To overcome host iron restriction, the human fungal pathogen Cryptococcus neoformans uses several iron uptake mechanisms to cause disease. In this study....
Keywords: ChIP-seq, Cryptococcus neoformans, iron, transcription factor, transcriptional network
Abstract
Iron is essential for the growth of the human fungal pathogen Cryptococcus neoformans within the vertebrate host, and iron sensing contributes to the elaboration of key virulence factors, including the formation of the polysaccharide capsule. C. neoformans employs sophisticated iron acquisition and utilization systems governed by the transcription factors Cir1 and HapX. However, the details of the transcriptional regulatory networks that are governed by these transcription factors and connections to virulence remain to be defined. Here, we used chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) and transcriptome analysis (RNA-seq) to identify genes directly regulated by Cir1 and/or HapX in response to iron availability. Overall, 40 and 100 genes were directly regulated by Cir1, and 171 and 12 genes were directly regulated by HapX, under iron-limited and replete conditions, respectively. More specifically, we found that Cir1 directly controls the expression of genes required for iron acquisition and metabolism, and indirectly governs capsule formation by regulating specific protein kinases, a regulatory connection not previously revealed. HapX regulates the genes responsible for iron-dependent pathways, particularly under iron-depleted conditions. By analyzing target genes directly bound by Cir1 and HapX, we predicted the binding motifs for the transcription factors and verified that the purified proteins bind these motifs in vitro. Furthermore, several direct target genes were coordinately and reciprocally regulated by Cir1 and HapX, suggesting that these transcription factors play conserved roles in the response to iron availability. In addition, biochemical analyses revealed that Cir1 and HapX are iron-containing proteins, implying that the regulatory networks of Cir1 and HapX may be influenced by the incorporation of iron into these proteins. Taken together, our identification of the genome-wide transcriptional networks provides a detailed understanding of the iron-related regulatory landscape, establishes a new connection between Cir1 and kinases that regulate capsule, and underpins genetic and biochemical analyses that reveal iron-sensing mechanisms for Cir1 and HapX in C. neoformans.
IRON is a key micronutrient that is essential for various cellular processes including mitochondrial respiration, DNA repair, and gene expression. The metal serves as an electron donor or acceptor and is required as a cofactor for a large number of enzymes. Excessive iron levels, however, are detrimental to cells because of the generation of toxic hydroxyl radicals via Haber-Weiss/Fenton chemistry, and this potential toxicity necessitates tight regulation of cellular iron homeostasis (Halliwell and Gutteridge 1984). Furthermore, iron plays an important role in the survival of pathogenic fungi within the vertebrate host environment, where iron availability is extremely limited. In particular, iron is sequestered and withheld from pathogens by iron-binding proteins, such as transferrin, lactoferrin, and ferritin in vertebrate hosts. This phenomenon is called nutritional immunity and inhibits the growth of invading pathogenic microbes (Wang and Cherayil 2009; Kehl-Fie and Skaar 2010; Ganz and Nemeth 2015). In response, pathogenic microbes have evolved sophisticated iron acquisition strategies regulated by iron-responsive transcription factors to adapt to environments in which iron is scarce.
The opportunistic fungal pathogen Cryptococcus neoformans is the causative agent of fungal meningoencephalitis in immunocompromised individuals, e.g., AIDS/HIV patients. C. neoformans is able to evade the host immune response via distinct virulence processes, including formation of a polysaccharide capsule, synthesis of the black pigment melanin, and the ability to grow at host body temperature (Casadevall and Perfect 1998). Interestingly, the elaboration of these virulence factors is closely linked to iron availability, indicating that iron homeostasis and pathogenesis are interconnected in C. neoformans (Jung et al. 2006; Liu and Nizet 2009; Kumar et al. 2011; Kronstad et al. 2012). To overcome iron sequestration within the host, C. neoformans utilizes sophisticated iron acquisition systems, including reductive iron transport, and the use of siderophore and heme uptake systems. Ferric reductases are also required for reductive iron transport. These enzymes reduce insoluble ferric iron to ferrous iron, which is subsequently oxidized by the ferroxidase Cfo1 and transported into the cell by the iron permease Cft1. Furthermore, the reductive iron uptake system (particularly, high-affinity reductive iron transport) is required for full virulence in a mouse model of cryptococcosis (Jung et al. 2008; Jung et al. 2009; Han et al. 2012; Saikia et al. 2014). Although C. neoformans does not possess the genes for siderophore biosynthesis, the fungus produces at least six siderophore transporters (Sit1–6) and can use exogenous siderophore-bound iron (Tangen et al. 2007). Furthermore, C. neoformans uses heme, which is the most abundant iron resource in the mammalian host, as a sole iron source and employs uptake mechanisms involving the putative hemophore Cig1, the ESCRT complexes containing Snf7, Vps20, Vps22, and Vps23, as well as clathrin-mediated endocytosis (Cadieux et al. 2013; Hu et al. 2013; Hu et al. 2015; Bairwa et al. 2019).Regulatory proteins that control iron uptake and homeostasis have been identified in various fungi. Examples include the GATA-type transcription factors Fep1, SreA, and Sfu1 in Schizosaccharomyces pombe, Aspergillus fumigatus, and Candida albicans, respectively. Additional regulators include the bZIP iron transcription factors, Php4, HapX, and Hap43, also known as the regulatory subunits of the CCAAT-binding complex (CBC) in S. pombe, A. fumigatus, and C. albicans, respectively (Pelletier et al. 2002; Mercier et al. 2006; Pelletier et al. 2007; Schrettl et al. 2008, 2010; Hsu et al. 2011). During iron starvation, the CBC negatively regulates the expression of genes involved in mitochondrial respiration, iron-sulfur cluster (ISC), and heme biosynthesis, as well as genes encoding iron-containing proteins, to reduce iron use (Mercier et al. 2006; Hortschansky et al. 2007; Schrettl et al. 2010). In addition, upon iron deprivation, the CBC represses the expression of the GATA-type iron transcription factor, which negatively regulates the expression of genes required for iron uptake under iron-replete conditions, and allows the activation of iron acquisition (Mercier et al. 2006; Schrettl et al. 2008). By contrast, under iron-replete conditions, the GATA-type iron transcription factors negatively regulate HapX, the regulatory subunit of CBC (Pelletier et al. 2003; Lan et al. 2004; Schrettl et al. 2008). Additional regulatory mechanisms that influence the activity of GATA-type iron transcription factors and the CBC have been proposed. For example, the Zn2Cys6 zinc knuckle transcription factor Sef1 in C. albicans participates in the regulatory network with Sfu1 and Hap43, and plays a role in activation of iron uptake systems, allowing fungal cells to respond to either iron deprivation or iron toxicity (Chen et al. 2011; Noble 2013). In S. pombe, the monothiol glutaredoxin Grx4 binds to Php4, along with a [2Fe–2S] cluster, and regulates the function and localization of Php4 in response to iron availability (Dlouhy et al. 2017). Collectively, it is evident that in most fungi, a GATA-type iron transcription factor and the CBC play key regulatory roles in iron uptake and homeostasis, although genome-wide information of their downstream transcriptional circuits remains to be elucidated (Haas 2012; Labbé et al. 2013; Brault et al. 2015).
Our previous studies in C. neoformans revealed that the GATA-type iron transcription factor Cir1 and the regulatory subunit of the CBC, HapX, play roles in the regulation of genes involved in iron acquisition and homeostasis, as well as virulence (Jung et al. 2006; Jung et al. 2010). In response to iron availability, Cir1 exhibits dual regulatory roles as an activator and a repressor (Jung et al. 2006; Jung and Kronstad 2011). For instance, transcriptome data from microarray analysis revealed that transcript levels for the CFO1 and CFT1 genes of the high-affinity iron uptake system are activated by Cir1, whereas the protein repressed the expression of CFO2 and CFT2, which are putative low-affinity iron transporters, and the laccase-encoding gene LAC1. Furthermore, a mutant lacking CIR1 shows a deficiency in the expression of virulence factors, such as capsule formation, and is avirulent in a murine model of cryptococcosis, suggesting that iron regulation by Cir1 is critical for the pathogenesis of C. neoformans. HapX is also implicated in iron metabolism in C. neoformans (Jung et al. 2010). In particular, HapX not only negatively regulates genes involved in amino acid synthesis, mitochondrial respiration, and ISC and heme synthesis, but also positively regulates the genes required for siderophore uptake for adaptation to iron deprivation (Jung et al. 2010). Although extensive data have been obtained to better understand the roles of Cir1 and HapX, the studies to date all rely on transcriptome analysis of mutants lacking CIR1 and HAPX. Hence, it is still largely unknown which genes are the direct regulatory targets of each iron transcriptional regulator and under which conditions.
In the current study, we employed genome-wide chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to directly identify downstream target genes of Cir1 and/or HapX in C. neoformans. The ChIP-seq profiles of Cir1 and HapX were then integrated with RNA sequencing (RNA-seq) data obtained for the cir1Δ and hapXΔ mutants to increase the stringency of selection of the target genes. Furthermore, ChIP-seq analysis was used to identify nucleotide motifs within the promoter regions of genes directly regulated by Cir1 and/or HapX. In addition, our analysis revealed that Cir1 and HapX are iron-containing proteins, with iron most likely present in the form of an Fe–S cluster. Importantly, our analysis identified a new regulatory circuit by which Cir1 directly controls the expression of two protein kinases that are negative regulators of capsule formation.
Materials and Methods
Strains and media
C. neoformans strains used in the current study are listed in Supplementary Material, Table S1. The strains were routinely grown in yeast extract-bacto peptone medium supplemented with 2.0% (wt/vol) glucose (YPD) or yeast nitrogen base (YNB; Difco) with 2.0% (wt/vol) glucose. The YNB low-iron medium (YNB-LIM) was prepared as previously described (Jung et al. 2010). Iron-replete medium was prepared by supplementing YNB-LIM with 100 μM FeCl3. Strains producing Cir1 or HapX proteins with the 3× FLAG epitope tag were constructed using primers listed in Table S2. To construct the strain producing the Cir1-FLAG fusion protein, the gene encoding Cir1 (CNAG_04864) was amplified by polymerase chain reaction (PCR) from the wild-type genomic DNA using primers Cir1_F and Cir1_R_KpnI. The obtained product was digested with KpnI. The digested DNA fragment was cloned into the KpnI site of pWH109, which contains 3× FLAG sequence, GAL7 terminator, and a neomycin resistance gene (Do et al. 2015). The resulting plasmid, pWH112, was digested with XmaI and introduced into the wild-type strain by biolistic transformation. Positive transformants were selected by PCR using primers Cir1_F and NATterm 2F, and protein production was confirmed by western blotting. To construct the strain producing the HapX-FLAG fusion protein, the HAPX gene (CNAG_01242) was PCR-amplified from the wild-type genomic DNA using primers HapX_F and HapX_R. The amplified DNA fragment was digested with BamHI and BglII. The digested DNA fragment was cloned into the BamHI site of pWH109. The resulting plasmid, pWH185, was digested with NheI and introduced into the hapX mutant by biolistic transformation. Positive transformants were selected by PCR using primers ConfirmF_HapX and NATterm 2F, and protein production was confirmed by western blotting. Strains containing the CCP1 promoter with the designated point-mutations were constructed using DNA fragments obtained using primers listed in Table S2. The promoter region of CCP1 was PCR-amplified using primers CCP1_F_re and CCP1_R_re, digested with BamHI and HindIII, and introduced into pJAF1 (Fraser et al. 2003) to construct pWH250. Neomycin resistance gene (NEO) was amplified using primers M13_F and Muta_CCP1_R, with pWH250 as a template. CCP1 promoters with designated mutations were amplified using primers Fusion_F and CCP1_R_re, with plasmid containing the synthesized CCP1 promoter used as a template. The subsequent DNA fragments were used as a DNA template for PCR amplification using primers NAT-up and Final_R and introduced into C. neoformans wild-type strain. Positive strains were then confirmed by PCR. The functionality and expression of Cir1-FLAG and HapX-FLAG in the constructed stains were confirmed by determination of cell surface reductase activities using triphenyltetrazolium chloride, by analyzing heme use, and by western blot analysis as described previously (Figure S1) (Jung et al. 2006; Jung et al. 2010).
RNA extraction
Three biological replicates for each strain (wild type, and cir1Δ and hapXΔ mutants) were grown in 50 ml of YPD, overnight at 30°. The cells were washed twice with iron-chelated water, and then grown in 50 ml of YNB-LIM for 16 hr to eliminate iron that had accumulated intracellularly during growth on the rich medium. Cells were harvested and diluted to 4.0 × 107 cells in 50 ml of YNB low-iron medium with or without 100 μM FeCl3. Cultures were incubated at 30° for 6 hr, and total RNA was extracted by using RiboPure-Yeast kit (Life Technologies) and treated with DNase I (Life Technologies), following the manufacturer’s instructions.
RNA library construction and sequencing
RNA was quantified using NanoDrop 2000 (ThermoFisher Scientific) and the integrity was checked using 2100 Bioanalyzer (Agilent Technologies). RNA library preparation, sequencing reactions, and bioinformatics analysis were conducted at GENEWIZ, Inc. Illumina TruSeq RNA Sample Preparation Kit was used, following the manufacturer’s recommendations (Illumina). Sequencing was performed using Illumina HiSeq 2000 instrument with 50 bp single end platform, according to the manufacturer’s instructions. Sequence reads were trimmed to remove possible adapter sequences and poor-quality nucleotides on the 3′-end. The trimmed reads were aligned with the reference genome of C. neoformans var. grubii H99, which is available in GenBank under project accession PRJNA411 (Janbon et al. 2014), and gene hit counts were calculated. All analyses were performed using CLC Genomics Workbench version 7.0.4 with the default parameters (Qiagen, Valencia, CA). Comparisons of gene expression between various sample pairs was conducted with the DESeq2 package (Love et al. 2014), and the related code is supplied in the Supplemental Material with sample data.
Chromatin extraction
Strains producing Cir1-FLAG or HapX-FLAG were grown in 50 ml of YPD overnight at 30°. The cells were harvested, washed twice with chelated water, and transferred to 50 ml of YNB-LIM. Cultures were grown overnight at 30°. Then, 1 × 107 cells/ml of each strain was transferred to 50 ml of YNB-LIM with or without 100 μM FeCl3, and incubated for 6 hr at 30°. We employed the 6 hr-incubation time because the cells grown in the iron-depleted condition displayed the highest abundance of CFO1 transcripts, which encodes a ferroxidase in the iron uptake system and is regulated by iron levels in C. neoformans (Figure S2) (Jung et al. 2008, 2009).
For in vivo cross-linking, formaldehyde (37%) was added to the culture medium (1% final concentration), and the culture was incubated for 5 min at room temperature. To quench the cross-linking reactions, samples were treated with 2.5 M glycine (300 mM final concentration) for 5 min at room temperature. Cells were harvested, washed twice with ice-cold phosphate-buffered saline (pH 7.5) and aliquoted into 2-ml screw-cap tubes. To extract chromatin, 400 μl of FA lysis buffer containing 50 mM HEPES KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, and protease inhibitor cocktail (Sigma, St. Louis, MO), and 400 μl of glass beads was added to the cells, which were then lysed using a bead-beater. The lysed cells were centrifuged and resuspended in 400 μl of FA lysis buffer. The samples were sonicated by using an ultrasonicator (Ulsso Hi-tech, Korea; 30 s ON and 30 s OFF, five times). They were then centrifuged at 14,000 rpm at 4° for 10 min, and the supernatant was transferred to new tubes. Finally, 20 μl of chromatin sample was obtained as input control, and DNA was purified using a DNA purification kit (Qiagen).
Chromatin immunoprecipitation
For the experiment, 60 μl of protein A Dynabeads (Invitrogen) were placed in a 1.5-ml tube and washed twice with 1 ml of ice-cold blocking buffer composed of phosphate-buffered saline and 5 mg/ml bovine serum albumin. The beads were resuspended in 60 μl of the blocking buffer, and anti-FLAG antibody (Abcam) was added. Each sample was incubated at 4°, overnight, with rotation. The beads conjugated with the antibody were then washed twice with the blocking buffer and aliquoted into four tubes. They were then incubated with the extracted chromatin at 4°, overnight, with rotation. After incubation, the beads were washed three times with 1 ml of FA lysis buffer using a magnet stand (Invitrogen) and washed three times with 1 ml of FA lysis buffer containing 0.5 M of NaCl. Finally, 1 ml of TE (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) was added to each sample, and the beads were removed using the magnet stand. DNA was eluted from the beads by incubating with the elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 1% SDS) at 65° for 15 min. Then, 100 μl of the eluted samples were incubated with 15 μl of 10% SDS, 1 μl of RNase H, and 3 μl of RNase cocktail (Invitrogen) at 65°, overnight. Each sample was treated with 0.14 mg of proteinase K at 65° for 1.5 hr. Reverse-cross-linked DNA samples were purified using a PCR purification kit (Qiagen).
ChIP-seq library construction and sequencing
Fragmented DNA samples were processed for the construction of the sequencing library using NEBNext Ultra II DNA Library Prep kit for Illumina (New England Biolabs, Hitchin, UK), following manufacturer’s instructions. Briefly, end-repair, dA-tailing, and adapter ligation of 1 ng of sample DNA were performed. The final library was prepared via 12 cycles of PCR amplification. Sequencing was performed by using Illumina HiSeq 2500 with 50 bp single end platform, according to the manufacturer’s instructions. Generated raw reads were quality-checked and trimmed by FastQC v0.11.4 (Andrews 2010) and Trimmomatic v0.35 with default parameters (Bolger et al. 2014). Cleaned reads were mapped to the reference genome (Janbon et al. 2014) using bowtie v2.2.6 (Langmead and Salzberg 2012) with -very-sensitive option. Peak calling was conducted using MACS2 v2.1.0 (Zhang et al. 2008) with default parameters except for the q-value cutoff, which was 0.01.
Manual curation of ChIP-seq data and bioinformatics for binding motif prediction
Peaks called by MACS were manually curated by comparison of each input and immunoprecipitated sample using Integrative Genome Viewer (Robinson et al. 2011) to remove false positives and dubious binding peaks (i.e., low peak signal in both input and immunoprecipitated sample, and/or unusual peak shape) (Rye et al. 2011). The manually curated peaks were used for the identification of target genes of each transcription factor by integrating with the data on genes that were twofold differentially expressed in the mutant lacking Cir1 or HapX compared to the wild type based on the transcriptome analysis. Promoter sequences of the target genes were analyzed by searching binding-site motifs using MEME-ChIP v4.12.0 (Machanick and Bailey 2011). Briefly, 200-bp DNA sequences from the summit of each peak were extracted and analyzed using the zero-or-one-occurrence-per-sequence constraint. The functional enrichment of FunCat was determined using webserver-based FungiFun 2.2.8 beta (https://sbi.hki-jena.de/fungifun/) (Priebe et al. 2015). Transcriptional regulatory networks were reconstructed using Cytoscape version 3.4.0 (Shannon et al. 2003).
Protein purification
Plasmids pWH065 (encoding the wild-type Cir1), pWH077 (Cir1 with C180A and C183A substitutions), pWH078 (Cir1 with C308A and C311A substitutions), and pWH079 (Cir1 with C320A and C321A substitutions) were constructed previously (Jung and Kronstad 2011). The cDNA of wild-type and mutated CIR1 sequences were PCR-amplified using primers Cir1_full_F and Cir1_full_R. The DNA fragments were digested with NheI and BamHI and cloned into the Escherichia coli expression vector pET28 (Merck, Germany). To construct a plasmid for HapX production, HAPX cDNA was PCR-amplified using primers HapX_F_KpnI and HapX_R_HindIII. The amplified DNA fragment was digested with KpnI and HindIII and cloned into pET28. To construct a plasmid for Hap2 production, HAP2 cDNA was PCR-amplified using primers Hap2_F_NdeI and Hap2_R_HindIII. The amplified DNA fragment was digested with NdeI and HindIII and cloned into pET28. To construct a plasmid for Hap3 production, HAP3 cDNA was PCR-amplified using primers Hap3_F_NdeI and Hap3_R_HindIII. The amplified DNA fragment was digested with NdeI and HindIII and cloned into pET28. To construct a plasmid for Hap5 production, HAP5 cDNA was PCR-amplified using primers Hap5_F_NdeI and Hap5_R_HindIII. The amplified DNA fragment was digested with NdeI and HindIII and cloned into pET28. All cloned DNA sequences were confirmed by nucleotide sequencing.
To overexpress the His-tagged recombinant proteins in E. coli, each plasmid was used to transform the E. coli BL21-CodonPlus (DE3)-RIL strain (Agilent Technologies). The cells were grown in LB medium at 37° to OD600 of 0.6. Gene expression was then induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and the incubation continued at 37° for additional 3 hr. The cells were lysed by ultrasonication (Ulsso Hi-tech, Korea) and then centrifuged at 6000 rpm at 4° for 15 min. Cell lysates were loaded onto His GraviTrap columns (GE Healthcare Life Science, Chalafont, St. Giles, UK), and the protein was eluted.
Electrophoretic mobility shift assay
The specific DNA binding motif of recombinant Cir1 was analyzed by electrophoretic mobility shift assay (EMSA) as previously described (Pelletier et al. 2002), with minor modifications. The binding reaction with the specific DNA probe was performed in 1× binding buffer containing 12.5 mM HEPES (pH 7.9), 75 mM NaCl, 4 mM MgCl2, 10% glycerol, 4 mM Tris-HCl (pH 7.9), 0.6 mM dithiothreitol, 1 μg poly (dI-dC)2, 25 μM ZnSO4, and 25 μM FeCl3, unless otherwise stated. The DNA probe (5ʹ-GCACCGAAGCATGAGTTTCGATCAAAGCCACAAGTTTAAAAAAGACTTGCCAAGAATTAATTTTCGGGCCCGCGCGCAGCGAGGAAGGTATCTGATAACTGGGCTAGGCACGCCGCTAAATGCACCGGTTCTCTCCGGTTCGCGCGCATCTGCAGCAGGATCTCGCTTTTTTCACTGGCTCAGGCAGCCACACCTACCGATTTCCCTTACCGATCTATGGTCCCACCAATCT-3ʹ), which contains the predicted binding motif 5ʹ-GATAA-3ʹ, was PCR-amplified using primers EMSA_CFO1_F and EMSA_CFO1_R, and then labeled with 32P-dATP using T4 polynucleotide kinase (Enzynomics). The reaction mixture containing recombinant Cir1 and 32P-end-labeled double-stranded DNA probe was incubated at room temperature for 30 min. The samples were then resolved on a 5% native polyacrylamide gel that had been pre-electrophoresed for 30 min in 0.5× TB buffer (44.5 mM Tris and 44.5 mM borate) at room temperature. The DNA-protein complex was separated by electrophoresis at room temperature for 1 hr. To analyze the specific DNA binding motif of HapX, EMSA was performed as previously described (Kato et al. 1997), with minor modifications. The binding reaction with the DNA probe, i.e., the wild-type CCP1 promoter region containing the predicted binding motif, 5′-CGAAT-3′, was performed in 1× binding buffer containing 25 mM HEPES (pH 7.9), 60 mM KCl, 1 mM EDTA (pH 8.0), 5 mM MgCl2, 1 mM DTT, 1 μg poly (dI-dC)2, and 10% glycerol, unless otherwise stated. The DNA probe (5ʹ-CGAGTAGTGTCAAAAGTGGGCGGTCACATGACATGTTTGAAGCCGCTTTCCGAAAAGGGCAAAAATGGAGAAAACTCGCGTGAATCACAAGGCTAAGTTGATATGTCGCCGACGAATCACGGCCCGCCCATTTCCCAGGCAGATCCCGCTCTATATATACTTGGCCCCCCCCTCATTGTTCATAT-3ʹ) was PCR-amplified using primers EMSA_CCP1_F and EMSA_CCP1_R, and then labeled with 32P-dATP using T4 polynucleotide kinase. The reaction mixtures were incubated at room temperature for 30 min, and then resolved on a 6% native polyacrylamide gel (that had been pre-electrophoresed in 0.5× TB at room temperature for 30 min) at room temperature for 40 min. The gel was dried and exposed to a Phosphor screen (PerkinElmer). The bands were detected by using Packard cyclone phosphor imager (PerkinElmer).
Northern blot analysis
The wild-type strain was grown in 50 ml of YPD overnight at 30°. The cells were transferred into 50 ml of YNB-LIM and incubated overnight at 30°. The following day, the cells were transferred to 50 ml of YNB-LIM with or without 100 μM FeCl3, and incubated at 30° for 6 hr. Total RNA was extracted and northern blot analysis was performed as previously described (Sambrook 2001).
Iron content assay
Iron content of the purified proteins was determined using the iron chelator ferrozine, as previously described (Nakashige et al. 2015). Briefly, the purified recombinant proteins were treated with 200 μl of 1.5 mM ascorbic acid and 200 μl of 0.4 M trichloroacetic acid, and then heated at 98° for 10 min. The samples were then centrifuged at 13,000 rpm at room temperature for 10 min and transferred to new microcentrifuge tubes. The samples were treated with 400 μl of 1.3 M ammonium acetate and 200 μl of 6.17 mM ferrozine and incubated at room temperature for 5 min. Sample absorbance was measured at 562 nm using a spectrophotometer.
DNase I footprinting
For the experiment, the 232-bp of the CFO1 promoter region was PCR-amplified by using a forward primer (5ʹ-AGGGGTCAAAAAATGCCTCAGAAACG-3ʹ) labeled at the 5ʹ-terminus with 6-carboxyfluorescein (FAM) and a reverse primer (5ʹ- GCTTTGATCGAAACTCATGCTTCGGTGC-3ʹ). The FAM-labeled DNA probe was purified using a PCR purification kit (Qiagen), according to the manufacturer’s instruction. The protein-DNA binding reactions were performed as described above for EMSA, and the samples were then treated with 0.025 U of DNase I at room temperature for 1 min. The reaction was stopped by the addition of 50 μl of stop solution containing 200 mM NaCl, 1% SDS, and 30 mM EDTA. The digested DNA fragments were extracted using phenol-chloroform and precipitated using ethanol. The samples were analyzed using ABI 3730 DNA analyzer (Life Technologies).
Western blot analysis
The strains were grown in 50 ml of YPD, overnight at 30°, and harvested. Cell pellets were washed twice with chelated water and resuspended in YNB-LIM, overnight at 30°. The cultures were then transferred to YNB-LIM with or without FeCl3, and incubated at 30° for 6 hr. The cells were harvested and lysed as previously described (Do et al. 2015). Equal amounts of protein were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto a nitrocellulose membrane using a semi-dry transfer system (Hoefer). Western blot analysis was performed using an anti-DDDDK polyclonal rabbit antibody (Abcam) and a goat anti-rabbit IgG horseradish peroxidase conjugate (Santa Cruz). Protein signals were visualized by using chemiluminescence.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The ChIP-seq and transcriptome data have been deposited at the Gene Expression Omnibus database of the National Center for Biotechnology Information under the SuperSeries record accession no. GSE126314. The accession number for the ChIP-seq data are GSE126312, and that for the RNA-seq data are GSE126313. R code and sample data for DESeq2 analysis can be found in Supplemental Materials. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12151674.
Results
ChIP-seq analysis identifies the overall numbers and distribution of Cir1 and HapX binding sites throughout the genome
We initially constructed strains producing functional Cir1-FLAG or HapX-FLAG proteins to enable identification by ChIP-seq of target genes directly bound by the transcription factors (see details in Materials and Methods). After isolation of the target DNA fragments bound by Cir1-FLAG or HapX-FLAG (obtained by ChIP), sequencing was performed and total sequence reads were mapped to the C. neoformans var. grubii genome, followed by computational analysis. In addition, we performed a visual inspection of the ChIP-seq data across the entire genome to distinguish false-positive peaks from the true peaks by comparison with the base-line noise. This highly stringent analysis of the ChIP-seq data revealed that the number of peaks for Cir1 and HapX, representing the number of binding sites, was 328 and 579 under low-iron conditions, respectively; and 530 and 497 under high-iron conditions, respectively (Table 1).
Table 1. The number of peaks identified in the ChIP-seq datasets.
| Cir1 | HapX | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Low- and high-irona | Unique: low-iron | Unique: high-iron | Total | Low- and high-irona | Unique: low-iron | Unique: high-iron | Total | ||
| Promoter | 234 | 1 | 124 | 359 | 215 | 264 | 177 | 656 | |
| Terminator | 38 | 0 | 32 | 70 | 8 | 19 | 41 | 68 | |
| Intergenic | 26 | 1 | 15 | 42 | 14 | 36 | 15 | 65 | |
| Exon | 21 | 4 | 30 | 55 | 6 | 14 | 15 | 35 | |
| Intron | 3 | 0 | 7 | 10 | 1 | 2 | 5 | 8 | |
| Total | 322 | 6 | 208 | 536 | 244 | 335 | 253 | 832 | |
These peaks were observed in the dataset from cells grown in low iron medium and in the dataset from cells grown in high iron medium. For Cir1, the promoter peaks present in both conditions plus the peaks unique to the low or high iron conditions equal 235 (e.g., 234 + 1) and 358 (234 + 124), respectively. For HapX, the corresponding numbers are 479 and 392.
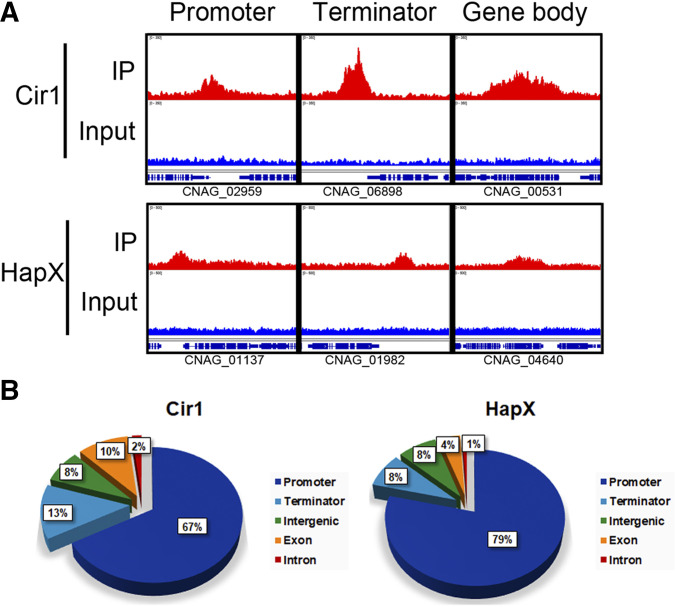
As mentioned, peak position (the binding locations of Cir1 and HapX in the genome) was also determined by visual inspection. Cir1 and HapX appeared to mainly bind the putative promoter regions of protein-coding genes: 72% (235/328) and 83% (479/579) in low-iron medium, respectively; and 67% (358/530) and 79% (392/497) in high-iron medium, respectively. However, the analysis also revealed that Cir1 binds not only at promoter regions but also at various other locations throughout the genome, including putative transcription termination sites, intergenic regions, and exon and intron sequences in the gene body. Similarly, a small portion of the HapX binding targets was found in regions of the genome outside of putative promoters (Figure 1). The overall numbers and distribution of Cir1 and HapX binding sites are consistent with previous findings indicating that Cir1 and HapX are key transcriptional regulators of the response to iron in C. neoformans (Jung et al. 2006, 2010). Specifically, our previous microarray analysis of genes with altered transcript levels in cir1 and hapX mutants identified a regulatory impact on genes encoding proteins for iron acquisition and iron-dependent functions (Jung et al. 2010). However, the regulatory mechanisms may be more complicated than initially anticipated given the observed binding of the proteins outside promoter regions.
Figure 1.
Analysis of the genomic distribution of the sequences bound by Cir1 and HapX. (A) Peak distributions determined by ChIP-seq, as analyzed using Integrative Genomics Viewer. Upper panels (red) show the enrichment of binding of each transcription factor at genomic loci and lower panels (blue) show input controls. (B) Pie charts of genomic distribution of Cir1-and HapX-binding peaks determined by ChIP-seq analysis. IP, immunoprecipitate.
Transcriptome analysis identifies genes whose expression is regulated by Cir1 and HapX
Enrichment bias at highly expressed loci has been reported to result in the inclusion of false-positive binding hits and the misinterpretation of ChIP-seq results (Park et al. 2013; Teytelman et al. 2013). The use of transcriptome profiles has been suggested as an approach to justify the enrichment bias of ChIP-seq results (Qin et al. 2014; Merhej et al. 2016), and we therefore performed transcriptome analysis of the cir1Δ and hapXΔ mutants grown under conditions identical to those used for the ChIP-seq experiments (Figure 2A). We conducted the transcriptome analysis in a high-throughput RNA-seq format, with three biological replicates. Differentially expressed genes in the mutant cells were sorted based on a specific selection criterion, i.e., that the relative transcript levels of genes in the mutant displayed an over twofold statistically significant up- or down-regulation (P < 0.05) compared with the wild type.
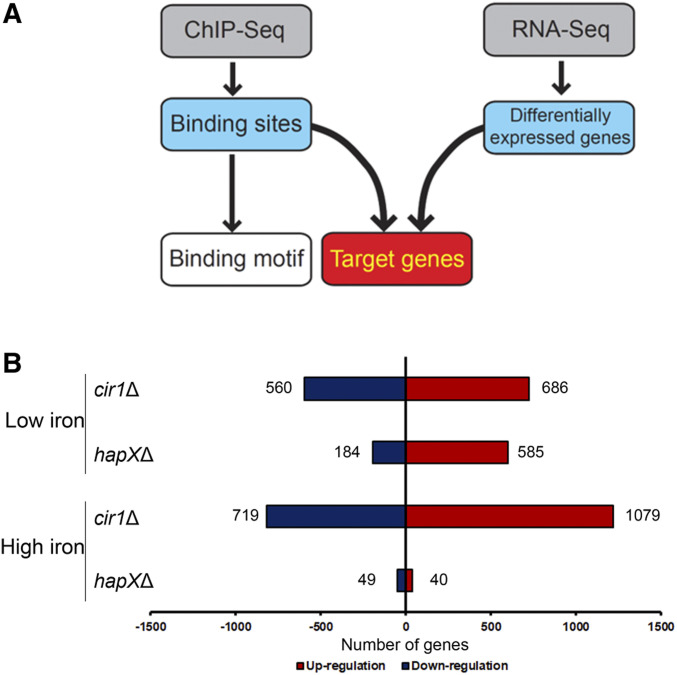
Figure 2.
Transcriptome changes in strains lacking CIR1 or HAPX. (A) Experimental strategy for defining the direct gene targets of Cir1 and HapX, and their binding motifs. (B) Overview of the numbers of differentially expressed genes in strains lacking CIR1 or HAPX in response to iron availability. The histogram shows the number of genes differentially expressed in the mutants, with at least twofold changes and statistical significance (P < 0.05), compared to the wild type.
The transcriptome analysis revealed that a large number of genes were differentially expressed in the mutants lacking CIR1 or HAPX. In the cir1Δ mutant grown in a low-iron medium, 560 and 686 genes were downregulated and upregulated, respectively, compared with the wild type. Interestingly, the number of differentially expressed genes increased in the same cells grown in a high-iron medium, to 719 and 1079 for downregulated and upregulated genes, respectively (Figure 2B). A number of genes were also differentially regulated in the hapXΔ mutant. Overall, 184 and 585 genes were downregulated and upregulated in the mutant, respectively, compared with the wild type in a low-iron medium. However, the number of differentially expressed genes was greatly reduced in the same mutant grown in a high-iron medium, with 49 and 40 downregulated and upregulated genes, respectively, compared with the wild type. This result suggests that HapX mainly plays a role under iron-limited conditions. Taken together, the transcriptome analysis demonstrated that the transcriptional responses of a number of genes to iron availability are governed mainly by Cir1 and HapX. Furthermore, the data implied that Cir1 and HapX might play reciprocal roles, with Cir1 regulating the expression of genes upon iron repletion and HapX regulating the genes upon iron deprivation in C. neoformans. Of note, the RNA-seq transcriptome profiles obtained in the current study were similar to our previously published microarray analysis of the transcriptomes in the cir1Δ and hapXΔ mutants (Jung et al. 2010).
Identification of genes directly regulated by Cir1
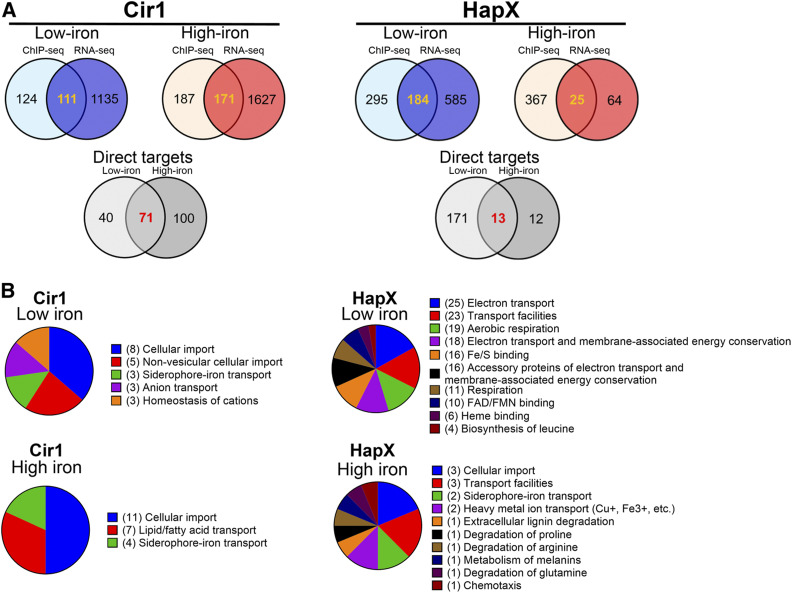
We hypothesized that transcript levels of genes whose promoter regions were identified by the ChIP-seq experiments as the binding targets of Cir1 and HapX would be altered in the strain lacking the corresponding transcription factor in comparison with the wild type, as determined by the transcriptome data. Therefore, the ChIP-seq data were combined with the transcriptome data to identify genes that fit the criteria of a putative binding target promoter by ChIP-seq and differential expression based on the transcriptome analysis. Promoter regions of 235 genes were identified by ChIP-seq as the binding targets of Cir1 in cells grown in low-iron medium (Table 1). Among these genes, 40 were differentially regulated in the cir1Δ mutant under low-iron conditions, suggesting that they are directly regulated by Cir1 upon iron deprivation (Figure 3A, left panel). Under high-iron conditions, the promoter regions of 358 genes were identified as the binding targets of Cir1 (Table 1). Among these genes, 100 were differentially regulated in the cir1Δ mutant in the high-iron condition, suggesting that they are directly regulated by Cir1 upon iron repletion (Figure 3A, left panel). Interestingly, the promoter regions of 71 genes were identified as binding targets in ChIP-seq experiments and exhibited differential expression between mutant and wild-type cells that was independent of iron levels in the growth medium. This result suggests that Cir1 also participates in iron-independent regulation of these genes (Figure 3A, left panel; Table S3). For the iron-dependent genes, functional category analysis using FunCat revealed that the genes involved in cellular import, nonvesicular cellular import, siderophore-iron transport, anion transport, and homeostasis of cations were enriched under iron-limited conditions (Figure 3B). In addition, genes involved in cellular import, lipid/fatty acid transport, and siderophore-iron transport appeared to be significantly enriched in response to iron-replete conditions (Figure 3B).
Figure 3.
Integration of transcriptome changes with ChIP-seq data. (A) Integration of the transcriptome data with ChIP-seq data yielded a list of genes directly regulated by Cir1 and HapX. The colored Venn diagrams indicate the number of genes identified by ChIP-Seq, as listed in Table 1, as well as the overlap with the number of regulated genes identified by RNA-Seq. The overlap in the diagrams indicate the numbers for candidate direct target genes and the numbers are further separated for low and high iron in the gray Venn diagrams. (B) Functional categories of gene targets of Cir1 and HapX analyzed using FunCat available from the webserver FungiFun2. Values in parentheses indicate the number of genes in each category.
Integrating the transcriptome data with the ChIP-seq data not only enabled a highly stringent selection of direct target genes for Cir1, but also revealed regulatory patterns for each transcription factor (i.e., positive or negative regulation) in the transcriptional regulatory networks. Based on this analysis, we constructed a comprehensive transcriptional regulatory network governed by Cir1 upon iron availability (Figure 4A). In the transcriptional regulatory network, Cir1 positively regulates 19 genes and negatively regulates 92 genes upon iron deprivation, demonstrating that the protein plays a dual regulatory role (Figure 4A). The genes directly and positively regulated by Cir1 include CFO1, SIT1 and SIT2, and CIG1, which are required for the reductive high-affinity iron uptake, siderophore uptake, and heme acquisition, respectively. This result suggested that, despite of the dual regulatory role, a positive regulatory role of Cir1 is required for iron uptake in C. neoformans under iron-limited conditions. The 92 genes that were directly and negatively regulated by Cir1 included LAC1, which encodes the laccase for melanin production. This result is consistent with previous data that laccase activity is constitutively derepressed in the cir1Δ mutant in a DOPA (L3–4 dihydroxyphenylalanine)-containing medium (Jung et al. 2006).
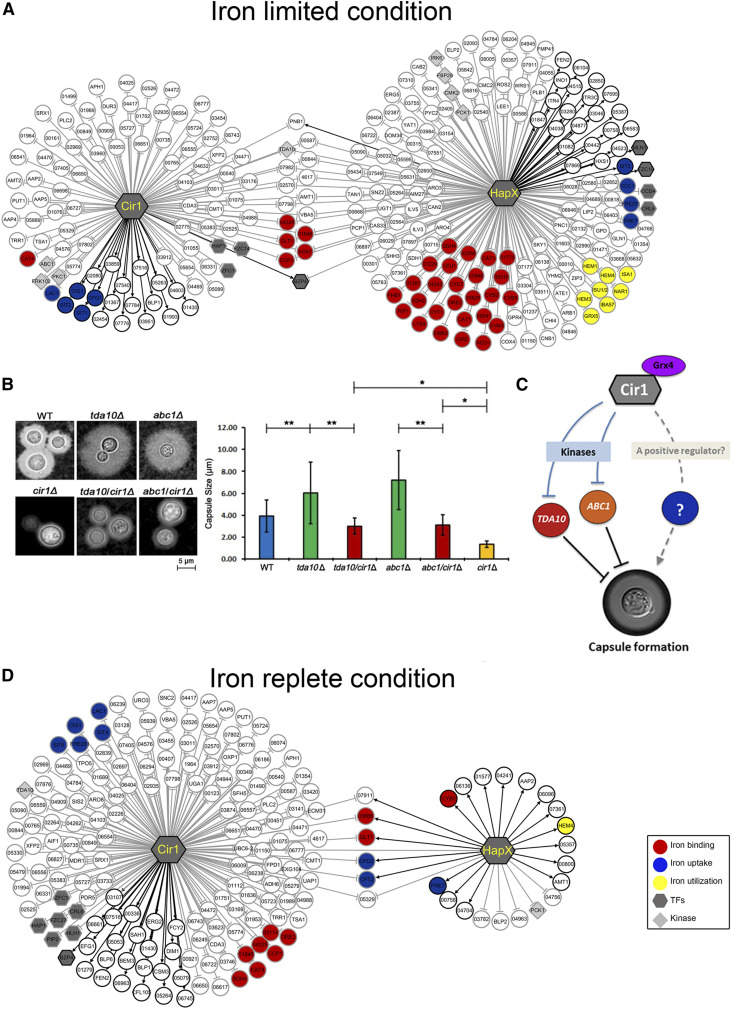
Figure 4.
The transcriptional networks of Cir1 and HapX. (A) Transcriptional regulatory networks are depicted to show the target genes of each transcription factor under iron-limited conditions. Genes regulated by Cir1 and HapX in an iron-independent manner are also included. (B) Capsules of C. neoformans strains grown in 1/10 sabouraud media were observed using a DCM310 microscope with India ink. Capsule sizes measured with 50 individual cells of each strain are shown in the graph. Note that the capsule size of the cir1Δ mutant is 1.36 ± 0.27 μm in these conditions. *P < 0.001; **P < 0.01. (C) Model for a possible regulatory mechanism of capsule formation based on direct regulation of Abc1 and Tda10 expression by Cir1. In the scenario shown, Cir1 indirectly regulates capsule formation by directly controlling expression of the genes encoding the protein kinases Tda10 and Abc1 which are negative regulators of capsule formation (Lee et al. 2016). A yet unknown regulatory factor, which might be a downstream of Cir1, may also play a positive regulatory role in capsule formation in C. neoformans. Additional regulatory scenarios are presented in the Discussion. (D) Transcriptional regulatory network depicting the target genes of each transcription factor under iron-replete conditions. Genes regulated by Cir1 and HapX in an iron-independent manner are also included. Black lines indicate positive regulation, and gray lines indicate negative regulation by the corresponding transcription factor.
We have previously demonstrated that deletion of CIR1 results in a deficiency in capsule formation, one of the well-known virulence factors of C. neoformans (Jung et al. 2006). However, the underlying mechanisms are not entirely clear. In the current study, we found that Cir1 directly represses expression of the protein kinases Abc1 and Tda10, which were recently found to negatively regulate capsule formation (Lee et al. 2016). To further understand how a deficiency in the cir1 mutant could potentially influence capsule formation, we deleted the coding region of CIR1 in the mutants lacking either ABC1 or TDA10 (generated in a previous high-throughput screening study (Lee et al. 2016)), and analyzed the capsule formation of the mutants. As previously reported (Lee et al. 2016), the mutants lacking either ABC1 or TDA10 display significantly increased capsule diameters, and the cir1Δ mutant has a barely detectable capsule (Jung et al. 2006) (Figure 4B). We found that the sizes of the capsules were reduced when CIR1 was deleted in the abc1Δ or tda10Δ mutant backgrounds, in comparison with the capsule sizes of the single mutants (Figure 4B). These results confirmed that ABC1 and TDA10, as downstream targets of Cir1, are indeed negative regulators of capsule formation. We also noted that the double mutants had reduced capsule sizes relative to WT cells but still produce larger capsule sizes than the cir1Δ mutant (Figure 4B). This observation suggests that as yet unknown regulatory factors, which might be downstream of Cir1, could play a positive regulatory role in capsule formation in C. neoformans (Figure 4C).
The transcriptional regulatory network defined herein also indicated that a number of genes were directly regulated by Cir1 in the iron-replete condition. Cir1 positively regulated 23 genes and negatively regulated 148 genes under high-iron conditions (Figure 4D). Under high-iron conditions, more genes were negatively regulated by Cir1 than those positively regulated by the protein, suggesting that Cir1 acts mainly as a repressor upon iron repletion. Indeed, the regulatory network clearly showed that a number of genes involved in iron uptake, such as CIG1, FRE201, SIT4, and SIT6, were repressed by Cir1 upon iron repletion. In this condition, we also observed binding of Cir1 to the HAPX promoter and negative regulation of transcription (Table S3). This result reinforces the idea that Cir1 plays an important role in the regulation of iron homeostasis by directly controlling the expression of iron uptake systems and, indirectly, iron-requiring functions.
Identification of genes directly regulated by HapX
Our ChIP-seq analysis also identified promoter regions of 479 genes as the binding targets of HapX under low-iron conditions (Table 1). Among them, 171 genes were differentially regulated in the hapXΔ mutant under low-iron conditions, suggesting that they are directly regulated by HapX upon iron deprivation (Figure 3A, right panel). Under high-iron conditions, the promoter regions of 392 genes were identified as the binding targets of HapX (Table 1). Among these genes, 12 were differentially regulated in the hapXΔ mutant under high-iron conditions, suggesting that they are directly regulated by HapX upon iron repletion (Figure 3A, right panel). Furthermore, the promoter regions of 13 genes were identified as binding targets by ChIP-seq experiments and showed differential expression that was independent of iron levels in the medium. This implied that HapX might also play an iron-independent role as found for Cir1 (Figure 3A (right panel), Table S4). The functional category analysis revealed that genes encoding proteins requiring iron, e.g., those involved in electron transport and aerobic respiration, were significantly enriched under iron-limited conditions (Figure 3B). By contrast, genes regulated by HapX under iron-replete conditions were involved in cellular import, siderophore-iron transport, and heavy-metal ion transport (Figure 3B). These results suggested that HapX plays an important role in the transcriptional regulation of genes involved in iron-requiring pathways upon iron deprivation. Furthermore, the data suggested that a small set of genes encoding iron transporters (mainly those involved in low-affinity reductive iron uptake and nonreductive iron transport) were directly regulated by HapX.
Under iron-limited conditions, HapX positively regulated 25 genes and negatively regulated 159 genes. These results also suggest that HapX, like Cir1, is a dual regulator. Furthermore, the number of genes that were negatively affected by HapX was larger than the number of positively regulated genes, implying that the transcription factor acts mainly as a repressor upon iron deprivation (Figure 4A). One notable finding was that the genes negatively regulated by HapX were closely related to iron-dependent metabolism. Examples include GLT1, LEU1, and LYS4 for amino acid synthesis as well as AOX1, CCP1, CYB2, CYC7, CYT1, RIP1, and SDH2 for mitochondrial respiration. Furthermore, genes required for ISC synthesis (IBA57, ISU1, and NAR1) and heme synthesis (HEM1, HEM3, and HEM4) were directly and negatively regulated by HapX. This suggested that HapX acts as a repressor of genes involved in iron use, which is a well conserved regulatory activity of the HapX orthologs in other fungi (Mercier et al. 2006; Hortschansky et al. 2007).
Only a few genes were directly regulated by HapX under high-iron conditions. Specifically, 19 genes were positively regulated, and 6 genes were negatively regulated by HapX (Figure 4D). The relatively small number of target genes of HapX in cells grown in a high-iron medium compared with cells grown in a low-iron medium suggested that the regulatory role of the protein is minimal upon iron repletion. However, we noticed CFO2 and CFT2 among the genes that were positively regulated by HapX. CFO2 and CFT2 are particularly interesting because they have been predicted to play a role in low-affinity iron uptake under high-iron conditions (Jung et al. 2008).
Cir1 and HapX reciprocally regulate downstream target genes
The comprehensive transcriptional regulatory network presented here suggested that Cir1 and HapX reciprocally regulate downstream target genes that are involved in multiple pathways of iron uptake and metabolism. In particular, the data suggested that, upon iron depletion, one of the main regulatory roles of Cir1 is to activate iron uptake systems, while that of HapX is to block expression of iron-dependent pathways. An additional role of Cir1 is, by contrast, to reduce the expression of iron uptake system genes under iron repletion. A specific example includes coordinated regulation by Cir1 and HapX of CFO2 and CFT2 which, as mentioned above, encode a putative low-affinity iron uptake system in C. neoformans. In the iron-replete condition, the expression of CFO2 and CFT2 was directly and negatively regulated by Cir1, but was positively regulated by HapX (Figure 4D).
We also note that characterization of Cir1 and HapX transcriptional regulatory networks revealed that genes encoding several transcription factors and kinases are directly regulated by these transcription factors (Tables 2 and 3). Therefore, the regulation of downstream transcription factors by Cir1 and/or HapX suggests a broader impact on the expression of genes involved in various metabolic pathways, and that analysis of the broader regulatory roles of Cir1 and HapX in C. neoformans is warranted (Jung et al. 2015). One or more of these factors may contribute to the positive regulation of capsule formation, as postulated earlier (Figure 4C).
Table 2. Transcription factors directly regulated by Cir1 or HapX.
| Gene ID | TF family | Gene name | Low-iron | High-iron | ||
|---|---|---|---|---|---|---|
| Cir1 | HapX | Cir1 | HapX | |||
| CNAG_06818 | Zn2Cys6 | HAP1 | –a | – | ||
| CNAG_03914 | Zn2Cys6 | FZC14 | – | |||
| CNAG_02603 | C2H2 zinc finger | ZFC1 | – | – | ||
| CNAG_03346 | bZIP | BZP4 | +b | – | + | |
| CNAG_04837 | bHLH | MLN1 | + | |||
| CNAG_04836 | Zn2Cys6 | FZC10 | + | |||
| CNAG_03279 | Zn2Cys6 | CCD4 | – | |||
| CNAG_07797 | Negative transcriptional regulator | CRL6 | – | – | ||
| CNAG_00830 | Zn2Cys6 | FZC38 | – | |||
| CNAG_00039 | C2H2 zinc finger | ZFC6 | – | |||
| CNAG_05049 | Zn2Cys6 | PIP201 | – | |||
| CNAG_04594 | Zn2Cys6 | FZC27 | – | |||
| CNAG_00791 | bHLH | HLH1 | – | |||
(–) indicates direct and negative regulation by either Cir1 or HapX.
(+) indicates direct and positive regulation by either Cir1 or HapX.
Table 3. Kinases directly regulated by Cir1 or HapX.
| Gene ID | Kinase family | Gene Name | Low-iron | High-iron | ||
|---|---|---|---|---|---|---|
| Cir1 | HapX | Cir1 | HapX | |||
| CNAG_01845 | Cysteine-rich domain | PKC1 | –a | |||
| CNAG_01061 | Protein kinase-like (PK-like) | FRK102 | – | |||
| CNAG_06632 | Protein kinase-like (PK-like) | ABC1 | – | |||
| CNAG_07779 | P-loop containing nucleoside triphosphate hydrolases | TDA10 | – | – | – | |
| CNAG_01704 | Sm-like ribonucleoproteins | IRK6 | – | |||
| CNAG_04221 | P-loop containing nucleoside triphosphate hydrolases | FBP26 | – | |||
| CNAG_04272 | Protein kinase-like (PK-like) | CMK2 | – | |||
(–) indicates direct and negative regulation by either Cir1 or HapX.
Prediction of the Cir1 binding motifs
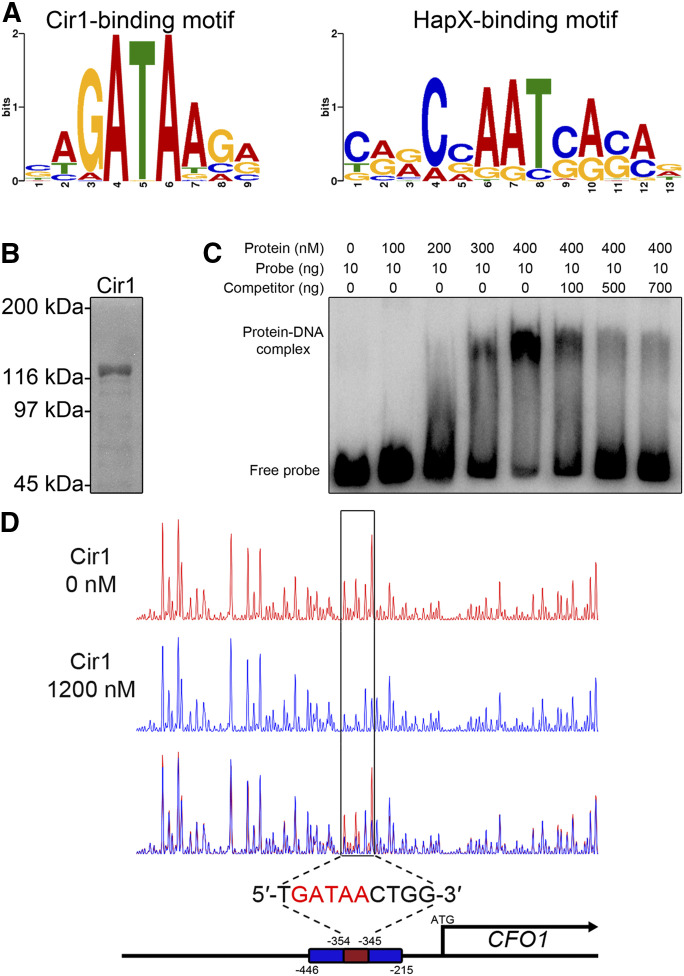
We next used the list of target genes directly regulated by each transcription factor to define the DNA-binding motifs within the target promoter region. We identified the binding motifs using the motif-prediction tool MEME-ChIP. The MEME-ChIP analysis revealed that the 5′-(C/G)(A/T/C)GATAA(G/C/A)(A/G/C)-3′ binding motif was highly enriched among the Cir1-bound target promoters, with an E-value of 3.2e−026 (Figure 5A). To confirm that Cir1 binds the predicted motif, a recombinant full-length Cir1 protein was expressed in E. coli and purified (Figure 5B). We then performed an EMSA with the purified Cir1 protein and a radioactively labeled probe representing the putative binding DNA target, i.e., the CFO1 promoter region containing the binding motif. We observed a concentration-dependent mobility shift for samples containing the purified Cir1 and labeled probe, compared with samples containing the probe but no protein. A competition assay further confirmed the binding of Cir1 to the predicted motif in the same experiment (Figure 5C). In addition to EMSA, we also performed a DNase I footprinting analysis to verify the binding of Cir1 to the predicted motif. A probe end-labeled with 6-FAM was incubated with the purified Cir1 protein, followed by DNase I digestion. The subsequent DNA fragments were analyzed by capillary electrophoresis and visualized. The DNase I footprinting suggested that Cir1 protects a 10-nt region (5′-TGATAACTGG-3′) spanning positions −354 to −345 relative to the CFO1 start codon (Figure 5D). These findings further confirmed that Cir1 binds a specific sequence motif, 5′-GATAA-3′, in the promoter region of target genes.
Figure 5.
Cir1 binds a GATA motif in the gene promoter region. (A) The binding motifs of Cir1 (left) and HapX (right) were identified using MEME analysis. (B) Full-length Cir1 protein was produced in the heterologous host E. coli, purified, and confirmed. (C) EMSA of DNA-protein binding was performed on a native polyacrylamide gel. The DNA probe was amplified from the CFO1 promoter region containing the predicted binding motif as a template. (D) DNase I footprinting using 6-FAM–labeled CFO1 promoter region was performed with (blue peaks) or without (red peaks) the purified Cir1 protein and analyzed by capillary electrophoresis. The position of the Cir1-binding motif was determined to span from −354 to −345 nt relative the transcription start site.
Prediction of the HapX binding motifs
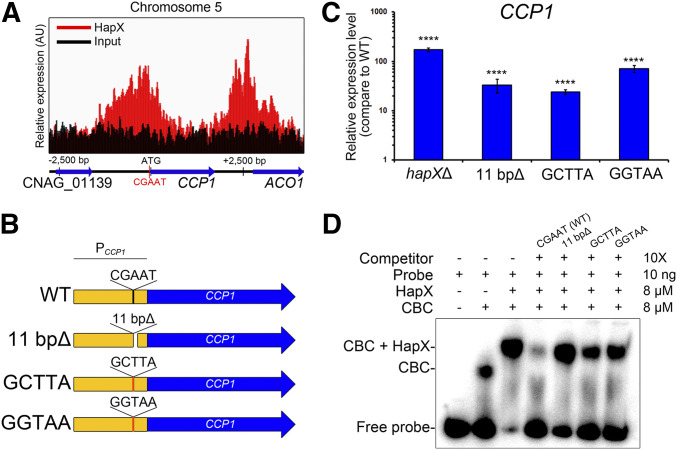
The 5′-(C/T/G)(A/G)(G/A)C(C/G/A)AAT(C/G)(A/G)(C/G)(A/C)-3′ motif was predicted as the binding motif of HapX with an E-value of 3.4e−008 (Figure 5A). To test whether HapX binds the predicted binding motif in the promoter region of target genes, we first analyzed the expression of CCP1 alleles with wild type or mutated versions of the binding motif. CCP1 encodes cytochrome c peroxidase involved in mitochondrial respiration, and, in the current study, we discovered that the gene is a binding target of HapX under low-iron conditions, with significant derepression in the hapXΔ mutant, as indicated by the transcriptome data. A CCP1 promoter region, −122 to −118 bp from the start codon, harbors the CGAAT motif, and the ChIP-seq analysis suggested that HapX occupies the region under iron-limited conditions (Figure 6A). For the expression analysis, a C. neoformans mutant strain lacking 11 bp (positions −122 to −113, including the CGAAT motif) within the promoter region of CCP1 was constructed and employed to investigate whether the predicted motif is essential for binding under low-iron conditions. Two additional C. neoformans strains containing the mutated promoter region of CCP1 (5′-GCTTA-3′ or 5′-GGTAA-3′ instead of 5′-CGAAT-3′) were also constructed and included in the analysis (Figure 6B). The wild type, hapXΔ mutant, and the strains containing the mutated CCP1 promoter regions were grown under iron-limited conditions. Then, quantitative reverse-transcription PCR was performed to evaluate the CCP1 transcript levels. The analysis revealed that the expression of CCP1 was de-repressed in the hapXΔ mutant, which agreed well with the transcriptome data. Similar to the hapXΔ mutant, the CCP1 transcript levels were elevated in the strain expressing CCP1 under a promoter with the CGAAT motif deleted. The expression of CCP1 in strains with the CGAAT motif mutated was also elevated. These observations suggested that the CGAAT motif is required for a negative regulation of CCP1 by HapX under iron-limited conditions (Figure 6C).
Figure 6.
HapX binds the CGAAT motif in the gene promoter region. (A) ChIP-seq analysis revealed that HapX binds to the CGAAT motif in the CCP1 promoter region on chromosome 5. Red color indicates enrichment profiles of the HapX-binding peaks, and black color indicates profiles of the input DNA. (B) Schematic representation of the constructs of the CCP1 promoter region with designated mutations, analyzed by EMSA. (C) The CCP1 transcript levels in strains harboring mutated CCP1 promoter regions, evaluated by quantitative reverse-transcription PCR. Data were normalized to TEF2 expression as an internal control. Three biological replicates were analyzed. ****P < 0.0001. (D) EMSA, performed using a native polyacrylamide gel. The DNA probe was amplified using the CCP1 promoter region containing the CGAAT sequence. The indicated competitors were added to observe competition with the probe. CCAAT-binding complex (CBC) was reconstituted by mixing equal amounts of Hap2, Hap3, and Hap5 that had been produced in the heterologous host E. coli and purified.
To further confirm the binding of HapX to the CGAAT motif, we used EMSA with a purified full-length HapX protein expressed in E. coli and a radioactively labeled probe representing the CCP1 promoter region containing the binding motif. It has been suggested that HapX forms a complex with the heterotrimeric CBC (consisting of HapB, HapC and HapE) in Aspergillus nidulans, and that the physical interaction between HapX and the CBC is essential for the binding of HapX to the promoter region of target genes (Hortschansky et al. 2007). Hap2, Hap3, and Hap5 are the C. neoformans homologs of A. nidulans HapB, HapC, and HapE, respectively (Jung et al. 2010). Therefore, in addition to HapX, we expressed these proteins in the heterologous host E. coli, purified them (Figure S3), and included them in the EMSA mixture. EMSA revealed that (1) the CBC binds to the CCP1 promoter region containing the predicted CGAAT binding motif; and (2) the addition of purified HapX triggers a supershift of the CBC-probe binding complex. Furthermore, a competition assay indicated that an unlabeled probe lacking the CGAAT motif, or one in which the motif was replaced with 5′-GCTTA-3′ or 5′-GGTAA-3′, no longer competed with the labeled probe containing the wild-type sequence (Figure 6D). These observations further confirmed that HapX binds the CGAAT motif, as predicted, via the formation of the CBC-HapX complex.
Cir1 and HapX are iron-binding proteins
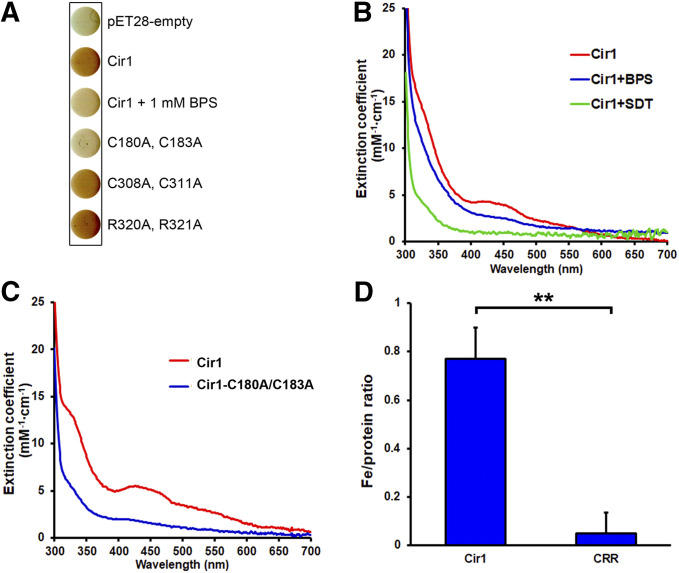
During the Ni-affinity purification of recombinant Cir1 produced in E. coli, we observed that the eluted protein was reddish brown in color (data not shown), similar to what has been reported for the iron-regulatory transcription factors Fep1, Sre1, and SRE in S. pombe, Histoplasma capsulatum, and Neurospora crassa, respectively (Harrison and Marzluf 2002; Chao et al. 2008; Encinar del Dedo et al. 2015; Kim et al. 2016). This led us to hypothesize that Cir1 might bind directly to iron as a potential mechanism to sense intracellular iron levels. Furthermore, bacterial colonies producing recombinant Cir1 protein were reddish brown, compared with cells harboring the empty vector. We reasoned that the addition of the iron chelator bathophenanthrolinedisulfonate (BPS) to the growth medium would influence the bacterial colony color because of the reduced availability of iron for Cir1 binding. As anticipated, the intensity of the reddish-brown color of bacterial colonies producing Cir1 grown in a BPS-containing medium was appreciably reduced (Figure 7A).
Figure 7.
Cir1 is an iron-containing protein. (A) E. coli cells were transformed with pET28 derivatives encoding Cir1Wild type (WT), Cir1C180A, C183A, Cir1C308A, C311A, or Cir1R320A, R321A. Protein production was induced by the addition of 1 mM of IPTG for 3 hr at 37°. The cultures were centrifuged and cell pellets were photographed. (B) Cir1 purified from cells grown in a medium containing 1 mM IPTG (red), or in one containing 1 mM IPTG and 1 mM BPS (blue) was analyzed using UV-visible spectrophotometer. Cir1 purified from cells grown in the presence of 1 mM IPTG and 1 mM BPS, and treated with sodium dithionite (SDT; green) was also analyzed. (C) UV-visible absorption spectra of proteins purified from strains producing Cir1WT (red) or Cir1C180A, C183A (blue). (D) Iron content of the purified Cir1WT and Cir1C180A, C183A proteins was determined. Three biological replicates were analyzed. **P < 0.01.
To examine the incorporation of iron into Cir1 more closely, E. coli strains producing full-length Cir1 with amino acid substitutions (C180A and C183A in the cysteine-rich region; and C308A and C311A, and R320A and R321A in the zinc-finger domain) were constructed. Interestingly, introduction of the C180A/C183A substitutions in the cysteine-rich region of Cir1 fully abrogated the reddish-brown color of the harboring cells, while the color of cells producing Cir1 with the C308A/C311A or R320A/R321A variants of the zinc-finger domain remained unchanged (Figure 7A). Collectively, these observations suggested that the cysteine-rich region of Cir1 participates in iron binding.
We next acquired the UV-visible absorption spectra of the purified Cir1 protein and its C180A/C183A variant. It has been shown that S. pombe Fep1 and N. crassa SRE are iron-sulfur containing proteins, with spectral absorption peaks at 320 and 420 nm, and an extended shoulder at ∼520 nm, which are common characteristics of Fe–S cluster binding proteins (Harrison and Marzluf 2002; Kim et al. 2016). In agreement with the studies on Fep1 and SRE, the absorption peak at 420 nm, and shoulders at ∼320 and 520 nm, were apparent in the UV-visible absorption spectrum of the purified Cir1 protein (Figure 7B). By contrast, these peaks and shoulders were not observed in the absorption spectrum of Cir1 that had been purified from E. coli cells grown in a BPS-containing low-iron medium. Furthermore, the absorption spectrum of Cir1 incubated with the reducing agent sodium dithionite (SDT) lacked these peaks and shoulders, supporting the notion that Cir1 is a Fe–S cluster-containing protein (Figure 7B). The analysis of UV-visible absorption spectrum also confirmed that the cysteine-rich region of Cir1 is responsible for the Fe–S cluster binding. Specifically, the spectral characteristics at 320, 420, and 520 nm disappeared in the Cir1 variant with the C180A/C183A substitutions (Figure 7C). These findings were further confirmed by measuring the iron content of the purified proteins. The iron‒to‒protein ratio of the purified wild type Cir1 protein was 0.77 ± 0.05, while that of the C180A/C183A variant of Cir1 was significantly reduced to 0.13 ± 0.09, thus confirming that the iron binding of Cir1 was dependent on the cysteine-rich region (Figure 7D).
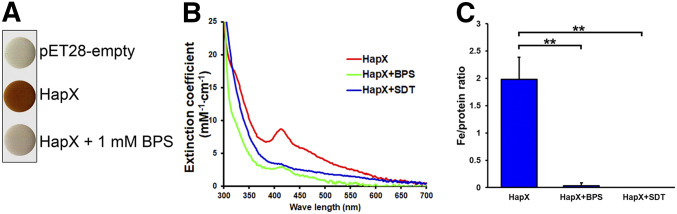
C. neoformans HapX contains six cysteine-rich regions, and we hypothesized that at least one of them binds the Fe–S cluster, similar to Yap5 in Saccharomyces cerevisiae. Yap5 is a transcription factor which plays a HapX-like role in S. cerevisiae, although it does not share homology with HapX in other pathogenic fungi. However, Yap5 has a similar structure with a basic-leucine zipper domain and cysteine-rich regions. In particular, the N-terminal cysteine-rich region of Yap5 binds an Fe–S cluster (Rietzschel et al. 2015). In addition, the cysteine-rich region (CRR-B) in HapX in A. fumigatus and A. nidulans is implicated in iron detoxification under iron excess, suggesting that the region is responsible for sensing intracellular iron status in fungi (Gsaller et al. 2014). Similar to E. coli cells producing Cir1, the bacterial cells producing HapX were brownish, indicating the possibility that the protein indeed harbors iron (Figure 8A). The color was abolished by the addition of 1 mM BPS to the growth medium, implying that the state of the HapX protein is influenced by iron availability. Subsequently, we monitored the UV-visible absorption spectrum of purified HapX. Accordingly, we noted the presence of an absorption peak at 420 nm, and shoulders at ∼320 and 520 nm, implying that the protein contains a Fe–S cluster (Figure 8B). The spectral peaks were noticeably less pronounced in a spectrum of HapX purified from E. coli cells grown in the presence of BPS, and that of a purified protein that had been incubated with SDT (Figure 8B). This result further suggests that the protein is an Fe–S–binding protein. Furthermore, determination of the iron content of the HapX protein revealed that it was significantly reduced when purified from E. coli cells grown in the presence of BPS (iron‒to‒protein ratio of 0.03 ± 0.05 vs. 1.98 ± 0.41 in a protein purified from cells grown the absence of the chelator) (Figure 8C).
Figure 8.
HapX is an iron-binding protein. (A) E. coli cells were transformed with pET28 encoding HapXWT and cultured in a medium containing 1 mM IPTG (red), with or without 1 mM BPS, for 2 hr at 37°. Cells were centrifuged and cell pellets were photographed. (B) HapX purified from cells grown in a medium containing 1 mM IPTG (red), or in a medium containing 1 mM IPTG and 1 mM BPS (blue) was analyzed using the UV-visible spectrophotometer. HapX purified from cells grown in the presence of 1 mM IPTG and 1 mM BPS was treated with sodium dithionite (SDT; green), and also analyzed. (C) Iron content of HapX purified from cells grown in a medium containing 1 mM IPTG (HapX), or 1 mM IPTG and BPS (HapX+BPS). The iron content of HapX purified from cells grown in the presence of 1 mM IPTG and treated with SDT (HapX+SDT) was also determined. Three biological replicates were analyzed. **P < 0.01.
Collectively, the data present here suggested that both Cir1 and HapX bind iron directly, very likely in the form of an Fe–S cluster, and that the cysteine-rich regions of these proteins are responsible for metal binding. Therefore, we speculate that Cir1 and HapX might directly sense intracellular iron levels perhaps as part of a mechanism to coordinately regulate the expression of iron-responsive target genes.
Discussion
ChIP-seq analysis in combination with transcriptome analysis is a well-established approach to identify genes that are directly regulated by a specific transcription factor and its binding motif (Chung et al. 2014; Leach et al. 2016; Liu et al. 2016). In this study, we used ChIP-seq and transcriptome analysis to identify the direct target genes and binding motifs of the major iron regulatory transcription factors Cir1 and HapX. We also successfully constructed transcriptional regulatory network models for iron uptake and metabolism in C. neoformans. The data demonstrated and confirmed that upon iron deprivation, Cir1 controls the expression of genes encoding iron uptake functions, including the proteins for high-affinity reductive iron uptake, siderophore transport, and heme utilization. In addition to genes involved in iron uptake, Cir1 also governs genes required for various other biological pathways that may support the proliferation of C. neoformans within the host. Examples include the AAP2, AAP4, AAP5, and AAP7 genes which encode amino acid permeases required for amino acid transport, and which are known to contribute to the virulence of C. neoformans (Fernandes et al. 2015; Martho et al. 2016; Calvete et al. 2019). Enhanced expression of amino acid transporters is also observed in C. neoformans in the central nervous system of an infected rabbit (Steen et al. 2003). Furthermore, macrophages trigger cellular amino acid deprivation by upregulating amino acid degradation pathways to limit the growth of intracellular pathogenic microbes (Weiss and Schaible 2015). Thus, the previously reported and current observations suggest that Cir1 may play an essential role in the acquisition of amino acids in host niches, a role crucial for fungal survival. Our results also imply that there is a link between amino acid uptake and iron metabolism.
The AMT1 and AMT2 genes encoding ammonium transporters are also directly regulated by Cir1, thus suggesting an important regulatory role for the factor in nitrogen acquisition. Interestingly, nitrogen availability may be important for the adaptation of C. neoformans to a primary ecological niche, pigeon guano, which contains uric acid, xanthine, urea, and creatinine. Additionally, nitrogen availability is critical for fungal survival in the mammalian host environment in which nitrogen sources may be greatly limited (Staib et al. 1976, 1978; Casadevall and Perfect 1998). Nitrogen metabolism is known to play a key role in the virulence of C. neoformans, and Gat1, another GATA-type transcription factor, positively regulates genes involved in the use of various nitrogen sources. Interestingly, Gat1 also regulates the expression of amino acid permeases and ammonium transporters, implying that the coordinate regulation by both Cir1 and Gat1 may govern nitrogen acquisition (Kmetzsch et al. 2011; Lee et al. 2011, 2012).
We have previously demonstrated that deletion of CIR1 results in a deficiency in capsule formation, one of the well-known virulence factors of C. neoformans, but the underlying mechanisms are not entirely clear (Jung et al. 2006). In a key discovery from our current study, we found that Cir1 appears to directly repress the expression of genes encoding Abc1 and Tda10, protein kinases known to negatively regulate capsule formation (Lee et al. 2016). Interestingly, our finding of an intermediate capsule size for the abc1Δ cir1Δ and tda10Δ cir1Δ double deletion mutants relative to WT and cir1Δ cells suggests the possibility that Cir1 plays a positive regulatory role in capsule formation by negatively regulating Abc1 and Tda10. Additionally, it is possible that one or more additional positive regulators may be regulated by Cir1 and function in capsule formation. Other regulatory scenarios are possible including a positive regulatory influence of Cir1 on capsule formation independent of Abc1 and Tda10, as well as activities of Tda10 in the abc1Δ cir1Δ mutant, or Abc1 in the tda10Δ cir1Δ mutant, that regulate capsule size. Further analysis is needed to explore these possibilities. Overall, the major regulatory role of Cir1 in iron uptake and capsule formation together would go a long way toward explaining the avirulent phenotype of a mutant lacking CIR1 in a murine model of cryptococcosis (Jung et al. 2006). The positive regulatory role of Cir1 in iron uptake and capsule formation upon iron deprivation is unique for the GATA-type iron regulators of pathogenic fungi, and suggests that C. neoformans possesses an iron regulatory mechanism that is more broadly integrated into the pathobiology of the fungus, compared with other pathogenic fungi.
In our study, we also defined the transcriptional regulatory network of HapX in detail. The analysis revealed that HapX plays a crucial role under iron-limited conditions by transcriptionally repressing the expression of genes involved in iron-dependent pathways (e.g., mitochondrial respiration, amino acid biosynthesis, and ISC and heme biosynthesis). The negative regulatory roles of HapX in iron-dependent pathways seem to be highly conserved among orthologs from other fungi including HapX in the Aspergillus species, Hap43 in C. albicans, and Php4 in S. pombe, indicating that, generally, fungi rely on HapX orthologs for the adaptation to iron deprivation (Mercier et al. 2006; Hortschansky et al. 2007; Schrettl et al. 2010; Hsu et al. 2011).
A recent study using ChIP-seq analysis suggested that A. fumigatus HapX is required for the binding of the CBC to the promoter region of the cyp51A gene, which encodes 14-α sterol demethylase needed for ergosterol biosynthesis, thus implicating the HapX in fungal susceptibility to azole antifungal drugs (Gsaller et al. 2016). Similarly, we previously showed that deletion of HAPX results in a reduced susceptibility of C. neoformans to fluconazole (Kim et al. 2012). In the current study, we discovered that HapX directly binds to the promoter region of ERG5, which encodes C-22 desaturase involved in the ergosterol biosynthesis pathway, and negatively regulates the expression of the gene. The involvement of ERG5 in azole resistance has been observed in a number of fungi, e.g., N. crassa and Fusarium verticillioides (Sun et al. 2013). In addition, Erg5 of S. cerevisiae exhibits high affinity to azole drugs (Kelly et al. 1997). Hence, the observations presented here further support the role of HapX in the regulation of genes involved in ergosterol biosynthesis in C. neoformans, and confirm the notion that HapX-mediated alteration of azole susceptibility might be common in fungi.
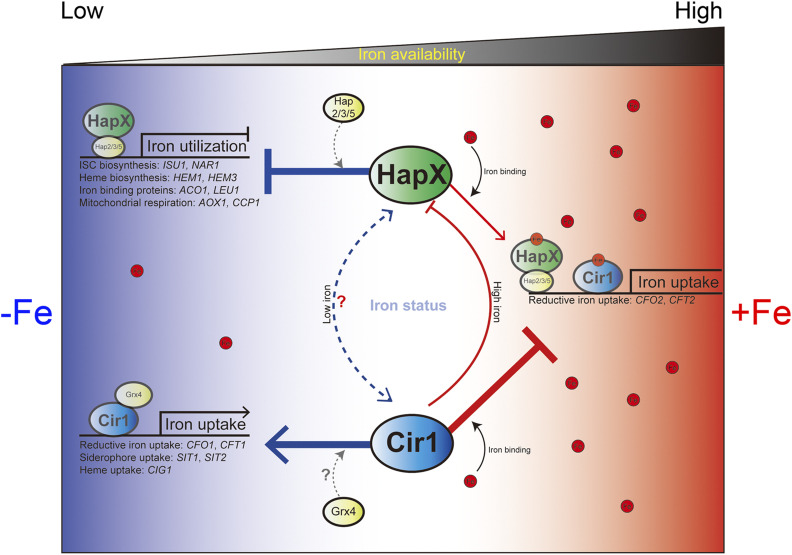
The A. fumigatus and S. pombe homologs of Cir1, SreA, and Fep1, respectively, inhibit the expression of HapX homologs under iron-replete conditions, while the HapX homologs inhibit the expression of Sre1 and Fep1 under iron-limited conditions, suggesting that the two major iron regulatory proteins reciprocally regulate one other (Haas 2012; Labbé et al. 2013). However, such reciprocal regulation is slightly different in other fungi. For example, in C. albicans, Sef1, a unique regulatory protein, intercalates between the Cir1 homolog Sfu1 and the HapX homolog Hap43, and coordinates iron regulation (Chen et al. 2011). The comprehensive data sets generated in the course of the current study revealed that the reciprocal regulation of Cir1 and HapX in response to iron availability in C. neoformans is slightly different from that in A. fumigatus, S. pombe, and C. albicans. Unlike in A. fumigatus and S. pombe, no direct binding or impact of HapX on the expression of Cir1 was observed in C. neoformans, although an indirect influence may account for our earlier finding of a 3.5 fold higher CIR1 transcript level in the hapXΔ mutant in low iron (Jung et al. 2010). By contrast, we showed here that the inhibition of HAPX transcription by Cir1 occurs directly in C. neoformans, as observed in other fungi (Figure 9).
Figure 9.
Regulatory roles of Cir1 and HapX in response to iron availability. The possible regulatory roles of Cir1 and HapX are indicated under iron-limited and iron-replete conditions. During iron deprivation, HapX forms a heteromeric complex with Hap2, Hap3, and Hap5, and represses genes involved in iron utilization (e.g., for ISC biosynthesis, heme biosynthesis, mitochondrial respiration, and iron-containing proteins). Under the same conditions, Cir1, together with Grx4, positively governs the expression of genes required for iron uptake, including reductive iron uptake, siderophore uptake, and heme uptake systems. Under iron-replete conditions, HapX incorporates iron and positively regulates reductive iron uptake genes, while iron-containing Cir1 represses the genes encoding iron uptake systems. The regulatory interactions between HapX and Cir1 are also indicated and include direct and negative control of HAPX transcription by Cir1 under the high iron condition. Direct interactions were not observed under low iron conditions, although modest and indirect regulation of Cir1 by HapX may occur (indicated by a dashed line), as suggested by previous microarray analyses (Jung et al. 2010)
We previously reported additional regulatory mechanisms at the level of protein activity, especially for Cir1 (Jung and Kronstad 2011). Specifically, we observed that Cir1 is regulated mainly by post-translation modifications and hypothesized that incorporation of an iron molecule might be critical for the stability of the protein. Moreover, we showed that CIR1 transcription is not significantly affected by iron levels (Jung and Kronstad 2011). A recent global profiling analysis of the C. neoformans phosphoproteome also showed that Cir1 is phosphorylated at amino acid positions S377, S532, S664, and S678, suggesting that protein stability and/or activity may be impacted by an as yet unknown protein kinase (Selvan et al. 2014). Our observations of the expression of the Cir1-FLAG protein are consistent with potential post-translational modification of the protein. In addition, the monothiol glutaredoxin Grx4 may also contribute to regulatory mechanisms controlled by Cir1 in C. neoformans compared to other fungi such as S. pombe. We recently observed that Grx4 binds to Cir1 under iron-depleted conditions in C. neoformans, as is observed for Grx4 and Fep1 in S. pombe. However, Grx4 relocates to the cytoplasm upon iron repletion to presumably release Cir1 to repress genes involved in iron transport, in contrast to no change in Grx4 protein localization in S. pombe (Attarian et al. 2018). Taken together, the data presented in the current and previous studies imply that a unique regulatory mechanism governs the expression and stability of Cir1, especially at the post-transcriptional level, and that a partnership with Grx4 likely contributes to the response to the availability of environmental iron in C. neoformans.
Some additional differences between the protein sequences of C. neoformans Cir1 and its homologs in other fungi are apparent. C. neoformans Cir1 contains only one N-terminal zinc-finger motif, whereas S. pombe Fep1, N. crassa SRE, A. nidulans SreA, and Ustilago maydis Urbs1 all possess two zinc-finger motifs, designated as ZnF1 and ZnF2, at their N- and C-termini, respectively (An et al. 1997; Haas et al. 1999; Zhou and Marzluf 1999; Pelletier et al. 2005). Mutational analysis of Fep1 revealed that ZnF2 is essential for DNA binding, while ZnF1 is auxiliary and increases the protein’s binding affinity to DNA (Pelletier et al. 2005; Jbel et al. 2009). In agreement with the findings for Fep1, ZnF2 rather than ZnF1 plays the major role in DNA binding in SreA, SRE, and Urbs1 (An et al. 1997; Haas et al. 1999; Zhou and Marzluf 1999; Harrison and Marzluf 2002; Pelletier et al. 2005). As mentioned, Cir1 lacks the N-terminal zinc-finger motif corresponding to ZnF1 of Cir1 homologs in other fungi. However, Cir1 contains the C-terminal zinc-finger motif corresponding to ZnF2, and, in the current study, our EMSA results showed that the motif is sufficient for binding to the promoter region of target genes. The absence of one of the zinc-finger motifs in Cir1 might shape the distinct regulatory role of the protein in C. neoformans in comparison with other fungi, although additional structural analysis is required to investigate this idea.
To summarize, we used ChIP-seq and transcriptome analysis to construct the transcriptional regulatory networks governed by the major iron regulatory transcription factors Cir1 and HapX in C. neoformans in response to iron availability. We showed that Cir1 and HapX orchestrate an iron-responsive regulatory network with the following key features. Cir1 controls the expression of genes required for iron acquisition and metabolism, as well as virulence factors, while HapX regulates the genes responsible for iron-dependent pathways, particularly under iron-depleted conditions (Figure 9). Importantly, we identified a new regulatory connection between Cir1 and the protein kinases Abc1 and Tda10 for the regulation of capsule formation. Furthermore, we identified the DNA-binding motifs of Cir1 and HapX and confirmed them by in vitro binding assays. Additionally, our biochemical analyses revealed that Cir1 and HapX are iron-containing proteins, implying that these proteins may directly sense the intracellular iron status of cells and that iron incorporation into Cir1 and HapX might impact the regulatory activities of these proteins. Further experimentation will be needed to assess the impact of iron binding on the regulatory interactions of Cir1 and HapX with target promoters. Taken together, the current study provides new insights into the regulatory networks of iron acquisition and homeostasis in C. neoformans.
Acknowledgments
We thank Aaron P. Mitchell for helpful comments and critical reading of the manuscript. This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT and Future Planning 2019R1F1A1061930 (W.H.J.), 2019R1A4A1024764 (W.H.J.), and the National Institute of Allergy and Infectious Diseases RO1 AI053721 (J.W.K.). J.W.K. is a Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology, and a Canadian Institute For Advanced Research (CIFAR) Fellow in the Fungal Kingdom: Threats & Opportunities Program. The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12151674.
Communicating editor: J. Stajich
Literature Cited
- Andrews S., 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- An Z., Zhao Q., McEvoy J., Yuan W. M., Markley J. L. et al. , 1997. The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis. Proc. Natl. Acad. Sci. USA 94: 5882–5887. 10.1073/pnas.94.11.5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attarian R., Hu G., Sanchez-Leon E., Caza M., Croll D. et al. , 2018. The monothiol glutaredoxin Grx4 regulates iron homeostasis and virulence in Cryptococcus neoformans. MBio 9: pii: e02377-18. 10.1128/mBio.02377-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa G., Caza M., Horianopoulos L., Hu G., and Kronstad J., 2019. Role of clathrin-mediated endocytosis in the use of heme and hemoglobin by the fungal pathogen Cryptococcus neoformans. Cell. Microbiol. 21: e12961 10.1111/cmi.12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault A., Mourer T., and Labbe S., 2015. Molecular basis of the regulation of iron homeostasis in fission and filamentous yeasts. IUBMB Life 67: 801–815. 10.1002/iub.1441 [DOI] [PubMed] [Google Scholar]
- Cadieux B., Lian T., Hu G., Wang J., Biondo C. et al. , 2013. The Mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J. Infect. Dis. 207: 1339–1347. 10.1093/infdis/jit029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete C. L., Martho K. F., Felizardo G., Paes A., Nunes J. M. et al. , 2019. Amino acid permeases in Cryptococcus neoformans are required for high temperature growth and virulence; and are regulated by Ras signaling. PLoS One 14: e0211393 10.1371/journal.pone.0211393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., and Perfect J. R., 1998. Cryptococcus neoformans. ASM press, Washington, DC: 10.1128/9781555818241 [DOI] [Google Scholar]
- Chao L. Y., Marletta M. A., and Rine J., 2008. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 47: 7274–7283. 10.1021/bi800066s [DOI] [PubMed] [Google Scholar]
- Chen C., Pande K., French S. D., Tuch B. B., and Noble S. M., 2011. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe 10: 118–135. 10.1016/j.chom.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D., Barker B. M., Carey C. C., Merriman B., Werner E. R. et al. , 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 10: e1004487 (erratum: PLoS Pathog. 10: e1004576). 10.1371/journal.ppat.1004487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlouhy A. C., Beaudoin J., Labbe S., and Outten C. E., 2017. Schizosaccharomyces pombe Grx4 regulates the transcriptional repressor Php4 via [2Fe-2S] cluster binding. Metallomics 9: 1096–1105. 10.1039/C7MT00144D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do E., Hu G., Caza M., Oliveira D., Kronstad J. W. et al. , 2015. Leu1 plays a role in iron metabolism and is required for virulence in Cryptococcus neoformans. Fungal Genet. Biol. 75: 11–19. 10.1016/j.fgb.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinar del Dedo J., Gabrielli N., Carmona M., Ayte J., and Hidalgo E., 2015. A cascade of iron-containing proteins governs the genetic iron starvation response to promote iron uptake and inhibit iron storage in fission yeast. PLoS Genet. 11: e1005106 10.1371/journal.pgen.1005106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J. D., Martho K., Tofik V., Vallim M. A., and Pascon R. C., 2015. The role of amino acid permeases and tryptophan biosynthesis in Cryptococcus neoformans survival. PLoS One 10: e0132369 10.1371/journal.pone.0132369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J. A., Subaran R. L., Nichols C. B., and Heitman J., 2003. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell 2: 1036–1045. 10.1128/EC.2.5.1036-1045.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T., and Nemeth E., 2015. Iron homeostasis in host defence and inflammation. Nat. Rev. Immunol. 15: 500–510. 10.1038/nri3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Beattie S. R., Klammer V., Tuppatsch K. et al. , 2014. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 33: 2261–2276. 10.15252/embj.201489468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Furukawa T., Carr P. D., Rash B. et al. , 2016. Correction: sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog. 12: e1006106 10.1371/journal.ppat.1006106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., 2012. Iron - a key nexus in the virulence of Aspergillus fumigatus. Front. Microbiol. 3: 28 10.3389/fmicb.2012.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas H., Zadra I., Stoffler G., and Angermayr K., 1999. The Aspergillus nidulans GATA factor SREA is involved in regulation of siderophore biosynthesis and control of iron uptake. J. Biol. Chem. 274: 4613–4619. 10.1074/jbc.274.8.4613 [DOI] [PubMed] [Google Scholar]
- Halliwell B., and Gutteridge J. M., 1984. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 219: 1–14. 10.1042/bj2190001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Do E., and Jung W. H., 2012. A human fungal pathogen Cryptococcus neoformans expresses three distinct iron permease homologs. J. Microbiol. Biotechnol. 22: 1644–1652. 10.4014/jmb.1209.09019 [DOI] [PubMed] [Google Scholar]
- Harrison K. A., and Marzluf G. A., 2002. Characterization of DNA binding and the cysteine rich region of SRE, a GATA factor in Neurospora crassa involved in siderophore synthesis. Biochemistry 41: 15288–15295. 10.1021/bi0204995 [DOI] [PubMed] [Google Scholar]
- Hortschansky P., Eisendle M., Al-Abdallah Q., Schmidt A. D., Bergmann S. et al. , 2007. Interaction of HapX with the CCAAT-binding complex–a novel mechanism of gene regulation by iron. EMBO J. 26: 3157–3168. 10.1038/sj.emboj.7601752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. C., Yang C. Y., and Lan C. Y., 2011. Candida albicans Hap43 is a repressor induced under low-iron conditions and is essential for iron-responsive transcriptional regulation and virulence. Eukaryot. Cell 10: 207–225. 10.1128/EC.00158-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Caza M., Cadieux B., Chan V., Liu V. et al. , 2013. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 81: 292–302. 10.1128/IAI.01037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Caza M., Cadieux B., Bakkeren E., Do E. et al. , 2015. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol. Microbiol. 96: 973–992. 10.1111/mmi.12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G., Ormerod K. L., Paulet D., Byrnes E. J. 3rd, Yadav V. et al. , 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10: e1004261 10.1371/journal.pgen.1004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbel M., Mercier A., Pelletier B., Beaudoin J., and Labbe S., 2009. Iron activates in vivo DNA binding of Schizosaccharomyces pombe transcription factor Fep1 through its amino-terminal region. Eukaryot. Cell 8: 649–664. 10.1128/EC.00001-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., and Kronstad J. W., 2011. Iron influences the abundance of the iron regulatory protein Cir1 in the fungal pathogen Cryptococcus neoformans. FEBS Lett. 585: 3342–3347. 10.1016/j.febslet.2011.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Sham A., White R., and Kronstad J. W., 2006. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 4: e410 10.1371/journal.pbio.0040410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Sham A., Lian T., Singh A., Kosman D. J. et al. , 2008. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 4: e45 10.1371/journal.ppat.0040045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Hu G., Kuo W., and Kronstad J. W., 2009. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot. Cell 8: 1511–1520. 10.1128/EC.00166-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Saikia S., Hu G., Wang J., Fung C. K. et al. , 2010. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 6: e1001209 10.1371/journal.ppat.1001209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. W., Yang D. H., Maeng S., Lee K. T., So Y. S. et al. , 2015. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 6: 6757 10.1038/ncomms7757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Aoyama A., Naruse F., Kobayashi T., and Tsukagoshi N., 1997. An Aspergillus nidulans nuclear protein, AnCP, involved in enhancement of Taka-amylase A gene expression, binds to the CCAAT-containing taaG2, amdS, and gatA promoters. Mol. Gen. Genet. 254: 119–126. 10.1007/s004380050399 [DOI] [PubMed] [Google Scholar]
- Kehl-Fie T. E., and Skaar E. P., 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14: 218–224. 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S. L., Lamb D. C., Baldwin B. C., Corran A. J., and Kelly D. E., 1997. Characterization of Saccharomyces cerevisiae CYP61, sterol delta22-desaturase, and inhibition by azole antifungal agents. J. Biol. Chem. 272: 9986–9988. 10.1074/jbc.272.15.9986 [DOI] [PubMed] [Google Scholar]
- Kim J., Cho Y. J., Do E., Choi J., Hu G. et al. , 2012. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet. Biol. 49: 955–966. 10.1016/j.fgb.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Lee K. L., Kim K. D., and Roe J. H., 2016. The iron uptake repressor Fep1 in the fission yeast binds Fe-S cluster through conserved cysteines. Biochem. Biophys. Res. Commun. 478: 187–192. 10.1016/j.bbrc.2016.07.070 [DOI] [PubMed] [Google Scholar]
- Kmetzsch L., Staats C. C., Simon E., Fonseca F. L., Oliveira D. L. et al. , 2011. The GATA-type transcriptional activator Gat1 regulates nitrogen uptake and metabolism in the human pathogen Cryptococcus neoformans. Fungal Genet. Biol. 48: 192–199. 10.1016/j.fgb.2010.07.011 [DOI] [PubMed] [Google Scholar]
- Kronstad J., Saikia S., Nielson E. D., Kretschmer M., Jung W. et al. , 2012. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot. Cell 11: 109–118. 10.1128/EC.05273-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Yang M., Haynes B. C., Skowyra M. L., and Doering T. L., 2011. Emerging themes in cryptococcal capsule synthesis. Curr. Opin. Struct. Biol. 21: 597–602. 10.1016/j.sbi.2011.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S., Khan M. G., and Jacques J. F., 2013. Iron uptake and regulation in Schizosaccharomyces pombe. Curr. Opin. Microbiol. 16: 669–676. 10.1016/j.mib.2013.07.007 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan C. Y., Rodarte G., Murillo L. A., Jones T., Davis R. W. et al. , 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53: 1451–1469. 10.1111/j.1365-2958.2004.04214.x [DOI] [PubMed] [Google Scholar]
- Leach M. D., Farrer R. A., Tan K., Miao Z., Walker L. A. et al. , 2016. Hsf1 and Hsp90 orchestrate temperature-dependent global transcriptional remodelling and chromatin architecture in Candida albicans. Nat. Commun. 7: 11704 10.1038/ncomms11704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. R., Chow E. W., Morrow C. A., Djordjevic J. T., and Fraser J. A., 2011. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 188: 309–323. 10.1534/genetics.111.128538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. R., Lim J. W., Ormerod K. L., Morrow C. A., and Fraser J. A., 2012. Characterization of an Nmr homolog that modulates GATA factor-mediated nitrogen metabolite repression in Cryptococcus neoformans. PLoS One 7: e32585 10.1371/journal.pone.0032585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. T., So Y. S., Yang D. H., Jung K. W., Choi J. et al. , 2016. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. 7: 12766 10.1038/ncomms12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. Y., and Nizet V., 2009. Color me bad: microbial pigments as virulence factors. Trends Microbiol. 17: 406–413. 10.1016/j.tim.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Bergenholm D., and Nielsen J., 2016. Genome-wide mapping of binding sites reveals multiple biological functions of the transcription factor Cst6p in Saccharomyces cerevisiae. MBio 7: pii: e00559-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machanick P., and Bailey T. L., 2011. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martho K. F. C., de Melo A. T., Takahashi J. P. F., Guerra J. M., Santos D. C. D. et al. , 2016. Amino acid permeases and virulence in Cryptococcus neoformans. PLoS One 11: e0163919 10.1371/journal.pone.0163919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier A., Pelletier B., and Labbe S., 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 5: 1866–1881. 10.1128/EC.00199-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej J., Thiebaut A., Blugeon C., Pouch J., Ali Chaouche Mel A. et al. , 2016. A network of paralogous stress response transcription factors in the human pathogen Candida glabrata. Front. Microbiol. 7: 645 10.3389/fmicb.2016.00645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashige T. G., Zhang B., Krebs C., and Nolan E. M., 2015. Human calprotectin is an iron-sequestering host-defense protein. Nat. Chem. Biol. 11: 765–771. 10.1038/nchembio.1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S. M., 2013. Candida albicans specializations for iron homeostasis: from commensalism to virulence. Curr. Opin. Microbiol. 16: 708–715. 10.1016/j.mib.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Lee Y., Bhupindersingh G., and Iyer V. R., 2013. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One 8: e83506 10.1371/journal.pone.0083506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier B., Beaudoin J., Mukai Y., and Labbe S., 2002. Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J. Biol. Chem. 277: 22950–22958. 10.1074/jbc.M202682200 [DOI] [PubMed] [Google Scholar]
- Pelletier B., Beaudoin J., Philpott C. C., and Labbe S., 2003. Fep1 represses expression of the fission yeast Schizosaccharomyces pombe siderophore-iron transport system. Nucleic Acids Res. 31: 4332–4344. 10.1093/nar/gkg647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier B., Trott A., Morano K. A., and Labbe S., 2005. Functional characterization of the iron-regulatory transcription factor Fep1 from Schizosaccharomyces pombe. J. Biol. Chem. 280: 25146–25161. 10.1074/jbc.M502947200 [DOI] [PubMed] [Google Scholar]
- Pelletier B., Mercier A., Durand M., Peter C., Jbel M. et al. , 2007. Expression of Candida albicans Sfu1 in fission yeast complements the loss of the iron-regulatory transcription factor Fep1 and requires Tup co-repressors. Yeast 24: 883–900. 10.1002/yea.1539 [DOI] [PubMed] [Google Scholar]
- Priebe S., Kreisel C., Horn F., Guthke R., and Linde J., 2015. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31: 445–446. 10.1093/bioinformatics/btu627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Hu Y., Xu F., Yalamanchili H. K., and Wang J., 2014. Inferring gene regulatory networks by integrating ChIP-seq/chip and transcriptome data via LASSO-type regularization methods. Methods 67: 294–303. 10.1016/j.ymeth.2014.03.006 [DOI] [PubMed] [Google Scholar]
- Rietzschel N., Pierik A. J., Bill E., Lill R., and Muhlenhoff U., 2015. The basic leucine zipper stress response regulator Yap5 senses high-iron conditions by coordination of [2Fe-2S] clusters. Mol. Cell. Biol. 35: 370–378. 10.1128/MCB.01033-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S. et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rye M. B., Saetrom P., and Drablos F., 2011. A manually curated ChIP-seq benchmark demonstrates room for improvement in current peak-finder programs. Nucleic Acids Res. 39: e25 10.1093/nar/gkq1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S., Oliveira D., Hu G., and Kronstad J., 2014. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect. Immun. 82: 839–850. 10.1128/IAI.01357-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., 2001. Molecular cloning: a laboratory manual / Joseph Sambrook, David W. Russell, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. [Google Scholar]
- Schrettl M., Kim H. S., Eisendle M., Kragl C., Nierman W. C. et al. , 2008. SreA-mediated iron regulation in Aspergillus fumigatus. Mol. Microbiol. 70: 27–43. 10.1111/j.1365-2958.2008.06376.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrettl M., Beckmann N., Varga J., Heinekamp T., Jacobsen I. D. et al. , 2010. HapX-mediated adaption to iron starvation is crucial for virulence of Aspergillus fumigatus. PLoS Pathog. 6: e1001124 10.1371/journal.ppat.1001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvan L. D., Renuse S., Kaviyil J. E., Sharma J., Pinto S. M. et al. , 2014. Phosphoproteome of Cryptococcus neoformans. J. Proteomics 97: 287–295. 10.1016/j.jprot.2013.06.029 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T. et al. , 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib F., Mishra S. K., Able T., and Blisse A., 1976. Growth of Cryptococcus neoformans on uric acid agar. Zentralbl. Bakteriol. Orig. A 236: 374–385. [PubMed] [Google Scholar]
- Staib F., Grave B., Altmann L., Mishra S. K., Abel T. et al. , 1978. Epidemiology of Cryptococcus neoformans. Mycopathologia 65: 73–76. 10.1007/BF00447178 [DOI] [PubMed] [Google Scholar]
- Steen B. R., Zuyderduyn S., Toffaletti D. L., Marra M., Jones S. J. et al. , 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2: 1336–1349. 10.1128/EC.2.6.1336-1349.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Wang W., Wang K., Yu X., Liu J. et al. , 2013. Sterol C-22 desaturase ERG5 mediates the sensitivity to antifungal azoles in Neurospora crassa and Fusarium verticillioides. Front. Microbiol. 4: 127 10.3389/fmicb.2013.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangen K. L., Jung W. H., Sham A. P., Lian T., and Kronstad J. W., 2007. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 153: 29–41. 10.1099/mic.0.2006/000927-0 [DOI] [PubMed] [Google Scholar]
- Teytelman L., Thurtle D. M., Rine J., and van Oudenaarden A., 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 110: 18602–18607. 10.1073/pnas.1316064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., and Cherayil B. J., 2009. Ironing out the wrinkles in host defense: interactions between iron homeostasis and innate immunity. J. Innate Immun. 1: 455–464. 10.1159/000210016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G., and Schaible U. E., 2015. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 264: 182–203. 10.1111/imr.12266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S. et al. , 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., and Marzluf G. A., 1999. Functional analysis of the two zinc fingers of SRE, a GATA-type factor that negatively regulates siderophore synthesis in Neurospora crassa. Biochemistry 38: 4335–4341. 10.1021/bi982543f [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. The ChIP-seq and transcriptome data have been deposited at the Gene Expression Omnibus database of the National Center for Biotechnology Information under the SuperSeries record accession no. GSE126314. The accession number for the ChIP-seq data are GSE126312, and that for the RNA-seq data are GSE126313. R code and sample data for DESeq2 analysis can be found in Supplemental Materials. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12151674.