Sas4 is a conserved basal body assembly protein. Here, Ruehle et al. describe a previously unknown link between basal bodies and the control of cell division by Hippo signaling molecules that depends on Sas4.
Abstract
Basal bodies (BBs) are macromolecular complexes required for the formation and cortical positioning of cilia. Both BB assembly and DNA replication are tightly coordinated with the cell cycle to ensure their accurate segregation and propagation to daughter cells, but the mechanisms ensuring coordination are unclear. The Tetrahymena Sas4/CPAP protein is enriched at assembling BBs, localizing to the core BB structure and to the base of BB-appendage microtubules and striated fiber. Sas4 is necessary for BB assembly and cortical microtubule organization, and Sas4 loss disrupts cell division furrow positioning and DNA segregation. The Hippo signaling pathway is known to regulate cell division furrow position, and Hippo molecules localize to BBs and BB-appendages. We find that Sas4 loss disrupts localization of the Hippo activator, Mob1, suggesting that Sas4 mediates Hippo activity by promoting scaffolds for Mob1 localization to the cell cortex. Thus, Sas4 links BBs with an ancient signaling pathway known to promote the accurate and symmetric segregation of the genome.
Introduction
Centrioles and basal bodies (BBs) are microtubule-organizing centers that are conserved across the eukaryotic lineage (Carvalho-Santos et al., 2011; Wickstead and Gull, 2011). As centrioles, they act in pairs to recruit pericentriolar material that nucleates cytoplasmic microtubules and the mitotic spindle apparatus (Brinkley, 1985). As BBs, these structures are positioned at the cell cortex to nucleate cilia that can function in both signaling and motility (Pala et al., 2017; Haimo and Rosenbaum, 1981). The conservation of these structures underscores the importance of their microtubule organizing functions in diverse organisms. The ciliate Tetrahymena thermophila harbors hundreds of BBs per cell organized into linear rows along the anterior-posterior cell axis to position motile cilia for cellular movement. During each cell cycle, new BBs are assembled and, like DNA replication, the number of BBs must double before cell division completes to ensure that both daughter cells have enough cilia for motility (Nanney et al., 1978; Galati et al., 2016). However, it is not understood how new BB assembly is coordinated with the timing of cell division. Premature cell division before an adequate number of BBs are produced would lead to a reductional loss of BBs and would therefore impair motility. It stands to reason that BB assembly and cell division are linked to guard against such outcomes. In the latter half of the cell cycle, the Tetrahymena micronuclear and macronuclear genomes replicate and segregate along the anterior-posterior cell axis (Flickinger, 1965). After nuclear segregation, the cell division furrow ingresses at an equatorial position perpendicular to the ciliary rows. Upon successful completion of cytokinesis, both BBs and the genomes are evenly distributed to the daughter cells.
Proper cell division is controlled, in part, by the conserved, eukaryotic Hippo signaling pathway (Misra and Irvine, 2018). In most organisms studied, environmental inputs signal to the core Hippo cassette, which includes Mst1 kinase, Lats kinase, and the coactivators Sav1 and Mob1. This serves to inhibit the downstream signaling effectors Yes-Associated Protein (YAP) and Transcriptional coactivator with PDZ-binding motif (TAZ) that control cell size and proliferation through transcriptional regulation (Misra and Irvine, 2018). However, in the case of cell division control, Hippo signaling is transcription-independent and instead regulates the timing and spatial position of the cell division furrow and cytokinesis (Florindo et al., 2012; Bui et al., 2016). Moreover, homologues of Hippo signaling factors in yeast are integral to the mitotic exit network and septation initiation network pathways that regulate mitotic spindle positioning and progression from mitosis to interphase (Hergovich and Hemmings, 2012; Simanis, 2015). The core Hippo and mitotic exit network/septation initiation network signaling factors localize to centrosomes and to spindle pole bodies, respectively, suggesting that centrosomes act as scaffolds to support Hippo signaling (Nishiyama et al., 1999; Morisaki et al., 2002; McPherson et al., 2004; Mardin et al., 2010; Campbell et al., 2020; Hergovich et al., 2009). Whether Hippo pathways act similarly in Tetrahymena is not clear. The known Hippo pathway molecules in Tetrahymena include CdaI (Mst kinase), Mob1, and Elo1 (Lats kinase), and they localize to BBs (Tavares et al., 2012; Jiang et al., 2017, 2019). Specifically, Hippo factors localize as an asymmetric gradient with the greatest levels of these proteins at the BBs of the cell’s posterior end. During cell division, a new gradient is established at an equatorial plane, thus defining the site of the division furrow and the new posterior and anterior of daughter cells. Tetrahymena Hippo pathway mutants exhibit mispositioned division furrows in either the anterior (mob1Δ and cdaI-1[Hippo/Mst]) or posterior (elo1-1 [Lats/Ndr]) direction. This suggests the Hippo pathway uses antagonistic activities to control the cortical pattern required for division furrow placement (Jiang et al., 2017, 2019; Tavares et al., 2012). It is not known how Hippo pathway proteins associate with BBs and if they depend on BB components for localization and proper division furrow position.
Centrioles and BBs are comprised of triplet microtubules arranged in a ninefold radial-symmetric array. In Tetrahymena, BB-appendage structures asymmetrically extend from the symmetric BB architecture to promote BB attachment to the cell cortex. The combined BB and BB-appendage structures form BB units of cortically attached, interconnected structures. How BB-appendages form around newly assembling BBs is an area of ongoing study (Junker et al., 2019). Plk4, Sas5/STIL, Sas6, Bld10/CEP135, and Sas4/CPAP are essential for centriole and BB assembly (Banterle and Gönczy, 2017). Of these, Sas4 is interesting because it acts as a slow and processive microtubule polymerase at plus-ends, but also interacts with Bld10 and Sas5 at the proximal end of centrioles and BBs where microtubule minus-ends are positioned. These data support the notion of a multifunctional role for Sas4 in centriole and BB biology to integrate centriole and BB assembly with the surrounding environment. Given the localization of Hippo molecules at BBs and centrosomes, we hypothesized that Sas4 might link BB and cortical structure replication to cell division and cytokinesis. Here, we investigate the role of Sas4 in T. thermophila BB assembly. We show that Sas4 not only functions in BB assembly and maintenance but also links cortical replication to cell division via the Hippo pathway.
Results and discussion
Sas4 localizes to BBs and BB-appendage structures
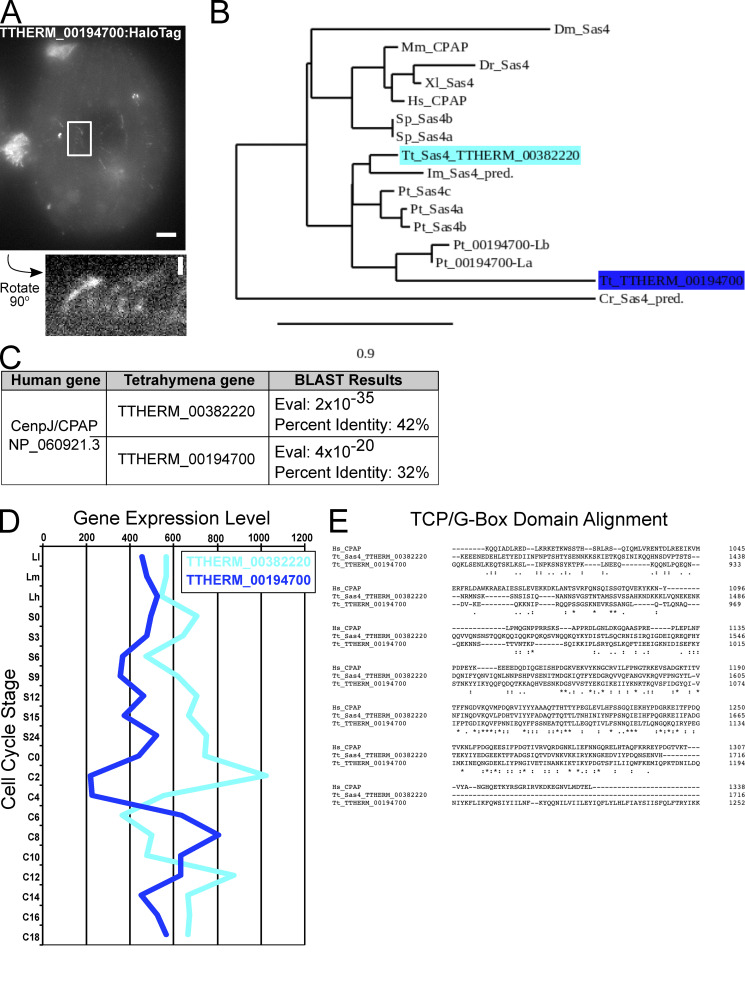
We identified a Tetrahymena SAS4 homologue using reciprocal Basic Local Alignment Search Tool (BLAST) against the human CPAP amino acid sequence (NCBI Protein database accession no. NP_060921.3). This revealed two candidate SAS4 genes, TTHERM_00382220 and TTHERM_00194700, with 42% and 32% identity, respectively, to human CPAP and strongest conservation in the C-terminal region that contains a TCP/G-box domain (Cottee et al., 2013; Zheng et al., 2014). Phylogenetic analysis revealed that TTHERM_00382220, compared with TTHERM_00194700, is more closely related to CPAP/SAS4 from human and other species (Fig. S1). We therefore focused our studies on TTHERM_00382220. To assess whether TTHERM_00382220 is a bona fide SAS4 homologue, we fused mCherry and HaloTag to the C terminus of the predicted open reading frame at the endogenous genomic locus. Fixed and live cell imaging revealed protein localization at both cortical and oral apparatus BBs, as well as other cortical structures (Fig. 1 A and Fig. S2, A–D). Thus, we conclude that TTHERM_00382220 is a BB protein and refer to it as Sas4.
Figure S1.
SAS4 genes in Tetrahymena. (A) Endogenous TTHERM_00194700:HaloTag (grayscale) localizes to oral apparatus and oral primordium cilia, cortical cilia, and to cortical rows. Scale bar, 5 µm. Enlarged panel is rotated 90° counter-clockwise and shows weak cortical row and cilia localization. Scale bar, 1 µm. (B) Phylogenetic analysis of Sas4 proteins, known and predicted (indicated by “pred.”). The SAS4 gene described in this paper (TTHERM_00382220) is closely related to the three previously studied Paramecium SAS4 paralogs, whereas TTHERM_00194700 is distantly related. Phylogenetic tree was created using phylogeny.fr in one-click mode. BLAST analysis of the TTHERM_00194700 protein sequence in the Paramecium genome revealed two additional Paramecium genes, which we identify as Pt_00194700-like(L). The phylogenetic tree shows that the TTHERM_00194700 and Pt_00194700-like proteins cluster together and are distantly related to the Paramecium Sas4 proteins. This supports the hypothesis that TTHERM_00382220 is a Sas4 homologue and that TTHERM_00194700 is not likely a true Sas4 homologue or a paralog to TTHERM_00382220. Dm, Drosophila melanogaster; Mm, Mus musculus; Dr, Danio rerio; Xl, Xenopus laevis; Hs, Homo sapiens; Sp, Strongylocentrotus purpuratus; Tt, T. thermophila; Im, Ichthyophthirius multifiliis; Pt, Paramecium tetraurelia; Cr, Chlamydomonas reinhardtii. (C) Table showing the E-values and percent identities of TTHERM_00382220 and TTHERM_00194700 aligned to the human CPAP protein sequence. (D) Gene expression levels for TTHERM_00382220 and TTHERM_00194700 in different stages of Tetrahymena growth, starvation, and mating (adapted from the Tetrahymena Functional Genomics Database, http://tfgd.ihb.ac.cn). Average of two replicate datasets is shown for each gene. (E) Alignment of human CPAP, TTHERM_00382220, and TTHERM_00194700 amino acid sequence in the C-terminal T complex protein 10 (TCP)/G-box region.
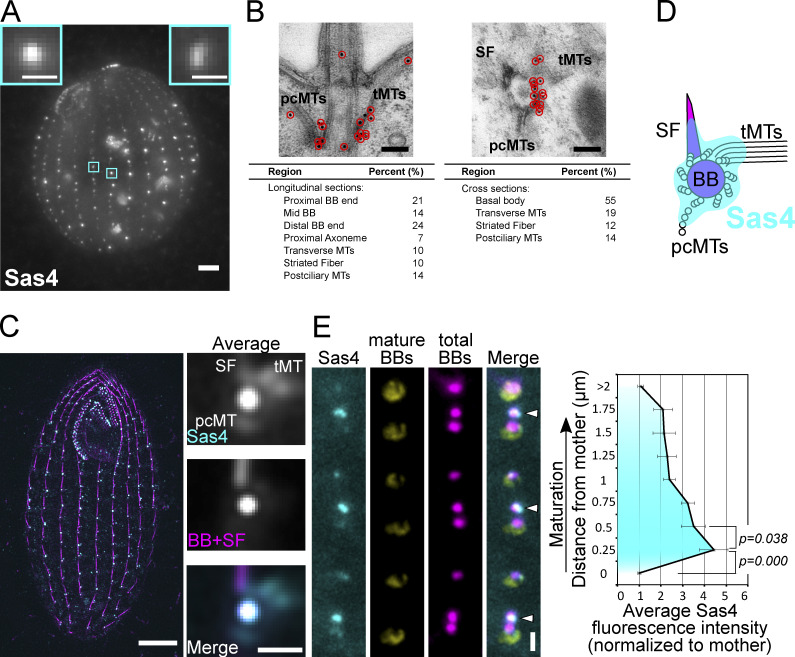
Figure 1.
Sas4 localizes to new BBs and to BB-appendage structures. (A) Sas4:HaloTag (grayscale; JF549) localizes to cortical and oral apparatus BBs. Scale bar, main image, 5 µm. Scale bars, insets, 1 µm. Insets denote individual BBs. (B) Sas4 immuno-electron microscopy localization to BBs, pcMTs, tMTs, and striated fibers. Scale bars, 200 nm. (C) SIM and image averaging of Sas4:HaloTag (JF549) localization at BBs and at proximal positions of BB-appendage structures (tMTs, pcMTs, and striated fibers). Cells were labeled with JF549 HaloTag ligand (cyan) and stained with an anti-Sas6A antibody (magenta) that labels BBs and SFs (Culver et al., 2009). Mature BBs were used for image averaging where each BB was centered on the centroid of the BB signal and oriented relative to the striated fiber. Right panels, average of 530 BBs. Scale bar (whole cell), 5 µm. Scale bar (right panels), 1 µm. (D) Model of Sas4 localization based on immuno-EM and SIM image averaging shown in B and C. (E) Sas4 is enriched at newly assembling daughter BBs. Sas4:HaloTag (cyan; JF549) was visualized relative to mature BBs (yellow; K-like antigen [Kl-Ag]; Williams et al., 1990) and all BBs (magenta; centrin). Arrowheads denote newly assembled BBs that are enriched for Sas4. Scale bar, 1 µm. Distance between peak centrin signals was used to calculate distance between the mother BB (Kl-Ag–positive) and daughter BBs (centrin). The average Sas4 fluorescence intensity is plotted as a function of distance from a mother BB. Data were collected from 10 cells per replicate, and three experimental replicates were performed, with a total of 702 daughter BBs analyzed. Error bars denote SEMs.
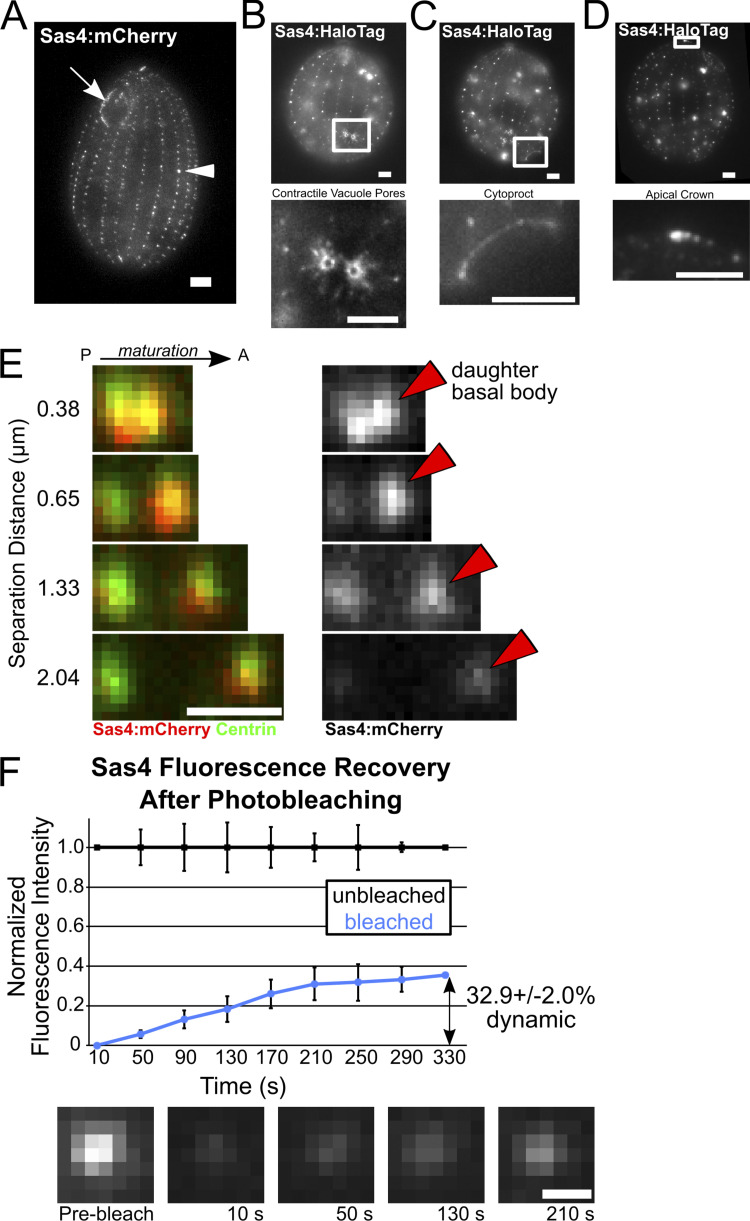
Figure S2.
Sas4 localization and dynamics. (A) Endogenous Sas4:mCherry (grayscale) localization to the cortical and oral apparatus BBs. All localizations described here (A–E) and in the main text are identical between HaloTag and mCherry fusions to Sas4. (B) Endogenous Sas4:HaloTag (grayscale; JF549) localizes to contractile vacuole pores (CVPs). Bottom panel, enlarged image of CVPs. CVPs are MT-based structures used for osmotic regulation (Organ et al., 1972). (C) Endogenous Sas4:HaloTag (grayscale; JF549) localizes to the cytoproct. Bottom panel, enlarged image of the cytoproct. Tetrahymena cytoprocts are sites of waste elimination (Allen and Wolf, 1974). (D) Endogenous Sas4:HaloTag (grayscale; JF549) localizes to the Tetrahymena apical crown. Bottom panel, enlarged image of the apical crown. The apical crown is a site of closely positioned doublet BBs (Jerka-Dziadosz, 1981). Scale bars, 5 µm. (E) Endogenous Sas4:mCherry (red at left, grayscale at right) is enriched at daughter BBs (red arrowheads). Costaining with centrin (green) to mark all BBs. Signal for mCherry is from the fluorescent protein itself, not from antibody staining against mCherry. P, posterior; A, anterior. Scale bar, 1 µm. (F) mCherry:Sas4 FRAP. Graph shows normalized signal for recovery of four bleached (blue) and four unbleached (black) BB. Error bars denote SD. Representative images of fluorescence recovery at a bleached BB. Scale bar, 500 nm.
Unlike other BB proteins, Sas4 diffusely localizes at and around BBs. The localization pattern of Sas4 at BBs varied between BBs of the same cell. Some basal bodies displayed a single strong focused spot of Sas4 signal, whereas others had multiple foci of signal. Cell cycle–dependent differences in localization were not observed. To precisely identify the location of Sas4 within the BB ultrastructure, we performed immuno-electron microscopy (Fig. 1 B). Sas4 localized along the length of the BB microtubule triplets and in the BB lumen. Sas4 also localized to the three BB-appendage structures: the striated fiber (SF), transverse microtubules (tMTs), and postciliary microtubules (pcMTs). These BB-appendages are distinct structures from the distal and subdistal centriole appendages of other eukaryotes (Bayless et al., 2016). Structured illumination microscopy (SIM) coupled with image averaging at mature BBs confirmed these BB-appendage structure localizations, indicating that, unlike other BB proteins required for new BB assembly (Sas6 and Bld10), Sas4 localizes to both BBs and to BB-appendages (Fig. 1 C; Bayless et al., 2012; Culver et al., 2009). Moreover, Sas4 localizes to the proximal end of the BB-appendage structures where the BB-appendage microtubule minus-ends are predicted to exist (Fig. 1 D).
Sas4 is enriched at nascent BBs
To measure the dynamics of Sas4 protein incorporation during new BB assembly and maturation, we quantified Sas4 levels relative to the maturation of BBs. Tetrahymena daughter BBs assemble immediately adjacent and anterior to mother BBs (Fig. 1 E; arrowheads). As daughter BBs mature, they migrate anteriorly along the ciliary row, away from the mother BB. Thus, the distance between mother and daughter BBs serves as a proxy for maturation stage (Allen, 1969; Bayless et al., 2012; Nanney, 1975). Using an independent marker for mother BBs (K-like antigen; Williams et al., 1990), we measured the intensity of Sas4:HaloTag signal at daughter BBs as a function of distance from their mother BBs (Fig. 1 E). Sas4 signal is greatest at close mother–daughter distances: daughter BBs have greater than fourfold the amount of Sas4 compared with mothers at 0.25 µm separation distance. This observation suggests that Sas4 plays an important role early during BB assembly. As daughter BBs mature, the intensity of Sas4 decreases but does not disappear (Fig. 1 E and Fig. S2 E). We conclude that a transient population of Sas4 is strongly recruited early during new BB assembly. This population likely assists in triplet microtubule elongation as has previously been shown for CPAP/Sas4 in other organisms (Sharma et al., 2016). Additionally, early Sas4 recruitment could contribute to BB-appendage microtubule elongation, which occurs simultaneously with BB maturation (Junker et al., 2019). However, a low level, persistent, or late-arriving population of Sas4 remains at BBs and BB-appendages through the life of the BB.
To determine whether Sas4 protein exchanges at its BB binding sites, FRAP of basal body localized mCherry:Sas4 was performed (Fig. S2 F). After photobleaching, Sas4 signal recovers to ∼33%. This indicates that 67% of the Sas4 protein at BBs stably associates with BBs during the time course of the experiment (330 s). It is not clear whether the dynamic Sas4 population resides at BBs or BB-appendages. Regardless, we conclude that, unlike other systems where Sas4 stably associates with centrioles without detectable exchange with the cytoplasm, Tetrahymena Sas4 has both stable and dynamic protein populations at BBs (Balestra et al., 2015; Kirkham et al., 2003).
Sas4 is required for BB assembly
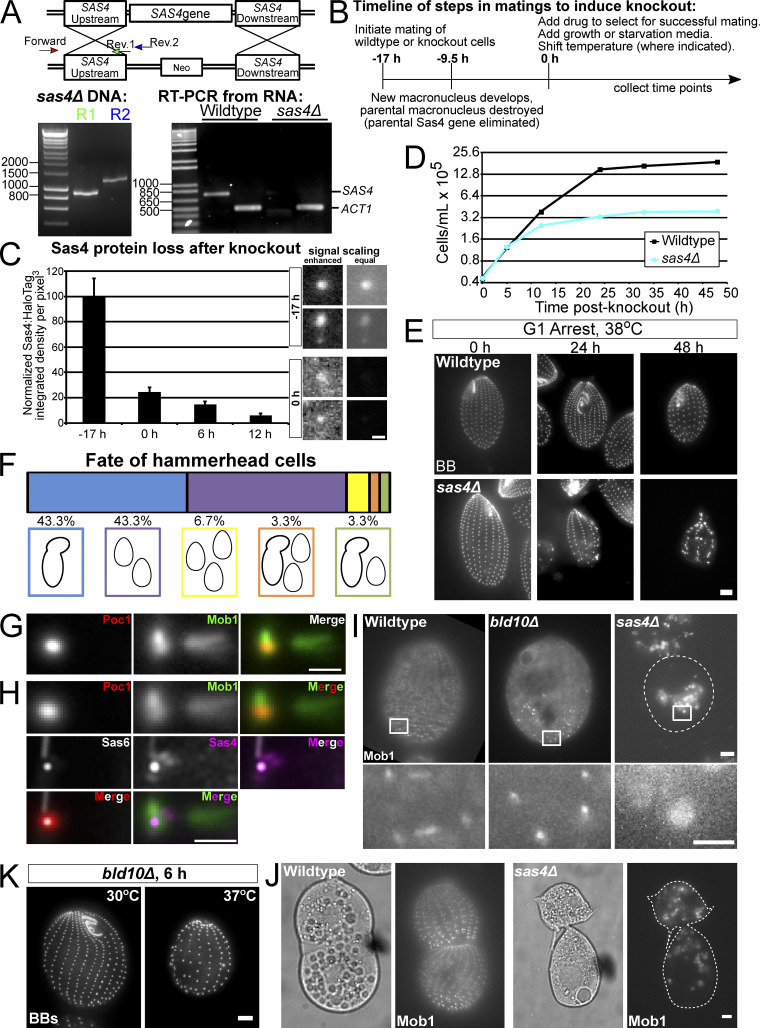
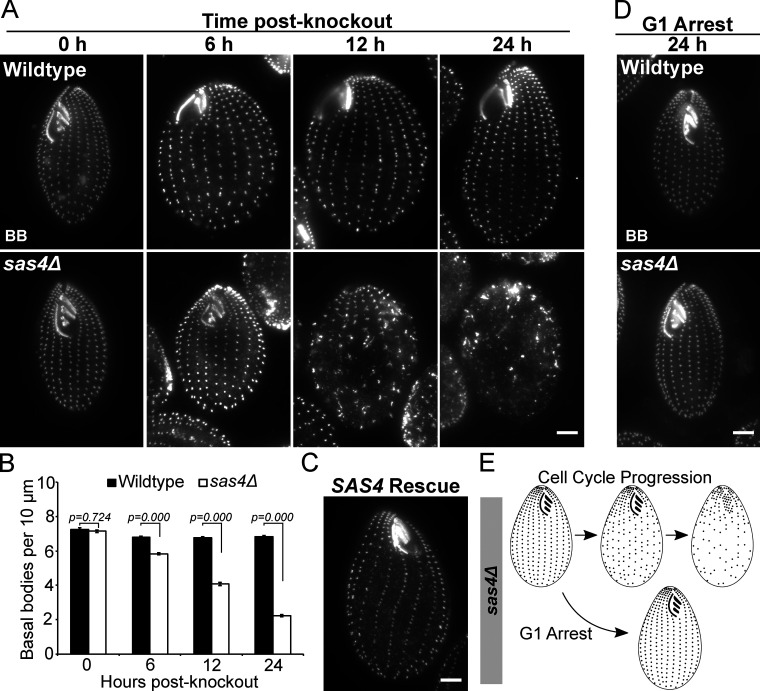
To determine whether Tetrahymena Sas4 is necessary for BB assembly, we generated a whole-genome knockout of SAS4 (Fig. S3 A). SAS4 knockout (sas4Δ) was induced through cell mating, and SAS4 loss leads to a rapid and severe decrease in cortical and oral apparatus BBs (Fig. 2 A). To quantify BB loss, the number of BBs in a ciliary row was quantified per 10 µm in the medial half of the Tetrahymena cell (Pearson et al., 2009). At 12 h post SAS4 knockout, 4.1 BBs per 10 µm were observed in sas4Δ cells, compared with 6.8 BBs per 10 µm in WT cells (Fig. 2 B). This 40% reduction in BB frequency occurs over approximately three cell divisions (Fig. S3 D). By 24 h post-knockout, the reduction increased to 67% compared with WT cells, leaving sas4Δ cells with only 2.2 BBs per 10 µm. Introduction of a WT copy of SAS4 to repair the endogenous SAS4 locus rescued the BB loss phenotype (Fig. 2 C). We conclude that Sas4 is essential for the normal frequency of BBs in Tetrahymena cells.
Figure S3.
SAS4 knockout, sas4Δ growth, and BB loss during G1 arrest at elevated temperature, and Mob1 localization. (A) Schematic of SAS4 knockout construct. The schematic shows that the cassette replaces the entire SAS4 locus with a neomycin resistance cassette. Bottom left panel, PCR of genomic DNA showing integration of the sas4Δ cassette at the SAS4 locus. Bottom right panel, RT-PCR from WT and sas4Δ cells showing loss of SAS4 transcript. ACT1 (actin) is a control for RNA quality. Numbering on molecular weight ladders is in base pairs. (B) Timeline shows mating timeline to induce SAS4 knockout. “0 h” is +17 h from time of mating initiation and is the time of drug and media addition in most experiments. “−9.5 h” is +7.5 h from mating initiation and is the time when most cells enter the second phase of macronuclear development, leading to the destruction of the parental macronucleus driving parental expression of SAS4 (Martindale et al., 1982). (C) Measurement of total cell Sas4 protein loss after SAS4 knockout. Protein levels were measured from at least six cells per time point. Error bars denote SEM. Representative images of two BBs at −17 h and two BBs at 0 h are shown at right. Enhanced scaling/contrast to allow for optimal visualization (left panels) shows that BBs can be identified at the cell cortex, while equal scaling between images (right panels) shows that the BBs at 0 h have drastically reduced Sas4 protein levels. (D) Representative cell growth curve of WT versus sas4Δ cells. Experiment was performed in triplicate. (E) WT and sas4Δ cells (BB, centrin, grayscale) in G1 arrest at 38°C for 24 h and 48 h after SAS4 knockout. Scale bar, 5 µm. (F) Chart showing the fate of 34 single hammerhead cells isolated at 24 h after SAS4 knockout and followed for an additional 24 h. (G) Image averaging of BBs from WT cells expressing Poc1:mCherry and GFP:Mob1. Each BB was centered on the centroid of the Poc1:mCherry signal and oriented relative to the BB just anterior in the ciliary row. Only BBs in the posterior half of the cell were used for analysis. Image is an average of 43 BBs. Note that Mob1 localizes strongly to BB-proximal regions of the striated fiber and distal regions of the transverse MTs, as well as to the BB itself. Scale bar, 1 µm. (H) Comparison of Sas4 and Mob1 average localization by overlay of averaged images from G and Fig. 1 C. BB alignment was determined using the centroid of either Sas6A or Poc1 signal. Merged image of Sas4 and Mob1 (magenta and green, respectively) shows that Mob1 localizes to BBs and striated fiber regions proximal to the BB, similar to where Sas4 localizes, and to distal regions of the transverse MTs. Sas4 also localizes to transverse MTs, but at a BB proximal location. (I) Images of GFP:Mob1 localization in nondividing WT, bld10Δ, and sas4Δ cells at 24 h post-knockout. Scale bar, 5 µm. Enlarged images show Mob1 localizes to the cell cortex in WT and bld10Δ cells but not in sas4Δ. Images are from the same experiments described in Fig. 4 F. Scale bar, 1 µm. (J) Left panels, DIC images of dividing WT and sas4Δ cells. Right panels, Mob1 localizes as a gradient to the posterior ends of both daughter cells in WT, but not in sas4Δ cells. Images are from the same experiments described in Fig. 4 F. Scale bar, 5 µm. (K) Cells at 6 h after BLD10 knockout at 30°C (left) or 37°C (right), stained for BBs (centrin, grayscale). Cells shifted to 37°C display greater BB loss. Scale bar, 5 µm.
Figure 2.
Sas4 is required for BB assembly. (A) WT and sas4Δ cells at 0, 6, 12, and 24 h post-knockout in cycling cells at 30°C (BB, centrin, grayscale). (B) Quantification of BB frequency in a 10-µm distance within the medial region of ciliary rows, based on centrin staining shown in A. Analysis was performed in triplicate on five ciliary rows per cell for 20 cells per condition (total of 300 rows per cell line per time point). P value for 0 h post-knockout between WT and sas4Δ is not significant. Error bars denote SEMs. (C) sas4Δ rescue by reintroduction of SAS4 gene. Image is representative of two independent experiments. (D) WT and sas4Δ cells (BB, centrin, grayscale) maintained in G1 arrest at 30°C for 24 h post-knockout. (E) Model of sas4Δ phenotypes. BBs are lost when cells progress through the cell cycle but are not lost if maintained in G1 arrest. Scale bars, 5 µm.
We envision three ways that BBs can be lost in sas4Δ cells. First, new BBs could fail to assemble as the cells proceed through the cell cycle. This would lead to a reductional loss of BBs at each cell division. Second, existing BBs could disassemble. This would cause BB loss independent of cell cycle progression. Third, BBs may not anchor properly at the cell cortex. This would cause BB loss by internalization of BBs. These possibilities are not mutually exclusive. To test the models of BB loss, we maintained sas4Δ cells in G1 arrest by nutrient starvation (Fig. 2 D; Kaczanowski, 1978). G1-arrested cells do not assemble new BBs or progress through the cell cycle; thus, any BB loss observed would result from the disassembly or cortical detachment of existing BBs. After G1 arrest, we did not observe BB loss as was observed in cycling conditions (Fig. 2, A and D). Therefore, Sas4 is responsible for new BB assembly during cell cycle progression, and not for stability or anchoring of existing BBs to the cell cortex during G1 arrest (Fig. 2 D). We did, however, observe loss of BBs and ciliary rows when G1-arrested sas4Δ cells were shifted to higher temperature, suggesting that Sas4 is required for proper cortical patterning during states of elevated ciliary force (Fig. S3 E; Allen, 1969; Dirksen, 1971; Pearson et al., 2009). This suggests that Sas4 functions to resist elevated ciliary force.
The specific role for Sas4 in BB assembly and not stability under normal force conditions is in stark contrast to the loss of the essential BB assembly and stability factor, Bld10/CEP135. In Tetrahymena bld10Δ cells, BB loss is observed, but it is not as rapid or severe as in sas4Δ cells (∼6 BBs/10 µm for bld10Δ versus 4.1 BBs/10 µm for sas4Δ at 12 h post-knockout; Fig. 2 B; Bayless et al., 2012). Further, while Bld10 is important for new BB assembly, it is also important for stabilizing existing BBs under normal force conditions (Bayless et al., 2012). Differences between bld10Δ and sas4Δ phenotypes may be reflected in their differences in protein incorporation dynamics during BB assembly. Bld10 gradually incorporates at BBs as they mature, whereas Sas4 is enriched at nascent BBs and lessens as they mature. In addition, the distinct sas4Δ and bld10Δ phenotypes could arise from differences in their localization within the BB ultrastructure. Bld10 is only found at BBs, whereas Sas4 also localizes to BB-appendage structures. These observations suggest distinct roles for each assembly factor that require further exploration to understand how they work together to build functional BBs.
Cell division arrest and anterior displacement of the cytokinetic furrow in sas4Δ cells
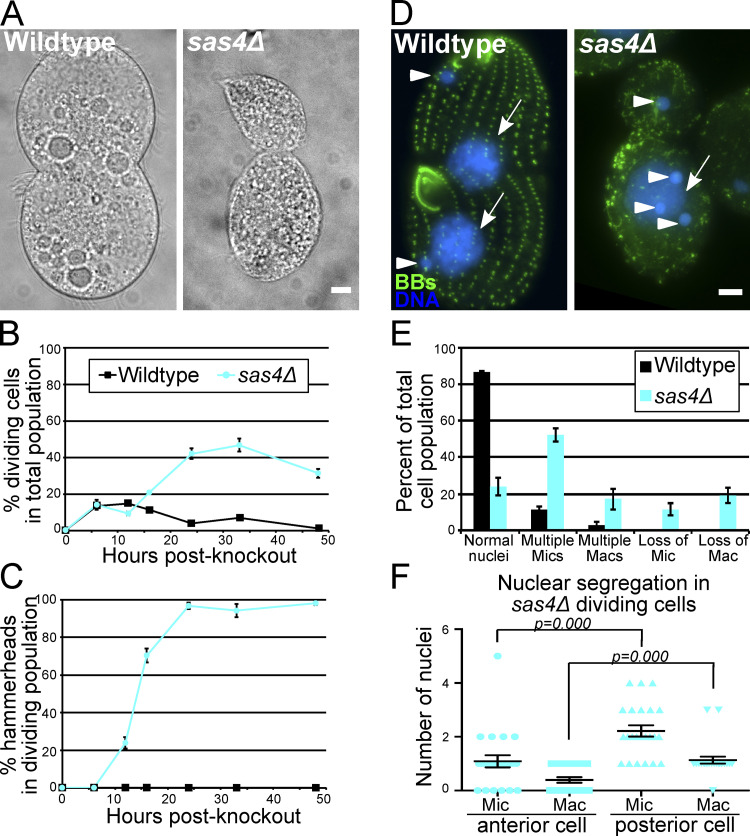
A prominent phenotype in sas4Δ cells is the appearance of cells with anteriorly positioned cytokinetic division furrows (Fig. 3 A). This was generally characterized by larger posterior cells and smaller anterior cells that were often tilted relative to the anterior–posterior axis. This cell division morphology is referred to as the “hammerhead” phenotype and is consistent with Tetrahymena mutants in the Hippo signaling pathway (Frankel, 2008; Jiang et al., 2017, 2019; Tavares et al., 2012). To understand the role that Sas4 plays in cell division, we quantified the percentage of cells with division furrows in WT and sas4Δ cells (Fig. 3 B). sas4Δ cells display a marked increase in the percentage of cells undergoing division. The hammerhead phenotype was not present within the first 6 h after SAS4 knockout (Fig. 3 C), corresponding to approximately two cell divisions (Fig. S3 D). By this time point, BB frequency has already dropped by ∼14% compared with WT (Fig. 2 B). This suggests while BB loss has initiated, the initial loss of Sas4 does not affect proper cell division furrow placement at the equatorial position, or, alternatively, that there is a parental population of Sas4 protein that supports normal cell division at these early time points. By 12 h after SAS4 knockout, when BB frequency was 58.8% of WT, 24% of the dividing cells displayed the hammerhead morphology. Conversely, when bld10Δ cells reach ∼60% BB frequency, they do not exhibit hammerhead phenotypes, suggesting that the hammerhead phenotype does not result from BB loss alone (Fig. 4 H; Bayless et al., 2012). At 24 h, nearly all of the dividing sas4Δ cells (98.6%) displayed the hammerhead phenotype (Fig. 3 C). This timing correlates with when the sas4Δ cell population stopped growing (Fig. S3 D), suggesting that cells that reach the hammerhead state arrest in cell division. To test this, individual hammerhead cells were isolated at 24 h post-knockout. Most hammerhead cells either never go on to complete cell division (43.3%) or only divide once, resolving the hammerhead into two daughter cells (43.3%, Fig. S3 F). Only 13.4% of hammerhead cells go on to attempt and/or complete another round of cell division, and none perform more than two cell divisions (Fig. S3 F). This is the first report that we are aware of in which disruption of a BB assembly protein results in cell division arrest and dysmorphology (Fig. 4 H).
Figure 3.
Sas4 is required for equal cell division and cell cycle progression. (A) Dividing WT and sas4Δ cells (DIC). In WT cells, the division plane is positioned equatorially, whereas sas4Δ cells have anteriorly positioned division furrows. (B) Percent of dividing cells in the total cell population in WT versus sas4Δ cells as a function of time after SAS4 knockout. Experiment was performed in triplicate, and at least 100 cells per cell line and time point were counted per experiment. Error bars denote SEM. (C) Percentage of dividing cells that exhibit the hammerhead phenotype as a function of time post-knockout. By 24 h post-knockout, nearly all dividing sas4Δ cells exhibit the hammerhead morphology. Experiment was performed in triplicate, and at least 100 cells per cell line and time point were counted per experiment. Error bars denote SEM. (D) BBs (green, centrin) and DNA (blue, Hoechst) in dividing WT and sas4Δ cells. Arrows and arrowheads denote macronuclei and micronuclei, respectively. (E) Quantification of the percentage of the total cell population that displayed nuclear gain or loss in WT or sas4Δ cells. Micronucleus, Mic; macronucleus, Mac. Experiment was performed in triplicate with between 24 and 35 cells analyzed per replicate per cell line. Error bars denote SEM. (F) Quantification of the number of micronuclei (Mic) and macronuclei (Mac) in dividing hammerhead sas4Δ cells in the anterior and posterior cell halves. 23 cells were analyzed. Error bars denote SEM. Scale bars, 5 µm.
Figure 4.
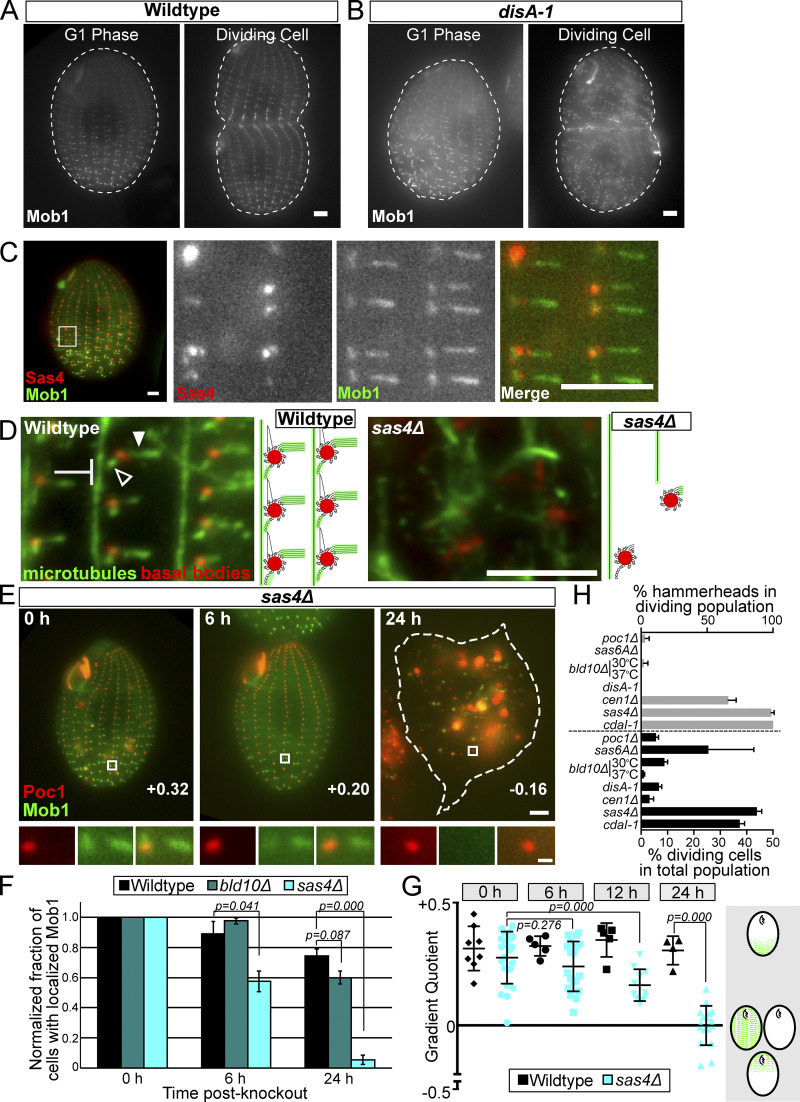
Sas4 is required for Hippo factor localization to the cell cortex. (A) GFP:Mob1 is asymmetrically localized as a gradient from the posterior to anterior of WT G1 phase cells and its localization is enriched above the division furrow in dividing WT cells. Mob1 localizes to BBs and tMTs. Scale bar, 5 µm. (B) GFP:Mob1 localization in G1 phase and dividing cells in the cortical disorganization mutant, disA-1. Mob1 localizes as a posterior to anterior gradient at BBs and tMTs, as in WT cells. Scale bar, 5 µm. (C) Sas4:HaloTag (JF549) and GFP:Mob1 colocalize at daughter basal bodies. Mob1 localizes along the length of tMTs, but not at the proximal ends adjacent to BBs. Conversely, Sas4 localizes to the proximal end of tMTs that are not labeled with Mob1. Scale bars, 5 µm. (D) Cortical microtubules are lost and disorganized in sas4Δ cells. WT and sas4Δ cells at 24 h post-knockout labeled for BBs (red, centrin) and MTs (green, α-tubulin). Images focus on the cortical microtubules: tMTs (filled arrowhead), pcMTs (empty arrowhead), and longitudinal MTs (blunted arrow). Scale bar, 5 µm. (E) sas4Δ cells expressing GFP:Mob1 and Poc1:mCherry at 0, 6, and 24 h post-knockout. Scale bar, 5 µm. White boxes indicate the enlarged region below. Scale bar, 500 nm. Gradient quotient (determined in G) for each image is shown. (F) Quantification of the percentage of the cell population with proper Mob1 localization. Mob1 fails to localize in sas4Δ cells starting at 6 h post-knockout and is reduced to less than 10% of the cell population by 24 h after SAS4 knockout. At least 100 cells per condition and time point were quantified, and the experiment was performed in triplicate. Error bars denote SEM. Representative images of WT, bld10Δ, and sas4Δ cells at 24 h are shown in Fig. S3 I. (G) Quantification of Mob1 gradient in WT and sas4Δ cells at 0 h, 6 h, 12 h, and 24 h post-knockout. The gradient quotient (see Materials and methods) is calculated such that positive numbers indicate Mob1 enrichment in the posterior half of the cell and negative numbers indicate enrichment in the anterior half of the cell. A zero value indicates no gradient, because of either equally high or equally low amounts of Mob1 in both halves of the cell. Experiment was performed in duplicate, and each point on the graph represents a single cell. Error bars denote SD. (H) Quantification of cell division percentages and hammerhead cells in BB, cortical (disA-1), and Hippo (cdaI-1) mutants. Data shown are the average number of hammerheads seen and percent dividing cells during knockout or temperature shift of the indicated gene. At least 100 cells were counted per cell line per experiment. Experiment was performed in duplicate for poc1Δ, sas6AΔ, and cen1Δ, in triplicate for sas4Δ, bld10Δ at 30oC, and cdaI-1, and in quadruplicate for disA-1 and bld10Δ at 37oC. Error bars denote SEM.
Nuclei are missegregated in sas4Δ cells
Because sas4Δ cells exhibit abnormal division furrow placement, we next asked whether the germline micronuclear and somatic macronuclear genomes properly segregate to daughter cells (Fig. 3 D). Using Hoechst 33342 to stain DNA, we assessed nuclear segregation in WT and sas4Δ cells at 24 h post-knockout. WT cells averaged 1.1 micronuclei and 1.0 macronuclei per cell, with little deviation from the normal configuration of one micronucleus and one macronucleus per cell. Conversely, sas4Δ cells averaged 1.8 micronuclei and 1.0 macronuclei per cell. The variation in nuclear configurations in sas4Δ cells was great with cells exhibiting both too many and too few nuclei (Fig. 3 E). Thus, sas4Δ cells have aberrant nuclear segregation where cells gain and lose nuclei.
To examine how nuclei missegregate in the hammerhead cells, we quantified nuclear frequencies in cells undergoing hammerhead cell divisions (Fig. 3 F). The anterior daughter cells frequently lacked a macronucleus, whereas the posterior daughter cells often had a macronucleus and multiple micronuclei. We observed diverse variations of nuclear gain and loss. These data suggest that the accurate mitotic micronuclear and amitotic macronuclear segregation is disrupted in sas4Δ cells. Because we observed bi- and multinucleated posterior cells, we suggest that failure to segregate micronuclei between anterior and posterior daughters in sas4Δ cells promotes the accumulation of multiple micronuclei. Importantly, we did not detect Sas4 at the spindle poles of dividing nuclei. These data show that Sas4, a cortically localized protein, is necessary for proper nuclear segregation.
Hippo signaling uses cortical structures to establish asymmetric subcellular localization
The hammerhead phenotype in dividing Tetrahymena cells was first discovered in forward genetic screens performed by Joseph Frankel (Frankel, 2008). One of these mutants was identified to be the Hippo signaling pathway factor, CdaI (Mst kinase; Jiang et al., 2017). Mutations in either CDAI or MOB1 produce anteriorly positioned cell division furrows and nuclear missegregation (Tavares et al., 2012; Jiang et al., 2017). Conversely, the elo1-1 (Lats kinase) mutant produces Tetrahymena cells with posteriorly positioned division furrows, suggesting antagonistic Hippo activities promote equal cell division (Jiang et al., 2019). The observation that sas4Δ cells phenocopy Hippo signaling mutants suggests they operate in the same pathway. This led us to investigate the relationship between Hippo signaling, BB assembly, and the cortical architecture.
That Sas4 and cortical organization could impact Hippo signaling is surprising for two reasons. First, bld10Δ cells, which are also defective in BB assembly and stability, do not exhibit a hammerhead phenotype (Bayless et al., 2012; Fig. 4 H). Second, the disorganized cortical architecture mutant, disA-1, also divides equatorially (Frankel, 1979). The latter observation led to the conclusion that cortical organization is dispensable for proper cell division (Jerka-Dziadosz et al., 1995). On the other hand, Tetrahymena Mob1, CdaI, and Elo1 localize to BBs and to BB-appendages in WT cells (Fig. 4, A and C; Tavares et al., 2012; Jiang et al., 2017, 2019). Additionally, the homologues of these Hippo factors localize to spindle pole bodies and centrosomes in other cell types, and to the cell cortex in the ciliate, Stentor coeruleus (Bolgioni and Ganem, 2016; Hergovich and Hemmings, 2012; Slabodnick et al., 2014). In Tetrahymena, the subcellular localization of Hippo factors is asymmetric (Fig. 4 A; Tavares et al., 2012; Jiang et al., 2017). In G1 cells, Hippo factors localize along an anterior-posterior gradient where they preferentially localize to BBs (and to tMTs in the case of Mob1 and CdaI) at the posterior end of the cell. In dividing cells, Hippo factors localize along two gradients, concentrating at what will become the posterior ends of both daughter cells (Fig. 4 A). The asymmetric distribution of Hippo factors around the division furrow may be necessary for proper division furrow placement, and these factors use BBs and other cortical features to effect their asymmetric localization (Tavares et al., 2012; Jiang et al., 2017, 2019).
To begin to understand how Hippo factors interact with the cortical architecture, we expressed GFP:Mob1 in disA-1 mutant cells, which have short SFs and disorganized and disoriented BBs (Jerka-Dziadosz et al., 1995; Galati et al., 2014). We found that despite the disorganized cortical architecture, Mob1 still localizes as an asymmetric gradient at the posterior end of the cell in G1 phase and above the equatorially positioned division furrow in dividing cells (Fig. 4 B). This supports the idea that Hippo signaling is not interrupted in this mutant and that a disorganized cortex is still capable of properly tethering Hippo signaling proteins.
We reasoned that if Sas4 is necessary for Hippo factor localization, we might observe colocalization of Mob1 at BBs and BB-appendage structures. To address this question, we coexpressed the Sas4:HaloTag with GFP:Mob1 in cycling Tetrahymena cells. Sas4 and Mob1 colocalize at BBs, but they did not colocalize at the appendage structures (Fig. 4 C). This is observed at the tMTs, where Sas4 localizes to the base or proximal side of tMTs, close to the BB, while Mob1 localizes distally along the length of the tMTs. To investigate this localization further, we created a cell line coexpressing the BB protein Poc1:mCherry and GFP:Mob1 and performed image averaging on BBs in the posterior half of the cell (Fig. S3 G). This revealed that Mob1 localizes to BBs and the distal regions of the tMTs, consistent with previous studies, as well as to the BB-proximal region of the SF (Tavares et al., 2012). To compare this localization with that of Sas4, we superimposed this averaged image with the averaged image of Sas4 shown in Fig. 1 C, using the centroid of Poc1 and BB-localized Sas6A signals to align the images (Fig. S3 H). This analysis revealed colocalization of Mob1 and Sas4 at the BB-proximal region of the SF, but no colocalization at the tMTs. These observations suggest that Mob1 localization to BBs and BB-proximal regions of SFs may depend on Sas4 either through a direct or indirect mechanism, whereas Mob1 localization to tMTs could depend on a Sas4-regulated loading mechanism.
Sas4 maintains cortical structure and Mob1 localization
Because Sas4 localizes to proximal ends of BB-appendage structures, notably the tMTs that Mob1 also localizes to, we hypothesized that loss of Sas4 might alter these appendage structures. To test this, we visualized microtubules (MTs) in sas4Δ cells at 24 h after SAS4 knockout, the time when sas4Δ cells exhibit the hammerhead phenotype. sas4Δ cells have fewer cilia, BBs, pcMTs, and tMTs, and what remains of them is severely disorganized (Fig. 4 D). This indicates that the cortical cytoskeletal architecture, as defined by these MT structures, is lost in the absence of Sas4. We conclude that Sas4 is required for the structural maintenance of cortical MTs.
A clear implication of the above findings is that, without BBs and specific cortical MTs, the Hippo pathway factors that are known to bind to these MTs may be mislocalized. If true, Hippo factors could not define the position of the cytokinetic furrow. To test this model, we expressed GFP:Mob1 in WT and sas4Δ cells and evaluated Mob1 localization following SAS4 knockout. In WT cells, Mob1 localized normally as a gradient (Fig. S3 I). Conversely, in sas4Δ cells as early as 6 h post-knockout, the number of cells with Mob1 localization to the cell cortex decreased by ∼50%, and by over 90% by 24 h post-knockout (Fig. 4 F). GFP:Mob1 signal was low or absent, and when low, it localized to intracellular bodies of varying size (Fig. 4, E and F; and Fig. S3 I). To test whether Mob1 mislocalization directly results from Sas4 loss or from loss of BBs and/or cortical MTs generally, we measured Mob1 loss in bld10Δ cells. Mob1 localizes normally in the majority of bld10Δ cells (Fig. 4 F and Fig. S3 I). Because bld10Δ also displays a BB loss phenotype, albeit less severe than sas4Δ, these data suggest that BB loss alone cannot explain the mislocalization of Mob1 in sas4Δ cells, and points to a more specific role for Sas4 in Hippo factor localization. We conclude that Sas4 is necessary for Mob1 localization.
To quantify the loss of Mob1 gradient localization, we calculated a gradient quotient to describe the strength of the Mob1 gradient during Sas4 loss (Fig. 4 G). Here, a positive value corresponds to enrichment of Mob1 at the posterior end of the cell, a negative value corresponds to anterior enriched Mob1, and a 0 value indicates no gradient. At 6 h post-knockout, the average Mob1 gradient quotient in sas4Δ cells decreased compared with sas4Δ cells at 0 h, and by 12 h after knockout this decrease was even stronger. At 24 h post-knockout, the gradient quotient was −0.001 ± 0.08 in sas4Δ, representing the complete loss of the Mob1 gradient due to the loss of Mob1 at all BBs (Fig. 4 E, 24 h). Because Mob1 mislocalization and gradient loss occurred at early time points before severe BB loss (6 h), this suggests that Sas4 is necessary for Mob1 tethering to the cell cortex, even when BBs are intact.
To further address whether the loss of Mob1 localization in sas4Δ cells is due to a specific role for Sas4 and not BBs generally, we tested whether Hippo-dependent asymmetric cell division could be forced by increasing BB loss using elevated temperature in bld10Δ cells (37°C; Fig. 4 H and Fig. S3 K). No hammerhead phenotypes were observed in dividing bld10Δ cells grown at 37°C (0/8 cells), suggesting that BB loss alone is not sufficient to induce the hammerhead phenotype. Fewer dividing cells were observed in bld10Δ cells at elevated temperature (8/915 cells). Importantly, cell division arrest is a hallmark of the Hippo mutants, and the lack of such an arrest in bld10Δ mutants suggests that they are refractory to the Hippo pathway arrest phenotype (Fig. 4 H; Tavares et al., 2012; Jiang et al., 2017, 2019). Hippo-associated phenotypes were also tested in additional BB mutants, the disA (striated fiber) mutant, and the cdaI-1 (Hippo) mutant as a positive control (Fig. 4 H; Culver et al., 2009; Stemm-Wolf et al., 2005; Pearson et al., 2009; Galati et al., 2014; Jerka-Dziadosz et al., 1995; Jiang et al., 2017; Frankel, 2008). Of the mutant cells tested, only cen1Δ knockout cells displayed an elevated fraction of dividing cells with hammerhead phenotypes (21/30 cells), although at a lower level than that in sas4Δ cells (263/267 cells). The lack of cell division arrest in cen1Δ mutants suggests that its interaction with the Hippo pathway is not as strong as in sas4Δ cells (Fig. 4 H). However, it is interesting to note that, like Sas4, Cen1 localizes to BB-proximal regions of the tMTs, indicating that this localization pattern may be critical for Hippo factor localization (Fig. 1 C; Stemm-Wolf et al., 2005). Together, these observations indicate that Mob1 requires Sas4 specifically, and not BBs generally, to promote its localization to the cell cortex and to properly position the cell division furrow.
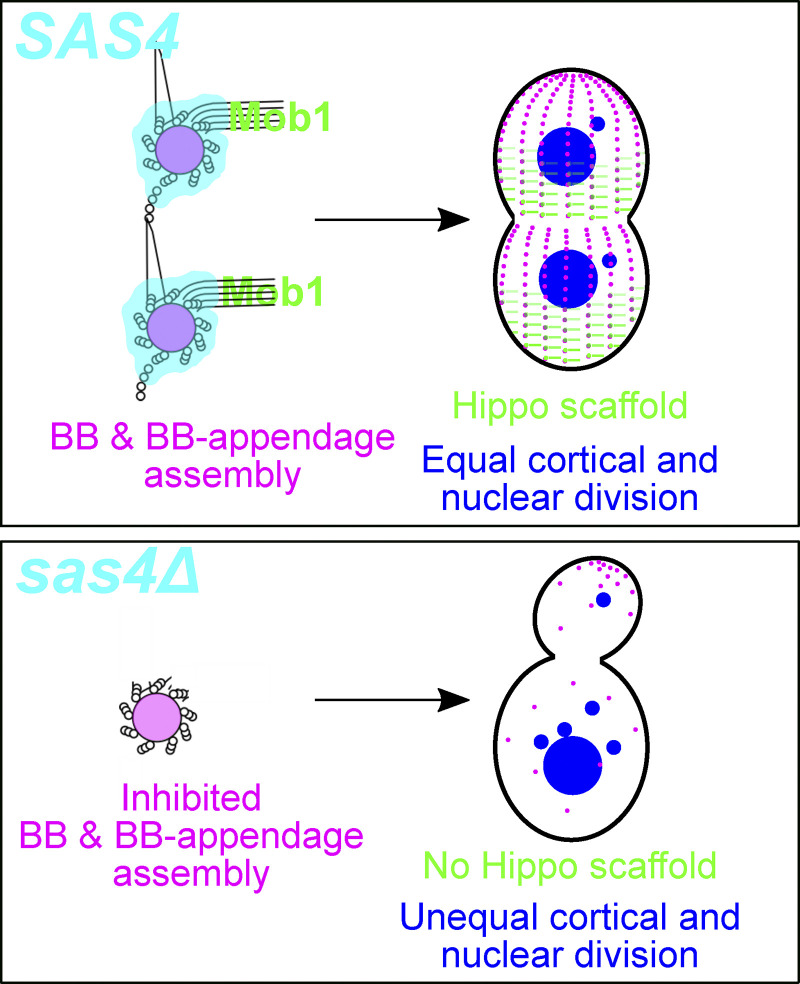
In summary, Tetrahymena Sas4 is a unique and multifunctional BB assembly protein. Sas4 localization and function are not restricted to the core BB structure, but also reside at proximal regions of BB-appendages. Because the ancestral role of centrioles and BBs is thought to be in cilium nucleation rather than at the centrosome, we speculate that the BB-appendage MT organization activity of Sas4 is an evolutionarily ancient function (Carvalho-Santos et al., 2011; Wickstead and Gull, 2011). This function is important to integrate BBs with their surrounding cytoplasmic and cortical environment. Coupling new centriole and BB assembly with MT organization in the activity of a single protein, Sas4, ensures that BBs integrate their assembly and cortical organization with high fidelity. This allows Sas4 to act as a link between BBs and Hippo signaling events for accurate cell division, both in regard to the placement of the cytokinetic furrow and the segregation of nuclei to daughter cells (Fig. 5). This is the first BB assembly protein mutant to exhibit a highly penetrant Hippo phenotype and suggests a specific role for Sas4 in coordinating the localization and function of Hippo signaling. We suggest that this is consistent with Sas4’s unique localization to the proximal ends of BB-appendages. In summary, our studies provide a link between BB assembly and cell cycle progression to explain the high degree of coordination between these two important cellular processes.
Figure 5.
Model of Sas4’s role in BB assembly, Hippo factor tethering, and cell division. Sas4 promotes BB assembly and maintains BB-appendages. Hippo signaling molecules (Mob1) localize to BBs and BB-appendages, and this is necessary to maintain proper Hippo gradients, nuclear segregation, and cytokinetic furrow position.
Materials and methods
Tetrahymena cell culture growth and media
T. thermophila strains SB1969 (TSC_SD00701), SB210 (TSC_SD00703), CU428.2 (TSC_SD00178), B* VI (TSC_SD00022), and B* VII (TSC_SD00023) were obtained from the Tetrahymena Stock Center at Cornell University. Cells were grown to mid-log phase at 30°C in 2% SPP (2% proteose peptone, 0.2% glucose, 0.1% yeast extract, and 0.003% Fe-EDTA) unless otherwise indicated. To arrest cells in the G1 phase of the cell cycle, cells were washed and resuspended in Tris starvation media (10 mM Tris-HCl, pH 7.4) or Dryl’s starvation media (2 mM sodium citrate, 1 mM NaHPO4, 1 mM NaH2PO4, and 1.5 mM CaCl2; used only in matings). For incubation at 38°C, cells were kept in a heated water bath. Cell densities were determined using a Coulter Z1 cell counter with size gating of 15–45 µm.
Plasmids
The Sas4:mCherry and Sas4:HaloTag strains were generated by transforming cells with p4T2-1:SAS4:mCherry or p4T2-1:SAS4:HaloTag. These cassettes integrate into the endogenous SAS4 locus and remain under control of the endogenous promoter. p4T2-1:SAS4:mCherry was generated by PCR amplification of a 1,062-bp fragment of SAS4 immediately upstream of the TGA termination codon (5′-GCgaattcATGCACATTTTCTGAGTTTTAATCAGG-3′ and 5′-GCggtaccTTCTGCTCATAAAAACGATAAAAAAAAATTGG-3′, lowercase letters denote restriction sites) that was cloned into p4T2-1-mCherryLAP to generate p4T2-1-SAS4US-mCherryLAP (Winey et al., 2012). A 1,009-bp fragment downstream of the TGA termination codon (5′-GCgagctcTAAATATTTAAATATCTATTTCTGATTCACCTAAGAAG-3′ and 5′-GCggatccGAAAGAGAGACTTGGCATATAACTAAGTG-3′) was then cloned into p4T2-1-SAS4US-mCherryLAP plasmid intermediate to create p4T2-1-SAS4:mCherry. This plasmid contains the NEO2 drug selection marker. p4T2-1:SAS4:HaloTag was created by restriction digestion of the p4T2-1:SAS4:mCherry vector with BstBI and XhoI to eliminate mCherry and replace it with a codon-optimized HaloTag flanked by BstBI and XhoI restriction sites. p4B2-1:SAS4:HaloTag harboring blasticidin resistance was created by restriction digest of p4T2-1:SAS4:HaloTag (NEOR) and p4B2-1 empty vector with XmaI and XbaI (Gaertig and Kapler, 1999). This releases the neomycin cassette from p4T2-1:SAS4:HaloTag (NEOR) backbone and the blasticidin resistance cassette from p4B2-1. The p4T2-1:SAS4:HaloTag backbone and the blasticidin resistance cassette were then gel isolated and ligated to produce p4B2-1:SAS4:HaloTag.
The p4T2-1:TTHERM_00194700:HaloTag plasmid was created by amplification of a 818-bp fragment of TTHERM_00194700 immediately upstream of the TGA termination codon (5′-CGCggtaccTTATTGATGGCTACTAAATTGTAATTTTTCC-3′ and 5′-CGCgaattcATTATTTAATTGCCATTTTTTGATATATCTTGTG-3′), and a 879-bp fragment downstream of the TGA termination codon (5′-CGCggatccATTTCAATAATAAAGATAAAATTACTTAGATCTTTC-3′ and 5′-CGCgagctcCAAGATACAATTCTTAAACTTATTAGACAAGC-3′) and cloning these fragments into a p4T2-1:HaloTag backbone using standard molecular cloning techniques.
Overexpression of GFP:Mob1 was achieved by transforming cells with pBS-MTT-GFP:MOB1. This vector was produced by generating MOB1 cDNA by RT-PCR (5′-CGAgaattcTTATGAGTTAGAAGACATATAAGCCTAAG-3′ and 5′-GCACtctagaTCATTGTTAAGTTTGAGGAACTTCTTTACG-3′), then cloning into the pENTR4 dual selection Gateway entry vector (Thermo Fisher Scientific) to create pENTR4:MOB1. This was subcloned into the pBS-MTT-GFP Gateway destination vector using the LR clonase reaction kit (Thermo Fisher Scientific) to create pBS-MTT-GFP:MOB1 (Matsuda et al., 2010). The vector was digested with BlpI before biolistic transformation and targeted to the rpL29 locus. Transformants were selected for by cycloheximide resistance. GFP:Mob1 expression was driven by the metallothionein (MTT1) promoter and was induced using 0.25–0.5 µg/ml CdCl2 (Shang et al., 2002).
The plasmid for SAS4 knockout was generated in the p4T2-1 vector with codon-optimized NEOR (Winey et al., 2012). A 747-bp fragment upstream of the SAS4 initiation codon (5′-GCggtaccTAATTTAGAACATATTTATATAAAATAAATAAGATAGAAAATAAATAGTAG-3′ and 5′-GCctcgagTCAAGTACCTTAAGACTAAGACTAATCTCCC-3′) and a 1,009-bp fragment downstream of the SAS4 termination codon (5′-GCgagctcTAAATATTTAAATATCTATTTCTGATTCACCTAAGAAG-3′ and 5′-GCggatccGAAAGAGAGACTTGGCATATAACTAAGTG-3′) were cloned into the p4T2-1 vector to create p4T2-1:SAS4Δ.
The plasmid for sas4Δ rescue was constructed as follows. A fragment beginning 817-bp upstream of the translational start site of SAS4 and including 1,495-bp of the 5′ portion of the open reading frame was amplified from genomic DNA (total size, 2,275 bp) using primers NotI_CPAP_Prom_5UTR_Nterm_F (5′-GCATTgcggccgcACCAACAACCAACACG-3′) and SpeI_CPAP_internal_R (5′-CATTTTATTAACATTTTactagtTTCTTGG-3′), and cloned into pBS-RPL29-polylinker using NotI and SpeI restriction endonuclease sites to create pBS-RPL29:SAS4_5′. Subsequently, a 1,943-bp fragment spanning the SpeI site and continuing to a region just upstream of the predicted Sas4 coiled-coil domain was amplified from genomic DNA using primers SpeI_CPAP_internal_F (5′-ccaagaaactagtaaaatgttaataaaatg-3′) and CPAP_PN23_MBD_R (5′-ggatccCTAATTGTTTCCTTTTTTGTTAAAATATTTTTAAACC-3′), then cloned into pBS-RPL29:SAS4_5′ using SpeI and BamHI restriction endonuclease cut sites, resulting in pBS-RPL29:SAS4ΔCC-Gbox. This 4,200-bp fragment was subsequently subcloned into pBlueScript II KS- using SacI and BamHI restriction sites, creating pBlueScript II:SAS4ΔCC-Gbox. A SAS4 fragment spanning the recognition sequences for BstBI and MfeI was amplified by PCR from genomic DNA with primers CPAP_BstBI_Fwd (5′-AATCTCttcgaaTGTAGATATTACTGAC-3′) and BamHI_CPAP_MfeI_Rev (5′-GATTggatccGCATCATCTACAATTGTTTTAGATGG-3′) and cloned into pBlueScript:SAS4ΔCC-Gbox using BstBI and BamHI to create pBlueScript:SAS4Δ3'. Finally, a SAS4 2,660-bp fragment spanning the MfeI site, including 714-bp of sequence downstream of the termination codon, was amplified from genomic DNA with primers CPAP_MfeI_Fwd (5′-CCATCTAAAAcaattgTAGATGATGC-3′) and BamHI_CPAP_DS_Rev (5′-GATTggatccACATTCGTTTAAATGGGAATTGAATC-3′), then cloned into pBlueScript:SAS4Δ3′ using MfeI and BamHI restriction enzymes to create pBlueScript:SAS4. The final construct is 7,659 bp and contains the entire SAS4 genomic locus plus 817 bp upstream and 714 bp downstream of the coding region. This provides ∼800 bp overlap upstream and ∼600 bp overlap downstream with the knockout construct such that there is sufficient overlap for homologous repair of the locus.
The plasmid for N-terminal fusion of mCherry to Sas4 (p4T2-1 Nterm mCherry:Sas4) was created by PCR amplification of a 646-bp region upstream of the SAS4 initiation codon (5′-GCggatccGCAGAACTATTTATTTTCCAAAATAAATTAAC-3′ and 5′-GCgagctcTAATTTAGAACATATTTATATAAAATAAATAAGATAGAAAATAAATAGTAG-3′) and a 945-bp region downstream of the SAS4 initiation codon (5′-GCgaattcATGGGAGATTAGTCTTAGTCTTAAGG-3′ and 5′-GCggtaccAAGTTACTGCTATTATTTCCAGTAATTTAAGG-3′). These fragments were inserted into a modified p4T2-1 vector using standard molecular cloning techniques. The modified vector targets the endogenous locus but inserts the MTT promoter followed by mCherry between the cloned genomic regions. After transformation, this construct disrupts the endogenous SAS4 promoter and drives expression through MTT promoter control once assorted to completion.
Tetrahymena strain production
Whole genome SAS4 knockout cells were produced using the genomic exclusion method (Hai and Gorovsky, 1997). Briefly, micronuclei of mating CU428 and B2086 cells were transformed using biolistic particle bombardment with linearized p4T2-1:SAS4Δ. Homologous recombination at the SAS4 locus removed the entire SAS4 gene and replaced the intervening sequence with a neomycin resistance cassette, conferring paromomycin resistance. Proper integration of the construct was verified by PCR (Fig. S3 A, forward primer: 5′-GTTTAAGTTTTGATCCTTTCTGAACC-3′, reverse primer 1: 5′-GCCTCGAGTCAAGTACCTTAAGACTAAGACTAATCTCCC-3′, reverse primer 2: 5′-GATTAATTACCTTCTAATAATTTGAAATAATTAATCC-3′). Cells that were transformed in the micronucleus were identified by paromomycin and 6-methyl purine resistance (inherited from the micronucleus of CU428). The progeny of this cross were grown vegetatively for approximately 4 wk to select for the loss of macronuclear chromosomes harboring paromomycin resistance and to bring cells to sexual maturity. These micronuclear heterozygous heterokaryon cells were then mated to a star strain (B* VI) to homozygose the sas4Δ allele in the micronucleus, producing homozygous heterokaryons, where the micronucleus is homozygous for the deletion and the macronucleus harbors WT SAS4. Homozygous heterokaryons were backcrossed to SB210 cells, and the process was repeated to produce a second set of homozygous heterokaryons, mating to B* VI and B* VII in the star strain crosses. These homozygous heterokaryon cells were then mated to induce whole-genome knockout of SAS4 in both nuclei of all progeny cells.
Macronuclear transformation was performed as previously described using DNA-coated particle bombardment (Bruns and Cassidy-Hanley, 1999). Transformed clones were selected using 100 µg/ml paromomycin to select for the NEO2 gene or 7.5 µg/ml cycloheximide to select for the CHX gene. To increase the copy number of the endogenously tagged constructs, cells were assorted using increasing concentrations of the appropriate drug.
Cell lines created in this work expressing fluorescent fusion proteins include Sas4:mCherry (in B2086 background), Sas4:HaloTag (in SB1969 background), mCherry:Sas4 (in SB1969 background), GFP:Mob1 (in SB1969, SB210, sas4Δ heterokaryons, bld10Δ heterokaryons, and disA-1 backgrounds), GFP:Mob1 + Sas4:HaloTag (SB1969 background and sas4Δ heterokaryons), GFP:Mob1 + Poc1:mCherry (in sas4Δ heterokaryons), and TTHERM_00194700:HaloTag (in SB1969 background).
RT-PCR
RNA was isolated from WT or sas4Δ cells using Trizol extraction followed by DNase treatment according to the manufacturer’s instructions (Invitrogen). cDNA was made using oligo dT primers in a first-strand synthesis reaction. PCR was performed from cDNA using primers within the coding region of SAS4 (5′-GCgaattcATGCACATTTTCTGAGTTTTAATCAGG-3′ and 5′-GCggtaccTTCTGCTCATAAAAACGATAAAAAAAAATTGG-3′), ACT1 (actin, 5′-GTTGAACAGAGAAAAGATGA-3′ and 5′-GAAGGTAAGTTCGTGGATAC-3′), or the entire MOB1 open reading frame (described above).
Cell mating and SAS4 knockout
Cells of different mating types were grown to 400,000 cells/ml in SPP and then starved in Dryl’s medium for 24 h at 30°C. Cells were mixed at 200,000 cells/ml and incubated at 30°C. Mating efficiency was assessed at 4 h post-mixing, and cells were refed with an equal volume of 2× SPP and treated with 200 µg/ml paromomycin at 17 h post-mixing to select for successful sas4Δ mating. The time of refeeding and drug addition is considered 0 h in all experiments. Cells were counted, fixed, and observed by brightfield microscopy at each of the indicated time points.
Analysis of dividing cells was performed by manually counting cells by brightfield microscopy (Fig. 3 C and Fig. 4 H). At least 100 cells per condition were counted. Cells were identified as dividing when the division furrow was visible. Generally, this underestimates the number of dividing cells since furrow ingression occurs later in the process of cell division. The experiment was performed in triplicate. For analysis of nuclear loss or gain (Fig. 3, E and F), nuclei were detected by Hoechst stain, and 35 cells were counted for each cell line per replicate. The experiment was performed in triplicate.
To visualize GFP:Mob1 in WT, sas4Δ, and bld10Δ cells, the appropriate heterokaryon cell lines (Tetrahymena strain production) were induced for GFP:Mob1 expression in mating cells at 4 h post-mixing by the addition of 0.5 µg/ml CdCl2. CdCl2 was washed out of the media at the time of SPP and drug addition (17 h post-mixing). GFP:Mob1 localization was assessed by widefield fluorescence microscopy. At least 100 cells total were counted per cell line at each time point. The fraction of each cell line’s population with localized Mob1 was normalized to its respective 0-h time point population to correct for differences in initial GFP:Mob1 induction.
Widefield fluorescence and differential interference contrast (DIC) microscopy
Fluorescence and DIC images were acquired using a Nikon Ti Eclipse inverted microscope with a Nikon 100× Plan-Apo objective, NA 1.4, at 23°C. Images were captured with a CMOS camera (Xyla 4.2, Andor Technology) using NIS Elements imaging software and analyzed using ImageJ image analysis software (National Institutes of Health; https://imagej.net). All images were acquired with exposure times between 50 and 500 ms, depending on the experiment and the channel of acquisition.
Immunofluorescence
Cells were washed in 10 mM Tris, pH 7.4, and then fixed in 3.2% paraformaldehyde and 0.25% Triton X-100 in PHEM buffer (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 10 min at 23°C. Cells were washed twice in 0.1% BSA-PBS, and either dried on a coverslip at 23°C or maintained in Eppendorf tubes for the remainder of the staining protocol. Blocking was performed in 1% BSA-PBS for 20 min, followed by incubation in primary antibody diluted in 1% BSA-PBS for 1 h at 23°C. Primary antibodies used in this study were mouse anti-Kl Ag (1:100; Williams et al., 1990), rabbit anti-TtCen1 (1:2,000; Stemm-Wolf et al., 2005), mouse anti-α-tubulin (1:200, 12G10, Developmental Studies Hybridoma Bank, University of Iowa; Wloga et al., 2010), and rabbit anti-TtSas6A (1:250; Culver et al., 2009). Cells were then washed twice with 0.1% BSA-PBS before incubation with secondary antibodies diluted in 1% BSA-PBS. Secondary antibodies used in this study were IgG derived from goat and conjugated to Alexa Fluor 488, 594, or 647 (Invitrogen) and diluted to 1:1,000. Hoechst 33342 DNA dye (Sigma-Aldrich) was used at 1:1,000. Cells were mounted in either Citifluor (Ted Pella) or Prolong Gold (Molecular Probes) mounting media. Coverslips were sealed with clear nail polish.
Sas4 incorporation dynamics analysis
Cells expressing Sas4:HaloTag were grown to mid-log phase at 30°C, then labeled with 100 µM JF549-HaloTag Ligand for 30 min at 37°C. Cells were returned to 30°C for 1 h. Cells were fixed as described for immunofluorescence and stained with Kl-Ag and centrin antibodies, and imaged. Maximum projected images were generated from four slices of 0.3-µm step size image volumes, using slices closest to the coverslip. Only mother–daughter BB pairs or “families” (one mother with multiple daughters) from the medial half of the cell were analyzed. Mothers were identified by the presence of Kl-Ag encircling a single centrin focus. Daughters were identified as centrin foci anterior to a mother. A 0.65 µm2 region of interest (ROI) was drawn over the Sas4 foci for each BB, and two 0.65-µm2 ROIs were placed adjacent to the initial ROI for local background subtraction. Sas4 intensity was measured as the mean intensity in the ROI minus the average of the mean intensities of the background ROIs. Distance between BBs was determined by calculating the distance between the BB (centrin) peak intensities using a line scan through the mother–daughter BB pair. Each mother–daughter BB pair was binned into 0.25-µm separation distances, and average Sas4 (JF549) intensity was obtained for each bin. SEM was calculated for each bin. Average intensities in each bin were then normalized by setting the average intensity in the 0.0 µm bin to 1.0, and the same normalization factor was applied to the SEM. The normalized intensities for each of three replicates were averaged and plotted, and error bars represent the average normalized SEM for each replicate. To ensure the results were not obscured by HaloTag labeling accessibility, the experiment was also performed with cells expressing Sas4:mCherry, using the native fluorescence from the mCherry protein for visualization (i.e., not using antibodies against mCherry, Fig. S2 E).
SIM and image averaging
SIM imaging was performed on the Nikon N-SIM system on a Ti2 inverted microscope equipped with a 100× CF160 Apochromat superresolution/TIRF NA 1.49 objective with correction collar (Nikon Instruments). Images were captured using a sCMOS camera (ORCA-Flash4.0, Hamamatsu). All images were collected at 25°C using NIS Elements software (Nikon). Raw SIM images were reconstructed using the image slice reconstruction algorithm (NIS Elements) with illumination modulation contrast: 0.0; high resolution noise suppression: 1.33; and out of focus blur suppression: 0.01. Reconstructed images were analyzed using ImageJ. ImageJ code for image alignment and averaging was kindly provided by A. Soh (University of Colorado Anschutz Medical Campus, Aurora, CO) and N. Galati (Western Washington University, Bellingham, WA; Galati et al., 2016). Briefly, maximum projected images of three 100-nm slices were created. Individual BBs were selected based on the BB-localized Sas6A signal, and the tip of the SF was selected based on the SF-localized Sas6A antibody signal (Culver et al., 2009). New images were created for each of the selected BBs with the centroid BB signal as the center position of the image and oriented with the tip of the SF at the top center. Each of the centered and oriented images was combined into a single stacked image and then Z-projected as an average to create the averaged image. The average image shown in Fig. 1 C was generated from 530 BBs.
Image averaging for Mob1 signal (Fig. S3, G and H) was performed using the same ImageJ code as above, except widefield images with 300-nm slices were used for analysis. In addition, BB selection was performed using Poc1:mCherry signal, and images were oriented by selecting the adjacent anterior BB. Only BBs in the posterior half of the cell were used for analysis. The average image shown in Fig. S3 G was generated from 43 BBs. To merge the averaged image of Poc1:mCherry and GFP:Mob1 with that of Sas6A and Sas4:HaloTag, the number of pixels from the average image of Poc1:mCherry and GFP:Mob1 was doubled with no interpolation using the Adjust>Size function in ImageJ. This is necessary because the pixel size in the SIM images is half the size of the widefield image pixel size (32.5 nm2 versus 65 nm2). These new images were then merged with the Sas6A and Sas4:HaloTag averaged image, using the centroid of Poc1:mCherry signal and the centroid of BB-localized Sas6A signal to align the images.
Immuno-electron microscopy
Cells expressing Sas4:mCherry were grown to mid-log and collected by centrifugation, then processed using high-pressure freezing and freeze substitution as previously described (Meehl et al., 2016; Pearson et al., 2009). Samples were sectioned at 70-nm thickness. Sas4:mCherry was localized by incubation with rabbit anti-mCherry primary antibody (gift from I. Cheesman, Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA) diluted to 1:500 followed by anti-rabbit secondary antibody conjugated to 15 nm gold particles. Grids were counterstained with uranyl acetate and lead citrate. Images were obtained on a Technai G2 electron microscope (FEI). Quantification was performed by classifying the location of gold particles on longitudinal and cross-section images. 136 gold particles in longitudinal section images and 42 gold particles in cross-section images were used in the analysis.
FRAP data collection and analysis
Cells were induced to express mCherry:Sas4 by treatment with 0.5 µg/ml CdCl2 for 2 h at 30°C. Live cells were immobilized by sandwiching between a microscope slide and coverslip. Individual BBs labeled with mCherry:Sas4 were photobleached with a 30-ms pulse of 561-nm laser light, and z-stacks were collected at 10-s intervals for a maximum of 330 s. On average, 93% of the total mCherry:Sas4 signal was photobleached to allow the same BB to be tracked over time with the remaining signal and to ensure that the laser did not ablate the BB structure.
Fluorescence measurements were performed on maximum projection images using 0.72-µm2 ROIs drawn around the photobleached spot and around a nearby unbleached mCherry:Sas4-labeled BB to serve as a control to correct for the photobleaching that occurs during the course of image acquisition. Four pairs of bleached and unbleached BBs were used in the analysis. Each pair of BBs analyzed was from a different cell. Background fluorescence was calculated using two equal-sized ROIs positioned adjacent to the bleached and unbleached BBs. Total integrated density was measured for all ROIs, and the average of the background ROIs was subtracted from the bleached and unbleached ROIs at each time point. These signals were then normalized such that the first time point collected after bleaching (10 s) was set to 0 for the bleached spot, and to 1 for the unbleached spot, binned into 40-s intervals, and averaged across four replicates. These data were then normalized such that the control signal is equal to 1 throughout the time course.
Sas4 protein loss quantification
sas4Δ heterokaryons expressing Sas4:HaloTag from the parental macronucleus were labeled with JaneliaFluor549 conjugated to Halo Ligand at the beginning of mating (T = −17 h). Dye was removed by washing three times in SPP at T = 0 h. Cells were fixed as described above and imaged using the same imaging parameters on the same day. Image slices encompassing the entire cell volume were sum-projected, and ROIs were drawn around whole cells using the freehand tool in ImageJ. Background fluorescence was calculated using two ROIs positioned adjacent to the cell being analyzed. Total integrated density was measured for all ROIs. These values were then divided by the corresponding ROI area and the number of slices used to create the sum projection image. This resulted in an integrated density per cubic pixel measurement. Data in Fig. S3 C were normalized to the Sas4 protein level at the beginning of mating. Protein levels were calculated from 6 cells (−17 h), 16 cells (0 h), 13 cells (6 h), and 10 cells (12 h).
Mob1 gradient analysis
Cells expressing Poc1:mCherry (Pearson et al., 2009) and GFP:Mob1 at 0 h, 6 h, 12 h, and 24 h post-knockout were imaged live and analyzed to determine the posterior enrichment of Mob1 signal, which we refer to as a “gradient quotient,” as a measure of the Mob1 gradient. 0.72 µm2 ROIs were drawn around four posterior BBs and four anterior BBs per cell, determined by Poc1:mCherry localization, with one equally sized ROI placed adjacent to each BB ROI to measure background. Mean intensities in the background ROIs were subtracted from the maximum pixel intensities in the BB ROIs to obtain a signal for each BB. The four posterior signals and the four anterior signals were each averaged, and the gradient quotient was determined as the average posterior signal divided by the total (anterior + posterior) signal − 0.5. This results in values theoretically ranging from +0.5 to −0.5, where a positive value indicates a posterior enriched gradient of Mob1, a negative value represents an anterior enriched gradient of Mob1, and a 0 value represents a uniform distribution of Mob1 signal throughout the cell (no gradient). Signals were determined using the BB maximum pixel intensity minus the background mean intensity to avoid obtaining negative values when the BB signal was nearly identical to the background signal (i.e., no Mob1 localization). This method overestimates the signal of BBs with low or no Mob1 localization and decreases the calculated gradient quotient of Mob1 signal. This is represented by WT cells having an average gradient quotient of +0.32 (and not a value closer to +0.5), even though the gradient is obvious.
Phylogenetic analysis
Phylogenetic analysis was performed using the Phylogeny.fr website in one-click mode (Dereeper et al., 2008, 2010). We thank S. Santini (CNRS/AMU IGS UMR7256) for maintenance of the website. Alignments were made using MUSCLE and curated using Gblocks v0.91b. Phylogeny was prepared by PhyML, and the trees were rendered using TreeDyn (Edgar, 2004; Castresana, 2000; Guindon and Gascuel, 2003; Anisimova and Gascuel, 2006; Chevenet et al., 2006).
Statistical analyses
All experimental datasets represent a minimum of three biological replicates. The total number of cells and structures analyzed for each dataset is described in the figure legends or in the Materials and methods. Statistical tests were performed in Prism8 (GraphPad Software). Normally distributed datasets were analyzed using Student’s t test. Non-normally distributed datasets were analyzed using the Mann–Whitney test. All P values are reported in the figures. All error bars denote SEM unless otherwise noted. All bars in bar graphs represent mean values.
Online supplemental material
Fig. S1 provides an analysis of putative SAS4 genes in T. thermophila. Fig. S2 shows additional localizations of Sas4 protein and Sas4 dynamics at BBs by FRAP. Fig. S3 describes the SAS4 knockout strategy, additional sas4Δ phenotypes, and Mob1 localization at WT BBs and in knockout cells.
Acknowledgments
The authors thank Adam Soh and Nick Galati for image averaging code. The authors would like to thank Garry Morgan and Courtney Ozzello for EM expertise. JF549-HaloTag ligand was a kind gift from the Luke Lavis laboratory at Janelia Farm Research Campus (Ashburn, VA). Anti-mCherry primary antibody was a kind gift from Iain Cheeseman (Whitehead Institute, Massachusetts Institute of Technolog). The authors would also like to thank the Tetrahymena Stock Center (Cornell University) for strains and information for culturing Tetrahymena.
The research was funded by National Institues of Health grant NIH-NIGMS R01GM099820, the American Cancer Society, the Linda Crnic Institute (C.G. Pearson), and National Institutes of Health grant NIH-NIGMS 5F32GM122239 (M.D. Ruehle).
The authors declare no competing financial interests.
Author contributions: M.C. Ruehle conceptualized and performed experiments, data acquisition, analysis, and interpretation. Alexander J. Stemm-Wolf performed molecular cloning. C.G. Pearson supervised and provided resources and funding. M.C. Ruehle wrote the manuscript; C.G. Pearson and Alexander J. Stemm-Wolf reviewed and edited the manuscript.
References
- Allen R.D. 1969. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 40:716–733. 10.1083/jcb.40.3.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R.D., and Wolf R.W.. 1974. The cytoproct of Paramecium caudatum: structure and function, microtubules, and fate of food vacuole membranes. J. Cell Sci. 14:611–631. [DOI] [PubMed] [Google Scholar]
- Anisimova M., and Gascuel O.. 2006. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 55:539–552. 10.1080/10635150600755453 [DOI] [PubMed] [Google Scholar]
- Balestra F.R., von Tobel L., and Gönczy P.. 2015. Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res. 25:642–644. 10.1038/cr.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banterle N., and Gönczy P.. 2017. Centriole Biogenesis: From Identifying the Characters to Understanding the Plot. Annu. Rev. Cell Dev. Biol. 33:23–49. 10.1146/annurev-cellbio-100616-060454 [DOI] [PubMed] [Google Scholar]
- Bayless B.A., Giddings T.H. Jr., Winey M., and Pearson C.G.. 2012. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol. Biol. Cell. 23:4820–4832. 10.1091/mbc.e12-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless B.A., Galati D.F., and Pearson C.G.. 2016. Tetrahymena basal bodies. Cilia. 5:1 10.1186/s13630-016-0022-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolgioni A.F., and Ganem N.J.. 2016. The interplay between centrosomes and the Hippo tumor suppressor pathway. Chromosome Res. 24:93–104. 10.1007/s10577-015-9502-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B.R. 1985. Microtubule organizing centers. Annu. Rev. Cell Biol. 1:145–172. 10.1146/annurev.cb.01.110185.001045 [DOI] [PubMed] [Google Scholar]
- Bruns P.J., and Cassidy-Hanley D.. 1999. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62:501–512. 10.1016/S0091-679X(08)61553-8 [DOI] [PubMed] [Google Scholar]
- Bui D.A., Lee W., White A.E., Harper J.W., Schackmann R.C.J., Overholtzer M., Selfors L.M., and Brugge J.S.. 2016. Cytokinesis involves a nontranscriptional function of the Hippo pathway effector YAP. Sci. Signal. 9:ra23 10.1126/scisignal.aaa9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I.W., Zhou X., and Amon A.. 2020. Spindle pole bodies function as signal amplifiers in the Mitotic Exit Network. Mol. Biol. Cell. 906–916. doi:. 10.1091/mbc.E19-10-0584 [DOI] [PMC free article] [PubMed]
- Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B., and Bettencourt-Dias M.. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 194:165–175. 10.1083/jcb.201011152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A.-L., Jacq B., and Christen R.. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 7:439 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottee M.A., Muschalik N., Wong Y.L., Johnson C.M., Johnson S., Andreeva A., Oegema K., Lea S.M., Raff J.W., and van Breugel M.. 2013. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. eLife. 2 e01071 10.7554/eLife.01071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver B.P., Meehl J.B., Giddings T.H. Jr., and Winey M.. 2009. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol. Biol. Cell. 20:1865–1877. 10.1091/mbc.e08-08-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server). W465-9 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Audic S., Claverie J.-M., and Blanc G.. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8 10.1186/1471-2148-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen E.R. 1971. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 51:286–302. 10.1083/jcb.51.1.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger C.J. 1965. The fine structure of the nuclei of Tetrahymena pyriformis throughout the cell cycle. J. Cell Biol. 27:519–529. 10.1083/jcb.27.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florindo C., Perdigão J., Fesquet D., Schiebel E., Pines J., and Tavares A.A.. 2012. Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. J. Cell Sci. 125:3085–3090. 10.1242/jcs.097147 [DOI] [PubMed] [Google Scholar]
- Frankel J. 1979. An analysis of cell-surface patterning in Tetrahymena. Determinants of Spatial Organization, Symp. Soc. Dev. Biol. 37:215–246. 10.1016/B978-0-12-612983-0.50017-1 [DOI] [Google Scholar]
- Frankel J. 2008. What do genic mutations tell us about the structural patterning of a complex single-celled organism? Eukaryot. Cell. 7:1617–1639. 10.1128/EC.00161-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., and Kapler G.. 1999. Transient and stable DNA transformation of Tetrahymena thermophila by electroporation. Methods Cell Biol. 62:485–500. 10.1016/S0091-679X(08)61552-6 [DOI] [PubMed] [Google Scholar]
- Galati D.F., Bonney S., Kronenberg Z., Clarissa C., Yandell M., Elde N.C., Jerka-Dziadosz M., Giddings T.H., Frankel J., and Pearson C.G.. 2014. DisAp-dependent striated fiber elongation is required to organize ciliary arrays. J. Cell Biol. 207:705–715. 10.1083/jcb.201409123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galati D.F., Abuin D.S., Tauber G.A., Pham A.T., and Pearson C.G.. 2016. Automated image analysis reveals the dynamic 3-dimensional organization of multi-ciliary arrays. Biol. Open. 5:20–31. 10.1242/bio.014951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., and Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hai B., and Gorovsky M.A.. 1997. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 94:1310–1315. 10.1073/pnas.94.4.1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimo L.T., and Rosenbaum J.L.. 1981. Cilia, flagella, and microtubules. J. Cell Biol. 91:125s–130s. 10.1083/jcb.91.3.125s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., and Hemmings B.A.. 2012. Hippo signalling in the G2/M cell cycle phase: lessons learned from the yeast MEN and SIN pathways. Semin. Cell Dev. Biol. 23:794–802. 10.1016/j.semcdb.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A., Kohler R.S., Schmitz D., Vichalkovski A., Cornils H., and Hemmings B.A.. 2009. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 19:1692–1702. 10.1016/j.cub.2009.09.020 [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M. 1981. Cytoskeleton-related structures in tetrahymena thermophila: microfilaments at the apical and division-furrow rings. J. Cell Sci. 51:241–253. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M., Jenkins L.M., Nelsen E.M., Williams N.E., Jaeckel-Williams R., and Frankel J.. 1995. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol. 169:644–661. 10.1006/dbio.1995.1176 [DOI] [PubMed] [Google Scholar]
- Jiang Y.-Y., Maier W., Baumeister R., Minevich G., Joachimiak E., Ruan Z., Kannan N., Clarke D., Frankel J., and Gaertig J.. 2017. The Hippo Pathway Maintains the Equatorial Division Plane in the Ciliate Tetrahymena. Genetics. 206:873–888. 10.1534/genetics.117.200766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-Y., Maier W., Baumeister R., Joachimiak E., Ruan Z., Kannan N., Clarke D., Louka P., Guha M., Frankel J., et al. 2019. Two Antagonistic Hippo Signaling Circuits Set the Division Plane at the Medial Position in the Ciliate Tetrahymena. Genetics. 211:651–663. 10.1534/genetics.118.301889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A.D., Soh A.W.J., O’Toole E.T., Meehl J.B., Guha M., Winey M., Honts J.E., Gaertig J., and Pearson C.G.. 2019. Microtubule glycylation promotes attachment of basal bodies to the cell cortex. J. Cell Sci. 132 jcs233726 10.1242/jcs.233726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczanowski A. 1978. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J. Exp. Zool. 204:417–430. 10.1002/jez.1402040313 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., and Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Mardin B.R., Lange C., Baxter J.E., Hardy T., Scholz S.R., Fry A.M., and Schiebel E.. 2010. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12:1166–1176. 10.1038/ncb2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D.W., Allis C.D., and Bruns P.J.. 1982. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp. Cell Res. 140:227–236. 10.1016/0014-4827(82)90172-0 [DOI] [PubMed] [Google Scholar]
- Matsuda A., Shieh A.W.-Y., Chalker D.L., and Forney J.D.. 2010. The conjugation-specific Die5 protein is required for development of the somatic nucleus in both Paramecium and Tetrahymena. Eukaryot. Cell. 9:1087–1099. 10.1128/EC.00379-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson J.P., Tamblyn L., Elia A., Migon E., Shehabeldin A., Matysiak-Zablocki E., Lemmers B., Salmena L., Hakem A., Fish J., et al. 2004. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 23:3677–3688. 10.1038/sj.emboj.7600371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl J.B., Bayless B.A., Giddings T.H. Jr., Pearson C.G., and Winey M.. 2016. Tetrahymena Poc1 ensures proper intertriplet microtubule linkages to maintain basal body integrity. Mol. Biol. Cell. 27:2394–2403. 10.1091/mbc.e16-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J.R., and Irvine K.D.. 2018. The Hippo Signaling Network and Its Biological Functions. Annu. Rev. Genet. 52:65–87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T., Hirota T., Iida S., Marumoto T., Hara T., Nishiyama Y., Kawasuzi M., Hiraoka T., Mimori T., Araki N., et al. 2002. WARTS tumor suppressor is phosphorylated by Cdc2/cyclin B at spindle poles during mitosis. FEBS Lett. 529:319–324. 10.1016/S0014-5793(02)03360-4 [DOI] [PubMed] [Google Scholar]
- Nanney D.L. 1975. Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 65:503–512. 10.1083/jcb.65.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney D.L., Chen S.S., and Meyer E.B.. 1978. Scalar constraints in Tetrahymena evolution. Quantitative basal body variations within and between species. J. Cell Biol. 79:727–736. 10.1083/jcb.79.3.727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y., Hirota T., Morisaki T., Hara T., Marumoto T., Iida S., Makino K., Yamamoto H., Hiraoka T., Kitamura N., et al. 1999. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 459:159–165. 10.1016/S0014-5793(99)01224-7 [DOI] [PubMed] [Google Scholar]
- Organ A.E., Bovee E.C., and Jahn T.L.. 1972. The mechanism of the water expulsion vesicle of the ciliate Tetrahymena pyriformis. J. Cell Biol. 55:644–652. 10.1083/jcb.55.3.644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pala R., Alomari N., and Nauli S.M.. 2017. Primary Cilium-Dependent Signaling Mechanisms. Int. J. Mol. Sci. 18:2272 10.3390/ijms18112272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Osborn D.P.S., Giddings T.H. Jr., Beales P.L., and Winey M.. 2009. Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 187:905–920. 10.1083/jcb.200908019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Song X., Bowen J., Corstanje R., Gao Y., Gaertig J., and Gorovsky M.A.. 2002. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA. 99:3734–3739. 10.1073/pnas.052016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Aher A., Dynes N.J., Frey D., Katrukha E.A., Jaussi R., Grigoriev I., Croisier M., Kammerer R.A., Akhmanova A., et al. 2016. Centriolar CPAP/SAS-4 Imparts Slow Processive Microtubule Growth. Dev. Cell. 37:362–376. 10.1016/j.devcel.2016.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanis V. 2015. Pombe’s thirteen - control of fission yeast cell division by the septation initiation network. J. Cell Sci. 128:1465–1474. 10.1242/jcs.094821 [DOI] [PubMed] [Google Scholar]
- Slabodnick M.M., Ruby J.G., Dunn J.G., Feldman J.L., DeRisi J.L., and Marshall W.F.. 2014. The kinase regulator mob1 acts as a patterning protein for stentor morphogenesis. PLoS Biol. 12 e1001861 10.1371/journal.pbio.1001861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A.J., Morgan G., Giddings T.H. Jr., White E.A., Marchione R., McDonald H.B., and Winey M.. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 16:3606–3619. 10.1091/mbc.e04-10-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares A., Gonçalves J., Florindo C., Tavares A.A., and Soares H.. 2012. Mob1: defining cell polarity for proper cell division. J. Cell Sci. 125:516–527. 10.1242/jcs.096610 [DOI] [PubMed] [Google Scholar]
- Wickstead B., and Gull K.. 2011. The evolution of the cytoskeleton. J. Cell Biol. 194:513–525. 10.1083/jcb.201102065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N.E., Honts J.E., and Kaczanowska J.. 1990. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 109:935–942. [DOI] [PubMed] [Google Scholar]
- Winey M., Stemm-Wolf A.J., Giddings T.H. Jr., and Pearson C.G.. 2012. Cytological analysis of Tetrahymena thermophila. Methods Cell Biol. 109:357–378. 10.1016/B978-0-12-385967-9.00013-X [DOI] [PubMed] [Google Scholar]
- Wloga D., Dave D., Meagley J., Rogowski K., Jerka-Dziadosz M., and Gaertig J.. 2010. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot. Cell. 9:184–193. 10.1128/EC.00176-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Gooi L.M., Wason A., Gabriel E., Mehrjardi N.Z., Yang Q., Zhang X., Debec A., Basiri M.L., Avidor-Reiss T., et al. 2014. Conserved TCP domain of Sas-4/CPAP is essential for pericentriolar material tethering during centrosome biogenesis. Proc. Natl. Acad. Sci. USA. 111:E354–E363. 10.1073/pnas.1317535111 [DOI] [PMC free article] [PubMed] [Google Scholar]