Abstract
Background
Mannose-binding lectin (MBL2) is considered to play a role in the human innate immune response to tuberculosis (TB) infections, and 4 common single nucleotide polymorphisms (SNPs) may be associated with pulmonary tuberculosis (PTB) risk. To examine these potential associations, we performed a comprehensive analysis to assess the relationships between MBL2 polymorphisms and PTB.
Methods
The PubMed, Embase, and SinoMed databases were searched for articles published prior to June 13, 2019. Odds ratios with 95% confidence intervals were calculated to evaluate the strength of the relationships.
Results
There were 37 case-control studies examining the effects of the four SNPs in MBL2 on PTB. A positive association between rs11003125 and PTB risk was observed in the hospital-based subgroup. Moreover, for the combined polymorphism and PTB risk, positive associations were detected not only in the total population but also in those with Asian origins across all source of control subgroups. No associations were found for rs7096206 or rs7095891.
Conclusions
Our current study indicated that several SNPs in MBL2 may be associated with susceptibility to PTB.
Keywords: Mannose-binding lectin, Pulmonary tuberculosis, Polymorphism, Meta-analysis, Susceptibility
Background
Tuberculosis (TB) is a global public health issue that poses serious threats to human health. It has been estimated that 1/3 of the world’s population may be infected with tubercle bacilli, but only 1/10 of individuals infected with Mycobacterium tuberculosis go on to develop TB [1], suggesting that there are inherent individual differences in susceptibility to TB that may be related to nutrition, constitution, specific and nonspecific resistance, and genetic susceptibility [2–6]. In fact, many studies have focused on the genetic variations within genes that increase the risk of TB [7, 8]. Previous case-control association studies have revealed that several human genes might be correlated with TB in certain populations. These genes include interferon-gamma (IFNG), vitamin D receptor (VDR), solute carrier family 11a member 1 (SLC11A1, which is also known as NRAMP1), and mannose-binding lectin (MBL2) [9–12].
The MBL2 gene, which is a member of the complement system, has been hypothesized to play a dual role in the innate immune response to infections by activating the classical lectin pathway and by phagocytosis [13, 14]. MBL and other soluble pattern recognition molecules [collectin-10, collectin-11, and ficolins (ficolin-1, ficolin-2, and ficolin-3)] act as mediators of host defense and participate in the maintenance of tissue homeostasis. They can bind to conserved pathogen-specific structures and altered self-antigens, and they form complexes with pentraxins to modulate innate immune functions. All these molecules exhibit distinct expressions in different tissue compartments, but all of them are found to varying degrees in the circulatory system. A common feature of these molecules is their ability to interact with a set of serine proteases named MASPs (MASP-1, MASP-2, and MASP-3) [15]. Human MBL is encoded by MBL2 on chromosome 10 (10q11.2-q21; OMIM 154545), which comprises four exons. MBL2 is reported to have several genetic polymorphisms that are commonly associated with MBL serum levels. Three point substitutions, located at codons 52, 54, and 57 in exon 1, are supposed to disrupt the assembly of MBL trimers or accelerate the degradation of the protein, thereby causing a decrease in the functional activity of MBL in the serum. These mutations are frequently referred to as variants D, B, and C, respectively, and they are collectively known as O, while A is the wild type. In addition, three other point substitutions have been reported in the nonstructural region: two at positions − 550 (H/L variants) and − 221 (X/Y variants) in the promoter region and one point mutation at position − 4 (P/Q variants) in the 5′-untranslated (UTR) region [16–18].
Many epidemiologic studies, including meta-analyses, suggest that there are relationships between MBL2 gene variations and pulmonary TB (PTB) risk [10, 16, 17, 19–47]. However, ambiguous conclusions have been reported; thus, it is necessary to perform an undated meta-analysis that includes a reanalysis of relevant studies.
Materials and methods
Search strategy and criteria
The PubMed, Embase, and SinoMed databases were searched for articles published prior to June 13, 2019, using the keywords “tuberculosis,” “TB,” “polymorphism,” and “mannose binding lectin 2 or MBL2”. A total of 163 papers were identified, 30 of which were consistent with our criteria. The inclusion criteria for papers were as follows: (i) examined the relationship between PTB susceptibility and MBL variations, (ii) case-control study, and (iii) contained a complete number of genotypes (MM + MW + WW) among cases and controls. The exclusion criteria were as follows: (i) no control group, (ii) incomplete genotype frequency data, (iii) duplicate publication, and (iv) controls did not meet the Hardy-Weinberg equilibrium (HWE) standards.
Data extraction
The essential data are listed as follows: first author name, publication year, original country, race, total samples of case/control, each genotype in cases/controls, source of control and genotype methods. Race was classified as Caucasian, Asian, African, or mixed. The source of control subgroups included population-based (PB) and hospital-based (HB) subgroups. The type of TB included total TB, PTB, and EPTB.
Quality score assessment (NOS)
The NOS was used to assess the quality of each study and to assess the various aspects of the methodology, including the selection of cases, the comparability of groups and the determination of exposure. The total score on the NOS ranges from 0 to 9 stars. Studies with scores greater than 7 are considered high-quality studies [48].
Statistical analysis
We used 95% CIs to measure the correlation between SNPs in MBL2 and PTB risk based on the genotype frequency of the case and control groups. The Z-test was used to determine the statistical significance of the correlations. The heterogeneity between the studies was evaluated using a Q-test based on the χ2 method. In the Q-test, a P value greater than 0.05 indicates that there is a lack of heterogeneity between the studies. Because the Q-statistic does not reveal the statistical significance of the heterogeneity, the I2 test was applied to better assess the extent of heterogeneity. As a guide, I2 values are divided into three categories (≤25%, 25–50%, ≥50%), corresponding to low risk, medium risk, and high risk, respectively [49]. If P ≤ 0.05 or I2 ≥ 50%, a random effects model was adopted; otherwise, a fixed effects model was used [50, 51]. We accessed the association between SNPs in MBL2 and PTB risk by testing the allelic contrast (X versus Y for rs7096206; L versus H for rs11003125; Q versus P for rs7095891; and O versus A for A/O combined SNP), heterozygote comparison (XY versus YY for rs7096206; LH versus HH for rs11003125; QP versus PP for rs7095891; and OA versus AA for A/O combined SNP), homozygote comparison (XX versus YY for rs7096206; LL versus HH for rs11003125; QQ versus PP for rs7095891; and OO versus AA for A/O combined SNP), recessive genetic model (XX versus XY + YY for rs7096206; LL versus LH + HH for rs11003125; and QQ versus QP + PP for rs7095891; OO versus OA + AA for A/O combined SNP) and dominant genetic model (XX + XY versus YY for rs7096206; LL + LH versus HH for rs11003125; QQ + QP versus PP for rs7095891; and OO + OA versus AA for A/O combined SNP). Sensitivity analysis was applied to assess the stability of the results. The HWE was evaluated by Pearson’s χ2 test, and P = 0.05 was considered statistically significant [52]. Publication bias was assessed by both Egger’s and Begg’s tests [53]. All statistical tests were carried out by version 11.0 of the Stata Software (StataCorp LP, College Station, TX, USA).
Genotyping methods
Methods for genotyping the SNPs in MBL2 were derived from the literature in Table 1.
Table 1.
Basic information of the association between 4 SNPs in MBL2 and TB, especially for PTB susceptibility
| First author | Year | Origin | Ethnicity | Source of | Type | Case | Control | Case | Control | Method | NOS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref No | Control | MM | MW | WW | MM | MW | WW | |||||||||
| rs7096206 | XX/GG | XY/GC | YY/CC | XX/GG | XY/GC | YY/CC | HWE | |||||||||
| Liu [10] | 2006 | China | Asian | PB | PTB | 141 | 212 | 6 | 44 | 91 | 7 | 54 | 151 | 0.43 | PCR-SSP/PCR-SSOP | 7 |
| Wu | 2017 | China | Asian | HB | PTB | 151 | 453 | 7 | 47 | 97 | 15 | 120 | 318 | 0.379 | PCR-RFLP/PCR-SSCP | 6 |
| Thye [41] | 2011 | Germany | Caucasian | PB | PTB | 1859 | 2180 | 26 | 396 | 1437 | 31 | 486 | 1663 | 0.503 | DASH-FRET | 7 |
| Feng [30] | 2016 | China | Asian | HB | PTB | 99 | 89 | 0 | 9 | 90 | 1 | 26 | 62 | 0.336 | Taqman | 5 |
| Liu | 2015 | China | Asian | HB | PTB | 112 | 120 | 11 | 35 | 66 | 2 | 40 | 78 | 0.215 | PCR-RFLP | 7 |
| Chen [24] | 2014 | China | Asian | PB | PTB | 205 | 216 | 5 | 77 | 123 | 8 | 49 | 159 | 0.098 | PCR-SSP | 7 |
| Wang [42] | 2009 | China | Asian | PB | PTB | 449 | 249 | 12 | 122 | 315 | 7 | 61 | 181 | 0.503 | AMLR | 7 |
| Alagarasu [19] | 2007 | India | Asian | HB | total | 109 | 146 | 3 | 44 | 62 | 13 | 61 | 72 | 0.987 | PCR-SSP | 6 |
| Chen [25] | 2015 | China | Asian | PB | total | 503 | 419 | 12 | 166 | 325 | 10 | 113 | 296 | 0.839 | PCR-SSP | 8 |
| Amiri [20] | 2017 | Iran | Asian | PB | PTB | 100 | 100 | 5 | 36 | 59 | 7 | 29 | 64 | 0.159 | PCR-SSP | 7 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | PTB | 119 | 148 | 4 | 40 | 75 | 6 | 32 | 110 | 0.076 | Sequencing | 6 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | EPTB | 36 | 148 | 1 | 9 | 26 | 6 | 32 | 110 | 0.076 | Sequencing | 6 |
| rs11003125 | LL | LH | HH | LL | LH | HH | HWE | |||||||||
| Liu [10] | 2006 | China | Asian | PB | PTB | 141 | 212 | 31 | 66 | 44 | 58 | 105 | 49 | 0.911 | PCR-SSP/PCR-SSOP | 7 |
| Thye [41] | 2011 | Germany | Caucasian | PB | PTB | 1843 | 2174 | 7 | 265 | 1571 | 9 | 287 | 1878 | 0.577 | DASH-FRET | 7 |
| Feng [30] | 2016 | China | Asian | HB | PTB | 99 | 89 | 28 | 45 | 26 | 21 | 36 | 32 | 0.092 | Taqman | 5 |
| Li [34] | 2011 | China | Asian | PB | PTB | 231 | 226 | 34 | 92 | 105 | 31 | 106 | 89 | 0.949 | PCR-SSP | 7 |
| Wu | 2017 | China | Asian | HB | PTB | 151 | 453 | 41 | 64 | 46 | 104 | 248 | 101 | 0.043 | PCR-RFLP/PCR-SSCP | 6 |
| Zhou [46] | 2011 | China | Asian | HB | PTB | 226 | 141 | 58 | 101 | 67 | 42 | 10 | 89 | < 0.01 | PCR-SSP | 6 |
| Zhang [44] | 2011 | China | Asian | HB | PTB | 220 | 213 | 29 | 75 | 116 | 51 | 76 | 86 | < 0.01 | PCR-SSP | 6 |
| Wang [42] | 2009 | China | Asian | PB | PTB | 449 | 249 | 91 | 235 | 123 | 60 | 108 | 81 | 0.046 | AMLR | 7 |
| Amiri [20] | 2017 | Iran | Asian | PB | PTB | 100 | 100 | 22 | 43 | 35 | 30 | 48 | 22 | 0.735 | PCR-SSP | 7 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | PTB | 119 | 148 | 66 | 45 | 8 | 68 | 61 | 19 | 0.367 | Sequencing | 6 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | EPTB | 36 | 148 | 16 | 18 | 2 | 68 | 61 | 19 | 0.367 | Sequencing | 6 |
| rs7095891 | QP | PP | QP | PP | HWE | |||||||||||
| Liu [10] | 2006 | China | Asian | PB | PTB | 141 | 212 | 1 | 22 | 118 | 2 | 39 | 171 | 0.891 | PCR-SSP/PCR-SSOP | 7 |
| Wu | 2017 | China | Asian | HB | PTB | 151 | 453 | 1 | 26 | 124 | 2 | 87 | 364 | 0.181 | PCR-RFLP/PCR-SSCP | 6 |
| Thye [41] | 2011 | Germany | Caucasian | PB | PTB | 1953 | 2230 | 308 | 920 | 725 | 319 | 1086 | 825 | 0.205 | DASH-FRET | 7 |
| Zhou [46] | 2011 | China | Asian | HB | PTB | 226 | 231 | 24 | 90 | 112 | 25 | 89 | 117 | 0.201 | PCR-SSP | 6 |
| Feng [30] | 2016 | China | Asian | HB | PTB | 99 | 89 | 7 | 34 | 58 | 0 | 24 | 65 | 0.141 | Taqman | 5 |
| Wang [42] | 2009 | China | Asian | PB | PTB | 449 | 249 | 3 | 114 | 332 | 2 | 64 | 183 | 0.155 | AMLR | 7 |
| Zhang [44] | 2011 | China | Asian | HB | PTB | 220 | 213 | 17 | 31 | 172 | 21 | 36 | 156 | < 0.01 | PCR-SSP | 6 |
| Amiri [20] | 2017 | Iran | Asian | PB | PTB | 100 | 100 | 3 | 21 | 76 | 5 | 26 | 69 | 0.233 | PCR-SSP | 7 |
| AA/AO/OO | OO | OA | AA | OO | OA | AA | HWE | |||||||||
| Garcia-Laorden [17] | 2006 | Spain | Caucasian | HB | total | 106 | 344 | 3 | 33 | 70 | 27 | 134 | 183 | 0.721 | PCR-RFLP | 6 |
| Søborg | 2003 | Denmark | Caucasian | PB |
total/ White |
59 | 250 | 4 | 18 | 37 | 7 | 86 | 157 | 0.235 | PCR-SSP | 8 |
| Søborg | 2003 | Denmark | Caucasian | PB |
total/ Nonwhite |
50 | 250 | 4 | 12 | 34 | 7 | 86 | 157 | 0.235 | PCR-SSP | 8 |
| Capparelli [22] | 2009 | Italy | Caucasian | HB | PTB | 274 | 288 | 61 | 158 | 55 | 10 | 112 | 166 | 0.087 | Sequencing | 6 |
| Garcıa-Gasalla [32] | 2014 | Spain | Caucasian | HB | total | 76 | 106 | 4 | 24 | 48 | 1 | 34 | 71 | 0.156 | PCR-SSP | 6 |
| Alagarasu [19] | 2007 | India | Asian | HB | total | 275 | 146 | 25 | 87 | 145 | 7 | 53 | 86 | 0.747 | PCR-SSP | 6 |
| Zhao [45] | 2014 | China | Asian | PB | PTB | 900 | 870 | 101 | 279 | 520 | 53 | 303 | 514 | 0.352 | PCR-RFLP | 7 |
| Li [34] | 2011 | China | Asian | PB | PTB | 231 | 226 | 3 | 57 | 171 | 3 | 37 | 186 | 0.461 | PCR-SSP | 7 |
| Li [33] | 2009 | China | Asian | HB | PTB | 141 | 152 | 6 | 56 | 79 | 8 | 38 | 106 | 0.075 | PCR-SSP | 6 |
| Liu [10] | 2006 | China | Asian | PB | PTB | 141 | 212 | 4 | 34 | 103 | 4 | 42 | 166 | 0.487 | PCR-SSP/PCR-SSOP | 7 |
| Zhou [47] | 2012 | China | Asian | HB | PTB | 226 | 231 | 14 | 106 | 106 | 5 | 80 | 146 | 0.114 | PCR-SSP | 6 |
| Liu | 2015 | China | Asian | HB | PTB | 112 | 120 | 3 | 29 | 80 | 2 | 22 | 96 | 0.576 | PCR-RFLP | 7 |
| Fang [29] | 2011 | China | Asian | HB | PTB | 100 | 100 | 1 | 25 | 74 | 0 | 25 | 75 | 0.153 | PCR-RFLP | 6 |
| Wu | 2017 | China | Asian | HB | PTB | 151 | 454 | 2 | 37 | 112 | 8 | 97 | 348 | 0.681 | PCR-RFLP/PCR-SSCP | 6 |
| Singla | 2011 | India | Asian | HB | PTB | 286 | 397 | 11 | 100 | 175 | 35 | 155 | 207 | 0.441 | PCR-RFLP | 6 |
| Singla | 2011 | India | Asian | HB | EPTB | 71 | 397 | 2 | 26 | 43 | 35 | 155 | 207 | 0.441 | PCR-RFLP | 6 |
| Özbaþ-Gerçeker | 2003 | Turkey | Caucasian | PB | PTB | 49 | 100 | 0 | 9 | 40 | 4 | 20 | 76 | 0.09 | PCR | 7 |
| Wit [28] | 2011 | South Africa | African | PB | total | 499 | 313 | 2 | 134 | 363 | 0 | 102 | 211 | < 0.01 | PCR-RFLP | 8 |
| Feng [30] | 2016 | China | Asian | HB | PTB | 381 | 267 | 14 | 177 | 190 | 12 | 176 | 79 | < 0.01 | Taqman | 5 |
| Wang [42] | 2009 | China | Asian | PB | PTB | 449 | 249 | 4 | 133 | 312 | 3 | 82 | 164 | 0.038 | AMLR | 7 |
| Thye [41] | 2011 | Germany | Caucasian | PB | PTB | 1893 | 1040 | 193 | 815 | 885 | 126 | 426 | 488 | 0.029 | DASH-FRET | 7 |
| Ceylan [23] | 2017 | Turkey | Caucasian | HB | total | 69 | 70 | 8 | 13 | 48 | 12 | 11 | 47 | < 0.01 | PCR-RFLP | 7 |
| Amiri [20] | 2017 | Iran | Asian | PB | PTB | 100 | 100 | 2 | 29 | 69 | 1 | 27 | 72 | 0.374 | PCR-SSP | 7 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | PTB | 119 | 148 | 7 | 41 | 71 | 6 | 34 | 108 | 0.129 | Sequencing | 6 |
| Cruz [27] | 2013 | Brazil | Caucasian | HB | EPTB | 36 | 148 | 1 | 14 | 21 | 6 | 34 | 108 | 0.129 | Sequencing | 6 |
| Selvaraj [37] | 1999 | India | Asian | PB | PTB | 202 | 109 | 22 | 73 | 107 | 2 | 39 | 68 | 0.175 | PCR-RFLP | 7 |
| Araújo | 2013 | Brazil | Caucasian | HB | PTB | 133 | 159 | 2 | 47 | 84 | 2 | 56 | 101 | 0.058 | PCR | 6 |
| Araújo | 2013 | Brazil | Caucasian | HB | EPTB | 34 | 159 | 1 | 15 | 18 | 2 | 56 | 101 | 0.058 | PCR | 6 |
| Fitness [31] | 2004 | UK | Caucasian | PB | total | 322 | 546 | 12 | 105 | 205 | 24 | 160 | 362 | 0.245 | fluorescence PCR/ARMS-PCR | 7 |
| Søborg | 2007 | Denmark | Caucasian | PB | PTB | 443 | 432 | 22 | 132 | 289 | 30 | 131 | 271 | 0.013 | PCR-RFLP/PCR-SSP | 7 |
HWE Hardy-Weinberg equilibrium; M Mutated allele; W Wide type allele; HB Hospital-based; PB Population-based; TB Tuberculosis; PTB: Pulmonary TB, EPTB Extra-pulmonary TB; PCR-FLIP Polymerase chain reaction and restrictive fragment length polymorphism; SSP Sequence specific primer; SSOP Sequence-specific oligonucleotide probe; SSCP Single-strand conformation polymorphism; DASH-FRET Dynamic allele-specific hybridization with fluorescence resonance energy transfer; AMLR Allelic-specific multiplex ligase-detection reaction; ARMS Amplification refractory mutation system; NOS Newcastle-Ottawa scale
Results
Study characteristics
A total of 163 articles were retrieved from the PubMed, Embase, and SinoMed databases by using various combinations of the abovementioned keywords. Fifty-three duplicate articles were removed after screening the titles, as shown in Fig. 1. Another 57 articles were removed because they did not contain relevant information. Next, the full texts of 53 articles were evaluated, and 23 additional articles were excluded because they contained duplicate data (4), they were meta-analyses/systematic reviews (10), they examined polymorphisms in other genes (2), or they were not case-control studies (7). Finally, 30 articles examining the association between the 4 SNPs in MBL2 and TB susceptibility were included (12 articles for rs7096206, 11 for rs11003125, 8 for rs7095891 and 30 for the A/O SNP). After filtering out studies that met our exclusion criteria, 9 different case-control studies were included for rs7096206, 6 for rs11003125, 7 for rs7095891, and 15 for the A/O SNP (Table 1). Overall, 37 case-control studies with 12,052 cases of PTB as well as 13,905 controls were included [10, 17, 19–47]. The controls were mainly healthy individuals.
Fig. 1.
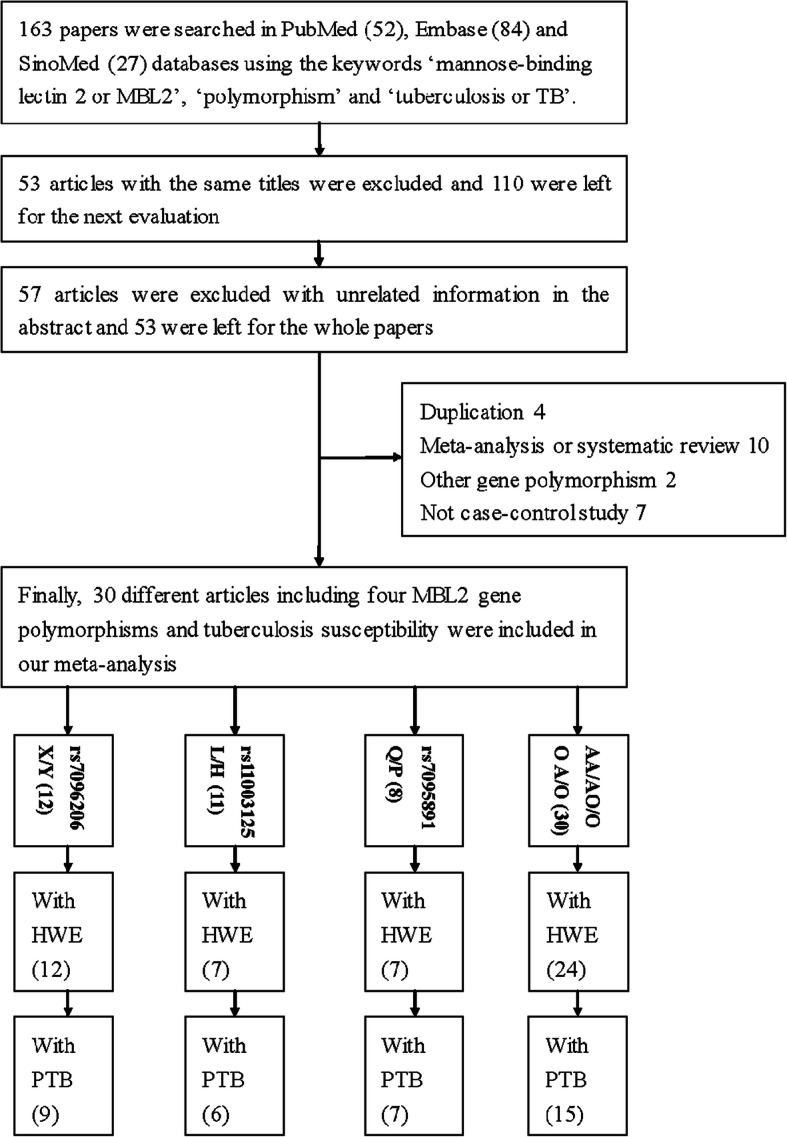
A flowchart illustrating the search strategy for identifying related studies. HWE: Hardy-Weinberg equilibrium; PTB: pulmonary tuberculosis
Quantitative synthesis
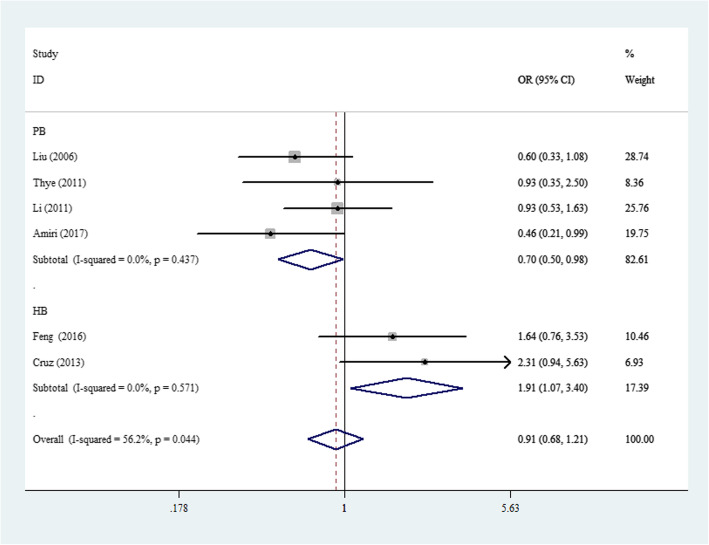
The associations between the 4 SNPs in MBL2 and PTB risk are shown in Table 2 and Figs. 2, 3, 4, and 5. For rs11003125, although negative associations were found in the total sample and in ethnic subgroups, a positive association was detected in the HB analysis (OR: 1.40, 95% CI: 1.06–1.85, P(heterogeneity): 0.755, P: 0.017, in the allelic contrast; OR: 1.91, 95% CI: 1.07–3.40, P(heterogeneity): 0.571, P: 0.029, in the homozygous comparison model (Fig. 2); and OR: 1.73, 95% CI: 1.05–2.86, P(heterogeneity): 0.633, P: 0.033 in the dominant genetic model).
Table 2.
Total and stratified subgroup analysis for 4 SNPs in MBL2 and PTB susceptibility
| Variables | N | Case/ | OR(95%CI) PhP I2(%) | OR(95%CI) PhP I2(%) | OR(95%CI) PhP I2(%) | OR(95%CI) PhP I2(%) | OR(95%CI) PhP I2(%) |
|---|---|---|---|---|---|---|---|
| Author/SNP | Control | M-allele vs. W-allele | MW vs. WW | MM vs. WW | MM + MW vs. WW | MM vs. MW + WW | |
| rs7096206(XX/XY/YY) | |||||||
| Total | 9 | 3235/3767 | 1.14(0.92–1.40)0.001 0.226 69.0 | 1.17(0.89–1.53)0.000 0.264 73.5 | 1.14(0.83–1.57)0.470 0.419 0.0 | 1.17(0.90–1.51)0.000 0.234 72.5 | 1.09(0.79–1.50)0.437 0.598 0.0 |
| Ethnicity | |||||||
| Asian | 7 | 1257/1439 | 1.13(0.86–1.49)0.005 0.366 68.0 | 1.13(0.80–1.61)0.002 0.482 71.8 | 1.30(0.84–1.99)0.308 0.238 16.0 | 1.15(0.83–1.60)0.002 0.412 70.7 | 1.20(0.79–1.84)0.264 0.395 21.7 |
| Caucasian | 2 | 1978/2328 | 1.11(0.75–1.64)0.081 0.608 67.1 | 1.24(0.65–2.37)0.022 0.504 80.9 | 0.97(0.60–1.58)0.922 0.908 0.0 | 1.20(0.68–2.11)0.035 0.531 77.6 | 0.96(0.59–1.56)0.804 0.864 0.0 |
| Source of control | |||||||
| HB | 4 | 481/810 | 1.01(0.58–1.76)0.001 0.973 82.4 | 0.93(0.48–1.83)0.001 0.840 82.5 | 1.77(0.96–3.27)0.159 0.069 42.0 | 0.97(0.51–1.87)0.001 0.935 82.9 | 1.67(0.91–3.06)0.147 0.099 44.1 |
| PB | 5 | 2754/2957 | 1.04(0.94–1.16)0.114 0.431 46.2 | 1.27(0.95–1.70)0.012 0.112 69.1 | 0.97(0.67–1.42)0.954 0.885 0.0 | 1.22(0.94–1.59)0.026 0.130 63.9 | 0.93(0.64–1.35)0.909 0.705 0.0 |
| rs11003125(LL/LH/HH) | |||||||
| Total | 6 | 2533/2949 | 0.99(0.80–1.22)0.011 0.900 66.2 | 0.93(0.70–1.25)0.049 0.649 55.1 | 0.95(0.60–1.49)0.044 0.808 56.2 | 0.96(0.71–1.29)0.013 0.716 65.3 | 1.01(0.80–1.28)0.309 0.930 16.3 |
| Ethnicity | |||||||
| Asian | 4 | 571/627 | 0.87(0.67–1.13)0.072 0.293 57.1 | 0.78(0.60–1.01)0.171 0.061 40.1 | 0.80(0.49–1.31)0.087 0.370 54.4 | 0.80(0.54–1.17)0.075 0.249 56.5 | 0.90(0.68–1.19)0.387 0.455 1.0 |
| Caucasian | 2 | 1962/2322 | 1.14(0.98–1.33)0.162 0.093 48.8 | 1.12(0.94–1.34)0.330 0.193 0.0 | 1.55(0.81–2.96)0.182 0.182 43.9 | 1.13(0.95–1.34)0.168 0.171 47.4 | 1.34(0.87–2.06)0.405 0.189 0.0 |
| Source of control | |||||||
| HB | 2 | 218/237 | 1.40(1.06–1.85)0.755 0.017 0.0 | 1.61(0.94–2.78)0.822 0.084 0.0 | 1.91(1.07–3.40)0.571 0.029 0.0 | 1.73(1.05–2.86)0.633 0.033 0.0 | 1.40(0.94–2.06)0.741 0.094 0.0 |
| PB | 4 | 2315/2712 | 0.87(0.70–1.08)0.047 0.217 62.6 | 0.82(0.59–1.13)0.048 0.219 62.1 | 0.70(0.50–0.98)0.437 0.039 0.0 | 0.80(0.57–1.11)0.026 0.179 67.7 | 0.84(0.62–1.13)0.634 0.204 0.0 |
| rs7095891(QQ/QP/PP) | |||||||
| Total | 7 | 3119/3524 | 1.02(0.95–1.11)0.166 0.552 34.3 | 0.97(0.87–1.08)0.713 0.589 0.0 | 1.10(0.93–1.31)0.586 0.262 0.0 | 0.99(0.90–1.10)0.389 0.912 4.9 | 1.12(0.96–1.32)0.641 0.161 0.0 |
| Ethnicity | |||||||
| Asian | 6 | 1166/1334 | 1.01(0.86–1.18)0.106 0.924 44.9 | 0.98(0.81–1.19)0.592 0.843 0.0 | 1.14(0.70–1.85)0.442 0.589 0.0 | 0.99(0.82–1.25)0.277 0.942 20.8 | 1.12(0.70–1.79)0.514 0.648 0.0 |
| Source of control | |||||||
| HB | 3 | 476/773 | 1.19(0.79–1.81)0.048 0.406 67.0 | 1.07(0.82–1.41)0.341 0.608 7.1 | 1.35(0.77–2.35)0.148 0.290 47.7 | 1.11(0.86–1.44)0.140 0.423 49.2 | 1.28(0.75–2.19)0.170 0.367 43.5 |
| PB | 4 | 2643/2791 | 1.01(0.93–1.10)0.495 0.808 0.0 | 0.95(0.84–1.07)0.817 0.404 0.0 | 1.08(0.90–1.30)0.787 0.406 0.0 | 0.97(0.87–1.09)0.670 0.638 0.0 | 1.11(0.94–1.31)0.815 0.236 0.0 |
| AA/AO/OO | |||||||
| Total | 15 | 3165/3665 | 1.33(1.05–1.70)0.000 0.020 85.4 | 1.37(1.06–1.77)0.000 0.018 79.8 | 1.82(0.94–3.51)0.000 0.073 79.2 | 1.41(1.06–1.86)0.000 0.017 84.3 | 1.61(0.94–2.77)0.000 0.083 69.2 |
| Ethnicity | |||||||
| Asian | 11 | 2590/2970 | 1.26(1.04–1.52)0.001 0.017 67.1 | 1.24(1.01–1.52)0.005 0.044 60.3 | 1.51(0.85–2.65)0.005 0.157 60.7 | 1.28(1.04–1.57)0.003 0.021 62.9 | 1.41(0.82–2.41)0.009 0.216 57.5 |
| Caucasian | 4 | 575/695 | 1.45(0.69–3.06)0.000 0.331 92.0 | 1.67(0.76–3.67)0.000 0.197 88.4 | 2.26(0.35–14.56)0.000 0.393 85.6 | 1.69(0.67–4.31)0.000 0.268 92.3 | 1.91(0.47–7.82)0.007 0.366 75.1 |
| Source of control | |||||||
| HB | 9 | 1425/2048 | 1.39(0.93–2.08)0.000 0.106 90.8 | 1.52(1.03–2.24)0.000 0.037 84.7 | 1.84(0.61–5.56)0.000 0.280 87.0 | 1.54(0.97–2.44)0.000 0.064 89.6 | 1.52(0.63–3.66)0.000 0.352 79.8 |
| PB | 6 | 1623/1617 | 1.24(1.10–1.40)0.283 0.001 19.9 | 1.05(0.90–1.23)0.221 0.543 28.6 | 1.94(1.42–2.56)0.340 0.000 11.7 | 1.16(1.00–1.34)0.339 0.050 11.9 | 1.97(1.45–2.68)0.348 0.000 10.6 |
M Mutated allele; W Wide type allele; HB Hospital-based; PB Population-based; Ph: value of Q-test for heterogeneity test; P: Z-test for the statistical significance of the OR
Fig. 2.
Forest plot of PTB risk associated with MBL2 rs11003125 polymorphism (LL vs. HH) in the subgroup about source of control. Square and horizontal lines correspond to specific OR or 95% CI. The area of the squares reflects the weight (inverse proportional variance). Diamonds represent the total OR or 95% CI
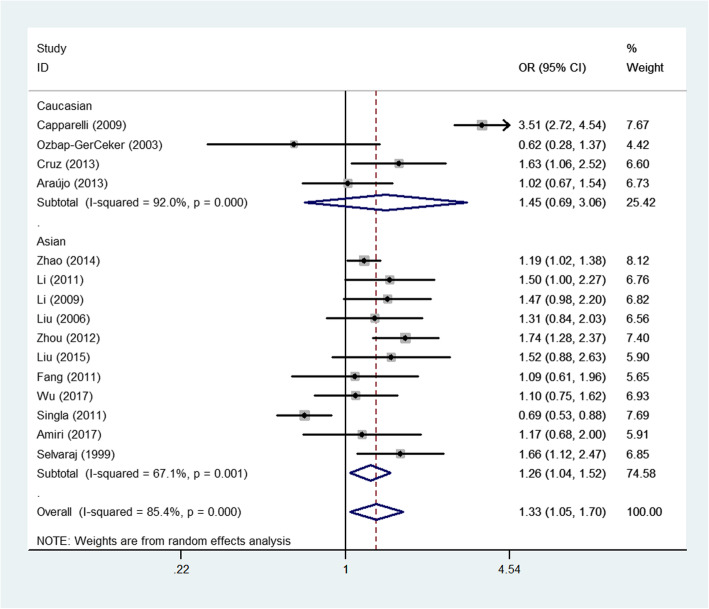
Fig. 3.
Forest plot of PTB risk associated with MBL2 A/O combined polymorphism (allelic contrast) by the whole samples and ethnicity
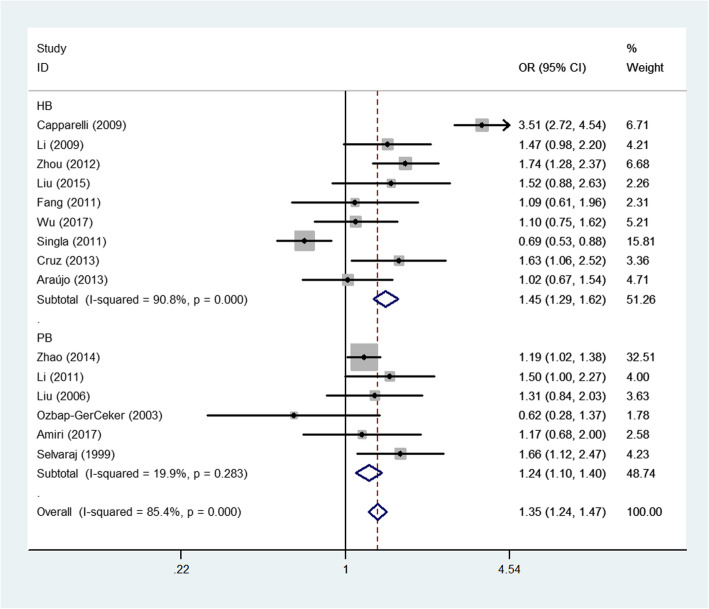
Fig. 4.
Forest plot of PTB risk associated with MBL2 A/O combined polymorphism (allelic contrast) by source of control
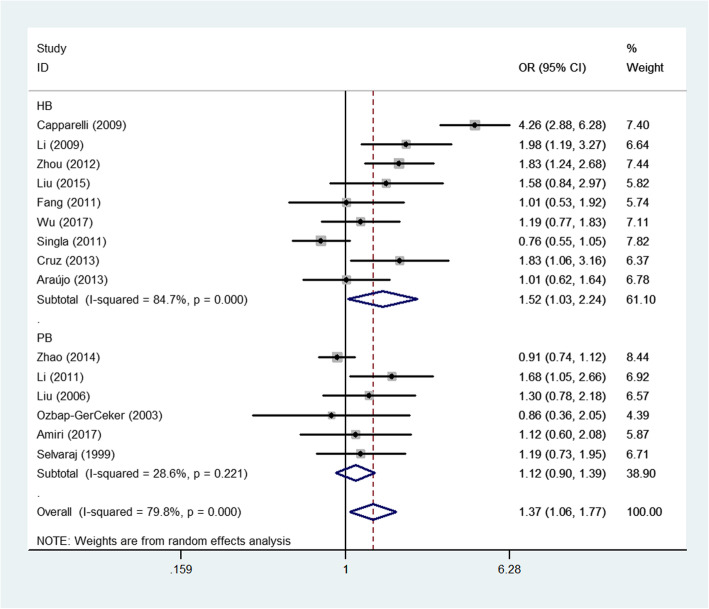
Fig. 5.
Forest plot of PTB risk associated with MBL2 A/O combined polymorphism (heterozygote comparison) by source of control
For the A/O combined SNP (AA/AO/OO) polymorphism, the O allele had a positive association with PTB risk in the total sample (heterozygote comparison: OR: 1.37, 95% CI: 1.06–1.7, P < 0.001 for heterogeneity, P: 0.018; dominant genetic model: OR: 1.41, 95% CI: 1.06–1.86, P < 0.001 for heterogeneity, P: 0.017; allelic contrast: OR: 1.33, 95% CI: 1.05–1.70, P < 0.001 for heterogeneity, P: 0.020, Fig. 3). In the subgroup analyses for different ethnicities, a similar significant association was detected for the Asian population (allelic contrast: OR: 1.26, 95% CI: 1.04–1.52, P: 0.001 for heterogeneity, P: 0.017, Fig. 3; heterozygote comparison: OR: 1.24, 95% CI: 1.01–1.52, P: 0.005 for heterogeneity, P: 0.044; dominant genetic model: OR: 1.28, 95% CI: 1.04–1.57, P: 0.003 for heterogeneity, P: 0.021, Fig. 3). Finally, in the subgroup analyses for different sources of control, PTB risk was significantly and positively associated with PB (e.g., allelic contrast: OR: 1.24, 95% CI: 1.10–1.40, P: 0.283 for heterogeneity, P: 0.001, Fig. 4) and HB studies (e.g., heterozygote comparison: OR: 1.52, 95% CI: 1.03–2.24, P < 0.001 for heterogeneity, P: 0.037, Fig. 5). In addition, no associations were observed for either rs7096206 or rs7095891, which indicated that heterogeneity might exist for these two SNPs (Table 2).
Publication bias and sensitivity analysis
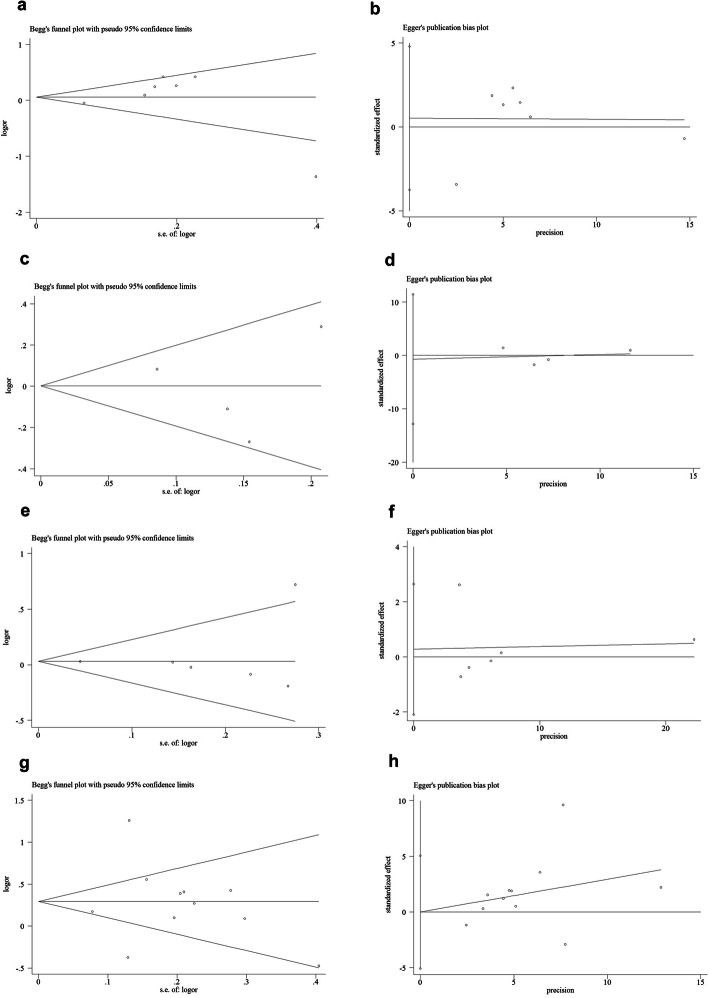
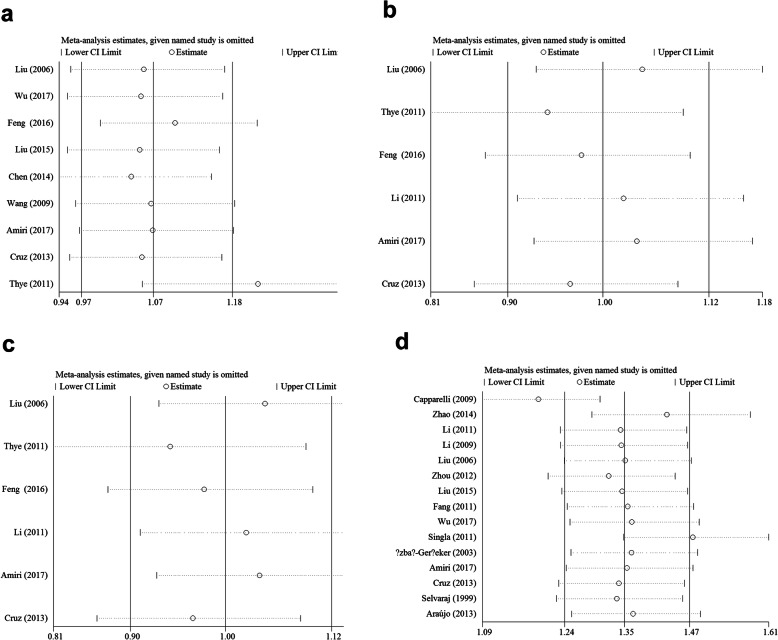
Begg’s test and Egger’s test were used to evaluate the publication bias of the included literature. The shape of the funnel plot did not show obvious asymmetry, and Egger’s test did not indicate publication bias (Fig. 6a-h, Table 3). We used sensitivity analysis to determine whether changes in a single study affected the outcome. For rs7096206 and O/A SNPs, two separate studies (Thye et al. for rs7096206, Fig. 7a and Capparelli et al. for O/A SNP, Fig. 7d) may have influenced the total OR according to the sensitivity analysis (data not shown).
Fig. 6.
Begg’s funnel plot for publication bias test (allelic contrast: a of rs7096206 [z = − 0.19, P = 0.851]; c of rs11003125 [z = − 0.21, P = 0.835]; e of rs 7,095,891 [z = − 1.65, P = 0.099] and g of A/O combined SNP [z = − 0.74, P = 0.458]). Each point represents a separate study for the indicated association. Log [OR], natural logarithm of OR. Horizontal line, mean effect size. Egger’s publication bias plot (allelic contrast: b of rs7096206 [t = 0.56, P = 0.592]; d of rs11003125 [t = − 0.25, P = 0.812]; f of rs 7,095,891 [t = − 0.12, P = 0.912] and h of A/O combined SNP [t = − 0.02, P = 0.988])
Table 3.
Publication bias tests (Begg’s funnel plot and Egger’s test for publication bias test) for 4 SNPs in MBL2.
| Egger’s test | Begg’s test | ||||||
|---|---|---|---|---|---|---|---|
| Genetic type | Coefficient | Standard error | t | P value | 95%CI of intercept | z | P value |
| rs7096206 | |||||||
| X-allele vs. Y-allele | 0.719 | 1.282 | 0.56 | 0.592 | (−2.312,3.751) | 0.1 | 0.917 |
| XY vs. YY | 0.597 | 1.084 | 0.55 | 0.599 | (−1.967,3.161) | −0.1 | 1 |
| XX vs. YY | 0.149 | 0.429 | 0.35 | 0.739 | (− 0.867,1.165) | 0.31 | 0.754 |
| XX + XY vs. YY | 0.625 | 1.12 | 0.56 | 0.594 | (−2.024,3.274) | −0.1 | 1 |
| XX vs. XY + YY | 0.147 | 0.434 | 0.34 | 0.744 | (−0.878,1.172) | 0.31 | 0.754 |
| rs11003125 | |||||||
| L-allele vs. H-allele | −0.597 | 2.35 | −0.25 | 0.812 | (−7.122,5.928) | 0 | 1 |
| LH vs. HH | −0.477 | 0.808 | −0.59 | 0.587 | (−2.721,1.768) | 0.38 | 0.707 |
| LL vs. HH | 1.899 | 1.558 | 1.22 | 0.29 | (−2.426,6.226) | 0.75 | 0.452 |
| LL + LH vs. HH | −0.495 | 0.899 | − 0.55 | 0.611 | (−2.993,2.002) | 0.75 | 0.452 |
| LL vs. LH + HH | −0.15 | 2.385 | −0.06 | 0.953 | (−6.772,6.472) | 0 | 1 |
| rs7095891 | |||||||
| Q-allele vs. P-allele | −0.09 | 0.781 | −0.12 | 0.912 | (−2.099,1.917) | 1.5 | 0.133 |
| QP vs. PP | −0.068 | 0.763 | −0.09 | 0.932 | (−2.031,1.893) | 1.2 | 0.23 |
| QQ vs. PP | −0.064 | 0.15 | −0.43 | 0.687 | (−0.451,0.322) | 0.3 | 0.764 |
| QQ + QP vs. PP | −0.077 | 0.775 | −0.1 | 0.924 | (−2.069,1.914) | 1.2 | 0.23 |
| QQ vs. QP + PP | −0.065 | 0.149 | −0.44 | 0.68 | (−0.448,0.317) | 0.3 | 0.764 |
| AA/AO/OO | |||||||
| A-allele vs. O-allele | −0.026 | 1.703 | −0.02 | 0.988 | (−3.706,3.653) | 0.69 | 0.488 |
| AO vs. OO | 0.469 | 1.407 | 0.33 | 0.744 | (−2.571,3.511) | 0.4 | 0.692 |
| AA vs. OO | 0.113 | 0.422 | 0.27 | 0.792 | (−0.798,1.025) | 1.39 | 0.166 |
| AA+AO vs. OO | 0.513 | 1.398 | 0.37 | 0.72 | (−2.507,3.533) | 0.59 | 0.533 |
| AA vs. AO + OO | 0.091 | 0.436 | 0.21 | 0.839 | (−0.852,1.033) | 1.39 | 0.166 |
Fig. 7.
Sensitivity analysis between 4 SNPs in MBL2 and PTB risk (allelic contrast: a for rs7096206; b for rs11003125; c for rs 7,095,891 and d for A/O combined SNP)
Discussion
Previous studies on the incidence of TB primarily focused on tubercle bacilli and the effects of environmental risk factors (such as sex, previous group TB, smoking status, drinking status, dominant status, age, group size, rainfall, immigration, number of eligible rovers, public health, economic, conservation importance). In recent decades, the effect of host susceptibility genes on TB has been increasingly recognized along with the development of genetic susceptibility. However, recent studies on the associations between SNPs in MBL2 and TB have produced different and even contradictory results. Some studies have indicated that mutations in the promoter and exon 1 of MBL2 may lead to the decline of MBL expression in the serum, while lower serum MBL levels can increase infections caused by tubercle bacilli [40, 54], indicating that polymorphisms in MBL2 may exert a protective effect against TB. Other studies have indicated that higher serum levels of MBL can reduce tubercle bacilli infections, which are associated with wild-type MBL2 alleles [5, 27, 55]. These studies suggest that MBL2 variants may increase the risk for TB.
Several meta-analyses have focused on the relationships between MBL2 polymorphisms and susceptibility to TB; however, each meta-analysis has its own conclusion and merits. Cao et al. analyzed 22 studies to assess the effect of MBL2 polymorphisms on TB risk. The rs1800451 polymorphism was associated with decreased TB risk in both the total sample and in some ethnic groups; in addition, A/O, rs7096206 and rs1800450 were likely only related to risk in some ethnic groups [56]. The analysis did not differentiate between the total sample and PTB subgroups. Tong et al. suggested that rs1800450 and rs5030737 polymorphisms were risk factors for susceptibility to TB; nevertheless, rs7095891 and rs1800451 polymorphisms acted as protective factors against TB [57]. Their study did not analyze the differences between the total sample and subgroups of TB. Denholm et al. [16] examined 12 case-control studies of HIV-negative patients and two studies of HIV-positive patients to determine the association of the MBL2 structural gene variants (B, C and D, referred to collectively as O, and A is the wild-type) with TB susceptibility. They did not find a significant association between the MBL2 genotype and PTB infection. By contrast, a meta-analysis of four studies examining MBL levels and susceptibility to TB found a significant association of high MBL levels with susceptibility to TB, although increased serum MBL levels due to the acute-phase reaction could not be ruled out. In addition, Areeshi et al. [58] found a statistically significant association of the C (rs1800451) alleles and genotypes with a reduced risk of TB in the overall population. No significant associations were observed in other variant sites (such as rs1800450, rs5030737, rs7096206, rs11003125, rs7095891 and combined rs1800450 O-alleles). Stratified analysis by ethnicity showed a decreased risk of TB in the African population for rs1800450 (B) and rs1800451 (C) alleles and genotypes. However, no association was observed between other MBL2 polymorphisms and TB risk in Asians. The results indicated a protective role of alleles B and C in TB infection. Finally, Shi et al. [59] indicated that individuals carrying the MBL2 codon 54 B allele had an increased risk of TB compared with AA homozygotes, whereas rs7095891 was possibly not associated with TB risk in Chinese.
To our knowledge, the current study is an updated systematic analysis exploring the relationships between MBL2 variants and PTB susceptibility. This analysis involved approximately 12,052 patients with PTB and 13,905 healthy samples. The most important finding of our study was that the rs11003125 L-allele and the A/O combined SNP were risk factors for PTB susceptibility in the HB subgroup, which was similar to findings from a previous meta-analysis. The O allele was also a risk factor for PTB in the Asian and PB subgroups. The aforementioned conclusions were novel concepts that have not been found in previously published meta-analyses.
The above contradictory results from previous meta-analyses further emphasize the controversy about the effect of MBL2 variants on susceptibility to TB. One possible explanation for this effect is that different polymorphisms may have different effects on gene function, resulting in changes in PTB susceptibility. Second, the complex interaction between several genetic and environmental factors may involve the development of PTB. We think these conflicting results among studies and different populations suggests linkage disequilibrium with other nearby genes (e.g., surfactant proteins A1, A2 and D [60] previously associated with TB) rather than a causative association between MBL2 variants and PTB. Third, it is now widely accepted that differences in ethnicities between cases and control measures may be a source of confusion in the compilation of studies. Fourth, research with “negative” results takes longer to publish due to the time-lag bias, and positive research results are published much faster. Fifth, small studies of with “negative” results have never been published, and small studies of similar quality with “positive” results will also be shown in the literature [61–63]. Sixth, rs7096206, rs11003125 and rs7095891 SNPs were not analyzed in the previous three meta-analyses; our study was the first to analyze these SNPs. Furthermore, we focused on PTB but not on total TB or extrapulmonary TB (EPTB), in contrast to previous meta-analyses.
Some limitations in our study should be noted. Initially, we collected all eligible studies; however, the sample size of these studies is not yet large enough, especially in certain ethnic groups. Therefore, not only is the likelihood of I/II type errors high, but there is insufficient statistical capacity to assess the correlations between the 7 SNPs and PTB risk. Second, serum MBL concentration was not assessed in our study, which would have been helpful for detecting and understanding the mechanism of SNPs in the MBL2 gene. Third, other factors such as age, sex, smoking, familial history, disease stage, specific environmental factors and lifestyles should be included. Fourth, only one article [19] included the subgroups of HIV- and HIV+, anti-TNF drugs, and DM; these groups were not evaluated in other included studies, so we could not analyze the associations within the above groups because of missing information. Fifth, the included studies had a high amount of heterogeneity. In addition, we cannot know whether patients had latent tuberculosis. Finally, all included studies were epidemiological surveys; there were no plausible biological hypotheses or mechanistic studies. We aimed to determine whether there is a relationship between MBL2 structural gene variants and susceptibility to PTB. Further studies should aim to overcome these limitations.
In summary, our study indicated that the rs11003125 and A/O-combined SNPs in MBL2 may be related to PTB risk. Larger sample sizes and additional gene-environment interactions should be considered in future studies.
Acknowledgements
Not applicable.
Abbreviations
- MBL
Mannose-binding lectin
- PTB
Pulmonary tuberculosis
- ORs
Odds ratios
- CIs
Confidence intervals
- SNPs
Single nucleotide polymorphisms
- HWE
Hardy-Weinberg equilibrium
Authors’ contributions
WS and LX conceived the study. YL searched the databases and extracted the data. DZ analyzed the data. WZ wrote the draft of the paper. WZ reviewed the manuscript. The author(s) read and approved the final manuscript.
Funding
This article was supported by Taizhou science and technology support plan (social development): TS202014.
Availability of data and materials
All the data generated in the present research is contained in this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenhao Shen, Li Xiao and Yang Li contributed equally to this work. Wei Zhang is the corresponding author, and Daming Zhou is the co-corresponding author.
Contributor Information
Daming Zhou, Email: zdmbxqarticle@sina.com.
Wei Zhang, Email: zhangweimeta@sina.com.
References
- 1.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186(10):1463–1468. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 2.Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80(10):3343–3359. doi: 10.1128/IAI.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo C, Zhang H, Gao Q, He D, Tang M, Liu S, Deng A, Wang Y, Lu S, Li J, et al. Monocyte chemoattractant protein-1 in spinal tuberculosis: -362G/C genetic variant and protein levels in Chinese patients. Diagn Microbiol Infect Dis. 2014;78(1):49–52. doi: 10.1016/j.diagmicrobio.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Yan J, Wan Q, Ye Q, Huang Y. The risk factors for tuberculosis in liver or kidney transplant recipients. BMC Infect Dis. 2014;14:387. doi: 10.1186/1471-2334-14-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narasimhan P, Wood J, Macintyre CR, Mathai D. Risk factors for tuberculosis. Pulm Med. 2013;2013:828939. doi: 10.1155/2013/828939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajaram MV, Ni B, Dodd CE, Schlesinger LS. Macrophage immunoregulatory pathways in tuberculosis. Semin Immunol. 2014;26(6):471–485. doi: 10.1016/j.smim.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis (Edinb) 2010;90(2):71–83. doi: 10.1016/j.tube.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Yim JJ, Selvaraj P. Genetic susceptibility in tuberculosis. Respirology. 2010;15(2):241–256. doi: 10.1111/j.1440-1843.2009.01690.x. [DOI] [PubMed] [Google Scholar]
- 9.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in west Africans. N Engl J Med. 1998;338(10):640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Zhang F, Xin ZT, Zhao QM, Wu XM, Zhang PH, de Vlas S, Richardus JH, Habbema JD, Yang H, et al. Sequence variations in the MBL gene and their relationship to pulmonary tuberculosis in the Chinese Han population. Int J Tuberc Lung Dis. 2006;10(10):1098–1103. [PubMed] [Google Scholar]
- 11.Wei Z, Wenhao S, Yuanyuan M, Yang L, Daming Z, Jiangchun X, Jijun J. A single nucleotide polymorphism in the interferon-gamma gene (IFNG +874 T/a) is associated with susceptibility to tuberculosis. Oncotarget. 2017;8(31):50415–50429. doi: 10.18632/oncotarget.17304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson RJ, Llewelyn M, Toossi Z, Patel P, Pasvol G, Lalvani A, Wright D, Latif M, Davidson RN. Influence of vitamin D deficiency and vitamin D receptor polymorphisms on tuberculosis among Gujarati Asians in west London: a case-control study. Lancet (London, England) 2000;355(9204):618–621. doi: 10.1016/S0140-6736(99)02301-6. [DOI] [PubMed] [Google Scholar]
- 13.Fujita T. Evolution of the lectin-complement pathway and its role in innate immunity. Nat Rev Immunol. 2002;2(5):346–353. doi: 10.1038/nri800. [DOI] [PubMed] [Google Scholar]
- 14.Santos IK, Costa CH, Krieger H, Feitosa MF, Zurakowski D, Fardin B, Gomes RB, Weiner DL, Harn DA, Ezekowitz RA, et al. Mannan-binding lectin enhances susceptibility to visceral leishmaniasis. Infect Immun. 2001;69(8):5212–5215. doi: 10.1128/IAI.69.8.5212-5215.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garred P, Genster N, Pilely K, Bayarri-Olmos R, Rosbjerg A, Ma YJ, Skjoedt MO. A journey through the lectin pathway of complement-MBL and beyond. Immunol Rev. 2016;274(1):74–97. doi: 10.1111/imr.12468. [DOI] [PubMed] [Google Scholar]
- 16.Denholm JT, McBryde ES, Eisen DP. Mannose-binding lectin and susceptibility to tuberculosis: a meta-analysis. Clin Exp Immunol. 2010;162(1):84–90. doi: 10.1111/j.1365-2249.2010.04221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Laorden MI, Pena MJ, Caminero JA, Garcia-Saavedra A, Campos-Herrero MI, Caballero A, Rodriguez-Gallego C. Influence of mannose-binding lectin on HIV infection and tuberculosis in a Western-European population. Mol Immunol. 2006;43(14):2143–2150. doi: 10.1016/j.molimm.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Madsen HO, Satz ML, Hogh B, Svejgaard A, Garred P. Different molecular events result in low protein levels of mannan-binding lectin in populations from southeast Africa and South America. J Immun. 1998;161(6):3169–3175. [PubMed] [Google Scholar]
- 19.Alagarasu K, Selvaraj P, Swaminathan S, Raghavan S, Narendran G, Narayanan PR. Mannose binding lectin gene variants and susceptibility to tuberculosis in HIV-1 infected patients of South India. Tuberculosis (Edinb) 2007;87(6):535–543. doi: 10.1016/j.tube.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Amiri A, Sabooteh T, Shahsavar F, Anbari K, Pouremadi F. Mannose-binding lectin (MBL) gene polymorphisms in susceptibility to pulmonary tuberculosis among the Lur population of Lorestan Province of Iran. Genomics Data. 2017;12:146–150. doi: 10.1016/j.gdata.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araujo MS, Graca ES, Azevedo VN, Cayres-Vallinoto I, Machado LF, Ishak MO, Ishak R, Vallinoto AC. No evidence of association between MBL2A/O polymorphisms and Mycobacterium tuberculosis infection in populations from the Brazilian Amazon region. Hum Immunol. 2013;74(1):82–84. doi: 10.1016/j.humimm.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Capparelli R, Iannaccone M, Palumbo D, Medaglia C, Moscariello E, Russo A, Iannelli D. Role played by human mannose-binding lectin polymorphisms in pulmonary tuberculosis. J Infect Dis. 2009;199(5):666–672. doi: 10.1086/596658. [DOI] [PubMed] [Google Scholar]
- 23.Ceylan E, Karkucak M, Coban H, Karadag M, Yakut T. Evaluation of TNF-alpha gene (G308A) and MBL2 gene codon 54 polymorphisms in Turkish patients with tuberculosis. J Infect Public Health. 2017;10(6):774–777. doi: 10.1016/j.jiph.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Deng J, Su C, Li J, Wang M, Abuaku BK, Hu S, Tan H, Wen SW. Impact of passive smoking, cooking with solid fuel exposure, and MBL/MASP-2 gene polymorphism upon susceptibility to tuberculosis. Int J Infect Dis. 2014;29:1–6. doi: 10.1016/j.ijid.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 25.Chen M, Liang Y, Li W, Wang M, Hu L, Abuaku BK, Huang X, Tan H, Wen SW. Impact of MBL and MASP-2 gene polymorphism and its interaction on susceptibility to tuberculosis. BMC Infect Dis. 2015;15:151. doi: 10.1186/s12879-015-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen MS: Impact of MBL/MASP-2 gene polymorphism and related factors on susceptibility to tuberculosis among Hans population in Hunan province. A doctor's degree thesis 2014.
- 27.da Cruz HL, da Silva RC, Segat L, de Carvalho MS, Brandao LA, Guimaraes RL, Santos FC, de Lira LA, Montenegro LM, Schindler HC, et al. MBL2 gene polymorphisms and susceptibility to tuberculosis in a northeastern Brazilian population. Infect Genet Evol. 2013;19:323–329. doi: 10.1016/j.meegid.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 28.de Wit E, van der Merwe L, van Helden PD, Hoal EG. Gene-gene interaction between tuberculosis candidate genes in a south African population. Mamm Genome. 2011;22(1–2):100–110. doi: 10.1007/s00335-010-9280-8. [DOI] [PubMed] [Google Scholar]
- 29.Fang GX, Yin CL. Research on relationship of polymorphism of mannose-binding lectin gene with susceptibility to tuberculosis in Zhuang population. Int J Respir. 2011;31(6):428–430. [Google Scholar]
- 30.Feng XY, Yang CE, Guan W, Hua M, Jiu T, Du FM, Cui JX, La Z, Li Y, An WJ, et al. Correlations between the mannose-binding lection (MBL) gene polymorphisms and the susceptibility to tuberculosis in Tibetan population in Qinghai province. Chin J Microbiol Immunol. 2016;36(11):838–842. [Google Scholar]
- 31.Fitness J, Floyd S, Warndorff DK, Sichali L, Malema S, Crampin AC, Fine PE, Hill AV. Large-scale candidate gene study of tuberculosis susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg. 2004;71(3):341–349. [PubMed] [Google Scholar]
- 32.Garcia-Gasalla M, Mila Llambi J, Losada-Lopez I, Cifuentes-Luna C, Fernandez-Baca V, Pareja-Bezares A, Mir-Villadrich I, Payeras-Cifre A. Mannose-binding lectin exon 1 and promoter polymorphisms in tuberculosis disease in a Mediterranean area. Int J Immunogenet. 2014;41(4):306–311. doi: 10.1111/iji.12126. [DOI] [PubMed] [Google Scholar]
- 33.Li Y: Association between NRAMP1, MBL SNPs and tuberculosis in the Ningxia HAIYUAN hui people. A master's degree thesis 2009.
- 34.Li Y, Wu F, Zhang Y, Zhang WJ. Association between polymorphism of MBL gene with susceptibility to tuberculosis in Han polymorphism in Xinjiang. Chin J Zoonoses. 2011;27(9):769–773. [Google Scholar]
- 35.Liu YD, Zheng RJ, Xiao HP, Sha W, Zhang Q, Wu FR, Sun H, Zhang ZS, Cui HY, Liu ZB, et al. Study on the correlation between polymorphisms of genes with susceptibility to tuberculosis and drug-resistant tuberculosis in Chinese Han population. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32(3):279–284. [PubMed] [Google Scholar]
- 36.Ozbas-Gerceker F, Tezcan I, Berkel AI, Ozkara S, Ozcan A, Ersoy F, Sanal O, Ozguc M. The effect of mannose-binding protein gene polymorphisms in recurrent respiratory system infections in children and lung tuberculosis. Turk J Pediatr. 2003;45(2):95–98. [PubMed] [Google Scholar]
- 37.Selvaraj P, Narayanan PR, Reetha AM. Association of functional mutant homozygotes of the mannose binding protein gene with susceptibility to pulmonary tuberculosis in India. Tuber Lung Dis. 1999;79(4):221–227. doi: 10.1054/tuld.1999.0204. [DOI] [PubMed] [Google Scholar]
- 38.Singla N, Gupta D, Joshi A, Batra N, Singh J, Birbian N. Association of mannose-binding lectin gene polymorphism with tuberculosis susceptibility and sputum conversion time. Int J Immunogenet. 2012;39(1):10–14. doi: 10.1111/j.1744-313X.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 39.Soborg C, Andersen AB, Range N, Malenganisho W, Friis H, Magnussen P, Temu MM, Changalucha J, Madsen HO, Garred P. Influence of candidate susceptibility genes on tuberculosis in a high endemic region. Mol Immunol. 2007;44(9):2213–2220. doi: 10.1016/j.molimm.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Soborg C, Madsen HO, Andersen AB, Lillebaek T, Kok-Jensen A, Garred P. Mannose-binding lectin polymorphisms in clinical tuberculosis. J Infect Dis. 2003;188(5):777–782. doi: 10.1086/377183. [DOI] [PubMed] [Google Scholar]
- 41.Thye T, Niemann S, Walter K, Homolka S, Intemann CD, Chinbuah MA, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, et al. Variant G57E of mannose binding lectin associated with protection against tuberculosis caused by Mycobacterium africanum but not by M. tuberculosis. PloS one. 2011;6(6):e20908. doi: 10.1371/journal.pone.0020908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang FF: Association between MBL gene polymorphism and the susceptibility to tuberculosis among Hans population and its potential mechanism. A doctor's degree thesis 2009.
- 43.Wu L, Deng H, Zheng Y, Mansjo M, Zheng X, Hu Y, Xu B. An association study of NRAMP1, VDR, MBL and their interaction with the susceptibility to tuberculosis in a Chinese population. Int J Infect Dis. 2015;38:129–135. doi: 10.1016/j.ijid.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, Deng XL, Wang X, Zhou J, Li Y, Wu F, Zhang WJ. A study on the association of polymorphisms of the MBL gene with the susceptibility to tuberculosis of Xinjiang Hazakhs. Land Reclamation Med. 2011;33(2):135–138. [Google Scholar]
- 45.Zhao YB, Zhu GY, Xhen DD, Yang YR. rs1800450 mannose-binding lectin gene polymorphisms and susceptibility to tuberculosis in Ningxia population. J Ningxia Med Univ. 2014;36(8):889–893. [Google Scholar]
- 46.Zhou J, Deng XL, Wang X, Wu F, Zhang J, Li Y, Zhang WJ. Study on the correlation between MBL-H/L, P/Q polymorphisms with susceptibility of TB in Xinjiang Uighurs population. Clin med Eng. 2011;18(7):976–978. [Google Scholar]
- 47.Zhou J, Zhang WJ. Correlation between MBL-A/B gene with the susceptibility of tuberculosis in Xinjiang Uighur population. Chin J Antibubere. 2012;34(7):445–451. [Google Scholar]
- 48.Wells G, Shea B, O'Connell D, Robertson J, Peterson J, Welch V, Losos M, Tugwell P: The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. In: Ottawa Health Research Institute. 21 Oct, 2011.
- 49.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed) 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 51.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 52.Napolioni V. The relevance of checking population allele frequencies and Hardy-Weinberg Equilibrium in genetic association studies: the case of SLC6A4 5-HTTLPR polymorphism in a Chinese Han Irritable Bowel Syndrome association study. Immunol Lett. 2014;162(1 Pt A):276–278. doi: 10.1016/j.imlet.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 53.Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol. 2005;15(6):235–243. doi: 10.2188/jea.15.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selvaraj P, Jawahar MS, Rajeswari DN, Alagarasu K, Vidyarani M, Narayanan PR. Role of mannose binding lectin gene variants on its protein levels and macrophage phagocytosis with live Mycobacterium tuberculosis in pulmonary tuberculosis. FEMS Immunol Med Microbiol. 2006;46(3):433–437. doi: 10.1111/j.1574-695X.2006.00053.x. [DOI] [PubMed] [Google Scholar]
- 55.You HL, Lin TM, Wang JC, Li CC, Chao TL, Liao WT, Eng HL. Mannose-binding lectin gene polymorphisms and mycobacterial lymphadenitis in young patients. Pediatr Infect Dis J. 2013;32(9):1005–1009. doi: 10.1097/INF.0b013e31829e6798. [DOI] [PubMed] [Google Scholar]
- 56.Cao Y, Wang X, Cao Z, Wu C, Wu D, Cheng X. Genetic polymorphisms of MBL2 and tuberculosis susceptibility: a meta-analysis of 22 case-control studies. Arch Med Sci : AMS. 2018;14(6):1212–1232. doi: 10.5114/aoms.2017.65319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong X, Wan Q, Li Z, Liu S, Huang J, Wu M, Fan H. Association between the mannose-binding lectin (MBL)-2 gene variants and serum MBL with pulmonary tuberculosis: An update meta-analysis and systematic review. Microb Pathog. 2019;132:374–380. doi: 10.1016/j.micpath.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 58.Areeshi MY, Mandal RK, Akhter N, Dar SA, Jawed A, Wahid M, Mahto H, Panda AK, Lohani M, Haque S. A meta-analysis of MBL2 polymorphisms and tuberculosis risk. Sci Rep. 2016;6:35728. doi: 10.1038/srep35728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi J, Xie M, Wang JM, Xu YJ, Xiong WN, Liu XS. Mannose-binding lectin two gene polymorphisms and tuberculosis susceptibility in Chinese population: a meta-analysis. J Huazhong Univ Sci Technol Med Sci. 2013;33(2):166–171. doi: 10.1007/s11596-013-1091-1. [DOI] [PubMed] [Google Scholar]
- 60.Sorensen GL, Husby S, Holmskov U. Surfactant protein a and surfactant protein D variation in pulmonary disease. Immunobiology. 2007;212(4–5):381–416. doi: 10.1016/j.imbio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Dickersin K, Min YI, Meinert CL. Factors influencing publication of research results. Follow-up of applications submitted to two institutional review boards. Jama. 1992;267(3):374–378. [PubMed] [Google Scholar]
- 62.Easterbrook PJ, Berlin JA, Gopalan R, Matthews DR. Publication bias in clinical research. Lancet (London, England) 1991;337(8746):867–872. doi: 10.1016/0140-6736(91)90201-y. [DOI] [PubMed] [Google Scholar]
- 63.Ioannidis JP. Effect of the statistical significance of results on the time to completion and publication of randomized efficacy trials. Jama. 1998;279(4):281–286. doi: 10.1001/jama.279.4.281. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated in the present research is contained in this manuscript.