Abstract
It is crucial to classify cervical lesions into high-grade squamous intraepithelial lesions (HSILs) and low-grade SILs (LSILs), as LSILs are conservatively treated by observation, based on an expectation of natural regression, whereas HSILs usually require electrosurgical excision. In the present study, peripheral blood gene expression profiles were analyzed to identify transcriptomic biomarkers distinguishing HSILs from LSILs. A total of 102 blood samples were collected from women with cervical SILs (66 HSIL and 36 LSIL) for microarray hybridization. Candidate gene signatures were identified using AdaBoost algorithms, and a predictive model was constructed using logistic regression to differentiate HSILs from LSILs. To correct for possible bias as a result of the limited sample size and to verify the stability of the predictive model, a two-fold cross validation and null set analysis was conducted over 1,000 iterations. The functions of the transcriptomic biomarkers were then analyzed to elucidate the pathogenesis of cervical SIL. A total of 10 transcriptomic genes (STMN3, TRPC4AP, DYRK2, AGK, KIAA0319L, GRPEL1, ZFC3H1, LYL1, ITGB1 and ARHGAP18) were identified. The predictive model based on the 10-gene panel exhibited well-discriminated power. A cross validation process using known disease status exhibited almost the same performance as that of the predictive model, whereas null-set analysis with randomly reassigned disease status exhibited much lower predictive performance for distinguishing HSILs from LSILs. These biomarkers were involved in the ‘Rho GTPase cycle’, ‘mitochondrial protein import’, ‘oncogenic MAPK signaling’, ‘integrin cell surface interaction’ and ‘signaling by BRAF and RAF fusions’. In conclusion, peripheral blood gene expression analysis is a promising method for distinguishing HSILs from LSILs. The present study proposes 10 candidate genes that could be used in the future as diagnostic biomarkers and potential therapeutic targets for cervical SILs. A simple, non-invasive blood test would be clinically useful in the diagnosis and classification of patients with cervical SILs.
Keywords: squamous intraepithelial lesion, cervical cancer, peripheral blood, transcriptome, biomarkers
Introduction
Precancerous cervical lesions are defined as localized, identifiable cervical lesions that carry an increased risk of developing into cancer, that are treatable and that can be eradicated to prevent the occurrence of cervical cancer. Large population-based screening programs for precursor lesions have been shown to be highly effective in the prevention of cervical cancer (1,2).
Over the past 100 years, however, the definition of ‘precancerous cervical lesions’ has been vague and variable, from carcinoma in situ (CIS) dysplasia to cervical intraepithelial neoplasia (CIN) (3). Furthermore, according to its potential to develop into cervical cancer, CIN can be divided into grades corresponding to mild, moderate or severe dysplasia and CIS (4,5).
In order to better understand the biology and epidemiology of cervical cancer and to improve the consistency of cervical biopsies, a two-tiered system of nomenclature for squamous intraepithelial lesion (SIL) was developed to replace the three-grade CIN classification (6–10). In this system, low-grade SILs (LSILs) equate to CIN1, and high-grade SILs (HSILs) equate to CIN3 and to most CIN2 lesions. p16-immunohistochemistry (IHC) was used to further classify CIN2 lesions into LSILs (p16-negative) and HSILs (p16-positive) (11–13). Since most LSILs are expected to regress naturally within ~2 years and are conservatively treated by observation, whereas HSILs usually require excision by electrosurgery, it is important to distinguish LSILs from HSILs (6).
Currently the diagnosis of HSILs and LSILs is based on histopathology. In total, 60% of LSILs will regress naturally without treatment, and the lesions are characterized by features that include hyperplasia of squamous epithelial basement and subbasal cells, mild nuclear disorder and mild atypia (6). LSILs are typically limited to the first one-third of the subepithelial layer and show an absence of p16 staining or positive scattered dots in the epithelium. HSILs, which may develop into infiltrating carcinoma, present with nuclear polarity disorder, an increased proportion of nucleoplasm (higher nuclear-to-cytoplasmic ratio) and increased cellular mitosis (6). Atypical cells extend to the subepithelial two-thirds of the epithelium or even throughout the whole epithelial layer with continuously positive p16 staining (6,14). SIL classification requires tissue biopsy, which is uncomfortable and invasive, and causes patients to be less compliant. Furthermore, it is difficult to distinguish human papilloma virus (HPV) infection from unequivocal LSIL, and HPV infection alone is increasingly being included in the category of LSIL by cytopathologists (15). Although HPV testing and the thinprep cytologic test (TCT) has improved the diagnosis of SIL (16), the final diagnosis still relies mainly on the histopathology. A non-invasive and simplified strategy to discriminate between HSILs and LSILs would greatly facilitate the diagnosis and treatment of these lesions.
A blood-based method to diagnose cancer by detecting circulating tumor cells or tumor DNA in peripheral blood has been proposed as a simpler, non-invasive strategy for cancer detection (17). This method, however, is not suitable for detecting precancerous lesions or cancer at early stages, as tumor-derived molecules released from a tiny cancer focus would rarely be detectable or would be undetectable. By contrast, we previously reported a novel ‘liquid biopsy’ (18) for cancer, using peripheral blood transcriptomic biomarkers in the diagnosis of various non-hematological disorders. As a blood-mRNA based rather than tumor dependent diagnostic, this technique is especially useful for detection in the early and pre-cancerous disease stages (19–22).
The present study uses peripheral blood transcriptome profiling to identify candidate genes for distinguishing HSILs from LSILs. The genes identified in this study may be clinically useful as the basis of a new blood test for the diagnosis of SILs, and may further promote our understanding of the pathophysiology of SILs and cervical cancer.
Materials and methods
Ethics
The present study was approved by the Ethics Committee of the Qingdao Women and Children's Hospital (Qingdao, China; approval no. QFELL-KY-2019-46). Sample acquisition for HSILs and LSILs was conducted between July 2019 and October 2019 at the Qingdao Women and Children's Hospital. All 102 participants, including 66 patients with HSILs and 36 with LSILs, were enrolled and provided written informed consent. The inclusion criteria were as follows: i) Ages from 20–65 years; and ii) HPV infection lasting longer than 6 months. The exclusion criteria were: i) Having autoimmune disease or immunodeficiency disease; ii) having cervical cancer or other malignancy; iii) pregnancy; and iv) having taken drugs affecting immune function within the previous 6 months. The age distribution of the patients is listed in Table SI. In order to verify the effectiveness of this method both for identifying transcriptomic biomarkers for LSIL and HSIL and for differentiating SIL from healthy populations, blood samples were also taken from 65 healthy female volunteers without cervical disease collected between July 2019 and October 2019 at the Qingdao Women and Children's Hospital. All healthy volunteers were recruited from patients who underwent a health examination in the hospital and who provided written informed consent.
Study population
A total of 102 blood samples were collected from the women before undergoing cervical tissue biopsy and before they had undergone any form of treatment, including hormone therapy, radio/chemo-therapy or surgery. SIL was categorized based on surgical pathological examination. LSIL was characterized as the proliferation of basal-like cells extending no more than one-third of the epithelial thickness and with normal mitoses. HSIL was characterized as the proliferative cell compartment extending into the middle one-third or the superficial one-third of the epithelium and with abnormal mitoses. All patients underwent HPV testing and TCT before cervical tissue biopsy.
Blood collection, RNA isolation and RNA quality control
Peripheral whole blood (2.5 ml) was collected in PaxGene Blood RNA tubes (PreAnalytix GmbH) and total RNA was isolated using an accessory PaxGene Blood RNA kit (PreAnalytix GmbH) according to the manufacturer's instructions. The isolated RNA quality was accessed using Agilent 2100 Bioanalyzer RNA 6000 Nano Chips (Agilent Technologies, Inc.). All the samples for microarray analysis met the following quality criteria: RNA integrity number ≥7.0 and 28S:18S rRNA ≥1.0. RNA quantity was determined by a NanoDrop 1000 UV–Vis spectrophotometer (Thermo Fisher Scientific, Inc.).
HPV test
A single cervical specimen was collected from each participant using a Rovers Cervex-Brush device (Rovers Medical Devices B.V.), and cells were suspended into BD SurePath collection vials containing preservative solution (Becton, Dickinson and Company), according to the manufacturer's instructions. HPV types, including 7 high-risk types 16/44/45/52/53/58/61, were detected with an Aptima HPV assay targeting E6/E7 mRNA (Aptima; Hologic, Inc.) according to the protocol previously described (23,24). The Aptima Auto Detect kit (Aptima; Hologic Inc.) was used to test these specimens with the Panther Fusion system (Hologic, Inc.), according to the manufacturer's instructions.
TCT test
Exfoliated cervical cells were collected from the ectocervix and endocervix with Rovers Cervex-Brush device (Rovers Medical Devices B.V.) and were analyzed using TCT tests according to the Bethesda classification system 2009 (25).
Microarray hybridization
Whole blood RNA from the 102 samples (66 HSILs and 36 LSILs) was analyzed using Gene Profiling Array cGMP U133 P2 microarray (Affymetrix; Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. Gene expression signal intensity was processed using Affymetrix Expression Console software (version 1.4.1; Affymetrix; Thermo Fisher Scientific, Inc.) and normalized by MAS5 normalization, in which the global signal intensity value was adjusted to 500 for each microarray to make it possible to compare profiling variations between microarrays.
Microarray data mining
To identify candidate genes that distinguish HSILs from LSILs, the entire 54,675 microarray probe sets were treated according to the following criteria: i) Only the probe sets reliably detected as ‘present’ in all of the samples were retained; and ii) the probe sets were included within the MicroArray Quality Control (MAQC) list from MAQC Consortium (26). The intensity values of gene expression signals were then log transformed to conform to a Gaussian distribution.
To select gene expression signatures, the ensemble learning strategy AdaBoost method was employed (27). This data mining method does not make restrictive assumptions on the training set, unlike traditional methods. Instead, AdaBoost creates a series of weak classifiers and then combines them into a single strong classifier by assigning each weak classifier its proper weight. In this way, AdaBoost outperforms existing methods in accuracy and training time (27). In the present study, the final 10 genes were selected using the AdaBoost method and then used to construct the predictive model via a logistic regression algorithm. To evaluate the performance of the predictive model for classifying the HSIL and LSIL groups, the model was characterized by the area under the receiver operating characteristic curve (ROC AUC), the sensitivity, the specificity and the accuracy.
As the sample size in this study was small, it was not practical to evaluate the performance of the predictive model by partitioning the entire group of samples into traditional training and test sets. Instead, as in our previously reported study, a 2-fold cross validation process was conducted to avoid data overfitting (28). Half of the LSIL and HSIL samples were randomly selected as a training fold to generate the predictive model; the remainder of the samples were included into a test fold, for prediction over 1,000 iterations.
An additional problem is that the final 10 genes identified may, due to clinical bias and limited sample size, derive from random chance. To avoid this problem, a null set analysis was performed to verify that the results were not due to chance and to reduce the bias owing to the limited number of HSIL and LSIL samples. The cross validation process was tested once again; the model was used to predict the sample cohorts with disease status randomly reassigned (null set). The distributions of diagnostic parameters of sensitivity, specificity, accuracy and ROC AUC were then compared between the two cross validation processes.
Bioinformatics analysis
Gene Ontology (GO) annotations of the candidate biomarkers were queried from the COXPRESdb v7 database (http://coxpresdb.jp) (29). The proteins interacting with the candidate biomarkers were downloaded from the STRING database (https://string-db.org/) with total confidence ≥0.7. Reactome (https://reactome.org/) pathway enrichment analysis using the clusterProfiler R package (version 3.16.0) (30) was performed on signature genes and their correlative proteins. Reactome pathways were identified with a strict cut-off of adjusted P<0.05, corrected with the Benjamini-Hochberg method and with a false-discovery rate (FDR) of <0.05. The visualization of the protein-protein interaction network was performed with Cytoscape software (https://cytoscape.org/; version 3.8.0).
Results
Basic and clinicopathological characteristics of patients with HSILs and LSILs
In the present study, a total of 102 samples were collected, including 66 HSIL and 36 LSIL samples. The mean ages of the two groups were not statistically significantly different (two-tailed Student's t-test; P=0.0642). Most patients were distributed between the ages of 31 and 40 years (Table SI).
HPV test information for all patients was summarized, as shown in Table SII. In the HSIL group, HPV16 was the most prevalent HPV type and accounted for more than half of all HSIL patients (37/66, 56.1%). HPV16 also predominated in the LSIL group, but its HPV16-positive rate was only one-fifth that found in the LSIL group (8/36, 22.2%). Except for HPV16, other single HPV subtypes comprised <8% of the total.
Peripheral blood gene expression profiling
Gene expression profiling was performed for peripheral blood samples taken from the 66 patients with HSILs and 36 patients with LSILs. Genome-wide expression profiles generated with Affymetrix Gene Profiling Array cGMP U133 P2 microarray were analyzed and correlated as between HSIL and LSIL. Finally, 10 candidate genes were identified as distinguishing HSILs from LSILs: STMN3, TRPC4AP, DYRK2, AGK, KIAA0319L, GRPEL1, ZFC3H1, LYL1, ITGB1 and ARHGAP18. The corresponding gene symbols and names of the final 10 probe sets are listed in Table I, as well as the fold-change between the two cohorts.
Table I.
Final 10 candidate genes for distinguishing HSILs from LSILs.
| Probe set | Gene symbol | Gene name | Fold-change |
|---|---|---|---|
| 222557_at | STMN3 | Stathmin-like 3 | 1.23 |
| 212059_s_at | TRPC4AP | Transient receptor potential cation channel, subfamily C, member 4 associated protein | 1.19 |
| 202968_s_at | DYRK2 | Dual specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | 1.09 |
| 222132_s_at | AGK | Acylglycerol kinase | 1.09 |
| 222468_at | KIAA0319L | KIAA0319-like | 1.08 |
| 212434_at | GRPEL1 | GrpE-like 1, mitochondrial | 1.08 |
| 213065_at | ZFC3H1 | Zinc finger, C3H1-type containing | 1.04 |
| 210044_s_at | LYL1 | Lymphoblastic leukemia-associated hematopoiesis regulator 1 | −1.09 |
| 1553530_a_at | ITGB1 | Integrin β1 | −1.23 |
| 225173_at | ARHGAP18 | Rho GTPase activating protein 18 | −1.23 |
HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade SIL.
Model construction and performance estimation
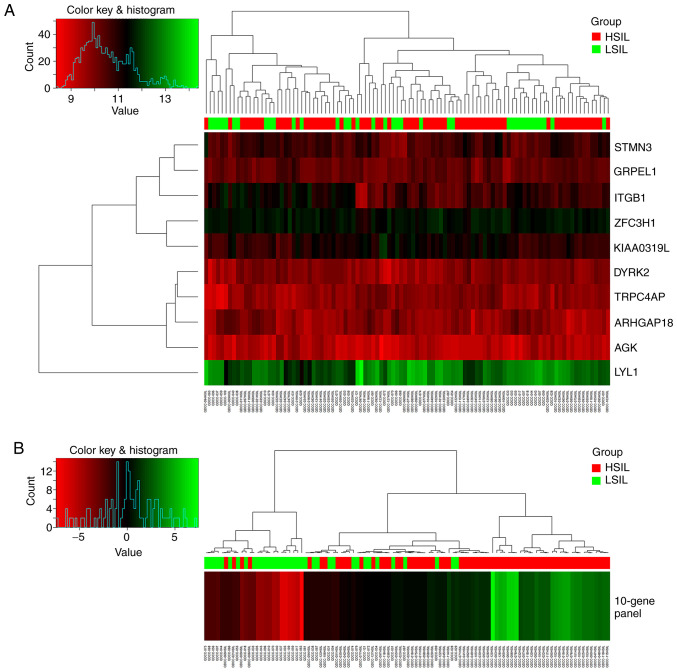
Based on the 10-gene panel identified, a predictive model was constructed for discriminating HSILs from LSILs using logistic regression. Fig. 1 shows the performance of the 10 candidate genes separately and the 10-gene panel as a whole for discriminating HSILs from LSILs for the total 102 samples. Use of the 10-gene panel showed better clustering of the HSIL samples.
Figure 1.
Heat map of gene expression and hierarchical cluster diagram showing the performance of 10 candidate genes and the 10-gene panel for clustering 102 samples in the training set, including 66 HSILs and 36 LSILs. (A) 10 separate candidate genes and (B) the 10-gene panel. The figure was generated using the ‘Heatmap’ function in R with default settings. HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade SIL.
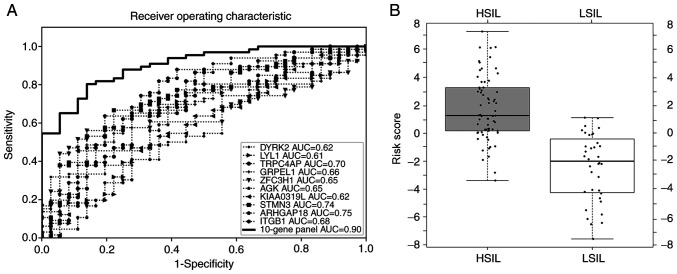
As the sample size in the present study was too small to be divided into a training set and a test set, the entire data set was used to construct the model. It was found that a 10-gene panel could discriminate HSILs from LSILs (sensitivity, 81.8%; specificity, 83.3%; accuracy, 82.4%). The 10-gene panel exhibited the highest ROC AUC of 0.90 as compared with that of each of 10 candidate genes (0.61–0.75) separately, as shown in Fig. 2A for the total 102 samples. The box-whisker plot (Fig. 2B) also illustrated a well-separated distribution of prediction scores of HSIL and the LSIL, based on the 10-gene panel and logistic regression algorithm.
Figure 2.
Model evaluation with ROC curve and box-whisker plot. (A) ROC curve for candidate genes of the training set. (B) Box-whisker plot to display the logistic regression scores in HSILs and LSILs in the training set. ROC, receiver operating characteristic; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade SIL; AUC, area under curve.
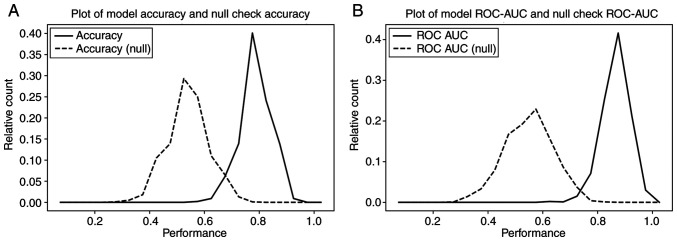
In order to validate the stability of the model, a 2-fold cross-validation method was performed, iterated 1,000 times. For each iteration, the samples were randomly and equally divided into training and test folds. The predictive model was constructed based on the samples in the training fold and then its performance was evaluated on the sample in the test fold. The distributions of accuracy and AUC ROC values over the 1,000 iterations exhibited good prediction performance, with a model mean accuracy of 82% (Fig. 3A) and ROC AUC of 0.9 (Fig. 3B) (solid line). To rule out the possibility that the final 10-gene panel was merely derived from chance and clinical bias, a null set analysis process was performed for 1,000 iterations, similar to the aforementioned 2-fold cross-validation process, but with HSIL/LSIL status randomly re-assigned. Comparing the prediction results with the known HSIL/LSIL status of the samples, it was found that the null set analysis results were significantly lower, with a model accuracy of 52% (Fig. 3A) and ROC AUC of 0.58 (Fig. 3B) (dashed line). The distribution of each analysis resulted in 2 well-separated curves with <5% overlap, from which it was concluded that the performance of the 10-gene panel for distinguishing HSILs from LSILs was unlikely to be merely the result of random chance.
Figure 3.
Two-fold cross-validation results over 1,000 iterations based on the known HSIL/LSIL disease status (true set, solid line) and re-assigned disease status (null set, dashed line) of the samples. This chart displays the distributions of (A) accuracy and (B) AUC ROC values over the 1,000 iterations, in which the model accuracy and ROC in the null set analysis were much lower than that in the known HSIL/LSIL status analysis. ROC, receiver operating characteristic; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade SIL; AUC, area under curve.
To further validate efficiency, this data mining method was performed in an analysis of blood-based gene expression profiles of women with cervical SILs and healthy volunteers. Transcriptomic biomarkers were identified and were found to exhibit good performance for differentiating cervical SIL patients from healthy volunteers. Detailed information is presented in Fig. S1 and Table SIII.
Protein-protein interaction and functional enrichment categorization
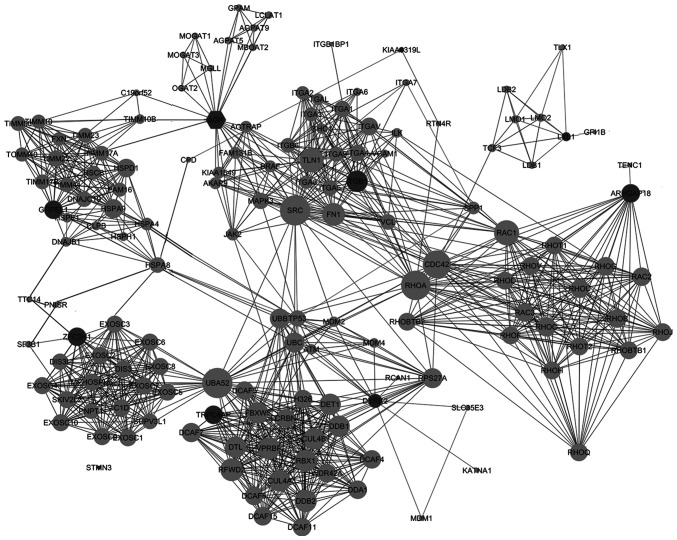
Proteins interacting among the 10 candidate genes were downloaded from the STRING database individually, with a total confidence ≥0.7, followed by the protein-protein interactions of the total 149 proteins thus identified. The protein-protein interaction network is shown in Fig. 4.
Figure 4.
Protein-protein interaction network of the 10 candidate genes for high-grade squamous intraepithelial lesions. Interaction map of 10 transcriptomic biomarkers (dark circles) and their interacting proteins (light circles), with total confidence ≥0.7.
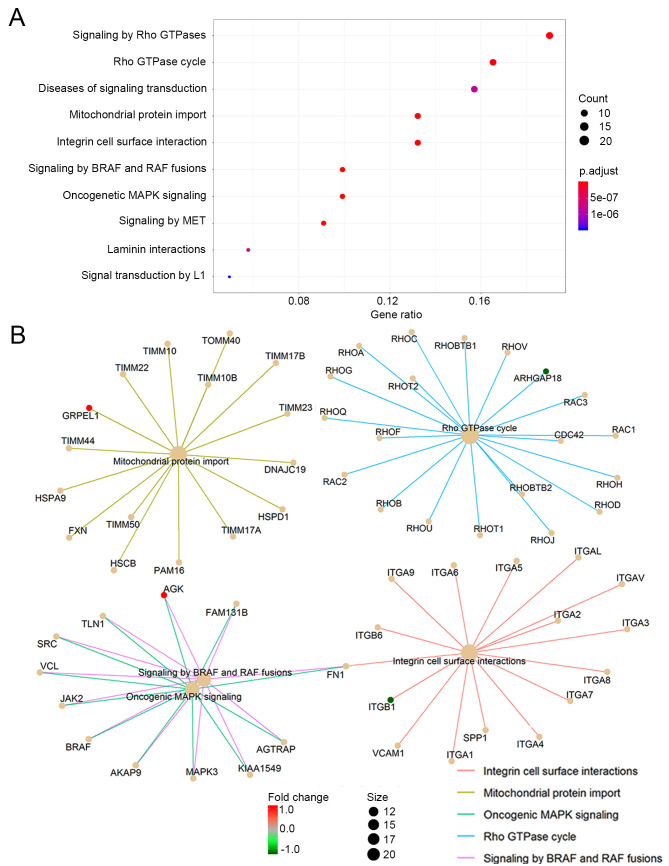
The genes corresponding to the selected 149 proteins were functionally categorized based on Reactome pathways. Reactome pathways were identified with a strict cut-off of P<0.05 and FDR of <0.05. When the top 10 significantly enriched pathways were analyzed, it was found that the 10 candidate genes and their interactive proteins were mainly engaged in the ‘Rho GTPase cycle’, ‘mitochondrial protein import’, ‘oncogenic MAPK signaling’, ‘integrin cell surface interaction’ and ‘signaling by BRAF and RAF fusions’, as shown in Fig. 5A. Since the Rho GTPase proteins act as binary switches by cycling between active GTP-bound and inactive GDP-bound conformations (Rho GTPase cycle) (31), the ‘signaling by Rho GTPase’ pathway mainly reflects the ‘Rho GTPase cycle’ and thus could be aligning to the pathway of ‘Rho GTPase cycle’ in the present study. The interactions of the engaged processes and the related candidate genes of each process are also indicated in Fig. 5B.
Figure 5.
Reactome pathway analysis of the 10 candidate genes. (A) The top 10 significantly enriched Reactome pathways. (B) The top 5 significantly enriched Reactome pathways and their associated genes (red, upregulated candidate genes; green, downregulated candidate genes; light orange, genes corresponding to the proteins interacting with the candidate genes).
Discussion
The present study was undertaken to explore a blood transcriptomic profiling method for distinguishing cervical HSILs from LSILs. Peripheral blood samples of patients with cervical HSILs and LSILs were collected, and blood transcriptomic features were identified using the AdaBoost algorithm. A panel of 10 candidate genes that successfully discriminated cervical HSILs from LSILs, with an accuracy of 82.4%, was identified. Functional enrichment analysis indicated that the candidate genes were mainly involved in the ‘Rho GTPase cycle’, ‘mitochondrial protein import’, ‘oncogenic MAPK signaling’, ‘integrin cell surface interaction’ and ‘signaling by BRAF and RAF fusions’. These preliminary results are promising; however, further research is needed to validate the findings in larger cohorts and in a multi-site validation clinical study.
Most types of cervical cancer are caused by HPV infections, and HPV is detected in ~99.7% of cervical cancer cases (32). Not all HPV infections, however, lead to cervical cancer. Most of these infections are temporary, resulting only in LSILs; it is only persistent HPV infection that may result in an HSIL (33). The comparative risks of progression to cervical cancer differ sharply between HSILs and LSILs (34). From a clinical perspective, the management of the two types of lesion is also different, with HSILs regarded as true cervical cancer precursor lesions, requiring colposcopy and treatment (35). Currently, the diagnosis of HSIL is mainly dependent on colposcopy and biopsy, but neither test is completely satisfactory. A non-invasive test would be welcome to distinguish HSILs from LSILs during the premalignant stage and to prevent cervical cancer.
In current clinical practice, the tools used for cervical cancer screening and diagnosis include HPV testing, cytology testing, colposcopy and cervical biopsy. Although combinations of different techniques have improved the screening and diagnosis of cervical cancer, the sensitivity, specificity, reliability and repeatability are still unsatisfactory due to sampling errors and/or screening errors, and the final diagnosis is still dependent on cervical biopsy (36). Thus, the development of a sensitive, non-invasive approach for the screening and diagnosis of HSILs and LSILs is essential to complement existing techniques.
The present study was based on our previous blood transcriptome profiling study (37) and on our previous reports in cancer research (19,21,22,37–39). In the present study, a 10-gene panel (Table I) was identified that can successfully distinguish HSILs from LSILs. A predictive model performed well in the training set (Figs. 1 and 2), with a sensitivity of 81.8%, specificity of 83.3% and accuracy of 82.4% (Table II). The 10 candidate genes (10-gene panel) used to construct the predictive model were: STMN3, TRPC4AP, DYRK2, AGK, KIAA0319L, GRPEL1, ZFC3H1, LYL1, ITGB1 and ARHGAP18. The last 3 genes (LYL1, ITGB1 and ARHGAP18) were downregulated in HSILs as compared with LSILs, while the other 7 genes were upregulated.
Table II.
Predictive performance of the training set.
| Performance | HSILs, n | LSILs, n | Sensitivity, % | Specificity, % | Accuracy, % | ROC AUC |
|---|---|---|---|---|---|---|
| Positive prediction | 54 | 6 | ||||
| Negative prediction | 12 | 30 | ||||
| Total | 66 | 36 | 81.8 | 83.3 | 82.4 | 0.9 |
HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade SIL; ROC, receiver operating characteristic; AUC, area under the curve.
To verify that this method is also efficient for identifying blood-based transcriptomic biomarkers suitable for discriminating HSILs/LSILs from healthy populations, the blood gene expression profiles of samples from healthy women and samples from women with cervical SIL were analyzed. It was found that the biomarkers identified by the present data mining method were able to differentiate SILs from healthy samples with relatively high accuracy, similar to the HSIL and LSIL classification results. Detailed information is provided in Fig. S1 and Table SIII. This result also confirmed the feasibility of applying blood-based transcriptomic biomarkers for SIL and cervical cancer screening.
To further explore the function of the 10-gene panel, the protein-protein interaction network was mapped and a total of 149 proteins were identified interacting with the candidate genes. The genes corresponding to the 149 interactive proteins were then functionally categorized into several biological processes, including the ‘Rho GTPase cycle’, ‘mitochondrial protein import’, ‘oncogenic MAPK signaling’, ‘integrin cell surface interaction’ and ‘signaling by BRAF and RAF fusions’, as shown in Fig. 5A. Of these, GRPEL1 is involved in ‘mitochondrial protein import’, ARHGAP18 in ‘Rho GTPase cycling’, ITGB1 in ‘integrin cell surface interaction’, and AGK in ‘oncogenic MAPK signaling’ and ‘BRAF and RAF fusions’, as indicated in Fig. 5B. These findings suggest that these processes may be associated with the progression of LSILs to HSILs.
Mitochondrial protein import is essential for maintaining cellular homeostasis. During this process, the mitochondria and nucleus co-ordinately communicate with each other, and the adaptive responses to stress depend on modulation of mitochondrial import (40). Dysfunction of this process plays a role in human disorders such as mitochondrial neuropathies, myopathies and cancer (41–43). It has been reported that cervical cancer HeLa cells can revert from an apoptotic state even after the induction of widespread mitochondrial-membrane permeabilization, and that mitochondrial protein import plays a role in the anastatic process of HeLa cells (44). Since most mitochondrial proteins are encoded in the nucleus and are imported into the matrix compartment where they are properly folded, the process is facilitated by mitochondrial heat shock protein 70 (mtHsp70) (45) GrpE-like 1 (GrpEL1), as a putative nucleotide exchange factors ortholog, acts as a hetero-oligomeric subcomplex to associate with mtHsp70 and regulate mtHsp70 function (46). Therefore, the GrpEL1-related mitochondrial protein import process may be a potential therapeutic target for cervical cancer and precancerous lesions.
The best characterized members of the Rho GTPase family, RhoA, Rac1 and Cdc42 (47), are associated with abnormalities in Rho GTPase function that have major consequences for tumorigenesis (48), cancer progression (49) and cancer immune suppression (50). The Rho GTPase member, Rac1, is expressed in the nucleus of epithelial cells in SIL and cervical cancer cell lines, and inhibition of Rac1 can decrease cellular proliferation, suggesting that Rho-GTPases may have a role in cervical cancer progression (51). One study demonstrated that stimulation of G12 proteins is capable of promoting cervical cancer invasion through RhoA/ROCK-JNK activation (52). RhoA has also shown a positive correlation with the progression and metastatic potential of cervical cancer, both in vivo and in vitro (53). ARHGAP18 is one of the crucial factors for the regulation of RhoA, whose knockdown and overexpression in HeLa cells in a previous study resulted in the inhibition and promotion of cell migration, respectively. Furthermore, it was also required for the polarization of cells for migration (54). Since ARHGAP18 was the most downregulated candidate gene in the present study, we speculate that this gene may be a biomarker associated with a good prognosis in cervical cancer, as has been reported in breast cancer (55).
Integrins belong to the family of heterodimeric cell surface receptors and trigger various cellular responses by forming physical crosstalk between the inside and outside of cells, and these receptors can bi-directionally control the signals for cell adhesion, migration, proliferation, survival and differentiation (56). Due to their crucial role in physiological functions, integrins also play a pivotal role in tumorigenesis (57). Integrins are heterodimers that include 8β and 18α subunits. ITGB1 protein, namely the integrin β1, which is a member of the β sub-family, forms dimers with different α subunits (α1–7 and αV) 55(t.). ITGB1 protein together with another integrin, ITGA6 protein, was found to be downregulated in Pap-cell smears of high-risk human papillomavirus-positive squamous cervical carcinoma in a previous study (58). The present results were consistent with this, as the gene expression of ITGB1 was also decreased in HSILs compared with that in LSILs. There was also a contradictory study, which suggested that ITGB1 was negatively mediated by miR-183-5p in cervical cancer cells; since miR-183-5p serves as a latent anti-oncogene, the investigators regarded ITGB1 as a metastasis-promoter gene (59). The precise role of ITGB1 in SILs and cervical cancer thus requires further research clarification.
Oncogenic MAPK signalling, as well as BRAF and RAF fusions with AGK, were the other biological processes identified in the present study. AGK protein, namely acylglycerol kinase, is a novel lipid kinase, which produces lysophosphatidic acid from monoacylglycerol (60). AGK is a cancer-related gene that is overexpressed in various human cancer types (61–64). Sun et al found that AGK protein and mRNA expression was significantly upregulated in cervical cancer cell lines and cancer tissues and demonstrated that the AGK protein expression level was an independent prognostic factor for the survival of patients with early-stage cervical squamous cell cancer (65). In the present study, it was also found that AGK was upregulated in HSILs, which suggested that AGK plays a role in the progression of LSILs to HSILs, and can be used as a prognostic marker for SIL and cervical cancer evolution.
On the journey of cancer as it progresses from precursor lesions towards full-scale malignancy, gene expression undergoes changes. Those genetic events in turn establish an environment permissive for neoplastic progression (66). We propose that the downregulation of ITGB1 and ARHGAP18 in women with HSILs leads to the dysfunction of cell adhesion and the promotion of cell migration. Dysfunction of cell adhesion then allows cells to migrate and to be more invasive, which occurs during the transformation of LSILs into HSILs. We also suggest that the upregulation of AGK and GRPEL1 in women with HSILs may disrupt the process of protein localization and transport (67,68), processes that establish and maintain proteins during oncogenesis. The present findings thus indicate that protein transport and cell migration may play important roles in the progression of LSILs to HSILs.
Conceptual progress for the hallmarks of cancer has been made over the past decade. In the present study, the downregulated genes, ITGB1 and ARHGAP18, were found to be involved in the integrin cell surface interaction and the Rho GTPase cycle pathways. Integrin, as one of the family of heterodimeric cell surface receptors, can mediate cell adhesion to the extracellular matrix, which is a network of macro-molecules. Integrins can bi-directionally control the signals for cell adhesion, migration, proliferation, survival and differentiation (56). The activation of Rho GTPases can trigger changes in the organization of the cytoskeleton, thereby regulating cell polarity and cell-cell junctions (47). RhoA has also shown a positive correlation with the progression and metastatic potential of cervical cancer (53). In brief, these two enriched pathways identified in the present study are associated with cell invasion and metastasis, two important hallmarks of cancer (69).
The upregulated genes, AGK and GRPEL1, are involved in the oncogenic MAPK signaling and mitochondrial protein import pathways. Accumulating evidence shows that the RAS/RAF/MAPK cascade is a critical pathway for cancer cell proliferation, differentiation and survival (70). Mitochondrial protein import is essential for maintaining cellular homeostasis (71). GRPEL1 acts as a hetero-oligomeric subcomplex to interact with mtHsp70, facilitating the process in which mitochondrial proteins are encoded and imported into the matrix (45), and regulating mtHsp70 function (46). In brief, these two enriched pathways identified in the present study are associated with selective growth and proliferative advantage, which are hallmarks of cancer (69,72).
A question arises as to any possible correlations that may be identified between blood gene expression and specific HPV types. In the present study, HPV16 was the most prevalent HPV type and accounted for more than half of all HSIL patients (37/66, 56.1%). A limitation of this study, however, is that only 20 samples in the HSIL cohort presented with other types of HPV, a number inadequate for statistically significant correlation analysis between blood gene expression and specific HPV type. This issue will be studied in future in larger cohorts.
Another notable future project would be to study variations in blood-based gene expression profiling between women with HSIL and women with cervical cancer. Such a study would be of clinical benefit for diagnosing cervical cancer.
In conclusion, the present study identified a panel of 10 blood transcriptomic biomarkers that discriminate cervical HSILs from LSILs, and which could form the basis of a novel blood test for SIL classification. Gene function investigation has indicated that these genes are mainly engaged in the ‘Rho GTPase cycle’, ‘mitochondrial protein import’, ‘oncogenic MAPK signaling’, ‘integrin cell surface interaction’ and ‘signaling by BRAF and RAF fusions’. We propose that these biological processes play important roles in the progression of LSILs to HSILs. We also suggest that these processes may be associated with cell invasion and metastasis, selective growth and proliferative advantage, which are important hallmarks of cancer. Moreover, the 10 candidate genes identified in this study (STMN3, TRPC4AP, DYRK2, AGK, KIAA0319L, GRPEL1, ZFC3H1, LYL1, ITGB1 and ARHGAP18) may participate in events leading to precancerous cervical lesions and require additional investigation. The genes identified here may also be used as diagnostic biomarkers and targeted therapy for cervical SILs.
Supplementary Material
Acknowledgements
The authors would like to thank Ms. Qian Shi, Shanghai Homeostasis Bio-Technology Inc., who conducted the Affymetrix microarray experiments and Ms. Isolde Prince, Shanghai Homeostasis Bio-Technology Inc., who helped with the editing of the original manuscript.
Funding
This study was sponsored by Shanghai Homeostasis Bio-Technology Inc. (Shanghai, China).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CZ was responsible for conception, study design and acquisition of data. YL was responsible for conceptualization, study design and writing the original draft of the manuscript. YC, CS, HL and JJ were involved in acquisition and analysis of data. MW was responsible for analysis and interpretation of data. RZ was responsible for analysis and interpretation of data. CC was involved in conception and design of the study, analysis and interpretation of data, drafting of the initial manuscript, revising the manuscript for important intellectual content and providing approval for the final version of the manuscript to be published. CCL was responsible for conceptualization and designing of the study, resources and revising the manuscript for important intellectual content. SZ was responsible for conception and design of the study, acquisition and analysis of data and revising the manuscript for important intellectual content. All authors have read and approved the manuscript.
Ethics approval
This study was approved by the Ethics Committee of the Qingdao Women and Children's Hospital (institutional review board no. QFELL-KY-2019-46). Sample acquisition for HSILs and LSILs was conducted between July 2019 and October 2019 at the Qingdao Women and Children's Hospital. All 102 participants, including 66 HSIL and 36 LSIL patients, were enrolled and provided written informed consent. All 65 healthy volunteers also provided informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors wish to declare the following competing interests: CC, YL, ME and RZ are employees of Shanghai Homeostasis Bio-Technology Inc., who sponsored this research. CCL was the scientific consultant at Shanghai Homeostasis Bio-Technology Inc., and the chair and founder of Golden Health Diagnostics. The authors declare that they have no competing interests.
References
- 1.Godoy-Vitorino F, Romaguera J, Zhao C, Vargas-Robles D, Ortiz-Morales G, Vázquez-Sánchez F, Sanchez-Vázquez M, de la Garza-Casillas M, Martinez-Ferrer M, White JR, et al. Cervicovaginal fungi and bacteria associated with cervical intraepithelial neoplasia and high-risk human papillomavirus infections in a hispanic population. Front Microbiol. 2018;9:2533. doi: 10.3389/fmicb.2018.02533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. WHO guidelines for screening and treatment of precancerous lesions for cervical cancer prevention. 2013 [PubMed] [Google Scholar]
- 3.Reagan JW, Seidemann IL, Saracusa Y. The cellular morphology of carcinoma in situ and dysplasia or atypical hyperplasia of the uterine cervix. Cancer. 1953;6:224–234. doi: 10.1002/1097-0142(195303)6:2<224::AID-CNCR2820060203>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 4.Richart RM. A theory of cervical carcinogenesis. Obstet Gynecol Surv. 1969;24:874–879. doi: 10.1097/00006254-196907001-00017. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman SR, Le T, Lockhart A, Sanusi A, Dal Santo L, Davis M, McKinney DA, Brown M, Poole C, Willame C, Smith JS. Patterns of persistent HPV infection after treatment for cervical intraepithelial neoplasia (CIN): A systematic review. Int J Cancer. 2017;141:8–23. doi: 10.1002/ijc.30623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voltaggio L, Cimino-Mathews A, Bishop JA, Argani P, Cuda JD, Epstein JI, Hruban RH, Netto GJ, Stoler MH, Taube JM, et al. Current concepts in the diagnosis and pathobiology of intraepithelial neoplasia: A review by organ system. CA Cancer J Clin. 2016;66:408–436. doi: 10.3322/caac.21350. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen HN, Nordqvist SR. The Bethesda system and evaluation of abnormal pap smears. Semin Surg Oncol. 1999;16:217–221. doi: 10.1002/(SICI)1098-2388(199904/05)16:3<217::AID-SSU4>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Solomon D, Ritu N, editors. The bethesda system for reporting cervical cytology: Definitions, criteria, and explanatory notes. Comprehensive cytopathology. 2008;10:77–90. [Google Scholar]
- 9.Darragh TM, Colgan TJ, Thomas Cox J, Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH, et al. The lower anogenital squamous terminology standardization project for HPV-associated lesions: Background and consensus recommendations from the College of American pathologists and the American society for colposcopy and cervical pathology. Int J Gynecol Pathol. 2013;32:76–115. doi: 10.1097/PGP.0b013e31826916c7. [DOI] [PubMed] [Google Scholar]
- 10.Stoler MH, Ronnett BM, Joste NE, Hunt WC, Cuzick J, Wheeler CM, New Mexico HPV Pap Registry Steering Committee The interpretive variability of cervical biopsies and its relationship to HPV status. Am J Surg Pathol. 2015;39:729–36. doi: 10.1097/PAS.0000000000000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charoonwatana T, Boonlikit S, Yanaranop M. Progression of precancerous cervical lesion predicted by p16 protein immunohistochemistry in Rajavithi Hospital. Asian Pac J Cancer Prev. 2019;20:1809–1815. doi: 10.31557/APJCP.2019.20.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Z, Chen J. Introduction of WHO classification of tumours of female reproductive organs fourth edition. Zhonghua Bing Li Xue Za Zhi. 2014;43:649–650. (In Chinese) [PubMed] [Google Scholar]
- 13.Liu Y, Alqatari M, Sultan K, Ye F, Gao D, Sigel K, Zhang D, Kalir T. Using p16 immunohistochemistry to classify morphologic cervical intraepithelial neoplasia 2: Correlation of ambiguous staining patterns with HPV subtypes and clinical outcome. Hum Pathol. 2017;66:144–151. doi: 10.1016/j.humpath.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wentzensen N, Bergeron C, Cas F, Eschenbach D, Vinokurova S, von Knebel Doeberitz M. Evaluation of a nuclear score for p16INK4a-stained cervical squamous cells in liquid-based cytology samples. Cancer. 2005;105:461–467. doi: 10.1002/cncr.21378. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Sigel K, Gaisa MM. Human papillomavirus genotypes predict progression of anal low-grade squamous intraepithelial lesions. J Infect Dis. 2018;218:1746–1752. doi: 10.1093/infdis/jiy463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CA, James D, Marzan A, Armaos M. Cervical cancer: An overview of pathophysiology and management. Semin Oncol Nurs. 2019;35:166–174. doi: 10.1016/j.soncn.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 18.Liew CC. Method for detection of gene transcripts in blood and uses thereof. 2009 [Google Scholar]
- 19.Shi J, Cheng C, Ma J, Liew CC, Geng X. Gene expression signature for detection of gastric cancer in peripheral blood. Oncol Lett. 2018;15:9802–9810. doi: 10.3892/ol.2018.8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omar H, Lim CR, Chao S, Lee MM, Bong CW, Ooi EJ, Yu CG, Tan SS, Abu Hassan MR, Menon J, et al. Blood gene signature for early hepatocellular carcinoma detection in patients with chronic hepatitis B. J Clin Gastroenterol. 2015;49:150–157. doi: 10.1097/MCG.0000000000000112. [DOI] [PubMed] [Google Scholar]
- 21.Mok CS, Kim JH, Skates SJ, Schorge JO, Cramer DW, Lu KH, Liew CC. Use of blood-based mRNA profiling to identify biomarkers for ovarian cancer screening. Gynecol Obstet (Sunnyvale) 2017;7:443. doi: 10.4172/2161-0932.1000443. [DOI] [Google Scholar]
- 22.Marshall KW, Mohr S, Khettabi FE, Nossova N, Chao S, Bao W, Ma J, Li XJ, Liew CC. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer. 2010;126:1177–1186. doi: 10.1002/ijc.24910. [DOI] [PubMed] [Google Scholar]
- 23.Monsonego J, Hudgens MG, Zerat L, Zerat JC, Syrjӓnen K, Halfon P, Ruiz F, Smith JS. Evaluation of oncogenic human papillomavirus RNA and DNA tests with liquid-based cytology in primary cervical cancer screening: The FASE study. Int J Cancer. 2011;129:691–701. doi: 10.1002/ijc.25726. [DOI] [PubMed] [Google Scholar]
- 24.Ratnam S, Coutlee F, Fontaine D, Bentley J, Escott N, Ghatage P, Gadag V, Holloway G, Bartellas E, Kum N, et al. Aptima HPV E6/E7 mRNA test is as sensitive as hybrid capture 2 assay but more specific at detecting cervical precancer and cancer. J Clin Microbiol. 2011;49:557–564. doi: 10.1128/JCM.02147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayar R, Wilbur DC, editors. 3rd. Springer International Publishing; Cham, Switzerland: 2015. The Bethesda system for reporting cervical cytology. Definitions, criteria, and explanatory Notes. [Google Scholar]
- 26.MAQC Consortium, corp-author. Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, et al. The microarray quality control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi Z, Meng F, Tian Y, Niu L, Shi Y, Zhang P. Adaboost-LLP: A boosting method for learning with label proportions. IEEE Trans Neural Netw Learn Syst. 2018;29:3548–3559. doi: 10.1109/TNNLS.2017.2727065. [DOI] [PubMed] [Google Scholar]
- 28.Chao S, Cheng C, Liew CC. Mining the dynamic genome: A method for identifying multiple disease signatures using quantitative RNA expression analysis of a single blood sample. Microarrays (Basel) 2015;4:671–689. doi: 10.3390/microarrays4040671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Obayashi T, Kagaya Y, Aoki Y, Tadaka S, Kinoshita K. COXPRESdb v7: A gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019;47(D1):D55–D62. doi: 10.1093/nar/gky1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 32.McCormack PL. Quadrivalent human papillomavirus (types 6, 11, 16, 18) recombinant vaccine (gardasil(®)): A review of its use in the prevention of premalignant anogenital lesions, cervical and anal cancers, and genital warts. Drugs. 2014;74:1253–1283. doi: 10.1007/s40265-014-0255-z. [DOI] [PubMed] [Google Scholar]
- 33.Stanley MA, Sterling JC. Host responses to infection with human papillomavirus. Curr Probl Dermatol. 2014;45:58–74. doi: 10.1159/000355964. [DOI] [PubMed] [Google Scholar]
- 34.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14:395–405. doi: 10.1038/nrc3728. [DOI] [PubMed] [Google Scholar]
- 35.Bierkens M, Wilting SM, Wieringen WN, van de Wiel MA, Ylstra B, Meijer CJ, Snijders PJ, Steenbergen RD. HPV type-related chromosomal profiles in high-grade cervical intraepithelial neoplasia. BMC Cancer. 2012;12:36. doi: 10.1186/1471-2407-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karia N, Van Loon A, Simoens C, Benoy I, Bogers J. The positive predictive value of high-grade squamous intraepithelial lesion on cytology for the histological diagnosis of cervical intraepithelial neoplasia 2 or higher: A systematic review. Acta Cytol. 2019;63:206–214. doi: 10.1159/000497110. [DOI] [PubMed] [Google Scholar]
- 37.Mohr S, Liew CC. The peripheral-blood transcriptome: New insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Osman I, Bajorin DF, Sun TT, Zhong H, Douglas D, Scattergood J, Zheng R, Han M, Marshall KW, Liew CC. Novel blood biomarkers of human urinary bladder cancer. Clin Cancer Res. 2006;12:3374–3380. doi: 10.1158/1078-0432.CCR-05-2081. [DOI] [PubMed] [Google Scholar]
- 39.Chao S, Ying J, Liew G, Marshall W, Liew CC, Burakoff R. Blood RNA biomarker panel detects both left- and right-sided colorectal neoplasms: A case-control study. J Exp Clin Cancer Res. 2013;32:44. doi: 10.1186/1756-9966-32-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicolas E, Tricarico R, Savage M, Golemis EA, Hall MJ. Disease-associated genetic variation in human mitochondrial protein import. Am J Hum Genet. 2019;104:784–801. doi: 10.1016/j.ajhg.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra F, Guaragnella N, Arbini AA, Bucci C, Giannattasio S, Moro L. Mitochondrial dysfunction: A novel potential driver of epithelial-to-mesenchymal transition in cancer. Front Oncol. 2017;7:295. doi: 10.3389/fonc.2017.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raimundo N. Mitochondrial pathology: Stress signals from the energy factory. Trends Mol Med. 2014;20:282–292. doi: 10.1016/j.molmed.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Madamba SM, Damri KN, Dejean LM, Peixoto PM. Mitochondrial ion channels in cancer transformation. Front Oncol. 2015;5:120. doi: 10.3389/fonc.2015.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seervi M, Sumi S, Chandrasekharan A, Sharma AK, Santhosh Kumar TR. Molecular profiling of anastatic cancer cells: Potential role of the nuclear export pathway. Cell Oncol (Dordr) 2019;42:645–661. doi: 10.1007/s13402-019-00451-1. [DOI] [PubMed] [Google Scholar]
- 45.Neupert W, Brunner M. The protein import motor of mitochondria. Nat Rev Mol Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 46.Srivastava S, Savanur MA, Sinha D, Birje A, R V, Saha PP, D'Silva P. Regulation of mitochondrial protein import by the nucleotide exchange factors GrpEL1 and GrpEL2 in human cells. J Biol Chem. 2017;292:18075–18090. doi: 10.1074/jbc.M117.788463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796:91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Jansen S, Gosens R, Wieland T, Schmidt M. Paving the Rho in cancer metastasis: Rho GTPases and beyond. Pharmacol Ther. 2018;183:1–21. doi: 10.1016/j.pharmthera.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Chaker M, Minden A, Chen S, Weiss RH, Chini EN, Mahipal A, Azmi AS. Rho GTPase effectors and NAD metabolism in cancer immune suppression. Expert Opin Ther Targets. 2018;22:9–17. doi: 10.1080/14728222.2018.1413091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendoza-Catalán MA, Cristóbal-Mondragón GR, Adame-Gómez J, del Valle-Flores HN, Coppe JF, Sierra-López L, Romero-Hernández MA, del Carmen Alarcón-Romero L, Illades-Aguiar B, Castañeda-Saucedo E. Nuclear expression of Rac1 in cervical premalignant lesions and cervical cancer cells. BMC Cancer. 2012;12:116. doi: 10.1186/1471-2407-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan B, Cui J, Wang W, Deng K. Gα12/13 signaling promotes cervical cancer invasion through the RhoA/ROCK-JNK signaling axis. Biochem Biophys Res Commun. 2016;473:1240–1246. doi: 10.1016/j.bbrc.2016.04.048. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Chen D, Liu G. Overexpression of RhoA promotes the proliferation and migration of cervical cancer cells. Biosci Biotechnol Biochem. 2014;78:1895–1901. doi: 10.1080/09168451.2014.943650. [DOI] [PubMed] [Google Scholar]
- 54.Maeda M, Hasegawa H, Hyodo T, Ito S, Asano E, Yuang H, Funasaka K, Shimokata K, Hasegawa Y, Hamaguchi M, Senga T. ARHGAP18, a GTPase-activating protein for RhoA, controls cell shape, spreading, and motility. Mol Biol Cell. 2011;22:3840–3852. doi: 10.1091/mbc.e11-04-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aleskandarany MA, Sonbul S, Surridge R, Mukherjee A, Caldas C, Diez-Rodriguez M, Ashankyty I, Albrahim KI, Elmouna AM, Aneja R, et al. Rho-GTPase activating-protein 18: A biomarker associated with good prognosis in invasive breast cancer. Br J Cancer. 2017;117:1176–1184. doi: 10.1038/bjc.2017.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Desgrosellier JS, Cheresh DA. Integrins in cancer: Biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan B, Guo J, Liao Q, Zhao Y. β1 and β3 integrins in breast, prostate and pancreatic cancer: A novel implication. Oncol Lett. 2018;15:5412–5416. doi: 10.3892/ol.2018.8076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manavi M, Hudelist G, Fink-Retter A, Gschwandtler-Kaulich D, Pischinger K, Czerwenka K. Gene profiling in Pap-cell smears of high-risk human papillomavirus-positive squamous cervical carcinoma. Gynecol Oncol. 2007;105:418–426. doi: 10.1016/j.ygyno.2006.12.032. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W, Zhang M, Liu L, Jin D, Wang P, Hu J. MicroRNA-183-5p inhibits aggressiveness of cervical cancer cells by targeting integrin subunit beta 1 (ITGB1) Med Sci Monit. 2018;24:7137–7145. doi: 10.12659/MSM.910295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bektas M, Payne SG, Liu H, Goparaju S, Milstien S, Spiegel S. A novel acylglycerol kinase that produces lysophosphatidic acid modulates cross talk with EGFR in prostate cancer cells. J Cell Biol. 2005;169:801–811. doi: 10.1083/jcb.200407123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nouh MA, Wu XX, Okazoe H, Tsunemori H, Haba R, Abou-Zeid AM, Saleem MD, Inui M, Sugimoto M, Aoki J, Kakehi Y. Expression of autotaxin and acylglycerol kinase in prostate cancer: Association with cancer development and progression. Cancer Sci. 2009;100:1631–1638. doi: 10.1111/j.1349-7006.2009.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Lin C, Zhao X, Liu A, Zhu J, Li X, Song L. Acylglycerol kinase promotes cell proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Mol Cancer. 2014;13:106. doi: 10.1186/1476-4598-13-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Ying Z, Lin X, Lin H, Wu J, Li M, Song L. Acylglycerol kinase augments JAK2/STAT3 signaling in esophageal squamous cells. J Clin Invest. 2013;123:2576–2589. doi: 10.1172/JCI68143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Y, Lin C, Wu Z, Liu A, Zhang X, Zhu J, Wu G, Wu J, Li M, Li J, Song L. AGK enhances angiogenesis and inhibits apoptosis via activation of the NF-κB signaling pathway in hepatocellular carcinoma. Oncotarget. 2014;5:12057–12069. doi: 10.18632/oncotarget.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun F, Xiong Y, Zhou XH, Li Q, Xiao L, Long P, Li LJ, Cai MY, Wei YX, Ma YL, Yu YH. Acylglycerol kinase is over-expressed in early-stage cervical squamous cell cancer and predicts poor prognosis. Tumour Biol. 2016;37:6729–6736. doi: 10.1007/s13277-015-4498-4. [DOI] [PubMed] [Google Scholar]
- 66.Gius D, Funk MC, Chuang EY, Feng S, Huettner PC, Nguyen L, Bradbury CM, Mishra M, Gao S, Buttin BM, et al. Profiling microdissected epithelium and stroma to model genomic signatures for cervical carcinogenesis accommodating for covariates. Cancer Res. 2007;67:7113–7123. doi: 10.1158/0008-5472.CAN-07-0260. [DOI] [PubMed] [Google Scholar]
- 67.Bauer NC, Detsch PW, Corbett AH. Mechanisms regulating protein localization. Traffic. 2015;16:1039–1061. doi: 10.1111/tra.12310. [DOI] [PubMed] [Google Scholar]
- 68.Kang Y, Fielden LF, Stojanovski D. Mitochondrial protein transport in health and disease. Semin Cell Dev Biol. 2018;76:142–153. doi: 10.1016/j.semcdb.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 69.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 70.Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006;1:7–9. doi: 10.1016/S1556-0864(15)31506-9. [DOI] [PubMed] [Google Scholar]
- 71.Kotiadis VN, Duchen MR, Osellame LD. Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim Biophys Acta. 2014;1840:1254–1265. doi: 10.1016/j.bbagen.2013.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7:1016–1036. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.