown to be required for acetate transport of acetate in Ms. mazei, and an AceP homolog is encoded in the genome of Methanothrix thermophila (Smith and Ingram-Smith, 2007; Welte et al., 2014). The transported acetate is converted to acetyl-CoA by acetate kinase (Ack) and phosphotransacetylase (Pta) in Methanosarcina, and by the AMP-forming acetyl-CoA synthetase (Acs) in Methanothrix (Berger et al., 2012). It was proposed that Ack and Pta were acquired by horizontal gene transfer from the genus Clostridium within the last 475 million years coinciding with evolution of aceticlastic pathways. This event resulted in a significant net increase of CH4 leading to climate change in agreement with that proposed for the end-Permian mass extinction (Fournier and Gogarten, 2008; Rothman et al., 2014).
The catalytic mechanism for Ack from Methanosarcina thermophila proceeds by nucleophilic attack of the carboxyl group of acetate on the γ-phosphate of ATP with direct in-line transfer to acetate producing acetyl phosphate (Buss et al., 2001; Miles et al., 2002; Ferry, 2011). The mechanism for Pta, also from Ms. thermophila, involves base-catalyzed abstraction of the thiol proton of HS-CoA followed by nucleophilic attack of the thiolate anion (–S-CoA) on the carbonyl carbon of acetyl phosphate forming acetyl-CoA (Iyer et al., 2004; Lawrence et al., 2006; Ferry, 2011). The crystal structure and biochemical characterization of Acs from Ms. acetivorans revealed the preference for medium chain substrates that excludes acetate, a result which indicates Acs functions other than activating acetate to acetyl-CoA (Ingram-Smith and Smith, 2007; Shah et al., 2009; Meng et al., 2010). The Acs of Methanothrix has a greater affinity for acetate than Ack of Ms. acetivorans which explains the dominance of Methanothrix in environments where acetate is in concentrations <0.1 mM (Berger et al., 2012). The acetyl-CoA is decarbonylated by the acetyl-CoA decarbonylase/synthase (ACDS) yielding a methyl group and CO. The methyl group is transferred to tetrahydrosarcinapterin (H4SPT) yielding CH3-H4SPT and CO is oxidized to CO2 with transfer of electrons to either ferredoxin (Fdx) or a novel flavodoxin (FldA) characterized from Ms. acetivorans (Prakash et al., 2019b).
The ACDS is predicted to be a component of the last universal common ancestor (LUCA) (Adam et al., 2018). Although of ancient origin and of central importance in the aceticlastic pathway, an atomic resolution structure of the intact ACDS complex from any methanogen is not reported. The enzymes from Methanosarcina and Methanothrix are known to have five subunits (αβγδε) based on the purified complexes and genomic analyses (Terlesky et al., 1986; Grahame and Demoll, 1996; Smith and Ingram-Smith, 2007). The β subunit catalyzes decarbonylation of acetyl-CoA while the αε subunits catalyze CO oxidation and the γδ subunits transfer the methyl group to H4SPT producing CH3-H4SPT (Murakami and Ragsdale, 2000). The crystal structure of the αε component of Ms. barkeri identified the active site in the α subunit comprised of a pseudocubane Ni-Fe3S4 cluster bridged to an exogenous iron atom (Gong et al., 2008). A mechanism was proposed wherein the CO bound to Ni, and the OH– bound to exogenous iron, H are coupled to form CO2. A role for the ε subunit was proposed in which bound FAD directs electrons from the α subunit to Fdx. This proposal fits with the possibility that FldA accepts electrons from the ε subunit of the ACDS from Ms. acetivorans at the proposed FAD site. Spectroscopic studies of the β subunit from Ms. thermophila indicate an active site Fe4S4 cluster bridged to a binuclear Ni–Ni site in analogy to the homolog from an acetogen of the domain Bacteria that synthesizes acetyl-CoA (Gu et al., 2003; Funk et al., 2004; Ragsdale, 2007). Kinetic and EPR spectroscopy results indicate that alterations in the Ni coordination environment of the active site cluster promote C–C bond cleavage dependent on conformational changes (Gencic and Grahame, 2008). The γδ component transfers the methyl group of acetyl-CoA to H4SPT involving a corrinoid coenzyme, although it is unknown which subunit interacts with H4SPT and a crystal structure is not available (Grahame, 1993).
Acetate-grown Ms. acetivorans up regulates a γ class carbonic anhydrase (Cam) for which the crystal structure and biochemical characterization of the homolog from Ms. thermophila revealed the catalytic mechanism involving an active-site iron (Kisker et al., 1996; Iverson et al., 2000; Macauley et al., 2009; Zimmerman et al., 2013). Although homologs are present in acetate grown Methanosarcina and Methanothrix, the physiological function is not established. A plausible function involves diffusion of cytoplasmic CO2 to the outer aspect of the membrane where AceP is located in a complex with Cam that hydrates CO2 to /H+ which supplies a local concentration of protons for symport of acetate by AceP (Figure 2). In this way, the proton gradient that drives ATP synthesis is not collapsed. The putative function for Cam is analogous to that reported for the α class carbonic anhydrase that supplies a proton for symport of lactate in mammalian cells (Peetz et al., 2014).
One-Carbon Reactions
The methyl group of CH3-H4SPT is transferred to coenzyme M (HS-CoM) coupled to sodium extrusion by a membrane bound methyltransferase (MtrABCDEFGH). The CH3-SCoM is reductively demethylated to CH4 by the methyl coenzyme M reductase (McrABG) requiring coenzyme B (HSCoB) as the reductant. Post-translational modified residues N1-methylhistidine (3-methylhistidine), 5-(S)-methylarginine, thioglycine, and S-methylcysteine are present in the active-sites of the catalytic McrA subunits from phylogenetically and metabolically diverse methanogenic and methanotrophic archaea (Grabarse et al., 2000; Kahnt et al., 2007). Mcr from Ms. acetivorans has emerged as a model for investigations of the modified residues. A unique radical SAM methyltransferase was shown required for methylation of the active-site arginine and concluded important for stability under imposed oxidative and heat stress (Deobald et al., 2018; Radle et al., 2019). Deletion of a homolog essential for arginine methylation in the obligate CO2-reducing methanogen Methanococcus maripaludis resulted in a 40–60% loss in the rate of methanogenesis consistent with partial loss of Mcr activity (Lyu et al., 2020). Deletion of two genes essential for thioglycine synthesis in McrA of Ms. acetivorans produced mutants severely impaired in the rate of growth with acetate and when exposed to thermal and oxidative stress, results supporting a role for thioglycine in stabilizing the McrA active-site although not essential. Combinatorial deletion of genes responsible for incorporation of 5-(S)-methylarginine, thioglycine and S-methylcysteine generated Ms. acetivorans mutants with phenotypes consistent with altered thermal stability of McrA (Nayak et al., 2020). The studies suggest that residue modifications of Mcr function in important ways although not essential for catalysis. The CoMS-SCoB product of Mcr is reduced by a membrane bound electron transport chain ending with heterodisulfide reductase (HdrE1D1) that regenerates sulfhydryl forms of the coenzymes.
Electron Transport and Energy Conservation
The electron transport pathways of all acetotrophic methanogens begin with the oxidation of Fdx and end with reduction of CoMS-SCoB by HdrE1D1 (Figure 2). As heterodisulfide is the terminal electron acceptor and generated internally, the process fits the definition of fermentative electron transport and energy conservation as opposed to respiration that requires an externally supplied electron acceptor. The aceticlastic pathways diverge in the mechanisms of membrane-bound electron transport that generates ion gradients driving ATP synthesis for growth (Figure 2). The H2 dependent pathway (Figure 2A) has been investigated in Ms. barkeri and Ms. mazei for which the understanding is well developed (Welte and Deppenmeier, 2014). Reduced Fdx donates electrons to Ech hydrogenase that pumps protons and also reduces protons to H2 that diffuses across the membrane where it is reoxidized at the outer aspect by the Vho hydrogenase, further contributing to the proton gradient (Welte and Deppenmeier, 2014; Kulkarni et al., 2018). Electrons from the oxidation of H2 by Vho are transferred to HdrE1D1 by the quinone-like electron carrier methanophenazine (MP) accompanied by the vectoral translocation of protons that supplements the proton gradient. The proton gradient, together with the Mtr imposed Na+ gradient, drives ATP synthesis.
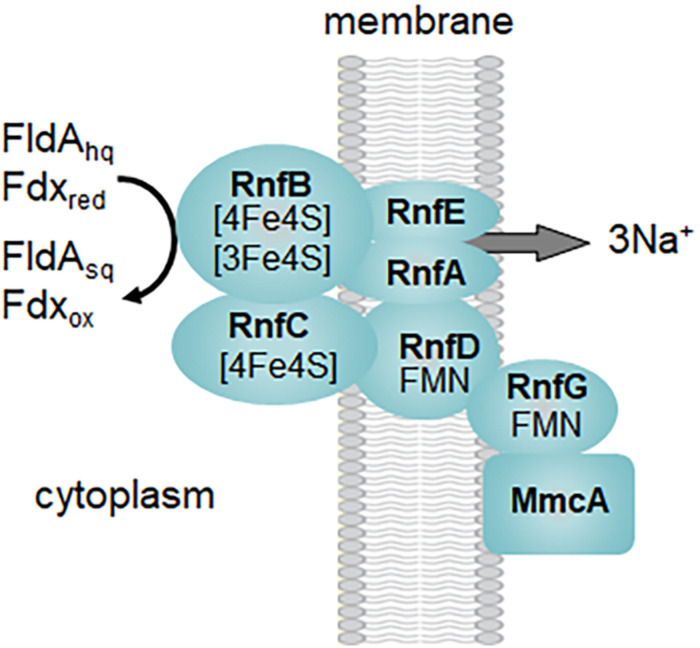
Several acetotrophic Methanosarcina lack Ech and Vho hydrogenases and are H2 independent (Zhilina, 1978; Ollivier et al., 1984; Sowers et al., 1984a; Zinder et al., 1985; Elberson and Sowers, 1997; Von Klein et al., 2002; Shimizu et al., 2011; Ganzert et al., 2014). Ms. acetivorans is typical of H2 independent Methanosarcina that instead utilize the membrane bound RnfCDGEAB complex to oxidize Fdx or FldA (Figure 2B; Li et al., 2006; Wang et al., 2011; Schlegel et al., 2012b; Prakash et al., 2019b). FldA accepts electrons from ACDS and is proposed to replace Fdx when growing in iron-limited environments (Prakash et al., 2019b). Fdx is an electron donor to the RnfB subunit of the Rnf complex (Suharti et al., 2014). It was further shown that the heterologously produced flavin-containing RnfG subunit is located on the outer aspect of the Escherichia coli membrane leading to the proposed model shown in Figure 3. Although MmcA is abundant in acetate-grown cells, its role in acetotrophic growth is questioned with the finding that a ΔmmcA mutant grows with acetate (Holmes et al., 2019). In contrast, the mutant is incapable of methanol-dependent respiratory growth with anthraquinone-2,6-disulfonate (AQDS), which suggests a role for MmcA in mediating electron transfer to external electron acceptors which fits the definition of respiratory electron transport and energy conservation. Rnf transfers electrons to MP for reduction of CoMS-SCoB by HdrE1D1 and pumps Na+ that thermodynamic considerations predict 3-4Na+/2 electrons (Schlegel et al., 2012b; Welte and Deppenmeier, 2014). Thus, electron transport generates H+ and Na+ gradients that, together with the Mtr-imposed Na+ gradient, drives ATP synthesis by the ATP synthase dependent on both H+ and Na+ (Schlegel et al., 2012a). It is proposed that the multi subunit Na+/H+ antiporter MrpABCDEFG adjusts the Na+/H+ ratio optimal for ATP synthesis (Jasso-Chavez et al., 2013, 2017). Although electron transport is remarkably different in Ms. barkeri and Ms. acetivorans, they have similar growth rates and yields in the absence of an exogenous electron acceptor which indicates that each conserve the same amount of energy (Sowers et al., 1984b). This result is consistent with equivalent H+ and Na+ gradients generated by electron transport and Mtr.
FIGURE 3.
Proposed organization of the Rnf complex from Methanosarcina acetivorans. Symbols: FldAsq, semiquinone of flavodoxin A; FldAhq, hydroquinone of flavodoxin A; Fdxox, oxidized ferredoxin; Fdxred, reduced ferredoxin. Modified (Suharti et al., 2014).
Methanosarcina acetivorans, Ms. barkeri and Ms. mazei each encode HdrE1D1, HdrA1B1C1, HdrD2, HdrA2, and HdrC2B2. HdrE1D1 was shown to function in acetotrophic growth of Ms. acetivorans whereas HdrA1B1C1 is apparently specific for methylotrophic growth (Buan and Metcalf, 2010; Catlett et al., 2015). It is proposed that reduced Fdx, generated in the oxidative branch, donates electrons to HdrA1B1C1 that then reduces F420 at the expense of CoMS-SCoB reduction in an electron bifurcation reaction (Buan and Metcalf, 2010). With this mechanism, electrons from Fdx are directed to the Fpo complex which results in additional energy conservation. A mechanism is proposed for the catalytic subunit HdrD that is distinct from the catalytic HdrB of the electron bifurcating HdrABC of obligate CO2-reducing methanogens. Based on the crystal structure alone, a mechanism is proposed for HdrB involving two novel non-cubane 4Fe4S clusters (Wagner et al., 2017). This mechanism contrasts with that proposed for HdrD involving one conventional 4Fe4S cluster although based primarily on spectroscopic analyses (Walters and Johnson, 2004). However, both mechanisms propose that on reduction of CoMS-SCoB the sulfur atoms of the HSCoM and HSCoB are bound to iron in a five-coordinate manner. The electron pair for reduction of CoMS-SCoB derives from a membrane-bound electron transport chain that accepts electrons from either reduced Fdx or a flavodoxin (FldA) generated by ACDS (Figure 2B). The HdrE1 subunit contains a b-type cytochrome that accepts electrons from MP for transfer to HdrD1 (Welte and Deppenmeier, 2014).
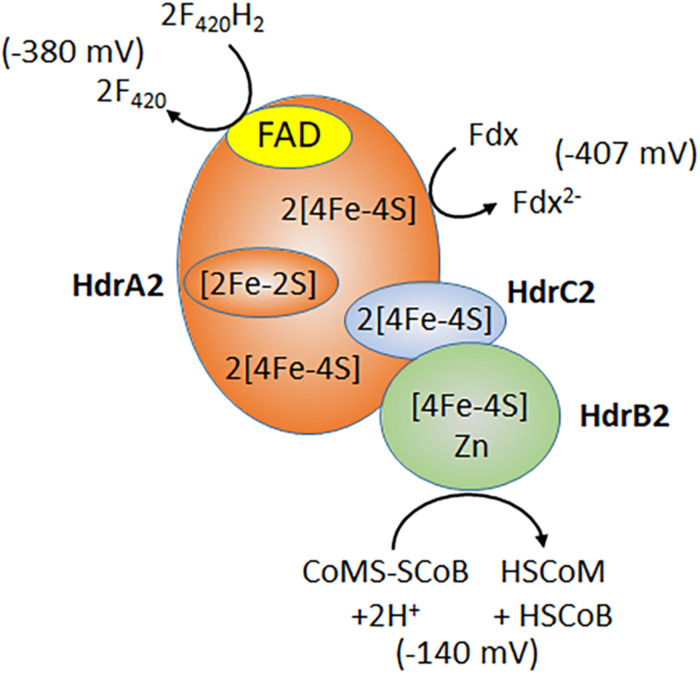
Subunits of the recently characterized electron bifurcating HdrA2B2C2 are up regulated in acetate-grown Ms. acetivorans consistent with a role in acetotrophic growth (Li et al., 2007; Buan and Metcalf, 2010; Rohlin and Gunsalus, 2010; Yan et al., 2017). Indeed, acetotrophic growth is impaired in a strain of Ms. acetivorans unable to synthesize HdrA2B2C2 (Buan and Metcalf, 2010). Expression of the individual HdrA2, HdrB2, and HdrB2C2 subunits in E. coli, and biochemical characterization of the reconstituted active HdrA2B2C2 complex, revealed a role for HdrA2 in the oxidation of reduced coenzyme F420 (F420H2) and FAD-dependent bifurcation of electrons that are transferred to Fdx and HdrC2 (Figure 4; Yan et al., 2017). The HdrC2 mediates electron transfer to HdrB2 for reduction of CoMS-SCoB. The thermodynamically unfavorable reduction of Fdx is driven by the more favorable reduction of CoMS-SCoB. Although up regulated in acetate grown cells, the role for HdrA2B2C2 in acetotrophic growth has not been established experimentally. It is postulated that the Rnf complex reduces coenzyme F420 that is oxidized by HdrA2B2C2 thereby recycling electrons to Fdx for oxidation by Rnf and an additional Na+ translocated, improving the thermodynamic efficiency (Buckel and Thauer, 2018). An unusual flavodoxin (FldA) can replace Fdx as electron donor to Rnf and acceptor for HdrA2B2C2 (Prakash et al., 2019b). FldA is a potential advantage in periods of oxidative stress that damage the iron-sulfur clusters of Fdx, or when iron is limiting in the environment (Prakash et al., 2019b).
FIGURE 4.
Electron bifurcation by HdrA2B2C2. F420, coenzyme F420; Fdx, ferredoxin; HSCoM, coenzyme M; HSCoB, coenzyme B. Redox potentials for F420 and CoMS-SCoB are published values (Thauer et al., 2008). The ferredoxin redox potential is determined for the 2(4Fe4S) ferredoxin from acetate grown Ms. thermophila (Clements et al., 1994). Modified (Yan et al., 2017).
Considerably less is known of electron transport and energy conservation in Methanothrix. The genomes are void of genes encoding Ech hydrogenase or Rnf and, instead, encode F420H2 dehydrogenase (FpoABCDHIJKLMNO) although lacking the gene encoding FpoF that in Methanosarcina is the input module oxidizing F420H2 (Zhu et al., 2012). Thus, it is postulated that Fpo accepts electrons directly from Fdx with MP-mediated reduction of HdrED that is encoded in Methanothrix genomes (Zhu et al., 2012). Thermodynamic considerations predict 3H+ translocated by Fpo for a total of seven ions contributing to the gradient driving ATP synthesis (Welte and Deppenmeier, 2014). Although equivalent to gradients generated by H2 dependent and H2 independent Methanosarcina (Figure 2), Methanothrix requires two ATP for activation of acetate compared to one for Methanosarcina which predicts lower growth yields. However, this thermodynamic disadvantage is at least partially compensated by the ability of Methanothrix to metabolize acetate at lower concentrations compared to Methanosarcina (Jetten et al., 1992).
Respiratory Energy Conservation
Methanosarcina acetivorans is capable of Fe(III)-dependent respiratory growth with acetate, a finding previously undocumented for acetotrophic methanogens (Prakash et al., 2019a). Growth and acetate consumption nearly doubles in the presence of ferrihydrite [Fe(OH)3], the metal oxide form of Fe(III) that is common in the environment. Ferric iron is stoichiometrically reduced to ferrous iron. The ATP/ADP ratio also doubles indicating a higher energetic state consistent with increased growth. However, CH4 is also produced indicating both fermentative and respiratory electron transport and energy conservation. The revised, ecologically relevant, pathway is shown in Figure 5. All one-carbon transformations leading to CH4 are the same as in Figure 2. Two Na+ are translocated for each Fe(III) reduced to Fe(II) in respiratory electron transport (Yan et al., 2018). Although further research is necessary, the present results indicate that productive Na+ translocation by the Rnf complex is dependent on electron transfer to MmcA that reduces an exogenous electron acceptor which fits the definition of respiratory electron transport. Respiratory electron transport is dependent on oxidation of the methyl group from CH3-H4SPT by reversal of reactions in the CO-dependent pathway of CO2 reduction to CH4 and acetate in Ms. acetivorans which generates reduced coenzyme F420 (F420H2) and additional reduced Fdx to enter the pool for both respiratory and fermentative electron transport (Lessner et al., 2006). The F420H2 dehydrogenase, essential for methylotrophic growth, is down regulated in acetate-grown cells leading to the proposal that oxidation of F420H2 is dependent on the electron bifurcating HdrA2B2C2 (Yan et al., 2017). As FldA can replace Fdx as electron acceptor for HdrA2B2C2, and donor to Rnf, either are available for initiating fermentative and respiratory electron transport (Prakash et al., 2019b). The combination of fermentative and respiratory electron transport generates both H+ and Na+ gradients that drive ATP synthesis by the ATP synthase dependent on both gradients (Schlegel et al., 2012a). It is proposed that the multi subunit Na+/H+ antiporter Mrp adjusts the Na+/H+ ratio optimal for ATP synthesis (Jasso-Chavez et al., 2013, 2017).
FIGURE 5.
The aceticlastic pathway proposed for growth of Ms. acetivorans in the presence of ferrihydrite. Respiratory electron transport is shown in bolded italicized red font. Modified (Prakash et al., 2019a).
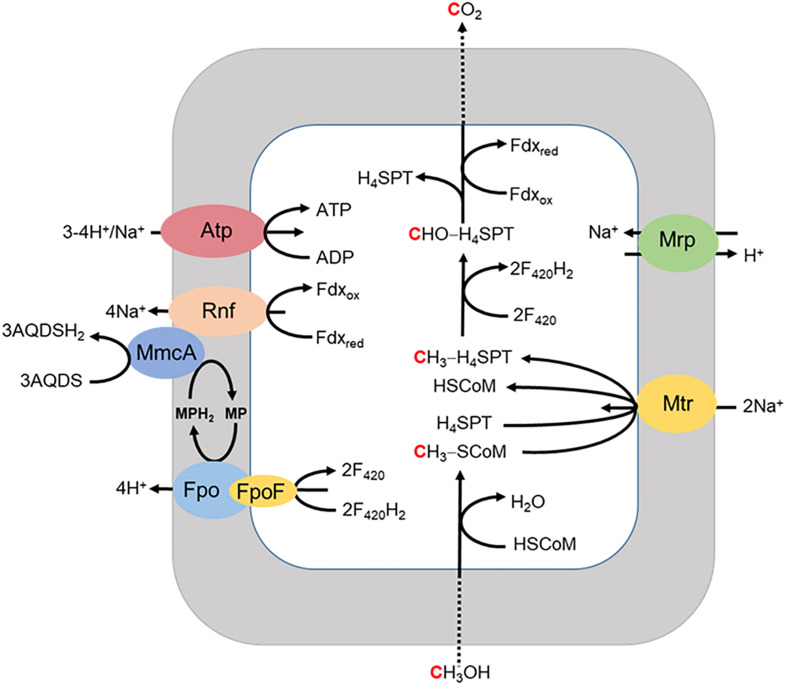
A respiratory pathway is also proposed for Ms. acetivorans grown with methanol when methanogenesis is inhibited by 2-bromoethanesulfonate (Figure 6; Holmes et al., 2019). The methyl group of methanol is oxidized to CO2 with reduction of Fdx and F420 for which the latter is reoxidized by the F420H2 dehydrogenase complex (Fpo and FpoF) that is up regulated in methanol grown cells. Fpo transfers the electrons to MP accompanied by the translocation of H+ which contributes to the ion gradient that drives ATP synthesis. Reduced MP transfers electrons to MmcA that reduces AQDS as the final electron acceptor. The reduced Fdx donates electrons to Rnf that also transfers electrons to MmcA with translocation of Na+ analogous to that proposed in the revised aceticlastic pathway (Figure 4). The imposed inhibition of methanogenesis precludes extrapolation to the environment although reinforces the discovery that Ms. acetivorans is capable of respiratory growth.
FIGURE 6.
Proposed model for extracellular electron transport to AQDS by Ms. acetivorans grown with methanol in the presence of the methanogenesis inhibitor 2-bromoethanesulfonic acid (BES). FpoF, input module to Fpo. Adapted (Holmes et al., 2019).
Ecology and Evolution
The revised aceticlastic pathway of Ms. acetivorans has important ecological and evolutionary implications. Without respiration, the amount of energy
| (1) |
| (2) |
available by methanogenesis alone, with equimolar reactants and products (Eq. 1), is barely enough to synthesize one ATP (Eq. 2). It is possible that growth by methanogenesis alone is only achievable in the laboratory with an abundant supply of acetate at optimal temperature, pH, and supply of nutrients whereas growth in the competitive and dynamic environment is dependent on additional energy gained by respiration. In environments where Fe(III) is limiting, energy conservation by methanogenic fermentation could afford an advantage over acetotrophic competitors that conserve energy only by respiration. Ms. acetivorans, and other Methanosarcina which are H2 independent, may have an advantage over H2 dependent Methanosarcina that are without multi-heme c-type cytochromes and incapable of respiratory growth.
Reverse Methanogenesis
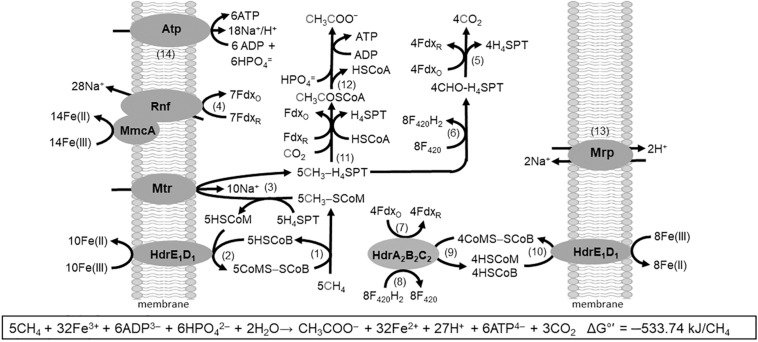
The discovery of respiratory energy conservation by Ms. acetivorans has impacted understanding of reverse methanogenesis, the CH4 cycle, and the iron cycle in nature. Previous models of the anaerobic oxidation of CH4 (AOM) involved anaerobic methanotrophic archaea (ANME) that oxidize CH4 by reversal of the CO2-reduction pathway of methanogens. The oxidation required a symbiosis with species utilizing reductant produced by ANME to make the overall reaction thermodynamically favorable. However, it was found that AQDS decouples CH4 oxidation from sulfate reduction which presented the possibility of independent respiratory methanotrophic growth by ANME. Ms. acetivorans is capable of trace CH4 oxidation during growth with methanogenic substrates (Moran et al., 2005, 2007). Furthermore, Ms. acetivorans is capable of Fe(III)-dependent AOM in the absence of methanogenic substrates when engineered with the Mcr gene derived from ANME-1 sediment (Soo et al., 2016). Biochemical investigations support a proposed AOM pathway for Ms. acetivorans anchored by Fe(III)-dependent mechanisms for energy conservation that drive endergonic reactions essential for methanotrophic growth (Figure 7) (Yan et al., 2018).
FIGURE 7.
Pathway proposed for Fe(III)-dependent CH4 oxidation, electron transport, and conservation of energy by Ms. acetivorans. See text for explanation of numbered steps. Not shown is AQDS-mediated reduction of Fe(III) at MmcA and HdrE1D1. Modified (Yan et al., 2018).
The AOM pathway postulates that CH4 is oxidized by Mcr producing CH3-SCoM (Rxn. 1) in analogy to that shown for the Mcr of obligate CO2-reducing methanogens (Scheller et al., 2010). The exergonic Fe(III)-dependent oxidation of HSCoM and HSCoB by HdrE1D1 (Rxn. 2) drives the endergonic oxidation of CH4 (Yan et al., 2018). The endergonic methyl transfer from CH3-SCoM to H4MPT by Mtr (Rxn. 3) is driven with the Na+ gradient generated by the Rnf complex (Rxn. 4) with a stoichiometry of 2Na+ translocated per electron transferred from Fdx to Fe(III) (Yan et al., 2018). Electrons are transferred from Rnf to MmcA that reduces Fe(III). Reduced Fdx is a product of the oxidation of the methyl group of CH3-H4SPT to CO2 (Rxn. 5) as is also F420H2 (Rxn. 6) that is oxidized by HdrA2B2C2 (Rxn. 7) with reduction of Fdx (Rxn. 8) and CoMS-SCoB (Rxn. 9). The CoMS-SCoB is regenerated (Rxn. 10) as for the Fe(III)-dependent oxidation of HSCoM and HSCoB by HdrE1D1 (Rxn. 2). Reactions oxidizing the methyl group of CH3-H4MPT to CO2 (Rxn. 5 and 6) are the reverse of reactions in the CO-dependent pathway of CO2 reduction to CH4 and acetate in Ms. acetivorans (Lessner et al., 2006). Reactions leading from CH3-H4MPT to acetate (Rxn. 11 and 12) are the reverse of reactions in the aceticlastic pathways (Figures 2, 4). The Na+/H+ antiporter Mrp is postulated to adjust the Na+/H+ ratio optimal for ATP synthesis by the Atp synthase dependent on both Na+ and H+ gradients (Rxn. 13 and 14) (Schlegel et al., 2012a; Jasso-Chavez et al., 2013, 2017). Not shown in Figure 7 is the requirement for AQDS to mediate electron transfer from HdrE1D1 to Fe(III) and MmcA to Fe(III). AQDS is an analog of humic substances that are proposed to replace AQDS in nature (Holmes et al., 2019).
The pathway resembles the AOM pathway predicted for an uncultured ANME-2a based on metagenomic analyses (Wang et al., 2014). However, it should be cautioned that the biochemistry of ANME is largely unknown and differences with methanogenic pathways are anticipated (Timmers et al., 2017). Nonetheless, the biochemical-based AOM pathway provides a working model for mechanistic understanding of the growing literature describing respiratory AOM by individual ANME using a variety of electron acceptors including Fe(III) (Raghoebarsing et al., 2006; Beal et al., 2009; Haroon et al., 2013; Ettwig et al., 2016; Cai et al., 2018; He et al., 2018; Liang et al., 2019; Luo et al., 2019; Aromokeye et al., 2020; Leu et al., 2020).
Ecology and Evolution
The realization of Fe(III)-dependent AOM has implications for understanding the CH4 and iron cycles, both past and present. It is postulated that symbiotic associations of ANME and sulfate-reducing species evolved from methanogenic species that first acquired the capacity to conserve energy by oxidizing CH4 and reducing metals (Scheller et al., 2016). Moreover, it is postulated that Fe(III)-dependent AOM was largely responsible for oxidizing all the CH4 produced on early Earth prior to the appearance of oxygen (Beal et al., 2009). It is further hypothesized that if only a small fraction of current global Mn(IV) and Fe(III) influx is used for AOM, it has the potential to consume a large amount of CH4 (Beal et al., 2009). Ms. acetivorans was isolated from off shore marine sediments near locations with CH4 seeps where single cells and aggregates of ANME are present and could play a role in non-symbiotic Fe(III)-dependent AOM (Sowers et al., 1984a; Orphan et al., 2002).
Conclusion
Acetotrophic methanogens utilize three aceticlastic pathways separated by mechanisms of electron transport and energy conservation that are well developed for the genus Methanosarcina and less so for Methanothrix. Ms. acetivorans is a model for H2 independent mechanisms whereas Ms. mazei and Ms. barkeri are models for the H2 dependent mechanisms. Recent developments establish respiratory energy conservation for Ms. acetivorans dependent on a multi-heme c-type cytochrome explaining growth in the environment and further separating H2 independent and H2 dependent Methanosarcina. However, gaps remain in our understanding of aceticlastic catabolism in Methanosarcina which include the mechanism of HdrED, a complete structure and mechanism for ACDS, and electron transport from multi-heme c-type cytochrome to exogenous electron acceptors.
Author Contributions
JF wrote the review.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. Research in the authors laboratory was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy through grant DE-FG02-95ER20198 and the Penn State Person Endowment.
References
- Adam P. S., Borrel G., Gribaldo S. (2018). Evolutionary history of carbon monoxide dehydrogenase/acetyl-CoA synthase, one of the oldest enzymatic complexes. Proc. Natl. Acad. Sci. U.S.A. 115 E1166–E1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aromokeye D. A., Kulkarni A. C., Elvert M., Wegener G., Henkel S., Coffinet S., et al. (2020). Rates and microbial players of iron-driven anaerobic oxidation of methane in methanic marine sediments. Front. Microbiol. 10:3041. 10.3389/fmicb.2019.03041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal E. J., House C. H., Orphan V. J. (2009). Manganese- and iron-dependent marine methane oxidation. Science 325 184–187. 10.1126/science.1169984 [DOI] [PubMed] [Google Scholar]
- Benedict M. N., Gonnerman M. C., Metcalf W. W., Price N. D. (2012). Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J. Bacteriol. 194 855–865. 10.1128/jb.06040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Welte C., Deppenmeier U. (2012). Acetate activation in Methanosaeta thermophila: characterization of the key enzymes pyrophosphatase and acetyl-CoA synthetase. Archaea 2012:315153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buan N. R., Metcalf W. W. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75 843–853. 10.1111/j.1365-2958.2009.06990.x [DOI] [PubMed] [Google Scholar]
- Buckel W., Thauer R. K. (2018). Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118 3862–3886. 10.1021/acs.chemrev.7b00707 [DOI] [PubMed] [Google Scholar]
- Buss K. A., Cooper D. R., Ingram-Smith C., Ferry J. G., Sanders D. A., Hasson M. S. (2001). Urkinase: structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 183 680–686. 10.1128/jb.183.2.680-686.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Leu A. O., Xie G. J., Guo J., Feng Y., Zhao J. X., et al. (2018). A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 12 1929–1939. 10.1038/s41396-018-0109-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett J., Ortiz A. M., Buan N. (2015). Rerouting cellular electron flux to increase the rate of biological methane production. Appl. Environ. Microbiol. 81 6528–6537. 10.1128/aem.01162-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements A. P., Kilpatrick L., Lu W.-P., Ragsdale S. W., Ferry J. G. (1994). Characterization of the iron-sulfur clusters in ferredoxin from acetate-grown Methanosarcina thermophila. J. Bacteriol. 176 2689–2693. 10.1128/jb.176.9.2689-2693.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deobald D., Adrian L., Schone C., Rother M., Layer G. (2018). Identification of a unique Radical SAM methyltransferase required for the sp(3)-C-methylation of an arginine residue of methyl-coenzyme M reductase. Sci. Rep. 8:7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elberson M. A., Sowers K. R. (1997). Isolation of an aceticlastic strain of Methanosarcina siciliae from marine canyon sediments and emendation of the species description for Methanosarcina siciliae. Int. J. Syst. Bacteriol. 47 1258–1261. 10.1099/00207713-47-4-1258 [DOI] [PubMed] [Google Scholar]
- Ettwig K. F., Zhu B., Speth D., Keltjens J. T., Jetten M. S., Kartal B. (2016). Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc. Natl. Acad. Sci. U.S.A. 113 12792–12796. 10.1073/pnas.1609534113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferry J. G. (2011). Acetate kinase and phosphotransacetylase. Methods Enzymol. 494 219–231. 10.1016/b978-0-12-385112-3.00011-1 [DOI] [PubMed] [Google Scholar]
- Fournier G. P., Gogarten J. P. (2008). Evolution of acetoclastic methanogenesis in Methanosarcina via horizontal gene transfer from cellulolytic Clostridia. J. Bacteriol. 190 1124–1127. 10.1128/jb.01382-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk T., Gu W. W., Friedrich S., Wang H. X., Gencic S., Grahame D. A., et al. (2004). Chemically distinct Ni sites in the A-cluster in subunit beta of the Acetyl-CoA decarbonylase/synthase complex from Methanosarcina thermophila: Ni L-edge absorption and x-ray magnetic circular dichroism analyses. J. Am. Chem. Soc. 126 88–95. 10.1021/ja0366033 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Nusbaum C., Roy A., Endrizzi M. G., Macdonald P., Fitzhugh W., et al. (2002). The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12 532–542. 10.1101/gr.223902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganzert L., Schirmack J., Alawi M., Mangelsdorf K., Sand W., Hillebrand-Voiculescu A., et al. (2014). Methanosarcina spelaei sp. nov., a methanogenic archaeon isolated from a floating biofilm of a subsurface sulphurous lake. Int. J. Syst. Evol. Microbiol. 64 3478–3484. 10.1099/ijs.0.064956-0 [DOI] [PubMed] [Google Scholar]
- Gencic S., Grahame D. A. (2008). Two separate one-electron steps in the reductive activation of the A cluster in subunit beta of the ACDS complex in Methanosarcina thermophila. Biochemistry 47 5544. 10.1021/bi7024035 [DOI] [PubMed] [Google Scholar]
- Gong W., Hao B., Wei Z., Ferguson D. J., Jr., Tallant T., Krzycki J. A., et al. (2008). Structure of the α2ε2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarbonylase/synthase complex. Proc. Natl. Acad. Sci. U.S.A. 105 9558–9563. 10.1073/pnas.0800415105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarse W. G., Mahlert F., Shima S., Thauer R. K., Ermler U. (2000). Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J. Mol. Biol. 303 329–344. 10.1006/jmbi.2000.4136 [DOI] [PubMed] [Google Scholar]
- Grahame D. A. (1993). Substrate and cofactor reactivity of a carbon monoxide dehydrogenase corrinoid enzyme complex. Stepwise reduction of iron sulfur and corrinoid centers, the corrinoid Co2+/1+ redox midpoint potential, and overall synthesis of acetyl-CoA. Biochemistry 32 10786–10793. 10.1021/bi00091a033 [DOI] [PubMed] [Google Scholar]
- Grahame D. A., Demoll E. (1996). Partial reactions catalyzed by protein components of the acetyl-CoA decarbonylase synthase enzyme complex from Methanosarcina barkeri. J. Biol. Chem. 271 8352–8358. 10.1074/jbc.271.14.8352 [DOI] [PubMed] [Google Scholar]
- Gu W. W., Gencic S., Cramer S. P., Grahame D. A. (2003). The A-cluster in subunit beta of the acetyl-CoA decarbonylase/synthase complex from Methanosarcina thermophila: Ni and Fe K-Edge XANES and EXAFS analyses. J. Am. Chem. Soc. 125 15343–15351. 10.1021/ja036602a [DOI] [PubMed] [Google Scholar]
- Haroon M. F., Hu S., Shi Y., Imelfort M., Keller J., Hugenholtz P., et al. (2013). Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500 567–570. 10.1038/nature12375 [DOI] [PubMed] [Google Scholar]
- He Z., Zhang Q., Feng Y., Luo H., Pan X., Gadd G. M. (2018). Microbiological and environmental significance of metal-dependent anaerobic oxidation of methane. Sci. Total Environ. 610-611 759–768. 10.1016/j.scitotenv.2017.08.140 [DOI] [PubMed] [Google Scholar]
- Holmes D. E., Ueki T., Tang H.-Y., Zhou J., Smith J. A., Chaput G., et al. (2019). A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer. mBio 10:e00789-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Smith C., Smith K. S. (2007). AMP-forming acetyl-CoA synthetases in Archaea show unexpected diversity in substrate utilization. Archaea 2 95–107. 10.1155/2006/738517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson T. M., Alber B. E., Kisker C., Ferry J. G., Rees D. C. (2000). A closer look at the active site of γ-carbonic anhydrases: High resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry 39 9222–9231. 10.1021/bi000204s [DOI] [PubMed] [Google Scholar]
- Iyer P. P., Lawrence S. H., Luther K. B., Rajashankar K. R., Yennawar H. P., Ferry J. G., et al. (2004). Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila. Structure 12 559–567. 10.1016/j.str.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Jasso-Chavez R., Apolinario E. E., Sowers K. R., Ferry J. G. (2013). MrpA functions in energy conversion during acetate-dependent growth of Methanosarcina acetivorans. J. Bacteriol. 195 3987–3994. 10.1128/jb.00581-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasso-Chavez R., Diaz-Perez C., Rodriguez-Zavala J. S., Ferry J. G. (2017). Functional role of MrpA in the MrpABCDEFG Na+/H+ antiporter complex from the archaeon Methanosarcina acetivorans. J. Bacteriol. 199:e00662-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten M. S. M., Stams A. J. M., Zehnder A. J. B. (1992). Methanogenesis from acetate. A comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88 181–198. [Google Scholar]
- Kahnt J., Buchenau B., Mahlert F., Kruger M., Shima S., Thauer R. K. (2007). Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS. J. 274 4913–4921. 10.1111/j.1742-4658.2007.06016.x [DOI] [PubMed] [Google Scholar]
- Kisker C., Schindelin H., Alber B. E., Ferry J. G., Rees D. C. (1996). A left-handed beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 15 2323–2330. 10.1002/j.1460-2075.1996.tb00588.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni G., Mand T. D., Metcalf W. W. (2018). Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. mBio 9:e01256-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence S. H., Luther K. B., Schindelin H., Ferry J. G. (2006). Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila. J. Bacteriol. 188 1143–1154. 10.1128/jb.188.3.1143-1154.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner D. J., Li L., Li Q., Rejtar T., Andreev V. P., Reichlen M., et al. (2006). An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U.S.A. 103 17921–17926. 10.1073/pnas.0608833103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu A. O., Cai C., Mcilroy S. J., Southam G., Orphan V. J., Yuan Z., et al. (2020). Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 14 1030–1041. 10.1038/s41396-020-0590-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li Q., Rohlin L., Kim U., Salmon K., Rejtar T., et al. (2007). Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6 759–771. 10.1021/pr060383l [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Karger B. L., Ferry J. G. (2005a). The proteome of Methanosarcina acetivorans. Part I, an expanded view of the biology of the cell. J. Proteome Res. 4 112–128. [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Karger B. L., Ferry J. G. (2005b). The proteome of Methanosarcina acetivorans. Part II, comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4 129–136. [DOI] [PubMed] [Google Scholar]
- Li Q., Li L., Rejtar T., Lessner D. J., Karger B. L., Ferry J. G. (2006). Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188 702–710. 10.1128/jb.188.2.702-710.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Wang Y., Sivan O., Wang F. (2019). Metal-dependent anaerobic methane oxidation in marine sediment: insights from marine settings and other systems. Sci. China Life Sci. 62 1287–1295. 10.1007/s11427-018-9554-5 [DOI] [PubMed] [Google Scholar]
- Luo J. H., Wu M., Liu J., Qian G., Yuan Z., Guo J. (2019). Microbial chromate reduction coupled with anaerobic oxidation of methane in a membrane biofilm reactor. Environ. Int. 130:104926. 10.1016/j.envint.2019.104926 [DOI] [PubMed] [Google Scholar]
- Lyu Z., Shao N., Chou C. W., Shi H., Patel R., Duin E. C., et al. (2020). Posttranslational methylation of arginine in methyl coenzyme M reductase has a profound impact on both methanogenesis and growth of Methanococcus maripaludis. J. Bacteriol. 202:e00654-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macauley S. R., Zimmerman S. A., Apolinario E. E., Evilia C., Hou Y., Ferry J. G., et al. (2009). The archetype γ-class carbonic anhydrase (Cam) contains iron when synthesized in vivo. Biochemistry 48 817–819. 10.1021/bi802246s [DOI] [PubMed] [Google Scholar]
- Mah R. A., Hungate R. E., Ohwaki K. (1977). “Acetate, a key intermediate in methanogenesis,” in Microbial Energy Conversion, eds Schlegel H. G., Barnea J. (Gottingen: E. Goltze; ), 97–106. 10.1016/b978-0-08-021791-8.50017-2 [DOI] [Google Scholar]
- Meng Y., Ingram-Smith C., Cooper L. L., Smith K. S. (2010). Characterization of an archaeal medium-chain acyl coenzyme A synthetase from Methanosarcina acetivorans. J. Bacteriol. 192 5982–5990. 10.1128/jb.00600-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles R. D., Gorrell A., Ferry J. G. (2002). Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila. J. Biol. Chem. 277 22547–22552. 10.1074/jbc.m105921200 [DOI] [PubMed] [Google Scholar]
- Moran J. J., House C. H., Freeman K. H., Ferry J. G. (2005). Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1 303–309. 10.1155/2005/650670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J. J., House C. J., Thomas B., Freeman K. H. (2007). Products of trace methane oxidation during nonmethyltrophic growth by Methanosarcina. J. Geophys. Res. 112:G02011 10.1029/2006JG000268 [DOI] [Google Scholar]
- Murakami E., Ragsdale S. W. (2000). Evidence for intersubunit communication during acetyl-CoA cleavage by the multienzyme CO dehydrogenase/acetyl-CoA synthase complex from Methanosarcina thermophila. Evidence that the beta subunit catalyzes C-C and C-S bond cleavage. J. Biol. Chem. 275 4699–4707. 10.1074/jbc.275.7.4699 [DOI] [PubMed] [Google Scholar]
- Nayak D. D., Liu A., Agrawal N., Rodriguez-Carerro R., Dong S. H., Mitchell D. A., et al. (2020). Functional interactions between posttranslationally modified amino acids of methyl-coenzyme M reductase in Methanosarcina acetivorans. PLoS Biol. 18:e3000507. 10.1371/journal.pbio.3000507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak D. D., Metcalf W. W. (2017). Cas9-mediated genome editing in the methanogenic archaeon Methanosarcina acetivorans. Proc. Natl. Acad. Sci. U.S.A. 114 2976–2981. 10.1073/pnas.1618596114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisbet E. G., Manning M. R., Dlugokencky E. J., Fisher R. E., Lowry D., Michel S. E., et al. (2019). Very strong atmospheric methane growth in the 4 Years 2014–2017: implications for the paris agreement. Glob. Biogeochem. Cycles 33 318–342. 10.1029/2018gb006009 [DOI] [Google Scholar]
- Ollivier B., Lombardo A., Garcia J. L. (1984). Isolation and characterization of a new thermophilic Methanosarcina strain (strain MP). Ann. Microbiol. 135b 187–198. 10.1016/s0769-2609(84)80026-5 [DOI] [PubMed] [Google Scholar]
- Orphan V. J., House C. H., Hinrichs K. U., Mckeegan K. D., Delong E. F. (2002). Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. U.S.A. 99 7663–7668. 10.1073/pnas.072210299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peetz J., Barros L. F., San Martin A., Becker H. M. (2014). Functional interaction between bicarbonate transporters and carbonic anhydrase modulates lactate uptake into mouse cardiomyocytes. Pflugers Arch. 467 1469–1480. 10.1007/s00424-014-1594-z [DOI] [PubMed] [Google Scholar]
- Peterson J. R., Labhsetwar P., Ellermeier J. R., Kohler P. R., Jain A., Ha T., et al. (2014). Towards a computational model of a methane producing archaeum. Archaea 2014:898453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash D., Chauhan S. S., Ferry J. G. (2019a). Life on the thermodynamic edge: respiratory growth of an acetotrophic methanogen. Sci. Adv. 5:eaaw9059. 10.1126/sciadv.aaw9059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash D., Iyer P. R., Suharti S., Walters K. A., Santiago-Martinez M. G., Golbeck J. H., et al. (2019b). Structure and function of an unusual flavodoxin from the domain Archaea. Proc. Natl. Acad. Sci. U.S.A. 116 25917–25922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radle M. I., Miller D. V., Laremore T. N., Booker S. J. (2019). Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. J. Biol. Chem. 294 11712–11725. 10.1074/jbc.ra119.007609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghoebarsing A. A., Pol A., Van De Pas-Schoonen K. T., Smolders A. J., Ettwig K. F., Rijpstra W. I., et al. (2006). A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440 918–921. 10.1038/nature04617 [DOI] [PubMed] [Google Scholar]
- Ragsdale S. W. (2007). Nickel and the carbon cycle. J. Inorg. Biochem. 101 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy V., Boucher O., Haigh J., Hauglustaine D., Haywood J., Myhre G., et al. (2001). “Radiative forcing of climate change,” in Climate Change 2001: The Scientific Basis. Contribution of Working Group I to the Third Assessment Report of the Intergovernmental Panel on Climate Change, eds Houghton J. T., Ding Y., Griggs D. J., Noguer M., Van Der Linden P. J., Dai X., et al. (Cambridge: Cambridge University Press; ), 349–416. [Google Scholar]
- Rhee T. S., Kettle A. J., Andreae M. O. (2009). Methane and nitrous oxide emissions from the ocean: a reassessment using basin-wide observations in the Atlantic. J. Geophys. Res. 114:D12304 10.1029/2008JD011662 [DOI] [Google Scholar]
- Ribas D., Soares-Silva I., Vieira D., Sousa-Silva M., Sa-Pessoa J., Azevedo-Silva J., et al. (2018). The acetate uptake transporter family motif “NPAPLGL(M/S)” is essential for substrate uptake. Fungal Genet. Biol. 122 1–10. 10.1016/j.fgb.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Rohlin L., Gunsalus R. P. (2010). Carbon-dependent control of electron transfer and central carbon pathway genes for methane biosynthesis in the Archaean, Methanosarcina acetivorans strain C2A. BMC Microbiol. 10:62. 10.1186/1471-2180-10-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman D. H., Fournier G. P., French K. L., Alm E. J., Boyle E. A., Cao C., et al. (2014). Methanogenic burst in the end-Permian carbon cycle. Proc. Natl. Acad. Sci. U.S.A. 111 5462–5467. 10.1073/pnas.1318106111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satish Kumar V., Ferry J. G., Maranas C. D. (2011). Metabolic reconstruction of the archaeon methanogen Methanosarcina acetivorans. BMC Syst. Biol. 5:28. 10.1186/1752-0509-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller S., Goenrich M., Boecher R., Thauer R. K., Jaun B. (2010). The key nickel enzyme of methanogenesis catalyses the anaerobic oxidation of methane. Nature 465 606–608. 10.1038/nature09015 [DOI] [PubMed] [Google Scholar]
- Scheller S., Yu H., Chadwick G. L., Mcglynn S. E., Orphan V. J. (2016). Artificial electron acceptors decouple archaeal methane oxidation from sulfate reduction. Science 351 703–707. 10.1126/science.aad7154 [DOI] [PubMed] [Google Scholar]
- Schlegel K., Leone V., Faraldo-Gomez J. D., Muller V. (2012a). Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U.S.A. 109 947–952. 10.1073/pnas.1115796109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel K., Welte C., Deppenmeier U., Muller V. (2012b). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279 4444–4452. 10.1111/febs.12031 [DOI] [PubMed] [Google Scholar]
- Shah M. B., Ingram-Smith C., Cooper L. L., Qu J., Meng Y., Smith K. S., et al. (2009). The 2.1 A crystal structure of an acyl-CoA synthetase from Methanosarcina acetivorans reveals an alternate acyl-binding pocket for small branched acyl substrates. Proteins 77 685–698. 10.1002/prot.22482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Upadhye R., Ishijima Y., Naganuma T. (2011). Methanosarcina horonobensis sp. nov., a methanogenic archaeon isolated from a deep subsurface Miocene formation. Int. J. Syst. Evol. Microbiol. 61 2503–2507. 10.1099/ijs.0.028548-0 [DOI] [PubMed] [Google Scholar]
- Smith K. S., Ingram-Smith C. (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol. 7 150–155. 10.1016/j.tim.2007.02.002 [DOI] [PubMed] [Google Scholar]
- Soo V. W., Mcanulty M. J., Tripathi A., Zhu F., Zhang L., Hatzakis E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Fact. 15:11 10.1186/s12934-12015-10397-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. (1984a). Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47 971–978. 10.1128/aem.47.5.971-978.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Nelson M. J. K., Ferry J. G. (1984b). Growth of acetotrophic, methane-producing bacteria in a pH auxostat. Curr. Microbiol. 11 227–230. [Google Scholar]
- Suharti S., Wang M., De Vries S., Ferry J. G. (2014). Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS One 9:e97966. 10.1371/journal.pone.0097966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlesky K. C., Nelson M. J. K., Ferry J. G. (1986). Isolation of an enzyme complex with carbon monoxide dehydrogenase activity containing a corrinoid and nickel from acetate-grown Methanosarcina thermophila. J. Bacteriol. 168 1053–1058. 10.1128/jb.168.3.1053-1058.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144 2377–2406. 10.1099/00221287-144-9-2377 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A. K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6 579–591. 10.1038/nrmicro1931 [DOI] [PubMed] [Google Scholar]
- Timmers P. H., Welte C. U., Kochorst J. J., Plugge C. M., Jetten M. S., Stams A. J. (2017). Reverse methanogenesis and respiration in methanotrophic Archaea. Archaea 2017:1654237. 10.1155/2017/1654237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine D. L. (2002). Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Antonie Van Leeuwenhoek 81 271–282. [DOI] [PubMed] [Google Scholar]
- Von Klein D., Arab H., Volker H., Thomm M. (2002). Methanosarcina baltica, sp. nov., a novel methanogen isolated from the Gotland Deep of the Baltic Sea. Extremophiles 6 103–110. 10.1007/s007920100234 [DOI] [PubMed] [Google Scholar]
- Wagner T., Koch J., Ermler U., Shima S. (2017). Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357 699–703. 10.1126/science.aan0425 [DOI] [PubMed] [Google Scholar]
- Walters E. M., Johnson M. K. (2004). Ferredoxin:thioredoxin reductase: disulfide reduction catalyzed via novel site-specific [4Fe-4S] cluster chemistry. Photosynth. Res. 79 249–264. 10.1023/b:pres.0000017195.05870.61 [DOI] [PubMed] [Google Scholar]
- Wang F. P., Zhang Y., Chen Y., He Y., Qi J., Hinrichs K. U., et al. (2014). Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J. 8 1069–1078. 10.1038/ismej.2013.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tomb J. F., Ferry J. G. (2011). Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165. 10.1186/1471-2180-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte C., Deppenmeier U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837 1130–1147. 10.1016/j.bbabio.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Welte C., Kroninger L., Deppenmeier U. (2014). Experimental evidence of an acetate transporter protein and characterization of acetate activation in aceticlastic methanogenesis of Methanosarcina mazei. FEMS Microbiol. Lett. 359 147–153. 10.1111/1574-6968.12550 [DOI] [PubMed] [Google Scholar]
- Yan Z., Ferry J. G. (2018). Electron bifurcation and confurcation in methanogenesis and reverse methanogenesis. Front. Microbiol. 9:1322. 10.3389/fmicb.2018.01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Joshi P., Gorski C. A., Ferry J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Wang M., Ferry J. G. (2017). A Ferredoxin- and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains Bacteria and Archaea. mBio 8:e02285-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhilina T. N. (1978). [Development of a pure Methanosarcina biotype 2 culture on acetate]. Mikrobiologiia 47 396–399. [PubMed] [Google Scholar]
- Zhu J., Zheng H., Ai G., Zhang G., Liu D., Liu X., et al. (2012). The genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS One 7:e36756. 10.1371/journal.pone.0036756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman S., Domsic J. F., Tu C., Robbins A. H., Mckenna R., Silverman D. N., et al. (2013). Role of Trp19 and Tyr200 in catalysis by the gamma-class carbonic anhydrase from Methanosarcina thermophila. Arch. Biochem. Biophys. 529 11–17. 10.1016/j.abb.2012.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Sowers K. R., Ferry J. G. (1985). Methanosarcina thermophila sp. nov., a thermophilic, acetotrophic, methane-producing bacterium. Int. J. Syst. Bacteriol. 35 522–523. 10.1099/00207713-35-4-522 [DOI] [Google Scholar]