Abstract
Organellar and secretory RNases, associated with different cellular compartments, are essential to maintain cellular homeostasis during development and in stress responses.
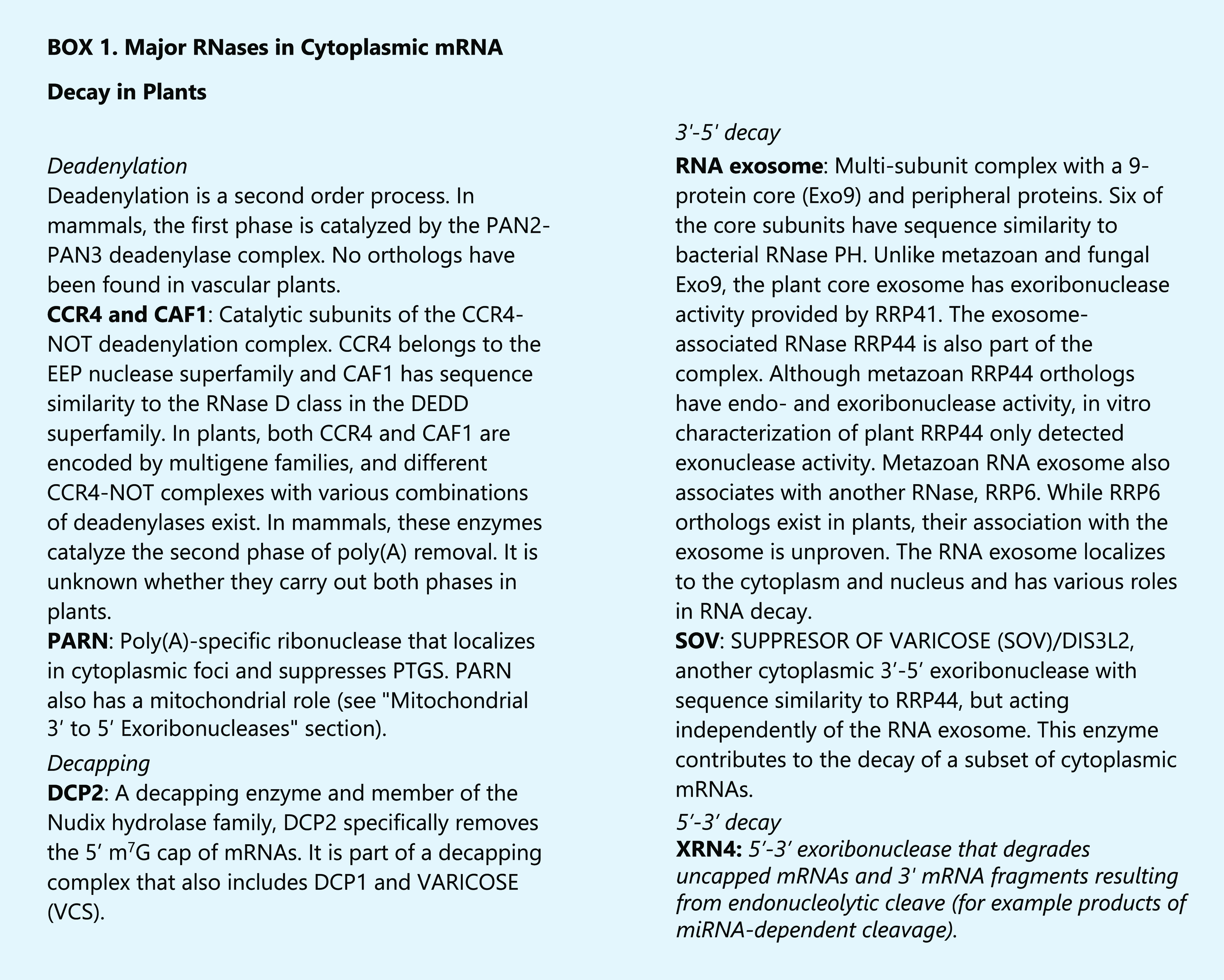
During the last 20 years there has been an intense focus on posttranscriptional regulation of gene expression, and the role played by ribonucleases (RNases) in this process. Most research has centered on cytoplasmic and nuclear RNA decay enzymes, particularly those involved in mRNA regulation, and in proteins mediating nucleus-localized coding and noncoding RNA processing. These efforts have created a well-defined picture of RNA homeostasis in eukaryotes, including plants (Schmid and Jensen 2018; Sieburth and Vincent 2018; Towler and Newbury, 2018; Matsui et al., 2019). Thus, cytoplasmic degradation of mRNAs in plants (see Box 1), which involves deadenylation, decapping, and degradation by 3′-5′ and 5′-3′ exoribonucleases, has been extensively studied and reviewed (e.g. Zhang and Guo 2017; Sieburth and Vincent 2018; Sorenson et al., 2018; Lange et al., 2019). Similarly, different roles for the nuclear exosome and other RNases in nuclear RNA processing and decay have been established (Lange et al., 2014; Sikorska et al., 2017; Tomecki et al., 2017; Sáez-Vásquez and Delseny 2019). RNA decay mediated by small RNAs associated to RNA-induced silencing complexes and the processing and decay of small RNAs (sRNAs) have also been the focus of intense research efforts (Fukudome and Fukuhara 2017; Yu et al., 2017; Wang et al., 2019).
On the other hand, RNases that do not reside in the cytoplasm or nucleus have received significant less attention, even though their activities are important for plant growth, development, and response to biotic and abiotic stimuli from the environment. As we will describe, organellar RNases are essential to maintain RNA homeostasis in chloroplasts and mitochondria. Plants also express members of the RNase T2 protein family, associated with the secretory pathway, which are involved in the maintenance of normal cellular homeostasis, with diverse roles, from stress responses to control of self-pollen rejection. This review will provide a summary of the present knowledge on these RNases, from enzymatic characterizations to biological functions, and highlight questions that should be addressed in the future.
SECRETORY RIBONUCLEASES
Secretory RNases are enzymes targeted to the cellular secretory system and thus localized in organelles or the extracellular space. In plants, characterized secretory RNases belong to the RNase T2 family, a group of enzymes typified by the fungal RNase T2 purified from Aspergillus oryzae (MacIntosh 2011). Enzymes from the T2 family are endoribonucleases without sequence or base specificity that degrade single-stranded RNA through a 2’,3′ cyclic phosphate intermediate, in a reaction catalyzed by two absolutely conserved His residues that define the active site (Irie 1999). It has been proposed that their enzymatic mechanism involves a two-step (transphosphorylation and hydrolysis) general acid–base catalysis (Irie 1999; Thorn et al., 2012). Interestingly, fungal enzymes can complete both steps and yield 3′ nucleotide monophosphates as final product, but bacterial enzymes end the reaction after the first step, resulting in the production of nucleotides (nt) with a 2’,3′ cyclic phosphate (Nicholson 1999; Fontaine et al., 2018). Although not many plant RNase T2 proteins have been characterized enzymatically, biochemical analyses of extracellular and vacuolar tomato enzymes indicated that plant RNase T2 enzymes generate 2’,3′ cyclic nucleotide monophosphates as primary product (Nürnberger et al., 1990; Löffler et al., 1992).
The RNase T2 family is conserved in almost all eukaryotes so far analyzed, with the exception of trypanosomes and Schizosaccharomyces pombe (Shang et al., 2018; Fricker et al., 2019) and is also found in a large number of bacteria and several viruses. In most nonplant organisms, only one gene belonging to this family is found in each genome (Hillwig et al., 2009; Ambrosio et al., 2014); however, in plants, the RNase T2 protein repertoire has expanded, and individual proteins have been adapted for a variety of functions. At least four genes are found in each seed plant genome that has been sequenced (MacIntosh et al., 2010; Ramanauskas and Igić 2017), and there is evidence of frequent duplications/gene losses that resulted in different number of RNase T2 gene in different species. In addition, gene expression and functional analyses have shown that these proteins have acquired a variety of biological roles, including participation in the cellular housekeeping salvage pathway, production of tRNA-derived sRNAs, defense activities, and a central role in gametophytic self-incompatibility in several plant families (Table 1). Phylogenetic analyses allowed separation of plant RNase T2 enzymes in three clades (Igic and Kohn 2001; MacIntosh et al., 2010; Ramanauskas and Igić 2017) that define three classes of proteins roughly associated with different functions: Class I are proteins associated with a variety of stress responses and show evidence of gene duplications and gene sorting that provide large variability to this class, with variable number of class I proteins in individual species; Class II enzymes are conserved in all seed plants, in general with only one gene per genome, and their function has been related to RNA salvage; and Class III enzymes are mainly associated with self-incompatibility, although other functions have also been assigned to this class (Table 1). Historically, Class III enzymes were named S-RNases, whereas other plant T2 proteins are referred to as S-like RNases (McClure et al., 1990; Taylor et al., 1993). Class I and II proteins are found in all land plants, whereas Class III proteins are found only in core eudicots (Ramanauskas and Igić, 2017). Some plant RNase T2 proteins in the three classes have lost their catalytic activity (MacIntosh et al., 2010; Ramanauskas and Igić 2017). A monocot-specific class I subclade of proteins without RNase activity is particularly conserved (MacIntosh et al., 2010). Some of these proteins, for example OsRNS4 (Table 1), seem to have significant stress-related functions (Zheng et al., 2014), although their specific biological activity is not known.
Table 1. Classification of plant RNase T2 proteins and proposed functions.
| Class and Functions | Examples | References |
|---|---|---|
| Class I | ||
| Defense, antiviral | RNS1 (Arabidopsis), ZnRNase II (Zinnia elegans) | Kim et al., 2019; Trifonova et al., 2012 |
| Defense, antimicrobial | RNase NE (Nicotiana tabacum) | Hugot et al., 2002 |
| Defense, wounding | RNS1 (Arabidopsis), RNase LE (Solanum lycopersicum), OsRNS4 and OsRNS5 (O. sativa) | Hillwig et al., 2008; Gross et al., 2004; MacIntosh et al., 2010 |
| Stress, drought/salinity | OsRNS4 (O. sativa), PvRNS3 (Phaseolus vulgaris) | Zheng et al., 2014; Diaz-Baena et al., 2020 |
| Stress, P-starvation | RNS1 (Arabidopsis), EsRNS1 (Eutrema salsugineum), RNase LX and RNase LE (S. lycopersicum) | Bariola et al., 1999; Velasco et al., 2016; Köck et al., 1995 |
| tsRNA synthesis | RNS1 and RNS3 (Arabidopsis) | Alves et al., 2017; Megel et al., 2019 |
| Development/PCD | RNase LX (S. lycopersicum), ZRNase I (Z. elegans) | Lehmann et al., 2001; Lers et al., 2006; Fukuda 2000 |
| Class II | ||
| RNA/nucleotide salvage | RNS2 (Arabidopsis) | Hillwig et al., 2011a |
| Stress, P-starvation | RNS2 (Arabidopsis) | Bariola et al., 1999 |
| Class III | ||
| Self-incompatibility | S-RNases (Solanaceae, Plantaginaceae, Rubiaceae, Rosaceae) | Ramanauskas and Igić, 2017 |
| Defense, antimicrobial | S1-RNase and SX-RNase (Petunia hybrida), SA2-RNase (Nicotiana alata) | Hillwig et al., 2011b; Silva et al., 2020 |
| Stress, P-starvation | NnSR1 and SX-RNase (N. alata) | Rojas et al., 2015; Rojas et al., 2018 |
For brevity, this review will focus on two plant RNase T2 functions for which significant advances have occurred in recent years: ribosomal RNA (rRNA) turnover and tRNA processing.
Housekeeping Role Recycling rRNA, the Ancestral Function of Eukaryotic RNase T2 Enzymes
The presence of RNase T2 enzymes in almost all eukaryote genomes suggests that these enzymes carry out an important biological function. Phylogenetic and gene expression analyses suggested that enzymes in Class II carry out a housekeeping function in plants, and that this role is likely the ancestral function of RNase T2 enzymes (Hillwig et al., 2009; MacIntosh et al., 2010). RNS2, the Class II enzyme present in Arabidopsis (Arabidopsis thaliana), is localized mainly in the vacuole (Hillwig et al., 2011a). Characterization of rns2 mutants showed that the enzyme is necessary for normal rRNA turnover, because the half-life of 28S rRNA and 18S rRNA almost doubled in the null mutant rns2-2 (Hillwig et al., 2011a), and rRNA accumulated in mutant vacuoles (Floyd et al., 2015). Plants lacking RNS2 activity showed a constitutive autophagy phenotype, indicating that housekeeping turnover of rRNA is an essential process to maintain cellular homeostasis (Hillwig et al., 2011a; Floyd et al., 2015). Characterization of rns2-1, a mutant expressing an active RNase missing the last 14 amino acids, determined that the C terminus of the protein contains a putative vacuolar localization signal, as the truncated RNS2 protein is mislocalized outside the vacuole and secreted outside of the cells. Moreover, rns2-1 mutants present a constitutive autophagy phenotype identical to the rns2-2 null mutant; thus, vacuolar activity of this enzyme is essential to maintain normal cellular functions (Floyd et al., 2017). Although the mechanism of ribosomal or rRNA transport from the cytoplasm to the vacuole is still not well understood, evidence suggests that a selective transport mechanism that uses part of the proteins necessary for the autophagy process, specifically ATG5 (Autophagy-Related Gene 5, a protein indispensable for autophagic vesicle formation; Le Bars et al., 2014), participates in the transport of ribosomes or rRNA to vacuoles for degradation (Floyd et al., 2015).
RNase T2-mediated RNA turnover is part of the salvage pathway that maintains normal cellular levels of nucleosides/nucleotides. Lack of vacuolar RNA turnover in the rns2 mutant causes metabolic changes, primarily in carbon flux through the pentose phosphate pathway, as a way to generate ribose-phosphate for de novo nucleotide synthesis (Morriss et al., 2017). A similar role for RNase T2 enzymes has been described in other organisms, including yeast (Saccharomyces cerevisiae; Huang et al., 2015), zebrafish (Danio rerio; Haud et al., 2011), and nematodes (Caenorhabditis elegans; Liu et al., 2018). Although RNS2, and potentially RNS2 orthologs in other plants, are necessary for normal rRNA turnover and homeostasis maintenance, rRNA still decays in rns2 mutants even if the rate is reduced (Hillwig et al., 2011a). This observation implies that other RNases can attack rRNA in vivo. Whether this turnover occurs in the vacuole or the cytoplasm is not known, but because rns2 plants have a constitutive autophagy phenotype, it is tempting to speculate that this process could transport extra RNase activities to the vacuole that could contribute to rRNA turnover. For example, in stress situations the presence of P-bodies, cytoplasmic complexes involved in mRNA translation repression and RNA decay, increases in plants. P-bodies contain several ribonucleases including the decapping complex, CCDR and CAF1 deadenylases and XRN4 (Chantarachot and Bailey-Serres, 2018), and it has been shown that P-bodies are processed through an autophagic process (granulophagy) in yeast and mammalian cells (Buchan et al., 2013; Frankel et al., 2017). It can thus be hypothesized that P-body-associated RNases could contribute to rRNA decay in the rns2 mutants.
As mentioned, it has been hypothesized that the role in RNA salvage (and perhaps P scavenging) is the ancestral function of RNase T2 enzymes (MacIntosh 2011). However, despite their almost absolute conservation, these enzymes are not strictly essential, although significant cellular and developmental phenotypes have been observed in plants and animals defective in RNase T2 activity (see above). It is still clear that the presence of T2 enzymes may provide a significant advantage to the organism that results in the conserved presence of these enzymes in eukaryotes. It is important to note that although the “ancestral” biological role has been assigned to RNS2 in Arabidopsis, neofunctionalization and gain/loss of gene duplications may have shifted the function to other T2 homologs in other species. For example, the Class II RNase T2 in tomato, RNase LER, seems to have a prominent role in guard cells, and it seems to localize exclusively to the ER (Köthke and Köck, 2011), whereas class I enzymes are found in the tomato vacuole and could be in charge of the RNA/nucleotide salvage function in this plant (Abel and Köck, 2001). For a more detailed analysis of phylogenetic relationships and evolution of the RNase T2 family in plants, the analysis by Ramanauskas and Igić, (2017) is recommended.
A Shared Role in the Biosynthesis of tRNA-Derived Small RNAs
Among the ever-growing collection of small RNA species found in living organisms, tRNA-derived small RNAs (tsRNAs) are a recent addition. tsRNAs vary in size, ranging from 19 to 35 nt, and are derived from different parts of tRNAs. Two main categories can be found in all eukaryotes: (1) 5′tsRNAs that are likely the result of cleavage in the tRNA d-loop and 3′tsRNAs produced by cleavage of the T-loop, commonly referred as tRNA-derived fragments (tRFs); and (2) 5′ tRNA halves and 3′ tRNA halves (tRHs) that result from processing at the anticodon loop (Dou et al., 2019). Note that nomenclature for these tsRNAs is not fully established, and some authors refer to all tsRNAs as “tRFs” and differentiate between “long tRFs” (∼30–35 nt) and “short tRFs” (<28 nt; Megel et al., 2015). Although the function of tsRNAs is less well understood than that of other sRNAs, their association with Argonaute (AGO) proteins indicate participation in posttranscriptional regulation of gene expression, with miRNA-like activity (Kumar et al., 2014). In addition, AGO-independent functions have been proposed, including regulation of both positive and negative regulation of translation, competitive binding to RNA-binding proteins, and modulation of mRNA secondary structures (reviewed by Dou et al., 2019). Given the significant conservation among tRNA sequences, it is not surprising that some tsRNAs are also highly conserved among organisms as divergent as mammals and bacteria (Kumar et al., 2014).
Analyses of tsRNA populations have been characterized in a variety of plants and relative abundance of individual tsRNAs is tissue specific and also modulated by biotic and abiotic stress conditions (reviewed by Zhu et al., 2018). The biosynthetic pathway(s) responsible for production of tRFs and tRHs is controversial. Recently, Martinez et al. (2017) described tRFs in the 18 to 25 nt range in Arabidopsis, and identified 19 nt 5′tRF species that accumulate preferentially in pollen. These 5′tRFs also accumulate in pollen from rice (Oryza sativa) and maize (Zea mays), and a similar phenomenon can be observed in the gametophyte/sporophyte of Physcomitrella patens (Martinez et al., 2017). The authors found a similar increase in 19 nt 5′tRFs in the ddm1 mutant that is defective in the DDM1 swi/snf family chromatin remodeler protein. Moreover, the accumulation of these 19 nt 5′tRFs depends on the Dicer RNase DCL1. These DCL1 products are found in AGO1 complexes and target transposon elements (TE). These results indicated that the tRFs are produced by a Dicer-dependent pathway and act in a miRNA-like manner to control TE expression. However, other authors have found that the overall population of 5′tRF and 3′tRF is only modestly affected in dcl1-4 Arabidopsis mutants (Nowacka et al., 2013; Alves et al., 2017).
On the other hand, Alves et al. (2017) found that several 5′tRF and 3′tRFs accumulate to higher levels in Arabidopsis plants overexpressing RNS1, a Class I RNase T2, suggesting that this enzyme could be involved in tRF production in plants. A more definitive demonstration of the role of plant RNase T2 enzymes in tsRNA production was recently presented by Megel et al. (2019), who used RNA sequencing (RNA-seq) and in vitro tRNA cleavage assays to demonstrate that tRF abundance is not affected in the quadruple Arabidopsis mutant dcl1234 and that protein extracts from this mutant are still able to process tRNAs in vitro to produce tRFs and tRHs. The same biochemical approach was used to demonstrate that RNS1 and RNS3, another class I RNase T2 protein from Arabidopsis, and to a minor extent RNS2, can produce tRF and tRHs in vitro. Expression analyses also indicated that rns1 mutants fail to accumulate tRFs and tRHs that are normally induced after Pi-starvation (Megel et al., 2019).
These conflicting results could indicate that specific tRFs and tRHs may be produced by different pathways, and, hypothetically, alternative biosynthetic pathways may be associated with different biological roles and mode of action, although both Dicer-dependent and -independent tRFs associate with Argonaute proteins in plants (Alves et al., 2017; Martinez et al., 2017). Also intriguing is the fact that a large number of plastid-derived tRFs accumulate in the cytoplasm and can associate with AGO1, although the RNases involved in their processing are not known (Cognat et al., 2017). Similarly, at least two pathways have been proposed for the formation of tsRNAs in human (Homo sapiens) cells. Cole et al. (2009) identified a large number of tsRNAs in HeLa cells, and determined that accumulation of a 19-nt 5′tsRNA derived from tRNAGln is Dicer dependent and showed that purified Dicer can cleave this tRNA in vitro. On the other hand, Yamasaki et al. (2009) identified stress-induced tRHs in human U2OS cells that depend of the activity of angiogenin, a member of the vertebrate-specific RNase A family, and overexpression of this enzyme increases accumulation of the tsRNAs in the absence of stress. Moreover, Rny1 is also responsible for the production of stress-induced tsRNAs in S. cerevisiae (Thompson and Parker 2009), several RNase T2 enzymes contribute to the production of tsRNAs in the ciliate Tetrahymena thermophila (Andersen and Collins 2012), and purified human RNASET2 can process tRNAs in vitro (Megel et al., 2019), suggesting that RNase T2 enzymes may have a conserved role in tsRNA production in eukaryotes.
Connecting Secretory RNases with their Substrates
Strong data support a role for plant RNase T2 proteins in the turnover of rRNA, and increasing evidence indicate that they also participate in the production of tsRNAs. However, the enzymes and their substrate are not normally found in the same subcellular location. Both tRNAs and rRNAs accumulate in the cytoplasm, but RNS2 is a vacuolar enzyme (Bariola et al., 1999; Hillwig et al., 2011a; Floyd et al., 2017) and RNS1 (and likely RNS3, given its similarity to RNS1) is mainly secreted outside of the cell (Bariola et al., 1999). As mentioned, autophagy-dependent mechanisms participate in the transport of rRNA to the vacuole where it can be attacked by RNS2, but data regarding the location where tRNA processing to produce tsRNAs takes place is missing. Whether tRNAs are transported to an organelle or whether the RNases are released into the cytoplasm to act on tRNAs (and/or rRNAs?) is not known. If the latter were true, the mechanisms that prevent cytotoxic effects of releasing RNases with little substrate specificity into the cytoplasm should also be investigated (see Outstanding Questions).
Many class I RNases are also associated with responses to abiotic and biotic stresses (Table 1). In some cases, it is possible to envision a substrate directly related to a biological function, for example, in defense against RNA viruses, or during phosphate starvation. But in other cases, their potential substrates and the specific role in stress responses is less evident. It is tempting to hypothesize that the role of RNase T2 enzymes in stress responses may also be connected to their ability to generate tsRNAs, thus having a regulatory role in these processes.
PLASTIDIAL RNASES
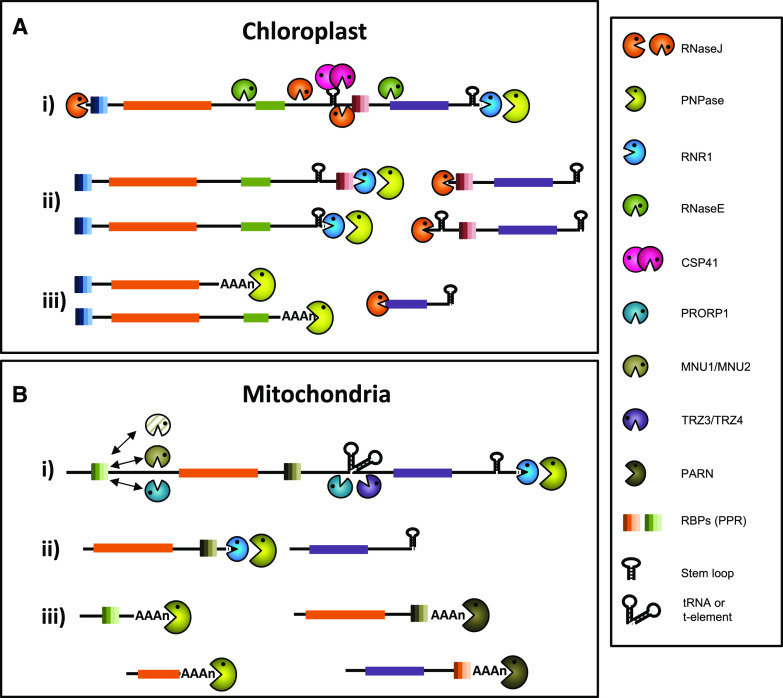
Both exo- and endo-ribonucleases have a dominant role in creating the chloroplast functional transcript population. They are responsible for intercistronic RNA cleavage along with 5′ and 3′ processing and RNA decay (Stoppel and Meurer 2012; Germain et al., 2013). Although seventeen chloroplastic RNases were predicted by mining the Arabidopsis genome for RNase annotations (Stoppel and Meurer, 2012), this figure certainly needs to be revised. Some predicted RNases were indeed shown to carry other functions (see, for example, the l-PSP/RidA enzyme; Niehaus et al., 2014), and new RNases keep being discovered in bacteria, some of them with a chloroplast homolog awaiting characterization (see, for example, the Rae1/YacP endoribonuclease; Condon et al., 2018). Indeed, as direct evidence of its cyanobacterial origin, nearly all chloroplast RNase homologs are present in bacteria (Stoppel and Meurer, 2012; Germain et al., 2013; dos Santos et al., 2018). Based on their chlorotic or embryo-lethal phenotypes and the associated disruption in RNA patterns, there are five major RNases whose putative roles are discussed below. Following transcription, the 5′-3′ exoribonuclease RNaseJ processes the 5′ ends, whereas two 3′-5′ exoribonucleases, polynucleotide phosphorylase (PNPase) and RNR1, cooperatively mature the 3′ ends. Intercistronic cleavage is performed by endoribonucleases, but the division of labor between RNaseJ, RNaseE, and CSP41 is unclear (Fig. 1A; Germain et al., 2013 for a model of chloroplast RNA maturation). These RNases are not specific and act on a wide variety of transcripts.
Figure 1.
The roles of ribonucleases in chloroplast and mitochondrial mRNA metabolism. A, (i) In chloroplasts, RNase J processes the 5′ end of polycistronic mRNAs, whereas PNPase and RNR1 cooperatively mature the 3′ ends. The transcript is also subject to intercistronic cleavage by several endoribonucleases, but the division of labor between RNase J, RNase E, and CSP41 is unclear. A, (ii) Another round of exoribonuclease digestion gives the final termini. Mature transcript ends are protected from exoribonuclease degradation by sequence-specific RNA binding proteins (for example, PPR proteins) and secondary structures. A, (iii) Illegitimate, partially, or misprocessed transcripts are then digested at the 3′ end by PNPase following the addition of a poly(A) tail. RNase J also participates in RNA degradation and surveillance. Its major role preventing the formation of double stranded RNA is not represented. B, (i) In mitochondria, endoribonucleases have a major role in defining 5′ ends through their interactions with PPR proteins that specifically bind RNA targets. Cleavage by PRORP1, MNU1/2, or yet unknown endoribonucleases occurs downstream of the PPR binding site. Alternatively, PRORP1 and TRZ3/4 can cleave tRNAs or t-elements between two genes and release mature or intermediate 5′ and 3′ ends. As in chloroplasts, PNPase and RNR1 cooperatively mature the 3′ ends. B, (ii) Mature 3′ ends are protected from exoribonuclease degradation by RBPs and/or secondary structures. B, (iii) Illegitimate, partially, or misprocessed transcripts are then digested at the 3′ end by PNPase, following the addition of a poly(A) tail. Additionally, the 3′-5′ exoribonuclease PARN appears to regulate the poly(A) status of correctly processed transcripts; however, the significance of this activity is unclear. RNase, RNase; PNPase, polynucleotide phosphorylase; RNR1, RNase R homolog 1; CSP41, chloroplast stem loop binding protein of 41 kD; PRORP1, PROTEINACEOUS RNASE P1; MNU, mitochondrial nuclease; TRZ, RNase Z; RBP, RNA binding protein;. Figure inspired by Germain et al. (2013).
Exoribonucleases
Absence of either of the two prominent exoribonucleases, PNPase and RNase R, leads to defects in 3′ end maturation for both mRNAs and rRNAs, and depletion of both causes embryo lethality in Arabidopsis (Germain et al., 2011; Germain et al., 2012).
PNPase
PNPase was the first discovered enzyme able to synthesize RNA in vitro (Grunberg-Manago et al., 1955). Its main role is, however, to degrade RNA through a phosphorolytic 3′-5′ exoribonuclease activity, and the balance between the two activities depends on the relative concentration of nucleoside diphosphates and Pi: the addition of Pi stimulates degradation, whereas nucleoside diphosphates promote degradation (Yehudai-Resheff et al., 2001). In vivo, the polymerase activity is responsible for the addition of short heteropolymeric A/U-rich tails to RNAs, but their biological role is still unclear (Germain et al., 2011). Different from the Escherichia coli enzyme, the active form purified from chloroplast is a complex of three PNPase monomers that does not seem to physically interact with other ribonucleases (Baginsky et al., 2001; Yehudai-Resheff et al., 2003). Extensive in vitro research has shown that PNPase is necessary to process RNA 3′ extensions and that its activity is inhibited by stem loops structures or tightly bound RBPs (Hayes et al., 1996; Yehudai-Resheff et al., 2001; Yehudai-Resheff et al., 2003; Prikryl et al., 2011). PNPase has a high affinity for poly(A) sequences, making it a major player in poly(A)-assisted RNA decay in chloroplast (Lisitsky et al., 1996; Yehudai-Resheff et al., 2003; Schuster and Stern, 2009). Arabidopsis PNPase mutants have chlorotic young leaves and are smaller than wild type but can still grow to full maturity (Marchive et al., 2009; Germain et al., 2011). Under phosphorus deprivation, Arabidopsis mutants fail to initiate lateral roots, whereas PNPase-depleted Chlamydomonas cells cannot survive in the same condition (Yehudai-Resheff et al., 2007; Marchive et al., 2009). PNPase was the first chloroplastic RNase whose influence on RNA maturation was investigated using RNA-seq (Hotto et al., 2011; Castandet et al., 2013; Castandet et al., 2019). This analysis, in association with traditional molecular biology techniques, showed that PNPase-deficient Arabidopsis plants accumulate numerous RNAs with 3′ extensions along with additional defects in RNA splicing and editing. Studies in Arabidopsis and maize determined that all types of RNAs are affected, including mRNAs, rRNAs, tRNA precursors, antisense RNAs, and spliced intron products (Walter et al., 2002; Marchive et al., 2009; Williams-Carrier et al., 2010; Germain et al., 2011; Hotto et al., 2011; Castandet et al., 2013; Ruwe et al., 2013; Castandet et al., 2019).
RNR1
In bacteria, members of the RNaseII/R 3′-5′ exoribonuclease superfamily mainly involved in mRNA metabolism are named RNaseII, whereas the ones displaying significant activity on rRNA are named RNR (Li and Deutscher, 2004). This interpretation of division of labor is, however, likely to be an oversimplification (Cheng and Deutscher, 2005). The Arabidopsis RNaseII/R homolog, RNR1, is dually localized to chloroplast and mitochondria (Kishine et al., 2004; Perrin et al., 2004b; Bollenbach et al., 2005) and combines the two activities described in bacteria (Germain et al., 2012). The rnr1 null mutants can only germinate on Suc, have white cotyledons that accumulate anthocyanins, and show retarded growth and pale green rosette leaves (Kishine et al., 2004; Bollenbach et al., 2005). Photosynthesis is impaired in the mutants, and electron microscopy revealed a lack of stacked thylakoids (Bollenbach et al., 2005). rnr1 mutants overaccumulate rRNA precursors at the expense of the mature forms (Kishine et al., 2004; Bollenbach et al., 2005; Germain et al., 2012). Additionally, mature rRNAs contain short 3′ extensions consistent with the ability of the recombinant enzyme to process the 3′ end of the rrn5S precursor in vitro (Bollenbach et al., 2005). It was noticed that the impaired maturation of the rrn5S transcript in the mutant is associated with the accumulation of an antisense RNA (asRNA), and this defective maturation was recapitulated in transplastomic plants artificially overexpressing the asRNA (Hotto et al., 2010; Sharwood et al., 2011b). Based on the ability of the E. coli enzyme to unwind RNA duplexes in vitro, it can be speculated that RNR1 might degrade the asRNA in vivo Sharwood et al., 2011b; Chu et al., 2017). A more thorough investigation of the rnr1 RNA patterns also demonstrated the essential role of the enzyme in mRNA 3′ maturation, in cooperation with PNPase. Most mRNAs in the rnr1 mutant accumulate short 3′ extensions, a feature probably explained by the ability of the enzyme to come closer than PNPase to the secondary structures and RBP binding sites marking the mature mRNA 3′ ends (Germain et al., 2012). Precursor mRNAs accumulate in nonpolysomal fractions and are more stable in the rnr1 mutant than wild type, suggesting a close link between RNA maturation and degradation (Germain et al., 2012).
The Endo- and Exoribonuclease RNase J
RNase J belongs to the metallo-β-lactamase family of ribonucleases (Condon and Gilet 2011), and its role in chloroplast RNA metabolism has recently been reviewed (Hotto et al., 2020). Initially described as an endoribonuclease in Bacillus subtilis, most members harbor both endo- and 5′-3′ exoribonuclease activity (Even et al., 2005; Mathy et al., 2007; Condon and Gilet 2011). As an illustration of its essential role in plant and chloroplast development, RNase J null mutants are embryo lethal (Tzafrir et al., 2004; Chen et al., 2015). Virus-induced gene silencing (VIGS) was therefore used to deplete RNase J in both Arabidopsis and tobacco chloroplasts (Sharwood et al., 2011a; Luro et al., 2013), and it showed that RNase J matures the 5′ end of several transcripts, with RBPs acting as a barrier to its activity (Luro et al., 2013). This result is in agreement with earlier in vitro studies that used commercial 5′-3′ exoribonucleases and recombinant RBPs (Prikryl et al., 2011; Hammani et al., 2012). The most striking effect in VIGS plants, however, was the accumulation of RNA species antisense to genic transcripts that could form duplexes preventing translation (Sharwood et al., 2011a). Because RNase J degrades preferentially single-stranded over double-stranded RNA in vitro, it is likely that its role in vivo is to degrade asRNAs before the formation of duplexes (Sharwood et al., 2011a; Halpert et al., 2019), making it an essential player in chloroplast RNA quality control. Interestingly, VIGS plants did not accumulate longer or unprocessed polycistronic precursors in place of mature transcripts, suggesting that RNase J does not mediate endonucleolytic processing (Sharwood et al., 2011a). This is in contrast with in vitro results where, similar to its bacterial homologs, Arabidopsis RNase J displays both activities (Halpert et al., 2019). On the other hand, although Chlamydomonas chloroplasts likely contain a 5′-3′ exoribonuclease activity (Drager et al., 1998; Drager et al., 1999), its RNase J acts only as an endoribonuclease in vitro (Liponska et al., 2018). One of the peculiarities of the plant enzyme is that it contains a C-terminal extension with high similarity to the GT-1 DNA binding domain usually found in transcription factors. It has been proposed that this domain could confer substrate specificity to the enzyme (Hotto et al., 2020). Finally, the possibility that chloroplast RNase J might also be involved in transcription termination of stalled RNA polymerase complexes, as recently described for the Bacillus enzyme (Šiková et al., 2020), has not been explored yet.
Endoribonucleases
CSP41
All photosynthetic organisms contain the CSP41 (Chloroplast Stem loop binding Protein of 41 kD) endoribonuclease whose elusive functions have recently been reviewed (Leister 2014). CSP41 proteins are highly abundant RNA binding proteins that display strong endoribonuclease activity on stem loops in vitro (Yang et al., 1996; Yang and Stern 1997; Bollenbach and Stern 2003a, b), a structure they preferentially bind (Yang et al., 1996). According to this characteristic, it was proposed that CSP41 cleaves the psbT-psbH transcript intergenic region (Chevalier et al., 2015). Chloroplasts contain two homologous proteins, CSP41A and CSP41B, which can form homo- and heterodimers (Bollenbach et al., 2009; Qi et al., 2012) and bind ribosomes (Beligni and Mayfield 2008; Bollenbach et al., 2009). Although the csp41a null mutant does not display any obvious phenotype, the csp41b null mutant shows mild chlorosis that is stronger under stress. Moreover, the absence of CSP41B also destabilizes CSP41A, resulting in similar phenotypes for the csp41b mutant and the csp41ab double mutant (Bollenbach et al., 2009; Qi et al., 2012). RNA immunoprecipitation-microarray (RIP-Chip) analyses more recently showed that CSP41 proteins are able to bind a large variety of transcripts and that their role is more likely to be RNA stabilization rather than cleavage (Qi et al., 2012). It has been proposed that the endoribonuclease activity could be regulated in vivo, making active forms behave like ribonucleases and inactive forms like stabilization factors (Leister 2014). If true, this hypothesis could reconcile the conflicting results obtained in vitro and in vivo.
RNase E
Chloroplast-localized homologs of E. coli RNase E can be found in most plants and green algae with the notable exception of Chlamydomonas (Schein et al., 2008). It is an essential endoribonuclease in E. coli, involved in both mRNA maturation and degradation (Aït-Bara and Carpousis, 2015). In vitro RNA cleavage assays confirmed that the plant enzyme is an endoribonuclease with a preference for single-stranded RNA with A/U rich sequences and 5′ monophosphate, a result in line with the described behavior of the bacterial enzyme (Mudd et al., 2008; Schein et al., 2008). RNase E is found in high Mr complexes in the soluble fraction of the chloroplast stroma, probably in an oligomeric state (Schein et al., 2008). Null rne mutants grow extremely slowly compared with wild type, are chlorotic, and contain smaller chloroplasts, demonstrating the essential role of RNase E in plants (Mudd et al., 2008; Walter et al., 2010). They accumulate very long transcripts precursors at the expense of processed, mature products, suggesting that the enzyme could be involved in intercistronic cleavage of the long mRNA precursors (Walter et al., 2010; Stoppel et al., 2012). It was indeed proposed that the strong phenotype observed for rne mutants was the consequence of the mutant inability to properly express some of the chloroplast-encoded ribosomal proteins transcripts (Walter et al., 2010). It is, however, important to note that mature transcripts still accumulate in the mutant, albeit at a lower extent than in wild type, suggesting that other enzymes can partially complement RNase E deficiency (Walter et al., 2010; Stoppel et al., 2012). Coimmunoprecipitation showed that RNase E forms a ∼800 kD complex with an RNA binding protein, RHON1, which has been hypothesized to confer endonucleolytic cleavage specificity (Stoppel et al., 2012). RHON1 seems particularly important for the processing of the rbcL-accD and rrn23S-4.5S intergenic area, but the exact role of the interaction with RNase E is still unclear. It was indeed shown that RHON1 is involved in transcription termination downstream of rbcL independently of RNase E (Chi et al., 2014) and that it interacts with the plastid-encoded RNA polymerase to prevent the formation of detrimental R-loops during transcription-replication-head-on conflicts (Yang et al., 2020).
Other Ribonucleases
Besides the five major RNases previously described, chloroplasts contain additional RNases that are more specific to their substrate. Chloroplast tRNAs are processed at the 5′ end by a protein-only RNase P enzyme named PRORP1 (PROTEINACEOUS RNASE P1; Gobert et al., 2010; Gutmann et al., 2012) that is dually localized to both the mitochondria and the chloroplast (see “Mitochondrial Endoribonucleases”) and at the 3′ by the RNase Z TRZ2 (Canino et al., 2009). Several other ribonucleases are involved in the processing of the rRNA transcriptional unit.
Mini-RNase III
Most plants contain one homolog of the Bacillus subtilis mini-III endoribonuclease. These enzymes are active on double-stranded RNA (dsRNA) and only contain the catalytic domain of the RNase III enzyme, without the dsRNA binding domain (Redko et al., 2008; Hotto et al., 2015). Arabidopsis plants contain two paralogs, RNC3 and RNC4, whose null mutants do not display any macroscopic phenotype. Double rnc3/4 mutants are, however, smaller than wild type and display reduced chlorophyll accumulation in mature leaves. Both gene-specific and genome-wide approaches (RNA-seq) demonstrated that mini-III processes the 5′ end of the 23S rRNA and the 3′ of the 4.5S rRNA. The enzyme is also involved in the processing of an RNA antisense to the 4.5S rRNA and plays a role in intron degradation following splicing (Hotto et al., 2015).
YbeY
In both E. coli and B. subtilis, rRNA 16S 3′ end is matured by an endoribonuclease of the YbeY family (Baumgardt et al., 2018). In vitro, the Arabidopsis homolog AtYbeY preferentially cleaves rRNA substrates but is not able to recapitulate the correct rRNA 16S 3′ end processing. Null mutants do not survive on soil, and plants with a reduced amount of YbeY grow slowly, have pale-green cotyledons and leaves, and completely altered chloroplast development. Mutants contain 5′ and 3′ extensions on the 16S, 23S, and 4.5S rRNA species, but mRNAs accumulate normally. AtYebY is therefore likely to be an endoribonuclease specific to the rRNA operon maturation (Liu et al., 2015).
ERIL1
ERI-1 proteins are conserved eukaryotic 3′-5′ exoribonucleases from the DEDDh family that participate in small RNA pathways (Thomas et al., 2014). The Arabidopsis homolog, ERIL1, is conserved in all sequenced plant genomes and targeted to the chloroplast. Knockdown mutants in Arabidopsis and tobacco showed mild chlorosis and smaller leaves compared with wild type. Recombinant ERIL1 possess a 3′-5′ exoribonuclease activity on single-stranded but not double-stranded RNA in vitro. The mutants showed incorrect processing of the 23S to 4.5S rRNA precursor and a decrease in mature 4.5S and 5S rRNA, but the exact molecular defects remain unclear (Mermigka et al., 2016).
PPR proteins
Present in all eukaryotes, the pentatricopeptide repeat (PPR) protein family has greatly expanded in plants and is involved in every step of plastidial gene expression (Barkan and Small, 2014). Molecular analyses of the crr2 (CHLORORESPIRATORY REDUCTION2) Arabidopsis mutant showed an accumulation of the unprocessed rps7-ndhB dicistronic transcript (Hashimoto et al., 2003). The CRR2 protein has 9 PPR domains and characteristic E and DYW domains found in many PPR proteins. Because some DYW domains (including the one present in CRR2) have endoribonucleolytic activity in vitro, it was proposed that CRR2 was a sequence-specific endoribonuclease responsible for the precise ndhB 5′ end processing, the specificity being conferred by the PPR repeats (Nakamura and Sugita 2008; Okuda et al., 2009). Recent analyses, however, suggest that CRR2 role in rps7-ndhB processing is probably to protect the ndhB 5′ end from exoribonucleases (Ruwe et al., 2019). The PPR SMR is another subfamily of PPR proteins that potentially contain sequence specific endoribonucleases (Zhang and Lu 2019). Arabidopsis mutant for the PPR SMR SOT1 (SUPPRESSOR OF THYLAKOID FORMATION1) protein has retarded growth, virescent leaves, and abnormal chloroplast structures, whereas maize mutants for the PPR53 ortholog are chlorotic and have an impaired NDH complex (Zoschke et al., 2016; Zhou et al., 2017). Molecularly, sot1 and ppr53 mutants accumulate incorrectly processed 23S to 4.5S rRNA precursor and mature forms, a possible explanation to the lower chloroplast protein synthesis observed (Wu et al., 2016; Zoschke et al., 2016; Zhou et al., 2017). Recombinant SOT1/PPR53 proteins can specifically bind the 5′ end of the 23S rRNA through their PPR domain, protecting the transcript from RNase degradation. Additionally, the SMR domain is necessary for correct rRNA maturation in vivo, and it can cleave the 5′ region of the 23S rRNA in vitro. Interestingly, the SMR domain expressed alone can cleave both Arabidopsis DNA and total RNA, whereas the full-length recombinant SOT1 specifically cleaves in the 5′ region of the 23S rRNA, around 19 to 20 nt downstream of its binding site (Zhou et al., 2017). It is, therefore, likely that the addition of a sequence-specific PPR domain to a nonspecific endonucleolytic SMR domain created a sequence-specific endoribonuclease in chloroplast. Arabidopsis chloroplasts contain at least five additional PPR SMR whose role as ribonucleases have not been explored yet (Liu et al., 2013; Zhang and Lu 2019).
Toward a Plastome View of RNase Action?
So far, analyses of RNA phenotypes for RNases mutants or silenced (VIGS) plants have been done with tedious and time-consuming gene by gene molecular techniques, leading to a general focus on a few heavily studied transcripts or clusters along with the rRNA operon. A consequence is that the knowledge has often been built on conflicting results from in vitro and/or in vivo analyses. For example, the relevance of strong in vitro endonuclease activity for CSP41 (Yang et al., 1996) does not easily reconcile with roles suggested by in vivo mutant analyses (Bollenbach et al., 2003; Beligni and Mayfield 2008; Bollenbach et al., 2009; Qi et al., 2012). Additionally, mutants for the endoribonuclease RNase E, its specificity partner RHON1, RNase J, and others, accumulate unique and apparently unprocessed transcripts, but the precise sites of cleavage are still unknown (Walter et al., 2010; Stoppel et al., 2012; Luro et al., 2013). These limitations have recently started to be overcome with the use of RNA-seq based approaches that have been instrumental in understanding the global role of two chloroplast ribonucleases, the exoribonuclease PNPase and the endoribonuclease mini-III (Hotto et al., 2011; Castandet et al., 2013; Ruwe et al., 2013; Hotto et al., 2015; Castandet et al., 2019). Combining a transcriptome-wide view to a single-nt resolution map of the chloroplast RNA processing sites is likely to speed up any future functional analysis of poorly studied chloroplastic ribonucleases whose RNA targets are still unknown.
MITOCHONDRIAL RNASES
Similar to chloroplast, 3′ ends of mitochondrial transcript are mainly matured through the cooperative action of the two 3′-5′ exoribonucleases, PNPase and RNR1. The major difference between the two organelle lies in the generation of mature 5′ ends: whereas the RNase J 5′-3′ exoribonuclease activity is essential in the chloroplast, there is no evidence of such an activity in plant mitochondria. Mature 5′ ends are, rather, generated through the action of various endoribonucleases whose identities have only begun to be revealed (Fig. 1B).
Mitochondrial 3′ to 5′ Exoribonucleases
As evidence of its essential role in plant and mitochondria physiology, mitochondrial PNPase null mutants are embryo lethal (Perrin et al., 2004b). In vivo studies using plants where the enzyme was down-regulated showed the accumulation of unprocessed transcripts containing long 3′ extensions (Perrin et al., 2004a; Perrin et al., 2004b). These plants also accumulate misprocessed, truncated, and cryptic polyadenylated transcripts that are not, or barely, detectable in wild-type plants. This is consistent with the known role of PNPase in the polyadenylation‐stimulated degradation pathway that is conserved from bacteria to organelles. No heteropolymeric tails could, however, be identified in these plants, suggesting that the mitochondrial PNPase might not function as a polymerase in vivo (Holec et al., 2006). Although plant mitochondria contain their own PNPase, RNR1 is dually targeted to both the chloroplast and mitochondria (Bollenbach et al., 2005; Perrin et al., 2004b). Mutants for RNR1 showed the accumulation of partially processed mitochondrial transcripts containing short 3′ tails that could be removed by the enzyme in vitro. This suggests that PNPase first removes large 3′ extensions, whereas RNR1 processes the remaining 3′ tails to produce mature 3′ ends. Like in chloroplast, secondary structures and RBPs can specify the correct site of processing by acting like barriers against 3′ to 5′ degradation (Forner et al., 2007; Haïli et al., 2013; Ruwe et al., 2016). It is important to note that such a model has so far only been validated for a few mitochondrial transcripts and still awaits global validation.
Plant mitochondria contain a third 3′ to 5′ exoribonuclease, the eukaryotic deadenylase PARN [poly(A)-specific RNase; Hirayama et al., 2013]. PARN contains three Exo domains characteristics of the RNase D family, and its homologs are widely distributed among eukaryotes (Chiba et al., 2004; Reverdatto et al., 2004). Null mutants are embryo lethal, and a weak allele is dwarf, with short roots, and displays hypersensitivity to the plant hormones abscisic acid and salicylic acid (Chiba et al., 2004; Reverdatto et al., 2004; Nishimura et al., 2005). The partially defective mutant accumulates 3′ polyadenylated mitochondrial mRNAs (Hirayama et al., 2013). It was shown in vitro that PARN only degrades the poly A tail and not the rest of a substrate RNA; however, it is unclear whether PARN is involved in vivo in the removal of poly A tract only or more largely in RNA degradation and/or processing (Reverdatto et al., 2004). Different from PNPase, the PARN mutant did not accumulate defective or misprocessed transcripts, suggesting that these two RNases have distinct roles in poly(A)-dependent RNA metabolism. The PARN role is, however, conserved in embryophytes as the enzyme from Marchantia polymorpha can complement the Arabidopsis mutant (Kanazawa et al., 2020).
Mitochondrial Endoribonucleases
Endoribonucleases have a major role in generating mitochondrial RNA termini, especially at the 5′ end (Binder et al., 2016). Termini mapping of mRNAs revealed that some 5′ and 3′ ends were located directly at the processing sites of adjacent tRNAs and t-elements (for tRNA-like elements), suggesting the involvement of RNase P and RNase Z endonucleases (Forner et al., 2007). The RNase P PRORP1 is a member of the PPR protein family and also contains a conserved NYN (for Nedd4-BP1, YacP Nuclease) metallonuclease domain (Matelska et al., 2017). Null mutants are lethal, but VIGS plants depleted for PRORP1 showed an altered mitochondrial structure with disorganized cristae. The enzyme can process in vitro a mRNA containing a t-element similarly to what is observed in vivo, and the same transcript is unstable in VIGS plants (Gobert et al., 2010; Gutmann et al., 2012). tRNAs and t-elements 3′ ends are processed by TRZ3 and TRZ4, two of the four RNase Z homologs encoded in the Arabidopsis genome. Single knock-out lines are viable but the double null mutant is lethal, pointing to a functional redundancy. Recombinant TRZ3 and TRZ4 correctly process tRNA and t-elements 3′ ends in vitro, at the exact mRNA site described in vivo (Forner et al., 2007; Canino et al., 2009).
Most mRNAs are, however, independent from tRNAs or t-elements and generated through the action of endoribonucleases. Mitochondrial nuclease1 (MNU1) and MNU2, two other members of the NYN family, also play a role in mitochondrial transcript maturation. Although single and/or double mutants did not display any obvious macroscopic phenotypes, large, 5′-extended mRNA precursors accumulated at the expense of some mature mRNAs in the mutants. This was, however, not true for most mRNAs and the endoribonuclease activities have not been tested in vitro (Stoll and Binder 2016). Coimmunoprecipitation and yeast two-hybrid assays recently showed that PRORP1 physically interacts with MNU2, and the latter also plays a role in tRNA accumulation (Bouchoucha et al., 2019). Because several sequence specific PPR proteins are known or predicted to bind upstream of mitochondrial mRNA 5′ mature ends (Ruwe et al., 2016), the current model is that they direct unspecific endoribonucleases to the correct site of processing (Binder et al., 2016). This mode of action is convincingly supported by an experiment with the RNA binding protein RPF2 that was altered to bind to a new transcript, different from its natural cognate. This induced a new endoribonuclease cleavage site downstream of the modified RPF2 binding site (Colas des Francs-Small et al., 2018). Similarly, binding of the PPR protein RFL2 to the mitochondrial orf291 transcript induces a cleavage by PRORP1 around 70 nt downstream of the binding site (Fujii et al., 2016).
CONCLUSION
RNA homeostasis is essential to maintain proper cellular function. In addition to the “usual suspects” that participate in cytoplasmic and nuclear RNA metabolism, it is clear that organellar and secretory RNases have an important role in this homeostatic regulation. RNases located in plastids and mitochondria maintain a functional transcriptome, affecting not only mRNA accumulation but also adequate synthesis of rRNA and other noncoding RNA species essential for their function. Similarly, secretory RNases connect RNA levels to primary metabolism, and have also been recruited for a variety of stress response mechanisms. Despite their importance, we still do not have a full understanding of their activities and the actual number of RNases that are located in organelles. More important, in some cases we do not know the in vivo substrate of these RNases and whether the substrates are recruited to the RNase location or RNases translocate to carry out their specific functions. Finally, although it is clear that many of these RNases have to coordinate their activity to achieve the desired RNA outcome, either in a synthetic or catabolic pathway, very little information exists on partners of organellar and secretory RNases, and posttranslational regulatory mechanisms that could control their activity (see Outstanding Questions). This is best exemplified by the unresolved question of the RNA quality control in chloroplast, where the same ribonucleases in charge of transcript processing also function in RNA degradation: How do they “know” whether to preserve or degrade RNA? In addition to the known polyadenylation-assisted degradation (Schuster and Stern, 2009; Germain et al., 2013) and the protective action of secondary structures and RNA binding proteins (Hayes et al., 1996; Perrin et al., 2004b; Manavski et al., 2018), three main hypotheses have been proposed. First, biochemical evidence showed that RNases do not act alone and are part of high Mr complexes, where they interact with multiple unknown partners that support their functions (Hayes et al., 1996; Stoppel et al., 2012). It is easy to speculate that some of these partners could play a role in cleavage specificity. Specific RNA binding proteins could also direct a RNase to a specific RNA target, similarly to what has been described in mitochondria. Second, proteomics studies have shown that RNases can be post translationally modified, for example through phosphorylation. As proposed for CSP41, this could represent a way to regulate the RNase specificity and/or activity (Leister 2014). Third, RNase activity could be regulated through the spatial localization of both enzymes and substrates, similarly to what has been described in bacteria (Redder 2016). It was for example proposed that the cooperation between PNPase and RNR1 could be based on their preferential localization in the chloroplast nucleoid and stroma, respectively (Germain et al., 2012). These questions should drive research in the near future if we are to have a systems understanding of RNA homeostasis in plants.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank all the members of our respective past and present laboratories for support.
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1714996 to G.C.M.) and Institute of Plant Sciences Paris-Saclay benefits from the Labex Saclay Plant Sciences (grant no. ANR–10–LABX–0040–SPS).
Articles can be viewed without a subscription.
References
- Abel S, Köck M(2001) Secretory acid ribonucleases from tomato, Lycopersicon esculentum Mill. Methods Enzymol 341: 351–368 [DOI] [PubMed] [Google Scholar]
- Aït-Bara S, Carpousis AJ(2015) RNA degradosomes in bacteria and chloroplasts: Classification, distribution and evolution of RNase E homologs. Mol Microbiol 97: 1021–1135 [DOI] [PubMed] [Google Scholar]
- Alves CS, Vicentini R, Duarte GT, Pinoti VF, Vincentz M, Nogueira FTS(2017) Genome-wide identification and characterization of tRNA-derived RNA fragments in land plants. Plant Mol Biol 93: 35–48 [DOI] [PubMed] [Google Scholar]
- Ambrosio L, Morriss S, Riaz A, Bailey R, Ding J, MacIntosh GC(2014) Phylogenetic analyses and characterization of RNase X25 from Drosophila melanogaster suggest a conserved housekeeping role and additional functions for RNase T2 enzymes in protostomes. PLoS One 9: e105444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen KL, Collins K(2012) Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol Biol Cell 23: 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S, Shteiman-Kotler A, Liveanu V, Yehudai-Resheff S, Bellaoui M, Settlage RE, Shabanowitz J, Hunt DF, Schuster G, Gruissem W(2001) Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA 7: 1464–1475 [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, MacIntosh GC, Green PJ(1999) Regulation of S-like ribonuclease levels in Arabidopsis. Antisense inhibition of RNS1 or RNS2 elevates anthocyanin accumulation. Plant Physiol 119: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I(2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Baumgardt K, Gilet L, Figaro S, Condon C(2018) The essential nature of YqfG, a YbeY homologue required for 3′ maturation of Bacillus subtilis 16S ribosomal RNA is suppressed by deletion of RNase R. Nucleic Acids Res 46: 8605–8615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beligni MV, Mayfield SP(2008) Arabidopsis thaliana mutants reveal a role for CSP41a and CSP41b, two ribosome-associated endonucleases, in chloroplast ribosomal RNA metabolism. Plant Mol Biol 67: 389–401 [DOI] [PubMed] [Google Scholar]
- Binder S, Stoll K, Stoll B(2016) Maturation of 5′ ends of plant mitochondrial RNAs. Physiol Plant 157: 280–288 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Lange H, Gutierrez R, Erhardt M, Stern DB, Gagliardi D(2005) RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res 33: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Sharwood RE, Gutierrez R, Lerbs-Mache S, Stern DB(2009) The RNA-binding proteins CSP41a and CSP41b may regulate transcription and translation of chloroplast-encoded RNAs in Arabidopsis. Plant Mol Biol 69: 541–552 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB(2003a) Divalent metal-dependent catalysis and cleavage specificity of CSP41, a chloroplast endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. Nucleic Acids Res 31: 4317–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach TJ, Stern DB(2003b) Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J Biol Chem 278: 25832–25838 [DOI] [PubMed] [Google Scholar]
- Bollenbach TJ, Tatman DA, Stern DB(2003) CSP41a, a multifunctional RNA-binding protein, initiates mRNA turnover in tobacco chloroplasts. Plant J 36: 842–852 [DOI] [PubMed] [Google Scholar]
- Bouchoucha A, Waltz F, Bonnard G, Arrivé M, Hammann P, Kuhn L, Schelcher C, Zuber H, Gobert A, Giegé P(2019) Determination of protein-only RNase P interactome in Arabidopsis mitochondria and chloroplasts identifies a complex between PRORP1 and another NYN domain nuclease. Plant J 100: 549–561 [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R(2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153: 1461–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canino G, Bocian E, Barbezier N, Echeverría M, Forner J, Binder S, Marchfelder A(2009) Arabidopsis encodes four tRNase Z enzymes. Plant Physiol 150: 1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B, Germain A, Hotto AM, Stern DB(2019) Systematic sequencing of chloroplast transcript termini from Arabidopsis thaliana reveals >200 transcription initiation sites and the extensive imprints of RNA-binding proteins and secondary structures. Nucleic Acids Res 47: 11889–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castandet B, Hotto AM, Fei Z, Stern DB(2013) Strand-specific RNA sequencing uncovers chloroplast ribonuclease functions. FEBS Lett 587: 3096–3101 [DOI] [PubMed] [Google Scholar]
- Chantarachot T, Bailey-Serres J(2018) Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176: 254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou W, Zhao J(2015) Ribonuclease J is required for chloroplast and embryo development in Arabidopsis. J Exp Bot 66: 2079–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z-F, Deutscher MP(2005) An important role for RNase R in mRNA decay. Mol Cell 17: 313–318 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Ghulam MM, Rondet D, Pfannschmidt T, Merendino L, Lerbs-Mache S(2015) Characterization of the psbH precursor RNAs reveals a precise endoribonuclease cleavage site in the psbT/psbH intergenic region that is dependent on psbN gene expression. Plant Mol Biol 88: 357–367 [DOI] [PubMed] [Google Scholar]
- Chi W, He B, Manavski N, Mao J, Ji D, Lu C, Rochaix JD, Meurer J, Zhang L(2014) RHON1 mediates a Rho-like activity for transcription termination in plastids of Arabidopsis thaliana. Plant Cell 26: 4918–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Johnson MA, Lidder P, Vogel JT, van Erp H, Green PJ(2004) AtPARN is an essential poly(A) ribonuclease in Arabidopsis. Gene 328: 95–102 [DOI] [PubMed] [Google Scholar]
- Chu L-Y, Hsieh T-J, Golzarroshan B, Chen Y-P, Agrawal S, Yuan HS(2017) Structural insights into RNA unwinding and degradation by RNase R. Nucleic Acids Res 45: 12015–12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognat V, Morelle G, Megel C, Lalande S, Molinier J, Vincent T, Small I, Duchêne A-M, Maréchal-Drouard L(2017) The nuclear and organellar tRNA-derived RNA fragment population in Arabidopsis thaliana is highly dynamic. Nucleic Acids Res 45: 3460–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Vincis Pereira Sanglard L, Small I(2018) Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun Biol 1: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G(2009) Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 15: 2147–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Gilet L(2011) The Metallo-β-Lactamase family of ribonucleases In Nicholson AW, ed, Ribonucleases. Springer Berlin, Heidelberg, pp 245–267 [Google Scholar]
- Condon C, Piton J, Braun F(2018) Distribution of the ribosome associated endonuclease Rae1 and the potential role of conserved amino acids in codon recognition. RNA Biol 15: 683–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Baena M, Galvez-Valdivieso G, Delgado-Garcia E, Pineda M, Piedras P(2020) Nuclease and ribonuclease activities in response to salt stress: Identification of PvRNS3, a T2/S-like ribonuclease induced in common bean radicles by salt stress. Plant Physiol Biochem 147: 235–241 [DOI] [PubMed] [Google Scholar]
- dos Santos RF, Quendera AP, Boavida S, Seixas AF, Arraiano CM, Andrade JM(2018) Major 3’-5’ exoribonucleases in the metabolism of coding and non-coding RNA. Prog Mol Biol Transl Sci 159: 101–155 [DOI] [PubMed] [Google Scholar]
- Dou S, Wang Y, Lu J(2019) Metazoan tsRNAs: Biogenesis, evolution and regulatory functions. Noncoding RNA 5: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drager RG, Girard-Bascou J, Choquet Y, Kindle KL, Stern DB(1998) In vivo evidence for 5→3′ exoribonuclease degradation of an unstable chloroplast mRNA. Plant J 13: 85–96 [DOI] [PubMed] [Google Scholar]
- Drager RG, Higgs DC, Kindle KL, Stern DB(1999) 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J 19: 521–531 [DOI] [PubMed] [Google Scholar]
- Even S, Pellegrini O, Zig L, Labas V, Vinh J, Bréchemmier-Baey D, Putzer H(2005) Ribonucleases J1 and J2: Two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res 33: 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, MacIntosh GC, Bassham DC(2015) Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11: 2199–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BE, Mugume Y, Morriss SC, MacIntosh GC, Bassham DC(2017) Localization of RNS2 ribonuclease to the vacuole is required for its role in cellular homeostasis. Planta 245: 779–792 [DOI] [PubMed] [Google Scholar]
- Fontaine BM, Martin KS, Garcia-Rodriguez JM, Jung C, Briggs L, Southwell JE, Jia X, Weinert EE(2018) RNase I regulates Escherichia coli 2′,3′-cyclic nucleotide monophosphate levels and biofilm formation. Biochem J 475: 1491–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J, Weber B, Thuss S, Wildum S, Binder S(2007) Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res 35: 3676–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel LB, Lubas M, Lund AH(2017) Emerging connections between RNA and autophagy. Autophagy 13: 3–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker R, Brogli R, Luidalepp H, Wyss L, Fasnacht M, Joss O, Zywicki M, Helm M, Schneider A, Cristodero M, Polacek N(2019) A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat Commun 10: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Suzuki T, Giegé P, Higashiyama T, Koizuka N, Shikanai T(2016) The Restorer-of-fertility-like 2 pentatricopeptide repeat protein and RNase P are required for the processing of mitochondrial orf291 RNA in Arabidopsis. Plant J 86: 504–513 [DOI] [PubMed] [Google Scholar]
- Fukuda H.(2000) Programmed cell death of tracheary elements as a paradigm in plants In Lam E, Fukuda H, and Greenberg J, eds, Programmed Cell Death in Higher Plants. Springer Netherlands, Dordrecht, pp 1–9 [DOI] [PubMed] [Google Scholar]
- Fukudome A, Fukuhara T(2017) Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J Plant Res 130: 33–44 [DOI] [PubMed] [Google Scholar]
- Germain A, Herlich S, Larom S, Kim SH, Schuster G, Stern DB(2011) Mutational analysis of Arabidopsis chloroplast polynucleotide phosphorylase reveals roles for both RNase PH core domains in polyadenylation, RNA 3′-end maturation and intron degradation. Plant J 67: 381–394 [DOI] [PubMed] [Google Scholar]
- Germain A, Hotto AM, Barkan A, Stern DB(2013) RNA processing and decay in plastids. Wiley Interdiscip Rev RNA 4: 295–316 [DOI] [PubMed] [Google Scholar]
- Germain A, Kim SH, Gutierrez R, Stern DB(2012) Ribonuclease II preserves chloroplast RNA homeostasis by increasing mRNA decay rates, and cooperates with polynucleotide phosphorylase in 3′ end maturation. Plant J 72: 960–971 [DOI] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P(2010) A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744 [DOI] [PubMed] [Google Scholar]
- Gross N, Wasternack C, Köck M(2004) Wound-induced RNaseLE expression is jasmonate and systemin independent and occurs only locally in tomato (Lycopersicon esculentum cv. Lukullus). Phytochemistry 65: 1343–1350 [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago M, Oritz PJ, Ochoa S(1955) Enzymatic synthesis of nucleic acidlike polynucleotides. Science 122: 907–910 [DOI] [PubMed] [Google Scholar]
- Gutmann B, Gobert A, Giegé P(2012) PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev 26: 1022–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H(2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41: 6650–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpert M, Liveanu V, Glaser F, Schuster G(2019) The Arabidopsis chloroplast RNase J displays both exo- and robust endonucleolytic activities. Plant Mol Biol 99: 17–29 [DOI] [PubMed] [Google Scholar]
- Hammani K, Cook WB, Barkan A(2012) RNA binding and RNA remodeling activities of the half-a-tetratricopeptide (HAT) protein HCF107 underlie its effects on gene expression. Proc Natl Acad Sci USA 109: 5651–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T(2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Haud N, Kara F, Diekmann S, Henneke M, Willer JR, Hillwig MS, Gregg RG, Macintosh GC, Gärtner J, Alia A, et al. (2011) rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci USA 108: 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R, Kudla J, Schuster G, Gabay L, Maliga P, Gruissem W(1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J 15: 1132–1141 [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC(2011a) RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Kanobe C, Thornburg RW, Macintosh GC(2011b) Identification of S-RNase and peroxidase in petunia nectar. J Plant Physiol 168: 734–738 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Lebrasseur ND, Green PJ, Macintosh GC(2008) Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Mol Genet Genomics 280: 249–261 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Rizhsky L, Wang Y, Umanskaya A, Essner JJ, MacIntosh GC(2009) Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals. BMC Evol Biol 9: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Matsuura T, Ushiyama S, Narusaka M, Kurihara Y, Yasuda M, Ohtani M, Seki M, Demura T, Nakashita H, et al. (2013) A poly(A)-specific ribonuclease directly regulates the poly(A) status of mitochondrial mRNA in Arabidopsis. Nat Commun 4: 2247. [DOI] [PubMed] [Google Scholar]
- Holec S, Lange H, Kühn K, Alioua M, Börner T, Gagliardi D(2006) Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol Cell Biol 26: 2869–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Castandet B, Gilet L, Higdon A, Condon C, Stern DB(2015) Arabidopsis chloroplast mini-ribonuclease III participates in rRNA maturation and intron recycling. Plant Cell 27: 724–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Huston ZE, Stern DB(2010) Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Schmitz RJ, Fei Z, Ecker JR, Stern DB(2011) Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 (Bethesda) 1: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto AM, Stern DB, Schuster G(2020) Plant ribonuclease J: An essential player in maintaining chloroplast RNA quality control for gene expression. Plants (Basel) 9: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kawamata T, Horie T, Tsugawa H, Nakayama Y, Ohsumi Y, Fukusaki E(2015) Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J 34: 154–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot K, Ponchet M, Marais A, Ricci P, Galiana E(2002) A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Mol Plant Microbe Interact 15: 243–250 [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR(2001) Evolutionary relationships among self-incompatibility RNases. Proc Natl Acad Sci USA 98: 13167–13171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie M.(1999) Structure-function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol Ther 81: 77–89 [DOI] [PubMed] [Google Scholar]
- Kanazawa M, Ikeda Y, Nishihama R, Yamaoka S, Lee N-H, Yamato KT, Kohchi T, Hirayama T(2020) Regulation of the poly(A) status of mitochondrial mRNA by poly(A)-specific ribonuclease is conserved among land plants. Plant Cell Physiol 61: 470–480 [DOI] [PubMed] [Google Scholar]
- Kim K, Yadav D, Cho M(2019) Multi-phased internalization of murine norovirus (MNV) in Arabidopsis seedlings and its potential correlation with plant defensive responses. Microb Pathog 135: 103648. [DOI] [PubMed] [Google Scholar]
- Kishine M, Takabayashi A, Munekage Y, Shikanai T, Endo T, Sato F(2004) Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis. Plant Mol Biol 55: 595–606 [DOI] [PubMed] [Google Scholar]
- Köck M, Löffler A, Abel S, Glund K(1995) cDNA structure and regulatory properties of a family of starvation-induced ribonucleases from tomato. Plant Mol Biol 27: 477–485 [DOI] [PubMed] [Google Scholar]
- Köthke S, Köck M(2011) The Solanum lycopersicum RNaseLER is a class II enzyme of the RNase T2 family and shows preferential expression in guard cells. J Plant Physiol 168: 840–847 [DOI] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A(2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Ndecky SYA, Gomez-Diaz C, Pflieger D, Butel N, Zumsteg J, Kuhn L, Piermaria C, Chicher J, Christie M, et al. (2019) RST1 and RIPR connect the cytosolic RNA exosome to the Ski complex in Arabidopsis. Nat Commun 10: 3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange H, Zuber H, Sement FM, Chicher J, Kuhn L, Hammann P, Brunaud V, Bérard C, Bouteiller N, Balzergue S, et al. (2014) The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet 10: e1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW(2014) ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun 5: 4121. [DOI] [PubMed] [Google Scholar]
- Lehmann K, Hause B, Altmann D, Köck M(2001) Tomato ribonuclease LX with the functional endoplasmic reticulum retention motif HDEF is expressed during programmed cell death processes, including xylem differentiation, germination, and senescence. Plant Physiol 127: 436–449 [PMC free article] [PubMed] [Google Scholar]
- Leister D.(2014) Complex(iti)es of the ubiquitous RNA-binding CSP41 proteins. Front Plant Sci 5: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lers A, Sonego L, Green PJ, Burd S(2006) Suppression of LX ribonuclease in tomato results in a delay of leaf senescence and abscission. Plant Physiol 142: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP(2004) Exoribonucleases and endoribonucleases. Ecosal Plus 1: 1. [DOI] [PubMed] [Google Scholar]
- Liponska A, Jamalli A, Kuras R, Suay L, Garbe E, Wollman F-A, Laalami S, Putzer H(2018) Tracking the elusive 5′ exonuclease activity of Chlamydomonas reinhardtii RNase J. Plant Mol Biol 96: 641–653 [DOI] [PubMed] [Google Scholar]
- Lisitsky I, Klaff P, Schuster G(1996) Addition of destabilizing poly (A)-rich sequences to endonuclease cleavage sites during the degradation of chloroplast mRNA. Proc Natl Acad Sci USA 93: 13398–13403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhou W, Liu G, Yang C, Sun Y, Wu W, Cao S, Wang C, Hai G, Wang Z, et al. (2015) The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol 168: 205–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Melonek J, Boykin LM, Small I, Howell KA(2013) PPR-SMRs: Ancient proteins with enigmatic functions. RNA Biol 10: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zou W, Yang P, Wang L, Ma Y, Zhang H, Wang X(2018) Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. eLife 7: e36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler A, Abel S, Jost W, Beintema JJ, Glund K(1992) Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiol 98: 1472–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luro S, Germain A, Sharwood RE, Stern DB(2013) RNase J participates in a pentatricopeptide repeat protein-mediated 5′ end maturation of chloroplast mRNAs. Nucleic Acids Res 41: 9141–9151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC.(2011) RNase T2 family: Enzymatic properties, functional diversity, and evolution of ancient ribonucleases In Nicholson AWW, ed, Ribonucleases, Vol Vol 26 Springer Berlin,, Heidelberg, pp 89–114 [Google Scholar]
- MacIntosh GC, Hillwig MS, Meyer A, Flagel L(2010) RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol Genet Genomics 283: 381–396 [DOI] [PubMed] [Google Scholar]
- Manavski N, Schmid L-M, Meurer J(2018) RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem 62: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Yehudai-Resheff S, Germain A, Fei Z, Jiang X, Judkins J, Wu H, Fernie AR, Fait A, Stern DB(2009) Abnormal physiological and molecular mutant phenotypes link chloroplast polynucleotide phosphorylase to the phosphorus deprivation response in Arabidopsis. Plant Physiol 151: 905–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK(2017) tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45: 5142–5152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matelska D, Steczkiewicz K, Ginalski K(2017) Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res 45: 6995–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathy N, Bénard L, Pellegrini O, Daou R, Wen T, Condon C(2007) 5′-To-3′ exoribonuclease activity in bacteria: Role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell 129: 681–692 [DOI] [PubMed] [Google Scholar]
- Matsui A, Nakaminami K, Seki M(2019) Biological function of changes in RNA metabolism in plant adaptation to abiotic stress. Plant Cell Physiol 60: 1897–1905 [DOI] [PubMed] [Google Scholar]
- McClure BA, Gray JE, Anderson MA, Clarke AE(1990) Self-incompatibility in Nicotiana alata involves degradation of pollen rRNA. Nature 347: 757–760 [Google Scholar]
- Megel C, Hummel G, Lalande S, Ubrig E, Cognat V, Morelle G, Salinas-Giegé T, Duchêne A-M, Maréchal-Drouard L(2019) Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res 47: 941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megel C, Morelle G, Lalande S, Duchêne A-M, Small I, Maréchal-Drouard L(2015) Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci 16: 1873–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermigka G, Helm JM, Vlatakis I, Schumacher HT, Vamvaka E, Kalantidis K(2016) ERIL1, the plant homologue of ERI-1, is involved in the processing of chloroplastic rRNAs. Plant J 88: 839–853 [DOI] [PubMed] [Google Scholar]
- Morriss SC, Liu X, Floyd BE, Bassham DC, MacIntosh GC(2017) Cell growth and homeostasis are disrupted in arabidopsis rns2-2 mutants missing the main vacuolar RNase activity. Ann Bot 120: 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd EA, Sullivan S, Gisby MF, Mironov A, Kwon CS, Chung W-I, Day A(2008) A 125 kDa RNase E/G-like protein is present in plastids and is essential for chloroplast development and autotrophic growth in Arabidopsis. J Exp Bot 59: 2597–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sugita M(2008) A conserved DYW domain of the pentatricopeptide repeat protein possesses a novel endoribonuclease activity. FEBS Lett 582: 4163–4168 [DOI] [PubMed] [Google Scholar]
- Nicholson AW.(1999) Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev 23: 371–390 [DOI] [PubMed] [Google Scholar]
- Niehaus TD, Nguyen TND, Gidda SK, ElBadawi-Sidhu M, Lambrecht JA, McCarty DR, Downs DM, Cooper AJL, Fiehn O, Mullen RT, et al. (2014) Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 26: 3010–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Kitahata N, Seki M, Narusaka Y, Narusaka M, Kuromori T, Asami T, Shinozaki K, Hirayama T(2005) Analysis of ABA hypersensitive germination2 revealed the pivotal functions of PARN in stress response in Arabidopsis. Plant J 44: 972–984 [DOI] [PubMed] [Google Scholar]
- Nowacka M, Strozycki PM, Jackowiak P, Hojka-Osinska A, Szymanski M, Figlerowicz M(2013) Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol Biol 83: 191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Abel S, Jost W, Glund K(1990) Induction of an extracellular ribonuclease in cultured tomato cells upon phosphate starvation. Plant Physiol 92: 970–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Chateigner-Boutin A-L, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T(2009) Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell 21: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Lange H, Grienenberger J-M, Gagliardi D(2004a) AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res 32: 5174–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R, Meyer EH, Zaepfel M, Kim Y-J, Mache R, Grienenberger J-M, Gualberto JM, Gagliardi D(2004b) Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J Biol Chem 279: 25440–25446 [DOI] [PubMed] [Google Scholar]
- Prikryl J, Rojas M, Schuster G, Barkan A(2011) Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc Natl Acad Sci USA 108: 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Armbruster U, Schmitz-Linneweber C, Delannoy E, de Longevialle AF, Rühle T, Small I, Jahns P, Leister D(2012) Arabidopsis CSP41 proteins form multimeric complexes that bind and stabilize distinct plastid transcripts. J Exp Bot 63: 1251–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanauskas K, Igić B(2017) The evolutionary history of plant T2/S-type ribonucleases. PeerJ 5: e3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redder P.(2016) How does sub-cellular localization affect the fate of bacterial mRNA? Curr Genet 62: 687–690 [DOI] [PubMed] [Google Scholar]
- Redko Y, Bechhofer DH, Condon C(2008) Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis. Mol Microbiol 68: 1096–1106 [DOI] [PubMed] [Google Scholar]
- Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA(2004) mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA 10: 1200–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas H, Floyd B, Morriss SC, Bassham D, MacIntosh GC, Goldraij A(2015) NnSR1, a class III non-S-RNase specifically induced in Nicotiana alata under phosphate deficiency, is localized in endoplasmic reticulum compartments. Plant Sci 236: 250–259 [DOI] [PubMed] [Google Scholar]
- Rojas HJ, Caspani C, Escobar EG, Quiroga R, Goldraij A(2018) NaPi/SX-RNase segregates as a functional S-RNase and is induced under phosphate deficiency in Nicotiana alata. Biol Plant 62: 261–268 [Google Scholar]
- Ruwe H, Castandet B, Schmitz-Linneweber C, Stern DB(2013) Arabidopsis chloroplast quantitative editotype. FEBS Lett 587: 1429–1433 [DOI] [PubMed] [Google Scholar]
- Ruwe H, Gutmann B, Schmitz-Linneweber C, Small I, Kindgren P(2019) The E domain of CRR2 participates in sequence-specific recognition of RNA in plastids. New Phytol 222: 218–229 [DOI] [PubMed] [Google Scholar]
- Ruwe H, Wang G, Gusewski S, Schmitz-Linneweber C(2016) Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res 44: 7406–7417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J, Delseny M(2019) Ribosome biogenesis in plants: From functional 45S ribosomal DNA organization to ribosome assembly factors. Plant Cell 31: 1945–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schein A, Sheffy-Levin S, Glaser F, Schuster G(2008) The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14: 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH(2018) Controlling nuclear RNA levels. Nat Rev Genet 19: 518–529 [DOI] [PubMed] [Google Scholar]
- Schuster G, Stern D(2009) RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci 85: 393–422 [DOI] [PubMed] [Google Scholar]
- Shang J, Yang Y, Wu L, Zou M, Huang Y(2018) The S. pombe mitochondrial transcriptome. RNA 24: 1241–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB(2011a) Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA 17: 2165–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharwood RE, Hotto AM, Bollenbach TJ, Stern DB(2011b) Overaccumulation of the chloroplast antisense RNA AS5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 17: 230–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Vincent JN(2018) Beyond transcription factors: Roles of mRNA decay in regulating gene expression in plants. F1000 Res 7: 1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorska N, Zuber H, Gobert A, Lange H, Gagliardi D(2017) RNA degradation by the plant RNA exosome involves both phosphorolytic and hydrolytic activities. Nat Commun 8: 2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šiková M, Wiedermannová J, Převorovský M, Barvík I, Sudzinová P, Kofroňová O, Benada O, Šanderová H, Condon C, Krásný L(2020) The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes. EMBO J 39: e102500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva F, Guirgis A, von Aderkas P, Borchers CH, Thornburg R(2020) LC-MS/MS based comparative proteomics of floral nectars reveal different mechanisms involved in floral defense of Nicotiana spp., Petunia hybrida and Datura stramonium. J Proteomics 213: 103618. [DOI] [PubMed] [Google Scholar]
- Sorenson RS, Deshotel MJ, Johnson K, Adler FR, Sieburth LE(2018) Arabidopsis mRNA decay landscape arises from specialized RNA decay substrates, decapping-mediated feedback, and redundancy. Proc Natl Acad Sci USA 115: E1485–E1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll B, Binder S(2016) Two NYN domain containing putative nucleases are involved in transcript maturation in Arabidopsis mitochondria. Plant J 85: 278–288 [DOI] [PubMed] [Google Scholar]
- Stoppel R, Manavski N, Schein A, Schuster G, Teubner M, Schmitz-Linneweber C, Meurer J(2012) RHON1 is a novel ribonucleic acid-binding protein that supports RNase E function in the Arabidopsis chloroplast. Nucleic Acids Res 40: 8593–8606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R, Meurer J(2012) The cutting crew–ribonucleases are key players in the control of plastid gene expression. J Exp Bot 63: 1663–1673 [DOI] [PubMed] [Google Scholar]
- Taylor CB, Bariola PA, delCardayré SB, Raines RT, Green PJ(1993) RNS2: A senescence-associated RNase of Arabidopsis that diverged from the S-RNases before speciation. Proc Natl Acad Sci USA 90: 5118–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MF, L’Etoile ND, Ansel KM(2014) Eri1: A conserved enzyme at the crossroads of multiple RNA-processing pathways. Trends Genet 30: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R(2009) The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn A, Steinfeld R, Ziegenbein M, Grapp M, Hsiao H-H, Urlaub H, Sheldrick GM, Gärtner J, Krätzner R(2012) Structure and activity of the only human RNase T2. Nucleic Acids Res 40: 8733–8742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Sikorski PJ, Zakrzewska-Placzek M(2017) Comparison of preribosomal RNA processing pathways in yeast, plant and human cells–focus on coordinated action of endo- and exoribonucleases. FEBS Lett 591: 1801–1850 [DOI] [PubMed] [Google Scholar]
- Towler BP, Newbury SF(2018) Regulation of cytoplasmic RNA stability: Lessons from Drosophila. Wiley Interdiscip Rev RNA 9: e1499. [DOI] [PubMed] [Google Scholar]
- Trifonova EA, Romanova AV, Sangaev SS, Sapotsky MV, Malinovsky VI, Kochetov AV(2012) Inducible expression of the gene of Zinnia elegans coding for extracellular ribonuclease in Nicotiana tabacum plants. Biol Plant 56: 571–574 [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco VME, Mansbridge J, Bremner S, Carruthers K, Summers PS, Sung WWL, Champigny MJ, Weretilnyk EA(2016) Acclimation of the crucifer Eutrema salsugineum to phosphate limitation is associated with constitutively high expression of phosphate-starvation genes. Plant Cell Environ 39: 1818–1834 [DOI] [PubMed] [Google Scholar]
- Walter M, Kilian J, Kudla J(2002) PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J 21: 6905–6914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Drechsel O, Weingartner M, Kudla J, Bock R(2010) Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J 64: 851–863 [DOI] [PubMed] [Google Scholar]
- Wang J, Mei J, Ren G(2019) Plant microRNAs: Biogenesis, homeostasis, and degradation. Front Plant Sci 10: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Stiffler N, Belcher S, Kroeger T, Stern DB, Monde R-A, Coalter R, Barkan A(2010) Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, et al. (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J 85: 607–621 [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Ivanov P, Hu GF, Anderson P(2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185: 35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Schuster G, Stern DB(1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell 8: 1409–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Stern DB(1997) The spinach chloroplast endoribonuclease CSP41 cleaves the 3′-untranslated region of petD mRNA primarily within its terminal stem-loop structure. J Biol Chem 272: 12874–12880 [DOI] [PubMed] [Google Scholar]
- Yang Z, Li M, Sun Q(2020) RHON1 co-transcriptionally resolves R-Loops for Arabidopsis chloroplast genome maintenance. Cell Rep 30: 243–256.e5 [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Hirsh M, Schuster G(2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol Cell Biol 21: 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G(2003) Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell 15: 2003–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB(2007) Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jia T, Chen X(2017) The ‘how’ and ‘where’ of plant microRNAs. New Phytol 216: 1002–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H(2017) mRNA decay in plants: Both quantity and quality matter. Curr Opin Plant Biol 35: 138–144 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu C(2019) The enigmatic roles of PPR-SMR proteins in plants. Adv Sci (Weinh) 6: 1900361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Wang Y, He Y, Zhou J, Li Y, Liu Q, Xie X(2014) Overexpression of an S-like ribonuclease gene, OsRNS4, confers enhanced tolerance to high salinity and hyposensitivity to phytochrome-mediated light signals in rice. Plant Sci 214: 99–105 [DOI] [PubMed] [Google Scholar]
- Zhou W, Lu Q, Li Q, Wang L, Ding S, Zhang A, Wen X, Zhang L, Lu C(2017) PPR-SMR protein SOT1 has RNA endonuclease activity. Proc Natl Acad Sci USA 114: E1554–E1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Ow DW, Dong Z(2018) Transfer RNA-derived small RNAs in plants. Sci China Life Sci 61: 155–161 [DOI] [PubMed] [Google Scholar]
- Zoschke R, Watkins KP, Miranda RG, Barkan A(2016) The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize. Plant J 85: 594–606 [DOI] [PMC free article] [PubMed] [Google Scholar]