Abstract
The MreB actin-like cytoskeleton assembles into dynamic polymers that coordinate cell shape in many bacteria. In contrast to most other cytoskeleton systems, few MreB-interacting proteins have been well characterized. Here, we identify a small protein from Caulobacter crescentus, an assembly inhibitor of MreB (AimB). AimB overexpression mimics inhibition of MreB polymerization, leading to increased cell width and MreB delocalization. Furthermore, aimB appears to be essential, and its depletion results in decreased cell width and increased resistance to A22, a small-molecule inhibitor of MreB assembly. Molecular dynamics simulations suggest that AimB binds MreB at its monomer-monomer protofilament interaction cleft and that this interaction is favored for C. crescentus MreB over Escherichia coli MreB because of a closer match in the degree of opening with AimB size, suggesting coevolution of AimB with MreB conformational dynamics in C. crescentus. We support this model through functional analysis of point mutants in both AimB and MreB, photo-cross-linking studies with site-specific unnatural amino acids, and species-specific activity of AimB. Together, our findings are consistent with AimB promoting MreB dynamics by inhibiting monomer-monomer assembly interactions, representing a new mechanism for regulating actin-like polymers and the first identification of a non-toxin MreB assembly inhibitor. Because AimB has only 104 amino acids and small proteins are often poorly characterized, our work suggests the possibility of more bacterial cytoskeletal regulators to be found in this class. Thus, like FtsZ and eukaryotic actin, MreB may have a rich repertoire of regulators to tune its precise assembly and dynamics.
Significance
The MreB actin-like cytoskeleton forms dynamic polymers that coordinate cell shape in many bacteria. In contrast to most other cytoskeleton systems, few MreB interacting proteins have been well characterized. Here, we identify Caulobacter crescentus assembly inhibitor of MreB (AimB). Using a combination of molecular dynamics simulations and biochemical assays, we demonstrate that AimB promotes MreB dynamics by inhibiting monomer-monomer assembly interactions, representing a new mechanism for regulating actin-like polymers. Because small proteins like AimB are often poorly characterized, we suggest that there may be additional bacterial cytoskeletal regulators to be found that are assembly inhibitors. Thus, like FtsZ and eukaryotic actin, our work suggests that MreB many have many accessory proteins that modulate its assembly and dynamics.
Introduction
Maintenance of proper cell size is an important physiological process for all organisms. Changes in cell size are often strongly coupled to cell fitness in laboratory evolution experiments (1), and mutations that affect cell size can be highly adaptive (2). Cell size is also dynamically regulated as in the example of rod-shaped bacteria whose dimensions are altered by environmental factors such as nutrient availability (3). A recent study developed a biophysical model in which cell size is determined by the relative rates of surface area and volume synthesis; upon nutrient upshift, the increased rate of cytoplasmic synthesis reduced the surface area-to-volume ratio via an increase in cell width (4). However, the molecular regulators of cell size remain largely unclear in most bacterial species.
Bacterial cell shape determination requires enzymes that directly synthesize and cross-link peptidoglycan chains in the periplasm and cytoskeletal factors that localize the activity of these enzymes. The actin homolog MreB serves this cytoskeletal function for cell elongation. Studies from Escherichia coli and Bacillus subtilis show that MreB forms filaments that localize and move (5, 6, 7) along the membrane based on the local cell geometry (8) and recruit cell wall enzymes to insert new cell wall material and change the shape of those sites, resulting in a feedback loop that establishes rod shape (9). In this model, MreB must dynamically assemble and disassemble to sample multiple cellular regions over time. Purified MreB filaments are quite stable in vitro (10), suggesting that MreB dynamics may be stimulated by accessory factors that have yet to be discovered.
For other well-characterized cytoskeletal systems such as eukaryotic actin and tubulin and bacterial FtsZ, there are multiple known regulators of filament dynamics (reviewed in (11, 12, 13)). By contrast, for MreB, RodZ is the only confirmed regulator, and it functions to increase the number of MreB filaments (14) and regulate filament properties (15), leaving MreB disassembly mysterious. There are several toxin-antitoxin systems whose toxins have been proposed to target MreB (16, 17, 18), but the degree to which these toxins are expressed and function under standard growth conditions remains unclear. The only other factor proposed to interact with MreB is MbiA, a small Caulobacter crescentus protein that interacts with MreB through an unknown mechanism (19). The effects of MbiA on MreB also remain unclear as its effects on MreB localization were characterized using a nonfunctional N-terminal fluorescent fusion to MreB (19).
Here, we address the lack of knowledge of MreB assembly inhibitors by directly screening for such factors with an overexpression library. We chose an overexpression approach because MreB and many of its known interactors are essential. To the best of our knowledge, our overexpression screen identified a previously uncharacterized factor that we named assembly inhibitor of MreB (AimB). Overexpression of AimB resulted in wider cells that resemble the loss of MreB, whereas depletion of AimB resulted in narrower cells that have increased resistance to a chemical inhibitor of MreB assembly, A22. To characterize the function of MreB, we developed a functional “sandwich” fusion of monomeric-superfolder GFP (msfGFP) to C. crescentus MreB (CcMreB) and found that AimB disrupts its proper localization. Genetic and biochemical studies confirmed that AimB directly interacts with MreB. To develop a model for how AimB inhibits the assembly of MreB, we used all-atom molecular dynamics (MD) simulations. Finally, we used site-specific cross-linking studies and analysis of the species specificity of AimB interactions to confirm specific predictions of our MD model. Together, these results support the model that AimB functions to inhibit the assembly of MreB by binding at MreB’s polymerization interface.
Materials and Methods
Bacterial growth conditions
C. crescentus wild-type strain NA1000 and its derivatives were grown at 30°C in peptone yeast extract medium (20). Escherichia coli strains were grown at 37°C in Lysogeny broth (LB) medium. When necessary, antibiotics were added at the following concentrations: kanamycin (Kan) 30 μg/mL in broth and 50 μg/mL in agar (abbreviated 30:50) for E. coli and 5:25 for C. crescentus; tetracycline (Tet) 1:2 for C. crescentus; chloramphenicol (Cm) 20:30 for E. coli; and carbenicillin (Carb) 50:100 for E. coli. Gene expression was induced in C. crescentus (0.03–0.3% w/v xylose; 0.5 mM vanillate) or E. coli (100 ng/mL anhydro-tetracycline (aTc); 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG)) as noted. Pharmacological inhibition of MreB was performed by adding 1–10 μg/mL A22 (methanol was used as the vehicle control). The strains, plasmids, and primers used in this study are described in Tables S1–S3, respectively.
CRISPR interference-mediated gene depletion
C. crescentus CRISPR interference (CRISPRi) was performed using the plasmids (Table S2) and methods developed in (21). Briefly, primers EK1003/1004 (Table S3), encoding the single guide RNA (sgRNA) mapping to the 5′-end of aimB, were phosphorylated and annealed. The annealed oligos were ligated into the BbsI site of plasmid psgRNA-Base. The resulting plasmid (pEK334) was transformed into a strain carrying a vanillate-inducible catalytically dead cas9 gene (CJW6270) to generate strain EK335 (ΔvanA::pV-dcas9hum-RBSmut1 with plasmid psgRNA-aimB). Gene depletion was initiated with 0.5 mM vanillate and monitored by quantitative reverse transcription PCR (qRT-PCR). Cells carrying psgRNA-Base (CJW5939) were used as controls.
High-throughput cloning and microscopy
Xylose-inducible plasmids for overexpression of conserved hypothetical proteins were generated using an in vivo Gateway strategy, as described previously (22,23). The resulting multicopy plasmids were conjugated into C. crescentus. Strains were induced with 0.3% xylose and imaged in high-throughput format using custom 48-pedestal agarose slides (22,23). Cell morphology was compared to wild-type controls to identify overexpression plasmids resulting in aberrant cell shape.
Fluorescence microscopy and image analysis
Cells were spotted onto pads made of 1% agarose with the corresponding growth medium. Fluorescence microscopy was performed on a Nikon Ti-E (Nikon Instruments, Melville, NY) inverted microscope equipped with a Lumen 220 PRO Illumination System (Prior Scientific, Rockland, MA), Zyla sCMOS 5.5-megapixel camera (Andor Technology, Concord, MA), CFI Plan Apochromat 100× oil immersion objective (NA 1.45, working distance 0.13 mm), and NIS Elements software for image acquisition. Images were segmented using Morphometrics (24). Cell width and length were calculated using custom MATLAB scripts (The MathWorks, Natick, MA). For time-lapse imaging, coverslips were sealed with VALAP (1:1:1 vaseline:lanolin:paraffin) to prevent drying of the agarose pad.
Cell growth measurements
For experiments up to 12 h, cells were grown in standard culture tubes, and aliquots were removed at the specified intervals for measurements of OD660 or colony forming units (CFUs). For experiments longer than 12 h, cells were aliquoted into a 96-well plate, and OD660 was measured on a CLARIOstar Plate Reader (BMG Labtech, Cary, NC) with shaking and temperature control.
Immunoblotting
Cell samples were normalized by optical density (1 mL of OD = 0.5) and lysed in 1× sodium dodecyl sulfate sample buffer. Samples were separated on a 4–20% gradient polyacrylamide gel, transferred to a polyvinylidene fluoride membrane, and blotted with antibodies against MreB (1:1000) (25), GFP (1:1000, ab6556; Abcam, Cambridge, U.K.), or FLAG (1:500, sc-166355; Santa Cruz Biotechnology, Dallas, TX). Horseradish peroxidase-conjugated secondary antibodies (1:5000) and enhanced chemiluminescence reagents (GE Healthcare, Chicago, IL) were used to detect the bands on a Bio-Rad ChemiDoc MP system (Bio-Rad Laboratories, Hercules, CA).
qRT-PCR
RNA was extracted from bacterial cultures using the QIAGEN RNeasy Kit (QIAGEN, Germantown, MD). After DNase digestion, RNA (5 ng/μL) was reverse transcribed using the High Capacity complementary DNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA). One microliter of complementary DNA was used as template in a 10 μL qRT-PCR reaction performed with Power SYBR reagent (Thermo Fisher Scientific). qRT-PCR was performed on an ABI QuantStudio 6 using the ΔΔCt method. rpoD expression was used as the loading control.
Molecular dynamics simulations
All simulations (Table S4) were performed using the MD package NAMD v. 2.10 (26) with the CHARMM27 force field, including CMAP corrections (27). Water molecules were described with the TIP3P model (28). Long-range electrostatic forces were evaluated by means of the particle-mesh Ewald summation approach with a grid spacing of <1 Å. An integration time step of 2 fs was used (29). Bonded terms and short-range, nonbonded terms were evaluated every time step, and long-range electrostatics were evaluated every other time step. Constant temperature (T = 310 K) was maintained using Langevin dynamics (30), with a damping coefficient of 1.0 ps−1. A constant pressure of 1 atm was enforced using the Langevin piston algorithm (31) with a decay period of 200 fs and a time constant of 50 fs. Setup, analysis, and rendering of the simulation systems were performed with the software VMD v. 1.9.2 (32).
Construction of homology models for AimB and E. coli MreB
The AimB and E. coli MreB (EcMreB) homology models were built using the Phyre2 web server (http://www.sbg.bio.ic.ac.uk/phyre2) (33). The amino acid sequences of AimB or EcMreB were submitted using the default settings of the web server. The AimB homology model was based on the Jannaschia sp. protein Jann_2546 (Protein Data Bank, PDB: 2KZC), and the EcMreB homology model was based on CcMreB (PDB: 4CZM).
Simulated systems
MD simulations performed in this study are described in Table S4. Simulations were initialized from the crystal structures of CcMreB (PDB: 4CZM) (34) or the EcMreB homology model. The bound nucleotide was replaced by ATP, and Mg2+-chelating ions were added for stability. The AimB homology model or the Jann_2546 2KZC crystal structure was docked to the MreB structure for AimB-MreB simulations. Water and neutralizing ions were added around each monomer or dimer, resulting in final simulation sizes of up to 89,000 atoms. All simulations were run for 100 ns. For mean values and distributions of measurements, only the last 30 ns were used. To ensure simulations had reached equilibrium, measurement distributions were fit to a Gaussian.
Analysis of opening angles
The centers of mass of the four subdomains of MreB were obtained using VMD. For each time step, we calculated one opening angle from the dot product between the vector defined by the centers of mass of subdomains IIA and IIB and the vector defined by the centers of mass of subdomains IIA and IA. Similarly, we calculated a second opening angle from the dot products between the vectors defined by the centers of mass of subdomains IA and IB and of subdomains IA and IIA. The opening angles we report are the average of these two opening angles. Subdomain definitions are as in (35).
In vitro cross-linking
A low-copy plasmid for the induction of MreB and AimB was constructed using the pZS2-123 vector backbone (36). The aTc-regulated CFP open reading frame (ORF) was removed by inverse PCR with primers EK644 and EK645 (Table S3). The arabinose-inducible RFP was replaced with AimB by Gibson assembly (primers EK646-649; Table S3). Wild-type MreB and a series of amber codon mutants (Table S1) were synthesized by GenScript Biotech (Piscataway, NJ) and used to replace the IPTG-inducible YFP to create a plasmid encoding Plac-MreB and Para-AimB (pMreBXL1-26). A C-terminal FLAG tag was introduced into AimB in a subset of amber codon mutants by inverse PCR using primers EK679-680. In vitro cross-linking of MreB and AimB was performed essentially as previously described (37). pMreBXL and pEVOL-pBpF (37) were co-transformed into strain NO36 (MC4100 ΔmreB) and grown overnight in LB containing kanamycin and chloramphenicol. Cells were diluted 1:100 into fresh LB with antibiotics along with inducers (1 mM IPTG and 0.1% w/v L(+)-arabinose) and 1 mM p-benzoylphenylalanine (pBPA) (Bachem, Torrance, CA). After 4 h, 1 mL of each culture was pelleted, resuspended in 50 μL cold PBS, and transferred to a white 96-well plate. The samples were irradiated under an ultraviolet (UV) bulb (Norman Lamps CFL15/UV/MED, Norman Lamps, St. Charles, IL) on ice for 15 min, and 50 μL 2× sodium dodecyl sulfate sample buffer was added to stop the reaction. Samples were boiled for 5 min and analyzed by immunoblotting.
Results
A C. crescentus protein overexpression screen identifies a novel cytoskeletal regulator
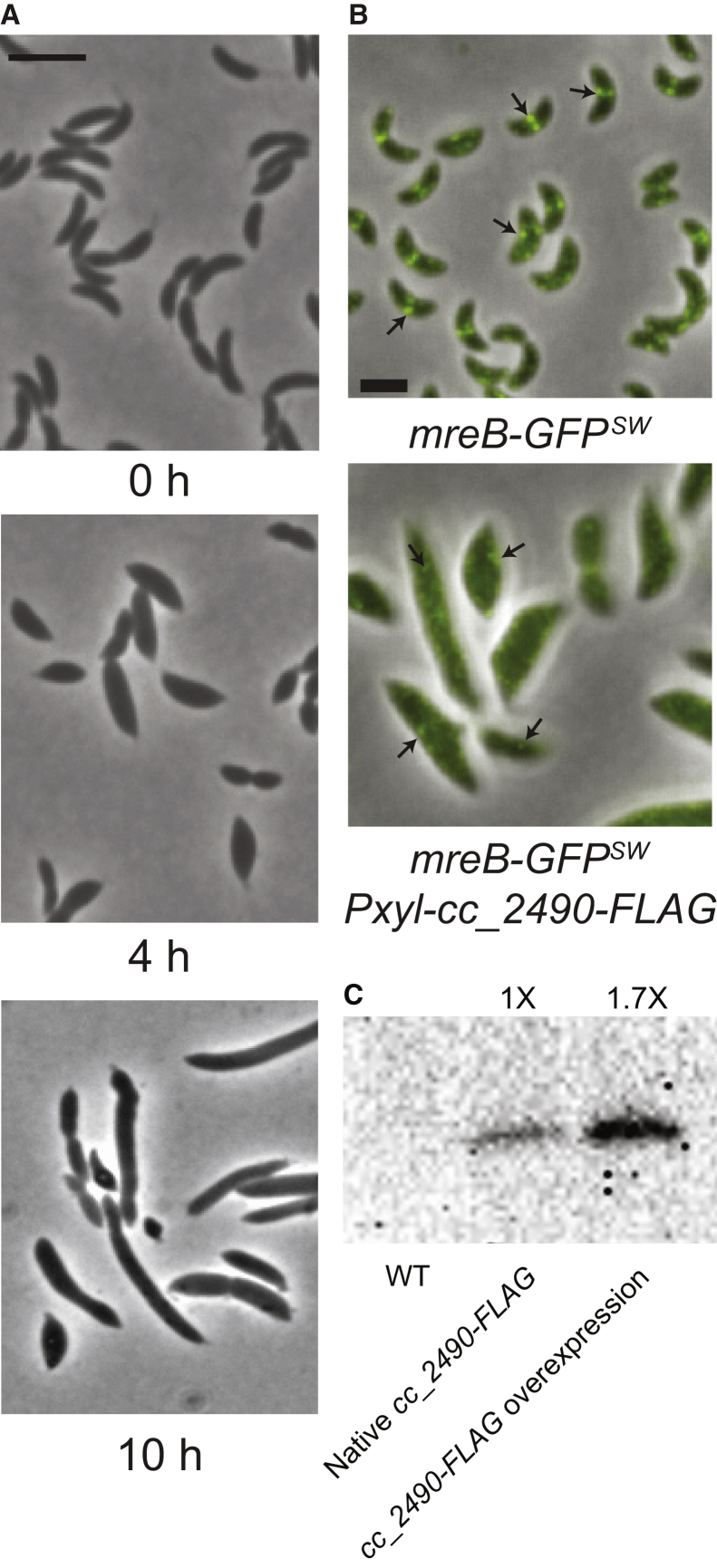
We previously constructed a C. crescentus Gateway entry vector library that includes 224 entry vectors containing ORFs encoding “conserved hypothetical” proteins (22). To identify candidate MreB regulators among these previously uncharacterized proteins, we transferred these ORFs into a xylose-inducible overexpression destination vector using an in vivo Gateway cloning system, conjugated these constructs into C. crescentus, and imaged the strains at the single-cell level (22). Among the various phenotypes observed, overexpression of cc_2490 resulted in a significant increase in cell width that was similar to that seen upon disruption of MreB assembly by the small-molecule inhibitor A22 (Fig. 1 A; (25)).
Figure 1.
Cell width and MreB localization are disrupted by CC_2490 overexpression. (A) CC_2490 expression was induced in wild-type C. crescentus for the indicated times. Phase-contrast images show disruption to cell width and cell shape in C. crescentus cells overexpressing CC_2490. Scale bars, 2 μm. (B) MreB-GFPsw cells with or without CC_2490 overexpression (Pxyl-cc_2490-FLAG) were imaged by phase and fluorescence microscopy at 9 h postinduction. MreB was delocalized in cells grown with CC_2490 overexpression. Arrows indicate examples of MreB localization. Scale bars, 2 μm. (C) Expression of CC_2490 was determined by Western blot using FLAG-tagged cc_2490 expressed from its native locus. Overexpression of CC_2490-FLAG was induced with 0.3% xylose for 9 h. Image quantification shows that xylose induction resulted in 1.7-fold overexpression. To see this figure in color, go online.
We expected that a factor that disrupts MreB assembly would have a strong effect on MreB localization. Because previous analyses of MreB localization in C. crescentus used N-terminal fluorescent fusions that we now know to be nonfunctional (38), we first developed a functional reporter for CcMreB localization. To this end, we inserted msfGFP, which is less prone to aggregation than most commonly used fluorescent proteins, into the same surface-exposed loop that tolerates functional fusion insertions in EcMreB (8). We replaced mreB at its native chromosomal locus under its native promoter to generate a strain in which the only copy of MreB is this new “sandwich” fusion (MreB-GFPsw). The MreB-GFPsw fusion does not affect proliferation rate (Video S1. Molecular Dynamics Simulation of CcMreB-AimB Interactions, Video S2. Molecular Dynamics Simulation of EcMreB-AimB Interactions, Document S2. Article plus Supporting Material), suggesting that it is functional with regards to regulating cell growth and division. As observed in the homologous msfGFP fusion in E. coli, C. crescentus cells expressing MreB-GFPsw were slightly wider and shorter than wild-type cells (Fig. S1, B and C).
Consistent with its effects on cell shape, overexpression of cc_2490 strongly disrupted MreB localization. Whereas wild-type cells showed MreB-GFPsw foci distributed in patches or at midcell in dividing cells, cc_2490 overexpression caused MreB-GFPsw to disperse and become diffuse or to accumulate at the poles (Fig. 1 B). Based on these morphological and MreB localization phenotypes, we renamed CC_2490 assembly inhibitor of MreB (AimB). AimB is a member of the domain-of-unknown function superfamily DUF1476 and is widely conserved among alphaproteobacteria but has no other known activity. RNA sequencing and ribosome profiling have shown that the expression levels of AimB and MreB are stable throughout the cell cycle (39). Ribosome profiling read counts are generally correlated with the number of protein molecules in the cell (40); thus, based on the reported data, the concentrations of AimB and MreB are comparable (39). Indeed, we readily detected native AimB expression in log-phase C. crescentus cell extracts (Fig. 1 C).
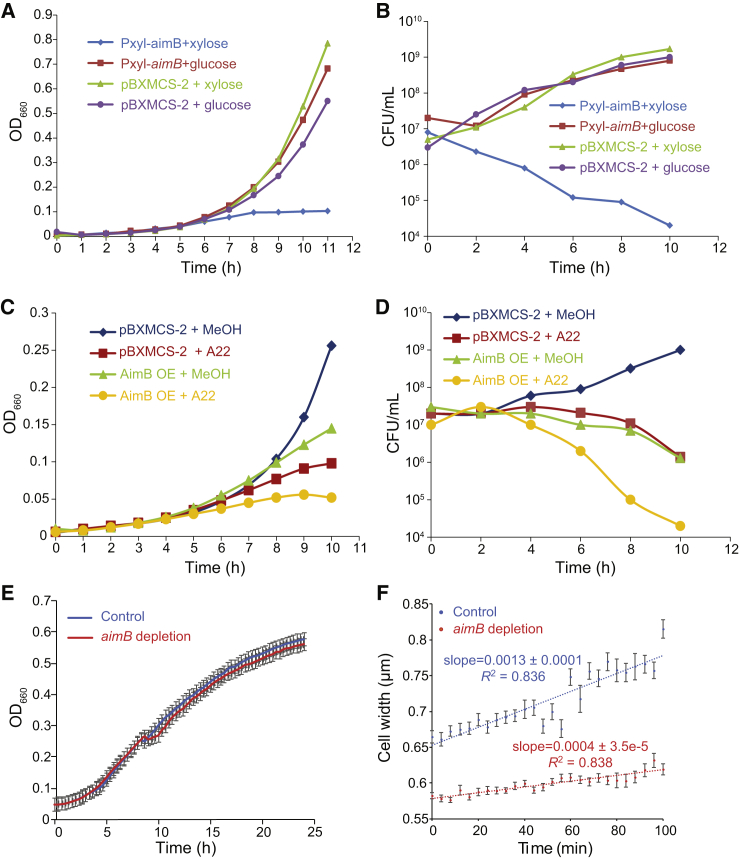
AimB and A22 have additive effects
Because A22 treatment is lethal to C. crescentus cells, we examined whether AimB overexpression is toxic. After only a few hours of overexpression, we observed a significant drop in growth rate as measured by optical density and CFUs, confirming that AimB overexpression is lethal (Fig. 2, A and B). Western blots for MreB showed no change in MreB protein levels when AimB is overexpressed (Fig. S2), demonstrating that AimB toxicity was not due to a reduction in MreB protein concentration. To compare the toxicity of AimB overexpression with that of A22, we measured growth with AimB overexpression and A22 treatment individually or in combination. Both treatments were toxic, and the combination of AimB overexpression and A22 treatment further enhanced lethality (Fig. 2, C and D).
Figure 2.
Regulation of AimB expression is critical for rapid C. crescentus growth. (A) AimB overexpression inhibited growth as measured by optical density. Cells containing either the empty vector (pBXMCS-2) or an AimB overexpression vector were grown with 0.03% xylose (induced) or 0.3% glucose (uninduced). (B) AimB overexpression was toxic to cells as measured by colony forming units (CFUs). Samples were removed every 2 h from the cultures in (A) and plated to measure CFUs. (C and D) AimB overexpression and A22 treatment synergistically resulted in toxicity as measured by optical density (C) and CFUs (D). In (C), cells containing either pBXMCS-2 or an AimB overexpression vector were grown in the presence of 0.03% xylose and either 10 μg/mL A22 or methanol (MeOH). Samples were removed every 2 h from the cultures in (C) and plated to measure CFUs (D). (E) Depletion of aimB messenger RNA using CRISPRi did not affect population growth as measured by optical density. dCas9 expression was induced with 0.5 mM vanillate in cells harboring either a control plasmid (psgRNA-base) or sgRNA-aimB (n = 2 biological replicates, each with three technical replicates; error bars are SEM). (F) The rate of width increase for cells grown on peptone yeast extract agarose pads containing 2.5 μg/mL A22 and 0.5 mM vanillate was higher in cells depleted for AimB than in wild-type cells (n > 24 cells per strain at each timepoint; error bars are SEM; error in slopes is the standard error of regression). To see this figure in color, go online.
The similarities between A22 treatment and AimB overexpression suggested that AimB functions to destabilize MreB. Thus, we hypothesized that loss of AimB would stabilize MreB filaments. Although aimB is annotated as nonessential (41), we were unable to generate a clean aimB deletion. We thus turned to conditional depletion of aimB using CRISPRi (21), which resulted in a 73.7 ± 2.0% (standard error of the mean; n = 3) knockdown of aimB messenger RNA without having an effect on cell growth (Fig. 2 E). In contrast to the increased cell width upon AimB overexpression, depletion resulted in narrower cells when compared to controls (Fig. 2 F, initial time point). If loss of AimB stabilizes MreB filaments, we would expect aimB-depleted cells to have increased resistance to A22 treatment. Time-lapse imaging of control and aimB-depleted cells grown on A22-containing agarose pads demonstrated that the rate of cell width increase was faster in the control cells (Fig. 2 F). Consistent with their opposing effects on MreB, aimB depletion doubled the minimal inhibitory concentration (MIC) of A22 compared to the control (control MIC = 2 μg/mL; aimB depletion MIC = 4 μg/mL). Thus, AimB modulates MreB function in a manner consistent with that of a negative regulator of MreB assembly.
AimB and MreB interact genetically
To identify the cellular targets of AimB, we performed a screen to identify suppressors of toxicity associated with AimB overexpression. Suppressors of overexpressed AimB-FLAG were isolated and subsequently screened by Western blot to filter out mutants with reduced AimB expression. This screening eliminated suppressors that decreased AimB production as well as nonsense and frameshift mutations in the aimB gene. For each isolated suppressor, we sequenced aimB from the overexpression vector and the chromosomal mreB gene. Three point mutations were identified in the overexpressed aimB that resulted in the residue changes V66M, L74Q, and A97P. Interestingly, 13 unique single point mutations were also found in mreB, demonstrating a genetic interaction between AimB and MreB.
To gain insight into the potential interaction between MreB and AimB, we mapped the altered residues in MreB and AimB suppressors onto structures of CcMreB and an AimB homolog from Jannaschia sp. (Fig. S3, A–C). Two mutations, MreBK236T and MreBT277A, were located at what is predicted to be the MreB longitudinal polymerization interface (34). These changes may suppress the effects of AimB overexpression by stabilizing MreB filaments via increasing the interaction strength between MreB monomers or disrupting MreB-AimB interaction. The remaining 11 mreB mutations map near the ATP binding pocket and are reminiscent of mutations that suppress the effects of A22 treatment (42).
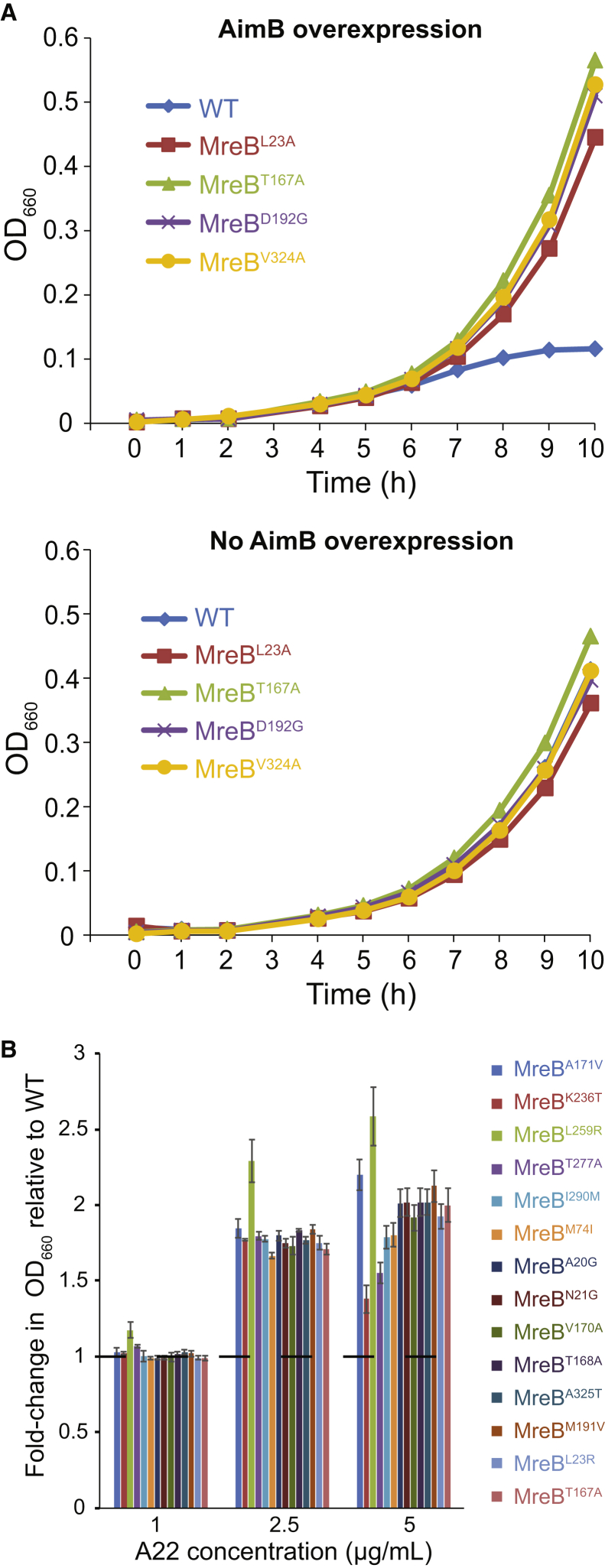
Because similar mreB mutations can confer resistance to A22 treatment and AimB overexpression, we tested previously characterized strains with A22-resistant point mutations in mreB (42) for their ability to suppress AimB overexpression. C. crescentus-producing chromosomally encoded MreBT167A, MreBL23A, MreBD192G, or MreBV324A were resistant to AimB overexpression (Fig. 3 A). These mutations are predicted to inhibit ATP hydrolysis, thereby stabilizing MreB filaments. Conversely, bacteria expressing AimB-resistant mreB mutants also exhibited increased resistance to A22 (Fig. 3 B). Interestingly, the two MreB mutations at the longitudinal polymerization interface (K236T and T277A) had the highest sensitivity to 5 μg/mL A22 of all the mutants. This variability in A22 sensitivity demonstrates that mutations in the ATP binding pocket likely suppress the effects of AimB overexpression by a different mechanism than the mutations involved in MreB subunit-subunit interactions. Thus, although A22 treatment and AimB overexpression both appear to destabilize MreB polymers, they may act through distinct molecular mechanisms, which would explain their additive effects on cell growth.
Figure 3.
MreB mutations confer increased resistance to AimB overexpression and A22. (A) A22 resistance mutations complemented the growth defect due to AimB overexpression. Shown are growth curves for wild-type and A22-resistant strains containing an AimB overexpression plasmid grown with 0.03% xylose (AimB overexpression) or 0.3% glucose (no AimB overexpression). (B) AimB overexpression-resistant strains exhibited higher resistance to A22 than wild-type. Overnight cultures were diluted and grown in media containing 1, 2.5, and 5 μg/mL A22 for 8 h, at which time OD660 readings were taken and standardized to the wild-type culture grown in that concentration of A22. Error bars are standard error of the mean (n = 3). To see this figure in color, go online.
The activity of AimB is specific to CcMreB
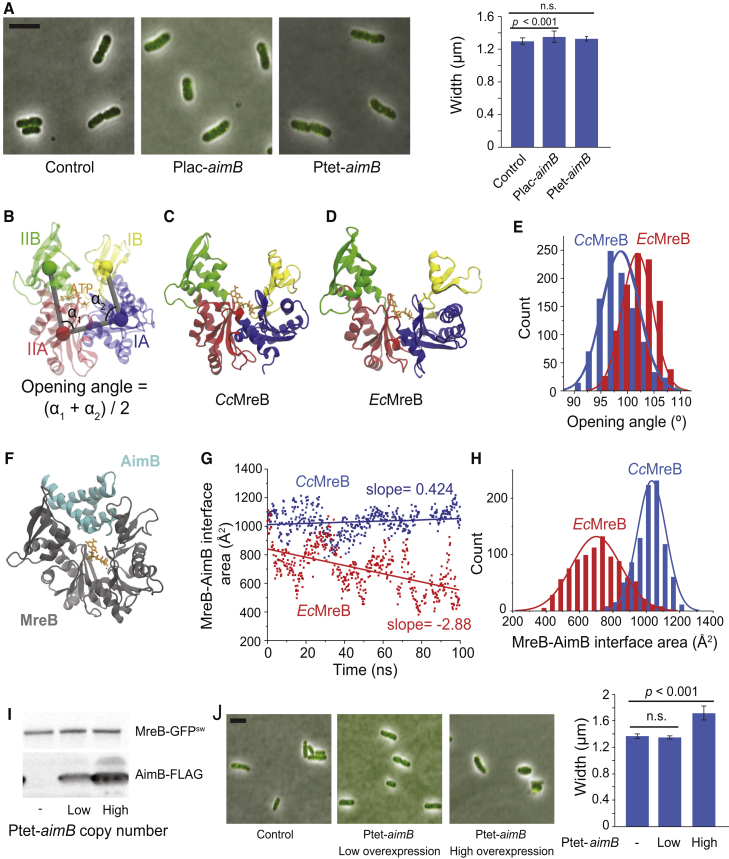
AimB is highly conserved among alphaproteobacteria but rarely found outside of this clade. Because the E. coli genome lacks an aimB homolog, we tested whether AimB alters cell shape and/or MreB localization in E. coli. Even when AimB was expressed at similar or slightly higher levels compared with those that have a strong impact in C. crescentus (Fig. S4), there was no effect on E. coli cell shape or on qualitative MreB localization (Fig. 4 A). This selectivity of AimB for CcMreB is particularly interesting given that the MreB orthologs in these organisms are 78% similar and 64% identical.
Figure 4.
AimB has low affinity for EcMreB, potentially because of differences in the binding pocket. (A) E. coli expressing MreB-GFPsw were transformed with low-copy plasmids for AimB-FLAG expression and induced with 1 mM IPTG or 100 ng/mL aTc for 6 h. Cells were back diluted 1:500 at 3 h to maintain log-phase growth. Cells were imaged by phase and fluorescence microscopy (overlay on left), and cell widths were analyzed (right). Minimal effects on cell width or shape were observed (n = 139 control, 106 Plac, 150 Ptet; error bars are SD; p-values are two-tailed t-test). Scale bars, 5 μm. (B) Definitions of opening angle for an MreB monomer are shown. The centers of mass of the four subdomains are shown as colored spheres. (C) Snapshot of an ATP-bound CcMreB (PDB: 4CZM) at the end of a 100-ns simulation is shown. (D) Shown is a snapshot of an ATP-bound EcMreB at the end of a 100-ns simulation, demonstrating a larger opening angle than CcMreB in (C). The initial EcMreB structure was a homology model of EcMreB built from the CcMreB crystal structure. (E) EcMreB exhibited larger opening angles than CcMreB in the last 30 ns of MD simulations. (F) Shown is the docking of a homology model of the Jannaschia sp. protein Jann_2546 (PDB: 2KZC), a homolog of AimB, to the equilibrated open structure from a CcMreB MD simulation. (G and H) The interfacial area between MreB and AimB showed that the docked heterodimer of CcMreB and AimB in (F) remained stable throughout 100 ns of MD simulation (Video S1), whereas the interfacial area of AimB docking to EcMreB decreased over time (G, Video S2). The distribution of interfacial areas over the course of the MD simulation demonstrates that AimB interacts more stably with CcMreB (H). (I and J) Substantial overexpression of AimB in E. coli disrupts cell width and MreB localization. EcMreB-GFPsw strains harboring low- or high-copy vectors for AimB-FLAG expression were induced as in (A). In (I), cell lysates (normalized to OD600) were analyzed by immunoblotting. In (J), cellular dimensions were quantified by phase and fluorescence microscopy (n = 30 control, 39 low-copy, 31 high-copy; error bars are SD; p-values are two-tailed t-test). Scale bars, 5 μm. To see this figure in color, go online.
Structural modeling suggests that AimB binds in the cleft of MreB
To develop a molecular hypothesis for how AimB may specifically affect CcMreB, we used MD simulations (35) to investigate whether the differential effects of AimB in E. coli and C. crescentus are due to different conformations of the two proteins. We performed all-atom MD simulations (see Materials and Methods) on ATP-bound EcMreB and CcMreB monomers; the EcMreB structure is a homology model we constructed based on the CcMreB structure (PDB: 4CZM, Materials and Methods) (34). We previously observed for Thermotoga maritima MreB monomers that the opening angle at the polymerization interface (Fig. 4 B) was polymerization dependent, with a larger value for monomers relative to the subunits of a dimer (35). Here, we found that the opening angle of an EcMreB monomer was significantly higher than that of CcMreB (Fig. 4, C–E). Thus, we hypothesized that AimB’s selectivity could be explained by binding within the gap formed at the MreB-MreB longitudinal polymerization interface. Specifically, binding of AimB when CcMreB monomers open would prevent binding by another MreB monomer and inhibit polymer assembly. Meanwhile, the larger opening in EcMreB would destabilize the binding of AimB, rendering it less active.
In support of our hypothesis, we were able to dock an AimB homology model of the Jannaschia sp. protein Jann_2546 (PDB: 2KZC; 51% identical and 64% similar) to the equilibrated open structure of our CcMreB MD simulations (Fig. 4 F), and this docked heterodimer remained stable throughout 100 ns of MD simulation (Fig. 4 G; Video S1). By contrast, after docking C. crescentus AimB to an EcMreB monomer with a similar opening angle to that of the equilibrated CcMreB-ATP structure, the AimB gradually dissociated during the simulation (Fig. 4 G; Video S2), coincident with further MreB opening. Quantification of the MreB-AimB interfacial area over 100 ns of simulation showed that AimB consistently had greater contact with CcMreB as compared to EcMreB (Fig. 4 H). To justify our use of a homology model to simulate AimB docking, we performed MD simulations using Jann_2546 and found that it also maintained a stable interaction with CcMreB, although the interfacial area was slightly lower than that of C. crescentus AimB (Fig. S5, A and B). Overexpression of Jann_2546 in C. crescentus resulted in an increase in cell width (Fig. S5 C), further supporting our use of this protein as the basis for a homology model of AimB. We note that overexpression of Jann_2546 had a relatively small effect on cell width, likely because of the fact that expression levels of Jann_2546 were only ∼25% of AimB when overexpressed from the same vector backbone despite codon optimization (Document S1. Supporting Materials and Methods, and Figs. S1–S6, and Tables S1–S4, Video S1. Molecular Dynamics Simulation of CcMreB-AimB Interactions, Video S2. Molecular Dynamics Simulation of EcMreB-AimB Interactions, Document S2. Article plus Supporting Material).
The interactions between AimB and CcMreB were simulated over 100 ns using molecular dynamics (see Materials and Methods). AimB remained stably associated with CcMreB.
The interactions between AimB and EcMreB were simulated over 100 ns using molecular dynamics (see Materials and Methods). AimB did not remain stably associated with EcMreB.
The species specificity of AimB function supports the validity of the MD model
Our MD simulations suggested that AimB can form a stable interaction within the opening cleft at the longitudinal polymerization interface of CcMreB, although AimB has a decreased affinity for EcMreB. Therefore, we hypothesized that the decreased affinity of AimB for EcMreB could be overcome by increasing its expression. Consistent with this prediction, when we expressed aimB from a high-copy E. coli expression vector, we observed an increase in E. coli cell width (Fig. 4, I and J), similar to the effects of sublethal A22 treatment (43). Importantly, the residues of CcMreB that likely interact with AimB (within 5 Å) are highly conserved in EcMreB (79% identical and 96% similar; Fig. S6). Thus, the relative affinities for CcMreB and EcMreB appear to be due to their opening angles rather than differences in binding site amino acids.
AimB and MreB interact directly through the regions predicted by the MD model
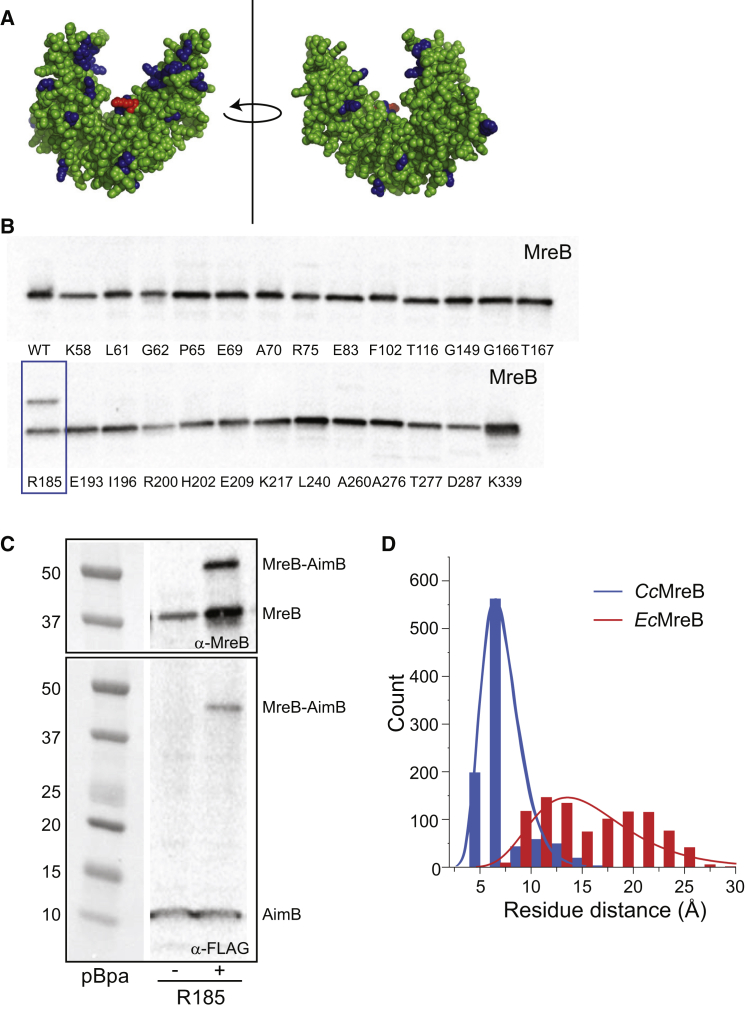
The isolation of AimB-resistant strains with MreB mutations suggested that MreB and AimB interact directly, and our MD docking simulations further predicted specific regions of the two proteins that may interact. To test these predictions, we used a photo-cross-linking assay. Specifically, we created an expression plasmid with CcMreB driven by the lac promoter and AimB driven by an arabinose-inducible promoter. Based on the CcMreB crystal structure, we selected 26 surface-accessible residues (Fig. 5 A) to probe for AimB interactions. Each of the 26 residues was individually mutated to the amber stop codon TAG to enable the incorporation of the unnatural amino acid pBPA. Each amber mutant plasmid was transformed into an E. coli ΔmreB strain carrying the plasmid pEVOL-pBpF, which encodes the transfer RNA synthase/transfer RNA pair for pBPA incorporation (37). We chose to use a ΔmreB strain so that the only potential MreB-AimB interaction would be that of the C. crescentus proteins. After cross-linking, an interaction was only observed when pBPA was incorporated at residue 185 of MreB (Fig. 5 B). Probing this interaction with an anti-FLAG antibody to detect AimB-FLAG confirmed the interaction (Fig. 5 C). The size of the shifted band indicated a 1:1 interaction stoichiometry between MreB and AimB. Strikingly, this position is at the base of the cleft where AimB and MreB are predicted to interact based on MD simulations; analysis of our simulations showed that the intermolecular distance between MreBR185 and AimBG64 remained small in CcMreB, whereas the distance was larger and more variable in EcMreB (Fig. 5 D). These cross-linking data provide evidence supporting our hypothesis that AimB directly interacts with MreB in vivo in a manner consistent with our MD simulations.
Figure 5.
AimB and MreB interact directly. (A) The location of the 26 residues of MreB that were mutated to the amber codon for in vitro cross-linking assays are highlighted in blue on the CcMreB crystal structure. Arginine 185 is highlighted in red. (B) In vitro cross-linking experiments were performed by incorporating the UV cross-linkable non-natural amino acid pBPA at various positions in MreB (Materials and Methods). Cross-linked samples were analyzed by immunoblotting for MreB. A cross-linked band was observed for position R185 (blue rectangle). (C) UV cross-linking of R185 was performed as in (A) using a FLAG-tagged AimB construct. Immunoblotting for MreB or the FLAG-tagged AimB showed similar cross-linked bands. (D) R185 is at the base of the cleft where AimB and MreB are predicted to interact. The intermolecular distance between MreBR185 and AimBG64, the nearest AimB residue, was quantified over the course of our CcMreB-AimB and EcMreB-AimB MD simulations. AimB interacts with CcMreB more stably compared with EcMreB, as shown by a smaller distance between the two residues. To see this figure in color, go online.
Discussion
As the number of sequenced bacterial genomes rapidly increases, a striking feature of virtually all genomes is the lack of comprehensive annotation, leading to an overwhelming number of “hypothetical genes” whose cellular functions are completely unknown. For even the best studied model organism, E. coli K-12, the fraction of hypothetical genes is >25% (UniProt “uncharacterized” or “putative” genes), roughly similar to other model organisms such as Pseudomonas aeruginosa PAO1 (39%), Vibrio cholera O1 El Tor (42%), B. subtilis 168 (43%), and C. crescentus (20%) (44). Moreover, these fractions are likely an underestimate because automated genome annotation pipelines have difficulty distinguishing bona fide small proteins from random unexpressed ORFs. Advanced transcriptomics and proteomics techniques, such as ribosome profiling (40), have improved our ability to robustly confirm the expression of small proteins (<50 residues), some of which are critical regulators of protein kinases, membrane bound enzymes, transport, cell division, or spore formation (reviewed in (45)). Using the C. crescentus genome as an example, of the 762 genes annotated as hypothetical proteins, there are 34 ORFs shorter than 50 codons and 172 ORFs with 50–100 residues. Here, we establish overexpression phenotypic screening as a rapid and robust platform to functionally characterize hypothetical proteins involved in the regulation of the bacterial cytoskeletal element MreB.
The turnover of eukaryotic actin filaments is accomplished by a variety of regulatory proteins that either sequester actin monomers or sever intact filaments (46,47). Although structural studies of actin-regulator interactions have yielded mechanistic insights into the modulation of actin polymerization, our understanding of MreB polymerization dynamics in general and polymer turnover in particular is quite limited. In C. crescentus, the protein MbiA binds directly to MreB, and its overexpression leads to a loss of proper cell shape and an increase in cell death (19). The E. coli toxins YeeV and CptA inhibit MreB polymerization in vitro (17,18); however, their roles in normal physiology are unclear. Importantly, the mechanism of action for all three proposed MreB inhibitors is unknown. Here, we identified AimB as a novel inhibitor of CcMreB and propose the first mechanistic model for MreB assembly inhibition. Specifically, in vivo cross-linking experiments (Fig. 5, A–D) coupled with MD simulations (Fig. 4, B–H) suggest a novel mechanism for the interaction between AimB and MreB in the cleft of open MreB subunits that blocks MreB dimerization.
Overexpression of AimB results in an increase in cell width (Fig. 1 A) and mislocalization of MreB (Fig. 1 B) in a manner similar to the MreB inhibitor A22. A screen for AimB overexpression suppressor mutants found mutations in MreB (Fig. 3 B), demonstrating a genetic interaction between MreB and AimB. To probe for a direct interaction between these proteins, we used a photo-cross-linking approach to discover that MreB residue 185 interacts with AimB (Fig. 5, B and C). These data support the model generated by our MD simulations that AimB binds to the longitudinal polymerization interface of MreB. In this model, AimB functions as a pointed-end competitive inhibitor of MreB-MreB dimerization. To the best of our knowledge, this model represents a novel mechanism for destabilizing actin-like filaments; thymosin-β4 sequesters G-actin monomers by stretching across the actin molecule and interacting with both the pointed and barbed ends (48), whereas twinfilin inhibits actin polymerization by binding G-actin barbed ends with high affinity (49).
In addition to explaining how AimB inhibits MreB assembly, our model can also explain the specificity of AimB for CcMreB as well as the additive effects of AimB overexpression and A22 treatment. Our simulations and cross-linking are consistent with AimB binding to the cleft that forms in MreB subunits at the longitudinal (intrapolymeric) polymer interface when the opening angle is large. Binding of AimB at this site would sterically prevent additional MreB monomers from adding to the polymer, thereby inhibiting MreB filament assembly. Furthermore, this binding site is conformationally distinct in C. crescentus and E. coli (Fig. 4, B–E), thereby explaining the species specificity. Finally, the structure of CcMreB filaments solved in the presence of A22 indicates that A22 disrupts the lateral (interpolymeric) MreB filament interface (34), which would explain how AimB and A22 use distinct mechanisms to inhibit MreB filament formation and therefore additively inhibit the growth rate.
MreB coordinates peptidoglycan insertion to regulate cellular elongation in a variety of species, including Gram-negative E. coli (7) and Gram-positive B. subtilis (5,6). Although mreB is found across a wide range of bacterial lineages, the aimB gene is restricted to alphaproteobacteria. Based on our overexpression studies and MD simulations, we suggest that AimB binds the longitudinal polymerization interface of CcMreB with a higher affinity than EcMreB. This species specificity is demonstrated by the ability of AimB to disrupt the localization of EcMreB only when highly overexpressed (Fig. 4 J). Indeed, recently published MD simulations with CcMreB, EcMreB, and Thermotoga maritima MreB exhibited qualitatively similar yet quantitatively distinct conformational dynamics, highlighting potential species-specific regulation of MreB (50). Species-specific regulation of a bacterial cytoskeletal protein is not unexpected given that the highly conserved tubulin ortholog FtsZ is regulated by a variety of divergent mechanisms. For example, placement of FtsZ and the divisome at midcell can be mediated by multiple distinct mechanisms. In E. coli and most Gram-negative bacteria, oscillations of the MinC/D complex are facilitated by MinE (51,52), whereas in B. subtilis and most Gram-positive bacteria, MinC/D restricts FtsZ to the midline via interactions with DivIVA (53). Similarly, nucleoid occlusion in E. coli and B. subtilis is directed by two different proteins, Noc and SlmA, respectively (54,55). Interestingly, C. crescentus does not use either the MinC/D or nucleoid occlusion mechanisms for FtsZ localization; instead, a gradient of MipZ antagonizes FtsZ polymerization closer to the poles, leading to midcell Z-ring formation (56). Thus, although the core MreB and FtsZ cytoskeletal proteins are widely conserved in bacteria, emerging evidence suggests that the regulation of these core cytoskeletons is largely performed by species-specific factors.
Author Contributions
J.N.W., H.S., J.H., K.C.H., Z.G., and E.A.K. designed the research. J.N.W., H.S., J.H., and E.A.K. performed research. J.N.W., H.S., J.H., K.C.H., Z.G., and E.A.K. analyzed data. E.A.K. wrote the manuscript. J.N.W., H.S., K.C.H., Z.G., and E.A.K. edited the manuscript.
Acknowledgments
We thank Anna Konovalova (Health Science Center, The University of Texas, Houston, TX) for helpful discussions and assistance with the photo-cross-linking assay and Christine Jacobs-Wager (Yale University, New Haven, CT) for providing CRISPRi reagents.
Funding was provided by National Science Foundation CAREER Award MCB-1149328, the Stanford University Center for Cancer Systems Biology under grant P50-GM107615, and the Allen Discovery Center at Stanford University on Systems Modeling of Infection (to K.C.H.); National Institutes of Health Ruth L. Kirschstein National Research Service Award 1F32GM100677 (to J.H.); an Agilent Fellowship, a Stanford Interdisciplinary Graduate Fellowship, and a James McDonnell Postdoctoral Fellowship (to H.S.); National Institutes of Health grant R01GM107384 (to Z.G.); and National Science Foundation CAREER Award MCB-1553004 (to E.A.K.). K.C.H. is a Chan Zuckerberg Investigator. All simulations were performed with computer time provided by the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant OCI-1053575, with allocation number TG-MCB110056 (to K.C.H.).
Editor: Margaret Cheung.
Footnotes
John N. Werner and Handuo Shi contributed equally to this work.
John N. Werner’s present address is Department of Biology, Wisconsin Lutheran College, Milwaukee, Wisconsin.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.04.029.
Supporting Citations
References (57, 58, 59, 60, 61, 62, 63, 64) appear in the Supporting Material.
Supporting Material
References
- 1.Lenski R.E., Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monds R.D., Lee T.K., Huang K.C. Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness. Cell Rep. 2014;9:1528–1537. doi: 10.1016/j.celrep.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaechter M., Maaløe O., Kjeldgaard N.O. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 4.Harris L.K., Theriot J.A. Relative rates of surface and volume synthesis set bacterial cell size. Cell. 2016;165:1479–1492. doi: 10.1016/j.cell.2016.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domínguez-Escobar J., Chastanet A., Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 6.Garner E.C., Bernard R., Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Teeffelen S., Wang S., Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ursell T.S., Nguyen J., Huang K.C. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. USA. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H., Bratton B.P., Huang K.C. How to build a bacterial cell: MreB as the foreman of E. coli construction. Cell. 2018;172:1294–1305. doi: 10.1016/j.cell.2018.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nurse P., Marians K.J. Purification and characterization of Escherichia coli MreB protein. J. Biol. Chem. 2013;288:3469–3475. doi: 10.1074/jbc.M112.413708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwal P., Zaidel-Bar R. Principles of actomyosin regulation in vivo. Trends Cell Biol. 2019;29:150–163. doi: 10.1016/j.tcb.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Maiato H., Sampaio P., Sunkel C.E. Microtubule-associated proteins and their essential roles during mitosis. Int. Rev. Cytol. 2004;241:53–153. doi: 10.1016/S0074-7696(04)41002-X. [DOI] [PubMed] [Google Scholar]
- 13.Ortiz C., Natale P., Vicente M. The keepers of the ring: regulators of FtsZ assembly. FEMS Microbiol. Rev. 2016;40:57–67. doi: 10.1093/femsre/fuv040. [DOI] [PubMed] [Google Scholar]
- 14.Bratton B.P., Shaevitz J.W., Morgenstein R.M. MreB polymers and curvature localization are enhanced by RodZ and predict E. coli’s cylindrical uniformity. Nat. Commun. 2018;9:2797. doi: 10.1038/s41467-018-05186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colavin A., Shi H., Huang K.C. RodZ modulates geometric localization of the bacterial actin MreB to regulate cell shape. Nat. Commun. 2018;9:1280. doi: 10.1038/s41467-018-03633-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller D.M., Tavag M., Hochschild A. CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with FtsZ and MreB. PLoS Genet. 2017;13:e1007007. doi: 10.1371/journal.pgen.1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda H., Tan Q., Inouye M. A novel membrane-bound toxin for cell division, CptA (YgfX), inhibits polymerization of cytoskeleton proteins, FtsZ and MreB, in Escherichia coli. FEMS Microbiol. Lett. 2012;328:174–181. doi: 10.1111/j.1574-6968.2012.02496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan Q., Awano N., Inouye M. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 2011;79:109–118. doi: 10.1111/j.1365-2958.2010.07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yakhnina A.A., Gitai Z. The small protein MbiA interacts with MreB and modulates cell shape in Caulobacter crescentus. Mol. Microbiol. 2012;85:1090–1104. doi: 10.1111/j.1365-2958.2012.08159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poindexter J.S. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irnov I., Wang Z., Jacobs-Wagner C. Crosstalk between the tricarboxylic acid cycle and peptidoglycan synthesis in Caulobacter crescentus through the homeostatic control of α-ketoglutarate. PLoS Genet. 2017;13:e1006978. doi: 10.1371/journal.pgen.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner J.N., Chen E.Y., Gitai Z. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werner J.N., Gitai Z. High-throughput screening of bacterial protein localization. Methods Enzymol. 2010;471:185–204. doi: 10.1016/S0076-6879(10)71011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ursell T., Lee T.K., Huang K.C. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. BMC Biol. 2017;15:17. doi: 10.1186/s12915-017-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitai Z., Dye N.A., Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackerell A.D., Jr., Feig M., Brooks C.L., III Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 29.Tuckerman M., Berne B.J., Martyna G.J. Reversible multiple time scale molecular dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 30.Brünger A., Brooks C.L., Karplus M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984;105:495–500. [Google Scholar]
- 31.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 32.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Kelley L.A., Mezulis S., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Ent F., Izoré T., Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colavin A., Hsin J., Huang K.C. Effects of polymerization and nucleotide identity on the conformational dynamics of the bacterial actin homolog MreB. Proc. Natl. Acad. Sci. USA. 2014;111:3585–3590. doi: 10.1073/pnas.1317061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox R.S., III, Dunlop M.J., Elowitz M.B. A synthetic three-color scaffold for monitoring genetic regulation and noise. J. Biol. Eng. 2010;4:10. doi: 10.1186/1754-1611-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chin J.W., Martin A.B., Schultz P.G. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc. Natl. Acad. Sci. USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gitai Z., Dye N., Shapiro L. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. USA. 2004;101:8643–8648. doi: 10.1073/pnas.0402638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrader J.M., Li G.W., McAdams H.H. Dynamic translation regulation in Caulobacter cell cycle control. Proc. Natl. Acad. Sci. USA. 2016;113:E6859–E6867. doi: 10.1073/pnas.1614795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G.W., Burkhardt D., Weissman J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christen B., Abeliuk E., Shapiro L. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dye N.A., Pincus Z., Theriot J.A. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol. Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tropini C., Lee T.K., Huang K.C. Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell Rep. 2014;9:1520–1527. doi: 10.1016/j.celrep.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storz G., Wolf Y.I., Ramamurthi K.S. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014;83:753–777. doi: 10.1146/annurev-biochem-070611-102400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 47.Sun H.Q., Kwiatkowska K., Yin H.L. Actin monomer binding proteins. Curr. Opin. Cell Biol. 1995;7:102–110. doi: 10.1016/0955-0674(95)80051-4. [DOI] [PubMed] [Google Scholar]
- 48.Xue B., Leyrat C., Robinson R.C. Structural basis of thymosin-β4/profilin exchange leading to actin filament polymerization. Proc. Natl. Acad. Sci. USA. 2014;111:E4596–E4605. doi: 10.1073/pnas.1412271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paavilainen V.O., Hellman M., Lappalainen P. Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc. Natl. Acad. Sci. USA. 2007;104:3113–3118. doi: 10.1073/pnas.0608725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H., Quint D.A., Huang K.C. Chiral twisting in a bacterial cytoskeletal polymer affects filament size and orientation. Nat. Commun. 2020;11:1408. doi: 10.1038/s41467-020-14752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Z., Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Mol. Microbiol. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- 52.Raskin D.M., de Boer P.A. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J. Bacteriol. 1999;181:6419–6424. doi: 10.1128/jb.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marston A.L., Thomaides H.B., Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernhardt T.G., de Boer P.A. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol. Cell. 2005;18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu L.J., Errington J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell. 2004;117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 56.Thanbichler M., Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 57.Amann E., Ochs B., Abel K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 58.Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ingerson-Mahar M., Briegel A., Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madeira F., Park Y.M., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon R., Priefer U., Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 62.Thanbichler M., Iniesta A.A., Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee T.S., Krupa R.A., Keasling J.D. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J. Biol. Eng. 2011;5:12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The interactions between AimB and CcMreB were simulated over 100 ns using molecular dynamics (see Materials and Methods). AimB remained stably associated with CcMreB.
The interactions between AimB and EcMreB were simulated over 100 ns using molecular dynamics (see Materials and Methods). AimB did not remain stably associated with EcMreB.