Abstract
The hyperactivation and overexpression of critical oncogenes is a common occurrence in multiple types of malignant tumors. Recently, the abnormal activation mechanism of an oncogene by a super-enhancer (SE) has attracted significant attention. A series of changes (insertion, deletion, translocation and rearrangement) in the genome occurring in cancer cells may generate new SEs, leading to the overexpression of SE-driven oncogenes. SEs are composed of typical enhancers densely loaded with mediator complexes, transcription factors, and chromatin regulators, and drive the overexpression of oncogenes associated with cellular identity and disease. Cyclin-dependent kinase 7 (CDK7) and bromodomain protein 4 (BRD4) are critical mediator complexes associated with SE-mediated transcription. Clinical trials have shown that emerging small-molecule inhibitors (CDK7 and BRD4 inhibitor), targeting the SE exert a notable effect on cancer treatment. Increasing evidences has illustrated that the SE and its associated complexes play a critical role in the development of various types of cancer. The present review discusses the composition, function and regulation of SEs and their contribution to oncogenic transcription. In addition, creative therapeutic approaches that target SE, their advantages and disadvantages, as well as the problems with their clinical application are discussed. It was found that targeting SE may be used in conventional treatment and establish more access for patients with cancer.
Keywords: super-enhancer, oncogene, small-molecular inhibitor, gene-editing, cancer therapy
1. Introduction
The first enhancer element was identified in a DNA sequence of the SV40 virus in the 1980s, and was found to have enhanced transcriptional activity of β-globin in Oryctolagus cuniculus (1). Since then, novel insights into the regulatory mechanism of the genome have been accumulatively gained, due to continuous exploration (2,3). It has been demonstrated that the transcriptional activation of genes is controlled by cell-type-specific proximal and distal regulatory elements, termed enhancers. As a non-coding regulatory element, an enhancer can activate gene expression through long-range chromatin interactions (4). Different from the typical enhancer (TE), super-enhancer (SE) can span dozens of kilo-base (kb) pairs compared with the dozens of base-pairs of the TE (5).
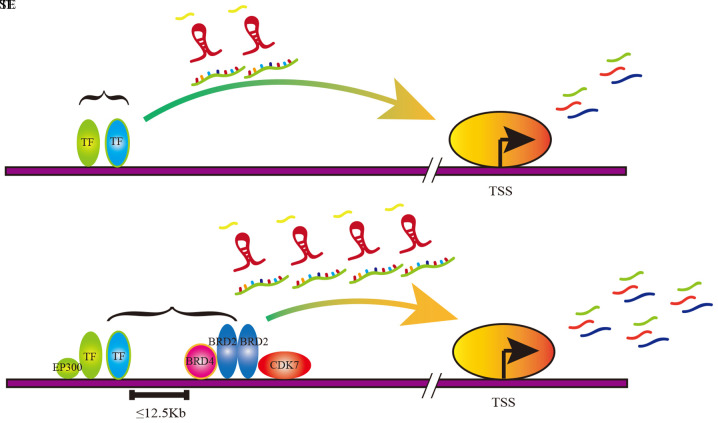
The regions of TEs and SEs are both occupied by enhancer-related molecules, including transcription factors (TFs), master cofactors, mediator complexes and RNA polymerase II (pol II) (6). However, compared with TEs, SEs are characterized by a wider span and more aggregation of SE-related molecules (Fig. 1) (7,8). Therefore, the SE can drive a higher level of gene transcription compared with TE, which is involved in various processes of tumorigenesis and progression (9). The focus of the present review includes the roles of the SE and its key complexes in cancer, with further discussion on the possible new therapies involving the SE in the targeted treatment of cancer.
Figure 1.
Schematic diagram of TE and SE function in transcriptional regulation. The TE promotes mRNA expression and the upregulation of miRNA and lncRNA, thereby promoting gene expression. The SE markedly promoted mRNA expression and the upregulation of miRNA and lncRNA, thereby increasing gene expression. TE, typical enhancer; SE, super-enhancer; miRNA, microRNA; lncRNA, long non-coding RNA; TSS, transcription start site.
2. SEs and cancer
Certain oncogenes exhibit low expression in normal cells and high expression in cancer cells via SE regulation, suggesting the significance of the SE function, particularly in the maintenance of cancer cell growth and survival (10–13). Compared with normal cells, cancer cells proactively construct SEs to drive the expression of oncogenes during tumorigenesis (14). Tumor mutations generally lead to the dysregulation of enhancers that normally control the signal-dependent expression of growth-associated genes, resulting in the uncontrolled proliferation of tumor cells (2). The results of genome wide association studies demonstrated that the majority of cancer-associated genetic variations are located outside the coding genome, and the mutation sites are often found in the putative enhancer enrichment regions (15). The early evidence supporting this view comes from studies in Burkitt lymphoma in which gene rearrangement caused the formation of MYC gene enhancer regions leading to MYC overexpression (16–19). Further studies have indicated that SE-driven MYC expression is critical for maintaining cell survival and proliferation in acute leukemia and neuroblastoma (20,21). The inhibition of SE assembly and maintenance inhibits carcinogenic transcription and tumor growth, which suggests that the SE is a promising anti-cancer target (22).
In 2017, Hnisz et al (23) proposed a model to explain the roles of SE through phase separation, without, however, adequate direct evidence. Phase separation was described as a dynamic process, where the single phase consisting of the originally miscible components is decomposed into two or more different phases. Thus, phase separation represents two or more distinct liquid phases produced from a single mixed liquid phase, such as oil, whose drops are separated from the water (24). In 2018, Sabari et al (25) confirmed the hypothesis of the phase separation model, reporting that the transcriptional coactivators bromodomain protein 4 (BRD4) and MED1 could phase separate to form droplets at SE regions. The droplets were recruited by various types of transcriptional machinery to gather in the vicinity of the SE regions to realize the compartmentalization reaction of the transcription process. The intrinsically disordered regions (IDRs) played a pivotal role in the phase separation process. IDRs are present in RNA granule proteins that trigger the disintegration of particulate matter by phosphorylation of self-association through the action of electrostatic, polar and hydrophobic action (26–28). The transcriptional coactivator forms phase-separated particle aggregates at the SE regions, and thus the transcription device can be compartmentalized and aggregated. This provides more detailed theoretical evidence to support that SE precisely regulates oncogene transcriptional activation and drives tumor progression.
The Assay for Transposase-Accessible Chromatin with high throughput sequencing and chromatin immunoprecipitation followed by sequencing, based on high-throughput whole-genome sequencing, have identified a series of SE-driven oncogenes in various tumor cells. SE can not only play a role in gene regulation in solid tumors, such as hepatocellular, breast, esophageal and gastric tumors, but can also elevate the expression of proteins in non-solid tumors, such as lymphocytic leukemia and myeloid leukemia (22). SE has been confirmed to regulate a variety of well-defined oncogenes, such as c-MYC and ETS-Variant Gene 6, and to affect multiple signaling pathways, such as the MAPK and Notch signaling pathways (29–32). It is generally believed that SE can promote tumorigenesis by upregulating SE-associated oncogenes. The histone H3 acetyl K27 (H3K27ac) and histone H3 methyl K4 (H3K4me1) peaks have been confirmed in the SE regions, since SE-driven gene requires an open chromatin microenvironment (33). However, extensive H3K4me3 peaks have also been confirmed to occur in the regions of the tumor suppressor genes (34). It was therefore hypothesized that if a non-specific demethylase intervenes the histone methylation modification, it may reduce the SE-driven oncogene as well as anti-oncogene expression. Therefore, targeting histone modification is challenging to use as a targeted drug.
Apart from their involvement in protein-coding gene regulation, SEs also play unique roles in RNA [including micro (mi)RNA, enhancer (e)RNA and long non-coding (lnc)RNA] regulation. Suzuki et al (35) demonstrated that SE may not only promote miRNA transcription, but also enhance the cell-specific miRNA production through Drosha/DGCR8 recruitment and pri-miRNA processing through genome editing, via clustered regularly interspaced short palindromic repeats (CRISPR/Cas9). Moreover, SE-miRNAs contain many highly cell-specific and vital miRNAs (such as ESC-, muscle-, neuron-, hematopoietic-, skin- and inflammation-associated miRNAs) compared with TE-miRNAs (35), indicating that SE can drive the corresponding miRNAs and affect their downstream pathways. Global run-on sequencing analysis revealed that abundant eRNAs were transcribed in SE regions in lymphoblastoid cell (LCL). In addition, the knockdown of these eRNAs was found to result in LCL cell growth arrest, with MYC expression significantly declining, following the knockdown of SE-associated eRNAs of MYC (36), illustrating that eRNA can affect not only gene expression, but also cell growth. The SE-associated lncRNA has also been demonstrated to play a fundamental role in the regulation of enhancer activity and gene programs in cardiovascular pathology (37). The disordered SE regulation associated with lncRNA causes a serious physiological and pathological deterioration, such as pathological stress, remodelling and failure in the cardiovascular system (37). It is possible to alleviate cardiovascular disease by targeting these non-coding RNAs in the future. In hepatocellular carcinoma (HCC), the novel lncRNA HCCL5 was identified as a key oncogene, which was driven by the SE and is significantly overexpressed in human HCC tissues (38). In addition, HCCL5 can promote the cell proliferation, regulate the G1-S phase transition and affect the invasive and metastatic ability of HCC cells. Therefore, the high expression of HCCL5 may be used as a biomarker for the poor overall survival of patients with HCC. Given the important role of SE in RNA regulation, it may be attempted to control the abnormal expression of RNAs by blocking the function of SE.
In conclusion, the modifications of H3K4me1 and H3K27ac often occur in the enhancer regions of cancer cells, representing the formation of open chromatin structures, both of which are absent in normal cells (39). Moreover, the SE-regulated mechanism affects abnormal regulation at both the transcriptional and post-transcriptional levels, resulting in malignant transformation.
3. Small-molecule inhibitors targeting SE for cancer treatment
The enhanced transcriptional activity of associated oncogenes caused by SE have been reported in various types of cancer. Therefore, blocking the SE is a viable anticancer therapy. The mechanism of action that describes the potential treatments, by targeting the SE-associated complexes, is shown in Fig. 2. However, transcription, as a biological process that occurs universally in vivo, cannot be inhibited as a whole, and a high degree of specificity in clinical antitumor therapy is required to inhibit it. It is therefore necessary to find a target for the possible intervention among these small molecules that are involved in the SE mechanism. Master TFs, cofactors and histone modification markers are difficult to select as targets due to their extensive effects, while mediator complexes such as cyclin-dependent kinase 7 (CDK7) and BRD4 are relatively characteristic. Decreased CDK7 expression or BRD4 expression suppresses cancer cell growth, confirming that cancer cells are sensitive to the dosage of SE-associated mediator complexes (40). The inhibition of mediator complexes is considered to be a promising approach. Studies have reported that the activity of SE must depend on the interaction of key TFs, cofactors and mediator complexes. Therefore, it is theoretically feasible to design inhibitors of mediator complexes to intervene in tumor cells (41). Targeted inhibitors can specifically block the interaction between SE regions and their corresponding complexes, thereby rescuing the upregulated oncogene. These types of small-molecule inhibitors have been found to exert significant inhibitory effects in animal experiments (42–44). At present, the small-molecule inhibitors designed for cancer treatments include the following three types: i) CDK7 inhibitor (THZ1); ii) BRD4 inhibitor (JQ1); and iii) other inhibitors, which are summarized in Table I (8,21,29,35,39,45–77).
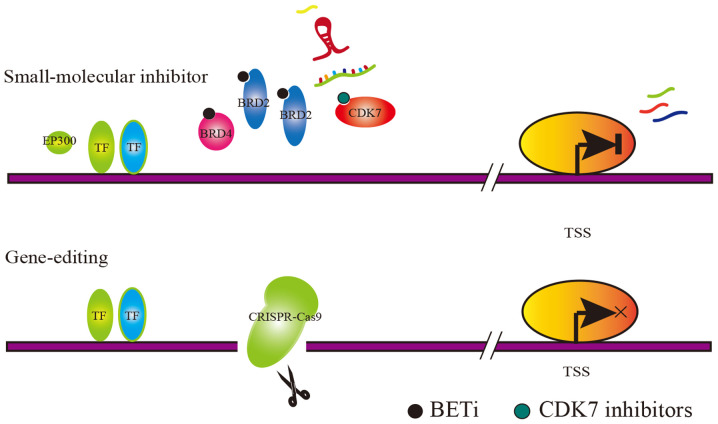
Figure 2.
Schematic diagram of two methods for targeting SE complexes, small-molecule inhibitor and gene-editing small-molecule inhibitor that can target SE-associated complexes (such as BRD4 and CDK7), thereby destroying SE function. Gene-editing technology can knock out regions targeted by SE-associated complexes, thereby disrupting the SE function. SE, super-enhancer; BRD4, bromodomain protein 4; CDK7, cyclin-dependent kinase 7.
Table I.
Examples of small-molecule inhibitors targeting super-enhancers in diseases.
| Small-molecule inhibitor | Target | Disease | Pubmed ID | (Refs.) |
|---|---|---|---|---|
| THZ1 | CDK7 | Osteosarcoma | 29644114 | (45) |
| THZ1 | CDK7 | Thyroid carcinoma | 11158054 | (46) |
| THZ1 | CDK7 | Neuroblastoma | 25416950 | (21) |
| THZ1 | CDK7 | Adult T-cell leukemia | 28978570 | (47) |
| THZ1 | CDK7 | Melanoma | 29408204 | (48) |
| THZ1 | CDK7 | T-cell acute lymphoblastic leukemia | 25043025 | (49) |
| THZ1 | CDK7 | Small cell lung cancer | 25490451 | (8) |
| THZ1 | CDK7, CDK12, CDK13 | Chordoma | 30664779 | (50) |
| THZ531 | CDK12, CDK13 | Chordoma | 30664779 | (50) |
| SY1365 | CDK7 | Ovarian and breast cancer | 31064851 | (51) |
| NVP-2/AT7519/dinaciclib/alvocidib | CDK9 | Chordoma | 30664779 | (50) |
| JQ1 | BRD4 | Cancer | 28283057 | (35) |
| JQ1 | BRD4 | Cervical cancer | 27624132 | (52) |
| JQ1 | BRD4 | Merkel cell carcinoma | 25941994 | (53) |
| JQ1 | BRD4 | Multiple myeloma | 23582323 | (54) |
| JQ1 | BRD4 | Prostate and breast cancer | 28359301 | (55) |
| JQ1 | BRD4 | Colon cancer | 28576751 | (29) |
| JQ1 | BRD4 | Diffuse large B-cell lymphoma | 24332044 | (56) |
| JQ1 | BRD4 | Hepatocellular carcinomas | 30659195 | (57) |
| JQ1 | BRD4 | Pancreatic cancer | 29533787 | (58) |
| JQ1 | BRD4 | Melanoma | 28991225 | (59) |
| JQ1 | BRD4 | Cancer | 27099234 | (60) |
| JQ1 | BRD4 | Colon cancer | 26983878 | (61) |
| JQ1 | BRD4 | Adenoid cystic carcinoma | 26829750 | (62) |
| JQ1 | BRD4 | Blastic plasmacytoid dendritic cell neoplasm | 27846392 | (63) |
| JQ1 | BRD4 | Breast cancer | 30518851 | (64) |
| JQ1 | BRD4 | Melanoma | 29149598 | (39) |
| JQ1 | BRD4 | Ovarian Cancer | 26877780 | (65) |
| JQ1 | BRD4 | Large B-cell lymphoma | 25165099 | (66) |
| JQ1 | BRD4 | Lymphoma | 23792448 | (67) |
| JQ1 | BRD4 | Acute myeloid leukemia | 26626481 | (68) |
| I-BET151 | BRD4 | Blastic plasmacytoid dendritic cell neoplasm | 27846392 | (63) |
| I-BET151 | BRD4 | Ovarian cancer | 26877780 | (65) |
| I-BET762 | BRD2, BRD3, BRD4 | Blastic plasmacytoid dendritic cell neoplasm | 27846392 | (63) |
| I-BET726 | BRD2, BRD3 BRD4 | Acute myeloid leukemia | 28595007 | (69) |
| OTX015 | BRD2, BRD3 BRD4 | Leukemia | 27063978 | (70) |
| OTX015 | BRD2, BRD3 BRD4 | Lymphoma | 27063978 | (70) |
| OTX015 | BRD2, BRD3 BRD4 | Myeloma | 27063978 | (70) |
| CPI0610 | BRD4 | Hematological malignancies | 26815195 | (71) |
| CPI0610 | BRD4 | Multiple myeloma | 27890933 | (72) |
| CPI0610 | BRD4 | T-cell lymphoma | 30529073 | (73) |
| Ribociclib (LEE011) | CDK4, CDK6 | Breast cancer | 28351928 | (74) |
| Ribociclib (LEE011) | CDK4, CDK6 | Melanoma | 28351928 | (74) |
| Ribociclib (LEE011) | CDK4, CDK6 | Neuroblastoma | 28351928 | (74) |
| Ribociclib (LEE011) | CDK4, CDK6 | Neuroblastoma | 28432176 | (75) |
| Ribociclib (LEE011) | CDK4, CDK6 | Malignant rhabdoid tumor | 28432176 | (75) |
| Ribociclib (LEE011) | CDK4, CDK6 | Breast cancer | 29229752 | (76) |
| Ribociclib (LEE011) | CDK4, CDK6 | Breast cancer | 28356261 | (77) |
CDK, cyclin-dependent kinase; BRD, bromodomain protein.
Cyclin-dependent kinases (CDKs) belong to the serine/threonine kinase family, and play a crucial role in the regulation of the cell cycle and transcription process (78). It has been reported that CDK7 not only affects the cell cycle, but is also associated with the regulation of SE-driven oncogenes (79). In addition, CDK7 has recently become an attractive anti-cancer target, since CDK7 inhibitors (CDK7i) can reduce the levels of oncogenic TFs that act on SE regions (51). The anticancer effects of CDK7i are attributed to their effects on gene transcription or interference with the function of the SE; THZ1 is one of the most effective small-molecule inhibitors (79,80). Despite the meaningful effects of THZ1 in a variety of tumors, studies have shown that THZ1 inhibits myogenic differentiation, suggesting the possible side effects of THZ1 on muscle function during treatment (81). CDK7i may also be combined with other anti-tumor drugs to improve efficacy and reduce side effects. In diffuse intrinsic pontine glioma, CDK7i has been found to disrupt transcriptional regulation in cancer cells; however, the sensitivity of cancer cells to CDK7i is notably increased when combined with histone deacetylase inhibitors (79).
The bromodomain and extra terminal (BET) family of bromodomain proteins (BRDs) consists of 4 members (BRD2, BRD3, BRD4 and BRDT) that may represent a promising new target for the discovery of small-molecule drugs (82). BRDs can recognize histone acetylation and promote the expression of corresponding genes, such as the MYC oncogene (83). Specifically, BET bromodomain inhibitor (BETi) mainly inhibits the binding function of BRD4, one of the mediator complexes of SE, thereby suppressing the expression of SE-driven oncogenes and attenuating the proliferation of cancer cells (84). However, it is still controversial whether the function of BETi depends on the expression of the MYC gene. Certain studies found BETi to preferentially affect the expression of the SE-driven MYC oncogene in multiple myeloma and colon cancer, suggesting that BETi sensitivity is significantly associated with c-MYC gene levels (54,61). However, other studies reported contrasting results, as they did not observe a significant correlation between JQ1 sensitivity and c-MYC expression in colon cancer (29). Therefore, further research should be focused on maximizing the efficacy of BETi. In a study on pancreatic cancer, KDM6A was reported to mediate the abnormal activation of the SE regions of MYC and RUNX3 (58). When patients with pancreatic cancer, accompanied by KDM6A deficiency, were treated with BETi simultaneously, the selective sensitivity was observed. Severe BETi side effects have been reported in multiple phase I clinical trials, and include heart toxicity, gastrointestinal toxicity, anemia, diarrhea, fatigue, nausea, neutropenia and thrombocytopenia (70,85,86). For example, a BETi, I-BET-151, may even reduce the right and left ventricular fractional shortening, resulting in impaired heart function (87). The cytotoxicity of BETi caused serious side effects in clinical trials; however, the combination of this drug with vitamin C may largely alleviate those side effects (88).
In addition to the above two types of markedly effective and widely applicable drugs, there are other small-molecule drugs involved in blocking the function of SE. Attempting to abolish the SE function, the dynamic conversion between RACK7/KDM5C gene deletion and the accumulation of two SE formation-dependent methylation modifications (H3K4me1 and H3K4me3) provided a new approach to breast cancer therapy (89). Similarly, acetylation modifications have also been reported to be involved in the course of SE action. The IKAROS gene was usually undetectable in B cell line acute lymphoblastic leukemia; however, it has been demonstrated that the artificial overexpression of IKAROS gene can inhibit the expression of the MYC gene by interfering with the H3K27ac3 modification at the SE regions of MYC (90). PAX3 and PAX5 are the key TFs of the SE in pulmonary alveolar rhabdomyosarcoma and chronic lymphocytic leukemia, and PAX inhibitors have been found to exert prominent antitumor effects (91,92). Another study proposed a novel paradigm for NF-κB-mediated gene inhibition, in which cofactors are redistributed and enriched through the accumulation of NF-κB at the SE regions (93). In addition to the key molecules mentioned above, other, novel small molecules that are considered to be SE-dependent in chordomas have gradually emerged, such as CDK9, CDK12 and CDK13 (50). In chordomas, the IRS4/IGF2 protein can directly interact with the SE sequence, thereby inhibiting the SE function detected by 4C-Seq assay (50). This phenomenon has been termed ‘enhancer hijacking’ and is widely reported in cancer. The development of these small-molecule inhibitors also provides promising therapeutic approaches against cancer.
Most of these drugs have expectant clinical application prospects, and some have entered the clinical trial phase (Table II).
Table II.
Clinical trial information on various small molecule inhibitors.
| Small-molecule inhibitor | Clinical phase (clinical trial no.) | Disease |
|---|---|---|
| SY-1365 | Phase 1 (NCT03134638) | Ovarian cancer, breast cancer, advanced solid tumors |
| OTX015 | Phase 1 (NCT01713582) | Acute myeloid leukemia |
| OTX015 | Phase 1 (NCT01713582) | Diffuse large B-cell lymphoma |
| OTX015 | Phase 1 (NCT01713582) | Acute lymphoblastic leukemia |
| OTX015 | Phase 1 (NCT01713582) | Multiple myeloma |
| OTX015 | Phase 1 (NCT02259114) | NUT midline carcinoma |
| OTX015 | Phase 1 (NCT02259114) | Triple negative breast cancer |
| OTX015 | Phase 1 (NCT02259114) | Non-small cell lung cancer with rearranged ALK gene/fusion protein or KRAS mutation |
| OTX015 | Phase 1 (NCT02259114) | Castrate-resistant prostate cancer |
| OTX015 | Phase 1 (NCT02259114) | Castration-resistant prostate cancer |
| OTX015 | Phase 1 (NCT02259114) | Pancreatic ductal adenocarcinoma |
| OTX015 | Phase 2 (NCT02296476) | Glioblastoma multiforme |
| OTX015 | Phase 1 Phase 2 (NCT02303782) | Acute myeloid leukemia |
| OTX015 | Phase 1 (NCT02698176) | NUT midline carcinoma |
| OTX015 | Phase 1 (NCT02698176) | Triple negative breast cancer |
| OTX015 | Phase 1 (NCT02698176) | Non-small cell lung cancer |
| OTX015 | Phase 1 (NCT02698176) | Castration-resistant prostate cancer |
| CPI0610 | Phase 2 (NCT02986919) | Peripheral nerve tumors |
| CPI0610 | Phase 1 (NCT01949883) | Lymphoma |
| CPI0610 | Phase 1 (NCT02157636) | Multiple myeloma |
| CPI0610 | Phase 2 (NCT02158858) | Leukemia, myelocytic, acute |
| LEE011 | Phase 2 (NCT02934568) | Advanced cancer, advanced solid tumors |
Previous studies on antitumor drugs have focused on genomics, while drug design mainly focuses on the abnormal activation of proteins caused by mutations; however, this treatment is not without limitations (94). Drug resistance and low mutation frequency should not be overlooked in cancer (95). In recent years, with the development of epigenomics, methylation in the promoter region has been reported to regulate gene expression. Due to the common epigenetic modification reported in subsequent studies, designing drugs for this epigenetic mechanism is of great significance, which is considered as a better target (96). In the future, both universality and specificity should be taken into consideration in the direction of new drug design, such as the key process of SE regulation.
4. Gene-editing targeting SE for cancer treatment
The abnormal base insertion, base deletion and chromatin rearrangement in cancer cells can result in the formation of SE. CRISPR/Cas9 technology could be applied to abolish the SE formation caused by the aforementioned reasons. In addition, the objective of anticancer treatment can be achieved through the CRISPR/Cas9 system.
The CRISPR/Cas9 knockout system targets specific SE regions to cut DNA sequences, causing non-homologous recombination repair, thereby abolishing the SE function. RUNX1 is a transcription factor that regulates normal and malignant blood cell production. The disruption the SE regions of RUNX1 gene by the CRISPR/Cas9 system was shown to increase apoptosis in acute leukemia cells, and subsequently alter the survival of mice with acute myeloid leukemia (AML) (97). It was observed that, in T-cell acute lymphoblastic leukemia (T-ALL) primary samples and cell lines, an indel mutation occurred at the hotspot 7.5 kb upstream the transcription initiation site of the T-cell acute leukemia 1 (TAL1) gene and contributed to the formation of the MYB binding site and SE, thereby resulting in the upregulation of oncogenes (98). Following the knockout of the abnormally inserted bases by the CRISPR/Cas9 system, SE formation and TAL1 gene overexpression were absent in ALL (98). The transcription activator-like effector and CRISPR/Cas9 genome editing systems may also be used to abolish the activation of abnormal enhancers in AML cells. Subsequent experiments confirmed that gene-editing acting on enhancer efficiently inhibited the expression of ecotropic viral integration site 1 and cancer growth (99).
The CRISPR/Cas9 knock-in system is also promising in the SE-driven oncogene expression pattern associated with single-nucleotide polymorphisms (SNPs). It has been demonstrated that most trait-associated SNPs occur in non-coding regions, with 64% occurring in the disease-associated SE regions defined by H3K27ac (7). Subsequent studies revealed one underlying mechanism through which SNPs located within the SE regions could affect gene expression (100,101). SNP rs6854845 is considered to be one of the risk factors for colon cancer (102). In colon cells, it was found that SNP rs6854845 formed in the SE region and affected the shifted enrichment of H3K4me1 and H3K27ac at the SE regions, further affecting the expression of SE-driven genes (103). The pathogenic SNP locus can be verified by expression quantitative trait locus, genomic chromatin interaction (high-throughput chromosome conformation capture), epigenetic annotation, and a series of functional assays (104). Therefore, the site-specific genome editing of SNPs by CRISPR/Cas9 can correct the multiple pathogenic changes in cells by reversing the interaction between SNPs and SE-associated genes.
Although the CRISPR technology is known for its effectiveness and versatility, it has two major drawbacks: The inability to arbitrarily edit bases and its off-target effects (105). The CRISPR/Cas9 system relies on the recognition of protospacer-adjacent motif (PAM) sites by single guide RNA to perform DNA shear, so the system can only edit DNA near the PAM site and cannot edit bases at any regions (106). As an important member of the CRISPR/Cas9 gene editing system, the Cas enzyme may cleave non-targeting sites after its introduction into cells, causing off-target effects (107). In addition, CRISPR/Cas9 gene editing relies on DNA double-stranded breaks, which may lead to an unpredictable disruption of cells following gene editing (108). These side effects of CRISPR/Cas9 limit their application in basic research and medicine, and may trigger safety issues.
A new precise gene editing tool, the Prime Editor, has been developed which can effectively convert all 12 single bases without relying on DNA templates and accurately insert or delete several bases (109). The Prime Editor system acts by combining Cas9 and reverse transcriptase into a complex, which is then brought to the specific DNA region to insert a new DNA sequence by the prime editing guide RNA (pegRNA) (109). This new method aims at improving the traditional cas9 enzyme and pegRNA to eliminate the 2 defects of the traditional CRISPR system. Through this new system, experiments in mouse cells have successfully repaired gene mutations that cause sickle cell anemia and Tay-Sachs disease (109). Sickle cell anemia is an autosomal dominant genetic disease, in which a single base mutation of A to T occurs in a gene encoding hemoglobin, resulting in the 6th amino acid glutamate of the hemoglobin β-peptide chain becoming valine, which makes the sickle hemoglobin replace the normal hemoglobin (110). Tay-Sachs disease is an autosomal recessive disorder in which the HEXA gene is mutated to an extra 4 bases, resulting in the inactivation of the lipolytic enzyme encoded by the HEXA gene, which in turn leads to the aggregation of gangliosides in the brain with ensuing toxicity (111). These genetic diseases could not be cured by traditional genome editing system, while the improved prime editor may prove more effective.
Although extensive animal and clinical experiments are required to further validate its safety and effectiveness, gene-editing therapy is already a promising method for tumor therapy. In conclusion, gene editing techniques are constantly being improved, and the prospects of the clinical application of these technologies in the near future are encouraging.
5. Discussion
SE is a controversial topic in current clinical and basic research, with great attention paid to its functions and potential therapeutic prospects in the clinic. With the improvement and breakthroughs made in next-generation sequencing technology, a more detailed and comprehensive understanding of the genome has emerged. By using the CRISPR/Cas9 system to interfere with the potential SE formation, the SE can be blocked to regulate the SE-driven oncogenes to avoid tumorigenesis. However, due to the off-target effects of the CRIPR/Cas9 system, it is impossible to accurately identify 100% of the human genome. Gene-editing at any non-target sites is associated with clinical risks. To date, the continuous improvement of CRISPR/Cas9 tools has not been effective in resolving the problem of off-target effects, causing great concerns regarding the safety of its clinical applications. The activation of SE is tightly linked to the interaction of certain key TFs. It has been reported that these key TFs play a critical role in cancer and further interference with small-molecule inhibitors also suppresses cancer cells (5). Since cancer cells are highly sensitive to SE-associated complexes, the inhibition of SE-derived gene expression is conceived as a safer and more promising therapeutic approach to cancer. The newly improved CAPTURE-Proteomics (ChIP-seq) technique is based on the core elements of CRISPR and adds biotin ligase BirA to improve the sensitivity and specificity of the experiment, which can more accurately detect the protein complex that binds to the SE site in order to find a new target for clinical small-molecule inhibitor therapy (112). Despite of the unclear mechanisms of SE and SE-associated complexes regulating the genome, the targeted therapy for SE-associated complexes remains one of the directions of treatment. With the persistent efforts focused on SE-associated complexes, greater breakthroughs are expected in the near future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81702789).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CZ conceived the study and was a major contributor to the manuscript. CZ and ML wrote the manuscript. HF revised the manuscript for important intellectual content. CZ and ML jointly collected relevant references. HF guided the research framework and approved the final version for publication. All the authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-X. [DOI] [PubMed] [Google Scholar]
- 2.Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 3.Rickels R, Shilatifard A. Enhancer Logic and mechanics in development and disease. Trends Cell Biol. 2018;28:608–630. doi: 10.1016/j.tcb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 7.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christensen CL, Kwiatkowski N, Abraham BJ, Carretero J, Al-Shahrour F, Zhang T, Chipumuro E, Herter-Sprie GS, Akbay EA, Altabef A, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang FH, Law CT, Tang TC, Cheng CL, Chin DW, Tam WV, Wei L, Wong CC, Ng IO, Wong CM. Aberrant super-enhancer landscape in human hepatocellular carcinoma. Hepatology. 2019;69:2502–2517. doi: 10.1002/hep.30544. [DOI] [PubMed] [Google Scholar]
- 10.Xie JJ, Jiang YY, Jiang Y, Li CQ, Lim MC, An O, Mayakonda A, Ding LW, Long L, Sun C, et al. Super-Enhancer-driven long non-coding RNA LINC01503, regulated by tp63, is over-expressed and oncogenic in squamous cell carcinoma. Gastroenterology. 2018;154:2137–2151. doi: 10.1053/j.gastro.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, Zhang C, Zhang Y, Lin Z, Li J, et al. Super-Enhancer-associated long noncoding RNA HCCL5 is activated by zeb1 and promotes the malignancy of hepatocellular carcinoma. Cancer Res. 2019;79:572–584. doi: 10.1158/0008-5472.CAN-18-0367. [DOI] [PubMed] [Google Scholar]
- 12.Lin L, Huang M, Shi X, Mayakonda A, Hu K, Jiang YY, Guo X, Chen L, Pang B, Doan N, et al. Super-enhancer-associated MEIS1 promotes transcriptional dysregulation in Ewing sarcoma in co-operation with EWS-FLI1. Nucleic Acids Res. 2019;47:1255–1267. doi: 10.1093/nar/gky1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Dang L, Zheng X, Lu Y, Lu Y, Ji R, Zhang T, Ruan X, Zhi J, Hou X, et al. Targeting super-enhancer-driven oncogenic transcription by cdk7 inhibition in anaplastic thyroid carcinoma. Thyroid. 2019;29:809–823. doi: 10.1089/thy.2018.0550. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Hu G. Remodeling super-enhancers and oncogenic transcription. Cell Cycle. 2016;15:3157–3158. doi: 10.1080/15384101.2016.1220715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma M, Ru Y, Chuang LS, Hsu NY, Shi LS, Hakenberg J, Cheng WY, Uzilov A, Ding W, Glicksberg BS, et al. Disease-associated variants in different categories of disease located in distinct regulatory elements. BMC Genomics. 2015;16(Suppl 8):S3. doi: 10.1186/1471-2164-16-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824–7827. doi: 10.1073/pnas.79.24.7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ar-Rushdi A, Nishikura K, Erikson J, Watt R, Rovera G, Croce CM. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983;222:390–393. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]
- 19.Erikson J, Ar-Rushdi A, Drwinga HL, Nowell PC, Croce CM. Transcriptional activation of the translocated c-myc oncogene in burkitt lymphoma. Proc Natl Acad Sci USA. 1983;80:820–824. doi: 10.1073/pnas.80.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubota S, Tokunaga K, Umezu T, Yokomizo-Nakano T, Sun Y, Oshima M, Tan KT, Yang H, Kanai A, Iwanaga E, et al. Lineage-specific RUNX2 super-enhancer activates MYC and promotes the development of blastic plasmacytoid dendritic cell neoplasm. Nat Commun. 2019;10:1653. doi: 10.1038/s41467-019-09710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sengupta S, George RE. Super-Enhancer-driven transcriptional dependencies in cancer. Trends Cancer. 2017;3:269–281. doi: 10.1016/j.trecan.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo H, Triandafillou C, Drummond DA. Cellular sensing by phase separation: Using the process, not just the products. J Biol Chem. 2019;294:7151–7159. doi: 10.1074/jbc.TM118.001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang JT, Smith J, Chen BC, Schmidt H, Rasoloson D, Paix A, Lambrus BG, Calidas D, Betzig E, Seydoux G. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. Elife. 2014;3:e4591. doi: 10.7554/eLife.04591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 28.Milin AN, Deniz AA. Reentrant phase transitions and non-equilibrium dynamics in membraneless organelles. Biochemistry. 2018;57:2470–2477. doi: 10.1021/acs.biochem.8b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura Y, Hattori N, Iida N, Yamashita S, Mori A, Kimura K, Yoshino T, Ushijima T. Targeting of super-enhancers and mutant BRAF can suppress growth of BRAF-mutant colon cancer cells via repression of MAPK signaling pathway. Cancer Lett. 2017;402:100–109. doi: 10.1016/j.canlet.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 30.Gunnell A, Webb HM, Wood CD, McClellan MJ, Wichaidit B, Kempkes B, Jenner RG, Osborne C, Farrell PJ, West MJ. RUNX super-enhancer control through the Notch pathway by Epstein-Barr virus transcription factors regulates B cell growth. Nucleic Acids Res. 2016;44:4636–4650. doi: 10.1093/nar/gkw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ke L, Zhou H, Wang C, Xiong G, Xiang Y, Ling Y, Khabir A, Tsao GS, Zeng Y, Zeng M, et al. Nasopharyngeal carcinoma super-enhancer-driven ETV6 correlates with prognosis. Proc Natl Acad Sci USA. 2017;114:9683–9688. doi: 10.1073/pnas.1705236114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dave K, Sur I, Yan J, Zhang J, Kaasinen E, Zhong F, Blaas L, Li X, Kharazi S, Gustafsson C, et al. Mice deficient of Myc super-enhancer region reveal differential control mechanism between normal and pathological growth. Elife. 2017;6:e23382. doi: 10.7554/eLife.23382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai B, Lee JE, Jang Y, Wang L, Peng W, Ge K. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 2017;45:6388–6403. doi: 10.1093/nar/gkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet. 2015;47:1149–1157. doi: 10.1038/ng.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki HI, Young RA, Sharp PA. Super-enhancer-mediated RNA processing revealed by integrative microrna network analysis. Cell. 2017;168:1000–1014.e15. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang J, Zhou H, Gerdt C, Tan M, Colson T, Kaye KM, Kieff E, Zhao B. Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc Natl Acad Sci USA. 2016;113:14121–14126. doi: 10.1073/pnas.1616697113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ounzain S, Pedrazzini T. Super-enhancer lncs to cardiovascular development and disease. Biochim Biophys Acta. 2016;1863:1953–1960. doi: 10.1016/j.bbamcr.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 38.Peng L, Jiang B, Yuan X, Qiu Y, Peng J, Huang Y, Zhang C, Zhang Y, Lin Z, Li J, et al. Super-Enhancer-associated long noncoding RNA HCCL5 Is activated by zeb1 and promotes the malignancy of hepatocellular carcinoma. Cancer Res. 2019;79:572–584. doi: 10.1158/0008-5472.CAN-18-0367. [DOI] [PubMed] [Google Scholar]
- 39.Fontanals-Cirera B, Hasson D, Vardabasso C, Di Micco R, Agrawal P, Chowdhury A, Gantz M, de Pablos-Aragoneses A, Morgenstern A, Wu P, et al. Harnessing BET inhibitor sensitivity reveals AMIGO2 as a melanoma survival gene. Mol Cell. 2017;68:731–744.e9. doi: 10.1016/j.molcel.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da SD, Caruso BT, Arefolov A, Fadeyi O, Christie AL, et al. Mediator kinase inhibition further activates super-enhancer- associated genes in AML. Nature. 2015;526:273–276. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.See YX, Wang BZ, Fullwood MJ. Chromatin interactions and regulatory elements in cancer: From bench to bedside. Trends Genet. 2019;35:145–158. doi: 10.1016/j.tig.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Enomoto K, Zhao L, Zhu YJ, Willingham MC, Meltzer P, Qi J, Cheng SY. Bromodomain and extraterminal protein inhibitor JQ1 suppresses thyroid tumor growth in a mouse model. Clin Cancer Res. 2017;23:430–440. doi: 10.1158/1078-0432.CCR-16-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augert A, MacPherson D. Treating transcriptional addiction in small cell lung cancer. Cancer Cell. 2014;26:783–784. doi: 10.1016/j.ccell.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Gerlach D, Tontsch-Grunt U, Baum A, Popow J, Scharn D, Hofmann MH, Engelhardt H, Kaya O, Beck J, Schweifer N, et al. The novel BET bromodomain inhibitor BI 894999 represses super-enhancer-associated transcription and synergizes with CDK9 inhibition in AML. Oncogene. 2018;37:2687–2701. doi: 10.1038/s41388-018-0150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, Zhao Z, Huang Z, Chen DC, Zhu XX, Wang YZ, Yan YW, Tang S, Madhavan S, Ni W, et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11. doi: 10.1038/s41413-018-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitazono M, Chuman Y, Aikou T, Fojo T. Construction of gene therapy vectors targeting thyroid cells: Enhancement of activity and specificity with histone deacetylase inhibitors and agents modulating the cyclic adenosine 3′,5′-monophosphate pathway and demonstration of activity in follicular and anaplastic thyroid carcinoma cells. J Clin Endocrinol Metab. 2001;86:834–840. doi: 10.1210/jcem.86.2.7196. [DOI] [PubMed] [Google Scholar]
- 47.Wong R, Ngoc P, Leong WZ, Yam A, Zhang T, Asamitsu K, Iida S, Okamoto T, Ueda R, Gray NS, et al. Enhancer profiling identifies critical cancer genes and characterizes cell identity in adult T-cell leukemia. Blood. 2017;130:2326–2338. doi: 10.1182/blood-2017-06-792184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eliades P, Abraham BJ, Ji Z, Miller DM, Christensen CL, Kwiatkowski N, Kumar R, Njauw CN, Taylor M, Miao B, et al. High MITF expression is associated with super-enhancers and suppressed by CDK7 inhibition in melanoma. J Invest Dermatol. 2018;138:1582–1590. doi: 10.1016/j.jid.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwiatkowski N, Zhang T, Rahl PB, Abraham BJ, Reddy J, Ficarro SB, Dastur A, Amzallag A, Ramaswamy S, Tesar B, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharifnia T, Wawer MJ, Chen T, Huang QY, Weir BA, Sizemore A, Lawlor MA, Goodale A, Cowley GS, Vazquez F, et al. Small-molecule targeting of brachyury transcription factor addiction in chordoma. Nat Med. 2019;25:292–300. doi: 10.1038/s41591-018-0312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu S, Marineau JJ, Rajagopal N, Hamman KB, Choi YJ, Schmidt DR, Ke N, Johannessen L, Bradley MJ, Orlando DA, et al. Discovery and characterization of SY-1365, a selective, covalent inhibitor of CDK7. Cancer Res. 2019;79:3479–3491. doi: 10.1158/0008-5472.CAN-19-0119. [DOI] [PubMed] [Google Scholar]
- 52.Dooley KE, Warburton A, McBride AA. Tandemly Integrated HPV16 Can Form a Brd4-dependent super-enhancer-like element that drives transcription of viral oncogenes. Mbio. 2016;7:e01446–e01416. doi: 10.1128/mBio.01446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sengupta D, Kannan A, Kern M, Moreno MA, Vural E, Stack BJ, Suen JY, Tackett AJ, Gao L. Disruption of BRD4 at H3K27Ac-enriched enhancer region correlates with decreased c-Myc expression in Merkel cell carcinoma. Epigenetics. 2015;10:460–466. doi: 10.1080/15592294.2015.1034416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuber V, Bettella F, Witoelar A, Andreassen OA, Mills IG, Urbanucci A. Bromodomain protein 4 discriminates tissue-specific super-enhancers containing disease-specific susceptibility loci in prostate and breast cancer. BMC Genomics. 2017;18:270. doi: 10.1186/s12864-017-3620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MG, Qi J, Rahl PB, Sun HH, Yeda KT, Doench JG, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong L, Wu F, Wu Q, Xu L, Cheung OK, Kang W, Mok MT, Szeto L, Lun CY, Lung RW, et al. Aberrant enhancer hypomethylation contributes to hepatic carcinogenesis through global transcriptional reprogramming. Nat Commun. 2019;10:335. doi: 10.1038/s41467-018-08245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andricovich J, Perkail S, Kai Y, Casasanta N, Peng W, Tzatsos A. Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to bet inhibitors. Cancer Cell. 2018;33:512–526.e8e0. doi: 10.1016/j.ccell.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gelato KA, Schöckel L, Klingbeil O, Rückert T, Lesche R, Toedling J, Kalfon E, Heroult M, Lejeune P, Mönning U, et al. Super-enhancers define a proliferative PGC-1alpha-expressing melanoma subgroup sensitive to BET inhibition. Oncogene. 2018;37:512–521. doi: 10.1038/onc.2017.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tasdemir N, Banito A, Roe JS, Alonso-Curbelo D, Camiolo M, Tschaharganeh DF, Huang CH, Aksoy O, Bolden JE, Chen CC, et al. BRD4 Connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6:612–629. doi: 10.1158/2159-8290.CD-16-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Togel L, Nightingale R, Chueh AC, Jayachandran A, Tran H, Phesse T, Wu R, Sieber OM, Arango D, Dhillon AS, et al. Dual targeting of bromodomain and extraterminal domain proteins and WNT or MAPK signaling, inhibits c-MYC expression and proliferation of colorectal cancer cells. Mol Cancer Ther. 2016;15:1217–1226. doi: 10.1158/1535-7163.MCT-15-0724. [DOI] [PubMed] [Google Scholar]
- 62.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJ, Kluk MJ, Carey CD, Rodig SJ, Sholl LM, Afrogheh AH, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet. 2016;48:265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ceribelli M, Hou ZE, Kelly PN, Huang DW, Wright G, Ganapathi K, Evbuomwan MO, Pittaluga S, Shaffer AL, Marcucci G, et al. A Druggable TCF4- and BRD4-Dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell. 2016;30:764–778. doi: 10.1016/j.ccell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Duan Z, Guo W, Zeng L, Wu Y, Chen Y, Tai F, Wang Y, Lin Y, Zhang Q, et al. Targeting the BRD4/FOXO3a/CDK6 axis sensitizes AKT inhibition in luminal breast cancer. Nat Commun. 2018;9:5200. doi: 10.1038/s41467-018-07258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Ma P, Jing Y, Yan Y, Cai MC, Zhang M, Zhang S, Peng H, Ji ZL, Di W, et al. BET Bromodomain Inhibition as a therapeutic strategy in ovarian cancer by downregulating FoxM1. Theranostics. 2016;6:219–230. doi: 10.7150/thno.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mottok A, Gascoyne RD. Bromodomain inhibition in diffuse large B-cell lymphoma-giving MYC a brake. Clin Cancer Res. 2015;21:4–6. doi: 10.1158/1078-0432.CCR-14-1651. [DOI] [PubMed] [Google Scholar]
- 67.Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. 2014;33:2928–2937. doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS, Suzuki Y, Pappin DJ, Joshua-Tor L, Vakoc CR. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell. 2015;60:847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan KH, Zengerle M, Testa A, Ciulli A. Impact of target warhead and linkage vector on inducing protein degradation: Comparison of bromodomain and extra-terminal (BET) degraders derived from triazolodiazepine (JQ1) and tetrahydroquinoline (I-BET726) BET inhibitor scaffolds. J Med Chem. 2018;61:504–513. doi: 10.1021/acs.jmedchem.6b01912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, et al. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: A dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol. 2016;3:e196–e204. doi: 10.1016/S2352-3026(16)00021-1. [DOI] [PubMed] [Google Scholar]
- 71.Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y, Nasveschuk CG, Bellon S, Bergeron L, Campbell R, et al. Identification of a Benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J Med Chem. 2016;59:1330–1339. doi: 10.1021/acs.jmedchem.5b01882. [DOI] [PubMed] [Google Scholar]
- 72.Siu KT, Ramachandran J, Yee AJ, Eda H, Santo L, Panaroni C, Mertz JA, Sims IR, Cooper MR, Raje N. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia. 2017;31:1760–1769. doi: 10.1038/leu.2016.355. [DOI] [PubMed] [Google Scholar]
- 73.Zhao L, Okhovat JP, Hong EK, Kim YH, Wood GS. Preclinical studies support combined inhibition of bet family proteins and histone deacetylases as epigenetic therapy for cutaneous t-cell lymphoma. Neoplasia. 2018;21:82–92. doi: 10.1016/j.neo.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tripathy D, Bardia A, Sellers WR. Ribociclib (LEE011): Mechanism of action and clinical impact of this selective cyclin-dependent kinase 4/6 Inhibitor in various solid tumors. Clin Cancer Res. 2017;23:3251–3262. doi: 10.1158/1078-0432.CCR-16-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geoerger B, Bourdeaut F, DuBois SG, Fischer M, Geller JI, Gottardo NG, Marabelle A, Pearson A, Modak S, Cash T, et al. A phase I study of the CDK4/6 inhibitor ribociclib (LEE011) in pediatric patients with malignant rhabdoid tumors, neuroblastoma and other solid tumors. Clin Cancer Res. 2017;23:2433–2441. doi: 10.1158/1078-0432.CCR-16-2898. [DOI] [PubMed] [Google Scholar]
- 76.Ribociclib extends survival in HR+ breast cancer. Cancer Discov. 2018;8:OF5. doi: 10.1158/2159-8290.CD-NB2017-175. [DOI] [PubMed] [Google Scholar]
- 77.Ribociclib approved for advanced breast cancer. Cancer Discov. 2017;7:OF3. doi: 10.1158/2159-8290.CD-NB2017-043. [DOI] [PubMed] [Google Scholar]
- 78.Cheng W, Yang Z, Wang S, Li Y, Wei H, Tian X, Kan Q. Recent development of CDK inhibitors: An overview of CDK/inhibitor co-crystal structures. Eur J Med Chem. 2019;164:615–639. doi: 10.1016/j.ejmech.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 79.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, Li M, Meng W, Ponnuswami A, Sun W, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 31:635–652. doi: 10.1016/j.ccell.2017.03.011. 2017.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cayrol F, Praditsuktavorn P, Fernando TM, Kwiatkowski N, Marullo R, Calvo-Vidal MN, Phillip J, Pera B, Yang SN, Takpradit K, et al. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat Commun. 2017;8:14290. doi: 10.1038/ncomms14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma X, Kuang X, Xia Q, Huang Z, Fan Y, Ning J, Wen J, Zhang H, Yan J, Zhang Q, et al. Covalent CDK7 inhibitor THZ1 inhibits myogenic differentiation. J Cancer. 2018;9:3149–3155. doi: 10.7150/jca.25395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ali I, Choi G, Lee K. BET inhibitors as anticancer agents: A patent review. Recent Pat Anticancer Drug Discov. 2017;12:340–364. doi: 10.2174/1574892812666170808121228. [DOI] [PubMed] [Google Scholar]
- 83.Henssen A, Althoff K, Odersky A, Beckers A, Koche R, Speleman F, Schafers S, Bell E, Nortmeyer M, Westermann F, et al. Targeting MYCN-Driven Transcription By BET-Bromodomain inhibition. Clin Cancer Res. 2016;22:2470–2481. doi: 10.1158/1078-0432.CCR-15-1449. [DOI] [PubMed] [Google Scholar]
- 84.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: Going beyond transcriptional regulation. Mol Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, et al. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. Lancet Haematol. 2016;3:e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 86.Stathis A, Zucca E, Bekradda M, Gomez-Roca C, Delord JP, de La Motte RT, Uro-Coste E, de Braud F, Pelosi G, French CA. Clinical response of carcinomas harboring the BRD4-NUT oncoprotein to the targeted bromodomain inhibitor OTX015/MK-8628. Cancer Discov. 2016;6:492–500. doi: 10.1158/2159-8290.CD-15-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Piquereau J, Boet A, Pechoux C, Antigny F, Lambert M, Gressette M, Ranchoux B, Gambaryan N, Domergue V, Mumby S, et al. The BET Bromodomain Inhibitor I-BET-151 induces structural and functional alterations of the heart mitochondria in healthy male mice and rats. Int J Mol Sci. 2019;20:1527. doi: 10.3390/ijms20071527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mustafi S, Camarena V, Qureshi R, Yoon H, Volmar CH, Huff TC, Sant DW, Zheng L, Brothers SP, Wahlestedt C, et al. Vitamin C supplementation expands the therapeutic window of BETi for triple negative breast cancer. Ebiomedicine. 2019;43:201–210. doi: 10.1016/j.ebiom.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen H, Xu W, Guo R, Rong B, Gu L, Wang Z, He C, Zheng L, Hu X, Hu Z, et al. Suppression of enhancer overactivation by a RACK7-histone demethylase complex. Cell. 2016;165:331–342. doi: 10.1016/j.cell.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katerndahl C, Heltemes-Harris LM, Willette M, Henzler CM, Frietze S, Yang R, Schjerven H, Silverstein K, Ramsey LB, Hubbard G, et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat Immunol. 2017;18:694–704. doi: 10.1038/ni.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, Schaefer B, Sen N, Song Y, Gualtieri A, et al. PAX3-FOXO1 establishes myogenic super enhancers and confers BET bromodomain vulnerability. Cancer Discov. 2017;7:884–899. doi: 10.1158/2159-8290.CD-16-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ott CJ, Federation AJ, Schwartz LS, Kasar S, Klitgaard JL, Lenci R, Li Q, Lawlor M, Fernandes SM, Souza A, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell. 2018;34:982–995.e7e0. doi: 10.1016/j.ccell.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schmidt SF, Larsen BD, Loft A, Nielsen R, Madsen JG, Mandrup S. Acute TNF-induced repression of cell identity genes is mediated by NF κB-directed redistribution of cofactors from super-enhancers. Genome Res. 2015;25:1281–1294. doi: 10.1101/gr.188300.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Biswas S, Rao CM. Epigenetics in cancer: Fundamentals and beyond. Pharmacol Ther. 2017;173:118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 95.Dzobo K, Senthebane DA, Thomford NE, Rowe A, Dandara C, Parker MI. Not everyone fits the mold: Intratumor and intertumor heterogeneity and innovative cancer drug design and development. Omics. 2018;22:17–34. doi: 10.1089/omi.2017.0174. [DOI] [PubMed] [Google Scholar]
- 96.Kelly AD, Issa JJ. The promise of epigenetic therapy: Reprogramming the cancer epigenome. Curr Opin Genet Dev. 2017;42:68–77. doi: 10.1016/j.gde.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 97.Mill CP, Fiskus W, DiNardo CD, Qian Y, Raina K, Rajapakshe K, Perera D, Coarfa C, Kadia TM, Khoury JD, et al. RUNX1 targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood. 2019;134:59–73. doi: 10.1182/blood.2018893982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gröschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman B, Erpelinck C, van der Velden V, Havermans M, Avellino R, van Lom K, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 100.Marsman J, Gimenez G, Day RC, Horsfield JA, Jones GT. A non-coding genetic variant associated with abdominal aortic aneurysm alters ERG gene regulation. Hum Mol Genet. 2020;29:554–565. doi: 10.1093/hmg/ddz256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kleinstern G, Yan H, Hildebrandt M, Vijai J, Berndt SI, Ghesquieres H, McKay J, Wang SS, Nieters A, Ye Y, et al. Inherited variants at 3q13.33 and 3p24.1 are associated with risk of diffuse large B-cell lymphoma and implicate immune pathways. Hum Mol Genet. 2020;29:70–79. doi: 10.1093/hmg/ddz228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He Y, Timofeeva M, Li X, Din F, Blackmur JP, Vaughan-Shaw P, Svinti V, Farrington SM, Campbell H, Dunlop MG, et al. A comprehensive study of the effect on colorectal cancer survival of common germline genetic variation previously linked with cancer prognosis. Cancer Epidemiol Biomarkers Prev. 2019;28:1944–1946. doi: 10.1158/1055-9965.EPI-19-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cong Z, Li Q, Yang Y, Guo X, Cui L, You T. The SNP of rs6854845 suppresses transcription via the DNA looping structure alteration of super-enhancer in colon cells. Biochem Biophys Res Commun. 2019;514:734–741. doi: 10.1016/j.bbrc.2019.04.190. [DOI] [PubMed] [Google Scholar]
- 104.Zhu DL, Chen XF, Hu WX, Dong SS, Lu BJ, Rong Y, Chen YX, Chen H, Thynn HN, Wang NN, et al. Multiple functional variants at 13q14 risk locus for osteoporosis regulate RANKL expression through long-range super-enhancer. J Bone Miner Res. 2018;33:1335–1346. doi: 10.1002/jbmr.3419. [DOI] [PubMed] [Google Scholar]
- 105.Eid A, Alshareef S, Mahfouz MM. CRISPR base editors: Genome editing without double-stranded breaks. Biochem J. 2018;475:1955–1964. doi: 10.1042/BCJ20170793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell. 2016;62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 108.Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fernandes Q. Therapeutic strategies in Sickle Cell Anemia: The past present and future. Life Sci. 2017;178:100–108. doi: 10.1016/j.lfs.2017.03.025. [DOI] [PubMed] [Google Scholar]
- 111.Vu M, Li R, Baskfield A, Lu B, Farkhondeh A, Gorshkov K, Motabar O, Beers J, Chen G, Zou J, et al. Neural stem cells for disease modeling and evaluation of therapeutics for Tay-Sachs disease. Orphanet J Rare Dis. 2018;13:152. doi: 10.1186/s13023-018-0886-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, Cao H, Ni M, Liu Y, Gu Z, et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170:1028–1043.e19e0. doi: 10.1016/j.cell.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.