Abstract
Peptide Nucleic Acid (PNA) are DNA/RNA synthetic analogs with 2‐([2‐aminoethyl] amino) acetic acid backbone. They partake unique antisense and antigene properties, just due to its inhibitory effect on transcription and translation; they also undergo complementary binding to RNA/DNA with high affinity and specificity. Hence, to date, many methods utilizing PNA for diagnosis and treatment of various diseases namely cancer, AIDS, human papillomavirus, and so on, have been designed and developed. They are being used widely in polymerase chain reaction modulation/mutation, fluorescent in‐situ hybridization, and in microarray as a probe; they are also utilized in many in‐vitro and in‐vivo assays and for developing micro and nano‐sized biosensor/chip/array technologies. Earlier reviews, focused only on PNA properties, structure, and modifications related to diagnostics and therapeutics; our review emphasizes on PNA properties and synthesis along with its potential applications in diagnosis and therapeutics. Furthermore, prospects in biomedical applications of PNAs are being discussed in depth.
Keywords: Biomedical application, Biosensors, Diagnosis, PNA properties , PNA, Therapeutics
Peptide nucleic acids are DNA/RNA synthetic analogs. They show unique stability and hybridization properties and have enormous biomedical applications for diagnostics (utilizing biosensors, microarray, and so on) and therapeutics. Hence, this review emphasizes the PNAs biomedical application along with its properties, synthesis, and prospects.
1. INTRODUCTION
Peptide nucleic acid (PNA) is a synthetic analog of DNA/RNA (Figure 1) with 2‐([2‐Aminoethyl] amino) acetic acid backbone, which results in achiral and uncharged mimic. It is regarded as DNA but has a neutral peptide backbone on the place of a negatively charged sugar‐phosphate backbone and due to their unique physical and chemical properties such as resistant to enzymatic degradation inside living cells have offered them with extensive consideration in various fields for application. 1 , 2 , 3 It has shown promising applications as a biomolecular tool, antisense and antigene agent, molecular probes, and biosensors. 4 Nielsen et al in 1991 designed a polyamide nucleic acid, by replacing the DNA backbone with an achiral polyamide backbone and found that in double‐stranded DNA these oligomers recognize their complementary target by strand displacement and the reason behind this displacement was due to the extremely high stability of the PNA‐DNA hybrids. 5 Though they still have stability complications 6 that can be improved by blocking or acetylating the N terminus, and apart from this, they are highly sequence‐dependent.
FIGURE 1.
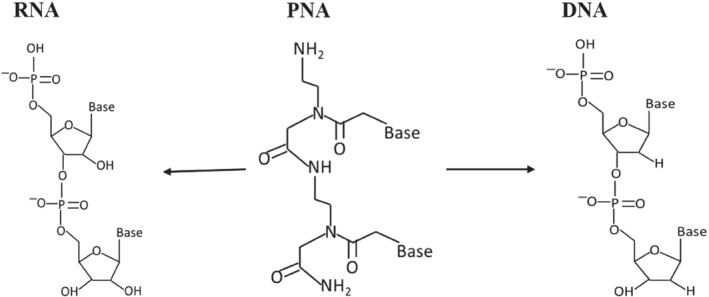
Structural comparison of PNA with RNA and DNA
PNA is chemically stable than its biological counterpart's DNA and RNA, as they are polyamide‐based synthetic molecules, which are highly stable in an acidic environment, and moderately stable in basic conditions as well at high temperatures. They are also biochemically stable as they do not act as a substrate for peptidases, proteases, and nucleases, but on the contrary, their native nucleic acids are highly sensitive to these enzymes; due to PNAs well‐known biological and chemical stability. It facilitates synthesis, storage, purification, and applications. They also show excellent stability over a wide range of pH, but DNA and RNA are sensitive to these changes, and it also discriminates DNA or RNA in a single‐point mismatch. Thus, it acts as a more specific sequence binder. DNA/RNA is negatively charged, on the other hand, PNA lacks charges, and as a result of this during hybridization of PNA with DNA or RNA strands, PNA lacks electrostatic repulsion, which offers PNA a greater affinity towards its target. Introducing positive charge in PNA could be more suitable for the formation and stability of triplex and duplex helices. At higher salt concentration DNA‐DNA duplexes are generally stable while PNA‐DNA duplexes are highly stable in this condition. Hence, PNA is better alternative for their native biological counterparts for wide range of applications. 7 , 8
PNA is composed of adenine, uracil, guanine, and cytosine‐N‐acetic acids which offers them to be a capable precursor to RNA, as there are a lot of problems that exist with present thinking of RNA as genetic material; as RNA is known to be the primary genetic molecule prior to DNA‐based organisms on earth over 3.5 billion years ago. But following the researches and ease in the synthesis of PNA components and possibility in the polymerization of AEG (N‐[2‐aminoethyl] glycine) supports the hypothesis that PNA may be the first genetic material and may have been used before RNA‐based organisms arose, to transmit genetic information in primitive life. 9 Thus, the study conducted by Banack et al 10 further confirmed that cyanobacteria produces PNA backbone, and they detected AEG in axenic strains as well as environmental samples of cyanobacteria. Hence, the production by diverse taxa of cyanobacteria of AEG suggests that it may be a primitive feature that resulted early in the development of life on earth.
PNA can recognize specific sequences of DNA and RNA by obeying the Watson‐Crick hydrogen bonding arrangement, and these hybrid complexes exhibit unique ionic strength and unexpected thermal stability. 2 These synthetic analogs display a wide range of applications in the biomedical domain. Along with a variety of applications, they also have few shortcomings such as issues with cellular uptake which can be dodged by modification of PNA or by making conjugates of PNA with cell‐penetrating peptides (CPPs). Thus, these modifications help them to improve their delivery inside the cells and offer them an application for targeted drug delivery. 11 Nucleic acid biosensors have proved applications in genetics and biomedicine; PNA specifically can be used in the development of high‐performance affinity biosensors for applicability in DNA genotyping, though little has been studied about sensitivity and specificity. 12 PNA was invented 28 years ago and has expanded briskly as helpful molecules in antisense technology and DNA hybridization. It is also used in diagnostics (in‐situ hybridization and polymerase chain reaction [PCR]‐based systems), and accompanies unique properties that have enlightened synthetic organic chemists to design and prepare other derivatives of these analogs with more refined properties. 13
PNAs have the potential to be used as novel antibiotics, gene‐activating agents, and also as molecular probes in techniques like fluorescent in‐situ hybridization (FISH) for imaging and in biosensors for diagnostic purposes. 14 PNA shows promising applications in detection techniques; wherein hybridization technologies based on FISH and novel approaches like matrix‐assisted laser desorption/lonization‐time of flight (MALDI‐TOF) utilizing PNA have gained lot of attention. PNA was a breakthrough among several efforts made to date for the development of gene therapeutic. Both in eukaryotes and prokaryotes, antisense activity of PNA as peptide conjugate has been reported with a positive result in the nerve cell‐line and in rat brain. 15 Furthermore, the significance of single‐cell biology in basic research and translational medicine is growing at a faster pace; in which efficacious detection and isolation of specific subclasses using intracellular markers (IcM) are key for understanding their functional heterogeneity. The use of peptide nucleic acid probes (PNPs) as an IcMs against certain microRNA for cancer surgical margin predictions are gaining attention. 16 Though nucleic acid analogs containing natural nucleobases on a modified backbone have been synthesized such as xeno‐nucleic acids, threose nucleic acids, locked nucleic acids (LNA), morpholinos, and so on, along with PNA, 17 , 18 , 19 , 20 , 21 among all these designed oligonucleotides, PNA is a remarkable nucleic acid mimic as it is neither a nucleic acids nor a peptide. 22 Furthermore, several reviews exist discussing PNA properties, 23 chemistry, 24 , 25 and its modifications 26 , 27 , 28 with details related to diagnostics and therapeutics, our review (Figure 2) specifically focuses on PNAs biomedical applications along with their properties, synthesis, and prospects.
FIGURE 2.
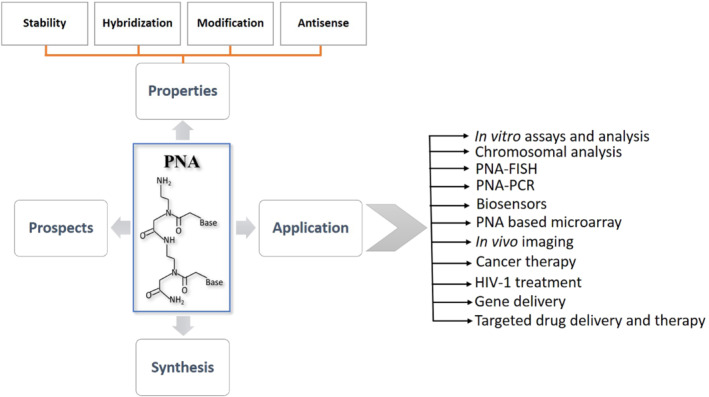
Diagrammatic illustration of details emphasized in this review
2. PROPERTIES
Properties of PNAs offer vast applications in the field of biomedical research, including diagnosis using in vivo imaging and biosensors; in therapeutics used for antisense and antigene therapies 11 which are elaborated in this section. Remarkable properties which are elaborated in this section.
2.1. Stability of PNAs
The complex structure of PNAs makes them more resistant to enzymes/chemicals such as DNases and proteinases, hence they do not degrade inside the cell and have high biostability which makes them a good candidate for in‐vivo study. Although its transportation within the cell is difficult due to its limited lipid membrane diffusion. 2 , 11 , 17 , 29 An experiment by Eriksson et al 6 suggests that PNA with thymine monomer at 11 pH with 37 days half‐life undergoes N‐acyl transfer rearrangement, and at pH 9 or above they exhibit sequential degradation and N‐acyl transfer reactions. They have a high binding affinity and are specific to DNA/RNA in targeting due to their complex structure and stability, this property provides PNA as a choice for gene therapy‐based applications. These modifications provide more stability, thus, chemical alterations in combination with nanotechnology can help PNAs to improve their intracellular delivery, which was previously a significant challenge. 30
2.2. Hybridization of PNA
PNA hybridize at low ion concentration which exhibit remarkable stability towards complimentary oligonucleotides due to the fact that it does not show electrostatic repulsion and has flexible and neutral backbone. 11 , 17 PNA was designed to undergo Hoogsteen base pairing with dsDNA (double stranded DNA), and is proven to show effective DNA mimic with ability to partake duplex and triplex hybridization. 31 Its hybrid complexes exhibit unique thermal stability and ionic strengths. 2 PNA‐DNA duplex, properties are mostly studied in H‐TGTACGTCACAACTAA‐NH2 (a penta‐decamer). 32 , 33 PNA‐PNA, PNA‐DNA, and DNA‐DNA have melting temperature (T m) of 67°C, 51°C, and 33.5°C respectively. Among these duplex hybrids, PNA‐PNA is most stable. 34 The DNA‐PNA or RNA‐DNA are more stable than the DNA‐DNA or RNA‐DNA duplexes due to the reason of high T m values; typically, T m value of 15‐mer PNA‐DNA duplex is approx. 70°C, and on the other hand T m of DNA‐DNA duplex is approx. 50°C at pH 7, and a general rule suggests that T m value of PNA‐DNA duplex is 1°C higher per base pair when compared with the T m values of DNA‐DNA duplex in 100 mmol dm−3 sodium chloride. Table 1 gives the comparison in a generalized manner about the T m values of 15‐mer RNA and DNA based duplex formation of PNA. 2 (PNA)2‐DNA is formed by homopyrimidine PNA oligomers/PNA with high pyrimidine: purine ratio, via binding with complementary DNA. These triplexes are very stable complexes with T m > 70°C (for decamer hybrid), binding of these complexes is governed by Hoogsteen and Watson‐crick base pairing (Figure 3). 5 , 35 , 36 , 37 , 38 PNA‐DNA duplex for forming PNA‐DNA‐PNA/(PNA)2‐DNA triplex structure can be performed by strand invasion, in which one DNA strand is replaced by PNA of duplex complex but the replaced strand of DNA exists as D‐loop (single stranded). 39
TABLE 1.
General example of T m values of 15‐mer duplex complexes formed by PNA, RNA, and DNA
| Hybrids | Formation of duplexes (15‐mer) | T m |
|---|---|---|
| RNA | RNA‐PNA | 72°C |
| RNA–DNA | 50°C | |
| DNA | DNA‐PNA | 69°C |
| DNA–DNA | 54°C |
FIGURE 3.
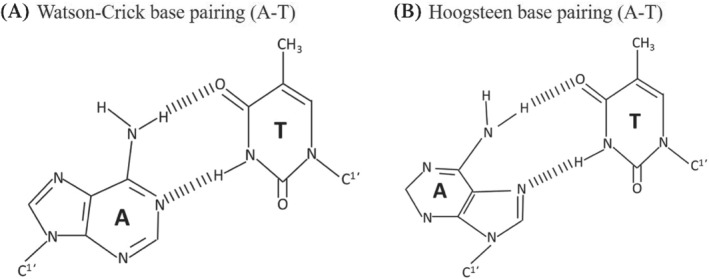
Difference between Watson‐crick and Hoogsteen base pairing in adenine–thymine (A‐T)
2.3. PNA modifications
Limited cellular uptake, poor membrane permeability, low aqueous solubility, and ambiguity in DNA binding orientation of PNA was a major challenge to use it as an antisense agent for in‐vivo/vitro studies. 40 Hence, chemical alterations in PNA backbone improved the antisense properties along with many other properties of PNA for application in diagnostics, medicine, and molecular biology. PNA modifications by using different strategies such as introducing chirality into achiral PNA backbone, adding cationic functional group (to improve aqueous solubility), modifying linker/nucleobase (to control DNA/RNA binding at physiological conditions), and so on, are being considered. Furthermore, these modifications will result in better solubility in aqueous, enhanced cellular uptake, and many other properties; which will make it a better tool for biomedical applications. 11 Apart from PNA backbone modifications, it can also be synthesized using artificial nucleobases, namely 2,6 diaminopurine, pseudoisocytosine, 2‐aminopurine, thiazole, hypoxanthine, thiouracil, and so on, which gives PNA unique feature such as increased affinity, selectivity for thymine, as a probe for detection of hybridization through fluorescence, and so on, these are just few examples with features of artificial nucleobases introduced in PNA. 41 , 42 , 43 , 44 , 45 PNA backbone and nucleobase modification will result in enhanced properties of PNA which will provide new applications to the PNA with several features (Table 2). Hnedzko et al 52 synthesized (M) modified PNA and found that, PNA are well‐suited ligands for dsRNA recognition in live cells and other biological systems. Hence, (M) modified PNA conjugates are promising probes, show excellent cellular uptake, and very less cytotoxicity; owing to wide range of biomedical applications of PNA.
TABLE 2.
Modification of PNA for enhancing their properties. (A) Backbone modified PNA, (B) nucleobase modified PNA
| S. No. | General name | Structure | Enhanced properties | Reference |
|---|---|---|---|---|
| (A) Backbone modified PNA | ||||
| 1 | Phosphono PNA |
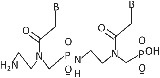
|
Antisense activity. | 46 |
| 2 | α‐Guanylated PNA |
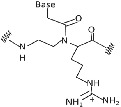
|
Cellular uptake properties while maintaining Watson‐Crick recognition with complementary DNA strand. | 47, 48 |
| 3 | α‐Amino methylene PNA |
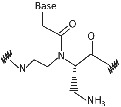
|
Cellular uptake. | 49, 50 |
| 4 | ϒ‐Amino methylene PNA |
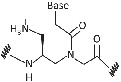
|
PNA‐DNA binding. | 49, 50 |
| 5 | Diethylene glycol PNA |
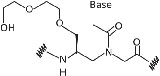
|
Water solubility and improved hybridization properties. | 51 |
| (B) Nucleobase modified PNA | ||||
| 1 | 2,6‐Diaminopurine |
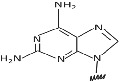
|
Affinity and selectivity for thymine. | 41 |
| 2 | Pseudoisocytosin |
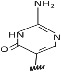
|
Mimics the C+ recognition pattern for triplex formation irrespective of surrounding pH. | 41 |
| 3 | 2‐Aminopurine |
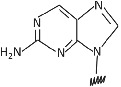
|
Hydrogen bond with uracil and thymine in the reverse Watson‐Crick mode and being inherently fluorescent, which can be used to study the kinetics of the hybridization process with complementary nucleic acids. | 42 |
| 4 | Thiazole |
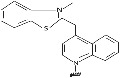
|
Forms PNA probe that fluoresced upon hybridization. | 43, 44 |
| 5 | Hypoxanthine |
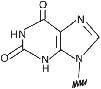
|
Form Watson‐Crick base pairs with A, C, T, and U and achieve multimutant specificity. | 45 |
| 6 | Thiouracil |
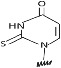
|
Invades dsDNA in antigene applications. | 41 |
2.4. PNA antisense properties
PNA exhibit several properties desirable to be a good antisense agent. 40 They identify DNA duplex homopurine sequences to which it binds by strand invasion, forming a very stable (PNA)2‐DNA triplex complex. In vitro studies indicate that PNA inhibits transcription and translation, and undergo complementary binding to RNA and DNA with high affinity and specificity; hence, these properties make PNA a “third generation” antisense and antigene agent. Owing to its superior properties, it can become a probe by replacing DNA for numerous investigation purposes. 2 , 3 , 53 Among several delivery strategies explored till date, to improve the antisense properties of PNA, nanotechnology‐based strategies hold enormous potential. From prior studies, advances in nanotechnology by using nanoparticle‐based on polymer, carbon, and so on, for the delivery of PNA will improve the therapeutic applications of PNA. Hence, nanoparticle delivery of PNA can be implied in gene targeting (antisense) based strategies. 30 The synthesized PNA exhibits enormous properties which make them a good antisense agent but they can be modified to enhance its antisense properties; one such example is Phosphono‐PNA (backbone modification), the added monomer gives the PNA improved antisense activity. 46
3. SYNTHESIS OF PNA
The pivotal requirement in PNA‐based research is its synthesis to form an oligomer. 7 , 54 PNA can be synthesized by self‐assembly and self‐organization process, and these processes are very versatile and easy, which produces a hybrid complex and also exhibits unique physicochemical and specific recognition properties. PNA monomer contains N‐protected AEG (Y), to which (X) protected nucleobase is attached (Figure 4A). Y and X are protecting groups in PNA and should be orthogonal that is X should be stable when Y is removed. 55 PNA monomers are synthesized can be performed by convergent method. 56 In this a suitable protected base and N‐protected AEG is initially synthesized, then their backbone is coupled with PNA nucleobase to get PNA monomer of interest (Figure 4B).
FIGURE 4.
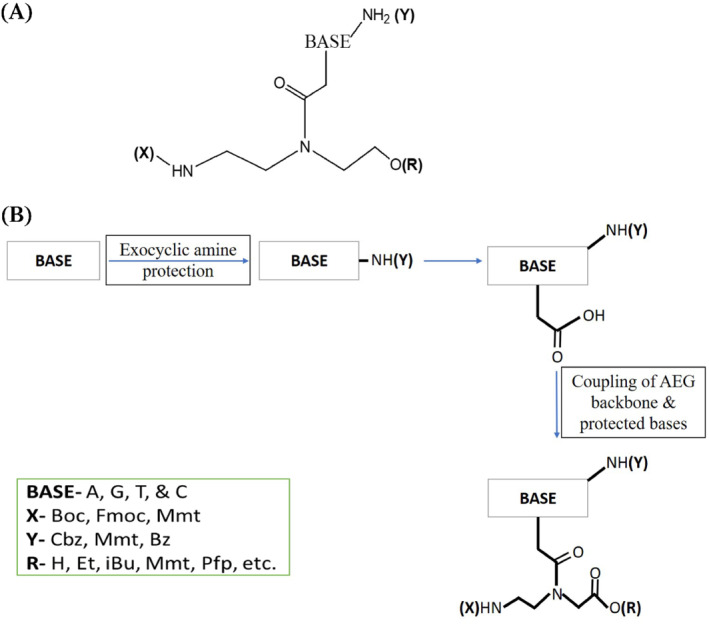
(A) General PNA monomer schematic illustration; (B) diagrammatic illustration explaining the synthesis procedure of PNA monomer
The protecting groups are always used in combination for the synthesis of PNA, to name a few group combinations are Boc‐Cbz, Fmoc‐Bhoc, Mmt‐Acyl, and Fmoc‐Mmt. Boc‐Cbz and Fmoc‐Bhoc PNA monomers are commercially available by Applied Biosystems (Foster City, United States) and can be used for PNA oligomer synthesis. Panageneⓒ is an international supplier of PNA since 2006, and the protocol utilizes a solid support via automated synthesis of PNA oligomer. Synthesis of PNA is a cyclic process and depends totally on the application for which the PNA is required. The synthesis of PNA oligomer starts by coupling of PNA monomers, then capping by acetic anhydride/lutidine, by concluding with deprotection (Figure 5). On the contrary, synthesis of Fmoc PNA monomer though was not cost‐effective, but face difficulties in mass production, needs a coupling reagent, and has purification complications leading to low yield. The solution to this was brought forward by Panageneⓒ by synthesizing Bts monomer, as the synthesis of this was cost‐effective, and does not require coupling reagent, has ease in mass production, and easy purification process leading to high yield. 57
FIGURE 5.
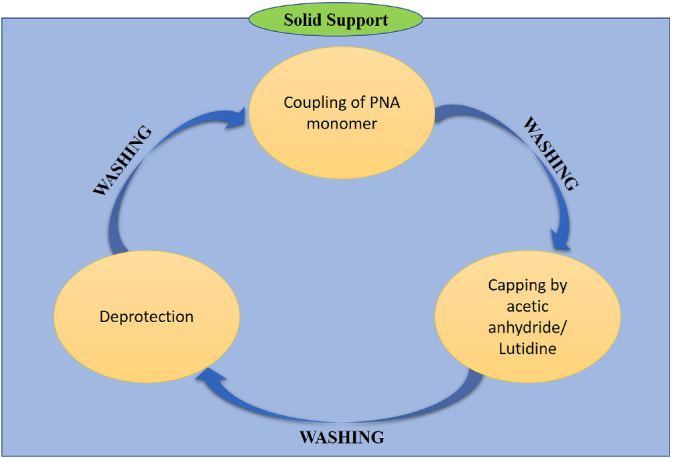
Diagrammatic illustration of basic solid‐phase PNA oligomer synthesis protocol
4. APPLICATIONS OF PNA
As PNA has better chemical, physical, and biological properties relative to nucleic acids, they find applications in various fields while showing promising development of diagnostic devices, therapeutic agent, molecular tool for nucleic acid manipulations, and as tool in biomedical applications since its discovery in 1991. 2 , 3 , 58 PNA is synthesized both at nano‐ and micro‐scale levels and have potential applications in various fields for making nanostructured devices for biochips, sensors, and microarrays. 59 Beavers et al 60 in their study reported the in‐situ synthesis of PNA from porous silicon (PSi) and confirmed the establishment of a novel PNA‐PSi platform with broad utility in drug delivery and biosensing. Ma et al 61 have explored CPPs‐PNAs by antisense inhibition of intracellular bacteria, and their results suggested that electroporation is the best approach for CPP‐based delivery. PNA exhibits potential antisense applications in biomedical field as they hybridize with neutral nucleic acids to form complexes and show enhanced biochemical properties. Thus, they have emerged as a tool for cancer diagnosis and therapy by utilizing a combination of nanobiotechnology with PNA biotechnology. PNAs have also proved to have potential applications in the cytogenetic and genetic diagnostic procedures. 22 Furthermore, PNA‐based nanoparticles can also be used to improve the efficiency of drug delivery in anticancer therapy. 8
4.1. PNA applications in diagnosis
4.1.1. In‐vitro assays and analysis
PNAs unique physicochemical properties have led to the development of in‐vitro diagnostic assays for advance routine clinical tests and environmental monitoring utilizing PNA technology. Apart from these applications PNAs also has an impact on other fields of cytogenetics and industrial microbiology. 62 Su et al 63 have developed an enzyme‐based colorimetric assay for nucleic acids utilizing PNA as a probe and have validated the assay on different DNA as well as on total RNA samples extracted from two human cancer cell lines of lung and HeLa cell line for detecting miRNA. This study signifies PNA assays have better detection and sensitivity, introduces additional robustness, and avoids labeling of sample, making them better assays over other colorimetric analyses. Furthermore, there are many more in‐vitro diagnosis techniques that utilize PNA, namely chromosomal analysis, PCR, FISH, microarray, and biosensors; that are elaborated in the below sections.
4.1.2. Chromosomal analyses
PNA is an excellent probe for molecular hybridization because of its unique hybridization properties. PNA‐based technology is developing and can replace FISH for human chromosomes detection in different tissues using in‐situ detection, which are reported by many researchers. PNA‐based hybridization technology utilizes PNA as a probe, having greater potential for chromosomal investigations. 29 , 64 , 65 Chen et al 66 synthesized 42 PNA probes for chromosome‐specific probe identification, which resulted in 10 PNAs were specific to human chromosomes 1, 2, 7, 9, 11, 17, 18, X, and Y. Their results also suggested that probes of PNA are very much capable of detecting trisomies in humans, and also are used for chromosome “bar‐coding” as sequence‐specific stains. FISH based on PNA is widely being investigated and has shown promising application for chromosomal analysis, as traditional techniques require denaturation of DNA by heat in formamide along with moderate temperature for longer hours to hybridize probe to its specific target. 67
4.1.3. PNA in PCR
In biomedical research PCR is a widely used technique for DNA amplification of targeted sequence, it generates thousands of copies for enormous applications. The methods based on PNA are used to modulate PCR, detecting genomic mutation/capturing nucleic acids. 22 Yu et al 68 designed a “modified PNA‐PCR method” to screen KRAS mutation in cancer patients, this mutation shows resistance to epidermal growth factor receptor (EGFR) target therapy. From the results it was evident that this modification in PCR using PNA was an accurate and sensitive method to screen pancreatic cancer patients with KRAS mutation, apart from screening, this method also has potential to quantify KRAS mutant DNA to envisage response of cancer patients towards therapy and monitoring of disease progression. PNA‐based PCR clamping is used for minor allele identification in blood chimerism, and on the other hand, normal PCR is not capable of identifying it. 69
PNA‐based real‐time PCR kits are highly sensitive and specific with very excellent detection limit, having no cross reactivity, stable reproducibility, and self‐life of more than 8 months, for the detection of carbapenemase genes. 70 Furthermore, PNA‐based quantitative PCR (Figure 6) are also used for rapid and sensitive detection of multiple sclerosis‐associated retrovirus, which is responsible for neuro‐inflammatory complications such as multiple sclerosis. 72 Nowadays, PNA‐based probes in PCR methods for highly sensitive detection of genetic drivers such as KRAS are in high demand, as normal quantitative PCR are not very sensitive for genetic mutation determination. 73 Clamping of PNA in PCR is used for diagnosis of McCune‐Albright syndrome which affects skin, endocrine tissues, and bones of effected person. 74 PNA clamping is ultrasensitive for detecting EGFR gene mutation when compared to traditional way of direct sequencing in non‐small cell lung cancer patients. 75 In addition to this, PNA‐LNA conjugated PCR clamping assay are being used to detect EGFR gene mutation from pleural fluid sample of non‐small cell lung cancer patients, and the results obtained suggest that these conjugate based PCR is highly sensitive than pre‐existing PCR techniques which are based on direct sequencing. 76 , 77
FIGURE 6.
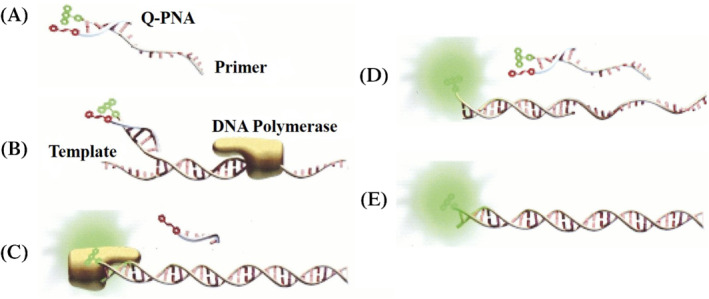
Illustration of basic Q‐PNA‐based PCR principle: (A) Q‐PNA labeled with quencher hybridized at 5′ end of the forward primer labeled with Fluor which will quench fluorescence. T m value of Q‐PNA‐primer duplex complex is ranged between temperature of PCR annealing and extension stages. (B) In initial stage of thermo‐cyclic reaction, denature DNA is hybridized by 3′ end of forward primer and DNA polymerase elongates the primer. (C) Reverse primer which is not illustrated in the figure, will displace the Q‐PNA and incorporate 13‐base‐Q‐PNA binding region into amplicon by initiating transcription of the reverse strand. (D) Later in annealing stage, forward primer fully anneals to reverse strand and report its presence in real‐time analysis because excess primer is quenched by the Q‐PNA. (E) This is end product: a double stranded piece of DNA labeled fluorescently at its one strand. Source: Reproduced from Fiandaca et al, Genome Research, 2001 71
4.1.4. PNA‐FISH
Fluorescent PNAs are an important tool in the biomedical domain for utilization in the FISH technique as a probe. Hnedzko et al 78 presented a simple protocol for labeling N‐terminus of PNA with HiLyte Fluor 488 in the last stage of PNA synthesis, which resulted significantly brighter and photostable in microscopy experiment when compared with PNA‐carboxyfluorescein. Santos et al 79 optimized the PNA probe (EUB338) targeting the bacteria by using flow cytometry for studying the three major factors responsible for hybridization that is time, temperature, and formamide concentration. Hence, their results signify that universal PNA‐FISH procedure does not apply to different microorganism's families as they require different hybridization parameters. In 2019 Huang et al 80 reported PNA‐FISH‐AFC (PNA‐FISH‐Acoustic Flow Cytometry) for bacterial detection in blood culture; as it was important to develop a rapid technique to diagnose bacteremia, which is a bacterial infection in the bloodstream caused by bacteria such as Escherichia coli, Klebsiella pneumoniae, or Pseudomonas aeruginosa, and so on, and conventional techniques like PCR‐based film array assay and MALDI‐TOF take 7‐24 hours for complete diagnosis. But PNA‐FISH‐AFC method was found to be more appropriate and had potential time advantages. PNAs are hydrophobic and can also penetrate the mycobacterial cell wall; considering this point, Kim et al 81 developed a FISH‐PNA probe method to differentially label Mycobacterium tuberculosis and non‐tuberculosis mycobacteria for detection in clinical respiratory specimens. The results obtained from their study suggest that dual‐color FISH with PNA probe was highly specific, but less sensitivity for detection and identification of mycobacterium. Hence, due to this reason, this method is best suited for culture confirmation.
Q fever endocarditis and vascular infections are very common infections caused by bacteria Coxiella burnetii, and FISH is used for diagnosis of this. Prudent et al 82 compared the conventional FISH method with PNA‐FISH for diagnosis of Q fever endocarditis and vascular infections in real‐samples collected from patients. Their result suggests that the FISH was approx. 43% sensitive and PNA‐FISH was approx. 60% sensitive. Hence, it suggests that PNA‐FISH is more sensitive than the FISH method for detecting C. burnetii. Naturally, biofilm is composed of a variety of species, 83 these multispecies biofilms can be held responsible for complications in different areas, namely drinking water distribution system biofouling, food processing environment contamination, biomedical complications, and infections from the device in humans. 84 , 85 , 86 Almeida et al quantified and visualize multispecies biofilm populations by utilizing PNA‐FISH and PNA‐FISH‐confocal laser scanning microscopy (CLSM) as a tool. The result suggested that PNA‐FISH was very efficient in quantification and microbial distribution of multiple species, and when this was combined with CLSM, it was more effective then PNA‐FISH. 87
4.1.5. PNA‐microarray
PNA microarray is more preferred over DNA chip in gene analysis, but a chemical synthesis of PNA microarray is a laborious process and very sophisticated art. Due to this reason, Wu et al 88 developed a fully automated synthesizer to fabricate the PNA microarray through photolithography. Yang et al 89 developed a PNA microarray that has improved performance when compared to DNA microarray. In their study, they have synthesized PNA and used it as building blocks for developing high‐density PNA microarrays and result suggest that the PNA microarray detected single and multiple base‐mismatches with high efficacy. DNA‐based microarray is very effective with a very few fallbacks, such as selectivity, sensitivity, and stability in varying conditions; the solution to this problem is PNA‐based probe due to its enormous properties. PNA‐based chips are very rapid, highly reproducible, accurate, reusable, and have long storage capacity. Figure 7, represents basic PNA‐based microarray, which illustrates PNA immobilization on the gold electrode chip surface. Then is hybridized with target DNA, and after target DNA gets hybridized, intercalator “Hoechst 33258” is introduced and incubated for binding with the PNA‐DNA complex. After incubation, the hybridization event is evaluated using the electrochemical analysis method. 91 , 92
FIGURE 7.
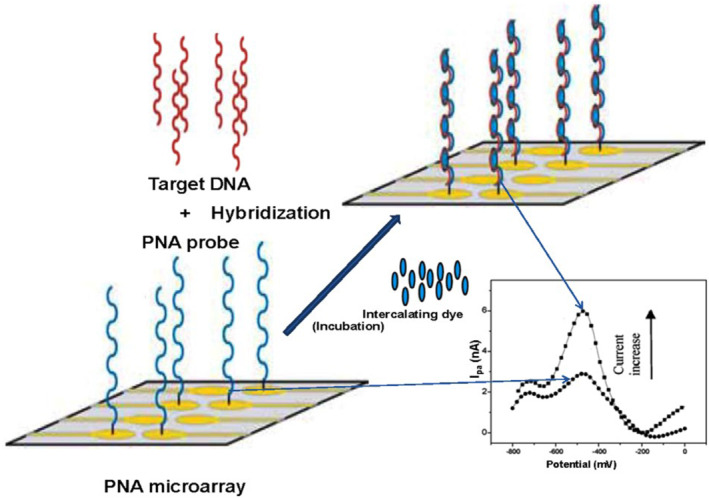
Diagrammatic representation of PNA‐based microarray basic set‐up for fabricating gold electrode based chip surface, to detect the hybridization events. Source: Reprinted by permission from Elsevier: Elsevier, Bioelectrochemistry, Application of peptide nucleic acid towards development of nanobiosensor arrays, Singh, R. P., Oh, B.‐K., & Choi, J.‐W. Bioelectrochemistry. 2010 90
Researchers have investigated a new rapid chip technology based on PNA‐microarray for cancer diagnosis, prognosis, and research. For this, they have hybridized unlabeled RNA on PNA microarray and further labeled this with pCp‐Cy3 by enzyme ligation on the chip. The designed chip was commercialized with trade name, PANArray™ miRNA (Figure 8), which demonstrated excellent reproducibility and low cross‐hybridization for let‐7 and the miR‐181 families of miRNA. Thus, the results of this study suggest that this microarray is rapid chip‐based technology for expression analysis with high fidelity. 93 Microarray technologies are widely used for biomedical studies. Endoh et al 94 prepared PNA‐based array and investigated it for capture ability of microRNAs, and showed that 10‐mer PNA probe microarray was highly sensitive than PNAs biological counterpart DNA as a probe for miRNAs targeting. For cancer diagnosis, miRNA plays a crucial role as a promising biomarker; low levels of miRNA in blood hampers their use for cancer diagnosis, and to overcome this issue development of the ultrasensitive analytical method is the need of current scenario. For the same, Jolly et al investigated PNA as probe immobilized on the gold electrode to capture miRNA, by utilizing dual‐mode detection that is electrochemical impedance spectroscopy and thiolated ferrocene. Their study suggests that the dual detection approach for miRNA was highly sensitive and were able to distinguish between mismatched target sequence of miRNA, and can be further exploited for the development of microarray platforms. 95
FIGURE 8.
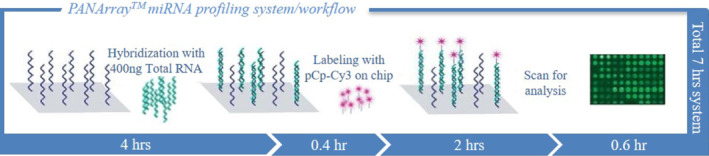
PANArray™ miRNA microarray system: In this system glass slide arrayed with miRNA specific PNA probes were hybridized with total RNA. Labeling with pCp‐Cy3 at 3′ end by enzyme ligation, to form a chip for analysis. Processing and analysis using standard software and scanning equipment. This system takes only 7 hours for the analytical process (form fabrication to analysis) to give an excellent result. Source: Reprinted by permission from Springer Nature: Springer Nature, BioChip Journal, A PNA microarray platform for miRNA expression profiling using on‐chip labeling technology, Kim, H., Choi, J., Cho, M., & Park, H. BioChip Journal. 2012 93
4.1.6. Biosensors
Biosensors based on nucleic acid have increasing applications in genetics and biomedical; for example, DNA genotyping and cancer biomarker detection by identifying single base mutations in a target nucleic acid at an extreme‐low concentration by using PNA‐based biosensors, which have an excellent affinity, sensitivity, and specificity. 12 , 96 Singh et al 90 reported applications of PNA for nano‐based biosensor (nanobiosensor) array, as PNA possess physicobiochemical properties which enables the establishment of new detection approach that is more reliable and faster than traditional biosensor arrays due to this reason PNA is used for the development of chips/sensors/arrays for the diagnosis and therapeutics. PNAs are used in sensing technology which has biomedical applications such as, diagnosis stratification of diseases, and prognosis, and is better as well as inexpensive than traditional technologies. Due to PNAs exceptional biophysical properties, fluorogenic‐PNA can be preferred over molecular beacons based on DNA for sensing in a cell‐free environment; hence, cellular uptake is not necessary. 97 Hamidi‐Asl et al 98 developed a genosensor (electrochemical detector) by labeling PNA with indigo carmine (IC) for detection of tumor suppressor gene p53 (15‐mer short sequence), using IC label in PNA for the first time and the result obtained from this study indicates that PNA sensors have a good logarithmic dependency.
Saadati et al 99 reported the applications of PNA in biosensor as a bioreceptor; PNA is used as a diagnostic probe by fabricating it on the surface of electrochemical/optical biosensors, which has application in identification of various analytes of interest; further these biosensors can be employed for recognizing DNA, RNA, bacteria, and so on. Bazin et al 100 reported in their review that PNA are attractive receptors, and recent developments in nanostructured and interface materials play a key role in future biosensor technology. Teengam et al 101 developed a paper‐based electrochemical PNA biosensor. In this, the PNA was labeled with anthraquinone‐labeled 28 pyrrolidinyl peptide nucleic acid probe (AQ‐PNA) and further, the electrode was modified by graphene‐polyaniline (G‐PANI) for the detection of human papillomavirus (HPV) type 16 DNA. The detection was done on real DNA samples, after PCR amplified DNA samples from the SiHa cell line. The result suggests that this device is inexpensive, can be incinerated with ease, and is reliable for detection of the primary stage cervical cancer by measuring the amount of HPV type 16 DNA.
Lab‐on‐chip based technologies are very high performing and cost‐effective; they are playing key role for advancing futuristic point‐of‐care diagnostic devices. Diagnostic work based on printed circuit board (PCB) that can be called as Lab‐on‐PCB, have recently reemerged as a promising technology for mass‐manufacturing of integrated microsystem technology. 102 , 103 Till date for genetic analysis microsystems, such as DNA, RNA, miRNA, and cRNA; are the main objectives. Keeping the above viewpoint, Jolly et al developed lab‐on‐chip‐PCB biosensor based on PNA as a probe for genetic diagnosis by DNA quantification. The result showed higher selectivity and sensitivity than the pre‐existing techniques for the same purpose. 104 Xuan et al 105 developed an economical and portable DNA recognition device that is an ultrasensitive DNA electrochemical sensor, which does not require immobilization of probe. There are two advantages of this method one is diffusivity between ferrocene‐PNA and ferrocene‐PNA hybridized to DNA, and the other one is the DNA polymerase activity of strand‐displacement. Thus, the results obtained from this study suggested 2‐3 folds higher detection sensitivity than other pre‐existing DNA biosensors, which are not based on immobilization. Hence, extraordinary sensitivity and ease in operating offered this method with a promising substitute for the development of miniaturized, integrated, cost‐effective, and portable biosensor devices for DNA detection.
4.1.7. In‐vivo imaging
Sonar et al 106 developed a method to detect KRAS2 mRNA hybridization using PNA‐fluorescence in lung cancer cells. They labeled PNA with thiazole orange (TO) dye at 5′ end for making PNA‐IGF1, this labeling with dye enhanced the fluorescence quantum yield. Their study showed that PNA‐IGF1 with TO shows less fluorescence, but hybridization of PNA‐IGF1 with KRAS2 RNA forms KRAS2 PNA‐IGF1, which has raised fluorescence multiple folds. Shigeto et al 107 designed a PNA‐DNA probe, in which PNA and DNA conjugated with fluorescein isothiocyanate for detection and Dabcyl as quencher probe respectively; for imaging analysis of EGFR mutation especially the following three EGFR mutations exon19del E746‐A750, T790M, and L858R in lung cancer cells. Chen et al 108 designed an assay based on PNA, tagged with a fluorophore for detection of BRAF V600 mutations with excess wild type background in melanoma samples. The tagged PNA act as both PCR clamp and sensor probe; the result obtained from this study has shown better sensitivity than traditional PCR plus Sanger sequencing. Hwang et al 3 developed a PNA probe labeled with fluorescent markers for convenient, rapid, and accurate detection of filaggrin (FLG) mutation. These mutations are of two types heterozygote and homozygote FLG mutations and are found in patients of atopic dermatitis (chronic inflammatory skin disease). Hence, these detection techniques will allow early prevention and will lower the incidence rate of atopic dermatitis.
4.2. Therapeutics utilizing PNA
PNA has a variety of properties such as antisense agent, antigene agent, versatility, affinity for targeting nucleic acids, and physiological stability. This offers PNA to be a potential therapeutic agent. Modifications in PNA have improved its potency for further developments in preclinical studies for drug delivery and has reached new statures. 109 , 110 It can be used as a research tool for gene regulation and targeting. The D‐loop formation during PNA hybridization to form a triplex complex has the ability to initiate transcription and induce gene expression. PNA‐based gene regulation will have important biomedical implications for the treatment of many deadly diseases such as cancer, genetic disorder, and age‐related diseases. 39 PNA in general cannot cross the blood‐brain barrier as it is permeable to only lipophilic molecules of molecular weight < 600 Da. Hence, conjugates of streptavidin (SA) and OX26 monoclonal antibody with PNA represent optimal antisense molecules for drug delivery to the brain for the treatment of Alzheimer's disease and cerebral AIDS. 111 Kaushik et al 112 reported that anti‐TAR PNA conjugation with membrane‐permeating peptide inhibits transactivation of the HIV‐1 (Human Immunodeficiency Virus Type 1) LTR (long terminal repeat), which results in reduced HIV‐1 virions production in chronically H9 cells infected from HIV‐1. Chaubey et al 113 designed PNATAR‐transportan conjugate, which has speedy cellular uptake and exhibits potential virucidal and antiviral properties. Neamine part from the aminoglycoside antibiotic neomycin B can be conjugated to a 16 mer PNA targeting HIV‐1 TAR RNA. The TAR RNA is the binding site of the viral protein TAT. Attachment of the neamine core allows cellular uptake of the PNA and results in effective inhibition of HIV‐1 replication. Neamine is a polycationic moiety which provides greater solubility to PNA and also gives it a unique property to cleave RNA, the conjugate is specific to its target site and is only functional at physiological concentrations of Mg2+. Hence, this conjugate of PNA with neamine has a potential therapeutic application. 114 Zeng et al 115 designed Tat‐PNA‐DR to target the direct repeat (DR) sequence of Hepatitis B virus (HBV); this was validated in‐vitro using HepG2.2.15 cells, and in‐vivo in acute hepatitis B mouse model by targeting LTR‐DR of HBV‐RNA. Targeting using this conjugate inhibited HBV replication in the in‐vitro study, and it also displays low cytotoxicity, less immunogenicity, and elevated stability in serum; an 80% decline in HBV‐DNA was observed in the in‐vivo studies. Hence, from these results, they have concluded that PNA conjugated drugs can be a promising candidate for HBV treatment.
Browne et al 116 reported for the first time, efficacy of PNA and their conjugates for the treatment of amyotrophic lateral sclerosis (ALS) by specific cellular pathologies, as ALS is a neurodegenerative disorder, which affects the spinal cord and nerve cells. Ricciardi et al 117 reported the details of PNA as a site‐specific gene‐editing tool due to its hybridization properties they form a stable triplex complex, which has potential for genome‐specific modification and DNA repair. PNA‐based strategies for gene editing can positively correct the human diseases caused due to mutations in preclinical studies. Bahal et al 118 reported γPNA‐mediated gene editing for correction of anemia in human β‐thalassemic mice model using nanoparticle‐based delivery system. This in‐vivo study result shows that the nanoparticle delivery system has low off‐target effects and stem cell factor, γPNAs and nano‐system based delivery will offer new treatment strategies for the genetic disorder related to the blood which can be treated safely by intravenous administration. Dorn et al 119 designed phosphonic ester PNA by modifying the side chain of PNA to interfere with gene function and to modulate cellular pathways according to the demand for therapy, as AEG PNA hampers in‐vivo applications. Their result suggests that PNA is a powerful antisense molecule which when directed against medaka (Oryzias latipes) six3 gene in fish embryos it was able to knock down the desired gene, which resulted in exact six3 phenotype.
5. FUTURE PROSPECTS AND CONCLUSIONS
PNA is a synthetic RNA/DNA analog with mimicking attributes of DNA; they exhibit unique features, such as higher specificity and affinity, poor solubility, can form hybridized complexes with high stability; its structure can be modified by targeting strand invasion and self‐tendency for aggregation. These distinctive features of PNA have given it a remarkable advantage over its corresponding analogs, to extend state‐of‐the‐art development of diagnostic and therapeutic tools in the biomedical domain after its synthesis in 1991. PNA is highly used for diagnostic applications such as in vivo and in vitro analysis. PNA biosensors have evolved recently and have shown improved detection of analytes when compared to conventional biosensors. Hence, we suggest researchers to utilize nanotechnology for enhancing clinical diagnosis attributes of PNA fabricated biosensors for detecting human diseases, which will open new avenues for future research in the making of cliniconanobiosensor and nanobiosensor arrays for clinical diagnosis. In vitro assays based on PNA are widely used for clinical diagnosis, but they have less detection sensitivity at nanomolar concentration and need improvement. 63 For this, we recommend that it can be achieved by employing nanobiotechnological applications. PNA‐based diagnosis has achieved great heights and apart from biomedical applications, they are also used for the detection of small‐size toxins in the environmental monitoring and food analysis industries.
PNA is a synthetic molecule that shows highly specific binding to target gene sequence due to which they are an appropriate candidate of interest in biomedical and biotechnological context. 3 Tagalakis et al 120 developed a nanovesicle (∼90 to 140 nm) complex made from giant unilamellar vesicle to efficiently deliver the drug to the target site of gene therapy by therapeutic nucleic acids, as targeted delivery is one of the challenging aspects in PNA‐based gene therapy. PNA in the therapeutic application was a challenge because of its intercellular delivery which is now improved utilizing nanoparticle‐based strategies for efficiently delivering the PNA conjugated drugs, to the cellular environment. 121 Furthermore, to extend the oligomeric family, many researchers are working to develop a new synthetic analog with properties more sophisticated than PNA, such an example is the development of AApeptides (a new class of peptidomimetics) which is based on chiral PNA backbone. 122 This class of peptide shows application in material science and biomedical domains because of their unique and few properties similar to PNA. A large number of transfection strategies for PNA to overcome intercellular uptake have been developed utilizing microinjection, electroporation, conjugation with lipophilic moieties, and so on, to date and is still developing at a faster pace. 53
The current situation of global viral pandemic (COVID‐19) is threatening the society; PNA and nanoparticle functionalized PNA can be used for the manufacturing of antiviral PPE (personal protective equipment) kits due to this reason PNA shows great potential as an antiviral agent. 123 PNA is not present in the human body, and the clearing mechanism of this from the human body is not known, and this may lead to systemic toxicity. Thus, there is an urgent need to lay attention towards the toxicological aspect of PNA and their conjugates. The search of synthetic nucleic acid with better properties such as higher biological stability, enhanced cellular uptake, and strong binding affinity with complementary DNA/RNA is still going on, but to date, PNA is the best synthetic nucleic acid analogs and can trigger the ideas for next‐generation novel technologies. 58 Hence, from a detailed study of PNA, we can suggest that researchers should lay their focus on engineering the PNA by functionalizing them with different nanoparticles (carbon‐based, quantum dots, metal and metal oxides, liposomal, and dendrimer nanoparticles) for improved and high‐end applications. Due to the higher prevalence of lifestyle diseases in the society 124 and PNA has well‐developed applications towards diagnostics and therapeutics. Hence, our review emphasizes research professional's attention towards the biomedical domain, for the treatment and diagnosis of deadliest lifestyle diseases, like cerebrocardiovascular diseases, cancer, diabetes, and so on, by utilizing PNA. Furthermore, PNAs plausible application potentialities towards the agriculture and environmental domain may be examined which are scantly reported or are in the infancy. To conclude our review gives a comprehensive picture on detailed biomedical application of PNA, along with their properties, synthesis, and prospects.
PEER REVIEW INFORMATION
Engineering Reports thanks Nuno Filipe Azevedo and other anonymous reviewers for their contribution to the peer review of this work.
AUTHOR CONTRIBUTIONS
Kshitij RB Singh contributed to the conceptualization, data curation, formal analysis, investigation, methodology, project administration, resources, validation, visualization, writing the original draft, review, and editing. Parikipandla Sridevi contributed to the conceptualization, resources, supervision, writing the review and editing. Ravindra Pratap Singh contributed to the conceptualization, project administration, resources, supervision, writing the original draft, review, and editing.
CONFLICT OF INTERESTS
The authors declare no potential conflict of interest.
ACKNOWLEDGMENTS
Authors are thankful to all faculty members of the Department of Biotechnology, Indira Gandhi National Tribal University, Amarkantak, M.P., India for their constant support. Special thanks to Hon'ble Vice‐chancellor, IGNTU, Amarkantak for providing financial assistance.
Singh KR, Sridevi P, Singh RP. Potential applications of peptide nucleic acid in biomedical domain. Engineering Reports. 2020;2:e12238. 10.1002/eng2.12238
REFERENCES
- 1. Nielsen PE, Haaima G. Peptide nucleic acid (PNA). A DNA mimic with a pseudopeptide backbone. Chem Soc Rev. 1997;26(2):73. 10.1039/cs9972600073. [DOI] [Google Scholar]
- 2. Shakeel S, Karim S, Ali A. Peptide nucleic acid (PNA) — a review. J Chem Technol Biotechnol. 2006;81(6):892‐899. 10.1002/jctb.1505. [DOI] [Google Scholar]
- 3. Hwang J, Lee S, Kim D, et al. Peptide nucleic acid (PNA) probe‐based analysis to detect filaggrin mutations in atopic dermatitis patients. Exp Dermatol. 2018;27(11):1304‐1308. 10.1111/exd.13765. [DOI] [PubMed] [Google Scholar]
- 4. Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr Issues Mol Biol. 1999;1:89‐104. [PubMed] [Google Scholar]
- 5. Nielsen P, Egholm M, Berg R, Buchardt O. Sequence‐selective recognition of DNA by strand displacement with a thymine‐substituted polyamide. Science. 1991;254(5037):1497‐1500. 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 6. Eriksson M, Christensen L, Schmidt J, Haaima G, Orgel L, Nielsen PE. Sequence dependent N‐terminal rearrangement and degradation of peptide nucleic acid (PNA) in aqueous solution. New J Chem. 1998;22(10):1055‐1059. 10.1039/a803214i. [DOI] [PubMed] [Google Scholar]
- 7. Braasch DA, Corey DR. Synthesis, analysis, purification, and intracellular delivery of peptide nucleic acids. Methods. 2001;23(2):97‐107. 10.1006/meth.2000.1111. [DOI] [PubMed] [Google Scholar]
- 8. Wu J, Meng Q, Ren H, Wang H, Wu J, Wang Q. Recent advances in peptide nucleic acid for cancer bionanotechnology. Acta Pharmacol Sin. 2017;38(6):798‐805. 10.1038/aps.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson KE, Levy M, Miller SL. Peptide nucleic acids rather than RNA may have been the first genetic molecule. Proc Natl Acad Sci. 2000;97(8):3868‐3871. 10.1073/pnas.97.8.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banack SA, Metcalf JS, Jiang L, Craighead D, Ilag LL, Cox PA. Cyanobacteria produce N‐(2‐Aminoethyl)glycine, a backbone for peptide nucleic acids which may have been the first genetic molecules for life on earth. PLoS One. 2012;7(11):e49043. 10.1371/journal.pone.0049043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta A, Mishra A, Puri N. Peptide nucleic acids: advanced tools for biomedical applications. J Biotechnol. 2017;259:148‐159. 10.1016/j.jbiotec.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Briones C, Moreno M. Applications of peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) in biosensor development. Anal Bioanal Chem. 2012;402(10):3071‐3089. 10.1007/s00216-012-5742-z. [DOI] [PubMed] [Google Scholar]
- 13. Ganesh NK, Nielsen EP. Peptide nucleic acids: Analogs and derivatives. Curr Org Chem. 2000;4(9):931‐943. 10.2174/1385272003375969. [DOI] [Google Scholar]
- 14. Malcher J, Wesoly J, Bluyssen H. Molecular properties and medical applications of peptide nucleic acids. Mini‐Rev Med Chem. 2014;14(5):401‐410. 10.2174/1389557514666140428114613. [DOI] [PubMed] [Google Scholar]
- 15. Nielsen PE. Applications of peptide nucleic acids. Curr Opin Biotechnol. 1999;10(1):71‐75. 10.1016/S0958-1669(99)80013-5. [DOI] [PubMed] [Google Scholar]
- 16. Abraham P, Maliekal TT. Single cell biology beyond the era of antibodies: relevance, challenges, and promises in biomedical research. Cell Mol Life Sci. 2017;74(7):1177‐1189. 10.1007/s00018-016-2382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lundin KE, Good L, Strömberg R, Gräslund A, Smith CIE. Biological activity and biotechnological aspects of peptide nucleic acid. Adv Genet. 2006;56:1‐51. 10.1016/S0065-2660(06)56001-8. [DOI] [PubMed] [Google Scholar]
- 18. Morihiro K, Kasahara Y, Obika S. Biological applications of xeno nucleic acids. Mol BioSyst. 2017;13(2):235‐245. 10.1039/C6MB00538A. [DOI] [PubMed] [Google Scholar]
- 19. Dunn MR, Chaput JC. An in vitro selection protocol for threose nucleic acid (TNA) using DNA display. Current Protocols Nucleic Acid Chem. 2014;2014:9.8.1‐9.8.19. 10.1002/0471142700.nc0908s57. [DOI] [PubMed] [Google Scholar]
- 20. Hagedorn PH, Persson R, Funder ED, et al. Locked nucleic acid: modality, diversity, and drug discovery. Drug Discov Today. 2018;23(1):101‐114. 10.1016/j.drudis.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 21. Summerton JE. Invention and early history of morpholinos: from pipe dream to practical products. In: Moulton HM, Moulton JD, eds. Morpholino Oligomers. Vol 1565. New York: Humana Press; 2017:1‐15. 10.1007/978-1-4939-6817-6_1. [DOI] [PubMed] [Google Scholar]
- 22. Pellestor F, Paulasova P. The peptide nucleic acids (PNAs), powerful tools for molecular genetics and cytogenetics. Eur J Hum Genet. 2004;12(9):694‐700. 10.1038/sj.ejhg.5201226. [DOI] [PubMed] [Google Scholar]
- 23. Järver P, Coursindel T, Andaloussi SEL, Godfrey C, Wood MJ, Gait MJ. Peptide‐mediated cell and in vivo delivery of antisense oligonucleotides and siRNA. Molecul Therapy ‐ Nucleic Acids. 2012;1:e27. 10.1038/mtna.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siddiquee S. A review of peptide nucleic acid. Adv Tech Biol Med. 2015;03(02):1‐10. 10.4172/2379-1764.1000131. [DOI] [Google Scholar]
- 25. Veldhoen S, Laufer S, Restle T. Recent developments in peptide‐based nucleic acid delivery. Int J Mol Sci. 2008;9(7):1276‐1320. 10.3390/ijms9071276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rozners E. Recent advances in chemical modification of peptide nucleic acids. J Nucleic Acids. 2012;2012:1‐8. 10.1155/2012/518162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sugiyama T, Kittaka A. Chiral peptide nucleic acids with a substituent in the N‐(2‐Aminoethy)glycine backbone. Molecules. 2012;18(1):287‐310. 10.3390/molecules18010287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moccia M, Adamo MFA, Saviano M. Insights on chiral, backbone modified peptide nucleic acids: properties and biological activity. Artif DNA PNA XNA. 2014;5(3):e1107176. 10.1080/1949095X.2015.1107176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paulasova P, Pellestor F. The peptide nucleic acids (PNAs): a new generation of probes for genetic and cytogenetic analyses. Ann Genet. 2004;47(4):349‐358. 10.1016/j.anngen.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 30. Malik S, Asmara B, Moscato Z, Mukker JK, Bahal R. Advances in nanoparticle‐based delivery of next generation peptide nucleic acids. Curr Pharm des. 2019;24(43):5164‐5174. 10.2174/1381612825666190117164901. [DOI] [PubMed] [Google Scholar]
- 31. Hyrup B, Nielsen PE. Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg Med Chem. 1996;4(1):5‐23. 10.1016/0968-0896(95)00171-9. [DOI] [PubMed] [Google Scholar]
- 32. Egholm M, Buchardt O, Christensen L, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson–crick hydrogen‐bonding rules. Nature. 1993;365(6446):566‐568. 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 33.Standard oligopeptide nomenclature is employed: H‐, the free N‐terminal amino group; T, C, A, and G, the T‐acetyl, C‐acetyl, A‐acetyl, and G‐acetyl N‐(2‐aminoethyl)glycinyl units, respectively; NH2, the C‐terminal Amido group.
- 34. Wittung P, Nielsen PE, Buchardt O, Egholm M, Nordén B. DNA‐like double helix formed by peptide nucleic acid. Nature. 1994;368(6471):561‐563. 10.1038/368561a0. [DOI] [PubMed] [Google Scholar]
- 35. Egholm M, Buchardt O, Nielsen PE, Berg RH. Peptide nucleic acids (PNA). Oligonucleotide analogs with an achiral peptide backbone. J Am Chem Soc. 1992;114(5):1895‐1897. 10.1021/ja00031a062. [DOI] [Google Scholar]
- 36. Egholm M, Nielsen PE, Buchardt O, Berg RH. Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA). J Am Chem Soc. 1992;114(24):9677‐9678. 10.1021/ja00050a068. [DOI] [Google Scholar]
- 37. Egholm M, Behrens C, Christensen L, Berg RH, Nielsen PE, Buchardt O. Peptide nucleic acids containing adenine or guanine recognize thymine and cytosine in complementary DNA sequences. J Chem Soc Chem Commun. 1993;(9):800‐801. 10.1039/c39930000800. [DOI] [Google Scholar]
- 38. Flensburg C, Egholm M. Ethyl 9‐adeninylacetate at 122 K. Acta Crystallograph Sect C Crystal Struct Commun. 1994;50(9):1480‐1482. 10.1107/S010827019400541X. [DOI] [Google Scholar]
- 39. Wang G, Xu XS. Peptide nucleic acid (PNA) binding‐mediated gene regulation. Cell Res. 2004;14(2):111‐116. 10.1038/sj.cr.7290209. [DOI] [PubMed] [Google Scholar]
- 40. Larsen HJ, Bentin T, Nielsen PE. Antisense properties of peptide nucleic acid. Biochimica et Biophysica Acta (BBA) ‐ Gene Struct Express. 1999;1489(1):159‐166. 10.1016/S0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 41. Haaima G, Hansen HF, Christensen L, Dahl O, Nielsen PE. Increased DNA binding and sequence discrimination of PNA oligomers containing 2,6‐diaminopurine. Nucleic Acids Res. 1997;25(22):4639‐4643. 10.1093/nar/25.22.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gangamani B, Kumar V. 2‐Aminopurine peptide nucleic acids (2‐apPNA): Intrinsic fluorescent PNA analogues for probing PNA–DNA interaction dynamics. Chem Commun. 1997(19):1913‐1914. 10.1039/a705539k1913. [DOI] [Google Scholar]
- 43. Köhler O, Seitz O. Thiazole orange as fluorescent universal base in peptide nucleic acids. Chem Commun. 2003;(23):2938‐2939. 10.1039/B308299G. [DOI] [PubMed] [Google Scholar]
- 44. Jarikote DV, Köhler O, Socher E, Seitz O. Divergent and linear solid‐phase synthesis of PNA containing Thiazole orange as artificial base. Eur J Org Chem. 2005;2005(15):3187‐3195. 10.1002/ejoc.200500201. [DOI] [Google Scholar]
- 45. Sanders JM, Wampole ME, Chen C‐P, et al. Effects of hypoxanthine substitution in peptide nucleic acids targeting KRAS2 oncogenic mRNA molecules: theory and experiment. J Phys Chem B. 2013;117(39):11584‐11595. 10.1021/jp4064966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shiraishi T, Hamzavi R, Nielsen PE. Subnanomolar antisense activity of phosphonate‐peptide nucleic acid (PNA) conjugates delivered by cationic lipids to HeLa cells. Nucleic Acids Res. 2008;36(13):4424‐4432. 10.1093/nar/gkn401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Katritzky AR, Narindoshvili T. Chiral peptide nucleic acid monomers (PNAM) with modified backbones. Org Biomol Chem. 2008;6(17):3171‐3176. 10.1039/b806141f. [DOI] [PubMed] [Google Scholar]
- 48. Gupta P, Muse O, Rozners E. Recognition of double‐stranded RNA by guanidine‐modified peptide nucleic acids. Biochemistry. 2012;51(1):63‐73. 10.1021/bi201570a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mitra R, Ganesh KN. PNAs grafted with (α/γ, R/S)‐aminomethylene pendants: Regio and stereo specific effects on DNA binding and improved cell uptake. Chem Commun. 2011;47(4):1198‐1200. 10.1039/C0CC03988H. [DOI] [PubMed] [Google Scholar]
- 50. Mitra R, Ganesh KN. Aminomethylene peptide nucleic acid ( am ‐PNA): synthesis, Regio‐/stereospecific DNA binding, and differential cell uptake of (α/γ, R / S ) am‐ PNA analogues. J Org Chem. 2012;77(13):5696‐5704. 10.1021/jo300860f. [DOI] [PubMed] [Google Scholar]
- 51. Sahu B, Sacui I, Rapireddy S, et al. Synthesis and characterization of Conformationally Preorganized, (R)‐diethylene glycol‐containing γ‐peptide nucleic acids with superior hybridization properties and water solubility. J Org Chem. 2011;76(14):5614‐5627. 10.1021/jo200482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hnedzko D, McGee DW, Karamitas YA, Rozners E. Sequence‐selective recognition of double‐stranded RNA and enhanced cellular uptake of cationic nucleobase and backbone‐modified peptide nucleic acids. RNA. 2017;23(1):58‐69. 10.1261/rna.058362.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA). Adv Drug Deliv Rev. 2003;55(2):267‐280. 10.1016/S0169-409X(02)00182-5. [DOI] [PubMed] [Google Scholar]
- 54. Nielsen PE. Peptide nucleic acid. a molecule with two identities. Acc Chem Res. 1999;32(7):624‐630. 10.1021/ar980010t. [DOI] [Google Scholar]
- 55. Slaitas A . Development of a New PNA Analogue as a Potential Antisense Drug and Tool for Life‐Science Studies. Stockholm, Sweden: Karolinka University; 2004. https://openarchive.ki.se/xmlui/handle/10616/39699. [Google Scholar]
- 56. Dueholm KL, Egholm M, Behrens C, et al. Synthesis of peptide nucleic acid monomers containing the four natural nucleobases: thymine, cytosine, adenine, and guanine and their oligomerization. J Org Chem. 1994;59(19):5767‐5773. 10.1021/jo00098a042. [DOI] [Google Scholar]
- 57. Lee H, Jeon JH, Lim JC, Choi H, Yoon Y, Kim SK. Peptide nucleic acid synthesis by novel amide formation. Org Lett. 2007;9(17):3291‐3293. 10.1021/ol071215h. [DOI] [PubMed] [Google Scholar]
- 58. Uhlmann E, Peyman A, Breipohl G, Will DW. PNA: synthetic polyamide nucleic acids with unusual binding properties. Angew Chem Int Ed. 1998;37(20):2796‐2823. . [DOI] [PubMed] [Google Scholar]
- 59. Bonifazi D, Carloni L‐E, Corvaglia V, Delforge A. Peptide nucleic acids in materials science. Artif DNA PNA XNA. 2012;3(3):112‐122. 10.4161/adna.21941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Beavers KR, Mares JW, Swartz CM, Zhao Y, Weiss SM, Duvall CL. In situ synthesis of peptide nucleic acids in porous silicon for drug delivery and biosensing. Bioconjug Chem. 2014;25(7):1192‐1197. 10.1021/bc5001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ma S, Schroeder B, Sun C, et al. Electroporation‐based delivery of cell‐penetrating peptide conjugates of peptide nucleic acids for antisense inhibition of intracellular bacteria. Integr Biol. 2014;6(10):973‐978. 10.1039/C4IB00172A. [DOI] [PubMed] [Google Scholar]
- 62. Porcheddu A, Giacomelli G. Peptide nucleic acids (PNAs), a chemical overview. Curr Med Chem. 2005;12(22):2561‐2599. 10.2174/092986705774370664. [DOI] [PubMed] [Google Scholar]
- 63. Su X, Teh HF, Lieu X, Gao Z. Enzyme‐based colorimetric detection of nucleic acids using peptide nucleic acid‐immobilized microwell plates. Anal Chem. 2007;79(18):7192‐7197. 10.1021/ac0709403. [DOI] [PubMed] [Google Scholar]
- 64. Pellestor F, Paulasova P. The peptide nucleic acids (PNAs): introduction to a new class of probes for chromosomal investigation. Chromosoma. 2004;112(8):375‐380. 10.1007/s00412-004-0282-8. [DOI] [PubMed] [Google Scholar]
- 65. Pellestor F. The peptide nucleic acids: a new way for chromosomal investigation on isolated cells? Hum Reprod. 2004;19(9):1946‐1951. 10.1093/humrep/deh386. [DOI] [PubMed] [Google Scholar]
- 66. Chen C, Wu B, Wei T, Egholm M, Strauss WM. Unique chromosome identification and sequence‐specific structural analysis with short PNA oligomers. Mamm Genome. 2000;11(5):384‐391. 10.1007/s003350010072. [DOI] [PubMed] [Google Scholar]
- 67. Cartwright IM. Modified PNA Telomere and Centromere FISH Protocols. Kato T., Wilson P., Radiation Cytogenetics. Methods in Molecular Biology. vol 1984. New York: Humana; 2019:101‐105. 10.1007/978-1-4939-9432-8_12. [DOI] [PubMed] [Google Scholar]
- 68. Yu S, Wu J, Xu S, Tan G, Liu B, Feng J. Modified PNA‐PCR method. Cancer Biol Ther. 2012;13(5):314‐320. 10.4161/cbt.19075. [DOI] [PubMed] [Google Scholar]
- 69. Sano R, Takahashi Y, Nakajima T, et al. ABO chimerism with a minor allele detected by the peptide nucleic acid‐mediated polymerase chain reaction clamping method. Blood Transfus. 2014;12(3):431‐434. 10.2450/2014.0162-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jeong S, Kim JO, Jeong SH, Bae IK, Song W. Evaluation of peptide nucleic acid‐mediated multiplex real‐time PCR kits for rapid detection of carbapenemase genes in gram‐negative clinical isolates. J Microbiol Methods. 2015;113:4‐9. 10.1016/j.mimet.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 71. Fiandaca MJ, Hyldig‐Nielsen JJ, Gildea BD, Coull JM. Self‐reporting PNA/DNA primers for PCR analysis. Genome Res. 2001;11(4):609‐613. 10.1101/gr.170401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Machnik G, Łabuzek K, Skudrzyk E, et al. A peptide nucleic acid (PNA)‐mediated polymerase chain reaction clamping allows the selective inhibition of the ERVWE1 gene amplification. Mol Cell Probes. 2014;28(5–6):237‐241. 10.1016/j.mcp.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 73. Lee B, Lee B, Han G, Kwon MJ, Han J, Choi Y‐L. KRAS mutation detection in non‐small cell lung cancer using a peptide nucleic acid‐mediated polymerase chain reaction clamping method and comparative validation with next‐generation sequencing. Korean J Pathol. 2014;48(2):100‐107. 10.4132/KoreanJPathol.2014.48.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Imanaka M, Iida K, Nishizawa H, et al. McCune‐Albright syndrome with acromegaly and fibrous dysplasia associated with the GNAS gene mutation identified by sensitive PNA‐clamping method. Intern Med. 2007;46(18):1577‐1583. 10.2169/internalmedicine.46.0048. [DOI] [PubMed] [Google Scholar]
- 75. Yoon S‐H, Choi Y‐D, Oh I‐J, et al. Peptide nucleic acid clamping versus direct sequencing for the detection of EGFR gene mutation in patients with non‐small cell lung cancer. Cancer Res Treat. 2015;47(4):661‐669. 10.4143/crt.2014.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Soh J, Toyooka S, Aoe K, et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer. 2006;119(10):2353‐2358. 10.1002/ijc.22190. [DOI] [PubMed] [Google Scholar]
- 77. Chen Y‐L, Lu C‐C, Yang S‐C, et al. Verification of wild‐type EGFR status in non–small cell lung carcinomas using a mutant‐enriched PCR on selected cases. J Mol Diagn. 2014;16(5):486‐494. 10.1016/j.jmoldx.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 78. Hnedzko D, McGee DW, Rozners E. Synthesis and properties of peptide nucleic acid labeled at the N‐terminus with HiLyte Fluor 488 fluorescent dye. Bioorg Med Chem. 2016;24(18):4199‐4205. 10.1016/j.bmc.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Santos RS, Guimarães N, Madureira P, Azevedo NF. Optimization of a peptide nucleic acid fluorescence in situ hybridization (PNA‐FISH) method for the detection of bacteria and disclosure of a formamide effect. J Biotechnol. 2014;187:16‐24. 10.1016/j.jbiotec.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 80. Huang XX, Urosevic N, Inglis TJJ. Accelerated bacterial detection in blood culture by enhanced acoustic flow cytometry (AFC) following peptide nucleic acid fluorescence in situ hybridization (PNA‐FISH). Plos One. 2019;14(2):e0201332. 10.1371/journal.pone.0201332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim N, Lee SH, Yi J, Chang CL. Evaluation of dual‐color fluorescence in situ hybridization with peptide nucleic acid probes for the detection of mycobacterium tuberculosis and non‐tuberculous mycobacteria in clinical specimens. Ann Lab Med. 2015;35(5):500‐505. 10.3343/alm.2015.35.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Prudent E, Lepidi H, Angelakis E, Raoult D. Fluorescence in situ hybridization (FISH) and peptide nucleic acid probe‐based FISH for diagnosis of Q fever endocarditis and vascular infections. J Clin Microbiol. 2018;56(9):e00542;18. 10.1128/JCM.00542-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83. Hall‐Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95‐108. 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 84. Murray TS, Egan M, Kazmierczak BI. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr. 2007;19(1):83‐88. 10.1097/MOP.0b013e3280123a5d. [DOI] [PubMed] [Google Scholar]
- 85. Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57(1):677‐701. 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 86. Bryers JD. Medical biofilms. Biotechnol Bioeng. 2008;100(1):1‐18. 10.1002/bit.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Almeida C, Azevedo NF, Santos S, Keevil CW, Vieira MJ. Discriminating multi‐species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS One. 2011;6(3):e14786. 10.1371/journal.pone.0014786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wu Y‐Q, Yang F‐P, Wang H‐Y, Liu J‐X, Liu Z‐C. Peptide nucleic acids (PNAs) patterning by an automated microarray synthesis system through photolithography. J Nanosci Nanotechnol. 2013;13(3):2061‐2067. 10.1166/jnn.2013.6890. [DOI] [PubMed] [Google Scholar]
- 89. Yang F, Dong B, Nie K, et al. Light‐directed synthesis of high‐density peptide nucleic acid microarrays. ACS Comb Sci. 2015;17(10):608‐614. 10.1021/acscombsci.5b00074. [DOI] [PubMed] [Google Scholar]
- 90. Singh RP, Oh B‐K, Choi J‐W. Application of peptide nucleic acid towards development of nanobiosensor arrays. Bioelectrochemistry. 2010;79(2):153‐161. 10.1016/j.bioelechem.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 91. Brandt O. PNA microarrays for hybridisation of unlabelled DNA samples. Nucleic Acids Res. 2003;31(19):119e‐119e. 10.1093/nar/gng120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Brandt O, Hoheisel JD. Peptide nucleic acids on microarrays and other biosensors. Trends Biotechnol. 2004;22(12):617‐622. 10.1016/j.tibtech.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 93. Kim H, Choi J, Cho M, Park H. A PNA microarray platform for miRNA expression profiling using on‐chip labeling technology. Biochip J. 2012;6(1):25‐33. 10.1007/s13206-012-6104-9. [DOI] [Google Scholar]
- 94. Endoh T, Kitamatsu M, Sisido M, Ohtsuki T. PNA arrays for miRNA detection. Chem Lett. 2009;38(5):438‐439. 10.1246/cl.2009.438. [DOI] [Google Scholar]
- 95. Jolly P, Batistuti MR, Miodek A, et al. Highly sensitive dual mode electrochemical platform for microRNA detection. Sci Rep. 2016;6(1):36719. 10.1038/srep36719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. D'Agata R, Giuffrida M, Spoto G. Peptide nucleic acid‐based biosensors for cancer diagnosis. Molecules. 2017;22(11):1951. 10.3390/molecules22111951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Choi Y, Metcalf G, Sleiman MH, Vair‐Turnbull D, Ladame S. Oligonucleotide‐templated reactions based on peptide nucleic acid (PNA) probes: concept and biomedical applications. Bioorg Med Chem. 2014;22(16):4395‐4398. 10.1016/j.bmc.2014.05.071. [DOI] [PubMed] [Google Scholar]
- 98. Hamidi‐Asl E, Raoof JB, Ojani R, Hejazi MS. Indigo carmine as new label in PNA biosensor for detection of short sequence of p53 tumor suppressor gene. Electroanalysis. 2013;25(9):2075‐2083. 10.1002/elan.201300155. [DOI] [Google Scholar]
- 99. Saadati A, Hassanpour S, de la Guardia M, et al. Recent advances on application of peptide nucleic acids as a bioreceptor in biosensors development. TrAC Trends Anal Chem. 2019;114:56‐68. 10.1016/j.trac.2019.02.030. [DOI] [Google Scholar]
- 100. Bazin I, Tria SA, Hayat A, Marty J‐L. New biorecognition molecules in biosensors for the detection of toxins. Biosens Bioelectron. 2017;87:285‐298. 10.1016/j.bios.2016.06.083. [DOI] [PubMed] [Google Scholar]
- 101. Teengam P, Siangproh W, Tuantranont A, Henry CS, Vilaivan T, Chailapakul O. Electrochemical paper‐based peptide nucleic acid biosensor for detecting human papillomavirus. Anal Chim Acta. 2017;952:32‐40. 10.1016/j.aca.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 102. Chin CD, Linder V, Sia SK. Commercialization of microfluidic point‐of‐care diagnostic devices. Lab Chip. 2012;12(12):2118‐2134. 10.1039/c2lc21204h. [DOI] [PubMed] [Google Scholar]
- 103. Jung W, Han J, Choi J‐W, Ahn CH. Point‐of‐care testing (POCT) diagnostic systems using microfluidic lab‐on‐a‐chip technologies. Microelectron Eng. 2015;132:46‐57. 10.1016/j.mee.2014.09.024. [DOI] [Google Scholar]
- 104. Jolly P, Rainbow J, Regoutz A, Estrela P, Moschou D. A PNA‐based lab‐on‐PCB diagnostic platform for rapid and high sensitivity DNA quantification. Biosens Bioelectron. 2019;123:244‐250. 10.1016/j.bios.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 105. Xuan F, Luo X, Hsing I‐M. Sensitive immobilization‐free electrochemical DNA sensor based on isothermal circular strand displacement polymerization reaction. Biosens Bioelectron. 2012;35(1):230‐234. 10.1016/j.bios.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 106. Sonar MV, Wampole ME, Jin Y‐Y, Chen C‐P, Thakur ML, Wickstrom E. Fluorescence detection of KRAS2 mRNA hybridization in lung cancer cells with PNA‐peptides containing an internal Thiazole Orange. Bioconjug Chem. 2014;25(9):1697‐1708. 10.1021/bc500304m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Shigeto H, Ohtsuki T, Iizuka A, Akiyama Y, Yamamura S. Imaging analysis of EGFR mutated cancer cells using peptide nucleic acid (PNA)–DNA probes. Analyst. 2019;144(15):4613‐4621. 10.1039/C9AN00725C. [DOI] [PubMed] [Google Scholar]
- 108. Chen T‐L, Chang JW‐C, Hsieh J‐J, Cheng H‐Y, Chiou C‐C. A sensitive peptide nucleic acid probe assay for detection of BRAF V600 mutations in melanoma. Cancer Genomics Proteomics. 2016;13(5):381‐386. http://www.ncbi.nlm.nih.gov/pubmed/27566656. [PMC free article] [PubMed] [Google Scholar]
- 109. Nielsen PE. Peptide nucleic acids as therapeutic agents. Curr Opin Struct Biol. 1999;9(3):353‐357. 10.1016/S0959-440X(99)80047-5. [DOI] [PubMed] [Google Scholar]
- 110. Quijano E, Bahal R, Ricciardi A, Saltzman WM, Glazer PM. Therapeutic peptide nucleic acids: principles, limitations, and opportunities. Yale J Biol Med. 2017;90(4):583‐598. http://www.ncbi.nlm.nih.gov/pubmed/29259523. [PMC free article] [PubMed] [Google Scholar]
- 111. Boado RJ, Tsukamoto H, Pardridge WM. Drug delivery of antisense molecules to the brain for treatment of Alzheimer's disease and cerebral AIDS. J Pharm Sci. 1998;87(11):1308‐1315. 10.1021/js9800836. [DOI] [PubMed] [Google Scholar]
- 112. Kaushik N, Basu A, Palumbo P, Myers RL, Pandey VN. Anti‐TAR polyamide nucleotide analog conjugated with a membrane‐permeating peptide inhibits human immunodeficiency virus type 1 production. J Virol. 2002;76(8):3881‐3891. 10.1128/JVI.76.8.3881-3891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chaubey B, Tripathi S, Ganguly S, Harris D, Casale RA, Pandey VN. A PNA‐transportan conjugate targeted to the TAR region of the HIV‐1 genome exhibits both antiviral and virucidal properties. Virology. 2005;331(2):418‐428. 10.1016/j.virol.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 114. Riguet E, Tripathi S, Chaubey B, Désiré J, Pandey VN, Décout J‐L. A peptide nucleic acid−Neamine conjugate that targets and cleaves HIV‐1 TAR RNA inhibits viral replication †. J Med Chem. 2004;47(20):4806‐4809. 10.1021/jm049642d. [DOI] [PubMed] [Google Scholar]
- 115. Zeng Z, Han S, Hong W, et al. A tat‐conjugated peptide nucleic acid tat‐PNA‐DR inhibits hepatitis B virus replication in vitro and in vivo by targeting LTR direct repeats of HBV RNA. Molecul Therap‐Nucleic Acids. 2016;5:e295. 10.1038/mtna.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Browne EC, Parakh S, Duncan LF, Langford SJ, Atkin JD, Abbott BM. Efficacy of peptide nucleic acid and selected conjugates against specific cellular pathologies of amyotrophic lateral sclerosis. Bioorg Med Chem. 2016;24(7):1520‐1527. 10.1016/j.bmc.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 117. Ricciardi A, Quijano E, Putman R, Saltzman W, Glazer P. Peptide nucleic acids as a tool for site‐specific gene editing. Molecules. 2018;23(3):632. 10.3390/molecules23030632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bahal R, Ali McNeer N, Quijano E, et al. In vivo correction of anaemia in β‐thalassemic mice by γPNA‐mediated gene editing with nanoparticle delivery. Nat Commun. 2016;7(1):13304. 10.1038/ncomms13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dorn S, Aghaallaei N, Jung G, et al. Side chain modified peptide nucleic acids (PNA) for knock‐down of six3 in Medaka embryos. BMC Biotechnol. 2012;12(1):50. 10.1186/1472-6750-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Tagalakis AD, Maeshima R, Yu‐Wai‐Man C, et al. Peptide and nucleic acid‐directed self‐assembly of cationic nanovehicles through giant unilamellar vesicle modification: targetable nanocomplexes for in vivo nucleic acid delivery. Acta Biomater. 2017;51:351‐362. 10.1016/j.actbio.2017.01.048. [DOI] [PubMed] [Google Scholar]
- 121. Gupta A, Bahal R, Gupta M, Glazer PM, Saltzman WM. Nanotechnology for delivery of peptide nucleic acids (PNAs). J Control Release. 2016;240:302‐311. 10.1016/j.jconrel.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bolarinwa O, Nimmagadda A, Su M, Cai J. Structure and function of AApeptides. Biochemistry. 2017;56(3):445‐457. 10.1021/acs.biochem.6b01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ndeboko B, Hantz O, Lemamy G, Cova L. Developments in cell‐penetrating peptides as antiviral agents and as vehicles for delivery of peptide nucleic acid targeting Hepadnaviral replication pathway. Biomol Ther. 2018;8(3):55. 10.3390/biom8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Singh KRB, Fernandes M, Sarkar T, Sridevi P. Assessment and analysis of lifestyle disease burden in tribes of central India. Infect Non Infect Diseas. 2019;4(1):1‐7. 10.24966/INID-8654/100027. [DOI] [Google Scholar]


