Abstract
A recent study reported that zinc finger protein (ZNF)281 is a tumor-suppressive long non-coding (lnc)RNA in glioma. The present study investigated the role of ZNF281 in non-small cell lung cancer (NSCLC). ZNF281 expression in paired cancer and non-cancerous tissues from patients with NSCLCs was analyzed by RNA extraction and reverse transcription-quantitative-PCR. A 5-year follow up on patients was performed to analyze the prognostic value of ZNF281 for NSCLC. Cell transfections of ZNF281 or phosphatase and tensin homolog (PTEN) expression vector and microRNA (miR)-221 mimic were performed to analyze the relationship between ZNF281, miR-221 and PTEN. Cell apoptosis and proliferation were analyzed using Cell Counting Kit-8 and flow cytometry, respectively. In patients with NSCLC, expression levels of ZNF281 were significantly lower in cancer tissues compared with in non-cancerous tissues, and lower levels of ZNF281 expression in cancerous tissues predicted poor survival. In NSCLC cells, ZNF281 overexpression resulted in upregulated PTEN and downregulated miR-221 expression, whereas cells with miR-221 overexpression exhibited downregulated PTEN expression and unaffected ZNF281 expression. In addition, ZNF281 and PTEN overexpression resulted in accelerated cell apoptosis and inhibited the cell proliferation of NSCLC cells. Notably, miR-221 overexpression exhibited an opposite effect and attenuated the functions of ZNF281 and PTEN overexpression. Therefore, ZNF281 may upregulate PTEN via downregulation of miR-221 in NSCLC, resulting in inhibition of cancer cell proliferation and the promotion of apoptosis.
Keywords: ZNF281, non-small cell lung cancer, microRNA-221, PTEN, survival
Introduction
Despite recent advances in cancer prevention, lung cancer remains the most common malignant cancer type and the leading cause of cancer-associated mortality worldwide (1). Overall, the 5-year survival rate of patients with lung cancer at all stages is ~15%, and this survival rate has not been significantly improved over previous years (2). The high mortality rate is primarily a result of the lack of tools available to facilitate early diagnosis and the lack of curative therapies (3). Non-small cell lung cancer (NSCLC) is the major subtype of lung cancer and accounts for >85% of all cases (4). Smoking is a major risk factor of NSCLC; however, this disease also affects non-smokers (5). In addition, smoking itself is insufficient to induce the occurrence of NSCLC (5).
Besides smoking, the tumorigenesis and progression of NSCLC are closely associated with certain genetic factors (6,7). Phosphatase and tensin homolog (PTEN) is a tumor-suppressive gene with pivotal roles in cell cycle regulation (8). In cancer biology, PTEN inhibits cancer cells from proliferating rapidly, predominantly via inhibition of the PI3K/AKT signaling pathway (9). Certain oncogenic microRNAs (miR/miRNA), such as miR-221, target PTEN to promote cancer progression (10). In a recent study, Li et al (11) identified a novel long non-coding RNA (lncRNA), zinc finger protein (ZNF)281, which serves a tumor-suppressive role in glioma via the inhibition of the NF-κB signaling pathway, and it has been revealed that the NF-κB signaling pathway interacts directly with miR-221 (12). Therefore, ZNF281 may interact with miR-221. The present study aimed to investigate the interactions between ZNF281 and miR-221 and the consequent effects on PTEN.
Materials and methods
Patients
A total of 66 patients were selected from the 182 patients with NSCLC admitted to Hiser Medical Center of Qingdao (Shandong, China) between January 2012 and April 2014. The present study was approved by the Review Board of Hiser Medical Center and the Qingdao Ethics Committee. The 66 patients with NSCLC comprised 30 cases of squamous cell carcinoma and 36 cases of adenocarcinoma. The inclusion criteria were as follows: i) Diagnosed for the first time; and ii) no treatment had been received before admission. The exclusion criteria were as follows: i) Recurrent NSCLC; ii) clinical disorders other than NSCLC were diagnosed; iii) therapy had already been initiated; and iv) patients who failed to complete the follow-up or who died from other diseases or accidents. According to the clinical findings, the 66 patients included 6, 19, 20 and 21 cases at clinical stage I–IV, respectively (13). According to cancer histologic grade, there were 14, 19, 20 and 13 cases at grade 1–4, respectively (14). All patients were informed of the principle of the present study. Written informed consent was provided by all 66 patients.
Follow-up
Starting from the day of admission, all 66 patients were followed-up for 5 years. Their survival conditions were monitored and recorded through monthly telephone calls.
NSCLC cells and tissues
The H1993 human NSCLC cell line (American Type Culture Collection) was used in the present study. Cells were cultured in a mixture of 90% RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) and 10% FBS (Sigma-Aldrich; Merck KGaA) supplemented with 1% penicillin-streptomycin (Sigma-Aldrich; Merck KGaA). Cell were cultured at 37°C with 5% CO2 and 95% humidity.
All 66 patients with NSCLC received lung biopsy. During biopsy, adjacent (≤2 cm from tumor) non-tumorous lung tissues and NSCLC tissues were obtained from each patient. Based on histopathological examination results, all non-tumor tissues contained <1% cancerous cells, and all NSCLC tissues contained >98% cancerous cells. Fresh tissues were stored in liquid nitrogen.
Cell transfections
Expression vectors of ZNF281 and PTEN were constructed using pcDNA3.1 (Sangon Biotech Co., Ltd.). Negative control (NC) miRNA (5′-UGUGGUUACGAUCGUGGGAACUG-3′) and miR-221 (5′-ACCUGGCAUACAAUGUAGAUUU-3′) were purchased from Guangzhou RiboBio Co., Ltd. Prior to transfections, H1993 cells were harvested at a confluency of 70–80%. Lipofectamine 2000® (Sangon Biotech Co., Ltd.) was used to transfect 40 nM miRNA (NC miRNA as NC group) or 10 nM vector (empty vector as NC group) into 1×106 cells. Untransfected cells were used as the control (C) group. Cells were harvested at 24 h post-transfection to perform all subsequent experiments.
RNA extraction and reverse transcription-quantitative (RT-q)PCR
H1993 cells were collected at 24 h post-transfection. Total RNA in 1×105 cells and 0.02 g tissue sample (ground in liquid nitrogen) was extracted using Ribozol reagent (Sigma-Aldrich; Merck KGaA). In order to harvest miRNAs, 80% ethanol was used to precipitate and wash RNA samples.
All RNA samples were digested with DNase I (Sigma-Aldrich; Merck KGaA) at 37°C for 2 h to remove genomic DNAs. All reverse transcriptions were performed using the PrimeScript RT Reagent kit (Takara Bio, Inc.) to synthesize cDNA, followed by preparation of qPCR mixtures using the QuantiTect SYBR-Green PCR kit (Qiagen) according to the manufacturer's instructions with GAPDH as an endogenous control to measure the expression levels of ZNF281 and PTEN mRNA.
To measure the expression levels of miR-221, both reverse transcriptions and qPCR mixture preparations were prepared using the All-in-One™ miRNA RT-qPCR Detection kit (GeneCopoeia, Inc.) with U6 as an endogenous control. The sequences of primers were: ZNF281 forward, 5′-GGACACATAGTGGAGAAAAG-3′ and reverse, 5′-GAGACAACACAGCCAGATTA-3′; PTEN forward, 5′-TGAGTTCCCTCAGCCGT-3′ and reverse, 5′-GAGGTTTCCTCTGGTCC-3′; and GADPH forward, 5′-GGATTTGGTCGTATTGG-3′ and reverse, 5′-GGAAGATGGTGATGGGAT-3′. The forward primer for miR-221 was: 5′-ACCUGGCAUACAAUGUAG-3′. Reverse primers for miR-221 and U6 primers were from the All-in-One™ miRNA RT-qPCR Detection kit (cat. no. QP015, GeneCopoeia, Inc.). The PCR conditions were as follows: 95°C for 1 min, and then 40 cycles of 95°C for 10 sec and 55°C for 60 sec. All experiments were performed in three technical replicates and the 2−ΔΔCq method was used to analyze data (15).
Cell apoptosis analysis
H1993 cells were collected at 24 h post-transfection and counted. Subsequently, 4×104 cells were mixed with 1 ml serum-free RPMI-1640 medium to prepare single-cell suspensions. Cells were cultivated in a 6-well cell culture plate (2 ml/well) for 48 h at 37°C. Subsequently, pre-cooled PBS was used to wash cells, followed by propidium iodide and Annexin V-FITC staining for 20 min at 4°C. Finally, flow cytometry was performed using CytoFLEX LX Flow Cytometer (Beckman Coulter) to separate apoptotic cells. CellQuest Pro v5.1 software (BD Biosciences) was used to analyze data.
Cell proliferation analysis
H1993 cells were collected at 24 h post-transfection and counted. Subsequently, 4×104 cells were mixed with 1 ml RPMI-1640 medium (10% FBS) to prepare single-cell suspensions. Cells were seeded in a 96-well cell culture plate (0.1 ml/well), followed by the addition of 10 µl Cell Counting Kit-8 (Dojindo Molecular Technologies, Inc.) for 4 h. Optical density was measured at 450 nm on a microplate reader.
Western blotting
H1993 cells were collected at 24 h post-transfection and counted. Total protein of 1×105 cells was extracted using RIPA solution (Sangon Biotech Co., Ltd.). A bicinchoninic acid kit (Sangon Biotech Co., Ltd.) was used to measure protein concentration. After denaturation in boiling water for 5 min, proteins (30 µg) were separated by 10% SDS-PAGE. Subsequently, proteins were transferred to PVDF membranes, followed by blocking for 1 h in 5% non-fat milk at room temperature. The membranes were first blotted with primary rabbit antibodies of PTEN (dilution, 1:1,500; cat. no. ab31392; Abcam) and GAPDH (dilution, 1:1,300; cat. no. ab37168; Abcam) for 18 h at 4°C, followed by blotting with horseradish peroxidase-cinjugated goat goat anti-rabbit immunoglobulin G secondary antibody (dilution, 1:1,500; cat. no. ab6721; Abcam) secondary antibodies at 24°C for 2 h. Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Inc.) was used to develop signals. Signals were processed using ImageJ v1.48 software (National Institutes of Health).
Statistical analysis
All data are presented as the mean values of three biological replicates. Evaluation of the significance of differences between non-tumor and NSCLC tissues was performed using the paired Student's t-test. Differences among cell transfection groups were analyzed using the Kruskal-Wallis test and post hoc Dunn's test. Associations were analyzed using linear regression. The 66 patients with NSCLC were grouped into high-(n=33) and low-ZNF281 (n=33) expression level groups according to its median expression level in NSCLC (2.28). GraphPad Prism 6 software (GraphPad Software, Inc.) was used for statistical analysis included the plotting of survival curves, and the log-rank test was used to compare survival curves. Differences in the clinical stage, cancer grade, subtypes, age and sex between two groups were analyzed using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
Low ZNF281 expression in NSCLC predicts poor overall survival rate
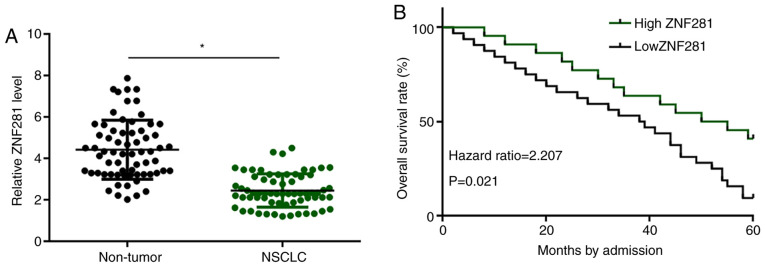
Levels of ZNF281 expression were measured using RT-qPCR and compared between two types of tissues (non-tumor vs. NSCLC) using the paired Student's t-test. Compared with non-tumor samples, expression levels of ZNF281 in NSCLC tissue samples were significantly lower (P<0.05; Fig. 1A). Patients were grouped into high- and low-expression groups according to the median expression level, and survival curves were plotted and compared. No significant differences in the clinical stage, cancer grade, subtype, age and sex were observed between the two groups (Table I). It was observed that the 5-year overall survival rate of patients in the high-ZNF281 expression group was significantly higher compared with that of patients in the low-ZNF281 expression group (Fig. 1B).
Figure 1.
High expression levels of ZNF281 in NSCLC predict poor survival. (A) ZNF281 expression was measured by reverse transcription-quantitative PCR and compared between non-tumor and NSCLC tissues using a paired Student's t-test. (B) A total of 66 patients with NSCLC were divided into high-(n=33) and low-ZNF281 (n=33) expression level groups according to its median expression level in NSCLC. GraphPad Prism 6 software was used to plot survival curves and the log-rank test was used to compare these. *P<0.05. ZNF281, zinc finger protein 281; NSCLC, non-small cell lung cancer.
Table I.
Comparison of clinical data between high- and low-ZNF281 expression level groups.
| Variable | High-ZNF281 expression, n | Low-ZNF281 expression, n | P-value |
|---|---|---|---|
| Cases | 33 | 33 | |
| Sex | |||
| Male | 23 | 21 | 0.60 |
| Female | 10 | 12 | |
| Age, years | |||
| >50 | 17 | 19 | 0.62 |
| ≤50 | 16 | 14 | |
| Subtype | |||
| Squamous cell carcinoma | 14 | 16 | 0.62 |
| Adenocarcinoma | 19 | 17 | |
| Grade | |||
| I | 6 | 8 | 0.79 |
| II | 9 | 10 | |
| III | 10 | 10 | |
| IV | 8 | 5 | |
| Stage | |||
| I | 2 | 4 | 0.81 |
| II | 9 | 10 | |
| III | 11 | 9 | |
| IV | 11 | 10 | |
ZNF281, zinc finger protein 281.
ZNF281 is significantly associated with miR-221 and PTEN mRNA in NSCLC tissues
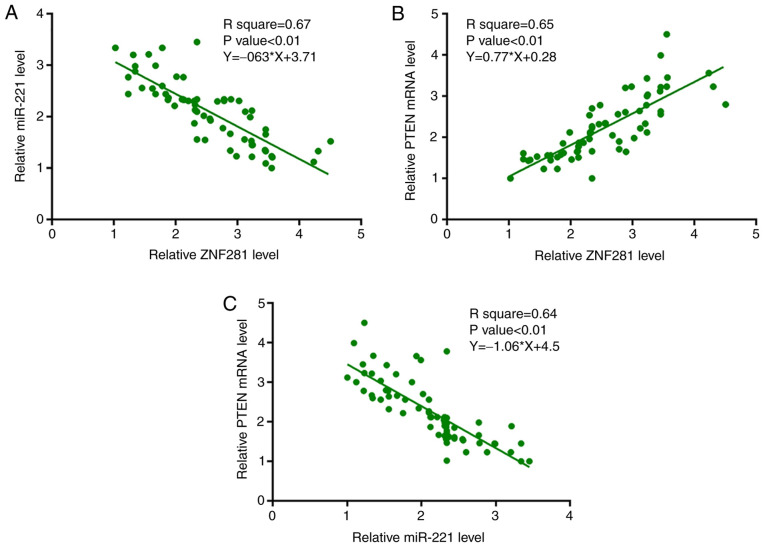
Furthermore, expression levels of miR-221 and PTEN mRNA in NSCLC tissues were measured by performing RT-qPCR. Associations between ZNF281 and miR-221/PTEN mRNA were analyzed using linear regression analysis. It was observed that the expression levels of ZNF281 were significantly negatively associated with miR-221 expression (Fig. 2A); however, the expression levels of ZNF281 and PTEN mRNA were significantly positively associated (Fig. 2B). Additionally, the association between miR-221 and PTEN mRNA expression was analyzed by linear regression analysis and it was observed that miR-221 and PTEN mRNA were significantly negatively associated (Fig. 2C).
Figure 2.
ZNF281 expression is significantly associated with miR-221 and PTEN mRNA levels in NSCLC tissues. Expression levels of miR-221 and PTEN mRNA in NSCLC tissues were measured by reverse transcription-quantitative PCR. Associations between ZNF281 and (A) miR-221 and (B) PTEN mRNA expression, and between (C) miR-221 and PTEN mRNA expression were analyzed using linear regression. ZNF281, zinc finger protein 281; NSCLC, non-small cell lung cancer; miR, microRNA; PTEN, phosphatase and tensin homolog.
ZNF281 upregulates PTEN mRNA and protein expression via the downregulation of miR-221 in H1993 cells
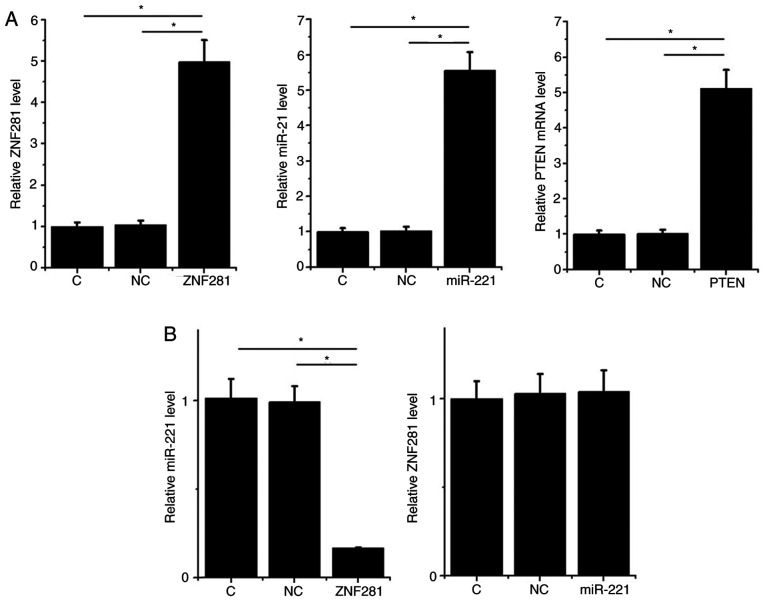
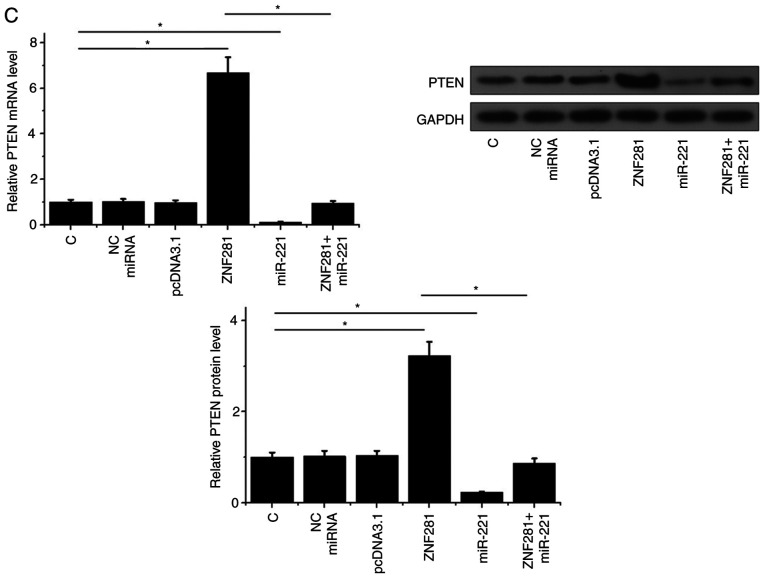
To analyze the interactions between ZNF281, PTEN and miR-221, expression vectors of ZNF281 and PTEN as well as a miR-221 mimic were transfected into H1993 cells. RT-qPCR results revealed that, compared with the NC and C groups, expression levels of ZNF281, PTEN mRNA and miR-221 were significantly increased at 24 h post-transfection (P<0.05; Fig. 3A). The effects of ZNF281 and miR-221 overexpression on mRNA and protein levels were analyzed by RT-qPCR and western blotting, respectively. Cells with ZNF281 overexpression exhibited downregulated miR-221 expression, whereas cells with miR-221 overexpression exhibited unaffected ZNF281 expression (P<0.05; Fig. 3B). Compared with the two control groups, cells with ZNF281 overexpression exhibited upregulated PTEN expression, whereas cells with miR-221 overexpression exhibited downregulated PTEN expression. Additionally, miR-221 overexpression reduced the effects of ZNF281 overexpression on PTEN expression at both mRNA and protein levels (P<0.05; Fig. 3C).
Figure 3.
ZNF281 upregulates PTEN via downregulation of miR-221 in H1993 cells. In order to analyze the interactions among ZNF281, PTEN and miR-221, expression vectors of ZNF281 and PTEN, and a miR-221 mimic were transfected into H1993 cells. (A) Overexpression of ZNF281, PTEN mRNA and miR-221 was confirmed by RT-qPCR. (B) Interaction between ZNF281 and miR-221 analyzed by RT-qPCR. (C) Effects of ZNF281 and miR-221 overexpression on PTEN expression were analyzed by western blotting and RT-qPCR. Data are presented as the means ± standard deviation. *P<0.05. C, control (untransfected cells); NC, negative control (cells transfected with empty vector or negative control miRNA); ZNF281, zinc finger protein 281; NSCLC, non-small cell lung cancer; miR, microRNA; PTEN, phosphatase and tensin homolog; RT-qPCR, reverse transcription-quantitative PCR.
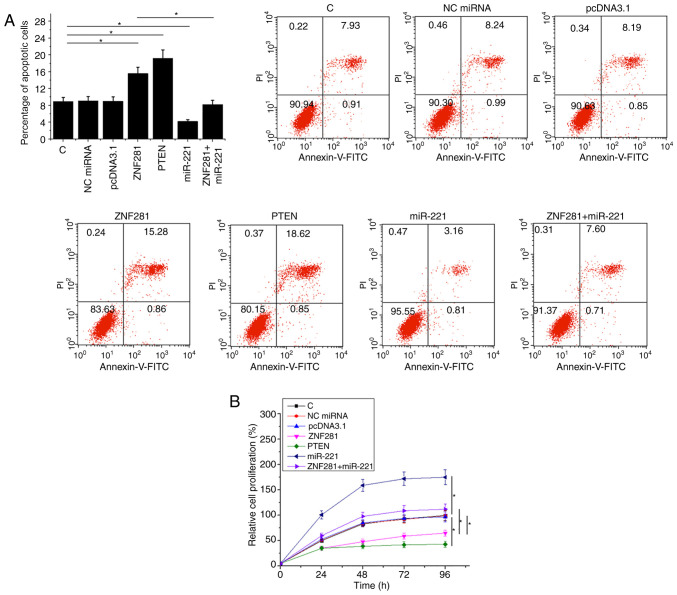
ZNF281 regulates H1993 cell apoptosis and proliferation via miR-221 and PTEN
Compared with the NC and C groups, ZNF281 and PTEN overexpression resulted in an increase in cell apoptosis (Fig. 4A) and inhibited cell proliferation (Fig. 4B) of NSCLC cells (P<0.05). Additionally, miR-221 overexpression partially attenuated the functions of ZNF281 (P<0.05).
Figure 4.
ZNF281 regulates H1993 cell apoptosis and proliferation via miR-221 and PTEN. Cell apoptosis and proliferation assays were performed to analyze the effects of ZNF281, miR-221 and PTEN overexpression on (A) cell apoptosis and (B) proliferation. Data are presented as the means ± standard deviation. *P<0.05. C, control (untransfected cells); NC, negative control (cells transfected with empty vector or negative control miRNA); ZNF281, zinc finger protein 281; miR, microRNA; PTEN, phosphatase and tensin homolog; PI, propidium iodide.
Discussion
The present study primarily investigated the role of ZNF281 in NSCLC, a major subtype of lung cancer. In NSCLC, ZNF281 was downregulated and could downregulate oncogenic miR-221 to upregulate PTEN, thereby promoting cancer cell apoptosis and inhibiting cancer cell proliferation.
To the best of our knowledge, the expression pattern and functionality of ZNF281 have only been investigated in glioma (11). In glioma, ZNF281 is downregulated and inhibits the stemness, proliferation and invasiveness of glioma cells via inactivation of the NF-κB signaling pathway (11), which is a well-characterized oncogenic signaling pathway that serves critical roles in numerous aspects of cancer biology (16,17). The present study was the first to report the downregulation of ZNF281 in NSCLC. In addition, ZNF281 overexpression resulted in the promotion of cell apoptosis and inhibition of cell proliferation. Therefore, the present data indicated that ZNF281 serves a tumor-suppressive role in NSCLC.
miR-221 is an oncogenic miRNA in numerous human cancer types, including liver cancer (18). However, in a previous study, miR-221 has been reported to have inhibitory effects on the proliferation of NSCLC cells (19), which was consistent with the results observed in the present study; for example, it was revealed that miR-221 overexpression resulted in the promotion of cancer cell proliferation and inhibited cancer cell apoptosis. This may be attributable to the different cell line used in the present study. In another study, miR-221 has been reported to directly target PTEN to inhibit gastric carcinoma cell proliferation (10). In the present study, PTEN was downregulated in NSCLC cells following miR-221 overexpression. Therefore, miR-221 may also target PTEN in NSCLC.
The present study demonstrated that ZNF281 downregulated miR-221 which resulted in the upregulation of PTEN. Additionally, it is known that both ZNF281 and miR-221 have direct interactions with NF-κB (11,12); therefore, NF-κB may mediate the interaction between ZNF281 and miR-221. However, further studies are required to investigate the mechanisms that mediate the interaction between ZNF281 and miR-221. Future study will include a higher sample number and in vivo experiments.
In conclusion, ZNF281 was downregulated in NSCLC and may downregulate miR-221, resulting in the upregulation of PTEN, thereby promoting cancer cell apoptosis and proliferation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XL and WJ designed the study. XL, BY and XW performed the experiments. FW, YL, NW and XY collected and analyzed the data. WJ drafted the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Review Board of Hiser Medical Center and the Qingdao Ethics Committee (approval no. 2011HC08LQ03). All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto H, Yatabe Y, Toyooka S. Inherited lung cancer syndromes targeting never smokers. Transl Lung Cancer Res. 2018;7:498–504. doi: 10.21037/tlcr.2018.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subramanian J, Govindan R. Molecular genetics of lung cancer in people who have never smoked. Lancet Oncol. 2008;9:676–682. doi: 10.1016/S1470-2045(08)70174-8. [DOI] [PubMed] [Google Scholar]
- 7.Brennan P, Hainaut P, Boffetta P. Genetics of lung-cancer susceptibility. Lancet Oncol. 2011;12:399–408. doi: 10.1016/S1470-2045(10)70126-1. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3, 4, 5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 10.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XT, Li JC, Feng M, Zhou YX, Du ZW. Novel lncRNA-ZNF281 regulates cell growth, stemness and invasion of glioma stem-like U251s cells. Neoplasma. 2019;66:118–127. doi: 10.4149/neo_2018_180613N391. [DOI] [PubMed] [Google Scholar]
- 12.Zhao D, Zhuang N, Ding Y, Kang Y, Shi L. miR-221 activates the NF-κB pathway by targeting A20. Biochem Biophys Res Commun. 2016;472:11–18. doi: 10.1016/j.bbrc.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 14.Sun Z, Aubry MC, Deschamps C, Marks RS, Okuno SH, Williams BA, Sugimura H, Pankratz VS, Yang P. Histologic grade is an independent prognostic factor for survival in non-small cell lung cancer: An analysis of 5018 hospital-and 712 population-based cases. J Thorac Cardiovasc Surg. 2006;131:1014–1020. doi: 10.1016/j.jtcvs.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Rinkenbaugh AL, Baldwin AS. The NF-κB pathway and cancer stem cells. Cells. 2016;5:16. doi: 10.3390/cells5020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, Zhang J, Arfuso F, Chinnathambi A, Zayed ME, Alharbi SA, Kumar AP, Ahn KS, Sethi G. NF-κB in cancer therapy. Arch Toxicol. 2015;89:711–731. doi: 10.1007/s00204-015-1470-4. [DOI] [PubMed] [Google Scholar]
- 18.Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, Sekido Y, Kondo M, Hasegawa Y. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4:551–564. doi: 10.1002/cam4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.